Abstract
Membrane-located monooxygenase systems, such as the Pseudomonas putida mt-2-derived xylene oxygenase, are attractive for challenging transformations of apolar compounds, including enantiospecific epoxidations, but are difficult to synthesize at levels that are useful for application to biotechnological processes. In order to construct efficient biocatalysis strains, we utilized the alkane-responsive regulatory system of the OCT plasmid-located alk genes of Pseudomonas oleovorans GPo1, a very attractive system for recombinant biotransformation processes. Determination of the nucleotide sequence of alkS, whose activated gene product positively regulates the transcription of the structural genes alkBFGHJKL, on a 3.7-kb SalI-HpaI OCT plasmid fragment was completed, and the N-terminal amino acid sequence of an AlkS-LacZ fusion protein was found to be consistent with the predicted DNA sequence. The alkS gene and the alkBp promoter were assembled into a convenient alkane-responsive genetic expression cassette which allowed expression of the xylene oxygenase genes in a recombinant Escherichia coli strain at a specific activity of 91 U per g (dry weight) of cells when styrene was the substrate. This biocatalyst was used to produce (S)-styrene oxide in two-liquid-phase cultures. Volumetric productivities of more than 2 g of styrene oxide per h per liter of aqueous phase were obtained; these values represented a fivefold improvement compared with previous results.
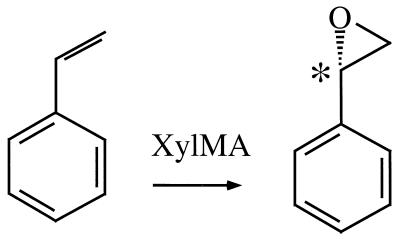
Pseudomonas putida mt-2-derived xylene oxygenase is encoded in the catabolic TOL plasmid pWW0 upper pathway operon, and together with a set of other enzymes it forms a catalytic cluster which degrades toluene and xylene to (substituted) benzoic acids (18, 30, 40). The ability of xylene oxygenase to hydroxylate methyl substituents on substituted benzenes or their heteroaromatic equivalents has made it an important biotechnological enzyme (24, 56). This enzyme consists of a membrane-bound component, XylM, which carries out the oxygenation step (50), and a cytoplasmic NADH:acceptor reductase component, XylA, which supplies reducing equivalents to XylM (45). The potential of this system for biological production of fine chemicals has already been exploited; wild-type cells of P. putida mt-2 are used by Lonza to produce heteroaromatic acids on a commercial scale (24). Furthermore, xylene oxygenase is selective for the si-face of prochiral vinyl functions on aromatic ring systems, which leads to the formation of optically active epoxides, such as (S)-styrene oxide (55) (Fig. 1). Escherichia coli recombinants carrying the genes for xylene oxygenase have produced (S)-styrene oxide from inexpensive styrene in a 2-liter reactor (54). Unfortunately, so far the productivities displayed by such recombinants have been insufficient to commercially exploit their synthetic potential (17, 55). A number of observations have indicated that expressing the xylene oxygenase genes via the alk regulatory system of Pseudomonas oleovorans GPo1 might provide suitable biocatalysis strains for two-liquid-phase cultures. P. oleovorans GPo1 degrades medium-chain-length alkanes with a set of enzymes encoded by two alk gene clusters on the catabolic OCT plasmid (Fig. 2A) (51). The first cluster contains the alkBFGHJKL operon, which contains all but one of the structural genes for conversion of alkanes to the corresponding alkanoic acids and coupling of these compounds to coenzyme A (Fig. 2B). The second cluster contains the remaining structural gene, alkT, and the gene which encodes the regulatory protein AlkS (11). Expression of the genes in the first cluster is under control of alkBp, the alk promoter, and is initiated in the presence of functional AlkS and alkanes or other, structurally nonrelated inducers, such as dicyclopropylketone (DCPK) (16, 49). DCPK is water soluble and hence is a convenient inducer in aqueous cultures, while alkanes are useful inducers in two-liquid-phase cultures which contain an organic phase. Expression of the alk genes in E. coli W3110 via the alk regulatory system from the low-copy-number RK2 derivative pGEc47 led to accumulation of membrane-located AlkB until it accounted for up to 10% of the total cell protein (35). This indicated that AlkS, together with its cognate promoter alkBp (26), could be a powerful general system to direct synthesis of recombinant proteins. In addition, the alk regulatory system is not subject to catabolite repression in E. coli (46, 58), which allows convenient utilization of cheap carbon sources, such as glucose, in cultures of recombinant strains. Because of these attractive features of the alk regulatory system we developed its components into an expression system for general use. In this paper we describe this system and its potential for efficient synthesis of xylene oxygenase for biotransformation of styrene to (S)-styrene oxide at high enantiomeric excess in a two-liquid-phase biotransformation system.
FIG. 1.
Conversion of inexpensive styrene to (S)-styrene oxide by xylene monooxygenase. The chiral carbon atom is indicated by an asterisk.
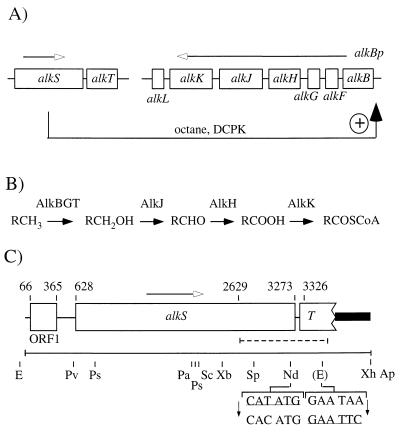
FIG. 2.
(A) Organization and regulation of the alk genes on the OCT plasmid in P. oleovorans GPo1. The regulatory protein AlkS is activated by octane or DCPK and induces transcription from the alkBp promoter. The directions of transcription are indicated by arrows with open arrowheads. Transcriptional regulation of alkT has not been described. (B) Alkane degradation by enzymes encoded by alk genes. Alkanes are converted to alkanols, alkanals, and the corresponding carboxylic acids, which are coupled to coenzyme A (CoA). R represents pentyl to undecyl residues. The functions of AlkF and AlkL are unclear. (C) Detailed structure of pBG11. The region sequenced is indicated by the solid line, while the previously described sequence is indicated by the dashed line. The coordinates (in the deposited sequence) are indicated at the top together with the direction of transcription of alkS. The solid bar indicates the portion of the plasmid that was derived from pJRD158. Restriction sites that were used for cloning in this work and sites which also occur in the pUC18 polylinker are indicated at the bottom. The EcoRI, XhoI, and ApaI sites were derived from the polylinker of pGEM-7Zf(+), which was the basis of pBG11. Two modifications introduced by site-directed mutagenesis are indicated at the bottom; the sequences of newly introduced (in parentheses) or removed restriction sites are underlined. Abbreviations: Ap, ApaI; E, EcoRI; Nd, NdeI; Pa, PacI; Ps, PstI; Pv, PvuII, Sc, SacI; Sp, SphI; Xb, XbaI; Xh, XhoI.
MATERIALS AND METHODS
Strains, media, and cultivation conditions.
