Abstract
VectorBase (VectorBase.org) is part of the VEuPathDB Bioinformatics Resource Center, providing free online access to multi-omics and population biology data, focusing on arthropod vectors and invertebrates of importance to human health. VectorBase includes genomics and functional genomics data from bed bugs, biting midges, body lice, kissing bugs, mites, mosquitoes, sand flies, ticks, tsetse flies, stable flies, house flies, fruit flies, and a snail intermediate host. Tools include the Search Strategy system and MapVEu, enabling users to interrogate and visualize diverse ‘omics and population-level data using a graphical interface (no programming experience required). Users can also analyze their own private data, such as transcriptomic sequences, exploring their results in the context of other publicly-available information in the database. Help Desk: help@vectorbase.org.
Introduction
As part of the Eukaryotic Pathogen, Vector and Host Informatics Resource (VEuPathDB.org) Bioinformatics Resource Center (BRC), VectorBase (VectorBase.org) is supported by the US National Institutes of Allergy and Infectious Diseases (NIAID) [1]. In addition to VectorBase [2], VEuPathDB [3] also supports eukaryotic pathogens (protists, fungi), selected mammalian host data, and provides resources for orthology determination and phylogenetic inference (OrthoMCL.org) [4]. Additional resources using the VEuPathDB model and infrastructure accommodate epidemiological (ClinEpiDB.org) [5] and microbiome data (MicrobiomeDB.org) [6].
Release 54 of VectorBase supports 53 vector genomes and integrates a wide range of other data types, including functional genomics and genetic variation data. The MapVEu geo-visualization tool displays different types of population data, including vector abundance, pathogen infection status, genetic variation, host blood meal source, and insecticide resistance phenotypes and genotypes, for ~470 species worldwide. Data are integrated from public repositories or directly from providers and analyzed with standard workflows using an ontology-driven framework to ensure data comparability. Expert knowledge from the community is also incorporated to improve genome annotation through an Apollo interface and in the form of User Comments. Here we present a general overview of the new VectorBase resource, including site use, data types, and tools, and finish with our future plans.
The new VectorBase: a merged BRC infrastructure
The rapid growth of genomic-scale datasets, increasing integration of scientific research, and funder mandates for improved efficiency have driven the development of VEuPathDB, coupling the Ensembl bioinformatic pipelines [7,8] long used by VectorBase, with the Genomics Unified Schema [9] and highly flexible Search Strategies [10] of EuPathDB. The net result offers improved scalability, flexibility, data flow, and overall user experience.
Web interface improvements
A redesigned common user interface provides convenient, consistent access to data, searches, and help infor- mation for all supported species. The home page (Figure 1) features a header (present on all pages), a main panel, an expandable ‘News & Tweets’ section (Figure 1d), and a footer with clickable icons to access other VEuPathDB resources (Figure 1f). The Site Search (Figure 1b) allows free text searches, returning categorized results; with filters allowing users to define categories or organisms of interest. Results (genes, SNPs, etc.) can then be exported to the ‘My Strategies’ system for further data mining, visualization, or download (see below). Educational materials, FAQs, virtual events, workshops, and methods are available under the ‘Help’ menu (Figure 1, arrow), and links to tutorials and exercises are at the bottom of the main panel (Figure 1e). Additional help is also available from the ‘Contact Us’ link (Figure 1b).
Figure 1.
Redesigned VectorBase home page. (a) Left-hand panel provides access to all available searches, categorized by datatype. (b) Header on all site pages, including Site Search box and access to My Strategies, Searches, Tools, My Workspace, Data, About, Help (educational materials, arrow) and Contact Us sections. Release date and version adjacent to the logo at left; social media, login, registration, and user profile links at right. (c) Central section provides an overview of resources and tools, including vignettes to help users get started on specific topics of likely interest. (d) News & Tweets section is an expandable tab (collapsed by default), providing access to recent announcements. (e) Links to more detailed step-by-step exercises. (f) Hyperlinked logos to other VEuPathDB components and affiliated sites, and community chat button enabling users to ask questions and share information.
The left sidebar provides access to all searches (Figure 1a; also accessible from the ‘Searches’ menu). Searches are organized into expandable categories containing configurable queries against the underlying data. Search results are returned as an expandable Search Strategy and are displayed in a dynamic table that can be configured by adding, removing, or moving columns. The central section of the main panel provides an overview of available resources and tools (Figure 1c).
