Abstract
The present study aims to find the effective natural enzyme inhibitors against alpha-amylase and alpha-glucosidase from the array of compounds identified in plants of the Simaroubaceae family using molecular docking and ADME/Toxicity studies. Among the 218 compounds docked against seven enzymes, buddlenol-A and citrusin-B showed the best binding energies (kcal/mol) of -7.830 and -7.383 against human salivary alpha-amylase and pancreatic alpha-amylase respectively. The other two compounds 9-hydroxycanthin-6-one and bruceolline-B had the best binding energy of -6.461 and -7.576 against N-terminal and C-terminal maltase glucoamylase respectively. Whereas the binding energy of prosopine (-6.499) and fisetinidol (-7.575) was considered as the best against N-terminal and C-terminal sucrase-isomaltase respectively. Picrasidine-X showed the best binding energy (-7.592) against yeast alpha-glucosidase. The study revealed that the seven compounds which showed the best binding energy against respective enzymes are considered as the ‘lead hit compounds’. Even though the ‘lead hit compounds’ are not obeying all the laws of ADMET, the drug-likeness properties of 9-hydroxycanthin-6-one, fisetinidol, picrasidine-X, and prosopine were considerable. Also, kaempferol-3-O-pentoside was the recent compound identified from the Simarouba glauca plant extract found to be one among the top five lead hit compounds against four enzymes. This study provides valuable insight into the direction of developing natural compounds as potential starch hydrolysing enzyme inhibitors for managing type 2 diabetes.
Keywords: Alpha-amylase, Alpha-glucosidase, ADME/Toxicity, Diabetes mellitus, Inhibitors, Molecular docking
Alpha-amylase; Alpha-glucosidase; ADME/Toxicity; Diabetes mellitus; Inhibitors; Molecular docking.
1. Introduction
It has been proved for ages that, even with the advancement of medical technology and lifestyle it will be a challenge for a human to escape from the epidemic or pandemic diseases. The frequent emerging of new diseases would motivate the scientific groups to discover novel drugs either from synthetic or from natural sources to eradicate them successfully. Irrespective of ample time, resources, manpower, and high cost, the clinical trial of novel drug molecules is one of the main barriers and ruins all the efforts taken during the entire drug discovery process in many instances (Hwang et al., 2016; Nicolaou, 2014). The high throughput screening (HTS) experiments that involve robotic technology are still an expensive process and require a vast array of targets and ligands. Due to such failures, many pharmaceutical companies are deviating from the routine screening of ligands towards particular targets and looking for alternative methods to increase the success rate (Subramaniam et al., 2008). Therefore, the computer-aided drug discovery (CADD) technique gained importance and attracted the pharmaceutical industries as well as academia in this field recently (Alagappan et al., 2016; Leelananda and Lindert, 2016).
Virtual high throughput screening (vHTS) is one of the alternative techniques to HTS that involves the molecular docking approach to screen in-silico compound database and to investigate the binding affinity of the active site of a macromolecule with the existing library of compounds (Shoichet, 2004). The main goal of the vHTS is to detect ‘hits’ (most active ligands), select the ‘leads’ (most likely ligand candidate for further assessment), and optimize the lead hits into suitable drug molecules by refining their physicochemical, pharmaceutical, and pharmacokinetic properties (Kapetanovic, 2008). Interestingly, the vHTS technique has been frequently applied in several drug development fields (Alam and Khan, 2018; Musyoka et al., 2016), and even in the area of recent SARS-CoV-2 outbreaks (Naik et al., 2020; Zhang et al., 2020). Besides, the vHTS technique has been applied in the development of starch hydrolysing enzyme inhibitor molecules to manage hyperglycaemia in type 2 diabetes (T2D) (Jhong et al., 2015; Park et al., 2008; Rasouli et al., 2017). Also, the success stories of vHTS through structure-based drug designing (SBDD) have been seen in the discovery of anti-HIV (Kim et al., 1995), anti-influenza drugs (Talele et al., 2010), etc. So, due to the feasibility of vHTS over HTS, it can be useful in the drug development process to treat a variety of diseases.
Over the past few decades, diabetes is one of the concerns along with other metabolic disorders. According to the recent reports by the International Diabetic Federation (IDF), diabetes is one of the reasons for the fastest emerging global crises in the 21st century (Saeedi et al., 2019). Among the several drug targets (Kahn et al., 2014), the concept of partial inhibition of starch hydrolysing enzymes (alpha-amylase and alpha-glucosidase) have been focused on for decades to regulate the hyperglycemia in T2D patients. In humans, there are six enzymes involved in the hydrolysis of complex starch-based food materials namely; salivary alpha-amylase (HSA), pancreatic alpha-amylase (HPA), both N-terminal (Nt) and C-terminal (Ct) maltase-glucoamylase (MGAM), both N-terminal (Nt) and C-terminal (Ct) sucrase-isomaltase (SI) (Sim et al., 2008). At the foremost, the HSA enzyme acts on the alpha 1, 4-glycosidic linkages of starch molecules in the mouth and breaks down into shorter chains. The complete hydrolysis of the remaining starch molecules will be carried out by the action of HPA , MGAM and SI in the stomuch and small intestine (Wahbeh and Christie, 2006). All the catalytic subunits of MGAM and SI will prefer the hydrolysis of alpha-1, 4-glycosidic linkages of the oligosaccharides. Additionally, the Nt-SI subunit has shown the specific action on alpha-1, 6 bonds of isomaltose and the Ct-SI subunit displays the specific activity on alpha-1, 2 bonds of sucrose (Sim et al., 2010b).
Acarbose, a pseudo-tetrasaccharide is the only drug available as an alpha-amylase inhibitor (AAI) so far. Acarbose has been used extensively for the treatment of T2D patients as an inhibitor of alpha-glucosidase (AG) enzyme along with miglitol and voglibose. Due to this, abdominal discomforts and other side effects have been noticed in the patients administered with these inhibitors (Campbell et al., 2000; Dabhi et al., 2013). Thus, several studies have been focused on identifying new alpha-amylase inhibitors (AAIs) and alpha-glucosidase inhibitors (AGIs) from natural sources (Lo Piparo et al., 2008; Sim et al., 2010a). Surprisingly, there are only two reports available on the inhibition of Ct-MGAM, Nt-MGAM, Ct-SI, and Nt-SI catalytic subunits by salacinol class of compounds and their derivatives so far (Jones et al., 2011; Sim et al., 2010a). Hence, there is a need for an effective starch hydrolysing enzyme inhibitor from natural sources as an alternative to the present drugs.
To date, there were more than 200 compounds have been identified and isolated from the plants of Simaroubaceae family (Simão et al., 1991). The discovery of ‘quassinoids’ revealed that this unique group of compounds has been attributed predominantly to the diversified biological activities of these plants (Dahar and Rai, 2019; Vieira and Braz-Filho, 2006). The plants from this family are traditionally known for the treatment of various diseases like malaria (Muhammad et al., 2004), cancer (Bedikian et al., 1979), and diseases caused by viruses and parasites (Silva et al., 2010). Surprisingly, the detailed studies on antidiabetic and starch hydrolysing enzyme inhibition properties of the Simaroubaceae family were lacking except for a few studies on Quassia amara (Husain et al., 2011), Brucea javanica (NoorShahida et al., 2009), and Simarouba glauca (Mugaranja and Kulal, 2020). Thus, in this study natural compounds identified in the Simaroubaceae family were used as ligands to find the ‘lead hit compounds’ as potent AAIs and/or AGIs using the molecular docking approach. The acarbose, miglitol, and voglibose which are standard inhibitors were used as controls during the study. The present study revealed that some of the ‘lead hit compounds’ including the recently identified compounds by our group have better binding affinity than the available drugs (Mugaranja and Kulal, 2020). Further, the drug likeliness properties of the ‘lead hit compounds’ evaluated by absorption, distribution, metabolism, excretion, and toxicity (ADMET) test provides additional evidence to consider them as effective starch hydrolysing enzyme inhibitors.
2. Experimental section
2.1. Selection of ligands
The natural compounds which were identified previously in the different genus of the Simaroubaceae family have been selected as ligands. In the present study total of 218 compounds have been docked against starch hydrolysing enzymes along with standard AAI and AGIs. Recently identified four compounds from Simarouba glauca (Mugaranja and Kulal, 2020) are also included in this study along with the compounds identified by others (Alves et al., 2014; Jiao et al., 2011; Kubo et al., 1993; Tan et al., 2012). The 2D structures of the majority of the compounds were retrieved from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) in sdf format. The compounds which were unavailable in the PubChem database were drawn using ChemDraw Pro 8.0 drawing software.
2.2. Homology modeling and Ramachandran plot
Since there are no crystal structures available in the database, the homology models were built for the Ct-SI and YAG enzymes. The protein sequences of Ct-SI (access code: P14410) and YAG (access code: P53341) enzymes were retrieved from UniProt (https://www.uniprot.org/) in FASTA format and submitted to RaptorX (http://raptorx.uchicago.edu/) homology modeling software to generate 3D structures. The quality of the homology models of both the protein structures was validated through Ramachandran plot analysis using PROCHECK software (https://servicesn.mbi.ucla.edu/PROCHECK/).
2.3. Selection of protein targets and active site residues
The crystal structures of starch hydrolysing enzymes such as HSA, HPA, Nt-MGAM, Ct-MGAM, and Nt-SI were retrieved from Protein Data Bank (RCSBPDB) with the protein ID; 3DHP, 2QV4, 2QMJ, 3TOP, and 3LPO respectively. The active site residues for HSA and HPA were selected by referring to the crystal structure solved by Ramasubbu et al., 1996, 2004. The structure of acarbose bound enzyme complex has referred for the selection of active site residues during the grid generation for Ct-MGAM and Nt-MGAM (Ren et al., 2011; Sim et al., 2008). The active site residues for the Nt-SI enzyme were selected by analysing the kotalanol-Nt-SI bound enzyme structure (Sim et al., 2010b). The active site residues for Ct-SI were designated by referring to the structure of Ct-MGAM in complex with acarbose (Ren et al., 2011). The active site residues for the YAG enzyme were selected by referring to the homology model.
2.4. Protein-ligand molecular docking studies
The docking study was performed against starch hydrolysing enzymes using the natural compounds which are previously identified in the different genera of the Simaroubaceae family as ligands by following the previously explained method with slight modifications (Aamir et al., 2018). All the protein structures were pre-processed before the docking analysis, where the water molecules beyond 5Å have deleted and the pH was set to 7 using the protein preparation module on Maestro with default settings. The sdf files of all the ligands were submitted to the Schrödinger, LLC, New York, NY, 2015, and 3D conformations for all the ligands were prepared with the default settings using the OPLS2005 force field. Wander walls scaling factor 1 and partial charge cut-off of 0.25 have been set in the LigPrep module. Also, the ligands with more than 500 atoms and 100 rotatable bonds have been excluded during the ligand preparation. A total of 732, 3D conformations of the ligands were generated and all the conformations were selected for the study. A docking grid of 20Å was generated around the active site pocket of each enzyme by providing the active site residues in the grid generation module. Docking was performed with standard precision (SP) and flexible mode in the Glide module on Maestro version 10.1 (Schrödinger, LLC, New York, NY, 2015). During the docking studies, the scaling factor was set as 0.8, and the number of poses/ligands was set as 5. Among the all docked poses, the protein-ligand complexes with the best pose were selected based on the Glide score, Glide energy, and Glide Emodel energy for further analysis. The following formula describes the calculation of Glide score,
where, vdW = van der Waals energy, Coul = Coulomb energy, Lipo = Lipophilic contacts, Hbond = Hydrogen-bonding, Metal = Metal-binding, BuryP = Penalty for buried polar groups, RotB = Penalty for freezing the rotatable bonds and, Site = Polar interactions with the residues in the active site. a = 0.065 is the coefficient constant of van der Waals energy and b = 0.130 is the coefficient constant of Coulomb energy (Aamir et al., 2018; Tripathi et al., 2013).
The docking analysis against each protein target was performed in duplicates (n = 2) by maintaining the same parameters. Also, the ligands which showed best poses with low binding energies were re-docked against respective protein targets to confirm the binding affinities. The 2D interactions of the ligands and active site residues were visually analysed with ligand-enzyme mode and, the 3D structures have drawn using Pymol software (https://pymol.org/2/).
2.5. ADME/toxicity evaluation
Absorption, distribution, metabolism, and excretion (ADME) properties including physicochemical, lipophilicity, water-solubility, pharmacokinetics, drug-likeness and medicinal chemistry properties have been evaluated for the ‘lead hit compounds’ found during the molecular docking study using SwissADME server (http://www.swissadme.ch/). The graphical outputs such as radar plots and Brain Or IntestinaL EstimateD permeation method (BOILED-Egg) have been implemented for the easy understanding of the drug-likeness property and, passive gastrointestinal absorption (GI) and blood-brain barrier penetrant (BBB) properties of the compounds respectively. The toxicity properties such as AMES toxicity, carcinogenic property, biodegradation property, acute oral toxicity, and Rat acute toxicity of the ‘lead hit compounds’ have been evaluated using a free online server; admetSAR (http://lmmd.ecust.edu.cn/admetsar1).
