Abstract
Maternal environment, including nutrition and microbiota, plays a critical role in determining offspring's risk of chronic diseases such as diabetes later in life. Heme iron requirement is amplified during pregnancy and lactation, while excessive dietary heme iron intake, compared to non-heme iron, has shown to trigger acute oxidative stress in the gut resulting from reactive aldehyde formation in conjunction with microbiota reshape. Given the immaturity of the antioxidant defense system in early life, we investigated the extent to which a maternal diet enriched with heme iron may have a lasting impact on gut homeostasis and glucose metabolism in 60-day-old C3H/HeN mice offspring.
As hypothesized, the form of iron added to the maternal diet differentially governed the offspring's microbiota establishment despite identical fecal iron status in the offspring. Importantly, despite female offspring was unaffected, oxidative stress markers were however higher in the gut of male offspring from heme enriched-fed mothers, and were accompanied by increases in fecal lipocalin-2, intestinal para-cellular permeability and TNF-α expression. In addition, male mice displayed blood glucose intolerance resulting from impaired insulin secretion following oral glucose challenge. Using an integrated approach including an aldehydomic analysis, this male-specific phenotype was further characterized and revealed close covariations between unidentified putative reactive aldehydes and bacterial communities belonging to Bacteroidales and Lachnospirales orders.
Our work highlights how the form of dietary iron in the maternal diet can dictate the oxidative status in gut offspring in a sex-dependent manner, and how a gut microbiota-driven oxidative challenge in early life can be associated with gut barrier defects and glucose metabolism disorders that may be predictive of diabetes development.
Keywords: Heme iron, Early life nutrition, Lipoperoxidation, Red meat, Gut barrier, Gut microbiota, Glucose metabolism
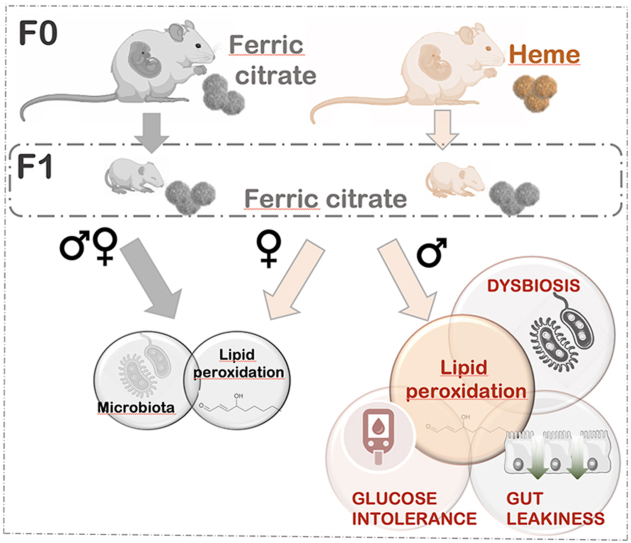
Graphical abstract
Highlights
-
•
Maternal heminic vs. non-heminic iron intake differentially and persistently imprints the offspring's fecal microbiota.
-
•
Males from heme-fed dams exhibit increased gut lumen reactive aldehydes in absence of direct dietary exposure to heme iron.
-
•
Some of the increased reactive aldehydes closely covariated with Orders belonging to Bacteroidales and Lachnospirales.
-
•
Maternal exposure to dietary heme iron impairs gut barrier and glucose tolerance in male offspring.
1. Introduction
Iron is a crucial mineral for numerous physiological processes during all stages of life, but its requirement is particularly critical during periods of rapid development, i.e. pregnancy and early infancy, for facilitating maternal blood volume expansion and ensuring adequate fetal iron stores [1,2]. However, due to its highly reactive nature, intracellular iron in excess has been shown to generate reactive oxygen species (ROS) [3,4]. Among them, highly reactive hydroxyl radicals by reacting notably with lipids can yield lipid peroxyl radicals that can further give rise to secondary lipoperoxidation products such as reactive aldehydes, among which malondialehyde (MDA) or 4-hydroxy-2-nonenal (4-HNE). Such iron-dependent ROS generation leading to increased oxidative stress have been shown to exert cytotoxicity resulting in cellular damage [5]. As such, tightly regulated mechanisms at both the systemic and cellular levels coordinate the uptake, recycling, transport and storage of iron to maintain balanced systemic iron levels and prevent related oxidative status and adverse health outcomes in adults [4,6]. However, in neonates, such iron regulation and antioxidant system defense would be immature [7,8].
Iron absorption in the gut lumen is mainly dependent on physiological requirements, but also dependent of its quantity and especially of its bioavailability in the diet [9]. Uptake of dietary iron is typically obtained from several forms: non-heme, heme and lactoferrin. For neonates, iron in breast milk is bound mainly to lactoferrin and is highly bioavailable [10]. As compared to the non-heme iron form, heme iron is the most bioavailable source of iron and is readily absorbed from animal-based diets, and particularly from red meat [11].
Similarly to the dual and versatile nature of iron, meat products provide on the one hand a highly nutritional value, but excessive consumption on the other hand promotes the development of non-communicable diseases such as cancer or diabetes and comorbidities such as cardiovascular diseases [[12], [13], [14]]. Even if several red meat constituents, as well as cooking and processing practices, or associated lifestyle contribute to enhanced risk for these chronic diseases, the relative contribution of heme iron appears to be central [15,16]: on the basis of cohort studies and nutritional studies in rodent, meat-induced promotion of colorectal cancer has shown to be at least in part explained by its heme iron content and especially its ability to catalyze lipoperoxidation of dietary polyunsaturated fatty acids in the gut lumen [[17], [18], [19]]. In the same way, results from epidemiological and intervention studies confirm that heme iron from red and processed meat, but not non-heme iron, is positively associated with an increased risk for diabetes [11,[20], [21], [22]]. Oxidative stress is known to participate in the development, progression and complications of diabetes [23]. Interestingly, increased concentration of thiobarbituric acid reactive substances (TBARS) both in plasma from diabetic patients [24] and in intestine of diabetic rats [25] have also been reported, suggesting the involvement of lipoperoxidation processes. In parallel, serum 4-HNE measurements in diabetic patients have also shown to be positively correlated with the progression of diabetes [26].
To meet the demand of iron during pregnancy, higher intake of iron-rich foods, especially red meat, are supposed to limit iron deficiency and related adverse maternal and birth/neonatal outcomes [1]. Prospective cohort studies as well as meta-analyses reported however that an excessive consumption of sources of heme iron, especially from red and processed meat, are significantly associated with pregnancy complications and higher risk of gestational diabetes [[27], [28], [29], [30], [31]]. Development of gestational diabetes has been associated with adverse birth and neonatal outcomes such as pre-eclampsia, prematurity or low body weight, but the long-term outcomes resulting from a maternal heme-enriched diet on the offspring remain largely unexplored.
Maternal diet during fetal and post-natal development is one of the key factors from the maternal environment that can determine both the immediate and future health of the offspring, by exerting a sustained influence on growth, metabolic phenotype, gut barrier maturation and gut microbiota establishment [[32], [33], [34], [35]]. While epigenetic processes has been evoked to explain the concept that chronic diseases in adulthood may originate in early life [36], other hypotheses related to microbiota have emerged [37]: gut microbiota diversity depletion over generations in response to the “western pattern diet”, as well as disruption within successive stages governing order of establishment of key bacterial early colonizers in response to neonatal environment have been reported [[38], [39], [40]].
In this context, we have previously shown that, at nutritional dose and in comparison to non-heme iron, short term exposure to dietary heme iron at adulthood resulted in alteration of the main components of the gut barrier, including microbiome through lipoperoxidation associated pathways [41]. Since we observed that Escherichia coli, a well-known maternally transmitted bacteria belonging to the first colonizers after birth, strongly covariated with luminal heme-induced lipoperoxidation products [41], we hypothesized whether a disruption of maternal gut homeostasis induced by heme-enriched diet during the perinatal period, may persistently impact (i) gut microbiota establishment, (ii) neoformation of lipid peroxidation-derived reactive aldehydes, (iii) gut barrier and (iv) glucose metabolism in mice offspring.
2. Materials and methods
2.1. Experimental diets
A purified powder-based AIN-76A diet was optimized to maintain iron bioavailability (calcium level limited to 0.09%) while taking care to meet pregnancy requirements in terms of protein and amino-acids (UE 1298, Sciences de l’Animal & de l’Aliment, INRAE). In order to prepare frozen portions that were daily distributed into cage feeders and thus, limit diet lipid peroxidation, the powdered form was further transformed into jellied mass cut into cubes by incorporating porcine gelatin (8%, Louis François) and completed with oil (5% safflower oil, Fischer scientific) and 1,4 μmol iron/g diet (0,094% hemine or 0,0036% ferric citrate, Sigma) before storage under vacuum at −20 °C.
2.2. Ethical approval
All experiments protocols were approved by the Animal Care Use Committee (Comité d’Ethique Pharmacologie-Toxicologie-Occitanie Toulouse, registered as n°86 at the Ministry of Research) under the authorization number [#12915–2018010514411433], and conducted in accordance with the European Union guidelines.
