Abstract
The reproducibility crisis in neuroimaging has led to an increased demand for standardized data processing workflows. Within the ENIGMA consortium, we developed HALFpipe (Harmonized Analysis of Functional MRI pipeline), an open‐source, containerized, user‐friendly tool that facilitates reproducible analysis of task‐based and resting‐state fMRI data through uniform application of preprocessing, quality assessment, single‐subject feature extraction, and group‐level statistics. It provides state‐of‐the‐art preprocessing using fMRIPrep without the requirement for input data in Brain Imaging Data Structure (BIDS) format. HALFpipe extends the functionality of fMRIPrep with additional preprocessing steps, which include spatial smoothing, grand mean scaling, temporal filtering, and confound regression. HALFpipe generates an interactive quality assessment (QA) webpage to rate the quality of key preprocessing outputs and raw data in general. HALFpipe features myriad post‐processing functions at the individual subject level, including calculation of task‐based activation, seed‐based connectivity, network‐template (or dual) regression, atlas‐based functional connectivity matrices, regional homogeneity (ReHo), and fractional amplitude of low‐frequency fluctuations (fALFF), offering support to evaluate a combinatorial number of features or preprocessing settings in one run. Finally, flexible factorial models can be defined for mixed‐effects regression analysis at the group level, including multiple comparison correction. Here, we introduce the theoretical framework in which HALFpipe was developed, and present an overview of the main functions of the pipeline. HALFpipe offers the scientific community a major advance toward addressing the reproducibility crisis in neuroimaging, providing a workflow that encompasses preprocessing, post‐processing, and QA of fMRI data, while broadening core principles of data analysis for producing reproducible results. Instructions and code can be found at https://github.com/HALFpipe/HALFpipe.
Keywords: fMRI, harmonization, image analysis, reproducibility, meta‐analysis pipeline, open source
HALFpipe is a user‐friendly software that facilitates reproducible analysis of fMRI data, including preprocessing, single‐subject, and group analysis. It provides state‐of‐the‐art preprocessing using fMRIPrep, but removes the necessity to convert data to the BIDS format. Common resting‐state and task‐based fMRI features can then be calculated on the fly using FSL and Nipype for statistics.
1. INTRODUCTION
The application of neuroimaging, in particular functional MRI (fMRI), has led to an explosion in knowledge about brain functions implicated in a range of human behaviors, cognitive processes, and emotions. Such research has been spurred by rapid advances in computationally intensive software required to perform complex algorithmic processing and statistical modeling of fMRI data. The resulting proliferation of software tools designed to fulfill various analytic functions has produced a large array of options for carrying out any given type of processing. Since the fMRI signal indirectly captures the neural processes of interest, a series of computational operations on fMRI data, referred to as the analysis pipeline, are necessary to arrive at interpretable results. In practice, each step is flexible and subject to a number of choices by the researcher, termed analytic flexibility (Poldrack et al., 2017). The steps in the analysis pipeline may be reordered, run with different parameters, or may be completely omitted in some cases. Understandably, users expect different tools performing the same function to generate (near) identical results when supplied with given input data. However, the multiplicity of tools has had the unintended consequence of generating inconsistent results from studies designed to answer the same research question, sometimes even when the same data is used as the starting point (Botvinik‐Nezer et al., 2020). Thus, analytic flexibility combined with the number of analysis steps, as well as the possible parameters for running each analysis step, has led to a vast multiplicity of methodologic variants and an equal number of possible results. This situation has contributed in part to what is widely hailed as a crisis of reproducibility, which now plagues the neuroimaging field (Gorgolewski et al., 2016; Poldrack et al., 2017).
One solution to improving reproducibility is to constrain the parameter space by limiting choices to default parameters established from empirically‐derived best practices (Grüning et al., 2018). For instance, established pipelines such as fMRIPrep (Esteban et al., 2019) and C‐PAC (Craddock et al., 2013) have automated many of these choices. An alternate approach is to run multiple analyses separately on the same input data with the same or different pipelines, but with different parameter selections for each analysis, and then compare the results. This second approach, termed multiverse analysis (Steegen, Tuerlinckx, Gelman, & Vanpaemel, 2016), has the advantage that results from multiple analyses may be compared and alternate solutions may be presented in published reports to promote increased transparency. However, multiverse analysis has the disadvantage that it may ultimately not be possible to determine the optimal or even the correct solution, as true effects in nonsimulated fMRI data are often unknown.
The reproducibility crisis has led to an increased demand for standardized workflows to conduct both the preprocessing and postprocessing stages of fMRI analysis. The recent introduction and widespread adoption of standardized pipelines for fMRI data preprocessing have provided the research community with much‐needed high‐quality tools that have improved reproducibility (Thompson et al., 2020). The four ingredients that are essential to data analysis and reproducible results are: (a) data and metadata availability, (b) code usage and transparency, (c) software installability, and (d) re‐creation of the runtime environment. Relative to other processing pipelines, fMRIPrep (Esteban et al., 2019) has grown in popularity due to its adoption of best practices, open‐source availability, favorable user experience, and glass‐box principles of transparency (Poldrack, Gorgolewski, & Varoquaux, 2019). However, fMRIPrep is limited to the minimal preprocessing steps of fMRI data analysis, while variability in parameter selection for further preprocessing (e.g., data cleaning) and subsequent postprocessing analysis steps (e.g., feature extraction, model specification) may compromise reproducibility.
The Enhancing neuro imaging genetics through meta analysis (ENIGMA) consortium has addressed the reproducibility crisis by pooling observational study data from structural and diffusion imaging (and more recently EEG and MEG), and by developing standardized pipelines, data harmonization methodology, and quality control protocols (Thompson et al., 2020). These workflows have successfully analyzed structural and diffusion MRI data aggregated from large numbers of small‐ and medium‐sized cohorts to accrue sufficient power to yield robust results on a wide range of neuropsychiatric conditions (e.g., Hoogman et al., 2020; Schmaal et al., 2020; van den Heuvel et al., 2020), However, until now the ENIGMA consortium has lacked the ability to reliably conduct consortium‐wide analyses on fMRI data. More recently, however, the ENIGMA task‐based (Veer, Waller, Lett, Erk, & Walter, 2019) and resting‐state fMRI (Adhikari et al., 2019) working groups have spurred initiatives to bring the ENIGMA framework to the functional domain.
To support these initiatives within ENIGMA, we developed a standardized workflow that encompasses the essential elements of task‐based and resting‐state fMRI analyses from raw data to group‐level statistics, builds on the progress and contributions of fMRIPrep developers, and extends its functionality beyond preprocessing steps to include additional preprocessing, post‐processing, and interactive tools for quality assessment. These extended features include: automatic and reliable conversion of fMRI data to BIDS format, spatial smoothing, temporal filtering, extended confounds regression, calculation of task‐based activations, and resting‐state feature extraction, including seed‐based functional connectivity, network‐template (dual) regression, atlas‐based functional connectivity matrices, regional homogeneity (ReHo) analysis, and fractional amplitude of low‐frequency fluctuations (fALFF). Although each of these post‐processing functions is available in other software packages and a few pipelines have incorporated a subset of these features, HALFpipe combines all these post‐processing tools from open‐source neuroimaging packages with the preprocessing steps performed by fMRIPrep (see Table 1). Furthermore, although HALFpipe provides recommended settings for each of the processing steps (see Table 3), it allows users to run any combinatorial number of these processing settings, thereby offering a streamlined infrastructure for pursuing multiverse analyses. Similar to other processing pipelines, HALFpipe is available as a containerized image, thereby offering full control over the computational environment. In this article, we provide a detailed description of HALFpipe. First, we explain the software architecture and implementation, followed by a walkthrough of the procedure for running the software, and finally a discussion of the potential applications of the pipeline.
TABLE 1.
Comparison to other neuroimaging pipelines
| HALFpipe | C‐PAC | fMRIPrep MRIQC FitLins | Conn toolbox | XCP toolbox | DPARSF DPABI | ||
|---|---|---|---|---|---|---|---|
| Quality assessment | Quality metrics | Yes | Yes | Yes | Yes | Yes | Yes |
| Visual quality assessment | Yes | Yes | Yes | Yes | Yes | Yes | |
| Features | Task‐based activation | Yes | No | Yes | No a | Yes | No |
| Seed‐based connectivity | Yes | Yes | No | Yes | Yes | Yes | |
| Dual regression | Yes | Yes | No | Yes | No | Yes | |
| Atlas‐based connectivity matrix | Yes | Yes | No | Yes | Yes | Yes | |
| ReHo | Yes | Yes | No | Yes b | Yes | Yes | |
| fALFF | Yes | Yes | No | Yes | Yes | Yes | |
| Group statistics | Yes | Yes | Yes | Yes | No | Yes |
Note: HALFpipe supports a number of different features that are also available in other pipelines such as the configurable pipeline for the analysis of connectome C‐PAC (Craddock et al., 2013), the Conn toolbox (Whitfield‐Gabrieli and Nieto‐Castanon 2012), the eXtensible Connectivity Pipeline XCP (Ciric et al. 2018) and the data processing and analysis of brain images toolbox DPABI (Yan et al. 2016). fMRIPrep (Esteban, Markiewicz, et al. 2019) in combination with Magnetic Resonance Imaging Quality Control tool (MRIQC) (Esteban et al., 2017) and FitLins (Markiewicz et al. 2016) allows users to construct an analysis pipeline fully within the Nipype ecosystem (Esteban, Ciric, et al. 2020).
Task‐based connectivity is supported.
As implemented with LCOR (local correlation).
TABLE 3.
Default values for preprocessing settings per feature
| Feature | ||||||||
|---|---|---|---|---|---|---|---|---|
| Preprocessed image | Task‐based activation | Seed‐based connectivity | Dual regression | Atlas‐based connectivity matrix | ReHo | fALFF | ||
| Preprocessing step | Spatial smoothing | 6 mm | 6 mm a | |||||
| Grand mean scaling | 10,000 | |||||||
| ICA‐AROMA | Yes | |||||||
| Temporal filter | Gaussian (128 s FWHM) | Frequency‐based (0.01–0.1 Hz) | ||||||
| Confound removal | None | None | ||||||
| Add confounds to model | None | |||||||
Note: Cells filled in gray indicate that this option cannot be selected in the user interface, all other settings can be adjusted by the user.
Done on the statistical maps after feature extraction.
