Abstract
Epichloë endophytes are a group of filamentous fungi that include both sexual (Epichloë) and asexual (Neotyphodium) species. As a group they are genetically diverse and form both antagonistic and mutualistic associations with temperate grasses. We report here on the development of a microsatellite-based PCR system for fingerprinting this group of fungi with template isolated from either culture or infected plant material. M13mp19 partial genomic libraries were constructed for size-fractionated genomic DNA from two endophyte strains. These libraries were screened with a mixture of DIG-labeled dinucleotide and trinucleotide repeat probes. Positive clones were sequenced, and nine unique microsatellite loci were identified. An additional microsatellite was serendipitously identified in the 3′ untranscribed region of the 3-hydroxy-3-methylglutaryl coenzyme A (HMG CoA) reductase gene from N. lolii Lp19. Primers were designed for each locus and a panel of endophytes, from different taxonomic groupings, was screened to determine the degree of polymorphism. On the basis of these results a multiplex assay was developed for strain identification with fluorescently labeled primers for five of these loci. Using this system the size of the products amplified can be precisely determined by automated analysis, and an allele profile for each strain can be readily generated. The assay was shown to resolve endophyte groupings to the level of known isozyme phenotype groupings. In a blind test the assay was used successfully to identify a set of endophytes in planta. A reference database of allele sizes has been established for the panel of endophytes examined, and this will be expanded as new strains are analyzed.
The Epichloë endophytes are a group of filamentous fungi comprised of the sexual Epichloë species, and their asexual derivatives the Neotyphodium species, which were formerly classified as Acremonium species (25, 52). This group of fungi infect cool season grasses (subfamily Pooideae), where they colonize the intercellular spaces of the leaves, culms, and in most cases the seeds. All form asymptomatic associations with the host during the vegetative phase of plant growth, but during flower development the sexual species are capable of emerging from the plant to form a stroma around the developing inflorescence which partially or completely sterilizes (chokes) the host. The sexual species are capable of contagious (horizontal) transmission either through ascospore or conidial infection of cut leaves and culms (63) or by the penetration of stigmata of florets of another plant and with subsequent seed transmission (13). In contrast, the asexual endophytes are seed (vertical) transmitted only and spend their entire life cycle within the host.
The Epichloë endophytes are important biological agents influencing the growth and persistence of temperate grasses (14, 16), since they can confer on the host protection from mammalian and insect herbivory (56), resistance to nematodes (32) and some fungal pathogens (29), drought tolerance (1), and greater field persistence (30, 62). An agricultural detriment of this association, however, is the production by the endophyte of metabolites that affect the health and productivity of grazing livestock. Ryegrass staggers (21) and fescue toxicosis (3) are syndromes usually associated with pastures containing endophyte-infected perennial ryegrass and tall fescue pastures (45). These syndromes are caused by ingestion of lolitrems (22) and ergopeptine alkaloids (2), respectively. These toxicoses are responsible for large economic losses to the sheep industry in New Zealand and to the beef and dairy cattle industry in the United States. Consequently, there is considerable interest in maximizing the beneficial aspects of this symbiotic association for pastoral agriculture.
As a group the Epichloë endophytes show considerable variation in morphology and physiology in culture (10–12). While these properties are useful in the identification of endophytes, used alone they are of limited value in defining distinct taxonomic groupings. However, when used in combination with molecular methods such as isozyme (37) and gene sequence (54) analysis, distinct taxonomic groupings can be defined and the phylogeny can be established. One such study of the strain variation for asexual endophytes isolated from perennial ryegrass, tall fescue, and meadow fescue by using isozyme analysis revealed the presence of several distinct taxonomic groups: three in tall fescue, FaTG-1 (N. coenophialum), FaTG-2, and FaTG-3; two in perennial ryegrass, LpTG-1 (N. lolii) and LpTG-2; and one in meadow fescue, FpTG-1 (N. uncinatum) (12). A comparison of the DNA sequences of the β-tubulin (tub2) and ribosomal (rrn) gene sequences of these same endophytes revealed that all, except N. lolii, are interspecific hybrids derived from Epichloë species (52, 53, 61), a conclusion supported by isozyme analysis (12, 37). These asexual hybrids are morphologically and genetically distinct from their proposed ancestors and comprise discrete taxa (12, 52, 53). However, both isozyme and DNA sequence analysis are rather lengthy processes for routine strain identification and require that the endophyte be first isolated from the grass host, a process which can take up to several weeks.
Several methods are available for the in planta detection of endophytes. These include histological staining (35), enzyme-linked immunosorbent assay (46), and tissue print-immunoblot (28). More recently, PCR-based methods, including both randomly amplified polymorphic DNA (RAPD) and microsatellite locus analysis, have been used for endophyte detection and quantification both in culture (27) and in planta (18, 26). PCR methods provide fast, sensitive, and specific amplification of target DNA sequences in complex DNA samples and thus are particularly amenable to endophyte detection in the plant background.
Microsatellites, also known as simple sequence repeats, are a class of repetitive DNA that is a ubiquitous component of eukaryotic genomes (59). Such loci are found scattered throughout the genome and are inherited in a Mendelian fashion. Microsatellites are composed of very short DNA motifs (1 to 10 nucleotides [nt]) and are found in tandem repeats, usually up to 100 bp (59). The number of tandemly repeated units has been shown to be highly polymorphic between individuals, and this is thought to be due to slippage of the DNA polymerase during the synthesis and repair of DNA (39, 59). The variation in the number of tandem repeats can be detected by PCR with primers designed for the conserved DNA sequences flanking the locus. The popularity of microsatellite markers as a tool for genetic analysis can be attributed to their utility in many applications, including studies of kinship analysis (15), population genetic structure (particularly in conservation genetics) (50), and genome mapping projects (particularly for plant breeding) (42). The majority of these studies have been carried out with animals and plants, where it has been found that the predominant microsatellite type in each group is (AC)n and (AT)n, respectively. Relatively little work has been done on fungal microsatellites, but DNA sequence database analysis suggests that (AT)n repeats are the predominant type in fungi (27).
In this study, we report on the development and application of a multilocus microsatellite-based PCR fingerprinting assay for the identification of Epichloë endophytes, both in culture and in planta. This assay combines the polymorphic properties of microsatellite loci, with the speed, sensitivity, and specificity of PCR. To automate the analysis of the microsatellite fingerprints, phosphoramidite dye-labeled primers were used to allow detection of the PCR products on polyacrylamide gels with a laser scanner and the appropriate computer software. A reference database of known endophyte microsatellite profiles has been established, and it has been shown that this assay is able to resolve endophyte groupings to the isozyme phenotype level as described by Christensen et al. (12).
MATERIALS AND METHODS
Fungal endophyte isolates and culture conditions.
Fungal endophyte isolates, their hosts, and other characteristics are listed in Table 1. Neotyphodium isolates were chosen which represent the range of variation of endophytes from perennial ryegrass (Lolium perenne), tall fescue (Festuca arundinaceae), and meadow fescue (F. pratensis) reported by Christensen et al. (12). In addition isolates of Epichloë typhina and Epichloë festucae were also included. The plant cultivars used in this study included Grasslands Roa tall fescue, Kentucky 31 tall fescue, an unnamed experimental L. perenne × L. multiflorum cultivar (seed line N1509), and a seed line (B3520) of a L. perenne × L. multiflorum population referred to as Coruna. All endophyte isolates were grown on 2.4% (wt/vol) potato dextrose (PD; Difco Laboratories, Detroit, Mich.) agar plates at 22°C. Liquid cultures were prepared by grinding a small amount of mycelium from a plate culture in 500 μl of PD broth and adding 100 μl of this to flasks containing 30 ml of PD broth. Cultures were incubated for 7 to 14 days at 22°C on a rotary shaker at 200 rpm. Outgroup fungal isolates of Claviceps purpurea and Echinodothis tuberiformis (American Type Culture Collection accession numbers 26245 and 201937, respectively) were cultured under the conditions described by Byrd et al. (9).
TABLE 1.
Fungal endophyte isolates used in this study
| Species or taxon | Isolate | Isozyme phenotypea | Host species | Origin | Source or referenceb |
|---|---|---|---|---|---|
| Neotyphodium sp. (LpTG-2) | Lp1 | LpA | Lolium perenne | France | 12 |
| Neotyphodium sp. (LpTG-2) | Lp2 | LpB | L. perenne | France | 12 |
| N. lolii (LpTG-1) | Lp5 | loA | L. perenne | New Zealand | 12 |
| N. lolii | Lp6 | loA | L. perenne | New Zealand | 12 |
| N. lolii | Lp13 | loA | L. perenne | France | 12 |
| N. lolii | Lp19 | loB | L. perenne | France | 12 |
| N. lolii | Lp20 | loB | L. perenne | France | 12 |
| N. lolii | Lp21 | loB | L. perenne | France | 12 |
| N. lolii | Lp7 | loD | L. perenne | Holland | 12 |
| N. lolii | Lp14 | loE | L. perenne | France | 12 |
| N. lolii | Lp9 | loF | L. perenne | Spain | 12 |
| N. lolii | AR1 | - | L. perenne | Spain | MFFGC NZ |
| N. coenophialum (FaTG-1) | Tf5 | coB | Festuca arundinacea | Spain | 12 |
| N. coenophialum | Tf27 | coB | F. arundinacea | Poland | 12 |
| N. coenophialum | Tf28 | coC | F. arundinacea | Portugal | 12 |
| Neotyphodium sp. (FaTG-2) | Tf13 | FaA | F. arundinacea | Spain | 12 |
| Neotyphodium sp. (FaTG-2) | Tf15 | FaC | F. arundinacea | Spain | 12 |
| Neotyphodium sp. (FaTG-2) | Tf20 | FaD | F. arundinacea | Algeria | MFFGC NZ |
| Neotyphodium sp. (FaTG-3) | Tf16 | FaF | F. arundinacea | Spain | 12 |
| Neotyphodium sp. (FaTG-3) | Tf18 | FaF | F. arundinacea | Algeria | 12 |
| N. uncinatum (FpTG-1) | Fp1 | unA | F. pratensis | Spain | 12 |
| N. uncinatum | Fp2 | unA | F. pratensis | Spain | 12 |
| N. uncinatum | Fp4 | unB | F. pratensis | New Zealand | 12 |
| E. typhina | E8 | - | L. perenne | Rhode Island | 54 |
| E. festucae | Fr1 | FF6 | F. rubra | New Zealand | 36 |
| E. festucae | Frc5 | - | F. rubra | Europe | MFFGC NZ |
| E. festucae | Frc7 | - | F. rubra | Europe | MFFGC NZ |
| E. festucae | Frr1 | - | F. rubra | New Zealand | MFFGC NZ |
| E. festucae | Fg1 | - | F. glauca | United States | MFFGC NZ |
| E. festucae | Fl1 | FF10 | F. longifolia | United States | 36 |
Preparation of fungal and plant genomic DNA.
