Abstract
Background
Although the antitumor efficacy of docetaxel (DTX) has long been attributed to the antimitotic activities, its impact on the tumor microenvironment (TME) has recently gained more attention. Macrophages are a major component of the TME and play a critical role in DTX efficacy; however, the underlying action mechanisms remain unclear.
Methods
DTX chemotherapeutic efficacy was demonstrated via both macrophage depletion and C–C motif chemokine ligand 3 (Ccl3)-knockout transgenic allograft mouse model. Ccl3-knockdown and Ccl3-overexpressing breast cancer cell allografts were used for the in vivo study. Combination therapy was used to evaluate the effect of Ccl3 induction on DTX chemosensitivity. Vital regulatory molecules and pathways were identified using RNA sequencing. Macrophage phagocytosis of cancer cells and its influence on cancer cell proliferation under DTX treatment were assessed using an in vitro coculture assay. Serum and tumor samples from patients with breast cancer were used to demonstrate the clinical relevance of our study.
Results
Our study revealed that Ccl3 induced by DTX in macrophages and cancer cells was indispensable for the chemotherapeutic efficacy of DTX. DTX-induced Ccl3 promoted proinflammatory macrophage polarization and subsequently facilitated phagocytosis of breast cancer cells and cancer stem cells. Ccl3 overexpression in cancer cells promoted proinflammatory macrophage polarization to suppress tumor progression and increase DTX chemosensitivity. Mechanistically, DTX induced Ccl3 by relieving the inhibition of cAMP-response element binding protein on Ccl3 via reactive oxygen species accumulation, and Ccl3 then promoted proinflammatory macrophage polarization via activation of the Ccl3–C-C motif chemokine receptor 5–p38/interferon regulatory factor 5 pathway. High CCL3 expression predicted better prognosis, and high CCL3 induction revealed better DTX chemosensitivity in patients with breast cancer. Furthermore, both the Creb inhibitor and recombinant mouse Ccl3 significantly enhanced DTX chemosensitivity.
Conclusions
Our results indicate that Ccl3 induced by DTX triggers proinflammatory macrophage polarization and subsequently facilitates phagocytosis of cancer cells. Ccl3 induction in combination with DTX may provide a promising therapeutic rationale for increasing DTX chemosensitivity in breast cancer.
Keywords: Breast Neoplasms, Macrophages, Phagocytosis, Tumor Microenvironment, Cytokines
WHAT IS ALREADY KNOWN ON THIS TOPIC
Docetaxel (DTX) exhibits chemoimmunomodulating property, but the underlying mechanisms remain unclear.
WHAT THIS STUDY ADDS
C–C motif chemokine ligand 3 (Ccl3) is indispensable for DTX-induced proinflammatory macrophage polarization.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Ccl3 induction combined with DTX may be a promising therapeutic strategy to overcome chemoresistance and achieve better clinical outcomes in patients with breast cancer.
Background
Breast cancer is a serious threat to women’s health,1 and chemotherapy remains among the main therapeutic approaches, especially for triple-negative breast cancer, but resistance frequently occurs.2 Therefore, novel, targeted therapeutic strategies or approaches to reverse chemoresistance are urgently needed.
Docetaxel (DTX) is a classic antimitotic chemotherapy drug widely used for treating cancers, including breast cancer,3 non-small cell lung cancer,4 and prostate cancer.5 In addition to the canonical mechanisms mediating the antitumor activity of DTX, it has been reported to exhibit chemoimmunomodulating property. However, current studies on immunomodulating effects of DTX have focused on myeloid-derived suppressor cells.6 Other studies have demonstrated that DTX enhances the differentiation of monocytes into proinflammatory macrophages, while the detailed mechanisms remain elusive.7 Another recent study on paclitaxel proposed that paclitaxel exerted antitumor effects partially by reprogramming macrophages toward the antitumor profile in a TLR4-dependent manner.8 These studies suggest that canonical chemotherapy drugs might exert effects through non-canonical pathways by remodeling the tumor microenvironment (TME), and the immune cells in the TME are supposed to participate in the therapeutic effects of chemotherapies.
Tumor immunomicroenvironment (TIME) plays important roles in tumor initiation, progression and therapeutic responses, in which macrophages constitute a major immune cell component.9 Generally, macrophages are inclined to exhibit an alternatively activated M2-like phenotype with anti-inflammatory and immunosuppressive characteristics.10 11 Correspondingly, the classically activated M1-like macrophages have been documented to demonstrate proinflammatory and antitumor immune activities.12 As macrophages are highly heterogeneous with notable plasticity, the antitumor therapies of inducing macrophage phenotypic transition or enhancing proinflammatory macrophage polarization have been demonstrated to be efficient.8 10 13–15 Classical lipopolysaccharide or interferon-γ induces M1-like macrophages, while interleukin 4 (IL4) drives macrophages into the M2-like state.16 Many other cytokines/chemokines or pathways have also been reported to affect the polarization balance of macrophages.10 14 Chemokines can be classified into four major subfamilies, including CXC, C–C, (X)C, and CX3C chemokines, according to the arrangement pattern of cysteines at the NH2-terminal.17 Notably, C–C motif chemokine ligand 3 (Ccl3), also known as macrophage inflammatory protein-1α, has been recognized as a proinflammatory macrophage polarization marker.18 However, little is known regarding whether Ccl3 functionally regulates proinflammatory macrophage polarization and chemosensitivity.
The present study used both in vitro and in vivo models to show that DTX induced Ccl3 expression in both macrophages and cancer cells through reactive oxygen species (ROS)-mediated cAMP-response element binding protein (Creb) inhibition and that Ccl3 then triggered proinflammatory macrophage polarization via the Ccl3–C-C motif chemokine receptor 5 (Ccr5) –p38/interferon regulatory factor 5 (Irf5) axis. DTX-educated macrophages showed enhanced phagocytic ability toward cancer cells, including cancer stem cells (CSCs). Creb inhibitor 666–15 and recombinant mouse Ccl3 (rmCcl3) enhanced DTX chemosensitivity in breast cancer, providing a promising therapeutic rationale to overcome DTX chemoresistance in breast cancer treatment by inducing Ccl3 expression.
Methods
Reagents
DTX stock solution was purchased from Hengrui (H20020543, Jiangsu, China) and diluted to indicated concentrations immediately before treatment. The commercial Creb inhibitor 666–15 (HY-101120, MedChemExpress) was used at 200 ng/mL in vitro and 10 mg/kg in vivo. The rmCcl3 (450-MA-050/CF, R&D) was used at 5 µg/mice in vivo. N-acetyl-L-cysteine (NAC) (S0077, Beyotime) was used at 10 mM for indicated time. Hydrogen dioxide (H2O2) (CN51001, Chinasun Specialty Products Co, Ltd) was administered at indicated concentrations.
Cell culture
Py8119 cells were cultured in Han’s F12 medium (21 700–075, Gibco) supplemented with 10% FBS (vs500T, Ausbian), 1% streptomycin/penicillin (C0222, Beyotime), 50 µg/mL gentamycin (G1397, Sigma), 1 µg/mL hydrocortisone (H110523, Aladdin), 10 ng/mL epidermal growth factor (AF-315–09, PeproTech), and 5 µg/mL insulin (BS901, Biosharp). 4T1 cells were maintained in RPMI1640 medium (31 800–022, Gibco) supplemented with 5% FBS, 4 µg/mL gentamycin, and 1% streptomycin/penicillin. Mvt1 cells were cultured in DMEM (12 800–017, Gibco) medium supplemented with 10% FBS and 1% streptomycin/penicillin. Immortalized bone marrow-derived macrophages (iBMMs) described previously19 were maintained in DMEM medium supplemented with 10% FBS and 1% streptomycin/penicillin. HEK293T cells were maintained in DMEM medium supplemented with 10% FBS and 1% streptomycin/penicillin. L929 cells were cultured in RPMI1640 medium supplemented with 10% FBS and 1% streptomycin/penicillin. All cells were cultured at 37°C with 5% carbon dioxide. Cells were routinely tested to avoid mycoplasma contamination.
Plasmid construction
The coding sequences of Ccl3 and Creb were amplified from the cDNA of iBMMs using the indicated primer pairs (online supplemental table S1) and cloned into the pSIN-EF1α-IRES-puro vector or inducible pTRIPZ vector using a one-step cloning kit (C112, Vazyme). The shRNA vectors used in this study were constructed following our previously published SuperSH method.20 The pLKO.1-puro vector was purchased from Addgene (8453). The primers used for shRNA vector construction are listed in online supplemental table S2.
jitc-2021-003793supp002.pdf (43.3KB, pdf)
Human breast cancer patient serum and tumor samples
Serum samples were collected at Fudan University Shanghai Cancer Center from patients receiving Taxane-containing neoadjuvant chemotherapy (TNC) at earlier chemotherapy cycles (the second or third cycle) and later chemotherapy cycles (the fifth or sixth cycle). Tumor samples were provided by Harbin Medical University Cancer Hospital. Paired tumor samples of patients before or after TNC were collected, and the patients were separated into response and non-response cohort according to the Miller-Payne (MP) score. The MP score of the non-response cohort was 1 or 2, and the MP score of the response cohort was 3, 4 or 5.
In vivo mouse models
Female C57BL/6, FVB and BALB/c mice (6–7 weeks old) were purchased from the Vital River (Beijing, China). Ccl3-knockout (Ccl3 −/−) and control wild type (WT) mice were self-bred. All the mice were housed in individually ventilated cages under specific pathogen-free conditions. Mice were orthotopically injected at the fourth pair of mammary fat pads with syngeneic mouse breast cancer cell lines. The injected cell numbers for Py8119, Mvt1, and 4T1 were 5×104, 1×105 and 3×104 cells per site, respectively. The indicated amounts of cells were suspended in 50 µL FBS and mixed with an equal volume of Matrigel (354234, Corning) for injection. Tumor size was measured at the indicated times, and the volume was calculated using the formula V=4π/3 × (length/2) × (width/2)2. For DTX chemotherapy in vivo, mice were randomly separated into the indicated groups with similar average tumor volumes when the average diameter reached approximately 2–3 mm. DTX (40 mg/kg for Py8119, 20 mg/kg for 4T1 and Mvt1) or vehicle (Veh) was administered by intraperitoneal injection every 7 days for the indicated time points.
Macrophage clearance in mice
Clodronate liposomes were used for macrophage clearance in mice. Clodronate liposomes (CLD-Lp) (40 337ES10) and control PBS liposomes (PBS-Lp) (40 338ES10) were developed by Professor Nico van Rooijen at Vrije University and were purchased from Yeasen Biotechnology Co, Ltd (Shanghai, China). The day before cancer cell injection, 200 µL of CLD-Lp (5 mg/mL) or control PBS-Lp was injected intraperitoneally. Subsequently, liposomes were given every 4 days for four consecutive time points.
Transgenic mice and genotyping
FVB and BALB/c Ccl3−/− mice generation
FVB and BALB/c Ccl3 −/− mice were generated by injecting the CRISPR/Cas9-sgRNA complex into zygotes at Bangyao Biotech (Shanghai, China). The following two sgRNA sequences were used to delete a 673bp fragment surrounding the start codon of Ccl3: FB_sgRNA1 (atgttgagcaggtgacaga) and FB_sgRNA2 (caaaatagtcaacgatgaat). All newborn mice were subjected to PCR-based genotyping using the primer pairs forward_1191/518bp (cactgtcctacctcctcctg) and reverse_1191/518bp (ctcactggcaccctttagat), which generate a 1191bp band in WT mice and a 518bp band in knockout mice, respectively.
