Summary
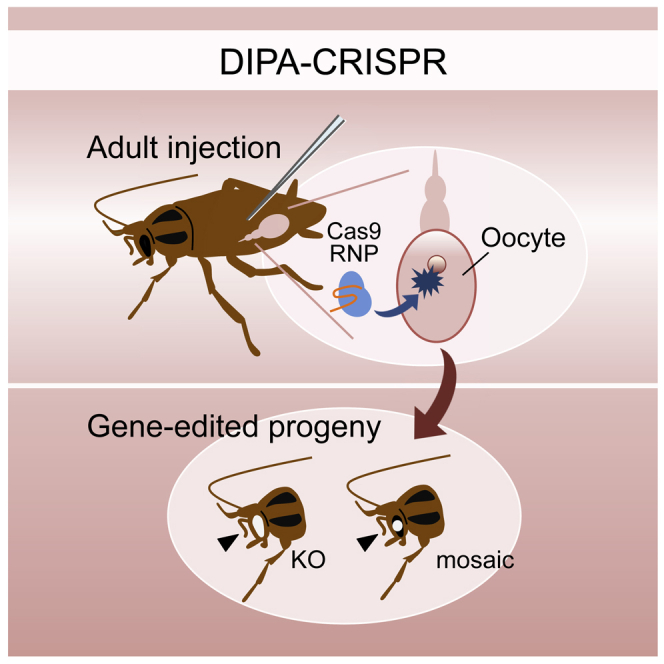
Current approaches for insect gene editing require microinjection of materials into early embryos. This severely limits the application of gene editing to a great number of insect species, especially to those whose reproduction systems preclude access to early embryos for injection. To overcome these limitations, we report a simple and accessible method for insect gene editing, termed “direct parental” CRISPR (DIPA-CRISPR). We show that injection of Cas9 ribonucleoproteins (RNPs) into the haemocoel of adult females efficiently introduces heritable mutations in developing oocytes. Importantly, commercially available standard Cas9 protein can be directly used for DIPA-CRISPR, which makes this approach highly practical and feasible. DIPA-CRISPR enables highly efficient gene editing in the cockroaches, on which conventional approaches cannot be applied, and in the model beetle Tribolium castaneum. Due to its simplicity and accessibility, DIPA-CRISPR will greatly extend the application of gene editing technology to a wide variety of insects.
Keywords: CRISPR-Cas9, DIPA-CRISPR, gene editing, genome editing, cockroaches, beetles
Graphical abstract
Highlights
-
•
A simple and efficient method for insect gene editing
-
•
Based on direct adult injection of Cas9 ribonucleoproteins
-
•
Readily implementable in non-specialist laboratories
-
•
Applicable to a wide diversity of non-model insects
Motivation
With over a million species described, insects are a treasure trove of diversity and represent boundless possibilities as research tools for answering fundamental questions in biology. Current approaches for insect gene editing require microinjection of materials into early embryos, which is highly challenging in most species. In this work, we established and optimized a simple and efficient method for insect gene editing by adult injection, which can be readily implemented in any laboratory and directly applied to a great diversity of non-model insect species.
Shirai et al. develop a simple and accessible method, DIPA-CRISPR, for insect gene editing by direct adult injection of Cas9 ribonucleoproteins. Using it, they successfully establish gene knockouts in cockroaches, in which conventional embryo microinjection cannot be applied. Given its simplicity and versatility, DIPA-CRISPR has the potential to greatly extend the application of gene editing technology to a wide diversity of insects.
Introduction
Recent advances of genome editing tools have enabled sophisticated engineering of insect genomes (Doudna and Charpentier, 2014; Gantz and Akbari, 2018; Matthews and Vosshall, 2020). However, current approaches rely on embryo injection, which requires expensive equipment, a specific experimental setup for each species, and highly skilled laboratory personnel (Matthews and Vosshall, 2020; Tamura et al., 2000). Furthermore, embryo injection must be completed in a small time window, from oviposition to preblastoderm stage, which is not applicable to species that give live birth rather than lay eggs (e.g., viviparous aphids and flies) or species in which an access to very early embryos is highly challenging (e.g., cockroaches, which encapsulate the eggs into a hard egg case or ootheca).
