Abstract
Background
Gestational diabetes mellitus (GDM) is a common pregnancy-specific disease and is growing at an alarming rate worldwide, which can negatively affect the health of pregnant women and fetuses. However, most studies are limited to one tissue, placenta or umbilical cord blood, usually with one omics assay. It is thus difficult to systematically reveal the molecular mechanism of GDM and the key influencing factors on pregnant women and offspring.
Results
We recruited a group of 21 pregnant women with GDM and 20 controls without GDM. For each pregnant woman, reduced representation bisulfite sequencing and RNA-seq were performed using the placenta and paired neonatal umbilical cord blood specimens. Differentially methylated regions (DMRs) and differentially expressed genes (DEGs) were identified with body mass index as a covariate. Through the comparison of GDM and control samples, 2779 and 141 DMRs, 1442 and 488 DEGs were identified from placenta and umbilical cord blood, respectively. Functional enrichment analysis showed that the placenta methylation and expression profiles of GDM women mirrored the molecular characteristics of “type II diabetes” and “insulin resistance.” Methylation-altered genes in umbilical cord blood were associated with pathways “type II diabetes” and “cholesterol metabolism.” Remarkably, both DMRs and DEGs illustrated significant overlaps among placenta and umbilical cord blood samples. The overlapping DMRs were associated with “cholesterol metabolism.” The top-ranking pathways enriched in the shared DEGs include “growth hormone synthesis, secretion and action” and “type II diabetes mellitus.”
Conclusions
Our research demonstrated the epigenetic and transcriptomic alternations of GDM women and offspring. Our findings emphasized the importance of epigenetic modifications in the communication between pregnant women with GDM and offspring, and provided a reference for the prevention, control, treatment, and intervention of perinatal deleterious events of GDM and neonatal complications.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-022-01289-5.
Keywords: Gestational diabetes mellitus, Placenta, Umbilical cord blood, Offspring, DNA methylation, Transcriptome
Introduction
Gestational diabetes mellitus (GDM) is a common pregnancy-specific disease, characterized by glucose intolerance. GDM usually occurs in the middle or late pregnancy [1–3]. Increased insulin resistance and impaired insulin secretion are the main causes of GDM [4]. Due to different lifestyles, ethnicities, diagnostic methods, the median (interquartile range) prevalence (%) of GDM varies significantly from the lowest in Europe (6.1, 1.8–31) to the highest in the Middle East and North Africa (15.2, 8.8–20) during 2005–2018 [5]. Independent studies show that factors affecting the occurrence of GDM include maternal weight, gestational weight gain, diet and exercise, ethnicity, maternal age, and family history of diabetes [6–11]. Women with GDM are prone to pregnancy-related complications or perinatal adverse events, such as hypertension, preeclampsia, preterm birth, shoulder dystocia, perinatal morbidity, and death [12]. Fetuses exposed to GDM have an increased risk of macrosomia, fetal distress, congenital malformations, and jaundice [13]. Meanwhile, the offspring of GDM women has an increased risk of metabolic diseases and cardiovascular diseases, such as obesity and type II diabetes. GDM is also related to immune function loss and cognitive impairment in the offspring [14]. The molecular mechanism of insulin resistance and insulin secretion defect in pregnant women is still under intensive investigation.
From a fertilized egg to the complete fetus, human genome remains relatively stable while the epigenome undergoes regulated reprogramming including dramatic DNA methylation changes and systemic histone modification alternations [15, 16]. Moreover, the epigenome of fetus is sensitive to the pregnant women’s intrauterine environment, especially high-glucose levels, which eventually leads to phenotypic changes [17]. How the exposure of fetal to maternal GDM contributes to increasing offspring risk remains to be explored. Results from animal studies suggest that the exposure to intrauterine high-glucose environment may cause disorders of hypothalamic neuropeptide neurons, impair blood glucose homeostasis and lead to abnormal renal development and islet β-cell dysfunction [18, 19]. Epidemiological studies have also confirmed that exposure of intrauterine high-glucose environment may cause the transfer of lipid matrix molecules to the fetus, resulting in disorders associated with adipocyte dysfunction and fatty acid accumulation [20]. Using bisulfite pyrosequencing assays, Houde and colleagues identified three candidate genes, LRP1B, BRD2 and CACNA1D, whose DNA methylation alterations in placenta and umbilical cord blood were related to energy metabolism and maternal glucose levels [21]. Through methylated DNA immunoprecipitation (MeDIP) assay and next-generation sequencing (NGS), Rong and colleagues revealed genome-wide changes in the placenta of GDM patients and transcriptional variations of four GDM-related genes [22]. However, the genes and pathways involved in GDM have not been systematically characterized and thus remain elusive. Therefore, genome-scale DNA methylation analysis and transcriptome studying of the placenta and paired umbilical cord blood from women with or without GDM can potentially uncover the underlying molecular mechanism.
Reduced representation bisulfite sequencing (RRBS) is an accurate and cost-effective method that can provide genome-wide mapping 5-methylcytosines, particularly enriching for coverage in promoter and enhancer regions that are crucial for transcriptional regulation [23, 24]. RNA-seq allows to profile thousands of gene expressions precisely and simultaneously [25]. Here, we employed the RRBS and RNA-seq methods to systematically analyze the genome-wide methylation and expression profiles of placenta and paired umbilical cord blood from a group of patients with GDM and control pregnant women without GDM. The aim of this study is to reveal critical epigenetic changes associated with GDM in pregnant women and to identify differentially expressed genes (DEGs) which may potentially contribute to GDM.
Results
Clinical characteristics of pregnant women and newborns
Forty-one pregnant women were recruited into this study, including 21 pregnant women with GDM and 20 without GDM as control. Those pregnant women delivered 41 babies, including 4 babies with macrosomia, 2 of which were from the GDM group. Anthropometric and metabolic parameters of pregnant women and newborns are shown in Table 1. We observed that both the maternal weight and body mass index (BMI) were significantly higher in the GDM group compared with those in the control group (Wilcoxon rank-sum test, P value < 0.05), while other characteristics, such as maternal age, gestational weight gain, maternal height, gestational weeks, and fetal birth weight, did not show significant differences between two groups. Given the significant differences of maternal BMI between GDM and control groups, BMI will be used as a covariate in the subsequent search for differentially methylated regions (DMRs) and DEGs.
Table 1.
Anthropometric and metabolic parameters of pregnant women and newborns
| Parameters | GDM (n = 21) | Control (n = 20) | P value | Difference |
|---|---|---|---|---|
| Maternal age (years) | 30.95 ± 3.8 | 30.05 ± 3.17 | 0.64 | n.s |
| Maternal birth weight (kg) | 73.56 ± 10.71 | 66.86 ± 6.08 | 0.029 | * |
| Gestational weight gain (kg) | 12.22 ± 3.01 | 14.25 ± 3.85 | 0.073 | n.s |
| Maternal height (cm) | 162.33 ± 5.36 | 161.35 ± 5.26 | 0.74 | n.s |
| BMI (kg/m2) | 22.61 ± 2.82 | 20.73 ± 1.92 | 0.021 | * |
| Gestational weeks (week) | 38.71 ± 1.06 | 39.15 ± 1.5 | 0.15 | n.s |
| 1 h post OGTT (mmol/L) | 9.17 ± 2.11 | 7.33 ± 1.09 | 7.6e-3 | ** |
| 2 h post OGTT (mmol/L) | 7.54 ± 1.26 | 6.21 ± 1.07 | 3.9e-3 | ** |
| Fasting cholesterol (mmol/L) | 5.83 ± 0.98 | 5.68 ± 1.2 | 0.62 | n.s |
| Fasting HDL-c (mmol/L) | 2.01 ± 0.44 | 1.93 ± 0.37 | 0.60 | n.s |
| Fasting LDL-c (mmol/L) | 2.76 ± 0.66 | 2.78 ± 0.91 | 0.94 | n.s |
| 24-week glycemia (mmol/L) | 7.54 ± 1.26 | 6.21 ± 1.07 | 3.96e-3 | ** |
| Fetal birth weight (g) | 3371.9 ± 507.8 | 3269 ± 433.89 | 0.566 | n.s |
| Fetal gender (male/female) | 11/10 | 12/8 | / | / |
Difference analysis was conducted by Wilcoxon rank-sum test, * P value < 0.05, ** P value < 0.01, n.s. P value > = 0.05
BMI Body mass index, OGTT Oral glucose tolerance test, HDL-c High-density lipoprotein cholesterol, LDL-c Low-density lipoprotein cholesterol
Placenta shows genome-wide methylation alterations associated with type II diabetes mellitus in GDM patients
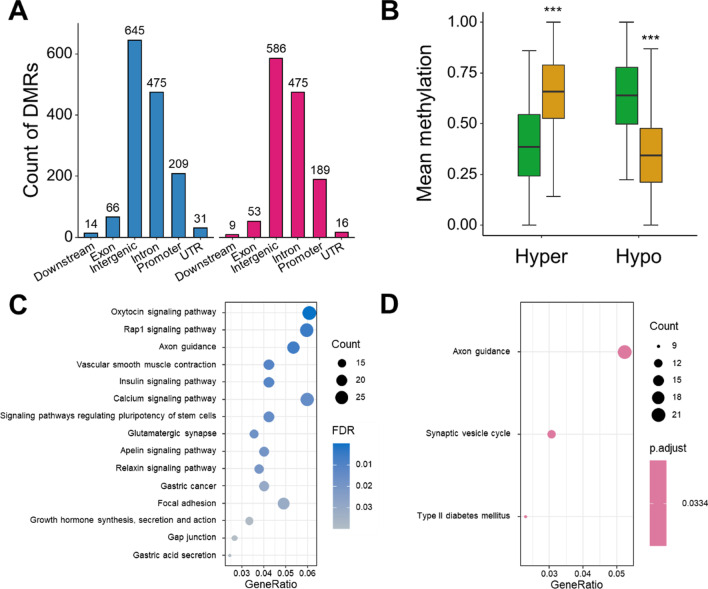
DMRs are genomic regions with differential methylation status across multiple CpG sites, which are considered to play an important role in gene imprinting and regulating the expression of nearby genes that were called differentially methylated genes (DMGs). In placenta samples, after excluding the effect of maternal BMI, a total of 2779 DMRs were identified, including 1446 hyper-methylated DMRs (hyper-DMRs) associated with 1293 hyper-methylated DMGs (hyper-DMGs) and 1333 hypo-methylated DMRs (hypo-DMRs) associated with 1197 hypo-methylated DMGs (hypo-DMGs) (Fig. 1A, Additional file 2: Table S2). Among them, 209 hyper-DMRs and 189 hypo-DMRs were located in promoter regions, respectively. The mean methylation of hyper-DMR and hypo-DMR in GDM and control groups is shown (Fig. 1B). As expected, compared with the control group, the distribution of mean methylation levels of all hyper-DMRs or hypo-DMRs in GDM group showed significant difference (Wilcoxon rank-sum test, P value < 0.05). Additionally, hyper-DMRs and hypo-DMRs widely spread across different chromosomes (Additional file 1: Fig. S1C), indicating that the placenta has extensive and significant methylation changes in a genome-wide manner.
