Abstract
Mis-expression of DUX4 in skeletal muscle causes facioscapulohumeral muscular dystrophy (FSHD). Human DUX4 and mouse Dux are retrogenes derived from retrotransposition of the mRNA from the parental DUXC gene. Primates and rodents have lost the parental DUXC gene, and it is unknown whether DUXC had a similar role in driving an early pluripotent transcriptional program. Dogs and other Laurasiatherians have retained DUXC, providing an opportunity to determine the functional similarity to the retrotransposed DUX4 and Dux. Here, we identify the expression of two isoforms of DUXC mRNA in canine testis tissues: one encoding the canonical double homeodomain protein (DUXC), similar to DUX4/Dux, and a second that includes an in-frame alternative exon that disrupts the conserved amino acid sequence of the first homeodomain (DUXC-ALT). The expression of DUXC in canine cells induces a pluripotent program similar to DUX4 and Dux and induces the expression of a similar set of retrotransposons of the ERV/MaLR and LINE-1 families, as well as pericentromeric satellite repeats; whereas DUXC-ALT did not robustly activate gene expression in these assays. Important for preclinical models of FSHD, human DUX4 and canine DUXC show higher conservation of their homeodomains and corresponding binding motifs compared with the conservation between human DUX4 and mouse Dux, and human DUX4 activates a highly similar transcriptional program in canine cells. Together, these findings show that retrotransposition resulted in the loss of an alternatively spliced isoform and that DUXC containing mammals might be good candidates for certain preclinical models ofFSHD.
Introduction
DUX4 and its mouse ortholog Dux are both retrogenes derived from the parental DUXC gene (1,2). These retrotranspositions are thought to have occurred separately in the primate and rodent lineages, and both primates and rodents have lost the intron-containing parental DUXC gene. Retrotransposition into a heterologous region of the genome results in the loss of many of the evolved enhancer and promoter elements of the parental gene and often results a non-functional pseudogene that accumulates mutations over time. However, occasionally retrotransposition into a genomic region that drives expression in different tissues or developmental periods can result in an evolved functional protein driving a new variation in phenotype that has selective advantage and can result in the rapid evolution of a new trait, such as the FGF4 retrotransposition driving the generation of short, chondrodysplastic legs in different dog breeds (3).
Another consequence of retrotransposition is the insertion of a single splice isoform from a gene that might have multiple different isoforms expressed at different times and in different tissues. In the case of mouse Dux and human DUX4, it remains unknown whether the retrotransposition occurred into the parental DUXC locus or at another region of the genome. The loss of the parental DUXC gene from both the mouse and primate lineages and the multi-copy array structure of the locus in cows and other Laurasiatheria suggests that the former may have occurred (1). However, since it is not known whether the parental DUXC gene produced alternative isoforms, the consequence of replacing the parental gene with a retrogene remains speculative.
In this study, we investigated DUXC in canine testis and other tissues. We identified two DUXC isoforms: one encodes a double homeodomain protein similar to Dux and DUX4, whereas an alternative splice isoform inserts an in-frame exon in the first homeodomain that is predicted to disrupt or alter its DNA binding specificity. We show that the canonical DUXC isoform, i.e. containing two intact homeodomains, has a DNA binding site similar to both human DUX4 and mouse Dux, and induces a similar transcriptional network, whereas the DUXC isoform with a disrupted first homeodomain while capable of binding a subset of the DUXC sites shows very little transcriptional activity in these assays. Surprisingly, human DUX4 expressed in canine cells more accurately induced the DUX4/pluripotent/facioscapulohumeral muscular dystrophy (FSHD) signature compared with human DUX4 expressed in mouse cells, suggesting that some DUXC containing mammals might be more appropriate than mice for preclinical models of FSHD that rely on mis-expression of humanDUX4.
Results
The canine DUXC gene resides in a repeated region at the most telomeric annotated region on chromosome 17 and produces different isoforms of DUXC
The current build (canFam4/GSD_1.0) (4) of the canine genome contains sequences predicted to encode a protein with high homology to the DUXC protein family at two nearby locations at the most distal region of chromosome 17 (see Materials and Methods). Because human DUX4 and mouse Dux are expressed in the testes, we used real-time reverse transcriptase-polymerase chain reaction (RT-PCR) based on the genomic sequence to determine the canine DUXC transcripts in canine testis RNA and discovered that canine DUXC produced two different splice isoforms from the DUXC gene: the predicted five exon transcript with two intact homeodomains and an alternative transcript that inserts an additional 174 nucleotide unannotated exon between the canonical exons 2 and 3 that is identical to NCI Reference Sequence XM_038453516.1 (Fig. 1A and Supplementary Material, Fig. S1A), which we will refer to as DUXC and DUXC-ALT, respectively. The insertion of this alternative exon changes the last 14 amino acids of HD1 and maintains the reading frame when spliced into exon 3, essentially creating a DUXC protein with amino acid substitutions in regions of HD1 previously shown to be critical for DUX4 DNA binding (5) and maintaining the sequence of the remainder of the protein, including the conserved region at the extreme c-terminus that characterizes the DUXC family (2), a region that has been shown to recruit p300 and function as the transcriptional activation domain for the DUX4 protein (6).
Figure 1.
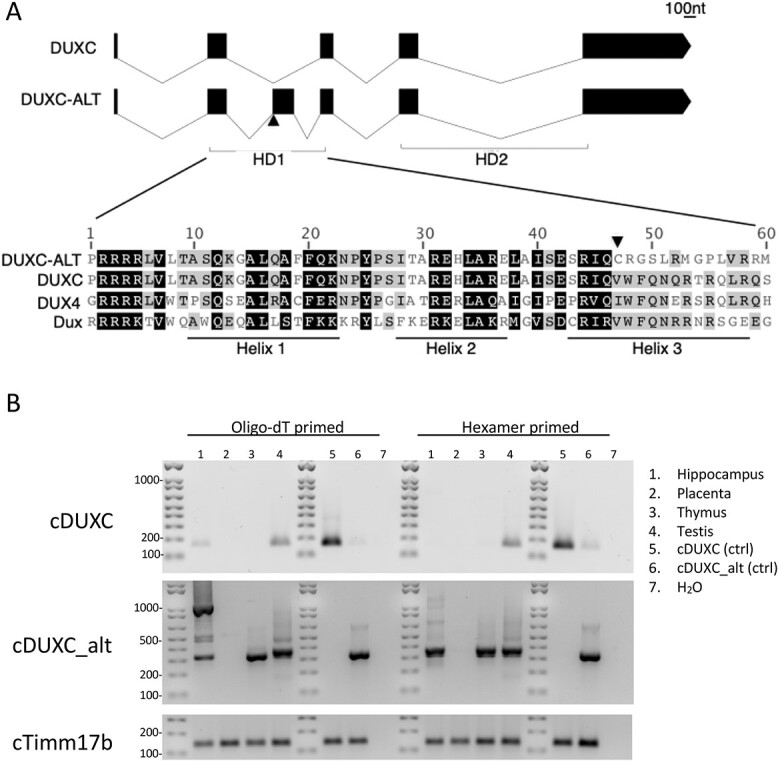
Canine DUXC locus makes two different isoforms. (A) Top panel shows a schematic of the exons encoding DUXC and DUXC-ALT and the bottom panel shows the alignment in the first homeodomain region with mouse Dux and Human DUX4 HD1. (B) Nested PCR primers specific to the DUXC (top panel) or DUXC-ALT (middle panel) mRNA isoforms show DUXC expression in testis a variably in hippocampus; DUXC-ALT is identified in testis, thymus and hippocampus.