2. METHODS
The HALFpipe software is containerized, similar to fMRIPrep or C‐PAC. This means that it comes bundled with all other software that is needed for it to run, such as fMRIPrep (Esteban, Markiewicz, et al., 2019), MRIQC (Esteban et al., 2017), FSL (Jenkinson, Beckmann, Behrens, Woolrich, & Smith, 2012), ANTs (Avants et al., 2011), FreeSurfer (Fischl & Dale, 2000), and AFNI (Cox, 1996; Cox & Hyde, 1997). As such, all users of one version of HALFpipe will be using identical versions of these tools, because they are packaged with the container. Thus, the containerization of HALFpipe software aids reproducibility across different researchers and computing environments.
We have provided the HALFpipe application in a Singularity container and a Docker container. Singularity or Docker, which are both freely available, must be installed prior to downloading the containerized HALFpipe application. Both Docker and Singularity perform so‐called operating‐system‐level virtualization, but are more efficient and require less resources than virtual machines. Running Docker containers on a Linux or macOS operating system usually requires administrator privileges. Singularity is typically run on a Linux operating system, and may be used without administrator privileges. Docker can be run on the Windows operating system, but compatibility issues may occur with respect to file systems.
Our HALFpipe development team adopted other software engineering best practices, which promoted faster development, and reduced code errors. These industry best practices, which have found their way into research applications (Das, 2018), involve writing code that is easy to read (albeit generally harder to write), the breakdown of complex systems into several simpler subsystems, dedicated effort toward thoughtful code design before implementation, and performing continuous integration via unit tests (Beck, 2000).
2.1. Ecosystem
HALFpipe has been developed as an open‐source project and is accepting contributions that offer new features, enhance functionality, or improve efficiency. All changes are tracked using the Git version control system, which is the de‐facto standard in the open‐source community. In addition, before inclusion in the source tree, changes will be reviewed and then undergo automated testing, which includes unit tests and also running an entire analysis for one subject of the OpenNeuro dataset ds000108 (Wager, Davidson, Hughes, Lindquist, & Ochsner, 2008). This way unexpected side effects and bugs will be caught and corrected before causing problems for users.
HALFpipe releases are made using semantic versioning as adapted for compatibility with Python (Coghlan & Stufft, 2013). This means that there will be feature releases that add new functionality and patch releases that make minor adjustments to solve specific issues or bugs. The development team takes great care that new patch releases of dependencies such as fMRIPrep are regularly incorporated into HALFpipe so that bug fixes are made available to users.
HALFpipe currently depends on the long‐term support release 20.2.x of fMRIPrep. For future releases containing new features, the developers will approve possible upgrades that are advantageous to ENIGMA consortium projects. We may also consider replacing tools used for specific processing steps or upgrading the standard brain template in future releases based on such considerations, which will be explained in the change log of each release.
2.2. Databases
To automatically construct a neuroimaging data processing workflow, the program needs to be able to fulfill queries such as “retrieve the structural image for subject x.” Many programs implement such queries using a database system. The queries also need to flexibly interface with the logic of neuroimaging and processing pipelines, which is relevant in the context of missing scans.
In the context of missing scans, HALFpipe always tries to execute the best possible processing pipeline based on the data that is available. For example, a field map may have been routinely acquired before each functional scan in a particular dataset. If one of these field maps is missing, HALFpipe flexibly assigns another field map, for example, one belonging to the preceding functional scan. However, HALFpipe will not use a field map from another scan session, as field inhomogeneities are likely to have changed. Finally, HALFpipe does not fail if a field map is missing, but simply omits the distortion correction step for that subject. Other examples include the ability of HALFpipe to match structural to functional images, and match task events to a functional scan. This strategy is used throughout the construction of processing workflows.
2.3. Metadata
Processing of neuroimaging data requires access to relevant metadata, such as temporal resolution, spatial resolution, and many others. Some elements of metadata, such as echo time (TE), are represented differently depending on scanner manufacturer and DICOM conversion software. The method for reading various types of data has been harmonized in HALFpipe using the following three methods.
First, metadata can be stored in BIDS format. This means that a JavaScript Object Notation (JSON) file accompanies each image file, which contains the necessary metadata. BIDS calls this file the sidecar, and common tools such as heudiconv (Halchenko et al., 2018) or dcm2niix (Li, Morgan, Ashburner, Smith, & Rorden, 2016) generate these files automatically. If these files are present, HALFpipe will detect and use them. Second, instead of sidecar files, some software tools store image metadata in the NIfTI header. The NIfTI format defines fields that can fit metadata, but depending on how the image file was created, this metadata may be missing. Some conversion programs also place the metadata in the description field in free‐text format. This description can also be parsed and read automatically. Third, information may be incorrectly represented due to user error, incompatible units of measurement, or archaic technical considerations. In such cases, HALFpipe provides a mechanism to override the incorrect values. For every metadata field, the user interface will prompt the user to confirm that metadata values have been read or inferred correctly. The user can choose to manually enter the correct values.
2.4. Interfaces
HALFpipe consists of different modules that need to pass data between each other, such as file pathnames and the results of quality assessment procedures. Developing an application as large and complex as HALFpipe requires establishing predictable interfaces, which prescribe data formats for communication within the application. An advantage of this approach is that knowledgeable users can write their own code to interface with HALFpipe.
HALFpipe uses the Python module marshmallow to implement interfaces, called schemas in the module's nomenclature. All schemas are defined in the halfpipe.schema module. When the user first starts the application, the user interface is displayed by HALFpipe. It asks the user a series of questions about the data set and the analysis plan, and stores the inputs in a configuration file called spec.json. The configuration file has predictable syntax and can be easily scripted or modified, which enables collaborative studies to harmonize analysis plans.
2.5. Workflow engine
To obtain reproducible results, a core requirement for HALFpipe was reproducible execution of the processing pipeline. As the ENIGMA consortium requires fMRI analysis of large datasets with several thousand samples, HALFpipe was designed to parallelize processing on multiple computers or processor cores. Both of these specifications were achieved by implementation in Nipype, NeuroImaging in Python: Pipelines, and Interfaces (Gorgolewski et al., 2011). Nipype is a workflow engine for neuroimaging that constructs an acyclic directed graph, in which nodes represent processing commands that need to be executed (the steps of the pipeline), while the edges represent inputs and outputs being passed between nodes (images or text files). In this formalization of a neuroimaging pipeline as a graph, the fastest order for execution across multiple processor cores can be determined.
The workflow graphs are modular and scalable, which means they can be nested and extended. HALFpipe uses the workflows defined by fMRIPrep and then connects these outputs to additional workflows. fMRIPrep itself is modular and divided into multiple workflows: sMRIPrep (Esteban, Markiewicz, Blair, Poldrack, & Gorgolewski, 2021), SDCFlows (Esteban, Markiewicz, Blair, Poldrack, & Gorgolewski, 2020), NiWorkflows (Esteban, Markiewicz, Esteban et al., 2021), and NiTransforms (Goncalves et al., 2021). The workflow graph facilitates saving and verifying intermediate results, and supports the user's ability to stop and later restart processing. HALFpipe also uses the graphs to determine which intermediate results files are not needed by subsequent commands by using a tracing garbage collection algorithm (Dijkstra, Lamport, Martin, Scholten, & Steffens, 1978). As such, intermediate files do not accumulate on the storage device. This feature is implemented as a plugin to Nipype.
Nipype forms the basis of fMRIPrep and C‐PAC, which are widely used in the neuroimaging community. However, it has several limitations that are relevant in the context of HALFpipe. HALFpipe is able to calculate features and statistical maps with different variations of preprocessing settings. To do this efficiently, intermediate results need to be re‐used whenever possible. An improved second version of Nipype is currently being developed, called Pydra (Jarecka et al., 2020), which will be able to automatically detect repetitive processing commands, and automatically re‐use outputs. Presently, until Pydra becomes available, HALFpipe calculates a four‐letter hash code that uniquely identifies each pre‐processing step. Before constructing a new pre‐processing command, HALFpipe checks whether its hash has already been added to the graph. If present, the existing command is re‐used, significantly reducing processing times in the context of multiverse analysis or pipeline comparison.
To illustrate the benefits of this approach participant 01 of a face matching task data set (Wakeman & Henson, 2015) was entered into HALFpipe and task contrasts were calculated with three pipelines. For the first pipeline, we used the recommended settings from Table 3. For the second and third pipelines, we used the same settings but disabled ICA‐AROMA. For the second pipeline we additionally added the motion parameters to the task model as confound time series. The third pipeline did not include any denoising or confound time series removal. The naive approach is to run HALFpipe three times, once for each pipeline. This approach is sub‐optimal, as many duplicate computations are performed. By default, HALFpipe combines all three pipelines using the hashing approach described above, making processing much faster. Processing was performed on an AMD Ryzen Threadripper 2950X 16‐core processor, and each run of HALFpipe was configured to use all cores. Table 2 shows the processing time (wall clock time) spent on each pipeline. For the naive approach, we also show the total time.
TABLE 2.
Efficient pipeline construction speeds up multiverse analyses
| Naive approach | HALFpipe approach (via hashing algorithm) | |
|---|---|---|
| Processing time (hh:mm) | 01:39 + 01:36 + 01:33 = 04:49 | 01:43 |
Another key requirement of HALFpipe was robust and flexible handling of missing data. For instance, a missing functional scan or statistical map does not cause HALFpipe to fail. In addition, HALFpipe defines inclusion and exclusion criteria for scans, such as the maximum allowed motion (mean framewise displacement) or a minimum brain coverage when extracting a brain region's average signal. Finally, depending on the data set, statistical maps may need to be aggregated across runs or sessions within single subjects before a group‐level model can be run. This means that the static graph has to be modified dynamically to adapt to the results of processing. HALFpipe solves this problem by defining a data structure that not only contains the file names of statistical maps, but also the tags and metadata that can be used to adjust processing on the fly. For example, using this data structure, design matrices can be constructed for group models based on the actual subjects that have statistical maps available.
2.6. Preprocessing
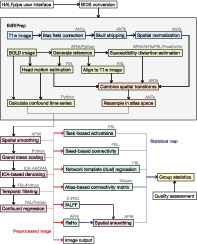
Main preprocessing is done in HALFpipe with fMRIPrep, which performs a consensus of preprocessing steps required for any fMRI study (Esteban, Markiewicz, et al., 2019). Consensus steps for structural images include skull stripping, tissue segmentation, and spatial normalization. Consensus steps for functional images include motion correction, slice time correction, susceptibility distortion correction, coregistration, and spatial normalization (Figure 1).