The strains and plasmids used and constructed in this study are shown in Table 1. We used Luria-Bertani (LB) complex medium (Difco Laboratories, Detroit, Mich.) or M9 mineral medium (44) supplemented with 1 ml of one of two trace element solutions per liter; when necessary the medium was solidified by adding 1.8% agarose (Difco). Trace element solution US contained 1 M hydrochloric acid and (per liter) 1.50 g of MnCl2 · 4H2O, 1.05 g of ZnSO4, 0.30 g of H3BO3, 0.25 g of Na2MoO4 · 2H2O, 0.15 g of CuCl2 · 2H2O, and 0.84 g of Na2EDTA · 2H2O. Trace element solution US* also contained (per liter) 4.87 g of FeSO4 · 7H2O and 4.12 g of CaCl2 · 2H2O. Alternatively, we used M9* mineral medium, which was identical to M9 mineral medium except that it contained three times more phosphate salts to increase the buffer capacity and did not contain calcium chloride. Glucose was added to each mineral medium at a concentration of 0.5% (wt/vol) as a carbon source or to complex media at a concentration of 1% (wt/vol) to bring about carbon catabolite repression of the lacZp promoter of cloning vectors and/or carbon catabolite repression of the synthesis of the tryptophanase of E. coli, which is involved in the formation of indole on complex media; indole can subsequently be converted to indigo by xylene oxygenase (32, 33). Antibiotics were added at the following concentrations: ampicillin, 150 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 30 μg/ml; and tetracycline, 12.5 μg/ml. When necessary, thiamine was added at a concentration of 1 mg per liter. The cloning and DNA modification protocols used have been described elsewhere (44). Cultivation for cloning procedures was carried out at 37°C. Cultures and precultures of the E. coli strains used to determine enzyme activities or for production were grown at 30°C. Unless mentioned otherwise, cultures were induced by adding 0.05% (vol/vol) DCPK (Aldrich, Buchs, Switzerland).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| E. coli strains | ||
| X7026 | F− Δ(gpt-lac)5 supE44 relA1 spoT1 | 23 |
| JM101 | supE thi-1 Δ(lac-proAB) F′[traD36 proAB+lacIqlacZΔM15] | 44 |
| CJ236 | dut-1 ung-1 thi-1 relA1/pCJ105(Cmr F′) | 29 |
| Plasmids | ||
| pUC18 | lacZα, cloning vehicle, Apr | 57 |
| pUC18Not | lacZα, cloning vector, polylinker flanked by NotI sites, Apr | 22 |
| pUC18Sfi | lacZα, cloning vector, polylinker flanked by SfiI sites, Apr | 22 |
| pHP45Ω | Source of the T4 transcriptional terminator, Smr Apr | 39 |
| pGEM-7Zf(+) | lacZα, cloning vector, phage f1ori, Apr | Promega |
| pGEM-7Zf(−) | Same as pGEM-7Zf(+) but f1 ori is inverted, Apr | Promega |
| mini-Tn5Km | Source of kanamycin interposon, Apr Kmr | 8 |
| pBR322 | Cloning vehicle, Apr Tcr | 3 |
| pGC2 | pBR322 derivative with M13ori, Apr | 34 |
| pMC1871 | pBR322 derivative with promoterless lacZ, Tcr | 6 |
| pCKO4 | Source of xylN, Cmr | 37 |
| pBG63 | Source of xylMA, f1ori, Apr | 55 |
| pGEc258 | pBR322/pJRD158 derivative, source of alkS, Apr | 11 |
| pGEc286 | Promoter probe vector with alkBp-CAT fusion, Apr | 25 |
| pGC2Mlu | pGC2 with additional MluI site, Apr | This study |
| pBG11 | pGEM7-Zf(+) with alkS, Apr | This study |
| pBG11lacZ | pBG11 derivative with alkS′-′lacZ fusion, Apr | This study |
| pBG4 | pUC18 with T4 transcriptional terminator of pHP45Ω, Apr | This study |
| pBG4ΔN | pBG4 without NdeI site, Apr | This study |
| pBG4NΔN | pBG4ΔN with xylN of pCKO4, Apr | This study |
| pBGL | pBG4NΔN with small new polylinker, Apr | This study |
| pGEMPalkN | Cloning vector with alkBp promoter and NdeI site on the ATG of alkB, Apr | This study |
| pBGPalk | pBGL with alkBp of pGEMPalkN, Apr | This study |
| pBG11EΔN | pBG11 with an additional EcoRI site and removed NdeI site internal to alkS, Apr | This study |
| pUC18NΔN | pUC18Not without NdeI, Apr | This study |
| pUC18NS* | pUC18NΔN with alkS* of pBG11EΔN, Apr | This study |
| pSPZ1(+) | pGEM-7Zf(+) with a new polylinker, Apr | This study |
| pSPZ1(−) | Same as pSPZ1(+) but from pGEM-7Zf(−), Apr | This study |
| pSPZ2Not | pUC18NS* with alkBP and T4t of pBGPalk, contains alkS* alkBp T4t as a NotI cassette, Apr | This study |
| pSPZ2Sfi | Same as pSPZ2Nde but SfiI sites instead of NotI sites, Apr | This study |
| pBG63N | pBG63 with NdeI site in xylM removed and new NdeI site on the ATG of xylM*, Apr | This study |
| pSPZ2MA | pSPZ2 with xylM*A of pBG63N under alkBp control, Apr | This study |
| pBRNS | pBR322 with rrnBT1 terminator and additional NotI and SfiI restriction sites, Apr | This study |
| pBRNSKm | pBRNS with kanamycin resistance of mini-Tn5Km replacing the bla gene, Kmr | This study |
| pBRNSKmΔ | pBRNSKm without remnants of the tet gene, Kmr | This study |
| pSPZ3 | pBRNSKmΔ with alkS* alkBp xylM*A of pSPZ2MA, Kmr | This study |
Nucleotide sequence analysis.
A 3.7-kb SalI-HpaI OCT plasmid fragment containing alkS was excised from plasmid pGEc258 together with a 340-bp portion of original cloning vector pJRD158 as a SalI-XhoI fragment. This fragment was inserted into the XhoI restriction site of pGEM-7Zf(+) so that the XhoI site that was reconstituted was oriented away from the lacZp promoter of the vector, while the destroyed SalI site was oriented towards the promoter. The resulting plasmid was designated pBG11. Unidirectional deletions from each end of the cloned fragment in pBG11 were generated by exonuclease III digestion as described previously (21). The collection of clones was completed by Sau3A and TaqI shotgun clones. The alkS sequence was determined by the Sanger dideoxy method. The subsequent data analysis was performed with the DNAstar software package (DNAstar, Madison, Wis.), the BlastP algorithm (1) of the GenBank database at the National Center for Biotechnology Information, and the analysis tools (42) available at the ExPASy Molecular Biology Server.
Determination of the N-terminal sequence of AlkS.
To construct an AlkS′-′LacZ fusion protein with the first 75 amino acids of AlkS and the ′lacZ gene product of plasmid pMC1871, we excised the ′lacZ gene as a PstI fragment and inserted it into the PstI site close to the 5′ end of alkS in pBG11, which generated the desired fusion in plasmid pBG11lacZ. E. coli X7026(pBG11lacZ) was grown in liquid LB medium and induced by adding 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) in the exponential growth phase. A crude cell extract was obtained 4 h later by sonication, and this extract was clarified by centrifugation. The supernatant was applied to a β-galactosidase affinity column containing p-aminobenzyl-1-thio-β-galactopyranoside agarose (Sigma Chemical Co., St. Louis, Mo.) (15) connected to a fast protein liquid chromatography system (Pharmacia, Dübendorf, Switzerland). The retained protein was eluted with 0.1 M sodium borate (pH 10). The fractions with the highest levels of β-galactosidase activity were pooled and desalted by fast protein liquid chromatography by using a Sephacryl column (Sigma). The resulting partially purified protein was subjected to sodium dodecyl sulfate gel electrophoresis and electroblotted onto polyvinylidene difluoride membranes as described previously (31), and the sequence of the first 15 amino acids of the fusion protein was determined by Edman degradation by using an Applied Biosystems automatic protein sequencer.
Site-directed mutagenesis.
