Abstract
Peripheral nerve injury (PNI) may lead to disability and neuropathic pain, which constitutes a substantial economic burden to patients and society. It was found that the peripheral nervous system (PNS) has the ability to regenerate after injury due to a permissive microenvironment mainly provided by Schwann cells (SCs) and the intrinsic growth capacity of neurons; however, the results of injury repair are not always satisfactory. Effective, long-distance axon regeneration after PNI is achieved by precise regulation of gene expression. Numerous studies have shown that in the process of peripheral nerve damage and repair, differential expression of non-coding RNAs (ncRNAs) significantly affects axon regeneration, especially expression of microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs). In the present article, we review the cellular and molecular mechanisms of axon regeneration after PNI, and analyze the roles of these ncRNAs in nerve repair. In addition, we discuss the characteristics and functions of these ncRNAs. Finally, we provide a thorough perspective on the functional mechanisms of ncRNAs in nervous injury repair, and explore the potential these ncRNAs offer as targets of nerve injury treatment.
Keywords: non-coding RNA, axon regeneration, peripheral nervous system, Schwann cells, peripheral nerve injury
Introduction
Injuries of the central and peripheral nervous system are common in clinical practice. Severe nerve injury has a destructive impact on patients' quality of life. Typical symptoms are sensory and motor function deficits, which can result in complete paralysis of the affected limb or development of intractable neuropathic pain 1. Neuropathic pain is caused by pathological changes or injury of the peripheral nerve or central sensory conduction pathway. After nerve injury, neuropathic pain is closely related to nerve regeneration and repair 2. Due to lack of an inherent ability to regenerate, as well as the inhibitory effect of the extracellular barrier, neurons in the central nervous system (CNS) are incapable of regeneration 3. In contrast, neurons in the peripheral nervous system (PNS) show strong regenerative capacity 4. In the PNS, functional recovery can often occur following injury since neurons have the capacity for self-repair and can reactivate intrinsic growth programs 5.
Currently, the treatment of choice for peripheral nerve injury (PNI) is advanced microsurgical end-to-end repair with tensionless epineurial sutures, autologous nerve grafting and nerve transfer where end-to-end anastomosis is not possible 6,7. Less than half of patients who undergo nerve repair after injury regain good to excellent motor or sensory function 8, and up to one-third of patients may have little or no recovery despite appropriate surgical intervention 9. Recently, growth factor (GF) supplementation has been developed as a new multifunctional treatment strategy which can promote nerve regeneration and functional recovery. However, due to the short half-life of GFs and their rapid inactivation in body fluids, administration of GFs alone is not sufficient for repairing PNI 10. Therefore, researchers urgently need to develop new therapeutic or adjunctive strategies to promote functional recovery in PNI patients. With the progress made by researchers in nerve regeneration after PNI, epigenetic regulation at the gene level has attracted greater attention.
The regulatory effect of non-coding RNAs (ncRNAs) on the expression of genes related to regeneration after PNI may provide a new target for axon regeneration after nerve injury. In this review, we summarize the cellular and molecular mechanisms of axon regeneration after PNI, introduce ncRNAs and focus on some ncRNAs that are critical for neuron regeneration. We also offer suggestions for future studies and treatment strategies in this field.
Molecular mechanisms of axon regeneration
After nervous system injury, the main internal factors that determine the inherent growth ability of axons are the expression and mobilization of axon regeneration-associated genes in nerve cells and the availability of cytoskeletal raw materials. The realization of axon regeneration requires the coordination of a variety of biological processes, including but not limited to recognition of cell damage, gene transcription, biological macromolecule synthesis, axoplasmic transport, cell energy homeostasis maintenance and cytoskeleton assembly. The transition to active neuronal regeneration is usually driven by the transcriptome, in which transcription factors (TFs) are responsible for coordinating the expression of multiple regeneration-associated genes 11. These TFs include ATF3, Smad1 and STAT3 among others 12-14. Regulation of these factors can effectively change neuronal transcription and promote regeneration. Epigenetic modification regulates gene transcription and can affect nerve regeneration by controlling ncRNAs, histone modification or DNA modification without altering the DNA sequence 15.
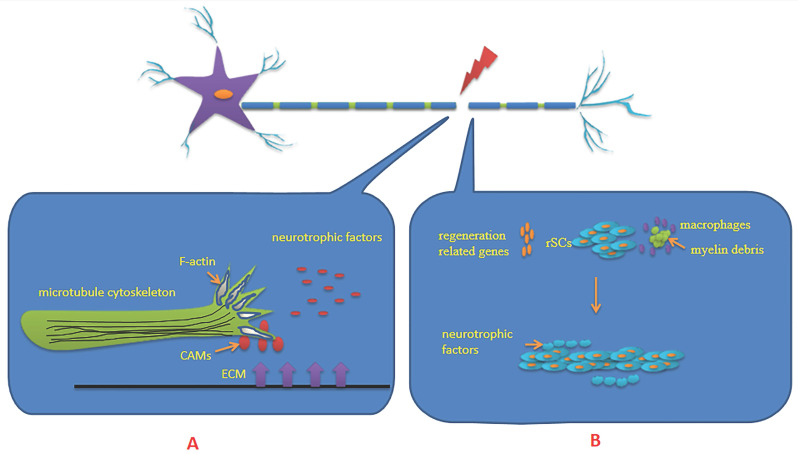
The PNS is mainly composed of neurons and axons that connect neurons. The axons are wrapped in a myelin sheath formed by Schwann cells (SCs). After PNI, the first step to ensure successful regeneration is the formation of a growth cone on the proximal segment of severed axons, which is a highly motile mechanosensitive structure at the tip of the axon. It receives multiple extracellular guidance cues through surface receptors and transduces this information into directional movements 16. The growth cone is primarily composed of microtubule cytoskeleton and filamentous actin (F-actin). Therefore, microtubule organization and dynamics, together with axonal transport, are essential for intrinsic axon regenerative capacity 17. The orientation of the advancing tip is guided by gradients of neurotrophic factors produced mainly by non-neuronal cells, which is also known as chemotaxis 18. These neurotrophic factors include nerve growth factor, glial cell line-derived neurotrophic factor, brain derived neurotrophic factor (BDNF) and neurotrophin 3, among others. The contribution of cytoskeletal proteins to axonal regeneration is crucial. The small GTPases of the Rho family integrate the upstream signaling cues for the downstream cytoskeleton rearrangements. Receptor complexes present at the growth cone can recruit GTPases, which are needed to control the contractile capability dependent on actin-myosin interactions 19. The extracellular matrix and cell adhesion molecules influence the state of the regenerating neurons and determines the response of the growth cone 20 (Figure 1).
Figure 1.
A: Growth cone is mostly made up of microtubule cytoskeleton and Filamentous actin (F-actin). The orientation of the advancing tip is guided by gradients of neurotrophic factors produced mainly by non-neuronal cells. The extracellular matrix (ECM) and cell adhesion molecules (CAMs) influence the state of the regenerating neurons and determine the response of the growth cone. B: Schwann cells (SCs) undergo dedifferentiation, proliferation, migration and remyelination after peripheral nerve injury. Differentiated Schwann cells transformed into repaired Schwann cells (rSCs). Proliferation of SCs can upregulate regeneration-associated genes, promote the aggregation of macrophages and clear myelin sheath debris. SCs form Bunger bands in the basement membrane tube and secrete a variety of neurotrophic factors. Under the support, guidance and stimulation of them, the growth cone extends from the proximal end to the distal target in the correct direction, and finally achieves nerve re-innervation.
Waller degeneration is initiated at the distal segment of severed axons, which involves a series of inflammatory and immune processes including axon degeneration, myelin degeneration, SCs proliferation, macrophage aggregation and cytokine secretion. Among them, the core of the regulation is provided by SCs. SCs undergo dedifferentiation, proliferation, migration and remyelination after PNI 21 (Figure 1). It is widely believed that the regenerated axons which arrive at their distal targets depend largely on dedifferentiation of SCs transitioning from a transmitted state toward an immature repair phenotype. Proliferation of SCs can activate monocytes/macrophages, clear myelin sheath debris and upregulate regeneration-associated genes including growth-associated protein-43, neurotrophic factors and their receptors, neuregulin and its receptors, and others. At the same time, they form Bungner bands in the basement membrane tube and secrete a variety of neurotrophic factors. Under the support and guidance of Bungner bands, the growth cone extends from the proximal end to the distal target in the correct direction, and nerve re-innervation is finally achieved 18,22,23. The guidance and intervention of SCs dedifferentiation, proliferation, remyelination and other physiological processes on axon regeneration also play an important role as the external cause of axon regeneration. Epigenetic regulation affects axon regeneration by regulating the dedifferentiation, proliferation and remyelination of SCs.
