Graphical abstract

Keywords: Antimicrobial peptides, Peptide self-assembly, Structural motifs, Physicochemical determinants, AMP evolution
Abbreviations: AMPs, Antimicrobial peptides; HDPs, Host defense peptides; MRSA, Methicillin-resistant Staphylococcus aureus; SUV, Single Unilamellar Vesicle; MUV, Multilamellar Vesicle; HD6, Human-α-defensin 6
Abstract
The burgeoning menace of antimicrobial resistance across the globe has necessitated investigations into other chemotherapeutic strategies to combat infections. Antimicrobial peptides, or host defense peptides, are a set of promising therapeutic candidates in this regard. Most of them cause membrane permeabilization and are a key component of the innate immune response to pathogenic invasion. It has also been reported that peptide self-assembly is a driving factor governing the microbicidal activity of these peptide candidates. While efforts have been made to develop novel synthetic peptides against various microbes, many clinical trials of such peptides have failed due to toxicity and hemolytic activity to the host. A function-guided rational peptide engineering, based on evolutionary principles, physicochemical properties and activity determinants of AMP activity, is expected to help in targeting specific microbes. Furthermore, it is important to develop a unified understanding of the evolution of AMPs in order to fully appreciate their importance in host defense. This review seeks to explore the evolution of AMPs and the physicochemical determinants of AMP activity. The specific interactions driving AMP self-assembly have also been reviewed, emphasizing implications of this self-assembly on microbicidal and immunomodulatory activity.
1. Introduction
Every living organism is in constant contact with a diverse array of infectious and pathogenic microorganisms. Despite this, the cornea or the buccal cavity of a human, or the gut of an insect is, on an average, largely devoid of any sign of infection. This is accomplished by the production of broad-spectrum peptides, which help in fending off harmful microorganisms. These evolutionarily ancient peptide weapons are referred to as antimicrobial peptides (AMPs), or host defense peptides (HDPs). These peptides are predominantly short cationic peptides with a diverse set of targets ranging from the microbial membrane to the translational machinery of the cell. Antibiotic resistance is a global public health menace, and there are chances of return to a pre-antibiotic era with the continued failure of last-line antibiotics [1]. This has led to intensive research into the field of AMPs, which have been enormously successful in retaining their activity despite continuous evolutionary conflict with various microbial species over the course of evolution.
AMPs are oligopeptides of 5 to 100 amino acids, predominantly with a net positive charge (typically +2 to +11) [2]. However, with the explosion in our knowledge of the diversity of AMPs, their has been increasing arbitrariness about their precise definition, particularly owing to their sheer structural and functional diversity [3]. Complement proteins like C9 which form ordered pores in microbial membranes have not been classified as AMPs due to the tight immune regulation of their formation, while large proteins like lactoferricin (molecular weight > 80 kDa) have been classified as AMPs due their ubiquitous presence in various body fluids and non-specific mode of action [3].
The panopoly of AMPs produced by various taxa are synthesized by two distinct routes – ribosomal and non-ribosomal. The former group of AMPs are synthesized by the usual process of translation executed by the ribosomes. The latter group comprises peptides that are end-product synthesized by enzyme complexes, which carry out various post-translational modifications like glycosylation and amidation or produce small peptides by carrying out proteolysis of larger proteins [4].
AMPs abound in all the kingdoms of the living world (Table 1) and constitute the first line of chemical sentinels in innate resistance to infection in most organisms. Among the prokaryotes (where the AMPs are associated with conferring competitive advantage to the host species over other microbes growing on the substratum and not strictly with immunity per se), the most commonly produced AMPs include bacteriocins, which are ribosomally synthesized. Among bacteriocins, lantibiotics are a widely studied group, members of which include nisin and cytolysin. These are small (<5 kDa) peptides containing the unusual amino acids lanthionine, β-methyllanthionine and a number of dehydrated amino acids [5]. Famous examples of non-lantibiotic bacteriocins include gramicidin S from Lactococcus lactis and Bacillus brevis [6] and colicins produced by E. coli. Bacteriocins are extremely potent AMPs, with some of them displaying activity against dreaded pathogens like Listeria monocytogenes [6]. Unlike the broad-spectrum effects of many natural AMPs, many bacteriocins display significant specificity and are targeted towards related strains. Bacteriocins are crucial for maintaining the structure of bacterial communities by defending against the invasion of other strains, enabling the invasion of a strain into an existing microbial community and even by regulating quorum sensing [7].
Table 1.
Antimicrobial peptide in all kingdoms of life.
| Kingdom | Peptide class | Examples | Example of producing organism | Target microbes | Ref |
|---|---|---|---|---|---|
| Archaebacteria | Sulfolobicins | SulA, SulB | Sulfolobus tokodaii DSM16993 | Members of Sulfolobus sp. | 1,2 |
| Halocins | Halocin A4 | Haloferax mediterranei R4 | Members of Halobacteriales like Halobacterium salinarium | 1, 3,4,5 | |
| Eubacteria | Circular bacteriocins | Acidocin B | Lactobacillus acidophilus | Listeria monocytogenes, Clostridium sporogenes | 6,7 |
| Lantibiotics | Nisin | Lactococcus lactis | Gram positive bacteria like Staphylococcus aureus, including MDR S. aureus biofilms | 8, 7 | |
| Fungi | Fungal defensin | Plectasin | Pseudoplectania nigrella | Streptococcus pneumoniae, MRSA | 9 |
| Plantae | Thionins | Purothionin | Triticum aestivum | Phytopathogens like Xanthomonas sp., Corynebacterium sp. | 10 |
| Knottin-type peptides | Psacotheasin | Psacothea hilaris | Fungal species like Candida albicans | 11 | |
| Animalia | Cathelicidins | LL-37 | Homo sapiens sapiens | Gram-positive (like Staphylococcus sp.) and Gram-negative (like Pseudomonas sp.) bacteria, as well as on the yeast C. albicans | 12 |
| Tachyplesins | Tachyplesin I | Tachypleus tridentatus | Gram-positive (like Staphylococcus sp.) and Gram-negative (like Salmonella sp.) | 13 |
1 Besse A, Peduzzi J, Rebuffat S, Carre-Mlouka A. 2015. Antimicrobial peptides and proteins in the face of extremes: lessons from archaeocins. Biochimie 118:344–355. https://doi.org/10.1016/j.biochi.2015.06.004.
2 A.F. Ellen, O.V. Rohulya, F. Fusetti, M. Wagner, S.-V. Albers, A.J.M. Driessen, The sulfolobicin genes of Sulfolobus acidocaldarius encode novel antimicrobial proteins, J. Bacteriol. 193 (2011) 4380e4387, http://dx.https://doi.org/10.1128/JB.05028-11.
3 I. Meseguer, F. Rodriguez-Valera, Production and purification of halocin H4, FEMS Microbiol. Lett. 28 (1985) 177 – 182.
4 A. Naor, P. Lapierre, M. Mevarech, R.T. Papke, U. Gophna, Low species barriers in halophilic archaea and the formation of recombinant hybrids, Curr. Biol. 22 (2012) 1444 – 1448, http://dx.https://doi.org/10.1016/j.cub.2012.05.056.
5 A. Naor, Y. Yair, U. Gophna, A halocin-H4 mutant Haloferax mediterranei strain retains the ability to inhibit growth of other halophilic archaea, Extrem. Life Extreme Cond. 17 (2013) 973 – 979, http://dx.https://doi.org/10.1007/s00792-013-0579-8.
6 Leer RJ, van der Vossen JM, van Giezen M, van Noort JM, Pouwels PH (1995) Genetic analysis of acidocin B, a novel bacteriocin produced by Lactobacillus acidophilus. Microbiology 141(Pt 7):1629–1635.
7 Drider, D., & Rebuffat, S. (2011). Prokaryotic antimicrobial peptides: From genes to applications. Springer.
8 Field D, O’ Connor R, Cotter PD, Ross RP and Hill C (2016) In Vitro Activities of Nisin and Nisin Derivatives Alone and In Combination with Antibiotics against Staphylococcus Biofilms. Front. Microbiol. 7:508. https://doi.org/10.3389/fmicb.2016.00508.
9 Mygind PH, Fischer RL, Schnorr KM, Hansen MT, Sonksen CP, Ludvigsen S, Raventos D, Buskov S, Christensen B, De Maria L, Taboureau O, Yaver D, Elvig-Jorgensen SG, Sorensen MV, Christensen BE, Kjaerulff S, Frimodt-Moller N, Lehrer RI, Zasloff M, Kristensen HH. (2005). Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature, 437, 975–980.
10 Fernandez de Caleya R, Gonzalez-Pascual B, García-Olmedo F, Carbonero P (1972) Susceptibility of phytopathogenic bacteria to wheat purothionins in vitro. Appl Microbiol 23:998–1000.
11 Hwang B, Hwang JS, Lee J, Lee DG (2010a) Antifungal properties and mode of action of psacotheasin, a novel knottin-type peptide derived from Psacothea hilaris. Biochem Biophys Res Commun 400:352–357.
