Abstract
The conventional drug discovery pipeline has proven to be unsustainable for rare diseases. Herein, we discuss the recent advances in biomedical knowledge mining applied to discovering therapeutics for rare diseases. We summarize current chemogenomics data of relevance to rare diseases and provide a perspective on the effectiveness of machine learning and biomedical knowledge graph mining in rare disease drug discovery. We illustrate the power of these methodologies using a chordoma case study. We expect that a broader application of knowledge graph mining and artificial intelligence approaches will expedite the discovery of viable drug candidates against both rare and common diseases.
Keywords: informatics, rare diseases, drug discovery, data mining, knowledge graphs
Teaser:
We describe how recent advances in biomedical knowledge graph mining and artificial intelligence could aid the discovery of viable drug candidates against rare diseases.
Introduction
Rare diseases are usually defined as conditions that affect fewer than 200,000 people in the United States and 1 in 2,000 people in the European Union.1 Unfortunately, there is currently no universal definition of what constitutes a rare disease. Several countries outside Europe and North America estimate rare disease prevalence ranging from 5 to 76 per 100,000 people, reaching, collectively, a conservative number of up to 446.2 million people affected worldwide, suffering from over 7,000 rare diseases.2 In Africa, access to diagnosis and advanced genetic testing has hindered the estimation of incidence.3 Some diseases are ultrarare, affecting less than 100 individuals worldwide and, in some cases, a single individual.4 In addition, many diseases, e.g., Malaria, Chagas disease, and sleeping sickness, uncommon in developed countries, are regarded as endemic in other geographical areas of the world. The World Health Organization calls these diseases “neglected” since big pharmaceutical companies often overlook them.5 Conversely, rare diseases are uncommon everywhere and they represent a substantial burden on individuals, families, and whole economies. As such, rare diseases demand disruptive and revolutionary drug discovery paradigms to promote the development of innovative therapies.6 Before 1983, there were only 34 treatments for rare diseases.7 In the last four decades, governmental incentives resulted in more than 600 therapies being approved as of 2021.8 Despite this progress, only a small fraction of patients can be treated with an approved medication.1
It is widely accepted that the conventional drug discovery pipeline is generally inadequate and unsustainable, as it usually takes 10–15 years and, on average, $2.6 billion in research and development costs to develop a single new drug.9 Inefficiencies in this pipeline are particularly problematic for rare diseases due to high research and development costs coupled with the potential for low revenue gains.10 Consequently, drug repurposing has become a trending topic among researchers.11 This approach allows researchers to identify new uses for already-approved or investigational drugs beyond the original therapeutic indication.12 Compared to new therapeutics, repurposed drugs can be approved for additional indications faster and at a reduced cost since FDA-approved medicines have already considered the effects of human exposure and can bypass additional expensive preclinical and Phase I safety studies.13
Drug repurposing studies are often only initiated after chance observations of unexpected or “off-label” effects.14 Typical examples include sildenafil (Viagra) that initially found unintended application as a treatment for erectile dysfunction,15 and zidovudine and thalidomide that were repurposed for HIV and multiple myeloma, respectively.16 Notably, these drugs were successfully repurposed serendipitously rather than systematically.
Computational approaches have emerged as practical solutions to accelerating drug discovery efforts and reducing drug development costs.17 Additionally, Literature-Based-Discovery (LBD), which seeks to unlock biological observations hidden within the existing information sources, such as published texts and manuscripts, has emerged as a promising approach.18 More recently, the exploration of biomedical knowledge graphs has found growing application as another promising approach.19 In such graphs, biomedical concepts are represented as nodes, and linkages between concepts are represented as edges connecting the respective nodes. The consolidation and integration of knowledge-driven drug discovery approaches toward developing a cost-effective, innovative drug discovery pipeline represents a formidable but potentially highly impactful challenge.
In this Perspective, we (i) review current data sources and computational approaches for the discovery of potential drug targets (e.g., genes or proteins) associated with rare diseases, (ii) describe different means to find information on a given disease and classify the diseases according to the putative treatment (small molecules, biologics, cell therapies, gene therapies, and nutraceuticals that are either approved, investigational, or experimental agents), (iii) propose a new approach to integrating existing data on drugs, rare diseases and imputed information from knowledge graphs to detect new targets and therapies; and, (iv) demonstrate the utility of this approach in a case study. We hope that this structured discussion and proof-of-concept studies will provide practical and useful guidance to enable many studies concerning drug discovery for rare diseases but also can be extended for neglected and common diseases.
Current therapeutics for treating rare diseases.
In contrast to many common diseases, the etiologies and pathogenesis for most rare diseases remain unknown. For example, amyotrophic lateral sclerosis is a rare disease that is long known as Lou Gehrig’s Disease and it still has an uncertain etiology.20 Phenotypic cell-based assays can identify potential drug candidates even if the disease pathophysiology is not well understood. In these assays, biological changes are observed when active compounds are present. However, when the disease etiology and pathophysiology are unknown, optimizing active compounds to improve efficacy, pharmacokinetics, and toxicity profiles is even more challenging. Still, thanks to the advances in these technologies, the number of new molecular entities or biologics license applications approved by the FDA for rare or orphan diseases increased from 5 in 2006 to 21 in 2015.21
Drug development for rare diseases is complicated because many such disorders are associated with multiple genotypic variations and phenotypic presentations.22 Furthermore, it is also challenging to collect sufficient safety and efficacy data with a naturally small number of patients recruited for clinical trials.23 Similarly, it is hard to find experts to help run these clinical trials. Finally, rare diseases most commonly develop in early childhood. Conducting clinical trials in children brings additional complications due to variable pharmacokinetics and pharmacodynamics of drugs, physiological differences, and ethical considerations.24
Despite the challenges, several therapeutics have been approved to treat rare diseases, especially those with higher incidence, such as multiple sclerosis, narcolepsy, primary biliary cholangitis, hemophilia, and cystic fibrosis. We summarize approved and investigational small molecules and biologics for some rare diseases in Table 1. Small molecules defined as chemical structures with molecular weight lower than 1 kDa and they constitute most of the approved drugs.25 Biologics are therapeutic modalities composed of large and complex structures derived from natural sources, such as proteins, antibodies, vaccines, nucleic acids (DNA, siRNA, mRNA, oligonucleotides), and cells.26 Some of these approaches can include replacing, altering, or introducing novel versions of genes, proteins, or cells in the patient’s body. In the last few years, these approaches have become promising for treating rare diseases (Table 1).21 For instance, growth hormone (e.g., somatropin for osteogenesis imperfecta27), stem-cell-based therapy (Cellavita HD for Huntington’s disease28), and gene therapy (e.g., ARU-1801 for sickle cell anemia29) have been explored. Furthermore, researchers use cell-based (normal or disease-affected) disease models, induced pluripotent stem cell models, and animal models to gain information about drug targets and treatment options.21
Table 1.
A summary of approved and investigational treatment modalities for some rare diseases.
| Disease State | Approved Treatments | Investigational Treatments |
|---|---|---|
| Multiple sclerosis | Ofatumumab (monoclonal antibody)127 | Fenebrutinib (small molecule)128 |
| Primary biliary cholangitis | Obeticholic acid (small molecule)129 | Elafibranor (small molecule)130 |
| Cystic Fibrosis | Elexacaftor-tezacaftor-ivacaftor (small molecules)40 | Approved CFTR modulators are being studies for non-approved cystic fibrosis genes.131 |
| Hemophilia | Emicizumab-kxwh (monoclonal antibody)132 | Valoctocogene roxaparvovec (gene therapy)133 |
| Narcolepsy | Pitolisant and Solriamfetol (small molecules)134 | FT218, JZP-258, reboxetine, and THN102 (small molecules).134 |
| Spinal muscular atrophy | Onasemnogene abeparvovec-xioi (gene therapy)37, risdiplam (small molecule)135, nusinersen (oligonucleotide)136 | SRK-015 (monoclonal antibody)137 |
| Osteogenesis imperfecta | Bisphosphonates (small molecules)134 | Somatropin (growth hormone)27 |
| Sickle cell anemia | Hydroxyurea (small molecule)134 | LentiGlobin BB305 (gene therapy), ARU-1801 (gene therapy)29 |
| Huntington Disease | Tetrabenazine (small molecule)134 | Cellavita HD (stem-cell therapy)28 |
| Amyotrophic lateral sclerosis |
Riluzole (small molecule)138 | Autologous MSC-NTF cells (stem-cell therapy)139 |
Despite some similarities to drug discovery for common diseases, rare disease drug discovery challenges are compounded by the lack of data and funding.30 In the pharmaceutical companies, success is often measured by the revenue from drug sales,31 which may explain the lack of interest in rare diseases. Measuring success by the revenue is a one-sided metric. In addition to not considering the opportunity to improve the quality of life of individuals, it does not consider the economic impact that finding the cure for a disease will have on the society. The profit-driven mentality hampers stakeholders to invest in research and development of therapeutics for these diseases, and often times, drug discovery for rare disease may receive significant funding from wealthy individuals only when they themselves or their loved ones are affected by that rare disease.32
Efforts have been made to increase the support for rare disease research by forming different consortia and centers.33 These organizations specialize in identifying undiscovered diseases and developing improved treatments for identified diseases. The International Rare Diseases Research Consortium spearheads these efforts in conjunction with public organizations such as the US National Institutes of Health and the European Commission.34 Consequently, one-third of new drug approvals for rare diseases occurred between 2010 to 2015.35 Currently, 230 rare diseases are being studied within the Rare Diseases Clinical Research Network.33 However, it is too soon to gauge the success of this endeavor as very few treatments have emerged thus far.
Gene therapy, protein/peptide replacement, and target-based small molecule discovery constitute existing approaches used to discover rare disease treatments.36 For example, onasemnogene abeparvovec-xioi (Zolgensma) was recently approved as gene therapy to treat spinal muscular atrophy caused by a mutation in the survival motor neuron 1 (SMN1) gene.37 The SMN1 was first described in 1995, allowing the development of a therapy to insert a functional SMN1 gene to prevent disease progression38, which eventually became available to patients only in 2019.37 Additionally, genetically validated drug targeting has become a popular approach for drug discovery.39 This approach involves identifying a gene that corresponds to a phenotype and creating small molecules or biologics to modulate the genetic target. It has been used for common diseases like hypercholesterolemia with the development of PCSK9 inhibitors and for cystic fibrosis, a rare disease resulting from mutations to the cystic fibrosis transmembrane conductance regulator (CFTR) gene.40 Vertex Pharmaceuticals, a biopharmaceutical company known for conducting drug discovery studies for cystic fibrosis, has developed the first genetically validated medication to treat the underlying cause of cystic fibrosis rather than manage cystic fibrosis symptoms.41 This medication, known as ivacaftor, acts as a potentiator of the CFTR channel targeting patients with a G551D gene mutation in the CFTR gene.42 Since this gene was discovered in 1989, studies of this gene have led to novel agents like elexacaftor-tezacaftor-ivacaftor (Trikafta), which is potent in patients with Phe508del-minimal function genotypes that did not respond to CFTR modulator therapies like ivacaftor.40
Current efforts in drug repurposing for rare diseases are often serendipitous and patient- family-driven.
