Abstract
Abstract
It has been about a century since the discovery of the first antibiotic, and during this period, several antibiotics were produced and marketed. The production of high-potency antibiotics against infections led to victories, but these victories were temporary. Overuse and misuse of antibiotics have continued to the point that humanity today is almost helpless in the fight against infection. Researchers have predicted that by the middle of the new century, there will be a dark period after the production of antibiotics that doctors will encounter antibiotic-resistant infections for which there is no cure. Accordingly, researchers are looking for new materials with antimicrobial properties that will strengthen their ammunition to fight antibiotic-resistant infections. One of the most important alternatives to antibiotics introduced in the last three decades is antimicrobial peptides (AMPs), which affect a wide range of microbes. Due to their different antimicrobial properties from antibiotics, AMPs can fight and kill MDR, XDR, and colistin-resistant bacteria through a variety of mechanisms. Therefore, in this study, we intend to use the latest studies to give a complete description of AMPs, the importance of colistin-resistant bacteria, and their resistance mechanisms, and represent impact of AMPs on colistin-resistant bacteria.
Key points
• AMPs as limited options to kill colistin-resistant bacteria.
• Challenge of antibiotics resistance, colistin resistance, and mechanisms.
• What is AMPs in the war with colistin-resistant bacteria?
Keywords: Antimicrobial peptides, Colistin-resistant bacteria, Bacterial infection, Multi-drug resistant
Introduction
It is a bitter scenario that the misuse and overuse of antibiotics has caused bacteria to become resistant very quickly in a short period of time, and humans are now in the age of multidrug-resistant (MDR), extensively drug-resistant (XDR), and drug-resistant (PDR) bacterial infectious diseases. It is very difficult for researchers and physicians to accept that modern-day humanity is likely to face the pre-antibiotic era and the era of antibiotics is coming to an end (Napier et al. 2013; Moghadam et al. 2020b). Today, high resistance antibiotics infections besides the treatment challenge in clinics are responsible for long hospital stays, high patient mortality, and increased health care costs (Shariati et al. 2020; Shahbandeh et al. 2020). If the life-threatening process of patients is preceded by antibiotic resistance without appropriate countermeasures, it can pose great challenges in performing many medical and surgical procedures as in the pre-antibiotic era. On the other hand, in these conditions, the possibility of death due to secondary infections of superbugs after COVID-19 (Coronavirus disease 2019) and influenza becomes more challenging than before (Lewies et al. 2019; Moghadam et al. 2021c; Dousari et al. 2020). In recent decades, with the emergence of high rates of infections caused by MDR and XDR bacteria, doctors have inevitably taken refuge in highly toxic antibiotics such as colistin, but recently resistance to these end-of-line antibiotics has also increased (Mousavi et al. 2021). One of the inevitable factors that has coincided with the crisis of antibiotic resistance is the production of new antibiotics, which has reached a dead end in recent years. At a glance, we can see that more than 50 new antimicrobial drug production programs were implemented to control infectious diseases between 1980 and 2000, but there have been less than 15 cases over the past decade (2013). Antibiotics do not generate good revenue compared to other drugs because they are only prescribed for short-term use and there is currently little incentive to invest in new antibiotics. However, if new antibiotics are developed, they will probably be used sparingly to maintain their effectiveness in the first few years, consequently the initial return will be small on the investment. Therefore, in this situation, humanity is facing a stubborn battle against antibiotic resistance as a serious threat to the effective treatment of infectious diseases, in which survival and victory in this battle requires the development of new strategies to target the weak point of resistance (Lewies et al. 2019; Kiaei et al. 2019; Chegini et al. 2020; Moghadam et al. 2020c; Sadeghi Dosari et al. 2016). Among the new strategies proposed by the researchers, antimicrobial peptides (AMPs) have been shown to be able to target the weaknesses of antibiotic resistance mechanisms. AMPs can not only strengthen humanity’s ability to withstand antibiotic resistance, but also have the potential to be an alternative to antibiotics, both in terms of treatment and prevention (Fox 2013; Hancock and Sahl 2006). Accordingly, in this context, we review some of the most promising data on the effect of antimicrobial peptides against colistin-resistant bacteria as bacteria resistant to last resort antibiotics to reframe efforts on the advances of AMPs toward clinical development.
Challenge of antibiotics resistance
Going back to about a century ago, one can concluded from the history of antibiotic production that after the discovery of penicillin in 1928, a strange development occurred in the production of a range of antibiotics and has revolutionized modern health care. Thus, the emergence of antibiotics as a ruthless weapon against common bacterial infections such as tuberculosis and pneumonia was effectively treated. Thus, antibiotics were not only responsible for the drastic reduction in mortality, but also formed the basis of the greatest advances in surgery and medicine in the twentieth century (Fleming 1929; Read and Woods 2014). It is a worrying fact that the successful use of antibiotics has made these drugs more of a lucrative commodity and therefore not reasonably managed as a valuable resource. Therefore, researchers and doctors encountered a huge flood of antibiotic resistance among many bacteria (Read and Woods 2014). It is predicted that by 2050, the outbreak of infectious diseases which are resistant to all antibiotics will devastate many other countries besides the USA, and an epidemic event similar to 1918, when the flu went hand in hand and devastated countries. At that time, the available antibiotics will not be able to fight infectious diseases, thus quarantining infected people to minimize infectious diseases, but millions of people die every year. In addition to the resurgence of previously treatable infectious diseases, a number of antibiotic-resistant microorganisms will become biological weapons, and several countries will fall victim to bioterrorism agents. This is not a portrayal of a Hollywood movie, but a bitter concept predicted by recent reports of antibiotic resistance (Lewies et al. 2019; O’Neill 2016; Shariati et al. 2019). Currently, humanity has entered the second dark age of post-antibiotic, an era in which besides humans have faced again with microbial diseases, these diseases also are resistant (Lewies et al. 2019). These resistances are not just the current characteristics of bacteria, but microorganisms that have adapted to the harsh environmental conditions for millions of years (Read and Woods 2014). It is noteworthy that bacteria inherit different genes from previous generations and easily pass them on to the next generation. Surprisingly, bacteria are one of the few organisms on Earth that have the potential to transmit genetic material and antibiotic resistance to other bacteria through horizontal gene transfer (Moradi et al. 2016; Taati Moghadam et al. 2016; Mohebi et al. 2016). In this case, the resistance is easily transmitted in bacterial communities and as well as mutation-inducing SOS response to certain environmental stressors gives these microorganisms an evolutionary counter to antibiotics, resulting in besides resistant genes, mutations able infer antibiotic resistance via various mechanisms such as (1) decreasing antibiotic drug uptake (preventing drug penetration of the outer and/or cytoplasmic membrane), (2) modifying the antimicrobial target (for example producing enzymes that inactivate the antibiotic), (3) universal changes in central biochemical pathways (evolutionary adapted mechanisms), and (4) activation of efflux mechanisms (active removal of antibiotics) (Cirz et al. 2005; Munita and Arias 2016). As a result, resistance to antimicrobials is so high today that it typically kills 10 million patients worldwide each year, highlighting isolates with high resistance to key antibiotics such as MDR, XDR, and PDR (Schwarz and Johnson 2016; Moghadam et al. 2021b, 2020a). It is estimated that MDR and XDR bacteria increase not only patient mortality and morbidity, but also the cost of medical care for patients in which the economic burden on health care by antibiotic resistance is estimated at 30 to 100 $ billion annually (Sileshi et al. 2016; Inchai et al. 2015). Thus, to treat these highly resistant bacteria, more toxic antibiotics such as colistin are used as the last available weapons. Unfortunately, a worrying rate of colistin resistance infections is reported to cause 25 to 71% mortality due to severe infections caused by these isolates concern (Moubareck et al. 2018; Menekşe et al. 2019). At present, the impact of antibiotic resistance in various areas of daily life, especially in medicine and the heavy economic burden on countries, is felt. The inability to cope with the onslaught of resistant bacterial infections is growing by the day, and suitable alternatives are becoming more vital every day.
