Abstract
Fertilization is the starting point for creating new progeny. At this time, the highly differentiated oocyte and sperm fuse to form one zygote, which is then converted into a pluripotent early embryo. Recent studies have shown that the lysosomal degradation system via autophagy and endocytosis plays important roles in the remodeling of intracellular components during oocyte-to-embryo transition. For example, in Caenorhabditis elegans, zygotes show high endocytic activity, and some populations of maternal membrane proteins are selectively internalized and delivered to lysosomes for degradation. Furthermore, fertilization triggers selective autophagy of sperm-derived paternal mitochondria, which establishes maternal inheritance of mitochondrial DNA. In addition, it has been shown that autophagy via liquid–liquid phase separation results in the selective degradation of some germ granule components, which are distributed to somatic cells of early embryos. This review outlines the physiological functions of the lysosomal degradation system and its molecular mechanisms in C. elegans and mouse embryos.
Keywords: endocytosis, autophagy, oocyte-to-embryo transition, fertilization
Introduction
The oocyte not only synthesizes maternal proteins and RNAs but also takes up nutrients for embryonic development. Oocyte growth is arrested at the first meiotic division in Caenorhabditis elegans and at the second meiotic division in mice. Upon receiving a signal from the sperm, egg maturation is initiated, followed by fertilization to form a zygote. The zygote then completes meiosis and starts mitotic cell division for embryogenesis. This is followed by large-scale degradation of maternal RNA and cytosolic proteins.1–3) In addition, translation of maternal mRNAs and subsequent zygotic gene expression drives protein synthesis to achieve intracellular remodeling during embryonic development.
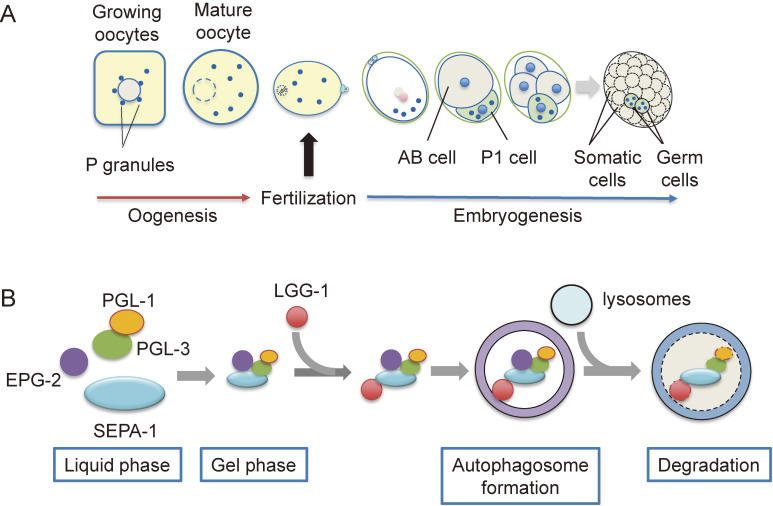
C. elegans has two sexes: hermaphrodites and males. It mainly lives as a self-fertilizing hermaphrodite, although approximately 0.1% of the progeny is male.4) C. elegans hermaphrodites have a pair of U-shaped gonad arms connecting to a spermatheca, which accommodates sperm (Fig. 1A). The first step in the oocyte-to-embryo transition is “meiotic maturation”. In C. elegans, the distal region of each gonad forms a syncytium that contains germ cell nuclei, and the cellularization of oocytes starts around the bend region of the gonad arm. Growing oocytes take up nutrients such as yolk proteins for embryogenesis by endocytosis as meiosis proceeds (see below). The oocytes continue to grow and gradually move toward the proximal region. In the C. elegans germ line, growing oocytes are arrested at the diakinesis stage of meiotic prophase I. This meiotic arrest is released by the major sperm proteins (MSPs) secreted from the sperm, leading to cortical rearrangement of the oocytes. A fully mature oocyte is ovulated into the spermatheca by gonadal sheath contraction and fertilized by a single sperm.5) Sperm entry triggers the second step in the oocyte-to-embryo transition, which is referred to as “egg activation”. Fertilized oocytes complete meiosis I and II, accompanied by polar body formation. During this process, C. elegans zygotes undergo cell polarization to form an anterior-posterior axis.6) The zygotes also form eggshells consisting of a chitin layer and a chondroitin proteoglycan layer.7) Chitin synthesis is driven by chitin synthase-1 (CHS-1) immediately after fertilization to prevent polyspermy.8) The chondroitin proteoglycan layer is formed by the exocytosis of cortical granules, including chondroitin proteoglycans and mucin glycoproteins.9–11)
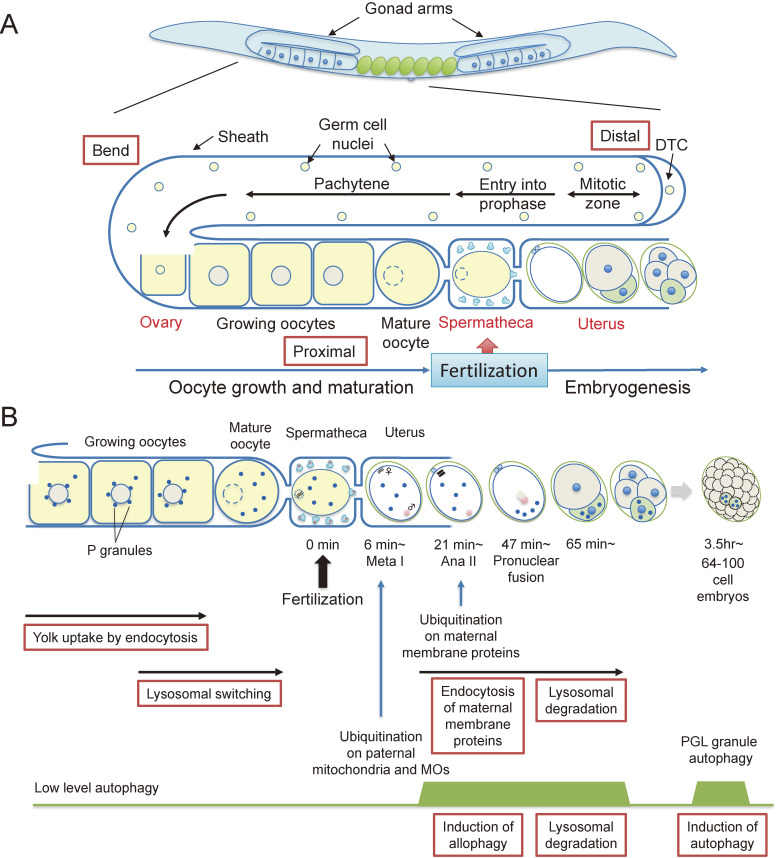
Figure 1.
(A) C. elegans germ line. The C. elegans hermaphrodite gonad contains a pair of U-shaped gonad arms containing a spermatheca, which accommodates sperms. A distal region of each gonad is composed of a syncytium that contains germ cell nuclei. Oocytes are formed by cellularization around the bend region of the gonad arm and growing oocytes move to the proximal region. In the most proximal region, the oocyte receives the signal from the sperm and undergoes meiotic maturation and cortical rearrangement. Mature oocytes are ovulated into the spermatheca which contains sperm and the oocytes are fertilized. The fertilized egg then moves to the uterus, completes meiosis I and II and starts embryogenesis. DTC, distal tip cell. (B) Multiple roles of endocytosis and autophagy during oocyte-to-embryo transition. Growing oocytes take up yolk components via receptor-mediated endocytosis in the proximal region. Oocytes receive a signal from the sperm and undergo lysosomal switching. Immediately after fertilization, sperm-derived paternal mitochondria and membranous organelles (MOs) are ubiquitinated at metaphase I and then eliminated by allophagy. Some populations of maternal membrane proteins are ubiquitinated at anaphase II and selectively endocytosed for degradation by the 2-cell stage. Induction of autophagy occurs at later embryonic stages again (64–100 cells). Autophagy of PGL granules also occurs in somatic cells of embryos.
