Abstract
Synapse formation is a process tightly controlled in space and time. How gene regulatory mechanisms specify spatial and temporal aspects of synapse formation is not well understood. In the nematode C.elegans, two subtypes of the D-type inhibitory motor neuron (MN) classes, the dorsal D (DD) and ventral D (VD) neurons, extend axons along both the dorsal and ventral nerve cords 1. The embryonically generated DD MNs initially innervate ventral muscles in the first (L1) larval stage and receive their synaptic input from cholinergic MNs in the dorsal cord. They rewire by the end of the L1 molt to innervate dorsal muscles and to be innervated by newly formed ventral cholinergic MNs 1. VD MNs develop after the L1 molt; they take over the innervation of ventral muscles and receive their synaptic input from dorsal cholinergic MNs. We show here that the spatiotemporal control of synaptic wiring of the D-type neurons is controlled by an intersectional transcriptional strategy in which the UNC-30 Pitx-type homeodomain transcription factor acts together in embryonic and early larval stages with the temporally controlled LIN-14 transcription factor to prevent premature synapse rewiring of the DD MNs and, together with the UNC-55 nuclear hormone receptor, to prevent aberrant VD synaptic wiring in later larval and adult stages. A key effector of this intersectional transcription factor combination is a novel synaptic organizer molecule, the single immunoglobulin domain protein OIG-1. OIG-1 is perisynaptically localized along the synaptic outputs of the D-type MNs in a temporally controlled manner and is required for appropriate selection of both pre- and post-synaptic partners.
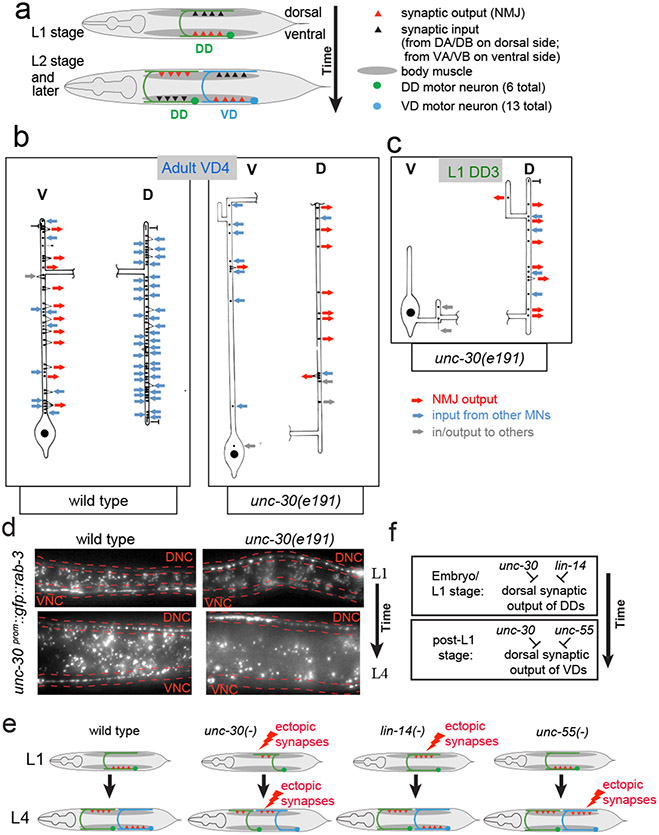
At the end of the first larval stage (L1), the synaptic outputs from the DD MNs to ventral muscle and their synaptic input from cholinergic DA/DB MNs is eliminated and, instead, synapses are formed onto dorsal muscle and synaptic input is received from cholinergic VA/VB MNs (Fig.1a)1. We sought to examine how the spatiotemporal specificity of this rewiring process is controlled and integrated with other aspects of D-type MN differentiation. To address this question, we examined the function of the C. elegans Pitx-type homeobox transcription factor UNC-30, which is known to control GABAergic neurotransmitter identity of the D-type MNs 2,3. The analysis of serial electron micrographs shows that the synaptic patterns of the DD and VD neurons are substantially disrupted in unc-30 null mutant animals. In adult unc-30(0) animals, VD MNs display ectopic synapses onto dorsal muscle and lack significant synaptic inputs from DA/DB on the dorsal side (Fig.1b; Extended Data Fig. 1). Furthermore, DD MNs which normally only synapse onto ventral muscle, show aberrant innervation of dorsal muscle in L1 stage unc-30 mutants (Fig.1c). These synaptic defects were confirmed with GFP-tagged RAB-3 protein, expressed specifically in D-type MNs (Fig.1d).
Figure 1: Loss of unc-30 disrupts the synaptic connectivity of the DD and VD MNs.
a: Schematic of DD rewiring 1.
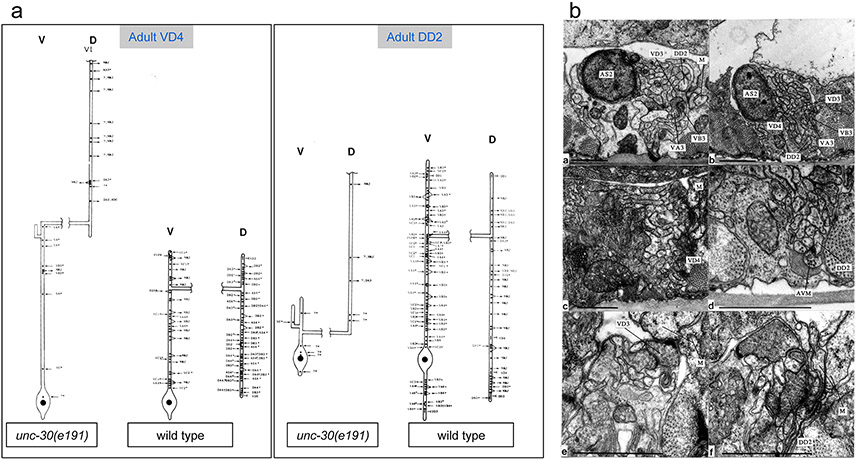
b: Reconstruction of a VD4 MN from an unc-30(e191) adult animal compared to the same neuron in a wild-type animal. Extended Data Fig. 1 shows a more detailed presentation of the EM data.
c: Reconstructed DD3 neuron from an unc-30(e191) L1 larva showing aberrant NMJs in the dorsal cord (D). Previous reconstructions of a wild-type L1 using the same techniques and personnel showed that a reconstructed DD3 made no NMJs on dorsal muscles and 9 NMJs on ventral muscles 1.
d: Presynaptic marker RAB-3 ectopically localizes mostly to ventral cord in wild-type L1 animals (16/20 animals), but ectopically in the dorsal nerve cord (DNC; outlined in red) in unc-30 mutant animals (19/20). At the L4 stage, in which presynaptic specialization are observed in both ventral nerve cord (VNC; outlined in red) and DNC (19/20 animals), unc-30 mutants show few specializations in the VNC (20/20 animals).
E,F: Summary of synapse formation defects in unc-30, lin-14, and unc-55 mutants (E) and genetic interpretation (F).
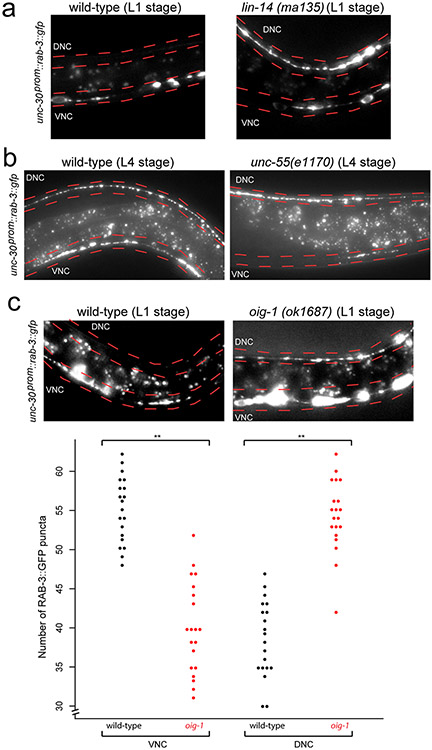
unc-30 is expressed in both DD and VD MNs at all stages 3, yet unc-30 inhibits dorsal DD synapses only in the L1 stage and not at later stages. However, at these later stages, unc-30 does inhibit dorsal synapses from VD neurons, but not the DD neurons. How can the temporal and spatial specificity of unc-30(e191) defects be explained? A potential answer to this question lies in the previously described mutant phenotype of two transcription factors, which recapitulate specific components of the cell-type specific, DD and VD synaptogenic defects of unc-30 mutants. In animals lacking the lin-14 transcription factor, whose expression is normally temporally restricted to embryonic and first larval stages in most tissues, including the D-type motorneurons 4,5, DD MNs form ectopic synapses in the dorsal cord in embryonic and L1 stages (Extended Data Fig. 2a; schematized in Fig.1e,f)4. These DD MN defects are similar to those that we observe in unc-30 mutants. The dorsal ectopic synapses in the VD neurons of unc-30 mutant animals (not observed in lin-14) are in turn recapitulated in animals lacking the unc-55 orphan nuclear receptor, in which VD MNs form aberrant synapses in the dorsal cord, as previously shown (Extended Data Fig. 2b; schematized in Fig.1e,f)6,7, while DD wiring at the L1 stage is normal. Taken together, the unc-30 phenotype in the DD and VD neurons can be viewed as a “composite” of the two individual phenotypes of lin-14 (DD neurons at L1 stage) and unc-55 (VD neurons at later stages) (Fig.1e,f schematic). One possible way to explain these concordances of phenotypes is that unc-30 may collaborate with lin-14 to control the expression of a molecule that acts in a temporally restricted manner in embryonic and L1 stages to inhibit dorsal synapse formation of the DD neurons. In the VD MNs, unc-30 may in turn collaborate with unc-55 to control expression of a molecule that acts in the VD neurons to inhibit dorsal synapse formation of these neurons.