Colistin resistance and mechanisms
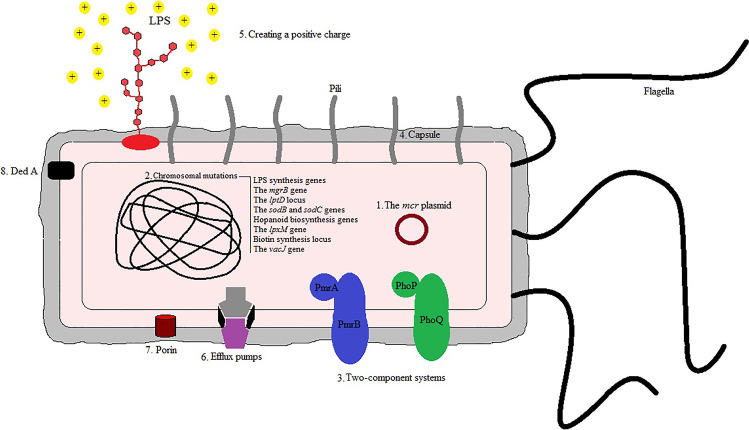
Colistin was first introduced as an antibiotic almost eight decades ago but due to the high side effects in patients and the discovery of less toxic antibiotics, doctors did not prescribe it for many years. Today, the emergence of a high rate of antibiotic resistance in bacteria such as MDR and XDR led to re-prescribe of colistin because of the lack of efficiency of other antibiotics (Moghadam et al.). Colistin is used as one of the main monotherapy-coping routes with MDR and XDR bacteria which are isolated from nosocomial infections and it has yet a good function on Gram-negative antibiotic resistance in despite of it has high side effects (Poulikakos et al. 2014). It was reported that nephrotoxicity (varies from 6 to 58%) was the common side effect after intravenous administration of colistin in infectious patients. Patients with abnormal kidney disease usually have a higher rate of nephrotoxicity (27–58%) than individuals with normal renal function (about 10% of cases) (Tigen et al. 2013; Falagas et al. 2010a). Irregular prescription of colistin in livestock in different areas of the world has gradually been responsible for the advent of transferable colistin-resistant bacteria from animals to humans through food. The use of colistin in livestock was interdicted with a significant increase in resistance in order to prevent further emergence of resistance to this antibiotic, especially in developed countries (Gwida et al. 2014; García-Meniño et al. 2019). Previous literatures have announced five mode of action for colistin such as (1) direct antibacterial activity of colistin which bind lipid A of LPS in the outer membrane (via electrostatic bonds) by its cationic diaminobutyric acid residues and resulting in cell lysis, (2) when colistin binds to LPS molecules exhibits anti-endotoxin activity by prohibiting the function of lipid A endotoxin which leads inhibiting shock induction via liberation of interleukin-8 (IL-8) and tumor necrosis factor-alpha (TNF-α) (Ahmed et al. 2016; El-Sayed Ahmed et al. 2020), (3) vesicle-vesicle contact pathway is another mode of action of colistin which binds to anionic phospholipid vesicles after transiting to the outer membrane, leading the fusion of the inner leaflet of the outer membrane with the outer leaflet of the cytoplasmic membrane, eventually resulting in the loss of phospholipids and Gram-negative bacteria death, (4) colistin in hydroxyl radical death pathway releases reactive oxygen species (ROS) in which the pathway is prominent to the Fenton reaction, consequently occurs bacterial death by DNA, lipids, and proteins damaging, and (5) prohibition of respiratory enzymes is final mode of action of colistin when events antibacterial function by interfering with fundamental respiratory enzymes (El-Sayed Ahmed et al. 2020). In addition to the severe side effects of colistin, the emergence of uncontrollable colistin resistance in bacteria has doubled the challenge of using it as last-line antibiotics for physicians. There has been report of colistin resistant in a wide range of Gram-negative isolates such as Klebsiella pneumoniae, Acinetobacter baumannii, Escherichia coli, Salmonella enterica serovar Typhimurium, Aeromonas spp., Pseudomonas aeruginosa, Enterobacter roggenkampii, Vibrio parahaemolyticus, Morganella morganii, Providencia spp., Burkholderia cepacia, Campylobacter, Serratia marcescens, Proteus spp., Vibrio cholera, Brucella, Edwardsiella spp., Legionella, Chromobacterium, Neisseria spp., and Stenotrophomonas spp. worldwide, especially MDR and XDR isolates (El-Sayed Ahmed et al. 2020; Kaye et al. 2016; Falagas et al. 2010b; Moghadam et al.). Colistin resistance in Gram-negative bacteria usually appears by chromosomal mutations or transmissible plasmid genes. The mcr genes including mcr-1 to mcr-10 are transmissible plasmid genes that are able to distribute colistin resistance via horizontal transfer among bacteria (Gharaibeh and Shatnawi 2019; Wang et al. 2020; Sun et al. 2018). The plasmid-borne mcr gene has a fascinating mechanism which reported various plasmids such as IncF, IncY, IncP, IncI2, IncX4, IncHI2, and ColE10-like ones can harbor mcr genes in bacteria (Madec and Haenni 2018). The genes are expressed in products which transfer phosphoethanolamine debris to the key target of colistin (lipid moiety of LPS) and responsible for changing in the LPS of Gram-negative bacteria, resulting in attenuating the bacterial affinity to colistin. In addition to colistin, it has been confirmed that the mcr gene can be a factor in resistance to other antibiotics (Andersson et al. 2016; Yang et al. 2017). It has been reported that integration of mcr into the bacterial membrane and altering in lipid A via its enzymatic activity occur when there is a high expression of the mcr gene which is responsible for alters in fitness, growth rate, and structural integrity of the outer membrane (Yang et al. 2017). Chromosomal mutation is another mechanism of colistin resistance which occurs in lipid A genes containing lpxA, lpxD, lpxO2, and lpxC, resulting in incomplete LPS. A high rate of resistance is occurs when the ISAba11 sequence is inserted within the LPS synthesis genes such as lpxC and lpxA because bacteria lose the potential of LPS production (Moffatt et al. 2011; Sherry and Howden 2018). Thus, this leads to a lack of LPS in bacteria and consequently a negative charge on Gram-negative bacteria, which causes a loss of affinity to react with colistin (Soon et al. 2011). Another chromosomal factor that contributes to intrinsic resistance to colistin are known as PhoPQ and PmrAB which are considered as part of two-component systems (Olaitan et al. 2014; Poirel et al. 2018). A response regulator (responds to environmental stimuli) and histidine kinase are the main parts of the two-component PmrAB system. There are different environmental circumstances, containing the presence of Mg2+, Al3+, and Fe3+ ions as well as various pH levels in which the PmrAB two-component system is able to sense and respond properly (Capra and Laub 2012; Willett 2012; Beceiro et al. 2011; Wösten et al. 2000). Impacting the expression of genes responsible for change of lipid A via the PmrAB system can contribute to colistin resistance in Gram-negative bacteria. On the other hand, point mutations in the pmrA and pmrB genes enhance the expression level of pmrAB resulting in change occurring in the bacterial outer membrane consequently diminishes membrane colistin entrance (Beceiro et al. 2011; Huang et al. 2020). PhoPQ activites through environmental agents including Mg2+ and Ca2+ as well as cationic antimicrobial peptides such as indolicidin, LL-37, and polymyxin. Thereby, this two-component system acts a crucial function in bacterial virulence and increased colistin resistance by changing in LPS (Minagawa et al. 2003; Devine and Hancock 2002; Regelmann et al. 2002; Huang et al. 2020; Cheung et al. 2008; Wi et al. 2017). Capsule formation in bacteria can be a main factor to attenuating polymyxin because the polysaccharides of the capsule react by anionic interactions with polymyxin and contributing to colistin resistance (Llobet et al. 2008). In addition, some efflux pumps such as KpnEF and PhoPQ are activated via regulators of capsule production including conjugative pilus expression (Cpx) and regulator of capsule synthesis (Rcs) and are responsible for colistin resistance. The KpnEF and PhoPQ were activated for Cpx and Rcs regulon, respectively (Olaitan et al. 2014; Llobet et al. 2008). Creating a positive charge on the surface of bacteria is responsible for changes in lipid A structure via the addition of compounds such as galactosamine (naxD), 4-amino-4-deoxy-L-arabinose (L-Ara4N, mediated by arnBCADTEF-ugd operon), and phosphoethanolamine (pEtN, mediated by chromosomally encoded eptA or plasmid-borne mcr) to lipid A. These created positive charges on the surface of Gram-negative bacteria not only reduce the potential of colistin for binding to bacterium, but also lead to the rupture of the outer membrane (Sato et al. 2018; Huang et al. 2020). Mutation of the chromosomal mgrB gene is responsible to activate PhoPQ two-component system by negative feedback of this system, consequently enhancing the expression of the arnBCADTEF operon and lead to colistin resistance (Baron et al. 2016). Various insertion sequences (e.g., IS5-like, IS102, IS5 family, IS3-like, and ISKpn14) can be located on mgrB genes and contribute to missense or nonsense mutations in clinical samples (Cannatelli et al. 2014, 2015; Bonura et al. 2015). Efflux pumps are one of the common forms of colistin resistance mechanism in Gram-negative bacteria which belong to the resistance nodulation cell division (RND) family. In the RND efflux pumps, adeR gene takes action as a regulator, adeA gene expresses to a membrane fusion protein, adeB gene becomes responsible to transports substrate from cytoplasm or phospholipid bilayer to the extracellular environment, and adeC gene act as the outer membrane protein channel (Magnet et al. 2001). Some of other efflux pumps also were reported contributing in colistin resistance but their mechanisms are not yet clear such as sapABCDF, MexXY–OprM, CarO, kpnEF, acrAB–tolC, and emrAB (Lin et al. 2017; Sundaramoorthy et al. 2019; Baron et al. 2016). Colistin heteroresistance is a mediocre estate that exhibits a determined phenotype through the existence of resistant subpopulations among a susceptible population; therefore, Gram-negative colistin-heteroresistant bacteria can acquire colistin resistance. Gram-negative colistin heteroresistance bacteria may be considered for the unaccountable therapeutic failure. Colistin heteroresistance phenotypes are mostly distinguished among MDR A. baumannii, K. pneumoniae, and P. aeruginosa isolates. Different mechanisms were attributed to colistin heteroresistance bacteria containing soxRS-regulated overexpression of the acrAB–tolC efflux pump, biofilm formation, putrescine/YceI communication, and activation of two-component regulatory systems PmrAB, PhoPQ, ParRS, CprRS, and ColRS, as well as mutations in lipid A biosynthesis genes (Lin et al. 2019; El-Sayed Ahmed et al. 2020). Recent studies by researchers to discover new mechanisms for resistance to colistin have shown that a number of “miscellaneous chromosomally encoded resistance genes” in different bacteria can cause resistance to this antibiotic. It was reported that lptD locus is an indispensable gene for locating the newly produced LPS within the outer membrane, the result of removing of this locus raising polymyxin resistance because bacteria completely lose LPS (Bojkovic et al. 2016). Another probable route of resistance to colistin has been reported through detoxification of reactive oxygen species in which this mechanism is formed by sodB and sodC genes (Mlynarcik and Kolar 2019). Two supposed hopanoid biosynthesis genes including Bmul_2133 and Bmul_2134 were detected in Burkholderia multivorans which were vital for the fixation of penetrance of outer membrane. Accordingly, these genes were attributed to polymyxin resistance via an independent of LPS-binding activity mechanism (El-Sayed Ahmed et al. 2020). Overexpression of an outer membrane protein as known OprH can bind to negatively charged phosphate groups of LPS, consequently preventing the binding of polymyxin to LPS. Intriguingly, downregulation of another outer membrane porin OprD is also contributed to polymyxin resistance in P. aeruginosa (El-Sayed Ahmed et al. 2020). The lpxM gene could be regulated by acylation of lipid A in Gram-negative bacteria resulting in colistin resistance which showed inactivation of lpxM gene can prevent L-Ara4N modifications and is responsible for reducing polymyxin resistance (Mills et al. 2017). Biotin synthesis locus acts a key role in polymyxin resistance which has demonstrated deletion mutation in the locus has been impressed lipid A production due to biotin an essential cofactor of lipid metabolism. Therefore, lower biotin levels contribute to the decreasing production of lipid A which can diminish colistin susceptibility (Hood et al. 2013; Whitfield and Trent 2014). A family of membrane transporter proteins called DedA was discovered in Burkholderia thailandensis which is related to colistin resistance. DedA was responsible for change in lipid A of LPS, resulting in colistin resistance (Panta et al. 2019). It is remarkable that a single mutation in vacJ gene attributes to the advent of colistin-resistant bacteria (Nhu et al. 2016). Figure 1 summarizes the mechanisms that Gram-negative bacteria use to become resistant to colistin.
