Abstract
As acquiring bigger data becomes easier in experimental brain science, computational and statistical brain science must achieve similar advances to fully capitalize on these data. Tackling these problems will benefit from a more explicit and concerted effort to work together. Specifically, brain science can be further democratized by harnessing the power of community-driven tools, which both are built by and benefit from many different people with different backgrounds and expertise. This perspective can be applied across modalities and scales and enables collaborations across previously siloed communities.
Keywords: computational, statistics, reference data, infrastructure
CHALLENGES IN BIG NEURAL DATA
The ability to collect and store brain data has grown exponentially over the past few decades. Petabytes (PBs) of anatomical, functional, and genetic data are being recorded with an ever-growing, ever-improving set of technologies. While some data have relatively specific needs, many large data sets, especially large imaging data sets, share a set of problems. This is true across imaging modalities, scales, and species. Therefore, there is an emerging opportunity to work together to efficiently develop the next generation of tools that are beneficial across subdisciplines and scales of brain science. To do so, however, will require overcoming a number of challenges. Our understanding of these challenges and opportunities can be informed by what big data means.
Computer Science Perspective
From a computer science perspective, the size of the data relative to the computer’s hardware is key to determining whether data are big. For example, if the data are small enough to fit into a computer’s main memory, but the calculations require more memory than the data (e.g., matrix inversion), then they are already big in a very practical way and require new analysis. If the data are so large that they cannot fit in the computer’s main memory, new visualization and analysis techniques are required; and if the data are too large to fit in a single computer’s storage drives, then new storage infrastructure is required. These scales of big data are context specific and depend on the particular hardware constraints (Figure 1). For example, for mobile computing devices, even a gigabyte might be big, whereas for servers, several terabytes (TB) would not yet be considered big. Regardless, at each scale, similar challenges must be overcome.
Figure 1.
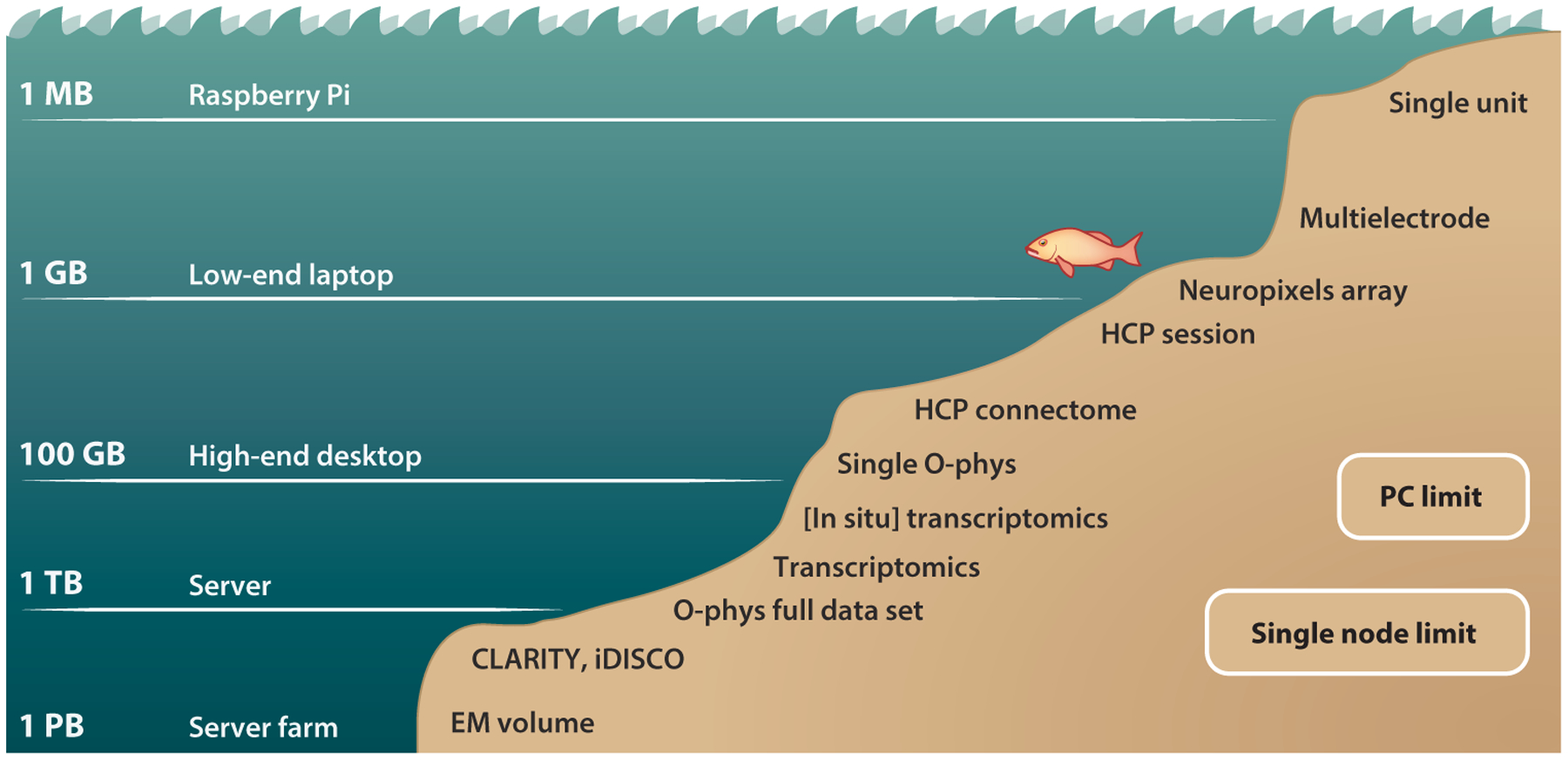
The big data deluge puts different pressure on different applications. At greater data sizes, more powerful systems are needed to operate in these ever–more challenging regimes. Most neuroscience data sets currently still reside at sizes computationally tractable on a single PC or, at worst, a single HPC node. All these modalities, however, are seeing a steady rise in data sizes. The methods that will enable neuroscientists to make use of these ever-richer data sets must be developed now.
The main challenge is in constructing computational infrastructure that can efficiently work with such large data. Whereas little data can be stored in standard file formats, visualized in typical desktop applications, analyzed using single file scripts, and accessed by double clicking, all of these functionalities break down for big data. Instead, we rely on databases for storage, Web-visualization tools, and distributed computing for analysis. Brain sciences are leveraging and modifying tools developed by other sciences and industries for some of these functionalities and developing others from scratch for our unique needs.
Statistics Perspective
From the perspective of statistics, data are big whenever the number of features (or dimensions) exceeds the number of samples. Such data, sometimes called wide data or high-dimensional low-sample-size data, suffer from the large p small n problem, also known as the curse of dimensionality. When data are big from a computational perspective, new computational tools are required, whereas when data are big from a statistical perspective, new statistical tools are required.
The main challenge here is that the traditional bedrock of statistical theory and practice has focused on asymptotic results that assume an effectively infinite number of samples relative to the number of features. For wide data, such assumptions fail to sufficiently restrict the set of potential answers to many quantitative questions. For example, linear regression on a single point permits an infinite number of solutions, but only an infinitesimally small number of them will be useful for predicting future points. Worse yet, as the number of dimensions increases, estimates of uncertainty require an exponentially increasing number of samples (unless they have strong prior knowledge). For these reasons, great care is required to arrive at valid and reproducible conclusions when leveraging wide data. Specifically, latent structures must be inferred, either explicitly or implicitly, and domain knowledge must restrict the search space to biologically sensible answers.
Approaches
For both sets of challenges, new ideas are required to overcome them, and new software is required to implement them. Both ideation and implementation greatly benefit from a global democratized science where anybody with interest can actively contribute. We have found that the most effective means for software development in this era of big data brain science is collaborative and open development (Vogelstein et al. 2018b). For this reason, we highlight a number of tools that are actively developed to overcome these challenges. We hope that this review will inspire others to contribute to these toolboxes rather than develop their own, typically poorly supported and non-sustainably developed, toolboxes. In our experience, community-developed toolboxes have a greater chance of providing more user support and sustainable development practices.
In the history of big data science, a common thread is that community standards emerge only after a data set of great community interest emerges (Burns et al. 2014). Two prominent examples are genetics and cosmology. When the first human genome was published, many in the field wanted to study it. This incentivized the development of tools specifically for operating on data in the format used for the first human genome. When other labs began generating other human genomes, they were incentivized to use the same format, as they had access to the previously developed tools. This pattern repeated as the format became a standard. Note that the incentive structure was internally rather than externally imposed. That is, the standard emerged because it immediately and obviously helped the researchers. Importantly, this included not just professors (who mostly do not do the work) but also graduate students and technicians (who mostly do the work). By aligning incentives, genetics (and also cosmology) was able to develop practices in support of open science and community standards, which greatly accelerated the field’s collective science. Of course, differences between fields, such as the wide variety of measurement modalities, scales, and taxa, create new challenges in neuroscience not seen before. Regardless, many of the same lessons hard learned in other domains stand to benefit how neuroscience approaches the big-data problem and accelerate solutions.
Brain sciences, for the most part, are yet to adopt community standards. We believe this is largely due to a lack of community-supported open data sets. Nonetheless, we are close. Here we first discuss the various subdisciplines that are facing these challenges. For each, we reference big data sets (Table 1) and tools (Table 2) that either already exist or could exist soon. For each modality, several spatial scales are considered. It is our hope that by understanding the shared challenges across scales and modalities, our community will further engage in community-led and community-developed data sets and analysis tools (Figure 2).
Table 1.
List of recommended data sets
| Data type | Data (source) | Limitations |
|---|---|---|
| Physiology | ||
| Micro (ephys) | International Brain Laboratory (https://www.internationalbrainlab.com/) | No data yet |
| Micro (opto) | Brain Observatory (http://observatory.brain-map.org/visualcoding) | Not community driven |
| Meso (fMRI) | Healthy Brain Network (http://fcon_1000.projects.nitrc.org/indi/cmi_healthy_brain_network/) | Only ~1,000 samples available now |
| Macro (behavior) | None | NA |
| Anatomy | ||
| Nano (EM) | TEMCA2 Data (FAFB) (https://www.temca2data.org) | Volumetric annotations not publicly available |
| Micro (LM) | MouseLight and Z Brain Atlas (http://mouselight.janelia.org/; https://engertlab.fas.harvard.edu/Z-Brain/home/) | Not integrated with other related data sets |
| Meso (sMRI & dMRI) | Healthy Brain Network (http://fcon_1000.projects.nitrc.org/indi/cmi_healthy_brain_network/) | Only ~1,000 samples available now |
| Genetics | ||
| Nano (in situ) | None | NA |
| Micro (scRNAseq) | None | NA |
| Meso (tissue) | Allen Brain Map (https://portal.brain-map.org) | NA |
Abbreviations: dMRI, diffusion MRI; EM, electron microscopy; ephys, electrophysiology; FAFB, full adult fly brain; fMRI, functional MRI; LM, light microscopy; NA, not applicable; opto, optical microscopy; scRNAseq, single-cell RNA sequencing; sMRI, structural MRI.