2.3. Experimental design
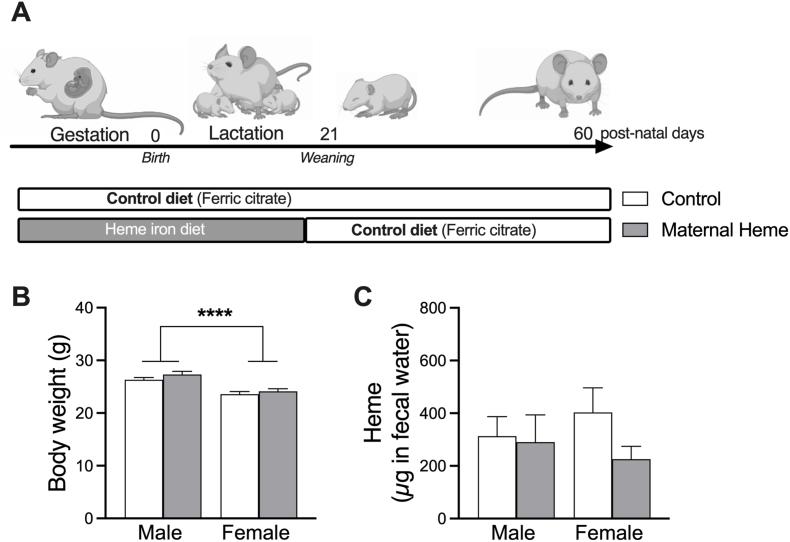
The experimental scheme is presented in Fig. 1A. Nulliparous female C3H/HeN mice were purchased at 8 weeks from Janvier Labs (Le Genest St Isle). Mice were kept at a constant temperature 22 °C on a 12-h light-dark cycle, and fed their experimental diet ad libitum daily just before their active period. During the 7 days of acclimatization and mating period with male C3H/HeN mice, mice were fed experimental Control diet supplemented with ferric citrate (inorganic iron). Female mice were mated by housing 3 females and 2 males per cage for 5 days, after which males were removed whereas females were randomly assigned to one of the 2 experimental groups until the pups were weaned: Control group (inorganic iron enriched diet) and Maternal Heme group (Heme iron-enriched maternal diet during pregnancy and lactation period). Group size was anticipated to generate a minimum of 5 pregnant females per group, and experimentation was repeated twice in order to minimize mother- and cage-specific divergence. A third independent experimentation devoted to repeating oral glucose tolerance tests and measuring insulin was performed. Pregnant females were housed individually five days before parturition. To avoid litter effect and resulting bias on body weight gain and oxidative status [42], litters were homogenized to 5–6 pups within the first 3 post-natal days (PND), while taking care to preserve male and female representation within each litter. Litter sizes less than 4 were excluded from the study. Weaning was performed at PND21, siblings were sex matched. From this stage, mice offspring were all fed the Control diet until PND60. Throughout the experiment, body weight was weekly monitored. Fecal samples stored at −80 °C were collected in F0 (pregnant and non-pregnant females) at PND05, PND15 and PND21 and in F1 (offspring) at PND21 and PND60.
Fig. 1.
In vivo effects of maternal heme iron exposure during gestation and lactation on the offspring. (A) Experimental design. Diet of females during gestation and lactation was enriched with Ferric citrate iron (Control) or Heme iron (Maternal Heme). After weaning, offspring were all fed the Control diet until the end of the experiment at post-natal day 60 (PND). At PND60 after maternal exposure to heme iron, measurement of (B) body weight (n = 25–30) and (C) fecal heme (n = 10–15). Values are presented as means ± S.E.M. Two-way ANOVA and Šídák's multiple comparison test: Fsex ****p < 0.0001.
One week before final plasma and tissue collection, 24-h fecal pellets and urine samples were collected for some mice under each metabolic cage to both quantify fecal and urinary biomarkers or perform fecal aldehydomic workflow [43].
2.4. High-throughput 16S rRNA gene amplicon analysis
Snap-frozen fecal samples and resulting purified genomic DNA were processed as described previously [41]. Briefly, fecal microbial DNA was purified on columns after mechanical lysis and hypervariable V3–V4 regions of the 16S rRNA gene were amplified using a two-step PCR. The resulting purified amplicon libraries were loaded onto the Illumina Miseq cartridge. The quality of the run and the lack of contamination were checked internally at each step in the sequencing facility (Get-PlaGe) and valid raw sequences were further processed using the FROGS pipeline (Galaxy Version 3.2.3) [44]. Each pair-end valid denoised sequences were filtered, merged and clustered with the swarm fastidious option using a maximum aggregation distance of 1 [45]. Once putative chimera were detected (vsearch) and removed, only clusters (i) whose abundance represented at least 0,005% of all sequences, (ii) presents in at least 5 times in a minimum of 5% of total samples with a prevalence threshold of 5% of all samples, were retained, yielding to 512 final clusters. The samples with fewer than 10,000 final valid sequences were removed from the statistical analysis. Clusters were affiliated to Operational Taxonomic Units (OTUs) using the silva 138.1_16S reference database and a taxonomic multi-affiliation procedure (Blast+with equal multi-hits that were manually verified and corrected if necessary). OTUs were agglomerated to examine differential abundances in response to maternal diet at the OTU and the family rank.
Community profiles (including α and β diversity) and differentially significant features were further analyzed using the R packages Phyloseq (v1.34.0) and DESeq2 (v1.30.1) respectively. Within sample community richness was assessed by the Chao-1 index and examined by two-way ANOVA analysis. Divergence of microbiota composition between samples was explored using the Unifrac distance matrices, visualized using multidimensional scaling (MDS) and assessed statistically using permutational multivariate analysis of variance (Adonis test with 9999 permutations).
Differences in relative abundances between groups were computed using the R’ DESeq2 package (v1.30.1) with LRT or Wald method according to the experimental factor design. Testings were corrected for false discovery rate (FDR) by Benjamini-Hochberg method, and adjusted p values less than 0.01 were considered statistically significant.
2.5. Fecal and urinary biomarkers
Fecal waters were prepared as previously described with 1 ml of distilled water and 50 μl of BHT in ethanol (butylated hydroxytoluene) for 0.5g of frozen feces [46]. Feces were grinded using Fast-Prep (MP Biomedicals) during 3 cycles of 30s at 6 m. with 1min breaks. After centrifugation for 20 min at 5.500g, supernatant was collected and kept at −20 °C until use.
Fecal heme was detected using a commercial kit (Sigma) according to the manufacturers protocol. Briefly, 96 wells flat plates (Greiner) were used to incubate fecal water sample and heme Calibrator (corresponding to 62.5 μM heme) with heme Reagent during 5 min. Absorbance measurement was perform (400 nm) using a microplate reader (Spark® Tecan Trading AG).
Fecal thiobarbituric reactive susbtances (TBARS) were measured according to Ohkawa et al. [47]. Briefly, 10 μl of fecal water were diluted in 90 μl of distilled water. A standard was performed with 1,1,3,3-tetramethoxypropane at 0 μM–1000 μM (TMOP). Sample and standard were incubated 1h at 95°c with 190 μl of 2-thiobarbituric acid replace by 190 μl of acetic acid (10%) for blank. The amount of TBARS was determined by measure of the absorbance at 532 nm.
The urinary DHN-MA (1,4-dihydroxynonene mercapturic acid) assay was performed using a competitive enzyme immunoassay as previously described [48].
2.6. Profiling of fecal reactive aldehydes through liquid chromatography-mass spectrometry
Free reactive aldehydes in fecal supernatants were stabilized and selectively labelled using a bromine atom according to Chevolleau et al. [43]. Briefly, derivatized aldehydes were selectively detected on the basis of their 79Br/81Br isotopic pattern through high performance liquid chromatography coupled to with high resolution mass spectrometry (HPLC-HRMS). Identification experiments were then realized via tandem mass spectrometry (MS/MS) using the iron trap mass analyzer of the LTQ-Orbitrap XL mass spectrometer.
Features corresponding to brominated compounds were extracted (XCMS software) and structural characterization of potential aldehydes was carried out by processing MSn spectra using Xcalibur QualBrowser (Thermo Scientific). In order to target discriminating potential aldehyde ions, both multivariate unsupervised principal component analysis (PCA) and supervised partial least squared discriminant analysis (PLS-DA) were used.
2.7. Oral glucose tolerance tests (OGTTs) and plasma analysis
OGTTs were performed in mice 6 h-fasted during day light on three independent experiments. Oral gavage was performed with glucose (2 mg glucose per g of bodyweight). Blood glucose levels were monitored from the blood of tail vein with a glucose meter (Accu-Chek® Performa) at −30, 0, 15, 30, 60, 90 and 120 min post-oral gavage. For plasma insulin measurement, blood samples from additional mice were collected from tail vein at 30 min after oral glucose gavage or obtained from cardiac puncture following mice euthanasia.
Plasma was prepared by centrifugation (1500g, 15 min, 4 °C) and stored at −80 °C until use. Insulin and GLP-1 were quantified using commercial ELISA kits (Merck Millipore).