Site-directed mutagenesis was performed as described previously (29) by using plasmid pGC2, plasmid pGC2Mlu (which is identical to pGC2 except that an additional MluI site is inserted by using oligonucleotide 6 [Table 2]), or one of the pGEM-7Zf plasmids as the source of the phage f1 origin. Successful mutagenesis was demonstrated by restriction analysis and functioning of the affected gene when possible. Silent mutations which removed NdeI sites were introduced into xylM and alkS by using oligonucleotides 1 and 2; the modified genes were designated xylM* and alkS*, respectively. New NdeI sites were placed on the ATG codons of xylM* and alkB by using oligonucleotides 3 and 4, and an additional EcoRI site was engineered 250 bp after the stop codon of alkS with oligonucleotide 5.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| 1 | 5′ TTG TTC CCA TGT GAT TCC ACG 3′ |
| 2 | 5′ TTT TCC TCA TGT GCC ACT TTA 3′ |
| 3 | 5′ GTG TCC ATA TGT CCA CCT ACT 3′ |
| 4 | 5′ GAG AAC ACC ATA TGC TTG AGA 3′ |
| 5 | 5′ ATG GTA ATA TTG GAA TTC GTA TAA AA 3′ |
| 6 | 5′ CTG GCA CGC GTT GGA CGC GCA 3′ |
| 7 | 5′ TAT GTT AAC GGC GCG CCC ATG 3′ |
| 8 | 5′ GGG CGC GCC GTT AAC A 3′ |
| 9 | 5′ AAT TCA TAA AAC GAA AGG CTC AGT CGA AAG ACT GGG CCT TTC GTT TTA TCT GTT GTT TGC GGC CGC GGC CGC CTA GGC C 3′ |
| 10 | 5′ AGC TGG CCT AGG CGG CCG CGG CCG CAA ACA ACA GAT AAA ACG AAA ACG AAA GGC CCA GTC TTT CGA CTG AGC CTT TCG TTT TAT G 3′ |
| 11 | 5′ GGC GCG CCT GCA GAA TTC TCG AGA AGC TTC CCG GGA TCC TAG GCG CGC CAT GCA 3′ |
| 12 | 5′ TGG CGC GCC TAG GAT CCC GGG AAG CTT CTC GAG AAT TCT GCA GGC GCG CCC GCC 3′ |
Oligonucleotides 1 to 6 were used as mutagenesis primers during site-directed mutagenesis (the mutagenic nucleotide[s] in each oligonucleotide is underlined). Oligonucleotides 7 to 12 were used for construction of the following polylinkers: oligonucleotides 7 and 8, NdeI-HpaI-AscI; oligonucleotides 9 and 10, EcoRI-rrnB T1-NotI-SfiI-kill HindIII; oligonucleotides 11 and 12, kill ApaI-AscI-PstI-EcoRI-XhoI-HindIII-SmaI-BamHI-AvrII-AscI-NsiI (Kill indicates that there were compatible cohesive ends which did not regenerate the restriction site indicated).
Construction of pSPZ2 plasmids.
Plasmid pBG4 was constructed by inserting the 272-bp phage T4 transcriptional terminator of pHP45Ω as an SphI-HindIII fragment into pUC18. NdeI sites were removed from plasmid pBG4 by digestion with NdeI, filling in with the Klenow polymerase, and religation leading to pBG4ΔN, and the plasmid received a segment that provided a new NdeI site necessary for subsequent cloning steps by transferring a 2.2-kb SalI-SphI fragment spanning xylN of pCKO4 into its polylinker. The resulting plasmid, pBG4NΔN, was digested with NdeI and SphI, which left the terminator intact but removed the 3′ part of xylN. The larger of the two fragments was ligated to a linker consisting of hybridized oligonucleotides 7 and 8, which introduced additional HpaI and AscI sites, eliminated the SphI site between the NdeI site and the terminator, and resulted in construct pBGL.
Plasmid pGEMPalkN contained the alkBp promoter as a 2.3-kb SmaI-HindIII OCT plasmid fragment with a new NdeI site on the ATG at the beginning of the alkB gene. The modified promoter was excised as a 273-bp SmaI-NdeI fragment and was inserted into pBGL digested in the same manner; this resulted in pBGPalk, from which the xylN gene was completely removed and in which the alkBp promoter pointed towards the small polylinker with NdeI, HpaI, and AscI sites and the terminator.
The alkS* gene was obtained as a 3.5-kb EcoRI fragment from plasmid pBG11EΔN and was inserted into pUC18NΔN, a pUC18Not derivative whose internal NdeI site had been removed as described above for pBG4ΔN. The resulting construct was designated pUC18NS* and contained the regulator gene in the orientation opposite that of the lacZp promoter of the vector. Plasmid pUC18NS* received the SmaI-HindIII fragment containing the alkBp promoter and the T4 transcriptional terminator of pBGPalk, which resulted in plasmid pSPZ2Not. An equivalent manipulation sequence based on the cloning vector pUC18Sfi resulted in construction of plasmid pSPZ2Sfi, which was identical to pSPZ2Not except that the segment of interest was flanked by SfiI sites rather than NotI sites.
Construction of expression plasmid pSPZ3.
Plasmid pBRNS was constructed by providing pBR322 with an rrnBT1 transcriptional terminator (4) and two new restriction sites by inserting a linker consisting of hybridized oligonucleotides 9 and 10 between its EcoRI and HindIII sites. In the second step, the bla gene was replaced by the kanamycin resistance gene from the kanamycin interposon of mini-Tn5Km; plasmid pBRNS was digested with EcoRI, filled in with T4 DNA polymerase, redigested with DraI, and ligated to the SmaI fragment of mini-Tn5Km with the kanamycin resistance gene (which eliminated one of the two phage T4 transcriptional terminators of the interposon) in such an orientation that the new resistance gene was flanked by a different transcriptional terminator on each side (the T4t terminator from the interposon and the rrnBT1 terminator from pBRNS). The resulting plasmid was designated pBRNSKm. The remaining portions of the tet gene of pBR322 were removed by digestion of pBRNSKm with EcoRV and AvaI, T4 DNA polymerase treatment, and religation. The remaining unique BamHI site upstream of the resistance gene (derived from the fragment containing the kanamycin resistance gene) in the resulting plasmid, pBRNSKmΔ, was made blunt by T4 DNA polymerase treatment. An identically treated NotI-AscI fragment of pSPZ2MA (see below) carrying alkS*, alkBp, and xylM*A but not the T4t terminator of pSPZ2MA was inserted, which yielded plasmid pSPZ3.
Determination of enzyme activities.
To determine enzyme activities by whole-cell assays, cells were grown in M9* mineral medium precultures supplemented with glucose, kanamycin, thiamine, and trace element solution US* and then inoculated into larger cultures growing in an identical medium. At an optical density at 450 nm of ca. 0.3, the cells were induced with 0.05% (vol/vol) DCPK; the cells were harvested by centrifugation at the appropriate time (see below) and were subjected to a whole-cell assay for styrene oxide formation under optimized conditions as described elsewhere (38). Briefly, cells were resuspended to a dry biomass concentration between 2 and 5 g per liter in 100 mM potassium phosphate buffer (pH 7.4) containing 1% (wt/vol) glucose and incubated for 5 min with 1.5 mM styrene, and the reaction was stopped by adding ice-cold ether. The biomass concentration was selected so that at least 0.3 mM styrene was left after the transformation. One unit of activity was defined as the amount of activity that produced 1 μmol of styrene oxide from styrene in 1 min. Specific activities were calculated from the measured transformation rates per unit of cell dry weight. The results given below are the averages based on three independent experiments.
Production of styrene oxide in two-liquid-phase cultures.