Classification and features of non-coding RNAs
There are many types of ncRNAs and classification methods. The two main groups are short ncRNAs and long ncRNAs, based on the length of the transcript. The former group includes small nuclear RNA, small nucleolar RNAs, miRNA, piwi-interacting RNA and small interfering RNA (siRNA). The latter group includes lncRNAs and other types. Based on the function of ncRNAs, there are housekeeping RNAs and regulatory RNAs. Ribosomal RNA, transfer RNA, small nucleolar RNA and small nucleolar RNAs are housekeeping RNAs, while miRNA, piwi-interacting RNA and siRNA are regulatory RNAs. According to subcellular location, ncRNAs can be divided into nuclear and cytoplasmic types. Many circRNAs are derived from protein-coding genes. However, circRNAs have not been proven to encode proteins, and so are also classified as non-coding RNAs. In this review, we focus on three types of ncRNAs: microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs), and describe in detail their functions and roles in axonal regeneration after PNI.
The tissue-specific expression and functional roles of ncRNAs in the nervous system under physiological conditions determine their putative involvement in the pathophysiological processes of neural injury, which include immune/inflammatory responses, glial scar formation, neuronal apoptosis, cell proliferation and migration, axonal regrowth, and target organ re-innervation 24. Notably, the differential expression of ncRNAs has been found in the regeneration of PNS neurons 25, which affects the survival and regeneration of neurons and regulates phenotypic changes of the SCs.
MiRNAs are a type of endogenous regulatory ncRNAs found in eukaryotes, which are about 22 nucleotides long 26. They are initially produced in the nucleus by RNA polymerase II as long primary transcripts (pri-miRNAs), which fold into hairpins to bind to Drosha and Dicer, the RNase III families of enzymes. Finally the mature miRNAs are produced 27. They regulate gene expression by directly binding to the 3'-UTR of the target mRNA, resulting in translational repression or degradation of the mRNA. MiRNAs function in the process of neurogenesis, involving pluripotent stem cell self-renewal capability, neural progenitor cell differentiation, synaptic plasticity and rearrangement, and the release of exosomes from neural cells delivering functional nucleic acids 28. These miRNA profiles may also be influenced by the heterogeneity of injury responses in different sensory neuron subtypes. It is of vital importance to determine how different neuronal subtypes respond to injury and how the regulation of translation in different populations of sensory neurons relates to their functional recovery 5. Recent studies have also found that miRNAs after PNI significantly affected the biological behaviors of neurons and SCs, affecting neuronal survival, axonal outgrowth and phenotype modulation of SCs 29. MiRNAs play a synergistic role in well-known molecular pathways of axonal growth, inflammation, apoptosis and remyelination 30.
LncRNAs are ncRNAs with a length of 200 nucleotides or longer 31. They were originally considered to be the by-products of RNA polymerase II transcription, which did not have clear biological functions. However, studies have found multiple types of lncRNAs according to their position in the genome relative to protein-coding genes, including sense, antisense, bidirectional, intronic and intergenic lncRNAs 32. They are weakly conserved in intergenic sequences and regulate gene expression at multiple levels including epigenetic, transcriptional and post-transcriptional levels, as well as by regulating miRNAs. LncRNAs are generated in the nucleus, and some of them are exported to the cytoplasm for clipping. In fact, different localizations of lncRNAs are closely associated with the regulatory mechanism in which they participate 33,34. The regulatory mechanisms of lncRNAs are complex and diverse, including miRNA sponges, regulation of protein activity and DNA interaction with binding proteins, chromatin modification, and others 35-37. Among them, lncRNAs that function as miRNA sponges are localized to the cytoplasm, while most other lncRNAs are localized to the nucleus, where they function in chromatin epigenetic modification and regulation of DNA-protein interactions 38. So it is beneficial to make clear the subcellular localization of lncRNAs to further explore their regulatory mechanisms. In the nervous system, lncRNAs are abundant and show precisely regulated temporal and spatial expression patterns 39, which have a significant impact on many neurological diseases, including brain tumors, neurodegenerative diseases and spinal cord injury 40. In addition, they have an important regulatory function in PNI 41. LncRNAs are key regulators of neuronal differentiation, development and regeneration in the nervous system 42.
The length of circRNAs varies from 200-2000 nucleotides, with the majority having lengths of about 500 nucleotides, which are generally located in the cytoplasm. The direct ligation of the exonic downstream 5' donor site with the upstream 3' acceptor site results in a closed loop structure, which makes circRNAs resistant to degradation by exonuclease RNaseR and helps them maintain high stability and conserved sequence 43. According to their origin, circRNAs may be composed of exons, introns and exon-intron structures (ElciRNA) 44. The circRNAs transcribed from exons contain miRNA binding site elements and have an miRNA sponging effect similar to that of lncRNAs. They regulate gene expression through competitive binding with corresponding miRNA site 45,46. The circRNAs transcribed from introns are mostly combined with RNA polymerase III to promote gene transcription. The circRNAs transcribed from introns and exons in cell nuclei can interact with small ribosomal U1 snRNP to form a complex with RNA polymerase II in the promoter region to promote gene expression 47. Moreover, circRNAs are efficiently translated in living human cells to produce abundant protein products by a rolling circle amplification mechanism 48. Through the above biological functions, circRNAs are thus able to regulate neuron regeneration and repair.
Non-coding RNAs in peripheral nerve regeneration
MiRNAs in peripheral nerve regeneration
Neuron survival and regeneration
The survival of injured neurons is a necessary prerequisite for activating their inherent regeneration ability. It has been determined that cell body damage or even cell death will occur around the lesion site after PNI 49. In addition to intrinsic neurotrophic factors, such as BDNF, ciliary neurotrophic factor and fibroblast growth factor-2, miRNAs also play important roles in the survival of injured neurons. Downregulated miR-192-5p can promote the expression of X-linked inhibitor of apoptosis protein (XIAP) to reduce apoptosis of nerve cells and promote the regeneration of sciatic nerve injury 50. Moreover, overexpression of miR-210 inhibited cell apoptosis by inhibiting its target ephrin-A3, thereby promoting sensory axon regeneration 51. Inhibition of endogenous miR-26a in sensory neurons impaired axon regeneration by increased expression of glycogen synthase kinase 3β. It was also found that a new miR-26a-glycogen synthase kinase 3β-Smad1 pathway could regulate axon regeneration 52. Smad1, a key member of the pathway, is a well-known axon regeneration-related transcription factor 53. During biological processes, epigenetic modification is frequently a pivotal approach to modulate gene expression. Researchers found that miR-9 targets FoxP1 directly and regulates mammalian axon regeneration post-injury. High level of endogenous miR-9 in sensory neurons inhibited axon regeneration 54.
Schwann cells
As mentioned above, SCs promote nerve repair through dedifferentiation, proliferation, migration, and remyelination after PNI. Therefore, it is necessary to discuss how miRNA regulates SCs. Damage or death of SCs induced by injuries adversely affects nerve regeneration. Research identified that miR-1b was downregulated following sciatic nerve injury and NDRG3 was verified as a direct target gene of miR-1b. Overexpression of miR-1b decreased the proliferation and migration as well as increased the apoptosis of SCs, while high expression of NDRG3 reversed the effect of miR-1b overexpression 55. In addition, other studies have found that overexpressed miR-1b inhibited the proliferation, migration, and promoted apoptosis of SCs by inhibiting BDNF 56.
Functional recovery after PNI relies on the reprogramming of differentiated SCs into repaired SCs. Transition into a repair phenotype requires expression of c-Jun and Sox2, which transcriptionally mediates inhibition of the differentiation program of myelination and activates a non-cell-autonomous repair program 4. Overexpression of miRNA-21 leads to downregulation of PTEN and activation of phosphatidylinositol-3 kinase (PI3K). Increased expression of miRNA-21 promotes axon regeneration and functional recovery after nerve injury 57.
