Abstract
Microglia have fundamental roles in health and disease; however, effects of age, sex, and genetic factors on human microglia have not been fully explored. We applied bulk and single‐cell approaches to comprehensively characterize human microglia transcriptomes and their associations with age, sex, and APOE. We identified a novel microglial signature, characterized its expression in bulk tissue and single‐cell microglia transcriptomes. We discovered microglial co‐expression network modules associated with age, sex, and APOE‐ε4 that are enriched for lipid and carbohydrate metabolism genes. Integrated analyses of modules with single‐cell transcriptomes revealed significant overlap between age‐associated module genes and both pro‐inflammatory and disease‐associated microglial clusters. These modules and clusters harbor known neurodegenerative disease genes including APOE, PLCG2, and BIN1. Meta‐analyses with published bulk and single‐cell microglial datasets further supported our findings. Thus, these data represent a well‐characterized human microglial transcriptome resource and highlight age, sex, and APOE‐related microglial immunometabolism perturbations with potential relevance in neurodegeneration.
Keywords: APOE, lipid metabolism, microglia, neurodegeneration, single cell, transcriptomics
Using bulk and single‐cell approaches, we identified microglial subclusters enriched for different biological functions. Activated microglia were more abundant with aging and lipid localization genes were upregulated. In APOE‐ε4 carriers, homeostatic microglia were reduced along with downregulation of carbohydrate metabolism genes, whereas cholesterol absorption genes were unregulated in macrophages and activated microglia.
Abbreviations
- AD
Alzheimer’s disease
- AMP‐AD
Accelerating Medicines Partnership Alzheimer’s Disease
- CNS
central nervous system
- CQN
Conditional Quantile Normalization
- DAM
disease‐associated microglia
- DNET
dysembryoplastic neuroepithelial tumor
- FACS
Fluorescence‐activated cell sorting
- GBM
glioblastoma multiforme
- GEM
gel beads‐in‐emulsion
- ID
identifier
- INF
Infinity
- MACS
Magnetic‐activated cell sorting
- MSBB
Mount Sinai brain bank
- MM
Module membership
- PCA
Principal components analysis
- RPKM
reads per kilo bases per million
- QC
quality control
- ROS‐MAP
Rush University Religious Orders Study and Memory and Aging Project
- scRNAseq
single cell RNA sequencing
- snRNAseq
single nuclei RNA sequencing
- UMAP
Uniform Manifold Approximation and Projection
- UMI
Unique Molecular Identifier
- WGCNA
weighted gene co‐expression network analysis
1. INTRODUCTION
Microglia are the resident macrophages of the central nervous system (CNS), responsible for clearance of cellular debris and pathological protein aggregates. In the healthy brain, they exist in a homeostatic state and can be induced to a reactive state in response to changes in the CNS microenvironment, such as inflammation and neuronal damage (Masuda et al., 2020). They are fundamental to maintaining brain homeostasis during development, aging, and disease; therefore, microglial dysfunction could ultimately lead to neurodegeneration (Li & Barres, 2018). Microglia are integral to the pathophysiology of neurodegenerative diseases, including Alzheimer's disease (AD) and multiple sclerosis, with chronic inflammation implicated as a contributing factor (Hickman et al., 2018; Keren‐Shaul et al., 2017; Krasemann et al., 2017).
Fresh human brain tissue studies are imperative to the characterization of the microglial transcriptome in health and disease; however, accessibility is limited. Although single nuclei studies using frozen tissue provide an easier alternative, recent studies have demonstrated limitations in detecting substantial populations of less abundant cell types (Del‐Aguila et al., 2019; Mathys et al., 2019). Additionally, it was recently reported that many microglial activation genes are expressed in the cytosol and therefore are likely to be missed by single nuclei RNA sequencing (snRNAseq; Thrupp et al., 2020). Recent single‐cell studies aiming to characterize microglial gene expression using fresh tissue have highlighted the heterogeneity in microglial phenotypes (Masuda et al., 2019; Olah et al., 2020; Sankowski et al., 2019). This has revealed that phenotypic changes are not binary but rather a spectrum of states in which microglia can simultaneously co‐exist during transition from homeostatic to more reactive states. Additionally, these different subsets could have specialized functions in brain homeostasis and dysfunction. Thus, it is increasingly important to characterize these heterogeneous subpopulations to understand their roles in health and disease. This could also help facilitate the design of novel therapeutic approaches to target‐specific subpopulations of cells and modulate their activity (Li & Barres, 2018).
Obtaining fresh human tissue from neurosurgeries allows us to study the mechanisms of microglial function in living cells. Unfortunately, this tissue is usually excised from surgical procedures for tumor resection or relieving temporal lobe epilepsy, rendering it difficult to distinguish between normal and disease‐affected tissue. Darmanis et al. (2017) investigated the effect of GBM tumors on CNS cell types and surrounding tissue, revealing that myeloid cells are greatly affected by the tumor microenvironment. They found that peri‐tumor myeloid populations were primarily pro‐inflammatory microglia compared to macrophages within the tumor core (Darmanis et al., 2017). In temporal lobe epilepsy, two distinct microglial phenotypes have been identified with microglia present in sclerotic areas with few neurons expressing markers of activation, including anti‐inflammatory cytokine IL10 (Kinoshita & Koyama, 2021; Morin‐Brureau et al., 2018). The other phenotype occurs transiently following a seizure, with secretion of interleukins CXCL8 and IL1B mediated by the NLRP3 inflammasome (Morin‐Brureau et al., 2018).
Microglial expression has also been shown to be affected by aging (Galatro et al., 2017; Olah et al., 2018); however, few studies have investigated the effects of sex and genetic factors on human microglia. Sex differences in microglia have been previously reported in mice, with females being predisposed to harboring more activated microglia than males (Frigerio et al., 2019; Nelson et al., 2017; Stephen et al., 2019). APOE, a lipoprotein of which the ε4 allele (APOE‐ε4) is a major risk factor for AD and also implicated in other neurodegenerative diseases (Yamazaki et al., 2019]), is upregulated in disease‐associated microglia (DAM) in mice and humans, but downregulated in astrocyte and oligodendrocyte subpopulations (Grubman et al., 2019; Hammond et al., 2019; Keren‐Shaul et al., 2017; Mathys et al., 2019). In microglia and neurons, APOE interacts with LDL receptors to facilitate endocytosis of cholesterol and phospholipids and modulate lipid homeostasis in the brain (Gamache et al., 2020). Such studies provide growing support for cell type‐specific functions of APOE; however, its effects on microglia remain to be fully elucidated. Thereby, identifying age, sex, and APOE‐associated pathways in microglia will provide greater insight into the functions of specific microglial subsets in relation to these risk factors. Interindividual variability and diversity in functional states makes targeting specific microglial subsets in disease challenging for modulating these cells (Li & Barres, 2018). Identifying the mechanisms regulating microglial homeostasis and activation can allow us to manipulate these cells for therapeutic purposes.
In this study, we leveraged both bulk and single‐cell approaches to provide a comprehensive characterization of the adult human microglial transcriptome. We obtained fresh intraoperative neurosurgical brain tissue and isolated an enriched population of microglial cells to investigate transcriptional changes associated with age, sex, and APOE‐ε4 in bulk microglia and further explored these in single microglial cells. Our findings support age‐, sex‐, and APOE‐related microglial transcriptome changes involving lipid and carbohydrate metabolic pathways and implicate microglial immunometabolism perturbations relevant to neurodegenerative diseases.
2. RESULTS
To uncover microglial transcriptional profiles and their associations with age, sex, and APOE, we performed microglial cell type‐specific and single‐cell RNA sequencing (scRNAseq) studies in fresh human brain tissue. We obtained neurosurgical tissue unaffected by the primary disease process from 19 human donors (Figure S1). Microglia were isolated by CD11b+ microbead selection followed by FACS sorting of cells expressing the CD11b+/CD45intermediate microglial signature to acquire a more purified population. These samples underwent bulk microglia RNAseq, with subsets of these and additional samples also undergoing 10x scRNAseq (n = 5) and bulk tissue RNAseq (n = 9; Figure 1a; Table 1). Validation of sorted microglia using qPCR showed the expected CD11b +/CD45 intermediate/P2RY12 + microglial signature (Li & Barres, 2018) with no expression of other cell type markers, indicating that we isolated a highly enriched microglial population (Figure S2).
FIGURE 1.
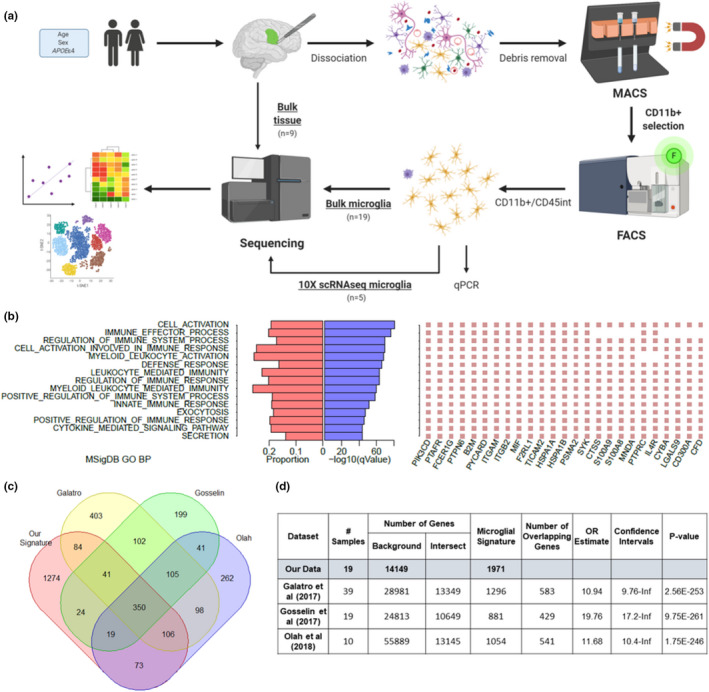
Characterization of our core human microglial signature. (a) Schematic illustrating our experimental approach for isolating microglial populations from fresh brain tissue and data analyses. [Created with BioRender.com] (b) MSigDB GO terms enriched in our microglial signature genes and top 25 genes for each. (c) Venn diagram showing number of overlapping genes between our microglial signature and those previously reported from Galatro et al. (2017), Gosselin et al. (2017) and Olah et al. (2018). (d) Hypergeometric tests of overrepresentation showing overlap with the published signatures
TABLE 1.
Demographics table of all samples included in the study and the associated demographics
| ID | Group | Sex | APOE genotype | Age | Ethnicity | Brain hemisphere | Brain region | Diagnosis | Bulk microglia | Bulk tissue | Single‐cell microglia | Surgical approach |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | F, APOEe4‐neg | F | 33 | 19 | White | Right | Temporal | Oligodendroglioma | X | Right temporal craniotomy for parahippocampal/brainstem low‐grade glioma | ||
| 13 | F, APOEe4‐neg | F | 33 | 35 | Black or African American | Left | Temporal lobe | Epilepsy | X | Left anterior temporal lobectomy | ||
| 1 | F, APOEe4‐neg | F | 33 | 37 | Hispanic or Latino | Right | Cerebellum | Metastatic carcinoma | X | Right craniotomy for tumor | ||
| 23 | F, APOEe4‐neg | F | 33 | 37 | White | Right | Frontal | Oligodendroglioma | X | Right awake craniotomy for perirolandic low‐grade glioma | ||
| 20 | F, APOEe4‐neg | F | 23 | 45 | White | Left | Temporal | GBM | X | Left awake craniotomy with cortical and subcortical mapping for 2 h for insular high‐grade glioma | ||
| 10 | F, APOEe4‐neg | F | 33 | 62 | White | Right | Temporal insular | GBM | X | X | X | Right temporal craniotomy for temporal insular high‐grade glioma with 5‐ALA |
| 24 | F, APOEe4‐neg | F | 33 | 64 | Hispanic or Latino | Left | Temporal | GBM | X | Left temporal craniotomy for high‐grade glioma | ||
| 12 | F, APOEe4‐neg | F | 33 | 65 | White | Left | Temporal | Astrocytoma | X | Left temporal craniotomy for low‐grade glioma | ||
| 21 | F, APOEe4‐neg | F | 33 | 66 | White | Left | Temporal | Meningioma | X | Left temporal craniotomy for tentorial meningioma | ||
| 4 | F, APOEe4‐neg | F | 33 | 71 | White | Right | Temporal lobe | Astrocytoma | X | Right temporal craniotomy for glioma | ||
| 17 | F, APOEe4‐pos | F | 24 | 37 | White | Left | Insula | Astrocytoma | X | Left awake craniotomy with cortical and subcortical mapping for insular glioma | ||
| 14 | F, APOEe4‐pos | F | 34 | 47 | White | Right | Parietal cortex | GBM | X | X | Right parietal craniotomy for parieto‐occipital high‐grade glioma with brain path tubular retractor | |
| 8 | F, APOEe4‐pos | F | 34 | 62 | White | Right | Temporal/occipital | GBM | X | Right temporal craniotomy for high‐grade glioma | ||
| 15 | F, APOEe4‐pos | F | 34 | 63 | White | Right | Temporal | Metastatic carcinoma | X | X | X | Right Temporal craniotomy for tumor resection |
| 9 | M, APOEe4‐neg | M | 33 | 19 | African American | Left | Temporal | DNET | X | Left temporal craniotomy for resection of mesiotemporal glioma with BrainPath retractor | ||
| 2 | M, APOEe4‐neg | M | 23 | 25 | White | Left | Mesial temporal lobe | Epilepsy | X | left anterior temporal lobectomy | ||
| 5 | M, APOEe4‐neg | M | 33 | 27 | Hispanic or Latino | Right | Mesial temporal cortex | Epilepsy | X | X | Right temporal lobectomy | |
| 6 | M, APOEe4‐neg | M | 33 | 27 | Hispanic or Latino | Right | Anterior temporal cortex | Epilepsy | X | Right temporal lobectomy | ||
| 26 | M, APOEe4‐neg | M | 33 | 27 | White | Left | Temporal | Astrocytoma | X | Left awake craniotomy for tumor resection, non‐IMRI | ||
| 19 | M, APOEe4‐neg | M | 33 | 31 | Hispanic or Latino | Right | Frontal | Oligodendroglioma | X | X | Right awake craniotomy with brain mapping and electrocorticography for tumor resection | |
| 3 | M, APOEe4‐neg | M | 33 | 34 | White | Right | Temporal insular | Oligodendroglioma | X | Right awake temporal craniotomy for temporal insular glioma | ||
| 7 | M, APOEe4‐neg | M | 23 | 55 | White | Right | Temporal lobe | GBM | X | Right temporal craniotomy for high‐grade glioma | ||
| 16 | M, APOEe4‐neg | M | 33 | 59 | White | Right | Frontal | Oligodendroglioma | X | Right awake craniotomy with cortical and subcortical mapping for low‐grade glioma | ||
| 18 | M, APOEe4‐neg | M | 33 | 66 | Middle Eastern | Left | Frontal | GBM | X | X | Left frontal craniotomy for tumor resection | |
| 22 | M, APOEe4‐pos | M | 34 | 67 | White | Left | Frontal | Meningioma | X | Left pterional craniotomy for tumor resection | ||
| 25 | M, APOEe4‐NA | M | NA | 63 | White | Left | Frontal | GBM | X | Left frontal craniotomy for opercular high‐grade glioma |
Samples were either included for bulk microglial RNAseq, bulk tissue RNAseq, 10× Genomics single cell RNAseq or on multiple platforms. Samples 5 and 6 were from two brain regions of the same person.
