Abstract
Ornithine decarboxylase (ODC) is the rate-limiting enzyme for the synthesis of polyamines (PAs). PAs are oncometabolites that are required for proliferation, and pharmaceutical ODC inhibition is pursued for the treatment of hyperproliferative diseases, including cancer and infectious diseases. The most potent ODC inhibitor is 1-amino-oxy-3-aminopropane (APA). A previous crystal structure of an ODC–APA complex indicated that APA non-covalently binds ODC and its cofactor pyridoxal 5-phosphate (PLP) and functions by competing with the ODC substrate ornithine for binding to the catalytic site. We have revisited the mechanism of APA binding and ODC inhibition through a new crystal structure of APA-bound ODC, which we solved at 2.49 Å resolution. The structure unambiguously shows the presence of a covalent oxime between APA and PLP in the catalytic site, which we confirmed in solution by mass spectrometry. The stable oxime makes extensive interactions with ODC but cannot be catabolized, explaining APA’s high potency in ODC inhibition. In addition, we solved an ODC/PLP complex structure with citrate bound at the substrate binding pocket. These two structures provide new structural scaffolds for developing more efficient pharmaceutical ODC inhibitors.
Introduction
ODC is a PLP-dependent enzyme that catalyzes the decarboxylation of ornithine to putrescine, the rate-limiting step in the biosynthesis of polyamines (PAs). PAs are required for proliferation and their synthesis and degradation are tightly regulated, including at the levels of ODC transcription, translation, degradation, and activity [1–6]. ODC is a transcriptional target of MYC that is frequently dysregulated in MYC-amplified tumors [7–10] and pharmacological inhibition of ODC is anti-proliferative [11, 12]. The most studied ODC inhibitor is α-difluoromethylornithine (DFMO), an ODC suicide inhibitor and FDA approved drug for the treatment of African sleeping sickness [13, 14] that has been in clinical trials as a cancer chemotherapeutic [15–18]. However, DFMO has a low ODC affinity [19], high renal clearance and a short half-life and thus requires an exceptionally high dose (up to 10.8 g/m2/day in adult patients [18] and 3 g/m2/day in pediatric patients [15]). In contrast, the potency of APA has been reported to be 3–4 orders of magnitude higher than that of DFMO [19–21], making APA a promising lead for the further development of anti-proliferative drugs.
ODC forms an obligate homodimer. Its catalytic center is located at the dimer interface and is formed by residues from both monomers. In the absence of a free amino acid, ODC covalently binds its cofactor PLP through formation of an internal Schiff base with the ε-amino group of ODC K69. A nucleophilic amino group nitrogen can attack the aldehyde group of PLP, which requires the amino group to be non-protonated. The α-amino group of a free amino acid such as ornithine has a lower pKa (~9.0) than an ε-amino group (~ 10.5). It is therefore more reactive (higher proportion non-protonated) and displaces the ε-amino group to form an external aldimine Schiff base with PLP. In turn, the ε-amino group of K69 rotates away from PLP and forms a charge interaction with the carboxyl group of D88, which further stabilizes the transamination reaction [22, 23]. The highly electrophilic Schiff base nitrogen promotes decarboxylation by functioning as sink for the free electron from the carboxylate group, followed by re-protonation through the weakly acidic SH group of C360 from the other monomer. Decarboxylation increases the pKa of the Schiff base nitrogen to allow displacement of the decarboxylation product, putrescine, by ODC K69 through transamination to close the catalytic cycle (Fig. 1).
Figure 1. ODC catalytic cycle and the inhibition mechanism by APA.
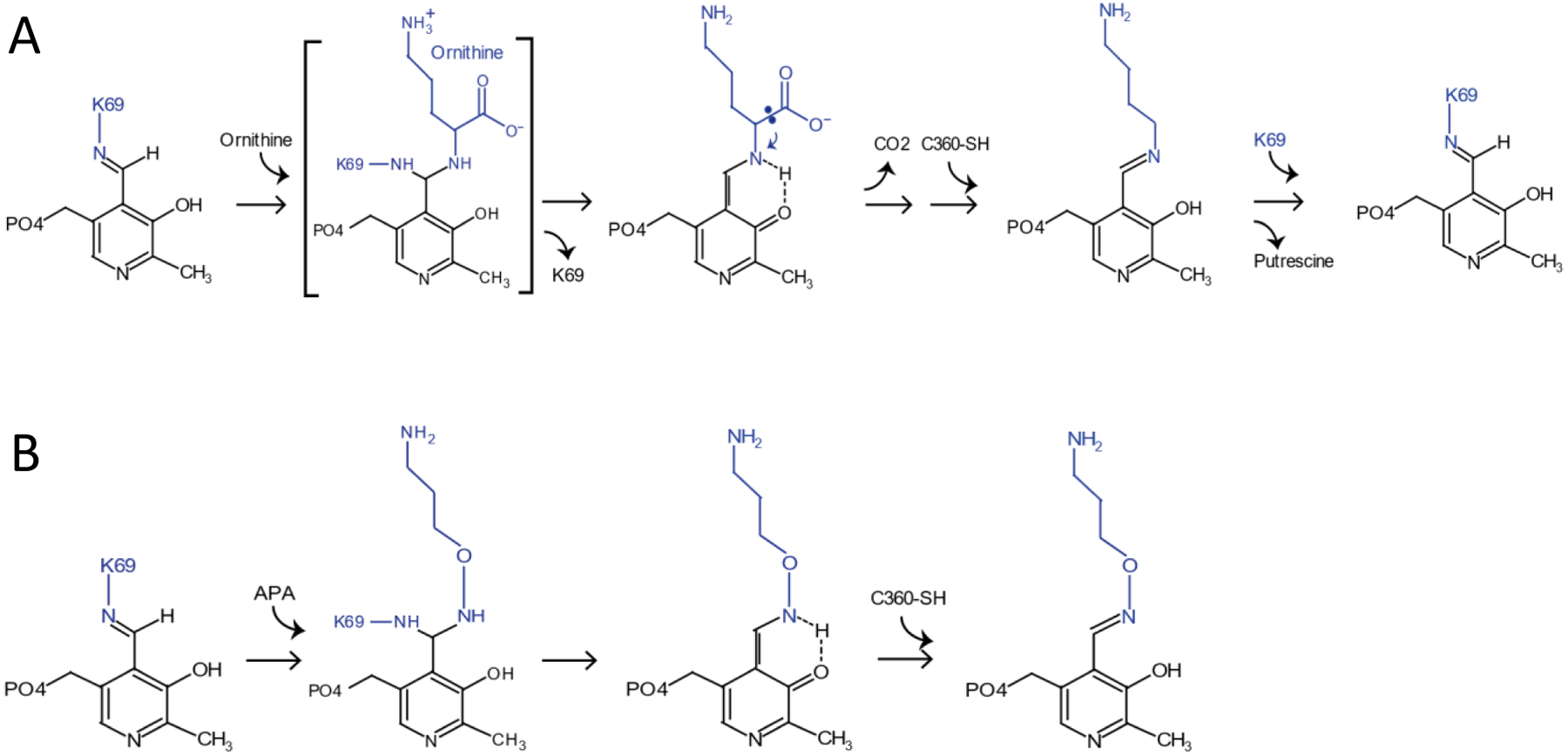
A, PLP binds to K69 of ODC through a Schiff base linkage with the ε-amino group of K69. The α-amino group of ornithine displaces the ε-amino group to form an external aldimine Schiff base with PLP. The highly electrophilic Schiff base nitrogen promotes decarboxylation of ornithine. The decarboxylation increases the pKa of the Schiff base nitrogen to allow displacement of the decarboxylation product, putrescine, by ODC K69 through transamination to close the catalytic cycle. B, APA mimics ornithine to bind to PLP and forms an oximine bond with PLP, which is much more stable than an aldimine bond, thus blocks the catalytic center from the access of ornithine.
APA is a cell-permeable isosteric analog of putrescine that inhibits ODC at low nanomolar concentrations [19–21] and has antiproliferative effects on tumor cells [24–26]. Its amino-oxy group has a pKa of ~5 and is therefore unprotonated at physiological pH, making it more reactive to forming a Schiff base with PLP than ornithine or putrescine. If APA is appropriately positioned in the ODC ornithine binding pocket to allow it to react with PLP, it would be expected to form a highly stable PLP adduct that cannot be catabolized by ODC as proposed previously [27]. This mechanism is notably analogous to the inhibition of mitochondrial aspartate amino transferase by hydroxylamine derivatives [28]. Unexpectedly, the published crystal structure of APA-bound ODC/PLP, which included the putrescine-related diamine cadaverine as crystallization additive, showed APA non-covalently bound in the pocket with the APA amino-oxy group in hydrogen bond distance to the PLP aldehyde group [19]. To explore APA as a scaffold for new ODC inhibitors, we revisited ODC binding and inhibition by APA, and solved a crystal structure of the ODC/PLP/APA complex with APA positioned at the substrate binding site and covalently bound to PLP through the formation of a Schiff base linkage. Surprisingly, we also obtained a citrate-bound ODC structure, in which citrate occupies a pocket at the catalytic center that is much larger than those occupied by the substrate ornithine and the inhibitor APA. The two structures provide new templates for developing more efficient inhibitors that block not only the catalytic center but the whole substrate binding pocket.
