Abstract
Lepidium meyenii or Maca is widely cultivated as a health care food supplement due to its nutritional and medicinal properties. Although there are a few in-depth studies evaluating Maca antihypertensive effects, the correlations between the chemical constituents and bioactivity of the plant have not been studied before. Thus, the roots were extracted using different solvents (aqueous, methanol, 50% methanol, and methylene chloride) and investigated for their antihypertensive and antioxidant activities through several in vitro assays. The methanolic extract exhibited the best renin and angiotensin converting enzyme (ACE) inhibitory activities with IC50 values of 24.79 ± 1.3 ng/mL and 22.02 ± 1.1 ng/mL, respectively, along with the highest antioxidant activity. In total, 120 metabolites from different classes, e.g., alkylamides, alkaloids, glucosinolates, organic acids, and hydantoin derivatives, were identified in the methanolic extract using ultrahigh-performance liquid chromatography/high-resolution mass spectrometry (UPLC/HRMS). Molecular docking simulations were used to investigate the potential binding modes and the intermolecular interactions of the identified compounds with ACE and renin active sites. Glucotropaeolin, β-carboline alkaloids, succinic acid, and 2,4-dihydroxy-3,5-cyclopentyl dienoic acid showed the highest affinity to target the ACE with high docking scores (S ranging from −35.32 to −22.51 kcal mol–1) compared to lisinopril (S = −36.64 kcal mol–1). Interestingly, macamides displayed the greatest binding affinity to the active site of renin with docking scores (S ranging from −22.47 to −28.25 kcal mol–1). Further, β-carbolines achieved docking scores comparable to that of the native ligand (S ranging from −13.50 to −20.06 kcal mol–1). Molecular dynamics simulations and MMPBSA were also carried out and confirmed the docking results. Additionally, the computational ADMET study predicted that the compounds attaining promising docking results had proper pharmacokinetics, drug-likeness characteristics, and safe toxicological profiles. Ultimately, our findings revealed that Maca roots could be considered a promising candidate as an antihypertensive drug.
1. Introduction
Maca (Lepidium meyenii Walp., Brassicaceae family), also known as Peruvian ginseng, is a well-known crop in Peru widely used as a health care food supplement and medicine.1,2 The Food and Agriculture Organization recommended Maca as a safe edible food in 1992.2 After that, it was considered one of the star products in the global health care market due to its various potential effects. The plant is well known for its nutrient compositions, such as minerals, proteins, starches, amino acids, sugars, and fatty acids, and commonly purchased in the form of capsules or powder worldwide as a nutritional supplement.3 Traditionally, L. meyenii root is used as an active component in oils, coffee, or chocolate to relieve fatigue, strengthen the body, and improve fertility and sexual function.4,5 Many pharmacological activities have been reported, such as aphrodisiac, antiproliferative, antioxidant, learning and memory enhancing, antidepressant, antirheumatic, UV radiation protection, hepatoprotective, antifatigue, immunomodulatory, neuroprotective, and fertility increasing.6 Additionally, it could be used for chemical and physical stress responses, premenstrual discomfort and menopausal symptoms, prostatic hyperplasia, osteoporosis, and locomotion.3,4 The main compounds found in L. meyenii are glucosinolates, isothiocyanates, (thio)hydantoins, macaenes, polysaccharides, fatty acids, alkaloids, macamides, and flavonols.6
Hypertension (HTN) is a growing public health concern where one in three adults worldwide has HTN, which is exceedingly more than 10% of the worldwide population.7 The prevalence of HTN has been shown to increase with age. Poor control of HTN is closely linked to the burden of cardiovascular diseases and high mortality rates.8 Several classes of HTN medications have been developed; however, many of these drugs have been reported to have undesirable adverse effects. It is known that plant extracts are less toxic, contributing to their preference over synthetic drugs. Moreover, plants consumed in diet have been found to be useful in the management of HTN.9 For this purpose, the investigation of plants used in traditional medicine for potential hypotensive effects is an important strategy for the development of new and safe drugs. Based on the strong relationship between oxidative stress and HTN, the use of a single agent with antioxidant and antihypertensive properties could provide an effective strategy for the prevention and treatment of HTN.10 Interestingly, many plants and functional foods displayed high antioxidant effects and a great ability to scavenge free radicals in oxidative stress.11
Liquid chromatography coupled with high-resolution mass spectrometry is a widely employed technique to identify chemical constituents in different plant extracts with high sensitivity and specificity.12 It allows the separation of many kinds of substances in a high throughput manner.13,14 The high-resolution mass analysis allowed accurate dereplication of monoisotopic masses of known natural products using commercial databases such as the Dictionary of Natural Products (DNP), LipidMaps, and KEGG.13
As a computer-assisted approach, molecular docking plays an important role in predicting the binding modes and abilities of small molecules toward the targeted enzymes and receptors. In addition, it has exhibited the efficient screening ability of bioactive components with multiple targets with a high degree of accuracy. Implementation of molecular docking methods facilitates the discovery of novel therapeutic drugs that can treat a variety of chronic diseases.15,16
Being a plant with many health effects, Maca has attracted a lot of attention as a potential antihypertensive agent.17 For this purpose, the objective of our study was to evaluate the antihypertensive effects of different Maca extracts through in vitro inhibition of Renin and ACE enzymes and to assess their antioxidant effects using several assays, i.e., total antioxidant capacity (TAC), 1,1-diphenyl-2-picrylhydrazyl (DPPH), 2,2′-azino-bis-3-ethylbenzthiazoline-6-sulfonic acid (ABTS), ferric-reducing antioxidant power (FRAP), and iron-reducing power. Furthermore, a comprehensive profiling of the active extract was performed through a sensitive and accurate UPLC/HRMS method to identify the bioactive compounds in the extract. The potential binding modes and intermolecular interactions of the identified compounds with ACE and Renin active sites were studied using molecular docking simulations to better investigate the mode of interaction underlying the inhibitory effects for the first time. In addition, molecular dynamics simulations and PBSA analysis were also carried out to further validate the binding of the most promising candidates as well as to examine the stability of the complexes formed. Finally, ADMET computational parameters were evaluated to predict the physicochemical properties and toxicity profiles of the tested compounds.
2. Results and Discussion
2.1. In Vitro Antioxidant Assays
There is a strong indication that many threatening pathophysiological processes such as cardiovascular diseases are associated with the accumulation of free radicals in the body. As innate antioxidant defense mechanisms are inefficient, the dietary intake of antioxidants is vital to protect our cells from damage by free radicals. To evaluate the antioxidant potential of the four extracts, five in vitro complementary assays were used. Some methods utilize radical producers, e.g., DPPH and ABTS, and some use metal ions for oxidation, e.g., iron-reducing power assay. Our results showed that the methanolic extract exhibited the most potent activity, as depicted in Table 1, with an IC50 value of 41.44 ± 2.44 μg/mL and EC50 of 60.05 ± 2.38 μg/mL in the DPPH and iron-reducing power assays, respectively, as compared to ascorbic acid with IC50 = 22.44 ± 1 μg/mL and EC50= 57.76 ± 4.75 μg/mL, respectively. In addition, the methanolic extract showed significantly higher total antioxidant capacity and FRAP and ABTS scavenging activities, 303.91 ± 6.5, 293.99 ± 2.5, and 1072.93 ± 7.77, respectively, calculated as μM ascorbic acid equivalent/g extract.
Table 1. In Vitro Antioxidant Activity of the Different Maca Root Extractsa.
| samples | DPPH IC50 (μg/mL) | ABTS (μM ascorbic acid equivalent/g extract) | FRAP (μM ascorbic acid equivalent/g extract) | TAC (μM ascorbic acid equivalent/g extract) | iron-reducing power EC50 (μg/mL) |
|---|---|---|---|---|---|
| aqueous extract | 1119.09 ± 6.1 | 508.06 ± 4.22 | 100.41 ± 1.89 | 111.85 ± 9.06 | 385.49 ± 4.75 |
| 50% methanolic extract | 208.44 ± 3.01 | 888.65 ± 4.64 | 155.11 ± 3.55 | 220.96 ± 7.43 | 135.99 ± 4.96 |
| dichloromethane extract | 500.93 ± 2.9 | 692.99 ± 8.11 | 127.33 ± 3.90 | 174.75 ± 7.99 | 204.87 ± 5.88 |
| methanolic extract | 41.44 ± 2.44 | 1072.93 ± 7.77 | 293.99 ± 2.5 | 303.91 ± 6.5 | 60.05 ± 2.38 |
| ascorbic acid | 22.44 ± 1 | - | - | - | 57.76 ± 4.75 |
All data are presented as mean ± SD.
2.2. In Vitro Antihypertensive Assays
The Renin–angiotensin system is considered an important determinant of blood pressure and fluid homeostasis in the human body. Renin catalyzes the first and rate-limiting step that initiates the production of angiotensin-I, which is subsequently converted by the action of ACE to angiotensin-II, a potent vasoconstrictor that contributes to blood pressure elevation. Therefore, simultaneous inhibition of Renin and ACE activities would efficiently regulate the Renin–angiotensin system, providing a new alternative way to treat hypertension efficiently.18,19
The results obtained from the inhibitory effects on Renin and ACE activities of Maca extracts (Table 2) showed that the methanolic extract exhibited the highest inhibitory effects compared to the other extracts (aqueous, 50% methanol, and methylene chloride). In our study, the IC50 values of the methanolic extract for Renin and ACE inhibitions were 24.79 ± 1.3 ng/mL and 22.02 ± 1.1 ng/mL, respectively, indicating a good hypotensive effect when compared to Quinapril with IC50 = 15.01 ± 0.8 ng/mL and 13.97 ± 0.7 ng/mL, respectively.
Table 2. In Vitro Antihypertensive Activity (Renin and ACE Inhibitory Activities) of the Different Maca Root Extractsa.
| IC50 (ng/mL) |
||
|---|---|---|
| samples | Renin | ACE |
| aqueous extract | 99.29 ± 5.4d | 63.14 ± 3.2d |
| 50% methanolic extract | 35.09 ± 1.9c | 22.76 ± 1.2b |
| dichloromethane extract | 42.78 ± 2.3c | 34.87 ± 1.8c |
| methanolic extract | 24.79 ± 1.3b | 22.02 ± 1.1b |
| quinapril | 15.01 ± 0.8a | 13.97 ± 0.7a |
All data are presented as (mean ± SD). Different superscripts letters for a given value within a column are significantly different from each other (Duncan’s significant difference multiple range post-hoc test, P < 0.05).
2.3. UPLC-HRMS Analysis of the Secondary Metabolites
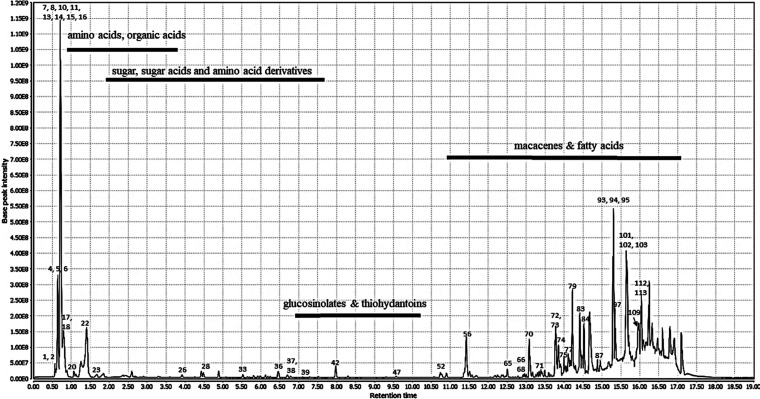
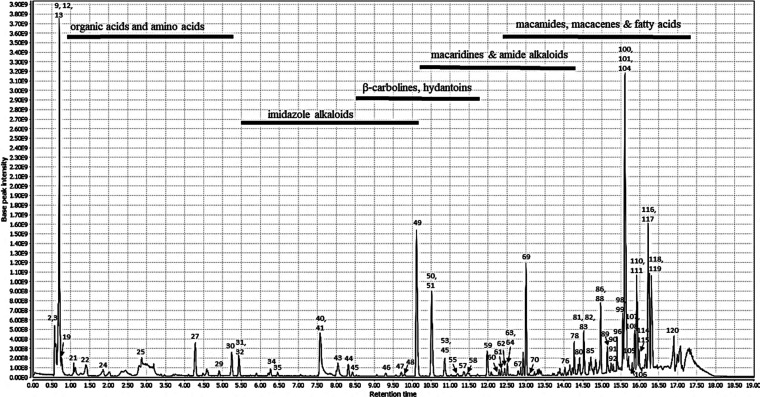
The chemical constituents of the active methanolic extract were analyzed via UPLC-HRMS in both positive and negative ionization modes to highlight possible different classes of bioactive compounds that may be correlated to the hypotensive effects of the Maca extract. All compounds characterized in the extract, including compound class, retention times (min), molecular ions (m/z), molecular formulas, fragment ions, as well as putative compounds, were summarized in Table 3. The representative LC/MS chromatograms of the methanolic extracts in both modes are shown in Figures 1 and 2. In total, 120 compounds were identified based on their accurate MS and fragment ions and by searching online databases, KEGG, lipid Maps, the dictionary of natural products (DNP, 2015), and available literature. Identified metabolites (Table 3) belonged to various classes including 19 macamides, 12 amide alkaloids, 8 β-carboline alkaloids, 2 macaridines, 13 imidazole alkaloids, 6 macacenes, 26 fatty acids, 7 organic acids, 1 glucosinolate, and 2 hydantoin derivatives along with 24 miscellaneous compounds. Alkaloids, macamides, and macaenes were the major chemical ingredients of Maca. The representative MS/MS spectra of selected compounds from each class were displayed in Figures S1–S20 (Supporting Information).