In C. elegans, the first large-scale degradation of maternal RNAs occurs immediately after fertilization.12) Several maternal proteins such as meiotic spindle formation protein (MEI)-1 and MEI-2, which are homologs of the microtubule-severing complex,13) are selectively degraded by the ubiquitin-proteasome system (UPS) after fertilization and before mitosis.14,15) Degradation of these meiotic cytosolic proteins is regulated by the dual-specificity tyrosine phosphorylation-regulated kinase (DYRK) family protein minibrain kinase (MBK)-2 and an E3 ubiquitin ligase, such as cullin (CUL)-3.16) MEI-1 and MEI-2 are required for meiotic cell division, but these preclude the transition of short meiotic spindles to large mitotic spindles and inhibit appropriate mitosis if they remain in embryos.14,15) Thus, UPS degradation of a subset of meiotic proteins is essential for normal embryonic development. In addition, recent studies have revealed that the lysosomal degradation system involving endocytosis and autophagy also play important roles in the remodeling of intracellular components during early development. This review will address the role of membrane trafficking in germline cells and recent developments in the understanding of lysosomal degradation systems, which mediate the degradation of intracellular membrane components and organelles during the oocyte-to-embryo transition, especially in C. elegans and mice.
1. Receptor-mediated endocytosis of yolk proteins by growing oocytes
In C. elegans, growing oocytes take up yolk components by receptor-mediated endocytosis and use them as nutrients for early development after fertilization (Fig. 1B, 2A). The yolk proteins, which are lipoproteins consisting of core proteins such as vitellogenin (VIT)-2 with neutral lipids and cholesterol, are secreted from intestinal cells into the body cavity. The secreted yolk proteins are then taken up by growing oocytes and are stored in yolk granules, which are lysosome-related organelles.17) By using a fusion protein of VIT-2 with green fluorescent protein (VIT-2-GFP) as a monitor protein, Grant et al. isolated 11 complementation groups of mutants, which were defective in the uptake of VIT-2-GFP by oocytes and referred to them as receptor-mediated endocytosis defective mutants (rme).17) Notably, many of the identified rme genes encoded novel genes that are conserved between C. elegans and mammals (Fig. 2A). The rme-2 gene encodes a homolog of the human low-density lipoprotein receptor RME-2, which functions as a yolk receptor in oocytes.17) The rme-3 gene encodes a clathrin heavy chain, suggesting that yolk uptake by oocytes is mediated by clathrin-mediated endocytosis.18) The rme-6 gene encodes a novel protein with a Vps9 domain that acts as a guanine nucleotide exchange factor for the small GTPase Rab5, and a Ras GTPase-activating protein-like domain.19) RME-6 interacts with α-adaptin and localizes mainly to clathrin-coated pits, suggesting that it activates Rab5 on clathrin-coated pits and transports cargo proteins in primary endocytic vesicles to endosomes. The rme-1 gene encodes an eps15-homology domain-containing protein, which localizes to recycling endosomes and functions during the recycling of the yolk receptor, RME-2 to the plasma membrane (PM).20) Recently, it was reported that RME-1 functions with amphiphysin-1 in the formation of transport intermediates from acidic lipid-rich recycling endosomes.21) The rme-5 and rme-4 genes encode a small GTPase, RAB-35 and a protein with a differentially expressed in normal and neoplastic cells (DENN) domain, a clathrin-binding box, and an AP-2 binding motif, respectively.22) It has been revealed that Connecdenn/DENND1, which is a human RME-4 homolog, acts as a guanine nucleotide exchange factor for Rab35.23) Both RAB-35 and RME-4 function in parallel with RAB-11 during the endocytic recycling of RME-2. Because RME-4 interacts with α-adaptin and localizes mainly to clathrin-coated pits, RME-4 activates RAB-35 on clathrin-coated pits, allowing for rapid recycling of the yolk receptor from early endosomes to the PM.22) RME-2 is recycled between the PM and endosomes for yolk uptake in growing oocytes, but it is delivered to lysosomes for degradation after fertilization,24,25) suggesting that switching of the transport pathway of RME-2 from a recycling route to a degradative route takes place during oocyte-to-embryo transition (Fig. 2, see below).
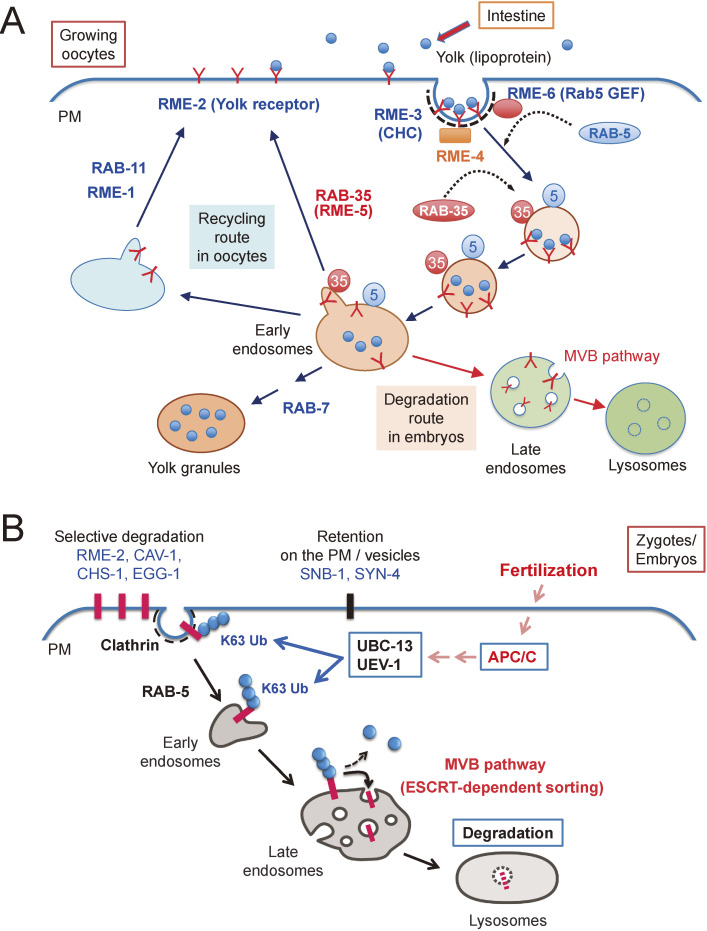
Figure 2.
(A) Receptor-mediated endocytosis of yolk components. Yolk components secreted from the intestine are endocytosed by the growing oocytes. RME-2 is a yolk receptor that is recycled between the plasma membrane and endosomes to deliver yolk components to yolk granules in growing oocytes. Many proteins, including RME proteins are involved in this process. After fertilization, RME-2 is delivered to lysosomes via the multivesicular body (MVB) pathway for degradation. CHC, clathrin heavy chain; PM, plasma membrane. (B) Selective degradation of maternal membrane proteins in embryos. Some of the maternal membrane proteins (RME-2, CAV-1, CHS-1, and EGG-1) are internalized by clathrin-dependent endocytosis and delivered to lysosomes via the MVB pathway. This process depends on meiotic cell cycle progression triggered by anaphase-promoting complex/cyclosome (APC/C). Ubiquitin-conjugating enzymes (E2) such as UBC-13 and its variants, UEV-1, are involved in the ubiquitination of maternal membrane proteins. By contrast, SNB-1 and SYN-4, which are general factors regulating membrane trafficking, remain after fertilization and continue to function in embryos. PM, plasma membrane.
2. Sperm signaling triggers a lysosomal switch to promote oocyte proteostasis
Recent studies have revealed that proteostasis in proximal oocytes is regulated by a lysosomal switching mechanism that converts the pH of lysosomal lumen from non-acidic to acidic and upregulates lysosomal functions in response to sperm signaling (Fig. 1B). In C. elegans, immature germ cells contain carbonylated proteins that are generated by oxidative stress.26) However, these proteins are eliminated from the cytosol of proximal oocytes. Because carbonylated proteins are not removed in the proximal oocytes of feminized mutants that lack a sperm, this process is thought to be signaled by MSPs secreted from sperm, which trigger the oocyte-to-embryo transition.26) To study the protein clearance mechanism in proximal oocytes, Kenyon and colleagues generated animals expressing GFP-tagged aggregation-prone proteins in the germ line.27) They found that these proteins formed immobile aggregates, which gradually accumulated in the proximal oocytes of young adult feminized animals, and that the elimination of these aggregates was triggered by the input of sperm signaling before fertilization, but not by fertilization itself. Using a protein-aggregate detection reagent, they observed that endogenous protein aggregates accumulated in the oocytes of feminized animals. They further sought sperm-responsive components that function in cytosolic aggregate elimination. They identified lysosomal V-ATPase as the downstream factor for aggregate clearance in proximal oocytes. Although the protein level of V-ATPase was relatively low in immature growing oocytes, the protein level of V-ATPase gradually increased and became localized to lysosomes as the oocyte moved to the proximal region and matured. This observation suggested that sperm-derived signals induce the expression of V-ATPase and trigger lysosomal acidification in proximal oocytes. The expression of V-ATPase subunits is repressed by germline development defective (GLD)-1 protein, which is a major translational repressor in the germ line,28) before oocyte maturation. When sperm-derived MSPs act on the proximal oocytes, proteasome-dependent degradation of GLD-1 is triggered to release translational repression of the mRNAs encoding V-ATPase subunits. The newly synthesized V-ATPase subunits are then targeted to lysosomes for acidification. Notably, the loss of macroautophagy-related genes did not affect aggregate clearance in the proximal oocytes. Instead, it was observed that cytosolic aggregates are surrounded by arm-like projections, which extend from lysosomes before their disappearance,27) suggesting the involvement of microautophagy in this process.29) Recently, a whole genome RNAi screen for genes regulating this process has identified many components involved in protein synthesis, cytoskeleton-associated processes, ER functions, lysosomal acidification, and vesicular transport.30) Further analysis will be required to understand the molecular mechanisms regulating the activation of proteostasis in the germ line during oocyte maturation.
