Abstract
The Hedgehog (Hh) signaling is one of the essential signaling pathways during embryogenesis and in adults. Hh signal transduction relies on primary cilium, a specialized cell surface organelle viewed as the hub of cell signaling. Protein kinase A (PKA) has been recognized as a potent negative regulator of the Hh pathway, raising the question of how such a ubiquitous kinase specifically regulates one signaling pathway. We reviewed recent genetic, molecular and biochemical studies that have advanced our mechanistic understanding of PKA’s role in Hh signaling in vertebrates, focusing on the compartmentalized PKA at the centrosome and in the primary cilium. We outlined the recently developed genetic and optical tools that can be harvested to study PKA activities during the course of Hh signal transduction.
Keywords: Hedgehog signaling, protein kinase A (PKA), primary cilium, centrosome, compartmentalized cell signaling
Introduction
Overview
The Hedgehog (Hh) signaling is an evolutionarily conserved pathway that regulates key events during embryonic development and in adults. During early embryonic development, it plays key instructive roles in pattern formation and cell fate determination in the telencephalon, developing spinal cord, and digits (Fuccillo et al., 2006; Sagner and Briscoe, 2019). Right after birth, Hh signaling controls cell proliferation in the developing cerebellum (Dahmane and Ruiz-i-Altaba, 1999; Wallace, 1999; Wechsler-Reya and Scott, 1999). In adults, it plays critical roles in the establishment and maintenance of stem cells in the nervous system and hair follicles in the skin (Han et al., 2008; Lee et al., 2016). Errors in Hh signaling lead to various birth defects, such as spina bifida, holoprosencephaly, polydactyly, skeletal malformations, heart and lung defects (Ruiz i Altaba et al., 2002; McMahon et al., 2003; Nieuwenhuis and Hui, 2005). Excessive Hh signaling leads to cancers, such as medulloblastoma, a malignant pediatric brain tumor caused by uncontrollable progenitor proliferation in the cerebellum (Stone et al., 1996; Pugh et al., 2012; Raleigh and Reiter, 2019). In adults, overactive Hh signaling leads to basal cell carcinoma, one of the most common skin cancers (Johnson et al., 1996; Xie et al., 1998).
The pathway originally got its name from the work of Nüsslein-Volhard and Wieschaus, who isolated Drosophila mutants to study body segmentation in the 1970s (Nüsslein-Volhard and Wieschaus, 1980). “Hedgehog” was named because the mutant embryos appeared stubby with denticle hairs pointing in random directions resembling a hedgehog. The mutated gene was later identified to be the eponymous ligand for the Hh pathway. The gene family of this ligand is expanded to three paralogs in amniotes: desert hedgehog (Dhh), Indian hedgehog (Ihh) and sonic hedgehog (Shh). Each ligand is expressed in distinct developing tissues (Bitgood and McMahon, 1995; Pathi et al., 2001). Shh is predominately expressed in tissue such as the central nervous system (CNS), limb, hair, gut, and is the best studied ligand (Goodrich et al., 1996). Shh may function as a mitogen to stimulate cell proliferation, such as the neural progenitor proliferation in the developing cerebellum (Stone et al., 1996; Pugh et al., 2012; Raleigh and Reiter, 2019). It may also function as a morphogen to induce cell fate determination by exposing target cells with different signaling strength and duration, such as the well-studied neuronal cell fate determination in the developing spinal cord (Sagner and Briscoe, 2019).
There are many similarities in the transduction mechanisms of Hh signaling between Drosophila and the vertebrate system (Ingham, 2018). In both systems, Hh signaling is transduced via alternative de-repression. When the signaling is off, Ptch inhibits Smo, and SuFu inhibits Gli transcription factors. Upon ligand binding, Ptch is inhibited, leading to the activation of Smo. Active Smo then suppresses SuFu to release SuFu’s inhibition on Gli transcription factors (Ci in Drosophila). However, details of signal transduction differ significantly in almost every step of the pathway, mainly because of the reliance on the primary cilium in vertebrates. Drosophila cells lack cilia, except in mechanical sensory neurons and sperm (Basto et al., 2006). The primary cilium provides a special cellular compartment where receptors and signal transducers are highly concentrated to effectively sense and transduce signaling cues from outside the cell into signaling cascades within the cell (Barr and Sternberg, 1999; Huangfu et al., 2003; Pazour and Witman, 2003). It also provides a privileged space to allow ubiquitous enzymes, such as cAMP-dependent protein kinase A (PKA), to regulate specific cell signaling, such as Hh signaling. A comprehensive review of biochemical events in Hh signaling can be found in the review by Kong et al. (2019). In this review, we will focus on the roles of PKA in Hh signal transduction in the vertebrate system. We will summarize previous discoveries from mouse genetics, cell biology and biochemical studies that implicate PKA as an essential regulator of the Hh pathway, and outline recent techniques that have been used to study PKA regulation of Hh signaling in the primary cilium.
Hh signal transduction relies on the primary cilium
Almost every vertebrate cell has a primary cilium. This specialized microtubule-based, membrane-enclosed organelle projects from the cell surface, and has been historically considered as an evolutionary vestige of the flagellum (Rosenbaum and Witman, 2002; Satir and Christensen, 2007). However, recent studies have revealed pivotal roles of this organelle in signal transduction. The very first clue that cilia may be involved in transducing cell signaling came from the seminal discovery by Kathryn Anderson. Mutations in genes encoding proteins involved in intraflagellar transport (IFT) cause defects in mammalian Hh signal transduction (Huangfu et al., 2003; Bangs and Anderson, 2017). Since then, almost all Hh signaling components have been found in the cilium, and Hh signal is now known to be transduced through a choreographed trafficking of signaling proteins into and out of the cilium (Mukhopadhyay et al., 2010; Bangs and Anderson, 2017).
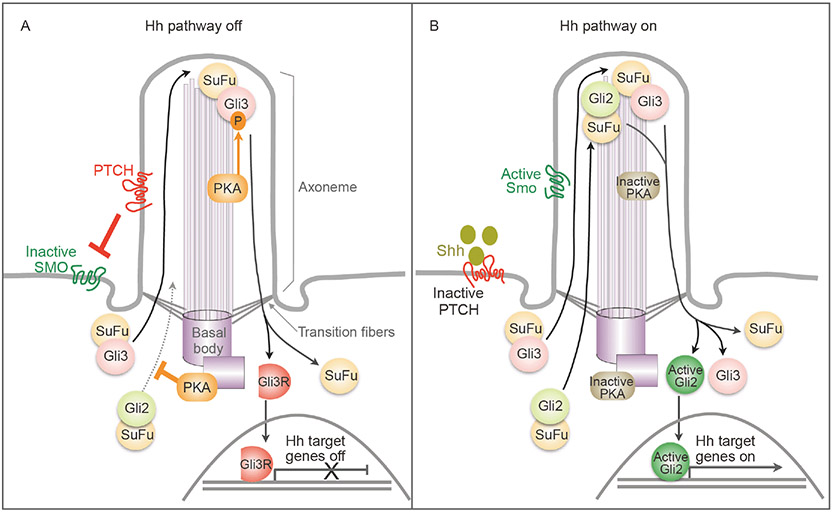
When the Hh pathway is off, the 12-transmembrane protein Ptch is enriched in cilia to inhibit Smo (Figure 1A). Recent results from cryo-EM suggest that the underlying mechanism may involve the local control of cholesterol composition in the ciliary membrane by Ptch, which structurally resembles a sterol transporter (Huang et al., 2018; Qi et al., 2018a; Qi et al., 2018b; Zhang et al., 2018; Deshpande et al., 2019; Rudolf et al., 2019). Sterol lipids such as cholesterol can activate Smo (Corcoran and Scott, 2006; Huang et al., 2016; Raleigh et al., 2018). The transcription factor Gli3, primed by PKA phosphorylation, is proteolytically processed into a transcription repressor Gli3R that translocates into the nucleus to keep Hh target genes off. PKA also prevents the activation of Gli2, a transcription activator. In the presence of Shh, Ptch binds to Shh and exits from the cilium (Figure 1B). Subsequently, Smo is activated and accumulates in the cilium. Active Smo antagonizes the inhibitory effects of PKA on Gli3 and Gli2. As a result, Gli3R production ceases, and Gli2 accumulates at the tip of the cilium where it is activated. Thus, inhibition from Gli3R is released, and active Gli2 enters the nucleus to turn on the transcription of Hh target genes. Not surprisingly, proteins involved in IFT, a bidirectional trafficking system along ciliary axonemal microtubules, are involved in the regulation of Hh signaling (Huangfu et al., 2003; Keady et al., 2012; Lechtreck, 2015; Eguether et al., 2018). These proteins include motor proteins, such as kinesin (Kif3a, Kif7) and dynein complexes (Dync2h1, Dync2li1, Wdr34), and IFT proteins (IFT172, IFT88 etc.). Comprehensive reviews about IFT in Hh signaling can be found in Lechtreck (2015), Bangs and Anderson (2017) and Eguether et al. (2018). Many steps in this process remain mysterious. For example, why do Hh signaling components need to transit to the primary cilium? How is this complicated protein trafficking coordinated in the cell? How does PKA, a ubiquitous housekeeping kinase, selectively regulate the Hh pathway? We will focus on the last question to summarize the specific regulation of Hh signaling by PKA in the primary cilium and at the centrosome. In ciliated cells, the mother centriole of the centrosome becomes the basal body to organize the cilium microtubules; for simplicity, hereafter we use centrosome to refer to both in this review.
Figure 1.
Overview of Hh signal transduction. Vertebrate Hh signaling relies on protein trafficking at the primary cilium, a centrosome organized, microtubule-based cell surface organelle. The membrane of the cilium is continuous with the cytoplasmic membrane, but the ciliary cytosol is separated from the cytoplasm via transition fibers that function as filter for material exchange between ciliary axoneme and the cytoplasm. A, When the Hh pathway is off, Ptch resides in the cilium and inhibits Smo. Active PKA phosphorylates Gli3, resulting in Gli3’s proteolytical processing into Gli3R, a transcription repressor. Active PKA also phosphorylates Gli2 and inhibits its trafficking to the cilium tip, preventing its activation. Note that both Gli proteins complex with SuFu, a negative regulator of the Hh pathway. B, Upon binding to the ligand Shh, Ptch exits from the cilium, and Smo is activated and accumulated in the cilium. PKA is inactivated and stops phosphorylating Gli proteins. As a result, both Gli proteins dissociate from SuFu. Gli3 remains in the full-length form and Gli2 is activated. Active Gli2 enters the nucleus to turn on the transcription of Hh target genes.
