Summary
Transposons significantly contribute to genome fractions in many plants. Although numerous transposon‐related mutations have been identified, the evidence regarding transposon‐derived genes regulating crop yield and other agronomic traits is very limited. In this study, we characterized a rice Harbinger transposon‐derived gene called PANICLE NUMBER AND GRAIN SIZE (PANDA), which epigenetically coordinates panicle number and grain size. Mutation of PANDA caused reduced panicle number but increased grain size in rice, while transgenic plants overexpressing this gene showed the opposite phenotypic change. The PANDA‐encoding protein can bind to the core polycomb repressive complex 2 (PRC2) components OsMSI1 and OsFIE2, and regulates the deposition of H3K27me3 in the target genes, thereby epigenetically repressing their expression. Among the target genes, both OsMADS55 and OsEMF1 were negative regulators of panicle number but positive regulators of grain size, partly explaining the involvement of PANDA in balancing panicle number and grain size. Moreover, moderate overexpression of PANDA driven by its own promoter in the indica rice cultivar can increase grain yield. Thus, our findings present a novel insight into the epigenetic control of rice yield traits by a Harbinger transposon‐derived gene and provide its potential application for rice yield improvement.
Keywords: rice, transposon‐derived gene, polycomb repressive complex 2, epigenetic regulation, grain yield
Introduction
Rice (Oryza sativa) is a staple food feeding more than 50% of the world’s population. Grain yield in rice is primarily determined by three components including panicle number, grain number and grain weight. A number of genes determining these traits separately have been well characterized and make it possible to improve one of the traits without compromising the other two (Li et al., 2018; Wang et al., 2018). These traits are usually negatively associated with each other (Sadras, 2007). Recently, more evidence suggests that genetic factors play roles in explaining the negative associations among yield components in plants. For example, GSN1 coordinates the trade‐off between grain number and grain size by integrating localized cell differentiation and proliferation (Guo et al., 2018). OsSHI1 can interact with IPA1 and modulates the transcriptional activity of two downstream genes, OsTB1 and OsDEP1, to coordinate panicle number and grain number (Duan et al., 2019). A fine gene network composed of microRNAs and transcription factors coordinates rice tiller formation and panicle branching (Wang et al., 2015). In addition to the extensively studied balance between panicle/tiller number and grain number, and between grain number and grain size, the balances between panicle number and grain size were also observed (Bai et al., 2017; Fu et al., 2010; Guo et al., 2018). However, the genetic mechanism underlying the coordination between panicle number and grain size is largely unknown.
Polycomb repressive complex 2 (PRC2) epigenetically represses gene expression by catalysing the trimethylation of lysine 27 on histone H3 (H3K27me3) (Bieluszewski et al., 2021). It was first discovered in Drosophila and then found to be functionally conserved in higher eukaryotes during normal growth and development and in response to environmental cues (Bieluszewski et al., 2021). The PRC2 complex consists of four canonical core components and large repertoires of accessory proteins (Bieluszewski et al., 2021). The core components are generally not DNA binding proteins, whereas some accessory proteins play roles in sequence‐specific recruitment of PRC2, which defines a selective and flexible target repression in association with the developmental plasticity and dynamics (Kassis and Brown, 2013; Qüesta et al., 2016; Xiao et al., 2017; Yuan et al., 2016, 2021; Zhou et al., 2017; Zhou et al., 2018). In rice, the paralog pairs OsCLF/OsSET1, OsEMF2a/b, OsFIE1/2 and OsMSI1/2, respectively correspond to the homologous counterparts of the core PRC2 components E(z), Su(z)12, ESC and p55 in Drosophila (Liu et al., 2014; Luo et al., 2009). A number of accessory proteins such as LC2, OsVIL2 and OsEMF1/DS1 modulate PRC2 recruitment by interacting with the core components of PRC2 (Calonje et al., 2008; Liu et al., 2018; Wang et al., 2013; Yang et al., 2013). The PRC2 complex has been extensively characterized in various organisms such as Drosophila, Arabidopsis and mammals, but it is still understudied in rice.
It has been documented that rice panicle number and grain size can be epigenetically controlled by the PRC2 complex. FIE1 negatively regulates grain width in response to heat stress (Dhatt et al., 2021). OsVIL2‐PRC2 suppresses OsTB1 expression by modifying the chromatin (Wang et al., 2013; Yang et al., 2013; Yoon et al., 2019). NGR5 drives the recruitment of PRC2 to repress the expression of D14 and OsSPL14 by interacting with LC2/OsVIL3, thereby subsequently promoting nitrogen‐induced tillering (Wu et al., 2020). RLB (RICE LATERAL BRANCH) physically binds to the PRC2 component OsEMF2b to regulate lateral branching through repressing the expression of OsCKX4 (Wang et al., 2021). These studies pinpoint a deep insight regarding a separate mode of regulation of panicle number or grain size by PRC2. However, it is still unknown whether PRC2 can synergistically regulate panicle number and grain size through common regulators in rice. In this study, we identified a rice epigenetic regulator, PANICLE NUMBER AND GRAIN SIZE, which fine‐tuned the balance between panicle number and grain size. The gene was identified to be a transposon‐derived gene with neofunctionalization, so we named it ‘PANDA’ (PANICLE NUMBER AND GRAIN SIZE), to indicate its nature as a living fossil such as the Giant panda (Ailuropoda melanoleuca). Mutation of PANDA reduced panicle number but increased grain size, while its overexpression plants reversed this balance. More importantly, we found that PANDA was neofunctionalized as an epigenetic regulator with an ability to bind PRC2, and the moderate overexpression of PANDA driven by its own native promoter can increase grain yield in indica rice cultivar.
Results
PANDA coordinates panicle number and grain size in rice
We isolated a natural mutant called panda with conspicuous changes in panicle number and grain size. Compared with its wild type parental japonica variety ‘Taibei 309’, panicle numbers in the panda mutant decreased by ~44.1%, but grain weight increased by ~42.7% in different growth environments (Figure 1; Figures S1 and S2A–C; Table S2). The larger grain size in the mutant panda was most likely caused by the increase in cell number rather than cell size in its glume, which is due to a number of the cell cycle‐related genes with significantly higher expression in the 2‐ to 3‐mm young panicles of the panda mutant than the wild type (Figure S2D–J). We did not observe consistent changes in spikelet number per panicle and seed‐setting rate in panda in multiple environments, indicative of environmental plasticity for these traits (Table S2). These changes in yield‐related traits in the panda mutant collectively led to reductions in its yield per plant in most cases (Table S2). In addition, compared with wild type, the panda mutant had increased plant height, larger and more drooping leaves, thicker culms, and sometimes longer awns and curly leaves (Figures S1 and S3).
Figure 1.

Map‐based cloning of PANDA. (a, b) Panicle numbers (a) and grain size (b) in wild type, the mutant panda and the complementary line (pPANDA::PANDA/panda) at maturity. (c) Fine mapping of PANDA to a 45‐kb region on the short arm of chromosome 7. (d) Sequence comparison of the candidate gene between wild type and panda. (e, f) Statistical analyses of panicle number (e) and grain size (f) in wild type, panda and pPANDA::PANDA/panda plants. (g, h) Panicle number (g) and grain size (h) in wild type and the PANDA‐overexpressing (OE) line at maturity. (i, j) Statistical analysis of panicle number (i) and grain size (j) in the wild type and three PANDA‐overexpressing lines at maturity. Different letters represent significant differences at the 5% level determined by Tukey’s test (n ≥ 8). Scale bars: a = 25 cm, b = 3 cm, g = 20 cm, h = 3 cm.
To dissect the genetic basis of the panda mutation, we obtained F1 plants by crossing panda with its wild type parent ‘Taibei 309’, and the F1 plants were indistinguishable from wild type in four measured traits (Figure S4). In the F2 population obtained by crossing panda with the wild type indica rice variety ‘Teqing’, the segregation ratio between the wild type and the mutant type with fewer tillers but larger grains was 3:1 (7437:2415, χ2 = 0.00042 < χ2 0.05,1 = 3.841, P < 0.05). These results indicated that the panda mutation was controlled by a single recessive gene.
To finely map the PANDA gene, we next conducted genetic mapping using the recombinants from the F2 population mentioned above, and finally narrowed the candidate interval down to a 45‐kilobase region, which is flanked by the simple sequence repeat marker (SSR) loci M5 and M7 on chromosome 7 (Figure 1c). We found that there are eight genes within this region, but only the LOC_Os07g07880 gene had a base change from G to A in the second exon, resulting in an amino acid change from Gly to Asp at the deduced residue 384 (Figure 1d). Thus, we considered this gene to be the candidate gene for PANDA.