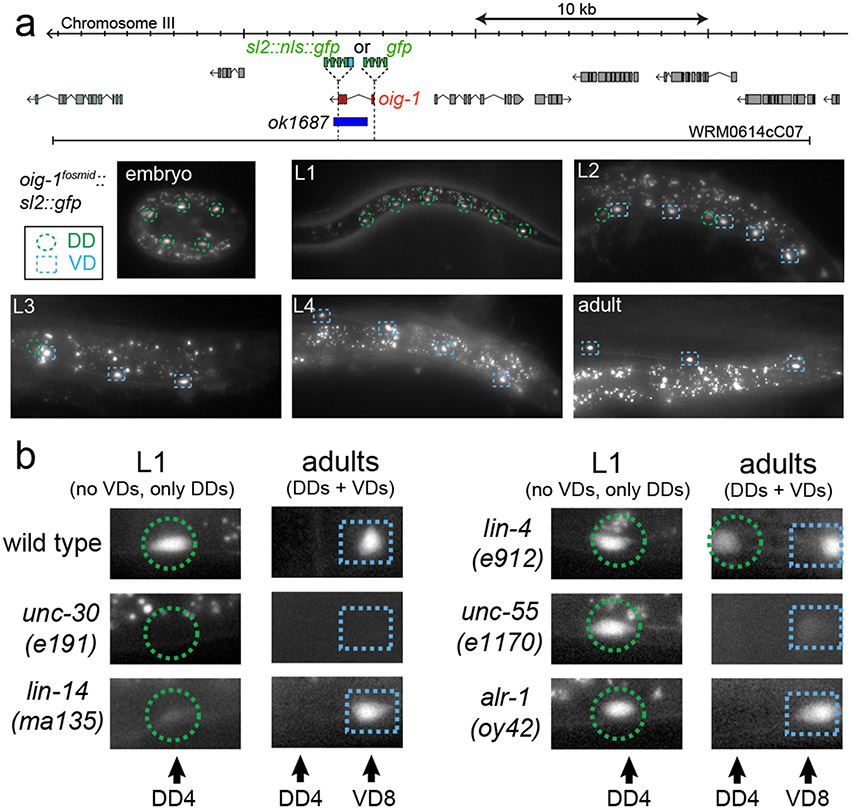
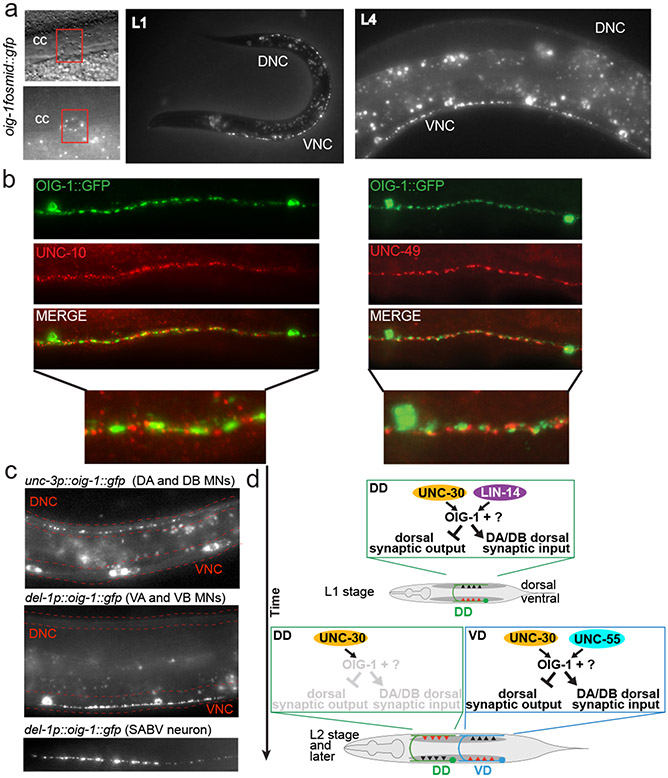
We sought to identify such potential effector molecule(s) through a candidate gene approach. In a survey of C. elegans immunoglobulin superfamily members, we had previously described a family of small proteins that are composed of a single Ig domain, the oig gene family 8. One of the oig family members, oig-1, encodes a 137aa long protein with a signal sequence, a single IgC2-type domain but no transmembrane domain or predicted GPI anchor. Transgenic animals carrying an oig-1 fosmid-based reporter construct showed expression both in the DD and VD MNs, but no other ventral nerve cord MNs (Fig.2a). Notably, expression of oig-1 in the D-type MNs is temporally controlled in a manner that correlates with the distinct periods of inhibition of dorsal muscle innervation exhibited by DD and VD neurons. oig-1 is transiently expressed in the DD neurons during the time when no dorsal synapses are formed (embryos and L1), but is downregulated in the DD neurons upon formation of their dorsal synapses (L2 and later; Fig.2a). In contrast, expression of oig-1 in the VD neurons, which have processes but no synaptic outputs in the dorsal cord, is continuously maintained throughout the life of the neuron (Fig.2a).
Figure 2: Expression of oig-1 correlates with the inhibition of dorsal synapse formation and is controlled by unc-30, lin-14 and unc-55.
a: oig-1 locus, oig-1 fosmid-based reporters (SL2::NLS::GFP fusion to assess gene regulation; GFP fusion after signal sequence to assess protein localization), deletion allele and oig-1fosmid::sl2::nls::gfp expression pattern (similar results with 2 independent lines).
b: oig-1 expression is regulated by the transcription factors unc-30 (0/20 animal express the reporter at any stage), lin-14 (3/20 L1 stage animals express the reporter but at reduced levels; scored in progeny from lin-14 null mothers carrying a lin-14 rescue array 23, n=20), and unc-55 (0/20 animal express the reporter in VD MNs; 20/20 animals express reporter in DD neurons), but not by alr-1 (20/20 animal express the reporter), in which VD identity but not synaptic wiring is affected 24. In lin-4 animals, oig-1 expression in DDs persists into the adult stage (20/20 adult animal express the reporter).
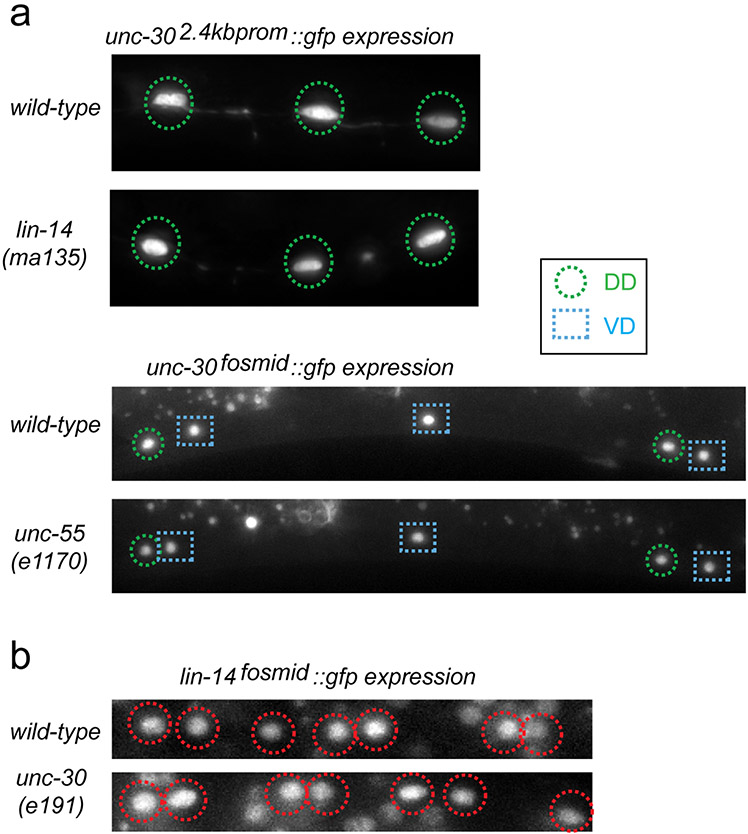
The transient expression in the DD and continuous expression in the VD neurons makes oig-1 a candidate effector gene for the unc-30, lin-14 and unc-55 transcription factors. Indeed, the oig-1fosmid::sl2::gfp reporter fails to be expressed in both DD- and VD-type MNs in unc-30 null mutants at all stages (Fig.2b). In lin-14 null mutant animals, transient expression of the oig-1 reporter in the L1 stage is diminished in the DD MNs (Fig.2b) and temporally prolonged expression of lin-14, achieved through genetic removal of a negative regulator of lin-14, the miRNA lin-4 9, results in prolonged expression of oig-1 in the DD MNs into the adult stage (Fig.2b). In animals lacking unc-55, which is normally expressed in VD, but not DD MNs 10, expression of oig-1 in the DD neurons at the L1 stage is unaffected but oig-1 expression in the VD neurons is absent at the adult stage (Fig.2b). The two distinct transcription factor combinations that control oig-1 expression in DD (unc-30 and lin-14) and in VD (unc-30 and unc-55) operate independently since the expression of each transcription factor is independent of the presence of the other transcription factor (Extended Data Fig. 3a,b)11.