Fungal genomic DNA was prepared by a modification of the method described by Byrd et al. (9). About 100 mg of freeze-dried mycelia from endophyte liquid culture was ground to a fine powder under liquid nitrogen and resuspended in 10 ml of extraction buffer (150 mM Na2EDTA, 50 mM Tris-HCl [pH 8.0], 1% [wt/vol] sodium lauroyl sarcosine, and 2 mg of proteinase K ml−1). This mixture was incubated at 37°C for 20 min and then centrifuged at 20,200 × g for 10 min to pellet the cellular debris. The supernatant was extracted successively with equal volumes of phenol, phenol-chloroform (1:1 [vol/vol]), and chloroform, and the DNA was precipitated with an equal volume of isopropanol. To remove polysaccharides from the sample, the pelleted DNA was resuspended in 5 ml of 1 M NaCl and then centrifuged at 20,200 × g for 5 min. DNA was precipitated from the supernatant with isopropanol and then pelleted. The DNA pellet was rinsed in 70% ethanol, air dried, and resuspended in 0.5 to 1.0 ml of sterile Milli-Q water.
Genomic DNA from freshly harvested endophyte-infected and endophyte-free plant material was isolated by using either the CTAB method of Doyle and Doyle (19) or a rapid miniprep method with the FastDNA Kit H (Bio 101, Inc., La Jolla, Calif.) according to the manufacturer’s instructions. To maximize the yield of endophyte DNA from plant material, the outer leaves were stripped from tillers, and the base 2 to 3 cm of tissue, where the endophyte is most abundant, was used for DNA extraction. A total of about 1 g of tissue was used in the CTAB method, and about 200 mg of tissue was used in the miniprep method.
DNA was quantified by Hoechst dye staining followed by fluorometry with a TKO100 fluorometer (Hoeffer Scientific Instruments, San Francisco, Calif.), which was calibrated by using known concentrations of calf thymus DNA. Alternatively, DNA concentrations were estimated by the intensity of ethidium bromide fluorescence of DNA samples compared to that of known concentrations of bacteriophage lambda DNA on agarose gels with an IS-1000 Digital Imaging System (Alpha Innotech Corp., San Leandro, Calif.).
Agarose gel electrophoresis and DNA hybridizations.
Restriction enzyme digestion of DNA with combinations of the restriction enzymes AluI, HaeIII, ThaI, and BamHI and agarose gel electrophoresis conditions for the separation of DNA were as described by Sambrook et al. (51). Genomic digests were transferred from agarose gels to positively charged nylon membranes (Boehringer GmbH, Mannheim, Germany) by capillary transfer (57) and fixed by UV cross-linking. Blots were probed with oligonucleotides of microsatellite sequences (CA)15, (GA)15, (CAA)10, (GAA)10, and (ATC)10 (Life Technologies, Gaithersburg, Md.), that were 3′ end labeled with DIG-11-ddUTP (Boehringer GmbH) by using terminal transferase (Boehringer GmbH) (4). Hybridizations were carried out in DIG standard hybridization buffer (5× SSC [1× SSC is 0.15 NaCl plus 0.015 M sodium citrate], 0.1% sodium lauroyl sarcosine, 0.02% sodium dodecyl sulfate, 1% blocking reagent) at either 60 or 50°C by using pools of end-labeled dinucleotide or trinucleotide repeat probes, respectively, at a final concentration of 2 pmol ml−1. After hybridization, blots were washed twice in 0.1× SSC for 10 min each at room temperature. A final stringency wash was performed for 2 min with a combination of temperature and salt concentration which, for low-stringency conditions was approximately 10 to 20°C below the Tm for the probe (calculated by using the formula of Bolton and McCarthy (5) and for high-stringency conditions was approximately 0 to 5°C below the Tm. Hybridized probes were detected by using the DIG chemiluminescent detection kit (4) with CSPD (Boehringer GmbH) as the substrate, followed by autoradiography for 30 to 120 min on RX X-ray film (Fuji Photo Film Co., Tokyo, Japan).
Construction and screening of endophyte partial genomic libraries.
Genomic DNA from E8 and Lp1 was digested to completion with combinations of the restriction enzymes AluI, HaeIII, ThaI, and BamHI and then separated by electrophoresis on 1.25% SeaPlaque agarose gels (FMC Bioproducts, Rockland, Maine) in 1× TAE buffer (40 mM Tris, 20 mM acetate acid, 1 mM Na2EDTA). Fragments in the size ranges of 200 to 500 bp (E8) and 100 to 1,000 bp (Lp1) were excised from these gels under long-wavelength UV light (366 nm), and DNA was isolated by phenol freeze extraction (60). BamHI linkers, constructed by the self-annealing of 5′ chemically phosphorylated 5′-CGGGATCCCG-3′ oligonucleotides (Life Technologies), were ligated in excess to the blunt-end fragments with T4 ligase (New England Biolabs, Beverly, Mass.). The fragments were digested with BamHI and ligated to calf alkaline phosphatase treated BamHI-digested M13mp19 vector (44). Ligation products were transformed into electrocompetent Escherichia coli cells of strain XL1-Blue (8) by electroporation and plated onto Luria-Bertani agar plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid), IPTG (isopropyl-β-d-thiogalactopyranoside), and tetracycline (15 μg/ml). Plaques were lifted onto Hybond N+ positively charged nylon membranes (Amersham, Buckinghamshire, United Kingdom) in duplicate and fixed by UV cross-linking.
Each library was screened by plaque hybridization by using low-stringency hybridization and the washing conditions described for the Southern blot hybridization, except for the screening of the E8 library with the trinucleotide repeat probes, which was conducted at high stringency. Plating and screening of the library was carried out by standard procedures (51). Single-stranded DNA was purified from positive plaques, and unique clones were identified by polyacrylamide gel electrophoresis (PAGE) of single deoxy- and dideoxynucleotide sequencing reactions prepared with an AmpliCycle sequencing kit (Perkin-Elmer Corp., Foster City, Calif.) and the M13 forward primer. Unique clones were then fully sequenced by using either the AmpliCycle sequencing kit according to the manufacturer’s instructions or a BigDye terminator cycle sequencing kit (PE Applied Biosystems, Foster City, Calif.) by using the M13 forward primer and the products separated by PAGE on an ABI Prism 377 DNA sequencer (Perkin-Elmer Corp.)
Nucleotide sequence accession numbers.
The sequences of the Epichloë microsatellite loci and flanking DNA have been deposited in the GenBank database and have the following accession numbers: B2 (AF063085), B3 (AF063086), B4 (AF063087), B5 (AF063088), B6 (AF063089), B7 (AF063090), B8 (AF063091), B9 (AF063092), B10 (AF063093), and B11 (AF063094).
Amplification of microsatellite loci by using PCR.
The primers used to amplify the microsatellite loci were designed for flanking sequences by using standard criteria (31) and are listed in Table 2. PCRs were performed in either 12.5- or 25-μl volumes containing 10 mM Tris-HCl, 1.5 mM MgCl2, and 50 mM KCl (pH 8.3) in the presence of 50 μM concentrations of each deoxynucleotide triphosphate (dATP, dCTP, dGTP, and dTTP), 200 nM of each primer (Life Technologies), 0.08 U of Taq DNA polymerase (Boehringer GmbH) μl−1, and either 40 pg of fungal genomic DNA or 400 pg of plant (plus or minus endophyte) genomic DNA μl−1. When PCR products were radiolabeled, 0.1 μCi of [α-33P]dCTP (Amersham) μl−1 was included in the PCR reaction mixture. Reactions were carried out in a PC-960 or FTS-960 thermocycler (Corbett Research, Mortlake, Australia) programmed for 30 cycles of 1 min at 94°C, 2 min at 65°C, and 1 min at 72°C followed by a final extension of 10 min at 72°C. Amplified products (2 μl) were fractionated by electrophoresis on 3% Nusieve gels (FMC Bioproducts) in 1× TBE buffer and visualized by ethidium bromide staining followed by UV fluorescence.
TABLE 2.