Results and Discussion
APA was prepared as shown in Fig. 2A. We determined its identity by 1H-NMR and mass spectrometry (see Methods), which is identical to that reported by others, prepared by an alternate route [29], and confirmed by its ability to inhibit the catalytic activity of ODC (Fig. 2B). Full length human ODC was expressed in E. coli as a His6-tagged protein and purified by Ni-affinity chromatography and size exclusion chromatography. Co-crystallization of ODC with PLP and APA yielded crystals under a number of different conditions, but the crystals diffracted only to moderate resolution, likely due to ODC’s highly flexible C-terminus. The C-terminus contains two degrons for ubiquitin-independent degradation of ODC by the 26S proteasome, which is promoted by the ODC-specific inhibitor protein, antizyme AZ1 [30]. Since the C-terminus of ODC is not required for its enzymatic function [31], we generated a construct lacking the C-terminal 38 amino acids. This region includes the C-terminal degron and was disordered in all structures of full length ODC, both in the presence and absence of AZ1 [32–34]. This allowed us to determine the structure of ODC(1–423) bound to PLP and APA at a resolution of 2.49 Å (Fig. 3A,C). We have also determined the structure of the PLP-bound ODC construct in the absence of APA at a resolution of 1.7 Å (Figs. 3B,D). The structure statistics are shown in Table 1.
Figure 2: APA synthesis and inhibitory function.
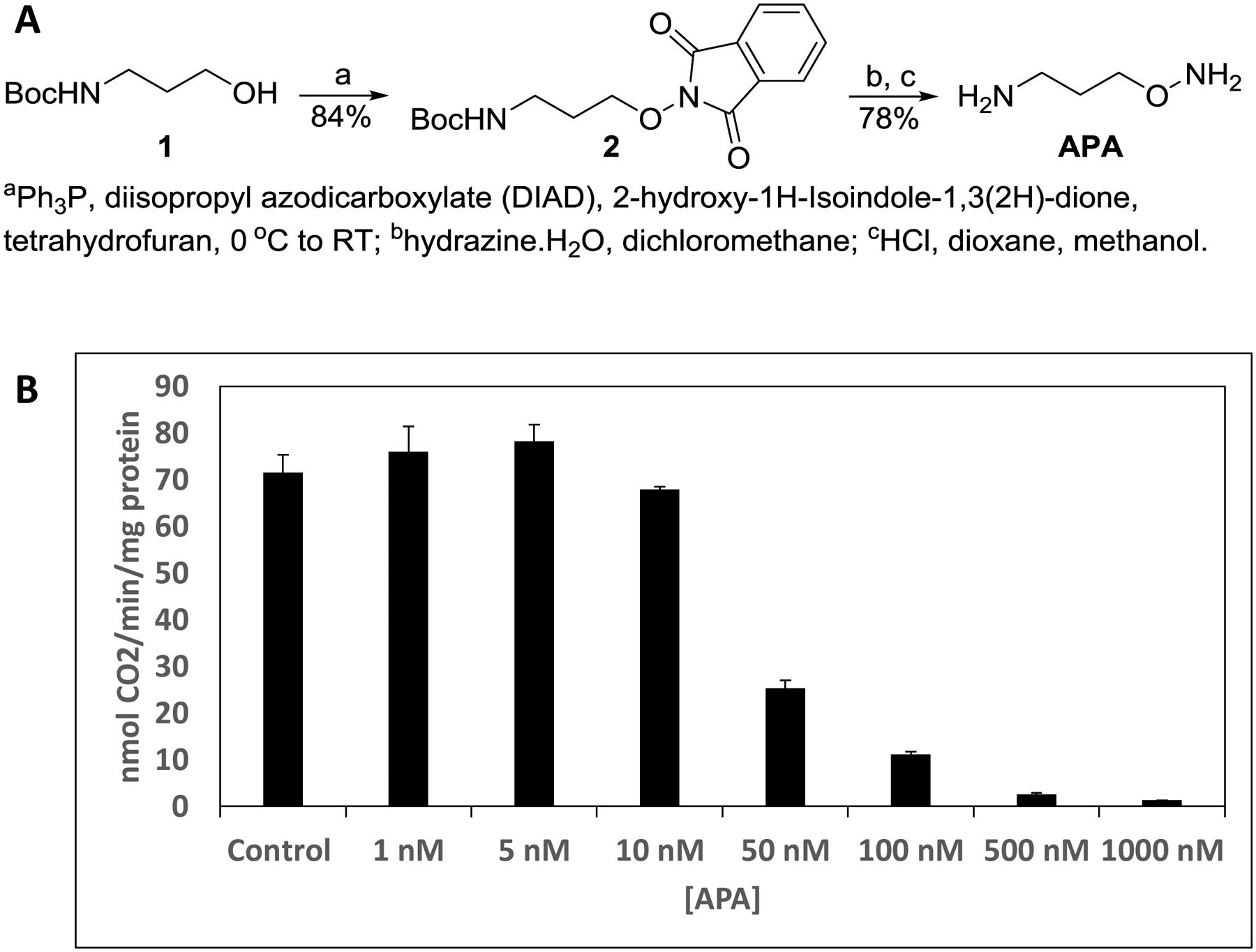
A. Synthesis scheme. B. Synthesized APA inhibits the enzymatic activity of ODC. Note that ODC in the enzymatic assay has a concentration of 40 nM, therefore APA at concentrations less than 40 nM can only inhibit a fraction of the activity.
Figure 3: Structures of apo and APA-bound ODC.
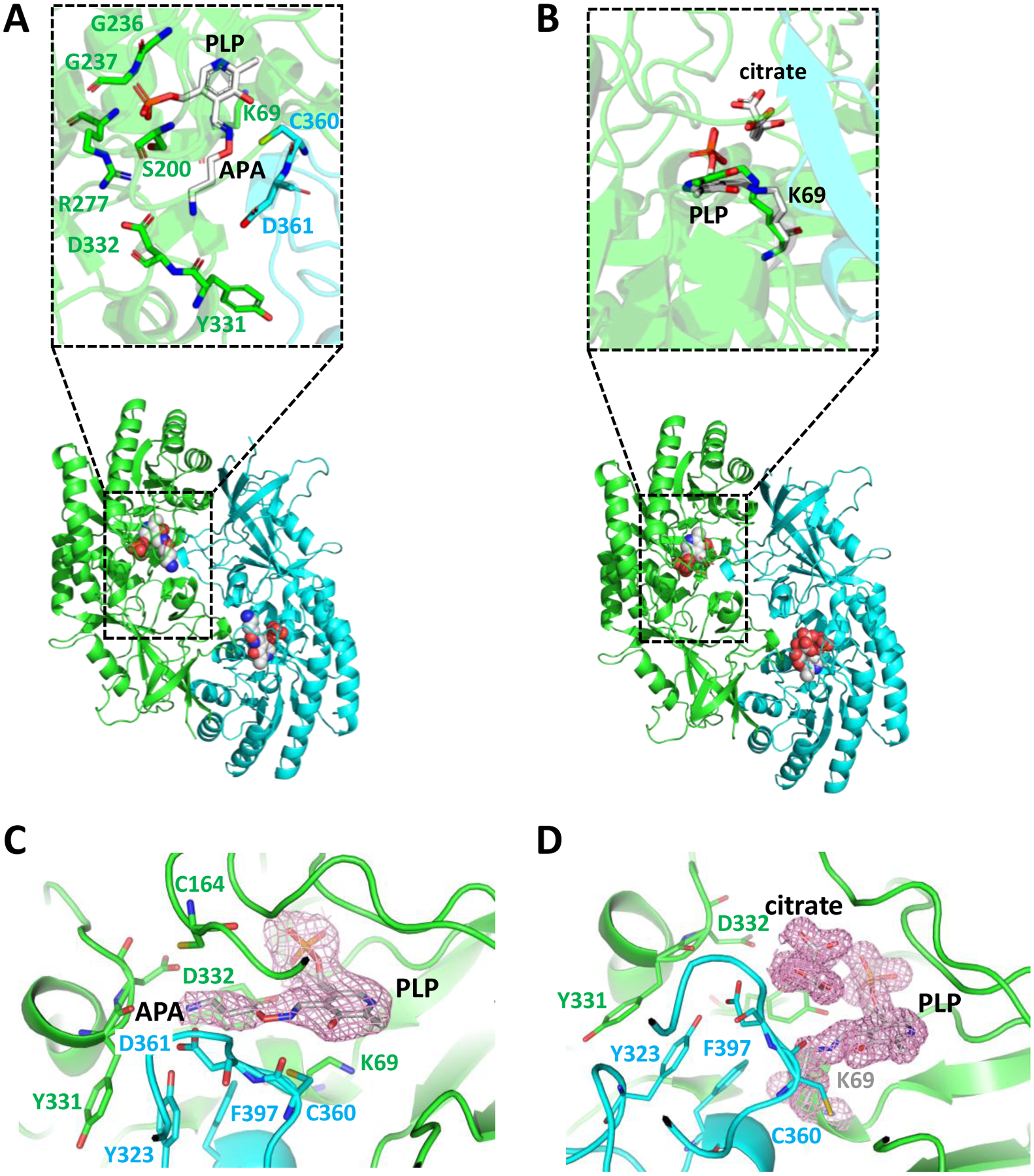
A,B. Overview ODC cartoon structures with catalytic site zoom-in in insets. A. APA-bound ODC/PLP. The amino-oxy group of APA forms a Schiff base with the aldehyde group of PLP. B. Apo-ODC/PLP with bound citrate. Citrate stabilizes the Schiff base bond between PLP and K69 of ODC. PLP and K69 are shown as white stick model in the inset, overlaid with structurally aligned PLP and K69 from apo ODC PDB 1D7K in green. The two monomers of the ODC homodimer are shown in green and cyan cartoon representation, respectively. PLP, PLP-APA, and citrate are shown in sphere representation in the overview cartoon models. C. Close-up of PLP-APA in the catalytic center of ODC with 2mFo-DFc density map contoured at 1 σ. D. Close-up of PLP and citrate in the catalytic center of ODC with 2mFo-DFc density map contoured at 1 σ. Key residues directly binding to APA are shown in stick presentation.
