Abstract
IMPORTANCE
The identification and validation of biomarkers in hidradenitis suppurativa (HS) has potential to improve the understanding and management of this chronic, burdensome disease.
OBJECTIVE
To systematically identify all known HS biomarkers, categorize them by biomarker type, and critically evaluate their validity according to established criteria.
EVIDENCE REVIEW
Eligibility criteria for this review (PROSPERO Registration 230830) included randomized clinical trials, uncontrolled clinical trials, cohort studies, case-control studies, and other observational studies with no restrictions of patient age, sex, race or ethnicity, or language of publication up until December 31, 2020. All articles were categorized into biomarker type, defined using the US Food and Drug Administration Biomarkers, Endpoints, and other Tools (BEST) glossary. Assessment of each identified biomarker was undertaken in line with the US Food and Drug Administration and European Medicines Agency guidelines for the validation of proposed biomarkers. Assessment of the strength of overall data regarding individual biomarkers was undertaken using the Grading of Recommendations, Assessment, Development, and Evaluations (GRADE) approach.
FINDINGS
A total of 3953 nonduplicate articles were screened, of which 1429 articles were retrieved based on the include/exclusion criteria applied. After full-text screen and data extraction, 106 articles were included in this review. The evidence of strength of 6 categories of biomarkers (susceptibility/risk, diagnostic, monitoring, predictive, prognostic, and pharmacodynamic/response biomarkers) was assessed using GRADE criteria. A total of 48 biomarkers were identified with a minimum GRADE rating of moderate.
Only 1 diagnostic (serum IL-2R), 1 monitoring (dermal Doppler vascularity), and 2 predictive biomarkers (epithelialized tunnels and positive family history of HS) achieved a GRADE rating of high. None of the identified biomarkers had sufficient clinical validity to be recommended for routine use in the clinical setting.
CONCLUSIONS AND RELEVANCE
Major barriers to the identification, validation, and introduction of routine biomarkers in the management of HS include lack of independent biomarker validation studies (especially assumption-free “omics”-based techniques); insufficient assessment of collinearity between identified or proposed biomarkers; and a lack of routine integration of biomarkers into the structure of clinical trials. International consensus among researchers, clinicians, and pharmaceutical stakeholders is required to standardize goals and methods and encourage biomarker integration into future HS clinical trials. This systematic review presents a number of priorities for near-term future research to overcome such barriers and limitations of biomarkers in HS.
Hidradenitis suppurativa (HS), also known as acne inversa, is a chronic autoinflammatory skin disease manifesting in painful nodules and abscesses, comedones, and draining malodorous tunnels.1,2 These lesions have a predilection to flexural areas of skin, namely the axillae, groin, and submammary regions, but can occur at any site.1,2 The disease is thought to occur owing to a combination of genetic and environmental factors (including bacterial dysbiosis) and, in a minority of individuals, has strong associations with inherited sequence variants in the gamma secretase complex.1-3 Hidradenitis suppurativa is associated with multiple inflammatory comorbidities, including obesity, metabolic syndrome, diabetes mellitus, and inflammatory bowel disease, among others.1-3 It is a heterogeneous disease with multiple phenotypes, clinical presentations, and disease trajectories. It has been proposed that the development of biomarkers may aid in the diagnosis, understanding, and management of this disease.4,5
A biomarker is a “defined characteristic that is measured as an indicator of normal biological processes, pathogenic processes, or responses to an exposure or intervention.”6(p4) There are several different types of biomarkers defined by the US Food and Drug Administration (FDA) Biomarkers, Endpoints, and other Tools (BEST) glossary (eMethods in the Supplement). The development, validation, and clinical use of biomarkers in HS would reap multiple benefits in the clinical assessment, diagnosis, and management of disease,6,7 as recently illustrated by the identification of various inflammatory endotypes in the context of atopic dermatitis and their subsequent therapeutic relevance.8 Susceptibility/risk and diagnostic biomarkers may aid in reversing the well-documented diagnostic delay in HS, while also identifying high-risk individuals who would benefit from closer monitoring for active disease. Diagnostic biomarkers may also complement the modified Dessau criteria as an objective measure to differentiate HS from other commonly misdiagnosed conditions, such as recurrent folliculitis, acne, or infection. Additionally, monitoring biomarkers (molecular biomarkers of disease activity) would allow for a deeper understanding of the associations between observed molecular findings in HS tissue and clinical disease activity over time. This may help define disease heterogeneity, refine drug repurposing, and identify novel therapeutic targets. Importantly, predictive biomarkers would allow for personalized medicine approaches with targeted selection of therapy to patients with greater reliability of response, rather than the relatively common trial-and-error approach currently used in HS.4
The aim of this review was to systematically identify all known HS biomarkers, categorize them by biomarker type, and critically evaluate their validity according to established criteria.6-8 Additionally, the strength of evidence supporting each biomarker was assessed using Grading of Recommendations, Assessment, Development, and Evaluations (GRADE) certainty criteria.9
Methods
This study was registered with PROSPERO (CRD42021230830) and conducted in line with the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) reporting guideline. Eligibility criteria for this review included randomized clinical trials, uncontrolled clinical trials, cohort studies, case-control studies, and other observational studies with no restrictions of patient age, sex, race or ethnicity, or language of publication up until December 31, 2020. Data collection was performed independently by 2 authors (S.D.S. and J.W.F.), with any disagreements regarding inclusion of citations being referred to a third author for mediation. All articles were categorized into biomarker type (eMethods in the Supplement). Biomarkers were defined using the FDA BEST glossary (https://www.ncbi.nlm.nih.gov/books/NBK338448/). Assessment of each identified biomarker was undertaken in line with the FDA and European Medicines Agency guidelines for the validation of proposed biomarkers. Assessment of the strength of overall data regarding individual biomarkers was undertaken using the GRADE approach. Detailed methods are presented in eMethods and eTable 1 in the Supplement.
Results
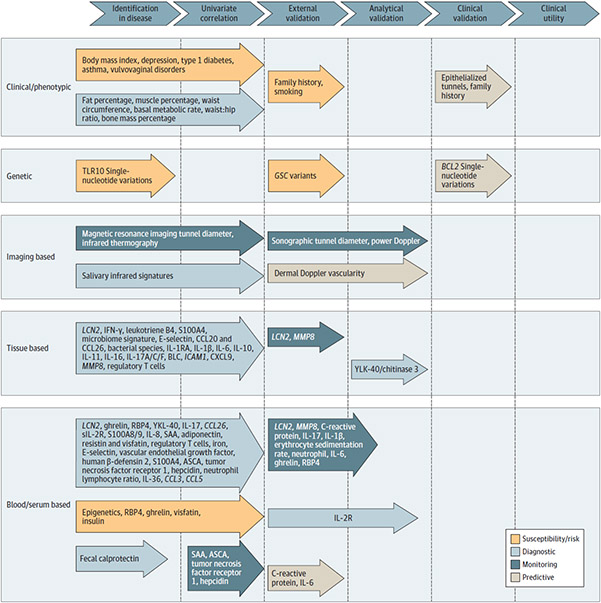
A total of 1429 articles were identified using the documented search strategy (eFigure in the Supplement). A total of 11 susceptibility/risk biomarkers, 85 diagnostic biomarkers, 39 monitoring biomarkers, and 20 predictive biomarkers were identified. Visual presentation of identified biomarkers with GRADE rating of moderate or high and their validity status is presented in the Figure. Biomarkers that met “moderate” or “high” GRADE criteria are listed in Tables 1, 2, 3, and 4; a complete list of all identified biomarkers with gradings (including those with GRADE ratings of very low or low) is presented in eTables 2-5 in the Supplement.
Figure. Biomarkers With GRADE Rating of Moderate or High and Degree of Biomarker Validation.
Multiple susceptibility/risk, diagnostic, and monitoring biomarkers were identified in this review, with lesser numbers of markers being independently validated in external cohorts. Predictive markers examining response to therapy were the only biomarkers that had undergone clinical validation in the setting of a clinical trial, and no biomarkers had assessment of clinical utility to recommend them for routine clinical use. The vast majority of these identified biomarkers met “moderate” GRADE criteria, and the only biomarkers that reached “high” GRADE criteria were serum IL-2R (diagnostic), dermal Doppler vascularity (monitoring), and epithelialized tunnels and positive family history of HS (predictive). Items were assessed based on criteria in line with the FDA biomarker definitions, GRADE criteria, and FDA/European Medicines Agency guidelines for the validation of proposed biomarkers as reported in eMethods in the Supplement. FDA indicates US Food and Drug Administration; GRADE, Grading of Recommendations, Assessment, Development, and Evaluations; GSC, gamma secretase complex; SAA, serum amyloid A.
Table 1.
Susceptibility/Risk Biomarkers (GRADE Moderate/High)
| Biomarker | Biomarker level |
Study type | Interpretation | Critical evaluation |
GRADE evidence profile |
References | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HS | OR | P value | External validation |
Analytical validation |
Clinical validation |
Clinical utility |
|||||
| Smoking | Increased | 14.87 | .001 | Case-control, regression modeling | Smoking is associated with a diagnosis of HS | Yes | No | No | No | Moderate | Akdogan et al,10 2018 |
| 1.90 | .01 | Garg et al,11 2018 | |||||||||
| FHX | Increased | NR | NR | Twin study/cross-sectional/heritability calculations | Narrow-sense heritability calculated at 77% (van Straalen et al,12 2020) | Yes | No | No | No | Moderate | van Straalen et al,12 2020; Schrader et al,13 2014; Molina-Leyva and Cuenca-Barrales,14 2019 |
| Fasting serum insulin | Increased | 1.09 | .03 | Case-control, regression modeling | Increased fasting serum insulin is associated with a diagnosis of HS | Yes | No | No | No | Moderate | Akdogan et al,10 2018; Vilanova et al,15 2018 |
| Genetic sequence variants in gamma secretase complex, POFUT1, PTSPIP1, etc | Present | NR | NR | Linkage analysis | Association but no analysis of predictive power or potential | Yes | No | No | No | Moderate | Reviewed in Frew et al,16 2017; Jfri et al,172019 |
Abbreviations: GRADE, Grading of Recommendations, Assessment, Development, and Evaluations; HS, hidradenitis suppurativa; NR, not reported; OR, odds ratio.
