Abstract
Chronic kidney disease (CKD) is defined by a reduced estimated glomerular filtration rate (eGFR). This failure can be related to a phenotype of accelerated aging. In this work, we considered 76 patients with end-stage renal disease (ESRD) and 83 healthy controls. We concomitantly evaluated for the first time two measures that can be informative of the rate of aging, i.e., whole blood DNA methylation using the Illumina Infinium EPIC array and plasma levels of a selection of inflammatory/immunological proteins using multiplex immunoassays. First of all, we demonstrated accelerated aging in terms of the most common epigenetic age estimators in CKD patients. Moreover, we developed a new clock/predictor of age based on the inflammatory/immunological profile (ipAGE) and identified the inflammatory/immunological biomarkers differentially expressed between cases and controls. IpAGE appeared to be more sensitive than epigenetic clocks in quantifying the accelerated aging phenotype of ESRD patients. Interestingly, we did not find any correlation between the age acceleration evaluated according to the epigenetic clocks and ipAGE in either the ESRD group or the control group. On the whole, our data show a consistent accelerated aging phenotype in ESRD patients, which is better appreciated by quantifying the underlying inflammatory processes (inflammaging) by ipAGE than by using epigenetic clocks.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-022-00540-4.
Keywords: Chronic kidney disease, Accelerated aging, DNA methylation, Inflammation
Introduction
The nature of aging and its key mechanisms is still a matter of debate [1]. However, there is no doubt that many pathologies, both spontaneously occurring and genetically determined, are age-related, to name cancer, endocrine and cardiovascular diseases, and growth and functional disorders of bones and joints.
Aging and age-related diseases share some basic mechanistic pillars that largely converge on inflammation. This has been conceptualized as inflammaging, the long-term result of the chronic physiological stimulation of the innate immune system, which can become damaging during aging [2, 3]. Inflammaging poses a highly significant risk factor for both morbidity and mortality in elderly people, as most if not all age-related diseases share inflammatory pathogenesis [4, 5].
Recently, Alpert, Davis, Shen-Orr, and colleagues have demonstrated the comprehensive remodeling of the immune cell composition with age and developed an aggregated score IMM-AGE, associated with both chronological and DNA methylation age, as well as cardiovascular disease risk [6]. Similarly, Sayed et al. recently proposed an inflammatory aging clock (iAge), based on circulating inflammatory proteins encompassing cytokines, chemokines, and growth factors that have been shown to be informative of multimorbidity, immunosenescence, and cardiovascular aging [7].
The inflammatory clocks are part of a larger family of biomarkers of aging [8], in which epigenetic clocks have been raised to fame [9]. In general, epigenetic clocks have a predictive power of chronological age higher than other biomarkers, including immunological ones. Numerous studies showed that the aging process and the development of age-related syndromes and diseases and syndromes are accompanied by epigenetic changes in DNA [10–18] and may depend on external factors such as alcohol, smoking, microbiome profile, region of residence, and diet [19–21]. Each of these factors contributes differently to epigenetic changes and age acceleration.
The premature aging in chronic kidney disease (CKD) patients is often associated with cellular senescence that contributes to chronic inflammation [22, 23]. The prevalence of CKD increased dramatically with age [24–26] and progressing to end-stage renal disease (ESRD) is considered a major issue for public health [27, 28]. Between hemodialysis sessions, the organism gets into the state of permanent intoxication, because the patients’ kidneys have completely lost their detoxification function. Intoxication emerging from impaired renal function can activate the processes of chronic inflammation. Similarly, to the development of oxidative stress and atherosclerosis, the self-proteins get modified by toxic metabolic products, lose their functional role, and become considered by the innate immune system as antigens, further stimulating the processes of inflammaging.
So far, some studies have reported accelerated aging phenotypes in ESRD [29–32], while a recent study demonstrated the association between epigenetic clocks and kidney disease, but it did not include ESRD patients [33].
In the present study, we investigated for the first time epigenetic and immunological/inflammatory biomarkers of age in ESRD patients.
Methods
Study design and participants
This is a case–control study involving patients with ESRD on hemodialysis and a control group of volunteers. The study participants, men and women aged 24 to 89, were recruited in the Nizhny Novgorod region, Russia, in 2019–2021. Participants with ESRD were recruited from hemodialysis centers “FESPHARM NN,” Nizhny Novgorod, Russia. All measurements were performed at the Laboratory of System Medicine for Healthy Aging, Nizhny Novgorod State University.
Features of the study procedure, possible inconveniences, and risks were explained to all study participants, each of whom signed an informed consent. Each participant filled out a consent to the processing of personal data, taking into account the principle of confidentiality (accessibility only to the research group and presentation of data in a common array). The study was approved by the local ethics committee of the Nizhny Novgorod State University. All study procedures were in accordance with the 1964 Declaration of Helsinki and its later amendments.
The exclusion criteria for the control group were the presence of chronic diseases in the acute phase, oncological diseases, and acute respiratory viral infections at the time of biomaterial donation, and pregnancy for women. Out of the 420 participants recruited for this study, the two groups were selected to maximize the similarity in age and sex. Additionally, the groups were compared against such characteristics as smoking, insomnia, regular nutrition, alcohol consumption, marital, and current employment status (Supplementary Table 1), to reveal the statistically significant differences (chi-squared test with Yates’ correction) only for nutrition and employment status, which for the apparent reason correlate with the health status (Supplementary Fig. 1).
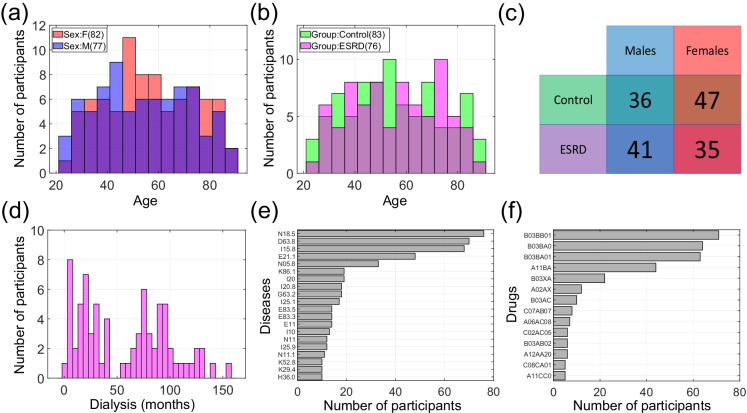
The selected dataset has an almost equal distribution by sex: 82 women and 77 men (Fig. 1a). The dataset includes 76 participants with 5-stage CKD (ESRD) and 83 controls (Fig. 1b). The age of participants ranges from 24 to 89 years. Figure 1c represents the ratio between controls and cases and males and females. Some of the participants in the dataset (mainly ESRD patients) have comorbidities. Figure 1e shows the most representative diseases in the group of participants with ESRD, coded according to the International Classification of Diseases (ICD-10) [34]. In addition to stage 5 chronic kidney disease (N18.5), which occurs in all patients in the respective group, the most common diseases are anemia (D63.8) in 69 participants, hypertension (I15.8) in 65 participants, and secondary hyperparathyroidism (E21.1) in 48 participants. Figure 1f shows the most representative drugs (codes in Anatomical Therapeutic Chemical Classification System [35]), which consume patients with ESRD. Most of them take folic acid (B03BB01) and cyanocobalamin (B03BA01). All biological data from the participants with the disease were taken within 30 min after the dialysis procedure. Supplementary Table 1 contains full information about the Russian population dataset (sheet “Main” provides information about age, sex, group, time on dialysis, sheet “Diseases” contains information about diseases, sheet “Drugs” contains information about drugs taken). We also obtained the inflammatory/immunological profiles for the additional 87 control participants (65 females and 22 males) to test ipAGE. Supplementary Table 2 contains the following information about these participants: age, ipAGE, ipAGE acceleration, 46 multiplex immunohistochemistry biomarkers.
Fig. 1.
Russian population dataset characteristics. a Distribution of men and women by age; b distribution of controls and participants with the ESRD by age; c Summary table by group and sex; d distribution participants with the disease by time on dialysis, measured in months; e the most representative diseases (ICD-10); f the most representative drugs (ATC classification system)
DNA methylation quantification and quality control
Phenol Chloroform DNA extraction was used. DNA was quantified using the DNA Quantitation Kit Qubit dsDNA BR Assay (Thermo Fisher Scientific) and 250 ng was bisulfite-treated using the EpiMark Bisulfite Conversion Kit (NEB) with case and control samples randomly distributed across arrays. The Illumina Infinium MethylationEPIC BeadChip [36] was used according to the manufacturer’s instructions. This platform allows measuring DNA methylation levels from a total number of 866,836 genomic sites, with single-nucleotide resolution. DNA methylation is expressed as β values, ranging from 0 for unmethylated to 1 representing complete methylation for each probe. Raw data were pre-processed as follows. First, probes with a detection p-value above 0.01 in at least 10% of samples were removed from the analysis. Second, probes with a beadcount less than three in at least 5% of samples were removed from the analysis. Third, all non-CpG probes were excluded from the results [37]. Fourth, SNP-related probes were removed from the analysis [38]. Fifth, multi-hit probes were removed [39]. Sixth, all probes located on chromosomes X and Y were filtered out. As a result, 733,923 probes remained for the analysis. All samples have less than 10% of probes with a detection p-value above 0.01. Functional normalization of raw methylation data was performed using the minfi [40] R package.
