Abstract
The CPP-like plant‐specific transcription factor has a prominent role in plant development and growth through cell division and differential activities. However, little information is available about the CPP gene family in Triticum aestivum L. Herein, we identified 37 and 11 CPP genes in the wheat and rice genome databases, respectively. The phylogeny of the CPP protein-like family members was further divided into five subfamilies based on structural similarities and phenotypic functional diversities. The in silico expression analysis showed that CPP genes are highly expressed in some tissues, such as shoot apex, shoot, leaf, leaf sheath, and microspore. Furthermore, the qRT-PCR found higher expression for TaCPP gene family members in leaf, leaf blade, young spike, mature spike, and differential expression patterns under abiotic stresses, including heat, drought, salt, and hormonal treatment, such as indole acetic acid and 1-aminocyclopropane-1 carboxylic acid. We found that CPP gene family members are mostly located in the nucleus after infiltrating the CPP5-1B-GFP and TaCPP11-3B-GFP into tobacco leaves. The overexpression of the TaCPP5-1D gene revealed that the CPP gene positively regulates the germanium, shoot, and root activities in Arabidopsis. The TaCPP5-1D-overexpressed plants showed less anti-oxidative sensitivity under drought stress conditions. These results demonstrated that TaCPP5-1D protein has a crucial contribution by interacting with TaCPP11-3B protein in maintaining stress homeostasis under the natural and unfavorable environmental conditions for growth, development, and stress resistance activities. Therefore, this study could be used as pioneer knowledge to further investigate the function of CPP genes in plant growth and development.
Keywords: wheat (Triticum aestivum L.), rice (Oryza sativa), CPP, IAA, ACC
1. Introduction
Previous studies reported that transcription factors (TFs) play a vital role in regulating gene expression at the mRNA transcript level. The CPP-like gene family is also known as cysteine-rich polycomb-like proteins, classified as important regulatory proteins mainly found in all plants and animals apart from yeast [1]. CPP TFs are derived from plants that consist of 1 or 2 cysteine-rich conserved motifs recognized as CXC domains (CXCX4CX3YCXCX6CX3CXCX2C) [1,2]. The CPP proteins are classified into two different categories based on their location composed of different amino acids, which controlled diverse functions in plants [3]. Furthermore, the CXC domain and the sequences that linked the CXC domains are highly conserved across all living organisms [4,5]. Furthermore, it has been reported that highly conserved inter-domain and domain sequences may help the CPP gene family to control gene expression by sticky DNA along with CXC motifs [6,7].
Recent studies confirmed the CXC domains’ functional role within CPP TFs to promote the expression of the target genes linked with plant growth, development, and stress responses [6,7]. For instance, the CXC domain-containing CBBP protein (also known as b1-repeat binding protein) may bind to a certain region within the b1 tandem repeat sequence to regulate para-mutation in maize [8]. Furthermore, the CPP-like TF played a vital role in cell division and flower development by negatively regulating the MYB3R1 in Arabidopsis [7,9,10]. CPP TFs also perform a diverse function, particularly to regulate cell division and the development of several tissues [9,10]. In addition, the first TF, namely TSO1, was identified in Arabidopsis by using map-based cloning, which showed high expression level in the flower ovule and microspore development, suggesting the contribution of the CPP genes in the development of reproductive organs [7,9,10,11,12]. It was also noted that STO1 could promote cellular expansion by altering the communication of the neighboring cell development, cytokinesis, and karyokinesis [7,11,12]. The CPP transcription-like protein, named CPP1, regulates the Gmlbc3 protein in root development in soybean [13].
Recent studies have reported that several TFs promote plant resistance against several adverse environmental conditions. For instance, it has been found that BBX and YABBY TFs perform a vital role in plant stress biology [14,15,16]. The biotic stresses greatly affect the function of CPP TFs in plant growth and developmental processes. For example, under heat stress, the transcript level of the CPP genes was promoted in the root [17]. The transcript level of the ZmCPP TF was increased under heat, cold, and drought stress in maize [18]. A recent study found that the CPP-like TFs displayed diverse expression patterns in different tissues in the tea plant, suggesting the involvement of CPP TFs in a wide range of physiological and developmental processes [2]. Therefore, we assumed that CPP regulatory proteins might potentially contribute to the plant developmental and growth processes in beneficial coordination with several abiotic stresses.
Undoubtedly, in agriculture, wheat and rice are regarded as utmost important cereal crops. Therefore, it is important to identify and study the TFs that could enhance the resistance against various unfavorable conditions. So far, the CPP-like TFs have been reported in maize, Arabidopsis, soybean, rice, cucumber, and tea plants [1,2,17,18]. However, no comprehensive study has investigated the CPP-like TF family members and their evolutionary relationship and expression profile under different hormonal and abiotic stresses in wheat. The present study evaluated the role of biological evolution in the expansion of the CPP-like TFs in rice, wheat, and Arabidopsis and the potential role of CPP proteins in the plant growth and development under various adverse environmental factors overexpressing the TaCPP5-1D gene in Arabidopsis. The results will provide a solid foundation for exploring the molecular mechanism of the CPP-like TFs in plants.
2. Materials and methods
2.1. Identification and physicochemical properties of CPP genes
To find CPP TFs in rice and wheat, we first retrieved the CPP TFs from the Arabidopsis genome (https://www.arabidopsis.org/). The identified Arabidopsis CPP proteins were then blasted in the rice genome database (http://rice.plantbiology.msu.edu/) and the wheat genome database (www.wheatgenome.org) using BlastP. To avoid the possible loss of the CPP gene due to the CPP incomplete domain, a local BLASTP was performed with a cutoff of 1 × 10−5. After that, all protein sequences were examined to confirm the presence of the CPP-conserved domain through SMART (http://smart.embl-heidelberg.de/) [19], Inter Pro Scan program (https://www.ebi.ac.uk/interpro/), Conservative Domain Database (CDD) (http://www.ncbi.nlm.nih.gov/cdd/), and Scan Prosite (https://prosite.expasy.org/scanprosite/). In the present study, the physicochemical characteristics, including isoelectric point (pI), molecular weight (MW), GRAVY, and the up–down strand of the identified CPP TFs, were carried out using ExPASy online server (https://www.expasy.org) [20].
2.2. Chromosomal location and multiple sequence alignment of CPP genes
Rice (http://rice.plantbiology.msu.edu/) and wheat genome databases (www.wheatgenome.org) were used to find information about the chromosome location of identified CPP TFs in wheat and rice. A diagram of the physical location of the identified CPP genes was constructed using MapDraw in excel 2007. The naming of the identified CPP genes was performed based on their location on the chromosome. Multiple sequence alignments were performed to identify the conserved regions in the CPP proteins using DNAMAN software (Lynnon Corporation, Canada, https://www.lynnon.com). The Logos of the conserved region were constructed through an online Web logo server (https://weblogo.berkeley.edu/logo.cgi) [21].
2.3. Phylogenetic relationships, gene structures, motif, and duplication analysis
To investigate the evolutionary relationship of the CPP proteins among the plant, the multiple sequence alignments of the rice, wheat, and Arabidopsis-identified CPP proteins were performed using the ClustalW v2.0 online package (http://www.ebi.ac.uk). Then, the aligned sequence of the identified CPP proteins of rice, wheat, and Arabidopsis was used for the phylogenetic analysis using iTOL (https://itol.embl.de/) [22] and MEGA software [23] via maximum likelihood methods with 1,000 bootstrap values. The genomic and coding sequence of the identified CPP genes was collected from their respective genome databases to evaluate the intron/exon distribution of the CPP genes through Gene Structure Display online Server (http://gsds1.cbi.pku.edu.cn/) [24]. The conserved motifs of CPP protein sequences were analyzed using the online Multiple Expectation Maximization for Motif Elucidation platform (version 4.12.0) (https://meme-suite.org/meme/doc/meme.html) [25], with default parameter. The maximum number of motifs was 10, and the optimum motif width was set to >6 and <200. Several studies have found that tandem and segmental gene duplication contributes significantly to the growth of genes in a living organism throughout evolution [15,16,26]. Therefore, the present study also explored the relationship between duplication and CPP TFs’ expansion in rice, and wheat using the Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/). The circus software (http://circos.ca/) [27] was used to draw the lines among the syntenic blocks that were obtained from the Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/).