3. Results and discussion
3.1. Homology model
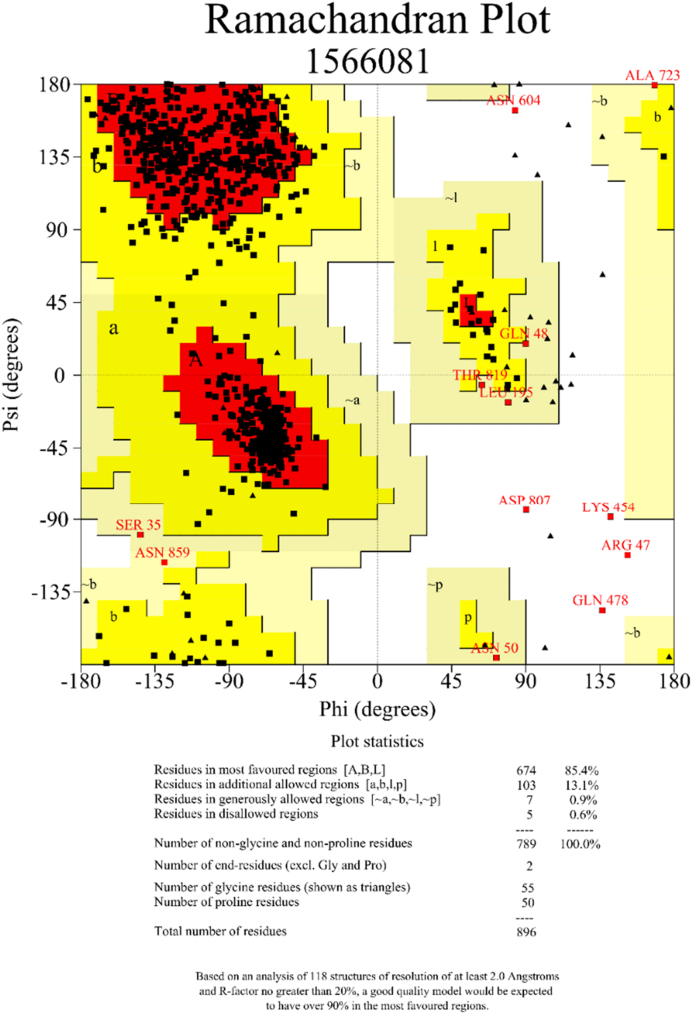
The homology model of Ct-SI was built using the crystal structures of Ct-MGAM (RCSB PDB: 3TON) and Nt-SI (RCSB PDB: 3LPO) as templates. The Ct-MGAM and Nt-SI have shown 59.16 and 38.87% sequence identity with the Ct-SI homology model respectively. Ramachandran plot analysis for the homology model of Ct-SI showed that there is 85.4% of residues were allocated in the most favoured regions (Figure 1). Also, 13.1, 0.9, and 0.6 % residues were allotted in additional allowed regions, generously allowed regions, and disallowed regions respectively. The Ramachandran plot analysis and the validation of yeast alpha-glucosidase (YAG) structure were made as explained in the earlier study (Mugaranja and Kulal, 2020). The authenticated homology models of Ct-SI and YAG enzymes along with HSA, HPA, Nt-MGAM, Ct-MGAM, and Nt-SI crystal structures were subjected to docking studies.
Figure 1.
Ramachandran plot for the homology model of Ct-SI. The homology model was built using RaptorX software. The authentication of homology model was done using Ramachandran plot analysis using PROCHECK software.
3.2. Active site residues of the enzyme targets
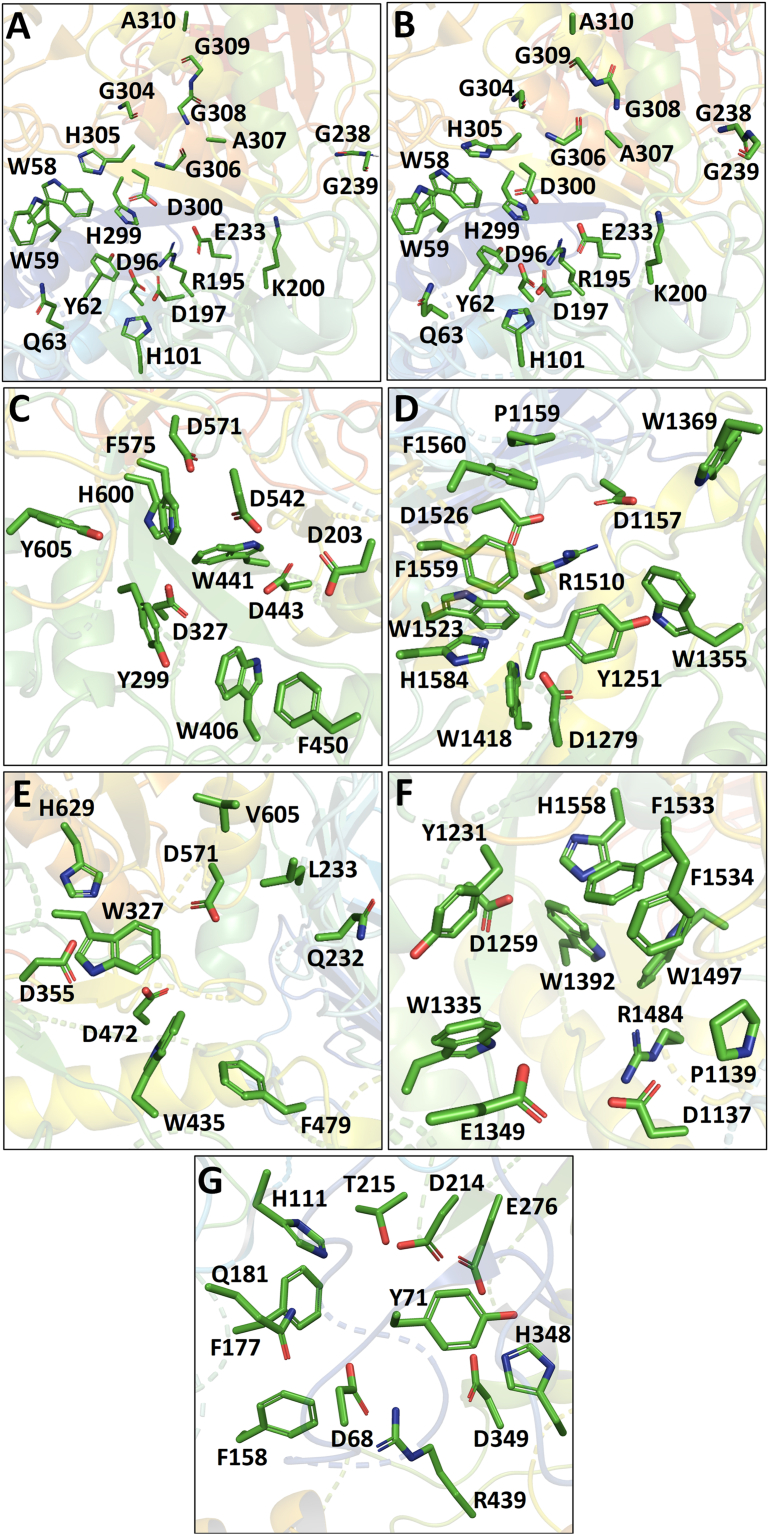
The active site residues of starch hydrolysing enzymes selected during the docking study were represented in the stick view model (Figure 2). There is 97% sequence similarity and 94% identity found in the catalytic domains of HSA and HPA. Thus, similar active site residues were annotated during the generation of the grid (Figure 2A and 2B) (Ramasubbu et al., 1996, 2004; Williams et al., 2012). The active site residues for Nt-MGAM, Ct-MGAM, and Nt-SI enzymes were represented in Figure 2C, 2D, and 2E respectively (Ren et al., 2011; Sim et al., 2008, 2010b). The MGAM and SI enzymes share 58.4% and 74.3% of sequence identity and similarity respectively. So, the active site residues for Ct-SI have been designated by referring to the structure of Ct-MGAM bound with acarbose (Figure 2F). The active site residues for YAG were assigned by referring to the homology model of YAG generated using RaptorX homology modeling software (Figure 2G).
Figure 2.
Active site residues of the protein targets. (A) HSA, (B) HPA, (C) Nt-MGAM, (D) Ct-MGAM, (E) Nt-SI, (F) Ct-SI, (G) YAG. The active site residues have represented in stick view and these residues were used for the grid generation during the docking study.
3.3. Molecular docking with HSA and HPA
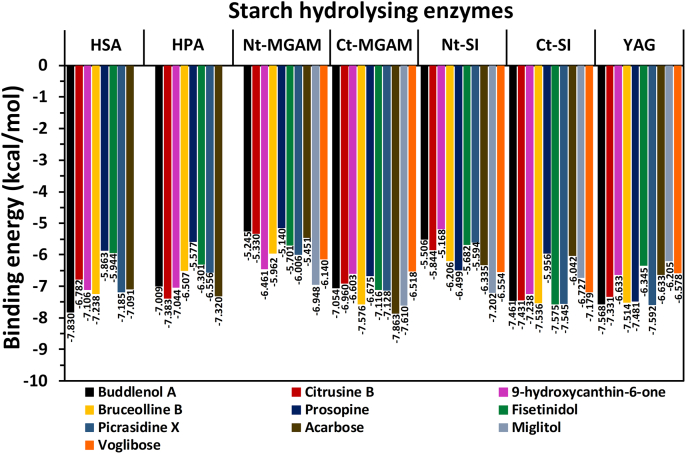
Among the 218 natural compounds docked, the compound buddlenol-A showed better binding energy with -7.830 kcal/mol against the HSA enzyme compared to the remaining compounds (Figure 3). The structure of buddlenol-A consists of an aryl glycerol portion attached to a dihydrobenzofuran neolignan moiety which was identified in Picrasma quassioides BENNET (Simaroubaceae) and it exhibited the inhibition of nitric oxide, tumour necrosis factor-α, and interleukin-6 production in a mouse model (Jiao et al., 2011). Also, this compound was identified in Hevea brasiliensis where it showed the antitumor activity against B-16 cancer cells (Ren et al., 2012). Another study showed the cytotoxic properties of buddlenol-A isolated from Solanum melongena L. against HepG2, Hela, and MCF-7 human tumour cell lines (Yang et al., 2020). However, there are no reports on this compound inhibiting starch hydrolysing enzymes so far.
Figure 3.
The binding energy of the ‘lead hit compounds’ and standard inhibitors against starch hydrolysing enzymes. The docking analysis of miglitol and voglibose was not performed against HSA and HPA enzymes. HSA; human salivary alpha-amylase, HPA; human pancreatic alpha-amylase, Nt-MGAM; N-terminal maltase glucoamylase, Ct-MGAM; C-terminal maltase glucoamylase, Nt-SI; N-terminal sucrase-isomaltase, Ct-SI; C-terminal sucrase-isomaltase and YAG; yeast alpha-glucosidase.
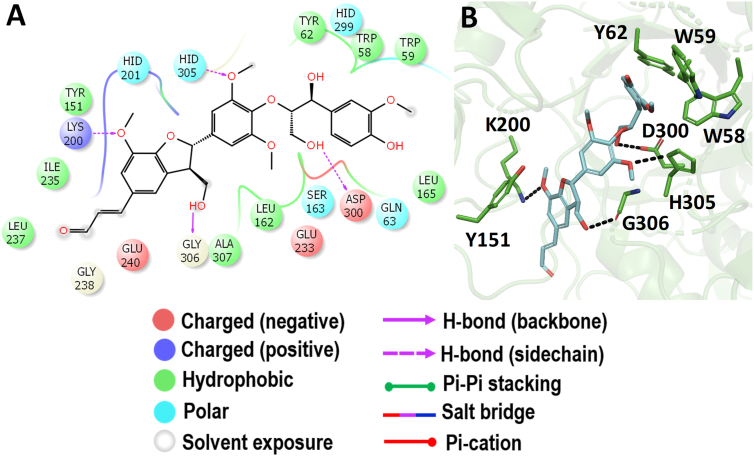
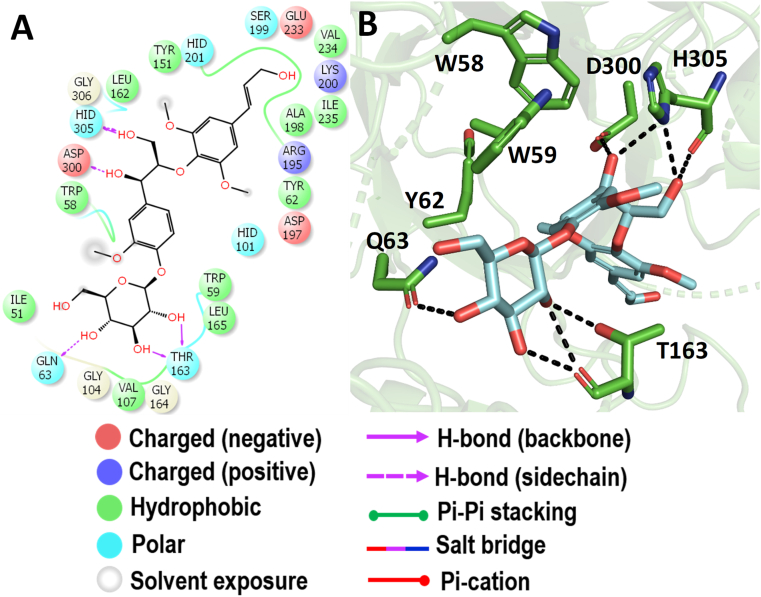
The interacting active site residues of HSA with buddlenol-A are represented in Figure 4. The dihydrobenzofuran neolignan unit of buddlenol-A showed the hydrogen bond interaction with the residues such as Q306, K200, and H305. The aryl glycerol portion showed a hydrogen bond interaction with the side chain of D300. Also, the aromatic residues such as W58, W59, Y62, and Y151 further stabilize the compound in the active site of HSA. This is the first evidence from the docking study on buddlenol-A inhibiting HSA with higher binding affinity than the known anti-diabetic compound acarbose.
Figure 4.
Molecular docking analysis of buddlenol-A. Interaction of buddlenol-A against HSA in 2D view (A) and 3D view (B). Buddlenol-A made a hydrogen bond interaction with D300 which acts as an acid/base catalyst during the hydrolysis of the substrates. In 3D view, the hydrogen bond interactions are represented in black coloured broken lines.
The second compound citrusin-B, showed the best binding energy of –7.383 kcal/mol against HPA among the other compounds as represented in Figure 3. The citrusin-B was identified in Picrasma quassioides (Yoshikawa et al., 1995) and Ailanthus altissima (Tan et al., 2012) plants that belong to the Simaroubaceae family. According to the previous studies, citrusin-B isolated from plants showed significant anti-inflammatory activity (Sun et al., 2014) as well as anti-bacterial activity (Yuan et al., 2007). Also, citrusin-B showed moderate inhibition activity against tobacco mosaic virus replication (Tan et al., 2012). Further, citrusin-B was identified in different plant extracts and reported for their liver-protective property (De Souza et al., 2019), anti-oxidant, and alpha-glucosidase inhibition properties (Saleem et al., 2019) along with other natural compounds. In this docking study, citrusin-B showed more activity towards inhibiting HPA than alpha-glucosidase.