FIGURE 1.
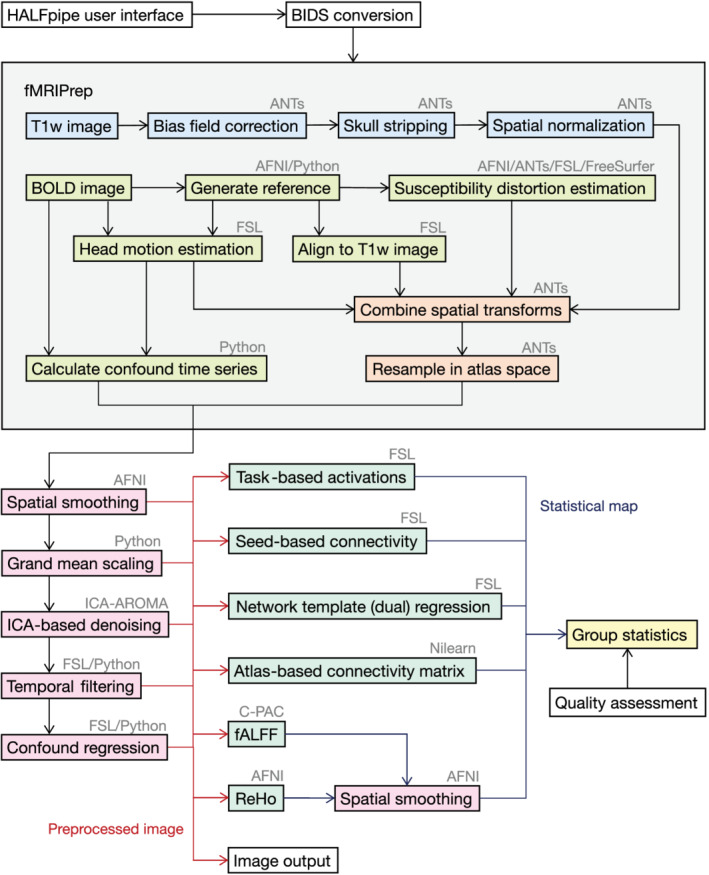
HALFpipe workflow. HALFpipe is configured in a user interface where the user is asked a series of questions about their data and the processing steps to perform. Data are then converted to BIDS format (Gorgolewski et al., 2016) to allow standardized processing (white). After minimal preprocessing of the structural (blue) and functional (green and orange) data with fMRIPrep (Esteban, Blair, et al., 2019), additional preprocessing steps can be selected (red). Using the preprocessed data, statistical maps can be calculated during feature extraction (turquoise). Finally, group statistics can be performed (yellow). Note that not all preprocessing steps are available for each feature, as is outlined in Table 3. The diagram omits this information to increase visual clarity
HALFpipe defines standard space as the MNI152NLin2009cAsym template, which is the most current and detailed template available (Horn, 2016). Note that the standard space template is not user‐configurable, so that any outputs generated by one version of HALFpipe can be easily compared to outputs generated by another version of HALFpipe.
Once the fMRI data have been processed with fMRIPrep and resampled into standard space, HALFpipe implements a number of additional preprocessing steps for denoising, filtering, and harmonizing the functional data (see also Figure 1):
-
ICA‐AROMA is an algorithm based on independent component analysis. It classifies components into those that contain signal and those that are noise (Pruim et al., 2015). To accomplish this, ICA‐AROMA relies on reference templates defined in MNI152NLin6Asym space, which is different from the standard space template MNI152NLin2009cAsym that is used by fMRIPrep. To allow ICA‐AROMA to run, it is thus necessary to provide the preprocessed image not just in the default template space, but also in the one required by ICA‐AROMA. By default, fMRIPrep will estimate a second normalization to this other template, apply it to the fMRI image in native space, and run ICA‐AROMA on the resulting image (Ciric et al., 2021). This approach effectively doubles the processor time spent on spatial normalization, and may require manually checking both spatial registrations.
To avoid this considerable effort, HALFpipe implements a different approach by using an existing warp between the two standard template spaces (Horn, 2016). This predefined warp is concatenated with the normalization that was already estimated by fMRIPrep, and then a second round of resampling is performed with fMRIPrep's bold_std_trans_wf. This way, only the resampling step needs to be run twice.
Finally, Independent component analysis‐based automatic removal Of motion artifacts (ICA‐AROMA) is run on the resulting fMRI image in MNI152NLin6Asym space using fMRIPrep's ica_aroma_wf workflow, which also includes spatial smoothing fixed to a 6 mm FWHM smoothing kernel. The resulting classifications are kept for step 4.
HALFpipeimplements spatial smoothing using AFNI's 3dBlurTo Full Width at Half Maximum (FWHM) (Friedman, Glover, Krenz, & Magnotta, 2006). Each voxel's signal is averaged with the signal of surrounding voxels weighted by an isotropic Gaussian kernel. At the edges of the brain, this kernel may include nonbrain voxels, so smoothing is constrained by the brain mask. This is equivalent to the procedure in the Minimal Preprocessing Pipelines for the Human Connectome Project (Glasser et al., 2013). In addition, 3dBlurToFWHM estimates the smoothness of the resulting image, and iteratively decreases the amount of smoothing so that the resulting smoothness matches the user setting. This way, differences in the intrinsic smoothness between datasets (e.g., due to different voxel sizes) can be harmonized.
Grand mean scaling sets the image mean, defined as the within‐scan mean across all voxels and time points, to a predefined value. The grand mean is closely related to scanner parameters such as RF power or amplifier gain but not to neural mechanisms (Gavrilescu et al., 2002). Adjusting the grand mean via scaling makes analysis results more interpretable and comparable across subjects, sessions, and sites. The scaling factor is calculated based on the masked functional image, and applied to both the fMRI data and the confound time series extracted by fMRIPrep.
If selected, the previously estimated ICA‐AROMA noise components are removed from the smoothed and grand‐mean‐scaled fMRI data. This is performed in a nonaggressive way to minimize removing variance that is shared between signal and noise components. ICA‐AROMA implements this step using the FSL command fsl_regfilt, which calculates an ordinary least squares regression for each voxel, where the design matrix includes both the signal and the noise components as regressors. This means that the resulting regression weights reflect the unique variance of the noise components (and not the shared variance with signal components). Then, the noise component regressors are multiplied by their regression weights and these products are added together to yield one time series of the noise. Subtracting the noise from the voxel time series yields a denoised time series (the regression residuals). This step is done using a custom re‐implementation of fsl_regfilt in HALFpipe using Numpy (Harris et al., 2020).
Temporal filtering can be selected to remove low‐frequency drift via a high‐pass filter, high‐frequency noise via a low‐pass filter, or both at the same time using a band‐pass filter. HALFpipe implements two approaches to temporal filtering, a frequency‐based approach (Jo et al., 2013) and a Gaussian‐weighted approach (Marchini & Ripley, 2000). The frequency‐based temporal filter is very exact in selecting frequencies to be kept or removed, and is commonly used to calculate fractional amplitude of LFF (fALFF) and regional homogeneity (ReHo). The Gaussian‐weighted temporal filter is the default used by FSL Feat (Jenkinson et al., 2012) and may have fewer edge effects at the start and end of the time series. However, its spectrum also has a more gradual roll‐off, meaning that it will be less aggressive in removing frequencies close to the chosen cutoff value.
Importantly, HALFpipe runs fMRIPrep with small modifications. For instance, we disabled fMRIPrep's experimental susceptibility distortion correction in the absence of field maps, because it is not yet validated. Furthermore, HALFpipe suggests default settings for each preprocessing step, which are outlined in Table 3. Note that some are selected based on best‐practices in the field (i.e., band‐pass temporal filter for ALFF and ReHo), whereas most default settings can be adjusted by the user. Last, HALFpipe does not output preprocessed and normalized functional images by default, because they use a lot of disk space. However, in the user interface users can manually choose to output a preprocessed functional image with their choice of preprocessing settings.
2.7. Confound time series removal
fMRIPrep not only outputs a preprocessed image in standard space but also a spreadsheet with confound time series named confounds.tsv. These include the (derivatives of) motion parameters (squared), aCompCor components (Behzadi, Restom, Liau, & Liu, 2007), white matter signal, CSF signal, and global signal. A key consideration needs to be made when using fMRIPrep confound time series in conjunction with the preprocessing steps outlined in the previous section: Using confound time series as nuisance regressors with data that was temporally filtered or denoised can re‐introduce the removed temporal or noise signals back into the voxel time series (Hallquist, Hwang, & Luna, 2013).
An example of this phenomenon may be regressing out a set of fMRIPrep confound time series after removing low‐frequency drift via temporal filtering. In practice, this means setting up a regression model for each voxel, where the voxel time series is the dependent variable and the regressors are the confound time series. The regression will yield a weight for each confound time series, so that the total model explains the maximum amount of variance (under assumption of normality). After multiplying the confound time series with these weights, their products are summed to one time series containing the confound‐related signal in that voxel. This time series is then subtracted from the original voxel time series to get the result (i.e., the regression residuals). However, if the confound time series happen to contain any low‐frequency drift, then their weighted sum likely will as well. It follows that subtracting a time series with temporal drift from the temporally filtered voxel data will re‐introduce temporal drift, independent of whether a temporal filter was applied before.
In HALFpipe, any (optional) denoising or filter applied to the voxel time series is also applied to the fMRIPrep confound time series. This way, previously removed variance is not re‐introduced accidentally, because it has been removed from both sides of the regression equation. For example, when the user chooses to perform ICA‐AROMA denoising, then the same denoising will be applied to the time fMRIPrep confound time series, and the same applies when using a temporal filter. Note that this means that the confound time series generated by HALFpipe will be different from the original fMRIPrep confound time series, and users should take care to use the appropriate file when running custom analyses.
2.8. Quality assessment
Assessing the quality of data and preprocessing is a laborious undertaking and often performed manually. Efforts to automate this process, either through predefined thresholds of image quality features (Alfaro‐Almagro et al., 2018) or machine learning (Esteban et al., 2017) are not yet ready to replace the eyes of a trained researcher checking the data. However, various approaches make this process easier. First, rather than viewing three‐dimensional neuroimaging files directly, generating and viewing reports containing two‐dimensional images offers a significant time savings. Second, tools such as slicesdir (in FSL), fMRIPrep, and MRIQC generate HTML files that contain multiple report images and can be explored in a web browser. MRIQC also provides an interactive widget to rate the quality of each image (Esteban, Blair, et al., 2019).