Table 3. Chemical Profiling of Maca Methanolic Extract Using LC-HR-ESI-MS/MS in Positive and Negative Ionization Modes.
| indicated
molecular ion (m/z) |
|||||||
|---|---|---|---|---|---|---|---|
| peak no. | RT (min) | (M – H)− | (M + H)+ | error (ppm) | elemental composition | identification | MS/MS (m/z) |
| Identified Imidazole Alkaloids | |||||||
| 40 | 7.51 | 227.1541 | –0.9 | C15H18N2 | lepidiline Ea | 174.9840, 135.0920, 113.9641 | |
| 41 | 7.57 | 225.1382 | –0.2 | C15H16N2 | 1-dibenzyl-2-propyne-4,5-dimethylimidaziliuma | 208.1135, 189.0699, 181.1024, 130.0651, 113.9644, 105.0341 | |
| 44 | 8.41 | 239.1543 | 0 | C16H18N2 | 1-dibenzyl-2-(1,3-butadiene)-4,5-dimethylimidaziliuma | 199.9881, 182.9850, 154.9903, 121.0644 | |
| 46 | 9.3 | 263.1542 | –0.4 | C18H18N2 | 1-dibenzyl-2-phenyl-4,5-dimethylimidaziliuma | 231.0294, 121.0644 | |
| 49 | 10.12 | 277.1700 [M – Cl]+ | 1.8 | C19H21N2 | lepidiline Aa | 185.1062 | |
| 50 | 10.51 | 307.1805 [M – Cl]+ | 1.5 | C20H23N2O | lepidiline Ca | 215.1188, 200.1296, 185.1083, 116.0713 | |
| 51 | 10.52 | 291.1855 [M – Cl]+ | –0.3 | C20H22N2 | lepidiline Ba | 200.1297, 199.1235 | |
| 54 | 10.86 | 321.1932 [M – Cl]+ | 1.8 | C21H25N2O | lepidiline Da | 246.2423, 199.1232, 161.0654 | |
| 60 | 12.08 | 339.1857 | 0.3 | C24H22N2 | 1,3-dibenzyl-2(1,3-glutaric alkynyl)-4,5-dimethylimidaziliuma | 319.2153, 248.1299, 231.0299, 199.1236, 184.0735, 171.0910, 136.0621, 121.0656, 113.9641 | |
| 62 | 12.35 | 353.2012 | –0.1 | C25H24N2 | 1,3-dibenzyl-2-phenyl-4,5-dimethylimidaziliuma | 262.1461, 226.2162, 184.0734, 171.0910, 136.0620, 121.0643 | |
| 63 | 12.43 | 333.2321 | –0.4 | C23H28N2 | 1,3-dibenzyl-2-butyl-4,5-dimethylimidaziliuma | 241.1693, 199.1232, 121.0654 | |
| 67 | 12.86 | 347.2482 | 0 | C24H30N2 | 1,3-dibenzyl-2-pentyl-4,5-dimethylimidaziliuma | 294.2201, 255.1865, 199.1233, 136.0620 | |
| 69 | 13.00 | 316.2846 | –8.5 | C21H35N2 | 1-dibenzyl-3-cyclohexyl-2-propenyl-4,5-dimethylimidaziliuma | 298.243, 280.2643, 262.2515, 245.2466, 184.0738, 109.1016 | |
| Identified β-Carboline Alkaloids | |||||||
| 27 | 4.28 | 233.1284 | –0.3 | C13H16N2O2 | 1-methyl tetrahydro-5,6-hydride carboline-3-carboxylic acid isomer Ia | 116.0711 | |
| 29 | 4.90 | 217.0972 | 0.1 | C12H12N2O2 | 1,2,3,4-tetrahydro-β-carboline-3-carboxylic acida | 144.0800, 113.9641 | |
| 30 | 5.24 | 233.1288 | 1.4 | C13H16N2O2 | 1-methyltetrahydro-5,6-hydride carboline-3-carboxylic acid isomer IIa | 116.0711 | |
| 31 | 5.34 | 231.1127 | –0.6 | C13H14N2O2 | 1-methyltetrahydro-β-carboline-3-carboxylic acida | 214.087, 168.0814, 158.0958, 143.0731, 130.0647, 113.9639 | |
| 34 | 6.27 | 273.1596 | –0.7 | C16H20N2O2 | 1-butyltetrahydro-β-5,6-hydridecarboline-3-carboxylic acida | 251.1186, 231.0293, 187.1239, 121.0644, 113.9641 | |
| 43 | 8.02 | 285.1596 | –0.6 | C17H20N2O2 | 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acida | 239.1545, 185.1061, 121.0645 | |
| 58 | 11.5 | 271.1440 | –0.5 | C16H18N2O2 | 1-butyltetrahydro-β-5,6-carboline-3-carboxylic acida | 121.0656 | |
| 59 | 12.03 | 211.0865 | –0.5 | C13H10N2O | 1-methyl-β-carboline-3-carbaldehydea | 193.0765, 184.0733, 169.0757, 121.0655 | |
| Identified Macaridines and Common Amide Alkaloids | |||||||
| 45 | 8.44 | 218.1176 | 0.1 | C13H15NO2 | 3-benzyl-1,2-dihydro-4-methoxy-N-hydroxypyridinea | 200.0005, 187.1241, 174.9839, 162.0914, 140.0699, 121.0644, 146.9619, 113.9641 | |
| 48 | 9.71 | 216.1019 | –0.1 | C13H13NO2 | 3-benzyl-1,2-dihydro-4-carbaldehyde-N-hydroxypyridine (macaridine)a | 198.0910, 170.0950, 141.0705, 128.0625, 121.0645, 113.9642 | |
| 21 | 1.08 | 136.0755 | –0.1 | C8H9NO | 2-phenylacetamidea | 119.0359, 112.0511 | |
| 25 | 3.19 | 166.0861 | 0.1 | C9H11NO2 | N-methyl-3-hydroxy-benzeneacetamidea | 121.0644 | |
| 32 | 5.43 | 136.0757 | –0.1 | C8H9NO | N-benzylformamidea | 113.9643 | |
| 35 | 6.45 | 150.0915 | –0.4 | C9H11NO | N-acetylbenzylaminea | 134.0603, 113.9642, 106.0656 | |
| 55 | 11.37 | 241.1334 | –0.7 | C15H16N2O | 1,3-dibenzylureaa | 227.1160, 205.1328, 121.0645, 113.9642 | |
| 57 | 11.46 | 212.1068 | –0.1 | C14H13NO | N-benzylbenzamidea | 136.0623 | |
| 89 | 15.2 | 280.2634 | –0.4 | C18H33NO | linoleamideb | 184.0728 | |
| 90 | 15.24 | 306.2789 | –0.9 | C20H35NO | N-ethyl-12-(2′-4′-)-cyclohexadiene-dodeceneamidea | 184.0732 | |
| 91 | 15.24 | 342.3002 | –0.3 | C20H39NO3 | N-cyclohexane-(3′-hydroxyl)-tridecane amidea | 184.0732, 109.1013, 104.0706 | |
| 92 | 15.25 | 300.2896 | –0.4 | C18H37NO2 | 16-hydroxyl-octadecanamidea | 282.1337, 121.0656 | |
| 96 | 15.52 | 256.2633 | –0.8 | C16H33NO | hexadecanamideb | 184.0732, 133.0861 | |
| 101 | 15.63 | 360.2912 | 362.3050 | 1.2 | C23H39NO2 | methanandamidea | 295.2281, 248.9612, 183.0110, 115.9951 |
| Identified Macamides | |||||||
| 61 | 12.29 | 382.274 | –0.5 | C25H35NO2 | N-(3-hydroxy-benzyl)-11Z,13Z,15Z,17Z-octadecatetrienamidea | 290.2130, 226.2166, 184.0736, 136.0622 | |
| 80 | 14.38 | 366.2791 | –0.2 | C25H35NO | N-benzyl-(9Z,12Z,15Z)-octadecatetraenamidea | 307.2653, 184.0730 | |
| 86 | 14.97 | 384.2895 | –0.6 | C25H37NO2 | N-benzyl-5-oxo-6E,8E-octadecadienamidea | 144.9823 | |
| 88 | 15.14 | 386.3055 | –0.5 | C25H39NO2 | N-benzyl-9-oxo-12E-octadecenamidea | 340.2638, 305.2472, 144.9818, 110.0203 | |
| 98 | 15.55 | 398.3053 | –0.2 | C26H39NO2 | N-(3-methoxybenzyl)-(9Z,12Z,15Z)-octadecatrienamidea | 121.0653 | |
| 99 | 15.61 | 368.294 | 0 | C25H37NO | N-benzyl-(9Z,12Z,15Z)-octadecatrienamidea | 345.2422, 108.0808 | |
| 100 | 15.62 | 308.2945 | –1 | C20H37NO | N-ethyl-(9E,11E)-octadecadienamidea | 290.2126, 1791300, 136.0756, 121.0643 | |
| 104 | 15.67 | 318.2791 | –0.2 | C21H35NO | N-benzyl-tetradeceneamidea | 184.0733, 115.0752 | |
| 107 | 15.81 | 344.2946 | –0.6 | C23H37NO | N-benzyl-4E-hexadecenamidea | 144.9822 | |
| 108 | 15.86 | 400.3210 | –0.1 | C26H41NO2 | N-(3-methoxybenzyl)-(9Z,12Z)-octadecadienamidea | 121.0651 | |
| 110 | 15.92 | 370.3104 | –0.2 | C25H39NO | N-benzyl-(9Z,12Z)-octadecadienamidea | 108.0808 | |
| 111 | 15.96 | 332.2947 | –0.3 | C22H37NO | N-benzyl pentadecanamidea | 317.2027, 284.2940, 184.0733, 108.0807 | |
| 114 | 16.14 | 406.331 | –1.5 | C25H43NO3 | N-(3,4-dimethoxybenzyl) hexadecanamidea | 286.2165, 184.0729 | |
| 115 | 16.16 | 376.3206 | –0.6 | C24H41NO2 | N-(3-methoxybenzyl) hexadecanamidea | 346.3087, 121.0653 | |
| 116 | 16.22 | 346.3102 | –0.8 | C23H39NO | N-benzyl hexadecanamide (macamide B)a | 121.0643 | |
| 117 | 16.31 | 372.326 | –0.3 | C25H41NO | N-benzyl-9Z-octadecenamidea | 190.1210, 121.9663, 108.0808 | |
| 118 | 16.41 | 360.326 | –0.3 | C24H41NO | N-benzyl heptadecanamidea | 121.9663 | |
| 119 | 16.43 | 404.3157 | –0.6 | C25H41NO3 | N-(3,4-dihydroxybenzyl)-oleamidea | 184.0735, 133.0863, 121.9664 | |
| 120 | 16.86 | 374.3417 | –0.2 | C25H43NO | N-benzyl octadecanamidea | 144.9821, 121.9664 | |
| Identified Macaenes and Fatty Acids | |||||||
| 52 | 10.58 | 215.1283 | –2.6 | C11H20O4 | undecanedioic acidb | 132.8669, 112.9846 | |
| 56 | 11.42 | 329.2332 | –0.4 | C18H34O5 | trihydroxy octadecenoic acidb | 293.2127, 248.9610, 29.1443, 211.1336, 171.1024, 139.1126, 112.9846 | |
| 64 | 12.5 | 274.2737 | –1.4 | C16H35NO2 | N-ethyl-tetradecene estera | 240.2310, 201.1021, 121.0655 | |
| 65 | 12.51 | 307.1922 | C18H28O4 | 7,12-dioxo-8,10-octadecadienoic acidb | 235.1351, 185.1186, 112.9847 | ||
| 66 | 12.79 | 331.2494 | 1.3 | C18H36O5 | 9,10,13-trihydroxy-octadecanoic acidb | 253.2176, 223.2058, 152.9950, 112.9846 | |
| 68 | 12.95 | 309.2077 | 1.9 | C18H30O4 | 7S,8S-dihydroxy-9Z,12Z,15Z-octadecatrienoic acidb | 209.1172, 152.9949, 112.9847 | |
| 70 | 13.08 | 311.2229 | 313.2388 | 0.4 | C18H32O4 | 9Z-octadecenedioic acidb | 223.1709, 152.9950, 112.9847 |
| 72 | 13.76 | 285.2073 | 0.7 | C16H30O4 | hexadecanedioic acidb | 265.1495, 223.2056, 152.9945, 112.9845 | |
| 73 | 13.86 | 315.2542 | 0.4 | C18H36O4 | 9,12-dihydroxy-octadecanoic acid | 293.2124, 116.9281 | |
| 74 | 13.91 | 293.2125 | 1 | C18H30O3 | 5-oxo-6E,8E-octadecadienoic acida | 279.2329, 185.1185, 152.9949, 116.928 | |
| 75 | 13.98 | 291.1968 | 0.9 | C18H28O3 | 5-oxo-6E,8E,10E-Octadecatrienoic acida | 277.2170, 152.9949, 112.9846 | |
| 76 | 14.13 | 284.3292 | 2.5 | C18H34O2 | (E)-octadec-9-enoic acid (elaidic acid)a | 184.0729 | |
| 77 | 14.22 | 295.228 | 0.5 | C18H32O3 | hydroxy-linoleic acidb | 281.2486, 255.2320, 183.1375, 152.9949, 116.9281 | |
| 78 | 14.22 | 283.2639 | 2.5 | C18H34O2 | oleic acida | 184.0732 | |
| 79 | 14.32 | 243.1965 | –0.2 | C14H28O3 | hydroxy myristic acidb | 183.9133, 130.0507, 110.0234 | |
| 81 | 14.50 | 363.2504 | –7.2 | C22H34O4 | dihydroxy-docosapentaenoic acidb | 184.0731, 119.0850 | |
| 82 | 14.53 | 277.2161 | –0.5 | C18H28O2 | stearidonic acidb | 184.0736, 107.0860 | |
| 83 | 14.53 | 293.2126 | 295.2266 | 1.4 | C18H30O3 | 9-oxo-10E,12E-octadecadienoic acida | 279.2331, 235.1693, 183.0106, 152.9949, 116.9281 |
| 84 | 14.53 | 317.2089 | –10.4 | C20H30O3 | 5-oxo-6E,8E,10E,12E-eicosatetraenoic acida | 277.2139, 184.0736, 107.0860 | |
| 85 | 14.72 | 297.2423 | –0.5 | C18H32O3 | hydroxy octadecadienoic acidb | 184.0732 | |
| 87 | 14.98 | 269.2124 | 0.8 | C16H30O3 | hydroxy hexadecenoic acidb | 152.9948, 116.9280 | |
| 93 | 15.3 | 271.2267 | 0.9 | C16H32O3 | hydroxy hexadecanoic acidb | 225.2212, 183.0106, 152.9948, 116.9280 | |
| 94 | 15.3 | 277.2174 | 0.4 | C18H30O2 | linolenic acidb | 225.2212, 116.9280 | |
| 95 | 15.45 | 349.2387 | 0.8 | C21H34O4 | dihydroxy eicosatetraenoic acid methyl esterb | 116.928 | |
| 97 | 15.55 | 397.3325 | 0.5 | C24H46O4 | tetracosanedioic acidb | 281.2484, 116.9280 | |
| 102 | 15.64 | 279.2331 | 0.6 | C18H32O2 | octadecadienoic acida | 183.0106, 152.9949, 116.9281 | |