Fig. 1.
Placenta shows genome-wide methylation alteration associated with glucose metabolism in GDM patients. A Genomic annotation of DMRs in placenta. Blue bar represents hyper-DMRs and red bar represents hypo-DMRs. B Differences of DMRs mean methylation levels between GDM and control group. Yellow box represents GDM samples and green box represents control samples. *** P value < 0.001, Wilcoxon rank-sum test. C KEGG pathway enrichment analysis of hyper-DMGs in placenta. D KEGG pathway enrichment analysis of hypo-DMGs in placenta
To further characterize DMRs identified, we performed functional enrichment analysis using Gene Ontology (GO) terms and identified 401 and 288 significantly enriched terms for hyper-DMGs and hypo-DMGs (FDR < 0.05), respectively (Additional file 2: Table S3, S5). For hyper-DMGs, the enriched pathways mainly included neural and developmental pathways, endocrine-, and immune-related pathways (Additional file 1: Fig. S1D). For hypo-DMGs, the enriched GO terms were similar to that in hyper-DMRs, but also included other ones, such as “cognition,” “long-term memory,” and epigenetic modification-related pathways (Additional file 1: Fig. S1E). Meanwhile, enrichment analysis with Kyoto Encyclopedia of Genes and Genomes (KEGG) database identified 31 and 3 pathways for hyper-DMGs and hypo-DMGs, respectively (Fig. 1C, D, Additional file 2: Table S4, S6, FDR < 0.05). Of the pathways associated with hyper-DMGs, “oxytocin signaling” (29 genes, FDR = 7.14e-05), “calcium signaling” (28 genes, FDR = 5.47e-06), and “insulin signaling” (19 genes, FDR = 8.63e-03) ranked in the top of the list (Fig. 1C). Interestingly, the “type II diabetes mellitus” pathway was significantly enriched in hypo-DMGs (FDR = 0.033), suggesting that GDM may have the epigenetic characteristics of type II diabetes mellitus (Fig. 1D).
Methylation contributes to expression change of genes associated with insulin signaling pathway
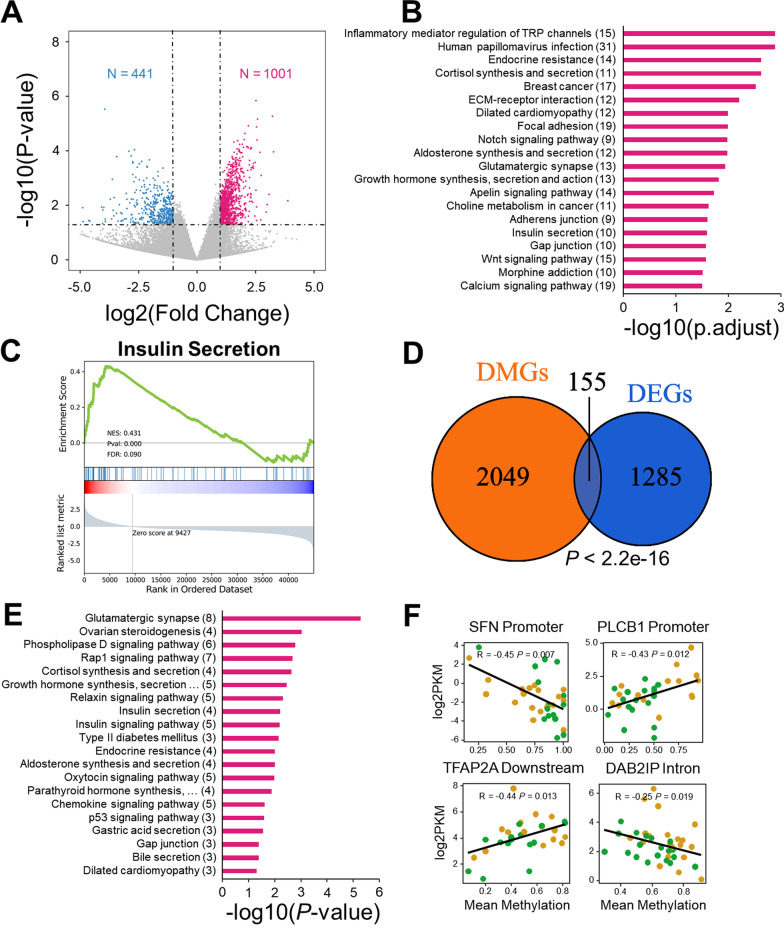
To further explore the pathogenesis of GDM, we next performed RNA-seq analysis using placenta samples from the GDM and control groups. After adjustments for multiple testing, we obtain 113 up-regulated and 9 down-regulated genes (FDR < 0.05). Given this number of DEGs is so small, it is challenging to interpret these genes in terms of biological processes and signaling pathways. However, this is not totally uncommon and has been observed in several previous type II diabetes mellitus-related studies [26, 27]. For this reason, researchers have developed the Gene Set Enrichment Analysis (GSEA) method, which was designed to detect modest but coordinate changes in the expression of groups of functionally related genes [28]. This also suggests that top-ranking genes are still good candidates for subsequent analysis, even though individual one of which is not statistically significant. For this reason, we used a P value-based threshold (|Fold changes|> 2, P value < 0.05) to select DEGs. With this threshold, 1001 genes were up-regulated, and 441 genes were down-regulated (Fig. 2A, Additional file 2: Table S7, S8). The up-regulated genes were annotated to multiple endocrine and metabolic-related pathways, including “endocrine resistance,” “cortisol synthesis, and secretion,” “dilated cardiomyopathy,” and “insulin secretion” (Fig. 2B, Additional file 2: Table S9 and S10). For down-regulated genes, 341 and 32 terms were enriched in GO and KEGG functional enrichment analysis, respectively (Additional file 2: Table S11 and S12). Examples of enriched KEGG terms include “antigen processing and presentation” (FDR = 8.89e-12), and “type I diabetes mellitus” (FDR = 2.81e-10) (Additional file 1: Fig. S2A). GO enrichment analysis showed that pathways related to immune regulation, cell proliferation, and glycolipid metabolism were most enriched (Additional file 1: Fig. S2B).
Fig. 2.
Methylation contributes to expression change of genes associated with insulin signaling pathway. A Volcano plot of placenta expression profile. B KEGG pathway enrichment analysis of up-regulated genes in placenta. The numbers in the brackets of the ordinate label represent the number of genes involved. C GSEA analysis of Insulin secretion pathway in placenta expression profile. D The Venn Diagram between DMGs and DEGs in placenta, Fisher’s Exact Test. E KEGG pathway enrichment analysis result of shared genes between DMGs and DEGs in placenta, the numbers in the brackets of the ordinate label represent the number of genes involved. F Correlation between gene methylation and gene expression in placenta, title of each panel shows the gene name and genomic location. Significance was tested by Spearman’s rank correlation test
We next conducted GSEA using expression profile of placenta to detect modest but coordinate changes in the expression of groups of functionally related genes. Of note, 28/40 and 31/32 of KEGG positive and negative enriched pathways were validated by GSEA results, respectively (FDR < 0.25), which show that DGE-based results are largely consistent with that from GSEA in the pathway-level (Additional file 2: Table S13, S14). Interestingly, we discovered two significant insulin-related pathways, “insulin secretion” (FDR = 0.0898, Fig. 2C) and “insulin resistance” (FDR = 0.111, Additional file 1: Fig. S2D). Other positively enriched pathways include the “notch signaling pathway,” “autophagy,” “growth hormone synthesis, secretion and action,” “prolactin signaling pathway,” and “aldosterone synthesis and secretion pathways” (Table 2). Most notably, pathway “type I diabetes mellitus” ranked first in the negative enriched pathways (FDR < 0.001, Additional file 1: Fig. S2E). Other negatively enriched pathways, such as “one carbon pool by folate” (FDR = 3.65e-3), “primary bile acid biosynthesis” (FDR = 6.96e-4), and “folate biosynthesis pathway” (FDR = 0.012), are listed in Table 3.
Table 2.
