Abstract
A State of the Art lecture entitled “Molecular Analysis of Vascular Gene Expression” was presented at the ISTH Congress in 2021. Endothelial cells (ECs) form a critical interface between the blood and underlying tissue environment, serving as a reactive barrier to maintain tissue homeostasis. ECs play an important role in not only coagulation, but also in the response to inflammation by connecting these two processes in the host defense against pathogens. Furthermore, ECs tailor their behavior to the needs of the microenvironment in which they reside, resulting in a broad display of EC phenotypes. While this heterogeneity has been acknowledged for decades, the contributing molecular mechanisms have only recently started to emerge due to technological advances. These include high‐throughput sequencing combined with methods to isolate ECs directly from their native tissue environment, as well as sequencing samples at a high cellular resolution. In addition, the newest technologies simultaneously quantitate and visualize a multitude of RNA transcripts directly in tissue sections, thus providing spatial information. Understanding how ECs function in (patho)physiological conditions is crucial to develop new therapeutics as many diseases can directly affect the endothelium. Of particular relevance for thrombotic disorders, EC dysfunction can lead to a procoagulant, proinflammatory phenotype with increased vascular permeability that can result in coagulopathy and tissue damage, as seen in a number of infectious diseases, including sepsis and coronavirus disease 2019. In light of the current pandemic, we will summarize relevant new data on the latter topic presented during the 2021 ISTH Congress.
Keywords: coagulation, endothelial cells, gene expression, high‐throughput sequencing, inflammation
Essentials.
Endothelial cells (ECs) play an important role in hemostasis and immunothrombosis.
ECs are highly heterogeneous with phenotypes tailored to the microenvironment in which they reside.
New technologies allow molecular analysis of ECs within their natural environment.
Understanding how ECs respond in (patho)physiology can help identify therapeutic targets.
1. INTRODUCTION
The vascular system extends to almost all tissues in the body, thereby providing oxygen and nutrients while removing waste products, via an intricate network of arteries and veins that are connected by capillaries. 1 , 2 , 3 Endothelial cells (ECs) form the inner lining of these vessels, where they not only provide a physical barrier between the blood and underlying cells but also act as a reactive interface to maintain tissue homeostasis. In addition to controlling vascular permeability and the extravasation of fluids and solutes, ECs regulate hemostasis and inflammation, as well as vasomotor tone, cell adhesion, and angiogenesis, and have been shown to participate in both innate and adaptive immune responses. 4 , 5 They also fulfill unique roles depending on their anatomic location along the vascular tree and the specific needs of the underlying tissue. To achieve all these functions, ECs display a remarkable heterogeneity. 1 , 2 , 3 While long recognized, the molecular mechanisms underlying this heterogeneity have only started to emerge in recent years, 2 , 3 , 6 , 7 as the EC’s microenvironmental interdependence, and interspersed and sparse distribution initially precluded direct in vivo analysis. As the integrity of the endothelium is essential to maintain tissue homeostasis, understanding how ECs function under physiological and pathological conditions is crucial in developing new therapeutics for many diseases, including thrombotic disorders.
In this review, we will first discuss ECs as an integral part of the blood coagulation system, and as an important mediator of the intricate connections between coagulation and inflammation, a topic that has received considerable attention due to the ongoing coronavirus disease 2019 (COVID‐19) pandemic. Next, we will provide an overview of methods to perform (endothelial) cell type‐specific molecular analyses. Here, we will not only highlight current approaches focused on transcriptomics, such as single‐cell RNA sequencing, but also discuss emerging techniques providing spatial information and combining different ‐omics approaches to further understand the complexity of biological systems such as the vasculature. Finally, we will present an update from the XXIX Congress of the ISTH, with a specific emphasis on the role of endothelial dysfunction in COVID‐19.
2. ENDOTHELIUM IN HEMOSTASIS
The endothelium plays a key role in hemostasis by maintaining blood in a fluid state under normal conditions, while facilitating rapid and localized thrombus formation at the site of vessel injury. 8 , 9 ECs achieve this by tightly regulating platelet adhesion and activation and controlling procoagulant, anticoagulant, and fibrinolytic processes (Figure 1).
FIGURE 1.
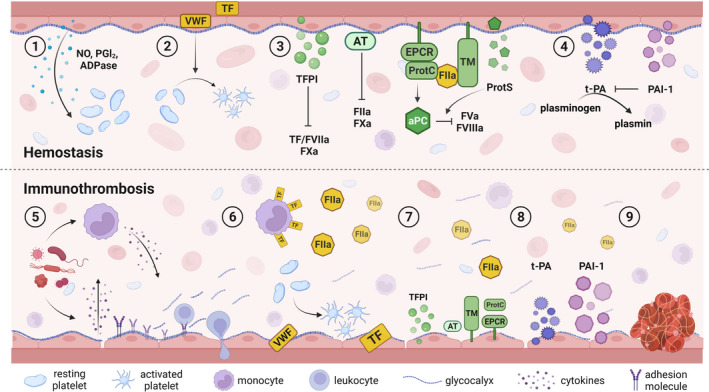
Endothelial contributions to hemostasis and immunothrombosis. ECs play a role in hemostasis via expression of (1) factors that prevent platelet aggregation, (2) procoagulant VWF, which recruits and activates platelets upon vessel injury, (3) several anticoagulant factors that limit thrombin formation and (4) pro‐ and antifibrinolytic factors that are important for thrombus resolution. (5) Pathogen exposure activates immune cells and ECs, which stimulates cytokine release that induce expression of adhesion molecules leading to leukocyte recruitment and extravasation, shedding of the glycocalyx, and vascular leakage. Cytokines also (6) upregulate tissue factor expression on immune cells and activation of ECs leading to thrombin generation, while at the same time causing (7) a decrease in production of anticoagulant factors and (8) a shift toward inhibition of fibrinolysis, thereby resulting in (9) thrombus formation containing the pathogen. ADPase, adenophosphatase; aPC, activated protein C; AT, antithrombin; ECs, endothelial cells; EPCR, endothelial protein C receptor; FIIa, activated factor II; FVa, activated factor V; FVIIa, activated factor VII; FVIIIa, activated factor VIII; FXa, activated factor X; NO, nitric oxide; PAI‐1, plasminogen activator inhibitor 1; PGI2, prostacyclin; TF, tissue factor; TFPI, tissue factor pathway inhibitor; VWF, von Willebrand factor
Healthy endothelium is covered by the glycocalyx, which contains glycosaminoglycans known for their anticoagulant properties. 10 Furthermore, ECs produce nitric oxide and prostacyclin, and express ecto‐adenophosphatase on their cell surface that keep platelets in a resting state. 11 However, platelet recruitment and thrombus formation upon vessel injury is essential to limit blood loss and tissue damage. This process starts with perivascular tissue factor (TF) exposure that binds and activates factor VII (FVII) to initiate the coagulation cascade. In addition, ultra‐large von Willebrand factor (VWF) is released from the endothelium and forms a bridge between platelets and extracellular matrix components. This latter interaction mediates shear stress–dependent platelet activation and aggregation, which further promotes coagulation and provides a surface for thrombus formation.