Omics and population data sets
VectorBase release 54 includes 492 datasets relating to vector species. Bimonthly releases incorporate new data and functionality into the site; the latest data can be found on the datasets page under the ‘Data’ menu (located in the header) (https://vectorbase.org/vectorbase/app/search/dataset/AllDatasets/result).
Forty-one vector genomes represent ‘reference’ strains for distinct species, while 12 are additional strains or resequencing of already available strains. Gene set predictions are available for all reference species (and most additional strains), including 12 with chromosomal map-pings. Other datasets, including transcriptomes, pro-teomes, genetic variation, and orthology profiles are aligned or cross-referenced to reference genomes, and genomes are also cross-referenced with ~20 external databases, including Chemical Entities of Biological Interest (ChEBI) [11], Kyoto Encyclopedia of Genes and Genomes (KEGG) [12,13] and Gene Ontology (GO) [14,15]. All omics datasets are also available for download or use with the site tools accessible under the ‘Tools’ menu in the header.
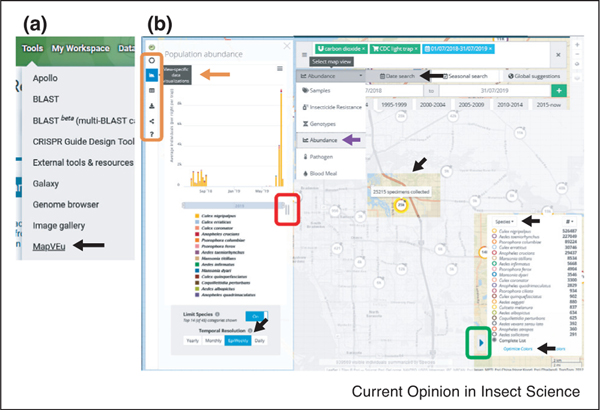
Population datasets include records for ~470 taxonomic groups, from field-collected samples divided into differ- ent map ‘views’ and/or data types, including >21 000 and >17 000 insecticide resistance phenotype and genotype assays respectively, >187 000 pathogen infection status assays, >12 000 blood meal source assays, >15 000 chromosomal inversions, >15 000 microsatellites, >2600 bar-codes, and >25 million population abundance records, among others. The MapVEu tool (Figure 2) is used for visualization, search, analysis, and raw data download. Specialized representations are also available; the bar graph in Figure 2b indicates species abundance counts for the geographic region shown.
Figure 2. Population data in MapVEu, a tool for visualizing, analyzing, and downloading geographic data. (a) Select MapVEu from the Tools menu on the home page (arrow). (b) Select Abundance ‘view’ from the dropdown menu below the map search bar (violet arrow). Select sampling location on the map or, enter into the search bar and select from the autocomplete menu (Manatee County used in this example). Date Search set = July 2018 to July 2019. Open/collapse blue arrowhead in legend panel (green box) to set: Collection Protocol = CDC light trap & Attractant = carbon dioxide, in the same panel select Species & Optimize Colors options. A point was selected (indicated in color, center of the map), to explore it in more detail. The icon at the left (orange arrow) defines graph type (bars in this example); ‘EpiWeekly’ is set as temporal resolution. Left panel size can be adjusted (red box). The orange box indicates other ‘view’ specific data visualizations, metadata details, raw data download, and so on. Login to VectorBase and follow this link to recreate the image shown: https://tinyurl.com/InsectGx2021VectorBaseFig2.
New and improved tools and resources
Genome and protein browsers
Genome browsing is facilitated by the JBrowse genome browser [16], an open-source platform allowing users to select tracks displaying aligned transcriptomic, proteomic, epigenomic, and variation data. Variation data sets (SNP calls) are available via Variant Call Format (VCF) files aligned to reference genomes. Protein Browser tracks include transmembrane domains (TMHMM predictions) [17], protein domains (InterPro predictions) [18], and synteny views across multiple genomes.
Gene pages
Gene pages, now with a new design, compile all the available data about a particular gene into a single webpage. Aligning of orthologs and paralogs are identified using OrthoMCL [4], and Clustal Omega [19] can be launched for multiple sequence alignments. New representations facilitate exploration of transcriptomics data, protein features and properties, use of functional prediction tools, and assessment of metabolic pathways.