Positive enriched pathways of GSEA result for placenta transcriptome (top20)
| Pathway | Size | NES | P value | FDR |
|---|---|---|---|---|
| Cortisol synthesis and secretion | 64 | 0.50 | < 0.001 | 0.16 |
| Adherens junction | 71 | 0.49 | < 0.001 | 0.097 |
| Notch signaling pathway | 59 | 0.48 | < 0.001 | 0.083 |
| Autophagy | 32 | 0.47 | < 0.001 | 0.085 |
| Growth hormone synthesis, secretion and action | 118 | 0.46 | < 0.001 | 0.087 |
| ERBB signaling pathway | 85 | 0.46 | < 0.001 | 0.075 |
| Snare interactions in vesicular transport | 33 | 0.45 | 5.52e-3 | 0.075 |
| Lysine degradation | 63 | 0.43 | < 0.001 | 0.10 |
| Ovarian steroidogenesis | 50 | 0.43 | < 0.001 | 0.093 |
| Insulin secretion | 84 | 0.43 | < 0.001 | 0.089 |
| Alpha-linolenic acid metabolism | 24 | 0.42 | 0.0175 | 0.086 |
| GNRH signaling pathway | 92 | 0.41 | < 0.001 | 0.10 |
| Prolactin signaling pathway | 69 | 0.41 | < 0.001 | 0.094 |
| Inositol phosphate metabolism | 73 | 0.41 | < 0.001 | 0.091 |
| Chronic myeloid leukemia | 76 | 0.41 | < 0.001 | 0.087 |
| Aldosterone synthesis and secretion | 97 | 0.41 | < 0.001 | 0.082 |
| Phosphatidylinositol signaling system | 96 | 0.40 | < 0.001 | 0.092 |
| Hedgehog signaling pathway | 56 | 0.40 | < 0.001 | 0.087 |
| Endometrial cancer | 58 | 0.40 | < 0.001 | 0.089 |
| Acute myeloid leukemia | 67 | 0.40 | < 0.001 | 0.086 |
Size represents the number of genes enriched in the corresponding pathway
NES Normalized enrichment score, FDR False discovery rate
Table 3.
Negative enriched pathways of GSEA result for placenta transcriptome (top20)
| Pathway | Size | NES | P value | FDR |
|---|---|---|---|---|
| Type I diabetes mellitus | 40 | − 0.78 | < 0.001 | < 0.001 |
| Allograft rejection | 34 | − 0.76 | < 0.001 | < 0.001 |
| Ribosome | 135 | − 0.74 | < 0.001 | < 0.001 |
| Graft-versus-host disease | 36 | − 0.74 | < 0.001 | < 0.001 |
| Intestinal immune network for IGA production | 44 | − 0.72 | < 0.001 | < 0.001 |
| Autoimmune thyroid disease | 41 | − 0.70 | < 0.001 | 1.14e-5 |
| Staphylococcus aureus infection | 87 | − 0.69 | < 0.001 | 9.77e-5 |
| Asthma | 28 | − 0.69 | < 0.001 | 8.55e-5 |
| Leishmaniasis | 74 | − 0.66 | < 0.001 | 4.64e-4 |
| Primary bile acid biosynthesis | 17 | − 0.65 | 2.63e-3 | 6.96e-5 |
| Systemic lupus erythematosus | 127 | − 0.64 | < 0.001 | 1.00e-3 |
| Inflammatory bowel disease | 60 | − 0.63 | < 0.001 | 1.09e-3 |
| Rheumatoid arthritis | 87 | − 0.63 | < 0.001 | 1.01e-3 |
| Thiamine metabolism | 15 | − 0.6 | 2.67e-3 | 1.38e-3 |
| Coronavirus disease | 220 | − 0.62 | < 0.001 | 1.84e-3 |
| Viral myocarditis | 57 | − 0.61 | < 0.001 | 1.81e-3 |
| Complement and coagulation cascades | 83 | − 0.60 | < 0.001 | 2.15e-3 |
| Antigen processing and presentation | 69 | − 0.60 | < 0.001 | 2.11e-3 |
| Pertussis | 76 | − 0.59 | < 0.001 | 2.99e-3 |
| One carbon pool by folate | 20 | − 0.58 | 2.55e-3 | 3.65e-3 |
Size represents the number of genes enriched in the corresponding pathway
NES Normalized enrichment score
Next, to investigate whether methylation will affect the expression of genes that may attribute to GDM, we combined methylation data and transcriptome data for an integrated analysis. The analysis indicated that 155 genes showed both methylation changes and expression alternations between the GDM and the control group (P value < 2.2e-16, odds ratio = 4.69, Fisher’s Exact Test, Fig. 2D). Though not statistically significant after adjustment for multiple comparisons, the top-ranking pathways enriched in the overlapping genes include “Glutamatergic synapse” (ADCY5/ADCY7/GRIK4/PLA2G4A/PLCB1/SHANK2/SHANK3/SLC1A6, P value = 5.32e-06), “cortisol synthesis and secretion” (ADCY5/ADCY7/LDLR/PLCB1, P value = 2.29e-03), “insulin secretion” (ADCY5/ADCY7/PLCB1/RYR2, P value = 6.28e-3), “insulin signaling pathway” (HK2/PRKCZ/SH2B2/SHC2/SOCS3, P value = 6.34e-3), “type II diabetes” (HK2/PRKCZ/SOCS3, P value = 7.10e-3) and “bile secretion” (ADCY5/ADCY7/LDLR, P value = 0.040) (Fig. 2E). We then selected 10 genes of interest and showed their mean methylation difference, expression change, and associated pathways in Table 4. Besides, from the 155 overlapped gene, we screened out four gene, SFN, PLCB1, TFAP2A and DAB2IP, whose gene expression level and gene mean methylation level show significant correlation and functional for glycolipid metabolism (Fig. 2F). Meanwhile, using the GSEA method, placenta CpG methylation generally had a negative regulatory effect on gene expression (P value < 0.001, NES = -0.255, Additional file 1: Fig. S2F). In summary, we found that some genes involved in glycolipid metabolism, insulin secretion, and insulin resistance had been epigenetically modified, which might contribute directly or indirectly to GDM.
Table 4.
Annotation of overlapping genes of placenta DMGs and DEGs
| Gene | Delta | log2FC | Pathway |
|---|---|---|---|
| ADCY5 | 0.5 | 1.93 | Ovarian steroidogenesis |
| Cortisol synthesis and secretion | |||
| Insulin secretion | |||
| Bile secretion | |||
| B3GALT5 | 0.12 | − 1.64 | Glycosphingolipid biosynthesis—globo and isoglobo series |
| GADD45A | − 0.25 | − 1.5 | p53 signaling pathway |
| Chronic myeloid leukemia | |||
| HK2 | − 0.67 | − 2.92 | Insulin signaling pathway |
| Type II diabetes mellitus | |||
| Fructose and mannose metabolism | |||
| JAG2 | 0.25 | 1.81 | Endocrine resistance |
| LDLR | − 0.63 | 1.31 | Cortisol synthesis and secretion |
| Aldosterone synthesis and secretion | |||
| Bile secretion | |||
| PLA2G4A | 0.4 | − 1.25 | Glutamatergic synapse |
| Ovarian steroidogenesis | |||
| Oxytocin signaling pathway | |||
| RYR2 | − 0.12 | 1.86 | Insulin secretion |
| Oxytocin signaling pathway | |||
| Dilated cardiomyopathy] | |||
| SH2B2 | 0.49 | 1.43 | Insulin signaling pathway |
| SOCS3 | − 0.5 | − 1.04 | Growth hormone synthesis, secretion and action |
| Insulin signaling pathway | |||
| Type II diabetes mellitus |
delta DNA methylation difference, log2FC log2(Fold Change), Pathway KEGG pathway annotation
Alterations for umbilical cord blood were related to insulin secretion and resistance
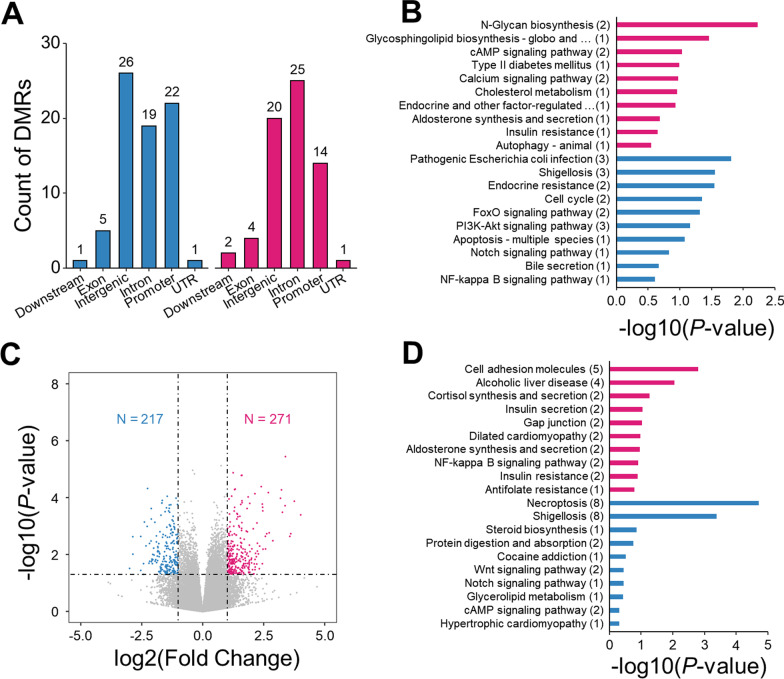
To explore whether the exposure to increased glucose level could cause epigenetic changes in fetus, we next conducted RRBS and RNA-seq analyses using the umbilical cord blood paired with placenta described above. Due to the existence of placental barrier, the changes of fetal epigenome were subtler comparing with that in placenta. Genome-wide methylation profiling by RRBS identified far less numbers of DMRs in contrast to that from the paired placenta. Specifically, after excluding the effect of maternal BMI, we detected 75 hyper-DMRs and 66 hypo-DMRs which annotated to 74 hyper-DMGs and 65 hypo-DMGs, respectively (Fig. 3A, B; Additional file 1: Fig. S3A). Next, KEGG pathway enrichment analysis was conducted to identify pathways of differentially methylated genes. Though not statistically significant after adjustment for multiple comparisons, some enlightening pathways were identified in hypo-DMRs (Fig. 3B, red bar), including “cAMP signaling pathway” (ATP2B3/DRD5), “type II diabetes mellitus” (PRKCD), “cholesterol metabolism” (OSBPL5), “aldosterone synthesis and secretion” (ATP2B3), “insulin resistance” (PRKCD). On the other hand, pathways “endocrine resistance” (JAG2/RB1), “PI3K-Akt signaling pathway” (BCL2L11/LPAR2/TCL1A), and “bile secretion” (SLC10A2) were found in hyper-methylated genes (Fig. 3B, blue bar).