Multiplex Assay Kits
The analysis was performed on plasma using the K3-EDTA anticoagulant, without hemolysis and lipemia. Plasma from matched participants was thawed, spun (3000 rpm, 10 min) to remove debris, and 25 µl was collected in duplicate. Plasma with antibody-immobilized beads was incubated with agitation on a plate shaker overnight (16–18 h) at 2–8 °C. The Luminex® assay [41] was run according to the manufacturer’s instructions, using a custom human cytokine 46-plex panel (EMD Millipore Corporation, HCYTA-60 K-PX48). The panel included the following: sCD40L (CD40LG), EGF, Eotaxin (CCL11), FGF-2, FLT-3L, Fractalkine (CX3CL1), M-CSF (CSF1), GROα (CXCL1), IFNα2, IFNγ (IFNG), IL-1α, IL-1β, IL-1RA, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12 (p40), IL-12 (p70) (IL12Bp70), IL-13, IL-15, IL-17A, IL-17E/IL-25 (IL25), IL-17F, IL-18, IL-22, IL-27, IP-10 (CXCL10), MCP-1 (CCL2), MCP-3 (CCL7), M-CSF, MDC (CCL22), MIG (CXCL9), MIP-1α (CCL3), MIP-1β (CCL4), PDGF-AA, PDGF-AB/BB (PDGFB), TGFα, TNFα, TNFβ (LTA), VEGF-A. Assay plates were measured using a Magpix (Milliplex MAP). Data acquisition and analysis were done using a standard set of programs MAGPIX®. Data quality was examined based on the following criteria: The standard curve for each analyte has a 5P R2 value > 0.95. To pass assay technical quality control, the results for two controls in the kit needed to be within the 95% of CI (confidence interval) provided by the vendor for > 40 of the tested analytes. No further tests were done on samples with results out of range low (< OOR). Samples with results that were out of range high (> OOR) or greater than the standard curve maximum value (SC max) were not tested at higher dilutions without further request. Supplementary Table 3 contains multiplex immunohistochemistry biomarkers data.
Biochemical markers
Two types of anticoagulants (K3-EDTA and Li-heparin) were used. To identify possible reasons for the difference between biological and chronological ages, the biological age estimation using Levine’s model [42] was carried out in all the samples obtained. For this purpose, a CBC was performed using the Abacus Junior 30 semi-automatic analyzer and a set of biochemical indicators using the analyzer StatFax 3300 and diagnostic kits DIAKON-DS (Russia). Supplementary Table 4 stores Phenotypic Age and the blood biochemistry markers which were used for calculating Phenotypic Age [42].
To investigate the effect of dialysis on the values changing of blood biochemistry parameters, which were used to calculate the Phenotypic Age, we considered data for 5 people who are not included in the main dataset (Supplementary Fig. 2, Supplementary Table 5). Creatinine shows the highest change after dialysis.
Age acceleration
Four estimators of epigenetic age are considered: DNAmAgeHannum [43], DNAmAge [44], DNAmPhenoAge [42], DNAGrimAge [45]. The DNAmAgeHannum quantitative aging model measures the aging rate of human methylome in whole blood. The DNAmAge multi-tissue age predictor allows estimating the age of DNA methylation in various tissues and cells. The biomarker of aging DNAmPhenoAge was developed considering composite clinical measures of phenotypic age [42], while the DNAmGrimAge is a composite biomarker based on DNAm surrogates of seven plasma proteins and of smoking history. Data for all models are obtained using Horvath’s calculator [46] and presented in Supplementary Table 6.
To calculate the acceleration of the epigenetic age of the participants considered in the study, first, for each type of epigenetic age, a linear regression model which estimates chronological age was built only on the group of control participants. Epigenetic age acceleration is a residual from this linear model. All models were built using statsmodels [47] package in the Python programming language. Mann–Whitney U test (two-sided) from the scipy [48] Python package was applied to analyze the statistically significant difference of epigenetic age acceleration between controls and participants with the disease. The Benjamini–Hochberg FDR correction was applied to the list of p-values. The same approach was used to analyze the age acceleration of the Phenotypic Age, the biological age estimator based on biochemical and hematological blood parameters together with chronological age [42].
We also calculated a measure of intrinsic epigenetic age acceleration (IEAA), which characterizes “pure” epigenetic aging effects without the influence of differences in blood cell count [15, 16]. IEAA is the residual resulting from a multivariate regression model of DNAm age on chronological age and blood immune cell counts (naïve CD8 + T cells, exhausted CD8 + T cells, plasma B cells, CD4 + T cells, natural killer cells, monocytes, and granulocytes). The model was built only on the group of control participants using the statsmodels [47] package in the Python programming language. We have calculated a measure of extrinsic epigenetic age acceleration (EEAA), which characterizes epigenetic aging in immune-related components [15, 16]. EEAA is the residual resulting from a univariate model that regressed the weighted average of the epigenetic age measure from Hannum and three estimated measures of blood cells (naïve (CD45RA + CCR7 +) cytotoxic T cells, exhausted (CD28-CD45RA-) cytotoxic T cells, plasma B cells on chronological age. The model was built only on the group of control participants using the statsmodels package in the Python programming language. The distributions of blood cell counts, obtained from Horvath’s calculator [46], are in Supplementary Fig. 3. IEAA and EEAA values are presented in Supplementary Table 6.
Immunology clock
Using immunology multiplex assays, we applied the elastic net regression model (sklearn [49] package in Python) where chronological age was regressed on 46 multiplex biomarkers to select variables for inclusion in our immunology age score (ipAGE). K-fold cross-validation (K = 5) was employed to select the optimal parameter alpha, which multiplies the penalty terms (lambda parameter in glmnet [50] R package). The search for the optimal value of this parameter was performed on an equidistant logarithmic grid: . Parameter l1_ratio, a merge weight of L1 and L2 regularization (corresponding to alpha in the glmnet R package), was taken as 0.5. Thus, for each set of parameters, the entire dataset was divided 5 times into training (80%) and test (20%) sets (sliding window of the test set). To calculate the ipAGE acceleration for each model, we firstly build a linear regression which estimates chronological age only on controls. After that, acceleration is calculated as a residual from this model. Mann–Whitney U test (two-sided) was applied to check the difference in age accelaration of ipAGE between the groups of participants. There are three groups of participants: controls, which were used for building the model; additional controls for testing the ipAGE model (Supplementary Table 2); ESRD group. The Mann–Whitney U test was performed pairwise for control vs controls (test), control vs ESRD, controls (test) vs ESRD.
To compare the results of the ipAGE clock, we compare their results from other phenotypic and epigenetic clocks by calculating the Pearson’s correlation coefficient between corresponding ages, as well as between accelerations relative to chronological age. The list of pairwise correlation p-values was FDR corrected using the Benjamini–Hochberg approach.
Associations of immunology biomarkers
To find the most disease-associated immunology biomarkers, the Mann–Whitney U test (scipy [48] Python package) was applied to the values of the biomarker, divided into groups (controls vs ESRD). The Benjamini–Hochberg FDR correction was applied to the list of p-values. To find biomarkers associated with chronological, phenotypic, epigenetic, and immune ages, the Pearson’s correlation test was performed (scipy [48] Python package). As a criterion for a statistically significant relationship between biomarkers and the selected age types, the Pearson’s correlation p-value (Benjamini–Hochberg FDR-corrected) of the corresponding biomarkers is used; the values of which are less than 0.05 indicate the presence of a relationship. The same approach was used to find associations between biomarkers and different types of age acceleration, which were defined as residuals from the chronological age.
