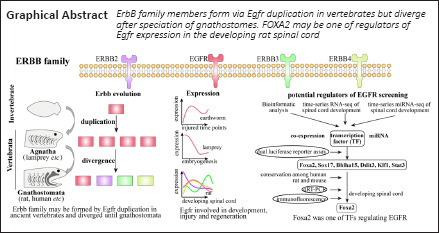
Key Words: co-expression, Egfr, evolution, FOXA2, gene expression, miRNA, spinal cord, transcription factor
Abstract
Egfr, a member of the ErbB gene family, plays a critical role in tissue development and homeostasis, wound healing, and disease. However, expression and regulators of Egfr during spinal cord development remain poorly understood. In this study, we investigated ErbB evolution and analyzed co-expression modules, miRNAs, and transcription factors that may regulate Egfr expression in rats. We found that ErbB family members formed via Egfr duplication in the ancient vertebrates but diverged after speciation of gnathostomes. We identified a module that was co-expressed with Egfr, which involved cell proliferation and blood vessel development. We predicted 25 miRNAs and nine transcription factors that may regulate Egfr expression. Dual-luciferase reporter assays showed six out of nine transcription factors significantly affected Egfr promoter reporter activity. Two of these transcription factors (KLF1 and STAT3) inhibited the Egfr promoter reporter, whereas four transcription factors (including FOXA2) activated the Egfr promoter reporter. Real-time PCR and immunofluorescence experiments showed high expression of FOXA2 during the embryonic period and FOXA2 was expressed in the floor plate of the spinal cord, suggesting the importance of FOXA2 during embryonic spinal cord development. Considering the importance of Egfr in embryonic spinal cord development, wound healing, and disease (specifically in cancer), regulatory elements identified in this study may provide candidate targets for nerve regeneration and disease treatment in the future.
Introduction
Epidermal growth factor receptor (EGFR, also known as ErbB1) is a member of the receptor tyrosine kinase (RTK) superfamily. The other three EGFR family members include ErbB2, ErbB3, and ErbB4 (Ayati et al., 2020). EGFR was the first receptor discovered in this family. Until now, at least seven primary ligands can activate EGFR, including epidermal growth factor (EGF), transforming growth factor-α, heparin-binding EGF, β-cellulin, amphiregulin, epiregulin, and epigen (Knudsen et al., 2014). The ErbB family plays a critical role in vital signaling pathways, such as the phosphatidylinositol 3-kinase/Akt, Ras/Raf/MEK/ERK1/2, and phospholipase C pathways, which are essential for cellular proliferation, differentiation, and survival (Schlessinger, 2002). Each member of the ErbB family contains an EGF-like domain (a conserved three-loop structure). EGFR consists of an extracellular ligand-binding domain, single transmembrane domain, and cytoplasmic domain. After ligand binding, EGFR can homo- or hetero-dimerize, and the EGFR-ErbB2 dimer forms the most active receptor (Klapper et al., 2000). After dimerization, specific tyrosine residues are phosphorylated and new binding sites are formed for adaptor proteins with Src homology 2 domains, which enable the activation of downstream signaling pathways.
EGFR plays a crucial role during vertebrate development. In mammals, EGFR improves embryo implantation and placenta development during embryogenesis (Chia et al., 1995). EGFR is involved in the development of many organs, including the brain, spinal cord, heart, bone, and liver (Chen et al., 2016). EGFR can be detected in cells of the central nervous system (CNS) during development, such as neurons, astrocytes, oligodendrocytes, and progenitor cells of the subventricular zone (Seroogy et al., 1994; Kornblum et al., 1997). However, the expression of EGFR is reduced in the adult brain, including in the nigrostriatal system, and the highest levels of expression have been detected during the neonatal stage (Seroogy et al., 1994). In the peripheral nervous system, EGFR is also found in the neurons of dorsal root ganglia (DRG), Schwann cells, and satellite glial cells (Morris et al., 1999). Sibilia et al. (2007) reported that EGFR-null mice displayed serious neural defects. Hence, EGFR participates in the development, differentiation, maintenance, and regeneration of various organs and the nervous system. Understanding the regulation of Egfr expression is of great significance for comprehending the development of the nervous system and exploring potential targeted therapies for related diseases and such spinal cord injury.
Gene expression is regulated by several factors, including chromatin modification, transcription factors (TFs), and non-coding RNAs (Kouzarides, 2007; Patil et al., 2014). In addition, accumulating studies showed that activation of G protein-coupled receptor (GPCR) by agonists (e.g. angiotensin II) could promote EGFR activity via transactivation (Forrester et al., 2016). TFs recognize cis-regulatory regions to correctly initiate gene transcription. MicroRNAs (miRNA, 20–30 NT) regulate gene expression post-transcriptionally by interacting with the 3’ untranslated region (3’-UTR) of target mRNAs (Filipowicz et al., 2008). TFs and miRNAs are important regulators of gene expression in eukaryotes. Several TFs (Sp1, EVI1, ETF) have been shown to activate Egfr (Kageyama et al., 1988; Mizuguchi et al., 2019; Tsai et al., 2019). Chromatin immunoprecipitation (ChIP)-seq technology has accelerated the discovery of TF binding sites (Mundade et al., 2014), and the majority have been studied in humans and mice. MiRNAs, such as miR-34a (Li et al., 2017) and miR-133a (Guo et al., 2018), and long non-coding RNAs, such as LINC00265 (Ge et al., 2019) and EGFR-AS1 (Wang et al., 2019a), have been validated as regulators of Egfr expression in cancers. In our previously sequenced transcription profile of rat spinal cord development (Yang et al., 2021), we demonstrated that Egfr was highly expressed in the late embryonic period (embryonic day 18, E18d) until postnatal week 1 (P1w) with low expression in adults (postnatal weeks 4–8), suggesting dynamic regulation of Egfr during spinal cord development. However, limited studies have explored potential regulators of Egfr expression in the spinal cord.
In this study, we performed an evolutionary analysis of the ErbB gene family in metazoans, screened a module containing genes that were co-expressed with Egfr, investigated GPCRs that may be involved in EGFR transactivation, and predicted miRNAs that targeted Egfr and TFs that regulated Egfr expression based on our previous transcription profiling of spinal cord development. We also validated the direct binding between TFs and Egfr using a dual luciferase reporter assay in vitro. Finally, we investigated the expression of candidate TFs during spinal cord development at the mRNA and protein level using qRT-PCR and immunohistochemistry, respectively, in rats ranging in age from embryonic to adult.
Materials and Methods
Animals and tissue preparation
The study was approved by the Animal Care and Use Committee of Nantong University and was reported in accordance with the ARRIVE 2.0 guidelines (Animal Research: Reporting of In Vivo Experiments) (Percie du Sert et al., 2020). Specific pathogen-free Sprague-Dawley rats, including pregnant rats (body weights: 300–330 g; n = 2), male pups (postnatal day 1 [P1d] and postnatal week 1 [P1w]; body weights: 7 g and 20 g, respectively; n = 4/group), and adult males (postnatal week 8 [P8w] and 12 [P12w]; average body weights: 200 g and 320 g, respectively; n = 4/group) were used for sample collection at different developmental stages. All rats were purchased from the Experimental Animal Center of Nantong University, Nantong, China (license No. SCXK (Su) 2019-0001). Four rats were housed per cage, and all rats were allowed access to food and water ad libitum. The animal room temperature was maintained at 23 ± 1°C. Spinal cord tissue samples were collected from embryos of pregnant rats at embryo day 11 (E11d), 13 (E13d), and 14 (E14d) and from pups/adults at P1d, P1w, and P8w. The sampling time was 6:30 a.m. The pregnant rats were deeply anesthetized by intraperitoneal injection of mixed narcotics (4.25 g chloral hydrate, 2.12 g magnesium sulfate, 886 mg sodium pentobarbital, 14.25 mL ethanol, 33.8 mL propylene glycol, and distilled water to a final volume of 100 mL) and the embryos were removed. The embryos were placed in a pre-prepared, oxygen-saturated artificial cerebrospinal fluid (ACSF, ice-water mixture; ACSF composition: 130 mM NaCl, 5 mM KCl, 2 mM KH2PO4, 1.5 mM CaCl2, 6 mM MgSO4, 10 mM glucose, 10 mM HEPES, pH 7.2, 305 mOsm osmotic pressure). The neural tubes of E11d rat embryos displayed a translucent, soft gel state. The strip outline of the neural tube and its dark boundary with surrounding tissues were observed under a microscope (ZEISS Type: Stemi 508, Jena, Germany). The neural tubes were carefully isolated using micro scissors and micro tweezers. For E13d, 14d, and E18d rat embryos, the cartilaginous spinal canals were carefully cut open with micro scissors, and the spinal cords were isolated using micro tweezers. Adult rats (P8w) were anesthetized by intraperitoneal injection of mixed narcotics, placed on an ice bag, and decapitated at the neck with a specialized decapitator. The spinal columns were separated immediately and washed twice with pre-prepared, oxygen-saturated ACSF. The spines were dissected in ACSF (ice-water mixture), and the muscles, bones, dura mater, arachnoid membranes, pia mater, and large blood vessels were carefully removed. All spinal cords were immediately quick-frozen and preserved in liquid nitrogen (–180°C).
Evolutionary analysis of the ErbB gene family in 32 metazoan species
We used BLAST (v2.2.26, https://ftp.ncbi.nlm.nih.gov/blast/) (Altschul et al., 1990) to identify potential ErbB gene family members from the proteomes of 32 species. The longest protein sequence was used if multiple isoforms existed. The phylogenetic relationship of 32 species was inferred by combining published literature. To understand the evolution of ErbB genes in the 32 species, we performed phylogenetic analysis as follows. Multiple sequence alignments were performed using MUSCLE (v3.8.31, https://www.ebi.ac.uk/Tools/msa/muscle/) (Edgar, 2004) and poor alignment was further trimmed using TrimAl (v1.2, http://trimal.cgenomics.org) (Capella-Gutierrez et al., 2009) with the parameter “-automated1”. Finally, IQ-TREE (v1.6.7, http://www.iqtree.org) (Nguyen et al., 2015) was used to reconstruct the phylogenetic tree using the maximum-likelihood algorithm. The iTOL tool (https://itol.embl.de/) (Letunic and Bork, 2019) was used for gene tree visualization. Synteny analysis was performed using JCVI (https://github.com/tanghaibao/jcvi) (Tang et al., 2015). MEGA-X (https://www.megasoftware.net/) (Kumar et al., 2018) was employed to analyze let-7 family evolution.