Recently, an alternative method that can bypass the requirement of embryo injection has been developed in the mosquito Aedes aegypti (Chaverra-Rodriguez et al., 2018). In this method, called Receptor-Mediated Ovary Transduction of Cargo (ReMOT), peptide ligands derived from yolk protein precursors are fused to Cas9 protein, and the complex of the engineered Cas9 and single-guide RNAs (sgRNAs) is injected into female adults to introduce mutations in developing oocytes. ReMOT-mediated targeted mutagenesis has been successfully used in a few other species, such as the mosquito Anopheles stephensi (Macias et al., 2020), the jewel wasp Nasonia vitripennis (Chaverra-Rodriguez et al., 2020), the red flour beetle Tribolium castaneum (Shirai and Daimon, 2020), and the silverleaf whitefly Bemisia tabaci (Heu et al., 2020). Although these examples are encouraging, the ReMOT approach appears to have a number of limitations. For example, the results reported so far indicate that the peptide ligands that are fused to Cas9 should be specifically tuned to a target species for efficient gene editing (Chaverra-Rodriguez et al., 2018; Heu et al., 2020; Shirai and Daimon, 2020), which may be a barrier for non-specialist laboratories.
Interestingly, however, the above studies, together with the study in the spider mite Tetranychus urticae (Dermauw et al., 2020), also showed that a very small number of gene-edited individuals could be recovered by adult injection of non-tagged Cas9 (i.e., Cas9 without a ligand sequence). Thus, these studies suggest that, although the addition of an appropriate ligand tag to Cas9 could increase gene editing efficiency, its addition is not essential for gene editing by adult injection.
During the course of our work aiming at developing a novel peptide tag for Cas9 that can cover a broad range of insect groups (i.e., a fragment of yolk protein precursor vitellogenin [Li et al., 2003; Murakami et al., 2019; discussed in Shirai and Daimon, 2020]), our group found that female adult injection of non-tagged Cas9 can efficiently introduce heritable mutations in developing oocytes of the German cockroach Blattella germanica, to which conventional approaches (i.e., embryo injection) are not feasible. As we used commercially available standard Cas9 protein (i.e., the one sold for general genome editing experiments in animals and cultured cells), such “non-tagged Cas9 approach” would become a more generalized method for insect gene editing by adult injection. To further explore this possibility, we here optimized this method and established it as an accessible technology for insect gene editing, which we named “direct parental” CRISPR (DIPA-CRISPR).
After exploring different optimizing conditions of DIPA-CRISPR in B. germanica, we demonstrated that gene editing efficiency (GEF; the proportion of edited individuals out of the total number of individuals hatched) could reach as high as 21.8%, which easily enabled the first establishment of knockout cockroach lines. Furthermore, we tested DIPA-CRISPR in the red flour beetle T. castaneum, in which GEF reached over 50%, a percentage comparable with the efficiency reached in conventional approaches (Gilles et al., 2015). Furthermore, we were able to generate gene knockin beetles by co-injecting single-stranded oligonucleotides (ssODNs) and Cas9 RNPs. The successful application of DIPA-CRISPR in the two evolutionarily distant insect species gives an idea of its generalizability. Without the need of custom-engineering of Cas9 or the use of special reagents that have been considered to facilitate the ovary uptake of injected Cas9 ribonucleoproteins (RNPs) (Chaverra-Rodriguez et al., 2018, 2020), DIPA-CRISPR could be readily implemented in any laboratory, so that it would greatly extend the application of gene editing to a wide diversity of insect species.
Results and discussion
Cockroach gene editing with DIPA-CRISPR
In general, cockroach females ovulate the oocytes into the genital atrium, where they are fertilized and then encapsulated into a hard egg case, or ootheca, where they will remain for days or weeks until egg hatching (Figure 1A) (Cornwell, 1968). Because of this unique reproduction system, it is impracticable to inject materials into very early embryos, thus genetic manipulation of cockroaches (i.e., transgenesis or gene editing) has not been achieved so far. To investigate whether adult injection of Cas9 RNPs enables cockroach gene editing, we tested non-tagged Cas9 in B. germanica, a global urban pest whose genome has been sequenced (Harrison et al., 2018), by targeting the autosomal eye color gene cinnabar (Figure S1), which is involved in the biosynthesis of ommochrome pigments (Lorenzen et al., 2002; Quan et al., 2002).
