Abstract
Ultraviolet-visible (UV-Vis) absorption spectra are routinely collected as part of high-performance liquid chromatography (HPLC) analysis systems and can be used to identify chemical reaction products by comparison to reference spectra. Here, we present UV-adVISor as a new computational tool for predicting UV-Vis spectra from a molecule’s structure alone. UV-Vis prediction was approached as a sequence-to-sequence problem. We utilized Long-Short Term Memory and attention-based neural networks with Extended Connectivity Fingerprint diameter 6 or molecule SMILES to generate predictive models for UV-spectra. We have produced two spectrum datasets (Dataset I, N = 949 and Dataset II, N = 2222) using different compound collections and spectrum acquisition methods to train, validate, and test our models. We evaluated the prediction accuracy of the complete spectra by the correspondence of wavelengths of absorbance maxima and with a series of statistical measures (the best test set median model parameters are in parentheses for Model II), including RMSE (0.064), R2 (0.71), and dynamic time warping (DTW, 0.194) of the entire spectrum curve. Scrambling molecule structures with experimental spectra during training resulted in a degraded R2, confirming the utility of the approaches for prediction. UV-adVISor is able to provide fast and accurate predictions for libraries of compounds.
Keywords: Attention-based Neural Network, Machine learning, Long-Short Term Memory, UV-VIS Spectrum Prediction, Dynamic Time Warping
Graphical Abstract
INTRODUCTION
Molecules absorb ultraviolet (UV) and visible (Vis) light with excitation of their electrons to higher energy molecular orbitals. The intensity of absorption varies as a function of wavelength, with greatest absorption corresponding to wavelengths having the energies of allowed electronic transitions. This variation, the absorption spectrum,1 underpins UV-Vis spectroscopy, a commonly used technique to characterize and quantify a variety of analytes, including solutions of macromolecules, conjugated organic compounds, and transition metal ions2.
Because the UV-Vis spectrum of a compound is sensitive to its structure, UV-Vis spectroscopy can be used to identify molecules with reliability comparable to that of low-resolution MS-MS4. Thus, UV-Vis spectroscopy is useful as a rapid, inexpensive, and non-destructive confirmatory tool in chemical synthesis and purification and natural product isolation. Routine analysis of compounds by high-performance liquid chromatography (HPLC) often involves a photodiode array (PDA) detector that measures UV-Vis spectra continuously during a chromatographic separation. UV-Vis spectroscopy is also used to monitor chemical reactions in situ, such as in flow reactors3. However, identification of a compound from its UV-Vis spectrum requires comparison to an experimental or predicted reference spectrum.
Development of dyes for biotechnology, genomics, immunoassays, and drug discovery utilizes different chromophores and makes frequent use of the UV-Vis absorbance spectra of molecules. A predicted spectrum for novel dyes would accelerate this process with value in automated molecular design and synthesis and analytical research.
The UV spectrum of a compound is also valuable for predicting other important optical and chemical properties, such as phototoxicity, which must be evaluated for potential drugs prior to Phase III clinical trials4. The ability to accurately predict UV spectra at the earliest stages of drug discovery, before compound synthesis, would be highly beneficial and cost-effective, versus embarking on a compound that might later be identified with this toxicity liability. Recent efforts have compiled data on compounds known to be phototoxic in in vitro assays, used for machine learning with quantum chemical descriptors producing accuracies between 83–85%4. Predicting the UV-Vis spectrum of a compound before synthesis and experimental testing also offers advantages in terms of avoiding molecules that interfere with high throughput assays5 and benefits in terms of cost of manufacture and speed.
Ab initio time dependent-density functional theory (TD-DFT) calculations are often used to predict electronic absorption spectra1 or the wavelengths of maximum absorbance (λmax) for compounds to aid in numerous applications4, 6–13. Such quantum chemistry approaches have been developed for decades with only modest success in spectrum prediction (Table S1). Hence, efficient and accurate UV spectrum prediction is still an unsolved problem. Alternative approaches to predicting a UV-Vis spectrum from molecular structure alone without resorting to quantum mechanical calculations would offer a quicker, and potentially more informative route for large collections of molecules. Recently, course-grained models have been developed for predicting absorption spectra for optoelectronic polymers using recurrent neural networks14. However, machine learning approaches to predicting the UV-Vis spectra for small molecules has not been described.
A challenge in the application of machine learning to prediction of UV-Vis absorption spectra is the paucity of available training data. There are few open-source databases of UV-Vis absorption spectra, and most focus only on λmax rather than the full spectrum within a useful wavelength range. The currently available databases with UV-Vis spectra include the Max Weaver dye library15, NIST Chemistry Webbook16, PhotochemCAD17, UV/Vis+ photochemistry database18 and the DSSC Database19 ranging from hundreds to several thousand molecules20. Few of these databases provide full spectra for use in machine learning, and most are biased toward specific classes of molecular structures, particularly dyes. PhotochemCAD provides spectra of ~339 entries for download; however, the wavelength range over which the spectra are measured varies, making compilation for machine learning purposes difficult17. Commercial UV-Vis spectral databases are also available including the KnowItAll UV-Vis spectral database collection from Wiley including over 30,000 spectra, with over 60% of them covering a narrow range 200–350 nm21.
These technical needs motivated us to develop our own datasets of spectra for a diverse collection of small molecules (Figure 1A, Supporting Information datasets I and II). We have now used these data with multiple machine learning approaches to reliably predict spectra for new molecules (UV-adVISor). We have used multiple measures to compare predicted to experimental spectra, including root mean square error (RMSE), R2, mean absolute error (MAE), RMSE of derivative spectra, and dynamic time warping (DTW), which is a distance measure technique that allows a non-linear mapping between two signals by minimizing the distance between them22. Altogether, our approach does not require time-intensive quantum chemistry calculations and provides accurate, multiple-wavelength spectrum predictions (across a complete spectrum rather than just the λmax), comparable to or better than currently used models.
Figure 1.
