Abstract
Epithelial organoids are already demonstrating their transformative impact in basic and translational research. However, formed through uncontrolled self-organization, they are heterogeneous and irreproducible, which limits their use in the lab and clinic. We describe methodologies for spatially and temporally controlling organoid formation, rendering a stochastic process more deterministic. Bioengineered stem cell microenvironments are used to specify the initial geometry of intestinal organoids, which in turn controls their patterning and crypt formation. We leveraged the reproducibility and predictability of the culture to identify the underlying mechanisms of epithelial patterning, which may contribute to reinforcing intestinal regionalization in vivo. We thus introduce next-generation, controlled organoid culture, and demonstrate how it can be used to answer questions not readily addressable with standard organoid and mouse models.
One Sentence Summary:
Synthetic stem cell niches promote highly stereotyped organoid development.
Stem cell-derived organoids are in vitro tissue and organ mimetics that hold promise as models of human organ development and disease, platforms for drug discovery and design of personalized therapies, and as a means to repair diseased and damaged tissue in the clinic (1–7). The self-organization processes through which organoids are generated can be credited for the previously unmatched architectural and functional sophistication of these structures. However, these processes are simultaneously responsible for introducing variability, and a general lack of reproducibility and control in organoid formation. For instance, intestinal organoid formation is largely stochastic and the resulting structures differ from the native organ in multiple aspects. Notably, the location and number of crypt-like domains cannot be controlled, and neither can the shape, size and cellular composition of the overall organoid. Indeed, the high variability has been listed as one of the principal limitations of current organoid models, as it introduces high levels of unpredictability, thus posing a significant challenge in basic and translational organoid-based research (8, 9). Although the field of stem cell-based organoids was conceived at least a decade ago and the range of organoid types is continuously expanding, it is only very recently that researchers have introduced methods to control organoid formation (10–15). Specifically, we have used microfabrication and microfluidics to control the macroscopic shape of intestinal organoids, which ultimately enabled the establishment of long-lived and perfused structures (13). This approach used fabrication to form the crypt-villus system, rather than relying on intrinsic morphogenetic programs, such as evagination and budding, which drive both organoid formation and intestinal development in vivo (16). Furthermore, the mechanisms whereby the macroscopic organoid shape can ultimately pattern the crypt-villus system were not elucidated. In this study, we set out to devise strategies for exerting extrinsic control over intestinal organoid symmetry breaking and crypt formation for the first time, and used these models to shed light on the mechanisms by which tissue geometry can regulate intestinal morphogenesis.
We have previously shown that the transformation of a round intestinal stem cell (ISC) colony into a crypt-containing organoid within a synthetic hydrogel requires matrix softening (17). The global matrix softening approach that we employed previously, however, resulted in stochastic and spatially uncontrolled budding, just like in the conventional organoid cultures based on native extracellular matrix (ECM)-derived 3D matrices (18, 19). We postulated that by introducing localized matrix softening, thus restricting the regions permissive to budding, we may achieve spatially controlled crypt formation.