Table 2.
List of recommended code bases and limitations to address each of the data modality specific and general challenges enumerated in the text
| Step | Code (source) | Current hurdles |
|---|---|---|
| Physiology | ||
| Micro (ephys) | Open Ephys (https://open-ephys.org/) | Limited analysis |
| Micro (opto) | CaImAn (https://github.com/flatironinstitute/CaImAn) | Requires manual fine-tuning |
| Meso (fMRI) | C-PAC (https://fcp-indi.github.io) | Single institution developing |
| Macro (behavior) | None | NA |
| Anatomy | ||
| Nano (EM) | NeuroData Cloud (https://neurodata.io/nd_cloud/) | Centralized |
| Micro (LM) | TeraSticher (https://abria.github.io/TeraStitcher/) | Only does linear registration |
| Meso (sMRI & dMRI) | DiPy (https://dipy.org) | No pipelines |
| Genetics | ||
| Nano (in situ) | None | NA |
| Micro (scRNAseq) | None | NA |
| Meso (Tissue) | None | NA |
| Systems | ||
| Storage | CloudVolume (https://github.com/seung-lab/cloud-volume) | Not yet widely adopted |
| Compression | Brotli (https://github.com/google/brotli) | Not yet widely adopted |
| Pipelines | Docker (https://www.docker.com) | Complex to set up |
| Visualization | NeuroGlancer (https://github.com/google/neuroglancer) | Lacks annotation support |
| Statistics | ||
| Tabular | Scikit-Learn (https://scikit-learn.org/stable/) | Parallel execution is weak |
| Images | Scikit-Image (https://scikit-image.org/) | Lacks sophisticated methods |
| Time series | StatsModels (https://www.statsmodels.org/stable/index.html) | Lacks sophisticated methods |
| Networks | NetworkX (https://networkx.github.io) | Lacks sophisticated methods |
Abbreviations: C-PAC, configurable pipelines for the analysis of connectomes; CaImAn, calcium imaging analysis; dMRI, diffusion MRI; EM, electron microscopy; ephys, electrophysiology; fMRI, functional MRI; LM, light microscopy; NA, not applicable; opto, optical microscopy; scRNAseq, single-cell RNA sequencing; sMRI, structural MRI.
Figure 2.
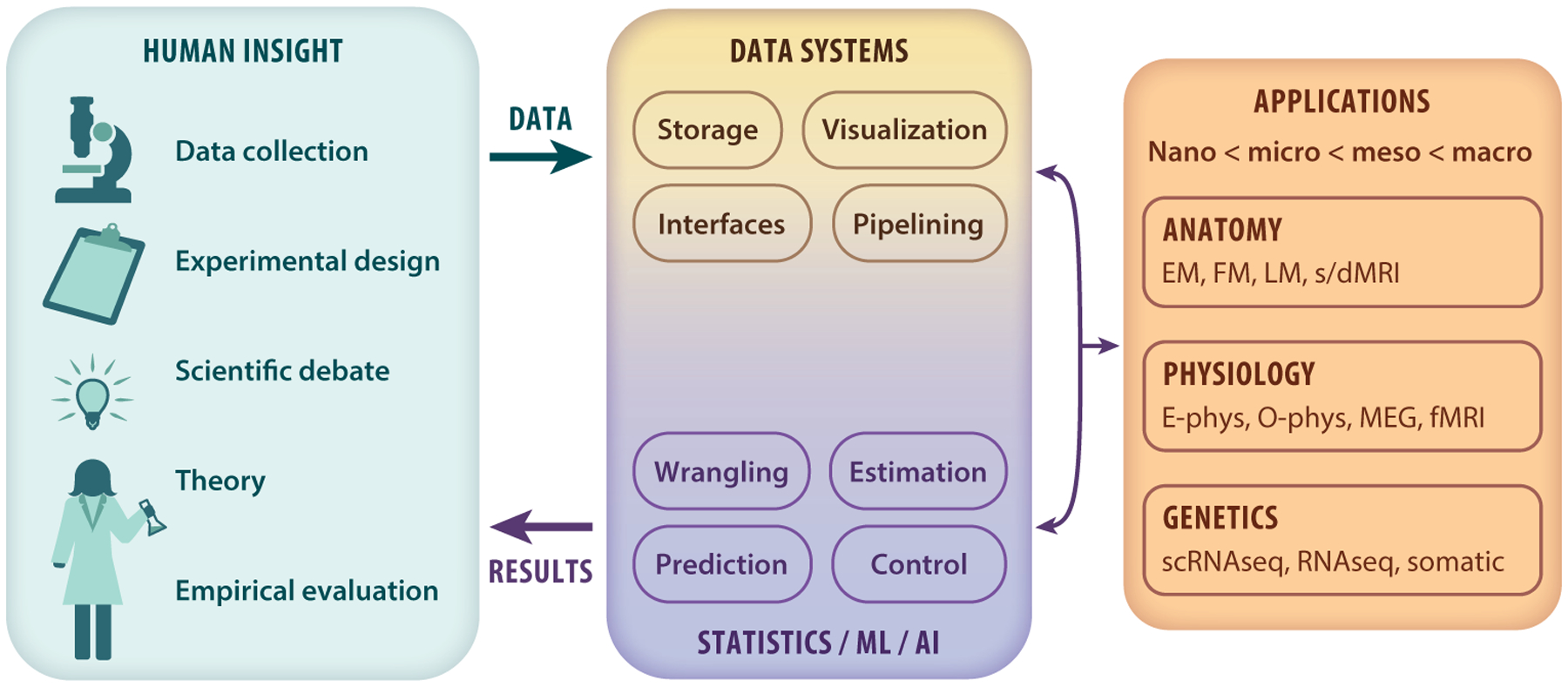
Big data brain science is the result of ingenious advances in recording technology and large-scale collaborations (left box). To maximally utilize the resulting data, we must determine how to convert the data coming from these new experimental paradigms into statistical conclusions on scientific questions (right box).
BIG DATA
We partition the kinds of data acquired in brain science into physiology (dynamics), anatomy (structure), and genetics (blueprint). The boundaries between these three areas are admittedly fuzzy. The structure of one’s brain is dynamic over one’s life span. Moreover, gene expression varies both spatially and temporally across an individual’s brain. Nonetheless, existing experimental methods and data sets for the most part investigate just one of the three areas. Because all three share certain challenges, building tools to address any one of them could accelerate discovery in the others, or better, unify the study across all three.
For each experimental modality, we list a potential reference data set. Reference data sets are data sets that are (a) of great interest to a large fraction of the community, (b) expensive to acquire, and (c) community driven (rather than top-down, i.e., a large fraction of the community collectively decided that such a data set would be highly valuable rather than a single institution producing such a data set). There are counterexamples to each of these; nonetheless, we find the above three principles to be helpful guidelines and provide a summary table of useful references across data types (Table 1). As this review is limited in space, we provide more complete discussions and reference lists in the Supplemental Appendix 1.
Big Dynamics
Physiological data are rapidly growing with advances in recording technologies across scales, including electrophysiology, optophysiology (voltage and calcium imaging), and magnetophysiology [i.e., functional MRI (fMRI)]. Current calculations even demonstrate a feasible path to simultaneous recording of every neuron in the brain (Marblestone et al. 2013). The particulars of each modality have led to different tools for different scales, often without considering their applicability to other scales (Figure 3). We believe developing shared methods would accelerate discovery across modalities.
Figure 3.
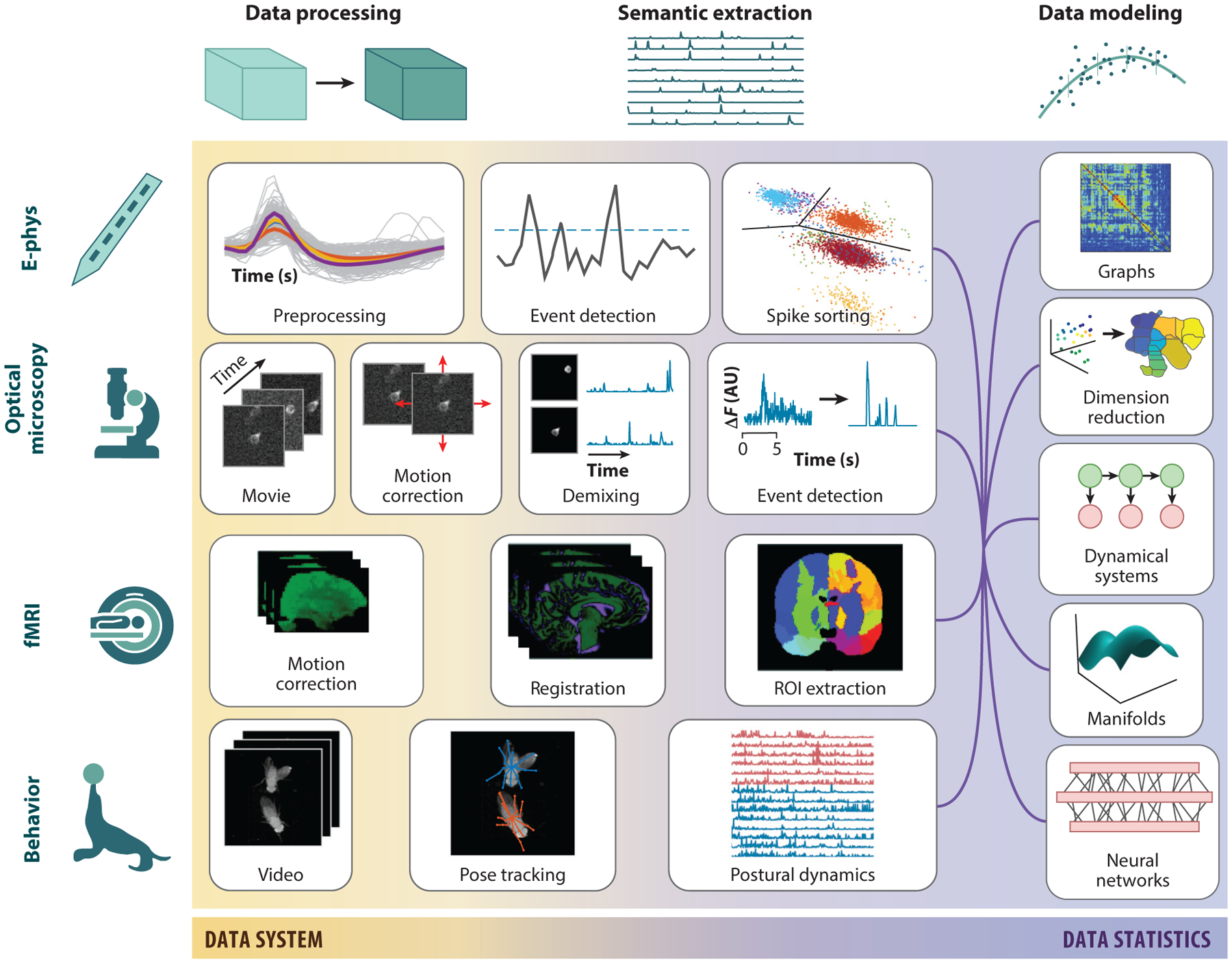
Physiology pipelines across scales. Pipelines have been independently developed for different brain data to transform the raw data through semantic information extraction and into a plethora of statistical analysis results. The raw data (left) are typically preprocessed via registration to a common space (e.g., motion correction). Next, semantic information, e.g., the regions of interest, individual neural traces, or animal poses, is extracted from the data. These are the variables used in final hypothesis generation or estimation.