The conventional drug discovery pipeline involves target identification and validation to detect molecules that may affect a disease state; these phases benefit most from bioinformatics and cheminformatics approaches. In the discovery process, preclinical research includes in vitro and in vivo efficacy, safety, and pharmacokinetic profiles, followed by clinical trials to establish safety and effectiveness in human subjects.43 The development of treatments for rare diseases also needs to follow this general workflow. However, there need to be additional knowledge-based innovative solutions to accelerate progress depending on the disease’s severity and rarity. In many cases, rare disease patients and their families undergo a multi-year diagnosis odyssey as data, knowledge, and physicians specializing in their conditions are all hard to find.44,45 Researchers and physicians often develop their understanding of the disease working with new patients, especially for ultra-rare diseases. Parents and rare disease patients have frequently started foundations connecting patients, clinicians, and researchers that focus on fundraising to assist with studies that may help develop treatments.21,30 The National Organization for Rare Disorders (NORD) (http://rarediseases.org/) provides recommendations for such organizations.
We briefly review drug repurposing case studies below to highlight the need to develop a systematized rare disease drug discovery pipeline. Lessons learned in these efforts can be translated into a reliable and reproducible workflow. One of the most famous cases is Augusto and Michaela Odone’s story, parents of Lorenzo Odone, who dedicated their lives to discovering a treatment for their son’s rare disease, adrenoleukodystrophy (ALD). Augusto and Michaela never had any formal medical training, but they found a cure for ALD and founded the Myelin Project, a non-profit research organization.46 ALD is a genetic disorder that causes the demyelination of neural fibers and degeneration of the adrenal gland, resulting in neurological instability and, ultimately, death.47 ALD causes the accumulation of saturated, long-chain fatty acids in the brain and adrenal cortex and leads to demyelination.47 Augusto and Michaela, with the help of researchers, eventually developed a treatment to break down these long-chain fatty acids by extracting acids from olive and rapeseed oils. This treatment was termed “Lorenzo’s Oil.” A study published in 2005 showed that, in some instances, ALD patients could positively benefit from treatment with Lorenzo’s Oil that may prevent the progression of the disease.48
More recently, Prof. Matthew Might, a computer scientist and father of a child with a rare disease involving NGLY1 deficiency, had to transition into a specialist in precision medicine and drug repurposing to find a treatment for his son’s rare disease. He discovered that, due to the NGLY1 deficiency, his son also lacked N-acetylglucosamine, a vital amino sugar. Further research proposed that NGLY1 deficiency could potentially be treated with endo-β-N-acetylglucosaminidase (ENGase) inhibitors.49 A structure-based screening of a drug database and an electrophoretic mobility shift assay revealed that several drugs, most notably proton pump inhibitors, could potentially be repurposed to treat the NGLY1 deficiency.49 These studies provided a direction for drug development and discovery for NGLY1 deficiency and possibly suggested a generalizable avenue for drug repurposing for rare diseases. Indeed, recent studies in Dr. Might’s group led to developing a systematic knowledge mining approach termed mediKanren50 that the researchers have started using effectively to propose personalized pharmacotherapy.51
Accelerating drug repurposing for rare diseases.
Drug repurposing is a strategy to identify novel uses for approved or investigational drugs beyond the original therapeutic indication.12 Many drugs considered for repurposing have already been characterized with respect to their safety and pharmacokinetic profile. Therefore, these drugs are less likely to fail in clinical trials than a new molecule. For instance, researchers found out-of-pocket clinical costs per approved orphan drug to be 43% cheaper than non-orphan drugs.52 In addition, the expected reduction in cost and time is essential not only to bring therapeutical options faster to patients but also to provide accessible treatment for people in economically disadvantaged areas of the world. Moreover, exploring a drug for potential repurposing may lead to the discovery of new targets.14 Drug repurposing is becoming a more widely used drug development method, with repurposed drugs making up about 30% of all drugs approved by the FDA in recent years.53
With recent technological advances, drug repurposing strategies have shifted from serendipitous observations to rational, computer-assisted methods.54 Computational approaches for drug repurposing include genetic association, pathway mapping, retrospective clinical analysis, molecular docking, virtual screening, signature patching, and LBD.14 Each of these approaches benefits from large-scale databases made publicly available in recent years. Even with technological advances, challenges for drug repurposing exist, especially for rare diseases.53 Fewer datasets exist for rare diseases since they have smaller markets and are not traditionally pursued by big pharmaceutical companies.11 However, collaboration among different biotech companies, academia, and private and government organizations may streamline the process to compile more information about rare diseases to help with treatment development.45
Literature-based discovery presents an exciting way of fueling testable drug repurposing hypotheses. Using a bibliometric approach, Baker et al.55 showed that more than 60% of all approved drugs and drug candidates (ca. 35,000 molecules) had been studied in more than one disease and 189 drugs were each tested in more than 300 diseases. More than 30% of approved drugs have been tested during their lifetime for at least one additional indication. More recently, we performed an additional bibliometric analysis of drug repurposing for rare diseases.56 In this analysis, we mined PubMed using earlier text-mining work57 to identify articles where a chemical entity was described in terms of its therapeutic association with a rare disease. We merged the rare disease list available in MalaCards58 with all the indications of drugs available in DrugBank59, which classifies drugs as approved, experimental (i.e., drugs that are at the preclinical or animal testing stage), and investigational (i.e., drugs that are in human clinical trials).60 As of 2021, there are more than 600 therapies approved for rare diseases.8 Our analysis integrating DrugBank and a combined list of rare diseases compiled from multiple sources showed 754 approved therapeutics associated with rare diseases (Table 2). This analysis involves drugs approved for common diseases in the investigational or experimental stage for rare diseases. Therefore, there is a rough estimate of 100–150 therapeutics approved for common diseases under investigation for rare diseases. In total, there were 1,421 rare diseases associated with these treatments. However, considering that many of the approved treatments only ease these diseases’ symptoms, there is a strong need to expand research on developing novel therapies for rare diseases.
Table 2.
Drugs associated with 1421 rare diseases and their DrugBank label in 2021 found using Abstract Sifter.
| Status | Drug modality | |
|---|---|---|
| Small molecules | Biologics | |
| Approved only | 350 | 27 |
| Approved and investigational | 335 | 17 |
| Approved and experimental | 18 | 0 |
| Approved, experimental, and investigational | 6 | 1 |
| Investigational | 133 | 18 |
| Experimental | 135 | 6 |
| Withdrawn | 11 | 0 |
Many diseases share common targets. For instance, patients with Niemann-Pick Disease Type C (NPC), a rare disease, are deficient in either the NPC1 or NPC2 gene. As a result of this autosomal recessive genetic mutation, individuals afflicted with NPC cannot transport cholesterol or lipids inside of cells. Simultaneously, those with NPC1 gene mutations cannot contract filoviruses like Ebola, as NPC1 is integral for cellular entry of filoviruses.61 There are enumerable potential interactions between genes and phenotypes of common and rare diseases. Using computational approaches, a deeper understanding of the biological networks and mechanisms underlying health and disease pathophysiology can be achieved.
Figure 1 summarizes major types of, and relationships between, basic and respective translational research that enable progress toward clinical intervention to provide care to rare disease patients. Key areas of basic research involve genotyping, clinical and pharmacological phenotyping, and knowledge mining, including computational modeling of biological targets and drug data. The respective translational tools include gene therapy, quality of life interventions, and (personalized, or precision) pharmacotherapy using repurposed or de novo developed drugs, including biologics. More specifically, the identification of rare disease genotypes allows the development of gene therapy medication. Establishing shared clinical and/or functional phenotypes between common and rare diseases can provide insights to propose both pharmacological and non-pharmacological interventions to improve the quality of life of rare disease patients. Computational approaches can be employed to connect the co-occurrence of phenotypes between common and rare diseases to identify possible novel targets for therapeutical intervention. This analysis can also identify approved drugs that can normalize disease phenotypes even at the level of individual patients (i.e., offer personalized pharmacotherapy), e.g., shift gene expression profile characteristic of the disease to those observed in healthy individuals, which can alleviate disease symptoms.
Figure 1.
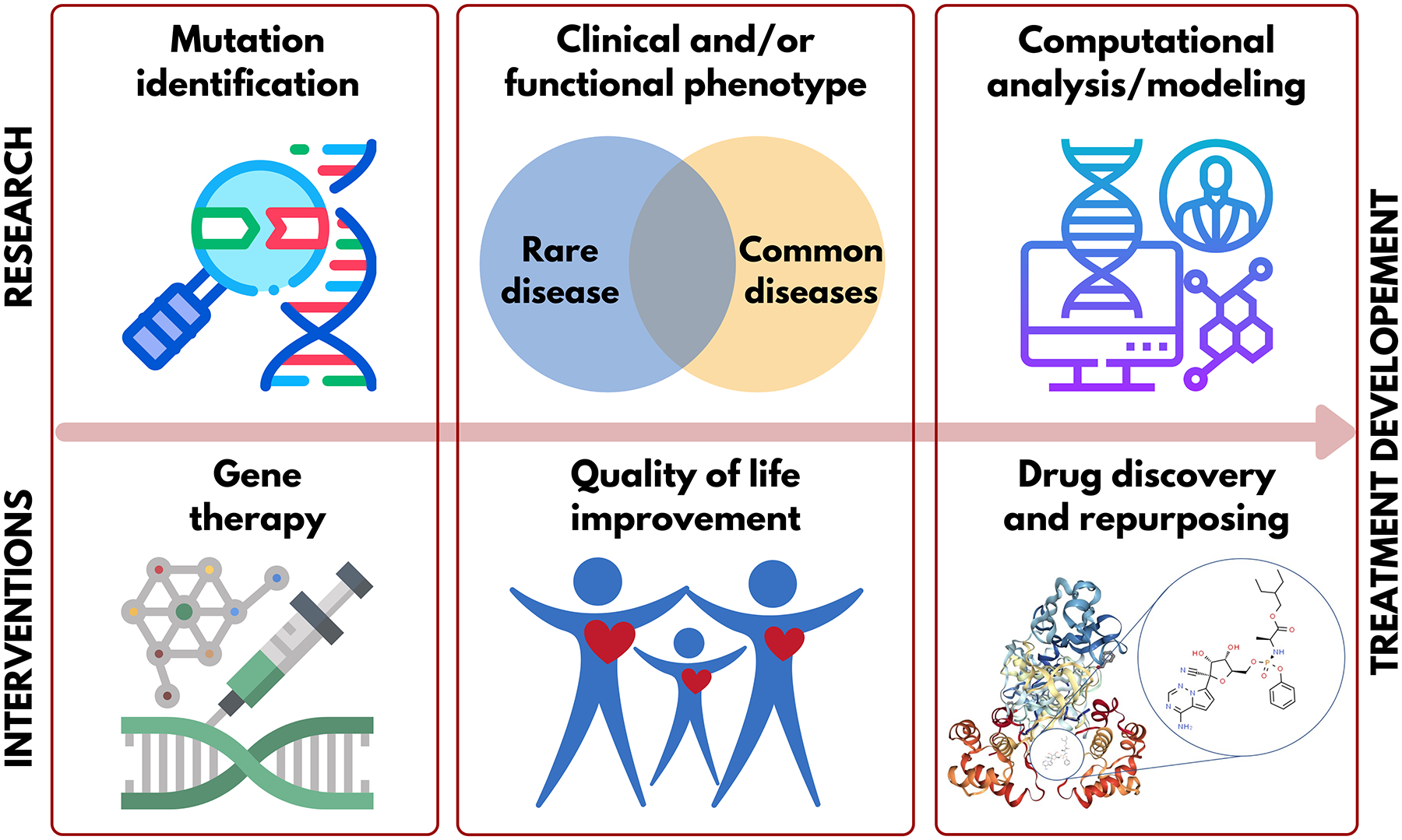
Key types of interrelated basic and translational research to enable clinical interventions for rare disease patients.