2.4. Developmental expression profile of CPP genes
The GENEVESTIGATOR V3 database (https://genevestigator.com/gv/) [28] was used to assess the transcript level of the CPP gene family at different developmental stages and tissues. The gene IDs of the identified CPP genes were pasted in the search area of these available microarray data search engines, and the data were collected in the form of a heat map. The different colors were used to distinguish the upregulated and downregulated genes in the heat map. The wheat plants were grown in the field of Hazara University for the transcript evaluation of CPP TFs in the diverse organs using semi-quantitative real-time polymerase chain reaction (qRT-PCR). The tissues, including leaf, root, shoot, stem, node, internode, leaf sheath, and leaf flag, were harvested at several stages, such as the booting stage, seedling stage, and heading stage. The tissues were stored at −80 for further analysis.
2.5. Expression profile analysis of CPP genes under different abiotic and hormonal stresses
The biological materials, such as seeds, were obtained from the Department of Genetics, Hazara University. The uniform and healthy seeds were grown in the field or the growth chamber in pods under optimum environmental conditions. The optimum condition for wheat growth was set as follows: daytime 16 h and night 8 h, the temperature should be between 21 and 25°C, light intensity should be 800 µmol m−2 s−1, and the relative humidity should be 60–70%. The hormonal application and abiotic stresses were performed according to our previously published paper in environmental research [29]. Briefly, for the heat stress treatment, the seedlings at the four-leaf stage were subjected to 40°C temperature with 60% humidity and a 16 h photoperiod in the growth chamber under fluorescent light for 24 h. For the dehydration of 20% polyethylene glycol (PEG 6000), the solution was purified through an ion-exchange column to remove any impurities and filtered using Miracloth (22–25 µm; Thomas Scientific, Swedesboro, NJ, USA). Salt (200 mM NaCl) was prepared from stock solution by dissolving in water. Then, seedlings were submerged in 20% PEG 6000 and 200 mM NaCl solution for drought and salt treatments, respectively. The final hormonal concentration of 1-aminocyclopropane-1 carboxylic acid (ACC) deaminase (50 µM) and indole acetic acid (IAA) (50 µM) was prepared from stock solutions; after the addition of wetting agent Tween-20 at 0.05% (v/v), the individual hormone was sprayed on 2-week-old wheat plants. The samples were collected at different time points for expression analysis.
2.6. Quantitative PCR analysis
The total RNA was extracted from all the samples by using the cetyltrimethylammonium bromide method [30]. The samples were run on 2% agarose gels to examine the intensity of ribosomal RNA bands, degraded products, and gDNA contamination. The residual genomic DNA was removed by treating the RNA samples with RNase-free DNase. The cDNA was synthesized through the PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio, Shiga, Japan) following the manufacturer’s instructions. All the primers were designed from wheat CPP sequences for real-time PCR using primer 6.0 (Table S2). Each primer pair was examined through standard real-time polymerase chain reaction (RT-PCR) to confirm the size of the amplified product through 1% agarose gel electrophoresis. Real-time PCR was carried out in an iCycler iQ Real-Time PCR Detection System (Bio-Rad). Each reaction consisted of 5 μL of SYBR Premix ExTaq (Takara, Kyoto, Japan), 2 μL of cDNA samples, and 0.5 μL of each primer (10 μM) and 2 μL of ddH2O in a reaction system of 10 μL. The thermal cycle was as follows: 95°C for 3 min, followed by 40 cycles at 94°C for 15 s, 62°C for 20 s, and 72°C for 20 s. The melting curve analysis was performed directly after real-time PCR to verify the presence of gene-specific PCR products. This analysis was done at 94°C for 15 s, followed by a constant increase from 60 to 95°C at a 2% ramp rate. The wheat actin gene was used as an internal control and served as a standard gene for normalizing all mRNA expression levels. The relative amount of template present in each PCR amplification mixture was evaluated using the 2−ΔΔCt method.
2.7. Sub-cellular location and protein–protein interaction
The sub-cellular location was performed by cloning the CDS sequences of the TaCPP5-1D and TaCPP11-3B genes into the pCAMBIA-1302 vector. The vectors were then transformed into Agrobacterium tumefaciens using the electric shock method. The vectors were then infiltrated into tobacco leaves, and the result was checked using the confocal microscope. The full-length CDS region of the TaCPP5-1D and TaCPP11-3B was ligated into different N-terminal and C-terminal of the luciferase reporter gene LUC, respectively. The constructed vectors were then co-infiltrated into tobacco leaves for firefly luciferase complementation imaging (LCI) assay. The co-expressed leaves were analyzed for the LUC activity at 60 h after co-infiltration through a plant living molecular marker imaging system (Lumazone Pylon 2048B, Princeton, USA). The primers used in the present study are listed in Table S2.
2.8. Construction of TaCPP5-1D Arabidopsis transgenic plants
The CDS region of the TaCPP5-1D gene was ligated into pCAMBIA-1302, and then the constructed vector was transformed into A. tumefaciens. The agrobacterium-mediated floral dipping method was used to transform the constructed vector into Arabidopsis wild-type (WT) Columbia. The ½ Murashige and Skoog (MS) medium containing 30 mg L−1 hygromycin was used to screen the T0 transgenic Arabidopsis plant. The seedlings with true green leaves were considered transformants. A total of three independent homozygous T3 lines (OE-1, OE-2, and OE-3) were used for further study. The transcript level of the three independent homozygous T3 lines (L1, L2, and L3) is presented in Figure 8a. All the primers used in this study are listed in Table S3.
Figure 8.
(a) Transcript level of TaCPP5-1D in the three independent homozygous T3 lines. (b) Transcript level of TaCPP5-1D gene in overexpressed plants under drought stress. (c) The antioxidant activities and contents of the antioxidant enzymes in Arabidopsis. The WT sample represents wild plants, and OE samples represent the TaCPP5-1D-overexpressed plants. Three-week-old Arabidopsis plants were exposed to drought stresses for the evaluation of the antioxidant analysis.
2.9. Evaluation of germination, root, shoot, and antioxidant activities of overexpressed and WT plants
For germination, root, and shoot activities, the transgenic seeds were grown on the ½ MS medium containing 250 mM inositol. The germination rate was counted based on the appearance of an embryonic axis protrusion. For root and shoot activities, 2-week-old plants were used. The drought stress was performed by withholding the water for 2 weeks, and the samples were collected for antioxidant analysis. The superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), and ascorbate peroxidase (APX) activities and malonaldehyde (MDA) content were evaluated by the previously described method [31].
2.10. Data analysis
The SPSS software was used to perform the statistical analysis. The mean and standard error were used for the graphs. The GraphPad Prism was used to construct the graphs.