Fig. 1.
Besides the colistin-heteroresistant, which is indicated by multiple pathways for colistin resistance, numbers 1 to 8 summarize the main reported colistin-resistant mechanisms
What is AMPs?
The AMPs are various classes of bioactive small proteins which are ubiquitous part of the first line of defense system of organisms (bacteria, fungi, plants, animals, and mammalian species) against pathogens (Haney et al. 2019; Jenssen et al. 2006). Although lysozyme was discovered by Alexander Fleming about a century ago, the pathway for identifying AMP was paved in the mid-1990s (Lemaitre et al. 1996). The mechanisms of AMPs are distinct from traditional antibiotics, resulting in having vigorous potential to affect a wide spectrum of microbes and even drug-resistant bacteria. So far, thousands of AMPs (natural or synthetic) have been discovered in which the most are cationic, water soluble, and act the key antimicrobial roles. AMPs are induced via particular external factors and expressed by the specific genes as well as usually consist a combination rich in hydrophobic and cationic amino acids resulting in they have positively charge (cationic) and both hydrophilic and hydrophobic (amphiphilic) characteristics (Lei et al. 2019). The AMPs frequently have short amino acid sequences (less than 100 amino acid residues) with helical form and with excessive amounts of lysine and arginine which are responsible for positive charge (Rathinakumar and Wimley 2010). The amphiphilic AMPs are α-helices which divide into two hydrophobic and hydrophilic parts and represent amphiphilicity when attaching to bacterial membranes. Thus, bacterial membranes undergo membrane conformational change when positive charge of AMPs lead to interacting negative charge of membranes via electrostatic interactions. AMPs attach to bacterial surfaces with their hydrophobic halves anchored in the hydrophobic lipid of the membrane bilayer (Chen et al. 2007; Som et al. 2008). The C-terminal of AMPs has hydrophobicity function and N-terminals are rich in essential amino acids with strong alkaline. The amount of pure cationic charges and hydrophobicity of AMPs correspond to their antibacterial and hemolytic activities, respectively. The synthetic AMPs are chemically modified and extend their half-life as far as some of them preserve biological activity at high temperature or are stable with the hydrolysis of pepsin and trypsin (Lei et al. 2019).
Advantages of AMPs
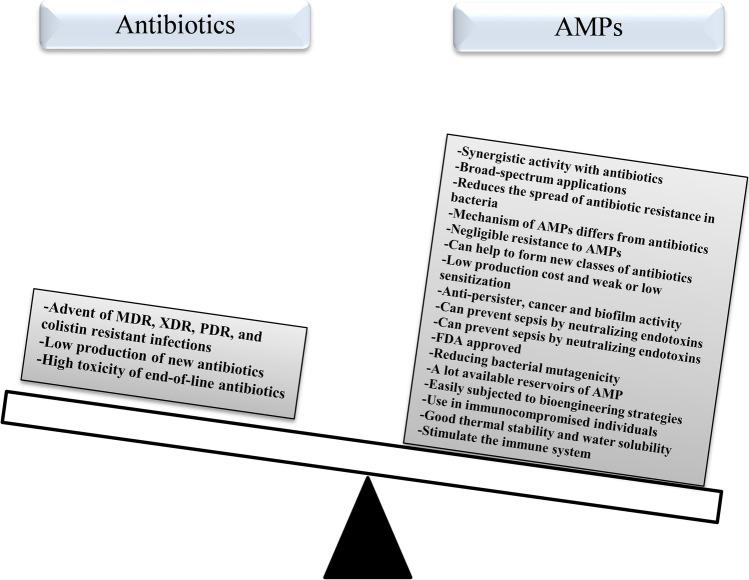
As mentioned earlier, the mechanism of AMPs differs from that of traditional antibiotics because they interact with the bacterial membrane by neutralizing the charge. As a result, they penetrate the cell membrane and cause the death of bacteria, which reduces the likelihood of the spread of antibiotic resistance in bacteria (Som et al. 2008; Mahlapuu et al. 2016). AMPs compared to antibiotics have broad-spectrum applications (antiviral, antibacterial, anticancer, and antiparasitic activities) while antibiotics usually display narrow spectrum function (only on bacteria). Resistance to AMPs is negligible compared to antibiotics (Marr et al. 2006). AMPs in low bactericidal concentration have great potential for rapid germ-killing and are effective on antibiotic-resistant bacteria. Although AMPs have been reported to stimulate the immune system, they can prevent sepsis by neutralizing endotoxins. This is when antibiotic treatment leads to the release of LPS from the cell wall of Gram-negative bacteria and eventually septic shock. Hence, AMPs represent synergistic activity with antibiotics as promising proxies to preventing septic shock by neutralizing lipopolysaccharide (LPS or endotoxin) (Bahar and Ren 2013; Mahlapuu et al. 2016; Prins et al. 1995). AMPs can be used for bacterial infections and problems in which antibiotics have not been successful such as septicemia and infection in immunocompromised individuals who are unable to provide an adequate immune system for antibiotic treatment (Hancock 2015). One of the important factors of AMPs is that they also do not activate the bacterial stress pathway such as SOS and rpoS compared to antibiotics, thus reducing bacterial mutagenicity (Rodríguez-Rojas et al. 2014). Surprisingly, good thermal stability and good water solubility are two other factors that are proven in AMPs. Ribosomally synthesized AMPs are easily subjected to bioengineering strategies to increase activity against resistant bacteria. For example, although nisin was only effective against Gram-positive bacteria, a nicin was produced using bioengineering technique called nisin A which was effective against Gram-positive bacteria even methicillin-resistant Staphylococcus aureus (MRSA) and a wider range of Gram-negative bacteria such as E. coli, Salmonella enterica serovar Typhimurium, and Cronobacter sakazakii (Raguse et al. 2002; Dehsorkhi et al. 2014; Field et al. 2012). AMPs are considered as small molecules with weak or low sensitization and simple structure–activity relationship (Raguse et al. 2002; Lee and Hodges 2003). Structural and mechanical studies of AMPs can help these antimicrobial compounds to form new classes of antibiotics because peptidomimetics are amphiphilic compounds modulated by the biological and structural properties of host defense peptides in order to be considered as a new chemical class of antibiotics (Wright 2016). It has been shown that AMPs have a greater effect on cancer cells than normal cells and easily enter adipose lipid membranes and create channels or pores. As a result, leakage of cell contents occurs and kills cancer cells (Lei et al. 2019). On the other hand, AMPs are with less or no side effects, and hard to lead drug resistance in bacteria compared to antibiotics. AMP reservoir remains high due to an abundance of uncommon sources of AMPs, containing marine bacteria, unculturable soil, and procedures available to produce vast libraries of derivatives. Not only are the AMPs used for treating infections, but also can be safely applied to therapeutics in aquaculture, food preservatives, food additives for livestock, and control plant diseases (Magana et al. 2020). A database of AMPs specifically tested against microbial biofilms (http://www.baamps.it/) was first published in 2015. Biofilms are accumulations of bacteria that form at different surfaces and are resistant to antibiotics due to the polysaccharide matrix that surrounds them. In addition, biofilms are notoriously recalcitrant to clearance by the host immune system and many antibiotics are unable to penetrate this form of resistance. Plethora AMPs have illustrated function in killing cells in biofilms by different modes of action such as inhibiting quorum sensing (QS)-dependent biofilm formation, preventing microbial adhesion when used to coat medical implants, and interfering with extracellular polymeric substance (EPS) production and stability (Batoni et al. 2021; Moghadam et al. 2021a). Importantly, in contrast to the majority of antibiotics which affect active cell processes, AMPs may target persisters that form biofilms in high frequencies, and are responsible for major contributors to the relapsing of many infections (Grassi et al. 2017; de Breij et al. 2018). Although many synthesized or natural antimicrobial compounds are not yet Food and Drug Administration (FDA) approved, many of AMPs have FDA approved. Imagine if we put the benefits of AMPs on one side of the scales and on the other side of the scales the limitations of using antibiotics, we will find that AMPs are good alternatives to antibiotics, co-administration with antibiotics, and even the production of new antibiotics (Fig. 2). All of the above benefits have attracted the attention of researchers to have a special look at AMPs and their use in the clinic in the future.
Fig. 2.
On one side of the scales are the benefits and important factors of using AMPs (right) and on the other side are the limitations of antibiotics (left), which weighs heavier on the benefits of AMPs
Mode of action of AMPs
Conventionally, there is a need to modify and synthesize the long-acting AMPs analogs for possible clinical applications because natural AMPs are not stable with a short half-life. Accurate identification of the mechanisms of AMPs is vital in designing a new generation of synthetic and effective AMPs. The three-dimensional structures and specific amino acid components of AMPs are not the main factors in their antimicrobial function, but the main antimicrobial functions are related to their surface activity. Thus, the physical–chemical interactions and surface characteristics considered essential factors to define the biological functions of AMPs with the membrane-permeabilizing and membrane-destabilizing potentials (Graf and Wilson 2019; Aisenbrey et al. 2019; Lei et al. 2019). According to membranes of eukaryotic cells consisting of uncharged neutral phospholipids such as cholesterol and sphingomyelins, the positive charges of α-helix on the surface of AMPs lead to interact with negatively charged membranes of microbes (due to negatively charged lipid groups such as phosphatidylglycerol and cardiolipin), and AMPs show antimicrobial activity without harming normal cells (Bahar and Ren 2013; Zanetti 2004). So far, several hypothetical modes of action are assumed to AMPs, containing the intracellular bactericidal mechanism, antimicrobial effect via participating in immune regulation, the damage of the organelles to cause DNA fragmentation, inhibition of enzyme activity, cell membrane damage, and inhibition of the synthesis of macromolecules (Bahar and Ren 2013; Som et al. 2008). Among the mentioned mechanisms, the most common was membrane permeability or interaction of cationic AMPs with cell membranes which impacted microbes through enhancing cytoplasmic membrane permeability. Scientists have suggested various presumptive patterns of pore formation for definition of action of AMPs, including the circular model, the concave barrel model, the blanket model, and the wormhole model. The electrostatic interaction between positive charge of AMPs and negative charge of microbial membranes is the first step of membrane permeability. Consequently, membrane damage occurs via pores which are caused by the blockage of membrane-coupled respiration and leakage of ions, metabolites, and biosynthesis and eventually responsible for the death of microbes (Lei et al. 2019). The effect of AMPs on cellular physiological processes occurs when they penetrate into bacterial cells and attack DNA and RNA, and inhibit of the synthesis of cell wall and protein without the permeability of bacterial membranes. This penetration led to accumulation of AMPs inside bacteria resulting in blocking bacterial functions and then enforce cell death by interacting with intracellular RNAs and DNAs (Moravej et al. 2018; Graf and Wilson 2019; Aisenbrey et al. 2019). Finally, the effect of AMPs on macromolecules is such that AMPs are able to interact with protein macromolecules associated with bacterial DNA replication, thus preventing DNA replication, and ultimately show a bactericidal function (Lei et al. 2019). These diverse mechanisms of action are one of the important potentials of AMPs, enabling them to be used against a wide range of microbes, even those with high drug resistance, because these mechanisms create a golden condition separate from the mechanisms of antibiotics to fight drug-resistant microorganisms.