3. Selective degradation of maternal membrane proteins after fertilization
In fertilized eggs, a subset of maternal PM proteins derived from oocytes is selectively internalized by endocytosis and degraded in lysosomes (Fig. 2).9,10,25,31) In the second meiotic phase, approximately 25 min after fertilization, several maternal membrane proteins such as RME-2, CAV-1, CHS-1, and EGG-1 are internalized and transported to endosomes (Fig. 2B).9,10,24,25,32) CAV-1, which is a caveolin homolog, localizes to cortical granules as well as the PM in oocytes and is transported to the PM immediately after fertilization.9) CHS-1 mainly localizes to the PM in the proximal oocytes and functions in the formation of the chitin layer to prevent polyspermy.33) EGG-1 was originally identified as a putative sperm receptor.34) These internalized maternal membrane proteins are then delivered to lysosomes for degradation and they disappear in the 2-cell stage (about 65–80 minutes after fertilization). In contrast, some PM proteins such as synaptobrevin-1 homolog (SNB-1) and syntaxin-4 homolog (SYN-4), which are generally required for membrane trafficking, are not degraded after fertilization and remain even in late-stage embryos, suggesting that this degradation is a substrate-selective process.
It is generally known that lysine 63 (K63)-linked ubiquitination is involved in selective endocytosis of membrane proteins.35) Indeed, a strong accumulation of K63-linked ubiquitin signals was observed on endosomes during the second meiotic anaphase (Fig. 1B and Fig. 2).25) In addition, the size of endosomes also increased significantly compared with that before fertilization, suggesting that endosomal activation takes place during fertilization. The accumulation of K63-linked ubiquitin signals on endosomes is transient and disappears at the 2-cell stage. These observations imply that considerable amounts of maternal membrane proteins are transiently ubiquitinated and transported to endosomes during this period.25) The ubiquitinated membrane proteins are then sorted by the ESCRT complex from the endosomal membrane into intraluminal vesicles via the multivesicular body (MVB) pathway36) and delivered to the lysosomal lumen (Fig. 2B).25)
We screened for factors that control the ubiquitination-mediated selective degradation of maternal membrane proteins in embryos.25) We found that ubiquitin-conjugating enzyme E2, UBC-13, and its variants, and UEV-1 are involved in this process. It has been reported that UBC-13 and UEV-1 form a complex that specifically affects the formation of K63-linked ubiquitin chains.37) In ubc-13 or uev-1-deficient mutants, the degradation of maternal membrane proteins such as CAV-1 and RME-2 is significantly inhibited, and the accumulation of K63-linked ubiquitin chains on endosomes that occurs after fertilization is strongly suppressed. In these mutants, maternal membrane proteins are transported to endosomes but recycled back to the PM, suggesting that the sorting of maternal membrane proteins on endosomes into the MVB pathway is impaired in the absence of K63-linked ubiquitination. Because the PM is the place for nutrient uptake, cell-cell communication, and various signal transduction pathways, selective degradation of maternal membrane proteins via endocytosis is a mechanism for converting the protein composition of the PM from the oocyte-type to the embryo-type.
As described above, maternal membrane proteins play a role in growing oocytes, but they are readily transported to lysosomes for degradation after fertilization. Therefore, there may be a signaling mechanism that triggers the degradation of maternal membrane proteins after fertilization. One of the upstream factors driving this mechanism is the anaphase-promoting complex/cyclosome (APC/C).10,25) APC/C is an E3 ubiquitin ligase required for the release of mature oocytes from their arrest at meiotic prophase I, and its loss drastically inhibits the endocytosis of GFP-CHS-1 from the PM after fertilization.25) These observations suggested that the endocytosis of maternal PM proteins after fertilization depends on APC/C-mediated meiotic cell cycle progression (Fig. 2B). Further elucidation of the signaling pathway that links developmental events and intracellular transport is required.
4. Selective degradation of maternal membrane proteins in mouse embryos
We also studied the fate of maternal PM proteins during mouse embryogenesis (Fig. 3).38) During mouse oogenesis, the growth of primary oocytes, which differentiate from primordial germ cells, is arrested at the prophase of meiosis I.39) Subsequently, oocytes resume meiosis I at the germinal vesicle (GV) stage and then undergo maturation. The matured oocytes arrest their growth at the metaphase of meiosis II to become metaphase II oocytes during ovulation. After fertilization, the fertilized oocytes complete meiosis II, resulting in the initiation of embryonic development.
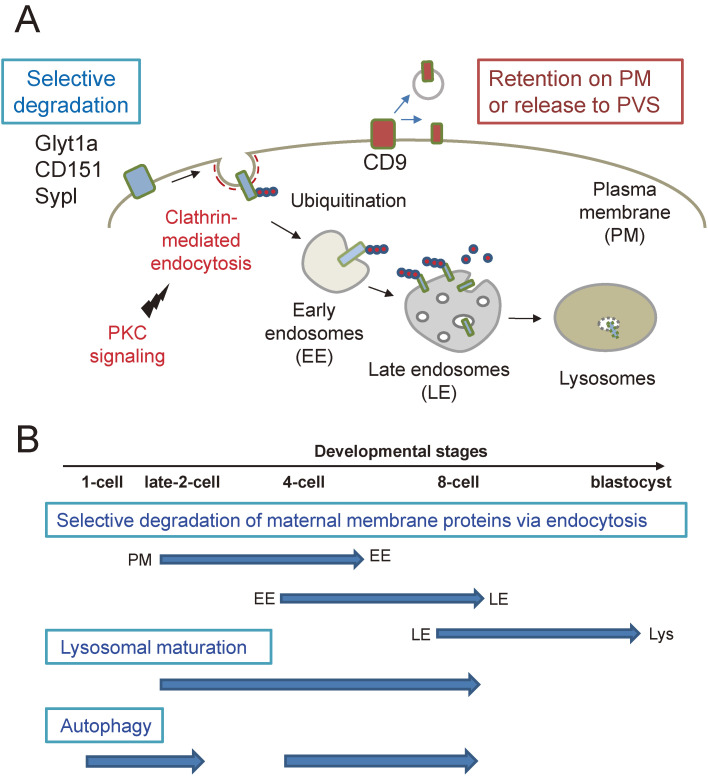
Figure 3.
Selective degradation of maternal membrane proteins in mouse embryos. (A) Some of the maternal membrane proteins (Glyt1a CD151, and Sypl) are selectively internalized by clathrin-dependent endocytosis by the late 2-cell stage (40 hours post fertilization) and slowly delivered to lysosomes for degradation by the 8-cell stage (54 hours post fertilization). In contrast, CD9 is partly retained on the plasma membrane or released into the perivitelline space (PVS). (B) Autophagy is induced immediately after fertilization and later stages. Lysosomal maturation appears to be initiated around the 2-cell stage.