Microphysiology: electrophysiology and optical microscopy.
On the micron scale, electrical and optical recordings provide systems neuroscience with rich access to population-level activity at single-neuron resolution. While electrophysiology remains the most established modality at this scale, even major advances in electrode densities have not yet brought these data sets to the scale of big data (Jun et al. 2017). Large-scale collaborations [e.g., the International Brain Lab (Int. Brain Lab. 2017)] and theorized technologies (Marblestone et al. 2013), however, promise a future for big electrophysiology. Optical imaging, on the other hand, and calcium imaging in particular have recently crossed the big data threshold and are continuing to grow with new technologies (Beaulieu et al. 2018, Hillman et al. 2018, Song et al. 2017) and chronic recording. Recently, both algorithmic toolboxes (Giovannucci et al. 2019, Pachitariu et al. 2017) and open data sets (e.g., Neurofinder) have been disseminated. The most promising public data set for community reference is the Allen Institute Mouse Brain Observatory; however, it remains to be seen if the top-down nature of the experimental design will impair widespread adoption. Nonetheless, recent activity and investment from prominent institutions and large-scale collaborations promise community-driven pipelines and analysis in the near future.
Mesophysiology: functional MRI.
There is a long history in fMRI of data availability, reference data sets, open resources, and code. The increase in quality, scale, and diversity of data acquisition has been paralleled by extensive algorithmic and infrastructure development as well as progress in social and scientific norms for reproducibility and knowledge representation. A few existing open data sets have the potential to become widely used and catalyze analytical synergy (Bycroft et al. 2018, Mueller et al. 2005, Van Essen et al. 2013). There remain, however, challenges in the fMRI community. Perhaps the most severe challenge is a lack of standard pipelines (Bridgeford et al. 2019, Kiar et al. 2018), which hampers reproducibility and validation and is further exacerbated by the difficulty of harmonizing data across experiments (Yu et al. 2018). For these reasons, the Configurable Pipeline for the Analysis of Connectomes (C-PAC) incorporates many pipelines. While not community driven, it incorporates community-developed algorithms and is gaining in prominence and flexibility.
Macrophysiology: behavior.
Interest in quantifying animal behavior, particularly in naturalistic settings, has recently grown due to its promise as a low-cost and noninvasive measurement of structure and variability in nervous systems (Gomez-Marin et al. 2014, Krakauer et al. 2017). Facilitated by open source assembly schematics and cheap fabrication techniques, behavioral monitoring systems collecting big data sets are becoming a staple in neuroscience laboratories. Traditionally limited by subjective and laborious manual scoring, the signals in behavioral data can now be automated and more objectively quantified due to advances in computer vision and machine learning (Graving et al. 2019, Pereira et al. 2019). These methods enable general purpose pose estimation (Graving et al. 2019, Mathis et al. 2018, Pereira et al. 2019), permitting the inference of postural dynamics necessary for understanding behavior in the context of the brain.
Interpretation of large-scale movement data with respect to the brain, however, remains a challenge (Gomez-Marin et al. 2014). Recent approaches using either unsupervised or supervised approaches have begun to succeed in mapping behavioral patterns to their neural substrates (Markowitz et al. 2018, Vogelstein et al. 2014). Despite this progress, there are no behavioral data sets poised to become reference data sets nor any community reference pipelines, presenting a significant opportunity for development.
Big Anatomy
The study of brain anatomy actually predates that of brain physiology, as the macroscale structure of the brain is readily measurable using relatively simple technology. Yet, the quantitative study of physiology, at least at the cellular scale, predates computational anatomy by nearly 50 years (Miller et al. 2015). Perhaps this is because physiology data can be one-dimensional (a single electrode’s time series), whereas anatomy is fundamentally three-dimensional (3D). Or perhaps it is because signal processing exploded as a computational discipline in the 1950s with Shannon’s communication theory, whereas the spatial statistics community remained relatively small and focused on geostatistics. In either case, in the twenty-first century there is an abundance of big anatomy data available across scales.
Nanoanatomy: electron microscopy.
Studying brain circuitry at nanoscale resolution using electron microscopy (EM) allows for the recovery of a complete wiring diagram of chemical synapses and, with higher resolution, also gap junctions and intracellular organelles. This level of detail also requires extremely large image volumes. A few large EM data sets are poised to become community reference data sets [Eichler et al. 2017, Knott et al. 2008; MICrONS (https://www.iarpa.gov/index.php/research-programs/microns)], but only one is currently properly supported: a data set of a full adult Drosophila brain (Zheng et al. 2018). Each of these data sets must be painstakingly aligned and segmented (Figure 4), both tasks being active areas of research, with no one tool being easy or automatic to use. For nonlinear registration, an elastic tool in Fiji is most popular, though it currently does not adequately scale (Saalfeld et al. 2012). We suspect that the EM community can benefit from advances in nonlinear registration developed by the MRI community, specifically in large deformation diffeomorphic metric mapping (LDDMM) (A.B. Miller et al. 2018), preliminary implementations of which are available using SciPy, ITK, and PyTorch. For segmentation and identification of subcellular structures (e.g., synapses), the most promising approaches leverage convolutional neural networks (CNNs). None of these tools, however, are plug-and-play yet. A further challenge for EM data is sustainability: Somebody must fund the continued storage of these PB-sized data sets for community analysis tools to be developed around them.
Figure 4.
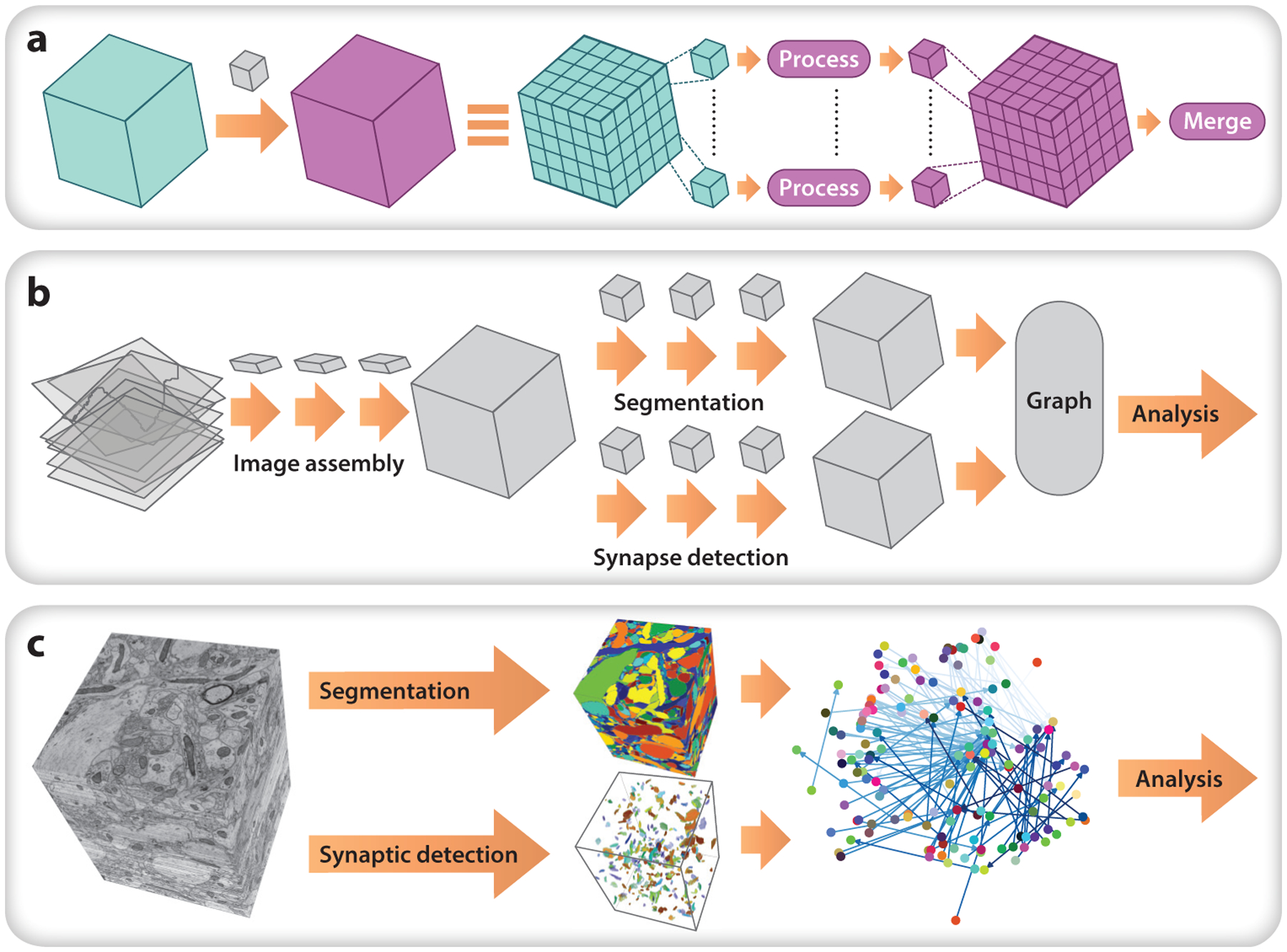
An example data pipeline for nanoscale anatomy. (a) A parallel chunk-processing motif used during processing. A large volume is broken into chunks, each of which is processed and merged. This involves shuttling data from cloud storage or other backends to a computational cluster and tracking process completion and handling failures. The chunk regions depicted here can be anisotropic (e.g., a few wide slices). Each task outside of ovals is handled by data system pipeline software. (b) Representation overview for an example serial section transmission electron microscopy pipeline, showing how the data system implements computational tasks. (c) Computational tasks exemplified on a small cutout of the open data set of Kasthuri et al. (2015).
