Abstract
The recently emerged B.1.1.529 (Omicron) severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant has a highly divergent spike (S) glycoprotein. We compared the functional properties of B.1.1.529 BA.1 S with those of previous globally prevalent SARS-CoV-2 variants, D614G and B.1.617.2. Relative to these variants, B.1.1.529 S exhibits decreases in processing, syncytium formation, virion incorporation, and ability to mediate infection of cells with high TMPRSS2 expression. B.1.1.529 and B.1.617.2 S glycoproteins bind ACE2 with higher affinity than D614G S. The unliganded B.1.1.529 S trimer is less stable at low temperatures than the other SARS-CoV-2 Ss, a property related to its more “open” S conformation. Upon ACE2 binding, the B.1.1.529 S trimer sheds S1 at 37°C, but not at 0°C. B.1.1.529 pseudoviruses are relatively resistant to neutralization by sera from patients with coronavirus disease 2019 (COVID-19) and vaccinees. These properties of the B.1.1.529 S glycoprotein likely influence the transmission, cytopathic effects, and immune evasion of this emerging variant.
Keywords: SARS-CoV-2, B.1.617.2, B.1.1.529, variants, Omicron, stability, S1 shedding, ACE2 affinity, neutralization sensitivity
Graphical abstract
Wang et al. show that, compared with spike glycoproteins of other prevalent SARS-CoV-2 variants, the B.1.1.529 (Omicron) BA.1 spike exhibits decreased processing, syncytium formation, virion incorporation, infection of cells with high TMPRSS2 levels, stability at 0°C, and sensitivity to antibodies elicited by natural infection or a vaccine.
Introduction
The continuing coronavirus disease 2019 (COVID-19) pandemic has stimulated the implementation of a number of countermeasures, including vaccines, therapeutic antibodies, and antiviral agents. Vaccines that elicit neutralizing antibodies against the spike (S) glycoprotein of the etiologic agent of COVID-19, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), have effectively reduced the probability of infection and death from this virus (Baden et al., 2021; Dai and Gao, 2021; Polack et al., 2020; Sadoff et al., 2021). However, the emergence of a SARS-CoV-2 variant in Botswana and South Africa in mid-November 2021, coupled with its rapid spread throughout the world, has raised concerns (Grabowski et al., 2022; Pulliam et al., 2022; Scott et al., 2021). The high rate of transmission has caused the World Health Organization to classify this variant as a variant of concern (VOC), and it has been termed Omicron (formally B.1.1.529), now found in over 70 countries (WHO, 2021).
B.1.1.529 was found to have an unprecedented level of divergence, with more than 55 mutations compared with the ancestral Wuhan-Hu-1 strain. Of particular importance to antibody-mediated protection, more than 30 of these mutations alter the sequence of viral S glycoprotein (Scott et al., 2021). Several of these changes in S, such as K417N and N501Y, have been previously demonstrated to confer resistance to antibody neutralization in other SARS-CoV-2 variants (Wang et al., 2021a, 2021b). Residue Glu 484, which is altered to Lys in the B.1.351 and P.1 variants, is an Ala in B.1.1.529, resulting in a similar loss of sensitivity to a subset of antibodies (Liu et al., 2022). These features suggest that the B.1.1.529 virus may be highly adapted to resist antibodies directed against the ancestral SARS-CoV-2.
Recent reports have confirmed this prediction. Several in vitro studies have demonstrated that B.1.1.529 evades several monoclonal antibodies, including some used clinically, and is less effectively neutralized by antibodies elicited by SARS-CoV-2 infection and vaccines (Garcia-Beltran et al., 2022; Gruell et al., 2022; Hoffmann et al., 2022; Liu et al., 2022; Pajon et al., 2022; Planas et al., 2022). The degree of B.1.1.529 resistance to antibodies seems to be greater than that observed in any variant to date, and consequently, many current vaccinees may be at risk of breakthrough infections or disease (Lu et al., 2021; Zeng et al., 2021). Data from the United Kingdom and South Africa indicate a substantial decline in the efficacy of the BNT162b2 vaccine (Pfizer), contextualizing the implications of the experimentally measured resistance (Abu-Raddad et al., 2021; Madhi et al., 2021; Sadoff et al., 2021).
Given the altered epidemiology of B.1.1.529 relative to other SARS-CoV-2 variants, we investigated the functional properties of the S glycoprotein of B.1.1.529. By comparison to the current predominant strain, B.1.617.2 (Delta variant), and its ancestor D614G (Wuhan-Hu-1 strain with a D614G change in S), we find that the alterations in B.1.1.529 specify interesting S phenotypes. Notably, B.1.1.529 has significantly reduced processing of its S glycoprotein into S1 and S2 subunits. Apparent consequences of poor S processing are reductions in syncytium formation and incorporation of S glycoproteins into pseudovirus particles, compared with these properties of D614G and B.1.617.2. The D614G, B.1.617.2, and B.1.1.529 S glycoproteins mediate pseudovirus infection of cells expressing different levels of the ACE2 receptor with comparable efficiency. By contrast, pseudoviruses with the D614G and B.1.617.2 S glycoproteins infect cells with high levels of TMPRSS2 more efficiently than B.1.1.529 pseudoviruses. For target cells with low TMPRSS2 expression, pseudovirus infection was mediated equivalently by the three S glycoproteins. Like other viruses descended from D614G, B.1.1.529 exhibits a higher affinity of its S for the ACE2 receptor. The B.1.1.529 S resists soluble ACE2-induced shedding of the S1 exterior glycoprotein at lower temperatures. After extended incubation at 0°C, the B.1.1.529 S glycoprotein spontaneously sheds the S1 subunit and loses its ability to support virus entry. The spontaneous shedding of S1 and the cold inactivation of the virus S are related to the conformation of the receptor-binding domains (RBDs) on the S glycoprotein. We confirm that the B.1.1.529 S glycoprotein is less sensitive to neutralization by sera from patients convalescing from COVID-19 and recipients of two doses of the mRNA-1273 vaccine (Moderna). The binding of the sera to recombinant S trimers indicates that many antibodies in the convalescent patient sera bind conserved epitopes on the B.1.1.529 S that are not available as neutralization targets. Serum binding to the D614G and B.1.1.529 RBDs correlated better with the virus neutralization activity of the sera. Collectively, our study reveals biological properties of the divergent B.1.1.529 S that may contribute to its rapid transmission, altered pathogenesis, and immune escape.
Results
S glycoprotein expression and post-translational processing
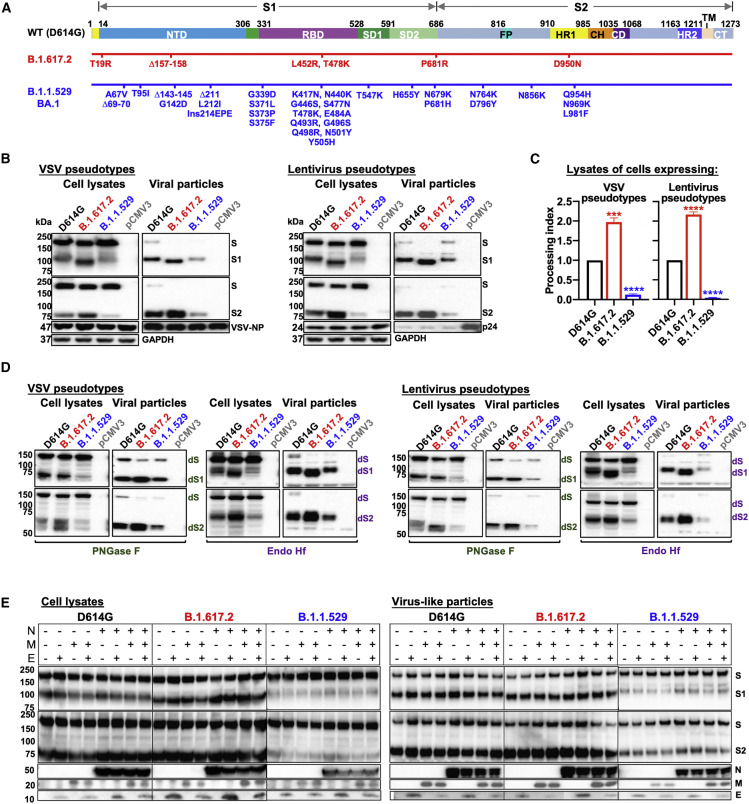
The significant number of changes in the B.1.1.529 BA.1 S glycoprotein (Figure 1A) led us to compare its properties with those of S glycoproteins from two other globally prevalent SARS-CoV-2 strains, D614G and B.1.617.2. We studied the expression and processing of the D614G, B.1.617.2, and B.1.1.529 S glycoproteins. We generated vesicular stomatitis virus (VSV) and lentivirus (HIV-1)-based pseudovirus particles as previously described and examined the incorporated S glycoproteins by western blot (Schmidt et al., 2020; Wang et al., 2021c). In both pseudoviral systems, relative to the D614G S, the B.1.617.2 S glycoprotein was proteolytically processed more efficiently, whereas the B.1.1.529 S glycoprotein exhibited significantly less cleavage (Figures 1B and 1C). For all these SARS-CoV-2 S glycoproteins, the cleaved (S1 and S2) glycoproteins were preferentially incorporated into pseudovirus particles. However, the decrease in B.1.1.529 S glycoprotein cleavage in the expressing cells was associated with reduced incorporation of S glycoproteins into pseudovirus particles (Figure 1B).
Figure 1.
SARS-CoV-2 variant S glycoprotein expression, post-translational processing, and incorporation into virus particles
(A) A schematic representation of the wild-type (D614G) S glycoprotein is shown in the upper figure. The N-terminal domain (NTD), receptor-binding domain (RBD), and subdomains 1 and 2 (SD1 and SD2) of the S1 glycoprotein are indicated. The fusion peptide (FP), heptad repeat regions (HR1 and HR2), central helical region (CH), connector domain (CD), transmembrane region (TM), and cytoplasmic tail (CT) of the S2 glycoprotein are indicated. The amino acid changes in the B.1.617.2 and B.1.1.529 BA.1 Ss, relative to the D614G S glycoprotein, are shown in the diagrams beneath the schematic.
(B) HEK293T cells producing VSV (left panels) and lentiviral (right panels) particles pseudotyped with the indicated SARS-CoV-2 S glycoproteins were used to prepare cell lysates and viral particles, which were western blotted and probed with rabbit anti-S1 and anti-S2 antibodies. The samples were also probed with anti-VSV NP, anti-HIV p17/p24/p55, and anti-GAPDH antibodies. The western blots shown are representative of those obtained in three independent experiments. Cells transfected with the pCMV3 vector were used as a negative control.
(C) Processing indices were calculated from western blots of cell lysates from three experiments. Data are presented as means ± SEM. A Student's unpaired t test was used to compare the values with those obtained for the wild-type D614G S glycoprotein (∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001).
