Summary
Human immunodeficiency virus type 1 (HIV-1) vaccination of cows has elicited broadly neutralizing antibodies (bNAbs). In this study, monoclonal antibodies (mAbs) are isolated from a clade A (KNH1144 and BG505) vaccinated cow using a heterologous clade B antigen (AD8). CD4 binding site (CD4bs) bNAb (MEL-1872) is more potent than a majority of CD4bs bNAbs isolated so far. MEL-1872 mAb with CDRH3 of 57 amino acids shows more potency (geometric mean half-maximal inhibitory concentration [IC50]: 0.009 μg/mL; breadth: 66%) than VRC01 against clade B viruses (29-fold) and than CHO1-31 against tested clade A viruses (21-fold). It also shows more breadth and potency than NC-Cow1, the only other reported anti-HIV-1 bovine bNAb, which has 60% breadth with geometric mean IC50 of 0.090 μg/mL in this study. Using successive different stable-structured SOSIP trimers in bovines can elicit bNAbs focusing on epitopes ubiquitous across subtypes. Furthermore, the cross-clade selection strategy also results in ultra-potent bNAbs.
Keywords: HIV-1, envelope glycoprotein, neutralization, cows, CDRH3, antibody, vaccine, antiretroviral
Graphical abstract
Highlights
-
•
Sequential vaccine with different SOSIP trimers could elicit bNAbs
-
•
Cross-clade B-cell-sorting probe could select ultra-potent bNAbs
-
•
Bovine CD4bs monoclonal antibody neutralizes HIV-1 isolates potently
Heydarchi et al. report that cows vaccinated with HIV-1 envelope can induce ultra-potent neutralizing antibodies targeting the key conserved sites on HIV-1 envelope. Isolated anti-HIV-1 bovine neutralizing antibodies are broader and more potent than a majority of anti-HIV-1 human broadly neutralizing antibodies.
Introduction
An effective human immunodeficiency virus type 1 (HIV-1) vaccine is considered the best way to halt the ongoing HIV-1 infections that have caused a major impact on social and economic development. Producing a broadly effective vaccine against HIV-1 has proven difficult, principally due to the HIV-1 high sequence variability and the limitations of the human immunoglobulin repertoire to develop broadly neutralizing antibodies (bNAbs).1 An effective vaccine will optimally present conserved epitopes represented2 on many strains in a manner where B-lymphocyte recognition elicits potent bNAbs against many strains. But to achieve this during natural infection, most bNAbs require a high level of affinity maturation over several years.3,4 Although vaccination of human5 and animals6,7 has yielded no or strain-specific neutralizing antibodies, cows have been able to induce bNAbs in a short period.8, 9, 10 Bovine antibodies naturally have some unique features, such as ultra-long CDRH311 or presence of certain amino acids12 that are similar to those playing roles in neutralization by human bNAbs. bNAbs prevent HIV-1 infection by targeting conserved epitopes on the envelope glycoprotein (Env),13 and a stabilized soluble immunogen must mimic native membrane-bound pre-fusion Env. Although HIV-1 Envs truncated to remove the trans-membrane domain were initially stabilized by mutating the gp120 furin cleavage site,14,15 resulting in uncleaved (Unc) Env gp140, this version had an open conformation and was antigenically distinct from proteolytically cleaved native pre-fusion Env on virions.16,17 The development of a stabilized cleavable version of Env, BG505 SOSIP, the prototype of native-like Env trimer, so far facilitated authentic presentation of the pre-fusion Env epitopes for multiple bNAbs with very few epitopes for non-neutralizing antibodies, thus avoiding immunological disruptions.18
Although BG505 SOSIP Env antigenically mimics HIV-1 Env, initial studies showed limited success in rabbits and non-human primates with induction of mainly autologous neutralizing antibodies with very little heterologous neutralizing activity.6,7 Optimization of BG505 SOSIP and germ-line-targeting priming antigens have resulted in broader neutralizing antibodies against most tier 1, autologous tier 2, and some heterologous tier 2 viruses.19 Despite the small success in eliciting tier 2 neutralizing responses in commonly used animal models, vaccination of cows with SOSIP BG505 Env18 resulted in elicitation of potent bNAbs. Analysis of cow B-cell clones led to isolation of NC-Cow1 bNAb, which was more potent than the majority of human bNAbs isolated so far.10 While NC-Cow1 showed the potential advantages of the bovine immune system in eliciting bNAbs against HIV-1, such advantages are not limited to SOSIP trimers. Indeed, our results showed that vaccinated cows could produce bNAbs in a much shorter time than human or other animals, with breadth achieved when cows were exposed to only non-native HIV-1 AD8 Unc gp140 HIV-1 Env vaccine without needing sequential immunogen modifications to cultivate an important germline lineage.8, 9, 10 These immunoglobulin Gs (IgGs) were able to neutralize diverse HIV-1 viruses and inhibit Env binding of anti-CD4 binding site human bNAbs8,9 and V3-loop-binding human bNAb.
Currently, there is a focus on therapeutic antibodies to provide immediate passive immunity to prevent or treat HIV-1 disease. Although there are numerous approved drugs against HIV-1 infection,20 passive antibody prophylaxis and immunotherapy could hold a valuable place in both prevention and treatment due to the prospect of high stability and long bio-availability of Fc-engineered monoclonal antibodies (mAbs). Passive transfer of some human bNAbs demonstrated both pre-exposure prophylaxis and treatment in macaque,21 humanized mouse models,10,11 and human.22 The result from antibody-mediated therapy to evaluate the safety, tolerability, and efficacy of the VRC01 antibody in preventing HIV-1 infection among men, transgender persons, and at-risk women showed proof of concept23 while emphasizing the need to develop bNAb combinations to achieve optimal potency and breadth.
Surprisingly, treatment with bNAbs could also reduce cell-associated HIV-1 DNA and aid the development of HIV-1-specific T cell immunity,25,26 which suggests a vaccine-like effect of bNAb immunotherapy.27 Thus, in this study, we aimed to study anti-HIV-1 bovine bNAbs as an advanced approach to design HIV-1 therapeutic and prophylactic agents and to discover potential neutralizing sites on HIV-1 Env to aid rational vaccine design. Here, we immunized cows with different forms and strains of HIV-1 Env and showed that a well-ordered trimer (SOSIP gp140) elicited bovine antibodies and, using a cross-clade selection strategy, could successfully select broadly active and ultra-potent monoclonal bNAbs with ultralong CDRH3 regions.
Results
Serum binding and neutralization
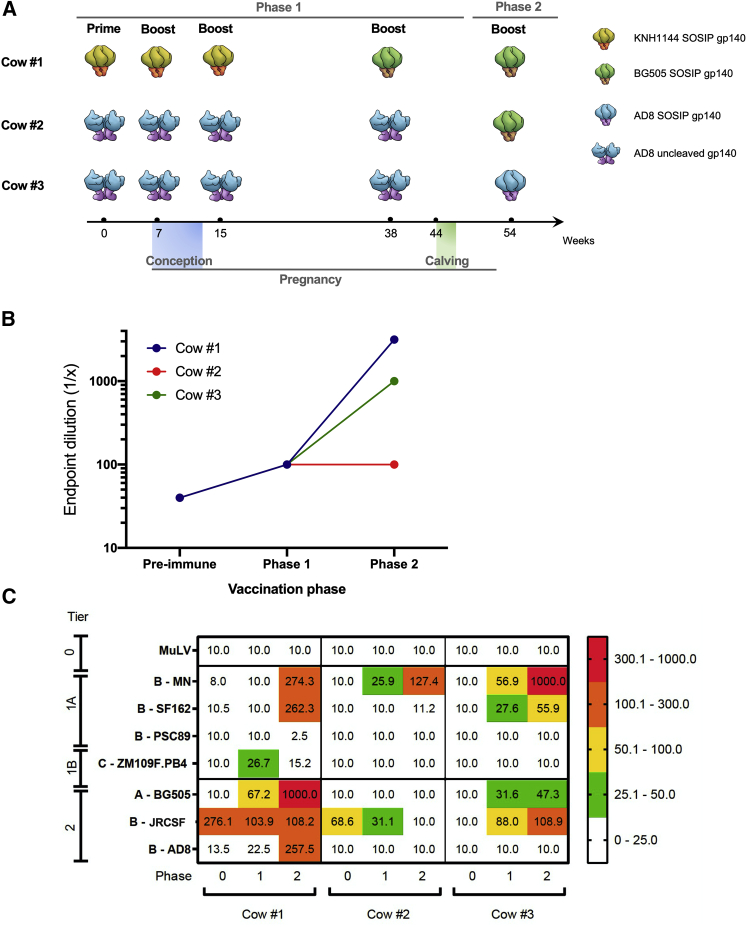
Female Holstein cattle were vaccinated prior to, during, and after pregnancy with different HIV Env proteins. To evaluate the antibody responses against HIV-1 vaccines, sera from different time points were collected and binding of bovine IgGs to Env immunogen was measured by direct ELISA (Figure 1B). Pre-immune sera presented low binding, while all samples at phase 1 presented endpoint titers of 100. At phase 2, re-vaccination with BG505 SOSIP in cow no. 1 increased binding to above 3,000, while changing the immunogen in cow no. 3 (from AD8 Unc gp140 on phase 1 to AD8 SOSIP on phase 2) showed a moderate increase in antibody titer (1,000). Cow no. 2, on the other hand, did not show an improved antibody titer despite changing the immunogen from AD8 Unc gp140 at phase 1 to BG505 SOSIP at phase 2. Neutralizing activity of sera from vaccinated animals was also investigated in a neutralization assay against a panel of seven pseudoviruses, including autologous Envs (Figure 1C). Cows no. 1 and no. 2 showed neutralization against JRCSF in pre-immunization phase, while neutralization decreased post-immunization. Neutralization against other tested viruses was induced only after immunization. Cow no. 1 showed the highest neutralizing activity against two pseudoviruses at phase 1 (ZM109F.PB4 and BG505 with median infectious dose [ID50] values of 26.7 and 67.2, respectively), and both potency and breadth increased at phase 2 with neutralizing activity against pseudoviruses MN, SF162, BG505, and AD8 (ID50 values of 274.3, 262.3, 1,000, and 257.5, respectively).
Figure 1.
HIV Env binding and neutralization of serum samples collected from immunized cows
(A) Vaccination of cows during pregnancy. Cow no. 1 was vaccinated with KNH1144 SOSIP gp140 and BG505 gp140, while cows no. 2 and no. 3 were vaccinated with AD8 uncleaved gp140 followed by BG505 uncleaved gp140 and AD8 SOSIP gp140, respectively.
(B) Env binding of bovine IgGs in sera of vaccinated cows. Binding was measured against Env vaccine (cow no. 1: BG505 SOSIP; cow no. 2 and cow no. 3: AD8 SOSIP) through direct ELISA.
(C) Neutralization assays were performed against seven pseudoviruses from clades A, B, and C and tiers 1A, 1B, and 2; as negative control, MuLV pseudovirus was used. The values show ID50. Heatmap scale shows no neutralization from a value of ID50 = 10 (white values) to the highest neutralization achieved at ID50 = 1,000 (red values). ELISA assays were performed in duplicate with two independent biological replicates.
Although both cows no. 2 and no. 3 vaccinated with AD8 Unc gp140 were unable to induce an autologous neutralization, they induced neutralizing antibodies against multiple pseudoviruses. On phase 1, cow no. 2 induced neutralization against MN (ID50 of 25.9), and this increased in phase 2 for MN (ID50 of 127.4). Cow no. 3 showed neutralization against MN, SF162, BG505, and JRCSF at phase 1 (ID50s of 56.9, 27.6, 31.6, and 88, respectively), and the neutralizing activity enhanced against all mentioned viruses at phase no. 2 (ID50 values of 1,000, 55.9, 47.3, and 108.9, respectively).