Another opportunity to accelerate the drug discovery and repurposing for rare diseases relies on polypharmacology, i.e., the effect observed when a therapeutic agent act on multiple targets or disease pathways, is often referred to as an innovative, potentially significant approach to design more effective and safer therapeutic agents for complex diseases.62 Several marketed drugs have been shown to have polypharmacological activity in cancer (e.g., sunitinib) and central nervous system diseases (e.g., clozapine).63 The rationale here is that a drug interacting with multiple key targets, with synergistic effects on biological pathways linked to a disease, may have higher efficacy at a lower dosage, limiting drawbacks arising from using a combination of various drugs. For this reason, multi-target drug design is posed as a promising approach for the discovery and development of drugs for rare diseases.64 Pierzynowska et al.65 suggested that the development of a therapeutic agent acting on multiple targets for several rare diseases may help economically for the development of drugs.
Data sources for rare diseases
In recent years, there has been an explosion of different types of data for rare diseases, including chemical, biological, and health care data. We summarize several of these sources in Table 3. There is an FDA database consisting of 1055 FDA-approved drugs with their orphan indications and chemical structures (https://www.accessdata.fda.gov/scripts/opdlisting/oopd/). A growing number of biomedical databases have added rare diseases registries, such as MalaCards58, Pharos66, ClinVar67, the Online Mendelian Inheritance in Man (OMIM)68, and others69. The GeneCards database has over 267,000 entries. We have examined the data in MalaCards database containing 12,863 rare disease names (many are subtypes of the same disease) and found that only about half of those (6,054 diseases) have genetic information in GeneCards. While efforts have been made to promote the sharing of information between multidisciplinary collaborations70, there is still a need to curate and adequately integrate all of this data.71
Table 3.
A summary of data sources for rare diseases.
| Dataset | Description | URL |
|---|---|---|
| FDA Orphan Drug Designations and Approvals | Collection of Drugs and their respective FDA Orphan Designations. | https://www.accessdata.fda.gov/scripts/opdlisting/oopd/ |
| MalaCards | Database of human diseases, with ontological descriptors, associated genes, and other related diseases. | https://www.malacards.org/ |
| Pharos | Targets associated with diseases. | https://pharos.nih.gov/ |
| ClinVar | Collection of gene variations and conditions known to be associated with. | https://www.ncbi.nlm.nih.gov/clinvar/ |
| Online Mendelian Inheritance in Man (OMIM) | Collection of both human genes and genetic disorders cross-linked by association. | https://omim.org/ |
| GeneCards | Collection of information on genes, including summaries and genomics data. | https://www.genecards.org |
| National Organization for Rare Disorders (NORD) | Summary, early signs, and symptoms for rare diseases. | https://rarediseases.org/ |
| Rare Disease InfoHub | Symptoms of rare diseases, including experts and funding opportunities. | https://rarediseases.oscar.ncsu.edu/ |
| Genetic and Rare Diseases Information Center (GARD) | Collection of synonyms, summary, and symptoms for rare diseases. | https://rarediseases.info.nih.gov/ |
Many organizations seek to collaborate and integrate data on rare diseases. For example, The NORD provides education to patients and caregivers and supports research initiatives for rare diseases.21 The Rare Disease InfoHub (https://rarediseases.oscar.ncsu.edu/) portal is currently under development to provide information about rare diseases to patients and help researchers diagnose patients with rare diseases. The InfoHub addresses the critical knowledge gap for rare disease patients described via web scraping, data mining, bioinformatics data linkages, and various other computational processes. The InfoHub provides its users with: (i) aggregation of disease information about specific conditions from various biomedical databases; (ii) access to communities formed around both specific and general rare diseases; (iii) biomedical inference tools tailored to specific rare diseases; (iv) and access to names and locations of providers and research specialists, including the ability to use geographic data to locate therapists. In addition, InfoHub acts as a data platform, where it provides inference on drug/disease relationships and others in the community may access current data through an application programming interface.
Several translational research centers help driving research and development for rare diseases, including the National Center for Advancing Translational Sciences (NCATS) and the International Rare Diseases Research Consortium21. NCATS, specifically, has a program called the Genetic and Rare Diseases (GARD) Information Center, which aims to help all individuals who may be affected by rare diseases, including patients, doctors, researchers, and patient advocates, by providing information about rare disease treatments, clinical trials, and other resources.
Another potential source of data for rare diseases is social media. Social media mining, also called “social listening,” has been used for pharmacovigilance72 as well as for estimating trends in disease outbreaks and symptoms.73 Therefore, it is a potential resource for drug repurposing hypotheses. In addition, social media may facilitate communications between providers, patients, and families. Data can be collected over social media through online surveys and information posted by users can promote fundraising and general awareness of rare diseases,34,45 which could bring these disorders to drug developers’ attention and identify potential study participants. Using social media to connect individuals with rare diseases across the world may expand clinical trial registries and spur research breakthroughs like determining the etiology and treatments for rare diseases.74
Data collection, curation, and integration
The growth of data for rare diseases is expected to boost respective computational drug discovery and development research. The collection and integration of data from disparate, heterogeneous sources create numerous challenges that could impact the content and knowledge discovery in the integrated datasets.75 When integrating multiple databases, the user must be aware of how to handle database collisions, i.e., when the same entry exists in both databases. Additionally, there is always concern over the validity of data. This concern can be addressed by data curation focusing on common types of errors found in many databases. The most common error is data duplication, which is often caused by differences in the naming nomenclature used by different original data sources (e.g., different names used for the same drug or the same gene), or it could also be different experimental measurements of the same property for the same compound (e.g., specific bioactivity or toxicity), or protein, or gene. The specifics of curation will vary based on the type of data presented.
The approaches and protocols for biological and chemical curation of chemogenomics datasets have been proposed and extensively discussed by our group.71 Briefly, chemical structure curation includes several steps, including structural normalization of specific chemotypes, such as aromatic and nitro groups, and removal of inorganic salts and organometallic compounds. Structural standardization enables chemical duplicate identification and removal. In the process, concordance of bioactivity (or any reported quantity) and intra- and inter-laboratory variability of the reported properties are examined and data from unreliable sources are excluded.
Biomedical knowledge graph databases
In recent years, significant advances in data integration, visualization, and knowledge generation have been made through the use of graph databases.76 A traditional relational database collects and stores data based on predefined properties. So, an entry in a disease database may have information for name, symptoms, prognosis, genes, etc. To link databases, unique identifiers of other tables must be added as another column in the relational database. At run time, to find links between these databases, both datasets must be searched. On the other hand, graph databases treat linkages between data points as first-class objects.76 A data point is mainly defined by the other data points to which it is linked. The benefits of this system are evident once datasets grow to the order of billions. For instance, searching for which genes are linked to a disease will only require to be executed once. Defining an upper-level ontology allows the avoidance of ambiguities and permits better knowledge derivation.77
Extracting meaningful information is a topic of particular concern for biomedical graph databases. Inferring meaningful relationships between two nodes in a graph could lead to new drugs or the discovery of previously unknown mechanisms of action. Unfortunately, these inferences are often extremely difficult to find due to the scarcity of data and the complexity of possible relationships that could be inferred from the graph.78
Embeddings, or techniques that seek to reduce the dimensionality of a graph by finding patterns in data, are particularly useful for database analytics.78 Once embedded, graph nodes can then easily be clustered. Clustering methods are unsupervised machine learning methods where algorithms are trained without a dependent variable to identify patterns in data sets based only on the features. This approach contributes to the identification of homogeneous subgroups among a heterogeneous dataset.79 Some well-known clustering techniques such as principal component analysis can be employed, but many are too computationally demanding to be useful in graphs with millions of data points. Many embedding algorithms that scale linearly with the number of nodes have been constructed. Of note, Node2Vec produces its embeddings by generating stochastic paths. These paths are then used to find a data point that is close to other data points.80 Leveraging these nearest neighbor calculations, inferences may be made on how various datapoints are interrelated. However, it is essential to acknowledge that the paucity of data in rare diseases may restrict the use of this approach.