The canine DUXC first homeodomain (HD1) protein sequence shows an overall 75% similarity to the human DUX4 HD1, whereas the DUXC-ALT isoform disrupts the highly conserved third helical region and shows an overall 58% similarity to human DUX4. RT-PCR, using primers that amplify both isoforms of DUXC, identified predominantly the DUXC-ALT isoform in testis and in the thymus (Supplementary Material, Fig. S1B), tissues previously shown to express DUX4 (7,8). Nested primers that specifically amplify the canonical DUXC isoform identified low amounts of this isoform in the testis and lower levels in the hippocampus, whereas the DUXC-ALT isoform was detected in RNA from the hippocampus, thymus and testis (Fig. 1B).
DUXC activates an early pluripotent program similar to human DUX4 and mouse Dux
To determine the transcriptomes of DUXC and DUXC-ALT and their relation to the DUX4 regulated transcriptome in FSHD muscle, we used a system that parallels tissue culture models of the human DUX4 transcriptome: forced expression of these transcription factors in cultured skeletal muscle cells (9). Specifically, we generated four stable canine myoblast cell lines with a doxycycline-inducible canine DUXC, canine DUXC-ALT, human DUX4 or Luciferase (as a control). It was necessary to establish inducible cell lines because the constitutive expression of DUXC, but not DUXC-ALT, in canine myoblasts resulted in cell death between 24 and 48 h of expression, similar to the toxicity of DUX4 expressed in human cells (5,10). The expression of canine DUXC in canine myoblasts changed the expression of 1753 genes compared with cells expressing Luciferase (adjusted P-value < 0.05 corresponding to 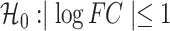 ; Fig. 2A; Supplementary Material, Table S1). The vast majority of these genes significantly increased in expression (1562 up versus 191 down), consistent with previous work showing both human DUX4 (9) and mouse Dux are transcriptional activators (11). Similarly, the expression of DUX4 in canine myoblasts caused 1784 genes to be differentially expressed (1553 up versus 231 down) (Fig. 2B, Supplementary Material, Table S1). There was a high correlation (Pearson = 0.803) between the genes induced by DUXC and DUX4 in canine myoblasts (Fig. 2C). In contrast to DUXC and DUX4, the expression of DUXC-ALT showed very little gene induction with only 20 genes increased compared with 114 decreased (Fig. 2D, Supplementary Material, Table S1).
; Fig. 2A; Supplementary Material, Table S1). The vast majority of these genes significantly increased in expression (1562 up versus 191 down), consistent with previous work showing both human DUX4 (9) and mouse Dux are transcriptional activators (11). Similarly, the expression of DUX4 in canine myoblasts caused 1784 genes to be differentially expressed (1553 up versus 231 down) (Fig. 2B, Supplementary Material, Table S1). There was a high correlation (Pearson = 0.803) between the genes induced by DUXC and DUX4 in canine myoblasts (Fig. 2C). In contrast to DUXC and DUX4, the expression of DUXC-ALT showed very little gene induction with only 20 genes increased compared with 114 decreased (Fig. 2D, Supplementary Material, Table S1).
Figure 2.
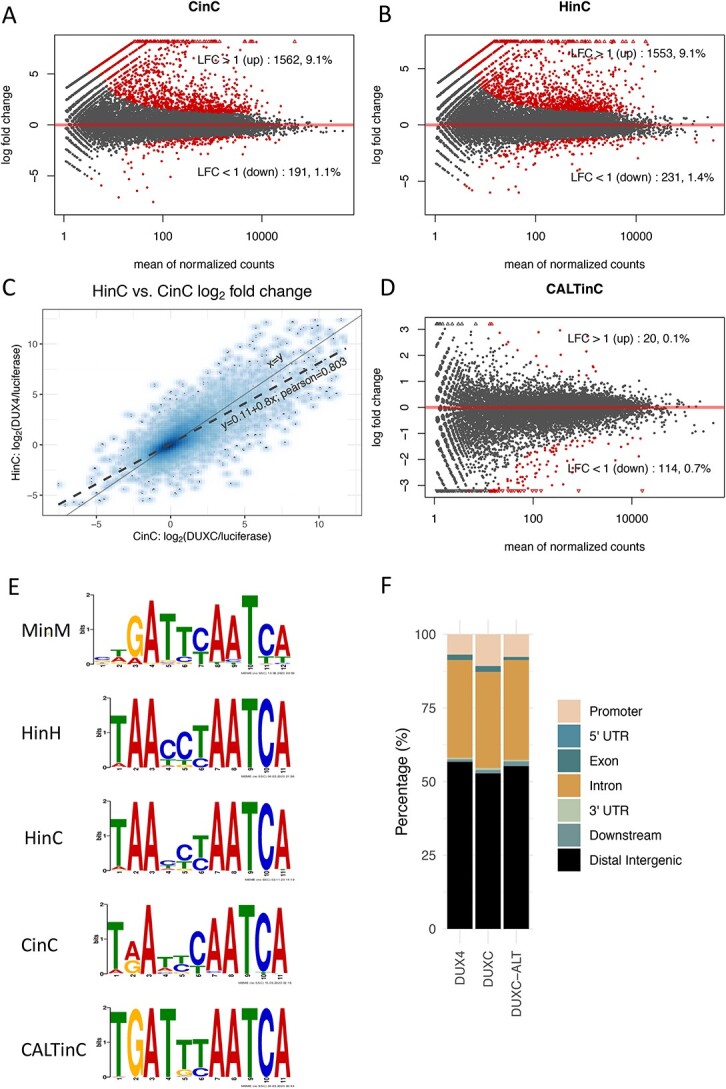
Transcriptome and DNA binding sites of DUXC, DUX4 and DUXC-ALT in canine skeletal muscle cells. (A) A MA plot of gene expression in canine muscle cells expressing DUXC (CinC) compared with the control luciferase expression vector. The x axis is the mean of normalized counts on each gene between two conditions (A value), and the y axis is the 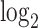 fold change (M value). Red dots represent the differentially expressed genes with adjusted P-values < 0.05 corresponding to the hypothesis
fold change (M value). Red dots represent the differentially expressed genes with adjusted P-values < 0.05 corresponding to the hypothesis 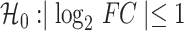 . (B) Similar to (A) but with comparison of the cells expressing DUX4 (HinC). (C) A smoothed scatter plot to show the linear relationship of the
. (B) Similar to (A) but with comparison of the cells expressing DUX4 (HinC). (C) A smoothed scatter plot to show the linear relationship of the 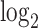 fold change between the CinC (x axis) and HinC (y axis) comparison models. The Pearson correlation between the two sets of
fold change between the CinC (x axis) and HinC (y axis) comparison models. The Pearson correlation between the two sets of 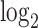 fold change is 0.803. The gray dashed line is the linear regression,
fold change is 0.803. The gray dashed line is the linear regression, 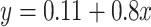 , and the solid gray line is
, and the solid gray line is 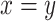 . (D) Similar to (A) but with the comparison of the cells expressing DUXC-ALT (CALTinC). (E) De novo motif discovery determined binding motifs for mouse Dux expressed in mouse myoblasts (MinM), human DUX4 expressed in human myoblasts (HinH), human DUX4 expressed in canine myoblasts (HinC), and canine DUXC and DUXC-ALT expressed in canine myoblasts (CinC and CALTinC). The HinH and MinM binding motifs were derived from published ChIP-seq data GSE33838 and GSE87279, respectively. (F) Genomic distribution of ChIP-seq peaks for DUX4, DUXC and DUXC-ALT.
