Summary
Mechanisms governing how immune cells and their derived molecules impact homeostatic brain function are still poorly understood. Here, we elucidate neuronal mechanisms underlying T-cell effects on synaptic function and episodic memory. Depletion of CD4 T cells led to memory deficits and impaired long-term potentiation. Severe combined immune-deficient mice exhibited amnesia, which was reversible by repopulation with T cells from wild-type but not from IL-4-knockout mice. Behaviors impacted by T cells were mediated via IL-4 receptors expressed on neurons. Exploration of snRNAseq of neurons participating in memory processing provided insights into synaptic organization and plasticity-associated pathways regulated by immune cells. IL-4Rα knockout in inhibitory (but not in excitatory) neurons was sufficient to impair contextual fear memory, and scRNAseq from these mice pointed to IL-4-driven regulation of synaptic function in promoting memory. These findings provide new insights into complex neuroimmune interactions at the transcriptional and functional levels in neurons under physiological conditions.
eTOC Blurb
In this article, Herz et al. demonstrate how T cells and their derived IL-4 regulate contextual fear memory in mice. They show that interleukin-4 plays an active role in supporting synapse function and identify expression of IL-4 receptor on GABAergic neurons to be essential for fear memory.
Graphical Abstract
Introduction
The nervous system integrates ongoing environment-derived information with previously stored experiences, thereby guiding behaviors and allowing environmental adaptation. Concomitantly the immune system, based on experience-derived memory, protects the host against microorganisms, allowing its better adaptation and survival. Despite similarities in principles guiding both systems, their communication in support of host adaptation is still elusive.
Episodic memory is a recollection of past experiences conveying distinct temporal and spatial information (Tulving, 2002). In the processing of such “mental time travel”, both in animal models and in humans, the hippocampus has a key role (Bird and Burgess, 2008; Burgess et al., 2002; Eichenbaum et al., 1999; Frisk and Milner, 1990; Hitti and Siegelbaum, 2014). The mechanisms of episodic memory have been dissected in numerous studies at multiple levels, including actively involved brain regions (Chen et al., 1996; Frisk and Milner, 1990), neuronal circuits (Herry et al., 2008; Roy et al., 2017), associated synaptic strength changes (Mahan and Ressler, 2012; McKernan and Shinnick-Gallagher, 1997), and molecular mechanisms (Johansen et al., 2011).
The immune system has been linked to cognitive function in a series of studies (Baruch et al., 2014; Castellano et al., 2017; Derecki et al., 2010; Kipnis et al., 2004; Villeda et al., 2014), mostly employing complex prolonged behavioral tasks in devices such as the Morris water maze (MWM), albeit without full understanding of the mechanisms invoked. Immune-deficient mice exhibited impaired spatial memory performance in the MWM, and reconstitution of splenocytes reversed the cognitive deficits (Derecki et al., 2010; Kipnis et al., 2004; Ziv et al., 2006). Similar memory deficits were also observed in mice deficient in interleukin (IL)-4, IL-4 receptor or IL-13 (cytokines mainly produced by Th2 lymphocytes, mast cells and ILC2s (Brown et al., 1987; Fallon et al., 2006; Neill et al., 2010; Plaut et al., 1989; Seder and Paul, 1994)), and an elusive mechanism mediated through astrocytes was suggested to account for it (Brombacher et al., 2021; Brombacher et al., 2020; Brombacher et al., 2017; Derecki et al., 2010). However, whether and how T cells and their derived cytokines affect episodic memory processing is not yet known.
A recently emerging new line of evidence suggests that cytokines can act as neuromodulators, thereby possibly altering neuronal responses and modifying neuronal function by signaling via their neuronally expressed cognate receptors (Chen et al., 2017; Choi et al., 2016; de Lima et al., 2020; Filiano et al., 2016). This intriguing novel indication of cytokine-induced potential communication between the immune and the nervous systems prompted us to revisit the roles of T cells and their derived IL-4 in episodic memory. Our aim was to acquire a better mechanistic insight into whether and how episodic memory is promoted by IL-4.
Here, we show that contextual fear conditioning (CFC) memory is impaired after depletion of CD4 T cells in wild-type (WT) mice. Consistently with the behavioral findings, CD4 T-cell depletion also impaired long-term synaptic potentiation in the hippocampal dentate gyrus (DG). We further found that memory deficits in immune-deficient mice could be restored by wild type T cells but not by T cells deficient in IL-4, and that conditional knockout of IL-4Rα from neurons, but not from microglia, recapitulated memory deficits. Single-nuclei RNA sequencing (snRNAseq) of neurons responding to a memory task revealed that compared to WT, immune-deficient mice exhibited dysregulation of synapse-related gene expression, and that this can be largely reversed by repopulation with T cells. Our study thus shows that CD4 T cells and their derived IL-4 regulate episodic memory, providing a mechanistic view that T cells and IL-4 alter synaptic transmission, synaptic plasticity and gene expression in task-activated neurons.
Results
T cells regulate episodic memory through interleukin-4
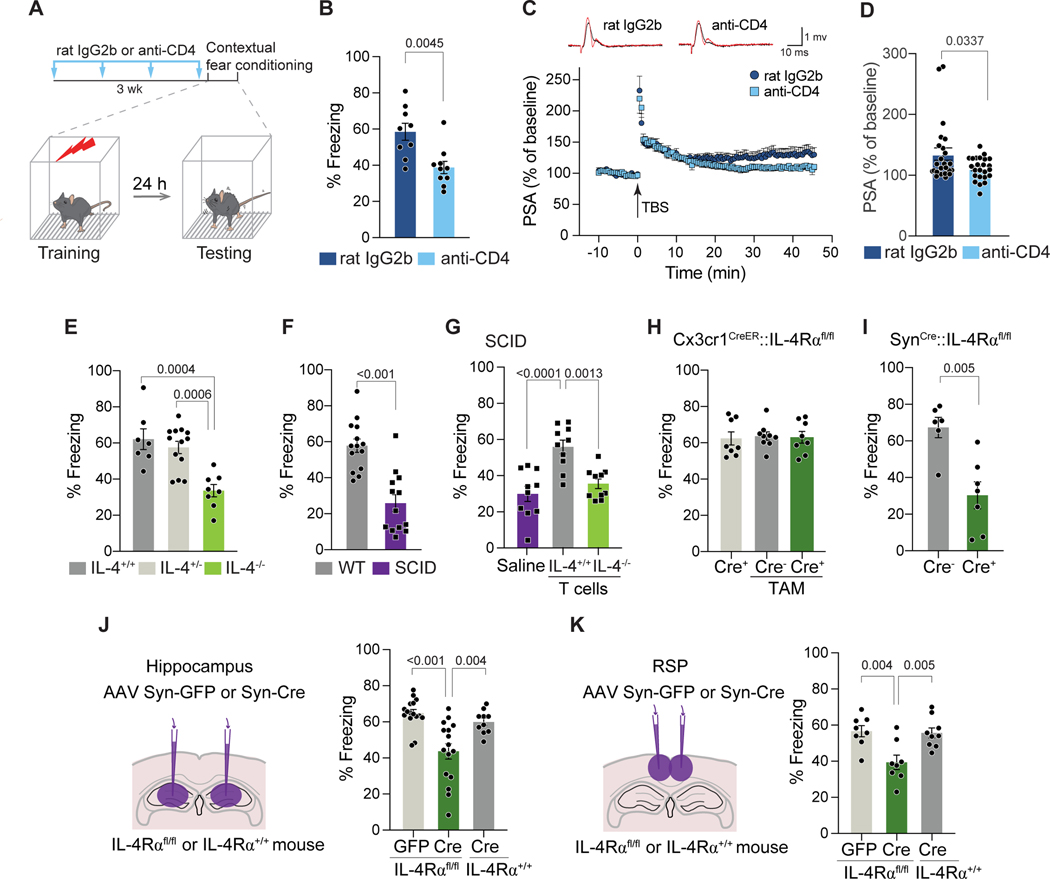
To explore the potential neuro-immune interactions during episodic memory processing, we employed the CFC memory paradigm, a robust memory task with well-studied circuitry (Goshen et al., 2011; Liu et al., 2012; Maren, 2001). In this task, mice are introduced to a new environment (new context) and concomitantly exposed to an unconditioned stimulus of foot shock (Maren, 2001; Phillips and LeDoux, 1992). They are then moved to a home cage for 24 hours, and upon reintroduction to the “shock context” but without the stimulus, their immobility (freezing) in anticipation of a shock is quantified. The time that freezing lasts is indicative of contextual memory (Figure 1A). We first determined whether CD4 T cells and IL-4 are required for CFC memory regulation. Acute depletion of CD4 T cells from WT mice for 3 weeks (Supplemental Figure S1A, B) resulted in their reduced freezing times during the memory test when compared to mice injected with control antibodies (Figure 1B). No differences were found before training and 1 hour after training (Supplemental Figure S1C). CD4 T cell depletion led to a significantly lower population spike amplitude (PSA) in a theta-burst stimulation induced long-term potentiation (LTP) (Figure 1C, D), decreased neuronal excitability (Supplemental Figure S1D) but not spontaneous excitatory and inhibitory postsynaptic currents (Supplemental Figure S1E-G). These data support the notion that mice lacking T cells show impaired contextual fear memory and neuronal function.
Figure 1. T cell-derived IL-4 promotes contextual fear memory through neuronal IL-4Rα.
A, Schematic diagram of the behavioral experimental procedure. Mice were injected i.p. with either 200 ug rat IgG2b isotype control (clone LTF-2) or anti-CD4 antibody (clone GK1.5) followed by 100 ug weekly injections for three weeks. Mice were then trained with contextual fear conditioning and subjected to the same cage (context) for testing 24 hours later. B, Percent freezing time during 3 minutes of testing of mice treated with control antibody (n=9) or depleted of CD4 T cells (n=10). Two-tailed unpaired Mann-Whitney U-test. Representative experiment is shown out of 2 independent experiments performed. C, Long-term potentiation of population spike amplitude (PSA) in the dentate gyrus of hippocampal slices after CD4 T cell depletion. Upper panel: representative traces of WT mice treated with control rat IgG2b or anti-CD4 antibody after three weeks. Lower panel: Average traces of PSA obtained before and after LTP induction by theta-burst stimulation (TBS). N=24 slices from 8 mice per group. D, Normalized PSA (% of baseline) for the last 5 minutes. Two-tailed unpaired Mann-Whitney U-test. E, Percent freezing time of IL-4+/+ (n=7), IL-4−/+ (n=13) and IL-4−/− (n=8) mice. One-Way ANOVA with Tukey post hoc test. Two independent experiments combined are presented. F, Percent freezing time of WT (n=14) and SCID (n=13) mice 24 hours after training. Two-tailed unpaired Mann-Whitney U-test. Representative experiment is shown out of 3 independent experiments performed. G, Percent freezing time of SCID mice injected with saline, 2 × 106 splenic IL4+/+ or IL4−/− T cells. Mice underwent training and fear memory testing 4 weeks after repopulation (n=10 mice per group). One-Way ANOVA with Tukey post hoc test. Representative experiment is shown out of 2 independent experiments performed. H, Percent freezing time of mice with microglia-specific knockout of IL-4Rα after 3 weeks of tamoxifen (TAM) treatment (n=8 – 9 mice per group) and I, neuron-specific knockout of IL-4Rα (n=6 – 7 mice per group) for 24-hour contextual fear memory testing. One-Way ANOVA with Tukey post hoc test (H; non-significant) and two-tailed unpaired Mann-Whitney U-test (I). J, Percent freezing time 24 hours after testing of IL-4Rα fl/fl mice injected bilaterally with AAV-Syn-GFP (n=16) or AAV-Syn-Cre (n=16) and IL-4Rα +/+ mice injected with AAV-Syn-Cre (n=10) into the hippocampus 4 weeks prior to behavioral testing. One-Way ANOVA with Tukey post hoc test. K, Percent freezing time 24 hours after testing of IL-4Rα fl/fl mice injected with AAV-Syn-GFP (n=8) or AAV-Syn-Cre (n=8) and IL-4Rα +/+ mice injected with AAV-Syn-Cre (n=9) into the retrosplenial cortex. One-Way ANOVA with Tukey post hoc test. Data are presented as mean ± SEM. See also Figure S1 and S2.
