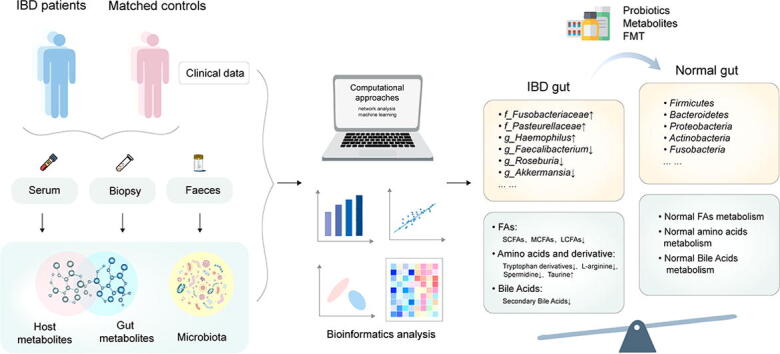
Graphical abstract
Abbreviations: AMPs, Antimicrobial peptides; BAs, Bile acids; BC, Bray Curtis; CD, Crohn’s disease; CDI, Clostridioides difficile infection; DC, Diversion colitis; DCA, Deoxycholic acid; DSS, Dextran sulfate sodium; FAs, Fatty acid; FMT, Fecal microbiota transplantation; FODMAP, Fermentable oligosaccharide, disaccharide, monosaccharide, and polyol; GC–MS, Gas chromatography-mass spectrometry; HDAC, Histone deacetylase; IBD, Inflammatory bowel disease; LCA, Lithocholic acid; LCFAs, Long-chain fatty acids; LC-MS, Liquid chromatography-mass spectrometry; MS, Mass spectrometry; MCFAs, Medium-chain fatty acids; MD, Mediterranean diet; NMR, Nuclear magnetic resonance; PBAs, Primary bile acids; SBAs, Secondary bile acids; SCD, Special carbohydrate diet; SCFAs, Short-chain fatty acids; TNBS, 2,4,6-trinitro-benzene sulfonic acid; UC, Ulcerative colitis; UDCA, Ursodeoxycholic acid; UU, Unweighted UniFrac; UPLC-MS, ultraperformance liquid chromatography coupled to mass spectrometry; WMS, Whole-metagenome shotgun
Keywords: Inflammatory bowel diseases, Metagenomics, Gut microbiota, Metabolomics, Metabolite, Microbial therapeutics
Abstract
Inflammatory bowel disease (IBD), comprising Crohn’s disease (CD) and ulcerative colitis (UC), is a set of clinically chronic, relapsing gastrointestinal inflammatory disease and lacks of an absolute cure. Although the precise etiology is unknown, developments in high-throughput microbial genomic sequencing significantly illuminate the changes in the intestinal microbial structure and functions in patients with IBD. The application of microbial metabolomics suggests that the microbiota can influence IBD pathogenesis by producing metabolites, which are implicated as crucial mediators of host-microbial crosstalk. This review aims to elaborate the current knowledge of perturbations of the microbiome–metabolome interface in IBD with description of altered composition and metabolite profiles of gut microbiota. We emphasized and elaborated recent findings of several potentially protective metabolite classes in IBD, including fatty acids, amino acids and derivatives and bile acids. This article will facilitate a deeper understanding of the new therapeutic approach for IBD by applying metabolome-based adjunctive treatment.
1. Introduction
IBD is known as a chronic nonspecific gastrointestinal inflammatory disease with unknown etiology, including CD and UC, and both of them are marked by unpredictable clinical course with alternating periods of exacerbation and remission[1], [2]. There is no absolute cure for IBD and existing treatments can offer only temporary but not lasting relief to many people[3]. It is well known that the development of IBD is associated with genetic predisposition and environmental factors. In particular, perturbations of gut microbiota are proposed to play important roles in IBD pathogenesis[4], which has been illustrated by germ-free mice[5], [6], [7].
Many studies have reported the imbalance of intestinal microorganisms in IBD. Profiling of intestinal microbial communities by high throughput sequencing showed substantial differences in composition among CD, UC, and non-IBD control subjects. IBD patients shared similar microbial patterns with lower microbial diversity and overgrowth of facultative anaerobes such as Proteobacteria phylum and relative reductions in obligate anaerobes such as Firmicutes phylum, defined as dysbiosis[8], [9].
Base on high-throughput analytical techniques and platforms such as targeted and untargeted fecal metabolomics, differences between the intestinal metabolite fingerprint of patients with active IBD and healthy people are also described in human cohorts[8], [9], [10], [11]. Gut metabolic profiles are made up of those molecules derived from bacterial metabolism of diet or directly from microbiota[12]. Changes in many small molecules like short-chain fatty acids (SCFAs), microbial tryptophan catabolites, and bile acids have been identified in patients with IBD[13], [14], [15]. Those microbial derived compounds act as signaling molecules, mediating the host-microbiota dialogue and regulating immune homeostasis.
From established steroids and anti-inflammation agents to the latest biological drugs, IBD patients are benefiting from a growing number of treatments. However, there is no complete cure, these treatments are sometimes accompanied by side effects. When all medications eventually fail to obtain disease control effectively, patients with IBD even have to undergo surgery, which can cause a series of complications and also destabilizes the microbiome[16], so it is needed to develop novel therapeutic approaches. The intestinal microbiota and its derivative metabolites are being expected to be the future targets for precision therapeutic for their important roles in IBD. In this article, we selectively reviewed the recent applications of metagenomics and metabolomics in IBD studies. Then we summarized the potential protective bacterial species and metabolites within the intestinal microenvironment to IBD. Finally, we reviewed the novel therapeutic approaches to provide the promising strategy to alleviate this inflammatory disease by targeting the gut microbiome–metabolome axis.
2. Gut microbiome in IBD
Next-generation sequencing, including 16S rRNA sequencing and the whole-metagenome shotgun sequencing (WMS), has contributed to a fast development in understanding human gut microbial communities. The 16S rRNA sequencing measurement process includes isolation of total DNA from collected microbiome samples, PCR amplification of selectively 16S gene regions in prokaryotes and high-throughput sequencing of amplicons, followed by gene annotation of sequence data to characterize microbial communities[17]. Species commonly are clustered into the same OTUs (Operational Taxonomic Units) if they have greater than 97% identity in the 16S rRNA gene[18]. The gastrointestinal tract, especially in the distal ileum and colon, is colonized by the largest number of microbes. Over 99% of them are of bacterial origin, and greater than 90% of all the phylotypes are Bacteroidetes and Firmicutes, which dominate the distal gut microbiota[19]. Normally, microbial diversity and abundance of individual’s fecal microbiota vary widely with the change of intestinal niches, while at longer time scales the gut microbiota generally keep quite stable[20], [21]. The homeostatic coexistence of commensal microbial communities can be perturbed in the context of some diseases, including IBD, in which the inflammation occurs in association with increased bacterial exposure and the loss of local tolerance for microbial or dietary antigens[4]. Based on 16S rRNA sequencing or WMS of biological samples, many studies have determined the significant differences of gut microbiota composition between objects with and without IBD. Collectively, these studies found reduced diversity in fecal and mucosal microbes, shown of imbalance between beneficial and aggressive bacteria[22], [23], [24], [25], [26], [27]. Due to limitation of sequencing region and the lack of taxonomic resolution, the taxonomic assignment with 16S rRNA sequencing was only up to the genus level but rarely species level. What’s more, 16S rRNA sequence cannot offer functional information of specific genes to better interpret the host–microbe interactions. As the cost of sequencing falling, WMS sequencing is playing an increasingly important part in characterizing microbial communities. WMS sequencing extends the sequencing information to the entire DNA content present in a microbiome sample and profiles the taxonomic composition and identifies functional potential of microbes by detecting functional genes[28], [29]. Several large human cohort studies have utilized WMS sequencing to characterize the species and strain-level differences in patients with IBD[8], [24], [30], [31]. In these studies, species from the Proteobacteria are generally increased in IBD, including (adherent-invasive) Escherichia coli, Enterobacteriaceae, Klebsiella, and Proteus spp., which have been reported enhancing inflammatory response and the Proteus mirabilis can significantly aggravate the colitis of mice induced by dextran sulfate sodium (DSS) [32], [33], [34], [35], [36]. Also, an enrichment of Fusobacteria was observed in IBD patients. Fusobacterium nucleatum, Fusobacterium Varium and Fusobacterium spp., members of Fusobacteria, have been demonstrated to be positively associated with colitis[37], [38], [39]. In addition, the imbalanced gut microbiota in IBD is almost always accompanied by a sustained decrease in Faecalibacterium and Roseburia genera[31]. Faecalibacterium prausnitzii, the only representative member of genus Faecalibacterium, can inhibit the formation and growth of biofilm of Candida albicans by inducing the antimicrobial peptides (AMPs) production and attenuate DSS-induced colitis[40]. The genus Roseburia, confirmed as negatively associated with the IBD genetic risk score[41]. Roseburia intestinalis has shown to be able to ameliorate DSS- and 2,4,6-trinitro-benzene sulfonic acid (TNBS)-induced colitis by inhibiting the immune response via different mechanisms[42], [43], [44]. Overall, large cohort analyses based on metagenomics data have provided a detailed characterization and substantial changes in intestinal microbiota structure in IBD, which emphasized the potency to dig the intestinal microbiome data as a modality to classify IBD patients. At the same time, these findings enable us to gain a more profound comprehension of the role of microorganisms in the pathogenesis of IBD as well. However, one of the most outstanding problems still existing in both methods is the lack of accurate resolution of strain-level variation. To better characterize microbial communities in more accurate resolution, the metagenome sequencing tools are continually improving. Recently, Lars Snipen et al. revealed the Reduced Metagenome Sequencing (RMS) is a good choice to full WMS sequencing when analyzing microbial community at strain level[45]. In addition, to understand the mechanism of species interaction, it is very important to image the microbiota in situ and map its spatial distribution. Ravi U. Sheth et al. used the method of metagenomic plot sampling by sequencing (MaPS-seq) and revealed the heterogeneous microbial distributions in the mouse intestine and showed the close associations between Bacteroidales taxa in intestinal compartments[46]. Hao Shi et al. combined fluorescence in situ hybridization (HiPR-FISH) with super-resolution imaging and distinguished hundreds of bacteria and obtained the spatial distribution map of microbiota at single-cell resolution[47]. In conclusion, understanding the functions and the relationship between specific strains with the host can offer biotechnological promise in microbial therapeutic discovery in the future. In order to gain a mechanistic insight into how gut microbiota influence host health, we need to focus our attention on microbial metabolome as well.
3. Gut metabolome in IBD
Although most of dietary intake is digested and absorbed in the small intestine, some dietary components not digested by the small intestine enter the colon and are converted into multifarious free metabolites including gases and toxic molecules by different microbial species known as fermentation[48]. Similar to the field of metagenome, technologies for detecting small molecular compounds greatly increase our knowledge of the metabolites in human intestinal tract. As an emerging member of ‘-omics’ technologies following genomics, transcriptomics, and proteomics, metabolomics allows us to characterize and quantify small-molecular compounds (≤1500 Da) in biological samples using technologies like nuclear magnetic resonance (NMR) spectroscopy or mass spectrometry (MS)[49], [50]. NMR is usually used to detect hydrogen-containing molecules in samples and gives a 1H NMR spectrum. An advantage of it is that metabolites will not be damaged before detection. Therefore, NMR is particularly useful for detecting metabolite levels in biopsy samples[49]. MS is generally coupled with liquid or gas chromatographic separation techniques, known as liquid chromatography-mass spectrometry (LC-MS), ultraperformance liquid chromatography coupled to mass spectrometry (UPLC-MS) and gas chromatography-mass spectrometry (GC–MS). Each of these methods has its own strengths and limitations and can complement each other[51], [52]. With the advances of metabolic profiling platforms and mathematical integration approaches, metabolomics has been applied extensively to understand the host-microbe interactions through analyzing intestinal microbiota metabolism and host-microbiota co-metabolisms[53]. Many studies have identified altered metabolite levels in stool, gut mucosa or serum of patients with IBD relative to controls by performing targeted or untargeted metabolomics (Table 1). Furthermore, owing to the relationships between several metabolite classes and intestinal inflammation, they have been the focus of intense researches in vitro or vivo, which potentially aid in the containment or in modulating the severity of IBD through various mechanisms.
Table 1.