(D) The cell lysates and viral particle preparations from (B) were treated with PNGase F and Endo Hf, and the deglycosylated samples were western blotted and probed with rabbit anti-S1 and anti-S2 antibodies. Cells transfected with the pCMV3 vector were used as a negative control. The deglycosylated forms of the S glycoprotein precursor (dS), S1 subunit (dS1), and S2 subunit (dS2) are indicated. The western blots shown are representative of those obtained in two independent experiments.
(E) Effect of SARS-CoV-2 membrane (M), envelope (E), and nucleocapsid (N) proteins on the processing of S glycoprotein variants. 293T cells were transfected with plasmids expressing the indicated SARS-CoV-2 S glycoproteins and with plasmids expressing one or more of the SARS-CoV-2 M, E, and N proteins. Cell lysates and supernatant pellets were western blotted and probed with antibodies against S1, S2, the N protein, and the Strep tag of the M and E proteins.
Next, we examined the glycosylation of these three SARS-CoV-2 S glycoproteins in the two pseudovirus systems. Cell lysates and pseudovirus particles were treated with either PNGase F or Endo Hf and then visualized by western blotting (Figure 1D). The S glycosylation patterns of the VSV and lentivirus pseudotypes were similar. As previously observed for SARS-CoV-2 S glycoproteins (Nguyen et al., 2020; Wang et al., 2021c; Zhang et al., 2021), the vast majority of the uncleaved S glycoprotein in cell lysates contained only high-mannose and hybrid glycans, whereas the S1 and S2 glycoproteins on pseudovirus particles were highly modified by complex carbohydrates. Following PNGase F digestion, the S2 glycoproteins incorporated into pseudovirus particles were more homogeneous than those in cell lysates; post-translational modifications of S2 other than N- or O-linked glycosylation account for this difference (Zhang et al., 2021). The untreated and the Endo Hf-treated S1 from B.1.617.2 migrated slightly faster than those of D614G and B.1.1.529, which may be due to a missing N-linked glycosylation site resulting from the T19R change in the N-terminal domain.
We also evaluated the effects of coexpression of the SARS-CoV-2 membrane (M), envelope (E), and nucleocapsid (N) proteins on D614G, B.1.617.2, and B.1.1.529 S glycoprotein processing. We examined cell lysates and particulate fractions concentrated from the supernatants of 293T cells transfected with plasmids expressing the variant S glycoproteins along with plasmids expressing the M, E, and N proteins, alone or in combination. Coexpression of the M, E, and N proteins, alone or in combination, did not significantly affect S glycoprotein processing (Figure 1E).
S-glycoprotein-mediated viral infection and cell-cell fusion
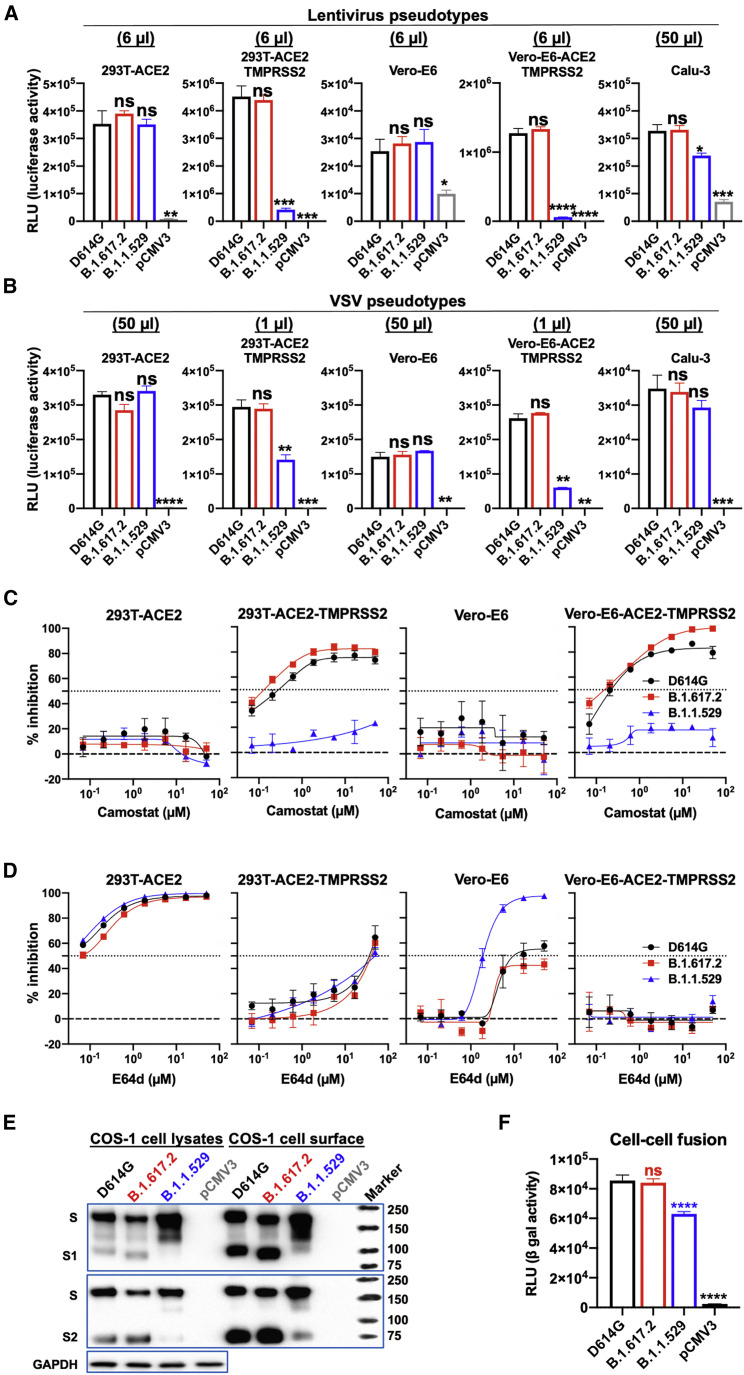
To determine whether the reduced cleavage and virion incorporation of the B.1.1.529 S affected viral entry, we quantified the infectivity of the pseudoviruses on target cells expressing different levels of ACE2 and TMPRSS2. The B.1.1.529 pseudoviruses infected 293T cells transiently expressing different levels of human ACE2 comparably to the D614G and B.1.617.2 pseudoviruses (Figure S1). The effect of TMPRSS2 overexpression on the efficiency of infection mediated by the D614G, B.1.617.2, and B.1.1.529 S glycoproteins was evaluated in 293T-ACE2, 293T-ACE2-TMPRSS2, Vero-E6, and Vero-E6-ACE2-TMPRSS2 target cells. No significant differences were observed in the infectivity of lentiviruses or VSV pseudotyped by the three SARS-CoV-2 S glycoproteins for 293T-ACE2 or Vero-E6 cells (Figures 2A and 2B). The D614G and B.1.617.2 S glycoproteins mediated 10- to 50-fold higher levels of infection of the corresponding TMPRSS2-expressing cells. Notably, both lentivirus and VSV vectors pseudotyped by the B.1.1.529 S glycoprotein exhibited significantly lower infectivity for the TMPRSS2-overexpressing cells compared with the D614G and B.1.617.2 pseudoviruses. Collectively, although the B.1.1.529 S glycoprotein exhibits reduced processing and decreased S incorporation into pseudovirus particles, its ability to mediate virus infection of cells with low levels of TMPRSS2 expression is not compromised. However, unlike D614G and B.1.617.2 S glycoproteins, which can mediate efficient viral entry by utilizing high levels of target cell TMPRSS2, B.1.1.529-S-glycoprotein-mediated viral entry is minimally enhanced by TMPRSS2 overexpression in target cells.
Figure 2.
S-glycoprotein-mediated viral infection and cell-cell fusion
(A) The infectivity of lentivirus vectors pseudotyped by the indicated SARS-CoV-2 S glycoproteins was determined on 293T-ACE2 cells, 293T-ACE2-TMPRSS2 cells, Vero-E6 cells, Vero-E6-ACE2-TMPRSS2 cells, and Calu-3 cells. The pCMV3-transfected cells serve as a negative control. The target cells were inoculated with the indicated volume of each virus, and the infectivity was quantified by measuring the NanoLuc luciferase activity in the target cells 2 days after inoculation. The results shown are representative of those obtained in two independent experiments. Data are presented as means ± SEM from triplicate samples. See also Figure S1. RLU, relative light unit.
(B) The infectivity of VSV vectors pseudotyped by the indicated SARS-CoV-2 S glycoproteins was determined on 293T-ACE2 cells, 293T-ACE2-TMPRSS2 cells, Vero-E6 cells, Vero-E6-ACE2-TMPRSS2 cells, and Calu-3 cells. The pCMV3-transfected cells serve as a negative control. The target cells were inoculated with the indicated volume of each virus, and the infectivity was quantified by measuring the luciferase activity in the target cells 20 h after inoculation. The results shown are representative of those obtained in three independent experiments. Data are presented as means ± SEM from triplicate samples. See also Figure S1.
(C) The effect of camostat on the infection of VSV particles pseudotyped by the indicated SARS-CoV-2 S glycoproteins in 293T-ACE2 cells, 293T-ACE2-TMPRSS2 cells, Vero-E6 cells, and Vero-E6-ACE2-TMPRSS2 cells was examined. The results shown are representative of those obtained in three independent experiments. Data are presented as means ± SEM from triplicate samples.
(D) The effect of E64d on the infection of VSV particles pseudotyped by the indicated SARS-CoV-2 S glycoproteins in 293T-ACE2 cells, 293T-ACE2-TMPRSS2 cells, Vero-E6 cells, and Vero-E6-ACE2-TMPRSS2 cells was examined. The results shown are representative of those obtained in three independent experiments. Data are presented as means ± SEM from triplicate samples.
(E) The transient expression of the indicated SARS-CoV-2 S glycoproteins in transfected COS-1 cell lysates (left panel) and on the cell surface (right panel) was examined. COS-1 cells were transiently transfected with plasmids expressing ⍺-gal and the indicated S glycoproteins. To evaluate the S glycoproteins on the COS-1 cell surface, the cells were incubated with sulfo-NHS-SS-biotin and the biotinylated cell-surface proteins were precipitated with NeutrAvidin Agarose. SARS-CoV-2 S glycoproteins precipitated from the cell surface and in the cell lysates were western blotted and probed with rabbit anti-S1 and anti-S2 antibodies. The western blots of the cell lysate samples were also probed with anti-GAPDH antibodies. The western blots shown are representative of those obtained in two independent experiments. Cells transfected with the pCMV3 vector were used as a negative control.
(F) Cell-cell fusion efficiency between COS-1 effector cells transiently expressing ⍺-gal and the indicated S glycoprotein and 293T target cells transiently expressing ω-gal and human ACE2 was determined by quantitation of β-galactosidase activity in fused cells. The COS-1 effector cells and 293T target cells were cocultivated at 37°C for 4 h. The cell-cell fusion activity of each SARS-CoV-2 variant S glycoprotein was compared with that of D614G. Cells transfected with pCMV3 serve as negative controls. The results shown represent the means and standard deviations derived from three independent experiments.