Prediction of regulatory miRNAs targeting Egfr
Four transcripts of the Egfr gene were present in rat reference annotation (RefSeq accession: GCF_000001895.5, rn6). The sequence of the longest 3’-UTR was extracted from the rat Egfr gene (accession No. XM_008770416.2). Mature miRNA sequences for rats were retrieved from the miRbase database (http://www.mirbase.org/) (Kozomara et al., 2019). The miRanda software (v3.3a, https://cbio.mskcc.org/miRNA2003/miranda.html) (Enright et al., 2003) was used to predict miRNAs targeting Egfr with the criterion of “-sc 140 -en -20”. To avoid multiple predictions, we also employed two other widely used programs: PITA (http://genie.weizmann.ac.il/pubs/mir07) (Kertesz et al., 2007) and TargetScan (http://www.targetscan.org/vert_72/) (Agarwal et al., 2015) using default parameters. MiRNAs with experimentally validated interactions in Tarbase (v8.0, http://www.microrna.gr/tarbase) (Karagkouni et al., 2018) were also collected. MiRNAs predicted by at least two programs were chosen for downstream analyses. Pearson’s correlation of expression between Egfr and miRNAs in rats was calculated using the function “cor.test” in the R program (https://www.r-project.org/) (Ripley, 2001) based on our previous RNA-seq data of rat spinal cord development. Significant negative correlations were used to further select candidate regulatory miRNAs (P < 0.05).
Prediction of potential TFs that regulate the expression of Egfr
We extracted a 5 kb sequence upstream of rat Egfr transcriptional start site and predicted potential promoter regions using the webserver Promoter 2.0 (http://www.cbs.dtu.dk/services/Promoter/) (Knudsen, 1999). Then, we predicted and screened potential regulatory TFs and binding sites by combining four methods: 1) JASPAR database (https://jaspar.genereg.net/) (Mathelier et al., 2016); 2) TRANSFAC database (https://genexplain.com/transfac/) (Matys et al., 2003) (v8.3); 3) animal TFDB database (http://bioinfo.life.hust.edu.cn/AnimalTFDB) (Zhang et al., 2015); and 4) MAST algorithms (https://meme-suite.org/meme/, from The MEME Suite) (Bailey and Gribskov, 1998). We focused on predicted TFs with Gene Ontology annotation by assigning “angiogenesis” and high expression in the early embryonal stage. ChIP-seq evidence of TFs targeting Egfr in humans and mice was also evaluated using ChIPBase (v2.0, https://rna.sysu.edu.cn/chipbase/) (Zhou et al., 2017).
Validation of direct interaction between TFs and the Egfr promoter in rats by dual-luciferase reporter assay
To further confirm nine predicted TFs, we employed a dual-luciferase reporter assay (Sherf et al., 1996). Sequences of nine TFs and Egfr promoter region were retrieved from the Genbank database and synthesized by GeneChem (Shanghai, China). GV141-TF (Foxa2, Sox17, Bhlha15, Klf4, Sp1, Ddit3, Arid3b, Klf1, and Stat3) over-expression vectors were constructed by GeneChem. A GV141-empty (control) vector was also provided by GeneChem. The plasmids individually containing full-length coding sequences of the nine TFs were used as the effectors. The plasmid containing the Egfr promoter was used as the reporter. Human embryonic kidney 293T cells (Cell bank of Shanghai Chinese Academy of Sciences, Shanghai, China, RRID:19375.09.3101HUMSCSP502) were transfected with these vectors (an effector and the reporter) using X-tremeGENE HP transfection reagent (Roche, Basel, Switzerland) in accordance with the manufacturer’s instructions. After 48 hours, the cells were harvested, and a dual-luciferase assay kit (Promega Biotech Co., Ltd.; Beijing, China) was used to detect LUC and REN luciferase activity via a Luminoskan Ascent Microplate Luminometer (Thermo Scientific, Rockford, IL, USA) in accordance with the manufacturer’s instructions. The transcriptional activity of each TF after binding to the rat Egfr promoter was estimated using the LUC/REN value. At least three independent experiments were performed for each TF-promoter pair.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) validation
The mRNA expression of Egfr and six candidate TFs (Bhlha15, Ddit3, Foxa2, Klf1, Sox17, and Stat3) in the developing rat spinal cord was examined by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Total RNA was separately extracted from the spinal cord of E13d, E18d, P1w, and P8w rats using TRIzol (Gibco; Carlsbad, CA, USA) in accordance with the manufacturer’s protocol. Egfr expression was analyzed at the additional time points E11d, E14d, P1d, and P12w. Total RNA (1 μg) was used to synthesize cDNA using the PrimeScriptTM RT reagent kit (TaKaRa Biotechnology CO., Ltd.; Dalian, Liaoning Province, China). The qRT-PCR was performed using the TB Green® Premix Ex TaqTM (TaKaRa). The expression of target genes was normalized to Gapdh expression using the ΔCt method (2–ΔΔCt). The primers used in this study are listed in Table 1.
Table 1.
Primers for quantitative reverse transcription-polymerase chain reaction used in this study
| Gene | Sequence (5’–3’) | Product size (bp) |
|---|---|---|
| Gapdh | Forward: ACA GCA ACA GGG TGG TGG AC | 20 |
| Reverse: TTT GAG GGT GCA GCG AAC TT | 20 | |
| Egfr | Forward: CCA CCA AGA CAG GCG ACG | 18 |
| Reverse: AGT AGC TTG GTT CTC GCA GTC | 21 | |
| Foxa2 | Forward: GGT GGG TAG CCA GAA AAA GGC | 21 |
| Reverse: CAC GGC TCC CAG CAT ACT T | 19 | |
| Sox17 | Forward: TTC AGC CGT CCT ATT TCC CC | 20 |
| Reverse: CCA CCA CCT CTC CTT TCA CC | 20 | |
| Bhlha15 | Forward: ACA ACC TCG TGG CTC TTT GT | 20 |
| Reverse: AGG ATC ACC CTG GTT GGA ACA | 21 | |
| Ddit3 | Forward: TCA TAC ACC ACC ACA CCT GAA | 21 |
| Reverse: AGG GAT GCA GGG TCA AGA GT | 20 | |
| Klf1 | Forward: CAC GCA CAC CGG AGA GAA G | 19 |
| Reverse: GAG TGG GAC CAT CCT TTG GG | 20 | |
| Stat3 | Forward: CCG GAG CTG CAG TTT AGA CA | 20 |
| Reverse: CAC TGT CTC TGG GGC TGA AG | 20 |
Immunofluorescence staining
Immunofluorescence staining was performed on tissues from E11d, E14d, E18d, P1d, P1w, and P8w rats to determine the expression and cellular localization EGFR. Immunofluorescence staining was performed on tissues from E13d, E18d, P1w, and P8w rats to evaluate FOXA2 expression and cellular localization during spinal cord development. For embryos (E11d–E18d) and neonatal rats (P1d, P1w), the spinal cord tissue was dissected in ice-cold phosphate-buffered saline (PBS) after a 75% ethanol wipe. Animal anesthesia method was the same with description above. To obtain the adult spinal cord (P8w), the animals were transcardially perfused with normal saline followed by 4% paraformaldehyde under deep anesthesia via intraperitoneal injections of mixed narcotics as described above. All collected spinal cord tissues were post-fixed in 4% paraformaldehyde at 4°C for 8 hours, dehydrated in sequential 10–20–30% sucrose solutions, and sectioned on a Leica cryostat (Leica, Wetzlar, Germany) into 12-μm-thick sections. Immunofluorescence procedures were performed as follows. The sections were placed in blocking buffer (Beyotime; Shanghai, China), incubated with the primary antibody at 4°C for 12 hours, and then treated with the secondary antibody at room temperature for one hour. Subsequently, the nuclei were counterstained with Hoechst 33342 in PBS (1:2000) for 10 minutes at room temperature. Images were observed using fluorescence microscopy (Leica). To quantify protein expression, the “integrated density” of the target channels (EGFR and FOXA2 only) with the same area at each developmental stage was calculated using ImageJ Fiji software (https://imagej.net/software/fiji/) (Schindelin et al., 2012) and then normalized to the first developmental stage (E11d for EGFR and E13d for FOXA2). Nestin was used as a marker of neural stem cells in the embryonic rat spinal cord, and NeuN was used as the marker of neuronal nuclei in the postnatal rat spinal cord. Primary and secondary antibodies used in this study are listed as follows: rabbit anti-EGFR (1:100, Proteintech, Rosemont, IL, USA, Cat# 18986-1-AP, RRID: AB_10596476), rabbit anti-FOXA2 (1:300, Abcam, Cambridge, UK, Cat# ab108422, RRID: AB_11157157), mouse anti-Nestin (1:100, Millipore, Bedford, MA, USA, Cat# MAB353, RRID: AB_94911), mouse anti-NeuN (1:100, MilliporeSigma, Burlington, MA, USA, Cat# MAB377A5, RRID: AB_2814948), Alexa Fluor 488 goat anti-mouse IgG (H+L) (1:2000, Thermo Fisher Scientific, Waltham, MA, USA, Cat# A-11001, RRID: AB_2534069), and Cy3-conjugated goat anti-rabbit IgG (H+L) (1:400, Proteintech, Cat# SA00009-2, RRID: AB_2890957).
Statistical analysis
No statistical methods were used to predetermine sample sizes; however, our sample sizes were similar to those reported in a previous publication (Zha et al., 2016). Histological analyses were blinded to experimental conditions. Data from the dual-luciferase reporter assays are presented as mean ± SD and were analyzed using one-way analysis of variance (ANOVA) followed by the Dunnett multiple comparison test. Quantitative data from qRT-PCR and immunofluorescence experiments are presented as means ± SEM after analysis using Student’s t-tests. All statistical analyses were performed using GraphPad Prism (v8.3, GraphPad Software Inc., San Diego, CA, USA). P-values < 0.05 were considered statistically significant.