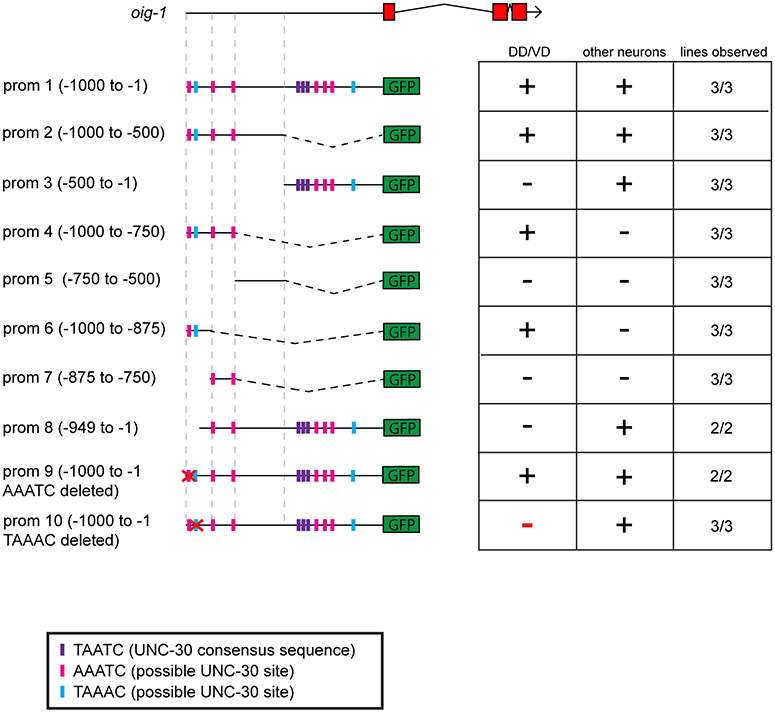
The transcriptional nature of oig-1 regulation is corroborated by the finding that 1kb of 5’ sequences of oig-1 conveys the same spatiotemporal regulation as the oig-1 fosmid reporter (Extended Data Fig. 4), Chromatin immunoprecipitation-sequencing data from the modEncode project shows binding of UNC-55 to this 1kb fragment of the oig-1 locus 12, suggesting direct regulation. A 125bp element that still recapitulates spatiotemporal control in the D-type MN expression contains two sites with partial match to the UNC-30 binding site 13 and one is required for DD MN expression (Extended Data Fig. 4).
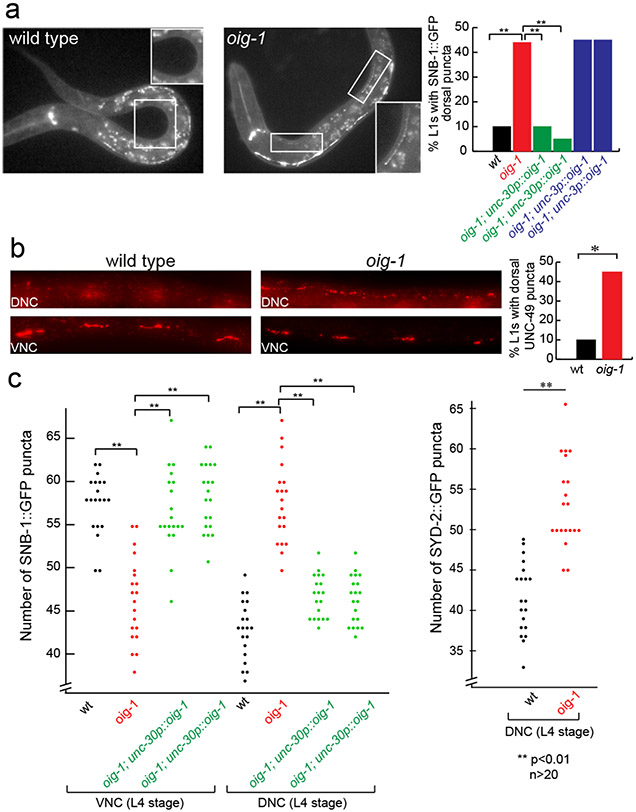
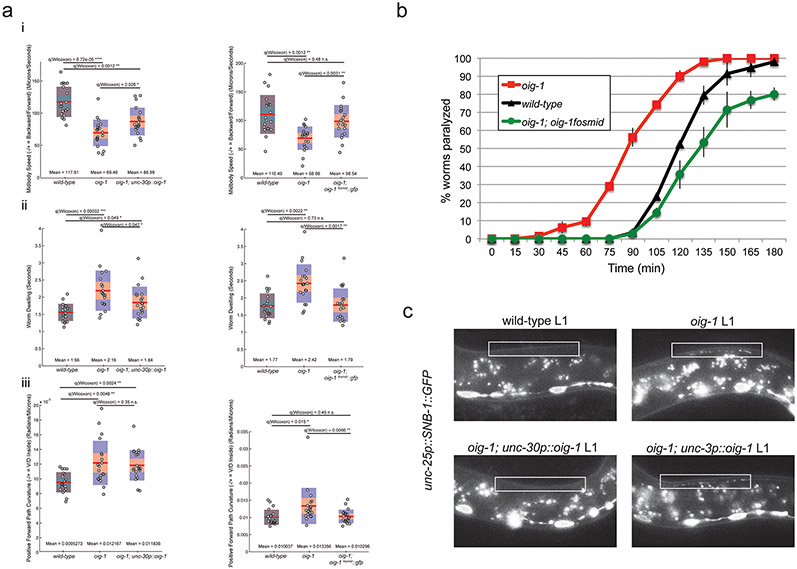
The expression pattern of oig-1 and its regulation by transcription factors that regulate synapse formation make oig-1 a candidate to be involved in the synapse-organizing activity of these transcription factors. Animals that carry an oig-1 deletion allele (Fig.2a) are viable and fertile, but display locomotory defects and hypersensitivity to the drug aldicarb (Extended Data Fig. 5a,b), which is characteristic of abnormalities in GABAergic signaling 14. In embryonic and L1 stages when only the DD MNs are present, the presynaptic vesicle protein SNB-1 and RAB-3 are aberrantly clustered along the dorsal nerve cord of oig-1 null mutants (Fig.3a; Extended Data Fig. 2c). Moreover, the postsynaptic GABA receptor UNC-49 15, which normally clusters on ventral muscle at the L1 stage, clusters ectopically along the dorsal nerve cord (Fig.3b). Therefore, oig-1 – like its upstream regulators unc-30 and lin-14 - is required to prevent premature DD synapse formation in the dorsal nerve cord.
Figure 3: Aberrant D-type MN synapse formation in oig-1 mutants.
a: Ectopic DD synapses in the dorsal nerve cord of oig-1 mutant L1s.
b: Ectopic UNC-49 localization in oig-1 mutant L1s, as assessed by UNC-49 antibody staining. UNC-17 staining was used as a control to identify the ventral and dorsal nerve cords of the animals (not shown).
c: Ectopic VD synapses in the dorsal nerve cord of L4 staged oig-1 mutants. For each marker in each strain, puncta in the posterior of L4 animals from DD4 to VD11 were scored.
Examining the synapses of the VD MN that normally exclusively form synapses on ventral muscle, we observed more puncta of three presynaptic markers (SNB-1 and RAB-3 proteins and the Liprin-α protein SYD-2) in the dorsal nerve cord and less in the ventral nerve cord of oig-1 mutants at post-L1 larval stages (Fig.3c; Extended Data Fig. 2c). This suggests the VD MNs have aberrant synaptic specializations in the dorsal nerve cord in oig-1 mutants; while less severe, this phenotype is similar to those of the VD synaptic defects of unc-30 and unc-55 mutants.