HIV-1-specific bovine monoclonal antibodies
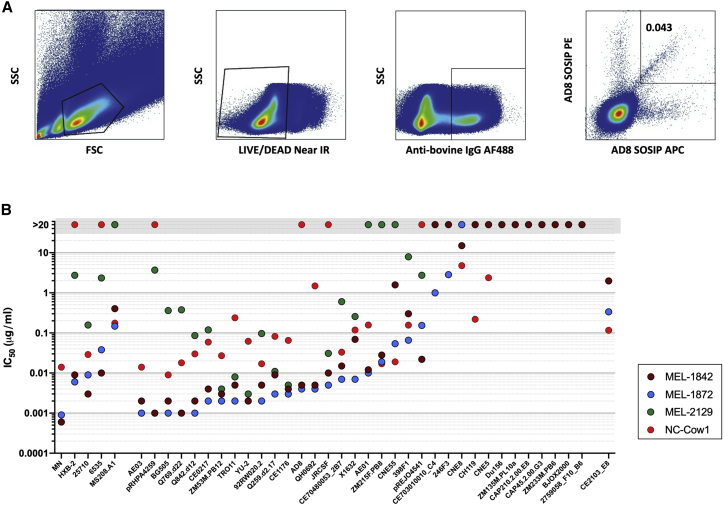
To isolate bovine B cells producing bNAbs, we developed a stabilized cleavable version of AD8 Env, AD8 SOSIP, to present the pre-fusion Env epitopes for multiple bNAbs with very few epitopes for non-neutralizing antibodies. The produced AD8 SOSIP was affinity purified using the 2G12 antibody followed by size exclusion chromatography (SEC) on a Superdex S200 16/600 column and characterized in reducing and non-reducing SDS gel, blue native (BN)-PAGE analysis, and capture ELISA (Figures S1C and S1D). Characterization of AD8 SOSIP confirmed that the proteins were predominantly trimeric and exposed epitopes of human bNAbs. Negative-stain electron microscopy (Figures S1G and S1H) also confirmed the trimeric structure, which appeared similar to BG505 SOSIP and closely resembled native Env spikes. AD8 SOSIP gp140 v4.1 binding to human bNAbs was confirmed in capture ELISA. HIV-1 Env-specific single B cells were sorted by fluorescence-activated cell sorting (FACS) from peripheral blood mononuclear cells (PBMCs) of cows no. 1, no. 2, and no. 3 using AD8 SOSIP. AD8 strain (clade B) is a tier 2 HIV-1 clade B virus, and we used this Env as bait to isolate B cells from AD8-vaccinated animals (cows no. 2 and no. 3). In addition, we used this bait to isolate B cells producing cross-clade anti-HIV-1 antibodies from the animal vaccinated with clade A Env (cow no. 1). HIV-1 Env-specific single B cells (IgG+ and AD8 SOSIP+) were sorted from PBMCs of vaccinated cows (Figure 2A), and after antibody variable gene amplification and further cloning of such genes into human antibody constant region expression vector, 47 chimeric mAbs were successfully produced from cow no. 1, from which 27 showed binding to AD8 SOSIP, four of which isolated from this cow showed neutralization against AD8 pseudovirus. Out of 87 chimeric mAbs constructed from cow no. 2, 46 mAbs could bind to AD8 SOSIP gp140 v4.1 trimer in capture ELISA (Figure S2). However, none of these mAbs from cow no. 2 neutralized autologous HIV-1 AD8 pseudovirus. From cow no. 3, 60 chimeric mAbs were produced, and although only 19 mAbs showed binding to SOSIP AD8, two of these antibodies showed autologous neutralization against AD8 pseudovirus in TZM-bl neutralization assay. The mAbs isolated from cow no. 1, MEL-1842 and MEL-1872, showed higher Env binding against AD8 SOSIP than VRC01 antibody. It is worth mentioning that, since the variable light gene was not rescued from MEL-1842 and MEL-1872 mAbs, these antibodies were produced using the light chain of MEL-2129 mAb. Three mAbs (MEL-1842, MEL-1872, and MEL-2129) belonged to the same antibody clonal family with ultralong CDRH3 length of 58 amino acids for MEL-2129 mAb and 57 amino acids for MEL-1842 and MEL-1872 mAbs. Sequences of the isolated antibody variable genes are deposited in GenBank and listed in Figure S3 and Table S3.
Figure 2.
Isolation of bNAbs from HIV-1 vaccinated cows
(A) Cow PBMCs were sorted for IgG+ cells that bound to biotinylated AD8 SOSIP-AviTag conjugated to phycoerythrin (PE) and antigen-presenting cell (APC) fluorophores. FSC, forward scatter; SSC, side scatter.
(B) Bovine bNAbs showed potent cross-clade neutralization against tier 1 and tier 2 viruses with low geometric mean IC50. Neutralization assays were performed in duplicates with two independent biological replicates.
Anti-HIV-1 bovine MEL-1872 mAb is extraordinarily potent
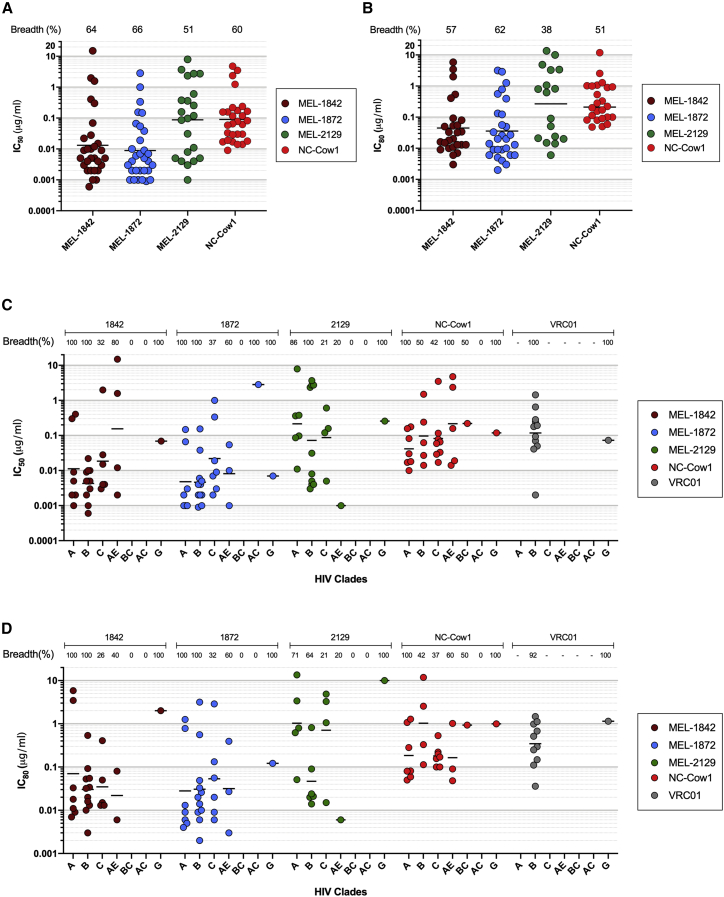
To evaluate the neutralization properties of AD8-SOSIP-binding antibodies, TZM-bl neutralization was performed using a virus panel including HIV-1 12-virus global panel and several heterologous HIV-1 viruses. Anti-HIV-1 mAbs isolated from cow no. 2 showed a narrow breadth, but mAbs of cow no. 3 showed a moderate breadth against clade B viruses (Figure S4; Tables S5 and S6). Most of the isolated bNAbs from all three cows neutralized <50% of HIV-1 viruses with geometric mean IC50 of above 0.09 μg/mL (Figure S5A), while three mAbs of MEL-1842, MEL-1872, and MEL-2129 isolated from cow no. 1 showed the highest breadth (64%, 66%, and 51%, respectively). Among them, MEL-1842 and MEL-1872 mAbs demonstrated the greatest potency with geometric mean IC50 of 0.013 μg/mL and 0.009 μg/mL and IC80 of 0.045 μg/mL and 0.033 μg/mL, respectively (Tables S5 and S6). Among mAbs isolated from cow no. 1, there was a correlation between low geometric mean IC50 and high breadth (Figure S5B). However, for AD8 neutralizing mAbs, there was a lack of correlation between IC50 and half-maximal effective concentration (EC50) for this strain (Figure S5C). Antibody MEL-1842, MEL-1872, and MEL-2129 all showed cross-clade neutralization against tier 1 and tier 2 viruses, with MEL-1872 neutralizing most of tier 2 viruses with IC50 value of 0.1 μg/mL (Figure 2B).
Antibodies MEL-1842 and MEL-1872 demonstrated broader and more potent HIV-1 neutralizing activity compared with NC-Cow1 (60% breadth with IC50 of 0.09 μg/mL) against tested viruses (Figures 3A and 3B). MEL-1842 and MEL-1872 showed the greatest potency against clade B viruses with geometric mean IC50 of 0.004 μg/mL followed by clade A viruses with geometric mean IC50 of 0.011 μg/mL and 0.005 μg/mL, respectively (Figures 3C and 3D). Antibody MEL-1872 was 29-fold more potent than VRC01 (geometric mean IC50 of 0.117 μg/mL) against tested clade B viruses (Figure S4) and 21-fold more potent than CHO1-31 (mixture of two CHO-1 [PG9-like] and CHO-31 [VRC01-like] bnAbs) against tested clade A viruses (geometric mean IC50 of 0.042 μg/mL).
Figure 3.
Neutralization profile of bNAb MEL-1842, MEL-1872, and MEL-2129
Comparison of IC50 (A) and IC80 (B) values of isolated mAbs with bNAb NC-COW1. Categorization and comparison of neutralization activity against different HIV clades using IC50 (C) and IC80 (D) values is shown. The black lines represent the geometric mean IC50 and IC80. Neutralization assays were performed in duplicates with two independent biological replicates.
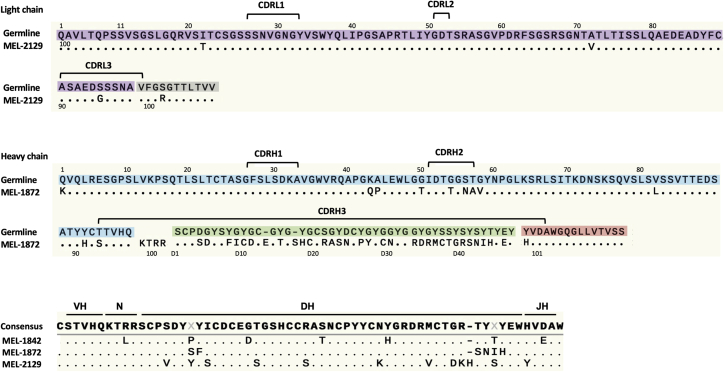
The alignment of germline genes with the heavy-chain and light-chain gene of MEL-1872 mAbs is shown in Figure 4. Sequences were then annotated with IMGT High V-Quest (http://www.imgt.org/HighV-QUEST/; Table S4). Conventional Kabat numbering was used for antibody light-chain and for heavy-chain amino acids (H1-100b and H101), but CDRH3 residues encoded by the IGHD8-2 gene segment (between H100b and H101 and its connection to the J chain) were numbered using a “D” identifier sequentially.28 IMGT was able to annotate both VH and JH germline genes for the bovine mAbs while failing to assign the germline genes for ultralong CDRH3. IgDiscover29 and AbMining Toolbox30 were used for identification of DH germline genes. As shown, there are limited hypersomatic mutations in the light gene and VH and JH of the heavy gene while, compared with germline DH (IGHD8-2∗01), there are significant mutations in CDRH3 region of MEL-1872 mAb.
Figure 4.
Amino acid alignment of heavy- and light-chain sequences of MEL-1872 mAb with the germline genes
Purple: IGLV1-47∗01 germline gene; gray: IGLJ4∗01 germline gene; blue: IGHV1-7∗02 germline gene; green: IGHD8-2∗01 germline gene; red: IGHJ2-4∗01 germline gene.
