Abstract
High immune-cell infiltration in glioblastomas (GBMs) leads to immunotherapy resistance. Emerging evidence has shown that zinc finger Asp-His-His-Cyc-type (ZDHHC) palmitoyl transferases participate in regulating tumor progression and the immune microenvironment. In the present study, a large cohort of patients with gliomas from The Cancer Genome Atlas (TCGA) and Rembrandt databases was included to perform omics analysis of ZDHHCs in gliomas. CCK-8, flow cytometry, quantitative real-time PCR, western blotting, and transwell assays were performed to determine the effects of ZDHHC inhibition on glioma cells and microglia. We found that five (ZDHHC11, ZDHHC12, ZDHHC15, ZDHHC22, and ZDHHC23) out of 23 ZDHHCs were aberrantly expressed in gliomas and might play their roles through the phosphatidylinositol 3-kinase/protein kinase B (PI3K/AKT) signaling pathway. Further results indicated that inhibition of ZDHHCs with 2-bromopalmitate (2-BP) suppressed glioma-cell viability and autophagy, as well as promoted apoptosis. Targeting ZDHHCs also promoted the sensitivity of glioma cells to temozolomide (TMZ) chemotherapy. In addition, the inhibition of ZDHHCs weakened the migratory ability of microglia induced by glioma cells in vitro and in vivo. Taken together, our findings suggest that the inhibition of ZDHHCs suppresses glioma-cell viability and microglial infiltration. Targeting ZDHHCs may be promising for glioma treatments.
Keywords: MT: Bioinformatics, ZDHHCs, immune-cell infiltration, gliomas, post-translational modification, signaling pathway
Graphical abstract
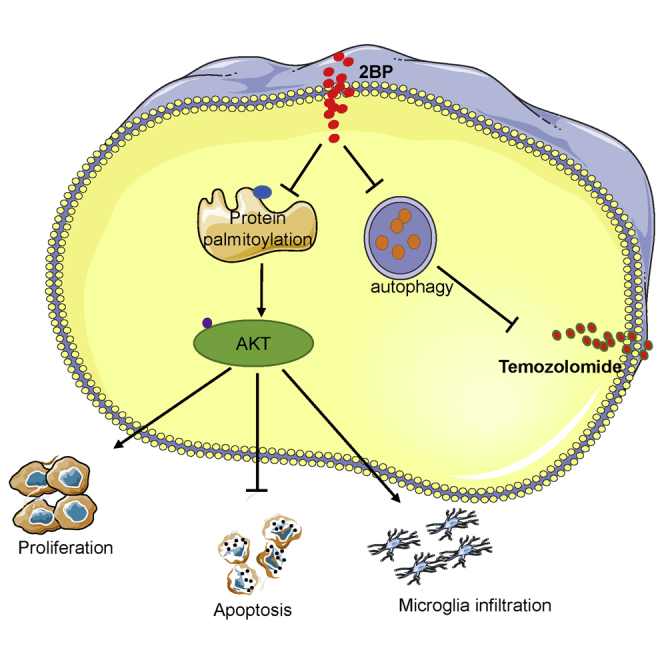
Glioma tissues exhibit aberrant protein palmitoylation. We found that several palmitoyl transferases are differentially expressed in gliomas and participate in regulating tumor progression and immune infiltration. Palmitoyl transferases inhibitor suppresses glioma-cell viability and microglial infiltration and may be promising for glioma treatments.
Introduction
Gliomas are the most lethal and common tumors in the central nervous system. Following surgical interventions and aggressive chemoradiotherapeutic treatments, many patients with glioblastomas (GBMs) still exhibit poor prognoses.1,2 In recent years, immune-checkpoint inhibitors, such as programmed cell death-1/programmed cell death-ligand 1 (PD-1/PD-L1) and cytotoxic-T-lymphocyte-associated antigen-4 (CTLA-4) inhibitors, have been used as cancer immunotherapies and have made a breakthrough in improving clinical treatments.3 In a phase I clinical trial, intracerebral administration of CTLA-4 inhibitor ipilimumab and PD-1 inhibitor nivolumab in combination with intravenous administration of nivolumab following maximal safe resection of recurrent GBMs was feasible, safe, and associated with encouraging overall survival.4 In a single-arm phase II clinical trial, a presurgical dose of nivolumab followed by postsurgical nivolumab could remodel the tumor immune microenvironment in resectable GBMs.5 However, high immune-cell infiltration mediated by GBM cells establishes an immunosuppressive tumor microenvironment, which can induce immunotherapy resistance.6,7 Hence, it is critical to identify novel targets associated with high immune-cell infiltration in gliomas.
S-palmitoylation is a reversible post-translational lipid modification, which is a universal feature of human cells that controls the localization, stability, and function of various proteins.8 Protein palmitoylation involves the covalent attachment of fatty acyl chains, typically a palmitate (C16:0), to internal cysteine residues of a protein via labile thioester linkages, and palmitoyl transferases catalyze this reaction.9,10 Palmitoyl transferases are a zinc finger Asp-His-His-Cys-type (ZDHHC) family containing 23 distinct genes (named ZDHHC1–ZDHHC24, excluding ZDHHC10) in mammals.11,12
Emerging evidence has shown that ZDHHC-mediated protein palmitoylation participates in regulating tumor progression and the immune microenvironment. For example, loss of ZDHHC9 has been shown to prevent the oncogene, N-Ras, from transforming bone marrow cells in a ZDHHC9-knockout mouse model.13 In colorectal cancer, ZDHHC3 has been found to be upregulated and to promote protein PD-L1 palmitoylation, which weakens T cell immune responses against tumors. Furthermore, inhibition of PD-L1 palmitoylation can activate anti-cancer immunity.14 A recent study showed that IFNGR1 palmitoylation stabilized IFNGR1 to drive immune evasion and immunotherapy resistance in tumors.15 In the central nervous system, compared with that in normal brain tissues, glioma tissues exhibit aberrant protein palmitoylation.16 Previous studies have found that several ZDHHCs indeed participated in regulating glioma cell survival, transition of stem cells, glycolysis, chemotherapy resistance, and progression. For example, ZDHHC18 and ZDHHC23 were distributed in the context of their specific niches in different GBM subsets to regulate the cellular plasticity of these subtypes, which contribute to the transition of glioma stem cells in GBM and cell survival under the stressful tumor microenvironment.17 ZDHHC17 could target JNK and p38 mitogen-activated protein kinase (MAPK) to drive GBM chemotherapy resistance.18 Moreover, ZDHHC5-mediated EZH2 palmitoylation was shown to drive p53-mutant glioma malignant development and progression.19 A recent study reported that GLUT1 palmitoylation-mediated DHHC9 promotes GBM glycolysis and tumorigenesis.20 Nevertheless, the expression profiles and potential biological functions of ZDHHCs, as well as whether ZDHHCs may represent a drug target in gliomas, have yet to be well elucidated.
In the present study, we first investigated the expression profiles of ZDHHCs through corresponding public datasets. Next, we explored the correlation between the differential expression of ZDHHCs and survival times of patients with glioma. In addition, we assessed the potential roles and mechanisms of ZDHHCs in gliomas via performing Gene Ontology (GO) enrichment analysis, immune-infiltration analysis, and gene set enrichment analysis (GSEA). Finally, CCK-8, flow cytometry, quantitative real-time PCR, western blotting, transwell, and immunofluorescence assays were also performed.
Results
Expression profiles of ZDHHCs in patients with gliomas
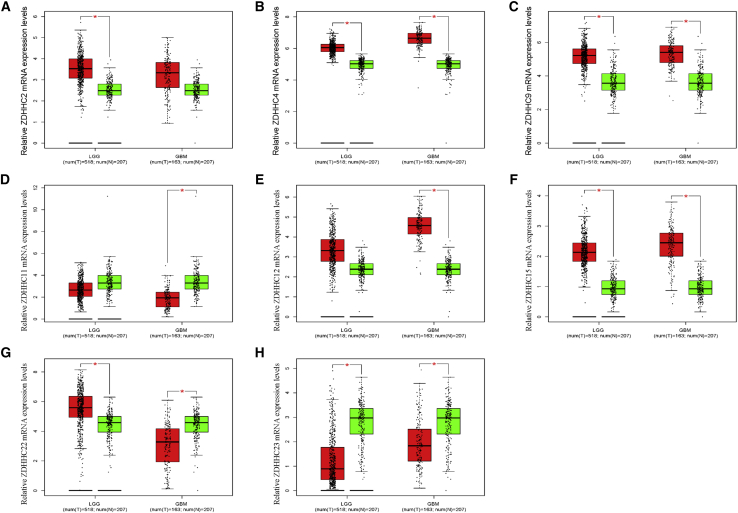
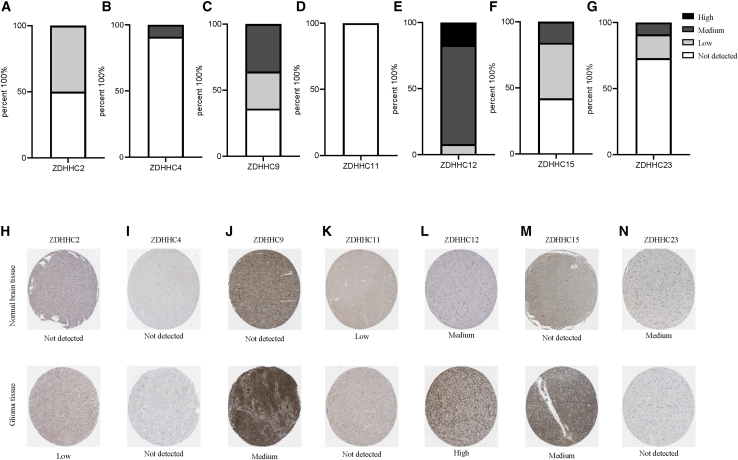
The details of what we have done were summarized and presented in Figure S1 and Table 1. The ZDHHCs are a gene family comprising 23 distinct genes. We first analyzed Pearson correlations among these genes. In lower grade gliomas (LGGs) (grades II and III), most ZDHHCs were positively associated with each other. Only ZDHHC12 was negatively correlated with other ZDHHCs (except for ZDHHC5 and ZDHHC19) in LGGs (Figure S2A). In GBMs (grade IV), Pearson correlations among ZDHHCs were lower than those in LGGs (Figure S1B). Next, we compared transcriptional levels of ZDHHCs between glioma tissues and normal brain tissues in The Cancer Genome Atlas (TCGA) database. As shown in Figure 1 and Table 1 (column 1), five ZDHHCs (ZDHHC2, ZDHHC4, ZDHHC9, ZDHHC12, and ZDHHC15) were upregulated and two ZDHHCs (ZDHHC11 and ZDHHC23) were downregulated in glioma tissues. Interestingly, ZDHHC22 was upregulated in LGGs, whereas it was downregulated in GBMs (Figure 1G). Microarray data from the Rembrandt database presented a similar result (Figure S3). However, we did not observe differential expression levels of ZDHHC9 between normal brain tissues and glioma tissues in the Rembrandt database (Figure S3C). In addition to gliomas, data from the TIMER (https://cistrome.shinyapps.io/timer/) dataset showed that ZDHHC2, ZDHHC4, ZDHHC9, ZDHHC11, ZDHHC12, ZDHHC15, ZDHHC22, and ZDHHC23 were also aberrantly expressed in the majority of tumors (Figure S4).21 In gliomas, compared with those in LGGs, the expression levels of six ZDHHCs (ZDHHC2, ZDHHC4, ZDHHC9, ZDHHC12, ZDHHC15, and ZDHHC23) were higher in GBMs. In contrast, ZDHHC11 and ZDHHC22 expression levels in GBMs were lower than those in LGGs (Figures S5A–S5H). With the exceptions of ZDHHC4 and ZDHHC15, there was a difference in ZDHHC mRNA expression among the three molecular subtypes of gliomas (Figures S5I–S5P). Moreover, isocitrate dehydrogenase (IDH) wild-type and mutant gliomas also presented differential expression levels of ZDHHCs (Figures S5Q–S5X). We also evaluated these ZDHHC protein levels through the Human Protein Atlas (HPA) database. There were no data related to ZDHHC22 protein levels in this database. As shown in Figure 2 and Table 1 (column 2 and column 3), compared with normal brain tissues, ZDHHC2, ZDHHC9, ZDHHC12, and ZDHHC15 protein levels were upregulated in glioma tissues, while ZDHHC11 and ZDHHC23 protein levels were downregulated. The protein levels of the five ZDHHCs (ZDHHC2, ZDHHC11, ZDHHC12, ZDHHC15, and ZDHHC23) were consistent with their corresponding mRNA levels.