Accurate path-finding is essential for axon growth. Ntn1 and its receptor Dcc have profound functions in axon guidance. Researchers have found that miR-9 decreases the levels of Dcc and inhibits SCs migration. miR let-7 and miR-9 decrease the protein levels of Ntn1 and Dcc, respectively, thereby inhibiting axonal regeneration. In addition, a potential regulatory network has been identified, involving let-7, miR-9, Ntn1, Dcc and other related molecules, such as RNA binding protein Lin-28 homologous protein A, SRC proto-oncogene nonreceptor tyrosine kinase and the transcription factor NF-κB 58. A recent study found that ulinastatin could significantly inhibit the expression of let-7 and increase the expression of nerve growth factor, thereby enhancing the proliferation and migration of SCs and reducing oxidative stress. Ulinastatin is a serine protease inhibitor with various anti-inflammatory and antioxidant biological activities. It can upregulate nerve growth-associated protein-43 and NF200 and myelin formation-related proteins MAG and PMP22 59. These findings suggest a possible approach for the treatment of PNI. In addition, miR-34a and miR-148b overexpression evidently inhibited the proliferation and migration of SCs by targeting contactin-2 and calreticulin, respectively 60,61. At the same time, a large number of miRNAs have been shown to promote the proliferation and migration of SCs. The upregulated miR-sc14 acts on fibroblast growth factor receptor 2 and miR-3099 acts on multiple targets including Aqp4, St8sia2, Tnfsf15 and Zbtb16 to promote the proliferation and migration of SCs 62,63. Mir-29a-3p inhibits proliferation and migration of SCs by negatively regulating peripheral myelin protein 22 (PMP22), which is closely correlated with myelin sheath formation 64.
Remyelination after PNI is a key step in axon regeneration. A study found that miR-485-5p overexpression targets Cdc42 and rac1 to inhibit SCs proliferation and myelination 65. Upregulation of miR-221-3p inhibited the myelination of SCs by target inhibiting Nab1 66. The enhanced expression of miR-30c significantly increased the amount of myelin-associated protein in co-cultured dorsal root ganglion (DRG) and SCs, suggesting that miR-30c functions to promote SCs myelination after PNI 67 (Table 1).
Table 1.
Summary of the functions and mechanisms of miRNAs in peripheral nerve injury.
| Human orthologs | Injury model | Expression | Target | Function | Reference | |
|---|---|---|---|---|---|---|
| miRNA-192-5p | Rat sciatic nerve crush | Upregulation | XIAP | Inhibition of miRNA-192-5p promote neurite outgrowth and decrease neurons apoptosis | 50 | |
| miRNA-210 | Mouse sciatic nerve crush | EFNA3 | miRNA-210 overexpression promote axon regeneration and inhibit apoptosis | 51 | ||
| miRNA-26a | Human | Mouse sciatic nerve crush |
GSK3β | Inhibition of miRNA-26a reduce axon regeneration | 52 | |
| miRNA-9 | Human | Mouse sciatic nerve crush |
FoxP1 | miRNA-9 overexpression inhibit axon regeneration | 54 | |
| miRNA-1b | Human | Rat sciatic nerve transection | Downregulation | NDRG3 | miRNA-1b overexpression inhibit proliferation, migration, and promote apoptosis of SCs | 55 |
| miRNA-1b | Human | Rat sciatic nerve transection | Upregulation | BDNF | miRNA-1b overexpression inhibit proliferation, migration, and promote apoptosis of SCs | 56 |
| miRNA let-7 | Rat sciatic nerve crush | Ntn1 | miRNA let-7 inhibit axon regeneration | 58 | ||
| miRNA-9 | Human | Rat sciatic nerve crush | Dcc | miRNA-9 inhibit SCs migration and axon regeneration | 58 | |
| miRNA-34a | Human | Rat sciatic nerve crush | Downregulation | CNTN2 | miRNA-34a inhibit SCs proliferation and migration | 60 |
| miRNA sc-14 | Rat sciatic nerve transection | Differential expression | FGFR2 | miRNA sc-14 overexpression promote SCs proliferation and migration |
62 | |
| miRNA-3099 | Rat sciatic nerve crush | Upregulation | Aqp4/ St8sia2/ Tnfsf15 /Zbtb16 | miRNA-3099 overexpression promote SCs proliferation and migration |
63 | |
| miRNA-29a-3p | Human | Rat sciatic nerve crush | Upregulation | PMP22 | miRNA-29a-3p overexpression inhibit SCs proliferation and migration |
64 |
| miRNA-485-5p | Human | Mouse sciatic nerve crush |
Downregulation | Cdc42 and Rac1 | miRNA-485-5p overexpression inhibit SCs proliferation and myelination |
65 |
| miRNA-221-3p | Human | Rat sciatic nerve resect | Nab1 | miRNA-221-3p overexpression inhibit SCs myelination | 66 | |
| miRNA-30c | Rat sciatic nerve transection | Differential expression | miRNA-30c promote SCs myelination | 67 |
LncRNAs in peripheral nerve regeneration
Neuron survival and regeneration
In recent years, studies have found that a large number of lncRNAs are differentially expressed after PNI and play an important role in peripheral nerve regeneration. It was found that silencing BC089918 and uc.217 by siRNA significantly promoted DRG neurite outgrowth 68,69. Sox11 is involved in the regulation of multiple genes including adhesion molecules, cytoskeleton elements, growth factors, cytokines, neuropeptides and other molecules associated with regeneration. Silc1 is an lncRNA with a conserved sequence and is highly expressed in neuronal tissues. Experiments indicated that Silc1 regulated nerve regeneration by cis-activating Sox11 70.
LncRNA-mRNA-miRNA co-expression network analysis has become an important research tool. Researchers found that the expression of lncRNA-GM4208 and lncRNA-GM30085 was upregulated in the injured mouse sciatic nerve, while the expression of miRNA-138, miRNA-500, miRNA-6538 and miRNA7239 was downregulated. It was hypothesized that the lncRNA, miRNA and mRNA formed a competing endogenous RNA (ceRNA) network to regulate neuron regeneration after sciatic nerve injury 71. LncRNA-mRNA-miRNA co-expression network analysis suggested lncRNA Arrl1 inhibited axon regeneration. A potential mechanism was proposed in which Arrl1 is a ceRNA acting as an miRNA molecular sponge which absorbs miR-761, an inhibitor of cyclin-dependent kinase inhibitor 2B (Cdkn2b), and thus, upregulates Cdkn2b (the inhibition of Cdkn2b significantly facilitates DRG axon regeneration) 72.
Schwann cells
The proliferation and migration of SCs have a profound impact on axon regeneration after PNI. Studies have shown that lncRNAs can act on SCs after PNI and play an important role in peripheral nerve regeneration. LncRNA snoRNA hostgene16 (SNHG16) is located on human chromosome 17 17q25.1 (13,14). Studies have reported that up-regulated SNHG16 promotes the expression of STAT3, which is a target of miR-93-5p, through endogenous competitive sponge adsorption of miR-93-5p, thus increasing SCs proliferation and migration 73. Similarly, up-regulated lncRNA MALAT1 elevates BDNF by sponging miR-129-5p 74. LncRNA Loc680254 was significantly upregulated after PNI, inducing SCs proliferation, thereby promoting axon regeneration and functional recovery. It may play a regulatory role by acting as an miRNA sponge to affect the expression of Psrc1 and Ska1. Psrc1 and Ska1 cooperate with each other to affect the cell cycle and proliferation 75. NEAT1 is a lncRNA that is upregulated in many tumor tissues. It promotes tumor cell proliferation and migration. MiR-34a is a tumor suppressor targeting NEAT1 in SCs. Satb1, an oncogene, is also the target gene of miR-34a, and there is a negative regulatory effect between Satb1 and miR-34a. Studies have shown that NEAT1 is upregulated after PNI. As a ceRNA of miR-34a, NEAT1 negatively regulates the expression of miR-34a, further increasing the expression of Satb1, and promoting the proliferation and migration of SCs, as well as peripheral nerve regeneration 76. Microarrays were used to screen the upregulated lncRNAs after sciatic nerve injury. It was found that lncRNA BC088259 showed the most significant upregulation. Further study found that BC088259 could interact with vimentin and regulate the migration of SCs after sciatic nerve injury. Vimentin is a major intermediate filament protein involved in cell development, attachment and migration. Silencing the expression of vimentin significantly inhibited the migration of SCs 77. Knockdown of lncRNA Bc088327 was found to inhibit the cell viability and induce cell apoptosis and S-phase arrest in the SCs. LncRNA Bc088327 may also synergize with Hereglin-1β, which participates in nerve regeneration 78. In addition to the above lncRNAs, the downregulation of some lncRNAs can also enhance the proliferation and migration of SCs. For instance, the downregulation of lncRNA MEG3 increased the proliferation and migration of SCs and facilitated nerve regeneration and functional recovery through the PTEN/PI3K/AKT pathway 79. Silencing lncRNA TNXA-PS1 could induce SCs migration to promote axon regeneration. A likely mechanism is that TNXA-PS1 may attenuate the inhibitory effect of miR-24-3p/ miR-152-3p on dual-specificity phosphatase 1 (DUSP1) by sponging miR-24-3p/miR-152-3p. After nerve injury, the downregulation of TNXA-PS1 decreased expression of DUSP1 and promoted the migration of SCs 80. It has been confirmed that overexpression of lncRNA MSTRG.24008.1 activates nucleotide-binding oligomerization domain-like pyrin domain containing 3 (NLRP3) and myelin and lymphocyte protein (MAL) via miR-331-3p to inhibit the proliferation of SCs at the injured site and further reduce the regeneration of the axon 81 (TABLE 2).