Glyt1, encoded by the Slc6a9 gene, is a membrane protein with 12-transmembrane domains, which localizes to the PM and functions as a glycine transporter in oocytes and embryos.40,41) This protein imports glycine into oocytes and early embryos in response to osmotic changes in the extracellular environment.42,43) It has been reported that mouse early embryos import glycine as an osmolyte to protect themselves from osmotic stress.44,45) In fact, it has been shown that the intracellular concentration of free glycine increases in metaphase II stage oocytes, its level is maintained during the 4–8-cell embryonic stage and then decreases in later stages.42) Notably, endogenous Glyt1 proteins have been reported to mainly localize to the PM in oocytes and also in early embryos but disappear after the late 8-cell stage of embryogenesis.42) To study the fate of Glyt1a during early embryogenesis, we observed the behavior of fluorescent protein-tagged Glyt1a in mouse embryos using a non-invasive live imaging system (Fig. 3).38) Whereas maternal membrane proteins are endocytosed immediately after fertilization in C. elegans, fluorescent protein-tagged Glyt1a was internalized from the PM to early endosomes at the late 2-cell stage and then transported to late endosomes and lysosomes for degradation during the 4–8-cell stage. An accumulation of a large amount of ubiquitinated proteins in endosomes was observed during this process. We also found that other maternal PM proteins such as CD151, which belongs to the tetraspanin family, and synaptophysin-like protein (Sypl), are also endocytosed by the late 2-cell stage for degradation as well as Glyt1a. Notably, CD9, which is a tetraspanin family protein that localizes to microvilli on oocytes, remains on the PM and is partly released into the perivitelline space between the PM and Zona pellucida in the 8-cell stage embryos,38,46) suggesting that maternal membrane protein degradation is a selective process. Treatment of 2-cell stage embryos with a clathrin inhibitor, Pitstop2, completely blocked the internalization of a fluorescent protein-tagged Glyt1a from the PM, suggesting that this process is not only mediated by ubiquitination but also by clathrin-mediated endocytosis. The endocytosis of Glyt1a appears to be mediated by a protein kinase C (PKC)-dependent mechanism because the activation and inhibition of PKC triggers the internalization of Glyt1a even in 1-cell stage embryos and strongly blocks Glyt1a endocytosis at the late 2-cell stage, respectively. Notably, Pitstop2 treatment also inhibited the cell division of 2-cell stage embryos and caused cell division defects in later stage embryos, suggesting the importance of clathrin-mediated endocytosis during early embryogenesis. These observations suggest that selective degradation of maternal membrane proteins is a conserved mechanism between C. elegans and mice, although the timing of endocytosis is different.
5. Paternal organelle degradation by autophagy and maternal inheritance of mitochondrial DNA
After fertilization, sperm-derived paternal organelles and cytosolic components enter the cytoplasm of the zygote. Among them, sperm-derived centrioles, which are platforms for centrosome assembly, persist during embryogenesis in C. elegans and some other species.47) It has been reported that approximately 10% of total RNA in early embryos is of paternal origin, which is derived from the sperm.48) On the other hand, paternal mitochondria and their mitochondrial DNA (mtDNA) are known to disappear after fertilization, resulting in maternal inheritance of mtDNA in many species,49,50) although the mechanisms involved remain largely unknown. Thus, we and other groups attempted to elucidate the mechanism of maternal inheritance of mtDNA in C. elegans (Fig. 4).51,52) We stained sperm-derived mitochondria (paternal mitochondria) by soaking males in a mitotracker-containing solution and observed their dynamics in fertilized eggs. We found that paternal mitochondria entered fertilized eggs but gradually disappeared from the 2–8-cell stage during embryogenesis. Next, we examined whether autophagy, which delivers cytoplasmic proteins and organelles to lysosomes for degradation via autophagosome formation, is involved in the degradation of paternal mitochondria in embryos. We analyzed the localization of autophagy-related factors such as LGG-1, which is a C. elegans homolog of the autophagosome marker Atg8/LC3, during the oocyte-to-embryo transition. Because LGG-1 localizes to small spherical structures scattered in the cytoplasm of growing oocytes, a low level of autophagy may take place in the process of oocyte maturation.51) In contrast, LGG-1 strongly accumulates on autophagosomes, which surround sperm-derived paternal mitochondria and post-Golgi organelles called membranous organelles (MOs) in fertilized eggs, leading to a local induction of autophagy (Fig. 4). Paternal mitochondria and MOs are then delivered to lysosomes for degradation. As paternal components derived from sperm are selectively eliminated from embryos by autophagy, we refer to this autophagy as allophagy (allogeneic [non-self] organelle autophagy).53) Loss of autophagy-related factors causes a defect in the degradation of paternal mitochondria, leading to the transmission of paternal mtDNA to the offspring. These findings clearly indicated that selective degradation of paternal mitochondria by autophagy ensures maternal inheritance of mtDNA in C. elegans. In addition to allophagy, autophagy is induced again in the 64–100-cell stage, and many LGG-1-positive punctate structures appear throughout the embryo (Fig. 1B). Although the substrates have not yet been identified, this suggests that there are multiple time points when autophagy activity increases during embryogenesis. In addition, most lgg-1 deficient animals suffer lethality in the late embryonic development stage or the L1 larval stage immediately after hatching, suggesting that autophagic activity is necessary for normal embryonic development.
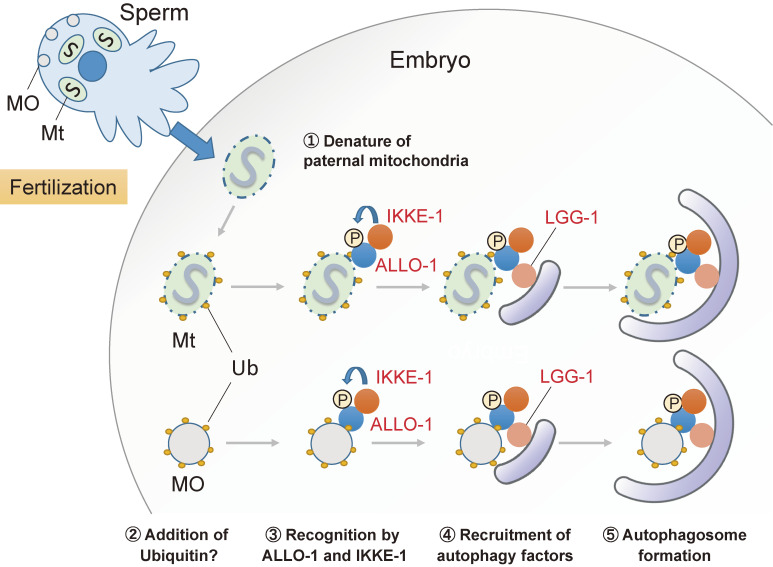
Figure 4.
Allophagy of paternal mitochondria and MOs in C. elegans embryos. In C. elegans embryos, sperm-derived paternal mitochondria and membranous organelles (MOs) are degraded by selective autophagy called allophagy. Paternal mitochondria seem to be denatured after fertilization and are ubiquitinated at a low level. An autophagy adaptor, ALLO-1 and its binding partner IKKE-1, localize to the paternal mitochondria and recruit the autophagy factor LGG-1 for the formation of autophagy. IKKE-1 is a kinase that phosphorylates ALLO-1. MOs are heavily ubiquitinated and eliminated by allophagy in the same manner.
We also studied the mechanisms underlying the recognition of paternal organelles during autophagy (Fig. 4). In fertilized zygotes, strong K48-linked and K63-linked ubiquitination occurs on MOs prior to autophagy, whereas a weak GFP-ubiquitin signal was observed on paternal mitochondria, suggesting a ubiquitin-mediated regulation of autophagy.51,54) However, it is unclear whether a ubiquitin monomer or a part of K48 or K63-linked ubiquitin chains is responsible for the weak GFP-ubiquitin signal on paternal mitochondria. In the ubiquitin-dependent selective autophagy pathway in mammals, ubiquitination of the substrates destined for degradation serves as a signal, which binds to an autophagy adaptor containing a ubiquitin-interacting motif. The autophagy adaptors then bind to autophagy-related factors such as LC3 via the LC3-interacting region (LIR), thereby promoting a local formation of autophagosomes around the substrates.55,56) Recently, we identified a novel factor, ALLO-1, which contains an LIR motif and functions as an autophagy adaptor in allophagy.54) ALLO-1 localizes to paternal organelles immediately after fertilization, and its localization is impaired by the loss of UBA-1, which is the sole ubiquitin-activating protein in C. elegans. In addition, ALLO-1 directly binds to LGG-1 via the LIR motif, and mutations in its LIR domain leads to defects in autophagosome formation around paternal organelles. From these observations, we concluded that ALLO-1 is an autophagy adaptor that promotes autophagosome formation around paternal organelles after fertilization. Although ALLO-1 is a nematode-specific factor based on its amino acid sequence, its function as an autophagy adaptor appears to be well conserved across species. We also identified IKKE-1, which is a homolog of the mammalian TBK1/IKKε kinase, as an ALLO-1-binding partner essential for allophagy. TBK1 is known to phosphorylate autophagy adaptors such as optineurin (OPTN), NDP52, and p62 in mammalian xenophagy and PINK1/Parkin-dependent mitophagy, thereby enhancing the binding ability of the adaptors to LC3 or ubiquitin.57) In fact, the phosphorylation of ALLO-1 is partly dependent on IKKE-1, and mutations in potential phosphorylation sites on ALLO-1 reduced the efficiency of allophagy. Thus, the mechanism of the selective autophagy pathway is conserved across species and targets. On the other hand, it has also been suggested that IKKE-1 has phosphorylation substrates other than ALLO-1, and IKKE-1 can promote the formation of a local autophagosome membrane by phosphorylating multiple targets simultaneously.