C57BL/6 Ccl3−/− mice generation
C57BL/6 Ccl3 −/− mice were generated by injecting the CRISPR/Cas9-sgRNA complex into zygotes at Xiamen University. The following two sgRNA sequences were used to delete a 62bp fragment surrounding the start codon of Ccl3: B6_sgRNA1 (ctgccggtttctcttagtc) and B6_sgRNA2 (caagaatacatcactgacc). All newborn mice were subjected to PCR-based genotyping using the primer pairs forward_274/212bp (aaccacagaggaagtcaga) and reverse_274/212bp (aacagcttataggagatggag), which generate a 274bp band in WT mice and a 212bp band in knockout mice, respectively.
Genotyping
Mouse tissues were boiled in 50 mM NaOH solution for 30 min. After a brief centrifugation, the supernatant was used for subsequent genotyping. PCR was performed according to the manufacturer’s recommendations (P213, Vazyme), and the product was separated in 2.0% agarose gel.
Quantitative real-time PCR (qRT-PCR) and RNA sequencing (RNA-Seq)
Total RNA was extracted with TRIzol reagent (9109, Takara), and cDNA was synthesized using HiScript II 1st Strand cDNA Synthesis Kit (R211-02, Vazyme). qRT-PCR was conducted using AceQ Universal SYBR qPCR Master Mix (Q511, Vazyme), and the signal was collected using a 7300Plus Real-Time PCR System (Applied Biosystems). The primers used for qRT-PCR are listed in online supplemental table S3. For RNA-Seq analysis, specific RNA libraries were prepared with standard procedure and sequenced at Singleron Biotechnology (Nanjing, China).
Western blotting and ELISA
Cells were lysed in RIPA lysis buffer (P0013C, Beyotime) on ice for 30 min and then centrifuged for 15 min (12 000 g, 4°C). The supernatant was quantified using the BCA kit (23227, Thermo) according to the manufacturer’s instructions. Equal amounts of total protein were boiled in 5× loading buffer for 10 min. The boiled protein samples were separated by SDS-PAGE and transferred to PVDF membrane (IPVH00010, Millipore). The membrane was blocked and incubated with the indicated primary antibodies overnight at 4°C, followed by washing in 1× TBST buffer. After secondary antibody incubation and washing, the signal was detected using chemiluminescence substrate (WBKLS0500, Millipore) on a chemiluminescence apparatus (SAGE, Beijing, China). The primary antibodies: Ccl3 (1:100, AF-450-NA, R&D), Gapdh (1:2000, HC301, TransGen), iNos (1:1000, 13120, CTS), Cox2 (1:1000, 160106, Cayman), p38 (1:1000, 9212, CST), p-p38 (1:1000, 4631, CST), Erk (1:1000, 4695, CST), p-Erk (1:1000, 9101, CST), Jnk (1:1000, 9252, CST), p-Jnk (1:1000, 9251, CST), Irf5 (1:1000, 10 547-1-AP, Proteintech), Creb (1:1000, 9197, CST), and p-Creb (1:1000, 9198, CST). The secondary antibodies: peroxidase-conjugated donkey antigoat (1:5000, SA00001-3, Proteintech), peroxidase-conjugated goat antirabbit (1:5000, HS101-01, TransGen), and peroxidase-conjugated goat antimouse (1:5000, HS201-01, TransGen). The uncropped blots of the western blotting experiments are shown in online supplemental figures S9–12. The CCL3 levels in patient sera were determined and calculated using an ELISA kit (ELH-MIP1a, RayBio) according to standard instructions.
jitc-2021-003793supp012.pdf (582.5KB, pdf)
jitc-2021-003793supp013.pdf (540.4KB, pdf)
jitc-2021-003793supp014.pdf (318.7KB, pdf)
jitc-2021-003793supp015.pdf (401.8KB, pdf)
Bone marrow-derived macrophages (BMDMs) induction and reconstitution
BM cells of mice were isolated and induced into BMDMs as previously described.21 Briefly, female mice (6–7 weeks old) were sacrificed and sterilized in 75% alcohol for 10 min. Subsequently, the BM in the femur and tibia was flushed out and treated with red blood cell lysis buffer (00-4333-57, Invitrogen). BM cells were then cultured in 60% RPMI1640, 10% FBS, 30% L929 conditioned medium (CM; with macrophage colony stimulating factor) and 1% streptomycin/penicillin at a density of 3×106 per 6 cm dish for 7 days, and the medium was changed every 2–3 days. After 7 days of induction, the obtained BMDMs were used for subsequent experiments. For BMDM reconstitution in Ccl3 −/− mice, 1×106 BMDMs derived from WT or Ccl3 −/− mice were adoptively transferred by intravenous injection the day before DTX administration, which was repeated every 3 days for six consecutive time points.
Flow cytometry
A total of 0.5 or 1 million cells were used for fluorescence-conjugated antibody staining or aldehyde dehydrogenase (ALDH) activity analysis. Specifically, for digested tumor tissue cells, as immune cells were a minority in the whole cell population, 2 million cells were used for fluorescence-conjugated antibody staining. For immune cell staining, antimouse CD16/32 (1:100, 101302, BioLegend) was used for blocking prior to the indicated antibody staining. Antibody staining was performed on ice for 30 min. ALDH activity analysis was conducted using the ALDEFLUOR kit (01700, STEMCELL Technologies), following the manufacturer’s instructions. After staining, the cells were suspended in 1× PBS containing 2% FBS with 4′,6-diamidino-2-phenylindole (DAPI) (D9564, Sigma) for detection. A Moflo Astrios or CytoFlex (Beckman Coulter) instrument was used for flow cytometry analysis, and a Moflo Astrios instrument was used for fluorescence-activated cell sorting. The following fluorescence-conjugated antibodies were purchased from BioLegend: antimouse CD45-FITC (1:100, 103108), antimouse F4/80-APC (1:80, 123116), antimouse MHCII-APC/Cy7 (1:80, 107628), antimouse CD11b-PE (1:80, 101208), antimouse CD140b-PE (1:100, 136006), antimouse CD19-PE (1:80, 115507) and antimouse CD3-APC/Cy7 (1:80, 100221). The following fluorescence-conjugated antibodies were purchased from BD Biosciences: antimouse CD31-PE (1:100, 553373) and antimouse CD45-PE (1:100, 553081). Intracellular ROS levels were determined using 2′,7′-dichlor-odihydrofluorescein diacetate probe (287810, Sigma-Aldrich) according to the manufacturer’s instructions. Further data analysis was performed using FlowJo software.
Lentiviral package and infection
Lentiviruses were produced in HEK293T cells in 6 cm culture dishes. Plasmids (8 µg) containing 2 µg psPAX2 (12260, Addgene), 2 µg pMD2.G (12259, Addgene) packaging plasmids, and 4 µg core lentiviral vector plasmids were mixed with 24 µg polyethylenimine (23966, Polysciences) in 300 µL Opti-MEM medium (31 985–062, Gibco) at room temperature (RT) for 20 min. The mixture was then added to HEK293T cells. The medium containing lentiviral particles was filtered through 0.45 µm filters 48 hours after transfection. Target cells were infected by incubation with virus-containing supernatant for 24 hours in the presence of polybrene (8 µg/mL, H9268, Sigma).
In vitro coculture
A total of 0.25 million cancer cells (Py8119-mCherry, Mvt1-mCherry or 4T1-mCherry) and 0.25 million iBMMs were preseeded together in 6 cm culture dishes to allow adherence overnight. DTX (30 nM for Py8119, 10 nM for Mvt1, and 5 nM for 4T1) was added to the coculture system for 2 days. Cells were counted, and 0.5 million cells were used for flow cytometry analysis. The absolute number of cancer cells after coculture with DTX was calculated according to the total cell number and the percentage of cancer cells in the flow cytometry analysis. For cancer cells cocultured with BMDMs, BMDMs were obtained by following the standard procedure for BMDM induction. Afterwards, 0.3 million cancer cells (Py8119, Mvt1 or 4T1) were seeded into BMDMs following overnight adherence. The subsequent experimental procedures were the same as cancer cell and iBMM coculture system, except that the mixed cells after coculture were stained with antimouse CD45-FITC to distinguish cancer cells and BMDMs.
DTX chemosensitivity assay
A total of 1000 4T1 cells were seeded in 96-well culture plates for overnight adherence. The next day, medium containing a series of concentrations of DTX diluted in gradient was added. The maximal DTX concentration for 4T1 was 270 nM, with threefold dilution for nine different concentrations. The cells were cultured for 3 days and viability was determined by MTT assay.
Immunohistochemistry (IHC) staining
Tumors were fixed in 4% formaldehyde for 1 day. After dehydration with gradient alcohol, paraffin embedding was performed. For IHC staining, paraffin sections were dewaxed in xylene for three times and rehydrated in graded alcohol. Endogenous peroxidase was inactivated using 3% H2O2 diluted in methanol at 37°C for 15 min. Antigen retrieval was then performed, and the tissues were blocked with non-immune serum containing 0.3% Triton X-100 (A600198, Sangon) for 20 min at RT. The primary antibody was incubated at 4°C overnight, followed by washing three times in 1× PBS for 5 min. The secondary antibody was incubated at RT for 20 min, and then washed in the same way. Positive staining was visualized using DAB kit (DAB-0031, MaxVision), and cell nuclei were stained with hematoxylin (ZLI-9610, ZSGB-BIO). The slides were dehydrated with gradient alcohol and xylene, then sealed with neutral balsam (BL085A-100g, Biomiky). A microscope (OLYMPUS BX43, Japan) with a 40× lens was used for observation and photograph. CCL3 expression was scored semiquantitatively using a manual histo-score (H-score) methodology based on staining intensity and the percentage of positive tumor cells or immune cells. Strongly staining scored 3, moderately staining scored 2, weakly staining scored 1, and negative staining scored 0. The H-score of CCL3 expression was obtained using the following formula: (3× percentage of strongly stained +2× percentage of moderately stained +percentage of weakly stained)×100. The primary antibodies used for IHC staining: CD68 (1:50, sc-9139, Santa Cruz), Cox2 (1:300, 160106, Cayman), and CCL3 (1:50, GTX52606, GeneTex). The secondary antibodies: peroxidase-conjugated goat antimouse/rabbit (KIT-5010, MaxVision). For HE staining, the sections were dewaxed, rehydrated, and stained with hematoxylin and eosin.
iBMM chemotaxis assay
Cancer cells or CM from cancer cells was used to actuate iBMM chemotaxis and placed in the lower well of a 24-well culture plate. A total of 2×105 cancer cells or 600 µL CM were used. iBMMs (3×104) suspended in serum-free DMEM were seeded in the upper chambers (8 µM, 353097, Corning). After 36 hours, the iBMMs were fixed with 4% formaldehyde at RT for 15 min and subsequently stained with 1% crystal violet.
Phagocytosis assay
iBMMs were pretreated with DTX (10 nM) for 2 days. Then DTX was washed out, and cells were deprived of FBS for 3 hours. Total, sorted ALDH+ and ALDH− 4T1 or Py8119 cells were labeled with carboxyfluorescein diacetate-succinimidyl ester (CFDA-SE) (C0051, Beyotime) and added to the prepared iBMMs. After 6 hours of coculture, cells were dissociated and stained with F4/80-APC to label iBMMs for flow cytometry analysis. For direct visualization by confocal microscope (TCS SP5 II, Leica), GFP-labeled iBMMs or mCherry-labeled cancer cells were used. At the end of the experiment, cells were fixed and stained with DAPI. Phagocytosis is represented as the percentage of CFDA-SE+APC+ or GFP+mCherry+ cells in all macrophages and normalized to the control. The ratios of macrophages and total or sorted cancer cells were 2:1 and 5:1, respectively.