Fig. 3.
Alterations for umbilical cord blood were related to insulin secretion and resistance. A Genomic annotation of DMRs in umbilical cord blood. Blue bar represents hyper-DMRs and red bar represents hypo-DMRs. B KEGG pathway enrichment analysis of hypo-DMGs (red bar) and hyper-DMGs (blue bar) of umbilical cord blood. C Volcano plot of umbilical cord blood expression profile. D KEGG pathway enrichment analysis of up-DEGs (red bar) and hyper-DEGs (blue bar) in umbilical cord blood
RNA-seq analyses of umbilical cord blood samples revealed 271 up-regulated genes and 217 down-regulated genes (|Fold change|> 2, P value < 0.05, Additional file 2: Table S15, S16) by comparing GDM patients to the controls (Fig. 3C). KEGG enrichment analysis showed that the up- and down-regulated genes were associated with 21 and 6 pathways, respectively (P value < 0.05, Fig. 3D). Interestingly, two pathways, “human papillomavirus infection” and “ovarian steroidogenesis” were overlapped for up-regulated genes between placenta and umbilical cord blood, showing that there is a certain relationship of expression profiles between pregnant women and offspring.
Due to minor changes in gene expression levels, GSEA was then performed as a cutoff-free method for pathway enrichment analysis (Additional file 2: Table S17, S18). In details, “collecting duct acid secretion” (FDR = 0.235), “FC gamma R-mediated phagocytosis” (FDR = 0.244), and “staphylococcus aureus infection” (FDR = 0.249) were significantly activated. By contrast, “systemic lupus erythematosus” (FDR = 0.0816), “one carbon pool by folate” (FDR = 0.0844), “fatty acid biosynthesis” (FDR = 0.203), and “primary bile acid biosynthesis” (FDR = 0.239) were significantly repressed. It is worth noting that pathway “one carbon pool by folate” were also enriched in the placenta samples. These results preliminarily show that there is a strong correlation between the current genomic changes and the offspring, and these changes are related to glucose metabolism and insulin, suggesting the possibility of metabolic diseases in the offspring.
In umbilical cord blood, we found one overlapping gene ZNF423 (DNA methylation delta = − 0.4, log2(Fold Change) = − 2.15), whose gene methylation and expression were both altered. Expression of ZNF423 has been reported directly to be proportional to the size of adipocytes [29]. The gene product of ZNF423 promotes the transformation of non-adipocytes into adipocytes and inhibits subcutaneous adipogenesis, leading to adipocyte hypertrophy and inflammation, and finally developing to obesity and insulin resistance [30]. The significant down-regulation of ZNF423 in umbilical cord blood may be related to the higher possibility of suffering obesity in offspring exposed to GDM.
Significant correlation between the changes of placenta and umbilical cord blood
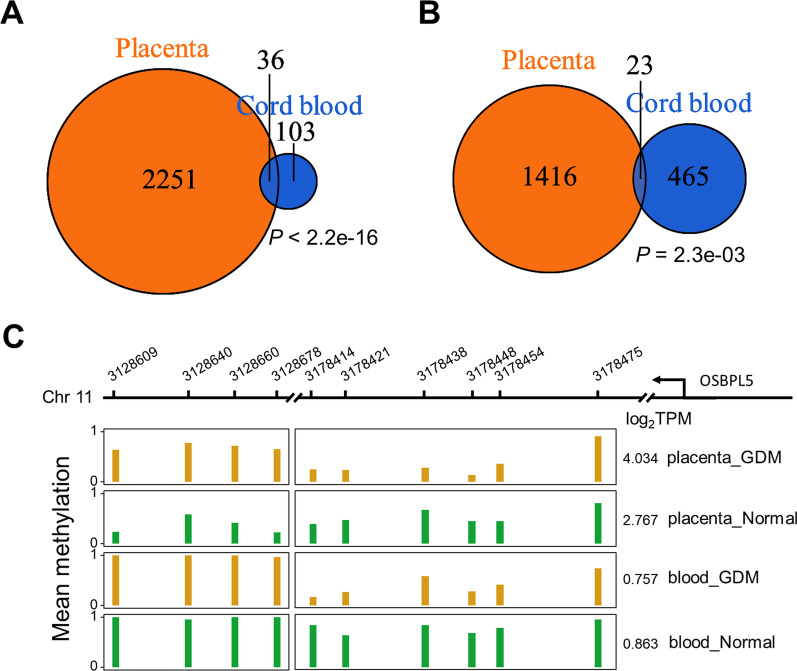
After discovering altered pathways shared by umbilical cord blood and placenta samples, we further explored the correlation between placenta and umbilical cord blood through an integrated analysis. The analysis revealed that 36 DMRs co-existed in the placentas and paired umbilical cord bloods (P value < 2.2e-16, odds ratio = 9.04, Fisher’s exact test, Fig. 4A). The top-ranking pathways enriched in the shared DMRs include “glycosphingolipid biosynthesis-globo and isoglobo series” (A4GALT, P value = 0.012), “N-glycan biosynthesis” (ALG10, P value = 0.042) and “cholesterol metabolism” (OSBPL5, P value = 0.042).
Fig. 4.
Significant correlation between the changes of placenta and umbilical cord blood, and involves glycolipid metabolism. A The Venn Diagram of DMGs in placenta and umbilical cord blood. Significance was tested by Fisher’s Exact Test. B The Venn Diagram of DEGs in placenta and umbilical cord blood. Significance was tested by Fisher’s Exact Test. C Mean methylation level of OSBPL5 DMR for placenta and umbilical cord blood in different group, log2TPM of each group is annotated on the right of each panel. Yellow bar represents GDM samples and green bar represents control samples
We then investigated altered gene expressions in placentas and paired umbilical cord blood samples by comparing GDM patients to the controls. The results demonstrated that 23 genes were overlapped between placenta and paired umbilical cord blood (P value = 2.3e-03, odds ratio = 2.04, Fisher’s exact test, Fig. 4B). The top-ranking pathways enriched in the shared genes include “ECM-receptor interaction” (FRAS1/SPP1, P value = 4.92e-03), “growth hormone synthesis, secretion and action” (ADCY5/SOCS3, P value = 8.84e-03) and “type II diabetes mellitus” (SOCS3, P value = 0.055).
The gene OSBPL5 encode a protein which belongs to the oxysterol-binding protein family, and proteins in this family are critical to maintenance of cholesterol balance in human body [31]. Comparison of the methylation levels of OSBPL5 across multiple specimens is illustrated in Fig. 4C. The DMR in placenta was annotated into one of the exons in the OSBPL5 gene, including four CpG sites (DNA methylation delta = 0.35; Fig. 4C left panel). Likewise, the DMR in umbilical cord blood located in an intron, including six CpG sites (DNA methylation delta = − 0.38; Fig. 4C right panel). Profiling of transcriptome showed that expression of OSBPL5 (log2TPM) in the placenta of GDM patient was 2.4-fold of that in the control group (GDM 4.034 vs. control 2.767), but the difference was small in umbilical cord blood (GDM 0.757 vs. control 0.863). These results suggest that the methylation and expression profile of umbilical cord blood are closely related to that in placenta, and the genes in glucose metabolism-related pathway are more likely to be regulated by DNA methylation.
Discussion
To the best of our knowledge, this is the first study that use epigenomic and transcriptomic assays to characterize placenta and the paired umbilical cord blood samples from GDM patients and controls. Our data demonstrated that placenta undergoes extensive methylation changes in genomic regions that are related to glucose metabolism-related pathways. Remarkably, methylation-altered genes in umbilical cord blood were associated with pathways insulin resistance and insulin secretion. And we also found that DMGs and DEGs were significantly overlapped between placenta and umbilical cord blood, indicating that the GDM conditions could affect fetus.
An early study found that insulin resistance in GDM patients led to a compensatory increase of the synthesis and secretion of insulin, which can promote the absorption and metabolism of blood glucose in islet β-cells [32]. However, when insulin resistance increases to a certain level, over-secreted insulin fails to maintain blood glucose levels in a normal range [33] and GDM patients could display hyperglycemia and hyperinsulinemia simultaneously [34]. GSEA result of placental RNA-seq data showed that several genes associated with the insulin secretion and the insulin resistance pathways were significantly up-regulated. More importantly, genes with both DNA methylation alternations and expression changes were identified, including PRKCZ, ADCY5, CACNA1C, PLCB1. The DNA methylation alternations in those genes might contribute to expression changes of these gene, which further aggravates insulin resistance and insulin secretion.
