Abstract
Primary cilia function as cells’ antennas to detect and transduce external stimuli and play crucial roles in cell signaling and communication. The vast majority of cilia genes that are causally linked with ciliopathies are also associated with neurological deficits, such as cognitive deficits. Yet, the roles of cilia dysfunctions in the pathogenesis of psychiatric disorders have not been studied. Our aim is to identify patterns of cilia gene dysregulation in the four major psychiatric disorders: schizophrenia (SCZ), autism spectrum disorder (ASD), bipolar disorder (BP), and major depressive disorder (MDD). For this purpose, we acquired differentially expressed genes (DEGs) from the largest and most recent publicly available databases. We found that 42%, 24%, 17%, and 15% of brain cilia genes were significantly differentially expressed in SCZ, ASD, BP, and MDD, respectively. Several genes exhibited cross-disorder overlap, suggesting that typical cilia signaling pathways’ dysfunctions determine susceptibility to more than one psychiatric disorder or may partially underlie their pathophysiology. Our study revealed that genes encoding proteins of almost all sub-cilia structural and functional compartments were dysregulated in the four psychiatric disorders. Strikingly, the genes of 75% of cilia GPCRs and 50% of the transition zone proteins were differentially expressed in SCZ.
The present study is the first to draw associations between cilia and major psychiatric disorders, and is the first step toward understanding the role that cilia components play in their pathophysiological processes, which may lead to novel therapeutic targets for these disorders.
Keywords: Cilia, Compartments, Schizophrenia, Autism, Bipolar disorder, Depression, DEGs
1. Introduction
Primary cilia are microtubule-based organelles that protrude from the surface of almost all cell types including neurons [1–3]. Cilia, functioning as cells’ antennas, detect and transduce external stimuli, thus playing crucial roles in cell communication and signaling. Cilia are essential sites for the Sonic Hedgehog (Shh) signaling pathway, and therefore, play a vital role in neurogenesis, cell differentiation, and migration [4, 5].
The sensory functions and signal transduction of primary cilia are achieved through specific receptors and downstream signaling components including receptor tyrosine kinases (RTK), hedgehog, WNT, ion channels, and G-protein-coupled receptors (GPCRs) [6–15]. Examples of neuronal cilia GPCRs are somatostatin receptor 3 (SSTR3), serotonin receptor 6 (5HTR6), melanin-concentrating hormone receptor 1 (MCHR1), melanocortin receptors MC4R, and dopamine receptors (DRD1, DRD2 and DRD5), GPR88, prolactin-releasing hormone receptor (PRLHR), and neuropeptide FF receptor 1 (NPFFR1) [6, 16–20].
Mutations or deletions of primary cilia genes cause disruptions in ciliary structure and functions and result in neurodevelopment defects. Of the 187 cilia genes that were causally associated with ciliopathies, several genes have been associated with neurological deficits such as cognitive deficits [21–23].
Since primary cilia’s main function of is to receive extracellular signals, dysfunctional cilia can cause cellular defects in response to extracellular cues. On the other hand, neuronal GPCRs play essential roles in regulating emotions, memory, and cognition (see [24–29] for reviews). Therefore, defects in the cilia GPCRs are expected to correlate with psychiatric disorders
Given the background information, our premises indicate that cilia structure/function has critical roles in cognitive brain functions. Thus, we hypothesize that the dysfunctions of cilia components are associated with psychiatric disorders.
In support of this hypothesis, the causative gene for the ciliopathy Joubert Syndrome, AHI1 which is involved in the ciliogenesis [30], and PCM1 and TSGA14, which are necessary for cilia assembly, have been linked with schizophrenia (SCZ) and autism spectrum disorder (ASD) [31–38]. Olfactory neural progenitor (ONP) cells derived from paranoid SCZ and bipolar disorder (BD) patients display a deficit in cilia assembly and maintenance [39].
However, a comprehensive study on the conclusive role of cilia in psychiatric disorders has never been conducted yet. Therefore, we examined here the changes in gene expression of cilia associated proteins in four psychiatric disorders including SCZ, ASD, BP, and MDD. We used publicly available databases of RNA sequencing (RNA-seq) and RNA-microarray in the largest cohorts of cases and controls to acquire differentially expressed genes (DEGs) in the four psychiatric disorders.
2. Methods
2.1. Compilation of brain cilia genes list
Genes experimentally probed and identified to be involved in cilia structure, ciliogenesis or ciliary function were collected from different studies and databases including Nauli et. al. [40], SysCilia Gold Standard (SCGS) version 1 database [41], CilDB database [42, 43] and CiliaCarta [44]. Search criteria included Homo sapiens species cilia genes. CiliaCarta database contains about 1000 putative cilia genes, including genes from Syscilia (302 genes) and Gene Ontology (GO) annotated genes (677), with 232 genes overlapped, as well as bayesian predictions (285 genes). Our initial list included the Sycilia and GO genes but not bayesian predictions. GPCRs (G-protein coupled receptor) localized on cilia were acquired from Schou et. al [45]. Ciliary evidence was then verified using CilDB. It is important to disclose that lists of cilia genes are continuously being updated, and one will never be ‘complete’ as novel cilia genes might always be identified. Thus, our list is a working list of cilia genes that will be updated as needed.
This list of 455 candidate ciliary genes were then compiled and further analyzed using Genotype-Tissue Expression (GTEx) https://gtexportal.org/home/. GTEx database was queried for cross tissue comparisons to determine each of the cilia genes’ brain location. Cilia genes were located in the following brain regions: amygdala, caudate, cerebral hemisphere, cortex, frontal cortex, hippocampus, hypothalamus, nucleus accumbens, putamen, and substantia nigra and expressed as the median transcripts per million (TPM). Cilia genes not located in the brain were removed from the list. Therefore, the final list of cilia genes totaled 281.
2.2. Transcriptomic analysis of cilia genes in SCZ, ASD, BP, and MDD
We screened publically available datasets for gene expression of genes encoding cilia associated proteins. We obtained the gene expression data of prefrontal cortex tissue samples (559 SCZ, 51 ASD, 222 BP, and 936 controls) from the study of Gandal et al [46], which retrieved RNA-seq data from PsychEncode (PEC, Brain-GVEX study) and CommonMind (CMC) datasets [46–49]. The DEGs data of cortical brain tissue samples (87 MDD, and 293 controls) were obtained from the study of Gandal et al [50], which performed meta-analyses of microarray data that were obtained from the Gene Expression Omnibus (GEO), ArrayExpress, or from the study authors directly conducted [50, 51]. Cilia differentially expressed genes (DEGs) in SCZ, ASD, BP, and MDD (P < 0.05) were identified from the transcriptomic data and underwent Fisher exact test to examine the enrichment of differentially expressed cilia genes in SCZ, ASD, BP, and MDD. The overlap of DEGs across the psychiatric disorders was computed using Fisher’s exact test. Cilia genes were then categorized based on their localization to the cilium, including basal body, axoneme, IFTA, IFTB, kinesin, dynein, transition zone, bbsome, golgi, nucleus, cytosol and the ciliary membrane. If unknown, the genes were placed in one group designated as other.
3. Results
3.1. Brain Cilia list
Using SysCilia, CilDB, and CiliaCarta databases, we identified and verified 455 candidate ciliary genes. Using GTEX, 281 genes out of these cilia genes were confirmed to be expressed in the brain’s following regions: amygdala, caudate, cerebral hemisphere, cortex, frontal cortex, hippocampus, hypothalamus, nucleus accumbens, putamen, and substantia nigra. The heatmap shows GTEx expression profiles in median TPMs of the cilia genes (Fig. 1, Table S1). Genes were considered if the median TPM was greater than zero. Although few genes had considerably low median TPMs, they were included in the list, as they are not vastly localized in other non-brain tissues in the body.
Figure 1. Identification of cilia gene expression across specific brain tissues in GTEx.