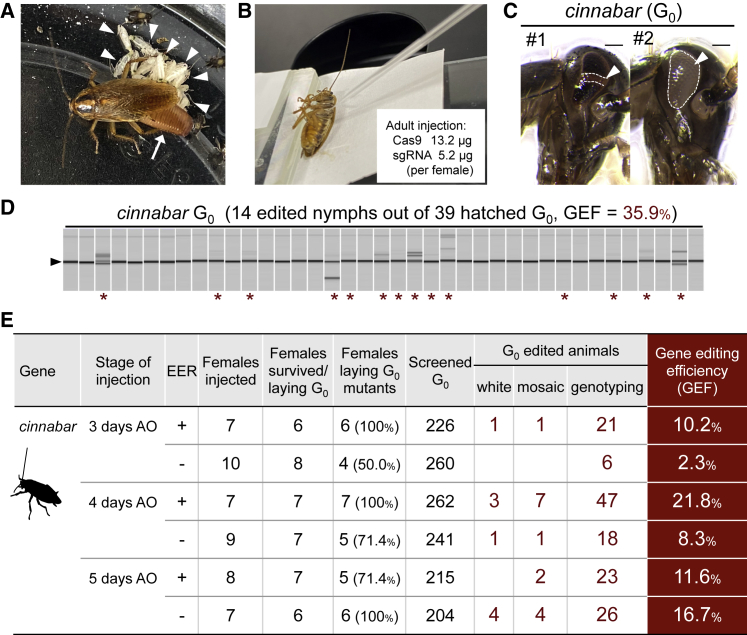
Figure 1.
Cockroach gene editing with DIPA-CRISPR
(A) Newly hatched nymphs (arrowheads) from the ootheca (arrow) of Blattella germanica.
(B) Adult injection in B. germanica.
(C) The eyes of cinnabar mosaic G0 nymphs with loss of black eye pigments. Scale bars, 100 μm.
(D) A representative result of cinnabar G0 genotyping. G0 nymphs with normal eyes hatched from a single ootheca were analyzed by heteroduplex mobility assay (HMA). Individuals having edited alleles (asterisks) and the homoduplex bands (156 bp, arrowhead) are indicated. See also Figure S1 for the detailed results.
(E) Gene editing efficiency (GEF) of DIPA-CRISPR. G0 mutants were first screened by phenotypes (white or mosaic), and then those without phenotypes were analyzed by HMA (genotyping). White, having phenotypes in entire regions of both eyes; AO, after ootheca drop; EER, presence or absence of chloroquine (2 mM) in injection solution.
We first injected commercial Cas9 RNPs into 16 fully matured females not carrying oothecae (Figure 1B). We presumed that they were undergoing a vitellogenic cycle, thus the injected Cas9 RNPs might be non-selectively incorporated into the growing oocytes with vitellogenins by receptor-mediated endocytosis (Ciudad et al., 2006; Cooper and Hausman, 2007; Dermauw et al., 2020; Raikhel and Dhadialla, 1992). Notably, 31% (5 out of 16) of the injected females that produced an ootheca after the injection yielded gene-edited G0 (generation zero) progeny. Of the nine edited G0 nymphs recovered from 385 hatchlings (GEF = 2.3%), two were eye color mosaics (Figure 1C), and the other seven carried edited alleles as judged by genotyping experiments (see Figure 1D for representative results). As cinnabar is an autosomal gene, the mosaic phenotypes observed in the G0 insects indicate that the incorporated Cas9 RNPs might persist in the oocyte for several days (i.e., from injection to fertilization) and then disrupted paternal alleles after fertilization, yielding cells having biallelic mutations.
The results obtained in this first experiment encouraged us to explore optimal conditions of our method, DIPA-CRISPR, by using carefully staged adults. We tested females at selected days of their reproductive cycle (Cornwell, 1968; Pascual et al., 1992; Treiblmayr et al., 2006) and found that injecting 4 days after ootheca drop and eggs hatching resulted in the highest efficiency (Figures 1E and S1C). In this condition, all the seven injected females produced gene-edited G0 nymphs, and they yielded 57 edited nymphs from 262 hatchlings in total (GEF = 21.8%) (Figure 1E). Notably, some G0 nymphs had “white” eyes (i.e., the entire surface of both eyes was white), suggesting that biallelic mutations were introduced at a very early stage of embryogenesis. When the GEF values of injected females were individually measured, some females had values exceeding 40% (Figure S1C). Collectively, our results show that highly efficient cockroach gene editing can be achieved by simple adult injection.