To this end, we embedded ISC colonies within RGD- and laminin-1-containing photosensitive poly(ethylene glycol) (PEG)-based hydrogels (20), which undergo degradation and softening when exposed to 405 nm light (Fig. S1). Localized light exposure allowed us to introduce softening at pre-defined regions within the hydrogel surrounding the colony (Fig. 1A). Importantly, the initial stiffness of the hydrogel matched the value which we previously found to support ISC colony formation but not budding (17) (Fig. S1), whereas the light dose supplied to specified regions was chosen to effect a drop of stiffness previously identified as crucial for organoid budding (17) (Fig. 1B). Shortly (<10 min) after photo-patterning, the epithelium adjacent to the softened regions underwent an evagination-like event (Supp. Movie 1), which we believe to be an attempt to establish mechanical balance. These pseudo-buds continued to extend over the next 72 h, forming structures that morphologically resembled intestinal crypts (Fig. 1C–E). Whereas crypt-like buds were frequent within softened regions, they were completely absent outside of these regions. Thus, we were able to control and predict the sites of bud formation with high (84±6%) fidelity (Fig. 1F).
Figure 1: Spatiotemporal control over organoid crypt formation through photopatterning.
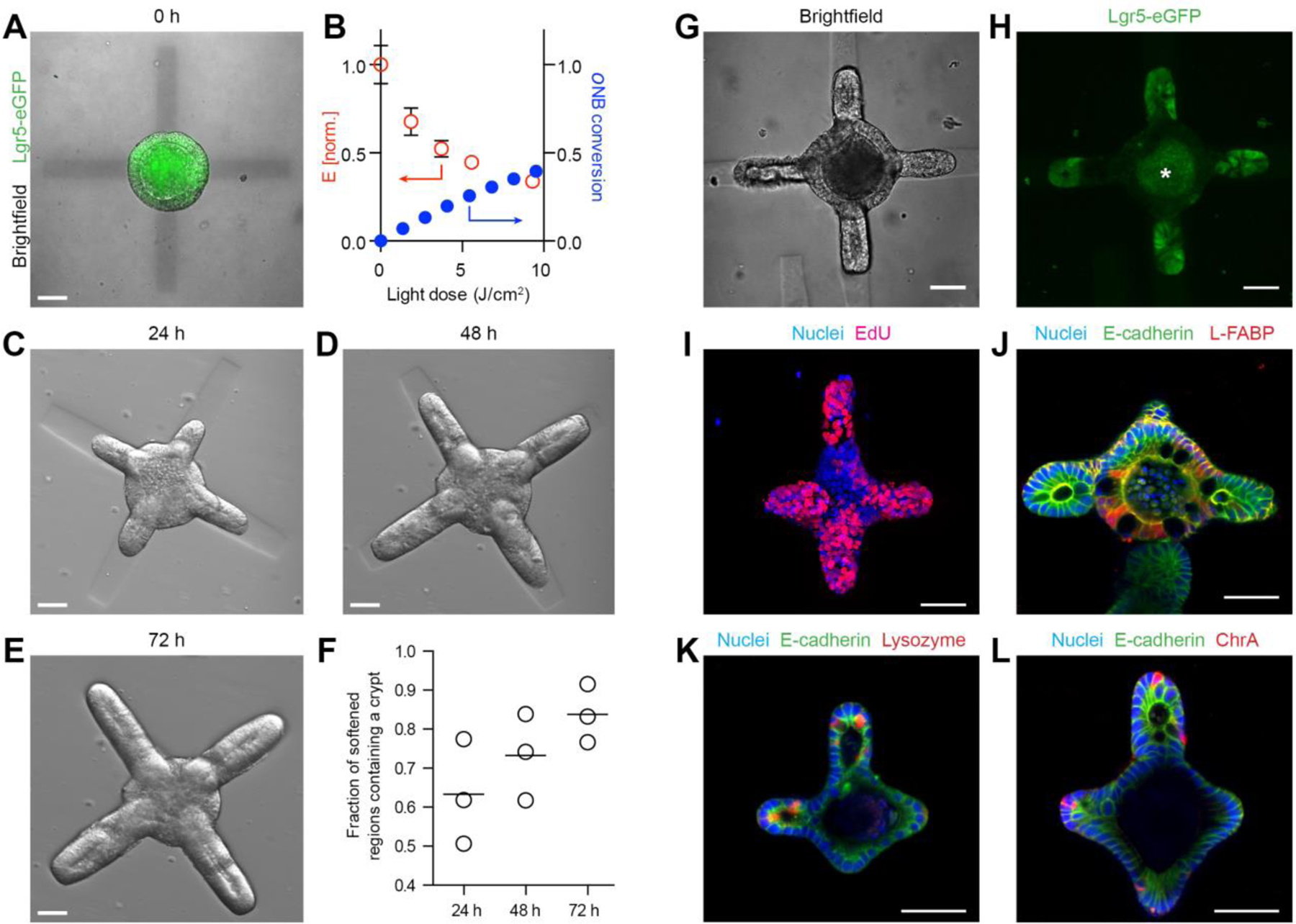
(A) Composite image showing Lgr5-GFP expression in a symmetric colony and photopattern visible with transmitted 405 nm light immediately after spatially restricted light exposure. (B) Mechanical characterization of gel regions with atomic force microscopy corresponds to conversion of photocleavable moieties within the gel. (C-E) Spatially defined crypt formation within photopatterned gels 24 h (C), 48 h (D) and 72 h (E) after light-induced softening. (F) Quantification of fraction of photo-softened gel regions containing a crypt. Individual data points and mean are shown. Lgr5-eGFP expression (G, H) and proliferation (I) are localized within the buds extending into the softened regions. Enterocytes are found in the central regions (J) of the organoids Paneth cells are restricted to the buds (K) of the organoids. Enteroendocrine cells (L) are also present. Scale bars, 30 µm.
To ensure that the buds were bona fide crypts formed through epithelial symmetry breaking and patterning, rather than merely by differential growth, we considered the distribution of ISCs and differentiated intestinal cells throughout the structure. Lgr5-expressing ISCs were present exclusively at the end of the buds, and absent in the central epithelial cyst (Fig. 1G,H). Cell division, indicated by incorporation of 5-ethynyl-2’-deoxyuridine (EdU) was similarly localized to crypt structures (Fig. 1I). Likewise, ISC-supporting Paneth cells were present within the buds, whereas enterocytes were confined to the central region of the cyst (Fig. 1J,K). Enteroendocrine cells were also found within the structures (Fig. 1L). Thus, we used light-mediated matrix softening to control organoid symmetry breaking and direct crypt formation. Of note, the timing of matrix softening is important, since crypt formation is substantially reduced when softening is performed two days after the induction of differentiation (Fig. S2).
In applying photo-mediated softening to control intestinal organoid formation, we noticed that the symmetry breaking and epithelial patterning were preceded by a change in epithelial shape (Supp. Movie 1). Specifically, following the rapid evagination-like event that produced nascent buds, the colonies were uniformly composed of ISCs. The budded structure was patterned and transformed into an organoid in the following 24 h. Bearing in mind that the appearance of the bud preceded the molecular symmetry breaking, we postulated that the bud shape of the crypt epithelium itself represents an integral part of the ISC niche, helping to restrict the ISC zone and establish the crypt-villus axis. Indeed, our recent work demonstrated that ISCs grown in crypt-shaped cavities within microfluidic scaffolds can be maintained in long-term cultures (13), suggesting that tissue shape directly influences patterning. To test this hypothesis, we set out to build intestinal tissues of pre-defined size and geometry that mimic those of the crypt, and monitor how the initial shape affects the spatial distributions of ISCs and the various differentiated cell types within the tissue (Fig. 2).
Figure 2: Geometrically controlled symmetry-breaking and epithelial patterning within intestinal organoids.