PKA is a strong negative regulator of the Hh pathway
PKA subunits and its activation
The PKA holoenzyme is a heterotetramer consisting of two catalytic (PKA-C) subunits and two regulatory (PKA-R) subunits. When cAMP binds to the R subunits, the PKA holoenzyme dissociates to release active C subunit (Turnham and Scott, 2016). In mice, two genes encode catalytic subunits (Cα and Cβ) and four genes encode regulatory subunits (RIα, RIβ, RIIα and RIIβ) (Taylor et al., 1990; Torres-Quesada et al., 2017). The expression pattern of these isoforms shows some degree of tissue specificity revealed by in situ hybridization in mouse embryos and adults (Cadd and McKnight, 1989; Guthrie et al., 1997). Within the cell, different regulatory subunits may localize to distinct subcellular sites (Ilouz et al., 2017). RI subunits are found in the primary cilium (Mick et al., 2015), whereas RII subunits localize to the centrosome (Barzi et al., 2010).
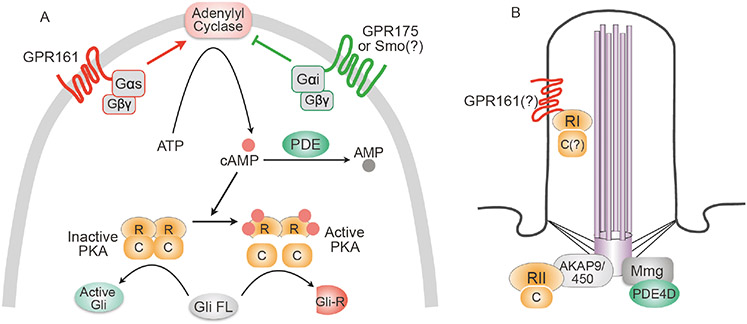
PKA activity is primarily controlled by the levels of the second messenger cAMP, which directly binds to and activates PKA (Figure 2A). cAMP levels are modulated by signaling cascades initiated by GPCRs. Upon activation by ligands, GPCRs activate heterotrimeric G-proteins by catalyzing the exchange of GDP for GTP on the Gα subunit, resulting in the dissociation of Gα from Gβ-γ subunits. Gαs activates adenylyl cyclases (AC) that synthesize cAMP, whereas Gαi subunits reduce cAMP levels by inhibiting AC. cAMP levels are also controlled by phosphodiesterases (PDEs) that hydrolyze cAMP (Beavo and Brunton, 2002) (Figure 2A). In addition to cAMP, PKA activity is also modulated by phosphorylation in an intra- or inter-molecular manner (Keshwani et al., 2012; Torres-Quesada et al., 2017). C subunits are phosphorylated on T197 and S338. Phosphorylation of T197 in the kinase activation loop is a characteristic of active PKA-C, and is presumably carried out by PDK1 (Taylor et al., 2013). Phosphorylation of the PKA-RII subunits affects the PKA holoenzyme assembly and impacts the dissociation of C subunits from the R subunits upon cAMP binding (Zhang et al., 2015). More details about PKA activity regulation and its implications in pathological processes can be found in these reviews (Gold et al., 2013; Röck et al., 2015; Torres-Quesada et al., 2017).
Figure 2.
Regulation of PKA activity and its compartmentalized localization in the cell. A, AC is activated by Gαs-coupled GPCR (such as GPR161), and inhibited by Gαi-coupled GPCR (GPR175 or potentially Smo; the mechanism of how Smo activates Gαi remains to be determined). AC converts ATP into the 2nd messenger cAMP, which binds to PKA-R and dissociates it from PKA-C. Active PKA-C suppresses Hh signaling by promoting the formation of Gli repressor. Inactive PKA allows the formation of active Gli, resulting in activation of Hh signaling. PDEs hydrolyze cAMP to inhibit PKA. B, PKA is sequestered to discrete subcellular compartments by binding to A kinase anchoring proteins (AKAPs). PKA-RII subunit is anchored to the centrosome by AKAP9 and AKAP450 (for simplicity, only one subunit is shown). PKA-RI subunit is found to be present in the cilium, potentially via binding to the C-terminus of GPR161. Some isoforms of PDE4D are anchored to the centrosome via myomegalin (Mmg), and degrade local cAMP to attenuate PKA activity.
PKA is involved in myriad cellular processes in development, metabolism, memory formation and proliferation. It is an intriguing question how a specific cell signaling pathway is selectively regulated by such a ubiquitous kinase. An emerging view proposes that the precise spatiotemporal control is achieved via compartmentalization of PKA activity in the cell. A-kinase anchoring proteins (AKAPs), multi-domain scaffold proteins, bind to PKA-R and anchor the holoenzyme to discrete subcellular compartments (Wong and Scott, 2004; Baillie et al., 2005). AKAPs serve as scaffolds to deposit PKA in the vicinity of its substrates together with other enzymes such as phosphatases, phosphodiesterases, or other components of the specific signaling pathways, thereby providing tailored PKA signaling units (aka, signalosomes) (Torres-Quesada et al., 2017). PKA activity at the centrosome and in the cilium is directly involved in the Hh signaling. AKAP9, AKAP450 and Myomegalin target PKA to the centrosome (Verde et al., 2001; Uys et al., 2011; Terrin et al., 2012), and GPR161 is reported to anchor PKA to the primary cilium (Bachmann et al., 2016) (Figure 2B). As detailed in section 3, manipulating local PKA activity at the centrosome and in the cilium, even though the global PKA activity is not impacted, is sufficient to control Hh signaling.
Genetic evidence of PKA as a potent suppressor of the Hh pathway
The involvement of PKA in Hh signaling was first observed in Drosophila. Loss of PKA activity leads to activation of Hh target genes in embryonic tissues such as eye, leg and wing imaginal discs (Jiang and Struhl, 1995; Lepage et al., 1995; Li et al., 1995; Pan and Rubin, 1995; Strutt et al., 1995). Subsequent studies in zebrafish showed that expression of dominant negative or constitutively active forms of PKA in the zebrafish embryo led to transcriptional activation or inhibition of Shh target genes, respectively (Concordet et al., 1996; Hammerschmidt et al., 1996). Pharmacological approaches that increase PKA activity were found to block Shh-mediated induction of dopaminergic neurons (Fan et al., 1995; Hynes et al., 1995).
A transgenic mouse model was generated by Epstein et al. (1996) in which a dominant-negative PKA (dn-PKA) was expressed in the developing CNS. This dnPKA encodes a PKA regulatory subunit that is insensitive to cAMP, thereby preventing the activation of PKA-C. The dnPKA transgenic mouse exhibited high levels of Hh signaling activity, including increased Hh-target expression and mis-patterning of the embryonic CNS (Epstein et al., 1996). Huang et al. (2002) generated PKA-deficient mice that retain only one functional PKA-C. Decreased PKA activity led to neural tube patterning deficits that resemble the phenotypes of overactive Hh signaling. Tuson et al. (2011) generated a mouse model that lacks both PKA-Cα and PKA-Cβ, and found that the double mutant caused complete ventralization of the neural tube to the same extent as Ptch1- and SuFu-null embryos (Goodrich et al., 1997; Cooper et al., 2005; Svärd et al., 2006). These results suggest that inhibiting PKA leads to maximal activation of the Hh pathway.
PKA activity is controlled by cAMP levels. Therefore, disturbing cAMP production in mutants along the axis of GPCR-G protein-AC also impairs Hh signaling in relevant tissues during embryogenesis. Mukhopadhyay et al. (2013) found that loss of GPR161, which is coupled to Gαs, inhibits PKA in the primary cilium and increases Shh signaling in the developing neural tube. Conversely, loss of GPR175, a Gαi-coupled receptor, increases cAMP/PKA activity and suppresses Hh signaling (Singh et al., 2015). Regard et al. (2013) reported that loss of Gαs upregulates Hh signaling via decreased cAMP/PKA activity, leading to ectopic ossification, like what is observed in osseous heteroplasia (POH). In the developing cerebellum and neural tube, overexpressing AC5 and AC6 represses the Hh pathway, whereas their knockdown results in the activation of Hh signaling (Vuolo et al., 2015). Recently, Somatilaka et al. identified that Ankmy2 is responsible for the maturation and trafficking of multiple ACs to the primary cilium, and Ankmy2 knockout mouse exhibits neural tube ventralization, to the similar degree as in Ptch1- and PKA-null mouse (Goodrich et al., 1997; Tuson et al., 2011; Somatilaka et al., 2020). Furthermore, the action of Ankmy2 is independent of Smo and directly controls Gli2 and Gli3 processing (Somatilaka et al., 2020). In summary, multiple lines of genetic evidence strongly implicate PKA as an essential and potent suppressor of the Hh pathway.
Gli transcription factors are targets of PKA in Hh pathway
The Gli homolog in Drosophila is Ci (Cubitus interruptus). In the absence of Hh ligand, Ci is phosphorylated by a protein complex that includes Cos2, PKA, GSK3, and Fused (Fu) (Robbins et al., 1997; Sisson et al., 1997; Zhang et al., 2005). Phosphorylated Ci is proteolytically processed to an inactive form. The presence of Hh ligand stabilizes fly Smo which competes for Cos2 association and prevents Cos2-Kinase complex formation. This prevents Ci from being phosphorylated, allowing it to remain in its full-length active form (Ruel et al., 2003), resulting in activation of the Hh pathway. In vertebrates, the Ci protein expands into three isoforms, Gli1/2/3, making Gli protein processing more complex (Ruppert et al., 1988). Mouse genetics studies show that Ci’s role seems to be unevenly distributed between the three Gli proteins, with Gli1 and Gli2 playing predominantly activating roles and Gli3 playing a repressive role (Ruppert et al., 1988; Ding et al., 1998; Matise et al., 1998; Bai and Joyner, 2001; Bai et al., 2002; Persson et al., 2002; Rallu et al., 2002). Gli1 is a Hh-target gene and acts to amplify Hh signaling (Park et al., 2000), whereas the ON and OFF switch of Hh signaling is determined by the balance between the Gli2 activator and Gli3 repressor.
In the absence of Shh, Gli3 is phosphorylated at six sites by PKA, which primes Gli3 for further phosphorylation by glycogen synthase kinase 3-beta (GSK3b) and casein kinase 1 (CK1) (Wang et al., 2000; Tempe et al., 2006; Wang and Li, 2006). The phosphorylated residues provide a high-affinity binding site for β-TrCP, which then recruits the SCF (Skp1/Cullin1/F-box) ubiquitin ligase complex (Wang and Li, 2006). Ubiquitinated Gli3 gets cleaved by the proteasome, converting the 190-kDa Gli3 full length (Gli3FL) protein into an 83-kD Gli3 repressor form (Gli3R) (Wang and Li, 2006; Pan and Wang, 2007; Wen et al., 2010). Gli3R enters the nucleus and suppresses the transcription of Hh target genes. Activation of Hh signaling represses this processing, via mechanisms including the inhibition of PKA activity, resulting in the existence of Gli3 predominantly in its full-length form. In contrast, Gli2 protein is minimally processed, and the vast majority of Gli2 is present in the full-length form (Pan et al., 2006). However, phosphorylation by PKA facilitates Gli2 degradation, as Gli2 protein lacking PKA phosphorylation sites is more stable than wild type Gli2, and is constitutively active in mouse embryos (Pan et al., 2009; Niewiadomski et al., 2014; Liu et al., 2015). Further, in PKA-null cells, Gli2 accumulates at the cilium tip, suggesting that PKA normally restricts Gli2 transportation into the cilium (Tuson et al., 2011).
