Abstract
RNA modifications affect many aspects of RNA metabolism and are involved in the regulation of many different biological processes. Mono-methylation of adenosine in the N1 position, N1-methyladensoine (m1A), is a reversible modification that is known to target rRNAs and tRNAs. m1A has been shown to increase tRNA structural stability and induce correct tRNA folding. Recent studies have begun to associate the dysregulation of epitranscriptomic control with age-related disorders such as Alzheimer’s disease. Here, we applied the newly developed m1A-quant-seq approach to map the brain abundant m1A RNA modification in the cortex of an Alzheimer’s disease mouse model, 5XFAD. We observed hypomethylation in both mitochondrial and cytosolic tRNAs in 5XFAD mice compared with wild type. Furthermore, the main enzymes responsible for the addition of m1A in mitochondrial (TRMT10C, HSD17B10) and cytosolic tRNAs (TRMT61A) displayed decreased expression in 5XFAD compared with wild-type mice. Knockdown of these enzymes results in a more severe phenotype in a Drosophila tau model, and differential m1A methylation is correlated with differences in mature mitochondrial tRNA expression. Collectively, this work suggests that hypo m1A modification in tRNAs may play a role in Alzheimer’s disease pathogenesis.
Introduction
RNA modifications are post-transcriptional changes at the RNA level that provide another means by which RNA function can be regulated. They tend to be conserved and have been implicated in many critical biological processes including development, disease, circadian rhythm and embryonic stem cell fate transition (1–3). Within mRNAs, N6-methyladenosine (m6A) is a predominant modification that is well studied and has been mapped transcriptome-wide using various approaches (4). Multiple studies have shown that m6A affects various aspects of RNA metabolism including mRNA degradation, mRNA translation and RNA splicing (5–11). Recently, greater emphasis has been placed on mapping and understanding the consequence of other RNA modifications.
Mono-methylation of adenosine in the N1 position, N1-methyladensoine (m1A), has been the focus of recent studies. m1A is a reversible modification that is known to modify rRNAs and tRNAs. In tRNAs, m1A occurs at positions 9, 14 and 58. Position 58 is found in cytoplasmic tRNAs and is catalyzed by TRMT61A and TRMT6 (12). Similar to m6A, m1A58 is a dynamic modification and is specifically removed by the human AlkB homolog 1 (ALKBH1) demethylase (13). Furthermore, human mitochondrial tRNAs are modified with m1A at positions 9 and 58 (12). The enzymes responsible for these modifications are TRMT10C and TRMT61B, respectively (14,15). HSD17B10 is a dehydrogenase and is required for TRMT10C activity; it likely acts as a scaffold protein for the formation of the complex (15). In humans, m1A is also found at position 1322 of 28S rRNA, which is installed by the human nucleolar protein nucleomethylin (16). In rRNAs, m1A can affect ribosome biogenesis, whilst within tRNAs, m1A can promote correct folding and structural stability (17,18). Initial transcriptome-wide studies using an antibody that enriches for m1A-marked targets coupled with next generation sequencing identified m1A as an abundant modification in mRNAs enriched in the 5′ untranslated region (19,20). However, since then, additional studies have suggested that m1A is rare (if it exists at all) in mRNAs (21,22). m1A is also known to disturb the formation of the Watson-Crick base pair and is therefore classified as a ‘hard stop’ modification. This results in stalling of strand elongation or nucleotide misincorporation during the reverse transcription reaction. This feature of m1A has been taken advantage of in the detection of this modification (23–26). m1A levels are highest in the brain, and in this context, there is limited knowledge surrounding its function.
Here we employed the newly developed m1A antibody independent approach (m1A-quant-seq) to gain an unbiased map of m1A. This approach takes advantage of an evolved reverse transcriptase enzyme that allows for optimal read-through and high mutation rates at the specific m1A site. We investigated the possible role of m1A in Alzheimer’s disease, and show that decreased expression of m1A tRNA writers are correlated with hypo m1A methylation, a more severe phenotype in Alzheimer’s disease fly tau model, and a decrease in mature mitochondrial tRNA expression which is suggested to contribute to Alzheimer’s disease etiology.
Results
Mitochondrial and cytoplasmic tRNAs are hypo m1A methylated in 5XFAD mice
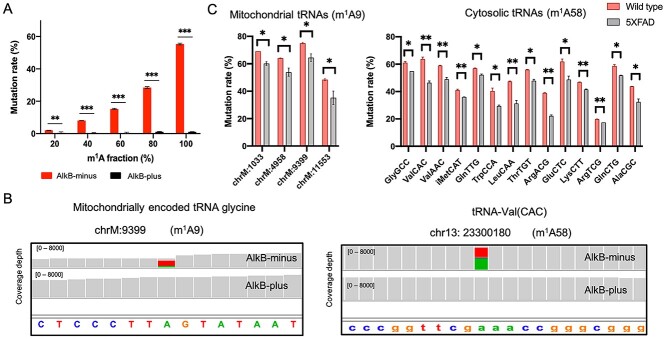
Briefly, in the m1A-quant-seq approach, half the RNA sample is treated with AlkB (an alpha-ketoglutarate-dependent hydroxylase), which removes the m1A methylation, whilst the other half is left untreated (27). The approach also employs an engineered reverse transcriptase that results in an A-to-T misincorporation at the m1A site in the AlkB-minus sample during reverse transcription. In this way, the presence of m1A can be reliably detected when compared to the AlkB-plus control (where no A-to-T misincorporation is observed). In addition, we spiked five synthetic RNAs containing increasing m1A levels as was previously described (27). Sensitivity to AlkB treatment is robustly observed with the spike-in RNAs, where increasing m1A levels correlates with higher observed mutation rate (Fig. 1A).