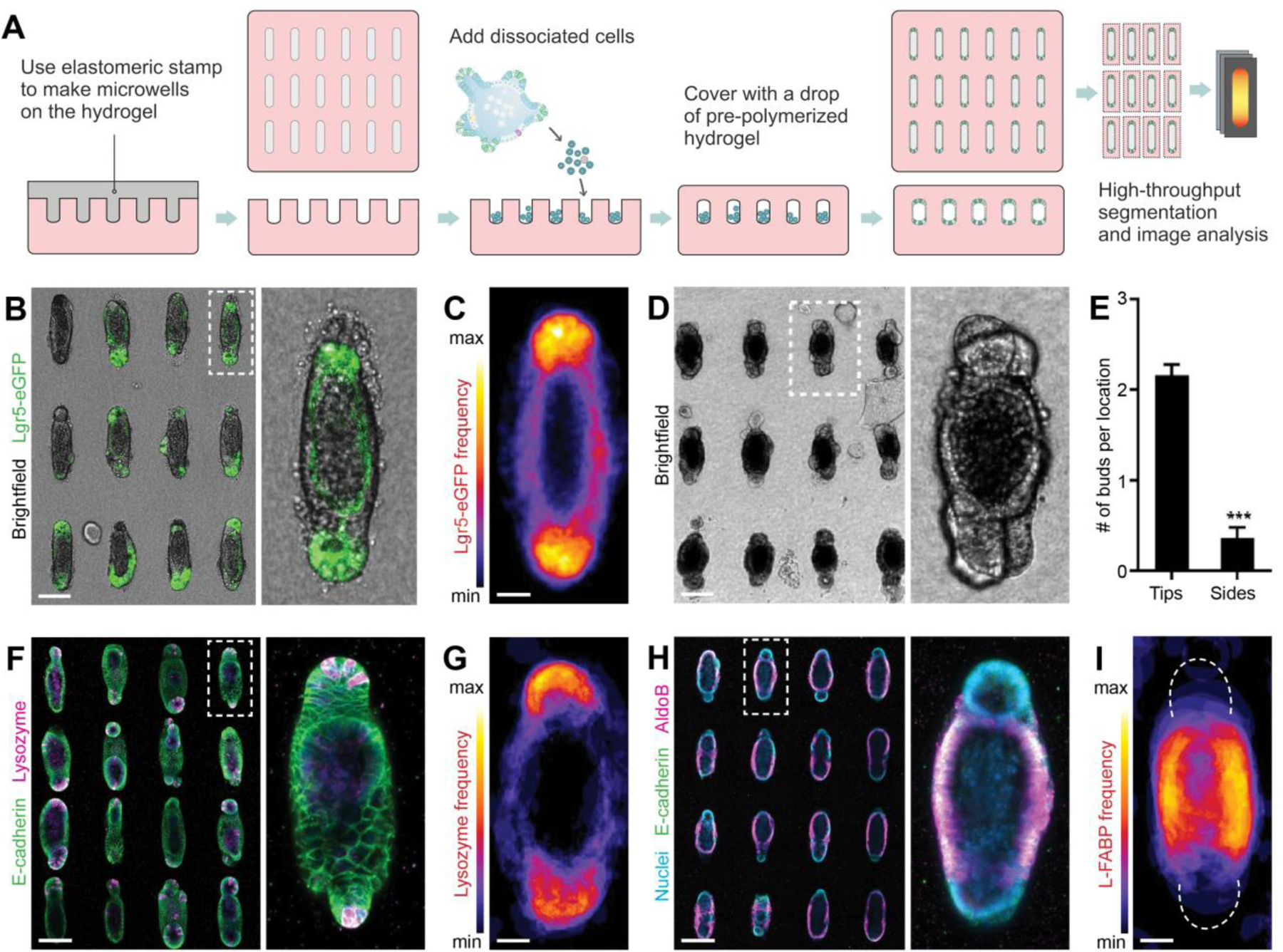
(A) Schematic depicting the generation of microfabricated tissues of controlled size and shape. (B) An array of intestinal organoids formed from engineered intestinal tissues of rod-like geometry and magnification. (C) Frequency map, showing average Lgr5 expression over ~80 tissues. (D) An array of intestinal organoids at day 5 and (E), quantification of the average number of buds per location within tubular intestinal tissues. (F) Paneth cells staining by lysozyme in the array of intestinal organoids and (G) average Paneth cell distribution. (H) AldoB-expressing enterocytes within rod-shaped organoids and (I) average enterocyte distribution. (B, D, F, H) Scale bars, 100 μm. (C, G, I) Scale bars, 25 μm.
We used soft lithography to microstructure a 3D hydrogel (composed of type I collagen (3 mg/ml) and 25% (v/v) Matrigel) with cavities of defined size and shape (21, 22), which were subsequently filled with purified Lgr5-eGFP+ ISCs (Fig. 2A). Initially randomly dispersed, the stem cells began to form contacts with each other and the surrounding matrix, and within 48 h self-organized into a lumenized epithelial tissue conforming to the shape of the pre-existing cavity. We used this approach to form 3D intestinal tissues of arbitrary sizes and shapes (Fig. S3A–C). Next, we sought to determine whether the differentiation of the engineered intestinal tissues followed a stereotypical pattern or occurred randomly. The method described above generates hundreds of regularly spaced tissues of identical size and shape, which permits rapid imaging of fluorescent markers or proteins visualized by immunofluorescence analysis, as well as quantification by automated image segmentation and analysis (Fig. S3D). Stacking images of a high number (>80) of individual tissues in registration provides information about the average distribution of the molecular marker of interest, with high statistical confidence (Fig. S3D). Although the tissues were formed from a cell suspension uniformly expressing Lgr5-eGFP, we found that within 4 days of culture, the signal became restricted to the curved ends of the tissues (Fig. 2B,C), indicating that ISCs are confined to these regions, in a pattern reminiscent to that of the native crypt. The microfabricated tissues proceeded to extend crypt-like buds with a spatial bias reflecting that of Lgr5 expression – the curved ends of the tissues were significantly more likely to extend buds than the flat sides (Fig. 2D,E). The spatial patterning also extended to differentiated intestinal cell types. Immunofluorescence analysis for Paneth cells and enterocytes revealed that the former are preferentially localized to the same end-locations as the ISCs (Fig. 2F,G), whereas the latter were on average excluded from the ends and confined to the middle of the tissue (Fig. 2H,I), suggesting that the spatial distribution of ISCs and differentiated cells within the engineered micro-tissues reflects the patterning of the crypt-villus axis in vivo (23). Thus, we are able to predict and control the site of budding and crypt formation within the organoids, by merely manipulating their initial geometry.
To elucidate how the initial epithelial geometry might dictate the patterning of intestinal tissue, we monitored the process using time-lapse microscopy (Supp. Movie 2, Fig. 3A–D). Interestingly, Lgr5 expression at the time of seeding and shortly thereafter (<2 h) appeared uniformly low. As the organoids formed in the crypt-like space, Lgr5 was re-expressed strongly at the ends of tissues, remaining low elsewhere (Fig. 3C–D). Preceding the Lgr5 regionalization, we observed a striking morphological difference between the curved end regions and the flat sides. Within 24 h of culture, and owing likely to crowding due to proliferation within a limited space at the ends, the cells in these regions became more packed, whereas cells in the lateral regions remained spread and flattened (Fig. 3E). Indeed, measuring the inter-nuclear distance at the different regions revealed a significant difference in cell packing (Fig. 3F).