The processing and activation of Gli2/3 requires the primary cilium and involves two other proteins, SuFu and Kif7 (Haycraft et al., 2005; Chen et al., 2009; Cheung et al., 2009; Endoh-Yamagami et al., 2009; Kim et al., 2009; Liem et al., 2009). SuFu binds to Gli proteins, preventing their degradation, but also inhibits their activation. Dissociation of Gli/SuFu complexes in cilia is a critical step in the activation of the vertebrate Hh pathway (Humke et al., 2010; Tukachinsky et al., 2010). In the current model, without Shh stimulation, Gli2 and Gli3 complex with SuFu and are primed by PKA at the base of the cilium. The complexes then enter the cilium, where Gli proteins are further phosphorylated by CK1 and GSK3b. After exiting the cilium, Gli3 is converted to Gli3R by the proteasome and Gli2 remains inactive. Upon Shh stimulation, Hh signaling cascade decreases PKA activity at the cilium base, preventing it from phosphorylating Gli proteins. The Gli/SuFu complex enters the cilium, where Gli is not fully phosphorylated by CK1 and GSK3b and the complex dissociates (via unknown mechanisms). Gli3R production ceases, and free Gli2 exits from the cilium and acts as the transcription activator to turn on transcription of Hh target genes in the nucleus.
The subcellular pool of PKA at the centrosome and in the cilium regulates Hh signaling pathway
Accruing evidence from pharmacological, biochemical and genetic studies suggest the inverse relationship between PKA and Hh signaling: increasing PKA activity suppresses Hh transduction, whereas inhibiting PKA activity enhances the strength of Hh signaling (Hammerschmidt et al., 1996; Huang et al., 2002; Barzi et al., 2010; Humke et al., 2010; Tukachinsky et al., 2010; Tuson et al., 2011; Niewiadomski et al., 2013; Vuolo et al., 2015; Williams et al., 2015; Ge et al., 2015; Bachmann et al., 2016; Moore et al., 2016; Kong et al., 2019). It is tempting to speculate that the reverse paradigm is also true: PKA activity must be suppressed in order for the Shh pathway to be activated; in other words, active Hh signaling must suppress PKA activity. However, early attempts at detecting global PKA activity in the entire cell failed to verify such assumptions (Niewiadomski et al., 2013). In fact, under some special circumstances, high total cell PKA levels can occur in parallel with Hh target gene transcription (Niewiadomski et al., 2013). Thus, global PKA levels are not good indicators of Hh signaling levels. Instead, studies in the field support a more precise model that core Hh signaling regulates compartmentalized pools of PKA at discrete subcellular sites, such as the centrosome and the primary cilium.
Signaling outcomes that change PKA activity at the centrosome regulate Hh signaling
Early immunocytochemical studies showed that PKA subunits are concentrated at the centrosome and manipulating PKA activity at this local site is sufficient to alter Hh signaling (Barzi et al., 2010; Tuson et al., 2011; Ge et al., 2015). In granule neural precursors (GNPs) from the developing cerebellum, the PKA holoenzyme is anchored to the basal body via an interaction between RII subunit and AKAP proteins. When this interaction is disrupted by a peptide St-HT31, GNP proliferation is inhibited (Barzi et al., 2010). In the developing neural tube, PKA-C immunofluorescence intensity concentrates at the centrosome and PKA-C knockout results in an increased Gli2 accumulation at the cilium tips, leading to the hypothesis that PKA controls Gli2 trafficking at the centrosome (Tuson et al., 2011). Local cAMP level is also controlled by phosphodiesterase 4D (PDE4D), the enzyme that converts cAMP into AMP. When PDE4D is recruited to the cytoplasmic membrane where most cAMP is produced, cAMP is more effectively degraded, leading to enhanced Hh signaling (Ge et al., 2015; Williams et al., 2015). In addition, a signalosome that includes PDE4D3 and the scaffold protein Myomegalin was identified at the centrosome (Verde et al., 2001); Myomegalin loss dislocates PDE4D3 from the centrosome, which subsequently promotes local PKA activity and suppresses Hh signaling (Peng et al., 2021).
PKA in the cilium specifically regulates Hh signaling
Since all Hh pathway components are found in the cilium, it is highly conceivable that PKA resides and signals inside the cilium, although early attempts failed to confirm so (Tuson et al., 2011). However, most Hh pathway transducers in the axis of GPCR-AC-cAMP reside in the primary cilium, such as GPR161 and AC3/5/6 (Mukhopadhyay et al., 2013; Somatilaka et al., 2020). Some G proteins and cAMP specific PDEs are also reported to be present in the cilium (Choi et al., 2011; Ishikawa et al., 2012). Therefore, the cAMP levels inside the cilium must be tightly controlled, and subtle changes in cAMP levels can only be perceived by local PKA in the cilium. Finally, PKA substrates Gli2 and Gli3 traffic through the cilium, and their transition in the cilium is crucial for their stability and activation (Haycraft et al., 2005; May et al., 2005; Chen et al., 2009; Wen et al., 2010). Hence, the critical phosphorylation events must happen in the cilium. In a recent proximity-based study, the subunits of PKA-RIα, -RIIβ and -Cα were discovered as candidate proteins in the cilium (Mick et al., 2015). When RIα was fused with GFP, the tagged proteins were observed in the axoneme of the primary cilium (Mick et al., 2015). The GFP-tagged PKA-C was also discovered to be present in the cilium (Arveseth et al., 2021; Truong et al., 2021). However, so far direct evidence showing that endogenous PKA is localized to the cilium, such as immunostaining results, is still missing. The endogenous PKA in the cilium is difficult to detect presumably because it rapidly transits through the cilium and the level is under current detection limits, or because of its lability for conventional fixation methods.
The validation of PKA subunits in the cilium indicates that the phosphorylation of Gli2/3 could take place directly in the cilium. Indeed, with an antibody that can recognize PKA’s phosphorylated residues in Gli2/3, Li et al. (2017) found that the levels of phosphorylated Gli2 and Gli3 in the cilium are inhibited by Hh signaling. Moreover, in a genome-wide RNAi screen, Jacob et al. (2011) reported that ablation of PKA-RI compromises Hh signaling. Finally, when Mick et al. (2015) targeted a PKA inhibitor, PKI, to the cilium to specifically inhibit local PKA activity, the processing of Gli3 into Gli3R was reduced. Therefore, it is proposed that high basal activity of cAMP/PKA in the cilium contributes to the phosphorylation of Gli3 and promotes its proteolytic processing into Gli3R. Once the Hh pathway is activated, cAMP levels in the cilium decease. Subsequent PKA inhibition reduces Gli3 processing and allows Gli2 to be activated. This model explains the dynamic regulation of Gli3 processing by local PKA in the cilium; however, how Gli2 activation is controlled by cilium localized PKA still remains enigmatic.
The signaling cascade that links Hh signaling with PKA activity
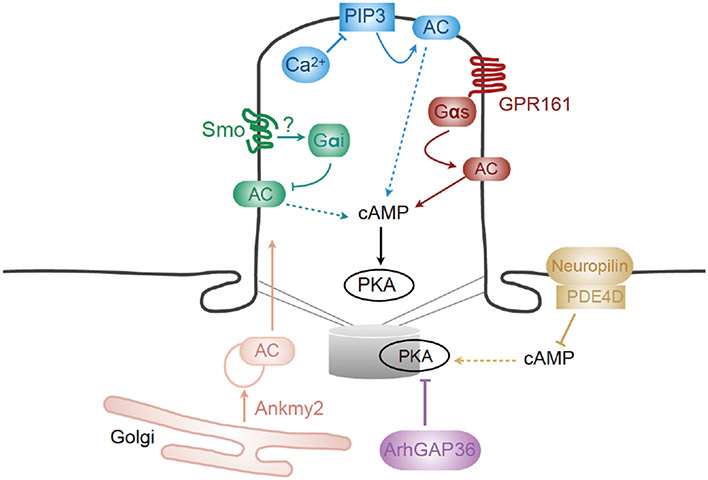
Based on the inverse correlation between PKA activity and Hh signaling, it is believed that the cascade of Hh signaling eventually abrogates PKA activities in the vicinity of the primary cilium, subsequently promoting the activation of Gli. Nevertheless, the specific molecular and biochemical mechanisms that link active Hh signaling to PKA remain controversial. Previous studies have implicated GPCRs, PDEs, and adenylyl cyclase-trafficking proteins in this process. In this section, we will summarize these studies and outline our current understanding of PKA regulation during Hh transduction in the vertebrate system (Figure 3).
Figure 3.
Current understanding of how PKA activity is regulated in the vicinity of the cilium. Each pathway is colored coded. Green path: Smo may activate Gαi via hitherto unknown mechanisms, which inhibits adenylyl (AC) and reduces cAMP levels to inhibit PKA activity. Red path: GPR161 is present at the cilium before Hh activation, where it activates Gαs to increase PKA activity. Blue path: High Ca2+ levels in the cilium inhibits PIP3-mediated AC activation, and contributes to the reduction of cAMP/PKA activity. This path is observed only after prolonged (>24 h) Hh agonist stimulation. Yellow path: Neuropilin recruits PDE4D to the cytoplasmic membrane to effectively degrade cAMP, and subsequently reduces PKA activity at the centrosome. Pink path: Ankmy2 is responsible for the maturation and trafficking of multiple ACs to the primary cilium; loss of Ankmy2 leads to marked elevation of Hh signaling in vitro and in vivo. Purple path: The Rho GTPase-activating protein ArhGAP36 blocks PKA activity by acting as a PKA pseudo-substrate or inducing PKA degradation. Dashed lines indicate that activation of this pathway will suppress cAMP production and PKA activity.
From Smo to PKA
Smo belongs to the family of class F (Frizzled) GPCRs. Some evidence in Drosophila suggest that Smo may act as a conventional GPCR to activate the trimeric G protein, Gαi (Ogden et al., 2008; Ayers and Thérond, 2010; Qi et al., 2019). As Gαi inhibits AC to reduce cAMP levels, its activation is predicted to reduce PKA activity, thereby promoting Hh signaling (Figure 3). However, contradicting results were revealed in other studies. Li et al. found that active PKA phosphorylates Smo and promotes Smo’s membrane localization to activate the Hh pathway (Li et al., 2014; Li et al., 2016). Further, a constitutively active PKA-C can substitute for endogenous PKA-C to govern the proper patterning during Drosophila organogenesis (Jiang and Struhl, 1995; Li et al., 1995). These results suggest a possible dual role of PKA in Hh signaling in Drosophila, indicating that the link between Smo and PKA may not be as simple as originally proposed.