M = male, F = female, GBM = glioblastoma multiforme, DNET = dysembryoplastic neuroepithelial tumor, males and APOE‐ε4 carriers are shown in bold.
2.1. Identification of a core human microglial transcriptional signature
To define a core human microglial signature, we calculated log2 fold change and q‐values of differential expression for each gene between bulk microglia RNAseq data in our study and bulk brain RNAseq data from 7 AMP‐AD datasets provided by Mayo Clinic (Allen et al., 2016]), Mount Sinai Brain Bank (Wang et al., 2018), and Rush University Religious Orders Study and Memory and Aging Project (ROSMAP; De Jager et al., 2018) representing 6 brain regions from 515 human samples. Using a cutoff of 4‐fold greater expression in our bulk microglia and a q‐value threshold of 0.05, we identified 1971 genes (Table S1–S6). These genes were expressed at significantly greater levels in our bulk microglial transcriptome data in comparison with each of the bulk brain transcriptome datasets. Therefore, we considered these 1971 genes as the core microglial signature in our dataset. This signature comprises several known marker genes, with 12.7% of the genes being BRETIGEA (McKenzie et al., 2018) microglial genes, suggesting that it also likely harbors novel microglial markers of interest (Table S7). GO enrichment using MSigDB showed that this signature was enriched for genes involved in immune‐related and inflammatory response pathways as would be expected, and leukocyte‐mediated immunity (Figure 1b).
To determine the ability of bulk brain tissue data to capture microglial genes, we assessed the expression levels of our microglial signature genes in each of the 7 AMP‐AD bulk brain RNAseq datasets. Of the 1971 microglial signature genes in our study, 37–47% were captured in these bulk brain datasets (Figure S3A‐B). Our microglial signature genes comprised 3.6–4.5% of the expressed bulk brain transcriptome, consistent with prior estimations (Mathys et al., 2019; Wang et al., 2020). We next compared bulk microglia RNAseq transcript levels to that obtained from bulk tissue RNAseq of neurosurgical fresh brain tissue samples. Bulk fresh brain tissue does not capture all microglial marker genes, as demonstrated by the low correlation between bulk tissue and bulk microglia data (R = 0.46, p = 0.94; Figures S3C, S4, Table S6). This reiterates the need for complementary single‐cell type data to deconvolute cell type‐specific expression. We provide the list of microglial signature genes that are also expressed at high levels in bulk brain tissue data (Table S5), which can serve as a validated resource for microglial signature gene markers in bulk RNAseq datasets.
To determine how the microglial signature in this study compared to previously published signatures, we performed hypergeometric tests of overrepresentation with Galatro et al. (2017), Gosselin et al. (2017) and Olah et al. (2018) studies. Significant overlap was observed across all datasets, with 350 genes common to all datasets (Figure 1c‐d, Tables S3–S4, S8). This comprised several established microglial marker genes, including P2RY12, TMEM119, and CX3CR1. The most significant overlap was shared with Gosselin, et al. (Gosselin et al., 2017) signature [OR = 19.6 (17.0‐Inf) p = 3.8E‐261], where 49.7% of their genes were also present in our signature, and 22% of ours in their signature. Gosselin et al. (2017) samples were also obtained from neurosurgical tissue resections like our cohort and are unlike Galatro et al. (2017) and Olah et al. (2018) samples that were harvested during autopsy. Although there appears to be a common set of microglial genes consistent across signatures, each also harbors many unique genes, which could be due to study or individual specific differences.
2.2. Transcriptional profiling of microglia discovers co‐expression networks and implicates lipid and carbohydrate metabolism pathways associated with age, sex, and APOE
We generated gene co‐expression networks using WGCNA (Langfelder & Horvath, 2008) to reduce number of tests and increase power to detect genetic associations with age, sex, and APOE. We identified 7 modules with significant associations (Figure 2; Figure S5; Table S9). Modules ME14 and ME34 associated with age, however, in opposite directions. ME14 was enriched for genes involved in the lipid localization pathway that were upregulated with age (R = 0.50, p = 0.03; Figure 2a‐c). ME34, enriched for DNA endoreduplication genes, had negative association with both age (R = −0.55, p = 0.01) and APOE‐ε4 (R = −0.50, p = 0.03), indicating that microglial transcripts involved in this pathway are downregulated with aging and in APOE‐ε4 carriers (Figure 2a). Several other modules also associated with APOE‐ε4, in either direction. The only module associated with sex was ME26, which was downregulated in females (R = −0.54, p = 0.02), and enriched for genes involved in cholesterol absorption and lipid digestion. This module also had the most significant association with APOE, in the positive direction with presence of APOE‐ε4 (R = 0.66, p = 0.002; Figure 2a,b,e). Of the APOE‐associated modules, ME23 had the second most significant association (R = −0.61, p = 0.006) and was enriched for carbohydrate metabolism genes (Figure 2a,b,d). Given recent discoveries in microglial immunometabolism (Bernier et al., 2020; Chausse et al., 2020; Loving & Bruce, 2020; Marschallinger et al., 2020), we focused on ME14, ME23, and ME26 that are enriched for lipid and carbohydrate metabolism genes.
FIGURE 2.
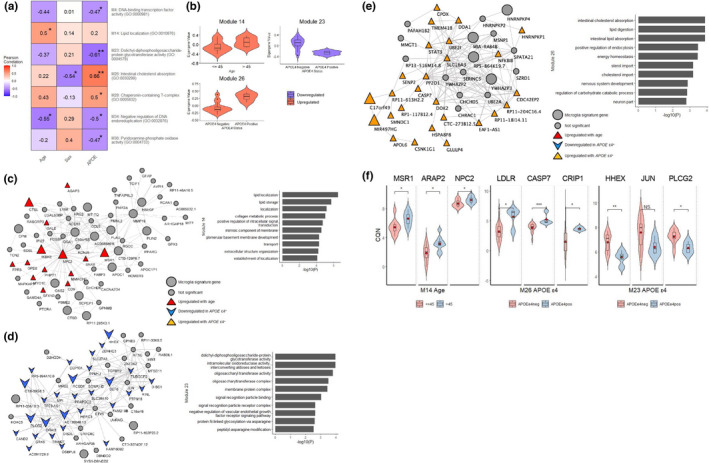
Age, sex and APOE ε4 pathway correlations in bulk microglia. (a) Heatmap showing correlation of age, sex and APOE ε4 status with WGCNA module eigengenes (MEs) significantly associated (p < 0.05) with traits, with top GO terms listed for each module. (b) Module eigengenes stratified by age or APOE ε4 status. (c) Module M14 gene co‐expression network, with genes of interest highlighted according to the key. Genes upregulated with age shown in red triangle ( ). Bar plot of top 10 significant GO terms (p < 0.05) for this module. (d) Module 23 gene co‐expression network, with genes downregulated in APOE ε4 carriers shown in blue arrow (
). Bar plot of top 10 significant GO terms (p < 0.05) for this module. (d) Module 23 gene co‐expression network, with genes downregulated in APOE ε4 carriers shown in blue arrow ( ). Bar plot of top 10 significant GO terms (p < 0.05) for this module. (e) Module 26 gene co‐expression network, with genes upregulated in APOE ε4 carriers shown in orange triangle (
). Bar plot of top 10 significant GO terms (p < 0.05) for this module. (e) Module 26 gene co‐expression network, with genes upregulated in APOE ε4 carriers shown in orange triangle ( ). Bar plot of top 10 significant GO terms (p < 0.05) for this module. (f) Violin plots showing expression of key genes in modules, stratified by age or APOE. *p < 0.05; **p < 0.01; ***p < 0.001
). Bar plot of top 10 significant GO terms (p < 0.05) for this module. (f) Violin plots showing expression of key genes in modules, stratified by age or APOE. *p < 0.05; **p < 0.01; ***p < 0.001
ME14 co‐expression network (Figure 2c) hub genes NPC2, MSR1, and PLAU are also microglial signature genes in our study and known to be involved in microglial functions (Butovsky & Weiner, 2018; Colombo et al., 2021; Cunningham et al., 2009; DePaula‐Silva et al., 2019; El Khoury et al., 1998]; Mehra et al., 2016). Several disease‐associated microglial (DAM) markers are also present in this network, including CD9, ARAP2, and MYO1E (Keren‐Shaul et al., 2017; Rangaraju et al., 2018a; Sobue et al., 2021) that are increased with aging, implicating activated microglial lipid localization pathways in aging (Figure 2f). Several genes in this module were also previously linked to neurodegeneration, including MYO1E (Gerrits et al., 2021]; Rangaraju et al., 2018b), CTSL, (Cermak et al., 2016) and UNC5B (Ahn et al., 2020; Xu et al., 2016). Due to the nature of the tissue, we compared expression levels of the key module genes within tumor and epilepsy samples (Figure S6). We observed significant differences due to diagnosis in NPC2, PLAU, APOC1, and IKBKE; therefore, it is important to note that some of their associations may be confounded due to the disease state.
Our microglial signature (Tables S1–S3) had significant overrepresentation of the age‐associated ME14 genes (Table S9; OR = 1.55 [95% CI = 1.23‐INF], p = 0.001), highlighting age‐related increases in microglial signature genes. Galatro et al. (2017) and Olah et al. (2018) also reported age‐related microglial signatures. Comparison of ME14 genes revealed significant overlap with Olah et al. (2018) (OR = 1.34 [95% CI = 1.05‐INF] p = 0.03), but not with Galatro et al. (2017) microglial aging signature genes (OR = 1.09 [95% CI = 0.81‐INF] p = 0.33).
ME26 cholesterol metabolism pathway genes exhibited reduced expression in males and were elevated in APOE‐ε4 carriers (Figure 2a,b). This module contains known microglial genes LDLR, CD36, and CRIP1 (Figure 2e,f). Assessment of individual ME26 network genes revealed C17orf49, RP11‐589P10.7, and MIR497HG to be the only microglial signature genes in this network to be associated with both sex and APOE (Figure 2e). Other microglial signature genes in ME26 associated with only sex or only APOE, suggesting that these traits may have independent effects on expression of some microglial genes. Several APOE‐associated genes in ME26 were previously implicated in AD, including CASP7 (Ayers et al., 2016; Zhang et al., 2019) and LDLR (Katsouri & Georgopoulos, 2011; Lämsä et al., 2008; Figure 2f).
Carbohydrate metabolism gene enriched module ME23 is downregulated in APOE‐ε4 carriers (Figure 2a,b,d). AD risk genes BIN1 (Crotti et al., 2019) and PLCG2 (Sims et al., 2017) are present in this network, which have both been implicated in microglial dysfunction in neurodegeneration (Figure 2d).
2.3. Single‐cell transcriptome reveals specific subtypes of microglia
To uncover distinct microglial subtypes, a subset of sorted microglial samples from neurosurgical brain tissue underwent single‐cell expression profiling. We obtained 26,558 cells from 5 unique individuals, including one individual who underwent epilepsy surgery and had samples from two brain regions (Table 1). Analysis of the scRNAseq data from these samples revealed 13 distinct cell clusters, which were annotated using established neuronal and glial marker genes from the literature (Darmanis et al., 2015; Keren‐Shaul et al., 2017; Masuda et al., 2019; Mathys et al., 2019; Olah et al., 2020; Rangaraju et al., 2018a; Sankowski et al., 2019; Zhou et al., 2020; Figure 3a, Tables S10‐S16). Myeloid markers (AIF1, PTPRC, and C1QA) were detected in all clusters except cluster 12 which expressed oligodendrocyte markers (PLP1, MBP, and MOBP). Cluster 9 expressed macrophage‐specific markers (VCAN, FCN1, CRIP1, and S100A8). These two clusters comprised <3% of all cells, indicating that our sorted samples represent a very pure microglial population. Each myeloid cluster had cellular contributions from all samples, albeit with some variability in their proportions, likely due to intrinsic differences between individuals (Figure 3b, Table S11). For these samples, the most marked difference was observed for macrophages (cluster 9) and homeostatic microglia (cluster 2), which had greater contributions from the mesiotemporal and anterior temporal regions, respectively. This could be due to the proximity of the mesiotemporal sample to the disease‐affected region.
FIGURE 3.