My strategies
Searches in VectorBase can be integrated into a Search Strategy, allowing users to integrate diverse results into a multistep in silico experiment (Figure 3). Multistep strategies (for example, find Aedes kinases expressed in a particular time or place, and conserved in species of interest) are built one step at a time, bringing together several searches by union (Figure 3c, step 2), intersection (Figure 3c, step 3), or subtraction operations. Strategies are extended by clicking ‘Add a step’ in the graphic panel (Figure 3c). Options for extending a strategy include ‘Combine’ with similar records’, ‘Transform’ to related records, and Genomic Colocation. Results can be transformed into orthologs, metabolic pathways, or compounds. Additionally, the genomic location can be exploited to search for additional features. Search Strategies can be saved, copied, revised, or shared with others using a private link. The Strategy System replaces BioMart functionalities, including the ability to download genome-wide information available from gene pages (e.g. homologs, expression values, GO terms, etc.).
Figure 3.
Search strategies as in silico experiments. (a) Site Search, a box that can be accessed from the header, returns site-wide hits, in the results page filters can be applied. Search strategies (for Genes, Organisms, Pathways, etc.) can be run from the Searches pull-down menu or the Search for . . . panel at the left. (b) Search filter can help to locate searches of interest, for example, identifying ‘text’ searches of Genes or Compounds. (c) Search results constitute one Step in a Search Strategy, which can be combined with other searches (+ Add a step; green arrow) using Boolean operators (e.g. union, intersection, subtraction). Results may be downloaded (black arrow), and searches edited, saved, shared, or published (icons at right; blue arrow). To retrieve this sample search for A. gambiae proteases expressed in the midgut with a specific promotor region DNA motif, see: https://vectorbase.org/vectorbase/app/workspace/strategies/import/bc4d101022805435. Publicly shared strategies are available from the menu at the top (orange arrow; https://vectorbase.org/vectorbase/app/workspace/strategies/public).
Enrichment analysis
Functional enrichment of gene results includes statistically valid gene ontology (GO) [14,15] and metabolic pathway enrichment results (also available as a word cloud). GO enrichment data can also be exported to REVIGO [20], facilitating data visualization using a variety of interactive tools.
Community annotation
VectorBase continues to support manual gene annotation with Apollo [21], which allows users to create & edit structural annotation, update product names, descriptions and symbols, and so on. For some species, VEuPathDB staff may integrate these annotations as part of the official gene set once several annotations have been submitted. Users can request that a specific genome be made available in Apollo for annotation by contacting the help desk. The ‘User Comments’ tool available on Gene Pages is new to VectorBase, allowing users to submit comments about specific genes, which are immediately integrated into the database and become searchable.
Homology predictions
VectorBase has historically been used to predict putative gene function, resolve evolutionary questions, and provide comparative genomic analyses using the Ensembl Compara pipeline. This functionality is now provided by OrthoMCL [4], but Compara can still be accessed via Ensembl Metazoa [7,8] (https://metazoa.ensembl.org/index.html) using the genome browser gene pages and BioMart [22,23].
Galaxy
Computationally intensive analysis of user-provided data (e.g. RNA-seq datasets, SNP calling, etc.) continues to be provided via a user-friendly front end to a cloud-based Galaxy pipeline [24], allowing users to privately analyze their own data. Output files can be exported for interrogation in the context of all other data in VectorBase.
Registration and citation
VectorBase does not require registration for use, but an account provides additional features including email alerts about new data sets, the ability to save and share BLAST jobs, Search Strategies, output results from Galaxy, gene annotations in Apollo, and more.
Much of the data in VectorBase is provided by independent researchers, and citation information is included for each VectorBase record, including publications or other attribution details for unpublished datasets, allowing users to cite primary data sources when relevant. A FAQ (https://vectorbase.org/vectorbase/app/static-content/faq.html) and the ‘About’ section provides information on how to cite VectorBase, and when appropriate, users are encouraged to include the VectorBase logo, tables, figures, and images in their original research presentations and publications.