Results
Epigenetic age measures in ESRD patients
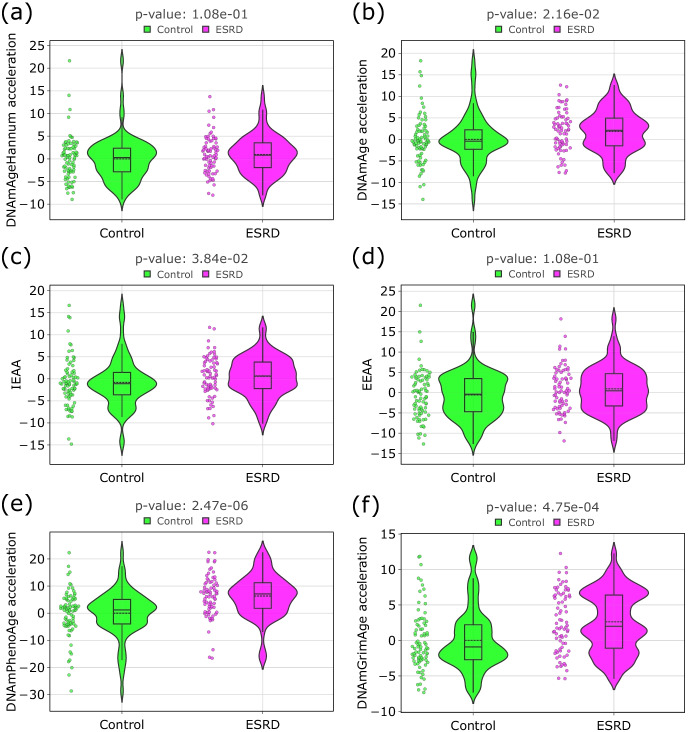
To identify the relationship between ESRD and aging processes, the 5 most common epigenetic clocks DNAmAgeHannum [43], IEAA [15, 16], EEAA [15, 16], DNAmAge [44], DNAmPhenoAge [42], DNAGrimAge [45] were analyzed (Fig. 2). Our results demonstrate that three out of four epigenetic clocks (DNAmAge [44], DNAmPhenoAge [42], DNAGrimAge [45]) show statistically significant (Mann–Whitney U test FDR-corrected p-value below 0.05) age acceleration in the group of participants with ESRD (Fig. 2b, e, and f), as well as IEAA [15, 16] (Fig. 2c), with DNAmAgeHannum [43] and EEAA [15, 16] as exceptions (Fig. 2a and d). On average, epigenetic age acceleration in the ESRD group was about 3 years. DNAmAgeHannum showed the smallest increase of 0.6 years (Q1 = − 1.9, Q3 = 3.5, p-value = 1.08e-1); the highest age-related acceleration of 7.0 years (Q1 = 1.7, Q3 = 11.2, p-value = 2.47e-6) was recorded for DNAmPhenoAge.
Fig. 2.
Violin plots for age acceleration differences in controls and participants with the ESRD: a DNAmAgeHannum [43]; b DNAmAge [44]; c IEAA [15, 16]; d EEAA [15, 16]; e DNAmPhenoAge [42]; f DNAmGrimAge [45]. The black line on the boxplot corresponds to the median value, dashed line—to mean value. For each type of epigenetic age, a linear regression model which estimates chronological age was built only on the group of control participants. Epigenetic age acceleration was calculated as a residual from this linear model. IEAA was calculated as the residual from a multivariate regression model of DNAm age on chronological age and blood immune cell counts only on the control group. EEAA is the residual resulting from a univariate model that regressed the weighted average of the epigenetic age measure from Hannum and three estimated measures of blood cells (naïve (CD45RA + CCR7 +) cytotoxic T cells, exhausted (CD28-CD45RA-) cytotoxic T cells, plasma B cells) on chronological age (only on control group). Mann–Whitney U test was applied to analyze the statistically significant difference of epigenetic age acceleration between controls and participants with ESRD. The resulting p-values were FDR-corrected with the Benjamini–Hochberg approach
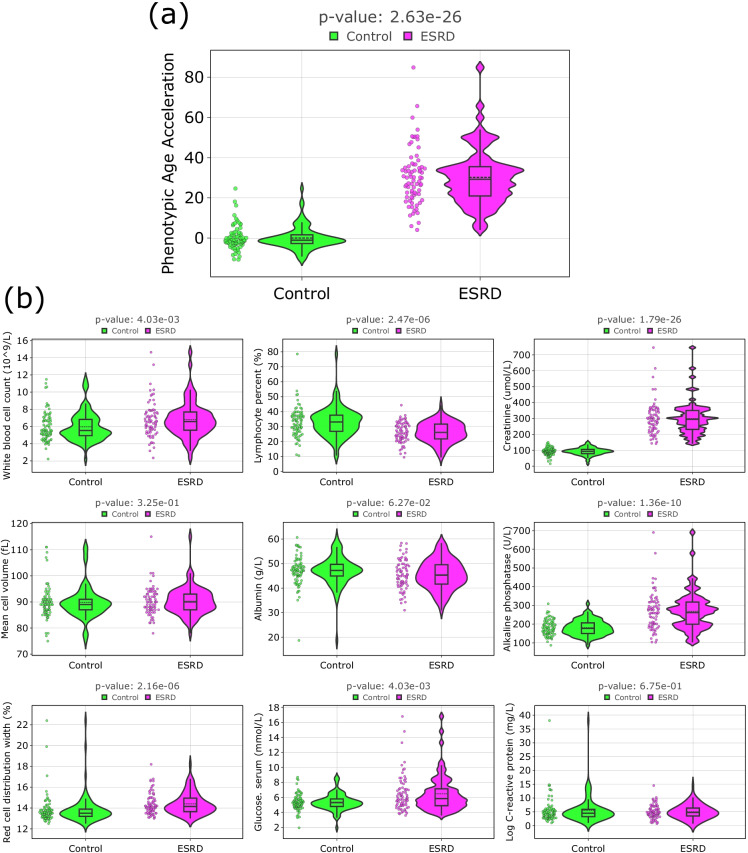
Next, we considered Phenotypic Age [42] a biological age estimator based on biochemical and hematological blood parameters, which could be expected to be more sensitive to the pathological changes in the organism. On the one hand, the employed markers are more responsive to various cues than epigenetic ones, not necessarily related to the pathology; on the other hand, it potentially allows to catch earlier the deficiencies of the body homeostatic regulation caused by the pathology. Besides, Phenotypic Age includes biomarkers, which are potentially associated with the disease (for example, creatinine). As a result, the Phenotypic Age obtained from blood biomarkers proved to manifest age acceleration in patients with ESRD substantially more pronounced than epigenetic age estimators (Fig. 3a). To get a deeper insight, we plot the distributions of biomarkers involved in these clocks for both the control group and patients with ESRD in Fig. 3b. It follows that almost all biomarkers in the ESRD group display significantly different values from the control group.
Fig. 3.
Comparison of Phenotypic Age [42] metrics in controls and cases (violin plots). a Age acceleration differences in controls and participants with the ESRD for Phenotypic Age. Age acceleration was calculated as a residual from a linear regression model which estimates chronological age only on controls; b distribution of components of the Phenotypic Age estimator in controls and participants with the ESRD. The black line on the box plot corresponds to the median value, dashed line—to mean value. Mann–Whitney U test was applied to analyze the statistically significant difference of Phenotypic Age metrics between controls and participants with ESRD. The resulting p-values of components of the Phenotypic Age estimator were FDR-corrected with the Benjamini–Hochberg approach
Inflammatory/immunological profile clock (ipAGE)
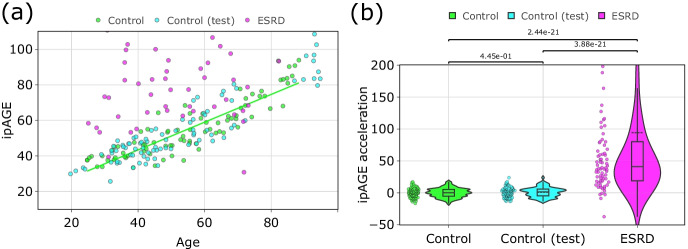
We evaluated 46 inflammatory/immunological markers in plasma from the participants involved in the study. Applying the elastic net regression model in the group of healthy controls (see “Methods”), we built a model that estimates age based on these biomarkers, termed inflammatory/immunological profile age (ipAGE). The model included 38 out of 46 biomarkers and yielded a determination coefficient of 0.79, mean absolute error (MAE) 6.82 years, root mean squared error (RMSE) 8.17 years in the control group. Supplementary Table 7 reports the list of the proteins included in the ipAGE clock, their model coefficients and the comparison with the iAge from Sayed et al. In particular, 27 plasma proteins are shared between the two predictors. ipAGE values for the considered cohort are listed in Supplementary Table 8. Figure 4a illustrates the resulting fit in the control group.
Fig. 4.
Characteristics of ipAGE. a Association between ipAGE and chronological age; b ipAGE acceleration in controls, test controls, and participants with the ESRD. ipAGE was built with the elastic net regression model where chronological age was regressed on multiplex biomarkers. Age acceleration was calculated as a residual from the linear regression model which estimates chronological age only on controls (green line). Mann–Whitney U test was applied to analyze the pairwise statistically significant difference of ipAGE acceleration between the groups of controls, test controls, and participants with ESRD
As for the epigenetic clocks, we calculated age acceleration as the residuals of the linear regression between ipAGE and chronological age (green line in Fig. 4a). Importantly, we found a statistically significant age acceleration in the ESRD group compared to controls, as illustrated in Fig. 4b (p-value = 2.44e-21). The group of participants with ESRD exhibited also a large ipAGE variance. To test the ipAGE model, we considered additional control participants (Supplementary Table 2). There is no statistically significant difference in ipAGE acceleration between controls, which were used to build the model, and test controls (p-value = 0.445). The ipAGE model demonstrated mean absolute error (MAE) 7.27 years and root mean squared error (RMSE) 8.33 years in the test control group. At the same time, we found a statistically significant age acceleration in the ESRD group compared to test controls (p-value = 3.88e-21) (Fig. 4b).