| 103 | 15.64 | 369.3013 | 0.8 | C22H42O4 | docosanedioic acidb | 339.2009, 279.2332, 172.9534, 116.9281 | |
| 105 | 15.78 | 325.2734 | –1.1 | C20H36O3 | 5-oxo-6E- eicosanoic acida | 307.2625, 279.2319, 144.9822, 110.0202 | |
| 106 | 15.79 | 457.3512 | –2.6 | C26H48O6 | byrsonic acidb | 426.2997, 413.3223, 378.2995, 344.2948, 327.1997, 283.1740, 239.1468, 177.1123, 133.0862 | |
| 109 | 15.91 | 299.2594 | 0.8 | C18H36O3 | hydroxy-octadecanoic acidb | 355.1583, 255.2320, 183.0106, 116.9280 | |
| 112 | 16.03 | 255.233 | 0.3 | C16H32O2 | palmitic acida | 183.0106, 134.8941, 116.9280, 100.9250 | |
| 113 | 16.06 | 327.2543 | 0.7 | C19H36O4 | nonadecandioic acidb | 229.1443, 211.1336, 171.1024, 112.9847 | |
| Identified Glucosinolates | |||||||
| 26 | 3.92 | 408.0435 | 1.7 | C14H19O9NS2 | benzyl glucosinolate (glucotropaeolin)a | 341.1110, 267.9721, 183.9132, 146.9383, 112.9846 | |
| Identified Hydantoins and Thiohydantoins | |||||||
| 47 | 9.57 | 307.0724 | 309.0869 | –0.3 | C14H16N2O2S2 | (+)-meyeniin Ca | 263.0815, 248.9609, 233.0729, 205.0779, 191.0604, 183.9132, 174.9554, 146.9383, 134.8644 |
| 53 | 10.77 | 261.1234 | 0 | C14H16N2O3 | meyeniihydantoin Aa | 231.0294, 184.0737, 121.0644 | |
| Organic Acids and Miscellaneous Compounds | |||||||
| 1 | 0.57 | 131.0817 | –6.6 | C5H12N2O2 | ornithineb | 112.9846 | |
| 2 | 0.57 | 173.1038 | 175.1198 | –3.9 | C6H14N4O2 | argeninea | 131.082 |
| 3 | 0.57 | 162.0761 | 0 | C6H11NO4 | N-methyl-l-glutamic acidb | 116.0712 | |
| 4 | 0.58 | 131.0453 | –6.8 | C4H8N2O3 | asparagineb | 101.0232 | |
| 5 | 0.59 | 335.1577 | 1.5 | C12H24N4O7 | hexosyl argeninea | 115.9201 | |
| 6 | 0.6 | 132.0293 | –6.8 | C4H7NO4 | aspartic acidb | 112.0394, 104.0337 | |
| 7 | 0.64 | 195.0505 | –2.6 | C6H12O7 | gluconic acida | 152.9950, 129.0188, 118.0503, 101.0233 | |
| 8 | 0.64 | 179.0554 | –3.8 | C6H12O6 | hexosea | 129.0188, 118.0503, 101.0233 | |
| 9 | 0.64 | 104.071 | 3.6 | C4H9NO2 | 4-amino-butanoic acidb | 102.0553 | |
| 10 | 0.65 | 113.0233 | –9.6 | C5H6O3 | 3-oxo-4-pentenoic acidb | 112.9846, 100.9250 | |
| 11 | 0.65 | 193.0349 | –2.3 | C6H10O7 | glucuronic acidb | 144.0653, 118.0502 | |
| 12 | 0.65 | 118.0865 | 1.9 | C5H11NO2 | l-valineb | 104.1068 | |
| 13 | 0.69 | 114.055 | 116.0713 | 2.4 | C5H9NO2 | prolinea | 104.0709 |
| 14 | 0.7 | 172.0607 | –4.7 | C7H11NO4 | carboxy methyl prolineb | 121.0287, 112.9847 | |
| 15 | 0.7 | 341.1084 | 1.1 | C12H22O11 | dihexosidea | 276.1084, 179.0558, 114.0549, 101.0232 | |
| 16 | 0.71 | 133.0135 | –3.9 | C4H6O5 | 2-hydroxy succinic acida | 128.0347, 116.0706, 103.9192 | |
| 17 | 0.71 | 467.1174 | –4.4 | C21H24O12 | catechin-4-ol 3-O-hexosideb | 387.1141, 341.1110, 276.1084, 179.0558, 133.0134, 101.10232 | |
| 18 | 0.71 | 618.2258 | 1.2 | C23H41NO18 | proline trihexosidea | 341.1108, 276.1083, 179.0557 | |
| 19 | 0.73 | 130.0863 | 0.9 | C6H11NO2 | N-methyl prolinea | 112.0511 | |
| 20 | 1.07 | 549.1679 | 1.2 | C19H34O18 | trihexosidea | 341.1109, 179.0558, 114.0549, 101.0232 | |
| 22 | 1.4 | 128.0347 | 130.0495 | –4.6 | C5H7NO3 | 5-oxo-prolinea | 116.0707, 103.9193 |
| 23 | 1.67 | 117.0183 | –8.5 | C4H6O4 | succinic acida | 103.9193 | |
| 24 | 1.85 | 132.1021 | 1.3 | C6H13NO2 | leucinea | 113.9642 | |
| 28 | 4.49 | 143.0342 | –5.3 | C6H8O4 | 2,4-dihydroxy-3,5-cyclopentyl dienoic acida | 112.9846, 103.9192 | |
| 33 | 5.53 | 210.0767 | –1.7 | C10H13NO4 | 3-hydroxy-5-methyl-l-tyrosineb | 183.9132, 124.0401 | |
| 35 | 6.7 | 132.0446 | –6.5 | C8H7NO | benzyl isocyanatea | 112.9846 | |
| 36 | 6.46 | 150.0552 | –5.5 | C8H9NO2 | phenyl glycineb | 112.9846, 109.0157 | |
| 38 | 6.78 | 206.0821 | –0.7 | C11H13NO3 | N-acetyl-l-phenylalanineb | 183.9132, 164.0711, 112.9846 | |
| 39 | 7.29 | 237.0408 | 1.5 | C11H10O6 | malic acid benzoatea | 183.9133, 121.087 | |
| 42 | 7.98 | 187.0972 | –1.9 | C9H16O4 | azelaic acidb | 141.0914, 125.0965, 112.9846 | |
| 71 | 13.37 | 194.0817 | –2.8 | C10H13NO3 | l-homotyrosineb | 152.9949, 112.9846 | |
Figure 1.
LC-MS chromatogram of Maca root (Lepidium meyenii) extract obtained in ESI negative mode.
Figure 2.
LC-MS chromatogram of Maca root (Lepidium meyenii) extract obtained in ESI positive mode.
2.3.1. Identification of Alkaloids in Maca Roots
Imidazole alkaloids, β-carboline alkaloids, macaridines, and common amide alkaloids are the most characteristic alkaloids in Maca.2 In total, 35 alkaloids (Table 3 and Figure 1) were tentatively identified in the Maca extract in the positive ionization mode.
2.3.1.1. Identified Imidazole Alkaloids
Many imidazole alkaloids such as lepidilines A–G were previously isolated and characterized from Maca roots.20,21 They are tertiary and quaternary alkaloids derived from histidine with the 1,3-dibenzylimidazolium nucleus.20,21 Lepidiline alkaloids were reported to increase fertility and inhibit human ovarian, bladder, and pancreatic cancer cells.22 Thirteen imidazole alkaloids were identified based on their MS/MS fragmentation and reported literature.2,22
Lepidiline A (compound 49, Table 3 and Figure S1) showed a precursor ion [M – Cl]+ at m/z 277.1700 and a product ion at m/z 185.1062 generated by splitting of the C7H7 group from the parent ion.
Lepidiline C (compound 50, Table 3 and Figure S2) had a molecular ion at m/z 307.1805 and gave the same characteristic ion as lepidiline A (m/z 185.1062) besides a fragment ion at m/z 121.0645, indicating the formation of a methoxy benzyl ion (C8H10O)+.
For compound 51 (lepidiline B, Table 3 and Figure S3), the [M – Cl]+ ion was at m/z 291.1855, and the base peak was at m/z 199.1232 (C13H15N2)+, which resulted from the loss of C7H7.
Lepidiline D (compound 54, Table 3 and Figure S4) displayed a molecular ion at m/z 321.1932 and produced a characteristic product ion due to the loss of phenyl, methoxy, benzyl, and methoxybenzyl groups. Thus, fragment ions at m/z 246.2423, 199.1232, and 121.0654 were obvious in the MS2 spectrum of lepidiline D (Figure S4). Following the same dissociation pathways, the remaining imidazole alkaloids were assigned.
2.3.1.2. Identified β-Carboline Alkaloids
Among the identified alkaloids, eight β-carboline alkaloids were detected and identified in the LC/MS chromatogram from the positive mode (Figure 1). Tetrahydro-β-carbolines are biologically active alkaloids naturally occurring in some fruit and fruit juices.23 They have antioxidants and monoamine oxidase (MAO) inhibitory activities.24
According to the literature,2,25 the characteristic fragment ions for β-carboline alkaloids are produced by the loss of C2H2 [M + H – 26]+, C2H4 [M + H – 28]+, CH2 [M + H – 14]+, and C2O2 [M + H – 56]+ groups from the precursor ions. Other ions are formed by the rearrangement of the molecular ion, leading to the loss of NH3, CH3, and C2H3N groups and the generation of [M + H – 17]+, [M + H – 15]+, and [M + H – 41]+ ions, respectively.26
The mass spectra of compounds 27 and 30 (Table 3 and Figures S5–6) showed the same [M + H]+ ion of m/z 233.1287 at retention times of 4.28 and 5.24 min, respectively. In the MS/MS fragmentation, both gave a diagnostic base peak at m/z 116.0711 [M + H – H2O]+ generated from the successive loss of C2H2 [M + H – 26]+, C2H4 [M + H – 28]+, CH2 [M + H – 14]+, and C2O2 [M + H – 56]+ groups from the precursor ions. Thus, compounds 27 and 30 were assigned as 1-methyl tetrahydro-β-5,6-hydride carboline-3-carboxylic acid isomer I and 1-methyltetrahydro-β-5,6-hydride carboline-3-carboxylic acid isomer II, which were consistent with the reported data.2,26
Compound 29 (1,2,3,4-tetrahydro-β-carboline-3-carboxylic acid, Table 3 and Figure S7) exhibited a molecular ion at m/z 217.0972 and a base peak ion at m/z 144.0800, corresponding to (C10H9N)+ dominated by the retro-Diels–Alder rearrangement of the parent trahydro-β-carboline ion.26
1-Methyltetrahydro-β-carboline-3-carboxylic acid (compound 31, Table 3 and Figure S8) displayed an abundant molecular ion at m/z 231.1127 and a major fragment ion at m/z 158.0958 (C11H12N)+ characteristic of the 1-methyl trahydro-β-carbolines.25,27 Further loss of CO2 and then NH3 groups from the molecular ions occurred and resulted in ions at m/z 187.1237 and 168.0814, respectively. Moreover, fragment ions corresponding to (C7H9N)+, (C10H9N)+, and (C9H7N)+ were detected at m/z values of 143.0731, 130.0647, and 121.0654, respectively, as shown in Figure S8.
The mass spectrum of compound 43 was identified as 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid (Table 3 and Figure S9) based on its MS/MS data and previously reported literature.2 It showed a [M + H]+ ion at m/z 285.1596 and two fragments at m/z 239.1545 (C16H20N2)+ and m/z 121.0645 (C7H9N)+, which were generated from the multiple breakages of phenyl, pentyl, and carboxylic groups as demonstrated in Figure S9.
Following the same fragmentation pathway, compound 58 was assigned as butyltetrahydro-β-5,6-carboline-3-carboxylic acid (Table 3 and Figure S10). It demonstrated a parent ion at m/z 271.1429 and two major peaks at m/z 136.0621 and m/z 121.0656 corresponding to (C8H12N2)+ and (C7H9N)+ fragments ions, respectively (Figure S10).
Two more β-carboline alkaloids (peak nos. 34 and 59, Table 3) were in accordance with the aforementioned fragmentation pathways and reported literature.2 Compound 34 was identified as 1-butyltetrahydro-β-5,6-hydridecarboline-3-carboxylic acid, while compound 59 was assigned as 1-methyl-β-carboline-3-carbaldehyde.
2.3.1.3. Identified Macaridines and Common Amide Alkaloids
Macaridines are N-hydroxypyridine derivatives previously reported in Maca roots.2,28 In this study, two macaridines were identified (compounds 45 and 48, Table 3 and Figures S11–12) in the positive mode.