The limitation of therapeutic use of AMPs and strategies to overcome them
Despite all the benefits mentioned for AMPs, these antimicrobial compounds also have limitations, the most important of which are discussed in this section. So far, one of the biggest limitation for the clinical use of AMPs is the high cost of their large-scale synthesis, especially ribosomally synthesized AMPs (Lewies et al. 2019). The cost of producing synthetic peptides was reported 5 to 20 times higher than that of antibiotics and the cost of using the currently available AMPs was estimated at about $ 50 to $ 400 per gram for each treatment of a patient’s infection at about 1 mg/kg body weight per day (Marr et al. 2006). Therefore, to resolve this issue, sufficient progress to production of recombinant DNA techniques has been performed for the cost-effective synthesis and purification of AMPs with enhanced yields for therapeutic usage in individuals. But the large-scale commercial possibility of these techniques still requires to be estimated via the accomplishment of commercial enterprises. It is thought that in the future, it is likely that pharmaceutical companies will increasingly present peptide drugs in production lines and it will be possible to produce cost-effective and high-quality peptides (Lewies et al. 2019; Giuliani et al. 2007). Bioavailability is an important issue for peptide-based drugs which needs to be addressed; therefore, the pharmacokinetics and pharmacodynamics of AMPs must be focused on defining the therapeutic dosage in order to avoid probable toxic side effects (Bell and Gouyon 2003). In general, peptides are sensitive to proteolytic degradation, so unfavorable pharmacokinetics and rapid clearance of peptides due to proteolytic degradation lead to their limitation. There are suggestions to address this defect through combining AMPs with negatively charged or lipid-friendly proteins that can protect against proteolytic degradation of AMPs in vivo. On the other hand, to enhance peptide half-life, many strategies are feasible such as ways of administration, several formulations, and different levels of chemical modification. The amidation at the N-terminus, introduction of D-amino acids, peptide cyclization, and non-natural amino acids are the most general plans to enhance peptide stability (Giuliani et al. 2007). Therefore, as mentioned, AMPs are part of the natural innate immunity of organisms, and monitoring their administration is a special issue because the emergence of cross-resistance to these compounds would be detrimental. Although monotherapy of AMPs has been observed cross-resistance between innate AMPs of the body and therapeutic AMPs, in solving this problem, the combination of therapeutic AMPs with conventional antibiotics has been helpful in restricting the development of cross-resistance and resistance (Bell and Gouyon 2003; Fleitas and Franco 2016). Given the few limitations reported for AMPs, there are solutions to each of these limitations. On the other hand, the many benefits of these antimicrobial compounds are a good option to replace or use them simultaneously with antibiotics against drug-resistant microorganisms, especially resistance to end-of-line antibiotics.
AMP in the war with colistin-resistant bacteria
As mentioned, humanity is in an unequal war against antibiotic-resistant infections, especially resistant to last-line treatment, and needs new weapons such as AMPs to win. Colistin is the last resort for physicians in the treatment of many infections and is one of the most effective antibiotics against MDR isolates and carbapenem-resistant Enterobacteriaceae. Colistin-resistant bacteria are usually resistant to most or all of the other antibiotics, so fighting them with new compounds such as AMPs is critical. In 2021, Jahangiri et al. in a study examined the synergistic effect of two AMPs (Nisin and P10), conventional antibiotics against XDR, and colistin-resistant Gram-negative isolates. The results of this study showed that nisin in the minimum inhibitory concentration (MIC) 64–256 μg/ml and P10 in the MIC 8–32 μg/ml have potential antibacterial activity against the resistant bacteria. It was also found that two studied AMPs, alone and in combination with antibiotics, showed the ability to kill XDR A. baumannii and colistin-resistant P. aeruginosa isolates. Among the combination therapy of AMPs and antibiotics, synergism of Nisin + colistin and P10 + ceftazidime/doripenem shows considerable therapeutic worthiness as antibacterial drugs against colistin-resistant isolates (Jahangiri et al. 2021). In another study, six cationic α-helical frog skin-derived AMPs including CPF-AM1, PGLa-AM1, B2RP-ERa, [E4K] alyteserin-1c, [D4K] B2RP, and [G4K] XT-7 against colistin-resistant Acinetobacter isolates were examined. All AMPs were effective on colistin-susceptible and colistin-resistant clinical isolates of A. baumannii and Acinetobacter nosocomialis in MIC ≤ 2 µg/mL and MIC ≥ 64 µg/mL, respectively. The [D4K] B2RP and [E4K] alyteserin-1c in MIC = 4–16 µg/mL showed the best antibacterial activity against colistin-resistant isolates (Conlon et al. 2012). In a study, Lin et al. examined the effect of LL37 as natural AMPs and WLBU2 as engineered AMPs on six colistin-resistant isolates containing 3 isolates of K. pneumoniae, 2 isolates of A. baumannii, and 1 isolate of P. aeruginosa in planktonic form and biofilm. Although LL37 showed significant antimicrobial properties on colistin-resistant A. baumannii isolates, it did not affect K. pneumoniae and P. aeruginosa isolates. On the other hand, WLBU2 was effective on colistin-resistant isolates of A. baumannii and K. pneumoniae but was not able to kill P. aeruginosa. Although both AMPs showed the ability to degrade biofilms, WLBU2 was more effective than LL37 (Lin et al. 2018). In 2019, the impact of AA139 and SET-M33 as two novel AMPs was tested against three colistin-resistant and MCR-producing isolates. The two AMPs were effective against colistin-resistant strains in MIC ≥ 16 µg/L and can be promising novel antimicrobial agents for the treatment of colistin-resistant MDR K. pneumoniae infections (van der Weide et al. 2019). In 2020, the combination of colistin and AMPs (MSI-78 and OTD-244) was examined to circumvent colistin-resistant bacteria such as E. coli, K. pneumoniae, A. baumannii, and P. aeruginosa. Not only did the MSI-78 alone showed great antibacterial activity against colistin-resistant bacteria (MIC 2.5–10 µg/mL), but also when combined with the colistin, it decreased more than fourfold for 75% of colistin-resistant isolates. The MIC for OTD-244 for all bacteria was > 500 µg/mL, and observed limited antibacterial effect against the all studied Gram-negative bacteria (Witherell et al. 2020). In another study, the effect of three AMPs including protamine, lysozyme, and lactoferrin on colistin-resistant K. pneumoniae and Enterobacter asburiae showed that lactoferrin did not any killing function but protamine and lysozyme exposition caused a 40% and 87% reduction of CFU on colistin-resistant K. pneumoniae isolate, respectively. All three AMPs had no effect on colistin-resistant E. asburiae and high-level tolerance was observed against these three AMPs (Kádár et al. 2015). Hashemi et al. evaluated the susceptibility of colistin-resistant clinical isolates of K. pneumoniae to a series of AMPs containing LL-37, cecropin A, magainin 1, CSA-13, CSA-44, CSA-131, CSA-138, and CSA-142. The result showed that there is approximately no cross-resistance between colistin and AMPs and AMPs kill colistin-resistant K. pneumoniae via lipid A modifications (Hashemi et al. 2017). Cirioni et al. combat colistin-resistant P. aeruginosa isolates using AMPs (s-thanatin) alone with colistin in vitro and in vivo. S-thanatin alone showed significant antimicrobial properties at MICs of 16 mg/L as well as observed a synergy interaction with colistin, and was able to prevent colistin-resistant P. aeruginosa isolates. On the other hand, the combination of s-thanatin with colistin showed the highest efficacy in an animal model of P. aeruginosa sepsis (Cirioni et al. 2011). In 2015, two engineered cationic AMPs (WLBU2 and WR12) and a natural AMPs (LL37) were applied to overcome MDR pathogenic bacteria. Although 25% of colistin-resistant strains were also susceptible to LL37, the inhibition of 80 to 86% of colistin- and LL37-resistant isolates was observed by two engineered cationic AMPs (Deslouches et al. 2015). LS-sarcotoxin and LS-stomoxyn as two insect-derived AMPs were profiled to assess their suitability for colistin-resistant MDR Gram-negative bacteria. Results showed that LS-sarcotoxin and LS-stomoxyn have selective and potent activities against MDR bacteria and no cross-resistance with antibiotics. LS-sarcotoxin in MIC 2 to 16 mg/L and LS-stomoxyn in MIC 2 to 128 mg/L have killing ability against colistin-resistant E. coli, E. cloacae, E. aerogenes, K. pneumoniae, S. enterica, A. baumannii, and P. aeruginosa (Hirsch et al. 2019). Antibacterial activity of ten α-helical AMPs containing D87(Lys1-6 Arg-1), D84(Lys1-6 Lys-1), D85(Lys1-6 Orn-1), D86(Lys1-6 Dab-1), D105(Lys1-6 Dap-1), D101(Lys1Ser26-5 Lys-1), D102(Lys1Ser26-5 Dab-1), D85(K13A/K16A)-(Lys1-6 Orn-1), D86(K13A/K16A)-(Lys1-6 Dab-1), and D105(K13A/K16A)-(Lys1-6 Dap-1) was measured against 7 polymyxin B and colistin-resistant A. baumannii strains. All of the studied AMPs presented excellent antimicrobial activity on polymyxin B- and colistin-resistant A. baumannii strains in MIC 0.5 to 2 μM (Mant et al. 2019). Kao at el. reported that LL-37, RL-37, LL-29, LL-29 V, LL-29V2, CAP-11, CAP-11V1, CAP-11V2, CAP-11V3, SMAP-29, SMAP-29 V, SMAP-29B, SMAP-29D, BMAP-27, BMAP-27A, BMAP-27B, and BMAP-27C as cathelicidins AMPs have killing activity against Gram-negative and Gram-positive strains. Among them, BMAP-27B and SMAP-29D have potent bactericidal activity against mcr-harboring and colistin-resistant E. coli in MIC 1.6 μM (Kao et al. 2016). In 2020, in a research study, although EtCec1-a and EtCec2-a displayed antimicrobial activity against the colistin-resistant isolates, EtCec2-a showed lower activity. EtCec1-a observed bactericidal activity on colistin-resistant E. coli, E. cloacae, E. aerogenes, K. pneumoniae, S. enterica, and A. baumannii in MIC 4 to 32 but MIC 64 to 128 on S. maltophilia. On the other hand, EtCec2-a showed antibacterial function on colistin-resistant E. coli and A. baumannii in MIC 4 to 32, but majority of colistin-resistant E. cloacae, E. aerogenes, K. pneumoniae, S. enterica, and S. maltophilia were susceptible in MIC 32 to 128 (Hirsch et al. 2020). Mourtada et al. designed stapled AMPs for overcoming MDR colistin-resistant E. coli and A. baumannii. Drug sensitivity test of a lead double-stapled AMP named Mag(i + 4)1,15(A9K,B21A,N22K,S23K) showed potential bactericidal activity on colistin-resistant E. coli and A. baumannii in MIC 1.56 to 3.12 g/ml. Result also showed that AMP without renal injury or hemolysis was able to kill MDR Gram-negative pathogen colistin-resistant A. baumannii in a murine peritonitis-sepsis model (Mourtada et al. 2019). The set of studies whose results were presented in this section (Table 1) demonstrated the effect of different AMPs on colistin-resistant Gram-negative bacteria, so AMPs can be considered as new weapons in the fight against these end-of-line antibiotic-resistant bacteria.