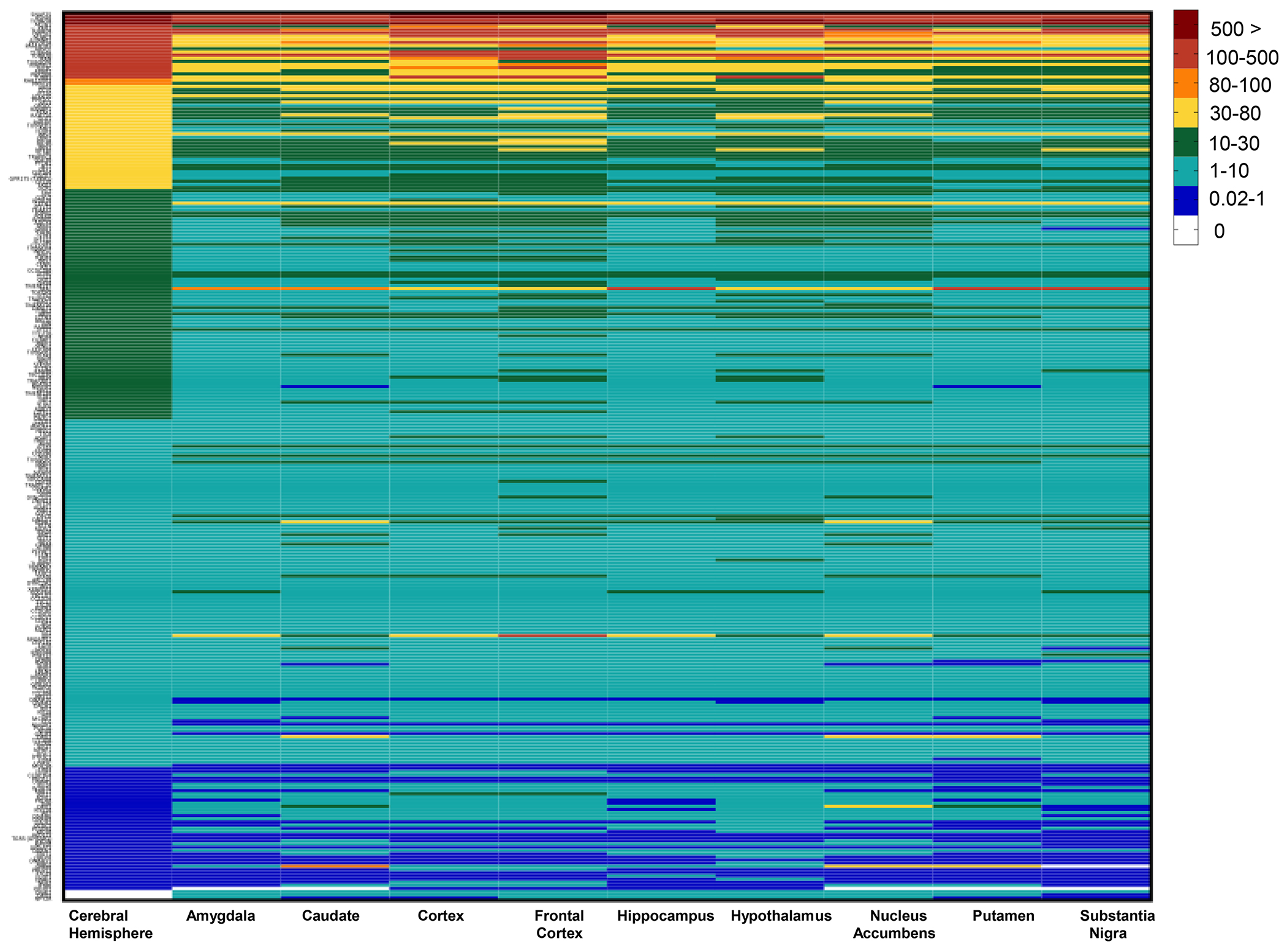
Heatmap of gene expressions of cilia genes in the GTEx brain tissues using median TPMs from the GTEx dataset. Columns denote brain regions and rows represent cilia genes. Genes are shown and categorized in median Transcripts Per Million (TPM).
3.2. The expressions of cilia genes are altered in psychiatric disorders
The RNA-Seq data for 268 genes of the 281 cilia genes in our compiled list were extracted from the largest TWAS study of psychiatric disorders [46, 50]. Out of the 268 cilia genes, 113 were differentially expressed in SCZ, (48 downregulated and 65 upregulated) compared to 7726 out of 25502 for non-cilia genes (Fig. 2a). Of these genes, 97 genes passed false discovery rate (FDR) < 0.1, and 74 genes passed FDR < 0.05. Exact fisher analysis revealed significant enrichment of differentially expressed cilia genes in SCZ (Odd ratio OR = 1.67, P < 0.0001, Fig. 2b). Strikingly, out of the 20 known cilia-associated GPCRs, 15 were differentially expressed in SCZ. Fisher analysis of cilia GPCRs revealed significant enrichment of cilia GPCRs in SCZ (OR: 5.35, P = 0.0003, Fig. 2c).
Figure 2. Cilia differentially expressed genes in SCZ, ASD, BP, and MDD.
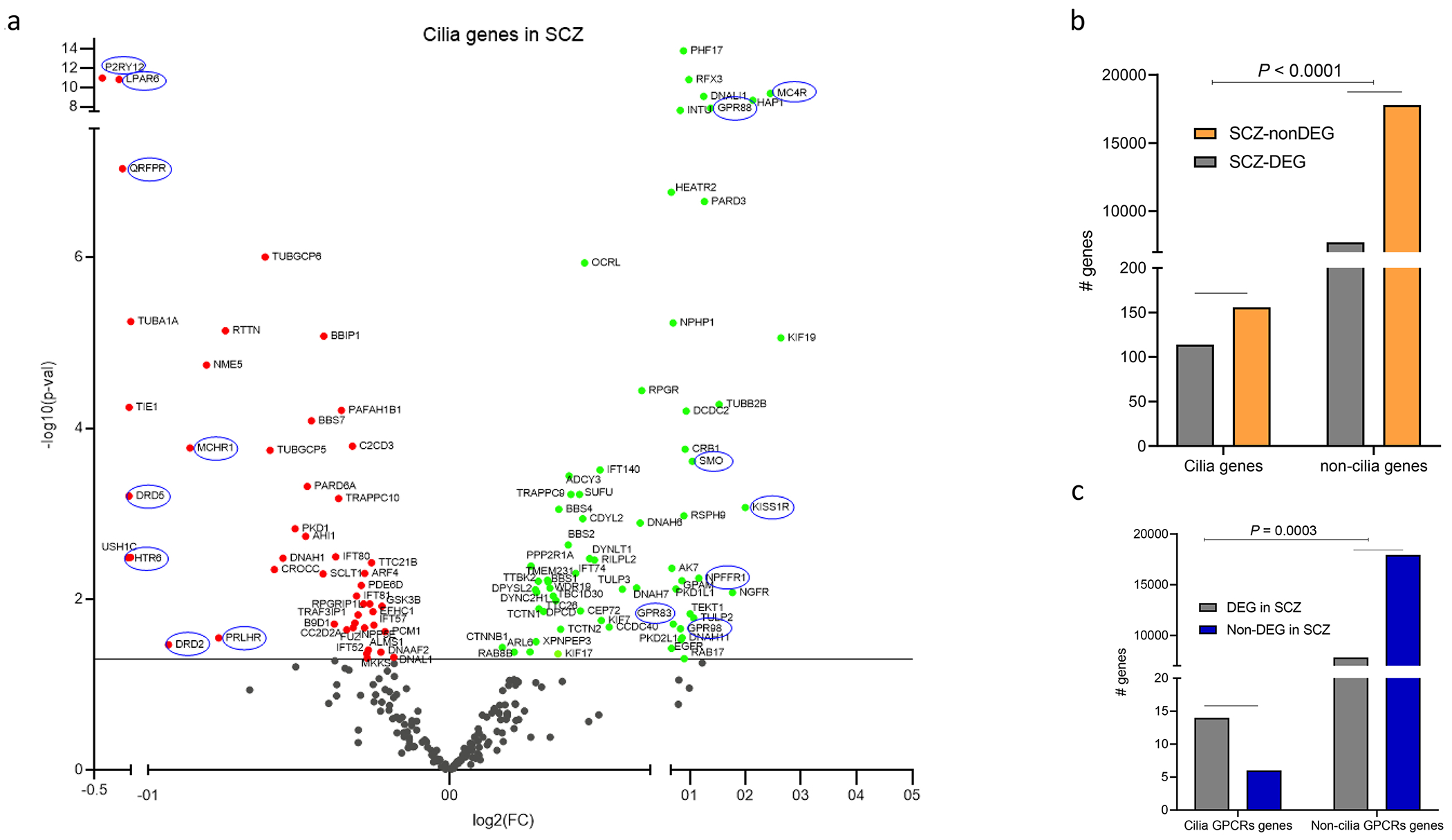
a. Volcano plot showing DEGs (P <0.05). Scattered points represent cilia genes: the x axis is the fold change while the y axis is the -log p value. Green and red dots refer to upregulated and downregulated genes. Genes circled in blue represent GPCRs that localize to the cilium.
b. Histogram showing the cilia genes and non-cilia genes that are differentially expressed in SCZ; Exact Fisher test OR = 1.681, P < 0.0001.
c. Histogram showing cilia GPCR genes and non-cilia GPCR genes that are differentially expressed in SCZ; Exact Fisher test: OR: 6.873, P = 0.0001.
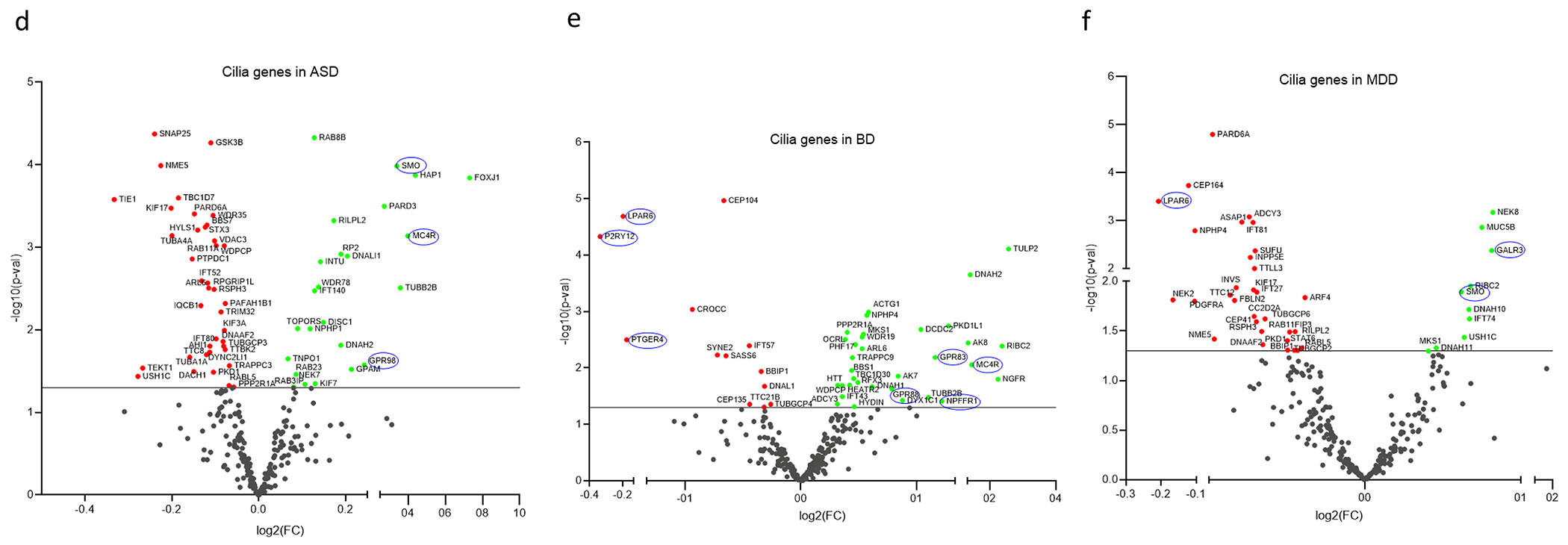
d-f. Volcano plots of cilia DEGs in (d) ASD, (e) BP, and (f) MDD.
