Abstract
As one of the most common endocrine disorders affecting women, polycystic ovary syndrome (PCOS) is associated with serious conditions including anovulation, endometrial cancer, infertility, hyperandrogenemia, and an increased risk for obesity and metabolic derangements. One contributing etiology to the pathophysiology of hyperandrogenemia associated with PCOS is an intrinsic alteration in ovarian steroidogenesis, leading to enhanced synthesis of androgens including testosterone. Studies have suggested that the increased testosterone synthesis seen in PCOS is driven in part by increased activity of CYP17A1, the rate-limiting enzyme for the formation of androgens in the gonads and adrenal cortex, which represents a critical factor driving enhanced testosterone secretion in PCOS. In this work, we evaluated the hypothesis that dysregulation of the noncoding RNA H19 results in aberrant CYP17 and testosterone production. To achieve this, we measured Cyp17 in ovarian tissues of H19 knockout mice, and quantified serum testosterone levels, in comparison with wild-type controls. We also evaluated circulating and ovarian H19 expression and correlated results with the presence or absence of PCOS in a group of women undergoing evaluation and treatment for infertility. We found that the loss of H19 in a mouse model results in decreased ovarian Cyp17, along with decreased serum testosterone in female mice. Moreover, utilizing serum samples and cumulus cells from women with PCOS, we showed that circulating and ovarian levels of H19 are increased in women with PCOS compared to controls. Findings from our multimodal experimental strategy, involving both a mouse model of dysregulated H19 expression and clinical serum and ovarian cellular samples from women with PCOS, suggest that the loss of H19 may disrupt androgen production via a Cyp17-mediated mechanism. Conversely, excess H19 may play a role in the pathogenesis of PCOS-associated hyperandrogenemia.
Keywords: H19, Noncoding RNA, ncRNA, PCOS
Introduction
Steroid hormones have broad physiologic roles and are essential for maintenance of reproductive capacity for both men and women. Multiple reproductive disease states can be characterized by aberrations in steroid hormone levels, and the mechanisms underlying these aberrations is in many cases still not well-understood. One disorder characterized by dysregulated steroid hormone production is the polycystic ovary syndrome (PCOS). PCOS affects up to 10% of reproductive-aged women [1]. As one of the most common endocrine disorders affecting women, PCOS is associated with serious conditions including irregular menstrual cycles and anovulation, endometrial cancer, infertility, hyperandrogenism, and hirsutism, and an increased risk for central adiposity, glucose intolerance, hyperinsulinemia, and development of the metabolic syndrome [2–4].
One contributing etiology to the pathophysiology of hyperandrogenemia associated with PCOS is an intrinsic alteration in ovarian steroidogenesis, leading to enhanced synthesis of androgens including testosterone. Elevated serum testosterone is a predominant clinical feature of PCOS [5]. Studies have suggested that the increased testosterone synthesis seen in patients with PCOS is driven in part by an increased activity of CYP17A1, the rate-limiting enzyme for the formation of androgens in the gonads and adrenal cortex [6] which represents a critical factor driving enhanced testosterone (T) secretion in PCOS [7]. In support of this theory, dysregulation of CYP17A1 mRNA expression has been described in ovarian theca cells from women with PCOS [8]. However, the study of the pathophysiology of PCOS is made more complex by the possible contribution of genetic and epigenetic alterations and environmental factors. Because of this, research into novel genetic areas which might have implications for diagnosis or treatment is important.
One of these potential areas of study is in the field of noncoding RNAs. The “central dogma” of molecular biology, that is, that DNA blueprints encode messenger RNAs which serve as intermediaries for protein translation, has been challenged by the discovery that a significant portion of the genome encodes functional RNA that is transcribed from DNA but not translated into protein [9]. These noncoding RNAs (ncRNAs) play critical roles in a wide variety of biological processes [10]. Over the past decade, a growing body of evidence has shown that ncRNAs can regulate the physiologic mechanisms of sex steroid biosynthesis and secretion [11]. Given their diverse roles in biological process, interest has grown in the role of ncRNAs in the pathogenesis of PCOS. We previously showed that the long ncRNA (lncRNA) H19 can regulate steroid hormone production via post-transcriptional control of the rate-limiting step of steroidogenesis [12]. In this work, we evaluated the hypothesis that the dysregulation of the noncoding RNA H19 results in aberrant CYP17 and testosterone production. To achieve this, we utilized a multimodal experimental strategy involving both a mouse model of dysregulated H19 expression and clinical serum and ovarian cellular samples from women with PCOS. We measured Cyp17 in ovarian tissues of H19 knockout mice, and quantified serum testosterone levels, in comparison with wild-type controls. We also evaluated circulating and ovarian H19 expression and correlated results with the presence or absence of PCOS in a group of women undergoing evaluation and treatment for infertility. Our findings suggest that loss of H19 may disrupt androgen production via a Cyp17-mediated mechanism, and that conversely, excess H19 may play a role in the pathogenesis of PCOS-associated hyperandrogenemia.
Materials and Methods
Mouse Strains and Animal Care
Homozygous H19Δ3 loss-of-function mice were used for this study as a model system for the dysregulation of H19. The strain is characterized by the deletion of the 3-kb transcription unit upstream of the H19 gene itself [13, 14]. Controls for H19KO females were homozygous wild-type (WT) littermates. Animals used in these studies were maintained and euthanized according to principles and procedures described in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. These studies were approved by the Yale University Institutional Animal Care and Use Committee and conducted in accordance with the Society for the Study of Reproduction’s specific guidelines and standards.
Reverse Transcription, Real-time PCR, and Western Blot
We chose to evaluate Cyp17 in gonadal tissues of H19KO mice based on a number of studies which have noted a possible linkage between Cyp17 polymorphisms, hyperandrogenism, and PCOS. To evaluate Cyp17 expression, ovarian tissue was collected from 8-week H19 knockout (H19KO) and wild-type (WT) female mice (n = 5 per group). RNA extraction was performed on tissue samples using the RNeasy Mini Kit (Qiagen Cat. #74,004, Hilden, Germany) according to manufacturer’s instructions, and reverse-transcribed using the iScript cDNA synthesis kit (Bio-Rad Cat. #1,708,891, Hercules, CA). One microgram RNA was combined with 19 μg of reaction mixture containing 5 μL iScript reaction mix, nuclease-free water, and iScript reverse transcriptase. Qualitative polymerase chain reaction (qPCR) was performed to assess Cyp17 in ovarian tissue from H19KO and WT mice. Twenty-five-microliter qPCR reactions were prepared containing 0.5 to 1.5 μl of cDNA and iQSYBRGreen (Bio-Rad Cat. #1,708,880, Hercules, CA) in a Bio-Rad iCycler. qPCR was performed by initial denaturation at 95 °C for 5 min, followed by 40 cycles for 30 s at 95 °C, 30 s at 60 °C, and 30 s at 72 °C. Specificity was verified by melting curve analysis and agarose gel electrophoresis. The threshold cycle values of each sample were used for post-PCR data analysis, and the ΔΔ threshold cycle method was used to calculate mRNA levels. The PCR primers for the indicated mouse genes are listed below. All primers had an efficiency of > 90%. Primers were purchased from Real Time Primers (USA).
H19 forward: 5′-CCTCAAGATGAAAGAAATGGTGCTA-3′
H19 reverse: 5′-TCAGAACGAGACGGACTTAAAGAA-3′
β-actin forward: 5′-AAGAGCTATGAGCTG CCTGA-3′
β-actin reverse: 5′-TACGGATGTCAACGTCACAC-3′
Cyp17α forward: 5′-TAT GCA TGC CAA CTT CTT CA-3′
Cyp17α reverse: 5′-CAA GAG GCC TAG AGT CAC CA-3′
Western Blot Analysis
We then performed Western blot analysis to measure protein levels in a mouse ovary [15]. For the assessment of protein levels, 8-week female H19KO and WT mice were euthanized, and ovarian tissues were harvested. Prior to tissue collection, mouse estrus cycle staging was evaluated by a single trained laboratory member via assessment of vaginal smear cytology [16]. Tissue lysates were prepared using the Nuclear Extract Kit (Activemotif, Cat. #40,010, Carlsbad, CA) according to the manufacturer’s protocol. Equal volumes of protein were electrophoresed through 4 to 15% polyacrylamide gels (Bio-Rad, Cat. #4,568,084) at 100 V for 60 min and then transferred onto Immun-Blot poly-vinylidene difluoride membranes (Bio-Rad, Cat. #1,620,177) in a transfer buffer (25 mmol/L Tris, 192 mmol/L glycine, and 20% methanol) at 100 V for 1 h. After incubation in blocking buffer (1 × PBS, 0.2% Tween 20, and 5% milk), the membrane was incubated individually with either rabbit monoclonal CYP17 antibody (Novus Biologicals, Cat. #OTI3F12, USA) diluted 1:500 or rabbit monoclonal GAPDH antibody (Cell Signaling, Cat. #14C10, USA) diluted 1:1000 overnight at 4 °C. After washing, the membranes were incubated with rabbit anti-mouse secondary antibody (Abcam, Cat. # Ab205718, USA) diluted in blocking buffer (3.5 mg/mL) at room temperature for 1 h. Western blot quantifications were performed using ImageJ.