Table 1.
Omics analysis of ZDHHCs in gliomas
| Genes | RNA expression levels (tumor versus normal) | Protein expression levels (percent account for total included glioma cases) | Protein expression levels (normal cerebral cortex) | Methylation levels (IDH mutant versus wild type) | Mutation frequency |
|---|---|---|---|---|---|
| ZDHHC1 | no significance | not detected | not detected | hypermethylation | 0 |
| ZDHHC2 | upregulated | not detected: 50% low: 50% |
not detected | hypermethylation | 0.6% |
| ZDHHC3 | no significance | not detected: 25% low: 8% medium: 58% high: 9% |
medium | hypermethylation | 1.4% |
| ZDHHC4 | upregulated | not detected: 91% medium: 9% |
not detected | hypermethylation | 1.2% |
| ZDHHC5 | no significance | not detected: 25% low: 58% medium: 9% high: 8% |
low | hypermethylation | 0 |
| ZDHHC6 | no significance | not detected: 50% low: 17% medium: 33% |
not detected | hypermethylation | 1.7% |
| ZDHHC7 | no significance | not detected | not detected | hypermethylation | 0.2% |
| ZDHHC8 | no significance | not detected: 46% low: 54% |
not detected | hypermethylation | 0.6% |
| ZDHHC9 | upregulated | not detected: 36% low: 28% medium: 36% |
low | no significance | 1.4% |
| ZDHHC11 | downregulated | not detected: 100% | low | hypermethylation | 1.6% |
| ZDHHC12 | upregulated | low: 8% medium: 75% high: 17% |
medium | hypermethylation | 1% |
| ZDHHC13 | no significance | not detected: 84% low: 8% medium: 8% |
not detected | hypermethylation | 1.6% |
| ZDHHC14 | no significance | not detected: 20% low: 50% medium: 20% high: 10% |
medium | hypermethylation | 2.1% |
| ZDHHC15 | upregulated | not detected: 42% low: 42% medium: 16% |
not detected | no significance | 2.9% |
| ZDHHC16 | no significance | not detected | not detected | hypermethylation | 0.4% |
| ZDHHC17 | no significance | low: 18% medium: 73% high: 9% |
high | hypermethylation | 0.6% |
| ZDHHC18 | no significance | not detected: 27% low: 18% high: 55% |
not detected | hypermethylation | 0.2% |
| ZDHHC19 | no significance | not available | not available | hypermethylation | 2.9% |
| ZDHHC20 | no significance | not detected: 50% low: 42% medium: 8% |
not detected | no significance | 1.9% |
| ZDHHC21 | no significance | low: 8% medium: 50% high: 42% |
medium | hypomethylation | 2.3% |
| ZDHHC22 | upregulated in LGGs downregulated in GBM |
not available | not available | hypomethylation | 0.8% |
| ZDHHC23 | downregulated | not detected: 73% low: 18% medium: 9% |
medium | hypomethylation | 1.4% |
| ZDHHC24 | no significance | not detected: 58% low: 42% |
not detected | hypomethylation | 0 |
Figure 1.
(A–H) Relative mRNA levels of the dysregulated ZDHHCs in glioma tissues and normal brain tissues. ∗p < 0.05 ; ∗∗p < 0.01
Figure 2.
Relative protein levels of seven ZDHHCs in normal brain tissues and glioma tissues
(A–G) Histograms of ZDHHC expression levels in glioma samples from the Protein Atlas. In total, 11 to 12 samples were analyzed for ZDHHCs. Immunohistochemical (IHC) staining was evaluated as high, medium, or low staining or not detected. No information could be retrieved for ZDHHC22. (H–N) Representative IHC staining for ZDHHCs in normal brain tissues and glioma tissues is shown.
DNA methylation of the CpG island of a gene promoter is the most common mechanism that results in the repression of gene expression.22 We explored the methylation levels of ZDHHC promoters via the MethSurv (https://biit.cs.ut.ee/methsurv/) and Xena Functional Genomics Explorer (https://xenabrowser.net/heatmap/) tools.23,24 Apart from ZDHHC11, ZDHHC22, and ZDHHC23, we noticed that other ZDHHCs (e.g., ZDHHC18 and ZDHHC19) exhibited low mRNA levels in both glioma tissues and normal brain tissues. Next, we investigated the methylation levels of ZDHHC promoters in gliomas. As shown in Figure S6, the promoters of ZDHHC7, ZDHHC11, ZDHHC18, and ZDHHC19 presented high methylation levels, which might account for the low expression levels of these genes in gliomas. However, there were no high methylation levels of the promoters of ZDHHC22 and ZDHHC23. In addition to that, we also compared methylation levels between IDH wild-type and mutant glioma samples. With respect to IDH wild-type gliomas, the majority of ZDHHCs presented higher methylation levels in IDH mutant glioma samples (Figure S7; Table 1, column 5).
Gene mutations frequently occur during tumor development. Many patients with LGGs harbor IDH gene-family mutations, which were consequently included in the classification of newly diagnosed gliomas by the World Health Organization in 2016.2 Next, we investigated genomic alterations of each ZDHHC gene in gliomas by using data available from the cBioPortal (http://www.cbioportal.org/datasets).25,26 Results presented in Figure S8 and Table 1 (column 6) showed that 21 LGGs and 22 GBMs out of all ZDHHCs harbored at least one type of genomic alteration (i.e., missense mutation, amplification, or deep deletion). The overall mutation frequency of ZDHHCs in LGGs was higher than that in GBMs. Among the 23 genes, ZDHHC14, ZDHHC15, ZDHHC19, and ZDHHC21 were the most frequently altered genes (2.1%, 2.9%, 2.9%, and 2.1%, respectively) in LGGs. However, there was no significant difference in survival times between altered and non-altered groups in gliomas (data not shown).
Prognostic values of ZDHHCs in patients with glioma
After obtaining the expression profiles of ZDHHCs, we then searched for an association between ZDHHC expression levels and survival times of patients with glioma. We extracted hazard ratios (HRs) from TCGA and Rembrandt databases by using an optimized algorithm via the Gliovis tool. As shown in Table 2, patients with glioma and with high expression levels of ZDHHC1, ZDHHC4, ZDHHC5, ZDHHC12, ZDHHC13, ZDHHC15, ZDHHC18, ZDHHC23, and ZDHHC24 had a poor overall survival. In contrast, data from the Rembrandt database showed that high expression levels of ZDHHC1, ZDHHC3, ZDHHC4, ZDHHC5, ZDHHC7, ZDHHC8, ZDHHC9, ZDHHC12, ZDHHC15, and ZDHHC23 were negatively associated with survival times of patients with glioma (Table 2). In TCGA database, patients with low expression levels of ZDHHC8, ZDHHC11, ZDHHC16, ZDHHC17, ZDHHC19, ZDHHC20, ZDHHC21, and ZDHHC22 showed poor survival times. However, only ZDHHC11, ZDHHC16, ZDHHC18, and ZDHHC22 expression levels were positively correlated with the overall survival of patients with glioma in the Rembrandt database (Table 2).
Table 2.