Microanatomy: light microscopy.
At the micron scale, labeling of individual cells plays a key role in brain science research. Unlike in EM, fluorescence probes can be designed based on genetic and environmental input to answer very specific questions about cellular function and connectivity. While traditional imaging methods involve slicing tissue and examining a small number of regions of interest under a standard light microscope, new technologies are allowing larger volumes of tissue or entire brains to be analyzed.
Like EM, several emerging data sets could become community references. The Mouse Brain Architecture Project (Bohland et al. 2009) provides hundreds of such samples for injection-based tractography such that, across the population, the injections cover the whole brain. The MouseLight project (Winnubst et al. 2019) uses cleared tissue (Chung & Deisseroth 2013, Renier et al. 2014) in combination with a block-face serial two-photon tomography strategy to achieve the best of both worlds, imaging a sparsely labeled set of cells at very high spatial resolution and avoiding registration problems (Narasimhan et al. 2017, Kim et al. 2017). Finally, the Allen Institute for Brain Science has a large portal with a wide variety of reference data sets. To date, there are no widely adopted pipelines for analysis of these data, though there are some disparate tools. The most widely used linear stitching tool is TeraStitcher (Bria & Iannello 2012). Like EM data, light microscopy data could benefit from the registration tools developed for MRI, if the tools could be scaled up.
Macroanatomy: structural and diffusion MRI.
At the macroscopic scale, anatomical analysis will generally involve segmenting the brain into well-characterized and often hierarchical regions of interest and quantifying their structure and function. Reference data sets for macroscale anatomy are the same for macroscale physiology, since experiments tend to acquire both. Similar to fMRI, there are many widely used tools and no community standards. DiPy is emerging as a suite of algorithms (Garyfallidis et al. 2014), and ndmg is a newly available optimized pipeline (Kiar et al. 2018). However, choosing the optimal pipeline (Bridgeford et al. 2019) and harmonizing across data sets (Fortin et al. 2017, Mirzaalian et al. 2017) are important open problems.
Big Genetics
A third kind of big data in brain science is genomics and transcriptomics. Both fields have recently exploded due to the exponential drop of DNA sequencing costs over the last two decades and are rapidly generating multitudes of sequencing data sets of ever-increasing size. Specifically, single-cell RNA sequencing (scRNAseq) that queries gene expression on a cell-by-cell level has rapidly spread in brain science (Svensson et al. 2017). In contrast to Big Anatomy and Big Dynamics applications, genomics and transcriptomics have wrestled with questions of big data storage, efficient analysis, and data sharing for many years, as sequencing data sets have been widespread and existed in the shifting domain of big data for some time. As a result, the majority of steps in the processing of sequencing data are relatively standardized and efficient, at least from the end user perspective.
Nanoscale: in situ transcriptomics.
In addition to Illumina-based single-cell transcriptomics, a rapidly evolving set of in situ transcriptomic techniques (Chen et al. 2015, Lee et al. 2014, Lein et al. 2017, Wang et al. 2018) is appearing on the big data landscape of brain science. These methods probe gene expression levels with subcellular resolution inside tissue sections, providing spatial information lost in traditional single-cell sequencing. Any experiment probing even a small fraction of a brain can generate several TB of raw data, and data volumes will only increase. These images must be registered across sequencing cycles, and messenger RNA signals need to be detected, segmented, and finally decoded to produce a table of gene identity and position in the imaged area. In contrast to the relatively established pipelines for Illumina sequencing data, however, the challenges of how to best handle big data challenges, including sharing, processing, and archiving, in in situ transcriptomics are unresolved and will require new, unified analysis pipelines.
Microscale: single-cell RNA sequencing.
In contrast to standardized preprocessing of sequencing data sets, work is currently underway to improve the analysis of the count matrix from scRNAseq. While from a big data perspective the count matrix of a scRNAseq experiment is relatively small, the ever-increasing number of cells profiled—already exceeding 105 and soon 106 cells—poses analysis challenges. These challenges are very similar to other big matrix challenges, including normalization procedures (Hafemeister & Satija 2019), matrix completion (Andrews & Hemberg 2019, Huang et al. 2018, van Dijk et al. 2018), dimensionality reduction, clustering, and data integration across experiments (Stuart et al. 2019, Welch et al. 2019). Many of these analyses are bundled in R or Python suites [e.g., Seurat (Stuart et al. 2019) and Scanpy (Wolf et al. 2018)], or as stand-alone packages. In addition, an HDF5-based, single-cell-specific data format (Loom) allows efficient access to the count matrix such that it need not be held in memory and is beginning to be integrated in analysis suites.
Mesoscale: tissue-specific gene expression profiles.
In genomics, as we coarsen the spatial scale, the data get increasingly smaller. Once we move beyond single-cell data to tissue-level data, the data are no longer particularly big, and the tools are fairly standard from the genomics and transcriptomics communities, so we refer the reader to standard treatments from those fields.
BIG DATA SYSTEMS
To support big data storage, visualization, and analysis requires extensive software developments and modifications. Many independent efforts have begun to support the analysis of big data, including at least 40 projects known to the authors. However, only a few have adopted best practices for developing tools that are widely adopted with long-term use and support. The key, in our experience, to developing such tools is doing so in an open and collaborative environment (Vogelstein et al. 2018b). As more people use the resource, the developers get more feedback for improvement, so they are not left relying on their gut judgments for how to improve. Moreover, when the code is open source, well documented (including developer documentation), and purposefully designed, other developers can easily contribute. Maintaining an active user and developer community is more of a soft skill that is often underappreciated in hard sciences. Nonetheless, if our community is to fully capitalize on big data, we will need to support, both intellectually and financially, individuals to build communities around tools.
Storage
Small data can be stored on a single hard drive or on a single workstation with multiple hard drives. Because hard drives only come in fixed sizes and workstations only have a limited number of hard drive bays, when data are larger than the maximum storage capacity from the maximum number of hard drives, researchers must consider other options. Our experience with the growth of the Open Connectome Project is a case in point (Burns et al. 2014). Originally, we began storing a 10-TB data set (Bock et al. 2011) on our local cluster. As other researchers began contributing additional data, our lab’s resources were overwhelmed, and we moved to using our institutional resources (which are available in many wealthy institutions). Even though the Institute for Data Intensive Engineering and Sciences had a multi-PB data center, the Open Connectome Project eventually required computing ill adapted to institutional resources. Specifically, we required elastic, on-demand, scale up and scale down computing. These computing features were important, as the MouseLight project could generate 40-TB data sets every two weeks (Economo et al. 2016), and the ingest process of writing data to our infrastructure was too slow. This, in combination with the upcoming MICrONS (https://www.iarpa.gov/index.php/research-programs/microns) data set destined to be multiple PBs in size, catalyzed us to port our infrastructure to the commercial cloud. Specifically, in collaboration with the Applied Physics Laboratory, we developed the NeuroData Cloud (NDCloud), a centralized cloud infrastructure (Lillaney et al. 2018). NDCloud currently stores data in NeuroGlancer precomputed file format (https://github.com/google/neuroglancer/tree/master/src/neuroglancer/datasource/precomputed), which both compresses the data and makes visualization fast and easy. CloudVolume (https://github.com/seung-lab/cloud-volume) enables fast and parallel data access (Silversmith 2018). This infrastructure hosts the current Open Connectome Project, available from Amazon Web Services registry as OpenNeuro (https://registry.opendata.aws/openneuro/), which serves approximately 50 TB of open access data, spanning data from 30 labs, several species, and multiple modalities (Vogelstein et al. 2018b).
This system is an evolution from previous systems in that it is now fundamentally decentralized, and data can be stored remotely and in a distributed fashion, without requiring a single gatekeeper as in our previous designs. This transition mirrors a similar conversion in other sectors, such as media sharing, which adopted a peer-to-peer (or P2P) strategy. Now, anybody who would like to host data can, and the Open Connectome Project simply serves as an archival site.
Pipelines
When neuroimaging data fit in memory, they can be simply processed and visualized using scripts or interactive analysis tools like Jupyter notebooks. Better code and algorithms can stretch the capability of single core analysis; however, as the data volume increases and faster execution times are sought, parallel processing such as Apache spark (Zaharia et al. 2016), Apache Beam (https://beam.apache.org/), and Dask (Rocklin 2015) and improved hardware (RAM, persistent storage, parallel CPUs, GPUs, or networking) can meet the increased demands.
To illustrate the potential issues, we discuss the implications of processing a petascale image, as per our experience processing most of a cubic millimeter of EM images. Before any image processing, the images are aligned and stitched into a 3D volume. TeraStitcher is capable of linear stitching and aligning light-sheet data, but it cannot address large, nonlinear deformations (Bria & Iannello 2012, Bria et al. 2019); LDDMM algorithms are instead desired (M.I. Miller et al. 2018). Then, data must be organized in a fashion that is amenable to efficient access and visualization. This is accomplished by splitting the image into a regular grid, saving each grid location as a separate file (called chunks), and recursively downsampling and compressing each file for storage. Other software packages have likewise been developed to organize and manage large scientific databases, e.g., DataJoint (Yatsenko et al. 2015).
Compression remains a complicated, unsolved problem. We would like a compression algorithm to have low compression ratios, high compression and decompression speeds, and native browser support (so browsers can open the compressed images directly, rather than decompressing first). JPEG 2000, while popular, lacks widespread browser support. We instead recommend brotli, a new general, lossless compression from Google with widespread browser support and better compression ratios but worse compression speeds (we do not currently use it for the Open Connectome Project because it is not yet compatible with NeuroGlancer, our visualization engine of choice).