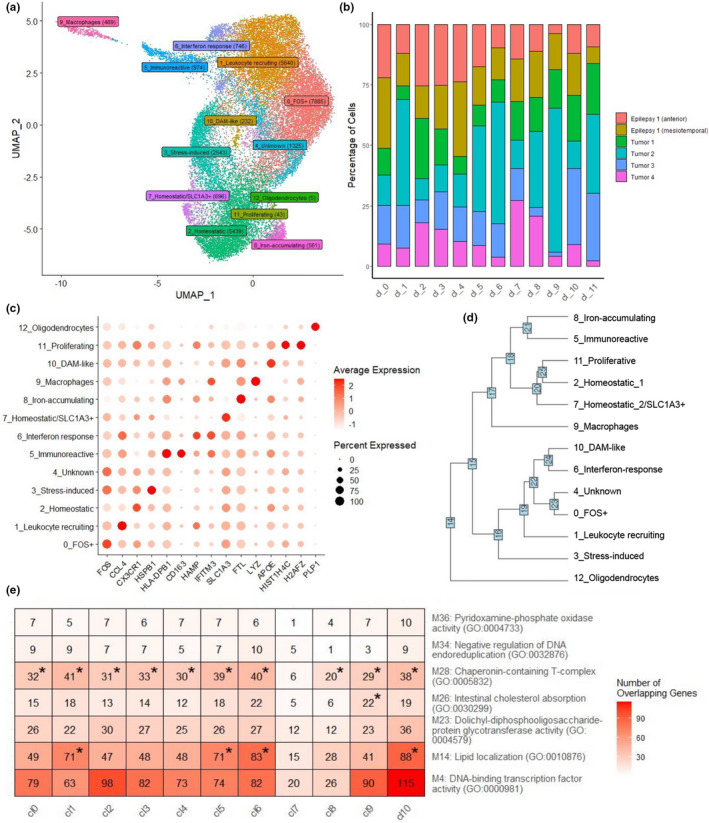
Single‐cell microglial data. (a) UMAP of clustered cells annotated with putative subtypes using cell type markers from the literature. (b) Stacked bar plot showing the distribution of cells across the clusters. (c) Dot plot showing the expression of key significant module genes across clusters. (d) Hierarchical clustering to highlight relationships between clusters. (e) Hypergeometric distribution of enrichment between module genes and clusters, showing number of overlapping genes. * Represents module genes that were significantly enriched in the cluster (p < 0.05)
We characterized the microglial clusters by their expression of established microglial subtype markers (Figure 3c, Figure S7) and their most significant marker genes (Figure S8). Homeostatic (TMEM119, P2RY12, and CX3CR1; Masuda et al., 2019; Rangaraju et al., 2018a; Sankowski et al., 2019; Zhou et al., 2020), pro‐inflammatory (CCL2, CCL4, Masuda et al., 2019; Sankowski et al., 2019) and DAM markers (APOE, C1QA, and C1QB; Hammond et al., 2019; Keren‐Shaul et al., 2017; Olah et al., 2020; Sankowski et al., 2019) were observed in clusters 2, 1/6, and 10, respectively. Cluster marker genes are defined as those expressed in at least 70% of the cells in the cluster with log fold change >0.6 and q < 0.05 in comparison to all other clusters. Expression levels of the top marker genes per cluster are shown (Figure 3c; Figure S8; Table S12). Most of these markers are distinct to a single cluster, although some clusters appeared to have similarities in their marker expressions. To define the proximity of their transcriptional profiles, we performed hierarchical clustering of the microglial clusters (Figure 3d). We determined that the homeostatic microglia cluster 2 was transcriptionally closest to clusters 7 and 11, which may represent subtypes of homeostatic microglia. Cluster 11 is enriched for markers of cell proliferation (STMN1, H2AFZ, PCNA, and MKI67), some of which were also observed by Olah et al. (2020), suggesting that these could be proliferating microglia. Clusters 1 and 6 both expressed inflammatory chemokines CCL2 and CCL4 and anti‐inflammatory molecule EGR2; however, cluster 6 was more closely related to DAM, whereas cluster 1 (named as Leukocyte‐recruiting cluster) represented a more pro‐inflammatory signature with greater expression of IL6 and TNFα. Cluster 6 (named as interferon‐response cluster) also highly expressed interferon‐related marker IFITM3 and ISG15, also observed in a cluster by Olah et al. (2020), which they defined as an interferon response‐enriched subset. The upregulation of chemokines and interleukins in these clusters suggest that they could be involved in recruitment of other immune cells. These findings highlight different transcriptional profiles for these two inflammatory clusters that may represent distinct activated microglia subtypes. Cluster 3 highly expressed heat shock protein HSPA1A, an immediate early gene (Schmunk et al., 2020) reportedly involved in antigen processing (Aung et al., 2012), response to stress and injury and exhibiting decreased gene expression in multiple sclerosis patients (Gandhi et al., 2010; Satoh et al., 2005). Several were upregulated in this cluster, suggesting that this may represent cells that underwent dissociation‐induced stress (Sankowski et al., 2019). Several of the clusters did not express well known existing cell type markers. Clusters 5/8 and FOS+0/4 were transcriptionally closest to one another (Figure 3d). Cluster 5 has distinct expression of immunoreactive marker CD163, which was not observed in other subsets except macrophages. Several HLA genes are also highly expressed in this cluster, suggesting that these may be immunoreactive microglia (Hendrickx et al., 2017). Cluster 8 marker FTL has recently been used to characterize iron‐accumulating microglia (Kenkhuis et al., 2021). Our findings highlight transcriptional profiles for known microglial clusters, describe the transcriptional proximity of these clusters and suggest that less well‐defined clusters could potentially represent novel or intermediate transcriptional states of microglia. Our microglial signature was significantly enriched in more clusters expressing more activated markers (Table S13), implicating this as the dominant expression profile within our samples. However, there is also enrichment of homeostatic cluster 2, demonstrating that we have not only captured activated cells as might be expected due to the nature of the tissue.
To determine whether the bulk microglial co‐expression networks (Figure 2a,c‐e, Figure S5) were representative of microglial subtypes, we performed enrichment analyses of the module genes within the myeloid clusters with sufficient cell numbers (Figure 3e; full enrichment statistics provided in Table S15). Age‐associated co‐expression network ME14, implicated in lipid metabolism, was significantly enriched in interferon‐response (cluster 6) and DAM (cluster 10) clusters. Genes within module 28, which was significantly upregulated with APOE‐ε4, had statistically significant enrichment in all clusters except cluster 7. There was no statistically significant enrichment for any of the other microglial modules that had significant age, sex, or APOE associations, suggesting that these factors may have ubiquitous effects on most microglial subtypes. Some of the remaining microglial co‐expression networks had distinct patterns of cluster enrichment (Figure S9), suggesting that some but not all networks could be representative of distinct microglial subtypes.
2.4. Meta‐analyses with published datasets support age, sex, and APOE associations
Each dataset was individually processed through the same MAPR‐seq pipeline for quality control to minimize variability due to data processing. Meta‐analysis of WGCNA results was performed by coercing the external dataset co‐expression networks onto our existing co‐expression networks. The forest plot in Figure 4a highlights the individual and combined associations for networks of interest across the datasets where age, sex, or APOE information was available (Table S17). For module ME14 associations with age, Galatro et al. (2017) also exhibited a similar direction of effect (R = 0.30, p = 0.06), whereas Srinivasan et al. (2020) did not show any association (R = −0.003, p = 0.99), likely due to a higher median age of individuals in the study. When meta‐analyzed, sex‐associated ME26 genes were significantly downregulated in males in all datasets, supporting our finding. This module was also significantly associated with APOE e4 carriers in the Srinivasan dataset (R = 0.42, p = 0.04). However, Srinivasan samples were inversely correlated with APOE for ME23 in comparison to our data (R = 0.61, p = 0.07). Srinivasan et al. (2020) was inversely correlated with age and APOE for ME23, but not with sex or APOE for ME26. Overall, it appears that the direction of effects was similar for correlation with these traits in most datasets (Figure S10).
FIGURE 4.
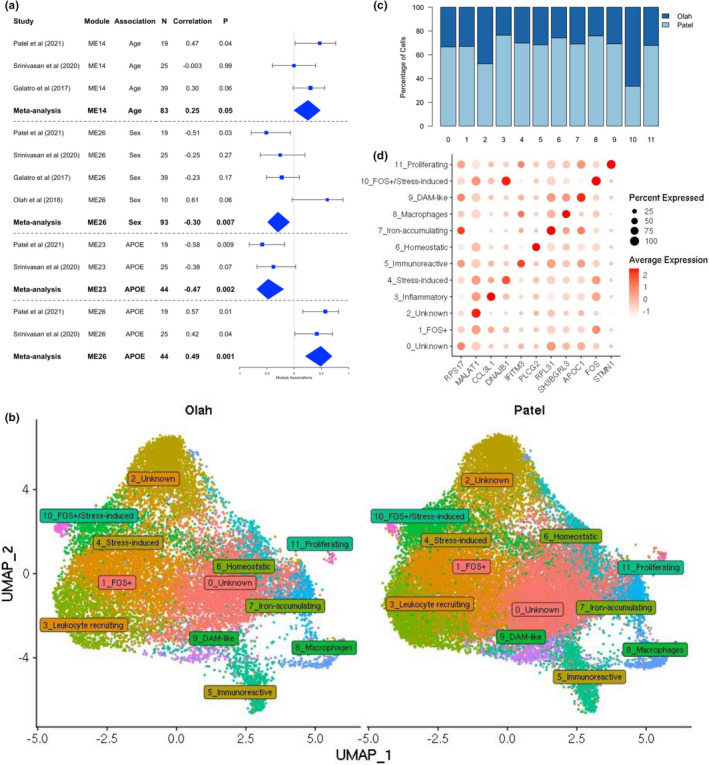
Meta‐analysis with published datasets. (a) Forest plots of module eigengene correlations across datasets and meta‐analyzed. (b) Integrated UMAP of our and Olah et al. (2020) single‐cell data, split by dataset. (c) Stacked bar plot showing the distribution of cells across the clusters. (d) Dot plot showing the expression of key significant module genes across clusters
We subsequently integrated microglial single‐cell data from Olah et al. (2020) (n = 9, 15,819 cells) with our samples (n = 6, 26,856 cells) to show how these single‐cell datasets compared. The UMAP in Figure 4b is split by study and shows relatively even contributions to each cluster from both datasets, also observed in the stacked bar plot in Figure 4c. However, a greater number of Olah cells were observed in cluster 10, which had similar expression patterns to our original FOS+ cluster 0. Hypergeometric tests of enrichment showed significant overlap between several of the original and integrated clusters, showcasing the high congruence between our data and Olah et al (Tables S18–S20). To further identify where our individual cells clustered when combined with the Olah dataset, we overlaid the cell IDs from the original clusters to the integrated dataset and calculated the percentage overlap (Table S20). A larger proportion of cells from Olah et al expressed markers of activation than homeostatic genes, indicating a greater enrichment for activated microglia. Many clusters retained their identity, as evidenced by the dot plot (Figure 4d) which shows some of the same top marker genes to that in Figure 3c. Additionally, enrichment of our microglial signature in the integrated single‐cell dataset was observed in several clusters, many of which highly expressed markers of activated microglia (Table S18). This is similar to what we observed in our single‐cell data (Table S13), supporting the notion that our microglial signature is enriched for activated cells.
3. DISCUSSION
Given their critical functions in maintaining homeostasis in the central nervous system (CNS) in health and their multifaceted roles during neurological diseases (Hickman et al., 2018; Li & Barres, 2018), understanding the biology of microglia and characterizing microglial subtypes is essential. Large‐scale studies in bulk brain tissue (Allen et al., 2016]; De Jager et al., 2018; Wang et al., 2018) have been instrumental in establishing transcriptional profiles in health and neurodegenerative diseases. Although these studies yielded information on brain expression signatures and uncovered perturbed pathways and molecules implicated in Alzheimer's disease and other neurological disorders (Allen et al., 2018a, 2018b; Mostafavi et al., 2018; Neff et al., 2021), they are limited in their ability to provide cell type‐specific transcriptional outcomes, especially for less abundant CNS cells such as microglia (Wang et al., 2020). Analytic deconvolution approaches began to leverage these bulk tissue transcriptome datasets to estimate cell type‐specific expression profiles (McKenzie et al., 2018; Wang et al., 2020), but the accuracy of these methods relies on the availability of high‐quality single cell‐type datasets. Such microglia‐specific transcriptome datasets are gradually emerging (Galatro et al., 2017; Gosselin et al., 2017; Olah et al., 2018, 2020), although the numbers of unique samples assessed remain limited given the arduous nature of collecting fresh human brain tissue. Additionally, comparative assessment of bulk brain vs. single cell‐type bulk microglia vs. single‐cell microglia studies are still rare (Alsema et al., 2020; Olah et al., 2020; Srinivasan et al., 2020). To our knowledge, there are no studies that evaluate human microglial transcriptome using all three approaches, as in our study. Further, investigations on effects of genetic and other factors on microglial transcriptional signatures in humans is likewise sparse, with the exception of age‐related effects assessed in a few studies (Galatro et al., 2017; Gosselin et al., 2017; Olah et al., 2018). Finally, unlike in bulk tissue studies (Allen et al., 2018a, 2018b; McKenzie et al., 2018; Mostafavi et al., 2018; Neff et al., 2021), microglia‐specific co‐expression networks, their molecular signatures and functional implications have not been evaluated.
In this study, we sought to overcome these knowledge gaps by characterizing the transcriptome of sorted bulk and single‐cell microglial populations isolated from fresh human brain tissue. We identified a robust microglial signature comprising 1971 genes enriched for immune‐related functions. These signature genes were selected due to their consistently higher expression levels in our sorted bulk microglial transcriptome in comparison with 7 different bulk brain tissue datasets from 6 different regions (Allen et al., 2016; De Jager et al., 2018; Wang et al., 2018). We also compared sorted bulk microglia to bulk fresh brain tissue and identified transcripts that are expressed in both. The microglial signature genes that are also reliably detected in bulk brain tissue represent a validated list of microglial markers that can be utilized in bulk brain tissue transcriptome analytic deconvolution studies (McKenzie et al., 2018; Wang et al., 2020).
Our microglial signature significantly overlapped with other signatures from bulk microglia previously reported by Galatro et al. (2017), Gosselin et al. (2017) and Olah et al. (2018), implicating a core set of genes consistently expressed in this cell type. However, there were additional genes unique to each signature, likely to be driven by factors such as patient demographics or study differences. Galatro et al. (2017) and Olah et al. (2018) both also reported age‐related microglial expression signatures. We found significant overlap of our age‐associated microglial gene expression module ME14 genes with the latter, which was also enriched for our microglial signature. This indicates that bulk microglial profiles can effectively capture genes affected by aging in microglia.
We leveraged the co‐expression network structure of sorted bulk microglia to further explore whether microglial subsets were associated with age, sex, or APOE‐ε4. To our knowledge, sex differences in microglial transcriptome were previously studied only in mice (Frigerio et al., 2019; Nelson et al., 2017; Stephen et al., 2019); however, APOE genotype‐specific microglial interactions with amyloid plaques have been previously observed in mice (Shi et al., 2019; Stephen et al., 2019) and humans (Nguyen et al., 2020). We identified two network modules associated with age, one with sex and six with APOE‐ε4. We observed that two modules, ME14 that is positively associated with increased age; and ME26 that is positively associated with both APOE‐ε4 and female sex, were both enriched for lipid metabolism biological terms (Chausse et al., 2020; Loving & Bruce, 2020; Marschallinger et al., 2020). Module ME14 included genes involved in lipid localization and storage pathways (PLIN2, IL6, LPL, MSR1, ENPP1, PPARG, PTPN2, SOAT1, IKBKE) and ME26 had lipid digestion/cholesterol transport pathway genes (CD36, LDLR). Both modules harbored known microglial genes (LDLR, CD36, CRIP1, NPC2, MSR1, PLAU) and those that are included in our microglial signature (PLIN2, IL6, MSR1, SOAT1, IKBKE, NPC2, PLAU).