To prevent pathological thrombosis, coagulation is tightly regulated by 3 main anticoagulant systems aimed at limiting thrombin generation, which are driven by tissue factor pathway inhibitor (TFPI), antithrombin (AT), and the protein C pathway. 12 TFPI is a factor X–dependent inactivator of TF/activated FVII (FVIIa), and ECs express both the secreted TFPIα and the membrane‐anchored TFPIβ isoform. 13 While AT is synthesized by hepatocytes and is present in plasma, its function is strongly enhanced upon interaction with heparan sulfates present in the glycocalyx covering the endothelium. 14 In addition to its main targets, thrombin (activated factor II [FIIa]) and activated factor X, AT can also inactivate other serine proteases within the coagulation cascade. Activation of the protein C pathway is initiated by binding of thrombin to thrombomodulin (TM), which has several consequences: not only does this binding reduce the amount of FIIa in the circulation, thus limiting further fibrin formation, it also activates thrombin‐activatable fibrinolysis inhibitor, which stabilizes fibrin clots. 15 Finally, by binding to TM, thrombin is converted into a protein with anticoagulant properties via protein C activation, a process augmented in the presence of the endothelial protein C receptor (EPCR). Activated protein C, together with EC‐derived protein S, can subsequently suppress coagulation via proteolysis of activated factor V and activated factor VIII.
As the vessel is repaired, clot resolution occurs through fibrinolysis via tissue‐type plasminogen activator (t‐PA) that is expressed by ECs and converts plasminogen into plasmin, leading to subsequent fibrin degradation. This process is inhibited by plasminogen activator inhibitor 1 (PAI‐1), which is produced by several cell types, including ECs. 16
3. ENDOTHELIUM IN IMMUNOTHROMBOSIS
Given its unique location between the blood and underlying parenchyma, ECs are often referred to as “gatekeepers” that maintain normal tissue homeostasis. 2 , 3 , 17 In addition to their role in coagulation, the endothelium has immunological functions, and both systems work together in host defense responses. 5 , 18 , 19 , 20 This is particularly evident in immunothrombosis, where inflammation triggers coagulation as part of the host’s reaction to pathogen invasion to contain and eliminate the threat, thereby preventing its dissemination throughout the vasculature and limiting tissue damage (Figure 1). 18 , 21
Recognition and binding of pathogens by traditional immune cells and endothelial cells triggers their activation and release of proinflammatory cytokines and chemokines. 4 , 5 Well known for their roles in regulating the immune response and inflammation, these mediators can further affect ECs in several ways. 22 , 23 First, they lead to upregulated expression of cell adhesion molecules necessary for leukocyte recruitment and extravasation into the underlying tissue. Cytokines also induce degradation of the glycocalyx, which not only affects its anticoagulant properties 10 but also contributes to increased vascular permeability, which is further enhanced by breakdown of junction proteins that interconnect the endothelium. In addition, cytokines stimulate the release of microvesicles, trigger the formation of neutrophil extracellular traps and activate the complement system. 19 , 21 All these components can directly influence both coagulation and inflammation, highlighting the extensive crosstalk between these two processes.
Cytokines can alter levels of coagulation and fibrinolytic factors, thereby shifting the endothelium from an anticoagulant to a procoagulant state. 18 , 19 , 21 Central in this process is the cytokine‐induced upregulation of TF on immune cells, which together with the release of VWF from activated ECs stimulates coagulation by increasing platelet activation and thrombin generation. Not only are platelets increasingly recognized as active participants in the immune response, 11 thrombin can also directly promote inflammation via cleavage of protease‐activated receptors (PARs), particularly PAR1 present on ECs. 24 Additionally, cytokines downregulate the expression of anticoagulant factors, many of which also have anti‐inflammatory properties and are involved in maintaining the integrity of the endothelial barrier. 13 , 25 , 26 Therefore, this downregulation not only shifts the coagulation balance further toward a procoagulant state but simultaneously enhances inflammation. Finally, cytokines cause a decrease in t‐PA while increasing PAI‐1 levels, thus suppressing clot resolution.
Together, the interaction between inflammation and coagulation leads to robust thrombus formation to contain and remove the pathogen from the circulation. However, it is important to note that as with hemostasis, immunothrombosis requires careful regulation, as an exaggerated procoagulant response without sufficient suppression by natural inhibitors can lead to thromboinflammation. 18 , 21 The latter can result in significant organ injury, potentially leading to organ failure and even death, as can be observed in severe sepsis and COVID‐19. 22 , 23 , 27 , 28
4. EC HETEROGENEITY
ECs are highly heterogeneous across organs, but also along different segments of the vascular tree. 1 , 2 , 3 As a result, ECs have distinct responses to stimuli depending on the vascular bed in which they reside. With respect to thrombosis, alterations in circulating levels of pro‐ or anticoagulant factor may result in site‐specific manifestations, for example, as observed in thrombotic microangiopathies such as thrombotic thrombocytopenic purpura (TTP). 29 However, while linked to abnormal VWF homeostasis, TTP does not lead to a general disseminated thrombotic phenotype, but instead predominantly affects the kidney and central nervous system. The idea of a tissue‐specific response is further reinforced by observations in mice deficient in (anti)coagulant factors, as they also display lesions associated with distinct segments of the vascular tree. 30
It has long been speculated that vascular site‐specific thrombosis phenotypes may be explained by unique expression patterns of EC‐derived coagulation factors. 31 However, demonstrating EC heterogeneity on a molecular level has been challenging since ECs not only form an integral part of a tissue, but they typically represent a small percentage of the total number of cells in an organ. 32 Therefore, early studies tried to assess gene expression by isolating ECs and evaluating them in vitro. While these studies have yielded important information, we and others have shown that extracting ECs from their native microenvironment and expanding them in culture induces phenotypic drift. 32 , 33 , 34 This drift leads to changes in expression profiles and the loss of tissue‐specific traits, thus not forming a true representative for their in vivo counterparts. Recognizing the importance of the natural EC environment, past approaches used histology‐based methods such as immunohistochemistry or in situ hybridization assays to determine expression patterns, showing distinct patterns for TFPI, EPCR, and t‐PA expression, whereas TM is ubiquitously expressed. 35 , 36 , 37 , 38 The distribution of VWF across the vascular tree has been the focus of several research groups, demonstrating by histology as well as the generation of transgenic mouse models, that VWF is more abundant in ECs of large vessels as compared to the microvasculature. 39 , 40 , 41 , 42 Furthermore, VWF is predominantly expressed in the venous rather than the arterial system. Although these latter methods provide information on the spatial distribution of proteins and RNA, they are limited by the ability to study only one or a few genes at the same time.
The introduction of high‐throughput approaches, particularly transcriptomic analysis, has vastly improved our understanding of cellular and molecular biology over the past two decades. 43 However, microarray or bulk RNA sequencing (RNASeq) data from intact tissues provides an average representation of transcript levels originating from all cell types present, and does not permit deconvolution of cell‐specific expression profiles. As ECs typically form only a minor fraction of the total cell content, variations in expression profiles are likely masked by differences in more abundant cell types. To overcome these limitations, several methods can be used to enrich for ECs prior to high‐throughput analyses, via either nongenetic or genetic methods requiring transgenic animals.
