Abstract
Cancer has been always considered as one of the main human health challenges worldwide. One of the main causes of cancer-related mortality is late diagnosis in the advanced stages of the disease, which reduces the therapeutic efficiency. Therefore, novel non-invasive diagnostic methods are required for the early detection of tumors and improving the quality of life and survival in cancer patients. MicroRNAs (miRNAs) have pivotal roles in various cellular processes such as cell proliferation, motility, and neoplastic transformation. Since circulating miRNAs have high stability in body fluids, they can be suggested as efficient noninvasive tumor markers. MiR-96 belongs to the miR-183-96-182 cluster that regulates cell migration and tumor progression as an oncogene or tumor suppressor by targeting various genes in solid tumors. In the present review, we have summarized all of the studies that assessed the role of miR-96 during tumor progression. This review clarifies the molecular mechanisms and target genes recruited by miR-96 to regulate tumor progression and metastasis. It was observed that miR-96 mainly affects tumorigenesis by targeting the structural proteins and FOXO transcription factors.
Introduction
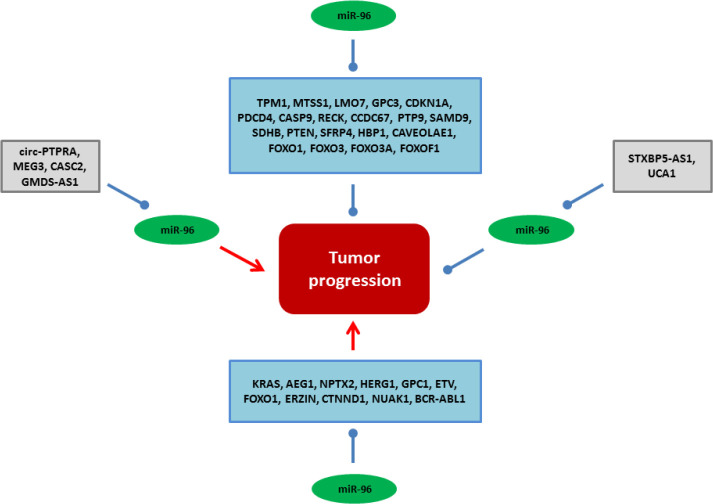
Cancer is still one of the main causes of death and remains an important health challenge globally. Although, improving the quality of life and increasing the life expectancy of cancer patients are the main goals of cancer therapy, the low efficacy of routine treatment modalities highlights the need to introduce novel specific therapeutic strategies (1). MicroRNAs (miRNAs) are a class of short non-coding RNAs (~22nt) involved in post-transcriptional regulation via mRNA degradation or translational suppression. They regulate cell proliferation, apoptosis, migration, and malignant transformation (2). Therefore, deregulation of miRNAs can be associated with tumor progression (3). They function as oncogene or tumor suppressors during neoplastic transformation. Regarding the tissue-specificity of miRNA expressions, there are specific miRNA signatures in different tumors (4). Non-invasive or minimally invasive markers are required for early-stage tumor detection to improve the patient’s survival. Circulating miRNAs have high stability in body fluids which can be suggested as efficient noninvasive tumor markers (5, 6). MiR-96 belongs to the miR-183-96-182 family that regulates cell motility and tumor progression as an oncogene or tumor suppressor by targeting various genes in solid tumors (7, 8). It can be regulated by ZEB1 (9), β‐catenin (10), and epidermal growth factor receptor (EGFR) (11). In the present review, we discussed all of the miR-96 based tumor reports to clarify the molecular mechanisms of miR-96 during tumor progressions and metastasis (Figure 1) (Table 1).
Figure 1.
All of the molecular mechanisms and interactions of miR-96 during tumor initiation, progression, and metastasis
Table 1.
Molecular mechanisms of miR-96 during tumor progression
| GUO [7] | 2012 | Bladder | miR-96 | FOXO1 | Tumor suppressor | 40 patients T24 cell line |
| tsai [11] | 2017 | Prostate | miR-96 | ETV | Tumor suppressor | 69 patients PC3 and RasB1 cell lines |
| yao [13] | 2018 | Osteosarcoma | miR-96 | EZRIN | Tumor suppressor | 4 patients MG-63 cell line |
| yu [14] | 2015 | Renal | miR-96 | EZRIN | Tumor suppressor | 63 patients CaKi-1 and 786-O cell lines |
| wu [17] | 2017 | Lung | miR-96 | LMO7 | Oncogene | BEAS-2B, A549, PC9, and H1299 cell lines |
| gao [18] | 2020 | Breast | miR-96 | CTNND1 | Tumor suppressor | 155 patients MDA-MB-231,MDA-Mb-246, T47D, and ZR-75-30 cell lines |
| yin [22] | 2020 | Cholangiocarcinoma | miR-96 | MTSS1 | Oncogene | 115 patients HuCCT1, HuH28, and RBE cell lines |
| XIE [23] | 2018 | Breast | miR-96 | MTSS1 | Oncogene | 5 patients MCF7 and MDA-MB-231 cell lines |
| XU [24] | 2016 | Prostate | miR-96 | MTSS1 | Oncogene | PC3 and LNCaP cell lines |
| ZHANG [28] | 2019 | Glioma | MEG3 | miR-96 | Oncogene | 30 patients GSC11, M059J, and D54 cell lines |
| liu [33] | 2018 | Bladder | MEG3 | miR-96 | Oncogene | 45 patients RT4, RT112, and T24 cell lines |
| ge [34] | 2020 | Colorectal | miR-96 | TPM1 | Oncogene | 40 patients SW480 cell lines |
| li [36] | 2014 | Pancreatic | miR-96 | GPC1 | Tumor suppressor | 38 patients Panc-1, AsPC-1, and BxPC-3 cell lines |
| FEI [39] | 2018 | Lung | miR-96 | GPC3 | Oncogene | 57 patients A549 and H460 cell lines |
| liu [44] | 2019 | Ovarian | miR-96 | CAVEOLAE1 | Oncogene | 23 patients A2780, OVCAR3, SKOV3, and CAOV3 cell lines |
| FENG [51] | 2014 | Pancreatic | miR-96 | HERG1 | Tumor suppressor | 20 patients PANC-1, SW1990, HPAC, CFPAC-1, and BxPC-3 cell lines |
| gao [53] | 2018 | Breast | CASC2 | miR-96 | Oncogene | 35 patients MCF7 and MDA-MB-231 cell lines |
| zhao [57] | 2020 | Lung | GMDS-AS1 | miR-96 | Oncogene | A549, SPCA-1, PC-9, H1975, and H838 cell lines |
| wei [58] | 2019 | Lung | CIRC-PTPRA | miR-96 | Oncogene | 34 patients H23, H1755, and H522 cell lines |
| liu [59] | 2019 | Thyroid | miR-96 | CCDC67 | Oncogene | 78 patients TPC-1, K-1, BCPAP, and 8505c cell lines |
| song [72] | 2015 | Thyroid | miR-96 | FOXO1 | Oncogene | 60 patients TPC-1 and K-1 cell lines |
| yang [73] | 2019 | Hepatocellular | miR-96 | FOXO1 | Oncogene | 60 patients HepG2 cell line |
| yang [75] | 2020 | Cervical | miR-96 | FOXO1 | Oncogene | 83 patients C41, C33A, HeLa, CaSki, MS751, and HT-3 cell lines |
| YU [77] | 2014 | Prostate | miR-96 | FOXO1 | Oncogene | 13 patients LNCaP and PC3 cell lines |
| LIN [79] | 2010 | Breast | miR-96 | FOXO3A | Oncogene | 23 patients MCF7, ZR-75-30, BT549, Bcap37, MDA-MB-453, and T47D cell lines |
| zhou [80] | 2018 | Pancreatic | UCA1 | miR-96 | Tumor suppressor | 36 patients PANC-1, SW1990, and AS-PC1 cell lines |
| li [81] | 2015 | Lung | miR-96 | FOXO3 | Oncogene | 68 patients A549 and SPC-A-1 cell lines |
| YIN [82] | 2020 | Breast | miR-96 | FOXO3 | Oncogene | MCF7 and T47D cell lines |
| gao [83] | 2015 | Colorectal | miR-96 | FOXO1, FOXO3A | Oncogene | 20 patients SW480 and SW620 cell lines |
| wang [89] | 2020 | Oral | miR-96 | FOXOF1 | Oncogene | 30 patients SCC1, SCC4, Tca8113, Cal-27, and HSC-2 cell lines |
| YAN [93] | 2014 | Glioma | miR-96 | HBP1 | Oncogene | U-87MG, U-251MG, U-373MG, and M059 cell lines |
| zhang [95] | 2020 | Cervical | miR-96 | SFRP4 | Oncogene | 60 patients HeLa, SiHa, Me180, and MS751 cell lines |
| guo [101] | 2020 | Pancreatic | UCA1 | miR-96 | Tumor suppressor | 46 patients PANC-1, BxPC-3, Aspc-1, SW1990, and HUVECs cell lines |
| FENG [108] | 2018 | Glioblastoma | miR-96 | AEG1 | Tumor suppressor | U251 cell line |
| xiang [111] | 2020 | Renal | miR-96 | NPTX2 | Tumor suppressor | 8 patients RenCa, 786-O, and HEK293T cell lines |
| shao [112] | 2019 | Cervical | STXBP5-AS1 | miR-96 | Tumor suppressor | 37 patients C33A, MS751, SiHa, HeLa, ME180, and CaSki cell lines |
| vahabi [113] | 2019 | Head and neck | miR-96 | PTEN | Oncogene | Cal27, FaDu, and H1299 cell lines |
| zhao [118] | 2019 | Thyroid | miR-96 | SDHB | Oncogene | 28 patients BCPAP, K-1, and TPC-1 cell lines |
| WU [120] | 2016 | Lung | miR-96 | SAMD9 | Oncogene | H358 and H23 cell lines |
| yu [126] | 2010 | Pancreatic | miR-96 | KRAS | Tumor suppressor | PaCa-2, PANC-1, BxPC-3, and HeLa cell lines |
| HONG [132] | 2016 | Breast | miR-96 | PTPN9 | Oncogene | 10 patients MCF7 and MDA-MB-468 cell lines |
| ma [133] | 2018 | Cervical | miR-96 | PTPN9 | Oncogene | HeLa cell line |
| huang [137] | 2014 | Pancreatic | miR-96 | NUAK1 | Tumor suppressor | 10 patients PaCa-2 and PANC-1 cell lines |
| huang [139] | 2019 | CML | miR-96 | BCR-ABL1 | Tumor suppressor | 50 patients K562 and MEG01 cell lines |
| xia [147] | 2014 | Esophageal | miR-96 | RECK | Oncogene | 145 patients TE-1, ECa-109, and EC-9706 cell lines |
| ZHANG [148] | 2014 | Breast | miR-96 | RECK | Oncogene | 38 patients MDA-MB-231, MCF7, MDA-MB-468, and T47D cell lines |