Prognostic values of ZDHHCs in gliomas
| Genes |
TCGA |
Rembrandt |
||||
|---|---|---|---|---|---|---|
| Hazard ratio | p value | Prognostic value | Hazard ratio | p value | Prognostic value | |
| ZDHHC1 | 0.28 (0.21–0.38) | <0.01 | poorer prognosis | 0.61 (0.48–0.76) | <0.01 | poorer prognosis |
| ZDHHC2 | 1.27 (0.99–1.65) | >0.05 | no significance | 0.94 (0.75–1.17) | >0.05 | no significance |
| ZDHHC3 | 0.87 (0.68–1.13) | >0.05 | no significance | 0.64 (0.52–0.81) | <0.05 | poorer prognosis |
| ZDHHC4 | 0.36 (0.28–0.48) | <0.01 | poorer prognosis | 0.52 (0.41–0.65) | <0.05 | poorer prognosis |
| ZDHHC5 | 0.3 (0.23–0.40) | <0.01 | poorer prognosis | 0.62 (0.5–0.78) | <0.05 | poorer prognosis |
| ZDHHC6 | 1.11 (0.86–1.43) | >0.05 | no significance | 1.22 (0.98–1.53) | >0.05 | no significance |
| ZDHHC7 | 0.84 (0.65–1.09) | >0.05 | no significance | 0.67 (0.53–0.83) | <0.01 | poorer prognosis |
| ZDHHC8 | 1.31 (1.01–1.7) | <0.05 | better prognosis | 1.24 (0.99–1.55) | >0.05 | no significance |
| ZDHHC9 | 0.8 (0.62–1.04) | >0.05 | no significance | 1.04 (0.83–1.3) | >0.05 | no significance |
| ZDHHC11 | 1.47 (1.13–1.91) | <0.01 | better prognosis | 1.62 (1.3–2.03) | <0.01 | better prognosis |
| ZDHHC12 | 0.22 (0.17–0.3) | <0.01 | poorer prognosis | 0.43 (0.34–0.54) | <0.01 | poorer prognosis |
| ZDHHC13 | 0.43 (0.33–0.65) | <0.01 | poorer prognosis | 1.04 (0.84–1.3) | >0.05 | no significance |
| ZDHHC14 | 0.93 (0.72–1.19) | >0.05 | no significance | 1.15 (0.92–1.44) | >0.05 | no significance |
| ZDHHC15 | 0.5 (0.38–0.65) | <0.01 | poorer prognosis | 0.66 (0.53–0.83) | <0.01 | poorer prognosis |
| ZDHHC16 | 1.48 (1.14–1.91) | <0.01 | better prognosis | 1.61 (1.29–2.01) | <0.01 | better prognosis |
| ZDHHC17 | 2.96 (2.24–3.9) | <0.01 | better prognosis | 0.89 (0.71–1.11) | >0.05 | no significance |
| ZDHHC18 | 0.24 (0.18–0.32) | <0.01 | poorer prognosis | 1.59 (1.27–1.99) | <0.01 | better prognosis |
| ZDHHC19 | 1.49 (1.16–1.93) | <0.01 | better prognosis | 0.87 (0.70–1.09) | >0.05 | no significance |
| ZDHHC20 | 1.33 (1.03–1.72) | <0.05 | better prognosis | 1.04 (0.84–1.3) | >0.05 | no significance |
| ZDHHC21 | 2.39 (1.83–3.11) | <0.01 | better prognosis | 1.68 (1.34–2.11) | <0.01 | better prognosis |
| ZDHHC22 | 6.56 (4.79–9.07) | <0.01 | better prognosis | 3.03 (2.4–3.84) | <0.01 | better prognosis |
| ZDHHC23 | 0.28 (0.21–0.37) | <0.01 | poorer prognosis | 0.76 (0.61–0.94) | <0.01 | poorer prognosis |
| ZDHHC24 | 0.4 (0.31–0.53) | <0.01 | poorer prognosis | 0.93 (0.75–1.06) | >0.05 | no significance |
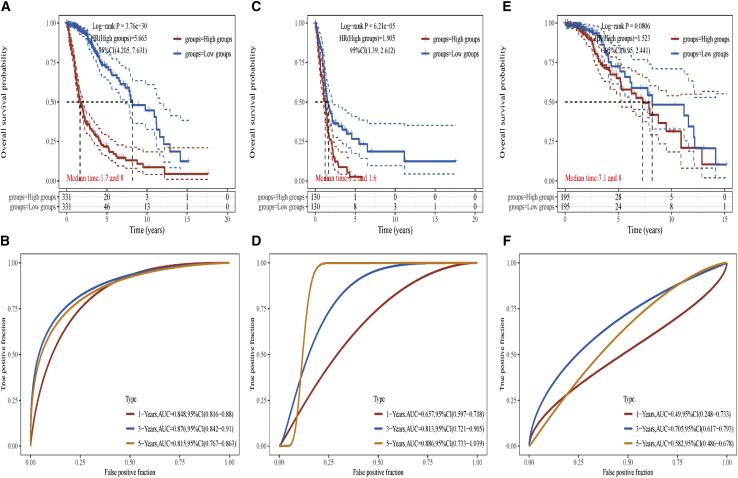
Combined with the above results, we noticed that there were five ZDHHCs (ZDHHC11, ZDHHC12, ZDHHC15, ZDHHC22, and ZDHHC23) that were not only differentially expressed in gliomas but were also associated with the overall survival of patients with glioma. Therefore, we used these five ZDHHCs to establish a ZDHHC-related risk signature via the assistance of a clinical bioinformatics tool (https://www.aclbi.com/static/index.html#/).27,28 By using the “glmnet” package in R, the least absolute shrinkage and selection operator (Lasso) regression model was selected to minimize overfitting and to identify the most significant survival-associated ZDHHCs in gliomas, which were found to be ZDHHC12, ZDHHC15, ZDHHC22, and ZDHHC23 (Figure S9). Risk scores were calculated for each sample (). As shown in Figure S9, ZDHHC12, ZDHHC15, and ZDHHC23 expression levels were higher in the high-risk group, while ZDHHC22 expression was higher in the low-risk group. Kaplan-Meier analysis showed that patients with glioma in the high-risk group had poorer survival times compared with those of the low-risk group (Figure 3A). To determine the prognostic significance of our ZDHHC-related risk signature, time-dependent receiver operating characteristic (ROC) curve analysis was performed. The area under the curve (AUC) values for the 1-, 3-, and 5-year ROC curves each reached 0.8, indicating that our ZDHHC-related risk signature yielded a good sensitivity and specificity for predicting the prognoses of patients with glioma (Figure 3B). We also evaluated prognostic values of the ZDHHC-related risk signature in separated IDH wild-type and mutant gliomas. As shown in Figures S10, 3C, and 3D, ZDHHC-related risk signature also presented an acceptable sensitivity and specificity for predicting the prognoses of patients with IDH wild-type gliomas. However, it seems that there were no values of ZDHHC-related risk signature for predicting the prognoses of patients with IDH mutant gliomas (Figures 3E, 3F, and S11).
Figure 3.
ZDHHC-related risk assessment model for prognostic prediction in gliomas
(A and B) Kaplan-Meier analysis and time-dependent ROC analysis of the ZDHHC-gene signature in gliomas (including IDH wild type and mutant). (C and D) Kaplan-Meier analysis and time-dependent ROC analysis of the ZDHHC-gene signature in IDH wild-type gliomas are shown. (E and F) Kaplan-Meier analysis and time-dependent ROC analysis of the ZDHHC-gene signature in IDH mutant gliomas are shown.
ZDHHCs are related to immune-cell infiltration and tumor-cell progression in gliomas
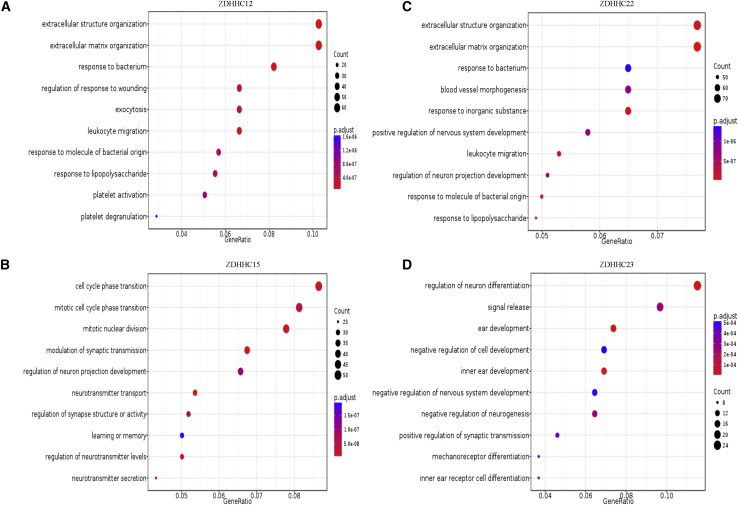
The microarray data from the Rembrandt dataset were used to perform GO enrichment analysis. Each ZDHHC was divided into high- and low-expression groups according to the corresponding expression levels. The top-50 differentially expressed genes between the high- and low-expression groups of each ZDHHC are shown in the heatmaps in Figures S12A and S13. We found that ZDHHC11 might regulate cell extracellular matrix organization (Figure S12B). We observed that ZDHHC12 and ZDHHC22 were related to leukocyte migration (Figures 4A and 4C). In contrast, ZDHHC15 and ZDHHC23 were mainly involved in regulating the cell cycle and development (Figures 4B and 4D).
Figure 4.
GO enrichment of ZDHHC12, ZDHHC15, ZDHHC22, and ZDHHC23 in gliomas
(A) ZDHHC12 is associated with extracellular structure organization, extracellular structure organization, and leukocyte migration. (B) ZDHHC15 is correlated with cell cycle phase transition, mitotic nuclear division, and neurotransmitter transport. (C) ZDHHC22 is related to extracellular structure organization, extracellular matrix organization, blood vessel morphogenesis, and leukocyte migration. (D) ZDHHC23 is involved in the regulation of neuron differentiation, signal release, and cell development.