Table 1.
Data collection and refinement statistics (molecular replacement)
| ODC1/PLP/Citrate | ODC1/PLP/APA | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 74.5, 86.5 154.6 | 75.0, 86.9, 154.5 |
| α, β, γ () | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 56–1.66 (1.70–1.66) * | 46–2.49 (2.55–2.49) |
| Rsym or Rmerge | 0.06 (1.16) | 0.08 (0.90) |
| I / σI | 19.2 (1.7) | 11.6 (1.7) |
| Completeness (%) | 100 (100) | 99.7 (100) |
| Redundancy | 8.2 (8.2) | 5.2 (5.3) |
| Refinement | ||
| Resolution (Å) | 42–1.66 (1.68–1.66) | 36–2.49 (2.56–2.49) |
| No. reflections | 118,397 (3597) | 35957 (2583) |
| Rwork / Rfree | 17.8/19.7 (26.8/27.2) | 18.0/20.8 (28.6/34.2) |
| No. residues/atoms | ||
| Protein | 808/6349 | 816/6387 |
| Ligand | 4/56 | 2/42 |
| Water | 501 | 183 |
| B-factors | ||
| Protein | 26.8 | 68.6 |
| Ligand | 18.9 | 69.9 |
| Water | 33.0 | 61.2 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.013 | 0.006 |
| Bond angles (°) | 1.315 | 0.801 |
| Ramachandran plot (%) | ||
| Favored | 98.4 | 98.3 |
| Outliers | 0.0 | 0.0 |
| Clash score | 2.12 | 3.10 |
| Molprobity score | 1.03 | 1.10 |
Values in parentheses are for highest-resolution shell.
As expected, our APA-free structure looks very similar to the previously determined structure of PLP-bound ODC (PDB 1D7K) [34] with PLP covalently bound to K69 (Figs. 3B). The two structures are superimposable with an RMSD value of 1.94 Å. However, in our structure, citrate from the crystallization solution (100 mM tri-sodium citrate) was found above the substrate binding pocket. Citrate binding shifted the position of the Schiff base carbon atom by 0.6 Å, allowing a more stable Schiff base double bond that is in the same plane as the PLP aromatic ring (Fig. 3B inset and Fig. 3D). Since the position of citrate partially overlaps with the binding site of ornithine, citrate would be expected to function as a competitive inhibitor and does indeed inhibit ODC at high concentrations (Fig. 4). In the structure, the citrate occupies most of the substrate binding pocket (which is about 350 Å2), much more than those occupied by APA, putrescine, or ornithine. This structure, therefore, provides a new scaffold for developing more efficient inhibitors that block not only the catalytic center but also the extended substrate binding pocket of ODC. Interestingly, bacterial decarboxylases from the same class as human ODC (e.g., Francicella FslC and Staphylococcus SaSbnH) use the adducts of amino acids and citrate as decarboxylation substrates for the synthesis of siderophores [35, 36]. In the corresponding structures (e.g., PDB 6KNH and 6KNK), the citryl moiety inserts into a groove at the dimer interface below the corresponding position of ornithine and interacts with both subunits. However, this groove is not related to the wide citrate-binding pocket that we have identified in human ODC, in which citate adopts a position parallel to PLP.
Figure 4: Citrate weakly inhibits ODC activity.
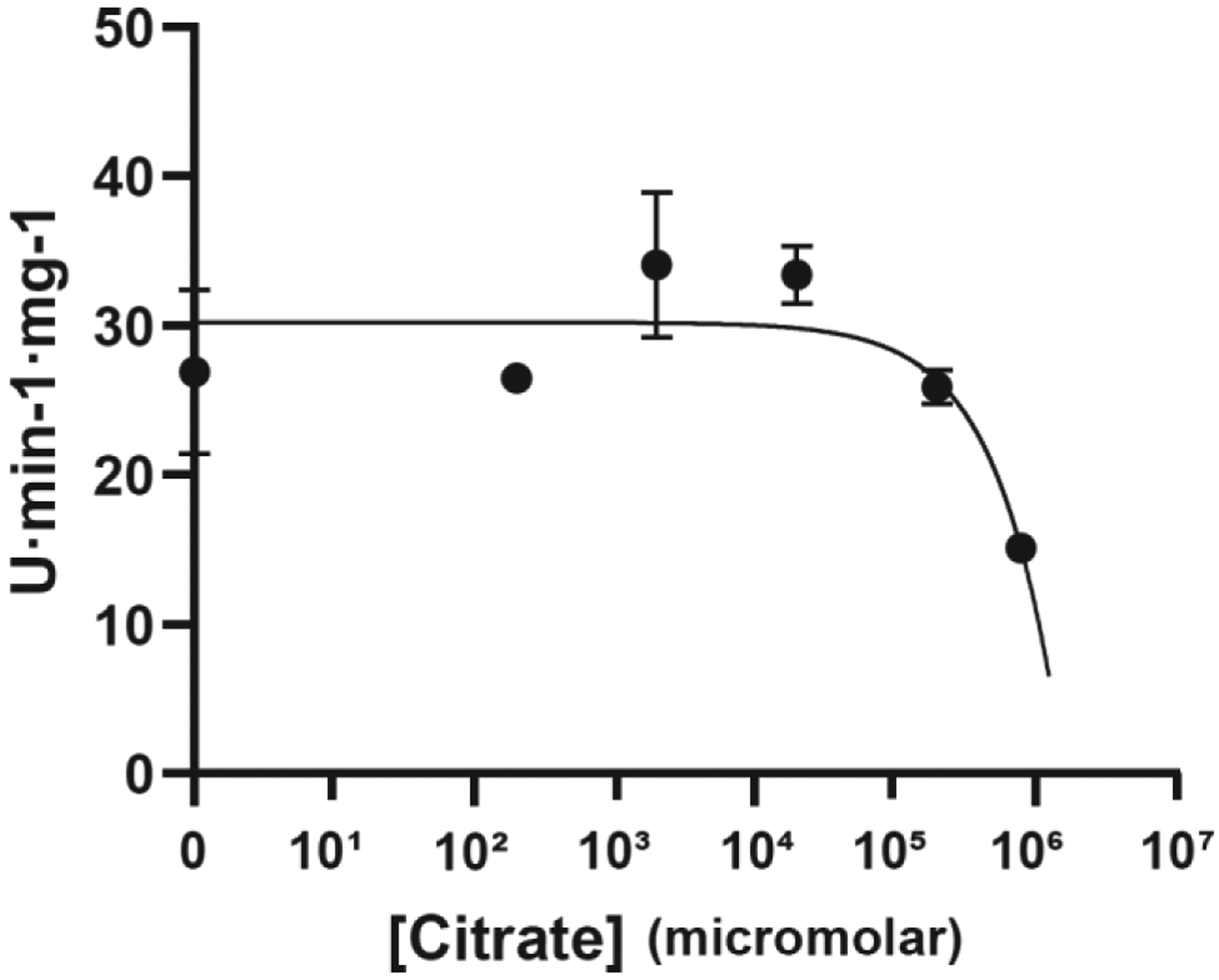
ODC activity in the presence of different concentrations of citrate.