3. Results
3.1. Identification of CPP genes in Triticum aestivum L. and Oryza sativa
In the present study, 37 CPP protein sequences were retrieved from the T. aestivum L. genome database. The nomenclature of all the CPP genes was performed based on their chromosome location and was named TaCPP1 to TaCPP17 (Table 1). We also found a total of 11 CPP genes in the rice genome, which were named OsCPP1 to OsCPP11 (Table 1). The family-specific domains, such as the CXC domain of the CPP-like gene family, were confirmed through the conserved domain, Pfam, and SMART databases. Based on the genome size, we found that TaCPP15-5D (9585 bp) was the largest, whereas TaCPP14-4D (963 bp) was the smallest CCP gene (Table 1). Based on the CDS and amino acid, the TaCPP7-1D and TaCPP14-4D were the largest and smallest CPP members, respectively (Table 1). We found that most of the identified CPP gene family members were located on the reversed strand in the rice and wheat genome. The physicochemical characteristics including MW, pI, amino acid composition, gravy, aliphatic index (AI), and instability index (II) of CPP-like gene family members were studied through the EXPASY PROTOPARAM http://www.expasy.org/tools/protparam.html online tool (Table S1). The range of MW was 14.28 (TaCPP11-3A) to 98.82 kDa (TaCPP3-1A) for the identified CPP-like proteins in rice and wheat. Most of the CPP gene family members were basic because their pI was higher than 7 (Table S1). The S (Ser) is the most abundant amino acid of the CPP proteins, followed by C (Cys), P (Pro), and A (Ala). The GRAVY value of all the identified rice and wheat CPP proteins is negative, suggesting that all the CPP proteins are non-polar and hydrophobic (Table S1). Moreover, the AI ranged from 30.31 (OsCPP8) to 75.95 (TaCPP9-2A) for the identified CPP proteins (Table S1). Based on the II, we found that the II value of all the CPP proteins was higher than 40, signifying that all the CPP proteins are unstable apart from TaCPP3-1B, TaCPP7-1D, and TaCPP8-1B.
Table 1.
Identification and nomenclature of the identified CPP proteins in rice and wheat genome.
| Name | Gene ID | Gene location | AA | CDS | Genomic | Strand |
|---|---|---|---|---|---|---|
| TaCPP1-1A | TraesCS1A02G327100.1 | 1A:516780762-516783619 | 372 | 1512 | 4058 | R |
| TaCPP1-1B | TraesCS1B02G340500.1 | 1B:568536481-568539365 | 374 | 1522 | 4085 | R |
| TaCPP1-1D | TraesCS1D02G329200.1 | 1D:420546981-420549825 | 374 | 1460 | 4045 | R |
| TaCPP2-1B | TraesCS1B02G411800.1 | 1B:637913058-637915448 | 362 | 789 | 3591 | R |
| TaCPP3-1A | TraesCS1A02G427700.1 | 1A:581879170-581880757 | 202 | 609 | 2788 | F |
| TaCPP3-1B | TraesCS1B02G423800.1 | 1B:646682762-646687533 | 219 | 1293 | 5972 | F |
| TaCPP4-1D | TraesCS1D02G393200.1 | 1D:462965510-462969112 | 180 | 1218 | 4803 | R |
| TaCPP5-1B | TraesCS1B02G423900.1 | 1B:646775147-646776631 | 228 | 898 | 2685 | F |
| TaCPP5-1D | TraesCS1D02G400400.1 | 1D:466604136-466605574 | 233 | 904 | 2639 | F |
| TaCPP6-1A | TraesCS1A02G428000.2 | 1A:581957739-581959345 | 303 | 912 | 2807 | F |
| TaCPP6-1B | TraesCS1B02G462600.1 | 1B:674816626-674821589 | 316 | 1530 | 6164 | F |
| TaCPP7-1B | TraesCS1B02G462800.1 | 1B:674933401-674934839 | 235 | 918 | 2639 | F |
| TaCPP7-1D | TraesCS1D02G437400.1 | 1D:484793121-484797317 | 230 | 1414 | 5397 | F |
| TaCPP8-1B | TraesCS1B02G462900.1 | 1B:675227333-675228275 | 136 | 411 | 2144 | R |
| TaCPP9-2A | TraesCS2A02G122300.2 | 2A:71665769-71671190 | 588 | 2324 | 6621 | R |
| TaCPP9-2B | TraesCS2B02G144400.1 | 2B:110479459-110485095 | 589 | 2394 | 6640 | R |
| TaCPP9-2D | TraesCS2D02G125000.1 | 2D:73099327-73104463 | 588 | 2213 | 6336 | R |
| TaCPP10-2D | TraesCS2D02G563300.1 | 2D:634645178-634647593 | 264 | 795 | 3616 | F |
| TaCPP11-3A | TraesCS3A02G219300.1 | 3A:402135204-402136682 | 242 | 729 | 2680 | R |
| TaCPP11-3B | TraesCS3B02G249700.1 | 3B:397677642-397679106 | 261 | 786 | 2666 | R |
| TaCPP11-3D | TraesCS3D02G233200.1 | 3D:321869369-321873110 | 243 | 2034 | 4442 | F |
| TaCPP12-3A | TraesCS3A02G304800.1 | 3A:540959694-540968654 | 615 | 2518 | 963 | F |
| TaCPP12-3B | TraesCS3B02G331700.1 | 3B:536986657-536994307 | 615 | 2485 | 8852 | R |
| TaCPP12-3D | TraesCS3D02G297000.1 | 3D:410666651-410674769 | 615 | 2445 | 9320 | R |
| TaCPP13-4A | TraesCS4A02G284600.1 | 4A:590993901-590999432 | 793 | 2841 | 6733 | R |
| TaCPP13-4B | TraesCS4B02G028500.1 | 4B:21187118-21192530 | 789 | 2711 | 6611 | F |
| TaCPP13-4D | TraesCS4D02G026100.1 | 4D:11364255-11369752 | 787 | 2799 | 6694 | F |
| TaCPP14-4D | TraesCS4D02G184600.2 | 4D:322849931-322857603 | 890 | 3204 | 8873 | F |
| TaCPP15-5A | TraesCS5A02G053800.1 | 5A:48916968-48924858 | 765 | 2812 | 9089 | F |
| TaCPP15-5B | TraesCS5B02G062400.1 | 5B:70090784-70098944 | 765 | 2962 | 9360 | R |
| TaCPP15-5D | TraesCS5D02G065300.1 | 5D:60631878-60640264 | 765 | 3047 | 9585 | F |
| TaCPP16-5A | TraesCS5A02G054200.1 | 5A:50743216-50746791 | 357 | 1656 | 4774 | F |
| TaCPP16-5B | TraesCS5B02G062200.1 | 5B:69855996-69859508 | 360 | 1298 | 4712 | R |
| TaCPP16-5D | TraesCS5D02G065500.1 | 5D:60896691-60899936 | 359 | 1345 | 4444 | F |
| TaCPP17-7A | TraesCS7A02G341100.1 | 7A:501056605-501062654 | 486 | 2071 | 7250 | F |
| TaCPP17-7B | TraesCS7B02G242300.1 | 7B:450563457-450569975 | 485 | 2185 | 7719 | F |
| TaCPP17-7D | TraesCS7D02G338500.1 | 7D:431930000-431936466 | 486 | 2147 | 7668 | R |
| OsCPP1 | LOC_Os01g55580.1 | 1: 32016622-32022401 | 619 | 1860 | 6980 | R |
| OsCPP2 | LOC_Os02g17460.1 | 2: 10042478-10049191 | 497 | 1494 | 7914 | R |
| OsCPP3 | LOC_Os03g43730.1 | 3: 24454953-24461662 | 757 | 2274 | 7910 | R |
| OsCPP4 | LOC_Os04g09560.1 | 4: 5130826-5132553 | 147 | 444 | 2928 | R |
| OsCPP5 | LOC_Os05g43380.1 | 5: 25224822-25227403 | 374 | 1125 | 3725 | R |
| OsCPP6 | LOC_Os05g51040.1 | 5: 29292436-29293646 | 194 | 585 | 1557 | F |
| OsCPP7 | LOC_Os06g22670.1 | 6: 13182291-13189126 | 518 | 1557 | 2411 | R |
| OsCPP8 | LOC_Os07g07974.1 | 7: 4,020265-4,027484 | 782 | 2349 | 8036 | F |
| OsCPP9 | LOC_Os08g28214.1 | 8: 17214129-17221249 | 598 | 1796 | 8420 | R |
| OsCPP10 | LOC_Os12g41210.1 | 12: 25553542-25557389 | 407 | 1224 | 5048 | R |
| OsCPP11 | LOC_Os12g41230.1 | 12: 25561750-25569052 | 670 | 2283 | 8503 | R |
AA, amino acid; CDS, coding sequence.
