Abstract
The topic of this work is the detection of antimicrobial resistance to Staphylococcus warneri strains and the genes encoding staphylococcal enterotoxins. It is considered a potential pathogen that can cause various—mostly inflammatory—diseases in immunosuppressed patients. The experimental part of the paper deals with the isolation of individual isolates from meat samples of Oryctolagus cuniculus, Oncorhynchus mykiss, Scomber scombrus, chicken thigh, beef thigh muscle, pork thigh muscle, and bryndza cheese. In total, 45 isolates were obtained and subjected to phenotypic (plasma coagulase activity, nuclease, pigment, hemolysis, lecithinase, and lipase production) and genotypic analyses to confirm the presence of the S. warneri species. The presence of genes encoding staphylococcal enterotoxins A (three isolates) and D (six isolates) was determined by PCR. Using the Miditech system, the minimum inhibitory concentration for various antibiotics or antibiotics combinations was determined, namely for ampicillin; ampicillin + sulbactam; oxacillin; cefoxitin; piperacillin + tazobactam; erythromycin; clindamycin; linezolid; rifampicin; gentamicin; teicoplanin; vancomycin; trimethoprim; chloramphenicol; tigecycline; moxifloxacin; ciprofloxacin; tetracycline; trimethoprim + sulfonamide; and nitrofurantoin. Resistance to ciprofloxacin and tetracycline was most common (73%). At the same time, out of a total of 45 isolates, 22% of the isolates were confirmed as multi-resistant. Isolates that showed phenotypic resistance to β-lactam antibiotics were subjected to mecA gene detection by PCR.
Keywords: antimicrobial resistance, enterotoxins, food, mecA, Staphylococcus warneri
1. Introduction
Staphylococcus warneri is a coagulase-negative staphylococcal (CNS) commonly present in the microbiota of human epithelium and mucous membranes. In the last two decades, S. warneri, like other species of the CNS, has been reported as a new emerging pathogen that can cause serious infections, usually in connection with the presence of implanted materials but sometimes even in the absence of a foreign body and in patients considered immunocompetent. At present, there is still a lack of scientific data on the pathogenesis and epidemiology of this species [1]. According to Bhardwaj et al. [2], Staphylococcus warneri is a common saprophyte on human skin, present in approximately 50% of the healthy adult population.
S. warneri creates a biofilm on the surface of various materials (e.g., air, walls, floors, and medical equipment). In addition, it possesses various virulence factors, such as adhesion to polymeric surfaces and metabolic changes in various situations. The ability to form biofilms in foreign bodies and medical devices allows this bacterium to be resistant to antimicrobials used to treat infections [3].
The CNS, including S. warneri, were previously considered food-related saprophytes [4,5]. S. warneri, as well as other CNSs, may possess or acquire mobile pathogenic factors (e.g., exotoxins, enterotoxins, adherence factors, leukocidins, and antibiotic resistance) in the form of transposons, pathogenicity islands (PIs), plasmids, and phages similar to those of Staphylococcus aureus, which may lead to a potential health risk for the consumer [5,6]. The most common contamination of food with this type of staphylococcus is secondary and comes from processing at food companies as well as from the poor handling of food during the production process [5,6,7]. Most recently, S. warneri was detected in chicken meat, ready-for-consumption fish dishes, and raw dairy products [8,9,10]. This species is also reported as a potential source of food enterotoxicosis [11,12].
The antibiotic resistance of microorganisms is a major health, social, and economic problem for all of us today. As consumers of animal products, humans should also be aware of the risks associated with consuming products that contain bacteria resistant to some antibiotics (chloramphenicol, streptomycin, enrofloxacin). When consuming such products, there is the possibility of transferring resistance genes to the microbial population in the human intestinal tract and possible future complications in the treatment of bacterial diseases, not to mention the spread of resistance genes further in the human population and environment [13].
Antimicrobial resistance in bacteria is not a new phenomenon. Estimates for the emergence of biosynthetic pathways have been published by Wright, showing that such resistance began hundreds of millions of years ago [14]. It has also been documented that bacteria share a pool of antimicrobial-resistant genes (AMRG) conferring antibiotic resistance before human discovery and the widespread use of antibiotics. In the permafrost of 30,000 years ago, genes encoding β-lactam resistance and resistance to other antibiotics were documented by D’Costa [15]. There are self-transferring plasmids carrying antibiotic resistance genes (ARGs) that can infect many phylogenetically distinct bacteria, forming a group of genes that can be shared [16]. Methicillin-resistant coagulase-negative staphylococci (MR-CoNS) are a major cause of infectious diseases [3].
The CNS, including S. warneri, are usually resistant to multiple drugs, have limited treatment options, cause incurable diseases, and become reservoirs of resistant strains in hospitals. The cause of antimicrobial resistance is multi-factorial, ranging from a lack of infection prevention to the irrational use of antimicrobials by health-care professionals and patients. Recent studies suggest that CNSs were highly resistant to penicillin, methicillin, vancomycin, oxacillin, and erythromycin. CNSs were less resistant to cephalosporins, aminoglycosides, and quinolones [17].
A significant proportion of S. warneri isolates belonging to the CNS group have a high degree of genetic diversity, indicating a high predisposition to antimicrobial resistance [18]. Biofilm isolates can carry multiple genes encoding resistance to beta-lactams, aminoglycosides, and macrolide–lincosamide antibiotics. Antibiotic resistance genes were found to be more common in biofilm-positive than biofilm-negative isolates of S. warneri. Biofilm structure, due to cell aggregation, may be ideal for horizontal gene transfer and thus facilitate the spread of antibiotic resistance [18].
A study by Chaves et al. [19] shows that CNS are capable of producing enterotoxins, confirming data previously published by other authors. Although current legislation only recommends counting coagulase-positive staphylococci, the study points to the possibility of reformulating existing microbiological standards. In addition, a new approach that correlates the levels and intervals of enterotoxin production could be used for a new and safer food policy.
The aim of our study was to point out the presence of resistant and enterotoxigenic isolates of S. warneri from various food commodities of animal origin. In particular, the study focused on the presence of genes encoding for the production of staphylococcal enterotoxins A–E. At the same time, the aim was to determine the MIC for different groups of antibiotics used in human and veterinary medicine and subsequently to detect methicillin-resistant staphylococci carrying the mecA gene.
2. Materials and Methods
Food samples, 4 from each species, investigated in this study derived from the following areas: Atlantic mackerel muscle (Scomber scombrus) originating in FAO 27 (Ireland), rainbow trout muscle (Oncorhynchus mykiss) originating in the South Bohemian Region (Czech Republic), wild rabbit muscle (Oryctolagus cuniculus) originating from a joint hunt on the grounds of the University facility for breeding and diseases of game, fish, and bees in Rozhanovce (Košice, Slovakia), samples of beef thigh muscle, pork thigh muscle (Sus scrofa domesticus) and thigh muscles from chickens (Gallus gallus domesticus) were provided from slaughterhouses located in eastern Slovakia. The samples of full-fat bryndza cheese were provided from two dairy farms located in central Slovakia.