To confirm the function of the candidate gene, we performed a genetic complementation test by transforming the entire gene including the coding sequence, the 2.6‐kb promoter region and the 1.0‐kb downstream region into the panda mutant using agrobacterium‐mediated transformation. All of the independent T3‐generation transgenic lines were restored to the wild type based on panicle number and grain size traits (Figure 1a,b,e,f; Figure S5). We also characterized PANDA‐overexpressing transgenic plants in which PANDA was driven either by the maize Ubiquitin promoter in the japonica rice cultivar ‘Taipei 309’ background or by its own promoter in the indica rice cultivar ‘Genit’ background. Compared with the control, the expression level of PANDA in the Ubiquitin promoter‐driving transgenic lines increased by ~50 folds, the number of panicles increased by ~35.4%, and the grain weight decreased by ~14.9% (Figure 1g–j). In addition, the transgenic lines decreased in plant height, spikelet per panicle and seed‐setting rate (Figure S6). All these changes collectively resulted in a decrease by 13.9% in grain yield per plant in these transgenic lines (Figures S6 and S8). In the PANDA promoter‐driving overexpression lines, the PANDA expression level increased by 6.7 folds, panicle number increased by 76.9%, while grain weight decreased by 5.3% (Figures S7 and S8). The moderate increase of PANDA expression level in these lines has less negative effects on spikelet number per panicle and seed‐setting per cent, altogether resulting in an increase in yield per plant by 27.7% (Figures S7 and S8). Taken together, these results confirm that LOC_Os07g07880 is the gene that we named PANDA. It is responsible for the balance between panicle number and grain size in rice, and its moderate overexpression has yield increasing potential in rice.
PANDA is expressed broadly and encodes a nuclear‐localized protein
According to the gene expression database CREP (http://crep.ncpgr.cn), the PANDA gene is expressed broadly in all rice tissues, such as roots, stems, leaves, young panicles and glumes (Figure S9A). The extensive expression pattern of the PANDA gene was verified by qRT‐PCR (Figure S9B). To further confirm this, we transformed the panda mutant with a construct consisting of the reporter GUS gene fused in frame with the PANDA gene driven by of 2.6‐Kb PANDA promoter region (pPANDA::PANDA‐GUS). The panda mutant phenotypes were restored to the wild type in the transgenic plants, suggesting that the fusion protein is functional (Figure S10A–C). We then examined the spatial–temporal glucuronidase (GUS) staining patterns in these plants and observed that the blue GUS stain signal was detectable in most of the rice tissues, such as leaves, glumes and roots (Figure S10D–K).
The PANDA protein is predicted to be a nuclear‐localized protein when analysed with ProtComp 9.0 software (http://linux1.softberry.com). To validate the computational prediction and investigate the subcellular localization of PANDA, we transiently expressed PANDA fused with green fluorescent protein (GFP) (PANDA‐GFP) in rice protoplasts and N. benthamiana leaves. We observed that the PANDA‐GFP fluorescent emissions completely overlapped the NLS‐mCherry signals in the nuclei of rice protoplasts and also in the nuclei of Nicotiana benthamiana leaf cells (Figure S11). Taken together, this indicates that the biological function of the PANDA protein can be achieved by nuclear localization and being expressed broadly in rice.
PANDA is a functionally conserved Harbinger transposon‐derived gene
Searching against GenBank and the Repbase database (https://www.girinst.org/repbase), the most comprehensive transposon database including all transposon superfamilies reported thus far (Bao et al., 2015), we found that PANDA had significant sequence similarity to the PIF/Harbinger transposons. The protein sequence of PANDA showed 36.1%, 31.1% and 23.4% similarity to the PIF/Harbinger transposons, Harbinger‐4_CMy_1p, Harbinger‐2_AMi_1p and Ping, with E values of 3.0E‐47, 3.0E‐34 and 4.0E‐9, respectively (Figure S12A,B). The nucleotide sequence of PANDA had a degree of similarity with Harbinger‐4_CMy_1p or Harbinger‐2_AMi_1p, but almost no similarity with Ping. For example, the conserved domain‐encoding nucleotide sequence of PANDA showed 68.1% identity with the corresponding part of Harbinger‐2_AMi_1p (Figure S12C). Based on comparisons of their protein sequences, rice PANDA and its Arabidopsis homolog ALP1 (Antagonist of Like Heterochromatin Protein1) showed closely evolutionary relationships to Harbinger‐2_AMi_1p and Harbinger‐4_CMy_1p than the Ping transposon (Figure S12D). It should be noted that ALP1 was derived from the evolutionary exaptation of Harbinger transposon (Velanis et al., 2020). Transposons have terminal inverted repeats and flanking target site duplication, such as Ping (Figure S12E), which are the typical features of transposons, while PANDA lacked these typical features, suggesting that PANDA is a gene without capability for movement in the genome. Additionally, PIF/Harbinger transposons can be present in high copy numbers in plant genomes (Casola et al., 2007; Grzebelus et al., 2007; Jiang et al., 2003), while there were only two homologs with high similarity to PANDA, Os11g0702700 and Os01g0838900 (Figure S13). The two homologs were knocked out using CRISPR/Cas9 system, and the resulting mutants did not exhibit any visible altered phenotypes, such as panicle number and grain size (data not shown), suggesting that the two homologs cannot regulate panicle number and grain size like PANDA. Given that PANDA showed significant sequence similarity to Harbinger transposons and the reported domesticated gene (Velanis et al., 2020), the results indicate that PANDA is a Harbinger transposon‐derived gene.
The homologs of PANDA were identified in a wide range of organisms including many important crops such as barley (Hordeum vulgare), maize (Zea mays), soybean (Glycine max) and rapeseed (Brassica napus) (E value <1 × 10−80). Impressively, PANDA also showed significant sequence similarity with the proteins in animals including HARBI1, which represents a domesticated PIF/Harbinger transposon in mammals (Kapitonov and Jurka, 2004). To understand the evolutionary relationships between PANDA and other homologous proteins, we conducted a phylogenetic analysis using these homologs and the three PIF/Harbinger transposons mentioned above. PANDA and other plant homologs such as ALP1 in Arabidopsis were grouped into a clade, while HARBI1 and other animal homologs were grouped into another clade. Neither clade contained the three transposons (Figure 2a). The wide distributions of the PANDA homologs in plants and their distant phylogenetic relationship with their animal homologs and the three transposons indicated that PANDA may be a conserved transposon‐derived gene in rice with neofunctionalization.
Figure 2.
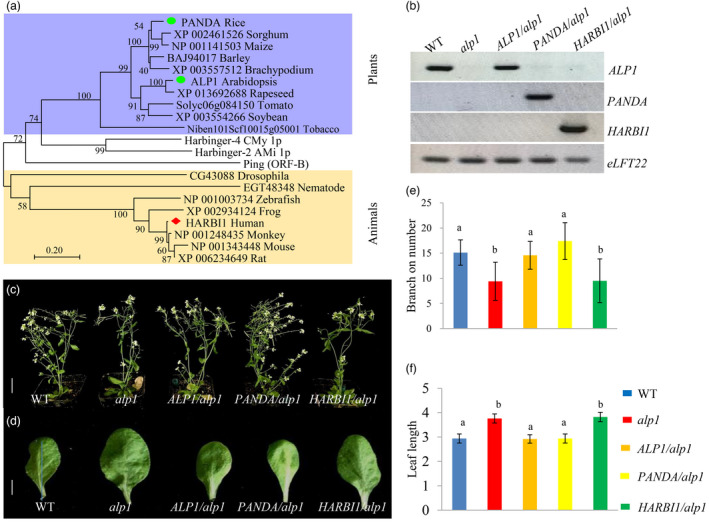
PANDA is a functionally conserved Harbinger transposon‐derived gene. (a) Phylogenetic tree of PANDA and its homologous proteins in some animals and plants. (b) Semi‐qRT‐PCR analysis of the transgenic lines expressing PANDA and two homologous genes in the Arabidopsis alp1 mutant. (c, d) Branch numbers (c) and leaf size (d) in the transgenic Arabidopsis alp1 lines expressing PANDA homologs. (e, f) Analysis of branch numbers (e) and leaf size (f) in the transgenic lines expressing PANDA homologs in the alp1 mutant background. Scale bars: c = 5 cm, d = 0.75 cm.
Since PANDA and its homologs distributed widely in plants, we performed genetic analyses to determine whether they are functionally conserved in plants. We first phenotyped the Arabidopsis homolog mutant alp1, which has not been well characterized before. Similar to panda mutant plants, alp1 plants showed increases in the sizes of the leaves, flowers and seeds, but had fewer branch numbers compared with the wild‐type Ler (Figure 2b–f; Figure S14 and S15). We transformed the rice PANDA gene, the Arabidopsis ALP1 gene and the control animal HARBI1 gene individually into the Arabidopsis mutant alp1 (Figure 2b). After phenotyping the transgenic lines that showed stable expression, we observed that both the ALP1/alp1 and PANDA/alp1 transgenic lines restored the wild type phenotypes, while the HARBI1/alp1 transgenic lines did not (Figure 2c–f; Figures S14, S15 and S18). Therefore, PANDA and ALP1 are neofunctionalized and functionally conserved Harbinger transposon‐derived genes in plants, which can balance the branch number and organ size in transgenic Arabidopsis. However, PANDA and HARBI1 showed divergent functions.