Results
ErbB gene family members are duplicated in vertebrates but diverge in Gnathostomata
We analyzed ErbB family genes in 32 species from Eukaryota (Placozoa, Cnidaria, Protostomia, and Deuterostomia) to understand the evolution of this gene family. The results indicated that ErbB genes occurred in species from Metazoa except for the phyla Placozoa and Cnidaria (Figure 1A). Multiple copies were observed in most species from vertebrate lineages but only in some invertebrate species, such as those in Platyhelminthes and in the earthworm Eisenia andrei (Figure 1A). According to the annotation in the public database, the ErbB gene in invertebrates was Egfr, while Egfr, Erbb2, Erbb3, and Erbb4 were found in vertebrates except for species from Agnatha which showed the best hit with Erbb4 (termed Erbb4-like). Phylogenetic analysis of ErbB genes indicated that Egfr was duplicated in the last common ancestor of Schistosoma mansoni and Schmidtea mediterranea and subsequently underwent independent expansion (Figure 1B). Egfr in E. andrei underwent species-specific expansion with a total of 11 copies. Interestingly, although duplication of ErbB genes was observed in vertebrates, divergence of ErbB genes occurred in Gnathostomata. Five to six ErbB genes in three species belonging to Agnatha were not diverged, but four diverged genes were present in the closest species, i.e., cartilaginous fish (shark), and remaining gnathostomes, such as humans and mice (Figure 1B). ErbB genes in species from Agnatha were clustered together with Erbb3-Erbb4 and divided into two subgroups before speciation (Figure 1B). Six ErbB genes in lampreys were located in six different scaffolds. Synteny analysis between humans and lampreys showed that blocks containing four ErbB genes in humans had multiple orthologs and cross relationships in lampreys (Figure 1C). These data suggested that the ErbB gene family may have originated via small-scale duplication of ancient Egfr in vertebrates. Gene duplication is an important source of novelty; however, most duplicates are eventually lost, and the remains may be preserved by neofunctionalization and subfunctionalization (Gout and Lynch, 2015). Transcriptomic information provides more details about the potential fate of duplicated genes. Planarian flatworms (S. mediterranea) have powerful abilities of regeneration after amputation owing to abundant stem cells (neoblasts) throughout the body (Adler et al., 2014). RNA interference (RNAi) knockdown that inhibited the expression of Egfr homologues smed-egfr-1–6 in S. mediterranea showed expression divergence in tissues with different copies even in the same group and one copy in each group mainly contributed to the phenotype (Barberan et al., 2016). It was also shown that RNAi specific for smed-egfr-1 and smed-egfr-3 influenced regeneration (Barberan et al., 2016). Nine out of 11 copies of Egfr in E. andrei were consistently up-regulated at 6 hours after injury, suggesting that most expanded Egfr copies were possibly a gene dosage response to injury and regeneration (Shao et al., 2020) (Figure 1D). In vertebrates, time-series transcriptome analysis of sea lamprey embryogenesis showed that most ErbB genes were upregulated in the early (1–2.5 days) period and decreased later (3–5 days) post-fertilization (Bryant et al., 2016) (Figure 1E). Expression divergence of different ErbB genes was observed in our previous rat spinal cord development dataset (Yang et al., 2021) (Figure 1F).
Figure 1.
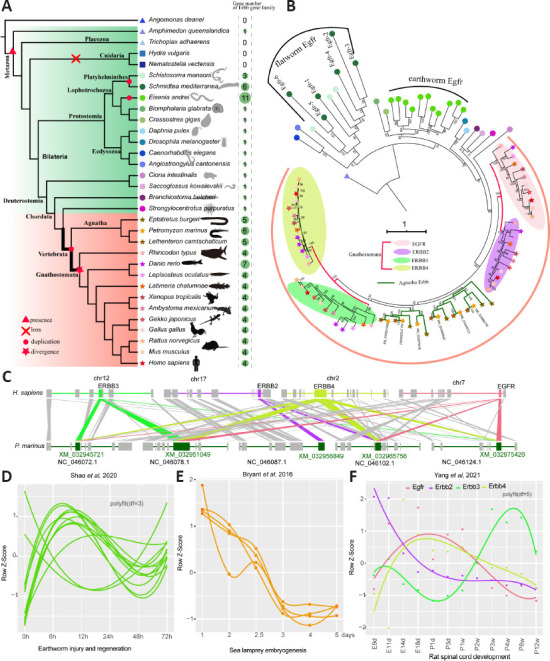
Identification and evolution of the ErbB gene family in Metazoa.
(A) Number of genes from the ErbB gene family that were found in 32 metazoan species. (B) The phylogeny of ErbB gene family members in 32 species. Four ellipses were used to label four typical members of the ErbB family in Gnathostomata. Egfr from A. queenslandica was used as a root. Leaf shapes and colors are the same as in (A) that depict different species. (C) Syntenic blocks containing ErbB genes that compare humans and lampreys. Shaded lines depict syntenic blocks of different ErbB genes in humans compared with that from lampreys. (D) Gene expression patterns of Efgr during the regeneration process of earthworms post-injury. Expression profiles were retrieved from the study (Shao et al., 2020). Smooth fit was performed using polyfit with df = 3. (E) Gene expression patterns of ErbB genes during lamprey embryogenesis. The gene expression profile was reanalyzed from the data (Bryant et al., 2016). Low expression levels or undetected genes are not shown. (F) Gene expression patterns of ErbB genes during rat spinal cord development from the embryonic stage to adulthood. Gene expression was retrieved from our previous study (Yang et al., 2021). Smooth fit was performed using polyfit with df = 5. Colors depict different members of the ErbB gene family.
Spatiotemporal expression of EGFR during spinal cord development
Egfr and Erbb2 from the ErbB gene family have important roles in tumor etiology and progression that are characterized by expression or activation of Egfr and altered expression of Erbb2 (Hynes and Lane, 2005; Tebbutt et al., 2013). In addition to cancer research, Egfr has been reported to be substantially upregulated in proximal nerve segments on days 4 and 7 after sciatic nerve transection (Gu et al., 2015). In this study, we focused on Egfr and its regulatory elements in the spinal cord. Egfr expression patterns during spinal cord development have not been systematically reported. Our previous RNA-seq data showed that Egfr expression was low in the early embryonic stage (E9d, E11d) and high in the later embryonic and neonatal stages (E18d–P1w). Expression decreased in adolescence and adulthood (P2w–P12w). These data were consistent with the qRT-PCR results for the developmental stages ranging from E11d to P8w (Figure 2A). Next, we investigated EGFR protein expression and cellular localization by immunohistochemistry. Consistent with the observation at the transcriptional level, immunostaining results showed low levels of EGFR protein at E11d (Figure 2B and C). At E14d, we detected EGFR-positive signals located approximately in the ependymal cells around the central canal of the rat spinal cord (Figure 2B). An obvious increase in EGFR protein was detected at E18d and was diffusely distributed within the spinal cord tissue. A relatively strong positive signal was observed in the anterior horn of gray matter (Figure 2B). In the P1d–P8w rats, EGFR protein levels were relatively stable and mostly confined to the ventral horn of the spinal cord with some co-localization with neurons (possibly motoneurons) (Figure 2B).
Figure 2.
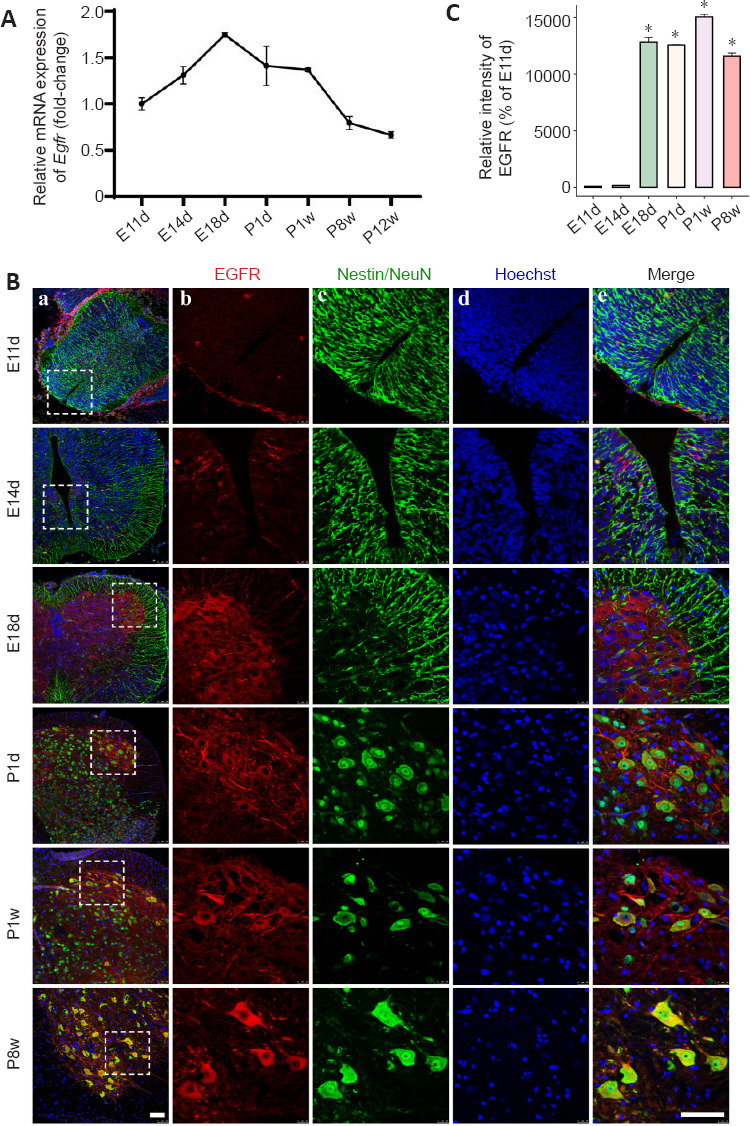
Spatiotemporal dynamics of EGFR expression during spinal cord development.
(A) qRT-PCR analysis of Egfr mRNA expression during spinal cord development. Data are expressed as means ± SEM (n = 3). The relative expression of Egfr from embryo day 11 (E11d) to postnatal week 12 (P12w). Expression levels were normalized to Gapdh. (B) Immunostaining for EGFR during spinal cord development. Colors depict EGFR-positive cells (red), neural stem cells (Nestin, green), or neuron (NeuN, green), and Hoechst 33342+ nuclei (blue). Individual stains (b–d) and merged (e) staining of markers within white boxed regions (a) shown on the left. Staining was performed at different time points during spinal cord development. E11d and E14d: embryo day 11 and 14, respectively; P1d: postnatal day 1; P1w and P8w: postnatal week 1 and 8, respectively. Left (a) and right (b-e) scale bars represent 100 μm. (C) Histogram depicts the relative fluorescence intensity of EGFR. All values are presented as the mean ± SEM (n = 3). *P < 0.05 vs. E11d (Student’s t-test).
Co-expression of Egfr-related genes inferred from rat spinal cord development data
Gene co-expression network analysis aids in identifying gene modules with potential biological functions and has been widely applied in biomedical research. Based on RNA-seq data from our previous rat spinal cord development study, we first constructed co-expression modules using weighted correlation network analysis. We identified a total of 27 modules of which Egfr and Erbb3 were contained in the blue module (Figure 3A). Integrating co-expression modules and interaction relationships retrieved from the STRING database, we extracted Egfr-neighboring subnetworks and labeled these genes as Egfr-related (Figure 3B) with a total of 67 genes. Gene ontology enrichment analysis of the 67 Egfr-related genes demonstrated that terms related to cell proliferation, blood vessel development, and ErbB signaling pathway were greatly enriched (Figure 3C). Gene expression profiles consisted of 65 genes with fragments per kilobase of exon per million (FPKM) greater than 1.0. The gene profiles showed that the majority of Egfr-related genes were upregulated in later development of the spinal cord (Figure 3D). We also examined the correlation (Pearson’s correlation coefficient r) of these genes with Egfr using our previous time-series transcriptome dataset from post-injury rat spinal cord (Yu et al., 2019). Forty-six out of 65 (71%) genes showed strong negative or positive correlations with Egfr expression (Figure 3D and Additional Table 1 (115.3KB, pdf) ; P < 0.05); for example, the correlation for Erbb3 was r = 0.73 (P = 2.94 × 10–17).
Figure 3.
