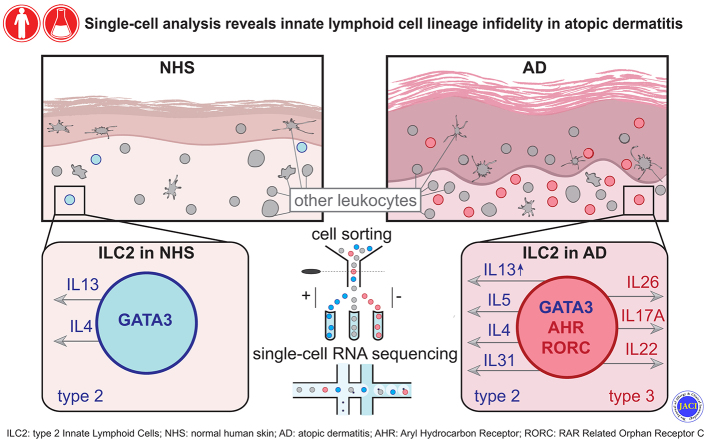
Abstract
Background
Although ample knowledge exists about phenotype and function of cutaneous T lymphocytes, much less is known about the lymphocytic components of the skin’s innate immune system.
Objective
To better understand the biologic role of cutaneous innate lymphoid cells (ILCs), we investigated their phenotypic and molecular features under physiologic (normal human skin [NHS]) and pathologic (lesional skin of patients with atopic dermatitis [AD]) conditions.
Methods
Skin punch biopsies and reduction sheets as well as blood specimens were obtained from either patients with AD or healthy individuals. Cell and/or tissue samples were analyzed by flow cytometry, immunohistochemistry, and single-cell RNA sequencing or subjected to in vitro/ex vivo culture.
Results
Notwithstanding substantial quantitative differences between NHS and AD skin, we found that the vast majority of cutaneous ILCs belong to the CRTH2+ subset and reside in the upper skin layers. Single-cell RNA sequencing of cutaneous ILC-enriched cell samples confirmed the predominance of biologically heterogeneous group 2 ILCs and, for the first time, demonstrated considerable ILC lineage infidelity (coexpression of genes typical of either type 2 [GATA3 and IL13] or type 3/17 [RORC, IL22, and IL26] immunity within individual cells) in lesional AD skin, and to a much lesser extent, in NHS. Similar events were demonstrated in ILCs from skin explant cultures and in vitro expanded ILCs from the peripheral blood.
Conclusion
These findings support the concept that instead of being a stable entity with well-defined components, the skin immune system consists of a network of highly flexible cellular players that are capable of adjusting their function to the needs and challenges of the environment.
Key words: Innate lymphoid cells, atopic dermatitis, human skin, single-cell RNA sequencing
Abbreviations used: AD, Atopic dermatitis; AHR, Aryl hydrocarbon receptor; GATA3, GATA binding protein 3; HC, Healthy control; ILC, Innate lymphoid cell; ILC1, Group 1 innate lymphoid cell; ILC2, Group 2 innate lymphoid cell; ILC3, Group 3 innate lymphoid cell; NHS, Normal human skin; PB, Peripheral blood; PE, Phycoerythrin; RORA, RAR related orphan receptor A; RORC, RAR related orphan receptor C; scRNA-seq, Single-cell RNA sequencing; TSLP, Thymic stromal lymphopoietin
Graphical abstract
T cells are the predominant, but not the only lymphocyte subpopulation within skin. Apart from a few reports on B cells,1 natural killer (NK) cells,2 and NKT cells,3 particular attention has recently focused on innate lymphoid cells (ILCs). These cells exhibit a lymphoid morphology, but lack antigen-specific receptors. Currently, ILCs are phenotypically defined as CD45+Lin–CD3–CD161+CD127+ cells and are subdivided into 3 subsets, namely, group 1 ILCs (ILC1s) (CD117–CRTH2–), group 2 ILCs (ILC2s) (CRTH2+), and group 3 ILCs (ILC3s) (CD117+CRTH2–).4,5
As compared to unperturbed tissues where they are present in small numbers only, ILCs are substantially increased upon inflammation. At the moment, whether these cells assemble in skin as a consequence of inflammation or, alternatively, serve as an initiator of this process is unclear. Studies in mice revealed that cutaneous overexpression of certain cytokines and growth factors such as IL-25 and IL-33,6,7 as well as thymic stromal lymphopoietin (TSLP),8 results in the expansion of IL-13–producing ILCs and the appearance of AD-like skin lesions. Interestingly, when appropriately stimulated, murine ILCs may cause skin inflammation in the absence of T lymphocytes.6,9,10 Another facet of ILC function is in maintaining skin homeostasis by modulating the equilibrium of skin commensal bacteria.11
Not that many data on human skin ILCs are available. Using flow cytometry, several groups of investigators demonstrated an increase in the numbers of ILC3s in patients with psoriasis12,13 and of ILC2s in patients with atopic dermatitis (AD).6 Even less is known about their in situ phenotype, localization, and relationship to other cell types. Immunofluorescence microscopy revealed the occurrence of ILCs in human skin and increased numbers of Lin–CD25+IL33R+ ILCs, presumably ILC2s, in lesional AD skin.14 In another study, all 3 ILC subclasses were identified by in situ immunostaining, with elevated numbers of GATA binding protein 3 (GATA3)-positive (GATA3+) and aryl hydrocarbon receptor (AHR)-positive (AHR+) ILCs in AD skin.15
In this study, we combined immunolabeling techniques and single-cell RNA sequencing (scRNA-seq) to investigate ILCs and their subsets in normal and perturbed human skin. We found that (i) ILCs and NK cells are present in small numbers in normal human skin (NHS), but their numbers are substantially expanded in lesional AD skin; (ii) ILC2s are the predominant ILC subset in both healthy and diseased skin; (iii) ILC2s from lesional AD skin and, to a much lesser extent, from normal skin exhibit substantial heterogeneity in their immunoprofile; and (iv) when appropriately stimulated, ILCs from either skin or peripheral blood (PB) of healthy persons rapidly change their molecular, immunophenotypic, and functional profile, making these cells versatile sentinels in host defense and, under different circumstances, disease-promoting vehicles.
Methods
Study approval
The study was approved by the ethics committee of the Medical University of Vienna, Austria (approval no. EK 071/2005). In accordance with the Declaration of Helsinki, written informed consent was received from each participant before inclusion in the study.
Skin and blood donors
Human skin samples were obtained from healthy individuals undergoing skin reduction surgery (mostly for aesthetic purposes), and in the form of punch biopsies, from lesional skin of patients with AD. Heparinized blood was collected by venipuncture from patients with AD and healthy persons (healthy controls [HCs]). All patients showed active disease, had AD since childhood, and did not receive any topical (for 2 weeks) or systemic (for 4 weeks) therapy before skin or blood sampling. Summarized clinical data as well as detailed demographics of the HCs and patients with AD can be found in Tables E1 and E2 (available in this article's Online Repository at www.jacionline.org).
Processing of skin tissue specimens
Enzymatic isolation of cells
Samples (punch biopsies or split-thickness skin sheets) were processed as described previously with minor modifications.15 Briefly, dermatomed 0.8-mm-thick NHS sheets (≥10 × 10 cm2) and 6-mm punch biopsy samples, collected from either NHS or the lesional skin areas of patients with AD, were fragmented and immersed in 500 U/mL of collagenase IV (Worthington Biochemical, Lakewood, NJ) and 50 U/mL of DNAse I (Sigma-Aldrich, St Louis, Mo) in RPMI 1640 medium (Gibco, Thermo Fisher Scientific, Waltham, Mass) and incubated at 37°C for 40 minutes. The digested tissue was sieved using a 70-μm cell strainer (Thermo Fisher Scientific). Cells were pelleted, resuspended in RPMI 1640 with 10% FCS (Gibco), and, in the case of NHS, further purified using Ficoll-Paque PLUS density gradient media (GE Healthcare, Chicago, Ill). The interphase was collected, pelleted, and stained with fluorescently labeled antibodies for further fluorescence-activated cell sorting. When necessary, skin tissue remnants containing nondigested epidermis were further processed by an additional incubation in 0.25% trypsin-EDTA (Gibco) for 10 minutes at 37°C, sieved using a 70-μm cell strainer, and stained for the sorting of CD45– EpCAM+/E-cadherin+ HLA-ABC+ keratinocytes.
Explant cultures of NHS
NHS tissue was dermatomed, mechanically fragmented (see Fig E4, A in the Online Repository at www.jacionline.org), and then incubated in culture flasks with RPMI 1640 growth medium, 10% FCS, Penicillin-Streptomycin (Gibco; 100 U/mL of penicillin, 100 μg/mL of streptomycin) and 2 mM l-glutamine (Gibco) at 37°C and 5% CO2 for a time line of 2, 4, 6, 8, and 10 days. At each time point, material was collected from a flask, sieved through 70-μm cell strainers, and stained with fluorescent-labeled antibodies for flow cytometry. To analyze ILCs in the nascent state, the skin obtained immediately after plastic surgery (day 0) was subjected to enzymatic tissue digestion (see the earlier section on enzymatic isolation of cells). The conditions for this procedure were optimized so that the major epitopes required for the detection of total ILCs were not digested.
Immunoperoxidase and immunofluorescence microscopy and automated analysis of the stainings
For the microscopic analyses, 4- to 6-mm punch biopsy samples of human skin were embedded in optimal cutting temperature (O.C.T.) compound (Tissue-Plus, Scigen Scientific, Gardena, Calif), deep-frozen in liquid nitrogen, and stored at –80°C until further processing, as previously described.15 Optimal cutting temperature compound–embedded tissue samples from the patients with AD and HCs (listed in Table E2) and from our biobank (AD patients A13-A21 [see Table E2]) were cut into 7-μm sections and mounted on SuperFrost Plus adhesion slides (Thermo Fisher Scientific). After 30 minutes of air-drying, the cryostat sections were fixed in ice-cold acetone (Sigma-Aldrich) for 10 minutes and stored at –20°C. For a detailed description of the immunoperoxidase and immunofluorescence procedures, please refer to the Methods section of the Online Repository (available at www.jacionline.org).