Compound 45 (Table 3 and Figure S11) was identified as 3-benzyl-1,2-dihydro-4-methoxy-N-hydroxypyridine. It exhibited a molecular [M + H]+ ion at m/z 218.1176. MS2 data showed fragments at m/z 200.0005, 187.1241, 140.0699, and 121.0644, corresponding to the loss of H2O, OCH3, C6H5, and C7H7 groups, respectively, from the precursor ion.2 Furthermore, successive loss of CH and CO groups was monitored from the hydroxypyridine ring, leading to the formation of fragments at m/z 174.9839, 162.0914, and 146.9619 (Figure S11).
Compound 48 (3-benzyl-1,2-dihydro-4-carbaldehyde-N-hydroxypyridine) showed a molecular ion [M + H]+ at m/z 216.1019 and fragment ions at m/z 198.0910 and m/z 170.0960 formed by the loss of a H2O molecule and HCO group, respectively. The further neutral loss of the NH3 group from the latter ion (m/z 170.0960) generated the fragment at m/z 153.0703 (Figure S12). In addition, the cleavage of C6H5, C9H6NO2, C7H7, and C7H7NO2 groups from the precursor ion was observed.2
Furthermore, common amide alkaloids such as 2-phenylacetamide, N-methyl-3-hydroxy-benzeneacetamide, and N-benzylformamide (Table 3) were deduced by comparing their fragmentation pattern together with the literature. The diagnostic fragment ions of amide alkaloids were produced by the cleavage of an amide bond, splitting of H2O, and successive loss of CH2 groups.2,10,29,30
2.3.2. Identification of Alkylamides in Maca
Macamides are a distinct class of compounds present in Maca roots.4 They are benzylalkylamides, which exhibit many biological functions such as improving sexual potential and neuroprotective, anti-inflammatory, anticancer, and antifatigue effects.31−35 They are better ionized and detected in the positive ionization mode.36 The molecular ions, fragment ions, and retention times for 19 macamides detected in the methanolic extract (Table 3 and Figure 1) were identical to the previously reported chemical profiling of Maca roots.2,37,38 All macamides showed a high intensity of protonated molecular ions [M + H]+ in their mass spectra, while the intensity of other fragment ions corresponding to an alkyl chain was relatively low in the spectra due to the relative instability of the long-chain aliphatic group in the mass spectrometer.51N-(3-Methoxybenzyl)-(9Z,12Z)-octadecadienamide and N-benzyl-(9Z,12Z)-octadecadienamide (compounds 108 and 110) are two representatives of the identified macamides (Table 3 and Figures S13–14).
Compound 108 exhibited a molecular ion at m/z 400.3185 and fragment ions at m/z 121.0651 and m/z 138.0906 characteristic of the N-(3-methoxybenzyl)-type macamides.39,40
Compound 110 showed a molecular ion at m/z 370.3127 beside the product ion at m/z 108.0808 characteristic of the N-benzyl-type macamides.39 The remaining macamides (Table 3) were identified following the same dissociation pathways which include the formation of the distinct ions at m/z values of 108, 121, and 138 which corresponded to the formation of N-benzyl, 3-methoxybenzyl, and N-3-methoxy benzyl ions, respectively.21,27
2.3.3. Identification of Macaenes and Fatty Acids in Maca
Macaenes are characteristic polyunsaturated fatty acids found in Maca roots.21 In the present study, six macaenes were observed mainly in the negative ionization mode (Figure 2). Among them, 55-oxo-6E,8E-octadecadienoic acid (compound 74, Table 3) was demonstrated as an example. Compound 74 demonstrated an obvious [M – H]− ion at m/z 293.2129 in the mass spectrum (Figure S15). The fragments ions at m/z 279.2331 and 152.9949 indicated the loss of CH2 and C10H18 groups, respectively, from the molecular ion (Figure S15). The fragment at m/z 125.0964 was generated by the cleavage of a CO group from the fragment ion at m/z 152.9949, while the product ion at m/z 185.1174 was attributed to the loss of a C9H16 group from the parent ion.2 The MS behaviors of the other identified macacenes were in line with that of compound 74 (Table 3).
Twenty-six fatty acids, mainly polyunsaturated and hydroxylated, such as oleic acid, linolenic acid, and hydroxy-linoleic acid, were tentatively identified in Maca extract (Table 3). The diagnostic fragment ions generated by loss of carbon dioxide and water molecules were predominant in their MS/MS spectra.41 Product ions were generated by loss of C6H6, C5H6O2, and C8H8O2 groups from the molecular ions.42
2.3.4. Identification of Glucosinolates in Maca
Glucosinolates are common secondary metabolites present in Maca and are responsible for the pungent flavor of the roots. Glucosinolates are assumed to exhibit many biological effects such as cancer chemoprotective, antioxidant, and apoptosis-promoting activities.29 Furthermore, glucosinolates can modulate insulin resistance and improve metabolic disorders.43 Glucotropaeolin, the most abundant glucosinolate in Maca, was observed in the negative mode at a retention time of 3.92 min (compound 26, Table 3 and Figure 2). The mass spectrum of glucotropaeolin (Figure S16) showed a molecular [M – H]− ion at m/z 408.0435 and a glucosinolate specific fragment ion at m/z 259.0134 corresponding to the neutral loss of R-N=C=S from the parent ion. Moreover, another diagnostic peak at m/z 195.0338, corresponding to the formation of a d-thioglucose fragment (C6H11O5S)− was observed in the spectrum.44 Two product ions at m/z 328.0877 (C14H19O6NS)− and 166.0322 (C8H7ONS)− were assigned as the loss of the SO3– group and further loss of glucose. Three fragment ions were noticed at the following m/z values, 274.9896 (C6H12O8S2), 241.0049 (C6H10O8S)−, and 212.8781 (C8H7O4NS)−, which were generated by the intramolecular rearrangements through which the sulfate group is transferred to the thioglucose moiety.2
2.3.5. Identification of Hydantoins and Thiohydantoins in Maca
One hydantoin derivative, meyeniihydantoin A, was detected in the positive ion mode at a retentoin time of 10.77 min (compound 53, Table 3 and Figure S17). It produced a protonated molecular ion at m/z 261.1234. MS2 data showed fragments at m/z 231.0294, 184.0737, and 121.0644, indicating loss of OCH3, C6H5, and C6H8N2O2 groups, respectively, from the molecular ion.6,45
Meyeniin C (compound 47, Table 3), a thiohydantoin derivative, that was recently isolated from Maca roots46 has been detected in both ionization modes. In the negative ion mode at a retention time of 9.57 min (Figure 2), meyeniin C revealed a parent ion with m/z 307.0724 (Figure S18). The successive breakdown of the hexahydroimidazo[1,5-c]thiazole ring in MS2 experiment was responsible for the fragmentation behavior of meyeniin C (Figures S18–19).6,29 In the MS/MS analysis of meyeniin C, the characteristic fragment ions at m/z 263.0815 (C13H15N2O2S)−, 233.0729 (C11H9N2O2S)−, and 183.9132 (C9H13NOS)− were yielded by neutral loss of the following groups, C=S, C3H6S, and C5H2NOS, respectively, from the precursor ion. The further loss of a methyl group from (C13H15N2O2S)− yielded the product ion at m/z 248.9609 (C12H13N2O2S)− which in turn produced the fragment at m/z 205.0779 (C11H13N2O2)− through the cleavage of the C=S group. Another fragment ion appeared at m/z 191.0604 (C10H11N2O2)− due to the splitting of the CH2 group from the ion at m/z 205.0779 (C11H13N2O2)−. Furthermore, two fragments at m/z 146.9383 and 134.8644 were generated after the cleavage of CH4NO and C2H2NO units, respectively, from the ion at m/z 191.0604 (Figures S18–19).
2.3.6. Identification of Organic Acids and Miscellaneous Compounds in Maca
Acids undergo better fragmentation and give higher abundant ions in the negative ionization mode. The fragmentation patterns of identified organic acids were consistent with the previously reported data.2,47 In agreement with the previous literature, the loss of CH2, CO2, and H2O groups was considered as the representative fragmentation pathway in acids (Table 3). Based on the accurate mass measurements and MS/MS data, seven organic acids have been identified, such as succinic acid, hydroxy succinic acid, and 2,4-dihydroxy-3,5-cyclopentyl dienoic acid (Table 3).
Finally, 24 miscellaneous compounds were identified in Maca extract including amino acids, organic esters, sugars, and benzyl isocyanate (Table 3). Their identities were confirmed by comparing their retention times and fragmentation patterns with data given in the literature,48,49 besides searching the available online databases.
2.4. In Silico Studies
2.4.1. Molecular Docking Studies
Although the in vitro ACE inhibition activity of Maca roots has been previously studied,17 the correlations between Maca chemical constituents and enzyme inhibitions have not yet been studied. Therefore, in this study ACE and renin inhibitory properties of different extracts of Maca were evaluated followed by metabolite profiling of the active extract using UPLC/HRMS. Moreover, the possible mechanisms of action based on structure–activity relationships and molecular docking were investigated.
To examine the possible ligand-structure/enzyme-binding relationships and to confirm the experimental data, selected compounds from each class, viz., lepidilines, β-carbolines, macaridines, macamides, glucosinolates, etc., were docked into the active site of ACE and renin.
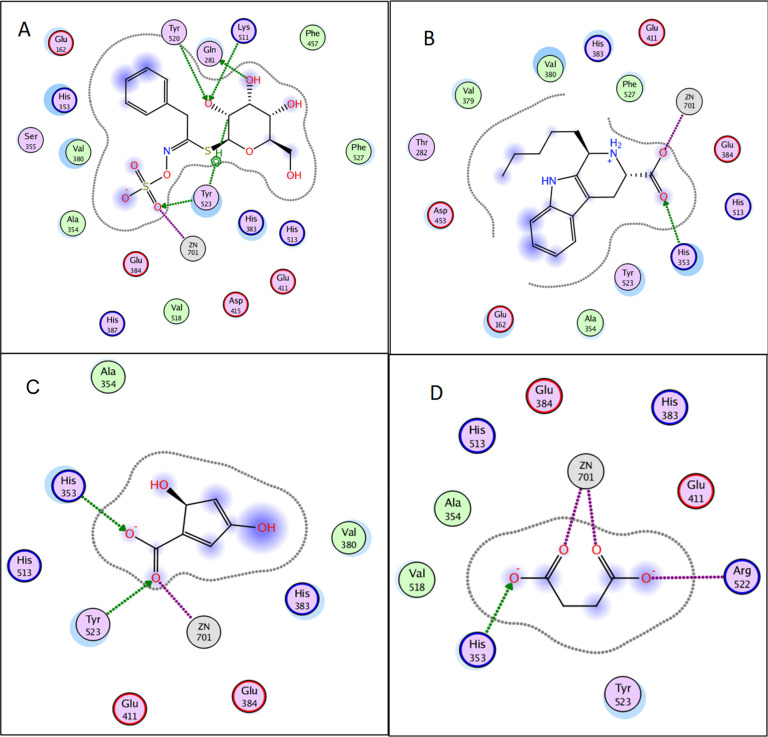
First, docking of the identified compounds into the three-dimensional X-ray crystallographic structure of the angiotensin converting enzyme (PDB code: 1O86) in complex with lisinopril was accomplished using Molecular Operating Environment (MOE) 2010.10. The antihypertensive drug, lisinopril, forms hydrogen bonds with Tyr 520, Lys 511, His 353, and Tyr 523 of ACE through its carbonyl and carboxylate groups, in addition to ionic bonds with Glu384 and Zn701 via charged amino and carboxylate groups (Figure S20). Interestingly, zinc is an essential catalytic component of ACE. Thus, interaction with the zinc-binding motif is crucial for ACE inhibition.
To confirm the accuracy of the used docking process, validation was achieved by redocking of the cocrystallized ligand into the active site of ACE. The validation results showed that the docking protocol is suitable for the anticipated docking study. This is confirmed by the perfect alignment between the coordinates of lisinopril and its self-docked pose with root-mean-square deviation (RMSD) of 0.811 Å and docking score (S) of −36.64 kcal mol–1. Moreover, the self-docked pose was able to attain the same binding interactions shown by the cocrystallized ligand with the key amino acids in the binding site of ACE.
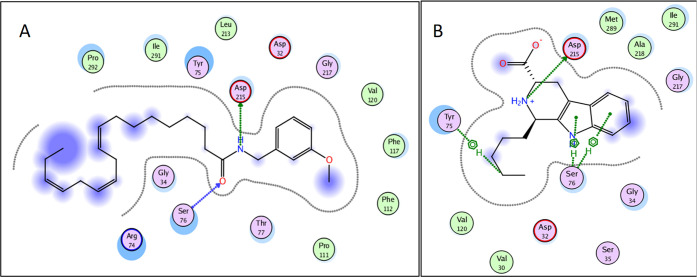
The results of the docking study in the active site of ACE showed that out of all studied compounds, ten compounds showed promising results. Glucotropaeolin, succinic acid, 2,4-dihydroxy-3,5-cyclopentyl dienoic acid exhibited high affinity to the target enzyme in descending order. Interestingly, the identified β-carboline alkaloids except for (1R,3S)-1-methyl-β-carboline-3-carbaldehyde (compound 59) showed high binding affinity to ACE active site. The affinity of the compounds to the target enzyme was judged by comparing the binding free energy (docking score) and the interactions displayed by the docked compounds with that of the cocrystallized ligand (Table 4). Upon docking in the active site of ACE, all the mentioned compounds showed ionic bond interactions with the zinc-binding motif (Figures 3 and 4). Additionally, glucotropaeolin with the highest docking score (S = −35.32 kcal mol–1) displayed multiple hydrogen bond interactions, with the amino acids Lys511, Tyr 520, Gln281, and Tyr523 achieving a binding mode like that of the native ligand lisinopril, which can be attributed to the presence of ionized sulfonic acid and hydroxyl groups (Figures 3A and 4A). Furthermore, the β-carbolines showed comparable docking scores to that of the cocrystallized ligand and demonstrated the same interactions with Zn701, His353, and/or Tyr 523 (Figures 3B and 4B). However, (1R,3S)-1-Methyl-β-carboline-3-carbaldehyde exhibited minimal binding affinity to the ACE active site which is probably due to the lack of the carboxyl group possessed by the other β-carbolines. In the same vein, the two organic acids, succinic acid and 2,4-dihydroxy-3,5-cyclopentyl dienoic acid, attained similar binding interactions in the ACE active site with comparable docking scores S = −34.56 and −31.39 kcal mol–1, respectively (Figures 3C–D and 4C–D).