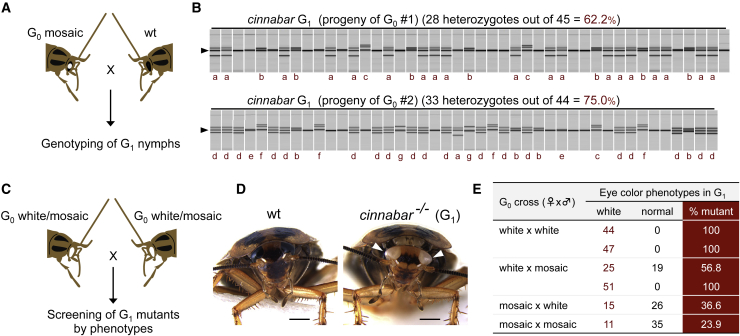
Inheritance of edited alleles in cockroaches
To test if edited alleles in G0 cockroaches are inherited to the next generation, we performed crossing experiments (Figure 2). When the two mosaic nymphs obtained in the first experiment (Figure 1C) were reared to adults and crossed to wild type (Figure 2A), 62.2% and 75.0% of their respective progenies (G1) were heterozygous mutants (Figure 2B), showing very high germline mutation rates in these mosaics. To further confirm this result, G0 adults with eye color phenotypes (white or mosaic) were crossed and their G1 progenies were screened for knockout phenotypes (Figure 2C). Notably, when G0 adults with both eyes white were crossed, all of their G1 progeny became white-eyed knockout insects (Figures 2D and 2E). This suggests that cinnabar was disrupted in all (or practically all) cells in the white-eyed G0 individuals. Similarly, very high germline mutation rates were also exhibited in mosaic-eyed G0 adults, which were roughly estimated to be >50% (Figure 2E). Together, our results demonstrate that DIPA-CRISPR is a powerful method that easily enabled the first establishment of knockout cockroaches.
Figure 2.
Inheritance of edited alleles in cockroaches
(A and B) A mating scheme (A) and the results of genotyping (B) of G1 individuals. The two cinnabar mosaics shown in Figure 1C (G0 #1 and #2) were crossed to wild type, and all the G1 progeny were individually analyzed by heteroduplex mobility assay. The mutant alleles are indicated below the panels (a–g, seven alleles in total). See Figure S1D for their nucleotide sequences.
(C) A mating scheme for screening of G1 knockout insects. G0 insects with eye color phenotypes were crossed to obtain G1 progenies.
(D) A cinnabar knockout G1 adult (right). Arrowheads indicate the white eyes. Scale bars, 1 mm.
(E) Phenotypes in G1 insects. Each row indicates the result of a single-pair mating of G0.
DIPA-CRISPR in beetles
To demonstrate potential for broad use, we applied DIPA-CRISPR to much more evolutionarily modified species, the red flour beetle T. castaneum (Figure 3). For these experiments, we targeted cardinal, an eye color gene on the X chromosome, as their mutant phenotypes are easily visible in hemizygous G0 males without genotyping (females = XX and males = XY in T. castaneum) (Grubbs et al., 2015; Shirai and Daimon, 2020).
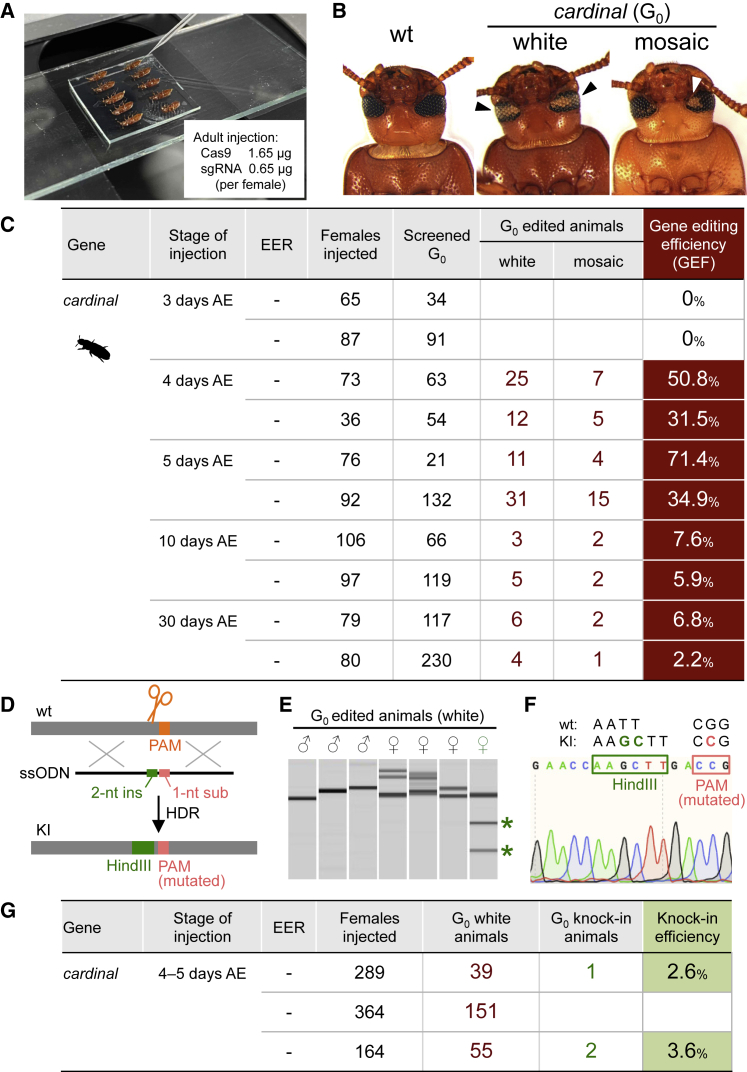
Figure 3.