Processing of blood specimens
Mononuclear cells (PBMCs) from peripheral blood samples were isolated by using Ficoll-Paque PLUS density gradient media (GE Healthcare), stained with ILC markers, and then analyzed by flow cytometry and/or subjected to fluorescence-activated cell sorting (FACS). In the experiments in which we expanded ILCs in culture, PBMCs were depleted of lineage marker–positive cells if necessary (see the Online Repository materials at www.jacionline.org).
Flow cytometry and cell sorting
Cells isolated from skin or blood specimens were analyzed/sorted with a 13-color flow cytometer (FACSAria III, BD Biosciences, Franklin Lakes, NJ). The following antibodies to human proteins were used: purified anti–glycophorin A (11E4B-7-6 [KC16], Beckman Coulter, Brea, Calif), anti-CD41 (P2, Beckman Coulter), Alexa Fluor 488–conjugated anti-TCRα/β (IP26, Biolegend, San Diego, Calif), fluorescein isothiocyanate–conjugated anti-TCRγ/δ (B1, Biolegend), anti-CD19 (4G7, BD Biosciences), anti-CD16 (NKP15, BD Biosciences), anti-CD15 (HI98, Biolegend), anti-CD14 (MϕP9, BD Biosciences), anti-CD11c (Bu15, Biolegend), anti-CD94 (DX22, Biolegend), anti-CD303 (BDCA-2) (201A, Biolegend), anti-FcεRIα (AER-37 [CRA-1]), Biolegend), anti-CD1a (HI149, BD Biosciences), anti-CD34 (8G12 [HPCA2], BD Biosciences), anti–HLA-ABC (W6/32, Bio-Rad, Hercules, Calif), phycoerythrin (PE)-conjugated anti-CD3 (SK7 (Leu-4), BD Biosciences), anti-CD94 (DX22, Biolegend), anti-CLA (HECA-452, Miltenyi Biotec, Bergisch Gladbach, Germany), anti-CD194 (CCR4) (L291H4, Biolegend), anti-CD196 (CCR6) (G034E3, Biolegend), anti-CD69 (FN50, Biolegend), anti-EpCAM (1B7, eBioscience, San Diego, Calif), anti–E-cadherin (67A4, Biolegend), PE-CF594–conjugated anti-CD45 (HI30, BD Biosciences), Alexa Fluor 647–conjugated anti-CD294 (CRTH2) (BM16, Biolegend), allophycocyanin/cyanine 7–conjugated anti-CD161 (HP-3G10, Biolegend), PE–cyanine 7–conjugated anti-CD127 (IL-7Rα) (A019D5, Biolegend); Brilliant Violet 421–conjugated anti-CD117 (c-kit) (104D2, Biolegend), Brilliant Violet 605–conjugated anti-CD3 (SK7, Biolegend), Brilliant Violet 711–conjugated anti-CD3 (SK7, Biolegend), Brilliant Violet 650–conjugated anti-CD45RA (HI100, Biolegend), and Brilliant Violet 785–conjugated anti-CD45RO (UCHL1, Biolegend). Viability was assessed by using 7-aminoactinomycin D. The signal from the lineage antibodies in fluorescein isothiocyanate or Alexa Fluor 488 channels was amplified with second-step Alexa Fluor 488–conjugated goat anti-mouse IgG (H+L, Thermo Fisher Scientific).
For a detailed description of the expansion of blood-derived ILCs, real-time PCR, and intracellular cytokine stainings, please refer to Online Repository materials at www.jacionline.org.
Statistics
Biologic repeats, acquired with flow cytometry and in situ immunolabeling, were compared either individually or (when available) by using the mean of technical repeats. The indicated populations were assessed for normal gaussian distribution with the D’Agostino and Pearson omnibus normality test and then analyzed by the nonparametric 2-tailed Mann-Whitney U test using Prism version 8.0.1 (GraphPad Software, La Jolla, Calif). Median values are given with interquartile ranges. P values less than .05 were considered significant.
Droplet-based scRNA-seq
Preparation of libraries and sequencing
Cells sorted from NHS and AD skin biopsies after mechanical and enzymatic isolation were used to generate libraries for scRNA-seq. Such libraries were generated by using the Chromium Controller and Single Cell 3’ Library and Gel Bead Kit, version 2 and version 3 (10x Genomics, Pleasanton, Calif), according to the manufacturer’s protocol and as previously described.16 For processing of scRNA-seq data, please refer to the Methods section of the Online Repository.
10x Genomics data sets were made available via Gene Expression Omnibus (GSE180885) subsequent to acceptance of this article.
Results
Phenotypic characterization of cutaneous ILCs by flow cytometry
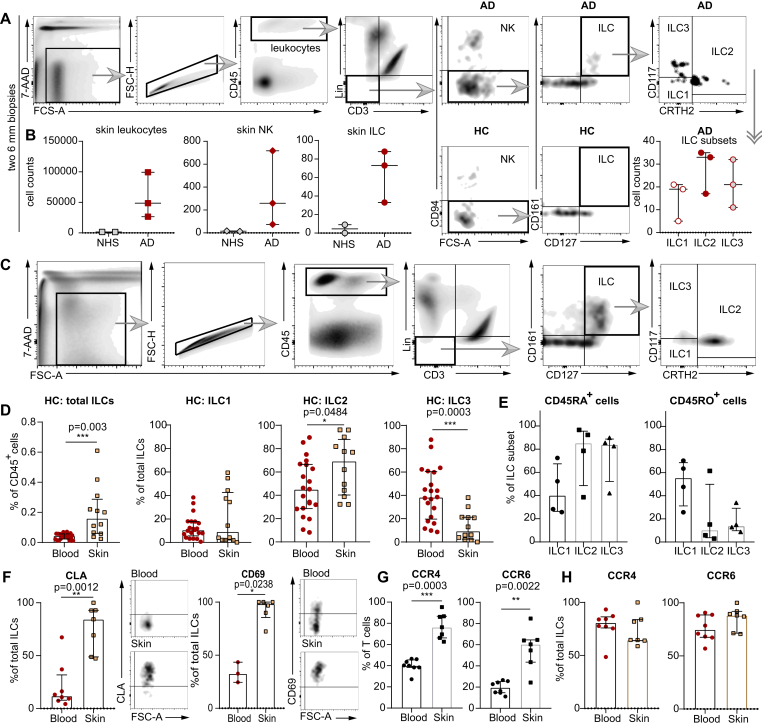
Comparative quantification of skin leukocytes/leukocyte subsets isolated from two 6-mm punch biopsies from either lesional AD skin or NHS showed that AD skin contains about 30-fold more CD45+ leukocytes than NHS does (Fig 1, A and B). Among these CD45+ AD leukocytes, approximately 0.5% could be identified as CD94+ NK cells and 0.15% as Lin–CD3–CD94–CD127+CD161+ ILCs. The latter consisted, in decreasing order, of CRTH2+ ILC2s, CD117+CRTH2– ILC3s and CD117–CRTH2– ILC1s (Fig 1, A and B). In contrast, the numbers of ILCs and also NK cells in NHS were at the limit of detection and, thus, did not allow for a reliable quantification.
Fig 1.
Characterization of ILCs with flow cytometry. A, Gating strategy for ILCs from skin punch biopsies. B, Numeric differences in leukocytes, NK cells, and ILCs between NHS (n = 2) and AD skin (n = 3) as well as ILC subpopulations detectable in AD skin samples (Lin for [A] and [B] without CD94 and CD16). C, Gating strategy for ILCs and ILC subsets extracted from NHS sheets and PB. D, Total ILCs and ILC subsets from NHS sheets (n = 12) and PB of HCs (n = 21). E, CD45RA+ and CD45RO+ ILC subsets in NHS (n = 4). F, Expression of CLA and CD69 on ILCs from NHS and PB of HCs (NHS [n = 7] and PB [n = 8] for CLA; NHS [n = 6] and PB [n = 3] for CD69). CD45+CD3+ T cells (G) and ILCs (H) expressing CCR4 and CCR6 in NHS (n = 7) and PB (n = 8). Dot plots represent medians with interquartile ranges (IQRs). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. 7-AAD, 7-Aminoactinomycin D, FSC-A/H, forward scatter area/height; Lin, lineage cocktail (TCRα/β, TCRγ/δ, CD19, CD16, CD15, CD14, CD94, CD11c, BDCA-2, FcεRIα, glycophorin A, CD41, CD1a, and CD34).
To address this issue, ie, to gain a clue about frequency and phenotype of ILCs and their subsets in NHS, we analyzed cells extracted from larger-sized (≥10 × 10 cm2) dermatomed NHS specimens. We then gated the total ILCs and their subpopulations (NK cells were excluded by adding anti-CD94 and anti-CD16 antibodies to the lineage cocktail) and compared them with those from the PB of healthy individuals (Fig 1, C). Results obtained showed that ILCs represent a minor subgroup of leukocytes (<<1% of all CD45+ cells) in both skin and blood. Interestingly though, the percentage of total ILCs among all CD45+ cells was considerably lower in the PB than in the skin (Fig 1, D). Furthermore, the distribution of ILC subsets within total ILCs was different between skin and blood. While the ratio of ILC1s was comparable between these 2 compartments, the percentage of ILC2s among total ILCs in the skin was one-third higher than that in the blood; in contrast, the percentage of ILC3s in the skin was far below that in the blood (Fig 1, D). Notably, the 2 CD45 isoforms, which in T cells signify different degrees of antigenic experience, are unevenly distributed among skin ILC subsets: while ILC2s and ILC3s predominantly exhibited the CD45RA+ phenotype, the percentage of CD45RO+ ILC1s exceeded that of CD45RA+ ILC1s (Fig 1, E).
One of the reasons why the percentage of ILCs among CD45+ cells is lower in the blood than in the skin could be immigration of blood ILCs into the skin. We therefore evaluated the presence of molecules, which are known from T-cell biology to be involved in skin homing and tissue residency,17, 18, 19 on skin and blood ILCs from healthy individuals. About 90% of skin ILCs were found to be CLA+, whereas only 10% of blood ILCs expressed this marker (Fig 1, F). Almost all of the skin ILCs were positive for CD69, whereas in the case of blood ILCs, the rate of CD69 positivity was approximately only 30%. In contrast to T cells, both skin and blood ILCs expressed equally high levels of other molecules presumably involved in skin homing, such as CCR4 and CCR6 (Fig 1, G and H).