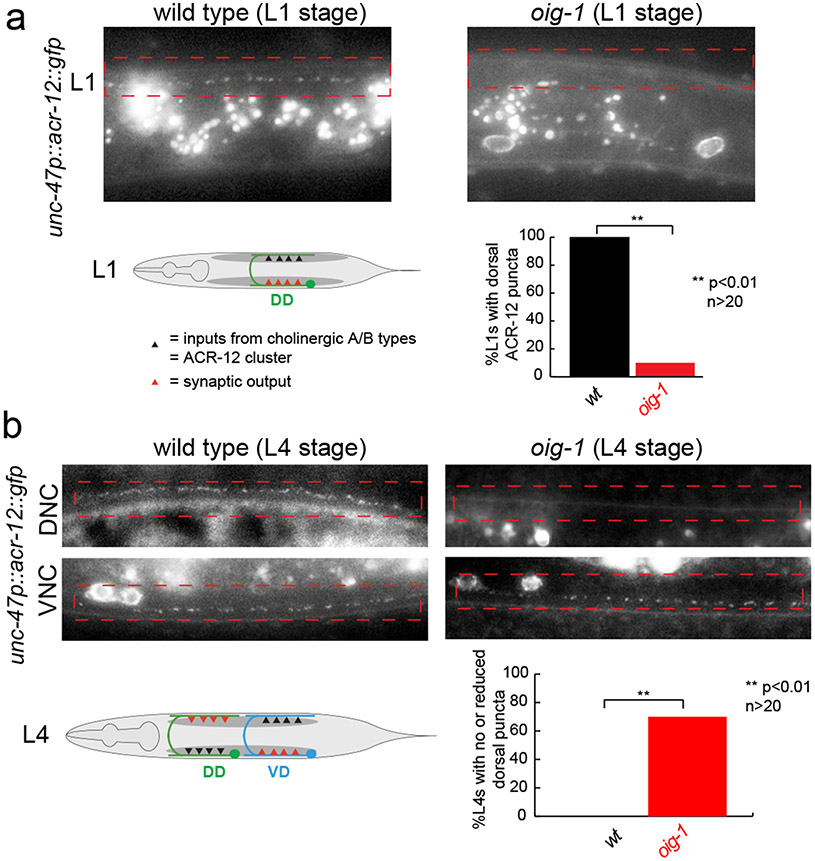
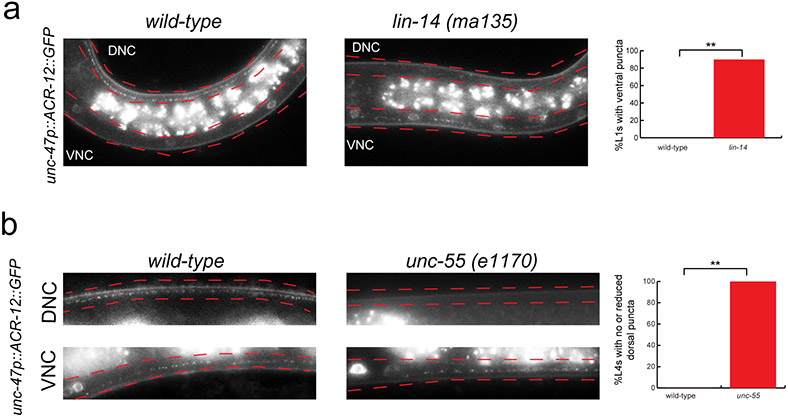
We next examined whether oig-1 function is restricted to controlling the synaptic output of D-type neurons or whether oig-1 may also affect localization of their synaptic input. Innervation of the DD and VD neurons from the cholinergic A- and B-type neurons can be visualized with GFP-tagged ACR-12 protein 16 which localizes to puncta in the DD neurons in the dorsal nerve cord in the L1 stage, indicative of the cholinergic input from the DA/DB MNs (Fig.4a). Strikingly, in oig-1 mutants, these dorsal puncta are not observed (Fig.4a). In post L1-stage wild-type animals, ACR-12 protein normally labels synapses from DA/DB to the VD neurons in the dorsal cord and synapses from VA/VB to the DD neurons in the ventral cord (Fig.4b). In oig-1 mutants, the dorsal, ACR-12(+) synaptic inputs in the VD neurons also do not form properly (Fig.4b). The coincidence of synaptic input and synaptic output defects in oig-1 mutants indicates that the localization of synaptic in- and outputs are coordinated and that this coordination requires the OIG-1 protein. As expected, the oig-1 defects in synaptic innervation, as determined by ACR-12 clustering, are mirrored by loss of the temporal (lin-14) and spatial (unc-55) specificity regulators of oig-1 expression (Extended Data Fig. 6a,b).
Figure 4: OIG-1 also controls cholinergic innervation into the DD and VD neurons.
Cholinergic innervation to the D-type MNs is visualized with an unc-47p::ACR-12::GFP reporter transgene, maintained in an acr-12(ok367) mutant background 16, in L1 stage animals (panel a) and L4 stage animals (panel b).
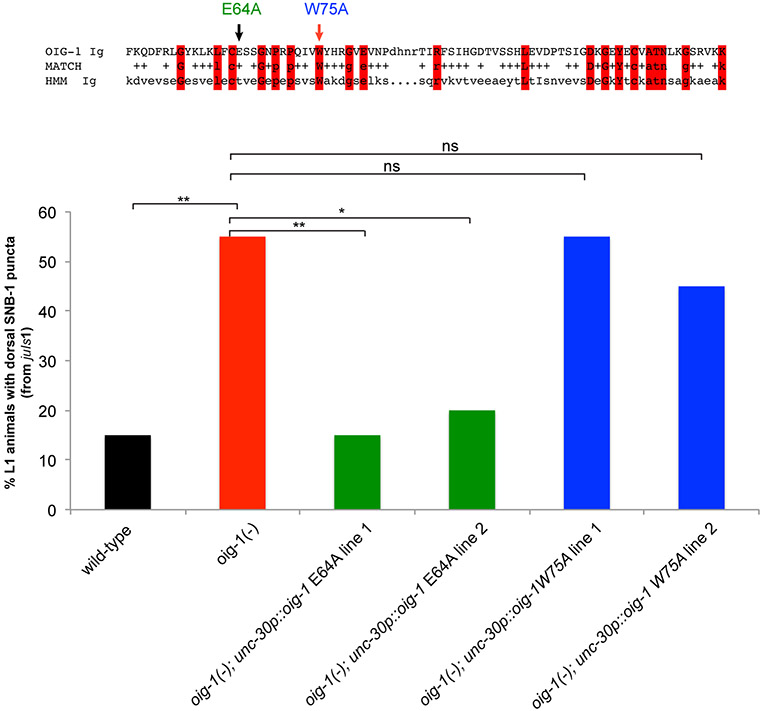
The synaptic defects (as well as the locomotory defects) of oig-1 mutants can be rescued by expressing oig-1 specifically in the D-type MN under control of the unc-30 promoter whereas expression under control of a cholinergic A- and B-type MN promoter does not rescue (Fig.3a,c Extended Data Fig. 5a,c). Since OIG-1 is predicted to encode a secreted protein, the lack of rescue of oig-1 with a cholinergic promoter suggests that OIG-1 protein functions cell-autonomously in/on the GABAergic DD and VD classes of MNs and argues against a long-range, diffusible function of OIG-1. Consistent with this autonomy, oig-1 mutants show no defects in the localization of synapses of the adjacent cholinergic MNs (data not shown). The rescuing activity of oig-1 critically depends on the integrity of the IgC2 domain (Extended Data Fig. 7). Forced expression of oig-1 in the D-type MNs under control of two promoters that are not downregulated in the D-type motor neurons (unc-25 and unc-30 promoters) is not sufficient to prevent the formation of dorsal synaptic outputs of the DD neurons (data not shown), indicating that oig-1 collaborates with other factors to regulate synapse formation.
The synaptic wiring defects in oig-1 mutants suggested the OIG-1 protein might be localized in a spatially restricted manner in the DD and VD MNs. A fosmid-based reporter in which OIG-1 protein is fused to GFP (and which rescues oig-1 locomotory defects and the aldicarb hypersensitivity; Fig.2a; Extended Data Fig. 5a,b) shows punctate localization along the processes of the D-type MNs (Fig.5a). The punctate pattern of OIG-1 in D-type MNs revealed a surprising localization pattern along the D-type processes. At the L1 stage, OIG-1 is not localized along the dorsal processes (in which the synaptic wiring defects are observed), but is localized along the ventral cord. After the generation of the VD MNs (and extinction of OIG-1 expression in the DD MNs, as described above), OIG-1 protein also localized in the VD neurons along the ventral cord. Co-labeling with the presynaptic RIM protein UNC-10 and the postsynaptic GABA receptors UNC-49 demonstrates that these puncta correspond to the perisynaptic region of synapses that D-type MNs form onto ventral muscle (Fig.5b). When ectopically expressed in excitatory cholinergic VNC or head MNs, OIG-1::GFP is also targeted to presynaptic specializations (Fig.5c), demonstrating that OIG-1 localization is not dependent on GABAergic specific synaptic features, but rather contains presynaptic targeting properties that are independent of the type of synapse. Taken together, the perisynaptic localization pattern of OIG-1 indicates that a highly localized, synaptic organizer protein is capable of orchestrating the wiring properties of an entire neuron, by promoting synaptic input and preventing ectopic synaptic output in a distal portion of the neuronal process (Fig.5b). Punctate OIG-1 protein is also observed in a few head neurons (Extended Data Fig. 8).
Figure 5: OIG-1 localization and summary.
a: OIG-1 localization as assessed by with an oig-1 translational reporter fusion protein shown in Fig.2a. cc = coelomocytes, which take up secreted proteins, including synaptically localized proteins 25.
b: OIG-1::GFP localizes perisynaptically as indicated by co-staining with UNC-10 (left panels) and UNC-49 (right panels) antibodies.
c: OIG-1 is targeted to presynaptic specializations when ectopically expressed in cholinergic neurons.
d: Summary.