Out of the 268 cilia genes, 63 genes including 3 GPCRs (39 genes downregulated and 24 upregulated), and 46 cilia genes including 7 GPCRs (13 genes downregulated and 33 genes upregulated) were differentially expressed in ASD and BP respectively (Fig. 2d,e). Exact fisher test revealed no significant enrichment of the cilia genes as a cluster in ASD (OR = 1.20, P = 0.2) or BP (OR = 0.91, P = 0.6). In MDD, 41 cilia genes including 3 GPCRs were differentially expressed, of which 31 genes were downregulated and only 10 were upregulated (Fig. 2f). Exact fisher test revealed no significant enrichment of the cilia genes in MDD (OR = 0.81, P = 0.23).
Several cilia DEGs overlapped across the four disorders, particularly between SCZ and each of the three other disorders (Fig. 3a–j). Fisher exact test revealed significant DEGs overlap between SCZ and BP (P = P = 0.0009), and a trend in overlap between SCZ and ASD (P = 0.0795), and between the down regulated genes in SCZ and MDD (P = 0.07, Fig. 3a).
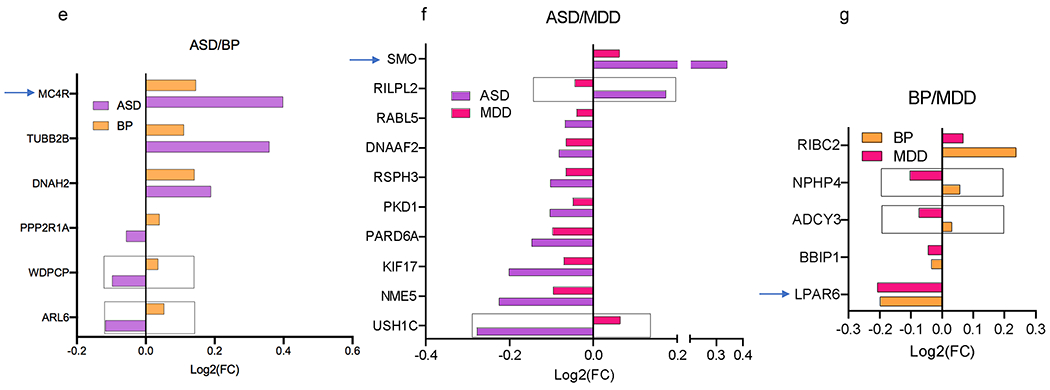
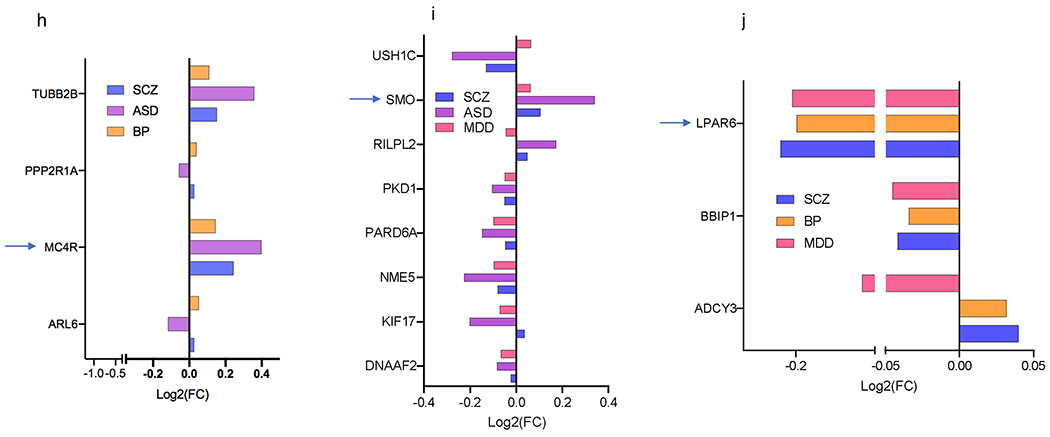
Figure 3. Cilia DEGs overlap across the four psychiatric disorders.
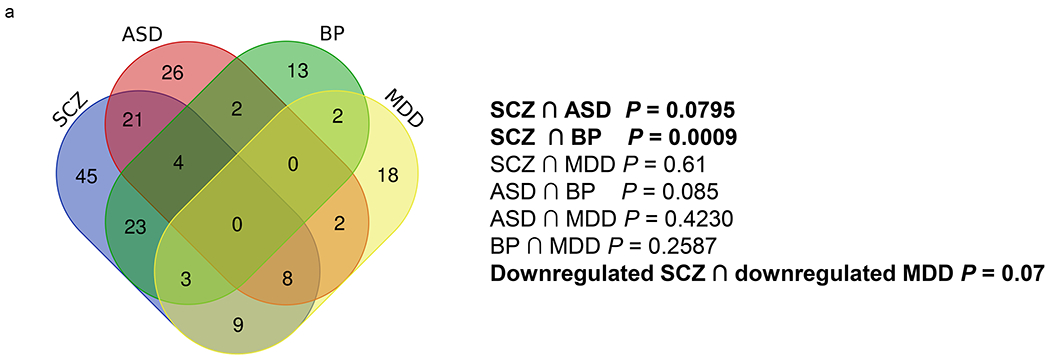
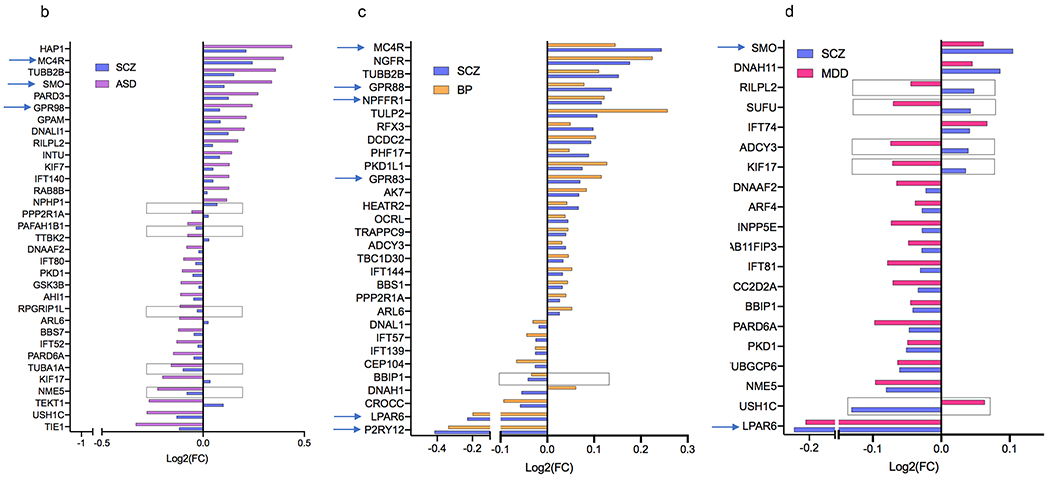
a. Venn diagram conveying overlap between significant DEGs (P<0.05) within 4 psychiatric disorders, SCZ, ASD, BP, and MDD.
b-d. Cilia DEGs genes overlapping between SCZ with (b) ASD, (c) BP, (d) and MDD. Genes that are differentially expressed in opposing directions are denoted with a box. GPCRs are identified by arrows.
e-g. Overlap of DEG between two disorders: (e) ASD and BP, (f) ASD and MDD, (g) BP and MDD.
h-j. Overlap of DEG among three disorders: (h) SCZ, ASD, and BP, (i) SCZ, ASD, and MDD, (j) SCZ, BP, MDD. The x axis shows Log2FC and along the y axis are the cilia genes. Genes that are differentially expressed in opposing directions are denoted with a box. GPCRs are identified by arrows.
Approximately 50% of ASD cilia DEGs (33 genes) including all the three differential GPCRs in ASD (MC4R, SMO, and GPR98) were also differentially expressed in SCZ (Fig. 3b). Around two-third of BP cilia DEGs (30 genes) including 6 GPCRs: MC4R, GPR83, GPR88, NPFFR1, and P2RY12, LPAR6 were differentially expressed in SCZ (Fig. 3c), and 20 MDD cilia DEGs overlapped with SCZ (Fig. 3d).