Statistical analysis
All data are presented as mean±SEM. In vitro experiments were independently repeated at least for two to three times. Differences between two groups were analyzed using two-tailed Student’s t-test. Two-way analysis of variance was used for multiple comparisons. Kaplan-Meier survival curves were analyzed using two-sided log-rank test. Statistical significance was set at p<0.05, and the p values are indicated with asterisks as follows: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; NS, no significance. All data were analyzed using GraphPad Prism V.6 software.
Results
Ccl3 is significantly induced in macrophages by DTX
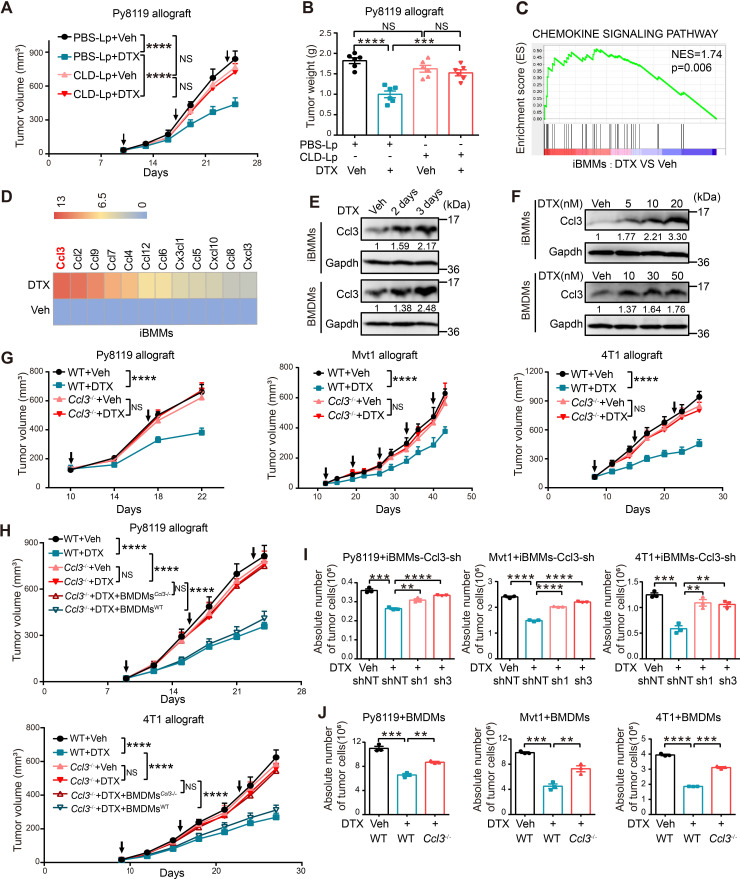
Emerging studies have demonstrated that chemotherapies stimulate macrophage infiltration and polarization.8 22 To explore the role of macrophages in DTX efficacy, we depleted macrophages in C57BL/6 mice using CLD-Lp, with PBS-Lp as control. Then, syngeneic mouse breast cancer cell line Py8119 was orthotopically injected into the mice, followed by DTX treatment (online supplemental figure S1A). Effective macrophage clearance was verified by IHC staining of CD68, a pan-macrophage marker (online supplemental figure S1B). Notably, the chemotherapeutic efficacy of DTX was almost abolished on macrophage clearance, although DTX significantly inhibited tumor growth in control mice (figure 1A, B, online supplemental figure S1C).
Figure 1.
Ccl3 induced by DTX in macrophages is indispensable for its chemotherapeutic efficacy. (A–B) C57BL/6 mice with macrophage depletion were subjected to DTX treatment experiments (n=5). (A) Tumor volume was measured, and tumor growth curves are shown. The arrows indicate the administration time of DTX. (B) After mouse sacrifice, the tumors were collected and weighed. (C) GSEA based on RNA-Seq results of iBMMs treated with Veh and DTX (10 nM). The normalized enrichment score (NES) and p value are shown. (D) Significantly upregulated chemokines in RNA-Seq analysis were screened and shown as heatmap. The order was rearranged by variation based on the fold change. (E) Western blotting was performed to detect Ccl3 expression after DTX (10 nM for iBMMs and 30 nM for BMDMs) treatment for 2 and 3 days in iBMMs and BMDMs. (F) Western blotting was performed to detect Ccl3 expression after DTX treatment at indicated concentrations in iBMMs and BMDMs. (G) WT or Ccl3 −/− mice of three individual strains, including C57BL/6, FVB and BALB/c, were orthotopically transplanted with syngeneic breast cancer cell line (Py8119, Mvt1 and 4T1, respectively) for DTX treatment experiments (n=8). The tumor volume was measured, and tumor growth curves are shown. The arrows indicate the time of DTX administration. (H) WT or Ccl3 −/− C57BL/6 and BALB/c mice were orthotopically transplanted with syngeneic breast cancer cell line (Py8119 and 4T1, respectively) for DTX treatment experiments (n=5). Ccl3 −/− mice with DTX treatment were reconstituted with BMDMs from syngeneic WT or Ccl3 −/− mice. The tumor volume was measured, and tumor growth curves are shown. The arrows indicate the time of DTX administration. (I) Py8119-mCherry, Mvt1-mCherry or 4T1-mCherry were cocultured with control iBMMs or iBMMs knocking down Ccl3 under DTX treatment for 2 days. (J) Py8119, Mvt1 or 4T1 were cocultured with BMDMs derived from WT or Ccl3 −/− mice under DTX treatment for 2 days. The cells were collected and stained with CD45-FITC for flow cytometry analysis. Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) was used as the loading control. Data are presented as mean±SEM. **P<0.01, ***p<0.001, ****p<0.0001; NS, no significance. BMDMs, bone marrow-derived macrophages; BMDMs Ccl3−/− , BMDMs derived from Ccl3 −/− mice; BMDMsWT, BMDMs derived from WT mice; Ccl3, C–C motif chemokine ligand 3; CLD-Lp, clodronate liposomes; DTX, docetaxel; NES, normalized enrichment score; Gapdh, glyceraldehyde-3-phosphate dehydrogenase; GSEA, gene set enrichment analysis; iBMM, immortalized bone marrow-derived macrophages; PBS-Lp, PBS liposomes; RNA-Seq, RNA sequencing; sh, short hairpin RNA; shNT, short hairpin RNA of non-target; Tbp, TATA-binding protein; Veh, vehicle; WT, wild type.
jitc-2021-003793supp004.pdf (5.1MB, pdf)
We then treated murine iBMMs with DTX and performed RNA-Seq to explore the underlying mechanisms. Since chemokines are important message mediators between immune cells23 and are responsive to chemotherapies,24 25 we studied whether DTX treatment changed the profile of chemokine expression. Gene set enrichment analysis (GSEA) revealed that DTX treatment activated the chemokine signaling pathway in iBMMs (figure 1C). Specifically, all significantly upregulated chemokines in RNA-Seq were screened. As immune cells secrete large amounts of chemokines, we rearranged the order of upregulated chemokines by variation in iBMMs based on the fold change to eliminate possible bias for the heatmap. Intriguingly, Ccl3 was the most upregulated chemokine in iBMMs after DTX treatment (figure 1D). Ccl3 upregulation was then confirmed in iBMMs and BMDMs at both protein and mRNA levels, where both a time-dependent and concentration-dependent induction patterns of Ccl3 by DTX were observed (figure 1E, F, online supplemental figure S1D, E).
Ccl3 is a chemokine involved in inflammation as well as antiviral and antibacterial infections whose function is strongly associated with macrophages.26 The expression levels of Ccl3 and its two main receptors Ccr1 and Ccr527 were analyzed in the sorted total cells (total, cancer cells and immune cells) and different immune cell subpopulations from tumors, including CD45+ (all immune cells), CD45+F4/80+ (macrophages, Mac), CD45+CD19+ (B cells), and CD45+CD3+ (T cells). We observed that Ccl3 and its receptors were mainly expressed in macrophages, indicating a possible autocrine role for Ccl3 in macrophages. In addition, T cells showed slightly lower Ccl3 expression than macrophages (online supplemental figure S1F). Collectively, these results demonstrated that the chemotherapeutic efficacy of DTX in vivo was dependent on macrophages and that Ccl3 was the most significantly upregulated chemokine in macrophages on DTX treatment.
Ccl3 enhances chemotherapeutic efficacy of DTX in breast cancer
To further elucidate the role of Ccl3 in DTX chemotherapeutic efficacy in breast cancer in vivo, we generated Ccl3 −/− mice of three distinct backgrounds: C57BL/6, FVB and BALB/c. Knockout efficiency was confirmed for all three mouse strains (online supplemental figure S2A). Ccl3 deficiency had no obvious influence on the expression of other C–C chemokines, except Ccl17 (online supplemental figure S2B), which was not induced by DTX (figure 1D). Murine breast cancer cell lines Py8119, Mvt1 and 4T1 were orthotopically injected into syngeneic WT (Ccl3 +/+) and Ccl3 −/− mice (C57BL/6, FVB and BALB/c, respectively), followed by DTX treatment. Our data showed that DTX remarkably suppressed tumor growth in WT mice, whereas this antitumor effect was largely abolished when Ccl3 was knocked out (figure 1G, online supplemental figure S2C–E). HE staining of paraffin-embedded tumor sections showed no obvious differences between WT and Ccl3−/− mice, except that the necrotic areas of tumors were larger in the WT mice with DTX treatment (online supplemental figure S2F). Reconstitution of BMDMs from WT mice to Ccl3 −/− mice largely restored the antitumor efficacy of DTX (figure 1H, online supplemental figure S2G, H). Furthermore, iBMMs with Ccl3 knockdown (online supplemental figure S2I) or BMDMs with Ccl3 knockout were cocultured with Py8119, Mvt1 or 4T1 cells in the presence of DTX. The inhibitory effect of DTX on cancer cell proliferation was remarkably decreased when Ccl3 expression was interfered in iBMMs or BMDMs (figure 1I, J). Taken together, these results indicated that Ccl3 induced by DTX in macrophages was indispensable for DTX chemotherapeutic efficacy in breast cancer.