In HALFpipe, we use a fixed set of processing steps for quality assessment. While slicesdir allows the researcher to easily compare the same image type across different subjects, it cannot be used to generate reports for all types of images. By contrast, fMRIPrep/MRIQC HTML files have a broad range of information and quality report images included, but one HTML file is always specific to one subject. As such, examining multiple processing steps in many subjects can be cumbersome.
To overcome these issues, HALFpipe provides an interactive web app that is contained in a single HTML file. The app dynamically loads reports with images, and can handle datasets up to thousands of images without a performance penalty. The images can be sorted both by subject, as is done by fMRIPrep/MRIQC, or by image type, as is performed in slicesdir. Each image can be rated as either good, uncertain, or bad. Predefined logic automatically converts these ratings into inclusion/exclusion decisions for HALFpipe's group statistics. In addition, tagging images as uncertain enables users to efficiently retrieve and discuss these with a colleague or collaborator, after which a definitive decision on image quality can be made.
2.9. Group statistics
HALFpipe uses FSL FMRIB Local Analysis of Mixed Effects (FLAME, Woolrich, Behrens, Beckmann, Jenkinson, & Smith, 2004) for group statistics, because FLAME considers the within‐subject variance of lower level estimates in its mixed‐effects models. In addition, its estimates are conservative, which means they offer robust control of the false positive rate (Eklund, Nichols, & Knutsson, 2016).
A common issue in fMRI studies is that the spatial extent of brain coverage may differ between subjects. A common choice is to restrict higher‐level statistics to only those voxels that were acquired in every subject. However, with a large variation in brain coverage, which is to be expected when pooling multi‐cohort data, sizable portions of the brain may ultimately be excluded from analysis. To circumvent this issue, HALFpipe uses a re‐implementation of FLAME in Numpy (Harris et al., 2020). In this implementation, a unique design matrix is generated for every voxel so that only subjects who have a measurable value for a given voxel are included. Then, the model is estimated using the FLAME algorithm. This list‐wise deletion approach depends on the assumption that voxels are missing completely at random (MCAR), meaning that the regressors (and thus statistical values) are independent of scanner coverage.
For group models, users can specify flexible factorial models that include covariates and group comparisons. By default, missing values for these variables are handled by list‐wise deletion as well, but the user may alternatively choose to replace missing values by zero in the demeaned design matrix. The latter approach is equivalent to imputation by the sample mean. Design matrices for the flexible factorial models are generated using the Python module Patsy (Smith et al., 2018). Contrasts between groups are specified using the lsmeans procedure (Lenth, 2016).
2.10. Running on a high‐performance cluster
Deploying Nipype to perform computations on multiple nodes, such as on a high‐performance cluster (HPC) is particularly challenging. By default, Nipype submits a separate job to the cluster queue for each processing command (graph node) regardless of the amount of time required to execute the command. A watcher process running on the head node collects outputs from completed commands and submits the next processing command. This process can be inefficient on some HPCs because computational resources need to be allocated and deallocated continually. We implemented a more efficient approach for HALFpipe that partitions the processing graph into many independent subgraphs, which the user may submit as separate jobs. The smallest granularity available is one subgraph per subject that is invoked automatically with the command line flag—use‐cluster. A Nipype workflow is created and validated for all subjects before the pipeline starts running. In a cluster setting, the most efficient resource utilization is to submit each subject as a separate job and to run each job on two CPU cores.
2.11. Procedure
HALFpipe starts up as a terminal‐based user interface that prompts the user with a series of questions about the dataset being analyzed and the desired analysis plan. The main stages of HALFpipe analysis, which are detailed below, include loading data, preprocessing with fMRIPrep, quality assessment, feature extraction, and group‐level statistics. Users have the flexibility to specify the settings for each processing stage at one time or separately at each stage. If HALFpipe is stopped and resumed at an intermediate stage, HALFpipe will detect which stages have been completed and ask the user to indicate further analyses that are desired. For instance, the user can request preprocessing and feature extraction, but not group‐level statistics, and later resume processing specifying group‐level statistics only.
2.12. Loading data
A major advantage of HALFpipe is that it accepts input data organized in various formats without the need for file naming conventions or a specific directory structure. Using the terminal interface, the user is asked to provide the location of the T1‐weighted and fMRI BOLD image files, which are required for preprocessing, as well as field maps and task event files if available or applicable. However, HALFpipe requires additional information linking the image files to run in an automated fashion, such as information specifying which set of images belong to the same subject.
Through the use of path templates, HALFpipe can handle a wide range of folder structures and data layouts. The syntax for path templates is adapted from C‐PAC's data configuration (Giavasis, Pellman, Clucas, & Lurie, 2020). Instead of manually adding each input file for each subject separately, as is performed in the SPM or FSL user interfaces, the template describes the pattern used for naming files. That pattern can match many file names, thereby reducing the amount of manual work for the user. For example, when placing the tag {subject} in the file path {subject}_t1.nii.gz, all files of which the name ends in _t1 and have the extension .nii.gz will be selected. The part of the filename that comes before _t1 is now interpreted by the parsing algorithm as the subject identifier. When multiple files from different modalities have the same subject identifier, or session number, and so on, they will be matched automatically by these tags. Automated processing workflows can then be constructed around the resulting data structure.
In contrast to C‐PAC's data configuration syntax, HALFpipe path templates use BIDS tags (Gorgolewski et al., 2016). HALFpipe path templates can be further specified by adding a colon and a regular expression after the tag name (as in standard Python regular expression syntax). For example, {subject:[0–9]} will only match subject identifiers that contain only digits. This can be useful for more complex data layouts, such as when multiple datasets are placed in the same directory, and only a single subset is to be used. For more examples, see Table 4.
TABLE 4.
Examples of path template syntax
| Example 1 | Example 2 | |
|---|---|---|
| Path template | data/{subject}/bold_rest.nii.gz | data/subject{subject}/bold_{task}.Nii.Gz |
| Matches these files |
data/subject01/bold_rest.nii.gz data/subject02/bold_rest.nii.gz data/subject03/bold_rest.nii.gz data/phantom/bold_rest.nii.gz |
data/subject01/bold_rest.nii.gz data/subject02/bold_rest.nii.gz data/subject03/bold_rest.nii.gz data/subject01/bold_task.nii.gz |
| Does not match |
Data/subject01/bold_task.Nii.Gz Data/subfolder/subject01/bold_rest.nii.gz |
data/phantom/bold_rest.nii.gz data/subfolder/subject01/bold_rest.nii.z |
Note: The undelined text shows the part of the file name that is responsible for matching/not matching.
In the HALFpipe user interface, the user receives feedback on how many and which files are matched, so that the path templates can be entered interactively. Importantly, after finishing the configuration process via the user interface, all files are internally converted into the standardized BIDS structure, which is a prerequisite for running fMRIPrep. However, no copies of files are made, the conversion is based entirely on symbolic links (aliases) to the original files. If the data are already in BIDS format, HALFpipe will still carry out this conversion for consistency. The resulting dataset in BIDS format is then stored in the working directory in a subfolder called rawdata.
2.13. Quality assessment
Quality assessment can be performed in an interactive, browser‐based user interface (see Figure 2). HALFpipe provides a detailed user manual for quality assessment that is linked on the web page. The web app shows report images of several preprocessing steps such as T1 skull stripping and normalization, BOLD tSNR, motion confounds, ICA‐based artifact removal, and spatial normalization (see the methods section on Quality assessment). These images can be visually inspected and rated by the viewer as either good, uncertain, or bad.
FIGURE 2.
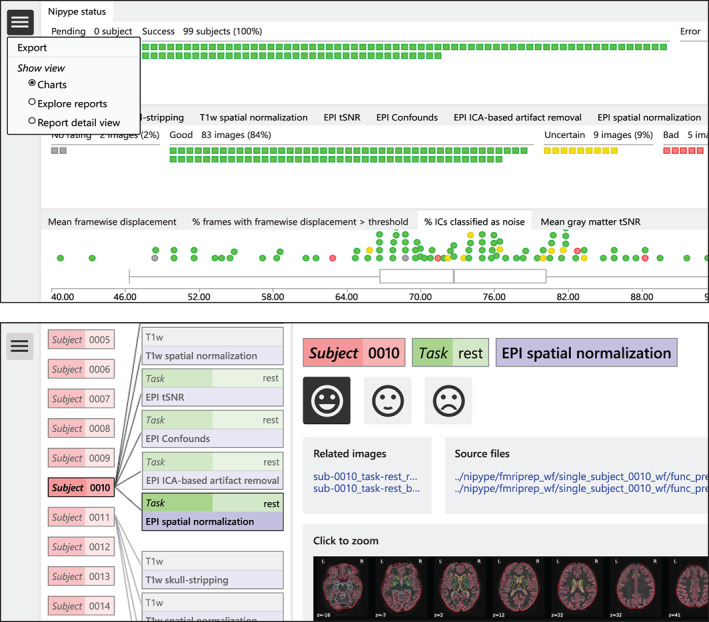
Quality assessment user interface. The top panel shows the charts view, containing one chart for processing status, one for quality ratings and one for image quality metrics. In the top left corner, the navigation menu is open, which shows the option to export ratings for use in group statistics. The bottom panel contains a screenshot of the explorer view that allows the user to navigate across subjects and image types. The explorer view shows the currently selected report image on the right, along with its rating, related images, and the source files that were used to construct it. By clicking on the image, or selecting the report detail view in the navigation menu, the image can be zoomed and panned using the mouse
Ratings will be saved in the local browser storage. Once completed, they can be downloaded in JSON format to be read by HALFpipe. If placed in the working directory, ratings will be automatically detected by HALFpipe and used to exclude subjects for group‐level statistics. In addition, HALFpipe will automatically detect all other JSON files whose names start with exclude, to accommodate quality assessment by multiple researchers. In the case of conflicts between ratings, the lower rating will be used.
HALFpipe will include as much data as possible while excluding all scans rated as “bad.” Ratings of “good” and “uncertain” will be included for group analysis. A “bad” rating for any report image related to structural/anatomical processing will exclude the entire subject. A “bad” rating for any report image related to functional image processing will only exclude the specific functional scan. This means that if a subject has one “bad” scan, its other scans may still be included for group statistics.