Microsatellite sequences and the primers used to amplify these loci
| Locusa | Microsatellite sequence | Type | Primer Sequence | Tmb | Product length (bp) | Isolate |
|---|---|---|---|---|---|---|
| B1c | (AAG)18 | B1.1 | 5′-CGC ACA ATA CGT CAG CTA GGA ATG | 55 | 325 | NF 62A |
| B1.2 | 5′-CCT GAA TCA ACT TTG CTA TCA GGC | 54 | ||||
| B2 | (AC)18 | B2.1 | 5′-TCC GTA ATG CTC TGC TTT GGC AGG | 57 | 163 | E8 |
| B2.2 | 5′-CTG CTG ACA GTA GGT GGA TGA TGG | 57 | ||||
| B3 | C12 and (GA)7 | B3.1 | 5′-TGC TGT CCG TTC TCG TGG CAT TCG | 59 | 115 | E8 |
| B3.2 | 5′-CGG TGT AGA AGG ACC TGC AGT TTG | 57 | ||||
| B4 | (CA)3GCG(CA)3ACG(CA)3A(CA)4 | B4.1 | 5′-TGG ACT CGA CTT GCC CTC TCT CAG | 59 | 124 | E8 |
| B4.2 | 5′-TGC GAG CAG CGT TTG CGT GTG CGT | 61 | ||||
| B5 | (CATCTCATCA)5 | B5.1 | 5′-CAA CTC CAA CAA ACT CAA CCA GCG | 55 | 107 | E8 |
| B5.2 | 5′-GGA CTC GTG CAA AGC TTC GGA TGG | 59 | ||||
| B6 | (CAT)2CAC(CAT)3 | B6.1 | 5′-GGC ATG GTA TGG GCA ATG AGT GTC | 57 | 173 | E8 |
| B6.2 | 5′-CTG CTG CGA TGT TTT GTA CTG TGG | 55 | ||||
| B7 | (AG)5A(AG)3 | B7.1 | 5′-AAC TTG AAT TAA CTA GGG GCT AGG | 52 | 111 | Lp1 |
| B7.2 | 5′-TTA ATT CGC TAC CCT CTC TTT TGC | 52 | ||||
| B8 | (AC)8T6 | B8.1 | 5′-CTA ACA CGC TCA AGG GAG ATT GCC | 57 | 142 | Lp1 |
| B8.2 | 5′-GGC GAG GTG CAA CCC TGT TAA TGG | 59 | ||||
| B9 | G5AG9 and (GAGAG)2C(GAGGA)2 | B9.1 | 5′-AAT CGT TGT GCG AGC CAT TCT GGC | 57 | 167d | Lp1 |
| B9.2 | 5′-TCC ATC TCC GCA ATC TGC ATG TCC | 57 | ||||
| B9.4 | 5′-GCC CCG TCA TGC ATT ATC TCC TTG | 57 | 246e | Lp1 | ||
| B10 | (AGC)3CG(CAT)3(CAA)5 | B10.1 | 5′-CGC TCA GGG CTA CAT ACA CCA TGG | 59 | 179 | Lp1 |
| B10.2 | 5′-CTC ATC GAG TAA CGC AGG CGA CG | 59 | ||||
| B11 | (GACA)18 | B11.1 | 5′-CAT GGA TGG ACA AGA GAT TGC ACG | 55 | 173 | Lp19 |
| B11.4 | 5′-TTC ACT GCT ACA ATT CTG TCC AGC | 54 |
B2, B3, B4, B7, B8, and B9 were isolated by using the dinucleotide probe mixture of (CA)15 and (GA)15. B5, B6, and B10 were isolated by using the trinucleotide probe mixture of (CAA)10, (GAA)10, and (ATC)10.
Tm is calculated by using as follows: Tm = 81.5 + 16.6(log10[Na+]) + 0.41(%G + C) − 600/N (5), where [Na+] is 0.05 M and N is the length of the oligonucleotide.
Expected product size for B9.1-B9.2 primer pair.
Expected product size for B9.1-B9.4 primer pair.
Where necessary, PCR assay conditions were optimized by adjusting the following conditions: annealing temperature, Mg2+ concentration, enzyme concentration, and hotstart PCR with AmpliWax beads (Perkin-Elmer Corp.).
Conditions for multiplex PCR of the microsatellite loci B4, B6, B9, B10, and B11 were the same as those used for a single locus amplification (see above), with each primer added to a final concentration of 200 nM. For automated analysis, one of the primers at each locus was 5′ end labeled with a fluorescent phosphoramidite dye (Life Technologies). Specifically, primers B4.1 and B6.1 were labeled with 6-carboxyfluorescein (6-FAM), B9.1 and B10.1 were labeled with 4,7,2′,7′-tetrachloro-6-carboxyfluorescein (TET), and B11.1 was labeled with 4,7,2′,4′,5′,7′-hexachloro-6-carboxyfluorescein (HEX). Where amplifications were poor, a problem particularly noticeable for plant tissue, the five-locus multiplex reaction was partitioned into three separate reactions, comprising locus B4 with B6, locus B9 with B10, and locus B11 alone. The products of these PCR reactions were pooled and concentrated under vacuum (SpeedVac; Savant Instruments, Inc., Farmingdale, N.Y.) to a final volume of about 5 μl and then run on Nusieve agarose or polyacrylamide gels.
Polyacrylamide gel separation of microsatellite PCR products.
Radiolabeled microsatellite PCR products (2 μl) were fractionated by PAGE on 6% sequencing polyacrylamide gels (41) in 1× sequencing TBE (134 mM Tris, 45 mM boric acid, 2.5 mM EDTA; pH 8.8) at a constant voltage of 1,500 V for 2 to 4 h. Gel running times were adjusted to obtain optimal separation of PCR products for each microsatellite locus. The sizes of the PCR products were determined by reference to the sequence of a control DNA, pBSMB, that was included with the AmpliCycle sequencing kit. Gels were dried on a model 583 Gel Dryer (Bio-Rad, Richmond, Calif.) and exposed to RX X-ray film (Fuji Photo Film Co.) for 16 to 40 h and then developed.
Fluorescently labeled microsatellite PCR products (1.5 μl of a 1/5 dilution) were added to 2.5 μl of formamide, 0.5 μl of 5% blue dextran, and 0.5 μl of GS-500 TAMRA (Perkin-Elmer Corp.), an internal lane size standard. A 2-μl portion of this mixture was fractionated by PAGE (4.25%) on an ABI Prism 377 DNA sequencer. The best estimates of sizes of the alleles were measured from their electrophoretic mobility through the gel relative to the internal size standard, as indicated by GeneScan 2.1 analysis software (Perkin-Elmer Corp.). The accuracy of the size estimates, expressed to a fraction of a nucleotide unit, is specific to the electrophoretic separation conditions. A database of allele sizes for each microsatellite locus of each endophyte isolate was compiled in ClarisWorks 4.0 (Claris Corp., Santa Clara, Calif.).
RESULTS
Abundance of microsatellites in Epichloë endophyte genomes.
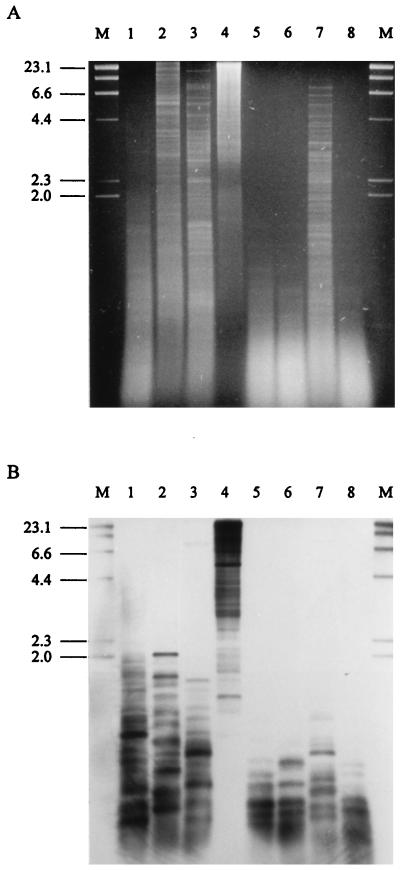
To determine the presence and relative abundance of microsatellites in the genomes of endophyte strains E8 and Lp1, genomic DNA was digested to completion with combinations of 4-bp blunt-end-cutting restriction enzymes and BamHI, and Southern blots of these digests were probed for the presence of microsatellite sequences. These digests resulted in large populations of small DNA fragments of a size suitable for the construction of small insert libraries (Fig. 1A). Hybridization of Lp1 digests with microsatellite probes, under conditions of low stringency, revealed up to 25 bands for dinucleotide repeat probes (results not shown) and up to 20 bands for the trinucleotide repeat probes (Fig. 1B). This represents an average frequency of one (CA)n or (GA)n microsatellite per 2 Mb of DNA or one (CAA)n, (GAA)n, or (ATG)n microsatellite per 2.5 Mb of DNA, assuming a genome size of 50 Mb for Lp1 (43). Genomic digests of strain E8 were also screened with di- and trinucleotide repeat probes, and similar results were obtained (results not shown).
FIG. 1.
Hybridization of microsatellite probes to the Lp1 genome. (A) Ethidium bromide-stained agarose (1.25%) gel of Lp1 genomic DNA (1 μg) digested with AluI (lane 1); ThaI (lane 2); HaeIII (lane 3); BamHI (lane 4); AluI, ThaI, and BamHI (lane 5); AluI, HaeIII, and BamHI (lane 6); ThaI, HaeIII, and BamHI (lane 7); and AluI, ThaI, HaeIII, and BamHI (lane 8). (B) Autoradiograph of Southern blot of gel from panel A, which has been probed at low stringency with DIG-labeled (CAA)10, (GAA)10, and (ATC)10 oligonucleotides, followed by chemiluminescent detection. The numbers on the left of the figures indicate the sizes in kilobases of the λ-HindIII size markers in lane M.
Cloning of microsatellite loci.
On the basis of the above results, M13mp19 partial genomic libraries were constructed for strains E8 and Lp1 by using size-fractionated DNA inserts of between 200 and 500 bp and between 100 and 1,000 bp, respectively. Approximately 110,000 plaques of the E8 library were screened by plaque hybridization with a mixture of DIG-labeled dinucleotide probes at a low stringency. Under these conditions 1% of the plaques hybridized, and 51 of these were taken through to the second round of screening. When the same filters were screened with a mixture of DIG-labeled trinucleotide probes under conditions of high stringency 0.1% of the plaques hybridized, and 30 of these were taken through a second round of screening. Screening of 120,000 plaques of the Lp1 library was carried out under conditions of low stringency for both pools of di- and trinucleotide probes. Under these conditions 0.4% of the plaques hybridized with each set of probes, and 24 and 12 plaques, respectively, were taken through to second-round screening. Of the total of 117 positive plaques that were picked from both libraries, 88 gave positive signals after the second round of screening.