Similar results were obtained in mice deficient in IL-4; while control and IL-4 knockout mice showed similar freezing times before training (Supplemental Figure S1H), the freezing response of the IL-4 knockout mice during testing was impaired compared to that of their IL-4−/+ or IL-4+/+ littermates (Figure 1E). In addition, we also examined the performance of the severe combined immune-deficient (SCID) mice, lacking adaptive immunity in the memory task. WT and SCID mice showed similar freezing behavior when assessed before training and 1 hour after training (Supplemental Figure S1I, J); however, compared to their WT counterparts, significantly reduced freezing times were evident in both male and female SCID mice when assessed at 24 hours and at 4 weeks post-training (Figure 1F; Supplemental Figure S1I, J).
To find out whether memory deficit is a result of immunodeficiency, we adoptively transferred T cells from WT donors or from IL4−/− donors into young adult SCID mice, in which CFC memory was tested 4 weeks after reconstitution. Compared to their non-repopulated counterparts, the SCID mice showed significantly longer freezing behavior when repopulated with T cells from WT but not from IL-4−/− mice (Figure 1G), suggesting that T cells and their derived IL-4 are required for normal CFC memory. As expected, immunosufficient WT mice showed no changes in freezing behavior when repopulated with T cells from either WT or IL-4−/− mice (Supplemental Figure S1K).
IL-4 regulates CFC memory through neuronal IL-4Rα
To better understand the IL-4-signaling processes underlying these changes, IL-4Rα was conditionally deleted from macrophages and microglia using Cx3cr1CreER mice crossed to IL4Rα fl/fl. No memory deficits or impaired locomotor and exploratory activities were observed in Cx3cr1CreER::IL4Rα fl/fl mice (Figure 1H; Supplemental Figure S1L, M). However, conditional deletion of IL-4Rα from neurons, using SynCre::IL4Rα fl/fl mice, resulted in impaired memory function (Figure 1I). Notably, SynCre+::IL4Rα fl/fl mice travelled more distance in the arena and spend less time in the center of the open field test when compared to SynCre-::IL4Rα fl/fl littermates (Supplemental Figure S1N). Confirmation of IL-4Rα deletion from neurons (both GABAergic and glutamatergic) and microglia was assessed by realtime PCR or RNA-fluorescence in situ hybridization (Supplemental Figure S2A-C). To further explore the regional specificity of IL-4 function, we deleted IL-4Rα from hippocampal or retrosplenial cortex (RSP) neurons via stereotactic AAV-hSyn1-eGFP-Cre (AAV Syn-Cre) delivery in IL-4Rα fl/fl mice and tested the mice for freezing behavior 24 hours after fear training. We found that deletion of IL-4Rα from hippocampal or RSP neurons (Supplemental Figure S2D) was sufficient to impair memory (Figure 1J, K). With AAV-hSyn1-eGFP (AAV Syn-GFP) used as a control vector, neither injection of AAV Syn-GFP into IL-4Rα fl/fl mice nor injection of AAV Syn-Cre virus into IL-4Rα +/+ mice altered freezing behavior (Figure 1J, K). These data suggest that neuronal expression of IL-4Rα is involved in CFC memory processing.
Rescue of synapse-related gene expression in CFC-activated neurons by T cells and IL-4
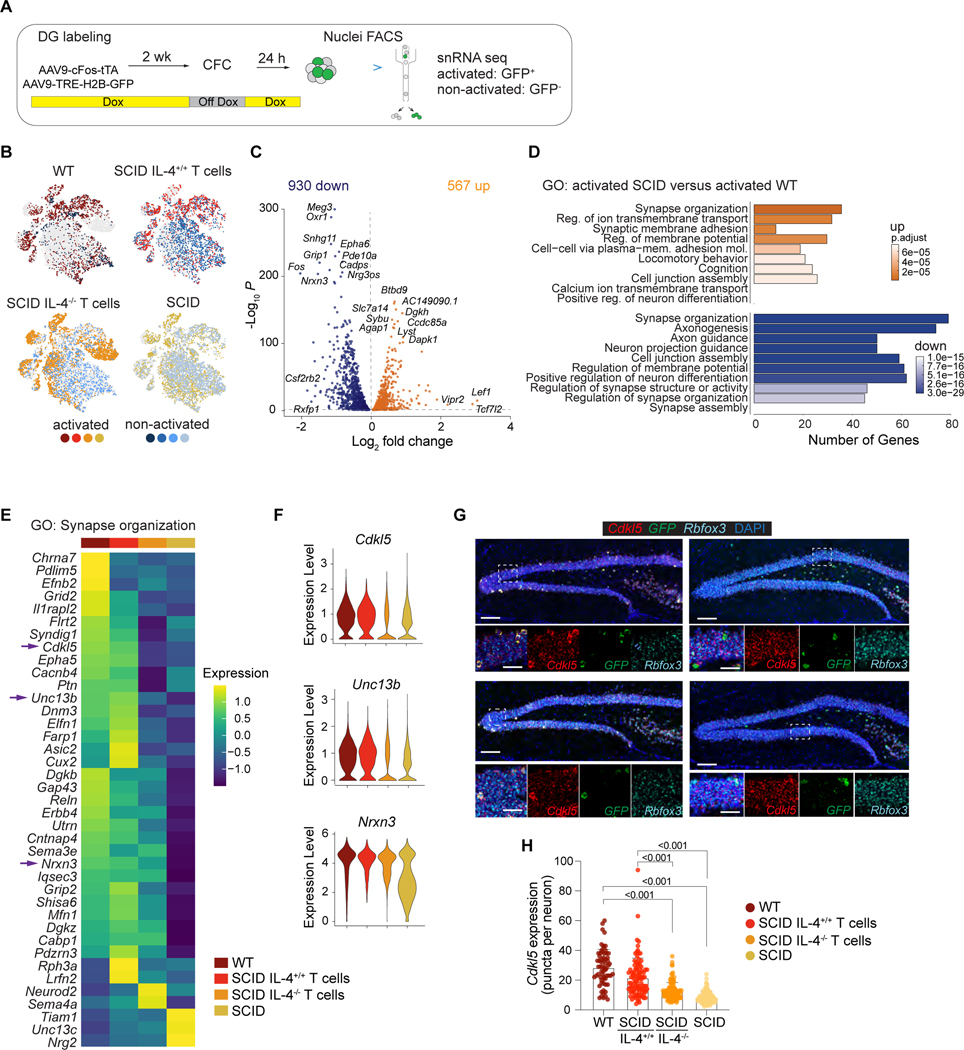
Peripheral immune cells have no physical contact with neurons in young-adult healthy mice. However, peripheral T cells, along with other immune cells are densely populating meningeal spaces (Cugurra et al., 2021; Kipnis, 2016; Louveau et al., 2015; Rustenhoven et al., 2021). We hypothesized that cytokines released into the cerebrospinal fluid (CSF) by meningeal immune cells diffuse through the brain parenchyma (Da Mesquita et al., 2018) and target their receptors on neurons and/or glia to influence memory. To determine how immune signals change neuronal function, we labeled with AAV9-cFos-tTA and AAV9-TRE-H2B-GFP viruses in the hippocampal DG (Figure 2A, Supplemental Figure S3A−D) (Liu et al., 2012). Labeled activated neuronal nuclei from WT, SCID and T-cell-repopulated SCID mice were isolated 24 hours after fear conditioning training, and snRNAseq was performed (Figure 2B). Analysis of neuronal nuclei from the DGs of WT, SCID and T-cell-repopulated SCID mice revealed unique clusters, distinctively falling into glutamatergic or GABAergic subsets (Supplemental Figure S3E) and eight neuronal subtypes (Supplemental Figure S3F, G). Of note, the cluster identified as dorsal CA3 pyramid cells was significantly increased compared to WT proportions in activated SCID neurons reconstituted with T cells, potentially linking these neurons with pro-cognitive effects of T cells in immunodeficient conditions (Supplemental Figure S3H). To gain an understanding of the molecular differences in task-activated neurons between WT and SCID mice we first examined the differential gene expression between the two groups, identifying 1,497 significant changes, of which 930 were significantly downregulated and 567 upregulated. (Figure 2C). Of the 930 down-regulated genes, the number of genes rescued by IL-4+/+ T cells was 513 and IL-4–/– T cell 93. Of the 567 up-regulated genes in SCID versus WT comparisons, 385 were abrogated with IL-4+/+ T cells and 496 with IL-4–/– T cells complementation. Among these changes, genes annotated to the Gene Ontology (GO) terms for “synapse organization”, “regulation of ion transmembrane transport”, and “synaptic membrane adhesion” were included in the top 10 of the most statistically significant observed (Figure 2D).
Figure 2. T cell-derived IL-4 orchestrates transcriptomic changes in synapse-related genes.
A, Schematic representation of the experimental design. WT and SCID mice were injected with AAV9c-Fos-tTA and AAV9-TRE-H2B-GFP and maintained on doxycycline (DOX) chow. The dentate gyrus (DG) was labeled two weeks prior to training. To specifically label training activated neurons, mice were taken off DOX food 24 hour before training and feed DOX chow immediately after training. Activated (GFP+) and non-activated (GFP-) neuronal nuclei from the DG were sorted by flow cytometry and subjected to single-nuclei RNA sequencing. B, t-SNE showing the distribution of nuclei from each sample (WT, SCID, SCID with IL-4+/+ T cells, SCID with IL-4−/− T cells) of activated and non-activated neurons across clusters. C, Volcano plot showing differentially expressed genes in activated WT compared to activated SCID neurons. Top differentially expressed genes are annotated with text. D, Gene ontology analysis (GO) of top 10 pathways by significance comparing differentially expressed genes in activated GFP+ neuronal nuclei from SCID and WT mice. E, Heat map showing the average scaled expression of significant differentially expressed genes in GO: Synapse organization (0050808) between activated WT, SCID with IL-4+/+ T cells, SCID with IL-4−/− T cells and SCID of neuronal nuclei. Mean of n = 3 – 4 biological samples. F, Violin plots showing the distribution of expression of Cdlk5, Unc13b and Nrxn3 in activated neuronal nuclei. G, Representative images of multi-color RNAscope experiments showing Cdkl5 gene expression of activated (GFP+) neurons (Rbfox3+) in the dentate gyrus for conditions shown in E and F. Scale bars = 200 μm or 40 μm. H, Quantifications of Cdkl5 gene expression in activated neurons (GFP+ Rbfoxp+) of WT (n=64 neurons), SCID with IL4+/+ T cells (n= 95 neurons), SCID with IL-4−/− T cells (n=116 neurons) and SCID mice (n=116 neurons) from 3–4 mice per group. Data are presented as mean ± SEM. Two-way ANOVA with Bonferroni post hoc test. See also Figure S3 and S4.
We next determined those genes that are involved in the enriched “synapse organization” pathway because we observed substantial downregulation in activated SCID versus WT neurons. Interestingly, upon repopulation of SCID mice with T cells (independent of IL-4 expression), we found that T cell reconstitution largely reversed the differential gene expression between WT and SCID (Figure 2E). We identified genes involved in positive regulation of synapse plasticity such as growth associated protein 43 (Gap43) and glutamate receptor-interacting protein 2 (Grip2). Violin plots of gene expression showed that in SCID mice, synapse-related genes such as cyclin-dependent kinase-like 5 (Cdkl5) and UNC-13 homolog B (Unc13b) were restored to WT levels by T-cell-repopulation with WT but not with IL-4−/− T cell repopulation (Figure 2F). Notably, expression of several genes such as neurexin 3-alpha (Nrxn3) were rescued by both WT T cell and IL-4−/− T cell repopulation (Figure 2F). Cdkl5, in particular, has been reported to regulate synaptic function and memory (Ricciardi et al., 2012; Tang et al., 2017; Zhu et al., 2013). We therefore collected brains, 24 hours after exposure to a foot shock, to further verify whether T cells and their derived IL-4 regulate Cdkl5 expression in CFC-activated neurons. Using in-situ hybridization methods, we quantified Cdkl5 expression on Rbfox3+- and GFP+-tagged neurons under four different conditions (Figure 2G). Consistently with our snRNAseq findings, RNAScope showed a significant increase in the number of activated Cdlk5+ neurons in the DG of mice reconstituted with T cells, compared to mice that remained without T cells (Figure 2H). Between non-activated WT and SCID groups, genes associated with synapse-related GO terms were also differentially expressed, suggesting that the adaptive immune system may regulate synapse-related gene expression under homeostatic conditions (Supplemental Figure S3I-K).