Freshly transformed E. coli JM101 cells harboring plasmid pSPZ3 were inoculated into a 5-ml LB medium preculture supplemented with kanamycin and 1% glucose, grown overnight at 30°C, and diluted 100-fold with 100 ml of M9 mineral medium supplemented with 0.5% glucose, kanamycin, and thiamine. The resulting culture was incubated for approximately 10 h (during which time the cells entered the stationary phase) on a horizontal shaker at 30°C and then was used as an inoculum for the biotransformation reaction mixture. The biotransformation reaction was carried out in a stirred tank reactor with two Rushton turbine impellers, four baffles, and a total volume of 3 liters. Seals and O-rings were made out of solvent-resistant Viton. The reactor contained 1.025 liters of a mineral medium which contained (per liter) 8.82 g of KH2PO4, 10.85 g of K2HPO4, 8.82 g of Na2HPO4, 1.0 g of NH4Cl, 0.5 g of NaCl, 0.49 g of MgSO4 · 7H2O, 14.7 mg of CaCl2 · 2H2O, 2.78 mg of FeSO4 · 7H2O, 5 g of glucose, 100 μl of polypropylene glycol 2000 (Fluka, Buchs, Switzerland), 0.5 ml of trace element solution US, thiamine, and kanamycin. The pH was maintained at 7.1 by adding 25% NH4OH and 25% phosphoric acid. The NH4OH also served as the source of nitrogen. After inoculation with 100 ml of the preculture, the reactor was aerated at a rate of 1 liter per min and was stirred at 1,500 rpm for ca. 12 h (overnight), which resulted in a culture in the stationary phase which contained approximately 2.2 g (dry weight) of cells per liter. The medium was then supplemented with 6 mg of thiamine per liter, 8 ml of trace elements solution US per liter, and 8.2 mg of FeSO4 · 7H2O per liter. Subsequently, the culture was fed at a rate of 10 ml · h−1 with an aqueous solution containing (per liter) 450 g of glucose, 50 g of yeast extract (Difco), and 9 g of MgSO4 · 7H2O. One hour later, 375 ml of a mixture containing hexadecane, 1% (vol/vol [organic phase]) octane (Sigma, Buchs, Switzerland), and 2% (vol/vol [organic phase]) styrene (Fluka) was added to the reactor; this mixture constituted an organic second liquid phase that was dispersed in the aqueous phase. Concomitantly the stirrer speed was increased to 2,500 rpm and the aeration rate was reduced to 0.67 liter per min to limit foam formation. The time when the organic phase was added to the reactor was the point of induction. Furthermore, before air entered the reactor, it was passed through a wash flask containing 280 ml of the organic phase. In additional experiments, the styrene concentration in the organic liquid added varied between 0.5 and 2% (vol/vol). Iron (9 mg/liter) was added 2.5 h after induction as described above. The dissolved oxygen tension (DOT) was always kept above 40% saturation by varying the aeration rate. Iron sulfate was added in pulses during cultivation until the concentration was 58 μM in order to compensate for the reduced amount of yeast extract compared to our previous experiments (13, 54); the nonheme iron monooxygenase iron requirements, which were around 3 μmol per g (dry weight) of cells (47), were taken into account. The preparation started to produce large amounts of foam within 1 h after induction; the foam was controlled by repeatedly adding silicone oil-based antifoam agent 289 (Sigma). Cultivation was carried out twice, and nearly identical results were obtained with the two preparations; the results obtained with one of the preparations are shown below.
Analytic procedures.
The DOT was determined with an autoclavable amperometric probe (Mettler Toledo, Greifensee, Switzerland). Cell dry weights and octane, styrene, and styrene oxide concentrations in the organic phase, as well as the styrene oxide concentrations in the aqueous phase of the reactor, were monitored over time. To do this, 10-ml samples were withdrawn from the reactor at regular intervals and centrifuged to separate the phases. The position of the interphase was marked, and the organic phase was removed, dried over sodium sulfate, diluted 50-fold with diethyl ether supplemented with dodecane as an internal standard, and analyzed by gas chromatography to determine the styrene, styrene oxide, and octane contents as described previously (38). The aqueous supernatant of each centrifuged sample was also completely removed and extracted with an equal volume of diethyl ether supplemented with dodecane. The ether phase was then dried and analyzed as described above. The remaining cell pellet was resuspended in aqueous 25 mM MgSO4, and aliquots were distributed into preweighed reaction tubes and centrifuged. The supernatants were discarded, and the tubes were incubated at 80°C until the weights were constant. The observed volumetric productivities and the observed styrene oxide formation rates were calculated as averages for intervals between two data points, taking into account volume corrections for sampling and feeding; the term “observed” refers to the fact that the activities which we measured in these biotransformation preparations were limited by substrate availability in some cases. As a result, the transformation rates given below may be underestimates of the available intrinsic enzyme activity.
Nucleotide sequence accession numbers.
The DNA sequence of the 3.7-kb OCT plasmid fragment has been deposited in the GenBank database under accession no. X52935. The sequences of plasmids pSPZ1(+), pSPZ2Not, and pSPZ3 have been deposited under accession no. AF118920, AF118921, and AF118922, respectively.
RESULTS
Gene for the AlkS regulatory protein.
Previous experiments showed that the regulatory protein AlkS is encoded on a 3.7-kb SalI-HpaI OCT plasmid fragment (11, 12). We determined the rest of the sequence of this fragment on plasmid pBG11 and found that it contained two open reading frames (ORFs). ORF 2, from coordinate 628 to coordinate 3276 of the sequence deposited in the database (Fig. 2C), encodes a protein having a molecular mass of 99,833 Da, which is very similar to the estimated molecular mass of AlkS, 99 kDa (11), and contains the previously determined 3′ part of the nucleotide sequence (12). Consequently, alkS was assigned to ORF 2. To verify the N terminus of the encoded protein, we constructed a ′lacZ gene fusion to alkS on plasmid pBG11lacZ and found that the first 15 amino acids of the partially purified AlkS′-′LacZ fusion protein were in perfect agreement with the amino acids predicted from the nucleotide sequence (MKIIINNDFPVAKVG). The hydropathy profile of AlkS indicated that it is a soluble protein (41, 42).
Assembling the alk regulatory elements for expression.
We developed an easily excisable cassette containing the alkS regulatory gene and the alkBp promoter (Fig. 3B). Since it has been found that the untranslated alkB-mRNA leader structure affects the efficiency of gene expression (25), we designed the system as an ATG expression vector with an NdeI site on the alkB start codon, which allowed precise insertion of a recombinant gene. Using restriction enzyme NdeI (instead of the frequently used enzyme NcoI) left the second codon of an inserted gene unaffected, while the leader mRNA sequence remained nearly unchanged (CAA ATG changed to CAT ATG). This cassette was assembled into the polylinkers of plasmids pUC18Not and pUC18Sfi, from which the internal NdeI sites had been removed previously; this resulted in plasmids pSPZ2Not and pSPZ2Sfi, each of which contained an alkS* gene lacking internal NdeI sites, the alkBp promoter, and a phage T4 transcriptional terminator to prevent readthrough to regions outside the cassette. Recombinant genes could be inserted because of the NdeI site and two additional unique restriction sites, an HpaI site and an AscI site. The whole segment was flanked either by NotI sites or by SfiI sites, which made the cassettes compatible with a variety of cloning and transposon vectors that have been developed over the past decade (8, 9, 22, 28).
FIG. 3.
Structure of the central genetic elements constructed in this study. (A) pSPZ1 helper plasmid with polylinker flanked by AscI sites. (B) pSPZ2Not. We assembled in the NotI-flanked polylinker of pUC18Not (i) the 2.7-kb OCT plasmid fragment with alkS* as an EcoRI fragment, (ii) the 275-bp OCT fragment with alkBp from the upstream SmaI site to the transcription start site of alkB, (iii) an artificial polylinker, and (iv) the 272-bp SphI-HindIII pHP45Ω fragment with the phage T4 transcriptional terminator. The mutated base pair is in parentheses. Useful restriction sites are indicated, as are the alkB Shine-Dalgarno (alkB SD) sequence and the translation start site. For a more detailed restriction map of alkS* see Fig. 2C. (C) pSPZ2MA carrying the xylene oxygenase expression cassette. The modification of xylM (to xylM*) which removed the NdeI site (underlined) is shown at the bottom. The T4t transcriptional terminator is indicated by the stem-loop structure (D) pSPZ3. Part of the xylene oxygenase expression plasmid is shown. The kanamycin resistance gene is flanked by two different transcriptional terminators. Abbreviations: As, AscI; B, BamHI; Hc, HincII; Hd, HindIII; Hp, HpaI; K, KpnI; No, NotI, Ns, NsiI; Sm, SmaI; Xh, XhoI, E, EcoRI; Ps, PstI; Pa, PacI; Sc, SacI; Nd, NdeI; Sf, SfiI.
