Summary
Calcium (Ca2+) is a universal second messenger involved in synaptogenesis and cell survival; consequently, its regulation is important for neurons. ATPase plasma membrane Ca2+ transporting 1 (ATP2B1) belongs to the family of ATP-driven calmodulin-dependent Ca2+ pumps that participate in the regulation of intracellular free Ca2+. Here, we clinically describe a cohort of 12 unrelated individuals with variants in ATP2B1 and an overlapping phenotype of mild to moderate global development delay. Additional common symptoms include autism, seizures, and distal limb abnormalities. Nine probands harbor missense variants, seven of which were in specific functional domains, and three individuals have nonsense variants. 3D structural protein modeling suggested that the variants have a destabilizing effect on the protein. We performed Ca2+ imaging after introducing all nine missense variants in transfected HEK293 cells and showed that all variants lead to a significant decrease in Ca2+ export capacity compared with the wild-type construct, thus proving their pathogenicity. Furthermore, we observed for the same variant set an incorrect intracellular localization of ATP2B1. The genetic findings and the overlapping phenotype of the probands as well as the functional analyses imply that de novo variants in ATP2B1 lead to a monogenic form of neurodevelopmental disorder.
Keywords: ATP2B1, de novo, development delay, intellectual disability, neurodevelopmental disorder, calcium homeostasis, abnormal behavior, seizure
Main text
Global developmental delay (DD), often leading to intellectual disability (ID), has a high prevalence of about 2% and is among the most frequent indications for genetic testing.1 Despite its frequency and importance, a genetic cause can only be found in 27% to 42% of routine diagnostic cases.2,3 One reason for this inadequate clarification rate is the extreme genetic heterogeneity of DD. According to the SysNDD4 database, pathogenic variants in 1,497 (gene statistics from July 29, 2021) different genes are an established cause of a neurodevelopmental disorder. Furthermore, several genes are under investigation to identify their association with DD/ID.
Calcium (Ca2+) is a second messenger that is involved in the regulation of signal transduction, gene expression, cell metabolism, and cell survival. Ca2+ homeostasis is especially important for neurons, as it controls the release of neurotransmitters and regulates neurite outgrowth, cell membrane depolarization, and depression of synaptic transmission, which are important processes for learning and memory consolidation (for a review, see Brini et al.5). The dysregulation of Ca2+ homeostasis by pathogenic variants in Ca2+ channel genes, such as CACNA1A (MIM: 183086), is a common cause for several forms of ataxia. Moreover, other genes whose protein products control intracellular levels of free Ca2+, such as CACNA1B (MIM: 618497),6 CAMK4,7 ANXA11,8 and MICU2,9 have been reported to be associated with DD/ID.
ATPase plasma membrane Ca2+ transporting 1 (ATP2B1, formerly known as plasma membrane Ca2+ pump isoform 1; PMCA1; MIM: 108731) encodes an ATP-driven calmodulin-dependent Ca2+ pump that maintains the homeostasis of intracellular Ca2+ by removing it from the cytosol. Moreover, Ca2+ pumps are considered to have a crucial role on neuronal function (for a review, see Calì et al.10) and certain variants in isoforms of ATP2B1 have been reported to be pathogenic. Cali and colleagues11 reported one individual with DD, generalized hypotonia, cerebellar ataxia, and a hemizygote missense variant in ATP2B3 (MIM: 302500). In another case report, Li and colleagues12 describe four members of Chinese family with progressive spastic paraplegia and a heterozygous missense variant in ATP2B4.
In this study, we report 12 individuals with an overlapping phenotype of mild to moderate DD/ID, each harboring a different variant in ATP2B1. Table 1 presents an outline of the clinical symptoms of each individual. Additionally, a comprehensive clinical description of each individual is in Table S1 and in the supplemental notes. Written consents for genetic testing and its publication, as well as the publication of photographs, were obtained from all subjects studied or their legal representatives after counseling and information about risks and benefits in accordance with national ethical standards and laws as confirmed by the responsible local committees. This study was approved by the ethics committee of the University of Leipzig (approval code: 402/16-ek).
Table 1.