Staphylococcus identification
2.1. Detection of the Phenotypic Properties of the Isolates of Isolation of Strains
Free coagulase detection
Staphylococcal colonies were inoculated from the blood–agar surface with sterile bacterial ashes into test tubes with 10 mL of broth containing brain–heart infusion (BHI broth; OXOID, Hampsire, UK). After incubation for 18–24 h at 37 °C, 0.1 mL of multiplied broth culture of the tested strains was added to test tubes with 1 mL of sterile rabbit plasma (StaphyloPK test, IMUNA, Šarišské Michaľany, Slovakia). The inoculated plasma was incubated at 37 °C. The results were read after 1, 2, 3, 6, and 24 h to capture the delayed positive reaction that may occur within 24 h.
Deoxyribonuclease production
DNase agar (OXOID, Hampshire, UK) containing DNA was used to detect nuclease activity. Isolates with positive nuclease activity hydrolyzed the DNA contained in the agar after 24 h incubation at 37 °C. Positive nuclease activity manifested itself on the surface of DNase agar after pouring the surface of the medium with 1 N hydrochloric acid solution as a transillumination zone. If no DNA hydrolysis occurred near the inoculated strains, the soil became cloudy due to HCl.
Other phenotypic properties of isolates
Pigment production as well as hemolysis of individual isolates were evaluated after 24 h of incubation on the agar surface with a 2% addition of lamb blood. Baird–Parker selective diagnostic medium was used to detect the formation of lecithinase and lipase on the surface of which the identified staphylococcal strains were inoculated with sterile bacterial ashes. The inoculated petri dishes were incubated at 37 °C/24 h. After incubation, in the positive case, a precipitation zone around the inoculated colonies in the agar medium in the presence of lecithinase can be detected. This is due to the hydrolysis of the lecithin phospholipid to 1.2-diglyceride and phosphorylcholine [20].
In the presence of the enzyme lipase, a brightening zone was created around the colonies. This is due to the cleavage of triacylglycerides to triacylglycerol and fatty acids, which are present in Baird–Parker agar medium.
2.2. Isolation DNA from Staphylococcal Isolates
Total genomic DNA was isolated according to Sharma et al. [21] from staphylococcal isolates grown in BHI broth. The pellet obtained after centrifugation of 1.5 mL of expanded staphylococcal culture (12,500× g/5 min) was resuspended in 100 μL of 0.5% Triton X-100 (Koch-Light Lab., Suffolk, VA, USA) and centrifuged again (12,500× g/5 min). Subsequently, the sediment was resuspended in 200 µL of 0.5% Triton X-100. The samples thus prepared were incubated at 95 °C/10 min and centrifuged again (12,500× g/5 min). The obtained supernatant was used as a source of DNA in PCR reactions.
2.3. Genotypic Analysis of Staphylococcal Isolates
The identification of individual staphylococcal strains was performed using the PCR method. The identification of individual staphylococcal strains was performed by using specific primers the SwarF (TGTAGCTAACTTAGATAGTGTTCCTTCT) and SwarR (CCGCCACCGTTATTTCTT) synthesized by Amplia s.r.o. (Bratislava, Slovakia) according to Iwas et al. [22]. Commercially produced Hot FIREPol® MasterMix (Amplia s.r.o., Bratislava, Slovakia) was used in PCR reactions. The total reaction mixture volume of 20 µL contained 5 ng/µL of template DNA and each primer at a concentration of 10 pmol. The finished mixture was heated at 95 °C for 12 min at initial denaturation, then heated over 30 amplification cycles: 95 °C/30 s, annealing 60 °C/30 s and 72 °C/2 min, using a thermal cycler (TECHNE TC-512, London, UK) with an extension of 10 min/72 °C. The 65 bp PCR product, which was amplified, was collected for 5 μL for analysis on a 1.5% agarose gel in TBE buffer (Tris-borate-EDTA). GelRed TM (Biotium, Fremont, CA, USA) was added to the agarose gel to visualize the PCR fragments using a Mini Bis Pro® reader (DNR Bio-Imaging Systems Ltd., Neve Yamin, Israel). The obtained PCR products were sequenced at the European Custom Sequencing Center GATCBiotech AG (Cologne, Germany) and compared with the nucleotide sequences of the reference strains in GenBank NCBI. The reference strain S. warneri CCM 2730T (Czech Collection of Microorganisms, Brno, Czech Republic) was used as a positive control for PCR strain identification.
2.4. Detection of Genes Encoding Staphylococcal Enterotoxins
To detect genes encoding staphylococcal enterotoxin (SE) production (sea, seb, sec, sed, and see), a multiplex PCR assay, according to Sharma et al. [21] was performed by using universal forward primer and five specific reverse primers for each staphylococcal enterotoxin to detect each of the five enterotoxins.
The reaction mixture used and the reaction conditions were the same as described above (except for the annealing temperature, which was 45 °C). The sizes of the PCR products are shown in Table 1.
Table 1.
Primers used in multiplex PCR to detect staphylococcal enterotoxins [21].
| Gene | Primer | Nucleotide Sequence 5′-3′ | Product Size (bp) |
|---|---|---|---|
| universal | Sauni-F | TGTATGTATGGAGGTGTAAC | |
| sea | SAA -R | ATTAACCGAAGGTTCTGT | 270 |
| seb | SAB -R | ATAGTGACGAGTTAGGTA | 165 |
| sec | SAC -R | AAGTACATTTTGTAAGTTCC | 102 |
| sed | SAD -R | TTCGGGAAAATCACCCTTAA | 306 |
| see | SAE -R | GCCAAAGCTGTCTGAG | 213 |
bp—base pairs.
The following reference strains of Staphylococcus aureus (CCM, Brno, Czech Republic) were used as a positive control for this multiplex PCR: CCM 5756 (sea gene), CCM 5757 (seb gene), CCM 5984 (sec gene), CCM 5973 (sed gene), and CCM 5972 (see gene).
2.5. Detection of Antimicrobial Resistance
Eighteen isolates of S. warneri were analyzed for their antibiotic susceptibility. Minimal inhibitory concentrations (MIC) were determined according to CLSI document [23] and EUCAST document [24], using a Miditech system (Bratislava, Slovakia) with interpretive reading of MIC. The antibiotics used in this study were as follows: ampicillin + sulbactam (SAM), piperacillin + tazobactam (TZP), oxacillin (OXA), erythromycin (ERY), clindamycin (CLI), teicoplanin (TEC), vancomycin (VAN), rifampicin (RIF), gentamicin (GEN), linezolid (LNZ), ciprofloxacin (CIP), moxifloxacin (MFX), tetracycline (TET), tigecycline (TGC), chloramphenenicol (CHL), trimethoprim (TMP), trimethoprim + sulfonamide (COT), and nitrofurantoin (NIT).
In isolates showing phenotypic resistance to oxacillin, the presence of the mecA gene was detected by PCR according to Poulsen et al. [25]. The primers used in this PCR reaction were MecA1 (GGGATCATAGCGTCATTATTC) and MecA2 (AACGATTGTGACACGATAGCC) (Amplia sro, Bratislava, Slovakia). The PCR reaction conditions were the same as for the species identification of staphylococci mentioned above. The resulting size of the PCR product was 527 bp. The reference isolate S. aureus CCM 4750 (Czech Collection of Microorganisms, Brno, Czech Republic) was used as a reference strain for the PCR and for detection of MIC in this study.