Construction of pSPZ1 helper plasmids.
To facilitate the introduction of a particular gene into the polylinker of one of the pSPZ2 plasmids, we constructed the helper plasmids pSPZ1(+) and pSPZ1(−) by digesting pGEM-7Zf(+) and pGEM-7Zf(−) with ApaI and NsiI and ligating them to a linker consisting of hybridized oligonucleotides 11 and 12, which resulted in a new polylinker flanked by AscI sites (Fig. 3A). AscI is a rarely cutting restriction enzyme with an octameric recognition sequence and is readily available from commercial sources. Thus, as an intermediate step, any fragment can be introduced into one of the pSPZ1 plasmids with the polylinker, be subjected to the necessary modifications with the phage f1 origin of replication provided by the pGEM vectors, and subsequently be excised as an NdeI-AscI fragment and inserted into pSPZ2Not or pSPZ2Sfi. Alternatively, the desired gene can be directly amplified by PCR by using extended primers that introduce the necessary restriction sites.
Xylene oxygenase expression controlled by the alk regulatory system.
Plasmid pBG63N is a pBG63 derivative with a 2.3-kb SmaI-HindIII fragment of the catabolic TOL plasmid pWW0 spanning the xylene oxygenase genes, which have been engineered to remove an NdeI site internal to xylM and to place another NdeI site on the ATG of xylM*. xylM*A was excised from this plasmid as an NdeI-SmaI fragment and was inserted into NdeI-HpaI-digested pSPZ2Not, which yielded pSPZ2MA. Thus, the whole DNA segment of interest, which consisted of alkS*, alkBp, and xylM*A plus the terminator, was available as a NotI fragment (Fig. 3C). We placed the cassette containing the desired elements on a medium-copy-number pBR322 derivative, which resulted in plasmid pSPZ3 (Fig. 3D). This plasmid contained a kanamycin resistance gene which was shielded by two different transcriptional terminators in order to limit elements of symmetry. The alkBp promoter pointed towards the phage T4 terminator, and the kanamycin resistance gene pointed towards the rrnB T1 terminator. Parts of the tetracycline resistance gene of pBR322 critical to segregational plasmid stability were removed (27). Two additional restriction sites, a NotI site and an SfiI site, were introduced, and these sites allow further easy modification of the plasmid (for instance, introduction of plasmid maintenance functions [14] or proteins that extend the range of pMB1 plasmids to other bacterial genera [52]).
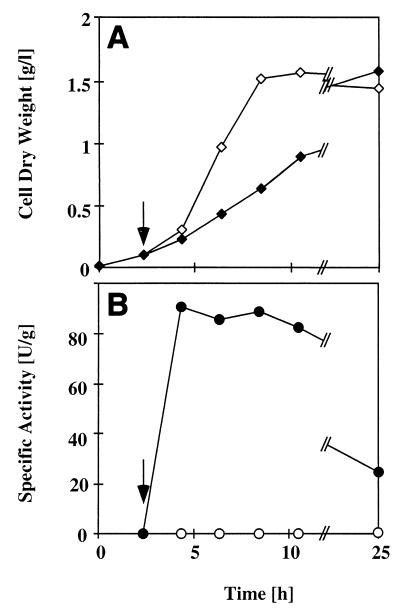
E. coli JM101 recombinants carrying plasmid pSPZ3 grown on mineral medium containing glucose as the carbon source accumulated xylene oxygenase at high specific enzyme activities (91 U · g [dry weight] of cells−1) (Fig. 4). Induction was rapid, and the growing cells exhibited the maximal specific xylene oxygenase activities 2 h after the inducer was added. Glucose did not repress translation from the alkBp promoter in the growth phase, which is consistent with previous observations (46, 58).
FIG. 4.
Induction kinetics of styrene monooxygenase activity in E. coli JM101(pSPZ3) in shaking flask experiments. (A) Growth curves with and without addition of DCPK at a cell density of approximately 0.09 g per liter. (B) Styrene monooxygenase activities with and without DCPK. Symbols: ◊ and ○, no DCPK added; ⧫ and ●, DCPK added at the time indicated by the arrow.
Production of (S)-styrene oxide by E. coli JM101(pSPZ3) in two-liquid-phase cultures.
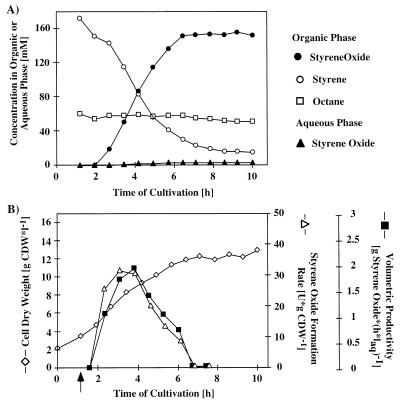
To investigate the efficacy of E. coli JM101(pSPZ3) as a biocatalyst under production conditions, we used this strain to produce (S)-styrene oxide in a two-liquid-phase fed-batch cultivation experiment analogous to our previous experiments performed with xylene oxygenase (54). E. coli JM101(pSPZ3) was first grown in a 3-liter reactor containing 1.025 liters of aqueous phase in batch mode in a mineral medium containing glucose as the carbon source. After the carbon source was exhausted, we initiated a linear feed which provided (per hour) 4.5 g of glucose and 0.5 g of yeast extract. One hour later, 375 ml of the organic second liquid phase was added; this phase consisted of the carrier n-hexadeane, 2% (vol/vol) styrene, and 1% (vol/vol) n-octane (an inducer). Approximately 2.5 h after the organic phase was added, a sharp increase in the DOT indicated a glucose limitation. To verify this, 10 ml of 50% glucose was added, which resulted in an immediate decrease in the DOT; this indicated that glucose had indeed been limiting. Approximately 1.5 h after the glucose was added, the DOT level started to increase again, and it kept increasing in an irregular fashion until the end of the experiment. Adding any of the components of the medium could not change this tendency. The biomass concentration was about 13 g (dry weight) of cells per liter of aqueous phase at the end of the experiment.
As shown in Fig. 5A, growth continued after the organic phase was added without any significant interruption. Styrene oxide formation began about 2 h later, which confirmed that the short induction period also observed in the shaking flask experiments was valid (Fig. 4). Styrene oxide was formed at rates between 20 and 35 U · g (dry weight) of cells−1 for 3 h, during which most of the styrene was consumed (Fig. 5B). The calculated bioconversion rate corresponded to a peak volumetric productivity of 2 g of styrene oxide per liter of aqueous phase per h, which was equivalent to a total transformation activity of at least 270 U per liter of aqueous phase. At the end of the experiment approximately 15 mM styrene was left in the organic phase, while the styrene oxide concentration was 155 mM. The styrene oxide concentration in the aqueous phase reached 3.1 mM during cultivation, while the styrene level in the aqueous phase decreased from 150 μM at the beginning of cultivation to levels below the detection level (5 μM).