Studies in vertebrates also revealed mixed results. Early biochemical assays in vertebrate cells overexpressing both Smo and Gαi indicate that Smo may directly activate Gαi (DeCamp et al., 2000; Shen et al., 2013). However, further biochemical and genetic studies failed to fully corroborate this notion. Biochemical studies in mammalian cells revealed that deleting the C-tail in Smo, the sequence required for Gli activation, has no impact on Gαi activation (Riobo et al., 2006). Since the Gαi family in vertebrates are encoded by multiple genes (Gnai1, Gnai2, Gnai3, Gnao1 and Gnaz) that are redundant to each other, many genetic studies employed pertussis toxin (Ptx), an uncoupler of all Gαi proteins (except Gαz) from upstream GPCRs. Ptx locks Gαi proteins in inactive states, resulting in the activation of adenylyl cyclase and increase in cAMP/PKA activity. Expressing Ptx in zebrafish embryos only partially impacted the ventral specification in the nervous system, but not other Hh-dependent patterning (Hammerschmidt and McMahon, 1998). Electroporation of dominant negative Gαi or pertussis toxin into the developing chicken neural tube failed to induce discernible phenotypes that are related to Hh defects (Low et al., 2008). Finally, genetic expression of Ptx in mouse embryos did not impair Hh-dependent limb patterning and skeletal development (Regard et al., 2013). Therefore, the evidence is inconsistent for Smo to act as a Gαi-coupled GPCR to control cAMP/PKA activity.
One recent study hinted at an alternative connection between Smo and PKA (Arveseth et al., 2021). PKA-C can directly bind to the C-terminal tail of active Smo, triggered by GPCR kinase 2 (GRK2)-mediated phosphorylation of the Smo intracellular domain. This sequestration of PKA prevents its phosphorylation of Gli proteins, releasing PKA’s inhibition on Gli. However, these results were obtained in non-ciliated 293T cells—whether the same mechanism applies to Smo-PKA interaction in the cilium needs to be confirmed.
GPR161 and PKA activity in the primary cilium
GPR161 is a cilium localized GPCR that negatively regulates Hh signaling (Mukhopadhyay et al., 2013). GPR161 resides in the cilium and activates Gαs to produce basally high cAMP levels within the cilium. Upon Hh activation, GPCR161 is cleared from the cilium within two hours, which dampens local PKA activity to promote Hh signaling (Figure 3). GPR161 knockout mice are lethal by E10.5 with ventralized neural tubes, resembling the phenotypes of higher Shh signaling in Ptch1- and PKA-null mice (Goodrich et al., 1997; Tuson et al., 2011; Mukhopadhyay et al., 2013). The knockout mice also exhibit morphogenesis defects in other tissues that involve Hh signaling, such as the limbs and skeleton (Hwang et al., 2018), as well as medulloblastoma formation in the developing cerebellum (Shimada et al., 2018). Consistent with its role in promoting cAMP/PKA activity, GPR161 regulates the proteolytic processing of Gli proteins. In GPR161 mutants, Gli3R levels are reduced, presumably due to reduced PKA activity, as well as the levels of full-length Gli2 and Gli3 (Mukhopadhyay et al., 2013). However, the link between GPCR and cAMP production needs G proteins, and so far, neither Gαi or Gαs has been reproducibly validated to be present in cilia, raising the question of how GPR161 regulates the local enzymatic activity of ACs and PKA.
Further studies elucidated the trafficking mechanisms of GPR161 into and out of the cilium. The intraflagellar transport A (IFT-A) core complex and the tubby family protein Tulp3 coordinate Gpr161’s trafficking to cilia (Badgandi et al., 2017). Indeed, IFT-A or Tulp3 mutants exhibit high Shh signaling, which is opposite to other mutants that affect the cilia (Goetz and Anderson, 2010). Removal of GPR161 from the cilium requires active GRK2 that recruits β-arrestin to the cilium. Active Smo promotes β-arrestin-GPR161 interaction, and the subsequent clathrin mediated endocytosis clears GPR161 from the cilium (Pal et al., 2016). GPR161 may also function as an AKAP to anchor the subunit of PKA-RI into the cilium with its C-terminal tail (Bachmann et al., 2016). In line with this, a recent study by May et al. (2021) proposed that upon Shh stimulation, PKA-RI reunites with PKA-C, and the holoenzyme piggybacks on GPCR161 to exit the cilium. However, this model assumes that PKA-C localizes to the cilium, which has not yet been verified.
A recent study reported that GPR161 functions as an attenuator for Hh signaling, rather than a negative regulator like Ptch and SuFu. Loss of the negative regulators leads to constitutive activation of the pathway in the absence of ligand; this is not observed in GPR161-null NIH3T3 cells (a ciliated cell line that expresses all components of the Hh pathway) (Pusapati et al., 2018). Instead, Shh and Smo agonists can still activate Hh signaling in GPR161-null cells, although with higher potency and efficacy than wild-type cells. The role of GPR161 as a signaling attenuator may explain why the phenotypes of Gpr161 mutant mice are much milder than these of PKA-null mutants (Tuson et al., 2011). Thus, GPR161 may function by modulating the cell’s sensitivity to Shh, a morphogen that drives cell fate determination and patterning of developing organs, such as the spinal cord, limb bud and paraxial mesoderm (Pusapati et al., 2018).
Control of compartmentalized cAMP/PKA activity by PDE
Within the cell, cAMP levels are controlled by two enzymes: production is regulated by ACs and degradation is regulated by cAMP-specific phosphodiesterases (PDE4,7,8). While most recent studies focus on local cAMP production by AC, PDEs have been long studied to play a pivotal role in shaping the spatial and temporal dimension of cAMP “pulses” in the cell to allow precise control of signal transduction. The concept of compartmentalized signaling is rooted in studies of PDE families in the early 1980s on cardiomyocytes (Hayes et al., 1980; Scotland et al., 1998; Rich et al., 2001; Houslay and Adams, 2003; Houslay, 2010). Visualization of cAMP levels in cardiomyocytes with cAMP reporters revealed strips of cAMP “hotspots” when β-adrenergic receptors (A Gαs-coupled GPCR) were activated. Co-treatment with a PDE inhibitor and the receptor agonist, however, abolished these “hotspots”, producing uniform and global increases in cAMP and concomitant activation of all cAMP effectors (Zaccolo and Pozzan, 2002). This led to the concept that cAMP levels are compartmentalized in the cell, and local cAMP levels are precisely sculped by PDEs that anchor to specific subcellular compartments (Wong and Scott, 2004). In other words, PDEs act as local cAMP “sinks”, allowing cAMP to accumulate in areas devoid of PDEs to form cAMP “hotspots”. In this way, local cAMP gradients can gate the activation of effector proteins, such as PKA, that are sequestered at the same subcellular compartments. Among all PDE proteins, the best characterized is the PDE4D subfamily that contains 11 isoforms derived from alternative splicing. Each isoform of PDE4D has an identical C-terminal catalytic domain and a unique N-terminal domain. The N-terminus anchors the enzyme to distinct subcellular sites (Scotland et al., 1998; Lefkowitz et al., 2002; Baillie et al., 2005; Lynch et al., 2006), where PDE4D selectively regulates specific signaling pathways (Scott and Pawson, 2009; McCormick and Baillie, 2014).
The effect of compartmentalized PDE4D on Hh signaling was first demonstrated by Ge et al. (2015). This study showed that Semaphorin 3-Neuropilin signaling recruits PDE4D to the juxtamembrane region right underneath the cytoplasmic membrane where cAMP is produced by AC. Due to this close proximity, PDE4D effectively degrades cAMP, resulting in an overall decrease in cAMP levels in the cell. This subsequently inhibits PKA at the centrosome and promotes Hh signaling (Hillman et al., 2011; Ge et al., 2015) (Figure 3). The regulation of Hh signaling by Neuropilin-PDE4D signaling was further corroborated by other studies (Hayden Gephart et al., 2013; Snuderl et al., 2013; Williams et al., 2015; Pinskey et al., 2017). Through a chemical genetic screen in zebrafish, Williams et al. (2015) discovered PDE4D as a positive regulator of the Hh pathway, and identified a small molecule inhibitor that selectively suppresses PDE4D thereby elevating PKA activity. Pinskey et al. (2017) reported that Neuropilin 1 is present in the cilium to promote Hh signaling via a previously uncharacterized cytoplasmic motif. Blocking Neuropilins’ activity via shRNA or function-blocking antibodies suppresses the growth of Hh-related tumors (Hayden Gephart et al., 2013; Snuderl et al., 2013). A more recent study showed that one PDE4D isoform, PDE4D3, is anchored to the centrosome by Myomegalin (aka. PDE4DIP), a centrosomal and Golgi scaffolding protein. Myomegalin knockout dislocates PDE4D3 from the centrosome, elevates local cAMP/PKA activity and attenuates Hh signaling (Peng et al., 2021). In renal epithelial cells, a protein complex comprising AC5/6, AKAP150, PKA and PDE4C was found to locate in the primary cilium (Choi et al., 2011). Whether and how this complex regulates Hh signaling remain to be identified. Since extensive arrays of PDE4D inhibitors have been developed for respiratory and inflammation diseases in the pharmaceutical industry (Spina, 2008; Page and Spina, 2012; Zaccolo et al., 2021), the identification of compartmentalized PDE4D activity and its local regulation on Hh signaling may provide opportunities to repurpose these inhibitors for pathway specific therapeutics.
Ankmy2, adenylyl cyclase and PKA activity in the cilium
Ankmy2, an ankytin-repeat and MYND-domain containing protein, binds to multiple ACs (AC3/5/6) and determines their maturation and trafficking to the primary cilium (Somatilaka et al., 2020). The lack of ACs in the cilium minimizes local cAMP levels and PKA activity, thereby promoting Hh signaling (Figure 3). Ankmy2-null mouse exhibits full activation of the Hh signaling in the neural tube, with the ventralization to the similar degree as in Ptch1- or PKA-null mice (Goodrich et al., 1997; Tuson et al., 2011). The levels of Gli3FL/Gli3R and Gli2 accumulation at the cilium tips are also similar to those in PKA-null mice. Intriguingly, genetic studies show that the phenotypes of Ankmy2−/−, Smo−/− mice resemble that of Ankmy2−/− mice, suggesting Ankmy2 is fully epistatic to Smo. This result indicates that PKA’s effect on Gli activation can be independent of Smo.
Other factors that impact PKA activity in Hh signaling
GPR175 is reported to reside in the cilium in a Hh-dependent manner in NIH3T3 cells. GPR175 moves to the cilium upon Shh stimulation, where it activates Gαi and subsequently inhibits AC and cAMP production. This in turn dampens PKA activity and Gli3R formation, maximizing Hh signal intensity (Singh et al., 2015). So far there is no genetic evidence of roles of GPR175 in Hh signaling.
ArhGAP36, a Rho GTPase-activating protein, was identified as a positive Hh regulator in a genome-wide screen for Hh agonists (Rack et al., 2014). ArhGAP36 overexpression leads to Smo-independent Hh pathway activation. A later study shows that ARHGAP36 antagonizes PKA activity via two mechanisms: blocking PKA-C activity via its pseudosubstrate motif, like a PKI, and targeting PKA-C for ubiquitin-mediated lysosomal degradation (Eccles et al., 2016; Zhang et al., 2019). Consistent with its role as a PKA inhibitor, overexpressing ArhGAP36 is sufficient to reduce Gli3R formation and promote Gli2 transportation to the cilium tip (Rack et al., 2014).