Bioinformatics
Bioinformatics corresponds to the application of informatics techniques to analyze and model biological data.81 It has been estimated that more than 70% of the 6,172 rare diseases cataloged in Orphanet are genetic2 and, therefore, bioinformatics is a core application science for analyzing and extracting knowledge from these data.82,83 The genetic roots of a rare disease can serve as invaluable information for finding a cure or useful therapeutics. By way of example, a treatment for spinal muscular atrophy was developed by inserting a functional SMN1 gene.37 These types of therapies require an explicit understanding of disease/gene relationships. The disease’s genetic roots are uncovered using two methods: candidate genome analysis (CGA) and genome scans.84 Candidate genome analysis attempts to find an association between a genetic variation and the presence of a disease. The apparent issue with this method is that a researcher must have a genetic variance and disease before an analysis can be run. This limits the functionality of new hypotheses. The other method for relating genetics to disease is whole-genome searches. This entails collecting the entire genome of multiple individuals afflicted with the disease, with no preference for specific sections of the gene. Once these genomes are assembled, statistical analysis can be performed to find areas of the genome that most likely correlate with disease activity. An issue with both methods is that the exact relation of genes to biological function is unknown for many genes. The GeneCards database identified only 19,168 genes out of the 332,121 entries in their database to be associated with diseases.85
Cheminformatics
Cheminformatics is an interdisciplinary field of science that uses computer and information science resources to solve chemistry problems.86 More specifically, it deals with (i) representation, visualization, manipulation, and processing of chemical structures; (ii) organization of chemical structure databases; and, (iii) studies of quantitative structure-activity relationships (QSAR).86,87
QSAR modeling is a fundamental computational approach widely employed by pharma and academia for the discovery of novel compounds with desired properties and for those lacking experimental activity. Nowadays, QSAR studies mainly employ machine learning algorithms to develop predictive models. The major challenge of applying this approach to discovering novel chemical probes for rare diseases is the lack of experimental chemogenomics data for most of the conditions.88 However, as data becomes available in the literature or in online repositories such as ChEMBL or PubChem, these data can be used to develop QSAR models. Once a target is known, and an experimental protocol is established, a large amount of data can be produced by applying High Throughput Screening (HTS) campaigns.89 Ekins et al.11 summarize several drugs discovered through HTS campaigns, such as riluzole, used to treat amyotrophic lateral sclerosis. The genesis of a chemical dataset with biological data obtained either from HTS or low-throughput screening may allow the development of QSAR models. For instance, after the Ebola virus outbreak, an original series of compounds blocking the entry of the Ebola virus into human cells were identified.90 Subsequently, Capuzzi et al.91 used this library to generate QSAR models and employed virtual screening of more than 17 million compounds. They identified 14 compounds with IC50 values under 10 μM, including several sub-micromolar inhibitors and more than 10-fold selectivity against host cytotoxicity. In another study, a database of FDA-approved compounds with activity against Ebola92 was used to generate a QSAR model that identified compounds to possess in vivo activity in mice93. A recent example from our group used repurposing for a rare disease which used published datasets for chordoma to build a Bayesian machine learning model that was used to score clinical candidates for testing and identified several compounds with promising in vitro activity.94
Health informatics
Health informatics, often called biomedical informatics95, uses computer and information science resources to collect, process, analyze, model, and interpret clinical data.96 Data-driven approaches are posed to help diagnose and develop interventions to address patients’ needs better and evaluate the impact of those interventions.97 The diagnosis of a rare disease may take several years.98 Patients and physicians might benefit from an integrated computational analysis of electronic health records (EHRs) to forecast the diagnosis of rare diseases.99 By running queries in the EHR database, it is possible to filter patients meeting specific parameters (e.g., phenotypic, laboratory, genotypic, treatment, etc.).100 Furthermore, patient registries for rare diseases exist through several consortiums or centers like The Orphan Disease Center (https://orphandiseasecenter.med.upenn.edu/) and Pulse Infoframe (https://www.pulseinfoframe.com/). These patient registries are used to collect demographic, clinical, laboratory, and outcomes data from patients with rare diseases to help researchers better understand rare diseases.
Employing deep learning for rare diseases
Deep learning (DL) refers to any supervised, semi-supervised, or unsupervised machine learning system composed of neural networks with multiple layers of non-linear processing capable of learning data representations.101 The architecture of this algorithm is represented by “neurons” or interconnected nodes that compute input data and release transformed output data. DL methods have been around for decades, but overfitting problems and data dependency often render them unused.102 In the last decade, DL has become a hot topic in biomedical research due to its availability as easier-to-model algorithms and faster computers with GPU acceleration, combined with the availability of more extensive data sets.103
In particular, DL has become popular in cheminformatics for the generation of QSAR models, which, as any other machine learning algorithm, can be used to predict the bioactivity of compounds lacking experimental data.17,87 In addition, due to its particular approach to learning representations of data with multiple levels of abstraction, DL has been employed to help diagnose genetic disorders.104 A phenotype-based machine learning system named The Rare Diseases Auxiliary Diagnosis system can help clinicians diagnose rare diseases using four diagnostic models.105 More recently, a study reported an improvement in rare disease diagnoses by 20–89% by employing a deep convolutional neural network trained on more than 17,000 patient images combined with genetic and patient data.106 DL can also drive the implementation of precision medicine.107
Integrative mining approaches
Mining of available biomedical data allows knowledge extraction to identify and develop new or repurposed drug candidates.55 The underlying biological pathways of diseases and potential drug treatments are described primarily in the biomedical literature.108 Text mining of published studies could confirm connections between drugs, their targets, underlying biological pathways, and diseases, including enabling new inferences of such connections.109 Swanson’s ABC approach was the first attempt to systematically extract novel hypotheses from existing literature.110 The proposed methods look for connections between two unlinked concepts, referred to as A and C, through some additional concept, called B. By looking at paper citations, Swanson formed links between papers. To ensure linkages had not already been hypothesized and debunked, no articles about concept A should have cited any papers about concept C. Similarly, no papers about C should have cited A. Further processes of ensuring no linkages are discussed in this paper. Examples of Swanson’s ABC method have been generated for many diseases, such as Parkinson’s disease111 and cancer112. This approach has also been used to elucidate adverse drug effects.18
This method is far from perfect, however. Finding valid candidates for A, B, and C is left up to manual curation. As stated, “[the trial and error] search strategy proposed is neither a recipe nor an algorithm. Success depends entirely on the knowledge and ingenuity of the searcher in forming hypotheses about potential logical connections”.110 An additional issue comes in the form of the number of connections to explore. As of 2021, approximately thirty-two million publications have been published in PubMed, accounting for approximately 1015 potential connections.
Inspired by the Swanson’s ABC method, our group designed and built Chemotext113 to help users find connections by mining existing biomedical literature. This tool uses every article indexed by PubMed with a Medical Subject Headings (MeSH) term. MeSH terms provide a systematic summary of the essential topics discussed in the publication. Chemotext’s functionality can be viewed as an inverse of Swanson’s ABC method. In Chemotext, the user selects two MeSH terms, viewed as concepts A and C, which they seek to connect. Chemotext then returns a list of possible intermediate terms, viewable as Swanson Method’s B concept. But unlike the Swanson’s method, which disregards B connections that relate A-C as already explored and therefore uninteresting, Chemotext weighs interconnected ideas higher. Intermediate connections are ranked by the total number of papers, including A, B, and C as MeSH terms.
Swanson’s method makes two problematic assumptions: (1) all medical professionals are completely up to date on every breakthrough and publication in their field and (2) an existing connection between two topics is tantamount to well-known and well-explored knowledge. Unfortunately, these two assumptions are very often untrue for large fields with thousands of publications per year. Enabling inference and exploration of already-existing-but-underexplored connections is, thus, a valuable contribution.
To expand Chemotext capabilities, our group at UNC has partnered and collaborated with other groups to develop ROBOKOP (Reasoning Over Biomedical Objects linked in Knowledge Oriented Pathways)114, an specialized knowledge graph database that develops and deploys data science cyberinfrastructure. ROBOKOP generates nodes (biomedical concepts) and edges (linkages between the concepts). Various sources must be consulted and standardized across fields. These genes, chemicals, and biomedical databases are extracted from medical publication abstracts, which could ultimately be used to explore a disease’s knowledge landscape and propose interesting new targets.
Graph databases belong to the category of non-relational, or NoSQL, databases.115 This unique structure allows storing extensive data with complex interrelationships as nodes (objects) and edges (known relationships between objects). ROBOKOP is a recent example of a biomedical knowledge graph that was developed utilizing the Neo4J graph database management system.114 The underlying system enables users to insert and query stored data systemically. By running queries against the established ROBOKOP database, the user can request dynamic and complex pattern queries from the database. An example of this would be the query “What are the genes associated with chordoma?”. This query will search the database for all genes associated with the condition chordoma and find all biological processes related to those genes.
All databases suffer from incomplete and incorrect information, especially as our scientific knowledge grows and evolves, and Knowledge Graphs are no exception. It is unavoidable that errors may occur within a robust graph database leveraging the data from dozens of external sources. These errors may come from outdated or false information in primary sources. In addition, integrating the sources is challenging due to an incomplete graph ontology, which hampers the ability to map all the incoming knowledge. These errors can be mitigated in a number of ways. Several algorithms have been proposed to complete and correct information within the database.116,117 In addition, it is important to assign confidence level to graph edges. These confidence values can be derived from various sources, such as experiments or the number of publications supporting an edge.
Case study
To demonstrate the utility of the approaches mentioned above, we have highlighted a case study in which we search for novel drug repurposing hypotheses aimed at treating chordoma. Chordoma is an umbrella term for carcinomas of the spinal cord. As a whole, chordomas represent a unique class of rare diseases, for which there are limited standardized treatment protocols established.118 Metformin is an antihyperglycemic drug used to control glycemic levels in type II diabetes patients.119 Its use has been investigated to treat multiple diseases, including several types of cancer.120 Metformin is currently in clinical trials for treating different types of sarcomas121,122 and similar reasoning could be put forward as a hypothesis for metformin as a potential treatment for chordoma.
Using the terms “metformin” and “cancer” to query ROBOKOP, we found that there were numerous links between this drug and chordoma. To investigate this hypothesis, we first queried ROBOKOP for “metformin” and “chordoma”, but no published literature directly described these connections. Then, we queried for graphs connecting “drugs” to “genes” to “chordoma” in ROBOKOP using the template question “Find a drug to treat chordoma by finding treatable diseases sharing genetics”. This query yielded multiple unique subgraphs, consisting of various combinations of 21 drugs connected to KIT and TSC1 genes linked to “chordoma” in the ROBOKOP database. Still, no direct connection between metformin and chordoma was found. The next approach was to query ROBOKOP using a more complex question: “What is the clinical outcome pathway for metformin and chordoma?”. Figure 2 shows the relationships between metformin and chordoma. Figure 2A details multiple connections between metformin, genes, pathways, cells, and chordoma. We focused on the catalase (CAT) gene as a prime example since it was related to only two types of cells (osteoblast and somatic cell). The edges highlighted in Figure 2B provide links to publish papers linking the two terms in the nodes. Analyzing these references, we found evidence that metformin increases CAT activity in mice without increasing its expression.123 CAT has shown to be essential for osteoblast growth.124 Chordoma is characterized by uncontrolled proliferation and maintenance of undifferentiated osteoblasts and proper osteoblast differentiation was found to reduce the development of chordoma cells.125
Figure 2.