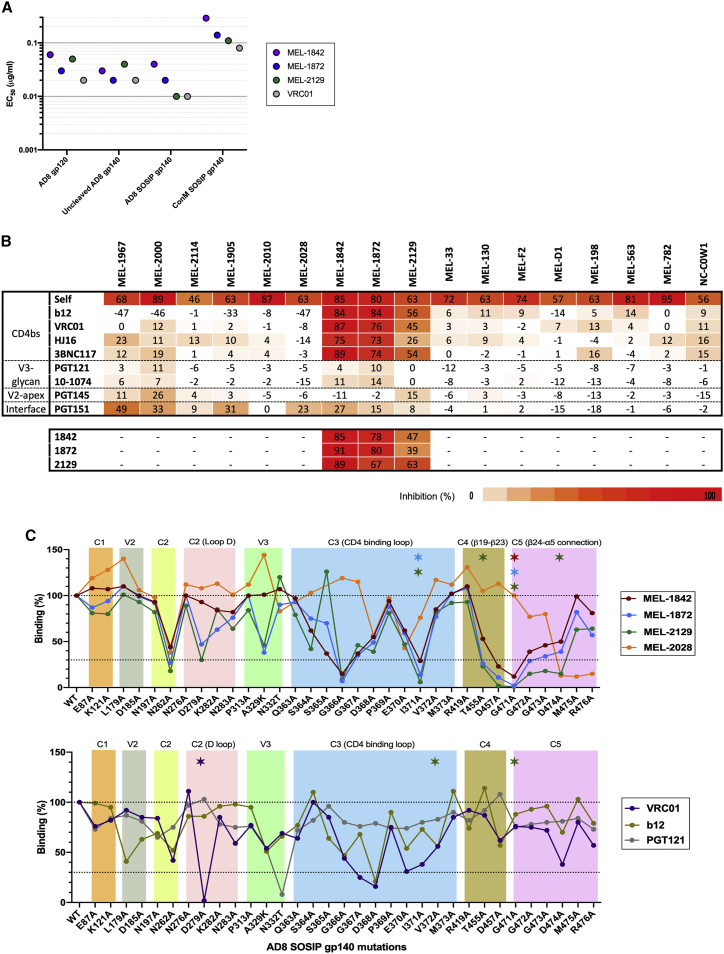
Bovine bNAbs bind to non-trimer-specific epitopes on HIV-1 Env
As shown in Figure 5A, bovine bNAbs displayed binding to monomeric AD8 gp120, Unc HIV-1 gp140, and AD8 SOSIP gp140 v4.1, confirming that their epitopes were present on gp120 monomers of both cleaved and Unc Env trimers. These mAbs could also bind to ConM SOSIP, which is an Env trimer based on a consensus sequence of all HIV-1 group M isolates. This trimer displays most bNAb epitopes and is made to minimize clade-specific and strain-specific antigenic determinants.31
Figure 5.
Epitope mapping of bovine monoclonal antibodies
(A) Binding of bovine bNAbs to HIV Envs. Bovine bNAbs were tested in direct ELISA assays to evaluate their binding to different forms of Env (monomeric gp120, uncleaved gp140, and SOSIP gp140) as well as ConM SOSIP, which is an Env trimer based on a consensus sequence of all HIV-1 group M isolates.
(B) Competition of bovine bNAbs with human bNAb for Env binding. The table shows the competition ELISA assay with values demonstrating Env binding inhibition (percentage) of human bNAbs by bovine antibodies. Competition ELISA between antibodies MEL-1842, MEL-1872, and MEL-2129 showed these antibodies bind to the same or proximate epitopes. Higher competition is shown in red, and lower inhibition values are in increasingly pale shades of orange. ELISA assays were performed in duplicates with two independent biological replicates.
(C) The effect of alanine mutagenesis on binding of bovine antibodies to AD8 gp120 captured from lysed virions. ELISA assay was performed using a constant half-maximal effective concentration (EC50) of each antibody to AD8 WT Env. Stars also show significant neutralization IC50 increase of each antibody against mutated virus compared with WT virus (5-fold for all mAbs, except 198). PGT121 (V3-glycan epitope) and b12 and VRC01 (CD4bs epitope) were included for comparison. ELISA and neutralization assays were performed in duplicate with two independent biological replicates.
Bovine bNAbs bind to CD4bs on HIV-1 Env
A competition ELISA with reference human bNAbs targeting four known epitopes was performed to evaluate interference with bovine mAbs binding to AD8 SOSIP. Bovine mAbs 2129, 1842, and 1872 inhibited Env binding of human CD4bs bNAbs b12, VRC01, HJ16, and 3BNC117, except for 2129, which showed incomplete inhibition of HJ16 binding (Figure 5B). Other bovine mAbs demonstrated partial competition (25%–50%) with V2-apex human bNAbs (PGT145) and gp120-gp41 interface human bNAbs (PGT151). There was also strong competition between MEL-1842 and MEL-1872 mAbs, demonstrating that these mAbs may share a common or proximate binding site(s). On the other hand, MEL-2129 mAb showed lower inhibitory effect on Env binding of MEL-1842 and MEL-1872 mAbs.
Epitope mapping of bovine bNAbs with HIV-1 Env mutants
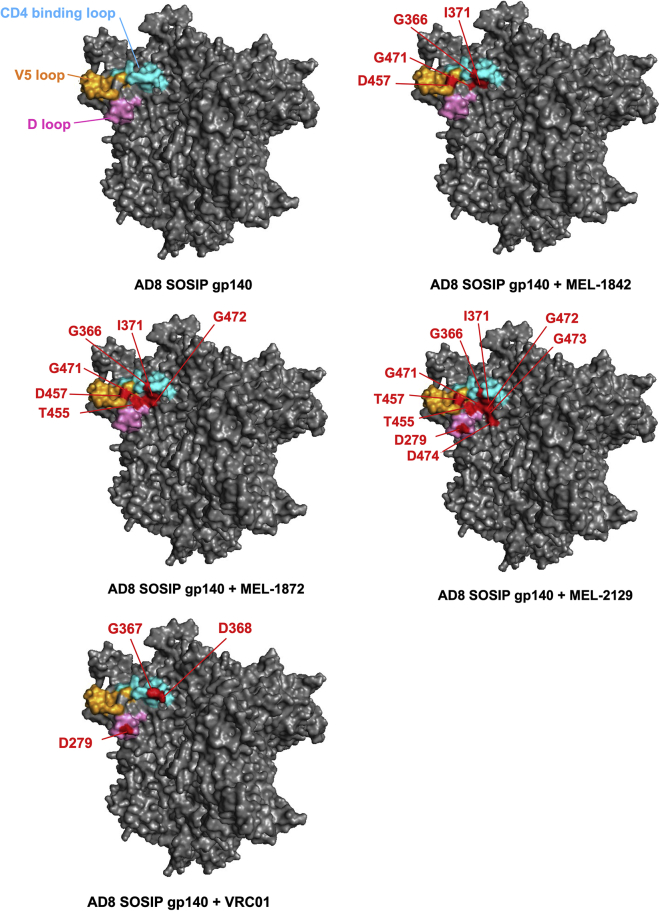
Affinity binding of bovine bNAbs to a panel of 33 AD8 Env mutants showed that mutations located primarily in the CD4bs, C4, and C5 regions of Env impeded the binding of these mAbs (Figures 5C and S6; Table S7). Using Swiss Modeller,24 the binding site of bovine bNAbs is shown in Figure 6. For MEL-1842, MEL-1872, and MEL-2129 mAbs (which are from the same clonal lineage antibody family), the most significant loss of binding was observed for mutations G366A, I371A, D457A, and G471A, which resulted in ≤30% Env binding compared with wild-type (WT) Env. In previous studies, alanine mutation in G366, I371, and D457 was shown to prevent binding of human CD4bs bNAbs, such as N6, VRC01, VRC-PG04, and VRC27 to JRCSF gp120.32 The mutations N262A, T455A, and G472A also inhibited the binding of MEL-1872 and MEL-2129 mAbs. Mutation in N262 was shown to inhibit binding of CD4bs bNAbs 3BNC117, VRC01, VRC-PG04, and VRC027, while substitution of G472 with alanine only hindered binding of VRC27.32 In addition, mutations D279A, G473A, and D474A resulted in a substantial loss of binding for MEL-2129 (all ≤30% Env binding compared with WT AD8 Env). VRC01 and b12 were used as controls and exhibited the lowest binding percentage to mutations introduced in the CD4bs, as expected.32 VRC01 showed a significant decrease in Env binding in response to mutations to the N-linked glycan at position 279 and the residues G367A and G368A (≤30% binding). b12 also demonstrated a similarly low decrease in binding to G368A (≤30% binding). For PGT121, the mutation of the glycan at position 332 resulted in the most substantial decrease in binding.
Figure 6.
Binding of bovine bNAbs to CD4bs
Highlighted on the BG505 SOSIP.664 trimer (top left) are residues from the D loop (275–283: SNFTDNAKN, pale pink), CD4 binding loop (362–375: NQSSGGDPEIVMHS, cyan), and V5 loop (458–469: GGNNHNNDTETFR, pale orange). Binding site of antibodies MEL-1842, MEL-1872, MEL-2129, and VRC01 to AD8 SOSIP V4.1 are shown with red color, indicating the residues that binding of bovine antibodies to them was reduced through mutagenesis. Homology model of AD8 SOSIP was obtained using Swiss Modeller.
Neutralization activity of bovine bNAbs was also assessed in a TZM-bl neutralization assay using a panel of 27 mutated AD8 pseudoviruses. As shown in Figure 5C, the G471A mutation resulted in the most significant effect on neutralization of MEL-1842, MEL-1872, and MEL-2129 mAbs and increased IC50 and IC80 values to ≥4 μg/mL. I371A mutation also obstructed the neutralization activity of MEL-2129 and MEL-1872 mAbs effectively, while MEL-1842 mAb was insensitive to this mutation. G471 and I371 are also epitope residues for most of human CD4bs bNAbs, including HJ16, 8ANC131, CH103, and b12,33 demonstrating the overlap between epitopes of bovine and human CD4bs bNAbs. In agreement with ELISA data and in addition to the mentioned mutations, mutations in C4 (T455A) and C5 (D474A) showed neutralization impediment for bovine MEL-2129 mAb. As expected, D279A mutation rendered VRC01 unable to neutralize AD8 mutated pseudovirus. In addition to G471A mutation, G366A and V372A mutations also inhibited neutralization activity of b12. PGT121 was affected by E370A and N332T mutations.
A neutralization assay with multi-clade panel of viruses and their corresponding CD4bs mapping mutations (N279A, N280D, and G458Y) showed evidence of CD4bs specificity for MEL-1842, MEL-1872, and MEL-2129 with at least three of the viruses (Table S8).
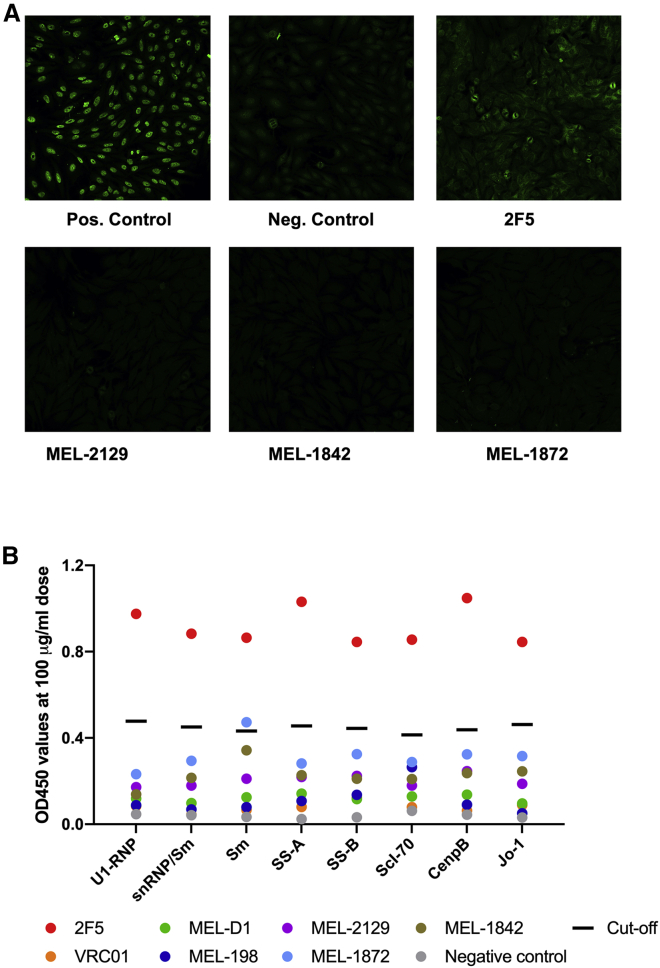
Bovine bNAbs are not polyreactive
Autoreactivity and polyreactivity of MEL-2129, MEL-1842, and MEL-1872 mAbs were evaluated in HEp-2 staining and ELISA assay against several human autoantigens. These bovine bNAbs showed no autoreactivity or polyreactivity against tested antigens (Figures 7A and 7B).