Figure 3: Tissue geometry controls organoid patterning through cell shape-mediated regulation of YAP and Notch signaling.
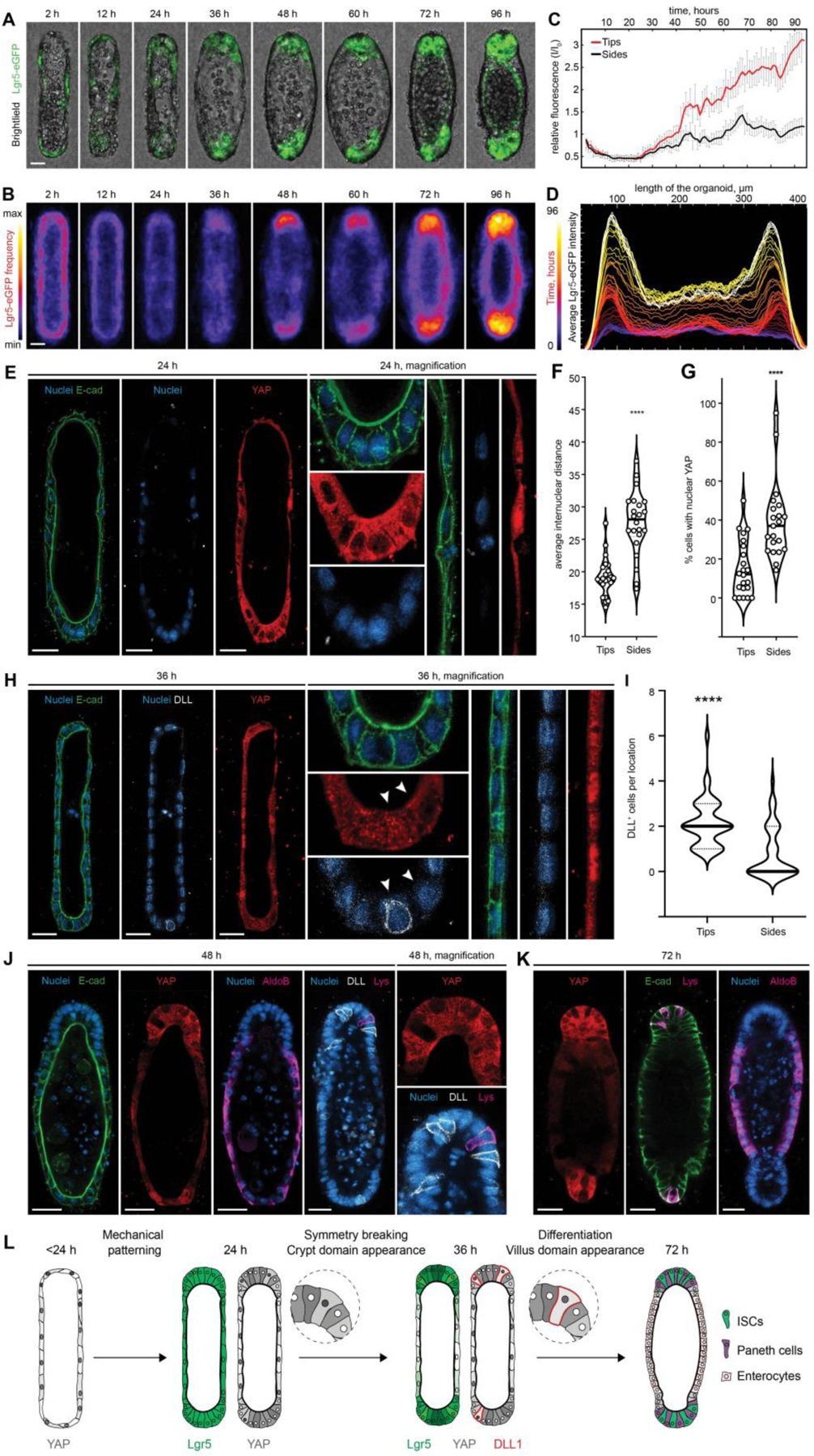
(A) Brightfield and Lgr5-eGFP time-lapse imaging of the representative organoid development and (B) frequency maps, showing average Lgr5 expression over ~80 tissues. (C) Relative changes in the Lgr5-eGFP expression in curved ends and flat sides of the organoids over time and (D) Lgr5-eGFP localization along length of the averaged tissue over time. (E) Immunofluorescence images showing the difference in internuclear distance, cell shape and the subcellular distribution of YAP between cells of the end and the side regions, 24 hours after cell loading. (F) Quantification of internuclear distance within the end and side regions of the tissues. Individual points, which represent the distance between neighboring nuclei, and means are shown. **** P < 0.0001 (G) Quantification of the nuclear localization of YAP within cells of the different organoid regions. Individual points and means are shown. **** P < 0.0001. (H) Immunofluorescence images showing the difference in the subcellular distribution of YAP cells between cells of the end and the side regions and appearance of the first DLL+, 36 hours after cell loading. (I) Quantification of DLL+ cells localization. **** P < 0.0001 (J, K) Immunofluorescence images showing YAP expression and localization of the enterocytes (AldoB), Paneth cells (Lys) and DLL+ cells in the representative organoids. (L) Schematic illustration summarizing the proposed mechanism of the geometry driven organoids patterning. Scale bars, 25 μm.