3. Results
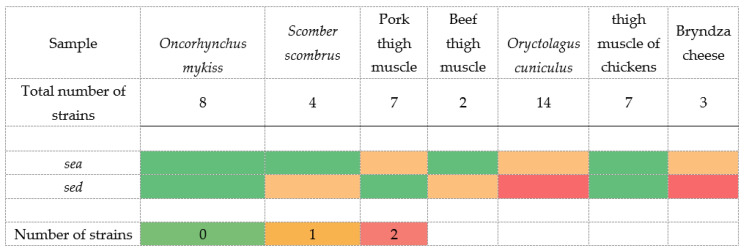
Typical and atypical colonies were harvested from the surface of the Baird–Parker agar medium after incubation for the initial culture screening of individual samples. A total of 560 staphylococcal isolates were obtained and subjected to further identification. All isolates obtained were subjected to detection of coagulase activity. In the detection of this phenotypic trait, for 200 isolates the result of coagulase assay was negative, showing no coagulase activity. Therefore, these isolates were classified as CNS. Subsequently, in this group of isolates, the detection of other phenotypic characteristics was carried out for a better characterization of the S. warneri strains as well as for detection of virulent strains: nuclease activity, pigment, hemolysis, lecithinase, and lipase production (Table 2). In isolates included in the CNS, genotypic identification was performed using the PCR method, where 45 isolates were identified (22% of the CNS group) as S. warneri. It was retrospectively evaluated that 8 isolates derived from brown trout, 4 isolates from Atlantic mackerel, 7 isolates from pork thigh muscle, 2 isolates from beef thigh muscle, 14 isolates of S. warneri came from wild rabbit, 7 isolates from chicken thigh muscle and, 3 isolates came from bryndza cheese.
Table 2.
Virulence phenotypic properties of S. warneri isolates.
| Number of Strains | Pigment | Hemolysis | Lecithinase | Lipase | Nuclease | ||||
|---|---|---|---|---|---|---|---|---|---|
| White (without Pigment) | Gray | Gray-White | Yellow-White | ||||||
| Oncorhynchus mykiss | 8 | 0 | 2 | 6 | 0 | 0 | 3 | 7 | 0 |
| Scomber scombrus | 4 | 0 | 2 | 1 | 1 | 3 (α) | 2 | 3 | 0 |
| Pork thigh muscle | 7 | 0 | 0 | 6 | 1 | 0 | 3 | 1 | 0 |
| Beef thigh muscle | 2 | 1 | 1 | 0 | 0 | 0 | 1 | 5 | 0 |
| Oryctolagus cuniculus | 14 | 0 | 3 | 7 | 4 | 5 (α) | 4 | 12 | 0 |
| thigh muscle of chickens | 7 | 1 | 0 | 4 | 2 | 1 (α) | 1 | 0 | 0 |
| Bryndza cheese | 3 | 1 | 1 | 1 | 0 | 1 (α) | 3 | 3 | 0 |
| Total | 45 | 9 | 6 | 23 | 7 | 10 | 17 | 31 | 0 |
(α)—α-hemolysis.
As shown in Table 2, gray pigment was observed most frequently on blood agar. The production of a yellowish-white coloration of S. warneri colonies also appeared. The production of α-hemolysis was also confirmed in these isolates. β-hemolysis was not confirmed in any of the isolates. Similarly, nuclease production on DNAse agar was not confirmed in any isolate. However, lipase production was confirmed in a large number of S. warneri isolates. Lipase production was manifested by a transillumination zone in Baird–Parker agar medium around the overgrown colony. In contrast, a smaller number of isolates produced the enzyme lecithinase. Its production appeared as a precipitating ring around the overgrown colony on Baird–Parker agar medium. The detection of lecithinase and lipase production confirmed their current production in three isolates of Oncorhynchus mykiss, two isolates from Oryctolagus cuniculus, and one isolate each from Scomber scombrus, pork thigh muscle, and Bryndza cheese.
Subsequently, after the detection of individual phenotypic manifestations, genes encoding the staphylococcal enterotoxins SEA, SEB, SEC, SED, and SEE were detected. As shown in Figure 1, S. warneri isolates confirmed the presence of genes encoding the production of SEA and SED enterotoxins. The presence of the sea gene was confirmed in three isolates and the sed gene in six isolates. Of these, the co-presence of the sea and sed genes was detected in two isolates. It was an isolate isolated from a sample of Oryctolagus cuniculus and a sample of Bryndza cheese. At the same time, these two isolates phenotypically showed α-hemolysis and lecithinase and lipase production.
Figure 1.
Heatmap demonstration of the presence and absence of genes encoding staphylococcal enterotoxins.
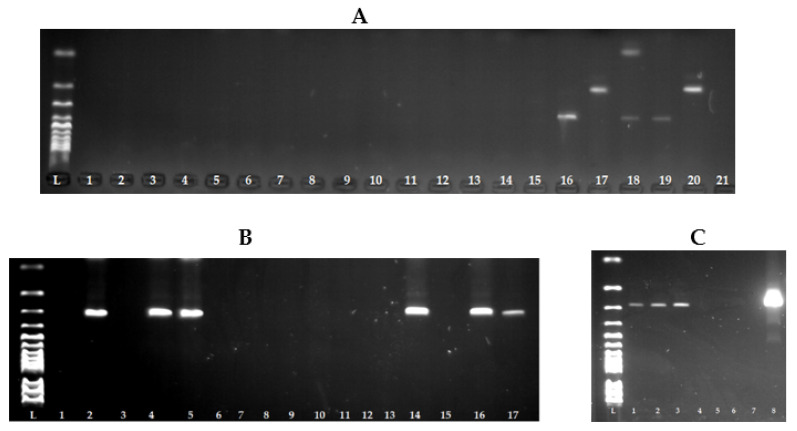
As seen in Figure 2A, we were unable to detect the presence of genes encoding enterotoxin production in S. warneri isolates by multiplex PCR, only in the reference strains. For this reason, we approached the detection of individual genes separately using primers from multiplex PCR. As seen in Figure 2B,C, the presence of the sea and sed genes was subsequently confirmed.
Figure 2.
Detection of genes encoding the staphylococcal enterotoxins of Staphyoccocus warneri using the PCR method. (A) L: 100 bp ladder; lines 1–15: isolates S. warneri without genes encoding the staphylococcal enterotoxins; line 16: reference strain for sea gene CCM 5756 S. aureus (270 bp); line 17: reference strain for seb gene CCM 5757 S. aureus (165 bp); line 18: reference strain for sec gene CCM 5984 S. aureus (102 bp); line 19: reference strain for sed gene CCM 5973 S. aureus (306 bp); line 20: reference strain for see gene CCM 5972 S. aureus (213 bp); line 21: negative control. (B) L: 100 bp ladder; line 1: negative control; line 2: reference strain for sed gene CCM 5973 S. aureus (306 bp); lines 4, 5, 14, 16, 17: isolates S. warneri with sed gene (306 bp). (C) L: 100 bp ladder; lines 1–3: isolates S. warneri with sea gene (270 bp); line 7: negative control; line 8: reference strain for sea gene CCM 5756 S. aureus (270 bp).