Figure 7.
Assessment of bovine bNAb polyreactivity
(A) Polyreactivity test in Hep-2 cells. Bovine bNAbs MEL-1842, MEL-1872, MEL-2129, and MEL-198 did not show any polyreactivity against human Hep-2 cells. 2F5 is human anti-HIV bNAb with polyreactivity, while anti-HIV bNAb PGT121 is not polyreactive.
(B) Assessment of antibody polyreactivity against human antigens. ELISA assay was performed against human antigens using constant amount of 100 μg/mL from tested mAbs. Black line indicates cutoff values as indicated by the manufacturer. Each assay was repeated twice. snRNP, small nuclear ribonucleoprotein particle.
Discussion
Despite decades of research, vaccination using HIV-1 Env formulations in humans and animal models have mostly yielded no or merely autologous, narrow-focused, neutralizing antibodies. In contrast, bovine antibodies naturally have some unique features, such as ultralong CDRH3 regions11 or the presence of key amino acids12 that are similar to those often observed in human bNAbs and promote development of bNAbs to HIV-1 Env.8, 9, 10 Previous studies have shown that the bovine immune system does not require a long period of affinity maturation to induce bNAbs.9,10 In this study, we vaccinated pregnant cows with different HIV-1 Env antigens, which induced bNAbs following vaccination. The pre-immune samples of bovines with no prior experience of HIV vaccines showed a low binding activity, despite no exposure to HIV antigen before the vaccination. We previously showed8,9 that the genetic property of ultralong CDRH3 immunoglobulin domains close to the unmuted germ line in cows was mediating a low-level background of anti-HIV activity in unvaccinated cows.
In agreement with previous studies,10 BG505 SOSIP vaccine was able to induce bNAbs. Although the serum samples showed low potency due to the transfer of bovine antibodies from serum to colostrum milk during pregnancy, we were able to isolate potent cross-clade bNAbs from this cow. While cell-mediated alloantigen rejection is suppressed at the maternal-fetal interface, further investigation is required to reveal whether these immunomodulatory effects impact on the attainment of anti-HIV bNAbs here after immunization of cows during pregnancy. In addition, different cell lines were used to produce HIV Env antigens in this study, and despite reports that glycosylation patterns of recombinant gp120 monomer from four HIV-1 clades produced in different cell lines revealed similar glycosylation profiles, we cannot exclude these small differences’ impact on binding of antibodies targeting key immunological epitopes on gp120 monomer and on protein immunogenicity.34
Neutralizing mAbs to BG505 SOSIP have been reported in guinea pigs,35 rabbits,33 cows,8, 9, 10 and rhesus macaques.36 In BG505-SOSIP.664-immunized rabbits eliciting autologous neutralizing antibodies, the dominant epitope was the 241/289-glycan hole in approximately 50% of the animals, while the C3/465 epitope was frequent in 25% of sera.7 On the other hand, the 241/289-glycan hole was targeted only in a minority of BG505-SOSIP.664-vaccinated rhesus macaques7,19 and the dominant potent autologous neutralizing epitopes were V1/V3 and C3/V5 epitopes.37 In this study, we evaluated binding and neutralizing activity of several mAbs to a panel of alanine mutants for HIV-1 AD8 isolate. The MEL-1872 mAb showed binding to CD4bs, and like CD4 and other CD4bs antibodies,38 mutagenesis of conserved gp120 residues (366–368 from the CD4 binding loop and 473 from the β24-α5 region) reduced Env binding of this antibody. The mutations I371A, T455A, D457A, G471A, and G472A were also found to be of importance for resistance towards MEL-1872 mAb in AD8 strain. All mutated positions are part of, or in close vicinity to, the CD4bs, and most are highly conserved.
Among reported anti-HIV-1 neutralizing antibodies, CAP256-VRC26.25 (targeting V1V2 apex) has been the most potent antibody with geometric mean IC50 of 0.012 μg/mL (median IC50 = 0.006 μg/mL; breadth 59%) thus far.39 The CDRH3 of CAP256-VRC26.25 comprises 36 amino acids and is one of the longest human CDRH3s identified.39 As shown in previous studies,39,40 there is a correlation between neutralization potency and CDRH3 length. CAP256-VRC26.25 has a protruding CDRH3 comprising a two-stranded antiparallel β sheet that is stabilized with a disulfide bond. Although CAP256-VRC26.25 has approximately 70% breadth on clade C viruses,41, 42, 43 it shows limited breadth (<30%) on clade B viruses39 due to the relative rarity of acidic and basic residues recognized by the antibody at positions 164 and 169, respectively.39,41 NC-Cow1 is also a potent anti-HIV-1 bovine CD4bs bNAb with a protruding CDRH3 of 60 amino acids and broad neutralization against clade A viruses but moderate neutralization against clade B and C viruses.10 The bovine antibody isolated in this study (MEL-1872) showed higher potency with geometric mean IC50 of 0.009 μg/mL (median IC50 = 0.006 μg/mL) and breadth (66%) than CAP256-VRC26.25. In addition, it showed more breadth and potency compared with NC-Cow1 against tested viruses (66% versus 60% breadth; geometric mean IC50 of 0.009 μg/mL versus 0.090 μg/0 mL). Although MEL-1872 and NC-Cow1 mAbs showed similar breadth against clade A and C viruses, the former antibody showed more potency (8-fold and 4-fold, respectively). MEL-1872 mAb also neutralized clade B viruses with more potency (above 23-fold) and breadth (100% versus 50%) than NC-Cow1. Using BG505 SOSIP as a sorting bait in a BG505-SOSIP-vaccinated cow resulted in isolation of one bnAb (NC-Cow1) out of seven strong Env-binding bnAbs (14%),10 while use of AD8 SOSIP as the sorting bait in this study resulted in isolation of three bnAbs out of nine strong Env-binding bnAbs (33%) from cow no. 1, which was vaccinated with BG505 SOSIP. Thus, although like NC-Cow1, MEL-1872 mAb was isolated from a BG505 SOSIP (clade A)-vaccinated cow, the usage of a tier-2 HIV-1 Env with different clade (AD8 SOSIP; clade B) for B cell sorting might be the explanation of why MEL-1872 neutralization was broader and more potent against tested viruses than NC-Cow1 and other CD4bs antibodies that were isolated using Env-binding-based, HIV-1-specific B cell selection.56,44 The approach of using mis-matched antigens has been used with other viral glycoproteins to isolate broad and potent antibodies. Sauer et al.45 immunized mice and screened antigen-specific B cells using multiple variants of coronavirus Spike glycoproteins, which resulted in isolation of a broad coronavirus bNAb with high affinity.
mAbs isolated from patient or vaccinated animals may guide vaccine design in an approach known as reverse vaccinology.34,46 This has become a productive avenue for HIV-1 vaccine research.46 A high-resolution structure of mAb-antigen complexes can facilitate immunogen design by disclosing the important details of epitopes.47 A compelling HIV vaccine is required to induce bNAbs against conserved epitopes,2 and many bNAbs target the CD4bs as a conserved region on HV Env.56,48 Immunization of guinea pigs with epitope-focused antigenic domain of the CD4bs VRC01 antibody, called EAD-VRC01, induced CD4bs neutralizing antibodies against subtype B viruses.49 Structure of MEL-1872 in complex with HIV Env would help reveal its mechanism of potent neutralization, and this could be useful in design of epitope vaccines.
Although there are numerous approved drugs against HIV-1 infection, they are limited to wealthy nations, and lifelong treatment requires diligent adherence and can be associated with some toxicity and economic cost.50 Long-term use of anti-retroviral drugs could also introduce drug-resistant escape mutants.51 Passive antibody prophylaxis and immunotherapy could hold a valuable place in both the prevention and treatment of HIV-1 infection. Disulfide bonds in bovine CDRH3 results in a rigid structure that might survive the acidic environment of mucosal environment better than human bNAbs.10 This rigid structure in bovine CDRH3 provides an excellent opportunity to design small-molecule drug inhibitors accessing deep recessed epitopes on HIV-1 Env more efficiently than human bNAbs. Bovine bNAbs with broad diagnostic, therapeutic, and prophylactic potential can be used not only against HIV-1 but also to control or prevent other diseases. There is evidence showing the effectiveness of bovine IgG isolated from colostrum to improve the management of patients with HIV-1-associated diarrhea,52 to boost the immune response in HIV-1-positive children,53 to neutralize respiratory syncytial virus (RSV) in vitro,54 or to prevent influenza infection in healthy volunteers.55 In a clinical trial, polyclonal IgG antibody produced from Middle East respiratory syndrome (MERS)-immunized cows was safe and well tolerated in healthy participants (up to 50 mg/kg). With the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), there has been an urgent need for therapeutic and prophylactic treatments. In this regard, vaccination of cows against SARS-CoV-2 spike protein can induce bNAbs against recessed epitopes, and since bovine IgG-derived colostrum has been reported to be resistant to proteolysis, consumption of SARS-CoV-2 hyperimmune colostrum or immune milk could offer short-term protection to individuals until effective and safe vaccines are more widely available worldwide.
Studying bovine antibody responses in HIV-1-vaccinated cows will be very valuable, as the induced bNAbs may introduce new neutralizing epitopes and/or sites on Env protein that are not easily accessible to human antibodies. Anti-HIV-1 bovine antibodies, particularly those that share similarities to human bNAbs in terms of target epitopes and antibody molecular features, will provide extremely important research output in advancing rational design of HIV-1 vaccines and antibody-based therapeutics and prophylactics.