Ca2+ has also been shown to regulate Hh signaling in the cilium via PKA. Studies with Patch-clamp and Ca2+ imaging techniques showed that the Ca2+ level is seven-fold higher in the cilium than the cytosol (DeCaen et al., 2013; Delling et al., 2013; Su et al., 2013). This high Ca2+ concentration is maintained by a heteromeric TRP channel, PKD1L1-PKD2L1 (DeCaen et al., 2013; Delling et al., 2013). In addition, prolonged treatment with Hh agonist increased PKD1L1-PKD2L1 levels in the cilium, leading to further elevation of ciliary Ca2+ levels. Moore et al. reported that the high Ca2+ level in the cilium inhibits AC5/6 to reduce local cAMP levels in the cilium. The subsequent PKA inhibition promotes Hh signaling (Moore et al., 2016). However, knocking out Ca2+ ion channels in the cilium (PKD1L1 & PKD2L1) only produces very mild Hh defects in mice (DeCaen et al., 2013; Delling et al., 2013), and the requirement of prolonged treatment with Hh agonist may not be relevant to the rapid change in PKA activity during Hh activation.
Detection of localized PKA activities at specific subcellular sites
Based on the robust inverse relationship between PKA and Hh signaling, it is believed that activation of Hh signaling suppresses PKA activity in the vicinity of the cilium. Since this subcellular pool of PKA only contributes to a small fraction of global PKA activity, conventional biochemical methods failed to detect this change. Over the years, multiple generations of probes for cAMP have been applied to monitor local cAMP levels in the centrosome and at the cilium (DeCaen et al., 2013; Delling et al., 2013; Moore et al., 2016). Nevertheless, direct monitoring of dynamic PKA activity in the cilium has not been reproducibly achieved. We will summarize recent studies on the detection of 2nd messenger levels in the cilium, as well as currently available tools for PKA activity, to inspire the ultimate direct measurement of PKA activity in the vicinity of the cilium.
Most cAMP sensors are based on FRET (Forster resonance energy transfer), which allows for direct monitoring of cAMP levels in live cells. The majority of cAMP sensors are built with the cyclic nucleotide-binding domain (CNBD) of Epac, a family of cAMP-binding effector proteins (Nikolaev et al., 2004; Ponsioen et al., 2004). cAMP binds to these sensors and induces a protein conformation change, resulting in FRET. Moore et al. (2016) targeted one such cAMP probe, cADDis, to the cilium after fusing it to 5-HT6 receptor. They found that the cilium cAMP level is five times higher than whole-cell levels; Shh reduces ciliary cAMP levels without changing the cAMP concentration in the whole cell. Other studies with a similar Epac-based FRET biosensor also revealed dynamic cAMP changes in the primary cilium in response to activation of Gαs-coupled GPCRs, such as β-adrenergic receptors, and dopamine receptor D1R (Marley et al., 2013; Mukherjee et al., 2016). With Epac-S, a 4th generation Epac-based FRET probe, Stoufflet et al. (2020) detected cAMP hotspots at the centrosome of migrating neurons in the embryonic, postnatal and adult brains. Ablation of the cilium or AC3 abolished the hotspots, leading to neuronal migratory defects. Within the cell, cAMP levels regulate multiple effector proteins, including Epac, cyclic nucleotide-gated (CNG) ion channels, and PKA (Beavo and Brunton, 2002). Therefore, the cAMP levels may not faithfully reflect bona fide local PKA activities. Sensitive biosensors that can directly report local PKA activities are needed.
One direct PKA sensor is A-Kinase activity reporter (AKAR) developed by Jin Zhang (Zhang et al., 2001). In this probe, a FRET pair (CFP and YFP) is connected by a linker sequence that contains PKA phosphorylation sites and a phosphoamino acid binding domain (PAABD). PKA phosphorylation induces conformational changes in the linker, bringing the FRET pair in close proximity to efficiently produce FRET. When AKAR3 is targeted to subcellular sites such as the plasma membrane, cytoplasm, nucleus and mitochondria, PKA activity changes in response to related cell signaling were successfully detected (Allen and Zhang, 2006). However, despite the success in these subcellular sites, direct detection of PKA dynamics in the cilium has so far not been achieved, owing to the low sensitivity of the current PKA sensors. The sensitivity is greatly improved in the most recent generation of excitation-ratiometric PKA activity reporter (ExRai-AKAR2) (Mehta et al., 2018; Zhang et al., 2021). Different from its predecessors, ExRai-AKAR is based on single fluorophore cpGFP from GCaMP3 (Tian et al., 2009), in combination with the PKA phosphorylation sequence and PAABD. Phosphorylation by PKA leads to opposite changes in the emission fluorescence intensity at two excitation wavelengths. The resulting ratiometric biosensor greatly increases the dynamic range and the sensitivity to subtle changes in PKA activity within the cell (Zhang et al., 2021). ExRai-AKAR2 may provide hope for the direct detection of PKA dynamics in the primary cilium during the course of Hh signal transduction.
Conclusions
The primary cilium is recognized as a privileged cellular compartment in which many signaling components can interact. 2nd messengers, such as cAMP, Ca2+, and phosphatidylinositol (PIP2, PIP3), have been found to be specially regulated within the cilium, generating a unique signaling environment. The concept of compartmentalized cAMP/PKA signaling in the cell has emerged decades ago. Earlier studies revealed a diverse array of PDEs that can be differentially sequestered to specific subcellular sites to sculpt local cAMP levels. Recent research uncovered the control of cAMP levels by the axis of GPCR-G protein-AC in the vicinity of the primary cilium. Both PDEs and components of GPCR axis are present in the cilium. It remains intriguing to decipher how the two systems cooperate to fine tune the local cAMP levels.
PKA sits at a crucial position in the Hh pathway. Hh signaling is believed to culminate at PKA activity that determines the On/Off switch of the pathway via Gli proteins. Several mechanistic questions remain to be answered regarding PKA’s role in this process. What are the molecular and biochemical mechanisms of PKA inhibition in the vicinity of the cilium by Hh signaling? PKA controls the activation and processing of Gli2 and Gli3; why is cilium trafficking required for the formation of Gli activators and suppressors? Where and how does PKA phosphorylate Gli and modulate the SuFu-Gli interaction? Finally, direct monitoring of PKA activity in the cilium has yet to be achieved. Application of the most recent technological advances in super resolution imaging (Milenkovic et al., 2015; Yang et al., 2019), proximity labeling (Mick et al., 2015; May et al., 2021), and nanobody-based optogenetic tools (Hansen et al., 2020) are essential to understanding mechanistic details on how a ubiquitous enzyme such as PKA specifically regulates individual signaling pathways over space and time.
Acknowledgements
This work was supported by the National Institutes of Health (CA235749). We apologize to the investigators whose work we were unable to cite owing to space constraints and the investigators working in the many important areas of Hh signaling we were unable to cover. We thank Dr. Aaron Gitler for comments on the manuscript.
Footnotes
Compliance and ethics The author(s) declare that they have no conflict of interest.
References
- Allen MD, and Zhang J (2006). Subcellular dynamics of protein kinase A activity visualized by FRET-based reporters. Biochem Biophys Res Commun 348, 716–721. [DOI] [PubMed] [Google Scholar]
- Arveseth CD, Happ JT, Hedeen DS, Zhu JF, Capener JL, Klatt Shaw D, Deshpande I, Liang J, Xu J, Stubben SL, et al. (2021). Smoothened transduces hedgehog signals via activity-dependent sequestration of PKA catalytic subunits. PLoS Biol 19, e3001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers KL, and Thérond PP (2010). Evaluating Smoothened as a G-protein-coupled receptor for Hedgehog signalling. Trends Cell Biol 20, 287–298. [DOI] [PubMed] [Google Scholar]
- Bachmann VA, Mayrhofer JE, Ilouz R, Tschaikner P, Raffeiner P, Röck R, Courcelles M, Apelt F, Lu TW, Baillie GS, et al. (2016). Gpr161 anchoring of PKA consolidates GPCR and cAMP signaling. Proc Natl Acad Sci USA 113, 7786–7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badgandi HB, Hwang SH, Shimada IS, Loriot E, and Mukhopadhyay S (2017). Tubby family proteins are adapters for ciliary trafficking of integral membrane proteins. J Cell Biol 216, 743–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai CB, Auerbach W, Lee JS, Stephen D, and Joyner AL (2002). Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 129, 4753–4761. [DOI] [PubMed] [Google Scholar]
- Bai CB, and Joyner AL (2001). Gli1 can rescue the in vivo function of Gli2. Development 128, 5161–5172. [DOI] [PubMed] [Google Scholar]
- Baillie GS, Scott JD, and Houslay MD (2005). Compartmentalisation of phosphodiesterases and protein kinase A: Opposites attract. FEBS Lett 579, 3264–3270. [DOI] [PubMed] [Google Scholar]
- Bangs F, and Anderson KV (2017). Primary cilia and mammalian Hedgehog signaling. Cold Spring Harb Perspect Biol 9, a028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr MM, and Sternberg PW (1999). A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature 401, 386–389. [DOI] [PubMed] [Google Scholar]
- Barzi M, Berenguer J, Menendez A, Alvarez-Rodriguez R, and Pons S (2010). Sonic-hedgehog-mediated proliferation requires the localization of PKA to the cilium base. J Cell Sci 123, 62–69. [DOI] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, and Raff JW (2006). Flies without centrioles. Cell 125, 1375–1386. [DOI] [PubMed] [Google Scholar]
- Beavo JA, and Brunton LL (2002). Cyclic nucleotide research—still expanding after half a century. Nat Rev Mol Cell Biol 3, 710–717. [DOI] [PubMed] [Google Scholar]
- Bitgood MJ, and McMahon AP (1995). Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev Biol 172, 126–138. [DOI] [PubMed] [Google Scholar]
- Cadd G, and McKnight GS (1989). Distinct patterns of cAMP-dependent protein kinase gene expression in mouse brain. Neuron 3, 71–79. [DOI] [PubMed] [Google Scholar]
- Chen MH, Wilson CW, Li YJ, Law KKL, Lu CS, Gacayan R, Zhang X, Hui C, and Chuang PT (2009). Cilium-independent regulation of Gli protein function by SuFu in Hedgehog signaling is evolutionarily conserved. Genes Dev 23, 1910–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung HOL, Zhang X, Ribeiro A, Mo R, Makino S, Puviindran V, Law KKL, Briscoe J, and Hui CC (2009). The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian Hedgehog signaling. Sci Signal 2, ra29. [DOI] [PubMed] [Google Scholar]
- Choi YH, Suzuki A, Hajarnis S, Ma Z, Chapin HC, Caplan MJ, Pontoglio M, Somlo S, and Igarashi P (2011). Polycystin-2 and phosphodiesterase 4C are components of a ciliary A-kinase anchoring protein complex that is disrupted in cystic kidney diseases. Proc Natl Acad Sci USA 108, 10679–10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concordet JP, Lewis KE, Moore JW, Goodrich LV, Johnson RL, Scott MP, and Ingham PW (1996). Spatial regulation of a zebrafish patched homologue reflects the roles of sonic hedgehog and protein kinase A in neural tube and somite patterning. Development 122, 2835–2846. [DOI] [PubMed] [Google Scholar]
