Abstract
Background:
Thymic stromal lymphopoietin (TSLP) is an epithelial-derived cytokine important in initiation of allergic inflammation. Single nucleotide polymorphisms (SNPs) in TSLP are associated with asthma, yet studies have shown inconsistent associations between circulating TSLP and asthma. Studies that integrate the combined effects of TSLP genotype, TSLP mRNA, circulating TSLP levels, and asthma outcome are lacking.
Objective:
To recruit a novel cohort based on asthma-relevant TSLP SNPs and determine their impact on TSLP mRNA expression and TSLP circulating protein levels, and their individual and combined effects on asthma.
Methods:
We developed an algorithm to prioritize TSLP SNPs and recruited 51 carriers and non-carriers based on TSLP genotypes. We quantified TSLP mRNA in nasal epithelial cells and circulating TSLP levels in plasma. We determined associations of defined TSLP risk genotypes and/or TSLP mRNA and protein levels with asthma.
Results:
TSLP mRNA expression, but not circulating TSLP, was significantly increased in asthmatics compared to non-asthmatics (P=0.007; OR=1.44). Notably, 90% of children with the defined TSLP risk genotypes and high nasal TSLP mRNA expression (top tertile) had asthma compared to 40% of subjects without risk genotypes and with low TSLP expression (bottom tertile) (P=0.024). No association between circulating TSLP and asthma was observed.
Conclusion:
Collectively, these data suggest childhood asthma is modified by the combined effects of TSLP genotype and TSLP expression in the nasal epithelium. The increased asthma risk likely manifests when genetic variation enables eQTL in the TSLP locus to elevate TSLP. It is important to consider both biomarkers when factoring asthma risk.
Clinical Implications:
TSLP genotype and nasal mRNA expression may be useful biomarkers to aid asthma prediction and/or identify individuals who would benefit from therapy targeting this pathway.
Keywords: TSLP, asthma, genotype, gene, biomarker, gene expression, mRNA
Capsule Summary:
Asthma risk is modified by the combined effects of TSLP genotype and TSLP expression in the nasal epithelium rather than circulating TSLP. These results suggest asthma risk is increased only when genetic variation enables eQTL in the TSLP locus to elevate TSLP mRNA expression.
INTRODUCTION
Thymic stromal lymphopoietin (TSLP) is an epithelial cell-derived immune cytokine that regulates T helper (Th) inflammatory 2 responses(1, 2). TSLP contributes to the initiation and maintenance of the allergic immune response, is a marker for defects in epidermal barrier differentiation, and is strongly implicated in the pathogenesis of atopic dermatitis (AD) and asthma(1, 3–5). Accordingly, a drug targeting TSLP as a therapeutic target in clinical trials demonstrated decreased airway inflammation and allergen sensitivity(6–9).
Genetic variation in TSLP modifies risk for allergic disease among patients carrying risk alleles in skin barrier genes(3, 10). Numerous SNPs in TSLP have been associated with AD and asthma(3, 10–24). The impact of TSLP genetic variation may be context dependent. Our group demonstrated that asthma risk partly results from independent associations of variants and epistasis between skin-related genes, TSLP and SPINK5, and that this effect differs by race(10). A recent study found that TSLP SNPs increased the risk of asthma among children with AD, especially among those who also had allergic sensitization(20). TSLP mRNA expression in skin and serum protein levels are increased in AD(25–27). TSLP mRNA expression is increased in nasal epithelial and bronchial airway tissues in asthma(28, 29). Disease-associated expression quantitative trait locus (eQTL) in the TSLP locus correlate with TSLP expression(14). These studies have been helpful in highlighting the allergic role of TSLP and in identifying potentially relevant genetic variants; however, which SNPs are the most informative in contributing to allergic disease pathogenesis and prevalence remains unclear.
Despite the strong association of TSLP genetic variation and epithelial expression with asthma and AD(14, 28–30), studies have failed to find a consistent association of circulating TSLP (a biomarker of epidermal TSLP production) with asthma and allergic disease(27, 31–34). In fact, one study of circulating TSLP found no positive association with recurrent wheezing, aeroallergen sensitization, or AD(35). This may be due to several factors. Local TSLP expression in skin cells at mucosal surfaces may be more relevant to disease than circulating levels, which may not accurately reflect local tissue levels(25, 31, 36). Genetic variation may have an impact beyond circulating TSLP protein expression, including on other genes or pathways as we demonstrated with SPINK5(10), which may impact asthma development or persistence. Thus, it is critical to simultaneously examine TSLP genotype, local expression, and circulating TSLP and determine their individual and combined impact on asthma.
Herein, we developed an unbiased algorithm to prioritize SNPs affecting TSLP gene expression levels. We then recruited a novel cohort of carriers with at least one TSLP risk allele from the identified TSLP SNPs and non-carriers with none of the identified TSLP risk alleles. Next, we systematically evaluated the individual and combined impact of TSLP genotypes, nasal mRNA expression, and circulating blood level on asthma to determine the functional and disease impact of these TSLP SNPs. We found that short form (sfTSLP) and long form TSLP (lfTSLP) expression in airway nasal epithelial cells, but not circulating TSLP levels, were increased in children with asthma compared with non-asthmatic children, suggesting local regulation of TSLP expression in the airway is important in asthma pathogenesis. Importantly, we found evidence of a combined effect of the identified TSLP risk alleles and upper airway TSLP expression such that increased sfTSLP and lfTSLP expression only associate with childhood asthma when the TSLP risk variants are present demonstrating a novel genotype-dependent contribution of TSLP expression to asthma risk. Collectively, these results suggest that TSLP SNPs and local airway expression of TSLP have synergistic contributions to asthma. The increase in asthma risk is likely manifest only when genetic variation enables eQTL in the TSLP locus to elevate lfTSLP.
METHODS
Study population
Fifty-one participants aged 6 to 18 years from the greater Cincinnati, Ohio metro area who were initially enrolled in the Greater Cincinnati Pediatric Clinic Repository (GCPCR)(37) or the Cincinnati Childhood Allergy and Air Pollution Study (CCAAPS)(38) were recruited from May to August 2019 using genotyping information available from these cohorts, as previously described(39–41), for TSLP SNPs rs2289277 and rs11466750. These SNPs were selected using the methods described in the Supplemental Materials. The GCPCR includes over 7500 patients and CCAAPS includes 762 participants. Carriers had at least one copy of the risk allele for TSLP rs2289277 or rs11466750; non-carriers had zero copies of the risk allele for both SNPs. Carriers and non-carriers were 1-to-1 matched on age (±2 years) to account for potential developmental changes. History of ever asthma and AD diagnosis were defined by medical chart review and patient report of history of doctor diagnosis. Individuals were excluded if they were born at gestational age <36 weeks, current age <6 years, or dependent on oral steroids or immunosuppressive drugs. All participants, or their legally authorized representatives (LAR), provided written informed consent for participation in the study. During the study visit, subjects/LARs completed our validated New Visit Questionnaire(37), which collects AD, allergic rhinitis, food allergy, asthma and allergy symptoms; medical history, family history of allergic disease, medications, diet, home characteristics and environmental exposures. Our Demographic Data Questionnaire(37) captured participants’ age, sex, race (American Indian/Alaskan Native, Asian, Black, Native Hawaiian, White, Other, Don’t Know), parental ethnicity, and socioeconomic status indicators. Race was categorized as Black and White. This study was approved by the Cincinnati Children’s Hospital Medical Center Institutional Review Board.
Statistical analyses were performed in R program. A p-value ≤ 0.05 was considered significant due to the correlation between TSLP measures. The full methods are available in the Supplementary Materials.
RESULTS
Prioritization of TSLP SNPs for analysis
A flow chart of SNP selection is outlined in Figure 1A and described in detail in the Supplemental Materials. Of the 670 SNPs located in the human TSLP gene based on the 1000 Genomes Project data, there were 26 SNPs with a MAF ≥0.10. We performed LD expansion (r2>0.9) to evaluate all genetic association candidates in the defined genomic region and they were all included in the 26 SNPs. Next, we selected 3 SNPs that were associated with allergic disease (including asthma, allergen sensitization, AD, and allergic rhinitis) in a published GWAS(21–24, 42–45) and 11 SNPs that were significant eQTLs in lung, skin, or esophageal mucosal tissues(46, 47) based on data accessed on or prior to March 1, 2019 (Figure 1B, Table 1). We performed genomic functional annotation for these 14 TSLP SNPs to prioritize those likely to impact TSLP expression.