At the beginning of pregnancy, the placenta releases hormones into the blood of pregnant women, raising the blood glucose concentration to ensure adequate nutrition for the fetus [35, 36]. Our data showed that many genes related with hormone production and secretion were highly expressed in placenta of GDM patients, which involved in “cortisol synthesis and secretion,” “growth hormone synthesis and secretion” and “aldosterone synthesis and secretion pathway” (Table 2). However, it is reported that excessive cortisol levels in pregnant women’s blood can promote anxiety [37], increase the risk of obesity in offspring [38] and reduce children’s cognitive ability [39]. Our investigation discovered decreased methylation levels in multiple genes of the cortisol synthesis and secretion pathway, suggesting that cortisol levels could be altered in GDM patients through placenta. This hypothesis is further supported by the GSEA result of RNA-seq data, which listed pathway “cortisol synthesis and secret” as one of the most positively enriched pathways (Additional file 1: Fig. S2C). The genes of which both methylation statuses and expression levels varied in this pathway included CACNA1C, LDLR, and ADCY9. The CACNA1C gene encodes an alpha-1 subunit of a voltage-dependent calcium channel and is related to the exocytosis of many hormones. Xu et al. [40] reported that miR-153 worked as a negative regulator for the expression of CACNA1C, which further increased the secretion of insulin. An independent GWAS study revealed that CACNA1C gene is associated with diabetic cataract [41]. Besides, the co-binding of low-density lipoprotein receptor (LDLR) and insulin receptor (IR) reduces the ability of LDLR to clean up LDLR in blood, and insulin can destroy this co-binding [42].
Moreover, compared with control pregnancy, GDM patients had significantly lower serum total bile acids at 24 weeks of gestation (Additional file 1: Fig. S4A) and at 40 weeks of gestation (Additional file 1: Fig. S4B) (Wilcoxon rank-sum test, P value < 0.05). Previous studies have shown that bile acids can activate glucose induced insulin secretion [43] and have a strong correlation with insulin resistance and nonalcoholic fatty liver [44]. Based on the technique of liquid chromatography, Li et al. reported that lower levels of serum bile acids in early pregnancy were independently associated with an increased risk of GDM in Chinese pregnant women [45]. At the same time, our GSEA analysis also showed that several genes associated with the primary bile acid biosynthesis pathway were significantly down-regulated in placental tissues (Additional file 1: Fig. S4C). Expression of CH25H, CYP7B1 and HSD3B7 were significantly down-regulated in core enrichment genes, which played an important role in catalytic synthesis of bile acids. CYP7B1 is also involved in the synthesis of dehydroepiandrosterone and pregnenolone [46]. Both dehydroepiandrosterone and pregnenolone are related to hippocampus-associated memory and learning, which may reveal the relationship between GDM patients and the risk of memory decline. We believe that this is the first report to connect bile acid metabolism with GDM using placental high-throughput sequencing data. Notably, bile acid has been reported for the treatment of patients with obesity and diabetes [47], suggesting that bile acids can potentially be used in pregnant women to prevent and treat GDM in the future.
In placental tissues, 155 genes were differentially methylated and differentially expressed (partially displayed in Table 4), and some of these genes were involved in key metabolic pathway. From those genes, we manifested that the expression of three critical genes in the type II diabetes pathway was altered, including HK2 (down-regulated), PRKCZ (up-regulated) and SOCS3 (down-regulated). HK2 is the key gene of glycolysis, and its down-regulation has been shown directly leading to the reduction of glucose metabolism [48]. SOCS3 can inhibit insulin secretion, and down-regulation of this gene promote insulin secretion, resulting in insulin resistance [49]. The activation of PRKCZ depends on insulin, and over-expression of this gene affects the expression of the insulin-like growth factor 1 receptor (IGF1R) gene [50]. Additionally, previous research show that genes illustrated in Fig. 2F play key roles in insulin secretion, lipid droplet formation and are associated with type II diabetes [51–53]. Taken together, those results suggested that the pathogenesis of GDM may be similar to type II diabetes, characterized with excessive insulin secretion and insulin resistance, and DNA methylation plays an important role in the process.
Previous studies have indicated that the methylation levels of metabolism-related genes or genes involved in pathways could be changed after exposing to persistent high-glucose levels in pregnant women with GDM, which is termed “metabolic programming” [54–56]. In present results, several pathways showed descending methylation levels in placenta of GDM patients, including the “aldosterone synthesis and secretion pathway,” “insulin resistance” and “type II diabetes mellitus pathway”. Besides, for up-regulated genes in placenta of GDM patients, enriched pathways included “EGFR tyrosine kinase inhibitor resistance,” “insulin secretion and “growth hormone synthesis, secretion and action”. Our results indicated that the glucose and lipid metabolism of offspring were probably affected by intrauterine exposure to GDM, both in epigenetic and expression levels. Furthermore, there was a significant relevance between the changes of placenta and umbilical cord blood, regardless differentially methylated genes (Fig. 4A) or differentially expressed genes (Fig. 4B). Most notably, genes in the one carbon pool by folate pathway were significantly down-regulated in placentas and umbilical cord bloods, indicating that both pregnant women and fetuses with GDM may have the characteristics of folate deficiency. However, a meta-analysis regarding to the correlation between folic acid and the pathogenesis of GDM provides controversial results [57]. Larger cohort studies are necessary to explore the correlations. The above results show that there are apparent associations between the methylome and the transcriptome in placenta and umbilical cord blood, which share the common characteristics of insulin resistance and type II diabetes.
Still, there are few limitations in this study. Firstly, due to the complexity of placenta dissection structure, it is inevitable to obtain a variety of tissues and cell types during sampling. Using single-cell sequencing technology or spatial transcriptome technology will make a more in-depth study on the pathogenesis and mother–child interaction of GDM comparing with traditional bulk RNA-seq. In addition, in differentially gene expression analysis, we did not correct P value by FDR method, because very few genes (placenta 122, umbilical cord blood 0) had significant differences with FDR cutoff of 5%. Therefore, we used GSEA method to support our point of view, which results are largely consistent with DGE-based results. Finally, as confounding factor, BMI weights much in present research, and we did not find a suitable tool to exclude the influence of BMI during calling DMRs. Instead, we divided samples into two groups with equal number of samples, BMI-high and BMI-low, and used DSS to identify BMI-related DMRs, which were removed from subsequent analysis.
Conclusions
In summary, our study systematically profiled the methylome and transcriptome of placenta and umbilical cord blood of pregnant women with or without GDM, and investigated the relationship between methylation and gene expression. Our results provide detailed molecular changes among GDM patients compared with the controls, which pave the way for the further investigation of GDM pathogenicity.
Materials and methods
Patient recruitment
Pregnant women were recruited at Hangzhou Women’s Hospital (Hangzhou Maternity and Child Health Care Hospital, China) from January 1, 2020 to May 31, 2020. All participants signed a written consent forms and this study was approved by the ethics committee of Hangzhou Women’s Hospital. GDM is diagnosed according to the International Association of Diabetes and Pregnancy Study Groups (IADPSG) guidelines. Specifically, at least one of the following conditions must be met: 1. Fasting blood glucose ≥ 5.1 mmol/L; 2. Blood glucose ≥ 10.0 mmol/L one hour after the oral glucose tolerance test (OGTT); 3. Blood glucose ≥ 8.5 mmol/L two hours post the OGTT [58]. Pregnant women who met the following conditions were enrolled in this study: (1) age 24–39; (2) singleton pregnancy; (3) full-term birth (37–40 weeks of gestation); (4) detailed information for the OGTT; (5) records showing that the perinatal examination, delivery and infant physical examination were conducted in our hospital. Pregnant women with normal blood glucose and no history of adverse diseases were recruited in the control group. We excluded the pregnant women with the following conditions from our study: (1) having chronic diabetes complicated with pregnancy or preeclampsia. (2) suffering from liver or kidney dysfunctions, or other chronic diseases which required long-term drug treatment; (3) showing a mental disorder or a serious infection; (4) smoking, drinking and drug abuse during pregnancy; (5) fetal chromosome abnormalities; (6) lacking registration information in the Women’s and Children’s Insurance Systems. The difference of physiological parameters of pregnant women and newborns between GDM group and control group was performed by Wilcoxon rank-sum test.
Placenta and umbilical cord blood sampling
Within 15 min after delivery, 5 ml of neonatal umbilical cord blood was collected using two PAXGenRNA sampling tubes (2.5 ml/tube), and collected samples were stored at – 80 ℃ [59]. The placental tissues were collected from both the maternal and fetal side. Specifically, using a clean scalpel, one full-thick placental tissue was incised 5 cm from the perimeter of the placenta (a size of about 1 × 1 × 2 cm, including maternal and fetal side). Then, the tissues were rinsed with phosphate buffer (PBS) or normal saline until no visible blood and stored at – 80 ℃ in a cryopreservation tube [60]. DNA or RNA was extracted after homogenizing the tissues together.