| guo [149] | 2014 | Lung | miR-96 | RECK | Oncogene | 28 patients A549, SK-MES-1, and H1299 cell lines |
| iwai [150] | 2018 | Hepatocellular | miR-96 | CASP9 | Oncogene | SNU387, SNU449, and HLF cell lines |
| guo [152] | 2018 | Glioblastoma | miR-96 | PDCD4 | Oncogene | U-87MG, U-251MG, and A172 cell lines |
| li [157] | 2020 | AML | UCA1 | miR-96 | Tumor suppressor | U937 and HL60 cell lines |
| wu [159] | 2015 | Bladder | miR-96 | CDKN1A | Oncogene | 60 patients T24 and EJ cell lines |
Structural proteins
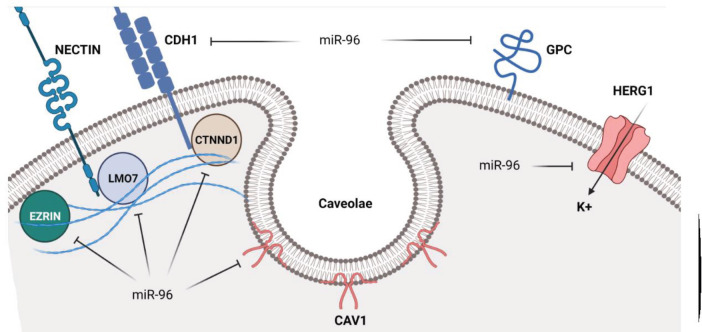
Structural proteins are involved in cell migration, adhesion, and ion homeostasis that can be regulated by miR-96 in tumor cells (Figure 2). EZRIN is a linker protein between the actin cytoskeleton and plasma membrane proteins participating in cell adhesion and migration. Its deregulation has been reported in metastatic tumors (12). It has been reported that miR-96 suppressed osteosarcoma (OS) cell invasion and proliferation while increasing apoptosis through EZRIN targeting. There was also miR-96 down-regulation in OS tissues in comparison with normal tissues (13). There was an inverse correlation between the levels of miR-96 expressions and the invasive capability of renal cell carcinoma (RCC) cells. MiR-96 reduced the RCC cells invasion through EZRIN targeting (14). LMO7 belongs to the PDZ/LIM domain-containing family of proteins participating in protein-protein interactions by actin binding (15). It maintains the epithelial architecture by regulation of actin cytoskeleton in normal cells. However, various studies have reported LMO7 up-regulation during carcinogenesis in different tumor types (16). It has been shown that there was miR-96 up-regulation in lung cancer patients that was directly correlated with high-grade and metastatic lymph node tumors. Silencing of miR-96 reduced lung tumor cell migration and cisplatin resistance by LMO7 up-regulation (17). CTNND1 is a critical cell adhesion regulator through binding with the juxta-membrane region of the cadherin cytoplasmic tails. There was miR-96-5p down-regulation in BC tissues and cell lines. MiR-96-5p down-regulation was also associated with lower overall survival, TNM stage, and distant metastasis. MiRNA-95-5p reduced BC cell proliferation and metastasis via suppression of CTNND1 mediated WNT signaling (18).
Figure 2.
Role of miR-96 in regulation of structural proteins during tumor progressions
MTSS1 regulates cytoskeletal dynamics and promotes membrane ruffle formation by interaction with actin and Rac proteins (19). It is an inhibitor of tumor metastasis by interaction with actin cytoskeleton associated with tumor progression in different organs (20, 21). There was significant miR-96 up-regulation in cholangiocarcinoma (CCA) tissues in comparison with normal margins that was significantly correlated with advanced TNM stage, shorter overall survival, lymph node metastasis, and poor differentiation. MiR-96 induced CCA cell proliferation and motility through MTSS1 targeting (22). There was a significantly higher serum miR96 level in breast cancer (BC) cells in comparison with controls that were reduced in patients with chemotherapy. MiR-96 down-regulated and up-regulated CDH1 and CDH2 in BC cells, respectively. It also inhibited BC cell migration through MTSS1 targeting (23). There was also a negative association between the serum levels of miR-96 and MTSS1 that was associated with survival in prostate cancer (PCa) patients (24). Long non-coding RNAs (lncRNAs) are involved in tumor progression and metastasis (25). Maternally expressed 3(MEG3) is a tumor suppressor lncRNA that is down-regulated in many cancers (26, 27). LncRNA MEG3 suppressed glioma cell proliferation and migration via miR96-5p sponging that resulted in MTSS1 up-regulation. Moreover, there was significant MEG3 down-regulation in glioma cells and tissues (28). MEG3 suppresses cell proliferation via p53 induction and TGF-β targeting (29, 30). Tropomyosin (TPM) is an actin-binding protein involved in inhibition of cellular transformation (31). TPM1 regulates the actin-myosin interaction (32). It has been reported that MEG3 inhibited bladder cancer (BCa) progression by regulating the miR-96/TPM1 axis. There was MEG3 down-regulation in BCa tissues that was correlated with high-grade and muscular invasion. MEG3 inhibited BCa proliferation while inducing apoptosis by miR-96 sponging and subsequent TPM1 up-regulation (33). There was miR-96 up-regulation in colorectal cancer (CRC) tissues. Suppression of miR96 increased oxaliplatin sensitivity in CRC cells by Bcl-2 down-regulation and the TPM1 and BAX up regulations (34).
Glypican-1 (GPC1) is a membrane-bound proteoglycan that is involved in organ development via regulation of extracellular growth factors, tumorigenesis, and angiogenesis (35). There was a significant miR-96-5p down-regulation in pancreatic cancer (PC) tissues that was associated with larger tumor sizes, poorer differentiation, and reduced survival. MiR-96 inhibited PC cell proliferation via GPC1 targeting (36). GPC3 belongs to the integral membrane proteoglycan family that is anchored with the membrane by a glycosylphosphatidylinositol. It is an important member of the extracellular matrix (ECM) involved in regulation of heparin-binding growth factor (37, 38). There was miR-96 up-regulation in non-small cell lung carcinoma (NSCLC) tissues in comparison with normal margins. MiR-96 increased NSCLC cell migration through GPC3 suppression (39). Caveolae1 (CAV1) is a structural component of caveolae involved in signal transduction and vesicular trafficking (40, 41). It has anti-tumor or oncogenic functions in different cancers (42, 43). There were significant miR-96-5p up-regulations in ovarian cancer (OC) tissues and cell lines in comparison with normal margins and cell lines. MiR-96-5p induced OC cell proliferation and migration via CAV1 targeting. It also up-regulated CCND1 by phosphorylation of AKT (44).
Ion channels are important structural proteins during tumor progression (45, 46). HERG1 belongs to the family of voltage-gated potassium channels that are up-regulated in various cancers (47, 48). It regulates cell proliferation and apoptosis (49, 50). HERG1 up-regulations in pancreatic tumor tissues were significantly associated with lymph node metastasis, TNM stage, and grade of differentiation. MiR-96 significantly inhibited pancreatic tumor cell proliferation and migration via HERG1 targeting (51).
SYVN1 is an E3 ubiquitin-protein ligase involved in elimination of unfolded proteins that are accumulated during ER stress. It promotes unfolded protein transportation to the cytoplasm where it uses the ubiquitin-proteasome process for protein degradation (52). There was significant CASC2 up-regulation in BC tissues compared with normal margins. CASC2 promoted apoptosis while inhibiting BC cell migration by miR-96-5p sponging that resulted in SYVN1 up-regulation (53).
As a deubiquitinase, CYLD has pivotal roles in microtubule stabilization by alpha-tubulin acetylation that is associated with cell proliferation, cytokinesis, migration, and angiogenesis (54-56). It has been reported that GMDS-AS1 inhibited lung tumor cell proliferation while inducing apoptosis. GMDS-AS1 up-regulated CYLD through miR-96-5p sponging (57).
CircPTPRA is a circular RNA transcribed from PTPRA. RASSF8 is a tumor suppressor with pivotal roles in maintenance of adherence junctions in epithelial cells and migration. There was circPTPRA down-regulation in NSCLC tumors compared with controls that was associated with invasive tumors and shorter survival rates. It sponged miR-96-5p to inhibit EMT in NSCLC cells through RASSF8 up-regulation (58). CCDC67 is a component of the deuterostome complex that is involved in centriole amplification in multi-ciliated cells. There were significant miR-96-5p up-regulations in papillary thyroid cancer (PTC) tissues and cell lines compared with normal margins and cells. MiR-96-5p induced PTC cell proliferation and migration by CCDC67 targeting (59).