Figure 1. TSLP locus and SNP prioritization strategy.
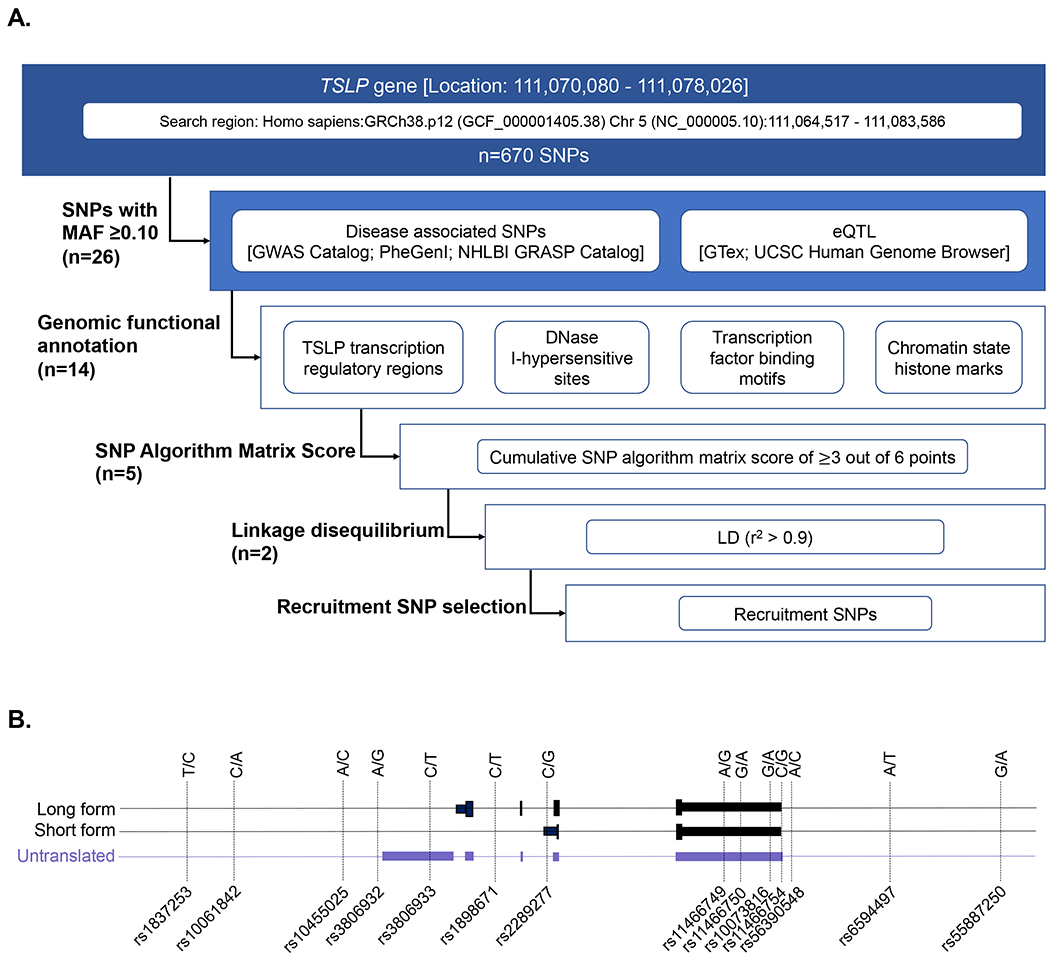
A) Flow chart of the SNP selection strategy. B) Schematic of TSLP locus and 14 SNPs of interest analyzed in discrete genomic functional annotation analyses. Genomic localization (GRCh38/hg38) of the human TSLP gene on chromosome 5q22.1 with mapped SNPs (MAF≥10%).
Table 1.
SNP algorithm analysis to calculate a cumulative score to select functionally important SNPs (n=14).
| TSLP SNP ID: | rs1837253 | rs10061842 | rs10455025 | rs3806932 | rs3806933 | rs1898671 | rs2289277 | rs11466749 | rs11466750 | rs10073816 | rs11466754 | rs56390548 | rs6594497 | rs55887250 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GWAS 1 | 1 | 1 | 1 | |||||||||||
| eQTL 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| TSLP transcription regulatory regions 3 | 1 | 1 | 1 | 1 | 1 | |||||||||
| DNase I-hypersensitive sites 4 | 1 | 1 | ||||||||||||
| Transcription factor binding motifs 5 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||
| Chromatin state histone marks 6 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||
| Total Score (out of 6) | 2 | 2 | 2 | 5 | 3 | 2 | 5 | 3 | 3 | 2 | 2 | 2 | 1 | 1 |
Includes all SNPs with MAF >0.10 in the human TSLP gene (within 5.6 kb of the 5’ flanking region of the promoter and 5.6 kb of the 3’ flanking region of the last exon) that were associated with allergic diseases (including asthma, allergy, allergen sensitization, atopic dermatitis, and allergic rhinitis) in a GWAS with a P-value <5x10−8 or that were eQTL in lung, skin (not sun-exposed), and esophageal mucosal tissues (n=14) as of March 1, 2019. SNP matrix scores ranged from 1-5, with a mean of 2.5 points (standard deviation=1.2) out of 6 points possible. rs2289277 and rs3806932 had the highest score of 5 points; rs11466749, rs11466750, rs3806933 had the second highest score of 3 points as noted with bold font.
GWAS; SNPs identified with genome-wide association with allergic disease phenotype in GWAS catalogue, Phenotype-Genotype Integrator (PheGenl), and NHLBI GRASP catalog at significance of P-value <5x10−8.
eQTL; eQTL effects with a P-value <0.001 in GTex for lung, skin (not sun-exposed), and esophageal mucosal tissues.
TSLP transcription regulatory regions; SNPs identified as being in accessible genomic regions and dynamic FAIRE-seq chromatin peaks within the TSLP gene during epidermal development and dysfunction(49, 50), and allergic disease associated SNPs in high LD (r2 = 0.8-1.0) with SNPs located in dynamic FAIRE peaks.
DNAse I hypersensitive sites; SNPs located in regions of DNase I sensitivity identified using HaploReg v4.1.
Transcription factor binding and regulation motifs; Assessment of SNPs that may modify or disrupt transcription factor regulation motifs using HaploReg v4.1 and SNP2TFBS databases.
Chromatin state histone modification; Chromatin state analysis to determine whether the identified SNPs overlapped with the major histone modifications (H3K4me1 and H3K27ac for enhancer; H3K4me3 and H3K9ac for promoter) in asthma relevant tissues using HaploReg v4.1.
TSLP transcription regulatory regions
TSLP is induced during epidermal barrier dysfunction and is involved in pathogenesis of asthma and AD(1, 48). FAIRE analyses performed by our colleagues, Lander et al., identified regulatory regions likely to regulate human TSLP expression in response to epidermal mechanical disruption or genetic dysfunction(49, 50). We performed an analysis of dynamic barrier-specific FAIRE peaks by identifying regions of open chromatin that correlated with lfTSLP expression. To select functional TSLP SNPs associated with increased asthma prevalence that are likely associated with TSLP expression, we prioritized SNPs that were located within these FAIRE peaks. TSLP rs2289277 is located in intron 2 of lfTSLP and in the sfTSLP promoter (Figure 2A).
Figure 2. Selection and use of disease-associated TSLP SNPs to define risk genotype carriers for TSLP cohort.