jitc-2021-003793supp005.pdf (2.2MB, pdf)
Ccl3 promotes DTX-induced proinflammatory macrophage polarization by activating Ccr5–p38/Irf5 signaling pathway
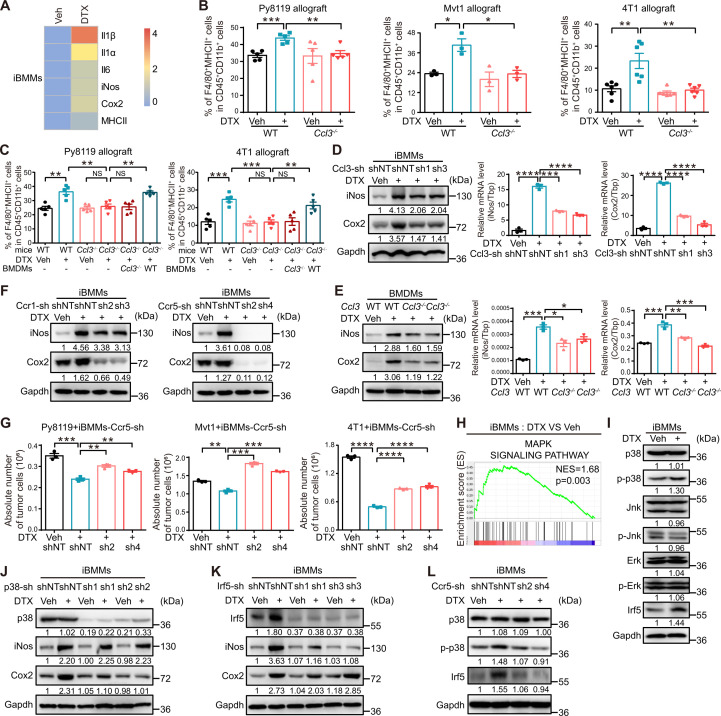
As the previous results demonstrated that macrophages were indispensable for the antitumor activity of DTX in vivo, we proposed that DTX might favor proinflammatory polarization to facilitate its chemotherapeutic efficacy. We observed that proinflammatory polarization markers, including major histocompatibility complex class II (MHCII), cyclooxygenase 2 (Cox2), inducible nitric oxide synthase (iNos), interleukin 6 (Il6), interleukin 1α (Il1α), and interleukin 1β (Il1β), were significantly upregulated in the RNA-Seq data of iBMMs treated with DTX, indicating an enriched proinflammatory profile after DTX treatment (figure 2A). Furthermore, F4/80+MHCII+ macrophages derived from Py8119, Mvt1 and 4T1 allograft tumors in both WT and Ccl3 −/− mice with or without DTX treatment were analyzed by flow cytometry, as F4/80+MHCII+ has been widely used to mark proinflammatory macrophages.28 A significant increase in the percentage of F4/80+MHCII+ macrophages after DTX treatment was observed in WT mice, but not in Ccl3 −/− mice (figure 2B, online supplemental figure S3A). This observation was further confirmed by IHC staining of paraffin-embedded tumor sections with two other proinflammatory polarization markers, CD68 and Cox2 (online supplemental figure S3B). The proinflammatory macrophage inducing ability of DTX was largely restored in Ccl3 −/− mice by reconstitution of BMDMs derived from WT mice (figure 2C). Furthermore, Ccl3-knockdown iBMMs were treated with DTX, and we found that the induction of both iNos and Cox2, two typical proinflammatory polarization markers, in the presence of DTX was diminished by Ccl3 knockdown (figure 2D). Similar results were obtained in BMDMs from Ccl3−/− mice (figure 2E). These findings suggested that DTX promoted proinflammatory macrophage polarization in a Ccl3-dependent manner.
Figure 2.
Ccl3 promotes DTX-induced proinflammatory macrophage polarization by activating Ccr5–p38/Irf5 signaling pathway. (A) Upregulated proinflammatory macrophage markers in RNA-Seq analysis of iBMMs treated with DTX (10 nM) are shown as heatmap. (B–C) Flow cytometry was used to analyze the percentage of F4/80+MHCII+ proinflammatory macrophages in cells isolated from tumors in figure 1G and H, respectively. (D) iBMMs with Ccl3 knockdown were subjected to DTX (10 nM) treatment for 2 days. Western blotting and qRT-PCR were performed to detect iNos and Cox2 expression. (E) BMDMs derived from FVB WT or Ccl3 −/− mice were subjected to DTX (50 nM) treatment for 2 days. Western blotting and qRT-PCR were performed to detect iNos and Cox2 expression. (F) iBMMs with Ccr1 or Ccr5 knockdown were subjected to DTX (10 nM) treatment for 2 days to determine iNos and Cox2 expression via western blotting. (G) Py8119-mCherry, Mvt1-mCherry or 4T1-mCherry were cocultured with iBMMs knocking down Ccr5 under DTX treatment for 2 days. (H) GSEA based on RNA-Seq results of iBMMs treated with Veh and DTX (10 nM). The NES and p value are shown. (I) MAPK family and Irf5 expression levels were determined via western blotting in iBMMs treated with DTX (10 nM) for 2 days. (J–K) iBMMs with p38 or Irf5 knockdown were subjected to DTX (10 nM) treatment for 2 days. The cells were collected and lysed for western blotting to detect iNos and Cox2 expression. (L) iBMMs with Ccr5 knockdown were subjected to DTX (10 nM) treatment for 2 days. The cells were collected for western blotting to detect p38 and Irf5 expression. Gapdh was used as the loading control. TATA-binding protein (Tbp) was used as the internal control. Data are presented as mean±SEM. *P<0.05, **p<0.01, ***p<0.001, ****p<0.0001; NS, no significance. BMDMs, bone marrow-derived macrophages; Ccl3, C–C motif chemokine ligand 3; Cox2, cyclooxygenase; Ccr, C–C motif chemokine receptor; DTX, docetaxel; Erk, extracellular-regulated protein kinases; Gapdh, glyceraldehyde-3-phosphate dehydrogenase; GSEA, gene set enrichment analysis; iBMM, immortalized bone marrow-derived macrophages; Il1α, interleukin 1α; Il1β, interleukin 1β; Il4, interleukin 4; Il6, interleukin 6; iNos, inducible nitric oxide synthase; Irf5, interferon regulatory factor 5; Jnk, c-Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; MHCII, major histocompatibility complex class II; NES, normalized enrichment score; qRT-PCR, quantitative real-time PCR; RNA-Seq, RNA sequencing; sh, short hairpin RNA; shNT, short hairpin RNA of non-target; Tbp, TATA-binding protein; Veh, vehicle; WT, wild type.
jitc-2021-003793supp006.pdf (4.7MB, pdf)
As a classic chemokine, Ccl3 initiates downstream signal transduction by binding to its cognate receptors. Hence, stable iBMM cell lines with Ccr1 or Ccr5 knockdown were established (online supplemental figure S3C) and treated with DTX. Ccr5 knockdown almost completely blocked DTX-induced upregulation of iNos and Cox2, while Ccr1 knockdown only slightly restrained the upregulation (figure 2F). Coculture experiments were performed using three breast cancer cell lines with Ccr5-knockdown iBMMs in the presence of DTX. Ccr5 knockdown reversed the antiproliferative effect of DTX on cancer cells (figure 2G), which was consistent with our previous results obtained in Ccl3-knockdown iBMMs (figure 1I).
To further explore the downstream mechanisms of Ccl3–Ccr5 signaling, RNA-Seq data of DTX-treated iBMMs were analyzed, revealing that DTX activated the mitogen-activated protein kinase (MAPK) signaling pathway (figure 2H). MAPK family consists of extracellular-regulated protein kinases (Erk), p38 and c-Jun N-terminal kinase (Jnk),29 and activation of p38 and Jnk has been reported to play prominent roles in proinflammatory macrophage polarization.30 31 In addition, transcriptional factor Irf5, which is critical for proinflammatory macrophage polarization,32 was in the upregulated gene list in RNA-Seq data. Hence, the expression levels of total and phosphorylated p38 (p-p38), Jnk (p-Jnk) and Erk (p-Erk), as well as total Irf5, were determined by western blotting in iBMMs after DTX treatment. P-p38 and total Irf5 were significantly upregulated after DTX treatment (figure 2I).
To determine the roles of p38 and Irf5 in DTX-induced proinflammatory macrophage polarization, p38 or Irf5 was individually knocked down in iBMMs (figure 2J, K). On DTX treatment, the protein levels of iNos and Cox2 were determined, and our data showed that the upregulation of Cox2 and iNos induced by DTX was dependent on p38 (figure 2J) and Irf5 (figure 2K), respectively. Furthermore, when Ccr5 was knocked down in iBMMs, the DTX-induced upregulation of p-p38 and Irf5 was reversed (figure 2L). These results indicated that p38 and Irf5 were the downstream effectors of Ccl3–Ccr5 signaling in DTX-induced proinflammatory macrophage polarization.
Ccl3 is indispensable for DTX-enhanced macrophage phagocytosis of cancer cells, including CSCs
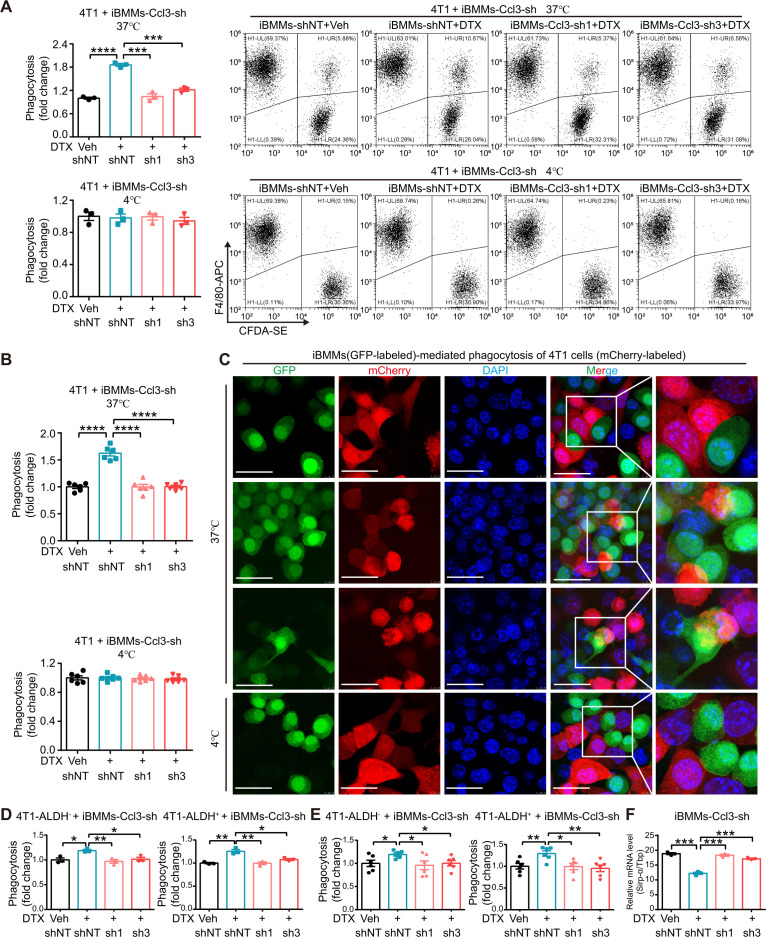
Proinflammatory macrophages have been documented to exhibit stronger phagocytic ability.33 Cancer cells, especially CSCs, have evolved corresponding mechanisms to avoid being engulfed.34 CSCs represent a subpopulation with self-renewal and multidifferentiation potential abilities, which are profoundly engaged in tumor metastasis, therapeutic resistance, and recurrence.35 ALDH has been widely recognized as a CSC marker in breast cancer.36 To evaluate the phagocytic ability of DTX-induced proinflammatory macrophages of bulk cancer cells and CSCs, we performed phagocytosis assay using both flow cytometry analysis and directly visualized confocal microscope photography by coculturing DTX-pretreated iBMMs with total, sorted ALDH− and ALDH+ cancer cells of 4T1 and Py8119. DTX promoted iBMM-mediated phagocytosis of total (figure 3A–C, (online supplemental figure S4A–C), ALDH− and ALDH+ (figure 3D, E, online supplemental figure S4D, E) cancer cells, where the increase in phagocytosis of ALDH+ cancer cells was slightly higher than that of ALDH− cancer cells (figure 3D, E, online supplemental figure S4D, E). Ccl3 knockdown in iBMMs dramatically reversed this effect (figure 3A–E, online supplemental figure S4A–E). The phagocytosis assay was conducted in parallel at 4°C (figure 3A–C, online supplemental figure S4A–C, lower panel). Signal regulatory protein-α (Sirp-α) is a critical inhibitory immune checkpoint on macrophages, and its activation by binding to CD47 on the surface of cancer cells suppresses macrophage phagocytosis.37 Surprisingly, Sirp-α was inhibited by DTX administration, and this inhibition was abolished by Ccl3 knockdown (figure 3F). These data indicated that Ccl3 was indispensable for DTX-enhanced macrophage-mediated phagocytosis of cancer cells, including CSCs.
Figure 3.