12 Dürr UH, Sudheendra US, Ramamoorthy A. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim Biophys Acta. 2006 Sep;1758(9):1408–25. https://doi.org/10.1016/j.bbamem.2006.03.030. Epub 2006 Apr 4. PMID: 16716248.
13 Nakamura T, Furunaka H, Miyata T, Tokunaga F, Muta T, Iwanaga S, Niwa M, Takao T, Shimonishi Y. Tachyplesin, a class of antimicrobial peptide from the hemocytes of the horseshoe crab (Tachypleus tridentatus). Isolation and chemical structure. J Biol Chem. 1988 Nov 15;263(32):16709–13. PMID: 3141410.
In plants and fungi, which lack an adaptive immune system, AMPs assume a profoundly significant role. Like in many other kingdoms, plants produce cysteine-rich AMPs. This enables the formation of disulfide bonds in the peptide, contributing to its physicochemical stability. Some widely studied plant AMPs include thionins, defensins, snakins, cyclotides, hevein-like peptides and knottin-like peptides [8]. A few AMPs have been reported from members of the fungal kingdom, plectasin being the most celebrated one among them [9]. Another very well-studied fungal AMP is alamethicin, which is member of a general family of secondary metabolites called peptaibols. ALM exhibits a wide spectrum of biological activities including antibacterial and antifungal effects, elicitation of systemic plant-defense responses and cytolytic activity towards mammalian cells [10].
Most of the natural AMPs reported in databases like APD3 [11] and DRAMP [12] come from animal sources (Fig. 1), owing primarily to the extensive research undertaken in this area to develop novel therapeutics. Insects and other arthropods produce many AMPs in their fat bodies. The most extensively studied arthropod-derived AMPs include cecropins, diptericins, tachyplesins and the big defensins [13]. Another exceptionally well-characterized AMP belonging to a different phylum is magainin, which comes from the skin secretions of frogs of the Pipidae family [13]. Mammals produce copious amounts of defensins and cathelicidins. Mammalian defensins can be subdivided into three categories based on the number and position of disulfide bonds – α, β and θ. Furthermore, the milk produced by mammals is rich in other AMPs like lactoferrin, lactoferricin B and lactoglobulin, some of which play well-documented promiscuous roles in other physiological processes [14].
Fig. 1.
Sources of AMPs based on the data available in APD3 database [1], [2], [3].
While the preceding discussion has been largely limited to domains bacteria and eukarya, the AMPs of Archaea are only beginning to be characterized. The AMPs produced by archaebacteria are called archaeocins, in analogy with the bacteriocins discussed previously. Halocins and sulfolobicins are the archaeocins most studied to date and have been discussed elaborately elsewhere [15], [16]. Archaeocins, particularly halocins, are showing promising therapeutic applications, including their suggested ability to treat cardiac ischemia [17] as they bind to the archaeal Na+/H+ exchanger, dysfunctions of whose human homolog has been implicated in cardiac diseases. The cherry on top of the cake is the exceptional stability of the archaeocins, owing to the extreme environments inhabited by the archaebacteria.
2. Evolution of AMPs
“Nothing in biology makes sense except in the light of evolution”, so said T. Dobzhansky. In order to develop a holistic picture of the diversity of AMPs and their roles in host defense and to fully harness their therapeutic potential, it is necessary to account for their evolution.
Genes of the immune system are often the most rapidly evolving genes in the genome. This is often attributed to the co-evolution of hosts and their pathogens. Following Woolhouse et al. [18], we define co-evolution as “the process of reciprocal, adaptive genetic change in two or more species”. Co-evolution can lead to varying outcomes on the genetic structure of the population. Two commonly known models of host-pathogen co-evolutionary dynamics are the so-called ‘Red Queen’ dynamics and the ‘trench warfare’ or ‘arms race’ dynamics. (We stick to these terminologies despite their incorrect and confusing interchangeable use in common parlance.).
In the Red Queen dynamics, dynamic polymorphisms occur. This is basically a natural selection regime in which multiple alleles are maintained at a genetic locus, albeit with (often cyclically) changing frequencies with time. Red Queen dynamics usually signify a widespread balancing selection acting in the population, which might be due to frequency-dependent selection or heterozygote advantage. On the contrary, in ‘trench warfare’ or ‘arms race’ dynamics, the population is under strong positive selection at a genetic locus, resulting in successive bouts of selective sweeps resulting in fixation of a particular allele in the population. A selective sweep in the host population is followed by another sweep in the pathogen population and vice versa. These two models of selection are most relevant in the interaction of a single host species with a single pathogenic species. Of course, this is far from reality, and typically a ‘diffuse’ co-evolution is seen [19].
Furthermore, since the beginning of the research on AMPs, the general understanding in the community has been that AMPs are typically broad-spectrum, and the diversity of AMPs produced by the hosts is functionally redundant [20]. This leads to the immediate conclusion that it should ideally be possible to knockout or knockdown any one of the specific classes of AMPs in an organism and yet prevent infection by providing other AMPs, provided that the latter is administered up to a threshold concentration rapidly enough to prevent infection during the time to attainment of the threshold.
However, such an understanding of the biochemical and physiological nature of AMPs is not entirely correct and the conclusions derived have been made too general. Research over the past half a decade has demonstrated that many AMPs display remarkable specificity [21]. For example, Diptericin A produced by Drosophila melanogaster is highly specific against Providencia rettgeri bacterium and fruit flies with null alleles for the gene encoding Diptericin A show acute susceptibility to infection by this bacterium [22]. Furthermore, it has also been found that a single Ser to Arg substitution in the AMP leads to remarkable sensitivity to P. rettgeri infection, and this mutation has been naturally found in multiple species in the Drosophila genus (insert citation to reference 16). These and other similar findings posit a direct challenge to our conventional wisdom on AMPs.
The preceding discussion thus leads to multiple questions immediately. What is the selection regime that AMPs (primarily the ribosomally synthesized AMPs) are subjected to? Given the significance of AMPs in the immune system (particularly invertebrates that lack an adaptive immune system), what explains the duplication and pseudogenization of AMP encoding genes in multiple organisms belonging to distinct families [23]? Why do some organisms encode for functionally redundant AMPs?
The opinion of the scientific community on the selection regime influencing AMPs has mainly been fractured. The idea predominantly prevailing until half a decade ago was that AMPs, immune genes, are primarily under positive selection. Based on the ratio of non-synonymous to synonymous mutations (dN/dS ratio), it was seen that AMPs are under moderate to strong positive selection across various taxa. This body of work has been reviewed elsewhere [24]. However, recent work [25], [26] has challenged this hypothesis. In cases where there is no evident signature of positive selection acting on AMPs, evidence for the polymorphism is still found, particularly in vertebrate AMPs from an array of taxa. Recently, Unckless and Lazzaro [26] have also placed irrefutable proof of balancing selection and adaptive maintenance of polymorphism in insect AMPs by evaluating and analyzing data available from Drosophila and revisiting the data available from other insect taxa. They have also documented trans-species polymorphism and recurrent convergent evolution across species in the Drosophila genus. Coupled with the available phenotypic effects of the polymorphisms, this strongly hints at adaptive maintenance of polymorphism owing to spatial and/or temporal variation of selective pressure on the AMP genes. Such variation in pressure has been indeed demonstrated [25], [27].
The older body of literature suggesting positive selection might have been due to the swamping of the selection signatures of AMP genes by that of other more prominent immune-defense genes like those encoding for signaling pathways or receptors. It might also be that different AMPs or even regions of the same AMP are subjected to different selective pressures; the exact nature of selection is determined by the function of the corresponding AMP or that particular domain. We cannot tell which of these is vastly closer to reality. Questions about the role of purifying selection on AMP evolution also remain unaddressed. Further studies and a re-investigation of the available data are needed to arrive at any less equivocal conclusion in this regard.
The repeated pseudogenization of AMP genes across various taxa might have to do with the evolutionary costs of immunity [28]. In the opinions of many ecologists, maintaining a functional immune system and mounting an appropriate immune response is a metabolically and nutritionally demanding process. This necessitates trade-off decisions between the competing nutritional and metabolic demands for growth, reproduction, thermoregulation and immunity. Details about these trade-offs and the cost of immunity have been reviewed elsewhere [29]. In this model, it might therefore be expected that AMPs are ‘evolutionary liabilities in the absence of infection’ [28]. Consequently, hosts with strong pathogen pressures would evolve to increase the net AMP production, while hosts with little pathogen pressures would evolve to reduce their AMP production, either at the transcriptional level or by way of deletions and/or pseudogenization. Loss of entire immune pathways have indeed been reported in pea aphids, which rely on a sterile food resource – plant phloem for subsistence [30]. Indeed, Hanson and co-workers have analysed genomes and transcriptomes of various Drosophila species and have presented evidence which offers a tell-tale signature that AMP genes are lost/pseudogenized in species with more sterile life history [28]. Furthermore, AMP alleles that are more effective against pathogens also tend to carry a higher autoimmune cost and overexpression of AMPs have significant fitness costs [31], [32], [33]. Another factor driving the loss of AMP genes might also be the tuning of the host immune system by symbiotic microflora. Duplication of AMP genes might be due to relaxed constraint on the duplicated genes, thereby paving the way for their neofunctionalization and diversification without being restrained to retain original functions to enable adaptation to suites of pathogens [23], [34], [35].