Table 2.
Diagnostic Biomarkers (GRADE Moderate/High)
| Biomarker | Statistical association |
Study type | Interpretation | Critical evaluation of biomarkers |
GRADE evidence profile |
References | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Population of comparison |
Significancea | External validation |
Analytical validation | Clinical validation |
Clinical utility |
|||||
| Serum IL-2R | HC | PPV, 0.68; P < .001 | Observational case-control | Elevation associated with HS | Yes | Yes (age, race, and sex matched in Matusiak et al,182015) | No | No | High | Wieland et al,19 2013 |
| HC | P < .001 | Matusiak et al,20 2009 | ||||||||
| HC | PPV, 0.8; P = .001 | Matusiak et al,182015 | ||||||||
| Serum hepcidin | HC | P < .01 | Observational case-control | Decreased levels associated with HS | Yes | No, insufficient info on controls in Ponikowska et al,21 2020 | No | No | Moderate | Ponikowska et al,21 2020 |
| HC | P = .03 | Ghias et al,22 2019 | ||||||||
| Combination of serum markers (E-selectin + VEGF + hBD2) | HC/IBD/staph infection | PPV, 85.3% | Multivariate analysis | Combination of markers identifies individuals with HS | Yes | Yes | No | No | Moderateb | Argyropoulou et al,23 2019b |
| Serum YLK-40/chitinase 3 | HC | PPV, 0.8; P < .001 | Observational case-control | Elevation associated with HS | No | Yes, matched for age, race, and sex | No | No | Moderate | Matusiak et al,18 2015 |
| Serum amyloid A | HC | P < .001 | Observational case-control | Elevation associated with HS | Yes | No, no matching for age or sex in Witte-Handel et al,24 2019 Matched for age and sex in Akdogan et al,25 2020 | No | No | Moderate | Witte-Händel et al,24 2019 |
| P = .008 | Akdogan et al,25 2020 | |||||||||
| Serum CRP | HC | P < .001 | Observational case-control | Elevation associated with HS | Yes | No, matched for age but not sex or BMI in Jiménez-Gallo et al,26 2017 No matching for age, sex or BMI in Jiménez-Gallo et al,27 2018 Matched for age and sex in Akdogan et al,25 2020 | No | No | Moderate | Jiménez-Gallo et al,26 2017 |
| P < .05 | Jiménez-Gallo et al,27 2018 | |||||||||
| P = .01 | Akdogan et al,25 2020 | |||||||||
| Serum IL-8 | HC | P < .01 | Observational case-control | Elevation associated with HS | Yes | No | No | No | Moderate | Jiménez-Gallo et al,27 2018 |
| Upregulated | Witte-Händel et al,24 2019 | |||||||||
| Serum IL-6 | HC | P < .01 | Observational case-control | Elevation associated with HS | Yes | No | No | No | Moderate | Witte-Händel et al,24 2019 |
| P < .001 | Jiménez-Gallo et al,27 2018 | |||||||||
| Serum adiponectin | HC | P < .001 | Observational case-control | Decreased levels associated with HS | Yes | No | No | No | Moderate | González-López et al,28 2020; Özkur et al,29 2020 |
| Serum resistin | HC | OR, 1.02; P = .02 | Elevation associated with HS | Yes | No | No | No | Moderate | ||
| Serum visfatin | HC | OR, 2.21; P < .001 | Elevation associated with HS | Yes | No | No | No | Moderate | ||
| Serum IL-17 | HC | P < .001 | Observational case-control | Elevation associated with HS | Yes | No | No | No | Moderate | Reviewed in Frew et al,4 2021 |
| P < .001 | Matusiak et al,30 2017 | |||||||||
| Serum S100A8/A9 | HC | PPV, 0.81; P < .001 | Observational case-control | Elevation associated with HS | No | Yes | No | No | Moderate | Wieland et al,19 2013 |
| Tissue and serum miRNAs | HC | miRNA-155-5p (P = .005); miRNA-223-p (P < .001); miRNA-31-5p (P = .04); miRNA-21–5p(P < .001); miRNA-146a-5p (P = .01) |
Observational case-control | Elevation associated with HS | Yes | No | No | No | Moderate | Radhakrishna et al,312019; Hessam et al,32 2017 |
| Nonlesional | miRNA-155-5p (P = .01); miRNA-223-5p (P = .01); miRNA-31-5p (P = .048); miRNA-21-5p (P = .01); miRNA-146a-5p (P = .04); miRNA-125b-5p (P = .04) |
Decrease associated with HS | ||||||||
| miRNA 155, 223, and 31 | miRNA-155-5p | P = .01 (L vs NL) | Observational case-control | Elevation associated with HS | Yes | No | No | No | Moderate | Radhakrishna et al,31 2019; Hessam et al,32 2017 |
| miRNA-223-5p | P = .01 | |||||||||
| miRNA-31-5p | P = .048 | |||||||||
| miRNA-21-5p | P = .01 | |||||||||
| miRNA-146a-5p | P = .04 | |||||||||
| miRNA-125b-5p | P = .04 | Decrease associated with HS | ||||||||
| Bacterial species | Corynebacterium (HC) | P < .001 | Observational case-control | Elevation associated with HS | Yes | No | No | No | Moderate | Ring et al,33 2017; Guet-Revillet et al,34 2017; Naik et al,35 2019; Ring et al,36 2019; Schneider et al,37 2020 |
| Acinetobacter/Moraxella (HC) | P = .10 | Yes | No | No | No | Moderate | ||||
| Staphylococcus epidermidis | Not given | Yes | No | No | No | Moderate | ||||
| Porphyromonas/Peptoniphilus (HC) | P = .02 | Yes | No | No | No | Moderate | ||||
| Propionibacterium acnes (NLT) | P < .001 | Yes | No | No | No | Moderate | ||||
| Tissue IL-17 | HC | P < .01 | Observational case-control | Elevation associated with HS | Yes | No, no matching for Witte-Händel et al,24 2019 | No | No | Moderate | Kelly et al,38 2015 |
| P < .001 | Wolk et al,39 2011 | |||||||||
| P < .01 | Hessam et al,40 2018 | |||||||||
| P < .001 | Witte-Händel et al,24 2019 | |||||||||
| Tissue IL-17A | NLT | P = .004 | Observational case-control | Elevation associated with HS | Yes | No | No | No | Moderate | Wolk et al,39 2011 |
| P = .05 | Kelly et al,38 2015 | |||||||||
| P < .001 | Witte-Händel et al,24 2019 | |||||||||
| P < .01 | Hotz et al,41 2016 | |||||||||
| P < .01 | Navrazhina et al,42 2020 | |||||||||
| Tissue IL-17F | NLT | P < .01 | Observational case-control | Elevation associated with HS | Yes | No | No | No | Moderate | Witte-Händel et al,24 2019; Navrazhina et al,42 2020 |
| Tissue IL-1B | HC | P < .01 | Observational case-control | Elevation associated with HS | Yes | No, no matching for age or sex in Witte-Händel et al,24 2019 | No | No | Moderate | Kelly et al,38 2015 |
| HC | P < .001 | Witte-Händel et al,24 2019 | ||||||||
| HC | P < .001 | Wolk et al,39 2011 | ||||||||
| Tissue IL-1B | NLT | 31-Fold increase, P = .003 | Observational case-control | Elevation associated with HS | Yes | No | No | No | Moderate | Wolk et al,39 2011 |
| P < .001 | Kelly et al,38 2015 | |||||||||
| P < .001 | van der Zee et al,43 2011 | |||||||||
| Tissue S100A7 | HC | P < .001 | Observational case-control | Elevation associated with HS | Yes | No | No | No | Moderate | Wolk et al,39 2011 |
| P = .002 | Batycka-Baran et al,44 2021 | |||||||||
| Tissue HBD3 | NLT | P < .05 | Observational case-control | Elevation associated with HS | Yes | No | No | No | Moderate | Wolk et al,39 2011 |
| P < .05 | Emelianov et al,45 2012 | |||||||||
| P < .01 | Multivariate logistic regression | Akdogan et al,25 2020 | ||||||||
| Tissue IFN-γ | NLT | P < .001 | Observational case-control | Elevation associated with HS | Yes | No | No | No | Moderate | Wolk et al,39 2011 |
| P < .01 | Hotz et al,412016 | |||||||||
| Tissue citrullinated H3 protein | HC | P = .03 | Observational case-control | Correlation with disease severity | Yes | No | No | No | Moderate | Byrd et al,46 2019 |
| P < .01 | Lowe et al,47 2020 | |||||||||
| Fecal calprotectin | Inactive disease | P < .001 | Observational case-control | Elevation associated with HS | Yes | No | No | No | Moderate | Eşer et al,48 2020 |
| Descriptive data only | Lloyd-McLennan et al,49 2021 | |||||||||
Abbreviations: BMI, body mass index; CRP, C-reactive protein; GRADE, Grading of Recommendations. Assessment, Development, and Evaluations; HC, healthy controls; HS, hidradenitis suppurativa; IBD, inflammatory bowel disease; L, lesional; miRNA, microRNA; NL, nonlesional; NLT, nonlesional tissue; OR, odds ratio; PPV, positive predictive value; staph, staphylococcus.