RRBS and RNA-seq libraries preparation and sequencing
Genomic DNA of placenta and umbilical cord blood specimens was extracted by using the TIANamp genomic DNA kit (Cat. No. DP304-02, TIANGEN Biotech CO. LTD., Beijing, China) per the manufacture’s recommendations. 50–300 ng of purified genomic DNA was utilized to generate RRBS libraries as previously described [61]. Indexed RRBS libraries were pooled accordingly and sequenced in a NovaSeq 6000 sequencer (Illumina, USA) with 100-base paired-end reads. Similarly, total RNAs were isolated using a miRNeasy Mini kit (QIAGEN, Germany) according to the manufacture’s recommendations. We used the Ribo-off rRNA depletion kit (Cat. No. N406-01, Vazyme Biotech Co. Ltd, Nanjing, China) to remove ribosomal RNA from total RNAs following the manufacturer’s instructions. To generate RNA-seq libraries, 10–100 ng rRNA-depleted RNAs were utilized by using the VAHTS total RNA-seq (H/M/R) library prep kit for Illumina (Cat. No. NR603-01, Vazyme Biotech Co. Ltd, Nanjing, China) per the manufacturer’s instructions. We sequenced the RNA-seq libraries in the NovaSeq 6000 sequencer with 100-base paired-end reads.
The sequencing data volume of a single sample is around 8 gigabases (Gb) and the average sequencing depth of the target region of the enzyme digestion fragment is > 30 × , the coverage proportion of CpG region is > 40%, and the total data volume is 100 Gb. Similarly, transcriptome profile for each placenta and umbilical cord blood sample was also conducted. VAHTSTM Total RNA-seq (H/M/R) Library Prep Kit for Illumina® based on Ribo-zero method was used to constructed transcriptome RNA library and high-throughput sequencing was performed subsequently. 10 Gb volume of transcriptome data for each sample was necessary.
Data processing
For RRBS data, FastQC (version 0.11.9) [62] software was used to evaluate the quality of the original RRBS sequencing reads, and TrimGalore (version 0.6.5) [63] was further applied to trim adapters and low-quality bases. The trimmed reads were then aligned to the hg19 genome using BSMAP [64] software. Methylation calling of BAM file obtained in the previous step was conducted by MethylDackel [65]. R package DSS [66] was used to discover de novo DMRs. In this research, DMRs were defined as regions whose mean methylation level across CpG sites increased or decreased > 0.1, width > 50 bp, CpG number > 3, and P value < 0.05. Specifically, statistical tests for differential methylation at each CpG site were performed by function “DMLtest” with default parameters, which output was then processed using function “callDMR” (parameters: delta = 0.1, p.threshold = 0.05) to call DMRs-GDM by comparing GDM group with control group. Meanwhile, to eliminate the influence of confounding factors, we divided samples into two groups with equal number of samples, BMI-high and BMI-low, and used DSS to identify BMI-related DMRs, which were removed from subsequent analysis. Finally, DMRs were annotated using R package ChIPseeker [67] with default parameters to find differentially methylated genes (DMGs) and genomic distribution of DMRs.
As for RNA-seq data, quality control and base trimming were similarly conducted by using FastQC and TrimGalore, respectively. STAR (2.6.1d) [68] was used to map the quality-controlled reads to the hg19 genome. Then, the transcripts were quantified by using software kallisto [69], and the expression abundance of transcripts was generated accordingly. RNASeQC is used to count the quality of genome alignment. Subsequently, using the output of kallisto, gene expression levels were estimated from the abundance of transcription level by function “tximport” of R package tximport [70]. Gene differential expression analysis between GDM group and control group was performed by function DESeq of R package DESeq2 [71] using default parameters. Briefly, dispersion was estimated by fitting a dispersion-mean relation via a robust gamma-family GLM (General linear model); library size was estimated by the standard median ratio method introduced in DESeq; finally, the GLM coefficients were tested for significance using Wald test. Meanwhile, BMI was added as a covariate in the difference analysis to exclude the influence of confounding factors.
The unique mapping ratio of RRBS and RNA-seq data were illustrated in Additional file 1: Fig. S1A, B, respectively. All quality control table of RRBS and RNA-seq data were listed in Additional file 2: Table S1.1–S1.4.
Functional enrichment analysis
The genomic annotation of DMRs was conducted by R package ChIPseeker [67] and consequently were grouped to 6 classes: promoter, exon, intron, intergenic, UTR (untranslated region) and downstream regions. To investigate the enriched functional pathways of DMGs and DEGs, functional enrichment analysis of GO (Gene Ontology) biological process and KEGG pathway annotation was performed by the R package clusterProfiler [72] (parameters: pvalueCutoff = 0.05, qvalueCutoff = 0.05, pAdjustMethod = “fdr”).
Gene Set Enrichment Analysis (GSEA) was used to detect modest but coordinate changes in the expression of groups of functionally related genes [73]. Java command line tool gsea (http://www.gsea-msigdb.org/gsea/msigdb/download_file.jsp?filePath=/resources/software/gsea2-2.2.4.jar) was used to find the key pathways in gene expression profiles of placenta and umbilical cord blood (parameters: xtools.gsea.GseaPreranked -scoring_scheme weighted -collapse true -mode Max_probe -norm None -nperm 1000 -include_only_symbols true make_sets true -plot_top_x 20 -rnd_seed timestamp -set_max 3000 -set_min 15 -zip_report false -gui false). Specifically, all genes in gene expression profile were firstly sorted by Z-scores, which were converted from P values and associated log2 fold change using function qnorm. Then, enrichment significance test of each KEGG pathway was performed by GSEA, and P values of all pathways were adjusted for multiple testing. According to official advice, pathways with FDR < 0.25 were considered statistically significant.
Integrating analysis
To explore the effect of DNA methylation on gene expression under GDM environment, we compared differentially methylated genes and differentially expressed genes in a tissue of interest, including placenta and umbilical cord blood. A gene was counted as an overlap if it was differentially expressed and near DMR. The statistical significance of overlapping was tested by Fisher’s exact test with all genes in gene expression profile as a background. Similarly, we compared DMGs in placenta and umbilical cord blood, as well as DEGs in placenta and umbilical cord blood. Spearman’s rank correlation test was used to assess the significance of correlation between mean methylation level of DMGs and mean gene expression level of DEGs across samples. The biological function of overlapped genes was then evaluated by GO/KEGG functional enrichment analysis.
Supplementary Information
Additional file1: Fig. S1 Placenta shows genome-wide methylation alteration associated with glucose metabolism in GDM patients. Unique mapping ratio of placenta and blood samples for A RRBS data and B RNA-seq data. C Genomic distribution of DMRs in placenta. GO pathway enrichment result of placenta for D hyper-DMGs and E hypo-DMGs. The number in brackets represents the number of enriched genes. Fig. S2 Methylation contributes to expression change of genes associated with insulin signaling pathway. A KEGG pathway enrichment analysis of down-DMGs in placenta. B “Cortisol synthesis and secretion” C “Adherens junction,” D “Insulin resistance” and E “Type I diabetes mellitus” pathway enrichment result of placenta expression profile GSEA analysis. F GSEA analysis of placenta DMGs to placenta expression profile. Fig. S3 Alterations for umbilical cord blood were related to insulin secretion and resistance A Genomic distribution of DMRs in umbilical cord blood. B Differences of DMRs mean methylation levels between GDM and control umbilical cord blood samples. Yellow box represents GDM samples and green box represents control samples. *** P value < 0.001, Wilcoxon rank-sum test. Fig. S4 Differential characteristics of primary bile acid synthesis and autophagy between GDM and control samples. Total bile acid concentration between GDM and control sample in A 24 gestational week and B 40 gestational week. Wilcoxon rank-sum test. C ‘primary bile acid biosynthesis’ and D ‘Autophagy’ pathway enrichment result of placenta expression profile GSEA analysis.
Additional file2: Table S1.1 Quality control table of placenta RRBS data. Table S1.2 Quality control table of umbilical cord blood RRBS data. Table S1.3 Quality control table of placenta RNA-seq data. Table S1.4 Quality control table of umbilical cord blood RNA-seq data. Table S2 The 2779 differential methylation regions of placenta. Table S3 GO pathway enrichment of hyper-DMGs of placenta. Table S4 KEGG pathway enrichment of hyper-DMGs of placenta. Table S5 GO pathway enrichment of hypo-DMGs of placenta. Table S6 KEGG pathway enrichment of hypo-DMGs of placenta. Table S7 Up-regulated differentially expressed genes of placenta. Table S8 Down-regulated differentially expressed genes of placenta. Table S9 GO pathway enrichment of UP-DEGs of placenta. Table S10 KEGG pathway enrichment of UP-DEGs of placenta. Table S11 GO pathway enrichment of DN-DEGs of placenta. Table S12 KEGG pathway enrichment of DN-DEGs of placenta. Table S13 Positive enriched pathways of GSEA for placenta gene expression profile. Table S14 Negative enriched pathways of GSEA for placenta gene expression profile. Table S15 Up-regulated differentially expressed genes of umbilical cord blood. Table S16 Down-regulated differentially expressed genes of umbilical cord blood. Table S17 Positive enriched pathways of GSEA for umbilical cord blood gene expression profile. Table S18 Negative enriched pathways of GSEA for umbilical cord blood gene expression profile.
Acknowledgements
We thank all the participants of this research study and the staff helping with this study. We also thank the referees for making important suggestions for analysis of our data.
