Abstract
Background
COVLIAS 1.0: an automated lung segmentation was designed for COVID-19 diagnosis. It has issues related to storage space and speed. This study shows that COVLIAS 2.0 uses pruned AI (PAI) networks for improving both storage and speed, wiliest high performance on lung segmentation and lesion localization.
Method
ology: The proposed study uses multicenter ∼9,000 CT slices from two different nations, namely, CroMed from Croatia (80 patients, experimental data), and NovMed from Italy (72 patients, validation data). We hypothesize that by using pruning and evolutionary optimization algorithms, the size of the AI models can be reduced significantly, ensuring optimal performance. Eight different pruning techniques (i) differential evolution (DE), (ii) genetic algorithm (GA), (iii) particle swarm optimization algorithm (PSO), and (iv) whale optimization algorithm (WO) in two deep learning frameworks (i) Fully connected network (FCN) and (ii) SegNet were designed. COVLIAS 2.0 was validated using “Unseen NovMed” and benchmarked against MedSeg. Statistical tests for stability and reliability were also conducted.
Results
Pruning algorithms (i) FCN-DE, (ii) FCN-GA, (iii) FCN–PSO, and (iv) FCN-WO showed improvement in storage by 92.4%, 95.3%, 98.7%, and 99.8% respectively when compared against solo FCN, and (v) SegNet-DE, (vi) SegNet-GA, (vii) SegNet-PSO, and (viii) SegNet-WO showed improvement by 97.1%, 97.9%, 98.8%, and 99.2% respectively when compared against solo SegNet. AUC > 0.94 (p < 0.0001) on CroMed and > 0.86 (p < 0.0001) on NovMed data set for all eight EA model. PAI <0.25 s per image. DenseNet-121-based Grad-CAM heatmaps showed validation on glass ground opacity lesions.
Conclusions
Eight PAI networks that were successfully validated are five times faster, storage efficient, and could be used in clinical settings.
Keywords: COVID-19, Lung CT, Hounsfield units, Glass ground opacities, AI, Deep learning, Pruning, Lung segmentation
Acronym table
- ACC
Accuracy
- AI
Artificial Intelligence
- ARDS
Acute Respiratory Distress Syndrome
- BA
Bland-Altman
- CC
Correlation Coefficient
- CE
Cross-Entropy
- COVID
Coronavirus disease
- COVLIAS
COVID Lung Image Analysis System
- CroMed
Croatian data set
- CT
Computed Tomography
- DE
Differential Evolution
- DL
Deep Learning
- DS
Dice Similarity
- EA
Evolutionary Algorithm
- FoM
Figure of merit
- GA
Genetic Algorithm
- GGO
Glass Ground Opacities
- GT
Ground Truth
- HDL
Hybrid Deep Learning
- HU
Hounsfield Units
- JI
Jaccard Index
- NovMed
Italian data set
- NIH
National Institute of Health
- PAI
Pruned AI
- POAI
Pruned and optimized AI
- PSO
Particle Swarm Optimization
- RT-PCR
Reverse Transcription-Polymerase Chain Reaction
- SDL
Solo Deep Learning
- SegNet
Segmentation Network
- VGG
Visual Geometric Group
- WHO
World Health Organization
- WO
Whale Optimization
- μ
Mean
- σ
Standard Deviation
Cross-entropy-loss
Classifier probability
i Input gold standard label 1
Optimized fitness function
Original hidden neuron
Compressed hidden neuron
Mean intersection over union accuracy
- μ1 and μ2
Weight factors for the fitness function
- TP
True Positive
- TN
True Negative
- FN
False Positive
- FP
False Negative
- m
Variable for counting EA and takes the values 1, 2, 3, and 4
- EA(m)
Evolutionary algorithm - DE, GA, PSO, and WO
Dice Similarity for the base models FCN & EA (m)
Dice Similarity for the base models FCN & EA (m)
Jaccard Index for the base models FCN & EA (m)
Jaccard Index for the base models SegNet & EA (m)
Correlation Coefficient for the base models FCN & EA (m)
Correlation Coefficient for the base models SegNet & EA (m)
Dice Similarity for the MedSeg
Jaccard Index for the MedSeg
Correlation Coefficient for the MedSeg
Difference between FCN and MedSeg
Difference between SegNet and MedSeg
Difference between FCN and MedSeg
Difference between SegNet and MedSeg
Difference between FCN and MedSeg
Difference between SegNet and MedSeg
Mean of all the four EA using FCN as base model
Mean of all the four EA using SegNet as base model
- [d]
Datatype - CroMed and NovMed
Mean area for EA models for the datatype “d”
Mean area for EA models for the datatype “d”
- N
Total number of images
Figure-of-Merit for EA using datatype “d”
Area of the lungs in the CT image ‘n’
GT area of the lungs in the CT image ‘n’
- Σ
Summation
1. Introduction
COVID-19 (the novel coronavirus) that was proclaimed by the World Health Organization (WHO) on March 11, 2020 [1] as a global pandemic has been a rapidly developing disease with limited hospital resources worldwide. As of February 15, 2022, it has infected more than 410 million people and killed almost 5.8 million [2] worldwide. COVID-19 molecular pathways [3,4] are worse in people with comorbidities such as coronary artery disease [3,5,6], diabetes [7], atherosclerosis [8], and fetal programming [9]. Further, it also damages the vasa vasorum of the aorta, causing micro thrombosis and atherosclerotic plaque vulnerability [10,11]. It has also caused architectural deformation because of interactions between alveolar and vascular modifications [12] and affects daily activities such as nutrition [13]. Pathology revealed that vaccine-induced immune thrombotic thrombocytopenia (VITT) is generated even after vaccine inoculation (ChAdOx1 nCoV-19) [14]. Adults who are born tiny, a condition known as intrauterine growth restriction (IUGR), are also more likely to be infected with COVID-19 [9]. Due to the lack of adequate immunization or therapy, early and accurate detection of COVID-19 is critical and in need of immediate attention. Due to the lower specificity of reverse transcription-polymerase chain reaction (RT-PCR) tests [[15], [16], [17]], research has become more inclined to use image-based analysis for detecting and diagnosing COVID-19.
With the advancement of artificial intelligence (AI) technology, machine learning (ML) and deep learning (DL) techniques have been widely adopted for pneumonia detection and classification [[18], [19], [20], [21]]. However, these technologies possess a threat when it comes to real-time analysis [22]. An advanced stage of DL is hybrid DL (HDL) [[23], [24], [25], [26], [27], [28], [29], [30]]. Even though DL/HDL models offer higher accuracy and performance, the cost associated with these models are high since it takes time to train and predict results. One way to solve the processing problem is to use GPU or supercomputers [29,[31], [32], [33]]]. However, they are expensive and difficult to maintain in the long run.
Originally, LeCun et al. [34], in their paper "Optimal Brain Damage," published in 1989, was the first to bring the concept of pruning to the area of deep learning. Pruning is a term that refers to trimming or cutting away excess from a model or search area to remove the redundant or insignificant sections [35]. This pruning concept was extended to improve storage [36,37] and speed up the model training by choosing the correct/appropriate hyperparameters [38,39]. They have mostly been applied to X-ray non-COVID and COVID-19 imaging [40]. As a result, the proposed study hypothesizes that such pruning methods can significantly enhance storage and processing time for computed tomography (CT) lung segmentation and lesion localization while requiring no additional hardware and maintenance.
The proposed study is the first to introduce eight DL-based innovative pruning models based on evolutionary algorithms (EA). This is our prime contribution and innovation in COVID-19 based CT lung segmentation. The eight-novel pruning AI (PAI) models are (i) differential evolution (DE), (ii) genetic algorithm (GA), (iii) particle swarm optimization algorithm (PSO), and (iv) whale optimization algorithm (WO) with FCN and SegNet as base DL infrastructure. As a result, the following eight pruning models emerges (i) FCN-DE, (ii) FCN-GA, (iii) FCN–PSO, (iv) FCN-WO when using solo FCN and (v) SegNet-DE, (vi) SegNet-GA, (vii) SegNet-PSO, and (viii) SegNet-WO when using solo SegNet. To scientifically validate the aforesaid innovation and contributions, we used a multicenter paradigm in an unseen framework. This requires training with our DL-based EA on the high-GGO cohort, which is taken from Croatia (CroMed), and predicting the segmented lung using the data from Novara, Italy (NovMed). Note that these “Unseen NovMed” data were acquired utilizing different CT machines and patients from various geographies and ethnicities, resulting in the Unseen AI paradigm. We consider this as our second innovation for this study. To assert the power of our innovation, we designed a unique pipeline to accurately localize the COVID-19 lesions in segmented lungs, which are taken from PAI and converted into a classification framework by fusing DenseNet-121 and GRAD-cam to generate powerful and visual color heatmaps. This takes us to our third and most distinctive innovation: evolutionary-based lung segmentation with lesion localization. To authenticate our contributions, we benchmarked our evolutionary-based COVLIAS 2.0 against MedSeg ([41]), a web-based lung segmentation tool, as the first attempt in low-storage and high-speed infrastructure. This is the fourth major contribution we have made. All our data analysis is comprehensive and adapted to 360-degree assessments. This consists of (i) Dice Similarity (DS), (ii) Jaccard Index (JI) [42], (iii) Bland-Altman plots (BA) [43,44], (iv) Correlation coefficient (CC) plot [45,46], (v) lung area error (AE), and (vi) receiver operator characteristic (ROC). We consider this as the fifth contribution for this study. We put COVLIAS 2.0 through clinical and statistical testing to prove the reliability and stability of our hypothesis, which is considered as the sixth and final contribution.
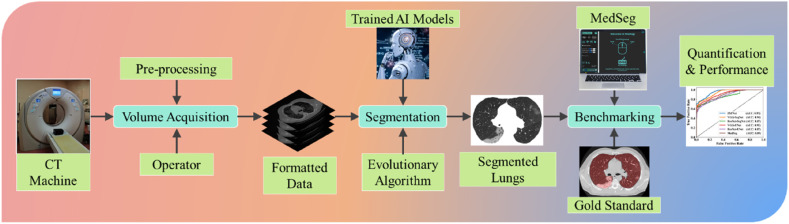
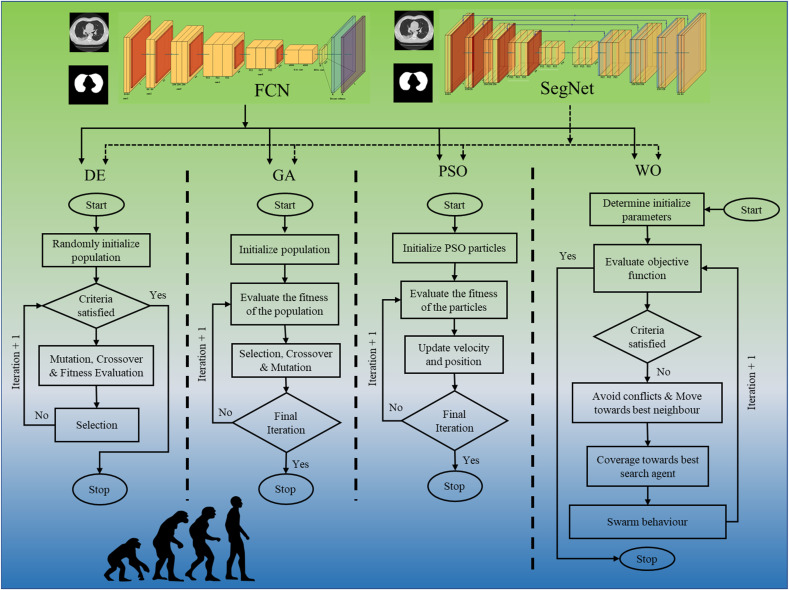
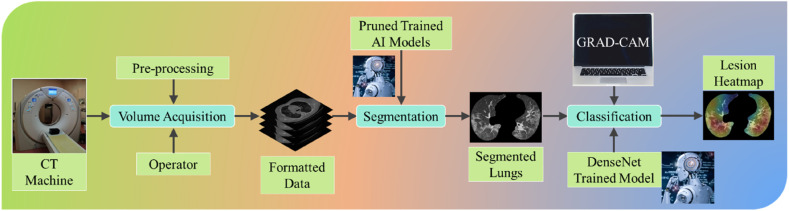
COVLIAS 2.0 system pipeline is presented in Fig. 1 , which demonstrates a universally applicable AI system for COVID-19 based lung segmentation that uses pruned and optimized AI (POAI) models for faster processing and less storage. It consists of volume acquisition, online lung segmentation, and benchmarking against MedSeg, as well as performance evaluation using “Unseen NovMed” data set.
Fig. 1.
COVLIAS 2.0 using pruned AI system for segmentation of CT lungs. The benchmarking is conducted using MedSeg. The performance is evaluated using manually delineated borders of lungs by radiologist. The scientific validation was conducted using Unseen NovMed data set.
Our suggested research is organized into seven sections. The background literature is presented in section 2, and the methodologies, demographics, image acquisition, and a brief explanation of the AI models employed in this work are presented in section 3. The findings of the models, as well as their performance evaluation, are presented in Section 4. The validation and statistical analysis of a different data set and MedSeg are shown in Section 5. Section 6 deals with discussion and benchmarking. Section 7 deals with the conclusion.
2. Literature survey
The art of segmentation in medical imaging has existed for several years [[47], [48], [49]], but it has recently been steered into an AI framework. Further, segmentation has caused problems in tissue characterization and classification in the early diagnosis of disease. Hence AI has begun to dominate that framework [19,20]. It started with ML, where it progressed to point-based models, such as problems related to diabetes [50,51], neonatal and infant mortality [52], and gene classification and analysis [53]. Moving from point-based to image-based machine learning for classification, several innovations have emerged which applied to lung segmentation [54], carotid plaque classification [[55], [56], [57], [58], [59]], thyroid [60], liver [61], stroke [62], coronary [63], ovarian [64], prostate [65], skin cancer [[66], [67], [68]], Wilson disease [32], and ophthalmology [69].
CT and X-ray play the most critical role in medical imaging for diagnosing COVID-19. CT has demonstrated high sensitivity and repeatability in the general diagnosis of COVID-19, body imaging [62,[70], [71], [72]], and the ability to detect various types of opacities such as ground-glass opacity (GGO), consolidation, crazy paving, and other opacities [[73], [74], [75]] that are mainly seen in COVID-19 patients [[76], [77], [78], [79], [80], [81]]. Jiang et al. presented COVID-19-based CT lung classification where the authors synthetically generated 1,186 CT images using CycleGAN [82]. The raw input lung cancer CT images were processed by the network taken from LUAN16 [83]. Each image patch had a size of 5122 pixels. A total of five AI models, namely, VGG16, ResNet-50, Inception ResNet_v2, Inception_v3, and DenseNet-169, were used to train on the synthetic data. The authors showed that Densenet-169 was the best performing model with an accuracy of 98.92%, while VGG16, ResNet-50, Inception ResNet_v2, and Inception_v3 had an accuracy of 94.19%, 94.83%, 96.55%, and 95.91%, respectively. There was no application of PAI in their research.
By analysing CT scan images, Kogilavani et al. [84] demonstrated the classification of CT images between COVID-19 and non-COVID-19. The author used six deep learning architectures, namely, VGG16 [85], DenseNet [86], MobileNet [87], Xception [88], NASNet [89], and EfficientNet [90]. All the models were trained on 3,873 CT images with an image resolution of 2242. While the authors used metrics like precision, recall, and F1-score for performance evaluation, the best-performing model was VGG16 with an accuracy of 97.68%. There was no application of PAI in their system design.
Paluru et al. [91] proposed a combination of UNet and ENet, the so-called AnamNet. It was designed to separate COVID-19-based lesions in segmented CT images of the lungs. The model was trained on a data set of 69 patients [92], in which the segmented lung was the input image to the AnamNet. The model was compared to ENet [93], UNet++ [94], SegNet, and LEDNet [95]. The pre-processing did not use any augmentation, and the dice similarity for lesion detection was 0.77. While the author did not use PAI, they deployed AnamNet on an edge device using an android application instead. The authors didn't compare lung area errors or create JI or BA plots.
Cai et al. [96] used the UNet model for lung and lesion segmentation, which resulted in a dice similarity of 0.77 using a ten-fold CV protocol with a database of 250 pictures taken from 99 patients. They also proposed a method for forecasting the length of an intensive care unit (ICU) stay based on the results of the lesion segmentation. The authors didn't compare lung area errors or create JI or BA plots.
Saood et al. [97] presented a COVID-19 based CT lung image segmentation system that used 100 COVID-19 lung CT images downscaled to 256 × 256. The scientists evaluated the findings of the two models, UNet and SegNet, and found they had similar DS scores of 0.73 and 0.74, respectively. The authors did not compare lung area errors or create JI or BA plots, and further, the authors did not apply PAI.
Our proposed study offers (i) eight pruning AI models demonstrating high speed and low storage, (ii) training of our pruning models using CroMed data set and performing unseen analysis using Unseen NovMed data set, (iii) classification framework to assert the COVID-19 lesions, (iv) MedSeg for benchmarking COVLIAS 2.0 to authenticate previous contributions, and finally, (v) conducting performance evaluation and statistical analysis to further provide the clinical evidence of our hypothesis.
3. Methodology
3.1. Patient demographics
3.1.1. CroMed Data set
The proposed study makes use of two distinct cohorts from separate nations. The first data set, also known as the experimental data set, consists of 80 CroMed COVID-19 positive individuals, of which 57 were male, and the rest were female. An RT-PCR test was conducted to confirm the positivity of COVID-19 in the selected cohort. The average value of ground-glass opacity (GGO), consolidation, and other opacities was ∼4. Out of the 80 CroMed patients who participated in this study, 83% had a cough, 60% had dyspnoea, 50% had hypertension, 8% were smokers, 12% had a sore throat, 15% were diabetic, and 3.7% had COPD. A total of 17 patients were admitted to the Intensive Care Unit (ICU), and 3 died due to a COVID-19 infection.
3.1.2. NovMed data set
The second data set, also known as the validation data set, consisted of 72 NovMed COVID-19 positive individuals, of which 47 were male, and the rest were female. An RT-PCR test was conducted to confirm the positivity of COVID-19 in the selected cohort. The average value of ground-glass opacity (GGO), consolidation, and other opacities was ∼2.4. Out of the 72 NovMed patients who participated in this study, 61% had a cough, 9% had a sore throat, 54% had dyspnoea, 42% had hypertension, 12% were diabetic, 11% had COPD, and 11% were smokers. Total 10 patients died due to COVID-19 infection.