2.8. Gut barrier analysis
2.8.1. Intestinal permeability in ussing chambers
Intestinal permeability measurement was performed as previously described [49]. Following mice euthanasia, jejunal fragments were immediately mounted in Ussing chambers (Physiologic Instruments) and incubated 2 h with Kreb's solution (Sigma) constantly oxygenated with carbogen (95% O2, 5% CO2) in presence of both Fluorescein Sodium Salt (FSS 376 Da, Sigma) and Horse Radish Peroxidase (HRP 4 kDA; Sigma) in the mucosal compartment. Gut integrity was continuously checked through electro-physiological measurements and electrical resistance was monitored (R, Ω.cm2). After 2h of incubation, the serosal compartment was sampled to follow the epithelial paracellular passage of FSS by measuring the fluorescence intensity (485nm/525 nm) using a microplate reader (Spark®). Epithelial permeability to HRP was determined by ELISA as previously described [49].
2.8.2. Genes expression
Total RNAs were extracted from jejunal frozen tissue using the Rneasy plus Mini kit (Qiagen) according to the manufacturer's instructions. Quantity and quality of extracted RNA samples were monitored using Nanodrop (Nanodrop™ 1000) and Bioanalyzer (Agilent) respectively. RNAs were reverse transcribed using enzyme IScript reverse transcription supermix (Biorad). The 384-well plates were completed by an Agilent Bravo Automated Liquid Handling Platform (Agilent Technologies, Santa Clara, CA, USA). All wells contain 5 μl of mix: 2.5 μl of ONEGreen® FAST qPCR Supermix (Ozyme), 1.5 μl of each primer set, and 1 μl of cDNA sample. Amplification was performed using a ViiA7 Real-Time PCR System (Applied Biosystems). Primers sequences and thermocycling conditions are listed in Supplemental Table 1. Raw data that passed quality control were normalized to HPRT1 mRNA levels and analyzed with LinRegPCR (version 2015.3).
2.8.3. Lipocalin-2 and TNF-α ELISA
Fecal lipocalin-2 and jejunal TNF-α concentrations were measured using commercial mouse Quantikine ELISA kits (R&D Systems), from homogenized feces and jejunal fragment suspended respectively in PBS Buffer 10% and in RIPA buffer (0.5% deoxycholate, 0.1% SDS and 1% Igepal in TBS) containing complete anti protease cocktail (Sigma). Fecal and jejunal protein concentrations was measured using BCA uptima kit (Interchim, Montlucon, France) and used to normalize assays.
2.9. Multi-omics analyses
Correlation structure between omics data sets was explored using the R package mixOmics DIABLO framework (Data Integration Analysis for Biomarker discovery using Latent cOmponents, v6.15.45) [50,51]. This integrative multivariate method is able to maximize correlations between different data-types and perform discriminant analysis with the aim of identifying a signature specific to the male offspring maternally exposed to heme iron. Only mice for whom both physiological traits (metadata without data associated with glucose challenge test), 16S rRNA gene amplicon data and aldehydomics data were available at PND60, were included in this framework (12 females Control, 10 females Maternal Heme, 11 males Control, 10 males Maternal Heme). Prior to fitting multi-block DIABLO model, 16S rRNA gene amplicon and aldehydomics datasets were preprocessed according to the mixMC framework and log-ratio transformed respectively [52], and independent PLS-DAs were built. A total of 10 physiological traits, 512 taxa and 1135 putative aldehydes were retained for analysis after data pre-processing.
2.10. Statistical analyses
GraphPad Prism version 9.2 was used for all remaining analyses displayed in graphs. Results are expressed as mean ± S.E.M unless otherwise stated. N refers to the number of mice per group used for in each experiment. The significance of differences between experimental groups was determined by two-way analysis of variance (ANOVA, 2 factors: iron, sex and their interaction were tested) with Šídák's multiple comparisons pos-test unless otherwise stated. For kinetics, a three-way Repeated Measures-ANOVA matching by time was performed (3 factors: iron, sex and time) with Šídák's multiple comparisons pos-test. Two-side analyses were used throughout, and a p value < 0.05 was considered significant.
3. Results
3.1. Impact of dietary iron form in maternal diet on offspring weight gain
During gestation and lactation, female mice were fed a modified semi-purified diet balanced either with ferric citrate (Control) or heme (Maternal Heme) as dietary iron source. At birth, the offspring sex ratio was not influenced by the iron form in the maternal diet during gestation (0.58 ± 0.04 males/litter from heme-fed mothers vs 0.58 ± 0.04 males/litter from mothers fed the Control diet, p > 0.05). At weaning, maternal heme exposure was discontinued and the offspring were all fed the Control diet until post-natal-day 60 (PND60, Fig. 1A). As expected, offspring female mice weighed less than males at PND60 (Fsex, p < 0.001, Fig. 1B), but their respective post-natal growth and final body weights were not affected by maternal exposure to heme iron as compared to non-heme iron (Firon, p > 0.05 at PND60, Fig. 1A, Fig. S1A). Early exposure to maternal dietary heme iron did not change basal detection of luminal heme iron in offspring at PND60 relative to the Control group (Fig. 1C).
3.2. Heme and non-heme iron in maternal diet differentially shape the fecal microbiota
Fecal microbiota analysis by sequencing the V3–V4 region of the 16S rRNA gene amplicon revealed that both the structure and the composition of microbiota was differentially affected according to the iron form added to the maternal diet.
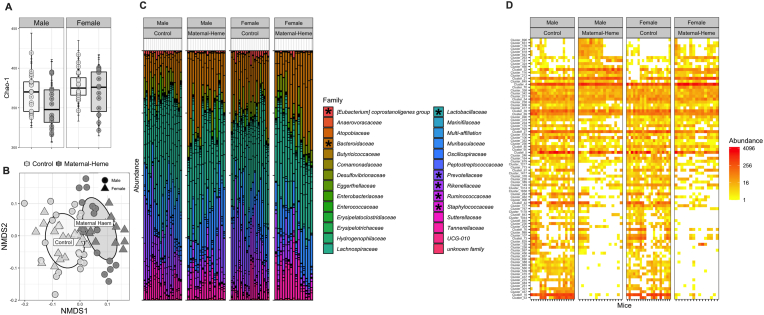
Microbiota richness at PND60, as indicated by the Chao-1 index (Fig. 2A), was indeed affected by both the maternal diet (Firon, p < 0.05), and the sex (Fsex, p < 0.05), revealing a more pronounced decrease in Operational Taxonomic Unit (OTUs) richness in males exposed to maternal heme enriched diet compared to females or Control males. Considering the importance of maternal microbiota inheritance during gut colonization stage, richness within fecal microbiota of adult female mice fed the heme-enriched diet (lactating or not, Supplemental Fig. 1B) and offspring at weaning stage (Supplemental Fig. 1C) was assessed. We showed that, even if lactation progressively enhanced richness whatever the diet (Fstage, p < 0.0001), heme-enriched diet was associated with a reduced Chao-1-index in the F0 generation (Firon, p = 0.05) that was passed on only to males in the F1 generation.
Fig. 2.
Effect of maternal exposure to heme iron as compared to non-heme iron on the fecal microbiota at PND60. The bacterial community diversity structure, distribution and composition of fecal microbiota was determined by 16S rRNA gene amplicon Illumina Miseq sequencing, at the OTU and family level (n = 16–20): (A) Richness (α-diversity) measured by Chao-1 index (ANOVA Firon = 0.035, Fsex = 0.0215) (B) UniFrac Non-metric Multidimensional Scaling (NMDS) ordination plot representing structural changes between diets (β-diversity, MANOVA Firon<0.0001, Fsex<0.0001, Firon xsex = 0.019) (C) Family Distribution; *p adj<0.05 using Differential abundance analysis (DESeq2) at the family level (D) Heatmap of OTUs significantly affected by the maternal diet; **p adj<0.01 using Differential abundance analysis (DESeq2) at the OTU level. Supplemental and related analyses, plots and taxonomic affiliations are presented in Supplemental Fig. 1,2 and Tables 2,3OTU, Operational Taxonomic Unit; PND, post-natal day.
Ordination based on UniFrac distances between samples at PND60 (Fig. 2B) revealed the existence of distinct bacterial communities that clustered mainly according to the maternal diet (Firon<0.0001) but also differentially according to the sex (Fsex<0.0001, Firon xsex = 0.019). This clustering of β-diversity based on the iron form added to the maternal diet probably stems from microbiota disruption observed early in the offspring (Supplemental Fig. 1C) and started initially in mothers (Supplemental Fig. 1B), as revealed by the relative abundance profile of affected OTUs (padj<0.05, Maternal Heme vs Control), which appeared to share similarities between mothers and their respective offspring at PND21 (Supplemental Fig. 1D and Table 2).
Regarding the microbiota composition at the family level of offspring from heme-enriched-fed mothers as compared to Control group at PND60 (Fig. 2C), 7 of the 28 families detected were significantly altered in response to the maternal diet (padj<0.05).
Among them, Bacteroidaceae was overabundant, whereas the Eubacterium coprostanoligenes group, Lactobacillaceae, Prevotellaceae, Rikenellaceae and Ruminococcaceae were underrepresented in mice early exposed to maternal heme diet compared to Control diet (Supplemental Fig. 2A). Changes in relative abundances for some of them in response to maternal diet were however more pronounced in males than in females (Bacteroidaceae, Lactobacillaceae, and Ruminococcaceae) whereas a divergent behavior of Staphylococcaceae relative abundances across sex was observed (less abundant in male and more in female). Deseq2 analysis at the OTU level allowed us to sort differentially abundant taxa at PND60 (a total of 88 out of 512 with padj<0.01, Fig. 2D, Supplemental Fig. 2B and Table 3), revealing the main driver effect of maternal diet on microbiota over the sex effect. By tracing the relative abundance of these selected significantly affected OTUs at earlier times (Supplemental Fig. 2C), we observed that they were not really impacted by the diet in mothers or offspring at PND21 (most of them were specific to the PND60 period, Supplemental Fig. 2D), suggesting that the bacterial profile associated with maternal heme-enriched diet at adulthood does not come directly from the maternal or post-natal microbiota imprint.