. (D) Similar to (A) but with the comparison of the cells expressing DUXC-ALT (CALTinC). (E) De novo motif discovery determined binding motifs for mouse Dux expressed in mouse myoblasts (MinM), human DUX4 expressed in human myoblasts (HinH), human DUX4 expressed in canine myoblasts (HinC), and canine DUXC and DUXC-ALT expressed in canine myoblasts (CinC and CALTinC). The HinH and MinM binding motifs were derived from published ChIP-seq data GSE33838 and GSE87279, respectively. (F) Genomic distribution of ChIP-seq peaks for DUX4, DUXC and DUXC-ALT.
DUXC binds motifs similar to both human DUX4 and mouse Dux
To determine the binding motifs of DUXC and DUXC-ALT, and their relation to human DUX4, we performed ChIP-seq of these three factors in cultured canine skeletal muscle. Because we do not have an antibody to DUXC or DUXC-ALT, we substituted the non-homeodomain C-terminal regions of these proteins with the human DUX4, i.e. maintaining the canine homeodomain region but replacing the C-terminal region with human DUX4 sequence. This allowed us to use the same antisera to the C-terminal region of DUX4 to perform ChIP on these chimeric proteins (as previously described for the comparison of human DUX4 and mouse Dux binding sites (11).
Using a peak calling algorithm (MACS2 (12)) at standard thresholds (q < 0.01), we observed 6031 peaks for DUXC and 581 peaks for DUXC-ALT. In cells expressing DUX4, we observed 85 825 peaks (Supplementary Material, Table S2). Using a de novo motif discovery algorithm (MEME), we determined the binding motif among the top peaks of each transcription factor and compared them with the binding motifs for DUX4 in human myoblasts and mouse Dux in mouse myoblasts (Fig. 2E). Together, the resulting data suggested that the canonical homeodomain isoform of canine DUXC binds a site that is a composite of the human DUX4 site and the mouse Dux site with either an A or G at position 2 compared with the G for mouse Dux and the A for human DUX4, whereas the DUXC-ALT favors a site similar to mouse Dux. All three factors had the majority of their peaks in intergenic regions; however, DUXC showed slightly greater enrichment in promoter regions compared with DUXC-ALT and DUX4 (Fig. 2F).
The DUXC transcriptome has high similarity to the FSHD transcriptome
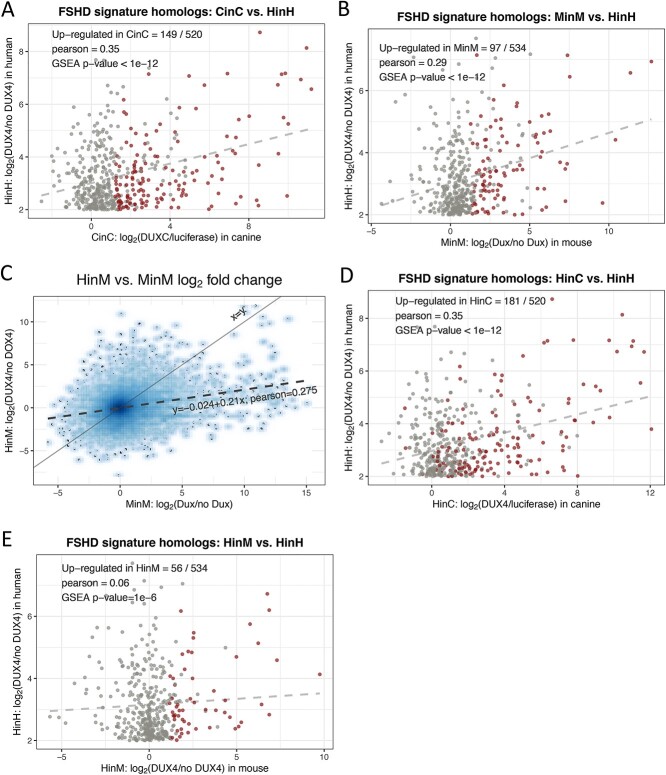
To determine whether DUXC induced a transcriptional program similar to the FSHD muscle cell transcriptome, we compared the DUXC transcriptome with the 666 genes identified as mis-expressed in several different model systems of FSHD (FSHD signature genes) (9). Of the subset of 520 genes that have a 1:1 homology between human and canine, 29% (146) were positively affected (adjusted P-value < 0.05 corresponding to 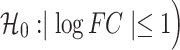 by the expression of canine DUXC in canine muscle cells [Pearson = 0.35; gene set enrichment analysis(GSEA) P-value <1e-12; Fig. 3A]. We then compared the previously described Dux transcriptome in mouse muscle cells (11) with the human DUX4 FSHD signature genes: of the 666 upregulated FSHD signature genes, 532 have 1:1 homology mapping to mouse and 97 of these (18%) were positively affected by the expression of mouse Dux in mouse muscle cells (Pearson = 0.29; GSEA P-value < 1e-12; Fig. 3B). Therefore, although mice and humans share a more recent common ancestor than dogs and humans (13), the expression of DUXC in canine muscle demonstrates an equally good, or slightly better, induction of the gene signature characteristic of human FSHD models.