DIPA-CRISPR in beetles
(A) Adult injection in Tribolium castaneum.
(B) G0 adults generated by DIPA-CRISPR targeting cardinal. Arrowheads indicate the loss of black eye pigments.
(C) The gene editing efficiency (GEF) in T. castaneum. See Figure S2 for the detailed results.
(D) A scheme of knockin using ssODNs as a template for homology-directed repair (HDR). ssODNs that contain 5′- and -3′-homology arms (96 nt each), a 2-nt insertion (producing a HindIII site) and a 1-nt substitution (mutating the PAM sequence) are knocked into the cardinal gene.
(E) A representative result of G0 genotyping. G0 adults with both eyes white were individually genotyped. The products of genomic PCR were digested with HindIII and analyzed by microchip electrophoresis. Asterisks indicate the HindIII-digested products.
(F) The nucleotide sequence of the recovered knockin (KI) allele.
(G) The efficiency of knockin through DIPA-CRISPR in three independent experiments. All the three knockin beetles carried the precise knockin allele shown in (F).
We injected Cas9 RNPs into females at selected days after adult emergence (Figures 3A and S2). We found that injection into 4- or 5-day-old adult females exhibited very high gene editing efficiency, with the GEF values being as high as 50.8% for 4-day-old females (32 out of 63 hatchlings) and 71.4% for 5-day-old females (15 out of 21) (Figures 3B and 3C), which is comparable with the efficiency in conventional embryo injection approaches (Gilles et al., 2015).
Cockroaches and beetles used in this study are evolutionarily very distant (Polyneopteran versus Endopterygote) (Harrison et al., 2018; Misof et al., 2014), show radically different modes of metamorphosis (hemimetabolan versus holometabolan) (Belles, 2020), and have different types of ovaries (panoistic in B. germanica versus telotrophic in T. castaneum) (McLaughlin and Bratu, 2015). Furthermore, previous ReMOT studies have shown that gene editing can be achieved by adult injection in wasps or mosquitos (Chaverra-Rodriguez et al., 2018, 2020; Macias et al., 2020), which have the most derived types of ovaries (i.e., polytrophic). Thus, these results point to DIPA-CRISPR as a generalizable approach for insect gene editing.
Gene knockin by DIPA-CRISPR in beetles
We next tested whether DIPA-CRISPR could be used to generate gene knockin insects, using T. castaneum as the experimental subject. We designed an ssODN having homology arms (96 nt each for 5′- and -3′-homology arms), a 2-nt insertion that introduces a novel HindIII restriction site, and a 1-nt substitution that mutates PAM, and used it as a template for homology-directed repair (HDR) (Figures 3D–3G). After injecting a mixture of Cas9 RNPs targeting cardinal and the ssODNs into female adults at optimized stages, we screened for white-eyed G0 adults. Genotyping of these adults showed that three adults (1.2%, 3 out of 245 in total) carried precise knockin alleles generated by HDR (Figures 3E and 3F). Although the efficiency is still low and should be further improved, our results indicate that the application of DIPA-CRISPR can be extended to knockin experiments.
Cas9 for DIPA-CRISPR
The direct use of commercial Cas9 protein for adult injection in the DIPA-CRISPR can eliminate time-consuming processes required in a similar adult injection approach (ReMOT) (Chaverra-Rodriguez et al., 2018), such as the development of a novel ligand that is tuned to target species, engineering of Cas9, and the expression and purification of recombinant Cas9 protein. Thus, the use of commercial Cas9 can enable gene editing in any non-specialist laboratory that cannot implement the above elaborated methods.
We also investigated and compared the performance of commercial Cas9 products from additional three companies in the market (Figure S3A). Although details were not disclosed to the authors, these Cas9 products should be engineered differently by manufacturers in many ways (e.g., the type, number, and location of nuclear localization signals [NLSs] or other epitope/purification tags). Nevertheless, their gene editing efficiencies were comparable and very high when tested in T. castaneum (GEF = 24%–32%; Figure S3A). This indicates that there is little requirement to use a particular Cas9 product, although the presence of NLSs should be essential for the delivery of Cas9 RNPs to the nucleus.