Table 2.
Summary of the functions and mechanisms of lncRNAs and circRNAs in peripheral nerve injury.
| Human orthologs | Injury model | Expression | Target | Function | Reference | |
|---|---|---|---|---|---|---|
| lncRNA BC089918/uc.217 | Rat sciatic nerve transection | Downregulation | Inhibition of lncRNA BC089918/uc.217 promote axon regeneration | 68,69 | ||
| lncRNA Silc1 | Human | Mouse sciatic nerve crush |
Upregulation | Sox11 | lncRNA Silc1 promote axon regeneration | 70 |
| lncRNA Arrl1 | Rat sciatic nerve crush | Downregulation | miRNA-761a/Cdkn2b | lncRNA Arrl1 inhibit neurite outgrowth | 72 | |
| lncRNA SNHG16 | Human | Rat sciatic nerve transection | Downregulation | miRNA-93-5p/STAT3 | lncRNA SNHG16 overexpression promote SCs proliferation and migration | 73 |
| lncRNA MALAT1 | Human | Mouse sciatic nerve crush |
Upregulation | miRNA-129-5p/BDNF | lncRNA MALAT1 overexpression promote SCs proliferation and migration | 74 |
| lncRNA Loc680254 | Rat sciatic nerve crush | Upregulation | Psrc1/Ska1 | lncRNA Loc680254 overexpression promote SCs proliferation | 75 | |
| lncRNA NEAT1 | Human | Mouse sciatic nerve crush |
Upregulation | miR-34a/Satb1 | lncRNA NEAT1 overexpression promote SCs proliferation, migration and axon regeneration |
76 |
| lncRNA BC088259 | Rat sciatic nerve crush | Upregulation | Vimentin | lncRNA BC088259 overexpression promote SCs migration | 77 | |
| lncRNA BC088327 | Rat sciatic nerve transection | Upregulation | Inhibition of lncRNA BC088327 suppress SCs proliferation and promote apoptosis | 78 | ||
| lncRNA MEG3 | Human | Rat sciatic nerve transection | Upregulation | PTEN/PI3K/AKT | Inhibition of lncRNA MEG3 promote SCs proliferation and migration | 79 |
| lncRNA TNXA-PS1 | Rat sciatic nerve crush | Downregulation | miRNA-24-3p/miRNA-152-3p/Dusp1 | Inhibition of lncRNA TNXA-PS1 promote SCs migration | 80 | |
| lncRNA MSTRG.24008.1 | Rat sciatic nerve crush | Upregulation | miRNA-331-3p/NLRP3/MAL | Inhibition of lncRNA MSTRG.24008.1 promote SCs proliferation | 81 | |
| circRNA-Spidr | Human | Rat sciatic nerve crush | Upregulation | PI3K-Akt | circRNA-Spidr promote axon regeneration | 83 |
| circRNA.2837 | Rat sciatic nerve crush | Downregulation | miR-34 | Inhibition of circRNA.2837 induce neurons autophagy | 84 | |
| circRNA-Ankib1 | Rat sciatic nerve crush | Downregulation | miR-423-5p/miR-485-5p/miR-666-3p/ Cyp26b1 | circRNA-Ankib1 inhibit SCs proliferation and axon regeneration | 85 |
CircRNAs in peripheral nerve regeneration
In recent years, circRNAs have been increasingly studied in nerve injury, which have a sponge-like effect on miRNAs. A bioinformatics database of circRNAs has not been completely established, so research progress on circRNAs has been relatively slow. It was found that circRNA 012142 was significantly upregulated after PNI. CircRNAs act as an miRNA sponge to inhibit miRNA activity through an interacting network with miRNAs. The circRNA/miRNA/mRNA network may be involved in multiple biological functions during axonal regeneration and degeneration 82. CircRNA-Spidr, which is derived from the Spidr gene, has its highly expression in the ovary and functions in DNA double-strand break repair through homologous recombination, thus helping to maintain the integrity of the genome. In addition, CircRNA-Spidr is a circRNA enriched in the cytoplasm of DRG neurons. Experiments have shown the downregulation of CircRNA-Spidr can regulate the PI3K-Akt signaling pathway to inhibit axon regeneration of DRG neurons after sciatic nerve injury 83. Researchers also found that downregulated circRNA.2837 alleviated sciatic nerve damage by inducing autophagy in a rat model of sciatic nerve injury. The mechanism may be that ceRNA regulates the autophagy of neurons by sponging members of the miR-34 family to protect neurons from injury 84.
CircRNA-Ankib1 is also associated with the proliferation of SCs. Overexpression of CircRNA-Ankib1 impairs SCs proliferation and axon regeneration after sciatic nerve injury. CircRNA-Ankib1 can directly bind with miR-423-5p, miR-485-5p, and miR-666-3p and function as their sponge to regulate Cyp26b1 expression 85 (Table 2).
Discussion
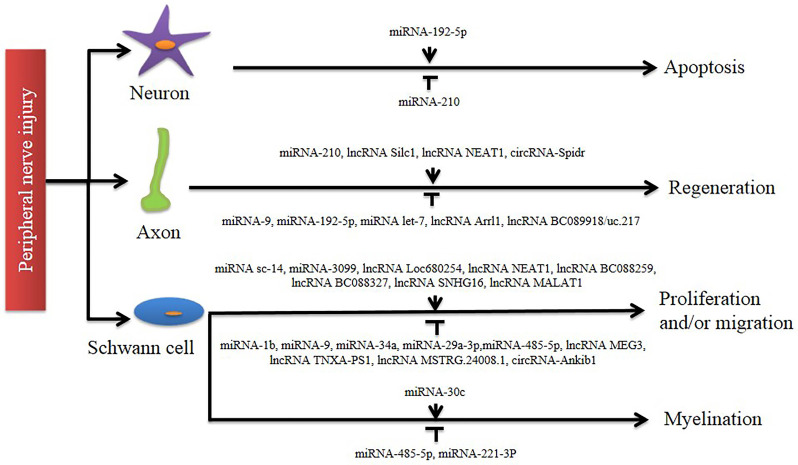
In this review, we first described the cellular and molecular mechanisms of axonal regeneration after PNI. A series of studies have shown that after PNI, a large number of ncRNAs become involved, and their differential expression suggests that they play an important role in the repair of the nervous system (Figure 2). Then, we explained the respective functions of miRNAs, lncRNAs and circRNAs in detail. In addition to the above, there are many ncRNAs that also play an important role, such as snRNA which is involved in splicing mRNA, snoRNA which guides post-transcriptional RNA modifications of other ncRNAs, piRNA which is important in maintaining genomic stability by targeting and inhibiting repetitive transposable elements, and siRNA which plays a role in inducing post-transcriptional gene silencing 86.
Figure 2.
Involvement of non-coding RNAs in peripheral nerve injury. A number of studies identify that different non-coding RNAs exert regulatory functions in axon regeneration and Schwann cell phenotype modulation (including proliferation, migration, and myelination). Their distinct roles in post-nerve-injury changes are indicated by an arrow line (for activation) and a T-shaped line (for inhibition), respectively.
Increasing evidence has shown that ncRNAs are involved in a variety of cellular processes and are associated with human diseases. This indicates that miRNAs, lncRNAs and circRNAs may be potential clinical diagnostic biomarkers and therapeutic targets. More importantly, these ncRNAs are easily detectable in body fluids. However, nerve injury repair is a very complex process involving multiple systems and multiple signaling pathways. Therefore, it's of vital importance to explore the source, characteristics, functions and regulatory mechanisms of ncRNAs in order to promote their clinical application. In previous studies, researchers have found that differentially expressed ncRNAs were derived from DRG neurons or SCs after sciatic nerve injury 68,69,87,88. In addition, some ncRNAs can also be carried by exosomes. Exosomes originating from SCs, macrophages and mesenchymal stem cells can promote peripheral nerve regeneration 89. However, more of them are endogenous, which is a significant factor in the regeneration process in PNS 52, 54.