Mouse Serum Steroid Hormone Quantification
For quantification of mouse serum steroid hormone levels, retroorbital blood collection was performed on 8-week female H19KO and WT mice (n = 5 per group). Prior to blood collection, mouse estrus cycle staging was evaluated by a single trained laboratory member via assessment of vaginal smear cytology [16]. Retroorbital blood collection was then performed at each estrus cycle stage, from alternating eyes at an interval of no less than 7 days per eye. Sera were prepared by clotting blood for 60–90 min at room temperature (RT). Supernatants were recovered by centrifugation at RT for 10–15 min at 2000 g. Sera samples were frozen at − 20 until quantification was performed. The samples were sent to the University of Virginia Ligand Assay and Analysis Core for measurement of testosterone, estradiol, and corticosterone levels. The UVA Core uses the following assays: the IBL America Testosterone ELISA (Cat. #IB79106, sensitivity 0.083 ng/mL; CV 11.6% (interassay); specificity: DHT 12.9%, 11b-hydroxytestosterone 3.3%, 19-nortestosterone 3.3%, progesterone < 0.1%, androstenedione 0.9%; other endogenous steroids tested were < 0.1%); the MP Biomedicals ImmuChem Double Antibody Corticosterone RIA kit (Cat. # 0,712,010-CF; sensitivity 25 ng/mL; CV 5.4% (interassay); specificity: deoxycorticosterone 0.34%, testosterone 0.10%, cortisol 0.05%, progesterone 0.02%; other endogenous steroids tested were < 0.1%), and the CalBiotech Estradiol ELISA (Cat. #E180S-100; sensitivity 3 pg/mL; CV 9.9% (interassay); specificity: progesterone, 0.0002%, androstenedione, 0.0001%, testosterone, 0.0002%, cortisol, 0.0001%; other endogenous steroids tested were < 0.0001% or undetectable).
Collection of Patient Samples and Quantification of H19 Expression
In order to evaluate circulating H19 expression and correlation with PCOS, discarded blood samples (2 mL) were collected from 39 patients undergoing evaluation for infertility at the Yale Fertility Center (New Haven, CT) (Table 1). Patients were divided into two groups: controls (women diagnosed with male and tubal factor infertility, n = 19) and women with hyperandrogenic PCOS evaluated by using Rotterdam criteria (n = 16). Patients were excluded if any of the following diagnoses were present: Cushing’s syndrome, hyperprolactinemia, adrenal hyperplasia, acromegaly, hypothalamic amenorrhea, hypothyroidism, or diabetes mellitus. This study was approved by the Yale University Institutional Review Board (IRB protocol # 1,606,017,946). Samples were collected in the early follicular phase (days 2–4 of the menstrual cycle).
Table 1.
Descriptive statistics for patients from whom serum samples were collected. There were no significant differences between patients with PCOS and non-PCOS patients in terms of age, BMI, day 3 FSH, or estradiol, or progesterone. Patients with PCOS had higher baseline luteinizing hormone (LH) (8.2 vs 5.7 mIU/mL; p < 0.05), anti-Mullerian hormone (AMH) (12.5 vs 2.5 ng/mL; p = 0.001) and total testosterone (51.3 vs 21.4 ng/dL; p < 0.01)
| PCOS (n = 16) | Male or tubal (n = 19) | p-value | |
|---|---|---|---|
| Age (years) | 33.3 ± 1.2 | 35.2 ± 1.2 | 0.30 |
| BMI (kg/m2) | 28.3 ± 1.6 | 29.3 ± 1.1 | 0.56 |
| FSH (mIU/mL) | 6.2 ± 0.5 | 6.1 ± 0.6 | 0.82 |
| LH (mIU/mL) | 8.2 ± 1.4 | 5.7 ± 0.8 | 0.04* |
| Estradiol (pg/mL) | 41.1 ± 8.4 | 30.5 ± 4.4 | 0.08 |
| Progesterone (ng/mL) | 0.3 ± 0.1 | 0.37 ± 0.1 | 0.70 |
| Testosterone (ng/dL) | 51.3 ± 26.5 | 21.4 ± 5.4 | 0.01** |
The serum was separated by centrifugation for 10 min at 12,000 rpm at 4 °C and stored at − 80 °C until RNA extraction [17–19]. RNA extraction from stored patient blood samples was performed using the Norgen Plasma/Serum RNA Purification Mini Kit (Cat. #55,000, Ontario, Canada). To purify RNA from cumulus cells, the Qiagen miRNeasy Mini Kit and RNeasy MinElute Cleanup Kits (Cat. #217,084, Germantown, MD, USA) were used according to the manufacturer’s instructions. RNA quantity and purity were determined using an ultraviolet spectrophotometer. Reverse transcription was carried out using the Bio-Rad iScript cDNA synthesis kit (Cat. #1,708,890, Hercules, CA, USA) in a 20 μL reaction mixture containing 0.5 μg of total RNA. For cDNA synthesis, the reaction mixtures were incubated at 16 °C for 30 min, at 42 °C for 30 min, and at then 85 °C for 5 min. The RNA was stored at − 80°C until qPCR.
qPCR was performed in a 25 μL reaction mixture containing 1.5 μl of cDNA using Bio-Rad QSYBRGreen in a Bio-Rad iCycler as outlined above. All SYBR green runs had dissociation curves to predict potential primer-dimers, and specificity was verified by melting curve analysis and agarose gel electrophoresis. The threshold cycle (Ct) values of each sample were used in the post-PCR data analysis, and the delta-delta Ct method was used to calculate mRNA expression. H19 serum and cumulus cell expression levels were normalized and expressed as fold change relative to that of β-actin. The PCR primers for the indicated genes are listed below; primers were purchased from Real Time Primers (Elkins Park, PA, USA).
Human H19 forward: 5′-GCACCTTGGACATCTGGAGT.
Human H19 reverse: 5′-TTCTTTCCAGCCCTAGCTCA.
β-actin forward: AAGAGCTATGAGCTGCCTGA.
β-actin reverse: TACGGATGTCAACGTCACAC.
We also sought to determine whether H19 is detectable in granulosa cells of women undergoing in vitro fertilization (IVF) with oocyte retrieval, and whether H19 expression correlates with the diagnosis of PCOS. For these studies, we utilized pooled cumulus cells collected from a total of 72 consecutive cycles in women undergoing infertility treatment with IVF at Gazi University School of Medicine IVF Center (Ankara, Turkey) (Table 2). For study analysis, patients were stratified by infertility diagnosis (PCOS (10 women) versus male/tubal factor infertility (29 women)). This study was approved by the Gazi University Institutional Review Board committee (IRB protocol #131/11.05.2011).
Table 2.