by the expression of canine DUXC in canine muscle cells [Pearson = 0.35; gene set enrichment analysis(GSEA) P-value <1e-12; Fig. 3A]. We then compared the previously described Dux transcriptome in mouse muscle cells (11) with the human DUX4 FSHD signature genes: of the 666 upregulated FSHD signature genes, 532 have 1:1 homology mapping to mouse and 97 of these (18%) were positively affected by the expression of mouse Dux in mouse muscle cells (Pearson = 0.29; GSEA P-value < 1e-12; Fig. 3B). Therefore, although mice and humans share a more recent common ancestor than dogs and humans (13), the expression of DUXC in canine muscle demonstrates an equally good, or slightly better, induction of the gene signature characteristic of human FSHD models.
Figure 3.
Comparisons to the FSHD transcriptome. (A) A scatter plot of 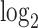 fold change of the 520 human-canine homologs of FSHD signature genes in the HinH and CinC transcriptome. Red dots indicate that the homologs are also upregulated in CinC. (B) Same as (A), but with the 534 human-mouse homologs in the HinH and MinM transcriptome, red dots indicate upregulation in MinM. (C) A smooth scatter plot to show the relationship of
fold change of the 520 human-canine homologs of FSHD signature genes in the HinH and CinC transcriptome. Red dots indicate that the homologs are also upregulated in CinC. (B) Same as (A), but with the 534 human-mouse homologs in the HinH and MinM transcriptome, red dots indicate upregulation in MinM. (C) A smooth scatter plot to show the relationship of 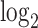 fold change between the HinH and MinM transcriptome. The Pearson correlation between these two sets is 0.275. Gray dashed line presents the linear regression line,
fold change between the HinH and MinM transcriptome. The Pearson correlation between these two sets is 0.275. Gray dashed line presents the linear regression line, 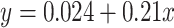 and the solid is
and the solid is 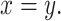 (D) Same as (A), but with 520 human-canine homologs in the cross-species HinC and HinH transcription, and red dots indicate upregulation in HinC. (E) Same as (A), but with 534 human-mouse homologs in the HinH and cross-species HinM transcriptome, and red dots indicate upregulation inHinM.
(D) Same as (A), but with 520 human-canine homologs in the cross-species HinC and HinH transcription, and red dots indicate upregulation in HinC. (E) Same as (A), but with 534 human-mouse homologs in the HinH and cross-species HinM transcriptome, and red dots indicate upregulation inHinM.
DUX4 expression in canine and murine cells
Several mouse models of FSHD have been made that introduce the human DUX4 locus or transgene into mice (14–18); however, human DUX4 does not robustly activate the same transcriptome in mouse muscle cells as in human muscle cells (11). The canine homeodomains show a higher similarity to human DUX4 than the comparison of human to mouse: canine HD1 shows 75% similarity to human, whereas mouse shows 60% similarity; canine HD2 shows 92% similarity to DUX4, whereas mouse shows 70% similarity to DUX4 (Supplementary Material, Fig. S2). This raises the possibility that canine DUXC and human DUX4 might retain a similar cross-species transcriptional program. As described above, the cross-species expression of human DUX4 in canine skeletal muscle cells positively regulated 1553 genes and there was a high correlation (Pearson = 0.803) between the genes induced by DUXC and DUX4 in canine myoblasts (see Fig. 2C). A similar comparison of our previous dataset expressing mouse Dux or human DUX4 in mouse myoblasts (11) showed a much lower correlation (Pearson = 0.275; Fig. 3C). Focusing on the set of FSHD signature genes, DUX4 regulated more of these genes in canine cells (181 out of the 520 homologs, 35%; Pearson = 0.35; GSEA P-value < 1e-12; Fig. 3D) than it regulated in mouse cells (56 out of 532 homologs, 10%; Pearson = 0.06; GSEA P-value = 1e-6; Fig. 3E).
The DUXC transcriptome compared with that of the early embryo
Given the correlation between human DUX4 and mouse Dux gene expression with the first wave of transcription in the early embryo (19), we asked whether the DUXC transcriptome is enriched for genes typically expressed in the early embryo. Since the transcriptome of early canine embryos is currently unavailable, we used Ensembl v88 homolog database to identify the canine homologs of the mouse 2C-like gene signature (20) and then performed GSEA by using hypergeometric tests. For this analysis, we used homologs with 1:1 canine-to-mouse mapping. Of the 466 genes that form the mouse 2C-like signature, 292 have 1:1 homology mapping to canine, and 58 of the 292 (20%) homologs were differentially upregulated by DUXC in canine cells. The GSEA inferred that the DUXC transcriptome is enriched with homologs of the mouse 2C-like gene signature (P-value = 7e-9).
DUXC and DUX4 activate retrotransposons
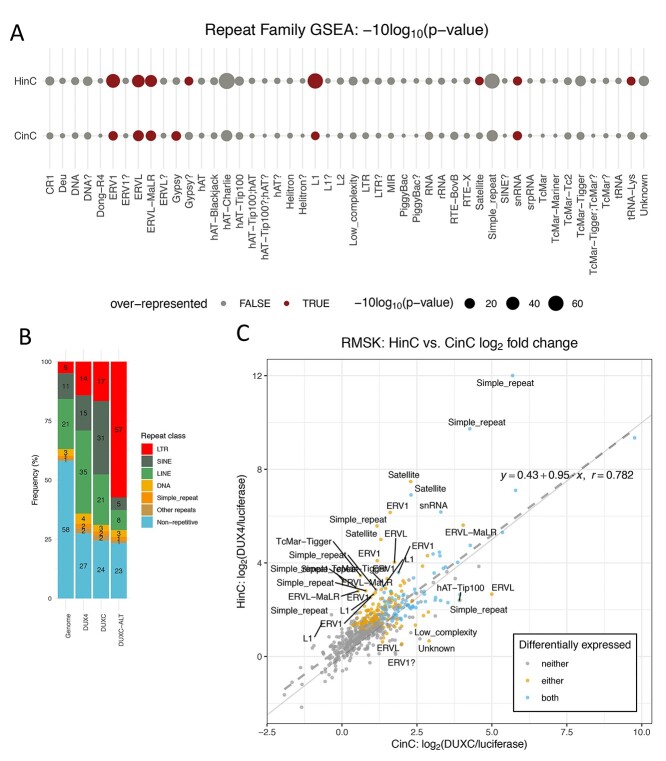
Human\DUX4 and mouse Dux activate similar sets of endogenous retrotransposons including MERV, HERV and MaLR families (11,21). In canine cells expressing canine DUXC or human DUX4, RNA-seq analysis (see Materials and Methods) detected elevated transcripts derived from many LINE-1 elements (L1s) and LTR subfamilies such as ERV1, ERVL and ERVL-MaLR, compared with Luciferase control (Supplementary Material, Table S3; Supplementary Material, Fig. S3A and B), although this analysis is based on a limited annotation of repeat elements in the canine genome. GSEA also confirmed the enrichment of these LTR subfamilies (Figs 4A and S3C–E). Moreover, DUXC and DUX4 bind to 70 and 95% of activated ERV1, ERVL and ERVL-MaLR elements, respectively, which suggested that the binding of DUX4 or DUXC directly control the activation of these LTR elements. Significantly higher than expected distributions of DUX4 (14%) and DUXC (17%) peak binding sites on LTRs also support the LTRs enrichment (Fig. 4B).