Table 4. Docking Scores as “S” (kcal mol–1) and Bond Interactions of the Docked Compounds Showing the Highest Affinities with the Zinc-Binding Motif of ACE and the Key Amino Acids of ACE and Renina.
|
S (kcal mol–1) |
amino
acids involved in hydrogen bonds |
|||
|---|---|---|---|---|
| compound | ACE | renin | ACE | renin |
| glucotropaeolin (compound 26) | –35.32 | –15.11 | Zn701 | |
| Lys511 | ||||
| Tyr520 | Ser219 | |||
| Gln281 | ||||
| Tyr523 | ||||
| 1,2,3,4-tetrahydro-β-carboline-3-carboxylic acid (compound 29) | –31.15 | –14.17 | Zn701 | Asp215 |
| His353 | ||||
| (1R,3S)-1-methyltetrahydro-β-carboline-3-carboxylic acid (compound 31) | –29.86 | –16.96 | Zn701 | Ser76 |
| His353 | Thr295 | |||
| Asp215 | ||||
| (1S,3S)-1-methyltetrahydro-β-5,6-hydride carboline-3-carboxylic acid (compound 27/30) | –34.56 | –14.13 | Zn701 | Ser219 |
| His353 | ||||
| Tyr523 | ||||
| (1R,3S)-1-butyltetrahydro-β-5,6-hydride carboline-3-carboxylic acid (compound 34) | –34.93 | –13.50 | Zn701 | Thr295 |
| His353 | Gln128 | |||
| (1R,3S)-1-methyltetrahydro-β-5,6-hydride carboline-3-carboxylic acid (compound 27/30) | –33.15 | –11.56 | Zn701 | Ser219 |
| His353 | ||||
| (1R,3S)-1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid (compound 43) | –33.90 | –20.06 | Zn701 | Asp215 |
| His353 | ||||
| (1R,3S)-1-butyltetrahydro-β-5,6-carboline-3-carboxylic acid (compound 58) | –32.37 | –18.45 | Zn701 | Asp215 |
| His353 | ||||
| (1R,3S)-1-methyl-β-carboline-3-carbaldehyde (compound 59) | –22.51 | –13.30 | - | Thr77 |
| succinic acid (compound 23) | –34.56 | –8.32 | Zn701 | Thr295 |
| His353 | Gln128 | |||
| 2,4-dihydroxy-3,5-cyclopentyl dienoic (compound 28) | –31.39 | –9.59 | Zn701 | Thr295 |
| His353 | Gln128 | |||
| Tyr523 | ||||
| N-(3-methoxybenzyl)-(9Z,12Z,15Z)-octadecatrienamide (compound 98) | –11.49 | –28.25 | - | Ser76 |
| Asp215 | ||||
| N-(3-methoxybenzyl)-(9Z,12Z)-octadecadienamide (compound 108) | - | –26.54 | - | Gly217 |
| N-(3,4-dihydroxybenzyl)-oleamide (compound 119) | - | –25.30 | - | Ser76 |
| Asp215 | ||||
| N-(3,4-dimethoxybenzyl)-hexadecanamide (compound 114) | - | –24.40 | - | Gln128 |
| Thr295 | ||||
| N-(3-hydroxybenzyl)-11Z,13Z,15Z,17Z-octadecatetrienamide (compound 61) | - | –22.47 | - | Ser76 |
| Thr295 | ||||
| methanandamide (compound 101) | - | –22.74 | - | Gly217 |
| Tyr14 | ||||
-: no detected interactions.
Figure 3.
2D diagrams of glucotropaeolin (A), 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid (B), succinic acid (C), and 2,4-dihydroxy-3,5-cyclopentyl dienoic (D) showing their interactions with the ACE active site.
Figure 4.
3D representation of glucotropaeolin (A), 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid (B), succinic acid (C), and 2,4-dihydroxy-3,5-cyclopentyl dienoic (D) (shown in magenta) demonstrating their interactions with the ACE active site.
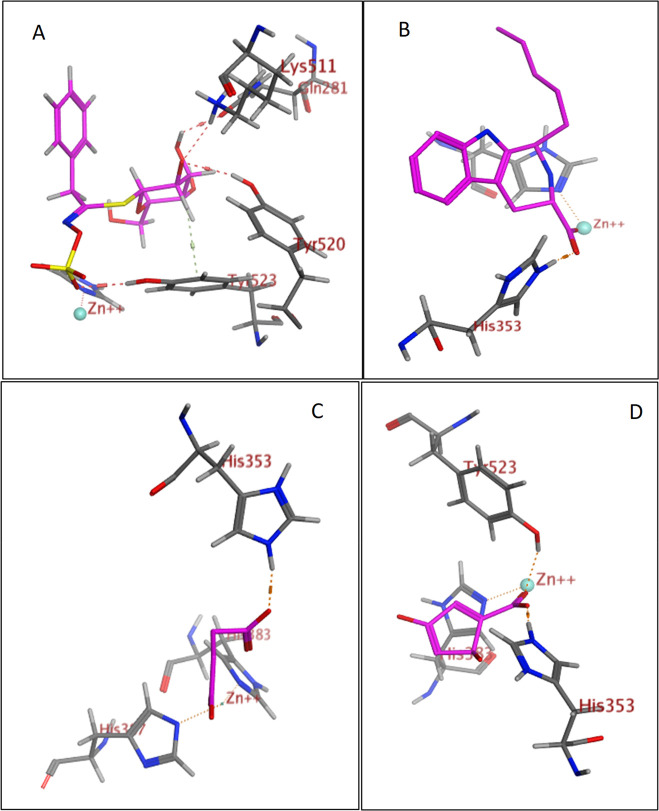
Second, docking of the identified compounds into the three-dimensional X-ray crystal structure of renin (PDB code: 2V0Z) complexed with aliskiren was performed. The cocrystallized ligand forms multiple hydrogen bonds with Arg74, Gly34, Asp32, Asp215, Ser76, and Tyr14 through its amino, hydroxyl, and methoxy groups, in addition to water-mediated hydrogen bonds with Thr295 and Gln128 (Figure S21). The validation results revealed a perfect alignment between the coordinates of the inhibitor and its self-docked pose with root-mean-square deviation (RMSD) of 1.33 Å and docking score (S) of −20.33 kcal mol–1.
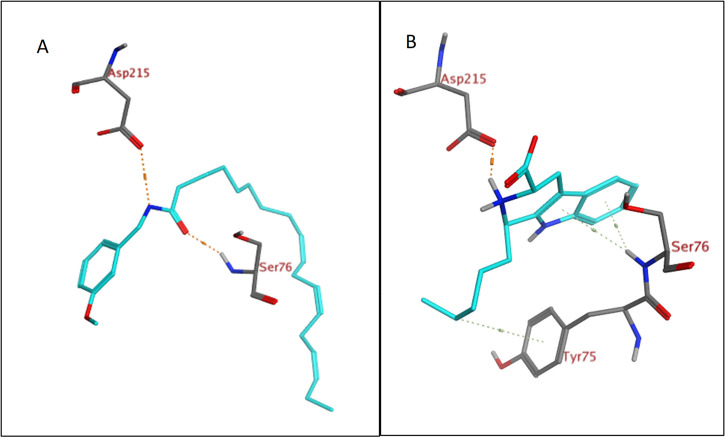
The obtained results revealed that 15 compounds showed good binding interactions. Macamides attained very poor docking scores toward ACE. However, upon docking of the different classes of the identified compounds into the active site of renin, several macamides (Table 4) displayed high docking scores (S ranging from −22.47 to −28.25 kcal mol–1), which are even higher than that of the native ligand (S = −20.33 kcal mol–1). N-(3-Methoxybenzyl)-(9Z,12Z,15Z)-octadecatrienamide interacted with two renin amino acid residues (Asp215 and Ser76), as illustrated in Figures 5A and 6A. Interesingly the eight identified β-carbolines have achieved docking scores comparable to that of the native ligand (S ranging from −13.50 to −20.06 kcal mol–1). For example, 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid displayed a hydrogen bond interaction with Asp215 in addition to three hydrogen−π interactions with Ser76 and Tyr75, as shown in Figures 5B and 6B. It is worth mentioning that renin contains two catalytic residues, namely, Asp32 and Asp215, located at its active site.50 In the present work, several mecamides and carbolines bound to Asp215, as shown in Table 4, suggesting that they can compete with the substrate for the active site.
Figure 5.
2D diagrams of N-(3-methoxybenzyl)-(9Z,12Z,15Z)-octadecatrienamide (A) and 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid (B) showing their interactions with the renin active site.
Figure 6.
3D representation of N-(3-methoxybenzyl)-(9Z,12Z,15Z)-octadecatrienamide (A) and 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid (B) (shown in light blue) demonstrating their interactions with the renin active site.
Finally, the molecular docking outcomes suggest that the promising inhibitory activity of the methanolic extract against ACE might be attributed to glucotropaeolin, β-carbolines, succinic acid, and 2,4-dihydroxy-3,5-cyclopentyl dienoic acid. In the same vein, macamides and β-carbolines have the highest affinity toward renin which could also justify the promising antihypertensive activity of the extract.
2.4.2. Molecular Dynamics and Binding Free Energy Calculation
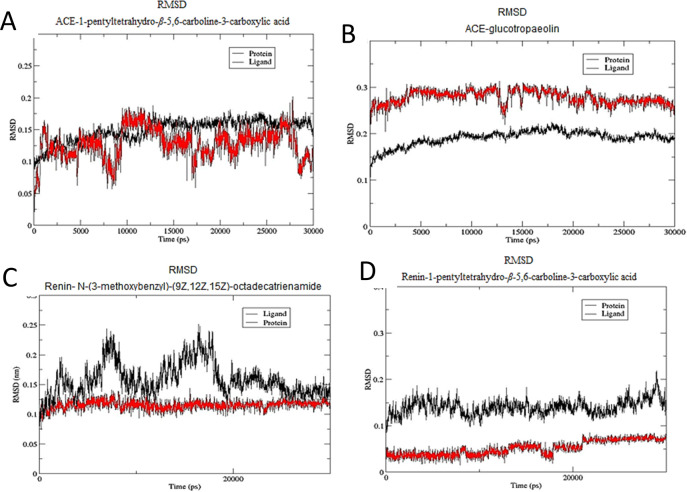
The stability of the ligands within the binding pockets of the enzyme was examined through the comparison of the RMSD of the ligand with the protein (Figure 7a–d). The RMSD of the ACE complexes with both 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid and glucotropaeolin showed flucctuations of 0.1 Å. This is slightly higher than the fluctuations seen in the renin complexes with N-(3-methoxybenzyl)-(9Z,12Z,15Z)-octadecatrienamide and 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid, which was found to be 0.05 Å. However, both values are small, suggesting that the ligand–protein complex is stable. Moreover, the trajectories resulting from the molecular dynamics simulations were visually examined to further study the stability of the ligands within the binding site.
Figure 7.
RMSD plot of (A) ACE shown in black and 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid shown in red, (B) ACE shown in black and glucotropaeolin shown in red, (C) renin enzyme shown in black and N-(3-methoxybenzyl)-(9Z,12Z,15Z)-octadecatrienamide shown in red, and (D) renin enzyme shown in black and 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid.
Additionally, the number of hydrogen bonds formed between the enzymes and the ligands throughout the simuation were also examined (Figure 8). The ACE glucotropaeolin was found to form an average of four hydrogen bonds throughout the simulation, while both the 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid complex with ACE and N-(3-methoxybenzyl)-(9Z,12Z,15Z)-octadecatrienamide complex with a renin enzyme were found to form an average of two hydrogen bonds throughout the simulation. Finally, the renin–1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid complex was found to form only an average of one hydrogen bond throughout the simulation. This is in agreement with the results observed in the docking analysis.
Figure 8.
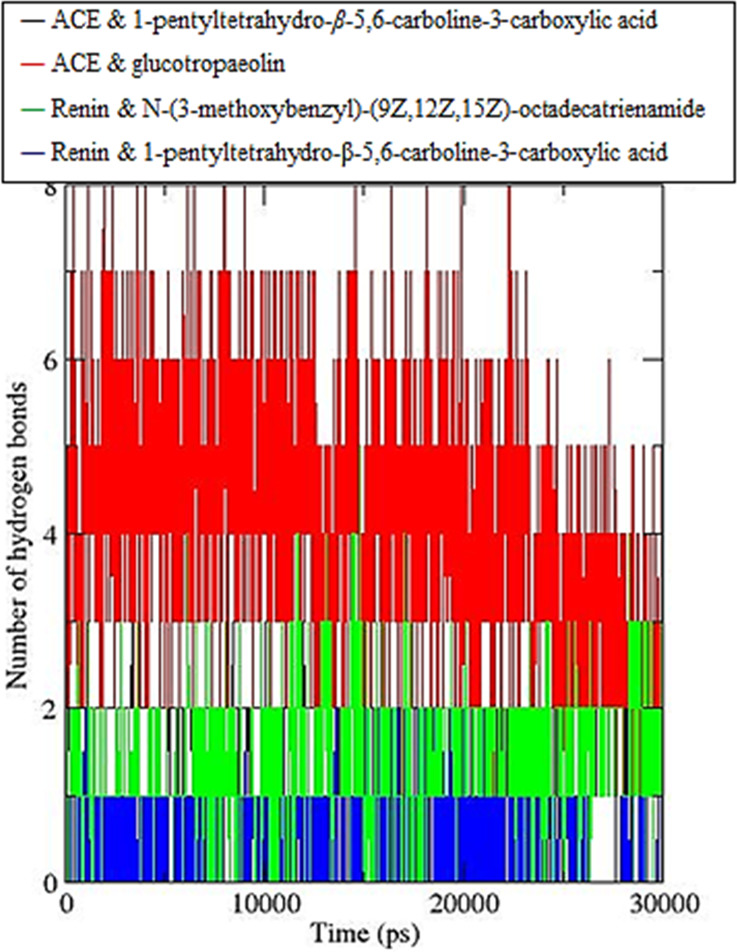
Number of hydrogen bonds calculated over 30 ns molecular dynamics, where the ACE and 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid simulation is shown in black, the ACE and glucotropaeolin simulation is shown in red, the renin enzyme and N-(3-methoxybenzyl)-(9Z,12Z,15Z)-octadecatrienamide simulation is shown in green, and the renin enzyme 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid simulation is shown in blue.