- Cooper AF, Yu KP, Brueckner M, Brailey LL, Johnson L, McGrath JM, and Bale AE (2005). Cardiac and CNS defects in a mouse with targeted disruption of suppressor of fused. Development 132, 4407–4417. [DOI] [PubMed] [Google Scholar]
- Corcoran RB, and Scott MP (2006). Oxysterols stimulate Sonic hedgehog signal transduction and proliferation of medulloblastoma cells. Proc Natl Acad Sci USA 103, 8408–8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmane N, and Ruiz-i-Altaba A (1999). Sonic hedgehog regulates the growth and patterning of the cerebellum. Development 126, 3089–3100. [DOI] [PubMed] [Google Scholar]
- DeCaen PG, Delling M, Vien TN, and Clapham DE (2013). Direct recording and molecular identification of the calcium channel of primary cilia. Nature 504, 315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCamp DL, Thompson TM, de Sauvage FJ, and Lerner MR (2000). Smoothened activates Gαi-mediated signaling in frog melanophores. J Biol Chem 275, 26322–26327. [DOI] [PubMed] [Google Scholar]
- Delling M, DeCaen PG, Doerner JF, Febvay S, and Clapham DE (2013). Primary cilia are specialized calcium signalling organelles. Nature 504, 311–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande I, Liang J, Hedeen D, Roberts KJ, Zhang Y, Ha B, Latorraca NR, Faust B, Dror RO, Beachy PA, et al. (2019). Smoothened stimulation by membrane sterols drives Hedgehog pathway activity. Nature 571, 284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Motoyama J, Gasca S, Mo R, Sasaki H, Rossant J, and Hui CC (1998). Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development 125, 2533–2543. [DOI] [PubMed] [Google Scholar]
- Eccles RL, Czajkowski MT, Barth C, Müller PM, McShane E, Grunwald S, Beaudette P, Mecklenburg N, Volkmer R, Zühlke, et al. (2016). Bimodal antagonism of PKA signalling by ARHGAP36. Nat Commun 7, 12963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguether T, Cordelieres FP, and Pazour GJ (2018). Intraflagellar transport is deeply integrated in hedgehog signaling. Mol Biol Cell 29, 1178–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh-Yamagami S, Evangelista M, Wilson D, Wen X, Theunissen JW, Phamluong K, Davis M, Scales SJ, Solloway MJ, de Sauvage FJ, et al. (2009). The mammalian Cos2 Homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr Biol 19, 1320–1326. [DOI] [PubMed] [Google Scholar]
- Epstein DJ, Marti E, Scott MP, and McMahon AP (1996). Antagonizing cAMP-dependent protein kinase A in the dorsal CNS activates a conserved Sonic hedgehog signaling pathway. Development 122, 2885–2894. [DOI] [PubMed] [Google Scholar]
- Fan CM, Porter JA, Chiang C, Chang DT, Beachy PA, and Tessier-Lavigne M (1995). Long-range sclerotome induction by sonic hedgehog: Direct role of the amino-terminal cleavage product and modulation by the cyclic AMP signaling pathway. Cell 81, 457–465. [DOI] [PubMed] [Google Scholar]
- Fuccillo M, Joyner AL, and Fishell G (2006). Morphogen to mitogen: The multiple roles of hedgehog signalling in vertebrate neural development. Nat Rev Neurosci 7, 772–783. [DOI] [PubMed] [Google Scholar]
- Ge X, Milenkovic L, Suyama K, Hartl T, Purzner T, Winans A, Meyer T, and Scott MP (2015). Phosphodiesterase 4D acts downstream of Neuropilin to control Hedgehog signal transduction and the growth of medulloblastoma. eLife 4, e07068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, and Anderson KV (2010). The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet 11, 331–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold MG, Gonen T, and Scott JD (2013). Local cAMP signaling in disease at a glance. J Cell Sci 126, 4537–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich LV, Johnson RL, Milenkovic L, McMahon JA, and Scott MP (1996). Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev 10, 301–312. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Milenković L, Higgins KM, and Scott MP (1997). Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277, 1109–1113. [DOI] [PubMed] [Google Scholar]
- Guthrie CR, Skålhegg BS, and McKnight GS (1997). Two novel brain-specific splice variants of the murine Cp gene of cAMP-dependent protein kinase. J Biol Chem 272, 29560–29565. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt M, Bitgood MJ, and McMahon AP (1996). Protein kinase A is a common negative regulator of Hedgehog signaling in the vertebrate embryo. Genes Dev 10, 647–658. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt M, and McMahon AP (1998). The effect of pertussis toxin on zebrafish development: A possible role for inhibitory G-proteins in Hedgehog signaling. Dev Biol 194, 166–171. [DOI] [PubMed] [Google Scholar]
- Han YG, Spassky N, Romaguera-Ros M, Garcia-Verdugo JM, Aguilar A, Schneider-Maunoury S, and Alvarez-Buylla A (2008). Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nat Neurosci 11, 277–284. [DOI] [PubMed] [Google Scholar]
- Hansen JN, Kaiser F, Klausen C, Stüven B, Chong R, Bönigk W, Mick DU, Möglich A, Jurisch-Yaksi N, Schmidt FI, et al. (2020). Nanobody-directed targeting of optogenetic tools to study signaling in the primary cilium. eLife 9, e57907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, and Yoder BK (2005). Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet preprint, e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden Gephart MG, Su YRS, Bandara S, Tsai FC, Hong J, Conley N, Rayburn H, Milenkovic L, Meyer T, and Scott MP (2013). Neuropilin-2 contributes to tumorigenicity in a mouse model of Hedgehog pathway medulloblastoma. J Neurooncol 115, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JS, Brunton LL, and Mayer SE (1980). Selective activation of particulate cAMP-dependent protein kinase by isoproterenol and prostaglandin El. J Biol Chem 255, 5113–5119. [PubMed] [Google Scholar]
- Hillman RT, Feng BY, Ni J, Woo WM, Milenkovic L, Hayden Gephart MG, Teruel MN, Oro AE, Chen JK, and Scott MP (2011). Neuropilins are positive regulators of Hedgehog signal transduction. Genes Dev 25, 2333–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houslay MD (2010). Underpinning compartmentalised cAMP signalling through targeted cAMP breakdown. Trends Biochem Sci 35, 91–100. [DOI] [PubMed] [Google Scholar]
- Houslay MD, and Adams DR (2003). PDE4 cAMP phosphodiesterases: Modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem J 370, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Nedelcu D, Watanabe M, Jao C, Kim Y, Liu J, and Salic A (2016). Cellular cholesterol directly activates smoothened in Hedgehog signaling. Cell 166, 1176–1187.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Zheng S, Wierbowski BM, Kim Y, Nedelcu D, Aravena L, Liu J, Kruse AC, and Salic A (2018). Structural basis of smoothened activation in Hedgehog signaling. Cell 174, 312–324.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Roelink H, and McKnight GS (2002). Protein kinase A deficiency causes axially localized neural tube defects in mice. J Biol Chem 277, 19889–19896. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, and Anderson KV (2003). Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426, 83–87. [DOI] [PubMed] [Google Scholar]
- Humke EW, Dorn KV, Milenkovic L, Scott MP, and Rohatgi R (2010). The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev 24, 670–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SH, White KA, Somatilaka BN, Shelton JM, Richardson JA, and Mukhopadhyay S (2018). The G-protein-coupled receptor Gpr161 regulates forelimb formation, limb patterning and skeletal morphogenesis in a primary cilium-dependent manner. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes M, Porter JA, Chiang C, Chang D, Tessier-Lavigne M, Beachy PA, and Rosenthal A (1995). Induction of midbrain dopaminergic neurons by Sonic hedgehog. Neuron 15, 35–44. [DOI] [PubMed] [Google Scholar]
- Ilouz R, Lev-Ram V, Bushong EA, Stiles TL, Friedmann-Morvinski D, Douglas C, Goldberg JL, Ellisman MH, and Taylor SS (2017). Isoform-specific subcellular localization and function of protein kinase a identified by mosaic imaging of mouse brain. eLife 6, e17681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW (2018). From Drosophila segmentation to human cancer therapy. Development 145. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Thompson J, Yates Iii JR, and Marshall WF (2012). Proteomic analysis of mammalian primary cilia. Curr Biol 22, 414–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob LS, Wu X, Dodge ME, Fan CW, Kulak O, Chen B, Tang W, Wang B, Amatruda JF, and Lum L (2011). Genome-wide RNAi screen reveals disease-associated genes that are common to Hedgehog and Wnt signaling. Sci Signal 4, ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, and Struhl G (1995). Protein kinase A and hedgehog signaling in Drosophila limb development. Cell 80, 563–572. [DOI] [PubMed] [Google Scholar]
- Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, Quinn AG, Myers RM, Cox DR, Epstein Ervin H, J., et al. (1996). Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 272, 1668–1671. [DOI] [PubMed] [Google Scholar]
- Keady BT, Samtani R, Tobita K, Tsuchya M, San Agustin JT, Follit JA, Jonassen JA, Subramanian R, Lo CW, and Pazour GJ (2012). IFT25 links the signal-dependent movement of Hedgehog components to intraflagellar transport. Dev Cell 22, 940–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshwani MM, Klammt C, von Daake S, Ma Y, Kornev AP, Choe S, Insel PA, and Taylor SS (2012). Cotranslational cisphosphorylation of the COOH-terminal tail is a key priming step in the maturation of cAMP-dependent protein kinase. Proc Natl Acad Sci USA 109, E1221–E1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kato M, and Beachy PA (2009). Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc Natl Acad Sci USA 106, 21666–21671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong JH, Siebold C, and Rohatgi R (2019). Biochemical mechanisms of vertebrate hedgehog signaling. Development 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF (2015). IFT-cargo interactions and protein transport in cilia. Trends Biochem Sci 40, 765–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RTH, Zhao Z, and Ingham PW (2016). Hedgehog signalling. Development 143, 367–372. [DOI] [PubMed] [Google Scholar]
- Lefkowitz RJ, Pierce KL, and Luttrell LM (2002). Dancing with different partners: protein kinase a phosphorylation of seven membrane-spanning receptors regulates their G protein-coupling specificity. Mol Pharmacol 62, 971–974. [DOI] [PubMed] [Google Scholar]
- Lepage T, Cohen SM, Diaz-Benjumea FJ, and Parkhurst SM (1995). Signal transduction by cAMP-dependent protein kinase A in Drosophila limb patterning. Nature 373, 711–715. [DOI] [PubMed] [Google Scholar]