Limitation of the study
Different cell lines were used to produce HIV Env antigens in this study, and despite reports that glycosylation patterns of recombinant gp120 monomer from four HIV-1 clades produced in different cell lines revealed similar glycosylation profiles, we cannot exclude these small differences’ impact on binding of bovine polyclonal antibodies targeting key immunological epitopes on gp120 monomer and on protein immunogenicity. While using cross-clade, B-cell-sorting probe resulted in isolation of potent bNAb, one limitation of the study is the small number of animals used. Vaccination of animals with HIV-1 Env and B cell sorting with different cross-clade antigen probes in future can lead to robust statistical analyses of the results. In addition, although MEL-1872 showed robust HIV-1 neutralization, the efficacy of protection in vivo needs to be further studied.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PGT121 | AIDS Reagent Program, Division of AIDS, NIAID, NIH. | Cat#ARP-12343; RRID:AB_2491041 |
| VRC01 | AIDS Reagent Program, Division of AIDS, NIAID, NIH. | Cat#ARP-12033; RRID:AB_249101 |
| b12 | AIDS Reagent Program, Division of AIDS, NIAID, NIH. | Cat#ARP-2640 |
| 2G12 | AIDS Reagent Program, Division of AIDS, NIAID, NIH. | Cat#ARP-1476 |
| CHO1-31 | Rosenberg et al., 2015 | N/A |
| PGMD1400 | Sok et al., 2014 | N/A |
| 3BNC117 | AIDS Reagent Program, Division of AIDS, NIAID, NIH. | Cat#ARP12474, RRID:AB_2491033 |
| HJ16 | AIDS Reagent Program, Division of AIDS, NIAID, NIH. | Cat#ARP12138, RRID:AB_2491032 |
| 10-1074 | AIDS Reagent Program, Division of AIDS, NIAID, NIH. | Cat#12477 , RRID:AB_2491062 |
| PGT145 | AIDS Reagent Program, Division of AIDS, NIAID, NIH. | Cat# ARP12703, RRID:AB_2491054 |
| PGT151 | Falkowska ET AL., 2014 | N/A |
| NC-Cow1 | Sok et al., 2017 | GenBank (MF167446.1 and MF167436.1) |
| HIVIG | AIDS Reagent Program, Division of AIDS, NIAID, NIH. | Cat#ARP-3957; RRID:AB_2890264 |
| Mouse Anti-Bovine IgG Monoclonal Antibody, Unconjugated, Clone BG-18 | Sigma-Aldrich | Cat# B6901, RRID:AB_258594 |
| D7324 Sheep anti-gp120 | Aalto Bio Reagents | Cat# D7324 |
| Goat anti-human IgG HRP | KPL | Cat# 474-1002 |
| 6X His tag® antibody | Abcam | Ab9108 |
| Sheep anti-bovine IgG-HRP | BioRad | Bio-Rad Cat# AAI23P, RRID:AB_323063 |
| Biological samples | ||
| Bovine PBMC samples | This paper | N/A |
| Bovine serum | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Monomeric AD8 gp120 | This paper | N/A |
| Uncleaved AD8 gp140 | This paper | N/A |
| AD8 SOSIP gp140. V4.1 | This paper | N/A |
| ConM SOSIP gp140 | This paper | N/A |
| AD8 6R SOSIP.664 | This paper | N/A |
| D7324 tagged- AD8 SOSIP gp140. V4.1 | This paper | N/A |
| Biotinylated AD8 SOSIP gp140. V4.1 | This paper | N/A |
| 6 His tagged- AD8 SOSIP gp140. V4.1 | This paper | N/A |
| Biotinylated MONOMERIC AD8 gp120 | This paper | N/A |
| KNH1144 SOSIP.v1 | Kang et al., 2009 | N/A |
| BG505 SOSIP.664 | Sanders et al., 2013 | N/A |
| BG505 Unc. SEKS | Ring et al., 2013 | N/A |
| Critical commercial assays | ||
| BirA Biotin-Protein Ligase Kit | Avidity | BirA-500 |
| Britelite plus Reporter Gene Assay System | PerkinElmer | Cat# 6066761 |
| RQ1 RNase-Free DNase | Promega | Cat# M6101 |
| LIVE/DEAD™ Fixable Aqua Dead Cell Stain | Thermofisher | Cat# L34965 |
| Streptavidin-APC | Life technologies | Cat# S868 |
| Streptavidin-PE | Life technologies | Cat# S866 |
| RNasin Plus Ribonuclease Inhibitor | Promega | Cat #N2615 |
| SuperScript™ III First-Strand Synthesis System | Thermofisher | Cat# 18080051 |
| MyTaq HS Red Mix | Bioline | Cat# BIO-25048 |
| EcoRI-HF Restriction enzyme | New England Biolabs | Cat# R3101L |
| NheI-HF Restriction enzyme | New England Biolabs | Cat# R3131LL |
| AvrII Restriction enzyme | New England Biolabs | Cat# R0174L |
| ExpiFectamine™ 293 Transfection Kit | Thermofisher | Cat# A14524 |
| Protein G Agarose Fast Flow | Millipore | Cat# A16266 |
| Phusion® High-Fidelity DNA Polymerase | New England Biolabs | Cat# M0530S |
| EZ-Link™ Sulfo-NHS-LC-Biotinylation Kit | Thermofisher | Cat# 21435 |
| SureBlue™ TMB 1-Component Microwell Peroxidase Substrate | KPL | Cat# 5120-0075 |
| ANA HEp-2 Standard Kit | Aesku Diagnostics | Cat # 51.101.US |
| AESKULISA ANA-8Pro | Aesku Diagnostics | Cat# 3101-AES |
| Montanide ISA206 | Seppic | N/A |
| Ficol paque™ plus | GE Healthcare | Cat# Cytiva 17-1440-02 |
| Casein Blocking Buffer 10x | Sigma-Aldrich | Cat# B6429 |
| TMB | Sigma-Aldrich | Cat# T5525 |
| Deposited data | ||
| Heavy variable genes | This paper | GenBank: OM331742-OM331759 |
| Light variable genes | This paper | GenBank: OM331760-OM331774 |
| Experimental models: Cell lines | ||
| HeLA-derived TZM-bl cells | NIH AIDS Reagent Program | Cat#8129; RRID:CVCL_B478 |
| HEK293T | ATCC | Cat#CRL-3216; RRID:CVCL_0063 |
| Expi293F cells | ThermoFisher Scientific Inc | Cat# A14525 |
| Hela | ATCC | Cat#CCL-2 |
| Oligonucleotides | ||
| Primer for mAb generation | This study | Table S1 |
| Primers for site directed mutagenesis | This study | Table S2 |
| Recombinant DNA | ||
| pFUSE2ss-CLIg-hL2 | Invivogen | Cat#pfuse2ss-hcll2 |
| pFUSEssCHIg-hG1 | Invivogen | Cat# pfusess-hchg1 |
| MN Env expression vector | AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat# ARP900 |
| 6535 Env expression vector | AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat# ARP11017 |
| HXB-2 Env expression vector | AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat# ARP11069 |
| QH0692 Env expression vector | AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat# ARP11018 |
| pREJO4541 Env expression vector | AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat# ARP11035 |
| pRHPA4259 Env expression vector | AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat# ARP11036 |
| AD8 Env expression vector | Purcell lab | N/A |
| JRCSF Env expression vector | Purcell lab | N/A |
| YU-2 | Purcell lab | N/A |
| ZM53M.PB12 | AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat# ARP11313 |
| BG505 | Purcell lab | N/A |
| 92RW020.2 | Montefiori lab | N/A |
| Q259.d2.17 | Montefiori lab | N/A |
| Q769.d22 | Montefiori lab | N/A |
| Q842.d12 | Montefiori lab | N/A |
| MS208.A1 | Montefiori lab | N/A |
| Du156 | Montefiori lab | N/A |
| ZM135M.PL10a | Montefiori lab | N/A |
| CAP210.2.00.E8 | Montefiori lab | N/A |
| CAP45.2.00.G3 | Montefiori lab | N/A |
| So431_C1_1 | AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat# ARP-13367 |
| 2969249 | AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat# ARP-13361 |
| 3728.v2.c6 | AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat# ARP-13362 |
| CE703010010_C4 | AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat# ARP-13364 |
| CE704810053_2B7 | AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat# ARP-13365 |
| ZM215F.PB8 | AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat# ARP-13360 |
| CE2103_E8 | AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat# ARP-13363 |
| ZM233M.PB6 | AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat# ARP-13359 |
| 2759058_F10_B6 | AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat# ARP-13366 |
| Ko243_H6.3 | AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat# ARP-13370 |
| CAP382.2.00.D7.19 | AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat# ARP-13368 |
| B005582-7_G7.8 | AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat# ARP-13369 |
| AE01 | Montefiori lab | N/A |
| AE03 | Montefiori lab | N/A |
| CNE5 | Montefiori lab | N/A |
| X2278 | deCamp et al., 2014 | N/A |
| TRO11 | deCamp et al., 2014 | N/A |
| 25710 | deCamp et al., 2014 | N/A |
| 398F1 | deCamp et al., 2014 | N/A |
| BJOX2000 | deCamp et al., 2014 | N/A |
| CE1176 | deCamp et al., 2014 | N/A |
| CE0217 | deCamp et al., 2014 | N/A |
| CH119 | deCamp et al., 2014 | N/A |
| CNE8 | deCamp et al., 2014 | N/A |
| CNE55 | deCamp et al., 2014 | N/A |
| 246F3 | deCamp et al., 2014 | N/A |
| X1632 | deCamp et al., 2014 | N/A |
| Mulv | Purcell lab | N/A |
| Software and algorithms | ||
| IMGT | International ImMunoGeneTics Information System | http://www.imgt.org |
| GraphPad Prism Software | GraphPad Prism Software, Inc. | SCR_002798 |
| FlowJo software v.10 | BD biosciences | SCR_008520 |
| PyMOL | Schrödinger, LLC | RRID: SCR_000305 |
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Prof. Damian Purcell (dfjp@unimelb.edu.au).
Materials availability
Material transfer agreements with standard academic terms will be established to document reagent sharing by the lead contact’s institution.
Experimental model and subject details
Cell lines and primary cells
The common laboratory cell lines HEK293T (ATCC CRL-11268), and Hela (ATCC CCL2) were originally purchased from ATCC. The source of other cell lines were: Expi293F cells (Thermo Fisher Scientific); and TZM-bl cells (AIDS Research and Reference Reagents Program, Division of AIDS ARP-8129). Master seed stocks of all cell lines were confirmed for identity through Short Tandem Repeat profiling and were negative for mycoplasma and were routinely tested bi-monthly. All cell lines were cultivated in complete Dulbecco’s modified Eagle’s medium (DMEM) media and 10% fetal bovine serum (FBS), except for Expi293F cells that were cultured in Expi293 Expression Medium (Gibco).
Primary bovine B-lymphocytes were prepared from bovine PBMCs obtained by centrifugation of cow blood on a Ficoll-Plaque PLUS step gradient (GE Healthcare).
Experimental animals
The Holstein Friesian cattle (Bos Taurus) used for the study were all females in the prime years for conception, 4–7 years of age. Cows were kept on a certified experimental open-gazing farm holding a Scientific Premises License from the Department of Economic Development, Jobs, Transport and Resources (DEDJTR) of the Victorian State Government that blocked any animal products from entering the food chain. Animal welfare was continuously evaluated and supervised by the Ellinbank Agricultural Research Farm Animal Husbandry Committee.
Ethics
This work was conducted under animal ethics approval 2015-17 from the Victorian State Government DEDJTR Research and Extension Animal Ethics Committee.
Method details
HIV-1 Env production, expression and purification
Vaccine proteins including HIV-1 NL AD8 (AD8) Unc gp140 Env (clade B) were expressed using stably-transfected Hela cell line and AD8 6R SOSIP.664 was produced by transient transfection of Expi293 cells. KNH1144 SOSIP.v1 (clade A),57 and BG505 SOSIP.664 (clade A) were produced as previously described.18
AD8 SOSIP.v4.1 was produced using an Env-expression plasmid encoding the Env from AD858 modified according to the “v4.1” mutations described previously59 (Figure S1A) with a C-terminal D7324 epitope-tag, 6His-tag or Avi-tag. Proteins were expressed in Expi293 cells by co-transfecting the Env-expression plasmid with a human furin protease expression plasmid. AD8 SOSIP Envs were purified from culture supernatant using a 2G12-sepharose affinity resin and eluted using 3M MgCl2 (Figure S1B). The Env was immediately buffer exchanged into PBS and trimeric Env was further purified by size exclusion chromatography using a HiLoad 16/600 Superdex prep grade column (GE Healthcare Life Sciences). Avi-Tagged trimers were biotinylated using BirA enzyme according to manufacturer’s instructions (Avidity, LLC). Monomeric AD8 gp120 was also produced as described previously.60, 61, 62 BG505 Unc. SEKS (uncleaved gp140) was produced as described above for AD8 SOSIP gp140 protein, except that the Env expression plasmid63 was not co-transfected with a furin expression plasmid.
Cow immunization
Three female Holstein cattle (Bos taurus) were vaccinated subcutaneously into the flank, using Seppic Montanide (ISA206) adjuvant. Cows were immunized prior to and during pregnancy and revaccinated after calving (Figure 1A). Cow #1 received 100μg KNH1144 SOSIP and BG505 SOSIP Env trimer in phase 1 and was revaccinated with 50μg BG505 SOSIP in phase 2. Cow #2 and #3 received 500μg AD8 Unc gp140 trimer during pregnancy and were revaccinated after calving with 50μg BG505 Unc gp140 and 100μg AD8 SOSIP. R6. 664, respectively. Sera samples were collected after calving at week 54 (phase 1) and week 59 (phase 2). Peripheral blood mononuclear cells (PBMC) from each cow were also isolated from bloods collected after phase 1 and phase 2 as described previously.12
Serum ELISA and neutralization of sera samples
IgG titres in sera against autologous Env vaccine antigens were measured by direct ELISA, with incubations performed at room temperature (RT) except when stated. Casein buffer 1x (Sigma) was used as sample diluent for each step. Briefly, 96-well plates were coated with 1 μg/mL recombinant Env gp140 proteins (BG505 SOSIP, AD8 Unc gp140) in coating buffer (200 mM Tris-HCl, 100 mM NaCl, pH 8.8) overnight at 4°C. The plate was washed four times with PBS+ 0.1% Tween and four times with PBS then blocked with casein buffer 1x (Sigma). Sera samples were added in half-log10 dilutions and incubated for 3 h at RT. Afterward, 1/1000 dilution of HRP-conjugated sheep-anti-bovine IgG (BioRad, #AAI23P) was loaded and incubated for 1 h at RT. Finally, color development was performed using TMB (Sigma, cat no: T5525) according to manufacturer instructions and the reaction was stopped using 1M H2SO4. Absorbance was measured at 450 nm against a reference of 690 nm.