In addition, the mean framewise displacement, percentage of frames with a framewise displacement above a specified threshold, percentage of the independent components that were classified as noise, and mean gray matter tSNR from all subjects is displayed in box plots. Next to the report images, links to the source images are shown so that these can be inspected in more detail by opening them in a preferred image viewer (e.g., fsleyes).
Images can be zoomed by clicking them. For faster operation by advanced users, rating, and navigation are accessible not just via user interface buttons, but also via keyboard shortcuts based on the WASD keys. Pressing the A goes back one image and D goes ahead, whereas W, S, and X rate an image as good, uncertain, or bad, respectively. The web app offers an overview chart that indicates subjects preprocessed successfully and subjects with errors, a chart with quality ratings, and box plots reflecting the sample distributions for motion, noise components, and temporal signal‐to‐noise ratio (tSNR). All three are implemented so that users can hover over chart elements with their cursor to view meta‐information, such as the subject identifier, and click to navigate to the associated report images. The HTML file is built as a frameworkless web app using TypeScript. Source code is available at https://github.com/HALFpipe/QualityCheck.
HALFpipe shows two report images for each subject on structural/anatomical processing and four additional images for each type of functional scan. Detailed explanations may be found in the quality assessment manual at https://github.com/HALFpipe/HALFpipe#quality-checks.
T1w skull stripping shows the bias‐field corrected anatomical image overlaid with a red line that outlines the brain mask. The user must check that no brain regions are missing from the mask, and that portions of the skull or head are not included in the mask.
T1w spatial normalization shows the anatomical image resampled to standard space overlaid with a brain atlas in standard space. The user needs to check whether the regions of the atlas closely match the resampled image.
Echo planar imaging (EPI) tSNR shows the temporal signal‐to‐noise ratio of the functional image after preprocessing using fMRIPrep. The user must check that signal recovery is distributed uniformly throughout the brain, and exclude scans with asymmetry, distortions, localized signal drop‐out, or striping artifacts.
EPI Confounds shows the carpet plot (Aquino, Fulcher, Parkes, Sabaroedin, & Fornito, 2020; Power, 2017), generated by fMRIPrep. A carpet plot is a two‐dimensional plot of time series within a scan, with time on the x‐axis and voxels on the y‐axis. Voxels are grouped into cortical gray matter (blue), subcortical gray matter (orange), cerebellum (green), and white matter and cerebrospinal fluid (red). Above the carpet plot are time courses (x‐axis) of the magnitude (y‐axis) of framewise displacement (FD), global signal (GS), global signal in CSF (GSCSF), global signal in white matter (GSWM), and DVARS, which is the temporal change in root‐mean‐square intensity (D being the temporal derivative of time courses and VARS the root‐mean‐square variance over voxels). The user must look for changes in heatmap/intensity in relation to motion and signal changes above. Abrupt changes in the carpet plot may correspond to motion spikes, whereas extended signal changes may indicate acquisition artifacts caused by defective scanner hardware.
EPI ICA‐based artifact removal shows the time course of the mean signal extracted from each ICA‐component and its classification as either signal (green) or noise (red). This figure is generated by fMRIPrep. For each component, there is a spatial map (left), the time series (top right) and the power spectrum (bottom right). The user must check that components classified as noise do not contain brain networks or temporal patterns that are known to be signal.
EPI spatial normalization shows the functional image after preprocessing using fMRIPrep overlaid with a brain atlas in standard space. As before, the user must check whether the regions of the atlas closely match the resampled image.
2.14. Feature extraction
Following preprocessing, HALFpipe can extract several features that are commonly used in resting‐state and task‐based analysis. These include various ways of examining functional connectivity between brain regions (seed‐based connectivity, network‐template (or dual) regression, atlas‐based connectivity matrices), as well as measures of local activity (ReHo, fALFF). HALFpipe allows the user to choose several region‐of‐interest masks (seeds), template networks, and atlases, for which a threshold indicates the minimum overlap the user requires between seeds, template networks, or atlas regions and the subjects' fMRI data. For each feature, the user can change the default settings for spatial smoothing and temporal filtering, and choose the confounds to be removed. The user is offered the option to extract the same feature multiple times, each time varying the preprocessing, confound, and denoising settings to explore the impact of analytical decisions in a multiverse analysis. Of note, for selected features, some options are not available. For example, spatial smoothing is disabled for atlas‐based connectivity matrices (Alakörkkö, Saarimäki, Glerean, Saramäki, & Korhonen, 2017), or performed after ReHo and fALFF have been calculated (see Table 3).
A brief description of the features is provided in Box 1.
Box 1. Overview of HALFpipe features.
Task‐based activations A first‐level general linear model (GLM) is run for event‐related or block designs. GLM regressors describing the stimulus presentations for each of the task conditions are convolved with a double Gamma HRF and the overall model is fit for each voxel in the brain using FSL FILM (Woolrich, Ripley, Brady, & Smith, 2001). Contrasts of interest are tested, which results in a whole‐brain task activation map for comparisons between task conditions.
Seed‐based connectivity Average BOLD time series are extracted from a seed region of interest (ROI), which is defined by a binary mask image. This time series is used as a regressor in a first‐level GLM, where the model is fit for each voxel in the brain using fsl_glm. This results in a whole‐brain functional connectivity map that represents the connectivity strength between the ROI and each voxel in the brain.
Network‐template (or dual) regression Subject‐specific representations of connectivity networks (e.g., default mode, salience, task‐positive networks) are generated using dual regression (Beckmann, Mackay, Filippini, & Smith, 2009) with fsl_glm. In a first regression model, the set of network template maps is regressed against the individual fMRI data, which generates time series for each of the template networks. Next, a second regression model is run, regressing the network time series against the individual fMRI data. This generates subject‐specific spatial representations of each of the template networks, which can be considered to represent the voxelwise connectivity strength within each of the networks.
Atlas‐based connectivity matrix Average time series are extracted from each region of a brain atlas of choice using custom code inspired by Pypes (Savio, Schutte, Graña, & Yakushev, 2017) and Nilearn (Abraham et al., 2014). From these, a pairwise connectivity matrix between atlas regions is calculated using Pearson product‐–moment correlations using Pandas (McKinney, 2010), which represent the pairwise functional connectivity between all pairs of regions included in the atlas.
Regional homogeneity (ReHo) Local similarity (or synchronization) between the time series of a given voxel and its nearest neighboring voxels is calculated using Kendall'’s coefficient of concordance (Zang, Jiang, Lu, He, & Tian, 2004) using FATCAT's 3dReHo, which is distributed with AFNI (Taylor & Saad, 2013).
Fractional amplitude of low frequency fluctuations (fALFF) Variance in amplitude of low frequencies in the BOLD signal is calculated, dividing the power in the low frequency range (0.01–0.1 Hz) by the power in the entire frequency range (Zou et al., 2008) with a customized version of the C‐PAC implementation of fALFF.
2.15. Group‐level statistics
Group‐level statistics on individual features can be performed with FSL's FLAME algorithm. Subjects who had poor quality data in the interactive quality assessment are excluded. In addition, subjects can be excluded based on movement by selecting the maximum allowed mean framewise displacement (FD) and percentage of outlier frames (i.e., frames with motion higher than the specified FD threshold).
For group‐level statistics, users can choose to calculate the intercept only (i.e., mean across all subjects) or run flexible factorial models. For the latter, HALFpipe prompts the user to specify the path to a covariates file (multiple file formats are supported) containing subject IDs, group membership, and other variables, and to specify whether these are continuous or categorical. Missing values in the covariates file can be handled with either listwise deletion or mean substitution. The user can specify main effects and interactions between variables, while within‐group regressions against a continuous variable (e.g., symptom severity) is also possible.
2.16. Outputs
After computation finishes, all outputs are accessible in the working directory. The outputs of fMRIPrep are stored in derivatives/fmriprep folders, similar to when fMRIPrep had been run outside of HALFpipe. The derivatives folder also contains the folder halfpipe which contains any preprocessed images and features that were generated. Just like the fmriprep folder, the halfpipe folder conforms to the BIDS standard for derived datasets (BIDS contributors, 2021). This means that all file names contain structured information such as subject ID sub‐01 or the name given to the feature feature‐seedConn in a standardized way that may be different from the original file naming. Underscores, dashes, and other nonalphanumeric characters are removed from the subject ID, session IDs, and so on. for compliance with BIDS. HALFpipe's standardized output file naming means that additional analysis steps can be automated easily.
The outputs from group statistics are placed in a similar folder structure that make it easy to share summary statistics for collaborative meta‐analysis projects.
3. DISCUSSION
Large samples are essential for recent neuroimaging applications, such as imaging‐genetics association studies, training of complex machine learning models, and even unsupervised learning. This demand has stimulated efforts to pool data from multiple observational studies, which typically incur greater bias than studies designed a priori to address a specific scientific question. Within ENIGMA, we developed HALFpipe to support the harmonization of task‐based and resting‐state fMRI data analysis and quality assessment across multiple labs and cohorts. HALFpipe bundles all software tools, library functions, and other dependencies by containerizing the requisite components in a Singularity (Kurtzer, Sochat, & Bauer, 2017) and Docker (Docker Inc.) release. Containerization ensures that all software dependencies and the runtime environment are provided. Therefore, containerized software such as HALFpipe can run reliably regardless of the computing environment where it is installed, be it a laptop, computational cluster, or cloud computing service (Grüning et al., 2018).
The design, implementation, and testing of the HALFpipe workflow resulted in its 1.0 version release in early 2021. Several thousand resting‐state fMRI datasets from 29 ENIGMA PTSD consortium sites have already been analyzed as part of the first published report to employ HALFpipe (Weis, 2020), while analyses of other large multi‐site datasets are currently underway in several ENIGMA working groups, including the ENIGMA task‐based fMRI working group (Veer et al., 2019). Running HALFpipe requires approximately 8–20 GB of RAM per computer or cluster node and 6–10 hours to complete on a single processor core. The exact resource usage depends on voxel resolution and the number of volumes in the fMRI data. The number of features the user chooses has a negligible impact on processing time.
The HALFpipe user experience includes an interactive user interface to facilitate rapid analysis prototyping while preserving the ability to script automated analyses of large datasets via configuration files in JSON format with detailed prescriptions of the dataset, analyses steps, and input parameters. Importantly, HALFpipe accommodates concurrent harmonized processing of task‐based and resting‐state fMRI data, which facilitates cross‐modal comparisons between the two fMRI modalities (e.g., Kerestes, Chase, Phillips, Ladouceur, & Eickhoff, 2017).