Clinical spectrum of the ATP2B1-related disorder
| Individual | Variant | Inheritance | Sex | Agea | DD | ID | Behavior | Seizures | Other |
|---|---|---|---|---|---|---|---|---|---|
| 1 | c.716A>G (p.Asp239Gly) | de novo | F | 6 years | yes | moderate | normal | yes | secundum atrial septal defect, toe clindactyly, facial dysmorphism, hypotonia |
| 2 | c.791C>T (p.Thr264Ile) | de novo | M | 8 years | yes | mild | ASD | no | transposition of large vessels, low set ears |
| 3 | c.1274C>A (p.Thr425Lys) | de novo | M | 9 years | yes | mild | N/A | pathological EEG | cerebral cavernous malformation, sparse hair |
| 4 | c.1376A>G (p.His459Arg) | de novo | M | 17 years | yes | mild | ASD | yes | marfanoid habitus, arachnodactyly, scoliosis, hypermobile thumb |
| 5 | c.2288G>C (p.Arg763Pro) | de novo | M | 21 years | yes | mild | ASD | no | facial dysmorphism |
| 6 | c.2365C>T (p.Arg789Cys) | de novo | M | 3 years | yes | unclassified | sleeping difficulties | yes | facial dysmorphism, hypotonia |
| 7 | c.2470G>A (p.Glu824Lys) | de novo | F | 22 years | yes | moderate | compulsive behavior | yes | N/A |
| 8 | c.2570A>G (p.Gln857Arg) | de novo | F | 3 years | yes | unclassified | normal | no | brachycephaly, facial dysmorphism, clinodactyly of the 5th finger |
| 9 | c.2972G>A (p.Arg991Gln) | unknown | M | 51 years | yes | mild | ASD | no | marfanoid habitus, aortic root dilation, pectus carinatum, scoliosis, arachnodactyly, facial dysmorphism |
| 10 | c.2632C>T (p.Gln878∗) | de novo | M | 6 years | yes | mild | hyperactivity | no | facial dysmorphism |
| 11 | c.458G>A (p.Trp153∗) | unknown | F | 6 years | yes | moderate | N/A | no | short stature |
| 12 | c.1789C>T (p.Arg597∗) | unknown | M | 5 years | yes | unclassified | ASD | infantile spasms | short stature, pectus excavatum, plagiocephaly |
Abbreviations: ID, intellectual disability; DD, development delay; ASD, autism spectrum disorder; N/A, not available; M, male; F, female. Variant descriptions based on GenBank: NM_001001323.2.
Age of last examination.
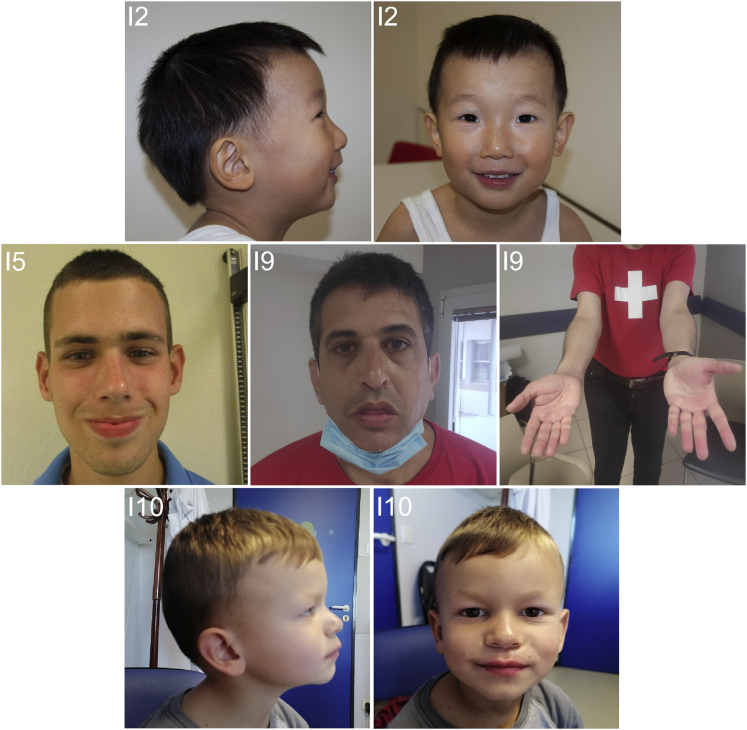
All individuals showed clear signs of DD. These included speech development delay in all individuals with a median age of first words of 26 months (spectrum of 12 to 36 months) and motor development delay in most individuals (median age at first walking 20 months; 16 to 36 months). The DD/ID was of variable degree; borderline to mild: 7/12, 58%; moderate: 3/12, 25%; moderate to severe: 2/12, 17%. Furthermore, five individuals (5/11, 45%, no information on one proband) were diagnosed with an autism spectrum disorder (ASD) and three additional individuals showed other behavior abnormalities. Dissimilar forms of seizures were reported in six individuals (6/12, 50%). Noteworthy, abnormal brain MRI findings were reported in three individuals without overlapping aspects. Individuals 1 and 6 had hypotonia. Other neurological symptoms, such as ataxia, were not reported. Individuals 11 and 12 had short stature, while all other individuals had normal growth development. Of note, two individuals, 4 and 9, exhibited a marfanoid habitus. Although minor facial dysmorphisms have been reported by six individuals (individuals 1, 5, 6, 8, 9, and 10), there was no apparent shared facial gestalt nor specific malformations (Figure 1, Table S1). Additionally, four individuals had anomalies of digits (individuals 1, 4, 8, and 9).
Figure 1.
Photos of individuals with ATP2B1 variants
No shared dysmorphic features in four individuals (for individual numbering, see Table 1).