The PANDA protein consists of 441 amino acids (UniPro No.: Q8H572), while Arabidopsis ALP1 is 397 amino acids in length (UniProtKB No.: Q94K49‐1). Rice PANDA has an additional 44 amino acid sequence at the N‐terminal end compared with APL1 (Figure S16). In order to determine whether the additional 44 amino acids affect the function of PANDA, we performed genetic complementation by transforming the entire 1,326 base pair PANDA ORF or a truncated PANDA ORF with a length of 1,194 base pairs fused with the 35S promoter into the rice panda and Arabidopsis alp1 mutants using Agrobacterium‐mediated transformation. The T3‐generation transformed lines for each transformation combination were characterized, such as PANDA441aa /panda, PANDA397aa /panda, PANDA441aa /alp1 and PANDA397aa /alp1. In rice, the transgenic PANDA441aa /panda lines were completely restored to the wild type phenotype, while PANDA397aa /panda plants were showed only partial recovery of the wild type phenotypes, such as complete recovery on panicle number, but partial recovery on grain size and plant height (Figure S17). In Arabidopsis, both PANDA441aa /alp1 and PANDA397aa /alp1 can restore the alp1 mutant to wild type in terms of branch number and organ size (Figure 2c–f; Figures S14, S15 and S18). Taken together, these results showed that the additional N‐terminal 44 amino acid sequence in the PANDA protein is necessary for the biological function of PANDA in rice but not in Arabidopsis, suggesting that the sequence divergence between PANDA and ALP1 is likely arisen after the evolutionary split from the common ancestor.
PANDA interacts with polycomb repressive complex 2 (PRC2) and regulates H3K27me3 deposition in the rice genome
ALP1, a homologous protein of PANDA, in Arabidopsis, has been reported to be an accessory protein associated with the MSI1 of PRC2 that is responsible for H3K27me3 formation (Liang et al., 2015). This prompted us to assess whether the PANDA protein interacts with the components of PRC2 in rice. An in vitro pull‐down assay was performed between PANDA and each of the components of PRC2. As shown in Figure 3a,b, The PANDA‐GST fusion protein can be immunoprecipitated with OsMSI1‐His or OsFIE2‐His in vitro, respectively. Bimolecular fluorescence complementation (BiFC) assays and the CoIP test were then performed to confirm the interaction in vivo. As shown in Figure 3c,d, a strong fluorescence signal was observed in the nucleus in both rice protoplasts and N. benthamiana leaves, which co‐expressed the C‐terminal half of YFP fused to the PANDA protein (PANDA‐cYFP) and the N‐terminal half of YFP fused to OsMSI1 (OsMSI1‐nYFP) or OsFIE2 (OsFIE2‐nYFP). The PANDA protein fused to the Flag tag co‐precipitated with the OsMSI1 or OsFIE2 protein with the MYC tag in rice protoplasts (Figure 3e,f). Taken together, these results simply that PANDA interacts with the PRC2 in rice by binding to both OsMSI1 and OsFIE2, the components of the PRC2. Additionally, we also assessed whether the mutation (G384D) of PANDA affects its interaction with the components of PRC2. The results of pull‐down assay showed that both the wild‐type and the mutant PANDA can bind to OsMSI1 and OsFIE2 (Figure S19A,B), suggesting that the mutation of PANDA with amino acid change from Gly to Asp at the position 384 does not affect its binding ability with PRC2.
Figure 3.
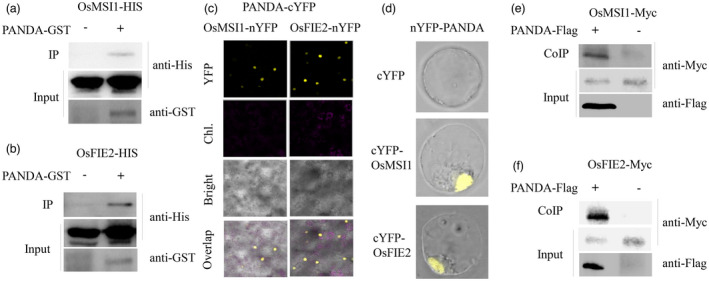
Protein–protein interactions between PANDA and PRC2 components. (a, b) Pull‐down analyses between PANDA and two components of PRC2, OsMSI1 and OsFIE2. (c, d) BiFC analyses between PANDA and OsMSI1 or OsFIE2 in leaf cells of Nicotiana benthamiana (c) or rice protoplasts (d). (e, f) CoIP analyses between PANDA and OsMSI1 (e) and PANDA and OsFIE2 (f) in rice protoplasts.
Polycomb repressive complex 2 has been reported to epigenetically repress gene expression through the deposition of H3K27me3 in the rice genome (Bieluszewski et al., 2021). To assess the effects of PANDA on gene expression, we firstly carried out RNA‐seq and found that there were 762 up‐regulated genes and 1,236 down‐regulated genes in the panda mutant compared with the wild type control (Figure 4a). To assess the repressive role of H3K27me3 in regulating gene expression through PANDA, we also performed anti‐H3K27me3 ChIP‐seq between wild type and panda (Figure S20). We found that 2,079 genomic loci exhibited significantly reduced H3K27me3 enrichment levels and 576 loci had significantly induced H3K27me3 enrichment levels in panda compared with wild type control (Figure S20A,B). To investigate the relationship between H3K27me3 enrichment levels and the expression levels of all genes in WT and panda, we plotted normalized read counts of H3K27me3 ChIP‐seq reads across ±1 kb of TSSs and TTSs of genes with different expression levels (high, medium or low FPKM value) and observed an overall negative correlation between H3K27me3 enrichment levels and the expression levels of all expressed genes in WT and panda (Figure S20C,D), which is consistent with repressive roles of H3K27me3 in gene expression. We then analysed the correlation between the change of H3K27me3 enrichment levels and expression levels for all genes between WT and panda. We found that the changes of H3K27me3 enrichment level did not exhibit a significant correlation with the expression changes of all genes (Figure S20E). A plausible explanation is that change of H3K27me3 enrichment level may affect expression level of a subset of genes. To test this possibility, we further analysed the relationship for both H3K27me3 and genes with significant changes between WT and panda. Indeed, we observed a negative correlation between changes of H3K27me3 enrichment level and expression levels in 978 genes with significant changes both in H3K27me3 enrichment and in expression levels (rho = −0.36, P < 0.01, Figure 4b). Therefore, the results reflected the repressive role of H3K27me3 in regulating gene expression (Liang et al., 2015). In detail, among the up‐regulated genes in panda, 63.6% of them had no significant differences in H3K27me3 enrichment levels between panda and wild type, while 24.0% and 12.3% of them had reduced and increased H3K27me3 enrichment levels in panda compared with wild type, respectively. Among the down‐regulated genes in panda, 55.5% had no significant differences in H3K27me3 enrichment levels between two genotypes, while 37.9% and 11.6% of them had increased and reduced H3K27me3 enrichment levels in panda relative to wild type, respectively (Figure 4c,d). These genes with a negative correlation between the levels of H3K27me3 enrichment and transcription may be the target genes epigenetically regulated by PANDA. Other differently expressed genes with no differences in H3K27me3 enrichment levels between genotypes may be the genes indirectly regulated by PANDA. All the above analyses suggest that PANDA affects the deposition of H3K27me3 in rice genome.
Figure 4.

PANDA regulates the deposition of H3K27me3 in the rice genome. (a) Differential expression of genes in leaves between wild type and the panda mutant. (b) Correlation analysis between RNA levels and H3K27me3 enrichment levels. (c) Percentages of the different H3K27me3 enrichment types of the down‐regulated genes in panda (up arrow, higher H3K27me3 in panda than wild type; down arrow, lower H3K27me3 in panda than wild type; N.S., no significant differences between two genotypes). (d) Percentage of the different H3K27me3 enrichment types of the up‐regulated genes in panda (the same as above). (e) Overlapping loci with deposition by H3K27me3 and PANDA‐Flag. (f) The motif enriched in the peaks with deposition of PANDA.
In order to find the direct PANDA‐regulating target genes associated with panicle number and grain size, we carried out anti‐Flag ChIP‐seq analysis using the T1‐generation transgenic plants overexpressing the PANDA‐Flag fusion gene, which restored the wild type phenotype in the panda mutant (Figure 1g–j; Figure S6). We detected a total of 2170 sites with PANDA‐Flag enrichment in the rice genome and 241 loci co‐localized with H3K27me3 enrichment (Figure 4e; Tables S5 and S6). Among the 241 loci, we identified 233 genes, including 52 and 33 genes highly expressed in WT and panda, respectively, and 148 genes without significant expression changes between WT and panda (Table S7). After conducting a motif enrichment assay, we detected a GCC motif that was significantly enriched in the loci bound by PANDA‐FLAG (Figure 4f). After combining anti‐H3K27me3 and anti‐Flag ChIP‐seq with RNA‐seq data, we found that the growth‐ and development‐related genes bound by PANDA, including OsMADS22, OsMADS55 and OsEMF1, had lower H3K27me3 enrichment level but higher transcription levels in the panda mutant relative to the wild type (Figure S21, Table S3, S5 and S6). Therefore, we speculate that these growth‐ and development‐related genes are directly epigenetically regulated by the binding of PANDA‐PRC2. In addition, there were other growth‐ and development‐related genes without PANDA deposition but with changes in both the levels of H3K27me3 enrichment and transcription (Table S3, S5 and S6). For example, compared with the wild type, the gene OsERF112 had higher H3K27me3 deposition but a lower transcription level in the panda mutant (Figure S26), suggesting that the gene may be indirectly regulated by PANDA.