Besides, the solvent-acceptable surface area (SASA) (Figure 9) was also alnalyzed for the four complexes, which could indicate changes in the teritiary structure of the protein. The results showed that the SASA of the four complexes showed small fluctuations throughout the simulations, suggesting that there was no change in the structure of the protein during the simulation. The estimation of the binding energies is shown in Table 5. The binding of 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid with ACE showed the most promising result, with the binding energy estimate calculated at −27.03 ± 5.8 kcal mol–1, with a significant contribution by electrostatic and van der Waals interactions. The binding energy for the glucotropaelin–ACE complex was estimated to be −25.78 ± 9.6 kcal mol–1, which is also promising with a significant contribution by van der Waals. On the other hand, the results obtained from the interaction energy analysis for the complexes with a renin enzyme showed less promising results, where the binding energy estimate for N-(3-methoxybenzyl)-(9Z,12Z,15Z)-octadecatrienamide with renin was −7.20 ± 5.6 kcal mol–1, whereas the binding energy estimate for 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid with renin was found to be 17.5406 ± 7.04 kcal mol–1. The poor results for the renin complexes may be attributed to the high free solvation energy.
Figure 9.
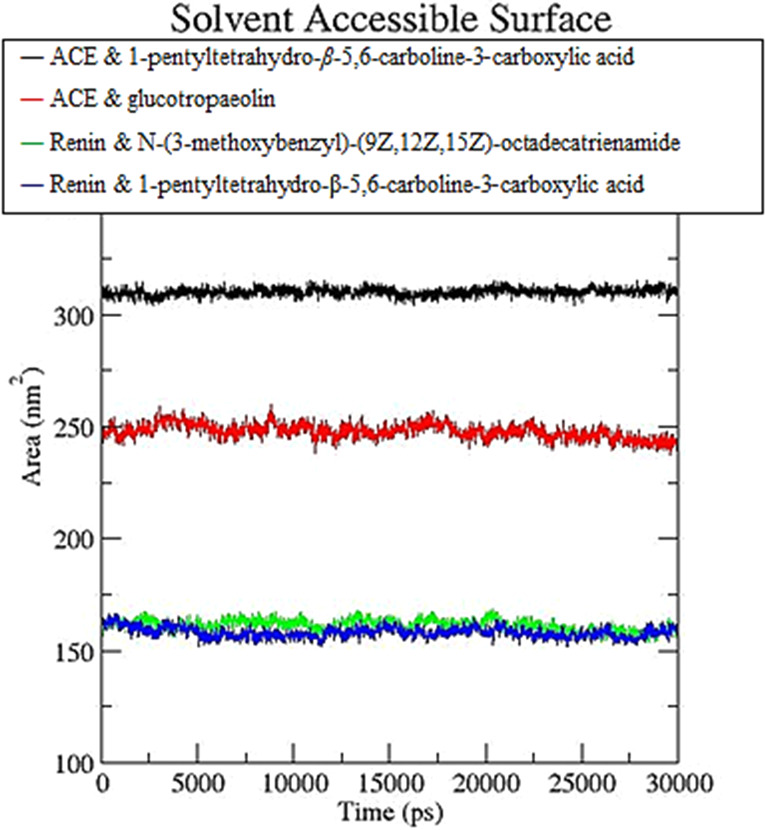
SASA analysis calculated over 30 ns molecular dynamics, where the ACE and 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid simulation is shown in black; the ACE and glucotropaeolin simulation is shown in red; the renin enzyme and N-(3-methoxybenzyl)-(9Z,12Z,15Z)-octadecatrienamide simulation is shown in green; and the renin enzyme and 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid simulation is shown in blue.
Table 5. Binding Energy Estimation.
| Compound | ||||
|---|---|---|---|---|
| ACE |
renin |
|||
| interaction (kcal/mol) | 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid | glucotropaeolin | 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid | N-(3-methoxybenzyl)-(9Z,12Z,15Z)-octadecatrienamide |
| VDWAALS | –26.44 | –36.44 | –23.86 | –27.02 |
| EEL | –57.5 | 3.71 | –13.54 | 5.32 |
| EGB | 60.97 | 12.56 | 59.26 | 19.27 |
| ESURF | –4.06 | –5.62 | –4.3194 | –4.77 |
| ΔTOTAL | –27.03 | –25.79 | 17.5406 | –7.2 |
2.4.3. In Silico ADMET Study
2.4.3.1. Pharmacokinetics and Drug-Likeness Properties
The prediction of pharmacokinetics and drug-likeness aspects of the compounds glucotropaeolin, (1R,3S)-1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid, and N-(3-methoxybenzyl)-(9Z,12Z,15Z)-octadecatrienamide was done using the freely available web server Swiss ADME (http://www.swissadme.ch/).30 These compounds were selected to perform the in silico study as they attained the most promising results in the docking study. The results of the Swiss ADME prediction of drug likeness of these compounds are shown in Table 6.
Table 6. ADME Properties of the Phytoleading Compounds Predicted Using the Swiss ADME Servera,b.
| property | glucotropaeolin | 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid | N-(3-methoxybenzyl)-(9Z,12Z,15Z)-octadecatrienamide |
|---|---|---|---|
| MW | 409.43 | 274.36 | 397.59 |
| consensus log Po/w | –0.63 | 1.28 | 6.75 |
| log S (ESOL) | very soluble | very soluble | moderately soluble |
| #rotatable bonds | 7 | 4 | 17 |
| #H-bond acceptors | 10 | 3 | 2 |
| #H-bond donors | 5 | 3 | 1 |
| MR | 91.57 | 86.37 | 126.01 |
| TPSA | 199.79 | 61.36 | 38.33 |
| Lipinski violations | 0 | 0 | - |
| Ghose violations | 0 | 0 | - |
| Veber violations | 1 | 0 | - |
| Egan violations | 1 | 0 | - |
| Muegge violations | 1 | 0 | - |
| bioavailability score | 0.11 | 0.55 | 0.166 |
| PAINS alerts | 0 | 0 | 0 |
| Brenk alerts | 4 | 0 | 0 |
| leadlikeness violations | 1 (MW > 350) | 0 | - |
| GI absorption | low | high | high |
| BBB permeant | no | yes | no |
| P-gp susbstrate | yes | yes | no |
| CYP1A2 inhibitor | no | no | no |
| CYP2C19 inhibitor | no | no | no |
| CYP2C9 inhibitor | no | no | no |
| CYP2D6 inhibitor | no | no | no |
| CYP3A4 inhibitor | no | no | yes |
BBB: blood–brain barrier; GI: gastrointestinal; MR: molar refractivity; P-gp: P-glycoprotein; PAINS: pan assay interfering substances; TPSA: topological polar surface area.
-: not determined.
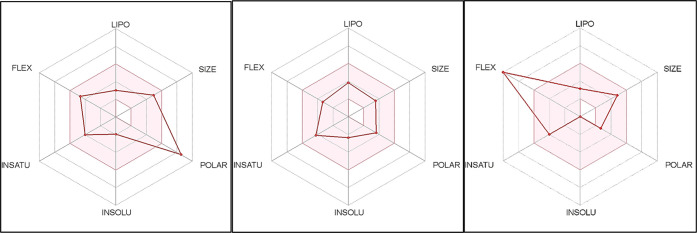
Human gastrointestinal absorption (HIA), blood–brain barrier (BBB) penetration, substrate or nonsubstrate for glycoprotein (P-gp) permeability, and the interaction of molecules with cytochrome P450 isomers (CYP) were predicted. Also, bioavailability scores were calculated based on the Lipinski rule of five51 and other models. Moreover, the oral bioavailability radar chart of the studied compound indicated their good predicted oral bioavailability and promising pharmacokinetic properties (Figure 10).
Figure 10.
Bioavailability radar chart of the phytoleading compounds (glucotropaeolin, 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid, and N-(3-methoxybenzyl)-(9Z,12Z,15Z)-octadecatrienamide, respectively). The pink area represents the range of the optimal property values for oral bioavailability, and the red line is the compound’s predicted properties. Saturation (INSATU), size (SIZE), polarity (POLAR), solubility (INSOLU), lipophilicity (LIPO), and flexibility (FLEX).
2.4.3.2. Toxicity Prediction
The selected compounds were further passed through one more web server, Osiris Property Explorer (http://www.organic-chemistry.org/prog/peo/), to predict their potential toxicities. Prediction through this program depends on the functional group resemblance of the tested compounds with the widely in vitro and in vivo studied compounds comprised in its database. The output data are demonstrated as color coded: red, green, and yellow. The green color suggests low toxic potential; yellow proposes mild toxicity; and red displays a high probability of toxicity.52,53 The outcomes revealed that all the examined compounds are expected to be safe, displaying no toxicity concerning mutagenicity, tumorigenicity, and irritant effects (Table S1).
3. Conclusion
In this study, we established an effective strategy based on the metabolite profiling and molecular docking for screening and determining compounds with ACE and renin inhibitory effects in Maca extract, whereas the docking simulations predicted the molecular interactions and revealed the binding modes of 17 compounds within the putative binding pockets of ACE and renin. The results of the molecular dynamics simulations further investigated the interactions formed between the most promising candidates and the ACE and renin enzymes. Moreover, the molecular dynamincs simulations also investigated the binding energies and stability of the complexes. The outcomes of the MD simulations suggested that the screened compounds have a higher potential to act as ACE inhibitors rather than renin inhibitors, due to the higher binding energy values estimated by the PBSA calculations. Proper drug likeness properties and safe toxicological profiles were attained by some selected compounds, which was illustrated by the computational ADMET study. Finally, our results specified Maca compounds as potential bioactive markers that could lay a foundation for the development of new hypotensive drugs derived from natural sources.
4. Methods
4.1. Plant Material and Extraction
Maca roots were purchased from Haraz Herbal Store in March 2020 and authenticated by Mrs. Teresa Labib, Head of the Taxonomists at Orman Botanic Garden. A herbarium voucher was deposited at Herbarium of the Department of Pharmacognosy, College of Pharmacy, Cairo University, Egypt (No. 7.3.2020). For extraction, four different solvents (water, methanol, 50% methanol, and methylene chloride) were used to extract the chemical components from Maca roots. The powders (100 g each) were extracted separately using an Ultra Turrax T50 homogenizer (IKA, Janke & Kunkel, Germany). Solvents (methanol, 50% methanol, and methylene chloride) were removed with a rotary evaporator to obtain the extracts in the yield of 1.67%, 1.89%, and 0.95% (w/w), respectively. The water extract was dried in a freeze-drier to obtain an extract in the yield of 1.50% (w/w).
4.2. Chemicals and Reagents
The solvents used were of analytical grade (El-Gomhouria Co., Cairo, Egypt). Renin enzyme, renin substrate (Arg-Glu(EDANS)-Ile-His-Pro-Phe-His-Leu-Val-Ile-His-Thr-Lys (Dabcyl)-Arg), ACE extracted from rabbit lung, ACE substrate (Hippuryl-histidyl-leucine, HHL), Quinapril, ABTS (2,2′-azino-bis-3-ethylbenzthiazoline-6-sulfonic acid), and ascorbic acid were all obtained from Sigma-Aldrich Chemicals, Germany. Sodium phosphate, ammonium molybdate, potassium persulfate, potassium ferricyanide, and ferric chloride were purchased from El-Nasr Company for Pharmaceutical Chemicals, Egypt. Phosphate buffer was bought from Bio diagnostic, Egypt. HPLC-grade acetonitrile, trichloroacetic acid, and methanol were purchased from SDFCL Fine-Chem Limited, Mumbai, India. Round bottom and black microplates (96 wells) were obtained from Corning Incorporated (USA).
4.3. Antioxidant Assays
DPPH, ABTS radical scavenging, TAC, and the iron-reducing power assays were assessed as described in previous works.54 Ascorbic acid was used as a standard.
In DPPH: The DPPH radical scavenging activity (%) was calculated as follows: IC50 = [(Ac – As)/Ac] × 100, where Ac was the absorbance of control (DPPH solution without a test sample) and As was the absorbance of the sample [DPPH solution + sample (extract/standard)]. The IC50 is defined as the concentration of substrate that causes 50% reduction of the DPPH color.
In iron-reducing power, the EC50 value (μg/mL) is the effective concentration giving an absorbance of 0.5 for reducing power and was obtained from linear regression analysis.
In FRAP, ABTS, and TAC, total antioxidant was expressed as ascorbic acid equivalent (AAE) (μmol/g of extract) after construction of the ascorbic acid calibration curve.
4.4. Renin and ACE Inhibition Assay
The renin inhibitory potentials of the samples and ACE were tested according to the method described in the previous literature.55 The inhibitory activities of the extracts were represented as percentages of inhibition in a concentration range of 10–150 ng/mL. An antihypertensive agent, Quinapril, was used as a standard at a concentration of 1–20 ng/mL. All experiments were performed 3 times.
4.5. Statistical Analysis
Each experiment was done in triplicate (n = 3). Statistical analysis was performed using SAS software ver. Sixteen (SAS Institute Inc., Cary, NC, USA). One-way ANOVA was performed, and the resultant values at P ≤ 0.05 were considered significant. Different letters indicate the significance according to Duncan’s multiple range test. EXCEL 2016 and SPSS software were used for data analysis.