For statistical analysis of the data in (A), (B), and (F), a Student’s unpaired t test was used to compare the values with those obtained for the wild-type D614G S glycoprotein (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001; ns, not significant).
SARS-CoV-2 can achieve entry into cells via two routes, either at the cell surface following TMPRSS2 proteolytic activation or from the endosome following proteolytic activation by the endosomal proteases cathepsin B or L (Peacock et al., 2021). The above infectivity data suggest that viruses with the B.1.1.529 S glycoproteins do not utilize the former TMPRSS2-dependent pathway as efficiently as viruses with the D614G and B.1.617.2 glycoproteins. To test this hypothesis, we examined the sensitivity of pseudovirus infections of cells expressing low and high levels of TMPRSS2 to camostat, a TMPRSS2 inhibitor, and E64d, a cathepsin inhibitor. In 293T-ACE2 and Vero-E6 cells, all three VSV vectors pseudotyped by the D614G, B.1.617.2, and B.1.1.529 S glycoproteins were totally resistant to camostat but inhibited by E64d (Figures 2C and 2D). Compared with the D614G and B.1.617.2 pseudoviruses, the B.1.1.529 pseudovirus was more sensitive to E64d inhibition in the Vero-E6 cells. In the target cells overexpressing TMPRSS2, the D614G and B.1.617.2 VSV pseudotypes were sensitive to camostat inhibition, whereas the B.1.1.529 pseudovirus infection was minimally affected by camostat. Pseudoviruses with the D614G, B.1.617.2, and B.1.1.529 S glycoproteins apparently can utilize the cathepsin-mediated endosomal entry pathway with comparable efficiency. However, whereas the D614G and B.1.617.2 pseudoviruses can utilize TMPRSS2 to enter cells, the pseudoviruses with the B.1.1.529 S glycoprotein are much less efficient at doing so.
To evaluate the efficiency of cell-cell fusion mediated by the D614G, B.1.617.2, and B.1.1.529 S glycoproteins, we cocultivated COS-1 cells expressing the S glycoproteins with 293T cells expressing human ACE2. The expression and processing of the D614G, B.1.617.2, and B.1.1.529 S glycoproteins in the COS-1 cell lysates and cell surface are shown in Figure 2E. The levels of the cleaved S1 and S2 glycoproteins in the cell lysates and on the cell surface were comparable for the D614G and B.1.617.2 variants. Although the B.1.1.529 S glycoprotein was expressed well, the level of processed S1 and S2 glycoproteins in cell lysates and on the cell surface was relatively low. To evaluate the efficiency of cell-cell fusion mediated by the D614G, B.1.617.2, and B.1.1.529 S glycoproteins, the COS-1 cells expressing these S glycoproteins and α-gal were cocultivated with 293T cells expressing ACE2 and ω-gal for 4 h at 37°C. The formation of syncytia between the S-expressing cells and ACE2-expressing cells results in the activation of β-galactosidase. Cell-cell fusion mediated by the B.1.1.529 S glycoprotein was mildly reduced compared with the levels observed for the D614G and B.1.617.2 S glycoproteins (Figure 2F). This result is consistent with the expectation that the process of syncytium formation is mediated by cleaved S1/S2 glycoprotein trimers on the surface of the expressing cell (Nguyen et al., 2020). The decreased processing of the B.1.1.529 S glycoprotein results in lower levels of mature S1/S2 trimers in expressing cells, compared with the levels of the D614G and B.1.617.2 S glycoproteins, likely contributing to the observed reduction in cell-cell fusion.
ACE2 binding and soluble ACE2-induced S1 shedding of variant spikes
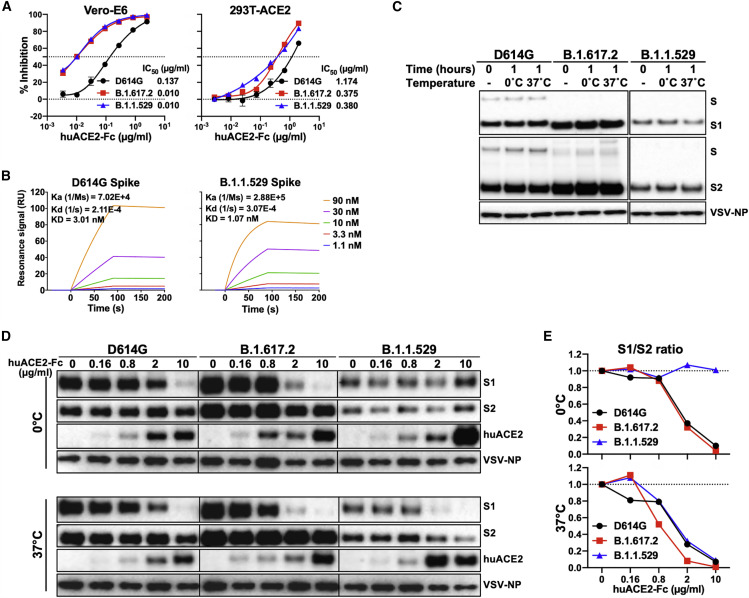
We investigated the interaction of the D614G, B.1.617.2, and B.1.1529 S variants with ACE2. We tested the inhibition of infection of D614G, B.1.617.2, and B.1.1.529 pseudotypes by soluble ACE2 (huACE2-Fc) and observed enhanced inhibition of B.1.617.2 and B.1.1.529 pseudotypes relative to D614G viruses in both Vero-E6 and 293T-ACE2 cells (Figure 3A). As expected for the lower level of ACE2 expression in Vero-E6 cells compared with that in 293T-ACE2 cells (Wang et al., 2021c), the differences in huACE2-Fc sensitivity between D614G and the other viruses were more pronounced in the former cells. No significant differences were observed between the B.1.617.2 and B.1.1.529 pseudotypes in these assays. Using a surface plasmon resonance assay, we found that the KD value for huACE2-Fc binding to the B.1.1.529 S (1.07 nM) was around 3-fold lower than that of the D614G S (3.01 nM) (Figure 3B). The higher affinity of huACE2-Fc for the B.1.1.529 S glycoprotein likely contributes to the enhanced huACE2-Fc inhibition of B.1.1.529 pseudovirus infection relative to that of D614G pseudoviruses (Figure 3A).
Figure 3.
Interaction of soluble ACE2 with the S glycoproteins of D614G, B.1.617.2, and B.1.1.529
(A) Neutralization activity of soluble huACE2-Fc against VSV vectors pseudotyped with the indicated S glycoproteins on Vero-E6 (left panel) and 293T-ACE2 (right panel) cells. The results shown in the graphs are the means ± SEM from triplicate measurements and are representative of those obtained in two independent experiments. The huACE2-Fc concentrations resulting in 50% inhibition of infectivity (IC50) were calculated and are presented to the right of the graphs.
(B) Binding of soluble huACE2-Fc to the S2P trimers tested by surface plasmon resonance (SPR). The Ka, Kd, and KD values are shown.
(C) VSV particles pseudotyped with the variant SARS-CoV-2 S glycoproteins were freshly prepared (0 time point) or were incubated for 1 h at 0°C or 37°C. Pelleted VSV particles were analyzed by western blotting with antibodies against S1, S2, and VSV-NP.
(D) VSV particles pseudotyped with the variant SARS-CoV-2 S glycoproteins were incubated with the indicated concentrations of soluble huACE2-Fc for 1 h at 0°C (upper panel) and 37°C (lower panel). Pelleted VSV particles were analyzed by western blotting with antibodies against S1, S2, huACE2-Fc, and VSV NP.
(E) The intensities of the S1 and S2 glycoprotein bands in (D) were measured, and the S1/S2 ratios for each concentration of sACE2 are shown.
We compared soluble ACE2-induced shedding of S1 from the S trimers of the D614G, B.1.617.2, and B.1.1.529 pseudoviruses. As differences in soluble ACE2-induced shedding of S1 among some SARS-CoV-2 strains were revealed at 0°C (Wang et al., 2021c), we performed these experiments at 0°C and 37°C. In the absence of huACE2-Fc, the D614G, B.1.617.2, and B.1.1.529 Ss on the pseudovirus particles were stable for at least 1 h at both temperatures (Figure 3C). Each variant pseudovirus was incubated with increasing concentrations of huACE2-Fc at either 0°C or 37°C for 1 h, and then, S1 shedding was quantified by western blotting (Figure 3D). While S1 shedding was similar for all three pseudoviruses at 37°C, the B.1.1.529 S was significantly more stable at 0°C, with minimal S1 shedding compared with those of the D614G or B.1.617.2 Ss (Figure 3E). The observed differences in S1 shedding were not explained by differences in huACE2-Fc binding by the variant S glycoproteins. These results indicate that the ACE2-bound B.1.1.529 S trimer resists the disruptive effects of incubation at 0°C better than the D614G and B.1.617.2 S trimers.
Stability of the variant SARS-CoV-2 spikes at different temperatures
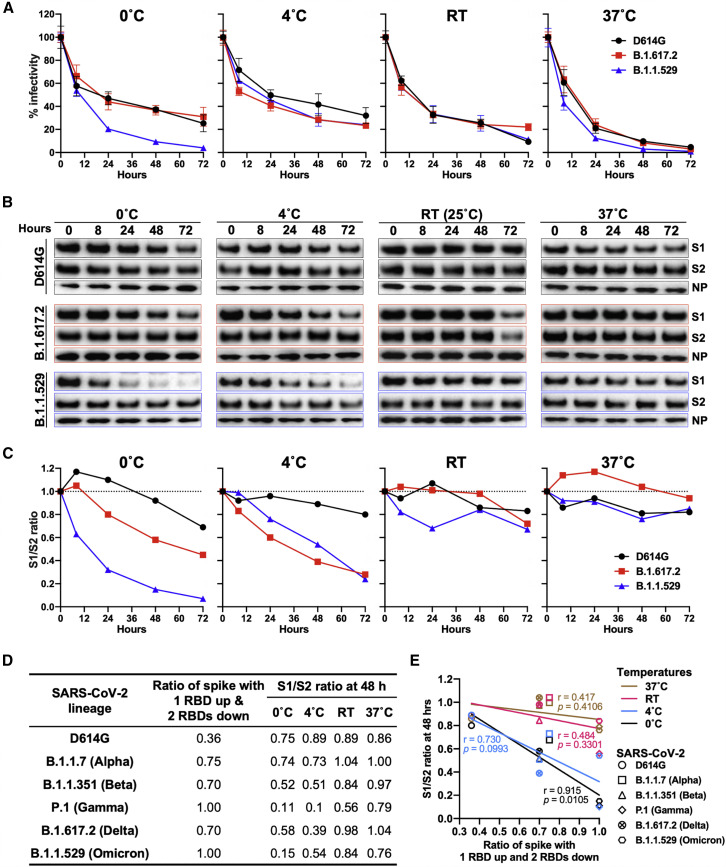
Extended periods of incubation at near freezing temperatures can reveal differences in the stability of the S glycoprotein trimers of SARS-CoV-2 variants (Nguyen et al., 2020; Wang et al., 2021c). We evaluated the effect of temperature on the infectivity of VSV pseudotyped by the D614G, B.1.617.2, and B.1.1.529 S glycoproteins. We incubated VSV pseudotypes at 0°C, 4°C, room temperature (RT), or 37°C for various periods of time before measuring their infectivity on Vero-E6 cells. No differences between the SARS-CoV-2 variants were observed at 4°C, RT, or 37°C; the infectivity of all these variants declined slightly faster at 37°C than at the other temperatures (Figure 4A). Notably, at 0°C, the infectivity of the B.1.1.529 pseudotype decayed faster than the infectivities of the D614G or B.1.617.2 pseudotypes.