The evolution of individual AMP classes in various taxa have been studied in substantially more detail than the selection which drives their evolution. Among vertebrates, the evolution of defensins have been very well studied [36], [37]. All three classes of vertebrate defensins are believed to have evolved from a single precursor. The identification of defensins in lower eukaryotes suggests that the ancestral gene of these peptides existed before the divergence of the fungal and arthropod lineages and hence might be at least 1 billion years old [38], [39]. In fact, defensin-like peptides (like laterosporulin) have also been isolated in prokaryotes [40]. The evolutionary relationship of vertebrate and invertebrate defensins in unclear. Nevertheless, it had been proposed that the vertebrate defensins may have evolved from the evolutionary conserved big defensins in invertebrates [41], whose evolutionary lineage has been traced to Bilateria. Furthermore, current evidence indicates that the entire array of vertebrate and invertebrate defensins consist of two analogous superfamilies, one composed of the cysteine-stabilized αβ proteins from plants, fungi, and invertebrates and the other consisting of the vertebrate α-, β-, θ-, and invertebrate big defensins [42]. Extensive convergent evolution has been suggested to be the source of their similarities [42]. Structural and functional similarities between laterosporulin10, a defensin-like bacteriocin and human defensins [43] also hint at evolutionary connections between them, which are yet to be explored. The evolutionary history of the repertoire of AMP genes in insects has also been studied [34] against the well-resolved and dated phylogeny of insects. Detailed phylogenetic reconstruction has also been done for anuran AMPs, which have displayed somewhat unique features as compared to insects including showing features of coordinated evolution to conserve net charge of the peptide [44], [45]. Greater insights into the evolution and ecology of AMPs will provide further clues about the divergence of these peptides and enable the creation of novel peptide-based translational technologies to combat gargantuan antimicrobial resistance.
3. Physicochemical determinants of AMP activity and selectivity
Modulating the physicochemical determinants of the AMP activity is a key step to the realization of their therapeutic potential. Few important physicochemical and structural parameters relevant to the activity of membranolytic AMPs include hydrophobicity, helicity, amphipathicity, polar angle and charge [46]. With rapid advances in techniques of peptide synthesis and combinatorial screening, there has been an explosion of (often contradictory) information in these regards. The apparent contradictions between different studies are often due to the fact that while some use live cells, the other use membrane mimics like single unilamellar vesicle (SUV) and multilamellar vesicle (MUV). It is also worth mentioning that single amino acid substitutions would change many (if not all) above-mentioned physicochemical parameters, which makes it difficult to investigate each of these parameters individually. It is noteworthy that most of these studies inherently assume that membranolytic activity necessary for microbicidal activity, while many AMPs have been shown to involve intracellular targets like metabolic pathways or nucleic acids [47]. Furthermore, even within studies utilizing membrane mimics, there is a broad spectrum of alternatives available for the phospholipids, their relative concentrations and the assays used. A nuanced discussion of the intracellular vs membranolytic activity of AMPs is available in literature [47] and is not pursued further in the current review. Further discussion in the ensuing section is thus limited to membrane-active AMPs.
3.1. α-helical AMPs
Most studies on the role of physicochemical parameters in modulating microbicidal activity have been restricted to α-helical AMPs. Like most AMPs, these remain largely unstructured in solution and attain an α-helical conformation upon coming in contact with the membrane. Mutation scan, spectroscopic techniques like CD and solid state NMR spectroscopy have provided a wealth of information about the influence of various parameters on their structure and concomitantly, their activity, some of which are discussed below.
3.1.1. Hydrophobicity
Hydrophobicity refers to the relative propensity of the peptide to partition into a hydrophobic phase from the aqueous phase. The partitioning of AMPs into the membranes is largely controlled by van der Waal’s interactions, conformational transitions and electrostatic interactions. Optimal microbicidal activity would require a trade-off in hydrophobicity – very low hydrophobicity would lead to poor lipidic affinity and consequent inefficient membrane-permeabilizing effect while too high hydrophobicity would cause peptide aggregation and poor selectivity, leading to significant destruction of host membranes and haemolysis. Several studies concur the existence of two hydrophobicity thresholds [48] – one which must be reached so that the peptides possess sufficient membrane binding and insertion effects and another which must not be exceeded because binding to the electrically neutral host membranes would become significant thereafter [49], [50], [51], [52]. However, hydrophobicity affects binding to neutral membranes more than to charged bilayers and haemolysis more than bactericidal activity. For example, a strong correlation between haemolysis and the retention times in a reversed phase-HPLC, and hence hydrophobicity, was found for analogues of the frog peptide brevinin 1E [53]. The explanation for this comes from the non-additivity of the hydrophobic and electrostatic effects [54]. The correlation of haemolytic activity with hydrophobicity can also be explained phenomenologically using a ‘membrane discrimination model’, based on the premise that AMPs interact via a ‘barrel-stave’ mechanism in eukaryotic cells having zwitterionic membranes and via a ‘carpet’ mechanism in prokaryotic cells with charged membranes [55]. Peptides with higher hydrophobicities will penetrate deeper into the hydrophobic core of the erythrocyte membrane, causing stronger haemolysis by forming pores. Furthermore, similar influence of hydrophobicity on haemolytic activity has been demonstrated for a 26-residue synthetic peptide V13KL [51], [54]. The peptide also showed a decrease in microbicidal activity outside an optimal hydrophobicity window. An explanation for the decreased antimicrobial activity at high peptide hydrophobicity could be that the strong peptide self-association which prevents the peptide from passing through the cell wall in prokaryotic cells, whereas increased peptide self-association has no effect on peptide access to eukaryotic membranes.
3.1.2. Amphipathicity
Amphipathicity measures the degree of asymmetry in the distribution of polar and hydrophobic residues. This property is quantified by the hydrophobic moment, which is defined (analogous to an electric dipole moment) as the vector sum of the hydrophobicities of the individual amino acids. However, two things need to be kept in mind while correlating hydrophobic moment with activity [56]. In many AMPs, hydrophobic and hydrophilic residues are not uniformly distributed. Furthermore, hydrophobic moment is calculated assuming an ideal helix. Despite this, some trends can be made out. Studies with analogues of the antibacterial magainin 2 (which is naturally obtained from the skin secretions of Xenopus laevis) amide indicated that slight enhancement of the hydrophobic moment by a few conservative amino acid substitutions substantially increased the antimicrobial activity but also increased haemolytic activity [57]. Furthermore, studies using a model peptide have revealed that amphipathicity is far more important than hydrophobicity in determining antimicrobial activity and partition free energy [58]. It has also been shown by De Grado et al that amphipathicity is alone sufficient to induce bactericidal activity in a helical peptide [59]. The general trend of the influence of amphipathicity on AMP activity resembles that for hydrophobicity - increasing amphipathicity above a critical threshold result in strong interaction with neutral membranes present in the host, leading to toxicity [60].
3.1.3. Helicity
The propensity of a peptide to form an α-helix has been found to be more closely related to toxicity than antimicrobial activity. An increased helicity, attained by either deleting Gly or substituting it with Leu in the N-terminal helix of melittin (obtained from the venom of Apis mellifera), correlated well with an enhanced haemolytic and antibacterial activity of the peptide [61]. Substitution of helix-destabilizing Gly or Pro residues present close to the centre of the sequence of natural, selective AMPs that attain a helical conformation in membranes by helix promoting residues like Ala lead to significant enhancement of antimicrobial and haemolytic activity and a very poor selectivity [62], [63]. It has been hypothesized that destabilization of the helical forms (which are partially populated even when the peptide is in solution) allows the peptide to fold on itself, thereby concealing the non-polar side chains and reducing the effective hydrophobicity, thereby diminishing its tendency to bind neutral membranes and leading to alleviated toxicity [48].
3.1.4. Charge
Most natural AMPs are cationic, which facilitates electrostatic interactions with the surface of microbial membranes. There is no general correlation of the total peptide charge with the antimicrobial and haemolytic activity, with regression analyses having been largely unfruitful (P > 0.05) [64]. It has been showed that increasing net charge of magainin 2 from +4 to +5 increased antimicrobial activity; further increase to +7 did not alter the maximal activity observed at +5, however, the haemolytic activity increased [65]. Furthermore, studies with the V13KL [66] model peptide have also demonstrated that decreasing the net charge to a lower level (<+4) made V13KL analogues antimicrobially and haemolytically inactive. Systematically increasing the net charge from +4 to +8 made them antimicrobially more potent and maintained low level haemolytic activity [57]. However, though an increase in net charge to +9 and +10 further improved antimicrobial activity it had a dramatic effect on increasing unwanted haemolytic activity, thereby suggesting that there is an optimal window of peptide charge for maximum antimicrobial activity. In fact, the majority of AMPs available on databases have charges in the range of +4 to +6 [67], which might be the optimal charge required for maximum activity [68].