Descriptive statistics for patients from whom cumulus cell samples were collected. There were no significant differences between patients with PCOS and non-PCOS patients in terms of age (29.8 vs 33.6), day 3 FSH (6.38 vs 6.96) or maximum estradiol level (1758 pg/mL vs 1441.5 pg/mL). Non-PCOS patients received higher total doses of gonadotropins (2617.5 vs 3394.6 units, < 0.05)
| PCOS (n = 10) | Male or tubal (n = 29) | p-value | |
|---|---|---|---|
| Age (years) | 29.8 ± 4.73 | 33.6 ± 5.94 | 0.48 |
| FSH (mIU/mL) | 6.4 ± 3.0 | 7.0 ± 2.3 | 0.30 |
| Total units FSH | 2617.5 ± 1015.2 | 3394.6 ± 806.1 | 0.01** |
| Peak estradiol (pg/mL) | 1758.3 ± 840.4 | 1441.5 ± 423.1 | 0.30 |
For patients undergoing agonist cycles, treatment was initiated by gonadotropin-releasing hormone (GnRH) agonists during the luteal phase of the preceding cycle. Stimulation with gonadotropins was initiated after downregulation had been achieved (estradiol level < 50 pg/mL in the absence of ovarian cysts on transvaginal sonography). For patients undergoing antagonist cycles, treatment with gonadotropins was initiated on cycle day 3 when serum progesterone was < 1 ng/mL and transvaginal sonography-confirmed absence of ovarian cysts. A GnRH antagonist was added for pituitary suppression after 5 days of gonadotropin stimulation. Stimulation protocols included 150–300 IU/day of gonadotropins, either recombinant follicle-stimulating hormone (FSH, GonalF, Merck) or FSH in combination with human menopausal gonadotropin (hMG, Menopur, Ferring). Patients received human chorionic gonadotropin (hCG, Ovidrel 250 μg, Merck) when two or more follicles > 18 mm in diameter were present with an adequate estradiol response. Oocytes were collected 36 h after hCG injection. Retrieved cumulus-oocyte complexes (COCs) were placed in a culture medium (VitroLife G-MOPS, Cat. #10,129, Sweden), and cumulus cells were dissected from the oocyte mechanically in the absence of hyaluronidase. Cumulus cells from each patient were pooled in a single Eppendorf tube. Samples were washed twice with 0.5 mL 1 × phosphate-buffered saline and centrifuged at 1000 × g for 1 min with the supernatant removed after each wash. After the final wash, the cell pellet was resuspended in 50 μL SideStep™ Lysis and Stabilization Buffer (Stratagene, Cat. #400,900, La Jolla, CA). Samples were stored at − 80 °C until analysis, at which time RNA extraction and qRT-PCR were performed as outlined above.
Statistical Analysis
All data are presented as mean ± SD. Data were analyzed using two-tailed Student t test, with the exception of ELISA data, which was analyzed using t-test or one-way ANOVA with correction for multiple comparisons, as appropriate. P values at 0.05 or less were considered significant. All statistical analysis was performed using the GraphPad software.
Results
Loss of H19 Results in Decreased Cyp17α in Female Mouse Ovary
We first evaluated steroidogenic enzyme levels in female H19KO mouse ovary. In order to achieve this goal, we performed RT-qPCR for Cyp17α in the ovary of H19KO and WT female mice. Cyp17α expression in the ovary of H19KO female mice was decreased compared with that of wild-type mice (Fig. 1a). We then evaluated whether the loss of H19 in the ovary of female H19KO mice affected CYP17α protein production. We found that CYP17α was decreased in the ovary of H19KO female mice (Fig. 1b). The quantification of Western blot was performed and confirmed a decreased CYP17α protein in H19KO female ovary compared to that in WT (Fig. 1c).
Fig. 1.
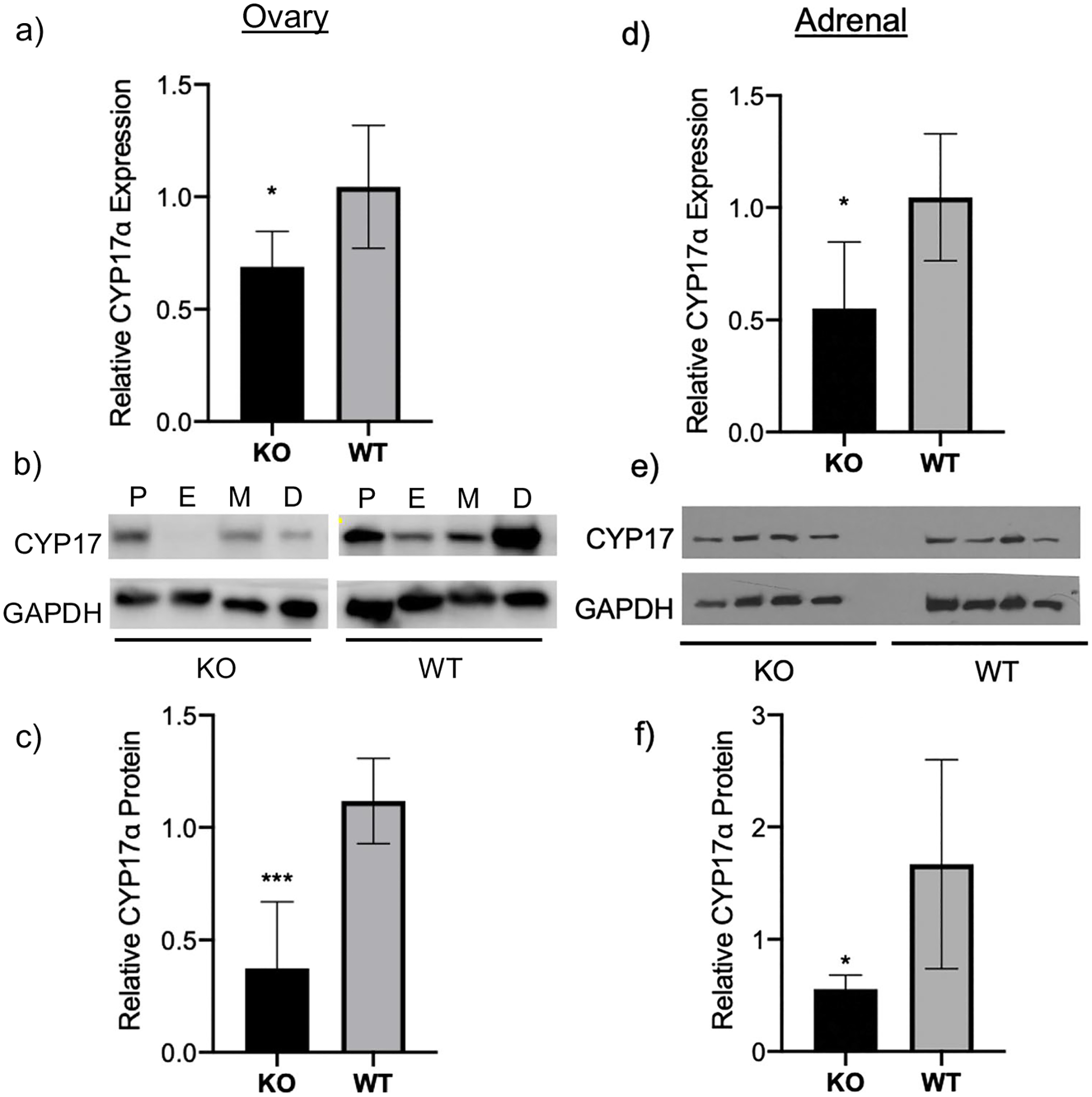
Decreased CYP17α levels in ovaries of H19KO female mice. To evaluate CYP17α expression, RNA extraction, cDNA synthesis, and RT-qPCR were performed on ovarian tissue from 8-week-old H19KO and WT female mice (n = 5 per group). a Mean relative Cyp17α expression levels by real-time PCR are shown. Cyp17α expression was decreased in ovaries from H19KO female mice compared to that from WT. Beta-actin was used as a control. b Western blot analysis was performed to measure CYP17α protein levels, using GAPDH as control. c Quantification of Western blot showing decreased CYP17α protein in ovary compared to that in WT. All data were analyzed using Student’s t-test. *p = 0.05; ***p < 0.001. Error bars represent one SEM. P, proestrus; E, estrus; M, metestrus; D, diestrus
Female H19KO Mice Have Decreased Serum Testosterone Levels
We then evaluated steroid hormone profiles in male and female H19KO and WT mice via retroorbital blood collection and quantification of serum hormone levels. Serum hormone quantification results from H19KO and WT mice were stratified by estrus cycle stage. Serum testosterone levels were lower in the estrus stage in female H19KO mice compared to those in WT (Fig. 2; 20.66 ng/dL vs 33.42 ng/dL; p < 0.01). Serum corticosterone concentrations were similar across estrus cycle stage in H19KO and WT mice (Fig. 2). Serum estradiol levels were not quantified in this study as these were previously evaluated [20].
Fig. 2.