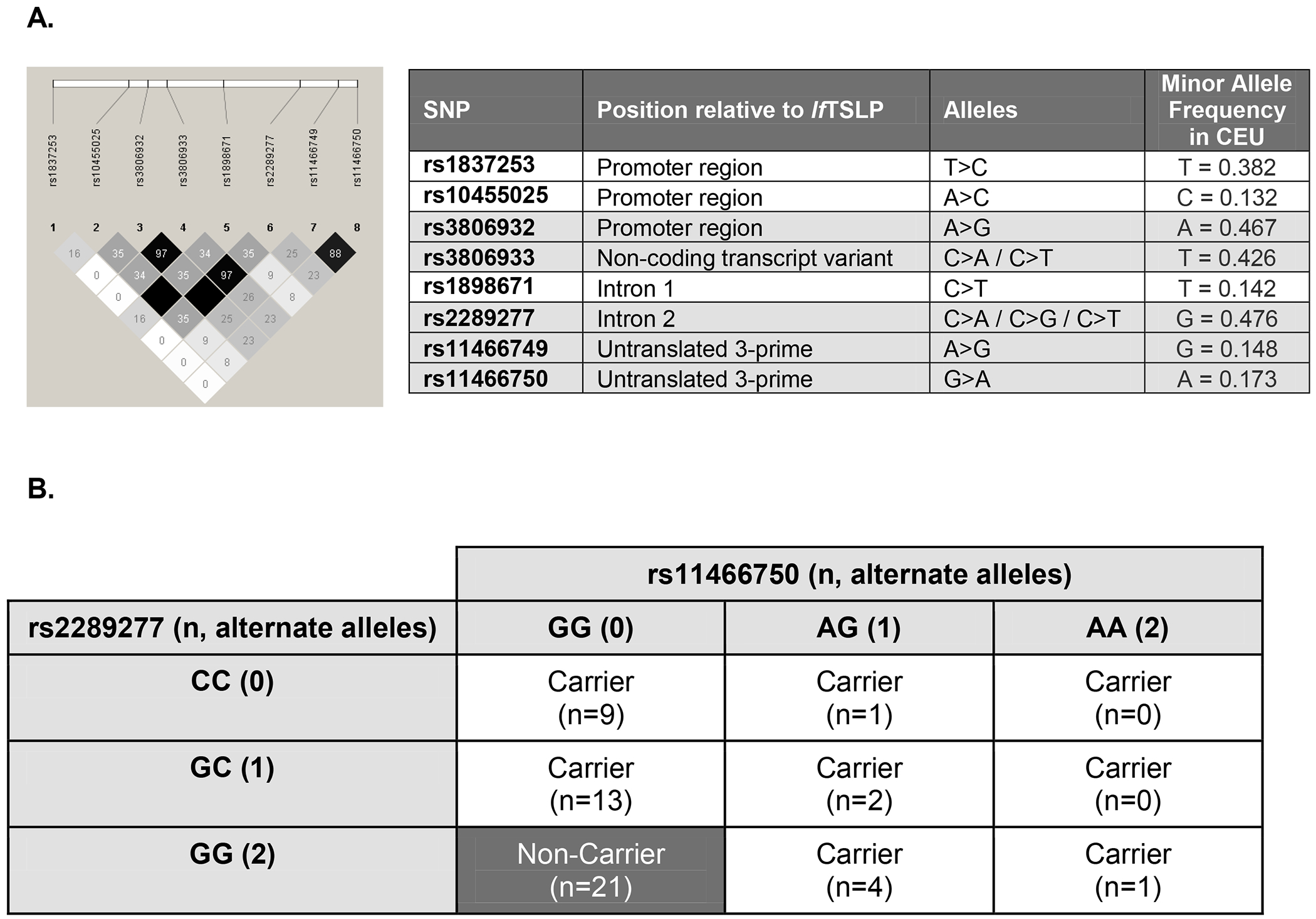
A) Linkage disequilibrium (r2) among 5 SNPs with highest scores in the SNP matrix discrete analysis and GWAS associations performed in the human TSLP gene in European descent (CEU) populations. Among these 8 SNPs, the median D’=1.0. Table includes data from dbSNP and MAF reported from 1000 Genomes. Gray shaded SNPs in the table indicate those that scored the highest in our SNP matrix analyses. B) Disease-associated TSLP SNPs rs2289277 and rs11466750 define carriers with the risk genotype associated with TSLP expression in the TSLP cohort (n=51). Of these, non-carriers (n=21) were defined using GTex data for each genotype such that rs2289277 ‘GG’ (alternate allele) and rs11466750 ‘GG’ (reference allele) have the lowest expected TSLP expression. There were n=30 carriers with a TSLP risk genotype. Carriers with the risk genotype had at least one copy of the rs2289277 (‘C’) reference allele and at least one copy of the rs11466750 (‘A’) alternate allele. The rs2289277 ‘CC’ genotype allele is associated with increased risk of allergic disease; the rs11466750 ‘AA’ genotype is associated with increased risk of allergic disease.
Haploreg in silico analyses
Histone modifications are important markers of function and chromatin state. Variants that affect chromatin at distal regulatory sites frequently also direct changes in gene expression at associated promoters(51). HaploReg analysis predicted overlap of 8 TSLP SNPs (including rs10061842, rs10455025, rs3806932, rs3806933, rs1898671, rs2289277, rs11466749, rs11466750) that might affect the histone mark of enhancers and promoters in asthma relevant tissues (see Table E1 in the Online Repository)(52).
SNPs that alter transcription factor binding motifs have been associated with multiple human diseases and may influence transcription factor binding affinity and alter gene expression and regulation. HaploReg analysis predicted altered motifs for 6 TSLP variants (rs1837253, rs3806932, rs2289277, rs10073816, rs11466754, rs56390548)(52). SNP2TFBS analysis predicted altered FOXF2 and Sox motifs associated with TSLP (rs1837253), and altered NR2C2 binding associated with rs2289277(53).
DNase I sensitivity is an indicator of open chromatin. DNase I hypersensitive sites typically mark regions that are associated with active histone marks and transcription factor binding, and often overlap with FAIRE peaks(51, 54). HaploReg identified DNase marks associated with TSLP rs2289277 and rs3806932 in asthma relevant tissues (52).
SNP algorithm scores
Using a simple algorithm that takes the above information into account, we calculated a cumulative score for 14 TSLP SNPs (Figure 1). SNPs were assigned one point for each of the functional characteristics examined (Table 1). The scores ranged from 1 to 5 [median=2 points; IQR: 2-3] out of 6 points possible. Figure 2A shows the LD among 5 TSLP SNPs with the highest scores in our SNP matrix analysis and 3 SNPs identified in GWAS associations. There is strong LD in the TSLP locus and among the 8 SNPs in Figure 2A in European descent (CEU) populations (median D’ = 1.0) suggesting shared information among these variants. Figure E1 shows the LD structure derived from African descent (ASW) populations, which exhibited weaker LD. Two SNPs (rs2289277 and rs3806932) scored 5 points and were strongly linked (r2 = 0.97). Three SNPs (rs11466749, rs11466750, rs3806933) scored 3 points. TSLP rs3806933 is in strong LD with rs2289277 and rs3806932 (r2 = 0.97 and 1.0, respectively), and rs11466749 and rs11466750 are strongly linked (r2 = 0.88). Within these two LD blocks, rs2289277 and rs11466750 each have the highest minor allele frequency. On this basis, we prioritized rs2289277 and rs11466750 in our recruitment strategy. Both of these SNPs are eQTLs and have been associated with asthma and AD(10, 13, 14, 49). TSLP rs2289277 is located in intron 2 of lfTSLP and in the promoter of sfTSLP, while rs11466750 is located in the 3’ untranslated region of lfTSLP and sfTSLP.
Demographics and clinical attributes of the TSLP cohort
To investigate the impact of genetic variation on TSLP expression and asthma diagnosis, we recruited individuals with 1 or 2 alternate alleles for TSLP disease-associated SNPs rs2289277 and/or rs11466750 as carriers using defined risk genotypes and age-matched non-carriers who had 2 copies of the reference allele at both SNPs (n=51, Figure 2B). The cohort included individuals of median age 16.1 years [range: 10.5-17.9 years], majority male sex (54.9%) and white race (58.8%) (Table 2). Carriers with the TSLP risk genotypes had a nominally higher prevalence of asthma diagnosis (p=0.064) and significantly higher prevalence of having both asthma and AD diagnosis (p=0.022). Carriers and non-carriers did not differ significantly by age, sex, or race.
Table 2.