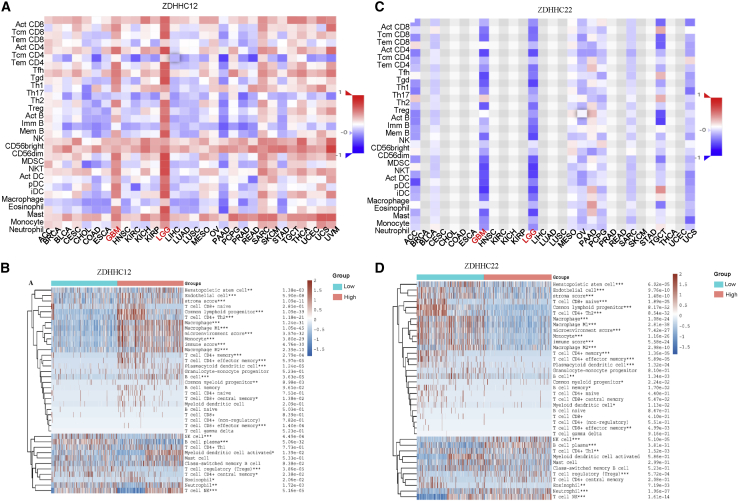
To validate whether ZDHHC12 and ZDHHC22 were indeed associated with infiltration of immune cells, we performed immune-cell infiltration analysis. We noticed that ZDHHC12 expression was positively related to immune-cell infiltration, while ZDHHC22 expression was negatively related to immune-cell infiltration in pan-cancer (Figures 5A and 5C). Furthermore, compared with other tumors, ZDHHC12- or ZDHHC22-associated immune-cell infiltration showed a high specificity for LGGs and GBMs (Figures 5A and 5C). In gliomas, higher macrophages and microglia, memory CD4+ T cell, B cell, regulatory T cell, myeloid-derived suppressor cell, and natural killer T cell infiltration were presented in ZDHHC12 high-expression group compared with that in low-expression group. In contrast, ZDHHC22 expression was negatively related to infiltration of these immune cells (Figures 5B and 5D). Considering that leukocyte migration is mediated by chemokines, we then analyzed the correlations between ZDHHC expression levels and related chemokines. As shown in Figure S14, ZDHHC12 was positively associated with most chemokines in gliomas, while ZDHHC22 was negatively associated with the majority of the chemokines analyzed. These results further support that ZDHHC12 and ZDHHC22 regulated leukocyte migration via chemokines. Importantly, immune-checkpoint-blockade-mediated cancer immunotherapies are promising strategies for clinical treatments.29 We found that ZDHHC12 was positively associated with the immune checkpoints, PD-L1, LAG3, and CTLA-4, while ZDHHC22 was negatively associated with these markers (Figure S15).
Figure 5.
Immune-cell infiltration analysis of ZDHHC12 and ZDHHC22 in tumors
(A) Pan-cancer analysis of the correlation between ZDHHC12 and immune-cell infiltration. (B) The differences of immune-cell infiltration between ZDHHC12 high- and low-expression group is shown. (C) Immune-cell infiltration analysis of ZDHHC22 in pan-cancer is shown. (D) The differences of immune-cell infiltration between ZDHHC22 high- and low-expression group is shown.
Next, we performed a GSEA to explore related biological mechanisms. We found that four out of five ZDHHCs were enriched in the oncogenic phosphatidylinositol 3-kinase/protein kinase B (PI3K/AKT) signaling pathway (Figure S16). Low expression levels of ZDHHC11 and ZDHHC22 were associated with the activation of PI3K/AKT signaling pathway (Figures S16A and S16D). In addition, high expression levels of ZDHHC12 and ZDHHC15 were related to the activation of PI3K/AKT signaling pathway (Figures S16B and S16C). In contrast, no enriched pathway was related to the expression of ZDHHC23.
2-BP suppresses cell proliferation and autophagy, as well as promotes apoptosis, in gliomas
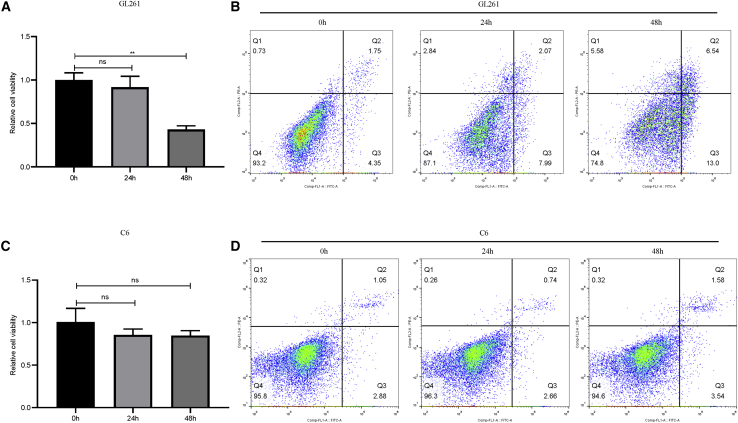
To determine whether ZDHHCs may represent a potential target for glioma treatment, we treated the GL261 and C6 glioma cell lines with the ZDHHC-specific inhibitor, 2-bromopalmitate (2-BP). Relative expression of five ZDHHCs in these two cell lines was presented in Figures S17A and S17B. Compared with ZDHHC11, ZDHHC12 and ZDHHC15 expression was higher in both cell lines, which are consistent with the result of our bioinformatics analysis. Nevertheless, there was differently relative expression of ZDHHC22 and ZDHHC23 between GL261 and C6 cells (Figures S17A and S17B). Interestingly, when these two cell lines were treated with 2-BP for indicated times, GL261 cell viability was inhibited in a dose-dependent manner, and the half-maximal inhibitory concentration (IC50) was approximately 50 μM (Figures S17C and S17E). However, C6 cell viability was almost not altered following 2-BP treatment (Figures S17D and S17F). It seems that C6 glioma cells were not sensitive to 2-BP (i.e., C6 glioma cells were thus defined as 2-BP-insensitive cells). To determine whether 2-BP inhibited glioma-cell viability in a time-dependent manner, glioma cells were treated with 50 μM of 2-BP. As shown in Figures 6A and 6B, 2-BP suppressed GL261 cell viability and promoted apoptosis in a time-dependent manner. By contrast, 2-BP still showed no influence on cell viability or apoptosis in 2-BP-insensitive cells (Figures 6C and 6D). The cell-proliferation-related protein, PCNA, and the anti-apoptosis-related proteins, Bcl-2 and Bcl-xl, were also decreased following 2-BP treatment in GL261 cells, whereas the pro-apoptosis protein, cleaved-caspase-3, was increased. In 2-BP-insensitive cells, no such result was observed (Figures S18A–S18E).
Figure 6.
Impact of 2-BP on glioma-cell viability and apoptosis
(A) Gl261 cells were treated with 50 μM 2-BP for indicated times, and cell viability was measured (∗∗p < 0.01; not significant [ns], p > 0.05 compared with the control group). (B) Gl261 cells were treated with 50 μM 2-BP for indicated times, and effects of 2-BP on GL261 apoptotic rate (Q2+Q3) at 0 h (6.10%), 24 h (10.06%), and 48 h (19.54%) were evaluated. (C) C6 cells were treated with 50 μM 2-BP for indicated times, and cell viability was measured (ns; p > 0.05 compared with the control group). (D) C6 cells were treated with 50 μM 2-BP for indicated times, and effects of 2-BP on C6 apoptotic rate (Q2+Q3) at 0 h (3.93%), 24 h (3.40%), and 48 h (5.12%) were evaluated.
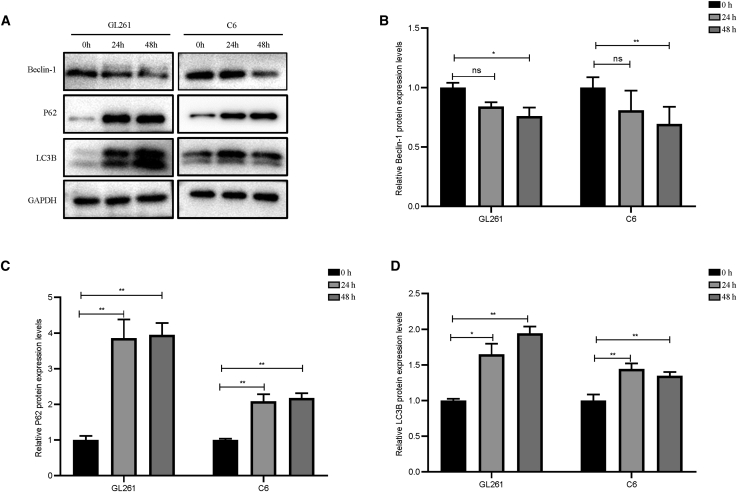
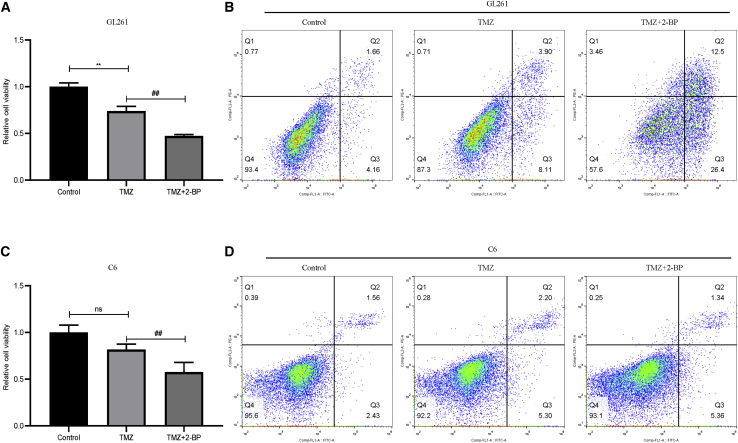
Moreover, as shown in Figure 7, compared with those in the control group, 2-BP decreased beclin-1 protein levels and increased P62 and LC3B protein levels in both GL261 and C6 cells. This result suggested that 2-BP treatment decreased autophagy levels in glioma cells. In glioma chemotherapy, temozolomide (TMZ)-induced autophagy is one of the common mechanisms that promote drug resistance.30,31 Hence, we hypothesized that 2-BP-mediated inhibition of autophagy may promote glioma-cell sensitivity to TMZ. We treated glioma cells with 200 μM of TMZ combined with 50 μM of 2-BP for 48 h. As shown in Figures 8A and 8B, TMZ inhibited cell viability and promoted apoptosis in GL261 cells compared with that of the control group. Compared with TMZ alone, 2-BP in combination with TMZ strengthened the effects of TMZ on GL261 cells. Interestingly, we found that C6 cells were also insensitive to 200 μM of TMZ. However, C6 cells treated with 2-BP combined with TMZ inhibited cell viability compared with that of TMZ alone (Figure 8C). We observed no difference in apoptosis among the three groups (Figure 8D).
Figure 7.
2-BP inhibits glioma-cell autophagy
(A) Autophagy-related key proteins of GL261 and C6 cells were detected by Western blotting assay. (B–D) Relative Beclin-1,P62, LC3B protein levels between control and 2-BP treated groups in GL261 and C6 cells. ∗p < 0.05; ∗∗p < 0.01; ns; p > 0.05 compared with the control group
Figure 8.