3.2. Image acquisition and data preparation
3.2.1. CroMed and NovMed dataset
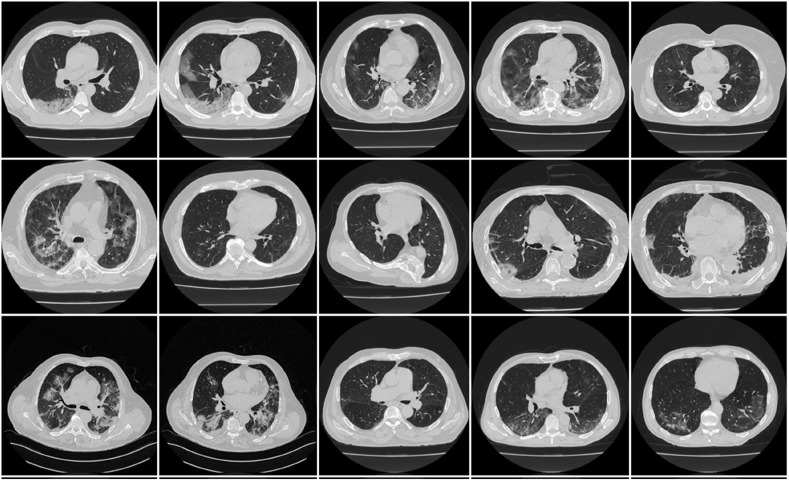
A CroMed data of 80 COVID-19 positive individuals (Fig. 2 ) was employed in this suggested investigation. The retrospective cohort study was conducted at the University Hospital for Infectious Diseases in Zagreb, Croatia, from March 1 to December 31, 2020. All patients over the age of 18, who agreed to take part in the study, had a positive RT-PCR test for the SARS-CoV-2 virus and underwent thoracic MDCT during their hospital stay. The patients met at least one of the following criteria: hypoxia (oxygen saturation 92%), tachypnea (respiratory rate 22 per minute), tachycardia (pulse rate >100), or hypotension (systolic blood pressure 100 mmHg). The UHID Ethics Committee approved the proposal. The scanner used was a 64-detector FCT Speedia HD (from Fujifilm Corporation, Tokyo, Japan, 2017), and the acquisition protocol for collecting the CT scan was a single full inspiratory breath-hold in the craniocaudal direction.
Fig. 2.
Sample CT scans taken from raw CroMed data set.
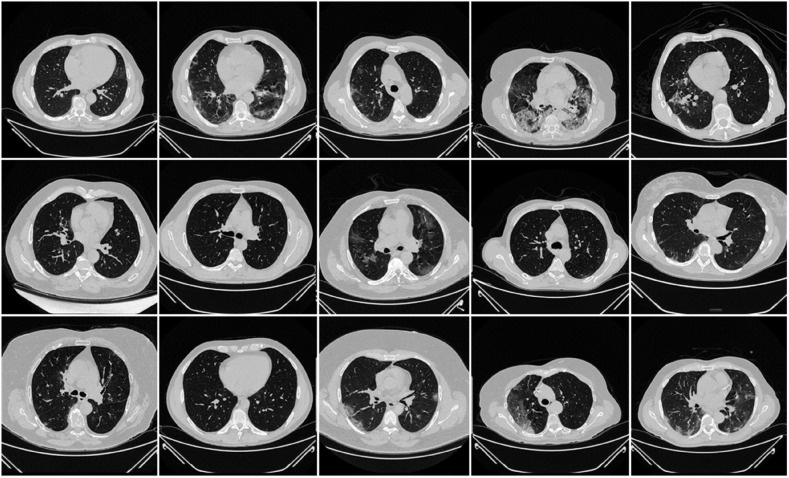
The NovMed dataset taken from Italy consisted of 72 patients (Fig. 3 ), were positioned supine, and chest CT scans were taken in a full inspiratory breath-hold using a 128 slice multidetector-row CT scanner (Philips Ingenuity Core, by Philips Healthcare). There was no intravenous or oral administration of the contrast agent. A soft tissue kernel with a lung kernel of a 768 × 768 matrix (lung window) was used to reproduce a one mm thick slice. The CT scans were performed using a 120 kV, 226 mAs/slice (using Philips' automatic tube current modulation – Z-DOM), a 1.08 spiral pitch factor, and a 0.5-s gantry rotation time 64 × 0.625 detector setup.
Fig. 3.
Sample CT scans taken from raw unseen NovMed data set.
3.3. Artificial intelligence models
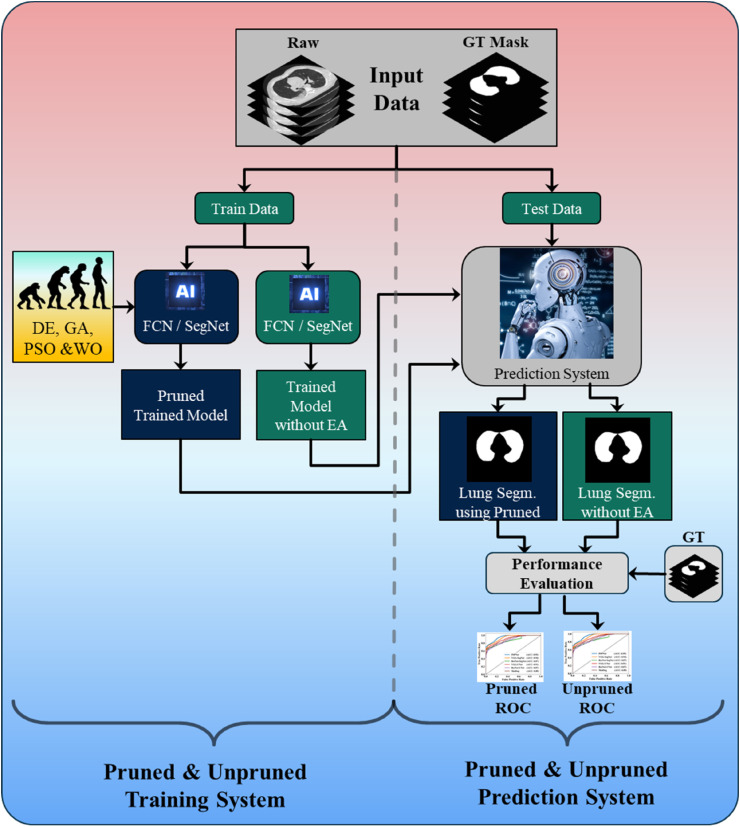
3.3.1. Overall system for pruned AI and unpruned AI
Fig. 4 shows the local pruned AI system for CT lung segmentation. It consists of two parts, the training and testing systems. The training system has two parts. Part A uses a conventional system of two types of base models such as FCN or SegNet for training the AI model without EA, the so-called unpruned AI (UnPAI). It uses raw grayscale pre-processed images and gold standard. In part B, the training system uses PAI system that utilizes evolutionary algorithms, such as DE, GA, PSO, and WO to generate the PAI models with EA. Next, we discuss the base DL models, which are well known in the DL industry.
Fig. 4.
Local COVLIAS 2.0 system for pruned and unpruned AI models.
3.3.2. Base model 1: Fully Convolutional Networks
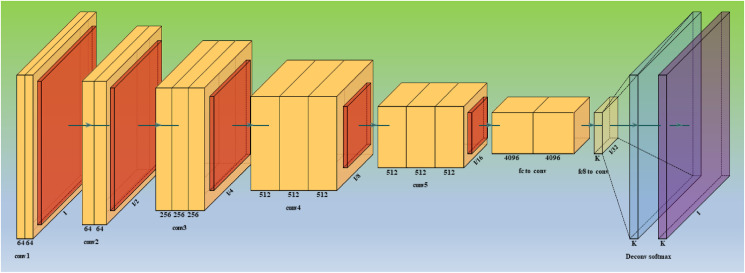
Fully Convolutional Networks, or FCNs (Fig. 5 ) are a type of architecture mainly utilized for semantic segmentation. They only use locally connected layers for convolution, pooling, and up sampling. Since they avoid the dense layers, it results in fewer parameters, which will make the networks faster to train. It also implies that an FCN can work with varying image sizes if all connections are local. The model learns the essential features in an image using a CNN feature extractor, referred to as the model's encoder. The image gets down sampled as it goes through convolutional layers. The output is then transmitted to the model's decoder portion, consisting of additional convolutional layers.
Fig. 5.
Architecture for base model one - FCN.
The decoder layers step-by-step up samples the image to its original dimensions, resulting in pixelwise labeling, also known as pixel mask or segmentation mask of the original image [98]. Some standard versions of FCNs are FCN-32, FCN-16, and FCN-8.
3.3.3. Base model 2: segmentation network
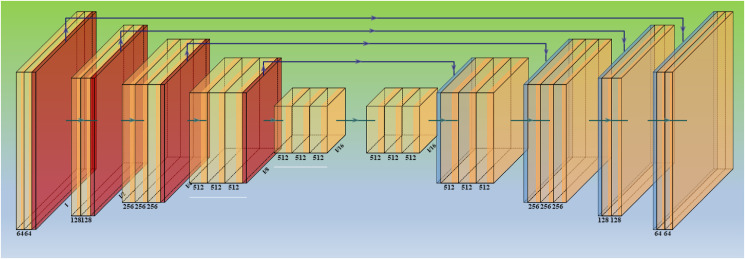
SegNet is a semantic segmentation model (Fig. 6 ) and has been implemented before [99]. However, we have fused evolutionary strategies with this base model, is a truly innovative component of this study, and it has been used for the first time in CT lung segmentation. SegNet is a trainable segmentation architecture that consists of (i) an encoder network (the left half of Fig. 6) topologically identical to the 13 convolutional layers, (ii) a corresponding decoder network, followed by (the right half of Fig. 6) (iii) a pixel-wise classification layer (the last block of Fig. 6). The encoder network's architecture down-samples the encoder output using a technique that requires storing the max-pooling (filter size of 2 × 2) indices. On the encoder side, the max-pooling layer is added at the end of each block, doubling the depth of the following layer (64–128 – 256–512). Similarly, up-sampling occurs in the second half of the design, where the layer depth is reduced by two (512–256 – 128–64). The max-pooling indices at the relevant encoder layer are recalled during the up-sampling operation. Finally, a K-class SoftMax classifier is used to predict the class for each pixel in the conclusion. This provides reasonable performance while also saving space. Typical SegNet examples for segmentation for other applications can be seen here [23,25,28].
Fig. 6.
Architecture for base model two – SegNet.
3.3.4. Design of eight PAI models
As discussed earlier, the overall structure of the interface between base FCN/SegNet models and the adaptation of four EA methods can be seen in Fig. 7 . Note that the input to the entire system is the raw grayscale lung CT scans along with the binary gold standard. These two base DL networks (FCN/SegNet) get optimized using the four EA methods. These four times two constitute the eight pruning techniques. The eight novel AI model pruning techniques are (i) DE, (ii) GA, (iii) PSO, and (iv) WO, with FCN and SegNet as base DL models. This yields the following eight pruning models: (i) FCN-DE, (ii) FCN-GA, (iii) FCN–PSO, (iv) FCN-WO when using solo FCN and (v) SegNet-DE, (vi) SegNet-GA, (vii) SegNet-PSO, and (viii) SegNet-WO when using solo SegNet.
Fig. 7.
Four pruning techniques (DE, GA, PSO, and WO) leading to eight system: FCN-DE, FCN-GA, FCN–PSO, FCN-WO, SegNet-DE, SegNet-GA, SegNet-PSO, and SegNet-WO.
Note that the EA methods are fundamentally compressing the base networks (FCN/SegNet). The steps are broken down as follows: (i) This compression process starts from 1st hidden convolutional layer and forms a vector equal to number of hidden neurons in that layer, comprising of random 0s and 1s. The trained FCN/SegNet model hidden neurons corresponding to 0 position of vector are temporarily removed and fitness criterion value is calculated. Fitness value is dual objective function with accuracy and compressed nodes ratio as the two objectives. Note that we need to maximize a weighted sum of both the objectives. After several EA iterations (say 20–30), we find the best vector with the best set of neurons that can be dropped to get the best compression ratio and accuracy after compression. (ii) The above process is repeated for all the hidden layers till we remove redundant filters from all the hidden convolutional layers. (iii) A new compressed model is created with each hidden layer having those number of neurons equal to sum of 1s in the EA vector for that hidden layer. (iv) Weights are then copied from original trained FCN/SegNet to the newly created compressed model. This is done such that each original model neuron's weight, which is at vector position that has a value 1, is copied to the corresponding compressed model neuron. (v) The compressed model is further trained for around 10–50 epochs to get the accuracy back which was reduced due to the nodes discarding process. After this, the accuracy and compressed model size is recorded. (vi) If the accuracy of the compressed model is within an acceptable value, i.e., not less than 1% of the original accuracy, then this compressed model is treated as the original uncompressed model and is further compressed by going back to step 1. By doing this, we get large compression after several compression iterations (say ∼10). The four evolutionary algorithms work similarly by making vectors of random 0s and 1s of an initial population (around 50 individuals). Only the intermediate steps are different to form new child vectors. Finally, the best vector is retained with the best fitness function value. This best vector helps to remove redundant neurons from the trained FCN/SegNet.
DE is a reproduction process that uses distance and orientation information via unit vectors, improving solutions through evolutionary processes [100,101]. The process uses mutation recombination to form new vectors [102,103]. These algorithms make minimal assumptions about the underlying optimization problem and explore enormous design spaces rapidly. In second EA method so-called GA, which originated from Darwin's Theory of Evolution [104], maintains a population of individuals who differ from one another. Those who are better adapted to their environment have a higher chance of surviving, reproducing, and passing on their traits to future generations (survival of the fittest). GA uses the process of selection, crossover, and mutation to produce optimized solutions [105,106]. The third EA method, so-called PSO was originally proposed by Kennedy and Eberhart [107] in 1995, originated from this concept that imitates a flock of birds or fishes which learn from each other to find the best position having food [108,109]. The position vector is formed in this scenario by having random 0 and 1. The vector with the highest fitness is assumed to be the position of food. It has a set of equations to find new position vectors in the next iterations. Finally, the last EA method so-called WO was inspired by the meta-heuristic optimization algorithm [110]. It originated from humpback whale behavior of encircling its prey in a spiral fashion [111,112]. The best vector with the highest fitness is assumed to be the position of prey, so the algorithm converges to that position. It also has a different set of equations to find new position vectors in the next iterations. These algorithms are shown in the diagrammatic form in Fig. 7(a–d) where (a), (b), (c), and (d) represent DA, GA, PSO, and WO.
3.4. Loss function for AI base models
The new models adopted the cross-entropy (CE)-loss functions during the model generation. If represented the CE-loss function, represents classifier's probability used in the AI model, i represents the input gold standard label 1, (1- i) represents the gold standard label 0. The loss function can be expressed mathematically as shown in Eq. (1).
| (1) |
Here represents the product of the two terms.
3.5. Fitness function for AI EA models
The main foundation of EA is fitness function which helps us to achieve the best possible solution set to maximize or minimize the objective fitness criterion. In each iteration the value of fitness function is compared with the previous best and solution space updated depending on its value. In all the evolutionary algorithms in this study, a dual objective fitness function has been used which depends on mean intersection over union (mIoU) accuracy () of test set and nodes compression ratio (Cr) [113,114]. If be the original hidden neuron and be the compressed hidden neuron, then the optimized fitness function can be mathematically given by the equation Eq. (2) below:
| (2) |
Where μ1 and μ2 are weight factors that helps to give more emphasis on one of the objectives. In our implementations, we have chosen these weights as 0.5. If we choose combinations such as 0.2 and 0.8, more compression can be achieved, but accuracy may be compromised and vice-versa.
3.6. Experimental protocol
A standardized cross-validation (CV) technique was used to train the AI models. Our team has created several CV-based protocols of various types using the AI framework. The K5 technique was used because the data was moderate. The data-consisted of 80% training data (4,000 CT scans) and 20% testing data (1,000 CT images). Five folds were designed in such a way that each fold has its own test set. Internal validation was included in the K5 protocol, with 10% of data being examined for validation.
The accuracy of an AI system is measured by comparing the expected output to ground truth pixel values. These readings were interpreted as binary (0 or 1) numbers because the output lung mask was only black or white. Finally, the total number of pixels in the image is divided by the sum of these binary integers. If TP, TN, FN, and FP stand for true positive, true negative, false negative, and false positive, respectively, then Eq. (3) can be used to calculate the AI system's accuracy.
| (3) |
4. Results and performance evaluation
The results have been compartmentalize using eight sets as proposed by COVLIAS 2.0, and eight pruning and optimization techniques. The first one uses (i) first base model FCN and four optimization techniques with FCN such as (ii) FCN-DE, (iii) FCN-GA, (iv) FCN–PSO, and (v) FCN-WO. The second one uses (ii) second base model SegNet and four optimization techniques with SegNet such as (ii) SegNet-DE, (iii) SegNet-GA, (iv) SegNet-PSO, and (v) SegNet-WO. This would enhance the speed and reduce the storage of the final AI models.