Comparing the sorted bulk microglial network modules to scRNAseq microglial clusters, we determined that ME14 genes were significantly over‐represented in interferon‐response cluster 6 and disease‐associated microglia (DAM) cluster 10. In our study, DAM cluster 10 included APOE, APOC1, ASAH1, and CTSD. Of these APOE (Leduc et al., 2010; Loving & Bruce, 2020; Yamazaki et al., 2019), APOC1 and ASAH1 (Paciotti et al., 2020) are involved in lipid metabolism and neurodegenerative diseases. APOE (Hammond et al., 2019; Keren‐Shaul et al., 2017; Krasemann et al., 2017), APOC1(Hammond et al., 2019 ), and CTSD (Keren‐Shaul et al., 2017) were also signature genes in mouse models of neurodegenerative diseases (Keren‐Shaul et al., 2017; Krasemann et al., 2017) or aging (Hammond et al., 2019). Our interferon‐response cluster 6 also included genes associated with mice microglial neurodegenerative (FTH1 Keren‐Shaul et al. (2017)) or aging signatures (CCL4 Hammond et al. (2019)), as well as IFITM3 (Marschallinger et al., 2020) and GOLGA4 (Marschallinger et al., 2020), previously shown to be upregulated in aging lipid droplet accumulating microglia (Marschallinger et al., 2020). Our findings that integrate human sorted bulk RNAseq and scRNAseq data, support a model where aging human microglia transition to a pro‐inflammatory and disease‐associated transcriptional profile, which is also associated with perturbations in lipid metabolism in these cells.
There is increasing evidence that tightly controlled lipid metabolism is essential to the functions of microglia during development and homeostatic functions of adulthood and may be disrupted in aging and disease (Chausse et al., 2020; Loving & Bruce, 2020). The complex interactions between microglial lipid metabolism and its cellular functions rely on lipid sensing by microglial receptors such as CD36 and TREM2 and uptake of lipids, including LDL and APOE (Chausse et al., 2020; Loving & Bruce, 2020). These interactions are necessary for microglia to become activated and perform functions including phagocytosis of myelin (Nugent et al., 2020) and misfolded proteins like amyloid ß (Yeh et al., 2016), cytokine release, migration and proliferation (Bernier et al., 2020; Chausse et al., 2020). Studies primarily focused on in vitro and animal models suggest disruption of the microglial immunometabolism and assumption of a pro‐inflammatory phenotype with aging (Hammond et al., 2019; Koellhoffer et al., 2017; Marschallinger et al., 2020; Norden & Godbout, 2013) and diseases including multiple sclerosis (MS) and Alzheimer's disease (Keren‐Shaul et al., 2017; Krasemann et al., 2017; Ulland et al., 2017). Interestingly, microglial lipid droplet accumulation has been demonstrated under all these conditions (Chausse et al., 2020; Loving & Bruce, 2020; Marschallinger et al., 2020; Nugent et al., 2020) and lipid droplet accumulating microglia in aging mice were shown to have a unique transcriptional state (Marschallinger et al., 2020). Our findings in sorted cells from fresh human brain tissue provide transcriptional evidence for immunometabolism changes and pro‐inflammatory phenotype with microglial aging, thereby contributing essential complementary data from humans for this cell type.
Besides module ME14, we determined that ME26 is also enriched for lipid metabolism genes. ME26 module expression is higher in both APOE‐ε4 and female sex; however, we note that in our sorted bulk microglia RNAseq samples, there were no male APOE‐ε4 carriers. Therefore, the distinct influence of sex and APOE on the expression of this module remains to be established. APOE‐ε4, a major risk factor for Alzheimer's disease, has the lowest lipid binding efficiency compared with other APOE isoforms (Chausse et al., 2020). Increased cholesterol accumulation has been reported in both iPSC‐driven astrocytes from APOE‐ε4 carriers (Lin et al., 2018) and in Apoe‐deficient microglia (Nugent et al., 2020). These findings collectively support a role for APOE‐ε4 associated microglial transcriptional changes and disrupted cholesterol metabolism. Using our sorted microglia RNAseq data, we identified five additional modules that associate with APOE‐ε4, one in a positive direction (ME28) and four negatively (ME4, ME23, ME34, and ME36). Of these, module ME23 had the second most significant APOE‐ε4 association after ME26. Interestingly, ME23 was enriched for carbohydrate metabolism biological processes, which are also tightly regulated in microglia (Bernier et al., 2020). Module ME23 harbors known AD risk genes BIN1 and PLCG2, where the latter is a microglial gene that modulates signaling through TREM2 (Andreone et al., 2020) and also a hub gene in this module. ME23 genes BIN1, JUN, and TGFBR2 were found to be reduced in a mouse microglial neurodegenerative phenotype gene signature (Krasemann et al., 2017). These findings further demonstrate the consistency of our human microglial data with that from mouse models and supports perturbed microglial immunometabolism as a potential pathogenic mechanism in neurodegeneration. Many of the meta‐analysis results comprising published datasets from Galatro et al. (2017), Olah et al. (2018), and Srinivasan et al. (2020) supported the directions of effects we had observed in our study despite small sample numbers.
In addition to analyzing gene expression modules from sorted bulk microglia, we also identified microglial clusters from sorted microglial scRNAseq data. To our knowledge, there are only two prior publications of scRNAseq characterizations on human microglia (Masuda et al., 2019; Olah et al., 2020). Masuda et al. (2019) analyzed 1,602 microglia isolated from 5 control and 5 MS patient brains, compared their findings to those from mice demonstrating clusters that are common and others that are species‐specific. Olah et al. assessed 16,242 microglia from 17 individuals and characterized subclusters of microglia from patients with mild cognitive impairment, AD, and epilepsy (Olah et al., 2020). Our scRNAseq dataset is from 5 unique individuals comprising 26,558 cells, 99.98% of which have myeloid markers. A small cluster highly enriched for mature oligodendrocyte and some neuronal marker genes were observed (Table S11). As there were only 5 cells in this cluster, this is likely to denote contamination from the sorting procedures. We identified microglial clusters that share characteristics of those previously reported in mice (Keren‐Shaul et al., 2017) and humans (Nguyen et al., 2020; Olah et al., 2020), such as DAM. We also uncovered clusters that had not been previously characterized in the literature, including cluster 7, exhibiting high expression levels of astrocytic marker gene SLC1A3. Microglial expression of SLC1A3 was previously shown to occur in mice and humans especially in disease states (Chrétien et al., 2004; Grassivaro et al., 2020; Wilhelmsson et al., 2017). We also leveraged these scRNAseq data to further characterize the sorted bulk microglial expression modules. Integration of our samples with Olah et al. allowed us to explore microglial subtypes at single‐cell resolution with increased power. We found that even in this larger dataset, our microglial signature was similarly enriched in the more activated subtype clusters, demonstrating the robustness of this signature across datasets. The homogeneity in clustering highlighted shared expression patterns between cells from both datasets, providing further support for the putative microglial subtypes we identified here. Hence, our microglial scRNAseq data contribute further to the emerging single‐cell landscape of this cell type.
Activated microglia are a hallmark in both brain cancers and temporal lobe epilepsy in response to the inflammation (Darmanis et al., 2017; Kaminska et al., 2021; Kinoshita & Koyama, 2021; Morin‐Brureau et al., 2018; Ochocka et al., 2021). We acknowledge that the tissue used in this study is sourced from tumor or epileptogenic tissue adjacent regions in individuals with diagnoses of brain tumors or epilepsy, meaning it is not entirely healthy. Therefore, it is possible that the activated subtypes we identify here may be due to the nature of the tissue. Additionally, sex‐specific expression differences have been observed in glioma‐activated microglia (Ochocka et al., 2021). Nonetheless, we did not observe significant differences in most genes we highlighted here due to diagnosis (Figure S5). There is also a large cluster of homeostatic microglia present in these samples, indicating that not all cells are activated. While the focus of our work is AD, these findings also have potential relevance in the fields of cancers, epilepsy, and other diseases.
We recognize that our study has several limitations, primarily owing to the difficulty in obtaining high‐quality neurosurgical brain tissue, which leads to limited sample size and variability in tissue, diagnoses, and patient demographics. Even though we have utilized tissue surgically separated from disease tissue, the samples are from epilepsy and various brain tumor patients representing multiple diagnoses. We isolated microglia using an approach which should minimize activation; however, we cannot definitively rule out stress‐induced transcriptomic changes during isolation. Despite these caveats, we could identify microglial co‐expression modules and subclusters with multiple features that are consistent with prior publications from model systems (Hammond et al., 2019; Keren‐Shaul et al., 2017; Krasemann et al., 2017; Marschallinger et al., 2020). Our scRNAseq clusters have contributions from both tumor and epilepsy samples, suggesting that our findings are unlikely to be driven by any one diagnoses. Furthermore, there are few published studies using fresh brain tissue to study microglia and thus our integrated single‐cell data showing a homogenous cluster of microglia highlights the robustness of the methodology.
In summary, our study on sorted bulk microglia RNAseq and scRNAseq from fresh brain tissue yield several key findings. We identify a microglial gene signature from sorted bulk microglia, characterize its expression in bulk brain RNAseq across 7 datasets comprising 6 regions, in bulk fresh brain RNAseq and in microglial scRNAseq subtype clusters. This signature provides a well‐characterized resource which can be utilized in analytic deconvolution studies of bulk transcriptome data (McKenzie et al., 2018; Wang et al., 2020). We uncovered microglial gene expression modules associated with age, sex and/or APOE‐ε4. Modules with age and APOE‐ε4‐associated transcriptional changes implicate microglial lipid and carbohydrate metabolism perturbations and microglial activation. Microglial scRNAseq data highlight the transcriptional complexity of this cell type, reveal both known and novel cell types, and demonstrate utility of this data in characterizing sorted bulk RNAseq data. These findings provide support for the emerging microglial immunometabolism (Bernier et al., 2020; Chausse et al., 2020) pathway as a plausible therapeutic target in aging‐related disorders; and provide a well‐characterized human transcriptome resource for the research community on this cell type with central roles in health and disease (Masuda et al., 2020).
4. MATERIALS AND METHODS
4.1. Patient samples
Fresh human brain tissue was obtained from patients undergoing neurosurgical procedures for epilepsy or tumor resection. Tissues determined to be grossly unaffected by the primary disease process were utilized for the present study (Figure 1). Patient samples were transported from the operating room to the laboratory in 1X DPBS (Thermo Fisher; 14287080) for processing within 1–2 h of resection. Human tissue was collected with informed consent prior to surgery and all procedures were approved by the Mayo Clinic Institutional Review Board and are HIPAA compliant.
4.2. Tissue dissociation
Tissue was dissected to remove necrotic tissue, white matter and excess vascular tissue, to retain only cortical gray matter. The remaining tissue was cut into sagittal slices and weighed before being processed using the Adult Brain Dissociation Kit (Miltenyi; 130‐107‐677) as per the manufacturer's protocol. Debris removal (Miltenyi; 130‐109‐398) and red blood cell lysis (Miltenyi; 130‐094‐183) were also performed. All procedures were carried out on ice. The resulting homogenate was filtered through a 70‐µm filter before proceeding.
4.3. Magnetic‐activated cell sorting (MACS)
The cell suspension was first enriched for CD11b+ cells by incubating with anti‐CD11b microbeads (Miltenyi; 130–049–601 clone M1/70) for 15 minutes according to manufacturer's recommendation. This was then washed with PB buffer (0.5% BSA, 1X PBS Ca2+/Mg2+ free pH 7.4) and filtered through a 70µm cell strainer before being applied to a large separation column (Miltenyi; 130‐042‐401) in a QuadroMACS separator magnet (Miltenyi; 130‐090‐976). The CD11b+ fraction was collected and resuspended in sterile filtered FACS staining buffer (1X PBS Ca2+/Mg2+ free, 0.5% BSA, 2% FBS, 3mM EDTA) for antibody staining.
4.4. Fluorescence‐activated cell sorting (FACS)
MACS sorted CD11b+ cells subsequently underwent FACS sorting to further purify the microglial population. The cell suspension was incubated in Human TruStain FcX blocking solution (1:20, Biolegend; 422302) at room temperature for 10 min. Subsequently, cells were stained with anti‐CD11b PE/Cy7 (1:100, Biolegend; 101206, M1/70) and anti‐CD45 Alexa Fluor 647 (1:100, Biolegend; 304056, HI30) antibodies for 30 min on ice. Following two washes with FACS staining buffer, SYTOX Green viability dye (1:1000, Thermo Fisher; S7020) was added for an additional 20 minutes. Single‐cell suspensions were filtered through a 40µm cell strainer (Falcon; 352235) before sorting on a BD FACS Aria II (BD Biosciences). CD11b+/CD45intermediate/SYTOX green− cells were sorted directly into FACS staining buffer. Independently CD11b and CD45 are not microglial specific markers; however, gating cells with a CD11b+/CD45intermediate signature allowed us to differentiate microglia from peripheral myeloid cells, such as macrophages, which are expected to be CD45high. An example of our FACS gating strategy is provided in Figure S2A. Briefly, we used cell lines expressing the markers of interest as positive controls to independently set the gates for CD11b (RAW 264.5 mouse macrophage cells) and CD45 (Jurkat cells) expression. CD45 intermediate cells were determined by selecting for mid‐level fluorescence within this gate. This was used to sort cells for both bulk and single‐cell RNAseq data generation to reduce the likelihood of peripheral myeloid cell contamination in the samples.
4.5. RNA isolation and sequencing
RNA from sorted microglial cells was isolated using the miRNeasy Serum/Plasma Kit (QIAGEN; 217184) and quantified on the Agilent BioAnalyzer 2100. cDNA libraries were generated using SMARTSeq2 v4 and Nextera Low Input Library Prep Kit. Samples were multiplexed and sequenced on the Illumina HiSeq 4000.
RNA from frozen bulk tissue was isolated using Trizol and chloroform, followed by DNase and clean up using the Rneasy Kit (QIAGEN; 74106). Libraries were generated using the TruSeq Stranded mRNA Library Prep Kit. Samples were multiplexed and sequenced on the Illumina HiSeq 4000. Base‐calling of all sequence data was performed using Illumina's RTA v2.7.7.
4.6. 10X Single cell 3’ v3 library preparation of sorted microglia
Viability of MACS plus FACS sorted cells was assessed by Trypan blue (Gibco; 15250061) exclusion and cell density was determined using a hemocytometer prior to adjustment to target 4000–5000 cells. Cells were loaded onto a 10X Chromium chip and run on the GemCode Single Cell Instrument (10X Genomics) to generate single‐cell gel beads‐in‐emulsion (GEMs). Single‐cell RNAseq libraries were prepared using the Chromium Single Cell 3’ Gel Bead and Library Kit v2 and v3 (10X Genomics; 120237) and the Chromium i7 Multiplex Kit (10X Genomics; 120262) according to the manufacturer's instructions. Quality of cDNA libraries was determined using a BioAnalyzer 2100 DNA High Sensitivity assay (Agilent; 5067–4626) prior to sequencing one per lane on an Illumina HiSeq 4000.