3.2. Evolutionary relationship, motifs analysis, and gene structure of CPP gene family members
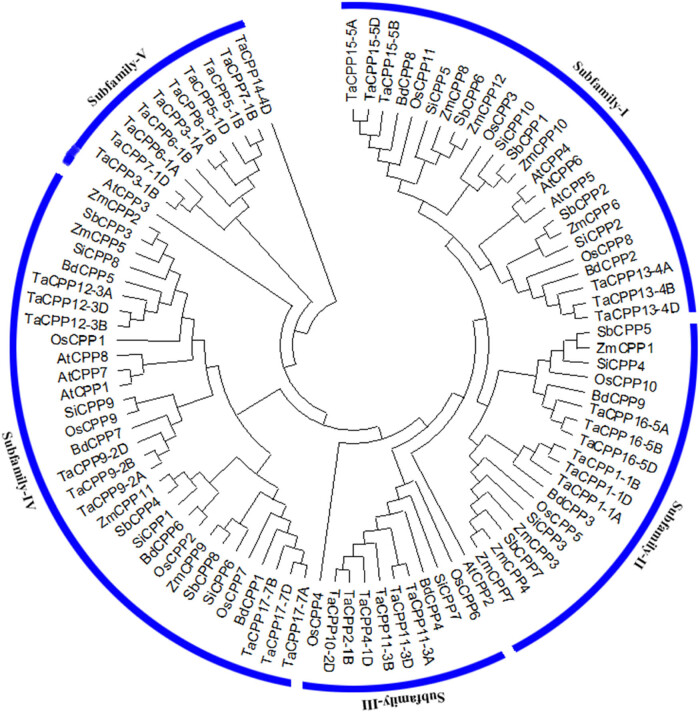
The identified CPP proteins from Arabidopsis, wheat, rice, maize, millet, sorghum, and stiff brome were used to construct a maximum likelihood phylogenetic tree (Figure 1), which provides more insight and knowledge about the evolutionary history CPP gene family in plants. The phylogenetic relationship divided the identified CPP proteins into five subfamilies (Figure 1). The highest number of CPP proteins was found in subfamily-IV, followed by subfamily-I and -II. The lowest number of CPP proteins was detected in subfamily-V and -III (Figure 1). We found that the distributions of CPP proteins into different subfamilies mainly occurred due to their physicochemical properties, intron–exon distribution, and motif arrangement. For instance, we found that the larger size CPP gene family members were clustered in subfamily-I, whereas the medium size CPP gene family members were found in subfamily-IV (Figure S1). Similarly, the smallest size CPP gene family members were noted in subfamily-II, -III, and -V. Based on the intron–exon distribution, we found that the CPP gene family members with a high number of exons (ten or more exons) were divided into subfamily-I. Most of the CPP genes possessing eight exons were found in subfamily-III. Similarly, six and five exons containing CPP genes were observed in subfamily-II and -IV, respectively (Figure S1). In subfamily-V, two to five exons were observed for most of the CPP gene family members apart from TaCPP14-4D. Furthermore, the CPP gene family members shared a quite similar motif distribution in each subfamily. For example, the CPP proteins possessing six to seven motifs were found in subfamily-I and -III, the CPP proteins having five motifs were clustered into subfamily-II, four motifs containing CPP proteins were detected in subfamily-IV, and two to three motifs possessing CPP proteins were divided into subfamily-V (Figure S1). Based on these results, we hypothesized that CPP gene family members shared conserved gene structure and common ancestor during biological evolution in plants. The intron–exon distribution was diverse among the identified CPP proteins (Figure S1). The highest numbers of exons (seventeen exons) were noted for TaCPP14-4D genes, whereas the lowest numbers of exons were observed for the TaCPP6-1B gene. Similarly, we found a total of ten motifs in the identified CPP proteins, which were named motif-1–10 (Figure S1). Motif-2 was the frequently repeated motif, followed by motif-7 and motif-3 in the identified CCP proteins in the studied plants. Moreover, motif-5 was the least repeated motif, followed by motif-6 and motif-8, respectively.
Figure 1.
The phylogenetic tree of the CPP proteins between Arabidopsis, rice, wheat, maize, stiff brome, millet, and sorghum. The phylogenetic analysis divided the CPP gene family members into five subfamilies based on their physiochemical properties, intron–exon, and motif arrangement.
3.3. Chromosomal location and duplication of CPP gene family members
The CPP genes are unevenly distributed on the chromosomes in the genome of the wheat and rice based on the chromosome location and annotation information. The identified 11 CPP gene family members were located on 9 out of the 12 chromosomes in the rice genome. Among them, chromosomes 1, 2, 3, 4, 6, 7, and 8 each had one OsCPP gene, whereas chromosomes 5 and 12 shared two CPP genes in the rice genome (Figure S2a). In wheat, the identified CPP genes were distributed on all the chromosomes apart from chromosomes 6A, B, and D (Figure S2b). The highest number of CPP genes is seven CPP gene family members detected on chromosome-1B in the wheat genome. Only one CPP gene was distributed on each chromosome-2A, -4A, -7A, -2B, -4B, -7B, and -7D in the wheat genome. Similarly, we found two CPP genes on each chromosome-3A, -5A, -3B, -5B, -2D, -3D, -4D, and -5D (Figure S2b). The second-highest number of CPP gene (four) family members was observed on chromosome-1D, followed by chromosome-1A (shared three CPP gene family members) in the wheat genome.
It has been reported that the plant genomes were widely composed of several segmental and duplicated genes. Therefore, we conducted a duplications analysis of the identified CPP gene family members in the studied plants to evaluate the role of duplication in expanding the CPP gene family in plants (Figure 2). We found several segmental duplicated pairs in the wheat genome for the CPP gene family members. A total of three segmental duplicated CPP genes were detected in the rice genome. Similarly, three segmental duplicated CPP pairs were observed in the Arabidopsis genome. All these results suggest that segmental duplication processes may play an important role in expanding the CPP gene family during biological evolution in plants.
Figure 2.
Genomic distribution of TaCPP, OsCPP, and AtCPP genes and gene homology analysis in rice, wheat, and Arabidopsis. The duplicated CPP gene pairs were found on the different chromosomes in rice, wheat, and Arabidopsis.