The positive plaques selected were amplified in E. coli and both single- and double-stranded M13 DNA was isolated from each clone. The double-stranded DNA was digested with BamHI and separated by agarose gel electrophoresis to determine the insert size. Many of the inserts appeared to be the same size and therefore were possibly clones of one another. This was confirmed by single dideoxy- and deoxynucleotide sequencing reactions of single-stranded DNA templates. Unique clones identified by this method were then completely sequenced by using the M13 forward primer. Nine different microsatellite types were identified; five from E8 (B2 to B6) and four from Lp1 (B7 to B10). The microsatellite sequences identified are listed in Table 2. Of these microsatellites, five (B2, B3, B5, B8, and B10) contain perfect repeats, and the remaining four (B4, B6, B7, and B9) contain imperfect repeats. Additional single-nucleotide microsatellite loci were identified in B3, B8, and B9. For B8, the single-nucleotide locus was immediately adjacent to the dinucleotide repeat locus as a compound microsatellite. For B5 and B9, the repeat motifs identified were only partially complementary to the probes used (tri- and dinucleotide mixtures, respectively), suggesting that they were identified as positives because of the low-stringency hybridization conditions employed in the screening process. Table 2 also includes two additional endophyte microsatellite sequences used in this study; a locus (referred to here as B1) identified by Groppe et al. (27) in an undescribed Epichloë sp. from Bromus erectus (mating population VI [55]) and B11, a locus serendipitously identified 3′ of the untranscribed region of the 3-hydroxy-3-methylglutaryl coenzyme A (HMG CoA) reductase gene from N. lolii strain Lp19 (17).
Primer design and PCR optimization.
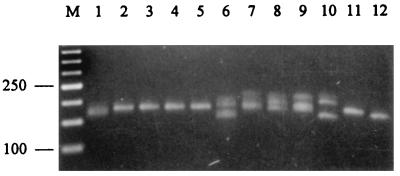
To establish PCR assays which amplify the microsatellite loci identified, primers were designed for the sequences flanking each of the microsatellites (Table 2). A particular consideration in the design of the primers was for all to have a similar length and GC content to facilitate potential multiplexing of all loci. Primer sequences for the B1 locus were those used previously by Groppe et al. (27). Initial evaluation of primer pairs (Bx.1-Bx.2 for loci B2 to B10 and B11.1-B11.4 for locus B11) was performed by screening a panel of 12 isolates, representative of seven taxonomic groups of endophytes, under a standard set of conditions. All loci, apart from B3, gave specific products with high yield. The results for amplification of locus B10 are shown in Fig. 2. All strains amplify well with this primer set, yielding a product(s) of a size characteristic of the allele amplified. The presence of multiple bands in some samples (lanes 1 and lanes 6 to 10) is consistent with the known hybrid nature of these particular endophytes. Amplification of the B3 locus resulted in the production of a number of nonspecific bands, despite attempts to optimize the PCR amplification conditions as described in Materials and Methods. Given the difficulty of redesigning primers to this locus, because of the lack of available flanking sequences, further analysis of locus B3 was discontinued. To test the specificity of the assay for the Epichloë endophytes, two outgroups from the Clavicipitaceae family, including Claviceps purpurea and Echinodothis tuberiformis, were also tested, but no products were obtained with any of these primer sets (results not shown).
FIG. 2.
Amplification of Epichloë endophyte microsatellite loci. Ethidium bromide-stained agarose (3% NuSieve) gel of PCR products amplified with primers B10.1 and B10.2 by using genomic DNA (40 pg μl−1) from Lp1 (lane 1), Lp5 (lane 2), Lp13 (lane 3), Lp19 (lane 4), Lp7 (lane 5), Tf27 (lane 6), Tf28 (lane 7), Tf13 (lane 8), Tf18 (lane 9), Fp2 (lane 10), E8 (lane 11), and Fr1 (lane 12). The numbers on the left of the figures indicate the sizes in base pairs of the 50-bp ladder in lane M.
In planta detection and identification of Epichloë endophytes.
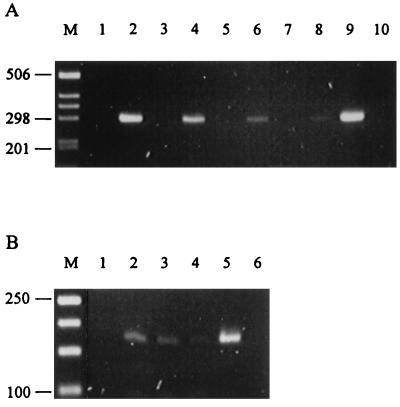
To determine the specificity of the microsatellite locus primers for detecting Epichloë endophyte DNA in planta, total DNA extracted from endophyte-infected grass tissue by using the CTAB method (19) was used as a template in PCR reactions. The results for AR1-infected L. perenne × L. multiflorum hybrid ryegrass with primers to locus B1 are shown in Fig. 3A. A product of approximately 298 bp was amplified from genomic DNA isolated from endophyte-infected plant material (lanes 2, 4, 6, and 8), but no product was detected in endophyte-free material (lanes 1, 3, 5, and 7). The DNA sequence of this product was identical to that from AR1 alone, thus confirming the specificity of the assay for detecting endophyte sequences in a template mixture that is predominantly of plant origin. The best yield of product (lane 2) was obtained by using a template concentration (2 ng μl−1) that was 50-fold higher than that required to amplify the corresponding fungal DNA (lane 9), presumably reflecting the relative abundance of fungal DNA to plant DNA in this particular association. Products of sizes characteristic of the corresponding AR1 alleles were also amplified from the same plant material by using primer sets for the loci B4 (100 bp), B6 (188 bp), B9 (246 bp), B10 (178 bp), and B11 (150 bp). With the same set of primers and PCR assay conditions, genomic DNAs from Tf15-infected tall fescue (Kentucky 31) and Fl1-infected tall fescue (Grasslands Roa) were screened, and products of a size characteristic of these endophytes were found. The sizes of these products were 298 (B1), 101 (B4), 187 (B6), 248 (B9), 172 and 178 (B10), and 129 and 165 bp (B11) for Tf15 and 310 (B1), 100 (B4), 187 (B6), 272 (B9), 178 (B10), and 117 bp (B11) for Fl1. No products were amplified from endophyte-free tall fescue samples.
FIG. 3.
Amplification of Epichloë microsatellite loci from endophyte-infected plant material. Ethidium bromide-stained agarose (3% NuSieve) gels of PCR products amplified from genomic DNA isolated from grass tissue by using primers to microsatellite loci B1 (A) and B10 (B). (A) PCR products generated with DNA prepared from AR1-infected L. perenne × L. multiflorum hybrid ryegrass (lanes 2, 4, 6, and 8) and endophyte-free hybrid ryegrass (lanes 1, 3, 5, and 7) by the CTAB method (18) at total DNA concentrations of 2 ng μl−1 (lanes 1 and 2), 400 pg μl−1 (lanes 3 and 4), 200 pg μl−1 (lanes 5 and 6), and 40 pg μl−1 (lanes 7 and 8); at an AR1 fungal genomic DNA concentration of 40 pg μl−1 (lane 9); and with no DNA (lane 10). Lane M, 1-kb ladder. (B) PCR products generated with DNA prepared from AR1-infected L. perenne × L. multiflorum hybrid ryegrass by the FastDNA Kit H minipreparation method at total DNA concentrations of 2 ng μl−1 (lane 1), 400 pg μl−1 (lane 2), 80 pg μl−1 (lane 3), and 16 pg μl−1 (lane 4); at an AR1 fungal genomic DNA concentration of 40 pg μl−1 (lane 5); and with no DNA (lane 6). Lane M, 50-bp ladder. The numbers on the left of the figures indicate the sizes in base pairs of the DNA size markers.
To improve the rapidity with which in planta detection of endophyte strains could be carried out, microsatellite loci were amplified with DNA extracted from plant material by the FastDNA Kit H miniprep method. The results for AR1-infected L. perenne × L. multiflorum hybrid ryegrass material amplified with primers to locus B10 are shown in Fig. 3B. A product of the size (178 bp) characteristic of this locus was obtained but only when DNA concentrations in the range of 16 to 400 pg μl−1 were used (lanes 2 to 4). At a DNA concentration of 2 ng μl−1 (lane 1), no product was generated, presumably as a result of the presence of inhibitors in DNA prepared by this method.
Degree of polymorphism of microsatellite loci.
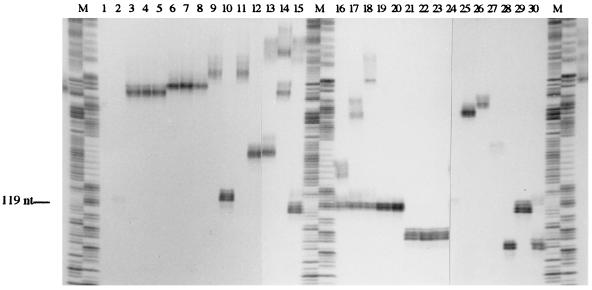
Once the standard conditions for PCR amplification of Epichloë endophyte microsatellite loci were established, all thirty fungal isolates listed in Table 1 were screened with primers to loci B1, B2, and B4 to B11. Products were labelled with [α-33P]dCTP incorporation and separated on polyacrylamide gels to determine the size of the single-stranded products generated at each locus. Figure 4 shows a representative gel of endophyte products amplified by primers to the B11 locus. Isolates sharing like-banding patterns were readily identifiable (e.g., lanes 3 to 5 and lanes 6 to 8) and fingerprinting trends between different taxonomic groups were readily recognized, though the absolute size of individual alleles was difficult to determine since (i) in most cases for each microsatellite allele amplified there are additional single-stranded DNA bands, corresponding to slippage by Taq polymerase during PCR, and (ii), because of the ability of this polymerase to add an additional deoxyadenylate at the end of the product, it was difficult to size bands run some distance from the sequencing ladder due to the variation in electrophoretic mobility across the gel. Despite these limitations it was possible to establish allele groupings, including the identification of hybrids which contain multiple alleles.
FIG. 4.
Autoradiograph of radiolabeled microsatellite products amplified from Epichloë genomic DNA. Fungal genomic DNA (40 pg μl−1) was amplified by PCR in the presence of [α-33P]dCTP with primers to the B11 microsatellite locus. These products were separated on a 6% denaturing polyacrylamide gel. Lanes: M, 1-bp ladder (pBSMB sequence) combining A and G reactions, and C with T; 1, Lp1; 2, Lp2; 3, Lp5; 4, Lp6; 5, Lp13; 6, Lp19; 7, Lp20; 8, Lp21; 9, Lp7; 10, Lp14; 11, Lp9; 12, AR1; 13, Tf5; 14, Tf27; 15, Tf28; 16, Tf13; 17, Tf15; 18, Tf20; 19, Tf16; 20, Tf18; 21, Fp1; 22, Fp2; 23, Fp4; 24, E8; 25, Fr1; 26, Frc5; 27, Frc7; 28, Frr1; 29, Fg1; 30, Fl1.