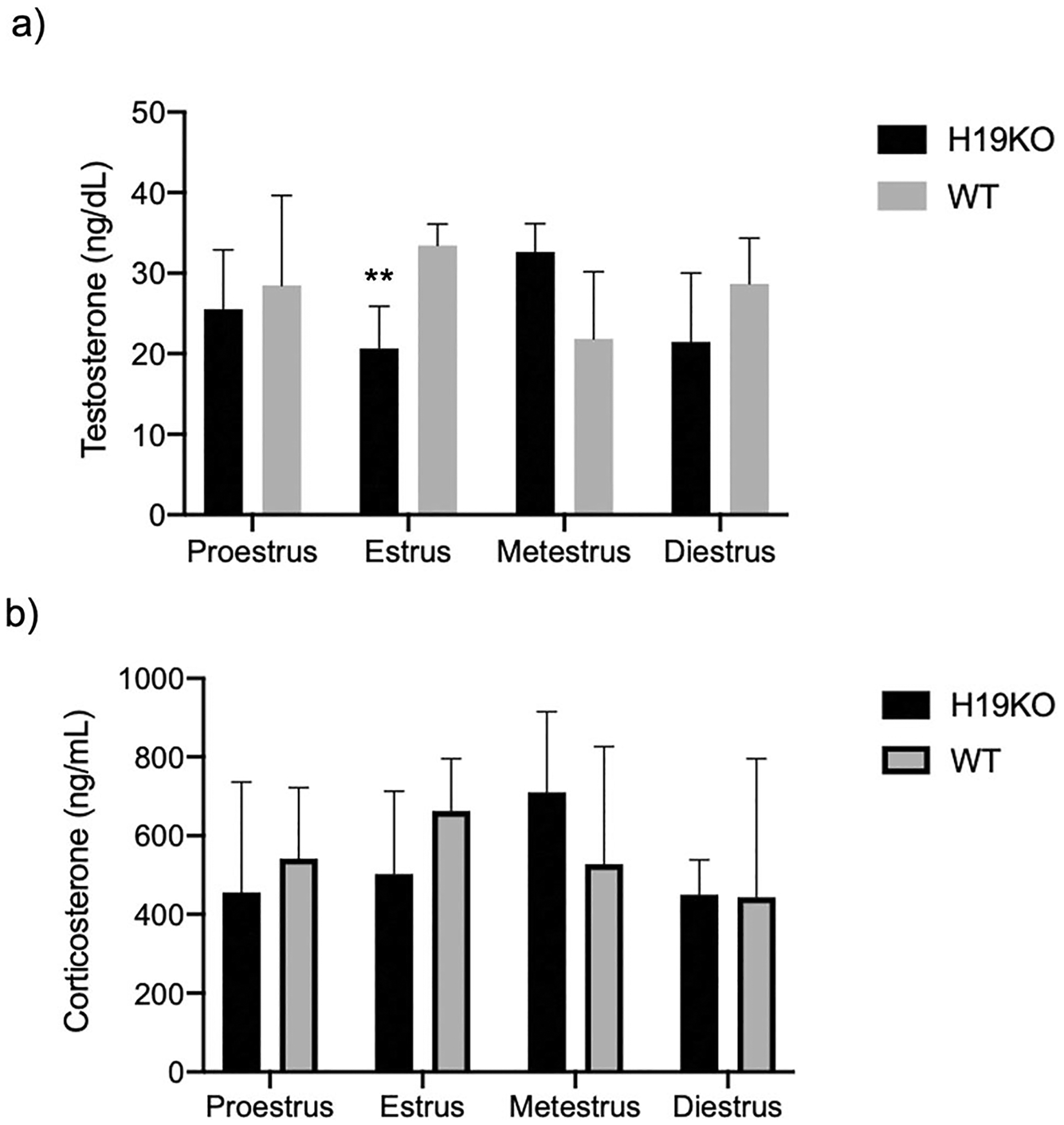
Serum quantification of hormone levels in female mice. Serum hormone levels were quantified via retroorbital blood collection in H19KO and WT female mice at each estrus cycle stage. Serum testosterone (T) levels were lower in the estrus stage in female H19KO mice compared to those in WT mice (Fig. 2a; 20.66 ng/dL vs 33.42 ng/dL; p < 0.01). Serum corticosterone concentrations were similar across estrus cycle stage in H19KO and WT mice. All data were analyzed using two-way ANOVA with correction for multiple comparisons. **p < 0.005. Error bars represent one standard error of the mean (SEM)
H19 Expression in Patient Samples
Women with PCOS presenting for baseline fertility evaluation were not significantly different from male and tubal factor control patients with respect to age, body mass index (BMI), day 3 FSH or estradiol, or progesterone (Table 1). Patient with PCOS had higher baseline LH (8.2 vs 5.7 mIU/mL; p < 0.05) and total testosterone (51.3 vs 21.4 ng/dL; p < 0.01) (Table 1). In a subset of 17 women with PCOS who had documentation of the presence or absence of insulin resistance (which is diagnosed in our clinic based on the presence of validated surrogate clinical features of hyperinsulinemia; e.g., acanthosis nigri-cans, metabolic syndrome [21, 22]), 11 women with PCOS had insulin resistance, while 5 were not insulin resistant (p = 0.17). Serum H19 expression was increased 2.5-fold in women with PCOS relative to women without PCOS (e.g., male and tubal factor controls) (p < 0.0005) (Fig. 3). There were no significant differences between women with PCOS undergoing controlled ovarian hyperstimulation for infertility and control women (tubal/male factor infertility) in terms of age (mean 29.8 in PCOS patients vs 33.6 in non-PCOS patients), day 3 FSH (6.38 vs 6.96), or maximum estradiol level (1758.3 vs 1441.5 pg/mL) (Table 2). Non-PCOS patients received higher total dose of gonadotropins over the course of a controlled ovarian hyperstimulation cycle (2617.5 vs 3394.6 IU; p = 0.02) (Table 2). H19 expression was increased 2.8-fold in cumulus cells of PCOS patients collected at the time of oocyte retrieval, compared to cumulus cells from non-PCOS control women (p < 0.05) (Fig. 4).
Fig. 3.
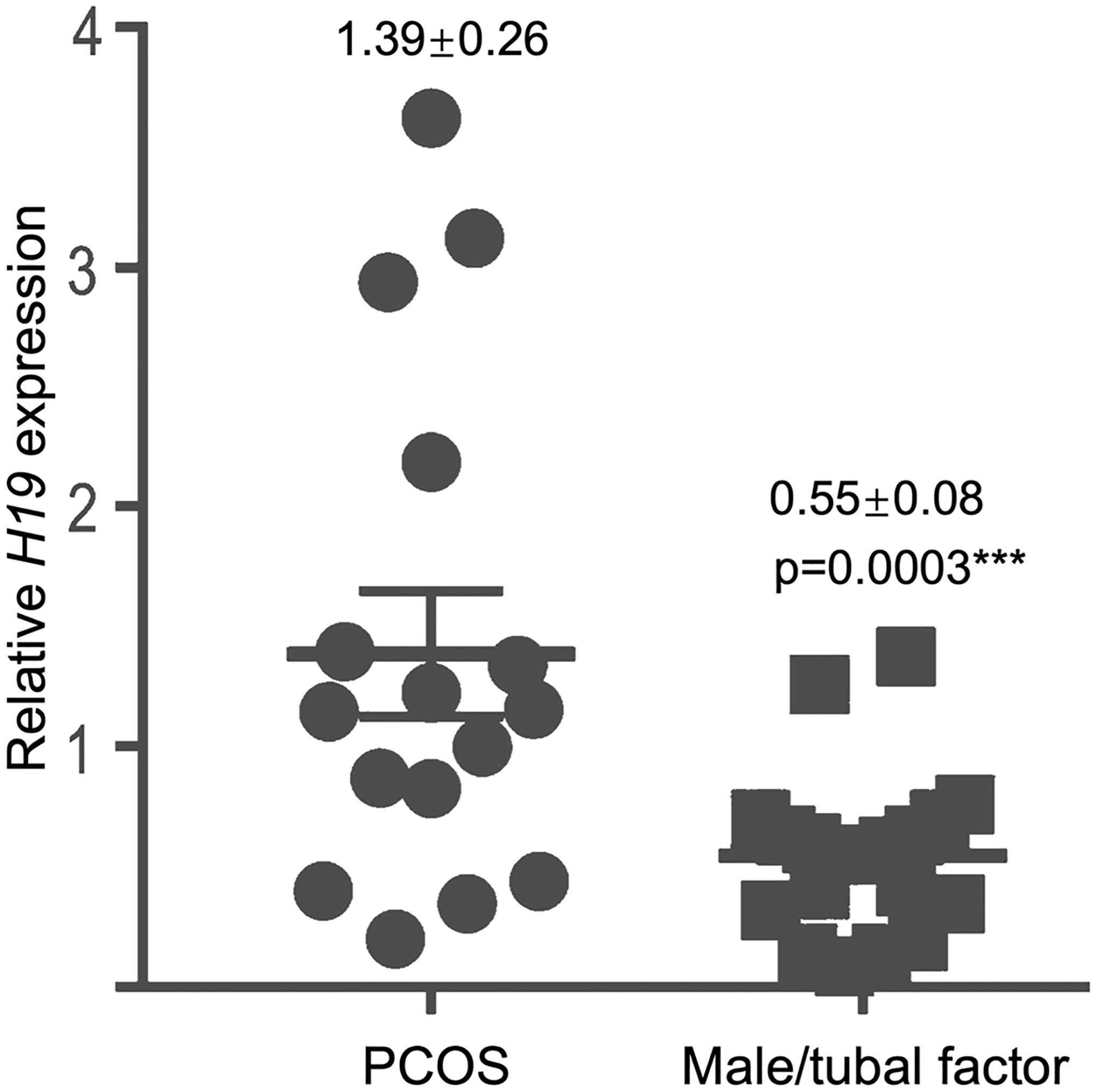
Serum H19 expression is increased in women with PCOS. Blood samples from 35 patients (19 controls diagnosed with male and/or tubal factor infertility and 16 women with hyperandrogenic PCOS) undergoing infertility evaluation were collected in the early follicular phase (days 2–4 of the menstrual cycle) and evaluated for H19 expression. The bar graph shows the expression level of H19 in women with PCOS, presented as fold change relative to women without PCOS (e.g., male and tubal factor controls). Relative H19 expression increased 2.5-fold in women with PCOS compared to that in women with male/tubal factor infertility (p < 0.0005)
Fig. 4.