To run workloads within a framework with appropriate execution guarantees, we recommend using containers run using a framework such as Docker or Singularity. These containers are a lightweight alternative to machine images that can be executed by a virtual operating system. Containerization forces the author to document the exact steps needed to create a functioning execution environment and generates a file that can be downloaded and run on various platforms. Once generated, containers are usually shared via online repositories. There are many container orchestration engines to choose from, which allow you to run containerized programs on your cluster and set resource limits (Bernstein 2014). Containerizing each step, even with container orchestration, is insufficient. Probably the largest bottleneck to widespread adoption of containers in brain science is that the overhead can be substantial, especially if it includes instructions for deployment. Several emerging approaches aim to mitigate these issues, including Boutiques and CodeOcean, which facilitate executing containers across different distributed environments, and Gigantum, which automatically builds docker containers. These approaches, while promising, remain unproven.
Visualization
Small, two-dimensional (2D) data are trivially visualized with local computer applications, (e.g., OSX’s Preview). Even before data get large, if they are 3D+, other software is required. Two of the most popular and widely supported 3D image visualization and annotation software tools are ITK-SNAP (Yushkevich et al. 2016) and Fiji (Schindelin et al. 2012). Both have a wealth of brain imaging–related plug-ins, although ITK-SNAP is largely geared at the macroscale (MRI), whereas Fiji is more geared to the nano- and microscales (EM and light microscopy). For example, BigDataViewer is a browser designed for multiview light-sheet microscopy that integrates with Fiji. When the data are too large to fit locally, other tools are required.
Of the many available 3D image viewers, such as NeuroGlancer, CATMAID, DVID, and Knossos, we currently recommend NeuroGlancer due to its more diverse community of contributors that includes industry, academia, and nonprofits. Moreover, NeuroGlancer enables sharing direct views via a URL so that collaborators can visualize precisely the same image.
NeuroGlancer has support for multichannel data and off-axis viewing. Yet NeuroGlancer also has several shortcomings, most of which are shared by the other web-based visualization tools. First, NeuroGlancer lacks support for manual volumetric labeling and error correcting. This step is crucial for any image processing, and therefore NeuroGlancer is not yet a one-stop shop for visualizing big image data. Second, NeuroGlancer stores all the image metadata in the URL because it is stateless. But, as visualizations get complicated, URLs can get quite long, sometimes hundreds or thousands of characters. For this reason, we developed a prototype service that converts long URLs into short ones. Third, although NeuroGlancer supports visualizing overlaid annotations, it natively does not provide annotation metadata. We have a deployment of NeuroGlancer that includes labels for the Allen Reference Atlas 3.0 labels, but it is not yet generalized to other parcellations, images, or species. As more groups adopt NeuroGlancer, we hope they will contribute such expansions.
Other, smaller kinds of data have their own limitations. For example, meshes enable the display of objects that span a large spatial distance and allow for more natural modes of interaction to understand how they lie in space. They represent 3D surface geometries as a network of vertices represented by three floating point numbers and a set of faces or edges. These structures are typically much lighter than dense voxel representations at sufficient surface area to volume ratios. Displaying meshes in 3D with a lighting model and reasonable frame rates typically requires GPUs. While meshes can be displayed across great spatial extents, they cannot in general be calculated in a single process, as the amount of voxel data required to contain the object is very large. There are techniques for reducing the data requirement, such as using lower resolution mip levels and packed binary representations of single objects, but to efficiently compute millions of objects, it is necessary to treat all labels in a bounding box at once. Open source libraries containing meshing algorithms are available, such as Zmesh (Zlateski & Silversmith 2019), which performs marching cubes (Lorensen & Cline 1987) on all labels in one pass with on-demand mesh simplification.
Mesh simplification is required to reduce the size of the mesh, which after marching cubes has a vertex located at every surface voxel of an object. Mesh-based extraction is also under development, e.g., based on mesh contraction (Au et al. 2008) and TEASAR-like approaches in MeshParty (Dorkenwald et al. 2020). Mesh-based approaches have the benefit of complete context and are especially suited for skeletonizing selected labels. While there are many techniques for simplifying, smoothing, and tuning meshes, Mcell is widely used in brain science for dynamics simulations, which require extremely detailed meshes.
BIG STATISTICS
Beyond the storage and management of big data is the litany of challenges in scientifically valid analyses. Even the most basic of operations, e.g., principal components analysis or linear regression, presents complications for standard single-core and in-memory numerical libraries when the data cannot fit in memory. This extreme regime has necessitated new classes of methods (Bzdok et al. 2019; D. Zheng et al. 2015; Mhembere et al. 2017a,b, 2019a,b; Wang et al. 2016; Zheng et al. 2016, 2018). The broader statistical issue is that brain data sets can have extremely small sample sizes with extremely large numbers of dimensions. To take the most extreme example, IARPA’s MICrONS project (https://www.iarpa.gov/index.php/research-programs/microns) will yield one mouse and has already amassed 2 PB in EM images alone. It is thus imperative that brain sciences remain at the forefront of statistical inference for wide data. We highlight here four types of high-dimensional data common in big brain science: unstructured feature matrices, multivariate time series, images, and networks. Each setting requires different developments; however, all benefit from the ability to learn latent structures from data. To do so effectively requires designing methods that use state-of-the-art knowledge of both brain science and data science. Collaborations between experts in those fields, as well as interdisciplinary training of young researchers, will therefore catalyze progress.
Big Unstructured Data
Unstructured data contain features with no known relationships with one another. For example, RNAseq experiments can generate up to 10,000 features, each corresponding to the measured quantity of RNA in a biological sample. As there is no natural ordering of gene products, these data are unstructured. Unstructured data has perhaps the longest and richest history in data science, with its modern revolution due to Ronald A. Fisher’s books on statistical methods and experimental design in the 1920s and 1930s (Fienberg 1992). Twenty-first century statistics has focused more on solving the large p small n problem with the spread of sparse modeling (Tibshirani 1996) and manifold learning (Lee & Verleysen 2007, Roweis & Saul 2000, Tenenbaum et al. 2000). More recently, theory has been catching up to explain how, when, and why such approaches work (Vershynin 2018, Wainwright 2019). These non-asymptotic theoretical results are often based on concentration inequalities, where guarantees can be provided even with finite data, in contrast to classical statistical theory. Complementary developments focus on algorithmic improvements, e.g., recursive least squares (Haykin 1996), batch-based updating via stochastic gradient descent (Xu & Yin 2015), and optimization (Cevher et al. 2014, Slavakis et al. 2014, Wahlberg et al. 2012).
Although many tools and programming languages are available for these kinds of operations, the neuroscience community is converging on using Python, specifically the NumPy and SciPy packages (Virtanen et al. 2020). Nonetheless, Python still has severe limitations that hinder even more widespread use. First, it is more difficult to install and configure than MATLAB or R. While cloud notebook environments such as binder and Gigantum are mitigating this issue, it remains problematic. Second, many statisticians and signal processing researchers still develop tools in R and MATLAB, respectively, so Python often lags in state-of-the-art tools. The community can combat this by shifting development to Python and working with the standard packages to ensure standardization and accessibility, as we have done with recent work on hypothesis testing (Vogelstein et al. 2019), dimensionality reduction (Vogelstein et al. 2018a), and classification (Tomita et al. 2020).
Big Images
Statistical theory for images is much more complex and difficult than for unstructured data. This is in part because representing an image requires an index for both where and what the feature is (i.e., magnitude). But the where and what are difficult to jointly model. Moreover, brain images can have very long dependence structures, requiring huge amounts of labeled data to effectively estimate. One of the most promising theories for images, the stochastic grammar of images (Zhu 2006), was promoted by David Mumford but never quite took off, likely due to its complexity and ineffectiveness. One realm of image analysis that has gained widespread use is the analysis of shape (Younes 2019), perhaps because shape effectively ignores magnitudes and only considers relative locations.
Instead of leveraging theory, in image analysis, engineering rules. The leading image analysis tools are CNNs (Goodfellow et al. 2016), despite scant theoretical understanding of their effectiveness (Zhang et al. 2018). More interpretable approaches, e.g., decision forests, may return and even overtake CNNs for image processing by virtue of them incorporating convolution and locality operators (Criminisi et al. 2012, Perry et al. 2019), but these are very preliminary ideas. Numerous articles have been published debunking various theories for why such methods work (Ba & Caruana 2014), and subsets of those authors publish papers debunking the debunking (Urban et al. 2017).
Nonetheless, many packages are widely available and supported for image analysis. Scikit-Image is one of the leading packages for basic image analysis (van der Walt et al. 2014), although it is yet to incorporate many standard techniques in brain imaging, such as nonlinear image registration (Kutten et al. 2016, 2017). For more sophisticated image analysis, various deep learning frameworks are available (e.g., TensorFlow and PyTorch). While TensorFlow holds more clout in industry, PyTorch has seen greater adoption in the academic community, particularly within the computer vision discipline (He 2019). The large-scale adoption of these open source frameworks thus holds promise that solutions to common image analysis tasks (e.g., object detection or segmentation) will become increasingly accessible to researchers hoping to apply new approaches to brain imaging data.
Big Time
Physiology data are fundamentally multivariate time series. In contrast to unstructured and image data, there is significantly less development for time series data. This is despite the fact that signal processing has been to a large extent focused on analysis of dynamics (Kay 1993). A key counterexample highlights the severe limitations faced with big time series data. There is no “scikit-time” for time series. Rather, perhaps the most widely used Python time series toolbox is Prophet. Developed by Facebook, it is explicitly designed to incorporate seasonal trends (e.g., holiday effects) and is therefore not appropriate for brain signals. Neural network frameworks support time series analysis, such as Long Short-Term Memory and other recurrent neural networks (RNNs) (Goodfellow et al. 2016), but like CNNs, it is not clear how or when RNNs work. A few recent theoretical works extended Kalman filters, the workhorse of much linear and nonlinear time series modeling, to big data regimes (Chen et al. 2017, Zipunnikov et al. 2011). Efficient implementations of these tools, however, are not yet available. Brain science has pushed latent process models (Smith & Brown 2003, Pakman et al. 2018, Sharma et al. 2018), but like the Kalman models, open source and community-developed packages for these tools do not exist. Many of the classical tools for time series analyses are incorporated into statsmodels, a Python package that includes many regression techniques and basic time series techniques. The need for time series packages yields an opportunity for the community to create, either anew or by expanding the current statsmodels, the toolbox that will fill that void.