Figure 1.
Detecting m1A in mouse cortex using m1A-quant-seq. (A) m1A mutation rates for the RNA spike-ins using m1A-quant-seq. (B) Integrated genomics viewer coverage traces displaying the mutational signatures of a detected m1A9 and m1A58 site. A-to-T misincorporation is observed in the AlkB-minus sample, whereas this is nonexistent in the AlkB-plus sample. (B) Bar graph displaying the wild type and 5XFAD mutation rate for the statistically significant hypo methylated mitochondrial and cytosolic m1A sites. * denotes a P-value ≤ 0.05, ** denotes a P-value ≤ 0.01. Error bars correspond to ±1 SD.
We generated small RNA libraries, and importantly, obtained abundant coverage per sample (Supplementary Material, Table S1). We defined a set of parameters to call high confidence sites: (1) the base must be covered by at least 50 reads in both AlkB treated and untreated samples, (2) show a signature A to T transition in the AlkB-minus untreated sample (at least 10% mutation rate) and (3) show a negligible mutation rate in the AlkB-plus treated sample across triplicates. Using these parameters, we only detected high confidence m1A sites at position 9 in mitochondrial tRNAs and at position 58 in cytosolic tRNAs, which is in agreement with recent studies (21,22). Presently, we observed robust A-to-T mutation signatures, again which is common for m1A-induced misincorporations (23,28), and mutation rates were virtually non-existent upon AlkB-plus treatment (Fig. 1B), confirming the quality of the data. In total, we detected m1A at position 9 in nine mitochondrial tRNAs and m1A at position 58 in 86 cytosolic tRNA isodecoders that were summed by their respective anticodon sequence into 30 genomically encoded anticodon classes (Table 1). Interestingly, 4 of the 9 mitochondrial tRNA sites and 14 out of the 30 cytosolic tRNAs anticodon classes show a statistically significant lower mutation rate in 5XFAD mice compared with wild-type mice (Fig. 1C), including the initiator tRNAMetCAT.
Table 1.
A-to-T mutation signatures for cytosolic and mitochondrial tRNAs in wild type and 5XFAD mice
| tRNA | Mean mutation rate of wild type | Mean mutation rate of 5XFAD | P-value |
|---|---|---|---|
| LysTTT | 45.03 | 35.73 | 0.083789 |
| LeuCAG | 50.35 | 40.94 | 0.09772 |
| GlyGCC | 61.03 | 54.88 | 0.023474 |
| GluCTC | 61.92 | 48.79 | 0.049736 |
| ArgTCT | 64.84 | 63.23 | 0.3794 |
| AsnGTT | 40.95 | 39.95 | 0.502964 |
| ArgCCT | 28.36 | 30.06 | 0.545949 |
| ArgTCG | 19.83 | 17.44 | 0.002312 |
| ValCAC | 63.74 | 46.42 | 0.012823 |
| ThrTGT | 55.96 | 47.81 | 0.019573 |
| ValAAC | 58.99 | 49.04 | 0.018121 |
| IleAAT | 35.77 | 28.82 | 0.174341 |
| SerAGA | 42.18 | 33.91 | 0.221755 |
| TrpCCA | 40.37 | 29.46 | 0.038817 |
| ThrCGT | 12.48 | 11.21 | 0.377434 |
| GlnTTG | 56.98 | 52.15 | 0.020528 |
| ArgCCG | 57.98 | 58.2 | 0.772954 |
| TyrGTA | 28.65 | 17.21 | 0.176075 |
| AlaAGC | 26.4 | 19.86 | 0.28752 |
| LysCTT | 46.83 | 41.47 | 0.00706 |
| iMetCAT | 41.1 | 36.01 | 0.015174 |
| PheGAA | 26.26 | 22.31 | 0.132256 |
| LeuAAG | 20.45 | 18.6 | 0.353735 |
| GlnCTG | 59.29 | 52.73 | 0.05007 |
| LeuCAA | 47.38 | 31.3 | 0.018676 |
| AlaCGC | 43.77 | 32.43 | 0.034584 |
| ArgACG | 38.94 | 22.21 | 0.002146 |
| IleTAT | 37.82 | 29.34 | 0.258774 |
| GlyCCC | 34.62 | 37.31 | 0.74946 |
| CysGCA | 65.17 | 63.75 | 0.857471 |
| chrM:9 | 72.43 | 70.99 | 0.330872 |