The interactions between citrusin-B and HPA enzyme was shown in Figure 5. The aromatic polycyclic compound, citrusin-B belongs to a class of lignan glycosides where the lignan moiety is linked to the carbohydrate unit through a glycosidic bond (Yoshikawa et al., 1995). The active site residues such as D300 and H305 showed the hydrogen bond with the lignin moiety of the citrusin-B. Further, the glycoside unit was stacked through the hydrogen bond interaction with T163 and Q63. The aromatic residues have also been involved in the stabilization of citrusin-B through hydrophobic interactions.
Figure 5.
Molecular docking analysis of citrusin-B: Interaction of citrusin-B against HPA in 2D view (A) and 3D view (B). Citrusin-B showed the hydrogen bond interaction with D300 which acts as an acid/base catalyst during the hydrolysis of the substrates. In 3D view, the hydrogen bond interactions are represented in black coloured broken lines.
The V-shaped active site pocket of the alpha-amylase enzyme has been divided into +1, +2, +3 subsites (aglycone binding sites) and -1, -2, -3, -4 subsites (glycone binding sites). Studies revealed that the acarviosine unit of the acarbose resides in the -1 and +1 subsites where generally the hydrolysis of substrates would occur. Most importantly, D197, D300, and E233 of domain A have been considered as the key catalytic residues of the alpha-amylase enzymes (Ramasubbu et al., 1996, 2004). It has been noted that D197 acts as a nucleophile as well as D300 and E233 either independently or mutually act as acid/base catalysts during the hydrolysis of the substrates. The crystal structure of alpha-amylase along with acarbose revealed that the binding of D197, D300, and E233 is believed to be involved in the strong inhibition of the enzyme (Brayer et al., 2000; Rydberg et al., 2002).
In the present study, the buddlenol-A believed to occupy the -1, -2, and -3 subsites of the active site cleft of HSA, because it shows the hydrogen bond with D300 (interacts with -1 subsite) as well as aromatic stabilization with W58 and W59 (interacts with -2 and -3 subsites). The binding of citrusin-B with HPA is involved in the hydrogen bond as well as aromatic stabilization through the residues such as D300, H305, T163, G63, W58, and W59 in the glycone binding site. Also, the recent docking studies on some of the synthetic compounds (Ganesan et al., 2020; Yousuf et al., 2020) and natural compounds derived from plants (Lo Piparo et al., 2008; Miao et al., 2015) showed relatively similar interactions against alpha-amylase enzymes. In the present docking study, both buddlenol-A and citrusin-B have shown better binding energies with amylase compared to the positive control acarbose. Thus, both the compounds could be potent alternatives to the acarbose as alpha-amylase inhibitors, which need further study.
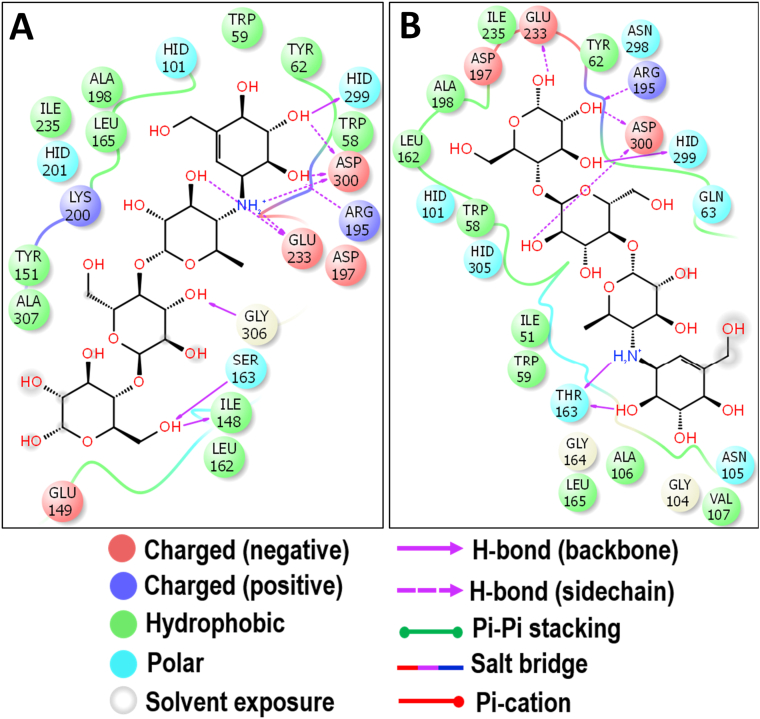
The present docking study of acarbose showed the binding energy of -7.081 and -7.320 kcal/mol against HSA and HPA enzymes respectively (Figure 3). The side chain of E233 of HSA sowed hydrogen bond with the –NH group of the acarviosine unit. Additionally, three more hydrogen bond interactions were observed between acarviosine units and D300, R195, and H299. The glycone ring showed three hydrogen bonds with the backbone of G306, I148, and S163 (Figure 6A). The interaction of acarbose and the HPA enzyme showed that the –NH and –OH groups of the acarviosine unit had the hydrogen bond interaction with T163. Also, the hydrogen bond interactions of the glycone rings have been observed through the active site residues D300, R195, H299, and E233 (Figure 6B).
Figure 6.
Molecular docking analysis of acarbose. Interaction of acarbose against HSA (A) and HPA (B) in 2D view. Acarbose showed the hydrogen bond interactions with the crucial active site residues D300, R195 and H299 of both HAS and HPA.
3.4. Molecular docking with Nt-MGAM
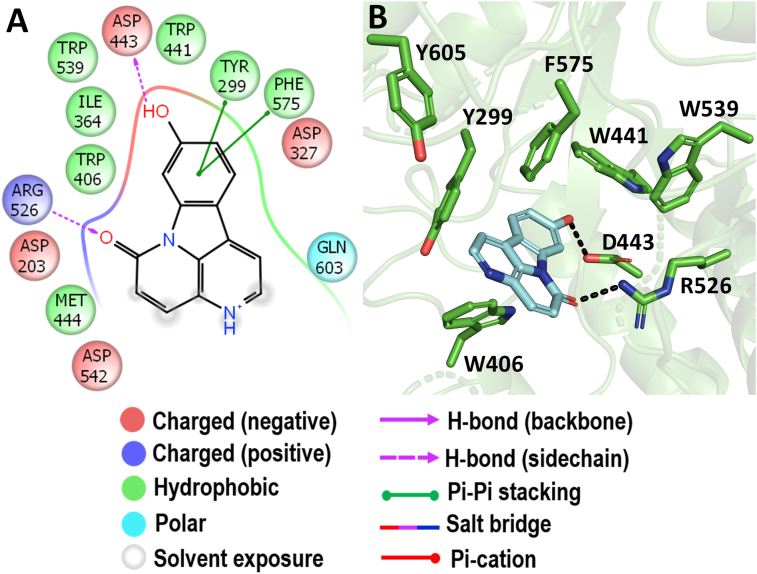
Among the Simaroubaceae compounds, the binding energy of 9-hydroxycanthin-6-one was found to be better with -6.461 kcal/mol against the Nt-MGAM enzyme as depicted in Figure 3. The compound 9-hydroxycanthin-6-one is a beta-carboline alkaloid identified in the several genera of Simaroubaceae such as Picrasma quassioides (Jiao et al., 2010), Ailanthus altissima (Jeong et al., 2018), Eurycoma longifolia (Ohishi et al., 2015), Simarouba berterona (Devkota et al., 2014), etc. The active site residues D443 and R526, which are in line with the -1 and +1 sugar-binding subsite of Nt-MGAM respectively (Sim et al., 2008) have been involved in the hydrogen bond interaction with the indole ring of 9-hydroxycanthin-6-one (Figure 7). Also, the indole ring has shown the Pi-Pi interaction with Y299 and F575 residues (−1 subsite) of the active site. This indicates that the compound may be held firmly within the -1 subsite of Nt-MGAM active site cleft.
Figure 7.
Molecular docking analysis of 9-hydroxycanthin-6-one. Interaction of 9-hydroxycanthin-6-one against Nt-MGAM in 2D view (A) and 3D view (B). 9-hydroxycanthin-6-one showed the hydrogen bond interaction with D443 which acts as a nucleophile during the hydrolysis of the substrates. In 3D view, the hydrogen bond interactions are represented in black coloured broken lines.
The docking study of acarbose showed that the residues such as D203, D443, D542, Q603, and Y605 of Nt-MGAM were involved in the hydrogen bond interactions with the acarviosine unit, which are comparatively similar as observed in the crystal structure of acarbose and Nt-MGAM complex (Sim et al., 2008). Further, the single hydrogen bond formed between R334 and glycone ring and the hydrophobic residue W406 which line the +1 active site of Nt-MGAM may help in further stacking of the acarbose (Figure 8A). The present docking study of miglitol with Nt-MGAM also showed similar interactions through the residues D327, D443, D542, R526, and H600 (Figure 8B) as found in the crystal structure of Nt-MGAM and miglitol complex (Sim et al., 2010a). The lack of hydrogen bond between miglitol and D203 is in line with the crystal structure of the Nt-MGAM-miglitol complex, and unlike acarbose, the miglitol was not extended towards the +1 subsite of the active site cleft (Sim et al., 2010a). Interestingly, the interaction of voglibose through the residues D203, D327, D542, R526, and H600 (Figure 8C) was comparatively similar as observed in Nt-MGAM- acarbose and Nt-MGAM-salacinol complexes (Sim et al., 2010a). Further, the 9-hydroxycanthin-6-one and all the AGIs (standards) were stabilized in the active site due to hydrophobic interactions with aromatic residues.
Figure 8.
Molecular docking analysis of acarbose (A), miglitol (B) and voglibose (C) against Nt-MGAM. Interactions are represented in 2D view. All the three standards have shown the interactions with D443 and D542 which acts as a nucleophile and an acid/base catalyst during the hydrolysis of substrates.
One of the recent studies by Miran Jeong et al., 9-hydroxycanthin-6-one isolated from A. altissima was effective in inducing apoptosis on human ovarian cancer cell lines (Jeong et al., 2018). In another study, the inhibition of the Wnt signaling pathway showed the importance of 9-hydroxycanthin-6-one as a therapeutic agent to control a wide range of human diseases (Ohishi et al., 2015). The effective inhibition of nitric oxide production in lipopolysaccharide-induced RAW 264.7 macrophages showed the anti-inflammatory activity of 9-hydroxycanthin-6-one (Kim et al., 2016). Further, 9-hydroxycanthin-6-one had a cytotoxic effect on the variety of human cancer cell lines (Devkota et al., 2014; Kardono et al., 1991; Kuo et al., 2003). There are no reports on studies related to the inhibition of glucosidase enzyme by this compound. Interestingly, the present study of 9-hydroxycanthin-6-one showed a better binding affinity against Nt-MGAM when compared to acarbose and voglibose. The docking study indicates that the compound 9-hydroxycanthin-6-one can be developed into a potent inhibitor of Nt-MGAM on a further detailed investigation.
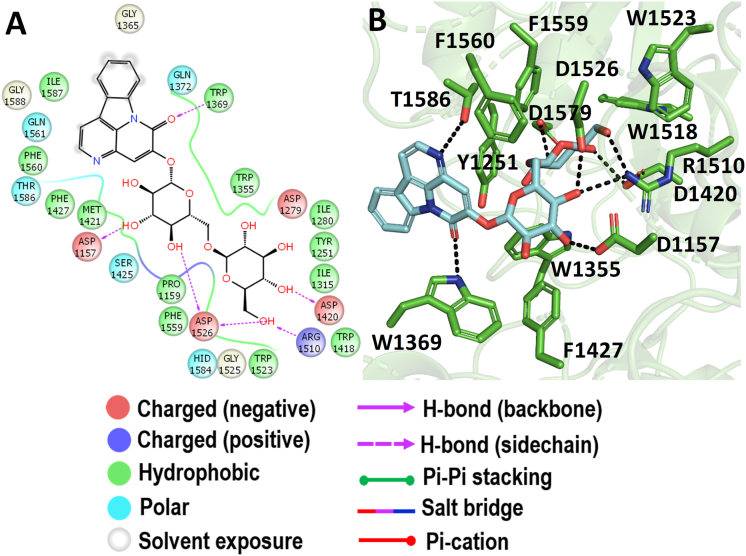
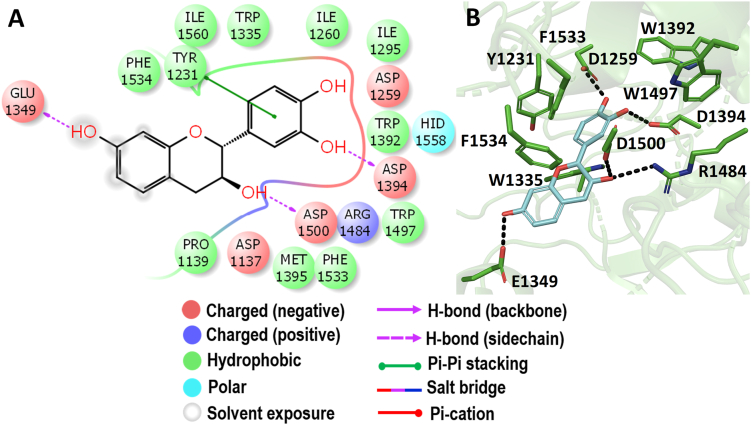
3.5. Molecular docking with Ct-MGAM
When the c-terminal of MGAM was docked with the Simaroubaceae compounds, bruceolline-B showed promising binding energy of -7.576 kcal/mol (Figure 3). Structurally, bruceolline-B is an alkaloidal glycoside consist of a glycone unit and a 5-hydroxycanthine-6-one unit which was identified in Brucea javanica. The compound bruceolline-B was analysed for its antimalarial property and found to be the only study reported on its biological activity so far (Kitagawa et al., 1994). Figure 9 represents the interaction of bruceolline-B with the active pocket, where the glycone unit showed the hydrogen bond interaction through the active site residues D1157, D1579, D1420, D1526, and R1510 of Ct-MGAM. Whereas, only two residues T1586 and W1427 have interacted through hydrogen bonds with the 5-hydroxycanthine-6-one. In the crystal structure of the Ct-MGAM-acarbose complex, the acarbose occupies all the sugar-binding subsites (−1, +1, +2, and +3) of the Ct-MGAM active site cleft (Ren et al., 2011). The bruceolline-B showed the hydrogen bond interactions as well as hydrophobic stabilization similar to the acarbose binding to Ct-MGAM. Hence, one could predict that the bruceolline-B can occupy the active site cleft of the Ct-MGAM comparatively similar mode as that of the acarbose.