What happens to paternal organelles after fertilization? Recently, detailed electron microscopic observation of C. elegans sperms and fertilized eggs has revealed that the membrane structure of paternal mitochondria changes drastically after fertilization.58) Paternal mitochondria in sperm have normal internal membrane structures before fertilization. In contrast, paternal mitochondria contain collapsed cristae and electron-dense aggregates in fertilized eggs. In addition, parts of the paternal mitochondria have a ruptured outer membrane and show a decrease in their membrane potential. As such changes occur before paternal mitochondria are completely engulfed by the autophagosome and not on maternal mitochondria, these seem to occur spontaneously only in paternal mitochondria (Fig. 4). On the other hand, a recent study has reported that paternal mitochondria remain polarized before they are engulfed by autophagosomes in fertilized eggs and that they never fuse with maternal mitochondria.59) The timing of paternal mitochondrial degeneration requires further investigation.
One of the candidates involved in the degeneration of paternal mitochondria after fertilization is CPS-6, a mitochondrial endonuclease G homolog, since it was reported that the loss of CPS-6 delays the degeneration of paternal mitochondria and the formation of autophagosomal membranes around paternal mitochondria.58) Based on these observations, it has been proposed that the degradation of mtDNA by CPS-6 leads to the structural breakdown of paternal mitochondria, and these mitochondria are specifically eliminated by allophagy. An interesting topic to be elucidated is how the fertilization signal is transmitted to the paternal mitochondria leading to their breakdown. Paternal mitochondrial degeneration may trigger ubiquitination and recognition by ALLO-1. In mammalian cells, a decrease in mitochondrial membrane potential triggers PINK1 and Parkin-dependent mitophagy. However, allophagy occurs normally in PINK1 and Parkin-deficient strains, suggesting the involvement of other ubiquitin ligases in this process.54) The mechanisms for regulating paternal mitochondrial degeneration and their recognition by ALLO-1 remain to be elucidated.
6. Autophagy-dependent degradation of P granule constituents in C. elegans
The P granule is a C. elegans germ cell-specific non-membrane bound organelle that is a type of germ granule widely observed in animal germ cells (Fig. 5).60) P granules are considered to be involved in the translational regulation of mRNA and are essential for the formation of functional germ cells.61) In addition to germ granules, nucleoli, P-bodies, and stress granules found in somatic cells are non-membrane bound organelles, but it remains unclear how such structures are formed without being separated by a membrane. However, recent analyses of P granules in C. elegans have revealed that these organelles form droplets, in which specific proteins and RNA are assembled by liquid–liquid phase separation.62,63) The P granule derived from the oocyte is transmitted and maintained only in the P cell lineage, which will differentiate into germ cells in the embryo (Fig. 5A).61) PGL-1 and its paralog PGL-3 form a scaffold for the formation of P granules by liquid–liquid phase separation (Fig. 5B).61,62,64,65) These proteins have an RGG domain, which is an intrinsically disordered region, and this region plays an important role in liquid–liquid phase separation. Recently, the intrinsically disordered proteins, MEG-3 and MEG-4, have been shown to be involved in the asymmetric localization of P granules in embryos.66,67)
Figure 5.
PGL granule autophagy in C. elegans embryos. (A) P granule is a type of germ granule that is selectively distributed to the germ cell lineage during embryogenesis. P granule components in somatic cells form PGL granules (PGL-1-positive granules) and are selectively removed by autophagy. (B) SEPA-1 is an autophagy adaptor for P granule components such as PGL-1 and PGL-3. EPG-2 localizes to PGL granules via SEPA-1 and changes the property of PGL granules from a soft liquid phase to a gel state for autophagy.
In addition to these mechanisms, when PGL-1 and PGL-3 are distributed to somatic cells, these proteins are selectively degraded by autophagy (Fig. 5).68) In the somatic cells of autophagy-deficient embryos, PGL-1 and PGL-3 exist in granular structures, which are referred to as PGL granules (PGL-1-positive granules) because their composition is different from that of P granules.68) In order to identify factors involved in autophagy, Zhang et al. attempted to isolate mutants that suppress PGL granule formation in autophagy-deficient mutants and isolated the suppressor of ectopic P granule in autophagy (sepa)-1 mutants.68) In the sepa-1 mutant, GFP::PGL-1 was present in somatic cells in embryos, but PGL granules were no longer formed. SEPA-1 is a self-associating protein with a helical structure and a KIX domain, and it functions as an autophagy adaptor in the degradation of PGL granules by interacting with both PGL-3 and the autophagy factor LGG-1.
Zhang et al. also isolated several ectopic PGL granules (epg) mutants, which showed abnormal localization of P-granule components in somatic cells, by performing a genetic screen using GFP-PGL-1 and identified many factors required for the degradation of PGL granules.69) Among these factors, EPG-1 and EPG-9 are Atg13 and ATG101 homologs, respectively, and form a complex with UNC-51 (Atg1 homolog) and act upstream of autophagy.70)
EPG-2 functions as a scaffold protein that localizes to PGL granules via SEPA-1 and further binds to LGG-1.69) EPG-2 also changes the properties of PGL granules from a soft liquid phase to a gel state destined for autophagy (Fig. 5B).71)
The epg-3 and epg-4 genes encode homologs of human vacuolar membrane protein-1 and etoposide-induced gene 24/p53-induced gene 8 (EI24/PIG8), which are membrane proteins localized in the ER.69) These mutants accumulate isolation membranes of omegasomes and autophagosomes formed from the ER, suggesting that EPG-3 and EPG-4 are factors involved in the formation and closure of isolation membranes on the ER.69,72) EPG-6 is a WIPI4 homolog, which is involved in omegasome formation by regulating the localization of ATG-9 vesicles together with ATG-2.73) In contrast, the epg-5 gene encodes a novel factor, EPG-5 that is conserved in mammals, and in epg-5 mutants, PGL granules are trapped in autophagosomes but not degraded. EPG-5 is thought to be involved in the formation of autolysosomes that occur in the late stages of autophagy.69) Recently, it has been revealed that human EPG5 functions as an effector of the small GTPase RAB7 to regulate the fusion specificity of autophagosomes and late endosomes/lysosomes,74) and mutations in EPG5 cause Vici syndrome.75)
7. Essential roles of autophagy in mouse embryogenesis
Autophagy is essential for early mouse development because Atg5-deficient oocytes fertilized with Atg5-deficient sperm arrest at 4- and 8-cell stages (Fig. 3B).76) Autophagy activity was relatively low in mouse oocytes prior to fertilization. In contrast, autophagy activity increased drastically approximately 4 h after fertilization. Although autophagy activity then decreases slightly before and after the 2-cell stage, it remains at a high level until the 4–8-cell stage. In fertilized mouse eggs, oocyte-derived proteins and mRNAs are rapidly degraded after fertilization, while embryonic gene transcription is initiated, and protein synthesis becomes active in the 4–8-cell stage. Because reduction of amino acid levels and decreased synthesis of new proteins are remarkable features in Atg5-deficient embryos, fertilization-triggered autophagy is thought to be essential for supplying amino acids to new protein synthesis driven by zygotic gene expression. mTOR is known to be a negative regulator of autophagy, and the phosphorylation of one of its substrates, S6 kinase, was reduced in fertilized eggs.76) However, the treatment of mouse embryos with an mTORC1 inhibitor did not induce autophagy by itself, suggesting that fertilization-triggered autophagy occurs independently of mTORC1.77) Treatment with strontium, which can artificially induce intracellular calcium release, induces autophagy even in unfertilized oocytes; raising the possibility that autophagy is triggered by calcium oscillation after fertilization. Further analysis of the signaling pathways that drive autophagy after fertilization is required.