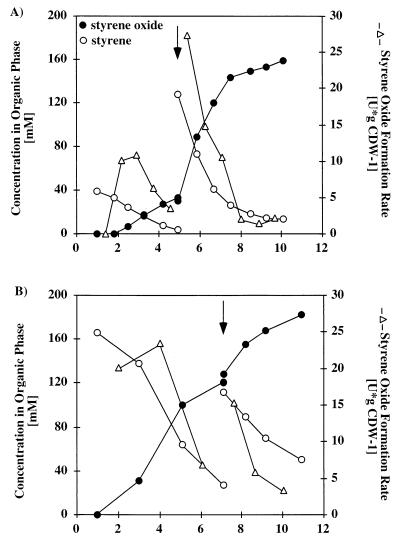
FIG. 5.
Production of (S)-styrene oxide by E. coli JM101(pSPZ3). The culture was grown in a two-liquid-phase medium which contained 25% (vol/vol) hexadecane containing 1% (vol/vol) octane as an inducer and 2% (vol/vol) styrene as the substrate. The organic phase was added 1 h after initiation of feeding. (A) Concentrations of styrene, octane, and styrene oxide in the hexadecane phase and of styrene oxide in the aqueous phase. (B) Formation of E. coli JM101(pSPZ3) biomass and development of productivities. Styrene oxide formation was calculated by determining the total rate of styrene oxide formation per gram (dry weight) of cells (CDW). Volumetric productivity was calculated by determining the mass of styrene oxide formed per hour per liter of aqueous phase (laq).
In order to investigate the reasons for the decrease in the rate of styrene oxide formation towards the end of the experiment, we carried out several additional experiments, in which organic material was added twice to the cultures. First, styrene was added together with 1% (vol/vol) octane and hexadecane. Later, fresh styrene without octane and hexadecane was added after nearly all of this substrate had been consumed. Figure 6A shows the effect of adding an organic phase which contained 0.5% (vol/vol) styrene instead of the 2% (vol/vol) styrene used for the experiment shown in Fig. 5. As Fig. 6A shows, cells were rapidly induced, as expected, but the rate of styrene oxide formation was lower and decreased as the styrene concentration in the organic phase decreased. The second addition of styrene, which resulted in a cumulative concentration of 2% (vol/vol) in the organic phase 4 h after induction, led to an immediate increase in the rate of product formation from 3.5 to 27.5 U · g (dry weight) of cells−1, which indicated that the system was substrate limited when the styrene concentration fell below 100 mM in the organic phase and 91 μM in the aqueous phase. After the second substrate addition the styrene oxide formation rate followed the same pattern as the pattern observed after the first addition; it decreased from the maximal value (ca. 28 U · g [dry weight] of cells−1) to ca. 0.5 U · g (dry weight) of cells−1 as the organic phase styrene concentration decreased to 15 mM and the aqueous styrene concentration decreased to 14 μM. The aqueous styrene oxide concentration after the pulse increased from 0.7 to 3.2 mM. A similar effect was observed when styrene was added until the cumulative concentration was 3% (vol/vol) 6 h after transformation had been started by the first addition of the organic phase containing 2% (vol/vol) styrene (Fig. 6B). In this case, the rate of styrene oxide formation increased from 7 to 15.5 U · g (dry weight) of cells−1 and then decreased. The aqueous styrene oxide concentration increased from 2.1 to 3.2 mM. In both experiments, cell growth remained very similar to cell growth in the first culture for the first 6 h after induction. When 3% (vol/vol) styrene was added, the cells ceased to grow after the second addition of styrene.
FIG. 6.
Behavior of E. coli JM101(pSPZ3) in two-liquid-phase cultures which received styrene pulses. Styrene was added at the times indicated by the arrows. Samples were taken immediately before and after the additions. (A) Culture started with 0.5% (vol/vol) styrene in the organic phase. The cumulative concentration of styrene was 2% (vol/vol). (B) Culture started with 2% (vol/vol) styrene in the organic phase. Enough styrene was added so that the cumulative concentration was 3% (vol/vol). CDW, cell dry weight.
DISCUSSION
Sequence of the alkS regulatory gene.
Previous AlkB induction experiments localized alkS, which encodes the positive regulator of the alkBFGHJKL operon, on a 3.7-kb SalI-HpaI OCT plasmid fragment (11). Induction occurred irrespective of the orientation of a somewhat larger 4.9-kb SalI fragment harboring the smaller fragment in pJRD158, suggesting that a transcriptionally autonomous unit was present. Furthermore, a 99-kDa translation product was attributed to the 3.7-kb fragment in minicell experiments (11). Sequencing of a region upstream of the alkT gene, which was also present on the 4.9-kb SalI fragment, had revealed the presence of the 3′-terminal part of alkS (12). These data were confirmed by the presence of a large ORF on the 3.7-kb SalI-HpaI subfragment harboring a 2,649-bp gene encoding a protein with a predicted molecular weight of 99,833. The 3′ part of this ORF contains the sequence upstream of alkT which had already been determined. In addition, in previous experiments Owen mapped the mutations of a number of pleiotropic Tn7S insertion mutants and nitrosoguanidine mutants and found that they are located at or very close to the location of this ORF (36). Given the limited accuracy of traditional mapping procedures, we believe that the large ORF 2 represents the gene, alkS which encodes the positive regulator of the alkBFGHJKL operon. The translation start of the gene could be verified by N-terminal sequencing of a partially purified fusion protein. A recent report localized the alkSp promoter at a position approximately 100 nucleotides upstream of the alkS initiation codon, suggesting that the DNA fragment used for cloning contains all of the necessary elements for alkS function (5).
alk regulatory system on pSPZ3 as an expression system.
Previous studies did not identify highly active recombinant biocatalysts that synthesize xylene monooxygenase. One reason for this might be that the biocatalysts used were inadequately designed; high-copy-number plasmids like pGEM-derived pBG63 have been shown to have detrimental effects on host physiology and are particularly unstable when inefficient antibiotic selection is used (2, 43, 54), and heat shock induction might interfere with protein function and plasmid stability (20, 56). Consequently, we designed plasmid pSPZ3 in order to avoid detrimental influences on gene expression based on predictable structural or segregational instability while a simple and effective regulatory system for gene expression is utilized. Our procedure included the use of transcriptional terminators to shield transcriptionally active regions (7, 48), elimination of elements of symmetry in the transcriptional terminators, deletion of known critical regions from the plasmid (27), and utilization of a kanamycin resistance gene instead of the ampicillin resistance gene (19). Use of a medium-copy-number plasmid based on pBR322 in combination with an efficient regulatory system should allow construction of a highly active biocatalyst while the physiological burden of multicopy plasmid DNA replication is reduced. The alk regulatory system has been used previously to express the membrane component of alkane hydroxylase, AlkB, at levels equivalent to up to 10% of total cell protein (35). It appeared that this regulatory system could also work well for functional expression of the xylMA genes. Furthermore, induction by octane is easy to achieve in a two-liquid-phase culture, and the system is not subject to catabolite repression by glucose in E. coli, which is why we used this regulatory system for expression. The resulting plasmid, pSPZ3, could be used very efficiently in shaking flask experiments for xylene oxygenase synthesis; we could express xylM*A in E. coli JM101 hosts at a specific activity of 91 U · g (dry weight) of cells−1 on a minimal medium containing glucose as the carbon source (Fig. 4), which is a substantial improvement over the values for recombinant strains reported previously (55).
Kinetic properties of xylene oxygenase.