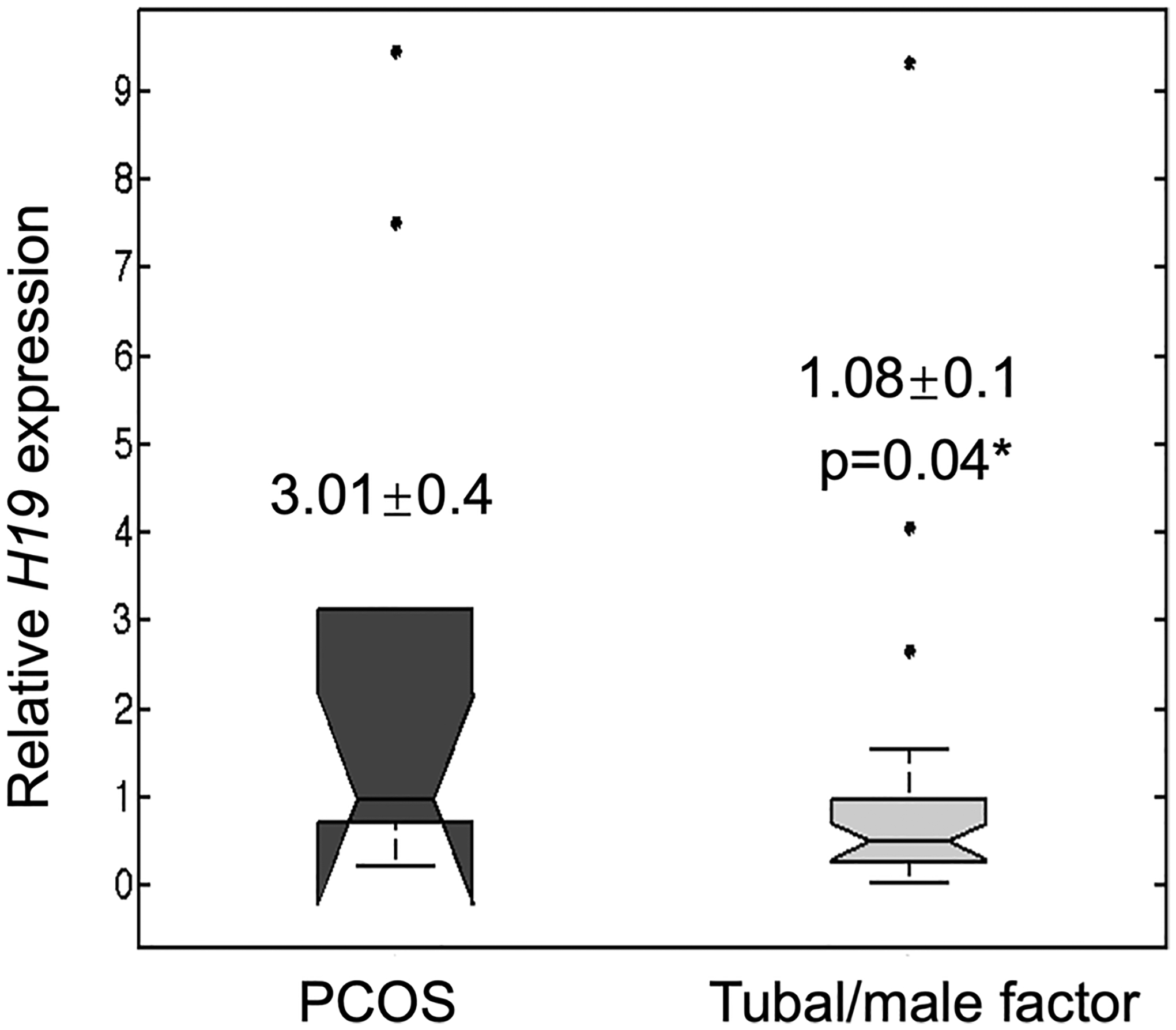
Cumulus cell expression of H19 in women with PCOS versus controls (male/tubal factor). A total of 39 women undergoing in vitro fertilization (29 non-PCOS women undergoing IVF for male factor infertility and 10 women with PCOS) were evaluated. At the time of oocyte retrieval, cumulus cells were collected, total RNA was isolated, and cDNA was synthesized by reverse transcription. RNA levels were analyzed using quantitative real-time PCR. H19 RNA levels were normalized to those of β-actin and presented as relative expression levels using the comparative CT method. Box plot demonstrates increased relative H19 expression (2.8-fold) in cumulus cells of PCOS patients undergoing IVF, compared to non-PCOS patients (p < 0.05)
Discussion
In this study, we show that loss of H19 in a mouse model results in a decreased ovarian Cyp17, along with decreased serum testosterone in female mice. Moreover, we have provided a clinical correlation by utilizing serum samples and cumulus cells from women with PCOS, showing that in these women, circulating and ovarian levels of H19 are increased compared to those in controls. These findings suggest that loss of H19 may disrupt androgen production via a Cyp17-mediated mechanism, and that conversely, excess H19 may play a role in the pathogenesis of PCOS-associated hyperandrogenemia.
Hyperandrogenemia is a recognized diagnostic criterion for PCOS. The molecular characterization of PCOS has focused in part on increased expression of steroidogenic enzymes involved in androgen biosynthesis, including CYP17A1, a critical component of the steroidogenic pathway. This enzyme converts pregnenolone and progesterone to their major products, including testosterone. CYP17A1 is responsible for both adrenal and gonadal steroid biosynthesis in humans. In the female mouse, CYP17 expression is primarily restricted to the androgen-producing theca cells of the ovary, as well as of the placenta [23]. The regulation of CYP17 is a significant factor in the expression of hyperandrogenism. Cyp17a1 knockout mice exhibit severe disruptions of steroidogenesis, including lack of testosterone synthesis and significantly elevated corticosterone levels (in XY mice) and a minor decrease in testosterone and accumulation of corticosterone in XX mice [24]. In a large study of 394 Kashmiri women with PCOS and 306 healthy controls, PCOS patients with a Cyp17 homozygous polymorphism had significantly higher levels of total testosterone than those without a Cyp17 polymorphism [25]. A case–control study of 50 Kurdish patients with PCOS and 109 controls found that patients with PCOS had a higher frequency of the CYP17 T-34C polymorphism than controls (30% vs 15.6%) [26]; this CYP17 polymorphism is significantly associated with increased testosterone [27–29]. A case–control study of 204 Pakistani PCOS patients and 100 controls noted similar findings (CYP17 polymorphism frequency 54.9% among PCOS patients vs 12% among controls) [30]. In vivo, RNA interference–based silencing of CYP17 in a rat ovary results in a significantly decreased testosterone production [31]. Of note, we observed decreased serum testosterone levels in female H19KO mice during the estrus stage, but not in other stages; this may be related to unidentified effects of loss of H19 on other gene targets related to testosterone production.