Figure 4.
Repeat element transcriptome shows retrotransposon activation by DUXC in canine skeletal muscle cells. (A) Repeat family enrichment analysis for DUX4 (HinC) and DUXC (CinC) expressed in canine skeletal muscle cells. Each dot represents the GSEA statistics for each repeat family with size reflecting the scaled P-value, −10log2 (P-value). Red dots indicate the family is over-represented with P-values < 0.1 and the number of significant genes in the family is higher than expected. (B) The distribution of peaks of DUX4, DUXC, DUXC-ALT transcription factors in classes of repeat elements versus the distribution of the repeat elements in the whole genome. (C) A scatter plot to show the relationship of 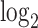 fold change of the HinC and CinC transcriptome. Each dot represents a repeat element, and the color coding presents the differential expression status. Dashed line is the linear regression line
fold change of the HinC and CinC transcriptome. Each dot represents a repeat element, and the color coding presents the differential expression status. Dashed line is the linear regression line 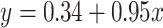 , whereas the solid is
, whereas the solid is 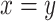 . A repeat element’s family name is labeled if the difference between two models is >1.5.
. A repeat element’s family name is labeled if the difference between two models is >1.5.
Comparing DUX4 and DUXC induction of retroelements in canine cells, the correlation of the repeat transcripts was high (Pearson = 0.782; Fig. 4C). (The ERVL-MaLR elements in canine are categorized as MLT1 (by Repeat Masker), which represent mammalian LTRs that populated the genome before the split of canine and primate lineages.) As for non-LTR retrotransposons, DUX4 upregulated 54% of L1s (52/97) and 28.5% of DUX4 binding sites overlapped with L1s. In comparison, DUXC had weaker effects on L1s—only 16% of L1s (16/97) showed differential expression (Supplementary Material, Fig. S4), and 15.6% of DUXC peaks overlap withL1s.
The canine genome has 5 major satellites and satellite-like family: Bs, Carnivore Satellite (CarSat1, CarSat2), Satellite Canis Familiaris (SAT1_CF, SAT2_CF, SAT3_CF, SAT4_CF, and SAT6_CF), (CATTC)n, (GAATG)n and SUBTEL_sa. Three satellites—Bs, CarSat1 and SAT1_CF—were differentially upregulated in cells expressing DUX4, whereas in cells expressing DUXC, only Bs satellite repeats were significantly upregulated, and CarSat1 was moderately affected (adjusted P-value = 0.14).
Discussion
This study shows that the canonical canine DUXC protein activates a program similar to human DUX4 and mouse Dux (11,22), including the pluripotent signature genes characteristic of the first wave of zygotic gene activation (19). Therefore, even prior to retrotransposition in the primate and rodent lineages, the DUXC family likely had a role in early embryonic development. Although we were able to show the expression of DUXC or DUXC-ALT in canine testis, thymus and hippocampus, we were unable to obtain RNA from cleavage-stage canine embryos and have yet to confirm DUXC expression at this stage. Therefore, we can only speculate that the activation of this early embryonic program by DUXC in canine myoblasts suggests that it might also have this role in the early embryo. Like DUXC, DUX4 is also expressed in the testis and thymus (7,8). In the testis, immunohistochemistry shows DUX4 expression in periluminal cells consistent with the spermatogonia and many of the genes known to be regulated by DUX4 in the early embryo and in FSHD skeletal muscle are also expressed in the testis (7,22). Dux knock-out mice remain fertile (23–26), and therefore any role in spermatogenesis is not essential since the reduced liter size in Dux knock-out mice appears to occur post-implantation (26). The cell type(s) expressing DUX4 or DUXC in the thymus remain unknown, as is the cell type(s) expressing DUXC in the hippocampus, although in the latter case it is interesting that DUX4 expressed in mouse ES cells induces neurogenesis (27),
Our prior studies comparing human DUX4 and mouse Dux not only showed their shared regulation of this early developmental program (11,19), but also highlighted a rapid divergence in their DNA binding motifs. Because of this, expressing human DUX4 in mouse cells or mouse Dux in human cells showed a highly degraded activation of the pluripotent transcription program (11). In this regard, it is interesting that the expression of human DUX4 in canine cells largely maintains the pluripotent transcriptional signature. On a practical level, this has implications for preclinical models of FSHD that are based on mis-expression of human DUX4, such as transgenic models using an inducible DUX4 or the human D4Z4 region containing the DUX4 retrogene (14–16). It is possible that species other than mice might be more suitable preclinical models for FSHD that utilize DUX4 expression.
Although human DUX4 and mouse Dux have been increasingly studied over the last decade, there have been few studies of the parental DUXC genes. In this study, one of the important findings is that the most abundant isoform of the DUXC mRNA in the testis and some other canine tissues includes an in-frame exon that results in a disruption of the amino acid sequence of the first homeodomain. Consequently, DUXC-ALT binds to the near palindromic TGAT[T/c][T/c]AATCA sequence rather than the more asymmetric T[A/G]A[t/a][t/c][C/T]AATCA motif of the canonical DUX4/DUXC protein. The changed sequence specificity might be due to the altered amino acid sequence of the first homeodomain region of the DUXC-ALT protein or might suggest that the first homeodomain has lost DNA binding and the DUCX-ALT binds as a homodimer. If the latter is correct, it raises the interesting possibility that the evolutionary precursor to the DUX family was a single homeodomain protein that functioned as a dimer and the duplication of the homeodomain region allowed a monomer to bind the double homeodomain motifs that regulated the transcriptional program of the ancestral precursor single homeodomain protein. This is an attractive hypothesis because of a current paradox in DUX4/Dux biology. Human DUX4 and mouse Dux both regulate a portion of the first wave of zygotic gene activation in the early embryo (11,19), suggesting a fundamental role in early development. Yet the DUX family arose in placental mammals through the duplication of the homeodomain region of the ancestral gene (1,2). If the ancestral single HD protein bound as a dimer, either as a homodimer or as a heterodimer with a yet to be identified homeodomain protein, then the intragenic HD duplication would allow a monomeric protein to regulate the genes with evolved double homeodomain motifs that previously required a homo- or heterodimer.