Schematic diagrams of each gel were compiled to show different allele groupings, and a summary of these is given in Table 3. With the microsatellite locus B2, only LpTG-2, FaTG-3, and E. typhina were amplified, with each giving a unique fingerprint indicative of separate taxonomic groupings. The B5 primers generated strong bands for LpTG-2, N. uncinatum, and E. typhina and faint bands for samples of N. coenophialum and some strains of N. lolii of isozyme phenotype grouping loA (12). However, only two alleles were identified in this group, making it a relatively uninformative locus. Primers to the B7 locus amplified LpTG-2, FaTG-3, and E. typhina but, like B5, this locus was relatively uninformative. The B8 primers amplified LpTG-2, N. lolii, FaTG-2, and E. festucae isolates Fg1 and Fl1, but the different taxonomic groups were not completely distinguishable from one another.
TABLE 3.
Endophyte allele groupings identified by microsatellite PCR assays
| Locus | Size range (bp) of PCR products | No. of fingerprint patternsa | Expected product size (bp)b | Isolates amplified |
|---|---|---|---|---|
| B1 | 300–340 | 8 | 325 | All except FaTG-3 and E. typhina |
| B2 | 95–170 | 3 | 163 | LpTG-2, FaTG-3, and E8 |
| B4 | 105–125 | 6 | 124 | All |
| B5 | 70–110 | 2 | 107 | LpTG-2, N. lolii (loA), N. coenophialum, N. uncinatum, and E8 |
| B6 | 150–200 | 7 | 173 | All |
| B7 | 70–120 | 2 | 111 | LpTG-2, FaTG-3, and E8 |
| B8 | 130–150 | 4 | 142 | LpTG-2, N. lolii, FaTG-2, Fg1, Fl1 |
| B9c | 145–190 | 10 | 157 | All except E8 |
| B9d | 235–280 | 7 | 246 | All except FaTG-3, N. uncinatum, and E8 |
| B10 | 145–200 | 16 | 179 | All |
| B11 | 115–240 | 20 | 169 | All |
For 30 isolates screened.
For allele cloned and sequenced (see Table 2).
With primer pair B9.1-B9.2.
With primer pair B9.1-B9.4.
Primers to the remaining six loci (B1, B4, B6, B9, B10, and B11) amplified almost all of the isolates tested and, between them, were able to distinguish isolates to at least their isozyme phenotype grouping (as described by Christensen et al. [12]). Locus B11 was particularly informative, with 20 different fingerprints distinguishable across the 30 isolates surveyed. This was the only locus that was able to distinguish most of the different isozyme phenotypes within the N. lolii group. Loci B9 and B10 proved to be particularly informative in identifying known hybrid endophytes, such as the tall fescue endophytes, that are characterized by the presence of multiple alleles. Multiple products were amplified from these same loci in N. uncinatum, providing evidence for a hybrid origin for this group as well.
Strategy for the automated analysis of microsatellite PCR products.
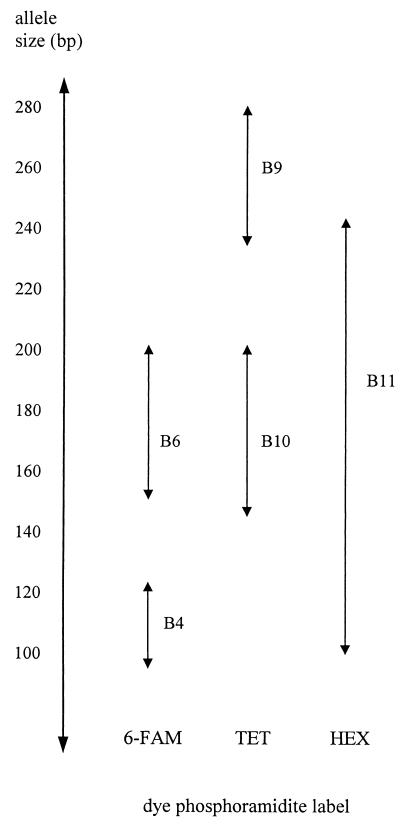
Given that primers to microsatellite loci B4, B6, B9, B10, and B11 amplified almost all of the isolates tested, these primer sets were selected for the development of an automated DNA fingerprinting system utilizing fluorescently labeled primers and the laser detection technology associated with the ABI automatic sequencer. The size range of alleles generated for each of these microsatellite loci are summarized in Table 3. With reference to these size ranges, a primer dye-labeling strategy that utilizes the three available phosphoramidite dyes, 6-FAM, TET, and HEX, was formulated to allow the simultaneous size analysis of the products of all five microsatellite loci on a single lane of the automatic sequencer (Fig. 5). As microsatellite B11 had such a wide range of PCR product sizes, it was assigned its own dye, HEX. Primers to loci B4 and B6 were both labeled with 6-FAM. A new primer (B9.4) was designed to pair with B9.1 to increase the expected product size by 89 bp, so that the products of this locus (235 to 280 bp) would not overlap with those from locus B10 (145 to 200 bp), thereby enabling both loci to be labeled with the third dye, TET. A consequence of this change was that the B9.1-B9.4 combination failed to amplify Tf16, N. uncinatum, and E. typhina and only weakly amplified Tf18, although very good amplification was obtained for all of the other isolates (Table 3). However, sufficiently informative fingerprints were generated for these isolates at the other four loci to resolve these groups.
FIG. 5.
Strategy for the dye labeling of primers to microsatellite loci. The strategy shows how the products for microsatellite loci B4, B6, B9, B10, and B11 can be amplified by multiplex PCR and analyzed in a single lane of the automatic sequencer. Double-headed arrows indicate the observed size range, as determined by separation on polyacrylamide gels, of microsatellite PCR products generated for the Epichloë endophytes analyzed. Primers used for each locus were Bx.1 and Bx.2, Exception for B9, which uses B9.1 and B9.4.
Multiplex PCR of informative microsatellite loci.
Primers to the five informative microsatellite loci identified above (B4, B6, B9, B10, and B11) were tested (as described in Materials and Methods) in a single multiplex reaction for their ability to amplify all loci, thus economizing on reagents, time, and labor. A comparison of the reaction products generated for the five-locus multiplex reaction and for individual locus amplifications by using template DNA prepared by the modified Byrd et al. (9) method from endophytes Tf13 and Fr1 is shown in Fig. 6. All products amplified in single-locus reactions were present in the multiplex reaction. This was confirmed by carrying out the same reactions in the presence of [α-33P]dCTP followed by separation of the products on polyacrylamide gels and detection by autoradiography. The products of the single-locus amplifications matched exactly the size of the products of the multiplex reaction (results not shown). The success of the multiplex reaction was, however, dependent on the quality of the template DNA used. Multiplex PCR of DNA prepared from plant material by the CTAB method (19) gave consistently stronger signals than that prepared with the FastDNA kit; presumably, this was a consequence of the presence of inhibitors in the latter.
FIG. 6.
Multiplex amplification of Epichloë endophyte microsatellite loci. Ethidium bromide-stained agarose (3% NuSieve) gel of the products generated from either a single or a multiplex PCR for endophytes Tf13 (lanes 1 to 6) and Fr1 (lanes 7 to 12). Lanes: M, 50-bp ladder; 1, multiplex; 2, B4; 3, B6; 4, B9; 5, B10; 6, B11; 7, multiplex; 8, B4; 9, B6; 10, B9; 11, B10; and 12, B11.
Automated analysis of microsatellite PCR products.
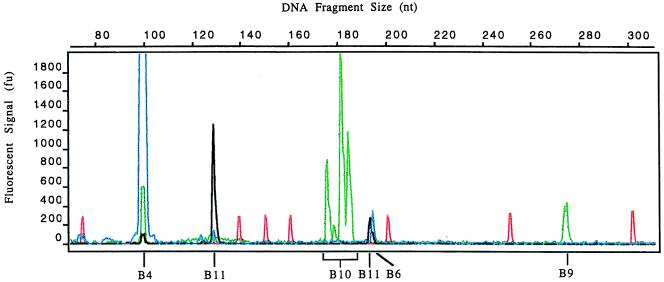
A limitation of the radiolabeling method described above is the inability to accurately determine both the size and the number of alleles present in a given sample. This problem was particularly acute in samples from hybrid endophytes with multiple alleles. To overcome these problems, samples were amplified by using fluorescently labeled primers, and the products were analyzed on an ABI Prism 377 DNA sequencer with the GeneScan Analysis 2.1 software. The electropherogram for one such analysis, that for Tf28, is shown in Fig. 7, and the allele sizes, in nucleotide units, for this sample are shown in Table 4.
FIG. 7.
Electropherogram of an endophyte microsatellite fingerprint. Genomic DNA from Tf28 was amplified in the presence of fluorescent primers, and the products were separated by PAGE. GeneScan 2.1 software was used to generate the electropherogram, and the sizes of the products were determined by reference to GS-500 TAMRA internal size standards. Peaks in red, blue, green, and black correspond to the size standards: 6-FAM (B4 and B6), TET (B9 and B10), and HEX (B11)-labeled products, respectively.
TABLE 4.