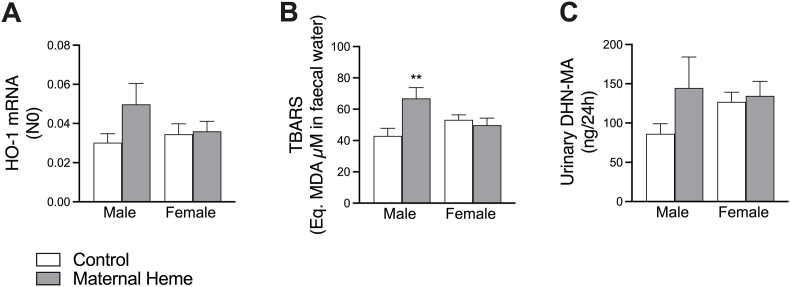
3.3. Maternal heme iron intake promotes luminal lipoperoxidation in male offspring
Although the levels of fecal heme detected were equivalent (Fig. 1C), a slight induction of heme oxygenase 1 expression was noticed in jejunal tissues of males early exposed to the maternal heme-enriched diet compared with the other groups (Fig. 3A), signing an oxidative insult. This male-specific induction was accompanied by higher lipoperoxidation activity, as evidenced by fecal TBARS levels representing the luminal side (Firon<0.05, Firon x sex<0.01, padjmale iron<0.01, Fig. 3B), and by substantial augmentation of 1,4-dihydroxynonene mercapturic acid (DHN-MA) levels detected in urines (padjmale iron = 0.085 males Maternal Heme vs Control, Fig. 3C).
Fig. 3.
Dietary maternal heme iron exposure during perinatal period influences lipoperoxidation status in gut offspring in a sex-dependent manner. Measurements at PND60 of (A) Expression of heme oxygenase 1 gene (HO-1 also known as Heat-Shock Protein-32) in jejunal tissues (n = 8–10), (B) TBARS in fecal waters (Firon = 0.047, Firon x sex = 0.009, n = 10–12), (C) Urinary DHN-MA (n = 10–12). Values are presented as means ± S.E.M. Two-way ANOVA and Šídák's multiple comparison test: **p < 0.01. DHN-MA, 1,4-dihydroxynonene mercapturic acid; HO-1, Heme Oxygenase 1; TBARS, Thiobabituric Reactive Substances.
Unlike males, none of these biomarkers was changed in female offspring. Furthermore, measurements of TBARS in male mice transiently exposed to heme-enriched diet during adulthood (Supplemental Fig. 3A) showed that heme-induced lipoperoxidation was completely normalized 30 days after discontinuation of heme exposure (Supplemental Fig. 3B), suggesting that maternal heme iron exposure during the perinatal period triggers specific neoformation of lipoperoxidation products in male offspring.
3.4. Maternal heme iron intake promotes gut barrier impairments in male offspring
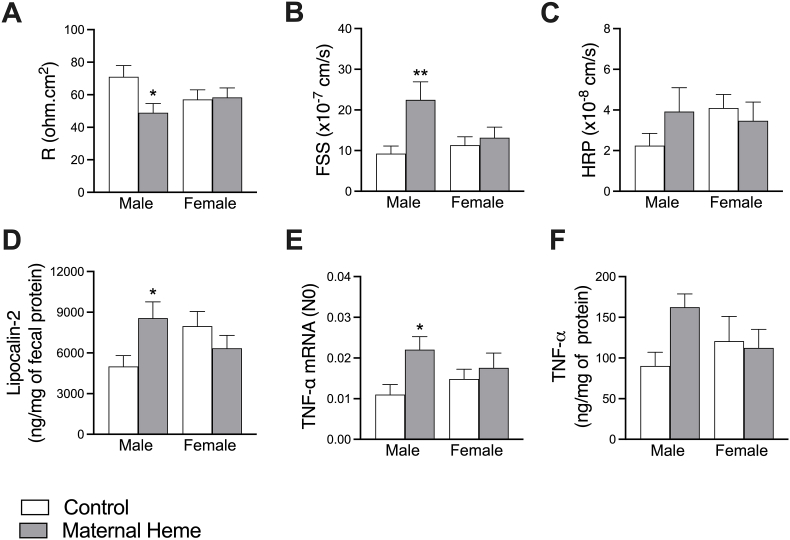
Next, we wished to know whether the neoformation of lipoperoxidation products in male offspring might lead to gut leakiness. To this end, jejunal tissues were mounted in Ussing chambers and permeability parameters were compared in offspring at PND60. Interestingly, male offspring, and not females, from heme-enriched fed mothers displayed reduced electrical resistance (R, Firon x sex<0.05, padjmale iron<0.05, Fig. 4A), associated with increased paracellular permeability to Fluorescein Sodium Salt (FSS, Firon<0.01, Firon x sex<0.05, padjmale iron<0.01, Fig. 4B) compared to Control groups or in comparison to males that were transiently exposed to heme iron as adults (Supplemental Fig. 3C). No difference in transcellular permeability was noticed (Horse Radish Peroxidase, HRP, Fig. 4C).
Fig. 4.
Dietary maternal heme iron exposure during perinatal period influences gut barrier homeostasis in offspring in a sex-dependent manner. Assessment at PND60 of intestinal epithelial permeability using Ussing chambers ex-vivo experiment on jejunal segments (n = 18–20) through the measurements of: (A) Electrical resistance (R, Firon x sex<0.05), (B) paracellular epithelial permeability to FSS (Firon0<0.01, Firon x sex<0.05), (C) transcellular epithelial permeability to HRP. Inflammation status (n = 7–11) was explored through measurements of (D) fecal lipocalin-2 (Firon x sex<0.05), (E) jejunal TNF-α gene (Firon<0.05) and (F) ileal TNF-α concentration. Values are presented as means ± S.E.M. Two-way ANOVA and Šídák's multiple comparaison test: *padj<0.05, **padj<0.01. FSS, Fluorescein Sodium Salt; HRP, Horse Radish Peroxidase; TNF-α, Tumor Necrosis Factor alpha.
In male offspring maternally exposed to heme-enriched diet, paracellular permeability defect was associated with both an increase of fecal inflammatory marker lipocalin-2 (Firon x sex<0.05, padjmale iron <0.05, Fig. 4D), and an induction of the pro-inflammatory cytokine TNF-α (gene and protein expression, Firon<0.05, padjmale iron<0.05 and Firon x sex = 0.08, padjmale iron = 0.06 respectively, Fig. 4E–F). Unlike males, none of these biomarkers was changed in female offspring in response to the maternal exposure to heme iron.
3.5. Glucose tolerance in male offspring exposed to maternal heme iron is impaired
Having shown that gut barrier function was compromised in male offspring from heme iron-enriched fed mothers, and that such leaky gut is often associated with systemic pathological outcomes including metabolic disorders [53], we next monitored glucose homeostasis in offspring.
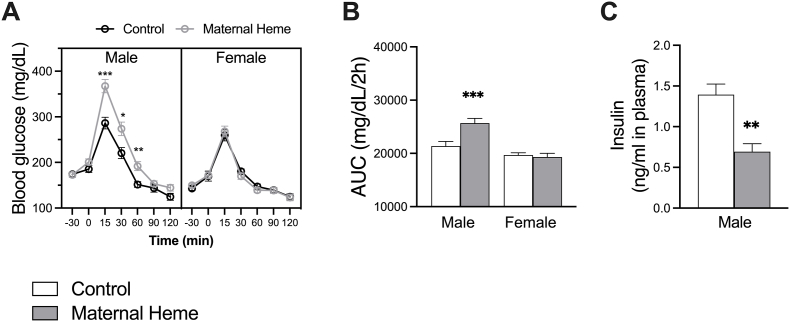
Fasting or not, blood glucose levels as well as plasma insulin and Glucagon Peptide 1 (GLP-1) levels were not affected by the form of the iron added in maternal diet (Supplemental Fig. 4). Fifteen min after the glucose challenge however, male mice from the Maternal Heme group peaked at 367 mg/dL ±15 glucose and showed impaired blood glucose disposal as compared to male mice in the Control group (286 mg/dL ±13, padj<0.001, Fig. 5A). Calculation of the area under the curve (AUC) above baseline glucose confirmed occurrence of glucose intolerance in males but not in females (Firon<0.05, Fsex<0.0001, Firon x sex<0.01, padjmale iron<0.001, Fig. 5B) or in adult males transiently exposed to heme iron (Supplemental Fig. 3D). Accordingly, glucose intolerance observed in males was associated to impaired insulin secretion 30 min after glucose challenge (unpaired t-test p < 0.01, Fig. 5C).
Fig. 5.