Figure 9.
Molecular docking analysis of bruceolline-B. Interaction of bruceolline-B against Ct-MGAM in 2D view (A) and 3D view (B). Bruceolline-B showed the hydrogen bond interactions with D1420 and D1526 which acts as a nucleophile and an acid/base catalyst during the hydrolysis of substrates. In 3D view, the hydrogen bond interactions are represented in black coloured broken lines.
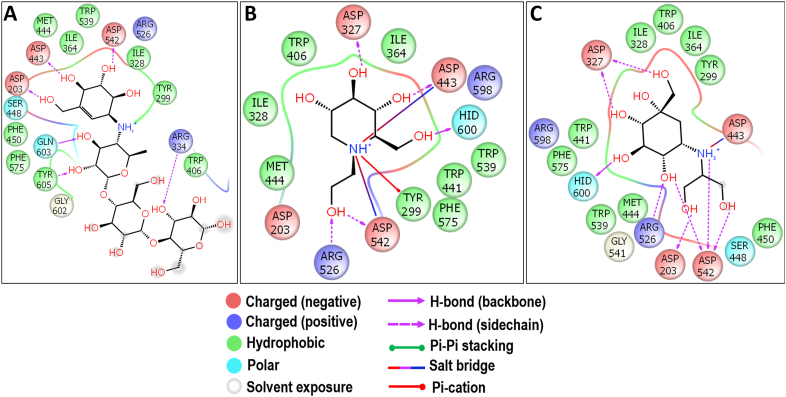
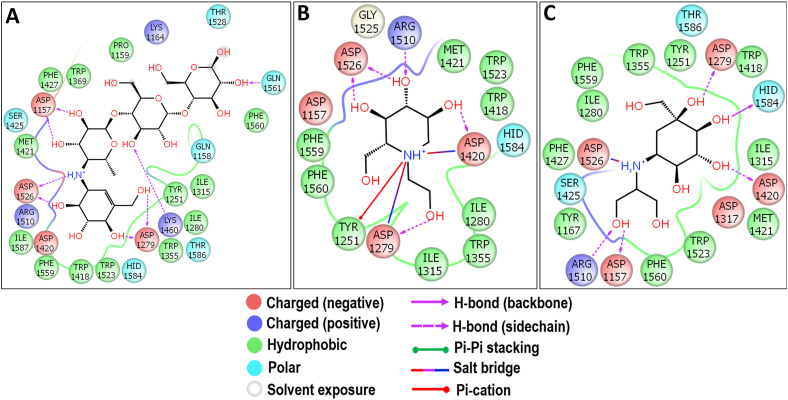
The molecular docking study of acarbose had the binding energy of -7.863 kcal/mol and showed similar interactions as that of the crystal structure of Ct-MGAM–acarbose complex (Figure 10A). Also, the docking study of miglitol and voglibose with Ct-MGAM had the binding energies of -7.610 and -6.518 kcal/mol respectively. Almost similar residues were involved in the hydrogen bond interactions of miglitol (Figure 10B) and voglibose (Figure 10C) with the Ct-MGAM, except D1157 and H1584 were the additional residues observed in the case of voglibose. It is notable that, unlike acarbose, the miglitol and voglibose have occupied the -1 and +1 subsite of Ct-MGAM active site cleft. Further, bruceolline-B, acarbose, miglitol, and voglibose were stabilized in the active site pocket of Ct-MGAM with the help of aromatic residues. The present study revealed that the binding energy of bruceolline-B can be comparable to the standard AGIs. Due to the similar mode of interactions with Ct-MGAM like acarbose, the bruceolline-B could exhibit an effective inhibition like acarbose which needs further detailed study.
Figure 10.
Molecular docking analysis of acarbose (A), miglitol (B) and voglibose (C) against Ct-MGAM. Interactions are represented in 2D view. All the three standards have shown the interactions with D1420 and D1526 which acts as a nucleophile and an acid/base catalyst during the hydrolysis of substrates.
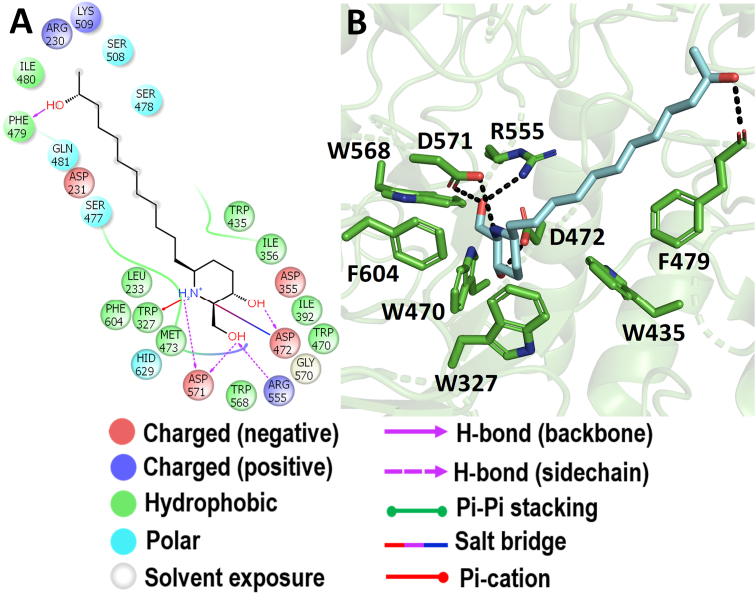
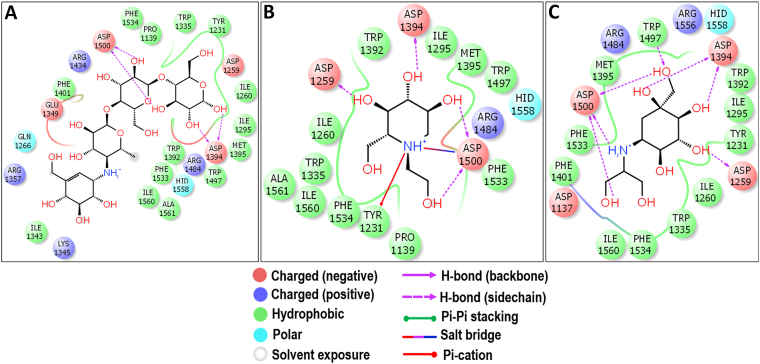
3.6. Molecular docking with Nt-SI
Among the docked compounds against Nt-SI, the prosopine showed moderate binding energy of -6.499 kcal/mol when compared to the binding energy of acarbose (−6.355 kcal/mol), miglitol (−7.202 kcal/mol), and voglibose (−6.554 kcal/mol) (Figure 3). The prosopine is a hydroxyflavan and isolated from Castelu tortuosa (Simaroubaceae) (Kubo et al., 1993), however, they have not studied its biological activity. The anti-cholinesterase activity, acaricide activity, and multiple organ toxicity were found in the alkaloid-rich extracts of Prosopis juliflora where prosopine was found to be one of the compounds identified after characterization (Lima et al., 2020; Singh and Verma, 2016). During the docking study, it was observed that prosopine was interacting through the salt bridge, hydrogen bond, hydrophobic and Pi-Pi interactions in the active site of Nt-SI as represented in Figure 11. The catalytic nucleophile, D472 interacted through a salt bridge with the –NH group and a hydrogen bond with piperidin-3-ol moiety of the prosopine which is also observed with the acarbose (Figure 12A), miglitol (Figure 12B), and voglibose (Figure 12C). Also, the –NH group of prosopine formed a Pi-Pi interaction with an aromatic residue W327 and a hydrogen bond with the acid/base catalyst D571. Similarly, all the AGIs had the hydrogen bond interaction with D571 depicts that the prosopine might have resided similar to the standard drugs in the active site cleft of Nt-SI. Additionally, the 2-hydroxymethyl moiety was stabilized through the hydrogen bond interactions of D571 and R555 along with the aromatic residue F479 which could help the stacking of 6-(11-hydroxydodecyl) moiety of prosopine through the hydrogen bond interaction in the active site of Nt-SI.
Figure 11.
Molecular docking analysis of prosopine. Interaction of prosopine against Nt-SI in 2D view (A) and 3D view (B). Prosopine showed the hydrogen bond interactions with D472 and D571 which acts as a nucleophile and an acid/base catalyst during the hydrolysis of substrates. In 3D view, the hydrogen bond interactions are represented in black coloured broken lines.
Figure 12.
Molecular docking analysis of acarbose (A), miglitol (B) and voglibose (C) against Nt-SI. Interactions are represented in 2D view. All the three standards have shown the interactions with D472 and D571 which acts as a nucleophile and an acid/base catalyst during the hydrolysis of substrates.
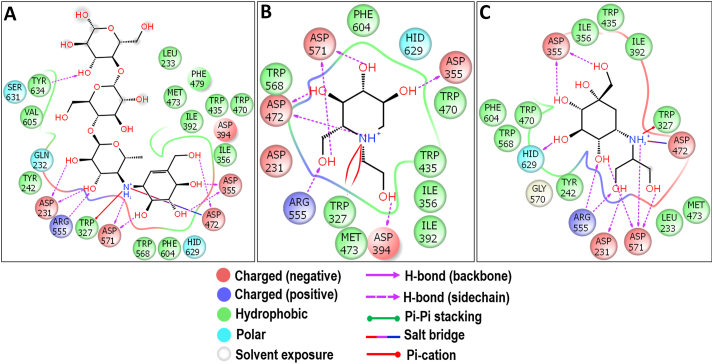
The comparison of the kotalanol binding structure of Nt-MGAM and Nt-SI revealed that both enzymes share similar sugar-binding subsites (−1 and +1) in their active site cleft. But a significant difference can be observed between +1 subsite that it is wide and open in Nt-MGAM when compared to the narrow groove of Nt-SI. Except for W327 which has the extension of its side chain towards +1 subsite, all the remaining residues involved in the interactions were lining the -1 subsite, which depicts that the prosopine might have extended towards the +1 subsite of the Nt-SI active site cleft (Sim et al., 2010b). Also, the perfect stacking of prosopine, acarbose, miglitol, and voglibose in the active site of Nt-SI may be due to the strong hydrophobic interaction of some aromatic residues.
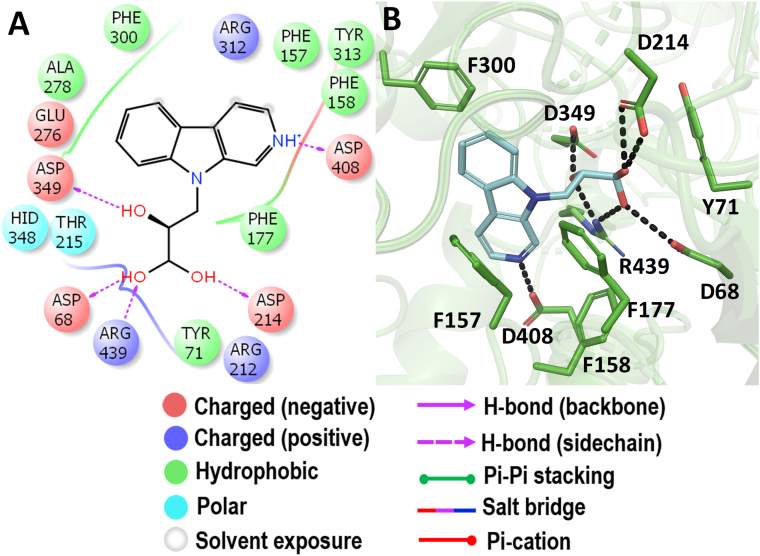
3.7. Molecular docking with Ct-SI
The molecular docking analysis of the Ct-SI enzyme was represented in Figure 13. Among the docked compounds of the Simaroubaceae family, fisetinidol showed a better binding energy of -7.575 kcal/mol (Figure 3). Since there is no crystal structure available for Ct-SI, we have compared the docking results of Ct-MGAM-bruceolline-B with Ct-S fisetinidol to know whether there is any similarity present in the active site residues that involved in the binding of bruceolline-B and fisetinidol against respective enzymes. Interestingly, the active site residues D1259, D1394, D1500, R1484, and E1349 which were involved in the hydrogen bond interaction of fisetinidol with Ct-SI have also been observed between bruceolline-B and Ct-MGAM. Further, all the active site residues except D1394 of Ct-SI–fisetinidol complex were also observed in the crystal structure of the Ct-MGAM –acarbose complex. Due to the high sequence similarity between Ct-MGAM and Ct-SI, one can predict that the Ct-SI could have a similar active site pocket and have -1, +1, +2, and +3 sugar-binding subsites as that of Ct-MGAM. Also, the comparison of the active site residues of Ct-SI-fisetinidol with Ct-MGAM-acarbose crystal structure and Ct-MGAM -bruceolline-B complex, is predictable that the fisetinidol could occupy the -1, +1, +2 subsites of Ct-SI and D1500 could act as an acid/base catalytic residue. Further, the slight difference noticed in the interaction of active site residues of the acarbose (Figure 14A), miglitol (Figure 14B), and voglibose (Figure 14C) when compared to the Ct-SI-fisetinidol complex indicates that the standard AGIs might occupied the -1 and +1 subsite of Ct-SI. Also, some of the aromatic residues have been involved in the further stabilization of fisetinidol and AGIs through hydrophobic interactions.
Figure 13.
Molecular docking analysis of fisetinidol. Interaction of fisetinidol against Ct-SI in 2D view (A) and 3D view (B). Fisetinidol showed the hydrogen bond interactions with D1394 and D1500 which acts as a nucleophile and an acid/base catalyst during the hydrolysis of substrates. In 3D view, the hydrogen bond interactions are represented in black coloured broken lines.
Figure 14.
Molecular docking analysis of acarbose (A), miglitol (B) and voglibose (C) against Ct-SI. Interactions are represented in 2D view. All the three standards have shown the interactions with D1394 and D1500 which acts as a nucleophile and an acid/base catalyst during the hydrolysis of substrates.