Transcription factors
Forkhead transcription factors are critical factors during embryogenesis (60). They are mainly considered tumor suppressors during tumor progressions (61). The Forkhead box O (FOXO) protein family has a critical role in regulation of PI3K/AKT signaling that mediates cell proliferation, differentiation, and tumorigenesis (62). FOXO proteins are involved in cell cycle regulation by up-regulations of p21Cip1 and p27Kip1 cell-cycle inhibitors, while CCND1 down-regulation results in G1/S arrest (63, 64). They also promote apoptosis by regulation of proapoptotic factors such as Bim and Fas ligand (65, 66). FOXO phosphorylation can result in DNA release and nucleus to cytoplasm translocation via 14-3-3 chaperones (67). FOXO1 is known as a tumor suppressor in the majority of cancers (68, 69), while it has an oncogenic function in female genital tract tumors (70). FOXO1 is mainly regulated by the PI3K/AKT pathway during thyroid tumorigenesis (71). There was miR-96 up-regulation in PTC tissues in comparison with normal samples. MiR-96 significantly induced PTC cell proliferation, while reducing 5FU mediated apoptosis by FOXO1 targeting that resulted in suppression of AKT/FOXO1/ Bim axis (72). There was miR-96 up-regulation in hepatocellular carcinoma (HCC) tissues and cell lines. MiR-96 induced AKT/GSK-3β/β-catenin axis via FOXO1 suppression in HCC cells. FOXO1 decreased tumor cell proliferation, invasion, and in vivo growth promoted by miR-96 in HCC. MiR-96 increased HCC progression via FOXO1 targeting. It also promoted β-catenin nuclear translocation (73). MiR-96 induced apoptosis via FOXO1 targeting in BCa cells (7), while suppressing camptothecin-induced apoptosis by FOXO1 targeting in PCa cells (74). p21 and p27 are critical factors involved in cell cycle regulation. MiR-96 was shown to induce cervical cancer (CC) cell proliferation through FOXO1 targeting. There were significant miR-96 up-regulations in CC tissues and cell lines that were associated with stage, grade, and lymph node invasion. MiR96 induced G1/S transition and cell proliferation in CC cells. Suppression of miR-96 increased apoptosis via p21 and p27 up-regulations (75). Another study showed that miR-96 was up-regulated by chemotherapeutic treatment that reduced the levels of FOXO1 expression and subsequent p21 down-regulation in GC cells (76). MiR-96 up-regulation was observed in PCa tissues in comparison with normal samples. MiR-96 induced PCa cell proliferation and clonogenicity through FOXO1 targeting (77).
FOXO3 belongs to the forkhead transcription factors involved in apoptosis, metabolism, and DNA repair (78). MiR-96 induced BC cell proliferation through FOXO3a targeting that reduced the levels of p27Kip1 and p21Cip1 expressions, while up-regulating CCND1. A significant miR-96 up-regulation was also reported in BC tissues compared with normal specimens (79). There was Urothelial Cancer Associated 1 (UCA1) up-regulation in PC tissues that was inversely associated with miR-96 expression. Silencing of UCA1 or FOXO3 inhibited pancreatic tumor cell invasion while promoting apoptosis. UCA1 increased pancreatic tumor cell invasion by miR-96 sponging that resulted in FOXO3 up-regulation (80). There was a significant miR-96 up-regulation in NSCLC tissues in comparison with normal specimens that was associated with TNM stage, grade of differentiation, and lymph node involvement. MiR-96 also induced NSCLC growth by FOXO3 targeting (81). The miRNA-96-5p up-regulation was also shown in BC tissues that induced cell proliferation through FOXO3 inhibition (82). There was miR-96 up-regulation in CRC tissues compared with corresponding normal samples. MiR-96 also induced CRC cell proliferation through FOXO1 and FOXO3a targeting (83). FOXF2 has tumor suppressor function in a variety of tumors such as breast, gastric, and colorectal cancers (84-86).
Proteolytic enzymes with ECM degradation ability are also essential factors during tumor cell invasion (87). Matrix metalloproteinases (MMPs) are a family of proteases that degrade ECM during angiogenesis and invasion (88). There was miR-96-5p up-regulation in oral squamous cell carcinoma (OSCC) tissues in comparison with normal margins. MiR-96-5p significantly induced OSCC cell proliferation via CDK4 and CCND1 up-regulations while p27 down-regulation. MiR-96-5p also reduced the levels of MMP-2 and MMP-9 expressions, while up-regulating TIMP-1 in OSCC cells. It induced the EMT of OSCC cells through CDH1 down-regulation and CDH2 up-regulation. MiR-96-5p exerted all of the oncogenic functions in OSCC cells by FOXF2 targeting (89). There was also miR-96 up-regulation in PCa tissues in comparison with normal specimens which was associated with lymph node involvement, distant metastasis, and high PSA levels. MiR-96 increased PCa cell proliferation via FOXF2 targeting. Moreover, miR-96 down-regulation reduced PCa cell proliferation by suppressing the expressions of CCNA1, CDK2, and CDK4 (90).
Signaling pathways
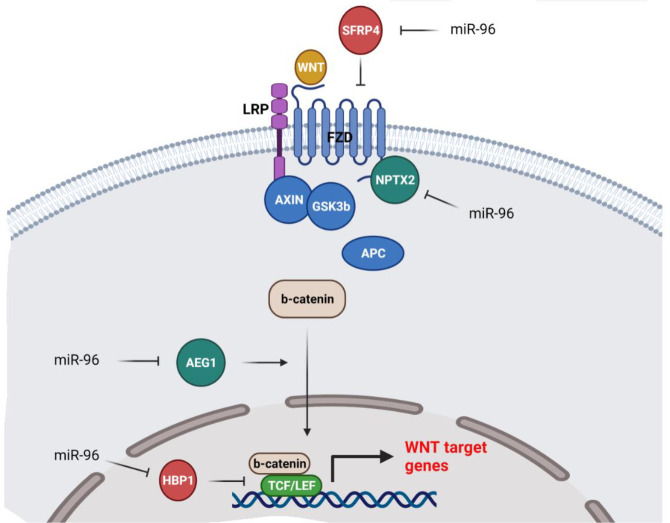
WNT is a developmental signaling pathway that has critical roles in normal tissue homeostasis and neoplastic transformation (91, 92). MiR-96 is involved in regulation of the WNT signaling pathway during tumor initiation and metastasis (Figure 3). It has been shown that miR-96 induced the WNT pathway and glioma cell growth by suppression of HBP1 as an inhibitor of the WNT pathway (93). SFRP4 belongs to the secreted frizzled related proteins (SFRPs) associated with regulation of cell proliferation and motility through the WNT signaling pathway (94). There was miR-96-5p up-regulation in CC tissues that was significantly associated with clinical stages and lymph node invasion. MiR-96-5p induced cell migration, while inhibiting apoptosis in CC cells via SFRP4 targeting (95). CTNNB1 is the main downstream effector of the canonical WNT pathway. It forms a complex with AXIN/APC/GSK3B in the absence of WNT ligands that results in CTNNB1 phosphorylation and subsequent degradation by the ubiquitin-proteasome system. However, WNT-FZD interaction prevents CTNNB1 degradation, and its nuclear accumulation causes the activation of TCF/LEF transcription factors and regulation of WNT target genes (96, 97).
Figure 3.
MiR-96 is involved in tumor progression and invasion through regulation of the WNT signaling pathway
Hypoxia is a pivotal feature of the tumor microenvironment during tumor progression (98). It promotes adaptive mechanisms by activation of hypoxia-inducible transcription factors that up-regulate various genes associated with angiogenesis, metastasis, and immune evasion (99, 100). AMOTL2 is an anti-angiogenic factor that regulates the actin cytoskeleton. It also suppresses the WNT signaling pathway via prevention of CTNNB1 nuclear translocation. It has been reported that there were higher levels of serum exosomal UCA1 in PC patients compared with healthy controls that were significantly associated with poor prognosis. Hypoxic pancreatic tumor cell-derived exosomal UCA1 promoted angiogenesis through regulation of miR-96-5p/AMOTL2/ERK1/2 axis (101).
During the EMT process, epithelial cells lose their polarity to convert to invasive mesenchymal cells (102). Astrocyte elevated gene-1 (AEG-1) is the activator of EMT-related signaling pathways such as SHH, TGF-b, and WNT (103-105). PI3K/AKT induction by AEG-1 can result in chemoresistance by MDR1 subsequent up-regulation (106). AEG-1 also increases the levels of LEF-1 expression as one of the critical WNT transcription factors. Moreover, AEG-1 promotes CTNNB1 nuclear translocation. Since, CTNNB1 phosphorylation by GSK3β results in its proteasomal degradation, AEG-1 can also induce CTNNB1 nuclear accumulation via GSK3β phosphorylation and inactivation (107). MiR-96 inhibited EMT and cell proliferation while inducing apoptosis via AEG-1 targeting in GBM cells (108). Neuronal pentraxin 2 (NPTX2) has a pivotal role in synapse formation and immune response (109). It also induces CRC cell proliferation and metastasis by a direct interaction with FZD6 that results in activation of the WNT signaling pathway (110). There was miR‐96 down-regulation in RCC samples. MiR‐96 reduced RCC cell invasion and proliferation via NPTX2 down-regulation (111).
PTEN is a protein phosphatase that suppresses the PI3K/AKT pathway by dephosphorylation of phosphoinositides. A significant STXBP5-AS1 up-regulation was observed in CC patients that was associated with poor prognosis. STXBP5-AS1 also inhibited CC cell proliferation by miR-96-5p sponging that resulted in PTEN up-regulation. Therefore, STXBP5-AS1 suppressed PI3K/AKT pathway via PTEN up-regulation in CC cells (112). There was miR-96-5p up-regulation in head and neck squamous cell carcinoma (HNSCC) specimens in comparison with normal margins. There were significantly higher levels of miR-96-5p expressions in HNSCC patients with TP53 mutations compared with wild types. MiR-96-5p up-regulation in mutant p53 carriers induced cell migration and chemo-resistance via PTEN targeting and activation of PI3K-AKT pathway in HNSCC (113). Succinate dehydrogenase (SDH) is a complex comprising SDHA-D components that is involved in the citric acid cycle of mitochondria by oxidation of succinate to fumarate (114, 115). It has also an important role in electron transport (116). Deregulation of SDHB has been associated with reduced oxidative phosphorylation and tumor progression in which SDHB deficiency results in increased cell migration and invasion (117). SDHB is also involved in regulation of the AKT/mTOR signaling pathway by inhibition of AKT. There was miR-96-3p up-regulation in PTC tissues in comparison with benign tissues. The advanced stage PTC patients had higher levels of miR-96-3p in tumor tissues compared with normal margins. MiR96-3p induced PTC cell invasion and migration through regulation of the SDHB/AKT/mTOR axis (118).