Yes-associated protein 1 (YAP) is an important regulator of ISC fate (24–27), which is strongly influenced by cell shape and mechanics (28, 29). To ascertain whether the differences in cell morphology between the different regions correlated with differences in YAP activity, we analyzed the subcellular distribution of YAP. Shortly after seeding, YAP was uniformly nuclear throughout the tissue, except in some cases where cell crowding was observed early in the curved regions due to (stochastic) variations in cell density (Fig. S4A). Between 12 and 24 h post-seeding, corresponding to the time when spatial differences in cell shape appear, and preceding the patterning of Lgr5, nuclear YAP localization became restricted to the lateral regions of the tissue. At the ends of the tissues, cytoplasmic translocation and, therefore, inactivation was observed (Fig. 3E,G). This geometry-dependent regionalization of YAP activity was again lost at later time points (>36 h) when cells throughout the tissue became uniformly packed (Fig. S4A). To test whether differential cell spreading (and, conversely, cell crowding) is sufficient to drive differences in YAP activity, we cultured ISCs at equal cell seeding density in microcavities of small (50 μm) and large (100 μm) diameter, resulting in a packed or a spread system (Fig. S4B–D). YAP activity strongly correlated with cell spreading: nuclear translocation was significantly more frequent within 100-μm wells compared with 50-μm wells, suggesting that tissue geometry per se controls the spatially patterned activation of YAP through differential cell spreading.
Several recent studies have implicated YAP activation in the repression of canonical ISC signatures, including Lgr5, Olfm4 and EphB3, during intestinal regeneration and cancer (24, 27, 30, 31). Given the spatial and temporal correlation between YAP induction and Lgr5 suppression, we hypothesized that the patterning of this system is driven by geometrically- and mechanically-established gradients in YAP activity. In line with this model, abrogating spatial gradients in YAP activity by uniform induction (via treatment with the MST1/2 signaling inhibitor XMU-MP-1 (32)) or inhibition (via treatment with verteporfin (33, 34)) both resulted in loss of intestinal tissue patterning (Fig. S4E–G). Furthermore, blocking cell spreading and the formation of mechanical gradients by treating with the contractility inhibitor blebbistatin (29, 35) abolished the patterns of both YAP activity and intestinal cell fate (Fig. S5). It is noteworthy that a blebbistatin treatment delayed by 36 hours did not interfere with Lgr5 patterning (Fig. S5A), while it was most efficient only in a short time window during the 36 hours in which the geometry-controlled cell spreading results in differential YAP activation (Fig. S5B,C). Thus, these gain-and-loss-of-function experiments show that an early geometrically induced YAP pre-pattern is necessary to promote later tissue regionalization. Previous studies by us and others have hinted that spatial heterogeneities in YAP are required for intestinal tissue morphogenesis (17, 30). Here, we show that these heterogeneities can be governed by geometrically-established gradients in cell mechanics to control downstream tissue patterning.
After determining that symmetry breaking in the system likely occurs through YAP-mediated spatial restriction of ISC maintenance, we wanted to further explore how the ISC niche at the ends is established. Multiple studies have emphasized that Paneth cells provide essential support to ISCs within intestinal organoids, and that their appearance is crucial for the establishment of the ISC niche (18, 19, 30, 36). Recent work by Serra et al. revealed a YAP-Notch symmetry breaking mechanism which is responsible for Paneth cell appearance and crypt initiation in classical intestinal organoids (30). In particular, it was found that Paneth cell differentiation and crypt initiation occur exactly at the sites where YAP ON cells are adjacent to YAP OFF cells. Bearing in mind the pattern of YAP activity described above, we reasoned that the ends of the tissue are the regions where YAP OFF and YAP ON cell pairs are more likely to coexist, triggering YAP-Notch symmetry breaking. In this model, an early indication of Paneth cell differentiation is the expression of the Notch ligand delta-like ligand 1 (DLL1), in cells featuring inactive Notch and active YAP. Strikingly, we observed the appearance of precisely this cell type at the end of the tissue around 36 h (Fig. 3H,I), along with localized Notch activation, as evidenced by the presence of the Notch intracellular domain (NICD) (Fig. S6A,B). Consistently with this model, Paneth cells were observed at these regions within 48 h of culture (Fig. 3J,K). Localized Notch activation and Paneth cell appearance were required for durable tissue patterning, as evidenced by the uniform inhibition (by treatment with the gamma secretase inhibitor DAPT) and global activation of Notch (by valproic acid (37)), both of which abolished patterns in Lgr5 and Paneth cell differentiation (Fig. S6C–F). Taken together, these data show that the tissue shape leads to a mechanically controlled pre-patterning of epithelial cells with different YAP activity, which in turn results in i) suppression of crypt cell fates at the side of the tissues and ii) a stereotypical Notch-DLL1 lateral inhibition event at the ends, driving the formation of the first Paneth cell that constitutes the niche (Supp. Movie 3; proposed mechanism summarized in Fig. 3L). Of note, in addition to the mechanism introduced, we also considered others that had previously been described for the regionalization of intestinal and other epithelia. Investigating the potential role of differential Wnt signaling in pattern establishment (Fig. S7), geometrically established inhibitory gradients in sonic hedgehog (SHH) (38) or transforming growth factor β (TGFβ) (21) (Fig. S8), as well as patterning through cell-cell repulsive interactions (39–41) (Fig. S9), we found that, whereas these mechanisms may contribute to or reinforce the patterning, they are likely not responsible for the initial symmetry breaking.
Next, intrigued by the possibility that differences in the packing of cells inside and outside the intestinal crypt may help to locate ISCs at the bottom of the crypt and establish a crypt-villus-like axis, we used a simplified intestinal surface with indentations that mimic the size and shape of the crypts, surrounded by flat regions that approximate the much larger surface of the villi (Fig. S10). Moving from the bottom of the engineered crypts to the exterior surfaces (at 24–36 h after cell loading), we observed a gradual increase in cell spreading and YAP activation (Supp. Movie 4). Conversely, Lgr5 expression was maximal at the bottom of the crypts, and was virtually absent outside of the cavities, indicating that stem cells were only preserved at the bottom of the crypts, in the familiar in vivo-like pattern (Fig. S10B), despite a homogeneous cocktail of soluble cell fate-determining factors.