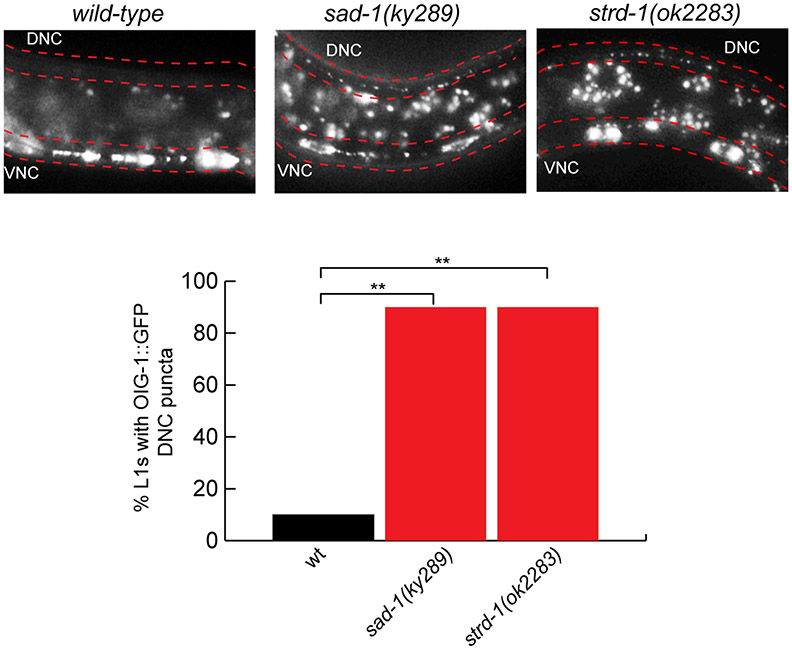
The mutant phenotype of oig-1, specifically the aberrant formation of synaptic output from dorsal DD and VD axons, resembles the mutant phenotype observed upon removal of the SAD-1 kinase 17-19. In animals lacking sad-1 or lacking strd-1/STRADα, a pseudokinase required for SAD-1 localization 20, OIG-1 clusters ectopically along the dorsal nerve cord (Extended Data Fig. 9). Conversely, loss of oig-1 does not affect localization of SAD-1 (data not shown). What sets OIG-1 apart from these molecules is that in contrast to the panneuronally expressed SAD-1 and STRADα , OIG-1 appears to operate as a spatiotemporally controlled nexus of this pathway that determines the spatiotemporal specificity of SAD-1 protein function in the context of the D-type MNs. The ability of ventrally and presynaptically localized OIG-1 to organize distally located synaptic inputs and outputs on the dorsal neurite, suggests that OIG-1 may trigger a cascade of downstream signaling events or anchor factors on the ventral side which would otherwise contribute to synapse organization on the dorsal side.
In conclusion, we have shown here that three different transcription factors cooperate in an intersectional manner in defined spatial and temporal contexts to control the expression of a perisynaptically-localized organizer molecule, OIG-1, which orchestrates the localization of synaptic outputs and inputs of two different neuron types (Fig.5d). unc-30 needs to cooperate with other transcription factors and these collaborators confer spatiotemporal specificity. In embryonic and L1 stages, spatially but not temporally restricted unc-30 cooperates with temporally, but not spatially controlled lin-14 to prevent DD MN synapse assembly at the inappropriate location via induction of oig-1 expression. After the L1 stage, the DD/VD-expressed unc-30 collaborates with the subtype (VD)-specific unc-55 transcription factor to restrict oig-1 expression to the VD neurons where it organizes synaptic in- and outputs (Fig.5d).
Our findings demonstrate that the localization of synaptic in- and outputs of a neuron are coordinated and that this coordination is apparently achieved, at least in part, by the OIG-1 organizer protein. It will be interesting to examine whether similar synaptic organizer functions can be ascribed to any of the multiple small, 1-Ig domain proteins, some secreted, some transmembrane, encoded in the C. elegans genome 21, but also in vertebrate genomes 22.
METHODS
C. elegans strains
Worms were grown at 20°C on nematode growth media (NGM) plates seeded with bacteria (E.coli OP50) as a food source. L1 animals were obtained by hypochlorite-treating gravid adult animals and letting embryos hatch and arrest in M9 for 16-18 hours.
Mutant alleles used in this study: LGI: unc-55(e1170) , LGII: lin-4(e912), LGIII: oig-1(ok1687), strd-1(ok2283), LGIV: unc-30(191), LGX: lin-14(ma135), alr-1(oy42), sad-1(ky289), acr-12(ok367)
Transgenes
otEx5663 (unc-302.4kbprom::GFP::RAB-3::unc-103’UTR) used in Fig.1a, Extended Data Fig. 2, otEx4816 (oig-1fosmid::sl2::gfp), otIs450 (oig-1fosmid::sl2::gfp) used in Fig.2., otEx5651 (unc-302.4kbprom::gfp) used in Extended Data Fig. 3a, wgIs395 (unc-30fosmid::TY1::EGFP::3xFLAG) used in Extended Data Fig. 3a, otEx5765 (lin-14fosmid::gfp) used in Extended Data Fig. 3b, juIs1 (unc-25p::snb-1::GFP)4 used in Fig.3a,c, Extended Data Fig. 5c,7, otEx4955 (unc-302.4kbprom::oig-1 line 1) used in Fig.3a,c, Extended Data Fig. 5a,c otEx4956 (unc-302.4kbprom::oig-1 line 2) used in Fig3a,c otEx4941 (unc-3prom::oig-1 line 1) used in Fig.3a,c Extended Data Fig. 5c, otEx4942 (unc-3prom::oig-1 line 2) used in Fig. 3a,c hpIs3 (unc-25p::syd-2::GFP)26 used in Fig.3c, ufIs92 (unc-47prom::ACR-12::GFP)16 used in Fig.4, Extended Data Fig. 6, otEx5664 (oig-1fosmid::gfp) used in Fig.5a,b, Extended Data Fig. 5a,b,8,9, otEx5858 (unc-3558bpprom::OIG-1::GFP) used in Fig.5c, otEx5859 (del-1488bpprom::OIG-1::GFP) used in Fig.5c, otEx6212 (unc-302.4kbprom::oig-1E64A line 1) used in Extended Data Fig. 7, otEx6213 (unc-302.4kbprom::oig-1E64A line 2) used in Extended Data Fig. 7, otEx6214 (unc-302.4kbprom::oig-1W75A line 1) used in Extended Data Fig. 7, otEx6215 (unc-302.4kbprom::oig-1W75A line 2) used in Extended Data Fig. 7.
oig-1 promoter bashing constructs (all used in Extended Data Fig. 4): otEx5993 (oig-1prom1::NLS::gfp line 1), otEx5994 (oig-1prom1::NLS::gfp line 2), otEx5995 (oig-1prom1::NLS::gfp line 3), otEx5996 (oig-1prom2::NLS::gfp line 1), otEx5997 (oig-1prom2::NLS::gfp line 2), otEx5998 (oig-1prom2::NLS::gfp line 3), otEx6003 (oig-1prom3:NLS::gfp line 1), otEx6004 (oig-1prom3::NLS::gfp line 2), otEx6005 (oig-1prom3::NLS::gfp line 3), otEx6006 (oig-1prom4::NLS::gfp line 1), otEx6007 (oig-1prom4::NLS::gfp line 2), otEx6008 (oig-1prom4::NLS::gfp line 3), otEx6009 (oig-1prom5::NLS::gfp line 1), otEx6010 (oig-1prom5::NLS::gfp line 2), otEx6011 (oig-1prom5::NLS::gfp line 3), otEx6034 (oig-1prom6::NLS::gfp line 1), otEx6035 (oig-1prom6::NLS::gfp line 2), otEx6036 (oig-1prom6::NLS::gfp line 3), otEx6037 (oig-1prom7::NLS::gfp line 1), otEx6038 (oig-1prom7::NLS::gfp line 2), otEx6039 (oig-1prom7::NLS::gfp line 3), otEx6060 (oig-1prom8::NLS::gfp line 1), otEx6061 (oig-1prom8::NLS::gfp line 2), otEx6121 (oig-1prom9::NLS::gfp line 1), otEx6122 (oig-1prom9::NLS::gfp line2), otEx6079 (oig-1prom10::NLS::gfp line 1), otEx6080 (oig-1prom10::NLS::gfp line 2), otEx6081 (oig-1prom10::NLS::gfp line 3)
Generation of oig-1 transgenes
The oig-1fosmid::sl2::gfp reporter (shown in Fig.2) was generated by fosmid recombineering using the fosmid WRM0614cC07 and a SL2-based, nuclear-localized gfp cassette, pBALU9 27. The inclusion of the SL2 sequence results in the production of nuclear localized GFP protein. The reporter was injected at 10ng/μl with rol-6(su1006) at 2ng/μl and sonicated OP50 genomic DNA at 120ng/μl. An extrachromosomal array (otEx4816) was integrated to yield otIs450 IV.