IL-4 regulates CFC memory through IL-4R in GABAergic neurons
We next focused on identifying the specific neuronal subsets responsible for promoting fear memory following IL-4 signaling. Previous reports have shown expression of IL-4Rα in axons (colocalized with SMI-31) of cortical and hippocampal neurons (Vogelaar et al., 2018). We also detected IL-4Rα in several brain regions in published data sets (mousebrain.org) (Supplemental Figure S4A), and identified Il4rα expression, albeit at low level, in our RNAseq data (Supplemental Figure S4B). Analysis of additional published RiboTag and RNAseq data sets (Furlanis et al., 2019; Paul et al., 2017) revealed expression of Il4rα1 and Il13rα1 but also their signaling molecules Janus kinase 3 (Jak3) and signal transducer and activator of transcription 6 (Stat6) in both excitatory and inhibitory neurons (Supplemental Figure S4C, D). We therefore investigated IL-4 signaling and expression of both, the JAK-STAT and IRS-PI3K pathways in fear-activated neurons (Supplemental Figure S4E, F). Interestingly, Gap43, Pik3c3 and Irs1 gene expression was increased in WT nuclei upon activation, but not in SCID. Of note, reduced gene expression of Gap43 in SCID mice could be rescued with IL-4+/+ but not IL-4–/– T cells, which further suggests functional effects of IL-4 in neurons via IL-4Rα signaling (Supplemental Figure S4E, F). While the percentages of cells expressing each receptor varied across tissues and datasets (Supplemental Figure S4A-F), we confirmed that both receptors are expressed in both excitatory and inhibitory neurons using in situ hybridization assays (Supplemental Figure S2B, Supplemental Figure S5A, B).
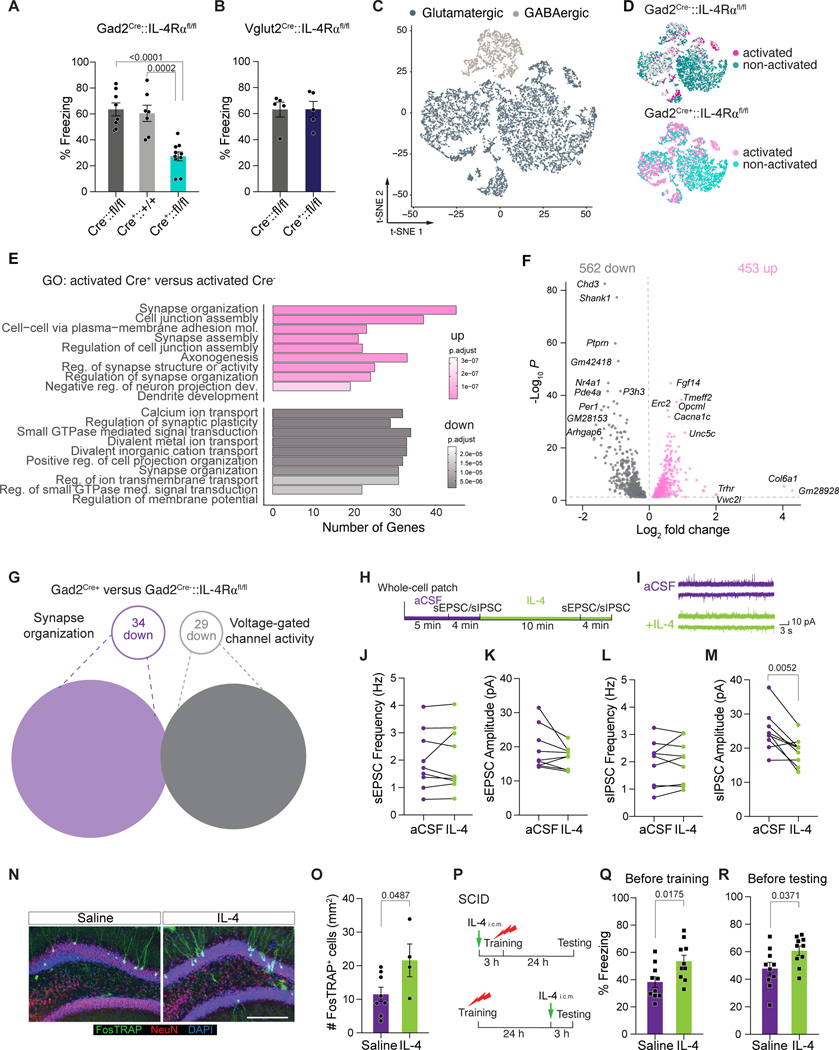
To determine the function of IL-4Rα in excitatory and inhibitory neurons for CFC memory regulation, we conditionally ablated IL-4Rα from either GABAergic (Gad2Cre+::IL-4Rα fl/fl) or glutamatergic (Vglut2Cre+::IL-4Rα fl/fl) neurons (Supplemental Figure S5A, B). Mice with IL-4Rα deficiency in GABAergic neurons (Figure 3A), but not in glutamatergic subsets (Figure 3B), exhibited significantly decreased freezing times, suggesting that in affecting CFC memory, IL-4 signals mainly through inhibitory neurons. No differences were found in baseline freezing (Supplemental Figure S5CD) or total distance traveled during the habituation phase in the fear chamber (Supplemental Figure S5E) and in the open field assay (Supplemental Figure S5F).
Figure 3. IL-4Rα on GABAergic neurons regulates contextual fear memory.
A, Percent freezing time of mice with GABAergic neuron-specific knockout of IL-4Rα . Gad2Cre-::IL-4Rα fl/fl (n=8), Gad2Cre+::IL-4R+/+ (n=7), and Gad2Cre+::IL-4Rα fl/fl (n=10) mice. One-Way ANOVA with Tukey post hoc test. B, Percent freezing level of mice with glutamatergic neuron-specific knockout of IL4Rα . Vglut2Cre-::IL-4Rα fl/fl and Vglut2Cre+::IL-4Rα fl/fl mice (n=5 mice per group). Two-tailed unpaired Mann-Whitney U-test; no significant difference was found between the groups. C, t-SNE visualization of nuclei from the dentate gyrus of Gad2Cre::IL-4Rα fl/fl mice, colored by glutamatergic and GABAergic neurons. D, t-SNE visualization of activated (GFP+) and non-activated (GFP-) nuclei in Gad2Cre-::IL-4Rα fl/fl mice and Gad2Cre+::IL-4Rα fl/fl mice. E, Gene ontology analysis (GO) of differently expressed genes that were identified between activated neuronal nuclei from Gad2Cre+::IL-4Rα fl/fl mice versus Gad2Cre-::IL-4Rα fl/fl mice. F, Volcano plots showing differentially expressed genes in activated neuronal nuclei in Gad2Cre+::IL-4Rα fl/fl versus Gad2Cre-::IL-4Rα fl/fl mice. Top significantly altered genes are annotated. G, List of synapse organization related (GO:0050808) and voltage-gated ion channels (calcium-, sodium-, potassium channels, GO:0022843 and GO:0022844) that are differentially expressed in activated glutamatergic neurons from Gad2Cre+::IL-4Rα fl/fl mice and Gad2Cre-::IL-4Rα fl/fl mice. H, Representative electrophysiological recording showing spontaneous excitatory and inhibitory post synaptic currents (sEPSC and sIPSC) in dentate gyrus neurons before and after perfusion with 100 ng/ml IL-4. I Example traces of sEPSC and sIPSC recordings before and after IL4 exposure. J - K, Statistical analysis of changes in sEPSC frequency (J) and amplitude (K) before and after IL-4 perfusion (n=10 neurons from 4 mice). Two-tailed paired t-test. L - M, Statistical analysis of changes in sIPSC frequency (J) and amplitude (K) before and after IL-4 perfusion form the same neurons shown in J and K. Twotailed paired t-test. N, Representative micrographs showing neuronal labeling in the dentate gyrus (DG) with FosTRAP mice subjected to saline or IL-4 intra-cisterna magna injection. Scale bar = 200 μm. O, Quantifications of FosTRAP+ neurons upon IL-4 injection. Two-tailed unpaired t-test. P, Experimental schematics showing SCID mice either injected with saline or 100 units of IL-4 (IL-4) through the magna (i.c.m.) 3 hours before training or testing. R - Q, Percent freezing time of SCID mice injected with saline or IL-4, 3 hours before training (Q) or testing (R). Two-tailed unpaired Mann-Whitney U-test. Data are presented as mean ± SEM. See also Figure S5 and S6.
Because our data on IL-4Rα depletion pointed to involvement of GABAergic neurons in CFC memory regulation, we conducted snRNAseq experiments on CFC-activated neurons from the DG using Gad2Cre–::IL-4Rα fl/fl and Gad2Cre+::IL-4Rα fl/fl mice (Figure 3C−G). Most strikingly, loss of IL-4Rα in Gad2+ (inhibitory) neurons led to changes in expression of genes of relevance to GO terms, such as “synapse organization”, “regulation of synaptic plasticity”, and “regulation of synapse structure or activity” (Figure 3E, F), indicating that IL-4 may regulate synaptic function through IL-4Rα on inhibitory neurons. Moreover, loss of IL-4Rα in Gad2+ (inhibitory) neurons also led to lower expression of genes related to voltage-gated potassium, sodium, and calcium channels in glutamatergic neurons in the DG (Figure 3G), suggesting that IL-4 may also regulate neuronal excitability.
To understand whether IL-4Rα signaling can generally change ion channel behavior in neurons, we used whole patch-clamp on neurons before and after application of IL-4 to simultaneously record alterations in spontaneous excitatory and inhibitory postsynaptic currents (Figure 3H-M). The mean frequency of spontaneous (s)EPSC and sIPSC and amplitude of sEPSC was not changed, however, sIPCS amplitude upon IL-4 treatment was lower than the respective control (aCSF) condition. The lack of effect on sEPSC but decreased sIPSC amplitude suggests a possibly favor of inhibition in dentate gyrus neurons upon IL-4 treatment. Additionally, we recorded AMPA-type glutamate-receptor-mediated miniature excitatory postsynaptic currents (mEPSC) and GABAA receptor-mediated miniature inhibitory postsynaptic currents (mIPSC) in excitatory and inhibitory neurons in the DGs of acutely prepared brain slices from postnatal day 11–20 WT mice (Supplemental Figure 6A−M). IL-4 perfusion significantly decreased mIPSC amplitude and frequency in excitatory neurons (Supplemental Figure 6B−D). In excitatory neurons, IL-4 perfusion did not change mEPSC amplitude or frequency (Supplemental Figure S6E−G), or mIPSC and mEPSC amplitude or frequencies in inhibitory neurons (Supplemental Figure S6H−M) suggests an active role of IL-4 disinhibiting DG excitatory neurons, although patching nonIL4Rα expressing neurons cannot entirely be excluded. Lastly, IL-4 perfusion was found to decrease the latency of the minimum injection of current required for an action potential (rheobase) in glutamatergic neurons, suggesting that IL-4 can increase glutamatergic neuronal excitability (Supplemental Figure 6N-Q).
IL-4 regulates synaptic transmission and restores memory deficits in SCID mice
Finally, we focused on the functional consequences of recombinant (r)IL-4 treatment in vivo. To enable activated neurons to be labeled in vivo and during a specific time window, we used mice that express CreER under the control of an activity-dependent c-Fos promoter (FosTRAP mice (Guenthner et al., 2013)) crossed with Ai6 mice (Rosa-CAG-LSL-ZsGreen1) to capture neurons that were active during IL-4 administration. Relative to saline-injected controls, injection of IL-4 into the CSF significantly increased the number of activated FosTRAP+ neurons in the DG (Figure 3N, O).
We next examined whether IL-4 plays a role in the behavioral reversal of a deficit in CFC task (Figure 3P-R). Remarkably, a single injection of recombinant IL-4 into the CSF of SCID mice 3 hours before CFC training improved memory deficits (Figure 3Q). Administration of IL-4 following task acquisition and 3 hours before memory retrieval also led to an increased freezing time (Figure 3R). However, freezing behavior was not changed by injection of rIL-4 into the CSF of WT mice before either training or testing (Supplemental Figure S6R-T), suggesting that the IL-4-mediated effects on memory are homeostatic, i.e., that the process is exacerbated in absence of IL-4, but is not affected by boosting it beyond physiological levels. Collectively, these data demonstrate that interference with CD4 T cells and their derived IL-4-mediated functions fosters transcriptional dysregulation, synaptic weakening in neurons, and an inability to fully operate during fear memory.