Tumor necrosis factor-alpha (TNF-α) is a cytokine involved in apoptosis and inflammation. TNF-α activates the TNF receptors which subsequently triggers various signal transduction pathways including MAPK, ERK, and JNK (119). SAMD9 as an effector of TNF-α signaling is involved in inflammatory responses. It has been shown that miR-96 suppressed cisplatin-induced apoptosis through SAMD9 in NSCLC cells (120). KRAS is a GTPase belonging to the RAS oncogene family that is involved in cell growth and differentiation (121). It induces pancreatic tumorigenesis by activation of PI3K/AKT, ERK, and NF-κB signaling pathways (122-125). It has been reported that miR-96 targeted KRAS oncogene in pancreatic tumor cells. MiR-96 inhibited AKT signaling and cell migration while inducing apoptosis via KRAS down-regulation (126).
Phosphatases, kinases, and matrix metalloproteinases
Protein tyrosine phosphatases (PTPs) are considered pivotal regulators of different cellular processes and signal transductions. PTPN9 belongs to the classic PTPs involved in different cellular processes (127). It is involved in dephosphorylation and suppression of EGFR, ErbB2, and STAT3 (128, 129). Since, these factors have important functions during BC progression (130, 131); PTPN9 can be suggested as a tumor suppressor via inhibition of ErbB and STAT3 signaling pathways. A significant miR-96 up-regulation was shown in BC tissues. MiR-96 promoted breast tumor in vivo growth, proliferation, and motility by PTPN9 sponging (132). There was miR-96 up-regulation in CC tissues that was inversely related to the levels of PTPN9 expression. MiR-96 increased CC cellular proliferation via PTNP9 targeting (133).
NUAK1 belongs to the AMPK-related kinase (ARK) family that is activated by AKT. It regulates cell survival during glucose deprivation and also inhibits apoptosis mediated by nutrient starvation (134). It regulates death receptors via CASP8 and pro-CASP6 suppressions (135, 136). MiR-96 repressed PC cell proliferation and migration through NUAK1 targeting (137). Chronic myeloid leukemia (CML) is a neoplastic transformation associated with (9; 22) translocation that generates a constitutively active BCR-ABL1 tyrosine kinase (138). ABL1 is a proto-oncogene involved in cell proliferation, migration, and stress response. MiR-96 suppressed the BCR-ABL1 oncogene in CML (139). EGFR is another tyrosine kinase receptor that is activated by EGF that triggers several signal transduction pathways such as MAPK and AKT. Therefore, EGFR is a pivotal regulator of cell growth and migration. Aberrant EGFR signaling is commonly observed during tumor progression (140). ETV6 has a tumor suppressor function by TWIST1 down-regulation in metastatic PCa (141). It has been reported that there was a converse association between the levels of ETV6 and miR-96 expressions in PCa tissues in which nuclear EGFR was correlated with ETV6 down-regulation. Nuclear EGFR increased the levels of miR-96 expressions that resulted in ETV6 targeting in PCa cells (11). EphrinA5 is a ligand for Eph receptors of tyrosine kinases that have important roles in regulation of angiogenesis and cell motility. It is an inhibitor of EGFR via inducing c-CBL binding and ubiquitylation (142). A significant miR-96 up-regulation was shown in HCC tissues in comparison with normal controls. MiR-96 increased HCC cell invasion and proliferation through ephrinA5 targeting (143).
MMPs are calcium-dependent proteases that have important roles in tissue remodeling, angiogenesis, and tumor cell metastasis. RECK is an MMP inhibitor involved in tumor metastasis by regulation of MMP-2 and MMP-9 (144, 145). Reduced levels of RECK expression are related to poor survival in different tumors (146). MiR-96 up-regulations were reported in esophageal cancer (EC) tissues and cell lines which was associated with clinical tumor stage and depth of invasion. MiR-96 reduced drug or irradiation responses in EC cells by RECK targeting (147). Significant miR-96 up-regulation was also shown in BC tissues in comparison with corresponding normal tissues. It also increased BC cell motility and proliferation by RECK inhibition (148). MiR-96 induced NSCLC cell growth and migration via RECK targeting. There was also miR-96 up-regulation in NSCLC tissues in comparison with normal tissues. Moreover, suppression of miR-96 reduced NSCLC cell proliferation (149).
Apoptosis and cell cycle regulation
Apoptosis and autophagy are the main cellular processes involved in regulation of the cell fate by protein and organelle turnovers. CASP9 is an initiator caspase required for the intrinsic apoptosis pathway. Due to the intracellular apoptotic stimuli, cytochrome c is released by mitochondria that form apoptosome by binding with Apaf-1. Then CASP9 will be activated by apoptosome that can subsequently activate executioner caspases to trigger apoptosis. A significant miR-96-5p up-regulation was observed in HCC tissues in comparison with normal samples. MiR-96-5p reduced the levels of FOXO1 expression in HCC cells. It increased doxorubicin resistance while inhibiting apoptosis through CASP9 targeting in HCC tumor cells (150). Programmed Cell Death 4 (PDCD4) is a tumor suppressor that activates BAX pro-apoptotic factors followed by cytochrome C mediated apoptosis (151). There was a significant miR-96 up-regulation in GBM cells. MiR-96 reduced GBM radiosensitivity through PDCD4 targeting (152). Autophagy is responsible for the maintenance of cellular homeostasis by elimination of dysfunctional cellular components using a lysosomal-mediated pathway. Therefore, this process can be associated with various biological processes and diseases (153, 154). Normally autophagy is an anti-apoptotic mechanism that maintains cell survival. Therefore, pro-apoptotic proteins should be inhibitors of autophagy. ATG7 belongs to the E1-like enzymes that suppress CASP9 translocation to the apoptosome resulting in apoptosis blocking (155, 156). It has been observed that UCA1 increased AML cell proliferation and autophagy by miR-96-5p sponging that resulted in ATG7 up-regulation (157). CDKN1A is a pivotal regulator of cell cycle progression that suppresses Cyclin/CDK2 complexes to mediate growth arrest due to DNA damages (158). There was miR-96 up-regulation in BCa tissues. MiR-96 reduced BCa cell proliferation while inducing apoptosis through CDKN1A down-regulation (159).
Conclusion
In the present review, we discussed all of the reports that have assessed the role of miR-96 in tumor initiation, progression, and invasion. This review clarifies the molecular mechanisms that are recruited by miR-96 to regulate tumor progression and metastasis. MiR-96 mainly exerts its role during tumorigenesis through targeting the structural proteins and FOXO transcription factors. Indeed, this review suggests miR-96 as an efficient diagnostic tumor marker in different cancers. Based on miR-96 as an oncogene or tumor suppressor, miR-96 or its inhibitors can be also suggested as novel therapeutic agents in cancer therapy. However, it is still required to perform animal studies in future research to find the efficiency of miR-96 targeted tumor therapy. More studies are also required to assess the miR-96 delivery method and long-term safety prior to medical practice as a novel therapeutic modality in cancer patients.
Authors’ Contributions
MMoghbeli study conception and design; HRR and MMojarrad draft manuscript preparation; MMoghbeli critical revision of the paper; MMoghbeli supervision of the research; HRR, MMojarrad, and MMoghbeli Final approval of the version to be published.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Acknowledgment
The authors are thankful to Mashhad University of Medical Sciences (MUMS), Mashhad, Iran. No funding to declare.