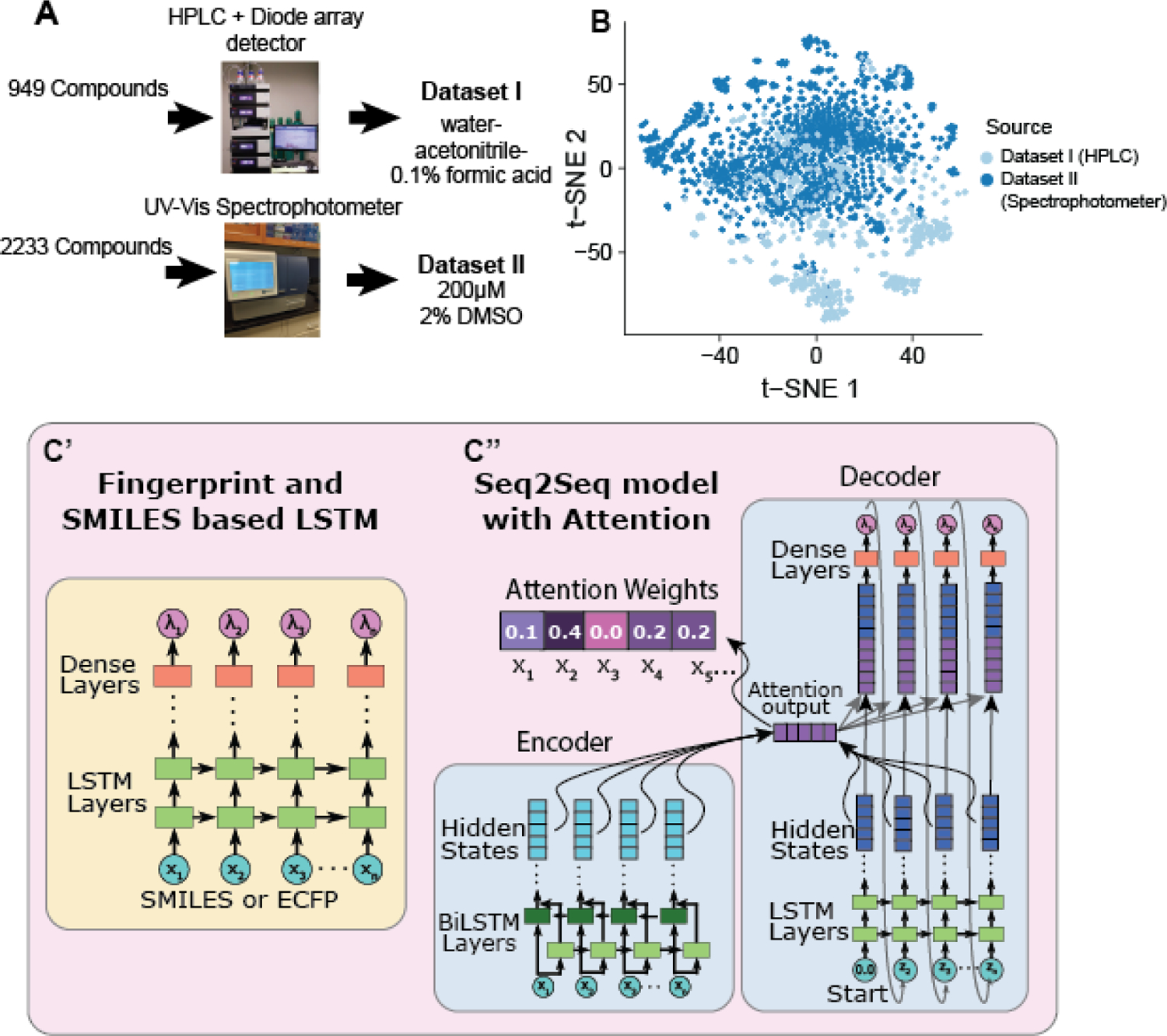
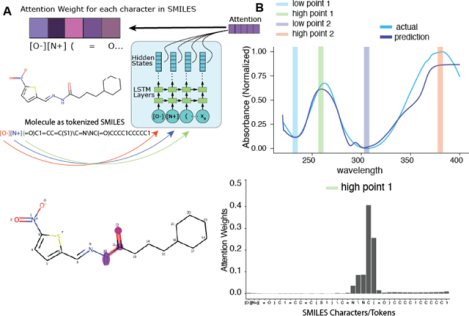
A. Overview of the experimental workflows for generating data with PDA or plate reader. B. t-distributed stochastic neighbor embedding (t-SNE) plot of chemical structure overlap between compounds generated by HPLC and spectrophotometer. Compounds that are structurally similar are close together in 2D space. No compounds are duplicated between the two datasets. C. Two LSTM architectures used for spectrum prediction. Left: LSTM model composed of LSTM layers followed by dense layers for the output, which takes in a SMILES string or ECFP6 as input. Right: Architecture of our Seq2Seq model with attention, which uses bi-directional LSTMs for the encoder and Luong attention and takes in SMILES strings.
EXPERIMENTAL
Compound libraries.
Absorbance spectra were acquired for a diverse set of compounds from SRI’s internal collection (393 compounds) and a collection of compounds purchased from OTAVA Chemicals (MMP2 Targeted Library, 596 compounds). Compounds were diluted to 200 μM with methanol or DMSO and arrayed in 96-well plates for analysis by HPLC with spectrum acquisition. The MicroSource Spectrum screening compound library of 2222 compounds (MicroSource Discovery Systems, Inc., Gaylordsville, CT, USA) was a generous gift from Dr. Ethan Perlstein, (Perlara).
UV-Vis Spectrum Acquisition.
Compounds for Dataset I were analyzed by HPLC using a Thermo Dionex Ultimate U3000 UPLC system equipped with a Thermo LCQ Fleet ion trap MS, a DAD-3000RS diode array detector (DAD), and a C18 column. The mobile phase was water-acetonitrile-0.1% formic acid, with an acetonitrile gradient.
The retention time for the compound of interest in each chromatographic run was determined from the extracted ion chromatogram (XIC). The XIC was scanned for the largest peak at the expected mass. When found, the peak was fit with a Gaussian and was accepted if it met constraints for lineshape (Gaussian FWHM < 0.1) and elution time greater than the void volume of 1.2 min. This process eliminated compounds that had no mass response or potential co-elution with sample impurities. It resulted in inclusion of spectra for 949 compounds from the starting set of 989. For each accepted chromatogram, an empirically determined time-offset was applied to extract the UV-Vis spectrum (200 nm to 800 nm) for that compound from the DAD data.
Background due to HPLC mobile phase absorption was subtracted from each spectrum. Due to the gradient in acetonitrile concentration, the background spectrum depended on the elution time of the analyzed compound. To assess the background at the relevant elution time for each compound, the minimum signal at each wavelength was extracted from the set of all spectra collected at that elution time for a given plate of compounds. The minimum signal from the set was taken to be the background without contribution from analytes or compound-specific impurities. The resulting inferred background spectrum for the relevant elution time was subtracted from the measured spectrum of each compound. The background-subtracted spectra were truncated (220 nm to 400 nm) and scaled by setting the minimum absorbance to zero and normalizing to a maximum absorbance of 1.0.
Compounds for Dataset II were obtained as 10 mM solutions in 100% DMSO. Each compound was diluted 50-fold (to 200 mM and 2% DMSO) with water and transferred to black, clear-bottom Greiner UV-STAR microplates. The UV absorption of each compound was read in a SpectraMax iD5 Multi-Mode Microplate spectrophotometer from 230nm to 400nm in 1 nm increments. The resulting spectra were scaled by setting the minimum absorbance to zero and normalizing to a maximum absorbance of 1.0.
Dataset preparation.
SMILES for the Dataset I compounds were exported from a CDD vault. Molecules were prepared as follows: Salts were removed and molecules were neutralized if possible. Molecules were converted into their canonical SMILES format using RDKit. Duplicates were then removed from the dataset.
Machine learning methods.
Spectrum prediction makes use of a Deep Learning Machine Learning algorithm called LSTM (Long-Short Term Memory) model23 (Figure 1). We use wavelength windows from 220 to 400nm for the spectra from Dataset I and 230 to 400nm for spectra from Dataset II (due to the wavelength limitations of the spectrophotometer). For input, we considered four different data representations: 1024-bit or 2048 bit ECFP6 fingerprint, a compressed fingerprint, and the tokenized SMILES string as parameters along with the full wavelength values for each molecule to build a model. Further details on the machine learning methods, server details, t-SNE visualization, clustering of spectra and spectrum comparison measures and can be found in the Supporting information Methods.
RESULTS
Overview of UV-adVISor.