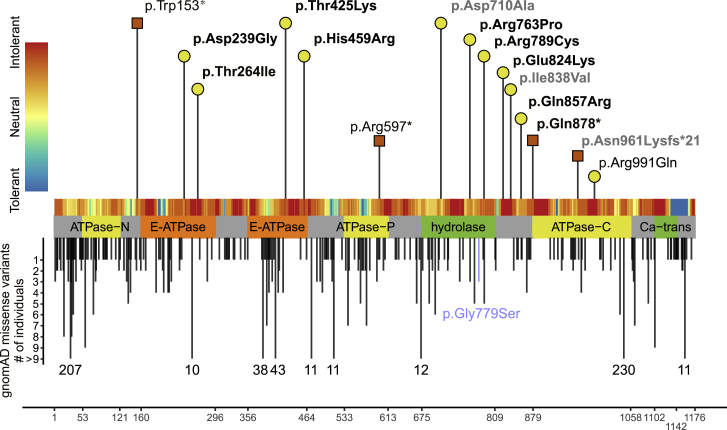
According to data from gnomAD,13 ATP2B1 contains a significantly reduced number of truncating and missense variants, indicating a selective constraint on both types of variants in a population without severe, early-onset phenotypes such as DD/ID (probability of being loss-of-function intolerant [pLI] score = 1.00; Z score missense = 5.29). In the present cohort, we identified 12 variants in ATP2B1, nine missense and three nonsense, that we consider of relevance. Nine (eight missense and one nonsense) were detected de novo, while for the remaining three individuals, parents were unavailable for segregation. All variants were absent from gnomAD, except the de novo variant from individual 6 (c.2365C>T [p.Arg789Cys]), which was detected in gnomAD once. The three truncating variants affect neither the last exon, the last 55 bp of the penultimate exon, nor the first 100 bp of ATP2B1; thus, nonsense-mediated mRNA decay is likely.14 The missense variants seem to be pathogenic in silico (conserved and predicted pathogenic by multiple tools) and have a combined annotation-dependent depletion (CADD)15 score between 25.6 and 32 (mean 28.5) and a rare exome variant ensemble learner (REVEL)16 score between 0.85 and 0.98 (mean 0.92, overview in Table S1). Additionally, seven out of nine missense variants of the cohort are located in established domains of ATP2B1 (Figure 2), which are critical for the protein’s function. The E1-E2 ATPase as well as the cation transporter ATPase domains, which are affected by six variants, are involved in Ca2+ binding, phosphoenzyme formation, and Ca2+ release.17 The haloacid dehalogenase-like hydrolase domain is affected by two variants and generates the energy for Ca2+ transport via hydrolysis of ATP to ADP. Two missense variants do not affect an established domain but are located in areas that are intolerant to missense variation, as proposed by MetaDome.18 Hence, an altered protein function of these seems plausible.
Figure 2.
Variants in ATP2B1
Location of missense and loss-of-function variant in ATP2B1 with respect to the domain structure of ATP2B1 (GenBank: NM_001001323.2). The x axis represents the corresponding amino acid position of ATP2B1. Variants reported in this study are labeled with the corresponding p-code and are indicated by a yellow circle (missense) or a red square (loss of function). Confirmed de novo variants are indicated in bold. Deciphering Developmental Disorders study variants with a lacking detailed phenotypic description are indicated in gray (see also Table S1 and supplemental methods). Missense variants in gnomAD with allele count are shown below the protein scheme. The gnomAD variant p.Gly779Ser, which was used as negative control for the Ca2+ imaging experiments, is blue color-coded. The tolerance landscape (MetaDome18) is shown color-coded above the protein scheme. Abbreviations: ATPase-N, cation transporter/ATPase, N terminus; ATPase-P, cation transport ATPase (P-type); ATPase-C, cation transporter/ATPase, C terminus; E-ATPase, E1-E2 ATPase; hydrolase, haloacid dehalogenase-like hydrolase; Ca-trans, plasma membrane calcium transporter ATPase C-terminal.
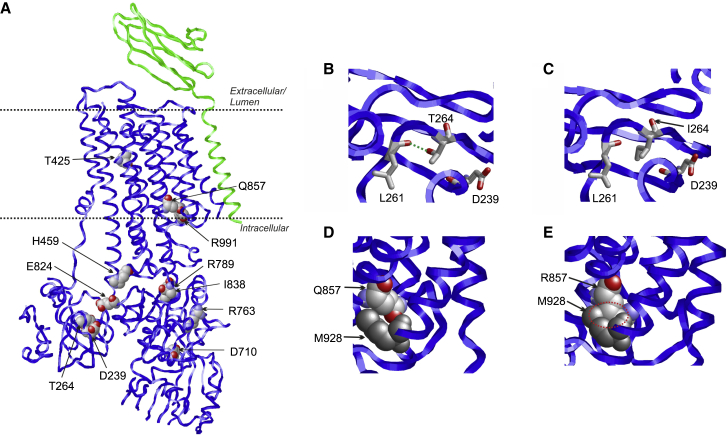
To better understand how missense variants affect ATP2B1 function, we investigated the effect of the variants by an in silico structural protein modeling. The availability of an experimental structure for ATP2B119 allows us to map the sequence positions of the variants in the 3D structure (Figure 3A). Three missense variants affect conserved amino acid (AA) residues in the transmembrane domain (Thr425, Gln857, and Arg991) that constitutes the Ca2+ exit pathway to the extracellular space. The remaining variants affect AA residues in the cytosolic protein part that harbors the active site. The observation that damaging variants are distributed over the structure is in line with the fact that ATP2B1 is a complex molecular machine that requires structural integrity of all domains for proper function.
Figure 3.
Structural effects in ATP21B1
(A) Structure of ATP2B1 (PDB: 6A6919) indicating the sites of genetic variants. The ATP2B1 structure is shown as blue ribbon and the neuroplastin subunit in green. Sites of genetic variants are shown in space-filled presentation (atom-type coloring) and labeled. The location of the membrane is indicated by two dotted lines.