5. (ENDOTHELIAL) CELL‐SPECIFIC ENRICHMENT FOR MOLECULAR ANALYSIS
5.1. Nongenetic approaches
An important advantage of methods not relying on genetic manipulations to characterize cells is that they can also be used in nonmodel species (Table 1). An example of such a method is laser microdissection (LMD), where cells of interest are identified based on morphological features and isolated directly from tissue sections. 44 Cells can then be processed for further downstream applications, including high‐throughput transcriptomic or proteomic analyses. 45 A limitation of this technique is that the identification of cells must be done manually to avoid contamination with neighboring cells. It is therefore very time consuming, and results are highly dependent on the expertise of the operator. Alternatively, immunostaining can be performed to help with this process, although these additional sample manipulations can affect RNA or protein integrity. 46 It is also important to note that even with these precautions, contamination can still occur, as cells below the dissection plane cannot be directly visualized. However, LMD allows for preservation of tissue architecture, which is highly valuable in evaluating spatial heterogeneity (Table 1), and it is therefore not surprising that LMD has been previously used to unmask molecular profiles of specific (micro)vascular segments. 45
TABLE 1.
Overview of molecular analysis technologies to profile cellular subsets
| LMD | FACS a | Vascular mapping | TRAP | INTACT | TU tagging | scRNASeq | FISSEQ / seqFISH | GeoMX | CosMX | MERFISH | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Spatial information | ✓ | X | X | X | X | X | X | ✓ | ✓ | ✓ | ✓ |
| Omics analysis | E, T, P | E, T, P | P | T | E | T | T | T | T, P | T, P | T |
| Purity isolated cells /Cell specificity | + | ++ | + | + | ++ | ++ | +++ | +++ | + | ++++ | ++++ |
| Specialized equipment required | ✓ | ✓ | X | X | X | X | ✓ | ✓ | ✓ | ✓ | ✓ |
| Nonmodel species compatible | ✓ | ✓ | ✓ | X | X | X | ✓ | ✓ | ✓ b | ✓ b | ✓ b |
| Tissue dissociation | None | Enzymatic | Mechanical | Mechanical | Mechanical | Mechanical | Enzymatic | None | None | None | None |
| Assay output | Variable: user dependent | Variable: user dependent | Protein | Translating mRNA | DNA | Nascent RNA | Total mRNA pool | Total RNA pool | Targeted mRNA subset | Targeted mRNA subset | Targeted mRNA subset |
Abbreviations: E, epigenome; FACS, fluorescence‐activated cell sorting; FISSEQ, fluorescence in situ hybridization sequencing; INTACT, isolation of nuclei tagged in specific cell types; LMD, laser microdissection; MERFISH, multiplexed error‐robust fluorescence in situ hybridization; P, proteome; scRNASeq, single‐cell RNA sequencing; seqFISH, sequential fluorescence in situ hybridization; T, transcriptome; TRAP, translating ribosome affinity purification; TU, thiouracil.
Cell labeling via exogenous antibody, transgenic or intravital labeling.
Provided that reference genome is available for probe design.
Another approach not relying on transgenic animals per se, is flow sorting by fluorescence‐activated cell sorting (FACS). Here, tissues are enzymatically dissociated to obtain a single cell suspension, followed by incubation with a fluorescently labeled antibody against a cell surface marker specific for the cell type of interest. While labeling of cells often occurs ex vivo, the unique position of the endothelium also allows for direct in vivo staining prior to tissue dissociation via intravital labeling, a method employed to show that tissue‐specific ECs establish specialized vascular niches. 47 An important caveat for this approach is that flow sorting requires the preparation of single cell suspensions. The resulting disruption of cell‐cell contacts, especially for ECs, may lead to stress‐induced transcriptional changes. 32 , 34 In addition, exposure to rapid flow may cause mechanical damage, further affecting gene expression or even result in cell death, thus introducing a potential selection bias. 43 Despite these limitations, FACS is widely used for selecting large numbers of cells for a specific population in an automated fashion, with minimal contamination (Table 1).
Many studies have used RNASeq analyses to evaluate the isolated cell types at a molecular level, as these provide genome‐wide coverage, great precision, and are accessible to many researchers. However, there is not always a direct correlation between mRNA and actual protein levels. 48 , 49 To directly assess protein levels in an unbiased manner, proteomic approaches can be applied. While these methods are often used to study intracellular protein levels, several groups have focused their attention on the luminal surface of the endothelium, which is in immediate contact with the blood. 50 , 51 , 52 Not surprisingly, these studies also identified (interactive regions of) proteins being present in organ‐specific patterns, and this knowledge could potentially be used to develop methods and therapeutics that target specific vascular beds.
5.2. Genetic approaches
As noted above, FACS is widely used to identify and isolate cell types of interest. However, its success is dependent on the availability of reliable, validated antibodies, which are still often lacking for (tissue‐specific) endothelial subsets. As an alternative, genetic means can be used to express fluorescent proteins on the surface of a specific cell type 53 (Figure 2). Although the in vivo labeling in these reporter animals has advantages, the downstream analyses are not free from the previously discussed limitations associated with FACS (Table 1).
FIGURE 2.
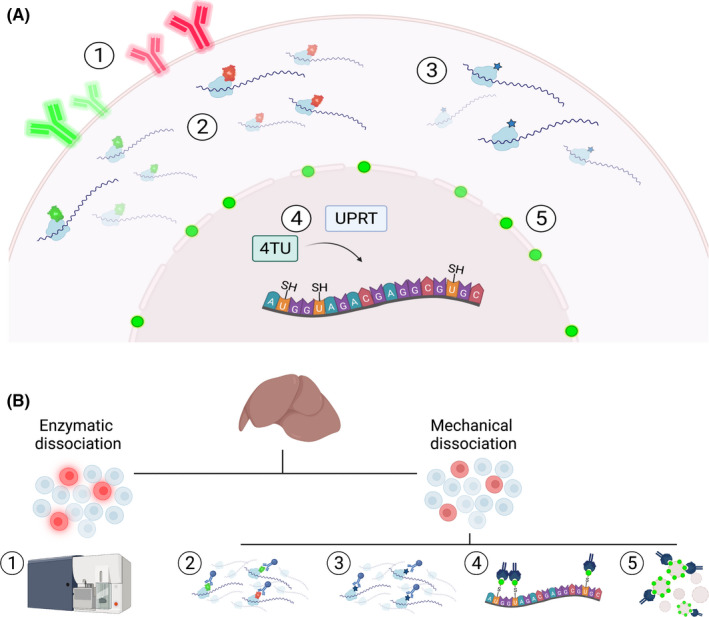
Genetic approaches allowing cell type‐specific enrichment for molecular analyses. (A) In vivo labeling of cells can be achieved by (1) expression of a fluorescent surface protein, (2) tagging of a ribosomal protein with a fluorescent or (3) hemagglutin epitope tag, (4) TU tagging, or (5) via expression of a biotinylated nuclear envelope protein. (B) Ex vivo sample processing of (1) fluorescently labeled cells includes enzymatic dissociation followed by FACS sorting to isolate cells of interest. (2, 3) Tagged ribosomal proteins are incorporated into polysomal complexes, thereby enabling the isolation of actively translating mRNA from mechanically dissociated tissues via TRAP. (4) Administration of TU leads to production of thioRNA in cells that express UPRT. Nascent thioRNA is subsequently conjugated with biotin and isolated via streptavidin immunoprecipitation. (5) Streptavidin immunoprecipitation is also used to select nuclei containing a biotinylated nuclear envelope protein to evaluate epigenetic landscapes in a cell‐specific manner. FACS, fluorescence‐activated cell sorting; TRAP, translating ribosome affinity purification; TU, thiouracil; UPRT, uracil phosphoribosyltransferase
To circumvent these problems, strategies have been developed to isolate cell‐specific RNA directly from the in vivo environment. One such approach is thiouracil (TU) tagging, which besides cell specificity also incorporates a temporal component, and has been used to identify intraorgan heterogeneity in nascent endothelial RNA levels (Figure 2). 54 An alternative approach to evaluate expression programs from distinct cell types is translating ribosome affinity purification (TRAP). Here, a ribosomal protein is labeled with either a fluorescent 55 , 56 , 57 or hemagglutinin epitope tag 32 , 58 , 59 in the cell population of interest (Figure 2). Incorporation of these tagged proteins into ribosomal complexes allows for immunoprecipitation of polysomes with their associated actively translating mRNA, which can be further processed for high‐throughput analyses. By combining in vivo perfusion using a translation inhibitor such as cycloheximide, with the mechanic disruption of frozen tissues, we have previously shown that TRAP provides an accurate snapshot of EC‐specific expression profiles, and identified a high degree of EC heterogeneity across organs in naïve mice as well as organ‐specific EC reactivity after lipopolysaccharide exposure. 32
The previously described genetic methods all focus on transcript levels, ranging from newly transcribed RNA (TU tagging) to actively translated RNA (TRAP models) (Table 1). The latter, also known as the translatome, has been demonstrated to correlate better to actual protein levels (proteome) as compared to the transcriptome, and might therefore be a better predictor for protein abundance. 48 , 49 However, it is also important to understand how regulatory programs control cellular and molecular heterogeneity, which can be done via epigenetic analyses. Although these can be performed on FACS‐sorted cells, 60 isolation of nuclei tagged in specific cell types (INTACT) provides a genetic model to label nuclear envelope proteins in vivo. 61 This method enables direct cell type–specific nucleus isolation via immunoprecipitation, which can then be used for downstream analyses (Figure 2). Recently, INTACT has been combined with TRAP, resulting in the nuTRAP (nuclear tagging and translating ribosome affinity purification) mouse. 62 , 63 Here, both the ribosomal and nuclear envelope protein are tagged, thus supporting the simultaneous analysis of genome‐wide transcript levels and chromatin features in distinct cell populations.