References
- 1.Chen J, Zhang K, Xu Y, Gao Y, Li C, Wang R, et al. The role of microRNA-26a in human cancer progression and clinical application. Tumour Biol. 2016;37:7095–7108. doi: 10.1007/s13277-016-5017-y. [DOI] [PubMed] [Google Scholar]
- 2.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng G. Circulating miRNAs: Roles in cancer diagnosis, prognosis and therapy. Adv Drug Deliv Rev. 2015;81:75–93. doi: 10.1016/j.addr.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA. MicroRNAome genome: A treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64:311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuk K, Zucknick M, Heil J, Madhavan D, Schott S, Turchinovich A, et al. Circulating microRNAs in plasma as early detection markers for breast cancer. Int J Cancer. 2013;132:1602–1612. doi: 10.1002/ijc.27799. [DOI] [PubMed] [Google Scholar]
- 6.Gorur A, Balci Fidanci S, Dogruer Unal N, Ayaz L, Akbayir S, Yildirim Yaroglu H, et al. Determination of plasma microRNA for early detection of gastric cancer. Mol Biol Rep. 2013;40:2091–2096. doi: 10.1007/s11033-012-2267-7. [DOI] [PubMed] [Google Scholar]
- 7.Guo Y, Liu H, Zhang H, Shang C, Song Y. miR-96 regulates FOXO1-mediated cell apoptosis in bladder cancer. Oncol Lett. 2012;4:561–565. doi: 10.3892/ol.2012.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, Wang Y. miR-96 targets SOX6 and promotes proliferation, migration, and invasion of hepatocellular carcinoma. Biochem Cell Biol. 2018;96:365–371. doi: 10.1139/bcb-2017-0183. [DOI] [PubMed] [Google Scholar]
- 9.Li XL, Hara T, Choi Y, Subramanian M, Francis P, Bilke S, et al. A p21-ZEB1 complex inhibits epithelial-mesenchymal transition through the microRNA 183-96-182 cluster. Mol Cell Biol. 2014;34:533–550. doi: 10.1128/MCB.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang X, Zheng D, Hu P, Zeng Z, Li M, Tucker L, et al. Glycogen synthase kinase 3 beta inhibits microRNA-183-96-182 cluster via the beta-Catenin/TCF/LEF-1 pathway in gastric cancer cells. Nucleic Acids Res. 2014;42:2988–2998. doi: 10.1093/nar/gkt1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai YC, Chen WY, Siu MK, Tsai HY, Yin JJ, Huang J, et al. Epidermal growth factor receptor signaling promotes metastatic prostate cancer through microRNA-96-mediated downregulation of the tumor suppressor ETV6. Cancer Lett. 2017;384:1–8. doi: 10.1016/j.canlet.2016.10.014. [DOI] [PubMed] [Google Scholar]
- 12.Hunter KW. Ezrin, a key component in tumor metastasis. Trends Mol Med. 2004;10:201–204. doi: 10.1016/j.molmed.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Yao Q, Pei Y, Zhang X, Xie B. microRNA-96 acts as a tumor suppressor gene in human osteosarcoma via target regulation of EZRIN. Life Sci. 2018;203:1–11. doi: 10.1016/j.lfs.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 14.Yu N, Fu S, Liu Y, Xu Z, Liu Y, Hao J, et al. miR-96 suppresses renal cell carcinoma invasion via downregulation of Ezrin expression. J Exp Clin Cancer Res. 2015;34 doi: 10.1186/s13046-015-0224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka-Okamoto M, Hori K, Ishizaki H, Hosoi A, Itoh Y, Wei M, et al. Increased susceptibility to spontaneous lung cancer in mice lacking LIM-domain only 7. Cancer Sci. 2009;100:608–616. doi: 10.1111/j.1349-7006.2009.01091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang S, Xu H, Duan X, Liu JJ, He Z, Yu F, et al. PCD1, a novel gene containing PDZ and LIM domains, is overexpressed in several human cancers. Cancer Res. 2000;60:5296–5302. [PubMed] [Google Scholar]
- 17.Wu H, Zhou J, Mei S, Wu D, Mu Z, Chen B, et al. Circulating exosomal microRNA-96 promotes cell proliferation, migration and drug resistance by targeting LMO7. J Cell Mol Med. 2017;21:1228–1236. doi: 10.1111/jcmm.13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao XH, Zhang YL, Zhang ZY, Guo SS, Chen XB, Guo YZ. MicroRNA-96-5p represses breast cancer proliferation and invasion through Wnt/beta-catenin signaling via targeting CTNND1. Sci Rep. 2020;10 doi: 10.1038/s41598-019-56571-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie F, Ye L, Ta M, Zhang L, Jiang WG. MTSS1: a multifunctional protein and its role in cancer invasion and metastasis. Front Biosci (Schol Ed) 2011;3:621–631. doi: 10.2741/s175. [DOI] [PubMed] [Google Scholar]
- 20.Huang XY, Huang ZL, Xu B, Chen Z, Re TJ, Zheng Q, et al. Elevated MTSS1 expression associated with metastasis and poor prognosis of residual hepatitis B-related hepatocellular carcinoma. J Exp Clin Cancer Res. 2016;35 doi: 10.1186/s13046-016-0361-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li XD, Zhang JX, Jiang LJ, Wang FW, Liu LL, Liao YJ, et al. Overexpression of maelstrom promotes bladder urothelial carcinoma cell aggressiveness by epigenetically downregulating MTSS1 through DNMT3B. Oncogene. 2016;35:6281–6292. doi: 10.1038/onc.2016.165. [DOI] [PubMed] [Google Scholar]
- 22.Yin X, Chai Z, Sun X, Chen J, Wu X, Yang L, et al. Overexpression of microRNA-96 is associated with poor prognosis and promotes proliferation, migration and invasion in cholangiocarcinoma cells via MTSS1. Exp Ther Med. 2020;19:2757–2765. doi: 10.3892/etm.2020.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie W, Sun F, Chen L, Cao X. miR-96 promotes breast cancer metastasis by suppressing MTSS1. Oncol Lett. 2018;15:3464–3471. doi: 10.3892/ol.2018.7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu L, Zhong J, Guo B, Zhu Q, Liang H, Wen N, et al. miR-96 promotes the growth of prostate carcinoma cells by suppressing MTSS1. Tumour Biol. 2016;37:12023–12032. doi: 10.1007/s13277-016-5058-2. [DOI] [PubMed] [Google Scholar]
- 25.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 26.Gordon FE, Nutt CL, Cheunsuchon P, Nakayama Y, Provencher KA, Rice KA, et al. Increased expression of angiogenic genes in the brains of mouse meg3-null embryos. Endocrinology. 2010;151:2443–2452. doi: 10.1210/en.2009-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kruer TL, Dougherty SM, Reynolds L, Long E, de Silva T, Lockwood WW, et al. Expression of the lncRNA maternally expressed gene 3 (MEG3) contributes to the control of lung cancer cell proliferation by the Rb pathway. PLoS One. 2016;11:e0166363. doi: 10.1371/journal.pone.0166363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang S, Guo W. Long noncoding RNA MEG3 suppresses the growth of glioma cells by regulating the miR965p/MTSS1 signaling pathway. Mol Med Rep. 2019;20:4215–4225. doi: 10.3892/mmr.2019.10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, et al. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461. doi: 10.1186/1471-2407-13-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 31.Qi L, Bart J, Tan LP, Platteel I, Sluis T, Huitema S, et al. Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer. 2009;9:163. doi: 10.1186/1471-2407-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nijak A, Alaerts M, Kuiperi C, Corveleyn A, Suys B, Paelinck B, et al. Left ventricular non-compaction with Ebstein anomaly attributed to a TPM1 mutation. Eur J Med Genet. 2018;61:8–10. doi: 10.1016/j.ejmg.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 33.Liu G, Zhao X, Zhou J, Cheng X, Ye Z, Ji Z. Long non-coding RNA MEG3 suppresses the development of bladder urothelial carcinoma by regulating miR-96 and TPM1. Cancer Biol Ther. 2018;19:1039–1056. doi: 10.1080/15384047.2018.1480279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ge T, Xiang P, Mao H, Tang S, Zhou J, Zhang Y. Inhibition of miR-96 enhances the sensitivity of colorectal cancer cells to oxaliplatin by targeting TPM1. Exp Ther Med. 2020;20:2134–2140. doi: 10.3892/etm.2020.8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S, Qiu Y, Bai B. The expression, regulation, and biomarker potential of glypican-1 in cancer. Front Oncol. 2019;9:614. doi: 10.3389/fonc.2019.00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C, Du X, Tai S, Zhong X, Wang Z, Hu Z, et al. GPC1 regulated by miR-96-5p, rather than miR-182-5p, in inhibition of pancreatic carcinoma cell proliferation. Int J Mol Sci. 2014;15:6314–6327. doi: 10.3390/ijms15046314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, et al. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 38.Cai Z, Grobe K, Zhang X. Role of heparan sulfate proteoglycans in optic disc and stalk morphogenesis. Dev Dyn. 2014;243:1310–1316. doi: 10.1002/dvdy.24142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fei X, Zhang J, Zhao Y, Sun M, Zhao H, Li S. miR-96 promotes invasion and metastasis by targeting GPC3 in non-small cell lung cancer cells. Oncol Lett. 2018;15:9081–9086. doi: 10.3892/ol.2018.8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chidlow JH Jr, Sessa WC. Caveolae, caveolins, and cavins: complex control of cellular signalling and inflammation. Cardiovasc Res. 2010;86:219–225. doi: 10.1093/cvr/cvq075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen AW, Hnasko R, Schubert W, Lisanti MP. Role of caveolae and caveolins in health and disease. Physiol Rev. 2004;84:1341–1379. doi: 10.1152/physrev.00046.2003. [DOI] [PubMed] [Google Scholar]