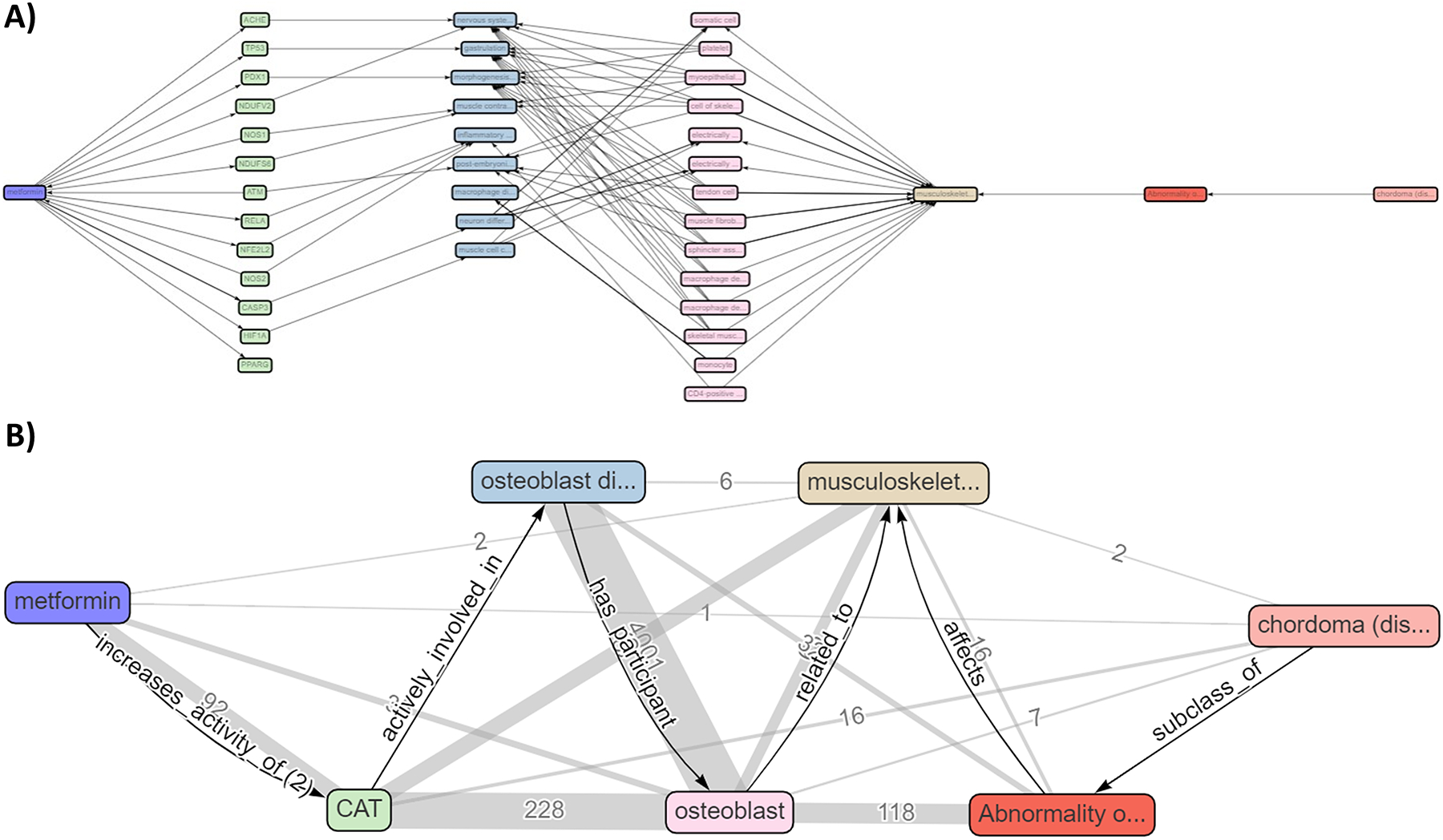
Knowledge graph illustrating the relationship of metformin and chordoma. A) Complete knowledge graph of the ROBOKOP query showing multiple relationships between metformin, genes, cells, and chordoma (https://bit.ly/3w3gNG6). B) Putative clinical outcome pathway linking metformin and chordoma showing that metformin increases CAT activity, which is involved in osteoblast differentiation in the musculoskeletal system.
Final remarks
Rare diseases are a global phenomenon, with some countries affected more than others by distinct rare diseases due to genetic variations between national populations worldwide. Therefore, we face a challenge to develop a unifying definition of what constitutes a rare disease since the widely used definitions promoted by the United States and the European Union are exclusive of their own population. Finding treatments for patients with rare diseases is much more severe in developing countries than in the developed world. Even though treatments for rare diseases might reach the United States and the European Union market, these therapies are likely out of financial reach for other countries until they come off patent.
The primary challenges facing rare diseases are related to discovering new treatments and making them readily available to patients. Global efforts to address this disparity are needed to make drugs available more broadly and cost-effectively. Over the past several decades, the scientific community has identified genes associated with many rare diseases, with more than 3,000 genes recently mapped against over 4,000 monogenic rare diseases.126 However, the number of FDA approved therapies remains very limited, roughly just over 600.8
Drug discovery for rare diseases continues to be challenging due to natural reasons such as the uniqueness of each disease, which requires years of focused research, prohibitively high cost of drug development, and lack of financial incentives given the small number of patients who can eventually benefit from such drugs. Consequently, the discovery and development of drugs for these diseases cannot be accomplished without a comprehensive approach integrating data and knowledge across genomics, chemogenomics, and EHRs.
The continuing accumulation and integration of such data coupled with the development of novel knowledge mining tools such as knowledge graph mining provide hope that at least some of the rare disease patients can benefit from drugs approved for other diseases, i.e., repurposed drugs. Furthermore, innovative knowledge mining and molecular modeling approaches can accelerate the discovery and characterization of novel targets for rare diseases and prioritize and expedite the development of respective drug candidates. We hope that the ongoing process of accumulating clinical and biomedical data and translating data to knowledge and knowledge to action in the form of data-driven and testable drug discovery hypotheses will enable the expedited and inexpensive development of new therapeutics for rare diseases in the near future. This challenge should not be left to the next generation of scientists. There is a huge opportunity to move the field and make scientific discoveries that can potentially make a remarkable impact on the lives of people suffering from rare diseases.
Acknowledgments
AT appreciates the financial support for studies in his group provided by the National Center for Advancing Translational Sciences (OT2TR003441), the National Institutes of Health (1U01CA207160), and the University of North Carolina Research Opportunity Initiative grant to support the Infohub for Rare Diseases project. SE acknowledges funding to Collaborations Pharmaceuticals Inc. from the National Institute of General Medical Sciences (GM122196-02A1), National Institute of Neurological Disorders and Stroke (1R43NS107079-01 and 3R43NS107079-01S1), and the FY2018 UNC Research Opportunities Initiative Award.
Biographies

Vinicius Alves received his Ph.D. in Pharmaceutical Sciences (2017) from the Federal University of Goias, Brazil. He joined UNC-Chapel Hill as a Postdoctoral Fellow in 2018 and NIEHS as a Research Fellow in 2020. Currently, he is a Research Assistant Professor at the UNC Eshelman School of Pharmacy. He has experience developing and implementing innovative cheminformatics and molecular modeling approaches for pharmaceutical and environmental research. He has authored or co-authored more than 30 peer-reviewed research papers, reviews, and book chapters. He is a recipient of the 2018 Lush Prize Young Researcher Award.

Anthony Hickey is a Distinguished Fellow at RTI International and Director of the UNC Catalyst for Rare Diseases of the UNC Eshelman School of Pharmacy. He obtained Ph.D. and D.Sc. degrees in pharmaceutical sciences from Aston University, Birmingham, UK. He is a Fellow of the Royal Society of Biology, the American Association of Pharmaceutical Scientists, the American Association for the Advancement of Science, and the Royal Society of Medicine. He founded Cirrus Pharmaceuticals, Inc., Oriel Therapeutics, Inc, and Astartein, LLC. He conducts multidisciplinary research in pulmonary therapy and vaccine delivery for various diseases and oversees research in rare and neglected diseases.

Alexander Tropsha, Ph.D. is the K. H. Lee Distinguished Professor and Associate Dean for Pharmacoinformatics and Data Science at the UNC Eshelman School of Pharmacy. He obtained his Ph.D. in Chemical Enzymology in 1986 from Moscow State University, Russia. His research interests are in Computer-Assisted Drug Design, Computational Toxicology, Cheminformatics, Materials Informatics, and Structural Bioinformatics. He has authored or co-authored more than 260 peer-reviewed research papers, reviews, and book chapters and co-edited two monographs. He is an Associate Editor of the ACS Journal of Chemical Information and Modeling and a Fellow of the American Institute for Medical and Biological Engineering.
Footnotes
Conflicts of interest
SE is CEO of Collaborations Pharmaceuticals Inc. and has numerous patents filed on rare and neglected diseases. The other authors declare no potential conflict of interest.