PANDA balances panicle number and grain size by epigenetic silencing of the target genes OsMADS55 and OsEMF1
To reveal how PANDA functions in the regulation of rice panicle number and grain size, we specifically analysed the growth‐ and development‐related target genes that were directly epigenetically regulated by PANDA. This group of genes, including OsMADS22, OsMADS55 and OsEMF1, exhibited lower H3K27me3 enrichment levels but increased expression levels in the panda mutant as compared to wild type (Figures S21 and S22, Table S3). Given that these target genes contain the GCC motif, we inferred that the motif is essential for involvement of PANDA in epigenetically suppressing the expression of the target genes. To prove this hypothesis, we first performed ChIP‐qPCR analysis using the PANDA‐Flag transgenic plants. The first exon of OsMADS55 containing two GCC motifs had significant levels of PANDA‐Flag enrichment, while the OsERF112 gene without GCC motifs had no significant deposition of PANDA‐Flag (Figure 5a,b). Then, we performed transient expression analyses using reporter genes fused with the first exon of OsMADS55 containing the GCC motifs in vivo (Figure 5c,d). Compared with the negative control, PANDA, its truncated version and its Arabidopsis homolog ALP1 significantly inhibited the expression of the fused GFP in leaf cells of N. benthamiana (Figure 5c). Similarly, we also conducted transient expression of the fused GFP reporter gene in the leaves of wild‐type (ALP1) Arabidopsis and the alp1 mutant. We observed that the expression level of GFP in the wild‐type leaves was significantly lower than that in the alp1 mutant (Figure 5d). When the motifs were mutated, the signals of GFP increased significantly as compared to the wild‐type motif (Figure S23). Therefore, we concluded that the GCC motif is essential for the functions of PANDA in epigenetically silencing its target genes. However, unfortunately, EMSAs failed to support the direct binding of PADNA to the GCC motif (Figure S24). A plausible explanation for this is that PANDA may indirectly bind to the GCC motif, or the epigenetic regulation of PANDA on the GCC motif‐containing target genes may require other protein or protein complex. Additionally, we also assessed the effects of two alleles of PANDA on repressing target gene expression using the GAL4BD system (Lu et al., 2013). The wild‐type PANDA, the mutant panda (G384D), the VP16 positive control and the Flag negative control were fussed to the GAL4 binding domain under the driver of the 35S promoter. Activities of firefly luciferase (LUC) driven by the GAL4 binding element UPSTREAM ACTIVATION SEQUENCE (UAS) were measured using plant in vivo imaging system. As compared to two controls, both GAL4BD‐PANDA and GAL4BD‐panda can inhibit the expression of LUC, but GAL4BD‐panda exhibited less inhibitory effects than GAL4BD‐PANDA (Figure S19C). Considering both genotypes binding to PRC2 as mentioned above, the results suggest that the mutation of PANDA (G384D) likely affects the expression of the target genes through the modified function of the complex.
Figure 5.

PANDA epigenetically regulates the expression of its downstream genes. (a) The distribution of the GCC motif and the fragments of ChIP‐qPCR in the OsMADS55 or OsERF112. (b) ChIP‐qPCR analyses in the PANDA‐Flag transgenic liens. (c) PANDA, its truncated form and ALP1 inhibited the expression of GFP fused with the GCC motif‐containing fragment from OsMADS55 in N. benthamiana leaf epidermal cells. (d) The expression of GFP fused with the GCC motif in the Arabidopsis alp1 mutant and wild type. (e) The combined analysis of the target gene OsMADS55 by multiple omics (RNA‐seq, anti‐H3K27me3 ChIP‐seq and anti‐PANDA‐Flag ChIP). (F‐I) Panicle numbers (f, h) and grain size (g, i) in the CRISPR/Cas9‐derived mutant Osmads55 and the wild type. (j) The working model of PANDA‐regulating panicle number and grain size. X, a yet‐to‐be‐identified protein or protein complex.
Through gene editing‐based functional analyses, we found that the PANDA target gene, OsMADS55, was involved in the regulation of the balance between panicle number and grain size. Both OsMADS55 and OsMADS22, which encode SVP‐group MADS‐box transcription factors, have been reported to act as negative regulators of brassinosteroid responses (Lee et al., 2008). Since it has been reported that the knockout of OsMADS22 did not result in any visible phenotypic changes (Lee et al., 2008), we then specifically analysed the CRISPR/Cas9‐mediated knockout of OsMADS55. Compared with the wild type control, plants of the Osmads55 mutant in the japonica rice cultivar ‘Zhonghua 11’ had a significant increase (60.3%) in panicle number, a significant decrease (7.5%) in grain weight and also a significant decrease (15.6%) in plant height (Figure 5e–i). The double gene mutations of OsMADS55 and PANDA were obtained by knocking out OsMADS55 in the panda mutant background. Compared with both panda and the wild type control, the pandaOsmads55 double‐mutant plants had more panicles but smaller grains and shorter stems (Figure S25), suggesting that OsMADS55 functions at the downstream of PANDA. In addition, the gene OsEMF1 bound by PANDA in the PANDA‐Flag overexpression plants also had reduced H3K27me3 enrichment level but higher transcription levels in the panda mutant relative to wild type (Figures S21 and S22, Table S3). OsEMF1 encodes a plant‐specific EMF1‐like protein and also has been reported to control rice architecture by regulation of brassinosteroid signalling. The Osemf1 mutant has been shown previously to have increases in the number of tillers but reduced grain weight (Liu et al., 2018; Yan et al., 2015; Zheng et al., 2015). These genetic analyses further confirmed that OsMADS55 and OsEMF1 were involved in the regulation of the balance between panicle number and grain size in rice (Figure 5j).
Additionally, we also genetically analysed the growth and development‐related genes that were indirectly regulated by PANDA. We found that OsERF112 was regulated by PANDA and related to grain size. OsERF112 is predicted to encode an AP2 domain‐containing protein, which functions as an ethylene response factor. This gene had no PANDA deposition in the rice genome, but was more enriched with H3K27me3 and down‐regulated in expression levels in the panda mutant relative to wild type (Figure S26A), suggesting that PANDA may promote transcription of OsERF112 by indirectly epigenetic regulation. The OsERF112 knockout plants did not show any significant change in panicle number, but exhibited a significant increase in grain size (Figure S26B–E), suggesting OsERF112 only negatively regulates grain size. Therefore, as shown in Figure S26F, PANDA promoted the transcription of OsERF112 indirectly by epigenetic regulation, and then, OsERF112 negatively regulated the grain size.
Taken together, these results show that PANDA epigenetically coordinates panicle number and grain size by directly silencing its target genes OsMADS55 and OsEMF1, and/or by indirectly regulating the transcription of other downstream genes that are independently responsible for panicle number or grain size.
Discussion
Both panicle number and grain size are important agronomic traits for rice yield. Considerable progress has been made on understanding the genetic and molecular bases of these two traits independently (Li et al., 2018; Wang et al., 2018). However, the genetic basis of the balance between panicle number and grain size in rice is still unclear (Fu et al., 2010; Guo et al., 2020). Dissection of the underlying molecular mechanism will provide a full understanding of the genetic control, which will benefit crop yield improvement. In this study, we characterized an epigenetic regulator PANDA balancing panicle number and grain size. The mutation of PANDA showed reduced panicle numbers and increased grain size (Figure 1a,b,e,f; Figures S1–S5), while PANDA overexpression lines showed an increase in panicle number but a decrease in grain size (Figure 1g–j; Figures S6 and S7). Analyses of genome‐wide histone modification, RNA‐seq and ChIP‐seq found that a set of genes were epigenetically regulated (Figure 4; Figure S20; Tables S3–S6). Knocking out of the growth‐ and development‐related target genes of PANDA, such as OsMADS55 and OsEMF1, reversed the mutant panda phenotype on panicle number and grain size (Figure 5f–i; Figure S25; Liu et al., 2018). OsMADS55 is a negative BR‐responsive gene (Lee et al., 2008). OsEMF1 can interact with OsARF11 and then bind to the OsBRI1 promoter to modulate BR signalling (Liu et al., 2018). The role of BR on the control of grain size has been shown in a number of BR biosynthesis and signalling mutants (Li et al., 2018). Recently, it was reported that BR is involved in rice panicle formation by regulating the stability of the D53‐OsBZR1 complex to regulate FC1 expression (Fang et al., 2020). Therefore, this suggests that PANDA balances panicle number and grain size through the BR‐dependent pathway. Consistently, an allele of PANDA, POW1, is involved in separable regulation of grain size and leaf angle development through the BR‐dependent signal pathway in rice (Zhang et al., 2021). However, we should note that other genetic factors, such as OsERF112, also contribute to the phenotypic changes in grain size independently. Knockout of OsERF112 partly phenocopied the panda mutant on grain size, but not on panicle number (Figure S26). Altogether, PANDA epigenetically coordinates panicle number and grain size in a BR‐dependent way, while BR‐independent factors that work downstream of PANDA also contribute to the phenotypic changes separately.