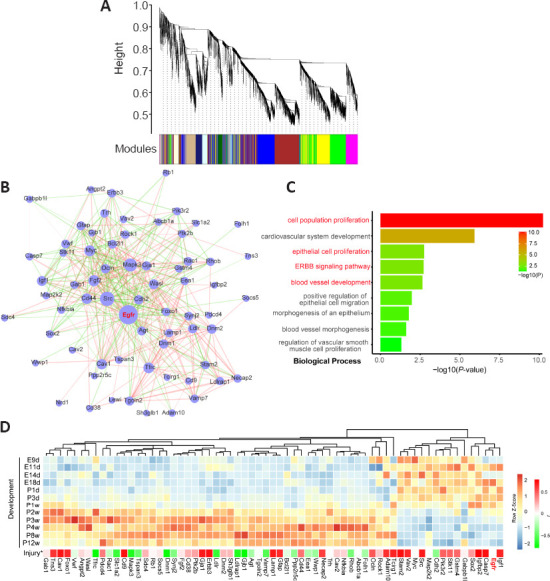
A co-expression module with Egfr inferred by weighted gene co-expression network analysis (WGCNA) based on an RNA-seq dataset of rat spinal cord development.
(A) Co-expressed modules inferred by the WGCNA method based on an RNA-seq dataset of rat spinal cord development and a blue module containing Egfr. (B) The subnetwork of first neighbors of Egfr and protein-protein interactions (PPIs) predicted from genes in the blue module. PPIs in the blue module were retrieved from the STRING database. (C) Gene ontology analysis of biological processes shows enrichment of genes in the subnetwork of first neighbors of Egfr. (D) Gene expression profile of genes in the subnetwork of first neighbors of Egfr. * Indicates the Pearson’s correlation (r) calculated based on the spinal cord injury dataset from Yu et al. (2019).
GPCR-mediated EGFR transactivation during spinal cord development
GPCRs have been reported to mediate transactivation of EGFR (Elliott et al., 2013; Forrester et al., 2016). GPCR-mediated transactivation of EGFR has been widely studied in cancers; however, data is limited for its role in spinal cord development. We found that five genes (Gpr153, Gpr21, Dear, Crcp, and Galr2) assigned to G protein-coupled receptor activity (GO:0004930) were clustered together with Egfr in the blue module. Previous studies have reported that some orphan receptors are involved in EGFR transactivation (Wang et al., 2010; Li et al., 2011; Fujiwara et al., 2012; Girgert et al., 2012; Ochiai et al., 2013; Cho-Clark et al., 2014). We determined that the two orphan receptor genes Gpr153 and Gpr21 showed positive correlations with the expression of Egfr (Pearson’s correlations of 0.87 and 0.56, respectively). Gpr153 was mainly expressed in the CNS (Fathi et al., 1998; Ruiz-Opazo et al., 1998). Mechanisms of GPCR-mediated EGFR transactivation include two types: the triple membrane passing signal (TMPS) and ligand-independent pathways. Our results showed that some important genes involved in the TMPS pathway were positively related to the expression of Egfr, such as Adam 1a, Adam4l1, Mmp16, Mmp17, Mmp23, Akt1, Sos2, Pik3cd, Raf1, and Map2k2 (Additional Table 2). In the ligand-independent pathway, three genes (Src, Fyn, and Lck) belonging to the Src family also showed a positive relationship with the expression of Egfr (Additional Table 2). The roles of these GPCR-related genes that are co-expressed with Egfr in rat spinal cord development require further experimental investigation.
Additional Table 2.
Key genes involved in TMPS pathway and ligand-independent pathway were positively
| GeneID | E9d | E11d | E14d | E18d | P1d | P3d | P1w | P2w | P3w | P4w | P8w | P12w | Pearson Correlation Coefficient with EGFR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adamla | 4.33 | 2.78 | 4.88 | 6.04 | 3.81 | 3.38 | 3.64 | 2.66 | 2.3 | 1.67 | 2.61 | 1.04 | 0.54 |
| Adam4l1 | 2.33 | 2.26 | 3.14 | 4.61 | 4.1 | 4.42 | 3.58 | 3.24 | 3.35 | 3.23 | 3.42 | 2.74 | 0.47 |
| Mmp16 | 9.36 | 11.77 | 13.45 | 18.54 | 16.72 | 14.01 | 13.42 | 9.78 | 7.46 | 5.11 | 6.11 | 5.99 | 0.78 |
| Mmp17 | 1.82 | 2.87 | 3.66 | 4.89 | 6.02 | 8.43 | 6.89 | 5.09 | 3.7 | 4.19 | 3.15 | 3.02 | 0.54 |
| Mmp23 | 0.55 | 1.47 | 0.57 | 0.26 | 0.35 | 0.32 | 0.44 | 0.35 | 0.3 | 0.16 | 0.13 | 0.12 | 0.41 |
| Aktl | 48.48 | 54.18 | 44.42 | 48.47 | 37.4 | 40.96 | 33.96 | 32.54 | 24.56 | 24.73 | 29.88 | 29.26 | 0.58 |
| Sos2 | 3.41 | 4.6 | 4.54 | 5.29 | 4.96 | 4.48 | 3.83 | 4.38 | 4.02 | 6.33 | 3.81 | 1.39 | 0.33 |
| Pik3cd | 0.7 | 1.58 | 1.75 | 1.6 | 0.81 | 0.76 | 0.86 | 0.66 | 0.49 | 0.4 | 0.83 | 1.44 | 0.33 |
| Rafl | 37.19 | 39.36 | 43.79 | 37.06 | 35.45 | 28.3 | 24.82 | 21.81 | 16.68 | 16.12 | 16.56 | 12.13 | 0.48 |
| Map2k2 | 33.52 | 36.83 | 32.8 | 33.97 | 33.75 | 34.66 | 28.06 | 22.92 | 19.25 | 19.79 | 29.77 | 28.88 | 0.5 |
| Src | 7.95 | 7.79 | 10.19 | 5.49 | 8.19 | 8.6 | 7.02 | 5.18 | 4.69 | 3.9 | 4.77 | 4.97 | 0.27 |
| Fyn | 23.67 | 22.22 | 49.59 | 52.92 | 65.71 | 69.96 | 77.95 | 76.94 | 59.5 | 39.67 | 40.52 | 26.04 | 0.36 |
| Lck | 0.59 | 0.07 | 1.4 | 1.21 | 1.3 | 1.31 | 1.31 | 0.39 | 0.23 | 0.08 | 0.23 | 0.46 | 0.43 |
| EGFR | 1.14 | 3.18 | 1.29 | 3.67 | 2.07 | 2.84 | 3 | 1.98 | 1.18 | 0.94 | 1.25 | 0.78 | - |
Prediction of miRNAs targeting Egfr
MiRNAs are a class of small, regulatory non-coding RNAs that normally bind to the 3’-UTR of target genes and influence gene expression at the post-transcriptional level (Bushati and Cohen, 2007). There are 59 human and 17 mouse miRNAs recorded in the TarBase database that are experimentally supported. Various miRNAs targeting humans and mouse Egfr have been widely investigated, but only miR-128-3p has been reported in rats. MiRNAs generally negatively regulate gene expression of their targets. Thus, we combined three widely applied prediction tools (PITA, miRanda, and TargetScan) and calculated correlations with Egfr expression to predict candidate miRNAs targeting Egfr in rats. In total, 120 miRNAs were supported by at least two programs of which 15 miRNAs were predicted by three of the programs (Figure 4A). To select miRNAs that may regulate Egfr during spinal cord development, we calculated Pearson’s correlation coefficients between 121 miRNAs, including miR-128-3p, and Egfr. Finally, 25 miRNAs showed a significant negative correlation with Egfr expression (Pearson’s correlation coefficients were –0.67 to –0.58; Figure 4B and Additional Table 3). Of the 25 candidates, three miRNAs (miR-34a-5p, miR-21-3p, and let-7b-5p) have been reported to target human EGFR and five miRNAs (miR-1843a-5p and let-7a, 7b, 7c, and 7e) have been reported to target mouse Egfr according to miRNA-target pairs deposited in Tarbase. Phylogenetic analysis of the let-7 family (six members: let-7a–f) in humans, mice, and rats demonstrated that five members (let-7b, 7c, 7d, 7e, and 7f) are conserved between these three species and, except for let-7e, they are conserved in their 5’ seed regions (nucleotides 2–8) (Figure 4C). These data suggested that let-7 in the rat may also target Egfr. Additionally, retrieval of publications deposited in PubMed showed that 13 out of 25 miRNAs have been reported to play a role in inhibiting cell proliferation (Wang et al., 2011, 2019b, c; Kim et al., 2012, 2015; Zhao et al., 2013; Xu et al., 2014; Yang et al., 2015, 2019; Lu et al., 2016, 2017; Fang et al., 2017; Wu et al., 2017; Sun et al., 2018) in humans or mice by targeting Egfr or other targets and include miR-330-5p, miR-29c-3p, and miR-34a-5p (Figure 4B).
Figure 4.
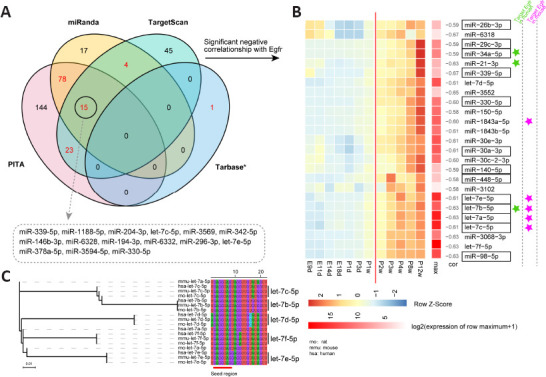
Prediction of miRNAs that target the 3’-UTR of rat Egfr.
(A) Venn diagram of predicted miRNAs targeting the 3’-UTR of rat Egfr using three programs and supported by experimentally validated data from the Tarbase database (indicated by *). Red numbers indicate miRNAs that were predicted by at least two programs or supported by experimental evidence. Fifteen miRNAs supported by three programs are shown in the dashed rectangle. (B) Expression of 25 candidate miRNAs that target Egfr in silico that were predicted based on the spinal cord development dataset. Boxed text indicates that miRNAs were reported to inhibit cell proliferation in humans or mice. MiRNAs with significant negative correlations (P < 0.05) with Egfr expression are shown. (C) Phylogenetic analysis of the let-7 family in humans, mice, and rats. Seed sequences of let-7c, 7b, 7d, 7f, and 7e are highlighted with a red line.
Additional Table 3.