Discussion
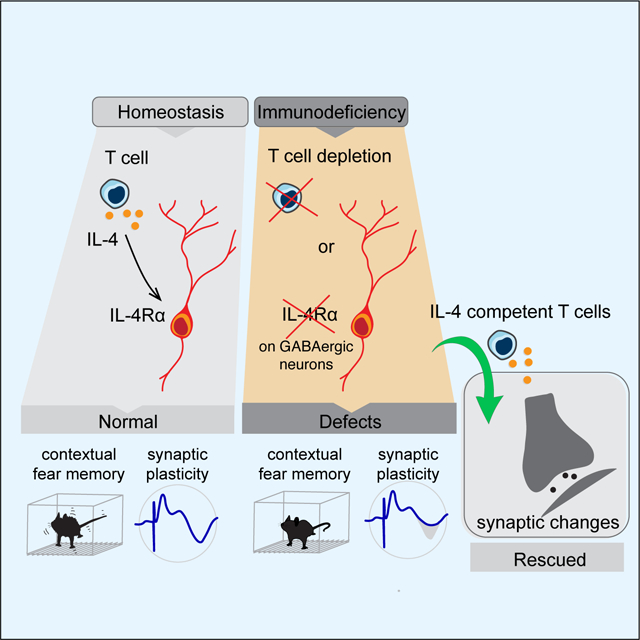
Appropriate defensive responses to previous experiences are vital in promoting survival when the individual is confronted with a potential threat. By studying an episodic memory task of CFC in mice, we showed here that T-cell-derived IL-4 plays a key role in regulating this type of memory. Mice depleted of CD4 T cells, or lacking adaptive immunity (SCID), as well as deficient in IL-4, exhibit memory deficits. In SCID mice, memory could be restored by reconstitution with T cells from WT donors but not with T cells from donors deficient in IL-4. Memory in SCID mice could also be improved by injection of recombinant IL-4 into the CSF either before acquisition of the CFC task or before memory retrieval. While memory function in the mice was not affected by conditional deletion of IL-4Rα from microglia and macrophages, deletion of IL-4Rα from their neurons was sufficient to recapitulate the cognitive deficits seen in IL-4-deficient mice. In-situ hybridization and snRNAseq data showed that IL-4Rα is expressed in both excitatory and inhibitory neurons, yet deletion of IL-4Rα from GABAergic neurons was required to recapitulate memory deficits. By comparing transcriptomic profiles of task-activated neurons in T-cell-sufficient and T-cell-deficient mice, we found that many of the differentially expressed genes were related to synaptic function.
The inhibitory inputs of dentate gyrus granule layer cells were decreased in the presence of IL-4, while the excitatory inputs are unchanged. This probably led hippocampal circuits to disinhibitory states and is consistent with our observation that neuronal firing activities were increased, and contextual memory enhanced. SnRNAseq of task-activated neurons from mice with specific IL-4Rα knockout in GABAergic neurons and their WT littermates showed that removal of IL-4Rα from GABAergic neurons alters the expression of genes associated with synapse organization and synaptic plasticity. This suggested that IL-4 could be an important contributory force in synapse reorganization, leading to strengthening of inter-neuron connections and improved cognition. It will be important to investigate in future studies whether IL4Rα is located in pre- or post-synaptic terminals and whether IL4Rα neuronal signaling might be therapeutically targeted.
The results presented here support the existence of a mechanistic link between adaptive immunity and neuronal function (de Lima et al., 2020; Reed et al., 2020), and extend those interactions to physiological homeostatic conditions. Our findings demonstrate a direct effect of peripheral T cells and their derived IL-4 on behavior modulation, neuronal transcription and synaptic function in CFC.
Many of the “immune” genes and pathways that are upregulated in activated neurons have previously been linked to neuronal function. As examples, TNF signaling was implicated in synaptic scaling (Stellwagen and Malenka, 2006), complement molecules were shown to modify synaptic function (Stevens et al., 2007), and MHCI was found to be expressed by neurons and to directly regulate synaptic pruning and function (Shatz, 2009). It is therefore plausible that “classical” immune molecules such as IL-4, which are expressed primarily by immune cells, can be used by the immune system for signaling to neurons about the state of the organism. Cytokine signaling would then modulate neuronal function, in part through “shared” immune molecules (such as TNF, MHCI and complement system-related molecules, all of which are expressed by neurons). Future studies should focus on cytokine-induced subcellular signaling to neurons and, more specifically, on the role of such shared immune molecules in mediating neuro-immune communications. Thus, for example, while it is likely that T cells affecting brain function are harbored in the meninges (Radjavi et al., 2014), in order to further address this communication enigma there is a critical need for new tools that would allow spatial targeting of T cells within such compartments and in tissues of interest.
Since the immune system is more easily accessible than the brain, modification of immune cells may indeed turn out to be a promising therapeutic option for disorders that are not classically characterized as neuroinflammatory, but in which an immune link is present.
STAR★Methods
Resources availability
Lead Contact
Further information and requests for resources and reagents should be directed to Jonathan Kipnis (kipnis@wustl.edu).
Materials Availability
This study did not generate new unique reagents.
Data and code availability
Fastq files and quantified gene counts for single cell sequencing are available at the Gene Expression Omnibus (GEO) and Sequence Read Archive (SRA) under accession numbers GSE143183.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Mice
Wild-type mice (C57BL/6J background) were either bred in-house or purchased from the Jackson Laboratory (JAX 000664). All mice were maintained in the animal facility for at least one week prior to the start of any experiment. The following strains were used: C57BL/6-Il4tm1Nnt/J (IL-4–/–, JAX 002518), B6.CB17-Prkdcscid/SzJ (SCID, JAX 001913), B6J.Cg-Gad2tm2(cre)Zjh/MwarJ (Gad2Cre, JAX 028867), B6J.129S6(FVB)-Slc17a6tm2(cre)Lowl/MwarJ (Vglut2Cre, JAX 028863), B6.129(Cg)-Fostm1.1(cre/ERT2)Luo/J (FosCreER, JAX 021882), B6.Cg-Gt(ROSA)26Sortm6(CAG-ZsGreen1)Hze/J (Ai6, JAX 007906), B6;129SGad2tm1.1Ksvo/J (Gad2-T2a-NLS-mCherry, JAX 023140), B6.Cg-Tg(Syn1-cre)671Jxm/J (SynCre, JAX 003966) and B6.129P2(Cg)-Cx3cr1tm2.1(cre/ERT2)Litt/WganJ (Cx3cr1CreER, JAX 021160). The Il4ratm2Fbb (IL-4Rα fl/fl) mice were kindly provided by Frank Brombacher (University of Cape Town). All mice were housed under standard 12 h light:dark cycle conditions (lights on at 7:00am) in rooms with controlled temperature and humidity. They were given standard rodent chow and sterilized tap water ad libitum unless stated otherwise. Male mice at 10 to 20 weeks of age were used for behavior experiments unless stated otherwise. All experiments were approved by the Institutional Animal Care and Use Committee of the University of Virginia and/or of the Washington University in St Louis.
Method details
Open Field assay
Adult male mice were transferred to the testing room with background white noise at least one hour before the initiation of the experiment. Mice were tested during the light cycle and placed individually in an eight-chambered arena constructed of white acrylic (35 cm by 35 cm each chamber) by a blinded experimenter. Mice were allowed to freely explore for 10 minutes. Tracking of mouse behavior was done using the EthoVision XT Tracking system (Noldus) and the percentage of time spent in the center (22 cm by 22 cm) and general activity assessed.
Contextual fear conditioning (CFC) training
The equipment was purchased from Coulbourn Instruments. During training, the test cage was scented with 0.25% benzaldehyde. Each mouse was put in an isolation cubicle test cage. After 3 min of habituation in the test cage (context B), electric foot-shocks (2 sec, 0.50 mA) were delivered during 198–200 sec, 258–260 sec, 318–320 sec. Mice were kept in the test cage for additional 30 sec after training and were put back to their home cage. Animal groups were blinded to the observer and video images analyzed offline and manually scored. Tracking of the total distance travelled in the fear chamber was done using the EthoVision XT Tracking system (Noldus). Freezing was defined as an event where the entire mouse was sitting still.
Tamoxifen treatment
Mice were feed tamoxifen diet for 3 weeks. On day 1 and 4, mice injected with 40 mg/kg 4-hydroxytamoxifen in a 1:4 mixture of castor oil (Sigma, 259853): sunflower oil (Sigma, S5007) intraperitoneally. All mice were returned to normal chow for 2 weeks before fear conditioning.
Intra-cisterna magna injection of IL-4
Mice were anesthetized with 1.5% isoflurane. The skin of the neck was shaved and sterilized with iodine and 70% ethanol. The head of the mouse was gently secured in a stereotaxic frame, a skin incision made, and the muscle layers were retracted to expose the cisterna magna. Using a Hamilton syringe with a 33-gauge needle, 3 μl of 100 U of murine IL-4 (eBioscience) or saline was injected into the cisterna magna compartment at a rate of ~2 ul/min. The syringe was left in place for an additional 2 minutes to minimize backflow of CSF. The skin was sutured, and the mice were allowed to recover for at least 3 hours before any behavioral experiments.
T cell depletion
Mice were injected i.p. with 200 ug anti-CD4 (clone GK1.5, BioXCell) or isotype control antibody (clone LTF-2, rat IgG2b, BioXCell). To ensure continuous depletion, mice were injected i.p. with 100 ug weekly.
TRAP labelling of activated cells
FosCreER mice were crossed to Ai6 mice to generate double heterozygous (FosTRAP) mice used for the labelling experiments. The mice were injected in the intra-cisterna magna (i.c.m.) compartment with 100 U of murine IL-4 (eBioscience) diluted in saline at a total volume of 3 μl. As control, mice were injected with the same volume of saline. After two hours, the mice were given an intraperitoneal injection of 10 mg kg−1 of 4-hydroxytamoxifen (4-OHT; Sigma) dissolved in a 1:4 mixture of castor oil (Sigma): sunflower oil (Sigma). The drug preparation has been previously described (Guenthner et al., 2013). After one week, mice were sacrificed, and the brains were harvested for further analysis.
Seizure induction
Mice were injected with 25 mg per kg kainic acid (Sigma-Aldrich) intraperitoneally and seizure behavior was monitored. Animals progressed at least to stage 3 on a Racine scale (continuous head bobbing and forepaws shaking) and were scarified 1 d after seizure induction.
Construction of the pAAV-TRE-H2B-GFP plasmid
The University of Massachusetts Viral Vector core constructed the pAAV-TRE-H2B-GFP plasmid. The H2B-GFP fragment was amplified from the CMV-H2B-GFP (Addgene) plasmid using the following primers: H2B-GFP-Forward: 5’- gct gcg gaa ttg tac ccg cgg ccg atc cac cgg tcg cca cca tgg atg cca gag cca gcg-3’ and H2B-GFP-Reverse: 5’- gca cag tcg agg ctg atc agc gag ctc tag tcg acg gta tcg att tac ttg tac agc tcg tcc atg ccg aga gtg a-3’.. The amplified fragment was cloned into the pAAV-TRE-EGFP backbone after restriction enzyme digestion with NcoI and ClaI to replace the original GFP sequence with H2B-GFP instead.
Preparation of adeno-associated virus constructs
The following plasmids were purchased from Addgene: pAAV-cFos-tTA, and CMV-H2B-GFP (Addgen). The plasmids were packaged with AAV9 coat proteins in the Viral Vector Core of the Gene Therapy Center at the University of Massachusetts Medical School. AAV9-TRE-H2B-GFP was constructed by the University of Massachusetts Viral Vector core. Viral titers were 2×1013 genome copies (GC) mL−1 for AAV9-cFos-tTA, 1.5×1013 GC mL−1 for AAV9-TRE-H2B-GFP.
Immunohistochemistry
Mice were given a lethal dose of anesthesia by intraperitoneal (i.p.) injection of euthasol (10% v/v in saline). They were transcardially perfused with ice-cold PBS with heparin (10 U ml−1) followed by 4% paraformaldehyde (PFA). Brains were dissected and kept in 4% PFA overnight at 4°C. The fixed brains were washed with PBS, cryoprotected in 30% sucrose at 4°C until sunk to the bottom of the vial, and frozen in Tissue-Plus OCT compound (Thermo Scientific). Frozen brains were sliced into 40 μm-thick free-floating coronal sections using a cryostat (Leica). Brain sections were blocked with 1% bovine serum albumin (BSA), 2% normal serum (either goat or chicken), and 0.2% Triton X-100in PBS for 1h at room temperature (RT). The blocking step was followed by incubation with rabbit anti-NeuN-Alexa Fluor 647 or 488 (1:500, Abcam, clone EPR12763) in PBS containing 1% BSA and 0.2% Triton X-100 overnight at 4°C. Sections were washed two times for 15 min at room temperature (RT) with PBS and nuclei were stained with 4’, 6-Diamidino-2-phenylindole (DAPI, 1:10,000, Sigma-Aldrich) at RT for 15 min. The tissues were washed with PBS once and were mounted with Prolong Gold (ThermoScientific).