P values are reported as they were in the original sources.
Abstract only.
Table 3.
Monitoring Biomarkers (GRADE Moderate/High)
| Biomarker | Disease severity association |
Study type | Interpretation | Critical evaluation |
GRADE evidence profile |
Reference | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Disease severity index |
Significance | External validation |
Analytical validation |
Clinical validation |
Clinical utility |
|||||
| Doppler dermal vascularity | Pain NRS | R, 0.98; P value, NS | Univariate and multivariate analysis | Correlation with disease severity | Yes | Yes | No | No | High | Nazzaro et al,50 2019 |
| Pain VAS | R, 0.67; P < .001 | Grand et al,51 2021 | ||||||||
| Fistula size | Size ≥25.6 mm associated with presence of Doppler signal: OR, 11.51; P = .01 and mixed vasculature distribution: OR, 3.84; P = .002 | Caposiena Caro et al,52 2018 | ||||||||
| Sartorius score | Score >19 associated with mixed vasculature distribution: OR, 6.65; P < .001 | Caposiena Caro et al,52 2018 | ||||||||
| Sonographic dermal tunnel diameter | HS-PGA | R, 0.75; P < .001 | Univariate | Correlation with disease severity | No | Yes | No | No | Moderate | Grand et al,51 2021 |
| BMI | Hurley stage | P < .001 | Univariate and multivariate regression modeling | Correlation with disease severity | Yes | No | No | No | Moderate | Theut Riis et al,53 2018 |
| PGA score | P < .001 | |||||||||
| Number of areas affected | P < .001 | |||||||||
| Patient-reported severity | P < .001 | |||||||||
| mHSS | R, 0.36; P = .002 for severe (mHSS ≥70) vs moderate and mild disease: OR, 1.12; P = .03 for severe and moderate (mHSS ≥40) vs mild disease: OR, 1.21; P < .001 | Hessam et al,54 2015 | ||||||||
| Hurley stage | Hurley III and II vs I: OR, 1.03; P = .01 | Schrader et al,13 2014 | ||||||||
| Hurley stage | Hurley III vs I and II: OR, 1.25; P = .03 | Alatas et al,55 2020 | ||||||||
| Serum ESR | Hurley stage | Kruskal-Wallis: P = .02; R, 0.30; P = .046 | Multivariate analysis and regression modeling | Correlation with disease severity | Yes | No | No | No | Moderate | Akdogan et al,25 2020 |
| mHSS | R, 0.6; P < .006 | Jiménez-Gallo et al,27 2018 | ||||||||
| HS-PGA | R, 0.6; P < .001 | Jiménez-Gallo et al,26 2017 | ||||||||
| Hurley stage | Kruskal-Wallis: P < .001 | Jiménez-Gallo et al,26 2017 | ||||||||
| Hurley stage | Only in male patients with HS: Hurley I vs III: P = .03; Hurley II vs III: P = .02 | Observational case-control | Matusiak et al,20 2009 | |||||||
| Number of skin areas involved by HS lesions | Only in male patients with HS: R, 0.41; P = .04 | Matusiak et al,20 2009 | ||||||||
| Male sex | Hurley stage | Hurley III and II vs I: OR, 2.11; P < .001 | Multivariate analysis and regression modeling | Correlation with disease severity | Yes | No | No | No | Moderate | Schrader et al,13 2014 |
| Hurley stage | Hurley III vs I and II: OR, 2.56; P = .02 | Alatas et al,55 2020 | ||||||||
| Serum SAA | Hurley stage | Kruskal-Wallis: P = .03; R, 0.31; P = .04 | Observational case-control | Correlation with disease severity | Yes | No | No | No | Moderate | Akdogan et al,25 2020 |
| Sartorius score | R, 0.32; P = .03 | Witte-Händel et al,24 2019 | ||||||||
| CRP | Hurley stage | Kruskal-Wallis: P = .003; R, 0.44; P = .003 | Observational case-control and multivariate regression | Correlation with disease severity | Yes | No | No | No | Moderate | Akdogan et al,25 2020 |
| mHSS | P < .001 | |||||||||
| Hurley stage | Kruskal-Wallis: P < .001 | Hessam et al,54 2015 | ||||||||
| Hurley stage | Kruskal-Wallis: P < .001 | Jiménez-Gallo et al,26 2017 | ||||||||
| Hurley stage | Hurley I vs III: P = .02 | Matusiak et al,20 2009 | ||||||||
| Hurley stage | Hurley I and II vs III: OR, 1.35; P = .001 | Alatas et al,55 2020 | ||||||||
| HS-PGA | R, 0.54; P < .001 | Jiménez-Gallo et al,26 2017 | ||||||||
| mHSS | R, 0.50; P < .001 | Hessam et al,54 2015 | ||||||||
| Serum IL-8 | mHSS | P = .02 | Observational case-control | Correlation with disease severity | Yes | No | No | No | Moderate | Jiménez-Gallo et al,27 2018 |
| HS-PGA | R, 0.41; P < .001 | Jiménez-Gallo et al,26 2017 | ||||||||
| Hurley stage | Kruskal-Wallis: P = .02 | Jiménez-Gallo et al,26 2017 | ||||||||
| Serum IL-17 | Hurley stage | Hurley I vs II: P = .01; Hurley I vs III: P = .005; R, 0.35; P < .001 | Observational case-control | Correlation with disease severity | Yes | No | No | No | Moderate | Matusiak et al,30 2017 |
| HS-PGA | R, 0.37; P = .001 | Observational case-control | Correlation with disease severity | Jiménez-Gallo et al,26 2017 | ||||||
Abbreviations: BMI, body mass index; GRADE, Grading of Recommendations, Assessment, Development, and Evaluations; HS, hidradenitis suppurativa; mHSS, Modified Hidradenitis Suppurativa Score; NRS, numeric rating scale; NS, not stated; OR, odds ratio; PGA, Physician Global Assessment; VAS, visual analog scale.
Table 4.
Predictive Biomarkers (GRADE Moderate/High)
| Biomarker | Response to therapy |
Study type | Interpretation | Critical evaluation |
GRADE evidence profile |
Reference | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Treatment | Outcome measure (significance) |
External validation |
Analytical validation |
Clinical validation |
Clinical utility |
|||||
| Epithelialized tunnels | Adalimumab | HiSCR (OR, 0.47; P = .01) | Multivariate analysis and regression modeling retrospective cohort | Tunnels decrease the odds of achieving a clinical response and increase the length of time to achieve a clinical response | Yes | No | Yes | No | High | Frew et al,56 2020 |
| Time to HiSCR (HR, 0.70; P = .03) | Frew et al,57 2021 | |||||||||
| Family history | Adalimumab | Time to HiSCR (HR, 2.01; P < .001) | Multivariate analysis and regression modeling; retrospective cohort | Family history increased time to loss of HiSCR | Yes | No | Yes | No | High | Frew et al,57 2021 |
| Sonographic vascularization and fibrosis | Adalimumab | Descriptive results only | Multivariate analysis and regression modeling; prospective cohort | Vascularization and fibrosis decrease with clinical response | Yes | No | No | No | Moderate | Nazzaro et al,502019 |
| rs59532114 BCL2 gene | Adalimumab | HiSCR (P < .001) | Genome-wide association study | BCL2 gene variant (increasing BCL2 mRNA) reduced response to adalimumab | No | No | Yes | No | Moderate | Liu et al,58 2020 |
Abbreviations: GRADE, Grading of Recommendations, Assessment, Development, and Evaluations; HiSCR, Hidradenitis Suppurativa Clinical Response; HR, hazard ratio; mRNA, messenger RNA; OR, odds ratio.
Susceptibility/Risk Biomarkers
Serum biomarkers, including fasting serum insulin,10,15 have been independently validated as associated with a diagnosis of HS (Table 1). Demographic variables, such as smoking status10,11 and positive family history,12-14 are also associated with disease (Table 1). Linkage analysis has demonstrated significant association of sequence variants in various components of the gamma secretase complex16 and other non-gamma secretase complex genes, including POFUT117; however, there is a lower prevalence of such variants in White populations with HS compared with East Asian populations, suggesting possible unidentified risk variants.59,60
Other serum risk biomarkers that had a low GRADE score and require future independent validation include serum RBP4,61 ghrelin,61 visfatin,10 TLR10 sequence variants,62 birthweight,63 childhood body mass index,63 and preceding conditions such as type 1 diabetes64 (eTable 2 in the Supplement). Prospective longitudinal studies are required to assess the clinical validity of such biomarkers and avoid selection and recall bias in existing retrospective studies.65
Diagnostic Biomarkers
Currently, the diagnosis of HS is based on clinical criteria defined by the modified Dessau definitions.40,66,67 A plethora of proposed biomarkers have been identified in tissue and serum (Figure) that differentiate between HS and healthy controls (Table 2). However, limited assessment of the analytical validity of such markers has been undertaken.