Big Networks
A convenient representation of many of the data sets described above is a network or graph. A graph is composed of a set of nodes and a set of edges between those nodes, where the edges represent some relationship between objects. With this representation we can ask, for example, what models best describe the network? How do the fit models vary across a population? How does the model relate to data sets in other modalities?
Graph representations can stem from many experimental modalities. EM circuit reconstruction has generated network representations, for example, in Caenorhabditis elegans (White et al. 1986), Ciona intestinalis (Ryan et al. 2016), larval and adult Drosophila (Eichler et al. 2017), zebrafish (Hildebrand et al. 2017), and mouse (Bock et al. 2011, Helmstaedter et al. 2013), with efforts underway to add zebra finch, nonhuman primates, octopus, and even humans. Connectomes can also be estimated via gene sequencing.
As graphs are much more concise descriptions of a brain than images (they can be stored as simple edge lists or adjacency matrices), storage is much more efficient. For the largest nanoscale connectomics, the final network should have approximately 100,000 nodes and one billion synapses [e.g., MICrONS (https://www.iarpa.gov/index.php/research-programs/microns)]. Even then, the network representation should fit easily in memory. The current issue is instead computational. Even for moderately sized networks, intuitive understanding of patterns in the graph is difficult. This makes visualization and dimensionality reduction (e.g., via eigendecompositions of the adjacency matrix) essential.
Specific graph analyses rely on the details of the node and edges in the graph. One model class assumes that each node in the graph is associated with a low-dimensional unobserved vector, or latent position (Athreya et al. 2017, Hoff 2007). The probability of two nodes connecting is a function of their latent positions. Under the random dot product graph (RDPG) model, this function is the inner product of the nodes’ positions (Sussman et al. 2011). Adjacency spectral embedding and omnibus embedding can estimate these latent positions from a single observed graph or a population of graphs, respectively (Fishkind et al. 2013, Levin et al. 2017). A further extension of the RDPG model assumes that the latent positions are distributed on a latent manifold, analogous to experimentally observed neural trajectory manifolds (Arroyo et al. 2019). RDPG and similar models provide a probability distribution, permitting inference on the modeled connectome. The standard tool in Python for analysis of networks, NetworkX (Hagberg et al. 2008), lacks many modern advances in statistical analysis of individual and populations of graphs. Graspy is an emerging tool that incorporates many developments in statistical learning from networks (Chung et al. 2019) but has not yet enjoyed widespread adoption.
DISCUSSION
Brain science is obtaining larger, more detailed data than ever before, and the pace of progress is only increasing. We have outlined here many of the challenges and current approaches to leveraging these rich data sets. While much progress has been made, there remain many open problems. The novelty and universality of many of these challenges creates a sequence of opportunities to unify the brain science community and best enable open sharing of data, methodologies, statistical analyses, and results.
One of the biggest missed opportunities in designing data infrastructure is connecting solutions for different data modalities. These solutions are often aimed at a current task facing a single group with a specific set of tools and scientific goals. Sharing good solutions with other domains that need or will need such advances is thus stymied.
The emerging challenge of simultaneous analysis of multiple data types, e.g., behavior and cellular recordings or fMRI and calcium imaging (Lake et al. 2018, Ma et al. 2016), is related. One interesting contrast between anatomical and functional data is the inertia of established methodology in the latter that has leveraged decades of small-data brain science. This creates a bias toward focusing on particular scientific questions rather than starting anew.
Paramount to progress is investment. Currently, pipelines are built and maintained by well-endowed institutions or large groups of labs collaborating under the new large-scale funding mechanisms. Access to these resources must be more widespread to permit new technologies and their benefits to have maximum impact. Endeavors to make standardized resources more accessible (e.g., the Allen Institute) are important and should be considered when creating new infrastructure. Unused time on these big-data systems is lost time that could have been used in the service of science.
An important aspect of operating at scale not discussed here is privacy. Certain applications, especially those with medical relevance such as genetics and MRI, necessitate infrastructure and statistics that are privacy preserving. Moreover, there are computational and statistical benefits to differential privacy-based algorithms that are often based on randomized statistics. Future designs and implementations should keep these considerations in mind, as they are often difficult to build post hoc.
One very promising area of advance is the ever-growing number of open reference data necessary to catalyze standardization and to broaden the pool of researchers with access to the data, which is needed to develop new infrastructure and statistical tools. Public data sets, through Collaborative Research in Computational Neuroscience, the Allen Institute, and others, are providing just those opportunities.
A related major opportunity in the short term is in the standardization of data files. Such standards can provide efficient storage by compressing the data in a way that is natural for brain data, create an open environment by providing universal data access data tools, and permit faster processing by allowing pipeline developers the freedom to optimize for a constant file structure. Earlier attempts [e.g., Neurodata Without Borders (Teeters et al. 2015)] have yet to take hold, which may be partially due to the constant evolution of data acquisition as well as competition from accessible standard image formats. Universality and flexibility to new imaging modalities are key in designing such a standard. Image standards have leveraged known natural image statistics to achieve this: No matter the camera, png will compress in a reasonable way. The statistics of brain data, rather than of the data modality, will similarly be key to successful standards.
Supplementary Material
SUMMARY POINTS.
Neuroscience is obtaining larger and more detailed recordings than ever before.
We have an opportunity to capitalize on cross-scale and cross-modality developments.
More investment in generalizable computational tools and reference data will accelerate discovery.
We need to use and build upon community-developed software, which can be widely accessed and based on the needs of researchers. If existing tools/data sets are not sufficient, we must add to them and build on them rather than starting something new.
ACKNOWLEDGMENTS
J.T.V. would like to acknowledge generous support from the National Science Foundation (NSF) under NSF award EEC-1707298. T.D.P. would like to acknowledge support from the NSF Graduate Research Fellowships Program under NSF award number DGE-1148900 and the Princeton Porter Ogden Jacobus Fellowship. W.S. and N.T. would like to acknowledge support by the Intelligence Advanced Research Projects Activity (IARPA) via Department of Interior/Interior Business Center (DoI/IBC) contract number D16PC0005, the National Institutes of Health (NIH)/National Institute of Mental Health (U01MH114824, U01MH117072, RF1MH117815), NIH/National Institute of Neurological Disorders and Stroke (U19NS104648, R01NS104926), NIH/National Eye Institute (R01EY027036), and ARO (W911NF-12-1-0594). S.S.G. was partially supported by and related to the intent of NIH awards R24MH117295, R01EB020740, P41EB019936, RF1MH12002101, and R01MH109682. The US Government is authorized to reproduce and distribute reprints for Governmental purposes notwithstanding any copyright annotation thereon. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of IARPA, DoI/IBC, or the US Government. The authors are grateful for assistance from Google, Amazon, and Intel.
Footnotes
DISCLOSURE STATEMENT
J.T.V. and R.B. are on the board of Gigantum.
LITERATURE CITED
- Andrews TS, Hemberg M. 2019. False signals induced by single-cell imputation. F1000Res. 7:1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo J, Athreya A, Cape J, Chen G, Priebe CE, Vogelstein JT. 2019. Inference for multiple heterogeneous networks with a common invariant subspace. arXiv:1906.10026 [stat.ME] [PMC free article] [PubMed] [Google Scholar]
- Athreya A, Fishkind DE, Tang M, Priebe CE, Park Y, et al. 2017. Statistical inference on random dot product graphs: a survey. J. Mach. Learn. Res 18(226):1–92 [Google Scholar]
- Au OK-C, Tai C-L, Chu H-K, Cohen-Or D, Lee T-Y. 2008. Skeleton extraction by mesh contraction. ACM Trans. Graph 27(3):1–10 [Google Scholar]
- Ba J, Caruana R. 2014. Do deep nets really need to be deep? In Advances in Neural Information Processing Systems 27, ed. Ghahramani Z, Welling M, Cortes C, Lawrence ND, Weinberger KQ, pp. 2654–62. San Diego, CA: NeurIPS [Google Scholar]
- Beaulieu DR, Davison IG, Bifano TG, Mertz J. 2018. Simultaneous multiplane imaging with reverberation multiphoton microscopy. arXiv:1812.05162 [physics.optics] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein D 2014. Containers and cloud: from LXC to Docker to Kubernetes. IEEE Cloud Comput. 1(3):81–84 [Google Scholar]
- Bock DD, Lee WC, Kerlin AM, Andermann ML, Hood G, et al. 2011. Network anatomy and in vivo physiology of visual cortical neurons. Nature 471(7337):177–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohland JW, Wu C, Barbas H, Bokil H, Bota M, et al. 2009. A proposal for a coordinated effort for the determination of brainwide neuroanatomical connectivity in model organisms at a mesoscopic scale. PLOS Comput. Biol 5(3):e1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bria A, Bernaschi M, Guarrasi M, Iannello G. 2019. Exploiting multi-level parallelism for stitching very large microscopy images. Front. Neuroinform 13:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bria A, Iannello G. 2012. TeraStitcher—A tool for fast automatic 3D-stitching of teravoxel-sized microscopy images. BMC Bioinform. 13:316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgeford EW, Wang S, Yang Z, Wang Z, Xu T, et al. 2019. Optimal experimental design for big data: applications in brain imaging. bioRxiv 802629. 10.1101/802629 [DOI] [Google Scholar]