Alteration of gut microbial metabolites in inflammatory bowel disease patients.
| Publication | Samples & Subjects | Methods | Major altered metabolites in IBD |
|---|---|---|---|
| Yang Z et al.2021[120] | Fecal sample from 32 UC, 23 controls | UPLC-MS/MS | • SBAs(LCA, DCA, glycol-deoxycholic acid, glycol-lithocholic acid, tauro-lithocholic acid↓ •PBAs(taurocholic acid, Cholic acid, taurochenodeoxycholic acid glycolchenodeoxycholic acid↑ |
| Wang Y et al.2021[129] | Fecal sample from 29 CD,20 controls | UPLC-MS/MS | • L-leucine, L-norleucine, methylmalonic acid, succinic acid↑; SCFAs (acetic acid, butyric acid, and propanol acid), BAs (deoxycholic acid, hyodeoxycholic acid, lithocholic acid) ↓ |
| Krzystek et al. 2020[95] | Large bowel tissues from 52 CD, 48 UC, 40 controls | LC-MS | •Arginine, Dimethylarginine ↓ •Citrulline, dimethylamine↑ |
| Bushman F et al. 2020[110] | Fecal samples from 28 IBD cases, 37 controls | UPLC-LC/MS | •Secondary bile acids(Deoxycholate, Lithocholate) ↓•Taurine, Primary bile acids (Chenodeoxycholate, Cholate) , Cadaverine, Kynurenine↑, Ceramide↑ |
| Diederen K et al. 2020[112] | Fecal samples from 43 CD,15 controls | 1HNMR, HPLC | •Arginine, Taurine, Glutamic acid ↓ •Primary bile acids, Trimethylamine, Cadaverine ↑ |
| Franzosa E et al. 2019[10] | Fecal samples from 68 CD,53 UC, 34 controls | Untargeted LC–MS | •SCFAs (Butyrate, Propionate), LCFA(2-hydroxymyristic acid), MCFA(Aprylic acid) SBAs (Lithocholate, Deoxycholate), Cholesterols, Phenylbenzodioxanes ↓ •PBAs(Cholate, Chenodeoxycholate), Sphingolipids, Cholesterylesters, Phosphatidylcholines, LCFAs (Arachidonic acid, Adrenic acid, Docosapentaenoic acid and Eicosatrienoic acid) ↑ |
| Scoville E et al. 2019[94] | Serum from 20 UC, 20 CD, and 20 non-IBD | HILIC/UPLC-MS/MS | •Arginine, LCFAs, MCFAs, Glutamine, Leucine, Lysine, Valine, Citrate, Conitate, α-ketoglutarate, Succinate, Fumarate, Malate ↓ |
| Lloyd et al. 2019[8] | Fecal samples | LS-MS/MS | •Butyrate, Propionate, Valerate/isovalerate, Indole-3-propionat, Secondary bile acids (Lithocholate, Deoxycholate), Arachidonoyl carnitine ↓ •Taurine, Free arachidonate, Uridine, Nicotinuric acid, Glycoche-nodeoxycholate ↑ |
| Weng Y et al.2019[220] | Fecal sample from 107 UC,173 CD,42 controls | GC/MS, LC/MS | • LCFAs(Arachidic acid, Oleic acid, Tridecanoic acid), MCFAs(Sebacic acid, Isocaproic acid), Bile acids (Lithocholic acid, Chenodeoxycholate, Taurolithocholic acid)↓ |
| Das P et al.2019[128] | Fecal sample from 25 IBD,14 controls | LC-MS | • Deoxycholate, Lithocholate ↓ • Cholate, Glycocholate, Taurocholate, Taurochenodeoxycholate↑ |
| Alghamdi A et al. 2018[110] | Fecal samples from 7 new-onset CD cases,11 controls | LC-MS | •Tyrosine, Ornithine isomer ↓• Taurine, Arachidonic acid, Eicosatrienoic acid, Docosatetraenoic acid, Kynurenine(Kyn) , Aspartate, Glycine, Tryptophan, Carnosine, Allantoin, Citrulline, Serine, Threonine, Ornithine, Creatine, Asparagine, Choline, Histidine, Phenylalanine, Alanine, Metanephrine ↑ |
| Nikolaus S et al. 2017[87] | Serum samples from 291 IBD cases,291 controls; | HPLC | •Tryptophan(Trp), TDO2, Picolinic acid↓ •Kyn/Trp ratio, IDO1, Anthranilic acid, Quinolinic acid ↑ |
| Santoru M et al. 2017[9] | Fecal samples from 82 UC, 50 CD, and 51 controls | 1HNMR, GC–MS, LC-QTOF-MS | •Putrescine, Cadaverine, Alanine, Beta-alanine, Phenylacetic acid, 4-hydroxyphenylacetic acid, Glyceric acid, Phenylethylamine ↑ •Nicotinic acid, Pantothenic acid, 3-methyladipic acid, 5β-coprostanol, 3-hydroxybutyric acid, Hydrocinnamic acid ↓ |
| Kolho K et al. 2017[109] | Fecal and serum samples from 69 IBD cases, 29 controls | UPLC-MS/MS | •L-Tryptophan, Kynurenic acid, Trimethylamine-N-oxide ↓ •Taurine, Kynurenine, Glycocholic acid, L-isoleucine, Symmetric dimethylarginine, Serine, Phosphoethanolamine, Proline, Hexanoylcarnitine↑ |
| Lamas B et al. 2016[88] | Fecal samples from 102 IBD cases,37 controls | HPLC, LC-MS | •Tryptophan, IAA, IAA/Trp ↓ •Kyn, Kyn/trp↑ |
| Coburn L et al. 2016[93] | Colonic tissues and Serum from 38 controls and 137 UC patients. | HPLC | •Tissue L- Arginine ↓ •Tissue L-citrulline ↑ •Serum L- Arginine:N |
| Bjerrum J et al. 2015[11] | Fecal samples from 48 UC,44 CD, 21 controls | 1HNMR | •Butyrate, Propionate ↓ •Taurine, Isoleucine, Leucine, Lysine, Phenylalanine, Valine ↑ |
| Lee T et al. 2016[221] | Fecal samples from 31 CD, 22 UC, 19 controls | High-resolution MS | •Pentadecanoic acid, Stearic acid, Hexadecadienoic acid↑ |
| De Preter V et al. 2015[73] | Fecal samples from 83 CD,68 UC, 16 controls | GC–MS | •MCFAs (Pentanoate, Hexanoate, Heptanoate, Octanoate, Nonanoate) ↓ |
| Jacobs J et al.2016[222] | Fecal sample from 26 CD,10 UC,54 controls | UPLC-MS | • Stercobilin, Acetyl-glutamic acid, Boldione↓ • Taurine, Tryptophan, Serinyl tryptophan, Omega-6 fatty acid (Adrenic acid), Bile acids (Cholic acid, 7-ketodeoxycholic acid, Chenodeoxycholic acid sulfate, 3-sulfodeoxycholic acid), Amino acid derivatives(Phenylethylamine, N-acetylcadaverine), ↑ |
Abbreviations: CD, Crohn’s disease; DCA, Deoxycholic acid; GC–MS, Chromatography-mass spectrometry; HILIC, Hydrophilic interaction liquid chromatography; HPLC, High-performance liquid chromatography; IAA, Indole-3-acetic acid; IBD, Inflammatory bowel disease; IDO1, Indoleamine-2,3-dioxygenase 1; Kyn, Kynurenine; LCA, Lithocholic acid; LCFAs, Long-chain fatty acids; LS-MS/MS, Liquid chromatography-triple quadrupole mass spectrometry; LC-MS, Liquid chromatography-mass spectrometry; LC-QTOF-MS, Liquid chromatography in combination with quadrupole time-of-flight mass spectrometry; MCFAs, medium-chain fatty acids; NMR, Nuclear magnetic resonance spectroscopy; PBAs, Primary bile acids; SBAs, Secondary bile acids; TDO2, Tryptophan-2,3-dioxygenase 2; Trp, Tryptophan; UC, Ulcerative colitis; UPLC-MS/MS, Ultra pressure liquid chromatography tandem mass spectrometry; ↑indicates increase; ↓indicates decrease.
3.1. Fatty acids
Triglycerides, commonly known as fats, are the second major source of dietary energy in human. Fatty acids are the important components of Triglycerides[54]. Both free and bound fatty acids play key roles in cellular functions and metabolism[55]. According to the carbon chain length, fatty acids can be divided into short-chain (≤6 carbons, SCFAs), medium-chain (7 ∼ 12 carbons, MCFAs) and long-chain fatty acids (more than 12 carbons, LCFAs)[56]. Among them, SCFAs are the major products, mainly derived from bacterial fermentative reactions of indigestible dietary fibers in intestines, including acetate, butyrate and propionate[57]. MCFAs and LCFAs derive mainly from dietary triglycerides in some animal fats and vegetable oils. Accumulating evidence suggest that the level of SCFAs in IBD patient's feces is decreased in varying degrees. For example, Julian R. Marches et al. found that acetic acid and butyric acid in feces of CD and UC patients were lower than those in the healthy group by performing 1H NMR spectroscopy[58]. Another cohort study revealed reduced level of propionic acid and acetic acid by carrying out GC–MS and low abundance of butyrate-producing bacteria in UC patient’s feces [59]. Nevertheless, SCFAs have shown to play significant roles in the immune response through host receptor signals. Recent study reported both microbiota-derived and administration of butyrate could activate G-protein receptor 41 and inhibit histone deacetylase (HDAC) to promote the production of IL-22 in human and mice ILCs and CD4+ T cells, and then suppressed intestinal inflammation[60]. Recently, another study by Li et al. revealed a new possible protective mechanism of SCFAs in TNFα-induced endothelial cells activation. This study showed that butyrate and propionate could inhibit IL-8 secretion and increase IL-33 secretion, and then halt the inflammatory responses in lesion sites[61]. Acetate, another SCFAs, can induce the production of T-cell-dependent IgA and further regulate the localization of commensal bacteria to maintain mucosal homeostasis[62]. Moreover, SCFAs can regulate the expression of epithelial genes involved in energy metabolism, support epithelial cell proliferation and enhance colonic epithelial barrier[63] and impact intestinal bacterial communication[64]. In addition to IBD, the possibility of SCFAs therapy for diversion colitis (DC) is being explored[65]. DC was first reported in 1974 as a nonspecific inflammation of diverted colon with fragile mucosa, aphthous ulcer, edema, erythema, etc.[66]. More and more studies have found that diversion colitis is associated with intestinal microbial dysbiosis such as decreased Bifidobacterium species and increased Proteobacteria[67], [68]. The decrease of SCFAs and immune dysfunction such as increased IgA have also been confirmed to be closely associated with the pathogenesis of DC[69], [70]. Luceri et al. found that the endoscopic score of DC patients treated with butyrate was significantly lower than that of DC patients treated with saline as placebo[71]. Another study conducted SCFAs treatment on 15 children with diversion proctocolitis and found that SCFAs could improve the disease symptoms, endoscopic and histopathological manifestations of children[72]. These provide evidence and explore possibilities for SCFAs to treat intestinal inflammatory diseases, including IBD.
At the same time, metabolomic profiling of cross-sectional stool samples from patients with CD and UC respectively showed that LCFAs were significantly depleted relative to controls[10], [73]. In another study, plasma lipid and metabolic profiles were quantified by UPLC-MS, the results revealed a decreased level of tetracosanoic acid, which belongs to LCFAs[74]. Clinical studies have shown that higher n-3 / n-6 PUFA intake was negatively correlated with IBD and LCFAs played a dual role in intestinal inflammation of IBD[75]. The n-6 unsaturated fatty acids are considered to be pro-inflammatory compounds, while n-3 unsaturated fatty acids have anti-inflammatory properties[76]. Jiwei Wang et al. proved that n-3PUFAs could improve the function of Paneth cells by activating IL-22 / Stat3 pathway and protect the intestinal barrier[77]. Recently, ulien Pujo et al. found certain bacteria such as Holdemanella biformis can produce high concentration of LCFA-3OH, which alleviated DSS-induced colitis in mice[78]. A recent randomized controlled trial has proved in mice and humans that monounsaturated LCFAs can improve endothelial cell function and reduce inflammatory factors such as TNF-α and IL-6 in plasma. Meanwhile, the intestinal microbial environment has changed. The authors found a decreased Firmicutes and/ or Bacteroidetes ratio in the monounsaturated LCFAs group, along with an increased abundance of Akkermansia, which changed the microbiota environment of SCFAs production, leading to the induction of glucagon-like peptide-1 secretion[79].
Besides, some case–control studies used GC–MS revealed the reduced level of MCFAs (pentanoate, hexanoate, heptanoate and octanoate etc.) in IBD patients[73], [80]. MCFAs have previously been shown to suppress inflammation and ameliorate experimental colitis in mice by activating the peroxisome proliferator activated receptor (PPAR)-γ[81]. Both capric acid and lauric acid have shown the anti-inflammatory and anti-bacterial properties[82]. Lauric acid, a compound containing 12 carbon atoms and mainly derived from coconut oil, recently has proved attenuating liver inflammation induced by lipopolysaccharide through inhibiting TLR4/MyD88 pathway[83]. These findings above indicate that lauric acid may have a protective role against IBD.
Of note, even if people begin to stress the role of FAs in the inflammatory regulation of intestinal diseases, clinical studies on them pay more attention to the relationship between their intake and inflammation, the specific pathway mechanism is still not fully clear. Metabolomics may provide more relevant factors for this and help develop the possibility of FAs in clinical treatment of IBD and other intestinal diseases.
3.2. Amino acids and derivatives
3.2.1. Tryptophan
As one of the nine essential dietary amino acids, tryptophan is the most complex aromatic amino acid. The absorption of dietary tryptophan within mammalian hosts mainly has the following four pathways: (a) the protein synthesis pathway; (b) the serotonin pathway; (c) the kynurenine pathway and (d) the microbial metabolic pathway[84], [85]. Among them, accumulating evidence implicates microbiota-derived tryptophan metabolites as crucial mediators of host-microbial cross-talk through serving as ligands of aryl hydrocarbon receptor or pregnane X receptor, two ligand-dependent transcription factors residing in the cytoplasm[86]. In many studies, targeted or untargeted metabolomics have been performed to reveal that tryptophan metabolism was disrupted and indoles and derivatives were significantly depleted in IBD patients [10], [87], [88]. In turn, tryptophan and its metabolites administration such as indole, indole‐3‐aldehyde, Indole‐3‐propionic acid and indole-3-acetic acid can reverse colitis-associated microbial dysbiosis, reduce colonic inflammation and protect the integrity of intestinal epithelium[13], [89], [90], [91]. Furthermore, dietary tryptophan deficiency was correlated with the exacerbation of colitis, and indole-3-carbinol treatment effectively reversed the alterations in microbial composition induced by TNBS and selectively increased the abundance of Roseburia, which is known as excellent butyrate producer as described above[90]. These findings suggest that microbiota-derived tryptophan metabolites hold promise in modulating the homeostasis of host immune response.