PANDA and its homolog ALP1 in Arabidopsis associate with PRC2 to modify the histone methylation and epigenetically repress expression of its target genes (Figures 3, 4, 5; Liang et al., 2015; Velanis et al., 2020). Our ChIP‐seq analyses showed a GCC motif that was significantly enriched in the regions of PANDA deposition (Figures 4f and 5a–d). However, it is still an open question as to how PANDA modulates the recruitment of the PRC2 complex to target loci containing this motif. A similar motif has been reported to be associated with H3K27me3 deposition in Arabidopsis (Xiao et al., 2017). This motif in rice can be recognized and bound by the transcription factor NGR5 to recruit PRC2 to modify the histone methylation and repress expression of its target genes (Wu et al., 2020). We did not obtain evidence showing a direct interaction of PANDA with NGR5 or its partner LC2 by yeast two‐hybrid assays (data not shown). Although PANDA has an HTH domain, which is a DNA‐recognition motif often involved in DNA binding (Zaveri et al., 2021), PANDA did not bind to the GCC motif through the HTH domain directly (Figure S24). The results suggest that the epigenetic regulation of PANDA on the GCC motif‐containing target genes, including OsMADS55 and OsEMF1, may require a yet‐to‐be‐identified protein or protein complex. Histone methylation can be either established by accessary protein‐mediated PRC2 deposition, or maintained and re‐established by PRC1 during chromosome duplication (Blackledge et al., 2014). ALP1 antagonizes LHP1, which is incorporated into the PRC1 complex functioning as a histone methylation reader and maintainer (Liang et al., 2015; Tao et al., 2019; Yuan et al., 2016). Both PANDA and LHP1 can bind to MSI1 in rice or in Arabidopsis (Figure 3; Derkacheva et al., 2013). As several independent PRC1 complexes have been reported to coordinate genome‐wide gene expression (Li et al., 2018), it is tempting to assume that PANDA incorporates in a PRC1 complex independent of OsLHP1 to modulate the PRC2 recruitment and gene silencing. Analysis of the PANDA interactome will provide a full picture of its genetic network and facilitate a deep understanding of epigenetic regulation in plants.
Transposons were often considered to be ‘junk DNA’ or ‘genome parasites’, but they can be ‘domesticated’ and evolve new cellular functions that benefit the host (Kapitonov and Jurka, 2004; Smit and Riggs, 1996). Thus far, most domesticated transposons were found in mammals, and only a few cases of transposon domestication have been reported in plants (McDowell and Meyers, 2013; Volff, 2006). Our results indicated PANDA showed significant sequence identity to Harbinger transposons and reported transposon‐related genes, suggesting that PANDA is likely derived from a Harbinger transposon but evolved into a new gene due to loss of its mobility and undergoing neofunctionalization that controls panicle number and grain size in rice. Moderate overexpression of PANDA under its own promoter in the indica background can significantly increase grain yield per plant (Figures S7 and S8). Therefore, PANDA is a potential gene for rice yield improvement in future. Homologs of PANDA were also present in many other important crops (Figure 2a) and showed high sequence similarity to PANDA. It is worthwhile to further address whether the homologs of PANDA in these crops also govern phenotypic traits similar to panicle number and grain size. Since the homologs of PANDA were found in diverse plants including both dicots and monocots, which split from a common ancestor over 150 million years ago (MYA) (Zeng et al., 2014), and PANDA can restore the wild type phenotype in the Arabidopsis alp1 mutant (Figure 2), this suggests that APL1 in Arabidopsis and PANDA in rice may be derived from the same ancient ancestor >150 MYA. The human HARBI1 protein cannot restore the wild type phenotype in Arabidopsis alp1 and was grouped into a different clade from APL1/PANDA (Figure 2b). It is possible that the HARBI1 and other homologous proteins in animals were domesticated independently. However, more analyses are necessary to further address the evolution and functional divergence of PANDA‐related genes in plants.
In conclusion, we have identified a transposon‐derived gene with neofunctionalization that epigenetically coordinates panicle number and grain size in rice. Our results also provide novel insights into the genetic control of rice yield traits and a potential way to improve crop yields associated with this gene.
Materials and methods
Plant materials and field experiments
The rice mutant ‘panda’ was a natural variant of the japonica rice variety ‘Taibei 309’. The F2 population used for map‐based cloning was obtained by crossing the mutant ‘panda’ with the indica rice variety ‘Teqing’, followed by self‐pollination of the F1 hybrid. The japonica rice cultivars ‘Taibei 309’ (TB309) and ‘Zhonghua 11’, and the indica rice cultivar ‘Genit’ were used for genetic transformation. Plants were cultivated in paddy fields following normal agricultural practices with a row spacing of 20 cm, a plant spacing of 15 cm, and 10 plants per row at our experimental farms in Sanya, Changsha and Beijing during the winter or the summer of 2014–2020. More than 10 plants of each line were used to dissect the yield‐related traits. The significant differences were determined by Student’s t‐test or Tukey’s honestly significant difference (HSD) test.
To test the functional conservation of PANDA in plants, we collected the mutant alp1 and its wild type (Ler) of Arabidopsis thaliana. The Arabidopsis plants were cultivated in a growth chamber (Percival Scientific, Perry, IA) under a 16‐h day/8‐h night photoperiod at temperatures of 23 °C (day) and 20 °C (night).
Map‐based cloning
In the F2 mapping population, we constructed two DNA pools composed of 30 wild‐type plants or 30 mutant‐type plants. The pools were genotyped using genome‐wide SSR markers for the primary mapping of PANDA. To fine‐map PANDA, 2,415 individual plants with the mutant phenotype were genotyped using the newly developed markers around the primary PANDA mapping interval. All genes in the fine‐mapped region were amplified by PCR, and the resulting fragments were subjected to Sanger DNA sequencing. The sequences from both wild type and the panda mutant were then compared using the Sequencher 5.0 software (Gene Codes Corporation, Ann Arbor, Michigan) to investigate sequence divergence and identify the candidate gene of PANDA.
Protein sequence analyses
The functional domains present in the PANDA protein were predicted using the Conserved Domain Database (https://www.ncbi.nlm.nih.gov/cdd). The proteins homologous to PANDA were identified using BLASTP searches against the NCBI database (https://www.ncbi.nlm.nih.gov) and the rice genome annotation database (http://rice.uga.edu). An alignment of the homologous protein sequences was performed using the software Clustal W, and a neighbour‐joining phylogenetic tree was constructed using MEGA 5.0 (https://www.megasoftware.net).
Plasmid construction and plant transformation
Gene fragments were amplified from genomic DNA of ‘Taibei 309’ and then cloned into the binary vectors based on homologous recombination technology using the ClonExpress Entry One Step Cloning Kit (Vazyme, Nanjing, China). The recipient vectors were pTCK303 for constitutive overexpression of PANDA driven by the maize Ubiquitin promoter (pUBI:PANDA), pCAMBIA1300 for the complementation assay in the panda mutant or moderate overexpression in the indica rice cultivar ‘Genit’ driven by its own promoter (pPANDA::PANDA), and pCAMBIA1301 for the analysis of the PANDA promoter activity (pPANDA::PANDA‐GUS). To obtain the CRISPR/Cas9 knockout lines, the Cas9 gene was driven by the CaMV 35S promoter, while the 20 bases upstream of the protospacer adjacent motifs (PAMs) were selected as target sequences, and their expression was driven by the OsU3 promoter in the binary vector of BGK03. The target regions in the genome of T0‐ and/or T1‐generation transgenic plants were sequenced using the related PCR products. All plasmid vectors were introduced into Agrobacterium tumefaciens strain EHA105 followed by Agrobacterium‐mediated transformation of japonica rice cultivars ‘Taibei 309’, ‘Zhonghua 11’, the mutant ‘panda’ or the indica rice cultivar ‘Genit’. For complementation assays in the Arabidopsis mutant alp1, the full‐length coding regions of ALP1, PANDA or HARBI1 were cloned into the plant binary vector pZH01 to generate 35S::PANDA, which was then used to transform the Arabidopsis mutant alp1 by Agrobacterium‐mediated transformation.
Subcellular localization
The vectors 35S::PANDA‐EGFP and 35S::EGFP, and the nuclear localization marker 35S::NLS‐mCherry were introduced into rice protoplast cells by PEG‐mediated transformation or into leaves of N. benthamiana by Agrobacterium‐mediated transformation. Green and red fluorescence signals were observed with a laser confocal microscope (LSM880, Zeiss Corporation, Oberkochen, Germany).
β‐Glucuronidase staining
Different tissues of the pPANDA::GUS transgenic plants were used for β‐GUS staining assays. The reaction solution was 100 mm sodium phosphate, 10 mm EDTA, 0.1% Triton X‐100 and 1 mm 5‐bromo‐4‐chloro‐3‐indolyl‐β‐glucuronic acid (Sigma‐Aldrich, St. Louis, Missouri), pH 7.0. Tissues were stained overnight at 37 °C and then cleared in 75% ethanol. The GUS‐stained signals were finally observed and photographed using a fluorescence microscope (BX51, Olympus Corporation, Tokyo, Japan).