The fact that DUXC-ALT binds DNA at all is surprising based on prior studies of the DUX4 homeodomains. Wallace et al. (5) showed that the HD1 residues in DUX4 that are disrupted by the DUXC-ALT splice-form were necessary for DUX4 DNA binding, transcriptional activation of target genes, and cell toxicity. Subsequent crystal structures of DUX4 bound to DNA further supported the role of these amino acids in DNA binding (28). The lack of transcriptional activity of the DUXC-ALT and the decreased DNA binding suggests that the disruption of the first homeodomain prevents high-affinity binding necessary for recruiting a stable transcription complex and for a strong ChIP-seq signal. An alternative, but not exclusive, possibility is that the DUXC-ALT is a less efficient pioneer transcription factor compared with the double homeodomain DUXC. DUX4 and Dux efficiently bind in areas of previously inaccessible chromatin and pioneer factor activity might require the extended motif of a double HD protein.
Our study also reveals an important evolutionary consequence of the retrotransposition of the DUXC gene to create human DUX4 and mouse Dux. The DUXC-ALT isoform is more easily detected in testis and some other canine tissues, whereas the DUXC isoform appears less abundant in these tissues. If the balance between these isoforms regulated the activity of the DUXC program, then the retrotransposition would ‘lock-in’ what appears to be the most transcriptionally active isoform and could result in the expression of some genes and retroelements at developmental timepoints where only the DUXC-ALT isoform had been expressed. This might have profound implications in the germline or early embryo if the retrotransposition induced retroelement activation. The activation of ERV, LINE1 and pericentromeric satellite repeats by both canine DUXC and human DUX4 in canine cells strongly supports the conclusion that this is a conserved evolutionary function of the DUXC family. It is notable that only the canonical double homeodomain isoform of DUXC activates these elements, suggesting that the canonical DUX4/DUXC isoform might have evolved for selective expression in specific tissues or at specific times that relied on retroelement expression. In this model, the DUXC retrotransposition in rodents and primates might have resulted in DUXC expression in the germline and other tissues that previously expressed predominantly the DUXC-ALT isoform. Because DUXC and DUX4 activate retroelements, it is possible that ‘locking-in’ the retroelement-activating isoform through retrotransposition conferred a partial benefit in the early embryo and germline and had strong evolutionary consequences. This model and further speculation regarding the relative biological roles of DUXC and DUXC-ALT will be greatly strengthened if similar splice forms of the DUXC gene are identified in other Laurasiatherians.
Materials and Methods
RT-PCR
Primers used that amplify both DUXC and DUXC-ALT were ATL-254a and ATL-265a. Forward nested primers used to specifically detect the DUXC isoform were ST304g nested with AB304i. Forward nested primers used to specifically detect the DUXC-ALT isoform were ATL-305b nested with ATL-305c. These forward primers were paired with ATL-265a nested with ATL265b. Primers for the housekeeping gene canine Timm17b were cTimm17b-F and cTimm17b-R.
ATL254a, GGCCTCCAGCAGCACCCCCG
ATL265a, GAGGATACTGGTTTGGGATG
ST340g, GAGAGCTGGCCATCTCCGAGTCTAGAATCCAGGTCTG
AB304i, GTCTAGAATCCAGGTCTGGTTCC
ATL-305b, AGAGGAAGCCTGCGCATGGGT
ATL-265b, GGATGCAGAAATGGATGTCC
cTimm17b-f, ATCAAGGGCTTCCGCAATG
cTimm17b-r, CACAGTCGATGGTGGAGAACAG
Cell lines, RNAseq and ChIPseq
For the RNA-seq and ChIP-seq, we transduced immortalized canine myoblasts (immortalized with hTERT and CDK4 expression vectors) with the pCW57.1 vector containing a doxycycline-inducible DUXC or DUXC-ALT, human DUX4, or Luciferase. After selection in puromycin, cells were treated with (or without) 1 μg/ml doxycycline for 24 h and then RNA harvested for RNA-seq. For the ChIP-seq, cells were similarly transduced with pCW57.1 containing a chimeric protein that substituted the human DUX4 C-terminal domain (CTD) for the canine CTD to permit the use of rabbit antisera generated to the human DUX4 CTD and the protocol previously used for human DUX4 in human cells (21,22).
ChIP-seq data analysis
We first filtered the low-quality reads based on FASTQC suggested procedure: removing reads with the filter flag = Y indicated in the sequence ID. We then used cutadapt to trim the adapter contamination and used BWA-0.7.10 to align the processed reads to reference genome canFam3. Peaks calling was done by MACS2-2.1.0 with Luciferase samples as negative controls; only peaks with q-value < 0.01 were considered. Last, de novo motif discovery was carried out by MEME-5.1.1 based on the top 1000 peaks, ranked by q-value, with flanking sequences 50 bps up/downstream from the peak summits.
RNA-seq data analysis
The RNA-seq reads derived from our canine skeletal muscle models were filtered to remove the unqualified and then aligned to reference genome canFam3 using Tophat-2.1.0/Bowtie2-2.2.6. The RNA-seq for the mouse (GSE87282) and human (GSE85461) myoblast models were collected from published GEO series; the reads were filtered and aligned to reference genome mm10 and hg38, respectively. The gene annotation for all canine, mouse and human were downloaded from Ensembl database. The gene expression for all mouse, human and canine models was quantified by counting the reads that were overlapped with exons of gene features using the summarizeOverlaps function of Bioconductor’s GenommicAlignments package. We applied DESeq2 (29) for gene expression normalization, log transformation and differential analysis, in which the P-value comparing two conditions was set to reflect the null hypothesis 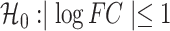 , and the threshold of the adjusted P-value for multiple testing was 0.05. To infer whether the DUXC and DUX4 transcriptomes in canine muscle cells are enriched for the FSHD and, 2C-like and human pluripotent homolog gene signatures, we applied hypergeometric tests as a practice of GSEA, which required four parameters—representing the numbers of (1) all annotated genes, (2) differentially upregulated genes in DUXC (or DUX4) expressed canine cells relative to the controls, (3) genes of a gene set of interest (FSHD, 2C-link or pluripotent) and (4) differentially upregulated genes included the gene set designated in reference (3).