Predicted miRNA targeting Egfr
| miRNA ID | Pearson’s correlation | Expression | Prediction | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||||
| cor | pval | E9d | E11d | E14d | E18d | P1d | P3d | P1w | P2w | P3w | P4w | P8w | P12w | PITA | miRanda | TargetScan | Number of support | |
| mo-let-7a-5p | -0.62625 | 0.029354 | 6477.333 | 6403.333 | 16388.67 | 21683.33 | 17401.33 | 21453 | 37934.67 | 53476.33 | 86457.33 | 118870.3 | 106036.3 | 124852.7 | 1 | 0 | 1 | 2 |
| mo-let-7b-5p | -0.627513 | 0.028932 | 223.6667 | 9.333333 | 554.3333 | 654.6667 | 620 | 868.6667 | 1761 | 2123.667 | 3526.667 | 4466.667 | 4260 | 5667.333 | 1 | 1 | 0 | 2 |
| rno-let-7c-5p | -0.611147 | 0.03475 | 7232.667 | 2547.667 | 20106.67 | 25500 | 18315 | 23583 | 39117 | 49975.33 | 75005.33 | 97705 | 85402.33 | 108606.3 | 1 | 1 | 1 | 3 |
| rno-let-7d-5p | -0.608652 | 0.035705 | 263 | 120.3333 | 586 | 878.6667 | 891.6667 | 1520 | 2732 | 4329 | 5791.333 | 6685.667 | 7998 | 9502 | 1 | 0 | 1 | 2 |
| rno-let-7e-5p | -0.609466 | 0.035391 | 3816 | 11203 | 8543 | 9899 | 9281 | 9472.333 | 19049 | 27465 | 43790.33 | 63673.33 | 44901.67 | 56654.33 | 1 | 1 | 1 | 3 |
| rno-let-7f-5p | -0.626573 | 0.029246 | 18170.33 | 12439.67 | 38217 | 68662.67 | 73109.67 | 78295.67 | 173574.3 | 251508 | 463744.3 | 561346.3 | 472311.3 | 526314.3 | 1 | 0 | 1 | 2 |
| rno-miR-140-5p | -0.590189 | 0.043365 | 10.66667 | 8.666667 | 6.333333 | 5.666667 | 3.333333 | 6.666667 | 11.33333 | 11.66667 | 11.33333 | 20.33333 | 28.33333 | 35 | 1 | 1 | 0 | 2 |
| rno-miR-150-5p | -0.582893 | 0.046689 | 33.66667 | 22.33333 | 78 | 139.6667 | 108.6667 | 243.3333 | 383.3333 | 707.6667 | 937 | 1075 | 1852 | 2634.333 | 1 | 1 | 0 | 2 |
| rno-miR-1843a-5p | -0.603038 | 0.037923 | 51.66667 | 39.33333 | 143.3333 | 246.3333 | 272.6667 | 560.6667 | 667 | 1032.667 | 1771.333 | 2236 | 3131.667 | 4342 | 1 | 1 | 0 | 2 |
| rno-miR-1843b-5p | -0.609339 | 0.03544 | 20 | 18.66667 | 58.33333 | 75.66667 | 99.66667 | 240.3333 | 299.6667 | 499.3333 | 793.3333 | 950.3333 | 1314 | 1811 | 1 | 1 | 0 | 2 |
| rno-miR-21-3p | -0.62683 | 0.02916 | 12.33333 | 14.33333 | 4.333333 | 1.666667 | 4 | 7 | 10 | 24.33333 | 33.33333 | 30.33333 | 38 | 73.33333 | 1 | 1 | 0 | 2 |
| rno-miR-26b-3p | -0.590143 | 0.043385 | 40 | 38.33333 | 16 | 8 | 6.666667 | 9 | 23.33333 | 32.66667 | 32.66667 | 38.66667 | 52 | 48.66667 | 1 | 1 | 0 | 2 |
| rno-miR-29c-3p | -0.586028 | 0.04524 | 2.333333 | 6 | 5.333333 | 12 | 11.33333 | 32.33333 | 37.66667 | 91.33333 | 157 | 229.3333 | 362 | 623.3333 | 1 | 1 | 0 | 2 |
| rno-miR-3068-3p | -0.631561 | 0.027609 | 149.6667 | 171.3333 | 171.3333 | 176.6667 | 203 | 288.3333 | 447 | 614 | 1067.333 | 1051 | 1090.333 | 1280.333 | 1 | 1 | 0 | 2 |
| rno-miR-30a-3p | -0.609427 | 0.035407 | 2825 | 3624.667 | 2524.333 | 1981 | 1386.333 | 2170 | 3178.333 | 4582.333 | 4628 | 5750 | 6397 | 6921 | 1 | 1 | 0 | 2 |
| rno-miR-30c-2-3p | -0.602544 | 0.038123 | 805.3333 | 884.3333 | 485.3333 | 532 | 364 | 486 | 642.6667 | 883.6667 | 1061 | 1433.667 | 1908.667 | 2270.667 | 1 | 1 | 0 | 2 |
| rno-miR-30e-3p | -0.609498 | 0.035379 | 3039 | 3778 | 2565.333 | 2113 | 1621.333 | 2840.667 | 3867 | 5414 | 5898 | 6972 | 7936 | 8478 | 1 | 1 | 0 | 2 |
| rno-miR-3102 | -0.579426 | 0.04833 | 0.666667 | 1 | 0 | 1 | 1 | 0 | 0.666667 | 0.666667 | 3.333333 | 3 | 2.333333 | 4 | 1 | 1 | 0 | 2 |
| rno-miR-330-5p | -0.604335 | 0.037402 | 76.66667 | 54.66667 | 62 | 61 | 74.33333 | 196 | 312.3333 | 696.6667 | 702.6667 | 833 | 1303 | 1552.667 | 1 | 1 | 1 | 3 |
| rno-miR-339-5p | -0.665811 | 0.018102 | 16 | 4.666667 | 2 | 2.333333 | 2.333333 | 3.333333 | 10 | 15.66667 | 16.66667 | 21.66667 | 26.33333 | 47.33333 | 1 | 1 | 1 | 3 |
| rno-miR-34a-5p | -0.594402 | 0.041523 | 3.666667 | 1 | 4 | 3.666667 | 11.66667 | 28.66667 | 52.33333 | 125.3333 | 239.6667 | 294 | 441 | 864 | 1 | 1 | 0 | 2 |
| rno-miR-3552 | -0.646476 | 0.02311 | 0 | 0 | 0 | 0.333333 | 1 | 1.333333 | 2.666667 | 9 | 11.33333 | 15 | 14.66667 | 22.66667 | 1 | 1 | 0 | 2 |
| rno-miR-448-5p | -0.580258 | 0.047933 | 1 | 0 | 0 | 0.666667 | 3.666667 | 6 | 6.666667 | 15 | 34.33333 | 15 | 18 | 31.33333 | 1 | 1 | 0 | 2 |
| rno-miR-6318 | -0.666447 | 0.017952 | 5.333333 | 4.333333 | 3.333333 | 0.333333 | 0.333333 | 0.333333 | 2.333333 | 4.666667 | 5.666667 | 6.333333 | 2.333333 | 5.666667 | 1 | 1 | 0 | 2 |
| rno-miR-98-5p | -0.631547 | 0.027614 | 535 | 221 | 1423.333 | 1830.333 | 1497.667 | 2475 | 4337.333 | 7388 | 12424 | 13643.33 | 13461.33 | 14301 | 1 | 0 | 1 | 2 |
Potential TFs regulating Egfr in silico and confirmed in vitro
TFs are the key cellular components that control gene expression via cis-regulatory activity. We predicted potential TFs in rats that regulate Egfr expression by combining multiple methods (Figure 5A). A total of nine TFs were identified as candidates (Figure 5B). A dual-luciferase reporter assay was used to determine the targeting relationship between the nine TFs and Egfr in vitro. Six out of nine TFs showed a significant change in Egfr reporter activity compared with that in the control (P < 0.05; Figure 5C). KLF1 and STAT3 significantly repressed Egfr reporter activity and FOXA2, SOX17, BHLHA15, and DDIT3 activated the Egfr reporter (P < 0.05; Figure 5C). These data suggested that these six TFs may be regulators of Egfr in rats. We also used the same methods to predict TFs that regulate mouse Egfr and human EGFR. Five TFs (FOXA2, KLF4, STAT3, SP1, and KLF1) were shared by the three species (Figure 5D). KLF4 and STAT3 were supported by ChIP-seq results for humans deposited in the ChIPBase database, and SP1 activation of EGFR has been experimentally validated (Tsai et al., 2019).
Figure 5.
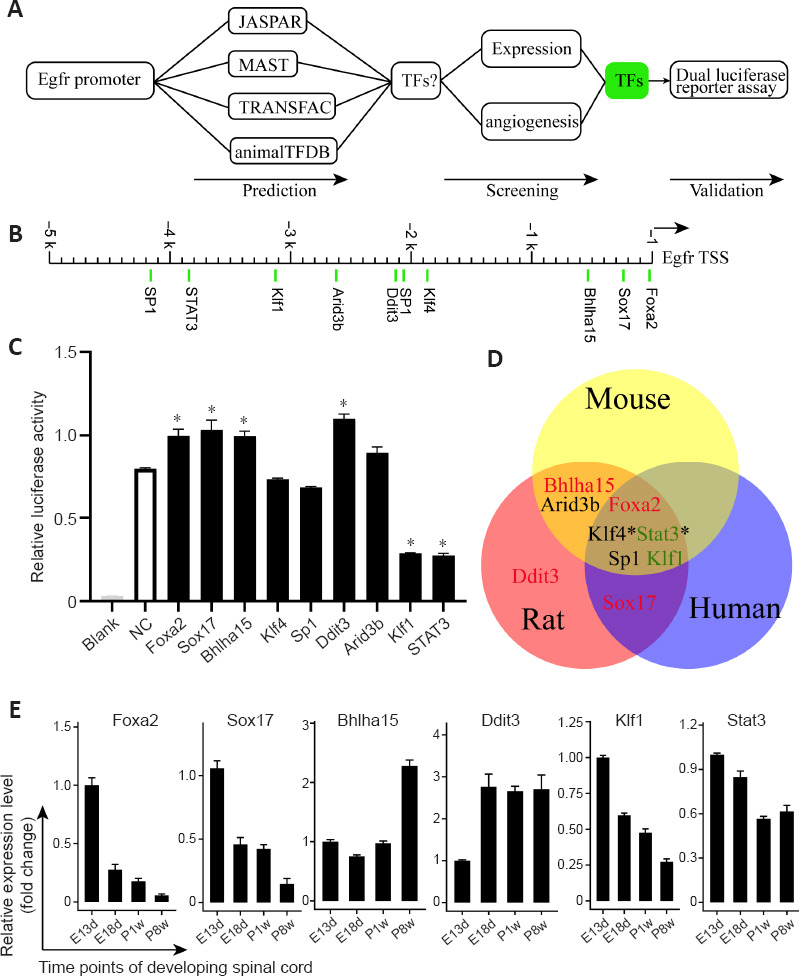
Prediction of transcription factors that regulate Egfr and the effect of nine predicted transcription factors on the Egfr promoter with bidirectional activity.