Fisetinidol is a tetrahydroxyflavone, identified in Castela tortuosa (Simaroubaceae) (Kubo et al., 1993). In the present study, the binding energy of fisetinidol was found to be significantly better than the standard AGIs. It is a well-known natural molecule that possesses antioxidants (Imai et al., 2008) and antibacterial activity (Da Silva et al., 2020). Additionally, fisetinidol is one of the main constituents of the acacia polyphenol which showed anti-obesity, anti-diabetic (Ikarashi et al., 2011b), lipase, and alpha-glucosidase inhibition effects (Ikarashi et al., 2011a). Thus, along with different biological activities observed in earlier studies, fisetinidol could also be considered as a potent inhibitor of Ct-SI after a thorough evaluation.
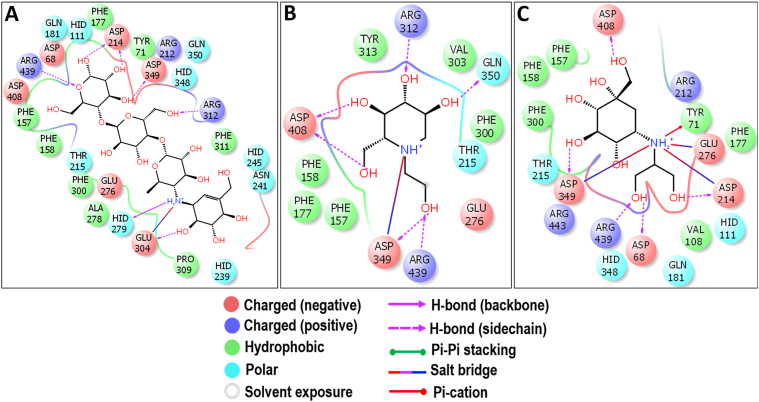
3.8. Molecular docking with YAG
Over the past few decades, demand for the AGIs have been immensely increased and many are trying to achieve this either synthetically or from natural sources. Since there is no crystal structure available for YAG, plenty of studies have been carried out by showing the inhibition against the homology models of YAG using synthetic and natural molecules in-silico (Peng et al., 2016; Rafique et al., 2020; Yan et al., 2014; Yousuf et al., 2020). The present study showed that the picrasidine-X had the best binding energy of -7.592 kcal/mol against the YAG enzyme (Figure 3). Picrasidine-X is a beta-carboline alkaloid isolated from Picrasma quassioides of the Simaroubaceae family (LI et al., 1993). The reports on the biological activity of picrasidine-X are available only from the single study where it showed the inhibitory activity against HepGP2 liver carcinoma cells (Zhao et al., 2019).
The present docking analysis revealed that the –NH group of the pyridine ring of picrasidine-X showed the hydrogen bond interaction with D408. Also, the additional hydrogen bond interactions were observed between the indole moiety and D68, D214, D349, and R439 (Figure 15). The active site residues of Saccharomyces cerevisiae isomaltase such as D215, 352, and E277 are believed to play a key role as the catalytic residues and, these correspond to D214, D349, and E276 in YAG (Tang et al., 2020). Interestingly, the picrasidine-X showed the hydrogen bond interactions with D214 and D349 making it difficult for the substrate molecules to bind to the YAG active site and thereby hinder the catalytic activity. Further, the acarbose (Figure 16A), miglitol (Figure 16B) and voglibose (Figure 16C) are also showed similar interactions with D214 and D349 along with other residues. These interactions indicate that picrasidine-X can also occupy the active site of YAG similarly to that of standard AGIs.
Figure 15.
Molecular docking analysis of picrasidine-X. Interaction of picrasidine-X against YAG in 2D view (A) and 3D view (B). Picrasidine-X showed the hydrogen bond interactions with D349, D408, D68, and D214 which could be the crucial active site residues of YAG. In 3D view, the hydrogen bond interactions are represented in black coloured broken lines.
Figure 16.
Molecular docking analysis of acarbose (A), miglitol (B) and voglibose (C) against YAG. Interactions are represented in 2D view. All the three standards have shown the interactions either with D349 or D408 or D68 or D214 which could be the crucial active site residues of YAG.
Several natural molecules such as luteolin (Yan et al., 2014), raspberry ketone (Xiong et al., 2018), and kaempferol (Peng et al., 2016) as well as some of the indole derivatives (Taha et al., 2021) and Indazole Schiff bases (Shamim et al., 2021) have stabilized in the active site pocket of YAG through hydrogen bond and hydrophobic interactions in a similar way as picrasidine-X observed in the present docking study. Further, few aromatic residues were involved in the hydrophobic interactions with picrasidine-X and standard AGIs. Interestingly, the binding energy of picrasidine-X was remarkably better than the acarbose (−6.633 kcal/mol), miglitol (−6.205 kcal/mol) and voglibose (−6.578 kcal/mol) indicate that the picrasidine-X can be a strong alternative to the available AGIs on further detailed studies.
Experimental Evidence: In the present study, the docking of Simaroubaceae compounds against the YAG enzyme resulted in a different ‘lead hit compound’ from that of the human enzymes. Interestingly, among the top 5 hit compounds of YAG enzyme, Buddlenol-A, citrusin-B, Bruceolline-B, and Prosopine were also the ‘lead hit compounds’ against HSA, HPA, Ct-MGAM, and Nt-SI respectively (Table 1). Recently, the phenolic extract from the S. glauca plant having alpha-glucosidase inhibition activity was purified and identified four compounds using the LC-MS method. Interestingly, among four identified compounds, kaempferol-3-O-pentoside was one among the top 5 ‘lead hit compounds’ against HSA, Nt-MGAM, Ct-MGAM, and Nt-SI enzymes (Mugaranja and Kulal, 2020). Another compound, kaempferol-3-O-glucoside identified by Mugaranja and Kulal (2020) was also very similar to kaempferol-3-O-pentoside in structure and binding energy as per the study. The latter compound was contributed a major portion of the active fraction and might be responsible for the highest AGI activity against YAG as seen in the in-vitro study (Mugaranja and Kulal, 2020). There was not much difference observed among the binding energy of ‘lead hit compounds’ which indicate that all these compounds could be effective in the inhibition of each enzyme docked in the present study. This is predictable that the alpha-amylase, alpha-glucosidase and YAG enzymes possess the retention of anomeric configuration as well as share the aspartic acid as a common catalytic nucleophile during the glycosidic bond hydrolysis (Davies and Henrissat, 1995; Frandsen et al., 2002). Further, during the docking study, all the ‘lead hit compounds’ were perfectly stacked and stabilized in the active site pocket of the respective enzymes which are shown in a surface representation view (Figure 17).
Table 1.
Top 5 ‘lead hit compounds’ against alpha-amylase and alpha-glucosidase enzymes.
| Enzymes | Compounds |
||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| HSA | Buddlenol-A (-7.830) | Bruceolline-B (-7.238) | Kaempferol-3-O-pentoside (-7.195) | Picrasidine-X (-7.185) | 2-hydroxy-canthin-6-one (-7.106) |
| HPA | Citrusine-B (-7.383) | Buddlenol-C (-7.102) | 2-hydroxy-canthin-6-one (-7.044) | Buddlenol-A (-7.009) | Picrasmalignane-A (-6.715) |
| Nt-MGAM | 9-hydroxycanthin-6-one (-6.461) | Picrasinoside-C (-6.25) | Kaempferol-3-O-pentoside (-6.026) | Picrasidine -X (-6.006) | Bruceolline-B (-5.962) |
| Ct-MGAM | Bruceolline-B (-7.576) | Kaempferol-3-O-pentoside (-7.563) | Buddlenol-C (-7.363) | Picrasidine-X (-7.128) | Fisetinidol (-7.116) |
| Nt-SI | Prosopine (-6.499) | Bruceolline-B (-6.206) | Buddlenol-A (-6.015) | Astragalin (-5.965) | Kaempferol-3-O-pentoside (-5.859) |
| Ct-SI | Fisetinidol (-7.575) | Picrasidin-X (-7.545) | Bruceolline-B (-7.536) | Luteolin (-7.528) | Buddlenol-A (-7.461) |
| YAG | Picrasidine-X (-7.592) | Buddlenol-A (-7.568) | Bruceolline-B (-7.514) | Prosopine (-7.481) | Citrusine-B (-7.331) |
∗Note: HSA; human salivary alpha-amylase, HPA; human pancreatic alpha-amylase, Nt-MGAM; N-terminal maltase glucoamylase, Ct-MGAM; C-terminal maltase glucoamylase, Nt-SI; N-terminal sucrase isomaltase, Ct-SI; C-terminal sucrase-isomaltase and YAG; yeast alpha-glucosidase. The binding energy (kcal/mol) for each compound was represented in the brackets.
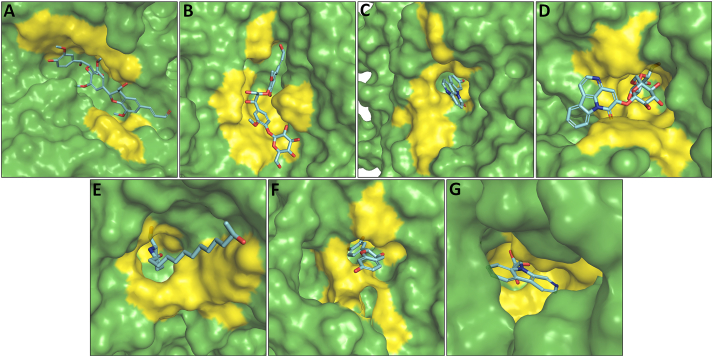
Figure 17.
Surface representation of the ‘lead hit compounds’ in the active site pocket of the enzymes: (A) Buddlenol-A, (B) Citrusin-B, (C) 9-hydroxycanthin-6-one, (D) Bruceolline-B, (E) Prosopine, (F) Fisetinidol and (G) Picrasidine-X in the active site pocket of HSA, HPA, Nt-MGAM, Ct-MGAM, Nt-SI, Ct-SI and YAG enzymes respectively. The ‘lead hit compounds’ have shown in a stick view. The yellow-coloured patches indicate the active site pocket of the enzymes.
3.9. ADME/toxicity evaluation
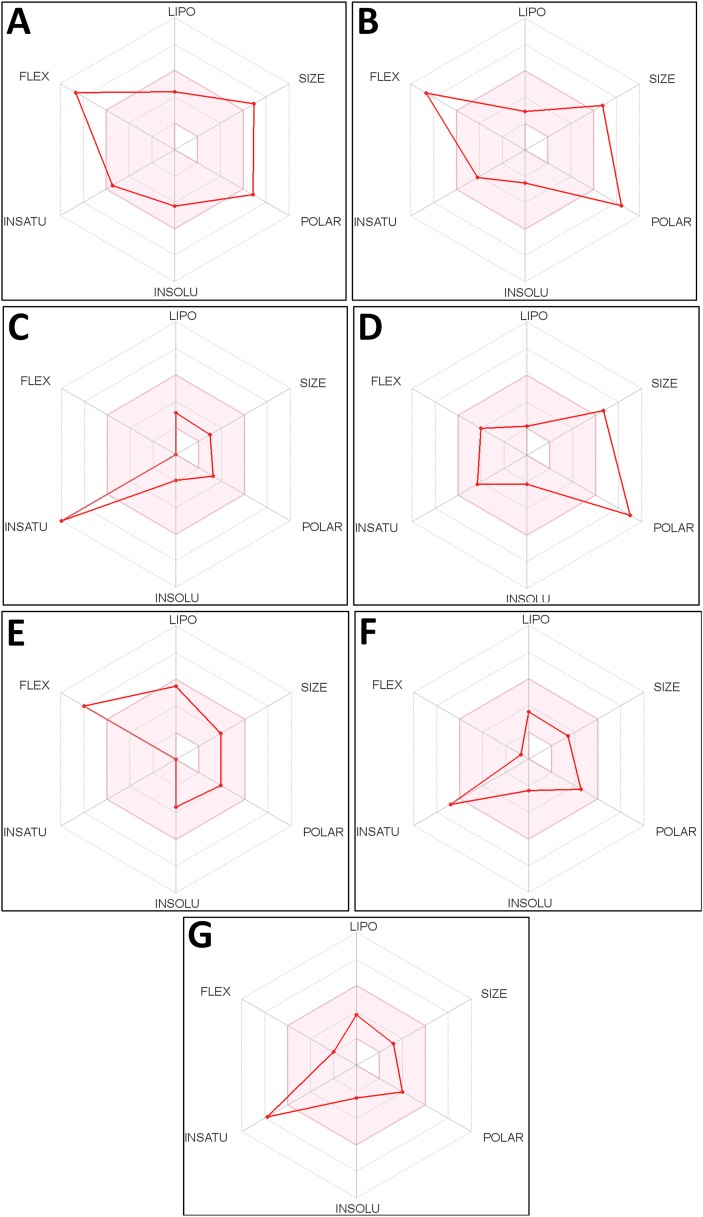
Evaluation of pharmacokinetics and pharmacodynamics properties of the ‘lead hit compounds’ before proceeding to in-vitro, in-vivo, or ex-vivo studies would help to know the real effect of the compounds, as well as it would increase the success rate of the drug discovery process. In the present study, all the seven ‘lead hit compounds’ were evaluated for ADME/Toxicity analysis. The physicochemical properties of ‘lead hit compounds’ were represented in Table 2. The evaluation of drug-likeness properties of the compounds shows that the compounds 9-hydroxycanthin-6-one, fisetinidol, and picrasidine-X have no violations for Lipinski's Rule of Five, Ghose, Veber, Egan, and Muegge criteria with the bioavailability score of 0.55, 0.55, and 0.56 respectively. The prosopine showed a bioavailability score of 0.55 with only one violation for Veber rules (Table 3). Also, the drug-likeness properties of the ‘lead hit compounds’ include lipophilicity (LIPO, consensus Log PO/W), molecular weight (SIZE), topological polar surface area (TPSA), solubility (INSOLU, Log S), saturation (INSATU), and flexibility (FLEX) can be easily understandable by analysing the Radar plots. The pink area of the Radar plots represents the optimum range of the drug-likeness properties. Interestingly, it was observed that the compounds 9-hydroxycanthin-6-one, fisetinidol, and picrasidine-X were obeyed all the properties except INSATU. The drug-likeness properties of the prosopine were also obeyed all the properties except FLEX (Figure 18, Table 3 and Table 4).