2-BP promotes the sensitivity of glioma cells to TMZ chemotherapy
(A) Gl261 cells were treated with 50 μM 2-BP and 200 μM TMZ for 48 h, and cell viability was measured (∗∗p < 0.01, compared with the control group; ##p < 0.01, compared with TMZ). (B) Gl261 cells were treated with 50 μM 2-BP and 200 μM TMZ for 48 h, and effects of 2-BP and TMZ on GL261 apoptotic rate (Q2+Q3) at 48 h were evaluated: control group (5.82%), TMZ (12.01%), and 2-BP + TMZ (38.90%). (C) C6 cells were treated with 50 μM 2-BP and 200 μM TMZ for 48 h, and cell viability was measured (ns; p > 0.05, compared with the control group; #p < 0.05, compared with TMZ). (D) C6 cells were treated with 50 μM 2-BP and 200 μM TMZ for 48 h, and effects of 2-BP and TMZ on C6 apoptotic rate (Q2+Q3) at 48 h were evaluated: control group (3.99%), TMZ (7.50%), and 2-BP + TMZ (6.70%).
2-BP inhibits GL261 cell-induced microglial migration
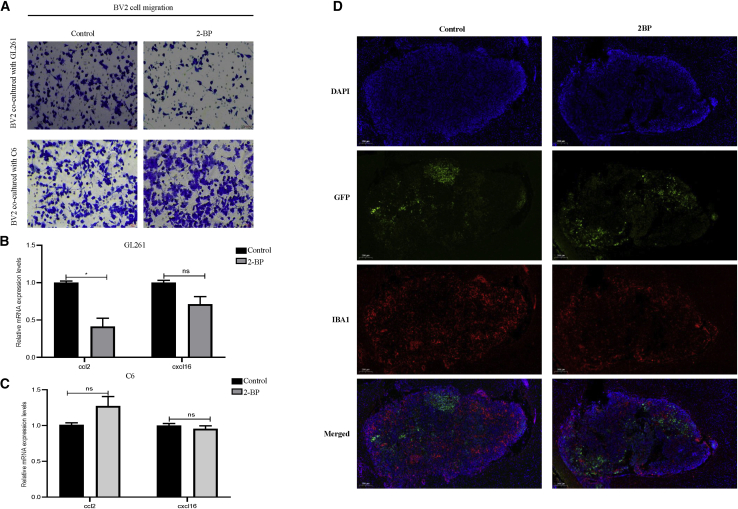
As shown in Figure 5, we found that ZDHHC expression was associated with immune-cell infiltration. Therefore, we explored the effects of ZDHHC inhibition on immune-cell migration. Considering that microglia are the most abundant immune cells in the central nervous system, we chose BV2 microglia for the following experiments. We pretreated GL261 and C6 cells with 2-BP for 48 h and then co-cultured them with microglia BV2 cells, respectively. Compared with that of the control group, GL261 cells pretreated with 2-BP exhibited reduced numbers of migratory BV2 cells (Figure 9A). Immune-cell migration is mediated by chemokines. Among these chemokines, we noticed that CCL2 and CXCL16, two chemokines associated with microglial infiltration, were highly related to both ZDHHC12 and ZDHHC22 in LGGs (Figure S14). We then investigated the effects of 2-BP on CCL2 and CXCL16 expression levels in gliomas. GL261 cells treated with 2-BP showed decreased mRNA levels of CCL2, but not CXCL16, compared with those of the control group (Figure 9B). Nevertheless, no such result was found in 2-BP-insensitive cells (Figures 9A and 9C). We have also performed an intracranial GFP-Gl261 cell implantation experiment in C57BL/6J mouse model. We found that mice treated with 2-BP presented lower microglia infiltration compared with those treated with PBS (Figures 9D and S19). These results suggest that 2-BP indeed inhibited microglial migration induced by glioma cells.
Figure 9.
Effects of 2-BP on glioma-induced microglia migration
(A) Glioma cells were pretreated with 2-BP for 48 h and then co-cultured with BV2 cells. Effects of 2-BP-treated GL261 and C6 cells on BV2 cell migration were evaluated. (B and C) Glioma cells were treated with 2-BP for 48 h, and CCL2 and CXCL16 expression was detected by qRT-PCR (∗p < 0.05; ns; p > 0.05; compared with the control group). (D) Effects of 2-BP on microglia migration in vivo are shown.
Discussion
In the present study, we first explored the expression profiles of ZDHHCs in glioma tissues. We found that five (ZDHHC2, ZDHHC11, ZDHHC12, ZDHHC15, and ZDHHC23) out of 23 ZDHHCs were aberrantly expressed at both transcriptional and translational levels in gliomas. Although no data could be retrieved for ZDHHC22 protein levels in the HPA database, aberrant ZDHHC22 mRNA levels were found in both TCGA and Rembrandt databases. Among these differentially expressed ZDHHCs, ZDHHC11, ZDHHC22, and ZDHHC23 were downregulated in gliomas. DNA methylation of the CpG islands of gene promoters represents the most common mechanism that leads to the repression of gene expression.22 Hence, we also explored the methylation levels of CpG sites of ZDHHC genes. Interestingly, only ZDHHC11 promoter regions presented high levels of methylation. In contrast, the promoters of ZDHHC22 and ZDHHC23 did not show high methylation levels, suggesting that the downregulated expression of these two genes was not due to hypermethylation of CpG islands. Furthermore, histone acetylation and N6-methyl adenosine also participate in modulating gene expression and/or mRNA-translational modifications.32,33 We speculated that histone acetylation and/or N6-methyl adenosine might be responsible for the low expression levels of ZDHHC22 and ZDHHC23. Apart from gliomas, ZDHHC2, ZDHHC11, ZDHHC12, ZDHHC15, ZDHHC22, and ZDHHC23 are also aberrantly expressed in the majority of tumors. In 2016, the World Health Organization released the latest classification of gliomas, which divided gliomas into IDH wild-type and IDH mutant gliomas according to the mutation status of the IDH gene family. In gliomas, the majority of patients with LGGs and 10% of patients with GBMs possess IDH gene-family alterations.2 Similar to the IDH gene family, our present results also showed that the mutation frequency of the ZDHHC gene family in patients with LGG was higher than that in patients with GBM. Nevertheless, unlike IDH gene-family mutations, global alterations of the ZDHHC gene family were not associated with survival times of patients with glioma in our present analysis.
Among these differentially expressed ZDHHCs (ZDHHC2, ZDHHC11, ZDHHC12, ZDHHC15, ZDHHC22, and ZDHHC23), patients with high expression levels of ZDHHC12 and ZDHHC15 exhibited poor prognoses, whereas ZDHHC11 and ZDHHC22 expression levels were positively correlated with the overall survival. Interestingly, downregulation of ZDHHC23 was negatively associated with survival times of patients with glioma. However, no correlation was observed between ZDHHC2 expression and the prognosis of patients with glioma in TCGA database. In gastric adenocarcinoma and hepatocellular carcinoma, patients with low ZDHHC2 expression have a poor prognosis.34,35 Patients with increased ZDHHC11 expression are predicted to have a superior progression-free survival rate in bladder cancer.36 In kidney renal clear-cell carcinoma, patients with decreased ZDHHC15 and ZDHHC23 expression exhibit poor overall survival.37 These studies reveal that the prognostic roles of ZDHHCs are tumor dependent. In our present study, we constructed a ZDHHC-related risk signature, and Kaplan-Meier analysis showed that patients with glioma in the high-risk group had a poorer survival time compared with those in the low-risk group. Moreover, the AUC values of 1-, 3-, and 5-year ROC curves reached 0.8, indicating that our ZDHHC-related risk signature possessed a good sensitivity and specificity for predicting the prognosis of patients with glioma.
After determining the profiles and prognostic values of ZDHHCs, we explored the potential roles of five ZDHHCs (ZDHHC11, ZDHHC12, ZDHHC15, ZDHHC22, and ZDHHC23). Results of GO enrichment analysis revealed that ZDHHC11, ZDHHC15, and ZDHHC23 were involved in regulating cell development and extracellular matrix organization. In gliomas, it has been reported that the inhibition of ZDHHC15 can impair glioma stem-cell proliferation and self-renewal via suppression of GP130 palmitoylation.38 Similar to ZDHHC15, ZDHHC23 also targets glioma stem cells to regulate cellular plasticity of these subtypes.17 These studies are also consistent with our present findings. Moreover, we found that ZDHHC12 and ZDHHC22 mainly participate in modulating leukocyte migration. Compared with other tumor types, ZDHHC12- and ZDHHC22-mediated immune-cell migration presented high specificities for gliomas. Further results suggested that these two genes were highly correlated with chemokines, which are known to induce immune-cell migration.39 To determine potential mechanisms of ZDHHCs in gliomas, we performed a GSEA. We found that ZDHHC11, ZDHHC12, ZDHHC15, and ZDHHC22 were enriched in the oncogenic PI3K/AKT signaling pathway. In our experiments, glioma GL261 treated with 50 μM 2-BP for indicated times indeed inhibited PI3K/AKT signaling pathway activation (Figures S18F–S18G). In gliomas, the activation of the PI3K/AKT pathway promotes tumor cell proliferation and invasion and glucose metabolism and inhibits apoptosis.40 Furthermore, the PI3K/AKT pathway also participates in modulating chemokine secretion and tumor immune evasion.41, 42, 43 Our present results showed that ZDHHC12 and ZDHHC15 were positively related to the PI3K/AKT pathway, suggesting that these two genes might play oncogenic roles in gliomas. On the contrary, ZDHHC22 might act as a tumor suppressor. These results suggest that ZDHHCs might play their roles in gliomas partially via modulating the PI3K/AKT pathway. Previous study reported that blocking EGFR palmitoylation suppresses PI3K/AKT signaling pathway.44 In addition, ZDHHC11 and ZDHHC22 expression was negatively correlated with EGFR, while ZDHHC12 and ZDHHC15 expression was positively correlated with EGFR expression in gliomas (data not shown). We speculated that EGFR might be one of the direct targets of palmitoylation modulated by ZDHHC inhibition, which need further exploration in our subsequent study.