It is of note that, unlike this study, very little gene editing was observed with non-tagged Cas9 in previous ReMOT studies. We speculate that the large differences in gene editing efficiency between this study and previous attempts are probably due to the difference in the preparation of Cas9 and doses of Cas9 injected. In previous studies, Cas9 was purified only by a single step of affinity chromatography, which might have led to a product contaminated with undesired materials. Although commercial Cas9 was tested in the jewel wasp N. vitripennis (Chaverra-Rodriguez et al., 2020), the dose used was much lower than that used in this study (0.36 versus 3.3 μg/μL in injection solution). To examine this view, we tested a serial dilution of Cas9 RNPs in T. castaneum and found a clear trend showing that decreased doses of Cas9 RNPs result in the decreased gene editing efficiency (Figures S3B and S3C). We thus propose using relatively high doses of commercial Cas9 when implementing our method to other species.
The use of endosomal escape reagents
It has been reported that the efficiency of ReMOT-mediated gene editing can be improved with the use of endosomal escape reagents (EERs) that facilitate the release of Cas9 RNPs from the endosome to the cytosol (Chaverra-Rodriguez et al., 2018, 2020; Heu et al., 2020; Macias et al., 2020). Similarly, in our experiments, there were cases where the use of EER increased the efficiency (see Figures 1E and S1C). However, the effect of EER was not always clear, especially at some time points (see Figures S1C and S2B). Furthermore, the number of the eggs laid and/or hatched decreased in some cases (Figure S2B). As EERs often reduce survival rates and/or fecundity of the injected females (Chaverra-Rodriguez et al., 2018, 2020; Heu et al., 2020; Macias et al., 2020), their use in new target species needs empirical optimization through multiple rounds of experiments. Thus, we propose to use only two components, Cas9 and sgRNA, in our DIPA-CRISPR approach, which greatly simplifies the procedures for gene editing experiments.
A key parameter for DIPA-CRISPR
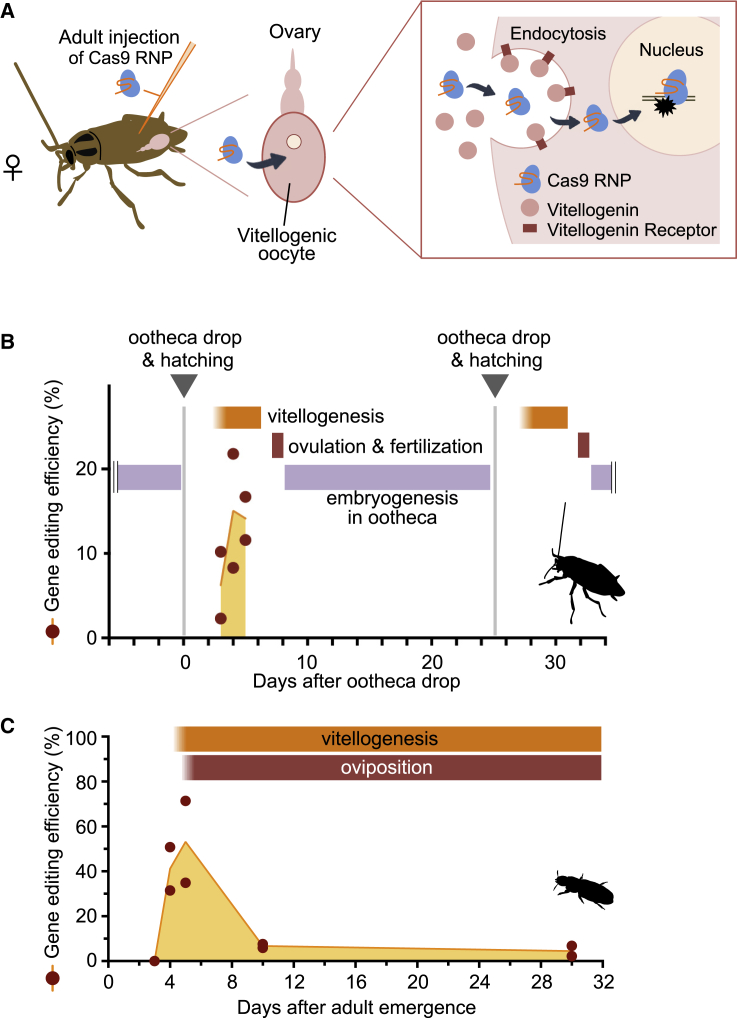
Our experiments demonstrated that the most critical parameter for successful gene editing by DIPA-CRISPR is the stage of the adult females injected (Figure 4), which is also shown in the previous ReMOT studies. In the species tested in this study, the highest GEF was achieved with females actively undergoing vitellogenesis (Cornwell, 1968; Parthasarathy et al., 2010; Pascual et al., 1992; Treiblmayr et al., 2006). This finding, together with the fact that endocytosis results in the non-selective uptake of extracellular materials (Cooper and Hausman, 2007), and that cultured insect ovaries can uptake and accumulate non-vitellogenin proteins (e.g., mouse IgG and bovine γ-globulin) (Kindle et al., 1988; Koller et al., 1989), suggests that the incorporation of Cas9 RNPs into vitellogenic oocytes occurs concomitantly with the massive uptake of vitellogenins from the hemolymph (Raikhel and Dhadialla, 1992) (Figure 4A). Thus, a good knowledge of the vitellogenesis process in the target species can be an important prerequisite for using DIPA-CRISPR.