Table 2.
Physicochemical properties of the ‘lead hit compounds’ determined using Swiss ADME server.
| Compounds | Molecular formula | Molecular weight (g/mol) | No. of heavy atoms | No. of aromatic heavy atoms | Fraction Csp3 | No. of rotatable bonds | No. of H-bond acceptor | No. of H-bond donor | Molar refractivity | TPSA (Å2) |
|---|---|---|---|---|---|---|---|---|---|---|
| Buddlenol-A | C31H34O11 | 582.6 | 42 | 18 | 0.32 | 13 | 11 | 4 | 152.25 | 153.37 |
| Citrusin-B | C27H36O13 | 568.57 | 40 | 12 | 0.48 | 13 | 13 | 7 | 139.04 | 196.99 |
| 9-hydroxycanthin-6-one | C14H8N2O2 | 236.23 | 18 | 16 | 0 | 0 | 2 | 1 | 70.49 | 54.34 |
| Bruceolline-B | C26H28N2O12 | 560.51 | 40 | 16 | 0.46 | 6 | 13 | 7 | 134.19 | 212.9 |
| Prosopine | C18H37NO3 | 315.49 | 22 | 0 | 1.0 | 12 | 4 | 4 | 96.73 | 72.72 |
| Fisetinidol | C15H14O5 | 274.27 | 20 | 12 | 0.2 | 1 | 5 | 4 | 72.31 | 90.15 |
| Picrasidine-X | C14H12N2O3 | 256.26 | 19 | 13 | 0.14 | 3 | 4 | 2 | 71.05 | 75.35 |
Table 3.
Drug-likeness properties of the ‘lead hit compounds’ determined using Swiss ADME server.
| Compounds | Lipinski violations | Ghose violations | Veber violations | Egan violations | Muegge violations | Bioavailability Score |
|---|---|---|---|---|---|---|
| Buddlenol-A | 2: MW > 500, NorO>10 | 3: MW > 480, MR > 130, #atoms>70 |
2: Rotors>10, TPSA>140 |
1: TPSA>131.6 |
2: TPSA>150, H-acc>10 |
0.17 |
| Citrusin-B | 3: MW > 500, NorO>10, NHorOH>5 |
4: MW > 480, WLOGP<-0.4, MR > 130, #atoms>70 |
2: Rotors>10, TPSA>140 | 1: TPSA>131.6 |
3; TPSA>150, H-acc>10, H-don>5 |
0.17 |
| 9-hydroxycanthin-6-one | 0 | 0 | 0 | 0 | 0 | 0.55 |
| Bruceolline-B | 3: MW > 500, NorO>10, NHorOH>5 |
3: MW > 480, WLOGP<-0.4, MR > 130 |
1: TPSA>140 |
1: TPSA>131.6 |
3: TPSA>150, H-acc>10, H-don>5 |
0.17 |
| Prosopine | 0 | 0 | 1: Rotors>10 |
0 | 0 | 0.55 |
| Fisetinidol | 0 | 0 | 0 | 0 | 0 | 0.55 |
| Picrasidine-X | 0 | 0 | 0 | 0 | 0 | 0.56 |
Figure 18.
Radar plot of the ‘lead hit compounds’ obtained after analysing the ADME properties using SwissADME server. (A) buddlenol-A, (B) citrusin-B, (C) 9-hydroxycanthin-6-one, (D) bruceolline-B, (E) prosopine, (F) fisetinidol, and (G) picrasidine-X. The radar plot determines the drug-likeness properties of the compounds. The optimum range of each property is represented by the pink area of the plot. LIPO = lipophilicity (consensus Log PO/W between -0.5 and 7), SIZE = molecular weight (between 150 and 500 g/mol), POLAR = topological polar surface area (TPSA should be between 20-130 Å2, INSOLU = solubility (Log S value does not exceed -6), INSATU = saturation (fraction of carbons in the sp3 hybridization should not less than 0.25), FLEX = flexibility (number of rotatable bonds should be less than 9).
Table 4.
Lipophilicity and Water solubility properties of the ‘lead hit compounds’ determined using Swiss ADME server.
| Compounds | Lipophilicity |
Water solubility |
|||||
|---|---|---|---|---|---|---|---|
| Consensus Log PO/W | Log S (ESOL) | ESOL Class |
Log S (Ali) | Ali Class |
Log S (SILICOS-IT) | SILICOS-IT Class |
|
| Buddlenol-A | 2.62 | -4.28 | Moderately soluble | -5.03 | Moderately soluble | -5.88 | Moderately soluble |
| Citrusin B | 0.02 | -2.5 | Soluble | -3.3 | Soluble | -1.87 | Soluble |
| 9-Hydroxycanthin-6-one | 1.55 | -1.96 | Very soluble | -0.68 | Very soluble | -4.83 | Moderately soluble |
| Bruceolline-B | -1.78 | -2.16 | Soluble | -2.29 | Soluble | -0.89 | Soluble |
| Prosopine | 3.10 | -3.58 | Soluble | -5.32 | Moderately soluble | -3.77 | Soluble |
| Fisetinidol | 1.22 | -2.37 | Soluble | -2.19 | Soluble | -2.72 | Soluble |
| Picrasidine-X | 1.18 | -2.46 | Soluble | -2.33 | Soluble | -3.02 | Soluble |
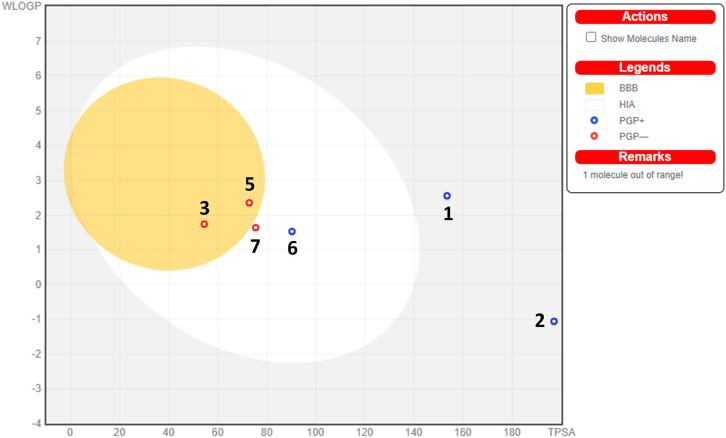
The crucial parameters of the ADME evaluation such as gastrointestinal absorption (GI), blood-brain barrier permeant (BBB), and permeability glycoprotein (P-gp) can be interpreted easily using the BOILED-Egg model (Figure 19). Even though the model relies solely on Log PO/W and TPSA, it will provide enough information for designing the drug in a variety of drug discovery settings (Daina et al., 2017). In the present study, the BOILED-Egg interpretation of the ‘lead hit compounds’ implies that the compounds 9-hydroxycanthin-6-one and prosopine have the properties of BBB permeation and acts as non-substrate for P-gp (PGP-). Further, compounds such as fisetinidol and picrasidine-X showed a high probability of GI absorption. Also, the fisetinidol acts as a substrate for P-gp (PGP+), and picrasidine-X act as a non-substrate for P-gp (PGP-). The buddlenol-A and citrusin-B which are observed in the grey region of the BOILED-Egg model and have blue coloured circles indicate that both the compounds are neither absorbed by GI nor a BBB permeant as well as they will be actively effluxes by P-gp (PGP+) (Figure 19 and Table 5).
Figure 19.
The BOILED-Egg model of the ‘lead hit compounds’ was obtained after analysing the ADME properties using the SwissADME server. The model helps to understand the pharmacokinetic properties such as gastrointestinal absorption (GI), blood-brain barrier permeant (BBB), and permeability glycoprotein (P-gp) of the compounds in an easy way. The white region represents that the molecules have a high probability of GI absorption and the yellow (yolk) region represents the compounds that have a high probability of BBB penetration. The compounds which have denoted as PGP + if they actively efflux by P-gp and are represented in a blue coloured circle. Otherwise, the compounds which have denoted as PGP- if they are the non-substrates for P-gp and represented in a red coloured circle. The compounds have been considered as not absorbed by GI and not a BBB permeant if they are located outside of the egg. The numbers inside the figure corresponds to the different compounds i.e., 1 (buddlenol-A), 2 (citrusin-B), 3 (9-hydroxycanthin-6-one), 5 (prosopine), 6 (fisetinidol) and 7 (picrasidine-X).
Table 5.
Pharmacokinetics properties of the ‘lead hit compounds’ determined using Swiss ADME server.
| Compounds | GI absorption | BBB permeant | P-gp substrate | Skin permeation (Log Kp) (cm/s) | CYP1A2 inhibitor | CYP2C19 inhibitor | CYP2C9 inhibitor | CYP2D6 inhibitor | CYP3A4 inhibitor |
|---|---|---|---|---|---|---|---|---|---|
| Buddlenol-A | Low | No | Yes | -8.31 | No | No | No | No | No |
| Citrusin-B | Low | No | Yes | -10.03 | No | No | No | No | No |
| 9-Hydroxycanthin-6-one | High | Yes | No | -7.75 | Yes | No | No | No | No |
| Bruceolline-B | Low | No | Yes | -10.9 | No | No | No | No | No |
| Prosopine | High | Yes | No | -5.32 | No | No | No | Yes | No |
| Fisetinidol | High | No | Yes | -7.46 | No | No | No | No | No |
| Picrasidine-X | High | No | No | -7.05 | No | No | No | No | No |
The cytochrome P450 (CYP) enzymes play a vital role in the processing of small molecules to improve the protection of tissues and organisms, metabolism of xenobiotic, and the elimination of drugs via metabolic transformation (van Waterschoot and Schinkel, 2011). Thus, the presence of an inhibitor or inducer molecules among the lead hits will hinder the normal action of CYP enzymes which results in the pharmacokinetics-related drug-drug interactions and leads to toxic or other unwanted adverse effects (Huang et al., 2008). Interestingly, among the seven ‘lead hit compounds’, only 9-hydroxycanthin-6-one (CYP1A2 inhibitor) and prosopine (CYP2D6 inhibitor) showed the probable inhibition of some of the CYP isoforms (Table 5).
The section of medicinal chemistry in the SwissADME server reveals whether or not the compounds have any substructures which show the false potent activity irrespective of the protein targets (PAINS alert) (Baell and Holloway, 2010) as well as the presence of problematic fragments with metabolically unstable, putative toxic, chemically reactive, or poor pharmacokinetic properties (Brenk alerts) (Brenk et al., 2008). Surprisingly, none of the compounds has violated the PAINS rule except fisetinidol (1 alert). But, among the 7 compounds buddlenol-A and fisetinidol have shown Brenk alerts. Also, no lead likeness (moieties suitable for optimization) violations have been observed for fisetinidol and picrasidine-X. Further, the predicted synthetic accessibility values for each compound present in the range of 2.02–5.98, which indicates that the compounds may have a moderately complex synthetic route (Table 6).
Table 6.
Medicinal chemistry properties of the ‘lead hit compounds’ determined using Swiss ADME server.
| Compounds | PAINS alerts | Brenk alerts | Leadlikeness violations | Synthetic Accessibility |
|---|---|---|---|---|
| Buddlenol-A | 0 | 2: aldehyde, michael_acceptor_1 | 2: MW > 350, Rotors>7 |
5.56 |
| Citrusin-B | 0 | 0 | 2: MW > 350, Rotors>7 |
5.98 |
| 9-hydroxycanthin-6-one | 0 | 0 | 1: MW < 250 |
2.02 |
| Bruceolline-B | 0 | 0 | 1: MW > 350 |
5.92 |
| Prosopine | 0 | 0 | 2: Rotors>7, XLOGP3>3.5 |
3.84 |
| Fisetinidol | 1: catechol_A | 1: catechol | 0 | 3.44 |
| Picrasidine-X | 0 | 0 | 0 | 2.44 |
Along with the ADME properties, the ‘lead hit compounds’ have been analysed for toxicity evaluation using the admetSAR server. No compounds have carcinogenic activity and, only the 9-hydroxycanthin-6-one showed positive for AMES toxicity. Also, all the compounds have been categorized under class III (LD50 > 500 mg/kg ≤ 5000 mg/kg) of the acute oral toxicity except fisetinidol which fall under class II (LD50 > 50 mg/kg ≤ 500 mg/kg) and all the compounds have been predicted as not readily biodegradable. Besides, all the compounds have shown relatively similar Rat acute toxicity where the prosopine (LD50, = 1.93 mol/kg) and buddlenol-A (LD50, = 2.68 mol/kg) showed highest and lowest Rat acute toxicity respectively when comparing to the remaining compounds (Table 7).
Table 7.
Toxicity evaluation of the ‘lead hit compounds’ determined using admetSAR server.
| Compound | AMES Toxicity | Carcinogens | Biodegradation | Acute Oral Toxicity (class) | Rat Acute Toxicity (LD50, mol/kg) |
|---|---|---|---|---|---|
| Buddlenol-A | No (0.77) | No (0.91) | Not readily biodegradable (0.97) | III (0.61) | 2.68 |
| Citrusin-B | No (0.79) | No (0.94) | Not readily biodegradable (0.74) | III (0.74) | 2.26 |
| 9-hydroxycanthin-6-one | Yes(0.71) | No (0.93) | Not readily biodegradable (0.96) | III (0.46) | 1.97 |
| Bruceolline-B | No (0.72) | No (0.94) | Not readily biodegradable (0.96) | III (0.53) | 2.24 |
| Prosopine | No (0.87) | No (0.97) | Not readily biodegradable (0.70) | III (0.71) | 1.93 |
| Fisetinidol | No (0.79) | No (0.93) | Not readily biodegradable (0.89) | II (0.33) | 2.48 |
| Picrasidine-X | No (0.65) | No (0.93) | Not readily biodegradable (0.89) | III (0.68) | 2.31 |
Taken together, none of the ‘lead hit compounds’ evaluated in the present study have exactly obeyed all the laws of ADME/Toxicity properties. Irrespective of the results obtained, there are always numerous routes to modify them for better efficacy against the respective protein targets. This study has given a glimpse of different properties of ‘lead hit compounds’ so that further studies would be required before considering them as therapeutics to treat T2D.