Single molecule multiplex fluorescent in situ hybridization with RNAscope
Euthanized mice were perfused with cold PBS containing heparin (10 U ml−1). Within 5 min, brains were embedded in Tissue-Plus OCT compound and were immediately frozen on dry ice. Coronal slices of 16 μm thickness were cut, mounted onto Superfrost plus slides and pretreated according to the manufacturer’s instructions for fresh frozen samples (RNAscope® Multiplex Fluorescent Reagent Kit v2 Assay). Briefly, sections were fixed in pre-chilled 10% neutral buffered formalin (Fisher Scientific) for 30 minutes at 4°C and were dehydrated using an ethanol series (50%, 70%, and 100% ethanol). The sections were pretreated with hydrogen peroxide for 10 min at RT and incubated with protease IV for 30 min at RT, provided in the kit. The hybridization and amplification steps were performed using Il4ra (Mm-IL4ra in C1), Gad1 (Mm-Gad1-C2), Gad2 (Mm-Gad2-C3), Cdkl5 (Mm-Cdkl5-C2), Rbfox3 (Mm-Cdkl5-C3), Vglut1 (Mm-Slc17a7-C3), Vglut2 (Mm-Slc17a6-C2) and Tmem119 (Mm-Tmem119-C2) RNAscope probes according to manufacturer’s instructions. Three-plex detection of Il4ra on glutaminergic and GABAergic neurons or microglia was achieved using Opal 520 (1:1000, Perkin Elmer), Opal 570 (1:1500, Perkin Elmer), and Opal 690 (1:1000, Perkin Elmer). Nuclei were stained with DAPI at RT for 30 sec and sections mounted with ProLong Gold antifade reagent (Invitrogen) were placed on the slide and covered with coverslips.
Confocal and widefield microscopy
Fluorescent images were acquired using a confocal microscope (Stellaris X8, Leica) or widefield slide scanner (VS200, Olympus), with a 20× objective with 0.70 NA. Quantitative analysis was performed using Fiji and CellProfiler by a blinded experimenter. Nuclei were segmented automatically in the DAPI channel and subsequently measured in each channel. For multiplex fluorescent image analysis, we used thresholding, watershed segmentation to identify and split overlapping nuclei. Adjust the size and circularity to count cells only and avoid counting fragments. Visually inspect all the detected particles and exclude the particles that do not look like a real signal. Expend the area of identified nuclei to a proportion and measure the puncta for target channel. For multiple channel measurement, repeat this method. For histocytometry, R was used and nuclear events were refined using size and DAPI mean fluorescent intensity, and cell type-specific markers (Gad1, Gad2, Vglut1, Vglut2, Tmem119) were plotted against Il4ra using ggplot2.
Stereotactic adeno-associated virus injections
Mice were anaesthetized by i.p. injection of ketamine (100 mg kg−1) and xylazine (10 mg kg−1) in saline solution. The head of the mouse was secured in a stereotaxic instrument with a digital display console (KOPF, Model 940) and a small hole made into the skull above the dorsal hippocampus or retrosplenial cortex. Viruses were injected using a glass micropipette (WPI, 1B120F-4) filled with mineral oil attached to Nanoliter Injector with SMARTouch Controller (WPI, NL2010MC2T). The needle was slowly lowered to the target brain region and was left in place for 5 min before starting the injection. The injection speed was set at 50 or 100 nl min-1.
For region-specific deletion of IL4Rα, IL-4Rα fl/fl mice received bilateral stereotactic injection of 1011 GC of AAV9-hSyn1-IRES-eGFP-WPRE or pENN.AAV.hSyn.HI.eGFP-Cre.WPRE.SV40 virus (300 nl per site) at a rate of 150 nl/min. For experiments targeting the hippocampus, mice were injected bilaterally in −2.06 anterior-posterior (AP), 1.33 medio-lateral (ML), and lowered 1.9 (DV). Retrosplenial cortex coordinates were −2.6 anterior-posterior, 0.25 medio-lateral, and lowered 1.0.
For activity labeling of neurons, mice were put on doxycycline food (ENVIGO, 40 mg kg−1) for 1 week. The mice were injected bilaterally with 300 nl of a 1:1 mixture of AAV9-cFos-tTA and AAV9-TRE-H2B-GFP into the retrosplenial cortex (AP: −2.06 mm; ML: ±0.4 mm; DV: 1.0 mm) or dentate gyrus (AP: −2.06 mm; ML: ±1.3 mm; DV: 1.9 mm). Two weeks after virus injection, mice were taken off Dox and were given standard rodent chow for 24 hours before the CFC training. After training, the mice were given doxycycline food. Twenty-four hours after training, mice were prepared for single nuclei sequencing.
After injection, the needle was left in place for an additional 10 min to minimize backflow. The mice were then injected subcutaneously with ketoprofen (2 mg kg−1) and allowed to recover on a heat pad until fully awake.
Field potential recording from dentate gyrus
After deeply anesthetized with isoflurane (Henry Schein, USA), mouse was quickly decapitated, and the head was dipped into ice-cold artificial cerebrospinal fluid (aCSF) briefly. The brain was taken out by swift dissection and submerged into ice-cold aCSF that was bubbled continuously with 5% CO2/95% O2. The aCSF was constituted as (in mM) NaCl 124.0, KCl 2.5, KH2PO4 1.2, CaCl2 2.4, MgSO4 1.3, NaHCO3 26.0 and glucose 10.0 (pH 7.4). The right hippocampus was taken out and cut into slices (400 μm thick) along its longitude axis with posterior part first using a tissue slicer (Stoelting Co., IL, USA). The left hippocampus was not used in this study. After at least 1 hour of incubation at room temperature in aCSF bubbled continuously with 5% CO2/95% O2, slices were recorded in submerged mode on the top of a chamber (Harvard Apparatus, USA) at room temperature with illumination underneath (Leica, Germany). Data were collected with an Axopatch- 2B amplifier and pClamp 10.4 program (Molecular Devices, US) through Digidata 1320A. Slices were perfused continuously with aCSF bubbling with 5% CO2/95% O2 at flow rate about 2 ml/min using two channels peristaltic pump (Dynamax, Rainin, USA). The recording electrode (resistance 1–3 MΩ) was fabricated with capillary glass tubing (Warner Instruments, Model No: G150F-3) and filled with aCSF. Biphasic current pulses (0.2 ms duration for one phase, 0.4 ms in total) were delivered in 10 s intervals using a concentric bipolar stimulating electrode (CBARC75, FHC, USA) through an isolator (ISO-Flex, AMPI, Israel). To record field population spikes (PS) in dorsal dentate gyrus (DG), the recording electrode was placed on inner part of granular cell layer and the stimulating electrode was placed right above hippocampal fissure to stimulate the bypassing perforant pathway fibers. To record field excitatory postsynaptic potential (EPSP) in DG the recording electrode was placed on the middle stratum above granular cell layer and stimulating electrode was placed above the hippocampal fissure.
Input/output curve was obtained for each slice by increasing intensities of 1 threshold (T, typically 0.02mA in CA1 and 0.05 mA in DG). The maximum intensity did not pass 1mA. Slices under following conditions were discarded to avoid experimental artifacts: 1) slices could not reach 0.5 mV/ms of the slope at the maximum input, 2) slices showing responses that were neither monotonic nor sigmodial shape, 3) slices showing non-zero postsynaptic responses when we give a stimulus close to zero. Initial Slope-Pop spike relationships were fitted y = a*(1-exp(−x/b)) + C graph.
To measure long-term potentiation, test pulse intensities were adjusted to evoke EPSP about 35–50% of maximal response. Theta burst (12 trains of 4 pulses at 100 Hz, delivered at 5 Hz, repeated three times at 0.1 Hz) and the amplitude of population spike and slope of EPSP were measured from the initial phase of negative wave with pClamp 10.4. Each data point was the average of 3 consecutive traces. LTP was plotted as the percentage to 10 min recording of baseline following high frequency stimulation. Representative trace is the average of 6 consecutive ones which last for 1 minute in span.
Patch-clamp recordings from dentate gyrus
For patch clamp recordings, C57BL/6J or B6.129S-Gad2tm1.1Ksvo/J mice at postnatal day 10 to 22 of both sexes were used and mice (wild type C57BL/6J or B6.129S-Gad2tm1.1Ksvo/J, specified otherwise, both sexes, at 10 – 22 postnatal days old, P10–P22), were decapitated under anesthesia with isoflurane. The animal’s brain was rapidly dissected and transferred to ice-cold (~4°C) oxygenated sucrose-substituted artificial cerebrospinal fluid (aCSF) solution. Coronal brain slices 250-μm thick were prepared with a Vibratome (Leica VT1000S) in oxygenated cold sucrose-substituted aCSF. The slices were transferred into an incubation chamber with normal aCSF, composing of (in mM) 130 NaCl, 1.2 NaH2PO4, 2.4 CaCl2, 2.5KCl, 1.3 MgSO4, 26 NaHCO3, 10 dextrose (pH 7.30, ~300 mOsm, oxygenated at 95% O2/5% CO2), for at least 1h at room temperature (25 ± 2 °C) prior to electrophysiological studies. Individual slices were transferred to a submersion recording chamber and were continuously perfused with 95% O2/5% CO2 bubbled with the aCSF. Recordings were made from neurons in dentate gyrus using a MultiClamp 700B amplifier (Molecular Devices, Sunnyvale, CA, USA). mIPSC or mEPSC recordings of hippocampal dentate gyrus neurons were performed for 10 min as baseline. The chamber was perfused with 100 ng/ml recombinant IL-4 (eBioscience) and the same neurons recorded for another 10 min. Additional 10 min were recorded under continuous perfusion with aCSF and are indicated as washout. Signals were low-pass filtered at 1 kHz and sampled at 10 kHz using Digidata 1440A (Molecular Devices). For mEPSC recordings, glass pipettes (resistance, 3–5 mΩ) were loaded with internal solution containing (in mM): 110 K-gluconate, 20 KCl, 20 HEPES, 5 MgCl2, 0.6 EGTA, 2 MgATP, 0.2 Na3GTP (pH 7.3, ~290 mOsm), and all neurons were held at −60 mV in voltage-clamp mode; 10 μM bicuculline methiodide and 1 μM tetrodotoxin (TTX) were added to aCSF to block GABAA receptor-mediated and voltage-gated Na+ currents, respectively. For mIPSC recordings, internal solution containing (in mM): 140 Cs-gluconate, 1 MgCl2, 11 EGTA, 1CaCl2 and 10 HEPES (pH 7.3, ~290 mOsm) was used, and cells were held at 0 mV in voltage-clamp mode; NBQX (30μM) and TTX (1 μM) were added to block AMPA receptor-mediated and Na+ currents, respectively. The spike frequency versus injected current experiments were performed in current-clamp mode. The internal solution contained (in mM): 110 K-gluconate, 20 KCl, 20 HEPES, 5 MgCl2, 0.6 EGTA, 2 MgATP, 0.2 Na3GTP (pH 7.3, 290 mOsm). The excitatory and inhibitory synaptic blockers (50 μM D-APV, 30 μM NBQX, and 10 μM bicuculline methiodide) were added to the aCSF. The action potential firing rate was averaged from 10 depolarizing current injections (500 ms in duration at an interval of 2 s). The maximum current injected in each experiment was below the current that induced spike frequency adaptation.