The decreases in the observed rates of styrene oxidation as the styrene concentrations in the organic phases of the 1.5-liter two-liquid-phase cultures decreased were due at least in part to kinetic properties of the xylene oxygenase rather than to physiological limitations of the biocatalysis strain. We reached this conclusion on the basis of the increase in styrene oxide formation when styrene was added (Fig. 6). In principle, this could have been due to one (or both) of two reasons; either the half-saturation constant of xylene oxygenase for styrene was rather high, or accumulation of product in the aqueous phase led to reversible product inhibition. Based on the experiments whose results are shown in Fig. 5 and 6, the organic styrene concentration at which product formation proceeded at the half-maximal rate appeared to be on the order of 45 mM. Since the partition coefficient for styrene in our organic phase-aqueous medium system was approximately 1,100 at the beginning of cultivation, this suggests that the half-saturation constant value was approximately 40 μM. For comparison, m-xylene-grown P. putida mt-2 cells synthesizing xylene oxygenase have been reported to have a half-saturation constant value of 8 μM for toluene. They have also been reported to exhibit product inhibition during conversion of m-xylene to 3-methylbenzylalcohol (10). Given that the aqueous styrene oxide concentrations at the times that styrene was added in the experiments shown in Fig. 6A and B were 0.7 and 2.1 mM, the possibility that reversible product inhibition occurred cannot be eliminated. However, it appears likely that toxification of the biocatalyst eventually also started to play a role in limiting the specific activity of the biocatalyst, as cell dry weight ceased to increase after the concentration of styrene added reached a cumulative amount of 3% (vol/vol) in the experiment shown in Fig. 6B, indicating that the combined influence of styrene and styrene oxide impaired cell growth.
Recombinant whole cells which synthesize xylene oxygenase as a suitable biocatalyst in two-liquid-phase cultures.
Expression of the xylene oxygenase genes with the alk regulatory system in recombinant strains resulted in whole-cell biocatalysts that had a very high specific activity, more than 90 U · g (dry weight) of cells−1, in shaking flask experiments, which is a remarkably high value for a recombinant monooxygenase. Furthermore, the activity was easy to induce during growth of the recombinants on inexpensive carbon sources like glucose. By studying the effects of improvements, we developed a simple two-liquid-phase fed-batch process for production of (S)-styrene oxide which converts 90% of the supplied styrene, and the observed volumetric productivity is more than 1 g of (S)-styrene oxide per liter of aqueous phase over a period of at least 4 h. Cells produced styrene oxide at a rate of at least 30 U · g (dry weight) of cells−1 when the culture contained 9 g (dry weight) of cells per liter of aqueous phase. This led to volumetric activity that was fivefold higher than the activity reported previously (54).
Two-liquid-phase bioprocesses, such as the one described here, represent a feasible technology with considerable economic potential for production of hydrophobic compounds in reactions when either the substrate or the product is toxic to cells (53). The expression tools described here should help integrate new challenging enzymes into these processes, thus resolving bottlenecks due to low biocatalyst activity.
ACKNOWLEDGMENTS
This work was supported by the Swiss Priority Program Biotechnology.
We are indebted to Víctor de Lorenzo for providing strains and ideas, to Fernánd Rojo for sharing results prior to publication, to Andreas Schmid, and Birgit Kessler for helpful discussions, and to Hans-Jürgen Feiten, Andrew Schmid, and Martina Röthlisberger for help with cultivation and sequencing.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Birnbaum S, Bailey J E. Plasmid presence changes the relative levels of many host cell proteins and ribosome components in recombinant Escherichia coli. Biotechnol Bioeng. 1991;37:736–745. doi: 10.1002/bit.260370808. [DOI] [PubMed] [Google Scholar]
- 3.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W, Crosa J H, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 4.Brosius J, Dulls T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 5.Canosa I, Yuste L, Rojo F. Role of the alternative sigma factor ςS in the expression of the AlkS regulator of the Pseudomonas oleovorans alkane degradation pathway. J Bacteriol. 1999;181:1748–1754. doi: 10.1128/jb.181.6.1748-1754.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casadaban M J, Martinez-Arias A, Shapira S K, Chou J. β-Galactosidase gene fusions for analyzing gene expression in Escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Morrison D A. Cloning of Streptococcus pneumoniae DNA fragments in Escherichia coli requires vectors protected by strong transcriptional terminators. Gene. 1987;55:179–187. doi: 10.1016/0378-1119(87)90278-2. [DOI] [PubMed] [Google Scholar]
- 8.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 10.Duetz W A, Wind B, van Andel J G, Barnes M R, Williams P A, Rutgers M. Biodegradation kinetics of toluene, m-xylene, p-xylene and their intermediates through the upper TOL-pathway in Pseudomonas putida (pWW0) Microbiology. 1998;144:1669–1675. doi: 10.1099/00221287-144-6-1669. [DOI] [PubMed] [Google Scholar]
- 11.Eggink G, Engel H, Meijer W G, Otten J, Kingma J, Witholt B. Alkane utilization in Pseudomonas oleovorans. Structure and function of the regulatory locus alkR. J Biol Chem. 1988;263:13400–13405. [PubMed] [Google Scholar]
- 12.Eggink G, Engel H, Vriend G, Terpstra P, Witholt B. Rubredoxin reductase of Pseudomonas oleovorans: structural relationship to other flavoprotein oxidoreductases based on one NAD and two FAD fingerprints. J Mol Biol. 1990;212:135–142. doi: 10.1016/0022-2836(90)90310-I. [DOI] [PubMed] [Google Scholar]
- 13.Favre-Bulle O, Witholt B. Biooxidation of n-octane by a recombinant Escherichia coli in a two-liquid-phase system: effect of medium components of cell growth and alkane oxidation activity. Enzyme Microb Technol. 1992;14:931–937. [Google Scholar]
- 14.Gerdes K. The parB (hok/sok) locus of plasmid R1: a general purpose plasmid stabilization system. Bio/Technology. 1988;6:1402–1405. [Google Scholar]
- 15.Germino J, Gray J, Charbonneau H, Vanaman T V, Bastia B. Use of gene fusions and protein-protein interactions in the isolation of a biologically active regulatory protein. The plasmid replication initiator protein of plasmid R6K. Proc Natl Acad Sci USA. 1983;80:6848–6852. doi: 10.1073/pnas.80.22.6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grund A, Shapiro J, Fennewald M, Bacha P, Leahy J, Markbreiter K, Nieder M, Toepfer M. Regulation of alkane oxidation in Pseudomonas putida. J Bacteriol. 1975;123:546–556. doi: 10.1128/jb.123.2.546-556.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harayama S, Leppik R A, Rekik M, Mermod N, Lehrbach P R, Reineke W, Timmis K N. Gene order of the TOL catabolic plasmid upper pathway operon and oxidation of both toluene and benzyl alcohol by the xylA product. J Bacteriol. 1986;167:455–461. doi: 10.1128/jb.167.2.455-461.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harayama S, Rekik M, Wubbolts M, Rose K, Leppik R A, Timmis K N. Characterization of five genes in the upper-pathway operon of TOL plasmid pWW0 from Pseudomonas putida and identification of the gene products. J Bacteriol. 1989;171:5048–5055. doi: 10.1128/jb.171.9.5048-5055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasenwinkle D, Jervis E, Kops O, Liu C, Lasnicki G, Haynes C A, Kilburn D G. Very high-level production and export in Escherichia coli of a cellulose binding domain for use in a generic secretion-affinity fusion system. Biotechnol Bioeng. 1997;55:854–863. doi: 10.1002/(SICI)1097-0290(19970920)55:6<854::AID-BIT4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 20.Hellmuth K, Korz D J, Sanders E A, Deckwer W-D. Effect of growth rate on stability and gene expression of recombinant plasmids during continuous and high cell density cultivation of Escherichia coli TG1. J Biotechnol. 1994;32:289–298. doi: 10.1016/0168-1656(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 21.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 22.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahn P L. Isolation of high-frequency recombining strains from Escherichia coli containing the V colicinogenic factor. J Bacteriol. 1968;96:205–214. doi: 10.1128/jb.96.1.205-214.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiener A. Enzymatic oxidation of methyl groups on aromatic heterocycles: a versatile method for the preparation of heteroaromatic carboxylic acids. Angew Chem Int Ed Engl. 1992;31:774–775. [Google Scholar]