, and the threshold of the adjusted P-value for multiple testing was 0.05. To infer whether the DUXC and DUX4 transcriptomes in canine muscle cells are enriched for the FSHD and, 2C-like and human pluripotent homolog gene signatures, we applied hypergeometric tests as a practice of GSEA, which required four parameters—representing the numbers of (1) all annotated genes, (2) differentially upregulated genes in DUXC (or DUX4) expressed canine cells relative to the controls, (3) genes of a gene set of interest (FSHD, 2C-link or pluripotent) and (4) differentially upregulated genes included the gene set designated in reference (3).
Repeat elements analysis
The repeat elements analysis applied to RNA-seq of our canine model included counting reads for repeat elements, normalization, differential analysis and repeat family/class enrichment analysis, all of which used the annotation from UCSC’s canFam3 Repeat Masker track. The challenge of counting reads for repeat elements was the potential over-estimation due to the multiple alignments of reads mapping to several instances of a repeat element. With this, the count of a read toward a repeat element was adjusted by the number of reported alignments in the BAM file (the NM column). Taking the size factor estimated by the known transcripts of the whole genome, we normalized the repeat element expression and then identified the differentially expressed elements by DESeq2 with adjusted P-value < 0.05 corresponding to the hypothesis 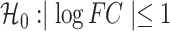 . Finally, we used the hypergeometric test, a conventional method for GSEA, to carry out the repeat family/class enrichment analysis.
. Finally, we used the hypergeometric test, a conventional method for GSEA, to carry out the repeat family/class enrichment analysis.
Identify DUXC copies on the canine genome
To find the copies of DUXC, we started with the canine genome reference canFam3. It has mature transcript and repeat element annotations, in which we used for our RNA-seq and ChIP-seq analysis. This version, however, has gaps in distal regions of chromosome 17, which hindered the identification of DUXC’s gene, one of the copies we found is in an uncharted region. Thus, we instead used the novel canFam4/GSD_1.0 build to map the DUX wild-type sequence. This version rescued most of the difficult-to-sequence regions (30) and enabled us to uncover the locations of two copies of DUXC gene: chr17/CM021978.1:65,040,493-65,045,279 (+) and chr17/CM021978.1:64,996,310-65,001,104 (−). With our DUXC and DUXC-ALT expressed canine RNA-seq, we verified the locations and structures of the expressed transcripts ofDUXC.
Software and packages
The downstream analyses for the RNA-seq data were conducted using the Bioconductor (v3.10) (31) and Tidyverse packages on R-3.6.2 environment.
Data and code availability
The ChIP-seq and RNA-seq of our canine models have been deposited to Genome Expression Omnibus (32) and are accessible through GEO accession number GSE188928. The RNA-seq and ChIP-seq data of the human DUX4 myoblast model were collected from published GEO series GSE85461 and GSE33838, and that of the mouse Dux myoblast model were from GSE87282. All shell scripts, R scripts and processed datasets in R/Bioconductor compatible format are available on our GitHub repository (https://github.com/FredHutch/canFam3.DuxFamily.git). We also host a GitHub page (https://FredHutch.github.io/canFam3.DuxFamily) that gives details of our analysis alone with readable, reproducible codes.
Supplementary Material
Acknowledgements
We thank B. Torok-Storb and the Animal Resource Core of the FHCRC Core Center for Excellence in Hematology (NIDDK P30DK056465) for providing RNA from the canine tissues used in this study.
Conflict of Interest statement. None declared.
Contributor Information
Chao-Jen Wong, Division of Human Biology, Fred Hutchinson Cancer Research Center, Seattle, WA 98109, USA.
Jennifer L Whiddon, Division of Human Biology, Fred Hutchinson Cancer Research Center, Seattle, WA 98109, USA.
Ashlee T Langford, Comparative Medicine, Fred Hutchinson Cancer Research Center, Seattle, WA 98109, USA.
Andrea E Belleville, Divisions of Public Health Sciences and Basic Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA 98109, USA.
Stephen J Tapscott, Division of Human Biology, Fred Hutchinson Cancer Research Center, Seattle, WA 98109, USA; Division of Clinical Research, Fred Hutchinson Cancer Research Center, Seattle, WA 98109, USA.
Funding
This work was supported by NIAMS R01AR045203 (S.J.T.), Friends of FSH Research (S.J.T.) and the Chris Carrino Foundation (S.J.T.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1. Leidenroth, A., Clapp, J., Mitchell, L.M., Coneyworth, D., Dearden, F.L., Iannuzzi, L. and Hewitt, J.E. (2012) Evolution of DUX gene macrosatellites in placental mammals. Chromosoma, 121, 489–497. [DOI] [PubMed] [Google Scholar]
- 2. Leidenroth, A. and Hewitt, J.E. (2010) A family history of DUX4: phylogenetic analysis of DUXA, B, C and Duxbl reveals the ancestral DUX gene. BMC Evol. Biol., 10, 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Parker, H.G., VonHoldt, B.M., Quignon, P., Margulies, E.H., Shao, S., Mosher, D.S., Spady, T.C., Elkahloun, A., Cargill, M., Jones, P.G. et al. (2009) An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science, 325, 995–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang, C., Wallerman, O., Arendt, M.L., Sundstrom, E., Karlsson, A., Nordin, J., Makelainen, S., Pielberg, G.R., Hanson, J., Ohlsson, A. et al. (2021) A novel canine reference genome resolves genomic architecture and uncovers transcript complexity. Commun Biol, 4, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wallace, L.M., Garwick, S.E., Mei, W., Belayew, A., Coppee, F., Ladner, K.J., Guttridge, D., Yang, J. and Harper, S.Q. (2011) DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann. Neurol., 69, 540–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choi, S.H., Gearhart, M.D., Cui, Z., Bosnakovski, D., Kim, M., Schennum, N. and Kyba, M. (2016) DUX4 recruits p300/CBP through its C-terminus and induces global H3K27 acetylation changes. Nucleic Acids Res., 44, 5161–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Snider, L., Geng, L.N., Lemmers, R.J., Kyba, M., Ware, C.B., Nelson, A.M., Tawil, R., Filippova, G.N., van der Maarel, S.M., Tapscott, S.J. et al. (2010) Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet., 6, e1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Das, S. and Chadwick, B.P. (2016) Influence of repressive histone and DNA methylation upon D4Z4 transcription in non-myogenic cells. PLoS One, 11, e0160022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jagannathan, S., Shadle, S.C., Resnick, R., Snider, L., Tawil, R.N., van der Maarel, S.M., Bradley, R.K. and Tapscott, S.J. (2016) Model systems of DUX4 expression recapitulate the transcriptional profile of FSHD cells. Hum. Mol. Genet., 25, 4419–4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kowaljow, V., Marcowycz, A., Ansseau, E., Conde, C.B., Sauvage, S., Matteotti, C., Arias, C., Corona, E.D., Nunez, N.G., Leo, O. et al. (2007) The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord., 17, 611–623. [DOI] [PubMed] [Google Scholar]