4. Conclusion
Despite the compounds including ‘quassinoids’ from different plants of Simaroubaceae which were reported for numerous biological properties, this is the first report on the in-silico analysis targeting the inhibition of starch hydrolysing enzymes. The binding affinity of buddlenol-A, citrusin-B, fisetinidol, and picrasidine-X against HSA, HPA, Ct-SI, and YAG respectively was found to be better than the acarbose, miglitol, and voglibose. The stacking of ‘lead hit compounds’ was found to be mainly due to the hydrogen bond interactions with the main active site residues as well as with the residues which act as the catalytic nucleophile and acid/base catalysts. Nonetheless, hydrophobic interactions, Pi-Pi stacking, Pi-cation interactions have also been helped in the stabilization of the compounds in the active site pocket of respective enzymes. It has been previously noticed that the direct correlation between in-silico and in-vitro or in-vivo analysis cannot always be established because there could be a considerable difference in the way that the ‘lead hit compounds’ could perform differently when subjected to in-vitro or in biological systems. Even though the ‘lead hit compounds’ satisfied some of the rules, it is always been important that the compounds should obey all the principles of ADME/Toxicity properties to proceed with them for further studies. Hence, the present study suggests that the ‘lead hit compounds’ identified from the array of Simaroubaceae compounds can be potent alpha-amylase and alpha-glucosidase enzyme inhibitors to treat T2D only after the necessary structural optimizations along with in-vitro and in-vivo evaluations.
Declarations
Author contribution statement
Kirana P Mugaranja: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Ananda Kulal: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
Kirana P Mugaranja was supported by Admar Mutt Education Foundation (AMEF).
Data availability statement
Data will be made available on request.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors of this manuscript would like to thank the Vision Group on Science and Technology (VGST), Govt. of Karnataka, India (GRD No.191 and GRD No. 535) and AMEF for the research facility.
References
- Aamir M., Singh V.K., Dubey M.K., Meena M., Kashyap S.P., Katari S.K., Upadhyay R.S., Umamaheswari A., Singh S. In silico prediction, characterization, molecular docking, and dynamic studies on fungal SDRs as novel targets for searching potential fungicides against Fusarium wilt in tomato. Front. Pharmacol. 2018;9:1038. doi: 10.3389/fphar.2018.01038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagappan M., Jiang D., Denko N., Koong A.C. Tumor Microenvironment. Vol. 899. Springer; 2016. A multimodal data analysis approach for targeted drug discovery involving topological data analysis (TDA) pp. 253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S., Khan F. Virtual screening, docking, ADMET and system pharmacology studies on Garcinia caged xanthone derivatives for anticancer activity. Sci. Rep. 2018;8(1):1–16. doi: 10.1038/s41598-018-23768-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves I.A., Miranda H.M., Soares L.A., Randau K.P. Simaroubaceae family: botany, chemical composition and biological activities. Rev. Brasil. Farmacogn. 2014;24(4):481–501. [Google Scholar]
- Baell J.B., Holloway G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010;53(7):2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- Bedikian A., Valdivieso M., Bodey G., Murphy W., Freireich E. Initial clinical studies with bruceantin. Cancer Treat Rep. 1979;63(11-12):1843–1847. [PubMed] [Google Scholar]
- Brayer G.D., Sidhu G., Maurus R., Rydberg E.H., Braun C., Wang Y., Nguyen N.T., Overall C.M., Withers S.G. Subsite mapping of the human pancreatic α-amylase active site through structural, kinetic, and mutagenesis techniques. Biochemistry. 2000;39(16):4778–4791. doi: 10.1021/bi9921182. [DOI] [PubMed] [Google Scholar]
- Brenk R., Schipani A., James D., Krasowski A., Gilbert I.H., Frearson J., Wyatt P.G. Lessons learnt from assembling screening libraries for drug discovery for neglected diseases. ChemMedChem. 2008;3(3):435. doi: 10.1002/cmdc.200700139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell L.K., Baker D.E., Campbell R.K. Miglitol: assessment of its role in the treatment of patients with diabetes mellitus. Ann. Pharmacother. 2000;34(11):1291–1301. doi: 10.1345/aph.19269. [DOI] [PubMed] [Google Scholar]
- Da Silva H.C., Leal A.L.A.B., de Oliveira M.M., Barreto H.M., Coutinho H.D.M., dos Santos H.S., Santiago G.M.P., de Freitas T.S., Lima I.K.C., Teixeira A.M.R. Structural characterization, antibacterial activity and NorA efflux pump inhibition of flavonoid fisetinidol. South Afr. J. Bot. 2020;132:140–145. [Google Scholar]
- Dabhi A.S., Bhatt N.R., Shah M.J. Voglibose: an alpha glucosidase inhibitor. J. Clin. Diagn. Res.: J. Clin. Diagn. Res. 2013;7(12):3023–3027. doi: 10.7860/JCDR/2013/6373.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahar D., Rai A. Screening of antimicrobial and antioxidant potentials of some medicinal plants from Simaroubaceae family. J. Pharmacogn. Phytochem. 2019;8(4):2576–2579. [Google Scholar]
- Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G., Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3(9):853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- De Souza G.R., De-Oliveira A.C.A., Soares V., Chagas L.F., Barbi N.S., Paumgartten F.J.R., da Silva A.J.R. Chemical profile, liver protective effects and analgesic properties of a Solanum paniculatum leaf extract. Biomed. Pharmacother. 2019;110:129–138. doi: 10.1016/j.biopha.2018.11.036. [DOI] [PubMed] [Google Scholar]
- Devkota K.P., Wilson J.A., Henrich C.J., McMahon J.B., Reilly K.M., Beutler J.A. Compounds from Simarouba berteroana which inhibit proliferation of NF1-defective cancer cells. Phytochem. Lett. 2014;7:42–45. doi: 10.1016/j.phytol.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen T.P., Palcic M.M., Svensson B. Substrate recognition by three family 13 yeast α-glucosidases: evaluation of deoxygenated and conformationally biased isomaltosides. Eur. J. Biochem. 2002;269(2):728–734. doi: 10.1046/j.0014-2956.2001.02714.x. [DOI] [PubMed] [Google Scholar]
- Ganesan M., Raja K.K., Narasimhan K., Murugesan S., Kumar B.K. Design, synthesis, α-amylase inhibition and in silico docking study of novel quinoline bearing proline derivatives. J. Mol. Struct. 2020;1208:127873. [Google Scholar]
- Huang S.M., Strong J.M., Zhang L., Reynolds K.S., Nallani S., Temple R., Abraham S., Habet S.A., Baweja R.K., Burckart G.J. New era in drug interaction evaluation: US Food and Drug Administration update on CYP enzymes, transporters, and the guidance process. J. Clin. Pharmacol. 2008;48(6):662–670. doi: 10.1177/0091270007312153. [DOI] [PubMed] [Google Scholar]
- Husain G.M., Singh P.N., Singh R.K., Kumar V. Antidiabetic activity of standardized extract of Quassia amara in nicotinamide–streptozotocin-induced diabetic rats. Phytother Res. 2011;25(12):1806–1812. doi: 10.1002/ptr.3491. [DOI] [PubMed] [Google Scholar]
- Hwang T.J., Carpenter D., Lauffenburger J.C., Wang B., Franklin J.M., Kesselheim A.S. Failure of investigational drugs in late-stage clinical development and publication of trial results. JAMA Intern. Med. 2016;176(12):1826–1833. doi: 10.1001/jamainternmed.2016.6008. [DOI] [PubMed] [Google Scholar]
- Ikarashi N., Takeda R., Ito K., Ochiai W., Sugiyama K. The inhibition of lipase and glucosidase activities by acacia polyphenol. Evid. base Compl. Alternative Med. 2011;2011 doi: 10.1093/ecam/neq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikarashi N., Toda T., Okaniwa T., Ito K., Ochiai W., Sugiyama K. Anti-obesity and anti-diabetic effects of acacia polyphenol in obese diabetic KKAy mice fed high-fat diet. Evid. base Compl. Alternative Med. 2011;2011 doi: 10.1093/ecam/nep241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T., Inoue S., Ohdaira N., Matsushita Y., Suzuki R., Sakurai M., de Jesus J.M.H., Ozaki S.K., Finger Z., Fukushima K. Heartwood extractives from the Amazonian trees Dipteryx odorata, Hymenaea courbaril, and Astronium lecointei and their antioxidant activities. J. Wood Sci. 2008;54(6):470–475. [Google Scholar]
- Jeong M., Kim H.M., Ahn J.-H., Lee K.-T., Jang D.S., Choi J.-H. 9-hydroxycanthin-6-one isolated from stem bark of Ailanthus altissima induces ovarian cancer cell apoptosis and inhibits the activation of tumor-associated macrophages. Chem. Biol. Interact. 2018;280:99–108. doi: 10.1016/j.cbi.2017.12.011. [DOI] [PubMed] [Google Scholar]
- Jhong C.H., Riyaphan J., Lin S.H., Chia Y.C., Weng C.F. Screening alpha-glucosidase and alpha-amylase inhibitors from natural compounds by molecular docking in silico. Biofactors. 2015;41(4):242–251. doi: 10.1002/biof.1219. [DOI] [PubMed] [Google Scholar]
- Jiao W.H., Gao H., Li C.Y., Zhou G.X., Kitanaka S., Ohmura A., Yao X.S. β-Carboline alkaloids from the stems of Picrasma quassioides. Magn. Reson. Chem. 2010;48(6):490–495. doi: 10.1002/mrc.2602. [DOI] [PubMed] [Google Scholar]
- Jiao W.H., Gao H., Zhao F., He F., Zhou G.X., Yao X.S. A new neolignan and a new sesterterpenoid from the stems of Picrasma quassioides Bennet. Chem. Biodivers. 2011;8(6):1163–1169. doi: 10.1002/cbdv.201000137. [DOI] [PubMed] [Google Scholar]
- Jones K., Sim L., Mohan S., Kumarasamy J., Liu H., Avery S., Naim H.Y., Quezada-Calvillo R., Nichols B.L., Pinto B.M. Mapping the intestinal alpha-glucogenic enzyme specificities of starch digesting maltase-glucoamylase and sucrase-isomaltase. Bioorg. Med. Chem. 2011;19(13):3929–3934. doi: 10.1016/j.bmc.2011.05.033. [DOI] [PubMed] [Google Scholar]
- Kahn S.E., Cooper M.E., Del Prato S. Pathophysiology and treatment of type 2 diabetes: perspectives on the past, present, and future. Lancet. 2014;383(9922):1068–1083. doi: 10.1016/S0140-6736(13)62154-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapetanovic I. Computer-aided drug discovery and development (CADDD): in silico-chemico-biological approach. Chem. Biol. Interact. 2008;171(2):165–176. doi: 10.1016/j.cbi.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardono L.B., Angerhofer C.K., Tsauri S., Padmawinata K., Pezzuto J.M., Kinghorn A.D. Cytotoxic and antimalarial constituents of the roots of Eurycoma longifolia. J. Nat. Prod. 1991;54(5):1360–1367. doi: 10.1021/np50077a020. [DOI] [PubMed] [Google Scholar]
- Kim E., Baker C., Dwyer M., Murcko M., Rao B., Tung R., Navia M. Crystal structure of HIV-1 protease in complex with VX-478, a potent and orally bioavailable inhibitor of the enzyme. J. Am. Chem. Soc. 1995;117(3):1181–1182. [Google Scholar]
- Kim H.M., Lee J.S., Sezirahiga J., Kwon J., Jeong M., Lee D., Choi J.-H., Jang D.S. A new canthinone-type alkaloid isolated from Ailanthus altissima Swingle. Molecules. 2016;21(5):642. doi: 10.3390/molecules21050642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa I., Mahmud T., Simanjunatak P., Hori K., Uji T., Shibuya H. Indonesian medicinal plants. VIII. Chemical structures of three new triterpenoids, bruceajavanin A, dihydrobruceajavanin A, and bruceajavanin B, and a new alkaloidal glycoside, bruceacanthinoside, from the stems of Brucea javanica (Simaroubaceae) Chem. Pharm. Bull. 1994;42(7):1416–1421. doi: 10.1248/cpb.42.1416. [DOI] [PubMed] [Google Scholar]
- Kubo I., Murai Y., Chaudhuri S.K. Castelalin, a quassinoid from Castela tortuosa. Phytochemistry. 1993;33(2):461–463. [Google Scholar]
- Kuo P.-C., Shi L.-S., Damu A.G., Su C.-R., Huang C.-H., Ke C.-H., Wu J.-B., Lin A.-J., Bastow K.F., Lee K.-H. Cytotoxic and antimalarial β-carboline alkaloids from the roots of Eurycoma longifolia. J. Nat. Prod. 2003;66(10):1324–1327. doi: 10.1021/np030277n. [DOI] [PubMed] [Google Scholar]
- Leelananda S.P., Lindert S. Computational methods in drug discovery. Beilstein J. Org. Chem. 2016;12(1):2694–2718. doi: 10.3762/bjoc.12.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.-Y., Koike K., Ohmoto T. New alkaloids, picrasidines W, X and Y, from Picrasma quassioides and X-ray crystallographic analysis of picrasidine Q. Chem. Pharm. Bull. 1993;41(10):1807–1811. [Google Scholar]
- Lima H.G.d., Santos F.O., Santos A.C.V., Silva G.D.d., Santos R.J.d., Carneiro K.d.O., Reis I.M.A., Estrela I.d.O., Freitas H.F.d., Bahiense T.C. Anti-tick effect and cholinesterase inhibition caused by Prosopis juliflora alkaloids: in vitro and in silico studies. Rev. Bras. Parasitol. Vet. 2020;29(2) doi: 10.1590/S1984-29612020036. [DOI] [PubMed] [Google Scholar]