Currently due to the structural complexity of the nervous system in humans which cannot be reproduced in vitro, researchers have employed the murine sciatic nerve injury model, which is the most used experimental paradigm for the pre-clinical investigation of peripheral nerve regeneration 90. The reason is that the size of sciatic nerve is large, which facilitates surgery, and data from previous comparable studies is readily available. However, due to limited size of the defects that can be created and the speed and quality of nerve regeneration, PNIs seen in the clinic are difficult to reproduce in murine sciatic nerve injury models. At present, there is no single experimental model that is superior to others. Hence, researchers need to carefully evaluate their specific requirements and expertise, choosing the most suited pre-clinical model 91.
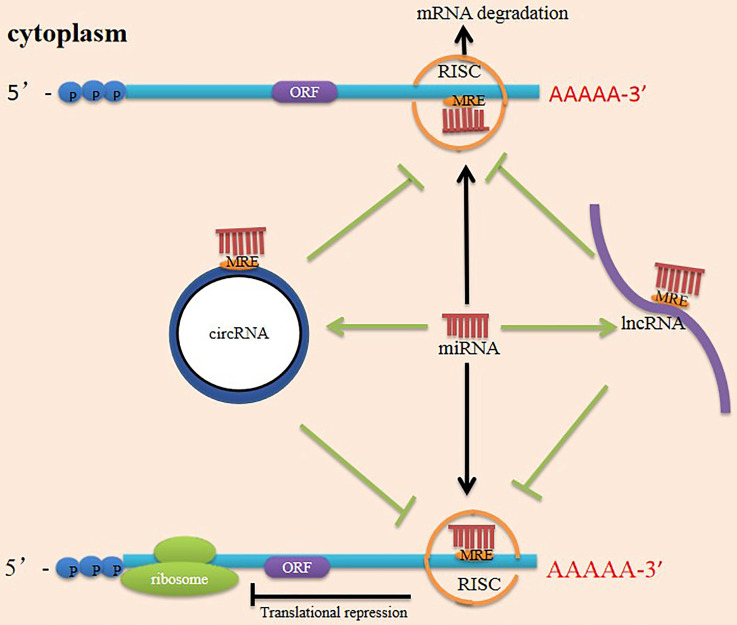
Currently, ceRNAs have attracted considerable attention, suggesting a new direction for future research. Researchers initially proposed the ceRNA hypothesis, positing that RNAs can engage in crosstalk with each other and manipulate biological functions independently of protein translation 92. MiRNAs recognize miRNA response elements (MREs) on the 3' UTR of mRNA and silence target genes by repressing mRNA translation or degrading the mRNA 93. LncRNAs and circRNAs contain MREs and can use MREs to bind miRNAs. They function as ceRNAs to sponge miRNAs and reduce miRNAs activity, impairing the interaction between miRNAs and their targeted genes 94 (Figure 3). The earliest ceRNA mechanisms and network construction have been reported in cancers. Numerous studies have reported that the ceRNA mechanism operates in various diseases and that ceRNA expression patterns vary under different tissue and cellular conditions 95. One study has connected differentially expressed lncRNAs with differentially expressed mRNAs after PNI to construct an endogenous competitive network of LIF and Hmox1 and describe the function of the mRNA-miRNA-lncRNA-ceRNA network following PNI 96. CircRNAs can also function as miRNA or lncRNA sponges to indirectly regulate mRNA expression through a complex RNA communication network in nerve regeneration 25. However, the interactions between these networks will require further documentation in PNI.
Figure 3.
Mechanism scheme of ceRNA network. There are multiple miRNA response elements (MRE) on the transcription of coding genes (mRNA), and miRNA can bind to mRNA through MRE, forming a RISC (RNA-induced silencing complex) in their 3' untranslated regions of the target mRNA. As a result, the miRNA silences the gene through mRNA degradation or transcriptional repression. Both lncRNAs and circRNAs have many MRE and serve as miRNA sponges in cells. They compete with mRNA for opportunities to bind with miRNAs. In this way, lncRNAs and circRNAs remove the miRNA's inhibition to the target mRNA, and the expression of the mRNA increases as a consequence. This mechanism is called competing endogenous RNA (ceRNA). ORF (open reading frame).
In recent years, more than 170 different modifications have been identified in RNA 97. N6-methyladenosine (m6A) is the most abundant form of methylation modification in mRNA, which is methylated at the N6 position of the adenosine base. It is dynamically regulated by methyltransferase and demethylase and binding proteins. M6A modification regulates cell proliferation, apoptosis and the cell cycle by promoting the maturation of miRNA, the translation and degradation of circRNA, and the stability of lncRNA 98. When the mammalian nervous system is damaged, m6A is involved in regulating protein synthesis and axon regeneration. M6A can control the translation process by regulating mRNA, through which axon regeneration and functional recovery is affected 99. To the best of our knowledge, the role of m6A methylation in regulating ncRNAs has not been examined in PNI, and therefore provides a possible direction for future research.
Exosomes are small secretory vesicles with a diameter of 30-150 nm that act as intercellular communication tools to transfer cytokines, mRNA, miRNA, DNA and proteins between cells, thereby causing biological reactions in recipient cells 100. During peripheral nerve repair, exosomes serve as important modulators to provide a suitable microenvironment for regeneration as well as regulate communication between different cell types. Exosomes have been shown in a variety of studies to bind to recipient cells through protein molecules on their surfaces, enter recipient cells and release contents such as miRNAs, or trigger signal transduction to affect the physiological functions of recipient cells 101. Emerging studies have shown that ncRNA-containing exosomes, which stem from sensory neurons, mesenchymal stem cells, SCs or macrophages, can mediate intercellular communication by transferring these types of cargo and stimulating axonal regeneration 102. It has been shown that after PNI, miR-21-5p is upregulated in DRG neurons. Sensory neuron-derived exosomes are readily phagocytosed by macrophages, and an increase in miR-21-5p expression promotes a pro-inflammatory phenotype, which contributes to sensory neuron-macrophage communication after damage to the peripheral nerve 103. Exosomal modification is an important part of the SCs repair program. Overexpression of miR-21 leads to downregulation of PTEN and activation of PI3K, which is responsible for the pre-regeneration ability of SC exosomes and promotes axon regeneration and functional recovery after nerve injury 57. The extracellular vesicles produced by mechanically stimulated SCs contain miR-23b-3p, whose overexpression inhibits Nrp1 expression and plays an active role in axon regeneration 104. This provides a new idea for the further study of axon regeneration in PNI. We need more exploration of ncRNAs to expand our thinking. This includes further exploration of the mechanism by which m6A methylation affects axon regeneration through the regulation of ncRNAs and exosomes carrying ncRNAs to complete intercellular transmission and participate in axon regeneration.
Although considerable progress has been made in understanding the underlying mechanisms of peripheral nerve regeneration and how these mechanisms can be harnessed to promote regeneration of the PNI, there are still many challenges to overcome in developing treatments that achieve complete regeneration and functional recovery of axons. At present, the clinical treatment of PNI mainly includes nerve repair, autologous nerve transplantation and artificial nerve conduits, among other approaches. These methods can improve the repair effect and reduce the formation of neuroma and neuropathic pain. However, functional recovery after PNI is often unsatisfactory 105,106. We will need to use combination therapy to improve the cure rate of patients. NcRNAs in the nervous system have tissue-specific expression and regulate the expression of regeneration-associated genes after PNI. Therefore, they have great potential for gene therapy in axonal regeneration after nerve injury. NcRNA-based therapeutics have rapidly emerged in recent years as a promising alternative strategy in neural regenerative medicine 28. The clinical value of ncRNAs in nerve regeneration therapy and a method to select the appropriate vector to ensure the successful delivery of ncRNAs to the desired target also need to be further explored. While we have made a good start, we still need to translate current animal studies into clinical applications as soon as possible.
In conclusion, ncRNAs provide us with further clues to understand the potential mechanism of peripheral nerve regeneration. More efforts are needed to promote the application of ncRNAs as a therapeutic target in PNI.
Acknowledgments
We would like to thank the study participants for taking part in this study.
Author Contributions
ML drafted the manuscript. PL and YJ discussed the manuscript. PL, QC and KZ made the figures and tables. JJ designed the study and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The work was supported by Natural Science Foundation of Liaoning Province (2019-MS-394) and 345 Talent Project of Shengjing Hospital.