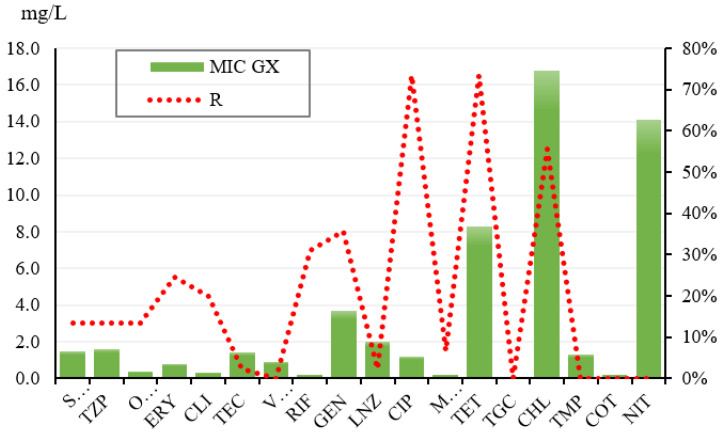
After detection of phenotypic characteristics and detection of toxinogenic isolates, MIC detection of antibiotics was performed using the Miditech system. In general, as shown in Table 3, the highest resistance to ciprofloxacin and tetracycline was confirmed. The MIC50 of ciprofloxacin was 2.0 mg/L and the MIC90 was 4.0 mg/L while the MIC of GX was 1.2 mg/L. For tetracycline, the MIC50 was 16.0 mg/L and the MIC90 was 32.0 mg/L. At the same time, the MIC GX of this antibiotic was 8.3 mg/L (Figure 3). At the same time, intermediate sensitivity was confirmed in all 45 isolates against trimethoprim + sulfonamide, and nitrofurantoin. The strains on vancomycin, trimethoprim, trimethoprim + sulfonamide, and nitrofurantoin appeared to be the most sensitive overall (Table 3).
Table 3.
Percentage of antimicrobial resistance.
| ATB | S | I | R | GX SI | MIC50 | MIC90 | Total |
|---|---|---|---|---|---|---|---|
| SAM | 86.67% | 0.00% | 13.33% | 0.9 | 1.00 | 64.00 | 45 |
| TZP | 86.67% | 0.00% | 13.33% | 0.8 | 1.00 | 128.00 | 45 |
| OXA | 86.67% | 0.00% | 13.33% | 0.2 | 0.25 | 8.00 | 45 |
| ERY | 75.56% | 0.00% | 24.44% | 0.3 | 0.50 | 16.00 | 45 |
| CLI | 89.00% | 0.00% | 20.00% | 0.1 | 0.25 | 8.00 | 45 |
| TEC | 97.78% | 0.00% | 2.22% | 1.3 | 1.00 | 4.00 | 45 |
| VAN | 100.00% | 0.00% | 0.00% | 0.9 | 1.00 | 2.00 | 45 |
| RIF | 42.22% | 26.67% | 31.11% | 0.1 | 0.50 | 2.00 | 45 |
| GEN | 64.44% | 0.00% | 35.56% | 0.4 | 0.50 | 256.00 | 45 |
| LNZ | 97.78% | 0.00% | 2.22% | 2.0 | 2.00 | 4.00 | 45 |
| CIP | 0.00% | 26.67% | 73.33% | 0.2 | 2.00 | 4.00 | 45 |
| MFX | 93.33% | 0.00% | 6.67% | 0.1 | 0.13 | 0.25 | 45 |
| TET | 26.67% | 0.00% | 73.33% | 0.4 | 16.00 | 32.00 | 45 |
| TGC | 100.00% | 0.00% | 0.00% | 0.1 | 0.06 | 0.13 | 45 |
| CHL | 44.44% | 0.00% | 55.56% | 5.3 | 16.00 | 64.00 | 45 |
| TMP | 0.00% | 100.00% | 0.00% | 1.3 | 2.00 | 2.00 | 45 |
| COT | 100.00% | 0.00% | 0.00% | 0.2 | 0.25 | 0.50 | 45 |
| NIT | 0.00% | 100.00% | 0.00% | 14.1 | 16.00 | 16.00 | 45 |
S—sensitive strain, I—intermediate sensitive strain, R—resistant strain; GX SI MIC—Geometric mean MIC of strains that can be treated with a given ATB (S + I). Mean MIC of treatable strains. MIC50—value which expresses the minimum inhibitory concentration of a given antibiotic at which at least 50% of the population is inhibited. MIC90—value which expresses the minimum inhibitory concentration of a given antibiotic at which at least 90% of the population is inhibited.
Figure 3.
Overview of MIC and MIC GX in resistant Staphylococcus warneri isolates. GX MIC—Geometric average MIC—the average MIC value at which the strains are inhibited.
Specifically, isolates isolated from the Oncorhynchus mykiss muscle (eight isolates) were the most frequently confirmed to be resistant to tetracycline (87.50%). Resistance to clindamycin and gentamicin (37.50%) was also among the common resistance detected in these isolates. Isolates isolated from Scomber scombrus muscle samples (four isolates) also showed the most common resistance to tetracycline (75.00%) and to gentamicin, ciprofloxacin, and chloramphenicol (50.00%). S. warneri strains isolated from the pork thigh muscle (7 isolates) and the Oryctolagus cuniculus muscle (14 isolates) showed the most common resistance to tertracycline (100.00%/92.86%). Isolates isolated from chicken thigh muscle (seven isolates) showed the highest resistance to ciprofloxacin (85.71%). In isolates derived from beef thigh muscle (two isolates), 100.00% resistance to clindamycin, rifampicin, and gentamicin was observed. The resistance of all S. warneri isolates (three isolates) to rifampicin (100.00%) was confirmed in the last examined bryndza samples.
Based on the antimicrobial resistance using the Miditech system, the various resistance mechanisms, which are shown in Table 4, were confirmed. As can be seen from the table, incomplete fluoroquinolone resistance was generally the most common. Specifically, this resistance mechanism was most commonly detected in isolates isolated from Oryctolagus cuniculus samples (13 isolates/92.86%). The presence of methicillin-resistant coagulase-negative staphylococci (MRCNS) in 13.33% of the examined isolates was also phenotypically confirmed.
Table 4.
Phenotypic detection of various mechanisms of resistance.
| Mechanisms of Resistance | Number | % |
|---|---|---|
| MRCNS | 6 | 13.33% |
| Aminogl.PH(2″)-AC(6′) | 15 | 33.33% |
| Fluoroq.incompl.resistance | 32 | 71.11% |
| Constitutive MLSB/c | 1 | 2.22% |
| Inducible MLSB/i | 0 | 0.00% |
| Multi-resistance | 10 | 22.22% |
MRCNS: methicillin-resistant coagulase-negative staphylococci; Amngl.PH (2″)-AC (6′): combined enzymatic resistance to GEN, TOB, and AMI; constitutive MLSB/c: constitutive resistance to macrolides, lincosamides, and streptogramin B; inducible MLSB/i: inducible resistance to macrolides, lincosamides, and streptogramin B; multi-resistance: current resistance in 3 or more unrelated ATB groups; fluoroq.incompl.resistanc: incomplete fluoroquinolone resistance, mutation with incomplete resistance to fluoroquinolones.
Based on the phenotypic expression of these isolates (six isolates), the mecA gene, which encodes one of the most common mechanisms of methicillin resistance, was detected. Based on the results of the PCR reaction and subsequent sequencing of the PCR products, the presence of the mecA gene was confirmed in four isolates (66.66%). These isolates were isolated from samples of Scomber scombrus, Oryctolagus cuniculus, thigh muscle of chickens, and Bryndza cheese. The isolates also exhibited virulence phenotypic properties such as α-hemolysis, lipase, and lecithinase production. All isolates carrying the mecA gene produced a yellow-white pigment. At the same time, the presence of the sea gene (1 isolate), the sed gene (1 isolate), and both genes (2 isolates) was confirmed in these isolates.