Finally, we sought to exploit these mechanistic insights to engineer macroscopic organoids with an in vivo tissue architecture (Fig. 4). We micro-fabricated hydrogel substrates resembling the native intestinal mucosa, with crypts at the bottom and villi protruding outwards (Fig. 4A–D). Within 48 h, ISCs seeded on these scaffolds expanded to establish a confluent monolayer of cells (Fig. 4E, Supp. Movie 5). Strikingly, induction of differentiation resulted in highly stereotyped organoid patterning, with stem cell-containing crypts (Fig. 4F) interspersed with villi composed of enterocytes and other differentiated cell types (Fig. 4G). These results demonstrate that the principles controlling cell fate patterning at the sub-tissue scale can be harnessed to engineer macroscopic intestinal surfaces that capture the cellular organization and periodicity of the crypt-villus system. It should be noted that previous attempts to engineer intestinal surfaces (10, 11) were dependent on complex chemical gradients to induce patterning, i.e. did not reveal or exploit tissue geometry as a patterning cue, and have not demonstrated recapitulation of the multicellular organization and periodicity of the in vivo crypt-villus system.
Figure 4: Bioengineered organoids with an in vivo-like tissue architecture.
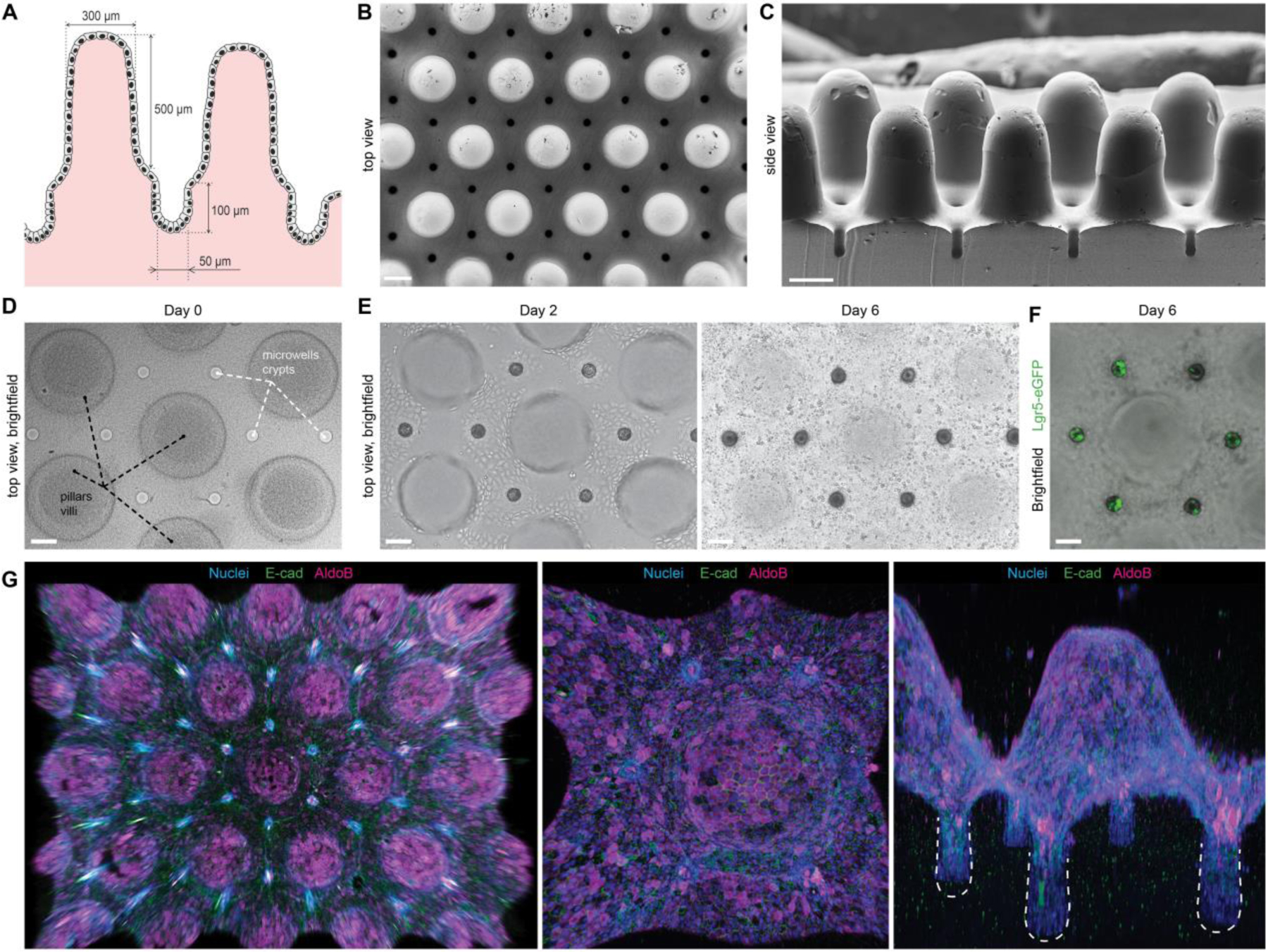
(A) Scheme of the designed topography resembling the native tissue with characteristic intestinal crypt-villus architecture. (B, C) SEM images of the poly(dimethylsiloxane) (PDMS) template used for fabricating bioengineered hydrogel substrates featuring a crypt-villus architecture, top and side view. Scale bars, 200 μm. (D) Top view of the hydrogel substrate shaped according to the topology of the native intestinal mucosa. (E) Brightfield time-course images of the intestinal epithelium development. (D,E) Extended depth of field for a z-stack ~600 μm. (F) Localization of the Lgr5+ stem cells in the engineered crypts. (D-F) Scale bars, 100 μm. (G) 3D reconstruction of the immunofluorescence images showing confluent monolayer of E-Cadherin expressing epithelial cells covering hydrogel substrates, harboring villi composed of enterocytes (AldoB) and other differentiated cell types.