(A) Workflow for screening potential transcription factors (TFs) that regulate Egfr expression. (B) Predicted TF binding loci within the rat Egfr promoter. (C) TF-mediated activation or repression of Egfr reporter activity using dual-luciferase assays. The reporters and effectors were co-expressed in 293T cells, and both REN and LUC activity were measured. The relative LUC activities normalized to the REN activities are shown. Results are presented as the mean ± SD from three independent experiments, each with three replicates. *P < 0.05 (one-way analysis of variance followed by the Dunnett multiple comparison test). (D) Three potential conserved transcription factors of Egfr in humans, mice, and rats. The star (*) indicates support from ChIP-seq data in humans. (E) Relative mRNA expression of six candidate TFs at embryonic days 18 (E18d) and postnatal week 1 (P1w) and 8 (P8w) was compared with the expression at embryonic day 13 (E13d). Expression levels of target genes were normalized to Gapdh. Data are expressed as means ± SEM (n = 3).
Expression of six candidate TFs during spinal cord development
We investigated whether FOXA2, SOX17, BHLHA15, DDIT3, KLF1, and STAT3 were expressed during spinal cord development at key developmental stages using qRT-PCR and immunohistochemistry. According to the expression pattern of Egfr mRNA during spinal cord development (Figure 2A), we selected several time points (E13d, E18d, P1w, and P8w). The qRT-PCR demonstrated that Foxa2, Klf1, Sox17, and Stat3 were relatively highly expressed during embryonic development (Figure 5E), which then gradually decreased, while Ddit3 expression was relatively low at E13d and increased by E18d. Bhlha15 showed relatively high expression in adult rats (P8w). The differences in expression of some TFs and their effects on EGFR promoter-luciferase activity may be caused by the detection of pooled mixed mRNA expression levels from different regions of the spinal cord. Immunostaining indicated that FOXA2 was highly expressed at E13d, and expression decreased by E18d and P1w and was undetectable at P8w (Figure 6A and B). Undetectable positive signals for the other five TFs are not shown. FOXA2 was co-localized within the cell nucleus, and FOXA2-positive cells were found in the floor plate suggesting the importance of FOXA2 in spinal cord development, which deserves further investigation.
Figure 6.
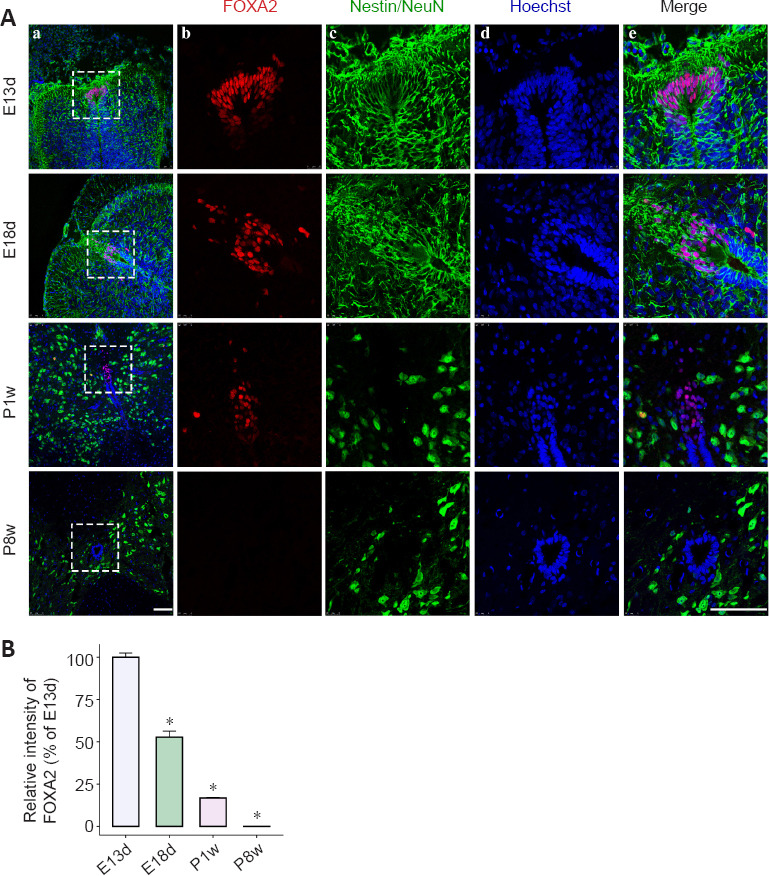
Spatiotemporal dynamics of FOXA2 expression during spinal cord development.
(A) Immunostaining for FOXA2 during spinal cord development. Colors depict FOXA2-positive cells (red), neural stem cells (Nestin) or neurons (NeuN) (green), and Hoechst 33342+ nuclei (blue). Individual stains (b–d) and merged staining (e) of markers within the white boxed regions shown on the left (a) at different time points during spinal cord development. Left scale bar and right scale bars (b–e) are 100 μm. FP: Floor plate. (B) Histogram depicts the relative fluorescence intensity of FOXA2. E13d and E18d: embryo day 13 and 18, respectively; P1w and P8w: postnatal week 1 and 8, respectively. All values are presented as means ± SEM (n = 3). *P < 0.05 vs. E13d (Student’s t-test).
Discussion
EGFR belongs to the receptor tyrosine kinase superfamily and plays an essential role in embryonic development and adult tissue homeostasis, including differentiation, growth, cell maintenance, and repair of damaged tissues. Egfr expression has been explored at specific stages of spinal cord development (Romano and Bucci, 2020); however, evaluation of Egfr expression patterns spanning developmental stages from neural tube formation to the adult spine has not been reported. RNA-seq and qPCR results demonstrated a bell-shaped Egfr expression pattern, which peaked near birth. Immunofluorescence staining showed that EGFR protein levels accumulated in and were confined to the ventral horn of the adult spine. EGFR showed inconsistent expression patterns at the mRNA and protein level, but FOXA2 showed consistent expression patterns. Protein abundance is affected by a series of linked processes, including transcription, post-transcriptional modification, translation, localization, post-translational modification, and protein degradation (Vogel and Marcotte, 2012). FOXA2 is a developmental transcription factor that is specifically expressed in the spine floor plate and is tightly controlled because knockdown or misexpression causes an abnormal floor plate (Bayly et al., 2012). EGFR has a complex structure with an extracellular ligand-binding domain, a single transmembrane domain, and a cytoplasmic domain. It can homo- or hetero-dimerize or form dimers with other proteins, activate multiple signal pathways, and is widely expressed in multiple organs and cells. We hypothesized that inconsistent Egfr mRNA and protein levels may be caused by protein modifications or estimates from pooled mixed mRNA expression levels from very different regions of the whole spinal cord. MiRNA-mediated gene regulation has been implicated in development and various diseases. Studies of the regulatory relationships between miRNAs and Egfr have been mainly focused on humans and mice, and only one miRNA has been reported in the rat. We predicted several miRNAs that may regulate Egfr expression during spinal cord development in rats, and some of these miRNAs have been shown to inhibit cell proliferation in other species. EGFR is widely distributed in the CNS and plays an important role in the maintenance of the pool of neural stem cells, maturation and function of astrocytes, oligodendrogenesis, and neurite growth. The contradictory effects of EGFR on nerve regeneration largely rely on the specific target cells. The changes in intracellular transport and signal transduction of EGFR are associated with the occurrence and progression of neurodegenerative diseases and injuries (Le Pichon et al., 2013; Chen et al., 2019), suggesting that regulating Egfr expression or EGFR-mediated signal transduction through upstream regulators may contribute to facilitating nerve regeneration and combating neurodegenerative diseases.
Because of the importance of the ErbB gene family, several studies (Stein and Staros, 2006; Liu et al., 2013; Barberan et al., 2016) have discussed the origin and evolution of ErbB genes in vertebrate and invertebrate animals. A vast amount of published genomic data is available to provide details regarding ErbB evolution in animals. The decoded genomes of flatworms and earthworms (Eisenia andrei) showed expansion of Egfr and revealed its relationship with regeneration (Barberan et al., 2016; Shao et al., 2020). Liu et al. (2013) analyzed many vertebrate species but lacked species from Agnatha, which are related to all vertebrates and represent the ancestral vertebrate. In this study, we added three species from Agnatha (Eptatretus burgeri, Petromyzon marinus, and Lethenteron camtschaticum) and the whale shark (Rhincodon typus) (Read et al., 2017), which is the sister group of the remaining gnathostomes. Our analysis indicated that Egfr was duplicated in ancient vertebrates via small-scale duplication but then diverged into other ErbB members after the speciation of gnathostomes. This was generally consistent with the previous study (Liu et al., 2013) but provided a more accurate node (Gnathostomata) for its divergence.
Egfr is not only involved in the development of the CNS but also participates in the biological process of CNS injury and repair (Barberan et al., 2016). It has been shown that low expression of Egfr normally occurs in astrocytes but high expression is observed after injury (Scholze et al., 2014). EGFR can activate astrocytes, which contributes to the formation of glial scars (Erschbamer et al., 2007). Understanding the regulators of Egfr expression is important in CNS development, injury, and regeneration. Transactivation of EGFR has been reported to be mediated by an expanding repertoire of GPCRs and promotes cell survival (Grisanti et al., 2017). We identified several factors that may be involved in transactivation. Several TFs that may regulate Egfr expression were identified in this study. FOXA2 opens compacted chromatin for other transcription-related proteins by displacing nucleosomal core histones and participates in embryonic development and regulation of tissue-specific gene expression (Lee et al., 2019). Our experiments showed that FOXA2 was tightly related to spinal cord embryonic development.
This study has limitations. First, functional experiments, such as RNA interference (RNAi) and over-expression, were not conducted in this study to specifically determine the roles of EGFR and other factors in spinal cord injury. Second, the expression of transcription factors screened by dual-luciferase reporter assay was not consistent with EGFR expression during spinal cord development, which may have resulted from pooling mixed expression levels from different regions of spinal cord tissue. Further studies will be required to confirm and determine the regulatory activities of the identified TFs and miRNAs on Egfr expression in vivo during spinal cord development and injury. In future studies, we plan to determine the expression of these regulators after spinal cord injury and compare expression to that during spinal cord development. Development-specific regulators will be over-expressed in the spinal cord after injury and effects on spinal cord repair will be evaluated.
In conclusion, we elucidated the long-term expression patterns of EGFR during rat spinal cord development and the evolutional complexity and diversity of the ErbB gene family in Metazoa. Initially screened candidate regulators that mediated Egfr expression were identified and require further investigation, including overexpression and RNAi studies as mentioned above. Our study provides new insight into the evolution of the ErbB gene family and regulators in Egfr expression at the transcriptional or post-transcriptional levels based on the long-term profile of spinal cord development. Given the importance of Egfr in the development and regeneration of the nervous system, the regulatory factors identified in this study may provide new candidate targets for CNS regeneration and disease treatment in the future.
Additional files:
Additional Table 1 (115.3KB, pdf) : Pearson’s correlation with Egfr expression based on spinal cord injury dataset.
Pearson's correlation with Egfr expression based on spinal cord injury dataset
Additional Table 2: Key genes involved in TMPS pathway and ligand-independent pathway were positively.
Additional Table 3: Predicted miRNA targeting Egfr.