Recently, Tsukamoto and colleagues revealed that autophagic activity in fertilized eggs is related to embryo viability and developmental ability.78) They found that the autophagic activity in aged eggs derived from 14–15-month-old mice was significantly reduced, probably due to a decreased activity of lysosomal degrading enzymes, compared with those from young mice. In addition, when they classified 4-cell stage embryos into two groups according to autophagic activity, the number of pups was significantly higher in females transplanted with embryos with high autophagy activity than those with embryos with low autophagy activity. Autophagic activity in embryos may be a good index for evaluating the quality of embryos and predicting their developmental ability.
8. Mechanisms to eliminate paternal mitochondria in mouse embryos
In mammals, sperm mitochondria are tightly coiled around an axoneme in a spiral shape at a site called the midpiece. In humans and mice, it has been reported that sperm-derived paternal mitochondria containing mtDNA enter eggs but disappear during embryogenesis.79) Ubiquitin signals were also detected in the paternal mitochondria of mouse embryos.79) Based on these observations, it has been thought that paternal mitochondria are somehow removed from embryos after fertilization. More recently, it has been reported that ubiquitinated paternal mitochondria are degraded by p62-mediated autophagy.80) In addition, the membrane potential of paternal mitochondria is reduced after fertilization, and the simultaneous knockdown of two ubiquitin ligases, PARKIN and MUL1, delays this degradation,80) suggesting that ubiquitination-mediated autophagy regulates the degradation of paternal mitochondria in mouse embryos. On the other hand, other groups have claimed that autophagy is dispensable for the elimination of paternal mitochondria in mouse embryos.81) They reported that paternal mitochondria are fragmented around the 4-cell stage, but they are not actively degraded and remain at least until the blastocyst stage, although autophagy factors such as LC3 are transiently localized in the vicinity of paternal mitochondria in 1-cell stage embryos. They also reported that mtDNA is already degraded during differentiation in approximately 65% of sperm, suggesting that this degradation is a major mechanism for maternal inheritance of mtDNA. The fate of paternal mitochondria in mouse embryos requires further verification.
Perspective
Recent studies have revealed multiple roles of endocytosis and autophagy during early development in C. elegans and mice. It has been shown that some populations of maternal membrane proteins are selectively degraded after fertilization in C. elegans and mouse embryos. However, the signaling mechanisms underlying these processes and the ubiquitination mechanism that determines substrate specificity have not yet been identified. Notably, the endocytosis of maternal PM proteins occurs immediately after fertilization (∼21 min after fertilization) and is quickly delivered to lysosomes for degradation by the 2-cell stage (∼65 min after fertilization) in C. elegans embryos.9,10,25) In contrast, maternal PM proteins in mouse embryos start to be internalized by the late-2-cell stage (40 hours post fertilization) and are slowly transported to lysosomes by the 8-cell (54 hours post fertilization) and later stages.38) This difference may be explained by the different timings of endosome/lysosome maturation in C. elegans and mouse embryos, as lysosomes in mouse embryos can be detected from the 2-cell stage by staining with LysoTracker Green, which labels acidic lysosomes.38,82) The physiological significance of the selective degradation of maternal membrane proteins after fertilization remains unclear. If the proteins that regulate this process are identified in the future, their physiological roles can be studied by analyzing the functions of the gene products and their deficient strains.
On the other hand, various roles of autophagy during oocyte-to-embryo transition have been elucidated. The sperm-derived paternal mitochondria are selectively degraded by allophagy, which has been shown to be essential for maternal inheritance of mtDNA. However, it is still unclear how allophagy is triggered and why only paternal mitochondria are selectively surrounded by autophagosomes and delivered to lysosomes for their degradation in embryos. In the future, it will be necessary to clarify the signaling pathways that initiate allophagy, identify changes occurring in paternal mitochondria before and after fertilization, and reveal the signals required to target ALLO-1 and other autophagy-related factors to paternal mitochondria. It is also unclear why paternal mitochondria need to be removed. Several hypotheses have been proposed in this regard. One model states that the mitochondria of sperm are exhausted by the flagellar movement and are dysfunctional at the time of fertilization, and another model states that mtDNA prefers to be in a homoplasmic state and mtDNA from other individuals is degraded to maintain this homoplasmic state. In fact, it has been reported that mice artificially carrying two types of mtDNA show metabolic abnormalities.83) If oocytes continue to accept mtDNA from sperm at each fertilization, mtDNA will be excessively heteroplasmic with each successive generation. In such a situation, it may be difficult to control multiple types of mitochondria with a different quality. Alternatively, if oocytes accept defective mitochondria that produce excessive reactive oxygen species even once, the survival of the species itself may be threatened. The physiological significance of paternal mitochondrial degradation is expected to become clearer through comparative analysis of development, growth, and effects on offspring using wild-type strains and mutants that retain paternal mitochondria, such as allo-1 and ikke-1 mutants. In mammals, autophagy is induced twice in early embryos, but the specific substrates for autophagy remain unclear. In the future, proteomic analysis of embryos from wild-type and autophagy-defective mice will enable us to identify the substrates of degradation by autophagy during early embryogenesis.
Acknowledgements
Thanks are given to Drs. Miyuki Sato and Yuhkoh Satouh for critically reading the manuscript and providing valuable advice. This study was supported by the Japan Society for the Promotion of Science KAKENHI (grant numbers 19H05711 and 20H00466 to K. Sato).
Abbreviations
- APC/C
anaphase-promoting complex/cyclosome
- CHS-1
chitin synthase-1
- CUL
cullin
- DENN
differentially expressed in normal and neoplastic cells
- DYRK
dual-specificity tyrosine phosphorylation-regulated kinase
- GFP
green fluorescent protein
- GLD
germline development defective
- GV
germinal vesicle
- LIR
LC3-interacting region
- MBK
minibrain kinase
- MEI
meiotic spindle formation protein
- MO
membranous organelle
- MSP
major sperm protein
- mtDNA
mitochondrial DNA
- MVB
multivesicular body
- OPTN
optineurin
- PKC
protein kinase C
- PM
plasma membrane
- PVS
perivitelline space
- rme
receptor-mediated endocytosis defective mutant
- SNB-1
synaptobrevin-1 homolog
- SYN-4
syntaxin 4 homolog
- Sypl
synaptophysin-like protein
- UPS
ubiquitin-proteasome system
- VIT
vitellogenin
Profile
Ken Sato was born in Oita Prefecture in 1968. He graduated from Kyusyu University in 1992. He then entered the Graduate School of Science of the University of Tokyo and received his Ph. D. degree in 1997 for studies on the mechanisms of ER protein localization by vesicle recycling under the supervision of Dr. Akihiko Nakano. He continued his study as a JSPS postdoctoral fellow in 1997 and became a Research Scientist of the Molecular Membrane Biology Laboratory at RIKEN in 1998. He joined Dr. Barth Grant’s laboratory at Rutgers University in the U.S. as a visiting scientist and initiated studies on the mechanisms and roles of receptor-mediated endocytosis in C. elegans. He became an Associate Professor in 2004 and a Professor in 2010 at Institute for Molecular and Cellular Regulation of Gunma University. He was appointed as Director of Institute for Molecular and Cellular Regulation in 2019. He discovered Rer1 as a sorting receptor to retrieve various ER membrane proteins from the Golgi and revealed that its functions are well conserved from yeast to mammals. Recently, he has studied the physiological functions and molecular mechanisms of membrane trafficking during development using C. elegans and mice. He and his colleagues have shown that sperm-derived paternal mitochondria are selectively eliminated by a fertilization-triggered autophagy named allophagy, which is essential for maternal inheritance of mitochondrial DNA in C. elegans. He was awarded the Young Investigator Award from the Japanese Biochemical Society in 2008, and Kihara Memorial Award in 2018.