Allele sizes for microsatellite loci of Tf28 from electropherogram of Fig. 7
| Color | Dye label | Size (nt U) | Peak ht (FU) | Deduced microsatellite locus |
|---|---|---|---|---|
| Blue | 6-FAM | 100.00 | 4,726 | B4 |
| Blue | 6-FAM | 193.75 | 348 | B6 |
| Green | TET | 175.14 | 909 | B10 |
| Green | TET | 176.15 | 382 | B10 |
| Green | TET | 180.93 | 2,054 | B10 |
| Green | TET | 181.93 | 990 | B10 |
| Green | TET | 183.81 | 1,191 | B10 |
| Green | TET | 184.81 | 632 | B10 |
| Green | TET | 272.23 | 407 | B9 |
| Green | TET | 273.22 | 447 | B9 |
| Yellow | HEX | 128.65 | 1,275 | B11 |
| Yellow | HEX | 192.65 | 273 | B11 |
| Yellow | HEX | 193.64 | 127 | B11 |
| Red | TAMRA | 75.00 | 292 | GS-500 TAMRA |
| Red | TAMRA | 100.00 | 113 | GS-500 TAMRA |
| Red | TAMRA | 139.00 | 298 | GS-500 TAMRA |
| Red | TAMRA | 150.00 | 296 | GS-500 TAMRA |
| Red | TAMRA | 160.00 | 301 | GS-500 TAMRA |
| Red | TAMRA | 200.00 | 304 | GS-500 TAMRA |
| Red | TAMRA | 250.00 | 337 | GS-500 TAMRA |
| Red | TAMRA | 300.00 | 366 | GS-500 TAMRA |
The peaks associated with each microsatellite locus were identified by reference to the dye label and the expected size range of products at each locus (Fig. 5). Many peaks occurred as a doublet, 1 nt U apart, corresponding to the labeled strand of the PCR product with or without the extra deoxyadenylate added at the 3′ end by Taq polymerase. In constructing a database of allele sizes for each endophyte strain, we have entered the size of the product without the additional deoxyadenylate. Where only a single peak was observed, the value for this was used. The allele sizes determined by this method were generally in good agreement with those generated by radiolabeled PCR (Table 3), though the ease of size determination was far greater for the automated multi-dye color system. In addition, automated analysis allows precise size determination of multiple products, including similar-sized products from different loci, in a single run. To check the reproducibility of allele size determinations by the automatic sequencer, dilutions of various fluorescently labeled PCR products of Tf13 were submitted for analysis. Allele sizes were found to be most consistent when the peak heights of the samples were less than 2,000 fluorescent units (FU). Working within this range resulted in an allele size variation of up to 0.4 nucleotide units.
Once the optimum conditions for automated analysis had been established, DNA from all endophyte isolates in Table 1 were amplified in the five-locus multiplex PCR assay and the sizes of the products generated were entered into a database. A summary of those results is given in Table 5. From these data we estimate that there are at least 5, 6, 3, 12, and 14 unique alleles at the loci B4, B6, B9, B10, and B11, respectively, with the allele groupings identified being the same as those shown in Table 3.
TABLE 5.
Allele sizes of Epichloë endophyte microsatellite loci as determined by automatic analysis
| Taxonomic group | Strain | Isozyme phenotypea | Allele size (nt U)b
|
||||
|---|---|---|---|---|---|---|---|
| B4 | B6 | B9 | B10 | B11 | |||
| LpTG-2 | Lp1 | LpA | 100.0 | 172.4 | 247.4* | 169.3* | 119.8 |
| 119.8 | 187.6* | 178.1* | |||||
| LpTG-2 | Lp2 | LpB | 100.0 | 172.4 | 247.3* | 169.3 | 119.6 |
| 120.0 | 187.5 | 178.1* | |||||
| LpTG-1 | Lp5 | loA | 100.0 | 187.7 | 247.3* | 178.1* | 177.0* |
| LpTG-1 | Lp6 | loA | 100.1 | 187.6 | 247.3* | 178.0* | 176.9* |
| LpTG-1 | Lp13 | loA | 100.0 | 187.7 | 247.4* | 178.0* | 177.0* |
| LpTG-1 | Lp19 | loB | 100.0 | 187.6 | 247.4* | 178.0 | 180.8 |
| LpTG-1 | Lp20 | loB | 100.0 | 187.6 | 247.4* | 178.1* | 180.9 |
| LpTG-1 | Lp21 | loB | 100.0 | 187.6 | 247.4* | 178.1 | 180.9 |
| LpTG-1 | Lp7 | loD | 100.0 | 187.6* | 247.4* | 178.1 | 188.9 |
| LpTG-1 | Lp14 | loE | 100.0 | 186.6* | 271.2* | 163.5 | 132.8 |
| LpTG-1 | Lp9 | loF | 100.1 | 187.6 | 248.3* | 178.0* | 188.8* |
| LpTG-1 | AR1 | 100.0 | 187.6 | 246.4* | 178.0* | 149.8 | |
| FaTG-1 | Tf5 | coB | 100.0 | 193.7 | 272.1* | 160.7* | 149.9 |
| 103.9 | 172.2* | 192.7* | |||||
| 186.7* | |||||||
| FaTG-1 | Tf27 | coB | 100.0 | 196.8 | 272.2* | 155.0* | 172.9* |
| 103.2 | 163.5* | 197.0* | |||||
| 180.9* | |||||||
| FaTG-1 | Tf28 | coC | 100.0 | 193.8 | 272.2* | 175.1* | 128.7 |
| 180.9* | 192.7* | ||||||
| 183.8* | |||||||
| FaTG-2 | Tf13 | FaA | 100.5 | 187.6 | 271.3* | 172.1* | 128.7* |
| 186.8* | 141.0 | ||||||
| FaTG-2 | Tf15 | FaC | 101.0 | 186.6* | 247.5* | 172.1* | 128.8* |
| 178.0* | 165.2* | ||||||
| FaTG-2 | Tf20 | FaD | 100.3 | 187.6 | 247.4* | 172.1* | 128.6 |
| 178.0* | 184.8* | ||||||
| FaTG-3 | Tf16 | FaF | 120.4 | 171.6* | –c | 172.2* | 128.6 |
| 180.9* | |||||||
| FaTG-3 | Tf18 | FaF | 120.3 | 172.6 | – | 172.2* | 128.6 |
| 180.9* | |||||||
| FpTG-1 | Fp1 | unA | 84.8 | 178.7 | – | 160.7* | 120.7 |
| 104.4 | 195.8* | ||||||
| FpTG-1 | Fp2 | unA | 84.7 | 178.7 | – | 160.6 | 120.7 |
| 103.7 | 195.7* | ||||||
| FpTG-1 | Fp4 | unB | 84.7 | 178.7 | – | 172.1* | 120.7 |
| 103.9 | 192.7 | ||||||
| E. typhina | E8 | 120.5 | 172.5 | – | 178.1 | – | |
| E. festucae | Fr1 | FF6 | 100.0 | 186.6* | 272.1* | 172.1* | 165.3 |
| E. festucae | Frc5 | 100.5 | 192.5* | 273.2* | 178.0* | 169.1* | |
| E. festucae | Frc7 | 100.0 | 187.7 | 272.1* | 178.0* | 149.9 | |
| E. festucae | Frr1 | 100.0 | 175.7 | 272.1* | 166.3* | 116.8 | |
| E. festucae | Fg1 | 100.0 | 187.7 | 279.0* | 195.7* | 128.7 | |
| E. festucae | Fl1 | FF10 | 100.0 | 187.5 | 272.2* | 178.0* | 116.8* |
Results of identification of endophytes in a blind test.
To test the robustness of this method for identifying endophyte strains in planta, 10 samples of plant material from endophyte-infected perennial ryegrass, tall fescue, and meadow fescue were submitted blindly by an independent researcher at AgResearch. DNA was isolated from these samples with the FastDNA Kit H and amplified in single-dye PCR reactions. The products from each reaction were pooled and submitted for automatic analysis. Alleles for each sample were identified and then compared with those entered into the database. In all cases a correct match was found with a taxonomic grouping that had been previously determined for that isolate by using a combination of morphological, biochemical, and genetic criteria (12, 38, 53, 61).
DISCUSSION
We describe here the molecular cloning of a set of microsatellite loci from Epichloë grass endophytes and the development of an automated PCR fingerprinting method based on sequences at those loci, a method that distinguishes the different groups of isolates to the level of their isozyme phenotype grouping. Surprisingly, there are few reports to date on the cloning of microsatellites from fungi (23, 27, 40) despite their widespread use for kinship, population structure, and mapping studies in plants and animals.
Based on Southern analysis, we found an average frequency of one (CA)n or (GA)n locus per 2 Mb of DNA and one (CAA)n, (GAA)n, or (ATC)n locus per 2.5 Mb of the Lp1 genome. By comparison with similar estimates for plants, (CA)n and (GA)n frequencies range from 1/270 kb in wheat (48) to 1/1.2 Mb in tomato plants (6). The frequency of the same classes of microsatellites in mammals is even greater (34). However, these figures should be regarded as a minimum, since a single hybridizing band in a Southern may correspond to more than one locus and, like the alternative approach of determining the number of positive clones in a library screen, they will also vary depending on the stringency of the hybridization used. The effect of the latter was evident here in that not all of the positive clones identified had microsatellite sequences that were perfect matches with the probes used. For example, the B5 and B9 loci only partially match the sequences of the tri- and dinucleotide repeat probes that were used for the isolation of these sequences.
It would also appear that the partial libraries made in M13mp19 were not as representative of the diversity of loci as the Southern blots had indicated. For example, the Southern analysis of Lp1 genomic DNA indicated that there were at least 20 (CA)n and (GA)n microsatellites in the size range of 100 to 1,000 bp used to construct the library, yet only three different microsatellite clone types were identified from the 24 clones screened. This was probably a consequence of preferential ligation of the smaller inserts into the M13mp19 vector, as all clones that were analyzed had inserts of between 100 and 300 bp. More accurate determinations of both the frequency and the diversity of microsatellites in fungi and other organisms will emerge from analysis of DNA sequence data generated from either partial or complete genome sequencing initiatives, such as that for Saccharomyces cerevisiae (20, 47).
A search by Groppe et al. (27) of fungal DNA sequences held in the GenBank and EMBL databases for all possible mono-, di-, and trinucleotide motifs (>20 bp) showed that the predominant microsatellite type found in fungi is (AT)n, representing 42% of the microsatellites identified. The other most frequently occurring microsatellite types were, in decreasing order of abundance: (A)n, (AAT)n, (AAC)n, (AAG)n, (ATC)n, (AC)n, and (AG)n. Screening for the abundant (AT)n repeat class was not carried out in this study, since these sequences are self-complementary, making it very difficult to isolate such sequences by solution hybridization methodology (33, 34).