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
Editor note: XSG is an Editorial Board member of Neural Regeneration Research. He was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal’s standard procedures, with peer review handled independently of this Editorial Board member and their research groups.
Availability of data and materials: RNA-Seq and miRNA-Seq data for different development stages of the rat spinal cord have been deposited in the NCBI database with BioProject accession tag PRJNA505253. All data generated or analyzed during this study are included in this published article and its supplementary information files.
Funding: This work was supported by the National Natural Science Foundation of China, No. 31730031 (to XSG), the Natural Science Foundation of Jiangsu Province, No. BK20202013 (to XSG), and the Jiangsu Provincial Key Medical Center and Priority Academic Program Development of Jiangsu High Education Institutions (PAPD) (to XSG).
C-Editor: Zhao M; S-Editor: Li CH; L-Editors: Zunino S, Song LP; T-Editor: Jia Y
References
- 1.Adler CE, Seidel CW, McKinney SA, Sanchez Alvarado A. Selective amputation of the pharynx identifies a FoxA-dependent regeneration program in planaria. Elife. 2014;3:e02238. doi: 10.7554/eLife.02238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Ayati A, Moghimi S, Salarinejad S, Safavi M, Pouramiri B, Foroumadi A. A review on progression of epidermal growth factor receptor (EGFR) inhibitors as an efficient approach in cancer targeted therapy. Bioorg Chem. 2020;99:103811. doi: 10.1016/j.bioorg.2020.103811. [DOI] [PubMed] [Google Scholar]
- 5.Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 6.Barberan S, Martin-Duran JM, Cebria F. Evolution of the EGFR pathway in Metazoa and its diversification in the planarian Schmidtea mediterranea. Sci Rep. 2016;6:28071. doi: 10.1038/srep28071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayly RD, Brown CY, Agarwala S. A novel role for FOXA2 and SHH in organizing midbrain signaling centers. Dev Biol. 2012;369:32–42. doi: 10.1016/j.ydbio.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryant SA, Herdy JR, Amemiya CT, Smith JJ. Characterization of somatically-eliminated genes during development of the sea lamprey (Petromyzon marinus) Mol Biol Evol. 2016;33:2337–2344. doi: 10.1093/molbev/msw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 10.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Zeng F, Forrester SJ, Eguchi S, Zhang MZ, Harris RC. Expression and function of the epidermal growth factor receptor in physiology and disease. Physiol Rev. 2016;96:1025–1069. doi: 10.1152/physrev.00030.2015. [DOI] [PubMed] [Google Scholar]
- 12.Chen YJ, Hsu CC, Shiao YJ, Wang HT, Lo YL, Lin AMY. Anti-inflammatory effect of afatinib (an EGFR-TKI) on OGD-induced neuroinflammation. Sci Rep. 2019;9:2516. doi: 10.1038/s41598-019-38676-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chia CM, Winston RM, Handyside AH. EGF, TGF-alpha and EGFR expression in human preimplantation embryos. Development. 1995;121:299–307. doi: 10.1242/dev.121.2.299. [DOI] [PubMed] [Google Scholar]
- 14.Cho-Clark M, Larco DO, Semsarzadeh NN, Vasta F, Mani SK, Wu TJ. GnRH-(1-5) transactivates EGFR in Ishikawa human endometrial cells via an orphan G protein-coupled receptor. Mol Endocrinol. 2014;28:80–98. doi: 10.1210/me.2013-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott KJ, Bourne AM, Takayanagi T, Takaguri A, Kobayashi T, Eguchi K, Eguchi S. ADAM17 silencing by adenovirus encoding miRNA-embedded siRNA revealed essential signal transduction by angiotensin II in vascular smooth muscle cells. J Mol Cell Cardiol. 2013;62:1–7. doi: 10.1016/j.yjmcc.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erschbamer M, Pernold K, Olson L. Inhibiting epidermal growth factor receptor improves structural, locomotor sensory and bladder recovery from experimental spinal cord injury. J Neurosci. 2007;27:6428–6435. doi: 10.1523/JNEUROSCI.1037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang Z, Yin S, Sun R, Zhang S, Fu M, Wu Y, Zhang T, Khaliq J, Li Y. miR-140-5p suppresses the proliferation, migration and invasion of gastric cancer by regulating YES1. Mol Cancer. 2017;16:139. doi: 10.1186/s12943-017-0708-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fathi Z, Battaglino PM, Iben LG, Li H, Baker E, Zhang D, McGovern R, Mahle CD, Sutherland GR, Iismaa TP, Dickinson KE, Zimanyi IA. Molecular characterization, pharmacological properties and chromosomal localization of the human GALR2 galanin receptor. Brain Res Mol Brain Res. 1998;58:156–169. doi: 10.1016/s0169-328x(98)00116-8. [DOI] [PubMed] [Google Scholar]
- 21.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 22.Forrester SJ, Kawai T, O'Brien S, Thomas W, Harris RC, Eguchi S. Epidermal growth factor receptor transactivation: mechanisms, pathophysiology and potential therapies in the cardiovascular system. Annu Rev Pharmacol Toxicol. 2016;56:627–653. doi: 10.1146/annurev-pharmtox-070115-095427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu X, Mao X, Wang Y, Ding X, Li Y. Let-7c-5p inhibits cell proliferation and induces cell apoptosis by targeting ERCC6 in breast cancer. Oncol Rep. 2017;38:1851–1856. doi: 10.3892/or.2017.5839. [DOI] [PubMed] [Google Scholar]
- 24.Fujiwara S, Terai Y, Kawaguchi H, Takai M, Yoo S, Tanaka Y, Tanaka T, Tsunetoh S, Sasaki H, Kanemura M, Tanabe A, Yamashita Y, Ohmichi M. GPR30 regulates the EGFR-Akt cascade and predicts lower survival in patients with ovarian cancer. J Ovarian Res. 2012;5:35. doi: 10.1186/1757-2215-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ge H, Yan Y, Yue C, Liang C, Wu J. Long noncoding RNA LINC00265 targets EGFR and promotes deterioration of colorectal cancer: a comprehensive study based on data mining and in vitro validation. Onco Targets Ther. 2019;12:10681–10692. doi: 10.2147/OTT.S227482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Girgert R, Emons G, Grundker C. Inactivation of GPR30 reduces growth of triple-negative breast cancer cells: possible application in targeted therapy. Breast Cancer Res Treat. 2012;134:199–205. doi: 10.1007/s10549-012-1968-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gout JF, Lynch M. Maintenance and loss of duplicated genes by dosage subfunctionalization. Mol Biol Evol. 2015;32:2141–2148. doi: 10.1093/molbev/msv095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grisanti LA, Guo S, Tilley DG. Cardiac GPCR-mediated EGFR transactivation: impact and therapeutic implications. J Cardiovasc Pharmacol. 2017;70:3–9. doi: 10.1097/FJC.0000000000000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu Y, Chen C, Yi S, Wang S, Gong L, Liu J, Gu X, Zhao Q, Li S. miR-sc8 inhibits Schwann cell proliferation and migration by targeting Egfr. PLoS One. 2015;10:e0145185. doi: 10.1371/journal.pone.0145185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo N, Zhao Y, Zhang W, Li S, Li S, Yu J. MicroRNA-133a downregulated EGFR expression in human non-small cell lung cancer cells via AKT/ERK signaling. Oncol Lett. 2018;16:6045–6050. doi: 10.3892/ol.2018.9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 32.Kageyama R, Merlino GT, Pastan I. A transcription factor active on the epidermal growth factor receptor gene. Proc Natl Acad Sci U S A. 1988;85:5016–5020. doi: 10.1073/pnas.85.14.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou D, Kavakiotis I, Maniou S, Skoufos G, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018;46:D239–D245. doi: 10.1093/nar/gkx1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 35.Kim BK, Yoo HI, Choi K, Yoon SK. miR-330-5p inhibits proliferation and migration of keratinocytes by targeting Pdia3 expression. FEBS J. 2015;282:4692–4702. doi: 10.1111/febs.13523. [DOI] [PubMed] [Google Scholar]
- 36.Kim SJ, Shin JY, Lee KD, Bae YK, Sung KW, Nam SJ, Chun KH. MicroRNA let-7a suppresses breast cancer cell migration and invasion through downregulation of C-C chemokine receptor type 7. Breast Cancer Res. 2012;14:R14. doi: 10.1186/bcr3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klapper LN, Kirschbaum MH, Sela M, Yarden Y. Biochemical and clinical implications of the ErbB/HER signaling network of growth factor receptors. Adv Cancer Res. 2000;77:25–79. [PubMed] [Google Scholar]
- 38.Knudsen S. Promoter2.0: for the recognition of PolII promoter sequences. Bioinformatics. 1999;15:356–361. doi: 10.1093/bioinformatics/15.5.356. [DOI] [PubMed] [Google Scholar]
- 39.Knudsen SL, Mac AS, Henriksen L, van Deurs B, Grovdal LM. EGFR signaling patterns are regulated by its different ligands. Growth Factors. 2014;32:155–163. doi: 10.3109/08977194.2014.952410. [DOI] [PubMed] [Google Scholar]