Dietary maternal heme iron exposure during perinatal period impaired blood glucose tolerance in male offspring. Oral glucose tolerance test (OGTT) was performed on 6h fasted mice (n = 18–23) and consisted in the measurements of (A) blood glucose (mg/dL) from t = −30min to t = 120 min after oral glucose gavage (t = 0 min), and the calculation of (B) Area under the Curve (AUC) of blood glucose 0–120 min (mg/dL/min). (C) Plasma insulin was determined in blood collected 30 min after oral glucose gavage (n = 5). Values are presented as means ± S.E.M. For kinetics in A, a three-way RM-ANOVA matching by time was performed: Firon<0.01, Fsex<0.0001, Firon x sex<0.01, Firon x sex x time<0.001 and Šídák's multiple comparison test: *padj<0.05, **padj<0.01, ***padj<0.001. In B, a two-way ANOVA was performed: Firon<0.05, Fsex<0.0001, Firon x sex<0.01, and Šídák's multiple comparison test: ***padj<0.001. In C, Unpaired t-test with Welch's correction was used: **p < 0.01. Related analyses and plots are presented in Supplemental Fig. 4.AUC, Area Under the Curve; OGTT, Oral Glucose Tolerance Test.
3.6. Identification of the fecal reactive aldehyde profile resulting from maternal dietary exposure to heme iron
Neoformation of lipoperoxidation products detected in feces of male offspring early exposed to maternal dietary heme iron-enriched diet (Fig. 3B) led us to continue their characterization using an untargeted omic approach [43]. To this aim, carbonyl compounds including reactive aldehydes present in fecal waters were selectively derivatized introducing a bromine atom, whose characteristic isotopic pattern is detectable using high-performance liquid chromatography coupled to high resolution mass spectrometry (HPLC-HRMS). Out of 6663 variables detected, 1135 variables displayed the expected brominated isotopic pattern. The corresponding PLS-DA score plot showed a main separation of males from females along the first component whereas separation according to maternal diet, and especially in males, appeared on the following components (Supplemental Fig. 5A).
In order to directly target both putative reactive aldehydes and bacterial taxa specifically associated with the leaky gut phenotype identified in males from heme iron-enriched fed mothers, a multiblock supervised model allowing datasets integration was built [50]. In contrast to aldehydomic data structuration, PLS-DA applied to microbiota dataset showed a separation of mice primarily by maternal diet and then by sex (Supplemental Fig. 5B), but connections between datasets were clearly found (Supplemental Fig. 5D) and allowed to capture signatures describing key features relevant to each mice group (Fig. 6A): One signature outlined the sex-associated features including mainly putative reactive unidentified aldehydes that were under-expressed in males and negatively correlated with body weight (Fig. 6B, Supplemental Table S4). Interestingly, correlations spanning the 3 block subspaces were primarily related to the phenotype encountered in males from heme iron-fed mothers (Fig. 6A–B): highest or lowest values in those males compared to others groups (visualized outside the circos plot in Fig. 6B), such as lipoperoxidation (TBARS) and permeability parameters (FSS, R), were strongly interconnected with numerous carbonyl compounds including reactive aldehydes and bacterial OTUs (Fig. 6B and Table S4). Among the closest features (Fig. 6C), interconnections between paracellular permeability values (FSS), 5 bacterial taxa (3 unknown Bacteroides, an unknown Muribaculaceae and an Lachnospiraceae belonging to A2 group or Lachnoclostridium) and 8 carbonyl compounds were highlighted (Supplemental Table S4). Using databases, 2 out of the 3 brominated compounds that correlated negatively with gut hyperpermeability were putatively annotated (namely cervonoyl ethanolamide and nutriacholic acid), but none of the other 5 candidates that could potentially explain the increase in fecal TBARS, corresponded to the previously identified lipoperoxidation products resulting from direct dietary heme iron uptake [43], suggesting a profile specific to the maternal exposure.
Fig. 6.
Identification of bacterial communities and putative reactive aldehydes that most covariated with leaky gut phenotype observed in male offspring from mothers fed dietary heme iron during perinatal period. Correlations between fecal aldehydomic (1135 variables in green) and fecal microbiota (512 OTUs in orange) datasets with gut metadata (in blue) available for 43 mice (n = 10–12) were maximized using the DIABLO framework: (A) Clustered image map representing variables selected in each block to characterize each mice sample. (B) Circos plot depicting the highest correlations (negatives and positives) between each block. “Expression” levels of each variable according to mice groups were plotted outside the circle. (C) corresponding relevance network with absolute correlations >0.68. Related analyses, plots and taxonomic affiliations are presented in Supplemental Fig. 5 and Table 4. DIABLO, Data Integration Analysis for Biomarker discovery using Latent variable approaches for Omics studies; FSS, Fluorescein Sodium Salt; R, Electrical Resistance; OTU, Operational Taxonomic Unit. . (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
In adulthood, it is established that dietary heme iron at nutritional dose catalyzes lipid oxidation, leading to gut luminal production of reactive aldehydes that have been associated with microbiota reshape [41,54,55], numerous detrimental alterations on the intestinal barrier function [41,[56], [57], [58]], including colon cancer promotion [15,17,59] and more putatively other systemic pathologies, including diabetes [16]. However to date, few studies have investigated the impact on offspring of maternal dietary heme iron intake during pregnancy and lactation, i.e. periods during which (i) iron requirement is greatly amplified, (ii) embryo and fetus have immature antioxidant defense system, and (iii) neonate is highly sensitive to its maternal environment.
Iron deficiency during pregnancy has shown to increase the risk of adverse birth/neonatal outcomes, including low birth weight, impaired body weight gain, neonatal iron deficiency and numerous associated long-term neurobehavioral effects [2,29,[60], [61], [62]]. Interestingly, maternal iron deficiency has shown however to improve glucose tolerance in 3-month-old rat offspring [63]. In our study, neurobehavioral abnormalities were not examined and the form of dietary iron did not alter offspring birth weight, weight gain or sex ratio, but we provide evidence that dietary heme iron, unlike non-heme iron, through its lipoperoxidation potential, drives impaired glucose tolerance in maternally exposed male offspring. Using an integrated approach, we showed that this male-dependent phenotype was linked with occurrence of a gut luminal pro-oxidative environment associated with a combination of interconnected factors well known to trigger diabetes, and particularly type 1 diabetes [64,65]: a specific microbiota pattern displayed reduced richness and close relationships with gut barrier defects in terms of epithelial permeability and low-grade inflammation.
In line with high concentrations of heme iron, red meat, and especially processed meat, is positively associated with type 1 and type 2 diabetes [11,13,20]. Although the underlying mechanism is not fully elucidated, the proposed hypothesis involves oxidative stress-induced ROS resulting from heme iron-induced lipoperoxidation products that may promote insulin resistance as well as detrimental activity on pancreatic beta cells [16]. Interestingly in our study, gut luminal reactive aldehydes detected in the male offspring did not result from higher concentration of heme iron in fecal content, and differed from these commonly detected in presence of dietary heme [43], suggesting a pattern of lipoperoxidation specific to the maternal exposure.
Therefore, this persistent pro-oxidant gut environment observed at PND60 raises questions about its origin. Gut luminal lipid peroxidation levels have shown to be influenced by both litter size and body weight gain during lactation [42] or following lipid overload [66], but litter size as well as dietary lipids intake were both controlled in our study, whereas body weight gain was not affected by the iron form added to the maternal diet. Numerous nutritional experiments conducted in animal models or human cohort studies (reviewed in Refs. [[67], [68], [69]]) revealed that a maternal pro-oxidant environment during the perinatal period may cause direct oxidative damage during development and promote nutritional programming to disease. In our study, only the male offspring from heme iron-fed mothers presented signs of oxidative insults, but remarkably, it has been widely observed that female neonates display an enhanced antioxidant defense system, probably due to estrogen metabolism [70]. Accordingly, the sex-dependent levels of luminal lipoperoxidation observed at PND60 in response to maternal heme iron-enriched diet may originate from the poor capacity of males to cope with oxidative stress during the neonatal period. In parallel, we can also presume that the heme iron-altered gut microbiota vertically transferred from mother to neonates may have an intrinsic ability to generate more specific reactive aldehydes. Indeed, Lee et al. demonstrated that some microbiota communities were able to limit or conversely to promote lipoperoxidation [71]. We also previously showed that depletion of gut microbiota by antibiotics prevents lipoperoxidation in heme iron-fed adult rats [54], suggesting that reactive aldehydes originate mainly from gut bacterial communities.
Even if we observed that heme iron-induced dysbiosis was reduced during lactation in mothers, we still noted that the alterations seen in offspring microbiota in response to maternal diet, mirrored those seen in their mothers at weaning stage. However, this heme iron-specific maternal imprint evolved after weaning and gave way in 60-days-old offspring, to a new combination of bacterial taxa differentially affected by the maternal diet. It remains to be determined whether the offspring microbiota at PND60 results from natural post-weaning trajectories in the maturation of the bacterial assembly based on two different maternal-inherited microbiota, from microbiota adaptation to its pro-oxidative gut environment, or from a combination of both. Interestingly in our study, putative reactive aldehydes produced mainly by males perinatally exposed to heme, covariated with some OTUs relative abundance. Among the most positively correlated OTUs, several belonged to the Bacteroides spp. i.e., a genus found to be vertically transmitted from mother to neonate, and a persistent colonizer in the infant gut [72]. Species belonging to Bacteroides require heme iron for growth, and consistently, they have shown to be well equipped to survive in an oxidative environment notably by detoxifying intracellularly peroxides [73]. Similarly to Bacteroides taxa, the unclassified OTU belonging to Muribaculaceae is described as a murine commensal genetically equipped to support oxidative stress [74]. Among Firmicutes, several multi-affiliated OTUs belonging to Lachnospiraceae were also found to covary with putative reactive aldehydes, but similarly to previous OTUs mentioned, these taxa are not known in the literature to directly generate lipoperoxidation products. Nonetheless, they all belong to communities previously described as being associated to gut barrier defects, as revealed by our circos plot, and/or predictive of diabetes development, in line with the observation in this study of glucose intolerance occurrence in male offspring [[75], [76], [77], [78], [79], [80], [81], [82], [83]].