- Lo Piparo E., Scheib H., Frei N., Williamson G., Grigorov M., Chou C.J. Flavonoids for controlling starch digestion: structural requirements for inhibiting human α-amylase. J. Med. Chem. 2008;51(12):3555–3561. doi: 10.1021/jm800115x. [DOI] [PubMed] [Google Scholar]
- Miao M., Jiang B., Jiang H., Zhang T., Li X. Interaction mechanism between green tea extract and human α-amylase for reducing starch digestion. Food Chem. 2015;186:20–25. doi: 10.1016/j.foodchem.2015.02.049. [DOI] [PubMed] [Google Scholar]
- Mugaranja K.P., Kulal A. Alpha glucosidase inhibition activity of phenolic fraction from Simarouba glauca: an in-vitro, in-silico and kinetic study. Heliyon. 2020;6(7) doi: 10.1016/j.heliyon.2020.e04392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad I., Bedir E., Khan S.I., Tekwani B.L., Khan I.A., Takamatsu S., Pelletier J., Walker L.A. A new antimalarial quassinoid from Simaba o rinocensis. J. Nat. Prod. 2004;67(5):772–777. doi: 10.1021/np030524n. [DOI] [PubMed] [Google Scholar]
- Musyoka T.M., Kanzi A.M., Lobb K.A., Bishop Ö.T. Structure based docking and molecular dynamic studies of plasmodial cysteine proteases against a South African natural compound and its analogs. Sci. Rep. 2016;6:23690. doi: 10.1038/srep23690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik B., Gupta N., Ojha R., Singh S., Prajapati V.K., Prusty D. High throughput virtual screening reveals SARS-CoV-2 multi-target binding natural compounds to lead instant therapy for COVID-19 treatment. Int. J. Biol. Macromol. 2020;160:1–7. doi: 10.1016/j.ijbiomac.2020.05.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaou K.C. Advancing the drug discovery and development process. Angew Chem. Int. Ed. Engl. 2014;53(35):9128–9140. doi: 10.1002/anie.201404761. [DOI] [PubMed] [Google Scholar]
- NoorShahida A., Wong T.W., Choo C.Y. Hypoglycemic effect of quassinoids from Brucea javanica (L.) Merr (Simaroubaceae) seeds. J. Ethnopharmacol. 2009;124(3):586–591. doi: 10.1016/j.jep.2009.04.058. [DOI] [PubMed] [Google Scholar]
- Ohishi K., Toume K., Arai M.A., Koyano T., Kowithayakorn T., Mizoguchi T., Itoh M., Ishibashi M. 9-Hydroxycanthin-6-one, a β-carboline alkaloid from Eurycoma longifolia, is the first Wnt signal inhibitor through activation of glycogen synthase kinase 3β without depending on casein kinase 1α. J. Nat. Prod. 2015;78(5):1139–1146. doi: 10.1021/acs.jnatprod.5b00153. [DOI] [PubMed] [Google Scholar]
- Park H., Hwang K.Y., Oh K.H., Kim Y.H., Lee J.Y., Kim K. Discovery of novel α-glucosidase inhibitors based on the virtual screening with the homology-modeled protein structure. Bioorg. Med. Chem. 2008;16(1):284–292. doi: 10.1016/j.bmc.2007.09.036. [DOI] [PubMed] [Google Scholar]
- Peng X., Zhang G., Liao Y., Gong D. Inhibitory kinetics and mechanism of kaempferol on α-glucosidase. Food Chem. 2016;190:207–215. doi: 10.1016/j.foodchem.2015.05.088. [DOI] [PubMed] [Google Scholar]
- Rafique R., Khan K.M., Chigurupati S., Wadood A., Rehman A.U., Karunanidhi A., Hameed S., Taha M., Al-Rashida M. Synthesis of new indazole based dual inhibitors of α-glucosidase and α-amylase enzymes, their in vitro, in silico and kinetics studies. Bioorg. Chem. 2020;94:103195. doi: 10.1016/j.bioorg.2019.103195. [DOI] [PubMed] [Google Scholar]
- Ramasubbu N., Paloth V., Luo Y., Brayer G.D., Levine M.J. Structure of human salivary α-amylase at 1.6 Å resolution: implications for its role in the oral cavity. Acta Crystallogr. Sect. D Biol. Crystallogr. 1996;52(3):435–446. doi: 10.1107/S0907444995014119. [DOI] [PubMed] [Google Scholar]
- Ramasubbu N., Ragunath C., Mishra P.J., Thomas L.M., Gyémánt G., Kandra L. Human salivary α-amylase Trp58 situated at subsite− 2 is critical for enzyme activity. Eur. J. Biochem. 2004;271(12):2517–2529. doi: 10.1111/j.1432-1033.2004.04182.x. [DOI] [PubMed] [Google Scholar]
- Rasouli H., Hosseini-Ghazvini S.M.-B., Adibi H., Khodarahmi R. Differential α-amylase/α-glucosidase inhibitory activities of plant-derived phenolic compounds: a virtual screening perspective for the treatment of obesity and diabetes. Food Funct. 2017;8(5):1942–1954. doi: 10.1039/c7fo00220c. [DOI] [PubMed] [Google Scholar]
- Ren F.-z., Chen S.-h., Li L.-h., Zhang X.-x., Zheng Z.-h., Dong A.-h. Chemical constituents of the seed shell of Hevea brasiliensis. Chin. J. New Drugs. 2012;19 [Google Scholar]
- Ren L., Qin X., Cao X., Wang L., Bai F., Bai G., Shen Y. Structural insight into substrate specificity of human intestinal maltase-glucoamylase. Protein Cell. 2011;2(10):827–836. doi: 10.1007/s13238-011-1105-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydberg E.H., Li C., Maurus R., Overall C.M., Brayer G.D., Withers S.G. Mechanistic analyses of catalysis in human pancreatic α-amylase: detailed kinetic and structural studies of mutants of three conserved carboxylic acids. Biochemistry. 2002;41(13):4492–4502. doi: 10.1021/bi011821z. [DOI] [PubMed] [Google Scholar]
- Saeedi P., Petersohn I., Salpea P., Malanda B., Karuranga S., Unwin N., Colagiuri S., Guariguata L., Motala A.A., Ogurtsova K. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: results from the international diabetes federation diabetes atlas. Diabetes Res. Clin. Pract. 2019;157:107843. doi: 10.1016/j.diabres.2019.107843. [DOI] [PubMed] [Google Scholar]
- Saleem H., Htar T.T., Naidu R., Ahmad I., Zengin G., Ahmad M., Ahemad N. Investigations into the therapeutic effects of aerial and stem parts of Buxus papillosa CK Schneid.: in vitro chemical, biological and toxicological perspectives. J. Pharmaceut. Biomed. Anal. 2019;166:128–138. doi: 10.1016/j.jpba.2019.01.007. [DOI] [PubMed] [Google Scholar]
- Shamim S., Khan K.M., Ullah N., Mahdavi M., Faramarzi M.A., Larijani B., Salar U., Rafique R., Taha M., Perveen S. Synthesis, in vitro, and in silico evaluation of indazole Schiff bases as potential α-glucosidase inhibitors. J. Mol. Struct. 2021;130826 [Google Scholar]
- Shoichet B.K. Virtual screening of chemical libraries. Nature. 2004;432(7019):862–865. doi: 10.1038/nature03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M.A.B.d., Melo L.V.L., Ribeiro R.V., Souza J.P.M.d., Lima J.C.S., Martins D.T.d.O., Silva R.M.d. Levantamento etnobotânico de plantas utilizadas como anti-hiperlipidêmicas e anorexígenas pela população de Nova Xavantina-MT, Brasil. Rev. Brasil. Farmacogn. 2010;20(4):549–562. [Google Scholar]
- Sim L., Jayakanthan K., Mohan S., Nasi R., Johnston B.D., Pinto B.M., Rose D.R. New glucosidase inhibitors from an ayurvedic herbal treatment for type 2 diabetes: structures and inhibition of human intestinal maltase-glucoamylase with compounds from Salacia reticulata. Biochemistry. 2010;49(3):443–451. doi: 10.1021/bi9016457. [DOI] [PubMed] [Google Scholar]
- Sim L., Quezada-Calvillo R., Sterchi E.E., Nichols B.L., Rose D.R. Human intestinal maltase–glucoamylase: crystal structure of the N-terminal catalytic subunit and basis of inhibition and substrate specificity. J. Mol. Biol. 2008;375(3):782–792. doi: 10.1016/j.jmb.2007.10.069. [DOI] [PubMed] [Google Scholar]
- Sim L., Willemsma C., Mohan S., Naim H.Y., Pinto B.M., Rose D.R. Structural basis for substrate selectivity in human maltase-glucoamylase and sucrase-isomaltase N-terminal domains. J. Biol. Chem. 2010;285(23):17763–17770. doi: 10.1074/jbc.M109.078980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão S.M., Barreiros E.L., Da Silva M.F.d.G., Gottlieb O.R. Chemogeographical evolution of quassinoids in Simaroubaceae. Phytochemistry. 1991;30(3):853–865. [Google Scholar]
- Singh S., Verma S.K. Synergistic effects of the alkaloids of Prosopis juliflora, causing multiple organ toxicity in mouse model. J. Biol. Active Prod. Nature. 2016;6(4):323–336. [Google Scholar]
- Subramaniam S., Mehrotra M., Gupta D. Virtual high throughput screening (vHTS)-A perspective. Bioinformation. 2008;3(1):14. doi: 10.6026/97320630003014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.N., Li W., Song S.B., Yan X.T., Yang S.Y., Kim Y.H. NF-κB inhibitory activities of phenolic and lignan components from the stems of Acanthopanax divaricatus var. albeofructus. Nat. Prod. Sci. 2014;20:232–236. [Google Scholar]
- Taha M., Alrashedy A.S., Almandil N.B., Iqbal N., Nawaz M., Uddin N., Chigurupati S., Wadood A., Rahim F., Das S., Venugopal V. Synthesis of indole derivatives as diabetics II inhibitors and enzymatic kinetics study of α-glucosidase and α-amylase along with their in-silico study. Int. J. Biol. Macromol. 2021;190:301–318. doi: 10.1016/j.ijbiomac.2021.08.207. [DOI] [PubMed] [Google Scholar]
- Talele T.T., Khedkar S.A., Rigby A.C. Successful applications of computer aided drug discovery: moving drugs from concept to the clinic. Curr. Top. Med. Chem. 2010;10(1):127–141. doi: 10.2174/156802610790232251. [DOI] [PubMed] [Google Scholar]
- Tan Q.-W., Ouyang M.-A., Wu Z.-J. A new seco-neolignan glycoside from the root bark of Ailanthus altissima. Nat. Prod. Res. 2012;26(15):1375–1380. doi: 10.1080/14786419.2011.587187. [DOI] [PubMed] [Google Scholar]
- Tang H., Huang L., Sun C., Zhao D. Exploring the structure–activity relationship and interaction mechanism of flavonoids and α-glucosidase based on experimental analysis and molecular docking studies. Food Funct. 2020;11(4):3332–3350. doi: 10.1039/c9fo02806d. [DOI] [PubMed] [Google Scholar]
- Tripathi S.K., Muttineni R., Singh S.K. Extra precision docking, free energy calculation and molecular dynamics simulation studies of CDK2 inhibitors. J. Theor. Biol. 2013;334:87–100. doi: 10.1016/j.jtbi.2013.05.014. [DOI] [PubMed] [Google Scholar]
- Van Waterschoot R.A., Schinkel A.H. A critical analysis of the interplay between cytochrome P450 3A and P-glycoprotein: recent insights from knockout and transgenic mice. Pharmacol. Rev. 2011;63(2):390–410. doi: 10.1124/pr.110.002584. [DOI] [PubMed] [Google Scholar]
- Vieira I.J.C., Braz-Filho R. Studies in Natural Products Chemistry. Vol. 33. Elsevier; 2006. Quassinoids: structural diversity, biological activity and synthetic studies; pp. 433–492. [Google Scholar]
- Wahbeh G.T., Christie D.L. Pediatric Gastrointestinal and Liver Disease. Elsevier; 2006. Basic aspects of digestion and absorption; pp. 11–23. [Google Scholar]
- Williams L.K., Li C., Withers S.G., Brayer G.D. Order and disorder: differential structural impacts of myricetin and ethyl caffeate on human amylase, an antidiabetic target. J. Med. Chem. 2012;55(22):10177–10186. doi: 10.1021/jm301273u. [DOI] [PubMed] [Google Scholar]
- Xiong S.-L., Yue L.-M., Lim G.T., Yang J.-M., Lee J., Park Y.-D. Inhibitory effect of raspberry ketone on α-glucosidase: docking simulation integrating inhibition kinetics. Int. J. Biol. Macromol. 2018;113:212–218. doi: 10.1016/j.ijbiomac.2018.02.124. [DOI] [PubMed] [Google Scholar]
- Yan J., Zhang G., Pan J., Wang Y. α-Glucosidase inhibition by luteolin: kinetics, interaction and molecular docking. Int. J. Biol. Macromol. 2014;64:213–223. doi: 10.1016/j.ijbiomac.2013.12.007. [DOI] [PubMed] [Google Scholar]
- Yang B.-Y., Yin X., Liu Y., Sun Y., Guan W., Zhou Y.-Y., Kuang H.-X. Terpenes and lignans from the roots of Solanum melongena L. Nat. Prod. Res. 2020;34(3):359–368. doi: 10.1080/14786419.2018.1533828. [DOI] [PubMed] [Google Scholar]
- Yoshikawa K., Sugawara S., Arihara S. Phenylpropanoids and other secondary metabolites from fresh fruits of Picrasma quassioides. Phytochemistry. 1995;40(1):253–256. [Google Scholar]
- Yousuf H., Shamim S., Khan K.M., Chigurupati S., Hameed S., Khan M.N., Taha M., Arfeen M. Dihydropyridines as potential α-amylase and α-glucosidase inhibitors: synthesis, in vitro and in silico studies. Bioorg. Chem. 2020;96:103581. doi: 10.1016/j.bioorg.2020.103581. [DOI] [PubMed] [Google Scholar]
- Yuan C.-S., Sun X.-B., Zhao P.-H., Cao M.-A. Antibacterial constituents from Pedicularis armata. J. Asian Nat. Prod. Res. 2007;9(7):673–677. doi: 10.1080/10286020601103171. [DOI] [PubMed] [Google Scholar]
- Zhang D.-h., Wu K.-l., Zhang X., Deng S.-q., Peng B. In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. J. Integr. Med. 2020;18(2):152–158. doi: 10.1016/j.joim.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W.-Y., Chen J.-J., Zou C.-X., Zhou W.-Y., Yao G.-D., Wang X.-B., Lin B., Huang X.-X., Song S.-J. 2019. Effects of Enantiomerically Pure β-carboline Alkaloids from Picrasma Quassioides on Human Hepatoma Cells. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.