Our implementation of HALFpipe enables users to tackle consortium analyses of multi‐cohort fMRI data with highly uniform application of methods. Specifically, we have established a standardized process and analysis methodology that involves a pre‐specified: (a) ensemble of software tools, (b) software version for each tool, (c) set of user‐defined parameters, (d) sequence of analytic steps, (e) quality assessment process, and (f) criteria for excluding substandard data. Thus, HALFpipe promotes the seamless implementation of a standardized process (preprocessing and feature extraction) at each site and/or cohort prior to initiating group level statistics. Such capabilities hold the promise of significantly advancing basic neuroscience, and particularly clinical neuroscience, by supporting the execution of multi‐site multi‐cohort studies of several hundred or several thousand samples—ultimately supporting harmonized cross‐disorder comparisons. While not part of the HALFpipe workflow, cross‐site/platform harmonization techniques for neuroimaging have recently experienced a dramatic increase (Fortin et al., 2018; Pezoulas, Exarchos, & Fotiadis, 2020; Wachinger et al., 2021). Much of this methodological innovation has arrived on the heels of earlier developments in cross‐platform harmonization of genetic data (Borisov et al., 2019; Haghverdi, Lun, Morgan, & Marioni, 2018; Johnson, Li, & Rabinovic, 2007; Pontikos et al., 2017). These advances in harmonization of neuroimaging data are expected to manifest synergy with standardized workflows such as HALFpipe, as both elements are essential to large‐scale imaging consortium efforts (Thompson, Jahanshad, et al., 2020).
The implementation of quality metrics for fMRI data has been an incremental process that has moved steadily toward establishing empirically‐informed best practices. Historically, quality criteria have been applied unevenly across research labs. Recent years have witnessed a heightened awareness about the essential role of applying systematic and principled quality metrics to minimize confounds, for example, motion artifacts (Murphy, Birn, & Bandettini, 2013; Power et al., 2014; Power, Barnes, Snyder, Schlaggar, & Petersen, 2012), and widespread fMRI signal deflections (Aquino et al., 2020). Automated quality control methods are being developed and adopted with increasing interest, such as the MRI Quality Control software MRIQC (Esteban et al., 2017). HALFpipe has adopted parts of the functionality of MRIQC with an enhanced user experience that generates quality reports via a web‐browser‐based interface to facilitate rapid viewing, screening, and selection of individual subject data for inclusion or exclusion. The application of uniform quality assessment procedures is particularly important when mega‐analyzing and even meta‐analyzing multi‐site/scanner data, as is performed in ENIGMA. That is, study variables that segregate by site are more likely to lead to confounds without the uniform implementation of quality assessment across sites (e.g., Wachinger et al., 2021). With its harmonized quality procedures, HALFpipe aims to minimize such effects.
3.1. Limitations
Computing platforms that are likely to differ between sites are known to introduce subtle differences in output attributable to operating systems and hardware (Gronenschild et al., 2012). Collecting raw multi‐site data at one central site prior to HALFpipe processing ensures that the same computing platform can be used to process all data. While optimal, this is often not practical due to restrictions on data sharing, even when the data is completely de‐identified (i.e., when linking data to protected health or other sensitive information is no longer possible).
HALFpipe offers harmonization through uniform processing of fMRI data, but other sources of nonuniformity are beyond its scope. Recent advances in cross‐site/platform harmonization may additionally correct for differences in site, scanner hardware, or computation on different processors (Fortin et al., 2018; Pezoulas et al., 2020; Wachinger et al., 2021). Such methods could be applied to extracted HALFpipe features, either centralized or through distributed computation using tools such as COINSTAC (Plis et al., 2016), to yield results that are potentially more generalizable.
4. CONCLUSION
HALFpipe provides a standardized workflow that encompases the essential elements of task‐based and resting‐state fMRI analyses, builds on the progress and contributions of fMRIPrep developers, and extends capabilities beyond preprocessing steps with a diverse set of post‐processing functions. HALFpipe represents a major step toward addressing the reproducibility crisis in functional neuroimaging by offering a workflow that maintains details of user options, steps performed in analyses, metadata associated with analyses, code transparency, containerized installation, and the ability to recreate the runtime environment, while implementing empirically‐supported best‐practices adopted by the functional neuroimaging community.
ACKNOWLEDGMENTS
This work has been supported by a Grant from the German Research Foundation to S.E. and H.W. (DFG ER 724/4–1, WA 1539/11–1). P.M.T. received research grant funding from Biogen, Inc., for research unrelated to this manuscript. L.S. received funding from the National Institutes of Health (NIH R01 MH117601 and NHMRC Career Development Fellowship 1140764). R.A.M received funding from National Institutes of Health (NIH R01 MH111671). I.M.V. and H.W. received funding from the European Union's Horizon 2020 research and innovation programme under under Grant Agreement number 777084 (DynaMORE project).
Open access funding enabled and organized by Projekt DEAL.
Waller, L. , Erk, S. , Pozzi, E. , Toenders, Y. J. , Haswell, C. C. , Büttner, M. , Thompson, P. M. , Schmaal, L. , Morey, R. A. , Walter, H. , & Veer, I. M. (2022). ENIGMA HALFpipe: Interactive, reproducible, and efficient analysis for resting‐state and task‐based fMRI data. Human Brain Mapping, 43(9), 2727–2742. 10.1002/hbm.25829
Rajendra A. Morey, Henrik Walter, and Ilya M. Veer contributed equally to this work.
Funding information German Research Foundation, Grant/Award Numbers: DFG ER 724/4‐1, WA 1539/11‐1; Biogen, Inc.; National Institutes of Health, Grant/Award Numbers: NIH R01, MH117601; NHMRC Career Development Fellowship, Grant/Award Number: 1140764; National Institutes of Health, Grant/Award Numbers: NIH R01, MH111671; European Union's Horizon 2020 research andinnovation programme, Grant/Award Number: 777084
DATA AVAILABILITY STATEMENT
This technical report does not rely on any data. Code and documentation are available at https://github.com/HALFpipe/HALFpipe.
REFERENCES
- Abraham, A. , Pedregosa, F. , Eickenberg, M. , Gervais, P. , Mueller, A. , Kossaifi, J. , … Varoquaux, G. (2014). Machine learning for neuroimaging with scikit‐learn. Frontiers in Neuroinformatics, 8, 14. 10.3389/fninf.2014.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari, B. M. , Jahanshad, N. , Shukla, D. , Turner, J. , Grotegerd, D. , Dannlowski, U. , … Kochunov, P. (2019). A resting state fMRI analysis pipeline for pooling inference across diverse cohorts: An ENIGMA rs‐fMRI protocol. Brain Imaging and Behavior, 13(5), 1453–1467. 10.1007/s11682-018-9941-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alakörkkö, T. , Saarimäki, H. , Glerean, E. , Saramäki, J. , & Korhonen, O. (2017). Effects of spatial smoothing on functional brain networks. European Journal of Neuroscience, 46(9), 2471–2480. 10.1111/ejn.13717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro‐Almagro, F. , Jenkinson, M. , Bangerter, N. K. , Andersson, J. L. , Griffanti, L. , Douaud, G. , … Smith, S. M. (2018). Image processing and quality control for the first 10,000 brain imaging datasets from UKbiobank. NeuroImage, 166, 400–424. 10.1016/j.neuroimage.2017.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquino, K. M. , Fulcher, B. D. , Parkes, L. , Sabaroedin, K. , & Fornito, A. (2020). Identifying and removing widespread signal deflections from fMRI data: Rethinking the global signal regression problem. NeuroImage, 212, 116614. 10.1016/j.neuroimage.2020.116614 [DOI] [PubMed] [Google Scholar]
- Avants, B. B. , Tustison, N. J. , Song, G. , Cook, P. A. , Klein, A. , & Gee, J. C. (2011). A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage, 54(3), 2033–2044. 10.1016/j.neuroimage.2010.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, K. (2000). Extreme programming explained: Embrace change. Reading, MA: Addison Wesley Longman, Inc. [Google Scholar]
- Beckmann, C. , Mackay, C. , Filippini, N. , & Smith, S. (2009). Group comparison of resting‐state FMRI data using multi‐subject ICA and dual regression. NeuroImage, 47, S148. 10.1016/s1053-8119(09)71511-3 [DOI] [Google Scholar]
- Behzadi, Y. , Restom, K. , Liau, J. , & Liu, T. T. (2007). A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. NeuroImage, 37(1), 90–101. 10.1016/j.neuroimage.2007.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIDS Contributors . (2021). The brain imaging data structure (BIDS) specification. doi: 10.5281/zenodo.4710751 [DOI]