| chrM:1033 | 69.41 | 59.12 | 0.016543 |
| chrM:4958 | 64.06 | 53.9 | 0.026689 |
| chrM:6950 | 62.4 | 49.88 | 0.379135 |
| chrM:7708 | 80.75 | 75.64 | 0.48222 |
| chrM:9399 | 75.45 | 64.5 | 0.014056 |
| chrM:9816 | 65.56 | 51.04 | 0.332473 |
| chrM:11553 | 45.86 | 30.19 | 0.025935 |
| chrM:15297 | 39.51 | 34.93 | 0.601065 |
Knockdown of m1A writers results in an enhanced Alzheimer’s disease-related phenotype
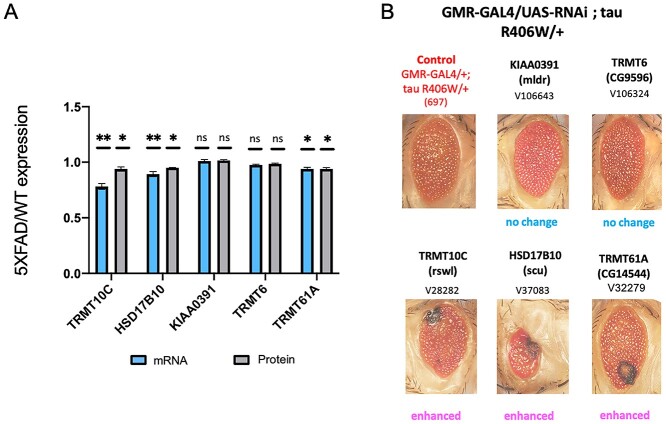
To begin to understand the role of the observed mitochondrial m1A tRNA hypomethylation in the 5XFAD Alzheimer’s disease mouse model, we first looked at the levels of m1A associated machinery in 5XFAD compared to control. We observed that TRMT10C, HSD17B10 and TRMT61A have significantly decreased expression in 5XFAD compared with wild-type mice using our RNA-seq data and publicly available protein mass spectrometry data generated using 5XFAD mice (Fig. 2A) (29). Additionally, a recent proteome-wide association study of human Alzheimer’s (ROS/MAP dataset) (30) revealed that the human TRMT10C protein has significantly lower expression in Alzheimer’s disease compared with controls.
Figure 2.
Characterizing the role of m1A writer proteins in the molecular pathogenesis of Alzheimer’s disease using a Drosophila tau model. (A) Eye phenotype following the knockdown of Drosophila TRMT10C (rswl), HSD17B10 (scu), KIAA0391 (mldr), TRMT6 (CG9596) and TRMT61A (CG14544) on the TauR406W background. In the case of knocking down TRMT10C, HSD17B10 and TRMT61A, the eye phenotype is aggravated compared with control, while there is no change in eye phenotype following knockdown of either KIAA0391 or TRMT6. (B) mRNA and protein expression levels of TRMT10C, HSD17B10, KIAA0391, TRMT6 and TRMT61A expressed as a ratio (5XFAD/WT). There is a statistically significant decrease in mRNA and protein levels in 5XFAD compared to WT for TRMT10C, HSD17B10 and TRMT61A. * denotes a P-value ≤ 0.05, ** denotes a P-value ≤ 0.01. Error bars correspond to ±1 SD.
Next, we investigated the impact of the reduced m1A modification on tau toxicity, a hallmark of Alzheimer’s disease. We employed a Drosophila transgenic Alzheimer’s disease model that specifically expresses the human tau gene with the R406W mutation in the eye using the gmr-GAL4 driver. Using the fly model allows us to determine the effects m1A players may have in the context of Alzheimer’s disease. This approach allows analysis of the results in an easy qualitative manner by visualizing changes in eye phenotype. To understand the effect of the m1A pathway on this Alzheimer’s disease fly model, we crossed RNAi lines of rswl, scu, mldr (Drosophila orthologs of the components that make up the m1A mitochondrial tRNA writer protein complex TRMT10C, HSD17B10 and KIAA0391, respectively) and cytoplasmic m1A tRNA writers CG9596, CG14544 (Drosophila orthologs of TRMT6, TRMT61A, respectively) with the tau fly. We found that loss of rswl, scu and CG14544 resulted in an enhanced eye phenotype compared to the control eye fly, suggesting that the loss of m1A may enhance tau toxicity (Fig. 2B). However, no changes in eye phenotype were observed after crossing the tau fly with mldr or CG9596 RNAi flies. Together, these results suggest that the reduction of TRMT10C, HSD17B10 and TRMT61A, and/or the concomitant hypomethylation observed in the 5XFAD mice may have a role in Alzheimer’s disease pathogenesis.