Figure 4.
Stability of SARS-CoV-2 D614G, B.1.617.2, and B.1.1.529 spikes at different temperatures
(A) VSV vectors pseudotyped by the variant SARS-CoV-2 S glycoproteins were incubated at 0°C, 4°C, room temperature (RT), and 37°C for various times. The infectivity of the viruses was measured on Vero-E6 cells. The infectivity at each time point is reported, relative to that observed at time 0 for each pseudovirus. Means ± SEM derived from three replicates are shown. The results are representative of those obtained in four independent experiments.
(B) VSV particles pseudotyped by the variant SARS-CoV-2 S glycoproteins were incubated at 0°C, 4°C, RT, and 37°C for various times. Pelleted VSV particles were analyzed by western blotting with antibodies against S1, S2, and VSV NP. The D614G, B.1.617.2, and B.1.1.529 S1 bands were exposed for 10 s, 10 s, and 30 s, respectively; the D614G, B.1.617.2, and B.1.1.529 S2 bands were exposed for 5 s, 2.5 s, and 15 s, respectively. The exposure time of the NP bands was 45 s. The results shown are representative of those obtained in two independent experiments.
(C) The intensities of the S1 and S2 glycoprotein bands in (B) were measured. The S1/S2 ratios are shown for each variant relative to the ratio for pseudoviruses at time 0.
(D) Relative populations of SARS-CoV-2 variant Ss with one RBD in the “up” position (Cerutti et al., 2022) and S1/S2 ratios after a 48-h incubation at 0°C, 4°C, RT, and 37°C (Wang et al., 2021c; see also C above).
(E) Correlation between the percentage of S populations with one RBD in the up position and the S1/S2 ratio after a 48-h incubation at 0°C, 4°C, RT, and 37°C for S glycoproteins from different SARS-CoV-2 variants. The r and p values for each curve were obtained by fitting the data with simple linear regression. A significant correlation between the percentage of spikes with one RBD up and the S1/S2 ratio was observed at the 0°C temperature.
As the observed reduction in infectivity of the B.1.1.529 pseudovirus at 0°C may be due to the shedding of the S1 glycoprotein from the S, we quantified S1 retention on the viral particles following various periods of incubation at different temperatures (Figures 4B and 4C). At 0°C, a higher rate of spontaneous S1 shedding from the B.1.1.529 pseudoviruses compared with the D614G and B.1.617.2 pseudoviruses was observed, suggesting that S disassembly may be part of the mechanism for the observed loss of infectivity at this temperature. The integrity of the B.1.617.2 and B.1.1.529 Ss decreased faster than that of D614G at 4°C; as the infectivity of these pseudoviruses differed only modestly at 4°C, mechanisms of S inactivation that do not result in S1 shedding are likely involved (Wang et al., 2021c).
In a previous study (Wang et al., 2021c), we found that, similar to the B.1.1.529 variant S glycoprotein, the S glycoprotein of the P.1 (Gamma) SARS-CoV-2 variant also exhibited an unusually high degree of S1 shedding after a 2-day incubation at 0°C. Interestingly, both P.1 and B.1.1.529 variant S trimers were reported to reside exclusively in a conformational state with only one RBD in the “up” position (Cerutti et al., 2022; Wang et al., 2021a). We found a correlation between the S1/S2 ratios of different SARS-CoV-2 variant S trimers after 48 h of incubation at 0°C (Wang et al., 2021c) and the population of the variant S with one RBD in the up position (Cerutti et al., 2022; Figures 4D and 4E).
SARS-CoV-2 variants are more resistant to convalescent patient and vaccinee sera
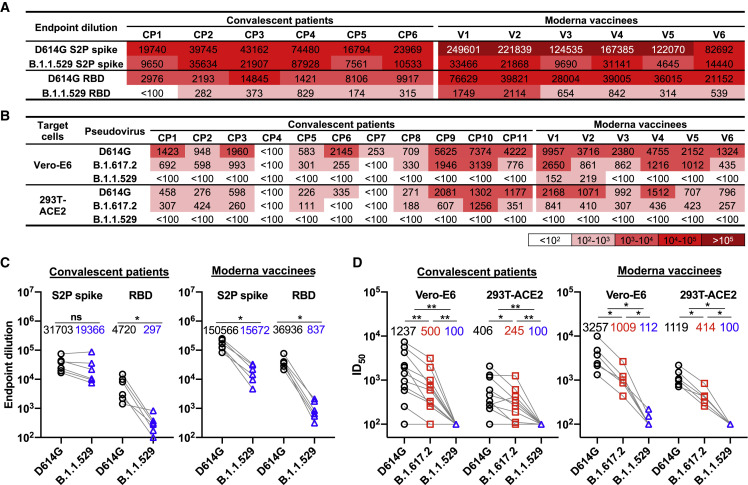
The B.1.1.529 variant has been reported to be significantly more resistant to antibody neutralization than previously characterized SARS-CoV-2 variants (Liu et al., 2022; Planas et al., 2022). We tested the binding of sera from patients convalescing from COVID-19 and vaccinees who had received two doses of mRNA-1273 (Moderna) to recombinant D614G and B.1.1.529 S trimers (S2P) and RBDs (Figures 5A, 5C, S2, and S3). Unexpectedly, we observed that, while binding to the B.1.1.529 S2P was lower than that to the D614G S2P for all serum samples, the magnitude of the decrease differed significantly between the groups; convalescent patient sera exhibited only a 1.6-fold decrease in binding titer, whereas sera from Moderna vaccinees exhibited a 9.6-fold decrease, on average. We also observed a decrease in binding to the B.1.1.529 RBD compared with that to the D614G RBD for both convalescent patient sera (15.9-fold decrease) and vaccinee sera (44.1-fold decrease); however, the magnitude of the decrease was more similar between the two groups of sera than that seen for binding to the D614G and B.1.1.529 S2P trimers. The vaccinees elicit higher titers of antibodies against the SARS-CoV-2 S and against the RBD region than the patients convalescing from COVID-19. Apparently, the S2P and RBD epitopes targeted by the vaccinee sera are less conserved between the D614G and B.1.1.529 S glycoproteins than the S2P epitopes recognized by the sera from patients convalescing from COVID-19.
Figure 5.
Binding and neutralization sensitivity of SARS-CoV-2 variants
(A) The serum-binding titers (endpoint dilution) of sera from patients convalescing from COVID-19 (CP1–CP6) and sera from Moderna vaccinees (V1–V6) to the SARS-CoV-2 D614G and B.1.1.529 S2P spike trimers and RBDs. The intensity of shading is proportionate to the titer of binding activity. See also Figures S2 and S3.
(B) The serum neutralization titers (ID50) of sera from patients convalescing from COVID-19 (CP1–CP11) and sera from Moderna vaccinees (V1–V6) against VSV particles pseudotyped by the variant SARS-CoV-2 S glycoproteins are reported. The infectivity of the viruses was measured on Vero-E6 cells and 293T-ACE2 cells. The intensity of shading is proportionate to the titer of neutralizing activity. See also Figure S3.
(C) Endpoint dilution values of the binding of sera from patients convalescing from COVID-19 (left) and sera from Moderna vaccinees (right) to the B.1.1.529 and D614G S2P trimers and RBDs are compared.
(D) Serum neutralization ID50 values of sera from patients convalescing from COVID-19 (left) and sera from Moderna vaccinees (right) against VSV particles pseudotyped by B.1.617.2 and B.1.1.529 S glycoproteins were compared with those against the D614G pseudovirus. The geometric means of endpoint dilutions and ID50 values of each group are listed in the graphs in (C) and (D), respectively.
Statistical analysis was performed using a paired t test (∗p < 0.05; ∗∗p < 0.01; ns, not significant).
To assess the functional consequence of the observed differences in antibody binding to the S glycoprotein trimers, we tested the capability of these sera to neutralize VSV pseudovirus infection of both Vero-E6 and 293T-ACE2 cells (Figures 5B, 5D, and S4). The B.1.617.2 pseudoviruses were neutralized less effectively than the D614G pseudoviruses by the convalescent patient sera and the vaccinee sera. Neutralization of the B.1.1.529 pseudoviruses by all samples decreased dramatically, with only two samples from Moderna vaccinees having a detectable titer in the Vero-E6 cells. These results indicate that the B.1.1.529 S glycoprotein is significantly less sensitive to neutralization by antibodies elicited to earlier SARS-CoV-2 variant S glycoproteins, either by vaccination or following natural infection.
A comparison of the binding of the convalescent patient and vaccinee sera to the S2P and RBD proteins with the neutralizing activity of these sera suggests differences in the targeted S glycoprotein epitopes. The vaccinees elicit higher titers of antibodies against the SARS-CoV-2 S and against the RBD region; although this binding correlates better with virus-neutralizing ability, the targeted sites are less conserved between the D614G and B.1.1.529 S glycoproteins. By contrast, many of the S2P epitopes recognized by the convalescent patient sera, although conserved between the D614G and B.1.1.529 S glycoproteins, are not available as neutralization targets on the functional virus S glycoprotein (Brewer et al., 2022).