Many of the genetic models rely on Cre recombinase to target specific cell populations. Several different Cre models have been developed to target the endothelium, all of which are based on well‐known EC markers such as Tie2 (Tek), vascular endothelial (VE)‐cadherin (Cdh5) or the vascular endothelial growth factor (VEGF) receptor (KDR) (reviewed in Payne et al 64 ). However, it is important to note that these genes exhibit differences in expression (and therefore activity) patterns, which can have a dramatic impact on the results. Furthermore, as ECs and hematopoietic cells share a similar embryonic origin, many constitutively expressed EC‐Cre transgenes also target blood cells. 64 , 65 To avoid this problem, investigators have used tamoxifen‐inducible Cre models that allow temporal expression. However, this latter requires careful optimizing of timing and dose as tamoxifen can directly affect EC gene expression and function. 66 , 67 An alternative approach potentially offering greater precision in targeting EC subtypes is provided by (sequential) intersectional genetics, where the expression of Cre is determined by the presence of two unique cell type–specific genes rather than one. This can be achieved via the use of an intermediate recombinase system such as Dre‐rox, which has been previously used to specifically target brain and coronary ECs 68 or via the split‐Cre system. 69
6. MORE THAN MEETS THE EYE: SINGLE‐CELL RNASeq
Although the above methods to study ECs have yielded valuable insights into organ specificity, they do not permit resolution at the individual cell level. The introduction of single‐cell RNA sequencing (scRNASeq) has revolutionized the field of cellular and molecular biology, and has proven particularly useful for studying cell types that are phenotypically diverse such as the endothelium. 6 , 7 , 70 For example, one of the first studies using scRNAseq to further evaluate vascular heterogeneity in the brain identified the transcriptional basis for the gradual changes in phenotypes along the arteriovenous axis, 71 and more recently it was shown that the alveolar endothelium consists of two distinct EC subtypes with each expressing different pro‐ and anticoagulant factors. 72 Furthermore, scRNASeq is being used to gain insights into the cellular and molecular mechanisms contributing to COVID‐19 pathogenesis, demonstrating differences in (endothelial) cell composition and gene expression profiles in the lung, as well as in other organs. 73 , 74 , 75
Whereas these examples already indicate the power of scRNASeq approaches, several potential limitations should be noted in addition to the previously mentioned limitations observed with FACS‐based analyses. First, even though scRNASeq analysis can differentiate between ECs and non‐ECs in a tissue based on expression profiles, the cost of sample preparation is currently often a limiting factor. With ECs forming a minor cell fraction within a tissue, enrichment strategies via FACS can be considered to make it more cost effective. However, this can be especially difficult for ECs, since there is no “one size fits all” protocol for this enrichment, as the embedding of ECs in different microenvironments requires optimized dissociation conditions to preserve sample integrity and avoid biases due to under‐ or overdigestion of the tissue sample. 6 , 7 , 43 , 70 , 76 Second, the increase in cellular resolution comes at the expense of decreased sensitivity in transcript detection, with only limited detection of less abundant transcripts, although this is highly dependent on the sequencing platform used. 7 , 76 Finally, with the rapid expansion of scRNASeq data sets, standardization of cell isolation protocols, library preparation, and data analysis are crucial to reduce data variability as each step can introduce bias and/or technical noise. 7 , 43
While efforts by large consortia are using scRNASeq to profile and categorize every cell in the body, 77 , 78 , 79 previous studies focused on generating cell atlases of particular organs or (endothelial) cell types. 80 , 81 , 82 , 83 All these studies aim to generate a platform that can eventually be used to evaluate disease‐induced changes in a systematic way, thereby providing molecular insights and identifying potential targets for therapeutic interventions, as previously illustrated for COVID‐19. 73 , 74 , 75 As these projects are ongoing, current work is directed at compiling curated scRNASeq data sets to provide scientists the opportunity to explore data from published scRNASeq studies and perform their own analyses. Examples include PanglaoDB, 84 which contains human and mouse data from a broad range of cells, and EndoDB, a database specifically focusing on the murine endothelium. 85
7. SPATIAL TRANSCRIPTOMICS
scRNASeq provides a powerful tool for identifying cellular heterogeneity at the molecular level, and defining tissue‐specific phenotypes and functions of vascular cells. However, it does not provide spatial information on the position of these cells within tissues (Table 1). In situ hybridization (ISH) assays do provide this information, particularly single‐molecule fluorescence ISH (smFISH). The design of the gene‐specific fluorescent probes makes it possible to visualize single RNA molecules while preserving tissue morphology, and therefore it has been widely used to validate sequencing data and further investigate cell‐cell interactions. 43 However, smFISH has been limited by the ability to only evaluate a few transcripts at the same time due to the overlapping spectra of fluorescent probes.
Therefore, early studies combined scRNASeq with smFISH, where transcripts unique to cellular subsets are identified by scRNASeq and validated by smFISH thereby providing information on the position of these cells within the tissue. 71 Conversely, differential expression of transcripts known to be spatially restricted, so‐called zonated landmark genes, can be used to identify cell clusters in scRNASeq data. 86 An alternative method is spatial sorting, where scRNASeq data are used to specifically identify surface markers that are unique to distinct cellular subsets. 87 This has the advantage that instead of single cells, single‐cell (sub)populations can be isolated, allowing for high‐resolution bulk multi‐omics analyses, including high‐sensitivity transcriptomics and proteomics analysis.