Differential mt-tRNA m1A methylation is associated with changes in mature tRNA expression but not with mitochondrial processing
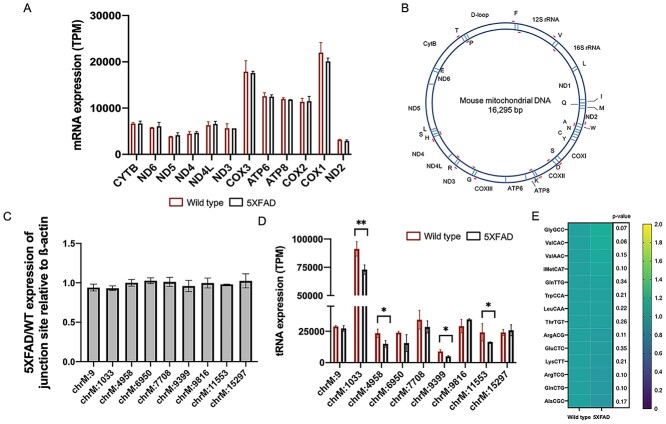
It has been suggested that mRNA translation is sensitive to even subtle changes in tRNA levels, and mutations in a number of genes involved in tRNA expression and processing are associated with neurodegenerative disease. Furthermore, TRMT10C has a known role in processing of the mitochondrial polycistronic transcript, and knockout of any of the proteins that make up the m1A writer complex have been shown to affect mitochondrial processing, resulting in the accumulation of precursor transcripts (31). Given that we observed differential levels of TRMT10C and differential m1A9 mitochondrial tRNA levels, we asked whether these differences could affect posttranscriptional substrate recognition and/or cleavage in the regions of the mitochondrial transcriptome in the 5XFAD model. To this end, we determined the expression levels of mitochondrial mRNAs including those immediately upstream from m1A-containing tRNAs in order to establish whether differences in m1A levels affect mitochondrial mRNA processing in the Alzheimer’s mouse model (Fig. 3A). We also performed RT-qPCR using primers that flank the mitochondrial tRNAs containing differential m1A methylation (Fig. 3B). This allowed us to detect ‘junction transcripts’ which correspond to unprocessed transcripts (Fig. 3C). Both analyses showed no difference in precursor transcript levels in 5XFAD compared with wild type, suggesting that in the context of the 5XFAD Alzheimer’s disease model, differences in TRMT10C/m1A levels do not alter mitochondrial processing compared with wild type.
Figure 3.
Assessing the functional consequence of the m1A9 hypomethylation in 5XFAD. (A) TPM expression levels of mitochondrial mRNAs including those upstream of tRNAs containing m1A. There are no differences in expression levels in these mRNAs between 5XFAD and WT. (B) Position of primers (in red) flanking the tRNAs that contain m1A9 sites as detected in this study. (C) RT-qPCR results showing no difference in ‘junction sites’ between 5XFAD and WT. (D) Mature tRNA expression correlates with the observed hypo m1A9 methylation. (E) No differences are observed in hypomethylated cytosolic tRNA abundances between 5XFAD and WT. * denotes a P-value ≤ 0.05 and ** denotes a P-value ≤ 0.01. Error bars correspond to ±1 SD.
As m1A methylation modifications are suggested to stabilize tRNA secondary structure, we next looked at whether differential methylation was associated with mature tRNA expression. Given that some RNA modifications, including m1A, are known to impede reverse transcription during library preparation, we made use of our AlkB-plus RNA-seq libraries (which removes the m1A modification) to determine accurate expression levels of the mature tRNAs, similar to the ARM-seq method (32). We observed a correlation between m1A methylation and mature tRNA levels. All of the hypomethylated tRNAs in 5XFAD were significantly less expressed compared with the wild- type control (Fig. 3D). Similar to what was reported in (33), tRNA expression is not significantly correlated with hypomethylation in the cytosolic tRNAs (Fig. 3E).
Discussion
RNA modifications represent an additional layer of control in the regulation of gene expression. They are found in both the nuclear and mitochondrial transcriptome, in various RNA types, and they have been implicated in many important roles including structural stability and translation efficiency. Here we show that m1A levels in mitochondrial and cytosolic tRNAs are modulated in the context of Alzheimer’s disease. We also demonstrate that the proteins responsible for installing m1A on mitochondrial and cytosolic tRNAs are expressed at lower levels in Alzheimer’s disease, and this correlates with the observed hypomethylation of particular m1A9 mitochondrial and m1A58 cytosolic tRNAs in the 5XFAD Alzheimer’s mouse model. Furthermore, loss of those m1A methyltransferases results in a more detrimental phenotype in a tau Drosophila model suggesting that perturbation of m1A may affect Alzheimer’s disease. Lastly, the hypomethylation in the Alzheimer’s disease model is associated with less mature mitochondrial tRNA expression, which in turn may affect mRNA translation, contributing to the known mitochondrial dysfunction observed in Alzheimer’s disease.