UV-adVISor is a new tool to enable a scientist to obtain predicted UV-Vis absorption spectra for input molecules using standard structure representations such as SDF24 and SMILES.25 Initially we tested several feed forward machine leaning models, however they all failed to converge (Figure S1). machine learning algorithm built from a Long Short-Term Memory (LSTM) network architecture to predict relative absorbance at wavelengths within a trained range (Figure 1) performed the best. To cover a wider range of applicability, we have trained two models, each with a different dataset which covers different chemical property space (Figure 1B). Dataset I was generated from a compound collection combining an internal chemical inventory and a commercial compound library. Spectra for these compounds were obtained with a PDA detector interfaced with a HPLC, elution time of each sample compound being judged by its initial detection with an in-line mass spectrometer. Dataset II was generated from a commercially obtained (MicroSource Spectrum) collection of drugs. Spectra for these compounds were obtained with a spectrophotometer using a multi-well plate format. Generating two datasets using two distinct methods allowed us to demonstrate the wider applicability of UV-Vis based models, as UV-Vis spectra can often be distinct based on conditions such as the solvent composition and the pH. The spectra in both data sets were baseline corrected (minimum value in wavelength range offset to 0) and normalized (maximum value in wavelength range set to 1). For each model, we used 70% of the compounds for training, 15% for validation, and 15% for testing. Our first set of models used LSTM layers to read SMILES sequences or an ECFP6 fingerprint (Figure 1C, left). We also used a second model architecture, taking advantage of recent advancements in using encoder-decoder architectures26, 27 with an attention mechanism for language translation (Figure 1C, right). This second network architecture is motivated by approaching spectrum prediction as a sequence to sequence (Seq2Seq) translation problem between a chemical structure (represented by SMILES string) and a wavelength sequence output. The final models are readily accessible through a web interface (https://www.collaborationspharma.com/uvadvisor), where the user can input a structure in 2D or SMILES format, and UV-AdVISor outputs the predicted spectrum as a graph or in .csv format.
UV-adVISor Enables Accurate Spectrum Predictions.
Models generated using Extended Connectivity Fingerprint Diameter 6 (ECFP6)28, 29 molecular representations as inputs to the LSTM network produced hiqh-quality predictions of spectra for test compounds. Representative examples from the model using Dataset I are shown in Figure 2. The full data set is available in Supporting Information data File 1. Many of the predictions accurately render absorption maxima, minima, and shoulders and good approximations of relative absorption across the wavelength range of the spectra. The best predicted spectrum had a RMSE of predicted versus measured spectra of 0.005 (SRI-1053215). Qualitatively, we assess RMSE values of less than 0.10 as “excellent”, values less than 0.20 as “good”, and anything at or above 0.25 as a “poor” prediction (Figure 2B). We obtained comparable prediction accuracy, as judged by RMSE (Table S2), with a model that used 2048 bit or 1024 bit ECFP6 descriptors (see Methods). The median RMSE for both sets of predictions is ~0.17. However, further compression of the fingerprint resulted in substantial degradation of the prediction quality (median RMSE = 0.21). Using tokenized SMILES as the molecular representation produced predictions of quality comparable to those produced with the uncompressed ECFP6 (median RMSE = 0.17). Using a Seq2Seq model resulted in the best predictive model (as judged by median RMSE = 0.15). Training the model with scrambled data, in which the compounds are paired randomly with spectra from the dataset, resulted in poor predictions as one would expect. The average median RMSE for predictions made with LSTM models trained with three randomly scrambled sets using 2048 bit ECFP6 was degraded to 0.25. Comparison of this performance metric with the that of the trained model with the correctly paired spectra and compounds confirms that the model has successfully learned structure-spectrum relationships. Certainly, there are other performance metrics which could be considered, for example peak-wavelength predictions.
Figure 2.
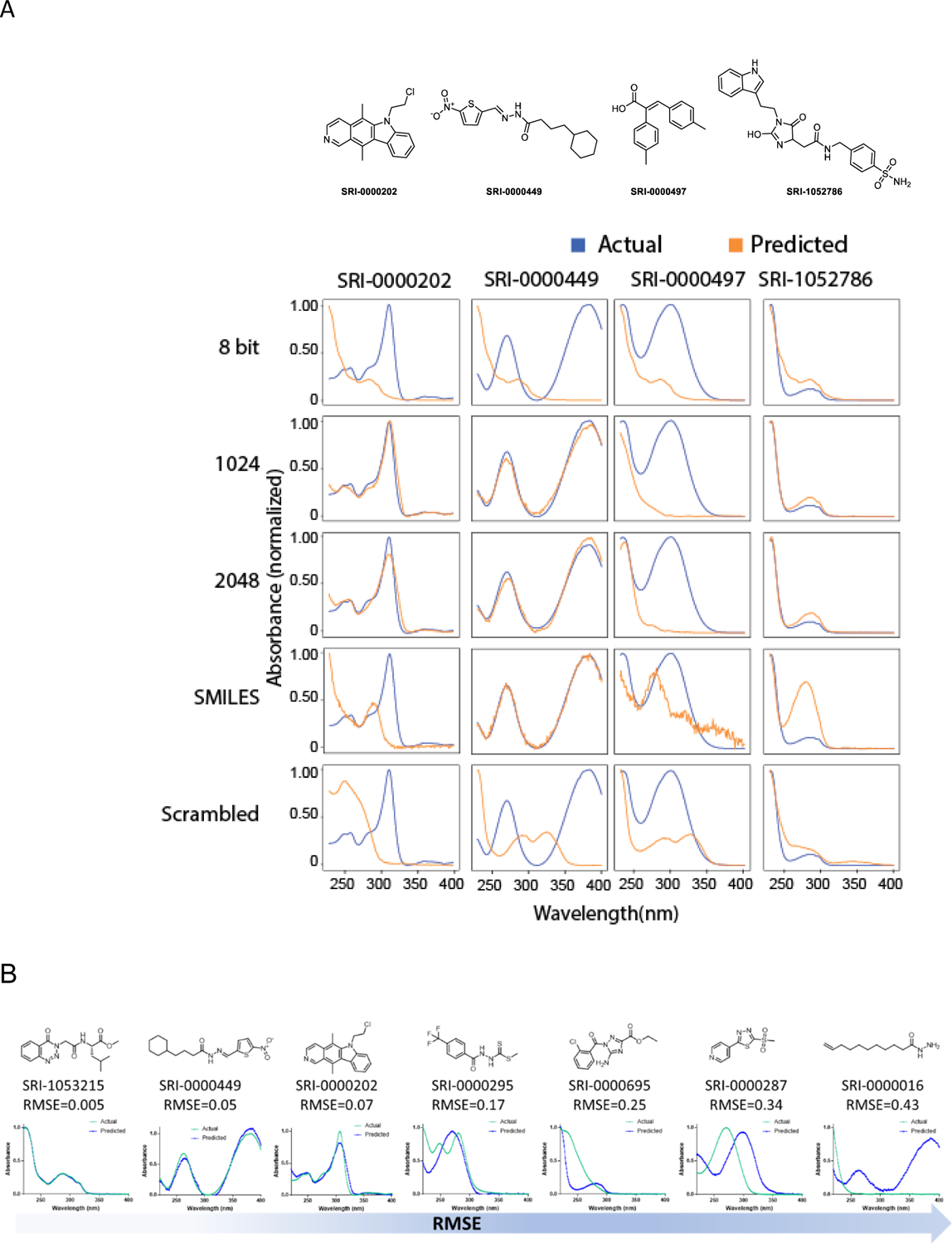
A. Comparison of different molecular descriptors to predict UV spectra for different representative molecules from Dataset I. B. Illustration of structures and spectra with varying qualities of prediction judged by RMSE.