3.4. Expression profiling of CPP genes in various tissues
Thirty-nine different tissues were selected to explore TaCPP gene family expression using the GENEVESTIGATOR V3 database (https://genevestigator.com/gv/) [28]. We found that all the homologs of TaCPP1, TaCPP12, TaCPP14, and TaCPP16 genes displayed high expression levels in the examined tissues (Figure S3). The TaCPP2, TaCPP3, TaCPP4, TaCPP5, TaCPP6, TaCPP7, and TaCPP8 genes were less expressed in the tissues apart from meiocyte, microspore, stamen, and anther. Extremely low expression was noted for TaCPP10-2D and TaCPP11-3A in the tissues except microspore (Figure S3). Most of the wheat CPP gene family members were detected with high transcript levels in meiocyte, microspore, stamen, and anther. The rice CPP gene family members were highly expressed in almost all the tissues (Figure S3). The expression of the rice CPP gene family was higher than wheat CPP gene family members in all the tissues. The (qRT-PCR) found that each CPP gene family member displayed a high transcript level of at least in more than two tissues. We found that most of the TaCPP genes were diversely expressed in all the tested tissues (Figure 3). In leaf, TaCPP gene family members had highly expressed apart from TaCPP3-1B, TaCPP4-1D, TaCPP9-2A, and TaCPP17-7B. Most of the TaCPP gene family members display high expression levels in seed except TaCPP3-1B and TaCPP17-7B. Similarly, most of the TaCPP gene family members exhibited diverse expression levels in root and shoot. The TaCPP1-1A, TaCPP3-1B, TaCPP5-1D, TaCPP9-2A, TaCPP10-2D, TaCPP11-3B, and TaCPP15-5B were noted with high transcript levels, whereas the remaining TaCPP genes were low expressed in stem (Figure 3). The transcript level of TaCPP2-1B, TaCPP3-1B, TaCPP9-2A, TaCPP13-4D, TaCPP16-5A, and TaCPP17-7B displayed low expression levels in node, internode, and mature spike. Furthermore, some TaCPP genes, such as TaCPP1-1B, TaCPP2-1B, TaCPP5-1D, TaCPP10-2D, and TaCPP15-5B, have highly expressed in leaf-related tissues. Based on the expression fold, we found that TaCPP11-3B was the most expressed, followed by TaCPP15-5B, TaCPP1-1B, and TaCPP5-1D, whereas TaCPP17-7B exhibited low expression (Figure 3).
Figure 3.
The expression analysis of TaCPP genes in different tissues, which were collected under normal conditions. Data of the quantitative RT-PCR analysis were presented in the form of the mean and standard deviation of three biological replicates of each biological sample. The y-axis is the relative expression level between two samples.
3.5. Expression analysis of TaCPP gene family under hormonal stresses
It has been reported that a few microbes and plants synthesize IAA, which plays a crucial role in shoot and root developments [32,33,34]. The ACC is recorded with function as a signal itself under stress conditions. Therefore, several mysterious questions are debatable about their function in extending the period. Here, we evaluated the transcript level of TaCPP gene family members under IAA and ACC stress to examine the role of TaCPP gene family members in growth and development (Figure 4). In response to exogenous ACC, we found that the transcript level of most of the TaCPP gene family members was promoted at different time points apart from the TaCPP17-7B gene. Most of the TaCPP gene family members were upregulated at the 24 h time point excluding the TaCPP11-3B gene. At 6 and 12 h time points, all the TaCPP gene family members showed high expression levels under ACC stress conditions (Figure 4). The transcript of the studied TaCPP gene family members was low at 1 and 3 h time points. Under exogenous IAA treatment, we found quite a mixed transcript level for the examined TaCPP gene family members (Figure 4). For instance, some TaCPP genes, such as TaCPP1-1A, TaCPP2-1B, TaCPP9-2A, TaCPP15-5B, and TaCPP16-5A, exhibited low transcript levels, while the remaining TaCPP gene family members showed high expression levels under IAA stress conditions. The transcript level of the TaCPP3-1B gene was high at all the time points and displayed high fold expression compared with other TaCPP gene family members in response to exogenous IAA treatment (Figure 4). The high expression for most of the TaCPP gene family members was noted at 24 h time points, followed by 1 and 6 h time points, respectively.
Figure 4.
The expression analysis of TaCPP genes under ACC and IAA treatments with extending period from 0 to 24 h. Data of the quantitative RT-PCR analysis were presented in the form of the mean and standard deviation of three biological replicates of each biological sample. The y-axis is the relative expression level between two samples.
3.6. Expression analysis of TaCPP gene family under abiotic stresses
It has been reported that gene expression analysis can provide important information regarding gene function; therefore, we used qRT-PCR to investigate the transcript level of the wheat CPP genes under diverse conditions, such as drought, cold, and salt, at different time points (Figure 5). Under treatment, more than a two-fold difference in the transcript level was considered to be true differences for a gene. Under salt treatment, we noted that most of the TaCPP gene family members displayed a low level of transcript level apart from TaCPP5-1D, TaCPP3-1B, TaCPP13-4D, TaCPP17-7B, and TaCPP10-2D genes. We found that most of the TaCPP gene family members were low expressed at all the time points, excluding the 1 h time point (Figure 5). We detected that the transcript level of TaCPP1-1A, TaCPP2-1B, TaCPP3-1B, TaCPP4-1D, TaCPP5-1D, TaCPP9-2A, TaCPP10-2D, TaCPP11-3B, TaCPP15-5B, and TaCPP16-5A was low, whereas the rest of the two CPP members was high under salt conditions. Only one CPP gene (TaCPP11-3B) had a high expression profile under drought stress compared to the 0 h sample (control), whereas TaCPP 13-4D and TaCPP5-1D had a high expression profile at all time points a low expression profile. Under drought stress, the expression of TaCPP2-1B, TaCPP4-1D, TaCPP11-3B, TaCPP, and TaCPP17-7B was high at 3 and 6 h time points, respectively, while the expression of other CPP members was upregulated at two or three points (Figure 5). Similarly, under heat stress, the transcript level of TaCPP1-1A, TaCPP9-2A, TaCPP10-2D, TaCPP13-4D, and TaCPP17-7B was high at all time points. Furthermore, several CPP members (TaCPP3-1B, TaCPP4-1D, TaCPP5-1D, TaCPP15-5B, and TaCPP16-5A) were downregulated, whereas the other four CPP genes were up- and downregulated at different points. Consequently, we identified that most wheat CPP member transcripts were greatly influenced by heat and salt stresses. Furthermore, we observed that CPP member transcripts were up- and downregulated at different time points during drought conditions. All these results showed the role of the CPP gene family in plant growth and development and their response to multiple stresses.
Figure 5.
The expression analysis of TaCPP genes under heat, drought, and NaCl treatments with extending the time point from 0 to 24 h. Data of the quantitative RT-PCR analysis were presented in the form of the mean and standard deviation of three biological replicates of each biological sample. The y-axis is the relative expression level between two samples.
3.7. Subcellular location and protein–protein interaction of CPP proteins
The full-length CDS of TaCPP5-1B and TaCPP11-3B genes were cloned into the pCAMBIA-1302 vector. The vectors were then infiltrated into tobacco leaves using A. tumefaciens. After 48 h, we checked the GFP signal under the confocal microscope. Several studies have reported that most of the TFs are located in the nucleus of the cell. Here, we found that TaCPP5-1B and TaCPP11-3B proteins produced strong GFP signals in the nucleus (Figure 6a), suggesting that CPP TFs are also located in the nucleus of the cell and perform a diverse role in plant growth and development, particularly under hormonal and abiotic stresses. We used the protein–protein interaction assay to investigate the interaction of CPP proteins with other CPP proteins (Figure 6b). The firefly LCI assay found that TaCPP5-1D promoted the luciferase activity when TaCPP5-1D was co-expressed with TaCPP11-3B proteins in the tobacco leaves (Figure 6b). The interaction of TaCPP5-1D with TaCPP11-3B protein was confirmed by the bimolecular fluorescence complementation (BiFC) assay. We observed that the co-infiltration of TaCPP5-1D and TaCPP11-3B proteins produced strong fluorescence signals in tobacco leaves (Figure 6c). The LCI and BiFC results were further evaluated by the yeast two-hybrid system. The TaCPP5-1D and TaCPP11-3B proteins were co-expressed in yeast cells (Figure 6d). The yeast cells containing TaCPP5-1D and TaCPP11-3B proteins displayed significant growth on S/D media compared with control, indicating that TaCPP5-1D regulates the plant growth and development by interacting with TaCPP11-3B protein.
Figure 6.
(a) The subcellular location of the CPP gene family members. The CDS region was ligated into the pCAMBIA-1302 vector and the constructed vector was then overexpressed in tobacco leaves. The GFP-fusion protein signals were detected using the confocal microscope. (b) The interaction of TaCPP5-1D and TaCPP11-3B proteins using firefly LCI assay. (c) The interaction of TaCPP5-1D and TaCPP11-3B proteins using the BiFC assay. (d) Yeast two-hybrid system confirmation of TaCPP5-1D and TaCPP11-3B proteins’ interaction.