3.1.5. Length
The antimicrobial activity, quite expectedly, depends on chain length. It has been reported that the antibacterial activity decreases with increasing chain length, while the reverse is true for haemolytic activity [69]. However, Liu et al. have reported that longer-chain linear peptides (RW)n-NH2 are more potent antibacterial agents [70]. In fact, the dependence of activity data on chain length suggested an almost biphasic behaviour with the two shortest chains being relatively inactive, while the three longer chains approximating a similar activity level. Mechanistic explanations or a unifying idea about the influence of chain length on antimicrobial activity are however lacking in literature. It may be that the greater conformational flexibility of longer peptides would lead to a greater entropic penalty upon binding to the microbial membrane, thereby leading to reduction of potency. However, this hypothesis has not yet been tested experimentally, nor have the enthalpic and entropic contributions to the energetics of interaction of AMPs to membranes been worked out.
3.2. β-hairpin AMPs
This class of peptides is characterized by the presence of an antiparallel β-sheet, which are generally stabilized by disulfide bond(s). Comparatively little information is available about these AMPs relative to their helical counterparts. Some of the well studied AMPs of this class include tachyplesins (isolated from the horseshoe crab Tachypleus tridentatus) and gomesins (which are produced by the hemocytes of the spider Acanthoscurria gomesiana). While helical AMPs are largely unstructured in solution, many β-hairpin AMPs largely retain their conformation even when detached from the target membrane. The physicochemical determinants of activity for these peptides are similar to α-helical AMPs, with hydrophobicity, amphipathicity and charge being the most significant. These are summarized briefly in the following paragraphs, without elaborate details, owing to the limited number of studies performed and the gross similarities with helical AMPs.
The influence of charge on polyphemusin I (obtained from the hemocytes of the American horseshoe crab Limulus polyphemus) derivatives has been investigated [71]. It has been revealed that introduction of an additional Arg residue, which increases the charge by +1, leads to an improved partitioning of the peptide into phosphatidylglycerol containing model membranes. However, it is unclear whether any optimal charge window exists for β-hairpin AMPs, particularly considering that the mode of action of many such peptides (including polyphemusin I) involves induction of non-bilayer phase transitions or lipid flip flop.
Most β-hairpin AMPs have high content of Tyr and Arg residues, which renders elements of positive charge and hydrophobicity to them [72]. However, the hydrophobic and hydrophilic residues of these peptides are more widely distributed throughout the AMP instead of being closely grouped together, as is the case with many helical AMPs. It has been demonstrated that most amphipathic and least hydrophobic β-hairpin peptides are the most bactericidal, but also most toxic, thereby displaying a poor therapeutic index profile [72]. In fact, amphipathicity has been directly correlated with haemolytic activity among a set of six β-hairpin AMPs.
An additional factor which assumes significance for β-hairpin AMPs is the role of disulfide bonds in antimicrobial activity. β-hairpin AMPs fall into two categories – (i) AMPs whose activity is independent of the presence of disulfide bonds and (ii) AMPs whose activity is significantly attenuated in the absence of disulfide bonds. While human β-defensin 3 belongs to the former category, bactenecin (found in bovine neutrophils) belongs to the latter category [73]. Surprisingly, for some peptides like tachyplesin 1, removal of all cysteine residues and complete abolition of disulfide bonds led to retention of antimicrobial activity but remarkable loss of haemolytic activity [74], [75]. In fact, the antimicrobial activity seems to depend on the ability of cysteine deleted/linearized analogues to retain the amphipathic structure [72]. Further details of the role of disulfide bond need to be probed, particularly with high resolution NMR techniques and calorimetric measurements to appreciate the interactions of cysteine deleted peptides with the membrane and the dependence on activity on the number or positions of disulfide bonds.
4. Role of self-assembly in AMP activity
Following Steed and Atwood, supramolecular self-assembly can be defined as “the spontaneous and reversible association of molecules or ions to form larger, more complex supramolecular entities according to the intrinsic information contained in the molecules themselves” [76]. Examples of self-assembled systems abound in the natural world – ranging from the double helix of DNA to microtubules.
Self-assembly is mediated by a variety of weak, non-covalent secondary interactions. These include hydrogen bond, cation-π interaction, anion-π interaction, van der Waal’s interaction, hydrophobic interactions, and π-π stacking interactions. Self-assembly usually proceeds towards a thermodynamic sink (leading to energy minimization for the entire system). However, kinetic considerations often modulate the outcome of self-assembly as well [77], [78].
Peptides are considered ideal building blocks for the construction of smart biological nanomaterials with well-ordered structures and diverse functions [79]. Peptides have been found to self-assemble into diverse nanostructures like nanofibers, nanospheres, nanonets, and hydrogels [80]. The degree of self-assembly and the details of the consequently formed nanostructure depend on the specific amino acid moieties, their spatial arrangement, and the external conditions like pH and polarity of the solvent. The ensuing section will thus examine briefly the chief interactions involved in AMP self-assembly before delving into details of self-assembled AMPs of natural and synthetic origin.
One of the simplest and most widely recognized motifs in peptide self-assembly is the diphenylalanine (or analogously, polyphenylalanine) motif, which is the most-minimalistic motif in driving self-assembly by π-π stacking interactions (Fig. 2) [81], [82]. It has also been demonstrated that FF (diphenylalanine) assembles into nanostructures which significantly inhibited the growth of E. coli, besides having biocidal properties against Listeria monocytogenes and Staphylococcus epidermidis [83]. FF also showed membrane permeation properties, led to transcriptional upregulation of stress-related genes, and caused abnormal bacterial morphology. Obviously, any chemical modification that increases aromaticity would increase the extent of π stacking, causing a concomitant increase in self-assembling propensity; for example, the increased self-assembling tendency of napthyl-capped peptides helps to eradicate up to 94% S. epidermidis biofilms [84].
Fig. 2.
Schematic of interactions involved in self-assembly of AMPs, shown with some representative amino acid residues. In the lower left corner of the image above, yellow boxes denote hydrophobic stretches of peptide while green boxes represent hydrophilic regions.
Hydrophobic interaction-mediated peptide self-aggregation to form supramolecular nanostructures also shows interesting antimicrobial properties. The peptide A9R (AAAAAAAAAR) has been demonstrated to be active against many Multi-Drug Resistant (MDR) pathogens while showing excellent self-assembling tendencies forming hydrogel nanostructures and stabilizing emulsions [85]. Peptide bola-amphiphiles, which contain a hydrophobic sequence capped at both ends by a hydrophilic residue, are also promising biocidal candidates in this class [86].
The two other types of interaction that can drive peptide self-assembly are ionic interactions and hydrogen bonding. Electrostatic interaction between oppositely charged lysine, arginine, aspartate, and glutamate residues within appropriate pH ranges has been extensively used to design potent AMPs with significantly reduced cytotoxicity like the peptide KLD-12 (KLDLKLDLKLDL) [87]. The intermolecular hydrogen bonding formed by glutamine has been the strongest among all secondary interactions in peptide-based supramolecular architectures [79]. Based on these first principles, Xu et al. have designed a self-assembling peptide K3W(QL)6K2, which shows excellent broad-spectrum biocidal activities with a MIC from 5 to 20 µM [88].
While the preceding paragraphs have largely focussed on physical principles driving self-assembly and have been strewn with handpicked examples from recent literature which have used these principles in molecular construction and peptide engineering, naturally occurring host defense peptides are replete with examples of self-assembling peptides. One of the most celebrated ones in this regard is the human α-defensin 6 (HD6). HD6 is a 32-amino acid cysteine-rich peptide produced by the Paneth cells in the small intestine [89], [90]. It is produced as an 81 amino-acid pro-peptide, which is co-packaged with proHD5 and trypsin in the secretory granules of Paneth cells [91]. Once released into the intestinal lumen, trypsin-catalyzed proteolysis (which may be activated by the luminal pH) leads to the formation of active HD6. HD6 self-assembles to form ‘nanonets’ mainly through hydrophobic and π stacking interactions [92], [93]. These nanonets have been demonstrated to physically entrap bacteria Gram-positive bacteria like L. monocytogenes and Gram-negative bacteria like Yersinia enterocolitica and S. Typhimurium and control their invasion into mammalian cells [92], [94]. Physical entrapment by peptide nanonets prevents bacterial invasion and leads to faster clearance by recruiting other immune cells or antimicrobial substances. HD6 also arrests biofilm formation in Candida albicans [95]. Furthermore, it has been proven that abolishing the ability of HD6 to self-assemble into nanonets by disrupting secondary interactions leads to the loss of this microbicidal activities [93].
Self-assembly has also been demonstrated for the core of human cathelicidin LL-37 (residues 17 to 29), which is the active core of the AMP, retaining the antimicrobial properties of the full-length protein [96]. The core of LL-37 (hLL-3717-29) self-assembles into a supramolecular fibril of densely packed protein helices. The surface has alternating hydrophobic and cationic zigzagged belts, which might be responsible for attachment to microbial membranes and concomitant membrane perturbation. Recent studies have also demonstrated that AMPs like dermaseptin S9 [97], [98] and protegrin [99] also self-assemble into amyloid-like fibrils. Recently, there have been multiple efforts to identify the inter-relationships between amyloids and AMPs due to the high degree of structural and sequence similarity between them, coupled with the observation that some amyloid fibrils possess concentration-dependent antimicrobial activity [100], [101], [102], [103], [104], [105].