Discussion
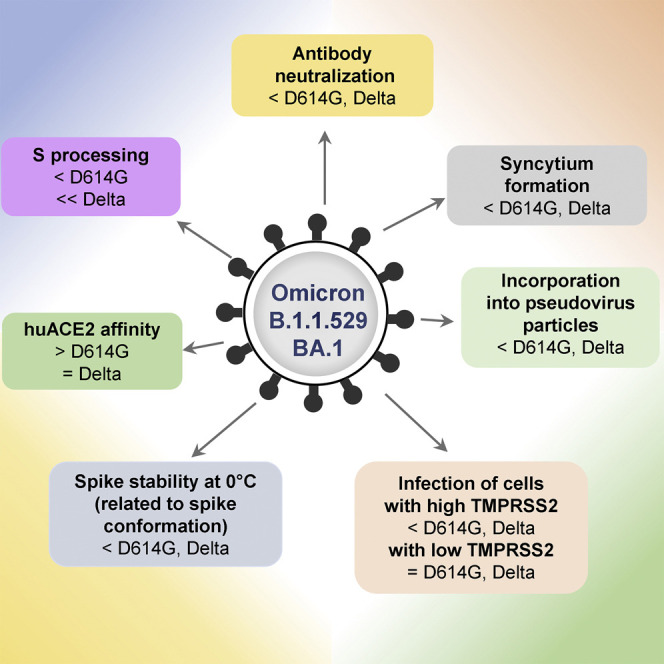
The recently emerged B.1.1.529 (Omicron) variant of SARS-CoV-2 has an unusually large number of changes in its S glycoprotein (Figure 1A). Although SARS-CoV-2 components other than S can potentially influence transmission, pathogenesis, and immune evasion, the concentration of variation in the B.1.1.529 S suggests its importance. Indeed, S changes render the virus less sensitive to some therapeutic antibodies and antibodies elicited by current vaccines (Grabowski et al., 2022; Pulliam et al., 2022; Scott et al., 2021). This has raised concerns about the potentially reduced efficacy of vaccines against B.1.1.529 as well as the increased risk for reinfection with this variant (Garcia-Beltran et al., 2022; Gruell et al., 2022; Hoffmann et al., 2022; Liu et al., 2022; Pajon et al., 2022; Planas et al., 2022). Insight into the biology of the novel B.1.1.529 variant would facilitate efforts to manage the COVID-19 pandemic. To this end, we compared the functional properties of the S glycoprotein of B.1.1.529 BA.1 with that of the globally dominant B.1.617.2 (Delta) variant and the ancestral D614G variant responsible for the early pandemic spread of SARS-CoV-2 (Figure 6 ).
Figure 6.
Summary of the phenotypes of the SARS-CoV-2 S glycoprotein variants
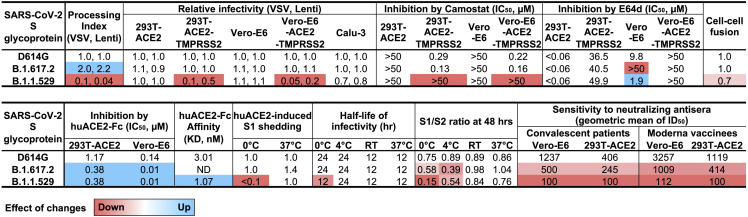
The primary data shown in Figures 1, 2, 3, 4, and 5 were used to calculate average values relative to those obtained for the D614G glycoprotein. Decreases in the phenotype relative to those of D614G are shaded red to a degree dependent on the level of the decrease. Increases in the phenotypes relative to those of D614G are shaded blue to a degree dependent on the level of the increase.
We observed significant differences in the proteolytic processing of the D614G, B.1.617.2, and B.1.1.529 S glycoproteins in expressing cells (Figure 1B). Cleavage of coronavirus S glycoproteins is a prerequisite for activation of their membrane-fusing activity in the infected cells, leading to the formation of short-lived syncytia. The S glycoprotein of the D614G variant, which supplanted the D614 variant responsible for the original SARS-CoV-2 outbreak, exhibited more efficient S glycoprotein cleavage compared with that of its D614 ancestor (Nguyen et al., 2020; Wang et al., 2021c). The S glycoprotein cleavage of most of the SARS-CoV-2 VOCs is not significantly different from that of the D614 variant (Wang et al., 2021c). Therefore, both the increased efficiency of B.1.617.2 S cleavage and the decreased efficiency of B.1.1.529 S cleavage, relative to that of the D614G S, are noteworthy (Figures 1B and 1C). Increases in the proteolytic processing of the B.1.617.2 S glycoprotein relative to the S glycoproteins of earlier SARS-CoV-2 variants have been reported (Mlcochova et al., 2021). The inefficient processing of the B.1.1.529 S glycoprotein on the surface of expressing cells likely contributes to its moderately decreased activity in mediating cell-cell fusion (Figures 2E and 2F). Syncytium formation represents a major form of the cytopathic effects of SARS-CoV-2, and the decreased cell-fusing capacity of the B.1.1.529 S glycoprotein could have implications with respect to pathogenesis. Very recent studies indicate that individuals infected with B.1.1.529 are at lower risk of hospitalization and have shorter hospital stays than individuals infected with other SARS-CoV-2 strains, including B.1.617.2 (Mahase, 2021). Although host variables, such as age and immune status, could potentially influence the observed differences in clinical outcome, intrinsic decreases in B.1.1.529 cytopathicity and pathogenicity may also play a role.
SARS-CoV-2 S glycoproteins incorporated into natural virions and pseudoviruses pass through the Golgi compartment. Relative to the total S glycoproteins synthesized in the producing cell, the virion S glycoproteins are enriched in cleaved glycoproteins modified extensively by complex glycans (Figure 1D; Nguyen et al., 2020; Zhang et al., 2021). This enrichment only partially compensates for the poor processing of the B.1.1.529 S glycoprotein in expressing cells. Nonetheless, despite the lower levels of the B.1.1.529 S on pseudovirus particles, the infectivity of these pseudoviruses was comparable to those of D614G and B.1.617.2 pseudoviruses on target cells with low levels of TMPRSS2 expression (Figures 2A and 2B). The ability of the B.1.1.529 S glycoprotein to support virus infection is likely aided by a high affinity for ACE2, a property shared by most emerging SAR-CoV-2 VOCs (Wang et al., 2021c). Relative to D614G pseudoviruses, both B.1.1.529 and B.1.617.2 pseudoviruses were neutralized more effectively by soluble ACE2 (Figure 3A). Direct measurement of the binding of soluble huACE2-Fc to the S glycoprotein trimers of D614G and B.1.1.529 confirmed the higher affinity of the latter S protein (Figure 3B).
Pseudoviruses with the D614G and B.1.617.2 S glycoproteins infected target cells expressing high levels of TMPRSS2 much more efficiently than the corresponding cells expressing lower levels of TMPRSS2 (Figures 2A and 2B). This was not the case for the B.1.1.529 pseudoviruses. E64d, an inhibitor of endosomal cathepsins, blocked infection of 293T-ACE2 and Vero-E6 cells by pseudoviruses with all three variant S glycoproteins. By contrast, only the D614G and B.1.617.2 pseudovirus infections and not the B.1.1.529 pseudovirus infections of TMPRSS2-overexpressing cells were inhibited by camostat mesylate, a serine protease (TMPRSS2) inhibitor (Figures 2C and 2D). These observations suggest that the D614G and B.1.617.2 S glycoproteins can efficiently utilize a high level of TMPRSS2 on the target cell surface to activate virus entry, whereas the B.1.1.529 S glycoprotein is much less able to do so. Thus, virus entry mediated by the B.1.1.529 S glycoprotein predominantly occurs through the use of endosomal cathepsins, consistent with other reports (Meng et al., 2022; Zhao et al., 2022).
Syncytium formation by SARS-CoV-2 S glycoproteins relies on cell-surface proteases like TMPRSS2 for activation of the membrane-fusing capacity of the S glycoprotein. The relatively poor ability of the B.1.1.529 S glycoprotein to utilize TMPRSS2 efficiently in support of virus entry contrasts with the rather modest reduction observed for cell-cell fusion mediated by the B.1.1.529 S glycoprotein. This may relate to the ability of SARS-CoV-2 S glycoproteins to use cell-surface proteases (e.g., matrix metalloproteases) other than TMPRSS2 to activate cell-cell fusion (Nguyen et al., 2020). Alternatively, utilization of TMPRSS2 by the B.1.1.529 S glycoprotein may be affected by the different contexts in which virus entry and cell-cell fusion occur.
The exposure of native SARS-CoV-2 S glycoproteins to 0°C can reveal strain-dependent differences in the stability and functional integrity of the S glycoprotein trimer. The infectivity of B.1.1.529 pseudoviruses decayed similarly to those of D614G and B.1.617.2 pseudoviruses at RT and 37°C but exhibited a relatively faster decay at 0°C (Figure 4A). This corresponded to an increase in the spontaneous shedding of the S1 glycoprotein from the B.1.1.529 S trimer during prolonged incubation at 0°C, compared with the D614G and B.1.617.2 Ss (Figures 4B and 4C). The cold resistance of enveloped viruses is related to the ability of their oligomeric envelope glycoproteins to resist the destabilizing effects of ice crystal formation (Privalov, 1990). The sensitivity of human immunodeficiency viruses (HIV-1) to inactivation at 0°C is related to the propensity of the viral envelope glycoprotein trimer to undergo conformational transitions from the pretriggered conformation to more “open” receptor-bound conformations (Kassa et al., 2009). Likewise, we found a correlation between cold sensitivity of SARS-CoV-2 variants and the occupancy of the S glycoprotein trimers in a conformation in which one RBD is in the up position and the other two RBDs are in the “down” position (Figures 4D and 4E). Thus, the cold sensitivity of the B.1.1.529 and P.1 variants may reflect the propensity of the “closed” S trimer to sample spontaneously the next more open state, a conformation with one RBD up. This propensity to undergo conformational changes could assist the triggering of conformational transitions in the B.1.1.529 S and help compensate for a lower S density on the virion. For the P.1 SARS-CoV-2 variant, which has a more typical S density, increased triggerability could account for the efficiency with which target cells with low levels of ACE2 can be infected (Wang et al., 2021c).
Relative to the D614G and B.1.617.2 Ss on pseudoviruses, the B.1.1.529 S demonstrated marked resistance to S1 shedding after exposure to soluble ACE2 at 0°C (Figures 3D and 3E). The B.1.1.529 S shares this property with the P.1 S, but not with the S glycoproteins of other SARS-CoV-2 variants studied (Cerutti et al., 2022; Wang et al., 2021a, 2021c). Apparently, in the more open ACE2-bound state, with RBDs in the up position, the B.1.1.529 and P.1 S trimers maintain a tighter association with S1. This property of the B.1.1.529 and P.1 Ss might be a secondary adaptation to an increased spontaneous sampling of the one-RBD-up conformation. During infection of cells, the prolonged stability of the receptor-bound S glycoprotein trimer could allow the requisite number of S-receptor interactions to be achieved, compensating for a lower density of virion Ss (in the case of B.1.1.529) or lower availability of ACE2 receptors (in the case of P.1). The temperature-dependent regulation of S conformational state could have value for respiratory viruses like SARS-CoV-2, because they infect both cells in the nasal passages that could be at cool temperatures, as well as cells in the lower respiratory tract that are at 37°C.
The B.1.1.529 pseudoviruses were remarkably resistant to neutralization by sera from patients convalescing from COVID-19 and sera from individuals who had received two doses of the Moderna vaccine (Figures 5B, 5D, and S4). These results indicate that most of the neutralizing antibodies generated to previous SARS-CoV-2 variants will be less effective against B.1.1.529. Resistance to neutralizing antibodies is clearly a major factor driving the evolution of SARS-CoV-2 variants (Choi et al., 2020). This conclusion is consistent with the increased number of vaccine breakthrough infections involving B.1.1.529 compared with earlier SARS-CoV-2 variants (Andrews et al., 2022; Kuhlmann et al., 2022; Pulliam et al., 2022). The B.1.1.529 S is apparently well adapted so that any negative consequences of an increased propensity to sample the one-RBD-up conformation, which potentially could result in greater exposure to neutralizing antibodies, are compensated during naturally encountered circumstances.