Ccl3 is indispensable for DTX-enhanced macrophage phagocytosis of cancer cells, including CSCs. (A) DTX-pretreated iBMMs were cocultured with total 4T1 cells at both 37°C (upper panel) and 4°C (lower panel) for in vitro phagocytosis assay and analyzed by flow cytometry. (B) DTX-pretreated iBMMs (GFP-labeled) were cocultured with total 4T1 cells (mCherry-labeled) at both 37°C (upper panel) and 4°C (lower panel) for in vitro phagocytosis assay and analyzed by directly visualized confocal microscope photography. (C) Representative images for figure 3B. The different stages of phagocytosis showing macrophages from contacting to internalizing cancer cells were observed at 37°C. Scale bar: 30 µm. (D–E) In vitro phagocytosis assay was performed using DTX-pretreated iBMMs cocultured with sorted ALDH− or ALDH+ 4T1 cells, and analyzed by flow cytometry analysis (D) and directly visualized confocal microscope photography (E). (F) iBMMs with Ccl3 knockdown were subjected to DTX (10 nM) treatment for 2 days to quantify Sirp-α expression by qRT-PCR. Tbp was used as the internal control. Data are presented as mean±SEM. *P<0.05, **p<0.01, ***p<0.001, ****p<0.0001. ALDH, aldehyde dehydrogenase; Ccl3, C–C motif chemokine ligand 3; CSCs, cancer stem cells; DAPI, 4′,6-diamidino-2-phenylindole; DTX, docetaxel; iBMMs, immortalized bone marrow-derived macrophages; qRT-PCR, quantitative real-time PCR; sh, short hairpin RNA; shNT, short hairpin RNA of non-target; Sirp-α, signal regulatory protein-α; Tbp, TATA-binding protein; Veh, vehicle.
jitc-2021-003793supp007.pdf (7.8MB, pdf)
Ccl3 is significantly induced in cancer cells by DTX and increases DTX chemotherapeutic efficacy
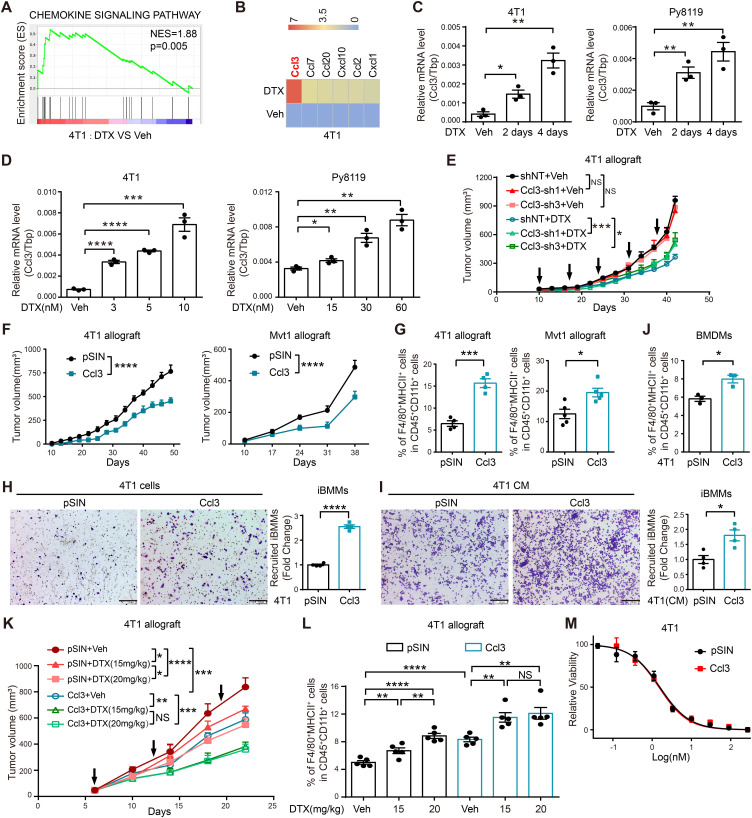
Cancer and immune cells are tightly tangled in the TME. Given that macrophages were remodeled by DTX and further inhibited tumor progression, we questioned whether DTX also provoked novel alterations in cancer cells. Therefore, 4T1 cells treated with DTX were subjected to RNA-Seq analysis. The results revealed that DTX activated the chemokine signaling pathway in cancer cells (figure 4A). Strikingly, Ccl3 was the most upregulated chemokine (figure 4B), and this upregulation was time and dose dependent (figure 4C, D). 4T1 stable cell line with Ccl3 knockdown was established (online supplemental figure S5A) and orthotopically injected into BALB/c mice, followed by DTX treatment. Ccl3 knockdown in cancer cells alone barely affected tumor growth in vivo but reduced the tumor-suppressing effect of DTX (figure 4E, online supplemental figure S5B), although this effect was weaker than that of Ccl3 −/− in TME (figure 1G). This observation indicated that Ccl3 of cancer cells also facilitated DTX chemotherapeutic efficacy.
Figure 4.
Ccl3 is significantly induced in cancer cells by DTX and increases DTX chemosensitivity in vivo. (A) GSEA based on RNA-Seq results of 4T1 treated with Veh and DTX (5 nM). The NES and p value are shown. (B) Significantly upregulated chemokines in RNA-Seq analysis were screened and presented as heatmap. The order was arranged by fold change. (C) qRT-PCR was performed to quantify Ccl3 expression after DTX (5 nM for 4T1 and 30 nM for Py8119) treatment for 2 and 4 days in 4T1 and Py8119. (D) qRT-PCR was performed to quantify Ccl3 expression after DTX treatment at indicated concentrations. (E) 4T1 cells with Ccl3 knockdown were orthotopically injected into BALB/c mice for DTX treatment experiments (n=5). The tumor volume was measured, and tumor growth curves are shown. The arrows indicate the time of DTX administration. (F) 4T1 or Mvt1 cells stably overexpressing Ccl3 and pSIN (Vector) control were orthotopically injected into BALB/c or FVB mice (n=5). Then, the tumor volume was measured, and tumor growth curves are shown. (G) Flow cytometry analysis of the F4/80+MHCII+ proinflammatory macrophage percentage in the tumors harvested in figure 4F. (H–I) Effects of Ccl3 overexpressing cancer cells (H) or the CM from Ccl3 overexpressing cancer cells (I) on iBMM migration were observed via chemotaxis experiment. Statistical graphs represent the fold change of recruited iBMMs. Scale bar: 200 µm. (J) BMDMs were cocultured with 4T1 overexpressing Ccl3 for 2 days and subsequently analyzed by flow cytometry for F4/80+MHCII+ proinflammatory macrophages. (K) 4T1 cells stably overexpressing Ccl3 and pSIN (Vector) control were orthotopically injected into BALB/c mice for different dosages of DTX treatment experiments (n=5). The tumor volume was measured, and tumor growth curves are shown. (L) Flow cytometry assay was used to analyze the percentage of F4/80+MHCII+ proinflammatory macrophages in the tumors harvested in figure 4K. (M) 4T1 overexpressing Ccl3 and pSIN control cell lines were subjected to DTX susceptibility assay. Tbp was used as the internal control. Data are presented as mean±SEM. *P<0.05, **p<0.01, ***p<0.001, ****p<0.0001; NS, no significance. BMDMs, bone marrow-derived macrophages; Ccl3, C–C motif chemokine ligand 3; CM, conditioned medium; DTX, docetaxel; GSEA, gene set enrichment analysis; iBMM, immortalized bone marrow-derived macrophages; MHCII, major histocompatibility complex class II; NES, normalized enrichment score; qRT-PCR, quantitative real-time PCR; RNA-Seq, RNA sequencing; sh, short hairpin RNA; shNT, short hairpin RNA of non-target; Tbp, TATA-binding protein; Veh, vehicle.
jitc-2021-003793supp008.pdf (10.1MB, pdf)
DTX-induced Ccl3 creates a positive feedback effect loop between macrophages and cancer cells
The simultaneous induction of Ccl3 by DTX treatment in both macrophages and cancer cells prompted us to explore their potential effects on each other via Ccl3. First, CM from cancer cells after DTX treatment facilitated Ccl3 expression and proinflammatory polarization of macrophages (online supplemental figure S6A). Then, CM from Ccl3-overexpressing 4T1 and Mvt1 cells (online supplemental figure S5C) was used to stimulate iBMMs Ccl3 expression and proinflammatory macrophage polarization of iBMMs were also enhanced (online supplemental figure S6B). Similarly, CM from DTX-treated iBMMs and Ccl3-overexpressing iBMMs induced Ccl3 expression in cancer cells (online supplemental figure S6C–E). These results suggested that Ccl3 expression between macrophages and cancer cells had a positive feedback amplification effect under DTX treatment, which might further augment the chemotherapeutic efficacy of DTX.
jitc-2021-003793supp009.pdf (277.6KB, pdf)
Ccl3 overexpression in cancer cells suppresses tumor progression and increases DTX chemosensitivity in vivo
Given that DTX also induced Ccl3 in cancer cells, we wondered whether Ccl3 overexpression in cancer cells inhibited tumor progression by promoting proinflammatory polarization of macrophages in the TIME. Hence, orthotopic transplantation with Ccl3-overexpressing 4T1 and Mvt1 cells was performed, where Ccl3 overexpression remarkably inhibited tumor growth in vivo (figure 4F, online supplemental figure S5D). Furthermore, analysis of the percentage of F4/80+MHCII+ macrophages in tumors revealed that Ccl3 overexpression in cancer cells upregulated the percentage of proinflammatory macrophages in the TIME (figure 4G). In addition, IHC staining demonstrated that proinflammatory-polarized macrophages (CD68+Cox2+) were upregulated in the TIME of Ccl3-overexpressing group (online supplemental figure S5E). In vitro macrophage chemotaxis experiments conducted with Ccl3-overexpressing 4T1 cells or their CM as the attractant demonstrated that Ccl3 overexpression in cancer cells enhanced macrophage recruitment (figure 4H, I). Coculturing assays with BMDMs and 4T1 cells revealed that Ccl3 overexpression in cancer cells promoted proinflammatory polarization of macrophages (figure 4J).
Since Ccl3 overexpression promoted the formation of a tumor-suppressive microenvironment, we wondered whether this effect could coordinate with DTX chemotherapy to inhibit tumor progression. Control or Ccl3-overexpressing 4T1 cells were transplanted into mice, and different doses of DTX were administered. Strikingly, although the antitumor effect of DTX was positively correlated with the doses in the control groups, Ccl3 overexpression resulted in a lower dose of DTX as effective as the higher dose (figure 4K, L). These results suggested that Ccl3 sensitized cancer cells to DTX in vivo, and this effect relied on TIME, as Ccl3 overexpression had no influence on DTX chemosensitivity in vitro (figure 4M). Collectively, Ccl3 overexpression in cancer cells promoted proinflammatory macrophage polarization in TIME to suppress tumor progression and increase DTX chemosensitivity.