UV-adVISor trained on different data sources.
Dataset I was produced on an HPLC-PDA system, modeling the type of analytical system used in a typical organic chemistry lab. Dataset II was directly read on a UV-Vis spectrophotometer, representing a faster data collection methodology, but without the chromatographic separation afforded by the HPLC-PDA system. Machine learning models trained using Dataset II were also found to provide accurate predictions (Supporting Information dataset 2), suggesting UV-adVISor is widely applicable to a variety of different detection methods. As with Dataset I, the median RMSE was comparable using the 2048 bit ECFP6 descriptor or the 1024 bit ECFP descriptor (Table S3). Using either descriptor, the median RMSE of the predictions was substantially lower than predictions using the model trained with Dataset I, (0.06–0.08 vs 0.17). The average median RMSE for predictions made with models trained with three randomly scrambled sets from Dataset II was 0.1, also substantially lower than the scrambled RMSE for Dataset I, which was 0.25.
Inspection of the datasets reveals that Dataset II, while derived from a diverse set of compounds, appeared to have a relatively low diversity of spectrum profiles in the training and test sets, with a large number of spectra having few or no features above ~240 nm. (Figure S2). To quantify this difference in diversity, we measured the average of the standard deviation of each wavelength value for both datasets. Dataset I had an average standard deviation of 0.23, while Dataset II had an average standard deviation of 0.08, indicating a lower diversity of spectra. Second, we used shape-based distance to divide the spectra into 25 distinct clusters. Dataset I exhibits a higher inter-cluster diversity compared to Dataset II (Figure S2A). Using the silhouette method30 (see Methods) to determine the optimal number of clusters, Dataset I is determined to have 4 major clusters, and Dataset II has 3 major spectrum clusters, consistent with the lower spectral diversity of Dataset II (Figure S2B). Because of this lower spectra diversity, the model trained and tested with Dataset II has a greater statistical probability of predicting the shape of the spectrum when trained with the actual data or the scrambled data (Table S3). This analysis again shows the importance of evaluating the model relative to a scrambled dataset, which captures the overall spectrum diversity for a given dataset. It also confirms that the model is able to learn structure-spectrum relationships for Dataset II. (Table S3).
Comparison of Measures of Prediction Accuracy.
To our knowledge no single measure of the difference between predicted and actual UV-Vis spectra has been previously adopted as an ideal metric for comparisons. Most comparisons of predicted spectra to measured spectra only consider λmax31, whereas our models predict the entire spectrum over a wavelength range. Therefore, we have applied a series of quality metrics to evaluate the predictions of UV-adVISor. In addition to RMSE, other commonly applied metrics are R2 and Mean Absolute Error (MAE). Applied to Dataset I, Median R2 was similar for 1024 bit ECFP6 and SMILES representations (~0.63) and lowest for the scrambled average (0.12). Median MAE was lowest for SMILES (0.10) and increased to 0.17 for the scrambled average for Dataset I (Table S2). A similar trend was observed using Dataset II, with stronger measures of concordance for both authentic and scrambled data (Table S3).
In addition, we have applied novel metrics aimed at emphasizing correct prediction of key features of a spectrum. DTW is an approach for comparing data series by finding the optimal match between the series. Applied to spectra, it allows comparison of spectrum shapes when features of the compared spectra are shifted in wavelength22. Thus, in principle, DTW is more robust than measures such as RMSE for comparing spectrum shapes and could also be used for shape-based classification32. We have generated DTW for the test spectra in each dataset and found it correlated with RMSE (R2 > 0.6, Figure S3). DTW therefore provides an interpretable method to compare predicted and observed spectra to assess machine learning prediction quality. For Dataset I, the median DTW showed considerable variability between 1024 bit, 2048 bit ECFP6 and SMILES representations (0.71–1.03, Table S2). Similarly, for Dataset II the median DTW shows a similar spread (0.194–0.232, Table S3) on a narrower scale, suggesting the error is generalizable, and therefore the 1024 bit ECFP6 was selected as the more favorable model in the latter case.
We have also applied the RMSE between the derivatives of the predicted and actual spectra to emphasize correct prediction of absorption maxima and minima (Table S3). The derivative is obtained using a forward finite-difference approximation applied to wave value at each wavelength:
where xt is value of the current wavelength, xt−1 is the value of the next nm wavelength measured, and δt is the difference between the two wavelengths (in our case, 1 nm for all spectra wavelength increments).
This measure was the lowest for the 1024 bit ECFP6 and highest for SMILES in Dataset I while being intermediate for the scrambled data. In contrast, SMILES and 2048 bit ECFP6 showed comparable RMSE. SMILES is an end-to-end model; using the encoded SMILES string as an input, whereas ECFP6 are features calculated from the molecule. It is possible that the end-to-end learning of SMILES, while requiring more data, is capable of learning a similar feature representation as fingerprints given a large enough dataset.
Based on the assessment of chemists in our group, none of these statistical measures adequately evaluates the utility of a predicted spectrum to the chemist’s task of identifying a compound or distinguishing a compound from others. We have therefore applied functional tests to the quality of spectra predicted with UV-adVISor based on the correspondence of peaks, i.e., wavelengths of local absorption maxima, with actual spectra. In one such test, the predicted spectrum is judged “useful” if 1) it has an equal number of local absorbance maxima within a defined wavelength range as the actual spectrum and 2) each of the peaks is within 15 nm of a corresponding peak in the actual spectrum (Table S4).
For the LSTM model trained with Dataset I (2048-bit ECFP6), 58 of 150 predictions (39%) meet these criteria. For the Seq2Seq model trained with Dataset I, 47 of 150 predictions (31%) meet these criteria. In contrast, spectra calculated using models trained with three random scrambles of Dataset I afford only 11, 15, and 17 of 150 predictions (7%, 10%, and 11%, respectively) that meet these criteria. For the model trained with Dataset II (2048-bit ECFP6), 235 of 330 predictions (71%) meet these criteria. Spectra calculated using models trained with three random scrambles of Dataset II afford 175, 181, and 184 of 330 predictions (53%, 55%, and 56%, respectively) that meet these criteria. As can be seen from these examples, the fraction of calculated spectra meeting this functional standard is roughly correlated with the median RMSE for the calculated spectra. However, at the level of individual spectra, this correlation is weak, because the functional criteria do not penalize a predicted spectrum for large deviations of absorption intensity from the actual spectrum; whereas, RMSE does penalize such deviations.
Spectrum Predictions for Additional Compounds.
After all model test sets were used for evaluation, a prediction was performed on a 17-compound external test set (Dataset III, Supporting information). Though there was no overlap of these compounds with Dataset I, 8 of the 17 were found in Dataset II. Therefore, we only made predictions using the model built with Dataset I. Similar to the test set, both the LSTM model trained with ECFP6 (1024) and the Seq2Seq model had comparable median RMSE (Table S5) for Dataset III (Supporting Information Data 3). Both models had a higher RMSE and lower Median R2 than the test or Dataset III which might be explained by the 17 compounds containing a variety of spectral shapes in comparison to the training, test, and validation sets, which had a number of similar spectrum peaks.