3.2.2. Arginine
Arginine is classified as conditionally essential amino acid and needs dietary requirement under several situations including early development, infection, inflammation and metabolic dysfunctions in kidney or intestine[92]. With no apparent difference in food intake of the L-arginine, decreased L-arginine levels in colonic tissue were observed in active UC and CD patients versus control subjects. Moreover, tissue L-arginine was negatively correlated with the disease activity index[93], [94], [95]. Arginase 1, one of arginase isoenzymes utilizing arginine as substrate, recently has been confirmed higher expression levels in intestinal tissues from IBD patients as well as DSS-induced colitis mouse model. The absence of arginase 1 and nutritional L-arginine exacerbated the severity colitis[96]. Besides, L-arginine supplementation significantly elevated the abundance of anti-inflammatory intestinal microbiota and reduced pro-inflammatory factor expression, which is partly attributed to the accumulation of polyamines in fecal[97], [98].
3.2.3. Polyamines
Polyamines are a class of bioactive chemicals derived from L-arginine and other polyamine precursors. The main source of fecal polyamines in colonic lumen is likely derived from intestinal microorganisms which metabolize the polyamine precursors to ornithine and feeds polyamine synthesis[99], [100], [101]. Polyamines mainly include putrescine, spermine, spermidine and cadaverine and the functions of them differ from each other. Among them, spermidine exhibits critical functions in maintaining cellular homeostasis. Dietary supplementation with spermidine ameliorated DSS and TNBS-induced colitis and improved gut barrier integrity[102], [103], as previously reported[104], [105], [106]. A recent study also demonstrated that putrescine derived from bacterium could increase the number of anti-inflammatory macrophages in the colon. They found colonization of germ-free mice with the wild-type, but not polyamine biosynthesis-deficient, E. coli accelerated epithelial renewal and microbial polyamines can affect the M1/M2 macrophage balance and play anti-inflammatory roles in a DSS-induced colitis model[107]. While in a clinical study, metabolomics and metagenomics approach have been used to analyze fecal samples from IBD patients and healthy people. The GC–MS analysis showed that the level of putrescine and cadaverine were significantly higher in CD and UC patients[9], which indicated putrescine and cadaverine may have adverse effects on IBD. In line with this, an imaging-based, quantitative, high-throughput screen was performed to identify the molecules which can disrupt intestinal barrier through CaCo-2 and T84 cells culture. They observed putrescine disrupted the epithelial tight junction in ex vivo and in vivo and putrescine administration exacerbated colon inflammation of mice. However, the addition of taurine blocked this effect and alleviated intestinal inflammation[108].
3.2.4. Taurine
Taurine is a sulfur-containing amino acid produced by endogenous oxidative cysteine metabolism. Besides, it can also be obtained from dietary sources. In different cross-sectional or longitudinal cohort studies about IBD, untargeted or targeted metabolomic based on GC/LC-MS were used to analyze fecal metabolome. Unlike several metabolites described above, serum or fecal taurine levels were significantly increased in IBD patients[8], [11], [109], [110], [111], [112], [113]. Even so, multiple studies have shown the protective effect of taurine on colitis[114], [115], [116], it can reduce colitis severity and inverse the dysbiotic microbiota by activating NLRP6-IL-18-AMPs pathway[117]. Another study shown elevated taurine in gut was metabolized by microbiota into sulfide and inhibited the pathogen respiration[118]. Besides, high level of taurine in the gut also enhanced tight junctions to increased intestinal epithelial integrity and reduced leaky gut and inflammation[119].
Herein, we reviewed the recent knowledge of the role potentially beneficial amino acids and derivatives, focusing on the interplay between bioactive compounds and host intestinal inflammation. Despite the role of some amino acids in IBD patients have been explored a lot, their functions may be controversial and need further research.
3.3. Secondary bile acids
Bile acids (BAs) are converted from cholesterol in the liver cells. When reaching the colon, primary bile acids (PBAs) are transformed into secondary bile acids (SBAs) by specific bacteria species. Previous works using MS-based metabolomics suggested that bile acid metabolism is disordered in IBD, shown of elevated PBAs and reduced SBAs. Recently, a case control study showed that the concentrations of fecal SBAs, including deoxycholic acid (DCA), lithocholic acid (LCA), were significantly reduced in UC patients, which were shown to be positively correlated with Roseburia, Clostridium IV, Butyricicoccus, and Faecalibacterium[120]. Henri Duboc et al. found that SBAs in serum and feces of patients with IBD were reduced, and sulfated LCA lost its anti-inflammatory effect[121]. Compared to familial adenomatous polyposis pouches, both LCA and DCA were strikingly reduced in UC group, and DCA and LCA supplementation mitigated inflammation in DSS-induced colitis [122]. In addition, a hydrolyzed protein diet increased secondary bile acids, reduced disease scores, regulated microbial dysbiosis, and relieved chronic enteropathy[123]. However, a study published in 2020 showed that exogenous supplementation of ursodiol, a commercial formulation of ursodeoxycholic acid (UDCA) approved by FDA, can alter the fecal bile acid profile, but has little effect on the microbiota[124]. Those results were consistent with previous studies[122], [125], [126], [127], [128], [129].
As one of the receptors of SBAs, the activation of FXR in small intestine has been proved to inhibit bacterial overgrowth and translocation[122], and then ameliorate intestinal inflammation[130]. Many studies have suggested that SBAs relieved inflammation and promoted colonic epithelial remission by downregulating the secretion of pro-inflammatory factors and inhibiting intestinal epithelial apoptosis[131], [132]. Intriguingly, S. Mroz et al. found that DCA inhibited colonic epithelial restitution in vivo[133]. As reported by Lotta K Stenman et al., high-fat diet significantly elevated fecal bile acid concentration and the decreased ratio of UDCA to DCA promoted the intestinal permeability of mice [134]. In addition, compared with the control group, DCA-treated mice had more inflammatory infiltrates of neutrophils in the lamina propria and higher intestinal inflammation score[135]. So far, studies have found that DCA has a dual function, there is still controversy about the role of DCA in IBD. Although there have been many studies on secondary bile acids, its role in IBD is still controversial, and further researches are needed.
4. Computational approaches
MS-based metabolomics has good sensitivity and ability to detect and quantify a large number of molecules produced by human microbiota, which greatly help us to understand the microbial community function in disease causality, yet the annotation of the signals identified in the data remains challenging[136]. Besides, the chemical-microorganism relationships within intestinal microenvironment are unclear, which is important for developing targeted therapies. To address this puzzle, James T. Morton et al. recommended to use mmvec (microbe-metabolite vectors) neural network to learn the interactions between microbes and metabolites. They not only confirmed the link between R. hominis and multiple carnitines in samples of IBD patients, but found high correlation of Klebsiella spp. with IBD status and several bile acids[137]. In addition, a recent meta-analysis based on statistical and machine learning found 97 metabolites could be robustly well-predicted by analyzing human intestinal microbiome composition with data processed from 1733 fecal samples from 10 independent studies[12]. Meanwhile, Shuo Han and co-workers have constructed an integrated mass-spectrometry pipeline centered on gut microbiome to provide the extended biochemical profiles of individual strains[138].
Meanwhile, despite the advent of novel technologies including high-throughput sequencing and targeted or untargeted metabolomics enable us to better understand the composition and functions of intestinal microbes, especially in IBD, they generate tremendous complex datasets of ever-increasing size. Therefore, choosing the appropriate computational approaches to analyze the datasets and determine the association of metabolites and microorganisms with disease is becoming a key discipline of this era, and a range of novel standardized bioinformatics pipelines are being developed and improved for raw data processing in microbiome research[139], [140], [141], [142]. In microbiome or metabolites sample clustering, machine learning methods are used more and more frequently compared with model-based methods. Among them, the weighted UniFrac distance, the unweighted UniFrac(UU) distance, the Bray Curtis(BC) dissimilarity metric, and the Aitchison distance are commonly used to characterize the distance metric of different samples and the incorporation of the UU metric and BC metric may perform more well[143]. Meanwhile, various data visualization tools are being developed for multi-omics data analysis, thus, users do not need to grasp technical expertise, they only need to upload raw data onto informatic software to get visualization results[144]. In addition, many data computational approaches are also being developed and used to interrogate multi-omics data, which greatly increased our further insights into IBD pathogenesis. In recent applications, two large studies integrated longitudinal multi-omics data to profile the temporal variations of gut microbial abundance or metabolite concentration in healthy controls and IBD patients between consecutive time points[8], [145]. Although the ‘omics’ technologies for integrating multi-omics datasets from IBD patients or mice models are still at an early stage, developments in systems biology and machine learning can drive this progress and then facilitate the translation of basic research towards clinical application[12]. Thus, identified microbial species and their secreted molecules could serve as potential biomarkers and diagnostic tools to help rapidly discriminate patients with IBD from individuals without IBD, and subsequently result in individualized assessments and guide precision medicine. Overall, the development of such tools applied to mine the links of gut microbiome-metabolites will help modulate metabolites to alleviate IBD by microbiome-based interventions and can also inform personalized medicine.
5. Therapeutics targeting metabolites for IBD
Due to the lack of mechanistic insights into IBD and the limit of existing treatment schemes, a challenge to the development of novel biotherapeutics based on microbiome and metabolite molecules is presented. The reduced diversity of the gut microbiota and the changes of microbial metabolism in IBD patients compared to healthy populations reveal the close relationships between microbiota and pathogenesis of IBD. While the properties of different members of microbiota in host intestine remain to be defined, it is possible that restoring the microbiota to its original state may be necessary to repair physiological functionality to our gut, which requires the administration of depleted taxa in combination with diminished metabolites. The proposed microbial therapies at present for IBD are described briefly below.
5.1. Antibiotics
Antibiotics are one of the traditional drugs for the treatment of IBD, which can control infection, reduce inflammation and alleviate the disease process by reducing the microbial concentration in the intestinal lumen and changing the composition of gut microbiota[146], [147]. Different antibiotics have different effects due to their characteristics. For example, metronidazole and ciprofloxacin have the best effect on the treatment of CD complications such as perianal fistula[148]. When IBD patients cannot use immunosuppressants immediately due to surgery or severe infection, the application of antibiotics is more important[149]. A meta-analysis which includes 15 randomized controlled trials showed that the general clinical remission rates were higher in CD patients using antibiotics than those in controls. However, the clinical remission rate of patients only treated with ciprofloxacin was not significantly different from that of the control group[150]. Another study randomly added tobramycin to 84 patients with acute recurrence of UC, and found that the clinical remission rate of patients using tobramycin was significantly higher than that of patients who didn’t used tobramycin[151]. At the same time, a large number of clinical data show that the side effects of antibiotics in the treatment of long-term recurrent IBD are obvious, such as patient intolerance, drug resistance, gastrointestinal dysfunction, peripheral neuropathy, tenosynovitis, etc.[148], [152].
With the development of omics technology, people have further insights into gut microbiota and its metabolites. More and more studies have shown that the effect of antibiotics on gut microbiota and its metabolites is not absolutely beneficial, and this complex effect may be closely related to the clinical outcome of IBD[153]. Nevertheless, another study on the effect of perinatal exposure to antibiotics on intestinal flora of premature infants showed that the application of β-lactam antibiotics could reduce the number of Firmicutes and Actinomycetes in the intestinal tract of newborns, along with the increased Proteobacteria. At the same time, SCFAs decreased in the intestinal tract detected by Gas-Chromatography Flame ionization / MS detection[154]. Current studies have shown that probiotics and Fecal microbiota transplantation (FMT) can restore the effects of antibiotics on intestinal microbiota and their metabolites, such as the recovery of SCFAs levels[155], [156].
At present, the effect of antibiotics combined with other emerging therapies is also gradually confirmed. A new study reviewed 28 studies in the database that treated IBD with FMT, 5 of which used antibiotics in advance. The results showed that pooled response rate of antibiotic pretreated group was significantly higher than that of untreated group, and antibiotic pretreatment improved the disease remission rate as well[157]. In general, the therapeutic effects and side effects of antibiotics on IBD and the effects of antibiotics on intestinal flora and its metabolites are also complex, which needs more clinical studies to explore it.
5.2. Dietary therapeutics
Given the diversity of molecules encoded within or produced by the microbiome, the interest in mining bioactive compounds and extracting drugs from microbiota is growing. The associations of the microbial metabolites with intestinal inflammation and IBD enable several metabolite classes to be the focus of microbial therapy research. As described above, SCFAs are the main players in the interplay between diet, microbiota, and health. Acetate, propionate, and butyrate have been proved to increase the intestinal barrier integrity and downregulate of inflammatory mediators, mainly by the HDAC inhibition mechanism, and are therefore of the potential to become therapeutic drugs for IBD[158]. However, although oral administration of SCFAs is able to get higher concentrations into the intestine, it is pharmacologically challenging. In a related study, treatment of primary monolayer intestinal epithelial cells of UC patients with sodium butyrate did not protect inflammation-induced barrier dysfunction[159], which revealed that local induction or supplementation with bioactive metabolites in therapy of IBD exists limitations to some extent. Besides, the optimal dose and indication are unknown. In the face of this dilemma, food may be helpful to solve this challenge. Intake of high-fiber diet can increase the level of SCFAs and improve intestinal dysbiosis and then increased quality of life in patient with ulcerative colitis[160]. Moreover, oral polyphenol ameliorated the colonic inflammation and enhanced colon barrier integrity by modulating intestinal microbial composition and increasing butyrate production[161], [162]. A recent study found novel diets containing increased levels of tryptophan, pectin and resistant starch helped improve active ulcerative colitis and achieved higher clinical remission and mucosal healing compared with single donor fecal transplantation[163]. Taken together, studies on dietary therapeutics targeting microbe-derived products are still in the early stages, and different patients may have different responses to the same nutrient, even so, it may be one of the most fruitful ways to develop an effective IBD therapeutic.