Only 1 diagnostic biomarker (serum IL-2R) achieved a GRADE rating of high based on multiple independent validation studies18-20 and preanalytical (with age-, sex-, and race-matched controls) and postanalytical validation (calculation of a positive predictive value). One combined panel of serum biomarkers23 (E-selectin/vascular endothelial growth factor [VEGF]/hBD2) initially achieved a GRADE rating of high based on independent validation and analytical validity as well as the ability to differentiate HS from inflammatory bowel disease and staphylococcal skin infection.23 However, this rating was downgraded to moderate because the publication was in abstract form and the study had not undergone peer review.23
Diagnostic biomarkers with validation in more than 1 study include tissue IL-17,24,38-40 IL-1B,24,38,39 serum hepcidin,21,22 E-selectin/VEGF/hBD2 combination,23,67 YLK-40,18 serum amyloid A (SAA),10,24 C-reactive protein (CRP),10,24,26,27 S100A7,39,41,44,45,68 IL-8,24,27 and IL-6.24,27 Additional markers include fecal calprotectin,48,49 serum visfatin,25 serum IL-17,30,41,69 microRNA,31,32 tissue bacterial species,33-37 IL-1B,24,39,43 IL-17A,24,27,39,70 IL-17F,24,42 hBD3,39,45 and IFN-γ.39,41,71,72 A wide range of other markers (eTable 3 in the Supplement) without external validation were also identified. The low GRADE assessments of these markers are a reflection of the lack of independent validation and observational univariate correlation (Table 2; eTable 3 in the Supplement).
Monitoring Biomarkers
The validation of proposed biomarkers to HS disease activity has typically been undertaken using validated clinical outcomes, such as the Sartorius score, HiSCR (Hidradenitis Suppurativa Clinical Response), and IHS4 (International Hidradenitis Suppurativa Severity Score System)11-13 (Table 3). However, validation is often undertaken at 1 time point rather than longitudinally. Monitoring biomarkers with moderate GRADE assessment include serum IL-17,30,38,69 SAA,24,25 CRP,24,25,27 IL-8,26,27 and sonographic measures including tunnel diameter51,73-75 (Table 3). Sonographic dermal vascularity was the only monitoring biomarker to achieve a GRADE rating of high owing to independent validation and assessment of analytical validity.50-52,73-75 Serum erythrocyte sedimentation rate,25-27 tissue citrullinated H3,46,47 and body mass index53,54 are also associated with disease severity using various measures (Table 3) and validated in independent studies. Other monitoring biomarkers identified are listed in eTable 4 in the Supplement.
Significant overlap between diagnostic and monitoring biomarkers was identified (eTables 3 and 4 in the Supplement) with a number of low GRADE-assessed diagnostic markers also having univariate association with disease severity. None of the proposed monitoring biomarkers have undergone analytical or clinical validation to demonstrate validity in a longitudinal setting. This is a particularly important aspect given the significant natural variability of clinical disease activity in HS76 and should be an aspect of future monitoring biomarker studies.
Predictive Biomarkers
Two predictive biomarkers (presence of epithelialized tunnels56,57 and positive family history57) achieved a GRADE assessment of high based on their independent validation and integration into the PIONEER 1 and PIONEER 2 phase 3 clinical trials of adalimumab in HS (Table 4). This indicates that particular morphological and clinical characteristics may be predictive of a response to adalimumab therapy as measured by HiSCR (Table 4). Additionally, BCL2 sequence variants (rs59532114)58 and elevated serum IL-6 levels have been associated with a decreased odds of clinical response to adalimumab58 and infliximab,77 respectively. It should be noted, however, that these results require external validation in future studies (eTable 5 in the Supplement). Given the recent insights into B cells being the site of action of adalimumab in HS,47,78 tissue cell markers such as BAFF and IL-1a47,78 are potential novel predictive markers that require validation in external studies (eTable 5 in the Supplement).
Prognostic, Pharmacodynamic/Response, and Safety Biomarkers
To our knowledge, no longitudinal studies have undertaken assessment of the risk of disease progression. Currently, to our knowledge, no studies have identified pharmacodynamic or safety markers in the setting of HS therapies.
Discussion
The identification, development, and validation of biomarkers for HS are vital aspects to improving clinical management of patients with this chronic, burdensome disease.4 Current biomarkers (Figure, Tables 1, 2, 3, and 4; eTables 2-5 in the Supplement) are primarily diagnostic biomarkers based on small patient cohorts, with few biomarkers integrated into large-scale clinical trials.79,80 Of 155 identified biomarkers, 44 achieved a GRADE rating of moderate, and only 4 biomarkers were rated high. Elevated serum IL-2R was associated with a diagnosis of HS,18-20 likely reflecting the systemic nature of HS inflammation. The monitoring biomarker dermal Doppler vascularity was associated with disease severity,50-52 which may be useful in future clinical and research studies. The presence of epithelialized tunnels in the setting of adalimumab treatment decreased the odds of achieving a clinical response56,57 and increases the length of time to achieve a clinical response.57 Similarly, a positive family history of HS increased time to loss of clinical response in adalimumab studies.57
Barriers and Limitations to Biomarker Identification and Validation
Limitations and proposed future directions of biomarker research in HS are summarized in eTable 6 in the Supplement. A major limitation to existing biomarker studies is the broad number of studies (including clinical/phenotypic markers, imaging-based markers, proteomics, transcriptomics, and genetic-based markers) with insufficient depth of biomarker validation. Additionally, given that (to our knowledge) there are no extant longitudinal studies evaluating the risk of developing HS, all susceptibility/risk biomarkers are based on cross-sectional studies of cases and controls. The identified susceptibility/risk biomarkers would require validation in longitudinal case-control studies to truly reflect the risk of developing HS. The majority of low GRADE-rated biomarkers were identified via univariate association with clinical outcomes (HiSCR, Sartorius score) and patient-reported outcomes (pain, Dermatology Life Quality Index) compared with healthy control participants. Such univariate associations require robust validation in large independent data sets. This should preferentially be undertaken via assumption-free (or -omics) methods to address known issues of selection bias and collinearity,81 rather than via reductive methods, such as examination of only a preselected range of markers.
Currently, as the majority of studies are based on reductive methods, collinearity is likely to exist within the compiled list of biomarkers with collinearity describing the linear association in a statistical regression model between 2 independent variables.81 Many examined markers have been identified based on previous studies in psoriasis vulgaris. Owing to this approach, many identified diagnostic biomarkers target mediators shared by psoriasis vulgaris and HS inflammatory pathways (including S100A7, IL-17A/F, and CXCL1/8). The lack of assumption-free methods leads to selection bias, which is only just beginning to be addressed by omics approaches.47,78 Identification of collinearity in an assumption-free data set enables the identification of the most specific or unique marker, and this may address the issues with identified markers not differentiating between HS and other inflammatory disorders. Addressing the issues of assumption-free biomarker identification and collinearity is important for future HS biomarker research.
Specific barriers to the identification and validation of susceptibility/risk biomarkers include the long-standing issue of diagnostic delay in HS16 and an incomplete understanding of the pathogenesis of disease.1,2,82 Particularly in the context of genomic risk biomarkers, large genome-wide association studies are required before reliable identification of novel risk biomarkers is likely to occur. Additionally, further basic research is needed to elucidate how such identified genetic loci translate to the observed molecular inflammatory pathways in HS, given the conflicting data regarding Notch, PI3K, and AKT signaling in HS.60,83,84 This will be important to understand how such risk loci translate into clinical disease.
Clinical outcomes used in the validation of monitoring biomarkers were largely confined to Hurley staging and the Modified Hidradenitis Suppurativa Score, with the HS Physician Global Assessment also used (Table 3). There is known variability and issues with reproducibility in Hurley staging85 and lesional counts.76 Further validation needs to be undertaken with other outcome measures, such as the IHS486 and HS Area and Severity Index.87
Additionally, recent mechanistic insights suggest that some tissue and serum monitoring biomarkers may be reflective of the different immunological pathways that are seen in individuals with and without tunnels.88,89 This is supported by the negative association of epithelialized tunnels as a predictive biomarker with clinical response to adalimumab.56,57 Further work is needed to clarify how such structures should be defined and identified, whether clinically, sonographically, or histologically, for such biomarkers to be relevant to the practicing physician. The continued integration of biomarkers into the design of future clinical trials is a vital step forward in translating biomarker research directly to the clinic. Currently, post hoc analyses of 2 clinical trials using complement inhibitors (vilobelimab90 and avacopan91) have identified epithelialized tunnels as a significant predictor of clinical response to this class of agents. Consideration of patient stratification by a validated predictive biomarker would aid the identification of more targeted therapeutics for subpopulations of individuals with HS (eg, those with and without epithelialized tunnels) as is done for fistulizing and nonfistulizing Crohn disease in the gastroenterology field. These data, combined with recent evidence suggesting that neutrophil- and B-cell–associated pathways are associated with these tunnels,78,88 further emphasize the need for evaluation of collinearity among predictive biomarkers in particular and validation with assumption-free methods.
The role of bacterial-based biomarkers requires further investigation given the established role of bacterial dysbiosis in HS.33-37 It is acknowledged that the inflammatory markers identified in multiple studies may be influenced by the presence or absence of specific microbionts,37 and hence, future studies correlating tissue and serum inflammatory profiles with microbiome studies are needed to examine the relevance and collinearity of bacterial dysbiosis to identified immunological alterations.