UV-adVISor predictions and molecule similarity to training set.
Chemical space is infinite33. Therefore, it would be unexpected for machine learning models trained with hundreds to thousands of molecules to correctly predict a UV-Vis spectrum for all possible new molecules. We discovered that UV-adVISor was capable of predicting near-identical spectral curves for some compounds but missed important features for others (Figure 2).The t-distributed stochastic neighbor embedding (t-SNE) plots34 (See Supporting Information Experimental) of structural similarity (based on ECFP6 fingerprints) suggests that predictive power is determined by training and test set overlap. Where the density of training examples is sparse in relation to the density of the test examples, the MAE of predictions is generally higher (Figure S4). This observation suggests that the reliability of predictions can be improved with sufficient representation in the training set of the model. The additional compounds (Dataset III) were also well distributed in the t-SNE plot for the Dataset I (Figure S5) suggesting they were likely within the applicability domain of this model.
Evaluating chemical-substructure contributions to spectrum prediction by exploiting model attention weights.
One of the advantages of using a Seq2Seq model with attention is the ability to visualize the attention mechanism27. In our Seq2Seq model, compounds are represented as tokenized SMILES strings. Upon generation of each wavelength value, a corresponding vector of weights over each character in the input is generated (Figure 3A). This vector of weights describes what parts of the input the model is “paying attention to” at each prediction step. Although caution must be used to not make direct inference from attention alone, we can exploit this mechanism to observe what part the compound structure the model is paying attention to and derive substructure importance from UV-Vis spectra. We chose a spectrum that was predicted with reasonable accuracy for example (Figure 3B). Here, we chose two “low points” and two “high points” and observed the attention weights for each. At the lowest wavelengths, the model’s attention is not focused on any part of the input (Figure 3C, top-left). During the first peak, however, the model is focused on the amide group. The second low point on the spectrum shows a focus on the thiophene ring, and the λmax indicates attention focus on the nitro group. This type of structure-spectrum analysis may also inform efforts to develop rules to calculate the λmax based on substructure features35, 36.
Figure 3:
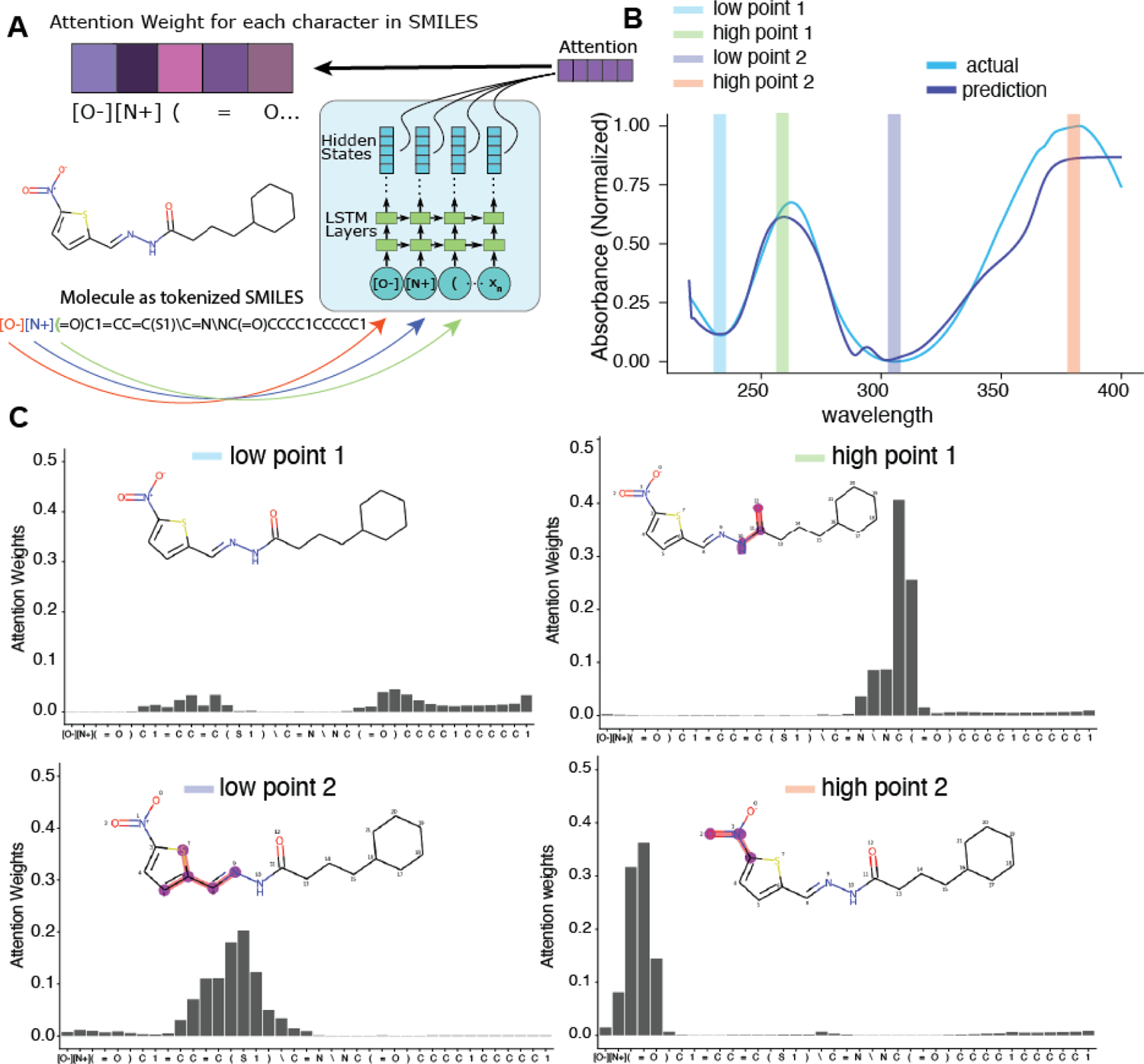
Exploration of the Seq2Seq model’s attention weights. A. Graphic showing the encoder side of Seq2Seq and the generation of an attention weight vector for each tokenized SMILES input. B. Example spectra and selected wavelengths at which the attention weights are visualized. C. Attention weights for each token SMILES input for each of the four chose wavelengths. At each prediction step, the attention weights focus on the most relevant SMILES input token as represented by the weight value.
DISCUSSION
In practice, UV-Vis spectra are most commonly used in reference to specific qualified standards or spectral libraries. The theoretical prediction of spectra has not achieved sufficient accuracy for routine use in chemistry labs, particularly for chemists analyzing mixtures of crude reaction products or extractions. In contrast, predictive tools for NMR and FT-IR spectra are used by almost all synthetic chemists in identification, characterization and structural elucidation of novel compounds (e.g. NMR predictor software, ACDlabs)37. Given that chemists routinely collect UV-Vis spectral data as part of standard HPLC analysis workflows, these data are essentially “free” and underutilized by them. The ability to accurately predict UV-vis spectra de novo would enable chemists to more easily identify compounds of interest without the need for qualified reference standards.