(B) Enlargement of the ATP2B1 structure showing the effect of the p.T264Ile and p.Gln857Arg variants. (B) T264 forms a hydrogen bond (green dotted line) to L261.
(C) In the p.Thr264Ile variant, this hydrogen bond cannot be formed by the nonpolar isoleucine sidechain.
(D) Q857 is in close spatial proximity to M928 (shown in gray).
(E) The bulkier R857 sidechain of the variant causes steric clashes (red dotted circle) with M928 leading to a destabilization of ATP2B1.
Most of the residues affected by the identified variants form tight interactions in the protein structure and are predicted to affect protein stability by a variety of structural mechanisms: for example, p.Thr264Ile results in a loss of a stabilizing hydrogen bond (Figures 3B and 3C). The p.Gln857Arg exchange causes protein destabilization by a steric clash between the Arg857 and Met928 sidechain in the altered protein (Figures 3D and 3E). The p.Arg991Gln exchange most likely causes the loss of a stabilizing interaction between the positively charged arginine and the negatively charged phospholipids. The p.Thr425Lys exchange in the transmembrane domain generates a positive charge close to the Ca2+ exit pathway, which most likely interferes with the transport of the positively charged Ca2+ ions. The p.Asp710Ala exchange disrupts a salt-bridge between the Asp710 sidechain and a conserved lysine (Lys476) of the active site.
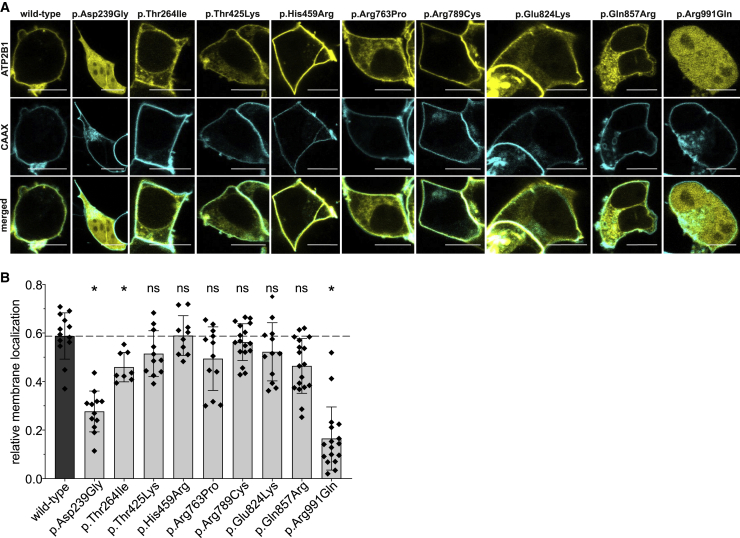
As the in silico predictions suggest that the variants would affect the function of ATP2B1, we tested this assumption experimentally. Therefore, we generated plasmids expressing ATP2B1 that harbor the variants of interest. Although the variants in our cohort affect several isoforms of ATP2B1, we decided to investigate the isoform (ATP2B1a) that is mainly detected in the human brain20 (GenBank: NM_001001323.2; Ensembl: ENST00000359142.7; compare also GTEx for expression data). As expected, we observed a membranous fluorescence after transfection of the ATP2B1-yellow fluorescent protein (YFP) wild-type expression plasmid in HEK293 cells (Figure 4A). More important, we also observed a significantly altered cellular localization of ATP2B1-YFP harboring the variants p.Asp239Gly, p.Thr264Ile, and p.Arg991Gln (Figures 4A and 4B). Of the remaining six missense variants, the variants p.Thr425Lys, p.Arg763Pro, p.Glu824Lys, and p.Gln857Arg also showed a shift of membranous to cytoplasmatic localization of ATP2B1. However, this effect was not statistically significant, as this was observable only in a proportion of analyzed cells (Figure S1). The variants p.His459Arg and p.Arg789Cys did not show a change and were comparable with the wild type.
Figure 4.
Subcellular localization of transiently overexpressed ATP2B1 variants
(A) Representative confocal laser scanning microscopy images of transfected HEK293 cells expressing YFP-fused ATP2B1 and a CAAX-box-modified cyan fluorescent protein. Scale bars: 10 μm.
(B) Quantification of relative membrane localization (for details, see supplemental methods). Data presented as mean and standard deviation from 8 to 17 independent analyzed cells. Data are shown as mean ± standard deviation represented by error bars. The results of a one-way ANOVA with the Games-Howell post-hoc test (each compared to wild type) is indicated as: ∗p < 0.05; ns: p > 0.05.