3.3. All cilia structural and functional compartments are associated with the four psychiatric disorders
We next identified the sub-cilia localization of cilia DEGs in the four disorders. We found that proteins encoded by the SCZ cilia DEGs were localized in almost all cilia sub-compartments, including the axoneme (core of the cilium), basal body, ciliary membrane proteins, intraflagellar transport cargo (IFTA and IFTB), cytoskeletal motor proteins (dynein and kinesin family), BBSome membrane protein trafficking complex, and the transition zone (Fig. 4a). We found that 50% of the transition zone genes (33% upregulated and 16% downregulated), 40% of the axoneme genes, 38% of the BBSome complex genes, and 30% of the basal body genes were DEGs in SCZ. While 40% of intraflagellar complex A (IFT-A) genes were DEGs in SCZ, only 6% of IFT-B genes displayed expression changes (upregulation) in this disorder. Along with the in IFT-A and IFT-B gene changes, 100% of kinesin DEGs compared with 33% of dynein DEGs exhibited upregulation. Strikingly, 75% of cilia GPCRs exhibited altered expression in SCZ.
Figure 4. Sub-cilia localization of the genes that exhibited variants association and/or differential gene expression in the four disorders.
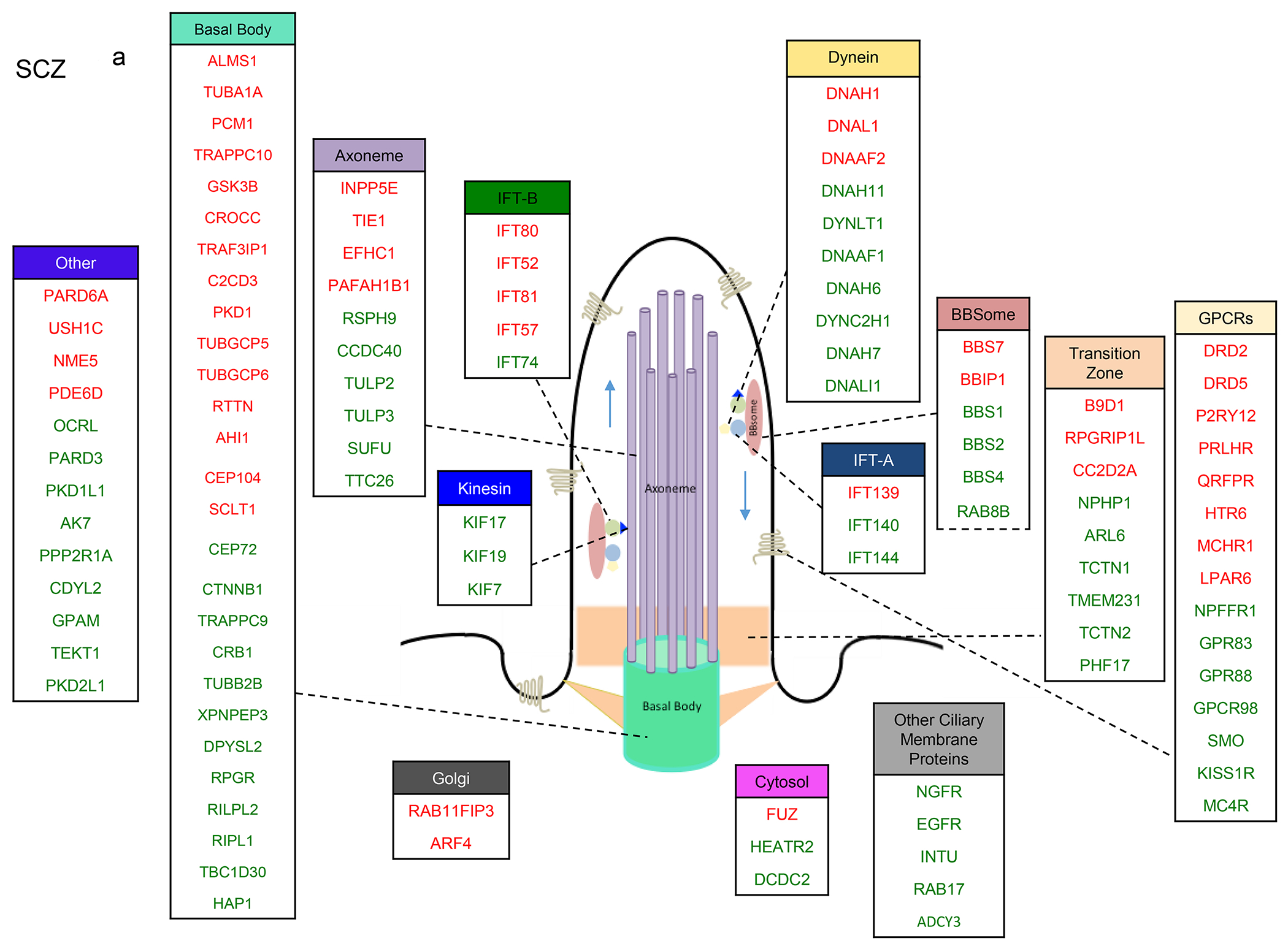
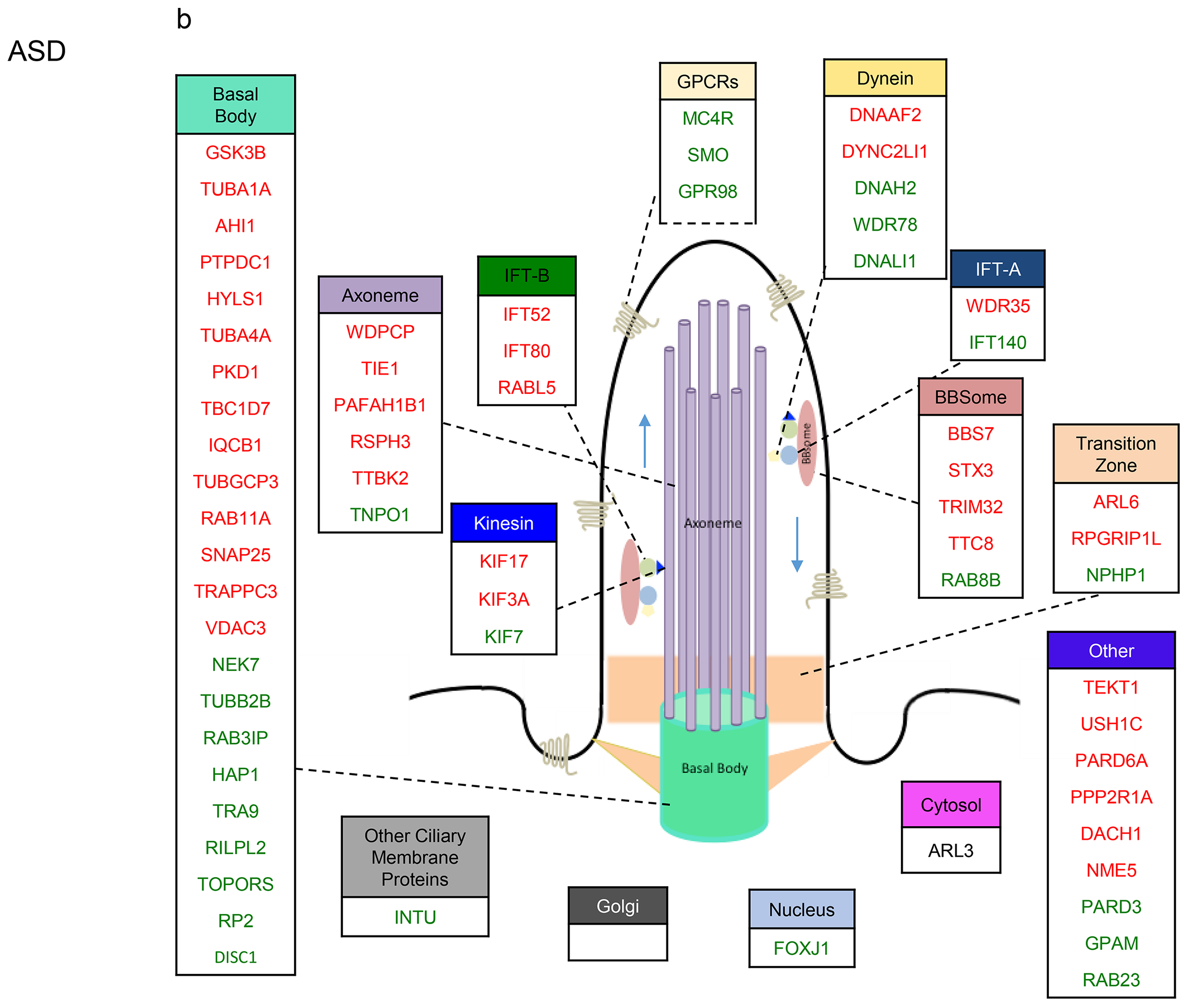
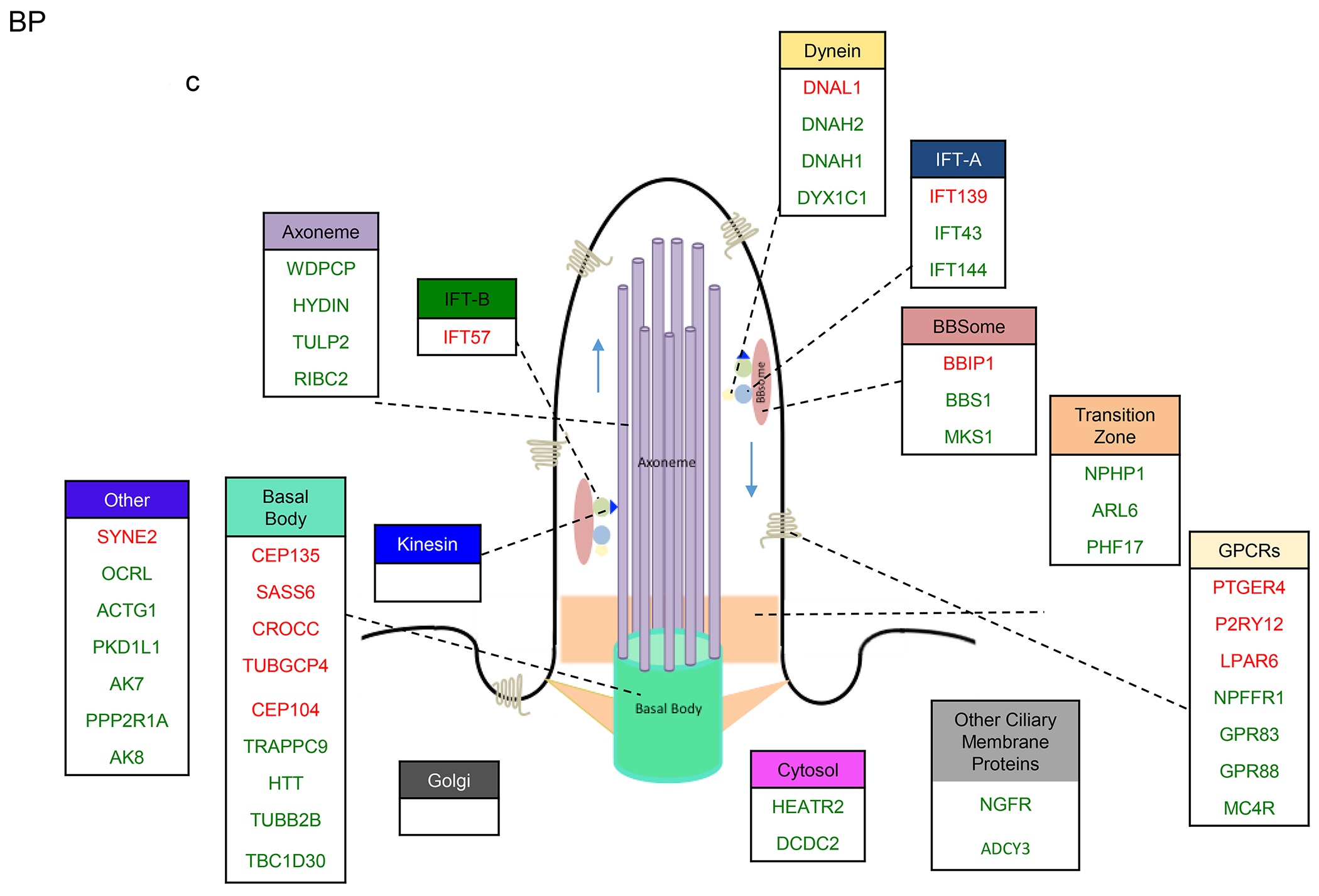
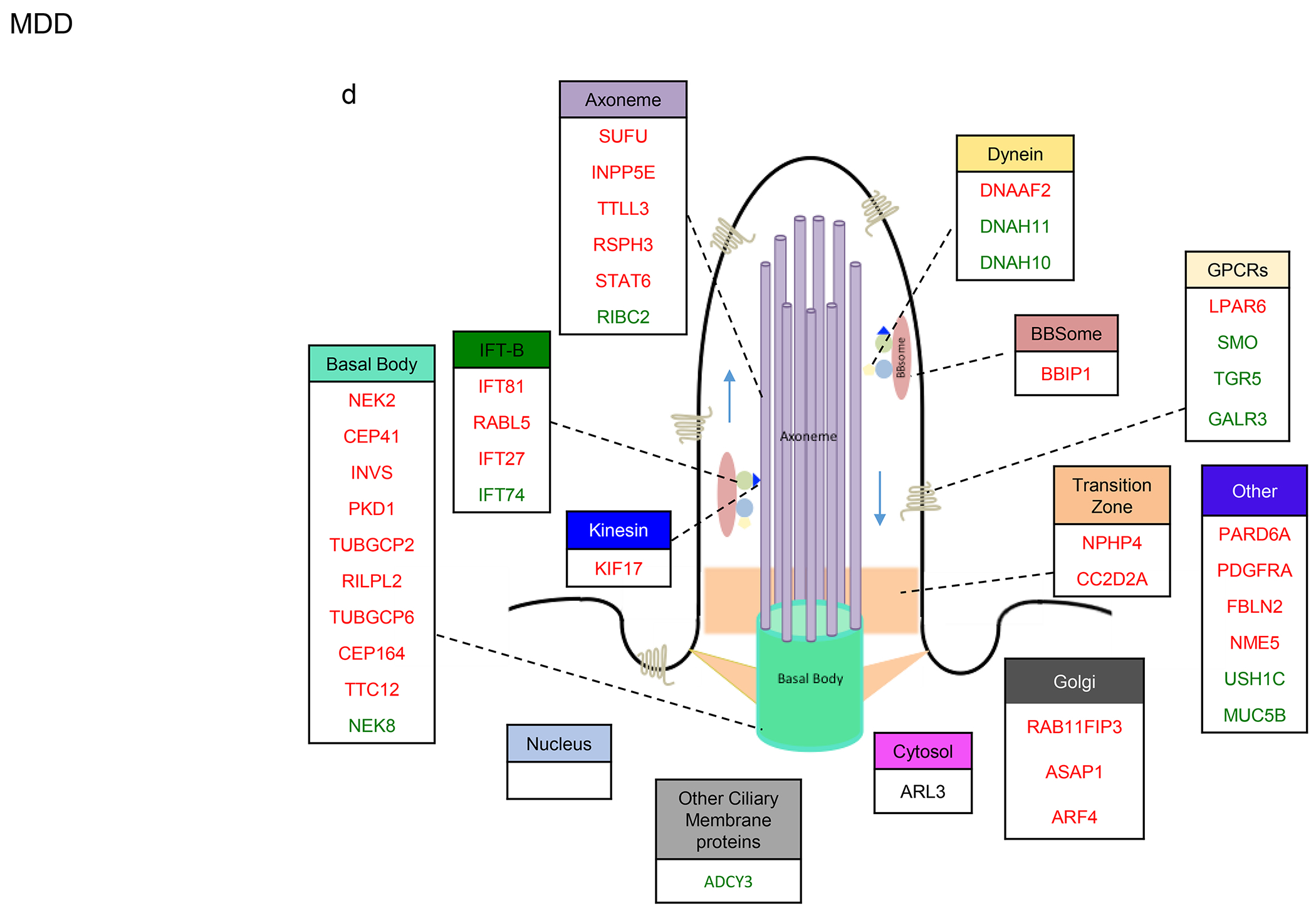
a-d. Sub-cilia localization of genes that exhibited differential gene expression and/or genetic variants in (a) SCZ, (b) ASD, (c) BP, (d) MDD. Upregulated and downregulated DEGs are in green and red respectively.
We found that 33% of IFT-A and 19% of IFT-B were DEGs in ASD, and that all IFT-B DEGs exhibited downregulation (Fig. 4b). While all Kinesin DEGs displayed downregulation, there were more upregulated dynein genes. Around 20% of genes localized to the cilia axoneme were DEGs in ASD, of which the vast majority exhibited downregulation. 29% of basal body genes were DEGs, and 20% of GPCRs were upregulated in ASD.