Differences between our structure of APA-bound ODC/PLP and the previously reported structure of ODC/PLP/APA/cadaverine (PDB 2OO0) were mainly found in loop regions, most notably in the position of the protease-sensitive loop (residues 197–205). The charged amino groups of APA in both structures, as well as in K69, ornithine, and putrescine in previously solved structures [22, 37, 38], are all anchored in the negatively charged pocket formed by D332, D361, and Y331 (Figs. 5 and 6). However, in contrast to PDB 2OO0, APA in our structure formed a cis oxime Schiff base with PLP as was previously predicted based on its chemistry [27, 28] (Fig. 3A inset). APA and the PLP ring are co-planar in the APA-PLP adduct (Fig. 3A inset and Fig. 3C), making extensive interactions with 18 ODC residues, 15 from the same monomer and 3 from the other monomer. These include direct hydrogen bonds with H197, S200, E274, G276, R277, Y331, D332, D361, and Y389 as well as water-mediated hydrogen bonds with F238, Y278, Y323, N327, and Y389 (Fig. 5A). These extensive interactions are in contrast to PDB 2OO0, in which APA makes interactions with only nine ODC residues and with PLP (Fig. 5B), significantly fewer than the residues that interact with the DFMO-PLP adduct in the structure of Trypanosoma brucei (Tb)-ODC/PLP/DFMO (PDB 2TOD) [39].
Figure 5: Interaction diagrams for APA-PLP (A) and APA (B).
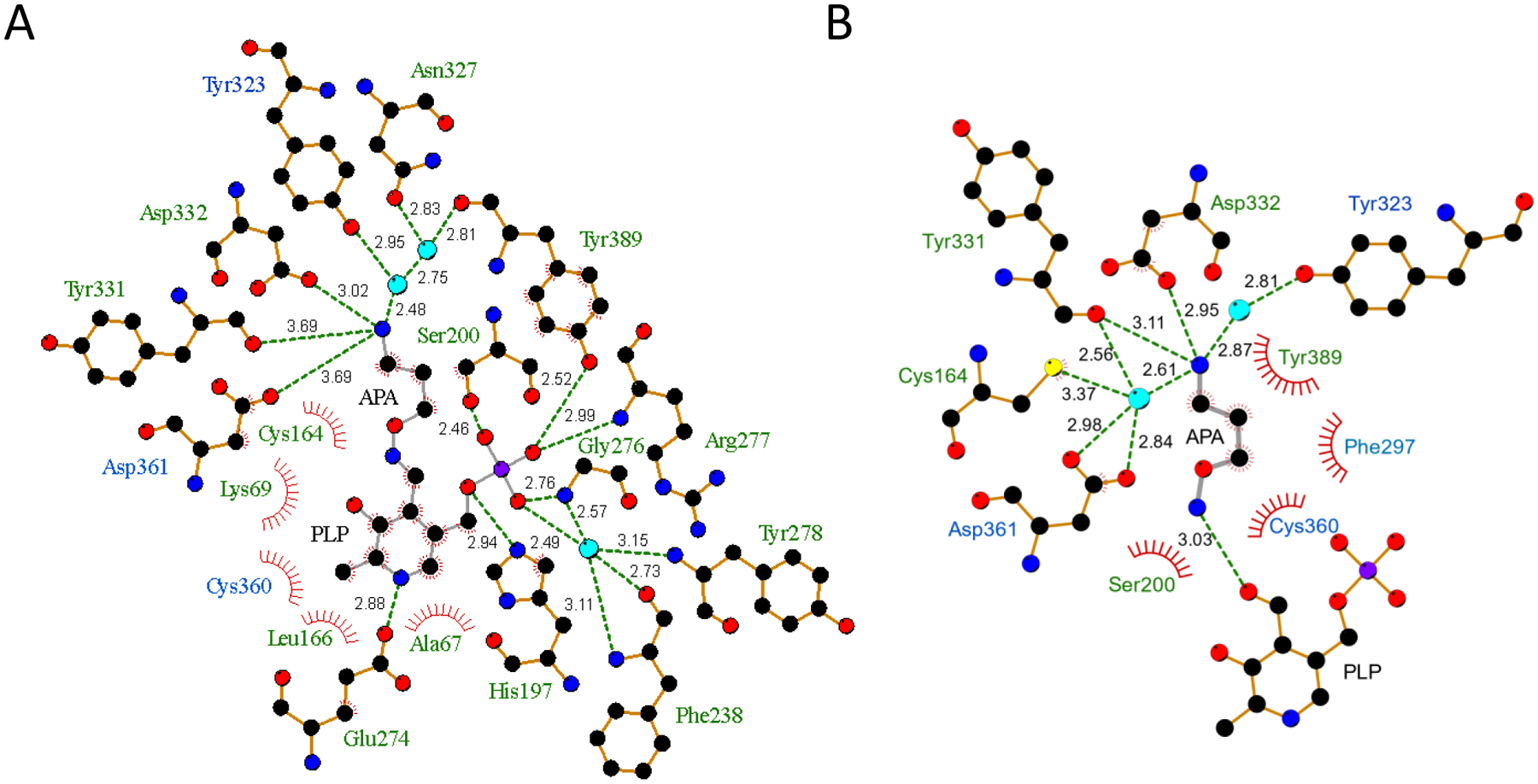
Diagrams were generated with LigPlot+. APA and APA-PLP are shown in grey stick representation, ODC residues that form hydrogen bonds are shown in orange stick representation, and structured water molecules as cyan spheres. Hydrogen bonds are shown as dashed lines with bond lengths labeled, and Van-der-Waals interactions as spoked arcs. Residues colored in green are from the same monomer as PLP and residues colored in blue are from the other monomer.
Figure 6: Structure close-ups of the catalytic center of different ODC complexes.
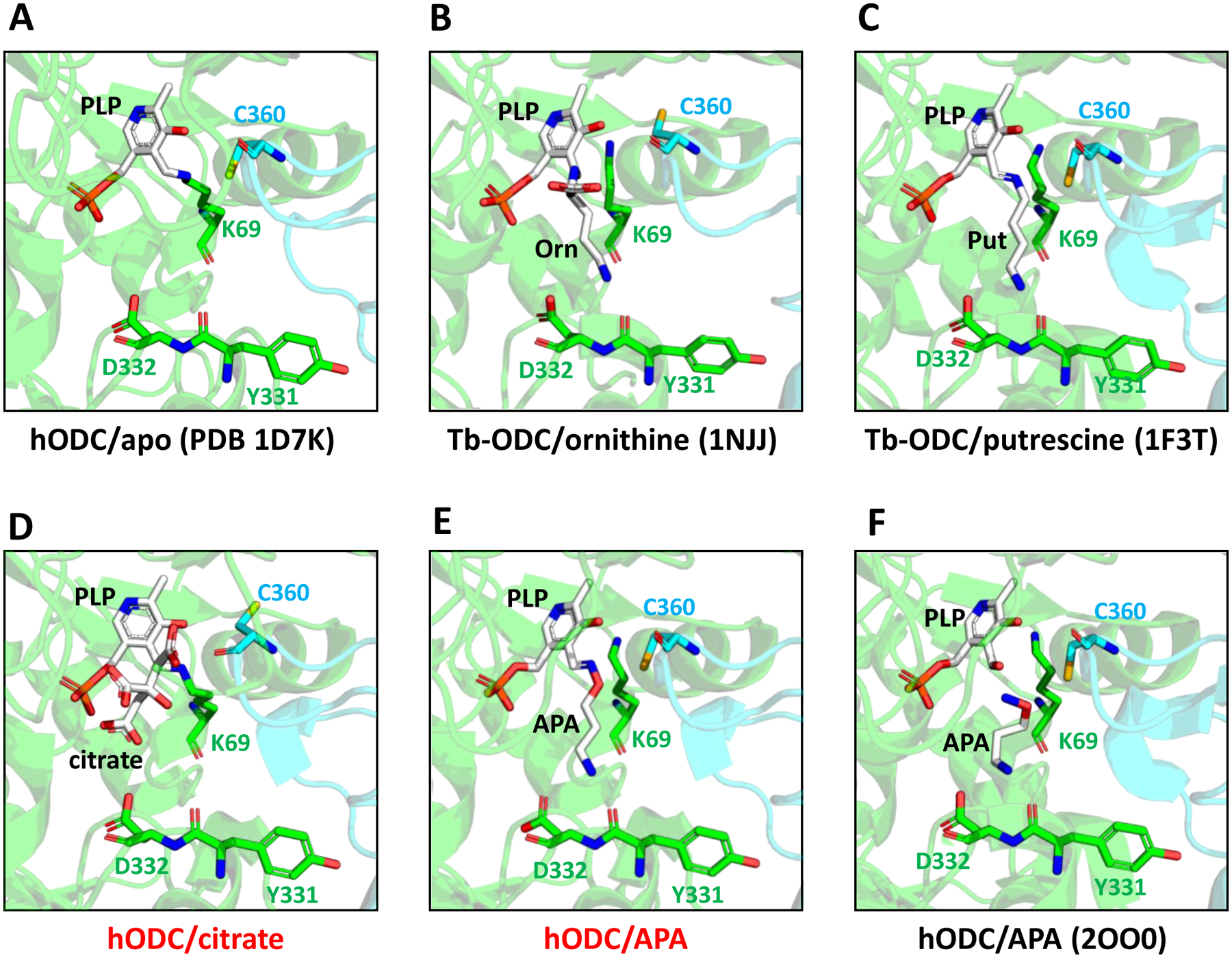
A. Human (h) apo-ODC. PLP (white) forms an internal aldimine with K69. The two monomers of the ODC homodimer are shown in green and cyan. B,C. Trypanosoma brucei (Tb) ODC bound to ornithine (B) and putrescine (C). D,E. hODC bound to citrate (D) and APA (E). F. Previously published structure (PDB 2OO0) of APA-bound hODC. PDB 2OO0 also contains cadaverine, but the position of cadaverine is outside of the binding pocket regions shown. Red letters: structures determined for this study; black letters: previously determined structures. Key residues are shown in stick representation.