We thus report novel means to engineer external guidance in stem cell-based organogenesis – a process otherwise fully driven by stochastic self-organization. We show that localized patterning of ECM mechanics and topographically structured hydrogel scaffolds can be used to build organoids of a controlled initial size and shape and to predict and influence the course of their development, in particular the breaking of symmetry and the number and location of crypt-like domains. We envision that this advance will help overcome the lack of reproducibility and control that are major limitations of organoids, undercutting their utility in basic and translational research. The uniquely controlled organoid development afforded by these approaches allowed us to identify a role for tissue geometry in intestinal tissue patterning, and dissect the underlying mechanisms. Our data suggest that anisometric tissue geometries drive stereotypic epithelial patterning by establishing reproducible local differences in cell packing and morphology. It is the heterogeneities in YAP activity that ultimately prescribe “villus” and “crypt” domains, by suppressing stem cell fates and localizing Notch-mediated Paneth cell differentiation, respectively. We believe that spatial variations in cell morphology alone are likely sufficient to induce symmetry breaking, and could be responsible for driving crypt initiation within classical organoids, where they would occur randomly. Here, we demonstrate that tissue geometry can be used as a means to regularize the spatial distribution of cell morphology and, consequently, cell fate. The notion that morphogenesis is self-referential, i.e. that tissue form can serve as an independent input into its further development is not new (21, 42). In the case of the native intestine, the shape of the villi has been shown to influence the restriction of the stem cell zones by introducing diffusion-based spatial heterogeneities in signals communicated between the epithelium and the mesenchyme (38). Here, we demonstrate that ISC restriction to the ends of crypt-like engineered tissue occurs in the absence of villi and mesenchyme, suggesting a complementary mechanism for intestinal regionalization, whereby the epithelial geometry per se allows for autonomous patterning of the tissue.
Supplementary Material
Acknowledgments:
We thank Celeste Nelson, Johan Auwerx and Jeffrey Hubbell for valuable feedback on the manuscript. We thank Freddy Radtke and Ute Koch for providing intestine from Hes1-GFP mice and Sylke Hoehnel for help with establishing intestinal crypt surfaces
Funding:
This work was funded by support from the Swiss National Science Foundation (SNSF) research grant 310030_179447, the EU Horizon 2020 Project INTENS (#668294–2), the PHRT - PM/PH Research Project Proposal 2017, Ecole Polytechnique Fédérale de Lausanne (EPFL), and the NIH (R01DK120921). N.G. was supported in part by an EMBO Long-Term Postdoctoral Fellowship. T.B. was supported by fellowships from the NSF GRFP and NIH (T32 GM-065103)
Footnotes
Competing interests: Ecole Polytechnique Fédérale de Lausanne (with M.P.L., N.G., M.N. and N.B.) has filed patent applications pertaining to organoid culture methods described in the paper
Data and materials availability: All data is available in the main text or the supplementary materials.
References and Notes:
- 1.Clevers H, Modeling Development and Disease with Organoids. Cell 165, 1586–97 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Drost J, Clevers H, Organoids in cancer research. Nat Rev Cancer 18, 407–418 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Drost J, van Boxtel R, Blokzijl F, Mizutani T, Sasaki N, Sasselli V, de Ligt J, Behjati S, Grolleman JE, van Wezel T, Nik-Zainal S, Kuiper RP, Cuppen E, Clevers H, Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 358, 234–238 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lancaster MA, Knoblich JA, Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 345, 1247125–1247125 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, Lampis A, Eason K, Huntingford I, Burke R, Rata M, Koh D-M, Tunariu N, Collins D, Hulkki-Wilson S, Ragulan C, Spiteri I, Moorcraft SY, Chau I, Rao S, Watkins D, Fotiadis N, Bali M, Darvish-Damavandi M, Lote H, Eltahir Z, Smyth EC, Begum R, Clarke PA, Hahne JC, Dowsett M, de Bono J, Workman P, Sadanandam A, Fassan M, Sansom OJ, Eccles S, Starling N, Braconi C, Sottoriva A, Robinson SP, Cunningham D, Valeri N, Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359, 920–926 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossi G, Manfrin A, Lutolf M, Progress and potential in organoid research. Nat Rev Genet 19, 671–687 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Sasai Y, Cytosystems dynamics in self-organization of tissue architecture. Nature 493, 318–26 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Huch M, Knoblich JA, Lutolf MP, Martinez-Arias A, The hope and the hype of organoid research. Dev Camb Engl 144, 938–941 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Advances in Organoid Technology: Hans Clevers, Madeline Lancaster, and Takanori Takebe. Cell Stem Cell 20, 759–762 (2017). [Google Scholar]
- 10.Wang Y, Gunasekara DB, Reed MI, DiSalvo M, Bultman SJ, Sims CE, Magness ST, Allbritton NL, A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials 128, 44–55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim R, Wang Y, Hwang S-HJ, Attayek PJ, Smiddy NM, Reed MI, Sims CE, Allbritton NL, Formation of arrays of planar, murine, intestinal crypts possessing a stem/proliferative cell compartment and differentiated cell zone. Lab Chip 18, 2202–2213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Kim R, Gunasekara DB, Reed MI, DiSalvo M, Nguyen DL, Bultman SJ, Sims CE, Magness ST, Allbritton NL, Formation of Human Colonic Crypt Array by Application of Chemical Gradients Across a Shaped Epithelial Monolayer. Cell Mol Gastroenterology Hepatology 5, 113–130 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nikolaev M, Mitrofanova O, Broguiere N, Geraldo S, Dutta D, Tabata Y, Elci B, Brandenberg N, Kolotuev I, Gjorevski N, Clevers H, Lutolf MP, Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature 585, 574–578 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Lancaster MA, Corsini NS, Wolfinger S, Gustafson EH, Phillips AW, Burkard