Based on the detection of resistance of S. warneri isolates, multi-drug resistance was also confirmed in 10 isolates (Table 4). Specifically, two isolates isolated from Oncorhynchus mykiss samples were confirmed to be resistant to four antimicrobial groups at the same time, namely RIF-GEN-CIP-TET. In isolates derived from Oryctolagus cuniculus samples, multi-drug resistance was confirmed in two isolates, namely GEN-CIP-TET. The other two isolates confirmed simultaneous resistance to CIP-TET-CHL-RIF-GEN, of which one isolate also carried the mecA gene. In isolates derived from pork thigh muscle, the co-resistance to GEN-CIP-TET was confirmed in three isolates. Multi-resistance was also confirmed in one isolate isolated from Bryndza cheese samples against OXA-ERY-CIP-CHL.
4. Discussion
The isolation of food samples produced 45 isolates that were subsequently tested for phenotypic and genotypic characteristics to confirm that they were Staphylococcus warneri, being catalase-positive, oxidase-negative, and coagulase-negative. This species is also a commensal bacterium that occurs as part of the skin microbiota in humans and animals. Although S. warneri accounts for less than 1% of the total staphylococcal population, it is responsible for a variety of human diseases, such as immune suppression, skin, eye, and urinary tract infections, nosocomial infections, and weakening the immune systems of patients and neonates [1]. However, in a study by Hoveida et al. [26], S. warneri accounted for up to 7.5%, similar to our study, where the incidence rate was up to 22% of the total identified CNS population. In our study, such a higher incidence of S. warneri may be due to a different origin of the samples. The samples came from bryndza, game, fish, and thigh muscles of livestock. The microbiota of these commodities can be affected by hygiene levels, where the role of food processors is crucial in determining the hygienic status of the final product, with mishandling leading to an increased likelihood of microbial contamination by humans, including multi-drug-resistant and/or enterotoxigenic staphylococci [27].
In our study, phenotypic and genotypic identification was used to identify this species. In terms of phenotypic characteristics, hemolytic activity was tested, which was demonstrated in 10 isolates in the form of α-hemolysis. This confirms the production of cytolytic alpha-toxin, which is also known as alpha-hemolysin. By binding to the cell surface, it causes necrotic cell death [28]. Alpha-hemolysin production was also confirmed in a study by Noumi et al. [29], in two tested S. warneri isolates.
Pigment formation was demonstrated in 36 isolates, where the most frequently produced was a gray-white pigment, to a lesser extent a yellow-white pigment. According to Becker et al. [4], most CNS species are usually unpigmented. Pigmented colonies are characteristic only for the species S. chromogenes, S. devriesei, S. lugdunensis, S. sciuri, S. vitulinus, S. warneri, and S. xylosus. They form a pigment of gray-white, gray-yellow, yellow, or yellow-orange color.
Other monitored properties were the production of hydrolytic enzymes (lecithinase and lipase). Hydrolytic enzymes are among the CNS virulence factors that contribute to soft-tissue degradation and aid in biofilm formation [30]. Lecithinase was observed in our study in 17 S. warneri isolates. It is a type of phospholipase that acts on lecithin [30]. Lipase production was confirmed in 35 S. warneri isolates. Staphylococcal lipases are usually released outside the cell, where they perform several functions, some of which are of hygienic importance. Staphylococcal extracellular lipases are overexpressed in connection with pathogenic events. They are also recognized as contributors to human and animal pathogens by improving bacterial nutrition on the skin, disrupting host cell membranes, disrupting the immune response, and host cell signaling [31]. Many studies have been conducted to examine staphylococcal lipase production, and these bacteria have been found to have high levels of lipase activity in meat and dairy production [32,33], as confirmed by our study. Lipolytic bacteria are classified into different genera based on their gene sequences and biochemical properties, and high lipase levels are also a common feature of staphylococci, including S. warneri [34]. The presence of the abovementioned hydrolytic properties was confirmed in S. warneri isolates by Noumi et al. [29].
Subsequently, the PCR method, which is currently considered to be the most accurate identification method, was used to accurately identify the S. warneri isolates obtained. The 16S rRNA gene is generally used as a target sequence to identify species in the genus Staphylococcus. However, this 16S rRNA gene in Staphylococcus epidermidis is very similar to the sequence of other 16S rRNAs in other CNS species. To solve this problem, it is possible to use an alternative target sequence that shows a greater sequence divergence than the 16S rRNA gene. Recently, a highly conserved, ubiquitous sodA gene was used that encodes manganese-dependent superoxide dismutase [35] In our study, 45 S. warneri isolates from 200 identified CNSs were confirmed using a specific sodA gene sequence. The presence of genes encoding the production of staphylococcal enterotoxins A to E was subsequently detected in the S. warneri isolates.
SE production is one of the most notable virulence factors in staphylococci. Staphylococcal enterotoxins (SE) are mainly divided into five classical serological types: SEA, SEB, SEC, SED, and SEE as well as the other recently discovered SEG, SHE, SEI, SER, SES, SET, and the enterotoxin-like proteins, such as SElK, SElN, SE10, SE1P, SE1Q, and SElU [36,37]. Reliable detection of SE genes has a dual function. First, it helps in the genotyping of coagulase-positive staphylococci (CPS) for epidemiological studies. Second, it provides an assessment of the possible occurrence of SE genes in CNS strains which pose a potential risk of staphylococcal enterotoxicosis to consumers [36]. In our study, the presence of the sea (6% of isolates) and sed (13% of isolates) genes was confirmed. De Freitas Guimarães et al. [38] also confirmed the presence of sea and sed genes in S. warneri isolates isolated from food of animal origin but in higher percentages than those confirmed in our study. In further studies, they confirmed the presence of other genes that encode enterotoxins, namely sec and she [29,39].
Banaszkiewicz et al. [39] and Hu et al. [40] also suggest the detection of the continuous transfer of elements containing SE genes from staphylococci which contain the stable enterotoxin genes of the CNS originating from wild animals, which was confirmed by our study.
Staphylococcal isolates of phenotypically and genotypically identified isolates were subsequently determined using the Miditech system, where the highest resistance was recorded against CIP and TET. Similarly, frequent resistance to TET was confirmed in the Hoveida et al. [41] study, which accounted for up to 50% of the resistance.
At the same time, our study confirmed the presence of S. warneri MRS in 13.33% of the S. warneri isolates. On the basis of the detection of the MRSCNS phenotypic expression, the mecA gene was detected in these isolates. Methicillin resistance is associated with the presence of the mecA gene, which encodes additional binding to a penicillin protein (PBP2A or PBP2’). This protein has a lower affinity for all beta-lactam antibiotics. The mecA gene is found on a mobile genetic element called the staphylococcal cassette chromosome mec (SCCmec) [42]. Another mechanism in staphylococcal resistance to beta-lactams is beta-lactamase production, encoded by the blaZ gene [43]. In our study, the presence of the mecA gene was confirmed in 66.66% of isolates (four isolates) that phenotypically appeared as MRSCNS (six isolates). Humphries et al. [44] confirmed the presence of the mecA gene in S. warneri isolates in a similarly high percentage. The mecA-positive S. warneri isolates accounted for up to 41.66% of the total of 48 isolates tested. Similarly, Hoveida et al. [41] confirmed the prevalence of mecA genes in S. warneri in 30% of the 40 isolates isolated from different types of food.