5.3. Probiotics
The recognition of the circulatory causal relationship between biological disorders and diseases urges people to look for comprehensive treatments that can deal with both host processes and microbiota. Probiotics intervention is one of the treatment tools[164]. Administration of probiotic cocktail can improve microbiota dysbiosis and relieve the intestinal inflammation both in in human and animal models[165]. Mechanistically, probiotic enhanced the activity of a microbial enzyme in feces, resulting in increased release of taurine[119]. Alternatively, the commensal bacteria with beneficial metabolites production in healthy people intestine are the promising candidates for next-generation probiotics, such as F. prausnitzii, Akkermansia muciniphila and Bacteroides fragilis[166]. F. prausnitzii has been proved to efficiently improve intestinal inflammation in animal models[167], [168], and its anti-inflammatory effects in intestinal epithelial cells were in part mediated by producing butyrate or anti-inflammatory protein[168], [169]. A. muciniphila is also a common resident of the human intestine, a clinical study with administration of A. muciniphila demonstrated its protective role against metabolic syndrome, showing the feasibility to administer A.muciniphila to humans[170]. Many recent studies also revealed A. muciniphila protected against animal models colitis[171], [172], one of the possible mechanisms is to increase the production of SCFAs[173]. However, the effect of A. muciniphila on IBD is still controversial and needs to be further confirmed in more human clinical trials[174]. In addition, gene recombination technologies are used to design the engineered probiotic bacteria such as E. coli Nissle 1917 (ECN), which can produce antimicrobial peptides and biological chemicals to modulate microenvironments of colon and further treat colitis[175], [176], [177]. Rationally designed engineered probiotics contribute to improving chronic immune-mediated colitis by acting as carriers of effector molecules[178]. In the future, probiotics targeting changes in specific microbial metabolites associated with a particular IBD phenotype may open the door to more personalized and personalized treatments.
5.4. Prebiotics/Synbiotics
Prebiotics, the energy and material basis of microecology, was defined as “a substrate that is selectively utilized by host microorganisms conferring a health benefit” by a panel of experts[179]. The prebiotics have been proved to be effective for IBD[180]. Specifically, studies showed that inulin, germinated barley foodstuff, and oat bran can enhance production of beneficial metabolites in IBD patients[181], [182], [183]. Among them, inulin can alleviate IBD by amending gut microbiota function, regulating gut microbiota, and increasing fecal SCFAs[184], [185]. Pectin relieved IBD through its side chain. It can inhibit pro-inflammatory cytokines and immunoglobulin production, and preserve gut flora diversity and then promote therapeutic effect of FMT[186], [187], [188]. Moreover, psyllium could modulate intestinal permeability and colitis severity to ameliorate colitis[189].
Likewise, some prebiotic foods also have potential therapeutic effect to IBD. For example, vaccinium macrocarpon can significantly increase the α-diversity of fecal microbial structure and beneficial bacteria abundance and then suppressed colonic inflammation in DSS-treated mice[190]. Recently, Goji berry and its functional constituents proved to have prebiotic effects, which can prevent gut microbiota dysbiosis associated with IBD[191]. In addition, the bioactive constituent of green tea, epigallocatechin-3-gallate, can ameliorate experimental colitis by increasing the SCFAs-producing bacteria and enrich butyrate production[161]. Furthermore, diet patterns can also be used to improve IBD, such as the Mediterranean diet (MD), the special carbohydrate diet (SCD), and the low fermentable oligosaccharide, disaccharide, monosaccharide, and polyol (FODMAP) diet[192]. FODMAP is a group of short-chain fermentable carbohydrates. Although dietary carbohydrates contained in FODMAP have prebiotic effects and promote the growth of beneficial bacteria[193], FODMAP diet can exacerbate functional gastrointestinal symptoms in IBD patients[194]. A study found that the low FODMAP diet group significantly improved gut symptoms and health-related quality of life in quiescent IBD patients, compared with control group[195]. Recently, a randomized trial indicated that both SCD and MD diet ameliorated disease activity index, pain, fatigue, and social isolation, comparing the efficacy of SCD and MD diet on Crohn's patients with mild to moderate symptoms[196].
In addition, synbiotics have been defined as “a mixture comprising live microorganisms and substrate(s) selectively utilized by host microorganisms that confers a health benefit on the host” in 2019[197]. Currently, a systematic review suggested that synbiotics could maintain IBD remission, reduce IBD disease activity index and prevent IBD recurrence[180]. Among them, the subgroup analysis showed that synbiotics may be more effective in inducing or maintaining IBD as compared to probiotic or prebiotics.
Despite the fact that prebiotic, synbiotics and prebiotic diet therapy are promising approaches for IBD treatment and maintenance, we also require more high-quality clinical research to further explore mechanisms by which they mitigate IBD.
5.5. Fecal microbiota transplantation
FMT can be delivered through the upper and lower gastrointestinal route. Examples include via capsules taken orally, nasoduodenal tube, colonoscopy, enema, etc.[198]. A randomized clinical trial showed that FMT given by oral capsules was similar to delivery by colonoscopy in the therapeutic effect[199]. Although colonoscopy delivery is more inconvenient, costly and invasive compared with oral capsules, it has great advantages in identifying alternative diagnoses. In turn, FMT delivery by oral capsules may decrease patient discomfort, reduce cost and wait time. Currently, FMT has been known as an established bacteriotherapy for recurrent Clostridioides difficile infection (CDI) and it is also used to treat CDI in patients with IBD[200], [201], [202], [203]. Meanwhile, several small scales, though not universal, cohort studies observed the effect of FMT on inflammatory bowel disease symptoms[39], [204], [205], [206]. Active UC patients received microbiota transplantation of blended homogenized stool from healthy donors obtained elevated microbial diversity, increased abundance of Roseburia inulivorans and Eubacterium hallii and higher levels of SCFAs and secondary bile acids[207]. Although some studies demonstrated promising results for FMT in inducing remission in IBD patients, it clearly poses complex challenges to clinicians, and its long-term effect remains unknown. Moreover, patients using FMT for active IBD are more prone to develop side effects compared to CDI treated with FMT. All UC patients in one clinical study developed fever and a transient elevation in C-reactive protein after FMT[208]. In addition, blood cultures were positive for the multidrug-sensitive E. coli strain in a 61-year-old CD patient twenty-four hours after FMT[209]. Meanwhile, fever was observed in CD patients[209], [210]. Though FMT treatment sometimes ameliorated clinical remission rate in UC patients, of 26 included studies, 23 precisely reported serious adverse events and 17 of them occurred in patients with IBD[198]. In general, FMT is a controversial treatment for IBD and is therefore still regarded as an experimental therapy.
5.6. Micro-nano conveying technology
Whether it is FMT or diet therapy, the current research is limited. There is not enough evidence to support the effectiveness and safety of these methods for IBD patients[211]. In order to avoid long-term immunosuppression, people need to find safer and more effective microbial targeted therapy strategy[157]. In addition to the treatment of antibiotics, diet therapy, probiotics and FMT mentioned above, many recent studies have shown that some nanoparticles can alter intestinal flora and metabolism in IBD patients. This may provide us with a new direction worth exploring for the development of IBD. Recently, a newly published editorial discussed the possibility of micro-nano conveying technology for the treatment of IBD associated with microbial disorders[212]. Among them, miRNA-loaded lipid NPs were proved to be able to bind to specific microorganisms and enter the bacteria to play a targeted role, such as achieveing a targeted downregulation of Lactobacillus rhamnosus GG SpaC gene[213]. Inorganic Ag NPs have been shown to target Fusoceaebacteria in the gut of IBD patients to achieve anti-inflammatory effects. In addition, micro- and nano-particles can target intestinal flora and affect its metabolites such as SCFAs, which are closely related[214], [215], [216]. In addition to targeting the regulation of intestinal microbiota and its metabolites, micro-nano technology can also repair intestinal mucosal injury and repair intestinal barrier. For example, hyaluronic acid-bilirubin nanoparticles have been proved to regulate intestinal flora, repair intestinal barrier, and play a strong anti-inflammatory role in acute colitis[217]. These are consistent with the pathogenesis of IBD, and the characteristics of targeted delivery make this treatment safer and more effective. With the gradual expansion of mass spectrometry data in microbial metabolism group, it may be possible to conduct metabolomics determination for IBD patients and a personalized treatment plan for them is customized by nanotechnology in the future. The new world of IBD treatment may be opened by targeting intestinal microorganisms and repairing intestinal barrier.
6. Summary and outlook
Increasing observational data in population-based researches have revealed the alterations in gut microbial structure and metabolic profiles in IBD patients. The advances of metagenomic sequencing and metabolomic analyses drive the identification and validation of disease-relevant microbiota and metabolites. As the output of host–microbiota co-metabolism, metabolites act as key regulators in the pathogenesis of IBD, and may hold promise for the treatment to this disease. Many fundamental questions nonetheless remain to be answered. Larger multinational cohorts of IBD patients and the channels for the detection of microbial members and microbial metabolites are needed. Moreover, owing to the composition and metabolism of microbiota changed dynamically on different temporal and spatial scales, more prospective longitudinal studies and new techniques are needed to explore high-resolution temporal and spatial profiling of microbiome and metabolite markers, such as sequential fluorescent in situ hybridization[218]. Furthermore, given the significant individual phenotypic differences in intestinal microbiota, even in the identical bacterial strains[219], it is crucial to further study the function of single strain and the mechanism of interaction between individual microbiota and the host. Ultimately, more metabolite-centered in ex vivo testing and in vivo clinical and transformational studies should be conducted to validate identified metabolic pathways and explore the dose effects as well as pharmacological features of metabolites.
7. Author statement
Mengfan Li, Lijiao Yang, Chenlu Mu were the major contributors. They prepared and revised the draft.
Mengfan Li: Writing-prepare most of original draft, Designing the graphical abstract, Writing – review & editing.
Lijiao Yang: Writing-Original draft preparation, Writing – review & editing.
Chenlu Mu: Writing-Original draft preparation, Writing – review & editing.
Yue Sun: Designing and drawing the graphical abstract, reviewing the draft.
Yu Gu and Danfeng Chen: Revising the draft critically.
Tianyu Liu: Reviewing the draft and put forward valuable suggestions for the manuscript.
Hailong Cao: Corresponding author; Involved in the study design and the critical revision of the manuscript.
8. Authors’ contributions
MFL, LJY and CLM were the major contributors. They participated in writing and collecting the related references. YS prepared the graphical abstract. YS, YG, DFC and TYL critically revised the manuscript and put forward valuable suggestions for the manuscript. HLC is involved in the study design and the critical revision of the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by grants (82070545 and 82100574) from the National Natural Science Foundation of China and the Key Project of Science and Technology Pillar Program of Tianjin (20YFZCSY00020).