To investigate the influence of additional characteristics of subjects on the resulting inflammatory/immunological age estimation, we built another model, based on the same 46 inflammatory/immunological markers and sex as an additional covariate. After elastic net optimization, the resulting model kept 39 features, and the sex variable did not remain among them. In other words, the sex covariate does not significantly affect the inflammatory/immunological age estimation. The obtained model demonstrates almost the same quality: RMSE = 8.13 and MAE = 6.78 in original controls and RMSE = 8.47 and MAE = 7.36 in test controls. Supplementary Fig. 4 illustrates the performance of this model: associations with chronological age and significance of age acceleration between the groups. Supplementary Table 9 reports the list of the proteins included in this model and estimated age values for the participants in all groups.
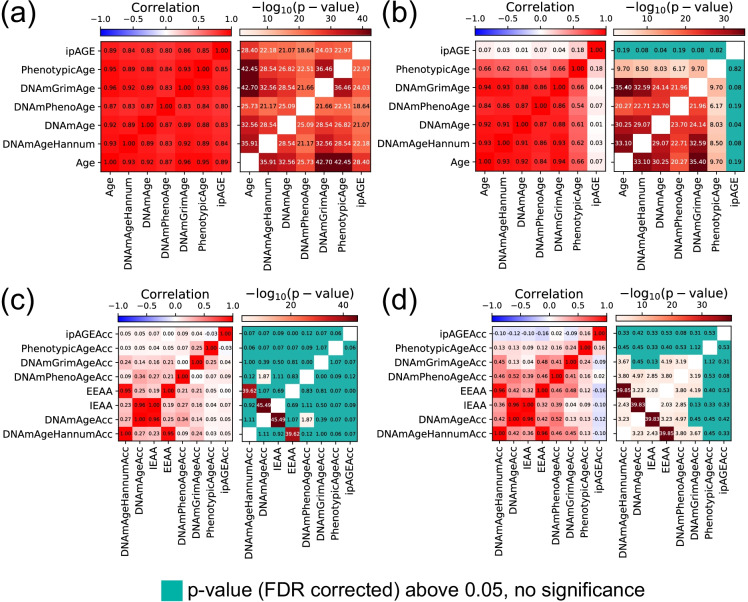
We then investigated the relationship between epigenetic clocks, phenotypic clock, and ipAGE. Expectedly, all the clocks showed a high correlation with chronological age in control participants (Fig. 5a) and between each other. The epigenetic age estimators maintained a high correlation with chronological age and among each other also in the ESRD patients, while in this group, the ipAGE was weakly correlated with chronological age and the other clocks. This is a consequence of the sensitive response of ipAGE to ESRD with respect to controls and significant intra-group differences between ESRD patients (Fig. 5b). Instructively, there was no significant correlation between age accelerations, except correlations between IEAA ~ DNAmAgeAcc and EEAA ~ DNAmAgeHannumAcc which are due to the calculation of IEAA and EEAA being based on DNAmAge and DNAmAgeHannum correspondingly (Fig. 5c and d). The effect was observed both for the control and ESRD groups, indicating that the same participant can have a positive age acceleration according to some clocks and negative by others.
Fig. 5.
Associations between different ages and age accelerations in controls and cases (separately). a Pearson’s correlation coefficient and corresponding logarithmic p-values of different ages for the control group; b—for the ESRD group. c Pearson’s correlation coefficient and corresponding logarithmic p-values of different age-accelerations for the control group; d—for the ESRD group. Age acceleration for each type of age estimator was calculated as a residual from the linear regression model which estimates chronological age only on controls. In each case, the list of pairwise correlation p-values was FDR corrected using the Benjamini–Hochberg approach
Association of immunology biomarkers with ESRD status
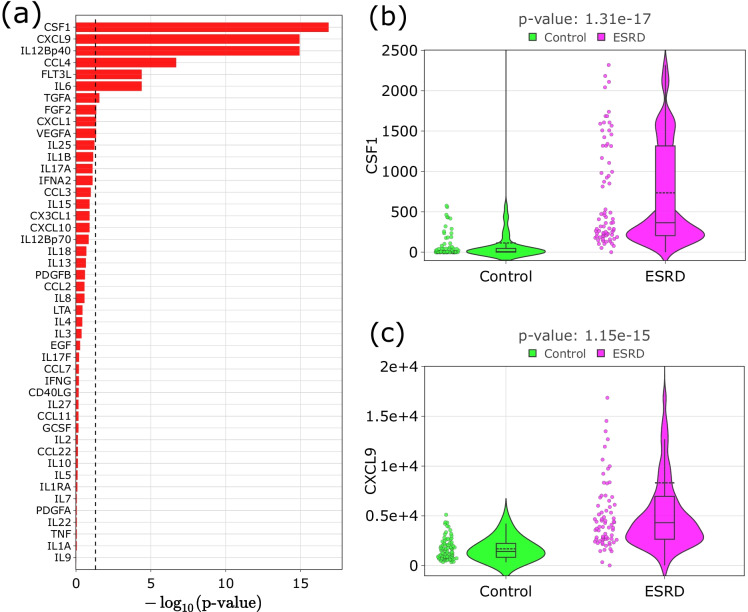
As a next step, we compared the 46 inflammatory/immunological markers between ESRD and controls. Ten out of 46 biomarkers (Supplementary Table 10) had a statistically significant association with ESRD (Mann–Whitney U test p-value with FDR correction below 0.05). Figure 6 illustrates the resulting p-values, manifesting the strongest association with the disease of CSF1, CXCL9, and IL12Bp40, and exemplifies distributions of the first two biomarkers in control and ESRD groups.
Fig. 6.
ESRD-associated inflammatory/immunological proteins. a FDR-corrected Mann–Whitney U test p-value (logarithmic), which identifies a significant difference of biomarkers values between groups. The dashed line corresponds to 0.05. b, c Violin plots for the two biomarkers with the highest association with ESRD: CSF1 and CXCL9. The black line on the boxplot corresponds to the median value, dashed line—to mean value
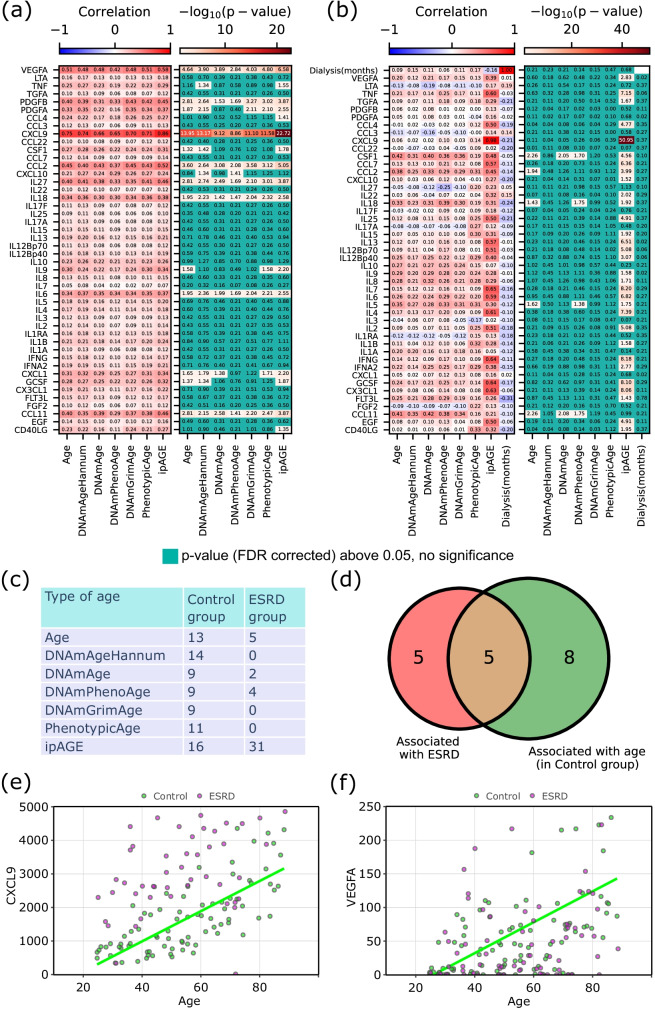
We also tested the individual proteins for their association with chronological age, epigenetic ages, phenotypic age, and ipAGE in the control and ESRD groups separately (Supplementary Table 10). The heatmaps of Pearson’s correlation coefficients, as well as corresponding FDR-corrected p-values for the considered biomarkers, are shown in Fig. 7a for the group of controls and in Fig. 7b for the patients with ESRD. Notably, some biomarkers implemented in immunological clocks do not have a significant correlation with age by themselves. Figure 7c illustrates the total number of significant biomarkers in both groups.