4.1. Results
-
(A)
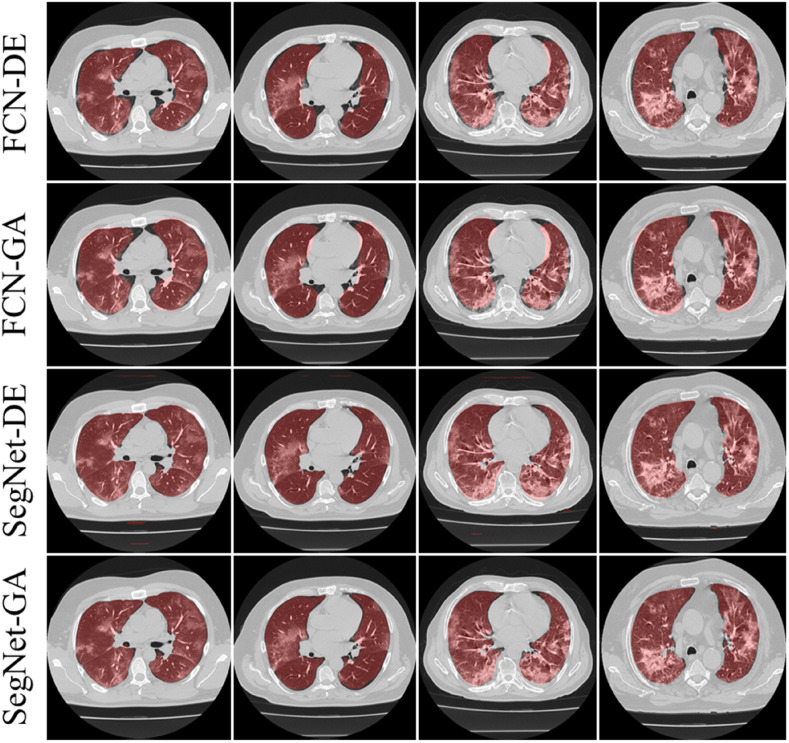
Visual Results using Pruning Models on CroMed Experimental Data set
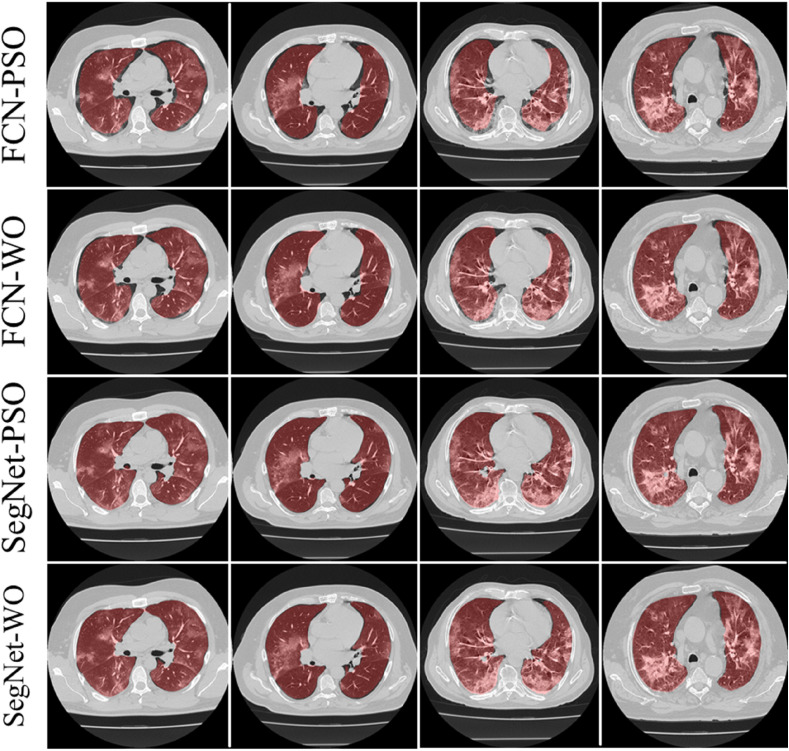
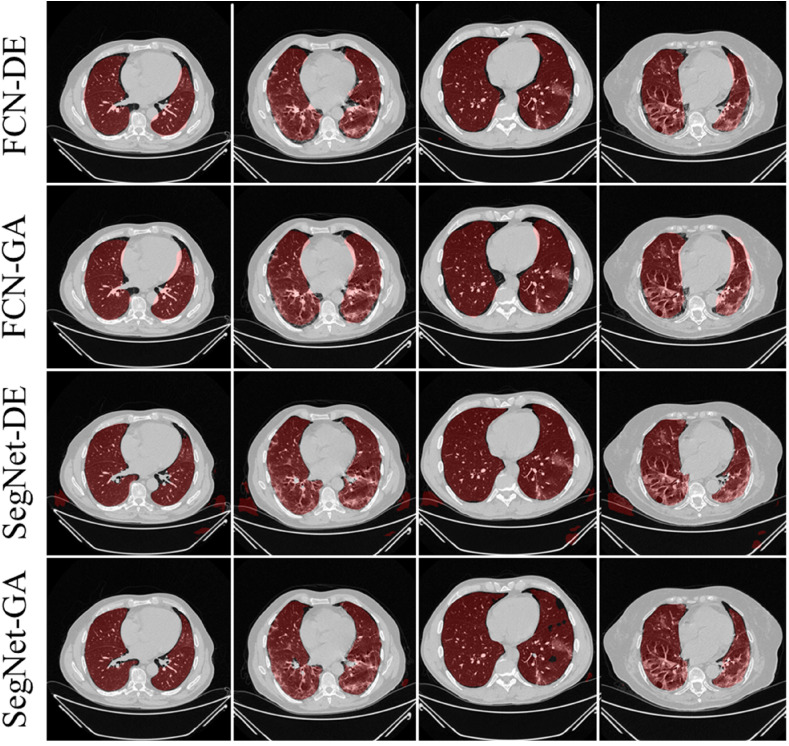
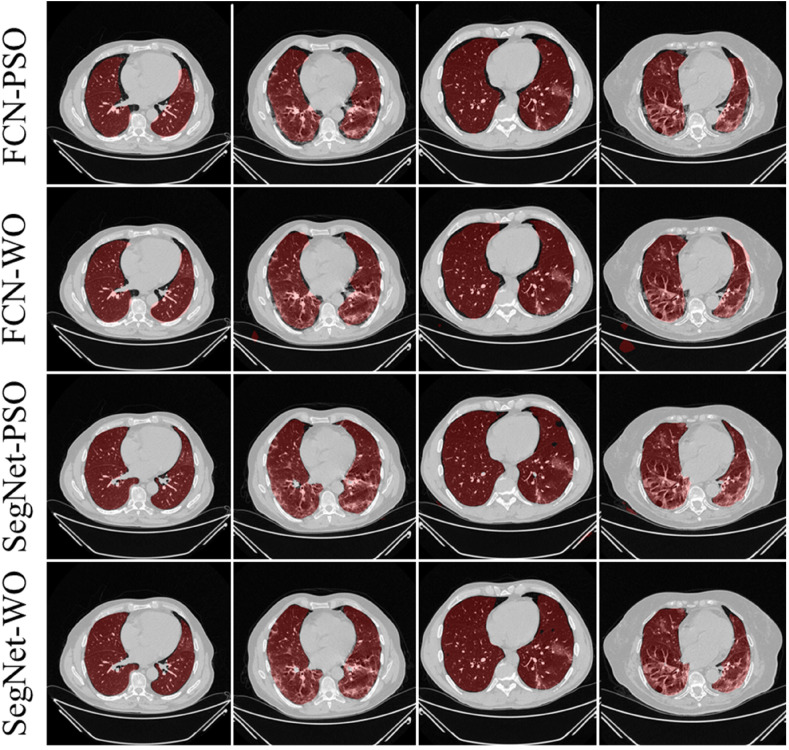
The training of the AI modes was done using the CroMed data set with 5,000 COVID-19 CT lung images, with one set of ground truth annotation from a senior radiologist. Fig. 8, Fig. 9 shows the overlay results of the segmented lungs with grayscale in the background. Fig. 8 is for the combinations FCN + DE, FCN + GA, SegNet + DE, and SegNet + GA and Fig. 9 for FCN + PSO, FCN + WO, SegNet + PSO, and SegNet + WO.
-
(B)
Percentage Storage Reduction using Pruning Models on CroMed Data set
Fig. 8.
Overlays for optimized pruning networks over raw grayscale CT scans using CroMed data set. Top: FCN-DE and FCN-GA pruning combination and Bottom: SegNet-DE and SegNet-GA pruning combination.
Fig. 9.
Overlays for optimized pruning networks over raw grayscale CT scans using CroMed data set. Top: FCN–PSO and FCN-WO pruning combination and Bottom: SegNet-PSO and SegNet-WO pruning combination.
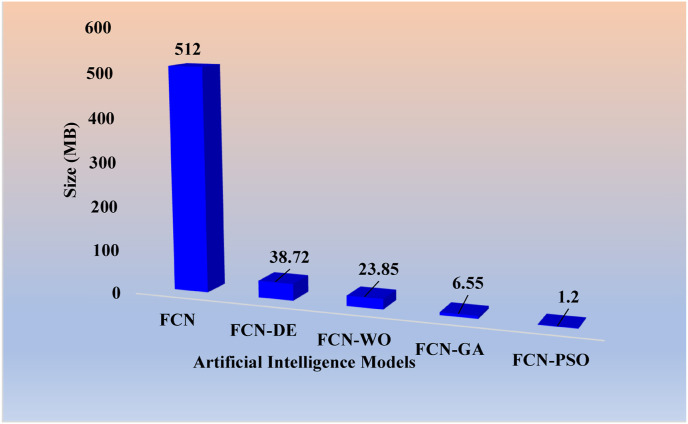
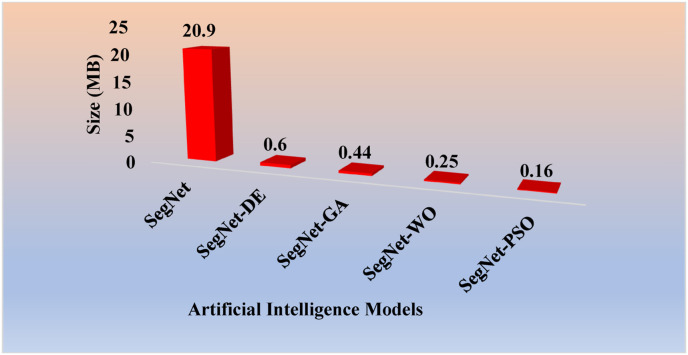
Table 1 shows the storage reduction for the eight pruning techniques using the percentage storage reduction (PSR) formula as shown in Eqs. [4], [5]).
| [4] |
where SFCN represents the storage of the FCN model and SFCN-EA represents the storage corresponding to the EA algorithm.
| [5] |
where SSegNet represents the storage of the SegNet model and SSegNet-EA represents the storage corresponding to the EA algorithm.
Table 1.
Percentage storage reduction (in MB) for the pruned AI models.
| Models | Size (MB) | PSR | Models | Size (MB) | PSR |
|---|---|---|---|---|---|
| FCN | 512 | – | SegNet | 20.9 | – |
| FCN-DE | 38.72 | 92.4% | SegNet-DE | 0.6 | 97.1% |
| FCN-WO | 23.85 | 95.3% | SegNet-GA | 0.44 | 97.9% |
| FCN-GA | 6.55 | 98.7% | SegNet-WO | 0.25 | 98.8% |
| FCN–PSO | 1.2 | 99.8% | SegNet-PSO | 0.16 | 99.2% |
For (i) FCN-DE, (ii) FCN-GA, (iii) FCN–PSO, and (iv) FCN-WO, the PSR using Eq. (4) was 92%, 95%, 99%, and 100%, respectively when compared against FCN and for (v) SegNet-DE, (vi) SegNet-GA, (vii) SegNet-PSO, and (viii) SegNet-WO, the PSR using Eq. (5) was 97%, 98%, 99%, and 99%, respectively when compared against SegNet. Thus, proving the hypothesis that pruning helps considerably reduce the size of the AI models, making the system fast, efficient, and most importantly, keeping the performance of the AI models in clinical standards. As per Table 1, the PSR values are made to increase along the rows. We observed that PSR was highest in FCN–PSO and SegNet-PSO pruning models. The lowest PSR was for FCN-DE and SegNet-DE models. The intermediate PSR models were FCN-WO and FCN-GA when FCN was used as a base. The intermediate models were SegNet-GA and SegNet-WO when SegNet was used as a model. Note that all the eight fused systems were five times faster compared to the base model FCN and SegNet.
PSR: Percentage Storage Reduction; DE: differential evolution; GA: genetic algorithm; PSO: particle swarm optimization algorithm; WO: whale optimization algorithm.