References
- 1.NIH. FAQs About Rare Diseases | Genetic and Rare Diseases Information Center (GARD) – an NCATS Program. https://rarediseases.info.nih.gov/diseases/pages/31/faqs-about-rare-diseases. Published 2021. Accessed June 7, 2021
- 2.Nguengang Wakap S, Lambert DM, Olry A, Rodwell C, Gueydan C, Lanneau V, et al. Estimating cumulative point prevalence of rare diseases: analysis of the Orphanet database. Eur J Hum Genet. 2020;28(2):165–73. doi: 10.1038/s41431-019-0508-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baynam GS, Groft S, van der Westhuizen FH, Gassman SD, du Plessis K, Coles EP, et al. A call for global action for rare diseases in Africa. Nat Genet. 2020;52(1):21–6. doi: 10.1038/s41588-019-0552-2 [DOI] [PubMed] [Google Scholar]
- 4.Crooke ST. A call to arms against ultra-rare diseases. Nat Biotechnol 2021 396. 2021;39(6):671–7. doi: 10.1038/s41587-021-00945-0 [DOI] [PubMed] [Google Scholar]
- 5.WHO. Neglected tropical diseases. https://www.who.int/news-room/q-a-detail/neglected-tropical-diseases. Accessed August 15, 2021
- 6.Kakkis ED, O’Donovan M, Cox G, Hayes M, Goodsaid F, Tandon P, et al. Recommendations for the development of rare disease drugs using the accelerated approval pathway and for qualifying biomarkers as primary endpoints. Orphanet J Rare Dis. 2015;10(1):16. doi: 10.1186/s13023-014-0195-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Institute of Medicine (US) Committee on Accelerating Rare Diseases Research and Orphan Product Development. Rare Diseases and Orphan Products. doi: 10.17226/12953 [DOI] [Google Scholar]
- 8.Developing products for rare diseases and conditions. FDA. https://www.fda.gov/industry/developing-products-rare-diseases-conditions. Accessed June 9, 2021 [Google Scholar]
- 9.PhRMA. Biopharmaceutical research industry profile. http://phrma.org/sites/default/files/pdf/biopharmaceutical-industry-profile.pdf. Published 2016. Accessed June 9, 2021
- 10.Baxter K, Horn E, Gal-Edd N, Zonno K, O’Leary J, Terry PF, et al. An end to the myth: there is no drug development pipeline. Sci Transl Med. 2013;5(171):171cm1. doi: 10.1126/scitranslmed.3003505 [DOI] [PubMed] [Google Scholar]
- 11.Ekins S, Williams AJ, Krasowski MD, Freundlich JS. In silico repositioning of approved drugs for rare and neglected diseases. Drug Discov Today. 2011;16(7–8):298–310. doi: 10.1016/j.drudis.2011.02.016 [DOI] [PubMed] [Google Scholar]
- 12.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3(8):673–83. doi: 10.1038/nrd1468 [DOI] [PubMed] [Google Scholar]
- 13.Nosengo N Can you teach old drugs new tricks? Nature. 2016;534(7607):314–6. doi: 10.1038/534314a [DOI] [PubMed] [Google Scholar]
- 14.Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18(1):41–58. doi: 10.1038/nrd.2018.168 [DOI] [PubMed] [Google Scholar]
- 15.Langtry HD, Markham A. Sildenafil. Drugs. 1999;57(6):967–89. doi: 10.2165/00003495-199957060-00015 [DOI] [PubMed] [Google Scholar]
- 16.Franks ME, Macpherson GR, Figg WD. Thalidomide. Lancet. 2004;363(9423):1802–11. doi: 10.1016/S0140-6736(04)16308-3 [DOI] [PubMed] [Google Scholar]
- 17.Ekins S, Puhl AC, Zorn KM, Lane TR, Russo DP, Klein JJ, et al. Exploiting machine learning for end-to-end drug discovery and development. Nat Mater. 2019;18(5):435–41. doi: 10.1038/s41563-019-0338-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henry S, McInnes BT. Literature Based Discovery: Models, methods, and trends. J Biomed Inform. 2017;74:20–32. doi: 10.1016/j.jbi.2017.08.011 [DOI] [PubMed] [Google Scholar]
- 19.Sang S, Yang Z, Wang L, Liu X, Lin H, Wang J. SemaTyP: a knowledge graph based literature mining method for drug discovery. BMC Bioinformatics. 2018;19(1):193. doi: 10.1186/s12859-018-2167-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiernan MC, Vucic S, Cheah BC, Turner MR, Eisen A, Hardiman O, et al. Amyotrophic lateral sclerosis. Lancet (London, England). 2011;377(9769):942–55. doi: 10.1016/S0140-6736(10)61156-7 [DOI] [PubMed] [Google Scholar]
- 21.Sun W, Zheng W, Simeonov A. Drug discovery and development for rare genetic disorders. Am J Med Genet A. 2017;173(9):2307–22. doi: 10.1002/ajmg.a.38326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.PhRMA. Biopharmaceutical Research and Development: The Process Behind New Medicines. http://phrma-docs.phrma.org/sites/default/files/pdf/rd_brochure_022307.pdf. Published 2015. Accessed June 9, 2021
- 23.Kempf L, Goldsmith JC, Temple R. Challenges of developing and conducting clinical trials in rare disorders. Am J Med Genet Part A. 2018;176(4):773–83. doi: 10.1002/ajmg.a.38413 [DOI] [PubMed] [Google Scholar]
- 24.Kern SE. Challenges in conducting clinical trials in children: approaches for improving performance. Expert Rev Clin Pharmacol. 2009;2(6):609–17. doi: 10.1586/ecp.09.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makurvet FD. Biologics vs. small molecules: Drug costs and patient access. Med Drug Discov. 2021;9:100075. doi: 10.1016/j.medidd.2020.100075 [DOI] [Google Scholar]
- 26.Kinch MS. An overview of FDA-approved biologics medicines. Drug Discov Today. 2015;20(4):393–8. doi: 10.1016/j.drudis.2014.09.003 [DOI] [PubMed] [Google Scholar]
- 27.Growth Hormone Therapy in Osteogenesis Imperfecta. ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT00001305. Published 2020. Accessed June 9, 2021
- 28.Safety Evaluation of Cellavita HD Administered Intravenously in Participants With Huntington’s Disease (SAVE-DH). ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT02728115. Published 2020. Accessed June 9, 2021
- 29.Hoban MD, Orkin SH, Bauer DE. Genetic treatment of a molecular disorder: gene therapy approaches to sickle cell disease. Blood. 2016;127(7):839–48. doi: 10.1182/blood-2015-09-618587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ekins S, Perlstein EO. Doing it All - How Families are Reshaping Rare Disease Research. Pharm Res. 2018;35(10):192. doi: 10.1007/s11095-018-2481-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.CBO. Research and Development in the Pharmaceutical Industry. https://www.cbo.gov/publication/57126. Published 2021. Accessed September 2, 2021
- 32.Tindera M A Billionaire’s Dying Wish: A $10 Million Prize To Fight Brain Diseases. Forbes. https://www.forbes.com/sites/michelatindera/2018/11/06/a-billionaires-dying-wish-a-10-million-prize-to-fight-brain-diseases. Published 2018. Accessed September 2, 2021 [Google Scholar]
- 33.Rare Disease Clinical Research Network. https://www.rarediseasesnetwork.org/diseases. Accessed August 15, 2021
- 34.Austin CP, Cutillo CM, Lau LPL, Jonker AH, Rath A, Julkowska D, et al. Future of Rare Diseases Research 2017–2027: An IRDiRC Perspective. Clin Transl Sci. 2018;11(1):21–7. doi: 10.1111/cts.12500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.PhrMA. A Decade of Innovation in Rare Diseases: 2005 to 2015. http://phrma-docs.phrma.org/sites/default/files/pdf/PhRMA-Decade-of-Innovation-Rare-Diseases.pdf. Published 2015. Accessed June 9, 2021
- 36.Al-Ali H. The evolution of drug discovery: from phenotypes to targets, and back. Medchemcomm. 2016;7(5):788–98. doi: 10.1039/C6MD00129G [DOI] [Google Scholar]
- 37.AveXis receives FDA approval for Zolgensma®, the first and only gene therapy for pediatric patients with spinal muscular atrophy (SMA). Novartis. https://www.novartis.com/news/media-releases/avexis-receives-fda-approval-zolgensma-first-and-only-gene-therapy-pediatric-patients-spinal-muscular-atrophy-sma. Published 2019. Accessed June 9, 2021 [Google Scholar]
- 38.Nurputra DK, Lai PS, Harahap NIF, Morikawa S, Yamamoto T, Nishimura N, et al. Spinal Muscular Atrophy: From Gene Discovery to Clinical Trials. Ann Hum Genet. 2013;77(5):435–63. doi: 10.1111/ahg.12031 [DOI] [PubMed] [Google Scholar]
- 39.Floris M, Olla S, Schlessinger D, Cucca F. Genetic-Driven Druggable Target Identification and Validation. Trends Genet. 2018;34(7):558–70. doi: 10.1016/j.tig.2018.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Middleton PG, Mall MA, Dřevínek P, Lands LC, McKone EF, Polineni D, et al. Elexacaftor-Tezacaftor-Ivacaftor for Cystic Fibrosis with a Single Phe508del Allele. N Engl J Med. 2019;381(19):1809–19. doi: 10.1056/NEJMoa1908639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vertex Pharmaceuticals. A Timeline: Vertex is Committed to Advances in Cystic Fibrosis (Infographic). https://www.vrtx.com/about-us/timeline-vertex-committed-advances-cystic-fibrosis-infographic/. Published 2021. Accessed June 9, 2021
- 42.Condren ME, Bradshaw MD. Ivacaftor: a novel gene-based therapeutic approach for cystic fibrosis. J Pediatr Pharmacol Ther. 2013;18(1):8–13. doi: 10.5863/1551-6776-18.1.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hughes JP, Rees S, Kalindjian SB, Philpott KL. Principles of early drug discovery. Br J Pharmacol. 2011;162(6):1239–49. doi: 10.1111/j.1476-5381.2010.01127.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ekins S Industrializing rare disease therapy discovery and development. Nat Biotechnol. 2017;35(2):117–8. doi: 10.1038/nbt.3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Litterman NK, Rhee M, Swinney DC, Ekins S. Collaboration for rare disease drug discovery research. F1000Research. 2014;3(0):261. doi: 10.12688/f1000research.5564.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.ALD Connect. The Myelin Project. https://aldconnect.org/about-us/the-myelin-project/. Accessed August 13, 2021