Recent science enabled by VectorBase data and tools
VectorBase data, tools, and analyses can demonstrably expedite basic discovery and translational research. For example, VectorBase genomes have been used to resolve questions involving individual genes [25–27], characterize gene families [28,29], and perform genome-wide analyses [30,31]. Genome resources have also been used to develop wet lab techniques, for example, for primer design [32] or a multilocus amplicon sequencing for simultaneous mosquito species identification and detection of parasite infection status [33]. BLAST [34,35], gene enrichment [36], and comparative genomic analyses among the same or different species, have been used for phylogenetic and homolog gene predictions [37•,38•,39•]. Genome assemblies have been improved, creating physical maps [40], karyotypes [41], and genome elements identified [42–44] using VectorBase files and tools. Researchers have also used VectorBase genome assembly and gene set files to perform analyses such as transcript differential expression [45,46] and peptide expression [47].
Transcriptomics and proteomics data sets deposited in VectorBase allow research groups to ask or test new hypotheses as described in this review paper on mosquito ‘omics [7•], now also possible using the Search Strategy system [12•]. Phenotype experiments, for example, insecticide susceptibility [50], have been analyzed using VectorBase genomes and the Ensembl Variation Effect Predictor (VEP), to interpret obtained genotypes (variant calling). The MapVEu tool has been used to generate meta-analyses, for example, with the population abundance view [51•], and/or facilitate reviews, for example, with the blood meal view [52•].
Summary and future perspectives
VEuPathDB provides consistent representation, interrogation, and visualization of data types and tools for hosts, vectors, parasites, and fungi species. Vector data can be accessed directly through VectorBase or through the VEuPathDB homepage. Infrastructure improvements resulting from the merger of VectorBase and EuPathDB allow for increased scalability, efficiency, and interoperability to incorporate the increasing quantities of data and new data types.
Future VectorBase releases are expected to provide organism preference parameters enabling customization of the user experience including the ability to select organisms across taxonomic groups (e.g. exploration of both Plasmodium parasites and Anopheles mosquito vectors). Development plans include tools for analysis and visualization of vector-pathogen interactions and systems biology research, resources for integrated exploration of VectorBase and the bacterial/viral BRC, improved visualizations and analyses for the MapVEu tool, an improved variant calling pipeline and associated searches, improved mechanisms for portability of data to other applications, and additional workflows using the VEuPathDB Galaxy instance.
Acknowledgements
To all the VEuPathDB team, including past and present VectorBase team members https://vectorbase.org/vectorbase/app/static-content/personnel.html. This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID) — National Institutes of Health (NIH) [Contract 75N93019C00077] and Wellcome Trust [grants 218288/Z/19/Z and 212929/Z/18/Z]. The authors also wish to thank members of the VEuPathDB communities for their continuous feedback to improve VectorBase, and for their data submission contributions.
Footnotes
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
- 1.Greene JM, Collins F, Lefkowitz EJ, Roos D, Scheuermann RH, Sobral B, Stevens R, White O, Di Francesco V: National Institute of Allergy and Infectious Diseases bioinformatics resource centers: new assets for pathogen informatics. Infect Immun 2007, 75:3212–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giraldo-Calderón GI, Emrich SJ, MacCallum RM, Maslen G, Dialynas E, Topalis P, Ho N, Gesing S: VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res 2015, 43:D707–D713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warrenfeltz S, Basenko EY, Crouch K, Harb OS, Kissinger JC, Roos DS, Shanmugasundram A, Silva-Franco F: EuPathDB: the eukaryotic pathogen genomics database resource. Methods Mol Biol 2018, 1757:69–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer S, Brunk BP, Chen F, Gao X, Harb OS, Iodice JB, Shanmugam D, Roos DS, Stoeckert CJ Jr: Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinform 2011:11–19. Chapter 6: Unit 6.