Our study uses a dedicated approach to determine m1A sites, i.e. an approach that is specifically designed to map m1A sites. It uses an evolved reverse transcriptase that allows for robust read-through and high mutation rates at m1A sites. This coupled with an AlkB treatment, which serves as a control, allows for robust, accurate detection of m1A sites. In addition, we generated dedicated small RNA libraries to get a clear picture of the m1A landscape in tRNAs in an Alzheimer’s disease mouse model. Thus, the approach taken here is a specialized one to accurately detect m1A methylation, specifically in tRNAs, rather than inferring m1A methylation from RNA-seq data, which has been done previously. It is important to note that it is difficult to distinguish between m1A-induced misincorporations, genetic variations such as single nucleotide polymorphisms, and sequencing errors in a standard RNA-seq. Furthermore, there would clearly be an under-representation of reads covering the pool of mature tRNA sequences in RNA-seq data. Therefore, any conclusions drawn from RNA-seq data may not accurately speak to differences in levels of methylated tRNAs.
There is little known regarding the specific role, if any, of m1A in disease, yet there is some evidence to suggest a role for this modification in neurodegenerative disease and cancer. For example, mutations in HSD17B10 have been linked with a disease that results in progressive neurodegeneration. These mutations have been shown to inhibit the interaction of HSD17B10 with TRMT10C (34). This is thought to reduce the levels of m1A modification, which may contribute to pathogenesis. Also, m1A9 tRNA methylation has been implicated in various cancers. In this context, hypermethylation was observed in cancer compared with normal (35). Overall, this suggests that perturbation of m1A levels could be involved in disease pathogenesis.
Presently, in the 5XFad mouse model, we find a reduction of m1A in mitochondrial and cytosolic tRNAs, and this observation is correlated with a reduced expression in m1A writers. Furthermore, our results were validated in vivo using a tau fly model. Knocking out m1A modifiers resulted in exacerbation of the eye phenotype, suggesting that either the loss of m1A writers and/or the loss of m1A methylation contributes to Alzheimer’s disease pathogenesis. Alzheimer’s disease is a multifactorial disease, and one factor that is known to contribute to AD etiology is mitochondrial dysfunction (36,37). It has been shown that increased oxidative stress contributes to the mitochondrial dysfunction and impaired energy metabolism in Alzheimer’s disease. Interestingly, though, Wang et al. (38) show that decreasing the m1A mitochondrial writer complex in Drosophila does not cause oxidative stress. However, as post-transcriptional processing can affect protein synthesis, perturbation of RNA modifications in mitochondrial tRNAs may contribute to the mitochondrial dysfunction in Alzheimer’s disease. m1A methylation at position 9 in mitochondrial tRNAs is thought to stabilize tRNA secondary structure. Currently, the hypo m1A9 mitochondrial tRNA methylation in 5XFAD may contribute to this mitochondrial dysfunction, likely by destabilizing the mitochondrial tRNAs, which could impact translation efficiency and downstream mitochondrial functions. On the other hand, in cytosolic tRNAs, we did not observe any correlation between hypomethylation and tRNA expression. On this note, studies in human cells report that m1A58 hypomodification in tRNAiMet does not decrease tRNA stability; however, the hypomethylation was found to decrease charging efficiency by the cognate aminoacyltransferase (33). Furthermore, they find that hypo m1A methylation does not affect tRNA stability of other tRNA classes but does affect their association with polysomes. This may suggest that the present differential m1A methylation in cytosolic tRNAs may impact translation efficiency. Altogether, the observed dysregulation of the m1A modification in mitochondrial and cytosolic tRNAs may contribute to the etiology of Alzheimer’s disease by ultimately affecting protein synthesis. In future studies, it would be interesting to determine the m1A landscape in other neurodegenerative diseases. Also, with the advent of long read direct RNA sequencing (such as nanopore sequencing), there is the potential to directly quantify RNA modification levels across multiple RNA modifications. This would definitely expand the current view on the role of RNA modifications and the biological significance of dynamic RNA modification regulation.
Materials and Methods
Animal care
Control wild type mice (C57BL/6J, Jackson Laboratory, Bar Harbor, ME, stock # 000664) and 5XFAD mice (generated on the same background, C57BL/6J 5XFAD, available from Jackson Laboratory, stock # 034840) were housed, maintained and euthanized according to the Emory University Institutional Animal Care and Use Committee guidelines. All animals were maintained and euthanized according to the Emory University Institutional Animal Care and Use Committee guidelines. The Institutional Review Board of Emory University approved the study.