References
- 1.Osborne NR, Anastakis DJ, Davis KD. Peripheral nerve injuries, pain, and neuroplasticity. J Hand Ther. 2018;31(2):184–194. doi: 10.1016/j.jht.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 2.Wei JH, Su WF, Zhao YY, Wei ZY, Hua YC, Xue P, Zhu X, Chen Y, Chen G. Maresin1 promotes nerve regeneration and alleviates neuropathic pain after nerve injury. J Neuroinflammation. 2022;19(1):32. doi: 10.1186/s12974-022-02405-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaudet AD, Mandrekar-Colucci S, Hall JC, Sweet DR, Schmitt PJ, Xu X. et al. miR-155 Deletion in Mice Overcomes Neuron-Intrinsic and Neuron-Extrinsic Barriers to Spinal Cord Repair. J Neurosci. 2016;36(32):8516–8532. doi: 10.1523/JNEUROSCI.0735-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nocera G, Jacob C. Mechanisms of Schwann cell plasticity involved in peripheral nerve repair after injury. Cell Mol Life Sci. 2020;77:3977–3989. doi: 10.1007/s00018-020-03516-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahar M, Cavalli V. Intrinsic mechanisms of neuronal axon regeneration. Nat Rev Neurosci. 2018;19(6):323–337. doi: 10.1038/s41583-018-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Modrak M, Talukder MA, Gurgenashvili K, Noble M, Elfar JC. Peripheral nerve injury and myelination: Potential therapeutic strategies. Journal of Neuroscience Research. 2020;98(5):780–795. doi: 10.1002/jnr.24538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raza C, Riaz HA, Anjum R, Shakeel N ul A. Repair strategies for injured peripheral nerve: Review. Life Sciences. 2020;243:117308. doi: 10.1016/j.lfs.2020.117308. [DOI] [PubMed] [Google Scholar]
- 8.Grinsell D, Keating CP. Peripheral Nerve Reconstruction after Injury: A Review of Clinical and Experimental Therapies. BioMed Research International. 2014;2014:698256. doi: 10.1155/2014/698256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanier ST, Hill JR, Dy CJ, Brogan DM. Evolving Techniques in Peripheral Nerve Regeneration. The Journal of Hand Surgery. 2021;46(8):695–701. doi: 10.1016/j.jhsa.2021.04.019. [DOI] [PubMed] [Google Scholar]
- 10.Li R, Li D, Zhang H, Wang J, Li X, Xiao J. Growth factors-based therapeutic strategies and their underlying signaling mechanisms for peripheral nerve regeneration. Acta Pharmacologica Sinica. 2020;41(10):1289–1300. doi: 10.1038/s41401-019-0338-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patodia S, Raivich G. Role of Transcription Factors in Peripheral Nerve Regeneration. Frontiers in Molecular Neuroscience. 2012;5:8. doi: 10.3389/fnmol.2012.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gey M, Wanner R, Schilling C, Pedro MT, Sinske D, Knöll B. Atf3mutant mice show reduced axon regeneration and impaired regeneration-associated gene induction after peripheral nerve injury. Open Biology. 2016;6(8):160091. doi: 10.1098/rsob.160091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saijilafu Hur E-M, Liu CM Jiao Z, Xu WL Zhou FQ. PI3K-GSK3 signalling regulates mammalian axon regeneration by inducing the expression of Smad1. Nature Communications. 2013;4(1):2690. doi: 10.1038/ncomms3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bareyre FM, Garzorz N, Lang C, Misgeld T, Buning H, Kerschensteiner M. In vivo imaging reveals a phase-specific role of STAT3 during central and peripheral nervous system axon regeneration. Proceedings of the National Academy of Sciences. 2011;108(15):6282–6287. doi: 10.1073/pnas.1015239108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palmisano I, Di Giovanni S. Advances and Limitations of Current Epigenetic Studies Investigating Mammalian Axonal Regeneration. Neurotherapeutics. 2018;15(3):529–540. doi: 10.1007/s13311-018-0636-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin M, Benzina O, Szabo V, Végh A-G, Lucas O, Cloitre T. et al. Morphology and Nanomechanics of Sensory Neurons Growth Cones following Peripheral Nerve Injury. PLoS ONE. 2013;8(2):e56286. doi: 10.1371/journal.pone.0056286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murillo B, Sousa MM. Neuronal intrinsic regenerative capacity: The impact of microtubule organization and axonal transport. Developmental Neurobiology. 2018;78(10):952–959. doi: 10.1002/dneu.22602. [DOI] [PubMed] [Google Scholar]
- 18.Allodi I, Udina E, Navarro X. Specificity of peripheral nerve regeneration: Interactions at the axon level. Progress in Neurobiology. 2012;98(1):16–37. doi: 10.1016/j.pneurobio.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Luo L. Actin Cytoskeleton Regulation in Neuronal Morphogenesis and Structural Plasticity. Annual Review of Cell and Developmental Biology. 2002;18(1):601–635. doi: 10.1146/annurev.cellbio.18.031802.150501. [DOI] [PubMed] [Google Scholar]
- 20.Kilinc D, Blasiak A, Lee GU. Microtechnologies for studying the role of mechanics in axon growth and guidance. Frontiers in Cellular Neuroscience. 2015;9:282. doi: 10.3389/fncel.2015.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viader A, Chang L, Fahrner T, Nagarajan R, Milbrandt J. MicroRNAs modulate Schwann cell response to nerve injury by reinforcing transcriptional silencing of dedifferentiation-related genes. J Neurosci. 2011;31(48):17358–17369. doi: 10.1523/JNEUROSCI.3931-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jessen KR, Mirsky R, Lloyd AC. Schwann Cells: Development and Role in Nerve Repair. Cold Spring Harbor Perspectives Biology. 2015;7(7):a020487. doi: 10.1101/cshperspect.a020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jessen KR, Mirsky R. The repair Schwann cell and its function in regenerating nerves. The Journal of Physiology. 2016;594(13):3521–31. doi: 10.1113/JP270874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou S, Ding F, Gu X. Non-coding RNAs as Emerging Regulators of Neural Injury Responses and Regeneration. Neurosci. Bull. 2016;32(3):253–264. doi: 10.1007/s12264-016-0028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao C, Yu B. Role of Long Noncoding RNAs and Circular RNAs in Nerve Regeneration. Front Mol Neurosci. 2019;12:165. doi: 10.3389/fnmol.2019.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalpachidou T, Kummer KK, Mitrić M, Kress M. Tissue Specific Reference Genes for MicroRNA Expression Analysis in a Mouse Model of Peripheral Nerve Injury. Front Mol Neurosci. 2019;12:283. doi: 10.3389/fnmol.2019.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartel DP. Metazoan MicroRNAs. Cell. 2018;173(1):20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang X, Sun C. The roles of microRNAs in neural regenerative medicine. Experimental Neurology. 2020;332:113394. doi: 10.1016/j.expneurol.2020.113394. [DOI] [PubMed] [Google Scholar]
- 29.Yu B, Zhou S, Yi S, Gu X. The regulatory roles of non-coding RNAs in nerve injury and regeneration. Progress in Neurobiology. 2015;134:122–139. doi: 10.1016/j.pneurobio.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Ghibaudi M, Boido M, Vercelli A. Functional integration of complex miRNA networks in central and peripheral lesion and axonal regeneration. Progress in Neurobiology. 2017;158:69–93. doi: 10.1016/j.pneurobio.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 32.Ponting CP, Oliver PL, Reik W. Evolution and Functions of Long Noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Chen L. Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci. 2016;41(9):761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Noh JH, Kim KM, McClusky WG, Abdelmohsen K, Gorospe M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA. 2018;9(3):e1471. doi: 10.1002/wrna.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–83. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 36.Ferrè F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17(1):106–16. doi: 10.1093/bib/bbv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nature reviews Molecular cell biology. 2021;22(2):96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021;220(2):e202009045. doi: 10.1083/jcb.202009045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Briggs JA, Wolvetang EJ, Mattick JS, Rinn JL, Barry G. Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution. Neuron. 2015;88(5):861–877. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- 40.Li P, Jia Y, Tang W, Cui Q, Liu M, Jiang J. Roles of Non-coding RNAs in Central Nervous System Axon Regeneration. Front Neurosci. 2021;15:630633. doi: 10.3389/fnins.2021.630633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Cai M, Feng Y, Yung B, Wang Y, Gao N. et al. Effect of lncRNA H19 on nerve degeneration and regeneration after sciatic nerve injury in rats. Developmental Neurobiology. 2022;82(1):98–111. doi: 10.1002/dneu.22861. [DOI] [PubMed] [Google Scholar]