Additional methods to measure RNA levels directly in tissue sections have been developed. 6 , 43 , 88 These are based on a combination of quantitative gene expression data and visualization of transcripts or proteins, thereby providing spatial information (Table 1). In general, these can be divided into targeted and untargeted approaches, with the latter including methods such as fluorescence in situ hybridization sequencing 89 and sequential fluorescence in situ hybridization. 90 , 91 Both of these are based on sequencing technology, where sequential rounds of hybridization of fluorescent nucleotides or probes, followed by signal imaging, creates a pattern that translates to an RNA molecule. Untargeted methods do not rely on gene‐specific probes and are thus unbiased, and in theory should be able to provide a genome‐wide analysis. However, with transcripts being densely packed in a cell, it has not (yet) been possible to resolve all individual RNA molecules within a single cell.
Targeted approaches, on the other hand, are based on specific target probes and therefore require prior knowledge of the (expected) expression profile. However, a major advantage is that there are several commercial platforms available for these probe‐based assays, which integrate the required high‐resolution microscopy with automated fluidics. For example, NanoString’s GeoMX Digital Spatial Profiler can detect RNA and protein within the same region of interest, 92 and has been recently used to generate a spatial atlas of lung samples from patients with COVID‐19. 73 , 74 The second‐generation CosMX Spatial Molecular Imager expands on this technique by providing (sub)single cell resolution. 93 Multiplexed error‐robust fluorescence in situ hybridization also provides direct detection of predefined RNA targets 94 and is commercially available in the Vizgen’s MERSCOPE platform. Since its unique barcoding method is able to detect and correct sequencing errors, this technique supports high sequencing precision and detection efficiency.
Many single‐cell studies are focused on transcriptional profiles, as unbiased proteomics on the single‐cell levels is not sensitive enough for mammalian cells. 95 Although currently limited to specific target panels, spatial ‐omics technologies have provided an avenue to visualize and correlate RNA levels and proteins in situ. The combination with approaches to identify epigenetic landscapes at single‐cell resolution such as Assay for Transposase‐Accessible Chromatin with high‐throughput sequencing (ATAC‐seq) to study open chromatin regions, will be vital for understanding cellular and molecular interactions in physiology and how these are affected in pathology.
8. ROLE OF ENDOTHELIAL CELLS IN COVID‐19: ISTH 2021 CONGRESS REPORT
Given the detrimental changes to the endothelium and coagulation‐related manifestations associated with severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infections, it is not surprising that a substantial number of abstracts presented at the XXIX Congress of the ISTH focused on the COVID‐19 patient population. Many of these studies involved clinical observations on plasma coagulation parameters, supporting previous data that COVID‐19 associated coagulopathy is different from coagulopathy associated with sepsis. 96 , 97 , 98 The latter can lead to disseminated intravascular coagulation, characterized by consumption of coagulation factors as evidenced by a prolonged activated partial thromboplastin time (aPTT) and prothrombin time (PT). Furthermore, sepsis patients show a typical thrombocytopenia and elevated D‐dimer levels. Patients with COVID‐19, on the other hand, are less likely to present with thrombocytopenia or changes in aPTT or PT levels. Instead, a stronger emphasis appears to be on circulating cytokines and immune cells, and because of the additional increase in VWF and factor VIII (FVIII) levels, an important role for EC dysfunction has been suggested. 27 , 28 Indeed, in addition to the hypercoagulable state, SARS‐CoV‐2 infection is associated with pathologic angiogenesis as an indicator of EC dysfunction. 99
While EC dysfunction has been acknowledged as part of COVID‐19–associated coagulopathy, especially in more severe cases, information is still lacking. To get a better understanding of EC dysfunction in relation to the increased thrombosis risk observed in patients with COVID‐19, several studies evaluated circulating fragments of EC surface markers as a proxy for endothelial stress. For example, Peralta and colleagues evaluated EC markers in plasma from over 150 patients with COVID‐19, ranging in disease severity. 100 They showed that patients who developed thrombosis or died had higher levels of not only cytokines, VWF, and FVIII, but also of t‐PA. Furthermore, SARS‐CoV‐2 infections resulted in increased levels of EC activation markers such as soluble intercellular adhesion molecule 1 (ICAM1), TEK tyrosine kinase (TIE2) and lymphatic vessel endothelial hyaluronan receptor 1 (LYVE1). 100 Additional studies identified greater abundance of soluble TM, soluble endothelial protein C receptor (sEPCR), and PAI‐1 as likely EC‐derived contributors to the hypercoagulable phenotype. 101 , 102 , 103 , 104 Although these latter studies often included smaller patient populations, some of these markers appear to have predictive value for (in‐hospital) mortality. 101 , 103 , 104 Others focused on identifying biomarkers that can be used to distinguish COVID‐19 from sepsis‐related coagulopathy. 101 , 102 Interestingly, SARS‐CoV‐2 infection seems to be associated with higher levels of soluble VCAM‐1 and sEPCR in blood, while PAI‐1, although higher than in controls, shows a less strong increase as compared to sepsis patients. 102
With changes in EC permeability playing an important role in immunothrombosis and thromboinflammation, Moraes et al 105 evaluated mediators of barrier disruption. They showed that markers such as angiopoietin 1 and 2, as well as their receptor TIE2, and VEGF‐A and VE‐cadherin were all elevated in patients with SARS‐CoV‐2 infection. Furthermore, levels of VEGF‐A were significantly associated with intensive care unit stay. These proteins are not only involved in barrier function, and increased levels may thus be a sign of barrier breakdown, but they also play a role in angiogenesis. 99 Therefore, these data could contribute to the understanding of the aberrant angiogenesis associated with severe COVID‐19. Another possible explanation for the altered angiogenesis came from a study that assessed endothelial colony‐forming cells (ECFCs) from recovered patients with COVID‐19, showing that patient‐derived ECFCs are immature with a reduced proliferative capacity and are not able to maintain or restore normal EC function. 106
While measuring circulating protein levels may be a useful indicator for the condition of the endothelium in general, it must be kept in mind that different vascular beds can exhibit distinct responses, as we and others have previously showed in systemic infections. 32 , 52 , 58 , 107 Even though this heterogeneity is difficult to recapitulate in vitro, studies using cultured cells have been crucial in providing mechanistic insights into the role of ECs in disease. The endothelium is being increasingly recognized as an integral part of hemostasis regulation, and ongoing efforts are being made to not only incorporate ECs into model systems for coagulopathy but also to generate more relevant in vitro models by using three‐dimensional systems that include extracellular matrix proteins relevant for the in vivo environment. 108 , 109
9. CONCLUSION AND FUTURE PERSPECTIVES
ECs play a key role in the coagulation system by producing both pro‐ and anticoagulant factors to ensure local hemostasis. As an integral component of the immune system, ECs also form an important mediator in the host defense response by connecting coagulation and inflammatory processes to contain and eliminate pathogens. This is a very delicate balance that can be easily disturbed and lead to coagulopathy as seen in sepsis and COVID‐19, a highly discussed topic at the ISTH 2021 Congress. Developing a better understanding of how ECs regulate these processes will not only provide insights into thrombosis and thromboinflammation, but could also help identify potential targets to prevent these pathological events.
Studying ECs is complicated by their extensive heterogeneity and high degree of dependence on the microenvironment in which they reside. Technological advances over the past decades have provided important information on the molecular mechanisms underlying this heterogeneity, both on an organ‐wide level and more recently at single‐cell resolution. The introduction of (commercially available) spatial transcriptomics platforms, as well as the growing possibilities to integrate multi ‐omics data sets will undoubtedly further contribute to this knowledge.