Fig. 7.
Association of inflammatory/immunological proteins with different types of age: a Pearson’s correlation coefficients and corresponding FDR-corrected p-values resulting from the correlation of protein levels and each type of age, in the control and b ESRD groups; c the number of proteins, which are significantly associated with chronological age, epigenetic ages, phenotypic age, and ipAGE (FDR-corrected Pearson’s correlation p-value below 0.05); d Venn diagram of ESRD-associated and chronological age-associated inflammatory/immunological proteins in the control group; e, f examples of biomarkers associated with chronological age
There are eight common biomarkers associated with chronological age, epigenetic ages, phenotypic age, and ipAGE in the control group (Fig. 7d, Supplementary Table 11). In the ESRD group, there are no biomarkers that are associated with all types of ages.
The time on dialysis, measured in months, appears as one of the variables in the ESRD group (Fig. 7b: upper row and right column). No correlation was found between time on dialysis and chronological age, epigenetic clocks, phenotypic age, and ipAGE. There are no biomarkers that have significant associations with time on dialysis.
Figure 7d illustrates the Venn diagram for disease-associated biomarkers and chronological age-associated biomarkers in the control group. Five cytokines lie in the intersection (CSF1, CXCL1, CXCL9, IL6, VEGFA). The most representative biomarkers associated with chronological age are shown in Fig. 7e and f.
We also considered associations between biomarkers and different types of age acceleration (Supplementary Fig. 5, Supplementary Table 10). In the control group as well as in the ESRD group, there are no biomarkers that are correlated with all types of age accelerations. However, in the ESRD group, most of the biomarkers are significantly associated with ipAGE acceleration. There are four biomarkers (CSF1, CXCL9, IL6, VEGFA) that are associated with the disease, chronological age in the control group, and ipAGE acceleration in the ESRD group.
Supplementary Figs. 6, 7, and 8 illustrate the dependencies of all biomarkers on chronological age, ipAGE, and ipAGE acceleration correspondingly. Associations between all immunological biomarkers in control and ESRD groups are separately shown in Supplementary Fig. 9.
Discussion
In this study, we evaluated biomarkers of age in patients with end-stage renal disease, focusing on epigenetic and inflammatory/immunological biomarkers.
So far, only a few studies have evaluated epigenetic changes related to kidney function and CKD [51–54]. More recently, Matías-García et al. investigated the association between epigenetic clocks and renal function [33], reporting that epigenetic age acceleration was associated with low renal function. The cohort assessed by Matías-García et al. did not include patients with ESRD or under dialysis, which on the contrary are the focus of our study. We found that most of the epigenetic clocks, in particular DNAmPhenoAge and GrimAge, detected age acceleration in the ESRD group, thus confirming and extending the results by Matías-García et al. in a more extreme phenotype. We also found that EEAA is not significantly different between ESRD and controls, suggesting that immunosenescence is not the main trigger of the accelerated aging in the ESRD group.
Phenotypic age revealed an even more marked age acceleration in ESRD patients, with most of its components showing significantly different values compared to the control group. This indicates that ESRD produces a significant impact on a variety of homeostasis regulation processes in the body, and its effect is not limited to an increase in creatinine, the renal filtration efficiency marker. Noteworthy, while chronic inflammation is viewed as one of the pathological mechanisms of CKD and ESRD development, the ESRD group did not show an increase in C-reactive protein, the main marker of inflammation in Phenotypic Age, and lymphocyte counts in patients with ESRD were statistically lower than the control values (p-value = 2.47e-06), suggesting the existence of more complex and systemic changes.
The development of CKD may be associated with chronic inflammatory reactions of an infectious nature (glomerulonephritis — 30% of cases) or non-infectious processes (type 2 diabetes mellitus — 55% of cases). Quite rarely, it is the result of genetically determined processes or toxic damage (15%). However, the process of CKD development is always associated with the so-called uremic inflammation. Uremic inflammation is related to the altering effects of the uremic milieu on the immune systems [55–57] and includes mechanisms of both immunoactivation and immunosuppression. As a rule, immunoactivation during the development of uremia is associated with an increase in the synthesis of IL-1, IL-6 and TNF [55, 58]. In ESRD, the innate immune system, which involves monocytes, macrophages, granulocytes, and also endothelial cell activation, is activated together with a depletion of natural regulatory T-cells, resulting in systemic inflammation and enhanced oxidative stress. These processes are connected with an adaptive immune deficiency due to the reduction of naïve and central memory T-cells and B-cells, dendritic cells, and altered functions of polymorphonuclear leukocytes and monocytes [59]. ESRD is characterized by systemic persistent inflammation that does not subside even after complete renal sclerosis. Inflammation becomes maladaptive, uncontrolled, and persistent [56]. This process recalls what is observed during inflammaging, the chronic, sterile, low-grade inflammation that develops during aging and contributes to the pathogenesis of age-related diseases [3]. Therefore, it calls for considering markers associated with inflammation and constructing novel (immunological) clocks on their basis that would potentially be more sensitive to disease in terms of manifested age acceleration.
We, therefore, developed an inflammatory/immunological clock that estimates chronological age in healthy participants and assesses the contribution of inflammaging to the processes of age acceleration. Implementation of machine learning algorithms allowed the selection of meaningful immunological biomarkers and to derive an ipAGE clock model that takes into account the complex relationships and redundancy of the cytokine network. The clock MAE is about 6.8 years, competing with epigenetic clocks in terms of accuracy. The components of the ipAGE clock largely overlap with the iAge clock, recently developed by Sayed et al. using alternative experimental and analytical approaches [7]. While the iAge predictor was shown to be associated with cardiovascular aging, here, we report that ipAGE successfully detects accelerated aging in ESRD patients.
Similar to the Phenotypic Age, ipAGE demonstrates a greater sensitivity in assessing the adaptive potential of an organism compared to epigenetic clocks, due to the higher blood parameters’ variation in comparison with DNA methylation changes. In addition, in analogy to Phenotypic Age, a higher variance was observed in ESRD patients. Unlike progressive CKD, ESRD is characterized by a state of “dynamic intoxication,” an increase in toxic low molecular weight products between hemodialysis sessions. Chronic dynamic intoxication is undoubtedly a unique phenomenon because of the permanent changes of milieu intérieur and dramatic changes in the concentrations of all low molecular weight compounds. Aging is a heterogeneous process of the body’s adaptation to the changing conditions of the external environment, or, as in the case of CKD, the internal one, when a variety of genetically determined mechanisms are triggered, determining the body’s individual response and an increase in the variance of the body’s metabolic parameters with age. Thus, an increase in the age-related variability of biological markers is further stimulated by impaired homeostasis under “dynamic intoxication” conditions.
We did not find any correlation between the age acceleration evaluated according to the epigenetic clocks and ipAGE in either the ESRD group or the control group. This is in line with previous results showing that different biomarkers of age can be informative of different aspects of aging [60, 61] and further suggests that currently available age estimators perform well for population data but are still not quite suited to characterize individuals, confirming the necessity of referring to several clocks to improve the reliability of results [12]. Interestingly, a positive correlation ipAGE with both pro-inflammatory and anti-inflammatory markers was found in the ESRD group, pointing out the extreme complexity of the processes of homeostatic regulation of the immune system. This explains the huge variance of indicators and ipAGE observed in the group of ESRD patients.
We also identified 10 plasma proteins that have significantly different levels between ESRD patients and healthy controls. Most of these proteins are part of the ipAGE clock, and 5 of them (CXCL9, VEGFA, IL6, CXCL1, CSF1) also show significant age association in healthy controls from our cohort. These findings indicate that a considerable number of immune system cytokines are both age and chronic kidney disease-associated, in accord with inflammaging theory.
A notable example is CXCL9 (MIG), which is associated with chronological, epigenetic, phenotypic age and with ipAGE, is one of the main components of the iAge clock from Sayed et al., and whose levels are significantly different between ESRD patients and healthy controls. For a long time, CXCL9 was not considered among the main markers of inflammaging. However, there is growing evidence of its key role in the development of age-related diseases, such as cardiovascular pathology [62], neurodegeneration [63], glaucoma [64], and depressed anti-tumor immunity [65]. CXCL9 is one of the chemokines that play a role in the induction of chemotaxis, promotes leukocyte differentiation and proliferation, and as a member of the IFN-γ-dependent cytokine family promotes the development of Th1-mediated pro-inflammatory responses. Despite the fact that the molecular and cellular action mechanisms of this chemokine are broadly known, its role in inflammaging requires further study.
Similar to CXCL9, also CSF1 is known to increase its concentration during aging and several age-related diseases, such as Alzheimer’s disease, cataracts, and cardiovascular disease [62, 66]. Other biomarkers (IL12Bp40, FGF2, FLT3L) that emerged from our study have been associated with inflammation processes only, but not with aging.