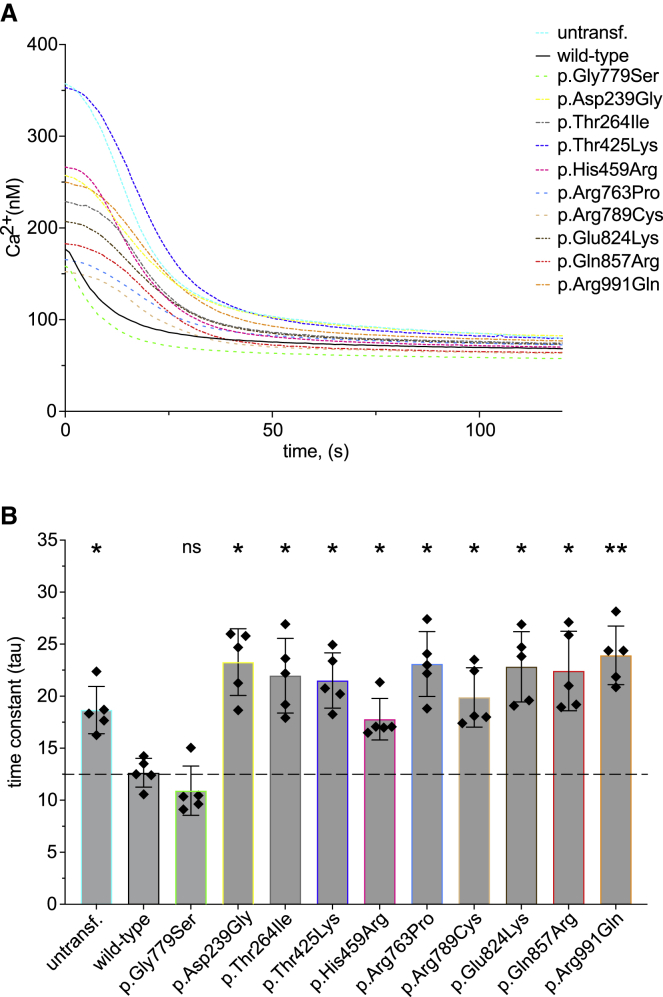
Next, we investigated whether the identified variants in ATP2B1 affect the Ca2+ transport function of ATP2B1. Therefore, we performed [Ca2+]i imaging in HEK293 cells after transfection of the corresponding expression plasmids. We chose this method21 because it allows a continuous measurement with a perfusion system to infuse the cells with different solutions. This makes it possible for us to investigate the cells in an approximate physiological environment and change of cellular Ca2+ gradients. The method allows even higher sensitivity because transfected cells can be separated from non-transfected ones by single-handedly picking the cells with a YFP signal, expressed through the plasmid. The result of this experiment is shown in Figure 5A. To investigate the ATP2B1 activity, we determined the relative Ca2+ extrusion rate. Therefore, after initial perfusion with EGTA, Ca2+ store depletion, and Ca2+ loading, we analyzed [Ca2+]i decline after perfusion with EGTA, which is represented by the time constant tau (see also Figure S2). Hence, a lower tau value indicates a higher ATP2B1 activity. Accordingly, we could detect a significantly reduced tau value after transfection of the wild-type ATP2B1 plasmid compared to untransfected HEK293 cells (Figure 5B). As an additional negative control, we analyzed a presumably non-pathogenic variant (p.Gly779Ser) that was detected three times in gnomAD. As expected, the [Ca2+]i imaging experiments revealed no statistically different Ca2+ extrusion rate of this variant compared to the wild type. The investigation of the identified variants in the present cohort revealed a significantly increased tau value after Ca2+ loading in all nine variants compared to the wild type (Figure 5B).
Figure 5.
De novo missense variants in ATP2B1 affect Ca2+ transport
(A) Fluorometric [Ca2+]i analysis (for details, see supplemental methods) in untransfected (untransf.) HEK293 cells and cells expressing wild-type or mutated ATP2B1. Data are shown as mean [Ca2+]i from five independent experiments after loading of the [Ca2+]i indicator dye fura-2/AM and the final addition of EGTA (for a representative complete sequence of the experiment, see Figure S2).
(B) In order to investigate the Ca2+ transport of transfected HEK293, time-dependent [Ca2+]i decline was analyzed after final addition of EGTA that is represented by the time constant tau. Data presented as mean and standard deviation from five independent experiments. As negative control served the likely non-pathogenic variant (3 times in gnomAD) p.Gly779Ser. The dashed line indicates the median tau value of ATP2B1 wild type. Data are shown as mean ± standard deviation represented by error bars. The results of a one-way ANOVA with the Games-Howell post-hoc test (each compared to wild type) is indicated as: ∗∗p < 0.005; ∗p < 0.05; ns: p > 0.05.
Interestingly, by ranking the variants on the basis of reduction of Ca2+ transport and on changes in cellular localization, we observe a significant correlation between the influence of a ATP2B1 variant on membrane localization and the tau value (p < 0.001; r2 = 0.768; Figure S3A). Of note, we also found a correlation between the REVEL score and the tau value (p < 0.02; r2 = 0.5; Figure S3B) on the basis of the rank. Although speculative, these observations indicate that the missense variants identified in this study reduce Ca2+ transport by disturbing the cellular trafficking and/or by inducing the degradation of abnormal ATP2B1 protein. A similar mechanism has also been reported for the creatine transporter SLC6A8.22
At this point, it should be noted that we identified another individual with DD and a de novo variant in ATP2B1 (c.1793T>C [p.Ile598Thr]; see supplemental note, “individual 16”). We have not included this individual into the clinical description of the present cohort, as the membrane localization and the tau value were indistinguishable from the wild type. It is possible that the variant causes a functional effect not measured by our assays. However, because the clinical relevance of this variant remains uncertain, we have excluded this individual from the phenotypic analysis.