References
- 1.Kobayashi T., Siegmund B., Le Berre C., Wei S.C., Ferrante M., et al. Ulcerative colitis. Nat Rev Dis Primers. 2020;6:74. doi: 10.1038/s41572-020-0205-x. [DOI] [PubMed] [Google Scholar]
- 2.Graham D.B., Xavier R.J. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature. 2020;578:527–539. doi: 10.1038/s41586-020-2025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisenstein M. Ulcerative colitis: towards remission. Nature. 2018;563:S33. doi: 10.1038/d41586-018-07276-2. [DOI] [PubMed] [Google Scholar]
- 4.Chang J.T. Pathophysiology of inflammatory bowel diseases. N Engl J Med. 2020;383:2652–2664. doi: 10.1056/NEJMra2002697. [DOI] [PubMed] [Google Scholar]
- 5.de Jong R.J., Ohnmacht C. Defining dysbiosis in inflammatory bowel disease. Immunity. 2019;50:8–10. doi: 10.1016/j.immuni.2018.12.028. [DOI] [PubMed] [Google Scholar]
- 6.Bryan P.F., Karla C., Edgar Alejandro M.T., Sara Elva E.P., Gemma F., et al. Sphingolipids as mediators in the crosstalk between microbiota and intestinal cells: implications for inflammatory bowel disease. Mediators Inflamm. 2016:9890141. doi: 10.1155/2016/9890141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britton G.J., Contijoch E.J., Mogno I., Vennaro O.H., Llewellyn S.R., et al. Microbiotas from Humans with Inflammatory Bowel Disease Alter the Balance of Gut Th17 and RORγt(+) Regulatory T Cells and Exacerbate Colitis in Mice. Immunity. 2019;50:212–224.e214. doi: 10.1016/j.immuni.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lloyd-Price J., Arze C., Ananthakrishnan A.N., Schirmer M., Avila-Pacheco J., et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santoru M.L., Piras C., Murgia A., Palmas V., Camboni T., et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci Rep. 2017;7:9523. doi: 10.1038/s41598-017-10034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franzosa E.A., Sirota-Madi A., Avila-Pacheco J., Fornelos N., Haiser H.J., et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4:293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjerrum J.T., Wang Y., Hao F., Coskun M., Ludwig C., et al. Metabonomics of human fecal extracts characterize ulcerative colitis, Crohn's disease and healthy individuals. Metabolomics. 2015;11:122–133. doi: 10.1007/s11306-014-0677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muller E., Algavi Y.M., Borenstein E. A meta-analysis study of the robustness and universality of gut microbiome-metabolome associations. Microbiome. 2021;9:203. doi: 10.1186/s40168-021-01149-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott S.A., Fu J., Chang P.V. Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proc Natl Acad Sci U S A. 2020 doi: 10.1073/pnas.2000047117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun M., Wu W., Liu Z., Cong Y. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J Gastroenterol. 2017;52:1–8. doi: 10.1007/s00535-016-1242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinken A., Ravcheev D.A., Baldini F., Heirendt L., Fleming R.M.T., et al. Systematic assessment of secondary bile acid metabolism in gut microbes reveals distinct metabolic capabilities in inflammatory bowel disease. Microbiome. 2019;7:75. doi: 10.1186/s40168-019-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang X., Vázquez-Baeza Y., Elijah E., Vargas F., Ackermann G., et al. Gastrointestinal surgery for inflammatory bowel disease persistently lowers microbiome and metabolome diversity. Inflamm Bowel Dis. 2021;27:603–616. doi: 10.1093/ibd/izaa262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnold J.W., Roach J., Azcarate-Peril M.A. Emerging Technologies for Gut Microbiome Research. Trends Microbiol. 2016;24:887–901. doi: 10.1016/j.tim.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen N.P., Warnow T., Pop M., White B. A perspective on 16S rRNA operational taxonomic unit clustering using sequence similarity. npj Biofilms Microbiomes. 2016;2:16004. doi: 10.1038/npjbiofilms.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faith J.J., Guruge J.L., Charbonneau M., Subramanian S., Seedorf H., et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caporaso J.G., Lauber C.L., Costello E.K., Berg-Lyons D., Gonzalez A., et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schirmer M., Denson L., Vlamakis H., Franzosa E.A., Thomas S., et al. Compositional and temporal changes in the gut microbiome of pediatric ulcerative colitis patients are linked to disease course. Cell Host Microbe. 2018;24:600–610.e604. doi: 10.1016/j.chom.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lepage P., Häsler R., Spehlmann M.E., Rehman A., Zvirbliene A., et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;141:227–236. doi: 10.1053/j.gastro.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 24.Gevers D., Kugathasan S., Denson L.A., Vázquez-Baeza Y., Van Treuren W., et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maldonado-Arriaga B., Sandoval-Jiménez S., Rodríguez-Silverio J., Lizeth Alcaráz-Estrada S., Cortés-Espinosa T., et al. Gut dysbiosis and clinical phases of pancolitis in patients with ulcerative colitis. Microbiologyopen. 2021;10 doi: 10.1002/mbo3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lo Sasso G., Khachatryan L., Kondylis A., Battey J.N.D., Sierro N., et al. Inflammatory Bowel Disease-Associated Changes in the Gut: Focus on Kazan Patients. Inflamm Bowel Dis. 2021;27:418–433. doi: 10.1093/ibd/izaa188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liguori G., Lamas B., Richard M.L., Brandi G., da Costa G., et al. Fungal Dysbiosis in Mucosa-associated Microbiota of Crohn's Disease Patients. J Crohns Colitis. 2016;10:296–305. doi: 10.1093/ecco-jcc/jjv209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quince C., Walker A.W., Simpson J.T., Loman N.J., Segata N. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017;35:833–844. doi: 10.1038/nbt.3935. [DOI] [PubMed] [Google Scholar]
- 29.New F.N., Brito I.L. what is metagenomics teaching us, and what is missed? Annu Rev Microbiol. 2020;74:117–135. doi: 10.1146/annurev-micro-012520-072314. [DOI] [PubMed] [Google Scholar]
- 30.Borren N.Z., Plichta D., Joshi A.D., Bonilla G., Sadreyev R., et al. Multi-“-Omics” profiling in patients with quiescent inflammatory bowel disease identifies biomarkers predicting relapse. Inflamm Bowel Dis. 2020;26:1524–1532. doi: 10.1093/ibd/izaa183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pittayanon R., Lau J.T., Leontiadis G.I., Tse F., Yuan Y., et al. Differences in gut microbiota in patients with vs without inflammatory bowel diseases: A systematic review. Gastroenterology. 2020;158:930–946.e931. doi: 10.1053/j.gastro.2019.11.294. [DOI] [PubMed] [Google Scholar]
- 32.Mukhopadhya I., Hansen R., El-Omar E.M., Hold G.L. IBD-what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol. 2012;9:219–230. doi: 10.1038/nrgastro.2012.14. [DOI] [PubMed] [Google Scholar]
- 33.Lupp C., Robertson M.L., Wickham M.E., Sekirov I., Champion O.L., et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Baldelli V., Scaldaferri F., Putignani L., Del Chierico F. The Role of Enterobacteriaceae in Gut Microbiota Dysbiosis in Inflammatory Bowel Diseases. Microorganisms. 2021;9 doi: 10.3390/microorganisms9040697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J., Hoedt E.C., Liu Q., Berendsen E., Teh J.J., et al. Elucidation of Proteus mirabilis as a Key Bacterium in Crohn's Disease Inflammation. Gastroenterology. 2021;160:317–330.e311. doi: 10.1053/j.gastro.2020.09.036. [DOI] [PubMed] [Google Scholar]
- 36.Atarashi K., Suda W., Luo C., Kawaguchi T., Motoo I., et al. Ectopic colonization of oral bacteria in the intestine drives T(H)1 cell induction and inflammation. Science. 2017;358:359–365. doi: 10.1126/science.aan4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strauss J., Kaplan G.G., Beck P.L., Rioux K., Panaccione R., et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011;17:1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 38.Geva-Zatorsky N., Sefik E., Kua L., Pasman L., Tan T.G., et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell. 2017;168:928–943.e911. doi: 10.1016/j.cell.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paramsothy S., Kamm M.A., Kaakoush N.O., Walsh A.J., van den Bogaerde J., et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet. 2017;389:1218–1228. doi: 10.1016/S0140-6736(17)30182-4. [DOI] [PubMed] [Google Scholar]
- 40.Mao X., Ma J., Jiao C., Tang N., Zhao X., et al. Faecalibacterium prausnitzii Attenuates DSS-Induced Colitis by Inhibiting the Colonization and Pathogenicity of Candida albicans. Mol Nutr Food Res. 2021 doi: 10.1002/mnfr.202100433. [DOI] [PubMed] [Google Scholar]
- 41.Imhann F., Vich Vila A., Bonder M.J., Fu J., Gevers D., et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut. 2018;67:108–119. doi: 10.1136/gutjnl-2016-312135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo W., Shen Z., Deng M., Li X., Tan B., et al. Roseburia intestinalis supernatant ameliorates colitis induced in mice by regulating the immune response. Mol Med Rep. 2019;20:1007–1016. doi: 10.3892/mmr.2019.10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu C.X., Song K.R., Shen Z.H., Quan Y.S., Tan B., et al. Roseburia intestinalis inhibits interleukin-17 excretion and promotes regulatory T cells differentiation in colitis. Mol Med Rep. 2018;17:7567–7574. doi: 10.3892/mmr.2018.8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quan Y.S., Song K.R., Zhang Y., Zhu C.X., Shen Z.H., et al. Roseburia intestinalis-derived flagellin is a negative regulator of intestinal inflammation. Biochem Biophys Res Commun. 2018;501:791–799. doi: 10.1016/j.bbrc.2018.05.075. [DOI] [PubMed] [Google Scholar]
- 45.Snipen L., Angell I.L., Rognes T., Rudi K. Reduced metagenome sequencing for strain-resolution taxonomic profiles. Microbiome. 2021;9:79. doi: 10.1186/s40168-021-01019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheth R.U., Li M., Jiang W., Sims P.A., Leong K.W., et al. Spatial metagenomic characterization of microbial biogeography in the gut. Nat Biotechnol. 2019;37:877–883. doi: 10.1038/s41587-019-0183-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi H., Shi Q., Grodner B., Lenz J.S., Zipfel W.R., et al. Highly multiplexed spatial mapping of microbial communities. Nature. 2020;588:676–681. doi: 10.1038/s41586-020-2983-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Evenepoel P., Claus D., Geypens B., Hiele M., Geboes K., et al. Amount and fate of egg protein escaping assimilation in the small intestine of humans. Am J Physiol. 1999;277:G935–943. doi: 10.1152/ajpgi.1999.277.5.G935. [DOI] [PubMed] [Google Scholar]
- 49.Nicholson J.K., Lindon J.C. Systems biology: Metabonomics. Nature. 2008;455:1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 50.Wishart D.S., Jewison T., Guo A.C., Wilson M., Knox C., et al. HMDB 3.0–The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amberg A., Riefke B., Schlotterbeck G., Ross A., Senn H., et al. In: Drug Safety Evaluation: Methods and Protocols. Gautier J.-.-C., editor. Springer; New York, New York, NY: 2017. NMR and MS Methods for Metabolomics; pp. 229–258. [Google Scholar]
- 52.Zhang A., Sun H., Wang P., Han Y., Wang X. Modern analytical techniques in metabolomics analysis. Analyst. 2012;137:293–300. doi: 10.1039/c1an15605e. [DOI] [PubMed] [Google Scholar]
- 53.Ursell L.K., Haiser H.J., Van Treuren W., Garg N., Reddivari L., et al. The intestinal metabolome: an intersection between microbiota and host. Gastroenterology. 2014;146:1470–1476. doi: 10.1053/j.gastro.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schoeler M., Caesar R. Dietary lipids, gut microbiota and lipid metabolism. Rev Endocr Metab Disord. 2019;20:461–472. doi: 10.1007/s11154-019-09512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calder P.C. Functional Roles of Fatty Acids and Their Effects on Human Health. JPEN J Parenter Enteral Nutr. 2015;39:18s–32s. doi: 10.1177/0148607115595980. [DOI] [PubMed] [Google Scholar]
- 56.Ratnayake W.M., Galli C. Fat and fatty acid terminology, methods of analysis and fat digestion and metabolism: a background review paper. Ann Nutr Metab. 2009;55:8–43. doi: 10.1159/000228994. [DOI] [PubMed] [Google Scholar]
- 57.Koh A., De Vadder F., Kovatcheva-Datchary P., Bäckhed F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 58.Marchesi J.R., Holmes E., Khan F., Kochhar S., Scanlan P., et al. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. 2007;6:546–551. doi: 10.1021/pr060470d. [DOI] [PubMed] [Google Scholar]
- 59.Machiels K., Joossens M., Sabino J., De Preter V., Arijs I., et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275–1283. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- 60.Yang W., Yu T., Huang X., Bilotta A.J., Xu L., et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat Commun. 2020;11:4457. doi: 10.1038/s41467-020-18262-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li M., van Esch B., Henricks P.A.J., Garssen J., Folkerts G. IL-33 Is Involved in the Anti-Inflammatory Effects of Butyrate and Propionate on TNFα-Activated Endothelial Cells. Int J Mol Sci. 2021;22 doi: 10.3390/ijms22052447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takeuchi T., Miyauchi E., Kanaya T., Kato T., Nakanishi Y., et al. Acetate differentially regulates IgA reactivity to commensal bacteria. Nature. 2021;595:560–564. doi: 10.1038/s41586-021-03727-5. [DOI] [PubMed] [Google Scholar]
- 63.Parada Venegas D., De la Fuente M.K., Landskron G., Gonzalez M.J., Quera R., et al. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front Immunol. 2019;10:277. doi: 10.3389/fimmu.2019.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hosseinkhani F., Heinken A., Thiele I., Lindenburg P.W., Harms A.C., et al. The contribution of gut bacterial metabolites in the human immune signaling pathway of non-communicable diseases. Gut Microbes. 2021;13:1–22. doi: 10.1080/19490976.2021.1882927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rolandelli R.H., Koruda M.J., Settle R.G., Rombeau J.L. Effects of intraluminal infusion of short-chain fatty acids on the healing of colonic anastomosis in the rat. Surgery. 1986;100:198–204. [PubMed] [Google Scholar]