RNA isolation, m 1A-quant-seq
Mice were sacrificed by cervical dislocation and the cortex was dissected. The tissues were dissolved in TriReagent (Thermo Fisher, Waltham, MA) using a mortar and pestle and total RNA was extracted according to the manufacturer’s instructions. Since our goal was to look at m1A in tRNAs, we specifically isolated small RNAs as our starting input. m1A-quant-seq was performed as described in (27). Half the sample was treated with AlkB, and the other half was left untreated. Libraries were then constructed from these samples as detailed in (27).
m1A-quant-seq analysis
We only used the R2 reads for our analysis due to the uncertainty in the position of the unique molecular identifier (UMI) in the R1 reads as shown in m1A-MAP (20). The method used for analysis is as described in (27). The R2 reads were first processed with the ‘clumpify.sh’ program in the BBMap package. PCR duplicates were removed by using ‘dedupe subs = 0.’ Adaptors were then trimmed using the ‘cutadapt’ program. Reads were filtered by quality and length using the options ‘-a AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGCCGTATCATT-q 20 -m 30.’ Reads were aligned to the mm10 genome using the hisat2 aligner with the option ‘—trim5 11’ to remove the UMI sequence. The default ‘soft-clipping’ option was used for alignment. Multi-mapping reads were discarded. Samtools mpileup was then used to generate text pileups from the BAM files (39). Finally, variant calling was performed using VarScan (40). m1A sites were called if they satisfied the following criteria: the site was covered by at least 50 reads, showed a signature A-to-T transition (>10% mutation rate) in the AlkB-minus sample and less than 1% mutation rate in the AlkB-plus sample across triplicates.
Drosophila genetics
We used the Drosophila model system as a genetic approach to identify the affect m1A players have on tau toxicity. This system has proven to be effective approach in gene discovery in Alzheimer’s disease (41). We used the transgenic fly line Tau R406W; this mutation is associated with neurotoxicity in Alzheimer’s disease. Drosophila RNAi lines were obtained from Bloomington Drosophila Stock Center or Vienna Drosophila Resource Center. The Drosophila RNAi lines were crossed with gmr-GAL4;Tau R406W and grown on standard medium at 25°C incubator. The progeny was collected, aged to 7 days, and the eye phenotype (such as reduced eye size, rough eye surfaces, and cell death) was observed using light microscopy.
RT-qPCR
Five micrograms of total RNA were treated with DNAse I (Promega), then reverse transcribed, using 200 U Superscript III (Invitrogen) and 50 μM of oligo dT primers (Invitrogen), according to the manufacturer’s instructions. cDNA was diluted 1:5 in nuclease-free water. Triplicate qPCR reactions (20 μl total volume) were performed using 50 ng of the diluted cDNA, 50 nm forward and reverse primers, 1× SYBR green Master Mix (Thermo Fisher Scientific). ß-actin was used for normalization.
Transcript quantification
We quantified reads mapped to mitochondrial tRNA genes in transcripts per kilobase million (TPM) using Kallisto (42). Fastq files of AlkB-plus treated libraries were used in order to gain an accurate estimate of tRNA abundance.
Statistical analyses
For statistical analyses, a Student t-test was performed using Prism. P-values are indicated in figures and figure legends.
Data availability
m1A-quant-seq datasets are available from NCBI’s Gene Expression Omnibus (GEO). The accession number for all the datasets reported in this paper is GSE168199. The 5XFAD proteomic data used in this study were sourced from (29) and are accessible from the PRIDE database (www.proteomexchange.org): with accession numbers of PXD007974 and PXD018590. The proteomic data from the ROS/MAP study (30) are available at https://www.synapse.org/#!Synapse:syn17015098. Scripts used throughout this study are available upon request.
Conflict of Interest statement. The authors declare no competing interests.
Supplementary Material
Contributor Information
Andrew M Shafik, Department of Human Genetics, School of Medicine, Emory University, Atlanta, GA 3032, USA.
Huiqing Zhou, Department of Chemistry, University of Chicago, Chicago, IL 60637, USA.
Junghwa Lim, Department of Human Genetics, School of Medicine, Emory University, Atlanta, GA 3032, USA.
Bryan Dickinson, Department of Chemistry, University of Chicago, Chicago, IL 60637, USA.
Peng Jin, Department of Human Genetics, School of Medicine, Emory University, Atlanta, GA 3032, USA.
Funding
National Institutes of Health (MH116441, HD104458, and NS111602 to P.J.).