- Borisov, N. , Shabalina, I. , Tkachev, V. , Sorokin, M. , Garazha, A. , Pulin, A. , … Buzdin, A. (2019). Shambhala: A platform‐agnostic data harmonizer for gene expression data. BMC Bioinformatics, 20(1), 66. 10.1186/s12859-019-2641-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botvinik‐Nezer, R. , Holzmeister, F. , Camerer, C. F. , Dreber, A. , Huber, J. , Johannesson, M. , … Schonberg, T. (2020). Variability in the analysis of a single neuroimaging dataset by many teams. Nature, 582(7810), 84–88. 10.1038/s41586-020-2314-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciric, R. , Rosen, A. F. G. , Erus, G. , Cieslak, M. , Adebimpe, A. , Cook, P. A. , … Satterthwaite, T. D. (2018). Mitigating head motion artifact in functional connectivity MRI. Nature Protocols, 13(12), 2801–2826. 10.1038/s41596-018-0065-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciric, R. , Lorenz, R. , Thompson, W. H. , Goncalves, M. , MacNicol, E. , Markiewicz, C. J. , … Esteban, O. (2021). TemplateFlow: A community archive of imaging templates and atlases for improved consistency in neuroimaging. bioRxiv, 430678. 10.1101/2021.02.10.430678 [DOI] [Google Scholar]
- Coghlan, N. , & Stufft, D. (2013). PEP 440 ‐ version identification and dependency specification [Python.org]. Retrieved from https://www.python.org/dev/peps/pep-0440/
- Cox, R. W. (1996). AFNI: Software for analysis and visualization of functional magnetic resonance Neuroimages. Computers and Biomedical Research, 29(3), 162–173. 10.1006/cbmr.1996.0014 [DOI] [PubMed] [Google Scholar]
- Cox, R. W. , & Hyde, J. S. (1997). Software tools for analysis and visualization of fMRI data. NMR in Biomedicine, 10(4–5), 171–178. [DOI] [PubMed] [Google Scholar]
- Craddock, C. , Sikka, S. , Cheung, C. , Khanuja, R. , Ghosh, S. S. , Yan, C. , … Milham, M. (2013). Towards automated analysis of Connectomes: The configurable pipeline for the analysis of Connectomes (C‐PAC). Frontiers in Neuroinformatics, 7. 10.3389/conf.fninf.2013.09.00042 [DOI] [Google Scholar]
- Das, R . (2018). Programming In Academia Vs Industry. Retrieved from https://medium.com/@rdasxy/programming-in-academia-vs-industry-5fb52852ea39
- Dijkstra, E. W. , Lamport, L. , Martin, A. J. , Scholten, C. S. , & Steffens, E. F. M. (1978). On‐the‐fly garbage collection: An exercise in cooperation. Communications of the ACM, 21(11), 966–975. 10.1145/359642.359655 [DOI] [Google Scholar]
- Eklund, A. , Nichols, T. E. , & Knutsson, H. (2016). Cluster failure: Why fMRI inferences for spatial extent have inflated false‐positive rates. Proceedings of the National Academy of Sciences, 113(28), 7900–7905. 10.1073/pnas.1602413113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban, O. , Birman, D. , Schaer, M. , Koyejo, O. O. , Poldrack, R. A. , & Gorgolewski, K. J. (2017). MRIQC: Advancing the automatic prediction of image quality in MRI from unseen sites. PLoS One, 12(9), e0184661. 10.1371/journal.pone.0184661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban, O. , Blair, R. W. , Nielson, D. M. , Varada, J. C. , Marrett, S. , Thomas, A. G. , … Gorgolewski, K. J. (2019). Crowdsourced MRI quality metrics and expert quality annotations for training of humans and machines. Scientific Data, 6(1), 30. 10.1038/s41597-019-0035-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban, O. , Ciric, R. , Finc, K. , Blair, R. W. , Markiewicz, C. J. , Moodie, C. A. , … Gorgolewski, K. J. (2020). Analysis of task‐based functional MRI data preprocessed with fMRIPrep. Nature Protocols, 15(7), 2186–2202. 10.1038/s41596-020-0327-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban, O. , Markiewicz, C. J. , Blair, R. , Poldrack, R. A. , & Gorgolewski, K. J. (2021). sMRIPrep: Structural MRI PREProcessing workflows. 10.5281/zenodo.4593442 [DOI] [Google Scholar]
- Esteban, O. , Markiewicz, C. J. , Blair, R. W. , Moodie, C. A. , Isik, A. I. , Erramuzpe, A. , … Gorgolewski, K. J. (2019). fMRIPrep: A robust preprocessing pipeline for functional MRI. Nature Methods, 16(1), 111–116. 10.1038/s41592-018-0235-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban, O. , Markiewicz, C. J. , Blair, R. W. , Poldrack, R. A. , & Gorgolewski, K. J. (2020). SDCflows: Susceptibility distortion correction workFLOWS. Zenodo. doi: 10.5281/zenodo.4025963 [DOI]
- Esteban, O. , Markiewicz, C. J. , Goncalves, M. , Blair, R. , Berleant, S. L. , Poldrack, R. A. , & Gorgolewski, K. J. (2021). NIWorkflows: NeuroImaging workflows. Zenodo. doi: 10.5281/zenodo.5544246 [DOI]
- Fischl, B. , & Dale, A. M. (2000). Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proceedings of the National Academy of Sciences, 97(20), 11050–11055. 10.1073/pnas.200033797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin, J.‐P. , Cullen, N. , Sheline, Y. I. , Taylor, W. D. , Aselcioglu, I. , Cook, P. A. , … Shinohara, R. T. (2018). Harmonization of cortical thickness measurements across scanners and sites. NeuroImage, 167, 104–120. 10.1016/j.neuroimage.2017.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, L. , Glover, G. H. , Krenz, D. , & Magnotta, V. (2006). Reducing inter‐scanner variability of activation in a multicenter fMRI study: Role of smoothness equalization. NeuroImage, 32(4), 1656–1668. 10.1016/j.neuroimage.2006.03.062 [DOI] [PubMed] [Google Scholar]
- Gavrilescu, M. , Shaw, M. E. , Stuart, G. W. , Eckersley, P. , Svalbe, I. D. , & Egan, G. F. (2002). Simulation of the effects of global normalization procedures in functional MRI. NeuroImage, 17(2), 532–542. 10.1006/nimg.2002.1226 [DOI] [PubMed] [Google Scholar]
- Giavasis, S. , Pellman, J. , Clucas, J. , & Lurie, D. (2020). Setting up a data configuration File (participant/subject list). https://fcp-indi.github.io/docs/latest/user/subject%5C_list%5C_config
- Glasser, M. F. , Sotiropoulos, S. N. , Wilson, J. A. , Coalson, T. S. , Fischl, B. , Andersson, J. L. , … Jenkinson, M. (2013). The minimal preprocessing pipelines for the human connectome project. NeuroImage, 80, 105–124. 10.1016/j.neuroimage.2013.04.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves, M. , Markiewicz, C. J. , Moia, S. , Ghosh, S. S. , Poldrack, R. A. , & Esteban, O. (2021). NiTransforms: A python tool to read, represent, manipulate, and apply n‐dimensional spatial transforms. Journal of Open Source Software, 6(65), 3459. 10.21105/joss.03459 [DOI] [Google Scholar]
- Gorgolewski, K. J. , Auer, T. , Calhoun, V. D. , Craddock, R. C. , Das, S. , Duff, E. P. , … Poldrack, R. A. (2016). The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Scientific Data, 3(1), 160044. 10.1038/sdata.2016.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski, K. J. , Burns, C. D. , Madison, C. , Clark, D. , Halchenko, Y. O. , Waskom, M. L. , & Ghosh, S. S. (2011). Nipype: A flexible, lightweight and extensible neuroimaging data processing framework in python. Frontiers in Neuroinformatics, 5, 13. 10.3389/fninf.2011.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronenschild, E. H. B. M. , Habets, P. , Jacobs, H. I. L. , Mengelers, R. , Rozendaal, N. , van Os, J. , & Marcelis, M. (2012). The effects of FreeSurfer version, workstation type, and Macintosh operating system version on anatomical volume and cortical thickness measurements. PLoS One, 7(6), e38234. 10.1371/journal.pone.0038234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüning, B. , Chilton, J. , Köster, J. , Dale, R. , Soranzo, N. , van den Beek, M. , … Taylor, J. (2018). Practical computational reproducibility in the life sciences. Cell Systems, 6(6), 631–635. 10.1016/j.cels.2018.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi, L. , Lun, A. T. L. , Morgan, M. D. , & Marioni, J. C. (2018). Batch effects in single‐cell RNA‐sequencing data are corrected by matching mutual nearest neighbors. Nature Biotechnology, 36(5), 421–427. 10.1038/nbt.4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halchenko, Y. , Goncalves, M. , Visconti di Oleggio Castello, M. , Ghosh, S. , Hanke, M. , Brett, M. , Salo, T. , Gorgolewski, K. J. , Stadler, J. , Lee John, J. , Lurie, D. , Pellman, J. , Poldrack, B. , Schiffler, B. , Szczepanik, M. , & Carlin, J. (2018). nipy/heudiconv: Heudiconv v0.5.1. 10.5281/zenodo.1306159 [DOI]
- Hallquist, M. N. , Hwang, K. , & Luna, B. (2013). The nuisance of nuisance regression: Spectral misspecification in a common approach to resting‐state fMRI preprocessing reintroduces noise and obscures functional connectivity. NeuroImage, 82, 208–225. 10.1016/j.neuroimage.2013.05.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, C. R. , Millman, K. J. , van der Walt, S. J. , Gommers, R. , Virtanen, P. , Cournapeau, D. , … Oliphant, T. E. (2020). Array programming with NumPy. Nature, 585(7825), 357–362. 10.1038/s41586-020-2649-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogman, M. , van Rooij, D. , Klein, M. , Boedhoe, P. , Ilioska, I. , Li, T. , … Franke, B. (2022). Consortium neuroscience of attention deficit/hyperactivity disorder and autism spectrum disorder: The ENIGMA adventure. Human Brain Mapping., 43, 37–55. 10.1002/hbm.25029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, A. (2016). MNI T1 6thGen NLIN to MNI 2009b NLIN ANTs transform. doi: 10.6084/m9.figshare.3502238.v2 [DOI]
- Jarecka, D. , Goncalves, M. , Markiewicz, C. J. , Esteban, O. , Lo, N. , Kaczmarzyk, J. , & Ghosh, S. (2020). Pydra—a flexible and lightweight dataflow engine for scientific analyses. In Proceedings of the 19th python in science conference (pp. 132–139). 10.25080/majora-342d178e-012 [DOI] [Google Scholar]
- Jenkinson, M. , Beckmann, C. F. , Behrens, T. E. , Woolrich, M. W. , & Smith, S. M. (2012). FSL. NeuroImage, 62(2), 782–790. 10.1016/j.neuroimage.2011.09.015 [DOI] [PubMed] [Google Scholar]