Over the last several years, long noncoding RNAs (lncRNAs, which are more than 200 nucleotides in length) have emerged as master regulators of tissue growth and differentiation. LncRNAs can interact with DNA, RNA, and proteins, and act in a variety of functions, including as molecular scaffolds [32], guides to a specific target locus [33], decoys or sponges [34], and enhancers of transcriptional activity [35]. Altered lncRNA levels have been associated with conditions as diverse as diabetes, insulin resistance, inflammation, and various cancers. The highly conserved H19 was discovered as the first lncRNA over three decades ago [36]. H19 is an imprinted gene, expressed exclusively from the maternal allele. It has been generally believed that H19 is abundantly expressed in the early stages of embryogenesis and repressed postnatally (except in skeletal muscle and heart), but more recently, this dogma has been challenged, as H19 expression has been observed in reproductive organs including the ovary and uterus [20, 37, 38].
Up to this point, attempts to link ncRNAs to steroid hormone production, including in women with PCOS, have focused primarily on the role of microRNAs (miRNAs), another class of noncoding RNAs that are able to regulate gene expression at the post-transcriptional level via direct translational repression of messenger RNAs (mRNAs) and/or mRNA destabilization [11, 39, 40]. In a study of letrozole-induced PCOS in rats, 158 lncRNAs were identified as differentially expressed between PCOS ovarian tissues and controls [41]; however, data on specific genes which were differentially expressed was not made available. In another study of lncRNA expression in cumulus cells isolated from PCOS patients, 623 lncRNAs were significantly upregulated or downregulated [42], but serum testosterone levels in patients with PCOS and controls were not provided. To our knowledge, this is the first report of a potential long ncRNA-mediated mechanism for the regulation of androgen production via Cyp17, and the first finding of a link between H19 and elevated testosterone levels in women with PCOS. Like Cyp17, prominent H19 expression has been observed in theca cells of mature preovulatory follicles [37], providing plausible mechanistic insight into H19-mediated regulation of testosterone production via Cyp17. Notably, lncRNAs can act as molecular “sponges” for miRNAs; we previously showed that H19 is a sponge for the miRNA let-7, a small molecule that predominantly functions to silence target genes [34] and which regulates glucose and insulin metabolism [43, 44]. Overexpression of let-7 in mice results in impaired glucose tolerance and reduced insulin secretion [44]. Moreover, hyperinsulinemia contributes to ovarian hyperandrogenism; in vitro studies have shown that insulin stimulates testosterone release from ovarian stroma [45]. H19, by binding and sequestering let-7, modulates its availability and leads to derepression of let-7 target genes. Thus, it is intriguing to consider the possibility that H19-mediated regulation of let-7 may be linked to adverse metabolic outcomes in women with PCOS. While fasting metabolic parameters were not available for our study population, Qin et al. demonstrated a significant positive correlation between H19 expression and fasting plasma glucose in women with PCOS [46]. Since our previous work, additional research has demonstrated H19’s broader role as a molecular sponge for other miRNAs, including miR-138, miR-200, and miR-152 [47–50], and shown that H19 can sponge these miRNAs in a context- and cell-specific manner and thus play contradictory roles in different cell types [48]; however, these miRNAs have not been evaluated in the context of PCOS.
Our findings regarding a role for H19 in the pathogenesis of PCOS may also have relevance with respect to treatment for this condition. Insulin resistance, which results in compensatory hyperinsulinemia, contributes to ovarian androgen production and further contributes to PCOS symptomatology [51]. The insulin-sensitizing medication metformin is used to treat type 2 diabetes mellitus and PCOS. In vitro, metformin attenuates granulosa cell apoptosis and improves insulin resistance [52]; in women with PCOS, treatment with metformin decreases androgen levels [53] and increases ovulation rates [54]. In addition to its role in the treatment of insulin resistance, metformin has direct effects on ovarian steroidogenesis [55, 56], including inhibition of androgen production in human theca cells [56]. Interestingly, metformin treatment also reduces H19 in endometrial cancer cell lines in vitro and in endometrium from women with endometrial cancer [57]. Moreover, metformin significantly blunts the effects of ACTH, a known stimulator of H19, on androgen production [58]. Finally, in a PCOS rat model, in which serum H19 expression was elevated at baseline, metformin was found to alleviate PCOS in part via a reduction of H19 expression [59]. These studies raise the intriguing possibility that metformin may exert its clinical effects on androgen production in part through H19, though this hypothesis warrants further study.
Our study is strengthened by the addition of clinical data both from patient serum samples and from cumulus cells collected at the time of oocyte retrieval. Noncoding RNAs, including miRNAs and lncRNAs, can be collected from tissues as well as bodily fluids including plasma, serum, and urine [60, 61]. While tissue-specific changes in H19 expression have been investigated as potential biomarkers in other conditions including breast [49] cancer, our laboratory was the first to report H19 expression in cumulus cells, which are easily accessible at the time of oocyte retrieval [62]. Moreover, our sample groups include a diverse population of women at a range of ages and ethnic backgrounds, lending generalizability to our results.
Limitations of our study include the small sample size and the fact that limited demographic data was available from the population of women from whom cumulus cells were obtained. However, while the finding of low H19 in women with PCOS may be correlative and requires further study, our findings in our H19KO mouse model regarding Cyp17 and testosterone levels in female mice lend support to H19 as a potential additive factor contributing to PCOS-related hyperandrogenemia. Future studies can further explore the relationship between H19 expression and PCOS subtypes (e.g., metabolic and reproductive phenotypes [63]). Additionally, while it has been established that imprinting of genes, including H19, is susceptible to alteration by assisted reproductive technologies [64–66], our findings of high H19 in women with PCOS are consistent across women undergoing controlled ovarian stimulation (COH) and those who presented prior to initiating COH, as well as across different testing modalities (serum and cumulus cells).
In conclusion, ncRNAs may represent new therapeutic targets for the treatment of PCOS and other disorders of steroidogenesis, and the use of circulating ncRNAs has also emerged in non-invasive diagnostic applications. H19 itself has been shown to be a promising biomarker for conditions including ischemic stroke and gastric cancer [17, 60]. The possibility that ovarian expression of H19 plays a role in follicular development, glucose metabolism, and/or androgen production is an intriguing one that warrants further study. Such a link may be particularly relevant to reproductive-aged women with PCOS, in whom associated impaired glucose tolerance has major implications for future cardiovascular health and fertility.
Acknowledgements
H19KO mice were provided by Luisa Dandolo, PhD, and Stefan Muljo, PhD. The University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core (used for serum steroid hormone analysis) is supported by the Eunice Kennedy Shriver NICHD Grant R24 HD102061. We thank Ms. Meirav Sela for her assistance with manuscript proofreading.
Funding
Dr Kallen received funding and research support provided by the NIH-NICHD (R01HD101475), the Reproductive Scientist Development Program (NIH-NICHD Project #2K12HD000849-26), the American Society for Reproductive Medicine, and the NIH Loan Repayment Program. Dr Kallen and Dr Xi received funding and support from the Milstein Medical Asian American partnership Foundation (MMAAPF). The University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core (used for serum steroid hormone analysis) is supported by the Eunice Kennedy Shriver NICHD Grant R24 HD102061.
Footnotes
Ethics Approvals
Studies involving mice were approved by the Yale University Institutional Animal Care and Use Committee (IACUC protocol #2021-20018). Studies utilizing patient samples were approved by the Gazi University Institutional Review Board committee (IRB protocol #131/11.05.2011) and the Yale University Institutional Review Board (IRB protocol #1606017946).
Conflict of Interest
The authors declare no competing interests.