Although both convalescent patient sera and vaccinee sera failed to neutralize B.1.1.529 pseudoviruses, the binding of the convalescent patient sera to recombinant S trimer (S2P) of B.1.1.529 was only modestly decreased compared with the binding to D614G S2P. By contrast, the difference in binding of the vaccinee sera to these two recombinant S trimers was much greater. The magnitude of the decrease in binding to D614G and B.1.1.529 RBDs between the convalescent patient sera and vaccinee sera was not as significant as that seen for binding to the S2P glycoproteins (Figures 5A and 5C). The vaccinee sera apparently recognize S2P and RBD epitopes that differ between D614G and B.1.1.529 but which represent targets for neutralization on the functional virus S. A significant fraction of antibodies in the sera of convalescent patients recognize S epitopes that are conserved between the D614G and B.1.1.529 S2P glycoproteins but are not available as neutralization targets on the native virus S. For example, Ss that shed S1 during natural infection could present S2 trimers to the host immune system; many of the antibodies directed against these S2 trimers would fail to bind intact S trimer (Brewer et al., 2022). By contrast, the S2P trimers used for vaccination are uncleaved and stabilized in a pre-fusion conformation, properties that may decrease trimer opening or exposure of epitopes for non-neutralizing antibodies.
In conclusion, we demonstrate that the B.1.1.529 S glycoprotein has a combination of properties differentiating it from previous SARS-CoV-2 S variants. In addition to the resistance of the B.1.1.529 S to antibodies reactive with other SARS-CoV-2 S glycoproteins, other notable features include the relatively low level of B.1.1.529 S processing and virus S density, decreased syncytium-forming ability, decreased ability to utilize TMPRSS2 to support virus entry, and the spontaneous and ACE2-regulated stability of the S trimer, which is temperature dependent and relates to S conformational states. A better understanding of the S-determined biological characteristics of this newly emerged SARS-CoV-2 variant should assist the design of vaccines and other interventions.
Limitations of the study
The assays conducted in our study used viruses pseudotyped by the SARS-CoV-2 S glycoproteins rather than authentic SARS-CoV-2 viruses. We used cell lines to study infection by the pseudoviruses and did not study natural target cells for SARS-CoV-2 infection. Although these experimental systems allow more precise control of variables, there may be differences from natural SARS-CoV-2 infection. In this study, we did not define the S glycoprotein determinants of the distinct B.1.1.529 S phenotypes.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-SARS-Spike S1 | SinoBiological | Cat# 40,591-T62 RRID: AB_2893171 |
| Rabbit anti-SARS-Spike S2 | SinoBiological | Cat# 40,590-T62 |
| Rabbit anti-p55/p24/p17 | Abcam | Cat# ab63917 RRID: AB_1139524 |
| Mouse anti-VSV-NP | Millipore | Cat# MABF2348 |
| Mouse anti-GAPDH | Millipore | Cat# CB1001 RRID: AB_2107426 |
| Rabbit anti-SARS-CoV-2 nucleocapsid antibody | GeneTex | Cat# GTX135357 RRID: AB_2868464 |
| HRP-conjugated anti-Rabbit antibody | Cytiva | Cat# NA934-1ML RRID: AB_2722659 |
| HRP-conjugated goat anti-mouse antibody | Jackson ImmunoResearch | Cat# 115-035-008 RRID:AB_2313585 |
| Strep•Tag® II Antibody HRP Conjugate | Sigma-Aldrich | Cat# 71,591 RRID: AB_10806716 |
| Peroxidase AffiniPure goat anti-human IgG (H + L) antibody | Jackson ImmunoResearch | Cat# 109-035-003 RRID: AB_2337577 |
| Bacterial and virus strains | ||
| VSV-G pseudotyped ΔG-luciferase | Kerafast | Cat# EH1020-PM |
| Biological samples | ||
| CP1-CP11 sera | (Wang et al., 2021a) | N/A |
| V1-V6 sera | (Wang et al., 2021a) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Human ACE2-Fc | SinoBiological | Cat# 10,108-H02H |
| SARS-CoV-2 D614G S2P | This paper | N/A |
| SARS-CoV-2 B.1.1.529 S2P | This paper | N/A |
| Critical commercial assays | ||
| Pseudotyped ΔG-luciferase (G∗ΔG-luciferase) rVSV | Kerafast | Cat# EH1025-PM |
| Luciferase Assay System | Promega | Cat# E4550 RRID: AB_1660600 |
| Nano-Glo® Luciferase Assay System | Promega | Cat# N1120 |
| Galacto-Star™ Reaction Buffer Diluent with Galacton-Star™ Substrate | ThermoFisher | Cat# T1056 |
| PNGase F | NEB | Cat# P0704L |
| Endo Hf | NEB | Cat# P0703L |
| QuikChange Lightning Site-Directed Mutagenesis Kit | Agilent | Cat# 210,518 |
| 3, 3′,5,5′-Tetramethylbenzidine Liquid Substrate, Supersensitive, for ELISA | Sigma-Aldrich | Cat# T4444 |
| Experimental models: Cell lines | ||
| 293T | ATCC | CRL-3216 |
| 293T-ACE2 | BEI | NR-5211 |
| Vero-E6 | ATCC | CRL-1586 |
| 293T-ACE2-TMPRSS2 | BEI | NR-55293 |
| Vero-E6-ACE2-TMPRSS2 | BEI | NR-54970 |
| Calu-3 | ATCC | HTB-55 |
| COS-1 | ATCC | CRL-1650 |
| Recombinant DNA | ||
| Plasmid: SARS-CoV-2 D614G spike | This paper | N/A |
| Plasmid: SARS-CoV-2 B.1.617.2 spike | This paper | N/A |
| Plasmid: SARS-CoV-2 B.1.1.529 spike | This paper | N/A |
| pHIV-1NL4-3 ΔEnv-NanoLuc | (Liu et al., 2020) | N/A |
| Plasmid: SARS-CoV-2 membrane (M) | Addgene | Cat# 141,386 |
| Plasmid: SARS-CoV-2 envelope (E) | Addgene | Cat# 141,385 |
| Plasmid: SARS-CoV-2 nucleocapsid (N) | (Zhang et al., 2020) | N/A |
| Software and algorithms | ||
| ImageJ | the National Institutes of Health | https://imagej.nih.gov/ij/ |
| Image lab | Bio-rad | https://www.bio-rad.com/en-us/product/image-lab-software?ID=KRE6P5E8Z |
| GraphPad Prism 9 | GraphPad Software Inc | https://www.graphpad.com/scientific-software/prism/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Joseph G. Sodroski (joseph_sodroski@dfci.harvard.edu).
Materials availability
All requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Joseph G. Sodroski (joseph_sodroski@dfci.harvard.edu). This includes selective cell lines, plasmids, antibodies, viruses, serum and proteins. All reagents will be made available on request after completion of a Material Transfer Agreement.
Experimental model and subject details
Human subjects
Plasma samples were obtained from patients (aged 27-85 years) convalescing from SARS-CoV-2 infection (Wang et al., 2021c). The patients were enrolled in an observational cohort study of patients convalescing from COVID-19 at the Columbia University Irving Medical Center (CUIMC) starting in the spring of 2020. The study protocol was approved by the CUIMC Institutional Review Board (IRB) and all participants provided written informed consent.
Sera were obtained from 6 participants in a phase-I clinical trial of the Moderna SARS-CoV-2 mRNA-1273 vaccine conducted at the NIH, under an NIH IRB-approved protocol (Andrews et al., 2022). The sera used in the present study were obtained after the participants had received two doses of mRNA-1273.
Cell lines
HEK293T, 293T-ACE2 (BEI), Vero-E6, 293T-ACE2-TMPRSS2 (BEI), Vero-E6-ACE2-TMPRSS2 (BEI) and COS-1 cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 mg/mL penicillin-streptomycin (Thermo Fisher Scientific, Cambridge, MA). Calu-3 cells (ATCC) were cultured in Eagle’s Minimum Essential Medium supplemented with 10% FBS and 100 mg/mL penicillin-streptomycin. Expi293 cells were maintained in suspension culture directly in Expi293TM Expression Medium, supplemented with penicillin and streptomycin, and were incubated at 37°C in a humidified atmosphere of 8% CO2 in air and on a shaker platform rotating at 125 rpm. HEK293T cells, 293T-ACE2 cells, 293T-ACE2-TMPRSS2 cells and Expi293 cells are of female origin. Vero-E6, Vero-E6-ACE2-TMPRSS2 and COS-1 cells are from African green monkey kidneys. Calu-3 epithelial cells were isolated from lung adenocarcinoma tissue derived from a 25-year-old, white male patient.
Method details
Plasmid constructs
The codon-optimized SARS-CoV-2 spike (S) gene (Sino Biological, Wayne, PA) encoding the S glycoprotein lacking 18 amino acids at the carboxyl terminus was cloned into the pCMV3 vector. The gene encoding the B.1.617.2 SARS-CoV-2 spike variant was made by introducing additional mutations into the wild-type (D614) S gene using a site-directed mutagenesis kit (Agilent, Santa Clara, CA). The B.1.1.529 S gene was generated by a high-throughput template-guided gene synthesis approach (Liu et al., 2022). Plasmids expressing the SARS-CoV-2 M (Cat# 141,386) and E (Cat# 141,385) proteins with 2XStrep tags at their C termini were from Addgene. The SARS-CoV-2 N gene cloned in pCAGFLAG was kindly provided by Dr. Pei-Hui Wang (Zhang et al., 2020). The studies conducted in this manuscript used the BA.1 subvariant of B.1.1.529. The genes encoding the SARS-CoV-2 D614G and B.1.1.529 S trimer ectodomains with 2P and furin cleavage-site changes, and containing a C-terminal 8x His tag (Wrapp et al., 2020), were synthesized and then cloned into the paH vector.
Expression and purification of protein reagents
The soluble SARS-CoV-2 S2P spike trimer proteins of the D614G and B.1.1.529 variants were generated by transfecting Expi293 cells with the trimer protein-expressing constructs using FectPRO DNA transfection reagent (Polyplus, New York, NY). The soluble S2P spike trimers were purified from cell supernatants 5 days later using Ni-NTA resin (Invitrogen), according to the manufacturer’s protocol (Liu et al., 2020).
Two μg of each S2P trimer and RBD protein was analyzed on a NuPAGE Bis-Tris protein gel (Invitrogen, Waltham, MA) run at 200 V using MES buffer, after which the gel was stained with Coomassie Blue dye.