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruhamyankaka E, Brunk BP, Dorsey G, Harb OS, Helb DA, Judkins J, Kissinger JC, Lindsay B, Roos DS, San EJ et al. : ClinEpiDB: an open-access clinical epidemiology database resource encouraging online exploration of complex studies. Gates Open Res 2019, 3:1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliveira FS, Brestelli J, Cade S, Zheng J, Iodice J, Fischer S, Aurrecoechea C, Kissinger JC, Brunk BP, Stoeckert CJ Jr et al. : MicrobiomeDB: a systems biology platform for integrating, mining and analyzing microbiome experiments. Nucleic Acids Res 2018, 46:D684–D691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howe KL, Contreras-Moreira B, De Silva N, Maslen G, Akanni W, Allen J, Alvarez-Jarreta J, Barba M, Bolser DM, Cambell L et al. : Ensembl genomes 2020-enabling non-vertebrate genomic research. Nucleic Acids Res 2020, 48:D689–D695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yates AD, Achuthan P, Akanni W, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R et al. : Ensembl 2020. Nucleic Acids Res 2020, 48:D682–D688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson SB, Crabtree J, Brunk B, Schug J, Tannen V, Overton GC, Stoeckert CJ: K2/Kleislit and GUS: experiments in integrated access to genomic data sources. IBM Syst J 2001, 40:512–531. [Google Scholar]
- 10.Fischer S, Aurrecoechea C, Brunk BP, Gao X, Harb OS, Kraemer ET, Pennington C, Treatman C, Kissinger JC, Roos DS et al. : The strategies WDK: a graphical search interface and web development kit for functional genomics databases. Database (Oxford) 2011, 2011:bar027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hastings J, Owen G, Dekker A, Ennis M, Kale N, Muthukrishnan V, Turner S, Swainston N, Mendes P, Steinbeck C: ChEBI in 2016: improved services and an expanding collection of metabolites. Nucleic Acids Res 2016, 44:D1214–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M: KEGG: integrating viruses and cellular organisms. Nucleic Acids Res 2021, 49:D545–D551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M, Sato Y, Kawashima M: KEGG mapping tools for uncovering hidden features in biological data. Protein Sci 2022, 31:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT et al. : Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000, 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The Gene Ontology Consortium et al. : The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res 2021, 49: D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buels R, Yao E, Diesh CM, Hayes RD, Munoz-Torres M, Helt G, Goodstein DM, Elsik CG, Lewis SE, Stein L et al. : JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol 2016, 17:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krogh A, Larsson B, von Heijne G, Sonnhammer EL: Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001, 305:567–580. [DOI] [PubMed] [Google Scholar]
- 18.Blum M, Chang H-Y, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Nuka G, Paysan-Lafosse T, Qureshi M, Raj S et al. : The InterPro protein families and domains database: 20 years on. Nucleic Acids Res 2021, 49:D344–D354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sievers F, Higgins DG: Clustal Omega for making accurate alignments of many protein sequences. Protein Sci 2018, 27:135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Supek F, Bosnjak M, Skunca N, Smuc T: REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 2011, 6:e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunn NA, Unni DR, Diesh C, Munoz-Torres M, Harris NL, Yao E, Rasche H, Holmes IH, Elsik CG, Lewis SE: Apollo: democratizing genome annotation. PLoS Comput Biol 2019, 15:e1006790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasprzyk A: BioMart: driving a paradigm change in biological data management. Database (Oxford) 2011, 2011:bar049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smedley D, Haider S, Durinck S, Pandini L, Provero P, Allen J, Arnaiz O, Awedh MH, Baldock R, Barbiera G et al. : The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res 2015, 43:W589598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Gruning BA et al. : The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 2018, 46: W537–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mysore K, Sun L, Roethele JB, Li P, Igiede J, Misenti JK, DumanScheel M: A conserved female-specific larval requirement for MtnB function facilitates sex separation in multiple species of disease vector mosquitoes. Parasit Vectors 2021, 14:338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vieira PH, Benjamim CF, Atella G, Ramos I: VPS38/UVRAG and ATG14, the variant regulatory subunits of the ATG6/Beclin1PI3K complexes, are crucial for the biogenesis of the yolk organelles and are transcriptionally regulated in the oocytes of the vector Rhodnius prolixus. PLoS Negl Trop Dis 2021, 15: e0009760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webster SH, Scott MJ: The Aedes aegypti (Diptera: Culicidae) hsp83 gene promoter drives strong ubiquitous DsRed and ZsGreen marker expression in transgenic mosquitoes. J. Med Entomol 2021, 58:2533–2537. [DOI] [PubMed] [Google Scholar]