Table 1.
A summary of the most important studies conducted in recent years on the effect of AMPs on colistin-resistant Gram-negative bacteria
| Name of author | Year | Name of AMPs | Organism | Result | Reference |
|---|---|---|---|---|---|
| Jahangiri et al | 2021 | Nisin and P10 | XDR A. baumannii and colistin-resistant P. aeruginosa isolates | AMPs, alone and in combination with antibiotics showed the ability to kill mentioned bacteria | (Jahangiri et al. 2021) |
| Conlon et al | 2012 | CPF-AM1, PGLa-AM1, B2RP-ERa, [E4K] alyteserin-1c, [D4K] B2RP, and [G4K] XT-7 | Colistin-resistant clinical isolates of A. baumannii and Acinetobacter nosocomialis | All six AMPs had an effect on colistin-resistant Acinetobacter isolates | (Conlon et al. 2012) |
| Lin et al | 2018 | WLBU2 and LL37 | Colistin-resistant isolates of K. pneumoniae, A. baumannii, and P. aeruginosa | The two studied AMPs showed a significant effect on colistin-resistant isolates of A. baumannii and K. pneumoniae but were not able to kill P. aeruginosa | (Lin et al. 2018) |
| Weide et al | 2019 | AA139 and SET-M33 | Colistin-resistant and mcr-producing isolates of K. pneumoniae | AMPs were effective against colistin-resistant strains in MIC ≥ 16 µg/L | (van der Weide et al. 2019) |
| Witherell et al | 2020 | MSI-78 and OTD-244 | Colistin-resistant E. coli, K. pneumoniae, A. baumannii, and P. aeruginosa | The MSI-78 alone and combination with colistin showed great antibacterial activity against colistin-resistant bacteria | (Witherell et al. 2020) |
| Kádár et al | 2015 | Protamine, lysozyme, and lactoferrin | Colistin-resistant K. pneumoniae and E. asburiae | Protamine and lysozyme were effective against colistin-resistant K. pneumoniae but all three AMPs were not able to eradicate colistin-resistant E. asburiae | (Kádár et al. 2015) |
| Hashemi et al | 2017 | LL-37, Cecropin A, Magainin 1, CSA-13, CSA-44, CSA-131, CSA-138, and CSA-142 | Colistin-resistant K. pneumoniae | AMPs kill colistin-resistant K. pneumoniae via lipid A modifications | (Hashemi et al. 2017) |
| Cirioni et al | 2011 | S-thanatin | Colistin-resistant P. aeruginosa | The s-thanatin alone and in combination with colistin showed the highest efficacy in vitro and in vivo | (Cirioni et al. 2011) |
| Deslouches et al | 2015 | WLBU2, WR12, and LL37 | Colistin-resistant MDR pathogenic bacteria | WLBU2 and WR12 as two engineered cationic AMPs display better antibacterial activity (80 to 86%) than LL37 (25%) as natural AMPs against colistin-resistant strains | (Deslouches et al. 2015) |
| Hirsch et al | 2019 | LS-sarcotoxin and LS-stomoxyn | Colistin-resistant MDR Gram-negative bacteria | LS-sarcotoxin and LS-stomoxyn have selective and potent activity against colistin-resistant MDR Gram-negative bacteria | (Hirsch et al. 2019) |
| Mant et al | 2019 | D87(Lys1-6 Arg-1), D84(Lys1-6 Lys-1), D85(Lys1-6 Orn-1), D86(Lys1-6 Dab-1), D105(Lys1-6 Dap-1), D101(Lys1Ser26-5 Lys-1), D102(Lys1Ser26-5 Dab-1), D85(K13A/K16A)-(Lys1-6 Orn-1), D86(K13A/K16A)-(Lys1-6 Dab-1), and D105(K13A/K16A)-(Lys1-6 Dap-1) | polymyxin B- and colistin-resistant A. baumannii strains | All of studied AMPs presented excellent antimicrobial activity on polymyxin B- and colistin-resistant A. baumannii strains | (Mant et al. 2019) |
| Kao et al | 2016 | LL-37, RL-37, LL-29, LL-29 V, LL-29V2, CAP-11, CAP-11V1, CAP-11V2, CAP-11V3, SMAP-29, SMAP-29 V, SMAP-29B, SMAP-29D, BMAP-27, BMAP-27A, BMAP-27B, and BMAP-27C | The mcr-harboring and colistin-resistant E. coli | BMAP-27B and SMAP-29D showed bactericidal activity against colistin-resistant E. coli | (Kao et al. 2016) |
| Hirsch et al | 2020 | EtCec1-a and EtCec2-a | Colistin-resistant E. coli, E. cloacae, E. aerogenes, K. pneumoniae, S. enterica, S. maltophilia and A. baumannii | Two AMPs displayed antimicrobial activity against the colistin-resistant isolates | (Hirsch et al. 2020) |
| Mourtada et al | 2019 | Mag (i + 4)1,15 (A9K,B21A,N22K,S23K) | Colistin-resistant E. coli and A. baumannii | AMP showed potential bactericidal activity on two studied colistin-resistant pathogens in vitro and colistin-resistant A. baumannii in a murine peritonitis-sepsis model | (Mourtada et al. 2019) |
Conclusion
At present, unfortunately, there is no powerful weapon in antibiotics that can target the strength of antibiotic resistance. It is a fact that in the face of the onslaught of antibiotic resistance, humanity must come up with new strategies to produce new weapons to deal effectively. AMPs are promising weapons in this regard due to their potency to directly kill bacteria (through multiple mechanisms), neutralize endotoxins, modulate host immunity to enhance the hosts’ ability to kill bacteria, and act synergistically with current antibiotics. High resistance bacteria, even those resistant to colistin, are not able to withstand a combination of AMPs and antibiotics, and the possibility of cross-resistance to AMPs is very low. Thus, the possibility is highlighted that adjuvant therapy with AMPs alone, combining several AMPs together, and in combination with antibiotics can manage the antibiotic resistance crisis. In addition, AMPs synthesized with genetically encoded ribosomes have considerable ability for development as antimicrobials because they have not only evolved with AMP-resistant mechanisms, but are also prone to bioengineering strategies to increase their activity and further circumvent bacterial resistance. Therefore, AMPs can be considered as agents of new antimicrobial agents in the near future, which can be considered a “coup de grace” for antibiotic resistance, especially colistin resistance, as one of the last lines of treatment antibiotics.
Author contribution
The core idea of this work came from MTM. RM was an advisor in the project. All of the authors contributed equally in writing the manuscript and approved it.