VSV pseudotyped by SARS-CoV-2 S glycoproteins
To generate VSV-based vectors pseudotyped with SARS-CoV-2 S glycoproteins, 6 X 106 HEK293T cells were plated in a 10-cm dish one day before transfection. Fifteen μg of the SARS-CoV-2 S glycoprotein plasmid was transfected into the HEK293T cells using Polyethylenimine (Polysciences, Warrington, PA). Twenty-four hours later, the cells were infected at a multiplicity of infection of 3–5 with rVSV-ΔG pseudovirus bearing a luciferase gene (Kerafast, Boston, MA) for 2 h at 37°C and then washed three times with PBS. Cell supernatants containing pseudoviruses were harvested after another 24 h, clarified by low-speed centrifugation (2000 rpm for 10 min) and filtered through a 0.45-μM filter. Viruses were then aliquoted and stored at −80°C until use (Liu et al., 2020; Wang et al., 2021a, 2021b).
Lentiviruses pseudotyped by SARS-CoV-2 S glycoproteins
To generate HIV-based vectors pseudotyped with SARS-CoV-2 S glycoproteins, 6 X 106 HEK293T cells were plated in a 10-cm dish one day before transfection. Then, 7.5 μg of SARS-CoV-2 S glycoprotein expressor plasmid and 7.5 μg pHIV-1NL4-3ΔEnv-NanoLuc reporter construct were cotransfected into the HEK293T cells using Polyethylenimine (Polysciences). Cell supernatants containing pseudoviruses were harvested 48 h after transfection, clarified by low-speed centrifugation (2000 rpm for 10 min) and filtered through a 0.45-μM filter. Viruses were then aliquoted and stored at −80°C until use.
S glycoprotein expression, processing and incorporation into pseudovirus particles
HEK293T cells were transfected to produce VSV- and HIV-based particles pseudotyped with SARS-CoV-2 S glycoprotein variants, as described above. To prepare viral particles, cell supernatants were collected, centrifuged at low speed (2000 rpm) to remove cell debris, filtered through a 0.45-μM filter and pelleted at 18,000 x g for 1 h at 4°C. In parallel with harvesting the pseudoviruses from the cell supernatants, cells were washed and lysed using 1.5% Cymal-5 at 4°C for 10 min. Cell lysates were then clarified by high-speed centrifugation (18,000 x g for 10 min). Cell lysates and virions were analyzed by Western blotting with the following primary antibodies: rabbit anti-SARS-Spike S1 (Sino Biological, Cat# 40,591-T62), rabbit anti-SARS-Spike S2 (Sino Biological, Cat# 40,590-T62), rabbit anti-p55/p24/p17 (Abcam, Cambridge, MA (Cat# ab63917)), mouse anti-VSV NP (Millipore, Burlington, MA (Cat# MABF2348)) or mouse anti-GAPDH (Millipore, Cat# CB1001). The Western blots were developed with the following secondary antibodies: HRP-conjugated anti-rabbit antibody (Cytiva, Marlborough, MA (NA934-1ML)) or HRP-conjugated goat anti-mouse antibody (Jackson ImmunoResearch, West Grove, PA (Cat# 115-035-008)).
S, S1, and S2 band intensities from unsaturated Western blots were calculated using ImageJ Software. S1/S2 ratios represent the ratios of the intensities of the S1 and S2 glycoprotein bands in the Western blots. The values for the processing index of the B.1.617.2 and B.1.1.529 variant S glycoproteins were calculated as follows:Processing index = (S1/S × S2/S)variant ÷ (S1/S × S2/S)D614G
For production of SARS-CoV-2 virus-like particles (VLPs), a plasmid expressing the variant SARS-CoV-2 S glycoprotein was transfected into 293T cells in 6-well plates in combination with equal amounts of plasmids expressing the SARS-CoV-2 M, E and N proteins. Two days after transfection, cell lysates and VLP pellets were prepared as described above. Samples were analyzed by Western blotting with antibodies against S1 and S2, as described above. Samples were also analyzed by Western blotting with rabbit anti-SARS-CoV-2 N antibody (GeneTex, Cat# GTX135357) and then developed with HRP-conjugated anti-rabbit antibody (Cytiva, Marlborough, MA (NA934-1ML)). Strep-tagged SARS-CoV-2 M and E proteins were analyzed by Western blotting with HRP-conjugated Strep tag antibody (Sigma-Aldrich, Cat# 71,591).
Deglycosylation of S glycoproteins
SARS-CoV-2 S glycoproteins in cell lysates or on pseudovirus particles were prepared as described above. Protein samples were boiled in 1 X denaturing buffer and incubated with PNGase F or Endo Hf (New England Biolabs, Ipswich, MA) for 1 h at 37°C according to the manufacturer’s protocol. The samples were then analyzed by SDS-PAGE and Western blotting as described above.
Virus infectivity and stability at different temperatures
The pseudoviruses were freshly prepared as described above, without freezing and thawing. The pseudovirus preparations were incubated with target cells seeded in 96-well plates at a density of 3 X 104 cells/well. For VSV-based pseudoviruses, target cells were cultured for 16-24 h after infection and then harvested to measure the luciferase activity (Promega, Madison, WI (Cat# E4550)). For HIV-1-based pseudoviruses, target cells were cultured for 2-3 days after infection and then cells were harvested to measure the NanoLuc luciferase activity (Promega, Cat# N1120).
To measure viral stability at different temperatures, pseudoviruses were incubated in an ice water bath (0°C), at 4°C, at room temperature and at 37°C for different lengths of time prior to measuring their infectivity in Vero-E6 cells.
To measure the infectivity of pseudovirus variants on target cells expressing different levels of human ACE2, serial dilutions (from 2 μg to 24.7 ng) of the human ACE2 expressor plasmid (Addgene, Watertown, MA (Cat# 1786)) were transfected into 293T cells in 12-well plates using 1 mg/mL PEI. Two days after transfection, cells were trypsinized and used as target cells for measuring the infectivity of VSV pseudotypes (Wang et al., 2021c).
Cell-cell fusion assay
For the alpha-complementation assay measuring cell-cell fusion, COS-1 effector cells were plated in black-and-white 96-well plates and then cotransfected with a plasmid expressing ⍺-gal and either pCMV3 or the SARS-CoV-1 S glycoprotein variant at a 1:1 ratio, using lipofectamine 3000 (Thermo Fisher) according to the manufacturer’s protocol. At the same time, 293T target cells in 6-well plates were cotransfected with plasmids expressing ω-gal and human ACE2 at a 1:1 ratio, using lipofectamine 3000. Forty-eight hours after transfection, target cells were scraped and resuspended in medium. Medium was removed from the effector cells, and target cells were then added to effector cells (one target-cell well provides sufficient cells for 50 effector-cell wells). Plates were spun at 500 x g for 3 min and then incubated at 37°C in 5% CO2 for 4 h. Medium was aspirated and cells were lysed in Tropix lysis buffer (Thermo Fisher Scientific). The β-galactosidase activity in the cell lysates was measured using the Galacto-Star Reaction Buffer Diluent with Galacto-Star Substrate (Thermo Fisher Scientific), following the manufacturer’s protocol.
To evaluate S glycoprotein expression in COS-1 cells, cells in 6-well plates were cotransfected with a plasmid expressing ⍺-gal and either pCMV3 or a plasmid expressing the SARS-CoV-2 S glycoprotein variant at a 1:1 ratio, using lipofectamine 3000 (Thermo Fisher) according to the manufacturer’s protocol. To evaluate the S glycoprotein in cell lysates, cells were lysed with 1.5% Cymal-5 containing a protease inhibitor two days after transfection. To evaluate the S glycoprotein on the cell surface, cells were incubated with EZ-link Sulfo-NHS-SS-Biotin. The biotinylated cell surface proteins were then precipitated by NeutrAvidin Agarose using a Pierce Cell Surface Protein Isolation Kit (Thermo Scientific, Cat# 89,881). The expression of the S glycoproteins in cell lysates and on the cell surface was analyzed by Western blotting as described above.
Surface plasmon resonance (SPR) assay
SPR assays for human ACE2-Fc (SinoBiological, Cat# 10,108-H02H) binding to SARS-CoV-2 D614G or B.1.1.529 spike trimers were performed using a Biacore T200 biosensor, equipped with a Series S CM5 chip (Cytiva, Cat# BR100530), at 25°C, in a running buffer of 10 mM HEPES pH 7.4, 150 mM NaCl, 0.2 mg/mL BSA and 0.01% (v/v) Tween 20 at 25°C. Each spike was captured through its C-terminal His6 tag on an anti-His6 antibody surface, generated using the His-capture kit (Cytiva, Cat# 28,995,056) according to the instructions of the manufacturer.
During a binding cycle, each spike was captured over individual flow cells at approximately 500-700 RU and an anti-His6 antibody surface was used as a reference flow cell. Human ACE2-Fc was prepared at six concentrations using a three-fold dilution series in running buffer, ranging from 1.1 to 90 nM. Samples were tested in order of increasing protein concentration. Blank buffer cycles were performed by injecting running buffer instead of the analyte, after two ACE2 injections. The association and dissociation rates were each monitored for 90s and 300s respectively, at 50 μL/min. Bound spike/ACE2 complexes were removed using a 10s pulse of 15 mM H3PO4 at 100 μL/min, thus regenerating the anti-His6 surface for a new cycle of recapturing of each spike, followed by a 60s buffer wash at 100 μL/min. The data were processed and fit to a 1:1 interaction model using the Scrubber 2.0 (BioLogic Software).
S1 shedding from spike trimers
VSV particles pseudotyped with S glycoprotein variants were prepared as described above. To evaluate spontaneous S1 shedding at different temperatures, the cell supernatants containing pseudoviruses were incubated in an ice water bath (0°C), at 4°C, room temperature and 37°C for different times. Virus particles were then pelleted at 18,000 x g for 1 h at 4°C. The pelleted samples were resuspended in 1 X LDS sample buffer (Invitrogen, Cat# NP0008) and analyzed by Western blotting.
To evaluate soluble huACE2-Fc-induced S1 shedding, the cell supernatants containing pseudovirus particles were incubated with soluble huACE2-Fc (Sino Biological Inc (cat# 10,108-H02H)) at different concentrations at 0°C or 37°C for 1 h. Afterward, virus particles were pelleted at 18,000 x g for 1 h at 4°C. The pelleted virus particles were washed once with cold PBS before the samples were resuspended in 1 X LDS sample buffer and analyzed by Western blotting. S1, S2 and NP were detected as described above; huACE2-Fc bound to the pseudovirus particles was detected with Peroxidase AffiniPure goat anti-human IgG (H + L) antibody (Jackson ImmunoResearch).
ELISA assays
Fifty nanograms of S2P spike trimer or RBD protein (Sino Biological Inc. (Cat: 40,592-V08H121 and 40,592-V08H)) was coated on each well of an ELISA plate at 4°C overnight. Then the plates were blocked with 300 mL of blocking buffer (1% BSA and 10% bovine calf serum in PBS) at 37°C for 2 h. Afterward, serially diluted convalescent serum or Moderna vaccinee serum was added and incubated at 37°C for 1 h. Next, 100 μL per well of 10,000-fold diluted Peroxidase AffiniPure goat anti-human IgG (H + L) antibody (Jackson ImmunoResearch) was added and incubated for 1 h at 37°C. Between each step, the plates were washed with PBST three times. Finally, the TMB substrate (Sigma, St. Louis, MO) was added and incubated for 5 min at room temperature before the reaction was stopped using 1 M sulfuric acid. Absorbance was measured at 450 nm.