Besides its structural and functional significance, the self-assembly of AMPs has interesting physiological (particularly immunomodulatory) consequences. It has been demonstrated that AMPs like LL37, mellitin, buforins, and β-defensins self-assemble with DNA via electrostatic interactions into grill-like nanostructures [106]. These nanostructures are spatially periodic and have regular spacing between the DNA molecules. The structures of these AMP-DNA supramolecular assemblies correlated well with their ability to activate plasmacytoid dendritic cells (pDCs) via TLR9 [107], [108]. Those nanostructures in which the inter-DNA spacing matched well with the steric size of TLR9 led to the maximum degree of cytokine production [109]. It is easy to imagine that such a nanostructure would lead to interdigitation of the DNA with a clustered, closely packed array of TLR9, leading to a ‘clustering’ of the immune ligands (Fig. 3) [110]. Such a clustered set of immune ligands would obviously lead to engagement of maximal number of receptors and cause a subsequent amplification of the immune response by enhanced downstream signalling, and subsequent strong induction of production of interferons and cytokines (Fig. 4). Monte-Carlo simulations based on such a model for AMP-RNA complexes have yielded similar results for engagement with TLR3 receptor, strongly affirming the validity of this simple and elegant mechanistic model [108]. Furthermore, self-assembly has important pathological consequences. The amyloid-β protein, which has been implicated in Alzheimer’s disease, has also been found to self-assemble into oligomers which have microbicidal activity [111], and can form transmembrane pores [83].
Fig. 3.
(A) (i) TLR9 dimer bound to its ligand – dsDNA (with unmethylated CpG nucleotides). The side view of the receptor is as shown in (ii), which has been schematically represented as in (iii) The amplification of the immune response depends on the distance between the DNA strands in the DNA-LL-37 nanostructure. (B) The distance between the DNA molecules in the spatially periodic nanostructure is same as the distance between TLR9 receptors in (ii), leading to amplified downstream signalling cascades, which is in contrast to the larger distance between the DNA molecules in (i), which does not match the inter-TLR9 distance, leading to weak immunological responses.
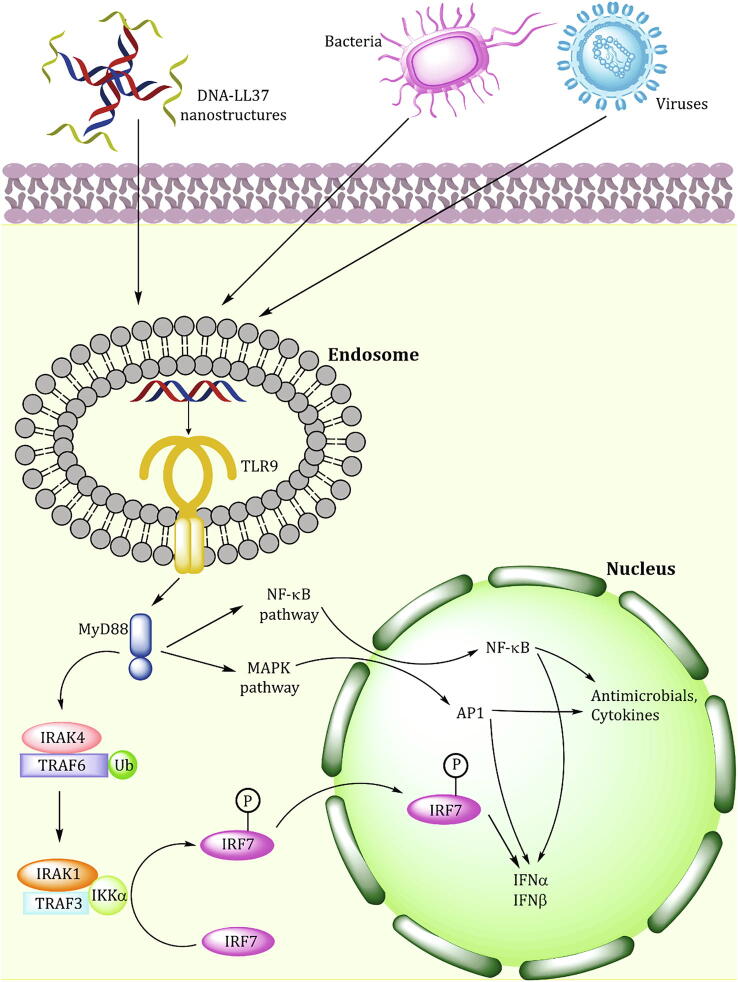
Fig. 4.
Signalling cascades activated upon binding of TLR9 to the DNA-AMP nanostructures. Other modes of TLR9 activation have also been shown for completeness, where MyD88 mediated activation of interferon signalling through nuclear factor kappa B (NF-kB) and Mitogen-activated protein kinase (MAPK). This interferon signalling further leads to production of AMPs and other cytokines.
While the role of self-assembly in determining antimicrobial properties of many natural HDPs has been thoroughly investigated, we lack a unified mechanistic understanding of its role. It is easy to imagine that despite there being multiple reports of self-assembly causing an increase in microbicidal potency, too large peptide aggregates would be sterically hindered from reaching the plasma membrane after crossing the outer membrane (in Gram negative bacteria) or cell wall (in Gram positive bacteria), which would lower antimicrobial activity. Peptides which self-assemble only at the biomembrane might solve this problem. Despite successful chemical synthesis of such peptides [112], it remains to be seen if such a strategy increases microbicidal ability of all self-assembling AMPs. What could be the optimum geometrical arrangement of the self-assembled nanostructure for optimal activity, or the optimal size of the supramolecular assemblies is still unknown. Furthermore, even in the formation of a barrel stave pore at the cell membrane, peptide self-assembly in a lipidic microenvironment is involved, which has not yet been investigated significantly.
5. Summary and outlook
AMPs have turned out to be promising candidates in our continuous exploration of newer chemotherapeutic strategies to combat infections caused by MDR pathogens. However, we need to exercise caution – the evolution of resistance may lead to AMPs meeting the same fate as conventional antibiotics. The early notion that bacteria are largely unable to evolve resistance to AMPs has been largely refuted [113]. Resistance to AMPs arises from a range of nonspecific mechanisms, including secretion of proteases, biofilm formation, activation of efflux pumps and two component sensors and modification of the cell envelope, including its charge [114]. An added challenge comes from the fact that most clinically tested AMPs are from human sources, and should resistance evolve in pathogens against these drug candidates, the entire innate defense system would be thrown into absolute disarray on a population-scale.
Nevertheless, the chances of evolution of AMP resistance are less than conventional antibiotics. This can be phenomenologically ascribed to the steep pharmacodynamic curves of AMPs and their high Hill coefficient, which lead to a small mutant selection window [115]. On a molecular scale, this has been attributed to the multiplicity of AMP targets and the synergistic action displayed by combinations of AMPs, either by amplification of downstream effects [116] or physical conjugation to form a supermolecule [117], [118]. It is to be noted, however, that we are very far from a definitive understanding of this issue.
Therefore, a logical way to combat evolution of resistance against AMPs is to use cocktails of judiciously chosen synergistic AMPs which show a rapid killing profile. In order to design such cocktails rationally, it is essential to be free from the constraints imposed by conventional template based AMP design (designing AMPs by tweaking on a natural template) and move towards function-guided design. The need of the hour is to unite the two apparently disconnected aspects of this review – structure and evolution of AMPs to develop a holistic understanding of this issue. Just as it is necessary to understand molecular determinants of AMP activity, it is also imperative to understand physicochemical features of AMPs susceptible to evolution of resistance. There has been extremely few studies undertaken in this area. In a recent study, Spohn et al. have shown that AMPs less susceptible to resistance during adaptive laboratory evolution displayed increased hydropathicity and contained fewer polar and positively charged amino acids [119]. Molecular reasons behind this are unclear. Such studies carrying out integrated evolutionary analysis are the need of the hour, as is the need to consciously consider the evolution of the AMPs in the host immune landscape in order to avoid dangerously playing with natural host innate defences while trying to develop chemotherapeutics. The principles outlined in this review would hopefully give a direction to such research.
This review systematically analyses the importance of various physicochemical parameters in modulating the microbicidal and haemolytic properties of AMPs. While there are multiple reviews detailing the tertiary structures, mechanisms of action and sequences of AMPs, a fuller picture would only emerge if basic physical principles are used to rationalize their activity. It is only by tweaking the structural determinants that we can design peptide resistance-resistant ‘silver bullets’ against deadly MDR pathogens. Furthermore, this review discusses the relevance of self-assembly in governing AMP activity – both in terms of synthetic microbicidal therapeutic candidates as well as in the natural context of the host. Since the details of immunomodulation by self-assembled nanostructures and the relevance of self-assembly in physical entrapment of pathogenic microbes is now known, it is hoped that efforts would be made to take advantage of this in engineering peptides. A detailed review of the physicochemical determinants and self-assembly properties would also make it easier to come up with quantitative structure activity relationships, which would make way for much easier design of AMPs with state-of-the-art microbicidal potential. Furthermore, such mechanistic and structural understanding of AMPs would make it easier to cast AMPs as a graph optimization problem [120] and computational language modelling problem [121], and thereby improve computational explorations for function-guided AMP designs.