As shown in Table 2, the nine microsatellite loci isolated contain a variety of sequence motifs, ranging from repeats of mononucleotides to decanucleotides. To determine the degree of length polymorphism associated with these microsatellites and those isolated by Groppe et al. (27) and Dobson (17), PCR assays were carried out with DNA from a collection of Epichloë endophytes representative of isolates from the forage grasses, perennial ryegrass, tall fescue, and meadow fescue.
Isolates from perennial ryegrass fall into two taxonomic groups: LpTG-1 (N. lolii) and LpTG-2, with the former being further resolved by isozyme analysis into six different phenotype groupings (12). While most of the microsatellite loci were unable to resolve the N. lolii isolates to the level of the isozyme phenotype, locus B11 was particularly informative, with five alleles being identified as unique to this species. While the other loci were less informative in resolving this relatively homogeneous group of isolates, strains Lp14 and Lp9 were frequently shown to be different from the other N. lolii isolates. There is now very good evidence from molecular phylogenetic studies that the asexual Neotyphodium spp. have evolved from the sexual Epichloë spp. (52). From these studies the most closely related sexual ancestors of the N. lolii group are the E. festucae (36, 53). Locus B4 is particularly informative in demonstrating this evolutionary connection, with both groups sharing a common allele.
The second taxonomic grouping within the perennial ryegrass isolates is represented in this study by just two isolates, Lp1 and Lp2 (12), and both are known to be interspecific hybrids between E. typhina and N. lolii (53). The hybrid nature of these isolates is supported here in that the two different alleles found in E. typhina and N. lolii for loci B4 and B6 were both amplified from the LpTG-2 isolates.
As outlined in the introduction, the endophyte isolates from tall fescue have been resolved into three taxonomic groups (12) and all are known to be interspecific hybrids (61). However, the proposed evolutionary origins of these hybrids is complex in that multiple hybridization events between N. uncinatum and the Epichloë species would be required to account for the diversity of tub2 and other genes that are found in this group (61). Loci B10 and B11 are particularly supportive of a hybrid origin for this group of isolates, since multiple alleles are present for all strains at locus B10 and for FaTG-1 and FaTG-2 at B11. All sexual species and the haploid N. lolii group have single alleles at these loci. Many common alleles were found between the tall fescue isolates and their proposed ancestors (52), which include N. uncinatum, E. festucae, and E. baconii for FaTG-1; E. festucae and E. baconii for FaTG-2; and E. typhina and E. baconii for FaTG-3.
The isolates from meadow fescue used in this study were readily identified from the other endophytes by the distinct alleles found at six loci (i.e., B1, B4, B6, B9, B10, and B11). While N. uncinatum has only a single copy of the β-tubulin gene (tub2), the phylogeny generated from sequences for this gene contradicts that for the rDNA, suggesting that N. uncinatum is also a hybrid (52). The hybrid origin of this group is supported by both isozyme analysis (36) and the multiple alleles generated at loci B4 and B10 (this study).
Epichloë festucae was the only group of sexual endophytes where we had several isolates available to study. These isolates, from very different hosts, showed a higher degree of polymorphism than the groups of asexual isolates, which would be expected for an out-crossing sexual species. However, it is interesting to note that the stability of microsatellite repeat tracts is similar both during meiosis and mitosis (58).
No microsatellite PCR products were obtained with any of the primer pairs with the outgroups Claviceps purpurea and Echinodothis tuberiformis, which are both from the same family as the Epichloë endophytes (Clavicipitaceae). This shows that these primers are highly specific for the Epichloë endophytes, demonstrating that the assay is not influenced by the presence of unrelated fungal endophytes or contaminating fungal species in the plant tissue.
While the microsatellite allele data generated by manual analysis of autoradiographs of 33P-labeled products was largely concordant with that generated by automated analysis of fluorescently labeled products, the latter method was superior in many respects. Automated analysis allows for precise PCR product size estimation over a wide size range in a single run, with a detection sensitivity that reduces the amount of product required for analysis. This becomes particularly important in the detection of endophytes in planta, where the fungal biomass can be very low. Dye-labeled primers and products are able to be stored for long periods of time without signal decay and are safer to use than radioisotopes. Automated analysis is also faster, since data is collected as the gel is running, thus eliminating the need for gel drying, film exposure, and development. The complexity of the banding pattern from a single product is reduced since only one strand of the product is labeled; this greatly simplifies the analysis of hybrid endophytes which have multiple alleles of similar size for some loci. The three-color dye system allows products from different loci to be identified simultaneously, even if their peaks overlap, as shown in Fig. 7. This cannot be done with manual methods, and separate gels are often required to analyze individual loci. Finally, the data are generated as the size of the DNA fragment, enabling comparisons to be readily made between laboratories, a task which is difficult to achieve with other typing methods, such as restriction fragment length polymorphism or RAPD analysis. Thus, microsatellite fingerprints may be generated at independent laboratories and submitted into a common database. However, as the estimated size of the fragment is dependent on its electrophoretic mobility, using a different separation matrix (such as non-cross-linked network polymer used in capillary electrophoresis) may alter the mobility of the DNA fragment, thus resulting in different size calling (49).
One problem encountered when analyzing fingerprints from the automated system was the inconsistent presence of split peak signals, 1 nt apart; this was a consequence of the inherent property of Taq polymerase to occasionally add a deoxyadenylate to the 3′ terminus of the PCR product. The primer dependence of this property of Taq polymerase (7) was reflected in the frequent occurrence of split peaks for amplifications at loci B9, B10, and B11. Single peaks predominated at loci B4 and B6 and were assumed to be the product without the addition of a deoxyadenylate. Methods that would potentially eliminate this problem would be the use of a DNA polymerase, such as Pwo or Tfl (which amplify only fragments with blunt ends), enzymatic removal of the overhang by T4 DNA polymerase (24), or use of the PIGtailing method (7).
In conclusion, we describe here the development of a microsatellite fingerprinting assay suitable for the rapid and accurate identification of endophytes both in culture and in planta. This assay should be a useful tool for grass breeders to rapidly determine both the identity of known endophytes in pasture and for monitoring the persistence in the field of endophytes in new grass associations. The assay will also be a useful tool for researchers to fingerprint strains and as an adjunct to the study of endophyte evolution, particularly for identifying the many hybrid asexual Epichloë endophytes.
ACKNOWLEDGMENTS
We thank Mike Christensen for donating many of the fungal strains and endophyte-infected grass tissues and for valuable discussion on the classification and identification of the endophytes. We also thank Joanne Dobson for providing sequence data for the B11 microsatellite, Lorraine Berry for the automated analysis of the microsatellite fingerprints, Carolyn Young for technical assistance, Walter Hollin for providing DNA from outgroup isolates, and Chris Schardl for critically reviewing the manuscript.
This research was supported by a grant from AgResearch, Grasslands Research Centre, Palmerston North, New Zealand.
REFERENCES
- 1.Arachevaleta M, Bacon C W, Hoveland C S, Radcliffe D E. Effect of the tall fescue endophyte on plant response to environmental stress. Agron J. 1989;81:83–90. [Google Scholar]
- 2.Bacon C W, Lyons P C, Porter J K, Robbins J D. Ergot toxicity from endophyte-infected grasses: a review. Agron J. 1986;78:106–116. [Google Scholar]
- 3.Bacon C W, Porter J K, Robbins J D, Luttrell E S. Epichloë typhina from toxic tall fescue grasses. Appl Environ Microbiol. 1977;34:576–581. doi: 10.1128/aem.34.5.576-581.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boehringer Mannheim GmbH. The DIG system user’s guide for filter hybridization. Mannheim, Germany: Boehringer Mannheim GmbH; 1995. [Google Scholar]
- 5.Bolton E T, McCarthy B J. A general method for the isolation of RNA complementary to DNA. Proc Natl Acad Sci USA. 1962;48:1390–1397. doi: 10.1073/pnas.48.8.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown P, Tanksley S D. Characterization and genetic mapping of simple repeat sequences in the tomato genome. Mol Gen Genet. 1996;250:39–49. doi: 10.1007/BF02191823. [DOI] [PubMed] [Google Scholar]
- 7.Brownstein M J, Carpten J D, Smith J R. Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. BioTechniques. 1996;20:1004–1010. doi: 10.2144/96206st01. [DOI] [PubMed] [Google Scholar]
- 8.Bullock W O, Fernandez J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–378. [Google Scholar]
- 9.Byrd A D, Schardl C L, Songlin P J, Mogen K L, Siegel M R. The β-tubulin gene of Epichloë typhina from perennial ryegrass (Lolium perenne) Curr Genet. 1990;18:347–354. doi: 10.1007/BF00318216. [DOI] [PubMed] [Google Scholar]
- 10.Christensen M J, Latch G C M. Variation among isolates of Acremonium endophytes (A. coenophialum and possibly A. typhinum) from tall fescue (Festuca arundinacea) Mycol Res. 1991;95:1123–1126. [Google Scholar]
- 11.Christensen M J, Latch G C M, Tapper B A. Variation within isolates of Acremonium endophytes from perennial rye-grasses. Mycol Res. 1991;95:918–923. [Google Scholar]