TR, Otani T, Livesey FJ, Knoblich JA, Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol 35, 659–666 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poling HM, Wu D, Brown N, Baker M, Hausfeld TA, Huynh N, Chaffron S, Dunn JCY, Hogan SP, Wells JM, Helmrath MA, Mahe MM, Mechanically induced development and maturation of human intestinal organoids in vivo. Nat Biomed Eng 2, 429–442 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zinner M, Lukonin I, Liberali P, Design principles of tissue organisation: How single cells coordinate across scales. Curr Opin Cell Biol 67, 37–45 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, Clevers H, Lutolf MP, Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560–564 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H, Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Sato T, Clevers H, Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Sci New York N Y 340, 1190–4 (2013). [DOI] [PubMed] [Google Scholar]
- 20.McKinnon DD, Brown TE, Kyburz KA, Kiyotake E, Anseth KS, Design and Characterization of a Synthetically Accessible, Photodegradable Hydrogel for User-Directed Formation of Neural Networks. Biomacromolecules 15, 2808–2816 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson CM, VanDuijn MM, Inman JL, Fletcher DA, Bissell MJ, Tissue Geometry Determines Sites of Mammary Branching Morphogenesis in Organotypic Cultures. Science 314, 298–300 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson CM, Inman JL, Bissell MJ, Three-dimensional lithographically defined organotypic tissue arrays for quantitative analysis of morphogenesis and neoplastic progression. Nat Protoc 3, 674–678 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clevers H, The Intestinal Crypt A Prototype Stem Cell Compartment. Cell 154, 274–284 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Gregorieff A, Liu Y, Inanlou MR, Khomchuk Y, Wrana JL, Yap-dependent reprogramming of Lgr5+ stem cells drives intestinal regeneration and cancer. Nature 526, 715–718 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Imajo M, Ebisuya M, Nishida E, Dual role of YAP and TAZ in renewal of the intestinal epithelium. Nat Cell Biol 17, 7–19 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Panciera T, Azzolin L, Cordenonsi M, Piccolo S, Mechanobiology of YAP and TAZ in physiology and disease. Nat Rev Mol Cell Bio 18, 758–770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yui S, Azzolin L, Maimets M, Pedersen MT, Fordham RP, Hansen SL, Larsen HL, Guiu J, Alves MRP, Rundsten CF, Johansen JV, Li Y, Madsen CD, Nakamura T, Watanabe M, Nielsen OH, Schweiger PJ, Piccolo S, Jensen KB, YAP/TAZ-Dependent Reprogramming of Colonic Epithelium Links ECM Remodeling to Tissue Regeneration. Cell Stem Cell 22, 35–49.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Digabel JL, Forcato M, Bicciato S, Elvassore N, Piccolo S, Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S, A Mechanical Checkpoint Controls Multicellular Growth through YAP/TAZ Regulation by Actin-Processing Factors. Cell 154, 1047–1059 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Serra D, Mayr U, Boni A, Lukonin I, Rempfler M, Meylan LC, Stadler MB, Strnad P, Papasaikas P, Vischi D, Waldt A, Roma G, Liberali P, Self-organization and symmetry breaking in intestinal organoid development. Nature 569, 66–72 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukonin I, Serra D, Meylan LC, Volkmann K, Baaten J, Zhao R, Meeusen S, Colman K, Maurer F, Stadler MB, Jenkins J, Liberali P, Phenotypic landscape of intestinal organoid regeneration. Nature 586, 275–280 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan F, He Z, Kong L-L, Chen Q, Yuan Q, Zhang S, Ye J, Liu H, Sun X, Geng J, Yuan L, Hong L, Xiao C, Zhang W, Sun X, Li Y, Wang P, Huang L, Wu X, Ji Z, Wu Q, Xia N-S, Gray NS, Chen L, Yun C-H, Deng X, Zhou D, Pharmacological targeting of kinases MST1 and MST2 augments tissue repair and regeneration. Sci Transl Med 8, 352ra108–352ra108 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Rosado-Olivieri EA, Anderson K, Kenty JH, Melton DA, YAP inhibition enhances the differentiation of functional stem cell-derived insulin-producing β cells. Nat Commun 10, 1464 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee S-J, Anders RA, Liu JO, Pan D, Genetic and pharmacological disruption of the TEAD–YAP complex suppresses the oncogenic activity of YAP. Gene Dev 26, 1300–1305 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, Chen CS, Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat Methods 7, 969–971 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farin HF, Jordens I, Mosa MH, Basak O, Korving J, Tauriello DVF, de Punder K, Angers S, Peters PJ, Maurice MM, Clevers H, Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 530, 340–343 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Yin X, Farin HF, van Es JH, Clevers H, Langer R, Karp JM, Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat Methods 11, 106–112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shyer AE, Huycke TR, Lee C, Mahadevan L, Tabin CJ, Bending Gradients: How the Intestinal Stem Cell Gets Its Home. Cell 161, 569–580 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holmberg J, Genander M, Halford MM, Annerén C, Sondell M, Chumley MJ, Silvany RE, Henkemeyer M, Frisén J, EphB Receptors Coordinate Migration and Proliferation in the Intestinal Stem Cell Niche. Cell 125, 1151–1163 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Genander M, Halford MM, Xu N-J, Eriksson M, Yu Z, Qiu Z, Martling A, Greicius G, Thakar S, Catchpole T, Chumley MJ, Zdunek S, Wang C, Holm T, Goff SP, Pettersson S, Pestell RG, Henkemeyer M, Frisén J, Dissociation of EphB2 Signaling Pathways Mediating Progenitor Cell Proliferation and Tumor Suppression. Cell 139, 679–692 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Batlle E, Henderson JT, Beghtel H, van den Born MMW, Sancho E, Huls G, Meeldijk J, Robertson J, van de Wetering M, Pawson T, Clevers H, β-Catenin and TCF Mediate Cell Positioning in the Intestinal Epithelium by Controlling the Expression of EphB/EphrinB. Cell 111, 251–263 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Nelson CM, Geometric control of tissue morphogenesis. Biochimica Et Biophysica Acta Bba - Mol Cell Res 1793, 903–910 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