Figure 4.
A schematic model and description of DIPA-CRISPR
(A) A schematic model of the uptake of Cas9 ribonucleoproteins (RNPs) by the oocyte. Vitellogenic oocytes massively uptake vitellogenins circulating in the hemolymph via receptor-mediated endocytosis (Raikhel and Dhadialla, 1992). Along with vitellogenin, injected Cas9 RNPs are likely non-selectively incorporated into the endosome of oocytes. Although the details of the process of endosomal escape are not clear, Cas9 RNPs then disrupt target genes in the nuclei of developing oocytes and/or fertilized embryos, producing gene-edited G0 insects.
(B and C) Relationship of gene editing efficiency and ovary development. Gene editing efficiencies are plotted against the stage of females of B. germanica (B) and T. castaneum (C). Each point represents the GEF value of each biological replication. Our results suggest that it is critical to set the right timing of adult injection, depending on the reproductive physiology of target species (i.e., continuous in T. castaneum versus discontinuous in B. germanica). The timetable of ovary development is inferred from Parthasarathy et al. (2010), Pascual et al. (1992), and Treiblmayr et al. (2006).
Like most insects, females of T. castaneum produce eggs continuously (i.e., they lay small number of eggs every day), whereas some insects such as cockroaches produce eggs in discrete batches (i.e., discontinuous reproductive cycle) (Figures 4B and 4C). Thus, vitellogenesis occurs almost throughout the adult stage in the former group, whereas at certain times in the latter group. Interestingly, we found a clear peak of GEF values in the former group. In T. castaneum, GEF values peaked on days 4 and 5 after emergence, after which the values decreased to a basal level (Figure 4C). Notably, this peak corresponds to the time of the onset of vitellogenesis that begins on day 4 after adult eclosion (Parthasarathy et al., 2010). The reason why these early stages give a very high efficiency is not clear, but our results would be helpful to design DIPA-CRISPR experiments in new target species.
DIPA-CRISPR as an accessible method for insect gene editing
Our method requires only minimal equipment for adult injection, such as a stereomicroscope and a micromanipulator commonly used for larval/nymphal RNAi (Linz et al., 2014; Posnien et al., 2009). Thus, it could be readily implemented in any laboratory. Furthermore, the minimal requirement of reagents (i.e., Cas9 protein and sgRNA) makes this method highly practical and feasible.
As adult injection requires a much larger amount of injection solution compared with embryo microinjection (μL versus nL scale per injection), the cost of reagents required for DIPA-CRISPR is expected to be higher than that for the conventional method. Thus, we calculated the cost of commercial Cas9 per recovered G0 edited insects (Figure S4). When high doses of Cas9 RNPs (3.3 μg/μL Cas9 in injection solution) were injected at optimized stages, the cost was calculated to be 2.0–7.4 USD for B. germanica (Figure S4A) and 4.1–10.5 USD for T. castaneum (Figure S4B). We consider that this cost is at an accessible level given the minimal requirement of equipment and no need to produce in-house Cas9. On the other hand, our results in T. castaneum suggested that the cost could not be reduced by decreasing the dose of Cas9 RNPs (Figure S4C), because this decreases the total number of recovered mutants (Figure S3C). Thus, injection of relatively high doses of Cas9 RNPs would be a reasonable option, as this optimizes the chances of success without largely increasing the cost of reagents.
Due to its simplicity and accessibility, DIPA-CRISPR will greatly extend the application of gene editing technology to a wide variety of model and non-model insects, including global/local agricultural and medical pests whose genomes have not been manipulated in any way.