6. Funding acknowledgment
We thank the financial support from Department of Biotechnology (DBT), Ministry of Science and Technology; Department of Science and Technology (DST), Ministry of Science and Technology. DC acknowledges DAE-SRC outstanding investigator award and funds and TATA Innovation fellowship. The authors jointly acknowledge the DBT-IISc partnership programme. Infrastructure support from ICMR (Center for Advanced Study in Molecular Medicine), DST (FIST), and UGC-CAS (special assistance), TATA fellowship is acknowledged. SC duly acknowledges the DST for the KVPY fellowship. RC duly acknowledges CSIR-SRF fellowship.
7. Disclosure statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
CRediT authorship contribution statement
Sukriyo Chakraborty: Conceptualization, Data curation, Formal analysis, Writing – original draft, Writing – review & editing. Ritika Chatterjee: Conceptualization, Data curation, Formal analysis, Writing – review & editing. Dipshikha Chakravortty: Conceptualization, Data curation, Formal analysis, Funding acquisition, Writing – review & editing.
References
- 1.World Health, O., Antimicrobial resistance: global report on surveillance. 2014, Geneva: World Health Organization.
- 2.Bin Hafeez A., et al. Antimicrobial peptides: an update on classifications and databases. Int J Mol Sci. 2021;22(21) doi: 10.3390/ijms222111691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phoenix, D.D.S.R.H.F., Antimicrobial peptides. 2013.
- 4.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415(6870):389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 5.McAuliffe O., Ross R.P., Hill C. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol Rev. 2001;25(3):285–308. doi: 10.1111/j.1574-6976.2001.tb00579.x. [DOI] [PubMed] [Google Scholar]
- 6.Drider, D.R.S., Prokaryotic antimicrobial peptides: from genes to applications. 2011.
- 7.Miller M.B., Bassler B.L. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 8.Li J., et al. Plant antimicrobial peptides: structures, functions, and applications. Bot Stud. 2021;62(1):5. doi: 10.1186/s40529-021-00312-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mygind P.H., et al. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature. 2005;437(7061):975–980. doi: 10.1038/nature04051. [DOI] [PubMed] [Google Scholar]
- 10.Leitgeb B., et al. The history of alamethicin: a review of the most extensively studied peptaibol. Chem Biodivers. 2007;4(6):1027–1051. doi: 10.1002/cbdv.200790095. [DOI] [PubMed] [Google Scholar]
- 11.Wang G., Li X., Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44(D1):D1087–D1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi G., et al. DRAMP 3.0: an enhanced comprehensive data repository of antimicrobial peptides. Nucleic Acids Res. 2022;50(D1):D488–D496. doi: 10.1093/nar/gkab651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huan Y., et al. Antimicrobial peptides: classification, design, application and research progress in multiple fields. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.582779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kell D.B., Heyden E.L., Pretorius E. The biology of lactoferrin, an iron-binding protein that can help defend against viruses and bacteria. Front Immunol. 2020;11:1221. doi: 10.3389/fimmu.2020.01221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makarova K.S., et al. Antimicrobial peptides, polymorphic toxins, and self-nonself recognition systems in archaea: an untapped armory for intermicrobial conflicts. mBio. 2019;10(3) doi: 10.1128/mBio.00715-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Besse A., et al. Antimicrobial peptides and proteins in the face of extremes: lessons from archaeocins. Biochimie. 2015;118:344–355. doi: 10.1016/j.biochi.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Lequerica J.L., et al. A halocin acting on Na+/H+ exchanger of haloarchaea as a new type of inhibitor in NHE of mammals. J Physiol Biochem. 2006;62(4):253–262. doi: 10.1007/BF03165754. [DOI] [PubMed] [Google Scholar]
- 18.Woolhouse M.E., et al. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genet. 2002;32(4):569–577. doi: 10.1038/ng1202-569. [DOI] [PubMed] [Google Scholar]
- 19.Janzen D.H. When is it coevolution? Evolution. 1980;34(3):611–612. doi: 10.1111/j.1558-5646.1980.tb04849.x. [DOI] [PubMed] [Google Scholar]
- 20.Lazzaro B.P., Zasloff M., Rolff J. Antimicrobial peptides: application informed by evolution. Science. 2020;368(6490) doi: 10.1126/science.aau5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unckless R.L., Howick V.M., Lazzaro B.P. Convergent balancing selection on an antimicrobial peptide in drosophila. Curr Biol. 2016;26(2):257–262. doi: 10.1016/j.cub.2015.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazzaro B.P. Natural selection on the Drosophila antimicrobial immune system. Curr Opin Microbiol. 2008;11(3):284–289. doi: 10.1016/j.mib.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tennessen J.A. Molecular evolution of animal antimicrobial peptides: widespread moderate positive selection. J Evol Biol. 2005;18(6):1387–1394. doi: 10.1111/j.1420-9101.2005.00925.x. [DOI] [PubMed] [Google Scholar]
- 24.Chapman J.R., Hill T., Unckless R.L. Balancing selection drives the maintenance of genetic variation in drosophila antimicrobial peptides. Genome Biol Evol. 2019;11(9):2691–2701. doi: 10.1093/gbe/evz191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Early A.M., et al. Survey of global genetic diversity within the drosophila immune system. Genetics. 2017;205(1):353–366. doi: 10.1534/genetics.116.195016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Unckless R.L., Lazzaro B.P. The potential for adaptive maintenance of diversity in insect antimicrobial peptides. Philos Trans R Soc Lond B Biol Sci. 2016;371(1695) doi: 10.1098/rstb.2015.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Behrman E.L., et al. Rapid seasonal evolution in innate immunity of wild Drosophila melanogaster. Proc Biol Sci. 2018;285(1870) doi: 10.1098/rspb.2017.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanson M.A., Lemaitre B., Unckless R.L. Dynamic evolution of antimicrobial peptides underscores trade-offs between immunity and ecological fitness. Front Immunol. 2019;10:2620. doi: 10.3389/fimmu.2019.02620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lochmiller R.L., Deerenberg C. Trade-offs in evolutionary immunology: just what is the cost of immunity? Oikos. 2000;88(1):87–98. [Google Scholar]
- 30.Gerardo N.M., et al. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 2010;11(2):R21. doi: 10.1186/gb-2010-11-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maneerat Y., et al. Increased alpha-defensin expression is associated with risk of coronary heart disease: a feasible predictive inflammatory biomarker of coronary heart disease in hyperlipidemia patients. Lipids Health Dis. 2016;15:117. doi: 10.1186/s12944-016-0285-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benachour H., et al. Association of human cathelicidin (hCAP-18/LL-37) gene expression with cardiovascular disease risk factors. Nutr Metab Cardiovasc Dis. 2009;19(10):720–728. doi: 10.1016/j.numecd.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Gilliet M., Lande R. Antimicrobial peptides and self-DNA in autoimmune skin inflammation. Curr Opin Immunol. 2008;20(4):401–407. doi: 10.1016/j.coi.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Mylonakis E., et al. Diversity, evolution and medical applications of insect antimicrobial peptides. Philos Trans R Soc Lond B Biol Sci. 2016;371(1695) doi: 10.1098/rstb.2015.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peschel A., Sahl H.G. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Microbiol. 2006;4(7):529–536. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- 36.Semple C.A., et al. The changing of the guard: Molecular diversity and rapid evolution of beta-defensins. Mol Divers. 2006;10(4):575–584. doi: 10.1007/s11030-006-9031-7. [DOI] [PubMed] [Google Scholar]
- 37.Hollox E.J., Armour J.A.L. Directional and balancing selection in human beta-defensins. BMC Evol Biol. 2008;8(1):113. doi: 10.1186/1471-2148-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu S. Discovery of six families of fungal defensin-like peptides provides insights into origin and evolution of the CSalphabeta defensins. Mol Immunol. 2008;45(3):828–838. doi: 10.1016/j.molimm.2007.06.354. [DOI] [PubMed] [Google Scholar]
- 39.Dimarcq J.L., et al. Cysteine-rich antimicrobial peptides in invertebrates. Biopolymers. 1998;47(6):465–477. doi: 10.1002/(SICI)1097-0282(1998)47:6<465::AID-BIP5>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 40.Singh P.K., et al. The intramolecular disulfide-stapled structure of laterosporulin, a class IId bacteriocin, conceals a human defensin-like structural module. FEBS J. 2015;282(2):203–214. doi: 10.1111/febs.13129. [DOI] [PubMed] [Google Scholar]