Since APA and putrescine are isosteric and form Schiff bases with PLP, they were expected to adopt very similar positions in the ODC ligand-binding pocket. Indeed, the position of putrescine in the structure of Tb-ODC (PDB 1F3T) closely overlays with the APA model in our structure (compare Figs. 6C and 6E), whereas the modeled APA in PDB 2OO0 adopted a bent conformation in which the amino-oxy group points away from PLP (Fig. 6F). PLP adopted only slightly varied positions in apo-, substrate-, product-, and inhibitor-bound structures (Figs. 5 and 6A–F). Its phosphate group is anchored in a positively charged pocket and forms hydrogen bonds with S200, G237, F238, G276, R277, and Y389, while its aromatic ring stacks against H197 and forms hydrogen bonds with R154 and D360, and Van-der-Waals interactions with A67 (Fig. 5). A key catalytic residue is C360. The C360 side chain points away from the negatively charged carboxyl groups of ornithine (Fig. 6B) and citrate (Fig. 6D) but rotates into the binding pocket upon substrate decarboxylation or when the pocket is occupied by molecules like APA that are non-charged at this position (Fig. 6E,F).
To test which form of APA is bound to ODC in solution, we incubated purified ODC with a five-fold molar excess of PLP and a ten-fold molar excess of APA as we had done for crystallization. Unbound APA and PLP were removed by first ultrafiltration, followed by gel filtration through a G-25 desalting column. Mass spectrometry revealed signals for APA, PLP, and APA-PLP. The APA-PLP compound ion intensity was 150 times higher than the APA ion intensity and 491 times higher than the PLP signal (Fig. 7). While differences in ionization efficiencies do not allow exact quantitation, the data clearly demonstrate that APA-PLP is the predominant APA form detected co-purifying with ODC. This is consistent with the high reactivity of APA toward aldehydes and explains the very high affinity of APA for ODC (more than three orders of magnitude lower IC50 for human ODC than DFMO) despite the comparatively small number of interactions of APA when not covalently bound to PLP (Fig. 5). In spite of its high affinity, the APA-PLP adduct binds ODC non-covalently and the mode of inhibition is competitive as shown by APA increasing the Km of the reaction without decreasing its Vmax (Fig. 8).
Fig. 7. Average MS1 of ODC bound APA-PLP.
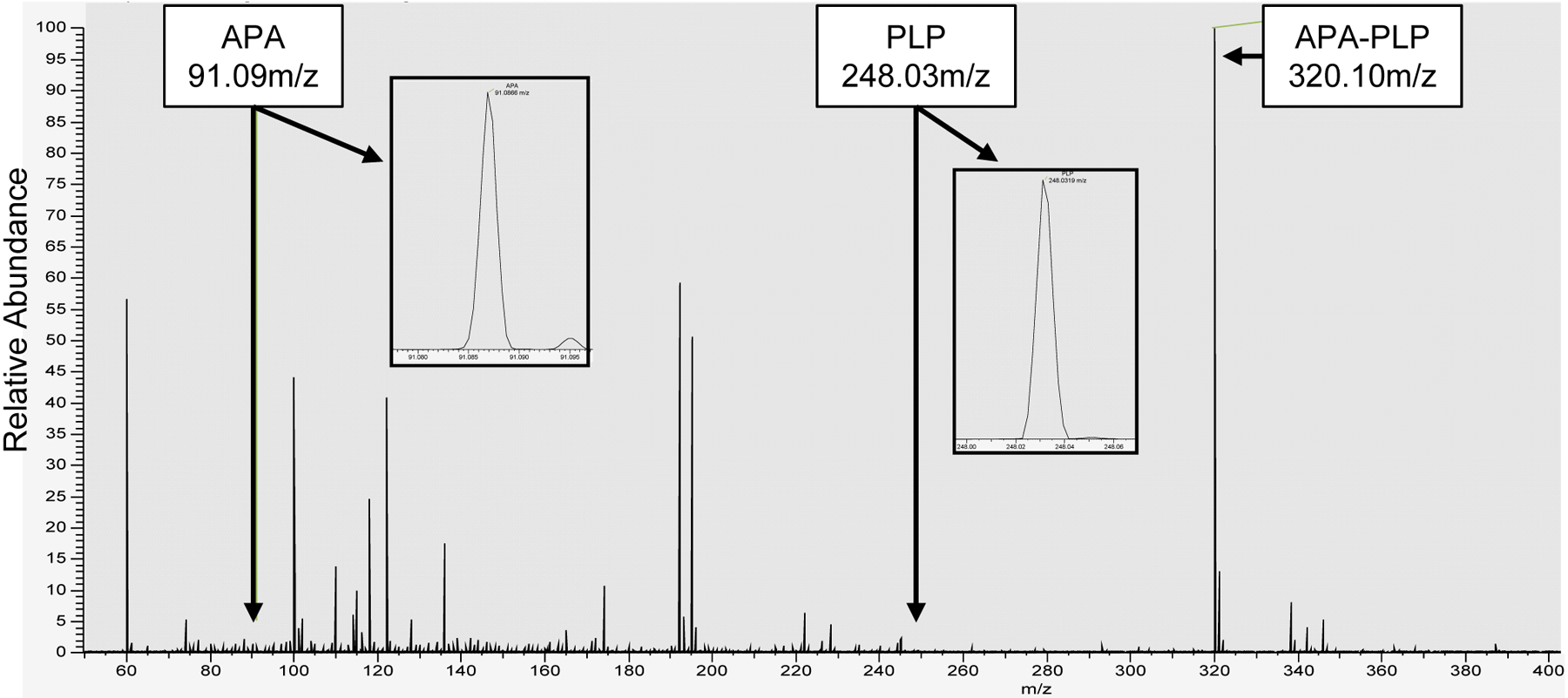
The mass spectrum shows that APA-PLP is the predominant detected ion (320.10 m/z) at normalized intensity (NL) of 4.40E7. This intensity is two orders of magnitude higher than that of the APA (91.09 m/z) and PLP (248.03 m/z) ion signals. Inserts display close-up spectra of the APA (NL: 3.01E5) and PLP (NL: 9.51E4) signals.
Figure 8: APA increases the KM of ODC without affecting Vmax.
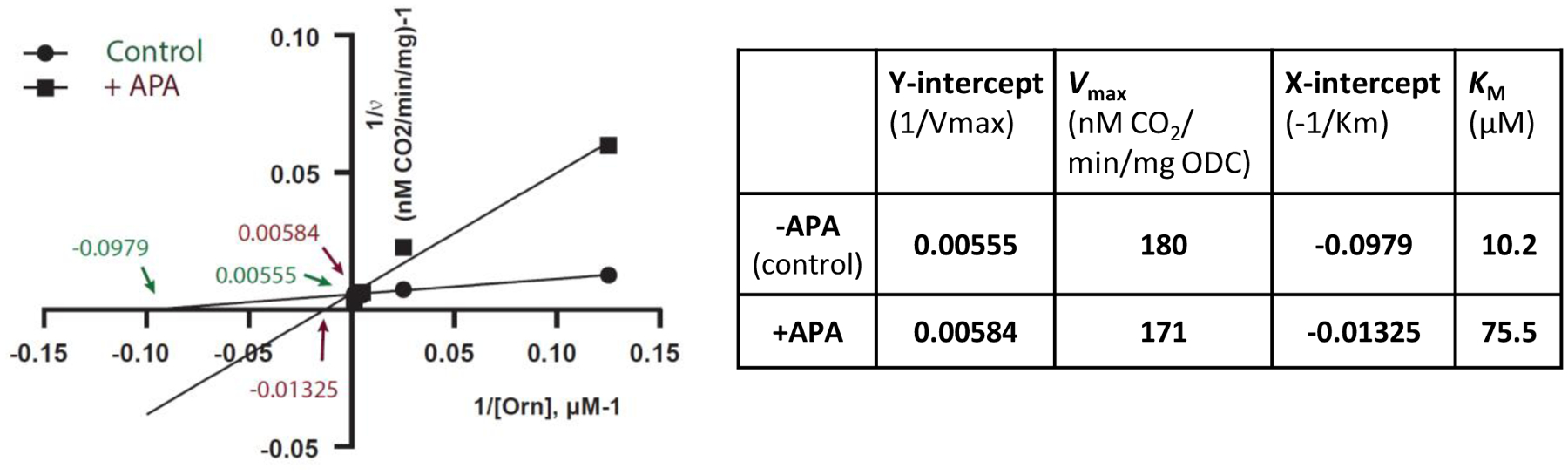
Lineweaver-Burk plot with Vmax and KM determined in the absence (control) or in the presence of the inhibitor APA (Y-intercept=1/Vmax; X-intercept=−1/KM). The results were confirmed by an independent repeat experiment (-APA: Vmax =177, KM =14.9; +APA: Vmax =173, KM =107.5).