In BP, 50% of IFT-A were DEGs (two-fold more upregulated genes than downregulated), and 6% of IFT-B were dysregulated (all downregulated) (Fig. 4c). Aligned with these, none of the kinesin genes and 19% dynein genes were DEGs.
In MDD, 25% of IFT-B genes, 13% of kinesin genes, 14% of dynein genes, 20% of axoneme genes, 11% of transition zone genes, and 15% of GPCRs were dysregulated (Fig. 4d). Interestingly, 75% of genes involved in the Golgi network were downregulated in MDD.
Across the disorders, several cilia DEGs overlapped within most of the cilia structure sub-compartments (Fig. S1a–f).
4. Discussion
This is the first study that describes the association between the brain cilia components at the transcriptomic and genomic and the cilia structural/compartmental level.
Despite the growing interest in cilia functions, very little is known about their potential involvement in psychiatric disorders. Remarkably, many of the cilia genes that have been causally associated with ciliopathies, which result in disruptions of ciliary structure and functions, are also associated with neurological defects, such as cognitive deficits [21–23]. Yet, the roles of cilia dysfunctions in the pathogenesis of psychiatric disorders have been largely overlooked.
Therefore, this study is the first step toward understanding the link between the dysregulation of the components of primary cilia and four major psychiatric disorders, namely schizophrenia, autism spectrum disorder, bipolar disorder, and major depressive disorder.
To examine whether the cilia genes are differentially expressed and identify the patterns of gene expression dysregulations in the four psychiatric disorders, we acquired transcriptomic data from the largest cohorts of cases and controls [46, 50]. We show that over 40% of the brain cilia genes are differentially expressed in SCZ, with significant enrichment of differentially expressed cilia genes in this disorder. The dysregulated cilia genes in SCZ encode proteins that are located in almost all cilia structural compartments including basal body, axoneme, cytoskeletal motor proteins (dynein and kinesin family), intraflagellar transport cargo (IFT-A and IFT-B), BBSome, and the transition zone.