- 41.Lehrer R.I., Lu W. alpha-Defensins in human innate immunity. Immunol Rev. 2012;245(1):84–112. doi: 10.1111/j.1600-065X.2011.01082.x. [DOI] [PubMed] [Google Scholar]
- 42.Shafee T.M., et al. The defensins consist of two independent, convergent protein superfamilies. Mol Biol Evol. 2016;33(9):2345–2356. doi: 10.1093/molbev/msw106. [DOI] [PubMed] [Google Scholar]
- 43.Baindara P., et al. Laterosporulin10: a novel defensin like Class IId bacteriocin from Brevibacillus sp. strain SKDU10 with inhibitory activity against microbial pathogens. Microbiology (Reading) 2016;162(8):1286–1299. doi: 10.1099/mic.0.000316. [DOI] [PubMed] [Google Scholar]
- 44.Duda T.F., Jr., Vanhoye D., Nicolas P. Roles of diversifying selection and coordinated evolution in the evolution of amphibian antimicrobial peptides. Mol Biol Evol. 2002;19(6):858–864. doi: 10.1093/oxfordjournals.molbev.a004143. [DOI] [PubMed] [Google Scholar]
- 45.Cortazar-Chinarro M., et al. Antimicrobial peptide and sequence variation along a latitudinal gradient in two anurans. BMC Genet. 2020;21(1):38. doi: 10.1186/s12863-020-00839-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dathe M., Wieprecht T. Structural features of helical antimicrobial peptides: their potential to modulate activity on model membranes and biological cells. Biochim Biophys Acta. 1999;1462(1–2):71–87. doi: 10.1016/s0005-2736(99)00201-1. [DOI] [PubMed] [Google Scholar]
- 47.Benfield A.H., Henriques S.T. Mode-of-action of antimicrobial peptides: membrane disruption vs. intracellular mechanisms. Front Med Technol. 2020;2 doi: 10.3389/fmedt.2020.610997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bobone S., Stella L. Selectivity of antimicrobial peptides: a complex interplay of multiple equilibria. Adv Exp Med Biol. 2019;1117:175–214. doi: 10.1007/978-981-13-3588-4_11. [DOI] [PubMed] [Google Scholar]
- 49.Mojsoska B., Zuckermann R.N., Jenssen H. Structure-activity relationship study of novel peptoids that mimic the structure of antimicrobial peptides. Antimicrob Agents Chemother. 2015;59(7):4112–4120. doi: 10.1128/AAC.00237-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glukhov E., Burrows L.L., Deber C.M. Membrane interactions of designed cationic antimicrobial peptides: the two thresholds. Biopolymers. 2008;89(5):360–371. doi: 10.1002/bip.20917. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y., et al. Rational design of alpha-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J Biol Chem. 2005;280(13):12316–12329. doi: 10.1074/jbc.M413406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saint Jean K.D., et al. Effects of hydrophobic amino acid substitutions on antimicrobial peptide behavior. Probiotics Antimicrob Proteins. 2018;10(3):408–419. doi: 10.1007/s12602-017-9345-z. [DOI] [PubMed] [Google Scholar]
- 53.Kwon M.Y., Hong S.Y., Lee K.H. Structure-activity analysis of brevinin 1E amide, an antimicrobial peptide from Rana esculenta. Biochim Biophys Acta. 1998;1387(1–2):239–248. doi: 10.1016/s0167-4838(98)00123-x. [DOI] [PubMed] [Google Scholar]
- 54.Wimley W.C. Energetics of peptide and protein binding to lipid membranes. Adv Exp Med Biol. 2010;677:14–23. doi: 10.1007/978-1-4419-6327-7_2. [DOI] [PubMed] [Google Scholar]
- 55.Chen Y., et al. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob Agents Chemother. 2007;51(4):1398–1406. doi: 10.1128/AAC.00925-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dathe M., Wieprecht T. Structural features of helical antimicrobial peptides: their potential to modulate activity on model membranes and biological cells. Biochim. Biophys. Acta (BBA) - Biomembranes. 1999;1462(1):71–87. doi: 10.1016/s0005-2736(99)00201-1. [DOI] [PubMed] [Google Scholar]
- 57.Wieprecht T., et al. Modulation of membrane activity of amphipathic, antibacterial peptides by slight modifications of the hydrophobic moment. FEBS Lett. 1997;417(1):135–140. doi: 10.1016/s0014-5793(97)01266-0. [DOI] [PubMed] [Google Scholar]
- 58.Fernandez-Vidal M., et al. Folding amphipathic helices into membranes: amphiphilicity trumps hydrophobicity. J Mol Biol. 2007;370(3):459–470. doi: 10.1016/j.jmb.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeGrado W.F., Kezdy F.J., Kaiser E.T. Design, synthesis, and characterization of a cytotoxic peptide with melittin-like activity. J Am Chem Soc. 1981;103(3):679–681. [Google Scholar]
- 60.Kindrachuk J., Napper S. Structure-activity relationships of multifunctional host defence peptides. Mini Rev Med Chem. 2010;10(7):596–614. doi: 10.2174/138955710791383983. [DOI] [PubMed] [Google Scholar]
- 61.Blondelle S.E., Houghten R.A. Hemolytic and antimicrobial activities of the twenty-four individual omission analogs of melittin. Biochemistry. 1991;30(19):4671–4678. doi: 10.1021/bi00233a006. [DOI] [PubMed] [Google Scholar]
- 62.Bobone S., et al. The importance of being kinked: role of Pro residues in the selectivity of the helical antimicrobial peptide P5. J Pept Sci. 2013;19(12):758–769. doi: 10.1002/psc.2574. [DOI] [PubMed] [Google Scholar]
- 63.Chen H.C., et al. Synthetic magainin analogues with improved antimicrobial activity. FEBS Lett. 1988;236(2):462–466. doi: 10.1016/0014-5793(88)80077-2. [DOI] [PubMed] [Google Scholar]
- 64.Dennison S.R., Harris F., Phoenix D.A. Factors determining the efficacy of alpha-helical antimicrobial peptides. Protein Pept Lett. 2003;10(5):497–502. doi: 10.2174/0929866033478663. [DOI] [PubMed] [Google Scholar]
- 65.Dathe M., et al. Optimization of the antimicrobial activity of magainin peptides by modification of charge. FEBS Lett. 2001;501(2–3):146–150. doi: 10.1016/s0014-5793(01)02648-5. [DOI] [PubMed] [Google Scholar]
- 66.Jiang Z., et al. Effects of net charge and the number of positively charged residues on the biological activity of amphipathic alpha-helical cationic antimicrobial peptides. Biopolymers. 2008;90(3):369–383. doi: 10.1002/bip.20911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Giangaspero A., Sandri L., Tossi A. Amphipathic alpha helical antimicrobial peptides. Eur J Biochem. 2001;268(21):5589–5600. doi: 10.1046/j.1432-1033.2001.02494.x. [DOI] [PubMed] [Google Scholar]
- 68.Tossi A., Sandri L., Giangaspero A. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers. 2000;55(1):4–30. doi: 10.1002/1097-0282(2000)55:1<4::AID-BIP30>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 69.Takuro N., et al. Effect of chain length of cationic model peptides on antibacterial activity. Bull Chem Soc Jpn. 2005;78(3):473–476. [Google Scholar]
- 70.Liu Z., et al. Length effects in antimicrobial peptides of the (RW)n series. Antimicrob Agents Chemother. 2007;51(2):597–603. doi: 10.1128/AAC.00828-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Powers J.P., et al. Solution structure and interaction of the antimicrobial polyphemusins with lipid membranes. Biochemistry. 2005;44(47):15504–15513. doi: 10.1021/bi051302m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edwards I.A., et al. Contribution of amphipathicity and hydrophobicity to the antimicrobial activity and cytotoxicity of β-hairpin peptides. ACS Infect Dis. 2016;2(6):442–450. doi: 10.1021/acsinfecdis.6b00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Powers J.P., Rozek A., Hancock R.E. Structure-activity relationships for the beta-hairpin cationic antimicrobial peptide polyphemusin I. Biochim Biophys Acta. 2004;1698(2):239–250. doi: 10.1016/j.bbapap.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 74.Saravanan R., et al. Structure, activity and interactions of the cysteine deleted analog of tachyplesin-1 with lipopolysaccharide micelle: mechanistic insights into outer-membrane permeabilization and endotoxin neutralization. Biochim Biophys Acta. 2012;1818(7):1613–1624. doi: 10.1016/j.bbamem.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 75.Ramamoorthy A., et al. Deletion of all cysteines in tachyplesin I abolishes hemolytic activity and retains antimicrobial activity and lipopolysaccharide selective binding. Biochemistry. 2006;45(20):6529–6540. doi: 10.1021/bi052629q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Self-Assembly, in Supramolecular Chemistry. 2009. p. 591-706.
- 77.Michaels T.C.T., et al. Kinetic constraints on self-assembly into closed supramolecular structures. Sci Rep. 2017;7(1):12295. doi: 10.1038/s41598-017-12528-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jung, S.H., M. Takeuchi, K. Sugiyasu, Chapter 10 - molecular self-assembly under kinetic control, in kinetic control in synthesis and self-assembly, M. Numata, S. Yagai, and T. Hamura, Editors. 2019, Academic Press. p. 205-229.