In addition to the increasing incidence of MRSCNS isolates, resistance to aminoglycosides has recently increased. Aminoglycoside resistance has increased, especially among methicillin-resistant strains carrying the mecA gene [45]. Inactivation of antibiotics by aminoglycoside-modifying enzymes (AME), which are encoded by genetic elements, is a major pathway for resistance [46]. The most important of these enzymes is aac (6′)/aph (2″), which alters aminoglycosides of medical importance, such as tobramycin and gentamicin [47]. This type of resistance to aminoglycosides was also confirmed in our investigated CNS isolates, specifically in 33.33% of the S. warneri isolates. Of these, MRSCNS was identified in three strains; they showed resistance to aminoglycosides using combined enzymatic resistance to tobramycin, gentamicin, and amikacin. Consistent with many other studies [47,48,49], the enzyme aac (6′)/aph (2″) is the most common, where 85% of AME-positive isolates were found.
Another resistance detected in the isolates we examined was constitutive resistance to macrolides, lincosamides and streptogramin B (cMLSB). However, studies mostly focus on the most common isolated staphylococci, i.e., S. aureus and S. epidermidis [50,51,52]. Recently, other staphylococcal species, such as S. hominis, S. haemolyticus, S. warneri and S. simulans, have emerged as etiological factors in serious human infections. The phenotypic expression of resistance to MLSB in these staphylococci may be inducible and manifest in clinical resistance to lincosamides and streptogramin B induced by 14- and 15-membered macrolides or constitutive, determining resistance to all MLSB antibiotics [53,54]. In our study, only constitutive macrolide resistance was phenotypically confirmed (2.22%). Similarly, a low percentage (10%) were confirmed cMLSB in S. warneri in a study by Szemraj et al. [55].
Incomplete fluoroquinolone resistance was also confirmed in the S. warneri isolates in our study, with up to 71.11%; in other words, this is a mutation with incomplete resistance to fluoroquinolones. However, little is known about the mechanisms involved in the development of fluoroquinolone resistance (through exogenous acquisition, de novo mutation, or selection of a minority-resistant mutant) in CNS [56]. However, previous studies have confirmed that methicillin-resistant strains developed resistance to fluoroquinolones more rapidly in S. aureus and the CNS than in methicillin-sensitive strains. This difference is partly explained by nosocomial transmission in some environments and thus the potential for co-selection with several antimicrobials (due to the common multi-drug resistance phenotype of MRS isolates [57]. Resistance to more than one antibiotic was also confirmed by Persson-Waller et al. [58], where multi-drug resistance occurred in 9% of the total 56 CoNS isolates. Nunes et al. [59] also confirmed 14 multi-drug-resistant CNS. Our demonstrated multi-drug resistance of CNS isolates, specifically S. warneri, is consistent with previous studies on coagulase-negative staphylococci, which found several resistant and multi-drug resistant staphylococcal isolates [60]. Our results are supported by the study of Senga et al. [61], who reported up to 80% resistance to several types of antibiotics in CNS isolates.
5. Conclusions
The results of the paper point to the ever-increasing incidence of resistant and multi-resistant and enterotoxigenic isolates of S. warneri in foodstuffs of animal origin, especially game and wild fish. These results point to the risk of the presence of enterotoxigenic and resistant isolates of S. warneri in the food industry, as they can serve as vectors for the transfer of resistance determinants into the genomes of bacteria inhabiting the consumer’s digestive tract. Therefore, the rational use of antibiotics, preventive measures in environmental hygiene, and the monitoring of antibiotic resistance are important prevention measures to prevent the spread of antimicrobial resistance. At the same time, the presence of enterotoxigenic isolates indicates a potential risk for staphylococcal enterotoxicosis in humans.
Author Contributions
Conceptualization: I.R.; methodology: I.R. and J.V.; software: G.G.; formal analysis: M.P.; investigation and data curation: P.J. and J.B.; writing—original draft preparation: I.R.; writing—review and editing: J.V.; supervision: J.V. and P.J.; project administration and funding acquisition: J.B. and F.Z. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
Additional financial support for the implementation of the study comes from Slovak grant KEGA no. 006UVLF-4-2020.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Azimi T., Mirzadeh M., Sabour S., Nasser A., Fallah F., Pourmand M.R. Coagulase-negative staphylococci (CoNS) meningitis: A narrative review of the literature from 2000 to 2020. New Microbes New Infect. 2020;37:100755. doi: 10.1016/j.nmni.2020.100755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhardwaj B., Bhatnagar U.B., Conaway D.G. An Unusual Presentation of Native Valve Endocarditis Caused by Staphylococcus warneri. Rev. Cardiovasc. Med. 2016;17:140–143. doi: 10.3909/ricm0823. [DOI] [PubMed] [Google Scholar]
- 3.Seng R., Kitti T., Thummeepak R., Kongthai P., Leungtongkam U., Wannalerdsakun S., Sitthisak S. Biofilm formation of methicillin-resistant coagulase negative staphylococci (MR-CoNS) isolated from community and hospital environments. PLoS ONE. 2017;12:e0184172. doi: 10.1371/journal.pone.0184172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker K., Heilmann C., Peters G. Coagulase-negative staphylococci. Clin. Microbiol. Rev. 2014;27:870–926. doi: 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fijalkowski K., Peitler D., Karakulska J. Staphylococci isolated from ready-toeat meat-identification, antibiotic resistance and toxin gene profile. Int. J. Food Microbiol. 2016;238:113–120. doi: 10.1016/j.ijfoodmicro.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Bhargava K., Zhang Y. Characterization of methicillin-resistant coagulasenegative staphylococci (MRCoNS) in retail meat. Food Microbiol. 2014;42:56–60. doi: 10.1016/j.fm.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 7.Incani R.N., Hernández M., Cortez J., González M.E., Dorel Salazar Y. Staphylococcus warneri meningitis in a patient with Strongyloides stercoralis hyperinfection and lymphoma: First report of a case. Rev. Inst. Med. Trop. São Paulo. 2010;52:169–170. doi: 10.1590/S0036-46652010000300011. [DOI] [PubMed] [Google Scholar]
- 8.Musharrafieh R., Tacchi L., Trujeque J., LaPatra S., Salinas I. Staphylococcus warneri, a resident skin commensal of rainbow trout (Oncorhynchus mykiss) with pathobiont characteristics. Vet. Microbiol. 2014;169:80–88. doi: 10.1016/j.vetmic.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H., Wang H., Bai Y., Xu X., Zhou G. Pathogenicity and antibiotic resistance of coagulase-negative staphylococci isolated from retailing chicken meat. LWT. 2018;90:152–156. doi: 10.1016/j.lwt.2017.12.029. [DOI] [Google Scholar]