Interestingly, all kinesin and kinesin-associated DEGs in SCZ were upregulated, yet most IFT-B DEGs were downregulated. Given the crucial role of the kinesin/IFT-B system in cilia anterograde transport, the dysregulation of this system may negatively affect base-to-tip transport of cilia proteins particularly cilia-associated GPCRs [52]. Studies have shown that a complete loss of any gene involved with the IFT-B complex or kinesin leads to shortening or cilia loss [53–61]. Also, the retrograde system consisting of dynein and IFT-A was also dysregulated, and more than 40% of dynein and IFT-A proteins, including IFT144 and IFT140, were dysregulated. IFT-A complex is associated with dynein motor proteins, and is required for cilia function, assembly, and trafficking within cilia [62]. Mutations in the IFT-A complex subunits and the dynein two retrograde motor result in shortened or swollen cilia with an accumulation of particles, including IFT-B proteins, and cause decreased retrograde transport velocities, increased ratio of anterograde to retrograde particles, and an accumulation of complex B proteins in the flagella [63–65]. This complex is particularly associated with ciliary retrograde transport, and it particularly promotes the trafficking of GPCRs [52], which were strikingly dysregulated SCZ (75% of cilia GPCRs were DEGs in SCZ).
TULP3, which encodes the axoneme protein TUB Like Protein 3 that interacts with IFT-A protein (particularly IFT144) [66], is also upregulated along with IFT144 in SCZ. TULP3 and IFT-A proteins negatively regulate Hedgehog signaling in embryonic development and brain formation [66–68]. Therefore, TULP3/IFT-A upregulation in SCZ suggests the role of cilia components in the abnormal neurodevelopment in this disorder [66].
The transcriptomic signature of the three other psychiatric disorders ASD, BP, and MDD included many cilia genes. Remarkably, 60% of cilia DEGs were downregulated in ASD, compared to only 30% in BP and 76% in MDD. The high rates of upregulated genes and downregulated genes in bipolar mood disorders and MDD (unipolar mood disorder) respectively are very interesting, given the similarities in the clinical presentations between MDD and the depressive episodes in BP. The differences observed in the trends and directions of cilia genes’ alterations in MDD and BP indicate that these two mood disorders, albeit their similar depressive symptoms, display distinct cilia transcriptomic signatures. These differences suggest discrete neuropathological mechanisms underlying these two disorders, and help develop potential classification tools, which might improve diagnostic accuracy and therapeutic precision.
Strikingly, over 50% of cilia DEGs overalp between ASD, BP, or MDD on one side with SCZ on the other side. ADCY3, which was largely upregulated in SCZ and BP, exhibited downregulation in MDD [46, 50]. Such large transcriptomic cross-disorder overlap suggests that dysfunctions of common cilia signaling pathways may partially underlie the pathophysiology of, or determine susceptibility to more than one psychiatric disorder. ADCY3 encodes the enzyme adenylate cyclase-3 that catalyzes the synthesis of cyclic adenosine monophosphate. ADCY3, known to be associated with ASD [69], was identified by the genome wide study (GWAS) of major depressive disorder (MDD) as one of the three top associated genes [70], and depressed patients display a reduction in platelet ADCY3 activity [71]. Interestingly, ADCY3 knockout mice exhibit depression like symptoms, more REM sleep, smaller hippocampus, less synaptic activity in the hippocampus and impaired sociability [72]. Another interesting cilia gene is TUBB2B, which is upregulated in SCZ, ASD, and BP. TUBB2B encodes tubulin beta-2B chain, and the dysregulation of this gene results in malformations of development of the cortex and basal ganglia [73–77]. These effects are thought to be caused by altering tubulin heterodimer or GTP binding, and disrupting the interaction between microtubule polymers with associated proteins (e.g. kinesin), ultimately leading to changes in the dynamic properties of microtubule polymers [78, 79].
The overlap observed in cilia DEGs between the four major psychiatric disorders is aligned with the consistent GWAS data on the shared genetic susceptibility across psychiatric disorders [50, 80–82]. Thus, our results suggest that shared cilia-related molecular and cellular mechanisms may partially underlie these disorders’ clinical phenotypes, particularly since most shared cilia DEGs encode products known to interact or function in common biochemical pathways. Our findings, however, acknowledge the need to construct new classifications of psychiatric disorders, considering the convergence and divergence of biological mechanisms and phenotypic presentations of these disorders.
PCM1, one of the top cilia DEGs in SCZ, has recently emerged as a credible candidate for severe schizophrenia [83]. Missense mutation and polymorphisms of PCM1, have been previously associated with schizophrenia [35, 84]. PCM1, which is a DISC1- interacting protein, is involved in the maintenance of centrosome integrity and regulation of microtubule cytoskeleton and dopamine transmission [85–87]. Suppression of PCM1 affects neuronal migration and plays a role in cortical development and genetic susceptibiity to schizophrenia [88], and Pcm1+/− mice display deficits in social interaction and show significant reduction in brain volume [89].
The striking alterations in the expressions of GPCRs in the four psychiatric disorders, particularly in SCZ (75% of cilia GPCRs) indicate a crucial role for the cilia GPCRs in the pathogenesis of these disorders. Cilia-associated GPCRs regulate cilia formation, structure, and ability to respond to external stimuli [90], and therefore, are essential for correct ciliary signal transduction. The high “druggability” of GPCRs, and the advancements in the cilia targeted drug delivery [91, 92] make these receptors of great interest as druggable sites, which might lead to novel treatments and/or prevention strategies for these psychiatric disorders.
In conclusion, we demonstrated gene dysregulations of almost all structural and functional components of primary cilia in four major psychiatric disorders, and identify large transcriptomic cross-disorder overlap. Our work provides strong evidence for the potential that primary cilia have as possible pathophysiological cause or therapeutic target in psychiatric disorders.
Supplementary Material
Highlights.
Around 90% of cilia genes’ mutations that result in disruptions of ciliary structure and functions and cause ciliopathies are also associated with neurological defect, such as cognitive dysfunctions
We demonstrate gene dysregulations of almost all structural and functional components of primary cilia in four major psychiatric disorders: schizophrenia, autism spectrum disorder, bipolar disorder, and major depressive disorder
We provide a strong evidence for the potential that primary cilia have as possible pathophysiological cause or therapeutic target in psychiatric disorders.
Acknowledgements
The work of AA was supported by the department of pharmaceutical sciences, UCI. The work of OC was supported by Eric L. and Lila D. Nelson Chair in Neuropharmacology. The work of SC and PB was supported in part by NIH grant GM123558.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare no competing interests.
Ethical Statement
We confirm that this work is original and has not been published elsewhere, nor is it currently under consideration for publication elsewhere.
References
- 1.Fuchs JL, Schwark HD: Neuronal primary cilia: a review. Cell biology international 2004, 28(2):111–118. [DOI] [PubMed] [Google Scholar]
- 2.Baudoin JP, Viou L, Launay PS, Luccardini C, Espeso Gil S, Kiyasova V, Irinopoulou T, Alvarez C, Rio JP, Boudier T et al. : Tangentially migrating neurons assemble a primary cilium that promotes their reorientation to the cortical plate. Neuron 2012, 76(6):1108–1122. [DOI] [PubMed] [Google Scholar]
- 3.Gerdes JM, Davis EE, Katsanis N: The vertebrate primary cilium in development, homeostasis, and disease. Cell 2009, 137(1):32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Louvi A, Grove EA: Cilia in the CNS: the quiet organelle claims center stage. Neuron 2011, 69(6):1046–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trulioff A, Ermakov A, Malashichev Y: Primary Cilia as a Possible Link between Left-Right Asymmetry and Neurodevelopmental Diseases. Genes 2017, 8(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Omori Y, Chaya T, Yoshida S, Irie S, Tsujii T, Furukawa T: Identification of G Protein-Coupled Receptors (GPCRs) in Primary Cilia and Their Possible Involvement in Body Weight Control. PloS one 2015, 10(6):e0128422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hilgendorf KI, Johnson CT, Jackson PK: The primary cilium as a cellular receiver: organizing ciliary GPCR signaling. Current opinion in cell biology 2016, 39:84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lancaster MA, Gleeson JG: The primary cilium as a cellular signaling center: lessons from disease. Current opinion in genetics & development 2009, 19(3):220–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen ST, Morthorst SK, Mogensen JB, Pedersen LB: Primary Cilia and Coordination of Receptor Tyrosine Kinase (RTK) and Transforming Growth Factor beta (TGF-beta) Signaling. Cold Spring Harbor perspectives in biology 2017, 9(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valencia-Gattas M, Conner GE, Fregien NL: Gefitinib, an EGFR Tyrosine Kinase inhibitor, Prevents Smoke-Mediated Ciliated Airway Epithelial Cell Loss and Promotes Their Recovery. PloS one 2016, 11(8):e0160216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans MJ, Fanucchi MV, Van Winkle LS, Baker GL, Murphy AE, Nishio SJ, Sannes PL, Plopper CG: Fibroblast growth factor-2 during postnatal development of the tracheal basement membrane zone. American journal of physiology Lung cellular and molecular physiology 2002, 283(6):L1263–1270. [DOI] [PubMed] [Google Scholar]