3.8. Overexpression of TaCPP5-1D genes positively regulates plant growth and development
To investigate the possible function of the CPP genes in plant growth and development, we overexpressed the TaCPP5-1D genes in Arabidopsis. We evaluate the function of the TaCPP-1D gene under osmotic stresses (250 mM inositol). In response to inositol, the TaCPP5-1D-overexpressed plants displayed a higher germination rate compared with WT plants (Figure 7a and b). The 1-week-old TaCPP5-1D-overexpressed plants showed a larger shoot size than WT plants (Figure 7c). However, no significant changes were observed in the number of leaves at this stage between 1-week-old TaCPP5-1D-overexpressed and WT plants (Figure 7d). Then, we evaluated the root and shoot activities of the 2-week-old TaCPP5-1D-overexpressed and WT plants under inositol stress conditions (Figure 7e). The TaCPP5-1D-overexpressed plants were noted with higher root activity than WT plants. The primary root length and number of TaCPP5-1D plant were higher than WT plants under 250 mM inositol condition (Figure 7f and g). Moreover, the number of leaves was more in TaCPP5-1D-overexpressed plants compared with WT plants at this stage (Figure 7h). Overall, we found that germination, root, and shoot activities were enhanced in TaCPP5-1D-overexpressed plants in response to an inositol stress condition, signifying that CPP proteins may positively regulate the plant growth and development under stress conditions.
Figure 7.
Phenotypic evaluation of Ta-CPP5-1D-overexpressed plants under 250 mM inositol. (a and b) Germination rate (%) of 1-week-old Ta-CPP5-1D-overexpressed and WT plants. (c) Shoot height of Ta-CPP5-1D-overexpressed and WT plants. (d) Average number of leaves per plant. (e) Root and shoot activities of 2-week-old Ta-CPP5-1D-overexpressed and WT plants. (f) Average primary root length. (g) Average primary root number. (h) Average number of leaves per plant.
3.9. Antioxidant activities and contents of antioxidant enzyme in Ta-CPP5-1D-overexpressed plants
Furthermore, we evaluated the expression level of the Ta-CPP5-1D gene in the overexpressed and WT plants. The transcript level of the TaCPP5-1D gene was upregulated in overexpressed plants compared with the WT plants in response to drought stress (Figure 8b). We further investigated the antioxidant activities and contents of the various antioxidant enzymes under drought stress conditions. The POD and SOD activities were high in WT plants as compared with TaCPP5-1D overexpressed plants (Figure 8c). In TaCPP5-1D-overexpressed plants, CAT and APX showed lower activities compared with WT plants in response to drought stress. Similarly, the MDA content was also higher in WT plants than in TaCPP5-1D-overexpressed plants apart from the OE1 line (Figure 8c). Overall, we found that WT plants displayed high oxidant activities and contents, suggesting the less sensitivity of the TaCPP5-1D-overexpressed plants in response to drought stress. Based on these results, we predicted that CPP genes might play important role in plant growth and development under various adverse environmental stress conditions by regulating the antioxidant enzymes and various other physiological pathways.
4. Discussion
Plants are continuously exposed to different kinds of environmental stimuli throughout their life span. Therefore, plants respond to environmental stimuli by utilizing the activities of TFs, families of genes coding for specific TF proteins. Hence, it is of utmost importance to have comprehensive knowledge about the functions of these TF families that regulate multiple responses against abiotic stresses in main staple crops such as wheat (T. aestivum L.), rice (O. sativa), and maize [35]. CPP (cysteine-rich polycomb-like protein) proteins, whose typical character is the presence of CXC domains, are a member of a small TF family, widely present in plants and animals [4]. The CPP gene family has a crucial role in controlling cell division and the development of reproductive tissues [7,11]. Therefore, it is crucial to comprehensively assess the CPP protein’s role under unfavorable environmental stresses and key regulator hormones. In the present study, we performed a genome-wide identification, characterization, and expression analysis of the CPP gene family members under various hormonal and abiotic stresses.
4.1. CPP proteins in various plant genomes and their evolutionary relationship
In the model plants Arabidopsis and rice, the investigation of the CPP gene family members has been performed. Previous studies reported a total of 8, 11, 20, 17, 5, and 8 CPP gene family members in maize, Arabidopsis, rice, soybean, cucumber, and tea plants, respectively [1,2,17,18,36]. Previous studies had found lineage-specific expansion to have a crucial role in the differentiation and growth of the proteomes of multicellular eukaryotes [37,38]. And 80% of these results are demonstrated in the model plant Arabidopsis thaliana [38]. Here, we found that the small CPP gene family comprised 37 members in the wheat genome (Table 1), which were scattered and distributed in the genome (Figure S1b). Our results demonstrated that most of the CPP gene family members in plants might share a common ancestor during biological evolution. The phylogenetic analysis found that each subfamily possessed quite similar intron–exon, motif distribution, and physiological properties. For instance, the larger size and more introns–exons with motif distribution CPP proteins were found in subfamily-I (Figure 1 and Figure S1), whereas the medium size and normal intron–exon with the same motif distribution CPP proteins were detected in subfamily-III, and the smaller size CPP proteins with less number of introns–exons were noted in subfamily-II, -IV, and -V (Figure 1 and Figure S1), suggesting that CPP gene family member may share common features and ancestor during evolution. Recently, it has been reported that some gene families may originate from the common ancestral genes and split into different monocot and dicot species [39,40]. Previous studies reported that gene duplication is an important source of raw materials for genesis, and gene duplication can be divided into three principal evolutionary patterns, such as tandem duplication, segmental duplication, and transposition events likewise, replicative and retroposition [41]. Previous successful work had witnessed three rounds of whole-genome duplication in Arabidopsis [42,43]. Here, we found a total of four segmentally duplicated pairs of the CPP genes in Arabidopsis and three duplicated pairs in rice (Figure 2). Furthermore, we observed several segmentally duplicated pairs for CPP gene family members in the wheat genome (Figure 2), further signifying the importance of duplication in the expansion of gene families in plants. We further evaluated the gene structure of the CPP gene family members using the intron–exon distribution and motif arrangement in rice, wheat, and Arabidopsis (Figure S1). There was highly inconsistency in the number of exons and introns and the motif arrangement in the CPP gene family members in the studied plants (Figure S1b), indicating that the variation in the gene structure may play a vital role in performing the diverse role of the CPP gene family members in the growth and development.