In situ immunolabeling of ILCs in NHS and AD skin biopsies
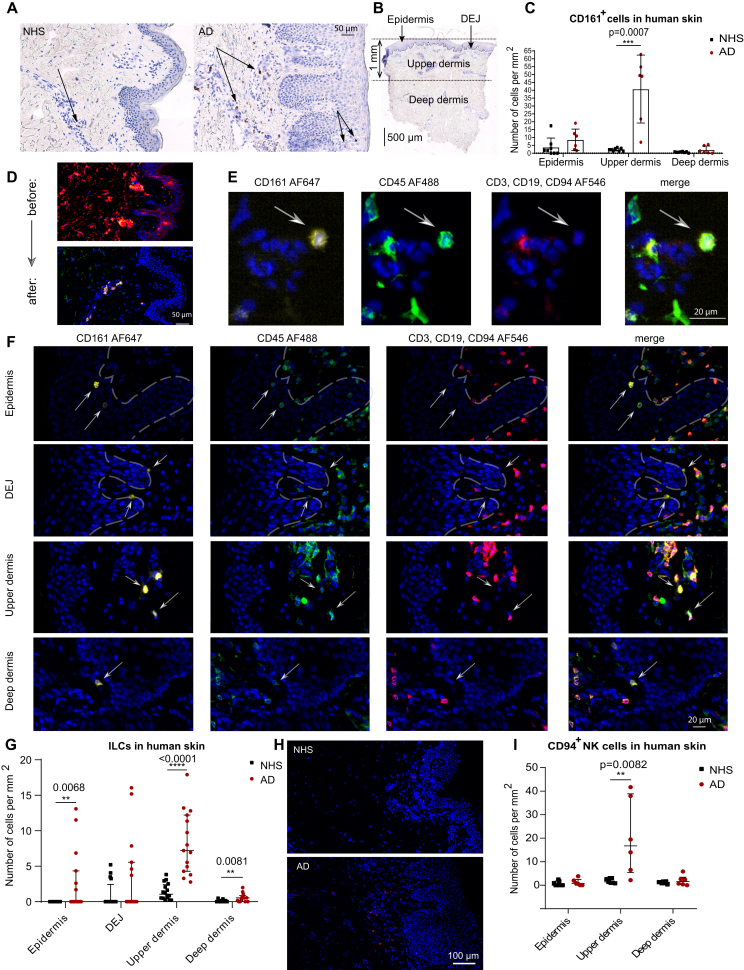
Characterizing ILCs in situ14,15,20 is challenging due to the paucity of these cells in NHS as well as to the absence of a single ILC-specific lineage marker. CD161, which is known to be present on NK cells, NKT cells, and T cells,21, 22, 23 is also expressed by human ILCs.24 This molecule is even considered to be one of the positive and uniform definition markers of total ILCs,5 provided that the co-occurrence of CD3 and CD94 has been excluded before. We therefore stained and enumerated CD161+ cells in cryosections from healthy and lesional AD skin (Fig 2, A-C) and found that AD skin contains higher numbers of CD161+ cells than the skin of healthy individuals does. These cells were primarily clustered in the upper dermis zone. To differentiate between CD161+ ILCs and non-ILCs, we performed additional immunofluorescence experiments. We found that in comparison with the isotype controls and positive stainings for CD3, CD161, and CD45 (see Fig E1, A and B in the Online Repository at www.jacionline.org), in human skin, CD161 is not coexpressed with any of the non-lymphocyte lineage markers, neither in situ nor by flow cytometry. (see Fig E1, C-E). With this in mind, we designed an immunofluorescence staining panel for the total set of ILCs that excluded T cells (CD3), B cells (CD19), and NK cells (CD94), but not the other lineage markers. This staining protocol led to a substantial reduction of the background noise, especially in the epidermis (Fig 2, D-F). Fig 2, G shows that the counts of CD161+Lin– ILCs per mm2 were higher in AD skin than in NHS, and that these cells were preferentially clustered in the upper dermis zone, often directly adjacent to the basement membrane (dermoepidermal junction). We also encountered a few ILCs in the epidermis of AD skin, but not in that of NHS. CD94+ NK cells were present at a slightly higher frequency than ILCs in AD lesional skin, and they were also located predominantly in the upper dermis zone (Fig 2, H and I). These observations are in line with our flow cytometry data (Fig 1, A and B).
Fig 2.
In situ immunolabeling of ILCs in NHS and AD skin biopsies. A, Anti-CD161 immunoperoxidase stainings of cryosections from healthy and AD skin. B, Subdivision of skin tissue sections into the following zones: epidermis, dermoepidermal junction (DEJ), upper dermis, and deep dermis. C, Summary of CD161 immunohistochemistry in NHS (n = 8) and AD skin (n = 6) sections. D, Demonstrative comparison of applying either all lineage antibodies (“before”) or reduced numbers of first-step antibodies (CD3, CD19, and CD94) (“after”). E, Immunofluorescence staining panel for total ILCs: CD161+, CD45+, and CD3–CD19–CD94–. F, Fluorescence images of cutaneous ILCs. G, Comparison of the numbers of ILCs per mm2 in various zones of sections of NHS (n = 16) and AD skin (n = 15) biopsy samples. H, Immunofluorescence labeling of CD94+ NK cells in NHS versus AD skin biopsy samples. I, Numeric comparison of NK cells between NHS (n = 7) and AD skin (n = 6). Dot plots represent medians with interquartile ranges (IQRs). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Characterization of cutaneous ILC-enriched samples by scRNA-seq
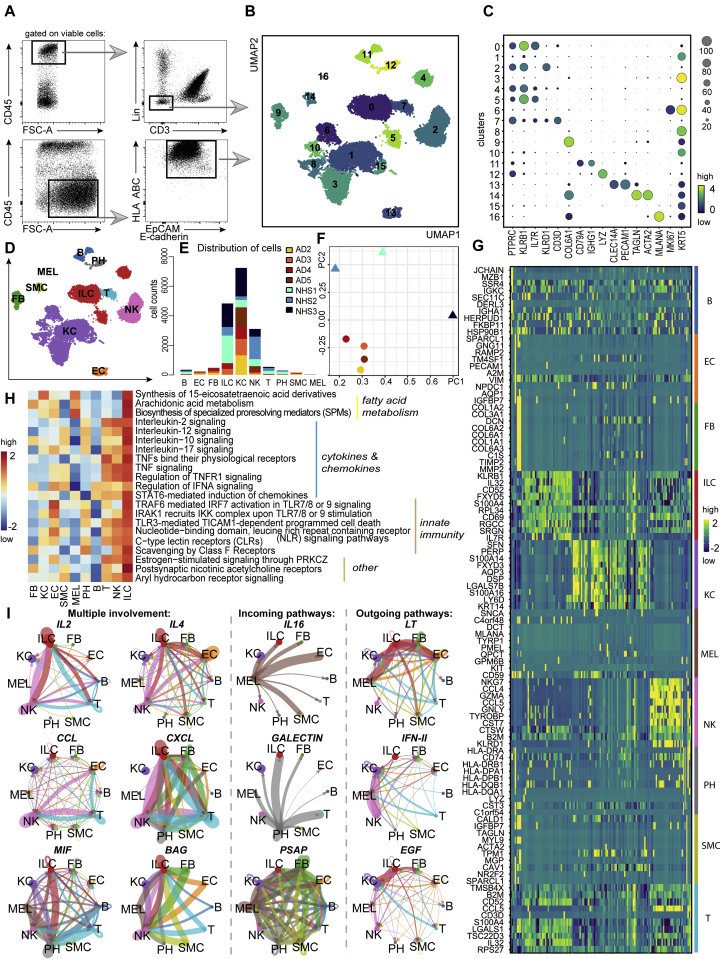
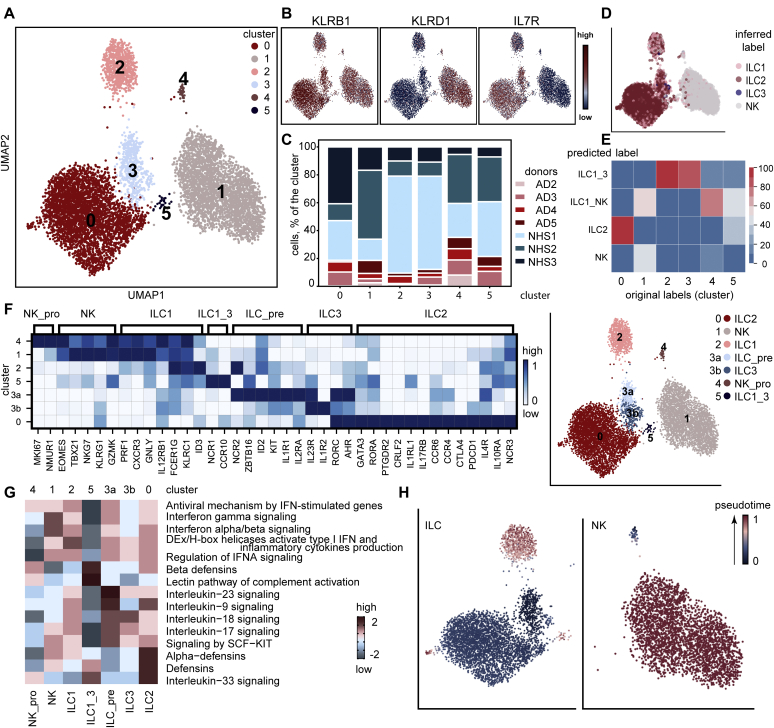
To determine the transcriptomic heterogeneity of ILCs, we performed scRNA-seq on cells from AD and healthy skin, as previously described.25,26 To maintain uniformity between the different sizes of tissue specimens (the dermatomed 10 × 10-cm2 NHS sheets were much larger than the 6-mm lesional AD skin punch biopsies) and to guarantee sufficient numbers of cells required for a sequencing run, we left the NK cells included when sorting viable CD45+Lin–CD3– cells (the lineage cocktail did not contain anti-CD94 and anti-CD16) and also supplemented samples with keratinocytes from corresponding specimens, as they belong to a histogenetically different cell lineage (Fig 3, A). In total, we profiled 3 NHS and 4 AD samples, which we integrated by using Harmony batch correction with good mixing between samples.27,28 We performed Leiden clustering (resolution 0.5) and obtained 17 clusters, which we annotated on the basis of differential gene expression (Fig 3, B): ILCs expressing PTPRC (CD45), KLRB1 (CD161), and IL7R (CD127) were located in clusters 0, 4, and 5; KLRD1 (CD94)+ NK cells were present in cluster 2; CD3D+ T cells predominated in cluster 7; KRT1+ and KRT5+ keratinocytes (KC) resided in clusters 1, 3, 6, 8, 10, and 15; a minor subset of CD79A- and IGHG1-expressing cells of B-cell lineage were found in cluster 11; LYZ+ phagocytes (PH) were present in cluster 12; CLEC14A- and PECAM1-expressing endothelial cells (EC) were located in cluster 13; TAGLN+ and ACTA2+ smooth muscle cells (SMC) were present in cluster 14; MLANA+ melanocytes (MEL) were found in cluster 16; and COL6A1+ fibroblasts (FB) were located in cluster 9 (Fig 3, C and D). Among the ILC clusters, around 83% of cells originated from NHS and about 17% came from AD. This was expected, as the differences in cell counts originate from the available sample size (more CD45+Lin–CD3– could be sorted from NHS sheets [>10 × 10-cm2] than from the two 6-mm punch biopsies from patients with AD) (Fig 3, E). Fig 3, F shows that probably because of differences in tissue processing (punch biopsies vs skin sheets), the AD skin samples but not the NHS ones cluster close together.