Tissue-based biomarker identification is prone to variability and heterogeneity in findings owing to natural immunological variability across cutaneous sites,92,93 as well as inconsistency in methods in identifying of lesional, perilesional, and nonlesional tissue.94 While recommendations for lesion definitions and biopsy sites have been published,94,95 implementation and further consensus regarding such definitions is required to standardize tissue-based biomarker investigations. Although outside the bounds of our search strategy, recent publications have identified serum IL-8, CCL-19, and CXCL9 as predictive biomarkers of adalimumab response in the PIONEER phase 3 randomized clinical trials.96 This study would have raised serum IL-8 level to a moderate GRADE rating and also provides independent validation of the importance of B-cell–associated chemokines in response to TNF-α blockade.47
The identification of prognostic biomarkers, particularly regarding progression to scarring/advanced disease or association with other complications such as squamous cell carcinoma,97-99 is an important area of need for future biomarker research. Biomarker research in the context of longitudinal registry efforts to understand the clinical course of HS, such as the multi-institutional Hidradenitis Suppurativa Prospective Observational Registry and Biospecimen Repository (HS PROGRESS), will be critical to developing robust prognostic biomarkers.100
Use of HS Biomarkers in Trials and the Clinic
The results of this systematic review suggest that in HS, no single biomarker has yet been adequately validated to recommend for routine clinical use. Identified biomarkers with the highest GRADE recommendations included serum IL-2R as a diagnostic biomarker,18-20 dermal doppler vascularity as a monitoring biomarker, and epithelialized tunnels and family history as predictive biomarkers.56,57
Identification and validation of high-quality predictive biomarkers are especially relevant given the number of mid-phase clinical trials that have failed to achieve primary outcome(s) owing to suspected disease heterogeneity.90,91 At present, the main focus of clinical trials is on clinical outcomes, while less emphasis is placed on analysis of serum or tissue-based biomarkers. Aside from the well-documented issues with outcome measures in HS clinical trials,86,87 the consideration of patient stratification by a validated predictive biomarker would aid the identification of more targeted therapeutics for subpopulations of individuals with HS (eg, those with and without epithelialized tunnels) as is done for fistulizing and nonfistulizing Crohn disease in the gastroenterology field. The first step to realize this would be the examination of collinearity between predictive biomarkers, given the theoretical underpinnings linking epithelialized tunnels, neutrophil activity, and B cells in HS.56-58
Limitations
Priorities for future biomarker studies should address the described limitations in the current literature as outlined previously (eTable 6 in the Supplement). More specifically, identified biomarkers require independent validation in an assumption-free omics setting in both tissue and whole blood/serum.81 Independent studies looking at the collinearity of identified clinical and inflammatory markers would complement feature selection algorithms81 to identify associations between biomarkers and the most appropriate biomarker for a given task (eg, diagnostic, monitoring). Current work in development and validation of clinical outcome measures (including the IHS4,86 Severity and Area Score for Hidradenitis,40 and HS Area and Severity Index87) will likely provide benefits in biomarker validation given that newer outcome measures take into account the variable morphologic characteristics (nodules, abscesses, and tunnels) that can contribute to disease activity in HS. This has particular relevance in the setting of predictive and monitoring biomarkers, where differential response may be seen in different disease morphologic characteristics (eg, nodules vs tunnels).
Conclusions
Studies in the extant literature provide an important step in the development of biomarkers for daily clinical use in HS, although no biomarker is yet at the point of established clinical utility. Biomarkers hold great potential to advance our understanding of disease pathophysiology as well as the clinical management of HS via identification of novel treatment targets. However, the process of biomarker validation requires multicenter and independent validation of findings and assumption-free methods to identify the most appropriate marker for clinical validation. Additionally, unique issues in the setting of HS include the need for assessment of biomarker stability over time as well as acknowledgment of the deficiencies in existing clinical outcome measures. Overall, to advance this field in HS and more robustly validate identified biomarkers, stakeholder consensus is required to outline and propose standardized methods for the identification, investigation, and validation of biomarkers in HS.
Supplementary Material
Key Points.
Question
What is the role of biomarkers in hidradenitis suppurativa (HS), and how are they validated?
Findings
In this systematic review, a total of 48 biomarkers were identified with a minimum Grading of Recommendations, Assessment, Development, and Evaluations rating of moderate; only 1 diagnostic (serum IL-2R), 1 monitoring (dermal Doppler vascularity), and 2 predictive biomarkers (epithelialized tunnels and positive family history of HS) achieved a high rating. None of the identified biomarkers had sufficient clinical validity to be recommended for routine use in the clinical setting; priorities were presented for near-term future research to overcome barriers and limitations of biomarkers in HS.
Meaning
The identification and validation of biomarkers in HS has potential to improve the understanding and management of this chronic, burdensome disease.
Footnotes
Conflict of Interest Disclosures: Dr Hessam reported serving on advisory boards for AbbVie and participating in trials for AbbVie and Novartis outside the submitted work. Dr Kirby reported receiving personal fees from AbbVie, ChemoCentryx, Incyte, Janssen, Novartis, and UCB Pharma outside the submitted work. Dr Lowes reported serving on advisory boards for AbbVie, InflaRx, Janssen, and Viela Bio; consulting for Almirall, BSN Medical, Incyte, Janssen, Kymera, Phoenicis, and XBiotech; and serving on the medical board of the Hidradenitis Suppurativa Foundation, a voluntary position. Dr Naik reported receiving personal fees (consulting) and grants from AbbVie, personal fees (consulting) from 23andMe and DAVA Oncology, and personal fees (advisory board) from Boehringer Ingelheim; serving as an investigator for Pfizer outside the submitted work; and serving as an unpaid board member of the US Hidradenitis Suppurativa Foundation. Dr Frew reported receiving personal fees from Janssen, Pfizer, Boehringer Ingelheim, AbbVie, Eli Lilly, and LEO Pharma and grants from Sun Pharma outside the submitted work. No other disclosures were reported.
Disclaimer: Dr Naik is an Associate Editor of JAMA Dermatology but was not involved in any of the decisions regarding review of the manuscript or its acceptance.
Contributor Information
Samuel Der Sarkissian, Department of Dermatology, Liverpool Hospital, Sydney, Australia.
Schapoor Hessam, Department of Dermatology, Venereology and Allergology, Ruhr-University Bochum, Bochum, Germany.
Joslyn S. Kirby, Department of Dermatology, Penn State Health Milton S. Hershey Medical Center, Hershey, Pennsylvania.
Michelle A. Lowes, Rockefeller University, New York, New York.
Dillon Mintoff, Department of Dermatology, Mater Dei Hospital, Msida, Malta.
Haley B. Naik, Department of Dermatology, University of California San Francisco; JAMA Dermatology.
Hans Christian Ring, Department of Dermato-Venereology & Wound Healing Centre, Bispebjerg Hospital, Copenhagen, Denmark.
Nisha Chandran Suyien, Division of Dermatology, Department of Medicine, National University Hospital, Singapore.
John W. Frew, Department of Dermatology, Liverpool Hospital, Sydney, Australia; University of New South Wales, Sydney, Australia, Laboratory of Translational Cutaneous Medicine, Ingham Institute of Applied Medical Research, Sydney, Australia.
REFERENCES
- 1.Goldburg SR, Strober BE, Payette MJ. Hidradenitis suppurativa: epidemiology, clinical presentation, and pathogenesis.J Am Acad Dermatol. 2020;82(5):1045–1058. doi: 10.1016/j.jaad.2019.08.090 [DOI] [PubMed] [Google Scholar]
- 2.Sabat R, Jemec GBE, Matusiak Ł, Kimball AB, Prens E, Wolk K. Hidradenitis suppurativa. Nat Rev Dis Primers. 2020;6(1):18. doi: 10.1038/s41572-020-0149-1 [DOI] [PubMed] [Google Scholar]