References
- 1).Tadros W., Lipshitz H.D. (2005) Setting the stage for development: mRNA translation and stability during oocyte maturation and egg activation in Drosophila. Dev. Dyn. 232, 593–608. [DOI] [PubMed] [Google Scholar]
- 2).Schier A.F. (2007) The maternal-zygotic transition: death and birth of RNAs. Science 316, 406–407. [DOI] [PubMed] [Google Scholar]
- 3).Merz E.A., Brinster R.L., Brunner S., Chen H.Y. (1981) Protein degradation during preimplantation development of the mouse. J. Reprod. Fertil. 61, 415–418. [DOI] [PubMed] [Google Scholar]
- 4).Marcello M.R., Singson A. (2010) Fertilization and the oocyte-to-embryo transition in C. elegans. BMB Rep. 43, 389–399. [DOI] [PubMed] [Google Scholar]
- 5).Miller M.A., Nguyen V.Q., Lee M.H., Kosinski M., Schedl T., Caprioli R.M., et al. (2001) A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science 291, 2144–2147. [DOI] [PubMed] [Google Scholar]
- 6).Gan W.J., Motegi F. (2020) Mechanochemical control of symmetry breaking in the Caenorhabditis elegans zygote. Front. Cell Dev. Biol. 8, 619869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Stein K.K., Golden A. (2018) The C. elegans eggshell. WormBook 2018, 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Olson S.K., Greenan G., Desai A., Muller-Reichert T., Oegema K. (2012) Hierarchical assembly of the eggshell and permeability barrier in C. elegans. J. Cell Biol. 198, 731–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Sato K., Sato M., Audhya A., Oegema K., Schweinsberg P., Grant B.D. (2006) Dynamic regulation of caveolin-1 trafficking in the germ line and embryo of Caenorhabditis elegans. Mol. Biol. Cell 17, 3085–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Sato M., Grant B.D., Harada A., Sato K. (2008) Rab11 is required for synchronous secretion of chondroitin proteoglycans after fertilization in Caenorhabditis elegans. J. Cell Sci. 121, 3177–3186. [DOI] [PubMed] [Google Scholar]
- 11).Bembenek J.N., Richie C.T., Squirrell J.M., Campbell J.M., Eliceiri K.W., Poteryaev D., et al. (2007) Cortical granule exocytosis in C. elegans is regulated by cell cycle components including separase. Development 134, 3837–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Stoeckius M., Grun D., Kirchner M., Ayoub S., Torti F., Piano F., et al. (2014) Global characterization of the oocyte-to-embryo transition in Caenorhabditis elegans uncovers a novel mRNA clearance mechanism. EMBO J. 33, 1751–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).McNally F.J., Vale R.D. (1993) Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell 75, 419–429. [DOI] [PubMed] [Google Scholar]
- 14).Dow M.R., Mains P.E. (1998) Genetic and molecular characterization of the Caenorhabditis elegans gene, mel-26, a postmeiotic negative regulator of mei-1, a meiotic-specific spindle component. Genetics 150, 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Srayko M., Buster D.W., Bazirgan O.A., McNally F.J., Mains P.E. (2000) MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 14, 1072–1084. [PMC free article] [PubMed] [Google Scholar]
- 16).Robertson S., Lin R. (2015) The maternal-to-zygotic transition in C. elegans. Curr. Top. Dev. Biol. 113, 1–42. [DOI] [PubMed] [Google Scholar]
- 17).Grant B., Hirsh D. (1999) Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell 10, 4311–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Sato K., Ernstrom G.G., Watanabe S., Weimer R.M., Chen C.H., Sato M., et al. (2009) Differential requirements for clathrin in receptor-mediated endocytosis and maintenance of synaptic vesicle pools. Proc. Natl. Acad. Sci. U.S.A. 106, 1139–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Sato M., Sato K., Fonarev P., Huang C.J., Liou W., Grant B.D. (2005) Caenorhabditis elegans RME-6 is a novel regulator of RAB-5 at the clathrin-coated pit. Nat. Cell Biol. 7, 559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Grant B., Zhang Y., Paupard M.C., Lin S.X., Hall D.H., Hirsh D. (2001) Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat. Cell Biol. 3, 573–579. [DOI] [PubMed] [Google Scholar]
- 21).Pant S., Sharma M., Patel K., Caplan S., Carr C.M., Grant B.D. (2009) AMPH-1/Amphiphysin/Bin1 functions with RME-1/Ehd1 in endocytic recycling. Nat. Cell Biol. 11, 1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Sato M., Sato K., Liou W., Pant S., Harada A., Grant B.D. (2008) Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME-4, a coated-pit protein. EMBO J. 27, 1183–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Allaire P.D., Marat A.L., Dall’Armi C., Di Paolo G., McPherson P.S., Ritter B. (2010) The Connecdenn DENN domain: a GEF for Rab35 mediating cargo-specific exit from early endosomes. Mol. Cell 37, 370–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Balklava Z., Pant S., Fares H., Grant B.D. (2007) Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat. Cell Biol. 9, 1066–1073. [DOI] [PubMed] [Google Scholar]
- 25).Sato M., Konuma R., Sato K., Tomura K. (2014) Fertilization-induced K63-linked ubiquitylation mediates clearance of maternal membrane proteins. Development 141, 1324–1331. [DOI] [PubMed] [Google Scholar]
- 26).Goudeau J., Aguilaniu H. (2010) Carbonylated proteins are eliminated during reproduction in C. elegans. Aging Cell 9, 991–1003. [DOI] [PubMed] [Google Scholar]
- 27).Bohnert K.A., Kenyon C. (2017) A lysosomal switch triggers proteostasis renewal in the immortal C. elegans germ lineage. Nature 551, 629–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Nousch M., Eckmann C.R. (2013) Translational control in the Caenorhabditis elegans germ line. Adv. Exp. Med. Biol. 757, 205–247. [DOI] [PubMed] [Google Scholar]
- 29).Oku M., Sakai Y. (2018) Three distinct types of microautophagy based on membrane dynamics and molecular machineries. BioEssays 40, e1800008. [DOI] [PubMed] [Google Scholar]
- 30).Samaddar M., Goudeau J., Sanchez M., Hall D.H., Bohnert K.A., Ingaramo M., et al. (2021) A genetic screen identifies new steps in oocyte maturation that enhance proteostasis in the immortal germ lineage. eLife 10, e62653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Sato M., Sato K. (2013) Dynamic regulation of autophagy and endocytosis for cell remodeling during early development. Traffic 14, 479–486. [DOI] [PubMed] [Google Scholar]
- 32).Audhya A., McLeod I.X., Yates J.R., Oegema K. (2007) MVB-12, a fourth subunit of metazoan ESCRT-I, functions in receptor downregulation. PLoS One 2, e956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Johnston W.L., Krizus A., Dennis J.W. (2010) Eggshell chitin and chitin-interacting proteins prevent polyspermy in C. elegans. Curr. Biol. 20, 1932–1937. [DOI] [PubMed] [Google Scholar]
- 34).Kadandale P., Stewart-Michaelis A., Gordon S., Rubin J., Klancer R., Schweinsberg P., et al. (2005) The egg surface LDL receptor repeat-containing proteins EGG-1 and EGG-2 are required for fertilization in Caenorhabditis elegans. Curr. Biol. 15, 2222–2229. [DOI] [PubMed] [Google Scholar]
- 35).Mukhopadhyay D., Riezman H. (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315, 201–205. [DOI] [PubMed] [Google Scholar]
- 36).Henne W.M., Buchkovich N.J., Emr S.D. (2011) The ESCRT pathway. Dev. Cell 21, 77–91. [DOI] [PubMed] [Google Scholar]
- 37).Hofmann R.M., Pickart C.M. (1999) Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96, 645–653. [DOI] [PubMed] [Google Scholar]
- 38).Morita A., Satouh Y., Kosako H., Kobayashi H., Iwase A., Sato K. (2021) Clathrin-mediated endocytosis is essential for the selective degradation of maternal membrane proteins and preimplantation development. Development 148, 199461. [DOI] [PubMed] [Google Scholar]
- 39).Wang J.J., Ge W., Liu J.C., Klinger F.G., Dyce P.W., De Felici M., et al. (2017) Complete in vitro oogenesis: retrospects and prospects. Cell Death Differ. 24, 1845–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Guastella J., Brecha N., Weigmann C., Lester H.A., Davidson N. (1992) Cloning, expression, and localization of a rat brain high-affinity glycine transporter. Proc. Natl. Acad. Sci. U.S.A. 89, 7189–7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Liu Q.R., Lopez-Corcuera B., Mandiyan S., Nelson H., Nelson N. (1993) Cloning and expression of a spinal cord- and brain-specific glycine transporter with novel structural features. J. Biol. Chem. 268, 22802–22808. [PubMed] [Google Scholar]
- 42).Richard S., Tartia A.P., Boison D., Baltz J.M. (2017) Mouse oocytes acquire mechanisms that permit independent cell volume regulation at the end of oogenesis. J. Cell. Physiol. 232, 2436–2446. [DOI] [PubMed] [Google Scholar]