For spontaneous EPSC and IPSC measurements, 3-month-old male C57BL/6 mice were used. Heads were decapitated under anesthesia with isoflurane, brains rapidly dissected and transferred to ice-cold oxygenated sucrose-substituted artificial cerebrospinal fluid (aCSF) solution. Coronal brain slices of 300μm thickness were prepared with a Vibratome (Leica VT1200S) in oxygenated cold dissection buffer containing (in mM) 212 sucrose, 25 NaHCO3, 5 KCl, 1.25 NaH2PO4, 0.5 CaCl2, 3.5 MgSO4, 10 DGlucose, 1.25 L-ascorbic acid and 2 Na-pyruvate. Slices were transferred into an incubation chamber with normal aCSF, composing of (in mM) 125 NaCl, 1.25 NaH2PO4, 2.5 CaCl2, 2.5 KCl, 1.3 MgCl2, 25 NaHCO3 and 10 D-glucose, and bubbled with 95% O2/5% CO2. After 20 minutes of recovery at 32 °C, slices were incubated at least 30 minutes at room temperature (25 ± 2 °C) prior to electrophysiological studies. A patch of membrane is subsequently ruptured by mild suction, monitored for 5 minutes, and EPSC (by holding −70 mV) and IPSC (by holding 0 mV) recorded in sequence. To check the direct effect of IL-4, the bath inlet was changed to aCSF containing 100 ng/ml IL-4 (R&D Systems), while the outlet was flowed through. After 5 minutes, the outlet bath was changed into IL-4 containing aCSF. After an additional 5 minutes, EPSC, and IPSC were recorded. Access resistance (Ra) was maintained during the whole experimental sequence and cells with an Ra change of more than 5 MΩ were discarded. The perfusion system was washed with aCSF for at least 10 minutes before soaking the next slice.
Adoptive transfer of T cells
Spleens, inguinal and cervical lymph nodes were collected. They were homogenized, and the cell suspension was passed through a 70-um cell strainer (Fisher Scientific). The red blood cells were lysed using ammonium-chloride-potassium (ACP) lysis buffer (Quality Biological) for 4 min at RT. The cells were washed once with RPMI (Gibco) supplemented with 5% fetal bovine serum (FBS). The single cell suspension was counted, pelleted, and adjusted so that the cell concentration was 1×108 cells per ml in sterile saline. At 4 weeks of age, SCID mice were reconstituted with 5×106 splenocytes or were given saline by intravenous (i.v.) injection. For T cell transfer, T cells were isolated from single-cell suspension from wild-type and IL-4–/– mouse spleens using the Pan T cell Isolation Kit II, LS columns, and a QuadroMACS Separator (Miltenyi Biotech) for manual cell enrichment. Purified T cells were resuspended in saline, and 2×106 cells (at a final volume of 100 μL) or sterile saline were injected into SCID recipients.
Flow cytometry
Mice were given a lethal dose of anesthesia by intraperitoneal (i.p.) injection of Euthasol (10% v/v in saline) and were transcardially perfused with ice-cold PBS containing 10 U mL−1 heparin. The spleen and dural meninges were dissected and digested in 1 mg mL−1 collagenase 8, D and 5 U mL−1 DNase I (Sigma Aldrich) at 37°C for 30 min. The digestion was stopped by adding EDTA to a final concentration of 10 mM. The cell suspension was passed through a 70-um cell strainer. Cells were washed with RPMI (Gibco), resuspended in PBS, and counted with a cell counter (Nexcelom). Cells were stained with Zombie Aqua™ dye (Biolegend) for 20 min on ice. Cells were washed with PBS containing 2% FBS (FACS buffer), and Fc-receptors were blocked with anti-CD16/32 antibody in 50 μl total volume. An equal volume of primary antibody mix was added containing anti-CD45.2-Alexa Fluor 700, anti-CD4-eFluor450, anti-CD8a-PE, anti-CD90.2-PE-Cy7, and anti-TCRβ-FITC. Cells were stained for 30 min on ice and washed twice with FACS buffer. The data was acquired on a flow cytometer (Gallios Beckman Coulter) and analyzed using FlowJo software (Tree Star, Inc.) or Cytobank Premium.
Single-nuclei isolations and RNA sequencing
Brains from mice injected with AAV9-cFos-tTA and AAV9-TRE-H2B-GFP in the retrosplenial cortex or dentate gyrus were rapidly extracted after perfusion with ice-cold artificial cerebrospinal fluid (aCSF). The brains were sectioned in aCSF with a vibratome (Leica) to obtain 500 um-thick slices. The retrosplenial cortex or dentate gyrus was micro-dissected, and tissues were pooled from 3–4 mice of the same group and experimental condition. The sections were then placed into working nuclei isolation medium (NIM; 0.25 M sucrose, 25 mM potassium, 5 mM magnesium chloride, 10 mM TrisCl, 100 mM dithiothreitol, 1x protease inhibitor, RNase inhibitor) and triturated with a 1000 μl wide-bore pipette tip. After adding 0.1% Triton-X-100, samples were homogenized using a 2 ml Dounce homogenizer with pestle A, followed by pestle B (Sigma), and filtered through a 70-um cell strainer (Fisher Scientific). The homogenate was spun at 1000 g for 8 min at 4oC, pellets, washed once, and resuspended in 500 μl 1% BSA in PBS for staining. Dissociated nuclei were stained by adding Hoechst 33342 (Invitrogen, 1:1000) and anti-NeuN-Alexa Fluor 647 antibody (Abcam, 1:500, ab190565, clone EPR12763) for 15 min on ice. Samples were then washed and resuspended in 1% BSA in PBS for FACS sorting using the Influx cell sorter (BD Biosciences). Single activity-labeled neuronal nuclei (Hoechst+ NeuN+ GFP+) or non-labeled nuclei (Hoechst+ NeuN+ GFP–) were directly deposited into 1.5 ml tubes containing 0.04% non-acetylated BSA in PBS, and diluted to 1000 nuclei per ul estimated from counting on a hemocytometer and Trypan blue staining. The sorted cortical neuronal nuclei (~2000) per sample were loaded onto a 10X Genomics Chromium platform to generate cDNAs carrying cell- and transcript-specific barcodes and sequencing libraries constructed using the Chromium Single Cell 3’ Library & Gel Bead Kit 2. Libraries were sequenced on the Illumina NextSeq using paired-end sequencing, resulting in 100,000 reads per cell.
Data analysis for single nuclei secquencing
WT and SCID with IL-4+/+ or IL-4−/− splenic T cell transfer single-nuclei analysis
Data Pre-processing.
Base call files were converted to Cellranger compatible Fastq files using the Illumina Bcl2fastq2 software. A reference pre-mRNA genome was created using the mkref functionality in the Cellranger software using the mm10 genome Fastq file and an edited GTF file with the feature type for each transcript set to exon to bypass the Cellranger exon filtering and to include intronic counts for further analysis. Reads were then aligned to the mm10 pre-mRNA genome using the count function of the Cellranger software pipeline (version 3.0.2) provided by 10x genomics. Base call files were converted to Cellranger compatible Fastq files using the Illumina Bcl2fastq2 software. A reference pre-mRNA genome was created using the mkref functionality in the Cellranger software using the mm10 genome Fastq file and an edited GTF file with the feature type for each transcript set to exon to bypass the Cellranger exon filtering and to include intronic counts for further analysis. Reads were then aligned to the mm10 premRNA genome using the count function of the Cellranger software pipeline (version 3.0.2) provided by 10x genomics. The resulting filtered gene by cell matrices of UMI counts for each sample were read into R using the read10xCounts function from the Droplet Utils package. Filtering was applied in order to remove low quality cells, excluding those with reads fewer than 2,000 or greater than 60,000, or unique genes fewer than 500 or greater than 8,000, or greater than 15% mitochondrial gene expression. Expression values for the remaining cells were then normalized using the scran and scater packages and the resulting log2 values were transformed to the natural log scale for compatibility with the Seurat (v3) pipeline (Butler et al., 2018; Lun et al., 2016; McCarthy et al., 2017).
Dimensionality Reduction and Clustering.
The filtered and normalized matrix was used as input to the Seurat pipeline and cells were scaled across each gene before the selection of the top 2,000 most highly variable genes using variance stabilizing transformation. Principal Components Analysis was conducted, and an elbow plot was used to select the first six principal components for tSNE analysis and clustering. Shared Nearest Neighbor (SNN) clustering optimized with the Louvain algorithm, as implemented by the Seurat FindClusters function was performed before manual annotation of clusters based on expression of canonical gene markers. Cell types not of interest to the analysis, namely glia, were removed and the remaining neuronal clusters were re-scaled, re-clustered, and identified as above. Clusters were then collapsed based on common cell types to result in two clusters.
Differential Expression.
For analysis of differentially expressed genes between conditions, Glutamatergic neurons were isolated and filtered to include genes that had at least 5 transcripts in at least 5 cells, then the top 2000 highly variable genes were determined and included for further analysis using the SingleCellExperiment modelGeneVar and getTopHVGs functions. After filtering, observational weights for each gene were calculated using the ZINB-WaVE zinbFit and zinbwave functions (Berge et al., 2018). These were then included in the edgeR model, which was created with the glmFit function, by using the glmWeightedF function (Robinson et al., 2009). Results were then filtered using a Benjamini-Hochberg adjusted p-value threshold of less than 0.05 as statistically significant. Volcano plots were created with the EnhancedVolcano package in R (Blighe et al., 2020).
Pathway Enrichment.
Over representation enrichment analysis with Fisher’s Exact test was used to determine significantly enriched Gene Ontology (GO) terms (adj. p < 0.05) for the sets of significantly differentially expressed genes between conditions. For each gene set, genes were separated into up- and down-regulated and separately (Hong et al., 2013) the enrichGO function from the clusterProfiler package was used with a gene set size set between 10 and 500 genes and p-values adjusted using the Benjamini-Hochberg correction (Yu et al., 2012, 2015).
Gad2Cre+::IL-4Rafl/fl and Gad2Cre-::IL-4Rafl/fl single-nuclei analysis
Data Pre-processing.
Base call files were converted to Cellranger compatible Fastq files using the Illumina Bcl2fastq2 software. A reference pre-mRNA genome was created using the mkref functionality in the Cellranger software using the mm10 genome Fastq file and an edited GTF file with the feature type for each transcript set to exon to bypass the Cellranger exon filtering and to include intronic counts for further analysis. Reads were then aligned to the mm10 pre-mRNA genome using the count function of the Cellranger software pipeline (version 3.0.2) provided by 10x genomics. The resulting filtered gene by cell matrices of UMI counts for each sample were read into R using the read10xCounts function from the Droplet Utils package. Filtering was applied in order to remove low quality cells, excluding those with reads fewer than 2,000 or greater than 60,000, or unique genes fewer than 500 or greater than 8,000, or greater than 15% mitochondrial gene expression. Expression values for the remaining cells were then normalized using the scran and scater packages and the resulting log2 values were transformed to the natural log scale for compatibility with the Seurat (v3) pipeline (Butler et al., 2018; Lun et al., 2016; McCarthy et al., 2017).
Dimensionality Reduction and Clustering.
The filtered and normalized matrix was used as input to the Seurat pipeline and cells were scaled across each gene before the selection of the top 2,000 most highly variable genes using variance stabilizing transformation. Principal Components Analysis was conducted and an elbow plot was used to select the first six principle components for tSNE analysis and clustering. Shared Nearest Neighbor (SNN) clustering optimized with the Louvain algorithm, as implemented by the Seurat FindClusters function was performed before manual annotation of clusters based on expression of canonical gene markers. Cell types not of interest to the analysis, namely glia, were removed and the remaining neuronal clusters were re-scaled, re-clustered, and identified as above. Clusters were then collapsed based on common cell types to result in two clusters.
Differential Expression.
For analysis of differentially expressed genes between conditions, Glutamatergic neurons were isolated and filtered to include genes that had at least 5 transcripts in at least 5 cells, then the top 2000 highly variable genes were determined and included for further analysis using the SingleCellExperiment modelGeneVar and getTopHVGs functions. After filtering, observational weights for each gene were calculated using the ZINB-WaVE zinbFit and zinbwave functions (Berge et al., 2018). These were then included in the edgeR model, which was created with the glmFit function, by using the glmWeightedF function (Robinson et al., 2009). Results were then filtered using a BenjaminiHochberg adjusted p-value threshold of less than 0.05 as statistically significant. Volcano plots were created with the EnhancedVolcano package in R (Blighe et al., 2020).
Pathway Enrichment.
Over representation enrichment analysis with Fisher’s Exact test was used to determine significantly enriched Gene Ontology (GO) terms (adj. p < 0.05) for the sets of significantly differentially expressed genes between conditions. For each gene set, genes were separated into up- and down-regulated and separately (Hong et al., 2013) the enrichGO function from the clusterProfiler package was used with a gene set size set between 10 and 500 genes and p-values adjusted using the Benjamini-Hochberg correction (Yu et al., 2012, 2015).