Interestingly, only one of the classic markers of uremic inflammation, namely IL6, correlates in our study with age-related acceleration in people with ESRD. The terminal stage of CKD has several features since the hemodialysis procedure aimed at detoxifying and maintaining the body’s homeostasis is stressful: it causes dynamic changes in low molecular weight metabolic products, most of which are toxic to the human body.
Particularly interesting is the fact that most of the interleukins associated with ipAGE in the ESRD group are modulators of the B-cell immune response, while those correlated with ipAGE, chronological age, and epigenetic ages in the control group are modulators of the Th1 cellular immune response. It is interesting to note the absence of correlations between individual immunomarkers and epigenetic or phenotypic age, in general, and satisfactory performance of ipAGE clocks requires 38 parameters. We conjecture that the observed variability of immunoreactivity is underpinned by the heterogeneity of the body’s adaptive responses.
The main limitation of the study is the moderate size of the cohort, which, however, did not preclude deriving a robust and powerful aging biomarker based on an inflammatory/immunological profile. Its precision and performance should improve with the further increase of the data set. Another potential confounding factor is the local nature of the cohort that should also be addressed in the future by scaling up the study. Lastly, the study is currently restricted to the case of ESRD, while probing the age acceleration in other pathologies with both epigenetic age estimators and ipAGE remains a challenge.
In conclusion, we demonstrated that epigenetic clocks, phenotypic clock, and a newly developed inflammatory/immune clock coherently detect an accelerated aging phenotype in ESRD patients, although they poorly correlate with each other. Future studies should integrate these different clocks, which are likely to grasp different aspects of human aging, to create new tools that could identify pathological deviations from aging-related trajectories. Furthermore, the possibility to include disease-specific parameters will allow using these new tools at the bedside, helping in the clinical management of highly prevalent conditions in the elderly like CKD.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We acknowledge the support of the Ministry of Science and Higher Education agreement No. 075-15-2021-639.
Author contribution
IY, EK, MGB, CF, MV, and MI contributed to the conception and design of the study. IY, EK, AK, MK, NL, and MV organized the datasets. IY, AK, MK, and MGB performed the statistical analysis. IY, EK, AK, MGB, CF, MV, and MI wrote the manuscript. All authors contributed to manuscript revision and read and approved the submitted version.
Data availability statement
All data (except DNA methylation) generated during this study are included in this published article (and its supplementary information files). DNA methylation data is available from the corresponding author on a reasonable request.
Declarations
Conflict of interest
The authors declare no competing interests.
Code availability statement
Source code for statistical analysis and building ipAGE clock can be found here: https://github.com/GillianGrayson/ipAGE
Footnotes
Maria Vedunova and Mikhail Ivanchenko are co-senior authors.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Igor Yusipov and Elena Kondakova contributed equally to this work.
References
- 1.Cohen AA, Kennedy BK, Anglas U, Bronikowski AM, Deelen J, Dufour F, Ferbeyre G, Ferrucci L, Franceschi C, Frasca D, Friguet B, Gaudreau P, Gladyshev VN, Gonos ES, Gorbunova V, Gut P, Ivanchenko M, Legault V, Lemaître J-F, Liontis T, Liu G-H, Liu M, Maier AB, Nóbrega OT, Olde Rikkert MGM, Pawelec G, Rheault S, Senior AM, Simm A, Soo S, Traa A, Ukraintseva S, Vanhaelen Q, Van Raamsdonk JM, Witkowski JM, Yashin AI, Ziman R, Fülöp T. Lack of consensus on an aging biology paradigm? A global survey reveals an agreement to disagree, and the need for an interdisciplinary framework. Mech Ageing Dev. 2020;191:111316. doi: 10.1016/j.mad.2020.111316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franceschi C, Capri M, Monti D, Giunta S, Olivieri F, Sevini F, Panourgia MP, Invidia L, Celani L, Scurti M, Cevenini E, Castellani GC, Salvioli S. Inflammaging and anti-inflammaging: a systemic perspective on aging and longevity emerged from studies in humans. Mech Ageing Dev. 2007;128:92–105. doi: 10.1016/j.mad.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 3.Franceschi C, Garagnani P, Parini P, Giuliani C, Santoro A. Inflammaging: a new immune–metabolic viewpoint for age-related diseases. Nat Rev Endocrinol. 2018;14:576–590. doi: 10.1038/s41574-018-0059-4. [DOI] [PubMed] [Google Scholar]
- 4.Furman D, Campisi J, Verdin E, Carrera-Bastos P, Targ S, Franceschi C, Ferrucci L, Gilroy DW, Fasano A, Miller GW, Miller AH, Mantovani A, Weyand CM, Barzilai N, Goronzy JJ, Rando TA, Effros RB, Lucia A, Kleinstreuer N, Slavich GM. Chronic inflammation in the etiology of disease across the life span. Nat Med. 2019;25:1822–1832. doi: 10.1038/s41591-019-0675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mittelbrunn M, Kroemer G. Hallmarks of T cell aging. Nat Immunol. 2021;22:687–698. doi: 10.1038/s41590-021-00927-z. [DOI] [PubMed] [Google Scholar]
- 6.Alpert A, Pickman Y, Leipold M, Rosenberg-Hasson Y, Ji X, Gaujoux R, Rabani H, Starosvetsky E, Kveler K, Schaffert S, Furman D, Caspi O, Rosenschein U, Khatri P, Dekker CL, Maecker HT, Davis MM, Shen-Orr SS. A clinically meaningful metric of immune age derived from high-dimensional longitudinal monitoring. Nat Med. 2019;25:487–495. doi: 10.1038/s41591-019-0381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sayed N, Huang Y, Nguyen K, Krejciova-Rajaniemi Z, Grawe AP, Gao T, Tibshirani R, Hastie T, Alpert A, Cui L, Kuznetsova T, Rosenberg-Hasson Y, Ostan R, Monti D, Lehallier B, Shen-Orr SS, Maecker HT, Dekker CL, Wyss-Coray T, Franceschi C, Jojic V, Haddad F, Montoya JG, Wu JC, Davis MM, Furman D. An inflammatory aging clock (iAge) based on deep learning tracks multimorbidity, immunosenescence, frailty and cardiovascular aging. Nat Aging. 2021;1:598–615. doi: 10.1038/s43587-021-00082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia X, Chen W, McDermott J, Han J-DJ (2017) Molecular and phenotypic biomarkers of aging. F1000Research 6:860 . 10.12688/f1000research.10692.1 [DOI] [PMC free article] [PubMed]
- 9.Oblak L, van der Zaag J, Higgins-Chen AT, Levine ME, Boks MP. A systematic review of biological, social and environmental factors associated with epigenetic clock acceleration. Ageing Res Rev. 2021;69:101348. doi: 10.1016/j.arr.2021.101348. [DOI] [PubMed] [Google Scholar]
- 10.Bacalini MG, D’Aquila P, Marasco E, Nardini C, Montesanto A, Franceschi C, Passarino G, Garagnani P, Bellizzi D. The methylation of nuclear and mitochondrial DNA in ageing phenotypes and longevity. Mech Ageing Dev. 2017;165:156–161. doi: 10.1016/j.mad.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19:371–384. doi: 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- 12.Bell CG, Lowe R, Adams PD, Baccarelli AA, Beck S, Bell JT, Christensen BC, Gladyshev VN, Heijmans BT, Horvath S, Ideker T, Issa J-PJ, Kelsey KT, Marioni RE, Reik W, Relton CL, Schalkwyk LC, Teschendorff AE, Wagner W, Zhang K, Rakyan VK. DNA methylation aging clocks: challenges and recommendations. Genome Biol. 2019;20:249. doi: 10.1186/s13059-019-1824-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, Gibson J, Henders AK, Redmond P, Cox SR, Pattie A, Corley J, Murphy L, Martin NG, Montgomery GW, Feinberg AP, Fallin MD, Multhaup ML, Jaffe AE, Joehanes R, Schwartz J, Just AC, Lunetta KL, Murabito JM, Starr JM, Horvath S, Baccarelli AA, Levy D, Visscher PM, Wray NR, Deary IJ. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16:25. doi: 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horvath S, Garagnani P, Bacalini MG, Pirazzini C, Salvioli S, Gentilini D, Di Blasio AM, Giuliani C, Tung S, Vinters HV, Franceschi C. Accelerated epigenetic aging in Down syndrome. Aging Cell. 2015;14:491–495. doi: 10.1111/acel.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai P-C, Roetker NS, Just AC, Demerath EW, Guan W, Bressler J, Fornage M, Studenski S, Vandiver AR, Moore AZ, Tanaka T, Kiel DP, Liang L, Vokonas P, Schwartz J, Lunetta KL, Murabito JM, Bandinelli S, Hernandez DG, Melzer D, Nalls M, Pilling LC, Price TR, Singleton AB, Gieger C, Holle R, Kretschmer A, Kronenberg F, Kunze S, Linseisen J, Meisinger C, Rathmann W, Waldenberger M, Visscher PM, Shah S, Wray NR, McRae AF, Franco OH, Hofman A, Uitterlinden AG, Absher D, Assimes T, Levine ME, Lu AT, Tsao PS, Hou L, Manson JE, Carty CL, LaCroix AZ, Reiner AP, Spector TD, Feinberg AP, Levy D, Baccarelli A, van Meurs J, Bell JT, Peters A, Deary IJ, Pankow JS, Ferrucci L, Horvath S (2016) DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging 8:1844–1865 . 10.18632/aging.101020 [DOI] [PMC free article] [PubMed]