The results of the present study imply that de novo missense variants in ATP2B1 cause a loss-of-function mechanism on the basis of the following observations. (1) The in silico structural modeling supports a reduced ATP2B1 activity resulting from changes of protein structure or stability. (2) The Ca2+ transport is significantly decreased by the missense variants. (3) The correlation between intracellular localization and tau value suggests the degradation of abnormal ATP2B1. (4) ATP2B1 exhibits a pLI score = 1.00, which supports a potential haploinsufficiency of this gene. (5) Three of the probands carry nonsense variants. This notion is further supported by the reports of seven independent individuals that harbor gross deletions, which affect ATP2B1 (summarized by Klein et al.23 and James et al.24). More important, these individuals exhibited an overlapping phenotype (DD, digit anomalies, facial dysmorphism, and growth deficiency) with the probands reported in the present study. We cannot exclude a dominant negative effect by the missense variants, even though this mechanism is unlikely. Instead of acting as monomers or homodimers, ATP2B1–4 form heteromeric complexes with neuroplastin or basigin.25 In accordance, none of the missense variants affect the interacting region of ATP2B1 and neuroplastin (Figure 3A).
Taken together, the overlapping phenotype of the probands and the experimental evidence strongly support that ATP2B1 is a DD/ID-associated gene. Furthermore, the role of ATP2B1 in neurodevelopment is strengthened by its expression pattern. While ATP2B1 is expressed at the early stages during development, the three other isoforms of the ATPase plasma membrane Ca2+ transporter family (ATP2B2–4) are expressed at later stages.26 The important role of ATP2B1 for neurological development is further corroborated by the fact that it can compensate for the absence of those other isoforms to a certain extent.27 ATP2B1 encodes different isoforms, which differ in calmodulin affinity and tissue expression, by alternative splicing (for a review, see Krebs28). Kenyon and colleagues found that Atp2b1b is nearly ubiquitously expressed in most tissues, while Aatp2ba is specifically expressed in the central nervous system. In rat brains, they found out that Atp2b1a is detected only in neurons, where it concentrates in the membrane of the soma, dendrites, and spines.20 It thus seems logical to consider ATP2B1 as a candidate gene for an early neurodevelopmental disorder as diagnosed in the probands of this study. Atp2b1–4 accumulate during the maturation of hippocampal neurons, which indicates the need of an increased Ca2+-exclusion capacity.26 Interestingly, Atp2b1–4 are transcriptionally regulated by Ca2+ itself.29 Likewise, studies with rats demonstrated the expression of ATP2B1b in the developing brain is later replaced by ATP2B1a, therefore further underlining its role in neurodevelopment.30
Thus, as ATP2B1 has a significant impact on Ca2+ homeostasis in the central nervous system31 during development on the basis of its expression pattern, it strongly supports that variants that affect ATP2B1’s function will impact neurons and therefore an individual’s neurodevelopment.
We also considered whether variants that affect a certain domain lead to a variable phenotype. However, we could observe neither a particular domain-specific correlation with the symptoms of the individuals nor a correlation between the severity of the symptoms and the impact of each variant on ATP2B1 activity. This may be due to the relatively small size of the herein reported cohort. Therefore, further studies with more individuals harboring a pathogenic variant in ATP2B1 may clarify a potential correlation.
In conclusion, the overlapping phenotype of 12 probands, the genetic findings, the in silico data, the structural modeling, the role of ATP2B1 in the central nervous system, and the pathogenicity of the variants prompt us to add ATP2B1 loss-of-function variants as a monogenetic cause of DD/ID.
Author contributions
M.J.R., R.J., M.W., and H.O. contributed to the concept and design and coordinated the study. Data curation and genetic analysis were performed by D.K., D.M., N.H., A.T., M.M., V.N., R.P., A.S., and T.B. Clinical investigations were performed by G.M., D.B., S.M., F.M., B.V., D.M., U.H., S.N., N.R., E.B., M. Sinnema, A.M., and I.D. The experiments were performed by M.J.R., N.U., and H.O. under the support of F.G. and M. Schaefer. H.S. performed the structural analysis and molecular modeling. M.J.R. and H.O. contributed to the acquisition, interpretation, statistical analysis, and visualization of the results. The original draft was written by M.J.R., R.J., and H.O., and all authors critically revised the manuscript and ensured the final approval of the manuscript for publication.
Acknowledgments
The authors would like to thank the proband’s families for their support and consent to the study. M.M. and V.N. were supported by the Telethon Undiagnosed Diseases Program (TUDP, GSP15001). The experiments were funded by budget resources of the Institute of Human Genetics (Leipzig, Germany), the Department of Neurosurgery (Leipzig, Germany), the Rudolf-Boehm-Institute of Pharmacology and Toxicology (Leipzig, Germany), and the Institute of Biochemistry (Erlangen, Germany). This work was generated within ITHACA: European Reference Network on Rare Congenital Malformations and Rare Intellectual Disability (M. Sinnema). We would also like to thank Patrick Yap and Polona Le Quesne Stabej from the University of Auckland, New Zealand for the fruitful discussions. Last but not least, we would like to thank the reviewers for the constructive comments that significantly improved the present work.
Declaration of interests
R.P. is an employee of GeneDx, Inc. All other authors declare no competing interests.
Published: March 30, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.03.009.
Data and code availability
The data of this study are available from the corresponding author with reasonable request. There was no code used for this study.