- Li J, Wang C, Wu C, Cao T, Xu G, Meng Q, and Wang B (2017). PKA-mediated Gli2 and Gli3 phosphorylation is inhibited by Hedgehog signaling in cilia and reduced in Talpid3 mutant. Dev Biol 429, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Ma G, Wang B, and Jiang J (2014). Hedgehog induces formation of PKA-smoothened complexes to promote smoothened phosphorylation and pathway activation. Sci Signal 7, ra62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Li S, Han Y, Tong C, Wang B, Chen Y, and Jiang J (2016). Regulation of smoothened phosphorylation and high-level Hedgehog signaling activity by a plasma membrane associated kinase. PLoS Biol 14, e1002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ohlmeyer JT, Lane ME, and Kalderon D (1995). Function of protein kinase A in hedgehog signal transduction and Drosophila imaginal disc development. Cell 80, 553–562. [DOI] [PubMed] [Google Scholar]
- Liem KF, He M, Ocbina PJR, and Anderson KV (2009). Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc Natl Acad Sci USA 106, 13377–13382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zeng H, and Liu A (2015). The loss of Hh responsiveness by a non-ciliary Gli2 variant. Development 142, 1651–1660. [DOI] [PubMed] [Google Scholar]
- Low WC, Wang C, Pan Y, Huang XY, Chen JK, and Wang B (2008). The decoupling of Smoothened from Gαi proteins has little effect on Gli3 protein processing and Hedgehog-regulated chick neural tube patterning. Dev Biol 321, 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MJ, Hill EV, and Houslay MD (2006). Intracellular targeting of phosphodiesterase-4 underpins compartmentalized cAMP signaling. In: Current Topics in Developmental Biology. New York: Academic Press. 225–259. [DOI] [PubMed] [Google Scholar]
- Marley A, Choy RWY, and von Zastrow M (2013). GPR88 reveals a discrete function of primary cilia as selective insulators of GPCR cross-talk. PLoS ONE 8, e70857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matise MP, Epstein DJ, Park HL, Platt KA, and Joyner AL (1998). Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development 125, 2759–2770. [DOI] [PubMed] [Google Scholar]
- May EA, Kalocsay M, D’Auriac IG, Schuster PS, Gygi SP, Nachury MV, and Mick DU (2021). Time-resolved proteomics profiling of the ciliary Hedgehog response. J Cell Biol 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May SR, Ashique AM, Karlen M, Wang B, Shen Y, Zarbalis K, Reiter J, Ericson J, and Peterson AS (2005). Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev Biol 287, 378–389. [DOI] [PubMed] [Google Scholar]
- McCormick K, and Baillie GS (2014). Compartmentalisation of second messenger signalling pathways. Curr Opin Genets Dev 27, 20–25. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Ingham PW, and Tabin CJ (2003). Developmental roles and clinical significance of Hedgehog signaling. In: Current Topics in Developmental Biology. New York: Academic Press. 1–114. [DOI] [PubMed] [Google Scholar]
- Mehta S, Zhang Y, Roth RH, Zhang JF, Mo A, Tenner B, Huganir RL, and Zhang J (2018). Single-fluorophore biosensors for sensitive and multiplexed detection of signalling activities. Nat Cell Biol 20, 1215–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick DU, Rodrigues RB, Leib RD, Adams CM, Chien AS, Gygi SP, and Nachury MV (2015). Proteomics of primary cilia by proximity labeling. Dev Cell 35, 497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic L, Weiss LE, Yoon J, Roth TL, Su YRS, Sahl SJ, Scott MP, and Moerner WE (2015). Single-molecule imaging of Hedgehog pathway protein Smoothened in primary cilia reveals binding events regulated by Patched1. Proc Natl Acad Sci USA 112, 8320–8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BS, Stepanchick AN, Tewson PH, Hartle CM, Zhang J, Quinn AM, Hughes TE, and Mirshahi T (2016). Cilia have high cAMP levels that are inhibited by Sonic Hedgehog-regulated calcium dynamics. Proc Natl Acad Sci USA 113, 13069–13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Jansen V, Jikeli JF, Hamzeh H, Alvarez L, Dombrowski M, Balbach M, Strünker T, Seifert R, Kaupp UB, et al. (2016). A novel biosensor to study camp dynamics in cilia and flagella. eLife 5, e14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Wen X, Chih B, Nelson CD, Lane WS, Scales SJ, and Jackson PK (2010). TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev 24, 2180–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Wen X, Ratti N, Loktev A, Rangell L, Scales SJ, and Jackson PK (2013). The ciliary G-protein-coupled receptor Gpr161 negatively regulates the sonic hedgehog pathway via cAMP signaling. Cell 152, 210–223. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis E, and Hui C (2005). Hedgehog signaling and congenital malformations. Clin Genets 67, 193–208. [DOI] [PubMed] [Google Scholar]
- Niewiadomski P, Zhujiang A, Youssef M, and Waschek JA (2013). Interaction of PACAP with Sonic hedgehog reveals complex regulation of the Hedgehog pathway by PKA. Cell Signal 25, 2222–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomski P, Kong JH, Ahrends R, Ma Y, Humke EW, Khan S, Teruel MN, Novitch BG, and Rohatgi R (2014). Gli protein activity is controlled by multisite phosphorylation in vertebrate hedgehog signaling. Cell Rep 6, 168–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev VO, Bünemann M, Hein L, Hannawacker A, and Lohse MJ (2004). Novel single chain cAMP sensors for receptor-induced signal propagation. J Biol Chem 279, 37215–37218. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, and Wieschaus E (1980). Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801. [DOI] [PubMed] [Google Scholar]
- Ogden SK, Fei DL, Schilling NS, Ahmed YF, Hwa J, and Robbins DJ (2008). G protein Gαi functions immediately downstream of Smoothened in Hedgehog signalling. Nature 456, 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page CP, and Spina D (2012). Selective PDE inhibitors as novel treatments for respiratory diseases. Curr Opin Pharmacol 12, 275–286. [DOI] [PubMed] [Google Scholar]
- Pal K, Hwang SH, Somatilaka B, Badgandi H, Jackson PK, DeFea K, and Mukhopadhyay S (2016). Smoothened determines β-arrestin-mediated removal of the G protein-coupled receptor Gpr161 from the primary cilium. J Cell Biol 212, 861–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, and Rubin GM (1995). cAMP-dependent protein kinase and hedgehog act antagonistically in regulating decapentaplegic transcription in Drosophila imaginal discs. Cell 80, 543–552. [DOI] [PubMed] [Google Scholar]
- Pan Y, Bai CB, Joyner AL, and Wang B (2006). Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol Cell Biol 26, 3365–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, and Wang B (2007). A novel protein-processing domain in Gli2 and Gli3 differentially blocks complete protein degradation by the proteasome. J Biol Chem 282, 10846–10852. [DOI] [PubMed] [Google Scholar]
- Pan Y, Wang C, and Wang B (2009). Phosphorylation of Gli2 by protein kinase A is required for Gli2 processing and degradation and the Sonic Hedgehog-regulated mouse development. Dev Biol 326, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HL, Bai C, Platt KA, Matise MP, Beeghly A, Hui CC, Nakashima M, and Joyner AL (2000). Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 127, 1593–1605. [DOI] [PubMed] [Google Scholar]
- Pathi S, Pagan-Westphal S, Baker DP, Garber EA, Rayhorn P, Bumcrot D, Tabin CJ, Blake Pepinsky R, and Williams KP (2001). Comparative biological responses to human Sonic, Indian, and Desert hedgehog. Mech Dev 106, 107–117. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, and Witman GB (2003). The vertebrate primary cilium is a sensory organelle. Curr Opin Cell Biol 15, 105–110. [DOI] [PubMed] [Google Scholar]
- Peng H, Zhang J, Ya A, Ma W, Villa S, Sukenik S, and Ge X (2021). Myomegalin regulates Hedgehog pathway by controlling PDE4D at the centrosome. Mol Biol Cell doi: 10.1091/mbc.E21-02-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson M, Stamataki D, te Welscher P, Andersson E, Böse J, Rüther U, Ericson J, and Briscoe J (2002). Dorsal-ventral patterning of the spinal cord requires Gli3 transcriptional repressor activity. Genes Dev 16, 2865–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinskey JM, Franks NE, McMellen AN, Giger RJ, and Allen BL (2017). Neuropilin-1 promotes Hedgehog signaling through a novel cytoplasmic motif. J Biol Chem 292, 15192–15204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponsioen B, Zhao J, Riedl J, Zwartkruis E, van der Krogt G, Zaccolo M, Moolenaar WH, Bos JL, and Jalink K (2004). Detecting cAMP-induced Epac activation by fluorescence resonance energy transfer: Epac as a novel cAMP indicator. EMBO Rep 5, 1176–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, et al. (2012). Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 488, 106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusapati GV, Kong JH, Patel BB, Gouti M, Sagner A, Sircar R, Luchetti G, Ingham PW, Briscoe J, and Rohatgi R (2018). G protein-coupled receptors control the sensitivity of cells to the morphogen Sonic Hedgehog. Sci Signal 11, eaao5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Schmiege P, Coutavas E, and Li X (2018a). Two patched molecules engage distinct sites on hedgehog yielding a signaling-competent complex. Science 362, eaas8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Schmiege P, Coutavas E, Wang J, and Li X (2018b). Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature 560, 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Liu H, Thompson B, McDonald J, Zhang C, and Li X (2019). Cryo-EM structure of oxysterol-bound human Smoothened coupled to a heterotrimeric Gi. Nature 571, 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rack PG, Ni J, Payumo AY, Nguyen V, Crapster JA, Hovestadt V, Kool M, Jones DTW, Mich JK, Firestone AJ, et al. (2014). Arhgap36-dependent activation of Gli transcription factors. Proc Natl Acad Sci USA 111, 11061–11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh DR, Sever N, Choksi PK, Sigg MA, Hines KM, Thompson BM, Elnatan D, Jaishankar P, Bisignano P, Garcia-Gonzalo FR, et al. (2018). Cilia-associated oxysterols activate smoothened. Mol Cell 72, 316–327.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh DR, and Reiter JF (2019). Misactivation of Hedgehog signaling causes inherited and sporadic cancers. J Clin Invest 129, 465–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallu M, Machold R, Gaiano N, Corbin JG, McMahon AP, and Fishell G (2002). Dorsoventral patterning is established in the telencephalon of mutants lacking both Gli3 and hedgehog signaling. Development 129, 4963–4974. [DOI] [PubMed] [Google Scholar]
- Regard JB, Malhotra D, Gvozdenovic-Jeremic J, Josey M, Chen M, Weinstein LS, Lu J, Shore EM, Kaplan FS, and Yang Y (2013). Activation of hedgehog signaling by loss of GNAS causes heterotopic ossification. Nat Med 19, 1505–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich TC, Fagan KA, Tse TE, Schaack J, Cooper DMF, and Karpen JW (2001). A uniform extracellular stimulus triggers distinct cAMP signals in different compartments of a simple cell. Proc Natl Acad Sci USA 98, 13049–13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riobo NA, Saucy B, Dilizio C, and Manning DR (2006). Activation of heterotrimeric G proteins by Smoothened. Proc Natl Acad Sci USA 103, 12607–12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins DJ, Nybakken KE, Kobayashi R, Sisson JC, Bishop JM, and Thérond PP (1997). Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell 90, 225–234. [DOI] [PubMed] [Google Scholar]