- 47.National Center for Biotechnology Information. Genes and Disease. Genes and Disease. https://www.ncbi.nlm.nih.gov/books/NBK22183/. Published 1998. Accessed June 9, 2021 [Google Scholar]
- 48.Moser HW, Raymond GV, Lu S-E, Muenz LR, Moser AB, Xu J, et al. Follow-up of 89 asymptomatic patients with adrenoleukodystrophy treated with Lorenzo’s oil. Arch Neurol. 2005;62(7):1073–80. doi: 10.1001/archneur.62.7.1073 [DOI] [PubMed] [Google Scholar]
- 49.Bi Y, Might M, Vankayalapati H, Kuberan B. Repurposing of Proton Pump Inhibitors as first identified small molecule inhibitors of endo-β-N-acetylglucosaminidase (ENGase) for the treatment of NGLY1 deficiency, a rare genetic disease. Bioorg Med Chem Lett. 2017;27(13):2962–6. doi: 10.1016/j.bmcl.2017.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patton M, Rosenblatt G, Byrd WE, Might M. mediKanren: A System for Bio-medical Reasoning. In: 25th ACM SIGPLAN International Conference on Functional Programming.; 2020:1–12. https://icfp20.sigplan.org/details/minikanren-2020-papers/10/mediKanren-A-System-for-Bio-medical-Reasoning [Google Scholar]
- 51.Diagnosis in 2.127 seconds: Solving a years-long vomiting mystery using AI, research and brain power. NewsWise. https://www.newswise.com/articles/diagnosis-in-2-127-seconds-solving-a-years-long-vomiting-mystery-using-ai-research-and-brain-power. Published 2020. Accessed June 9, 2021 [Google Scholar]
- 52.Jayasundara K, Hollis A, Krahn M, Mamdani M, Hoch JS, Grootendorst P. Estimating the clinical cost of drug development for orphan versus non-orphan drugs. Orphanet J Rare Dis. 2019;14(1):12. doi: 10.1186/s13023-018-0990-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Polamreddy P, Gattu N. The drug repurposing landscape from 2012 to 2017: evolution, challenges, and possible solutions. Drug Discov Today. 2019;24(3):789–95. doi: 10.1016/j.drudis.2018.11.022 [DOI] [PubMed] [Google Scholar]
- 54.Lee H-M, Kim Y Drug Repurposing Is a New Opportunity for Developing Drugs against Neuropsychiatric Disorders. Schizophr Res Treatment. 2016;2016:6378137. doi: 10.1155/2016/6378137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baker NC, Ekins S, Williams AJ, Tropsha A. A bibliometric review of drug repurposing. Drug Discov Today. 2018;23(3):661–72. doi: 10.1016/j.drudis.2018.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alves VM, Capuzzi SJ, Baker N, Muratov EN, Trospsha A, Hickey AJ. Mining Complex Biomedical Literature for Actionable Knowledge on Rare Diseases. In: Bizzarri M, ed. Approaching Complex Diseases. Human Perspectives in Health Sciences and Technology Springer, Cham; 2020:77–94. doi: 10.1007/978-3-030-32857-3_4 [DOI] [Google Scholar]
- 57.Baker NC, Hemminger BM. Mining connections between chemicals, proteins, and diseases extracted from Medline annotations. J Biomed Inform. 2010;43(4):510–9. doi: 10.1016/j.jbi.2010.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rappaport N, Twik M, Plaschkes I, Nudel R, Iny Stein T, Levitt J, et al. MalaCards: An amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2017;45(D1):D877–87. doi: 10.1093/nar/gkw1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–82. doi: 10.1093/nar/gkx1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36(Database issue):D901–6. doi: 10.1093/nar/gkm958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuroda M, Fujikura D, Nanbo A, Marzi A, Noyori O, Kajihara M, et al. Interaction between TIM-1 and NPC1 Is Important for Cellular Entry of Ebola Virus. García-Sastre A, ed. J Virol. 2015;89(12):6481–93. doi: 10.1128/JVI.03156-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Proschak E, Stark H, Merk D. Polypharmacology by Design: A Medicinal Chemist’s Perspective on Multitargeting Compounds. J Med Chem. 2018;62(2):420–44. doi: 10.1021/ACS.JMEDCHEM.8B00760 [DOI] [PubMed] [Google Scholar]
- 63.Anighoro MA, Rgen Bajorath J, Rastelli G. Polypharmacology: Challenges and Opportunities in Drug Discovery. Published online 2014. doi: 10.1021/jm5006463 [DOI] [PubMed] [Google Scholar]
- 64.Evox Therapeutics Signs Multi-Target Rare Disease Collaboration with Takeda - Global Genes. https://globalgenes.org/2020/03/26/evox-therapeutics-signs-multi-target-rare-disease-collaboration-with-takeda/. Accessed August 27, 2021
- 65.Pierzynowska K, Kamińska T, Węgrzyn G. One drug to treat many diseases: unlocking the economic trap of rare diseases. Metab Brain Dis 2020 358. 2020;35(8):1237–40. doi: 10.1007/S11011-020-00617-Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nguyen DT, Mathias S, Bologa C, Brunak S, Fernandez N, Gaulton A, et al. Pharos: Collating protein information to shed light on the druggable genome. Nucleic Acids Res. 2017;45(D1):D995–1002. doi: 10.1093/nar/gkw1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):D1062–7. doi: 10.1093/nar/gkx1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43(D1):D789–98. doi: 10.1093/nar/gku1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao M, Wei D-QQ. Rare Diseases: Drug Discovery and Informatics Resource. Interdiscip Sci Comput Life Sci. 2018;10(1):195–204. doi: 10.1007/s12539-017-0270-3 [DOI] [PubMed] [Google Scholar]
- 70.Kaufmann P, Pariser AR, Austin C. From scientific discovery to treatments for rare diseases - the view from the National Center for Advancing Translational Sciences - Office of Rare Diseases Research. Orphanet J Rare Dis. 2018;13(1):196. doi: 10.1186/s13023-018-0936-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fourches D, Muratov E, Tropsha A. Curation of chemogenomics data. Nat Chem Biol. 2015;11(8):535–535. doi: 10.1038/nchembio.1881 [DOI] [PubMed] [Google Scholar]
- 72.Tricco AC, Zarin W, Lillie E, Jeblee S, Warren R, Khan PA, et al. Utility of social media and crowd-intelligence data for pharmacovigilance: a scoping review. BMC Med Inform Decis Mak. 2018;18(1):38. doi: 10.1186/s12911-018-0621-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kagashe I, Yan Z, Suheryani I. Enhancing Seasonal Influenza Surveillance: Topic Analysis of Widely Used Medicinal Drugs Using Twitter Data. J Med Internet Res. 2017;19(9):e315. doi: 10.2196/jmir.7393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Milne C-P, Ni W. The Use of Social Media in Orphan Drug Development. Clin Ther. 2017;39(11):2173–80. doi: 10.1016/j.clinthera.2017.08.016 [DOI] [PubMed] [Google Scholar]
- 75.Roos M, López Martin E, Wilkinson MD. Preparing Data at the Source to Foster Interoperability across Rare Disease Resources. In: Posada de la Paz M, Taruscio D, Groft S, eds. Rare Diseases Epidemiology: Update and Overview. Advances in Experimental Medicine and Biology Springer, Cham; 2017:165–79. doi: 10.1007/978-3-319-67144-4_9 [DOI] [PubMed] [Google Scholar]
- 76.Robinson I, Webber J, Eifrem E. Graph Databases. “ O’Reilly Media, Inc.”; 2013. [Google Scholar]
- 77.Santana da Silva F, Jansen L, Freitas F, Schulz S. Ontological interpretation of biomedical database content. J Biomed Semantics. 2017;8(1):24. doi: 10.1186/s13326-017-0127-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goyal P, Ferrara E. Graph embedding techniques, applications, and performance: A survey. Knowledge-Based Syst. 2018;151:78–94. doi: 10.1016/j.knosys.2018.03.022 [DOI] [Google Scholar]
- 79.Melnykov V, Michael S. Clustering Large Datasets by Merging K-Means Solutions. J Classif. 2020;37(1):97–123. doi: 10.1007/s00357-019-09314-8 [DOI] [Google Scholar]
- 80.Grover A, Leskovec J. Node2vec: Scalable feature learning for networks. In: Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining.; 2016. doi: 10.1145/2939672.2939754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.López-López E, Bajorath J, Medina-Franco JL. Informatics for Chemistry, Biology, and Biomedical Sciences. J Chem Inf Model. 2021;61(1):26–35. doi: 10.1021/acs.jcim.0c01301 [DOI] [PubMed] [Google Scholar]
- 82.Bellgard MI, Sleeman MW, Guerrero FD, Fletcher S, Baynam G, Goldblatt J, et al. Rare Disease Research Roadmap: Navigating the bioinformatics and translational challenges for improved patient health outcomes. Heal Policy Technol. 2014;3(4):325–35. doi: 10.1016/J.HLPT.2014.08.007 [DOI] [Google Scholar]
- 83.Zhao M, Havrilla JM, Fang L, Chen Y, Peng J, Liu C, et al. Phen2Gene: rapid phenotype-driven gene prioritization for rare diseases. NAR Genomics Bioinforma. 2020;2(2). doi: 10.1093/nargab/lqaa032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.SCHORK NJ. Genetics of Complex Disease. Am J Respir Crit Care Med. 1997;156(4):S103–9. doi: 10.1164/ajrccm.156.4.12-tac-5 [DOI] [PubMed] [Google Scholar]
- 85.GeneCards Version 5.4. [August 13, 2021]. https://www.genecards.org/. Accessed.