The most commonly used method to date for UV-Vis spectrum prediction is TD-DFT (Table S1) using CAM-B3LYP functionals.1 This approach requires quantum chemistry software, significant computing resources, and expertise in their use and interpretation. Nevertheless, it has been used in hundreds of publications for diverse range of compounds. Most of these publications report studies of individual compounds or at most a few analogs, and the experimental data for the various studies have been generated in a variety of solvents, limiting their value as a spectrum database. Most measure agreement between prediction and experiment only at λmax, providing at best, a qualitative assessment of agreement for other spectral features. In many cases, the predicted values of λmax are significantly different than those observed.
Though the limitations of purely theoretical approaches to predicting UV-Vis spectra hinder the application of these approaches to compound identification and characterization in organic chemistry, chemists routinely use empirical rules to make qualitative or partial predictions of compounds’ UV-Vis absorbance behavior. The utility of such methods suggests the potential for data-driven approaches such as machine learning to prediction of UV-Vis spectra. Key issues that we have addressed to realize this objective are the availability of sufficient data for training, validating, and testing ML algorithms; the relationship between the content of training data and the reliability of predictions; machine readable (i.e., vector) representations of molecular structure that capture sufficient detail to generalize structure-spectrum relationships; network architectures that output predicted spectra that are continuous across a wavelength range; and useful metrics for assessing the predictive power of ML models.
Despite the routine nature of UV-Vis spectrum acquisition, assembly of a sizeable dataset from publicly available sources that meets the needs of training and testing for spectrum prediction was not possible. Existing publicly available datasets are inadequate because they lack full spectra across a consistent wavelength range (rather than λmax only or varying wavelength ranges), absorption values across the wavelength range (rather than plotted spectra only), consistent solvent environments (solvent composition and pH), or a diversity of molecular structures (e.g. the compound sets often being focused on an analogous series of compounds such as dyes). Data harvested piecewise from the literature suffered similar deficits. We sought to avoid these limitations in the construction of our own datasets.
The library of compounds we used to construct Dataset I comprised an internal collection aggregated from a variety of projects with a range of objectives undertaken at SRI. In addition to emulating the type of analytical system used in a typical organic chemistry lab, the HPLC methodology that we used for collection of Dataset I ensured that the spectra we analyzed were of pure compounds. The larger library of compounds in Dataset II from a commercial vendor comprised a wide range of drugs and natural products. By using these two datasets, we have created machine learning models that relate to a broader range of compound classes than literature datasets created primarily using dyes.
At the outset, it was unknown how much data would be required for machine learning models to learn structure-UV-VIS spectrum relationships to generalize to new molecules. We found that surprisingly small datasets can result in accurate predictions for new molecules. With less than 1000 molecules, we can obtain good levels of accuracy of prediction as judged by median statistics. Not surprisingly, it appears that the quality of spectrum prediction depends on the overlap between the chemical space of the training data and the compounds for which predictions are made (Figure 3). Similarly, we find that the accuracy of predictions depends on the similarity of spectral profiles between the training compounds and the compounds for prediction. Future work will expand our datasets to cover more chemical space, which we anticipate will improve the reliability of predictions. Understanding whether models for different solvent conditions are required or whether we can reliably extend datasets to create “generic UV-Vis spectrum models” will also be important to assess.
The LSTM network architectures we have employed are well-suited to the modeling of UV-Vis spectra. The recurrent structure of the LSTM architecture facilitates the modeling of spectra as continuous data series. Such models are particularly apt for UV-Vis spectra, which are typically smooth functions with broad features. The LSTM models described can be generated in minutes, and molecule predictions are processed in seconds. The Seq2Seq model with attention provided predictive accuracy comparable to the LSTM model that we tested and represents a novel method for visualizing what parts of the chemical substructure are most relevant to the prediction at hand. To our knowledge this is also the first use of an attention mechanism for probing substructure-UV-Vis prediction relevance. While interpretation of the attention weights must be done with care (we cannot, for example infer what atom centers contribute directly to what wavelengths), attention placed on certain substructures that appear repeatedly for specific wavelength peaks may indicate a chemical feature to investigate further and could the focus of further research. We can reasonably interpret the attention placed on each atom as importance to the predictive ability and use this information to refine the model by altering the training set.
We have demonstrated that UV-Vis spectra can be predicted from molecular structure alone (i.e., without additional physics-based information) represented by either ECFP6 descriptors or SMILES. We have previously demonstrated how compression of 1024 bit fingerprints to 8 bits of information could facilitate the use of machine learning approaches on a quantum computer38 and we reasoned this same approach could be used to assess how much structural information needed to be retained for accurate model spectra prediction. The reduction to 1024 bit fingerprints did not result in any significant information loss versus the 2048 bit fingerprints, while ECFP6 8 bit compression showed a dramatic loss based on degradation of statistical measures such as RMSE (Table S2).
Development of models to predict UV-Vis spectra requires metrics to evaluate the quality of predictions. Statistical measures such as RMSE, R2, and MAE are commonly used metrics of agreement between predicted and actual values, and we have applied them to evaluate our models, to test different input formats, and to test the effect of scrambling the structure-spectrum relationship during training. We have also used MAE as the metric of loss during training of our models. We find that these measures are generally in concurrence. To the extent they differ, RMSE agrees best with our qualitative assessment of prediction quality.
Many test set predictions were remarkably close to the observed spectra (e.g., spectra in Figure 2 and Figure S6), an agreement reflected in values for RMSE, R2, and MAE. However, other spectrum predictions of our models capture important and useful features of the observed spectra in ways that are not well-reflected in these common statistical measures. For example, a small shift in wavelength of a large absorption peak results in a large contribution to RMSE but will often have a small impact on the utility of the prediction for distinguishing between two compounds. Similarly, a discrepancy in the relative height of a peak in a predicted spectrum from an actual spectrum will degrade the RMSE but have a small impact on interpretation. To address this shortcoming of standard statistical measures we have applied additional measures of prediction quality to our models, DTW and derivative spectrum RMSE.
DTW is a distance measure technique that allows a non-linear mapping between two signals by minimizing the distance between them39. This method is flexible, allowing two data series that are similar but locally out of phase to align non-linearly. It is a well-known solution for time-series alignment 46. To our knowledge, it has not been used previously for comparisons of spectra. As a measure of agreement between predicted and actual spectra, it accommodates small shifts in wavelength between spectra of similar shape. DTW is correlated with RMSE for the predictions made with our test sets (Figure S3). Median DTW is also correlated with median RMSE (Tables S2 and S3).
Comparison of derivatives of predicted and observed spectra also allows comparison of the overall shapes of spectra, emphasizing agreement in the wavelength positions of peaks and valleys, where the value of the derivative is zero irrespective of the magnitude of the absorption at those wavelengths. Derivative spectroscopy is frequently used to visualize poorly resolved spectral features and to differentiate similar spectra40. We are not aware of its use in quantitative comparison of predicted and experimental spectra. As with DTW, the trend in median values for this measure mirrors that of median RMSE.
Functional tests for assessing the quality of spectrum predictions provide a practical and intuitive measure of predictive success. The test we have described, correspondence of peak wavelengths between predicted and experimental spectra, emphasizes the peak positions over other spectrum features. Though this approach is similar to the typical analysis of results of TD-DFT predictions, which judges success by prediction of λmax values only, the measure we have applied adds the rigor of requiring that no peaks are predicted that are not in the actual spectrum.