Note that in Eq. (2), Cr can have a minimum value of 1 and a maximum value equal to the total number of hidden neurons. To avoid unacceptable conditions, one must ensure that compressed hidden neurons never be zero. The maximum Cr obtained for FCN–PSO and SegNet-PSO after 10 iterations were 962.47 and 133.12, respectively. The maximum Cr obtained for FCN-GA and SegNet-GA after 4 iterations were 80.03 and 48.71, respectively. The Cr for FCN-DE, after 4 iterations was 13.54, while after 8 iterations for SegNet-DE it was 34.79. The Cr for FCN-WO, after 12 iterations was 31.99, while after 10 iterations for SegNet-WO it was 85.73. The further effort to compress led to degradation in performance, therefore it was not considered.
4.2. Performance evaluation
-
(A)
Performance Evaluation for eight EA models
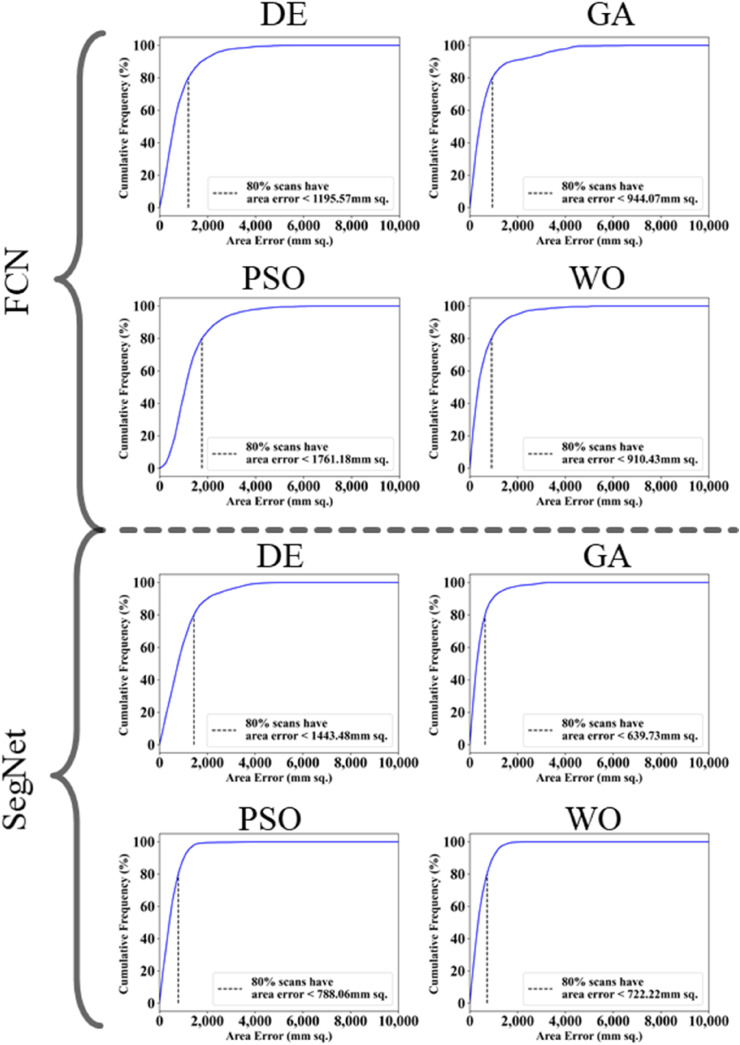
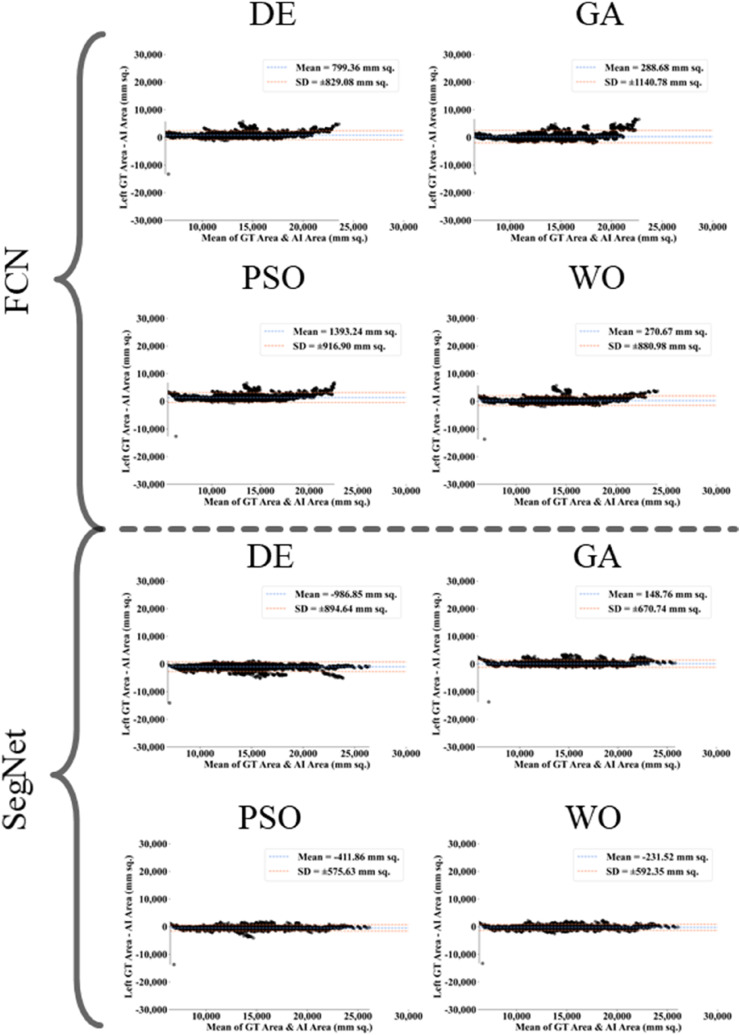
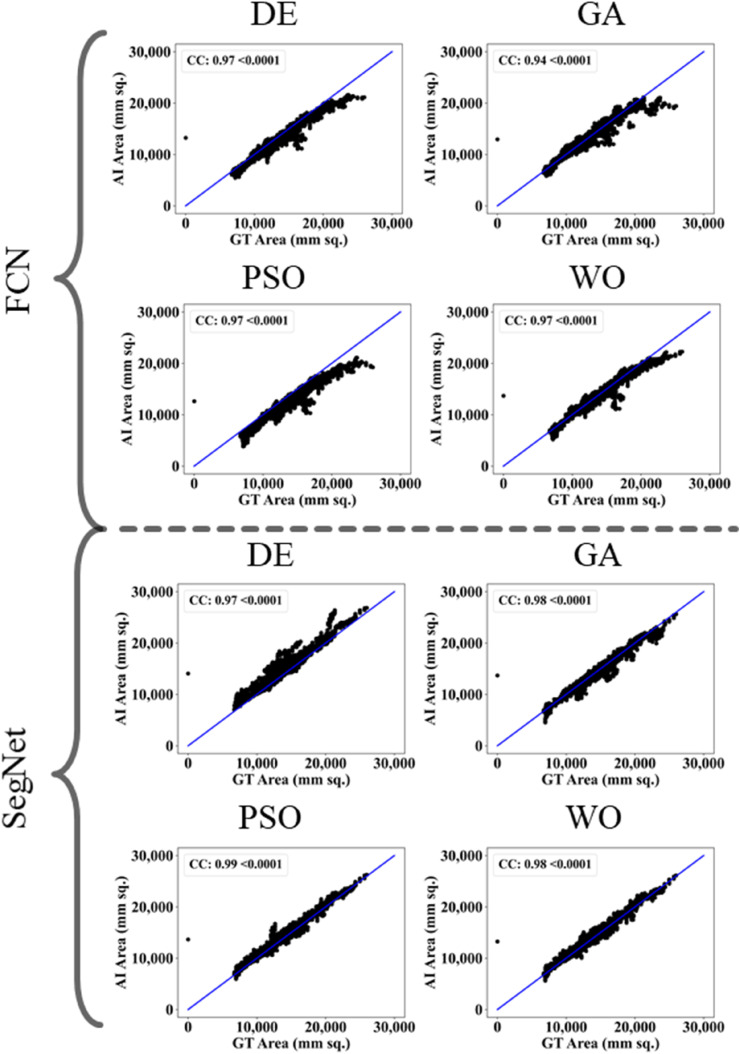
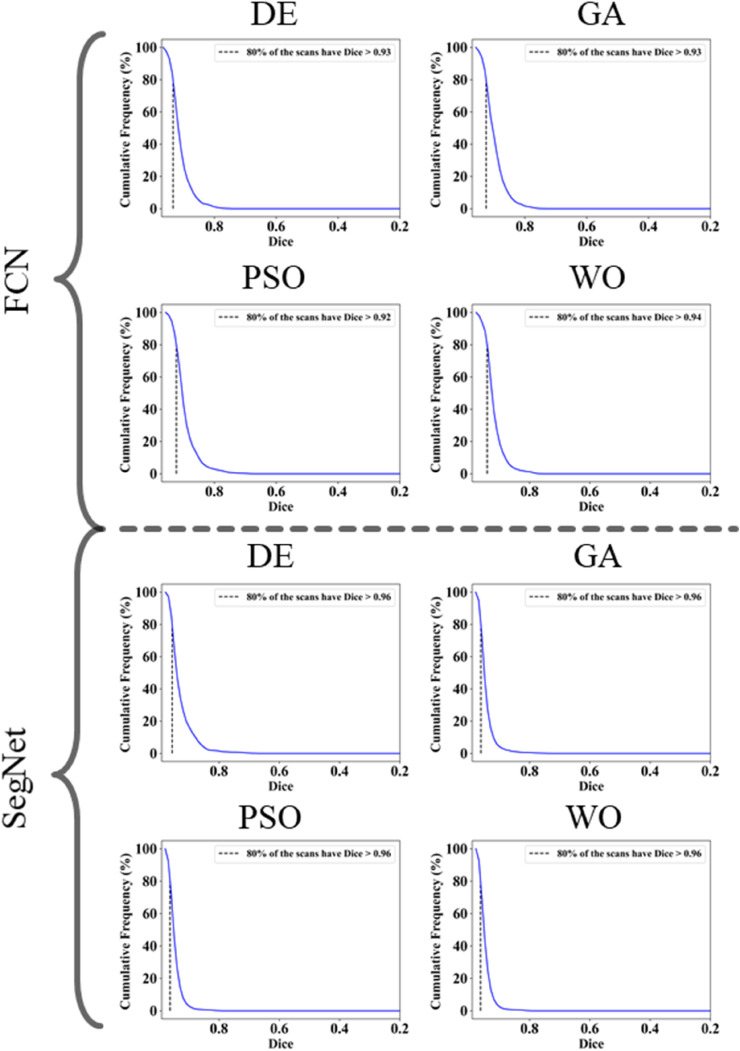
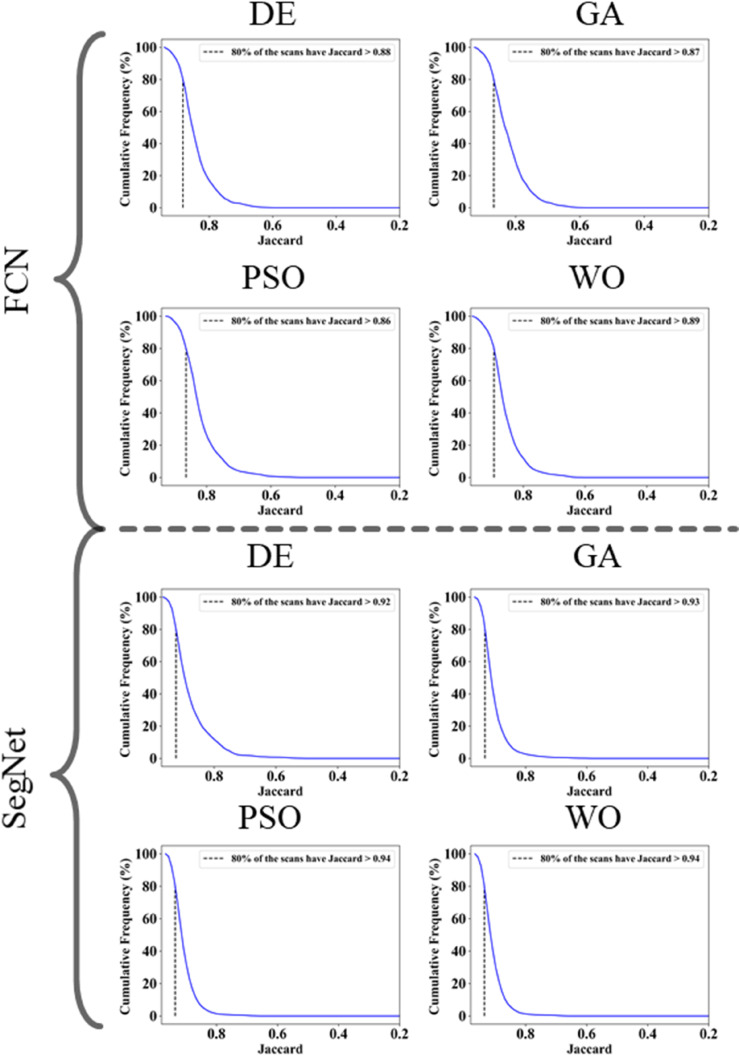
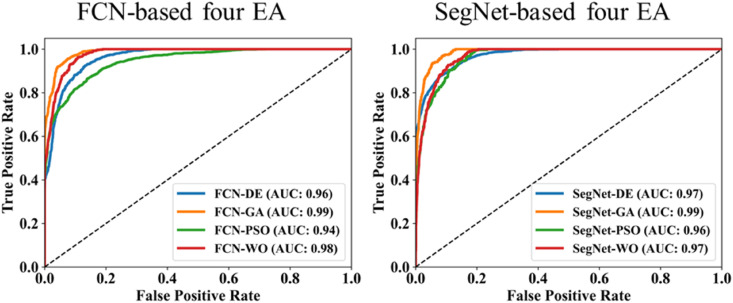
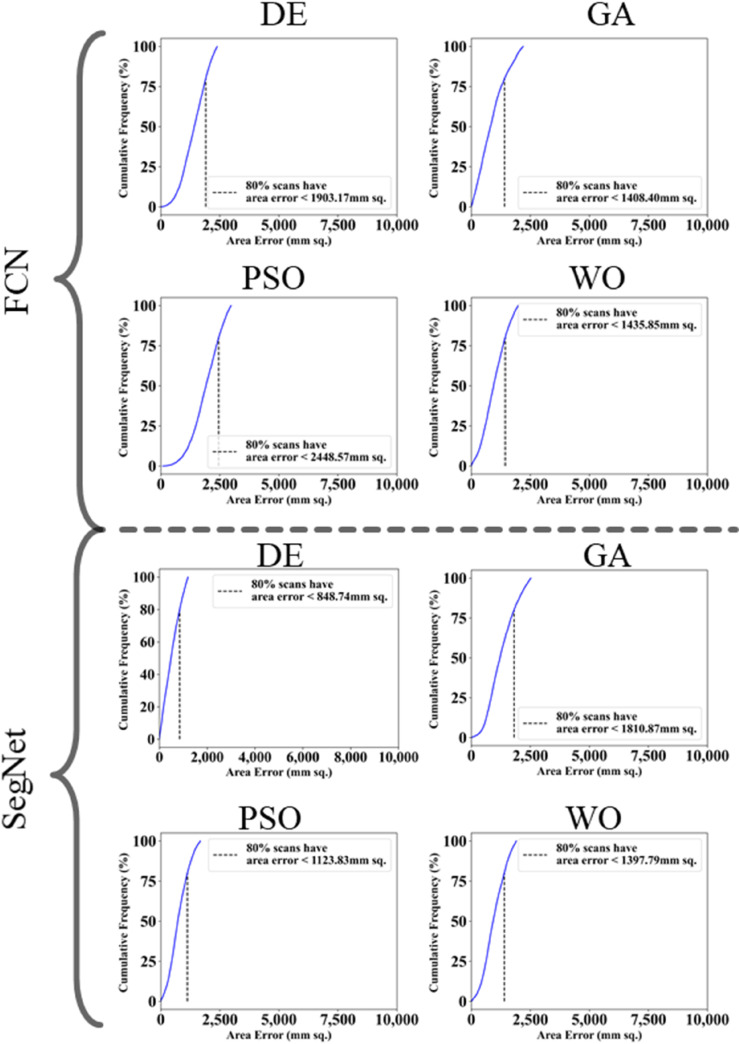
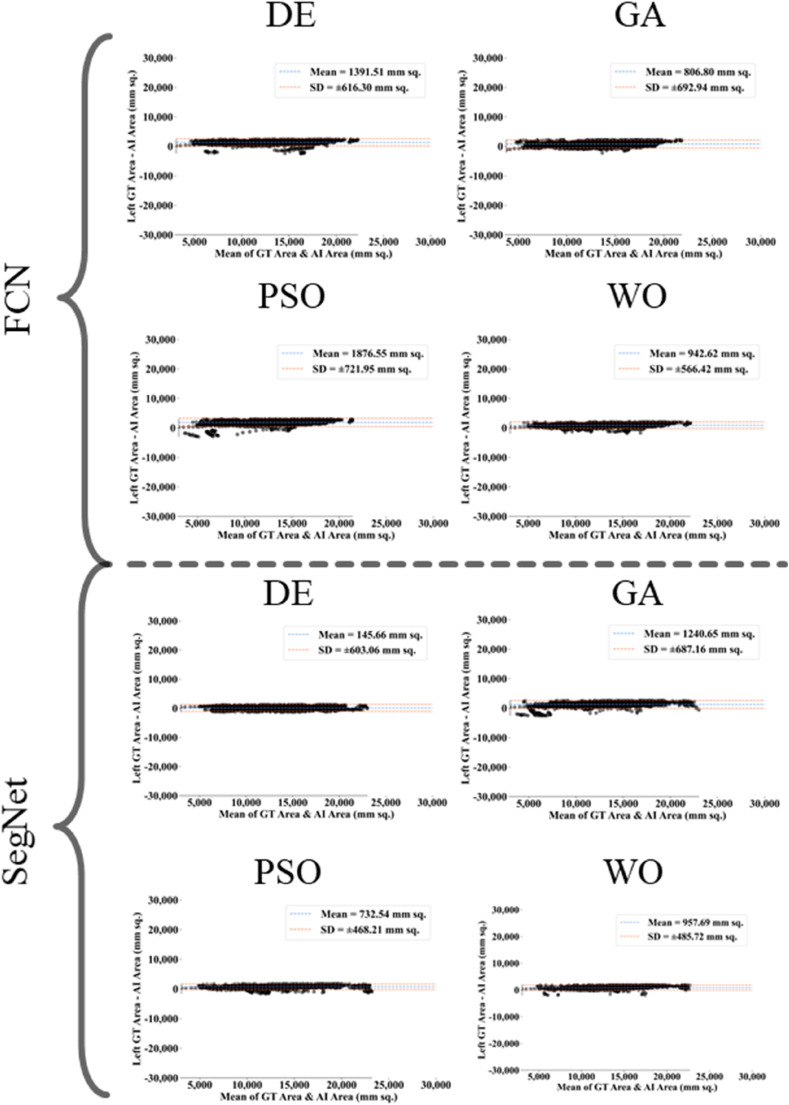
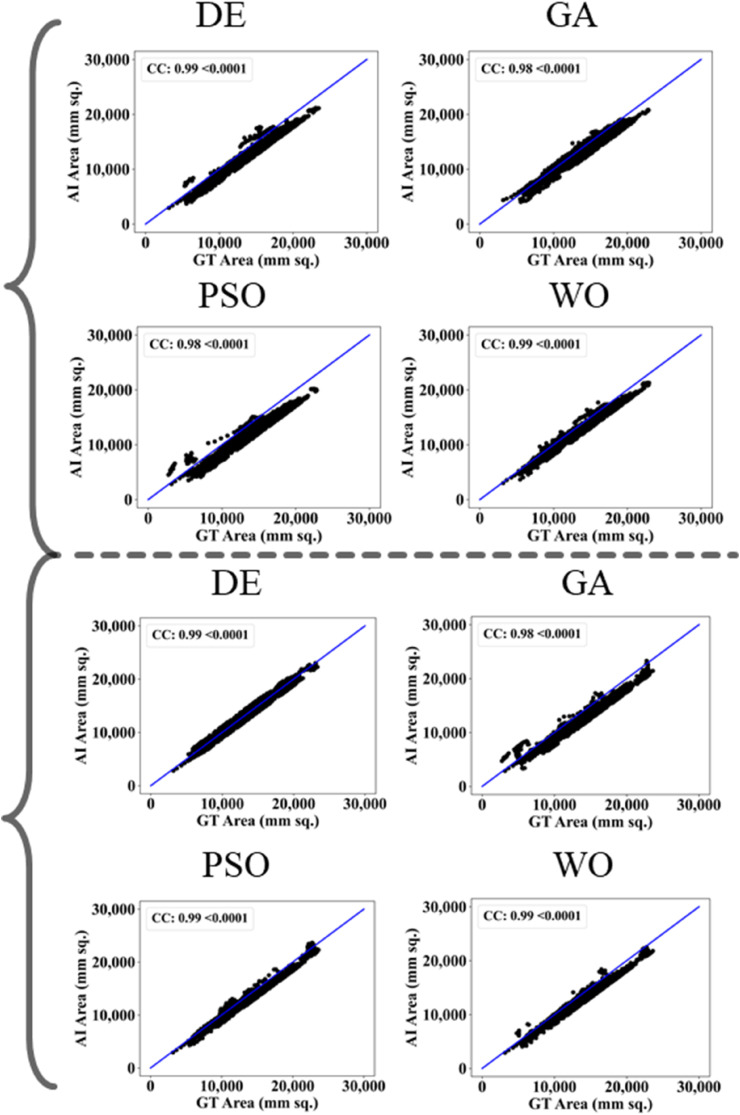
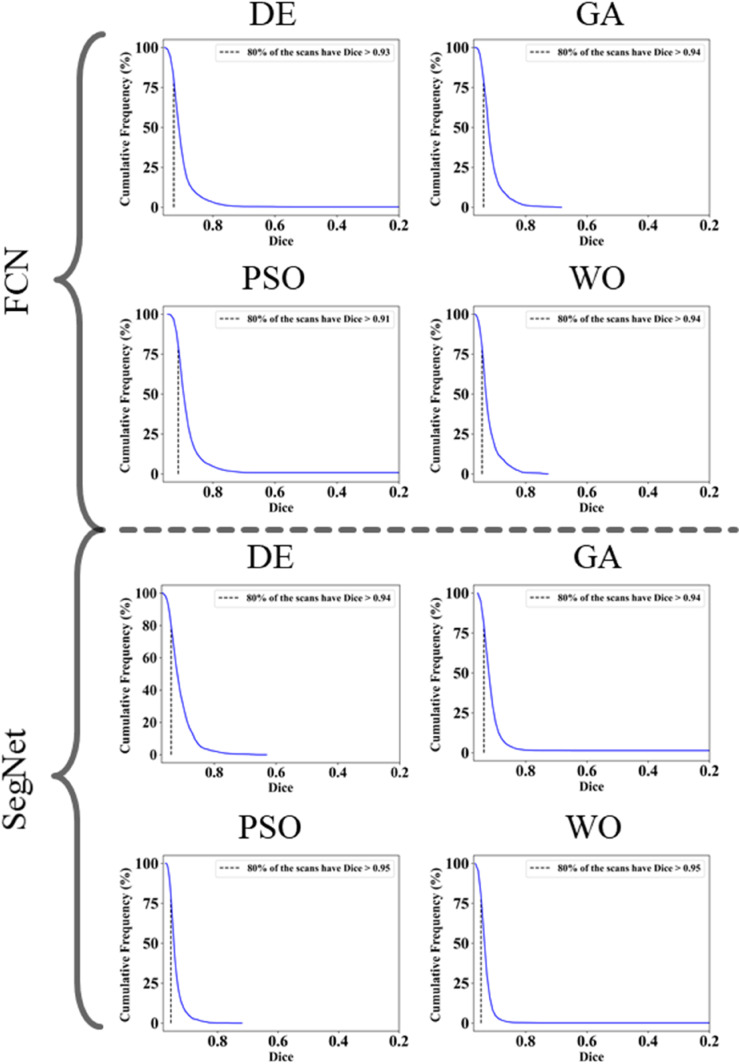
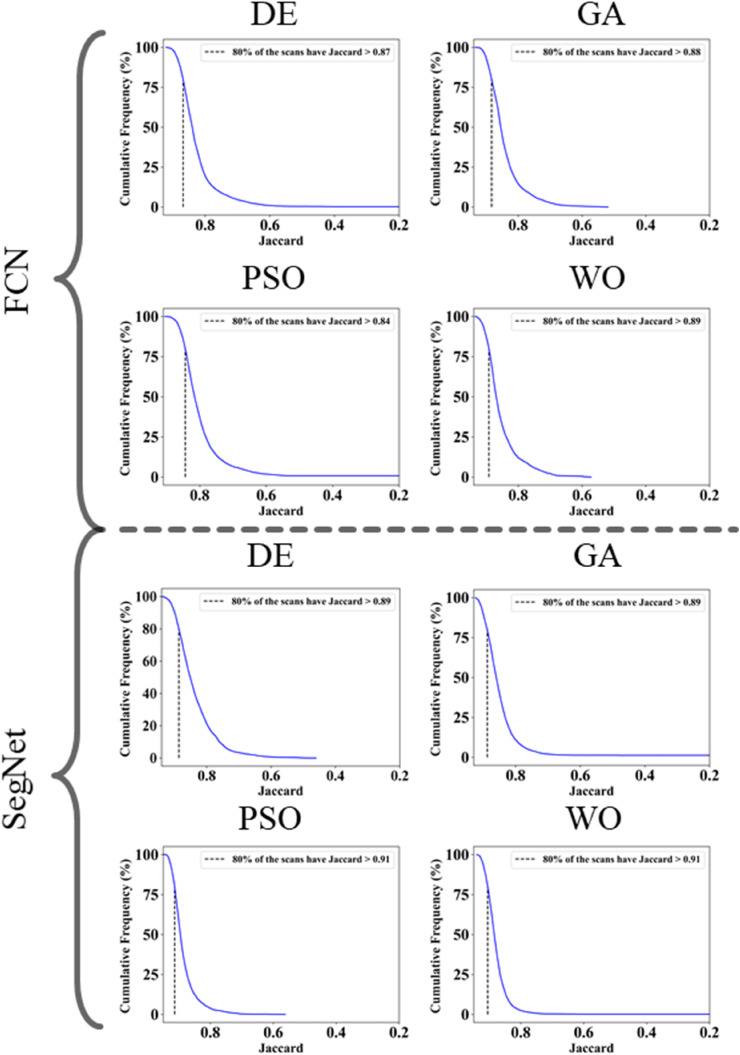
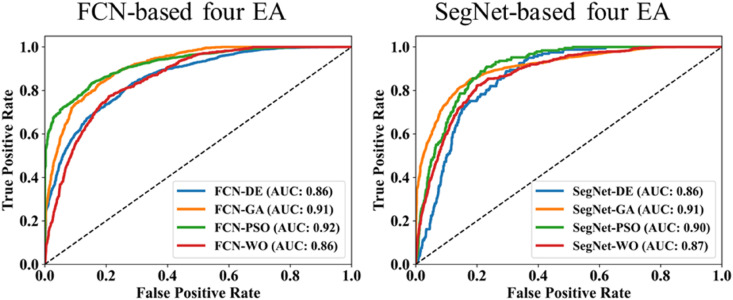
The proposed study makes use of mainly five kinds of performance evaluation metrics (i) AE, (ii) BA, (iii) CC, (iv) DS, (v) JI, and (vi) ROC to compare the performance of the pruned AI models. The cumulative frequency distribution (CFD) plot for AE is presented in Fig. 10 at a threshold cutoff of 80% for CroMed data. Fig. 11 shows the BA plot with mean and standard deviation (SD) line for the estimated lung area against the AI models and ground truth tracings for CroMed data. Similarly, CC plots with a cutoff of 80% are displayed in Fig. 12 . CFD plots for DS and JI at a threshold cutoff of 80% for the CroMed is presented in Fig. 13, Fig. 14 . The COVIAS 2.0 diagnostic performance can be calculated utilizing the variable threshold technique and ROC analysis. Fig. 15 shows the ROC curves using CroMed data set for FCN-based and SegNet-based four EA models, each having AUC values greater than ∼0.94 (p < 0.0001) and ∼0.96 (p < 0.0001), respectively.
-
(B)
DS, JI, and CC for eight EA models for CroMed and NovMed Data sets
Fig. 10.
Cumulative frequency plot for lung Area Error for CroMed data. Top: FCN with four pruning (DE, GA, PSO, WO) and Bottom: SegNet with four pruning (DE, GA, PSO, WO).
Fig. 11.
Bland-Altman plots for Area Error on Croatia data. Top: FCN with four pruning (DE, GA, PSO, WO) and Bottom: SegNet with four pruning (DE, GA, PSO, WO).
Fig. 12.
CC plots for Area Error on CroMed data. Top: FCN with four pruning (DE, GA, PSO, WO) and Bottom: SegNet with four pruning (DE, GA, PSO, WO).
Fig. 13.
Dice Similarity on CroMed data. Top: FCN with four pruning (DE, GA, PSO, WO) and Bottom: SegNet with four pruning (DE, GA, PSO, WO).
Fig. 14.
Jaccard Index plots for Area Error on CroMed data. Top: FCN with four pruning (DE, GA, PSO, WO) and Bottom: SegNet with four pruning (DE, GA, PSO, WO).
Fig. 15.
ROC using CroMed data set. Left: FCN-based four EA (DE, GA, PSO, WO) and Right: SegNet-based four EA (DE, GA, PSO, WO).