For neutralization assay of sera samples, HIV-1 Env-pseudotyped viruses were produced as described previously.9,64 Pseudoviruses were produced in HEK 293T cells by co-transfecting a backbone plasmid with one of Env expressing plasmids. TZM-bl neutralization assay was performed for sera samples collected pre-immunisation, at phase 1 (week 54) and phase 2 (week 59). The assay was performed as described previously.64 Briefly, 50 μL of pseudovirus in complete growth medium (DMEM +10% FBS) was mixed with serial dilutions of serum samples in a final volume of 150 μL in 96-well plates (Corning, flat bottom, non-pyrogenic), and incubated for 1 h at 37 °C. Thereafter, 104 TZM-bl cells (containing DEAE-Dextran at pre-determined optimal concentration (Sigma)) was added to each well and plates were incubated for 48 h at 37 °C. Inhibition of infection was calculated by measuring relative luminescence units (RLUs) using Britelite plus (PerkinElmer) in a VICTOR Multilabel Plate Reader (PerkinElmer). ID50 neutralizing antibody titers are expressed as the reciprocal of the sample dilution required to reduce RLU by 50%.
Single particle negative stain electron microscopy
Purified AD8 SOSIP (100 ng/μL) was placed on glow-discharged carbon coated copper mesh grids and stained with 1% uranyl formate. Grids were screened for appropriate stain thickness and particle distribution and images were collected using an FEI Talos L120C electron microscope. Images were collected at 730,00X magnification with a −1.8 μm defocus for a final magnified pixel size of 1.9 Å/pix. Negatively stained AD8 particles were then automatically picked based on an empirical evaluation of maximum particle radius of 110 Å , characteristic particle radius of 80 Å, and with threshold peak high of 5 standard deviation above the noise using cisTEM software version 1.0.0-beta.65 Further, a 2D classification was performed on 38,000 particles in cisTEM that resulted in 50 classes. The initial model was generated ab initio using the dataset and processed under the filter for particles from 20 Å to 8 Å in the first step of classification within cisTEM. Best 18 classes representing different orientations of AD8 were selected for further iterative 3D classification under C3 symmetry. This was followed by a local refinement and a final 3D refinement in cisTEM. UCSF Chimera was used to generate figures.66
Single cell sorting by fluorescence-activated cell sorting (FACS)
Sorting of bovine PBMCs was performed as described previously12,64 with minor modifications (Figure 2B). In brief, 2.5 million cryopreserved PBMCs were thawed and resuspended in 10 mL pre-warmed 37°C RPMI 1640 medium (Life technologies) (containing 10% FBS, 20 μg/mL or 10 U/mL DNaseI) for 5 min at RT followed by centrifugation at 500 x g for 10 min at 4°C. The cells were resuspended in chilled PBS and LIVE/DEAD™ Fixable Aqua Dead Cell Stain (Thermo Fisher Scientific) was added and incubated for 10 min on ice. PBMCs were then stained with Alexa-fluor 488 conjugated anti-bovine IgG (Sigma, B6901) and 50 nM biotinylated AD8 SOSIP-avi gp140 coupled to streptavidin-APC and PE (Life Technologies) in equimolar ratios. The cells were incubated for 1 h at 4°C in PBS containing 1 mM EDTA and 1% horse serum (Sigma). Then, live IgG + AD8 SOSIP PE+/AD8 SOSIP APC + cells were single-sorted into 96-well plates containing lysis buffer (3.7μL/well PBS, 10 mM DTT and 8 U RNasin Ribonuclease Inhibitor (Promega)) on an ARIA III sorter using BD FACSDiva Software and were immediately frozen at −80°C until further use. Data were analyzed using FlowJo software v.10.
Single cell cDNA synthesis, RT-PCR, and cloning
cDNA was synthesized from mRNA of single cells as follows: Sorted cells were incubated with 200ng Random Hexamer Primer, 4 μL 5x RT Buffer (Thermo Fisher Scientific), 1 μL dNTPs mix (10 mM, Promega), 1 μL DTT (Thermo Fisher Scientific), 8U RNasin Ribonuclease Inhibitor (Promega), 0.25 μL Superscript III (200 U/mL, Thermo Fisher Scientific), and 6.5 μL of RNase-free H2O at 42°C for 10 min, 25°C for 10 min, 50°C for 60 min, and 94°C for 5 min67 and antibody variable genes were amplified as described previously12 with minor modifications. Briefly, antibody heavy gamma (γ) and light lambda (λ) variable genes were amplified independently in nested PCR using MyTaq HS Red Mix (Bioline) according to the manufacturer’s instruction. The PCR reaction primers and conditions are listed in Table S1. PCR1 reactions were set up in 25 μL with 2.5μL cDNA, while PCR2 reaction volumes were 50μL using 5 μL PCR1 product. The PCR reaction cycle was performed as 94 °C 5 min, 50 cycles of 94°C for 45 s, 60°C for 45 s and 72°C for 45 s and final extension at 72 °C for 10 min. The annealing temperature of PCR1 for lambda gene was 58°C. The amplifid bovine VH/VL genes were cloned into the human constant heavy (CH) and constant light (CL) region expression vectors in pFUSEssCHIg-hG1 and pFUSE2ss-CLIg-hL2 (Invivogen), using EcoRI/NheI and EcoRI/AvrII restriction enzymes, respectively.
Antibody production and purification
Antibody plasmids containing heavy chain and light chain genes were co-transfected (2:3 ratio) into Expi293F cells using Expifectamine (Thermo Fisher Scientific) according to manufacturer instructions. Supernatants containing antibodies were harvested 4 days after transfection and filtered using 0.22 μm filters. NC-Cow1 antibody was produced as an anti-HIV-1 bovine antibody control by codon optimisation of genes available from the GenBank (MF167446.1 and MF167436.1). Antibody supernatants were purified using Protein G Agarose Fast Flow (Merck Millipore). Antibodies were eluted from chromatography columns using 50 mM glycine (pH 2.7) and immediately neutralised by addition of 1/10 volume of 1 M Tris (pH 8.0) before being buffer exchanged into PBS, concentration using Amicon 50 kDa spin membranes (Millipore) and sterilization using 0.22 μm filters.
Generation of HIV-1 AD8 pseudovirus mutant
Specific amino acid changes to HIV-1 AD8 gp160 Env were introduced using the following PCR reaction set up: 100ng Full-length AD8 gp160 Env plasmid, 5% Dimethyl sulfoxide (DMSO), 10 μL 5x Phusion Reaction buffer (New England BioLabs), 1 μL dNTP mix(10 mM, Promega), 3U Phusion HF DNA Polymerase (New England BioLabs, (MEL-2000 units/mL)), 0.5 μL from each forward and Reverse primer (20 μM) (Table S2) and nuclease-free H20 up to the total volume of 50 μL. The PCR reaction was performed as following: 95°C for 5 min, 30 cycles of 95°C for 30 s, 48°C for 30 s, 72°C for 8 min and final extension of 72°C 15 min. Mutations were confirmed by sequence analysis.
Neutralization assays of anti-HIV-1 monoclonal antibodies
The neutralization assay of TZM-bl was performed as described previously.12 In brief, pseudoviruses and serial dilutions of mAbs were mixed and incubated for 1 h at 37°C. Then, 104 TZM-bl cells supplemented with DEAE-dextran at a final concentration of 10 μg/mL were added and the plate was incubated for 72 h at 37°C. The infectivity of pseudoviruses was calculated according to luciferase relative light unit readout using either a Fluostar (BMG Labtech) or Victor XLight (Perkin-Elmer) luminometer. Human bNAbs were used as the controls in the assay. CH01-31 antibody was an equal concentration mixture of two bNAbs (CH01 and VRC-CH31).
ELISA assays of anti-HIV-1 monoclonal antibodies
To screen HIV-1 Env binding mAbs, ELISA plates were coated with 2 μg/mL D7324 Sheep anti-gp120 (Aalto Bio Reagents) at 4°C in 1X PBS overnight. Plates were washed four times with PBS +0.1% Tween and two times with PBS, then blocked with 5% Skim milk in 1× PBS at RT for 1 h. The plates were washed and 600 ng/mL D7324 tagged AD8 SOSIP trimer was added and incubated for 1 h at RT. Plates were washed and a 1/1000 dilution of goat anti-human IgG HRP (KPL Cat No. 474-1002) (pre-incubated with 2% normal sheep serum) was added to the wells. The plate was incubated at RT for 1 h, washed then developed by adding SureBlue TMB (KPL) according to the manufacturer’s instructions. The absorbance was measured at 450 nm against a reference of 690 nm.
To assess the binding of mAbs to mutated HIV-1 Env gp160, harvested pseudoviruses were lysed with 1% Triton X-100 detergent (Astral Scientific). ELISA plates were coated with D7324 Sheep anti-gp120, blocked with skim milk and the lysed pseudoviruses were then captured on ELISA for 2 h at 37°C. Next, serial dilutions of mAbs were added before the addition of goat anti-human IgG HRP.
For experiments involving antibody binding to untagged HIV-1 Env (monomeric AD8 gp120, AD8 Unc gp140, AD8 SOSIP and ConM SOSIP), plates were coated directly with 1 μg/mL Env proteins at 4°C in PBS overnight then washed and blocked before addition of mAbs and goat anti-human gamma HRP.
Competition ELISA
To investigate the epitopes of AD8 Env-binding mAbs, a competition ELISA was performed using competing antibodies that were biotinylated with EZ-Link Sulfo-NHS-LC-Biotin kit (Thermo Fisher Scientific). The plates were coated with 1/1000 dilution of anti-6X His antibody (Abcam, #9108) and incubated overnight at 4°C. Then, the plates were washed and blocked with 5% skim milk in PBS +0.1% Tween (0.1%) for 1 h at RT. Following washing, 500 ng/mL His-tagged AD8 SOSIP trimer was added, and the plates were incubated for 2 h at RT. Then, bovine mAbs in the following amounts were added: 1 μg/mL (MEL-1842, MEL-1872, MEL-2129, MEL-2000, MEL-2028 and MEL-782), 2 μg/mL (MEL-2010, MEL-33, MEL-130, MEL-563 and MEL-663) and 5 μg/mL (MEL-1905, MEL-1967, MEL-2114, MEL-F2, MEL-198 and MEL-D1). After washing, the biotinylated human mAbs were added in the following amounts to give sufficient signal: 1 μg/mL (PGT121, PGT145, 10-1074, PGT151), 2 μg/mL (VRC01 and 3BNC117) and 5 μg/mL (b12, HJ16). Then, a 1/1000 dilution of Streptavidin HRP was added and incubated for 1 h at RT followed by addition of SureBlue according to the manufacturer’s instructions.
For self-competition of bovine bNAbs, the assay was performed as above except using biotinylated bovine mAbs as the following amounts: 1 μg/mL for MEL-2028, MEL-2000, MEL-782, MEL-2129, MEL-1842, MEL-1872, 2 μg/mL for MEL-2010, MEL-33, MEL-130, MEL-563, 5 μg/mL for MEL-1905, MEL-1967, MEL-2114, MEL-F2, MEL-198 and MEL-D1.
Polyreactivity assay: HEp-2 cell staining assay
The HEp-2 cell-staining kit (Aesku Diagnostics) was used according to manufacturer’s instructions. In brief, 2.5 μg of mAbs and controls were added to HEp-2 cell containing wells and incubated in a moist chamber at RT for 30 min. Slides were washed with PBS then 25 μL FITC-conjugated goat anti-human IgG was applied with incubation of 30 min at RT. The slide was washed and mounted on coverslips using the provided mounting medium. Slides were viewed at 20x magnification and imaged on the Zeiss LSM780 confocal microscope. All images were captured with the following conditions: digital gain 800, laser power 2.0%. Samples showing fluorescence greater than the negative control (provided by the vendor) were considered positive for HEp-2 staining.