- Röck R, Mayrhofer JE, Bachmann V, and Stefan E (2015). Impact of kinase activating and inactivating patient mutations on binary PKA interactions. Front Pharmacol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, and Witman GB (2002). Intraflagellar transport. Nat Rev Mol Cell Biol 3, 813–825. [DOI] [PubMed] [Google Scholar]
- Rudolf AF, Kinnebrew M, Kowatsch C, Ansell TB, El Omari K, Bishop B, Pardon E, Schwab RA, Malinauskas T, Qian M, et al. (2019). The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism. Nat Chem Biol 15, 975–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruel L, Rodriguez R, Gallet A, Lavenant-Staccini L, and Thérond PP (2003). Stability and association of Smoothened, Costal2 and fused with Cubitus interruptus are regulated by Hedgehog. Nat Cell Biol 5, 907–913. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Palma V, and Dahmane N (2002). Hedgehog-GLI signaling and the growth of the brain. Nat Rev Neurosci 3, 24–33. [DOI] [PubMed] [Google Scholar]
- Ruppert JM, Kinzler KW, Wong AJ, Bigner SH, Kao FT, Law ML Seuanez HN, O’Brien SJ, and Vogelstein B (1988). The GLI-Kruppel family of human genes. Mol Cell Biol 8, 3104–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagner A, and Briscoe J (2019). Establishing neuronal diversity in the spinal cord: A time and a place. Development 146. [DOI] [PubMed] [Google Scholar]
- Satir P, and Christensen ST (2007). Overview of structure and function of mammalian cilia. Annu Rev Physiol 69, 377–400. [DOI] [PubMed] [Google Scholar]
- Scotland G, Beard M, Erdogan S, Huston E, McCallum E, MacKenzie SJ, Peden AH, Pooley L, Rena NG, Ross AH, et al. (1998). Intracellular compartmentalization of PDE4 cyclic AMP-specific phosphodiesterases. Methods 14, 65–79. [DOI] [PubMed] [Google Scholar]
- Scott JD, and Pawson T (2009). Cell signaling in space and time: Where proteins come together and when they’re apart. Science 326, 1220–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F, Cheng L, Douglas AE, Riobo NA, and Manning DR (2013). Smoothened is a fully competent activator of the heterotrimeric G protein Gi. Mol Pharmacol 83, 691–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada IS, Hwang SH, Somatilaka BN, Wang X, Skowron P, Kim J, Kim M, Shelton JM, Rajaram V, Xuan Z, et al. (2018). Basal suppression of the Sonic Hedgehog pathway by the G-protein-coupled receptor Gpr161 restricts medulloblastoma pathogenesis. Cell Rep 22, 1169–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Wen X, and Scales SJ (2015). The orphan G protein-coupled receptor Gpr175 (Tpra40) enhances Hedgehog signaling by modulating cAMP levels. J Biol Chem 290, 29663–29675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisson JC, Ho KS, Suyama K, and Scott MP (1997). Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell 90, 235–245. [DOI] [PubMed] [Google Scholar]
- Snuderl M, Batista A, Kirkpatrick ND, Ruiz de Almodovar C, Riedemann L, Walsh EC, Anolik R, Huang Y, Martin JD, Kamoun W, et al. (2013). Targeting placental growth factor/neuropilin 1 pathway inhibits growth and spread of medulloblastoma. Cell 152, 1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somatilaka BN, Hwang SH, Palicharla VR, White KA, Badgandi H, Shelton JM, and Mukhopadhyay S (2020). Ankmy2 prevents smoothened-independent hyperactivation of the Hedgehog pathway via cilia-regulated adenylyl cyclase signaling. Dev Cell 54, 710–726.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spina D (2008). PDE4 inhibitors: Current status. British J Pharmacol 155, 308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, Scott MP, Pennica D, Goddard A, Phillips H, et al. (1996). The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 384, 129–134. [DOI] [PubMed] [Google Scholar]
- Stoufflet J, Chaulet M, Doulazmi M, Fouquet C, Dubacq C, Metin C, Schneider-Maunoury S, Trembleau A, Vincent P, and Caillé I (2020). Primary cilium-dependent cAMP/PKA signaling at the centrosome regulates neuronal migration. Sci Adv 6, eaba3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt DI, Wiersdorff V, and Mlodzik M (1995). Regulation of furrow progression in the Drosophila eye by cAMP-dependent protein kinase A. Nature 373, 705–709. [DOI] [PubMed] [Google Scholar]
- Su S, Phua SC, DeRose R, Chiba S, Narita K, Kalugin PN, Katada T, Kontani K, Takeda S, and Inoue T (2013). Genetically encoded calcium indicator illuminates calcium dynamics in primary cilia. Nat Methods 10, 1105–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svärd J, Heby-Henricson K, Henricson KH, Persson-Lek M, Rozell B, Lauth M, Bergström A, Ericson J, Toftgård R, and Teglund S (2006). Genetic elimination of suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell 10, 187–197. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Buechler JA, and Yonemoto W (1990). cAMP-dependent protein kinase: Framework for a diverse family of regulatory enzymes. Annu Rev Biochem 59, 971–1005. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Zhang P, Steichen JM, Keshwani MM, and Kornev AP (2013). PKA: Lessons learned after twenty years. Biochim Biophys Acta 1834, 1271–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempe D, Casas M, Karaz S, Blanchet-Tournier MF, and Concordet JP (2006). Multisite protein kinase A and glycogen synthase kinase 3β phosphorylation leads to Gli3 ubiquitination by SCFβTrCP. Mol Cell Biol 26, 4316–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrin A, Monterisi S, Stangherlin A, Zoccarato A, Koschinski A, Surdo NC, Mongillo M, Sawa A, Jordanides NE, Mountford JC, et al. (2012). PKA and PDE4D3 anchoring to AKAP9 provides distinct regulation of cAMP signals at the centrosome. J Cell Biol 198, 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, et al. (2009). Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods 6, 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Quesada O, Mayrhofer JE, and Stefan E (2017). The many faces of compartmentalized PKA signalosomes. Cell Signal 37, 1–11. [DOI] [PubMed] [Google Scholar]
- Truong ME, Bilekova S, Choksi SP, Li W, Bugaj LJ, Xu K, and Reiter JF (2021). Vertebrate cells differentially interpret ciliary and extraciliary cAMP. Cell 184, 2911–2926.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukachinsky H, Lopez LV, and Salic A (2010). A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol 191, 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumharn RE, and Scott JD (2016). Protein kinase A catalytic subunit isoform PRKACA; History, function and physiology. Gene 577, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuson M, He M, and Anderson KV (2011). Protein kinase A acts at the basal body of the primary cilium to prevent Gli2 activation and ventralization of the mouse neural tube. Development 138, 4921–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uys GM, Ramburan A, Loos B, Kinnear CJ, Korkie LJ, Mouton J, Riedemann J, and Moolman-Smook JC (2011). Myomegalin is a novel A-kinase anchoring protein involved in the phosphorylation of cardiac myosin binding protein C. BMC Cell Biol 12, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde I, Pahlke G, Salanova M, Zhang G, Wang S, Coletti D, Onuffer J, Jin SLC, and Conti M (2001). Myomegalin is a novel protein of the Golgi/centrosome that interacts with a cyclic nucleotide phosphodiesterase. J Biol Chem 276, 11189–11198. [DOI] [PubMed] [Google Scholar]
- Vuolo L, Herrera A, Torroba B, Menendez A, and Pons S (2015). Ciliary adenylyl cyclases control the Hedgehog pathway. J Cell Sci 128, 2928. [DOI] [PubMed] [Google Scholar]
- Wallace VA (1999). Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr Biol 9, 445–448. [DOI] [PubMed] [Google Scholar]
- Wang B, Fallon JF, and Beachy PA (2000). Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 100, 423–434. [DOI] [PubMed] [Google Scholar]
- Wang B, and Li Y (2006). Evidence for the direct involvement of TrCP in Gli3 protein processing. Proc Natl Acad Sci USA 103, 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler-Reya RJ, and Scott MP (1999). Control of neuronal precursor proliferation in the cerebellum by sonic hedgehog. Neuron 22, 103–114. [DOI] [PubMed] [Google Scholar]
- Wen X, Lai CK, Evangelista M, Hongo JA, de Sauvage FJ, and Scales SJ (2010). Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Mol Cell Biol 30, 1910–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CH, Hempel JE, Hao J, Frist AY, Williams MM, Fleming JT, Sulikowski GA, Cooper MK, Chiang C, and Hong CC (2015). An in vivo chemical genetic screen identifies phosphodiesterase 4 as a pharmacological target for Hedgehog signaling inhibition. Cell Rep 11, 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W, and Scott JD (2004). AKAP signalling complexes: focal points in space and time. Nat Rev Mol Cell Biol 5, 959–970. [DOI] [PubMed] [Google Scholar]
- Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, Bonifas JM, Lam CW, Hynes M, Goddard A, et al. (1998). Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature 391, 90–92. [DOI] [PubMed] [Google Scholar]
- Yang TT, Tran MNT, Chong WM, Huang CE, and Liao JC (2019). Single-particle tracking localization microscopy reveals nonaxonemal dynamics of intraflagellar transport proteins at the base of mammalian primary cilia. Mol Biol Cell 30, 828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccolo M, and Pozzan T (2002). Discrete microdomains with high concentration of cAMP in stimulated rat neonatal cardiac myocytes. Science 295, 1711–1715. [DOI] [PubMed] [Google Scholar]
- Zaccolo M, Zerio A, and Lobo MJ (2021). Subcellular organization of the camp signaling pathway. Pharmacol Rev 73, 278–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Zhuang T, Lin Q, Yang B, Xu X, Xin G, Zhu S, Wang G, Yu B, Zhang T, et al. (2019). Patched1-ArhGAP36-PKA-Inversin axis determines the ciliary translocation of smoothened for sonic hedgehog pathway activation. Proc Natl Acad Sci USA 116, 874–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ma Y, Taylor SS, and Tsien RY (2001). Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering. Proc Natl Acad Sci USA 98, 14997–15002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JF, Liu B, Hong I, Mo A, Roth RH, Tenner B, Lin W, Zhang JZ, Molina RS, Drobizhev M, et al. (2021). An ultrasensitive biosensor for high-resolution kinase activity imaging in awake mice. Nat Chem Biol 17, 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Knape MJ, Ahuja LG, Keshwani MM, King CC, Sastri M, Herberg FW, and Taylor SS (2015). Single turnover autophosphorylation cycle of the PKA RIIβ holoenzyme. PLoS Biol 13, e1002192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhao Y, Tong C, Wang G, Wang B, Jia J, and Jiang J (2005). Hedgehog-regulated Costal2-kinase complexes control phosphorylation and proteolytic processing of cubitus interruptus. Dev Cell 8, 267–278. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Bulkley DP, Xin Y, Roberts KJ, Asarnow DE, Sharma A, Myers BR, Cho W, Cheng Y, and Beachy PA (2018). Structural basis for cholesterol transport-like activity of the Hedgehog receptor patched. Cell 175, 1352–1364.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]