The oig-1 translational fosmid gfp reporter, oig-1fosmid::gfp (shown in Fig.5) was generated by fosmid recombineering using the fosmid WRM0614cC07 and a gfp cassette pBALU25, modified from pBALU1 27. pBALU25 was created by mutating the coding sequence of gfp in pBALU1 to contain the amino acid changes F64L and S65T. This cassette was recombineered and inserted into the oig-1 fosmid immediately following the predicted signal peptide sequence (after the 72nd base pair of oig-1). This translational reporter was injected at 10ng/μl with ttx-3prom::mCherry at 3ng/μl and sonicated OP50 genomic DNA at 120ng/μl.
The unc-3558bpprom::OIG-1::GFP construct was generated by TOPO cloning a PCR fusion of 558bp upstream of the unc-3 translational start site with a fragment of the oig-1fosmid::gfp construct containing from the oig-1 translational start site to 500bp downstream of the stop codon. This construct was PCR-amplified from the start of the unc-3 promoter to 500bp downstream of the oig-1 locus and injected at 10ng/μl with ttx-3prom::mCherry at 3ng/μl and sonicated OP50 genomic DNA at 120ng/μl.
The del-1488bpprom::OIG-1::GFP construct was generated by TOPO cloning as the unc-3558bpprom::OIG-1::GFP with 488bp upstream of the del-1 translational start site. This construct was PCR-amplified from the start of the del-1 promoter to 500bp downstream of the oig-1 locus and injected at 10ng/μl with ttx-3prom::mCherry at 3ng/μl and sonicated OP50 genomic DNA at 120ng/μl.
The unc-30p::oig-1 construct was generated by cloning the 2.4kb unc-30 promoter into the EcoRV site of the first MCS of pPD49.26 and the oig-1 locus from the start to stop codon (1521bp) into the BamHI site of the second MCS. This construct was digested with PvuI and injected at 5ng/μl with myo-2p::gfp at 3ng/μl and OP50 genomic DNA at 120ng/μl. Site directed mutagenesis of this construct was used to generate unc-30p::oig-1E64A and unc-30p::oig-1W75A. These constructs were injected as described above.
The unc-3p::oig-1 construct was generated by cloning the 558bp unc-3 promoter into the EcoRV site of the first MCS of pPD49.26 and the oig-1 locus from the start to stop codon (1521bp) into the BamHI site of the second MCS. This construct was digested with PvuI and injected at 10ng/μl with myo-2p::gfp at 3ng/μl and OP50 genomic DNA at 120ng/μl.
The oig-1 promoter deletion constructs were generated by cloning the various promoter fragments into the HindIII and BamHI sties of the MSC of a 2XNLSGFP plasmid. Promoter constructs with potential UNC-30 binding sites deleted were generated using site-directed mutagenesis. These constructs were injected at 50ng/μl with rol-6 at 30ng/μl and rol-6(su1006) at 20ng/μl.
Wormtracker assays
Tracking assays were performed as previously described 28. Briefly, L4 animals were placed on an NGM plate seeded with 20 μl of OP50 bacteria in the center. Automated tracking was performed with the Worm Tracker 2.0 (WT2) which uses a camera to track and record individual worms. Twenty worms of each genotype were tracked for 5 minutes each at 20°C. Analysis was performed as previously described 28.
Aldicarb assays
Aldicarb assays were performed as previously described (Mahoney et al., 2006). Briefly, ~20 young adult animals (24hrs after L4 stage, blinded for genotype) were picked to freshly seeded NGM plates containing1mM aldicarb (ChemService). Worms were assayed for paralysis every 15 minutes by prodding with a platinum wire. A worm was considered paralyzed if it did not respond to prodding to the head and tail three times each at a given time point. Strains were grown and assayed at 20°C.
Antibody staining
Antibody staining was performed at previously described 29. Briefly, following a freeze-crack procedure, worms were fixed by a treatment in ice-cold acetone for 5 minutes and then ice-cold methanol for 5 minutes. Worms were collected in 1X PBS and centrifuged briefly. The PBS was removed and worms were incubated in a blocking solution (1XPBS, 0.2% gelatin, 0.25% Triton X-100) for 30 minutes at 20°C. After the blocking solution was removed, worms were incubated with primary antibodies diluted in PGT (1X PBS, 0.1% gelatin, 0.25% Triton X-100) overnight at 4°C. The anti-UNC-49 antibody (Gally and Bessereau 2003) was used at a 1:500 dilution. The anti-UNC-17 antibody 30 was used at a 1:3000 dilution. The anti-GFP antibody (Life Technologies A10262) was used at 1:1000. The anti-RIM2 (used to recognize UNC-10) was used at a 1:10 dilution (Developmental Studies Hybridoma Bank – University of Iowa). Worms were washed 5 times in wash solution (1XPBS, 0.25% Triton X-100) for 20 minutes each wash. Worms were then incubated with secondary antibodies diluted 1:1000 in PGT for 3 hours at 20°C. Alexa Fluor 488 goat anti-chicken (Invitrogen A11039) was used to detect the anti-GFP antibody. Alexa Fluor 594 donkey anti-mouse (Invitrogen A-21203) was used to detect the anti-RIM2 antibody. Alexa Fluor 555 donkey anti-rabbit (Invitrogen A-31572) was used to detect the anti-UNC-49 antibody. Alexa Fluor 488 donkey anti-mouse (Invitrogen A-21202) was used to detect the anti-UNC-17 antibody. Worms were then washed 5 times for 20 minutes each wash. Following the final wash, worms were mounted in Fluorogel with Tris buffer (Electron Microscopy Sciences).
Statistical Analysis
For results shown in Fig.3a,b, Fig.4a,b, 6a,c, 7, 9, we performed Fisher’s exact test, **p<0.01, *p<0.05. For results shown in Fig.3c, Extended Data Fig. 2c, we performed a Student’s t-test (2 sided, type 2), **p<0.01, *p<0.05. For WormTracker analysis in Extended Data Fig. 3a, we used Wilcoxon rank sum to test the differences between oig-1, wild-type, and rescued strains, **q<0.01, *q<0.05.
Extended Data
Extended Data Figure 1: Electron microscopical analysis of unc-30(e191) mutants.
a: Reconstructions of a VD4 and a DD2 MN from an unc-30(e191) animal compared to the same neurons in a wild-type animal. Cell bodies (large black dots) are all situated in the ventral cord. Processes emanate anteriorly (up on plots) from the cell body and run along the ventral cord. Lateral branches leave the ventral cord (v) and run round to the dorsal cord (d) as a circumferential commissure (broken horizontal process in the plots). Commissures from unc-30 mutant type-D neurons are situated in the same regions as those of their wild-type counterparts. However the cell bodies of DD neurons are often displaced anteriorly in the mutants, with the consequence that DD neurons have shorter processes in the ventral cord (c). Processes in the dorsal cord run anteriorly in mutant animals, whereas they branch with the main branch running posteriorly in wild-type animals. Neuromuscular junctions (NMJs) in unc-30 mutants are made predominantly in the dorsal cord by both the DDs and VDs, whereas in the wild type, only DD neurons innervate dorsal muscles. The synaptic inputs to the DD and VD neurons in mutant animals (inward pointing arrows for chemical synapses and "T" for gap junctions) are generally abnormal. The reconstructed DD2 neuron received synapses from several unidentified processes on the ventral side (depicted with a "?"). These processes do not belong to VA or VB neurons, the normal pre-synaptic partners of DD, as all the local VA and VB neurons were identified. From the location and synaptic behavior of these processes, it is probable that they belong to interneurons which span the length of the cord and do not usually innervate D-type neurons. An Asterisk ("*") indicates that a synapse has multiple post-synaptic elements. A total of six VDs and three DDs were reconstructed. Each reconstruction covered around 2000 EM sections corresponding to a length of 100μ along the body of the animal. The unc-30(e191) mutation does not affect MN cell body position or the synaptic behaviour of the DA/DB neurons, except in regard to their synapses to D-type neurones; this made it possible to unambiguously identify D type neurones from their positions and by eliminating other identified classes of MNs. Electron microscopy and reconstructions of micrographs of serial sections were performed as described in 31.
b: Electron micrographs. The processes of DD and VD neurons normally run subjacent to the bounding basal lamina of the ventral cord immediately dorsal to the axons of the VA and VB neurons (a). NMJs are made through the basal lamina onto muscle arms (m). In unc-30(e191) animals, the axons of the DD and VD neurons wander round the cord and do not run in defined locations; the configuration shown in (b) is typical but not stereotyped. Very few NMJs are made in the ventral cord by the DD or VD neurons in unc-30 mutants; those that are made look rather small (c). Atypical synapses (d) are often made onto DD or VD processes from neurons such as the touch receptor neuron AVM (6). It is probable that these synapses are not normally found as the processes of AVM and DD or VD do not normally run alongside each other in wild-type animals. Both the DD and VD MNs make NMJs to dorsal muscles in the dorsal cord of unc-30 mutants, whereas in wild-type animals, only DD neurons do so and VD neurons innervate ventral muscles in the ventral cord. (e) & (f) show NMJs made by VD (e) and DD (f) neurons in the dorsal cord of an unc-30(e191) mutant. Scale bars 1μ.