- 16.Horvath S, Gurven M, Levine ME, Trumble BC, Kaplan H, Allayee H, Ritz BR, Chen B, Lu AT, Rickabaugh TM, Jamieson BD, Sun D, Li S, Chen W, Quintana-Murci L, Fagny M, Kobor MS, Tsao PS, Reiner AP, Edlefsen KL, Absher D, Assimes TL. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016;17:171. doi: 10.1186/s13059-016-1030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durso DF, Bacalini MG, Sala C, Pirazzini C, Marasco E, Bonafé M, do Valle ÍF, Gentilini D, Castellani G, Faria AMC, Franceschi C, Garagnani P, Nardini C (2017) Acceleration of leukocytes’ epigenetic age as an early tumor and sex-specific marker of breast and colorectal cancer. Oncotarget 8:23237–23245 . 10.18632/oncotarget.15573 [DOI] [PMC free article] [PubMed]
- 18.Gensous N, Bacalini MG, Franceschi C, Meskers CGM, Maier AB, Garagnani P. Age-related DNA methylation changes: potential impact on skeletal muscle aging in humans. Front Physiol. 2019;10:996. doi: 10.3389/fphys.2019.00996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiorito G, Polidoro S, Dugué P-A, Kivimaki M, Ponzi E, Matullo G, Guarrera S, Assumma MB, Georgiadis P, Kyrtopoulos SA, Krogh V, Palli D, Panico S, Sacerdote C, Tumino R, Chadeau-Hyam M, Stringhini S, Severi G, Hodge AM, Giles GG, Marioni R, Karlsson Linnér R, O’Halloran AM, Kenny RA, Layte R, Baglietto L, Robinson O, McCrory C, Milne RL, Vineis P. Social adversity and epigenetic aging: a multi-cohort study on socioeconomic differences in peripheral blood DNA methylation. Sci Rep. 2017;7:16266. doi: 10.1038/s41598-017-16391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dugué P-A, Bassett JK, Joo JE, Baglietto L, Jung C-H, Wong EM, Fiorito G, Schmidt D, Makalic E, Li S, Moreno-Betancur M, Buchanan DD, Vineis P, English DR, Hopper JL, Severi G, Southey MC, Giles GG, Milne RL. Association of DNA methylation-based biological age with health risk factors and overall and cause-specific mortality. Am J Epidemiol. 2018;187:529–538. doi: 10.1093/aje/kwx291. [DOI] [PubMed] [Google Scholar]
- 21.Gensous N, Garagnani P, Santoro A, Giuliani C, Ostan R, Fabbri C, Milazzo M, Gentilini D, di Blasio AM, Pietruszka B, Madej D, Bialecka-Debek A, Brzozowska A, Franceschi C, Bacalini MG. One-year Mediterranean diet promotes epigenetic rejuvenation with country- and sex-specific effects: a pilot study from the NU-AGE project. GeroScience. 2020;42:687–701. doi: 10.1007/s11357-019-00149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schroth J, Thiemermann C, Henson SM. Senescence and the aging immune system as major drivers of chronic kidney disease. Front Cell Dev Biol. 2020;8:564461. doi: 10.3389/fcell.2020.564461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Figuer A, Bodega G, Tato P, Valera G, Serroukh N, Ceprian N, de Sequera P, Morales E, Carracedo J, Ramírez R, Alique M. Premature aging in chronic kidney disease: the outcome of persistent inflammation beyond the bounds. Int J Environ Res Public Health. 2021;18:8044. doi: 10.3390/ijerph18158044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevens LA, Viswanathan G, Weiner DE. Chronic kidney disease and end-stage renal disease in the elderly population: current prevalence, future projections, and clinical significance. Adv Chronic Kidney Dis. 2010;17:293–301. doi: 10.1053/j.ackd.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams ME, Sandeep J, Catic A. Aging and ESRD demographics: consequences for the practice of dialysis: aging and ESRD demographics. Semin Dial. 2012;25:617–622. doi: 10.1111/sdi.12029. [DOI] [PubMed] [Google Scholar]
- 26.Ravani P, Quinn R, Fiocco M, Liu P, Al-Wahsh H, Lam N, Hemmelgarn BR, Manns BJ, James MT, Joanette Y, Tonelli M. Association of age with risk of kidney failure in adults with stage IV chronic kidney disease in Canada. JAMA Netw Open. 2020;3:e2017150. doi: 10.1001/jamanetworkopen.2020.17150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levey AS, Coresh J. Chronic kidney disease. The Lancet. 2012;379:165–180. doi: 10.1016/S0140-6736(11)60178-5. [DOI] [PubMed] [Google Scholar]
- 28.McCullough K, Sharma P, Ali T, Khan I, Smith WCS, MacLeod A, Black C. Measuring the population burden of chronic kidney disease: a systematic literature review of the estimated prevalence of impaired kidney function. Nephrol Dial Transplant. 2012;27:1812–1821. doi: 10.1093/ndt/gfr547. [DOI] [PubMed] [Google Scholar]
- 29.Chiu Y-L, Tsai H-H, Lai Y-J, Tseng H-Y, Wu Y-W, Peng Y-S, Chiu C-M, Chuang Y-F. Cognitive impairment in patients with end-stage renal disease: accelerated brain aging? J Formos Med Assoc. 2019;118:867–875. doi: 10.1016/j.jfma.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Chiu Y-L, Shu K-H, Yang F-J, Chou T-Y, Chen P-M, Lay F-Y, Pan S-Y, Lin C-J, Litjens NHR, Betjes MGH, Bermudez S, Kao K-C, Chia J-S, Wang G, Peng Y-S, Chuang Y-F. A comprehensive characterization of aggravated aging-related changes in T lymphocytes and monocytes in end-stage renal disease: the iESRD study. Immun Ageing. 2018;15:27. doi: 10.1186/s12979-018-0131-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung H, Choi IY, Shin DW, Han K, Yoo JE, Chun S, Yi Y. Association between physical performance and incidence of end-stage renal disease in older adults: a national wide cohort study. BMC Nephrol. 2021;22:85. doi: 10.1186/s12882-021-02291-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang F, Chen R, Cao X, Shen B, Chen X, Ding X, Zou J. Premature aging of circulating T cells predicts all-cause mortality in hemodialysis patients. BMC Nephrol. 2020;21:271. doi: 10.1186/s12882-020-01920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matías-García PR, Ward-Caviness CK, Raffield LM, Gao X, Zhang Y, Wilson R, Gào X, Nano J, Bostom A, Colicino E, Correa A, Coull B, Eaton C, Hou L, Just AC, Kunze S, Lange L, Lange E, Lin X, Liu S, Nwanaji-Enwerem JC, Reiner A, Shen J, Schöttker B, Vokonas P, Zheng Y, Young B, Schwartz J, Horvath S, Lu A, Whitsel EA, Koenig W, Adamski J, Winkelmann J, Brenner H, Baccarelli AA, Gieger C, Peters A, Franceschini N, Waldenberger M. DNAm-based signatures of accelerated aging and mortality in blood are associated with low renal function. Clin Epigenetics. 2021;13:121. doi: 10.1186/s13148-021-01082-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.World Health Organization . The ICD-10 classification of mental and behavioural disorders: clinical descriptions and diagnostic guidelines. Geneva: World Health Organization; 1992. [Google Scholar]
- 35.Nahler G. Dictionary of Pharmaceutical Medicine. Vienna: Springer Vienna; 2009. Anatomical Therapeutic Chemical Classification System (ATC) pp. 8–8. [Google Scholar]