- 3.Kohorst JJ, Kimball AB, Davis MD. Systemic associations of hidradenitis suppurativa. J Am Acad Dermatol. 2015;73(5)(suppl 1):S27–S35. doi: 10.1016/j.jaad.2015.07.055 [DOI] [PubMed] [Google Scholar]
- 4.Frew JW, Marzano AV, Wolk K, et al. A systematic review of promising therapeutic targets in hidradenitis suppurativa: a critical evaluation of mechanistic and clinical relevance. J Invest Dermatol. 2021;141(2):316–324.e2. doi: 10.1016/j.jid.2020.06.019 [DOI] [PubMed] [Google Scholar]
- 5.Byrd AS, Dina Y, Okoh UJ, et al. Specimen collection for translational studies in hidradenitis suppurativa. Sci Rep. 2019;9(1):12207. doi: 10.1038/s41598-019-48226-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amur A. Biomarker terminology: speaking the same language. US Food and Drug Administration. Accessed October 12, 2020. https://www.fda.gov/files/BIOMARKER-TERMINOLOGY--SPEAKING-THE-SAME-LANGUAGE.pdf [Google Scholar]
- 7.Biomarker. European Medicines Agency. Accessed October 12, 2020. https://www.ema.europa.eu/en/glossary/biomarker#:~:text=A%20biological%20molecule%20found%20in,European%20Medicines%20Agency [Google Scholar]
- 8.Renert-Yuval Y, Thyssen JP, Bissonnette R, et al. Biomarkers in atopic dermatitis—a review on behalf of the International Eczema Council. J Allergy Clin Immunol. 2021;147(4):1174–1190.e1. doi: 10.1016/j.jaci.2021.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guyatt G, Oxman AD, Sultan S, et al. GRADE guidelines: 11. making an overall rating of confidence in effect estimates for a single outcome and for all outcomes. J Clin Epidemiol. 2013;66(2):151–157. doi: 10.1016/j.jclinepi.2012.01.006 [DOI] [PubMed] [Google Scholar]
- 10.Akdogan N, Alli N, Uysal PI, Topcuoglu C, Candar T, Turhan T. Visfatin and insulin levels and cigarette smoking are independent risk factors for hidradenitis suppurativa: a case-control study. Arch Dermatol Res. 2018;310(10):785–793. doi: 10.1007/s00403-018-1867-z [DOI] [PubMed] [Google Scholar]
- 11.Garg A, Papagermanos V, Midura M, Strunk A. Incidence of hidradenitis suppurativa among tobacco smokers: a population-based retrospective analysis in the USA. Br J Dermatol. 2018;178(3):709–714. doi: 10.1111/bjd.15939 [DOI] [PubMed] [Google Scholar]
- 12.van Straalen KR, Prens EP, Willemsen G, Boomsma DI, van der Zee HH. Contribution of genetics to the susceptibility to hidradenitis suppurativa in a large, cross-sectional Dutch twin cohort. JAMA Dermatol. 2020;156(12):1359–1362. doi: 10.1001/jamadermatol.2020.3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schrader AM, Deckers IE, van der Zee HH, Boer J, Prens EP. Hidradenitis suppurativa: a retrospective study of 846 Dutch patients to identify factors associated with disease severity. J Am Acad Dermatol. 2014;71(3):460–467. doi: 10.1016/j.jaad.2014.04.001 [DOI] [PubMed] [Google Scholar]
- 14.Molina-Leyva A, Cuenca-Barrales C. Adolescent-onset hidradenitis suppurativa: prevalence, risk factors and disease features. Dermatology. 2019;235(1):45–50. doi: 10.1159/000493465 [DOI] [PubMed] [Google Scholar]
- 15.Vilanova I, Hernández JL, Mata C, et al. Insulin resistance in hidradenitis suppurativa: a case-control study. J Eur Acad Dermatol Venereol. 2018;32(5):820–824. doi: 10.1111/jdv.14894 [DOI] [PubMed] [Google Scholar]
- 16.Frew JW, Vekic DA, Woods J, Cains GD. A systematic review and critical evaluation of reported pathogenic sequence variants in hidradenitis suppurativa. Br J Dermatol. 2017;177(4):987–998. doi: 10.1111/bjd.15441 [DOI] [PubMed] [Google Scholar]
- 17.Jfri AH, O'Brien EA, Litvinov IV, Alavi A, Netchiporouk E. Hidradenitis suppurativa: comprehensive review of predisposing genetic mutations and changes. J Cutan Med Surg. 2019;23(5):519–527. doi: 10.1177/1203475419852049 [DOI] [PubMed] [Google Scholar]
- 18.Matusiak Ł, Salomon J, Nowicka-Suszko D, Bieniek A, Szepietowski JC. Chitinase-3-like protein 1 (YKL-40): novel biomarker of hidradenitis suppurativa disease activity? Acta Derm Venereol. 2015;95(6):736–737. doi: 10.2340/00015555-2061 [DOI] [PubMed] [Google Scholar]
- 19.Wieland CW, Vogl T, Ordelman A, et al. Myeloid marker S100A8/A9 and lymphocyte marker, soluble interleukin 2 receptor: biomarkers of hidradenitis suppurativa disease activity? Br J Dermatol. 2013;168(6):1252–1258. doi: 10.1111/bjd.12234 [DOI] [PubMed] [Google Scholar]
- 20.Matusiak Ł, Bieniek A, Szepietowski JC. Soluble interleukin-2 receptor serum level is a useful marker of hidradenitis suppurativa clinical staging. Biomarkers. 2009;14(6):432–437. doi: 10.1080/13547500903075218 [DOI] [PubMed] [Google Scholar]
- 21.Ponikowska M, Matusiak L, Kasztura M, Jankowska EA, Szepietowski JC. Deranged iron status evidenced by iron deficiency characterizes patients with hidradenitis suppurativa. Dermatology. 2020;236(1):52–58. doi: 10.1159/000505184 [DOI] [PubMed] [Google Scholar]
- 22.Ghias M, Cameron S, Shaw F, et al. Anemia in hidradenitis suppurativa, hepcidin as a diagnostic tool. Am J Clin Pathol. 2019;152(suppl 1):S15. doi: 10.1093/ajcp/aqz112.029 [DOI] [Google Scholar]
- 23.Argyropoulou M, Grundhuber M, Kanni T, et al. A composite biomarker score for the diagnosis of hidradenitis suppurativa [8th European Hidradenitis Suppurativa Foundation Conference oral session abstract 028 OS06-01]. Exp Dermatol. 2019;28(suppl 2):18. doi: 10.1111/exd.13893 [DOI] [Google Scholar]
- 24.Witte-Händel E, Wolk K, Tsaousi A, et al. The IL-1 pathway is hyperactive in hidradenitis suppurativa and contributes to skin infiltration and destruction. J Invest Dermatol. 2019;139(6):1294–1305. doi: 10.1016/j.jid.2018.11.018 [DOI] [PubMed] [Google Scholar]
- 25.Akdogan N, Dogan S, Incel-Uysal P, et al. Serum amyloid A and C-reactive protein levels and erythrocyte sedimentation rate are important indicators in hidradenitis suppurativa. Arch Dermatol Res. 2020;312(4):255–262. doi: 10.1007/s00403-019-02014-8 [DOI] [PubMed] [Google Scholar]
- 26.Jiménez-Gallo D, de la Varga-Martínez R, Ossorio-García L, Albarrán-Planelles C, Rodríguez C, Linares-Barrios M. The clinical significance of increased serum proinflammatory cytokines, C-reactive protein, and erythrocyte sedimentation rate in patients with hidradenitis suppurativa. Mediators Inflamm. 2017;2017:2450401. doi: 10.1155/2017/2450401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiménez-Gallo D, de la Varga-Martínez R, Ossorio-García L, Collantes-Rodríguez C, Rodríguez C, Linares-Barrios M. Effects of adalimumab on T-helper-17 lymphocyte- and neutrophil-related inflammatory serum markers in patients with moderate-to-severe hidradenitis suppurativa. Cytokine. 2018;103:20–24. doi: 10.1016/j.cyto.2017.12.020 [DOI] [PubMed] [Google Scholar]
- 28.González-López MA, Vilanova I, Ocejo-Viñals G, et al. Circulating levels of adiponectin, leptin, resistin and visfatin in non-diabetics patients with hidradenitis suppurativa. Arch Dermatol Res. 2020;312(8):595–600. doi: 10.1007/s00403-019-02018-4 [DOI] [PubMed] [Google Scholar]
- 29.Özkur E, Erdem Y, Altunay IK, et al. Serum irisin level, insulin resistance, and lipid profiles in patients with hidradenitis suppurativa: a case-control study. An Bras Dermatol. 2020;95(6):708–713. doi: 10.1016/j.abd.2020.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matusiak Ł, Szczęch J, Bieniek A, Nowicka-Suszko D, Szepietowski JC. Increased interleukin (IL)-17 serum levels in patients with hidradenitis suppurativa: implications for treatment with anti-IL-17 agents. J Am Acad Dermatol. 2017;76(4):670–675. doi: 10.1016/j.jaad.2016.10.042 [DOI] [PubMed] [Google Scholar]
- 31.Radhakrishna U, Vishweswaraiah S, Ratnamala U, Bahado-Singh R, Saiyed N. DNA methylation and microRNA biomarkers for noninvasive detection of hidradenitis suppurativa [8th European Hidradenitis Suppurativa Foundation Conference, oral session 004OS01-02]. Exp Dermatol. 2019;28(suppl 2):6–7. doi: 10.1111/exd.13893 [DOI] [Google Scholar]
- 32.Hessam S, Sand M, Skrygan M, Gambichler T, Bechara FG. Expression of miRNA-155, miRNA-223, miRNA-31, miRNA-21, miRNA-125b, and miRNA-146a in the inflammatory pathway of hidradenitis suppurativa. Inflammation. 2017;40(2):464–472. doi: 10.1007/s10753-016-0492-2 [DOI] [PubMed] [Google Scholar]
- 33.Ring HC, Thorsen J, Saunte DM, et al. The follicular skin microbiome in patients with hidradenitis suppurativa and healthy controls. JAMA Dermatol. 2017;153(9):897–905. doi: 10.1001/jamadermatol.2017.0904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guet-Revillet H, Jais JP, Ungeheuer MN, et al. The microbiological landscape of anaerobic infections in hidradenitis suppurativa: a prospective metagenomic study. Clin Infect Dis. 2017;65(2):282–291. doi: 10.1093/cid/cix285 [DOI] [PubMed] [Google Scholar]