Extended Data Figure 2: RAB-3 is ectopically localized in lin-14, unc-55, and oig-1 mutants.
a: Presynaptic marker RAB-3 ectopically localizes to the dorsal nerve cord (DNC; marked in red) in lin-14 mutant animals. RAB-3::GFP puncta (from otEx5663, unc-30p::GFP::RAB-3) localize mostly to the ventral nerve cord (VNC; marked in red) in wild-type L1 animals (left). Ectopic RAB-3::GFP puncta localize mostly to the dorsal nerve cord in 95% of lin-14 L1 animals (right, scored in progeny from lin-14 null animals carrying a lin-14 rescue array 23. Ventral and dorsal nerve cords are indicated by red dotted lines. L1 animals were obtained by hypochlorite-treating gravid adult animals and letting embryos hatch and arrest in M9 for 16-18 hours. n>20 for each strain scored.
b: RAB-3 ectopically localizes to the dorsal nerve cord in unc-55 L4 mutant animals. RAB-3::GFP puncta localizes to both the ventral (VNC) and dorsal (DNC) nerve cord in 100% of wild-type L4 animals (left). RAB-3::GFP puncta localize mostly to the dorsal nerve cord in 100% of unc-55 L4 animals (right). Ventral and dorsal nerve cords are indicated by red dotted lines. Signals between the nerve cords are autofluorescence from the gut. n>20 for each strain scored.
c: RAB-3 ectopically localizes to the dosal nerve cord in oig-1 mutants. RAB-3 normally localizes to the ventral nerve cord (VNC, marked in red) in wild-type L1 animals (top left). Ectopic RAB-3::GFP puncta localize to the dorsal nerve cord in 55% of oig-1 L1 animals (top right, compared to 20% of wild-type animals). L1 animals were obtained by hypochlorite-treating gravid adult animals and letting embryos hatch and arrest in M9 for 16-18 hours. n>20 for each strain scored. In wild-type L4 animals, more RAB-3::GFP puncta are localized in the VNC than in the DNC of the animal (bottom, black dots). Conversely, in oig-1 mutants, more RAB-3::GFP puncta are localized in the DNC than in the VNC (bottom, red dots). **p<0.01, n=20 for each strain.
Extended Data Figure 3: Independence of transcription factor activities.
a: Expression of unc-30 is not affected by loss of lin-14 or unc-55. A 2.4kb unc-30 promoter gfp fusion reporter is expressed in the DD MNs (green circles) in wild-type L1 animals; this expression is not affected in lin-14(−) mutant animals (scored in progeny from lin-14 null mothers carrying a lin-14 rescue array 23). An unc-30 fosmid-based reporter, kindly provided by the Transgeneome project, is expressed in the DD (green circles) and VD (blue squares) MNs (DD4 to VD10 shown) in wild-type L4 animals; this expression is not affected in unc-55(e1170) L4 animals. n>20 for each genotype.
b: Expression of a lin-14 fosmid-based reporter construct 32 is unaffected by loss of unc-30. lin-14 is expressed in the DA, DB, and DD MNs in the VNC at the L1 stage (Average number of VNC cells=15); this expression is not affected in unc-30(e191) mutant L1s (Average number of VNC cells=15);. n>20 for each genotype. Loss of unc-30 also does not affect unc-55 expression, as shown by 11.
Extended Data Figure 4: Deletion of a putative UNC-30 binding site results in loss of oig-1 expression in the D-type neurons.
Regions of the oig-1 promoter were fused to gfp to analyze expression. (+) indicates robust expression of the reporter construct in the specified cell type, whereas (−) indicates loss of expression in the specified cell type. Twenty worms at both the L1 and L4 stage were scored for each line. Expression of a 1kb promoter reporter (prom 1) recapitulates expression of the oig-1fosmid::gfp reporter in the D-type MNs (see Fig.2). This region contained 3 elements that exactly match the UNC-30 consensus binding site (TAATC, purple box, 13) and multiple others that are a partial match to the UNC-30 binding site (magenta and blue boxes). Further deletion of this prom 1 defined a minimal 125bp element that is sufficient to drive oig-1 expression in the D-type MNs (prom 6). This element contains two sites that partially match the UNC-30 binding sequence. Deletion of the AAATC site in the context of the 1kb promoter (prom 9) has no effect on oig-1 expression in the D-type MNs. Deletion of the TAAAC site in the context of the 1kb promoter reporter (prom 10) results in complete loss of oig-1 expression specifically in the D-type MNs.
Extended Data Figure 5. oig-1 mutants defects and their rescue.
a: oig-1 mutants display locomotory defects. Locomotion of L4 animals was analyzed with tracking assays (Yemini et al 2013). The graphs on the left side of each panel correspond to assays comparing wild-type, oig-1 mutant, and oig-1; unc-30p::oig-1 animals. The graphs on the right side of each panel correspond to assays comparing wild-type, oig-1 mutant, and oig-1; oig-1fosmid::gfp animals. Twenty animals (each dot on a plot) were tracked for each genotype for both comparisons. Mean and Q values are indicated. Note that in a previously published analysis of a large panel of available mutants, the same set of locomotory defects that we describe here for oig-1 mutants were found to be affected in unc-55 mutants 28, albeit in a stronger manner than oig-1 mutants. Also note that the very strong locomotory defects unc-30 defects are qualitatively very different from oig-1 mutants, but this is to be expected since unc-30 mutant do not only show the synaptic defects that we describe here, but also lack the neurotransmitter GABA 3, thereby disabling any neuromuscular signaling.
i: The midbody speed of oig-1 mutant animals is significantly lower than that of wild-type animals. This defect is partially rescued (statistically different from oig-1 mutants but also from wild-type animals) by expressing unc-30p::oig-1 in oig-1 animals (left graph). The lower midbody speed of oig-1 mutants is completely rescued (statistically different from oig-1 mutants but not from wild-type animals) by expressing the oig-1fosmid::gfp in oig-1 mutants.
ii: oig-1 mutants exhibit more dwelling than wild-type L4 animals. This defect is partially rescued (statistically different from oig-1 mutants but also from wild-type animals) by expressing unc-30p::oig-1 in oig-1 animals (left graph). The increased dwelling of oig-1 mutants is completely rescued (statistically different from oig-1 mutants but from not wild-type animals) by expressing the oig-1fosmid::gfp in oig-1 mutants.
iii: oig-1 mutants exhibit an increased path curvature compared to wild-type animals. This defect is not rescued by expressing unc-30p::oig-1 in oig-1 animals (left graph). The increased path curvature of oig-1 mutants is completely rescued (statistically different from oig-1 mutants but not from wild-type animals) by expressing the oig-1fosmid::gfp in oig-1 mutants.
b: Aldicarb-sensitivity defects in oig-1 mutants. oig-1 mutant young adult animals (red squares), which display aberrant GABAergic synapses in both the ventral and dorsal cord, show hypersensitivity to aldicarb-induced paralysis compared to wild-type (black triangles). Expression of oig-1fosmid::gfp from a multicopy transgenic array (green circles) does not only rescue the oig-1 mutant phenotype, but even results in a slight hyposensitivity to aldicarb. Worms were tested every 15 minutes for paralysis by touching the head and tail three times each. n=20 for each strain, repeated 3 times.
c: Expression of oig-1 in the D-type neurons rescues ectopic DD synapses in the dorsal nerve cord. At the L1 stage when only the DD MNs are present, SNB-1::GFP (from juIs1-unc-25p::SNB-1::GFP) localizes to the ventral nerve cord (VNC) in wild-type animals (top left). Ectopic SNB-1::GFP puncta localize to the dorsal nerve cord (DNC) of oig-1 mutant L1s (top right). This phenotype is rescued by expressing oig-1 in the D-type MNs (unc-30p::oig-1, otEx4955, bottom left), but not by expressing oig-1 in the neighboring cholinergic MNs (unc-3p::oig-1, otEx4942, bottom right). White boxes indicate the dorsal nerve cord.
Extended Data Figure 6: ACR-12 is mislocalized in lin-14 and unc-55 mutants.
Cholinergic innervation to the D-type MNs is visualized with an unc-47p::ACR-12::GFP reporter transgene, maintained in an acr-12(ok367) mutant background. 16.
a: ACR-12 puncta localization is affected by loss of lin-14. In wild-type L1 animals, ACR-12 puncta are observed only in the DD neurons in the dorsal (DNC) nerve cord (left). In lin-14 mutant L1 animals (scored in progeny from lin-14 null animals carrying a lin-14 rescue array 23), ACR-12 puncta are detected in the ventral nerve cord (VNC) of the DD MNs. Quantification of this data is represented in the graph. Some dorsal puncta in the DD MNs were still observed in 83% of the lin-14 mutant L1s that had puncta in the ventral nerve cord. L1 animals were obtained by hypochlorite-treating gravid adult animals and letting embryos hatch and arrest in M9 for 16-18 hours. n > 20 for each strain scored, **p < 0.01.
b: ACR-12 puncta localization is affected by loss of unc-55. In wild-type L4 animals, ACR-12 puncta are observed in both the ventral (VNC) and dorsal (DNC) nerve cords (left). In unc-55 mutant L4 animals, ACR-12 puncta are observed mostly in the ventral nerve cord of unc-55 mutants. Ventral and dorsal nerve cords are marked by red dotted lines. Quantification of this data is represented in the graph. n>20 for each strain, **p < 0.01.