- 40.Kornblum HI, Hussain RJ, Bronstein JM, Gall CM, Lee DC, Seroogy KB. Prenatal ontogeny of the epidermal growth factor receptor and its ligand, transforming growth factor alpha in the rat brain. J Comp Neurol. 1997;380:243–261. doi: 10.1002/(sici)1096-9861(19970407)380:2<243::aid-cne7>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 41.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 42.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le Pichon CE, Dominguez SL, Solanoy H, Ngu H, Lewin-Koh N, Chen M, Eastham-Anderson J, Watts R, Scearce-Levie K. EGFR inhibitor erlotinib delays disease progression but does not extend survival in the SOD1 mouse model of ALS. PLoS One. 2013;8:e62342. doi: 10.1371/journal.pone.0062342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee K, Cho H, Rickert RW, Li QV, Pulecio J, Leslie CS, Huangfu D. FOXA2 is required for enhancer priming during pancreatic differentiation. Cell Rep. 2019;28:382–393. doi: 10.1016/j.celrep.2019.06.034. e387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256-259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li G, Deng X, Wu C, Zhou Q, Chen L, Shi Y, Huang H, Zhou N. Distinct kinetic and spatial patterns of protein kinase C (PKC)- and epidermal growth factor receptor (EGFR)-dependent activation of extracellular signal-regulated kinases 1 and 2 by human nicotinic acid receptor GPR109A. J Biol Chem. 2011;286:31199–31212. doi: 10.1074/jbc.M111.241372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li YL, Liu XM, Zhang CY, Zhou JB, Shao Y, Liang C, Wang HM, Hua ZY, Lu SD, Ma ZL. MicroRNA-34a/EGFR axis plays pivotal roles in lung tumorigenesis. Oncogenesis. 2017;6:e372. doi: 10.1038/oncsis.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, He W, Long J, Pang F, Xian L, Chen M, Wu Y, Hu Y. Natural selection and functional diversification of the epidermal growth factor receptor EGFR family in vertebrates. Genomics. 2013;101:318–325. doi: 10.1016/j.ygeno.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 50.Lu Y, Hu J, Sun W, Li S, Deng S, Li M. MiR-29c inhibits cell growth, invasion and migration of pancreatic cancer by targeting ITGB1. Onco Targets Ther. 2016;9:99–109. doi: 10.2147/OTT.S92758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, Zhang AW, Parcy F, Lenhard B, Sandelin A, Wasserman WW. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44:D110–115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matys V, Fricke E, Geffers R, Gössling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Münch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mizuguchi A, Yamashita S, Yokogami K, Morishita K, Takeshima H. Ecotropic viral integration site 1 regulates EGFR transcription in glioblastoma cells. J Neurooncol. 2019;145:223–231. doi: 10.1007/s11060-019-03310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morris JK, Lin W, Hauser C, Marchuk Y, Getman D, Lee KF. Rescue of the cardiac defect in ErbB2 mutant mice reveals essential roles of ErbB2 in peripheral nervous system development. Neuron. 1999;23:273–283. doi: 10.1016/s0896-6273(00)80779-5. [DOI] [PubMed] [Google Scholar]
- 55.Mundade R, Ozer HG, Wei H, Prabhu L, Lu T. Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism epigenetic marks and beyond. Cell Cycle. 2014;13:2847–2852. doi: 10.4161/15384101.2014.949201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ochiai S, Furuta D, Sugita K, Taniura H, Fujita N. GPR87 mediates lysophosphatidic acid-induced colony dispersal in A431 cells. Eur J Pharmacol. 2013;715:15–20. doi: 10.1016/j.ejphar.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 58.Patil VS, Zhou R, Rana TM. Gene regulation by non-coding RNAs. Crit Rev Biochem Mol Biol. 2014;49:16–32. doi: 10.3109/10409238.2013.844092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Percie du Sert N, Hurst V, Ahluwalia A, Alam S, Avey MT, Baker M, Browne WJ, Clark A, Cuthill IC, Dirnagl U, Emerson M, Garner P, Holgate ST, Howells DW, Karp NA, Lazic SE, Lidster K, MacCallum CJ, Macleod M, Pearl EJ, et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. PLoS Biol. 2020;18:e3000410. doi: 10.1371/journal.pbio.3000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Read TD, Petit RA, 3rd, Joseph SJ, Alam MT, Weil MR, Ahmad M, Bhimani R, Vuong JS, Haase CP, Webb DH, Tan M, Dove ADM. Draft sequencing and assembly of the genome of the world's largest fish, the whale shark: Rhincodon typus Smith 1828. BMC Genomics. 2017;18:532. doi: 10.1186/s12864-017-3926-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ripley BD. The R project in statistical computing. MSOR connections. 2001;1:23–25. [Google Scholar]
- 62.Romano R, Bucci C. Role of EGFR in the nervous system. Cells. 2020;9:1887. doi: 10.3390/cells9081887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruiz-Opazo N, Hirayama K, Akimoto K, Herrera VL. Molecular characterization of a dual endothelin-1/Angiotensin II receptor. Mol Med. 1998;4:96–108. [PMC free article] [PubMed] [Google Scholar]
- 64.Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- 65.Scholze AR, Foo LC, Mulinyawe S, Barres BA. BMP signaling in astrocytes downregulates EGFR to modulate survival and maturation. PLoS One. 2014;9:e110668. doi: 10.1371/journal.pone.0110668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seroogy KB, Numan S, Gall CM, Lee DC, Kornblum HI. Expression of EGF receptor mRNA in rat nigrostriatal system. Neuroreport. 1994;6:105–108. doi: 10.1097/00001756-199412300-00028. [DOI] [PubMed] [Google Scholar]
- 67.Shao Y, Wang XB, Zhang JJ, Li ML, Wu SS, Ma XY, Wang X, Zhao HF, Li Y, Zhu HH, Irwin DM, Wang DP, Zhang GJ, Ruan J, Wu DD. Genome and single-cell RNA-sequencing of the earthworm Eisenia andrei identifies cellular mechanisms underlying regeneration. Nat Commun. 2020;2656;11 doi: 10.1038/s41467-020-16454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sherf BA, Navarro SL, Hannah RR, Wood KV. Dual-luciferase reporter assay: an advanced co-reporter technology integrating firefly and Renilla luciferase assays. Promega Notes. 1996;57:2–8. [Google Scholar]
- 69.Sibilia M, Kroismayr R, Lichtenberger BM, Natarajan A, Hecking M, Holcmann M. The epidermal growth factor receptor: from development to tumorigenesis. Differentiation. 2007;75:770–787. doi: 10.1111/j.1432-0436.2007.00238.x. [DOI] [PubMed] [Google Scholar]
- 70.Stein RA, Staros JV. Insights into the evolution of the ErbB receptor family and their ligands from sequence analysis. BMC Evol Biol. 2006;6:79. doi: 10.1186/1471-2148-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun X, Liu H, Li T, Qin L. MicroRNA3395p inhibits cell proliferation of acute myeloid leukaemia by directly targeting SOX4. Mol Med Rep. 2018;18:5261–5269. doi: 10.3892/mmr.2018.9552. [DOI] [PubMed] [Google Scholar]
- 72.Tang H, Li J, Krishnakumar V. jcvi: JCVI utility libraries. Zenodo 2015 [Google Scholar]
- 73.Tebbutt N, Pedersen MW, Johns TG. Targeting the ERBB family in cancer: couples therapy. Nat Rev Cancer. 2013;13:663–673. doi: 10.1038/nrc3559. [DOI] [PubMed] [Google Scholar]
- 74.Tsai CN, Tsai CL, Yi JS, Kao HK, Huang Y, Wang CI, Lee YS, Chang KP. Activin A regulates the epidermal growth factor receptor promoter by activating the PI3K/SP1 pathway in oral squamous cell carcinoma cells. Sci Rep. 2019;9:5197. doi: 10.1038/s41598-019-41396-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang A, Bao Y, Wu Z, Zhao T, Wang D, Shi J, Liu B, Sun S, Yang F, Wang L, Qu L. Long noncoding RNA EGFR-AS1 promotes cell growth and metastasis via affecting HuR mediated mRNA stability of EGFR in renal cancer. Cell Death Dis. 2019a;10:154. doi: 10.1038/s41419-019-1331-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang CM, Wang Y, Fan CG, Xu FF, Sun WS, Liu YG, Jia JH. miR-29c targets TNFAIP3, inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma. Biochem Biophys Res Commun. 2011;411:586–592. doi: 10.1016/j.bbrc.2011.06.191. [DOI] [PubMed] [Google Scholar]
- 78.Wang S, Jin S, Liu MD, Pang P, Wu H, Qi ZZ, Liu FY, Sun CF. Hsa-let-7e-5p Inhibits the Proliferation and Metastasis of Head and Neck Squamous Cell Carcinoma Cells by Targeting Chemokine Receptor 7. J Cancer. 2019b;10:1941–1948. doi: 10.7150/jca.29536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Y, Wang F, He J, Du J, Zhang H, Shi H, Chen Y, Wei Y, Xue W, Yan J, Feng Y, Gao Y, Li D, Han J, Zhang J. miR-30a-3p targets MAD2L1 and regulates proliferation of gastric cancer cells. Onco Targets Ther. 2019c;12:11313–11324. doi: 10.2147/OTT.S222854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Z, Jin C, Li H, Li C, Hou Q, Liu M, Dong Xda E, Tu L. GPR48-induced keratinocyte proliferation occurs through HB-EGF mediated EGFR transactivation. FEBS Lett. 2010;584:4057–4062. doi: 10.1016/j.febslet.2010.08.028. [DOI] [PubMed] [Google Scholar]
- 81.Wu X, Yan L, Liu Y, Xian W, Wang L, Ding X. MicroRNA-448 suppresses osteosarcoma cell proliferation and invasion through targeting EPHA7. PLoS One. 2017;12:e0175553. doi: 10.1371/journal.pone.0175553. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 82.Xu H, Liu C, Zhang Y, Guo X, Liu Z, Luo Z, Chang Y, Liu S, Sun Z, Wang X. Let-7b-5p regulates proliferation and apoptosis in multiple myeloma by targeting IGF1R. Acta Biochim Biophys Sin (Shanghai) 2014;46:965–972. doi: 10.1093/abbs/gmu089. [DOI] [PubMed] [Google Scholar]
- 83.Yang G, Zhang X, Shi J. MiR-98 inhibits cell proliferation and invasion of non-small cell carcinoma lung cancer by targeting PAK1. Int J Clin Exp Med. 2015;8:20135–20145. [PMC free article] [PubMed] [Google Scholar]
- 84.Yang J, Zhao L, Yi S, Ding F, Yang Y, Liu Y, Wang Y, Liu M, Xue C, Xu L, Gong L, Wang X, Zhang Y, Yu B, Ming Gl, Gu X. Developmental temporal patterns and molecular network features in the transcriptome of rat spinal cord. Engineering. 2021 doi: 10.1016/j.eng.2021.10.001. [Google Scholar]
- 85.Yang X, Zhao H, Yang J, Ma Y, Liu Z, Li C, Wang T, Yan Z, Du N. MiR-150-5p regulates melanoma proliferation, invasion and metastasis via SIX1-mediated Warburg Effect. Biochem Biophys Res Commun. 2019;515:85–91. doi: 10.1016/j.bbrc.2019.05.111. [DOI] [PubMed] [Google Scholar]
- 86.Yu B, Yao C, Wang Y, Mao S, Wang Y, Wu R, Feng W, Chen Y, Yang J, Xue C, Liu D, Ding F, Gu X. The landscape of gene expression and molecular regulation following spinal cord hemisection in rats. Front Mol Neurosci. 2019;12:287. doi: 10.3389/fnmol.2019.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zha GB, Shen M, Gu XS, Yi S. Changes in microtubule-associated protein tau during peripheral nerve injury and regeneration. Neural Regen Res. 2016;11:1506–1511. doi: 10.4103/1673-5374.191227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang HM, Liu T, Liu CJ, Song S, Zhang X, Liu W, Jia H, Xue Y, Guo AY. AnimalTFDB 2.0: a resource for expression, prediction and functional study of animal transcription factors. Nucleic Acids Res. 2015;43:D76–81. doi: 10.1093/nar/gku887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao G, Guo J, Li D, Jia C, Yin W, Sun R, Lv Z, Cong X. MicroRNA-34a suppresses cell proliferation by targeting LMTK3 in human breast cancer mcf-7 cell line. DNA Cell Biol. 2013;32:699–707. doi: 10.1089/dna.2013.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou KR, Liu S, Sun WJ, Zheng LL, Zhou H, Yang JH, Qu LH. ChIPBase v2.0: decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic Acids Res. 2017;45:D43–50. doi: 10.1093/nar/gkw965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pearson's correlation with Egfr expression based on spinal cord injury dataset