Demographic and clinical characteristics of the TSLP cohort (n=51).
| Characteristics | All (n=51) | Non-Carriers (n=21) | Carriers (n=30) | P-value |
|---|---|---|---|---|
|
| ||||
| n (%) | n(%) | n(%) | ||
| Age (years), Median [IQR] | 16.1 [14.4-17.0] | 16.2 [13.8-17.0] | 16.0 [14.4-16.6] | 0.716 |
| Weight (kg), Median [IQR] | 65.8 [54.1-75.1] | 62.4 [53.8-70.7] | 68.8 [54.4-86.4] | 0.141 |
| Sex | ||||
| M | 28 (54.9%) | 10 (47.6%) | 18 (60.0%) | 0.387 |
| F | 23 (45.1%) | 11 (52.4%) | 12 (40.0%) | |
| Race | ||||
| White | 30 (58.8%) | 11 (52.4%) | 19 (63.3%) | 0.439 |
| Black | 21 (41.2%) | 10 (47.6%) | 11 (36.7%) | |
| Ever diagnosed with asthma | ||||
| Asthma− | 19 (37.3%) | 11 (52.4%) | 8 (26.7%) | 0.064 |
| Asthma+ | 32 (62.7%) | 10 (47.6%) | 22 (73.3%) | |
| Ever diagnosed with AD | ||||
| AD− | 8 (15.7%) | 5 (23.8%) | 3 (10.0%) | 0.186 |
| AD+ | 43 (84.3%) | 16 (76.2%) | 27 (90.0%) | |
| Ever diagnosed with Asthma and AD | ||||
| Asthma− AD− | 2 (3.9%) | 2 (9.5%) | 0 (0.0%) | 0.022 |
| Asthma+ or AD+ | 23 (45.1%) | 12 (57.1%) | 11 (36.7%) | |
| Asthma+ and AD+ | 26 (51.0%) | 7 (33.3%) | 19 (63.3%) | |
P-value comparing carriers with TSLP risk genotype and non-carriers using fisher’s exact test for categorical variables and the nonparametric Wilcoxon rank-sum test for continuous variables. Boldface indicates a p-value of less than 0.10.
TSLP risk genotype carriers have increased TSLP mRNA expression in nasal epithelial cells
In order to investigate the impact of genetic variation on TSLP expression at the local and systemic level, we examined TSLP expression in nasal epithelial cells (NEC) and circulating TSLP levels in plasma comparing carriers with the risk genotypes to non-carriers. Both sfTSLP and lfTSLP transcripts have been reported(30, 32, 55, 56), thus we examined expression of both forms. All subjects had detectable sfTSLP in NEC. Carriers had a significantly greater proportion of detectable lfTSLP in NEC than non-carriers (82.8% vs. 55.0%, p=0.036). Detection of total TSLP circulating in plasma did not significantly differ between carriers and non-carriers. Both sfTSLP and lfTSLP mRNA expression were significantly correlated with each other among carriers with the risk genotypes (rho=0.60, p=0.0005), but not among non-carriers (rho=0.31, p=0.20). Median lfTSLP expression in NECs was significantly higher (p=0.031) and median circulating total TSLP in plasma was nominally higher (p=0.056) in carriers than non-carriers (Figure 3A). Median sfTSLP expression in NECs was non-significantly increased in carriers compared to non-carriers.
Figure 3. Median TSLP expression in nasal epithelial cells and median circulating TSLP concentration in plasma.
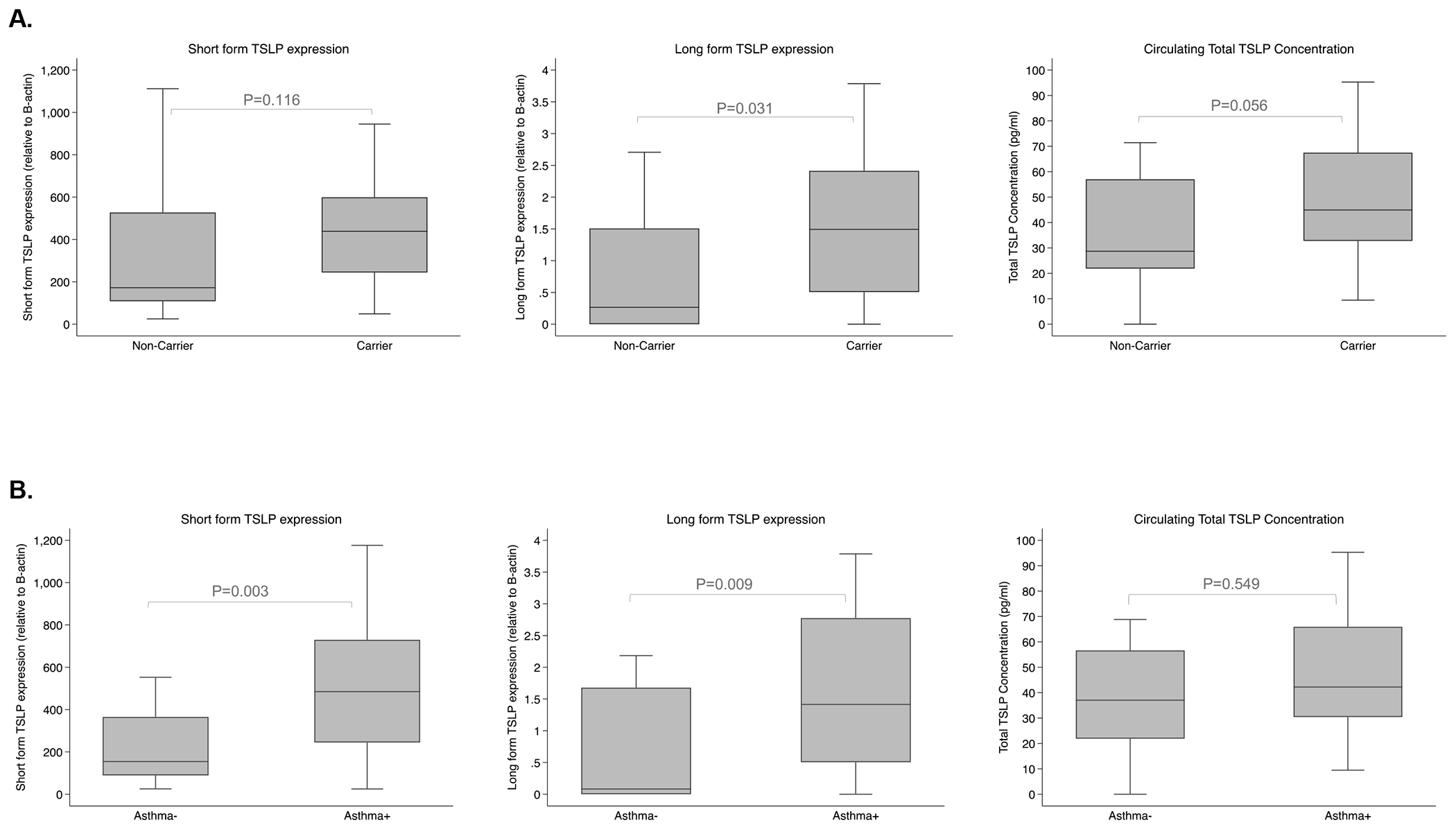
A) Median TSLP level by TSLP risk genotype. Differences in median long and short form TSLP expression (relative to B-actin) in nasal epithelium and total circulating TSLP concentration (pg/ml) levels in plasma comparing carriers with the TSLP risk genotype to non-carrier subjects without. P-value comparing TSLP levels in carriers and non-carriers using nonparametric Wilcoxon rank-sum test. Outliers (defined as 1.5 times the interquartile range above the upper quartile and below the lower quartile) are not shown for visualization purposes in this figure. Outliers were not removed from the analyses. B) Median TSLP level by asthma status. Differences in median short form and long form TSLP expression (relative to B-actin) in the nasal epithelium, total circulating TSLP concentration (pg/ml) levels in plasma comparing asthmatic and nonasthmatic children. P-value comparing asthmatic and nonasthmatic children using nonparametric Wilcoxon rank-sum test. Outliers (defined as 1.5 times the interquartile range above the upper quartile and below the lower quartile) are not shown for visualization purposes in this figure. Outliers were not removed from the analyses.