- 42.Ayala G, Morello M, Frolov A, You S, Li R, Rosati F, et al. Loss of caveolin-1 in prostate cancer stroma correlates with reduced relapse-free survival and is functionally relevant to tumour progression. J Pathol. 2013;231:77–87. doi: 10.1002/path.4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kannan A, Krishnan A, Ali M, Subramaniam S, Halagowder D, Sivasithamparam ND. Caveolin-1 promotes gastric cancer progression by up-regulating epithelial to mesenchymal transition by crosstalk of signalling mechanisms under hypoxic condition. Eur J Cancer. 2014;50:204–215. doi: 10.1016/j.ejca.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 44.Liu B, Zhang J, Yang D. miR-96-5p promotes the proliferation and migration of ovarian cancer cells by suppressing Caveolae1. J Ovarian Res. 2019;12:57. doi: 10.1186/s13048-019-0533-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fraser SP, Diss JK, Chioni AM, Mycielska ME, Pan H, Yamaci RF, et al. Voltage-gated sodium channel expression and potentiation of human breast cancer metastasis. Clin Cancer Res. 2005;11:5381–5389. doi: 10.1158/1078-0432.CCR-05-0327. [DOI] [PubMed] [Google Scholar]
- 46.Kunzelmann K. Ion channels and cancer. J Membr Biol. 2005;205:159–173. doi: 10.1007/s00232-005-0781-4. [DOI] [PubMed] [Google Scholar]
- 47.Afrasiabi E, Hietamaki M, Viitanen T, Sukumaran P, Bergelin N, Tornquist K. Expression and significance of HERG (KCNH2) potassium channels in the regulation of MDA-MB-435S melanoma cell proliferation and migration. Cell Signal. 2010;22:57–64. doi: 10.1016/j.cellsig.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 48.Shao XD, Wu KC, Guo XZ, Xie MJ, Zhang J, Fan DM. Expression and significance of HERG protein in gastric cancer. Cancer Biol Ther. 2008;7:45–50. doi: 10.4161/cbt.7.1.5126. [DOI] [PubMed] [Google Scholar]
- 49.Lastraioli E, Guasti L, Crociani O, Polvani S, Hofmann G, Witchel H, et al. herg1 gene and HERG1 protein are overexpressed in colorectal cancers and regulate cell invasion of tumor cells. Cancer Res. 2004;64:606–611. doi: 10.1158/0008-5472.can-03-2360. [DOI] [PubMed] [Google Scholar]
- 50.Pillozzi S, Brizzi MF, Bernabei PA, Bartolozzi B, Caporale R, Basile V, et al. VEGFR-1 (FLT-1), beta1 integrin, and hERG K+ channel for a macromolecular signaling complex in acute myeloid leukemia: role in cell migration and clinical outcome. Blood. 2007;110:1238–1250. doi: 10.1182/blood-2006-02-003772. [DOI] [PubMed] [Google Scholar]
- 51.Feng J, Yu J, Pan X, Li Z, Chen Z, Zhang W, et al. HERG1 functions as an oncogene in pancreatic cancer and is downregulated by miR-96. Oncotarget. 2014;5:5832–5844. doi: 10.18632/oncotarget.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kikkert M, Doolman R, Dai M, Avner R, Hassink G, van Voorden S, et al. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J Biol Chem. 2004;279:3525–3534. doi: 10.1074/jbc.M307453200. [DOI] [PubMed] [Google Scholar]
- 53.Gao Z, Wang H, Li H, Li M, Wang J, Zhang W, et al. Long non-coding RNA CASC2 inhibits breast cancer cell growth and metastasis through the regulation of the miR-96-5p/SYVN1 pathway. Int J Oncol. 2018;53:2081–2090. doi: 10.3892/ijo.2018.4522. [DOI] [PubMed] [Google Scholar]
- 54.Gao J, Huo L, Sun X, Liu M, Li D, Dong JT, et al. The tumor suppressor CYLD regulates microtubule dynamics and plays a role in cell migration. J Biol Chem. 2008;283:8802–8809. doi: 10.1074/jbc.M708470200. [DOI] [PubMed] [Google Scholar]
- 55.Gao J, Sun L, Huo L, Liu M, Li D, Zhou J. CYLD regulates angiogenesis by mediating vascular endothelial cell migration. Blood. 2010;115:4130–4137. doi: 10.1182/blood-2009-10-248526. [DOI] [PubMed] [Google Scholar]
- 56.Wickstrom SA, Masoumi KC, Khochbin S, Fassler R, Massoumi R. CYLD negatively regulates cell-cycle progression by inactivating HDAC6 and increasing the levels of acetylated tubulin. EMBO J. 2010;29:131–144. doi: 10.1038/emboj.2009.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao M, Xin XF, Zhang JY, Dai W, Lv TF, Song Y. LncRNA GMDS-AS1 inhibits lung adenocarcinoma development by regulating miR-96-5p/CYLD signaling. Cancer Med. 2020;9:1196–1208. doi: 10.1002/cam4.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei S, Zheng Y, Jiang Y, Li X, Geng J, Shen Y, et al. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine. 2019;44:182–193. doi: 10.1016/j.ebiom.2019.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu ZM, Wu ZY, Li WH, Wang LQ, Wan JN, Zhong Y. MiR-96-5p promotes the proliferation, invasion and metastasis of papillary thyroid carcinoma through down-regulating CCDC67. Eur Rev Med Pharmacol Sci. 2019;23:3421–3430. doi: 10.26355/eurrev_201904_17706. [DOI] [PubMed] [Google Scholar]
- 60.Kaufmann E, Knochel W. Five years on the wings of fork head. Mech Dev. 1996;57:3–20. doi: 10.1016/0925-4773(96)00539-4. [DOI] [PubMed] [Google Scholar]
- 61.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 62.Kousteni S. FoxO1: a molecule for all seasons. J Bone Miner Res. 2011;26:912–917. doi: 10.1002/jbmr.306. [DOI] [PubMed] [Google Scholar]
- 63.Kops GJ, Medema RH, Glassford J, Essers MA, Dijkers PF, Coffer PJ, et al. Control of cell cycle exit and entry by protein kinase B-regulated forkhead transcription factors. Mol Cell Biol. 2002;22:2025–2036. doi: 10.1128/MCB.22.7.2025-2036.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidt M, Fernandez de Mattos S, van der Horst A, Klompmaker R, Kops GJ, Lam EW, et al. Cell cycle inhibition by FoxO forkhead transcription factors involves downregulation of cyclin D. Mol Cell Biol. 2002;22:7842–7852. doi: 10.1128/MCB.22.22.7842-7852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dijkers PF, Medema RH, Lammers JW, Koenderman L, Coffer PJ. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr Biol. 2000;10:1201–1204. doi: 10.1016/s0960-9822(00)00728-4. [DOI] [PubMed] [Google Scholar]
- 66.Modur V, Nagarajan R, Evers BM, Milbrandt J. FOXO proteins regulate tumor necrosis factor-related apoptosis inducing ligand expression Implications for PTEN mutation in prostate cancer. J Biol Chem. 2002;277:47928–47937. doi: 10.1074/jbc.M207509200. [DOI] [PubMed] [Google Scholar]
- 67.Brunet A, Kanai F, Stehn J, Xu J, Sarbassova D, Frangioni JV, et al. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J Cell Biol. 2002;156:817–828. doi: 10.1083/jcb.200112059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim SJ, Winter K, Nian C, Tsuneoka M, Koda Y, McIntosh CH. Glucose-dependent insulinotropic polypeptide (GIP) stimulation of pancreatic beta-cell survival is dependent upon phosphatidylinositol 3-kinase (PI3K)/protein kinase B (PKB) signaling, inactivation of the forkhead transcription factor Foxo1, and down-regulation of bax expression. J Biol Chem. 2005;280:22297–22307. doi: 10.1074/jbc.M500540200. [DOI] [PubMed] [Google Scholar]
- 69.Machida S, Spangenburg EE, Booth FW. Forkhead transcription factor FoxO1 transduces insulin-like growth factor’s signal to p27Kip1 in primary skeletal muscle satellite cells. J Cell Physiol. 2003;196:523–531. doi: 10.1002/jcp.10339. [DOI] [PubMed] [Google Scholar]
- 70.Hou T, Ou J, Zhao X, Huang X, Huang Y, Zhang Y. MicroRNA-196a promotes cervical cancer proliferation through the regulation of FOXO1 and p27Kip1. Br J Cancer. 2014;110:1260–1268. doi: 10.1038/bjc.2013.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zaballos MA, Santisteban P. FOXO1 controls thyroid cell proliferation in response to TSH and IGF-I and is involved in thyroid tumorigenesis. Mol Endocrinol. 2013;27:50–62. doi: 10.1210/me.2012-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song HM, Luo Y, Li DF, Wei CK, Hua KY, Song JL, et al. MicroRNA-96 plays an oncogenic role by targeting FOXO1 and regulating AKT/FOXO1/Bim pathway in papillary thyroid carcinoma cells. Int J Clin Exp Pathol. 2015;8:9889–9900. [PMC free article] [PubMed] [Google Scholar]
- 73.Yang N, Zhou J, Li Q, Han F, Yu Z. miR-96 exerts carcinogenic effect by activating AKT/GSK-3beta/beta-catenin signaling pathway through targeting inhibition of FOXO1 in hepatocellular carcinoma. Cancer Cell Int. 2019;19 doi: 10.1186/s12935-019-0756-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fendler A, Jung M, Stephan C, Erbersdobler A, Jung K, Yousef GM. The antiapoptotic function of miR-96 in prostate cancer by inhibition of FOXO1. PLoS One. 2013;8:e80807. doi: 10.1371/journal.pone.0080807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang L, Liu L, Zhang X, Zhu Y, Li L, Wang B, et al. miR-96 enhances the proliferation of cervical cancer cells by targeting FOXO1. Pathol Res Pract. 2020;216:152854. doi: 10.1016/j.prp.2020.152854. [DOI] [PubMed] [Google Scholar]
- 76.Lang C, Xu M, Zhao Z, Chen J, Zhang L. MicroRNA-96 expression induced by low-dose cisplatin or doxorubicin regulates chemosensitivity, cell death and proliferation in gastric cancer SGC7901 cells by targeting FOXO1. Oncol Lett. 2018;16:4020–4026. doi: 10.3892/ol.2018.9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu JJ, Wu YX, Zhao FJ, Xia SJ. miR-96 promotes cell proliferation and clonogenicity by down-regulating of FOXO1 in prostate cancer cells. Med Oncol. 2014;31:910. doi: 10.1007/s12032-014-0910-y. [DOI] [PubMed] [Google Scholar]
- 78.Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 79.Lin H, Dai T, Xiong H, Zhao X, Chen X, Yu C, et al. Unregulated miR-96 induces cell proliferation in human breast cancer by downregulating transcriptional factor FOXO3a. PLoS One. 2010;5:e15797. doi: 10.1371/journal.pone.0015797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou Y, Chen Y, Ding W, Hua Z, Wang L, Zhu Y, et al. LncRNA UCA1 impacts cell proliferation, invasion, and migration of pancreatic cancer through regulating miR-96/FOXO3. IUBMB Life. 2018;70:276–290. doi: 10.1002/iub.1699. [DOI] [PubMed] [Google Scholar]