Data analysis of Publicly Available Datasets
The publicly available RiboTag data was downloaded from the GEO portal (GSE133291) and expression levels of genes of interest were replotted using ggplot2 (PMID:31451803). Publicly available count data (GSE92522) was downloaded from the Gene Expression Omnibus website and reanalyzed in R using the scran and scater packages for normalization and the Seurat workflow for further analysis including scaling, principal component analysis, clustering and tSNE. Violin plots were generated using the Seurat package and the percentage of Il4ra expression was calculated within each of the subpopulations identified in the provided metadata sample guide (PMID:2894923).
The level 6, taxonomy level 2 dataset for all CNS neurons was downloaded from the mousebrain.org website in loom format and analyzed in python to extract count information for cells isolated from the Dentate Gyrus, CA1 hippocampus (CA1), Hippocampus (HC), Cortex (Ctx1, Ctx1.5, Ctx2, and Ctx3), the SS Cortex, Amygdala, Thalamus, and Hypothalamus as well as information about the Linnarsson identified cluster to which each cell belongs (Zeisel et al., 2018). Within each region the percentage of Il4ra expressing cells (counts greater than 0) was calculated. Additionally, for each Linnarson identified cluster within each region, the percentage of Il4ra positive cells was determined and regions of interest were visualized with pie charts.
PCR for assaying IL-4Ra knockout mice
Whole hippocampi or cortical tissue from Il4rafl/fl mice was dissected from perfused mice and frozen on dry ice. Tissue was dissociated in 1 ml TRIzol reagent using silica beads and a homogenizer (Mini-beadbeater, Biospec Products). Total RNA was extracted from tissue using a RNA mini-prep kit (RNeasy kit, Qiagen) or from isolated cells using a micro-prep kit (RNAqueous-Micro Total RNA Isolation kit, Invitrogen) and converted into cDNA (High-Capacity cDNA Reverse Transcription kit, Applied Biosystems). One microliter of 50 ng/ul cDNA was diluted in 9 ul SyBR green reaction volume (iTaq Universal Supermix, BioRad) and gene expression levels of Il4ra and GAPDH were determined by qRTPCR (6). The following primers were used: Il4ra 5’-TCT GCA TCC CGT TGT TTT GC-3’ and 5’-GCA CCT GTG CAT CCT GAA TG-3’; GAPDH 5’-GGT CCT CAG TGT AGC CCA AG-3’ and 5’-ACC CAG AAG ACT GTG GAT GG-3’ (Integrated DNA Technologies).
Statistical Analysis
GraphPad Prism version 9.2.0, OriginPro 7.5 and MATLAB R2017b software was used for statistical analysis. Values are presented as dot plots with individual data points, violin plots or boxes and whiskers with mean interquartile range, the minimum, maximum and mean ± s.e.m. as indicated in the figure legends. N indicates the number of animals unless otherwise specified. For comparisons between two groups or more, data analyzed by t-test (Mann-Whitney or Student’s, two-tailed, unpaired). To test whether the response of neurons to cytokine differed in amplitude or frequency, two-tailed paired t-tests were applied. For comparisons of three groups or more data were analyzed by one- or two-way ANOVA followed by Tukey, Bonferroni or Dunn’s multiple comparisons post-hoc test, as appropriate. P value <0.05 were considered statistically significant unless Log2FC criteria were also considered, as specified in the figure legends. Experiments reported in this manuscript were repeated two to four times, using mice from different generations. Experimenters were blinded during data acquisition and analysis of the subject treatment and conditions.
Supplementary Material
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-CD16/32, mouse clone 93, purified | Biolegend | 101302 |
| anti-CD4, mouse clone RM4–5, eFluor450 | eBioscience | 48–0042 |
| anti-CD45.2, mouse clone Ly-5.2, Alexa Fluor 700 | Biolegend | 109822 |
| anti-CD8a, mouse clone 53–6.7, PE | eBioscience | 12–0081 |
| anti-CD90.2, mouse clone 53–2.1, PE-Cyanine7 | eBioscience | 25–0902 |
| anti-NeuN, monoclonal clone EPR12763, Alexa Fluor | Abcam | ab190195 |
| 4nti-NeuN, monoclonal clone EPR12763, Alexa Fluor | Abcam | ab190565 |
| 6nti-TCR-β, mouse clone 53–2.1, FITC | Biolegend | 109206 |
| InVivoMAb anti-CD4, rat monoclonal mouse clone | BioXCell | BE0003 |
| InVivoMAb rat IgG2b isotype control; anti-keyhole | BioXCell | BP0090 |
| Virus strains and plasmids | ||
| AAV9-cFos-tTA | this paper | N/A |
| AAV9-hSyn1 -IRES-eGFP-WPRE | Vectorbiolabs | N/A |
| AAV 9-TRE-H2B-GFP | this paper | N/A |
| CMV-H2B-GFP | Addgene | 11680 |
| pAAV-cFos-tTA | Addgene | 66794 |
| pAAV -TRE-EGFP | Addgene | 89875 |
| pENN.AAV.hSyn.HI.eGFP-Cre.WPRE.SV40 | Addgene | 105540-AAV9 |
| Chemicals, peptides, and recombinant proteins | ||
| (+) bicuculline | Sigma | 14340 |
| 4-hydroxytamoxifen | Sigma | H6278 |
| 6-Diamidino-2-phenylindole | Sigma-Aldrich | D9542 |
| ACK Lysing Buffer | Quality Biological | 118–156-101CS |
| artificial cerebrospinal fluid | Harvard Apparatus | 597316 |
| Benzaldehyde | Sigma-Aldrich | B1334 |
| Castor oil | Sigma | 259853 |
| Cs-gluconate | Hello Bio | HB4822 |
| D-AP5 | Tocris | 106 |
| Deoxyribonuclease I | Sigma-Aldrich | DN25–1G |
| DNQX | Sigma | D0540 |
| Doxycycline diet (40 mg / kg) | Envigo | TD.120240 |
| EGTA | Sigma | E3889 |
| Formalin, Phosphate Buffered (10%) | Fisher Scientific | SF100–4 |
| Heparin Sodium | Fisher Scientific | H19 |
| Hoechst 33342 | Sigma-Aldrich | 14533 |
| Interleukin-4, recombinant murine protein | eBioscience | 14–8041-80 |
| K-gluconate | Sigma | P1847 |
| Kainic acid | Sigma-Aldrich | 420318 |
| LS columns | Miltenyi Biotec | 130–042-401 |
| Opal 520 Reagent Pack | Perkin Elmer | FP1487001KT |
| Opal 570 Reagent Pack | Perkin Elmer | FP1488001KT |
| Opal 690 Reagent Pack | Perkin Elmer | FP1497001KT |
| Optiprep density gradient medium (Iodixonoal) | Sigma-Aldrich | D1556 |
| Papain, Suspension | Worthington | LS003126 |
| Paraformaldehyde, Aqueous, EM grade (16%) | Fisher Scientific | 50–980-487 |
| ProLong Gold | Thermo Fisher | P36934 |
| Proteose Inhibitor Tablets, Complete Mini, EDTA-free | Thermo Fisher | 4693159001 |
| RNAscope Target Probe EGFP-O3 | Sigma-Aldrich | 492011 |
| RNAscope Target Probe Mm-Cdkl5-C2 | Advanced Cell | 500851-C2 |
| RNAscope Target Probe Mm-Fos-C2 | Advanced Cell | 316921-C2 |
| RNAscope Target Probe Mm-Gad1-C2 | Advanced Cell | 400951 |
| RNAscope Target Probe Mm-Gad2-C3 | Advanced Cell | 439371-C3 |
| RNAscope Target Probe Mm-IL4ra-C1 | Advanced Cell | 520171 |
| RNAscope Target Probe Mm-Rbfox3-C3 | Advanced Cell | 313311-C3 |
| RNAscope Target Probe Mm-Sall1-C3 | Advanced Cell | 469661-C3 |
| RNAscope Target Probe Mm-Slc17a6-C2 | Advanced Cell | 319171-C2 |
| RNAscope Target Probe Mm-Slc17a7-C3 | Advanced Cell | 416631 |
| RNAscope Target Probe Mm-Tmem119-C2 | Advanced Cell | 472901-C2 |
| Sunflower oil | Sigma | S5007 |
| Superase-In | Invitrogen | AM2696 |
| Tamoxifen diet (250 mg / kg) | Envigo | TD.130856 |
| Tetrodotoxin citrate | Hello Bio | HB1035 |
| Tissue-Plus O.C.T. Compound, clear | Thermo Fisher | 23–730-571 |
| TRIzol reagent | Thermo Fisher | 15596026 |
| Zombie Aqua Fixable Viability dye | Biolegend | 423102 |
| Critical commercial assays | ||
| Chromium Single Cell 3’ Library & Gel Bead Kit v2 | 10x Genomics | GN- B20237 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | 4374966 |
| iTaq Universal SYBR Green Supermix | Bio-Rad | 1725121 |
| Pan T cell Isolation Kit II, mouse | Miltenyi Biotec | 130–095-130 |
| RNAqueous-Micro Total RNA Isolation kit | Invitrogen | AM1931 |
| RNAscope Multiplex Fluorescent Reagent Kit v2 | Advanced Cell | 323110 |
| RNeasy Mini kit | Qiagen | 74106 |
| Deposited data | ||
| Fastq files and quantified gene counts for single nuclei | Gene Expression | GSE143183 |
| Experimental models: Organisms/strains | ||
| Mus musculus/C57BL/6J | The Jackson | 000664 |
| C57BL/6-Il4tm1Nnt/J (IL-4-/-) | The Jackson | 002518 |
| B6.Cg-Prkdcscid/SzJ (SCID) | The Jackson | 001913 |
| B6.129P2(Cg)-Cx3cr1tm2.1(cre/ERT2)Litt/WgadJ | The Jackron | 021160 |
| B6.Cg-Tg(Synl-cre)671Jxm/J (Syn-cre) | The Jackron | 003966 |
| The Il4ratm2Fbb (IL-4ra fl/fl) | Frank Brombacher, | |
| B6J.Cg-Gad2tm2(cre)Zjh/MwarJ (Gad2-cre) | The Jackson | 028867 |
| B6J.129S6(FVB)-Slc17a6tm2(cre)Lowl/MwarJ (Vglut2- | Hie Jackron | 028863 |
| B6.129(Cg)-Fostml.l(cre/ERT2)Luo/J (FosCreET) | Hie Jackron | 021882 |
| B6.Cg-Gt(ROSA)26Sortm6(CAGZsGreen1)Hze/J (Ai6) | Hie Jackron | 007906 |
| B6;129S-Gad2tm1.1Ksvo/J (Gad2-T2a-NLS-mCherry) | Hie Jackron | 023140 |
| Software and algorithms | ||
| CellProfiler v4.2.1 | Broad Institute | https://cellprofiler.or |
| Cellranger software pipeline v2.20 and v.4.0.0 | 10X Genomics | https://support.10xge |
| Cytobank | Beckman Coulter | https://www.cytoban |
| Fiji package for ImageJ | NIH | https://imagej.net/Fij |
| FlowJo v10.3 | Tree Star | https://www.flowjo.c |
| Illumina Bcl2fastq software g | Illumina | https://support.illumi |
| ImageJ | NIH | https://imagej.nih.go |
| MATLAB R2017b | MathWorks | https://mathworks.co |
| OriginPro v7.5 | OriginLab | https:/www.orginla |
| Prism v9.2.0 | GraphPad | https://www.graphpa |
| R Studio v3 | R Studio | https://rstudio.com/ |
| Seurat pipeline for R studio v.2.3.4 and 3.0 | Butler et al., 2018 | https://satijalab.org/s |
| Other | ||
| Axopatch- 2B amplifier | Molecular Devices | https://www.molecul |
| Capillary glass tubing | Warner Instruments | G150F-3 |
| Concentric bipolar stimulating electrode | FHC | CBATC75 |
| Digidata 1320A | Molecular Devices | https://www.molecul |
| Digidata 1440A | Molecular Devices | https://www.molecul |
| Isolator | AMPI | ISO-Flex |
| Mini-Beadbeater | Biospec Products | C321001 |
| MultiClamp 700B amplifier | Molecular Devices | https://www.molecul |
| pClamp 10.4 program | Molecular Devices | https://www.molecul |
| Peristaltic pump | Rainin | RP-1 |
| Vibratome | Leica | VT1200S |
Highlights.