4.2. CPP proteins could be important in plant stress biology
As we know that the gene expression profile can give clues about their functional features within developing plants. Therefore, we investigated the expression pattern of the CPP gene family members in rice and wheat. It has been reported that the CPP gene family members have been examined with a distinct feature in several plant species [3,5,17]. The microarray results demonstrated that higher expression of CPP gene family members was recoded within vegetative tissues; shoot apex, shoot, and root in wheat (Figure S3). These results further verify the previous finding on the CPP gene family role during cell division, growth, and development [5,17]. A previous study reported that the CPP gene family displayed distinct expression profiles in different parts of the plant [3]. For instance, it was found that SOL2/TCX2 and TSO1/AtCPP5 were less expressed in pollen and carpel than in other plants tissues [4]. The present study also found a similar expression pattern for most of the genes. For instance, the transcript of TaCPP1-1A and TaCPP13-4D genes was highly regulated in leaf but less expressed in mature spike (Figure 3). A quite similar expression level was also noted for TaCPP16-5A and TaCPP13-4D, which were highly expressed in seeds but a low transcript was recorded in leaf flag and young spike. It has been reported that IAA and ACC are considered the main regulators of plant growth and development through cell division, elongation, and tissue differentiation [44,45]. However, various studies also reported the independent role of ACC as a growth regulator under stress conditions [45]. In the present study, the qRT-PCR found that most of the CPP gene family members were highly expressed in response to ACC, particularly at 24 h time points (Figure 4), signifying the important role of the CPP gene family in plant growth and development in response to the high dose of ACC. In contrast, IAA significantly reduces the expression profile of TaCPP1-1A, TaCPP2-1B, TaCPP4-1D, TaCPP9-2A, TaCPP10-2D, TaCPP11-3B, TaCPP13-4D, TaCPP15-5B, TaCPP16-5A, and TaCPP17-7B by extending the period until 24 h (Figure 4). It has been reported that IAA could reduce the growth activities in plants [44]. Salt, heat, and drought stresses comprise osmotic and ionic homeostasis, growth regulation, and detoxification. TFs, such as the CPP-like family, have a crucial role in growth and development through cell division [3]. It was found that TFs had a specific influence on gene expression by altering the activity of a protein by either suppressing or promoting its function [46]. A recent study reported that the CPP gene family members displayed diverse expression levels in response to various abiotic stimuli in maize [18]. For instance, it has been noticed that the ZmCPP7 gene displayed a high expression level under salt, heat, drought, and cold stresses suggesting the role of CPP genes in plant growth and development under stress conditions. In maize, most of the CPP gene family members were upregulated in response to heat stresses, further signifying the involvement of CPP genes in stresses [18]. Recently, it was noticed that the cucumber CPP genes were highly expressed in response to various abiotic and hormonal stresses [1]. It was observed that CsCPP1, CsCPP3, and CsCPP4 were upregulated in response to cold stresses [1]. In tea plants, it was observed that the promoter of the CPP genes possessed the stress-responsive cis-elements, which regulate the expression of CPP genes under stress conditions [2]. Moreover, except for GmCPP03 and GmCPP07, the remaining 18 GmCPP genes were all induced by heat shock under drought stress conditions, indicating that these genes are involved in the responses of soybean root systems to high-temperature stress and play important roles in regulating heat shock responses [17]. We found that most of the CPP gene family members were low expressed under salt stress apart from TaCPP3-1B, TaCPP15-1D, TaCPP13-4D, and TaCPP17-7B, whereas most of the CPP gene family members displayed high expression levels in response to drought and heat stresses (Figure 5), suggesting the important role of the CPP gene family in plant growth and development under unfavorable condition. Based on the previous and our current studies, we postulated that the CPP gene family may contribute to plant growth and development under various hormonal and abiotic stresses. Based on the previous and our current studies, we postulated that the CPP gene family may contribute to plant growth and development under various hormonal and abiotic stresses. Our results confirmed that CPP proteins bind with other CPP proteins to perform various functions in plant growth and development, particularly under adverse environmental stress conditions (Figure 6b–d). Furthermore, we found that the overexpression of TaCPP5-1D gene passivity regulates the germanium, shoot, and root activities in Arabidopsis (Figure 7). We further found that antioxidant activities and contents were lower in TaCPP5-1D-overexpressed plants compared with WT plants (Figure 8), signifying the less sensitivity of CPP genes and the importance of CPP genes in plant growth and development under various abiotic stresses.
5. Conclusion
In this study, we found a total of 37 and 11 CPP genes reported in wheat and rice, which were divided into five subfamilies based on the domain and structural distribution. Furthermore, the microarray and qRT-PCR analysis found the distinct expression pattern for most of the CPP gene family members in different tissues, suggesting that CPP gene family members may play a diverse role in plant growth and development. The qRT-PCR analysis of TaCPP genes under ACC, IAA, heat, drought, and NaCl treatments displayed varying transcript levels at different time points, indicating the vital role of the CPP gene family in plant growth and development under various abiotic and hormonal stresses. The overexpression of TaCPP5-1D detected that the CPP proteins might promote various plant developmental processes by targeting other CPP proteins under abiotic stress conditions. Thus, the present investigation provided important information for further functional studies on CPP genes in stress biology. Therefore, the current work could be used as primary knowledge to elucidate the regulation and pathway analysis of TaCPP TFs in plants.
Supplementary Material
Acknowledgments
We thank Izhar Muhammad (State Key Laboratory of Crop Stress Biology in Arid Areas, College of Life Sciences, Northwest A&F University) and Usman Aziz (College of Agronomy, Northwest A&F University, Yangling 712100, China) for their critical revision of the manuscript.
Footnotes
Funding information: Authors state no funding involved.
Author contributions: Conceptualization, U.U., Z.B. and A.S. data curation, U.U. and I.M. data analysis, U.U., A.D, and H. A. visualization, U.U., Z.B., A.S., and H.A. writing original draft preparation, U.U., A.Z.U., and H.A. writing review and editing, furthermore, all authors have read and agreed to the published version of the manuscript.
Conflict of interest: Authors state no conflict of interest.
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Contributor Information
Aziz Ud-Din, Email: geneticsaz@gmail.com.
Hamid Ali, Email: biotechdr.ali@gmail.com.
References
- [1].Zhou Y, Hu L, Ye S, Jiang L, Liu S. Genome-wide identification and characterization of cysteine-rich polycomb-like protein (CPP) family genes in cucumber (Cucumis sativus) and their roles in stress responses. Biologia. 2018;73(4):425–35.
- [2].Nan H, Lin Y, Wang X, Gao L. Comprehensive genomic analysis and expression profiling of cysteine-rich polycomb-like transcription factor gene family in tea tree. Horticul Plant J. 2021;7:469–78.
- [3].Lu T, Dou Y, Zhang C. Fuzzy clustering of CPP family in plants with evolution and interaction analyses. BMC Bioinforma. 2013;14(13):1–8. [DOI] [PMC free article] [PubMed]
- [4].Andersen SU, Algreen-Petersen RG, Hoedl M, Jurkiewicz A, Cvitanich C, Braunschweig U, et al. The conserved cysteine-rich domain of a tesmin/TSO1-like protein binds zinc in vitro and TSO1 is required for both male and female fertility in Arabidopsis thaliana. J Exp Botany. 2007;58(13):3657–70. [DOI] [PubMed]
- [5].Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290(5499):2105–10. [DOI] [PubMed]
- [6].Schmit F, Cremer S, Gaubatz S. LIN54 is an essential core subunit of the DREAM/LINC complex that binds to the cdc2 promoter in a sequence‐specific manner. The. FEBS J. 2009;276(19):5703–16. [DOI] [PubMed]
- [7].Hauser BA, He JQ, Park SO, Gasser CS. TSO1 is a novel protein that modulates cytokinesis and cell expansion in Arabidopsis. Development. 2000;127(10):2219–26. [DOI] [PubMed]
- [8].Brzeska K, Brzeski J, Smith J, Chandler VL. Transgenic expression of CBBP, a CXC domain protein, establishes paramutation in maize. Proc Natl Acad Sci. 2010;107(12):5516–21. [DOI] [PMC free article] [PubMed]
- [9].Sijacic P, Wang W, Liu Z. Recessive antimorphic alleles overcome functionally redundant loci to reveal TSO1 function in Arabidopsis flowers and meristems. PLoS Genet. 2011;7(11):e1002352. [DOI] [PMC free article] [PubMed]
- [10].Wang W, Sijacic P, Xu P, Lian H, Liu Z. Arabidopsis TSO1 and MYB3R1 form a regulatory module to coordinate cell proliferation with differentiation in shoot and root. Proc Natl Acad Sci. 2018;115(13):E3045–54. [DOI] [PMC free article] [PubMed]
- [11].Hauser BA, Villanueva JM, Gasser CS. Arabidopsis TSO1 regulates directional processes in cells during floral organogenesis. Genetics. 1998;150(1):411–23. [DOI] [PMC free article] [PubMed]
- [12].Song J-Y, Leung T, Ehler LK, Wang C, Liu Z. Regulation of meristem organization and cell division by TSO1, an Arabidopsis gene with cysteine-rich repeats. Development. 2000;127(10):2207–17. [DOI] [PubMed]
- [13].Cvitanich C, Pallisgaard N, Nielsen KA, Hansen AC, Larsen K, Pihakaski-Maunsbach K, et al. CPP1, a DNA-binding protein involved in the expression of a soybean leghemoglobin c3 gene. Proc Natl Acad Sci. 2000;97(14):8163–8. [DOI] [PMC free article] [PubMed]
- [14].Buttar ZA, Yang Y, Sharif R, Nan Wu S, Xie Y, Wang C. Genome wide identification, characterization, and expression analysis of YABBY-gene family in wheat (Triticum aestivum L.). Agronomy. 2020;10(8):1189.