Fig 3.
Characterization of RNA-sequenced cutaneous ILC-enriched samples. A, Gating strategy of ILC-enriched samples sorted for scRNA-seq as viable CD45+Lin–CD3– cells (NHS, n = 3; AD skin, n = 4) and supplemented with CD45–EpCAM/E-cadherin+HLA ABC+ keratinocytes (NHS, n = 1; AD skin, n = 4) (lineage cocktail without CD94 and CD16). B, Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) plot (17,307 cells) of ILC-enriched samples sorted from NHS sheets (n = 3) and AD skin biopsies (n = 4). C, Dot plot demonstrating mean loge normalized expression of cluster-specific marker genes. Color scale indicates mean expression, and circle size indicates fraction of cells in a group. D, Cell types present in the sequenced data set (NHS, n = 3; AD skin, n = 4) from (B) and represented by using a UMAP plot. E, Numeric distribution of cells among cell types depicted in (D) and color-coded on the basis of sample origin (see Table E5 [in the Online Repository at www.jacionline.org]). F, Principal component analysis of the sequenced samples. G, Heatmap, indicating the top 10 differentially expressed genes (P < .05) between annotated cell types from (D) (see Table E3). H, Major ILC-related pathways, as determined with the ReactomeGSA package; pathway expression levels are shown as z score normalized values. I, ILC-related interaction patterns, as revealed by using the CellChat toolkit.
On the basis of differential gene expression analysis (see Table E3 in the Online Repository at www.jacionline.org), we identified the top 10 genes characteristic for each of the attributed cell types, such as KLRB1 and IL7R for ILCs; KLRD1 and GNLY for NK cells; CD3D for T cells; JCHAIN and IGKC for the B-cell lineage; HLA-DQB1, LYZ, and CD74 for phagocytes; ACTA2 and TAGLN for smooth muscle cells; PMEL, MLANA, and KIT for melanocytes; RAMP2 and PECAM1 for endothelial cells; COL1A2 and COL6A2 for fibroblasts; and KRT14 for keratinocytes (Fig 3, G).
To determine particular signaling features that are characteristic for all ILCs as a cell group, we processed the object with assigned cell types by using the R package ReactomeGSA.29 Results obtained implied that ILCs are involved in fatty acid metabolism, cytokine- and chemokine-related pathways, innate immunity, and some other events such as AHR signaling (Fig 3, H and see Table E3).
Communication pathways of skin ILCs
We then applied the CellChat algorithm to explore possible relationships between the cell types shown in Fig 3, D.30 This analysis infers communication patterns, namely, connection of cell groups on the basis of signaling pathways in the context of outgoing signaling (cells are treated as sources) or incoming signaling (cells are treated as targets). Among both incoming and outgoing communication patterns we detected IL2, IL4, CCL, CXCL, MIF, and BAG (multiple involvement) as communication pathways between ILCs and various other cell types (mainly NK cells and phagocytes [Fig 3, I and see Fig E2, A and B in the Online Repository at www.jacionline.org]). Especially pronounced were ILC incoming pathways (eg, IL16 from melanocytes and GALECTIN from phagocytes). In turn, ILCs expressed several outgoing patterns such as LT, IFN-II, and EGF pathway components that mediate communication primarily with keratinocytes and melanocytes (Fig 3, I). These data are indicative of potential interactions between keratinocytes, melanocytes, phagocytes, and NK cells on the one hand and ILCs on the other.
Molecular profiles of skin ILCs/ILC subsets and NK cells
To further characterize the molecular heterogeneity between the cells of interest, we reclustered ILCs and NK cells identified in Fig 3, D. Among the 6 newly formed clusters, numbers 1 and 4 showed a pronounced KLRD1 (CD94) signal, indicating their belonging to the NK cell family (Fig 4, A and B31,32). Clusters 0, 2, 3, and 5 expressed KLRB1 (CD161) and IL7R (CD127) and, thus, could be attributed to an ILC phenotype. As shown in Fig 4, C, each cluster contained cells from healthy donors and patients with AD.
Fig 4.
Detailed analysis of cutaneous ILC and NK cell subsets. A, Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) plot (7949 cells) representing reclustered ILCs and NK cells identified in Fig 3, D. B, Feature plots showing the distribution of KLRB1+IL7R+ ILCs and KLRD1+ NK cells. C, Bar chart representing the portion of cells originating from each donor in every cluster of cells depicted in (A). D, UMAP plot with an overlap of annotations from published data of sequenced tonsil ILC subsets.31E, Logistic regression prediction probability: overlap of annotations from sequenced skin leukocytes,32 in which ILC1_3, ILC1_NK, ILC2 and NK cell subsets were used for the overlap with current data. F, Differences between NK cell and ILC clusters from (A) (NHS, n = 3; AD skin; n = 4). Genes were manually curated; scaled expression is shown. G, Pathways, determined with the R package ReactomeGSA, that are characteristic for clusters of cutaneous ILCs and NK cell subsets; pathway expression levels are shown as z score normalized values. H, Partition–based graph abstraction (PAGA) analysis overlaid onto UMAP coordinates of ILCs (left) and NK cells (right).
To juxtapose our clusters with known ILC subsets, we used the reference data on sequenced tonsil ILC subsets (ILC1, ILC2, ILC3, and NK cells).31 NK cells overlapped nicely with clusters 1 and 4; the majority of cells in cluster 0 were attributable to the ILC2 phenotype; ILC1s overlapped with clusters 2 and 5, and ILCs from cluster 3 showed certain features of ILC3s (Fig 4, D). We also performed a comparative prediction probability analysis of cutaneous ILC/NK cell phenotypes (ILC1_NK, ILC1_3, ILC2, and NK) based on a recently published data set32 and our ILC/NK cell clusters depicted in Fig 4, A. The ILC2 subset overlapped with cluster 0; NK cells showed a slight overlap with cluster 1; the ILC1_NK phenotype corresponded to signatures of clusters 1, 4 and 5; and the ILC1_3 group correlated with phenotypes of clusters 2 and 3 (Fig 4, E). Thus, cluster 0 could be attributed to ILC2s; clusters 1 and 4 to NK cells; cluster 2 to ILC1s; cluster 3 to ILC3s; and cluster 5 to ILC1s/ILC3s (ILC1_3).
Within cluster 4 we noticed the expression of MKI67 and NMUR1 and would conclude that they represent proliferating NK cells (NK_pro); EOMES+ and TBX21+ cells from cluster 1 were assigned to NK cells (NK), and FCER1G+ and KLRC1+ cells from cluster 2 were assigned to ILC1s. Cluster 5 ILCs, which expressed NCR1 (NKp46) and CCR10, demonstrated a mixture of type 1 and type 3 markers and were, thus, attributed to the ILC1_3 subset (Fig 4, F). Since ILC3s and ILC precursors are known to share marker genes such as KIT (CD117), IL1R1, and several others,33 we subdivided cluster 3 into the subgroups 3a and 3b to detect a potential precursor population within cluster 3. We observed indeed that the subset 3a demonstrated a combination of markers ZBTB16 (PLZF), ID2, KIT, and IL1R1, which are characteristic of ILC precursors (ILC_pre) (Fig 4, F).33, 34, 35 The subset 3b was assigned to ILC3s because these cells expressed IL23R, IL1R2 as well asRORC and AHR. Cluster 0 ILCs expressing GATA3, RORA, PTGDR2 (CRTH2), CRLF2 (TSLPR), IL1RL1 (ST2), and IL17RB were assigned to ILC2s.24,31,36, 37, 38 Differentially expressed genes for the ILC and NK subsets are listed in Table E3. Interestingly, only NK cells formed a separate proliferating MKI67+ cluster (number 4), whereas only individual ILCs showed the presence of this marker, particularly in ILC1/ILC2 clusters (see Fig E3, A in the Online Repository at www.jacionline.org). NMUR1 was reported to be expressed by ILC2s,39 and we were surprised by the observation that cutaneous NK cells rather than ILCs express this marker (see Fig E3, B). To substantiate our findings, we evaluated the NMUR1 expression in cutaneous ILC/NK subgroups from a published data set32 and also observed NMUR1 on NK cells rather than in ILC clusters (see Fig E3, C).
A comparative ReactomeGSA analysis between the ILC and NK clusters revealed interferon-related pathways for NK cells and ILC1s, IL-23 as well as SCF-KIT signaling for ILC3s and ILC precursors, and involvement of ILC2s in the IL-33 signaling pathway29 (Fig 4, G and see Table E3).
To learn more about the differentiation status of ILCs in various clusters, we applied the partition-based graph abstraction (PAGA) algorithm40 to ILCs and NK cells separately. The ILC precursor (ILC_pre) cluster and proliferating NK cells (NK_pro) were selected as the “roots” in diffusion pseudotime analysis for ILCs and NK cells, respectively. On the basis of this input, cells from ILC3 and ILC2 clusters were slightly more differentiated, whereas cells from ILC1 and ILC1_3 clusters showed the highest diffusion pseudotime values (Fig 4, H). As for the NK cell trajectory, the NK cell cluster also demonstrated higher pseudotime values compared to the NK_pro cluster.