Let and be the DS for the base models FCN and SegNet, corresponding to EA(m), where m can take the values 1, 2, 3, and 4, which implies DE, GA, PSO, and WO, respectively. Similarly, and be the JI for the base models FCN and SegNet, corresponding to EA(m), where m can take the values 1, 2, 3, and 4, which implies DE, GA, PSO, and WO, respectively.
Let and be the CC for the base models FCN and SegNet, corresponding to EA(m), where m can take the values 1, 2, 3, and 4, which implies DE, GA, PSO, and WO, respectively. Using the above notations, we can define and as given in Eq. (6), and as given in Eq. (7) and and as in Eq. (8).
| [6] |
| [7] |
| [8] |
Using Eqs. [6], [7], [8]) the one can compute the mean of all the four EA models for FCN and SegNet base models as defined in Eq. (9).
| [9] |
Table 2 summarizes the DS, JI, and CC values for the eight pruned AI models over CroMed (experimental) and Unseen NovMed (validation) data sets, along with the results achieved by the MedSeg. Table 3 presents the percentage difference for the DS over eight AI pruned models with FCN-DE, FCN-GA, FCN–PSO and FCN-WO on the left side and SegNet-DE, SegNet-GA, SegNet-PSO and SegNet-WO on the right side using Eq. (9). The mean dice for the four pruned FCN models is 3%, 8%, and 2%, while for four pruned SegNet models was 0%, 4%, and 1%, respectively for CroMed (experimental) and Unseen NovMed (validation) data sets using Eq. (9). Further, the mean percentage difference using Eq. (7) for JI values is presented in Table 4 , which is 5%, 15%, and 3% for pruned FCN models, and 1%, 8%, and 1% for pruned SegNet models, respectively for CroMed (experimental) and Unseen NovMed (validation) data sets. Similarly, the mean percentage difference for CC values using Eq. (8) is presented in Table 5 , which is 3%, 1%, and 1% for pruned FCN models, and 1%, 1%, and 0% for pruned SegNet models, respectively for CroMed (experimental) and Unseen NovMed (validation) data set.
-
(C)
Comparison between eight EA models using Figure-of-Merit
Table 2.
DS, JI, and CC values for experimental CroMed and Unseen NovMed.
| Models | CroMed |
Unseen NovMed |
||||
|---|---|---|---|---|---|---|
| DS | JI | CC | DS | JI | CC | |
| FCN-DE | 0.93 | 0.88 | 0.97 | 0.93 | 0.87 | 0.99 |
| FCN-GA | 0.93 | 0.87 | 0.94 | 0.94 | 0.88 | 0.98 |
| FCN–PSO | 0.92 | 0.86 | 0.97 | 0.91 | 0.84 | 0.98 |
| FCN-WO | 0.94 | 0.89 | 0.97 | 0.94 | 0.89 | 0.99 |
| SegNet-DE | 0.96 | 0.92 | 0.97 | 0.94 | 0.89 | 0.99 |
| SegNet-GA | 0.96 | 0.93 | 0.98 | 0.94 | 0.89 | 0.98 |
| SegNet-PSO | 0.96 | 0.94 | 0.99 | 0.95 | 0.91 | 0.99 |
| SegNet-WO | 0.96 | 0.94 | 0.98 | 0.95 | 0.91 | 0.99 |
| MedSeg | 0.96 | 0.92 | 0.99 | 0.95 | 0.90 | 0.99 |
Table 3.
Percentage difference in Dice Similarity (against GT) for the eight EA models when compared against MedSeg using experimental CroMed and Unseen NovMed.
| Models | CroMed | Unseen NovMed | Models | CroMed | Unseen NovMed |
|---|---|---|---|---|---|
| FCN-DE | 3% | 2% | SegNet-DE | 0% | 1% |
| FCN-GA | 3% | 1% | SegNet-GA | 0% | 1% |
| FCN–PSO | 4% | 4% | SegNet-PSO | 0% | 0% |
| FCN-WO | 2% | 1% | SegNet-WO | 0% | 0% |
| μ | 3% | 2% | μ | 0% | 1% |
| σ | 1% | 1% | σ | 0% | 1% |
Table 4.
Percentage Difference in Jaccard Index (against GT) for the eight EA models when compared against MedSeg using experimental CroMed and Unseen NovMed.
| Models | CroMed | Unseen NovMed | Models | CroMed | Unseen NovMed |
|---|---|---|---|---|---|
| FCN-DE | 4% | 3% | SegNet-DE | 0% | 1% |
| FCN-GA | 5% | 2% | SegNet-GA | 1% | 1% |
| FCN–PSO | 7% | 7% | SegNet-PSO | 2% | 1% |
| FCN-WO | 3% | 1% | SegNet-WO | 2% | 1% |
| μ | 5% | 3% | μ | 1% | 1% |
| σ | 1% | 2% | σ | 1% | 0% |
Table 5.
Percentage difference in Correlation Coefficient (against GT) for the eight EA models when compared against MedSeg using experimental CroMed and Unseen NovMed.
| Models | CroMed | Unseen NovMed | Models | CroMed | Unseen NovMed |
|---|---|---|---|---|---|
| FCN-DE | 2% | 0% | SegNet-DE | 2% | 0% |
| FCN-GA | 5% | 1% | SegNet-GA | 1% | 1% |
| FCN–PSO | 2% | 1% | SegNet-PSO | 0% | 0% |
| FCN-WO | 2% | 0% | SegNet-WO | 1% | 0% |
| μ | 3% | 1% | μ | 1% | 0% |
| σ | 2% | 1% | σ | 1% | 1% |
We compared the FCN and SegNet-based eight pruned AI models using standardized Figure-of-Merit (FoM) algorithm [25,[115], [116], [117]]. Mathematically, this can be represented as follows. Let represents mean area for EA models on a datatype [d], represents mean ground truth (GT) area for the datatype [d], both taken over ‘n’ images of the cohort datatype [d] having ‘N’ images, where ‘d’ representing CroMed, or NovMed. Using these notations, the FoM in % for the EA using datatype [d] can be represented as shown in Eq. (10).
| [10] |
where, and ,
represents the area of the image ‘n’ using the evolutionary algorithm EA (DE, GA, PSO, and WO). represents the GT area corresponding to image ‘n’, ‘Σ’ represents the summation of area of all the image in the cohort. Table 6, Table 7 shows the FoM for the CroMed, and NovMed data sets. Table 6 shows the FoM for CroMed corresponding to FCN and SegNet based EA models in the column 2 and 3. The difference of the two columns is shown in the column ‘Superior’. For the EA algorithm except DE, SegNet-based EA is superior in the range 1–10%. Overall mean for the four EA using the two base models are comparable, having a different of 2.39%. Table 7 shows the FoM for unseen NovMed corresponding to FCN and SegNet based EA models in the column 2 and 3. The difference of the two columns is shown in the column ‘Superior’. For the EA algorithm except GA and WO, SegNet-based EA is superior in the range 1–10%. Overall mean for the four EA using the two base models are comparable, having a difference of 5.45%.
Table 6.
FoM comparison for CroMed using eight EA models.
| FoM comparison for CroMed Seen Data | |||
|---|---|---|---|
| Models | FCN | SegNet | Superior |
| DE | 94.33 | 93.00 | 1.33 |
| GA | 97.95 | 98.94 | 0.99 |
| PSO | 90.12 | 97.08 | 6.96 |
| WO | 98.08 | 98.35 | 0.28 |
| μ | 95.12 | 96.84 | 2.39 |
| σ | 3.76 | 2.68 | 3.08 |
Table 7.
FoM comparison for Unseen NovMed using eight EA models.
| FoM comparison for NovMed Unseen Data | |||
|---|---|---|---|
| Models | FCN | SegNet | Superior |
| DE | 89.81 | 98.50 | 8.70 |
| GA | 93.21 | 89.25 | 3.96 |
| PSO | 85.62 | 94.32 | 8.70 |
| WO | 92.90 | 92.47 | 0.43 |
| μ | 90.38 | 93.64 | 5.45 |
| σ | 3.53 | 3.86 | 4.02 |
Finally, we can conclude from Table 6, Table 7 that the mean FoM for FCN based EA models over all three data sets (CroMed and NovMed) is ∼94%, while for SegNet it was ∼96%. This clearly demonstrated that the four EA models based on SegNet are better than the four EA models based on FCN. This is mainly because the number of layers present in the SegNet (Fig. 6) models are more than the layers in the FCN (Fig. 5) model. SegNet has skip connections where FCN lacks it. The results in which SegNet is superior to FCN are similar to previous applications [23,25,26,28].
5. Validation and statistical tests
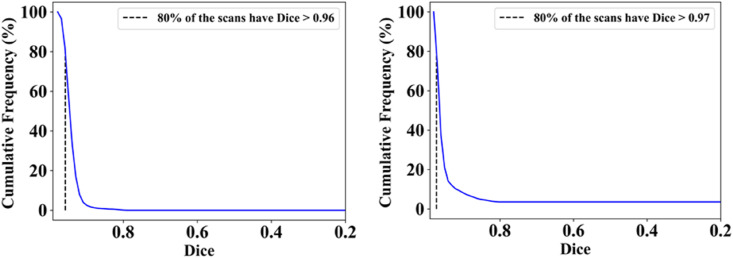
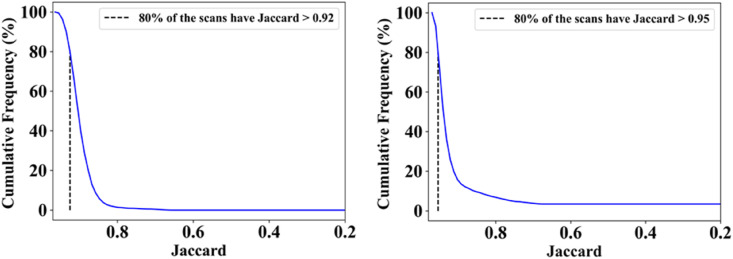
The proposed study presents a lung segmentation validation using (i) Unseen NovMed data set on the trained pruned AI models and (ii) compared the results against MedSeg, a web-based CT lung segmentation tool. Fig. 16, Fig. 17 shows the overlay results of the segmented lungs (red) with grayscale in the background using the Unseen NovMed data, respectively. Fig. 16 is a combination of FCN-DE, FCN-GA, SegNet-DE, and SegNet-GA. Fig. 17 is a combination of FCN–PSO, FCN-WO, SegNet-PSO, and SegNet-WO using the Unseen NovMed data, respectively. Further, note that “Unseen NovMed” data sets were taken to validate the results. Fig. 18, Fig. 19, Fig. 20, Fig. 21, Fig. 22 show the CFD plots of AE, BA, CC, DS, and JI, respectively, for the segmented lungs using COVLIAS 2.0 on the “Unseen NovMed” data set. Fig. 23 shows the ROC curves using Unseen NovMed for FCN-based and SegNet-based four EA models, having each AUC values more than ∼0.86 (p < 0.0001) and ∼0.86 (p < 0.0001), respectively.
Fig. 16.
Overlays (red) for optimized pruning networks over raw grayscale CT scans for NovMed data set. Top: FCN-DE and FCN-GA pruning combination and Bottom: SegNet-DE and SegNet-GA pruning combination.
Fig. 17.
Overlays (red) for optimized pruning networks over raw grayscale CT scans for NovMed data set. Top: FCN-DE and FCN-GA pruning combination and Bottom: SegNet-DE and SegNet-GA pruning combination.
Fig. 18.
Cumulative frequency plot for Area Error using NovMed data.
Fig. 19.
BA plot using NovMed data.
Fig. 20.
CC plot using NovMed data.
Fig. 21.
Cumulative frequency plot for Dice using NovMed data.
Fig. 22.
Cumulative frequency plot for Jaccard using NovMed data.
Fig. 23.
ROC using NovMed data set. Left: FCN-based four EA (DE, GA, PSO, WO) and Right: SegNet-based four EA (DE, GA, PSO, WO).
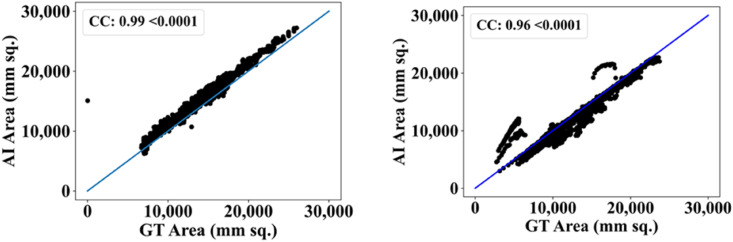
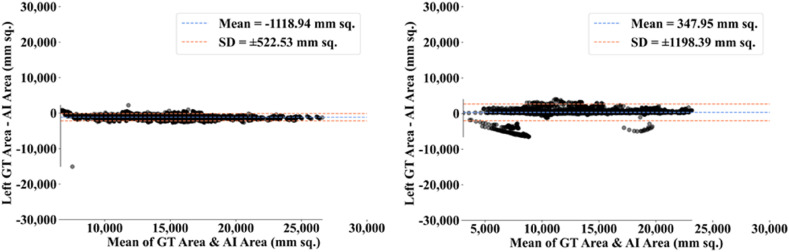
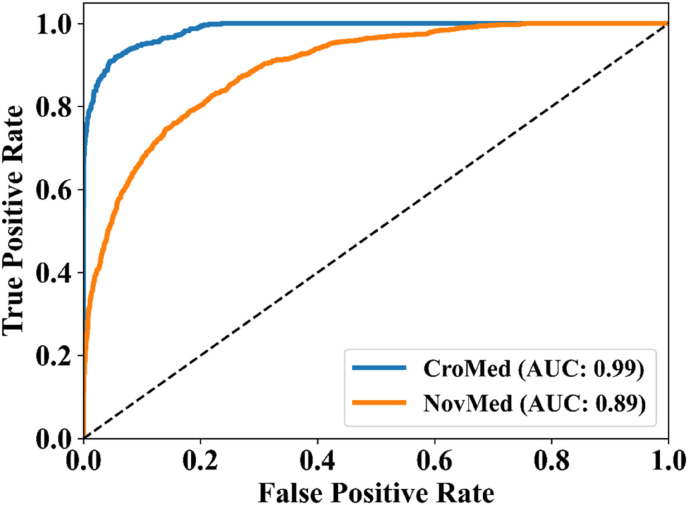
The MedSeg tool's results were compared to the gold standard tracings of the two data sets utilized in the study. CFD plot of DS, JI, and CC for the segmented lungs using the MedSeg tool for CroMed and “Unseen NovMed” data sets are shown in Fig. 24, Fig. 25, Fig. 26 , respectively. Similarly, Fig. 27 show the BA plot of the results from the MedSeg compared to the ground truth tracings of the two data sets (CroMed on the left and NovMed on the right). The percentage difference between the DS, JI, and CC score of the COVLIAS 2.0 AI models compared to MedSeg is <5%, thus proving the applicability of the proposed AI models in the clinical domain. Fig. 28 shows the ROC curve and AUC values for the MedSeg, with CroMes and Unseen NovMed having AUC values of 0.99 (p < 0.0001) and 0.89 (p < 0.0001), respectively. The size utilized by the models in decreasing order were FCN, FCN-DE, FCN-GA, FCN–PSO, and FCN-WO is shown in Fig. 29 and for SegNet, SegNet-DE, SegNet-GA, SegNet -PSO, and SegNet-WO is shown in Fig. 30 . The quantitative analysis using AE, DS, JI, BA, CC, and ROC on Unseen NovMed have similar behavior as Seen CroMed data.
Fig. 24.
Cumulative frequency plot of DS for MedSeg using CroMed (left) and Unseen NovMed (right) data sets.
Fig. 25.
Cumulative frequency plot of JI for MedSeg using CroMed (left) and Unseen NovMed (right) data sets.
Fig. 26.
CC plot for MedSeg vs. GT using CroMed (left) and Unseen NovMed (right) data sets.
Fig. 27.
BA plot for MedSeg vs. GT using CroMed (left) and Unseen NovMed (right) data sets.
Fig. 28.
ROC plot for MedSeg vs. GT using CroMed and Unseen NovMed data sets.
Fig. 29.
Size (in Megabyte) of the FCN-based models in descending order.
Fig. 30.
Size (in Megabyte) of the SegNet-based models in descending order.
We present a summary and percentage improvement for all eight pruned AI models for DS, JI, and CC values using experimental CroMed, validation Unseen NovMed data set in Table 2, Table 3, Table 4, Table 5, respectively. When comparing four pruning techniques against the base model (FCN and SegNet) for experimental and validation data, the pruning model performs far better than the base model for all three performance evaluation matrices. Finally, to prove the reliability of the AI-based segmentation system COVLIAS 2.0, statistical test such as Mann-Whitney, Paired t-Test, and Wilcoxon test were presented for using experimental CroMed (Table 8 ), validation Unseen NovMed (Table 9 ) analysis. All the above analysis was conducted by using MedCalc software (v18.2.1, Osteen, Belgium).
Table 8.
Statistical test for CroMed data set.
| Models | Paired t-Test | Mann-Whitney | Wilcoxon |
|---|---|---|---|
| FCN-DE | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| FCN-GA | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| FCN–PSO | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| FCN-WO | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| SegNet-DE | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| SegNet-GA | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| SegNet-PSO | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| SegNet-WO | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| MedSeg | P < 0.0001 | P < 0.0001 | P < 0.0001 |
Table 9.
Statistical test for NovMed data set.
| Models | Paired t-Test | Mann-Whitney | Wilcoxon |
|---|---|---|---|
| FCN-DE | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| FCN-GA | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| FCN–PSO | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| FCN-WO | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| SegNet-DE | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| SegNet-GA | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| SegNet-PSO | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| SegNet-WO | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| MedSeg | P < 0.0001 | P < 0.0001 | P < 0.0001 |
5.1. Lesion localization validation
Lesions have different characteristics such as texture, contrast, intensity variation, density changes, and etc. [118]. Fig. 31 presents the pipeline for lesion validation using heatmaps, where the input to the eight pruned segmentation model is the CT image, which produces the segmented lungs. This segmented lung goes to the DenseNet-121 for classification into two classes, i.e., COVID-19 and Controls.
Fig. 31.
COVLIAS 2.0 Lesion heatmap pipeline using DenseNet model using the infrastructure of pruned AI models during lung segmentation.
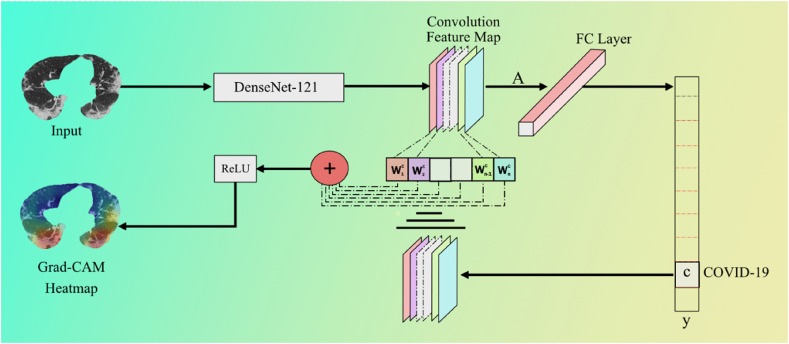
The Gradient-weighted Class Activation Mapping (Grad-CAM) (Fig. 32 ) algorithm is applied to produce the lesion heatmap. Grad-CAM employs the gradients of any target concept (example “COVID-19” in this classification network) to build a coarse feature map by showing the critical regions in the picture for predicting the concept. Grad-CAM produces a coarse localization map showing the essential regions in the image for predicting the idea by using the gradients of any target concept flowing into the final convolutional layer. From a high level, we take an image as input to the model for which a Grad-CAM heatmap is desired. Then this image is passed through the network following the normal prediction cycle (including the FC Layer) and generating the class probability scores followed by the loss calculation. Next, we calculate the gradient of our desired model layer's output concerning the model loss. Then we trim, resize, and rescale areas of the gradient that contribute to the prediction such that the heatmap can be overlaid with the original image. In this scenario, ReLU is the best option because it highlights qualities that positively impact the class of interest.