Depletion of CD4 T cell disrupts contextual fear memory in mice
T-cell derived IL-4 rescues memory deficits and synapse function in SCID mice
IL-4Rα depletion on GABAergic neurons is sufficient to impair contextual fear memory
IL-4 delivery into the cerebrospinal fluid ameliorates memory deficits in SCID mice
Acknowledgements:
We would like to thank S. Smith for editing the manuscript. We thank all the members of the Kipnis lab for their valuable comments and the staff of the University of Virginia Flow Cytometry Core for help with cell sorting and the Genome Analysis and Technology Core for library preparations and sequencing. This work was supported by grants from the National Institutes of Health (AT010416, AG034113, NS096967 and AG057496) to J.K.
Footnotes
Declaration of interests: J.K. is a member of a scientific advisory group for PureTech. XSX, BZ and NY are employed by AfaSci.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baruch K, Deczkowska A, David E, Castellano JM, Miller O, Kertser A, Berkutzki T, Barnett-Itzhaki Z, Bezalel D, Wyss-Coray T, et al. (2014). Aging. Aging-induced type I interferon response at the choroid plexus negatively affects brain function. Science 346, 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird CM, and Burgess N (2008). The hippocampus and memory: insights from spatial processing. Nature Reviews Neuroscience 9, 182–194. [DOI] [PubMed] [Google Scholar]
- Brombacher TM, Ajonijebu DC, Scibiorek M, Berkiks I, Moses BO, Mpotje T, and Brombacher F (2021). IL-4Ralpha deletion disrupts psychomotor performance and reference memory in mice while sparing behavioural phenotype associated with spatial learning. Brain Behav Immun 92, 157164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brombacher TM., Berkiks I., Pillay S., Scibiorek M., Moses BO., and Brombacher F. (2020). IL-4R alpha deficiency influences hippocampal-BDNF signaling pathway to impair reference memory. Sci Rep 10, 16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brombacher TM, Nono JK, De Gouveia KS, Makena N, Darby M, Womersley J, Tamgue O, and Brombacher F (2017). IL-13–mediated regulation of learning and memory. The Journal of Immunology 198, 2681–2688. [DOI] [PubMed] [Google Scholar]
- Brown MA, Pierce JH, Watson CJ, Falco J, Ihle JN, and Paul WE (1987). B cell stimulatory factor-1/interleukin-4 mRNA is expressed by normal and transformed mast cells. Cell 50, 809–818. [DOI] [PubMed] [Google Scholar]
- Burgess N, Maguire EA, and O’Keefe J (2002). The human hippocampus and spatial and episodic memory. Neuron 35, 625–641. [DOI] [PubMed] [Google Scholar]
- Castellano JM, Mosher KI, Abbey RJ, McBride AA, James ML, Berdnik D, Shen JC, Zou B, Xie XS, Tingle M, et al. (2017). Human umbilical cord plasma proteins revitalize hippocampal function in aged mice. Nature 544, 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Itakura E, Nelson GM, Sheng M, Laurent P, Fenk LA, Butcher RA, Hegde RS, and de Bono M (2017). IL-17 is a neuromodulator of Caenorhabditis elegans sensory responses. Nature 542, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Kim JJ, Thompson RF, and Tonegawa S (1996). Hippocampal lesions impair contextual fear conditioning in two strains of mice. Behavioral neuroscience 110, 1177. [DOI] [PubMed] [Google Scholar]
- Choi GB, Yim YS, Wong H, Kim S, Kim H, Kim SV, Hoeffer CA, Littman DR, and Huh JR (2016). The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 351, 933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cugurra A., Mamuladze T., Rustenhoven J., Dykstra T., Beroshvili G., Greenberg ZJ., Baker W., Papadopoulos Z., Drieu A., Blackburn S., et al. (2021). Skull and vertebral bone marrow are myeloid cell reservoirs for the meninges and CNS parenchyma. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Mesquita S, Fu Z, and Kipnis J (2018). The Meningeal Lymphatic System: A New Player in Neurophysiology. Neuron 100, 375–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima KA, Rustenhoven J, Da Mesquita S, Wall M, Salvador AF, Smirnov I, Cebinelli GM, Mamuladze T, Baker W, and Papadopoulos Z (2020). Meningeal γδ T cells regulate anxiety-like behavior via IL-17a signaling in neurons. Nature immunology 21, 1421–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derecki NC, Cardani AN, Yang CH, Quinnies KM, Crihfield A, Lynch KR, and Kipnis J (2010). Regulation of learning and memory by meningeal immunity: a key role for IL-4. The Journal of Experimental Medicine 207, 1067–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenbaum H, Dudchenko P, Wood E, Shapiro M, and Tanila H (1999). The hippocampus, memory, and place cells: is it spatial memory or a memory space? Neuron 23, 209–226. [DOI] [PubMed] [Google Scholar]
- Fallon PG, Ballantyne SJ, Mangan NE, Barlow JL, Dasvarma A, Hewett DR, McIlgorm A, Jolin HE, and McKenzie AN (2006). Identification of an interleukin (IL)-25-dependent cell population that provides IL-4, IL-5, and IL-13 at the onset of helminth expulsion. J Exp Med 203, 1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiano AJ, Xu Y, Tustison NJ, Marsh RL, Baker W, Smirnov I, Overall CC, Gadani SP, Turner SD, and Weng Z (2016). Unexpected role of interferon-γ in regulating neuronal connectivity and social behaviour. Nature 535, 425–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisk V, and Milner B (1990). The role of the left hippocampal region in the acquisition and retention of story content. Neuropsychologia 28, 349–359. [DOI] [PubMed] [Google Scholar]
- Furlanis E, Traunmuller L, Fucile G, and Scheiffele P (2019). Landscape of ribosome-engaged transcript isoforms reveals extensive neuronal-cell-class-specific alternative splicing programs. Nat Neurosci 22, 1709–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshen I, Brodsky M, Prakash R, Wallace J, Gradinaru V, Ramakrishnan C, and Deisseroth K (2011). Dynamics of retrieval strategies for remote memories. Cell 147, 678–689. [DOI] [PubMed] [Google Scholar]
- Guenthner CJ., Miyamichi K., Yang HH., Heller HC., and Luo L. (2013). Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron 78, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herry C, Ciocchi S, Senn V, Demmou L, Müller C, and Lüthi A (2008). Switching on and off fear by distinct neuronal circuits. Nature 454, 600–606. [DOI] [PubMed] [Google Scholar]
- Hitti FL, and Siegelbaum SA (2014). The hippocampal CA2 region is essential for social memory. Nature 508, 88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen JP, Cain CK, Ostroff LE, and LeDoux JE (2011). Molecular mechanisms of fear learning and memory. Cell 147, 509–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipnis J (2016). Multifaceted interactions between adaptive immunity and the central nervous system. Science 353, 766–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipnis J, Cohen H, Cardon M, Ziv Y, and Schwartz M (2004). T cell deficiency leads to cognitive dysfunction: implications for therapeutic vaccination for schizophrenia and other psychiatric conditions. Proc Natl Acad Sci U S A 101, 8180–8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, and Tonegawa S (2012). Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature 484, 381385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louveau A, Smirnov I, Keyes TJ, Eccles JD, Rouhani SJ, Peske JD, Derecki NC, Castle D, Mandell JW, Lee KS, et al. (2015). Structural and functional features of central nervous system lymphatic vessels. Nature 523, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahan AL, and Ressler KJ (2012). Fear conditioning, synaptic plasticity and the amygdala: implications for posttraumatic stress disorder. Trends in neurosciences 35, 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maren S (2001). Neurobiology of Pavlovian fear conditioning. Annu Rev Neurosci 24, 897–931. [DOI] [PubMed] [Google Scholar]
- McKernan M, and Shinnick-Gallagher P (1997). Fear conditioning induces a lasting potentiation of synaptic currents in vitro. Nature 390, 607–611. [DOI] [PubMed] [Google Scholar]
- Neill DR., Wong SH., Bellosi A., Flynn RJ., Daly M., Langford TK., Bucks C., Kane CM., Fallon PG., Pannell R., et al. (2010). Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 464, 1367–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A, Crow M, Raudales R, He M, Gillis J, and Huang ZJ (2017). Transcriptional Architecture of Synaptic Communication Delineates GABAergic Neuron Identity. Cell 171, 522–539 e520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips R, and LeDoux J (1992). Differential contribution of amygdala and hippocampus to cued and contextual fear conditioning. Behavioral neuroscience 106, 274. [DOI] [PubMed] [Google Scholar]
- Plaut M, Pierce JH, Watson CJ, Hanley-Hyde J, Nordan RP, and Paul WE (1989). Mast cell lines produce lymphokines in response to cross-linkage of Fc epsilon RI or to calcium ionophores. Nature 339, 64–67. [DOI] [PubMed] [Google Scholar]
- Radjavi A, Smirnov I, Derecki N, and Kipnis J (2014). Dynamics of the meningeal CD4+ T-cell repertoire are defined by the cervical lymph nodes and facilitate cognitive task performance in mice. Molecular psychiatry 19, 531–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed MD, Yim YS, Wimmer RD, Kim H, Ryu C, Welch GM, Andina M, King HO, Waisman A, Halassa MM, et al. (2020). IL-17a promotes sociability in mouse models of neurodevelopmental disorders. Nature 577, 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciardi S, Ungaro F, Hambrock M, Rademacher N, Stefanelli G, Brambilla D, Sessa A, Magagnotti C, Bachi A, and Giarda E (2012). CDKL5 ensures excitatory synapse stability by reinforcing NGL-1–PSD95 interaction in the postsynaptic compartment and is impaired in patient iPSCderived neurons. Nature cell biology 14, 911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy DS, Kitamura T, Okuyama T, Ogawa SK, Sun C, Obata Y, Yoshiki A, and Tonegawa S (2017). Distinct neural circuits for the formation and retrieval of episodic memories. Cell 170, 1000–1012. e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustenhoven J., Drieu A., Mamuladze T., de Lima KA., Dykstra T., Wall M., Papadopoulos Z., Kanamori M., Salvador AF., Baker W., et al. (2021). Functional characterization of the dural sinuses as a neuroimmune interface. Cell 184, 1000–1016 e1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seder RA, and Paul WE (1994). Acquisition of lymphokine-producing phenotype by CD4+ T cells. Annu Rev Immunol 12, 635–673. [DOI] [PubMed] [Google Scholar]
- Shatz CJ (2009). MHC class I: an unexpected role in neuronal plasticity. Neuron 64, 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen D, and Malenka RC (2006). Synaptic scaling mediated by glial TNF-α. Nature 440, 10541059. [DOI] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, and Stafford B (2007). The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178. [DOI] [PubMed] [Google Scholar]
- Tang S, Wang I-TJ, Yue C, Takano H, Terzic B, Pance K, Lee JY, Cui Y, Coulter DA, and Zhou Z (2017). Loss of CDKL5 in glutamatergic neurons disrupts hippocampal microcircuitry and leads to memory impairment in mice. Journal of Neuroscience 37, 7420–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulving E (2002). Episodic memory: From mind to brain. Annual review of psychology 53, 1–25. [DOI] [PubMed] [Google Scholar]
- Villeda SA, Plambeck KE, Middeldorp J, Castellano JM, Mosher KI, Luo J, Smith LK, Bieri G, Lin K, Berdnik D, et al. (2014). Young blood reverses age-related impairments in cognitive function and synaptic plasticity in mice. Nat Med 20, 659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelaar CF, Mandal S, Lerch S, Birkner K, Birkenstock J, Buhler U, Schnatz A, Raine CS, Bittner S, Vogt J, et al. (2018). Fast direct neuronal signaling via the IL-4 receptor as therapeutic target in neuroinflammation. Sci Transl Med 10. [DOI] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, Van Der Zwan J, Häring M, Braun E, Borm LE, and La Manno G (2018). Molecular architecture of the mouse nervous system. Cell 174, 999–1014. e1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y-C., Li D., Wang L., Lu B., Zheng J., Zhao S-L., Zeng R., and Xiong Z-Q. (2013). Palmitoylation-dependent CDKL5–PSD-95 interaction regulates synaptic targeting of CDKL5 and dendritic spine development. Proceedings of the National Academy of Sciences 110, 9118–9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Ron N, Butovsky O, Landa G, Sudai E, Greenberg N, Cohen H, Kipnis J, and Schwartz M (2006). Immune cells contribute to the maintenance of neurogenesis and spatial learning abilities in adulthood. Nat Neurosci 9, 268–275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Fastq files and quantified gene counts for single cell sequencing are available at the Gene Expression Omnibus (GEO) and Sequence Read Archive (SRA) under accession numbers GSE143183.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.