Molecular heterogeneity in ILC subsets in AD skin versus NHS
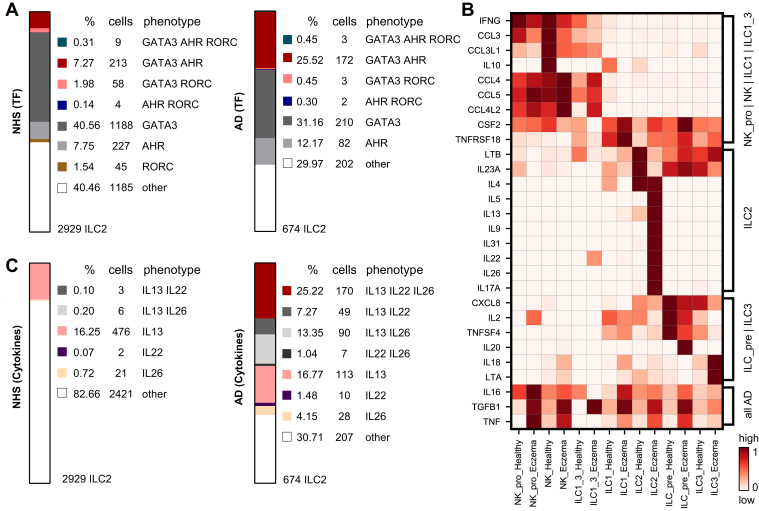
We were surprised to observe in the ILC2 cluster the coexpression of the type 2 marker GATA3, the type 3 marker RORC as well as considerable amounts of AHR, previously thought to be an indicator of ILC3s.15,41 Thus, we wondered whether the very same cells could coexpress different transcription factors (TF). We therefore calculated the percentages of GATA3, AHR, and RORC triple-, various double-, and single-positive cells in the ILC2 cluster. As can be seen in Fig 5, A, not only a sizable percentage of AD skin ILC2s (many more in donor A3 than in other patients), but also a small portion of NHS ILC2s coexpressed AHR and GATA3. We even detected a minor fraction of triple-positive cells as well as cells in which both GATA3 and RORC mRNA were found.
Fig 5.
Evidence for lineage infidelity in cutaneous ILCs in AD skin. A, Coexpression of various combinations of transcription factors GATA3, AHR, and RORC in NHS (left) and AD skin (right) ILCs from ILC2 cluster (cluster 0 in Fig 4, A). B, Chemokines and cytokines of various types detectable in ILC and NK cell clusters; scaled expression is shown. C, Coexpression of the cytokine transcripts IL13, IL22, and IL26 in either NHS (left) or AD skin (right) cells from the ILC2 cluster.
In our search of further biologic properties of skin innate lymphocytes, we looked at the expression of cytokines and chemokines from different families in ILC and NK cell clusters from AD skin and NHS donors. NK cells from healthy individuals showed higher expression of IFNG, CCL3, CCL3L1, and IL10 than did those from atopic individuals, whereas AD skin NK cells upregulated CCL4, CCL5, and CCL4L2 (Fig 5, B). NHS ILC2s showed abundant amounts of LTB, IL23A, and IL4, whereas AD skin ILC2s showed a comparatively higher expression of IL5, IL13, IL22, and IL26. These observations are in line with the previously reported potential of cell lines generated from normal skin CRTH2+ ILC2s to produce IL-13, IL-22, and IL-17A.13 AD skin ILC1s showed a slightly higher expression of CSF2 and TNFRSF18 than did NHS ILC1s; comparatively, cells in the ILC precursor cluster from NHS demonstrated a more abundant expression of CXCL8, IL2, and TNFSF4, whereas those from AD skin upregulated IL20. AD skin ILC3s exhibited higher levels of IL18 and LTA than NHS ILC3s. Cells from most of the clusters expressed higher levels of IL16, TGFB1, and TNF in AD skin as compared to NHS samples (Fig 5, B). Differentially expressed genes between AD skin and NHS samples are listed for each ILC and NK cluster in Table E4 (in the Online Repository at www.jacionline.org).
The presence of the type 2 cytokine IL13 and type 3 cytokines IL22 and IL26 made us wonder whether single cells could coexpress these markers in NHS and/or AD skin. Similar to what we observed with transcription factors in individual AD skin ILC2s, we identified cells with different cytokine gene combinations (IL13+IL22+IL26+, IL13+IL26+, and IL13+IL22+) primarily in ILC2s originating from AD skin (donor AD3 >> other patients), but almost no such cells in NHS (Fig 5, C).
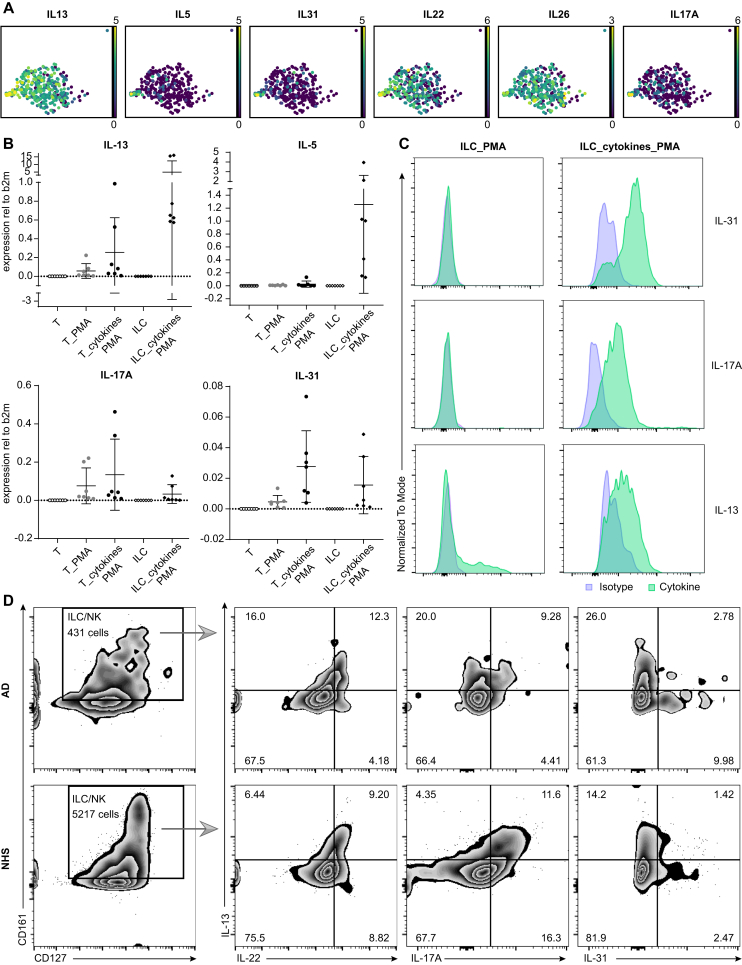
Moreover, individual ILC2s from 1 particular donor (AD3) demonstrated an even more “bizarre” combination of markers in that these cells simultaneously exhibited IL13, IL5, IL31, IL22, IL26, and IL17A (Fig 6, A). To determine whether ILCs can indeed express IL31, we sorted PBMC-derived ILCs from healthy donors and incubated them with a mixture of IL-2, IL-4, IL-7, IL-25, IL-33, and TSLP cytokines for 7 days. We then stimulated them with the Cell Activation Cocktail (Biolegend) containing phorbol 12-myristate-13-acetate (PMA), ionomycin, and brefeldin A for 4 hours and subsequently analyzed the cells by flow cytometric immunostaining and/or their RNA by RT-PCR. Fig 6, B shows that ILCs and T cells thus activated exhibited IL31 mRNA transcripts. In line with these findings, we detected cytoplasmic anti–IL-31 immunoreactivity in cultured, but not in freshly isolated ILCs stimulated with Cell Activation Cocktail (Fig 6, C).
Fig 6.
Exceptional features of ILCs. A, Individual ILC2s from patient AD3 show a mixed IL13+IL5+IL31+IL22+IL26+IL17A+ phenotype. B, Expression of cytokine transcripts (RT-PCR) in PBMC-derived ILCs and T cells (T/ILC, freshly isolated T cells/ILCs; T_PMA, freshly isolated T cells stimulated with Cell Activation Cocktail (Biolegend); T_cytokines_PMA/ILC_cytokines_PMA, T cells/ILCs expanded in culture with IL-2, IL-4, IL-7, IL-25, IL-33, and TSLP cytokine cocktail for 7 days and additionally stimulated with cell activation cocktail at day 7 for 4 hours). C, Protein expression of cytokines on either freshly isolated ILCs stimulated with a cell activation cocktail or an “ILC_cytokines_PMA” sample. D, Cell suspensions isolated from either two 6-mm AD skin punch biopsies (upper row) or NHS sheets (>10 × 10 cm2) (lower row) were incubated with cell activation cocktail containing phorbol 12-myristate-13-acetate (PMA), ionomycin, and brefeldin A for 4 hours and then immunolabeled for flow cytometry. CD45+Lin–CD3–CD127+CD161+ ILC/NK cells in NHS and AD skin coexpress IL-13 with IL-22/IL-17A/IL-31 at the protein level.