References
- 1. Batista, P.J., Molinie, B., Wang, J., Qu, K., Zhang, J., Li, L., Bouley, D.M., Lujan, E., Haddad, B., Daneshvar, K. et al. (2014) m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell, 15, 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fustin, J.M., Doi, M., Yamaguchi, Y., Hida, H., Nishimura, S., Yoshida, M., Isagawa, T., Morioka, M.S., Kakeya, H., Manabe, I. et al. (2013) RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell, 155, 793–806. [DOI] [PubMed] [Google Scholar]
- 3. Hsu, P.J., Shi, H. and He, C. (2017) Epitranscriptomic influences on development and disease. Genome Biol., 18, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhu, W., Wang, J.Z., Xu, Z., Cao, M., Hu, Q., Pan, C., Guo, M., Wei, J.F. and Yang, H. (2019) Detection of N6methyladenosine modification residues (review). Int. J. Mol. Med., 43, 2267–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coots, R.A., Liu, X.M., Mao, Y., Dong, L., Zhou, J., Wan, J., Zhang, X. and Qian, S.B. (2017) m(6)A facilitates eIF4F-independent mRNA translation. Mol. Cell, 68(504–514), e507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hsu, P.J., Shi, H., Zhu, A.C., Lu, Z., Miller, N., Edens, B.M., Ma, Y.C. and He, C. (2019) The RNA-binding protein FMRP facilitates the nuclear export of N (6)-methyladenosine-containing mRNAs. J. Biol. Chem., 294, 19889–19895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roundtree, I.A., Luo, G.Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., Sha, J., Huang, X., Guerrero, L., Xie, P. et al. (2017) YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife, 6, e31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang, X., Lu, Z., Gomez, A., Hon, G.C., Yue, Y., Han, D., Fu, Y., Parisien, M., Dai, Q., Jia, G. et al. (2014) N6-methyladenosine-dependent regulation of messenger RNA stability. Nature, 505, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang, X., Zhao, B.S., Roundtree, I.A., Lu, Z., Han, D., Ma, H., Weng, X., Chen, K., Shi, H. and He, C. (2015) N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell, 161, 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xiao, W., Adhikari, S., Dahal, U., Chen, Y.S., Hao, Y.J., Sun, B.F., Sun, H.Y., Li, A., Ping, X.L., Lai, W.Y. et al. (2016) Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell, 61, 507–519. [DOI] [PubMed] [Google Scholar]
- 11. Zhou, J., Wan, J., Gao, X., Zhang, X., Jaffrey, S.R. and Qian, S.B. (2015) Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature, 526, 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ozanick, S., Krecic, A., Andersland, J. and Anderson, J.T. (2005) The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA, 11, 1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu, F., Clark, W., Luo, G., Wang, X., Fu, Y., Wei, J., Wang, X., Hao, Z., Dai, Q., Zheng, G. et al. (2016) ALKBH1-mediated tRNA demethylation regulates translation. Cell, 167, 816, e816–e828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chujo, T. and Suzuki, T. (2012) Trmt61B is a methyltransferase responsible for 1-methyladenosine at position 58 of human mitochondrial tRNAs. RNA, 18, 2269–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vilardo, E., Nachbagauer, C., Buzet, A., Taschner, A., Holzmann, J. and Rossmanith, W. (2018) A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase - extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res., 46, 11126–11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Waku, T., Nakajima, Y., Yokoyama, W., Nomura, N., Kako, K., Kobayashi, A., Shimizu, T. and Fukamizu, A. (2016) NML-mediated rRNA base methylation links ribosomal subunit formation to cell proliferation in a p53-dependent manner. J. Cell Sci., 129, 2382–2393. [DOI] [PubMed] [Google Scholar]
- 17. Helm, M. (2006) Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res., 34, 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sharma, S., Hartmann, J.D., Watzinger, P., Klepper, A., Peifer, C., Kotter, P., Lafontaine, D.L.J. and Entian, K.D. (2018) A single N(1)-methyladenosine on the large ribosomal subunit rRNA impacts locally its structure and the translation of key metabolic enzymes. Sci. Rep., 8, 11904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li, X., Xiong, X., Wang, K., Wang, L., Shu, X., Ma, S. and Yi, C. (2016) Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat. Chem. Biol., 12, 311–316. [DOI] [PubMed] [Google Scholar]
- 20. Li, X., Xiong, X., Zhang, M., Wang, K., Chen, Y., Zhou, J., Mao, Y., Lv, J., Yi, D., Chen, X.W. et al. (2017) Base-resolution mapping reveals distinct m(1)A methylome in nuclear- and mitochondrial-encoded transcripts. Mol. Cell, 68, 993–1005 e1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grozhik, A.V., Olarerin-George, A.O., Sindelar, M., Li, X., Gross, S.S. and Jaffrey, S.R. (2019) Antibody cross-reactivity accounts for widespread appearance of m(1)A in 5'UTRs. Nat. Commun., 10, 5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Khoddami, V., Yerra, A., Mosbruger, T.L., Fleming, A.M., Burrows, C.J. and Cairns, B.R. (2019) Transcriptome-wide profiling of multiple RNA modifications simultaneously at single-base resolution. Proc. Natl. Acad. Sci. U. S. A., 116, 6784–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hauenschild, R., Tserovski, L., Schmid, K., Thuring, K., Winz, M.L., Sharma, S., Entian, K.D., Wacheul, L., Lafontaine, D.L., Anderson, J. et al. (2015) The reverse transcription signature of N-1-methyladenosine in RNA-Seq is sequence dependent. Nucleic Acids Res., 43, 9950–9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Limbach, P.A. and Paulines, M.J. (2017) Going global: the new era of mapping modifications in RNA. Wiley Interdiscip. Rev. RNA, 8, 10.1002/wrna.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Motorin, Y. and Helm, M. (2019) Methods for RNA modification mapping using deep sequencing: established and new emerging technologies. Genes, 10, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Motorin, Y., Muller, S., Behm-Ansmant, I. and Branlant, C. (2007) Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol., 425, 21–25. [DOI] [PubMed] [Google Scholar]