Abbreviations
- GDM
Gestational diabetes mellitus
- RRBS
Reduced representation bisulfite sequencing
- DMRs
Differentially methylated regions
- DEGs
Differentially expressed genes
- DMGs
Differentially methylated genes
- BMI
Body mass index
- MeDIP
Methylated DNA immunoprecipitation
- NGS
Next-generation sequencing
- OGTT
Oral glucose tolerance test
- HDL-c
High-density lipoprotein cholesterol
- LDL-c
Low-density lipoprotein cholesterol
- hyper-DMRs
Hyper-methylated DMRs
- hyper-DMGs
Hyper-methylated DMGs
- hypo-DMRs
Hypo-methylated DMRs
- hypo-DMGs
Hypo-methylated DMGs
- GO
Gene ontology
- FDR
False discovery rate
- KEGG
Kyoto encyclopedia of genes and genomes
- GSEA
Gene set enrichment analysis
- NES
Normalized enrichment score
- IADPSG
International association of diabetes and pregnancy study groups
- PBS
Phosphate buffer
- GLM
General linear model
- UTR
Untranslated region
Author contributions
WH contributed to study design; SL, NK, WH, JX, and YW contributed to data collection; JW, SH, HG, and JS involved in data analysis; WH and JS involved in study supervision; JW, SL, and HG contributed to manuscript writing. All authors have approved the submitted version of the manuscript. All authors agree to be personally accountable for the manuscript’s content. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (82173530), National Health Commission Scientific Research Fund–Major Science and Technology Program of Medicine and Health of Zhejiang Province (WKJ-ZJ-1911), Natural Science Foundation of Zhejiang Province (LQ21H040001), "Pioneer" and "Leading Goose" R&D Program of Zhejiang (2022C03102), Science and Technology Program of Medicine and Health of Hangzhou (ZD20200035 & OO2019054). Jiantao Shi is a recipient of the Hundred Talents Program Award of the Chinese Academy of Sciences.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
This study was performed in accordance with the Declaration of Helsinki and approved by the ethics committee of Hangzhou Women’s Hospital. All participants provided written informed consent. Ethics Number: No. 201800901.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sha Lu and Jiahao Wang contributed equally
Contributor Information
Jiantao Shi, Email: jtshi@sibcb.ac.cn.
Wensheng Hu, Email: huws@zju.edu.cn.
References
- 1.A American Diabetes. 2. Classification and diagnosis of diabetes: standards of medical care in diabetes-2020. 2020; 43: S14–31. [DOI] [PubMed]
- 2.A American Diabetes. Gestational diabetes mellitus. 2004; 27(Suppl 1): S88–90. [DOI] [PubMed]
- 3.Mack LR, Tomich PG. Gestational diabetes: diagnosis classification, and clinical care. Obstet Gynecol Clin. 2017;44:207–217. doi: 10.1016/j.ogc.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Plows JF, Stanley JL, Baker PN, et al. The pathophysiology of gestational diabetes mellitus. Int J Mol Sci. 2018;19(11):3342. doi: 10.3390/ijms19113342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McIntyre HD, Catalano P, Zhang C, et al. Gestational diabetes mellitus. Nat Rev Dis Prim. 2019;5:47. doi: 10.1038/s41572-019-0098-8. [DOI] [PubMed] [Google Scholar]
- 6.Alwash SM, McIntyre HD, Mamun A. The association of general obesity, central obesity and visceral body fat with the risk of gestational diabetes mellitus: evidence from a systematic review and meta-analysis. Obes Res Clin Pract. 2021;15(5):425–430. doi: 10.1016/j.orcp.2021.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Chuang YC, Huang L, Lee WY, et al. The association between weight gain at different stages of pregnancy and risk of gestational diabetes mellitus. J Diabetes Invest. 2021;13(2):359–366. doi: 10.1111/jdi.13648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Artal R, Catanzaro RB, Gavard JA, et al. A lifestyle intervention of weight-gain restriction: diet and exercise in obese women with gestational diabetes mellitus. App Physiol Nutr Metab. 2007;32:596–601. doi: 10.1139/H07-024. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz N, Nachum Z, Green MS. The prevalence of gestational diabetes mellitus recurrence–effect of ethnicity and parity: a metaanalysis. Am J Obstetr Gynecol. 2015;213:310–317. doi: 10.1016/j.ajog.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Ren X, He L, et al. Maternal age and the risk of gestational diabetes mellitus: a systematic review and meta-analysis of over 120 million participants. Diabetes Res Clin Pract. 2020;162:108044. doi: 10.1016/j.diabres.2020.108044. [DOI] [PubMed] [Google Scholar]
- 11.Moosazadeh M, Asemi Z, Lankarani KB, et al. Family history of diabetes and the risk of gestational diabetes mellitus in Iran: a systematic review and meta-analysis. Diabetes Metab Syndr Clin Res Rev. 2017;11(Suppl 1):S99–S104. doi: 10.1016/j.dsx.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 12.Johns EC, Denison FC, Norman JE, et al. Gestational diabetes mellitus: mechanisms treatment, and complications. Trends Endocrinol Metab. 2018;29:743–754. doi: 10.1016/j.tem.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Ruchat SM, Houde AA, Voisin G, et al. Gestational diabetes mellitus epigenetically affects genes predominantly involved in metabolic diseases. Epigenetics. 2013;8:935–943. doi: 10.4161/epi.25578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szmuilowicz ED, Josefson JL, Metzger BE. Gestational diabetes mellitus. Endocrinol Metab Clin. 2019;48:479–493. doi: 10.1016/j.ecl.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nature Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 16.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 17.Hanson M, Godfrey KM, Lillycrop KA, et al. Developmental plasticity and developmental origins of non-communicable disease: theoretical considerations and epigenetic mechanisms. Prog Biophys Mol Biol. 2011;106:272–280. doi: 10.1016/j.pbiomolbio.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 18.Franke K, Harder T, Aerts L, et al. ‘Programming’ of orexigenic and anorexigenic hypothalamic neurons in offspring of treated and untreated diabetic mother rats. Brain Res. 2005;1031:276–283. doi: 10.1016/j.brainres.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Amri K, Freund N, Duong Van Huyen JP, et al. Altered nephrogenesis due to maternal diabetes is associated with increased expression of IGF-II/mannose-6-phosphate receptor in the fetal kidney. Diabetes. 2001;50:1069–1075. doi: 10.2337/diabetes.50.5.1069. [DOI] [PubMed] [Google Scholar]
- 20.Kitajima M, Oka S, Yasuhi I, et al. Maternal serum triglyceride at 24–32 weeks’ gestation and newborn weight in nondiabetic women with positive diabetic screens. Obstetr Gynecol. 2001;97:776–780. doi: 10.1016/S0029-7844(01)01328-X. [DOI] [PubMed] [Google Scholar]
- 21.Houde AA, Ruchat SM, Allard C, et al. LRP1B, BRD2 and CACNA1D: new candidate genes in fetal metabolic programming of newborns exposed to maternal hyperglycemia. Epigenomics. 2015;7:1111–1122. doi: 10.2217/epi.15.72. [DOI] [PubMed] [Google Scholar]
- 22.Rong C, Cui X, Chen J, et al. DNA methylation profiles in placenta and its association with gestational diabetes mellitus. Exp Clin Endocrinol Diabetes. 2015;123:282–288. doi: 10.1055/s-0034-1398666. [DOI] [PubMed] [Google Scholar]
- 23.Gu H, Bock C, Mikkelsen TS, et al. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods. 2010;7:133–136. doi: 10.1038/nmeth.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu H, Smith ZD, Bock C, et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nature Protoc. 2011;6:468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Gerstein M, Snyder M. RNA-seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sreekumar R, Halvatsiotis P, Schimke JC, et al. Gene expression profile in skeletal muscle of type 2 diabetes and the effect of insulin treatment. Diabetes. 2002;51(1):913–920. doi: 10.2337/diabetes.51.6.1913. [DOI] [PubMed] [Google Scholar]
- 27.Yang X, Pratley RE, Tokraks S, et al. Microarray profiling of skeletal muscle tissues from equally obese, non-diabetic insulin-sensitive and insulin-resistant pima Indians. Diabetologia. 2002;45:1584–1593. doi: 10.1007/s00125-002-0901-y. [DOI] [PubMed] [Google Scholar]
- 28.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 29.Longo M, Raciti GA, Zatterale F, et al. Epigenetic modifications of the Zfp/ZNF423 gene control murine adipogenic commitment and are dysregulated in human hypertrophic obesity. Diabetologia. 2018;61:369–380. doi: 10.1007/s00125-017-4471-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghaben AL, Scherer PE. Adipogenesis and metabolic health. Nature Rev Mol Cell Biol. 2019;20:242–258. doi: 10.1038/s41580-018-0093-z. [DOI] [PubMed] [Google Scholar]
- 31.Lehto M, Laitinen S, Chinetti G, et al. The OSBP-related protein family in humans. J Lipid Res. 2001;42:1203–1213. doi: 10.1016/S0022-2275(20)31570-4. [DOI] [PubMed] [Google Scholar]
- 32.Tripathi BK, Srivastava AK. Diabetes mellitus: complications and therapeutics. Med Sci Monit. 2006;12:130–147. [PubMed] [Google Scholar]
- 33.Petersen MC, Shulman GI. Mechanisms of insulin action and insulin resistance. Physiol Rev. 2018;98:2133–2223. doi: 10.1152/physrev.00063.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee Y, Fluckey JD, Chakraborty S, et al. Hyperglycemia- and hyperinsulinemia-induced insulin resistance causes alterations in cellular bioenergetics and activation of inflammatory signaling in lymphatic muscle. FASEB J. 2017;31:2744–2759. doi: 10.1096/fj.201600887R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keller-Wood M, Feng X, Wood CE, et al. Elevated maternal cortisol leads to relative maternal hyperglycemia and increased stillbirth in ovine pregnancy. Am J Physiol Regul Integr Comp Physiol. 2014;307:R405–R413. doi: 10.1152/ajpregu.00530.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Cianni G, Miccoli R, Volpe L, et al. Intermediate metabolism in normal pregnancy and in gestational diabetes. Diabetes Metab Res Rev. 2003;19:259–270. doi: 10.1002/dmrr.390. [DOI] [PubMed] [Google Scholar]