4.7. Validation with quantitative real‐time PCR
Total RNA was extracted from sorted cells using the miRNeasy Serum/Plasma Kit (QIAGEN; 217184). Concentration and quality were assessed using the Agilent BioAnalyzer RNA 6000 Pico Kit (Agilent; 5067‐1514). RNA was normalized to 0.5ng/µl for cDNA synthesis using the SuperScript IV VILO Master Mix (Thermo Fisher; 11756050). TaqMan PreAmp Master Mix (Thermo Fisher; 4391128) was used to pre‐amplify the cDNA, followed by TaqMan Universal PCR Master Mix (Thermo Fisher; 4304437) with the following gene expression probes: MOG, AQP4, THY1, PTPRC, ITGAM, P2RY12, PECAM1, CD34, GAPDH (Thermo Fisher; Hs01555268_m1, Hs00242342_m1, Hs00174816_m1, Hs04189704_m1, Hs00355885_m1, Hs00224470_m1, Hs01065279_m1, Hs02576480_m1, Hs99999905_m1). RT‐qPCR was performed on a QuantStudio 7 Flex Real‐Time PCR System (Thermo Fisher) using a relative standard curve to quantify gene expression.
4.8. Validation with immunocytochemistry
Cultured cells were fixed with 4% paraformaldehyde (PFA) overnight at 4°C and blocked with blocking solution (10% BSA, 5% normal goat serum and 0.1% Triton‐X). Fixed cells were stained with anti‐TMEM119 (1:100, Biolegend; 853302) extracellular primary antibody with Goat anti‐mouse IgG secondary antibody conjugated to Alexa‐488 (1:100, Abcam; ab150113). Nuclei were stained with 1µg/ml DAPI (1:1000, Thermo Fisher; 62248) before mounting with AquaPoly Mount (Poly Sciences, 18606‐20). Images were acquired with a Zeiss LSM880 Confocal microscope using a Plan‐Apochromat 20x magnification and 0.8 objective at 1024 by 1024 pixels with a 0.5 microsecond pixel dwell time.
4.9. Data analysis
4.9.1. Bulk microglia RNAseq processing
The MAPR‐Seq pipeline (Kalari et al., 2014) was used to align reads to human reference genome hg38 using STAR (Dobin et al., 2013) and count reads using featureCounts (Liao et al., 2014). FastQC was used for quality control (QC) of raw sequence reads, and RseQC was used for QC of mapped reads. Quality measures were examined including base calling quality, GC content, mapping statistics and sex check to ensure consistency between the recorded and inferred sex from expression of chromosome Y genes. Raw read counts were normalized using Conditional Quantile Normalization (CQN) to generate log2 scaled expression values via the Bioconductor package cqn, accounting for sequencing depth, gene length and GC content. Normalized CQN expression values were assessed using Principal components analysis (PCA) to identify and remove outliers, defined as greater than 4 standard deviations from the mean of the first two principal components. In addition, RPKM (reads per kilo bases per million) values were calculated.
4.10. Identification of a core microglial signature from bulk microglia data
To define a core microglial signature, we compared our bulk microglia data to cognitively normal control samples from the AMP‐AD bulk tissue transcriptome data from 7 different datasets representing 6 brain regions (Synapse ID: syn2580853); Mayo Clinic (Allen et al., 2016]) (cerebellum and superior temporal gyrus), Mount Sinai Brain Bank (Wang et al., 2018) BM10 (frontal pole), BM22 (superior temporal gyrus), BM36 (parahippocampal gyrus), BM44 (inferior frontal gyrus), and Rush University Religious Order Study‐Memory and Aging Project (ROSMAP; De Jager et al., 2018) (dorsolateral prefrontal cortex). Raw gene counts and metadata (see Acknowledgments) were obtained from the AMP‐AD RNAseq Harmonization study which had performed alignment and processing of all datasets and brain regions through a consensus pipeline (Wan et al., 2020). Samples were removed that had inconsistent sex between that indicated in metadata and that inferred from RNAseq expression; a RIN <5; were identified as gene expression outliers based on principal component analysis (PCA) (>4 standard deviation (SD) from mean PC1 or PC2), or missing metadata. In addition, duplicates (lowest read count sample removed) and those with rRNA (>5%) were removed from the MSBB datasets. Furthermore, samples not meeting neuropathological criteria as Alzheimer's disease (AD; McKhann et al., 1984) or control were excluded. To generate the microglial expression signature, only control samples from the AMP‐AD datasets were included. Raw read counts were normalized using Conditional Quantile Normalization (CQN). Log2 fold change and q‐values between each bulk tissue brain region and the bulk microglia profiles were calculated for each gene via linear regression using log2 (RPKM) without correction for covariates. Genes were filtered using a cutoff of 4‐fold greater expression in bulk microglia compared to each bulk tissue region and q < 0.05. Genes that passed these criteria and were significant in comparisons with all 7 bulk brain datasets determined the microglial signature. These signature genes were assessed for GO term enrichment with biological pathways using MsigDB. REViGO (Supek et al., 2011) tree plots were generated in R using GO terms obtained from MsigDB.
4.11. Weighted gene co‐expression network analysis
The CQN normalized expression values from bulk microglia were input to R WGCNA (Langfelder & Horvath, 2008) package v1.69. This analysis included 14,149 expressed genes, that is, median (CQN) > 2. Modules were identified, their eigengenes were calculated and merged if correlation of eigengenes > 0.7. Genes in the 40 modules identified were tested for GO term enrichment via WGCNA. Module membership (MM) for each gene was calculated as the correlation between expression of each gene and its module eigengene. Genes with MM ≥ 0.7 are considered the hub genes for the network. Gene co‐expression network plots were generated in Cytoscape v3.8 (http://www.cytoscape.org/). Each module eigengene was tested for association with age, sex and APOE e4 carrier status independently using Pearson correlation. Co‐expression network genes were annotated if they were significantly associated (p < 0.05) with the tested trait.
4.12. Over‐representation and correlation analyses
Hypergeometric testing was performed in R to determine the enrichment of a select set of genes in previously reported signatures, bulk tissue expressed genes, WGCNA modules or 10X single‐cell clusters. Correlation between bulk tissue and bulk microglial normalized CQN data was calculated using Spearman's rank correlation. Concordant and discordantly correlated genes were determined using the upper and lower quartiles from each dataset.
4.13. Single‐cell data analysis
For single‐cell RNA samples, 10X Genomics Cell Ranger Single Cell Software Suite v3.1.0 (Zheng et al., 2017) was used to demultiplex raw base call files generated from the sequencer into FASTQ files. Raw reads were aligned to human genome build GRCh38. Reads aligned to gene transcript locus were counted to generate raw UMI counts per gene per barcode for each sample. The raw UMI matrices were filtered to only keep barcodes with >500 UMIs and those that were classified as cells by Cell Ranger's cell calling algorithm.
Quality control, normalization, clustering, and marker gene identification were performed with Seurat v3 (Stuart et al., 2019), followed by annotation of clusters using established cell type markers. We kept (1) barcodes with >10% of UMI mapped to mitochondrial genome; (2) barcodes with <400 or >8000 detected genes; (3) barcodes with <500 or >46,425 mapped UMIs; (4) genes that are detected in <5 cells. These thresholds were determined by UMI or gene distribution to identify undetectable genes and outlier barcodes that may encode background, damaged or multiple cells. UMI counts of remaining cells and genes were normalized using NormalizeData function, which gave natural log transformed expression adjusted for total UMI counts in each cell. The top 2000 genes whose normalized expression varied the most across cells were identified through FindVariableFeatures function with default parameters. Using those genes, cells from 6 samples were integrated using functions FindIntegrationAnchors and IntegrateData with default parameters. Principal components (PCs) of the integrated and scaled data were computed; and the first 31 PCs, which accounted for >95% variance, were used in clustering cells. Cell clustering was performed using FindNeighbors and FindClusters with default parameters. Marker genes were identified in each cluster using FindMarkers in Seurat. Marker genes on one cluster must (1) be present in >20% cells in the cluster; (2) the log(fold change) between expression in the cluster and other clusters must be >0.25; (3) the rank sum test p‐value (Bonferroni‐adjusted) between cells in the cluster and cells in other clusters <0.05.
4.14. Meta‐analysis of bulk microglia RNAseq datasets
We obtained sets of co‐expression genes, that is, WGCNA modules from our own data and identified several modules whose eigengenes were correlated with age/sex/APOE. To check whether these correlations hold for external datasets, we downloaded raw reads in FASTQ format for three external bulk microglia datasets listed in Table S17. Reads were mapped to human genome build hg38 and were counted and normalized in the same fashion as described previously. For each WGCNA module, we identified the central genes (i.e., genes whose Pearson correlation with module eigengene > 0.75). Using these central genes, module eigengenes were calculated in all datasets—ours and three external ones. We correlated module eigengenes with traits. Finally, we performed meta‐analysis to combine correlations from multiple datasets using metacor function (random effect model) in R meta package.
4.15. Meta‐analysis of single‐cell RNAseq datasets
Raw single‐cell RNAseq data from Olah et al. (2020) was downloaded through Synapse (syn21438358) and processed through Cell Ranger. Quality control was performed as described prior to integration with our samples using Seurat v4.0.4. Integration was performed to combine the datasets by individual rather than sample. A total of 9 samples and 15,819 cells were retained from the Olah data. Cells were annotated to include dataset of origin. Hypergeometric tests were performed to determine the enrichment of our microglial signature within each integrated cluster. Additionally, we also looked at overlap between integrated cluster genes and marker genes from our original single‐cell dataset. To identify where our original cells localized within the integrated dataset, we mapped the original cell IDs to the integrated clusters and calculated percentage overlap.
CONFLICT OF INTEREST
None.
AUTHOR CONTRIBUTIONS
TP and NET wrote the manuscript; NET and MA designed the study; TP, XW, and ZQ performed data analysis; JC consulted on statistical methods; TP, TPC, XW, YM, and RMA generated tables and figures; EM, CAG, SG, KC, RW, HGC, and AQH provided neurosurgical tissue samples; TP, TPC, LJLT, SJL, SL, FQTN, CCGH, KGM, and TN performed experimental procedures from blood and tissue samples. All authors read the manuscript and provided input and consultation. NET oversaw the study and provided direction, funding, and resources.
Supporting information
Fig S1‐S10
Table S1‐S20
ACKNOWLEDGMENTS
The authors thank the patients and their families for their participation, without whom these studies would not have been possible. We thank our colleagues in the neurosurgery team Christopher Louie, Karim ReFaey, and Ivan Segura Duran. We thank our colleagues at the Mayo Clinic Genome Analysis Core (GAC) for their collaboration, particular gratitude to Bruce Eckloff and Julie Lau. We also acknowledge the AMP‐AD RNAseq reprocessing team, in particular Dr. Kirsten Dang, Dr. Thanneer Perumal and Dr. Ben Logsdon at Sage Bionetworks. AMP‐AD RNASeq datasets: This study is a cross‐consortia project using RNAseq data generated through grants U01AG046152, U01AG046170, and U01AG046139. For the Mayo RNAseq study: The results published here are in whole or in part based on data obtained from the AD Knowledge Portal (https://adknowledgeportal.synapse.org/). Study data were provided by the following sources: The Mayo Clinic Alzheimer’s Disease Genetic Studies, led by Dr. Nilüfer Ertekin‐Taner and Dr. Steven G. Younkin, Mayo Clinic, Jacksonville, FL using samples from the Mayo Clinic Study of Aging, the Mayo Clinic Alzheimer’s Disease Research Center, and the Mayo Clinic Brain Bank. Data collection was supported through funding by NIA grants P50 AG016574, R01 AG032990, U01 AG046139, R01 AG018023, U01 AG006576, U01 AG006786, R01 AG025711, R01 AG017216, R01 AG003949, NINDS grant R01 NS080820, CurePSP Foundation, and support from Mayo Foundation. Study data includes samples collected through the Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona. The Brain and Body Donation Program is supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026 National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders), the National Institute on Aging (P30 AG19610 Arizona Alzheimer’s Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05‐901, and 1001 to the Arizona Parkinson's Disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research. For the ROSMAP study: The results published here are in whole or in part based on data obtained from the AD Knowledge Portal (https://adknowledgeportal.synapse.org). Study data were provided by the Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago. Data collection was supported through funding by NIA grants P30AG10161 (ROS), R01AG15819 (ROSMAP; genomics and RNAseq), R01AG17917 (MAP), R01AG30146, R01AG36042 (5hC methylation, ATACseq), RC2AG036547 (H3K9Ac), R01AG36836 (RNAseq), R01AG48015 (monocyte RNAseq) RF1AG57473 (single nucleus RNAseq), U01AG32984 (genomic and whole exome sequencing), U01AG46152 (ROSMAP AMP‐AD, targeted proteomics), U01AG46161(TMT proteomics), U01AG61356 (whole genome sequencing, targeted proteomics, ROSMAP AMP‐AD), the Illinois Department of Public Health (ROSMAP), and the Translational Genomics Research Institute (genomic). Additional phenotypic data can be requested at www.radc.rush.edu. For the MSBB study: The results published here are in whole or in part based on data obtained from the AD Knowledge Portal (https://adknowledgeportal.synapse.org/). These data were generated from postmortem brain tissue collected through the Mount Sinai VA Medical Center Brain Bank and were provided by Dr. Eric Schadt from Mount Sinai School of Medicine.
Patel, T. , Carnwath, T. P. , Wang, X. , Allen, M. , Lincoln, S. J. , Lewis‐Tuffin, L. J. , Quicksall, Z. S. , Lin, S. , Tutor‐New, F. Q. , Ho, C. C. G. , Min, Y. , Malphrus, K. G. , Nguyen, T. T. , Martin, E. , Garcia, C. A. , Alkharboosh, R. M. , Grewal, S. , Chaichana, K. , Wharen, R. , … Ertekin‐Taner, N. (2022). Transcriptional landscape of human microglia implicates age, sex, and APOE‐related immunometabolic pathway perturbations. Aging Cell, 21, e13606. 10.1111/acel.13606
Funding information
This study was supported by NIH funding U01 AG046193, RF1 AG051504, and R01 AG051504 to NET.