In PDB 2OO0, the APA amino-oxy group is in hydrogen bond distance (3.0 Å) to the PLP aldehyde group and is unobstructed to rearrange in the binding pocket to allow Schiff base formation. Why then did APA fail to form a Schiff base with PLP in PDB 2OO0? Crystallization required cadaverine as additive and Dufe et al. speculated that the binding of cadaverine in the vicinity of the active site (>7 Å distance to the phosphate group of PLP and > 9 Å distance to APA) might affect the interaction between PLP and APA [19]. However, it is not immediately clear how cadaverine binding would modulate the interaction. When we scrutinized the electron density, we found that ethylene glycol from the PEG precipitant of the crystallization solution would fit the density better than the modeled APA (Fig. 9). It is therefore possible that PDB 2OO0 does not contain APA at the substrate binding site.
Fig. 9. Ethylene glycol (EG) fits into the density at the catalytic center of PDB 2OO0.
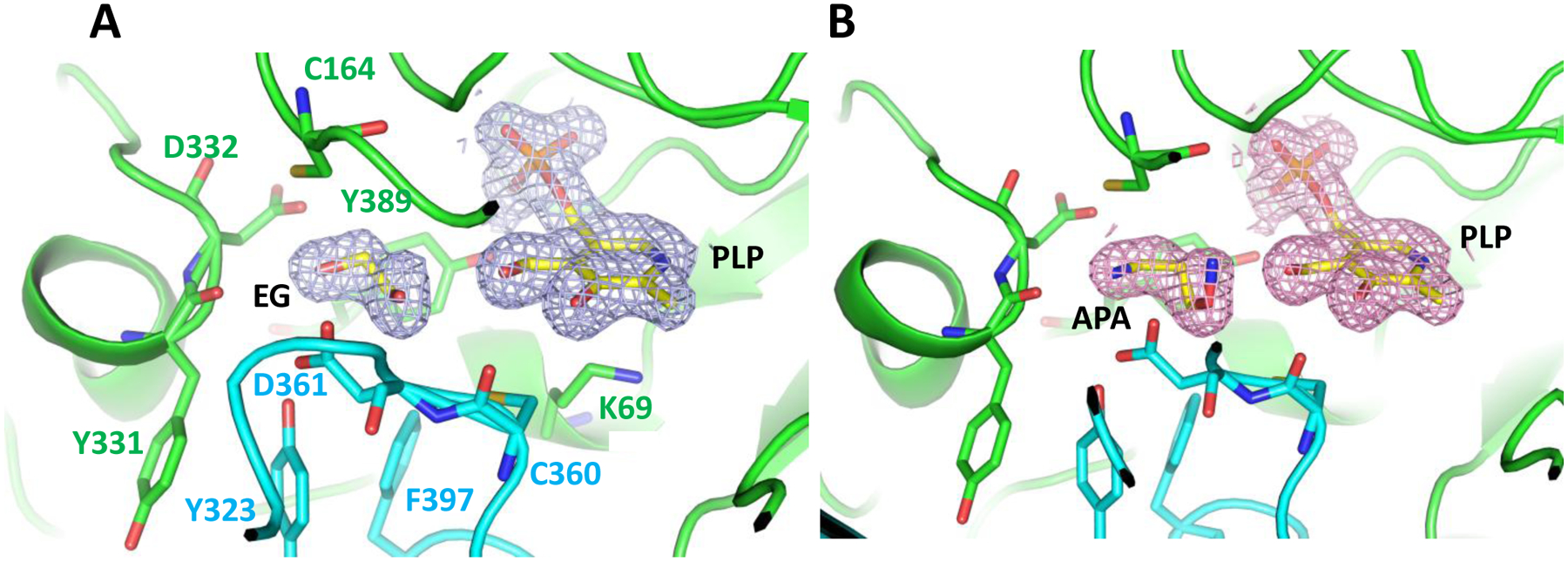
A. 2mFo-DFc density map calculated from the structure factors downloaded from PDB 2OO0 contoured at 1 σ with ethylene glycol used as model. B. Original 2OO0 model contoured at 1 σ with APA built in the substrate binding site.
Materials and Methods
Synthesis of tert-Butyl (3-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)oxy]propyl)[18]carbamate (compound 2 in Fig. 2).
A mixture of 3-(tert-butoxycarbonylamino)-1-propanol (1) (1.0 g, 5.71 mmol), N-hydroxyphthalimide (1.03 g, 6.28 mmol) and triphenylphosphine (1.60 g, 6.28 mmol) in dry tetrahydrofuran (20 mL), under an argon atmosphere, was stirred at 0 °C for 20 minutes. A solution of diisopropyl azodicarboxylate (1.30 g, 6.28 mmol) in tetrahydrofuran (10 mL) was then added dropwise and allowed to slowly warm to room temperature (2 hours). Upon completion, the reaction was concentrated in vacuo. The resulting residue was purified via medium pressure liquid chromatography (SiO2, 100% hexanes to 60% ethyl acetate / hexanes (1.54 g, 84% yield). 1H NMR (500 MHz, Chloroform-d) δ 7.84 (dd, J = 5.4, 3.1 Hz, 2H), 7.76 (dd, J = 5.5, 3.1 Hz, 2H), 4.27 (t, J = 5.9 Hz, 2H), 3.42 (q, J = 6.4 Hz, 2H), 1.95 (q, J = 6.1 Hz, 2H), 1.44 (s, 9H).
Synthesis of 3-(aminooxy)propan-1-amine dihydrochloride (APA).
To a solution of tert-butyl {3-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)oxy]propyl}carbamate (compound 2 in Fig. 2) (1.0 g, 3.13 mmol) in dry dichloromethane (15 mL), under an argon atmosphere was added hydrazine monohydrate (0.30 mL, 3.13 mmol) dropwise during 5 minutes. The solution turned into a suspension in 30 minutes. After 2 hours, the white precipitate was filtered off and washed with cold dichloromethane (10 mL). The filtrate was concentrated in vacuo and the resulting residue was used without further purification. The residue was taken up in methanol (5 mL) and treated dropwise with hydrochloric acid (4.0 N in dioxane, 4.0 mL, 16.0 mmol). The mixture was stirred at room temperature, under an argon atmosphere, for 2 hours and concentrated. The resulting residue was taken up in water (10 mL), washed with ethyl acetate (3 × 5 mL) and lyophilized to give the di-HCl salt of 3-(aminooxy)propan-1-amine (APA) as a fine white powder (0.40 g, 78% yield for two steps). 1H NMR (500 MHz, Deuterium Oxide) δ 4.03 (t, J = 5.9 Hz, 2H), 2.96 (t, J = 7.5 Hz, 2H), 1.91 (p, J = 6.4 Hz, 2H). HRMS (ESI) m/z calculated for C3H11N2O [M+H], 91.0871; found 91.0879.
Protein expression and purification.
Full length human ODC1 and ODC1(1–423) were expressed in pET28 (Novagen) with a C-terminal His6 tag. E. coli BL21(DE3) cells transformed with this expression plasmid were grown in LB broth at 16 °C to an OD600 of ~1 and induced with 0.1 mM IPTG. The next morning, cells were harvested, resuspended in 150 ml buffer A (20 mM Tris [pH8], 200 mM NaCl,10% glycerol) per 2 liters of cells, and passed three times through a French Press with pressure set at 1000 Pa. The lysate was centrifuged at 34,571 × g for 1 hour, and the supernatant was loaded on a 5 ml HisTrap FF column (GE Healthcare). The column was washed with 100 ml buffer A + 25 mM imidazole and eluted with 50 ml of Buffer B (20 mM Tris [pH8], 500 mM imidazole, 10% glycerol). The peak fractions were further purified by passing through at HiLoad 26/200 Superdex 200pg column (GE Healthcare) in 20 mM Tris [pH8], 200 mM NaCl, 2 mM DTT, 1 mM EDTA, 10% glycerol. A portion of the eluted protein was complexed with pyridoxal 5′-phosphate monohydrate (PLP) (Sigma-Aldrich) at a 5-fold molar excess, and APA at a 10-fold molar excess; another portion was complexed with a 5-fold molar excess of PLP in the absence of APA (“apo”) for 1 h at 4 ⁰C. The complexes were filter-concentrated to 16 mg/ml.