Declarations
Ethical approval
This manuscript is a review article that doesn’t require prior approval.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ahmaed SS, Alp E, Hopman J, Voss A (2016) Global epidemiology on colistin resistant Acinetobacter baumannii. Infect Dis Ther
- Aisenbrey C, Marquette A, Bechinger B (2019) The mechanisms of action of cationic antimicrobial peptides refined by novel concepts from biophysical investigations. Adv Exp Med Biol :33–64 [DOI] [PubMed]
- Andersson DI, Hughes D, Kubicek-Sutherland JZ. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist Update. 2016;26:43–57. doi: 10.1016/j.drup.2016.04.002. [DOI] [PubMed] [Google Scholar]
- Bahar AA, Ren D. Antimicrobial peptides. J Pharm. 2013;6:1543–1575. doi: 10.3390/ph6121543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron S, Hadjadj L, Rolain J-M, Olaitan AO. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int J Antimicrob Agents. 2016;48:583–591. doi: 10.1016/j.ijantimicag.2016.06.023. [DOI] [PubMed] [Google Scholar]
- Batoni G, Maisetta G, Esin S. Therapeutic potential of antimicrobial peptides in polymicrobial biofilm-associated infections. Int J Mol Sci. 2021;22:482. doi: 10.3390/ijms22020482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother. 2011;55:3370–3379. doi: 10.1128/AAC.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G, Gouyon P-H. Arming the enemy: the evolution of resistance to self-proteins. J Microbiol. 2003;149:1367–1375. doi: 10.1099/mic.0.26265-0. [DOI] [PubMed] [Google Scholar]
- Bojkovic J, Richie DL, Six DA, Rath CM, Sawyer WS, Hu Q, Dean CR. Characterization of an Acinetobacter baumannii lptD deletion strain: permeability defects and response to inhibition of lipopolysaccharide and fatty acid biosynthesis. J Bacteriol. 2016;198:731–741. doi: 10.1128/JB.00639-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonura C, Giuffre M, Aleo A, Fasciana T, Di Bernardo F, Stampone T, Giammanco A, Palma DM, Mammina C, Group M-GW (2015) An update of the evolving epidemic of bla KPC carrying Klebsiella pneumoniae in Sicily, Italy, 2014: emergence of multiple non-ST258 clones. PloS ONE 10:e0132936 [DOI] [PMC free article] [PubMed]
- Cannatelli A, Giani T, D’Andrea MM, di Pilato V, Arena F, Conte V, Tryfinopoulou K, Vatopoulos A, Rossolini GM. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother. 2014;58:5696–5703. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannatelli A, Santos-Lopez A, Giani T, Gonzalez-Zorn B, Rossolini GM. Polymyxin resistance caused by mgrB inactivation is not associated with significant biological cost in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2015;59:2898–2900. doi: 10.1128/AAC.04998-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012;66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centres for Disease Control and Prevention (2013) Antibiotic resistance threats in the United States. https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf. Accessed 17 July 2017
- Chegini Z, Khoshbayan A, Moghadam MT, Farahani I, Jazireian P, Shariati A. Bacteriophage therapy against Pseudomonas aeruginosa biofilms: A review. Ann Clin Microbiol. 2020;19:1–17. doi: 10.1186/s12941-020-00389-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Guarnieri MT, Vasil AI, Vasil ML, Mant CT, Hodges RS. Role of peptide hydrophobicity in the mechanism of action of α-helical antimicrobial peptides. Antimicrob Agents Chemother. 2007;51:1398–1406. doi: 10.1128/AAC.00925-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J, Bingman CA, Reyngold M, Hendrickson WA, Waldburger CD. Crystal structure of a functional dimer of the PhoQ sensor domain. J Biol Chem. 2008;283:13762–13770. doi: 10.1074/jbc.M710592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirioni O, Wu G, Li L, Orlando F, Silvestri C, Ghiselli R, Shen Z, Gabrielli E, Brescini L, Lezoche G. S-thanatin in vitro prevents colistin resistance and improves its efficacy in an animal model of Pseudomonas aeruginosa sepsis. Peptides. 2011;32:697–701. doi: 10.1016/j.peptides.2011.01.016. [DOI] [PubMed] [Google Scholar]
- Cirz RT, Chin JK, Andes DR, De Crecy-Lagard V, Craig WA, Romesberg FE. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 2005;3:e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon JM, Sonnevend A, PáL T, Vila-Farrés X. Efficacy of six frog skin-derived antimicrobial peptides against colistin-resistant strains of the Acinetobacter baumannii group. Int J Antimicrob Agents. 2012;39:317–320. doi: 10.1016/j.ijantimicag.2011.12.005. [DOI] [PubMed] [Google Scholar]
- De Breij A, Riool M, Cordfunke RA, Malanovic N, De Boer L, Koning RI, Ravensbergen E, Franken M, Van Der Heijde T, Boekema BK (2018) The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci Transl Med 10 [DOI] [PubMed]
- Dehsorkhi A, Castelletto V, Hamley IW. Self-assembling amphiphilic peptides. J Pept Sci. 2014;20:453–467. doi: 10.1002/psc.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslouches B, Steckbeck JD, Craigo JK, Doi Y, Burns JL, Montelaro RC. Engineered cationic antimicrobial peptides to overcome multidrug resistance by ESKAPE pathogens. Antimicrob Agents Chemother. 2015;59:1329–1333. doi: 10.1128/AAC.03937-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine DA, Hancock RE. Cationic peptides: distribution and mechanisms of resistance. Curr Pharm Des. 2002;8:703–714. doi: 10.2174/1381612023395501. [DOI] [PubMed] [Google Scholar]
- Dousari AS, Moghadam MT, Satarzadeh N. COVID-19 (Coronavirus disease 2019): a new coronavirus disease. Infect Drug Resist. 2020;13:2819. doi: 10.2147/IDR.S259279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed Ahmed MAE-G, Zhong L-L, Shen C, Yang Y, Doi Y, Tian G-B. Colistin and its role in the Era of antibiotic resistance: an extended review (2000–2019) Emerg Microbes & Infect. 2020;9:868–885. doi: 10.1080/22221751.2020.1754133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falagas ME, Rafailidis PI, Ioannidou E, Alexiou VG, Matthaiou DK, Karageorgopoulos DE, Kapaskelis A, Nikita D, Michalopoulos A. Colistin therapy for microbiologically documented multidrug-resistant Gram-negative bacterial infections: a retrospective cohort study of 258 patients. Int J Antimicrob Agents. 2010;35:194–199. doi: 10.1016/j.ijantimicag.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Falagas ME, Rafailidis PI, Matthaiou DK. Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist Update. 2010;13:132–138. doi: 10.1016/j.drup.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Field D, Begley M, O’Connor PM, Daly KM, Hugenholtz F, Cotter PD, Hill C, Ross RP. Bioengineered nisin A derivatives with enhanced activity against both Gram positive and Gram negative pathogens. PLOS One. 2012;7:10–22. doi: 10.1371/journal.pone.0046884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleitas O, Franco OL. Induced bacterial cross-resistance toward host antimicrobial peptides: a worrying phenomenon. Front Microbiol. 2016;7:381. doi: 10.3389/fmicb.2016.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Br J Exp Pathol. 1929;10:226. [PMC free article] [PubMed] [Google Scholar]
- Fox JL. Antimicrobial peptides stage a comeback: better understanding of the mechanisms of action, modification and synthesis of antimicrobial peptides is reigniting commercial development. Nat Biotechnol. 2013;31:379–383. doi: 10.1038/nbt.2572. [DOI] [PubMed] [Google Scholar]
- García-Meniño I, Díaz-Jiménez D, García V, de Toro M, Flament-Simon SC, Blanco J, Mora A. Genomic characterization of prevalent mcr-1, mcr-4, and mcr-5 Escherichia coli within swine enteric colibacillosis in Spain. Front Microbiol. 2019;10:2469. doi: 10.3389/fmicb.2019.02469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharaibeh MH, Shatnawi SQ. An overview of colistin resistance, mobilized colistin resistance genes dissemination, global responses, and the alternatives to colistin: a review. Vet World. 2019;12:1735. doi: 10.14202/vetworld.2019.1735-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliani A, Pirri G, Nicoletto S. Antimicrobial peptides: an overview of a promising class of therapeutics. Open Life Sci. 2007;2:1–33. doi: 10.2478/s11535-007-0010-5. [DOI] [Google Scholar]
- Graf M, Wilson DN (2019) Intracellular antimicrobial peptides targeting the protein synthesis machinery. Adv Exp Med Biol :73–89 [DOI] [PubMed]
- Grassi L, di Luca M, Maisetta G, Rinaldi AC, Esin S, Trampuz A, Batoni G. Generation of persister cells of Pseudomonas aeruginosa and Staphylococcus aureus by chemical treatment and evaluation of their susceptibility to membrane-targeting agents. Front Microbiol. 2017;8:1917. doi: 10.3389/fmicb.2017.01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwida M, Horzel H, Geue L, Tomaso H (2014) Occurrence of Enterobacteriaceae in raw meat and in human samples from Egyptian retail sellers. Int sch res notices [DOI] [PMC free article] [PubMed]
- HANCOCK RE. Rethinking the antibiotic discovery paradigm. EBioMedicine. 2015;2:629–630. doi: 10.1016/j.ebiom.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock RE, Sahl H-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- Haney EF, Straus SK, Hancock RE. Reassessing the host defense peptide landscape. Front Chem. 2019;7:43. doi: 10.3389/fchem.2019.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi MM, Rovig J, Weber S, Hilton B, Forouzan MM, Savage PB. Susceptibility of colistin-resistant, Gram-negative bacteria to antimicrobial peptides and ceragenins. Antimicrob Agents Chemother. 2017;61:e00292–e317. doi: 10.1128/AAC.00292-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch R, Wiesner J, Marker A, Pfeifer Y, Bauer A, Hammann PE, Vilcinskas A. Profiling antimicrobial peptides from the medical maggot Lucilia sericata as potential antibiotics for MDR Gram-negative bacteria. J Antimicrob Chemother. 2019;74:96–107. doi: 10.1093/jac/dky386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch R, Wiesner J, Bauer A, Marker A, Vogel H, Hammann PE, Vilcinskas A. Antimicrobial peptides from rat-tailed maggots of the drone fly eristalis tenax show potent activity against multidrug-resistant gram-negative bacteria. Microorganisms. 2020;8:626. doi: 10.3390/microorganisms8050626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood MI, Becker KW, Roux CM, Dunman PM, Skaar EP. Genetic determinants of intrinsic colistin tolerance in Acinetobacter baumannii. Infect Immun. 2013;81:542–551. doi: 10.1128/IAI.00704-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Li C, Song J, Velkov T, Wang L, Zhu Y, Li J. Regulating polymyxin resistance in Gram-negative bacteria: roles of two-component systems PhoPQ and PmrAB. Future Microbiol. 2020;15:445–459. doi: 10.2217/fmb-2019-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inchai J, Liwsrisakun C, Theerakittikul T, Chaiwarith R, Khositsakulchai W, Pothirat C. Risk factors of multidrug-resistant, extensively drug-resistant and pandrug-resistant Acinetobacter baumannii ventilator-associated pneumonia in a Medical Intensive Care Unit of University Hospital in Thailand. J Infect Chemother. 2015;21:570–574. doi: 10.1016/j.jiac.2015.04.010. [DOI] [PubMed] [Google Scholar]