- 27. Zhou, H., Rauch, S., Dai, Q., Cui, X., Zhang, Z., Nachtergaele, S., Sepich, C., He, C. and Dickinson, B.C. (2019) Evolution of a reverse transcriptase to map N(1)-methyladenosine in human messenger RNA. Nat. Methods, 16, 1281–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ryvkin, P., Leung, Y.Y., Silverman, I.M., Childress, M., Valladares, O., Dragomir, I., Gregory, B.D. and Wang, L.S. (2013) HAMR: high-throughput annotation of modified ribonucleotides. RNA, 19, 1684–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bai, B., Wang, X., Li, Y., Chen, P.C., Yu, K., Dey, K.K., Yarbro, J.M., Han, X., Lutz, B.M., Rao, S. et al. (2020) Deep multilayer brain proteomics identifies molecular networks in Alzheimer's disease progression. Neuron, 105, 975–991 e977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wingo, A.P., Fan, W., Duong, D.M., Gerasimov, E.S., Dammer, E.B., Liu, Y., Harerimana, N.V., White, B., Thambisetty, M., Troncoso, J.C. et al. (2020) Shared proteomic effects of cerebral atherosclerosis and Alzheimer's disease on the human brain. Nat. Neurosci., 23, 696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sanchez, M.I., Mercer, T.R., Davies, S.M., Shearwood, A.M., Nygard, K.K., Richman, T.R., Mattick, J.S., Rackham, O. and Filipovska, A. (2011) RNA processing in human mitochondria. Cell Cycle, 10, 2904–2916. [DOI] [PubMed] [Google Scholar]
- 32. Cozen, A.E., Quartley, E., Holmes, A.D., Hrabeta-Robinson, E., Phizicky, E.M. and Lowe, T.M. (2015) ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods, 12, 879–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saikia, M., Fu, Y., Pavon-Eternod, M., He, C. and Pan, T. (2010) Genome-wide analysis of N1-methyl-adenosine modification in human tRNAs. RNA, 16, 1317–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vilardo, E. and Rossmanith, W. (2015) Molecular insights into HSD10 disease: impact of SDR5C1 mutations on the human mitochondrial RNase P complex. Nucleic Acids Res., 43, 5112–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Idaghdour, Y. and Hodgkinson, A. (2017) Integrated genomic analysis of mitochondrial RNA processing in human cancers. Genome Med., 9, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iturria-Medina, Y., Carbonell, F.M., Sotero, R.C., Chouinard-Decorte, F., Evans, A.C. and Alzheimer's Disease Neuroimaging, I (2017) Multifactorial causal model of brain (dis)organization and therapeutic intervention: application to Alzheimer's disease. NeuroImage, 152, 60–77. [DOI] [PubMed] [Google Scholar]
- 37. Veitch, D.P., Weiner, M.W., Aisen, P.S., Beckett, L.A., Cairns, N.J., Green, R.C., Harvey, D., Jack, C.R., Jr., Jagust, W., Morris, J.C. et al. (2019) Understanding disease progression and improving Alzheimer's disease clinical trials: recent highlights from the Alzheimer's disease neuroimaging initiative. Alzheimers Dement., 15, 106–152. [DOI] [PubMed] [Google Scholar]
- 38. Wang, W., Zhao, F., Ma, X., Perry, G. and Zhu, X. (2020) Mitochondria dysfunction in the pathogenesis of Alzheimer's disease: recent advances. Mol. Neurodegener., 15, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G., Durbin, R. and Genome Project Data Processing, S (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koboldt, D.C., Chen, K., Wylie, T., Larson, D.E., McLellan, M.D., Mardis, E.R., Weinstock, G.M., Wilson, R.K. and Ding, L. (2009) VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics, 25, 2283–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shulman, J.M., Chipendo, P., Chibnik, L.B., Aubin, C., Tran, D., Keenan, B.T., Kramer, P.L., Schneider, J.A., Bennett, D.A., Feany, M.B. et al. (2011) Functional screening of Alzheimer pathology genome-wide association signals in Drosophila. Am. J. Hum. Genet., 88, 232–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bray, N.L., Pimentel, H., Melsted, P. and Pachter, L. (2016) Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol., 34, 525–527. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
m1A-quant-seq datasets are available from NCBI’s Gene Expression Omnibus (GEO). The accession number for all the datasets reported in this paper is GSE168199. The 5XFAD proteomic data used in this study were sourced from (29) and are accessible from the PRIDE database (www.proteomexchange.org): with accession numbers of PXD007974 and PXD018590. The proteomic data from the ROS/MAP study (30) are available at https://www.synapse.org/#!Synapse:syn17015098. Scripts used throughout this study are available upon request.
Conflict of Interest statement. The authors declare no competing interests.