- 42.Chen L, Feng P, Zhu X, He S, Duan J, Zhou D. Long non-coding RNA Malat1 promotes neurite outgrowth through activation of ERK/MAPK signalling pathway in N2a cells. J Cell Mol Med. 2016;20(11):2102–2110. doi: 10.1111/jcmm.12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao Y, Zhang R, Liu Q, Sun S, Qi M, Wang Y. et al. Functional predication of differentially expressed circRNAs/lncRNAs in the prefrontal cortex of Nrf2-knockout mice. Aging (Albany NY) 2021;13(6):8797–8816. doi: 10.18632/aging.202688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guerra B, Lima J, Araujo B, Torres L, Santos J, Machado D. et al. Biogenesis of circular RNAs and their role in cellular and molecular phenotypes of neurological disorders. Seminars in Cell and Developmental Biology. 2021;114:1–10. doi: 10.1016/j.semcdb.2020.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier, L. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 46.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 47.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 48.Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H. et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, Liu Y, Lu L. Emerging role of MicroRNAs in peripheral nerve system. Life Sciences. 2018;207:227–233. doi: 10.1016/j.lfs.2018.06.011. [DOI] [PubMed] [Google Scholar]
- 50.Liu X, Cui X, Guan G, Dong Y, Zhang Z. MicroRNA-192-5p is involved in nerve repair in rats with peripheral nerve injury by regulating XIAP. Cell Cycle. 2020;19(3):326–338. doi: 10.1080/15384101.2019.1710916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu Y, Jiang J, Yan G, Wang R, Tu G. MicroRNA-210 promotes sensory axon regeneration of adult mice in vivo and in vitro. Neurosci Lett. 2016;622:61–66. doi: 10.1016/j.neulet.2016.04.034. [DOI] [PubMed] [Google Scholar]
- 52.Jiang J, Liu C, Zhang B, Wang X, Zhang M, Saijilafu. et al. MicroRNA-26a supports mammalian axon regeneration in vivo by suppressing GSK3β expression. Cell Death Dis. 2015;6:e1865. doi: 10.1038/cddis.2015.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saijilafu Hur EM, Liu C Jiao Z, Xu W Zhou F. PI3K-GSK3 signalling regulates mammalian axon regeneration by inducing the expression of Smad1. Nat Commun. 2013;4:2690. doi: 10.1038/ncomms3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang J, Hu Y, Zhang B, Shi Y, Zhang J, Wu X. et al. MicroRNA-9 regulates mammalian axon regeneration in peripheral nerve injury. Mol Pain. 2017;13:1–9. doi: 10.1177/1744806917711612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y, Xu P, Guo C, Luo Z, Zhu J, Mou F. et al. miR-1b overexpression suppressed proliferation and migration of RSC96 and increased cell apoptosis. Neurosci Lett. 2018;687:137–145. doi: 10.1016/j.neulet.2018.09.041. [DOI] [PubMed] [Google Scholar]
- 56.Liu Y, Luo Z, Wang C, Cai H, Zhao T, Li H. et al. Electroacupuncture Promoted Nerve Repair After Peripheral Nerve Injury by Regulating miR-1b and Its Target Brain-Derived Neurotrophic Factor. Front Neurosci. 2020;14:525144. doi: 10.3389/fnins.2020.525144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.López-Leal R, Díaz-Viraqué F, Catalán RJ, Saquel C, Enright A, Iraola G. et al. Schwann cell reprogramming into repair cells increases miRNA-21 expression in exosomes promoting axonal growth. J Cell Sci. 2020;133:jcs239004. doi: 10.1242/jcs.239004. [DOI] [PubMed] [Google Scholar]
- 58.Wang X, Chen Q, Yi S, Liu Q, Zhang R, Wang P, The microRNAs let-7 and miR-9 down- regulate the axon-guidance genes Ntn1 and Dcc during peripheral nerve regeneration. J Biol Chem. 2019. 294(10) 3489-3500. [DOI] [PMC free article] [PubMed]
- 59.Zhang J, Zhang Y, Chen L, Rao Z, Sun Y. Ulinastatin Promotes Regeneration of Peripheral Nerves After Sciatic Nerve Injury by Targeting let-7 microRNAs and Enhancing NGF Expression. Dovepress. 2020;14:2695–2705. doi: 10.2147/DDDT.S255158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zou D, Zhou X, Liu J, Zhao Y, Jiang X. MiR-34a regulates Schwann cell proliferation and migration by targeting CNTN2. Neuroreport. 2020;31:1180–1188. doi: 10.1097/WNR.0000000000001539. [DOI] [PubMed] [Google Scholar]
- 61.Zhou Z, Zhang N, Shi P, Xie J. Mechanism of miR-148b inhibiting cell proliferation and migration of Schwann cells by regulating CALR. Artif Cells Nanomed Biotechnol. 2019;47(1):1978–1983. doi: 10.1080/21691401.2019.1609008. [DOI] [PubMed] [Google Scholar]
- 62.Ji X, Wang S, Cai X, Wang X, Liu Q, Wang P. et al. Novel miRNA, miR-sc14, promotes Schwann cell proliferation and migration. Neural Regen Res. 2019;14(9):1651–1656. doi: 10.4103/1673-5374.255996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Q, Miao Y, Wang X, Wang P, Cheng Z, Qian T. Increased levels of miR-3099 induced by peripheral nerve injury promote Schwann cell proliferation and migration. Neural Regen Res. 2019;14(3):525–531. doi: 10.4103/1673-5374.245478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shen Y, Cheng Z, Chen S, Zhang Y, Chen Q, Yi S. Dysregulated miR-29a-3p/PMP22 Modulates Schwann Cell Proliferation and Migration During Peripheral Nerve Regeneration. Molecular Neurobiology. 2022;59(2):1058–1072. doi: 10.1007/s12035-021-02589-2. [DOI] [PubMed] [Google Scholar]
- 65.Zhang Z, Li X, Li A, Wu G. miR-485-5p suppresses Schwann cell proliferation and myelination by targeting cdc42 and Rac1. Exp Cell Res. 2020;388(1):111803. doi: 10.1016/j.yexcr.2019.111803. [DOI] [PubMed] [Google Scholar]
- 66.Zhao L, Yuan Y, Li P, Pan J, Qin J, Liu Y. et al. miR-221-3p Inhibits Schwann Cell Myelination. Neuroscience. 2018;379:239–245. doi: 10.1016/j.neuroscience.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 67.Yi S, Wang Q, Zhao L, Qin J, Wang Y, Yu B. et al. miR-30c promotes Schwann cell remyelination following peripheral nerve injury. Neural Regen Res. 2017;12(10):1708–1715. doi: 10.4103/1673-5374.217351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu B, Zhou S, Hu W, Qian T, Gao R, Ding G. et al. Altered long noncoding RNA expressions in dorsal root ganglion after rat sciatic nerve injury. Neurosci Lett. 2013;534:117–122. doi: 10.1016/j.neulet.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 69.Yao C, Wang J, Zhang H, Zhou S, Qian T, Ding F. et al. Long non-coding RNA uc.217 regulates neurite outgrowth in dorsal root ganglion neurons following peripheral nerve injury. Eur J Neurosci. 2015;42:1718–1725. doi: 10.1111/ejn.12966. [DOI] [PubMed] [Google Scholar]
- 70.Perry RB, Hezroni H, Goldrich MJ, Ulitsky I. Regulation of Neuroregeneration by Long Noncoding RNAs. Mol Cell. 2018;72:553–567. doi: 10.1016/j.molcel.2018.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jia Y, Zhang M, Li P, Tang W, Liu Y, Hu Y. et al. Bioinformatics analysis of long non-coding RNAs involved in nerve regeneration following sciatic nerve injury. Mol Pain. 2020;16:1–10. doi: 10.1177/1744806920971918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang D, Chen Y, Liu M, Cao Q, Wang Q, Zhou S. et al. The long noncoding RNA Arrl1 inhibits neurite outgrowth by functioning as a competing endogenous RNA during neuronal regeneration in rats. J Biol Chem. 2020;295(25):8374–8386. doi: 10.1074/jbc.RA119.011917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen Y, Fan Z, Dong Q. LncRNA SNHG16 promotes Schwann cell proliferation and migration to repair sciatic nerve injury. Annals of Translational Medicine. 2021;9(16):1349. doi: 10.21037/atm-21-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu G, Li X, Li M, Zhang Z. Long non-coding RNA MALAT1 promotes the proliferation and migration of Schwann cells by elevating BDNF through sponging miR-129-5p. Experimental Cell Research. 2020;390(1):111937. doi: 10.1016/j.yexcr.2020.111937. [DOI] [PubMed] [Google Scholar]