- 66.Rodríguez-Padilla Á., Morales-Martín G., Pérez-Quintero R., Gómez-Salgado J., Ruiz-Frutos C. Serological Biomarkers and Diversion Colitis: Changes after Stimulation with Probiotics. Biomolecules. 2021;11 doi: 10.3390/biom11050684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neut C., Guillemot F., Gower-Rousseau C., Biron N., Cortot A., et al. Treatment of diversion colitis with short-chain fatty acids. Bacteriological study. Gastroenterol Clin Biol. 1995;19:871–875. [PubMed] [Google Scholar]
- 68.Baek S.J., Kim S.H., Lee C.K., Roh K.H., Keum B., et al. Relationship between the severity of diversion colitis and the composition of colonic bacteria: a prospective study. Gut Liver. 2014;8:170–176. doi: 10.5009/gnl.2014.8.2.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tominaga K., Kamimura K., Takahashi K., Yokoyama J., Yamagiwa S., et al. Diversion colitis and pouchitis: A mini-review. World J Gastroenterol. 2018;24:1734–1747. doi: 10.3748/wjg.v24.i16.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tominaga K., Tsuchiya A., Mizusawa T., Matsumoto A., Minemura A., et al. Evaluation of intestinal microbiota, short-chain fatty acids, and immunoglobulin a in diversion colitis. Biochem Biophys Rep. 2021;25 doi: 10.1016/j.bbrep.2020.100892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luceri C., Femia A.P., Fazi M., Di Martino C., Zolfanelli F., et al. Effect of butyrate enemas on gene expression profiles and endoscopic/histopathological scores of diverted colorectal mucosa: A randomized trial. Dig Liver Dis. 2016;48:27–33. doi: 10.1016/j.dld.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 72.Pal K., Tinalal S., Al Buainain H., Singh V.P. Diversion proctocolitis and response to treatment with short-chain fatty acids–a clinicopathological study in children. Indian J Gastroenterol. 2015;34:292–299. doi: 10.1007/s12664-015-0577-0. [DOI] [PubMed] [Google Scholar]
- 73.De Preter V., Machiels K., Joossens M., Arijs I., Matthys C., et al. Faecal metabolite profiling identifies medium-chain fatty acids as discriminating compounds in IBD. Gut. 2015;64:447–458. doi: 10.1136/gutjnl-2013-306423. [DOI] [PubMed] [Google Scholar]
- 74.Tefas C., Ciobanu L., Tanțău M., Moraru C., Socaciu C. The potential of metabolic and lipid profiling in inflammatory bowel diseases: A pilot study. Bosn J Basic Med Sci. 2020;20:262–270. doi: 10.17305/bjbms.2019.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma C., Vasu R., Zhang H. The Role of Long-Chain Fatty Acids in Inflammatory Bowel Disease. Mediators Inflamm. 2019;2019:8495913. doi: 10.1155/2019/8495913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Piotrowska M., Binienda A., Fichna J. The role of fatty acids in Crohn's disease pathophysiology - An overview. Mol Cell Endocrinol. 2021;538 doi: 10.1016/j.mce.2021.111448. [DOI] [PubMed] [Google Scholar]
- 77.Wang J., Tian F., Zheng H., Tian H., Wang P., et al. N-3 polyunsaturated fatty acid-enriched lipid emulsion improves Paneth cell function via the IL-22/Stat3 pathway in a mouse model of total parenteral nutrition. Biochem Biophys Res Commun. 2017;490:253–259. doi: 10.1016/j.bbrc.2017.06.032. [DOI] [PubMed] [Google Scholar]
- 78.Pujo J., Petitfils C., Le Faouder P., Eeckhaut V., Payros G., et al. Bacteria-derived long chain fatty acid exhibits anti-inflammatory properties in colitis. Gut. 2021;70:1088–1097. doi: 10.1136/gutjnl-2020-321173. [DOI] [PubMed] [Google Scholar]
- 79.Tsutsumi R., Yamasaki Y., Takeo J., Miyahara H., Sebe M., et al. Long-chain monounsaturated fatty acids improve endothelial function with altering microbial flora. Transl Res. 2021;237:16–30. doi: 10.1016/j.trsl.2021.03.016. [DOI] [PubMed] [Google Scholar]
- 80.Garner C.E., Smith S., de Lacy Costello B., White P., Spencer R., et al. Volatile organic compounds from feces and their potential for diagnosis of gastrointestinal disease. FASEB J. 2007;21:1675–1688. doi: 10.1096/fj.06-6927com. [DOI] [PubMed] [Google Scholar]
- 81.Bassaganya-Riera J., Viladomiu M., Pedragosa M., De Simone C., Carbo A., et al. Probiotic bacteria produce conjugated linoleic acid locally in the gut that targets macrophage PPAR γ to suppress colitis. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0031238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang W.C., Tsai T.H., Chuang L.T., Li Y.Y., Zouboulis C.C., et al. Anti-bacterial and anti-inflammatory properties of capric acid against Propionibacterium acnes: a comparative study with lauric acid. J Dermatol Sci. 2014;73:232–240. doi: 10.1016/j.jdermsci.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 83.Khan H.U., Aamir K., Jusuf P.R., Sethi G., Sisinthy S.P., et al. Lauric acid ameliorates lipopolysaccharide (LPS)-induced liver inflammation by mediating TLR4/MyD88 pathway in Sprague Dawley (SD) rats. Life Sci. 2021;265 doi: 10.1016/j.lfs.2020.118750. [DOI] [PubMed] [Google Scholar]
- 84.Lavelle A., Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease, Nature Reviews. Gastroenterology & Hepatology. 2020;17:223–237. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 85.Roager H.M., Licht T.R. Microbial tryptophan catabolites in health and disease. Nat Commun. 2018;9:3294. doi: 10.1038/s41467-018-05470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stockinger B., Shah K., Wincent E. AHR in the intestinal microenvironment: safeguarding barrier function. Nat Rev Gastroenterol Hepatol. 2021;18:559–570. doi: 10.1038/s41575-021-00430-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nikolaus S., Schulte B., Al-Massad N., Thieme F., Schulte D.M., et al. Increased Tryptophan Metabolism Is Associated With Activity of Inflammatory Bowel Diseases. Gastroenterology. 2017;153:1504–1516.e1502. doi: 10.1053/j.gastro.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 88.Lamas B., Richard M.L., Leducq V., Pham H.P., Michel M.L., et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cervantes-Barragan L., Chai J.N., Tianero M.D., Di Luccia B., Ahern P.P., et al. Lactobacillus reuteri induces gut intraepithelial CD4(+)CD8alphaalpha(+) T cells. Science. 2017;357:806–810. doi: 10.1126/science.aah5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Busbee P.B., Menzel L., Alrafas H.R., Dopkins N., Becker W., et al. Indole-3-carbinol prevents colitis and associated microbial dysbiosis in an IL-22-dependent manner. JCI Insight. 2020;5 doi: 10.1172/jci.insight.127551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zelante T., Iannitti R.G., Cunha C., De Luca A., Giovannini G., et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 92.Böger R.H., Bode-Böger S.M. The clinical pharmacology of L-arginine. Annu Rev Pharmacol Toxicol. 2001;41:79–99. doi: 10.1146/annurev.pharmtox.41.1.79. [DOI] [PubMed] [Google Scholar]
- 93.Coburn L.A., Horst S.N., Allaman M.M., Brown C.T., Williams C.S., et al. L-arginine availability and metabolism is altered in ulcerative colitis. Inflamm Bowel Dis. 2016;22:1847–1858. doi: 10.1097/MIB.0000000000000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Scoville E.A., Allaman M.M., Brown C.T., Motley A.K., Horst S.N., et al. Alterations in lipid, amino acid, and energy metabolism distinguish crohn's disease from ulcerative colitis and control subjects by serum metabolomic profiling. Metabolomics. 2018;14:17. doi: 10.1007/s11306-017-1311-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Krzystek-Korpacka M., Fleszar M.G., Bednarz-Misa I., Lewandowski Ł., Szczuka I., et al. Transcriptional and metabolomic analysis of l-arginine/nitric oxide pathway in inflammatory bowel disease and its association with local inflammatory and angiogenic response: preliminary findings. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21051641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Caldwell R.B., Toque H.A., Narayanan S.P., Caldwell R.W. Arginase: an old enzyme with new tricks. Trends Pharmacol Sci. 2015;36:395–405. doi: 10.1016/j.tips.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Baier J., Gansbauer M., Giessler C., Arnold H., Muske M., et al. Arginase impedes the resolution of colitis by altering the microbiome and metabolome. J Clin Invest. 2020;130:5703–5720. doi: 10.1172/JCI126923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Andrade M.E., Santos R.D., Soares A.D., Costa K.A., Fernandes S.O., et al. Pretreatment and Treatment With L-Arginine Attenuate Weight Loss and Bacterial Translocation in Dextran Sulfate Sodium Colitis. JPEN J Parenter Enteral Nutr. 2016;40:1131–1139. doi: 10.1177/0148607115581374. [DOI] [PubMed] [Google Scholar]
- 99.Madeo F., Hofer S.J., Pendl T., Bauer M.A., Eisenberg T., et al. Nutritional Aspects of Spermidine. Annu Rev Nutr. 2020;40:135–159. doi: 10.1146/annurev-nutr-120419-015419. [DOI] [PubMed] [Google Scholar]
- 100.Matsumoto M., Benno Y. The relationship between microbiota and polyamine concentration in the human intestine: a pilot study. Microbiol Immunol. 2007;51:25–35. doi: 10.1111/j.1348-0421.2007.tb03887.x. [DOI] [PubMed] [Google Scholar]
- 101.Tofalo R., Cocchi S., Suzzi G. Polyamines and Gut Microbiota. Front Nutr. 2019;6:16. doi: 10.3389/fnut.2019.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ma L., Ni L., Yang T., Mao P., Huang X., et al. Preventive and therapeutic spermidine treatment attenuates acute colitis in mice. J Agric Food Chem. 2021;69:1864–1876. doi: 10.1021/acs.jafc.0c07095. [DOI] [PubMed] [Google Scholar]
- 103.Ma L., Ni Y., Wang Z., Tu W., Ni L., et al. Spermidine improves gut barrier integrity and gut microbiota function in diet-induced obese mice. Gut Microbes. 2020;12:1–19. doi: 10.1080/19490976.2020.1832857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li L., Rao J.N., Bass B.L., Wang J.Y. NF-kappaB activation and susceptibility to apoptosis after polyamine depletion in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2001;280:G992–g1004. doi: 10.1152/ajpgi.2001.280.5.G992. [DOI] [PubMed] [Google Scholar]
- 105.Medina M.A., Urdiales J.L., Rodríguez-Caso C., Ramírez F.J., Sánchez-Jiménez F. Biogenic amines and polyamines: similar biochemistry for different physiological missions and biomedical applications. Crit Rev Biochem Mol Biol. 2003;38:23–59. doi: 10.1080/713609209. [DOI] [PubMed] [Google Scholar]
- 106.Carriche G.M., Almeida L., Stüve P., Velasquez L., Dhillon-LaBrooy A., et al. Regulating T-cell differentiation through the polyamine spermidine. J Allergy Clin Immunol. 2021;147:335–348.e311. doi: 10.1016/j.jaci.2020.04.037. [DOI] [PubMed] [Google Scholar]
- 107.Nakamura A., Kurihara S., Takahashi D., Ohashi W., Nakamura Y., et al. Symbiotic polyamine metabolism regulates epithelial proliferation and macrophage differentiation in the colon. Nat Commun. 2021;12:2105. doi: 10.1038/s41467-021-22212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Grosheva I., Zheng D., Levy M., Polansky O., Lichtenstein A., et al. High-throughput screen identifies host and microbiota regulators of intestinal barrier function. Gastroenterology. 2020;159:1807–1823. doi: 10.1053/j.gastro.2020.07.003. [DOI] [PubMed] [Google Scholar]
- 109.Kolho K.L., Pessia A., Jaakkola T., de Vos W.M., Velagapudi V. Faecal and serum metabolomics in paediatric inflammatory bowel disease. J Crohns Colitis. 2017;11:321–334. doi: 10.1093/ecco-jcc/jjw158. [DOI] [PubMed] [Google Scholar]
- 110.Bushman F.D., Conrad M., Ren Y., Zhao C., Gu C., et al. Multi-omic analysis of the interaction between clostridioides difficile infection and pediatric inflammatory bowel disease. Cell Host Microbe. 2020;28:422–433.e427. doi: 10.1016/j.chom.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Alghamdi A., Gerasimidis K., Blackburn G., Akinci D., Edwards C., et al. Untargeted metabolomics of extracts from faecal samples demonstrates distinct differences between paediatric crohn's disease patients and healthy controls but no significant changes resulting from exclusive enteral nutrition treatment. Metabolites. 2018;8 doi: 10.3390/metabo8040082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Diederen K., Li J.V., Donachie G.E., de Meij T.G., de Waart D.R., et al. Exclusive enteral nutrition mediates gut microbial and metabolic changes that are associated with remission in children with Crohn's disease. Sci Rep. 2020;10:18879. doi: 10.1038/s41598-020-75306-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sinniger V., Pellissier S., Fauvelle F., Trocmé C., Hoffmann D., et al. A 12-month pilot study outcomes of vagus nerve stimulation in Crohn's disease. Neurogastroenterol Motil. 2020;32 doi: 10.1111/nmo.13911. [DOI] [PubMed] [Google Scholar]
- 114.Son M., Ko J.I., Kim W.B., Kang H.K., Kim B.K. Taurine can ameliorate inflammatory bowel disease in rats. Taurine 3: Cell Regul Mech. 1998:291–298. doi: 10.1007/978-1-4899-0117-0_37. [DOI] [PubMed] [Google Scholar]
- 115.Son M.W., Ko J.I., Doh H.M., Kim W.B., Park T.S., et al. Protective effect of taurine on TNBS-induced inflammatory bowel disease in rats. Arch Pharmacal Res. 1998;21:531–536. doi: 10.1007/BF02975370. [DOI] [PubMed] [Google Scholar]
- 116.Zhao Z., Satsu H., Fujisawa M., Hori M., Ishimoto Y., et al. Attenuation by dietary taurine of dextran sulfate sodium-induced colitis in mice and of THP-1-induced damage to intestinal Caco-2 cell monolayers. Amino Acids. 2008;35:217–224. doi: 10.1007/s00726-007-0562-8. [DOI] [PubMed] [Google Scholar]
- 117.Levy M., Thaiss C.A., Zeevi D., Dohnalová L., Zilberman-Schapira G., et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell. 2015;163:1428–1443. doi: 10.1016/j.cell.2015.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stacy A., Andrade-Oliveira V., McCulloch J.A., Hild B., Oh J.H., et al. Infection trains the host for microbiota-enhanced resistance to pathogens. Cell. 2021;184:615–627.e617. doi: 10.1016/j.cell.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ahmadi S., Wang S., Nagpal R., Wang B., Jain S., et al. A human-origin probiotic cocktail ameliorates aging-related leaky gut and inflammation via modulating the microbiota/taurine/tight junction axis. JCI Insight. 2020;5 doi: 10.1172/jci.insight.132055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang Z.H., Liu F., Zhu X.R., Suo F.Y., Jia Z.J., et al. Altered profiles of fecal bile acids correlate with gut microbiota and inflammatory responses in patients with ulcerative colitis. World J Gastroenterol. 2021;27:3609–3629. doi: 10.3748/wjg.v27.i24.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Duboc H., Rajca S., Rainteau D., Benarous D., Maubert M.A., et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531–539. doi: 10.1136/gutjnl-2012-302578. [DOI] [PubMed] [Google Scholar]