CONCLUSION
The machine learning technique embodied in UV-AdVISor allows very large compound libraries to be scored more quickly than previous methods. Thus, it will enable chemists to more rapidly and reliably identify compounds with desirable UV-Vis spectra. It could have applications for new compound discovery (e.g. prediction of dye colors), organic chemistry reaction monitoring, phototoxicity prediction, and numerous other important chemistry applications6–13. We have also shown that alternative spectrum comparison measures such as DTW may help in assessment of observed and predicted spectra. These scores may be used in the future as elements of machine learning algorithm cost functions. Future work will include comparison of 2D and 3 descriptors, evaluation and optimization of additional machine learning algorithms41–43 as well as applying additional algorithms for selection of training and test sets. The algorithms used herein are also likely applicable to NMR and MS spectrum prediction. Generation of spectra for significantly larger training sets (tens to hundreds of thousands of molecules) will assist in broadening the scope of these computational models and be useful in training recurrent neural network models to assist in the de novo design of molecules44, 45 with a particular spectrum of interest for specific applications requiring ideal physicochemical or UV-Vis properties.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge Dr. Ethan Perlstein for providing the Microsource Spectrum library. Dr. John Byrnes is also thanked for helpful comments and Dr. Antony Williams is kindly acknowledged for discussions on spectra prediction.
Funding
DARPA (HR0011-19-C-0108; PI: P. Madrid). Distribution Statement “A” (Approved for Public Release, Distribution Unlimited). This research was developed with funding from the Defense Advanced Research Projects Agency (DARPA). The views, opinions, and/or findings expressed are those of the author and should not be interpreted as representing the official views or policies of the Department of Defense or the U.S. Government. We kindly acknowledge NIH funding to develop the software from R44GM122196-02A1 “Centralized assay datasets for modelling support of small drug discovery organizations” from NIGMS and NIEHS for 1R43ES031038-01 “MegaTox for analyzing and visualizing data across different screening systems”. “Research reported in this publication was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number R43ES031038. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.”
Footnotes
The supporting Information is available free of charge.
Supporting Information consists of: Methods, Datasets provided as a zip file, Tables S1–S6, Figures S1–S7 and References. We have also made the Supporting dataset files available on FigShare (https://doi.org/10.6084/m9.figshare.15217512) which consists of spectra and SMILES files for Datasets I–III, Data for Figures 1, 3 and 4 and data for Tables S2–S5.
Code availability statement
Code is available from the authors upon written request for non-commercial use.
Competing interests
S.E. is owner and K.B, F.U. and M.A.Z.H. work for Collaborations Pharmaceuticals, Inc. Others have no conflicts of interest. A provisional patent on this work has been filed.
REFERENCES
- 1.Gonzalez L; Escudero D; Serrano-Andres L, Progress and challenges in the calculation of electronic excited states. Chemphyschem 2012, 13 (1), 28–51. [DOI] [PubMed] [Google Scholar]
- 2.Perkampus HH, UV-VIS Spectroscopy and Its Applications. Springer-Verlag: Berlin, 1992. [Google Scholar]
- 3.Shen Y; Abolhasani M; Chen Y; Xie L; Yang L; Coley CW; Bawendi MG; Jensen KF, In-Situ Microfluidic Study of Biphasic Nanocrystal Ligand-Exchange Reactions Using an Oscillatory Flow Reactor. Angew Chem Int Ed Engl 2017, 56 (51), 16333–16337. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt F; Wenzel J; Halland N; Gussregen S; Delafoy L; Czich A, Computational Investigation of Drug Phototoxicity: Photosafety Assessment, Photo-Toxophore Identification, and Machine Learning. Chem Res Toxicol 2019, 32 (11), 2338–2352. [DOI] [PubMed] [Google Scholar]
- 5.Simeonov A; Davis MI, Interference with Fluorescence and Absorbance. In Assay Guidance Manual, Markossian S; Sittampalam GS; Grossman A; Brimacombe K; Arkin M; Auld D; Austin CP; Baell J; Caaveiro JMM; Chung TDY; Coussens NP; Dahlin JL; Devanaryan V; Foley TL; Glicksman M; Hall MD; Haas JV; Hoare SRJ; Inglese J; Iversen PW; Kahl SD; Kales SC; Kirshner S; Lal-Nag M; Li Z; McGee J; McManus O; Riss T; Saradjian P; Trask OJ Jr.; Weidner JR; Wildey MJ; Xia M; Xu X, Eds. Bethesda (MD), 2004. [Google Scholar]
- 6.Ghidinelli S; Longhi G; Abbate S; Hattig C; Coriani S, Magnetic Circular Dichroism of Naphthalene Derivatives: A Coupled Cluster Singles and Approximate Doubles and Time-Dependent Density Functional Theory Study. J Phys Chem A 2020. [DOI] [PubMed] [Google Scholar]
- 7.Anouar el H; Weber JF, Time-dependent density functional theory study of UV/vis spectra of natural styrylpyrones. Spectrochim Acta A Mol Biomol Spectrosc 2013, 115, 675–82. [DOI] [PubMed] [Google Scholar]
- 8.Martynov AG; Mack J; May AK; Nyokong T; Gorbunova YG; Tsivadze AY, Methodological Survey of Simplified TD-DFT Methods for Fast and Accurate Interpretation of UV-Vis-NIR Spectra of Phthalocyanines. ACS Omega 2019, 4 (4), 7265–7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daengngern R; Camacho C; Kungwan N; Irle S, Theoretical Prediction and Analysis of the UV/Visible Absorption and Emission Spectra of Chiral Carbon Nanorings. J Phys Chem A 2018, 122 (37), 7284–7292. [DOI] [PubMed] [Google Scholar]
- 10.Garcia RD; Maltarollo VG; Honorio KM; Trossini GH, Benchmark studies of UV-vis spectra simulation for cinnamates with UV filter profile. J Mol Model 2015, 21 (6), 150. [DOI] [PubMed] [Google Scholar]
- 11.Aguilar-Martinez M; Cuevas G; Jimenez-Estrada M; Gonzalez I; Lotina-Hennsen B; Macias-Ruvalcaba N, An Experimental and Theoretical Study of the Substituent Effects on the Redox Properties of 2-[(R-phenyl)amine]-1,4-naphthalenediones in Acetonitrile. J Org Chem 1999, 64 (10), 3684–3694. [DOI] [PubMed] [Google Scholar]
- 12.Blase X; Duchemin I; Jacquemin D; Loos PF, The Bethe-Salpeter Equation Formalism: From Physics to Chemistry. J Phys Chem Lett 2020, 11 (17), 7371–7382. [DOI] [PubMed] [Google Scholar]
- 13.Yahyaei H; Shahab S; Sheikhi M; Filippovich L; Almodarresiyeh HA; Kumar R; Dikusar E; Borzehandani MY; Alnajjar R, Anisotropy (optical, electrical and thermal conductivity) in thin polarizing films for UV/Vis regions of spectrum: Experimental and theoretical investigations. Spectrochim Acta A Mol Biomol Spectrosc 2018, 192, 343–360. [DOI] [PubMed] [Google Scholar]
- 14.Simine L; Allen TC; Rossky PJ, Predicting optical spectra for optoelectronic polymers using coarse-grained models and recurrent neural networks. Proc Natl Acad Sci U S A 2020, 117 (25), 13945–13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuenemann MA; Szymczyk M; Chen Y; Sultana N; Hinks D; Freeman HS; Williams AJ; Fourches D; Vinueza NR, Weaver’s historic accessible collection of synthetic dyes: a cheminformatics analysis. Chem Sci 2017, 8 (6), 4334–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talrose V; Yermakov AN; Leskin AN; Usov AA; Goncharova AA; Messineva NA; Usova NV; Efimkina MV; Aristova EV NIST chemistry webbook. https://webbook.nist.gov/chemistry/uv-vis/.