In our present study, we also explored the effects of ZDHHC inhibition or overexpression in gliomas. Take ZDHHC12, for example: overexpression of ZDHHC12 promoted glioma C6, GL261, and T98G cell viability (Figures S20A–S20C). Knockdown of ZDHHC12 inhibited glioma cell viability (Figure S20D). We then explored whether palmitoyl transferase could be a target for glioma treatment. We found that the palmitoyl transferase inhibitor, 2-BP, suppressed GL261 cell viability in a time- and dose-dependent manner. In addition, inhibition of protein palmitoylation suppressed glioma-cell autophagy. At present, surgical interventions and aggressive chemoradiotherapeutic treatments present standard treatments for patients with glioma.1,2 Despite TMZ being the most common chemotherapeutic drug used in the clinic, as it prolongs the survival time of patients with glioma, TMZ-induced autophagy can induce drug resistance.30 Interestingly, in our present study, we found that glioma cells treated with 2-BP increased cell sensitivity to TMZ. This phenomenon might partially be due to the inhibition of autophagy induced by 2-BP. Finally, we also explored the effects of 2-BP on immune-cell infiltration. Microglia are the resident immune cells in the central nervous system. However, only limited primary microglia can be obtained from 2- to 3-day-old mice or rats. Hence, we chose microglial BV2 cell lines in our present study. Our results showed that pretreatment of GL261 glioma cells with 2-BP co-cultured with microglia weakened microglial migration. Moreover, 2-BP decreased CCL2 secretion. Numerous studies have reported that CCL2 secreted by glioma cells is involved in establishing an immunosuppressive tumor microenvironment via the recruitment of microglia and other immune cells.45, 46, 47, 48, 49, 50 These results reveal that the inhibition of ZDHHCs not only suppresses glioma cell viability but also inhibits glioma-cell-mediated microglial migration. In contrast, no such results were found in 2-BP-insensitive C6 cells. One reason for this result may be due to the drug-uptake rate of C6 cells being approximately 10 times lower than that of GL261 cells.51 Another reason for this result may be due to high levels of S100 protein expression in C6 cells.52 In gliomas as well as other tumors, it is reported that S100 participated in promoting cell survival and preventing cell apoptosis.53, 54, 55, 56 Apart from that, the differential ZDHHC23 or other ZDHHCs expression between Gl261 and C6 cell lines may be also another cause.
In conclusion, this study comprehensively analyzed expression profiles and prognostic values of ZDHHCs in gliomas. Indeed, we found that five ZDHHCs (ZDHHC11, ZDHHC12, ZDHHC15, ZDHHC22, and ZDHHC23) were differentially expressed in gliomas and expression of these ZDHHCs were associated with prognoses of patients with glioma. Our results also demonstrated that ZDHHCs might regulate glioma progression and immune-cell infiltration via PI3K/AKT signaling pathway. In addition, inhibition of ZDHHCs suppressed cell proliferation and autophagy, as well as increased cell sensitivity to TMZ, in gliomas. More importantly, ZDHHC inhibition weakened microglial migration induced by glioma cells. These results indicate that ZDHHCs may represent promising therapeutic targets for glioma treatment. Nevertheless, the limitation of our study is that we did not provide more in vivo results, which is the field we will focus on in the subsequent study.
Materials and methods
Downloading of data from public databases
To explore the expression levels of ZDHHCs in glioma tissues and normal brain tissues, we used the online website Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/) tool.57 The HPA (https://www.proteinatlas.org/) was used to obtain the protein levels of ZDHHCs in glioma samples.58 We downloaded ZDHHC expression levels and corresponding clinical data of patients with glioma from TCGA and Rembrandt databases via the GlioVis (http://gliovis.bioinfo.cnio.es/) data portal.59 This analysis included 1,139 glioma samples (TCGA: 667 patients; Rembrandt: 472 patients).
Survival prognosis analysis
When we performed survival prognosis analysis, glioma samples were divided into two groups by the median of each ZDHHC expression. “Survival” module of GlioVis was applied to assess the clinical relevance of each ZDHHC expression, and the Cox proportional hazard model was used to evaluate the outcome significance of ZDHHC expression in gliomas.
Differential expression analysis and GO enrichment analysis
We performed differential expression analysis and GO enrichment analysis via the GlioVis data portal. First, we divided the microarray data from the Rembrandt database into low- and high-expression groups according to ZDHHC expression levels. Differentially expressed genes between the low- and high-ZDHHC-expression groups were selected when |logFC| ≥ 1 was combined with a p < 0.01. Subsequently, we analyzed related biological processes to gain insights into the roles of ZDHHCs in gliomas, according to the differentially expressed genes between the low- and high-ZDHHC-expression groups.
Immune-infiltration analysis
We performed immune-infiltration analysis through the TISIDB web server. TISIDB (http://cis.hku.hk/TISIDB/) is a web portal for assessing tumor and immune-system interactions, which integrates multiple heterogeneous data types.60 We evaluated the correlation of ZDHHC expression with the abundance of immune cells and related chemokines. To validate the result of immune-cell infiltration in gliomas, we applied xCELL methods to infer the infiltration fraction of different types of immune cells among gliomas. The xCell was a novel gene-signature-based method, which could infer 64 immune and stromal cell types.61
GSEA
GSEA was performed with the R software packages, clusterProfiler and enrichplot. A p < 0.05 was considered statistically significant.
Reagents
2-BP (18,263-25-7) was purchased from Merck (Darmstadt, Germany). TMZ (HY-17364) was purchased from MedChemExpress (New Jersey, USA). Antibodies against P62 (1:1,000; ab109012), LC3B (1:1,000; ab192890), and Bcl2 (1:1,000; ab196495) were purchased from Abcam (Cambridgeshire, England). Antibodies against Beclin-1 (1:1,000; no. 3495), PCNA (1:1,000; no. 13110), Bcl-xl (1:1,000; no. 2764), and caspase-3 (1:1,000; no. 9662) were purchased from Cell Signaling Technology (Massachusetts, USA). Antibodies against GAPDH (1:1,000; bs-0755R) were purchased from Bioss (Beijing, China). A goat anti-rabbit secondary antibody (1:10,000; lot: 70,100,200) was purchased from Biosharp (Wuhan, China).
Cell culture
C6 and T98G glioma cell lines were obtained from the American Type Culture Collection. The GL261 glioma cell line was kindly donated by Professor Jie Luo (Taihe Hospital, Hubei Province, China). BV2 microglia were purchased from the China Center for Type Culture Collection. All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, USA) and supplemented with 10% fetal bovine serum (FBS) (WISENT, Canada).
Cell transfection
Using Lipofectamine 3000 (Invitrogen, USA), the ZDHHC12 small interfering RNAs (siRNAs) (sense:5′-GCGCAACCACCCACUCUUUTT-3'; antisense:5'-AAAGAGUGGGUGGUUGCGCTT-3′) were transfected into glioma cells. The ZDHHC12 empty vector and ZDHHC12 overexpression plasmid were transfected into glioma cells according to the Lipofectamine 3000 manufacturer’s protocol.
CCK-8 assay
First, 8,000 glioma cells were seeded in a 96-well plate for 24 h and were then treated with corresponding drugs and equal volumes of DMSO for the indicated durations. Next, 10 μL of CCK-8 solution (HY-K0301; MCE) was added to each well and incubated for 1 h. The absorbance was measured at 450 nm. Each experiment was replicated three times.
Flow cytometry
Glioma cells were seeded in a six-well plate for 24 h and then treated with corresponding drugs and equal volumes of DMSO for the indicated durations. Finally, cells were harvested and stained with annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) for flow cytometry analysis via Cytoflex. Each experiment was replicated three times.
Transwell assay
First, 5 × 104 BV2 cells were plated onto upper transwell chambers, after which they were co-cultured with treated glioma cells for 24 h. The cells in the upper chamber were fixed with 4% paraformaldehyde for 15 min and were then stained with 0.1% crystal violet for 15 min at room temperature. Migrating cells were imaged by microscopy. Each experiment was replicated three times.
Quantitative real-time PCR
Total RNA of cells was extracted via Trizol reagent (Ambion, USA). Reverse transcription and quantitative real-time PCR were performed by using a high-quality cDNA reverse-transcription kit (TOYOBO, Japan) and SYBR Green Master Mix (Biomake, Japan), respectively. The relative expression of RNA was calculated according to the 2−ΔΔCT method. All primers used in this study are listed in Table 3.
Table 3.
List of primers used for quantitative PCR
| Gene | Primer sequence |
|---|---|
| GAPDH (mouse) | forward 5′-TGGAGAAACCTGCCAAGTATG-3′ reverse 5′-CATACCAGGAAATGAGCTTGAC-3′ |
| CCL2 (mouse) | forward 5′-GATGCAGTTAACGCCCCACT-3′ reverse 5′-GAGGGCCGGGGTATGTAACT-3′ |
| CXCL16 (mouse) | forward 5′-GGACCCTTGTCTCTTGCGTT-3′ reverse 5′-GATCCAAAGTACCCTGCGGT-3′ |
| ZDHHC11 (mouse) | forward 5′-AAGCCCAGGAAAACAAT GTGAA-3′ reverse 5′-GAACACC CCACCCATCACAATA-3′ |
| ZDHHC12 (mouse) | forward 5′-CGAGCCCACCCGATCAC-3′ reverse 5′-AGTAGTCACATAGCCGGGGT-3′ |
| ZDHHC15 (mouse) | forward 5′-AACCGGGAGTGGAGCTGTA-3′ reverse 5′-TAACCCATGGGCAGTGATGG-3′ |
| ZDHHC22 (mouse) | forward 5′-CTGGCACCCAAGTAGGAGGG-3′ reverse 5′-CATCCTGGATCACATTCCGAGC-3′ |
| ZDHHC23 (mouse) | forward 5′-TGGACCATCATTGTGTCTGGATA-3′ reverse 5′-ATGGTGTTCAAGGTCAGCGA-3′ |
| GAPDH (rat) | forward 5′-GAGTCAACGGATTTGGTCGTT-3′ reverse 5′-TTGATTTTGGAGGGATCTCG-3′ |
| CCL2 (rat) | forward 5′-CAGTTAATGCCCCACTCACCT-3′ reverse 5′-TGAAGACCATCCCTTGTTCTCG-3′ |
| CXCL16 (rat) | forward 5′-CGGGACTCTTAAACCTCACCC-3′ reverse 5′-GCAGGACATGAGCTGAATTGG-3′ |
| ZDHHC11 (rat) | forward 5′-AAGAAACTGCTACCACGCCC-3′ reverse 5′-CACCCCACCCATCACTAGTACA-3′ |
| ZDHHC12 (rat) | forward 5′-CGCTGCTCAACTCTGGGATG-3′ reverse 5′-CATTGACGCAGCTCGGTGT-3′ |
| ZDHHC15 (rat) | forward 5′-TCGTCACTTGCTGTGGAGTC-3′ reverse 5′-AGGATCTCTTGAGCTCGGGT-3′ |
| ZDHHC22 (rat) | forward 5′-CAGTCCCCGAAGCGAAGC-3′ reverse 5′-TCCTCGATTACATTCCCAGCTC-3′ |
| ZDHHC23 (rat) | forward 5′-TGGATCACCATTGTGTCTGGATA-3′ reverse 5′-TCCGTAAACCGAAGTGAGCA-3′ |
Western blotting
Total proteins of cells were extracted by using radioimmunoprecipitation assay (RIPA) (Beyotime Biotechnology) in combination with a phosphatase inhibitor cocktail (MCE, USA) and protease inhibitor cocktail (MCE, USA). Equal amounts of proteins of each group were separated by 10% sodium-dodecyl-sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and electrotransferred to a polyvinylidene fluoride (PVDF) membrane. The PVDF membrane was blocked with 5% BSA at room temperature for 1.5 h and then incubated with corresponding primary antibodies at 4°C overnight. Subsequently, the membrane was incubated with the secondary antibody at room temperature for 1.5 h. Finally, the membranes were visualized with an enhanced chemiluminescence (ECL) solution (34,094; Thermo Fisher Scientific) and detected by using a Protein Imager (Find-do×6; Tanon). Each experiment was replicated three times.