Dietary intake of heme iron on the one hand [13] and increased levels of TBARS, lipid hydroperoxides and lipoperoxides in plasma or urines on the other hand [24,26,84,85], have previously been associated with the risk of diabetes later in life, whereas the underlying mechanisms explaining these associations, including the generation of lipoperoxidation endproducts [13,16,84], have not been clearly demonstrated. Further, aberrant gut microbiota establishment with reduced diversity has been observed in newly diagnosed diabetic children [65,77,79], and most importantly, rescue of the gut barrier disruption by microbiota manipulation, has been shown to prevent low-grade intestinal inflammation, and more specifically, to prevent activation of islet-reactive T cells preceding their migration to the pancreas in diabetes-prone rats [86], suggesting a direct causal relationship between leaky gut and autoimmune disease aggravation.
Collectively, our results recapitulate the sequential events described above, starting in the gut with enhanced luminal lipoperoxidation sustained by an altered microbiota with reduced bacterial richness, and leading to impaired insulin secretion, as a result of altered epithelial barrier integrity and associated low-grade inflammation. By containing this pro-oxidant luminal environment, as observed in female offspring, this domino effect was not observed, suggesting that some metabolic disorders at adulthood may be prevented early in life. Both epidemiological and experimental studies in adulthood have shown that natural and dietary antioxidants, through their ability to limit the luminal formation of heme iron-induced reactive aldehydes, also limited disease risk without altering the luminal iron bioavailability [17,18]. Maternal interventions with antioxidant nutrients could be a strategy to meet nutritional iron requirements while avoiding the associated deleterious effects.
Author contribution statement
Conceptualization: MO with VT & FHFP, Methodology & Experimentations: AM, LM, CM, CL, VB, ML, IA, VAB, NN, CHT, SC, FG & MO, Formal analysis: AM, LM, VB, ML, LD, SC, FG, FHFP, VT & MO, Writing original draft preparation: AM & MO, Writing review and editing: LM, LD, SC, FG, FHFP & VT, All authors read and approved the final manuscript.
Fundings
This work was supported by the Human Nutrition and Food safety research division (AlimH) of the French National Research Institute for Agriculture, Food and Environment (INRAE), and the Région Occitanie.
Data availability statement
The raw data set (16S rRNA sequences) described in this project has been deposited in the MG-RAST database under accession code mgp100998. Further information and requests for resources and reagents will be fulfilled by the corresponding author, MO.
Declaration of competing interest
The authors state they have no competing interests.
Acknowledgments
The authors thank Xavier Blanc (UE 1298, Sciences de l’Animal & de l’Aliment, INRAE) for providing custom experimental diets and all members of the EZOP (Animal facility) for assistance with the animal experimentation. The authors thank the Get-PlaGe platform (Toulouse) for 16S rRNA gene libraries and sequencing and to the Genotoul bioinformatics platform Toulouse Occitanie and Sigenae group for providing help and storage resources thanks to Galaxy instance (https://galaxy-workbench.toulouse.inra.fr). The authors are grateful to Alyssa Bouville, Carla Orlandi and Jean-François Martin from the Metatoul-AXIOM analytical platform (Metabolomics and Toxicology) for their careful help with this project. All MS experiments were performed on the instruments of the MetaToul-AXIOM platform, partner of the national infrastructure of metabolomics and fluxomics: MetaboHUB [MetaboHUB-ANR-11-INBS-0010, 2011].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102333.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Sungkar A. The role of iron adequacy for maternal and fetal health. World Nutrition J. 2021;5:10–15. [Google Scholar]
- 2.Brannon P.M., Taylor C.L. Iron supplementation during pregnancy and infancy: uncertainties and implications for research and policy. Nutrients. 2017;9:1–17. doi: 10.3390/nu9121327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galaris D., Barbouti A., Pantopoulos K. Iron homeostasis and oxidative stress: an intimate relationship. Biochim. Biophys. Acta Mol. Cell Res. 2019;1866 doi: 10.1016/j.bbamcr.2019.118535. Elsevier. [DOI] [PubMed] [Google Scholar]
- 4.Puntarulo S. Iron, oxidative stress and human health. Mol. Aspect. Med. 2005;26:299–312. doi: 10.1016/j.mam.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Dixon S.J., Stockwell B.R. The role of iron and reactive oxygen species in cell death. Nat. Chem. Biol. 2014;10:9–17. doi: 10.1038/nchembio.1416. [DOI] [PubMed] [Google Scholar]
- 6.Gammella E., Recalcati S., Cairo G. Dual role of ROS as signal and stress agents: iron tips the balance in favor of toxic effects. Oxid. Med. Cell. Longev. 2016;2016 doi: 10.1155/2016/8629024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collard K.J. Iron homeostasis in the neonate. Pediatrics. 2009;123:1208–1216. doi: 10.1542/peds.2008-1047. [DOI] [PubMed] [Google Scholar]
- 8.Davis J.M., Auten R.L. Maturation of the antioxidant system and the effects on preterm birth. Semin. Fetal Neonatal Med. 2010;15:191–195. doi: 10.1016/j.siny.2010.04.001. Elsevier Ltd. [DOI] [PubMed] [Google Scholar]
- 9.Fairweather-Tait S., Sharp P., Iron Adv. Food Nutr. Res. 2021;96:219–250. doi: 10.1016/bs.afnr.2021.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Andreas N.J., Kampmann B., Mehring Le-Doare K. Human breast milk: a review on its composition and bioactivity. Early Hum. Dev. 2015;91:629–635. doi: 10.1016/j.earlhumdev.2015.08.013. Elsevier Ireland Ltd. [DOI] [PubMed] [Google Scholar]
- 11.Czerwonka M., Tokarz A. Iron in red meat–friend or foe. Meat Sci. 2017;123:157–165. doi: 10.1016/j.meatsci.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 12.Bastide N.M., Pierre F.H.F., Corpet D.E. Vol. 4. Cancer prevention research; Philadelphia, Pa: 2011. pp. 177–184. (Heme Iron from Meat and Risk of Colorectal Cancer: a Meta-Analysis and a Review of the Mechanisms Involved). [DOI] [PubMed] [Google Scholar]
- 13.Misra R., Balagopal P., Raj S., Patel T.G. Red meat consumption (heme iron intake) and risk for diabetes and comorbidities? Curr. Diabetes Rep. 2018;18 doi: 10.1007/s11892-018-1071-8. [DOI] [PubMed] [Google Scholar]
- 14.De Oliveira Mota J., Boué G., Guillou S., Pierre F., Membré J.M. Estimation of the burden of disease attributable to red meat consumption in France: influence on colorectal cancer and cardiovascular diseases. Food Chem. Toxicol. 2019;130:174–186. doi: 10.1016/j.fct.2019.05.023. Elsevier. [DOI] [PubMed] [Google Scholar]
- 15.Bastide N.M., Chenni F., Audebert M., Santarelli R.L., Taché S., Naud N., et al. A central role for heme iron in colon carcinogenesis associated with red meat intake. Cancer Res. 2015;75:870–879. doi: 10.1158/0008-5472.CAN-14-2554. [DOI] [PubMed] [Google Scholar]
- 16.White D.L., Collinson A. Red meat, dietary heme iron, and risk of type 2 diabetes: the involvement of advanced lipoxidation endproducts. Adv. Nutr. 2013;4:403–411. doi: 10.3945/an.113.003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin O.C.B., Naud N., Taché S., Debrauwer L., Chevolleau S., Dupuy J., et al. Vol. 11. Cancer Prevention Research; Philadelphia, Pa): 2018. pp. 569–580. (Targeting Colon Luminal Lipid Peroxidation Limits Colon Carcinogenesis Associated with Red Meat Consumption). [DOI] [PubMed] [Google Scholar]
- 18.Bastide N., Morois S., Cadeau C., Kangas S., Serafini M., Gusto G., et al. Heme iron intake, non-enzymatic antioxidant capacity, and risk of colorectal adenomas in a large cohort study of French women. Cancer Epidemiol. Biomarkers Prev. 2016;24:1–28. doi: 10.1158/1055-9965.EPI-15-0724. [DOI] [PubMed] [Google Scholar]
- 19.Pierre F.H.F., Martin O.C.B., Santarelli R.L., Taché S., Naud N., Guéraud F., et al. Calcium and α-tocopherol suppress cured-meat promotion of chemically induced colon carcinogenesis in rats and reduce associated biomarkers in human volunteers. Am. J. Clin. Nutr. 2013;98:1255–1262. doi: 10.3945/ajcn.113.061069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Talaei M., Wang Y.L., Yuan J.M., Pan A., Koh W.P. Meat, dietary heme iron, and risk of type 2 diabetes mellitus. Am. J. Epidemiol. 2017;186:824–833. doi: 10.1093/aje/kwx156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Z., Li S., Liu G., Yan F., Ma X., Huang Z., et al. Body iron stores and heme-iron intake in relation to risk of type 2 diabetes: a systematic review and meta-analysis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0041641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao W., Rong Y., Rong S., Liu L. Dietary iron intake, body iron stores, and the risk of type 2 diabetes: a systematic review and meta-analysis. BMC Med. 2012;10 doi: 10.1186/1741-7015-10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maritim A.C., Sanders R.A., Watkins J.B. Diabetes, oxidative stress, and antioxidants: a review. J. Biochem. Mol. Toxicol. 2003;17:24–38. doi: 10.1002/jbt.10058. [DOI] [PubMed] [Google Scholar]