ChIP assays
The anti‐H3K27me3 ChIP‐seq assay was performed using the panda mutant and its wild type cultivar ‘Taibei 309’. The PANDA ChIP‐seq assay was performed using plants expressing pUBI::PANDA‐Flag in the japonica rice cultivar ‘Taibei 309’. Samples of leaf tissue (~4 g) or ~1.0‐cm young panicles were cross‐linked in 1% formaldehyde under vacuum, and the cross‐linking was stopped by adding glycine to a final concentration of 0.125 m. The samples were ground to fine powders in liquid nitrogen for nuclei preparation. After chromatin preparation according to the method described previously (Zhao et al., 2020), anti‐H3K27me3 or anti‐Flag polyclonal antibody (#A16199, ABclonal, Wuhan, China; #14793, Cell Signaling Technology, Danvers, Massachusetts) was used to immunoprecipitate the corresponding protein–DNA complexes, and the precipitated DNA was recovered for library preparation followed by sequencing on the Illumina platform. ChIP‐seq data analysis was performed as previously described (Lu et al., 2013). The H3K27me3 or PANDA‐Flag ChIP‐qPCR assays were performed using plants expressing pUBI::PANDA‐Flag and the wild‐type japonica rice cultivar ‘Taibei 309’. Chromatin precipitated with normal mouse IgG was used as the negative control. The precipitated DNA was recovered and analysed by ChIP–qPCR with the primers listed in Table S1.
RNA sequencing and data analyses
Whole‐transcriptome comparison analyses between the panda mutant and the wild type were performed using the leaves of 3‐week‐old plants or about 1‐cm young panicles. Total RNA was extracted with TRIzol reagent (Sangon Biotech, Shanghai, China). The cDNA libraries were constructed following Illumina standard protocols and sequenced on an Illumina HiSeq instrument by Novogene Biotech Co. Ltd. in Beijing city of China. RNA‐seq reads were aligned to the rice reference genome (http://rice.uga.edu) using TopHat after filtering out low‐quality reads (lowest base score <20) using SeqPrep and Sickle (Trapnell et al., 2009). Gene expression levels were calculated and normalized to FPKM (fragments per kilobase of transcript per million mapped reads) with HTSeq (Anders et al., 2015). Differential gene expression levels were examined using the R package DEGSeq (Wang et al., 2010). The cut‐off for significant differential expression was set as log2 (fold‐change) ≥1 and FDR < 0.05.
qRT‐PCR
Total RNA was extracted from plant tissues using TRIzol reagent (Sangon, Shanghai, China), and the mRNA was reverse‐transcribed into cDNA using the M‐MLV reverse transcriptase kit (Vazyme, Nanjing, China) following the manufacturers’ instructions. Quantitative RT‐PCR (qRT‐PCR) was performed using SYBR I Premix ExTaq (Vazyme, Nanjing, China). The gene expression levels in at least three biological replicates were calculated using the ∆∆C t method. Student’s t‐test was used to determine significant differences between samples.
Pull‐down assay
The PANDA CDS and the CDS of its interacting protein gene were cloned into the vectors pGSTA or pHISK for fusion with the GST or His tag, respectively. The GST or His fusion proteins were expressed in Escherichia coli BL21 and purified using Glutathione Sepharose 4 FF (#175132‐01 GE Healthcare, Chicago, Illinois) and NI‐NTA SefinoseTM Resin (C600033‐0010 BBI South Wales, UK) kits. Pull‐down assays were performed with the Pierce™ GST Protein Interaction Pull‐Down Kit (#21516 Thermo Scientifc, Waltham, Massachusetts).
BiFC assay
For the BiFC assay, the CDS of PANDA or its interacting protein genes was amplified and cloned into the binary vectors pC1300S‐nYFP and pC2300‐cYFP for fusion with nYFP and cYFP, respectively. These vectors were then co‐transformed into rice protoplasts or into N. benthamiana leaves using the PEG or agrobacterium‐mediated methods for transient expression. The transformed cells were finally observed with a laser confocal microscope (LSM880, Zeiss Corporation, Oberkochen, Germany).
CoIP assay
The CDS of PANDA or its interacting proteins was cloned into the pC1300S‐Flag and pC2300S‐Myc vectors to produce Flag and Myc fusion protein. The vectors were co‐transformed into rice protoplasts for transient expression of these fusion proteins. For the CoIP assay, total protein was extracted from the rice protoplasts. Following the manufacturer’s instructions, 30 μL of the agarose‐conjugated anti‐Flag monoclonal antibody (Sangon, Shanghai, China) was added to 500 μL total extracted proteins and incubated at 4 °C for 3 h with gentle rotation. The beads were washed three times with 350 μL of extraction buffer, and the proteins were eluted with 30 μL SDS‐PAGE sample buffer. Immunoblotting was then performed as previously described (Lu et al., 2013). The antibodies we used are as follows: anti‐flag rabbit antibody (#D110005‐0100, Sangon, Shanghai, China), anti‐Myc rabbit antibody (#D110006‐0100, Sangon, Shanghai, China) and HRP‐conjugated goat anti‐rabbit IgG (#D110053‐0025, Sangon, Shanghai, China).
Conflict of interest
The authors declare no competing interests.
Author contributions
D.M., D.Z., W.Z. and C.C. designed the research; D.M., S.T., X.L., M.T., C.L., D.W., L.B., Z.H., X.W., L.Y., Y.Z. and D.Z. performed experiments; D.M., D.G. and S.T. analysed the data; and D.M., D.G., W.Z. and C.C. wrote the manuscript.
Supporting information
Figure S1 Phenotypic characteristics of the rice panda mutant.
Figure S2 Statistical analyses of the grain size traits and the related gene expression levels between wild type (WT) and the mutant panda.
Figure S3 Statistical analyses of leaf length, leaf width, and leaf angle between wild type (WT) and panda.
Figure S4 Statistical analysis of plant height, panicle number, grain number, seed setting rate, grain weight, and grain yield among the wild type japonica cultivar ‘TB309’, the mutant panda, and their hybrid F1.
Figure S5 Statistical analyses of panicle number, grain number, seed setting rate, grain weight, and grain yield between the wild type cultivar ‘TB309’, the mutant panda, and the transgenic pPANDA::PANDA/panda complementation lines in the panda genetic background.
Figure S6 Expression and phonotype analyses of the constitutive PANDA‐overexpression lines.
Figure S7 Expression and phonotype analyses of the moderate PANDA‐overexpression lines.
Figure S8 Comparison of changes in expression and yield‐related traits between two kinds of the PANDA overexpression lines.
Figure S9 Expression pattern of PANDA in rice tissues at various developmental stages.
Figure S10 GUS staining of pPANDA::PANDA‐GUS transgenic plants.
Figure S11 Subcellular localization of PANDA.
Figure S12 Homology analyses between PANDA and the PIF/Harbinger transposons.
Figure S13 Phylogenetic tree of PANDA homologous proteins in rice.
Figure S14 Genetic complementation in the Arabidopsis alp1 mutant by using the rice PANDA gene.
Figure S15 Seed sizes in wild‐type Arabidopsis, the alp1 mutant, and the 35S::PANDA transgenic line in the mutant alp1 background.
Figure S16 Protein structures of PANDA and ALP1.
Figure S17 The N‐terminal 44 amino acids are necessary for PANDA to completely rescue the mutant phenotype in rice.
Figure S18 Comparisons of the phenotypes of the whole plant (A), flowers (B), and leaves (C) in WT, alp1, and two transgenic lines overexpressing the truncated 397 aa‐length PANDA; PANDA397 /alp1 #1 and PANDA397 /alp1 #2. Scale bars in A = 3.5 cm, B = 4 cm, and C = 0.5 cm.
Figure S19 Function diversity between wild type and the mutant of PANDA.
Figure S20 anti‐H3K27me3 ChIP‐seq analysis between wild type and panda.
Figure S21 RNA‐seq, anti‐H3K27me3 ChIP‐seq, and anti‐Flag ChIP‐seq analyse of target genes of PANDA, such as OsMADS22, OsMADS55 and OsEMF1.
Figure S22 qRT‐PCR analyses of PANDA, OsMADS55, OsEMF1, OsMADS22 and OsERF112 in wild type (WT) and the panda mutant.
Figure S23 PANDA inhibit the expression of GFP fused with the first exon of OsMADS55 containing GCC motifs in N. benthamiana.
Figure S24 Electrophoretic Mobility Shift Assay (EMSA) of PANDA binding to the GCC motif.
Figure S25 Analyses of plants carrying the double mutation in PANDA and its target gene OsMADS55.
Figure S26 Genetic analyses of the relationship between PANDA and OsREF112.
Table S1 Primers used in this study.
Table S2 Phenotypic analysis of the rice panda mutant and the wild type in the different environments.
Table S3 List of all genes with changes in the level of RNA or H3K27me3 enrichment.
Table S4 List of the groups with different H3K27me3 enrichment levels in the genes regulated by PANDA.