Immunofluorescence assay
The immunofluorescence assay was performed as previously described.62 Briefly, the frozen slice of mouse glioma tissues were fixed in 4% paraformaldehyde for 20 min, permeabilized with 0.5% Triton X-100 for 20 min, sealed with 10% BSA (BSA) in room temperature for 30 min, and incubated with IBA1 antibody (1:500; Wako, Japan; cat. 019–19,741) in a wet box overnight at 4°C in turn. After that, samples were washed three times with PBST and incubated with R-phycoerythrin-conjugated secondary antibody (1:200; 111-116-144; Jackson Laboratory) for 1 h at 37°C. Finally, these samples were stained with DAPI for 5 min at room temperature, and images were acquired using a fluorescent-microscope imaging system.
Intracranial glioma-cell implantation
Six- to eight-week-old male C57BL/6J mice (n = 12) weighing 22–28 g were randomly allocated to two groups and underwent intracranial GFP-GL261-cell implantation. Seven days after implantation, mice were intraperitoneal administered daily with 2-BP (3 mg/kg) or equal PBS for 2 weeks.16 All animal experiments were approved by the Ethics Committee of Wuhan University Zhongnan Hospital.
Statistical analysis
In this study, Shapiro-Wilk tests were used to assess the distributions of variables. If the data followed a normal distribution, Student’s t test or one-way ANOVA was performed to analyze the data. Otherwise, Mann-Whitney tests were used to evaluate data that did not exhibit a normal distribution. SPSS 23.0, GraphPad Prism 8.0, and ImageJ software were used for statistical analysis. A p < 0.05 was considered statistically significant.
Acknowledgments
This research was supported by the National Health Commission of China (2018ZX-07S-011) and Translational Medicine Research Fund of Zhongnan Hospital of Wuhan University (ZLYNXM202011 and ZNLH201901). We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Author contributions
Z.-Q.L. and Z.-F.W. were involved in the conception or the design of the study. F.T., C.Y., and D.-H.Y. performed the experiments. F.T. and Z.-Y.P. participated in the acquisition and analyzation of the data. F.T. and F.-P.L. wrote the manuscript. Z.-Q.L. and Z.-F.W. reviewed the manuscript and made contributions for improvement.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.04.030.
Contributor Information
Ze-Fen Wang, Email: wangzf@whu.edu.cn.
Zhi-Qiang Li, Email: lizhiqiang@whu.edu.cn.
Supplemental information
References
- 1.Louis D., Ohgaki H., Wiestler O., Cavenee W., Burger P., Jouvet A., Scheithauer B., Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis D., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W., Ohgaki H., Wiestler O., Kleihues P., Ellison D. The 2016 World Health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 3.Wang H., Xu T., Huang Q., Jin W., Chen J. Immunotherapy for malignant glioma: current status and future directions. Trends Pharmacol. Sci. 2020;41:123–138. doi: 10.1016/j.tips.2019.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Duerinck J., Schwarze J., Awada G., Tijtgat J., Vaeyens F., Bertels C., Geens W., Klein S., Seynaeve L., Cras L., et al. Intracerebral administration of CTLA-4 and PD-1 immune checkpoint blocking monoclonal antibodies in patients with recurrent glioblastoma: a phase I clinical trial. J. Immunother. Cancer. 2021;9:e002296. doi: 10.1136/jitc-2020-002296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schalper K., Rodriguez-Ruiz M., Diez-Valle R., López-Janeiro A., Porciuncula A., Idoate M., Inogés S., de Andrea C., López-Diaz de Cerio A., Tejada S., et al. Neoadjuvant nivolumab modifies the tumor immune microenvironment in resectable glioblastoma. Nat. Med. 2019;25:470–476. doi: 10.1038/s41591-018-0339-5. [DOI] [PubMed] [Google Scholar]
- 6.Zheng Z., Zhang J., Jiang J., He Y., Zhang W., Mo X., Kang X., Xu Q., Wang B., Huang Y. Remodeling tumor immune microenvironment (TIME) for glioma therapy using multi-targeting liposomal codelivery. J. Immunother. Cancer. 2020;8:e000207. doi: 10.1136/jitc-2019-000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohme M., Neidert M., Regli L., Weller M., Martin R. Immunological challenges for peptide-based immunotherapy in glioblastoma. Cancer Treat. Rev. 2014;40:248–258. doi: 10.1016/j.ctrv.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 8.Ko P., Dixon S. Protein palmitoylation and cancer. EMBO Rep. 2018;19:e46666. doi: 10.15252/embr.201846666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linder M., Deschenes R. Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 10.Zhang M., Hang H. Protein S-palmitoylation in cellular differentiation. Biochem. Soc. Trans. 2017;45:275–285. doi: 10.1042/BST20160236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greaves J., Chamberlain L. DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends Biochem. Sci. 2011;36:245–253. doi: 10.1016/j.tibs.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Greaves J., Chamberlain L. New links between S-acylation and cancer. J. Pathol. 2014;233:4–6. doi: 10.1002/path.4339. [DOI] [PubMed] [Google Scholar]
- 13.Liu P., Jiao B., Zhang R., Zhao H., Zhang C., Wu M., Li D., Zhao X., Qiu Q., Li J., et al. Palmitoylacyltransferase Zdhhc9 inactivation mitigates leukemogenic potential of oncogenic Nras. Leukemia. 2016;30:1225–1228. doi: 10.1038/leu.2015.293. [DOI] [PubMed] [Google Scholar]
- 14.Yao H., Lan J., Li C., Shi H., Brosseau J., Wang H., Lu H., Fang C., Zhang Y., Liang L., et al. Inhibiting PD-L1 palmitoylation enhances T-cell immune responses against tumours. Nat. Biomed. Eng. 2019;3:306–317. doi: 10.1038/s41551-019-0375-6. [DOI] [PubMed] [Google Scholar]
- 15.Du W., Hua F., Li X., Zhang J., Li S., Wang W., Zhou J., Wang W., Liao P., Yan Y., et al. Loss of optineurin drives cancer immune evasion via palmitoylation-dependent IFNGR1 lysosomal sorting and degradation. Cancer Discov. 2021 doi: 10.1158/2159-8290.CD-20-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X., Li H., Fan X., Zhao C., Ye K., Zhao Z., Hu L., Ma H., Wang H., Fang Z. Protein palmitoylation regulates cell survival by modulating XBP1 activity in glioblastoma multiforme. Mol. Ther. Oncolytics. 2020;17:518–530. doi: 10.1016/j.omto.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X., Hu L., Yang H., Ma H., Ye K., Zhao C., Zhao Z., Dai H., Wang H., Fang Z. DHHC protein family targets different subsets of glioma stem cells in specific niches. J. Exp. Clin. Cancer Res. 2019;38:25. doi: 10.1186/s13046-019-1033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X., Hao A., Li X., Ye K., Zhao C., Yang H., Ma H., Hu L., Zhao Z., Hu L., et al. Activation of JNK and p38 MAPK mediated by ZDHHC17 drives glioblastoma multiforme development and malignant progression. Theranostics. 2020;10:998–1015. doi: 10.7150/thno.40076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X., Ma H., Wang Z., Zhang S., Yang H., Fang Z. EZH2 palmitoylation mediated by ZDHHC5 in p53-mutant glioma drives malignant development and progression. Cancer Res. 2017;77:4998–5010. doi: 10.1158/0008-5472.CAN-17-1139. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Z., Li X., Yang F., Chen C., Liu P., Ren Y., Sun P., Wang Z., You Y., Zeng Y., et al. DHHC9-mediated GLUT1 S-palmitoylation promotes glioblastoma glycolysis and tumorigenesis. Nat. Commun. 2021;12:5872. doi: 10.1038/s41467-021-26180-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T., Fan J., Wang B., Traugh N., Chen Q., Liu J., Li B., Liu X. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alhaji S., Ngai S., Abdullah S. Silencing of transgene expression in mammalian cells by DNA methylation and histone modifications in gene therapy perspective. Biotechnol. Genet. Eng. Rev. 2019;35:1–25. doi: 10.1080/02648725.2018.1551594. [DOI] [PubMed] [Google Scholar]
- 23.Modhukur V., Iljasenko T., Metsalu T., Lokk K., Laisk-Podar T., Vilo J. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics. 2018;10:277–288. doi: 10.2217/epi-2017-0118. [DOI] [PubMed] [Google Scholar]
- 24.Goldman M., Craft B., Hastie M., Repečka K., McDade F., Kamath A., Banerjee A., Luo Y., Rogers D., Brooks A., et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020;38:675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerami E., Gao J., Dogrusoz U., Gross B., Sumer S., Aksoy B., Jacobsen A., Byrne C., Heuer M., Larsson E., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J., Aksoy B., Dogrusoz U., Dresdner G., Gross B., Sumer S., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z., Lin E., Zhuang H., Xie L., Feng X., Liu J., Yu Y. Construction of a novel gene-based model for prognosis prediction of clear cell renal cell carcinoma. Cancer Cell Int. 2020;20:27. doi: 10.1186/s12935-020-1113-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin W., Wu S., Chen X., Ye Y., Weng Y., Pan Y., Chen Z., Chen L., Qiu X., Qiu S. Characterization of hypoxia signature to evaluate the tumor immune microenvironment and predict prognosis in glioma groups. Front. Oncol. 2020;10:796. doi: 10.3389/fonc.2020.00796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright J., Powers A., Johnson D. Endocrine toxicities of immune checkpoint inhibitors. Nat. Rev. Endocrinol. 2021;17:389–399. doi: 10.1038/s41574-021-00484-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johannessen T., Hasan-Olive M., Zhu H., Denisova O., Grudic A., Latif M., Saed H., Varughese J., Røsland G., Yang N., et al. Thioridazine inhibits autophagy and sensitizes glioblastoma cells to temozolomide. Int. J. Cancer. 2019;144:1735–1745. doi: 10.1002/ijc.31912. [DOI] [PubMed] [Google Scholar]