- 81.Li J, Li P, Chen T, Gao G, Chen X, Du Y, et al. Expression of microRNA-96 and its potential functions by targeting FOXO3 in non-small cell lung cancer. Tumour Biol. 2015;36:685–692. doi: 10.1007/s13277-014-2698-y. [DOI] [PubMed] [Google Scholar]
- 82.Yin Z, Wang W, Qu G, Wang L, Wang X, Pan Q. MiRNA-96-5p impacts the progression of breast cancer through targeting FOXO3. Thorac Cancer. 2020;11:956–963. doi: 10.1111/1759-7714.13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao F, Wang W. MicroRNA-96 promotes the proliferation of colorectal cancer cells and targets tumor protein p53 inducible nuclear protein 1, forkhead box protein O1 (FOXO1) and FOXO3a. Mol Med Rep. 2015;11:1200–1206. doi: 10.3892/mmr.2014.2854. [DOI] [PubMed] [Google Scholar]
- 84.Cai J, Tian AX, Wang QS, Kong PZ, Du X, Li XQ, et al. FOXF2 suppresses the FOXC2-mediated epithelial-mesenchymal transition and multidrug resistance of basal-like breast cancer. Cancer Lett. 2015;367:129–137. doi: 10.1016/j.canlet.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 85.Higashimori A, Dong Y, Zhang Y, Kang W, Nakatsu G, Ng SSM, et al. Forkhead box F2 suppresses gastric cancer through a novel FOXF2-IRF2BPL-beta-catenin signaling axis. Cancer Res. 2018;78:1643–1656. doi: 10.1158/0008-5472.CAN-17-2403. [DOI] [PubMed] [Google Scholar]
- 86.Zhang Y, Wang X, Wang Z, Tang H, Fan H, Guo Q. miR-182 promotes cell growth and invasion by targeting forkhead box F2 transcription factor in colorectal cancer. Oncol Rep. 2015;33:2592–2598. doi: 10.3892/or.2015.3833. [DOI] [PubMed] [Google Scholar]
- 87.Simpson-Haidaris PJ, Rybarczyk B. Tumors and fibrinogen The role of fibrinogen as an extracellular matrix protein. Ann N Y Acad Sci. 2001;936:406–425. [PubMed] [Google Scholar]
- 88.Vihinen P, Kahari VM. Matrix metalloproteinases in cancer: prognostic markers and therapeutic targets. Int J Cancer. 2002;99:157–166. doi: 10.1002/ijc.10329. [DOI] [PubMed] [Google Scholar]
- 89.Wang H, Ma N, Li W, Wang Z. MicroRNA-96-5p promotes proliferation, invasion and EMT of oral carcinoma cells by directly targeting FOXF2. Biol Open. 2020:9. doi: 10.1242/bio.049478. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 90.Wei WR, Zeng GJ, Liu C, Zou BW, Li L. Overexpression of miR-96 promotes cell proliferation by targeting FOXF2 in prostate cancer. Int J Clin Exp Pathol. 2017;10:7596–7602. [PMC free article] [PubMed] [Google Scholar]
- 91.Abbaszadegan MR, Riahi A, Forghanifard MM, Moghbeli M. WNT and NOTCH signaling pathways as activators for epidermal growth factor receptor in esophageal squamous cell carcinoma. Cell Mol Biol Lett. 2018;23 doi: 10.1186/s11658-018-0109-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moghbeli M, Abbaszadegan MR, Golmakani E, Forghanifard MM. Correlation of Wnt and NOTCH pathways in esophageal squamous cell carcinoma. J Cell Commun Signal. 2016;10:129–135. doi: 10.1007/s12079-016-0320-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yan Z, Wang J, Wang C, Jiao Y, Qi W, Che S. miR-96/HBP1/Wnt/beta-catenin regulatory circuitry promotes glioma growth. FEBS Lett. 2014;588:3038–3046. doi: 10.1016/j.febslet.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 94.Bafico A, Gazit A, Pramila T, Finch PW, Yaniv A, Aaronson SA. Interaction of frizzled related protein (FRP) with Wnt ligands and the frizzled receptor suggests alternative mechanisms for FRP inhibition of Wnt signaling. J Biol Chem. 1999;274:16180–16187. doi: 10.1074/jbc.274.23.16180. [DOI] [PubMed] [Google Scholar]
- 95.Zhang H, Chen R, Shao J. MicroRNA-96-5p facilitates the viability, migration, and invasion and suppresses the apoptosis of cervical cancer cells bynegatively modulating SFRP4. Technol Cancer Res Treat. 2020;19:1533033820934132. doi: 10.1177/1533033820934132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moghbeli M, Rad A, Farshchian M, Taghehchian N, Gholamin M, Abbaszadegan MR. Correlation between meis1 and msi1 in esophageal squamous cell carcinoma. J Gastrointest Cancer. 2016;47:273–277. doi: 10.1007/s12029-016-9824-6. [DOI] [PubMed] [Google Scholar]
- 97.Moghbeli M, Sadrizadeh A, Forghanifard MM, Mozaffari HM, Golmakani E, Abbaszadegan MR. Role of Msi1 and PYGO2 in esophageal squamous cell carcinoma depth of invasion. J Cell Commun Signal. 2016;10:49–53. doi: 10.1007/s12079-015-0314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rankin EB, Giaccia AJ. Hypoxic control of metastasis. Science. 2016;352:175–180. doi: 10.1126/science.aaf4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lu X, Kang Y. Hypoxia and hypoxia-inducible factors: master regulators of metastasis. Clin Cancer Res. 2010;16:5928–5935. doi: 10.1158/1078-0432.CCR-10-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shih JW, Chiang WF, Wu ATH, Wu MH, Wang LY, Yu YL, et al. Long noncoding RNA LncHIFCAR/MIR31HG is a HIF-1alpha co-activator driving oral cancer progression. Nat Commun. 2017;8:15874. doi: 10.1038/ncomms15874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guo Z, Wang X, Yang Y, Chen W, Zhang K, Teng B, et al. Hypoxic tumor-derived exosomal long noncoding RNA UCA1 promotes angiogenesis via miR-96-5p/AMOTL2 in pancreatic cancer. Mol Ther Nucleic Acids. 2020;22:179–195. doi: 10.1016/j.omtn.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 103.Karhadkar SS, Bova GS, Abdallah N, Dhara S, Gardner D, Maitra A, et al. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. 2004;431:707–712. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
- 104.Shin SY, Rath O, Zebisch A, Choo SM, Kolch W, Cho KH. Functional roles of multiple feedback loops in extracellular signal-regulated kinase and Wnt signaling pathways that regulate epithelial-mesenchymal transition. Cancer Res. 2010;70:6715–6724. doi: 10.1158/0008-5472.CAN-10-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Valcourt U, Kowanetz M, Niimi H, Heldin CH, Moustakas A. TGF-beta and the Smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol Biol Cell. 2005;16:1987–2002. doi: 10.1091/mbc.E04-08-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yoo BK, Chen D, Su ZZ, Gredler R, Yoo J, Shah K, et al. Molecular mechanism of chemoresistance by astrocyte elevated gene-1. Cancer Res. 2010;70:3249–3258. doi: 10.1158/0008-5472.CAN-09-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Robertson CL, Srivastava J, Rajasekaran D, Gredler R, Akiel MA, Jariwala N, et al. The role of AEG-1 in the development of liver cancer. Hepat Oncol. 2015;2:303–312. doi: 10.2217/hep.15.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Feng S, Yao J, Zhang Z, Zhang Y, Zhang Z, Liu J, et al. miR96 inhibits EMT by targeting AEG1 in glioblastoma cancer cells. Mol Med Rep. 2018;17:2964–2972. doi: 10.3892/mmr.2017.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chang S, Bok P, Tsai CY, Sun CP, Liu H, Deussing JM, et al. NPTX2 is a key component in the regulation of anxiety. Neuropsychopharmacology. 2018;43:1943–1953. doi: 10.1038/s41386-018-0091-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xu C, Tian G, Jiang C, Xue H, Kuerbanjiang M, Sun L, et al. NPTX2 promotes colorectal cancer growth and liver metastasis by the activation of the canonical Wnt/beta-catenin pathway via FZD6. Cell Death Dis. 2019;10:217. doi: 10.1038/s41419-019-1467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xiang W, Han L, Mo G, Lin L, Yu X, Chen S, et al. MicroRNA-96 is a potential tumor repressor by inhibiting NPTX2 in renal cell carcinoma. J Cell Biochem. 2020;121:1504–1513. doi: 10.1002/jcb.29385. [DOI] [PubMed] [Google Scholar]
- 112.Shao S, Wang C, Wang S, Zhang H, Zhang Y. LncRNA STXBP5-AS1 suppressed cervical cancer progression via targeting miR-96-5p/PTEN axis. Biomed Pharmacother. 2019;117:109082. doi: 10.1016/j.biopha.2019.109082. [DOI] [PubMed] [Google Scholar]
- 113.Vahabi M, Pulito C, Sacconi A, Donzelli S, D’Andrea M, Manciocco V, et al. miR-96-5p targets PTEN expression affecting radio-chemosensitivity of HNSCC cells. J Exp Clin Cancer Res. 2019;38:141. doi: 10.1186/s13046-019-1119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Iwashita H, Okudela K, Matsumura M, Yamanaka S, Sawazumi T, Enaka M, et al. Succinate dehydrogenase B-deficient renal cell carcinoma: A case report with novel germline mutation. Pathol Int. 2017;67:585–589. doi: 10.1111/pin.12587. [DOI] [PubMed] [Google Scholar]
- 115.Miettinen M, Lasota J. Succinate dehydrogenase deficient gastrointestinal stromal tumors (GISTs) - a review. Int J Biochem Cell Biol. 2014;53:514–519. doi: 10.1016/j.biocel.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fuhrmann DC, Wittig I, Brune B. TMEM126B deficiency reduces mitochondrial SDH oxidation by LPS, attenuating HIF-1alpha stabilization and IL-1beta expression. Redox Biol. 2019;20:204–216. doi: 10.1016/j.redox.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Loriot C, Domingues M, Berger A, Menara M, Ruel M, Morin A, et al. Deciphering the molecular basis of invasiveness in Sdhb-deficient cells. Oncotarget. 2015;6:32955–32965. doi: 10.18632/oncotarget.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhao X, Li Y, Zhou Y. MicroRNA-96-3p promotes metastasis of papillary thyroid cancer through targeting SDHB. Cancer Cell Int. 2019;19:287. doi: 10.1186/s12935-019-1003-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.van Horssen R, Ten Hagen TL, Eggermont AM. TNF-alpha in cancer treatment: molecular insights, antitumor effects, and clinical utility. Oncologist. 2006;11:397–408. doi: 10.1634/theoncologist.11-4-397. [DOI] [PubMed] [Google Scholar]