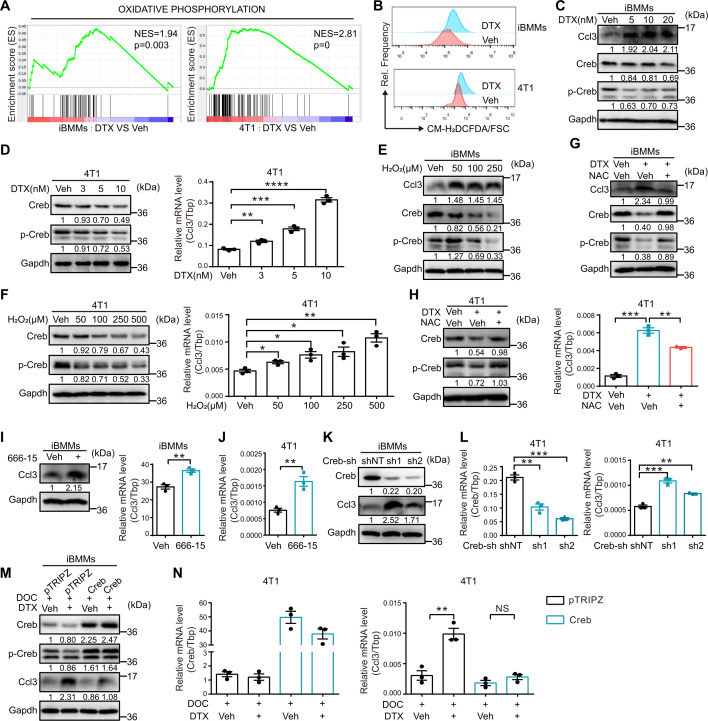
DTX induces Ccl3 by relieving the inhibitory effect of Creb via ROS accumulation
ROS and Creb have been reported to be involved in regulating Ccl3 expression.38 39 RNA-Seq analysis indicated that the oxidative phosphorylation pathway was enriched in both iBMMs and 4T1 cells treated with DTX (figure 5A), which was usually accompanied with ROS accumulation.40 Analysis of ROS levels in both macrophages (iBMMs and BMDMs) and breast cancer cells (4T1 and Py8119) treated with DTX revealed that the ROS levels significantly increased in response to DTX treatment (figure 5B, online supplemental figure S7A). Macrophages and cancer cells were treated with different concentrations of DTX to determine Ccl3 and Creb expression. An opposite expression pattern was observed between Ccl3 and Creb, in which Ccl3 was induced, while total Creb and p-Creb were inhibited by DTX (figure 5C, D, online supplemental figure S7B, C). The inhibitory effect of DTX on Creb was verified using qRT-PCR (online supplemental figure S7D).
Figure 5.
DTX induces Ccl3 by relieving the inhibitory effect of Creb via ROS accumulation. (A) GSEA based on RNA-Seq results of iBMMs and 4T1 cells treated with Veh and DTX (10 nM for iBMMs and 5 nM for 4T1). The NES and p value are shown. (B) ROS levels in iBMMs and 4T1 cells treated with DTX (10 nM for iBMMs and 5 nM for 4T1) for 2 days were determined via flow cytometry. (C) Western blotting was performed to detect the expression of Ccl3, Creb and p-Creb in iBMMs treated with different concentrations of DTX (0, 5, 10 and 20 nM) for 1 day. (D) Western blotting was used to detect the expression of Creb and p-Creb, whereas qRT-PCR was used to quantify Ccl3 expression in 4T1 treated with different concentrations of DTX (0, 3, 5 and 10 nM) for 1 day. (E) Various concentrations of H2O2 (0, 50, 100 and 250 µM) were used to induce ROS in iBMMs for 16 hours. Ccl3, Creb and p-Creb expression levels were determined via western blotting. (F) Various concentrations of H2O2 (0, 50, 100, 250 and 500 µM) were used to induce ROS in 4T1 for 16 hours. Western blotting was used to detect the expression of Creb and p-Creb, whereas qRT-PCR was used to quantify Ccl3 expression. (G) NAC (10 mM) was used to neutralize ROS in combination with DTX treatment (10 nM) for 1 day in iBMMs. Ccl3 and Creb expression levels were determined via western blotting. (H) NAC (10 mM) was used to neutralize ROS in combination with DTX treatment (5 nM) for 1 day in 4T1. Creb and p-Creb expression levels were determined via western blotting, whereas Ccl3 expression was quantified by qRT-PCR. (I) Creb inhibitor 666–15 (200 nM) was administered to iBMMs for 12 hours. The cells were collected for western blotting and qRT-PCR analysis to detect Ccl3 expression. (J) Creb inhibitor 666–15 (200 nM) was administered to 4T1 for 12 hours. qRT-PCR was performed to quantify Ccl3 expression. (K) Ccl3 expression was determined via western blotting in iBMMs knocking down Creb. (L) Ccl3 expression was quantified by qRT-PCR in 4T1 knocking down Creb. (M) iBMM stable cell line overexpressing Creb was subjected to DTX (10 nM) and doxycycline (DOC, 1 µg/mL) treatment for 2 days. Creb was cloned into an inducible pTRIPZ overexpression vector induced with DOC. The cells were collected and lysed for western blotting to detect Ccl3. (N) 4T1 stable cell line overexpressing Creb was subjected to DTX (5 nM) and DOC (1 µg/mL) treatment for 2 days. Creb was cloned into an inducible pTRIPZ overexpression vector induced with DOC. The cells were collected for qRT-PCR to quantify Ccl3 expression. Gapdh was used as the loading control. Tbp was used as the internal control. Data are presented as mean±SEM. *P<0.05, **p<0.01, ***p<0.001, ****p<0.0001; NS, no significance. Ccl3, C–C motif chemokine ligand 3; Creb, cAMP-response element binding protein; DOC, doxycycline; DTX, docetaxel; Gapdh, glyceraldehyde-3-phosphate dehydrogenase; GSEA, gene set enrichment analysis; H2O2, hydrogen dioxide; iBMMs, immortalized bone marrow-derived macrophages; NAC, N-acetyl-L-cysteine; NES, normalized enrichment score; qRT-PCR, quantitative real-time PCR; RNA-Seq, RNA sequencing; ROS, reactive oxygen species; sh, short hairpin RNA; shNT, short hairpin RNA of non-target; Tbp, TATA-binding protein; Veh, vehicle.
jitc-2021-003793supp010.pdf (789.5KB, pdf)
To explore whether the downregulation of Creb resulted from ROS accumulation, macrophages and cancer cells were treated with H2O2 at different concentrations. The results were similar to those after DTX treatment (figure 5E, F, online supplemental figure S7E, F), implying that ROS played a critical role in DTX-induced suppression of Creb. Furthermore, when NAC was used to neutralize ROS induced by DTX, Ccl3 induction and Creb inhibition were simultaneously reversed (figure 5G, H, online supplemental figure S7G, H). These results suggested that ROS accumulation on DTX treatment initiated downstream signaling.
Although Creb has been reported to be constitutively associated with the proximal promoter region of Ccl3 and to promote the transcription of Ccl3 in neutrophils,39 our results suggested a possible inhibitory role of Creb on Ccl3 expression in macrophages and breast cancer cell lines. To verify our hypothesis, macrophages and cancer cells were treated with 666–15, a commercial Creb inhibitor,41 and Ccl3 was significantly upregulated on Creb inhibition (figure 5I, J, online supplemental figure S7I, J). In addition, Creb knockdown upregulated Ccl3 expression (figure 5K,L, online supplemental figure S7K). In contrast, Creb overexpression significantly reversed Ccl3 upregulation induced by DTX (figure 5M, N, online supplemental figure S7L).
Collectively, ROS accumulation following DTX treatment in both macrophages and cancer cells inhibited Creb expression, which further relieved the inhibitory effect of Creb on Ccl3 and resulted in Ccl3 upregulation.
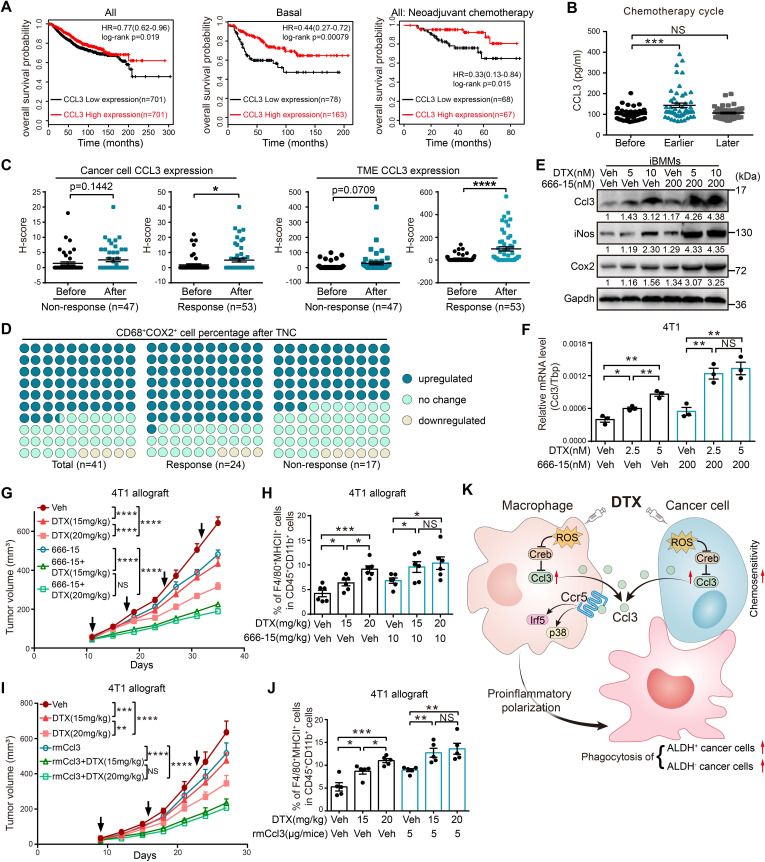
High CCL3 expression predicts better prognosis and DTX chemosensitivity in patients with breast cancer
We further investigated whether Ccl3 expression was relevant to clinical prognosis using the Kaplan-Meier database.42 The results showed that higher CCL3 expression indicated better prognosis, and it was more significant in patients with basal-type breast cancer and those receiving neoadjuvant chemotherapy (figure 6A). In addition, as determined by ELISA, CCL3 in the sera of patients receiving TNC was upregulated in the earlier chemotherapy cycles during which the chemotherapeutic effect was quite remarkable, but returned to the starting level in later chemotherapy cycles with no significant effects (figure 6B). IHC staining of CCL3 was performed using tumor sections from patients receiving TNC. The CCL3 expression levels in cancer cells and TME cells were both upregulated after TNC (online supplemental figure S8A), wherein CCL3 upregulation in TME cells was more obvious and vastly attributed to the response cohort (figure 6C, online supplemental figure S8B). Furthermore, the proportion of CD68+COX2+ proinflammatory macrophages after chemotherapy increased in 63.4% of patients receiving TNC, and this increase was more obvious in the response cohort (70.8%) than in the non-response cohort (52.9%) (figure 6D, online supplemental figure S8C). These observations suggested that upregulating CCL3 might provide new advantages to enhance DTX chemosensitivity in patients with breast cancer.
Figure 6.