- 11. Whiddon, J.L., Langford, A.T., Wong, C.J., Zhong, J.W. and Tapscott, S.J. (2017) Conservation and innovation in the DUX4-family gene network. Nat. Genet., 49, 935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang, Y., Liu, T., Meyer, C.A., Eeckhoute, J., Johnson, D.S., Bernstein, B.E., Nusbaum, C., Myers, R.M., Brown, M., Li, W. et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol., 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lunter, G. (2007) Dog as an outgroup to human and mouse. PLoS Comput. Biol., 3, e744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jones, T.I., Chew, G.L., Barraza-Flores, P., Schreier, S., Ramirez, M., Wuebbles, R.D., Burkin, D.J., Bradley, R.K. and Jones, P.L. (2020) Transgenic mice expressing tunable levels of DUX4 develop characteristic facioscapulohumeral muscular dystrophy-like pathophysiology ranging in severity. Skelet. Muscle, 10, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krom, Y.D., Thijssen, P.E., Young, J.M., den Hamer, B., Balog, J., Yao, Z., Maves, L., Snider, L., Knopp, P., Zammit, P.S. et al. (2013) Intrinsic epigenetic regulation of the D4Z4 macrosatellite repeat in a transgenic mouse model for FSHD. PLoS Genet., 9, e1003415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bosnakovski, D., Shams, A.S., Yuan, C., da Silva, M.T., Ener, E.T., Baumann, C.W., Lindsay, A.J., Verma, M., Asakura, A., Lowe, D.A. et al. (2020) Transcriptional and cytopathological hallmarks of FSHD in chronic DUX4-expressing mice. J. Clin. Invest., 130, 2465–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bosnakovski, D., Chan, S.S.K., Recht, O.O., Hartweck, L.M., Gustafson, C.J., Athman, L.L., Lowe, D.A. and Kyba, M. (2017) Muscle pathology from stochastic low level DUX4 expression in an FSHD mouse model. Nat. Commun., 8, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Giesige, C.R., Wallace, L.M., Heller, K.N., Eidahl, J.O., Saad, N.Y., Fowler, A.M., Pyne, N.K., Al-Kharsan, M., Rashnonejad, A., Chermahini, G.A. et al. (2018) AAV-mediated follistatin gene therapy improves functional outcomes in the TIC-DUX4 mouse model of FSHD. JCI Insight, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hendrickson, P.G., Dorais, J.A., Grow, E.J., Whiddon, J.L., Lim, J.W., Wike, C.L., Weaver, B.D., Pflueger, C., Emery, B.R., Wilcox, A.L. et al. (2017) Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet., 49, 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Akiyama, T., Xin, L., Oda, M., Sharov, A.A., Amano, M., Piao, Y., Cadet, J.S., Dudekula, D.B., Qian, Y., Wang, W. et al. (2015) Transient bursts of Zscan4 expression are accompanied by the rapid derepression of heterochromatin in mouse embryonic stem cells. DNA Res., 22, 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Young, J.M., Whiddon, J.L., Yao, Z., Kasinathan, B., Snider, L., Geng, L.N., Balog, J., Tawil, R., van der Maarel, S.M. and Tapscott, S.J. (2013) DUX4 binding to retroelements creates promoters that are active in FSHD muscle and testis. PLoS Genet., 9, e1003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Geng, L.N., Yao, Z., Snider, L., Fong, A.P., Cech, J.N., Young, J.M., van der Maarel, S.M., Ruzzo, W.L., Gentleman, R.C., Tawil, R. et al. (2012) DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell, 22, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. De Iaco, A., Planet, E., Coluccio, A., Verp, S., Duc, J. and Trono, D. (2017) DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet., 49, 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guo, M., Zhang, Y., Zhou, J., Bi, Y., Xu, J., Xu, C., Kou, X., Zhao, Y., Li, Y., Tu, Z. et al. (2019) Precise temporal regulation of dux is important for embryo development. Cell Res., 29, 956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen, Z. and Zhang, Y. (2019) Loss of DUX causes minor defects in zygotic genome activation and is compatible with mouse development. Nat. Genet., 51, 947–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bosnakovski, D., Gearhart, M.D., Ho Choi, S. and Kyba, M. (2021) Dux facilitates post-implantation development, but is not essential for zygotic genome activationdagger. Biol. Reprod., 104, 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dandapat, A., Hartweck, L.M., Bosnakovski, D. and Kyba, M. (2013) Expression of the human FSHD-linked DUX4 gene induces neurogenesis during differentiation of murine embryonic stem cells. Stem Cells Dev., 22, 2440–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee, J.K., Bosnakovski, D., Toso, E.A., Dinh, T., Banerjee, S., Bohl, T.E., Shi, K., Orellana, K., Kyba, M. and Aihara, H. (2018) Crystal structure of the double homeodomain of DUX4 in complex with DNA. Cell Rep., 25, 2955, e2953–e2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Love, M.I., Huber, W. and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang, C., Wallerman, O., Arendt M.L., Sundström, E., Karlsson, Å., Nordin, J., Mäkeläinen, S., Pielberg, G.R., Hanson, J., Ohlsson, Å. et al. (2021) A novel canine reference genome resolves genomic architecture and uncovers transcript complexity. Commun Biol., 4, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reimers, M. and Carey, V.J. (2006) Bioconductor: an open source framework for bioinformatics and computational biology. Methods Enzymol., 411, 119–134. [DOI] [PubMed] [Google Scholar]
- 32. Edgar, R., Domrachev, M. and Lash, A.E. (2002) Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res., 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ChIP-seq and RNA-seq of our canine models have been deposited to Genome Expression Omnibus (32) and are accessible through GEO accession number GSE188928. The RNA-seq and ChIP-seq data of the human DUX4 myoblast model were collected from published GEO series GSE85461 and GSE33838, and that of the mouse Dux myoblast model were from GSE87282. All shell scripts, R scripts and processed datasets in R/Bioconductor compatible format are available on our GitHub repository (https://github.com/FredHutch/canFam3.DuxFamily.git). We also host a GitHub page (https://FredHutch.github.io/canFam3.DuxFamily) that gives details of our analysis alone with readable, reproducible codes.