- 12.Christensen M J, Leuchtmann A, Rowan D D, Tapper B A. Taxonomy of Acremonium endophytes of tall fescue (Festuca arundinacea), meadow fescue (F. pratensis) and perennial rye-grass (Lolium perenne) Mycol Res. 1993;97:1083–1092. [Google Scholar]
- 13.Chung K-R, Schardl C L. Sexual cycle and horizontal transmission of the grass symbiont, Epichloë typhina. Mycol Res. 1997;101:295–301. [Google Scholar]
- 14.Clay K. Fungal endophytes, grasses, and herbivores. In: Barbosa P, Krischik V A, Jones C G, editors. Microbial mediation of plant-herbivore interactions. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 199–226. [Google Scholar]
- 15.Crawford A M, Buchanan F C, Swarbrick P A. Ovine dinucleotide repeat polymorphism at the MAF18 locus. Anim Genet. 1990;21:433–434. doi: 10.1111/j.1365-2052.1990.tb01990.x. [DOI] [PubMed] [Google Scholar]
- 16.Dahlman D L, Eichenseer H, Siegel M R. Chemical perspectives on endophyte-grass interactions and their implications to insect herbivory. In: Barbosa P, Kirschik L, Jones E, editors. Microbial mediation of plant-herbivore interactions. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 227–252. [Google Scholar]
- 17.Dobson J M. MSc. thesis. Palmerston North, New Zealand: Massey University; 1997. [Google Scholar]
- 18.Doss R P, Welty R E. A polymerase chain reaction-based procedure for detection of Acremonium coenophialum in tall fescue. Phytopathology. 1995;85:913–917. [Google Scholar]
- 19.Doyle J J, Doyle L L. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- 20.Field D, Wills C. Abundant microsatellite polymorphism in Saccharomyces cerevisiae, and the different distributions of microsatellites in eight prokaryotes and S. cerevisiae, result from strong mutation pressures and a variety of selective forces. Proc Natl Acad Sci USA. 1998;95:1647–1652. doi: 10.1073/pnas.95.4.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fletcher L R, Harvey I C. An association of a Lolium endophyte with ryegrass staggers. N Z Vet J. 1981;29:185–186. doi: 10.1080/00480169.1981.34839. [DOI] [PubMed] [Google Scholar]
- 22.Gallagher R T, Hawkes A D, Steyn P S, Vleggaar R. Tremorgenic neurotoxins from perennial ryegrass causing ryegrass staggers disorder of livestock: structure elucidation of lolitrem B. London, United Kingdom: Chemical Society (Chemical Communication); 1984. pp. 614–616. [Google Scholar]
- 23.Geistlinger J, Maqbool S, Kaiser W J, Kahl G. Detection of microsatellite fingerprint markers and their mendelian inheritance in Ascochyta rabiei. Mycol Res. 1997;101:1113–1121. [Google Scholar]
- 24.Ginot F, Bordelais I, Nguyen S, Gyapay G. Correction of some genotyping errors in automated fluorescent microsatellite analysis by enzymatic removal of one base overhangs. Nucleic Acids Res. 1996;24:540–541. doi: 10.1093/nar/24.3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glenn A E, Bacon C W, Price R, Hanlin R T. Molecular phylogeny of Acremonium and its taxonomic implications. Mycologia. 1996;88:369–383. [Google Scholar]
- 26.Groppe K, Boller T. PCR assay based on a microsatellite-containing locus for detection and quantification of Epichloë endophytes in grass tissue. Appl Environ Microbiol. 1997;63:1543–1550. doi: 10.1128/aem.63.4.1543-1550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Groppe K, Sanders I, Wiemken A, Boller T. A microsatellite marker for studying the ecology and diversity of fungal endophytes (Epichloë spp.) in grasses. Appl Environ Microbiol. 1995;61:3943–3949. doi: 10.1128/aem.61.11.3943-3949.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gwinn K D, Collins-Shepard M H, Reddick B B. Tissue print-immunoblot, an accurate method for the detection of Acremonium coenophialum in tall fescue. Phytopathology. 1991;81:747–748. [Google Scholar]
- 29.Gwinn K D, Gavin A M. Relationship between endophyte infestation level of tall fescue and seed lots and Rhizoctonia zeae seedling disease. Plant Dis. 1992;76:911–914. [Google Scholar]
- 30.Hill N S, Stringer W C, Rottinghaus G E, Belesky D P, Parrott W A, Pope D D. Growth, morphological, and chemical component responses of tall fescue to Acremonium coenophialum. Crop Sci. 1990;30:156–161. [Google Scholar]
- 31.Innis M A, Gelfand D H. Optimization of PCRs. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. A guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 3–12. [Google Scholar]
- 32.Kimmons C A, Gwinn K D, Bernard E C. Nematode reproduction on endophyte-infected and endophyte-free tall fescue. Plant Dis. 1990;74:757–761. [Google Scholar]
- 33.Kostia S, Varvio S-L, Vakkari P, Pulkkinen P. Microsatellite sequences in a conifer, Pinus sylvestris. Genome. 1995;38:1244–1248. doi: 10.1139/g95-163. [DOI] [PubMed] [Google Scholar]
- 34.Lagercrantz U, Ellegren H, Andersson L. The abundance of various polymorphic microsatellite motifs differs between plants and vertebrates. Nucleic Acids Res. 1993;21:1111–1115. doi: 10.1093/nar/21.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Latch G C M, Christensen M J, Samuels G J. Five endophytes of Lolium and Festuca in New Zealand. Mycotaxon. 1984;20:535–550. [Google Scholar]
- 36.Leuchtmann A. Isozyme relationships of Acremonium endophytes from twelve Festuca species. Mycol Res. 1994;98:25–33. [Google Scholar]
- 37.Leuchtmann A, Clay K. Isozyme variation in the Acremonium/Epichloë fungal endophyte complex. Phytopathology. 1990;80:1133–1139. [Google Scholar]
- 38.Leuchtmann A, Schardl C L, Siegel M R. Sexual compatibility and taxonomy of a new species of Epichloë symbiotic with fine fescue grasses. Mycologia. 1994;86:802–812. [Google Scholar]
- 39.Levinson G, Gutman G A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 40.Longato S, Bonfante P. Molecular identification of mycorrhizal fungi by direct amplification of microsatellite regions. Mycol Res. 1997;101:425–432. [Google Scholar]
- 41.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 42.Morgante M, Olivieri A M. PCR-amplified microsatellites as markers in plant genetics. Plant J. 1993;3:175–182. [PubMed] [Google Scholar]
- 43.Murray F R, Latch G C M, Scott D B. Surrogate transformation of perennial ryegrass, Lolium perenne, using genetically modified Acremonium endophyte. Mol Gen Genet. 1992;233:1–9. doi: 10.1007/BF00587554. [DOI] [PubMed] [Google Scholar]
- 44.Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 45.Raisbeck M F, Rottinghaus G E, Kendall J D. Effects of naturally occurring mycotoxins on ruminants. In: Smith J E, Henderson R S, editors. Mycotoxins and animal foods. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 647–677. [Google Scholar]
- 46.Reddick B B, Collins M H. An improved method for detection of Acremonium coenophialum in tall fescue plants. Phytopathology. 1988;78:418–420. [Google Scholar]
- 47.Richard G F, Dujon B. Distribution and variability of trinucleotide repeats in the genome of the yeast Saccharomyces cerevisiae. Gene. 1996;174:165–174. doi: 10.1016/0378-1119(96)00514-8. [DOI] [PubMed] [Google Scholar]
- 48.Röder M S, Plaschke J, König S U, Börner A, Sorrells M E, Tanksley S D, Ganal M W. Abundance, variability and chromosomal location of microsatellites in wheat. Mol Gen Genet. 1995;246:327–333. doi: 10.1007/BF00288605. [DOI] [PubMed] [Google Scholar]
- 49.Rosenblum B B, Oaks F, Menchen S, Johnson B. Improved single-strand DNA sizing accuracy in capillary electrophoresis. Nucleic Acids Res. 1997;25:3925–3929. doi: 10.1093/nar/25.19.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roy M S, Geffen E, Smith D, Ostrander E A, Wayne R K. Patterns of differentiation and hybridization in North American wolflike canids, revealed by analysis of microsatellite loci. Mol Biol Evol. 1994;11:553–570. doi: 10.1093/oxfordjournals.molbev.a040137. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 52.Schardl C L. Epichloë species: fungal symbionts of grasses. Annu Rev Phytopathol. 1996;34:109–130. doi: 10.1146/annurev.phyto.34.1.109. [DOI] [PubMed] [Google Scholar]
- 53.Schardl C L, Leuchtmann A, Tsai H-F, Collett M A, Watt D M, Scott D B. Origin of a fungal symbiont of perennial ryegrass by interspecific hybridization of a mutualist with the ryegrass choke pathogen, Epichloë typhina. Genetics. 1994;136:1307–1317. doi: 10.1093/genetics/136.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schardl C L, Liu J-S, White J F, Jr, Finkel R A, An Z-Q, Siegel M R. Molecular phylogenetic relationships of nonpathogenic grass mycosymbionts and clavicipitaceous plant pathogens. Plant Syst Evol. 1991;178:27–41. [Google Scholar]
- 55.Schardl C L, Phillips T D. Protective grass endophytes. Where are they from and where are they going? Plant Dis. 1997;81:430–438. doi: 10.1094/PDIS.1997.81.5.430. [DOI] [PubMed] [Google Scholar]
- 56.Siegel M R, Latch G C M, Bush L P, Fannin F F, Rowan D D, Tapper B A, Bacon C W, Johnson M C. Fungal endophyte-infected grasses: alkaloid accumulation and aphid response. J Chem Ecol. 1990;16:3301–3315. doi: 10.1007/BF00982100. [DOI] [PubMed] [Google Scholar]
- 57.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 58.Strand M, Prolla T A, Liskay R M, Petes T D. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 59.Tautz D, Renz M. Simple sequences are ubiquitous repetitive components of eukaryote genomes. Nucleic Acids Res. 1984;12:4127–4138. doi: 10.1093/nar/12.10.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thuring R W J, Sanders J P M, Borst P. A freeze-squeeze method for recovering long DNA from agarose gels. Anal Biochem. 1975;66:213–220. doi: 10.1016/0003-2697(75)90739-3. [DOI] [PubMed] [Google Scholar]
- 61.Tsai H-F, Liu J-S, Staben C, Christensen M J, Latch G C M, Siegel M R, Schardl C L. Evolutionary diversification of fungal endophytes of tall fescue grass by hybridization with Epichloë species. Proc Natl Acad Sci USA. 1994;91:2542–2546. doi: 10.1073/pnas.91.7.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.West C P, Izekor E, Oosterhuis D M, Robbins R T. The effect of Acremonium coenophialum on the growth and nematode infestation of tall fescue. Plant Soil. 1988;112:3–6. [Google Scholar]
- 63.Western J H, Cavett J J. The choke disease of cocksfoot (Dactylis glomerata) caused by Epichloë typhina. Trans Br Mycol Soc. 1959;42:298–307. [Google Scholar]