- 37.Murphy SE, Braithwaite EC, Hubbard I, et al. Salivary cortisol response to infant distress in pregnant women with depressive symptoms. Arch Womens Ment Health. 2015;18:247–253. doi: 10.1007/s00737-014-0473-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Entringer S, Buss C, Rasmussen JM, et al. Maternal cortisol during pregnancy and infant adiposity: a prospective investigation. J Clin Endocrinol Metab. 2017;102:1366–1374. doi: 10.1210/jc.2016-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LeWinn KZ, Stroud LR, Molnar BE, et al. Elevated maternal cortisol levels during pregnancy are associated with reduced childhood IQ. Int J Epidemol. 2009;38:1700–1710. doi: 10.1093/ije/dyp200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu H, Abuhatzira L, Carmona GN, et al. The Ia-2beta intronic miRNA, miR-153, is a negative regulator of insulin and dopamine secretion through its effect on the Cacna1c gene in mice. Diabetologia. 2015;58:2298–2306. doi: 10.1007/s00125-015-3683-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang C, Zhang K, Veluchamy A, et al. A genome-wide association study provides new evidence that CACNA1C gene is associated with diabetic cataract. Invest Ophthalmol Vis Sci. 2016;57:2246–2250. doi: 10.1167/iovs.16-19332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramakrishnan G, Arjuman A, Suneja S, et al. The association between insulin and low-density lipoprotein receptors. Diabetes Vasc Dis Res. 2012;9:196–204. doi: 10.1177/1479164111430243. [DOI] [PubMed] [Google Scholar]
- 43.Dufer M, Horth K, Wagner R, et al. Bile acids acutely stimulate insulin secretion of mouse beta-cells via farnesoid X receptor activation and K(ATP) channel inhibition. Diabetes. 2012;61:1479–1489. doi: 10.2337/db11-0815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei J, Qiu DK, Ma X. Bile acids and insulin resistance: implications for treating nonalcoholic fatty liver disease. J Dig Dis. 2009;10:85–90. doi: 10.1111/j.1751-2980.2009.00369.x. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Huo X, Cao YF, et al. Bile acid metabolites in early pregnancy and risk of gestational diabetes in Chinese women: a nested case-control study. EBioMedicine. 2018;35:317–324. doi: 10.1016/j.ebiom.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stiles AR, McDonald JG, Bauman DR, et al. CYP7B1: one cytochrome P450, two human genetic diseases, and multiple physiological functions. J Biol Chem. 2009;284:28485–28489. doi: 10.1074/jbc.R109.042168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chavez-Talavera O, Tailleux A, Lefebvre P, et al. Bile acid control of metabolism and inflammation in obesity, type 2 diabetes dyslipidemia, and nonalcoholic fatty liver disease. Gastroenterology. 2017;152:1679–1694. doi: 10.1053/j.gastro.2017.01.055. [DOI] [PubMed] [Google Scholar]
- 48.Tan VP, Miyamoto S. HK2/hexokinase-II integrates glycolysis and autophagy to confer cellular protection. Autophagy. 2015;11:963–964. doi: 10.1080/15548627.2015.1042195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang SJ, Xu CQ, Wu JW, et al. SOCS3 inhibits insulin signaling in porcine primary adipocytes. Mol Cell Biochem. 2010;345:45–52. doi: 10.1007/s11010-010-0558-7. [DOI] [PubMed] [Google Scholar]
- 50.Seto KK, Andrulis IL. Atypical protein kinase C zeta: potential player in cell survival and cell migration of ovarian cancer. PLoS ONE. 2015;10:e0123528. doi: 10.1371/journal.pone.0123528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khanom R, Nguyen CT, Kayamori K, et al. Keratin 17 Is induced in oral cancer and facilitates tumor growth. PLoS ONE. 2016;11:e0161163. doi: 10.1371/journal.pone.0161163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hwang HJ, Yang YR, Kim HY, et al. Phospholipase C-beta1 potentiates glucose-stimulated insulin secretion. FASEB J. 2019;33:10668–10679. doi: 10.1096/fj.201802732RR. [DOI] [PubMed] [Google Scholar]
- 53.Rahman MH, Peng S, Hu X, et al. A network-based bioinformatics approach to identify molecular biomarkers for type 2 diabetes that are linked to the progression of neurological diseases. Int J Environ Res Public Health. 2020;17(3):1035. doi: 10.3390/ijerph17031035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.El Hajj N, Pliushch G, Schneider E, et al. Metabolic programming of MEST DNA methylation by intrauterine exposure to gestational diabetes mellitus. Diabetes. 2013;62:1320–1328. doi: 10.2337/db12-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Desgagne V, Hivert MF, St-Pierre J, et al. Epigenetic dysregulation of the IGF system in placenta of newborns exposed to maternal impaired glucose tolerance. Epigenomics. 2014;6:193–207. doi: 10.2217/epi.14.3. [DOI] [PubMed] [Google Scholar]
- 56.Petropoulos S, Guillemin C, Ergaz Z, et al. Gestational diabetes alters offspring DNA methylation profiles in human and rat: identification of key pathways involved in endocrine system disorders insulin signaling, diabetes signaling, and ILK signaling. Endocrinology. 2015;156:2222–2238. doi: 10.1210/en.2014-1643. [DOI] [PubMed] [Google Scholar]
- 57.Wang L, Hou Y, Meng D, et al. Vitamin B12 and folate levels during pregnancy and risk of gestational diabetes mellitus: a systematic review and meta-analysis. Front Nutr. 2021;8:670289. doi: 10.3389/fnut.2021.670289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sacks DA, Hadden DR, Maresh M, et al. Frequency of gestational diabetes mellitus at collaborating centers based on IADPSG consensus panel-recommended criteria the hyperglycemia and adverse pregnancy outcome (HAPO) study. Diabetes Care. 2012;35:526–528. doi: 10.2337/dc11-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Landon MB, Gabbe SG. Gestational diabetes mellitus. Obstet Gynecol. 2011;118:1379–1393. doi: 10.1097/AOG.0b013e31823974e2. [DOI] [PubMed] [Google Scholar]
- 60.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 61.Charlton J, Downing TL, Smith ZD, et al. Global delay in nascent strand DNA methylation. Nat Struct Mol Biol. 2018;25:327–332. doi: 10.1038/s41594-018-0046-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.B Bioinformatics. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. 2019.
- 63.B Bioinformatics. https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/. 2019.
- 64.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinform. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.D Ryan. https://github.com/dpryan79/MethylDackel. 2017.
- 66.Feng H, Conneely KN, Wu H. A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res. 2014;42:e69. doi: 10.1093/nar/gku154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 68.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bray NL, Pimentel H, Melsted P, et al. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 70.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu G, Wang LG, Han Y, et al. Clusterprofiler: an R package for comparing biological themes among gene clusters. OMICS J Integr Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file1: Fig. S1 Placenta shows genome-wide methylation alteration associated with glucose metabolism in GDM patients. Unique mapping ratio of placenta and blood samples for A RRBS data and B RNA-seq data. C Genomic distribution of DMRs in placenta. GO pathway enrichment result of placenta for D hyper-DMGs and E hypo-DMGs. The number in brackets represents the number of enriched genes. Fig. S2 Methylation contributes to expression change of genes associated with insulin signaling pathway. A KEGG pathway enrichment analysis of down-DMGs in placenta. B “Cortisol synthesis and secretion” C “Adherens junction,” D “Insulin resistance” and E “Type I diabetes mellitus” pathway enrichment result of placenta expression profile GSEA analysis. F GSEA analysis of placenta DMGs to placenta expression profile. Fig. S3 Alterations for umbilical cord blood were related to insulin secretion and resistance A Genomic distribution of DMRs in umbilical cord blood. B Differences of DMRs mean methylation levels between GDM and control umbilical cord blood samples. Yellow box represents GDM samples and green box represents control samples. *** P value < 0.001, Wilcoxon rank-sum test. Fig. S4 Differential characteristics of primary bile acid synthesis and autophagy between GDM and control samples. Total bile acid concentration between GDM and control sample in A 24 gestational week and B 40 gestational week. Wilcoxon rank-sum test. C ‘primary bile acid biosynthesis’ and D ‘Autophagy’ pathway enrichment result of placenta expression profile GSEA analysis.
Additional file2: Table S1.1 Quality control table of placenta RRBS data. Table S1.2 Quality control table of umbilical cord blood RRBS data. Table S1.3 Quality control table of placenta RNA-seq data. Table S1.4 Quality control table of umbilical cord blood RNA-seq data. Table S2 The 2779 differential methylation regions of placenta. Table S3 GO pathway enrichment of hyper-DMGs of placenta. Table S4 KEGG pathway enrichment of hyper-DMGs of placenta. Table S5 GO pathway enrichment of hypo-DMGs of placenta. Table S6 KEGG pathway enrichment of hypo-DMGs of placenta. Table S7 Up-regulated differentially expressed genes of placenta. Table S8 Down-regulated differentially expressed genes of placenta. Table S9 GO pathway enrichment of UP-DEGs of placenta. Table S10 KEGG pathway enrichment of UP-DEGs of placenta. Table S11 GO pathway enrichment of DN-DEGs of placenta. Table S12 KEGG pathway enrichment of DN-DEGs of placenta. Table S13 Positive enriched pathways of GSEA for placenta gene expression profile. Table S14 Negative enriched pathways of GSEA for placenta gene expression profile. Table S15 Up-regulated differentially expressed genes of umbilical cord blood. Table S16 Down-regulated differentially expressed genes of umbilical cord blood. Table S17 Positive enriched pathways of GSEA for umbilical cord blood gene expression profile. Table S18 Negative enriched pathways of GSEA for umbilical cord blood gene expression profile.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.