- 75.Yao C, Wang Q, Wang Y, Wu J, Cao X, Lu Y. et al. Loc680254 regulates Schwann cell proliferation through Psrc1 and Ska1 as a microRNA sponge following sciatic nerve injury. Glia. 2021;69:2391–2403. doi: 10.1002/glia.24045. [DOI] [PubMed] [Google Scholar]
- 76.Liu X, Yu X, He Y, Wang L. Long noncoding RNA nuclear enriched abundant transcript 1 promotes the proliferation and migration of Schwann cells by regulating the miR-34a/Satb1 axis. J Cell Physiol. 2019;234:16357–16366. doi: 10.1002/jcp.28302. [DOI] [PubMed] [Google Scholar]
- 77.Yao C, Chen Y, Wang J, Qian T, Feng W, Chen Y. et al. LncRNA BC088259 promotes Schwann cell migration through Vimentin following peripheral nerve injury. Glia. 2020;68:670–679. doi: 10.1002/glia.23749. [DOI] [PubMed] [Google Scholar]
- 78.Wang H, Wu J, Zhang X, Ding L, Zeng Q. Microarray analysis of the expression profile of lncRNAs reveals the key role of lncRNA BC088327 as an agonist to heregulin-1β-induced cell proliferation in peripheral nerve injury. Int J Mol Med. 2018;41:3477–3484. doi: 10.3892/ijmm.2018.3571. [DOI] [PubMed] [Google Scholar]
- 79.Ma Y, Zhai D, Zhang W, Zhang H, Dong L, Zhou Y. et al. Down-regulation of long non-coding RNA MEG3 promotes Schwann cell proliferation and migration and repairs sciatic nerve injury in rats. J Cell Mol Med. 2020;24:7460–7469. doi: 10.1111/jcmm.15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yao C, Wang Y, Zhang H, Feng W, Wang Q, Shen D. et al. lncRNA TNXA-PS1 Modulates Schwann Cells by Functioning As a Competing Endogenous RNA Following Nerve Injury. J Neurosci. 2018;38(29):6574–6585. doi: 10.1523/JNEUROSCI.3790-16.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yin G, Peng Y, Lin Y, Wang P, Li Z, Wang R. et al. Long Non-coding RNA MSTRG.24008.1 Regulates the Regeneration of the Sciatic Nerve via the miR-331-3p-NLRP3/MAL Axis. Frontiers in Cell and Developmental Biology. 2021;9:641603. doi: 10.3389/fcell.2021.641603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sohn EJ, Park HT. Differential expression of circular RNAs in the proximal and distal segments of the sciatic nerve after injury. Neuroreport. 2020;31:76–84. doi: 10.1097/WNR.0000000000001371. [DOI] [PubMed] [Google Scholar]
- 83.Mao S, Huang T, Chen Y, Shen L, Zhou S, Zhang S. et al. Circ-Spidr enhances axon regeneration after peripheral nerve injury. Cell Death Dis. 2019;10:787. doi: 10.1038/s41419-019-2027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou Z, Niu Y, Huang G, Lu J, Chen A, Zhu L. Silencing of circRNA.2837 Plays a Protective Role in Sciatic Nerve Injury by Sponging the miR-34 Family via Regulating Neuronal Autophagy. Mol Ther Nucleic Acids. 2018;12:718–729. doi: 10.1016/j.omtn.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mao S, Zhang S, Zhou S, Huang T, Feng W, Gu X. et al. A Schwann cell-enriched circular RNA circ-Ankib1 regulates Schwann cell proliferation following peripheral nerve injury. Faseb j. 2019;33:12409–12424. doi: 10.1096/fj.201900965R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hombach S, Kretz M. Non-coding RNAs: Classification, Biology and Functioning. Advances in Experimental Medicine and Biology. 2016;937:3–17. doi: 10.1007/978-3-319-42059-2_1. [DOI] [PubMed] [Google Scholar]
- 87.Xia B, Gao J, Li S, Huang L, Zhu L, Ma T. et al. Mechanical stimulation of Schwann cells promote peripheral nerve regeneration via extracellular vesicle-mediated transfer of microRNA 23b-3p. Theranostics. 2020;10(20):8974–8995. doi: 10.7150/thno.44912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou S, Shen D, Wang Y, Gong L, Tang X, Yu B. et al. microRNA-222 Targeting PTEN Promotes Neurite Outgrowth from Adult Dorsal Root Ganglion Neurons following Sciatic Nerve Transection. PLoS ONE. 2012;7(9):e44768. doi: 10.1371/journal.pone.0044768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Qing L, Chen H, Tang J, Jia X. Exosomes and Their MicroRNA Cargo: New Players in Peripheral Nerve Regeneration. Neurorehabil Neural Repair. 2018;32(9):765–76. doi: 10.1177/1545968318798955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Savastano LE, Laurito SR, Fitt MR, Rasmussen JA, Gonzalez Polo V, Patterson SI. Sciatic nerve injury: A simple and subtle model for investigating many aspects of nervous system damage and recovery. Journal of Neuroscience Methods. 2014;227:166–180. doi: 10.1016/j.jneumeth.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 91.Geuna S. The sciatic nerve injury model in pre-clinical research. Journal of Neuroscience Methods. 2015;243:39–46. doi: 10.1016/j.jneumeth.2015.01.021. [DOI] [PubMed] [Google Scholar]
- 92.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kartha RV, Subramanian S. Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Frontiers in Genetics. 2014;5:8. doi: 10.3389/fgene.2014.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Moreno-García L, López-Royo T, Calvo AC, Toivonen JM, de la Torre M, Moreno-Martínez L. et al. Competing Endogenous RNA Networks as Biomarkers in Neurodegenerative Diseases. International Journal of Molecular Sciences. 2020;21(24):9582. doi: 10.3390/ijms21249582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cai Y, Wan J. Competing Endogenous RNA Regulations in Neurodegenerative Disorders: Current Challenges and Emerging Insights. Frontiers in Molecular Neuroscience. 2018;11:370. doi: 10.3389/fnmol.2018.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Qian T, Fan C, Liu Q, Yi S. Systemic functional enrichment and ceRNA network identification following peripheral nerve injury. Mol Brain. 2018;11:73. doi: 10.1186/s13041-018-0421-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science. 2018;361(6409):1346–1349. doi: 10.1126/science.aau1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tang F, Chen L, Gao H, Xiao D, Li X. m6A: An Emerging Role in Programmed Cell Death. Frontiers in Cell and Developmental Biology. 2022;10:817112. doi: 10.3389/fcell.2022.817112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Weng Y, Wang X, An R, Cassin J, Vissers C, Liu Y. et al. Epitranscriptomic m6A Regulation of Axon Regeneration in the Adult Mammalian Nervous System. Neuron. 2018;97(2):313–325. doi: 10.1016/j.neuron.2017.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Qing L, Chen H, Tang J, Jia X. Exosomes and Their MicroRNA Cargo: New Players in Peripheral Nerve Regeneration. Neurorehabilitation and Neural Repair. 2018;32(9):765–776. doi: 10.1177/1545968318798955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang J, Li S, Li L, Li M, Guo C, Yao J. et al. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genomics Proteomics Bioinformatics. 2015;13(1):17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bischoff JP, Schulz A, Morrison H. The role of exosomes in intercellular and inter-organcommunication of the peripheral nervous system. FEBS Letters. 2022;596(5):655–664. doi: 10.1002/1873-3468.14274. [DOI] [PubMed] [Google Scholar]
- 103.Simeoli R, Montague K, Jones HR, Castaldi L, Chambers D, Kelleher JH. et al. Exosomal cargo including microRNA regulates sensory neuron to macrophage communication after nerve trauma. Nature Communications. 2017;8(1):1778. doi: 10.1038/s41467-017-01841-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xia B, Gao J, Li S, Huang L, Zhu L, Ma T. et al. Mechanical stimulation of Schwann cells promote peripheral nerve regeneration via extracellular vesicle-mediated transfer of microRNA 23b-3p. Theranostics. 2020;10(20):8974–8995. doi: 10.7150/thno.44912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Burks SS, Levi DJ, Hayes S, Levi AD. Challenges in sciatic nerve repair: anatomical considerations. J Neurosurg. 2014;121:210–218. doi: 10.3171/2014.2.JNS131667. [DOI] [PubMed] [Google Scholar]
- 106.Gordon T. Peripheral Nerve Regeneration and Muscle Reinnervation. Int J Mol Sci. 2020;21(22):8652. doi: 10.3390/ijms21228652. [DOI] [PMC free article] [PubMed] [Google Scholar]