Table S5 List of all genes with deposition of PANDA‐Flag in the first ChIP‐seq.
Table S6 List of all genes with deposition of PANDA‐Flag in the second ChIP‐seq.
Table S7 List of the 241 PANDA‐FLAG binding sites co‐localized with H3K27me3 enrichment sites.
Acknowledgements
We thank the Arabidopsis Biological Resource Center (ABRC) at the Ohio State University for kindly providing Arabidopsis seeds. This work was supported by the National Natural Science Foundation of China (31371596, 32071995, 31071376), Youth Innovation Promotion Association of Chinese Academy of Sciences (2018398), Natural Science Foundation of Hunan Province for Distinguished Young Scholars (2021JJ10041) and Creative Research Groups (2019JJ10003), Key R&D Programs of Hunan Province (2020WK2023) and Hainan Yazhou Bay Seed Lab (B21HJ0108). Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Mao, D. , Tao, S. , Li, X. , Gao, D. , Tang, M. , Liu, C. , Wu, D. , Bai, L. , He, Z. , Wang, X. , Yang, L. , Zhu, Y. , Zhang, D. , Zhang, W. and Chen, C. (2022) The Harbinger transposon‐derived gene PANDA epigenetically coordinates panicle number and grain size in rice. Plant Biotechnol. J., 10.1111/pbi.13799
Contributor Information
Dechun Zhang, Email: zhangdechun@ctgu.edu.cn.
Wenli Zhang, Email: wzhang25@njau.edu.cn.
Caiyan Chen, Email: cychen@isa.ac.cn.
References
- Anders, S. , Pyl, P.T. and Huber, W. (2015) HTSeq‐‐a Python framework to work with high‐throughput sequencing data. Bioinform. 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, X. , Huang, Y. , Hu, Y. , Liu, H. , Zhang, B. , Smaczniak, C. , Hu, G. et al. (2017) Duplication of an upstream silencer of FZP increases grain yield in rice. Nat. Plants, 3, 885–893. [DOI] [PubMed] [Google Scholar]
- Bao, W. , Kojima, K.K. and Kohany, O. (2015) Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA, 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieluszewski, T. , Xiao, J. , Yang, Y. and Wagner, D. (2021) PRC2 activity, recruitment, and silencing: a comparative perspective. Trends Plant Sci. 19, S1360–1385. [DOI] [PubMed] [Google Scholar]
- Blackledge, N.P. , Farcas, A.M. , Kondo, T. , King, H.W. , McGouran, J.F. , Hanssen, L.L.P. , Ito, S. et al. (2014) Variant PRC1 complex‐dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell, 157, 1445–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calonje, M. , Sanchez, R. , Chen, L. and Sung, Z. (2008) EMBRYONIC FLOWER1 participates in Polycomb group‐mediated AG gene silencing in Arabidopsis . Plant Cell, 20, 277–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola, C. , Lawing, A.M. , Betrán, E. and Feschotte, C. (2007) PIF‐like transposons are common in drosophila and have been repeatedly domesticated to generate new host genes. Mol. Biol. Evol. 24, 1872–1888. [DOI] [PubMed] [Google Scholar]
- Derkacheva, M. , Steinbach, Y. , Wildhaber, T. , Mozgová, I. , Mahrez, W. , Nanni, P. , Bischof, S. et al. (2013) Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J. 32, 2073–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhatt, B.K. , Paul, P. , Sandhu, J. , Hussain, W. , Irvin, L. , Zhu, F. , Adviento‐Borbe, M.A. et al. (2021) Allelic variation in rice Fertilization Independent Endosperm 1 contributes to grain width under high night temperature stress. New Phytol. 229, 335–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, E. , Wang, Y. , Li, X. , Lin, Q. , Zhang, T. , Wang, Y. , Zhou, C. et al. (2019) OsSHI1 regulates plant architecture through modulating the transcriptional activity of IPA1 in rice. Plant Cell, 31, 1026–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Z. , Ji, Y. , Hu, J. , Guo, R. , Sun, S. and Wang, X. (2020) Strigolactones and Brassinosteroids antagonistically regulate the stability of the D53‐OsBZR1 complex to determine FC1 expression in rice tillering. Mol. Plant, 13, 586–597. [DOI] [PubMed] [Google Scholar]
- Fu, Q. , Zhang, P. , Tan, L. , Zhu, Z. , Ma, D. , Fu, Y. , Zhan, X. et al. (2010) Analysis of QTLs for yield‐related traits in Yuanjiang common wild rice (Oryza rufipogon Griff.). J. Genet. Genomics, 37, 147–157. [DOI] [PubMed] [Google Scholar]
- Grzebelus, D. , Lasota, S. , Gambin, T. , Kucherov, G. and Gambin, A. (2007) Diversity and structure of PIF/Harbinger‐like elements in the genome of Medicago truncatula . BMC Genom., 8, 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, T. , Chen, K. , Dong, N.Q. , Shi, C.L. , Ye, W.W. , Gao, J.P. , Shan, J.X. et al. (2018) GRAIN SIZE AND NUMBER 1 negatively regulates the OsMKKK10‐OsMKK4‐OsMPK6 cascade to coordinate the trade‐off between grain number per panicle and grain size in rice. Plant Cell, 30, 871–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, T. , Chen, K. , Dong, N.Q. , Ye, W.W. , Shan, J.X. and Lin, H.X. (2020) Tillering and small grain 1 dominates the tryptophan aminotransferase family required for local auxin biosynthesis in rice. J. Integr. Plant Biol. 62, 581–600. [DOI] [PubMed] [Google Scholar]
- Jiang, N. , Bao, Z. , Zhang, X. , Hirochika, H. , Eddy, S.R. , McCouch, S.R. and Wessler, S.R. (2003) An active DNA transposon family in rice. Nature, 421, 163–167. [DOI] [PubMed] [Google Scholar]
- Kapitonov, V.V. and Jurka, J. (2004) Harbinger transposons and an ancient HARBI1 gene derived from a transposase. DNA Cell Biol. 23, 311–324. [DOI] [PubMed] [Google Scholar]
- Kassis, J.A. and Brown, J.L. (2013) Polycomb group response elements in Drosophila and vertebrates. Adv. Genet. 81, 83–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. , Choi, S.C. and An, G. (2008) Rice SVP‐group MADS‐box proteins, OsMADS22 and OsMADS55, are negative regulators of brassinosteroid responses. Plant J. 54, 93–105. [DOI] [PubMed] [Google Scholar]
- Li, Z. , Fu, X. , Wang, Y. , Liu, R. and He, Y. (2018) Polycomb‐mediated gene silencing by the BAH‐EMF1 complex in plants. Nat. Genet. 50, 1254–1261. [DOI] [PubMed] [Google Scholar]
- Liang, S.C. , Hartwig, B. , Perera, P. , Mora‐García, S. , de Leau, E. , Thornton, H. , de Lima, A.F. et al. (2015) Kicking against the PRCs – a domesticated transposase antagonises silencing mediated by Polycomb group proteins and is an accessory component of Polycomb Repressive Complex 2. PLoS Genet. 11, e1005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Yang, C.Y. , Miao, R. , Zhou, C.L. , Cao, P.H. , Lan, J. , Zhu, X.J. et al. (2018) DS1/OsEMF1 interacts with OsARF11 to control rice architecture by regulation of brassinosteroid signaling. Rice (N Y), 11, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Zhou, C. , Zhao, Y. , Zhou, S. , Wang, W. and Zhou, D.X. (2014) The rice enhancer of zeste [E(z)] genes SDG711 and SDG718 are respectively involved in long day and short day signaling to mediate the accurate photoperiod control of flowering time. Front. Plant Sci. 5, 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Z. , Yu, H. , Xiong, G. , Wang, J. , Jiao, Y. , Liu, G. , Jing, Y. et al. (2013) Genome‐wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell, 25, 3743–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M. , Platten, D. , Chaudhury, A. , Peacock, W.J. and Dennis, E.S. (2009) Expression, imprinting, and evolution of rice homologs of the polycomb group genes. Mol. Plant, 2, 711–723. [DOI] [PubMed] [Google Scholar]
- McDowell, J.M. and Meyers, B.C. (2013) A transposable element is domesticated for service in the plant immune system. Proc. Natl Acad. Sci. USA, 110, 14821–14822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qüesta, J.I. , Song, J. , Geraldo, N. , An, H. and Dean, C. (2016) Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science, 353, 485–488. [DOI] [PubMed] [Google Scholar]
- Sadras, V.O. (2007) Evolutionary aspects of the trade‐off between seed size and number in crops. Field Crops Res. 100, 125–138. [Google Scholar]
- Smit, A.F. and Riggs, A.D. (1996) Tiggers and DNA transposon fossils in the human genome. Proc. Natl Acad. Sci. USA, 93, 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Z. , Hu, H. , Luo, X. , Jia, B. , Du, J. and He, Y. (2019) Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis. Nature Plants, 5, 424–435. [DOI] [PubMed] [Google Scholar]