The serum endpoint dilutions that achieved an OD450 value > 3-fold over background were calculated by fitting the data in five-parameter dose-response curves in GraphPad Prism 9 (GraphPad Software Inc., San Diego, CA).
Virus inhibition assay
To test the inhibition of virus infection by camostat and E64d, 293T-ACE2, 293T-ACE2-TMPRSS2, Vero-E6, or Vero-E6-ACE2-TMPRSS2 cells were pre-treated with serial dilutions of camostat or E64d in triplicate in 96-well plates for 1 h at 37°C prior to inoculation with VSV pseudotypes. The cultures were maintained for 16-24 h at 37°C before luciferase activity was measured as described above.
Sera were collected from March to June 2020 from New York City patients that recovered from COVID-19; six sera were collected from Moderna vaccinees (Wang et al., 2021b). Pseudovirus neutralization assays were performed by incubating VSV vectors pseudotyped by S glycoprotein variants with serial dilutions of huACE2-Fc (Cat# 10,108-H02H), sera from convalescent COVID-19 patients or vaccinee sera in triplicate in 96-well plates for 1 h at 37°C. Approximately 3 x 104 target cells (Vero- E6 or 293T-ACE2 cells) per well were then added. The cultures were maintained for an additional 16-24 h at 37°C before luciferase activity was measured as described above.
The % inhibition and neutralization values were calculated from the reduction in luciferase activity compared with mock-treated controls. The concentrations of camostat, E64d, huACE2-Fc and serum titers that inhibit 50% of infection (the IC50 and ID50 values, respectively) were determined by fitting the data in five-parameter dose-response curves in GraphPad Prism 9 (GraphPad Software Inc., San Diego, CA).
Quantification and statistical analysis
Evaluations of statistical significance were performed employing Student’s unpaired or paired t test using GraphPad Prism 9 software. Levels of significance are indicated as follows: ns, p > 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; and ∗∗∗∗p < 0.0001. Linear correlation was determined by fitting the data with simple linear regression. EC50, IC50 and ID50 values were determined by fitting the data in five-parameter dose-response curves in GraphPad Prism 9. Western blot data were analyzed by Image Lab and ImageJ software. All data presented is representative or mean data derived from at least two independent experiments.
Acknowledgments
We thank Elizabeth Carpelan for manuscript preparation and Hanh T. Nguyen for valuable suggestions on this study. This study was supported by a gift from William F. McCarty-Cooper.
Author contributions
Q.W. and J.G.S. designed the research. Q.W. and S.A. performed the experimental work. S.I., Y.G., L.L., and D.D.H. provided the convalescent and vaccinee sera and soluble S trimers. P.S.K. and L.S. generated the surface plasmon resonance (SPR) data. Q.W., S.A., and J.G.S. analyzed the data and wrote the manuscript. All authors reviewed and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: June 14, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110924.
Supplemental information
Data and code availability
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Data reported in this paper will be shared by the lead contact upon request.
This paper does not report original codes.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abu-Raddad L.J., Chemaitelly H., Butt A.A., National Study Group for C.-V. Effectiveness of the BNT162b2 Covid-19 vaccine against the B.1.1.7 and B.1.351 variants. N. Engl. J. Med. 2021;385:187–189. doi: 10.1056/nejmc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., Gower C., Kall M., Groves N., O'Connell A.M., et al. Covid-19 vaccine effectiveness against the omicron (B.1.1.529) variant. N. Engl. J. Med. 2022;386:1532–1546. doi: 10.1056/nejmoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/nejmoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer R.C., Ramadoss N.S., Lahey L.J., Jahanbani S., Robinson W.H., Lanz T.V. BNT162b2 vaccine induces divergent B cell responses to SARS-CoV-2 S1 and S2. Nat. Immunol. 2022;23:33–39. doi: 10.1038/s41590-021-01088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti G., Guo Y., Liu L., Liu L., Zhang Z., Luo Y., Huang Y., Wang H.H., Ho D.D., Sheng Z., Shapiro L. Cryo-EM structure of the SARS-CoV-2 omicron spike. Cell. Rep. 2022;38:110428. doi: 10.1016/j.celrep.2022.110428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., Solomon I.H., Kuo H.H., Boucau J., Bowman K., et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185:457–466.e4. doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski F., Kochanczyk M., Lipniacki T. The spread of SARS-CoV-2 variant omicron with a doubling time of 2.0-3.3 days can be explained by immune evasion. 2022;14:294. doi: 10.3390/v14020294. Viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruell H., Vanshylla K., Tober-Lau P., Hillus D., Schommers P., Lehmann C., Kurth F., Sander L.E., Klein F. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Nat. Med. 2022;28:477–480. doi: 10.1038/s41591-021-01676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kruger N., Schulz S., Cossmann A., Rocha C., Kempf A., Nehlmeier I., Graichen L., Moldenhauer A.S., Winkler M.S., et al. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell. 2022;185:447–456.e11. doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassa A., Finzi A., Pancera M., Courter J.R., Smith A.B., 3rd, Sodroski J. Identification of a human immunodeficiency virus type 1 envelope glycoprotein variant resistant to cold inactivation. J. Virol. 2009;83:4476–4488. doi: 10.1128/jvi.02110-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann C., Mayer C.K., Claassen M., Maponga T., Burgers W.A., Keeton R., Riou C., Sutherland A.D., Suliman T., Shaw M.L., Preiser W. Breakthrough infections with SARS-CoV-2 omicron despite mRNA vaccine booster dose. Lancet. 2022;399:625–626. doi: 10.1016/s0140-6736(22)00090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Iketani S., Guo Y., Chan J.F., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Lu L., Mok B.W.Y., Chen L.L., Chan J.M.C., Tsang O.T.Y., Lam B.H.S., Chuang V.W.M., Chu A.W.H., Chan W.M., Ip J.D., et al. Neutralization of SARS-CoV-2 Omicron variant by sera from BNT162b2 or Coronavac vaccine recipients. Clin. Infect. Dis. 2021:ciab1041. doi: 10.1093/cid/ciab1041. ciab1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., Padayachee S.D., Dheda K., Barnabas S.L., Bhorat Q.E., et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N. Engl. J. Med. 2021;384:1885–1898. doi: 10.1056/nejmoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahase E. Covid-19: hospital admission 50-70% less likely with omicron than delta, but transmission a major concern. BMJ. 2021;375:n3151. doi: 10.1136/bmj.n3151. [DOI] [PubMed] [Google Scholar]
- Meng B., Abdullahi A., Ferreira I.A.T.M., Goonawardane N., Saito A., Kimura I., Yamasoba D., Gerber P.P., Fatihi S., Rathore S., et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. 2022;603:706–714. doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlcochova P., Kemp S.A., Dhar M.S., Papa G., Meng B., Ferreira I.A.T.M., Datir R., Collier D.A., Albecka A., Singh S., et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599:114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H.T., Zhang S., Wang Q., Anang S., Wang J., Ding H., Kappes J.C., Sodroski J. Spike glycoprotein and host cell determinants of SARS-CoV-2 entry and cytopathic effects. J. Virol. 2021;95 doi: 10.1128/jvi.02304-20. e02304–02320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajon R., Doria-Rose N.A., Shen X., Schmidt S.D., O'Dell S., McDanal C., Feng W., Tong J., Eaton A., Maglinao M., et al. SARS-CoV-2 omicron variant neutralization after mRNA-1273 booster vaccination. N. Engl. J. Med. 2022;386:1088–1091. doi: 10.1056/NEJMc2119912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock T.P., Brown J.C., Zhou J., Thakur N., Newman J., Kugathasan R., Sukhova K., Kaforou M., Bailey D., Barclay W.S. The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry. Priprint at bioRxiv. 2021 doi: 10.1101/2021.12.31.474653. [DOI] [Google Scholar]
- Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalov P.L. Cold denaturation of protein. Crit. Rev. Biochem. Mol. Biol. 1990;25:281–306. doi: 10.3109/10409239009090612. [DOI] [PubMed] [Google Scholar]
- Pulliam J.R.C., van Schalkwyk C., Govender N., von Gottberg A., Cohen C., Groome M.J., Dushoff J., Mlisana K., Moultrie H. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science. 2022;376 doi: 10.1126/science.abn4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoff J., Gray G., Vandebosch A., Cardenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B., et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/nejmoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F., Weisblum Y., Muecksch F., Hoffmann H.H., Michailidis E., Lorenzi J.C.C., Mendoza P., Rutkowska M., Bednarski E., Gaebler C., et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 2020;217 doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L., Hsiao N.Y., Moyo S., Singh L., Tegally H., Dor G., Maes P., Pybus O.G., Kraemer M.U.G., Semenova E., et al. Track Omicron's spread with molecular data. Science. 2021;374:1454–1455. doi: 10.1126/science.abn4543. [DOI] [PubMed] [Google Scholar]
- Wang P., Casner R.G., Nair M.S., Wang M., Yu J., Cerutti G., Liu L., Kwong P.D., Huang Y., Shapiro L., Ho D.D. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell. Host. Microbe. 2021;29:747–751.e4. doi: 10.1016/j.chom.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wang Q., Nair M.S., Anang S., Zhang S., Nguyen H., Huang Y., Liu L., Ho D.D., Sodroski J.G. Functional differences among the spike glycoproteins of multiple emerging severe acute respiratory syndrome coronavirus 2 variants of concern. iScience. 2021;24:103393. doi: 10.1016/j.isci.2021.103393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2021. Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., Evans J.P., Qu P., Faraone J., Zheng Y.M., Carlin C., Bednash J.S., Zhou T., Lozanski G., Mallampalli R., et al. Neutralization and stability of SARS-CoV-2 omicron variant. bioRxiv. 2021 doi: 10.1101/2021.12.16.472934. Preprint at. [DOI] [Google Scholar]
- Zhang J., Cruz-Cosme R., Zhuang M.W., Liu D., Liu Y., Teng S., Wang P.H., Tang Q. A systemic and molecular study of subcellular localization of SARS-CoV-2 proteins. Signal. Transduct. Target. Ther. 2020;5:269. doi: 10.1038/s41392-020-00372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Go E.P., Ding H., Anang S., Kappes J.C., Desaire H., Sodroski J.G. Glycosylation and disulfide bonding of wild-type SARS-CoV-2 spike glycoprotein. J. Virol. 2021:JVI0162621. doi: 10.1128/jvi.01626-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Lu L., Peng Z., Chen L.L., Meng X., Zhang C., Ip J.D., Chan W.M., Chu A.W.H., Chan K.H., et al. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg. Microbes. Infect. 2022;11:277–283. doi: 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Data reported in this paper will be shared by the lead contact upon request.
This paper does not report original codes.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.