TSLP risk genotype is associated with increased asthma prevalence
Since a recent genetic study found that TSLP SNPs increased the risk of asthma among children with AD(20), we evaluated the associations of asthma and AD with TSLP risk genotypes. Carriers with the risk genotypes were 3-times more likely to be diagnosed with asthma compared with non-carriers after adjusting for race in logistic regression models (p=0.06; OR 3.05; Table 3). A similar association between risk genotypes and ever having a dual asthma and AD diagnosis was observed compared to non-carriers without the risk alleles after adjustment (p=0.04; OR 3.33). Models adjusting for sex did not yield significantly different effect estimates; thus, sex was not included as a covariate (data not shown).
Table 3.
Association of TSLP risk genotype with clinical outcomes.
| Outcome | All (n=51) | |
|---|---|---|
|
| ||
| Adjusted OR | P-value | |
| Ever diagnosed with Asthma | 3.05 (0.93 - 9.98) | 0.06 |
| Ever diagnosed with AD | 2.60 (0.53 - 12.70) | 0.25 |
| Ever diagnosed with Asthma and AD* | 3.33 (1.02 - 10.86) | 0.04 |
Multivariable logistic regression models to calculate odds ratios (OR) of the clinical outcome comparing carriers with risk genotype and non-carriers adjusted for race. Empiric p-values were derived by randomly permuting TSLP risk genotype 10,000 times. Due to sparseness of the data, confidence intervals from the regression model are reported. Boldface indicates a p-value of less than 0.10.
Subjects with asthma and AD were compared to all other subjects (includes individuals who had only asthma, only AD, or neither condition).
TSLP expression is increased among individuals with asthma
Previous reports suggest that TSLP expression is increased in asthma(28, 29) and disease-associated eQTL in TSLP correlate with TSLP expression(14). In our data, median sfTSLP and lfTSLP expression in NECs were significantly higher in subjects with asthma compared to non-asthmatics (p=0.003 and p=0.009, respectively; Figure 3B). Increased sfTSLP and lfTSLP expression in NECs remained significantly associated with increased asthma diagnosis after adjusting for race and sex in logistic regression models using all data (sfTSLP, p=0.011; lfTSLP, p=0.028; Table 4).
Table 4.
Association of TSLP levels with asthma stratified by TSLP risk genotype.
| Asthma Outcome | All (n=51) | Carriers with TSLP risk genotype (n=30) | Non-carriers without risk genotype (n=21) | Interaction term | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Adjusted OR | P-value | Adjusted OR | P-value | Adjusted OR | P-value | P-value | |
| Short form TSLP expression* | 1.33 (1.01 – 1.93) | 0.011 | 2.53 (1.11 -6.16) | 0.012 | 1.02 (0.56 - 1.83) | 0.91 | 0.005 |
| Long form TSLP expression | 1.47 (1.02 – 3.35) | 0.028 | 3.12 (1.19 – 70.3) | 0.006 | 1.20 (0.06 – 5.13) | 0.52 | 0.073 |
| Circulating Total TSLP | 1.00 (0.98 – 1.03) | 0.55 | 1.01 (0.94 - 1.08) | 0.68 | 0.99 (0.85 - 1.09) | 0.76 | 0.59 |
Multivariable logistic regression models to calculate odds ratios (OR) for asthma diagnosis using continuous TSLP expression data adjusted for race and sex. Models consider the full dataset as well as the data stratified by risk genotype. Given the differences in association between asthma and TSLP in carriers with the risk genotype and non-carriers, we performed multivariable logistic regression including a continuous TSLP level*TSLP genotype (carrier/non-carrier) interaction term (other variables in the model include continuous TSLP expression, genotype, race, and sex). TSLP levels measured long and short form TSLP expression (relative to B-actin) in nasal epithelium, and circulating total TSLP concentration (pg/ml) levels in plasma and were analyzed as continuous variables. Empiric p-values were derived by randomly permuting asthma diagnosis 10,000 times. Bootstrapping was used to estimate the confidence intervals for the odds ratio. Boldface indicates a p-value of less than 0.10.
Data for short form TSLP were rescaled per 100 units to calculate the adjusted odds ratio for ease of interpretation.
Individuals with the TSLP risk genotype and high TSLP expression are more likely to have asthma
Since (1) the TSLP risk genotypes were associated with increased asthma and (2) expression of sfTSLP and lfTSLP in NECs were increased among asthmatics, we sought to determine whether there was a combined effect of TSLP risk genotype and TSLP mRNA expression on asthma prevalence. We performed analyses of sfTSLP and lfTSLP expression in NECs and asthma prevalence stratified by TSLP risk genotypes (Figure 4). Among carriers with the defined risk genotypes, those with high TSLP expression (top tertile) had significantly greater asthma prevalence than those with low TSLP expression (bottom tertile; sfTSLP 100% vs. 29%, p=0.002; lfTSLP 90% vs 30%, p=0.013; Figure 4). Among non-carriers, high sfTSLP and lfTSLP expression were not associated with asthma prevalence. Carriers with high TSLP expression had significantly increased asthma prevalence compared to non-carriers with low TSLP expression (sfTSLP p=0.008; lfTSLP p=0.024). Multivariable logistic regression analysis using a 4-level variable for TSLP genotype and TSLP expression to account for combinatorial effects showed that asthma prevalence was significantly higher among carriers with the risk genotypes and high lfTSLP expression compared to non-carriers with low lfTSLP expression (aOR=3.415; p=0.024). However, individuals with only one risk factor (being a carrier or having elevated TSLP) were not appreciably different than those with neither risk factor. Similarly, multivariable logistic regression models evaluating the interaction of continuous lfTSLP and sfTSLP expression level with risk genotypes were significant (Table 4). Thus, Table 4 presents results stratified by TSLP genotype that demonstrate carriers with increased TSLP NEC expression had increased asthma prevalence (sfTSLP p=0.012; lfTSLP p=0.006; Table 4). Taken together, these data suggest that TSLP risk genotypes act in combination with sfTSLP and lfTSLP expression to influence asthma prevalence.
Figure 4. Association of asthma and TSLP mRNA expression stratified by TSLP risk genotype.

A) Short form TSLP. B) Long form TSLP. TSLP SNPs rs2289277 and rs11466750 define carriers with the risk genotypes and non-carriers without the risk genotype. Prevalence of asthma diagnosis among children with high and low (A) short form and (B) long form TSLP expression (High TSLP+ = high TSLP ≥ 66th percentile; TSLP− = low TSLP < 33th percentile) stratified by TSLP risk genotype (Risk genotype+ = Carriers with risk genotype; Risk genotype− = Non-carriers without risk genotype). P-values for pairwise comparisons using Fisher’s exact test.
DISCUSSION
Our study is distinct from previous studies because it simultaneously examines TSLP SNPs, TSLP local tissue mRNA expression (sfTSLP and lfTSLP), and circulating protein to determine their independent and combined impact on asthma prevalence. Further, our study used a novel algorithm to prioritize SNPs to define risk genotypes and then recruited a cohort based on selected TSLP genotypes, which served as the basis for our studies. Our data demonstrate that TSLP SNPs and local airway expression of TSLP have synergistic contributions to asthma. The increase in asthma risk is likely manifest only when genetic variation enables eQTL in the TSLP locus to elevate lfTSLP. First, our data demonstrate that children carrying one or more alleles for rs2289277 and/or rs11466750 have increased prevalence of detectable TSLP expression, correlation between TSLP isoforms in the upper airway, and marginally increased childhood asthma in carriers compared to non-carriers. Second, we demonstrate that TSLP expression in the upper airway, but not circulating total TSLP in plasma, is increased among children with asthma, suggesting that pathogenesis of asthma is driven by local rather than systemic TSLP secretion. Third, we identify the combined effect of TSLP rs2289277 and rs11466750 genotypes and TSLP (sfTSLP and lfTSLP) expression in NEC that associates with increased childhood asthma thereby demonstrating a novel genotype-dependent contribution of TSLP expression to asthma risk. This indicates that TSLP rs2289277 and rs11466750 genotypes and TSLP expression have non-redundant contributions to asthma.