- Burns R, Vogelstein JT, Szalay AS. 2014. From cosmos to connectomes: the evolution of data-intensive science. Neuron 83(6):1249–52 [DOI] [PubMed] [Google Scholar]
- Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, et al. 2018. The UK Biobank resource with deep phenotyping and genomic data. Nature 562(7726):203–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzdok D, Nichols TE, Smith SM. 2019. Towards algorithmic analytics for large-scale datasets. Nat. Mach. Intell 1(7):296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevher V, Becker S, Schmidt M. 2014. Convex optimization for big data: scalable, randomized, and parallel algorithms for big data analytics. IEEE Signal Process. Mag 31(5):32–43 [Google Scholar]
- Chen L, Vogelstein JT, Lyzinski V, Priebe CE. 2015. A joint graph inference case study: the C. elegans chemical and electrical connectomes. arXiv:1507.08376 [stat.AP] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Liu K, Yang Y, Xu Y, Lee S, et al. 2017. An M-estimator for reduced-rank system identification. Pattern Recognit. Lett 86:76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J, Pedigo BD, Bridgeford EW, Varjavand BK, Helm HS, Vogelstein JT. 2019. GraSPy: graph statistics in Python. J. Machine Learn. Res 20(158):1–7 [Google Scholar]
- Chung K, Deisseroth K. 2013. CLARITY for mapping the nervous system. Nat. Methods 10(6):508–13 [DOI] [PubMed] [Google Scholar]
- Criminisi A, Shotton J, Konukoglu E. 2012. Decision forests: a unified framework for classification, regression, density estimation, manifold learning and semi-supervised learning. Found. Trends Comput. Graph. Vis 7(2–3):81–227 [Google Scholar]
- Dorkenwald S, Schneider-Mizell C, Collman F. 2020. sdorkenw/MeshParty: v1.9.0 (Version v1.9.0). Software. 10.5281/zenodo.3710398 [DOI] [Google Scholar]
- Economo MN, Clack NG, Lavis LD, Gerfen CR, Svoboda K, et al. 2016. A platform for brain-wide imaging and reconstruction of individual neurons. eLife 5:e10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler K, Li F, Litwin-Kumar A, Park Y, Andrade I, et al. 2017. The complete connectome of a learning and memory centre in an insect brain. Nature 548(7666):175–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fienberg SE. 1992. A brief history of statistics in three and one-half chapters: a review essay. Stat. Sci 7(2):208–25 [Google Scholar]
- Fishkind DE, Lyzinski V, Pao H, Chen L, Priebe CE. 2013. Vertex nomination schemes for membership prediction. Ann. App. Stat 9(3):1510–32 [Google Scholar]
- Fortin J-P, Parker D, Tunç B, Watanabe T, Elliott MA, et al. 2017. Harmonization of multi-site diffusion tensor imaging data. NeuroImage 161:149–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garyfallidis E, Brett M, Amirbekian B, Rokem A, van der Walt S, et al. 2014. Dipy, a library for the analysis of diffusion MRI data. Front. Neuroinform 8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannucci A, Friedrich J, Gunn P, Kalfon J, Brown BL, et al. 2019. CaImAn an open source tool for scalable calcium imaging data analysis. eLife 8:e38173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Marin A, Paton JJ, Kampff AR, Costa RM, Mainen ZF. 2014. Big behavioral data: psychology, ethology and the foundations of neuroscience. Nat. Neurosci 17(11):1455–62 [DOI] [PubMed] [Google Scholar]
- Goodfellow I, Benigo Y, Courville A. 2016. Deep Learning. Cambridge, MA: MIT press [Google Scholar]
- Graving JM, Chae D, Naik H, Li L, Koger B, et al. 2019. DeepPoseKit, a software toolkit for fast and robust animal pose estimation using deep learning. eLife 8:e47994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C, Satija R. 2019. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20(1):296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg A, Schult D, Swart P. 2008. Exploring network structure, dynamics, and function using NetworkX. In Proceedings of the 7th Python in Science Conference, ed. Varoquaux G, Vaught T, Millman J, pp. 11–15. Pasadena, CA: SciPy [Google Scholar]
- Haykin S 1996. Adaptive Filter Theory. Upper Saddle River, NJ: Prentice-Hall. 3rd ed. [Google Scholar]
- He H 2019. The state of machine learning frameworks in 2019. The Gradient, Oct. 10. https://thegradient.pub/state-of-ml-frameworks-2019-pytorch-dominates-research-tensorflow-dominates-industry/ [Google Scholar]
- Helmstaedter MN, Briggman KL, Turaga SC, Jain V, Seung HS, Denk W. 2013. Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature 500(7461):168–74 [DOI] [PubMed] [Google Scholar]
- Hildebrand DGC, Cicconet M, Torres RM, Choi W, Quan TM, et al. 2017. Whole-brain serial-section electron microscopy in larval zebrafish. Nature 545(7654):345–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman EM, Voleti V, Patel K, Li W, Yu H, et al. 2018. High-speed 3D imaging of cellular activity in the brain using axially-extended beams and light sheets. Curr. Opin. Neurobiol 50:190–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff PD. 2007. Modeling homophily and stochastic equivalence in symmetric relational data. In Advances in Neural Information Processing Systems 20, ed. Platt JC, Koller D, Singer Y, Roweis ST. San Diego, CA: NeurIPS [Google Scholar]
- Huang M, Wang J, Torre E, Dueck H, Shaffer S, et al. 2018. SAVER: gene expression recovery for single-cell RNA sequencing. Nat. Methods 15(7):539–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Int. Brain Lab. 2017. An international laboratory for systems and computational neuroscience. Neuron 96(6):1213–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun JJ, Steinmetz NA, Siegle JH, Denman DJ, Bauza M, et al. 2017. Fully integrated silicon probes for high-density recording of neural activity. Nature 551(7679):232–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasthuri N, Hayworth KJ, Berger DR, Schalek RL, Conchello JA, et al. 2015. Saturated reconstruction of a volume of neocortex. Cell 162(3):648–61 [DOI] [PubMed] [Google Scholar]
- Kay SM. 1993. Fundamentals of Statistical Signal Processing, Volume I: Estimation Theory. Upper Saddle River, NJ: Prentice Hall. 1st ed. [Google Scholar]
- Kiar G, Bridgeford EW, Gray Roncal WR, Consort. Reliab. Reprod. (CoRR), Chandrashekhar V, et al. 2018. A high-throughput pipeline identifies robust connectomes but troublesome variability. bioRxiv 188706. 10.1101/188706 [DOI] [Google Scholar]
- Kim Y, Yang GR, Pradhan K, Venkataraju KU, Bota M, et al. 2017. Brain-wide maps reveal stereotyped cell-type-based cortical architecture and subcortical sexual dimorphism. Cell 171(2):456–469.e22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott G, Marchman H, Wall D, Lich B. 2008. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. J. Neurosci 28(12):2959–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakauer JW, Ghazanfar AA, Gomez-Marin A, MacIver MA, Poeppel D. 2017. Neuroscience needs behavior: correcting a reductionist bias. Neuron 93(3):480–90 [DOI] [PubMed] [Google Scholar]
- Kutten KS, Charon N, Miller MI, Ratnanather JT, Matelsky J, et al. 2017. A large deformation diffeomorphic approach to registration of CLARITY images via mutual information. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2017, ed. Descoteaux M, Maier-Hein L, Franz A, Jannin P, Collins DL, Duchesne S, pp. 275–82. Cham, Switz.: Springer [Google Scholar]
- Kutten KS, Vogelstein JT, Charon N, Ye L, Deisseroth K, Miller MI. 2016. Deformably registering and annotating whole CLARITY brains to an atlas via masked LDDMM. In Optics, Photonics and Digital Technologies for Imaging Applications IV, ed. Schelkens P, Ebrahimi T, Cristóbal G, Truchetet F, Saarikko P. Bellingham, WA: SPIE [Google Scholar]
- Lake EMR, Ge X, Shen X, Herman P, Hyder F, et al. 2018. Spanning spatiotemporal scales with simultaneous mesoscopic Ca2+ imaging and functional MRI: neuroimaging spanning spatiotemporal scales. bioRxiv 464305. 10.1101/464305 [DOI] [Google Scholar]
- Lee JA, Verleysen M. 2007. Nonlinear Dimensionality Reduction. New York: Springer [Google Scholar]
- Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, et al. 2014. Highly multiplexed subcellular RNA sequencing in situ. Science 343(6177):1360–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein E, Borm LE, Linnarsson S. 2017. The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science 358(6359):64–69 [DOI] [PubMed] [Google Scholar]
- Levin K, Athreya A, Tang M, Lyzinski V, Park Y, Priebe CE. 2017. A central limit theorem for an omnibus embedding of multiple random graphs and implications for multiscale network inference. arXiv:1705.09355 [stat.ME] [Google Scholar]
- Lillaney K, Kleissas D, Eusman A, Perlman E, Gray Roncal W, et al. 2018. Building NDStore through hierarchical storage management and microservice processing. In 2018 IEEE 14th International Conference on e-Science (e-Science), pp. 223–233. Los Alamitos, CA: IEEE [Google Scholar]
- Lorensen WE, Cline HE. 1987. Marching cubes: a high resolution 3D surface construction algorithm. In Proceedings of the 14th Annual Conference on Computer Graphics and Interactive Techniques, pp. 163–69. New York: ACM [Google Scholar]
- Ma Y, Shaik MA, Kim SH, Kozberg MG, Thibodeaux DN, et al. 2016. Wide-field optical mapping of neural activity and brain haemodynamics: considerations and novel approaches. Philos. Trans. R. Soc. Lond. B Biol. Sci 371(1705):20150360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marblestone A, Zamft BM, Maguire YG, Shapiro MG, Cybulski TR, et al. 2013. Physical principles for scalable neural recording. Front. Comput. Neurosci 7:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz JE, Gillis WF, Beron CC, Neufeld SQ, Robertson K, et al. 2018. The striatum organizes 3D behavior via moment-to-moment action selection. Cell 174(1):44–58.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis A, Mamidanna P, Cury KM, Abe T, Murthy VN, et al. 2018. DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci 21(9):1281–89 [DOI] [PubMed] [Google Scholar]
- Mhembere D, Zheng D, Priebe CE, Vogelstein JT, Burns R. 2017a. knor: a NUMA-optimized in-memory, distributed and semi-external-memory k-means library. arXiv:1606.08905 [cs.DC] [Google Scholar]
- Mhembere D, Zheng D, Priebe CE, Vogelstein JT, Burns R. 2017b. knor: a NUMA-optimized in-memory, distributed and semi-external-memory k-means library. In Proceedings of the 26th International Symposium on High-Performance Parallel and Distributed Computing, pp. 67–78. New York: Assoc. Comput. Mach. [Google Scholar]
- Mhembere D, Zheng D, Priebe CE, Vogelstein JT, Burns R. 2019a. clusterNOR: a NUMA-optimized clustering framework. arXiv:1902.09527 [cs.DC] [Google Scholar]