- Jahangiri A, Neshani A, Mirhosseini SA, Ghazvini K, Zare H, Sedighian H. Synergistic effect of two antimicrobial peptides, Nisin and P10 with conventional antibiotics against extensively drug-resistant Acinetobacter baumannii and colistin-resistant Pseudomonas aeruginosa isolates. Microb Pathog. 2021;150:104700. doi: 10.1016/j.micpath.2020.104700. [DOI] [PubMed] [Google Scholar]
- Jenssen H, Hamill P, Hancock RE. Peptide antimicrobial agents. Clin Microbiol Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kádár B, Kocsis B, Kristof K, Tóth Á, Szabó D. Effect of antimicrobial peptides on colistin-susceptible and colistin-resistant strains of Klebsiella pneumoniae and Enterobacter asburiae. Acta Microbiol Immunol Hung. 2015;62:501–508. doi: 10.1556/030.62.2015.4.12. [DOI] [PubMed] [Google Scholar]
- Kao C, Lin X, Yi G, Zhang Y, Rowe-Magnus DA, Bush K. Cathelicidin antimicrobial peptides with reduced activation of Toll-like receptor signaling have potent bactericidal activity against colistin-resistant bacteria. Mbio. 2016;7:e01418–e1516. doi: 10.1128/mBio.01418-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye KS, Pogue JM, Tran TB, Nation RL, Li J. Agents of last resort: polymyxin resistance. Infect Dis Clin North Am. 2016;30:391. doi: 10.1016/j.idc.2016.02.005. [DOI] [PubMed] [Google Scholar]
- Kiaei S, Moradi M, Nave HH, Hashemizadeh Z, Taati-Moghadam M, Kalantar-Neyestanaki D. Emergence of co-existence of bla NDM with rmtC and qnrB genes in clinical carbapenem-resistant Klebsiella pneumoniae isolates in burning center from southeast of Iran. Folia Microbiol. 2019;64:55–62. doi: 10.1007/s12223-018-0630-3. [DOI] [PubMed] [Google Scholar]
- Lee DL, Hodges RS. Structure–activity relationships of de novo designed cyclic antimicrobial peptides based on gramicidin S. J Pept Sci. 2003;71:28–48. doi: 10.1002/bip.10374. [DOI] [PubMed] [Google Scholar]
- Lei J, Sun L, Huang S, Zhu C, Li P, He J, Mackey V, Coy DH, He Q. The antimicrobial peptides and their potential clinical applications. Am J Transl Res. 2019;11:3919. [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Nicolas E, Michaut L, Reichhart J-M, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell J. 1996;86:973–983. doi: 10.1016/S0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Lewies A, du Plessis LH, Wentzel JF. Antimicrobial peptides: the Achilles’ heel of antibiotic resistance? Probiotics and Antimicrobial Proteins. 2019;11:370–381. doi: 10.1007/s12602-018-9465-0. [DOI] [PubMed] [Google Scholar]
- Lin M-F, Lin Y-Y, Lan C-Y. Contribution of EmrAB efflux pumps to colistin resistance in Acinetobacter baumannii. J Microbiol. 2017;55:130–136. doi: 10.1007/s12275-017-6408-5. [DOI] [PubMed] [Google Scholar]
- Lin Q, Deslouches B, Montelaro RC, Di YP. Prevention of ESKAPE pathogen biofilm formation by antimicrobial peptides WLBU2 and LL37. Int J Antimicrob Agents. 2018;52:667–672. doi: 10.1016/j.ijantimicag.2018.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Xu C, Fang R, Cao J, Zhang X, Zhao Y, Dong G, Sun Y, Zhou T. Resistance and heteroresistance to colistin in Pseudomonas aeruginosa isolates from Wenzhou, China. Antimicrob Agents Chemother. 2019;63:e00556–e619. doi: 10.1128/AAC.00556-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llobet E, Tomas JM, Bengoechea JA. Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. J Microbiol. 2008;154:3877–3886. doi: 10.1099/mic.0.2008/022301-0. [DOI] [PubMed] [Google Scholar]
- Madec J-Y, Haenni M. Antimicrobial resistance plasmid reservoir in food and food-producing animals. Plasmid. 2018;99:72–81. doi: 10.1016/j.plasmid.2018.09.001. [DOI] [PubMed] [Google Scholar]
- Magana M, Pushpanathan M, Santos AL, Leanse L, Fernandez M, Ioannidis A, Giulianotti MA, Apidianakis Y, Bradfute S, Ferguson AL (2020) The value of antimicrobial peptides in the age of resistance. Lancet Infect Dis [DOI] [PubMed]
- Magnet S, Courvalin P, Lambert T. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother. 2001;45:3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlapuu M, Håkansson J, Ringstad L, Björn C. Antimicrobial peptides: an emerging category of therapeutic agents. Front Cell Infect Microbiol. 2016;6:194. doi: 10.3389/fcimb.2016.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mant CT, Jiang Z, Gera L, Davis T, Nelson KL, Bevers S, Hodges RS. De Novo designed amphipathic α-helical antimicrobial peptides incorporating dab and dap residues on the polar face to treat the gram-negative pathogen, Acinetobacter baumannii. J Med Chem. 2019;62:3354–3366. doi: 10.1021/acs.jmedchem.8b01785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr AK, Gooderham WJ, Hancock RE. Antibacterial peptides for therapeutic use: obstacles and realistic outlook. Curr Opin Pharmacol. 2006;6:468–472. doi: 10.1016/j.coph.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Menekşe Ş, Çağ Y, Işık ME, Şahin S, Hacıseyitoğlu D, Can F, Ergonul O. The effect of colistin resistance and other predictors on fatality among patients with bloodstream infections due to Klebsiella pneumoniae in an OXA-48 dominant region. Int J Infect Dis. 2019;86:208–211. doi: 10.1016/j.ijid.2019.06.008. [DOI] [PubMed] [Google Scholar]
- Mills G, Dumigan A, Kidd T, Hobley L, Bengoechea JA (2017) Identification and characterization of two Klebsiella pneumoniae lpxL lipid A late acyltransferases and their role in virulence. Infect Immun 85 [DOI] [PMC free article] [PubMed]
- Minagawa S, Ogasawara H, Kato A, Yamamoto K, Eguchi Y, Oshima T, Mori H, Ishihama A, Utsumi R. Identification and molecular characterization of the Mg2+ stimulon of Escherichia coli. J Bacteriol. 2003;185:3696–3702. doi: 10.1128/JB.185.13.3696-3702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynarcik P, Kolar M (2019) Molecular mechanisms of polymyxin resistance and detection of mcr genes. Biomed Pap Med 163 [DOI] [PubMed]
- Moffatt JH, Harper M, Adler B, Nation RL, Li J, Boyce JD. Insertion sequence ISAba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob Agents Chemother. 2011;55:3022–3024. doi: 10.1128/AAC.01732-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadam MT, Amirmozafari N, Shariati A, Hallajzadeh M, Mirkalantari S, Khoshbayan A, Jazi FM. How phages overcome the challenges of drug resistant bacteria in clinical infections. Infect Drug Resist. 2020;13:45. doi: 10.2147/IDR.S234353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadam MT, Khoshbayan A, Chegini Z, Farahani I, Shariati A. Bacteriophages, a new therapeutic solution for inhibiting multidrug-resistant bacteria causing wound infection: lesson from animal models and clinical trials. Drug Des Devel Ther. 2020;14:1867. doi: 10.2147/DDDT.S251171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadam MT, Taati B, PaydarArdakani SM, Suzuki K. Ramadan fasting during the COVID-19 pandemic; observance of health, nutrition and exercise criteria for improving the immune system. Front Nutr. 2021;7:349. doi: 10.3389/fnut.2020.570235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadam M, Shariati A, Mirkalantari S, Karmostaji A (2020a) The complex genetic Region Conferring transferable antibiotics resistance in MDR and XDR Klebsiella pneumoniae clinical isolates. New Microbes New Infect, 100693 [DOI] [PMC free article] [PubMed]
- Moghadam MT, Mirzaei M, Moghaddam MFT, BabakhaniI S, Yeganeh O, Asgharzadeh S, Farahani HE, Shahbazi S (2021) The Challenge of global emergence of novel colistin-resistant Escherichia coli ST131. Microb Drug Resist (Larchmont, NY) [DOI] [PubMed]
- Moghadam MT, Chegini Z, Khoshbayan A, Farahani I, Shariati A (2021a) Helicobacter pylori Biofilm and New Strategies to Combat it. Curr Mol Med [DOI] [PubMed]
- Moghasdam MT, Chegini Z, Norouzi A, Dosari AS, Shariati A (2021b) Three-decade failure to the eradication of refractory Helicobacter pylori infection and recent efforts to eradicate the infection. Curr Pharm Biotechnol [DOI] [PubMed]
- Mohebi S, Hossieni Nave H, Norouzi A, KandehkarGharaman M, TaatiMoghadam M. Detection of extended spectrum beta lactamases on class I integron in Escherichia coli isolated from clinical samples. J Maz Univ Med Sci. 2016;26:66–76. [Google Scholar]
- Moradi M, Norouzi A, Taatimoghadam M. Prevalence of bla-CTX-M, bla-SHV, and bla-TEM genes and comparison of antibiotic resistance pattern in extended-spectrum β-lactamase producing and non-producing groups of Klebsiella pneumoniae isolated from clinical samples in Kerman hospitals. J Adv Med Biomed Sci. 2016;6:120–128. [Google Scholar]
- Moravej H, Moravej Z, Yazdanparast M, Heiat M, Mirhosseini A, MoosazadehMoghaddam M, Mirnejad R. Antimicrobial peptides: features, action, and their resistance mechanisms in bacteria. Microb Drug Resist. 2018;24:747–767. doi: 10.1089/mdr.2017.0392. [DOI] [PubMed] [Google Scholar]
- Moubareck CA, Mouftah SF, Pál T, Ghazawi A, Halat DH, Nabi A, AlSharhan MA, AlDeesi ZO, Peters CC, Celiloglu H. Clonal emergence of Klebsiella pneumoniae ST14 co-producing OXA-48-type and NDM carbapenemases with high rate of colistin resistance in Dubai, United Arab Emirates. Int J Antimicrob Agents. 2018;52:90–95. doi: 10.1016/j.ijantimicag.2018.03.003. [DOI] [PubMed] [Google Scholar]