Polyreactivity assay: single autoantigen reactivity
Single antigen ELISA assays were performed using AESKULISA ANA-8Pro (Aesku) for U1- ribonucleoprotein (RNP), SnRNP/Sm, Sm, SS-A, SS-B, Jo-1, ScI-70, and CenpB. The 96 wells were coated with these cellular and nuclear antigens for the qualitative detection of mAbs reactivity. The cut-off calibrator, negative control and positive control were provided by the manufacturer.
Quantification and statistical analysis
Statistics
Statistical analyses were performed with Graphpad Prism 9. Correlations between the neutralizing breadth and neutralization activity (IC50) of AD8 SOSIP binding antibodies were assessed using the non-parametric Spearman test.
Acknowledgments
This work was conducted under animal ethics approval 2015-17 from the Victorian State Government DEDJTR Research and Extension Animal Ethics Committee. Funding support was provided to D.F.J.P. from the Australian NHRMC - EU Horizons 2020 Grant (APP1115828), as a partner in the European AIDS Vaccine Initiative 2020 (EAVI 2020), from the Australian Centre for HIV-1 and Hepatitis Virology Research; to B.H. from the Melbourne HIV-1 Cure Consortium and the Australian Centre for HIV-1 and Hepatitis Virology Research; to A.S. from the University of Melbourne Early Career Research Grant; and to J.P.M. from grant P01 AI110657 and to D.M. from grant HSN272201800004C from the National Institutes of Health. This project has received funding from the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement no. 681137. We acknowledge contributions of Leanne Horstman, Greg Morris, and Lianne Dorling (Department Jobs, Precincts and Resources, Victoria) for veterinarian support and animal husbandry and George Lovrecs and Brian Muller for technical support.
Author contributions
Conceptualization and oversight of the experiment, B.H. and D.F.J.P.; antigen B cell sorting, B.H.; PCR and antibody cloning, B.H. and J.M.E.; antibody gene analysis, B.H. and N.A.S.-Q.; antibody expression and purification, B.H., J.M.E., and S.G.; antibody characterization, neutralization, and mutagenesis experiments, B.H., D.S.F., N.A.S.-Q., H.G., and D.M.; provided proteins for immunization, R.W.S., M.J.v.G., J.P.M., A.C., and C.A.G.; structural work and analysis, A.S.; polyreactivity assays, B.H. and T.E.A.; cow immunization, D.F.J.P., W.J.W., and C.M.; analysis, figures, and original draft of manuscript, B.H., D.S.F., A.S., and N.A.S.-Q.; review and editing of the manuscript, B.H., D.S.F., H.G., N.A.S.-Q., J.M.E., C.A.G., S.G., T.E.A., C.M., W.J.W., M.J.v.G., A.C., I.R., P.R.G., J.P.M., R.W.S., D.M., A.S., and D.F.J.P.; funding acquisition, D.F.J.P., B.H., and A.S.
Declaration of interests
B.H. and D.F.J.P. are inventors on a corresponding patent from University of Melbourne: HIV-1 antibodies PCT/AU2021/050593. The other authors declare no competing interests.
Published: May 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100635.
Supplemental information
The GenBank accession codes for the heavy variable genes: OM331742-OM331759; light variable genes: OM331760-OM331774 are annotated at the bottom right of each box.
IgDiscover and AbMining Toolbox were used for identification of DH germline genes.
Values in red color show low IC50 and better neutralization while, those in yellow color show high IC50 values and less neutralization activity. Neutralization assays were performed in duplicates with two independent biological replicates. ND: Not done.
Values in red color show low IC80 and better neutralization while, those in yellow color show high IC80 values and less neutralization activity. Neutralization assays were performed in duplicates with two independent biological replicates.
ELISA assay was performed using a constant half maximal effective concentration (EC50) of each antibody to AD8 WT Env. PGT121 (V3-glycan epitope) and b12 and VRC01 (CD4bs epitope) were included for comparison. Blue colour indicates a decrease in binding, and Red/orange shows an increase in binding compared to wild type. The amount of lysed virus/ amount of Env added was equilibrated according to 2G12 capture ELISA binding. IgG Polyclonal serum (NIH, #3957) was used for mutants that did not bind well to 2G12. ELISA assays were performed in duplicates with two independent biological replicates.
Values in yellow color show high IC50 and less neutralization activity. Neutralization assays were performed in duplicates with two independent biological replicates.
Data and code availability
-
•
All materials described in this manuscript are available via a material transfer agreement with the University of Melbourne. All data required to state the conclusions in the paper are present in the paper and/or the Supplementary data. Sequence data are provided in this paper or already published.15,25, 26, 27, 28, 29, 30, 31,56 The GenBank accession codes for the heavy variable genes: OM331742-OM331759; light variable genes: OM331760-OM331774 are annotated at the bottom right of each box in Table S3.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Burton D.R., Poignard P., Stanfield R.L., Wilson I.A. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Subbaraman H., Schanz M., Trkola A. Broadly neutralizing antibodies: what is needed to move from a rare event in HIV-1 infection to vaccine efficacy? Retrovirology. 2018;15:52. doi: 10.1186/s12977-018-0433-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohno S., Mori N., Matsunaga T. Antigen-binding specificities of antibodies are primarily determined by seven residues of VH. Proc. Natl. Acad. Sci. U S A. 1985;82:2945–2949. doi: 10.1073/pnas.82.9.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hraber P., Seaman M.S., Bailer R.T., Mascola J.R., Montefiori D.C., Korber B.T. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS. 2014;28:163–169. doi: 10.1097/qad.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Excler J.L., Michael N.L. Lessons from HIV-1 vaccine efficacy trials. Curr. Opin. HIV AIDS. 2016;11:607–613. doi: 10.1097/coh.0000000000000312. [DOI] [PubMed] [Google Scholar]
- 6.Sanders R.W., van Gils M.J., Derking R., Sok D., Ketas T.J., Burger J.A., Ozorowski G., Cupo A., Simonich C., Goo L., et al. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klasse P.J., Ketas T.J., Cottrell C.A., Ozorowski G., Debnath G., Camara D., Francomano E., Pugach P., Ringe R.P., LaBranche C.C., et al. Epitopes for neutralizing antibodies induced by HIV-1 envelope glycoprotein BG505 SOSIP trimers in rabbits and macaques. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heydarchi B., Center R.J., Gonelli C., Muller B., Mackenzie C., Khoury G., Lichtfuss M., Rawlin G., Purcell D.F. Repeated vaccination of cows with HIV Env gp140 during subsequent pregnancies elicits and sustains an enduring strong env-binding and neutralising antibody response. PLoS One. 2016;11 doi: 10.1371/journal.pone.0157353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kramski M., Center R.J., Wheatley A.K., Jacobson J.C., Alexander M.R., Rawlin G., Purcell D.F.J. Hyperimmune bovine colostrum as a low-cost, large-scale source of antibodies with broad neutralizing activity for HIV-1 envelope with potential use in microbicides. Antimicrob. Agents Chemother. 2012;56:4310–4319. doi: 10.1128/aac.00453-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sok D., Le K.M., Vadnais M., Saye-Francisco K.L., Jardine J.G., Torres J.L., Berndsen Z.T., Kong L., Stanfield R., Ruiz J., et al. Rapid elicitation of broadly neutralizing antibodies to HIV by immunization in cows. Nature. 2017;548:108–111. doi: 10.1038/nature23301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F., Ekiert D.C., Ahmad I., Yu W., Zhang Y., Bazirgan O., Torkamani A., Raudsepp T., Mwangi W., Criscitiello M.F., et al. Reshaping antibody diversity. Cell. 2013;153:1379–1393. doi: 10.1016/j.cell.2013.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heydarchi B., Center R.J., Bebbington J., Cuthbertson J., Gonelli C., Khoury G., Mackenzie C., Lichtfuss M., Rawlin G., Muller B., Purcell D. Trimeric gp120-specific bovine monoclonal antibodies require cysteine and aromatic residues in CDRH3 for high affinity binding to HIV Env. MAbs. 2017;9:550–566. doi: 10.1080/19420862.2016.1270491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Gils M.J., Sanders R.W. Broadly neutralizing antibodies against HIV-1: templates for a vaccine. Virology. 2013;435:46–56. doi: 10.1016/j.virol.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Sellhorn G., Caldwell Z., Mineart C., Stamatatos L. Improving the expression of recombinant soluble HIV Envelope glycoproteins using pseudo-stable transient transfection. Vaccine. 2009;28:430–436. doi: 10.1016/j.vaccine.2009.10.028. [DOI] [PubMed] [Google Scholar]
- 15.Yang X., Wyatt R., Sodroski J. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J. Virol. 2001;75:1165–1171. doi: 10.1128/jvi.75.3.1165-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dey A.K., David K.B., Lu M., Moore J.P. Biochemical and biophysical comparison of cleaved and uncleaved soluble, trimeric HIV-1 envelope glycoproteins. Virology. 2009;385:275–281. doi: 10.1016/j.virol.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pancera M., Wyatt R. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology. 2005;332:145–156. doi: 10.1016/j.virol.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 18.Sanders R.W., Derking R., Cupo A., Julien J.P., Yasmeen A., de Val N., Kim H.J., Blattner C., de la Pena A.T., Korzun J., et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pauthner M., Havenar-Daughton C., Sok D., Nkolola J.P., Bastidas R., Boopathy A.V., Carnathan D.G., Chandrashekar A., Cirelli K.M., Cottrell C.A., et al. Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Immunity. 2017;46:1073–1088.e6. doi: 10.1016/j.immuni.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson S.S., Sumner L.E., Finnegan M.A., Billings E.A., Huffman D.L., Rush M.A. A 35-year review of pre-clinical HIV therapeutics research reported by NIH ChemDB: influences of target discoveries, drug approvals and research funding. J. AIDS Clin. Res. 2020;11:11. [PMC free article] [PubMed] [Google Scholar]
- 21.Watkins J.D., Siddappa N.B., Lakhashe S.K., Humbert M., Sholukh A., Hemashettar G., Wong Y.L., Yoon J.K., Wang W., Novembre F.J., et al. An anti-HIV-1 V3 loop antibody fully protects cross-clade and elicits T-cell immunity in macaques mucosally challenged with an R5 clade C SHIV. PLoS One. 2011;6 doi: 10.1371/journal.pone.0018207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoofs T., Klein F., Braunschweig M., Kreider E.F., Feldmann A., Nogueira L., Oliveira T., Lorenzi J.C.C., Parrish E.H., Learn G.H., et al. HIV-1 therapy with monoclonal antibody 3BNC117 elicits host immune responses against HIV-1. Science. 2016;352:997–1001. doi: 10.1126/science.aaf0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corey L., Gilbert P.B., Juraska M., Montefiori D.C., Morris L., Karuna S.T., Edupuganti S., Mgodi N.M., deCamp A.C., Rudnicki E., et al. Two randomized trials of neutralizing antibodies to prevent HIV-1 acquisition. N. Engl. J. Med. 2021;384:1003–1014. doi: 10.1056/nejmoa2031738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shingai M., Nishimura Y., Klein F., Mouquet H., Donau O.K., Plishka R., Buckler-White A., Seaman M., Piatak M., Jr., Lifson J.D., et al. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature. 2013;503:277–280. doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barouch D.H., Whitney J.B., Moldt B., Klein F., Oliveira T.Y., Liu J., Stephenson K.E., Chang H.W., Shekhar K., Gupta S., et al. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013;503:224–228. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng C.T., Jaworski J.P., Jayaraman P., Sutton W.F., Delio P., Kuller L., Anderson D., Landucci G., Richardson B.A., Burton D.R., et al. Passive neutralizing antibody controls SHIV viremia and enhances B cell responses in infant macaques. Nat. Med. 2010;16:1117–1119. doi: 10.1038/nm.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanfield R.L., Wilson I.A., Smider V.V. Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies. Sci. Immunol. 2016;1 doi: 10.1126/sciimmunol.aaf7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corcoran M.M., Phad G.E., Bernat N.V., Stahl-Hennig C., Sumida N., Persson M.A.A., Martin M., Hedestam G.B.K. Production of individualized V gene databases reveals high levels of immunoglobulin genetic diversity. Nat. Commun. 