- 28.Waterhouse RM, Lazzaro BP, Sackton TB: Characterization of insect immune systems from genomic data. In Immunity in Insects. Edited by Sandrelli F, Tettamanti G. Springer US; 2020:334. [Google Scholar]
- 29.Feuda R, Goulty M, Zadra N, Gasparetti T, Rosato E, Pisani D, Rizzoli A, Segata N, Ometto L, Stabelli OR: Phylogenomics of opsin genes in diptera reveals lineage-specific events and contrasting evolutionary dynamics in Anopheles and Drosophila. Genome Biol Evol 2021, 13:evab170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campos M, Rona LDP, Willis K, Christophides GK, MacCallum RM: Unravelling population structure heterogeneity within the genome of the malaria vector Anopheles gambiae. BMC Genomics 2021, 22:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeRaad DA, Cobos ME, Alkishe A, Ashraf U, Ahadji-Dabla KM, Nuñez-Penichet C, Peterson AT: Genome-environment association methods comparison supports omnigenic adaptation to ecological niche in malaria vector mosquitoes. Mol Ecol 2021, 30:6468–6485. [DOI] [PubMed] [Google Scholar]
- 32.Montanez-Gonzalez R, Vallera AC, Calzetta M, Pichler V, Love RR, Guelbeogo MW, Dabire RK, Pombi M, Costantini C, Simard F et al. : A PCR-RFLP method for genotyping of inversion 2Rc in Anopheles coluzzii. Parasit Vectors 2021, 14:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Makunin A, Korlevic P, Park N, Goodwin S, Waterhouse RM, von Wyschetzki K, Jacob CG, Davies R, Kwiatkowski D, St Laurent B et al. : A targeted amplicon sequencing panel to simultaneously identify mosquito species and Plasmodium presence across the entire Anopheles genus. Mol Ecol Resour 2022, 22:28–44 • The authors developed a wet-lab tool, a multilocus amplicon sequencing approach, to simultaneously reveal both the mosquito species and whether that mosquito carries malaria parasites, which can be used in large-scale mosquito genetic surveillance and vector control. The initial selection of phylogenetically informative markers was based on the whole-genome alignment of 21 genomes from 19 species, available from VectorBase.
- 34.Speth Z, Kaur G, Mazolewski D, Sisomphou R, Siao DDC, Pooraiiouby R, Vasquez-Gross H, Petereit J, Gulia-Nuss M, Mathew D et al. : Characterization of Anopheles stephensi odorant receptor 8, an abundant component of the mouthpart chemosensory transcriptome. Insects 2021, 12:593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Brito TF, Coelho VL, Cardoso MA, Brito IAd A, Berni MA, Zenk FL, Iovino N, Pane A: Transovarial transmission of a core virome in the Chagas disease vector Rhodnius prolixus. PLoS Pathog 2021, 17:e1009780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gestuveo RJ, Royle J, Donald CL, Lamont DJ, Hutchinson EC, Merits A, Kohl A, Varjak M: Analysis of Zika virus capsid-Aedes aegypti mosquito interactome reveals pro-viral host factors critical for establishing infection. Nat Commun 2021, 12:2766. • The authors used VectorBase GO and metabolic pathway enrichment to identify mosquito interactions with the Zika virus capsid proteins. To construct the protein-protien interaction (PPI) networks, Uniprot IDs were converted to VectorBase gene IDs.
- 37. Cheng C, Kirkpatrick M: Molecular evolution and the decline of purifying selection with age. Nat Commun 2021, 12:2657. • The authors used as input data, in a molecular evolution model, ortholog genes extracted from VectorBase.
- 38. Kabaka JM, Wachira BM, Mang’era CM, Rono MK, Hassanali A, Okoth SO, Oduol VO, Macharia RW, Murilla GA, Mireji PO:Expansions of chemosensory gene orthologs among selected tsetse fly species and their expressions in Glossina morsitans morsitans tsetse fly. PLoS Negl Trop Dis 2020, 14:e0008341 • The authors used VectorBase for the analyses of a (chemosensory) gene group. Resources include genome files for species of interest and, Compara phylogentic trees (with homolog genes) and, Computational Analysis of gene Family Evolution, CAFÉ (gene tree with expansions/ contractions), both tools currently available in Ensembl Metazoa.