DATA AVAILABILITY STATEMENT
The data in this manuscript are available via the AD Knowledge Portal (https://adknowledgeportal.synapse.org). The AD Knowledge Portal is a platform for accessing data, analyses and tools generated by the Accelerating Medicines Partnership (AMP‐AD) Target Discovery Program and other National Institute on Aging (NIA)‐supported programs to enable open‐science practices and accelerate translational learning. The data, analyses, and tools are shared early in the research cycle without a publication embargo on secondary use. Data are available for general research use according to the following requirements for data access and data attribution (https://adknowledgeportal.synapse.org/DataAccess/Instructions). The raw data files are available through Synapse (https://www.synapse.org/#!Synapse:syn28450881.3/datasets/).
| Dataset | Data Type | Description | SynapseID | DoD |
|---|---|---|---|---|
| Mayo RNAseq TCX | Metadata | Individual human and RNAseq | syn5550404 | na |
| Mayo RNAseq CER | Metadata | Individual human and RNAseq | syn5550404 | na |
| Mayo RNAseq TCX | RNASeq Expression | Consensus processed RNASeq raw counts | syn8690799 | 10/2/2019 |
| Mayo RNAseq CER | RNASeq Expression | Consensus processed RNASeq raw counts | syn8690904 | 10/2/2019 |
| ROSMAP | Metadata | ID Key | syn3382527 | 10/2/2019 |
| ROSMAP | Metadata | Individual human | syn3191087 | 10/2/2019 |
| ROSMAP | Metadata | Assay RNAseq | syn21088596 | 1/2/2020 |
| ROSMAP | RNASeq Expression | Consensus processed RNASeq raw counts | syn8691134 | 10/2/2019 |
| MSBB | Metadata | Individual human | syn6101474 | 11/22/2019 |
| MSBB | Metadata | Assay RNAseq | syn6100548 | 10/2/2019 |
| MSBB | RNASeq Expression | Consensus processed RNASeq raw counts | syn8691099 | 10/2/2019 |
Data from AD knowledge portal utilized in this study. DoD = Date of download, “na” indicates data that were generated by study authors and shared within the AD knowledge portal. Data were obtained from the RNAseq Harmonization Study in the AD knowledge portal (DOI: https://doi.org/10.7303/syn9702085).
REFERENCES
- Ahn, E. H. , Kang, S. S. , Qi, Q. , Liu, X. , & Ye, K. (2020). Netrin1 deficiency activates MST1 via UNC5B receptor, promoting dopaminergic apoptosis in Parkinson’s disease. Proceedings of the National Academy of Sciences, 117, 24503–24513. 10.1073/pnas.2004087117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, M. , Carrasquillo, M. M. , Funk, C. , Heavner, B. D. , Zou, F. , Younkin, C. S. , Burgess, J. D. , Chai, H.‐S. , Crook, J. , Eddy, J. A. , Li, H. , Logsdon, B. , Peters, M. A. , Dang, K. K. , Wang, X. , Serie, D. , Wang, C. , Nguyen, T. , Lincoln, S. , … Ertekin‐Taner, N. (2016). Human whole genome genotype and transcriptome data for Alzheimer's and other neurodegenerative diseases. Scientific Data, 3, 160089. 10.1038/sdata.2016.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, M. , Wang, X. , Burgess, J. D. , Watzlawik, J. , Serie, D. J. , Younkin, C. S. , Nguyen, T. , Malphrus, K. G. , Lincoln, S. , Carrasquillo, M. M. , Ho, C. , Chakrabarty, P. , Strickland, S. , Murray, M. E. , Swarup, V. , Geschwind, D. H. , Seyfried, N. T. , Dammer, E. B. , Lah, J. J. , … Ertekin‐Taner, N. (2018). Conserved brain myelination networks are altered in Alzheimer's and other neurodegenerative diseases. Alzheimer's & Dementia: the Journal of the Alzheimer's Association, 14, 352–366. 10.1016/j.jalz.2017.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, M. , Wang, X. , Serie, D. J. , Strickland, S. L. , Burgess, J. D. , Koga, S. , Younkin, C. S. , Nguyen, T. T. , Malphrus, K. G. , Lincoln, S. J. , Alamprese, M. , Zhu, K. , Chang, R. , Carrasquillo, M. M. , Kouri, N. , Murray, M. E. , Reddy, J. S. , Funk, C. , Price, N. D. , … Ertekin‐Taner, N. (2018). Divergent brain gene expression patterns associate with distinct cell‐specific tau neuropathology traits in progressive supranuclear palsy. Acta Neuropathologica, 136, 709–727. 10.1007/s00401-018-1900-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsema, A. M. , Jiang, Q. , Kracht, L. , Gerrits, E. , Dubbelaar, M. L. , Miedema, A. , Brouwer, N. , Hol, E. M. , Middeldorp, J. , van Dijk, R. , Woodbury, M. , Wachter, A. , Xi, S. , Möller, T. , Biber, K. P. , Kooistra, S. M. , Boddeke, E. W. G. M. , & Eggen, B. J. L. (2020). Profiling microglia from Alzheimer’s disease donors and non‐demented elderly in acute human postmortem cortical tissue. Frontiers in Molecular Neuroscience, 13, 134. 10.3389/fnmol.2020.00134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreone, B. J. , Przybyla, L. , Llapashtica, C. , Rana, A. , Davis, S. S. , van Lengerich, B. , Karin Lin, J. U. , Shi, Y. M. , Astarita, G. , Di Paolo, G. , Sandmann, T. , Monroe, K. M. , & Lewcoc, J. W. (2020). Alzheimer's‐associated PLCgamma2 is a signaling node required for both TREM2 function and the inflammatory response in human microglia. Nature Neuroscience, 23, 927–938. [DOI] [PubMed] [Google Scholar]
- Aung, L. L. , Brooks, A. , Greenberg, S. A. , Rosenberg, M. L. , Dhib‐Jalbut, S. , & Balashov, K. E. (2012). Multiple sclerosis‐linked and interferon‐beta‐regulated gene expression in plasmacytoid dendritic cells. Journal of Neuroimmunology, 250, 99–105. 10.1016/j.jneuroim.2012.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers, K. L. , Mirshahi, U. L. , Wardeh, A. H. , Murray, M. F. , Hao, K. E. , Glicksberg, B. S. , Li, S. , Carey, D. J. , & Chen, R. (2016). A loss of function variant in CASP7 protects against Alzheimer's disease in homozygous APOE ε4 allele carriers. BMC Genomics, 17(S2), 445. 10.1186/s12864-016-2725-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier, L. P. , York, E. M. , & MacVicar, B. A. (2020). Immunometabolism in the brain: how metabolism shapes microglial function. Trends in Neurosciences, 43, 854–869. 10.1016/j.tins.2020.08.008 [DOI] [PubMed] [Google Scholar]
- Butovsky, O. , & Weiner, H. L. (2018). Microglial signatures and their role in health and disease. Nature Reviews Neuroscience, 19, 622–635. 10.1038/s41583-018-0057-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak, S. , Kosicek, M. , Mladenovic‐Djordjevic, A. , Smiljanic, K. , Kanazir, S. , & Hecimovic, S. (2016). Loss of Cathepsin B and L leads to lysosomal dysfunction, NPC‐like cholesterol sequestration and accumulation of the key Alzheimer's proteins. PLoS One, 11, e0167428. 10.1371/journal.pone.0167428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chausse, B. , Kakimoto, P. A. , & Kann, O. (2020). Microglia and lipids: How metabolism controls brain innate immunity. Seminars in Cell & Developmental Biology, 112, 137–144. 10.1016/j.semcdb.2020.08.001 [DOI] [PubMed] [Google Scholar]
- Chrétien, F. , Le Pavec, G. , Vallat‐Decouvelaere, A.‐V. , Marie‐Bernadette‐Delisle , Uro‐Coste, E. , Ironside, J. W. , Gambetti, P. , Parchi, P. , Créminon, C. , Dormont, D. , Mikol, J. , Gray, F. , & Gras, G. (2004). Expression of excitatory amino acid transporter‐1 (EAAT‐1) in brain macrophages and microglia of patients with prion diseases. Journal of Neuropathology & Experimental Neurology, 63, 1058–1071. 10.1093/jnen/63.10.1058 [DOI] [PubMed] [Google Scholar]
- Colombo, A. , Dinkel, L. , Müller, S. A. , Sebastian Monasor, L. , Schifferer, M. , Cantuti‐Castelvetri, L. , König, J. , Vidatic, L. , Bremova‐Ertl, T. , Lieberman, A. P. , Hecimovic, S. , Simons, M. , Lichtenthaler, S. F. , Strupp, M. , Schneider, S. A. , & Tahirovic, S. (2021). Loss of NPC1 enhances phagocytic uptake and impairs lipid trafficking in microglia. Nature Communications, 12, 1158. 10.1038/s41467-021-21428-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti, A. , Sait, H. R. , McAvoy, K. M. , Estrada, K. , Ergun, A. , Szak, S. , Marsh, G. , Jandreski, L. , Peterson, M. , Reynolds, T. L. , Dalkilic‐Liddle, I. , Cameron, A. , Cahir‐McFarland, E. , & Ransohoff, R. M. (2019). BIN1 favors the spreading of Tau via extracellular vesicles. Scientific Reports, 9, 9477. 10.1038/s41598-019-45676-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, O. , Campion, S. , Perry, V. H. , Murray, C. , Sidenius, N. , Docagne, F. , & Cunningham, C. (2009). Microglia and the Urokinase plasminogen activator receptor/uPA system in innate brain inflammation. Glia, 57, 1802–1814. 10.1002/glia.20892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis, S. , Sloan, S. A. , Croote, D. , Mignardi, M. , Chernikova, S. , Samghababi, P. , Zhang, Y. E. , Neff, N. , Kowarsky, M. , Caneda, C. , Li, G. , Chang, S. D. , Connolly, I. D. , Li, Y. , Barres, B. A. , Gephart, M. H. , & Quake, S. R. (2017). Single‐cell RNA‐Seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Reports, 21, 1399–1410. 10.1016/j.celrep.2017.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis, S. , Sloan, S. A. , Zhang, Y. E. , Enge, M. , Caneda, C. , Shuer, L. M. , Hayden Gephart, M. G. , Barres, B. A. , & Quake, S. R. (2015). A survey of human brain transcriptome diversity at the single cell level. Proceedings of the National Academy of Sciences, 112, 7285. 10.1073/pnas.1507125112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager, P. L. , Ma, Y. , McCabe, C. , Xu, J. , Vardarajan, B. N. , Felsky, D. , Klein, H.‐U. , White, C. C. , Peters, M. A. , Lodgson, B. , Nejad, P. , Tang, A. , Mangravite, L. M. , Yu, L. , Gaiteri, C. , Mostafavi, S. , Schneider, J. A. , & Bennett, D. A. (2018). A multi‐omic atlas of the human frontal cortex for aging and Alzheimer's disease research. Scientific Data, 5, 180142. 10.1038/sdata.2018.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del‐Aguila, J. L. , Li, Z. , Dube, U. , Mihindukulasuriya, K. A. , Budde, J. P. , Fernandez, M. V. , Ibanez, L. , Bradley, J. , Wang, F. , Bergmann, K. , Davenport, R. , Morris, J. C. , Holtzman, D. M. , Perrin, R. J. , Benitez, B. A. , Dougherty, J. , Cruchaga, C. , & Harari, O. (2019). A single‐nuclei RNA sequencing study of Mendelian and sporadic AD in the human brain. Alzheimer's Research & Therapy, 11, 71. 10.1186/s13195-019-0524-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePaula‐Silva, A. B. , Gorbea, C. , Doty, D. J. , Libbey, J. E. , Sanchez, J. M. S. , Hanak, T. J. , Cazalla, D. , & Fujinami, R. S. (2019). Differential transcriptional profiles identify microglial‐ and macrophage‐specific gene markers expressed during virus‐induced neuroinflammation. Journal of Neuroinflammation, 16, 152. 10.1186/s12974-019-1545-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A. , Davis, C. A. , Schlesinger, F. , Drenkow, J. , Zaleski, C. , Jha, S. , Batut, P. , Chaisson, M. , & Gingeras, T. R. (2013). STAR: ultrafast universal RNA‐seq aligner. Bioinformatics, 29, 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Khoury, J. , Hickman, S. E. , Thomas, C. A. , Loike, J. D. , & Silverstein, S. C. (1998). Microglia, scavenger receptors, and the pathogenesis of Alzheimer's disease. Neurobiology of Aging, 19, S81–84. 10.1016/S0197-4580(98)00036-0 [DOI] [PubMed] [Google Scholar]
- Frigerio, C. , Wolfs, L. , Fattorelli, N. , Thrupp, N. , Voytyuk, I. , Schmidt, I. , Mancuso, R. , Chen, W.‐T. , Woodbury, M. E. , Srivastava, G. , Möller, T. , Hudry, E. , Das, S. , Saido, T. , Karran, E. , Bradley Hyman, V. , Perry, H. , Fiers, M. , & De Strooper, B. (2019). The major risk factors for Alzheimer's disease: Age, sex, and genes modulate the microglia response to amyloid‐beta plaques. Cell Reports, 27, 1293–1306 e1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galatro, T. F. , Holtman, I. R. , Lerario, A. M. , Vainchtein, I. D. , Brouwer, N. , Sola, P. R. , Veras, M. M. , Pereira, T. F. , Leite, R. E. P. , Möller, T. , Wes, P. D. , Sogayar, M. C. , Laman, J. D. , den Dunnen, W. , Pasqualucci, C. A. , Oba‐Shinjo, S. M. , Boddeke, E. W. G. M. , Marie, S. K. N. , & Eggen, B. J. L. (2017). Transcriptomic analysis of purified human cortical microglia reveals age‐associated changes. Nature Neuroscience, 20, 1162–1171. 10.1038/nn.4597 [DOI] [PubMed] [Google Scholar]
- Gamache, J. , Yun, Y. , & Chiba‐Falek, O. (2020). Sex‐dependent effect of APOE on Alzheimer's disease and other age‐related neurodegenerative disorders. Disease Models & Mechanisms, 13, dmm045211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi, K. S. , McKay, F. C. , Cox, M. , Riveros, C. , Armstrong, N. , Heard, R. N. , Vucic, S. , Williams, D. W. , Stankovich, J. , Brown, M. , Danoy, P. , Stewart, G. J. , Broadley, S. , Moscato, P. , Lechner‐Scott, J. , Scott, R. J. , Booth, D. R. , Griffiths, L. , Slee, M. , … Bahlo, M. (2010). The multiple sclerosis whole blood mRNA transcriptome and genetic associations indicate dysregulation of specific T cell pathways in pathogenesis. Human Molecular Genetics, 19, 2134–2143. 10.1093/hmg/ddq090 [DOI] [PubMed] [Google Scholar]