We also detected (by flow cytometry) coexpression of the conventional type 2 cytokine IL-13 with (i) the itch-inducing type 2 cytokine IL-31 (IL-13 and IL-31 single-positive cells as well as IL-13/IL-31 double-positive ILCs) and (ii) the type 3 molecules IL-17A and IL-22 in a stimulated CD45+Lin–CD3–CD127+CD161+ ILC/NK cell population from both NHS and AD skin (Fig 6, D), thus supporting our transcriptional data. It therefore appears that there are no fixed borders between the individual ILC subsets and that cutaneous ILCs represent an immunologically flexible cell population.42, 43, 44
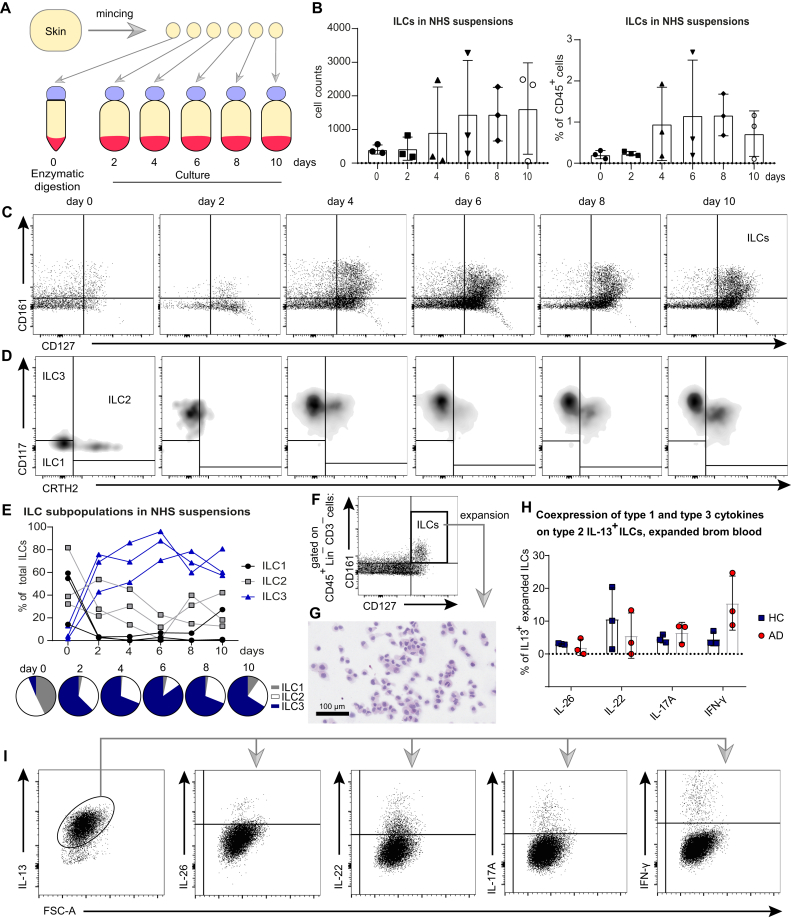
Behavior of ILCs in cultures: Evidence for molecular metamorphosis
Further support for the validity of the concept of ILC plasticity (as illustrated in Fig 5, A and C) came from a study on the behavior of ILCs in cultures of NHS in which we monitored the phenotype of cells that had been spontaneously released in the culture media over a time line of 10 days. A schematic representation of these experiments is depicted in Fig 7, A and Fig E4, A. Fig 7, B and C shows that the numbers of ILCs recovered from culture supernatants (expressed in absolute cell counts and as percentage of CD45+ cells) were increasing over time, and, more importantly, ILCs were undergoing a change in phenotype. While, as expected from what was said above, ILC2s and ILC1s were the predominant subpopulations on day 0, they soon became outnumbered by CD117+ cells (Fig 7, D and E). This transition occurred rather rapidly and did not result from an artifact stemming from CD117 epitope digestion, because a pronounced CD117+ signal was seen on Lin+ cells that had also been exposed to the very same enzyme mix (see Fig E4, B). These observations support and extend dermal crawl-out experiments on Lin–CD127+ ILCs from other investigators.13 Conceivably, this phenotypic shift may well be the consequence of a direct transformation of individual cells with a particular phenotype into those with another.
Fig 7.
Behavior of ILCs in cultures: evidence for molecular metamorphosis. A, Experimental design scheme. B, Absolute and relative numeric increase of viable CD45+Lin–CD3–CD161+CD127+ ILCs in skin explant cultures over 10 days (summary of 3 independent experiments). C, Flow cytometry plots indicating these quantitative differences. D and E, Shift in phenotype from primarily CRTH2+ ILC2s and CD117–CRTH2– ILC1s at day 0 (native skin state) toward a predominance of CD117+ ILC3s in skin explant cultures already starting from day 2 in experiments with skin explant cultures: flow cytometry plots and summary of 3 independent experiments. F, Blood ILCs, sorted as viable CD45+Lin–CD3–CD161+CD127+ and expanded in culture with a mixture of the cytokines IL-2, IL-4, IL-7, IL-33, and TSLP. G, Hematoxylin and eosin (H&E) staining of cytospinned ILCs, expanded in culture. H and I, Coexpression of type 1 and type 3 cytokines in blood ILCs, expanded in culture with type 2 inducing cytokines (IL-2, IL-4, IL-7, IL-33, and TSLP) (ie, bona fide type 2 IL13+ ILC2s): summary for 6 independent experiments (HCs, n = 3; patients with AD, n = 3) and flow cytometry plots. Dot plots represent means ± SDs, in which each dot is an individual donor.
The concept of ILC plasticity gained further support from our studies on the behavior of PB ILCs in culture. To that end, we sorted CD45+Lin–CD3–CD127+CD161+ ILCs consisting of CD117–CRTH2– ILC1s, CRTH2+ ILC2s, and CD117+ ILC3s from the PB of patients with AD and HCs and stimulated them with a type 2–promoting cytokine cocktail consisting of IL-2, IL-4, IL-7, IL-33, and TSLP (Fig 7, F). This resulted in clear signs of proliferative activity that could be maintained for several weeks. Expanded cells (Fig 7, G) consisted in their vast majority of Lin–CD3–CD161+CD127+ cells, which were almost uniformly CRTH2+ (see Fig E4, C). Interestingly enough, a proportion of these cells coexpressed CD117 (see Fig E4, D). In line with this phenotypic shift was the observation that within expanded cells from both healthy subjects and atopic donors, IL-13 was frequently coexpressed with varying levels of IFN-γ, IL-17A, IL-22, and IL-26 (Fig 7, H and I). This indicates that individual cells coexpress cytokines, which are known to belong to different subsets, not only at the mRNA levcl (as described for ILC2s in Figs 5, C and 6, A) but also at the protein level.
Discussion
Over the past decade, it became increasingly clear that the repertoire of cutaneous lymphocytes is more complex than previously appreciated. We now know that in addition to T cells and small numbers of B cells, NK cells, and NKT cells, human skin harbors a population of CD45+Lin–CD127+CD161+ cells termed ILCs. In this study, we used different methods to quantitatively and qualitatively characterize these cells, aiming for a better understanding of their biologic role under physiologic (healthy donors) and pathologic (lesional AD skin) conditions. Results obtained showed that (i) ILCs represent a minor leukocyte subset in both skin and blood, with numbers in AD skin clearly exceeding those in NHS; (ii) the vast majority of ILCs in both NHS and AD skin belong to the CRTH2+ ILC2 subset; (iii) CD117+ ILC3s represent an only minor fraction of skin ILCs, but comprise a sizable portion of blood ILCs; (iv) ILCs reside mainly in the epidermis and upper dermis, with their numbers in AD skin being much higher than those in NHS; (v) sort-enriched skin ILCs, when subjected to scRNA-seq, consist mainly of ILC2s but display different signs of activation (eg, expression of type 2-specific cytokines) in AD skin versus NHS; (vi) ILCs from AD skin frequently exhibit both type 2 and type 17/3 molecular features at the single-cell level; and (vii) a phenotypic shift, at the single-cell level, from a type 2 into a type 3/17 signature occurs in cytokine-stimulated blood ILCs and ILCs within skin explant cultures.
Confirming and extending previous investigations,15 we regularly detected small numbers of CD45+Lin–CD3–CD127+CD161+ cells (ie, bona fide ILCs5 in samples of NHS). By immunohistology, they were found to be located predominantly along either side of the dermoepidermal junction, but as opposed to what has been reported for mice,11 they could not be clearly associated with a particular skin-specific structure such as the pilosebaceous unit. According to the flow cytometry data, they comprised approximately 0.2 % of all CD45+ cells, which compares favorably with the percentage of ILCs among blood leukocytes (0.05 %). Multicolor immunolabeling revealed that in their vast majority, ILCs from NHS consisted of CRTH2+ ILC2s. CD117+ cells, by contrast, represented only a minor subset of the entire ILC population. These findings gained further support from our scRNA-seq experiments, which revealed a predominant expression of the transcription factor GATA3.
Both Villanova et al12 and Teunissen et al13 have reported on the occurrence of a cutaneous CD117+ ILC3 population that is devoid of natural cytotoxicity receptors (NCRs), including NKp44. Supposedly, when allowed to “crawl out” from dermal explant cultures, these cells do acquire NCR2/NKp44.13 We have also observed a phenotypic switch of ILCs in skin explant cultures, but have reasons to believe that CD117+ cells emerging under these conditions are derived from either CRTH2+ ILC2s or CD117– ILCs/putative ILC precursors. First, the phenotypic shift happened very rapidly and the numbers of viable cells recovered during the first 2 days of ex vivo culture were comparable. This excludes the possibility that the transition from ILC2 to ILC3 or ILC3-like cells has occurred because of a massive demise of one subset and replacement and/or overgrowth by another. Second, we have performed enzymatic digestion experiments showing that critical ILC3 moieties such as CD117 are not adversely affected by proteases and/or nucleases. The subcellular/molecular events responsible for the phenotypic transformation escape us at the present time, but they may be related to an altered microenvironment in skin explant cultures.
Issues that urgently need clarification relate to the derivation and function of skin ILCs under both physiologic and pathologic conditions. Regarding the former question, it has been reported that nonlymphoid tissues such as the skin are populated by ill-defined ILC precursors within the fetal liver and perhaps within the yolk sac.45 Following this reasoning, one would postulate that the microenvironment of the respective tissues would skew such precursors in one or the other direction. Within the skin, molecules such as IL-33, TSLP, and IL-25, and perhaps others, are probably responsible for the shift toward ILC2s46; by contrast, in the gut the predominance of ILC3s may be explained by factors such as IL-23 and IL-1β.47,48 Alternatively, one could also argue that certain ILCs and/or ILC precursors in the PB have the capacity to selectively home to peripheral tissues.33 Our finding of an inverse correlation between the numbers of ILC2s in the skin and in the PB as well as the expression of skin-homing molecules (CLA, CCR4, and CCR6) lends further support to this concept, but does not definitively prove it.
As far as the functional role of ILCs in NHS is concerned, our results obtained by scRNA-seq do suggest an important role of these cells in the maintenance of skin homeostasis,49 as exemplified by the expression of genes related to the circadian clock (NR1D1 and PER1) and receptors for neurotransmitters (ADRB2, CHRNA7, and VIPR1)50,51 (see Table E4). It remains to be seen whether ILC-dependent host defense mechanisms in experimental animals such as the regulation of the equilibrium of commensal bacteria11 or the initiation of wound healing52 are also operative in humans and, if so, which cellular and subcellular events are underlying these processes. In this context, it has recently been reported that the capacity of gut ILC3s to maintain intestinal homeostasis is controlled by the circadian machinery53 and by the feeding behavior of the animals.54
The situation in AD skin appears to be quite different. Although we found little, if any, evidence for the expression of proinflammatory molecules in the ILC2s of NHS, we observed a substantial upregulation of cytokine genes typically associated with type 2 inflammation (IL13, IL5, IL31, and CSF2) in ILCs of AD skin. The roles of IL-25, IL-33, TSLP, and PGD2 in this process are well established,6, 7, 8,55 but others that could trigger similar events may exist.