- 120.Wu L, Pu X, Wang Q, Cao J, Xu F, Xu LI, et al. miR-96 induces cisplatin chemoresistance in non-small cell lung cancer cells by downregulating SAMD9. Oncol Lett. 2016;11:945–952. doi: 10.3892/ol.2015.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hall A. The cellular functions of small GTP-binding proteins. Science. 1990;249:635–640. doi: 10.1126/science.2116664. [DOI] [PubMed] [Google Scholar]
- 122.Campbell PM, Groehler AL, Lee KM, Ouellette MM, Khazak V, Der CJ. K-Ras promotes growth transformation and invasion of immortalized human pancreatic cells by Raf and phosphatidylinositol 3-kinase signaling. Cancer Res. 2007;67:2098–2106. doi: 10.1158/0008-5472.CAN-06-3752. [DOI] [PubMed] [Google Scholar]
- 123.Kallifatidis G, Rausch V, Baumann B, Apel A, Beckermann BM, Groth A, et al. Sulforaphane targets pancreatic tumour-initiating cells by NF-kappaB-induced antiapoptotic signalling. Gut. 2009;58:949–963. doi: 10.1136/gut.2008.149039. [DOI] [PubMed] [Google Scholar]
- 124.McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta. 2007;1773:1263–1284. doi: 10.1016/j.bbamcr.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 126.Yu S, Lu Z, Liu C, Meng Y, Ma Y, Zhao W, et al. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer Res. 2010;70:6015–6025. doi: 10.1158/0008-5472.CAN-09-4531. [DOI] [PubMed] [Google Scholar]
- 127.He RJ, Yu ZH, Zhang RY, Zhang ZY. Protein tyrosine phosphatases as potential therapeutic targets. Acta Pharmacol Sin. 2014;35:1227–1246. doi: 10.1038/aps.2014.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Su F, Ren F, Rong Y, Wang Y, Geng Y, Wang Y, et al. Protein tyrosine phosphatase Meg2 dephosphorylates signal transducer and activator of transcription 3 and suppresses tumor growth in breast cancer. Breast Cancer Res. 2012;14:R38. doi: 10.1186/bcr3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yuan T, Wang Y, Zhao ZJ, Gu H. Protein-tyrosine phosphatase PTPN9 negatively regulates ErbB2 and epidermal growth factor receptor signaling in breast cancer cells. J Biol Chem. 2010;285:14861–14870. doi: 10.1074/jbc.M109.099879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gritsko T, Williams A, Turkson J, Kaneko S, Bowman T, Huang M, et al. Persistent activation of stat3 signaling induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin Cancer Res. 2006;12:11–19. doi: 10.1158/1078-0432.CCR-04-1752. [DOI] [PubMed] [Google Scholar]
- 131.Turkson J, Jove R. STAT proteins: novel molecular targets for cancer drug discovery. Oncogene. 2000;19:6613–6626. doi: 10.1038/sj.onc.1204086. [DOI] [PubMed] [Google Scholar]
- 132.Hong Y, Liang H, Uzair Ur R, Wang Y, Zhang W, Zhou Y, et al. miR-96 promotes cell proliferation, migration and invasion by targeting PTPN9 in breast cancer. Sci Rep. 2016;6:37421. doi: 10.1038/srep37421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ma X, Shi W, Peng L, Qin X, Hui Y. MiR-96 enhances cellular proliferation and tumorigenicity of human cervical carcinoma cells through PTPN9. Saudi J Biol Sci. 2018;25:863–867. doi: 10.1016/j.sjbs.2017.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Suzuki A, Kusakai G, Kishimoto A, Lu J, Ogura T, Lavin MF, et al. Identification of a novel protein kinase mediating Akt survival signaling to the ATM protein. J Biol Chem. 2003;278:48–53. doi: 10.1074/jbc.M206025200. [DOI] [PubMed] [Google Scholar]
- 135.Suzuki A, Kusakai G, Kishimoto A, Lu J, Ogura T, Esumi H. ARK5 suppresses the cell death induced by nutrient starvation and death receptors via inhibition of caspase 8 activation, but not by chemotherapeutic agents or UV irradiation. Oncogene. 2003;22:6177–6182. doi: 10.1038/sj.onc.1206899. [DOI] [PubMed] [Google Scholar]
- 136.Suzuki A, Kusakai G, Kishimoto A, Shimojo Y, Miyamoto S, Ogura T, et al. Regulation of caspase-6 and FLIP by the AMPK family member ARK5. Oncogene. 2004;23:7067–7075. doi: 10.1038/sj.onc.1207963. [DOI] [PubMed] [Google Scholar]
- 137.Huang X, Lv W, Zhang JH, Lu DL. miR96 functions as a tumor suppressor gene by targeting NUAK1 in pancreatic cancer. Int J Mol Med. 2014;34:1599–1605. doi: 10.3892/ijmm.2014.1940. [DOI] [PubMed] [Google Scholar]
- 138.Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5:172–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 139.Huang T, Fu Y, Wang S, Xu M, Yin X, Zhou M, et al. miR-96 acts as a tumor suppressor via targeting the BCR-ABL1 oncogene in chronic myeloid leukemia blastic transformation. Biomed Pharmacother. 2019;119:109413. doi: 10.1016/j.biopha.2019.109413. [DOI] [PubMed] [Google Scholar]
- 140.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 141.Tsai YC, Zeng T, Abou-Kheir W, Yeh HL, Yin JJ, Lee YC, et al. Disruption of ETV6 leads to TWIST1-dependent progression and resistance to epidermal growth factor receptor tyrosine kinase inhibitors in prostate cancer. Mol Cancer. 2018;17:42. doi: 10.1186/s12943-018-0785-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li JJ, Liu DP, Liu GT, Xie D. EphrinA5 acts as a tumor suppressor in glioma by negative regulation of epidermal growth factor receptor. Oncogene. 2009;28:1759–1768. doi: 10.1038/onc.2009.15. [DOI] [PubMed] [Google Scholar]
- 143.Wang TH, Yeh CT, Ho JY, Ng KF, Chen TC. OncomiR miR-96 and miR-182 promote cell proliferation and invasion through targeting ephrinA5 in hepatocellular carcinoma. Mol Carcinog. 2016;55:366–375. doi: 10.1002/mc.22286. [DOI] [PubMed] [Google Scholar]
- 144.Chiang CH, Hou MF, Hung WC. Up-regulation of miR-182 by beta-catenin in breast cancer increases tumorigenicity and invasiveness by targeting the matrix metalloproteinase inhibitor RECK. Biochim Biophys Acta. 2013;1830:3067–3076. doi: 10.1016/j.bbagen.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 145.Hirata H, Ueno K, Shahryari V, Tanaka Y, Tabatabai ZL, Hinoda Y, et al. Oncogenic miRNA-182-5p targets Smad4 and RECK in human bladder cancer. PLoS One. 2012;7:e51056. doi: 10.1371/journal.pone.0051056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhao H, Wang Y, Yang L, Jiang R, Li W. MiR-25 promotes gastric cancer cells growth and motility by targeting RECK. Mol Cell Biochem. 2014;385:207–213. doi: 10.1007/s11010-013-1829-x. [DOI] [PubMed] [Google Scholar]
- 147.Xia H, Chen S, Chen K, Huang H, Ma H. MiR-96 promotes proliferation and chemo- or radioresistance by down-regulating RECK in esophageal cancer. Biomed Pharmacother. 2014;68:951–958. doi: 10.1016/j.biopha.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 148.Zhang J, Kong X, Li J, Luo Q, Li X, Shen L, et al. miR-96 promotes tumor proliferation and invasion by targeting RECK in breast cancer. Oncol Rep. 2014;31:1357–1363. doi: 10.3892/or.2013.2934. [DOI] [PubMed] [Google Scholar]
- 149.Guo H, Li Q, Li W, Zheng T, Zhao S, Liu Z. MiR-96 downregulates RECK to promote growth and motility of non-small cell lung cancer cells. Mol Cell Biochem. 2014;390:155–160. doi: 10.1007/s11010-014-1966-x. [DOI] [PubMed] [Google Scholar]
- 150.Iwai N, Yasui K, Tomie A, Gen Y, Terasaki K, Kitaichi T, et al. Oncogenic miR-96-5p inhibits apoptosis by targeting the caspase-9 gene in hepatocellular carcinoma. Int J Oncol. 2018;53:237–245. doi: 10.3892/ijo.2018.4369. [DOI] [PubMed] [Google Scholar]
- 151.Matsuhashi S, Manirujjaman M, Hamajima H, Ozaki I. Control Mechanisms of the Tumor Suppressor PDCD4: Expression and Functions. Int J Mol Sci. 2019:20. doi: 10.3390/ijms20092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Guo P, Yu Y, Tian Z, Lin Y, Qiu Y, Yao W, et al. Upregulation of miR-96 promotes radioresistance in glioblastoma cells via targeting PDCD4. Int J Oncol. 2018;53:1591–1600. doi: 10.3892/ijo.2018.4498. [DOI] [PubMed] [Google Scholar]
- 153.Colhado Rodrigues BL, Lallo MA, Perez EC. The controversial role of autophagy in tumor development: A systematic review. Immunol Invest. 2020;49:386–396. doi: 10.1080/08820139.2019.1682600. [DOI] [PubMed] [Google Scholar]
- 154.Guo F, Liu X, Cai H, Le W. Autophagy in neurodegenerative diseases: pathogenesis and therapy. Brain Pathol. 2018;28:3–13. doi: 10.1111/bpa.12545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Han J, Hou W, Goldstein LA, Stolz DB, Watkins SC, Rabinowich H. A Complex between Atg7 and Caspase-9: A Novel mechanism of cross-regulation between autophagy and apoptosis. J Biol Chem. 2014;289:6485–6497. doi: 10.1074/jbc.M113.536854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zheng W, Xie W, Yin D, Luo R, Liu M, Guo F. ATG5 and ATG7 induced autophagy interplays with UPR via PERK signaling. Cell Commun Signal. 2019;17:42. doi: 10.1186/s12964-019-0353-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Li JJ, Chen XF, Wang M, Zhang PP, Zhang F, Zhang JJ. Long non-coding RNA UCA1 promotes autophagy by targeting miR-96-5p in acute myeloid leukaemia. Clin Exp Pharmacol Physiol. 2020;47:877–885. doi: 10.1111/1440-1681.13259. [DOI] [PubMed] [Google Scholar]
- 158.Bianco S, Jangal M, Garneau D, Gevry N. LRH-1 controls proliferation in breast tumor cells by regulating CDKN1A gene expression. Oncogene. 2015;34:4509–4518. doi: 10.1038/onc.2014.382. [DOI] [PubMed] [Google Scholar]
- 159.Wu Z, Liu K, Wang Y, Xu Z, Meng J, Gu S. Upregulation of microRNA-96 and its oncogenic functions by targeting CDKN1A in bladder cancer. Cancer Cell Int. 2015;15:107. doi: 10.1186/s12935-015-0235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]