- Gerrits, E. , Brouwer, N. , Kooistra, S. M. , Woodbury, M. E. , Vermeiren, Y. , Lambourne, M. , Mulder, J. , Kummer, M. , Möller, T. , Biber, K. , Dunnen, W. F. A. D. , De Deyn, P. P. , Eggen, B. J. L. , & Boddeke, E. W. G. M. (2021). Distinct amyloid‐β and tau‐associated microglia profiles in Alzheimer’s disease. Acta Neuropathologica, 141, 681–696. 10.1007/s00401-021-02263-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin, D. , Skola, D. , Coufal, N. G. , Holtman, I. R. , Schlachetzki, J. C. M. , Sajti, E. , Jaeger, B. N. , O’Connor, C. , Fitzpatrick, C. , Pasillas, M. P. , Pena, M. , Adair, A. , Gonda, D. D. , Levy, M. L. , Ransohoff, R. M. , Gage, F. H. , & Glass, C. K. (2017). An environment‐dependent transcriptional network specifies human microglia identity. Science, 356, eaal3222. 10.1126/science.aal3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassivaro, F. , Menon, R. , Acquaviva, M. , Ottoboni, L. , Ruffini, F. , Bergamaschi, A. , Muzio, L. , Farina, C. , & Martino, G. (2020). Convergence between microglia and peripheral macrophages phenotype during development and neuroinflammation. The Journal of Neuroscience, 40, 784–795. 10.1523/JNEUROSCI.1523-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubman, A. , Chew, G. , Ouyang, J. F. , Sun, G. , Choo, X. Y. , McLean, C. , Simmons, R. K. , Buckberry, S. , Vargas‐Landin, D. B. , Poppe, D. , Pflueger, J. , Lister, R. , Rackham, O. J. L. , Petretto, E. , & Polo, J. M. (2019). A single‐cell atlas of entorhinal cortex from individuals with Alzheimer's disease reveals cell‐type‐specific gene expression regulation. Nature Neuroscience, 22, 2087–2097. 10.1038/s41593-019-0539-4 [DOI] [PubMed] [Google Scholar]
- Hammond, T. R. , Dufort, C. , Dissing‐Olesen, L. , Giera, S. , Young, A. , Wysoker, A. , Walker, A. J. , Gergits, F. , Segel, M. , Nemesh, J. , Marsh, S. E. , Saunders, A. , Macosko, E. , Ginhoux, F. , Chen, J. , Franklin, R. J. M. , Piao, X. , McCarroll, S. A. , & Stevens, B. (2019). Single‐cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell‐state changes. Immunity, 50, 253–271 e256. 10.1016/j.immuni.2018.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx, D. A. E. , van Eden, C. G. , Schuurman, K. G. , Hamann, J. , & Huitinga, I. (2017). Staining of HLA‐DR, Iba1 and CD68 in human microglia reveals partially overlapping expression depending on cellular morphology and pathology. Journal of Neuroimmunology, 309, 12–22. 10.1016/j.jneuroim.2017.04.007 [DOI] [PubMed] [Google Scholar]
- Hickman, S. , Izzy, S. , Sen, P. , Morsett, L. , & El Khoury, J. (2018). Microglia in neurodegeneration. Nature Neuroscience, 21, 1359–1369. 10.1038/s41593-018-0242-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalari, K. R. , Nair, A. A. , Bhavsar, J. D. , O’Brien, D. R. , Davila, J. I. , Bockol, M. A. , Nie, J. , Tang, X. , Baheti, S. , Doughty, J. B. , Middha, S. , Sicotte, H. , Thompson, A. E. , Asmann, Y. W. , & Kocher, J.‐P. (2014). MAP‐RSeq: Mayo analysis pipeline for RNA sequencing. BMC Bioinformatics, 15, 224. 10.1186/1471-2105-15-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminska, B. , Ochocka, N. , & Segit, P. (2021). Single‐cell omics in dissecting immune microenvironment of malignant gliomas: Challenges and perspectives. Cells, 10, 2264. 10.3390/cells10092264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsouri, L. , & Georgopoulos, S. (2011). Lack of LDL receptor enhances amyloid deposition and decreases glial response in an Alzheimer's disease mouse model. PLoS One, 6, e21880. 10.1371/journal.pone.0021880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenkhuis, B. , Somarakis, A. , de Haan, L. , Dzyubachyk, O. , IJsselsteijn, M. E. , de Miranda, N. F. C. C. , Lelieveldt, B. P. F. , Dijkstra, J. , van Roon‐Mom, W. M. C. , Höllt, T. , & van der Weerd, L. (2021). Iron loading is a prominent feature of activated microglia in Alzheimer's disease patients. Acta Neuropathologica Communications, 9, 27. 10.1186/s40478-021-01126-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren‐Shaul, H. , Spinrad, A. , Weiner, A. , Matcovitch‐Natan, O. , Dvir‐Szternfeld, R. , Ulland, T. K. , David, E. , Baruch, K. , Lara‐Astaiso, D. , Toth, B. , Itzkovitz, S. , Colonna, M. , Schwartz, M. , & Amit, I. (2017). A unique microglia type associated with restricting development of Alzheimer's disease. Cell, 169, 1276–1290 e1217. 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Kinoshita, S. , & Koyama, R. (2021). Pro‐ and anti‐epileptic roles of microglia. Neural Regeneration Research, 16, 1369–1371. 10.4103/1673-5374.300976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koellhoffer, E. C. , McCullough, L. D. , & Ritzel, R. M. (2017). Old maids: Aging and its impact on microglia function. International Journal of Molecular Sciences, 18, 769. 10.3390/ijms18040769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann, S. , Madore, C. , Cialic, R. , Baufeld, C. , Calcagno, N. , El Fatimy, R. , Beckers, L. , O'Loughlin, E. , Yang, X. U. , Fanek, Z. , Greco, D. J. , Smith, S. T. , Tweet, G. , Humulock, Z. , Zrzavy, T. , Conde‐Sanroman, P. , Gacias, M. , Weng, Z. , Chen, H. , & … Butovsky, O. (2017). The TREM2‐APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity, 47, 566–581 e569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lämsä, R. , Helisalmi, S. , Herukka, S.‐K. , Tapiola, T. , Pirttilä, T. , Vepsäläinen, S. , Hiltunen, M. , & Soininen, H. (2008). Genetic study evaluating LDLR polymorphisms and Alzheimer's disease. Neurobiology of Aging, 29, 848–855. 10.1016/j.neurobiolaging.2006.12.009 [DOI] [PubMed] [Google Scholar]
- Langfelder, P. , & Horvath, S. (2008). WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics, 9, 559. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc, V. , Jasmin‐Belanger, S. , & Poirier, J. (2010). APOE and cholesterol homeostasis in Alzheimer's disease. Trends in Molecular Medicine, 16, 469–477. 10.1016/j.molmed.2010.07.008 [DOI] [PubMed] [Google Scholar]
- Li, Q. , & Barres, B. A. (2018). Microglia and macrophages in brain homeostasis and disease. Nature Reviews Immunology, 18, 225–242. 10.1038/nri.2017.125 [DOI] [PubMed] [Google Scholar]
- Liao, Y. , Smyth, G. K. , & Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30, 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lin, Y.‐T. , Seo, J. , Gao, F. , Feldman, H. M. , Wen, H.‐L. , Penney, J. , Cam, H. P. , Gjoneska, E. , Raja, W. K. , Cheng, J. , Rueda, R. , Kritskiy, O. , Abdurrob, F. , Peng, Z. , Milo, B. , Yu, C. J. , Elmsaouri, S. , Dey, D. , Ko, T. , … Tsai, L.‐H. (2018). APOE4 causes widespread molecular and cellular alterations associated with Alzheimer's disease phenotypes in human iPSC‐Derived brain cell types. Neuron, 98, 1141–1154 e1147. 10.1016/j.neuron.2018.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loving, B. A. , & Bruce, K. D. (2020). Lipid and lipoprotein metabolism in microglia. Frontiers in Physiology, 11, 393. 10.3389/fphys.2020.00393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschallinger, J. , Iram, T. , Zardeneta, M. , Lee, S. E. , Lehallier, B. , Haney, M. S. , Pluvinage, J. V. , Mathur, V. , Hahn, O. , Morgens, D. W. , Kim, J. , Tevini, J. , Felder, T. K. , Wolinski, H. , Bertozzi, C. R. , Bassik, M. C. , Aigner, L. , & Wyss‐Coray, T. (2020). Lipid‐droplet‐accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nature Neuroscience, 23(2), 194–208. 10.1038/s41593-019-0566-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda, T. , Sankowski, R. , Staszewski, O. , Böttcher, C. , Amann, L. , Sagar , Scheiwe, C. , Nessler, S. , Kunz, P. , van Loo, G. , Coenen, V. A. , Reinacher, P. C. , Michel, A. , Sure, U. , Gold, R. , Grün, D. , Priller, J. , Stadelmann, C. , & Prinz, M. (2019). Spatial and temporal heterogeneity of mouse and human microglia at single‐cell resolution. Nature, 566, 388–392. 10.1038/s41586-019-0924-x [DOI] [PubMed] [Google Scholar]
- Masuda, T. , Sankowski, R. , Staszewski, O. , & Prinz, M. (2020). Microglia heterogeneity in the single‐cell era. Cell Reports, 30, 1271–1281. 10.1016/j.celrep.2020.01.010 [DOI] [PubMed] [Google Scholar]
- Mathys, H. , Davila‐Velderrain, J. , Peng, Z. , Gao, F. , Mohammadi, S. , Young, J. Z. , Menon, M. , He, L. , Abdurrob, F. , Jiang, X. , Martorell, A. J. , Ransohoff, R. M. , Hafler, B. P. , Bennett, D. A. , Kellis, M. , & Tsai, L.‐H. (2019). Single‐cell transcriptomic analysis of Alzheimer's disease. Nature, 570, 332–337. 10.1038/s41586-019-1195-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie, A. T. , Wang, M. , Hauberg, M. E. , Fullard, J. F. , Kozlenkov, A. , Keenan, A. , Hurd, Y. L. , Dracheva, S. , Casaccia, P. , Roussos, P. , & Zhang, B. (2018). Brain cell type specific gene expression and co‐expression network architectures. Scientific Reports, 8, 8868. 10.1038/s41598-018-27293-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann, G. , Drachman, D. , Folstein, M. , Katzman, R. , Price, D. , & Stadlan, E. M. (1984). Clinical diagnosis of Alzheimer's disease: report of the NINCDS‐ADRDA work group under the auspices of department of health and human services task force on Alzheimer's disease. Neurology, 34, 939–944. 10.1212/WNL.34.7.939 [DOI] [PubMed] [Google Scholar]
- Mehra, A. , Ali, C. , Parcq, J. , Vivien, D. , & Docagne, F. (2016). The plasminogen activation system in neuroinflammation. Biochimica Et Biophysica Acta (BBA) ‐ Molecular Basis of Disease, 1862, 395–402. 10.1016/j.bbadis.2015.10.011 [DOI] [PubMed] [Google Scholar]
- Morin‐Brureau, M. , Milior, G. , Royer, J. , Chali, F. , Le Duigou, C. , Savary, E. , Blugeon, C. , Jourdren, L. , Akbar, D. , Dupont, S. , Navarro, V. , Baulac, M. , Bielle, F. , Mathon, B. , Clemenceau, S. , & Miles, R. (2018). Microglial phenotypes in the human epileptic temporal lobe. Brain, 141, 3343–3360. 10.1093/brain/awy276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi, S. , Gaiteri, C. , Sullivan, S. E. , White, C. C. , Tasaki, S. , Xu, J. , Taga, M. , Klein, H.‐U. , Patrick, E. , Komashko, V. , McCabe, C. , Smith, R. , Bradshaw, E. M. , Root, D. E. , Regev, A. , Yu, L. , Chibnik, L. B. , Schneider, J. A. , Young‐Pearse, T. L. , … De Jager, P. L. (2018). A molecular network of the aging human brain provides insights into the pathology and cognitive decline of Alzheimer's disease. Nature Neuroscience, 21, 811–819. 10.1038/s41593-018-0154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, R. A. , Wang, M. , Vatansever, S. , Guo, L. , Ming, C. , Wang, Q. , Wang, E. , Horgusluoglu‐Moloch, E. , Song, W.‐M. , Li, A. , Castranio, E. L. , Tcw, J. , Ho, L. , Goate, A. , Fossati, V. , Noggle, S. , Gandy, S. , Ehrlich, M. E. , Katsel, P. , … Zhang, B. (2021). Molecular subtyping of Alzheimer's disease using RNA sequencing data reveals novel mechanisms and targets. Science Advances, 7, eabb5398. 10.1126/sciadv.abb5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, L. H. , Warden, S. , & Lenz, K. M. (2017). Sex differences in microglial phagocytosis in the neonatal hippocampus. Brain, Behavior, and Immunity, 64, 11–22. 10.1016/j.bbi.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, A. T. , Wang, K. , Hu, G. , Wang, X. , Miao, Z. , Azevedo, J. A. , Suh, E. R. , Van Deerlin, V. M. , Choi, D. , Roeder, K. , Li, M. , & Lee, E. B. (2020). APOE and TREM2 regulate amyloid‐responsive microglia in Alzheimer’s disease. Acta Neuropathologica, 140, 477–493. 10.1007/s00401-020-02200-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden, D. M. , & Godbout, J. P. (2013). Review: microglia of the aged brain: primed to be activated and resistant to regulation. Neuropathology and Applied Neurobiology, 39, 19–34. 10.1111/j.1365-2990.2012.01306.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent, A. A. , Lin, K. , van Lengerich, B. , Lianoglou, S. , Przybyla, L. , Davis, S. S. , Llapashtica, C. , Wang, J. , Kim, D. J. , Xia, D. , Lucas, A. , Baskaran, S. , Haddick, P. C. G. , Lenser, M. , Earr, T. K. , Shi, J. U. , Dugas, J. C. , Andreone, B. J. , Logan, T. , … Di Paolo, G. (2020). TREM2 regulates microglial cholesterol metabolism upon chronic phagocytic challenge. Neuron, 105, 837–854 e839. 10.1016/j.neuron.2019.12.007 [DOI] [PubMed] [Google Scholar]