Also very noteworthy is our additional finding of coexpression of varying amounts of cytokine transcripts characteristic of type 17 and/or type 3 immunity (ie, IL17A, IL22, and IL26) within a given ILC2. This phenomenon was frequently present in AD skin ILC2s, most notably in donor AD3, but it was only rarely detected in the ILC2s of NHS. Conversely, ILC1 and ILC3 from AD skin were almost completely devoid of “foreign” subset transcripts. Together with our observations regarding crawl-out cells from ex vivo skin explant cultures and in vitro–stimulated ILCs from PB, this points to the occurrence of ILC “unfaithfulness” under pathologic conditions.26,56, 57, 58, 59, 60 It will be important to determine whether such a mixed marker constellation defines a particular disease subset. In this context, it should be remembered that the bulk analysis of AD skin biopsies at the mRNA and protein levels, although regularly showing the signs of type 2 inflammation, also revealed the presence of other cytokine gene signatures under certain conditions.61 IL-17A, for example, was detected in skin biopsies from small children, IFN-γ was present in chronic lesions, and IL-22 was observed throughout the different stages of the disease.62,63
It will be most interesting to determine, in a larger number of AD skin biopsy samples, not only the nature of the cellular and molecular signals leading to this diversity but also the cellular origin (ILCs vs T cells) of these transcripts. In fact, evidence exists that in certain animal models of AD10 and psoriasis64 innate circuits play a more important pathogenic role than classical T-cell immunity. Methodologies such as those used in this and many other studies now allow for similar investigations of other human skin diseases, hopefully resulting in an increasingly refined Human Cell Atlas and Human Skinatlas.65,66 Most importantly, these new technologies have taught us that the mechanisms operative in skin physiology and pathology are flexible processes and highly diverse between different persons and that a successful interference with these will require individualized approaches.
Clinical implications.
The phenotypic and molecular profile of ILCs in lesional AD skin makes them candidate players in the pathogenesis of this disease.
Acknowledgments
We thank Professor Wolfgang Weninger for continued support. We are also thankful to the teams of the core facilities Flow Cytometry and Imaging of the Medical University of Vienna as well as to the Biomedical Sequencing Facility at Center for Molecular Medicine of the Austrian Academy of Sciences for assistance with next-generation sequencing.
Footnotes
Supported by grants from the Austrian Science Fund (FWF; DK W 1248-B30) and the Medical University of Vienna, as well as by grant IF_2017_29 from the innovation fund of the Austrian Academy of Sciences (to G.S.) and, in part, by grant 18130 from the Medical Scientific Fund of the Mayor of the City of Vienna (to G.S.).
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Supplementary data
References
- 1.Nihal M., Mikkola D., Wood G.S. Detection of clonally restricted immunoglobulin heavy chain gene rearrangements in normal and lesional skin: analysis of the B cell component of the skin-associated lymphoid tissue and implications for the molecular diagnosis of cutaneous B cell lymphomas. J Mol Diagn. 2000;2:5–10. doi: 10.1016/S1525-1578(10)60609-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ebert L.M., Meuter S., Moser B. Homing and function of human skin gammadelta T cells and NK cells: relevance for tumor surveillance. J Immunol. 2006;176:4331–4336. doi: 10.4049/jimmunol.176.7.4331. [DOI] [PubMed] [Google Scholar]
- 3.Gober M.D., Fishelevich R., Zhao Y., Unutmaz D., Gaspari A.A. Human natural killer T cells infiltrate into the skin at elicitation sites of allergic contact dermatitis. J Invest Dermatol. 2008;128:1460–1469. doi: 10.1038/sj.jid.5701199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mjosberg J.M., Trifari S., Crellin N.K., Peters C.P., van Drunen C.M., Piet B., et al. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol. 2011;12:1055–1062. doi: 10.1038/ni.2104. [DOI] [PubMed] [Google Scholar]
- 5.Hazenberg M.D., Spits H. Human innate lymphoid cells. Blood. 2014;124:700–709. doi: 10.1182/blood-2013-11-427781. [DOI] [PubMed] [Google Scholar]
- 6.Salimi M., Barlow J.L., Saunders S.P., Xue L., Gutowska-Owsiak D., Wang X., et al. A role for IL-25 and IL-33-driven type-2 innate lymphoid cells in atopic dermatitis. J Exp Med. 2013;210:2939–2950. doi: 10.1084/jem.20130351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imai Y., Yasuda K., Sakaguchi Y., Haneda T., Mizutani H., Yoshimoto T., et al. Skin-specific expression of IL-33 activates group 2 innate lymphoid cells and elicits atopic dermatitis-like inflammation in mice. Proc Natl Acad Sci U S A. 2013;110:13921–13926. doi: 10.1073/pnas.1307321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim B.S., Siracusa M.C., Saenz S.A., Noti M., Monticelli L.A., Sonnenberg G.F., et al. TSLP elicits IL-33-independent innate lymphoid cell responses to promote skin inflammation. Sci Transl Med. 2013;5 [Google Scholar]
- 9.Roediger B., Kyle R., Yip K.H., Sumaria N., Guy T.V., Kim B.S., et al. Cutaneous immunosurveillance and regulation of inflammation by group 2 innate lymphoid cells. Nat Immunol. 2013;14:564–573. doi: 10.1038/ni.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imai Y., Yasuda K., Nagai M., Kusakabe M., Kubo M., Nakanishi K., et al. IL-33-induced atopic dermatitis-like inflammation in mice is mediated by group 2 innate lymphoid cells in concert with basophils. J Invest Dermatol. 2019;139:2185–2194.e3. doi: 10.1016/j.jid.2019.04.016. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi T., Voisin B., Kim D.Y., Kennedy E.A., Jo J.H., Shih H.Y., et al. Homeostatic control of sebaceous glands by innate lymphoid cells regulates commensal bacteria equilibrium. Cell. 2019;176:982–997.e16. doi: 10.1016/j.cell.2018.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villanova F., Flutter B., Tosi I., Grys K., Sreeneebus H., Perera G.K., et al. Characterization of innate lymphoid cells in human skin and blood demonstrates increase of NKp44+ ILC3 in psoriasis. J Invest Dermatol. 2014;134:984–991. doi: 10.1038/jid.2013.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teunissen M.B.M., Munneke J.M., Bernink J.H., Spuls P.I., Res P.C.M., Te Velde A., et al. Composition of innate lymphoid cell subsets in the human skin: enrichment of NCR(+) ILC3 in lesional skin and blood of psoriasis patients. J Invest Dermatol. 2014;134:2351–2360. doi: 10.1038/jid.2014.146. [DOI] [PubMed] [Google Scholar]
- 14.Kim B.S., Wang K., Siracusa M.C., Saenz S.A., Brestoff J.R., Monticelli L.A., et al. Basophils promote innate lymphoid cell responses in inflamed skin. J Immunol. 2014;193:3717–3725. doi: 10.4049/jimmunol.1401307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruggen M.C., Bauer W.M., Reininger B., Clim E., Captarencu C., Steiner G.E., et al. In situ mapping of innate lymphoid cells in human skin: evidence for remarkable differences between normal and inflamed skin. J Invest Dermatol. 2016;136:2396–2405. doi: 10.1016/j.jid.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 16.Rendeiro A.F., Krausgruber T., Fortelny N., Zhao F., Penz T., Farlik M., et al. Chromatin mapping and single-cell immune profiling define the temporal dynamics of ibrutinib response in CLL. Nat Commun. 2020;11:577. doi: 10.1038/s41467-019-14081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferran M., Santamaria-Babi L.F. Pathological mechanisms of skin homing T cells in atopic dermatitis. World Allergy Organ J. 2010;3:44–47. doi: 10.1097/WOX.0b013e3181d675f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tubo N.J., McLachlan J.B., Campbell J.J. Chemokine receptor requirements for epidermal T-cell trafficking. Am J Pathol. 2011;178:2496–2503. doi: 10.1016/j.ajpath.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szabo P.A., Miron M., Farber D.L. Location, location, location: tissue resident memory T cells in mice and humans. Sci Immunol. 2019;4 [Google Scholar]
- 20.de Boer O.J., Krebbers G., Mackaaij C., Florquin S., de Rie M.A., van der Wal A.C., et al. Comparison of two different immunohistochemical quadruple staining approaches to identify innate lymphoid cells in formalin-fixed paraffin-embedded human tissue. J Histochem Cytochem. 2020;68:127–138. doi: 10.1369/0022155419897257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi T., Dejbakhsh-Jones S., Strober S. Expression of CD161 (NKR-P1A) defines subsets of human CD4 and CD8 T cells with different functional activities. J Immunol. 2006;176:211–216. doi: 10.4049/jimmunol.176.1.211. [DOI] [PubMed] [Google Scholar]
- 22.Rai A.K., Thakur C.P., Seth T., Mitra D.K. Early activated Th-1 type and dominantly diverse natural killer T (CD3(+)CD161(+)Valpha24(-)) cells in bone marrow among visceral leishmaniasis patients. Int J Parasitol. 2011;41:1069–1077. doi: 10.1016/j.ijpara.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 23.Kurioka A., Cosgrove C., Simoni Y., van Wilgenburg B., Geremia A., Bjorkander S., et al. CD161 defines a functionally distinct subset of pro-inflammatory natural killer cells. Front Immunol. 2018;9:486. doi: 10.3389/fimmu.2018.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mjosberg J., Spits H. Human innate lymphoid cells. J Allergy Clin Immunol. 2016;138:1265–1276. doi: 10.1016/j.jaci.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 25.Zheng G.X., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rojahn T.B., Vorstandlechner V., Krausgruber T., Bauer W.M., Alkon N., Bangert C., et al. Single-cell transcriptomics combined with interstitial fluid proteomics defines cell type-specific immune regulation in atopic dermatitis. J Allergy Clin Immunol. 2020;146:1056–1069. doi: 10.1016/j.jaci.2020.03.041. [DOI] [PubMed] [Google Scholar]