- Trapnell, C. , Pachter, L. and Salzberg, S.L. (2009) TopHat: discovering splice junctions with RNA‐Seq. Bioinform. 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velanis, C.N. , Perera, P. , Thomson, B. , de Leau, E. , Liang, S.C. , Hartwig, B. , Förderer, A. et al. (2020) The domesticated transposase ALP2 mediates formation of a novel Polycomb protein complex by direct interaction with MSI1, a core subunit of Polycomb Repressive Complex 2 (PRC2). PLoS Genet. 16, e1008681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff, J.N. (2006) Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. BioEssays, 28, 913–922. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Feng, Z. , Wang, X. , Wang, X. and Zhang, X. (2010) DEGseq: an R package for identifying differentially expressed genes from RNA‐seq data. Bioinform. 26, 136–138. [DOI] [PubMed] [Google Scholar]
- Wang, B. , Smith, S.M. and Li, J. (2018) Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 69, 437–468. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Tong, X. , Tang, L. , Wang, Y. , Zhao, J. , Li, Z. , Liu, X. et al. (2021) RLB (RICE LATERAL BRANCH) recruits PRC2‐mediated H3K27 tri‐methylation on OsCKX4 to regulate lateral branching. Plant Physiol. 188, 460–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Hu, J. , Qian, Q. and Xue, H.W. (2013) LC2 and OsVIL2 promote rice flowering by photoperoid‐induced epigenetic silencing of OsLF. Mol. Plant, 6, 514–527. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Sun, S. , Jin, J. , Fu, D. , Yang, X. , Weng, X. , Xu, C. et al. (2015) Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl Acad. Sci. USA, 112, 15504–15509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K. , Wang, S. , Song, W. , Zhang, J. , Wang, Y. , Liu, Q. , Yu, J. et al. (2020) Enhanced sustainable green revolution yield via nitrogen‐responsive chromatin modulation in rice. Science, 367, eaaz2046. [DOI] [PubMed] [Google Scholar]
- Xiao, J. , Jin, R. , Yu, X. , Shen, M. , Wagner, J.D. , Pai, A. , Song, C. et al. (2017) Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat. Genet. 49, 1546–1552. [DOI] [PubMed] [Google Scholar]
- Yan, D. , Zhang, X. , Zhang, L. , Ye, S. , Zeng, L. , Liu, J. , Li, Q. et al. (2015) Curved chimeric palea 1 encoding an EMF1‐like protein maintains epigenetic repression of OsMADS58 in rice palea development. Plant J. 82, 12–24. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Lee, S. , Hang, R. , Kim, S.R. , Lee, Y.S. , Cao, X. , Amasino, R. et al. (2013) OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. Plant J. 73, 566–578. [DOI] [PubMed] [Google Scholar]
- Yoon, J. , Cho, L.H. , Lee, S. , Pasriga, R. , Tun, W. , Yang, J. , Yoon, H. et al. (2019) Chromatin interacting factor OsVIL2 is required for outgrowth of axillary buds in rice. Mol. Cells, 42, 858–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, L. , Song, X. , Zhang, L. , Yu, Y. , Liang, Z. , Lei, Y. , Ruan, J. et al. (2021) The transcriptional repressors VAL1 and VAL2 recruit PRC2 for genome‐wide Polycomb silencing in Arabidopsis. Nucleic Acids Res. 49, 98–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, W. , Luo, X. , Li, Z. , Yang, W. , Wang, Y. , Liu, R. , Du, J. et al. (2016) A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis . Nat. Genet. 48, 1527–1534. [DOI] [PubMed] [Google Scholar]
- Zaveri, A. , Bose, A. , Sharma, S. , Rajendran, A. , Biswas, P. , Shenoy, A.R. and Visweswariah, S.S. (2021) Mycobacterial STAND adenylyl cyclases: the HTH domain binds DNA to form biocrystallized nucleoids. Biophys. J. 120, 1231–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, L. , Zhang, Q. , Sun, R. , Kong, H. , Zhang, N. and Ma, H. (2014) Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat. Commun. 5, 4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Wang, R. , Xing, Y. , Xu, Y. , Xiong, D. , Wang, Y. and Yao, S. (2021) Separable regulation of POW1 in grain size and leaf angle development in rice. Plant Biotechnol. J. 19, 2517–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, L. , Xie, L. , Zhang, Q. , Ouyang, W. , Deng, L. , Guan, P. , Ma, M. et al. (2020) Integrative analysis of reference epigenomes in 20 rice varieties. Nat. Commun. 11, 2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, M. , Wang, Y. , Wang, Y. , Wang, C. , Ren, Y. , Lv, J. , Peng, C. et al. (2015) DEFORMED FLORAL ORGAN1 (DFO1) regulates floral organ identity by epigenetically repressing the expression of OsMADS58 in rice (Oryza sativa). New Phytol. 206, 1476–1490. [DOI] [PubMed] [Google Scholar]
- Zhou, Y. , Tergemina, E. , Cui, H. , Förderer, A. , Hartwig, B. , Velikkakam James, G. , Schneeberger, K. et al. (2017) Ctf4‐related protein recruits LHP1‐PRC2 to maintain H3K27me3 levels in dividing cells in Arabidopsis thaliana . Proc. Natl Acad. Sci. USA, 114, 4833–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , Wang, Y. , Krause, K. , Yang, T. , Dongus, J.A. , Zhang, Y. and Turck, F. (2018) Telobox motifs recruit CLF/SWN‐PRC2 for H3K27me3 deposition via TRB factors in Arabidopsis. Nat Genet, 50, 638–644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Phenotypic characteristics of the rice panda mutant.
Figure S2 Statistical analyses of the grain size traits and the related gene expression levels between wild type (WT) and the mutant panda.
Figure S3 Statistical analyses of leaf length, leaf width, and leaf angle between wild type (WT) and panda.
Figure S4 Statistical analysis of plant height, panicle number, grain number, seed setting rate, grain weight, and grain yield among the wild type japonica cultivar ‘TB309’, the mutant panda, and their hybrid F1.
Figure S5 Statistical analyses of panicle number, grain number, seed setting rate, grain weight, and grain yield between the wild type cultivar ‘TB309’, the mutant panda, and the transgenic pPANDA::PANDA/panda complementation lines in the panda genetic background.
Figure S6 Expression and phonotype analyses of the constitutive PANDA‐overexpression lines.
Figure S7 Expression and phonotype analyses of the moderate PANDA‐overexpression lines.
Figure S8 Comparison of changes in expression and yield‐related traits between two kinds of the PANDA overexpression lines.
Figure S9 Expression pattern of PANDA in rice tissues at various developmental stages.
Figure S10 GUS staining of pPANDA::PANDA‐GUS transgenic plants.
Figure S11 Subcellular localization of PANDA.
Figure S12 Homology analyses between PANDA and the PIF/Harbinger transposons.
Figure S13 Phylogenetic tree of PANDA homologous proteins in rice.
Figure S14 Genetic complementation in the Arabidopsis alp1 mutant by using the rice PANDA gene.
Figure S15 Seed sizes in wild‐type Arabidopsis, the alp1 mutant, and the 35S::PANDA transgenic line in the mutant alp1 background.
Figure S16 Protein structures of PANDA and ALP1.
Figure S17 The N‐terminal 44 amino acids are necessary for PANDA to completely rescue the mutant phenotype in rice.
Figure S18 Comparisons of the phenotypes of the whole plant (A), flowers (B), and leaves (C) in WT, alp1, and two transgenic lines overexpressing the truncated 397 aa‐length PANDA; PANDA397 /alp1 #1 and PANDA397 /alp1 #2. Scale bars in A = 3.5 cm, B = 4 cm, and C = 0.5 cm.
Figure S19 Function diversity between wild type and the mutant of PANDA.
Figure S20 anti‐H3K27me3 ChIP‐seq analysis between wild type and panda.
Figure S21 RNA‐seq, anti‐H3K27me3 ChIP‐seq, and anti‐Flag ChIP‐seq analyse of target genes of PANDA, such as OsMADS22, OsMADS55 and OsEMF1.
Figure S22 qRT‐PCR analyses of PANDA, OsMADS55, OsEMF1, OsMADS22 and OsERF112 in wild type (WT) and the panda mutant.
Figure S23 PANDA inhibit the expression of GFP fused with the first exon of OsMADS55 containing GCC motifs in N. benthamiana.
Figure S24 Electrophoretic Mobility Shift Assay (EMSA) of PANDA binding to the GCC motif.
Figure S25 Analyses of plants carrying the double mutation in PANDA and its target gene OsMADS55.
Figure S26 Genetic analyses of the relationship between PANDA and OsREF112.
Table S1 Primers used in this study.
Table S2 Phenotypic analysis of the rice panda mutant and the wild type in the different environments.
Table S3 List of all genes with changes in the level of RNA or H3K27me3 enrichment.
Table S4 List of the groups with different H3K27me3 enrichment levels in the genes regulated by PANDA.
Table S5 List of all genes with deposition of PANDA‐Flag in the first ChIP‐seq.
Table S6 List of all genes with deposition of PANDA‐Flag in the second ChIP‐seq.
Table S7 List of the 241 PANDA‐FLAG binding sites co‐localized with H3K27me3 enrichment sites.