Data Availability
The raw data used to support the findings of this study are available upon request.
References
- 1.Bozdag G, Mumusoglu S, Zengin D, Karabulut E, Yildiz BO. The prevalence and phenotypic features of polycystic ovary syndrome: a systematic review and meta-analysis. Hum Reprod. 2016;31(12):2841–55. 10.1093/humrep/dew218. [DOI] [PubMed] [Google Scholar]
- 2.Randeva HS, Tan BK, Weickert MO, et al. Cardiometabolic aspects of the polycystic ovary syndrome. Endocr Rev. 2012;33(5):812–41. 10.1210/er.2012-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gottschau M, Kjaer SK, Jensen A, Munk C, Mellemkjaer L. Risk of cancer among women with polycystic ovary syndrome: a Danish cohort study. Gynecol Oncol. 2015;136(1):99–103. 10.1016/j.ygyno.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Azziz R, Woods KS, Reyna R, Key TJ, Knochenhauer ES, Yildiz BO. The prevalence and features of the polycystic ovary syndrome in an unselected population. J Clin Endocrinol Metab. 2004;89(6):2745–9. 10.1210/jc.2003-032046. [DOI] [PubMed] [Google Scholar]
- 5.Lerchbaum E, Schwetz V, Rabe T, Giuliani A, Obermayer-Pietsch B. Hyperandrogenemia in polycystic ovary syndrome: exploration of the role of free testosterone and androstenedione in metabolic phenotype. PLoS ONE. 2014;9(10):e108263–e108263. 10.1371/journal.pone.0108263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenfield RL, Ehrmann DA. The pathogenesis of polycystic ovary syndrome (PCOS): the hypothesis of PCOS as functional ovarian hyperandrogenism revisited. Endocr Rev. 2016;37(5):467–520. 10.1210/er.2015-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson VL, Qin KN, Rosenfield RL, et al. The biochemical basis for increased testosterone production in theca cells propagated from patients with polycystic ovary syndrome. J Clin Endocrinol Metab. 2001;86(12):5925–33. 10.1210/jcem.86.12.8088. [DOI] [PubMed] [Google Scholar]
- 8.Wickenheisser JK, Quinn PG, Nelson VL, Legro RS, Strauss JF 3rd, McAllister JM. Differential activity of the cytochrome P450 17alpha-hydroxylase and steroidogenic acute regulatory protein gene promoters in normal and polycystic ovary syndrome theca cells. J Clin Endocrinol Metab. 2000;85(6):2304–11. 10.1210/jcem.85.6.6631. [DOI] [PubMed] [Google Scholar]
- 9.Cech TR, Steitz JA. Review The noncoding RNA revolution — trashing old rules to forge new ones. Cell. 2014;157(1):77–94. 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Yao R-W, Wang Y, Chen L-L. Cellular functions of long noncoding RNAs. Nat Cell Biol. 2019;21(5):542–51. 10.1038/s41556-019-0311-8. [DOI] [PubMed] [Google Scholar]
- 11.Zhang R, Wesevich V, Chen Z, Zhang D, Kallen AN. Emerging roles for noncoding RNAs in female sex steroids and reproductive disease. Mol Cell Endocrinol. 2020;518:110875. 10.1016/j.mce.2020.110875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Men Y, Fan Y, Shen Y, Lu L, Kallen AN. The steroidogenic acute regulatory protein (StAR) is regulated by the H19/let-7 axis. Endocrinology. 2017;158(2):402–9. 10.1210/en.2016-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabory A, Ripoche M-A, Le Digarcher A, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136(20):3413–21. 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- 14.Ripoche MA, Kress C, Poirier F, Dandolo L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 1997;11(12):1596–604. [DOI] [PubMed] [Google Scholar]
- 15.Men Y, Fan Y, Shen Y, Lu L, Kallen AN. The steroidogenic acute regulatory protein (StAR) is regulated by the H19/let-7 axis. Endocrinology. 2017;158(2):402–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caligioni CS. Assessing reproductive status/stages in mice. Curr Protoc Neurosci. 2009;(SUPPL. 48):1–11. 10.1002/0471142301.nsa04is48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Zhao H, Fan Z, et al. Long noncoding RNA H19 promotes neuroinflammation in ischemic stroke by driving histone deacetylase 1–dependent M1 microglial polarization. Stroke. 2017;48(8):2211–21. 10.1161/STROKEAHA.117.017387. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y, Wang Y-Q, Weng W-W, et al. A serum-circulating long noncoding RNA signature can discriminate between patients with clear cell renal cell carcinoma and healthy controls. Oncogenesis. 2016;5(2): e192. 10.1038/oncsis.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Yang K, Yuan W, Gao Z. Determination of serum exosomal H19 as a noninvasive biomarker for bladder cancer diagnosis and prognosis. Med Sci Monit. 2018;24:9307–16. 10.12659/msm.912018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin C, Xia X, Fan Y, et al. A novel, noncoding-RNA-mediated, post-transcriptional mechanism of anti-Mullerian hormone regulation by the H19/let-7 axis. Biol Reprod. 2018;0(August):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moller DE, Flier JS. Insulin resistance — mechanisms, syndromes, and implications. N Engl J Med. 1991;325(13):938–48. 10.1056/NEJM199109263251307. [DOI] [PubMed] [Google Scholar]
- 22.Lorenzo C, Haffner SM, Stančáková A, Laakso M. Relation of direct and surrogate measures of insulin resistance to cardiovascular risk factors in nondiabetic finnish offspring of type 2 diabetic individuals. J Clin Endocrinol Metab. 2010;95(11):5082–90. 10.1210/jc.2010-1144. [DOI] [PubMed] [Google Scholar]
- 23.Su AI, Cooke MP, Ching KA, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci. 2002;99(7):4465–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aherrahrou R, Kulle AE, Alenina N, et al. CYP17A1 deficient XY mice display susceptibility to atherosclerosis, altered lipidomic profile and atypical sex development. Sci Rep. 2020;10(1):1–11. 10.1038/s41598-020-65601-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashraf S, Rasool SUA, Nabi M, et al. CYP17 gene polymorphic sequence variation is associated with hyperandrogenism in Kashmiri women with polycystic ovarian syndrome. Gynecol Endocrinol Off J Int Soc Gynecol Endocrinol. 2021;37(3):230–4. 10.1080/09513590.2020.1770724. [DOI] [PubMed] [Google Scholar]
- 26.Rahimi Z, Mohammadi M Sc E. The CYP17 MSP AI (T-34C) and CYP19A1 (Trp39Arg) variants in polycystic ovary syndrome: a case-control study. Int J Reprod Biomed. 2019;17(3):201–8. 10.18502/ijrm.v17i3.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panda PK, Rane R, Ravichandran R, Singh S, Panchal H. Genetics of PCOS: A systematic bioinformatics approach to unveil the proteins responsible for PCOS. Genomics data. 2016;8:52–60. 10.1016/j.gdata.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Liu F, Luo S, Hu H, Li X-H, Li S-W. Polymorphism T→C of gene CYP17 promoter and polycystic ovary syndrome risk: A meta-analysis. Gene. 2012;495(1):16–22. 10.1016/j.gene.2011.12.048. [DOI] [PubMed] [Google Scholar]
- 29.Pusalkar M, Meherji P, Gokral J, Chinnaraj S, Maitra A. CYP11A1 and CYP17 promoter polymorphisms associate with hyperandrogenemia in polycystic ovary syndrome. Fertil Steril. 2009;92(2):653–9. 10.1016/j.fertnstert.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 30.Munawar Lone N, Babar S, Sultan S, Malik S, Nazeer K, Riaz S. Association of the CYP17 and CYP19 gene polymorphisms in women with polycystic ovary syndrome from Punjab, Pakistan. Gynecol Endocrinol Off J Int Soc Gynecol Endocrinol. September 2020:1–6. 10.1080/09513590.2020.1822803 [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Liang X, Wei L, et al. Study of RNA interference inhibiting rat ovarian androgen biosynthesis by depressing 17alpha-hydroxylase/17, 20-lyase activity in vivo. Reprod Biol Endocrinol. 2009;7:73. 10.1186/1477-7827-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(345):1240–1. 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA Hotair in mouse and human. PLoS Genet. 2011;7(5):1–10. 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kallen AN, Zhou XB, Xu J, et al. The imprinted H19 LncRNA antagonizes Let-7 microRNAs. Molefile///Users/amandakallen/Downloads/Gabory_et_al-2010-BioEssays.pdfcular Cell. 2013;52(1):101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ørom UA, Shiekhattar R. Minireview long noncoding RNAs usher in a new era in the biology of enhancers. Cell. 2013;154(6):1190–3. 10.1016/j.cell.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rachmilewitz J, Goshen R, Ariel I, Schneider T, de Groot N, Hochberg A. Parental imprinting of the human H19 gene. FEBS Lett. 1992;309(1):25–8. 10.1016/0014-5793(92)80731-U. [DOI] [PubMed] [Google Scholar]