- 12.Nauli SM, Pala R, Kleene SJ: Calcium channels in primary cilia. Current opinion in nephrology and hypertension 2016, 25(5):452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nachury MV: The molecular machines that traffic signaling receptors into and out of cilia. Current opinion in cell biology 2018, 51:124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu B, Chen S, Cheng D, Jing W, Helms JA: Primary cilia integrate hedgehog and Wnt signaling during tooth development. Journal of dental research 2014, 93(5):475–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.May-Simera HL, Kelley MW: Cilia, Wnt signaling, and the cytoskeleton. Cilia 2012, 1(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brailov I, Bancila M, Brisorgueil MJ, Miquel MC, Hamon M, Verge D: Localization of 5-HT(6) receptors at the plasma membrane of neuronal cilia in the rat brain. Brain research 2000, 872(1-2):271–275. [DOI] [PubMed] [Google Scholar]
- 17.Berbari NF, Lewis JS, Bishop GA, Askwith CC, Mykytyn K: Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proceedings of the National Academy of Sciences of the United States of America 2008, 105(11):4242–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Domire JS, Green JA, Lee KG, Johnson AD, Askwith CC, Mykytyn K: Dopamine receptor 1 localizes to neuronal cilia in a dynamic process that requires the Bardet-Biedl syndrome proteins. Cellular and molecular life sciences : CMLS 2011, 68(17):2951–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Handel M, Schulz S, Stanarius A, Schreff M, Erdtmann-Vourliotis M, Schmidt H, Wolf G, Hollt V: Selective targeting of somatostatin receptor 3 to neuronal cilia. Neuroscience 1999, 89(3):909–926. [DOI] [PubMed] [Google Scholar]
- 20.Varela L, Horvath TL: Neuronal Cilia: Another Player in the Melanocortin System. Trends in molecular medicine 2018, 24(4):333–334. [DOI] [PubMed] [Google Scholar]
- 21.Hartwig C, Monis WJ, Chen X, Dickman DK, Pazour GJ, Faundez V: Neurodevelopmental disease mechanisms, primary cilia, and endosomes converge on the BLOC-1 and BORC complexes. Developmental neurobiology 2018, 78(3):311–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valente EM, Rosti RO, Gibbs E, Gleeson JG: Primary cilia in neurodevelopmental disorders. Nature reviews Neurology 2014, 10(1):27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reiter JF, Leroux MR: Genes and molecular pathways underpinning ciliopathies. Nature reviews Molecular cell biology 2017, 18(9):533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung CCY, Wong YH: Role of G Protein-Coupled Receptors in the Regulation of Structural Plasticity and Cognitive Function. Molecules 2017, 22(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts AJ, Hedlund PB: The 5-HT(7) receptor in learning and memory. Hippocampus 2012, 22(4):762–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bassilana F, Nash M, Ludwig MG: Adhesion G protein-coupled receptors: opportunities for drug discovery. Nature reviews Drug discovery 2019, 18(11):869–884. [DOI] [PubMed] [Google Scholar]
- 27.Huang Y, Thathiah A: Regulation of neuronal communication by G protein-coupled receptors. FEBS letters 2015, 589(14):1607–1619. [DOI] [PubMed] [Google Scholar]
- 28.Catapano LA, Manji HK: G protein-coupled receptors in major psychiatric disorders. Biochimica et biophysica acta 2007, 1768(4):976–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Civelli O, Reinscheid RK, Zhang Y, Wang Z, Fredriksson R, Schioth HB: G protein-coupled receptor deorphanizations. Annual review of pharmacology and toxicology 2013, 53:127–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsiao YC, Tong ZJ, Westfall JE, Ault JG, Page-McCaw PS, Ferland RJ: Ahi1, whose human ortholog is mutated in Joubert syndrome, is required for Rab8a localization, ciliogenesis and vesicle trafficking. Human molecular genetics 2009, 18(20):3926–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alvarez Retuerto AI, Cantor RM, Gleeson JG, Ustaszewska A, Schackwitz WS, Pennacchio LA, Geschwind DH: Association of common variants in the Joubert syndrome gene (AHI1) with autism. Human molecular genetics 2008, 17(24):3887–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren Z, Qiu A, Zhang A, Huang L, Rao S: A cis-eQTL in AHI1 confers risk to schizophrenia in European populations. Neuroscience letters 2016, 632:130–135. [DOI] [PubMed] [Google Scholar]
- 33.Ingason A, Giegling I, Cichon S, Hansen T, Rasmussen HB, Nielsen J, Jurgens G, Muglia P, Hartmann AM, Strengman E et al. : A large replication study and meta-analysis in European samples provides further support for association of AHI1 markers with schizophrenia. Human molecular genetics 2010, 19(7):1379–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ingason A, Sigmundsson T, Steinberg S, Sigurdsson E, Haraldsson M, Magnusdottir BB, Frigge ML, Kong A, Gulcher J, Thorsteinsdottir U et al. : Support for involvement of the AHI1 locus in schizophrenia. European journal of human genetics : EJHG 2007, 15(9):988–991. [DOI] [PubMed] [Google Scholar]
- 35.Datta SR, McQuillin A, Rizig M, Blaveri E, Thirumalai S, Kalsi G, Lawrence J, Bass NJ, Puri V, Choudhury K et al. : A threonine to isoleucine missense mutation in the pericentriolar material 1 gene is strongly associated with schizophrenia. Molecular psychiatry 2010, 15(6):615–628. [DOI] [PubMed] [Google Scholar]
- 36.Gurling HM, Critchley H, Datta SR, McQuillin A, Blaveri E, Thirumalai S, Pimm J, Krasucki R, Kalsi G, Quested D et al. : Genetic association and brain morphology studies and the chromosome 8p22 pericentriolar material 1 (PCM1) gene in susceptibility to schizophrenia. Archives of general psychiatry 2006, 63(8):844–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider E, Mayer S, El Hajj N, Jensen LR, Kuss AW, Zischler H, Kondova I, Bontrop RE, Navarro B, Fuchs E et al. : Methylation and expression analyses of the 7q autism susceptibility locus genes MEST , COPG2, and TSGA14 in human and anthropoid primate cortices. Cytogenetic and genome research 2012, 136(4):278–287. [DOI] [PubMed] [Google Scholar]
- 38.Korvatska O, Estes A, Munson J, Dawson G, Bekris LM, Kohen R, Yu CE, Schellenberg GD, Raskind WH: Mutations in the TSGA14 gene in families with autism spectrum disorders. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics 2011, 156B(3):303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munoz-Estrada J, Lora-Castellanos A, Meza I, Alarcon Elizalde S, Benitez-King G: Primary cilia formation is diminished in schizophrenia and bipolar disorder: A possible marker for these psychiatric diseases. Schizophrenia research 2018, 195:412–420. [DOI] [PubMed] [Google Scholar]
- 40.Nauli SM SR, Reese CJ, Nauli AM: Mechanosensory and chemosensory primary cilia in ciliopathy and ciliotherapy. Hoboken, NJ: Wiley-Blackwell; 2017. [Google Scholar]
- 41.van Dam TJP, Townsend MJ, Turk M, Schlessinger A, Sali A, Field MC, Huynen MA: Evolution of modular intraflagellar transport from a coatomer-like progenitor. P Natl Acad Sci USA 2013, 110(17):6943–6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnaiz O, Cohen J, Tassin AM, Koll F: Remodeling Cildb, a popular database for cilia and links for ciliopathies. Cilia 2014, 3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arnaiz O, Malinowska A, Klotz C, Sperling L, Dadlez M, Koll F, Cohen J: Cildb: a knowledgebase for centrosomes and cilia. Database : the journal of biological databases and curation 2009, 2009:bap022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Dam TJP, Kennedy J, van der Lee R, de Vrieze E, Wunderlich KA, Rix S, Dougherty GW, Lambacher NJ, Li C, Jensen VL et al. : CiliaCarta: An integrated and validated compendium of ciliary genes. PloS one 2019, 14(5):e0216705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schou KB, Pedersen LB, Christensen ST: Ins and outs of GPCR signaling in primary cilia. EMBO Rep 2015, 16(9):1099–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, Won H, van Bakel H, Varghese M, Wang Y et al. : Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 2018, 362(6420). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, Ruderfer DM, Oh EC, Topol A, Shah HR et al. : Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nature neuroscience 2016, 19(11):1442–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parikshak NN, Swarup V, Belgard TG, Irimia M, Ramaswami G, Gandal MJ, Hartl C, Leppa V, Ubieta LT, Huang J et al. : Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 2016, 540(7633):423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G et al. : Spatio-temporal transcriptome of the human brain. Nature 2011, 478(7370):483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, Schork AJ, Appadurai V, Buil A, Werge TM et al. : Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 2018, 359(6376):693–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang LC, Jamain S, Lin CW, Rujescu D, Tseng GC, Sibille E: A conserved BDNF, glutamate- and GABA-enriched gene module related to human depression identified by coexpression meta-analysis and DNA variant genome-wide association studies. PloS one 2014, 9(3):e90980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mukhopadhyay S, Wen XH, Chih B, Nelson CD, Lane WS, Scales SJ, Jackson PK: TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Gene Dev 2010, 24(19):2180–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beales PL, Bland E, Tobin JL, Bacchelli C, Tuysuz B, Hill J, Rix S, Pearson CG, Kai M, Hartley J et al. : IFT80, which encodes a conserved intraflagellar transport protein, is mutated in Jeune asphyxiating thoracic dystrophy. Nat Genet 2007, 39(6):727–729. [DOI] [PubMed] [Google Scholar]