- 35.Naik HB, Nassif A, Ramesh MS, et al. Are bacteria infectious pathogens in hidradenitis suppurativa? debate at the Symposium for Hidradenitis Suppurativa Advances Meeting, November 2017. J Invest Dermatol. 2019;139(1):13–16. doi: 10.1016/j.jid.2018.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ring HC, Sigsgaard V, Thorsen J, et al. The microbiome of tunnels in hidradenitis suppurativa patients. J Eur Acad Dermatol Venereol. 2019;33(9):1775–1780. doi: 10.1111/jdv.15597 [DOI] [PubMed] [Google Scholar]
- 37.Schneider AM, Cook LC, Zhan X, et al. Loss of skin microbial diversity and alteration of bacterial metabolic function in hidradenitis suppurativa. J Invest Dermatol. 2020;140(3):716–720. doi: 10.1016/j.jid.2019.06.151 [DOI] [PubMed] [Google Scholar]
- 38.Kelly G, Hughes R, McGarry T, et al. Dysregulated cytokine expression in lesional and nonlesional skin in hidradenitis suppurativa. Br J Dermatol. 2015;173(6):1431–1439. doi: 10.1111/bjd.14075 [DOI] [PubMed] [Google Scholar]
- 39.Wolk K, Warszawska K, Hoeflich C, et al. Deficiency of IL-22 contributes to a chronic inflammatory disease: pathogenetic mechanisms in acne inversa. J Immunol. 2011;186(2):1228–1239. doi: 10.4049/jimmunol.0903907 [DOI] [PubMed] [Google Scholar]
- 40.Hessam S, Scholl L, Sand M, Schmitz L, Reitenbach S, Bechara FG. A novel severity assessment scoring system for hidradenitis suppurativa. JAMA Dermatol. 2018;154(3):330–335. doi: 10.1001/jamadermatol.2017.5890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hotz C, Boniotto M, Guguin A, et al. Intrinsic defect in keratinocyte function leads to inflammation in hidradenitis suppurativa. J Invest Dermatol. 2016;136(9):1768–1780. doi: 10.1016/j.jid.2016.04.036 [DOI] [PubMed] [Google Scholar]
- 42.Navrazhina K, Frew JW, Krueger JG. Interleukin 17C is elevated in lesional tissue of hidradenitis suppurativa. Br J Dermatol. 2020;182(4):1045–1047. doi: 10.1111/bjd.18556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Zee HH, de Ruiter L, van den Broecke DG, Dik WA, Laman JD, Prens EP. Elevated levels of tumour necrosis factor (TNF)-α, interleukin (IL)-1β and IL-10 in hidradenitis suppurativa skin: a rationale for targeting TNF-α and IL-1β. Br J Dermatol. 2011;164(6):1292–1298. doi: 10.1111/j.1365-2133.2011.10254.x [DOI] [PubMed] [Google Scholar]
- 44.Batycka-Baran A, Baran W, Nowicka-Suszko D, et al. Serum concentration and skin expression of S100A7 (psoriasin) in patients suffering from hidradenitis suppurativa. Dermatology. 2021;237(5):733–739. doi: 10.1159/000510689 [DOI] [PubMed] [Google Scholar]
- 45.Emelianov VU, Bechara FG, Gläser R, et al. Immunohistological pointers to a possible role for excessive cathelicidin (LL-37) expression by apocrine sweat glands in the pathogenesis of hidradenitis suppurativa/acne inversa. Br J Dermatol. 2012;166(5):1023–1034. doi: 10.1111/j.1365-2133.2011.10765.x [DOI] [PubMed] [Google Scholar]
- 46.Byrd AS, Carmona-Rivera C, O'Neil LJ, et al. Neutrophil extracellular traps, B cells, and type I interferons contribute to immune dysregulation in hidradenitis suppurativa. Sci Transl Med. 2019;11(508):eaav5908. doi: 10.1126/scitranslmed.aav5908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lowe MM, Naik HB, Clancy S, et al. Immunopathogenesis of hidradenitis suppurativa and response to anti-TNF-α therapy. JCI Insight. 2020;5(19):e139932. doi: 10.1172/jci.insight.139932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eşer E, Engin B, Yüksel P, et al. Relationship between fecal calprotectin level and disease activity in patients with hidradenitis suppurativa. Dermatol Ther. 2020;33(2):e13232. doi: 10.1111/dth.13232 [DOI] [PubMed] [Google Scholar]
- 49.Lloyd-McLennan AM, Ali S, Kittler NW. Prevalence of inflammatory bowel disease among pediatric patients with hidradenitis suppurativa and the potential role of screening with fecal calprotectin. Pediatr Dermatol. 2021;38(1):98–102. doi: 10.1111/pde.14417 [DOI] [PubMed] [Google Scholar]
- 50.Nazzaro G, Passoni E, Calzari P, et al. Color Doppler as a tool for correlating vascularization and pain in hidradenitis suppurativa lesions. Skin Res Technol. 2019;25(6):830–834. doi: 10.1111/srt.12729 [DOI] [PubMed] [Google Scholar]
- 51.Grand D, Frew JW, Navrazhina K, Krueger JG. Doppler ultrasound-based noninvasive biomarkers in hidradenitis suppurativa: evaluation of analytical and clinical validity. Br J Dermatol. 2021;184(4):688–696. doi: 10.1111/bjd.19343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caposiena Caro RD, Solivetti FM, Bianchi L. Power Doppler ultrasound assessment of vascularization in hidradenitis suppurativa lesions. J Eur Acad Dermatol Venereol. 2018;32(8):1360–1367. doi: 10.1111/jdv.14745 [DOI] [PubMed] [Google Scholar]
- 53.Theut Riis P, Saunte DM, Benhadou F, et al. Low and high body mass index in hidradenitis suppurativa patients-different subtypes? J Eur Acad Dermatol Venereol. 2018;32(2):307–312. doi: 10.1111/jdv.14599 [DOI] [PubMed] [Google Scholar]
- 54.Hessam S, Sand M, Gambichler T, Bechara FG. Correlation of inflammatory serum markers with disease severity in patients with hidradenitis suppurativa (HS). JAm Acad Dermatol. 2015;73(6):998–1005. doi: 10.1016/j.jaad.2015.08.052 [DOI] [PubMed] [Google Scholar]
- 55.Alatas ET, Biteker M, Alatas OD. Epicardial fat thickness is increased and associated with disease severity in hidradenitis suppurativa. Arch Dermatol Res. 2020;312(7):467–472. doi: 10.1007/s00403-019-02032-6 [DOI] [PubMed] [Google Scholar]
- 56.Frew JW,Jiang CS,Singh N, et al. Clinical response rates, placebo response rates, and significantly associated covariates are dependent on choice of outcome measure in hidradenitis suppurativa: a post hoc analysis of PIONEER 1 and 2 individual patient data. J Am Acad Dermatol. 2020;82(5):1150–1157. doi: 10.1016/j.jaad.2019.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frew JW, Jiang CS, Singh N, et al. Dermal tunnels influence time to clinical response and family history influences time to loss of clinical response in patients with hidradenitis suppurativa treated with adalimumab. Clin Exp Dermatol. 2021;46(2):306–313. doi: 10.1111/ced.14448 [DOI] [PubMed] [Google Scholar]
- 58.Liu M, Degner J, Georgantas RW, et al. A genetic variant in the BCL2 gene associates with adalimumab response in hidradenitis suppurativa clinical trials and regulates expression of BCL2. J Invest Dermatol. 2020;140(3):574–582.e2. doi: 10.1016/j.jid.2019.06.152 [DOI] [PubMed] [Google Scholar]
- 59.Duchatelet S, Miskinyte S, Delage M, et al. Low prevalence of GSC gene mutations in a large cohort of predominantly Caucasian patients with hidradenitis suppurativa. J Invest Dermatol. 2020;140(10):2085–2088.e14. doi: 10.1016/j.jid.2019.10.025 [DOI] [PubMed] [Google Scholar]
- 60.Frew JW. Differential profiles of gamma secretase Notch signalling in hidradenitis suppurativa: the need for genotype-endotype-phenotype analysis. Br J Dermatol. 2021;185(3):636–637. doi: 10.1111/bjd.19805 [DOI] [PubMed] [Google Scholar]
- 61.González-López MA, Ocejo-Viñals JG, Mata C, et al. Association of retinol binding protein4 (RBP4) and ghrelin plasma levels with insulin resistance and disease severity in non-diabetic patients with hidradenitis suppurativa. Exp Dermatol. 2020;29(9):828–832. doi: 10.1111/exd.14132 [DOI] [PubMed] [Google Scholar]
- 62.Ocejo-Vinyals JG, Irure-Ventura J, Guiterrez-Larranaga M, et al. Association of TLR polymorphisms with susceptibility to hidradenitis suppurativa in a Spanish Caucasian population [34th European Immunogenetics and Histocompatibility and 31st British Society for Histocompatibility and Immunogenetics Conference abstract]. HLA. 2020;95(4):410–411. doi: 10.1111/tan.13844 [DOI] [Google Scholar]
- 63.Jørgensen AR,Aarestrup J, Baker JL, Thomsen SF. Association of birth weight, childhood body mass index, and height with risk of hidradenitis suppurativa. JAMA Dermatol. 2020;156(7):746–753. doi: 10.1001/jamadermatol.2020.1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kjæsgaard Andersen R, Jørgensen IF, Reguant R, Jemec GBE, Brunak S. Disease trajectories for hidradenitis suppurativa in the Danish population. JAMA Dermatol. 2020;156(7):780–786. doi: 10.1001/jamadermatol.2020.1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Althubaiti A. Information bias in health research: definition, pitfalls, and adjustment methods. J Multidiscip Healthc. 2016;9:211–217. doi: 10.2147/JMDH.S104807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zouboulis CC, Del Marmol V, Mrowietz U, Prens EP,Tzellos T, Jemec GB. Hidradenitis suppurativa/acne inversa: criteria for diagnosis, severity assessment, classification and disease evaluation. Dermatology. 2015;231(2):184–190. doi: 10.1159/000431175 [DOI] [PubMed] [Google Scholar]
- 67.Revuz JE, Jemec GB. Diagnosing hidradenitis suppurativa. Dermatol Clin. 2016;34(1):1–5. doi: 10.1016/j.det.2015.08.009 [DOI] [PubMed] [Google Scholar]