- 36.Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, Van Djik S, Muhlhausler B, Stirzaker C, Clark SJ. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17:208. doi: 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCartney DL, Walker RM, Morris SW, McIntosh AM, Porteous DJ, Evans KL. Identification of polymorphic and off-target probe binding sites on the Illumina Infinium MethylationEPIC BeadChip. Genomics Data. 2016;9:22–24. doi: 10.1016/j.gdata.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou W, Laird PW, Shen H. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res. 2017;45:e22. doi: 10.1093/nar/gkw967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nordlund J, Bäcklin CL, Wahlberg P, Busche S, Berglund EC, Eloranta M-L, Flaegstad T, Forestier E, Frost B-M, Harila-Saari A, Heyman M, Jónsson ÓG, Larsson R, Palle J, Rönnblom L, Schmiegelow K, Sinnett D, Söderhäll S, Pastinen T, Gustafsson MG, Lönnerholm G, Syvänen A-C. Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol. 2013;14:r105. doi: 10.1186/gb-2013-14-9-r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luminex Assays & Instrumentation. https://www.rndsystems.com/products/luminex-assays-and-high-performance-assays
- 42.Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, Hou L, Baccarelli AA, Stewart JD, Li Y, Whitsel EA, Wilson JG, Reiner AP, Aviv A, Lohman K, Liu Y, Ferrucci L, Horvath S (2018) An epigenetic biomarker of aging for lifespan and healthspan. Aging 10:573–591 . 10.18632/aging.101414 [DOI] [PMC free article] [PubMed]
- 43.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan J-B, Gao Y, Deconde R, Chen M, Rajapakse I, Friend S, Ideker T, Zhang K. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, Hou L, Baccarelli AA, Li Y, Stewart JD, Whitsel EA, Assimes TL, Ferrucci L, Horvath S (2019) DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 11:303–327 . 10.18632/aging.101684 [DOI] [PMC free article] [PubMed]
- 46.DNA Methylation Age Calculator. https://dnamage.genetics.ucla.edu/home
- 47.Seabold S, Perktold J (2010) statsmodels: econometric and statistical modeling with python. In: 9th python in science conference
- 48.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, van der Walt SJ, Brett M, Wilson J, Millman KJ, Mayorov N, Nelson ARJ, Jones E, Kern R, Larson E, Carey CJ, Polat İ, Feng Y, Moore EW, VanderPlas J, Laxalde D, Perktold J, Cimrman R, Henriksen I, Quintero EA, Harris CR, Archibald AM, Ribeiro AH, Pedregosa F, van Mulbregt P, SciPy 1.0 Contributors. Vijaykumar A, Bardelli AP, Rothberg A, Hilboll A, Kloeckner A, Scopatz A, Lee A, Rokem A, Woods CN, Fulton C, Masson C, Häggström C, Fitzgerald C, Nicholson DA, Hagen DR, Pasechnik DV, Olivetti E, Martin E, Wieser E, Silva F, Lenders F, Wilhelm F, Young G, Price GA, Ingold G-L, Allen GE, Lee GR, Audren H, Probst I, Dietrich JP, Silterra J, Webber JT, Slavič J, Nothman J, Buchner J, Kulick J, Schönberger JL, de Miranda Cardoso JV, Reimer J, Harrington J, Rodríguez JLC, Nunez-Iglesias J, Kuczynski J, Tritz K, Thoma M, Newville M, Kümmerer M, Bolingbroke M, Tartre M, Pak M, Smith NJ, Nowaczyk N, Shebanov N, Pavlyk O, Brodtkorb PA, Lee P, McGibbon RT, Feldbauer R, Lewis S, Tygier S, Sievert S, Vigna S, Peterson S, More S, Pudlik T, Oshima T, Pingel TJ, Robitaille TP, Spura T, Jones TR, Cera T, Leslie T, Zito T, Krauss T, Upadhyay U, Halchenko YO, Vázquez-Baeza Y. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay É. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 50.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. doi: 10.18637/jss.v033.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chu AY, Tin A, Schlosser P, Ko Y-A, Qiu C, Yao C, Joehanes R, Grams ME, Liang L, Gluck CA, Liu C, Coresh J, Hwang S-J, Levy D, Boerwinkle E, Pankow JS, Yang Q, Fornage M, Fox CS, Susztak K, Köttgen A. Epigenome-wide association studies identify DNA methylation associated with kidney function. Nat Commun. 2017;8:1286. doi: 10.1038/s41467-017-01297-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qiu C, Hanson RL, Fufaa G, Kobes S, Gluck C, Huang J, Chen Y, Raj D, Nelson RG, Knowler WC, Susztak K. Cytosine methylation predicts renal function decline in American Indians. Kidney Int. 2018;93:1417–1431. doi: 10.1016/j.kint.2018.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morgado-Pascual JL, Marchant V, Rodrigues-Diez R, Dolade N, Suarez-Alvarez B, Kerr B, Valdivielso JM, Ruiz-Ortega M, Rayego-Mateos S. Epigenetic modification mechanisms involved in inflammation and fibrosis in renal pathology. Mediators Inflamm. 2018;2018:1–14. doi: 10.1155/2018/2931049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wing MR, Devaney JM, Joffe MM, Xie D, Feldman HI, Dominic EA, Guzman NJ, Ramezani A, Susztak K, Herman JG, Cope L, Harmon B, Kwabi-Addo B, Gordish-Dressman H, Go AS, He J, Lash JP, Kusek JW, Raj DS, for the Chronic Renal Insufficiency Cohort (CRIC) Study DNA methylation profile associated with rapid decline in kidney function: findings from the CRIC Study. Nephrol Dial Transplant. 2014;29:864–872. doi: 10.1093/ndt/gft537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ebert T, Pawelzik S-C, Witasp A, Arefin S, Hobson S, Kublickiene K, Shiels PG, Bäck M, Stenvinkel P. Inflammation and premature ageing in chronic kidney disease. Toxins. 2020;12:227. doi: 10.3390/toxins12040227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cobo G, Lindholm B, Stenvinkel P. Chronic inflammation in end-stage renal disease and dialysis. Nephrol Dial Transplant. 2018;33:iii35–iii40. doi: 10.1093/ndt/gfy175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kooman JP, Dekker MJ, Usvyat LA, Kotanko P, van der Sande FM, Schalkwijk CG, Shiels PG, Stenvinkel P. Inflammation and premature aging in advanced chronic kidney disease. Am J Physiol-Ren Physiol. 2017;313:F938–F950. doi: 10.1152/ajprenal.00256.2017. [DOI] [PubMed] [Google Scholar]
- 58.Sato Y, Yanagita M. Immunology of the ageing kidney. Nat Rev Nephrol. 2019;15:625–640. doi: 10.1038/s41581-019-0185-9. [DOI] [PubMed] [Google Scholar]
- 59.Diaz-Ricart M, Torramade-Moix S, Pascual G, Palomo M, Moreno-Castaño AB, Martinez-Sanchez J, Vera M, Cases A, Escolar G. Endothelial damage, inflammation and immunity in chronic kidney disease. Toxins. 2020;12:361. doi: 10.3390/toxins12060361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCrory C, Fiorito G, McLoughlin S, Polidoro S, Cheallaigh CN, Bourke N, Karisola P, Alenius H, Vineis P, Layte R, Kenny RA (2019) Epigenetic clocks and allostatic load reveal potential sex-specific drivers of biological aging. J Gerontol Ser A glz241. 10.1093/gerona/glz241 [DOI] [PubMed]
- 61.Li X, Ploner A, Wang Y, Magnusson PK, Reynolds C, Finkel D, Pedersen NL, Jylhävä J, Hägg S. Longitudinal trajectories, correlations and mortality associations of nine biological ages across 20-years follow-up. eLife. 2020;9:e51507. doi: 10.7554/eLife.51507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Altara R, Gu Y-M, Struijker-Boudier HAJ, Thijs L, Staessen JA, Blankesteijn WM. Left ventricular dysfunction and CXCR3 ligands in hypertension: from animal experiments to a population-based pilot study. PLoS ONE. 2015;10:e0141394. doi: 10.1371/journal.pone.0141394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koper O, Kamińska J, Sawicki K, Kemona H (2018) CXCL9, CXCL10, CXCL11, and their receptor (CXCR3) in neuroinflammation and neurodegeneration. Adv Clin Exp Med 27:849–856 . 10.17219/acem/68846 [DOI] [PubMed]
- 64.Chua J, Vania M, Cheung CMG, Ang M, Chee SP, Yang H, Li J, Wong TT. Expression profile of inflammatory cytokines in aqueous from glaucomatous eyes. Mol Vis. 2012;18:431–438. [PMC free article] [PubMed] [Google Scholar]
- 65.Gao H-F, Cheng C-S, Tang J, Li Y, Chen H, Meng Z-Q, Chen Z, Chen L-Y (2020) CXCL9 chemokine promotes the progression of human pancreatic adenocarcinoma through STAT3-dependent cytotoxic T lymphocyte suppression. Aging 12:502–517 . 10.18632/aging.102638 [DOI] [PMC free article] [PubMed]
- 66.Suehiro A, Imagawa T, Hosokawa H, Suehiro M, Ohe Y, Kakishita E. Age related elevation of serum macrophage colony stimulating factor (M-CSF) level. Arch Gerontol Geriatr. 1999;29:13–20. doi: 10.1016/S0167-4943(99)00015-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data (except DNA methylation) generated during this study are included in this published article (and its supplementary information files). DNA methylation data is available from the corresponding author on a reasonable request.