- 54.Brazelton WJ, Amundsen CD, Silflow CD, Lefebvre PA: The bld1 mutation identifies the Chlamydomonas osm-6 homolog as a gene required for flagellar assembly. Curr Biol 2001, 11(20):1591–1594. [DOI] [PubMed] [Google Scholar]
- 55.Follit JA, Tuft RA, Fogarty KE, Pazour GJ: The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell 2006, 17(9):3781–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haycraft CJ, Swoboda P, Taulman PD, Thomas JH, Yoder BK: The C-elegans homolog of the murine cystic kidney disease gene Tg737 functions in a ciliogenic pathway and is disrupted in osm-5 mutant worms. Development 2001, 128(9):1493–1505. [DOI] [PubMed] [Google Scholar]
- 57.Houde C, Dickinson RJ, Houtzager VM, Cullum R, Montpetit R, Metzler M, Simpson EM, Roy S, Hayden MR, Hoodless PA et al. : Hippi is essential for node cilia assembly and Sonic hedgehog signaling. Dev Biol 2006, 300(2):523–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huangfu DW, Liu AM, Rakeman AS, Murcia NS, Niswander L, Anderson KV: Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 2003, 426(6962):83–87. [DOI] [PubMed] [Google Scholar]
- 59.Pazour GJ, Dickert BL, Vucica Y, Seeleye ES, Rosenbaum JL, Witman GB, Cole DG: Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene Tg737, are required for assembly of cilia and flagella. Mol Biol Cell 2000, 11:369a–369a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qin HM, Rosenbaum JL, Barr MM: An autosomal recessive polycystic kidney disease gene homolog is involved in intraflagellar transport in C-elegans ciliated sensory neurons. Curr Biol 2001, 11(6):457–461. [DOI] [PubMed] [Google Scholar]
- 61.Tsujikawa M, Malicki J: Intraflagellar transport genes are essential for differentiation and survival of vertebrate sensory neurons. Neuron 2004, 42(5):703–716. [DOI] [PubMed] [Google Scholar]
- 62.Nakayama K, Katoh Y: Ciliary protein trafficking mediated by IFT and BBSome complexes with the aid of kinesin-2 and dynein-2 motors. Journal of biochemistry 2018, 163(3):155–164. [DOI] [PubMed] [Google Scholar]
- 63.Piperno G, Siuda E, Henderson S, Segil M, Vaananen H, Sassaroli M: Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J Cell Biol 1998, 143(6):1591–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iomini C, Li LY, Esparza JM, Dutcher SK: Retrograde Intraflagellar Transport Mutants Identify Complex A Proteins With Multiple Genetic Interactions in Chlamydomonas reinhardtii. Genetics 2009, 183(3):885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iomini C, Babaev-Khaimov V, Sassaroli M, Piperno G: Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J Cell Biol 2001, 153(1):13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mukhopadhyay S, Wen X, Chih B, Nelson CD, Lane WS, Scales SJ, Jackson PK: TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes & development 2010, 24(19):2180–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Legue E, Liem KF Jr.: Mutations in Ciliary Trafficking Genes affect Sonic Hedgehog-dependent Neural Tube Patterning Differentially along the Anterior-Posterior Axis. Neuroscience 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Norman RX, Ko HW, Huang V, Eun CM, Abler LL, Zhang Z, Sun X, Eggenschwiler JT: Tubby-like protein 3 (TULP3) regulates patterning in the mouse embryo through inhibition of Hedgehog signaling. Human molecular genetics 2009, 18(10):1740–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guan J, Cai JJ, Ji G, Sham PC: Commonality in dysregulated expression of gene sets in cortical brains of individuals with autism, schizophrenia, and bipolar disorder. Translational psychiatry 2019, 9(1):152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wray NR, Pergadia ML, Blackwood DH, Penninx BW, Gordon SD, Nyholt DR, Ripke S, MacIntyre DJ, McGhee KA, Maclean AW et al. : Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Molecular psychiatry 2012, 17(1):36–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hines LM, Tabakoff B, Trait WISS: Platelet adenylyl cyclase activity: A biological marker for major depression and recent drug use. Biol Psychiat 2005, 58(12):955–962. [DOI] [PubMed] [Google Scholar]
- 72.Chen XM, Luo J, Leng YH, Yang YM, Zweifel LS, Palmiter RD, Storm DR: Ablation of Type III Adenylyl Cyclase in Mice Causes Reduced Neuronal Activity, Altered Sleep Pattern, and Depression-like Phenotypes. Biol Psychiat 2016, 80(11):836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Keays DA, Tian G, Poirier K, Huang GJ, Siebold C, Cleak J, Oliver PL, Fray M, Harvey RJ, Molnar Z et al. : Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell 2007, 128(1):45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jaglin XH, Poirier K, Saillour Y, Buhler E, Tian GL, Bahi-Buisson N, Fallet-Bianco C, Phan-Dinh-Tuy F, Kong XP, Bomont P et al. : Mutations in the beta-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat Genet 2009, 41(6):746–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tischfield MA, Baris HN, Wu C, Rudolph G, Van Maldergem L, He W, Chan WM, Andrews C, Demer JL, Robertson RL et al. : Human TUBB3 Mutations Perturb Microtubule Dynamics, Kinesin Interactions, and Axon Guidance. Cell 2010, 140(1):74–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Breuss M, Heng JIT, Poirier K, Tian GL, Jaglin XH, Qu ZD, Braun A, Gstrein T, Ngo L, Haas M et al. : Mutations in the beta-Tubulin Gene TUBB5 Cause Microcephaly with Structural Brain Abnormalities. Cell Rep 2012, 2(6):1554–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cushion TD, Paciorkowski AR, Pilz DT, Mullins JGL, Seltzer LE, Marion RW, Tuttle E, Ghoneim D, Christian SL, Chung SK et al. : De Novo Mutations in the Beta-Tubulin Gene TUBB2A Cause Simplified Gyral Patterning and Infantile-Onset Epilepsy. Am J Hum Genet 2014, 94(4):634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Romaniello R, Arrigoni F, Fry AE, Bassi MT, Rees MI, Borgatti R, Pilz DT, Cushion TD: Tubulin genes and malformations of cortical development. Eur J Med Genet 2018, 61(12):744–754. [DOI] [PubMed] [Google Scholar]
- 79.Jaglin XH, Chelly J: Tubulin-related cortical dysgeneses: microtubule dysfunction underlying neuronal migration defects. Trends Genet 2009, 25(12):555–566. [DOI] [PubMed] [Google Scholar]
- 80.Cross-Disorder Group of the Psychiatric Genomics Consortium. Electronic address pmhe, Cross-Disorder Group of the Psychiatric Genomics C: Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell 2019, 179(7):1469–1482 e1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cross-Disorder Group of the Psychiatric Genomics C: Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013, 381(9875):1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao H, Nyholt DR: Gene-based analyses reveal novel genetic overlap and allelic heterogeneity across five major psychiatric disorders. Human genetics 2017, 136(2):263–274. [DOI] [PubMed] [Google Scholar]
- 83.Monroe TO, Garrett ME, Kousi M, Rodriguiz RM, Moon S, Bai Y, Brodar SC, Soldano KL, Savage J, Hansen TF et al. : PCM1 is necessary for focal ciliary integrity and is a candidate for severe schizophrenia. Nature communications 2020, 11(1):5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moens LN, Ceulemans S, Alaerts M, Van Den Bossche MJ, Lenaerts AS, De Zutter S, Norrback KF, Adolfsson R, Del-Favero J: PCM1 and schizophrenia: a replication study in the Northern Swedish population. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics 2010, 153B(6):1240–1243. [DOI] [PubMed] [Google Scholar]
- 85.Eastwood SL, Hodgkinson CA, Harrison PJ: DiSc-1 Leu607Phe alleles differentially affect centrosomal PCM1 localization and neurotransmitter release. Molecular psychiatry 2009, 14(6):556–557. [DOI] [PubMed] [Google Scholar]
- 86.Kamiya A, Tan PL, Kubo K, Engelhard C, Ishizuka K, Kubo A, Tsukita S, Pulver AE, Nakajima K, Cascella NG et al. : Recruitment of PCM1 to the centrosome by the cooperative action of DISC1 and BBS4: a candidate for psychiatric illnesses. Archives of general psychiatry 2008, 65(9):996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rampino A, Walker RM, Torrance HS, Anderson SM, Fazio L, Di Giorgio A, Taurisano P, Gelao B, Romano R, Masellis R et al. : Expression of DISC1-interactome members correlates with cognitive phenotypes related to schizophrenia. PloS one 2014, 9(6):e99892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gurling HMD, Critchley H, Datta SR, McQuillin A, Blaveri E, Thirumalai S, Pimm J, Krasucki R, Kalsi G, Quested D et al. : Genetic association and brain morphology studies and the chromosome 8p22 pericentriolar material 1 (PCM1) gene in susceptibility to schizophrenia. Arch Gen Psychiat 2006, 63(8):844–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zoubovsky S, Oh EC, Cash-Padgett T, Johnson AW, Hou Z, Mori S, Gallagher M, Katsanis N, Sawa A, Jaaro-Peled H: Neuroanatomical and behavioral deficits in mice haploinsufficient for Pericentriolar material 1 (Pcm1). Neuroscience research 2015, 98:45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Verleyen D, Luyten FP, Tylzanowski P: Orphan G-protein coupled receptor 22 (Gpr22) regulates cilia length and structure in the zebrafish Kupffer’s vesicle. PloS one 2014, 9(10):e110484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pala R, Mohieldin AM, Shamloo K, Sherpa RT, Kathem SH, Zhou J, Luan ZY, Zheng JG, Ahsan A, Nauli SM: Personalized Nanotherapy by Specifically Targeting Cell Organelles To Improve Vascular Hypertension. Nano Lett 2019, 19(2):904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pala R, Mohieldin AM, Sherpa RT, Kathem SH, Shamloo K, Luan ZY, Zhou J, Zheng JG, Ahsan A, Nauli SM: Ciliotherapy: Remote Control of Primary Cilia Movement and Function by Magnetic Nanoparticles. Acs Nano 2019, 13(3):3555–3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