- 25.Kok M. Ph.D. thesis. Groningen, The Netherlands: Rijksuniversiteit; 1988. [Google Scholar]
- 26.Kok M, Oldenhuis R, van der Linden M P G, Raatjes P, Kingma J, van Lelyveld P H, Witholt B. The Pseudomonas oleovorans alkane hydroxylase gene: sequence and expression. J Biol Chem. 1998;264:5435–5441. [PubMed] [Google Scholar]
- 27.Kolot M N, Kashlev M V, Gragerov A I, Khmel I A. Stability of the pBR322 plasmid as affected by the promoter region of the tetracycline-resistance gene. Gene. 1989;75:335–339. doi: 10.1016/0378-1119(89)90280-1. [DOI] [PubMed] [Google Scholar]
- 28.Kristensen C S, Eberl L, Sánchez-Romero J M, Givskov M, Molin S, de Lorenzo V. Site-specific deletions of chromosomally located DNA segments with the multimer resolution system of broad-host-range plasmid RP4. J Bacteriol. 1995;177:52–58. doi: 10.1128/jb.177.1.52-58.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 30.Marqués S, Ramos J L. Transcriptional control of the Pseudomonas putida TOL plasmid catabolic pathways. Mol Microbiol. 1993;9:923–929. doi: 10.1111/j.1365-2958.1993.tb01222.x. [DOI] [PubMed] [Google Scholar]
- 31.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 32.Mc Fall E, Newman E B. Amino acids as carbon sources. In: Neidhardt F C, editor. Escherichia coli and Salmonella. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 358–379. [Google Scholar]
- 33.Mermod N, Harayama S, Timmis K N. New route to bacterial production of indigo. Bio/Technology. 1986;4:321–324. [Google Scholar]
- 34.Meyers R M, Lerman L S, Maniatis T. A general method for saturation mutagenesis of cloned DNA fragments. Science. 1985;229:242–247. doi: 10.1126/science.2990046. [DOI] [PubMed] [Google Scholar]
- 35.Nieboer M, Kingma J, Witholt B. The alkane oxidation system of Pseudomonas oleovorans: induction of the alk genes in Escherichia coli W3110(pGEc47) affects membrane biogenesis and results in overexpression of alkane hydroxylase in a distinct cytoplasmic membrane subfraction. Mol Microbiol. 1993;8:1039–1051. doi: 10.1111/j.1365-2958.1993.tb01649.x. [DOI] [PubMed] [Google Scholar]
- 36.Owen D J. Molecular cloning and characterization of sequences from the regulatory cluster of the Pseudomonas plasmid alk system. Mol Gen Genet. 1986;203:64–72. doi: 10.1007/BF00330385. [DOI] [PubMed] [Google Scholar]
- 37.Panke S, Sánchez-Romero J M, de Lorenzo V. Engineering quasi-natural Pseudomonas putida strains for metabolism of toluene through an ortho-cleavage degradation pathway. Appl Environ Microbiol. 1998;64:748–751. doi: 10.1128/aem.64.2.748-751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panke S, Witholt B, Schmid A, Wubbolts M G. Towards a biocatalyst for (S)-styrene oxide production: characterization of the styrene degradation pathway of Pseudomonas sp. strain VLB120. Appl Environ Microbiol. 1998;64:2032–2043. doi: 10.1128/aem.64.6.2032-2043.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 40.Ramos J L, Marqués S, Timmis K N. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators. Annu Rev Microbiol. 1997;51:341–373. doi: 10.1146/annurev.micro.51.1.341. [DOI] [PubMed] [Google Scholar]
- 41.Rao M J K, Argos P. A conformational preference parameter to predict helices in integral membrane proteins. Biochim Biophys Acta. 1986;869:197–214. doi: 10.1016/0167-4838(86)90295-5. [DOI] [PubMed] [Google Scholar]
- 42.Rost B, Fariselli P, Casadio R. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 1996;5:1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryan W, Parulekar S J. Recombinant protein synthesis and plasmid instability in continuous cultures of Escherichia coli JM103 harboring a high copy number plasmid. Biotechnol Bioeng. 1991;37:415–429. doi: 10.1002/bit.260370504. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 45.Shaw J P, Harayama S. Purification and characterization of the NADH:acceptor reductase component of xylene monooxygenase encoded by the TOL plasmid pWW0 of Pseudomonas putida mt-2. Eur J Biochem. 1992;209:51–61. doi: 10.1111/j.1432-1033.1992.tb17260.x. [DOI] [PubMed] [Google Scholar]
- 46.Staijen I E, Marcionelli R, Witholt B. The PalkBFGHJKL promoter is under carbon catabolite repression control in Pseudomonas oleovorans but not in Escherichia coli alk+ recombinants. J Bacteriol. 1999;181:1610–1617. doi: 10.1128/jb.181.5.1610-1616.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Staijen I E, Witholt B. Synthesis of alkane hydroxylase of Pseudomonas oleovorans increases the iron requirement of alk+ bacterial strains. Biotechnol Bioeng. 1998;57:228–237. [PubMed] [Google Scholar]
- 48.Stassi D L, Lacks S A. Effect of strong promoters on the cloning in Escherichia coli of DNA fragments from Streptococcus pneumoniae. Gene. 1982;18:319–328. doi: 10.1016/0378-1119(82)90170-6. [DOI] [PubMed] [Google Scholar]
- 49.Sticher P, Jaspers M C M, Stemmler K, Harms H, Zehnder A J B, van der Meer J R. Development and characterization of a whole-cell bioluminescent sensor for bioavailable middle-chain alkanes in contaminated groundwater samples. Appl Environ Microbiol. 1997;63:4053–4060. doi: 10.1128/aem.63.10.4053-4060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki M, Hayakawa T, Shaw J P, Rekik M, Harayama S. Primary structure of xylene monooxygenase: similarities to and differences from the alkane hydroxylation system. J Bacteriol. 1991;173:1690–1695. doi: 10.1128/jb.173.5.1690-1695.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Beilen J B, Wubbolts M G, Witholt B. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation. 1994;5:161–174. doi: 10.1007/BF00696457. [DOI] [PubMed] [Google Scholar]
- 52.West S E H, Schweizer H-P, Dall C, Sample A K, Runyen-Janecky L J. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;128:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 53.Witholt B, de Smet M-J, Kingma J, van Beilen J B, Lageveen R G, Eggink G. Bioconversions of alipathic compounds by Pseudomonas oleovorans in multiphase bioreactors: background and economic potential. TIBTECH. 1990;8:46–52. doi: 10.1016/0167-7799(90)90133-i. [DOI] [PubMed] [Google Scholar]
- 54.Wubbolts M G, Favre-Bulle O, Witholt B. Biosynthesis of synthons in two-liquid-phase media. Biotechnol Bioeng. 1996;52:301–308. doi: 10.1002/(SICI)1097-0290(19961020)52:2<301::AID-BIT10>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 55.Wubbolts M G, Hoven J, Melgert B, Witholt B. Efficient production of optically active styrene epoxides in two-liquid phase cultures. Enzyme Microb Technol. 1994;16:887–893. [Google Scholar]
- 56.Wubbolts M G, Reuvekamp P, Witholt B. TOL plasmid-specified xylene oxygenase is a wide substrate range monooxygenase capable of olefin epoxidation. Enzyme Microb Technol. 1994;16:608–615. doi: 10.1016/0141-0229(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 57.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–109. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 58.Yuste L, Canosa I, Rojo F. Carbon-source-dependent expression of the PalkB promoter from the Pseudomonas oleovorans alkane degradation pathway. J Bacteriol. 1998;180:5218–5226. doi: 10.1128/jb.180.19.5218-5226.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]