- 122.Sinha S.R., Haileselassie Y., Nguyen L.P., Tropini C., Wang M., et al. Dysbiosis-Induced Secondary Bile Acid Deficiency Promotes Intestinal Inflammation. Cell Host Microbe. 2020;27:659–670 e655. doi: 10.1016/j.chom.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang S., Martins R., Sullivan M.C., Friedman E.S., Misic A.M., et al. Diet-induced remission in chronic enteropathy is associated with altered microbial community structure and synthesis of secondary bile acids. Microbiome. 2019;7:126. doi: 10.1186/s40168-019-0740-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Winston J.A., Rivera A.J., Cai J., Thanissery R., Montgomery S.A., et al. Ursodeoxycholic Acid (UDCA) Mitigates the Host Inflammatory Response during Clostridioides difficile Infection by Altering Gut Bile Acids. Infect Immun. 2020;88 doi: 10.1128/IAI.00045-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dong S., Zhu M., Wang K., Zhao X., Hu L., et al. Dihydromyricetin improves DSS-induced colitis in mice via modulation of fecal-bacteria-related bile acid metabolism. Pharmacol Res. 2021;171 doi: 10.1016/j.phrs.2021.105767. [DOI] [PubMed] [Google Scholar]
- 126.Ke J., Li Y., Han C., He R., Lin R., et al. Fucose ameliorate intestinal inflammation through modulating the crosstalk between bile acids and gut microbiota in a chronic colitis murine model. Inflamm Bowel Dis. 2020;26:863–873. doi: 10.1093/ibd/izaa007. [DOI] [PubMed] [Google Scholar]
- 127.Liu L., Yang M., Dong W., Liu T., Song X., et al. Gut Dysbiosis and Abnormal Bile Acid Metabolism in Colitis-Associated Cancer. Gastroenterol Res Pract. 2021;2021:6645970. doi: 10.1155/2021/6645970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Das P., Marcisauskas S., Ji B., Nielsen J. Metagenomic analysis of bile salt biotransformation in the human gut microbiome. BMC Genomics. 2019;20:517. doi: 10.1186/s12864-019-5899-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang Y., Gao X., Zhang X., Xiao F., Hu H., et al. Microbial and metabolic features associated with outcome of infliximab therapy in pediatric Crohn's disease. Gut Microbes. 2021;13:1–18. doi: 10.1080/19490976.2020.1865708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gadaleta R.M., van Erpecum K.J., Oldenburg B., Willemsen E.C., Renooij W., et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut. 2011;60:463–472. doi: 10.1136/gut.2010.212159. [DOI] [PubMed] [Google Scholar]
- 131.Ward J.B.J., Lajczak N.K., Kelly O.B., O'Dwyer A.M., Giddam A.K., et al. Ursodeoxycholic acid and lithocholic acid exert anti-inflammatory actions in the colon. Am J Physiol Gastrointest Liver Physiol. 2017;312:G550–G558. doi: 10.1152/ajpgi.00256.2016. [DOI] [PubMed] [Google Scholar]
- 132.Lajczak-McGinley N.K., Porru E., Fallon C.M., Smyth J., Curley C., et al. The secondary bile acids, ursodeoxycholic acid and lithocholic acid, protect against intestinal inflammation by inhibition of epithelial apoptosis. Physiol Rep. 2020;8 doi: 10.14814/phy2.14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mroz M.S., Lajczak N.K., Goggins B.J., Keely S., Keely S.J. The bile acids, deoxycholic acid and ursodeoxycholic acid, regulate colonic epithelial wound healing. Am J Physiol Gastrointest Liver Physiol. 2018;314:G378–G387. doi: 10.1152/ajpgi.00435.2016. [DOI] [PubMed] [Google Scholar]
- 134.Stenman L.K., Holma R., Korpela R. High-fat-induced intestinal permeability dysfunction associated with altered fecal bile acids. World J Gastroenterol. 2012;18:923–929. doi: 10.3748/wjg.v18.i9.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xu M., Cen M., Shen Y., Zhu Y., Cheng F., et al. Deoxycholic Acid-Induced Gut Dysbiosis Disrupts Bile Acid Enterohepatic Circulation and Promotes Intestinal Inflammation. Dig Dis Sci. 2021;66:568–576. doi: 10.1007/s10620-020-06208-3. [DOI] [PubMed] [Google Scholar]
- 136.Bauermeister A., Mannochio-Russo H., Costa-Lotufo L.V., Jarmusch A.K., Dorrestein P.C. Mass spectrometry-based metabolomics in microbiome investigations. Nat Rev Microbiol. 2021 doi: 10.1038/s41579-021-00621-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Morton J.T., Aksenov A.A., Nothias L.F., Foulds J.R., Quinn R.A., et al. Learning representations of microbe-metabolite interactions. Nat Methods. 2019;16:1306–1314. doi: 10.1038/s41592-019-0616-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Han S., Van Treuren W., Fischer C.R., Merrill B.D., DeFelice B.C., et al. A metabolomics pipeline for the mechanistic interrogation of the gut microbiome. Nature. 2021;595:415–420. doi: 10.1038/s41586-021-03707-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yang C., Chowdhury D., Zhang Z., Cheung W.K., Lu A., et al. A review of computational tools for generating metagenome-assembled genomes from metagenomic sequencing data. Comput Struct Biotechnol J. 2021;19:6301–6314. doi: 10.1016/j.csbj.2021.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shi Y., Zhang L., Peterson C.B., Do K.A., Jenq R.R. Performance determinants of unsupervised clustering methods for microbiome data. Microbiome. 2022;10:25. doi: 10.1186/s40168-021-01199-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Beniddir M.A., Kang K.B., Genta-Jouve G., Huber F., Rogers S., et al. Advances in decomposing complex metabolite mixtures using substructure- and network-based computational metabolomics approaches. Nat Prod Rep. 2021;38:1967–1993. doi: 10.1039/d1np00023c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhou Z., Tran P.Q., Breister A.M., Liu Y., Kieft K., et al. METABOLIC: high-throughput profiling of microbial genomes for functional traits, metabolism, biogeochemistry, and community-scale functional networks. Microbiome. 2022;10:33. doi: 10.1186/s40168-021-01213-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Park S.Y., Ufondu A., Lee K., Jayaraman A. Emerging computational tools and models for studying gut microbiota composition and function. Curr Opin Biotechnol. 2020;66:301–311. doi: 10.1016/j.copbio.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hernández-de-Diego R., Tarazona S., Martínez-Mira C., Balzano-Nogueira L., Furió-Tarí P., et al. PaintOmics 3: a web resource for the pathway analysis and visualization of multi-omics data. Nucleic Acids Res. 2018;46:W503–w509. doi: 10.1093/nar/gky466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Clooney A.G., Eckenberger J., Laserna-Mendieta E., Sexton K.A., Bernstein M.T., et al. Ranking microbiome variance in inflammatory bowel disease: a large longitudinal intercontinental study. Gut. 2021;70:499–510. doi: 10.1136/gutjnl-2020-321106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sartor R.B. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics. Gastroenterology. 2004;126:1620–1633. doi: 10.1053/j.gastro.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 147.Scott K.P., Antoine J.M., Midtvedt T., van Hemert S. Manipulating the gut microbiota to maintain health and treat disease. Microb Ecol Health Dis. 2015;26:25877. doi: 10.3402/mehd.v26.25877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nitzan O., Elias M., Peretz A., Saliba W. Role of antibiotics for treatment of inflammatory bowel disease. World J Gastroenterol. 2016;22:1078–1087. doi: 10.3748/wjg.v22.i3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Oliveira S.B., Monteiro I.M. Diagnosis and management of inflammatory bowel disease in children. BMJ. 2017;357 doi: 10.1136/bmj.j2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Su J.W., Ma J.J., Zhang H.J. Use of antibiotics in patients with Crohn's disease: a systematic review and meta-analysis. J Dig Dis. 2015;16:58–66. doi: 10.1111/1751-2980.12216. [DOI] [PubMed] [Google Scholar]
- 151.Burke D.A., Axon A.T., Clayden S.A., Dixon M.F., Johnston D., et al. The efficacy of tobramycin in the treatment of ulcerative colitis. Aliment Pharmacol Ther. 1990;4:123–129. doi: 10.1111/j.1365-2036.1990.tb00456.x. [DOI] [PubMed] [Google Scholar]
- 152.Banfi D., Moro E., Bosi A., Bistoletti M., Cerantola S., et al. Impact of Microbial Metabolites on Microbiota-Gut-Brain Axis in Inflammatory Bowel Disease. Int J Mol Sci. 2021;22 doi: 10.3390/ijms22041623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ianiro G., Tilg H., Gasbarrini A. Antibiotics as deep modulators of gut microbiota: between good and evil. Gut. 2016;65:1906–1915. doi: 10.1136/gutjnl-2016-312297. [DOI] [PubMed] [Google Scholar]
- 154.Arboleya S., Sánchez B., Solís G., Fernández N., Suárez M., et al. Impact of prematurity and perinatal antibiotics on the developing intestinal microbiota: a functional inference study. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17050649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mu C., Zhu W. Antibiotic effects on gut microbiota, metabolism, and beyond. Appl Microbiol Biotechnol. 2019;103:9277–9285. doi: 10.1007/s00253-019-10165-x. [DOI] [PubMed] [Google Scholar]
- 156.Merenstein D., Fraser C.M., Roberts R.F., Liu T., Grant-Beurmann S., et al. Bifidobacterium animalis subsp. lactis BB-12 Protects against Antibiotic-Induced Functional and Compositional Changes in Human Fecal Microbiome. Nutrients. 2021;13 doi: 10.3390/nu13082814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mocanu V., Rajaruban S., Dang J., Kung J.Y., Deehan E.C., et al. Repeated fecal microbial transplantations and antibiotic pre-treatment are linked to improved clinical response and remission in inflammatory bowel disease: a systematic review and pooled proportion meta-analysis. J Clin Med. 2021;10 doi: 10.3390/jcm10050959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Deleu S., Machiels K., Raes J., Verbeke K., Vermeire S. Short chain fatty acids and its producing organisms: An overlooked therapy for IBD? EBioMedicine. 2021;66 doi: 10.1016/j.ebiom.2021.103293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vancamelbeke M., Laeremans T., Vanhove W., Arnauts K., Ramalho A.S., et al. Butyrate does not protect against inflammation-induced loss of epithelial barrier function and cytokine production in primary cell monolayers from patients with ulcerative colitis. J Crohns Colitis. 2019;13:1351–1361. doi: 10.1093/ecco-jcc/jjz064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fritsch J., Garces L., Quintero M.A., Pignac-Kobinger J., Santander A.M., et al. Low-Fat, high-fiber diet reduces markers of inflammation and dysbiosis and improves quality of life in patients with ulcerative colitis. Clin Gastroenterol Hepatol. 2021;19:1189–1199.e1130. doi: 10.1016/j.cgh.2020.05.026. [DOI] [PubMed] [Google Scholar]
- 161.Wu Z., Huang S., Li T., Li N., Han D., et al. Gut microbiota from green tea polyphenol-dosed mice improves intestinal epithelial homeostasis and ameliorates experimental colitis. Microbiome. 2021;9:184. doi: 10.1186/s40168-021-01115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hu B., Yu S., Shi C., Gu J., Shao Y., et al. Amyloid-Polyphenol Hybrid Nanofilaments Mitigate Colitis and Regulate Gut Microbial Dysbiosis. ACS Nano. 2020;14:2760–2776. doi: 10.1021/acsnano.9b09125. [DOI] [PubMed] [Google Scholar]
- 163.Sarbagili Shabat C., Scaldaferri F., Zittan E., Hirsch A., Mentella M.C., et al. Use of Fecal transplantation with a novel diet for mild to moderate active ulcerative colitis: The CRAFT UC randomized controlled trial. J Crohns Colitis. 2021 doi: 10.1093/ecco-jcc/jjab165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sanders M.E., Merenstein D.J., Reid G., Gibson G.R., Rastall R.A. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol. 2019;16:605–616. doi: 10.1038/s41575-019-0173-3. [DOI] [PubMed] [Google Scholar]
- 165.Chen P., Xu H., Tang H., Zhao F., Yang C., et al. Modulation of gut mucosal microbiota as a mechanism of probiotics-based adjunctive therapy for ulcerative colitis, Microb. Biotechnol. 2020;13:2032–2043. doi: 10.1111/1751-7915.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.El Hage R., Hernandez-Sanabria E., Van de Wiele T. Emerging Trends in “Smart Probiotics”: Functional Consideration for the Development of Novel Health and Industrial Applications. Front Microbiol. 2017;8:1889. doi: 10.3389/fmicb.2017.01889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Martín R., Chain F., Miquel S., Lu J., Gratadoux J.J., et al. The commensal bacterium Faecalibacterium prausnitzii is protective in DNBS-induced chronic moderate and severe colitis models. Inflamm Bowel Dis. 2014;20:417–430. doi: 10.1097/01.MIB.0000440815.76627.64. [DOI] [PubMed] [Google Scholar]
- 168.Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermúdez-Humarán L.G., et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lenoir M., Martín R., Torres-Maravilla E., Chadi S., González-Dávila P., et al. Butyrate mediates anti-inflammatory effects of Faecalibacterium prausnitzii in intestinal epithelial cells through Dact3. Gut Microbes. 2020;12:1–16. doi: 10.1080/19490976.2020.1826748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Depommier C., Everard A., Druart C., Plovier H., Van Hul M., et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med. 2019;25:1096–1103. doi: 10.1038/s41591-019-0495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Qu S., Fan L., Qi Y., Xu C., Hu Y., et al. Akkermansia muciniphila Alleviates Dextran Sulfate Sodium (DSS)-Induced Acute Colitis by NLRP3 Activation. Microbiol Spectr. 2021 doi: 10.1128/Spectrum.00730-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang L., Tang L., Feng Y., Zhao S., Han M., et al. Akkermansia muciniphilaA purified membrane protein from or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8 T cells in mice. Gut. 2020 doi: 10.1136/gutjnl-2019-320105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhai R., Xue X., Zhang L., Yang X., Zhao L., et al. Strain-Specific Anti-inflammatory Properties of Two Akkermansia muciniphila Strains on Chronic Colitis in Mice. Front Cell Infect Microbiol. 2019;9:239. doi: 10.3389/fcimb.2019.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhang T., Ji X., Lu G., Zhang F. The potential of Akkermansia muciniphila in inflammatory bowel disease. Appl Microbiol Biotechnol. 2021;105:5785–5794. doi: 10.1007/s00253-021-11453-1. [DOI] [PubMed] [Google Scholar]