4.6. UPLC/HRMS Analysis
The extract was resuspended in methanol, and 2 μL was injected and separated on a reversed phase (RP) C18 column, using a UPLC system. The mass spectra were obtained in positive and negative ionization modes using a heated electrospray ionization (HESI) source in combination with an Exactive, Orbitrap-type HRMS.56
4.7. Data Mining and Processing
The RAW data were converted with ProteoWizard 3.0 into mzXML format and imported to MZmine 2.53 software. Mass ion peaks were isolated with a centroid mass detector threshold with the noise level set to 1.0 × 104 and an MS level of 1–2. The chromatogram builder was used with a minimum time span set to 0.001 min and the minimum height and m/z tolerance to 1 × 106 and 0.001 m/z or 5.0 ppm, respectively. Chromatogram deconvolution was performed using a baseline cutoff algorithm with minimum peak height of 1 × 106, peak duration range of 0–0.1 min, and baseline level of 1× 104. The separated peaks were then deisotoped using the function of isotopic peaks grouper (m/z tolerance: 0.001 m/z or 5.0 ppm, retention time tolerance: 0.05 absolute (min), maximum charge: 2, and representative isotope: most intense). The parameters for data filtering and gap filling were set to m/z tolerance 0.001 m/z or 5.0 ppm, retention time tolerance 0.05 absolute (min), and minimum standard intensity 1 × 104. An adduct search was performed for Na+, K+, NH+4, formate, and ACN+ (RT tolerance: 0.05 absolute (min), m/z tolerance: 0.001 m/z or 5.0 ppm, max relative adduct peak height: 50%). The processed data were then subjected to formula prediction by selecting atoms C, H, N, O, and S and adjusting parameters with a heuristic element count with all suboptions to get the isotope pattern filter working with all features with isotope peaks. Finally, peak identification was used to search for unidentified metabolites using KEGG and LipidMaps online databases or Dictionary of natural products, 2015 (DNP, 15).
4.8. In Silico Studies
4.8.1. Docking Study
All the molecular docking studies were achieved using Molecular Operating Environment (MOE, 2010.10) software. The X-ray crystal structure of the angiotensin converting enzyme (PDB code: 1O86),57 in complex with lisinopril and renin cocrystallized with aliskiren as an inhibitor (PDB code: 2V0Z),58 were imported from the protein data bank. Regarding ACE protein structure, it was prepared for docking by removal of all water molecules, whereas the cofactors, zinc, and two chloride ions were retained throughout the docking process. As for renin protein structure, the enzyme was prepared by removal of chain B and all ligands and water molecules that are not involved in the binding. The protein structures were prepared for docking study using the Protonate 3D protocol in MOE with default options. The structures of the compounds were constructed using ChemBioDraw Ultra 17.0, and their SMILES were copied to MOE. 3D Protonation of the compounds was carried out using the precise mode in “Protonate” tool, and the most prevalent ionized form was selected for subsequent steps. Energy minimization of the structures using the MMFF94x force field and a gradient of 0.05 was applied using the “Energy Minimize” tool. The partial charges were automatically calculated for each molecule. Conformational analysis was run using the default settings for a systematic search. The lowest energy conformer of each molecule was saved to another database to be docked into the binding site of the target enzymes. The Triangle Matcher placement method and London dG scoring function were used for the evaluation of the binding pattern and binding affinity of the ligands. The cocrystallized ligands were used to define the active site for docking. A docking protocol was first validated by redocking of the cocrystallized ligands in the active site of the enzymes with an energy score (S) = −36.64 kcal mol–1 and root-mean-square deviation (RMSD) of 0.811 Å for ACE and energy score (S) = −20.33 kcal mol–1 and root-mean-square deviation (RMSD) of 1.33 Å for renin. The validated docking protocol was then used to study the ligand–target interactions for the identified compounds in the active site of the target enzymes. This was performed to predict their binding modes and binding affinities to rationalize the promising antihypertensive activity.
4.8.2. Molecular Dynamics and Binding Free Energy Calculation
Molecular dynamics simulations were carried out to further validate the molecular docking results for the compounds showing the highest activity. The selected compounds were as follows: 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid which showed promising results for both enzymes, ACE and renin, in addition to N-(3-methoxybenzyl)-(9Z,12Z,15Z)-octadecatrienamide and glucotropaeolin, which showed promising results for renin and ACE, respectively. The best docked pose for each enzyme–ligand complex was selected and used for the 30 ns molecular dynamics simulation using GROMACS (Groningen Machine for Chemical Simulations).59 Both the topology and the coordinate file for the proteins were generated using the AMBER99SB force field,60 while the ligand parametrization was carried out using the ACPYPE (AnteChamber Python Parser Interface)61 and the automated topology builder (http://bio2byte.be/acpype/).62 The system was solvated using TIP3P water molecules, and 15932 and 15941 water molecules were added to the renin-N-(3-methoxybenzyl)-(9Z,12Z,15Z)-octadecatrienamide and renin-1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid systems, respectively. Similarly, 25 663 and 25 665 water molecules were added to the ACE–1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid and ACE-glucotropaeolin systems, respectively. Na+ and Cl– ions were added to the system to achieve a final concentration of 0.15 mM as well as to neutralize the system. For the renin system, 57 Na+ and 49 Cl– ions were added, while 93 Na+ and 79 Cl– were added to the ACE system. The systems were then equilibrized through energy minimization, followed by two restrained molecular dynamics simulations. Molecular dynamics simulations were carried out for 30 ns, at a constant temperature of 300 K and 1 bar. The resultant trajectories were analyzed using XMGrace63 and were visually inspected using VMD (Visual Molecular Dynamics).64 In addition, the binding free energy of the previously mentioned complexes was calculated using molecular Mechanic/Poisson–Boltzmann Surface Area (MM-PBSA). GMX_pbsa65 was used to calculate the binding free energy over the 30 ns of the MD simulation.
4.8.3. In Silico ADMET Study
4.8.3.1. Pharmacokinetic and Drug-Likeness Properties
SMILES notations of the studied compounds were inserted directly into the Swiss ADME (http://www.swissadme.ch/)66 web server followed by running the prediction of the pharmacokinetics and drug-likeness characteristics of the studied compounds.
4.8.3.2. Toxicity Prediction
The computational tool, Osiris Property Explorer (http://www.organic-chemistry.org/prog/peo/), was used for estimation of the potential toxicities, viz., mutagenicity, tumorigenicity, and irritant effect.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c01342.
MS/MS spectra obtained for lepidiline A (Figure S1), MS/MS spectra obtained for lepidiline C (Figure S2), MS/MS spectra obtained for lepidiline B (Figure S3), MS/MS spectra obtained for lepidiline D (Figure S4), MS/MS spectra obtained for 1-methyltetrahydro-β-5,6-hydride carboline-3-carboxylic acid isomer I (Figure S5), MS/MS spectra obtained for 1-methyltetrahydro-β-5,6-hydride carboline-3-carboxylic acid isomer II (Figure S6), MS/MS spectra obtained for 1,2,3,4-tetrahydro-β-carboline-3-carboxylic acid (Figure S7), MS/MS spectra obtained for 1-methyltetrahydro-β-carboline-3-carboxylic acid (Figure S8), MS/MS spectra obtained for 1-pentyltetrahydro-β-5,6-carboline-3-carboxylic acid (Figure S9), MS/MS spectra obtained for 1-butyltetrahydro-β-5,6-carboline-3-carboxylic acid (Figure S10), MS/MS spectra obtained for 3-benzyl-1,2-dihydro-4-methoxy-N-hydroxypyridine (Figure S11), MS/MS spectra obtained for 3-benzyl-1,2-dihydro-4-carbaldehyde-N-hydroxypyridine (Figure S12), MS/MS spectra obtained for N-(3-methoxybenzyl)(9Z,12Z)-octadecadienamide (Figure S13), MS/MS spectra obtained for N-benzyl-(9Z,12Z)-octadecadienamide (Figure S14), MS/MS spectra obtained for 5-oxo-6E,8E-octadecadienoic acid (Figure S15), MS/MS spectra obtained for glucotropaeolin (Figure S16), MS/MS spectra obtained for meyeniihydantoin A (Figure S17), MS/MS spectra obtained for meyeniin C (Figure S18), proposed fragmentation pathway of meyeniin C (Figure S19), 2D interaction diagram showing lisinopril interactions with the key amino acids in the ACE active site (Figure S20), and 2D interaction diagram showing aliskiren interactions with the key amino acids in the renin active site (Figure S21). Supplementary Table S1. Orisis calculated toxicity risks of the studied compounds (PDF)
Author Contributions
⊥ All authors contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Yábar E.; Pedreschi R.; Chirinos R.; Campos D. Glucosinolate content and myrosinase activity evolution in three maca (Lepidium meyenii Walp.) ecotypes during preharvest, harvest and postharvest drying. Food Chem. 2011, 127, 1576–1583. 10.1016/j.foodchem.2011.02.021. [DOI] [Google Scholar]
- Zhou Y.; Li P.; Brantner A.; Wang H.; Shu X.; Yang J.; Si N.; Han L.; Zhao H.; Bian B. Chemical profiling analysis of Maca using UHPLC-ESI-Orbitrap MS coupled with UHPLC-ESI-QqQ MS and the neuroprotective study on its active ingredients. Sc. Rep. 2017, 7, 1–14. 10.1038/srep44660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho F. V.; Fonseca Santana L.; da Silva V.; Costa S. L.; Zambotti-Villelae L.; Colepicolo P.; Ferraz C. G.; Ribeiro P. R. Combination of a multiplatform metabolite profiling approach and chemometrics as a powerful strategy to identify bioactive metabolites in Lepidium meyenii (Peruvian maca). Food Chem. 2021, 364, 130453. 10.1016/j.foodchem.2021.130453. [DOI] [PubMed] [Google Scholar]
- Uchiyama F.; Jikyo T.; Takeda R.; Ogata M. Lepidium meyenii (Maca) enhances the serum levels of luteinising hormone in female rats. J. Ethnopharmacol. 2014, 151, 897–902. 10.1016/j.jep.2013.11.058. [DOI] [PubMed] [Google Scholar]
- Miao H. The research on the impact of maca polypeptide on sport fatigue. Open Biomed. Eng. J. 2015, 9, 322. 10.2174/1874120701509010322. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Carvalho F. V.; Ribeiro P. R. Structural diversity, biosynthetic aspects, and LC-HRMS data compilation for the identification of bioactive compounds of Lepidium meyenii. Food Res. Int. 2019, 125, 108615. 10.1016/j.foodres.2019.108615. [DOI] [PubMed] [Google Scholar]
- Tata C. M.; Sewani-Rusike C. R.; Oyedeji O. O.; Gwebu E. T.; Mahlakata F.; Nkeh-Chungag B. N. Antihypertensive effects of the hydro-ethanol extract of Senecio serratuloides DC in rats. BMC Complement Altern. Med. 2019, 19, 1–10. 10.1186/s12906-019-2463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iman S.; Asif H.; Saleem M. Evaluation of antihypertensive potential of Ficus carica fruit. Pharm. Biol. 2017, 55, 1047–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baradaran A.; Nasri H.; Rafieian-Kopaei M. Oxidative stress and hypertension: Possibility of hypertension therapy with antioxidants, Journal of research in medical sciences. J. Isfahan Med. Sch. 2014, 19, 358. [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Zhu F. Chemical composition and health effects of maca (Lepidium meyenii). Food chem. 2019, 288, 422–443. 10.1016/j.foodchem.2019.02.071. [DOI] [PubMed] [Google Scholar]
- Lobo V.; Patil A.; Phatak A.; Chandra N. Free radicals, antioxidants and functional foods: Impact on human health. Pharmacognosy Rev. 2010, 4, 118. 10.4103/0973-7847.70902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Li C.; Gu W.; Qiu R.; Chao J.; Pei L.; Ma L.; Guo Y.; Tian R. Metabolomics analysis of dandelions from different geographical regions in China. Phytochem. Anal. 2021, 32, 899–906. 10.1002/pca.3033. [DOI] [PubMed] [Google Scholar]
- Chagas-Paula D. A.; Zhang T.; Da Costa F. B.; Edrada-Ebel R. A metabolomic approach to target compounds from the Asteraceae family for dual COX and LOX inhibition. Metabolites 2015, 5, 404–430. 10.3390/metabo5030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S.; Du H.; Tang B.; Xi C.; Chen Z. Non-target metabolomics based on high-resolution mass spectrometry combined with chemometric analysis for discriminating geographical origins of Rhizoma Coptidis. Microchem. J. 2021, 160, 105685. 10.1016/j.microc.2020.105685. [DOI] [Google Scholar]
- Tripathi A.; Misra K. Molecular docking: A structure-based drug designing approach. JSM Chem. 2017, 5, 1042–1047. [Google Scholar]