- 10.El-Ashker M., Gwida M., Monecke S., Ehricht R., Elsayed M., El-Gohary F., Reißig A., Müller E., Paul A., Igbinosa E.O., et al. Microarray-based detection of resistance genes in coagulase-negative staphylococci isolated from cattle and buffalo with mastitis in Egypt. Trop. Anim. Health Prod. 2020;52:3855–3862. doi: 10.1007/s11250-020-02424-1. [DOI] [PubMed] [Google Scholar]
- 11.Chajęcka-Wierzchowska W., Gajewska J., Wiśniewski P., Zadernowska A. Enterotoxigenic potential of coagulase-negative staphylococci from ready-to-eat food. Pathogens. 2020;9:734. doi: 10.3390/pathogens9090734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costanzo N., Ceniti C., Santoro A., Clausi M.T., Casalinuovo F. Foodborne pathogen assessment in raw milk cheeses. Int. J. Food Sci. 2020;2020:3616713. doi: 10.1155/2020/3616713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hleba L., Kačániová M. Antibiotická Rezistencia Čeľade Enterobacteriaceae vo Vzťahu k Potravinám. Slovenská Poľnohospodárska Univerzita v Nitre; Nitra, Slovenska: 2015. p. 93. [Google Scholar]
- 14.Wright G.D. The antibiotic resistome: The nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 2007;5:175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- 15.D’Costa V.M., King C.E., Kalan L., Morar M., Sung W.W., Schwarz C., Froese D., Zazula G., Calmels F., Debruyne R., et al. Antibiotic resistance is ancient. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 16.Schlüter A., Szczepanowski R., Pühler A., Top E.M. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol. Rev. 2007;31:449–477. doi: 10.1111/j.1574-6976.2007.00074.x. [DOI] [PubMed] [Google Scholar]
- 17.Deyno S., Fekadu S., Seyfe S. Prevelance and antimicrobial resistance of coagulase staphylococci clinical isolates from Ethiopia: A meta analysis. BMC Microbiol. 2018;18:43. doi: 10.1186/s12866-018-1188-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szczuka E., Krzyminska S., Kaznowski A. Clonality, virulance and the occurrence of genes encoding antibiotic resistance among Staphylococcus warneri from bloodstream infections. J. Med. Microbiol. 2016;65:828–836. doi: 10.1099/jmm.0.000287. [DOI] [PubMed] [Google Scholar]
- 19.Chaves R.D., Pradella F., Turatti M.A., Amaro E.C., da Silva A.R., dos Santos Farias A., Pereira J.S., Khaneghah A.M. Evaluation of Staphylococcus spp. in food and kitchen premises of Campinas, Brazil. Food Control. 2018;84:463–470. doi: 10.1016/j.foodcont.2017.09.001. [DOI] [Google Scholar]
- 20.Owens J.J. The egg yolk reaction produced by several species of bacteria. J. Appl. Bacteriol. 1974;37:137–148. doi: 10.1111/j.1365-2672.1974.tb00424.x. [DOI] [PubMed] [Google Scholar]
- 21.Sharma N.K., Rees C.E., Dodd C.E. Development of a single-reaction multiplex PCR toxin typing assay for Staphylococcus aureus strains. Appl. Environ. Microbiol. 2000;66:1347–1353. doi: 10.1128/AEM.66.4.1347-1353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwase T., Seki K., Shinji H., Mizunoe Y., Masuda S. Development of a real-time PCR assay for the detection and identification of Staphylococcus capitis, Staphylococcus haemolyticus and Staphylococcus warneri. J. Med. Microbiol. 2007;56:1346–1349. doi: 10.1099/jmm.0.47235-0. [DOI] [PubMed] [Google Scholar]
- 23.CLSI Document VET01-S2 Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2013. pp. 1–168. [Google Scholar]
- 24.EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance. Version 2.0. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2017. pp. 1–43. [Google Scholar]
- 25.Poulsen A.B., Skov R., Pallesen L.V. Detection of methicillin resistance in coagulase-negative staphylococci and in staphylococci directly from simulated blood cultures using the EVIGENE MRSA Detection Kit. J. Antimicrob. Chemother. 2003;51:419–421. doi: 10.1093/jac/dkg084. [DOI] [PubMed] [Google Scholar]
- 26.Hoveida L., Ataei B., Amirmozafari N., Noormohammadi Z. Species diversity and molecular analysis of Staphylococcus in confectioneries of a developing country, Iran. Infez. Med. 2018;26:148–154. [PubMed] [Google Scholar]
- 27.Hammad A.M., Watanabe W., Fujii T., Shimamoto T. Occurrence and characteristics of methicillin-resistant and -susceptible Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci from Japanese retail ready-to-eat raw fish. Int. J. Food Microbiol. 2016;156:286–289. doi: 10.1016/j.ijfoodmicro.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 28.Vandenesch F., Lina G., Henry T. Staphylococcus aureus hemolysins, bi-component leukocidins, and cytolytic peptides: A redundant arsenal of membrane-damaging virulence factors. Front. Cell. Infect. Microbiol. 2012;2:12. doi: 10.3389/fcimb.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noumi E., Merghni A., Alreshidi M., Del Campo R., Adnan M., Haddad O., De Feo V., Snoussi M. Phenotypic and Genotypic Characterization with MALDI-TOF-MS Based Identification of Staphylococcus spp. Isolated from Mobile Phones with their Antibiotic Susceptibility, Biofilm Formation, and Adhesion Properties. Int. J. Environ. Res. Public Health. 2020;17:3761. doi: 10.3390/ijerph17113761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisher K., Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology. 2009;155:1749–1757. doi: 10.1099/mic.0.026385-0. [DOI] [PubMed] [Google Scholar]
- 31.LEPIDI A. Pet-to-Man Travelling Staphylococci. Academic Press; Cambridge, MA, USA: 2018. Chapter 12—Staphylococcal Lipases; pp. 147–159. [Google Scholar]
- 32.Gundogan N., Ataol O., Torlak F.O. Determination of Some Virulence Factors in Staphylococcus aureus, Enterococcus faecalis and Enterococcus faecium Isolated from Meat and Milk Products. J. Food Saf. 2012;33:387–393. doi: 10.1111/jfs.12062. [DOI] [Google Scholar]
- 33.Gundogan N., Ataol O. Biofilm, protease and lipase properties and antibiotic resistance profiles of staphylococci isolated from various foods. Afr. J. Microbiol. Res. 2013;33:3582–3588. doi: 10.5897/AJMR2012.2316. [DOI] [Google Scholar]
- 34.Zhao J., Ma M., Zeng Z., Yu P., Gong D., Deng S. Production, purification and biochemical characterisation of a novel lipase from a newly identified lipolytic bacterium Staphylococcus caprae NCU S6. J. Enzym. Inhib. Med. Chem. 2021;36:248–256. doi: 10.1080/14756366.2020.1861607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poyart C., Quesne G., Boumaila C., Trieu-Cuot P. Rapid and accurate species-level identification of coagulase-negative staphylococci by using the sodA gene as a target. J. Clin. Microbiol. 2001;39:4296–4301. doi: 10.1128/JCM.39.12.4296-4301.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinchuk I.V., Beswick E.J., Reyes V.E. Staphylococcal enterotoxins. Toxins. 2010;2:2177–2197. doi: 10.3390/toxins2082177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sospedra I., Soriano J.M., Mañes J. Enterotoxinomics: The omic sciences in the study of staphylococcal toxins analyzed in food matrices. Food Res. Int. 2013;54:1052–1060. doi: 10.1016/j.foodres.2013.03.002. [DOI] [Google Scholar]
- 38.De Freitas Guimarães F., Nóbrega D.B., Richini-Pereira V.B., Marson P.M., de Figueiredo Pantoja J.C., Langoni H. Enterotoxin genes in coagulase-negative and coagulase-positive staphylococci isolated from bovine milk. J. Dairy Sci. 2013;96:2866–2872. doi: 10.3168/jds.2012-5864. [DOI] [PubMed] [Google Scholar]