With ECs forming a large therapeutic target, capitalizing on established molecular differences in EC reactivity under (patho)physiologic conditions can pave the way for developing effective antithrombotic therapies without inducing adverse effects such as an increased bleeding risk, an important unmet clinical need.
RELATIONSHIP DISCLOSURE
The authors have no conflicts of interest to disclose.
AUTHOR CONTRIBUTIONS
ME, DS, and AC wrote, edited, and approved the final version of the manuscript.
ACKNOWLEDGMENTS
The authors thank Dr David Ginsburg for careful reading of the manuscript. DS and AC are supported by grants from the National Institutes of Health (T32HL007622 and K12HD028820 to DS; R03AG070541 to AC). Figures were created using BioRender.
Van der Ent MA, Svilar D, Cleuren ACA. Molecular analysis of vascular gene expression. Res Pract Thromb Haemost. 2022;6:e12718. doi: 10.1002/rth2.12718
Handling Editor: Dr Henri Spronk
REFERENCES
- 1. Aird WC. Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ Res. 2007;100:158‐173. [DOI] [PubMed] [Google Scholar]
- 2. Augustin HG, Koh GY. Organotypic vasculature: from descriptive heterogeneity to functional pathophysiology. Science. 2017;357(6353):eaal2379. [DOI] [PubMed] [Google Scholar]
- 3. Potente M, Makinen T. Vascular heterogeneity and specialization in development and disease. Nat Rev Mol Cell Biol. 2017;18:477‐494. [DOI] [PubMed] [Google Scholar]
- 4. Mai J, Virtue A, Shen J, Wang H, Yang XF. An evolving new paradigm: endothelial cells–conditional innate immune cells. J Hematol Oncol. 2013;6:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shao Y, Saredy J, Yang WY, et al. Vascular endothelial cells and innate immunity. Arterioscler Thromb Vasc Biol. 2020;40:e138‐e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chavkin NW, Hirschi KK. Single cell analysis in vascular biology. Front Cardiovasc Med. 2020;7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Paik DT, Cho S, Tian L, Chang HY, Wu JC. Single‐cell RNA sequencing in cardiovascular development, disease and medicine. Nat Rev Cardiol. 2020;17:457‐473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bochenek ML, Schäfer K. Role of endothelial cells in acute and chronic thrombosis. Hamostaseologie. 2019;39:128‐139. [DOI] [PubMed] [Google Scholar]
- 9. Wang M, Hao H, Leeper NJ, Zhu L. Thrombotic regulation from the endothelial cell perspectives. Arterioscler Thromb Vasc Biol. 2018;38:e90‐e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sobczak AIS, Pitt SJ, Stewart AJ. Glycosaminoglycan neutralization in coagulation control. Arterioscler Thromb Vasc Biol. 2018;38:1258‐1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koupenova M, Clancy L, Corkrey HA, Freedman JE. Circulating platelets as mediators of immunity, inflammation, and thrombosis. Circ Res. 2018;122:337‐351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dahlbäck B. Blood coagulation and its regulation by anticoagulant pathways: genetic pathogenesis of bleeding and thrombotic diseases. J Intern Med. 2005;257:209‐223. [DOI] [PubMed] [Google Scholar]
- 13. Mast AE. Tissue factor pathway inhibitor: multiple anticoagulant activities for a single protein. Arterioscler Thromb Vasc Biol. 2016;36:9‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roemisch J, Gray E, Hoffmann JN, Wiedermann CJ. Antithrombin: a new look at the actions of a serine protease inhibitor. Blood Coagul Fibrinolysis. 2002;13:657‐670. [DOI] [PubMed] [Google Scholar]
- 15. Dahlbäck B, Villoutreix BO. The anticoagulant protein C pathway. FEBS Lett. 2005;579:3310‐3316. [DOI] [PubMed] [Google Scholar]
- 16. Urano T, Suzuki Y, Iwaki T, Sano H, Honkura N, Castellino FJ. Recognition of plasminogen activator inhibitor type 1 as the primary regulator of fibrinolysis. Curr Drug Targets. 2019;20:1695‐1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aird WC. Endothelial cell heterogeneity. Cold Spring Harb Perspect Med. 2012;2:a006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Engelmann B, Massberg S. Thrombosis as an intravascular effector of innate immunity. Nat Rev Immunol. 2013;13:34‐45. [DOI] [PubMed] [Google Scholar]
- 19. Foley JH, Conway EM. Cross talk pathways between coagulation and inflammation. Circ Res. 2016;118:1392‐1408. [DOI] [PubMed] [Google Scholar]
- 20. Keller TT, Mairuhu AT, de Kruif MD, et al. Infections and endothelial cells. Cardiovasc Res. 2003;60:40‐48. [DOI] [PubMed] [Google Scholar]
- 21. Stark K, Massberg S. Interplay between inflammation and thrombosis in cardiovascular pathology. Nat Rev Cardiol. 2021;18:666‐682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Joffre J, Hellman J, Ince C, Ait‐Oufella H. Endothelial responses in sepsis. Am J Respir Crit Care Med. 2020;202(3):361‐370. [DOI] [PubMed] [Google Scholar]
- 23. Lupu F, Kinasewitz G, Dormer K. The role of endothelial shear stress on haemodynamics, inflammation, coagulation and glycocalyx during sepsis. J Cell Mol Med. 2020;24(21):12258‐12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Willis Fox O, Preston RJS. Molecular basis of protease‐activated receptor 1 signaling diversity. J Thromb Haemost. 2020;18:6‐16. [DOI] [PubMed] [Google Scholar]
- 25. Esmon CT. Protein C anticoagulant system–anti‐inflammatory effects. Semin Immunopathol. 2012;34:127‐132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Levy JH, Sniecinski RM, Welsby IJ, Levi M. Antithrombin: anti‐inflammatory properties and clinical applications. Thromb Haemost. 2016;115:712‐728. [DOI] [PubMed] [Google Scholar]
- 27. Bonaventura A, Vecchié A, Dagna L, et al. Endothelial dysfunction and immunothrombosis as key pathogenic mechanisms in COVID‐19. Nat Rev Immunol. 2021;21:319‐329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iba T, Connors JM, Levy JH. The coagulopathy, endotheliopathy, and vasculitis of COVID‐19. Inflamm Res. 2020;69:1181‐1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Motto D. Endothelial cells and thrombotic microangiopathy. Semin Nephrol. 2012;32:208‐214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cleuren AC, van Vlijmen BJ, Reitsma PH. Transgenic mouse models of venous thrombosis: fulfilling the expectations? Semin Thromb Hemost. 2007;33:610‐616. [DOI] [PubMed] [Google Scholar]
- 31. Rosenberg RD, Aird WC. Vascular‐bed–specific hemostasis and hypercoagulable states. N Engl J Med. 1999;340:1555‐1564. [DOI] [PubMed] [Google Scholar]
- 32. Cleuren ACA, van der Ent MA, Jiang H, et al. The in vivo endothelial cell translatome is highly heterogeneous across vascular beds. Proc Natl Acad Sci USA. 2019;116:23618‐23624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Amatschek S, Kriehuber E, Bauer W, et al. Blood and lymphatic endothelial cell‐specific differentiation programs are stringently controlled by the tissue environment. Blood. 2007;109:4777‐4785. [DOI] [PubMed] [Google Scholar]
- 34. Durr E, Yu J, Krasinska KM, et al. Direct proteomic mapping of the lung microvascular endothelial cell surface in vivo and in cell culture. Nat Biotechnol. 2004;22:985‐992. [DOI] [PubMed] [Google Scholar]
- 35. Bajaj MS, Kuppuswamy MN, Manepalli AN, Bajaj SP. Transcriptional expression of tissue factor pathway inhibitor, thrombomodulin and von Willebrand factor in normal human tissues. Thromb Haemost. 1999;82:1047‐1052. [PubMed] [Google Scholar]