- Yang L.; Gao S.; Su Z.; Qin X.; Li Z. Identification of the constituents and the cancer-related targets of the fruit of Solanum nigrum based on molecular docking and network pharmacology. J. Pharm. Biomed. 2021, 200, 114067. 10.1016/j.jpba.2021.114067. [DOI] [PubMed] [Google Scholar]
- Ranilla L. G.; Kwon Y.-I.; Apostolidis E.; Shetty K. Phenolic compounds, antioxidant activity and in vitro inhibitory potential against key enzymes relevant for hyperglycemia and hypertension of commonly used medicinal plants, herbs and spices in Latin America. Bioresour. Technol. 2010, 101, 4676–4689. 10.1016/j.biortech.2010.01.093. [DOI] [PubMed] [Google Scholar]
- Olagunju A. I.; Omoba O. S.; Enujiugha V. N.; Alashi A. M.; Aluko R. E. Antioxidant properties, ACE/renin inhibitory activities of Pigeon pea hydrolysates and effects on systolic blood pressure of spontaneously hypertensive rats. Food Sci. Nutr. 2018, 6, 1879–1889. 10.1002/fsn3.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malomo S. A.; Niwachukwu I.; Girgih A. T.; Idowu A. O.; Aluka R.; Fagbemi T. N. Antioxidant and renin-angiotensin system inhibitory properties of cashew nut and fluted-pumpkin protein hydrolysates. Polym. Food Sci. Nutr. 2020, 70, 275–289. [Google Scholar]
- Le H. T.; Van Roy E.; Dendooven E.; Peeters L.; Theunis M.; Foubert K.; Pieters L.; Tuenter E. Alkaloids from Lepidium meyenii (Maca), structural revision of macaridine and UPLC-MS/MS feature-based molecular networking. Phytochemistry 2021, 190, 112863. 10.1016/j.phytochem.2021.112863. [DOI] [PubMed] [Google Scholar]
- Tafuri S.; Cocchia N.; Carotenuto D.; Vassetti A.; Staropoli A.; Mastellone V.; Peretti V.; Ciotola F.; Albarella S.; Del Prete C. Chemical analysis of Lepidium meyenii (Maca) and its effects on redox status and on reproductive biology in stallions. Molecules 2019, 24, 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.; Liu H.; Huang X.; Zhan L.; Luo P.; Xue J.; Chen R.; Nie Z. The metabolism and distribution of imidazole alkaloids from Lepidium meyenii (Maca) in mouse by matrix-assisted laser desorption/ionization mass spectrometry imaging. Int. J. Mass Spectrom. 2018, 434, 93–99. 10.1016/j.ijms.2018.09.014. [DOI] [Google Scholar]
- Herraiz T.; Galisteo J. Tetrahydro-β-carboline alkaloids occur in fruits and fruit juices. Activity as antioxidants and radical scavengers. J. Agric. Food Chem. 2003, 51, 7156–7161. 10.1021/jf030324h. [DOI] [PubMed] [Google Scholar]
- Herraiz T. β-Carboline alkaloids, Bioactive compounds in foods. Nutrients 2008, 11, 199–223. 10.1002/9781444302288.ch8. [DOI] [Google Scholar]
- Freire V. F.; Silva G. R.; Yariwake J. H. Targeted-Analysis of β-Carboline Alkaloids in Passion fruit (“Maracujá”) by SBSE (PDMS)-LC/Flu and UHPLC-MS. J. Braz. Chem. Soc. 2018, 29, 775–781. [Google Scholar]
- Zhao T.; Zheng S.-S.; Zhang B.-F.; Li Y.-Y.; Bligh S. A.; Wang C. H.; Wang Z. T. Metabolic pathways of the psychotropic-carboline alkaloids, harmaline and harmine, by liquid chromatography/mass spectrometry and NMR spectroscopy. Food chem. 2012, 134, 1096–1105. 10.1016/j.foodchem.2012.03.024. [DOI] [PubMed] [Google Scholar]
- Herraiz T. Analysis of the bioactive alkaloids tetrahydro-β-carboline and β-carboline in food. J. Chromatogr. A 2000, 881, 483–499. 10.1016/S0021-9673(99)01313-8. [DOI] [PubMed] [Google Scholar]
- Muhammad I.; Zhao J.; Dunbar D. C.; Khan I. A. Constituents of Lepidium meyenii ‘Maca’. Phytochemistry 2002, 59, 105–110. 10.1016/S0031-9422(01)00395-8. [DOI] [PubMed] [Google Scholar]
- Huang Y. J.; Peng X. R.; Qiu M. H. Progress on the chemical constituents derived from glucosinolates in maca (Lepidium meyenii). Nat. prod. bioprospect. 2018, 8, 405–412. 10.1007/s13659-018-0185-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.; Zhan L.; Liu C.; Fu L.; Chen R.; Nie Z. Mass spectrometry imaging of small molecule in situ in Lepidium meyenii (Maca) using gold nanoparticles matrix. Microchem. J. 2019, 150, 104190. 10.1016/j.microc.2019.104190. [DOI] [Google Scholar]
- Gugnani K. S.; Vu N.; Rondón-Ortiz A. N.; Böhlke M.; Maher T. J.; Pino-Figueroa A. J. Neuroprotective activity of macamides on manganese-induced mitochondrial disruption in U-87 MG glioblastoma cells. Toxicol. Appl. Pharmacol. 2018, 340, 67–76. 10.1016/j.taap.2017.12.014. [DOI] [PubMed] [Google Scholar]
- Yang Q.; Jin W.; Lv X.; Dai P.; Ao Y.; Wu M.; Deng W.; Yu L. Effects of macamides on endurance capacity and anti-fatigue property in prolonged swimming mice. Pharm. Biol. 2016, 54, 827–834. 10.3109/13880209.2015.1087036. [DOI] [PubMed] [Google Scholar]
- Pérez V. T.; Ticona L. A.; Serban A. M.; Gómez J. A.; Sánchez Á.R. Synthesis and biological screening of a library of macamides as TNF-α inhibitors. RSC Med. Chem. 2020, 11, 1196–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu Z.; Nicolussi S.; Rau M.; Lorántfy L.; Forgo P.; Hohmann J.; Csupor D.; Gertsch J. Identification of endocannabinoid system-modulating N-alkylamides from Heliopsis helianthoides var. scabra and Lepidium meyenii. J. Nat. Prod. 2014, 77, 1663–1669. 10.1021/np500292g. [DOI] [PubMed] [Google Scholar]
- Xia C.; Chen J.; Deng J.; Zhu Y.; Li W.; Jie B.; Chen T. Novel macamides from maca (Lepidium meyenii Walpers) root and their cytotoxicity. Phytochem. Lett. 2018, 25, 65–69. 10.1016/j.phytol.2018.03.001. [DOI] [Google Scholar]
- Chen S.; Li K.; Pubu D.; Jiang S.; Chen B.; Chen L.; Yang Z.; Ma C.; Gong X. Optimization of ultrasound-assisted extraction, HPLC and UHPLC-ESI-Q-TOF-MS/MS analysis of main macamides and macaenes from maca (cultivars of Lepidium meyenii Walp). Molecules 2017, 22, 2196. 10.3390/molecules22122196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.; Zhao Q.; Liu Y.; Gong P.; Cao L.; Wang X.; Zhao B. Macamides present in the commercial maca (Lepidium meyenii) products and the macamide biosynthesis affected by postharvest conditions. Int. J. Food Prop. 2017, 20, 3112–3123. 10.1080/10942912.2016.1274905. [DOI] [Google Scholar]
- Esparza E.; Hadzich A.; Kofer W.; Mithöfer A.; Cosio E. Bioactive maca (Lepidium meyenii) alkamides are a result of traditional Andean postharvest drying practices. Phytochemistry 2015, 116, 138–148. 10.1016/j.phytochem.2015.02.030. [DOI] [PubMed] [Google Scholar]
- Pan Y.; Zhang J.; Li H.; Wang Y.; Li W. Simultaneous analysis of macamides in maca (Lepidium meyenii) with different drying process by liquid chromatography tandem mass spectrometry. Food Anal. Methods 2016, 9, 1686–1695. 10.1007/s12161-015-0346-9. [DOI] [Google Scholar]
- Lin L.; Huang J.; Sun-Waterhouse D.; Zhao M.; Zhao K.; Que J. Maca (Lepidium meyenii) as a source of macamides and polysaccharide in combating of oxidative stress and damage in human erythrocytes. Int. J. Food Sci. 2018, 53, 304–312. 10.1111/ijfs.13586. [DOI] [Google Scholar]
- Yang K.; Zhao Z.; Gross R.; Han X. Identification and quantitation of unsaturated fatty acid isomers by electrospray ionization tandem mass spectrometry: a shotgun lipidomics approach. Anal. Chem. 2011, 83, 4243–4250. 10.1021/ac2006119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M.; Dunn S.; Altvater J.; Dove S.; Nette G. Rapid identification of long-chain polyunsaturated fatty acids in a marine extract by HPLC-MS using data-dependent acquisition. Anal. Chem. 2012, 84, 5976–5983. 10.1021/ac3006523. [DOI] [PubMed] [Google Scholar]
- Li A.; Liu J.; Ding F.; Wu X.; Pan C.; Wang Q.; Gao M.; Duan S.; Han X.; Xia K.; et al. Maca extracts regulate glucose and lipid metabolism in insulin-resistant HepG2 cells via the PI3K/AKT signalling pathway. Food Sci. Nutr. 2021, 9, 2894–2907. 10.1002/fsn3.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldi T.; Lelario F.; Orlando D.; Bufo S. Collision-induced dissociation of the A+ 2 isotope ion facilitates glucosinolates structure elucidation by electrospray ionization-tandem mass spectrometry with a linear quadrupole ion trap. Anal. Chem. 2010, 82, 5686–5696. 10.1021/ac100703w. [DOI] [PubMed] [Google Scholar]
- Geng H.; Yang D.; Chen X.; Wang L.; Zhou M.; Mei W. Meyeniihydantoins A–C, three novel hydantoin derivatives from the roots of Lepidium meyenii Walp. Phytochem. Lett. 2018, 26, 208–211. 10.1016/j.phytol.2018.06.010. [DOI] [Google Scholar]
- Zhou M.; Ma H.; Liu Z.; Yang G.; Du G.; Ye Y.; Li G. P.; Hu Q. F. (+)-Meyeniins A–C, novel hexahydroimidazo [1, 5-c] thiazole derivatives from the tubers of Lepidium meyenii: complete structural elucidation by biomimetic synthesis and racemic crystallization. J. Agric. Food Chem. 2017, 65, 1887–1892. 10.1021/acs.jafc.6b05805. [DOI] [PubMed] [Google Scholar]
- Sandín-España P.; Mateo-Miranda M.; López-Goti C.; De Cal A.; Alonso-Prados J. Development of a rapid and direct method for the determination of organic acids in peach fruit using LC–ESI-MS. Food chem. 2016, 192, 268–273. 10.1016/j.foodchem.2015.07.012. [DOI] [PubMed] [Google Scholar]
- Yang S.; Sun X.; Gao Y.; Chen R. Differentiation of Lepidium meyenii (Maca) from Different Origins by Electrospray Ionization Mass Spectrometry with Principal Component Analysis. ACS omega 2019, 4, 16493–16500. 10.1021/acsomega.9b02128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng P.; Sun J.; Chen P.; Brand E.; Frame J.; Meissner H.; Stewart J.; Gafner S.; Clark S.; Miller Characterization of maca (Lepidium meyenii/Lepidium peruvianum) using a mass spectral fingerprinting, metabolomic analysis, and genetic sequencing approach. Planta Med. 2020, 86, 674–685. 10.1055/a-1161-0372. [DOI] [PubMed] [Google Scholar]
- Sielecki A.; Hayakawa K.; Fujinaga M.; Murphy M.; Fraser M.; Muir A.; Carilli C.; Lewicki J.; Baxter J.; James M. Structure of recombinant human renin, a target for cardiovascular-active drugs, at 2.5 A resolution. Science 1989, 243, 1346–51. 10.1126/science.2493678. [DOI] [PubMed] [Google Scholar]
- Lipinski C.; Lombardo F.; Dominy B.; Feeney P. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 1997, 23, 3–25. 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- Sander T.Actelion’s Property explorer; Pharm. Ltd.: Gewerbestrasse, Allschwil, Switz., 2001; Vol. 16, p 4123. [Google Scholar]
- Sander T.; Freyss J.; von Korff M.; Reich J.; Rufener C. OSIRIS, an entirely in- house developed drug discovery informatics system. J. Chem. Inf. Model. 2009, 49, 232–246. 10.1021/ci800305f. [DOI] [PubMed] [Google Scholar]
- El-Shiekh R.; Abdelmohsen U.; Ashour H.; Ashour R. Novel antiviral and antibacterial activities of Hibiscus schizopetalus. Antibiotics 2020, 9, 756. 10.3390/antibiotics9110756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olarewaju O.; Alashi A.; Aluko R. Antihypertensive effect of aqueous polyphenol extracts of Amaranthusviridis and Telfairiaoccidentalis leaves in spontaneously hypertensive rats. JFB 2018, 1, 166–173. 10.31665/JFB.2018.1135. [DOI] [Google Scholar]
- Salem M.; Jüppner J.; Bajdzienko K.; Giavalisco P. Protocol: a fast, comprehensive and reproducible one-step extraction method for the rapid preparation of polar and semi-polar metabolites, lipids, proteins, starch and cell wall polymers from a single sample. Plant Methods. 2016, 12, 1–15. 10.1186/s13007-016-0146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natesh R.; Schwager S.; Sturrock E.; Acharya K. Crystal structure of the human angiotensin-converting enzyme–lisinopril complex. Nature 2003, 421, 551–554. 10.1038/nature01370. [DOI] [PubMed] [Google Scholar]
- Rahuel J.; Rasetti V.; Maibaum J.; Rüeger H.; Göschke R.; Cohen N.; Stutz S.; Cumin F.; Fuhrer W.; Wood J.; Grütter M. Structure-based drug design: the discovery of novel nonpeptide orally active inhibitors of human renin. Chem. Biol. 2000, 7, 493–504. 10.1016/S1074-5521(00)00134-4. [DOI] [PubMed] [Google Scholar]
- Pettersen E.; Goddard T.; Huang C.; Couch G.; Greenblatt D.; Meng E.; Ferrin T. UCSF Chimera-A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Berendsen H.; van der Spoel D.; van Drunen R. GROMACS: a message passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- Maier J.; Martinez C.; Kasavajhala K.; Wickstrom L.; Hauser K.; Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput.h 2015, 11, 3696–3713. 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refaey R.; El-Ashrey M.; Nissan Y. Repurposing of renin inhibitors as SARS-COV-2 main protease inhibitors: A computational study. Virology 2021, 554, 48–54. 10.1016/j.virol.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner P.XMGRACE, Version 5.1.19; Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology: Beaverton, OR, 2005. [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Valdés-Tresanco M.; Valdés-Tresanco M.; Valiente P.; Moreno E. Gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. 10.1021/acs.jctc.1c00645. [DOI] [PubMed] [Google Scholar]
- Daina A.; Michielin O.; Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.