- 68.Lima AL, Karl I, Giner T, et al. Keratinocytes and neutrophils are important sources of proinflammatory molecules in hidradenitis suppurativa. Br J Dermatol. 2016;174(3):514–521. doi: 10.1111/bjd.14214 [DOI] [PubMed] [Google Scholar]
- 69.Kanni T, Tzanetakou V, Savva A, et al. Compartmentalized cytokine responses in hidradenitis suppurativa. PLoS One. 2015;10(6):e0130522. doi: 10.1371/journal.pone.0130522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van der Zee HH, Laman JD, de Ruiter L, Dik W A, Prens EP. Adalimumab (antitumour necrosis factor-α) treatment of hidradenitis suppurativa ameliorates skin inflammation: an in situ and ex vivo study. Br J Dermatol. 2012;166(2):298–305. doi: 10.1111/j.1365-2133.2011.10698.x [DOI] [PubMed] [Google Scholar]
- 71.Thomi R, Schlapbach C, Yawalkar N, Simon D, Yerly D, Hunger RE. Elevated levels of the antimicrobial peptide LL-37 in hidradenitis suppurativa are associated with a Th1/Th17 immune response. Exp Dermatol. 2018;27(2):172–177. doi: 10.1111/exd.13482 [DOI] [PubMed] [Google Scholar]
- 72.Ten Oever J, van de Veerdonk FL, Joosten LA, et al. Cytokine production assays reveal discriminatory immune defects in adults with recurrent infections and noninfectious inflammation. Clin Vaccine Immunol. 2014;21(8):1061–1069. doi: 10.1128/CVI.00152-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grand D, Navrazhina K, Frew JWA. A scoping review of non-invasive imaging modalities in dermatological disease: potential novel biomarkers in hidradenitis suppurativa. Front Med (Lausanne). 2019;6:253. doi: 10.3389/fmed.2019.00253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wortsman X, Moreno C, Soto R, Arellano J, Pezo C, Wortsman J. Ultrasound in-depth characterization and staging of hidradenitis suppurativa. Dermatol Surg. 2013;39(12):1835–1842. doi: 10.1111/dsu.12329 [DOI] [PubMed] [Google Scholar]
- 75.Martorell A, Alfageme Roldán F, Vilarrasa Rull E, et al. Ultrasound as a diagnostic and management tool in hidradenitis suppurativa patients: a multicentre study. J Eur Acad Dermatol Venereol. 2019;33(11):2137–2142. doi: 10.1111/jdv.15710 [DOI] [PubMed] [Google Scholar]
- 76.Frew JW, Jiang CS, Singh N, Navrazhina K, Vaughan R, Krueger JG. Quantifying the natural variation in lesion counts over time in untreated hidradenitis suppurativa: implications for outcome measures and trial design. JAAD Int. 2020;1(2):208–221. doi: 10.1016/j.jdin.2020.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Montaudié H, Seitz-Polski B, Cornille A, Benzaken S, Lacour JP, Passeron T. Interleukin 6 and high-sensitivity C-reactive protein are potential predictive markers of response to infliximab in hidradenitis suppurativa. J Am Acad Dermatol. 2017;76(1):156–158. doi: 10.1016/j.jaad.2016.08.036 [DOI] [PubMed] [Google Scholar]
- 78.Gudjonsson JE, Tsoi LC, Ma F, et al. Contribution of plasma cells and B cells to hidradenitis suppurativa pathogenesis. JCI Insight. 2020;5(19):e139930. doi: 10.1172/jci.insight.139930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kimball AB, Okun MM, Williams DA, et al. Two phase 3 trials of adalimumab for hidradenitis suppurativa. N Engl J Med. 2016;375(5):422–434. doi: 10.1056/NEJMoa1504370 [DOI] [PubMed] [Google Scholar]
- 80.Zouboulis CC, Okun MM, Prens EP, et al. Long-term adalimumab efficacy in patients with moderate-to-severe hidradenitis suppurativa/acne inversa: 3-year results of a phase 3 open-label extension study. J Am Acad Dermatol. 2019;80(1):60–69.e2. doi: 10.1016/j.jaad.2018.05.040 [DOI] [PubMed] [Google Scholar]
- 81.Torres R, Judson-Torres RL. Research techniques made simple: feature selection for biomarker discovery. J Invest Dermatol. 2019;139 (10):2068–2074.e1. doi: 10.1016/j.jid.2019.07.682 [DOI] [PubMed] [Google Scholar]
- 82.Frew JW. Hidradenitis suppurativa is an autoinflammatory keratinization disease: a review of the clinical, histologic, and molecular evidence. JAAD Int. 2020;1(1):62–72. doi: 10.1016/j.jdin.2020.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hessam S,Gambichler T,Skrygan M, et al. Increased expression profile of NCSTN, Notch and PI3K/AKT3 in hidradenitis suppurativa. J Eur Acad Dermatol Venereol. 2021;35(1):203–210. doi: 10.1111/jdv.16962 [DOI] [PubMed] [Google Scholar]
- 84.Xiao X, He Y, Li C, Zhang X, Xu H, Wang B. Nicastrin mutations in familial acne inversa impact keratinocyte proliferation and differentiation through the Notch and phosphoinositide 3-kinase/AKT signalling pathways. Br J Dermatol. 2016;174(3):522–532. doi: 10.1111/bjd.14223 [DOI] [PubMed] [Google Scholar]
- 85.Sartorius K, Lapins J, Emtestam L, Jemec GB. Suggestions for uniform outcome variables when reporting treatment effects in hidradenitis suppurativa. Br J Dermatol. 2003;149(1):211–213. doi: 10.1046/j.1365-2133.2003.05390.x [DOI] [PubMed] [Google Scholar]
- 86.Zouboulis CC, Tzellos T, Kyrgidis A, et al. ; European Hidradenitis Suppurativa Foundation Investigator Group. Development and validation of the International Hidradenitis Suppurativa Severity Score System (IHS4), a novel dynamic scoring system to assess HS severity. Br J Dermatol. 2017;177(5):1401–1409. doi: 10.1111/bjd.15748 [DOI] [PubMed] [Google Scholar]
- 87.Goldfarb N, Lowes MA, Butt M, King T, Alavi A, Kirby JS. Hidradenitis Suppurativa Area and Severity Index Revised (HASI-R): psychometric property assessment. Br J Dermatol. 2021;184(5):905–912. doi: 10.1111/bjd.19565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Navrazhina K, Frew JW, Gilleaudeau P, Sullivan-Whelan M, Garcet S, Krueger JG. Epithelialized tunnels are a source of inflammation in hidradenitis suppurativa. J Allerg Clin Immunol. 2021;147(6):2213–2224. doi: 10.1016/j.jaci.2020.12.651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Navrazhina K, Garcet S, Gonzalez J, Grand D, Frew JW, Krueger JG. In-depth analysis of the hidradenitis suppurativa serum proteome identifies distinct inflammatory subtypes. J Invest Dermatol. 2021;141(9):2197–2207. doi: 10.1016/j.jid.2021.02.742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Giamarellos-Bourboulis EJ, Argyropoulou M, Kanni T, et al. Clinical efficacy of complement C5a inhibition by IFX-1 in hidradenitis suppurativa: an open-label single-arm trial in patients not eligible for adalimumab. Br J Dermatol. 2020;183(1):176–178. doi: 10.1111/bjd.18877 [DOI] [PubMed] [Google Scholar]
- 91.ChemoCentryx announces positive topline results of phase II AURORA clinical trial of avacopan in the treatment of hidradenitis suppurativa (HS). News release. Accessed March 1, 2021. https://ir.chemocentryx.com/news-releases/news-release-details/chemocentryx-announces-positive-topline-results-phase-ii-aurora [Google Scholar]
- 92.Jenei A, Dajnoki Z, Medgyesi B, et al. Apocrine gland-rich skin has a non-inflammatory IL-17-related immune milieu, that turns to inflammatory IL-17-mediated disease in hidradenitis suppurativa. J Invest Dermatol. 2019;139(4):964–968. doi: 10.1016/j.jid.2018.10.020 [DOI] [PubMed] [Google Scholar]
- 93.Béke G, Dajnoki Z, Kapitány A, et al. Immunotopographical differences of human skin. Front Immunol. 2018;9:424. doi: 10.3389/fimmu.2018.00424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Frew JW, Navrazhina K, Byrd AS, et al. Defining lesional, perilesional and unaffected skin in hidradenitis suppurativa: proposed recommendations for clinical trials and translational research studies. Br J Dermatol. 2019;181(6):1339–1341. doi: 10.1111/bjd.18309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Frew JW, Lowes MA, Goldfarb N, et al. Global harmonization of morphological definitions in hidradenitis suppurativa for a proposed glossary. JAMA Dermatol. 2021;157(4):449–455. doi: 10.1001/jamadermatol.2020.5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cao Y, Hong F, Conlon DM, et al. Potential predictive biomarkers of adalimumab response in patients with hidradenitis suppurativa. Br J Dermatol. 2021;185(4):804–814. doi: 10.1111/bjd.20097 [DOI] [PubMed] [Google Scholar]
- 97.Roy CF, Roy SF, Ghazawi FM, Patocskai E, Bélisle A, Dépeault A. Cutaneous squamous cell carcinoma arising in hidradenitis suppurativa: a case report. SAGE Open Med Case Rep. 2019;7:X19847359. doi: 10.1177/2050313X19847359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lapins J, Ye W, Nyrén O, Emtestam L. Incidence of cancer among patients with hidradenitis suppurativa. Arch Dermatol. 2001;137(6):730–734. [PubMed] [Google Scholar]
- 99.Frew JW, Jiang CS, Singh N, et al. Malignancy and infection risk during adalimumab therapy in hidradenitis suppurativa. Clin Exp Dermatol. 2020;45(7):859–865. doi: 10.1111/ced.14264 [DOI] [PubMed] [Google Scholar]
- 100.Naik HB, Lowes MA. A call to accelerate hidradenitis suppurativa research and improve care—moving beyond burden. JAMA Dermatol. 2019;155(9):1005–1006. doi: 10.1001/jamadermatol.2019.1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.