- [15].Shalmani A, Jing X-Q, Shi Y, Muhammad I, Zhou M-R, Wei X-Y, et al. Characterization of B-BOX gene family and their expression profiles under hormonal, abiotic and metal stresses in Poaceae plants. BMC Genomics. 2019;20(1):1–22. [DOI] [PMC free article] [PubMed]
- [16].Shalmani A, Fan S, Jia P, Li G, Muhammad I, Li Y, et al. Genome Identification of B-BOX gene family members in seven Rosaceae Species and their expression analysis in response to flower induction in Malus domestica. Molecules. 2018;23(7):1763. [DOI] [PMC free article] [PubMed]
- [17].Zhang L, Zhao HK, Wang YM, Yuan CP, Zhang YY, Li HY, et al. Genome-wide identification and expression analysis of the CPP-like gene family in soybean. Genet Mol Res. 2015;14(1):1260–8. [DOI] [PubMed]
- [18].Song X, Zhang Y, Wu F, Zhang L. Genome-wide analysis of the maize (Zea may L.) CPP-like gene family and expression profiling under abiotic stress. Genet Mol Res. 2016;15:(3):gmr.15038023. [DOI] [PubMed]
- [19].Schultz J, Milpetz F, Bork P, Ponting CPSMART. a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci. 1998;95(11):5857–64. [DOI] [PMC free article] [PubMed]
- [20].Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, De Castro E, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40(W1):W597–603. [DOI] [PMC free article] [PubMed]
- [21].Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–90. [DOI] [PMC free article] [PubMed]
- [22].Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–6. [DOI] [PMC free article] [PubMed]
- [23].Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evolution. 2013;30(12):2725–9. [DOI] [PMC free article] [PubMed]
- [24].Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–7. [DOI] [PMC free article] [PubMed]
- [25].Bailey TL, Johnson J, Grant CE, Noble WS. The MEME suite. Nucleic Acids Res. 2015;43(W1):W39–49. [DOI] [PMC free article] [PubMed]
- [26].Shalmani A, Muhammad I, Sharif R, Zhao C, Ullah U, Zhang D, et al. Zinc finger-homeodomain genes: evolution, functional differentiation, and expression profiling under flowering-related treatments and abiotic stresses in plants. Evolut Bioinforma. 2019;15:1176934319867930. [DOI] [PMC free article] [PubMed]
- [27].Naquin D, d’Aubenton-Carafa Y, Thermes C, Silvain M. CIRCUS: a package for Circos display of structural genome variations from paired-end and mate-pair sequencing data. BMC Bioinforma. 2014;15(1):1–6. [DOI] [PMC free article] [PubMed]
- [28].Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, et al. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma. 2008;2008:420747. [DOI] [PMC free article] [PubMed]
- [29].Shalmani A, Ullah U, Muhammad I, Zhang D, Sharif R, Jia P, et al. The TAZ domain-containing proteins play important role in the heavy metals stress biology in plants. Environ Res. 2021;197:111030. [DOI] [PubMed]
- [30].Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Report. 1993;11(2):113–6.
- [31].Shalmani A, Huang Y-B, Chen Y-B, Muhammad I, Li B-B, Ullah U, et al. The highly interactive BTB domain targeting other functional domains to diversify the function of BTB proteins in rice growth and development. Int J Biol Macromolecules. 2021;192:1311–24. [DOI] [PubMed]
- [32].Basse CW, Lottspeich F, Steglich W, Kahmann R. Two potential indole‐3‐acetaldehyde dehydrogenases in the phytopathogenic fungus Ustilago maydis. Eur J Biochem. 1996;242(3):648–56. [DOI] [PubMed]
- [33].Prusty R, Grisafi P, Fink GR. The plant hormone indoleacetic acid induces invasive growth in Saccharomyces cerevisiae. Proc Natl Acad Sci. 2004;101(12):4153–7. [DOI] [PMC free article] [PubMed]
- [34].Yamada T, Palm CJ, Brooks B, Kosuge T. Nucleotide sequences of the Pseudomonas savastanoi indoleacetic acid genes show homology with Agrobacterium tumefaciens T-DNA. Proc Natl Acad Sci. 1985;82(19):6522–6. [DOI] [PMC free article] [PubMed]
- [35].Kimotho RN, Baillo EH, Zhang Z. Transcription factors involved in abiotic stress responses in Maize (Zea mays L.) and their roles in enhanced productivity in the post genomics era. PeerJ. 2019;7:e7211. [DOI] [PMC free article] [PubMed]
- [36].Yang Z, Gu S, Wang X, Li W, Tang Z, Xu C. Molecular evolution of the CPP-like gene family in plants: insights from comparative genomics of Arabidopsis and rice. J Mol evolution. 2008;67(3):266–77. [DOI] [PubMed]
- [37].Jordan IK, Makarova KS, Spouge JL, Wolf YI, Koonin EV. Lineage-specific gene expansions in bacterial and archaeal genomes. Genome Res. 2001;11(4):555–65. [DOI] [PMC free article] [PubMed]
- [38].Lespinet O, Wolf YI, Koonin EV, Aravind L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002;12(7):1048–59. [DOI] [PMC free article] [PubMed]
- [39].Yang X, Tuskan GA, Cheng Z-M. Divergence of the Dof gene families in poplar, Arabidopsis, and rice suggests multiple modes of gene evolution after duplication. Plant Physiol. 2006;142(3):820–30. [DOI] [PMC free article] [PubMed]
- [40].Zhang S, Chen C, Li L, Meng L, Singh J, Jiang N, et al. Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol. 2005;139(3):1107–24. [DOI] [PMC free article] [PubMed]
- [41].Kong H, Landherr LL, Frohlich MW, Leebens‐Mack J, Ma H, DePamphilis CW. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 2007;50(5):873–85. [DOI] [PubMed]
- [42].Raes J, Vandepoele K, Simillion C, Saeys Y, Van de Peer Y. Investigating ancient duplication events in the Arabidopsis genome. J Struct Funct Genomics. 2003;3(1):117–29. [PubMed]
- [43].Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van, de Peer Y. The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci. 2002;99(21):13627–32. [DOI] [PMC free article] [PubMed]
- [44].Fu S-F, Wei J-Y, Chen H-W, Liu Y-Y, Lu H-Y, Chou J-Y. Indole-3-acetic acid: A widespread physiological code in interactions of fungi with other organisms. Plant Signal & Behav. 2015;10(8):e1048052. [DOI] [PMC free article] [PubMed]
- [45].Polko JK, Kieber JJ. 1-aminocyclopropane 1-carboxylic acid and its emerging role as an ethylene-independent growth regulator. Front Plant Sci. 2019;10:1602. [DOI] [PMC free article] [PubMed]
- [46].Wang H, Zhao S, Gao Y, Yang J. Characterization of Dof transcription factors and their responses to osmotic stress in poplar (Populus trichocarpa). PLoS One. 2017;12(1):e0170210. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.