High CCL3 expression predicts better prognosis, and Creb inhibitor or rmCcl3 increases DTX chemosensitivity in breast cancer. (A) Overall survival analysis of CCL3 in breast cancer using the Kaplan-Meier tool (https://kmplot.com/analysis/). HR and log-rank p value are as shown. (B) The serum CCL3 levels of TNC-receiving patients before chemotherapy (before, n=41), at earlier chemotherapy cycles (earlier, the second or third cycle, n=52) and at later chemotherapy cycles (later, the fifth or sixth cycle, n=51) were determined using ELISA. (C–D) IHC staining was performed using paired tumor sections of patients before or after TNC, and the patients were separated into response and non-response cohorts according to the MP score. (C) CCL3 expression levels of cancer cells and TME cells were separately calculated by H-score (non-response: n=47, response: n=53). (D) IHC staining of CD68 and COX2 were performed. The changes of CD68+COX2+ macrophages after TNC were defined as upregulated, no change and downregulated and presented as percentages of these three groups (non-response: n=17, response: n=24). (E) Indicated concentrations of 666–15 and DTX were combined to treat iBMMs for 20 hours. Western blotting was performed to determine the expression levels of Ccl3 and proinflammatory macrophage markers. (F) Indicated concentrations of 666–15 and DTX were combined to treat 4T1 for 20 hours, and qRT-PCR was performed to quantify Ccl3 expression. (G) 4T1 cells were orthotopically injected into BALB/c mice for the treatment of different dosed DTX in the presence or absence of 666–15 (n=5). The tumor volume was measured, and the tumor growth curves are shown. The arrows indicate the time of DTX and 666–15 administration. (H) Flow cytometry was used to analyze the percentage of F4/80+MHCII+ proinflammatory macrophages in cells isolated from tumors in figure 6G. (I) 4T1 cells were orthotopically injected into BALB/c mice for the treatment of different dosed DTX in the presence or absence of rmCcl3 (n=5). The tumor volume was measured, and the tumor growth curves are shown. The arrows indicate the time of DTX and rmCcl3 administration. (J) Flow cytometry was used to analyze the percentage of F4/80+MHCII+ proinflammatory macrophages in cells isolated from tumors in figure 6I. (K) Schematic diagram for the findings of the present work. DTX administration led to the intracellular ROS accumulation in both macrophages and cancer cells, which further upregulated Ccl3 by relieving the inhibitory effect of Creb on Ccl3. Then, the binding of Ccl3 to Ccr5 activated p38 and Irf5, resulting in the proinflammatory polarization of macrophages. These DTX educated macrophages suppressed tumor progression and augmented DTX chemosensitivity in breast cancer by enhancing phagocytosis of both bulk cancer cells and ALDH+ CSCs. Gapdh was used as the loading control. Tbp was used as the internal control. Data are presented as mean±SEM. *P<0.05, **p<0.01, ***p<0.001, ****p<0.0001; NS, no significance. ALDH, aldehyde dehydrogenase; Ccl3, C–C motif chemokine ligand 3; Ccr, C–C motif chemokine receptor; Cox2, cyclooxygenase 2; Creb, cAMP-response element binding protein; CSCs, cancer stem cells; TNC, Taxane-containing neoadjuvant chemotherapy; DTX, docetaxel; Gapdh, glyceraldehyde-3-phosphate dehydrogenase; H-score, histo-score; iBMMs, immortalized bone marrow-derived macrophages; IHC, immunohistochemistry; iNos, inducible nitric oxide synthase; MHCII, major histocompatibility complex class II; MP, Miller-Payne; qRT-PCR, quantitative real-time PCR; rmCcl3, recombinant Ccl3; ROS, reactive oxygen species; Tbp, TATA-binding protein; TME, tumor microenvironment.
jitc-2021-003793supp011.pdf (2.4MB, pdf)
Both Creb inhibitor and rmCcl3 increase DTX chemosensitivity in breast cancer
Commercially available 666–15 is a potent and selective Creb inhibitor.41 666–15 treatment upregulated Ccl3 expression, indicating that it might be a perfect candidate for combination with DTX. The combination of 666–15 and DTX prominently upregulated Ccl3 in both macrophages and cancer cells (figure 6E, F, online supplemental figure S8D) and promoted proinflammatory macrophage polarization (figure 6E). In addition, 666–15 made a lower dose of DTX comparable with a higher dose in Ccl3 induction and proinflammatory macrophage promotion (figure 6E, F, online supplemental figure S8D). Furthermore, the combination of 666–15 and DTX had a better tumor suppressive effect and enhanced DTX sensitivity, as the antitumor effect of DTX at a lower dose was comparable with that at a higher dose when combined with 666–15 (figure 6G, H, online supplemental figure S8E). We observed similar results in rmCcl3 combined with DTX (figure 6I, J, online supplemental figure S8F). These studies implied that Ccl3 induction might provide a novel strategy for improving DTX chemosensitivity in breast cancer.
Discussion
Previous studies have already proposed the notion that classic chemotherapeutic agents may elicit antitumor effects through immunomodulatory capability.43 In our study, we found that conventional antimicrotubule chemotherapy drug DTX could also exhibit therapeutic effects by provoking an antitumor immune response via enhanced proinflammatory macrophage polarization along with intensive phagocytosis of cancer cells, which depended on Ccl3 induction. The induction of Ccl3 by DTX was concomitant in both macrophages and cancer cells, and further formed a positive feedback loop to amplify the antitumor activity of DTX (figure 6K).
Considerable efforts have been devoted to restricting tumors by motivating the host antitumor immune response.44 Our study of macrophage depletion for DTX chemotherapy in tumor-bearing mice revealed that the therapeutic effect of DTX was indeed dependent on macrophages. Furthermore, DTX treatment promoted proinflammatory macrophage polarization in breast cancer TIME by inducing Ccl3 in macrophages. DTX-educated macrophages exhibited enhanced phagocytic ability of both bulk cancer cells and ALDH+ CSCs, thereby sensitizing cancer cells and CSCs to DTX and resulting in tumor recession. Furthermore, the therapeutic effects of DTX were abolished by Ccl3 knockout in TIME and to a less degree by Ccl3 knockdown in cancer cells. Recently, researchers have proposed that the proportion of cells in the active dividing cycle is small in the tumor mass. Thus, microtubule-targeting agents causing tumor regression might result from other non-mitotic-related mechanisms and factors, including interfering interphase cells, inducing cancer-cell-to-cancer-cell killing, targeting non-cancer cells in TME, and drug retention effect.45–47 In addition, Orth et al 48 developed high-resolution microscopy methods to visualize mitosis in vivo and demonstrated that the mitotic arrest ability of antimicrotubule drug paclitaxel in tumors was not as efficient as in culture. Although most cancer cells escaped from paclitaxel-induced mitotic arrest, tumor growth was inhibited in their model.48 These previous studies perfectly support our observation that the chemotherapy efficacy of DTX in vivo indispensably relies on proinflammatory macrophage polarization. Our results indicated the novel immunomodulatory competence of DTX via tumor immune remodeling and unveiled the underlying mechanisms. Given that T cells have a relatively high expression of Ccl3, whether DTX also influences T cells via Ccl3 deserves further in-depth study.
Skewing macrophage polarization state toward proinflammatory phenotype has proven to be a promising antitumor therapeutic strategy.8 10 13–15 Our in vivo and in vitro experimental results indicated that proinflammatory macrophage polarization was enhanced by DTX treatment. Furthermore, this effect was Ccl3 dependent and could be reversed by Ccl3 knockout or knockdown. By individually interfering with the two main Ccl3 receptors (Ccr1 and Ccr5), we determined that proinflammatory polarization induced by DTX was substantially Ccr5 dependent. Further studies indicated that in response to DTX, p38 and Irf5 played crucial roles in the induction of Cox2 and iNos, respectively. However, according to our present study, whether DTX can induce a macrophage phenotype change remains elusive and requires further exploration.
Ccl3 is a chemokine that has not been extensively studied in tumor biology. In the few available studies, Ccl3 has been reported to play contradictory roles in tumor progression. Ccl3 participates in the induction and maintenance of antitumor immune responses in vivo.49 However, it promotes angiogenesis50 and lung metastasis27 of tumor. The present study demonstrated that Ccl3 was induced by DTX in both macrophages and cancer cells. Mechanistically, accumulated ROS after DTX treatment initiated a signaling cascade by inhibiting Creb expression and further releasing Ccl3. Notably, as a transcriptional activating factor that recognizes the cAMP response element sequences in the promoter regions of target genes in most studies,29 the mechanism by which Creb inhibits Ccl3 needs further exploration. Furthermore, DTX-induced Ccl3 mediated the positive feedback loop between macrophages and cancer cells, functioning together to augment the chemotherapeutic effects of DTX. Either aspect disturbance would attenuate the antitumor activity of DTX, with macrophages being more important in this process. The administration of DTX combined with Creb inhibitor 666–15 or rmCcl3 achieved a prominent tumor inhibitory effect compared with individual application, as well as enhanced DTX chemosensitivity. Collectively, these results indicated that Ccl3 upregulation sensitized cancer cells to DTX, further augmenting its chemotherapeutic effects. Notably, the possibility of other antimitotic drugs taking effects via similar mechanisms cannot be excluded.
In summary, we discovered that DTX induced Ccl3 in both macrophages and cancer cells. Mechanistically, ROS accumulation after DTX treatment suppressed Creb expression and alleviated its inhibitory effects on Ccl3. Ccl3 upregulation triggered proinflammatory polarization of macrophages via Ccr5–p38/Irf5 pathway. DTX induced proinflammatory macrophages exhibited enhanced phagocytic ability. We concluded that the Ccl3-mediated proinflammatory macrophage polarization in TIME played indispensable roles in DTX chemotherapeutic efficacy by provoking innate antitumor immune responses. Ccl3 induction in combination with DTX may provide a promising therapeutic strategy to overcome chemoresistance and achieve better clinical outcomes in patients with breast cancer.
jitc-2021-003793supp001.pdf (1.3MB, pdf)
jitc-2021-003793supp003.pdf (112.5KB, pdf)
Acknowledgments
Many thanks to Dr Zhenyu Cai and Dr Tao Li for kindly providing Mvt1 and immortalized bone marrow-derived macrophage (iBMM) cell lines.
Footnotes
DS, WM and RZ contributed equally.
Correction notice: This article has been corrected since it was first published to correct the first affiliation.
Contributors: DS, WM, RZ, LZho, QD, JT, WC, FZ, NG, MD, DW, FL, XH, LZha, and SD conducted the experiments and analyzed the data. DS, WM, and SL designed the research. TL and YL collected tumor and serum samples of patients. DS and WM drafted the manuscript. SL, LZho, RZ, LZha, SD, and TL revised the manuscript. SL and TL supervised the work. SL is responsible for the overall content as the guarantor.
Funding: This work was supported by The National Key Research and Development Program of China (Stem Cell and Translational Research 2020YFA0112300); National Natural Science Foundation of China (81930075, 81772799, 81530075, 81773155, 82072903, 82073267); 'Ten Thousand Plan' – National High-Level Talents Special Support Plan WR-YK5202101; Program for Outstanding Medical Academic Leader in Shanghai (2019LJ04); Program of Shanghai Academic/Technology Research Leader 20XD1400700; The innovative research team of high-level local university in Shanghai; Shenzhen Science and Technology Innovation Commission Project, Shenzhen Municipal Government of China (KQTD20170810160226082); The Fudan University Research Foundation (IDH 1340042); The Research Foundation of the Fudan University Shanghai Cancer Center (YJRC1603); and Heilongjiang Province Natural Science Foundation Joint Guidance Project (LH2020H125).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by Fudan University Shanghai Cancer Center Institutional Review Board (050432-4-1212B) and Harbin Medical University Cancer Hospital Institutional Review Board (IRB: KY2019-08). Participants gave informed consent to participate in the study before taking part.