- 17.Taniguchi M; Du H; Lindsey JS, PhotochemCAD 3: Diverse Modules for Photophysical Calculations with Multiple Spectral Databases. Photochem Photobiol 2018, 94 (2), 277–289. [DOI] [PubMed] [Google Scholar]
- 18.Noelle A; Vandaele AC; Martin-Torres J; Yuan C; Rajasekhar BN; Fahr A; Hartmann GK; Lary D; Lee YP; Limao-Vieira P; Locht R; McNeill K; Orlando JJ; Salama F; Wayne RP, UV/Vis(+) photochemistry database: Structure, content and applications. J Quant Spectrosc Radiat Transf 2020, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venkatraman V; Raju R; Oikonomopoulos SP; Alsberg BK, The dye-sensitized solar cell database. J Cheminform 2018, 10 (1), 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beard EJ; Sivaraman G; Vazquez-Mayagoitia A; Vishwanath V; Cole JM, Comparative dataset of experimental and computational attributes of UV/vis absorption spectra. Sci Data 2019, 6 (1), 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anon KnowItAll UV-Vis Spectral Database Collection. https://sciencesolutions.wiley.com/solutions/technique/uv-vis/knowitall-uv-vis-collection/.
- 22.Keogh E; Ratanamahatana CA, Exact indexing of dynamic time warping. Knowledge and Information Systems 2004, 7, 358–386. [Google Scholar]
- 23.Greff K; Srivastava RK; Koutnik J; Steunebrink BR; Schmidhuber J, LSTM: A Search Space Odyssey. IEEE Trans Neural Netw Learn Syst 2017, 28 (10), 2222–2232. [DOI] [PubMed] [Google Scholar]
- 24.Dalby A; Nourse JG; Hounshell WD; Gushurst AKI; Grier DL; Leland BA; Laufer J, Description of several chemical structure file formats used by computer programs developed at Molecular Design Limited. Journal of Chemical Information and Computer Sciences 1992, 32 (3), 244–255. [Google Scholar]
- 25.Weininger D, SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. Journal of Chemical Information and Computer Sciences 1988, 28 (1), 31–36. [Google Scholar]
- 26.Sutskever I; Vinyals O; Le QV, Sequence to Sequence Learning with Neural Networks. arXiv:1409.3215v3 2014. [Google Scholar]
- 27.Luong T; Pham HT; Manning CD In Effective Approaches to Attention-based Neural Machine Translation, Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing, Lisbon, Portugal, Lisbon, Portugal, 2015; pp 1412–1421. [Google Scholar]
- 28.Rogers D; Brown RD; Hahn M, Using extended-connectivity fingerprints with Laplacian-modified Bayesian analysis in high-throughput screening follow-up. J Biomol Screen 2005, 10 (7), 682–6. [DOI] [PubMed] [Google Scholar]
- 29.Rogers D; Hahn M, Extended-connectivity fingerprints. J Chem Inf Model 2010, 50 (5), 742–54. [DOI] [PubMed] [Google Scholar]
- 30.Rousseeuw PJ, Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. Journal of Computational and Applied Mathematics 1987, 20, 53–65. [Google Scholar]
- 31.Shao Y; Mei Y; Sundholm D; Kaila VRI, Benchmarking the Performance of Time-Dependent Density Functional Theory Methods on Biochromophores. Journal of Chemical Theory and Computation 2020, 16 (1), 587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallwitz R; Baumann W, A new shape-oriented classification method for UV/VIS-spectra. Anal Bioanal Chem 1996, 354 (4), 385–91. [DOI] [PubMed] [Google Scholar]
- 33.Dobson CM, Chemical space and biology. Nature 2004, 432 (7019), 824–828. [DOI] [PubMed] [Google Scholar]
- 34.van der Maaten L; Hinton G, Visualizing Data using t-SNE. J Machine Learning Research 2008, 9, 2579–2605. [Google Scholar]
- 35.Woodward RB, Structure and the Absorption Spectra of α,β-Unsaturated Ketones. Journal of the American Chemical Society 1941, 63 (4), 1123–1126. [Google Scholar]
- 36.Fieser LF; Fieser M; Rajagopalan S, ABSORPTION SPECTROSCOPY AND THE STRUCTURES OF THE DIOSTEROLS. The Journal of Organic Chemistry 1948, 13 (6), 800–806. [DOI] [PubMed] [Google Scholar]
- 37.Moser A; Elyashberg ME; Williams AJ; Blinov KA; Dimartino JC, Blind trials of computer-assisted structure elucidation software. J Cheminform 2012, 4 (1), 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Batra K; Zorn KM; Foil DH; Minerali E; Gawriljuk VO; Lane TR; Ekins S, Quantum Machine Learning Algorithms for Drug Discovery Applications. J Chem Inf Model 2021, 61 (6), 2641–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berndt D; Clifford J In Using dynamic time warping to find patterns in time series, AAAI Workshop on Knowledge Discovery in Databases, 1994; pp 229–248. [Google Scholar]
- 40.Ojeda CB; Rojas FS, Recent developments in derivative ultraviolet/visible absorption spectrophotometry. Anal Chim Acta 2004, 518, 1–24. [Google Scholar]
- 41.Lane T; Russo DP; Zorn KM; Clark AM; Korotcov A; Tkachenko V; Reynolds RC; Perryman AL; Freundlich JS; Ekins S, Comparing and Validating Machine Learning Models for Mycobacterium tuberculosis Drug Discovery. Mol Pharm 2018, 15 (10), 4346–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russo DP; Zorn KM; Clark AM; Zhu H; Ekins S, Comparing Multiple Machine Learning Algorithms and Metrics for Estrogen Receptor Binding Prediction. Mol Pharm 2018, 15 (10), 4361–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zorn KM; Lane TR; Russo DP; Clark AM; Makarov V; Ekins S, Multiple Machine Learning Comparisons of HIV Cell-based and Reverse Transcriptase Data Sets. Mol Pharm 2019, 16 (4), 1620–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Segler MHS; Kogej T; Tyrchan C; Waller MP, Generating Focused Molecule Libraries for Drug Discovery with Recurrent Neural Networks. ACS Cent Sci 2018, 4 (1), 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gomez-Bombarelli R; Wei JN; Duvenaud D; Hernandez-Lobato JM; Sanchez-Lengeling B; Sheberla D; Aguilera-Iparraguirre J; Hirzel TD; Adams RP; Aspuru-Guzik A, Automatic Chemical Design Using a Data-Driven Continuous Representation of Molecules. ACS Cent Sci 2018, 4 (2), 268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bagnall A; Lines J; Bostrom A; Large J; Keogh E Data Min. Knowl. Disc 2017, 31, 606–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