Fig. 32.
Grad-CAM process using DenseNet-121 model utilized for lesion localization.
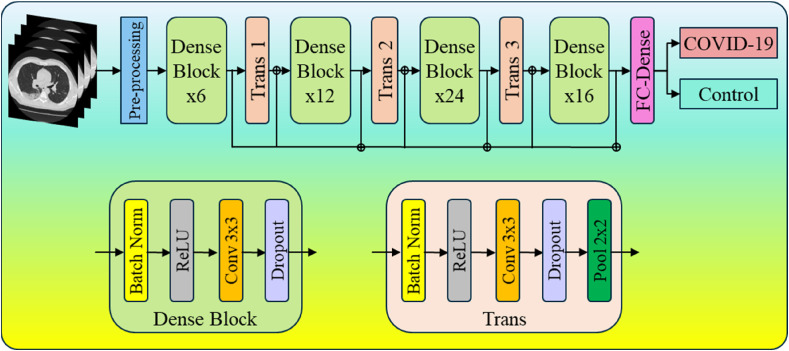
DenseNet-121(Fig. 33 ) is made up of 120 convolutions and 4 AvgPool layers. All layers, including those within the same dense block and transition layers, spread their weights across numerous inputs, allowing deeper layers to leverage characteristics collected earlier in the process. DenseNets result in more compact models, attain state-of-the-art performances, and better outcomes across competing data sets than their typical CNN or ResNet equivalents. This is because they require fewer parameters and allow feature reuse [[119], [120], [121]]. These critical regions that differentiate the COVID-19 CT scan from the Control CT scans are presented in Figs. Fig. 34, Fig. 35, Fig. 36 .
Fig. 33.
DenseNet-121 model utilized for lesion localization.
Fig. 34.
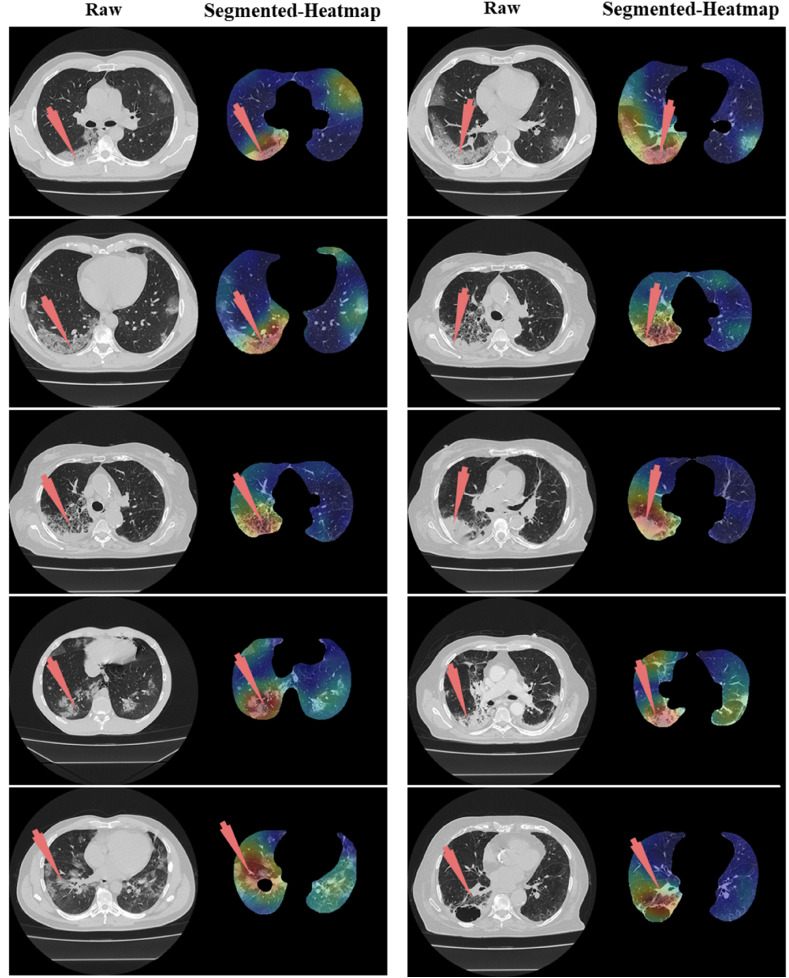
Left: Raw grayscale CT slice. Right: Segmented lungs with colored heatmap where red arrow indicates the right lung damage only.
Fig. 35.
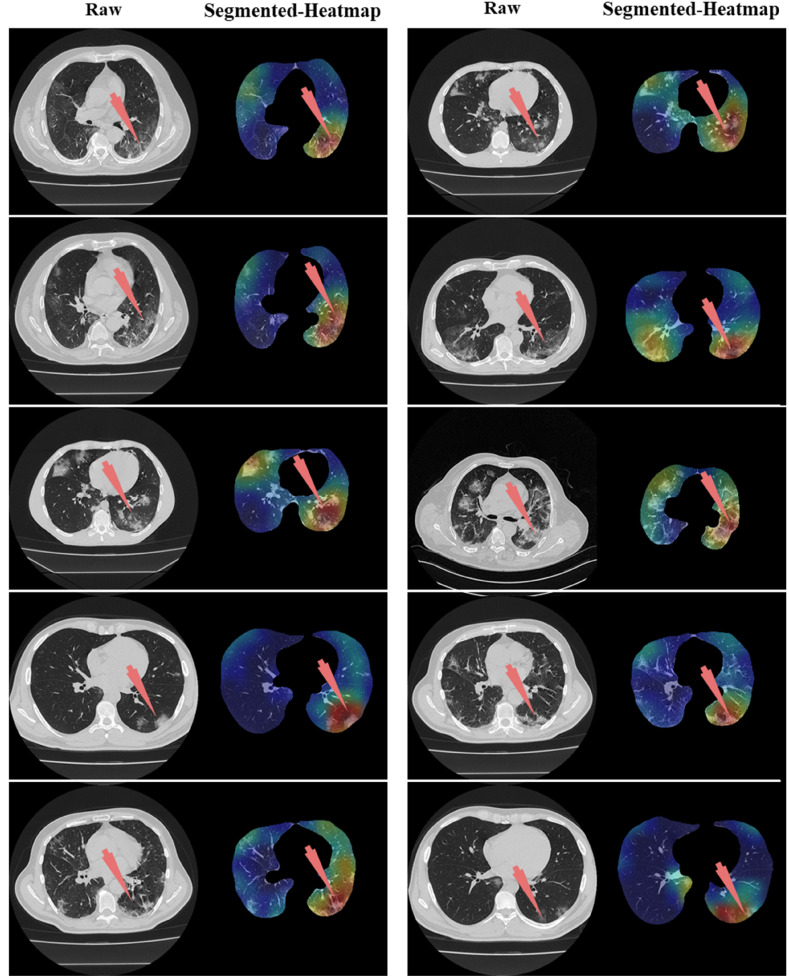
Left: Raw grayscale CT slice. Right: Segmented lungs with heatmap where red arrow indicated the left lung damage.
Fig. 36.
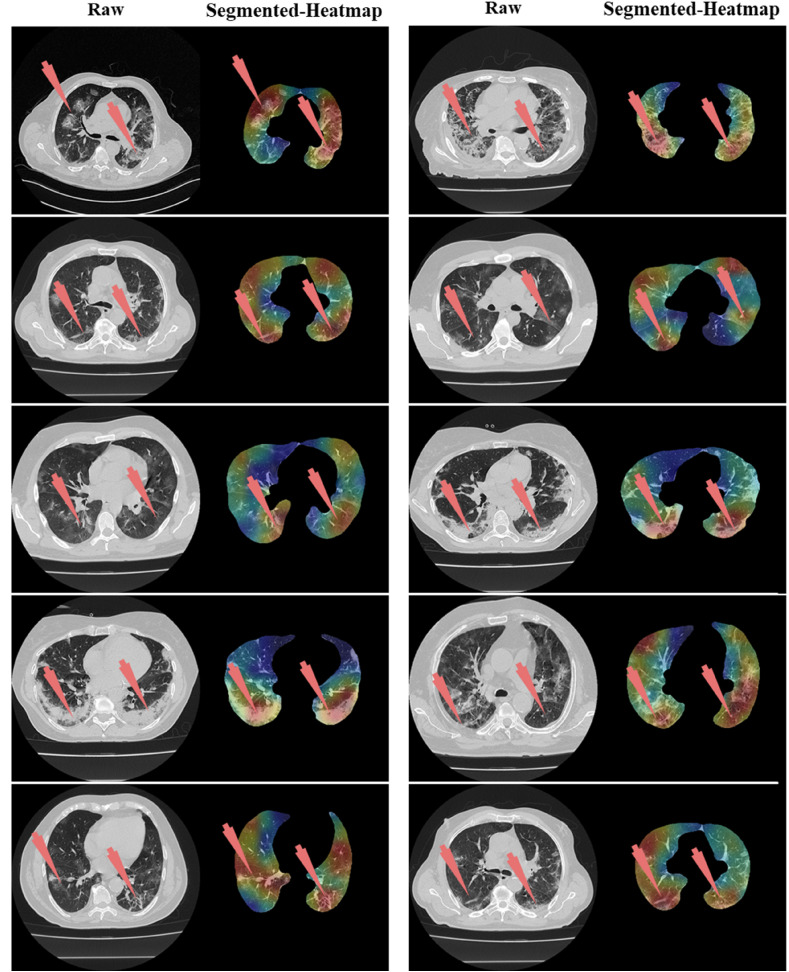
Left: Raw grayscale CT slice. Right: Segmented lungs with heatmap where red arrow indicated the damage in both left and right lungs.
6. Discussion
6.1. Major contributions and study findings
The proposed study presented eight pruned AI techniques, namely, (i) FCN-DE, (ii) FCN-GA, (iii) FCN–PSO, (iv) FCN-WO, (v) SegNet-DE, (vi) SegNet-GA, (vii) SegNet -PSO, and (viii) SegNet-WO for CT lung segmentation, namely, and this was our premier contribution of this study. The pruned AI models were trained on a CroMed data set, containing 5,000 COVID-19 CT images collected from 80 patients. The pre-processing involved in the CroMed data set consists of Hounsfield unit (HU) that was adjusted to highlight the lung region (1600, −400), making the model train efficiently. The second major contribution was the Unseen data analysis taken from Unseen NovMed data and it consists of validating the eight pruned AI models using Unseen NovMed data on 4,000 CT images from 72 patients. The two data sets CroMed and Unseen NovMed were annotated by senior radiologists. The third major innovation of our study is the design of the lesion localization that consists of the superposition of heatmaps on the pruned AI segmented lungs, displaying red color on the lesions. This innovation was first designed to develop the classification model using DensNet-121 and the trained model was then adapted for applying the Grad-CAM heatmap system on the grayscale lungs (Fig. 33, Fig. 34, Fig. 35).
Our fourth major contribution mainly deals with benchmarking our evolutionary-based COVLIAS 2.0 against MedSeg ([41], a web-based lung segmentation tool). This was the first time attempted in a low-storage and high-speed infrastructures. The percentage difference between the DS, JI, and CC score of the COVLIAS 2.0 AI models was compared to MedSeg is <5%, thus proving that the proposed AI models are clinically applicable. For DS, JI, and CC values, we presented a summary and percentage improvement for all eight pruned AI models in Table 3, Table 4, Table 5
The study revealed that the pruned AI models (i) FCN-DE, (ii) FCN-GA, (iii) FCN–PSO, and (iv) FCN-WO) had storage reductions of 92%, 95%, 99%, and 100%, respectively, when compared against the base FCN. When compared against base SegNet for (v) SegNet-DE, (vi) SegNet-GA, (vii) SegNet -PSO, and (viii) SegNet -WO, it showed a percentage storage reduction 97%, 98%, 99%, and 99%, respectively. Thus, we validate our hypothesis and demonstrate our superior clinical performance. We put COVLIAS 2.0 through clinical and statistical tests to ensure its reliability and stability to support our theory. All our data analysis was comprehensively adapted to 360-degree evaluations, and this consists of (i) DS, (ii) JI, (iii) BA (iv) CC, (v) AE and (vi) ROC plots. We consider this as our fifth contribution of our study.
6.2. Benchmarking
We couldn't identify any articles that used pruning techniques to segment CT lungs. There have been several articles that use pruning approaches on X-ray images to classify COVID-19 pneumonia. As a result, we chose to include papers that used CT lung segmentation in our benchmarking strategy (Table 10 ). This included Jiang et al. [82], Kogilavani et al. [84] that used VGG16 [85], DenseNet [86], MobileNet [87], Xception [88], NASNet [89], and EfficientNet [90] for COVID-19 lung classification. Paluru et al. [91] developed a CT-based COVID-19 detection model, named AnamNet, based on the data set in Ref. [92]. Other authors include Cai et al. [96] and Saood et al. [97]. The authors did not compare lung area errors or create JI or BA plots. The authors did not use any kind of model pruning technique to reduce the size of the training model, which helps to improve running time.
Table 10.
Benchmarking table.
| C1 |
C2 |
C3 |
C4 |
C5 |
C6 |
C7 |
C8 |
C9 |
C10 |
C11 |
C12 |
C13 |
C14 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R# | Author | # Patients | # Images | Image Dim | Model Types | Classification vs Segmentation |
Pruning | Model type | Dim | AE | DS | JI | BA | ACC |
| R1 | Jiang et al. | – | 1168 | 5122 | VGG16 ResNet-50 Inception v3 Inception ResNet v2 DenseNet-169 |
Classification | ✗ | SDL | 2D | ✗ | ✗ | ✗ | ✗ | 0.94 0.95 0.96 0.96 0.99 |
| R2 | Kogilavani et al. | – | ∼3873 | 2242 | VGG16 MobileNet Densenet121 Xception Efficientnet NASNet |
Classification | ✗ | SDL | 2D | ✗ | ✗ | ✗ | ✗ | 0.97 0.96 0.97 0.92 0.80 0.89 |
| R3 | Paluru et al. | 69 | ∼4339 | 5122 | AnamNet | Segmentation | ✗ | SDL | 2D | ✗ | 0.75 | ✗ | ✗ | 0.99 |
| R4 | Saood et al. | – | ∼100 | 2562 | UNet SegNet |
Segmentation | ✗ | SDL | 2D | ✗ | 0.73 0.74 |
✗ | ✗ | 0.91 0.95 |
| R5 | Cai et al. | 99 | ∼250 | – | UNet | Segmentation | ✗ | SDL | 2D | ✗ | 0.98 | 0.96 | ✗ | ✗ |
| R6 | Suri et al. (Proposed) | ∼152 | ∼9,000 | 5122 |
FCN FCN-DE FCN-GA FCN–PSO FCN-WO SegNet SegNet-DE SegNet-GA SegNet-PSO SegNet-WO |
Segmentation | ✓ | SDL | 2D | ✓ |
0.78 0.93 0.93 0.92 0.94 0.96 0.96 0.96 0.96 0.96 |
0.65 0.88 0.87 0.86 0.89 0.93 0.92 0.93 0.94 0.94 |
✓ |
0.96 0.97 0.97 0.97 0.98 0.98 0.98 0.98 0.98 0.99 |
#: number; AE: Area Error; DS: Dice Similarity; JI: Jaccard Index; BA: Bland-Altman; ACC: Accuracy; Dim: Dimension (2D vs. 3D).
R#: Row number; DE: Differential Evolution; GA: Genetic Algorithm; PSO: Particle swarm Optimization; WO: Whale Optimization.
6.3. Strengths, weakness, and extensions
The study shows that using the eight pruning techniques such as (i) FCN-DE, (ii) FCN-GA, (iii) FCN–PSO, (iv) FCN-WO, (v) SegNet-DE, (vi) SegNet-GA, (vii) SegNet-PSO, and (viii) SegNet-WO showed a reduction in storage by ∼97%. Thus, the hypothesis proves that pruning helps considerably reduce the size of the AI models, making the system fast, efficient, and most importantly, keeping the performance of the AI models in clinical standards. Our pruning techniques did not result in a decrease in the overall performance of the AI models. First, it was five times faster than the existing base models. Second, the system was validated using the “Unseen NovMed” data set. Third, the lesion localization showed powerful results and was graded by our expert radiologists. Fourth, the system has shown strong clinical statistical results.
The current pilot study is encouraging and yields promising results. However, we still need to explore the hybrid deep learning networks since it has proved to be efficient in classification and segmentation. We did not adapt mix-matching of the data for testing its abilities as done by our group earlier [122]. Further, we wish to explore statistical or non-statistical pruning and optimization techniques. The recent evolution of UNet has a potential in our application [23,26,33,123]. The current study can therefore be extended by comparing SegNet-EA and FCN-EA against UNet-EA. Big data framework can be a powerful paradigm for including data from multiple sources [124].
Note that we did the HU adjustments to enhance the lung region in the CT images in our case, which can possibly also use noise removal or a mixture of window-level with noise smoothing [125] One can also use conventional methods like level sets [126] or stochastic segmentation [47] to get lesion region and then modify the loss function. AI tends to give a higher accuracy with a lack of clinical validations, thus bringing bias in AI systems. One can therefore compute the AI bias using AP(ai)Bias [[127], [128], [129]].
7. Conclusions
The proposed study is the first pilot study that integrates eight kinds of evolution with two different DL paradigms. Thus, it accomplishes in designing eight different low storage and high-speed strategies for COVID-19 based CT lung segmentation. COVLIAS 2.0 demonstrated that the performance was retained while yielding the percentage storage reduction by ∼97%. Overall, on average, the speed of the eight pruning methods was almost five times faster than the base models. We compare our eight pruned AI models against the open-source web-based lung segmentation tools MedSeg. We also validated our hypothesis that the overall system error was under 6%, thus proving the clinical applicability. The online COVLIAS 2.0 takes ∼0.25 s for one CT slice. The COVLIAS 2.0 is reliable, accurate, and stable in clinical settings.
Eight different pruning techniques (i) differential evolution (DE), (ii) genetic algorithm (GA), (iii) particle swarm optimization algorithm (PSO), and (iv) whale optimization algorithm (WO) in two deep learning frameworks namely, (i) Fully connected network (FCN) and (ii) SegNet.
Declaration of competing interest
We respectfully want to declare that there was a conflict of interest between our senior authors (by working with them for over 10 years) and Dr. U. Rajendra Acharya and Dr. Filippo Molinari. We therefore humbly request you that they be not the reviewers or AE of this manuscript. We deeply appreciate your kind understanding.