Web resources
DECIPHER, https://www.deciphergenomics.org/
ENSEMBL, https://www.ensembl.org/
GeneMatcher, https://genematcher.org/
MetaDome, https://stuart.radboudumc.nl/metadome
MutationTaster, https://www.mutationtaster.org/
OMIM, http://www.omim.org/
STRING database, https://www.string-db.org/
SysNDD, https://www.sysid.dbmr.unibe.ch/
The Genotype-Tissue Expression (GTEx) Project, https://gtexportal.org/home/
The National Center for Biotechnology Information (NCBI), https://www.ncbi.nlm.nih.gov/
UCSC Cell Browser (human cerebral cortex), http://hgw1.soe.ucsc.edu/
UniProt database, https://www.uniprot.org/
Variant Effect Predictor (VEP) from ENSEMBL, https://www.ensembl.org/Tools/VEP
WebAutoCasC, https://autocasc.uni-leipzig.de/
Supplemental information
References
- 1.McRae J.F., Clayton S., Fitzgerald T.W., Kaplanis J., Prigmore E., Rajan D., Sifrim A., Aitken S., Akawi N., Alvi M., et al. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilissen C., Hehir-Kwa J.Y., Thung D.T., van de Vorst M., van Bon B.W., Willemsen M.H., Kwint M., Janssen I.M., Hoischen A., Schenck A., et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 3.Aspromonte M.C., Bellini M., Gasparini A., Carraro M., Bettella E., Polli R., Cesca F., Bigoni S., Boni S., Carlet O., et al. Characterization of intellectual disability and autism comorbidity through gene panel sequencing. Hum. Mutat. 2019;40:1346–1363. doi: 10.1002/humu.23822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kochinke K., Zweier C., Nijhof B., Fenckova M., Cizek P., Honti F., Keerthikumar S., Oortveld M.A.W., Kleefstra T., Kramer J.M., et al. Systematic Phenomics Analysis Deconvolutes Genes Mutated in Intellectual Disability into Biologically Coherent Modules. Am. J. Hum. Genet. 2016;98:149–164. doi: 10.1016/j.ajhg.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brini M., Calì T., Ottolini D., Carafoli E. Neuronal calcium signaling: function and dysfunction. Cell. Mol. Life Sci. 2014;71:2787–2814. doi: 10.1007/s00018-013-1550-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorman K.M., Meyer E., Grozeva D., Spinelli E., McTague A., Sanchis-Juan A., Carss K.J., Bryant E., Reich A., Schneider A.L., et al. Bi-allelic Loss-of-Function CACNA1B Mutations in Progressive Epilepsy-Dyskinesia. Am. J. Hum. Genet. 2019;104:948–956. doi: 10.1016/j.ajhg.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zech M., Lam D.D., Weber S., Berutti R., Poláková K., Havránková P., Fečíková A., Strom T.M., Růžička E., Jech R., Winkelmann J. A unique de novo gain-of-function variant in CAMK4 associated with intellectual disability and hyperkinetic movement disorder. Cold Spring Harb. Mol. Case Stud. 2018;4:a003293. doi: 10.1101/mcs.a003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nahm M., Lim S.M., Kim Y.-E., Park J., Noh M.-Y., Lee S., Roh J.E., Hwang S.-M., Park C.-K., Kim Y.H., et al. ANXA11 mutations in ALS cause dysregulation of calcium homeostasis and stress granule dynamics. Sci. Transl. Med. 2020;12:eaax3993. doi: 10.1126/scitranslmed.aax3993. [DOI] [PubMed] [Google Scholar]
- 9.Shamseldin H.E., Alasmari A., Salih M.A., Samman M.M., Mian S.A., Alshidi T., Ibrahim N., Hashem M., Faqeih E., Al-Mohanna F., Alkuraya F.S. A null mutation in MICU2 causes abnormal mitochondrial calcium homeostasis and a severe neurodevelopmental disorder. Brain. 2017;140:2806–2813. doi: 10.1093/brain/awx237. [DOI] [PubMed] [Google Scholar]
- 10.Calì T., Brini M., Carafoli E. The PMCA pumps in genetically determined neuronal pathologies. Neurosci. Lett. 2018;663:2–11. doi: 10.1016/j.neulet.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Calì T., Lopreiato R., Shimony J., Vineyard M., Frizzarin M., Zanni G., Zanotti G., Brini M., Shinawi M., Carafoli E. A Novel Mutation in Isoform 3 of the Plasma Membrane Ca2+ Pump Impairs Cellular Ca2+ Homeostasis in a Patient with Cerebellar Ataxia and Laminin Subunit 1α Mutations. J. Biol. Chem. 2015;290:16132–16141. doi: 10.1074/jbc.M115.656496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M., Ho P.W.-L., Pang S.Y.-Y., Tse Z.H.-M., Kung M.H.-W., Sham P.-C., Ho S.-L. PMCA4 (ATP2B4) mutation in familial spastic paraplegia. PLoS ONE. 2014;9:e104790. doi: 10.1371/journal.pone.0104790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindeboom R.G.H., Supek F., Lehner B. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat. Genet. 2016;48:1112–1118. doi: 10.1038/ng.