- Mourtada R, Herce HD, Yin DJ, Moroco JA, Wales TE, Engen JR, Walensky LD. Design of stapled antimicrobial peptides that overcome antibiotic resistance and in vivo toxicity. Nat Biotechnol. 2019;37:1186. doi: 10.1038/s41587-019-0222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi SM, Babakhani S, Moradi L, Karami S, Shahbandeh M, Mirshekar M, Mohebi S, Moghadam MT (2021) Bacteriophage as a novel therapeutic weapon for killing colistin-resistant multi-drug-resistant and extensively drug-resistant gram-negative bacteria. Curr Microbiol :1–14 [DOI] [PMC free article] [PubMed]
- Munita J, Arias C (2016) Mechanisms of antibiotic resistance. Microbiol Spectr 4 (2), 10.1128/microbiolspec. VMBF-0016–2015 10.1128/microbiolspec. VMBF-0016–2015 [DOI] [PMC free article] [PubMed]
- Napier BA, Burd EM, Satola SW, Cagle SM, Ray SM, McGann P, Pohl J, Lesho EP, Weiss DS. Clinical use of colistin induces cross-resistance to host antimicrobials in Acinetobacter baumannii. Mbio. 2013;4:e00021–e113. doi: 10.1128/mBio.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nhu NTK, Riordan DW, Nhu TDH, Thanh DP, Thwaites G, Lan NPH, Wren BW, Baker S, Stabler RA. The induction and identification of novel colistin resistance mutations in Acinetobacter baumannii and their implications. Sci Rep. 2016;6:1–8. doi: 10.1038/srep28291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J (2016) Review on antimicrobial resistance: tackling drug-resistant infections globally: final report and recommendations. Review on antimicrobial resistance: tackling drug-resistant infections globally: final report and recommendations
- Olaitan AO, Morand S, Rolain J-M. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panta PR, Kumar S, Stafford CF, Billiot CE, Douglass MV, Herrera CM, Trent MS, Doerrler WT. A DedA family membrane protein is required for Burkholderia thailandensis colistin resistance. Front Microbiol. 2019;10:2532. doi: 10.3389/fmicb.2019.02532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Madec JY, Lupo A, Scink AK, Kieffer N, Nordmann P, Schwarz S (2018) Antimicrobial resistance in Escherichia coli. Antimicrob Resist Bact Livest Compan Anim :289–316 [DOI] [PMC free article] [PubMed]
- Poulikakos P, Tansarli G, Falagas M. Combination antibiotic treatment versus monotherapy for multidrug-resistant, extensively drug-resistant, and pandrug-resistant Acinetobacter infections: a systematic review. Eur J Clin Microbiol Infect Dis. 2014;33:1675–1685. doi: 10.1007/s10096-014-2124-9. [DOI] [PubMed] [Google Scholar]
- Prins JM, Kuijper EJ, Mevissen M, Speelman P, van Deventer S. Release of tumor necrosis factor alpha and interleukin 6 during antibiotic killing of Escherichia coli in whole blood: influence of antibiotic class, antibiotic concentration, and presence of septic serum. Infect Immun. 1995;63:2236–2242. doi: 10.1128/iai.63.6.2236-2242.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raguse TL, Porter EA, Weisblum B, Gellman SH. Structure− activity studies of 14-helical antimicrobial β-peptides: probing the relationship between conformational stability and antimicrobial potency. J Am Chem Soc. 2002;124:12774–12785. doi: 10.1021/ja0270423. [DOI] [PubMed] [Google Scholar]
- Rathinakumar R, Wimley WC. High-throughput discovery of broad-spectrum peptide antibiotics. FASEB J. 2010;24:3232–3238. doi: 10.1096/fj.10-157040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read AF, Woods RJ. Antibiotic resistance management. Evol Med Public Health. 2014;2014:147. doi: 10.1093/emph/eou024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regelmann AG, Lesley JA, Mott C, Stokes L, Waldburger CD. Mutational analysis of the Escherichia coli PhoQ sensor kinase: differences with the Salmonella enterica serovar Typhimurium PhoQ protein and in the mechanism of Mg2+ and Ca2+ sensing. J Bacteriol. 2002;184:5468–5478. doi: 10.1128/JB.184.19.5468-5478.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Rojas A, Makarova O, Rolff J. Antimicrobials, stress and mutagenesis. PLoS Pathog. 2014;10:e1004445. doi: 10.1371/journal.ppat.1004445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi Dosari A, Norouzi A, Taati Moghadam M, Satarzadeh N (2016) Antimicrobial activity of Ephedra pachyclada methanol extract on some enteric gram negative bacteria which causes nosocomial infections by agar dilution method. Zahedan J Res Med Sci :18
- Sato T, Shiraishi T, Hiyama Y, Honda H, Shinagawa M, Usui M, Kuronuma K, Masumuri N, Takahashi S, Tamura Y (2018) Contribution of novel amino acid alterations in PmrA or PmrB to colistin resistance in mcr-negative Escherichia coli clinical isolates, including major multidrug-resistant lineages O25b: H4-ST131-H30Rx and Non-x. Antimicrob Agents Chemother 62 [DOI] [PMC free article] [PubMed]
- Schwarz S, Johnson AP. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother. 2016;71:2066–2070. doi: 10.1093/jac/dkw274. [DOI] [PubMed] [Google Scholar]
- Shahbandeh M, Moghadam MT, Mirnejad R, Mirkalantari S, Mirzaei M. The efficacy of AgNO3 nanoparticles alone and conjugated with imipenem for combating extensively drug-resistant Pseudomonas aeruginosa. Int J Nanomedicine. 2020;15:6905. doi: 10.2147/IJN.S260520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariati A, Asadian E, Fallah F, Azimi T, Hashemi A, Sharahi JY, Moghadam MT. Evaluation of Nano-curcumin effects on expression levels of virulence genes and biofilm production of multidrug-resistant Pseudomonas aeruginosa isolated from burn wound infection in Tehran. Iran Infect Drug Resist. 2019;12:2223. doi: 10.2147/IDR.S213200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariati A, Dadashi M, Moghadam MT, van Belkum A, Yaslianifard S, Darban-Sarokhalil D. Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: a systematic review and meta-analysis. Sci Rep. 2020;10:1–16. doi: 10.1038/s41598-020-69058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry N, Howden B. Emerging Gram negative resistance to last-line antimicrobial agents fosfomycin, colistin and ceftazidime-avibactam–epidemiology, laboratory detection and treatment implications. Expert Rev Anti Infect Ther. 2018;16:289–306. doi: 10.1080/14787210.2018.1453807. [DOI] [PubMed] [Google Scholar]
- Sileshi A, Tenna A, Feyissa M, Shibeshi W. Evaluation of ceftriaxone utilization in medical and emergency wards of Tikur Anbessa specialized hospital: a prospective cross-sectional study. BMC Pharmacol Toxicol. 2016;17:1–10. doi: 10.1186/s40360-016-0057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Som A, Vemparala S, Ivanov I, Tew GN. Synthetic mimics of antimicrobial peptides. Pept Sci. 2008;90:83–93. doi: 10.1002/bip.20970. [DOI] [PubMed] [Google Scholar]
- Soon RL, Nation RL, Cockram S, Moffatt JH, Harper M, Adler B, Boyce JD, Larson I, Li J. Different surface charge of colistin-susceptible and-resistant Acinetobacter baumannii cells measured with zeta potential as a function of growth phase and colistin treatment. J Antimicrob Chemother. 2011;66:126–133. doi: 10.1093/jac/dkq422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zhang H, Liu Y-H, Feng Y. Towards understanding MCR-like colistin resistance. Trends Microbiol. 2018;26:794–808. doi: 10.1016/j.tim.2018.02.006. [DOI] [PubMed] [Google Scholar]
- Sundaramoorthy NS, Suresh P, Ganesan SS, Ganeshprasad A, Nagarajan S. Restoring colistin sensitivity in colistin-resistant E. coli: Combinatorial use of MarR inhibitor with efflux pump inhibitor. Sci Rep. 2019;9:1–13. doi: 10.1038/s41598-019-56325-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TaatiMoghadam M, Hossieni Nave H, Mohebi S, Norouzi A. The evaluation of connection between integrons class I and II and ESBL-producing and Non-ESBL klebsiella pneumoniae isolated from clinical samples, Kerman. Iran J Med Microbiol. 2016;10:1–9. [Google Scholar]
- Tigen ET, Koltka EN, Dogru A, Orhon ZN, Gura M, Vahaboglu H. Impact of the initiation time of colistin treatment for Acinetobacter infections. J Infect Chemother. 2013;19:703–708. doi: 10.1007/s10156-013-0549-1. [DOI] [PubMed] [Google Scholar]
- van der Weide H, Vermeulen-de Jongh DM, van der Meijden A, Boers SA, Kreft D, Ten Kate MT, Falciani C, Pini A, Strandh M, Bakker-Woudenberg IA. Antimicrobial activity of two novel antimicrobial peptides AA139 and SET-M33 against clinically and genotypically diverse Klebsiella pneumoniae isolates with differing antibiotic resistance profiles. Int J Antimicrob Agents. 2019;54:159–166. doi: 10.1016/j.ijantimicag.2019.05.019. [DOI] [PubMed] [Google Scholar]
- Wang C, Feng Y, Liu L, Wei L, Kang M, Zong Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg Microbes & Infect. 2020;9:508–516. doi: 10.1080/22221751.2020.1732231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C, Trent MS. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem. 2014;83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- Wi YM, Choi J-Y, Lee J-Y, Kang C-I, Chung DR, Peck KR, Song J-H, Ko KS. Emergence of colistin resistance in Pseudomonas aeruginosa ST235 clone in South Korea. Int J Antimicrob Agents. 2017;49:767–769. doi: 10.1016/j.ijantimicag.2017.01.023. [DOI] [PubMed] [Google Scholar]
- Willett J (2012) Bacterial two-component systems share a common mechanism to regulate signaling and specificity
- Witherell KS, Price J, Bandaranayake AD, Olson J, Call DR. Circumventing colistin resistance by combining colistin and antimicrobial peptides to kill colistin-resistant and multidrug-resistant Gram-negative bacteria. J Glob Antimicrob Resist. 2020;22:706–712. doi: 10.1016/j.jgar.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wösten MM, Kox LF, Chamnongpol S, Soncini FC, Groisman EA. A signal transduction system that responds to extracellular iron. Cell J. 2000;103:113–125. doi: 10.1016/S0092-8674(00)00092-1. [DOI] [PubMed] [Google Scholar]
- Wright GD. Antibiotic adjuvants: rescuing antibiotics from resistance. Trends Microbiol. 2016;24:862–871. doi: 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]
- Yang Q, Li M, Spiller OB, Andrey DO, Hinchliffe P, Li H, Maclean C, Niumsup P, Powell L, Pritchard M. Balancing mcr-1 expression and bacterial survival is a delicate equilibrium between essential cellular defence mechanisms. Nat Commun. 2017;8:1–12. doi: 10.1038/s41467-017-02149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti M. Cathelicidins, multifunctional peptides of the innate immunity. J Leukoc Biol. 2004;75:39–48. doi: 10.1189/jlb.0403147. [DOI] [PubMed] [Google Scholar]