References
- 1.Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomedica Atenei Parmensis. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO coronavirus (COVID-19) dashboard. Janurary. 2022;24 https://covid19.who.int/ [Google Scholar]
- 3.Saba L., Gerosa C., Fanni D., Marongiu F., La Nasa G., Caocci G., Barcellona D., Balestrieri A., Coghe F., Orru G., Coni P., Piras M., Ledda F., Suri J.S., Ronchi A., D'Andrea F., Cau R., Castagnola M., Faa G. Molecular pathways triggered by COVID-19 in different organs: ACE2 receptor-expressing cells under attack? A review. Eur Rev Med Pharmacol Sci. 2020;24:12609–12622. doi: 10.26355/eurrev_202012_24058. [DOI] [PubMed] [Google Scholar]
- 4.Suri J.S., Puvvula A., Biswas M., Majhail M., Saba L., Faa G., Singh I.M., Oberleitner R., Turk M., Chadha P.S., Johri A.M., Sanches J.M., Khanna N.N., Viskovic K., Mavrogeni S., Laird J.R., Pareek G., Miner M., Sobel D.W., Balestrieri A., Sfikakis P.P., Tsoulfas G., Protogerou A., Misra D.P., Agarwal V., Kitas G.D., Ahluwalia P., Kolluri R., Teji J., Maini M.A., Agbakoba A., Dhanjil S.K., Sockalingam M., Saxena A., Nicolaides A., Sharma A., Rathore V., Ajuluchukwu J.N.A., Fatemi M., Alizad A., Viswanathan V., Krishnan P.R., Naidu S. COVID-19 pathways for brain and heart injury in comorbidity patients: a role of medical imaging and artificial intelligence-based COVID severity classification: a review. Comput Biol Med. 2020;124:103960. doi: 10.1016/j.compbiomed.2020.103960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cau R., Bassareo P.P., Mannelli L., Suri J.S., Saba L. Imaging in COVID-19-related myocardial injury. Int J Cardiovasc Imaging. 2021;37:1349–1360. doi: 10.1007/s10554-020-02089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Onnis C., Muscogiuri G., Bassareo P.P., Cau R., Mannelli L., Cadeddu C., Suri J.S., Cerrone G., Gerosa C., Sironi S., Faa G. Non-invasive coronary imaging in patients with COVID-19: a narrative review. Eur. J. Radiol. 2022;149:110188. doi: 10.1016/j.ejrad.2022.110188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viswanathan V., Puvvula A., Jamthikar A.D., Saba L., Johri A.M., Kotsis V., Khanna N.N., Dhanjil S.K., Majhail M., Misra D.P. Bidirectional link between diabetes mellitus and coronavirus disease 2019 leading to cardiovascular disease: a narrative review. World journal of diabetes. 2021;12:215. doi: 10.4239/wjd.v12.i3.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fanni D., Saba L., Demontis R., Gerosa C., Chighine A., Nioi M., Suri J.S., Ravarino A., Cau F., Barcellona D., Botta M.C., Porcu M., Scano A., Coghe F., Orru G., Van Eyken P., Gibo Y., La Nasa G., D'Aloja E., Marongiu F., Faa G. Vaccine-induced severe thrombotic thrombocytopenia following COVID-19 vaccination: a report of an autoptic case and review of the literature. Eur Rev Med Pharmacol Sci. 2021;25:5063–5069. doi: 10.26355/eurrev_202108_26464. [DOI] [PubMed] [Google Scholar]
- 9.Gerosa C., Faa G., Fanni D., Manchia M., Suri J., Ravarino A., Barcellona D., Pichiri G., Coni P., Congiu T. Fetal programming of COVID-19: may the barker hypothesis explain the susceptibility of a subset of young adults to develop severe disease? European Review for Medical Pharmacological Sciences. 2021;25:5876–5884. doi: 10.26355/eurrev_202109_26810. [DOI] [PubMed] [Google Scholar]
- 10.Congiu T., Demontis R., Cau F., Piras M., Fanni D., Gerosa C., Botta C., Scano A., Chighine A., Faedda E. Scanning electron microscopy of lung disease due to COVID-19-a case report and a review of the literature. European review for medical and pharmacological sciences. 2021;25:7997–8003. doi: 10.26355/eurrev_202112_27650. [DOI] [PubMed] [Google Scholar]
- 11.Suri J.S., Laxminarayan S. CRC press; 2003. Angiography and Plaque Imaging: Advanced Segmentation Techniques. [Google Scholar]
- 12.Faa G., Gerosa C., Fanni D., Barcellona D., Cerrone G., Orrù G., Scano A., Marongiu F., Suri J., Demontis R. Aortic vulnerability to COVID-19: is the microvasculature of vasa vasorum a key factor? A case report and a review of the literature. European review for medical and pharmacological sciences. 2021;25:6439–6442. doi: 10.26355/eurrev_202110_27018. [DOI] [PubMed] [Google Scholar]
- 13.Munjral S., Ahluwalia P., Jamthikar A.D., Puvvula A., Saba L., Faa G., Singh I.M., Chadha P.S., Turk M., Johri A.M. Nutrition, atherosclerosis, arterial imaging, cardiovascular risk stratification, and manifestations in COVID-19 framework: a narrative review. Frontiers in bioscience (Landmark edition) 2021;26:1312–1339. doi: 10.52586/5026. [DOI] [PubMed] [Google Scholar]
- 14.Congiu T., Fanni D., Piras M., Gerosa C., Cau F., Barcellona D., D'Aloja E., Demontis R., Chighine F., Nioi M. Ultrastructural findings of lung injury due to Vaccine-induced Immune Thrombotic Thrombo-cytopenia (VITT) following COVID-19 vaccination: a scanning electron microscopic study. European review for medical and pharmacological sciences. 2022;26:270–277. doi: 10.26355/eurrev_202201_27777. [DOI] [PubMed] [Google Scholar]
- 15.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296:E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dramé M., Teguo M.T., Proye E., Hequet F., Hentzien M., Kanagaratnam L., Godaert L. Should RT-PCR be considered a gold standard in the diagnosis of COVID-19? J. Med. Virol. 2020;92(11):2312–2313. doi: 10.1002/jmv.25996. Epub 2020 Jul 14. PMID: 32383182; PMCID: PMC7267274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao A.T., Tong Y.X., Zhang S. False negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: rather than recurrence. J. Medi. Virol. 2020;92(10):1755–1756. doi: 10.1002/jmv.25855. Epub 2020 Jul 11. PMID: 32270882; PMCID: PMC7262304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suri J.S., Agarwal S., Gupta S.K., Puvvula A., Biswas M., Saba L., Bit A., Tandel G.S., Agarwal M., Patrick A. A narrative review on characterization of acute respiratory distress syndrome in COVID-19-infected lungs using artificial intelligence. Computers in Biology Medicine. 2021:104210. doi: 10.1016/j.compbiomed.2021.104210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biswas M., Kuppili V., Saba L., Edla D.R., Suri H.S., Cuadrado-Godia E., Laird J.R., Marinhoe R.T., Sanches J.M., Nicolaides A., Suri J.S. State-of-the-art review on deep learning in medical imaging. Front Biosci (Landmark Ed) 2019;24:392–426. doi: 10.2741/4725. [DOI] [PubMed] [Google Scholar]
- 20.Saba L., Biswas M., Kuppili V., Cuadrado Godia E., Suri H.S., Edla D.R., Omerzu T., Laird J.R., Khanna N.N., Mavrogeni S., Protogerou A., Sfikakis P.P., Viswanathan V., Kitas G.D., Nicolaides A., Gupta A., Suri J.S. The present and future of deep learning in radiology. Eur J Radiol. 2019;114:14–24. doi: 10.1016/j.ejrad.2019.02.038. [DOI] [PubMed] [Google Scholar]
- 21.Kuppili V., Biswas M., Sreekumar A., Suri H.S., Saba L., Edla D.R., Marinhoe R.T., Sanches J.M., Suri J.S. Extreme learning machine framework for risk stratification of fatty liver disease using ultrasound tissue characterization. Journal of Medical Systems. 2017;41:1–20. doi: 10.1007/s10916-017-0797-1. [DOI] [PubMed] [Google Scholar]
- 22.Chen X., Tang Y., Mo Y., Li S., Lin D., Yang Z., Yang Z., Sun H., Qiu J., Liao Y. A diagnostic model for coronavirus disease 2019 (COVID-19) based on radiological semantic and clinical features: a multi-center study. European radiology. 2020;30:4893–4902. doi: 10.1007/s00330-020-06829-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain P.K., Sharma N., Giannopoulos A.A., Saba L., Nicolaides A., Suri J.S. Hybrid deep learning segmentation models for atherosclerotic plaque in internal carotid artery B-mode ultrasound. Comput Biol Med. 2021;136:104721. doi: 10.1016/j.compbiomed.2021.104721. [DOI] [PubMed] [Google Scholar]
- 24.Jena B., Saxena S., Nayak G.K., Saba L., Sharma N., Suri J.S. Artificial intelligence-based hybrid deep learning models for image classification: the first narrative review. Comput Biol Med. 2021;137:104803. doi: 10.1016/j.compbiomed.2021.104803. [DOI] [PubMed] [Google Scholar]
- 25.Suri J.S., Agarwal S., Pathak R., Ketireddy V., Columbu M., Saba L., Gupta S.K., Faa G., Singh I.M., Turk M. Covlias 1.0: lung segmentation in COVID-19 computed tomography scans using hybrid deep learning artificial intelligence models. Diagnostics. 2021;11:1405. doi: 10.3390/diagnostics11081405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain P.K., Sharma N., Saba L., Paraskevas K.I., Kalra M.K., Johri A., Nicolaides A.N., Suri J.S. Automated deep learning-based paradigm for high-risk plaque detection in B-mode common carotid ultrasound scans: an asymptomatic Japanese cohort study. International Angiology: a Journal of the International Union of Angiology. 2022;41(1):9–23. doi: 10.23736/S0392-9590.21.04771-4. Epub 2021 Nov 26. PMID: 34825801. [DOI] [PubMed] [Google Scholar]
- 27.Suri J.S., Agarwal S., Carriero A., Paschè A., Danna P.S., Columbu M., Saba L., Viskovic K., Mehmedović A., Agarwal S. COVLIAS 1.0 vs. MedSeg: artificial intelligence-based comparative study for automated COVID-19 computed tomography lung segmentation in Italian and Croatian cohorts. Diagnostics. 2021;11:2367. doi: 10.3390/diagnostics11122367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suri J.S., Agarwal S., Elavarthi P., Pathak R., Ketireddy V., Columbu M., Saba L., Gupta S.K., Faa G., Singh I.M., Turk M., Chadha P.S., Johri A.M., Khanna N.N., Viskovic K., Mavrogeni S., Laird J.R., Pareek G., Miner M., Sobel D.W., Balestrieri A., Sfikakis P.P., Tsoulfas G., Protogerou A., Misra D.P., Agarwal V., Kitas G.D., Teji J.S., Al-Maini M., Dhanjil S.K., Nicolaides A., Sharma A., Rathore V., Fatemi M., Alizad A., Krishnan P.R., Ferenc N., Ruzsa Z., Gupta A., Naidu S., Kalra M.K. Inter-variability study of COVLIAS 1.0: hybrid deep learning models for COVID-19 lung segmentation in computed tomography. Diagnostics (Basel) 2021;11:2025. doi: 10.3390/diagnostics11112025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skandha S.S., Nicolaides A., Gupta S.K., Koppula V.K., Saba L., Johri A.M., Kalra M.S., Suri J.S. A hybrid deep learning paradigm for carotid plaque tissue characterization and its validation in multicenter cohorts using a supercomputer framework. Computers in biology and medicine. 2022;141:105131. doi: 10.1016/j.compbiomed.2021.105131. [DOI] [PubMed] [Google Scholar]
- 30.Gupta N., Gupta S.K., Pathak R.K., Jain V., Rashidi P., Suri J.S. Human activity recognition in artificial intelligence framework: a narrative review. Artificial Intelligence Review. 2022:1–54. doi: 10.1007/s10462-021-10116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saba L., Biswas M., Suri H.S., Viskovic K., Laird J.R., Cuadrado-Godia E., Nicolaides A., Khanna N.N., Viswanathan V., Suri J.S. Ultrasound-based carotid stenosis measurement and risk stratification in diabetic cohort: a deep learning paradigm. Cardiovasc Diagn Ther. 2019;9:439–461. doi: 10.21037/cdt.2019.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agarwal M., Saba L., Gupta S.K., Johri A.M., Khanna N.N., Mavrogeni S., Laird J.R., Pareek G., Miner M., Sfikakis P.P. Wilson disease tissue classification and characterization using seven artificial intelligence models embedded with 3D optimization paradigm on a weak training brain magnetic resonance imaging datasets: a supercomputer application. Medical Biological Engineering Computing. 2021;59:511–533. doi: 10.1007/s11517-021-02322-0. [DOI] [PubMed] [Google Scholar]
- 33.Sanagala S.S., Nicolaides A., Gupta S.K., Koppula V.K., Saba L., Agarwal S., Johri A.M., Kalra M.S., Suri J.S. Ten fast transfer learning models for carotid ultrasound plaque tissue characterization in augmentation framework embedded with heatmaps for stroke risk stratification. Diagnostics. 2021;11:2109. doi: 10.3390/diagnostics11112109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeCun Y., Denker J., Solla S. Optimal brain damage. Advances in neural information processing systems. 1989:2. [Google Scholar]
- 35.Zhu M., Gupta S. 2017. To Prune, or Not to Prune: Exploring the Efficacy of Pruning for Model Compression. arXiv preprint arXiv:1710.01878. [Google Scholar]
- 36.Band S.S., Janizadeh S., Chandra Pal S., Saha A., Chakrabortty R., Shokri M., Mosavi A. Novel ensemble approach of deep learning neural network (DLNN) model and particle swarm optimization (PSO) algorithm for prediction of gully erosion susceptibility. Sensors. 2020;20:5609. doi: 10.3390/s20195609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brodzicki A., Piekarski M., Jaworek-Korjakowska J. The whale optimization algorithm approach for deep neural networks. Sensors. 2021;21:8003. doi: 10.3390/s21238003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashraf N.M., Mostafa R.R., Sakr R.H., Rashad M. Optimizing hyperparameters of deep reinforcement learning for autonomous driving based on whale optimization algorithm. Plos one. 2021;16 doi: 10.1371/journal.pone.0252754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Acharya U.R., Mookiah M.R., Vinitha Sree S., Yanti R., Martis R.J., Saba L., Molinari F., Guerriero S., Suri J.S. Evolutionary algorithm-based classifier parameter tuning for automatic ovarian cancer tissue characterization and classification. Ultraschall Med. 2014;35:237–245. doi: 10.1055/s-0032-1330336. [DOI] [PubMed] [Google Scholar]
- 40.Horry M., Chakraborty S., Pradhan B., Paul M., Zhu J., Loh H.W., Barua P.D., Arharya U.R. 2022. Debiasing Pipeline Improves Deep Learning Model Generalization for X-Ray Based Lung Nodule Detection. arXiv preprint arXiv:2201.09563. [Google Scholar]
- 41.MedSeg February 10, 2022. https://htmlsegmentation.s3.eu-north-1.amazonaws.com/index.html
- 42.Eelbode T., Bertels J., Berman M., Vandermeulen D., Maes F., Bisschops R., Blaschko M.B. Optimization for medical image segmentation: theory and practice when evaluating with dice score or jaccard index. IEEE Transactions on Medical Imaging. 2020;39:3679–3690. doi: 10.1109/TMI.2020.3002417. [DOI] [PubMed] [Google Scholar]
- 43.Giavarina D. Understanding bland altman analysis. Biochemia medica. 2015;25:141–151. doi: 10.11613/BM.2015.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dewitte K., Fierens C., Stockl D., Thienpont L.M. Application of the Bland–Altman plot for interpretation of method-comparison studies: a critical investigation of its practice. Clinical chemistry. 2002;48:799–801. [PubMed] [Google Scholar]
- 45.Asuero A.G., Sayago A., Gonzalez A. The correlation coefficient: an overview. Critical reviews in analytical chemistry. 2006;36:41–59. [Google Scholar]
- 46.Taylor R. Interpretation of the correlation coefficient: a basic review. Journal of diagnostic medical sonography. 1990;6:35–39. [Google Scholar]
- 47.El-Baz A., Gimel’farb G., Suri J.S. first ed. CRC Press; 2015. Stochastic Modeling for Medical Image Analysis. [Google Scholar]
- 48.El-Baz A., Jiang X., Suri J.S. CRC Press; Boca Raton: 2016. Biomedical Image Segmentation: Advances and Trends. [Google Scholar]
- 49.El-Baz A.S., Acharya R., Mirmehdi M., Suri J.S. ume 2. Springer Science & Business Media; Boca Raton: 2011. (Multi Modality State-Of-The-Art Medical Image Segmentation and Registration Methodologies). [Google Scholar]
- 50.Maniruzzaman M., Rahman M.J., Al-MehediHasan M., Suri H.S., Abedin M.M., El-Baz A., Suri J.S. Accurate diabetes risk stratification using machine learning: role of missing value and outliers. Journal of Medical Systems. 2018;42:1–17. doi: 10.1007/s10916-018-0940-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maniruzzaman M., Kumar N., Abedin M.M., Islam M.S., Suri H.S., El-Baz A.S., Suri J.S. Comparative approaches for classification of diabetes mellitus data: machine learning paradigm. Computer methods and programs in biomedicine. 2017;152:23–34. doi: 10.1016/j.cmpb.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 52.Maniruzzaman M., Suri H.S., Kumar N., Abedin M.M., Rahman M.J., El-Baz A., Bhoot M., Teji J.S., Suri J.S. Risk factors of neonatal mortality and child mortality in Bangladesh. Journal of global health. 2018;8 doi: 10.7189/jogh.08.010421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maniruzzaman M., Jahanur Rahman M., Ahammed B., Abedin M.M., Suri H.S., Biswas M., El-Baz A., Bangeas P., Tsoulfas G., Suri J.S. Statistical characterization and classification of colon microarray gene expression data using multiple machine learning paradigms. Comput Methods Programs Biomed. 2019;176:173–193. doi: 10.1016/j.cmpb.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 54.Noor N.M., Than J.C., Rijal O.M., Kassim R.M., Yunus A., Zeki A.A., Anzidei M., Saba L., Suri J.S. Automatic lung segmentation using control feedback system: morphology and texture paradigm. Journal of medical systems. 2015;39:1–18. doi: 10.1007/s10916-015-0214-6. [DOI] [PubMed] [Google Scholar]
- 55.Acharya R.U., Faust O., Alvin A.P., Sree S.V., Molinari F., Saba L., Nicolaides A., Suri J.S. Symptomatic vs. asymptomatic plaque classification in carotid ultrasound. J Med Syst. 2012;36:1861–1871. doi: 10.1007/s10916-010-9645-2. [DOI] [PubMed] [Google Scholar]
- 56.Acharya U.R., Faust O., S V.S., Alvin A.P., Krishnamurthi G., Seabra J.C., Sanches J., Suri J.S. Understanding symptomatology of atherosclerotic plaque by image-based tissue characterization. Comput Methods Programs Biomed. 2013;110:66–75. doi: 10.1016/j.cmpb.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 57.Acharya U.R., Faust O., Sree S.V., Alvin A.P.C., Krishnamurthi G., Sanches J., Suri J.S. 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE; 2011. Atheromatic™: symptomatic vs. asymptomatic classification of carotid ultrasound plaque using a combination of HOS, DWT & texture; pp. 4489–4492. [DOI] [PubMed] [Google Scholar]
- 58.Acharya U.R., Mookiah M.R., Vinitha Sree S., Afonso D., Sanches J., Shafique S., Nicolaides A., Pedro L.M., Fernandes E.F.J., Suri J.S. Atherosclerotic plaque tissue characterization in 2D ultrasound longitudinal carotid scans for automated classification: a paradigm for stroke risk assessment. Med Biol Eng Comput. 2013;51:513–523. doi: 10.1007/s11517-012-1019-0. [DOI] [PubMed] [Google Scholar]
- 59.Molinari F., Liboni W., Pavanelli E., Giustetto P., Badalamenti S., Suri J.S. 2007 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE; 2007. Accurate and automatic carotid plaque characterization in contrast enhanced 2-D ultrasound images; pp. 335–338. [DOI] [PubMed] [Google Scholar]