- 31.Huang T., Wan X., Alvarez A., James C., Song X., Yang Y., Sastry N., Nakano I., Sulman E., Hu B., et al. MIR93 (microRNA -93) regulates tumorigenicity and therapy response of glioblastoma by targeting autophagy. Autophagy. 2019;15:1100–1111. doi: 10.1080/15548627.2019.1569947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krug B., De Jay N., Harutyunyan A., Deshmukh S., Marchione D., Guilhamon P., Bertrand K., Mikael L., McConechy M., Chen C., et al. Pervasive H3K27 acetylation leads to ERV expression and a therapeutic vulnerability in H3K27M gliomas. Cancer Cell. 2019;35:782–797.e8. doi: 10.1016/j.ccell.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du J., Ji H., Ma S., Jin J., Mi S., Hou K., Dong J., Wang F., Zhang C., Li Y., et al. m6A regulator-mediated methylation modification patterns and characteristics of immunity and stemness in low-grade glioma. Brief Bioinform. 2021;22:bbab013. doi: 10.1093/bib/bbab013. [DOI] [PubMed] [Google Scholar]
- 34.Yan S., Tang J., Huang C., Xi S., Huang M., Liang J., Jiang Y., Li Y., Zhou Z., Ernberg I., et al. Reduced expression of ZDHHC2 is associated with lymph node metastasis and poor prognosis in gastric adenocarcinoma. PLoS One. 2013;8:e56366. doi: 10.1371/journal.pone.0056366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li S., Tang G., Zhou D., Pan Y., Tan Y., Zhang J., Zhang B., Ding Z., Liu L., Jiang T., et al. Prognostic significance of cytoskeleton-associated membrane protein 4 and its palmitoyl acyltransferase DHHC2 in hepatocellular carcinoma. Cancer. 2014;120:1520–1531. doi: 10.1002/cncr.28593. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto Y., Chochi Y., Matsuyama H., Eguchi S., Kawauchi S., Furuya T., Oga A., Kang J., Naito K., Sasaki K. Gain of 5p15.33 is associated with progression of bladder cancer. Oncology. 2007;72:132–138. doi: 10.1159/000111132. [DOI] [PubMed] [Google Scholar]
- 37.Liu Z., Liu C., Xiao M., Han Y., Zhang S., Xu B. Bioinformatics analysis of the prognostic and biological significance of ZDHHC-protein acyltransferases in kidney renal clear cell carcinoma. Front. Oncol. 2020;10:565414. doi: 10.3389/fonc.2020.565414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan X., Yang H., Zhao C., Hu L., Wang D., Wang R., Fang Z., Chen X. Local anesthetics impair the growth and self-renewal of glioblastoma stem cells by inhibiting ZDHHC15-mediated GP130 palmitoylation. Stem Cell Res. Ther. 2021;12:107. doi: 10.1186/s13287-021-02175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozga A., Chow M., Luster A. Chemokines and the immune response to cancer. Immunity. 2021;54:859–874. doi: 10.1016/j.immuni.2021.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao H., Wang J., Shao W., Wu C., Chen Z., To S., Li W. Recent advances in the use of PI3K inhibitors for glioblastoma multiforme: current preclinical and clinical development. Mol. Cancer. 2017;16:100. doi: 10.1186/s12943-017-0670-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Briand J., Nadaradjane A., Bougras-Cartron G., Olivier C., Vallette F., Cartron P. Diuron exposure and Akt overexpression promote glioma formation through DNA hypomethylation. Clin. Epigenetics. 2019;11:159. doi: 10.1186/s13148-019-0759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu-Ju Wu C., Chen C., Lin C., Feng L., Lin Y., Wei K., Huang C., Fang J., Chen P. CCL5 of glioma-associated microglia/macrophages regulates glioma migration and invasion via calcium-dependent matrix metalloproteinase 2. Neuro Oncol. 2020;22:253–266. doi: 10.1093/neuonc/noz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ellert-Miklaszewska A., Dabrowski M., Lipko M., Sliwa M., Maleszewska M., Kaminska B. Molecular definition of the pro-tumorigenic phenotype of glioma-activated microglia. Glia. 2013;61:1178–1190. doi: 10.1002/glia.22510. [DOI] [PubMed] [Google Scholar]
- 44.Kharbanda A., Walter D., Gudiel A., Schek N., Feldser D., Witze E. Blocking EGFR palmitoylation suppresses PI3K signaling and mutant KRAS lung tumorigenesis. Sci. Signal. 2020;13 doi: 10.1126/scisignal.aax2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H., Zhang L., Zhang I., Chen X., Da Fonseca A., Wu S., Ren H., Badie S., Sadeghi S., Ouyang M., et al. S100B promotes glioma growth through chemoattraction of myeloid-derived macrophages. Clin. Cancer Res. 2013;19:3764–3775. doi: 10.1158/1078-0432.CCR-12-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen R., Keoni C., Waker C., Lober R., Chen Y., Gutmann D. KIAA1549-BRAF expression establishes a permissive tumor microenvironment through NFκB-mediated CCL2 production. Neoplasia. 2019;21:52–60. doi: 10.1016/j.neo.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindemann C., Marschall V., Weigert A., Klingebiel T., Fulda S. Smac mimetic-induced upregulation of CCL2/MCP-1 triggers migration and invasion of glioblastoma cells and influences the tumor microenvironment in a paracrine manner. Neoplasia. 2015;17:481–489. doi: 10.1016/j.neo.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang A., Miska J., Wainwright D., Dey M., Rivetta C., Yu D., Kanojia D., Pituch K., Qiao J., Pytel P., et al. CCL2 produced by the glioma microenvironment is essential for the recruitment of regulatory T cells and myeloid-derived suppressor cells. Cancer Res. 2016;76:5671–5682. doi: 10.1158/0008-5472.CAN-16-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cherry J., Meng G., Daley S., Xia W., Svirsky S., Alvarez V., Nicks R., Pothast M., Kelley H., Huber B., et al. CCL2 is associated with microglia and macrophage recruitment in chronic traumatic encephalopathy. J. Neuroinflammation. 2020;17:370. doi: 10.1186/s12974-020-02036-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J., Sarkar S., Cua R., Zhou Y., Hader W., Yong V. A dialog between glioma and microglia that promotes tumor invasiveness through the CCL2/CCR2/interleukin-6 axis. Carcinogenesis. 2012;33:312–319. doi: 10.1093/carcin/bgr289. [DOI] [PubMed] [Google Scholar]
- 51.Watabe T., Kaneda-Nakashima K., Shirakami Y., Liu Y., Ooe K., Teramoto T., Toyoshima A., Shimosegawa E., Nakano T., Kanai Y., et al. Targeted alpha therapy using astatine (At)-labeled phenylalanine: a preclinical study in glioma bearing mice. Oncotarget. 2020;11:1388–1398. doi: 10.18632/oncotarget.27552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zimmer D., Van Eldik L. Levels and distribution of the calcium-modulated proteins S100 and calmodulin in rat C6 glioma cells. J. Neurochem. 1988;50:572–579. doi: 10.1111/j.1471-4159.1988.tb02949.x. [DOI] [PubMed] [Google Scholar]
- 53.Emberley E., Murphy L., Watson P.J.B. S100 proteins and their influence on pro-survival pathways in cancer. Biochem. Cell Biol. 2004;82:508–515. doi: 10.1139/o04-052. [DOI] [PubMed] [Google Scholar]
- 54.Szatmári T., Huszty G., Désaknai S., Spasokoukotskaja T., Sasvári-Székely M., Staub M., Esik O., Sáfrány G., Lumniczky K. Adenoviral vector transduction of the human deoxycytidine kinase gene enhances the cytotoxic and radiosensitizing effect of gemcitabine on experimental gliomas. Cancer Gene Ther. 2008;15:154–164. doi: 10.1038/sj.cgt.7701115. [DOI] [PubMed] [Google Scholar]
- 55.Jin T., Zhang Z., Yang X., Luo J. S100A4 expression is closely linked to genesis and progression of glioma by regulating proliferation, apoptosis, migration and invasion. Asian Pac. J. Cancer Prev. 2015;16:2883–2887. doi: 10.7314/apjcp.2015.16.7.2883. [DOI] [PubMed] [Google Scholar]
- 56.Chow K., Park H., George J., Yamamoto K., Gallup A., Graber J., Chen Y., Jiang W., Steindler D., Neilson E., et al. S100A4 is a biomarker and regulator of glioma stem cells that is critical for mesenchymal transition in glioblastoma. Cancer Res. 2017;77:5360–5373. doi: 10.1158/0008-5472.CAN-17-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uhlen M., Zhang C., Lee S., Sjöstedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F., et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 59.Bowman R., Wang Q., Carro A., Verhaak R., Squatrito M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol. 2017;19:139–141. doi: 10.1093/neuonc/now247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ru B., Wong C., Tong Y., Zhong J., Zhong S., Wu W., Chu K., Wong C., Lau C., Chen I., et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35:4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 61.Aran D., Hu Z., Butte A. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang F., Zhao Y., Zhang Q., Wei W., Tian S., Li C., Yao J., Wang Z., Li Z. Impact of beta-2 microglobulin expression on the survival of glioma patients via modulating the tumor immune microenvironment. CNS Neurosci. Ther. 2021;27:951–962. doi: 10.1111/cns.13649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.