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rentzsch P., Schubach M., Shendure J., Kircher M. CADD-Splice-improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 2021;13:31. doi: 10.1186/s13073-021-00835-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D., et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mangialavori I.C., Ferreira-Gomes M.S., Saffioti N.A., González-Lebrero R.M., Rossi R.C., Rossi J.P.F.C. Conformational changes produced by ATP binding to the plasma membrane calcium pump. J. Biol. Chem. 2013;288:31030–31041. doi: 10.1074/jbc.M113.494633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiel L., Baakman C., Gilissen D., Veltman J.A., Vriend G., Gilissen C. MetaDome: Pathogenicity analysis of genetic variants through aggregation of homologous human protein domains. Hum. Mutat. 2019;40:1030–1038. doi: 10.1002/humu.23798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong D., Chi X., Ren K., Huang G., Zhou G., Yan N., Lei J., Zhou Q. Structure of the human plasma membrane Ca2+-ATPase 1 in complex with its obligatory subunit neuroplastin. Nat. Commun. 2018;9:3623. doi: 10.1038/s41467-018-06075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenyon K.A., Bushong E.A., Mauer A.S., Strehler E.E., Weinberg R.J., Burette A.C. Cellular and subcellular localization of the neuron-specific plasma membrane calcium ATPase PMCA1a in the rat brain. J. Comp. Neurol. 2010;518:3169–3183. doi: 10.1002/cne.22409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lenz J.C., Reusch H.P., Albrecht N., Schultz G., Schaefer M. Ca2+-controlled competitive diacylglycerol binding of protein kinase C isoenzymes in living cells. J. Cell Biol. 2002;159:291–302. doi: 10.1083/jcb.200203048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salazar M.D., Zelt N.B., Saldivar R., Kuntz C.P., Chen S., Penn W.D., Bonneau R., Koehler Leman J., Schlebach J.P. Classification of the Molecular Defects Associated with Pathogenic Variants of the SLC6A8 Creatine Transporter. Biochemistry. 2020;59:1367–1377. doi: 10.1021/acs.biochem.9b00956. [DOI] [PubMed] [Google Scholar]
- 23.Klein O.D., Cotter P.D., Schmidt A.M., Bick D.P., Tidyman W.E., Albertson D.G., Pinkel D., Rauen K.A. Interstitial deletion of chromosome 12q: genotype-phenotype correlation of two patients utilizing array comparative genomic hybridization. Am. J. Med. Genet. A. 2005;138:349–354. doi: 10.1002/ajmg.a.30867. [DOI] [PubMed] [Google Scholar]
- 24.James P.A., Oei P., Ng D., Kannu P., Aftimos S. Another case of interstitial del(12) involving the proposed cardio-facio-cutaneous candidate region. Am. J. Med. Genet. A. 2005;136:12–16. doi: 10.1002/ajmg.a.30693. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt N., Kollewe A., Constantin C.E., Henrich S., Ritzau-Jost A., Bildl W., Saalbach A., Hallermann S., Kulik A., Fakler B., Schulte U. Neuroplastin and Basigin Are Essential Auxiliary Subunits of Plasma Membrane Ca2+-ATPases and Key Regulators of Ca2+ Clearance. Neuron. 2017;96:827–838.e9. doi: 10.1016/j.neuron.2017.09.038. [DOI] [PubMed] [Google Scholar]
- 26.Kip S.N., Gray N.W., Burette A., Canbay A., Weinberg R.J., Strehler E.E. Changes in the expression of plasma membrane calcium extrusion systems during the maturation of hippocampal neurons. Hippocampus. 2006;16:20–34. doi: 10.1002/hipo.20129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okunade G.W., Miller M.L., Pyne G.J., Sutliff R.L., O’Connor K.T., Neumann J.C., Andringa A., Miller D.A., Prasad V., Doetschman T., et al. Targeted ablation of plasma membrane Ca2+-ATPase (PMCA) 1 and 4 indicates a major housekeeping function for PMCA1 and a critical role in hyperactivated sperm motility and male fertility for PMCA4. J. Biol. Chem. 2004;279:33742–33750. doi: 10.1074/jbc.M404628200. [DOI] [PubMed] [Google Scholar]
- 28.Krebs J. The plethora of PMCA isoforms: Alternative splicing and differential expression. Biochim. Biophys. Acta. 2015;1853:2018–2024. doi: 10.1016/j.bbamcr.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 29.Guerini D., García-Martin E., Gerber A., Volbracht C., Leist M., Merino C.G., Carafoli E. The expression of plasma membrane Ca2+ pump isoforms in cerebellar granule neurons is modulated by Ca2+ J. Biol. Chem. 1999;274:1667–1676. doi: 10.1074/jbc.274.3.1667. [DOI] [PubMed] [Google Scholar]
- 30.Brandt P., Neve R.L., Kammesheidt A., Rhoads R.E., Vanaman T.C. Analysis of the tissue-specific distribution of mRNAs encoding the plasma membrane calcium-pumping ATPases and characterization of an alternately spliced form of PMCA4 at the cDNA and genomic levels. J. Biol. Chem. 1992;267:4376–4385. [PubMed] [Google Scholar]
- 31.Brini M. Plasma membrane Ca(2+)-ATPase: from a housekeeping function to a versatile signaling role. Pflugers Arch. 2009;457:657–664. doi: 10.1007/s00424-008-0505-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data of this study are available from the corresponding author with reasonable request. There was no code used for this study.