- 175.Mejía-Pitta A., Broset E., de la Fuente-Nunez C. Probiotic engineering strategies for the heterologous production of antimicrobial peptides. Adv Drug Deliv Rev. 2021;176 doi: 10.1016/j.addr.2021.113863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Praveschotinunt P., Duraj-Thatte A.M., Gelfat I., Bahl F., Chou D.B., Engineered E., et al. for the delivery of matrix-tethered therapeutic domains to the gut. Nat Commun. 1917;10(2019):5580. doi: 10.1038/s41467-019-13336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yan X., Liu X.Y., Zhang D., Zhang Y.D., Li Z.H., et al. Construction of a sustainable 3-hydroxybutyrate-producing probiotic Escherichia coli for treatment of colitis. Cell Mol Immunol. 2021;18:2344–2357. doi: 10.1038/s41423-021-00760-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wang L., Liao Y., Yang R., Zhu Z., Zhang L., et al. An engineered probiotic secreting Sj16 ameliorates colitis via Ruminococcaceae/butyrate/retinoic acid axis. Bioeng Transl Med. 2021;6 doi: 10.1002/btm2.10219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gibson G.R., Hutkins R., Sanders M.E., Prescott S.L., Reimer R.A., et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017;14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 180.Zhang X.F., Guan X.X., Tang Y.J., Sun J.F., Wang X.K., et al. Clinical effects and gut microbiota changes of using probiotics, prebiotics or synbiotics in inflammatory bowel disease: a systematic review and meta-analysis. Eur J Nutr. 2021;60:2855–2875. doi: 10.1007/s00394-021-02503-5. [DOI] [PubMed] [Google Scholar]
- 181.De Preter V., Joossens M., Ballet V., Shkedy Z., Rutgeerts P., et al. Metabolic profiling of the impact of oligofructose-enriched inulin in Crohn's disease patients: a double-blinded randomized controlled trial. Clin Transl Gastroenterol. 2013;4 doi: 10.1038/ctg.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mitsuyama K., Saiki T., Kanauchi O., Iwanaga T., Tomiyasu N., et al. Treatment of ulcerative colitis with germinated barley foodstuff feeding: a pilot study. Aliment Pharmacol Ther. 1998;12:1225–1230. doi: 10.1046/j.1365-2036.1998.00432.x. [DOI] [PubMed] [Google Scholar]
- 183.Hallert C., Björck I., Nyman M., Pousette A., Grännö C., et al. Increasing fecal butyrate in ulcerative colitis patients by diet: controlled pilot study. Inflamm Bowel Dis. 2003;9:116–121. doi: 10.1097/00054725-200303000-00005. [DOI] [PubMed] [Google Scholar]
- 184.Valcheva R., Koleva P., Martínez I., Walter J., Gänzle M.G., et al. Inulin-type fructans improve active ulcerative colitis associated with microbiota changes and increased short-chain fatty acids levels. Gut Microbes. 2019;10:334–357. doi: 10.1080/19490976.2018.1526583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Joossens M., De Preter V., Ballet V., Verbeke K., Rutgeerts P., et al. Effect of oligofructose-enriched inulin (OF-IN) on bacterial composition and disease activity of patients with Crohn's disease: results from a double-blinded randomised controlled trial. Gut. 2012;61:958. doi: 10.1136/gutjnl-2011-300413. [DOI] [PubMed] [Google Scholar]
- 186.Ishisono K., Mano T., Yabe T., Kitaguchi K. Dietary Fiber Pectin Ameliorates Experimental Colitis in a Neutral Sugar Side Chain-Dependent Manner. Front Immunol. 2019;10:2979. doi: 10.3389/fimmu.2019.02979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ye M.B., Lim B.O. Dietary pectin regulates the levels of inflammatory cytokines and immunoglobulins in interleukin-10 knockout mice. J Agric Food Chem. 2010;58:11281–11286. doi: 10.1021/jf103262s. [DOI] [PubMed] [Google Scholar]
- 188.Wei Y., Gong J., Zhu W., Tian H., Ding C., et al. Pectin enhances the effect of fecal microbiota transplantation in ulcerative colitis by delaying the loss of diversity of gut flora. BMC Microbiol. 2016;16:255. doi: 10.1186/s12866-016-0869-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Llewellyn S.R., Britton G.J., Contijoch E.J., Vennaro O.H., Mortha A., et al. Interactions Between Diet and the Intestinal Microbiota Alter Intestinal Permeability and Colitis Severity in Mice. Gastroenterology. 2018;154:1037–1046.e1032. doi: 10.1053/j.gastro.2017.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Cai X., Han Y., Gu M., Song M., Wu X., et al. Dietary cranberry suppressed colonic inflammation and alleviated gut microbiota dysbiosis in dextran sodium sulfate-treated mice. Food Funct. 2019;10:6331–6341. doi: 10.1039/c9fo01537j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sun Q., Du M., Kang Y., Zhu M.J. Prebiotic effects of goji berry in protection against inflammatory bowel disease. Crit Rev Food Sci Nutr. 2022:1–25. doi: 10.1080/10408398.2021.2015680. [DOI] [PubMed] [Google Scholar]
- 192.Oka A., Sartor R.B. Microbial-Based and Microbial-Targeted Therapies for Inflammatory Bowel Diseases. Dig Dis Sci. 2020;65:757–788. doi: 10.1007/s10620-020-06090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gibson P.R., Halmos E.P., Muir J.G. Review article: FODMAPS, prebiotics and gut health-the FODMAP hypothesis revisited. Aliment Pharmacol Ther. 2020;52:233–246. doi: 10.1111/apt.15818. [DOI] [PubMed] [Google Scholar]
- 194.Cox S.R., Prince A.C., Myers C.E., Irving P.M., Lindsay J.O., et al. Fermentable Carbohydrates [FODMAPs] Exacerbate Functional Gastrointestinal Symptoms in Patients With Inflammatory Bowel Disease: A Randomised. Double-blind, Placebo-controlled, Cross-over, Re-challenge Trial, J Crohns Colitis. 2017;11:1420–1429. doi: 10.1093/ecco-jcc/jjx073. [DOI] [PubMed] [Google Scholar]
- 195.Cox S.R., Lindsay J.O., Fromentin S., Stagg A.J., McCarthy N.E., et al. Effects of Low FODMAP Diet on symptoms, fecal microbiome, and markers of inflammation in patients with quiescent inflammatory bowel disease in a randomized trial. Gastroenterology. 2020;158:176–188.e177. doi: 10.1053/j.gastro.2019.09.024. [DOI] [PubMed] [Google Scholar]
- 196.Lewis J.D., Sandler R.S., Brotherton C., Brensinger C., Li H., et al. A Randomized trial comparing the specific carbohydrate diet to a mediterranean diet in adults with Crohn's disease. Gastroenterology. 2021;161:837–852.e839. doi: 10.1053/j.gastro.2021.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Swanson K.S., Gibson G.R., Hutkins R., Reimer R.A., Reid G., et al. The international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat Rev Gastroenterol Hepatol. 2020;17:687–701. doi: 10.1038/s41575-020-0344-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Green J.E., Davis J.A., Berk M., Hair C., Loughman A., et al. Efficacy and safety of fecal microbiota transplantation for the treatment of diseases other than Clostridium difficile infection: a systematic review and meta-analysis. Gut Microbes. 2020;12:1–25. doi: 10.1080/19490976.2020.1854640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kao D., Roach B., Silva M., Beck P., Rioux K., et al. Effect of oral capsule- vs colonoscopy-delivered fecal microbiota transplantation on recurrent clostridium difficile infection: a randomized clinical trial. JAMA. 2017;318:1985–1993. doi: 10.1001/jama.2017.17077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chen T., Zhou Q., Zhang D., Jiang F., Wu J., et al. Effect of faecal microbiota transplantation for treatment of clostridium difficile infection in patients with inflammatory bowel disease: a systematic review and meta-analysis of cohort studies. J Crohns Colitis. 2018;12:710–717. doi: 10.1093/ecco-jcc/jjy031. [DOI] [PubMed] [Google Scholar]
- 201.Jiang Z.D., Ajami N.J., Petrosino J.F., Jun G., Hanis C.L., et al. Randomised clinical trial: faecal microbiota transplantation for recurrent Clostridum difficile infection - fresh, or frozen, or lyophilised microbiota from a small pool of healthy donors delivered by colonoscopy. Aliment Pharmacol Ther. 2017;45:899–908. doi: 10.1111/apt.13969. [DOI] [PubMed] [Google Scholar]
- 202.Hvas C.L., Dahl Jørgensen S.M., Jørgensen S.P., Storgaard M., Lemming L., et al. Fecal microbiota transplantation is superior to fidaxomicin for treatment of recurrent clostridium difficile infection. Gastroenterology. 2019;156:1324–1332.e1323. doi: 10.1053/j.gastro.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 203.Allegretti J.R., Mullish B.H., Kelly C., Fischer M. The evolution of the use of faecal microbiota transplantation and emerging therapeutic indications. Lancet. 2019;394:420–431. doi: 10.1016/S0140-6736(19)31266-8. [DOI] [PubMed] [Google Scholar]
- 204.Rossen N.G., Fuentes S., van der Spek M.J., Tijssen J.G., Hartman J.H., et al. Findings From a Randomized Controlled Trial of Fecal Transplantation for Patients With Ulcerative Colitis. Gastroenterology. 2015;149:110–118.e114. doi: 10.1053/j.gastro.2015.03.045. [DOI] [PubMed] [Google Scholar]
- 205.Moayyedi P., Surette M.G., Kim P.T., Libertucci J., Wolfe M., et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology. 2015;149:102–109.e106. doi: 10.1053/j.gastro.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 206.Costello S.P., Hughes P.A., Waters O., Bryant R.V., Vincent A.D., et al. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: a randomized clinical trial. JAMA. 2019;321:156–164. doi: 10.1001/jama.2018.20046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Paramsothy S., Nielsen S., Kamm M.A., Deshpande N.P., Faith J.J., et al. Specific bacteria and metabolites associated with response to fecal microbiota transplantation in patients with ulcerative colitis. Gastroenterology. 2019;156:1440. doi: 10.1053/j.gastro.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 208.Angelberger S., Reinisch W., Makristathis A., Lichtenberger C., Dejaco C., et al. Temporal bacterial community dynamics vary among ulcerative colitis patients after fecal microbiota transplantation. Am J Gastroenterol. 2013;108:1620–1630. doi: 10.1038/ajg.2013.257. [DOI] [PubMed] [Google Scholar]
- 209.Quera R., Espinoza R., Estay C., Rivera D. Bacteremia as an adverse event of fecal microbiota transplantation in a patient with Crohn's disease and recurrent Clostridium difficile infection. J Crohns Colitis. 2014;8:252–253. doi: 10.1016/j.crohns.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 210.Cui B., Feng Q., Wang H., Wang M., Peng Z., et al. Fecal microbiota transplantation through mid-gut for refractory Crohn's disease: safety, feasibility, and efficacy trial results. J Gastroenterol Hepatol. 2015;30:51–58. doi: 10.1111/jgh.12727. [DOI] [PubMed] [Google Scholar]
- 211.Chande N., Costello S.P., Limketkai B.N., Parker C.E., Nguyen T.M., et al. Alternative and complementary approaches for the treatment of inflammatory bowel disease: evidence from cochrane reviews. Inflamm Bowel Dis. 2020;26:843–851. doi: 10.1093/ibd/izz223. [DOI] [PubMed] [Google Scholar]
- 212.Long D., Merlin D. Micro- and nanotechnological delivery platforms for treatment of dysbiosis-related inflammatory bowel disease. Nanomedicine (Lond) 2021;16:1741–1745. doi: 10.2217/nnm-2021-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Teng Y., Ren Y., Sayed M., Hu X., Lei C., et al. Plant-Derived Exosomal MicroRNAs Shape the Gut Microbiota. Cell Host Microbe. 2018;24:637–652.e638. doi: 10.1016/j.chom.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hsiao E.Y., McBride S.W., Hsien S., Sharon G., Hyde E.R., et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Pluznick J.L., Protzko R.J., Gevorgyan H., Peterlin Z., Sipos A., et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci U S A. 2013;110:4410–4415. doi: 10.1073/pnas.1215927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ghebretatios M., Schaly S., Prakash S. Nanoparticles in the Food Industry and Their Impact on Human Gut Microbiome and Diseases. Int J Mol Sci. 2021;22 doi: 10.3390/ijms22041942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lee Y., Sugihara K., Gillilland M.G., 3rd, Jon S., Kamada N., et al. Hyaluronic acid-bilirubin nanomedicine for targeted modulation of dysregulated intestinal barrier, microbiome and immune responses in colitis. Nat Mater. 2020;19:118–126. doi: 10.1038/s41563-019-0462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lohoff T., Ghazanfar S., Missarova A., Koulena N., Pierson N., et al. Integration of spatial and single-cell transcriptomic data elucidates mouse organogenesis. Nat Biotechnol. 2021 doi: 10.1038/s41587-021-01006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Zeevi D., Korem T., Godneva A., Bar N., Kurilshikov A., et al. Structural variation in the gut microbiome associates with host health. Nature. 2019;568:43–48. doi: 10.1038/s41586-019-1065-y. [DOI] [PubMed] [Google Scholar]
- 220.Weng Y.J., Gan H.Y., Li X., Huang Y., Li Z.C., et al. Correlation of diet, microbiota and metabolite networks in inflammatory bowel disease. J Dig Dis. 2019;20:447–459. doi: 10.1111/1751-2980.12795. [DOI] [PubMed] [Google Scholar]
- 221.Lee T., Clavel T., Smirnov K., Schmidt A., Lagkouvardos I., et al. Oral versus intravenous iron replacement therapy distinctly alters the gut microbiota and metabolome in patients with IBD. Gut. 2017;66:863–871. doi: 10.1136/gutjnl-2015-309940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Jacobs J.P., Goudarzi M., Singh N., Tong M., McHardy I.H., et al. A Disease-Associated Microbial and Metabolomics State in Relatives of Pediatric Inflammatory Bowel Disease Patients, Cell Mol. Gastroenterol Hepatol. 2016;2:750–766. doi: 10.1016/j.jcmgh.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