This study adds several new findings to the current knowledge. Previous studies demonstrated that increased TSLP expression is linked to the pathogenesis of Th2 inflammatory diseases, including asthma and AD(32, 57–59). TSLP is necessary and sufficient for the development of Th2 cytokine-associated inflammation of the airways in mice(1, 60). In human studies, airway biopsy samples of asthmatic patients reveal significantly increased expression of TSLP and Th2 cytokines in the airway epithelium, lamina propria, and bronchial submucosa(28, 61). A bronchial allergen challenge study in mild asthmatics resulted in substantially increased IL-25, IL-33, and TSLP expression in the bronchial epithelium and submucosa(59). Importantly, expression of lfTSLP, but not sfTSLP, is highly inducible by polyI:C, proinflammatory cytokines (IL-1B and TNF-a), and Th2 cytokines (IL-4 and IL-13) in bronchial epithelial cells, underscoring the importance of lfTSLP expression in patients with allergic inflammation(30). While lfTSLP is proinflammatory and expressed during inflammation, sfTSLP demonstrates anti-inflammatory and antimicrobial activities by affecting the capacity of PBMCs and dendritic cells to produce inflammatory cytokines(55, 56). sfTSLP mRNA is constitutively expressed in a variety of tissues, including bronchial and colonic epithelial cells, keratinocytes, and lung fibroblasts(30, 32, 55, 56), but its function in vivo remains unclear. One report demonstrated a modest biological effect of administered sfTSLP in vivo in a mouse house dust mite-induced airway inflammation model(62); however, there is no known receptor for the putative sfTSLP protein so it is less likely to be relevant and translated into a biologically active protein in vivo(32). Consistent with previous studies on small airway epithelial cells and NECs(29, 30), our data demonstrate that lfTSLP expression is increased in the nasal epithelium of asthmatic patients. We also demonstrate a simultaneous increase in sfTSLP expression that is correlated with lfTSLP, and context and genotype dependent despite their differing biological properties.
Genetic studies demonstrated associations between TSLP SNPs and asthma prevalence, including meta-analyses of GWAS in ethnically diverse populations(10, 21, 23, 24, 63–65). Our study confirmed an association between TSLP rs2289277 and/or rs11466750 risk genotypes and asthma. Previous studies also demonstrated associations of TSLP variants with TSLP mRNA expression and asthma, including TSLP rs2289277 and rs11466750 with AD(13), asthma(10), EoE(66, 67), IgE(13, 68), and regulation of TSLP expression in allergic disease(49). Other studies demonstrated associations of TSLP rs3806933, rs3806932, and rs11466749 with asthma(2, 10, 69) and EoE(66, 70, 71). TSLP rs1837253 was associated with asthma risk in three ethnic groups(24) and identified as a susceptibility locus in a Japanese cohort(22). We found that individuals with defined TSLP risk alleles and high levels of NEC sfTSLP and lfTSLP expression had increased odds of asthma. Other studies have illustrated cell-type-specific regulation of expression of TSLP and other asthma-related genes(14, 29), which taken with our results and the importance of local secretion of lfTSLP(31), may explain the inconsistent associations reported with circulating TSLP in plasma and asthma, recurrent wheeze or AD(27, 31–35). By using orthogonal pieces of evidence, surprisingly, the variants we identified with the strongest evidence of contributing to etiology were not the top variants from GWAS studies. This may be is due to the fact that GWAS is a marker for a region that harbors genetic association, but the strength of this association is likely multifactorial including the allele frequency and LD with causal variant(s). GWAS data is dynamic and changes over time. Our study reflects GWAS data that were available at the date of accession; however, new data have since been added. Thus, future studies may benefit from inclusion of TSLP expression in addition to TSLP genotype. Combined, these results suggest increased asthma risk is manifest only when genetic variation enables eQTL in the TSLP locus to elevate lfTSLP expression. There may be additional genetic variants not evaluated in the present study outside of TSLP that also impact TSLP expression and impact asthma.
To prioritize TSLP SNPs for this study, we developed a novel algorithm to identify SNPs that can be applied to other genes of interest. By using a targeted approach to select subjects enriched for specific TSLP genotypes, we ensured sufficient genetic variability and disease homogeneity(23, 72) that enabled us to rigorously examine the combined effects of TSLP genotype, TSLP secretion, and asthma prevalence. This is a highly innovative feature of this study as the subjects included were carefully phenotyped and genotyped. This study design may have value over larger population studies where the subject heterogeneity and population substructure can reduce power and lead to spurious results.
Our observation that TSLP rs2289277 and rs11466750 genotypes and TSLP mRNA (sfTSLP and lfTSLP) expression act in combination to confer asthma risk has important clinical implications. TSLP genotype alone is not sufficient and requires a modifier that enables expression from the allele, i.e. the eQTL to express. Indeed, we did not observe increased asthma prevalence among subjects without the TSLP risk genotypes when comparing those with high and low sfTSLP and lfTSLP expression. It is possible that NEC expression of TSLP is a surrogate for lfTSLP status in other tissues critical for promoting asthma when combined with the presence of risk alleles and a genome permissive for their effect. In any case, TSLP expression alone is not a suitable biomarker for predicting asthma risk despite the associations observed in the present study. Additional studies to identify other risk factors are warranted because the failure to carry the TSLP risk genotypes or to have high TSLP expression does not rule out asthma.
The observed combined effects between TSLP genotypes and TSLP (sfTSLP and lfTSLP) expression might elucidate differences in therapeutic effects of anti-TSLP therapy. Clinical trials of Tezepelumab demonstrated the therapeutic potential of inhibiting TSLP in patients with asthma and with AD. In a small proof-of-concept study of patients with moderate-to-severe AD, treatment with Tezepelumab resulted in a numerical but not statistically significant improvement in eczema-severity scores, potentially due to the use of ‘background medication’ during the trial(73). Based on our data, perhaps the efficacy would be most evident amongst those with both AD and asthma. In a bronchial allergen challenge study, treatment with Tezepelumab in patients with mild allergic asthma resulted in significant reductions of blood and sputum eosinophils and exhaled nitric oxide indicating TSLP has a key upstream role(8, 32). Treatment with Tezepelumab in a phase 2 randomized placebo-controlled trial of moderate-to-severe uncontrolled asthma was associated with a reduction in the annualized asthma exacerbation rate in patients with and without type 2 asthma(6, 7, 32). The observed improvements in disease control in patients who received Tezepelumab highlights the potential pathogenic role of TSLP across different asthma phenotypes(6, 7). The combined effects of TSLP genotypes and nasal TSLP expression in airway tissues, which are sensitive to increased TSLP secretion to drive allergic disease pathogenesis, might explain how treatments can be broadly effective against a variety of asthma and atopic diseases. TSLP may affect disease activity more broadly than inhibition of a single downstream pathway. Further, because TSLP expression acts in combination with TSLP genotypes, it is critical to genotype carrier and non-carrier subjects to minimize underlying heterogeneity that may impact the therapeutic response and disease pathogenesis.
Supplementary Material
Acknowledgment
We thank the individuals and their families who participated in this study. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. This work was supported by NIH R01AI127392 (GKKH, LBM, JMB, LJM, RK) and NIH U19AI1070235 (GKKH, JMB, LJM).
Funding Source:
Supported by NIH R01AI127392 (GKKH, LBM, JMB, LJM, RK) and NIH U19AI1070235 (GKKH, JMB, LJM).
Abbreviations:
- TSLP
Thymic stromal lymphopoietin
- SNP
Single nucleotide polymorphism
- NEC
Nasal epithelial cell
- Th
T helper
- AD
atopic dermatitis
- eQTL
expression quantitative trait locus
- MAF
minor allele frequency
- GTEx
Genotype-Tissue Expression
- FAIRE
formaldehyde-assisted isolation of regulatory elements
- TF
Transcription factor
- GCPCR
Greater Cincinnati Pediatric Clinic Repository
- CCAAPS
Cincinnati Childhood Allergy and Air Pollution Study
- LAR
legally authorized representatives
- ELISA
enzyme-linked immunosorbent assay
- LD
linkage disequilibrium
- sfTSLP
short form TSLP
- lfTSLP
long form TSLP
- CEU
Utah Residents (CEPH) with Northern and Western European Ancestry
- ASW
Americans of African Ancestry in South West USA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflicts of interest.