- 60.Acharya U., Vinitha Sree S., Mookiah M., Yantri R., Molinari F., Zieleźnik W., Małyszek-Tumidajewicz J., Stępień B., Bardales R., Witkowska A. Diagnosis of Hashimoto's thyroiditis in ultrasound using tissue characterization and pixel classification. Proceedings of the Institution of Mechanical Engineers, Part H: Journal of Engineering in medicine. 2013;227:788–798. doi: 10.1177/0954411913483637. [DOI] [PubMed] [Google Scholar]
- 61.Biswas M., Kuppili V., Edla D.R., Suri H.S., Saba L., Marinhoe R.T., Sanches J.M., Suri J.S. Symtosis: a liver ultrasound tissue characterization and risk stratification in optimized deep learning paradigm. Comput Methods Programs Biomed. 2018;155:165–177. doi: 10.1016/j.cmpb.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 62.Saba L., Sanagala S.S., Gupta S.K., Koppula V.K., Johri A.M., Khanna N.N., Mavrogeni S., Laird J.R., Pareek G., Miner M., Sfikakis P.P., Protogerou A., Misra D.P., Agarwal V., Sharma A.M., Viswanathan V., Rathore V.S., Turk M., Kolluri R., Viskovic K., Cuadrado-Godia E., Kitas G.D., Sharma N., Nicolaides A., Suri J.S. Multimodality carotid plaque tissue characterization and classification in the artificial intelligence paradigm: a narrative review for stroke application. Ann Transl Med. 2021;9:1206. doi: 10.21037/atm-20-7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banchhor S.K., Londhe N.D., Araki T., Saba L., Radeva P., Laird J.R., Suri J.S. Wall-based measurement features provides an improved IVUS coronary artery risk assessment when fused with plaque texture-based features during machine learning paradigm. Computers in biology and medicine. 2017;91:198–212. doi: 10.1016/j.compbiomed.2017.10.019. [DOI] [PubMed] [Google Scholar]
- 64.Acharya U.R., Saba L., Molinari F., Guerriero S., Suri J.S. 2012 Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE; 2012. Ovarian tumor characterization and classification: a class of GyneScan™ systems; pp. 4446–4449. [DOI] [PubMed] [Google Scholar]
- 65.Pareek G., Acharya U.R., Sree S.V., Swapna G., Yantri R., Martis R.J., Saba L., Krishnamurthi G., Mallarini G., El-Baz A. Prostate tissue characterization/classification in 144 patient population using wavelet and higher order spectra features from transrectal ultrasound images. Technology in cancer research treatment. 2013;12:545–557. doi: 10.7785/tcrt.2012.500346. [DOI] [PubMed] [Google Scholar]
- 66.Shrivastava V.K., Londhe N.D., Sonawane R.S., Suri J.S. Exploring the color feature power for psoriasis risk stratification and classification: a data mining paradigm. Computers in Biology and Medicine. 2015;65:54–68. doi: 10.1016/j.compbiomed.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 67.Shrivastava V.K., Londhe N.D., Sonawane R.S., Suri J.S. A novel and robust Bayesian approach for segmentation of psoriasis lesions and its risk stratification. Computer methods and programs in biomedicine. 2017;150:9–22. doi: 10.1016/j.cmpb.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 68.Shrivastava V.K., Londhe N.D., Sonawane R.S., Suri J.S. Reliable and accurate psoriasis disease classification in dermatology images using comprehensive feature space in machine learning paradigm. Expert Systems with Applications. 2015;42:6184–6195. [Google Scholar]
- 69.Acharya U.R., Kannathal N., Ng E., Min L.C., Suri J.S. 2006 International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE; 2006. Computer-based classification of eye diseases; pp. 6121–6124. [DOI] [PubMed] [Google Scholar]
- 70.Saba L., Suri J.S. CRC Press; Boca Raton: 2013. Multi-Detector CT Imaging: Principles, Head, Neck, and Vascular Systems. [Google Scholar]
- 71.Murgia A., Erta M., Suri J.S., Gupta A., Wintermark M., Saba L. CT imaging features of carotid artery plaque vulnerability. Annals of Translational Medicine. 2020;8 doi: 10.21037/atm-2020-cass-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saba L., di Martino M., Siotto P., Anzidei M., Argiolas G.M., Porcu M., Suri J.S., Wintermark M. Radiation dose and image quality of computed tomography of the supra-aortic arteries: a comparison between single-source and dual-source CT Scanners. Journal of Neuroradiology. 2018;45:136–141. doi: 10.1016/j.neurad.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 73.Wu J., Pan J., Teng D., Xu X., Feng J., Chen Y.-C. Interpretation of CT signs of 2019 novel coronavirus (COVID-19) pneumonia. European radiology. 2020;30:5455–5462. doi: 10.1007/s00330-020-06915-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.De Wever W., Meersschaert J., Coolen J., Verbeken E., Verschakelen J.A. The crazy-paving pattern: a radiological-pathological correlation. Insights into imaging. 2011;2:117–132. doi: 10.1007/s13244-010-0060-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Niu R., Ye S., Li Y., Ma H., Xie X., Hu S., Huang X., Ou Y., Chen J. Chest CT features associated with the clinical characteristics of patients with COVID-19 pneumonia. Annals of medicine. 2021;53:169–180. doi: 10.1080/07853890.2020.1851044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salehi S., Abedi A., Balakrishnan S., Gholamrezanezhad A. Coronavirus disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. AJR Am J Roentgenol. 2020;215:87–93. doi: 10.2214/AJR.20.23034. [DOI] [PubMed] [Google Scholar]
- 77.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical coronavirus disease 2019 (COVID-19) pneumonia: relationship to negative RT-PCR testing. Radiology. 2020;296:E41–E45. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gozes O., Frid-Adar M., Greenspan H., Browning P.D., Zhang H., Ji W., Bernheim A., Siegel E. 2020. Rapid Ai Development Cycle for the Coronavirus (Covid-19) Pandemic: Initial Results for Automated Detection & Patient Monitoring Using Deep Learning Ct Image Analysis. arXiv preprint arXiv:.05037. [Google Scholar]
- 79.Shalbaf A., Vafaeezadeh M. Automated detection of COVID-19 using ensemble of transfer learning with deep convolutional neural network based on CT scans. International journal of computer assisted radiology surgery. 2021;16:115–123. doi: 10.1007/s11548-020-02286-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang X., He X., Zhao J., Zhang Y., Zhang S., Xie P. 2020. COVID-CT-dataset: a CT Scan Dataset about COVID-19. arXiv preprint arXiv:.13865. [Google Scholar]
- 81.Cau R., Falaschi Z., Pasche A., Danna P., Arioli R., Arru C.D., Zagaria D., Tricca S., Suri J.S., Karla M.K., Carriero A., Saba L. Computed tomography findings of COVID-19 pneumonia in intensive care unit-patients. J Public Health Res. 2021;10 doi: 10.4081/jphr.2021.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang X., He X., Zhao J., Zhang Y., Zhang S., Xie P. 2020. COVID-CT-dataset: a CT Scan Dataset about COVID-19. arXiv preprint arXiv:2003.13865. [Google Scholar]
- 83.Setio A.A.A., Traverso A., De Bel T., Berens M.S., Van Den Bogaard C., Cerello P., Chen H., Dou Q., Fantacci M.E., Geurts B. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: the LUNA16 challenge. Medical image analysis. 2017;42:1–13. doi: 10.1016/j.media.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 84.Kogilavani S., Prabhu J., Sandhiya R., Kumar M.S., Subramaniam U., Karthick A., Muhibbullah M., Imam S.B.S. COVID-19 detection based on lung ct scan using deep learning techniques. Computational and Mathematical Methods in Medicine. 2022:2022. doi: 10.1155/2022/7672196. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 85.Simonyan K., Zisserman A. 2014. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv preprint arXiv: [Google Scholar]
- 86.Iandola F., Moskewicz M., Karayev S., Girshick R., Darrell T., Keutzer K. 2014. Densenet: Implementing Efficient Convnet Descriptor Pyramids. arXiv preprint arXiv:1404.1869. [Google Scholar]
- 87.Howard A.G., Zhu M., Chen B., Kalenichenko D., Wang W., Weyand T., Andreetto M., Adam H., Mobilenets . 2017. Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv preprint arXiv:1704.04861. [Google Scholar]
- 88.Chollet F. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Xception: deep learning with depthwise separable convolutions; pp. 1251–1258. [Google Scholar]
- 89.Zoph B., Vasudevan V., Shlens J., Le Q.V. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2018. Learning transferable architectures for scalable image recognition; pp. 8697–8710. [Google Scholar]
- 90.Tan M., Le Q. International Conference on Machine Learning, PMLR. 2019. Efficientnet: rethinking model scaling for convolutional neural networks; pp. 6105–6114. [Google Scholar]
- 91.Paluru N., Dayal A., Jenssen H.B., Sakinis T., Cenkeramaddi L.R., Prakash J., Yalavarthy P.K., Systems L. Anam-Net: anamorphic depth embedding-based lightweight CNN for segmentation of anomalies in COVID-19 chest CT images. IEEE Transact. Neural Networks Learn. Syst. 2021;32:932–946. doi: 10.1109/TNNLS.2021.3054746. 3rd ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.COVID-19 database. February. 2022;10 https://radiopaedia.org/articles/covid-19-4?lang=gb 2022. [Google Scholar]
- 93.Paszke A., Chaurasia A., Kim S., Culurciello E. 2016. Enet: A Deep Neural Network Architecture for Real-Time Semantic Segmentation. arXiv preprint arXiv:1606.02147. [Google Scholar]
- 94.Zhou Z., Siddiquee M.M.R., Tajbakhsh N., Liang J. UNet++: redesigning skip connections to exploit multiscale features in image segmentation. IEEE Transactions on Medical Imaging. 2020;39:1856–1867. doi: 10.1109/TMI.2019.2959609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Y., Zhou Q., Liu J., Xiong J., Gao G., Wu X., Latecki L.J. 2019 IEEE International Conference on Image Processing (ICIP) IEEE; 2019. Lednet: a lightweight encoder-decoder network for real-time semantic segmentation; pp. 1860–1864. [Google Scholar]
- 96.Cai W., Liu T., Xue X., Luo G., Wang X., Shen Y., Fang Q., Sheng J., Chen F., Liang T. CT quantification and machine-learning models for assessment of disease severity and prognosis of COVID-19 patients. Acad Radiol. 2020;27:1665–1678. doi: 10.1016/j.acra.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Saood A., Hatem I. COVID-19 lung CT image segmentation using deep learning methods: U-Net versus SegNet. BMC Medical Imaging. 2021;21:1–10. doi: 10.1186/s12880-020-00529-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Biswas M., Kuppili V., Saba L., Edla D.R., Suri H.S., Sharma A., Cuadrado-Godia E., Laird J.R., Nicolaides A., Suri J.S. Deep learning fully convolution network for lumen characterization in diabetic patients using carotid ultrasound: a tool for stroke risk. Medical Biological Engineering Computing. 2019;57:543–564. doi: 10.1007/s11517-018-1897-x. [DOI] [PubMed] [Google Scholar]
- 99.Badrinarayanan V., Kendall A., Cipolla R. SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39:2481–2495. doi: 10.1109/TPAMI.2016.2644615. [DOI] [PubMed] [Google Scholar]
- 100.Fleetwood K. Proceedings of Mathematics and Statistics of Complex Systems (MASCOS) One Day Symposium, 26th November. 2004. An introduction to differential evolution; pp. 785–791. Brisbane, Australia. [Google Scholar]
- 101.Price K.V. Springer; 2013. Differential Evolution, Handbook of Optimization; pp. 187–214. [Google Scholar]
- 102.Singh D., Kumar V., Kaur M. Classification of COVID-19 patients from chest CT images using multi-objective differential evolution–based convolutional neural networks. European Journal of Clinical Microbiology & Infectious Diseases. 2020;39:1379–1389. doi: 10.1007/s10096-020-03901-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Baştürk A., Günay E. Efficient edge detection in digital images using a cellular neural network optimized by differential evolution algorithm. Expert Systems with applications. 2009;36:2645–2650. [Google Scholar]
- 104.Ruse M. Charles Darwin's theory of evolution: an analysis. Journal of the History of Biology. 1975:219–241. doi: 10.1007/BF00130439. [DOI] [PubMed] [Google Scholar]
- 105.Kozek T., Roska T., Chua L.O. Genetic algorithm for CNN template learning. IEEE Transactions on Circuits and Systems I: Fundamental Theory and Applications. 1993;40:392–402. [Google Scholar]
- 106.Sun Y., Xue B., Zhang M., Yen G.G., Lv J. Automatically designing CNN architectures using the genetic algorithm for image classification. IEEE transactions on cybernetics. 2020;50:3840–3854. doi: 10.1109/TCYB.2020.2983860. [DOI] [PubMed] [Google Scholar]
- 107.Kennedy J., Eberhart R. Proceedings of ICNN'95-international Conference on Neural Networks. IEEE; 1995. Particle swarm optimization; pp. 1942–1948. [Google Scholar]
- 108.Navaneeth B., Suchetha M. PSO optimized 1-D CNN-SVM architecture for real-time detection and classification applications. Computers in biology and medicine. 2019;108:85–92. doi: 10.1016/j.compbiomed.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 109.Wang Y., Zhang H., Zhang G., cPSO-CNN An efficient PSO-based algorithm for fine-tuning hyper-parameters of convolutional neural networks. Swarm and Evolutionary Computation. 2019;49:114–123. [Google Scholar]
- 110.Mirjalili S., Lewis A. The whale optimization algorithm. Advances in engineering software. 2016;95:51–67. [Google Scholar]
- 111.Dixit U., Mishra A., Shukla A., Tiwari R. Texture classification using convolutional neural network optimized with whale optimization algorithm. SN Applied Sciences. 2019;1:1–11. [Google Scholar]
- 112.Rana N., Latiff M.S.A., Abdulhamid S.i.M., Chiroma H. Whale optimization algorithm: a systematic review of contemporary applications, modifications and developments. Neural Computing and Applications. 2020;32:16245–16277. [Google Scholar]
- 113.Yuan T., Liu W., Han J., Lombardi F. High performance CNN accelerators based on hardware and algorithm co-optimization. IEEE Transactions on Circuits and Systems I: Regular Papers. 2020;68:250–263. [Google Scholar]
- 114.Zhang J., Raj P., Zarar S., Ambardekar A., Garg S. CompAct: on-chip com pression of act ivations for low power systolic array based CNN acceleration. ACM Transactions on Embedded Computing Systems (TECS) 2019;18:1–24. [Google Scholar]
- 115.Saba L., Banchhor S.K., Londhe N.D., Araki T., Laird J.R., Gupta A., Nicolaides A., Suri J.S. Web-based accurate measurements of carotid lumen diameter and stenosis severity: an ultrasound-based clinical tool for stroke risk assessment during multicenter clinical trials. Comput Biol Med. 2017;91:306–317. doi: 10.1016/j.compbiomed.2017.10.022. [DOI] [PubMed] [Google Scholar]
- 116.Saba L., Than J.C., Noor N.M., Rijal O.M., Kassim R.M., Yunus A., Ng C.R., Suri J.S. Inter-observer variability analysis of automatic lung delineation in normal and disease patients. J Med Syst. 2016;40:142. doi: 10.1007/s10916-016-0504-7. [DOI] [PubMed] [Google Scholar]
- 117.Molinari F., Meiburger K.M., Saba L., Acharya U.R., Famiglietti L., Georgiou N., Nicolaides A., Mamidi R.S., Kuper H., Suri J.S. Springer; 2014. Automated Carotid IMT Measurement and its Validation in Low Contrast Ultrasound Database of 885 Patient Indian Population Epidemiological Study: Results of AtheroEdge® Software, Multi-Modality Atherosclerosis Imaging and Diagnosis; pp. 209–219. [Google Scholar]
- 118.Mirmehdi M. Imperial College Press; 2008. Handbook of Texture Analysis. [Google Scholar]
- 119.He G., Ping A., Wang X., Zhu Y. 2019 10th International Conference on Information Technology in Medicine and Education (ITME) IEEE; 2019. Alzheimer's disease diagnosis model based on three-dimensional full convolutional DenseNet; pp. 13–17. [Google Scholar]
- 120.Ouhami M., Es-Saady Y., Hajji M.E., Hafiane A., Canals R., Yassa M.E. Deep Transfer Learning Models for Tomato Disease Detection, International Conference on Image and Signal Processing. Springer; 2020. pp. 65–73. [Google Scholar]
- 121.Ruiz J., Mahmud M., Modasshir M., Shamim Kaiser M., Alzheimer’s Disease Neuroimaging Initiative f.t. 3D DenseNet Ensemble in 4-way Classification of Alzheimer's Disease, International Conference on Brain Informatics. Springer; 2020. pp. 85–96. [Google Scholar]
- 122.Saba L., Sanagala S.S., Gupta S.K., Koppula V.K., Laird J.R., Viswanathan V., Sanches M.J., Kitas G.D., Johri A.M., Sharma N. A multicenter study on carotid ultrasound plaque tissue characterization and classification using six deep artificial intelligence models: a stroke application. IEEE Transactions on Instrumentation Measurement. 2021;70:1–12. [Google Scholar]
- 123.Jain P.K., Sharma N., Saba L., Paraskevas K.I., Kalra M.K., Johri A., Laird J.R., Nicolaides A.N., Suri J.S. Unseen artificial intelligence—deep learning paradigm for segmentation of low atherosclerotic plaque in carotid ultrasound: a multicenter cardiovascular study. Diagnostics. 2021;11:2257. doi: 10.3390/diagnostics11122257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.El-Baz A., Suri J.S. CRC Press; Boca Raton: 2019. Big Data in Multimodal Medical Imaging. [Google Scholar]
- 125.Sudeep P., Palanisamy P., Rajan J., Baradaran H., Saba L., Gupta A., Suri J.S. Speckle reduction in medical ultrasound images using an unbiased non-local means method. Biomedical Signal Processing Control. 2016;28:1–8. [Google Scholar]
- 126.Suri J.S., Liu K., Singh S., Laxminarayan S.N., Zeng X., Reden L. Shape recovery algorithms using level sets in 2-D/3-D medical imagery: a state-of-the-art review. IEEE Transactions on information technology in biomedicine. 2002;6:8–28. doi: 10.1109/4233.992158. [DOI] [PubMed] [Google Scholar]
- 127.Suri J.S., Agarwal S., Gupta S., Puvvula A., Viskovic K., Suri N., Alizad A., El-Baz A., Saba L., Fatemi M., Naidu D.S. Systematic review of artificial intelligence in acute respiratory distress syndrome for COVID-19 lung patients: a biomedical imaging perspective. IEEE J Biomed Health Inform. 2021;25:4128–4139. doi: 10.1109/JBHI.2021.3103839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Paul S., Maindarkar M., Saxena S., Saba L., Turk M., Kalra M., Krishnan P.R., Suri J.S. Bias investigation in artificial intelligence systems for early detection of Parkinson's disease: a narrative review. Diagnostics. 2022;12:166. doi: 10.3390/diagnostics12010166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Suri J.S., Bhagawati M., Paul S., Protogeron A., Sfikakis P.P., Kitas G.D., Khanna N.N., Ruzsa Z., Sharma A.M., Saxena S. Understanding the bias in machine learning systems for cardiovascular disease risk assessment: the first of its kind review. Computers in Biology and Medicine. 2022:105204. doi: 10.1016/j.compbiomed.2021.105204. [DOI] [PubMed] [Google Scholar]