Extended Data Figure 7: The IgC2 domain is necessary for OIG-1 function.
At the L1 stage when only the DD MNs are present, ectopic SNB-1::GFP puncta (from juIs1-unc-25p::SNB-1::GFP) localize to the dorsal nerve cord (DNC) of oig-1 mutant L1s (red bar) but not in wild-type animal (black bar) (see Fig. 3A). Based on an alignment with the hidden Markov model (HMM) Ig domain (top), a highly conserved residue (W75) and a nonconserved residue (E64) in the OIG-1 Ig domain were mutated in the context of an unc-30p::oig-1 transgene that is able to rescue the L1 ectopic synapse defects (see Fig. 3A). The unc-30p::oig-1E64A transgenes (otEx6212, otEx6213) were still able to rescue the synaptic defects of oig-1 L1 animals (green bars), whereas the unc-30p::oig-1W75A transgenes (otEx6214,otEx6215) had no rescue ability (blue bars). L1 animals were obtained by hypochlorite-treating gravid adult animals and letting embryos hatch and arrest in M9 for 16-18 hours. n>20 for each strain scored, **p < 0.01, *p < 0.05.
Extended Data Figure 8: OIG-1 localization in other neuron types.
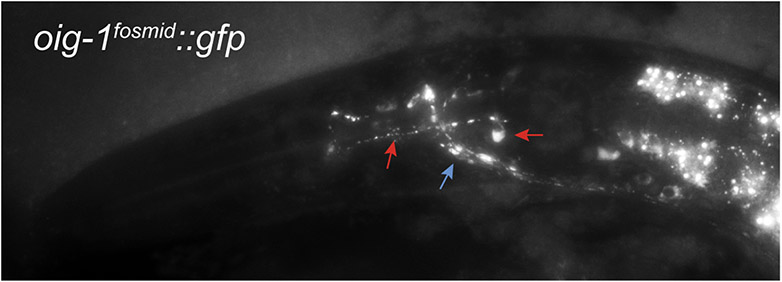
OIG-1::GFP (from oig-1fosmid::gfp) localizes in a punctate manner along axons in the nerve ring (blue arrow) and along a pair of neurons in the pharynx, tentatively identified as the M2 MNs (red arrows point to cell body and process). These neurons form synapses onto pharyngeal muscles along their processes, and these processes also show punctate localization of OIG-1.
Extended Data Figure 9: OIG-1 is mislocalized in sad-1 and strd-1 mutants.
OIG-1::GFP (from oig-1fosmid::gfp) localizes to the ventral nerve cord (VNC) of wild-type L1 animals (left). In sad-1 mutants (middle), OIG-1::GFP is ectopically localized to the dorsal side (DNC) of L1 animals. In strd-1 mutants, OIG-1::GFP is ectopically localized to the dorsal side of L1 animals. Quantification of the data is shown in graph. n > 20 for each strain scored. **p<.01.
ACKNOWLEDGEMENTS
We thank the Transgeneome project, the CGC, J.L. Bessereau, M. Gendrel, S.Y. Kerk for reagents, Q. Chen for microinjection, E. Southgate and N. Thomson for EM analsys, S. Brenner for first noting the neural defects of unc-30(e191), E. Yemini for assistance with worm tracking, and D.Miller, S.He, I. Greenwald and members of the Hobert lab for comments on this manuscript. This work was funded by the National Institutes of Health [R01NS039996-05 and R01NS050266-03], the Howard Hughes Medical Institute and the UK Medical Research Council.
Footnotes
DECLARATION
The authors declare no competing interests.
REFERENCES
- 1.White JG, Albertson DG & Anness MA Connectivity changes in a class of motoneurone during the development of a nematode. Nature 271, 764–6 (1978). [DOI] [PubMed] [Google Scholar]
- 2.Eastman C, Horvitz HR & Jin Y Coordinated transcriptional regulation of the unc-25 glutamic acid decarboxylase and the unc-47 GABA vesicular transporter by the Caenorhabditis elegans UNC-30 homeodomain protein. J Neurosci 19, 6225–34 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin Y, Hoskins R & Horvitz HR Control of type-D GABAergic neuron differentiation by C. elegans UNC-30 homeodomain protein. Nature 372, 780–3 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Hallam SJ & Jin Y lin-14 regulates the timing of synaptic remodelling in Caenorhabditis elegans. Nature 395, 78–82 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Ruvkun G & Giusto J The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature 338, 313–9 (1989). [DOI] [PubMed] [Google Scholar]
- 6.Walthall WW & Plunkett JA Genetic transformation of the synaptic pattern of a motoneuron class in Caenorhabditis elegans. J Neurosci 15, 1035–43 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedgecock EM, Culotti JG, Hall DH & Stern BD Genetics of cell and axon migrations in Caenorhabditis elegans. Development 100, 365–82 (1987). [DOI] [PubMed] [Google Scholar]
- 8.Aurelio O, Hall DH & Hobert O Immunoglobulin-domain proteins required for maintenance of ventral nerve cord organization. Science 295, 686–90 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Wightman B, Ha I & Ruvkun G Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–62 (1993). [DOI] [PubMed] [Google Scholar]
- 10.Zhou HM & Walthall WW UNC-55, an Orphan Nuclear Hormone Receptor, Orchestrates Synaptic Specificity among Two Classes of Motor Neurons in Caenorhabditis elegans. J Neurosci 18, 10438–10444 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shan G, Kim K, Li C & Walthall WW Convergent genetic programs regulate similarities and differences between related motor neuron classes in Caenorhabditis elegans. Dev Biol 280, 494–503 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Araya CL et al. Regulatory analysis of the C. elegans genome with spatiotemporal resolution. Nature 512, 400–5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cinar H, Keles S & Jin Y Expression profiling of GABAergic motor neurons in Caenorhabditis elegans. Curr Biol 15, 340–6 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Miller KG et al. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc Natl Acad Sci U S A 93, 12593–8 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bamber BA, Beg AA, Twyman RE & Jorgensen EM The Caenorhabditis elegans unc-49 locus encodes multiple subunits of a heteromultimeric GABA receptor. J Neurosci 19, 5348–59 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrash HA, Philbrook A, Haburcak M, Barbagallo B & Francis MM ACR-12 ionotropic acetylcholine receptor complexes regulate inhibitory motor neuron activity in Caenorhabditis elegans. J Neurosci 33, 5524–32 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crump JG, Zhen M, Jin Y & Bargmann CI The SAD-1 Kinase Regulates Presynaptic Vesicle Clustering and Axon Termination. Neuron 29, 115–129. (2001). [DOI] [PubMed] [Google Scholar]
- 18.Kim JS, Hung W & Zhen M The long and the short of SAD-1 kinase. Commun Integr Biol 3, 251–5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shelly M & Poo MM Role of LKB1-SAD/MARK pathway in neuronal polarization. Dev Neurobiol 71, 508–27 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Kim JS, Hung W, Narbonne P, Roy R & Zhen MC elegans STRADalpha and SAD cooperatively regulate neuronal polarity and synaptic organization. Development 137, 93–102 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Hobert O The neuronal genome of Caenorhabditis elegans. WormBook, 1–106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Letunic I, Doerks T & Bork P SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40, D302–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong Y, Lee RC & Ambros V Structure and function analysis of LIN-14, a temporal regulator of postembryonic developmental events in Caenorhabditis elegans. Mol Cell Biol 20, 2285–95 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melkman T & Sengupta P Regulation of chemosensory and GABAergic motor neuron development by the C. elegans Aristaless/Arx homolog alr-1. Development 132, 1935–49 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Rapti G, Richmond J & Bessereau JL A single immunoglobulin-domain protein required for clustering acetylcholine receptors in C. elegans. Embo J 30, 706–18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES FOR ONLINE MATERIAL
- 26.Yeh E, Kawano T, Weimer RM, Bessereau JL & Zhen M Identification of genes involved in synaptogenesis using a fluorescent active zone marker in Caenorhabditis elegans. J Neurosci 25, 3833–41 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tursun B, Cochella L, Carrera I & Hobert O A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS ONE 4, e4625 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yemini E, Jucikas T, Grundy LJ, Brown AE & Schafer WR A database of Caenorhabditis elegans behavioral phenotypes. Nat Methods 10, 877–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gendrel M, Rapti G, Richmond JE & Bessereau JL A secreted complement-control-related protein ensures acetylcholine receptor clustering. Nature 461, 992–6 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Duerr JS, Han HP, Fields SD & Rand JB Identification of major classes of cholinergic neurons in the nematode Caenorhabditis elegans. J Comp Neurol 506, 398–408 (2008). [DOI] [PubMed] [Google Scholar]
- 31.White JG, Southgate E, Thomson JN & Brenner S The structure of the nervous system of the nematode Caenorhabditis elegans. Philosophical Transactions of the Royal Society of London B. Biological Sciences 314, 1–340 (1986). [DOI] [PubMed] [Google Scholar]
- 32.Li J & Greenwald I LIN-14 inhibition of LIN-12 contributes to precision and timing of C. elegans vulval fate patterning. Curr Biol 20, 1875–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]