REFERENCES
- 1.Demehri S, Morimoto M, Holtzman MJ, Kopan R. Skin-derived TSLP triggers progression from epidermal-barrier defects to asthma. PLoS Biol. 2009;7(5):e1000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harada M, Hirota T, Jodo AI, Hitomi Y, Sakashita M, Tsunoda T, et al. Thymic stromal lymphopoietin gene promoter polymorphisms are associated with susceptibility to bronchial asthma. Am J Respir Cell Mol Biol. 2011;44(6):787–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Margolis DJ, Kim B, Apter AJ, Gupta J, Hoffstad O, Papadopoulos M, et al. Thymic stromal lymphopoietin variation, filaggrin loss of function, and the persistence of atopic dermatitis. JAMA dermatology. 2014;150(3):254–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han H, Xu W, Headley MB, Jessup HK, Lee KS, Omori M, et al. Thymic stromal lymphopoietin (TSLP)-mediated dermal inflammation aggravates experimental asthma. Mucosal Immunol. 2012;5(3):342–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soumelis V, Reche PA, Kanzler H, Yuan W, Edward G, Homey B, et al. Human epithelial cells trigger dendritic cell mediated allergic inflammation by producing TSLP. Nat Immunol. 2002;3(7):673–80. [DOI] [PubMed] [Google Scholar]
- 6.Menzies-Gow A, Corren J, Bourdin A, Chupp G, Israel E, Wechsler ME, et al. Tezepelumab in Adults and Adolescents with Severe, Uncontrolled Asthma. N Engl J Med. 2021;384(19):1800–9. [DOI] [PubMed] [Google Scholar]
- 7.Corren J, Parnes JR, Wang L, Mo M, Roseti SL, Griffiths JM, et al. Tezepelumab in Adults with Uncontrolled Asthma. N Engl J Med. 2017;377(10):936–46. [DOI] [PubMed] [Google Scholar]
- 8.Gauvreau GM, O’Byrne PM, Boulet LP, Wang Y, Cockcroft D, Bigler J, et al. Effects of an anti-TSLP antibody on allergen-induced asthmatic responses. N Engl J Med. 2014;370(22):2102–10. [DOI] [PubMed] [Google Scholar]
- 9.Van Rompaey D, Verstraete K, Peelman F, Savvides SN, Augustyns K, Van Der Veken P, et al. Virtual screening for inhibitors of the human TSLP:TSLPR interaction. Sci Rep. 2017;7(1):17211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biagini Myers JM, Martin LJ, Kovacic MB, Mersha TB, He H, Pilipenko V, et al. Epistasis between serine protease inhibitor Kazal-type 5 (SPINK5) and thymic stromal lymphopoietin (TSLP) genes contributes to childhood asthma. J Allergy Clin Immunol. 2014;134(4):891–9 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He JQ, Hallstrand TS, Knight D, Chan-Yeung M, Sandford A, Tripp B, et al. A thymic stromal lymphopoietin gene variant is associated with asthma and airway hyperresponsiveness. J Allergy Clin Immunol. 2009;124(2):222–9. [DOI] [PubMed] [Google Scholar]
- 12.Bunyavanich S, Melen E, Wilk JB, Granada M, Soto-Quiros ME, Avila L, et al. Thymic stromal lymphopoietin (TSLP) is associated with allergic rhinitis in children with asthma. Clin Mol Allergy. 2011;9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao PS, Rafaels NM, Mu D, Hand T, Murray T, Boguniewicz M, et al. Genetic variants in thymic stromal lymphopoietin are associated with atopic dermatitis and eczema herpeticum. J Allergy Clin Immunol. 2010;125(6):1403–7 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Hastie AT, Hawkins GA, Moore WC, Ampleford EJ, Milosevic J, et al. eQTL of bronchial epithelial cells and bronchial alveolar lavage deciphers GWAS-identified asthma genes. Allergy. 2015;70(10):1309–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong X, Wang G, Liu X, Kumar R, Tsai HJ, Arguelles L, et al. Gene polymorphisms, breast-feeding, and development of food sensitization in early childhood. The Journal of allergy and clinical immunology. 2011;128(2):374–81 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harada M, Hirota T, Jodo AI, Doi S, Kameda M, Fujita K, et al. Functional analysis of the thymic stromal lymphopoietin variants in human bronchial epithelial cells. Am J Respir Cell Mol Biol. 2009;40(3):368–74. [DOI] [PubMed] [Google Scholar]
- 17.Liu W, Xu LS, Liu QJ, Dong FZ, Qiu RF, Wen MC, et al. Two single nucleotide polymorphisms in TSLP gene are associated with asthma susceptibility in Chinese Han population. Exp Lung Res. 2012;38(8):375–82. [DOI] [PubMed] [Google Scholar]
- 18.Hunninghake GM, Soto-Quiros ME, Avila L, Kim HP, Lasky-Su J, Rafaels N, et al. TSLP polymorphisms are associated with asthma in a sex-specific fashion. Allergy. 2010;65(12):1566–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birben E, Sahiner UM, Karaaslan C, Yavuz TS, Cosgun E, Kalayci O, et al. The genetic variants of thymic stromal lymphopoietin protein in children with asthma and allergic rhinitis. International archives of allergy and immunology. 2014;163(3):185–92. [DOI] [PubMed] [Google Scholar]
- 20.Wang IJ, Wu LS, Lockett GA, Karmaus WJ. TSLP polymorphisms, allergen exposures, and the risk of atopic disorders in children. Ann Allergy Asthma Immunol. 2016;116(2):139–45 e1. [DOI] [PubMed] [Google Scholar]
- 21.Demenais F, Margaritte-Jeannin P, Barnes KC, Cookson WOC, Altmuller J, Ang W, et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune cell enhancer marks. Nat Genet. 2018;50(1):42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Doi S, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet. 2011;43(9):893–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363(13):1211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43(9):887–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoo J, Omori M, Gyarmati D, Zhou B, Aye T, Brewer A, et al. Spontaneous atopic dermatitis in mice expressing an inducible thymic stromal lymphopoietin transgene specifically in the skin. J Exp Med. 2005;202(4):541–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergot AS, Monnet N, Le Tran S, Mittal D, Al-Kouba J, Steptoe RJ, et al. HPV16 E7 expression in skin induces TSLP secretion, type 2 ILC infiltration and atopic dermatitis-like lesions. Immunol Cell Biol. 2015;93(6):540–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alysandratos KD, Angelidou A, Vasiadi M, Zhang B, Kalogeromitros D, Katsarou-Katsari A, et al. Increased affected skin gene expression and serum levels of thymic stromal lymphopoietin in atopic dermatitis. Annals of allergy, asthma & immunology : official publication of the American College of Allergy, Asthma, & Immunology. 2010;105(5):403–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shikotra A, Choy DF, Ohri CM, Doran E, Butler C, Hargadon B, et al. Increased expression of immunoreactive thymic stromal lymphopoietin in patients with severe asthma. J Allergy Clin Immunol. 2012;129(1):104–11 e1–9. [DOI] [PubMed] [Google Scholar]
- 29.Hui CC, Yu A, Heroux D, Akhabir L, Sandford AJ, Neighbour H, et al. Thymic stromal lymphopoietin (TSLP) secretion from human nasal epithelium is a function of TSLP genotype. Mucosal Immunol. 2015;8(5):993–9. [DOI] [PubMed] [Google Scholar]
- 30.Harada M, Hirota T, Jodo AI, Doi S, Kameda M, Fujita K, et al. Functional Analysis of the Thymic Stromal Lymphopoietin Variants in Human Bronchial Epithelial Cells. American Journal of Respiratory Cell and Molecular Biology. 2009;40(3):368–74. [DOI] [PubMed] [Google Scholar]
- 31.Han H, Xu W, Headley MB, Jessup HK, Lee KS, Omori M, et al. Thymic stromal lymphopoietin (TSLP)-mediated dermal inflammation aggravates experimental asthma. Mucosal Immunology. 2012;5(3):342–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corren J, Ziegler SF. TSLP: from allergy to cancer. Nat Immunol. 2019;20(12):1603–9. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura K, Tsuchida T, Tsunemi Y, Saeki H, Tamaki K. Serum thymic stromal lymphopoietin levels are not elevated in patients with atopic dermatitis. J Dermatol. 2008;35(8):546–7. [DOI] [PubMed] [Google Scholar]
- 34.Lee EB, Kim KW, Hong JY, Jee HM, Sohn MH, Kim KE. Increased serum thymic stromal lymphopoietin in children with atopic dermatitis. Pediatr Allergy Immunol. 2010;21(2 Pt 2):e457–60. [DOI] [PubMed] [Google Scholar]