- Mhembere D, Zheng D, Priebe CE, Vogelstein JT, Burns R. 2019b. Graphyti: a semi-external memory graph library for FlashGraph. arXiv:1907.03335 [cs.DC] [Google Scholar]
- Miller AB, Sheridan MA, Hanson JL, McLaughlin KA, Bates JE, et al. 2018. Dimensions of deprivation and threat, psychopathology, and potential mediators: a multi-year longitudinal analysis. J. Abnorm. Psychol 160–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MI, Arguillère S, Tward DJ, Younes L. 2018. Computational anatomy and diffeomorphometry: a dynamical systems model of neuroanatomy in the soft condensed matter continuum. Wiley Interdiscip. Rev. Syst. Biol. Med 10(6):e1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MI, Trouvé A, Younes L. 2015. Hamiltonian systems and optimal control in computational anatomy: 100 years since D’Arcy Thompson. Annu. Rev. Biomed. Eng 17:447–509 [DOI] [PubMed] [Google Scholar]
- Mirzaalian H, Ning L, Savadjiev P, Pasternak O, Bouix S, et al. 2017. Multi-site harmonization of diffusion MRI data in a registration framework. Brain Imaging Behav. 12(1):284–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, et al. 2005. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin. N. Am 15(4):869–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan A, Venkataraju KU, Mizrachi J, Albeanu DF, Osten P. 2017. Oblique light-sheet tomography: fast and high resolution volumetric imaging of mouse brains. bioRxiv 132423. 10.1101/132423 [DOI] [Google Scholar]
- Pachitariu M, Stringer C, Dipoppa M, Schröder S, Rossi LF, et al. 2017. Suite2p: beyond 10,000 neurons with standard two-photon microscopy. bioRxiv 061507. 10.1101/061507 [DOI] [Google Scholar]
- Pakman A, Wang Y, Mitelut C, Lee JH, Paninski L. 2018. Discrete neural processes. arXiv:1901.00409 [stat.ML] [Google Scholar]
- Pereira TD, Aldarondo DE, Willmore L, Kislin M, Wang SS, et al. 2019. Fast animal pose estimation using deep neural networks. Nat. Methods 16(1):117–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R, Tomita TM, Patsolic J, Falk B, Vogelstein JT. 2019. Manifold forests: closing the gap on neural networks. arXiv:1909.11799 [cs.LG] [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M. 2014. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159(4)896–910 [DOI] [PubMed] [Google Scholar]
- Rocklin M 2015. Dask: parallel computation with blocked algorithms and task scheduling. In Proceedings of the 14th Python in Science Conference, pp. 126–32. Austin, TX: SciPy [Google Scholar]
- Roweis ST, Saul LK. 2000. Nonlinear dimensionality reduction by locally linear embedding. Science 290(5500):2323–26 [DOI] [PubMed] [Google Scholar]
- Ryan K, Lu Z, Meinertzhagen IA. 2016. The CNS connectome of a tadpole larva of Ciona intestinalis (L.) highlights sidedness in the brain of a chordate sibling. eLife 5:e16962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saalfeld S, Fetter R, Cardona A, Tomancak P. 2012. Elastic volume reconstruction from series of ultra-thin microscopy sections. Nat. Methods 9(7):717–20 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9(7):676–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Johnson R, Engert F, Linderman S. 2018. Point process latent variable models of larval zebrafish behavior. In Advances in Neural Information Processing Systems 31, ed. Bengio S, Wallach H, Larochelle H, Grauman K, Cesa-Bianchi N, Garnett R, pp. 10942–53. San Diego, CA: NeurIPS [Google Scholar]
- Silversmith W 2018. CloudVolume: client for reading and writing to Neuroglancer precomputed volumes on cloud services. GitHub. https://github.com/seung-lab/cloud-volume [Google Scholar]
- Slavakis K, Giannakis GB, Mateos G. 2014. Modeling and optimization for big data analytics:(statistical) learning tools for our era of data deluge. IEEE Signal Process. Mag 31(5):18–31 [Google Scholar]
- Smith AC, Brown EN. 2003. Estimating a state-space model from point process observations. Neural Comput. 15(5):965–91 [DOI] [PubMed] [Google Scholar]
- Song A, Charles AS, Koay SA, Gauthier JL, Thiberge SY, et al. 2017. Volumetric two-photon imaging of neurons using stereoscopy (vTwINS). Nat. Methods 14(4):420–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, et al. 2019. Comprehensive integration of single-cell data. Cell 177(7):1888–902.e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman DL, Tang M, Fishkind DE, Priebe CE. 2011. A consistent adjacency spectral embedding for stochastic blockmodel graphs. J. Am. Stat. Assoc 107(499):1119–28 [Google Scholar]
- Svensson V, Natarajan KN, Ly LH, Miragaia RJ, Labalette C, et al. 2017. Power analysis of single-cell RNA-sequencing experiments. Nat. Methods 14(4):381–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeters JL, Godfrey K, Young R, Dang C, Friedsam C, et al. 2015. Neurodata without borders: creating a common data format for neurophysiology. Neuron 88(4):629–34 [DOI] [PubMed] [Google Scholar]
- Tenenbaum JB, de Silva V, Langford JC. 2000. A global geometric framework for nonlinear dimensionality reduction. Science 290(5500):2319–23 [DOI] [PubMed] [Google Scholar]
- Tibshirani R 1996. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B Stat. Methodol 58(1):267–88 [Google Scholar]
- Tomita TM, Browne J, Shen C, Chung J, Patsolic JL, et al. 2020. Sparse projection oblique randomer forests. J. Mach. Learn. Res In press [Google Scholar]
- Urban G, Geras KJ, Ebrahimi Kahou S, Aslan O, Wang S, et al. 2017. Do deep convolutional nets really need to be deep and convolutional? arXiv:1603.05691 [stat.ML] [Google Scholar]
- van der Walt S, Schönberger JL, Nunez-Iglesias J, Boulogne F, Warner JD, et al. 2014. scikit-image: image processing in Python. PeerJ 2:e453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, et al. 2018. Recovering gene interactions from single-cell data using data diffusion. Cell 174(3):716–29.e27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, et al. 2013. The WU-Minn Human Connectome Project: an overview. NeuroImage 80:62–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershynin R 2018. High-Dimensional Probability: An Introduction with Applications in Data Science. Cambridge, UK: Cambridge Univ. Press. 1st ed. [Google Scholar]
- Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, et al. 2020. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17:261–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein JT, Bridgeford E, Tang M, Zheng D, Burns R, Maggioni M. 2018a. Geometric dimensionality reduction for subsequent classification. arXiv:1709.01233 [stat.ML] [Google Scholar]
- Vogelstein JT, Bridgeford EW, Wang Q, Priebe CE, Maggiono M, et al. 2019. Discovering and deciphering relationships across disparate data modalities. eLife 8:e41690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein JT, Park Y, Ohyama T, Kerr RA, Truman JW, et al. 2014. Discovery of brainwide neural-behavioral maps via multiscale unsupervised structure learning. Science 344(6182):386–92 [DOI] [PubMed] [Google Scholar]
- Vogelstein JT, Perlman E, Falk B, Baden A, Gray Roncal W, et al. 2018b. A community-developed open-source computational ecosystem for big neuro data. Nat. Methods 15(11):846–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg B, Boyd S, Annergren M, Wang Y. 2012. An ADMM algorithm for a class of total variation regularized estimation problems. IFAC Proc. Vol 45(16):83–88 [Google Scholar]
- Wainwright MJ. 2019. High-Dimensional Statistics: A Non-Asymptotic Viewpoint. Cambridge, UK: Cambridge Univ. Press. 1st ed. [Google Scholar]
- Wang C, Chen M-H, Schifano E, Wu J, Yan J. 2016. Statistical methods and computing for big data. Stat. Interface 9(4):399–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, et al. 2018. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361(6400):eaat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JD, Kozareva V, Ferreira A, Vanderburg C, Martin C, Macosko EZ. 2019. Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell 177(7):1873–87.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci 314(1165):1–340 [DOI] [PubMed] [Google Scholar]
- Winnubst J, Bas E, Ferreira TA, Wu Z, Economo MN, et al. 2019. Reconstruction of 1,000 projection neurons reveals new cell types and organization of long-range connectivity in the mouse brain. Cell 179(1):268–81.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, Theis FJ. 2018. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Yin W. 2015. Block stochastic gradient iteration for convex and nonconvex optimization. SIAM J. Optim 25(3):1686–716 [Google Scholar]
- Yatsenko D, Reimer J, Ecker AS, Walker EY, Sinz F, et al. 2015. DataJoint: managing big scientific data using MATLAB or Python. bioRxiv 031658. 10.1101/031658 [DOI] [Google Scholar]
- Younes L 2019. Shapes and Diffeomorphisms. New York: Springer. 2nd ed. [Google Scholar]
- Yu M, Linn KA, Cook PA, Phillips ML, McInnis M, et al. 2018. Statistical harmonization corrects site effects in functional connectivity measurements from multi-site fMRI data. Hum. Brain Mapp 39(11):4213–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Yang G, Gerig G. 2016. ITK-SNAP: an interactive tool for semi-automatic segmentation of multi-modality biomedical images. In Proceedings of the 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, pp. 3342–45. Los Alamitos, CA: IEEE; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharia M, Xin RS, Wendell P, Das T, Armbrust M, Dave A, et al. 2016. Apache spark: a unified engine for big data processing. Comm. ACM 59(11):56–65 [Google Scholar]
- Zhang D, Yin J, Zhu X, Zhang C. 2018. Network representation learning: a survey. arXiv:1801.05852 [cs.SI] [Google Scholar]
- Zheng D, Mhembere D, Vogelstein JT, Priebe CE, Burns R. 2016. FlashMatrix: parallel, scalable data analysis with generalized matrix operations using commodity SSDs. arXiv:1604.06414v1 [cs.DC] [Google Scholar]
- Zheng D, Mhembere D, Burns R, Vogelstein J, Priebe CE, Szalay AS. 2015. FlashGraph: processing billion-node graphs on an array of commodity SSDs. In Proceedings of the 13th USENIX Conference on File and Storage Technologies, pp. 45–58. Santa Clara, CA: FAST [Google Scholar]
- Zheng Z, Lauritzen JS, Perlman E, Robinson CG, Nichols M, et al. 2018. A complete electron microscopy volume of the brain of adult Drosophila melanogaster. Cell 174(3):730–43.e22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M 2006. Discriminant analysis with common principal components. Biometrika 93(4):1018–24 [Google Scholar]
- Zipunnikov V, Caffo B, Yousem DM, Davatzikos C, Schwartz BS, Crainiceanu C. 2011. Multilevel functional principal component analysis for high-dimensional data. J. Comput. Graph. Stat 20(4):852–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlateski A, Silversmith W. 2019. Zmesh. GitHub. https://github.com/seung-lab/zmesh [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