References
- 1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2. Nedeljković M, Damjanović A. Mechanisms of chemotherapy resistance in triple-negative breast Cancer-How we can rise to the challenge. Cells 2019;8:957. 10.3390/cells8090957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saloustros E, Mavroudis D, Georgoulias V. Paclitaxel and docetaxel in the treatment of breast cancer. Expert Opin Pharmacother 2008;9:2603–16. 10.1517/14656566.9.15.2603 [DOI] [PubMed] [Google Scholar]
- 4. Matikas A, Georgoulias V, Kotsakis A. The role of docetaxel in the treatment of non-small cell lung cancer lung cancer: an update. Expert Rev Respir Med 2016;10:1229–41. 10.1080/17476348.2016.1240620 [DOI] [PubMed] [Google Scholar]
- 5. Vale CL, Burdett S, Rydzewska LHM, et al. Addition of docetaxel or bisphosphonates to standard of care in men with localised or metastatic, hormone-sensitive prostate cancer: a systematic review and meta-analyses of aggregate data. Lancet Oncol 2016;17:243–56. 10.1016/S1470-2045(15)00489-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kodumudi KN, Woan K, Gilvary DL, et al. A novel chemoimmunomodulating property of docetaxel: suppression of myeloid-derived suppressor cells in tumor bearers. Clin Cancer Res 2010;16:4583–94. 10.1158/1078-0432.CCR-10-0733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Millrud CR, Mehmeti M, Leandersson K. Docetaxel promotes the generation of anti-tumorigenic human macrophages. Exp Cell Res 2018;362:525–31. 10.1016/j.yexcr.2017.12.018 [DOI] [PubMed] [Google Scholar]
- 8. Wanderley CW, Colón DF, Luiz JPM, et al. Paclitaxel reduces tumor growth by reprogramming tumor-associated macrophages to an M1 profile in a TLR4-dependent manner. Cancer Res 2018;78:canres.3480.2017–900. 10.1158/0008-5472.CAN-17-3480 [DOI] [PubMed] [Google Scholar]
- 9. Larionova I, Cherdyntseva N, Liu T, et al. Interaction of tumor-associated macrophages and cancer chemotherapy. Oncoimmunology 2019;8:1596004. 10.1080/2162402X.2019.1596004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tan B, Shi X, Zhang J, et al. Inhibition of Rspo-Lgr4 facilitates checkpoint blockade therapy by switching macrophage polarization. Cancer Res 2018;78:4929–42. 10.1158/0008-5472.CAN-18-0152 [DOI] [PubMed] [Google Scholar]
- 11. Pathria P, Louis TL, Varner JA. Targeting tumor-associated macrophages in cancer. Trends Immunol 2019;40:310–27. 10.1016/j.it.2019.02.003 [DOI] [PubMed] [Google Scholar]
- 12. Wang Y, Smith W, Hao D, et al. M1 and M2 macrophage polarization and potentially therapeutic naturally occurring compounds. Int Immunopharmacol 2019;70:459–66. 10.1016/j.intimp.2019.02.050 [DOI] [PubMed] [Google Scholar]
- 13. Ubil E, Caskey L, Holtzhausen A, et al. Tumor-secreted PROS1 inhibits macrophage M1 polarization to reduce antitumor immune response. J Clin Invest 2018;128:2356–69. 10.1172/JCI97354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vidyarthi A, Khan N, Agnihotri T, et al. TLR-3 stimulation skews M2 macrophages to M1 through IFN-αβ signaling and restricts tumor progression. Front Immunol 2018;9:1650. 10.3389/fimmu.2018.01650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang J-C, Sun X, Ma Q, et al. Metformin's antitumour and anti-angiogenic activities are mediated by skewing macrophage polarization. J Cell Mol Med 2018;22:3825–36. 10.1111/jcmm.13655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Negreiros-Lima GL, Lima KM, Moreira IZ, et al. Cyclic AMP regulates key features of macrophages via PKA: recruitment, reprogramming and efferocytosis. Cells 2020;9:128. 10.3390/cells9010128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zlotnik A, Yoshie O. The chemokine superfamily revisited. Immunity 2012;36:705–16. 10.1016/j.immuni.2012.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vidyarthi A, Agnihotri T, Khan N, et al. Predominance of M2 macrophages in gliomas leads to the suppression of local and systemic immunity. Cancer Immunol Immunother 2019;68:1995–2004. 10.1007/s00262-019-02423-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hornung V, Bauernfeind F, Halle A, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol 2008;9:847–56. 10.1038/ni.1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou L, Sheng D, Deng Q, et al. Development of a novel method for rapid cloning of shRNA vectors, which successfully knocked down CD44 in mesenchymal triple-negative breast cancer cells. Cancer Commun 2018;38:57. 10.1186/s40880-018-0327-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang W-C, Zheng X-J, Du L-J, et al. High salt primes a specific activation state of macrophages, M(Na). Cell Res 2015;25:893–910. 10.1038/cr.2015.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rangasami VK, Samanta S, Parihar VS, et al. Harnessing hyaluronic acid-based nanoparticles for combination therapy: a novel approach for suppressing systemic inflammation and to promote antitumor macrophage polarization. Carbohydr Polym 2021;254:117291. 10.1016/j.carbpol.2020.117291 [DOI] [PubMed] [Google Scholar]
- 23. Galdiero MR, Marone G, Mantovani A. Cancer inflammation and cytokines. Cold Spring Harb Perspect Biol 2018;10:a028662. 10.1101/cshperspect.a028662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang D, Zhou J, Zeng T, et al. Serum chemokine network correlates with chemotherapy in non-small cell lung cancer. Cancer Lett 2015;365:57–67. 10.1016/j.canlet.2015.05.001 [DOI] [PubMed] [Google Scholar]
- 25. Ma Y, Adjemian S, Galluzzi L, et al. Chemokines and chemokine receptors required for optimal responses to anticancer chemotherapy. Oncoimmunology 2014;3:e27663. 10.4161/onci.27663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Menten P, Wuyts A, Van Damme J. Macrophage inflammatory protein-1. Cytokine Growth Factor Rev 2002;13:455–81. 10.1016/S1359-6101(02)00045-X [DOI] [PubMed] [Google Scholar]
- 27. Kitamura T, Qian B-Z, Soong D, et al. CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. J Exp Med 2015;212:1043–59. 10.1084/jem.20141836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Taniguchi S, Elhance A, Van Duzer A. Tumor-Initiating cells establish an IL-33-TGF-beta niche signaling loop to promote cancer progression. Science;2020:eaay1813. 10.1126/science.aay1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koga Y, Tsurumaki H, Aoki-Saito H, et al. Roles of cyclic AMP response element binding activation in the ERK1/2 and p38 MAPK signalling pathway in central nervous system, cardiovascular system, osteoclast differentiation and mucin and cytokine production. Int J Mol Sci 2019;20:1346. 10.3390/ijms20061346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alam S, Liu Q, Liu S, et al. Up-regulated cathepsin C induces macrophage M1 polarization through FAK-triggered p38 MAPK/NF-κB pathway. Exp Cell Res 2019;382:111472. 10.1016/j.yexcr.2019.06.017 [DOI] [PubMed] [Google Scholar]
- 31. Zhong J, Wang H, Chen W, et al. Correction: Ubiquitylation of MFHAS1 by the ubiquitin ligase praja2 promotes M1 macrophage polarization by activating JNK and p38 pathways. Cell Death Dis 2018;9:782. 10.1038/s41419-018-0828-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wei J, Tang D, Lu C, et al. Irf5 deficiency in myeloid cells prevents necrotizing enterocolitis by inhibiting M1 macrophage polarization. Mucosal Immunol 2019;12:888–96. 10.1038/s41385-019-0169-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. M-Y W. Autophagy and macrophage functions: inflammatory response and phagocytosis. Cells 2020;9:70. 10.3390/cells9010070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jaiswal S, Jamieson CHM, Pang WW, et al. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell 2009;138:271–85. 10.1016/j.cell.2009.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang L, Shi P, Zhao G, et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Target Ther 2020;5:8. 10.1038/s41392-020-0110-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu S, Cong Y, Wang D, et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Reports 2014;2:78–91. 10.1016/j.stemcr.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hu J, Xiao Q, Dong M, et al. Glioblastoma immunotherapy targeting the innate immune checkpoint CD47-SIRPα axis. Front Immunol 2020;11:593219. 10.3389/fimmu.2020.593219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jiang L, Zheng T, Huang J, et al. Association of semen cytokines with reactive oxygen species and histone transition abnormalities. J Assist Reprod Genet 2016;33:1239–46. 10.1007/s10815-016-0756-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mayer TZ, Simard FA, Cloutier A, et al. The p38-MSK1 signaling cascade influences cytokine production through CREB and C/EBP factors in human neutrophils. J Immunol 2013;191:4299–307. 10.4049/jimmunol.1301117 [DOI] [PubMed] [Google Scholar]
- 40. Sinenko SA, Starkova TY, Kuzmin AA, et al. Physiological signaling functions of reactive oxygen species in stem cells: from flies to man. Front Cell Dev Biol 2021;9:714370. 10.3389/fcell.2021.714370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sapio L, Salzillo A, Ragone A, et al. Targeting CREB in cancer therapy: a key candidate or one of many? an update. Cancers 2020;12:3166. 10.3390/cancers12113166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010;123:725–31. 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- 43. Aldarouish M, Su X, Qiao J, et al. Immunomodulatory effects of chemotherapy on blood lymphocytes and survival of patients with advanced non-small cell lung cancer. Int J Immunopathol Pharmacol 2019;33:205873841983959. 10.1177/2058738419839592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bhatia S, Miller NJ, Lu H, et al. Intratumoral G100, a TLR4 agonist, induces antitumor immune responses and tumor regression in patients with Merkel cell carcinoma. Clin Cancer Res 2019;25:1185–95. 10.1158/1078-0432.CCR-18-0469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mitchison TJ. The proliferation rate paradox in antimitotic chemotherapy. Mol Biol Cell 2012;23:1–6. 10.1091/mbc.e10-04-0335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang J, Yang Y, Zhou Shen'ao, et al. Membrane-bound TNF mediates microtubule-targeting chemotherapeutics-induced cancer cytolysis via juxtacrine inter-cancer-cell death signaling. Cell Death Differ 2020;27:1569–87. 10.1038/s41418-019-0441-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Komlodi-Pasztor E, Sackett D, Wilkerson J, et al. Mitosis is not a key target of microtubule agents in patient tumors. Nat Rev Clin Oncol 2011;8:244–50. 10.1038/nrclinonc.2010.228 [DOI] [PubMed] [Google Scholar]
- 48. Orth JD, Kohler RH, Foijer F, et al. Analysis of mitosis and antimitotic drug responses in tumors by in vivo microscopy and single-cell pharmacodynamics. Cancer Res 2011;71:4608–16. 10.1158/0008-5472.CAN-11-0412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schaller TH, Batich KA, Suryadevara CM, et al. Chemokines as adjuvants for immunotherapy: implications for immune activation with CCL3. Expert Rev Clin Immunol 2017;13:1049–60. 10.1080/1744666X.2017.1384313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu Y, Li Y-Y, Matsushima K, et al. CCL3-CCR5 axis regulates intratumoral accumulation of leukocytes and fibroblasts and promotes angiogenesis in murine lung metastasis process. J Immunol 2008;181:6384–93. 10.4049/jimmunol.181.9.6384 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-003793supp002.pdf (43.3KB, pdf)
jitc-2021-003793supp012.pdf (582.5KB, pdf)
jitc-2021-003793supp013.pdf (540.4KB, pdf)
jitc-2021-003793supp014.pdf (318.7KB, pdf)
jitc-2021-003793supp015.pdf (401.8KB, pdf)
jitc-2021-003793supp004.pdf (5.1MB, pdf)
jitc-2021-003793supp005.pdf (2.2MB, pdf)
jitc-2021-003793supp006.pdf (4.7MB, pdf)
jitc-2021-003793supp007.pdf (7.8MB, pdf)
jitc-2021-003793supp008.pdf (10.1MB, pdf)
jitc-2021-003793supp009.pdf (277.6KB, pdf)
jitc-2021-003793supp010.pdf (789.5KB, pdf)
jitc-2021-003793supp011.pdf (2.4MB, pdf)
jitc-2021-003793supp001.pdf (1.3MB, pdf)
jitc-2021-003793supp003.pdf (112.5KB, pdf)
Data Availability Statement
Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information.