- 37.Ariel I, Weinstein D, Voutilainen R, et al. The expression of the imprinted gene H19 in the human female reproductive organs. Diagnostic Mol Pathol. 1997;61(1):17–25. [DOI] [PubMed] [Google Scholar]
- 38.Khatib H, Schutzkus V. The expression profile of the H19 gene in cattle. Mamm Genome. 2006;17(9):991–6. 10.1007/s00335-006-0038-2. [DOI] [PubMed] [Google Scholar]
- 39.Dai A, Sun H, Fang T, et al. MicroRNA-133b stimulates ovarian estradiol synthesis by targeting Foxl2. FEBS Lett. 2013;587(15):2474–82. 10.1016/j.febslet.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 40.Chen B, Xu P, Wang J, Zhang C. The role of MiRNA in polycystic ovary syndrome (PCOS). Gene. 2019;706(March):91–6. 10.1016/j.gene.2019.04.082. [DOI] [PubMed] [Google Scholar]
- 41.Fu L lu, Xu Y, Li D dan, et al. Expression profiles of mRNA and long noncoding RNA in the ovaries of letrozole-induced polycystic ovary syndrome rat model through deep sequencing. Gene. 2018;657(February):19–29. 10.1016/j.gene.2018.03.002 [DOI] [PubMed] [Google Scholar]
- 42.Huang X, Hao C, Bao H, Wang M, Dai H. Aberrant expression of long noncoding RNAs in cumulus cells isolated from PCOS patients. J Assist Reprod Genet. 2016;33(1):111–21. 10.1007/s10815-015-0630-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu H, Shyh-Chang N, Segrè AV, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147(1):81–94. 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frost RJA, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci U S A. 2011;108(52):21075–80. 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barbieri RL, Makris A, Randall RW, Daniels G, Kistner RW, Ryan KJ. Insulin stimulates androgen accumulation in incubations of ovarian stroma obtained from women with hyperandrogenism. J Clin Endocrinol Metab. 1986;62(5):904–10. 10.1210/jcem-62-5-904. [DOI] [PubMed] [Google Scholar]
- 46.Qin L, Huang C, Yan X, Wang Y, Li Z, Wei X. Long non-coding RNA H19 is associated with polycystic ovary syndrome in Chinese women: a preliminary study. Endocr J. 2019. 10.1507/endocrj.ej19-0004. [DOI] [PubMed] [Google Scholar]
- 47.Ou L, Wang D, Zhang H, Yu Q, Hua F. Decreased expression of MiR-138–5p by LncRNA H19 in cervical cancer promotes tumor proliferation. Oncol Res Featur Preclin Clin Cancer Ther. 2017. 10.3727/096504017X15017209042610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou W, Ye X-L, Xu J, et al. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci Signal. 2017;10(483). 10.1126/scisignal.aak9557 [DOI] [PubMed] [Google Scholar]
- 49.Li Z, Li Y, Li Y, et al. Long non-coding RNA H19 promotes the proliferation and invasion of breast cancer through upregulating DNMT1 expression by sponging miR-152. J Biochem Mol Toxicol. 2017;31(e21933):1–9. 10.1002/jbt.21933. [DOI] [PubMed] [Google Scholar]
- 50.Imig J, Brunschweiger A, Brümmer A, et al. miR-CLIP capture of a miRNA targetome uncovers a lincRNA H19–miR-106a interaction. Nat Chem Biol. 2014;11(2):107–14. 10.1038/nchembio.1713. [DOI] [PubMed] [Google Scholar]
- 51.Dumesic DA, Oberfield SE, Stener-Victorin E, Marshall JC, Laven JS, Legro RS. Scientific statement on the diagnostic criteria, epidemiology, pathophysiology, and molecular genetics of polycystic ovary syndrome. Endocr Rev. 2015;36(5):487–525. 10.1210/er.2015-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Q, Shang J, Zhang Y, Zhou W. Metformin and sitagliptin combination therapy ameliorates polycystic ovary syndrome with insulin resistance through upregulation of lncRNA H19. Cell Cycle. 2019;18(19):2538–49. 10.1080/15384101.2019.1652036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Norman RJ, Dewailly D, Legro RS, Hickey TE. Polycystic ovary syndrome. Lancet (London, England). 2007;370(9588):685–97. 10.1016/S0140-6736(07)61345-2. [DOI] [PubMed] [Google Scholar]
- 54.Palomba S, Orio F, Falbo A, et al. Prospective parallel randomized, double-blind, double-dummy controlled clinical trial comparing clomiphene citrate and metformin as the first-line treatment for ovulation induction in nonobese anovulatory women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2005;90(7):4068–74. 10.1210/jc.2005-0110. [DOI] [PubMed] [Google Scholar]
- 55.Mansfield R, Galea R, Brincat M, Hole D, Mason H. Metformin has direct effects on human ovarian steroidogenesis. Fertil Steril. 2003;79(4):956–62. 10.1016/S0015-0282(02)04925-7. [DOI] [PubMed] [Google Scholar]
- 56.Attia GR, Rainey WE, Carr BR. Metformin directly inhibits androgen production in human thecal cells. Fertil Steril. 2001;76(3):517–24. 10.1016/S0015-0282(01)01975-6. [DOI] [PubMed] [Google Scholar]
- 57.Zhong T, Men Y, Lu L, et al. Metformin alters DNA methylation genome-wide via the H19/SAHH axis. Oncogene. 2017;36(17):2345–54. 10.1038/onc.2016.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.La Marca A, Morgante G, Paglia T, Ciotta L, Cianci A, De Leo V. Effects of metformin on adrenal steroidogenesis in women with polycystic ovary syndrome. Fertil Steril. 1999;72(6):985–9. 10.1016/S0015-0282(99)00407-0. [DOI] [PubMed] [Google Scholar]
- 59.Chen Z, Wei H, Zhao X, et al. Metformin treatment alleviates polycystic ovary syndrome by decreasing the expression of MMP-2 and MMP-9 via H19/miR-29b-3p and AKT/mTOR/autophagy signaling pathways. J Cell Physiol. 2019;234(11):19964–76. 10.1002/jcp.28594. [DOI] [PubMed] [Google Scholar]
- 60.Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5(May):1–10. 10.1038/srep11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hashad D, Elbanna A, Ibrahim A, Khedr G. Evaluation of the role of circulating long non-coding RNA H19 as a promising novel biomarker in plasma of patients with gastric cancer. J Clin Lab Anal. 2016;30(6):1100–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xia X, Burn M, Chen Y, Johnson J, Kallen A. The relationship between H19 and parameters of ovarian reserve. Reprod Biol Endocrinol. 2020;In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dapas M, Lin FTJ, Nadkarni GN, et al. Distinct subtypes of polycystic ovary syndrome with novel genetic associations: an unsupervised, phenotypic clustering analysis. PLoS Med. 2020;17(6):e1003132–e1003132. 10.1371/journal.pmed.1003132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sakian S, Louie K, Wong EC, et al. Altered gene expression of H19 and IGF2 in placentas from ART pregnancies. Placenta. 2015;36(10):1100–5. [DOI] [PubMed] [Google Scholar]
- 65.Li T, Vu TH, Ulaner GA, et al. IVF results in de novo DNA methylation and histone methylation at an Igf2-H19 imprinting epigenetic switch. Mol Hum Reprod. 2005;11(9):631–40. [DOI] [PubMed] [Google Scholar]
- 66.Shi X, Ni Y, Zheng H, et al. Abnormal methylation patterns at the IGF2/H19 imprinting control region in phenotypically normal babies conceived by assisted reproductive technologies. Eur J Obstet Gynecol Reprod Biol. 2011;158(1):52–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data used to support the findings of this study are available upon request.