- 24.Martín-Gallán P., Carrascosa A., Gussinyé M., Domínguez C. Biomarkers of diabetes-associated oxidative stress and antioxidant status in young diabetic patients with or without subclinical complications. Free Radic. Biol. Med. 2003;34:1563–1574. doi: 10.1016/s0891-5849(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 25.Maritim A.C., Moore B.H., Sanders R.A., Watkins J.B. Effects of melatonin on oxidative stress in streptozotocin-induced diabetic rats. Int. J. Toxicol. 1999;18:161–166. [Google Scholar]
- 26.Lou B., Boger M., Bennewitz K., Sticht C., Kopf S., Morgenstern J., et al. Elevated 4-hydroxynonenal induces hyperglycaemia via Aldh3a1 loss in zebrafish and associates with diabetes progression in humans. Redox Biol. 2020;37:101723. doi: 10.1016/j.redox.2020.101723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu C., Zhang C., Gelaye B., Enquobahrie D.A., Frederick I.O., Williams M.A. Gestational diabetes mellitus in relation to maternal dietary heme iron and nonheme iron intake. Diabetes Care. 2011;34:1564–1569. doi: 10.2337/dc11-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hajianfar H., Abbasi K., Azadbakht L., Esmaeilzadeh A., Mollaghasemi N., Arab A. The association between maternal dietary iron intake during the first trimester of pregnancy with pregnancy outcomes and pregnancy-related complications. Clinical Nutrition Res. 2020;9:52. doi: 10.7762/cnr.2020.9.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iqbal S., Ekmekcioglu C. Maternal and neonatal outcomes related to iron supplementation or iron status: a summary of meta-analyses. J. Matern. Fetal Neonatal Med. 2017;32:1528–1540. doi: 10.1080/14767058.2017.1406915. Informa UK Ltd. [DOI] [PubMed] [Google Scholar]
- 30.Marí-Sanchis A., Díaz-Jurado G., Basterra-Gortari F.J., de la Fuente-Arrillaga C., Martínez-González M.A., Bes-Rastrollo M. Association between pre-pregnancy consumption of meat, iron intake, and the risk of gestational diabetes: the SUN project. Eur. J. Nutr. 2018;57:939–949. doi: 10.1007/s00394-017-1377-3. [DOI] [PubMed] [Google Scholar]
- 31.Middleton P. Gestational diabetes: higher animal protein intake during pregnancy is associated with increased risk, and higher vegetable protein intake with decreased risk. Evid. Base Nurs. 2014;17 doi: 10.1136/eb-2013-101550. 75–75. [DOI] [PubMed] [Google Scholar]
- 32.Paul H.A., Bomhof M.R., Vogel H.J., Reimer R.A. Diet-induced changes in maternal gut microbiota and metabolomic profiles influence programming of offspring obesity risk in rats. Sci. Rep. 2016;6:1–14. doi: 10.1038/srep20683. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hussain A., Nookaew I., Khoomrung S., Andersson L., Larsson I., Hulthén L., et al. A maternal diet of fatty fish reduces body fat of offspring compared with a maternal diet of beef and a post-weaning diet of fish improves insulin sensitivity and lipid profile in adult C57BL/6 male mice. Acta Physiol. 2013;209:220–234. doi: 10.1111/apha.12130. [DOI] [PubMed] [Google Scholar]
- 34.Merrifield C.A., Lewis M.C., Berger B., Cloarec O., Heinzmann S.S., Charton F., et al. Neonatal environment exerts a sustained influence on the development of the intestinal microbiota and metabolic phenotype. ISME J. 2016;10:145–157. doi: 10.1038/ismej.2015.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hallam M.C., Reimer R.A. Impact of diet composition in adult offspring is dependent on maternal diet during pregnancy and lactation in rats. Nutrients. 2016;8 doi: 10.3390/nu8010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bianco-Miotto T., Craig J.M., Gasser Y.P., van Dijk S.J., Ozanne S.E. Epigenetics and DOHaD: from basics to birth and beyond. J. Develop. Origins Health Dis. 2017;8:513–519. doi: 10.1017/S2040174417000733. [DOI] [PubMed] [Google Scholar]
- 37.Stiemsma L.T., Michels K.B. The role of the microbiome in the developmental origins of health and disease. Pediatrics. 2018:141. doi: 10.1542/peds.2017-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonnenburg E.D., Smits S.A., Tikhonov M., Higginbottom S.K., Wingreen N.S., Sonnenburg J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature. Nature Publishing Group. 2016;529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blaser M.J. The theory of disappearing microbiota and the epidemics of chronic diseases. Nat. Rev. Immunol. 2017;17:461–463. doi: 10.1038/nri.2017.77. [DOI] [PubMed] [Google Scholar]
- 40.Dogra S., Sakwinska O., Soh S.E., Ngom-Bru C., Brück W.M., Berger B., et al. Rate of establishing the gut microbiota in infancy has consequences for future health. Gut Microb. 2015;6:321–325. doi: 10.1080/19490976.2015.1078051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin O.C.B., Olier M., Ellero-Simatos S., Naud N., Dupuy J., Huc L., et al. Haem iron reshapes colonic luminal environment: impact on mucosal homeostasis and microbiome through aldehyde formation. Microbiome. 2019;7:72. doi: 10.1186/s40168-019-0685-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.dos Santos-Junior E.F., Gonçalves-Pimentel C., de Araujo L.C.C., da Silva T.G., de Melo-Junior M.R., Moura-Neto V., et al. Malnutrition increases NO production and induces changes in inflammatory and oxidative status in the distal colon of lactating rats. Neuro Gastroenterol. Motil. 2016;28:1204–1216. doi: 10.1111/nmo.12820. [DOI] [PubMed] [Google Scholar]
- 43.Chevolleau S., Noguer-Meireles M.-H., Mervant L., Martin J.-F., Jouanin I., Pierre F., et al. Towards aldehydomics: untargeted trapping and analysis of reactive diet-related carbonyl compounds formed in the intestinal lumen. Antioxidants. 2021;10:1261. doi: 10.3390/antiox10081261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Escudié F., Auer L., Bernard M., Mariadassou M., Cauquil L., Vidal K., et al. FROGS: find, rapidly, OTUs with Galaxy solution. Berger B., editor. Bioinformatics. 2018;34:1287–1294. doi: 10.1093/bioinformatics/btx791. [DOI] [PubMed] [Google Scholar]
- 45.Mahé F., Rognes T., Quince C., de Vargas C., Dunthorn M. Swarm: robust and fast clustering method for amplicon-based studies. PeerJ. 2014;2:e593. doi: 10.7717/peerj.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pierre F., Tache S., Petit C.R., Van der Meer R., Corpet D.E. Meat and cancer: haemoglobin and haemin in a low-calcium diet promote colorectal carcinogenesis at the aberrant crypt stage in rats. Carcinogenesis. 2003;24:1683–1690. doi: 10.1093/carcin/bgg130. 2003/08/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohkawa H., Ohishi N., Yagi K. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal. Biochem. 1979;95:351–358. doi: 10.1016/0003-2697(79)90738-3. [DOI] [PubMed] [Google Scholar]
- 48.Gueraud F., Peiro G., Bernard H., Alary J., Creminon C., Debrauwer L., et al. Enzyme immunoassay for a urinary metabolite of 4-hydroxynonenal as a marker of lipid peroxidation. Free Radic. Biol. Med. 2006;40:54–62. doi: 10.1016/j.freeradbiomed.2005.08.011. 2005/12/13. [DOI] [PubMed] [Google Scholar]
- 49.Riba A., Olier M., Lacroix-Lamandé S., Lencina C., Bacquié V., Harkat C., et al. Early life stress in mice is a suitable model for Irritable Bowel Syndrome but do not predispose to colitis nor increase susceptibility to enteric infections. Brain Behav. Immun. 2018;73:403–415. doi: 10.1016/j.bbi.2018.05.024. Elsevier. [DOI] [PubMed] [Google Scholar]
- 50.Singh A., Shannon C., Gautier B., Rohart F., Vacher M., Tebbutt S., et al. Diablo - an integrative, multiomics, multivariate method for multigroup classification. Bioinformatics. 2019;35:3055–3062. doi: 10.1093/bioinformatics/bty1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rohart F., Gautier B., Singh A., Le Cao K.-A. mixOmics: an R package for ’omics feature selection and multiple data integration. PLoS Comput. Biol. 2017;13 doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lê Cao K.A., Costello M.E., Lakis V.A., Bartolo F., Chua X.Y., Brazeilles R., et al. MixMC: a multivariate statistical framework to gain insight into microbial communities. PLoS One. 2016;11 doi: 10.1371/journal.pone.0160169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bosi E., Molteni L., Radaelli M.G., Folini L., Fermo I., Bazzigaluppi E., et al. Increased intestinal permeability precedes clinical onset of type 1 diabetes. Diabetologia. 2006;49:2824–2827. doi: 10.1007/s00125-006-0465-3. [DOI] [PubMed] [Google Scholar]