- 86.Engel T, Gasteiger J, eds. Chemoinformatics : Basic Concepts and Methods. Wiley-VCH; 2018. [Google Scholar]
- 87.Gasteiger J. Chemistry in Times of Artificial Intelligence. ChemPhysChem. 2020;21(20):2233–42. doi: 10.1002/cphc.202000518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cherkasov A, Muratov EN, Fourches D, Varnek A, Baskin II, Cronin M, et al. QSAR modeling: where have you been? Where are you going to? J Med Chem. 2014;57(12):4977–5010. doi: 10.1021/jm4004285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ekins S, Clark AM, Swamidass SJ, Litterman N, Williams AJ. Bigger data, collaborative tools and the future of predictive drug discovery. J Comput Aided Mol Des. 2014;28(10):997–1008. doi: 10.1007/s10822-014-9762-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kouznetsova J, Sun W, Martínez-Romero C, Tawa G, Shinn P, Chen CZ, et al. Identification of 53 compounds that block Ebola virus-like particle entry via a repurposing screen of approved drugs. Emerg Microbes Infect. 2014;3(1):1–7. doi: 10.1038/emi.2014.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Capuzzi SJ, Sun W, Muratov EN, Martínez-Romero C, He S, Zhu W, et al. Computer-Aided Discovery and Characterization of Novel Ebola Virus Inhibitors. J Med Chem. 2018;61(8):3582–94. doi: 10.1021/acs.jmedchem.8b00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Madrid PB, Chopra S, Manger ID, Gilfillan L, Keepers TR, Shurtleff AC, et al. A Systematic Screen of FDA-Approved Drugs for Inhibitors of Biological Threat Agents. Yu X, ed. PLoS One. 2013;8(4):e60579. doi: 10.1371/journal.pone.0060579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lane TR, Comer JE, Freiberg AN, Madrid PB, Ekins S. Repurposing Quinacrine against Ebola Virus Infection In Vivo. Antimicrob Agents Chemother. 2019;63(9). doi: 10.1128/AAC.01142-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Anderson E, Havener TM, Zorn KM, Foil DH, Lane TR, Capuzzi SJ, et al. Synergistic drug combinations and machine learning for drug repurposing in chordoma. Sci Rep. 2020;10(1):12982. doi: 10.1038/s41598-020-70026-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bernstam EV, Smith JW, Johnson TR. What is biomedical informatics? J Biomed Inform. 2010;43(1):104–10. doi: 10.1016/j.jbi.2009.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Coiera Enrico. Guide to Health Informatics. 3rd ed. CRC Press; 2015. [Google Scholar]
- 97.Ravi D, Wong C, Deligianni F, Berthelot M, Andreu-Perez J, Lo B, et al. Deep Learning for Health Informatics. IEEE J Biomed Heal Informatics. 2017;21(1):4–21. doi: 10.1109/JBHI.2016.2636665 [DOI] [PubMed] [Google Scholar]
- 98.Blöß S, Klemann C, Rother A-K, Mehmecke S, Schumacher U, Mücke U, et al. Diagnostic needs for rare diseases and shared prediagnostic phenomena: Results of a German-wide expert Delphi survey. PLoS One. 2017;12(2):e0172532. doi: 10.1371/journal.pone.0172532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Colbaugh R, Glass K, Rudolf C, Tremblay Volv Global Lausanne Switzerland M. Learning to Identify Rare Disease Patients from Electronic Health Records. AMIA. Annu Symp proceedings AMIA Symp 2018;2018:340–7. http://www.ncbi.nlm.nih.gov/pubmed/30815073 [PMC free article] [PubMed] [Google Scholar]
- 100.Shen F, Liu S, Wang Y, Wen A, Wang L, Liu H. Utilization of Electronic Medical Records and Biomedical Literature to Support the Diagnosis of Rare Diseases Using Data Fusion and Collaborative Filtering Approaches. JMIR Med Informatics. 2018;6(4):e11301. doi: 10.2196/11301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–44. doi: 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 102.Ekins S The Next Era: Deep Learning in Pharmaceutical Research. Pharm Res. Published online 2016:2594–603. doi: 10.1007/s11095-016-2029-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Miotto R, Wang F, Wang S, Jiang X, Dudley JT. Deep learning for healthcare: Review, opportunities and challenges. Brief Bioinform. 2017;19(6):1236–46. doi: 10.1093/bib/bbx044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gurovich Y, Hanani Y, Bar O, Nadav G, Fleischer N, Gelbman D, et al. Identifying facial phenotypes of genetic disorders using deep learning. Nat Med. 2019;25(1):60–4. doi: 10.1038/s41591-018-0279-0 [DOI] [PubMed] [Google Scholar]
- 105.Jia J, Wang R, An Z, Guo Y, Ni X, Shi T. RDAD: A Machine Learning System to Support Phenotype-Based Rare Disease Diagnosis. Front Genet. 2018;9:587. doi: 10.3389/fgene.2018.00587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hsieh TC, Mensah MA, Pantel JT, Aguilar D, Bar O, Bayat A, et al. PEDIA: prioritization of exome data by image analysis. Genet Med. 2019;21(12):2807–14. doi: 10.1038/s41436-019-0566-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Uddin M, Wang Y, Woodbury-Smith M. Artificial intelligence for precision medicine in neurodevelopmental disorders. npj Digit Med. 2019;2(1):112. doi: 10.1038/s41746-019-0191-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hunter LE. Knowledge-based biomedical Data Science. Kuhn T, ed. Data Sci. 2017;1(1–2):1–7. doi: 10.3233/DS-170001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Przybyła P, Shardlow M, Aubin S, Bossy R, Eckart de Castilho R, Piperidis S, et al. Text mining resources for the life sciences. Database. 2016;2016. doi: 10.1093/database/baw145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Swanson DR. Medical literature as a potential source of new knowledge. BMLA. 1990;78(1):29–36. [PMC free article] [PubMed] [Google Scholar]
- 111.Kostoff RN, Briggs MB. Literature-Related Discovery (LRD): Potential treatments for Parkinson’s Disease. Technol Forecast Soc Change. 2008;75(2):226–38. doi: 10.1016/j.techfore.2007.11.007 [DOI] [Google Scholar]
- 112.Choi B-K, Dayaram T, Parikh N, Wilkins AD, Nagarajan M, Novikov IB, et al. Literature-based automated discovery of tumor suppressor p53 phosphorylation and inhibition by NEK2. Proc Natl Acad Sci. 2018;115(42):10666–71. doi: 10.1073/pnas.1806643115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Capuzzi SJ, Thornton TE, Liu K, Baker N, Lam WI, O’Banion C, et al. Chemotext: A Publicly Available Web Server for Mining Drug–Target–Disease Relationships in PubMed. J Chem Inf Model. 2018;58(2):212–8. doi: 10.1021/acs.jcim.7b00589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bizon C, Cox S, Balhoff J, Kebede Y, Wang P, Morton K, et al. ROBOKOP KG and KGB: Integrated Knowledge Graphs from Federated Sources. J Chem Inf Model. 2019;59(12):4968–73. doi: 10.1021/acs.jcim.9b00683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hogan A, Blomqvist E, Cochez M, D’Amato C, de Melo G, Gutierrez C, et al. Knowledge Graphs. arXiv. Published online March 4, 2020:3509–10. http://arxiv.org/abs/2003.02320. Accessed December 20, 2020 [Google Scholar]
- 116.Nguyen TA, Perkins WA, Laffey TJ, Pecora D. Knowledge-base verification. AI Mag. 1987;8(2):69–75. [Google Scholar]
- 117.Paulheim H Knowledge graph refinement: A survey of approaches and evaluation methods. Semant Web. 2017;8(3):489–508. doi: 10.3233/SW-160218 [DOI] [Google Scholar]
- 118.Frezza AM, Botta L, Trama A, Dei Tos AP, Stacchiotti S. Chordoma: update on disease, epidemiology, biology and medical therapies. Curr Opin Oncol. 2019;31(2):114–20. doi: 10.1097/CCO.0000000000000502 [DOI] [PubMed] [Google Scholar]
- 119.Bailey CJ. Metformin: historical overview. Diabetologia. 2017;60(9):1566–76. doi: 10.1007/s00125-017-4318-z [DOI] [PubMed] [Google Scholar]
- 120.Pryor R, Cabreiro F. Repurposing metformin: An old drug with new tricks in its binding pockets. Biochem J. 2015;471(3):307–22. doi: 10.1042/BJ20150497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Molenaar RJ, Coelen RJS, Khurshed M, Roos E, Caan MWA, Van Linde ME, et al. Study protocol of a phase IB/II clinical trial of metformin and chloroquine in patients with IDH1-mutated or IDH2-mutated solid tumours. BMJ Open. 2017;7(6). doi: 10.1136/bmjopen-2016-014961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Longhi A, Istituto Ortopedico Rizzoli. Metformin as Maintenance Therapy in Patients With Bone Sarcoma and High Risk of Relapse. ClinicalTrials.gov. [Google Scholar]
- 123.Dai J, Liu M, Ai Q, Lin L, Wu K, Deng X, et al. Involvement of catalase in the protective benefits of metformin in mice with oxidative liver injury. Chem Biol Interact. 2014;216(1):34–42. doi: 10.1016/j.cbi.2014.03.013 [DOI] [PubMed] [Google Scholar]
- 124.Schreurs AS, Torres S, Truong T, Moyer EL, Kumar A, Tahimic CGT, et al. Skeletal tissue regulation by catalase overexpression in mitochondria. Am J Physiol - Cell Physiol. 2020;319(4):C734–45. doi: 10.1152/ajpcell.00068.2020 [DOI] [PubMed] [Google Scholar]
- 125.Marie PJ, Fromigué O, Modrowski D. Chapter 4 - Deregulation of osteoblast differentiation in primary bone cancers. In: Heymann DBT-BC (Second E, ed. Bone Cancer (Second Edition). Academic Press; 2015:39–54. doi: 10.1016/B978-0-12-416721-6.00004-2 [DOI] [Google Scholar]
- 126.Ehrhart F, Willighagen EL, Kutmon M, van Hoften M, Curfs LMG, Evelo CT. A resource to explore the discovery of rare diseases and their causative genes. Sci Data 2021 81. 2021;8(1):1–8. doi: 10.1038/s41597-021-00905-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Novartis. FDA approves Novartis Kesimpta® (ofatumumab), the first and only self-administered, targeted B-cell therapy for patients with relapsing multiple sclerosis. https://www.novartis.com/news/media-releases/fda-approves-novartis-kesimpta-ofatumumab-first-and-only-self-administered-targeted-b-cell-therapy-patients-relapsing-multiple-sclerosis. Accessed September 3, 2021
- 128.Weber M, Harp C, Bremer M, Goodyear A, Crawford J, Johnson A, et al. Fenebrutinib Demonstrates the Highest Potency of Bruton Tyrosine Kinase Inhibitors (BTKis) in Phase 3 Clinical Development for Multiple Sclerosis (MS) (4437). Neurology. 2021;96(15 Supplement):4437. http://n.neurology.org/content/96/15_Supplement/4437.abstract [Google Scholar]
- 129.FDA. FDA approves Ocaliva for rare, chronic liver disease. https://www.fda.gov/news-events/press-announcements/fda-approves-ocaliva-rare-chronic-liver-disease. Published 2016. Accessed September 3, 2021
- 130.Schattenberg JM, Pares A, Kowdley KV, Heneghan MA, Caldwell S, Pratt D, et al. A randomized placebo-controlled trial of elafibranor in patients with primary biliary cholangitis and incomplete response to UDCA. J Hepatol. 2021;74(6):1344–54. doi: 10.1016/j.jhep.2021.01.013 [DOI] [PubMed] [Google Scholar]
- 131.Testing Drug Efficacy in Cystic Fibrosis Through N-of-1 Trials - Full Text View - ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT04580368. Published 2020. Accessed September 9, 2021
- 132.Scott LJ, Kim ES. Emicizumab-kxwh: First Global Approval. Drugs. 2018;78(2):269–74. doi: 10.1007/s40265-018-0861-2 [DOI] [PubMed] [Google Scholar]
- 133.Single-Arm Study To Evaluate The Efficacy and Safety of Valoctocogene Roxaparvovec in Hemophilia A Patients - Full Text View - ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT03370913. Accessed September 9, 2021
- 134.Dwan K, Phillipi CA, Steiner RD, Basel D. Bisphosphonate therapy for osteogenesis imperfecta. Cochrane Database Syst Rev. 2016;10(10):CD005088. doi: 10.1002/14651858.CD005088.pub4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.FDA. FDA Approves Oral Treatment for Spinal Muscular Atrophy. https://www.fda.gov/news-events/press-announcements/fda-approves-oral-treatment-spinal-muscular-atrophy. Published 2020. Accessed September 9, 2021
- 136.FDA. FDA approves first drug for spinal muscular atrophy. https://www.fda.gov/news-events/press-announcements/fda-approves-first-drug-spinal-muscular-atrophy. Accessed September 9, 2021
- 137.An Active Treatment Study of SRK-015 in Patients With Type 2 or Type 3 Spinal Muscular Atrophy. https://clinicaltrials.gov/ct2/show/NCT03921528. Accessed September 9, 2021
- 138.Miller RG, Mitchell JD, Moore DH. Riluzole for amyotrophic lateral sclerosis (ALS)/motor neuron disease (MND). Cochrane Database Syst Rev. Published online March 14, 2012. doi: 10.1002/14651858.CD001447.pub3 [DOI] [Google Scholar]
- 139.Phase 2, Randomized, Double Blind, Placebo Controlled Multicenter Study of Autologous MSC-NTF Cells in Patients With ALS (NurOwn). ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT02017912. Accessed October 13, 2020