- Jo, H. J. , Gotts, S. J. , Reynolds, R. C. , Bandettini, P. A. , Martin, A. , Cox, R. W. , & Saad, Z. S. (2013). Effective preprocessing procedures virtually eliminate distance‐dependent motion artifacts in resting state FMRI. Journal of Applied Mathematics, 2013, 1–9. 10.1155/2013/935154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, W. E. , Li, C. , & Rabinovic, A. (2007). Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics, 8(1), 118–127. 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- Kerestes, R. , Chase, H. W. , Phillips, M. L. , Ladouceur, C. D. , & Eickhoff, S. B. (2017). Multimodal evaluation of the amygdala's functional connectivity. NeuroImage, 148, 219–229. 10.1016/j.neuroimage.2016.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzer, G. M. , Sochat, V. , & Bauer, M. W. (2017). Singularity: Scientific containers for mobility of compute. PLoS One, 12(5), e0177459. 10.1371/journal.pone.0177459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenth, R. V. (2016). Least‐squares means: The R package lsmeans. Journal of Statistical Software, Articles, 69(1), 1–33. 10.18637/jss.v069.i01 [DOI] [Google Scholar]
- Li, X. , Morgan, P. S. , Ashburner, J. , Smith, J. , & Rorden, C. (2016). The first step for neuroimaging data analysis: DICOM to NIfTI conversion. Journal of Neuroscience Methods, 264, 47–56. 10.1016/j.jneumeth.2016.03.001 [DOI] [PubMed] [Google Scholar]
- Marchini, J. L. , & Ripley, B. D. (2000). A new statistical approach to detecting significant activation in functional MRI. NeuroImage, 12(4), 366–380. 10.1006/nimg.2000.0628 [DOI] [PubMed] [Google Scholar]
- Markiewicz, C. J. , De La Vega, A. , Wagner, A. , Halchenko, Y. O. , Finc, K. , Ciric, R. , … Gorgolewski, K. J. poldracklab/fitlins: v0.9.2. 10.5281/zenodo.5120201 [DOI]
- McKinney, W. (2010). Data structures for statistical computing in python. In Proceedings of the 9th python in science conference (pp. 56–61). 10.25080/majora-92bf1922-00a [DOI] [Google Scholar]
- Murphy, K. , Birn, R. M. , & Bandettini, P. A. (2013). Resting‐state fMRI confounds and cleanup. NeuroImage, 80, 349–359. 10.1016/j.neuroimage.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezoulas, V. , Exarchos, T. , & Fotiadis, D. (2020). Medical data sharing, harmonization and analytics. London, England: Academic Press. 10.1016/C2018-0-00523-X [DOI] [Google Scholar]
- Plis, S. M. , Sarwate, A. D. , Wood, D. , Dieringer, C. , Landis, D. , Reed, C. , … Calhoun, V. D. (2016). COINSTAC: A privacy enabled model and prototype for leveraging and processing decentralized brain imaging data. Frontiers in Neuroscience, 10, 365. 10.3389/fnins.2016.00365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack, R. A. , Baker, C. I. , Durnez, J. , Gorgolewski, K. J. , Matthews, P. M. , Munafò, M. R. , … Yarkoni, T. (2017). Scanning the horizon: Towards transparent and reproducible neuroimaging research. Nature Reviews Neuroscience, 18(2), 115–126. 10.1038/nrn.2016.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack, R. A. , Gorgolewski, K. J. , & Varoquaux, G. (2019). Computational and Informatic advances for reproducible data analysis in neuroimaging. Annual Review of Biomedical Data Science, 2(1), 1–20. 10.1146/annurev-biodatasci-072018-021237 [DOI] [Google Scholar]
- Pontikos, N. , Yu, J. , Moghul, I. , Withington, L. , Blanco‐Kelly, F. , Vulliamy, T. , … Plagnol, V. (2017). Phenopolis: An open platform for harmonization and analysis of genetic and phenotypic data. Bioinformatics, 33(15), 2421–2423. 10.1093/bioinformatics/btx147 [DOI] [PubMed] [Google Scholar]
- Power, J. D. (2017). A simple but useful way to assess fMRI scan qualities. NeuroImage, 154, 150–158. 10.1016/j.neuroimage.2016.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power, J. D. , Barnes, K. A. , Snyder, A. Z. , Schlaggar, B. L. , & Petersen, S. E. (2012). Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage, 59(3), 2142–2154. 10.1016/j.neuroimage.2011.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power, J. D. , Mitra, A. , Laumann, T. O. , Snyder, A. Z. , Schlaggar, B. L. , & Petersen, S. E. (2014). Methods to detect, characterize, and remove motion artifact in resting state fMRI. NeuroImage, 84, 320–341. 10.1016/j.neuroimage.2013.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim, R. H. , Mennes, M. , van Rooij, D. , Llera, A. , Buitelaar, J. K. , & Beckmann, C. F. (2015). ICA‐AROMA: A robust ICA‐based strategy for removing motion artifacts from fMRI data. NeuroImage, 112, 267–277. 10.1016/j.neuroimage.2015.02.064 [DOI] [PubMed] [Google Scholar]
- Savio, A. M. , Schutte, M. , Graña, M. , & Yakushev, I. (2017). Pypes: Workflows for processing multimodal neuroimaging data. Frontiers in Neuroinformatics, 11, 25. 10.3389/fninf.2017.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaal, L. , Pozzi, E. , Ho, T. C. , van Velzen, L. S. , Veer, I. M. , Opel, N. , … Veltman, D. J. (2020). ENIGMA MDD: Seven years of global neuroimaging studies of major depression through worldwide data sharing. Translational Psychiatry, 10(1), 172. 10.1038/s41398-020-0842-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, N. J. , Hudon, C. , Broessli, Seabold, S. , Quackenbush, P. , Hudson‐Doyle, M. , Humber, M. , Leinweber, K. , Kibirige, H. , Davidson‐Pilon, C. & Portnoy, A. (2018). pydata/patsy: v0.5.1. 10.5281/zenodo.1472929 [DOI]
- Steegen, S. , Tuerlinckx, F. , Gelman, A. , & Vanpaemel, W. (2016). Increasing transparency through a multiverse analysis. Perspectives on Psychological Science, 11(5), 702–712. 10.1177/1745691616658637 [DOI] [PubMed] [Google Scholar]
- Taylor, P. A. , & Saad, Z. S. (2013). FATCAT: (an efficient) functional and Tractographic connectivity analysis toolbox. Brain Connectivity, 3(5), 523–535. 10.1089/brain.2013.0154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, P. M. , Ching, C. R. K. , Dennis, E. L. , Salminen, L. E. , Turner, J. A. , van Erp, T. G. M. , & Jahanshad, N. (2020). Big data initiatives in psychiatry: Global neuroimaging studies. In Kubicki M. & Shenton M. E. (Eds.), Neuroimaging in Schizophrenia (pp. 411–426). Cham: Springer International Publishing. 10.1007/978-3-030-35206-6_21 [DOI] [Google Scholar]
- Thompson, P. M. , Jahanshad, N. , Ching, C. R. K. , Salminen, L. E. , Thomopoulos, S. I. , Bright, J. , … Zelman, V. (2020). ENIGMA and global neuroscience: A decade of large‐scale studies of the brain in health and disease across more than 40 countries. Translational Psychiatry, 10(1), 1–28. 10.1038/s41398-020-0705-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel, O. A. , Boedhoe, P. S. W. , Bertolin, S. , Bruin, W. B. , Francks, C. , Ivanov, I. , … Stein, D. J. (2020). An overview of the first 5 years of the ENIGMA obsessive–compulsive disorder working group: The power of worldwide collaboration. Human Brain Mapping, 43, 23–36. 10.1002/hbm.24972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veer, I. M. , Waller, L. , Lett, T. A. , Erk, S. , & Walter, H. (2019). ENIGMA task‐based fMRI: A workgroup studying the genetic basis of task‐evoked brain activity. In 25th Organization of Human Brain Mapping Annual Meeting. https://archive.aievolution.com/2019/hbm1901/index.cfm?do=doc.viewDocument&documentID=2746 [Google Scholar]
- Wachinger, C. , Rieckmann, A. , Pölsterl, S. , & for the Alzheimer's Disease Neuroimaging Initiative and the Australian Imaging Biomarkers and Lifestyle flagship study of ageing . (2021). Detect and correct bias in multi‐site neuroimaging datasets. Medical Image Analysis, 67, 101879. 10.1016/j.media.2020.101879 [DOI] [PubMed] [Google Scholar]
- Wager, T. D. , Davidson, M. L. , Hughes, B. L. , Lindquist, M. A. , & Ochsner, K. N. (2008). Prefrontal‐subcortical pathways mediating successful emotion regulation. Neuron, 59(6), 1037–1050. 10.1016/j.neuron.2008.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeman, D. G. , & Henson, R. N. (2015). A multi‐subject, multi‐modal human neuroimaging dataset. Scientific Data, 2(1), 150001. 10.1038/sdata.2015.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis, C. (2020). Data‐driven approach to dynamic resting state functional connectivity in post‐traumatic stress disorder. PhD dissertation, The University of Wisconsin Milwaukee . https://dc.uwm.edu/etd/2623
- Whitfield‐Gabrieli, S. , & Nieto‐Castanon, A. (2012). Conn: a functional connectivity toolbox for correlated and anticorrelated brain networks. Brain Connectivity, 2(3), 125–141. 10.1089/brain.2012.0073 [DOI] [PubMed] [Google Scholar]
- Woolrich, M. W. , Behrens, T. E. , Beckmann, C. F. , Jenkinson, M. , & Smith, S. M. (2004). Multilevel linear modelling for FMRI group analysis using Bayesian inference. NeuroImage, 21(4), 1732–1747. 10.1016/j.neuroimage.2003.12.023 [DOI] [PubMed] [Google Scholar]
- Woolrich, M. W. , Ripley, B. D. , Brady, M. , & Smith, S. M. (2001). Temporal autocorrelation in Univariate linear modeling of FMRI data. NeuroImage, 14(6), 1370–1386. 10.1006/nimg.2001.0931 [DOI] [PubMed] [Google Scholar]
- Yan, C.‐G. , Wang, X.‐D. , Zuo, X.‐N. , & Zang, Y.‐F. (2016). DPABI: Data Processing & Analysis for (Resting‐State) Brain Imaging. Neuroinformatics, 14(3), 339–351. 10.1007/s12021-016-9299-4 [DOI] [PubMed] [Google Scholar]
- Zang, Y. , Jiang, T. , Lu, Y. , He, Y. , & Tian, L. (2004). Regional homogeneity approach to fMRI data analysis. NeuroImage, 22(1), 394–400. 10.1016/j.neuroimage.2003.12.030 [DOI] [PubMed] [Google Scholar]
- Zou, Q.‐H. , Zhu, C.‐Z. , Yang, Y. , Zuo, X.‐N. , Long, X.‐Y. , Cao, Q.‐J. , … Zang, Y.‐F. (2008). An improved approach to detection of amplitude of low‐frequency fluctuation (ALFF) for resting‐state fMRI: Fractional ALFF. Journal of Neuroscience Methods, 172(1), 137–141. 10.1016/j.jneumeth.2008.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This technical report does not rely on any data. Code and documentation are available at https://github.com/HALFpipe/HALFpipe.