- 43).Van Winkle L.J., Haghighat N., Campione A.L., Gorman J.M. (1988) Glycine transport in mouse eggs and preimplantation conceptuses. Biochim. Biophys. Acta 941, 241–256. [DOI] [PubMed] [Google Scholar]
- 44).Anas M.K., Hammer M.A., Lever M., Stanton J.A., Baltz J.M. (2007) The organic osmolytes betaine and proline are transported by a shared system in early preimplantation mouse embryos. J. Cell. Physiol. 210, 266–277. [DOI] [PubMed] [Google Scholar]
- 45).Baltz J.M., Zhou C. (2012) Cell volume regulation in mammalian oocytes and preimplantation embryos. Mol. Reprod. Dev. 79, 821–831. [DOI] [PubMed] [Google Scholar]
- 46).Miyado K., Yoshida K., Yamagata K., Sakakibara K., Okabe M., Wang X., et al. (2008) The fusing ability of sperm is bestowed by CD9-containing vesicles released from eggs in mice. Proc. Natl. Acad. Sci. U.S.A. 105, 12921–12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Balestra F.R., von Tobel L., Gonczy P. (2015) Paternally contributed centrioles exhibit exceptional persistence in C. elegans embryos. Cell Res. 25, 642–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Stoeckius M., Grun D., Rajewsky N. (2014) Paternal RNA contributions in the Caenorhabditis elegans zygote. EMBO J. 33, 1740–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Sato K., Sato M. (2017) Multiple ways to prevent transmission of paternal mitochondrial DNA for maternal inheritance in animals. J. Biochem. 162, 247–253. [DOI] [PubMed] [Google Scholar]
- 50).Sato M., Sato K. (2013) Maternal inheritance of mitochondrial DNA by diverse mechanisms to eliminate paternal mitochondrial DNA. Biochim. Biophys. Acta 1833, 1979–1984. [DOI] [PubMed] [Google Scholar]
- 51).Sato M., Sato K. (2011) Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science 334, 1141–1144. [DOI] [PubMed] [Google Scholar]
- 52).Al Rawi S., Louvet-Vallee S., Djeddi A., Sachse M., Culetto E., Hajjar C., et al. (2011) Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 334, 1144–1147. [DOI] [PubMed] [Google Scholar]
- 53).Sato M., Sato K. (2012) Maternal inheritance of mitochondrial DNA: degradation of paternal mitochondria by allogeneic organelle autophagy, allophagy. Autophagy 8, 424–425. [DOI] [PubMed] [Google Scholar]
- 54).Sato M., Sato K., Tomura K., Kosako H., Sato K. (2018) The autophagy receptor ALLO-1 and the IKKE-1 kinase control clearance of paternal mitochondria in Caenorhabditis elegans. Nat. Cell Biol. 20, 81–91. [DOI] [PubMed] [Google Scholar]
- 55).Mizushima N., Komatsu M. (2011) Autophagy: renovation of cells and tissues. Cell 147, 728–741. [DOI] [PubMed] [Google Scholar]
- 56).Gubas A., Dikic I. (2022) A guide to the regulation of selective autophagy receptors. FEBS J. 289, 75–89. [DOI] [PubMed] [Google Scholar]
- 57).Yamano K., Matsuda N., Tanaka K. (2016) The ubiquitin signal and autophagy: an orchestrated dance leading to mitochondrial degradation. EMBO Rep. 17, 300–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58).Zhou Q., Li H., Li H., Nakagawa A., Lin J.L., Lee E.S., et al. (2016) Mitochondrial endonuclease G mediates breakdown of paternal mitochondria upon fertilization. Science 353, 394–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59).Rubio-Pena K., Al Rawi S., Husson F., Lam F., Merlet J., Galy V. (2021) Mitophagy of polarized sperm-derived mitochondria after fertilization. iScience 24, 102029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60).Strome S., Wood W.B. (1982) Immunofluorescence visualization of germ-line-specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 79, 1558–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61).Seydoux G. (2018) The P Granules of C. elegans: A genetic model for the study of RNA-protein condensates. J. Mol. Biol. 430, 4702–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62).Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., et al. (2009) Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732. [DOI] [PubMed] [Google Scholar]
- 63).Hyman A.A., Brangwynne C.P. (2011) Beyond stereospecificity: liquids and mesoscale organization of cytoplasm. Dev. Cell 21, 14–16. [DOI] [PubMed] [Google Scholar]
- 64).Kawasaki I., Amiri A., Fan Y., Meyer N., Dunkelbarger S., Motohashi T., et al. (2004) The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics 167, 645–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65).Kawasaki I., Shim Y.H., Kirchner J., Kaminker J., Wood W.B., Strome S. (1998) PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94, 635–645. [DOI] [PubMed] [Google Scholar]
- 66).Chen J.X., Cipriani P.G., Mecenas D., Polanowska J., Piano F., Gunsalus K.C., et al. (2016) In vivo interaction proteomics in Caenorhabditis elegans embryos provides new insights into P granule dynamics. Mol. Cell. Proteomics 15, 1642–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67).Wang J.T., Smith J., Chen B.C., Schmidt H., Rasoloson D., Paix A., et al. (2014) Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife 3, e04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68).Zhang Y., Yan L., Zhou Z., Yang P., Tian E., Zhang K., et al. (2009) SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell 136, 308–321. [DOI] [PubMed] [Google Scholar]
- 69).Tian Y., Li Z., Hu W., Ren H., Tian E., Zhao Y., et al. (2010) C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 141, 1042–1055. [DOI] [PubMed] [Google Scholar]
- 70).Liang Q., Yang P., Tian E., Han J., Zhang H. (2012) The C. elegans ATG101 homolog EPG-9 directly interacts with EPG-1/Atg13 and is essential for autophagy. Autophagy 8, 1426–1433. [DOI] [PubMed] [Google Scholar]
- 71).Zhang G., Wang Z., Du Z., Zhang H. (2018) mTOR regulates phase separation of PGL granules to modulate their autophagic degradation. Cell 174, 1492–1506.e22. [DOI] [PubMed] [Google Scholar]
- 72).Zhao Y.G., Chen Y., Miao G., Zhao H., Qu W., Li D., et al. (2017) The ER-localized transmembrane protein EPG-3/VMP1 regulates SERCA activity to control ER-isolation membrane contacts for autophagosome formation. Mol. Cell 67, 974–989.e6. [DOI] [PubMed] [Google Scholar]
- 73).Lu Q., Yang P., Huang X., Hu W., Guo B., Wu F., et al. (2011) The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev. Cell 21, 343–357. [DOI] [PubMed] [Google Scholar]
- 74).Wang Z., Miao G., Xue X., Guo X., Yuan C., Wang Z., et al. (2016) The vici syndrome protein EPG5 is a Rab7 effector that determines the fusion specificity of autophagosomes with late endosomes/lysosomes. Mol. Cell 63, 781–795. [DOI] [PubMed] [Google Scholar]
- 75).Cullup T., Kho A.L., Dionisi-Vici C., Brandmeier B., Smith F., Urry Z., et al. (2013) Recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder with defective autophagy. Nat. Genet. 45, 83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76).Tsukamoto S., Kuma A., Murakami M., Kishi C., Yamamoto A., Mizushima N. (2008) Autophagy is essential for preimplantation development of mouse embryos. Science 321, 117–120. [DOI] [PubMed] [Google Scholar]
- 77).Yamamoto A., Mizushima N., Tsukamoto S. (2014) Fertilization-induced autophagy in mouse embryos is independent of mTORC1. Biol. Reprod. 91, 1–7. [DOI] [PubMed] [Google Scholar]
- 78).Tsukamoto S., Hara T., Yamamoto A., Kito S., Minami N., Kubota T., et al. (2014) Fluorescence-based visualization of autophagic activity predicts mouse embryo viability. Sci. Rep. 4, 4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79).Sutovsky P., Moreno R.D., Ramalho-Santos J., Dominko T., Simerly C., Schatten G. (1999) Ubiquitin tag for sperm mitochondria. Nature 402, 371–372. [DOI] [PubMed] [Google Scholar]
- 80).Rojansky R., Cha M.Y., Chan D.C. (2016) Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. eLife 5, e17896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81).Luo S.M., Ge Z.J., Wang Z.W., Jiang Z.Z., Wang Z.B., Ouyang Y.C., et al. (2013) Unique insights into maternal mitochondrial inheritance in mice. Proc. Natl. Acad. Sci. U.S.A. 110, 13038–13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82).Tsukamoto S., Hara T., Yamamoto A., Ohta Y., Wada A., Ishida Y., et al. (2013) Functional analysis of lysosomes during mouse preimplantation embryo development. J. Reprod. Dev. 59, 33–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83).Sharpley M.S., Marciniak C., Eckel-Mahan K., McManus M., Crimi M., Waymire K., et al. (2012) Heteroplasmy of mouse mtDNA is genetically unstable and results in altered behavior and cognition. Cell 151, 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]