- 35.Demehri S, Yockey LJ, Visness CM, Jaffee KF, Turkoz A, Wood RA, et al. Circulating TSLP associates with decreased wheezing in non-atopic preschool children: data from the URECA birth cohort. Clin Exp Allergy. 2014;44(6):851–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saenz SA, Taylor BC, Artis D. Welcome to the neighborhood: epithelial cell-derived cytokines license innate and adaptive immune responses at mucosal sites. Immunol Rev. 2008;226:172–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butsch Kovacic M, Biagini Myers JM, Lindsey M, Patterson T, Sauter S, Ericksen MB, et al. The Greater Cincinnati Pediatric Clinic Repository: A Novel Framework for Childhood Asthma and Allergy Research. Pediatr Allergy Immunol Pulmonol. 2012;25(2):104–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LeMasters GK, Wilson K, Levin L, Biagini J, Ryan P, Lockey JE, et al. High prevalence of aeroallergen sensitization among infants of atopic parents. J Pediatr. 2006;149(4):505–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin LJ, Gupta J, Jyothula SS, Butsch Kovacic M, Biagini Myers JM, Patterson TL, et al. Functional variant in the autophagy-related 5 gene promotor is associated with childhood asthma. PLoS One. 2012;7(4):e33454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kovacic MB, Myers JM, Wang N, Martin LJ, Lindsey M, Ericksen MB, et al. Identification of KIF3A as a novel candidate gene for childhood asthma using RNA expression and population allelic frequencies differences. PLoS One. 2011;6(8):e23714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biagini Myers JM, Wang N, LeMasters GK, Bernstein DI, Epstein TG, Lindsey MA, et al. Genetic and environmental risk factors for childhood eczema development and allergic sensitization in the CCAAPS cohort. J Invest Dermatol. 2010;130(2):430–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferreira MA, Matheson MC, Tang CS, Granell R, Ang W, Hui J, et al. Genome-wide association analysis identifies 11 risk variants associated with the asthma with hay fever phenotype. J Allergy Clin Immunol. 2014;133(6):1564–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pickrell JK, Berisa T, Liu JZ, Segurel L, Tung JY, Hinds DA. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet. 2016;48(7):709–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferreira MAR, Mathur R, Vonk JM, Szwajda A, Brumpton B, Granell R, et al. Genetic Architectures of Childhood- and Adult-Onset Asthma Are Partly Distinct. Am J Hum Genet. 2019; 104(4):665–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Y, Byrne EM, Zheng Z, Kemper KE, Yengo L, Mallett AJ, et al. Genome-wide association study of medication-use and associated disease in the UK Biobank. Nat Commun. 2019;10(1):1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003; 100(16):9440–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oyoshi MK, Larson RP, Ziegler SF, Geha RS. Mechanical injury polarizes skin dendritic cells to elicit a T(H)2 response by inducing cutaneous thymic stromal lymphopoietin expression. J Allergy Clin Immunol. 2010;126(5):976–84, 84 e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lander JM, Supp DM, He H, Martin LJ, Chen X, Weirauch MT, et al. Analysis of chromatin accessibility in human epidermis identifies putative barrier dysfunction-sensing enhancers. PLoS One. 2017;12(9):e0184500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simon JM, Giresi PG, Davis IJ, Lieb JD. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat Protoc. 2012;7(2):256–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McVicker G, van de Geijn B, Degner JF, Cain CE, Banovich NE, Raj A, et al. Identification of genetic variants that affect histone modifications in human cells. Science. 2013;342(6159):747–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar S, Ambrosini G, Bucher P. SNP2TFBS - a database of regulatory SNPs affecting predicted transcription factor binding site affinity. Nucleic Acids Res. 2017;45(D1):D139–D44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Degner JF, Pai AA, Pique-Regi R, Veyrieras JB, Gaffney DJ, Pickrell JK, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482(7385):390–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bjerkan L, Schreurs O, Engen SA, Jahnsen FL, Baekkevold ES, Blix IJ, et al. The short form of TSLP is constitutively translated in human keratinocytes and has characteristics of an antimicrobial peptide. Mucosal Immunol. 2015;8(1):49–56. [DOI] [PubMed] [Google Scholar]
- 56.Fornasa G, Tsilingiri K, Caprioli F, Botti F, Mapelli M, Meller S, et al. Dichotomy of short and long thymic stromal lymphopoietin isoforms in inflammatory disorders of the bowel and skin. J Allergy Clin Immunol. 2015;136(2):413–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mack MR, Kim BS. The Itch-Scratch Cycle: A Neuroimmune Perspective. Trends Immunol. 2018;39(12):980–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han H, Roan F, Johnston LK, Smith DE, Bryce PJ, Ziegler SF. IL-33 promotes gastrointestinal allergy in a TSLP-independent manner. Mucosal Immunol. 2018;11(2):394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang W, Li Y, Lv Z, Chen Y, Li Y, Huang K, et al. Bronchial Allergen Challenge of Patients with Atopic Asthma Triggers an Alarmin (IL-33, TSLP, and IL-25) Response in the Airways Epithelium and Submucosa. J Immunol. 2018;201(8):2221–31. [DOI] [PubMed] [Google Scholar]
- 60.Ziegler SF, Artis D. Sensing the outside world: TSLP regulates barrier immunity. Nature Immunology. 2010;11(4):289–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ying S, O’Connor B, Ratoff J, Meng Q, Fang C, Cousins D, et al. Expression and cellular provenance of thymic stromal lymphopoietin and chemokines in patients with severe asthma and chronic obstructive pulmonary disease. J Immunol. 2008;181(4):2790–8. [DOI] [PubMed] [Google Scholar]
- 62.Dong H, Hu Y, Liu L, Zou M, Huang C, Luo L, et al. Distinct roles of short and long thymic stromal lymphopoietin isoforms in house dust mite-induced asthmatic airway epithelial barrier disruption. Sci Rep. 2016;6:39559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harada M, Hirota T, Jodo AI, Hitomi Y, Sakashita M, Tsunoda T, et al. TSLP Promoter Polymorphisms are Associated with Susceptibility to Bronchial Asthma. Am J Respir Cell Mol Biol. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hunninghake GM, Soto-Quiros ME, Avila L, Kim HP, Lasky-Su J, Rafaels N, et al. TSLP polymorphisms are associated with asthma in a sex-specific fashion. Allergy. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu M, Rogers L, Cheng Q, Shao Y, Fernandez-Beros ME, Hirschhorn JN, et al. Genetic variants of TSLP and asthma in an admixed urban population. PLoS One. 2011;6(9):e25099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martin LJ, He H, Collins MH, Abonia JP, Biagini Myers JM, Eby M, et al. Eosinophilic esophagitis (EoE) genetic susceptibility is mediated by synergistic interactions between EoE-specific and general atopic disease loci. J Allergy Clin Immunol. 2018;141(5):1690–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sherrill JD, Gao PS, Stucke EM, Blanchard C, Collins MH, Putnam PE, et al. Variants of thymic stromal lymphopoietin and its receptor associate with eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126(1):160–5 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song X, Ionita-Laza I, Liu M, Reibman J, We Y. A General and Robust Framework for Secondary Traits Analysis. Genetics. 2016;202(4):1329–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Olafsdottir TA, Theodors F, Bjarnadottir K, Bjornsdottir US, Agustsdottir AB, Stefansson OA, et al. Eighty-eight variants highlight the role of T cell regulation and airway remodeling in asthma pathogenesis. Nat Commun. 2020;11(1):393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kottyan LC, Davis BP, Sherrill JD, Liu K, Rochman M, Kaufman K, et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet. 2014;46(8):895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. 2010;42(4):289–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hattersley AT, McCarthy MI. What makes a good genetic association study? Lancet. 2005;366(9493):1315–23. [DOI] [PubMed] [Google Scholar]
- 73.Simpson EL, Parnes JR, She D, Crouch S, Rees W, Mo M, et al. Tezepelumab, an anti-thymic stromal lymphopoietin monoclonal antibody, in the treatment of moderate to severe atopic dermatitis: A randomized phase 2a clinical trial. J Am Acad Dermatol. 2019;80(4):1013–21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