- 36. Laszik Z, Mitro A, Taylor FB Jr, Ferrell G, Esmon CT. Human protein C receptor is present primarily on endothelium of large blood vessels: implications for the control of the protein C pathway. Circulation. 1997;96:3633‐3640. [DOI] [PubMed] [Google Scholar]
- 37. Levin EG, del Zoppo GJ. Localization of tissue plasminogen activator in the endothelium of a limited number of vessels. Am J Pathol. 1994;144:855‐861. [PMC free article] [PubMed] [Google Scholar]
- 38. Weiler‐Guettler H, Aird WC, Husain M, Rayburn H, Rosenberg RD. Targeting of transgene expression to the vascular endothelium of mice by homologous recombination at the thrombomodulin locus. Circ Res. 1996;78:180‐187. [DOI] [PubMed] [Google Scholar]
- 39. Guan J, Guillot PV, Aird WC. Characterization of the mouse von Willebrand factor promoter. Blood. 1999;94:3405‐3412. [PubMed] [Google Scholar]
- 40. Kawanami O, Jin E, Ghazizadeh M, et al. Heterogeneous distribution of thrombomodulin and von Willebrand factor in endothelial cells in the human pulmonary microvessels. J Nippon Med Sch. 2000;67:118‐125. [DOI] [PubMed] [Google Scholar]
- 41. Pusztaszeri MP, Seelentag W, Bosman FT. Immunohistochemical expression of endothelial markers CD31, CD34, von Willebrand factor, and Fli‐1 in normal human tissues. J Histochem Cytochem. 2006;54:385‐395. [DOI] [PubMed] [Google Scholar]
- 42. Yamamoto K, de Waard V, Fearns C, Loskutoff DJ. Tissue distribution and regulation of murine von Willebrand factor gene expression in vivo. Blood. 1998;92:2791‐2801. [PubMed] [Google Scholar]
- 43. Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet. 2019;20:631‐656. [DOI] [PubMed] [Google Scholar]
- 44. Decarlo K, Emley A, Dadzie OE, Mahalingam M. Laser capture microdissection: methods and applications. Methods Mol Biol. 2011;755:1‐15. [DOI] [PubMed] [Google Scholar]
- 45. Langenkamp E, Kamps JA, Mrug M, et al. Innovations in studying in vivo cell behavior and pharmacology in complex tissues–microvascular endothelial cells in the spotlight. Cell Tissue Res. 2013;354:647‐669. [DOI] [PubMed] [Google Scholar]
- 46. Mojsilovic‐Petrovic J, Nesic M, Pen A, Zhang W, Stanimirovic D. Development of rapid staining protocols for laser‐capture microdissection of brain vessels from human and rat coupled to gene expression analyses. J Neurosci Methods. 2004;133:39‐48. [DOI] [PubMed] [Google Scholar]
- 47. Nolan DJ, Ginsberg M, Israely E, et al. Molecular signatures of tissue‐specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev Cell. 2013;26:204‐219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535‐550. [DOI] [PubMed] [Google Scholar]
- 49. Wang ZY, Leushkin E, Liechti A, et al. Transcriptome and translatome co‐evolution in mammals. Nature. 2020;588:642‐647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Arap W, Kolonin MG, Trepel M, et al. Steps toward mapping the human vasculature by phage display. Nat Med. 2002;8:121‐127. [DOI] [PubMed] [Google Scholar]
- 51. Rajotte D, Arap W, Hagedorn M, Koivunen E, Pasqualini R, Ruoslahti E. Molecular heterogeneity of the vascular endothelium revealed by in vivo phage display. J Clin Investig. 1998;102:430‐437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Toledo AG, Golden G, Campos AR, et al. Proteomic atlas of organ vasculopathies triggered by Staphylococcus aureus sepsis. Nat Commun. 2019;10:4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Doh SJ, Yamakawa M, Santosa SM, et al. Fluorescent reporter transgenic mice for in vivo live imaging of angiogenesis and lymphangiogenesis. Angiogenesis. 2018;21:677‐698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gay L, Miller MR, Ventura PB, et al. Mouse TU tagging: a chemical/genetic intersectional method for purifying cell type‐specific nascent RNA. Genes Dev. 2013;27:98‐115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Heiman M, Schaefer A, Gong S, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738‐748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hupe M, Li MX, Gertow Gillner K, Adams RH, Stenman JM. Evaluation of TRAP‐sequencing technology with a versatile conditional mouse model. Nucleic Acids Res. 2014;42:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhou P, Zhang Y, Ma Q, et al. Interrogating translational efficiency and lineage‐specific transcriptomes using ribosome affinity purification. Proc Natl Acad Sci USA. 2013;110:15395‐15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jambusaria A, Hong Z, Zhang L, et al. Endothelial heterogeneity across distinct vascular beds during homeostasis and inflammation. Elife. 2020;9:e51413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell‐type‐specific isolation of ribosome‐associated mRNA from complex tissues. Proc Natl Acad Sci USA. 2009;106:13939‐13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sabbagh MF, Heng JS, Luo C, et al. Transcriptional and epigenomic landscapes of CNS and non‐CNS vascular endothelial cells. eLife. 2018;7:e36187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Deal RB, Henikoff S. The INTACT method for cell type‐specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat Protoc. 2011;6:56‐68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chucair‐Elliott AJ, Ocañas SR, Stanford DR, et al. Inducible cell‐specific mouse models for paired epigenetic and transcriptomic studies of microglia and astroglia. Commun Biol. 2020;3:693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Roh HC, Tsai LT, Lyubetskaya A, Tenen D, Kumari M, Rosen ED. Simultaneous transcriptional and epigenomic profiling from specific cell types within heterogeneous tissues in vivo. Cell Rep. 2017;18:1048‐1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Payne S, De Val S, Neal A. Endothelial‐specific Cre mouse models. Arterioscler Thromb Vasc Biol. 2018;38:2550‐2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gritz E, Hirschi KK. Specification and function of hemogenic endothelium during embryogenesis. Cell Mol Life Sci. 2016;73:1547‐1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Liu J, Willet SG, Bankaitis ED, Xu Y, Wright CV, Gu G. Non‐parallel recombination limits Cre‐LoxP‐based reporters as precise indicators of conditional genetic manipulation. Genesis. 2013;51:436‐442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. McNamara DA, Harmey J, Wang JH, Kay E, Walsh TN, Bouchier‐Hayes DJ. Tamoxifen inhibits endothelial cell proliferation and attenuates VEGF‐mediated angiogenesis and migration in vivo. Eur J Surg Oncol. 2001;27:714‐718. [DOI] [PubMed] [Google Scholar]
- 68. Pu W, He L, Han X, et al. Genetic targeting of organ‐specific blood vessels. Circ Res. 2018;123:86‐99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hirrlinger J, Scheller A, Hirrlinger PG, et al. Split‐Cre complementation indicates coincident activity of different genes in vivo. PLoS One. 2009;4:e4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gupta RK, Kuznicki J. Biological and medical importance of cellular heterogeneity deciphered by single‐cell RNA sequencing. Cells. 2020;9:1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vanlandewijck M, He L, Mae MA, et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018;554:475‐480. [DOI] [PubMed] [Google Scholar]