- Ochocka, N. , Segit, P. , Walentynowicz, K. A. , Wojnicki, K. , Cyranowski, S. , Swatler, J. , Mieczkowski, J. , & Kaminska, B. (2021). Single‐cell RNA sequencing reveals functional heterogeneity of glioma‐associated brain macrophages. Nature Communications, 12, 1151. 10.1038/s41467-021-21407-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olah, M. , Menon, V. , Habib, N. , Taga, M. F. , Ma, Y. , Yung, C. J. , Cimpean, M. , Khairallah, A. , Coronas‐Samano, G. , Sankowski, R. , Grün, D. , Kroshilina, A. A. , Dionne, D. , Sarkis, R. A. , Cosgrove, G. R. , Helgag, J. , Golden, J. A. , Pennell, P. B. , Prinz, M. , … De Jager, P. L. (2020). Single cell RNA sequencing of human microglia uncovers a subset associated with Alzheimer's disease. Nature Communications, 11, 6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olah, M. , Patrick, E. , Villani, A.‐C. , Xu, J. , White, C. C. , Ryan, K. J. , Piehowski, P. , Kapasi, A. , Nejad, P. , Cimpean, M. , Connor, S. , Yung, C. J. , Frangieh, M. , McHenry, A. , Elyaman, W. , Petyuk, V. , Schneider, J. A. , Bennett, D. A. , De Jager, P. L. , & Bradshaw, E. M. (2018). A transcriptomic atlas of aged human microglia. Nature Communications, 9, 539. 10.1038/s41467-018-02926-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciotti, S. , Albi, E. , Parnetti, L. , & Beccari, T. (2020). Lysosomal ceramide metabolism disorders: Implications in Parkinson's disease. Journal of Clinical Medicine, 9, 594. 10.3390/jcm9020594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju, S. , Dammer, E. B. , Raza, S. A. , Gao, T. , Xiao, H. , Betarbet, R. , Duong, D. M. , Webster, J. A. , Hales, C. M. , Lah, J. J. , Levey, A. I. , & Seyfried, N. T. (2018). Quantitative proteomics of acutely‐isolated mouse microglia identifies novel immune Alzheimer's disease‐related proteins. Molecular Neurodegeneration, 13, 34. 10.1186/s13024-018-0266-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju, S. , Dammer, E. B. , Raza, S. A. , Rathakrishnan, P. , Xiao, H. , Gao, T. , Duong, D. M. , Pennington, M. W. , Lah, J. J. , Seyfried, N. T. , & Levey, A. I. (2018). Identification and therapeutic modulation of a pro‐inflammatory subset of disease‐associated‐microglia in Alzheimer's disease. Molecular Neurodegeneration, 13, 24. 10.1186/s13024-018-0254-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankowski, R. , Böttcher, C. , Masuda, T. , Geirsdottir, L. , Sagar , Sindram, E. , Seredenina, T. , Muhs, A. , Scheiwe, C. , Shah, M. J. , Heiland, D. H. , Schnell, O. , Grün, D. , Priller, J. , & Prinz, M. (2019). Mapping microglia states in the human brain through the integration of high‐dimensional techniques. Nature Neuroscience, 22, 2098–2110. 10.1038/s41593-019-0532-y [DOI] [PubMed] [Google Scholar]
- Satoh, J.‐I. , Nakanishi, M. , Koike, F. , Miyake, S. , Yamamoto, T. , Kawai, M. , Kikuchi, S. , Nomura, K. , Yokoyama, K. , Ota, K. , Kanda, T. , Fukazawa, T. , & Yamamura, T. (2005). Microarray analysis identifies an aberrant expression of apoptosis and DNA damage‐regulatory genes in multiple sclerosis. Neurobiology of Diseases, 18, 537–550. 10.1016/j.nbd.2004.10.007 [DOI] [PubMed] [Google Scholar]
- Schmunk, G. , Kim, C. N. , Soliman, S. S. , Keefe, M. G. , Bogdanoff, D. , Tejera, D. , Ziffra, R. S. , Shin, D. , Allen, D. E. , Chhun, B. B. , McGinnis, C. S. , Winkler, E. A. , Abla, A. A. , Chang, E. F. , Gartner, Z. J. , Mehta, S. B. , Piao, X. , Hengen, K. B. , & Nowakowski, T. J. (2020). Human microglia upregulate cytokine signatures and accelerate maturation of neural networks. bioRxiv, 2020.2003.2024.006874. [Google Scholar]
- Shi, Y. , Manis, M. , Long, J. , Wang, K. , Sullivan, P. M. , Remolina Serrano, J. , Hoyle, R. , & Holtzman, D. M. (2019). Microglia drive APOE‐dependent neurodegeneration in a tauopathy mouse model. Journal of Experimental Medicine, 216, 2546–2561. 10.1084/jem.20190980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims, R. , van der Lee, S. J. , Naj, A. C. , Bellenguez, C. , Badarinarayan, N. , Jakobsdottir, J. , Kunkle, B. W. , Boland, A. , Raybould, R. , Bis, J. C. , Martin, E. R. , Grenier‐Boley, B. , Heilmann‐Heimbach, S. , Chouraki, V. , Kuzma, A. B. , Sleegers, K. , Vronskaya, M. , Ruiz, A. , Graham, R. R. , … Schellenberg, G. D. (2017). Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial‐mediated innate immunity in Alzheimer's disease. Nature Genetics, 49, 1373–1384. 10.1038/ng.3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobue, A. , Komine, O. , Hara, Y. , Endo, F. , Mizoguchi, H. , Watanabe, S. , Murayama, S. , Saito, T. , Saido, T. C. , Sahara, N. , Higuchi, M. , Ogi, T. , & Yamanaka, K. (2021). Microglial gene signature reveals loss of homeostatic microglia associated with neurodegeneration of Alzheimer’s disease. Acta Neuropathologica Communications, 9, 1. 10.1186/s40478-020-01099-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, K. , Friedman, B. A. , Etxeberria, A. , Huntley, M. A. , van der Brug, M. P. , Foreman, O. , Paw, J. S. , Modrusan, Z. , Beach, T. G. , Serrano, G. E. , & Hansen, D. V. (2020). Alzheimer's patient microglia exhibit enhanced aging and unique transcriptional activation. Cell Reports, 31, 107843. 10.1016/j.celrep.2020.107843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen, T. L. , Cacciottolo, M. , Balu, D. , Morgan, T. E. , LaDu, M. J. , Finch, C. E. , & Pike, C. J. (2019). APOE genotype and sex affect microglial interactions with plaques in Alzheimer's disease mice. Acta Neuropathologica Communications, 7, 82. 10.1186/s40478-019-0729-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, T. , Butler, A. , Hoffman, P. , Hafemeister, C. , Papalexi, E. , Mauck, W. M. , Hao, Y. , Stoeckius, M. , Smibert, P. , & Satija, R. (2019). Comprehensive integration of single‐cell data. Cell, 177, 1888–1902.e1821. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek, F. , Bosnjak, M. , Skunca, N. , & Smuc, T. (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One, 6, e21800. 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrupp, N. , Sala Frigerio, C. , Wolfs, L. , Skene, N. G. , Fattorelli, N. , Poovathingal, S. , Fourne, Y. , Matthews, P. M. , Theys, T. , Mancuso, R. , de Strooper, B. , & Fiers, M. (2020). Single‐nucleus RNA‐seq is not suitable for detection of microglial activation genes in humans. Cell Reports, 32, 108189. 10.1016/j.celrep.2020.108189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulland, T. K. , Song, W. M. , Huang, S.‐C. , Ulrich, J. D. , Sergushichev, A. , Beatty, W. L. , Loboda, A. A. , Zhou, Y. , Cairns, N. J. , Kambal, A. , Loginicheva, E. , Gilfillan, S. , Cella, M. , Virgin, H. W. , Unanue, E. R. , Wang, Y. , Artyomov, M. N. , Holtzman, D. M. , & Colonna, M. (2017). TREM2 maintains microglial metabolic fitness in Alzheimer's disease. Cell, 170, 649–663 e613. 10.1016/j.cell.2017.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, Y. W. , Al‐Ouran, R. , Mangleburg, C. G. , Perumal, T. M. , Lee, T. V. , Allison, K. , Swarup, V. , Funk, C. C. , Gaiteri, C. , Allen, M. , Wang, M. , Neuner, S. M. , Kaczorowski, C. C. , Philip, V. M. , Howell, G. R. , Martini‐Stoica, H. , Zheng, H. , Mei, H. , & Zhong, X. … Logsdon, B. A. (2020). Meta‐analysis of the Alzheimer's disease human brain transcriptome and functional dissection in mouse models. Cell Reports, 32, 107908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Beckmann, N. D. , Roussos, P. , Wang, E. , Zhou, X. , Wang, Q. , Ming, C. , Neff, R. , Ma, W. , Fullard, J. F. , Hauberg, M. E. , Bendl, J. , Peters, M. A. , Logsdon, B. , Wang, P. , Mahajan, M. , Mangravite, L. M. , Dammer, E. B. , Duong, D. M. , … Zhang, B. (2018). The Mount Sinai cohort of large‐scale genomic, transcriptomic and proteomic data in Alzheimer's disease. Scientific Data, 5, 180185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Allen, M. , Li, S. , Quicksall, Z. S. , Patel, T. A. , Carnwath, T. P. , Reddy, J. S. , Carrasquillo, M. M. , Lincoln, S. J. , Nguyen, T. T. , Malphrus, K. G. , Dickson, D. W. , Crook, J. E. , Asmann, Y. W. , & Ertekin‐Taner, N. (2020). Deciphering cellular transcriptional alterations in Alzheimer's disease brains. Molecular Neurodegeneration, 15, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsson, U. , Andersson, D. , de Pablo, Y. , Pekny, R. , Ståhlberg, A. , Mulder, J. , Mitsios, N. , Hortobágyi, T. , Pekny, M. , & Pekna, M. (2017). Injury leads to the appearance of cells with characteristics of both microglia and astrocytes in mouse and human brain. Cerebral Cortex, 27, 3360–3377. 10.1093/cercor/bhx069 [DOI] [PubMed] [Google Scholar]
- Xu, H. , Han, Y. , Liu, B. , & Li, R. (2016). Unc‐5 homolog B (UNC5B) is one of the key downstream targets of N‐α‐Acetyltransferase 10 (Naa10). Scientific Reports, 6, 38508. 10.1038/srep38508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, Y. , Zhao, N. , Caulfield, T. R. , Liu, C. C. , & Bu, G. (2019). Apolipoprotein E and Alzheimer disease: pathobiology and targeting strategies. Nature Reviews. Neurology, 15, 501–518. 10.1038/s41582-019-0228-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, F. L. , Wang, Y. , Tom, I. , Gonzalez, L. C. , & Sheng, M. (2016). TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of amyloid‐beta by microglia. Neuron, 91, 328–340. 10.1016/j.neuron.2016.06.015 [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Zhu, C. , Beecham, G. , Vardarajan, B. N. , Ma, Y. , Lancour, D. , Farrell, J. J. , & Chung, J. Alzheimer's Disease Sequencing Project ; Mayeux, R. , Haines, J. L. , Schellenberg, G. D. , Pericak‐Vance, M. A. , Lunetta, K. L. , & Farrer, L. A. (2019). A rare missense variant of CASP7 is associated with familial late‐onset Alzheimer's disease. Alzheimer's & Dementia: the Journal of the Alzheimer's Association, 15, 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, G. X. Y. , Terry, J. M. , Belgrader, P. , Ryvkin, P. , Bent, Z. W. , Wilson, R. , Ziraldo, S. B. , Wheeler, T. D. , McDermott, G. P. , Zhu, J. , Gregory, M. T. , Shuga, J. , Montesclaros, L. , Underwood, J. G. , Masquelier, D. A. , Nishimura, S. Y. , Schnall‐Levin, M. , Wyatt, P. W. , Hindson, C. M. , … Bielas, J. H. (2017). Massively parallel digital transcriptional profiling of single cells. Nature Communications, 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , Song, W. M. , Andhey, P. S. , Swain, A. , Levy, T. , Miller, K. R. , Poliani, P. L. , Cominelli, M. , Grover, S. , Gilfillan, S. , Cella, M. , Ulland, T. K. , Zaitsev, K. , Miyashita, A. , Ikeuchi, T. , Sainouchi, M. , Kakita, A. , Bennett, D. A. , Schneider, J. A. , … Colonna, M. (2020). Human and mouse single‐nucleus transcriptomics reveal TREM2‐dependent and TREM2‐independent cellular responses in Alzheimer's disease. Nature Medicine, 26, 131–142. 10.1038/s41591-019-0695-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S10
Table S1‐S20
Data Availability Statement
The data in this manuscript are available via the AD Knowledge Portal (https://adknowledgeportal.synapse.org). The AD Knowledge Portal is a platform for accessing data, analyses and tools generated by the Accelerating Medicines Partnership (AMP‐AD) Target Discovery Program and other National Institute on Aging (NIA)‐supported programs to enable open‐science practices and accelerate translational learning. The data, analyses, and tools are shared early in the research cycle without a publication embargo on secondary use. Data are available for general research use according to the following requirements for data access and data attribution (https://adknowledgeportal.synapse.org/DataAccess/Instructions). The raw data files are available through Synapse (https://www.synapse.org/#!Synapse:syn28450881.3/datasets/).
| Dataset | Data Type | Description | SynapseID | DoD |
|---|---|---|---|---|
| Mayo RNAseq TCX | Metadata | Individual human and RNAseq | syn5550404 | na |
| Mayo RNAseq CER | Metadata | Individual human and RNAseq | syn5550404 | na |
| Mayo RNAseq TCX | RNASeq Expression | Consensus processed RNASeq raw counts | syn8690799 | 10/2/2019 |
| Mayo RNAseq CER | RNASeq Expression | Consensus processed RNASeq raw counts | syn8690904 | 10/2/2019 |
| ROSMAP | Metadata | ID Key | syn3382527 | 10/2/2019 |
| ROSMAP | Metadata | Individual human | syn3191087 | 10/2/2019 |
| ROSMAP | Metadata | Assay RNAseq | syn21088596 | 1/2/2020 |
| ROSMAP | RNASeq Expression | Consensus processed RNASeq raw counts | syn8691134 | 10/2/2019 |
| MSBB | Metadata | Individual human | syn6101474 | 11/22/2019 |
| MSBB | Metadata | Assay RNAseq | syn6100548 | 10/2/2019 |
| MSBB | RNASeq Expression | Consensus processed RNASeq raw counts | syn8691099 | 10/2/2019 |
Data from AD knowledge portal utilized in this study. DoD = Date of download, “na” indicates data that were generated by study authors and shared within the AD knowledge portal. Data were obtained from the RNAseq Harmonization Study in the AD knowledge portal (DOI: https://doi.org/10.7303/syn9702085).