- 54.Martin O.C.B., Lin C., Naud N., Tache S., Raymond-Letron I., Corpet D.E., et al. Antibiotic suppression of intestinal microbiota reduces heme-induced lipoperoxidation associated with colon carcinogenesis in rats. Nutr. Cancer. 2015;67:119–125. doi: 10.1080/01635581.2015.976317. [DOI] [PubMed] [Google Scholar]
- 55.IJssennagger N., Derrien M., van Doorn G.M., Rijnierse A., van den Bogert B., Müller M., et al. Dietary heme alters microbiota and mucosa of mouse colon without functional changes in host-microbe cross-talk. PLoS One. 2012;7 doi: 10.1371/journal.pone.0049868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cindric M., Cipak A., Zapletal E., Jaganjac M., Milkovic L., Waeg G., et al. Stobadine attenuates impairment of an intestinal barrier model caused by 4-hydroxynonenal. Toxicol. Vitro. 2013;27:426–432. doi: 10.1016/j.tiv.2012.08.005. 2012/08/22. [DOI] [PubMed] [Google Scholar]
- 57.Ijssennagger N., Rijnierse A., de Wit N.J., Boekschoten M.V., Dekker J., Schonewille A., et al. Dietary heme induces acute oxidative stress, but delayed cytotoxicity and compensatory hyperproliferation in mouse colon. Carcinogenesis. 2013;34:1628–1635. doi: 10.1093/carcin/bgt084. 2013/03/05. [DOI] [PubMed] [Google Scholar]
- 58.Ijssennagger N., Belzer C., Hooiveld G.J., Dekker J., van Mil S.W.C., Müller M., et al. Gut microbiota facilitates dietary heme-induced epithelial hyperproliferation by opening the mucus barrier in colon. Proc. Natl. Acad. Sci. Unit. States Am. 2015;112 doi: 10.1073/pnas.1507645112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pierre F., Tache S., Guéraud F., Rerole A.L., Jourdan M.-L.L., Petit C. Apc mutation induces resistance of colonic cells to lipoperoxide-triggered apoptosis induced by faecal water from haem-fed rats. Carcinogenesis. 2007;28:321–327. doi: 10.1093/carcin/bgl127. [DOI] [PubMed] [Google Scholar]
- 60.Mégier C., Peoc’h K., Puy V., Cordier A.G. Iron metabolism in normal and pathological pregnancies and fetal consequences. Metabolites. 2022;12 doi: 10.3390/metabo12020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Georgieff M.K. Long-term brain and behavioral consequences of early iron deficiency. Nutr. Rev. 2011;69:S43–S48. doi: 10.1111/j.1753-4887.2011.00432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiegersma A.M., Dalman C., Lee B.K., Karlsson H., Gardner R.M. Association of prenatal maternal anemia with neurodevelopmental disorders. JAMA Psychiatr. 2019;76:1294–1304. doi: 10.1001/jamapsychiatry.2019.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lewis R.M., Petry C.J., Ozanne S.E., Hales C.N. Effects of maternal iron restriction in the rat on blood pressure, glucose tolerance, and serum lipids in the 3-month-old offspring. Metab., Clin. Exp. 2001;50:562–567. doi: 10.1053/meta.2001.22516. [DOI] [PubMed] [Google Scholar]
- 64.Vaarala O., Atkinson M.A., Neu J. The “perfect storm” for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes. 2008:2555–2562. doi: 10.2337/db08-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou H., Sun L., Zhang S., Zhao X., Gang X., Wang G. The crucial role of early-life gut microbiota in the development of type 1 diabetes. Acta Diabetol. 2021;58:249–265. doi: 10.1007/s00592-020-01563-z. Springer Milan. [DOI] [PubMed] [Google Scholar]
- 66.Serino M., Waget A., Marsollier N., Masseboeuf M., Payros G., Kabani C., et al. Lipid-induced peroxidation in the intestine is involved in glucose homeostasis imbalance in mice. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hsu C.-N., Tain Y.-L. The good, the bad and the ugly of pregnancy nutrients and developmental programming of adult disease. Nutrients. 2019;11:1–21. doi: 10.3390/nu11040894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strakovsky R.S., Pan Y.X. In utero oxidative stress epigenetically programs antioxidant defense capacity and adulthood diseases. Antioxidants Redox Signal. 2012;17:237–253. doi: 10.1089/ars.2011.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu C., Yan Y., Ji F., Zhou H. Maternal obesity increases oxidative stress in placenta and it is associated with intestinal microbiota. Front. Cell. Infect. Microbiol. 2021;11:1–11. doi: 10.3389/fcimb.2021.671347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lavoie J.C., Tremblay A. Sex-specificity of oxidative stress in newborns leading to a personalized antioxidant nutritive strategy. Antioxidants. 2018;7 doi: 10.3390/antiox7040049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee I.-A., Bae E.-A., Hyun Y.-J., Kim D.-H. Dextran sulfate sodium and 2,4,6-trinitrobenzene sulfonic acid induce lipid peroxidation by the proliferation of intestinal gram-negative bacteria in mice. J. Inflamm. 2010;7:7. doi: 10.1186/1476-9255-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lou Y.C., Olm M.R., Diamond S., Crits-Christoph A., Firek B.A., Baker R., et al. Infant gut strain persistence is associated with maternal origin, phylogeny, and traits including surface adhesion and iron acquisition. Cell Reports Medicine. 2021;2 doi: 10.1016/j.xcrm.2021.100393. Elsevier Company. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mishra S., Imlay J.A. An anaerobic bacterium, Bacteroides thetaiotaomicron, uses a consortium of enzymes to scavenge hydrogen peroxide. Mol. Microbiol. 2013;90:1356–1371. doi: 10.1111/mmi.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ormerod K.L., Wood D.L.A., Lachner N., Gellatly S.L., Daly J.N., Parsons J.D., et al. Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome. 2016;4:36. doi: 10.1186/s40168-016-0181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kostic A.D., Gevers D., Siljander H., Vatanen T., Hyötyläinen T., Hämäläinen A.-M., et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17:260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Siljander H., Honkanen J., Knip M. Microbiome and type 1 diabetes. EBioMedicine. The Authors. 2019;46:512–521. doi: 10.1016/j.ebiom.2019.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brown C.T., Davis-Richardson A.G., Giongo A., Gano K.A., Crabb D.B., Mukherjee N., et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6:1–9. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davis-Richardson A.G., Ardissone A.N., Dias R., Simell V., Leonard M.T., Kemppainen K.M., et al. Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Front. Microbiol. 2014;5:1–11. doi: 10.3389/fmicb.2014.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.De Goffau M.C., Fuentes S., Van Den Bogert B., Honkanen H., De Vos W.M., Welling G.W., et al. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia. 2014;57:1569–1577. doi: 10.1007/s00125-014-3274-0. [DOI] [PubMed] [Google Scholar]
- 80.Vatanen T., Kostic A.D., D'Hennezel E., Siljander H., Franzosa E.A., Yassour M., et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang J., Li W., Wang C., Wang L., He T., Hu H., et al. Enterotype Bacteroides is associated with a high risk in patients with diabetes: a pilot study. J. Diabetes Res. 2020;2020 doi: 10.1155/2020/6047145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 83.Kameyama K., Itoh K. Intestinal colonization by a Lachnospiraceae bacterium contributes to the development of diabetes in obese mice. Microb. Environ. 2014;29:427–430. doi: 10.1264/jsme2.ME14054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Davì G., Falco A., Patrono C. Lipid peroxidation in diabetes mellitus. Antioxidants Redox Signal. 2005;7:256–268. doi: 10.1089/ars.2005.7.256. [DOI] [PubMed] [Google Scholar]
- 85.Davì G., Chiarelli F., Santilli F., Pomilio M., Vigneri S., Falco A., et al. Enhanced lipid peroxidation and platelet activation in the early phase of type 1 diabetes mellitus: role of interleukin-6 and disease duration. Circulation. 2003;107:3199–3203. doi: 10.1161/01.CIR.0000074205.17807.D0. [DOI] [PubMed] [Google Scholar]
- 86.Sorini C., Cosorich I., Conte M.L., De Giorgi L., Facciotti F., Lucianò R., et al. Loss of gut barrier integrity triggers activation of islet-reactive T cells and autoimmune diabetes. Proc. Natl. Acad. Sci. U. S. A. 2019;116:15140–15149. doi: 10.1073/pnas.1814558116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data set (16S rRNA sequences) described in this project has been deposited in the MG-RAST database under accession code mgp100998. Further information and requests for resources and reagents will be fulfilled by the corresponding author, MO.