- 72. Gillich A, Zhang F, Farmer CG, et al. Capillary cell‐type specialization in the alveolus. Nature. 2020;586:785‐789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Delorey TM, Ziegler CGK, Heimberg G, et al. COVID‐19 tissue atlases reveal SARS‐CoV‐2 pathology and cellular targets. Nature. 2021;595:107‐113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pujadas E, Beaumont M, Shah H, et al. Molecular profiling of coronavirus disease 2019 (COVID‐19) autopsies uncovers novel disease mechanisms. Am J Pathol. 2021;191:2064‐2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang S, Yao X, Ma S, et al. A single‐cell transcriptomic landscape of the lungs of patients with COVID‐19. Nat Cell Biol. 2021;23:1314‐1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yasen A, Aini A, Wang H, et al. Progress and applications of single‐cell sequencing techniques. Infect Genet Evol. 2020;80:104198. [DOI] [PubMed] [Google Scholar]
- 77. HuBMAP Consortium . The human body at cellular resolution: the NIH human biomolecular atlas program. Nature. 2019;574:187‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Han X, Wang R, Zhou Y, et al. Mapping the mouse cell atlas by microwell‐seq. Cell. 2018;172:1091‐107.e17. [DOI] [PubMed] [Google Scholar]
- 79. Regev A, Teichmann SA, Lander ES, et al. The human cell atlas. Elife. 2017;6:e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tabula Muris Consortium , Overall Coordination ; Logistical Coordination , et al. Single‐cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367‐372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kalucka J, de Rooij L, Goveia J, et al. Single‐cell transcriptome atlas of murine endothelial cells. Cell. 2020;180:764‐79.e20. [DOI] [PubMed] [Google Scholar]
- 82. Paik DT, Tian L, Williams IM, et al. Single‐cell RNA sequencing unveils unique transcriptomic signatures of organ‐specific endothelial cells. Circulation. 2020;142:1848‐1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Feng W, Chen L, Nguyen PK, Wu SM, Li G. Single cell analysis of endothelial cells identified organ‐specific molecular signatures and heart‐specific cell populations and molecular features. Front Cardiovasc Med. 2019;6:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Franzén O, Gan L‐M, Björkegren JLM. PanglaoDB: a web server for exploration of mouse and human single‐cell RNA sequencing data. Database. 2019;2019:baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Khan S, Taverna F, Rohlenova K, et al. EndoDB: a database of endothelial cell transcriptomics data. Nucleic Acids Res. 2019;47:D736‐D744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Halpern KB, Shenhav R, Matcovitch‐Natan O, et al. Single‐cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542:352‐356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Inverso D, Shi J, Lee KH, et al. A spatial vascular transcriptomic, proteomic, and phosphoproteomic atlas unveils an angiocrine Tie‐Wnt signaling axis in the liver. Dev Cell. 2021;56:1677‐93.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Rad HS, Rad HS, Shiravand Y, et al. The Pandora's box of novel technologies that may revolutionize lung cancer. Lung Cancer. 2021;159:34‐41. [DOI] [PubMed] [Google Scholar]
- 89. Lee JH, Daugharthy ER, Scheiman J, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360‐1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lubeck E, Cai L. Single‐cell systems biology by super‐resolution imaging and combinatorial labeling. Nat Methods. 2012;9:743‐748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single‐cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014;11:360‐361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Merritt CR, Ong GT, Church SE, et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat Biotechnol. 2020;38:586‐599. [DOI] [PubMed] [Google Scholar]
- 93. He S, Bhatt R, Birditt B, et al. High‐Plex multiomic analysis in FFPE tissue at single‐cellular and subcellular resolution by spatial molecular imaging. bioRxiv. 2021. doi: 10.1101/2021.11.03.467020 [DOI] [Google Scholar]
- 94. Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348(6233):aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kelly RT. Single‐cell proteomics: progress and prospects. Mol Cell Proteomics. 2020;19:1739‐1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Iba T, Levy JH, Levi M, Thachil J. Coagulopathy in COVID‐19. J Thromb Haemost. 2020;18:2103‐2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Levi M, Iba T. COVID‐19 coagulopathy: is it disseminated intravascular coagulation? Intern Emerg Med. 2021;16:309‐312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Mackman N, Antoniak S, Wolberg AS, Kasthuri R, Key NS. Coagulation abnormalities and thrombosis in patients infected with SARS‐CoV‐2 and other pandemic viruses. Arterioscler Thromb Vasc Biol. 2020;40(9):2033‐2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Norooznezhad AH, Mansouri K. Endothelial cell dysfunction, coagulation, and angiogenesis in coronavirus disease 2019 (COVID‐19). Microvasc Res. 2021;137:104188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Peralta MR, Muczynski V, McVey J, et al. The link between inflammation, coagulation and endothelium damage in COVID‐19: evidence from an exploratory cross‐sectional study. Res Pract Thromb Haemost. 2021;5(suppl 2):OC 68.3. [Google Scholar]
- 101. Cani E, Dwivedi DJ, Liaw KL, et al. Immunothrombosis biomarkers for distinguishing COVID‐19 patients from non‐COVID septic pneumonia patients and for predicting ICU mortality. Res Pract Thromb Haemost. 2021;5(suppl 2):PB0138. [Google Scholar]
- 102. Fernández S, Moreno‐Castaño AB, Palomo M, et al. Distinctive biomarker features in the endotheliopathy of COVID‐19 and septic syndromes. Shock. 2022;57:95‐105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Juneja GK, Castelo M, Yeh CH, et al. Biomarkers of coagulation, endothelial function, and fibrinolysis in critically ill patients with COVID‐19: a single‐center prospective longitudinal study. J Thromb Haemost. 2021;19:1546‐1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Marchetti M, Gomez‐Rosas P, Sanga E, et al. Endothelium activation markers in severe hospitalized COVID‐19 patients: role in mortality risk prediction. TH Open. 2021;5:e253‐e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Moraes CRP, Lima F Borba Junior IT, et al. Association of the angiopoietin/Tie2 and VEGF‐A pathways with clinical and laboratory markers of disease severity in COVID‐19. Res Pract Thromb Haemost. 2021;5(suppl 2):PB0144. [Google Scholar]
- 106. Alvarado‐Moreno JA, Davila‐Moreno J, Dominguez‐Reyes V, et al. Morphological and functional alterations in endothelial colony‐forming cells from recovered COVID‐19 patients. Thromb Res. 2021;206:55‐59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Gunawardana H, Romero T, Yao N, et al. Tissue‐specific endothelial cell heterogeneity contributes to unequal inflammatory responses. Sci Rep. 2021;11:1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. McCafferty CLL, Cai T, Praporski S, et al. Fibrin clot characteristics and anticoagulant response in a SARS‐CoV‐2 infected endothelial cell model. Res Pract Thromb Haemost. 2021;5(suppl 2):PB0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Riddle RHK, Jennbacken K, Harper M. A 3D in vitro model of inflammation‐associated bleeding. Res Pract Thromb Haemost. 2021;5(suppl 2):PB1040. [Google Scholar]