- 27.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griss J., Viteri G., Sidiropoulos K., Nguyen V., Fabregat A., Hermjakob H. ReactomeGSA - efficient multi-omics comparative pathway analysis. Mol Cell Proteomics. 2020 mcp.TIR120.002155. [Google Scholar]
- 30.Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.H., et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bjorklund A.K., Forkel M., Picelli S., Konya V., Theorell J., Friberg D., et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat Immunol. 2016;17:451–460. doi: 10.1038/ni.3368. [DOI] [PubMed] [Google Scholar]
- 32.Reynolds G., Vegh P., Fletcher J., Poyner E.F.M., Stephenson E., Goh I., et al. Developmental cell programs are co-opted in inflammatory skin disease [abstract] Science. 2021;371 [Google Scholar]
- 33.Lim A.I., Li Y., Lopez-Lastra S., Stadhouders R., Paul F., Casrouge A., et al. Systemic human ILC precursors provide a substrate for tissue ILC differentiation. Cell. 2017;168:1086–1100.e10. doi: 10.1016/j.cell.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 34.Hams E., Bermingham R., Fallon P.G. Macrophage and innate lymphoid cell interplay in the genesis of fibrosis. Front Immunol. 2015;6:597. doi: 10.3389/fimmu.2015.00597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vacca P., Chiossone L., Mingari M.C., Moretta L. Heterogeneity of NK cells and other innate lymphoid cells in human and murine decidua. Front Immunol. 2019;10:170. doi: 10.3389/fimmu.2019.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roediger B., Kyle R., Tay S.S., Mitchell A.J., Bolton H.A., Guy T.V., et al. IL-2 is a critical regulator of group 2 innate lymphoid cell function during pulmonary inflammation. J Allergy Clin Immunol. 2015;136:1653–1663.e7. doi: 10.1016/j.jaci.2015.03.043. [DOI] [PubMed] [Google Scholar]
- 37.Salimi M., Xue L., Jolin H., Hardman C., Cousins D.J., McKenzie A.N., et al. Group 2 innate lymphoid cells express functional NKp30 receptor inducing type 2 cytokine production. J Immunol. 2016;196:45–54. doi: 10.4049/jimmunol.1501102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Moltke J., O'Leary C.E., Barrett N.A., Kanaoka Y., Austen K.F., Locksley R.M. Leukotrienes provide an NFAT-dependent signal that synergizes with IL-33 to activate ILC2s. J Exp Med. 2017;214:27–37. doi: 10.1084/jem.20161274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cardoso V., Chesne J., Ribeiro H., Garcia-Cassani B., Carvalho T., Bouchery T., et al. Neuronal regulation of type 2 innate lymphoid cells via neuromedin U. Nature. 2017;549:277–281. doi: 10.1038/nature23469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolf F.A., Hamey F.K., Plass M., Solana J., Dahlin J.S., Gottgens B., et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 2019;20:59. doi: 10.1186/s13059-019-1663-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J., Doty A., Glover S.C. Aryl hydrocarbon receptor signaling involves in the human intestinal ILC3/ILC1 conversion in the inflamed terminal ileum of Crohn's disease patients. Inflamm Cell Signal. 2016;3:e1404. doi: 10.14800/ics.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Golebski K., Ros X.R., Nagasawa M., van Tol S., Heesters B.A., Aglmous H., et al. IL-1beta, IL-23, and TGF-beta drive plasticity of human ILC2s towards IL-17-producing ILCs in nasal inflammation. Nat Commun. 2019;10:2162. doi: 10.1038/s41467-019-09883-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagasawa M., Heesters B.A., Kradolfer C.M.A., Krabbendam L., Martinez-Gonzalez I., de Bruijn M.J.W., et al. KLRG1 and NKp46 discriminate subpopulations of human CD117(+)CRTH2(-) ILCs biased toward ILC2 or ILC3. J Exp Med. 2019;216:1762–1776. doi: 10.1084/jem.20190490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lanier L.L. Plastic fantastic innate lymphoid cells. J Exp Med. 2019;216:1726–1727. doi: 10.1084/jem.20191183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Popescu D.M., Botting R.A., Stephenson E., Green K., Webb S., Jardine L., et al. Decoding human fetal liver haematopoiesis. Nature. 2019;574:365–371. doi: 10.1038/s41586-019-1652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Camelo A., Rosignoli G., Ohne Y., Stewart R.A., Overed-Sayer C., Sleeman M.A., et al. IL-33, IL-25, and TSLP induce a distinct phenotypic and activation profile in human type 2 innate lymphoid cells. Blood Adv. 2017;1:577–589. doi: 10.1182/bloodadvances.2016002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bernink J.H., Krabbendam L., Germar K., de Jong E., Gronke K., Kofoed-Nielsen M., et al. Interleukin-12 and -23 control plasticity of CD127(+) group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity. 2015;43:146–160. doi: 10.1016/j.immuni.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 48.Forkel M., Mjosberg J. Dysregulation of group 3 innate lymphoid cells in the pathogenesis of inflammatory bowel disease. Curr Allergy Asthma Rep. 2016;16:73. doi: 10.1007/s11882-016-0652-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kobayashi T., Ricardo-Gonzalez R.R., Moro K. Skin-resident innate lymphoid cells - cutaneous innate guardians and regulators. Trends Immunol. 2020;41:100–112. doi: 10.1016/j.it.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarker G., Larabee C.M., Domingos A.I. ILC3s gut rhythm. Nat Immunol. 2020;21:106–108. doi: 10.1038/s41590-019-0580-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klose C.S.N., Artis D. Innate lymphoid cells control signaling circuits to regulate tissue-specific immunity. Cell Res. 2020;30:475–491. doi: 10.1038/s41422-020-0323-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rak G.D., Osborne L.C., Siracusa M.C., Kim B.S., Wang K., Bayat A., et al. IL-33-dependent group 2 innate lymphoid cells promote cutaneous wound healing. J Invest Dermatol. 2016;136:487–496. doi: 10.1038/JID.2015.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Godinho-Silva C., Domingues R.G., Rendas M., Raposo B., Ribeiro H., da Silva J.A., et al. Light-entrained and brain-tuned circadian circuits regulate ILC3s and gut homeostasis. Nature. 2019;574:254–258. doi: 10.1038/s41586-019-1579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seillet C., Luong K., Tellier J., Jacquelot N., Shen R.D., Hickey P., et al. The neuropeptide VIP confers anticipatory mucosal immunity by regulating ILC3 activity. Nat Immunol. 2020;21:168–177. doi: 10.1038/s41590-019-0567-y. [DOI] [PubMed] [Google Scholar]
- 55.Xue L., Salimi M., Panse I., Mjosberg J.M., McKenzie A.N., Spits H., et al. Prostaglandin D2 activates group 2 innate lymphoid cells through chemoattractant receptor-homologous molecule expressed on TH2 cells. J Allergy Clin Immunol. 2014;133:1184–1194. doi: 10.1016/j.jaci.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liang S.C., Tan X.Y., Luxenberg D.P., Karim R., Dunussi-Joannopoulos K., Collins M., et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dambacher J., Beigel F., Zitzmann K., De Toni E.N., Goke B., Diepolder H.M., et al. The role of the novel Th17 cytokine IL-26 in intestinal inflammation. Gut. 2009;58:1207–1217. doi: 10.1136/gut.2007.130112. [DOI] [PubMed] [Google Scholar]
- 58.Fabre T., Molina M.F., Soucy G., Goulet J.P., Willems B., Villeneuve J.P., et al. Type 3 cytokines IL-17A and IL-22 drive TGF-beta-dependent liver fibrosis. Sci Immunol. 2018;3 [Google Scholar]
- 59.Bernink J.H., Ohne Y., Teunissen M.B.M., Wang J., Wu J., Krabbendam L., et al. c-Kit-positive ILC2s exhibit an ILC3-like signature that may contribute to IL-17-mediated pathologies. Nat Immunol. 2019;20:992–1003. doi: 10.1038/s41590-019-0423-0. [DOI] [PubMed] [Google Scholar]
- 60.Nomura T., Wu J., Kabashima K., Guttman-Yassky E. Endophenotypic variations of atopic dermatitis by age, race, and ethnicity. J Allergy Clin Immunol Pract. 2020;8:1840–1852. doi: 10.1016/j.jaip.2020.02.022. [DOI] [PubMed] [Google Scholar]
- 61.Nograles K.E., Zaba L.C., Shemer A., Fuentes-Duculan J., Cardinale I., Kikuchi T., et al. IL-22-producing "T22" T cells account for upregulated IL-22 in atopic dermatitis despite reduced IL-17-producing TH17 T cells. J Allergy Clin Immunol. 2009;123:1244–1252.e2. doi: 10.1016/j.jaci.2009.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Werfel T., Morita A., Grewe M., Renz H., Wahn U., Krutmann J., et al. Allergen specificity of skin-infiltrating T cells is not restricted to a type-2 cytokine pattern in chronic skin lesions of atopic dermatitis. J Invest Dermatol. 1996;107:871–876. doi: 10.1111/1523-1747.ep12331164. [DOI] [PubMed] [Google Scholar]
- 63.Brunner P.M., He H., Pavel A.B., Czarnowicki T., Lefferdink R., Erickson T., et al. The blood proteomic signature of early-onset pediatric atopic dermatitis shows systemic inflammation and is distinct from adult long-standing disease. J Am Acad Dermatol. 2019;81:510–519. doi: 10.1016/j.jaad.2019.04.036. [DOI] [PubMed] [Google Scholar]
- 64.Zenz R., Eferl R., Kenner L., Florin L., Hummerich L., Mehic D., et al. Psoriasis-like skin disease and arthritis caused by inducible epidermal deletion of Jun proteins. Nature. 2005;437:369–375. doi: 10.1038/nature03963. [DOI] [PubMed] [Google Scholar]
- 65.Villani A.C., Satija R., Reynolds G., Sarkizova S., Shekhar K., Fletcher J., et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356 [Google Scholar]
- 66.Dyring-Andersen B., Lovendorf M.B., Coscia F., Santos A., Moller L.B.P., Colaco A.R., et al. Spatially and cell-type resolved quantitative proteomic atlas of healthy human skin. Nat Commun. 2020;11:5587. doi: 10.1038/s41467-020-19383-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.