- 79.Lombardi L., et al. A new hope: self-assembling peptides with antimicrobial activity. Pharmaceutics. 2019;11(4):166. doi: 10.3390/pharmaceutics11040166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tian X., et al. Role of peptide self-assembly in antimicrobial peptides. J Pept Sci. 2015;21(7):530–539. doi: 10.1002/psc.2788. [DOI] [PubMed] [Google Scholar]
- 81.Yang Z., et al. Nanostructured antimicrobial peptides: crucial steps of overcoming the bottleneck for clinics. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.710199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cross E.R., et al. Unravelling the antimicrobial activity of peptide hydrogel systems: current and future perspectives. Soft Matter. 2021;17(35):8001–8021. doi: 10.1039/d1sm00839k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schnaider L., et al. Self-assembling dipeptide antibacterial nanostructures with membrane disrupting activity. Nat Commun. 2017;8(1):1365. doi: 10.1038/s41467-017-01447-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Laverty G., et al. Ultrashort cationic naphthalene-derived self-assembled peptides as antimicrobial nanomaterials. Biomacromolecules. 2014;15(9):3429–3439. doi: 10.1021/bm500981y. [DOI] [PubMed] [Google Scholar]
- 85.Castelletto V., et al. Peptide-stabilized emulsions and gels from an arginine-rich surfactant-like peptide with antimicrobial activity. ACS Appl Mater Interfaces. 2019;11(10):9893–9903. doi: 10.1021/acsami.9b00581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Edwards-Gayle C.J.C., et al. Self-assembly, antimicrobial activity, and membrane interactions of arginine-capped peptide bola-amphiphiles. ACS Appl Bio Mater. 2019;2(5):2208–2218. doi: 10.1021/acsabm.9b00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sun J., Zheng Q. Experimental study on self-assembly of KLD-12 peptide hydrogel and 3-D culture of MSC encapsulated within hydrogel in vitro. J Huazhong Univ Sci Technolog Med Sci. 2009;29(4):512–516. doi: 10.1007/s11596-009-0424-6. [DOI] [PubMed] [Google Scholar]
- 88.Xu D., et al. Designed supramolecular filamentous peptides: balance of nanostructure, cytotoxicity and antimicrobial activity. Chem Commun (Camb) 2015;51(7):1289–1292. doi: 10.1039/c4cc08808e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jones D.E., Bevins C.L. Paneth cells of the human small intestine express an antimicrobial peptide gene. J Biol Chem. 1992;267(32):23216–23225. [PubMed] [Google Scholar]
- 90.Schroeder B.O., et al. Paneth cell α-defensin 6 (HD-6) is an antimicrobial peptide. Mucosal Immunol. 2015;8(3):661–671. doi: 10.1038/mi.2014.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chairatana P., et al. Proteolysis triggers self-assembly and unmasks innate immune function of a human α-defensin peptide. Chem Sci. 2016;7(3):1738–1752. doi: 10.1039/c5sc04194e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chu H., et al. Human alpha-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science. 2012;337(6093):477–481. doi: 10.1126/science.1218831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chairatana P., Nolan E.M. Molecular basis for self-assembly of a human host-defense peptide that entraps bacterial pathogens. J Am Chem Soc. 2014;136(38):13267–13276. doi: 10.1021/ja5057906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chairatana P., Nolan E.M. Human α-Defensin 6: a small peptide that self-assembles and protects the host by entangling microbes. Acc Chem Res. 2017;50(4):960–967. doi: 10.1021/acs.accounts.6b00653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chairatana P., Chiang I.L., Nolan E.M. Human alpha-defensin 6 self-assembly prevents adhesion and suppresses virulence traits of Candida albicans. Biochemistry. 2017;56(8):1033–1041. doi: 10.1021/acs.biochem.6b01111. [DOI] [PubMed] [Google Scholar]
- 96.Engelberg Y., Landau M. The Human LL-37(17–29) antimicrobial peptide reveals a functional supramolecular structure. Nat Commun. 2020;11(1):3894. doi: 10.1038/s41467-020-17736-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Auvynet C., et al. Structural requirements for antimicrobial versus chemoattractant activities for dermaseptin S9. FEBS J. 2008;275(16):4134–4151. doi: 10.1111/j.1742-4658.2008.06554.x. [DOI] [PubMed] [Google Scholar]
- 98.Caillon L., et al. Biophysical investigation of the membrane-disrupting mechanism of the antimicrobial and amyloid-like peptide dermaseptin S9. PLoS ONE. 2013;8(10) doi: 10.1371/journal.pone.0075528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jang H., et al. Antimicrobial protegrin-1 forms amyloid-like fibrils with rapid kinetics suggesting a functional link. Biophys J. 2011;100(7):1775–1783. doi: 10.1016/j.bpj.2011.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang M., Zhao J., Zheng J. Molecular understanding of a potential functional link between antimicrobial and amyloid peptides. Soft Matter. 2014;10(38):7425–7451. doi: 10.1039/c4sm00907j. [DOI] [PubMed] [Google Scholar]
- 101.White M.R., et al. Alzheimer's associated beta-amyloid protein inhibits influenza A virus and modulates viral interactions with phagocytes. PLoS ONE. 2014;9(7) doi: 10.1371/journal.pone.0101364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Soscia S.J., et al. The Alzheimer's disease-associated amyloid beta-protein is an antimicrobial peptide. PLoS ONE. 2010;5(3) doi: 10.1371/journal.pone.0009505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hirakura Y., et al. Channel formation by serum amyloid A: a potential mechanism for amyloid pathogenesis and host defense. Amyloid. 2002;9(1):13–23. doi: 10.3109/13506120209072440. [DOI] [PubMed] [Google Scholar]
- 104.Last N.B., Miranker A.D. Common mechanism unites membrane poration by amyloid and antimicrobial peptides. Proc Natl Acad Sci U S A. 2013;110(16):6382–6387. doi: 10.1073/pnas.1219059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Allen Reish H.E., Standaert D.G. Role of alpha-synuclein in inducing innate and adaptive immunity in Parkinson disease. J Parkinsons Dis. 2015;5(1):1–19. doi: 10.3233/JPD-140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee E.Y., et al. Helical antimicrobial peptides assemble into protofibril scaffolds that present ordered dsDNA to TLR9. Nat Commun. 2019;10(1):1012. doi: 10.1038/s41467-019-08868-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lande R., et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449(7162):564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 108.Schmidt N.W., et al. Liquid-crystalline ordering of antimicrobial peptide-DNA complexes controls TLR9 activation. Nat Mater. 2015;14(7):696–700. doi: 10.1038/nmat4298. [DOI] [PubMed] [Google Scholar]
- 109.Lee E.Y., et al. Functional reciprocity of amyloids and antimicrobial peptides: rethinking the role of supramolecular assembly in host defense, immune activation, and inflammation. Front Immunol. 2020;11:1629. doi: 10.3389/fimmu.2020.01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee E.Y., et al. A review of immune amplification via ligand clustering by self-assembled liquid–crystalline DNA complexes. Adv Colloid Interface Sci. 2016;232:17–24. doi: 10.1016/j.cis.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kumar D.K., et al. Amyloid-beta peptide protects against microbial infection in mouse and worm models of Alzheimer's disease. Sci Transl Med. 2016;8(340):p. 340ra72. doi: 10.1126/scitranslmed.aaf1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shen Z., et al. Biomembrane induced in situ self-assembly of peptide with enhanced antimicrobial activity. Biomater Sci. 2020;8(7):2031–2039. doi: 10.1039/c9bm01785b. [DOI] [PubMed] [Google Scholar]
- 113.Perron G.G., Zasloff M., Bell G. Experimental evolution of resistance to an antimicrobial peptide. Proc Biol Sci. 2006;273(1583):251–256. doi: 10.1098/rspb.2005.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Joo H.S., Fu C.I., Otto M. Bacterial strategies of resistance to antimicrobial peptides. Philos Trans R Soc Lond B Biol Sci. 2016;371(1695) doi: 10.1098/rstb.2015.0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yu G., et al. Combination effects of antimicrobial peptides. Antimicrob Agents Chemother. 2016;60(3):1717–1724. doi: 10.1128/AAC.02434-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chongsiriwatana N.P., Wetzler M., Barron A.E. Functional synergy between antimicrobial peptoids and peptides against gram-negative bacteria. Antimicrob Agents Chemother. 2011;55(11):5399–5402. doi: 10.1128/AAC.00578-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Matsuzaki K., et al. Mechanism of synergism between antimicrobial peptides magainin 2 and PGLa. Biochemistry. 1998;37(43):15144–15153. doi: 10.1021/bi9811617. [DOI] [PubMed] [Google Scholar]
- 118.Brogden K.A. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3(3):238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 119.Spohn R., et al. Integrated evolutionary analysis reveals antimicrobial peptides with limited resistance. Nat Commun. 2019;10(1):4538. doi: 10.1038/s41467-019-12364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nagarajan D., et al., Omega76: A designed antimicrobial peptide to combat carbapenem- and tigecycline-resistant Acinetobacter baumannii. Sci Adv, 2019. 5(7): p. eaax1946. [DOI] [PMC free article] [PubMed]
- 121.Nagarajan D., et al. Computational antimicrobial peptide design and evaluation against multidrug-resistant clinical isolates of bacteria. J Biol Chem. 2018;293(10):3492–3509. doi: 10.1074/jbc.M117.805499. [DOI] [PMC free article] [PubMed] [Google Scholar]