- 39.Banaszkiewicz S., Wałecka-Zacharska E., Schubert J., Tabiś A., Król J., Stefaniak T., Węsierska E., Bania J. Staphylococcal Enterotoxin Genes in Coagulase-Negative Staphylococci—Stability, Expression, and Genomic Context. Int. J. Mol. Sci. 2022;23:2560. doi: 10.3390/ijms23052560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu D.L., Li S., Fang R., Ono H.K. Update on molecular diversity and multipathogenicity of staphylococcal superantigen toxins. Anim. Dis. 2021;1:7. doi: 10.1186/s44149-021-00007-7. [DOI] [Google Scholar]
- 41.Hoveida L., Ataei B., Amirmozafari N., Noormohammadi Z. Species variety, antibiotic susceptibility patterns and prevalence of enterotoxin genes in Staphylococci isolated from foodstuff in Central Iran. Iran. J. Public Health. 2020;49:96. doi: 10.18502/ijph.v49i1.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saber H., Jasni A.S., Jamaluddin T.Z.M.T., Ibrahim R. A review of staphylococcal cassette chromosome mec (SCCmec) types in coagulase-negative staphylococci (CoNS) species. Malays. J. Med. Sci. 2017;24:7–18. doi: 10.21315/mjms2017.24.5.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ballhausen B., Kriegeskorte A., Schleimer N., Peters G., Becker K. The mecA homolog mecC confers resistance against-lactams in Staphylococcus aureus irrespective of the genetic strain background. Antimicrob. Agents Chemother. 2014;58:3791–3798. doi: 10.1128/AAC.02731-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Humphries R.M., Magnano P., Burnham C.A.D., Dien Bard J., Dingle T.C., Callan K., Westblade L.F. Evaluation of surrogate tests for the presence of mecA-mediated methicillin resistance in Staphylococcus capitis, Staphylococcus haemolyticus, Staphylococcus hominis, and Staphylococcus warneri. J. Clin. Microbiol. 2020;59:e02290-20. doi: 10.1128/JCM.02290-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Al-Saadi D.A.A., Abd Al-Mayahi F.S. Antibiogram Susceptibility Patterns of Staphylococcus Aureus Harboring of MecA Gene and Prevalence Aminoglycoside Modifying Enzymes (AMEs) Genes in Iraq. IOP Conf. Ser. Earth Environ. Sci. 2021;923:012049. doi: 10.1088/1755-1315/923/1/012049. [DOI] [Google Scholar]
- 46.Perumal N., Murugesan S., Krishnan P. Distribution of genes encoding aminoglycoside-modifying enzymes among clinical isolates of methicillin-resistant staphylococci. Indian J. Med. Microbiol. 2016;34:350–352. doi: 10.4103/0255-0857.188339. [DOI] [PubMed] [Google Scholar]
- 47.Klingenberg C., Sundsfjord A., Rønnestad A., Mikalsen J., Gaustad P., Flægstad T. Phenotypic and genotypic aminoglycoside resistance in blood culture isolates of coagulase-negative staphylococci from a single neonatal intensive care unit, 1989–2000. J. Antimicrob. Chemother. 2004;54:889–896. doi: 10.1093/jac/dkh453. [DOI] [PubMed] [Google Scholar]
- 48.Ardic N., Sareyyupoglu B., Ozyurt M., Haznedaroglu T., Ilga U. Investigation of aminoglycoside modifying enzyme genes in methicillin-resistant staphylococci. Microbiol. Res. 2006;161:49–54. doi: 10.1016/j.micres.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 49.Emaneini M., Taherikalani M., Eslampour M.A., Sedaghat H., Aligholi M., Jabalameli F., Shahsavan S., Sotoudeh N. Phenotypic and genotypic evaluation of aminoglycoside resistance in clinical isolates of staphylococci in Tehran, Iran. Microb. Drug Resist. 2009;15:129–132. doi: 10.1089/mdr.2009.0869. [DOI] [PubMed] [Google Scholar]
- 50.Martineau F., Picard F.J., Lansac N., Ménard C., Roy P.H., Ouellette M., Bergeron M.G. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2000;44:231–238. doi: 10.1128/AAC.44.2.231-238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chaudhury A., Kumar A.G. In vitro activity of antimicrobial agents against oxacillin resistant staphylococci with special reference to Staphylococcus haemolyticus. Indian J. Med. Microbiol. 2007;25:50–52. doi: 10.1016/S0255-0857(21)02234-9. [DOI] [PubMed] [Google Scholar]
- 52.Goudarzi G., Tahmasbi F., Anbari K., Ghafarzadeh M. Distribution of genes encoding resistance to macrolides among staphylococci isolated from the nasal cavity of hospital employees in Khorramabad, Iran. Iran. Red Crescent Med. J. 2016;18:e25701. doi: 10.5812/ircmj.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roberts M.C., Sutcliffe J., Courvalin P., Jensen L.B., Rood J., Seppala H. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 1999;43:2823–2830. doi: 10.1128/AAC.43.12.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leclercq R. Mechanisms of resistance to macrolides and lincosamides: Nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 2002;34:482–492. doi: 10.1086/324626. [DOI] [PubMed] [Google Scholar]
- 55.Szemraj M., Czekaj T., Kalisz J., Szewczyk E.M. Differences in distribution of MLS antibiotics resistance genes in clinical isolates of staphylococci belonging to species: S. epidermidis, S. hominis, S. haemolyticus, S. simulans and S. warneri. BMC Microbiol. 2019;19:124. doi: 10.1186/s12866-019-1496-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Munier A.L., de Lastours V., Barbier F., Chau F., Fantin B., Ruimy R. Comparative dynamics of the emergence of fluoroquinolone resistance in staphylococci from the nasal microbiota of patients treated with fluoroquinolones according to their environment. Int. J. Antimicrob. Agents. 2015;46:653–659. doi: 10.1016/j.ijantimicag.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 57.Pegues D.A., Colby C., Hibberd P.L., Cohen L.G., Ausubel F.M., Calderwood S.B., Hooper D.C. The epidemiology of resistance to ofloxacin and oxacillin among clinical coagulase-negative staphylococcal isolates: Analysis of risk factors and strain types. Clin. Infect. Dis. 1998;26:72–79. doi: 10.1086/516270. [DOI] [PubMed] [Google Scholar]
- 58.Persson-Waller K., Aspán A., Nyman A., Persson Y., Grönlund Andersson U. CNS species and antimicrobial resistance in clinical and subclinical bovine mastitis. Vet. Microbiol. 2011;152:112–116. doi: 10.1016/j.vetmic.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 59.Nunes R.S.C., Del Aguila E.M., Paschoalin V.M.F. Safety evaluation of the coagulase-negative staphylococci microbiota of salami: Superantigenic Toxin Production and antimicrobial resistance. BioMed Res. Int. 2015;2015:483–548. doi: 10.1155/2015/483548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thomas D.Y., Jarraud S., Lemercier B., Cozon G., Echasserieau K., Etienne J., Gougeon M.L., Lina G., Vandenesch F. Staphylococcal enterotoxin-like toxins U2 and V, two new staphylococcal superantigens arising from recombination within the enterotoxin gene cluster. Infect. Immun. 2006;74:4724–4734. doi: 10.1128/IAI.00132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seng P., Boushab B.M., Romain F., Gouriet F., Bruder N., Martin C., Papazian L. Emerging role of Raoultella ornithinolytica in human infections: A series of cases and review of the literature. Int. J. Infect. Dis. 2016;45:65–71. doi: 10.1016/j.ijid.2016.02.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.