- 39. Kimenyi KM, Abry MF, Okeyo W, Matovu E, Masiga D, Kulohoma BW: Detecting bracoviral orthologs distribution in five tsetse fly species and the housefly genomes. BMC Res Notes 2020, 13:318. • The authors used VectorBase to retrieve sequences from bracoviruses in multiple Dipteran genomes. OrthoMCL DB was also used for data analyses.
- 40.Rafael MS, Bridi LC, Sharakhov IV, Marinotti O, Sharakhova MV, Timoshevskiy V, Guimaraes-Marques GM, Santos VS, da Silva CGN, Astolfi-Filho S et al. : Physical mapping of the Anopheles (Nyssorhynchus) darlingi genomic scaffolds. Insects 2021, 12:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love RR, Redmond SN, Pombi M, Caputo B, Petrarca V, Della Torre A, Anopheles gambiae Genomes C, Besansky NJ: In silico karyotyping of chromosomally polymorphic malaria mosquitoes in the Anopheles gambiae complex. G3 2019, 9:3249–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farley EJ, Eggleston H, Riehle MM: Filtering the junk: assigning function to the mosquito non-coding genome. Insects 2021, 12:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma Q, Srivastav SP, Gamez S, Dayama G, Feitosa-Suntheimer F, Patterson EI, Johnson RM, Matson EM, Gold AS, Brackney DE et al. : A mosquito small RNA genomics resource reveals dynamic evolution and host responses to viruses and transposons. Genome Res 2021, 31:512–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schember I, Halfon MS: Identification of new Anopheles gambiae transcriptional enhancers using a cross-species prediction approach. Insect Mol Biol 2021, 30:410–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mang’era CM, Khamis FM, Awuoche EO, Hassanali A, Ombura FLO, Mireji PO: Transcriptomic response of Anopheles gambiae sensu stricto mosquito larvae to Curry tree (Murraya koenigii) phytochemicals. Parasit Vectors 2021, 14:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gakii C, Bwana BK, Mugambi GG, Mukoya E, Mireji PO, Rimiru R: In silico-driven analysis of the Glossina morsitans morsitans antennae transcriptome in response to repellent or attractant compounds. PeerJ 2021, 9:e11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perdomo HD, Hussain M, Parry R, Etebari K, Hedges LM, Zhang G, Schulz BL, Asgari S: Human blood microRNA hsa-miR-21–5p induces vitellogenin in the mosquito Aedes aegypti. Commun Biol 2021, 4:856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ruzzante L, Reijnders MJMF, Waterhouse RM: Of genes and genomes: mosquito evolution and diversity. Trends Parasitol 2019, 35:32–51 • In this review paper the authors discussed how deposited data in VectorBase (e.g. genomes, transcritptomes, etc.) allows research groups to ask or test new hypotheses, especially for mosquito research. The advantages of having the Apollo tool for gene manual annotation are also discussed.
- 49. Kumar V, Garg S, Gupta L, Gupta K, Diagne CT, Misse D, Pompon J, Kumar S, Saxena V: Delineating the role of Aedes aegypti ABC transporter gene family during mosquito development and arboviral infection via transcriptome analyses. Pathogens 2021, 10:1127. • The authors used VectorBase to identify a family of genes with resources that include: extraction of list of genes, genome assembly and 42 RNAseq data sets for species of interest.
- 50.Denlinger DS, Hudson SB, Keweshan NS, Gompert Z, Bernhardt SA: Standing genetic variation in laboratory populations of insecticide-susceptible Phlebotomus papatasi and Lutzomyia longipalpis (Diptera: Psychodidae: Phlebotominae) for the evolution of resistance. Evol Appl 2021, 14:1248–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van Klink R, Bowler DE, Gongalsky KB, Swengel AB, Gentile A,Chase JM: Meta-analysis reveals declines in terrestrial but increases in freshwater insect abundances. Science 2020, 368:417–420 • In this meta-analysis the authors used MapVEu to download long term/ historical population surveillance data to evaluate insect population declines.
- 52. Bellekom B, Hackett TD, Lewis OT: A network perspective on the vectoring of human disease. Trends Parasitol 2021, 37:391–400 • In this review the authors used MapVEu to download long term/historical host blood meal source data to evaluate host vector interactions.