Crystallization.
The ODC apo crystals were grown in sitting drop wells with 0.2 μl of protein and 0.2 μl of well solution containing 100 mM tri-sodium citrate pH 5.0 and 30% polyethylene glycol monomethyl ether 550 (PEGmme550). The ODC1-PLP-APA crystals were grown in sitting drop wells with 0.2 μl of protein and 0.2 μl of well solution containing 2% tacsimate, 0.1 M sodium citrate tribasic dihydrate pH 5.6, and 16% w/v polyethylene glycol 3,350.
Crystal data collection, structure determination and analysis.
The crystals of ODC1/PLP/citrate and ODC1/PLP/APA formed in the P212121 space group. The datasets were collected with an EIGER 16M pixel array detector at LS-CAT, 21-ID-D at the Advanced Photon Source at Argonne National Laboratory (Argonne, IL). The data of ODC1/PLP/citrate were indexed to 1.66 Å and the data of ODC1/PLP/APA were indexed to 2.49 Å with MOSFLM [40] and scaled with AIMLESS in the ccp4 package (http://www.ccp4.ac.uk). The CCP4 program PHASER was used for molecular replacement, with the crystal structure of 2OO0 [19] as a search model. The initial model was manually built in COOT [41] and refined with the PHENIX program package [42]. Figure 5 was prepared using LigPlot+ [43], all other structure figures were prepared using PyMOL (DeLano Scientific, San Carlos, CA, http://www.pymol.org).
ODC activity assay.
In vitro ODC activity was measured using 100 ng (40 nM) of purified full length enzyme (Ray Biotech). ODC was incubated with increasing concentrations of APA (1 – 1000 nM) for 30 min at room temperature in a buffer containing 25 mM Tris and 0.1 mM EDTA. Each of the reactions was added to 200 μL of assay mix containing 6.25 mM Tris HCl (pH 7.5), 100 μM L-ornithine, 50 μM pyridoxal-5-phosphate, 1.56 mM DTT and 0.1 μCi [1–14C] L-ornithine (American Radiolabeled Chemicals, Inc., specific activity 55 mCi/mmol) in a microcentrifuge tube. The microcentrifuge tubes were then placed into scintillation vials containing a piece of filter paper saturated with 200 μL of 0.1 M NaOH to capture the release of radiolabeled carbon dioxide. The samples were incubated in a 37 °C incubator while shaking for 30 min. The enzymatic reaction was stopped by adding 250 μL of 5 M sulfuric acid to each sample and incubating at 37 °C while shaking for 30 min. The microcentrifuge tubes were removed from the scintillation vials, and 5 mL of scintillation fluid was added. Disintegrations per minute (DPM) of each sample were measured using a TriCarb liquid scintillation counter (PerkinElmer). The specific ODC activity is expressed as nmol CO2/min/mg protein.
Kinetic measurements.
To determine Michaelis-Menten kinetics, 100 ng/32 nM ODC was incubated for 30 min at room temperature both in the absence and presence of 100 nM APA. Reaction mixes with increasing concentrations of total ornithine (final concentrations of 15.2 μM, 47.2 μM, 207.2 μM, and 807.2 μM) were then added. ODC activities were determined after 1, 3, 5, and 10 minutes reaction time at 37°C for each ornithine concentration. Reaction velocities were determined by linear regression.
The curve of ODC inhibition by citrate was determined by curve fitting using GraphPad Prism. Since inhibition by citrate is incomplete even at the highest citrate concentration, confident calculations of IC50 and Ki values are not available.
Mass spectrometry.
Purified ODC protein (12.4 μM) was complexed with a 5-fold molar excess of PLP and a 10-fold molar excess of APA for 1 h at 4 ⁰C and was then spin concentrated to 125 μM using Amicon ultrafiltration with a 30 kDa molecular cutoff. The protein was frozen, two-fold diluted, and then buffer-exchanged against ammonium acetate using a PD MiniTrap G-25 desalting column (Cytivia).
100 μl of the buffer exchanged protein (at 125 μM) was diluted 1:1 with 100% methanol and 0.2% formic acid. Mass spectra were collected by direct infusion into a Q Exactive HF-X (ThermoFisher) via HESI II ionization source with an electrospray voltage of 3.9 kV. MS1 spectra were collected with alternating MS2 spectra of APA, PLP, and APA-PLP compound. Spectra were acquired over 10 minutes at a 5 μl/min flow rate. The ion capillary temperature was 280 °C and the RF level was 55.0. MS1 scans of m/z 300–1500 were acquired in the orbitrap with a resolution (M/ΔM) of 30,000 at 200 m/z, maximum injection time of 50 ms, and automatic gain control (AGC) target of 3e6. To confirm the molecular identity of the compounds, MS2 scans of APA, PLP, and the APA-PLP compound (91.0866 m/z, 248.0319 m/z, and 320.1006 m/z respectively) were acquired in the orbitrap with a resolution of 30,000 at 200 m/z and maximum injection time of 100 ms, AGC target of 2e5, and isolation width of 1/3 m/z. MS1 spectral intensities for each compound were extracted using Skyline (version 4.2.0). The median values were used for comparison.
Acknowledgments.
Research reported in this publication was supported by NIH grant R21 CA216867 (ASB) and by the Van Andel Institute (KM). We also thank staff members of the Life Science Collaborative Access Team of the Advanced Photon Source (APS) for assistance in data collection at the beam lines of sector 21. Use of the APS, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357. The authors also thank ChemAxon for the use of both Marvin Sketch and Marvin Space to support this work.
Footnotes
Declaration of Interests. The authors declare no competing interests.
Data availability.
The crystal structures have been deposited in the Protein Database (PDB) under accession numbers 7S3F (ODC/PLP/APA) and 7S3G (ODC/PLP/citrate). All other data needed to evaluate the conclusions in the paper are presented in the main text.
References
- 1.Abrahamsen MS, Li RS, Dietrich-Goetz W and Morris DR (1992) Multiple DNA elements responsible for transcriptional regulation of the ornithine decarboxylase gene by protein kinase A. J Biol Chem. 267, 18866–18873 [PubMed] [Google Scholar]
- 2.Reddy SG, McLlheran SM, Cochran BJ, Worth LL, Bishop LA, Brown PJ, Knutson VP and Haddox MK (1996) Multisite phosphorylation of ornithine decarboxylase in transformed macrophages results in increased intracellular enzyme stability and catalytic efficiency. J Biol Chem. 271, 24945–24953 [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg-Hasson Y, Strumpf D and Kahana C (1991) Mouse ornithine decarboxylase is phosphorylated by casein kinase-II at a predominant single location (serine 303). Eur J Biochem. 197, 419–424 [DOI] [PubMed] [Google Scholar]
- 4.Hayashi S, Murakami Y and Matsufuji S (1996) Ornithine decarboxylase antizyme: a novel type of regulatory protein. Trends Biochem Sci. 21, 27–30 [PubMed] [Google Scholar]
- 5.Heller JS, Fong WF and Canellakis ES (1976) Induction of a protein inhibitor to ornithine decarboxylase by the end products of its reaction. Proc Natl Acad Sci U S A. 73, 1858–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pegg AE (2006) Regulation of ornithine decarboxylase. J Biol Chem. 281, 14529–14532 [DOI] [PubMed] [Google Scholar]
- 7.Bachmann AS and Geerts D (2018) Polyamine synthesis as a target of MYC oncogenes. J Biol Chem. 293, 18757–18769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassiri H, Benavides A, Haber M, Gilmour SK, Norris MD and Hogarty MD (2015) Translational development of difluoromethylornithine (DFMO) for the treatment of neuroblastoma. Transl Pediatr. 4, 226–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bello-Fernandez C and Cleveland JL (1992) c-myc transactivates the ornithine decarboxylase gene. Curr Top Microbiol Immunol. 182, 445–452 [DOI] [PubMed] [Google Scholar]
- 10.Bello-Fernandez C, Packham G and Cleveland JL (1993) The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci U S A. 90, 7804–7808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casero RA Jr., Murray Stewart T and Pegg AE (2018) Polyamine metabolism and cancer: treatments, challenges and opportunities. Nat Rev Cancer. 18, 681–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casero RA Jr. and Marton LJ (2007) Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov. 6, 373–390 [DOI] [PubMed] [Google Scholar]