2016;7:13642. doi: 10.1038/ncomms13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D'Angelo S., Glanville J., Ferrara F., Naranjo L., Gleasner C.D., Shen X., Bradbury A.R., Kiss C. The antibody mining toolbox: an open source tool for the rapid analysis of antibody repertoires. MAbs. 2014;6:160–172. doi: 10.4161/mabs.27105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sliepen K., Han B.W., Bontjer I., Mooij P., Garces F., Behrens A.J., Rantalainen K., Kumar S., Sarkar A., Brouwer P.J.M., et al. Structure and immunogenicity of a stabilized HIV-1 envelope trimer based on a group-M consensus sequence. Nat. Commun. 2019;10:2355. doi: 10.1038/s41467-019-10262-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., O'Dell S., Walker L.M., Wu X., Guenaga J., Feng Y., Schmidt S.D., McKee K., Louder M.K., Ledgerwood J.E., et al. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J. Virol. 2011;85:8954–8967. doi: 10.1128/jvi.00754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCoy L.E., van Gils M.J., Ozorowski G., Messmer T., Briney B., Voss J.E., Kulp D.W., Macauley M.S., Sok D., Pauthner M., et al. Holes in the glycan shield of the native HIV envelope are a target of trimer-elicited neutralizing antibodies. Cell Rep. 2016;16:2327–2338. doi: 10.1016/j.celrep.2016.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rappuoli R., Bottomley M.J., D'Oro U., Finco O., De Gregorio E. Reverse vaccinology 2.0: human immunology instructs vaccine antigen design. J. Exp. Med. 2016;213:469–481. doi: 10.1084/jem.20151960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lei L., Yang Y.R., Yang Y.H.R., Tran K., Wang Y.M., Chiang C.I., Ozorowski G., Xiao Y.L., Ward A.B., Wyatt R.T., Li Y.X. The HIV-1 envelope glycoprotein C3/V4 region Defines a prevalent neutralization epitope following immunization. Cell Rep. 2019;27:586–598.e6. doi: 10.1016/j.celrep.2019.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cottrell C.A., van Schooten J., Bowman C.A., Yuan M., Oyen D., Shin M., Morpurgo R., van der Woude P., van Breemen M., Torres J.L., et al. Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates. PLoS Pathog. 2020;16:e1008753. doi: 10.1371/journal.ppat.1008753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao F.Z., Joyce C., Burns A., Nogal B., Cottrell C.A., Ramos A., Biddle T., Pauthner M., Nedellec R., Qureshi H., et al. Mapping neutralizing antibody epitope specificities to an HIV Env trimer in immunized and in infected rhesus macaques. Cell Rep. 2020;32:108122. doi: 10.1016/j.celrep.2020.108122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stanfield R.L., Berndsen Z.T., Huang R., Sok D., Warner G., Torres J.L., Burton D.R., Ward A.B., Wilson I.A., Smider V.V. Structural basis of broad HIV neutralization by a vaccine-induced cow antibody. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aba0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorman J., Chuang G.Y., Lai Y.T., Shen C.H., Boyington J.C., Druz A., Geng H., Louder M.K., McKee K., Rawi R., et al. Structure of super-potent antibody CAP256-VRC26.25 in complex with HIV-1 envelope reveals a combined Mode of trimer-apex recognition. Cell Rep. 2020;31:107488. doi: 10.1016/j.celrep.2020.03.052. [DOI] [PubMed] [Google Scholar]
- 40.Chuang G.Y., Zhou J., Acharya P., Rawi R., Shen C.H., Sheng Z., Zhang B., Zhou T., Bailer R.T., Dandey V.P., et al. Structural survey of broadly neutralizing antibodies targeting the HIV-1 Env trimer Delineates epitope categories and characteristics of recognition. Structure. 2019;27:196–206.e6. doi: 10.1016/j.str.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doria-Rose N.A., Bhiman J.N., Roark R.S., Schramm C.A., Gorman J., Chuang G.Y., Pancera M., Cale E.M., Ernandes M.J., Louder M.K., et al. New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J. Virol. 2016;90:76–91. doi: 10.1128/jvi.01791-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Julg B., Sok D., Schmidt S.D., Abbink P., Newman R.M., Broge T., Linde C., Nkolola J., Le K., Su D., et al. Protective efficacy of broadly neutralizing antibodies with incomplete neutralization activity against simian-human immunodeficiency virus in rhesus monkeys. J. Virol. 2017;91 doi: 10.1128/jvi.01187-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagh K., Bhattacharya T., Williamson C., Robles A., Bayne M., Garrity J., Rist M., Rademeyer C., Yoon H., Lapedes A., et al. Optimal combinations of broadly neutralizing antibodies for prevention and treatment of HIV-1 clade C infection. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang J., Kang B.H., Pancera M., Lee J.H., Tong T., Feng Y., Imamichi H., Georgiev I.S., Chuang G.Y., Druz A., et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 2014;515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sauer M.M., Tortorici M.A., Park Y.J., Walls A.C., Homad L., Acton O., Bowen J., Wang C., Xiong X., de van der Schueren W., et al. Structural basis for broad coronavirus neutralization. bioRxiv. 2021 doi: 10.1101/2020.12.29.424482. Preprint at. [DOI] [PubMed] [Google Scholar]
- 46.Burton D.R. Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2002;2:706–713. doi: 10.1038/nri891. [DOI] [PubMed] [Google Scholar]
- 47.Ward A.B., Wilson I.A. Insights into the trimeric HIV-1 envelope glycoprotein structure. Trends Biochem. Sci. 2015;40:101–107. doi: 10.1016/j.tibs.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J., Kang B.H., Ishida E., Zhou T., Griesman T., Sheng Z., Wu F., Doria-Rose N.A., Zhang B., McKee K., et al. Identification of a CD4-binding-site antibody to HIV that evolved near-Pan neutralization breadth. Immunity. 2016;45:1108–1121. doi: 10.1016/j.immuni.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H., Chen X., Wang D., Yao C., Wang Q., Xie J., Shi X., Xiang Y., Liu W., Zhang L. Epitope-focused immunogens against the CD4-binding site of HIV-1 envelope protein induce neutralizing antibodies against auto- and heterologous viruses. J. Biol. Chem. 2018;293:830–846. doi: 10.1074/jbc.m117.816447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oguntibeju O.O. Quality of life of people living with HIV and AIDS and antiretroviral therapy. HIV AIDS (Auckl) 2012;4:117–124. doi: 10.2147/hiv.s32321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iyidogan P., Anderson K.S. Current perspectives on HIV-1 antiretroviral drug resistance. Viruses. 2014;6:4095–4139. doi: 10.3390/v6104095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Floren C.H., Chinenye S., Elfstrand L., Hagman C., Ihse I. ColoPlus, a new product based on bovine colostrum, alleviates HIV-associated diarrhoea. Scand. J. Gastroenterol. 2006;41:682–686. doi: 10.1080/00365520500380817. [DOI] [PubMed] [Google Scholar]
- 53.Odong P.O., Angwech P.J., Obol J., Florén C.H. Management of HIV in children using a bovine colostrum-based food product— an observational field study. World J. AIDS. 2015;5:100–104. doi: 10.4236/wja.2015.52012. [DOI] [Google Scholar]
- 54.den Hartog G., Jacobino S., Bont L., Cox L., Ulfman L.H., Leusen J.H.W., van Neerven R.J. Specificity and effector functions of human RSV-specific IgG from bovine milk. PLoS One. 2014;9 doi: 10.1371/journal.pone.0112047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cesarone M.R., Belcaro G., Di Renzo A., Dugall M., Cacchio M., Ruffini I., Pellegrini L., Del Boccio G., Fano F., Ledda A., et al. Prevention of influenza episodes with colostrum compared with vaccination in healthy and high-risk cardiovascular subjects: the epidemiologic study in San Valentino. Clin. Appl. Thromb. Hemost. 2007;13:130–136. doi: 10.1177/1076029606295957. [DOI] [PubMed] [Google Scholar]
- 56.Scheid J.F., Mouquet H., Ueberheide B., Diskin R., Klein F., Oliveira T.Y.K., Pietzsch J., Fenyo D., Abadir A., Velinzon K., et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang Y., Andjelic S., Binley J.M., Crooks E.T., Franti M., Iyer S.P.N., Donovan G.P., Dey A.K., Zhu P., Roux K.H., et al. Structural and immunogenicity studies of a cleaved, stabilized envelope trimer derived from subtype A HIV-1. Vaccine. 2009;27:5120–5132. doi: 10.1016/j.vaccine.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Freed E.O., Englund G., Martin M.A. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J. Virol. 1995;69:3949–3954. doi: 10.1128/jvi.69.6.3949-3954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Taeye S.W., Ozorowski G., Torrents de la Pena A., Guttman M., Julien J.P., van den Kerkhof T.L., Burger J.A., Pritchard L.K., Pugach P., Yasmeen A., et al. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell. 2015;163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Center R.J., Earl P.L., Lebowitz J., Schuck P., Moss B. The human immunodeficiency virus type 1 gp120 V2 domain mediates gp41-independent intersubunit contacts. J. Virol. 2000;74:4448–4455. doi: 10.1128/jvi.74.10.4448-4455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Center R.J., Wheatley A.K., Campbell S.M., Gaeguta A.J., Peut V., Alcantara S., Siebentritt C., Kent S.J., Purcell D.F. Induction of HIV-1 subtype B and AE-specific neutralizing antibodies in mice and macaques with DNA prime and recombinant gp140 protein boost regimens. Vaccine. 2009;27:6605–6612. doi: 10.1016/j.vaccine.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 62.Gonelli C.A., Khoury G., Center R.J., Purcell D.F.J. HIV-1-based virus-like particles that Morphologically Resemble mature, infectious HIV-1 virions. Viruses. 2019;11:507. doi: 10.3390/v11060507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ringe R.P., Sanders R.W., Yasmeen A., Kim H.J., Lee J.H., Cupo A., Korzun J., Derking R., van Montfort T., Julien J.P., et al. Cleavage strongly influences whether soluble HIV-1 envelope glycoprotein trimers adopt a native-like conformation. Proc. Natl. Acad. Sci. U S A. 2013;110:18256–18261. doi: 10.1073/pnas.1314351110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Montefiori D.C. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. 2005 doi: 10.1002/0471142735.im1211s64. Chapter 12: Unit 12 11. [DOI] [PubMed] [Google Scholar]
- 65.Grant T., Rohou A., Grigorieff N. cisTEM, user-friendly software for single-particle image processing. Elife. 2018;7 doi: 10.7554/elife.35383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 67.Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The GenBank accession codes for the heavy variable genes: OM331742-OM331759; light variable genes: OM331760-OM331774 are annotated at the bottom right of each box.
IgDiscover and AbMining Toolbox were used for identification of DH germline genes.
Values in red color show low IC50 and better neutralization while, those in yellow color show high IC50 values and less neutralization activity. Neutralization assays were performed in duplicates with two independent biological replicates. ND: Not done.
Values in red color show low IC80 and better neutralization while, those in yellow color show high IC80 values and less neutralization activity. Neutralization assays were performed in duplicates with two independent biological replicates.
ELISA assay was performed using a constant half maximal effective concentration (EC50) of each antibody to AD8 WT Env. PGT121 (V3-glycan epitope) and b12 and VRC01 (CD4bs epitope) were included for comparison. Blue colour indicates a decrease in binding, and Red/orange shows an increase in binding compared to wild type. The amount of lysed virus/ amount of Env added was equilibrated according to 2G12 capture ELISA binding. IgG Polyclonal serum (NIH, #3957) was used for mutants that did not bind well to 2G12. ELISA assays were performed in duplicates with two independent biological replicates.
Values in yellow color show high IC50 and less neutralization activity. Neutralization assays were performed in duplicates with two independent biological replicates.
Data Availability Statement
-
•
All materials described in this manuscript are available via a material transfer agreement with the University of Melbourne. All data required to state the conclusions in the paper are present in the paper and/or the Supplementary data. Sequence data are provided in this paper or already published.15,25, 26, 27, 28, 29, 30, 31,56 The GenBank accession codes for the heavy variable genes: OM331742-OM331759; light variable genes: OM331760-OM331774 are annotated at the bottom right of each box in Table S3.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.