- 13.Alirol E, Schrumpf D, Amici Heradi J, Riedel A, de Patoul C, Quere M and Chappuis F (2013) Nifurtimox-eflornithine combination therapy for second-stage gambiense human African trypanosomiasis: Medecins Sans Frontieres experience in the Democratic Republic of the Congo. Clin Infect Dis. 56, 195–203 [DOI] [PubMed] [Google Scholar]
- 14.Priotto G, Kasparian S, Mutombo W, Ngouama D, Ghorashian S, Arnold U, Ghabri S, Baudin E, Buard V, Kazadi-Kyanza S, Ilunga M, Mutangala W, Pohlig G, Schmid C, Karunakara U, Torreele E and Kande V (2009) Nifurtimox-eflornithine combination therapy for second-stage African Trypanosoma brucei gambiense trypanosomiasis: a multicentre, randomised, phase III, non-inferiority trial. Lancet. 374, 56–64 [DOI] [PubMed] [Google Scholar]
- 15.Saulnier Sholler GL, Gerner EW, Bergendahl G, MacArthur RB, VanderWerff A, Ashikaga T, Bond JP, Ferguson W, Roberts W, Wada RK, Eslin D, Kraveka JM, Kaplan J, Mitchell D, Parikh NS, Neville K, Sender L, Higgins T, Kawakita M, Hiramatsu K, Moriya SS and Bachmann AS (2015) A Phase I Trial of DFMO Targeting Polyamine Addiction in Patients with Relapsed/Refractory Neuroblastoma. PLoS One. 10, e0127246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sholler GLS, Ferguson W, Bergendahl G, Bond JP, Neville K, Eslin D, Brown V, Roberts W, Wada RK, Oesterheld J, Mitchell D, Foley J, Parikh NS, Eshun F, Zage P, Rawwas J, Sencer S, Pankiewicz D, Quinn M, Rich M, Junewick J and Kraveka JM (2018) Maintenance DFMO Increases Survival in High Risk Neuroblastoma. Sci Rep. 8, 14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyskens FL Jr., McLaren CE, Pelot D, Fujikawa-Brooks S, Carpenter PM, Hawk E, Kelloff G, Lawson MJ, Kidao J, McCracken J, Albers CG, Ahnen DJ, Turgeon DK, Goldschmid S, Lance P, Hagedorn CH, Gillen DL and Gerner EW (2008) Difluoromethylornithine plus sulindac for the prevention of sporadic colorectal adenomas: a randomized placebo-controlled, double-blind trial. Cancer Prev Res (Phila). 1, 32–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachmann A and Levin V (2012) Clinical applications of polyamine-based therapeutics In Polyamine Drug Discovery. pp. 257–276 Royal Society of Chemistry, London [Google Scholar]
- 19.Dufe VT, Ingner D, Heby O, Khomutov AR, Persson L and Al-Karadaghi S (2007) A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 1-amino-oxy-3-aminopropane. Biochem J. 405, 261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khomutov RM, Denisova GF, Khomutov AR, Belostotskaia KM and Shlosman RB (1985) [Aminooxypropylamine--an effective inhibitor of ornithine decarboxylase in vitro and in vivo]. Bioorg Khim. 11, 1574–1576 [PubMed] [Google Scholar]
- 21.Stanek J, Frei J, Mett H, Schneider P and Regenass U (1992) 2-substituted 3-(aminooxy)propanamines as inhibitors of ornithine decarboxylase: synthesis and biological activity. J Med Chem. 35, 1339–1344 [DOI] [PubMed] [Google Scholar]
- 22.Jackson LK, Brooks HB, Osterman AL, Goldsmith EJ and Phillips MA (2000) Altering the reaction specificity of eukaryotic ornithine decarboxylase. Biochemistry. 39, 11247–11257 [DOI] [PubMed] [Google Scholar]
- 23.Osterman AL, Kinch LN, Grishin NV and Phillips MA (1995) Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase identified by alanine scanning mutagenesis. J Biol Chem. 270, 11797–11802 [DOI] [PubMed] [Google Scholar]
- 24.Hyvonen T, Alakuijala L, Andersson L, Khomutov AR, Khomutov RM and Eloranta TO (1988) 1-Aminooxy-3-aminopropane reversibly prevents the proliferation of cultured baby hamster kidney cells by interfering with polyamine synthesis. J Biol Chem. 263, 11138–11144 [PubMed] [Google Scholar]
- 25.Mett H, Stanek J, Lopez-Ballester JA, Janne J, Alhonen L, Sinervirta R, Frei J and Regenass U (1993) Pharmacological properties of the ornithine decarboxylase inhibitor 3-aminooxy-1-propanamine and several structural analogues. Cancer Chemother Pharmacol. 32, 39–45 [DOI] [PubMed] [Google Scholar]
- 26.Milovica V, Turchanowa L, Khomutov AR, Khomutov RM, Caspary WF and Stein J (2001) Hydroxylamine-containing inhibitors of polyamine biosynthesis and impairment of colon cancer cell growth. Biochem Pharmacol. 61, 199–206 [DOI] [PubMed] [Google Scholar]
- 27.Khomutov AR (2002) Inhibition of enzymes of polyamine biosynthesis by substrate-like O-substituted hydroxylamines. Biochemistry (Mosc). 67, 1159–1167 [DOI] [PubMed] [Google Scholar]
- 28.Markovic-Housley Z, Schirmer T, Hohenester E, Khomutov AR, Khomutov RM, Karpeisky MY, Sandmeier E, Christen P and Jansonius JN (1996) Crystal structures and solution studies of oxime adducts of mitochondrial aspartate aminotransferase. Eur J Biochem. 236, 1025–1032 [DOI] [PubMed] [Google Scholar]
- 29.Klee N, Wong PE, Baragana B, Mazouni FE, Phillips MA, Barrett MP and Gilbert IH (2010) Selective delivery of 2-hydroxy APA to Trypanosoma brucei using the melamine motif. Bioorg Med Chem Lett. 20, 4364–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mangold U (2005) The antizyme family: polyamines and beyond. IUBMB Life. 57, 671–676 [DOI] [PubMed] [Google Scholar]
- 31.Ghoda L, van Daalen Wetters T, Macrae M, Ascherman D and Coffino P (1989) Prevention of rapid intracellular degradation of ODC by a carboxyl-terminal truncation. Science. 243, 1493–1495 [DOI] [PubMed] [Google Scholar]
- 32.Wu D, Kaan HY, Zheng X, Tang X, He Y, Vanessa Tan Q, Zhang N and Song H (2015) Structural basis of Ornithine Decarboxylase inactivation and accelerated degradation by polyamine sensor Antizyme1. Sci Rep. 5, 14738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu HY, Chen SF, Hsieh JY, Chou F, Wang YH, Lin WT, Lee PY, Yu YJ, Lin LY, Lin TS, Lin CL, Liu GY, Tzeng SR, Hung HC and Chan NL (2015) Structural basis of antizyme-mediated regulation of polyamine homeostasis. Proc Natl Acad Sci U S A. 112, 11229–11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Almrud JJ, Oliveira MA, Kern AD, Grishin NV, Phillips MA and Hackert ML (2000) Crystal structure of human ornithine decarboxylase at 2.1 A resolution: structural insights to antizyme binding. J Mol Biol. 295, 7–16 [DOI] [PubMed] [Google Scholar]
- 35.Tang J, Ju Y, Gu Q, Xu J and Zhou H (2019) Structural Insights into Substrate Recognition and Activity Regulation of the Key Decarboxylase SbnH in Staphyloferrin B Biosynthesis. J Mol Biol. 431, 4868–4881 [DOI] [PubMed] [Google Scholar]
- 36.Li B, Deng X, Kim SH, Buhrow L, Tomchick DR, Phillips MA and Michael AJ (2021) Alternative pathways utilize or circumvent putrescine for biosynthesis of putrescine-containing rhizoferrin. J Biol Chem. 296, 100146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson LK, Goldsmith EJ and Phillips MA (2003) X-ray structure determination of Trypanosoma brucei ornithine decarboxylase bound to D-ornithine and to G418: insights into substrate binding and ODC conformational flexibility. J Biol Chem. 278, 22037–22043 [DOI] [PubMed] [Google Scholar]
- 38.Kern AD, Oliveira MA, Coffino P and Hackert ML (1999) Structure of mammalian ornithine decarboxylase at 1.6 A resolution: stereochemical implications of PLP-dependent amino acid decarboxylases. Structure. 7, 567–581 [DOI] [PubMed] [Google Scholar]
- 39.Grishin NV, Osterman AL, Brooks HB, Phillips MA and Goldsmith EJ (1999) X-ray structure of ornithine decarboxylase from Trypanosoma brucei: the native structure and the structure in complex with alpha-difluoromethylornithine. Biochemistry. 38, 15174–15184 [DOI] [PubMed] [Google Scholar]
- 40.Battye TG, Kontogiannis L, Johnson O, Powell HR and Leslie AG (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Lohkamp B, Scott WG and Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC and Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laskowski RA and Swindells MB (2011) LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 51, 2778–2786 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The crystal structures have been deposited in the Protein Database (PDB) under accession numbers 7S3F (ODC/PLP/APA) and 7S3G (ODC/PLP/citrate). All other data needed to evaluate the conclusions in the paper are presented in the main text.