Limitations of the study
DIPA-CRISPR requires a good knowledge of ovary development in target species, since injected Cas9 RNPs utilize the process of vitellogenesis to be internalized into the oocyte. Thus, a precise staging of vitellogenic females may be crucial. However, this can be challenging in some species, given the diverse life histories and reproductive strategies in insects. Also, DIPA-CRISPR would not be applicable to species in which oogenesis proceeds without apparent vitellogenesis (e.g., aphids undergoing parthenogenetic reproduction) (Miura et al., 2003), or species in which vitellogenesis is mainly an ovarian autosynthetic process. For example, in the fruit fly Drosophila melanogaster and some higher Diptera, the major source of yolk proteins is the follicle cells surrounding the oocytes, but not the extraovarian fat body (Brennan et al., 1982; Houseman and Morrison, 1986). Furthermore, the temporal and spatial range of “patency,” the opening of intercellular spaces between follicle cells that allows hemolymph-borne materials to reach the surface of oocytes (Raikhel and Dhadialla, 1992), is more limited in D. melanogaster than in most other species (Isasti-Sanchez et al., 2021; Row et al., 2021). To empirically establish the limits of our method in ovarian autosynthetic species, we tested it in D. melanogaster (Table S1), verifying that it does not work in them. Thus, our DIPA-CRISPR approach would not be directly applicable to some peculiar species that undergo a highly derived mode of vitellogenesis.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| T7 RiboMAX Express Large Scale RNA Production System | Promega | Cat#P1320 |
| Phenol:chloroform:isoamyl alcohol mixture | Sigma-Aldrich | Cat#77619 |
| Alt-R® S.p. Cas9 Nuclease V3 | Integrated DNA Technologies | Cat#1081059 |
| Cas9 Protein | Sigma-Aldrich | Cat#CAS9PROT |
| Cas9 Nuclease protein NLS | FUJIFILM Wako | Cat#316-08651 |
| GenomeCraft Cas9 | Fasmac | Cat#GE-005-S |
| Chloroquine Diphosphate | FUJIFILM Wako | Cat#038-17971 |
| Saponin Quilaja sp. | Sigma-Aldrich | Cat#S4521 |
| KOD FX Neo | TOYOBO | Cat#KFX-201 |
| Experimental models: Organisms/strains | ||
| B. germanica wildtype Japanese strain | Sumika Technoservice | N/A |
| T. castaneum wildtype Okinawa strain | Shirai and Daimon, 2020 | N/A |
| D. melanogaster wildtype Canton S strain | Gift from Christen Mirth | N/A |
| Oligonucleotides | ||
| PCR primers used in this study, see Method Details | This paper | N/A |
| ssODN (5′- to -3′) used as a HDR template: ACCCTTTATCCGAATTTAATGTCACTTGTA TGGAATTTGTGCGGTCGGCAAATGCCGC CACTTGTTGTCTGGGGCCCAGGGAACAG ATGAACCAAGCTTGACCGCGTTTATAGAC GGGTCGGTTATTTACGGGGTGGAGGAAA AGACAGTTGGGGCGCTCCGGACGATGTC AGGGGGTGAACTCGAAATGTTTG |
Integrated DNA Technologies | N/A (Ultramer® DNA Oligonucleotides) |
| Recombinant DNA | ||
| pDR274 | Hwang et al., 2013 | Addgene Plasmid #42250 |
| Software and algorithms | ||
| GraphPad Prism (v.6.0h) | GraphPad | RRID:SCR_002798 |
| Leica Application Suite X (LASX) | Leica Microsystems | https://www.leica-microsystems.com/ |
| Adobe Photoshop | Adobe | RRID:SCR_014199 |
| Other | ||
| Femtojet 4i Microinjector | Eppendorf | Cat#5252000021 |
| MultiNA Microchip Electrophoresis System | SHIMADZU | MCE-202 |
| Needle puller | Narishige | PC-100 |
| Borosilicate glass with filament | Sutter Instrument | BF-100-50-10 |
| Stereomicroscope | Leica Microsystems | M165FC |
| Digital microscope camera | Leica Microsystems | DFC7000T |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Takaaki Daimon (daimon.takaaki.7a@kyoto-u.ac.jp).
Materials availability
The knockout lines generated in this study is available on reasonable request to the lead contact.
Experimental model and subject details
Insects
A Blattella germanica colony derived from a Japanese population was maintained at 25 ± 1.5°C under a 16 h:8 h light:dark cycle with a constant supply of solid feed (MF, Oriental Yeast) and water. A Tribolium castaneum (Okinawa strain) colony was maintained on wheat flour containing 5% (w/w) brewer’s dry yeast at 30 ± 1°C and 50%–70% relative humidity as described previously (Shirai and Daimon, 2020). The wildtype Drosophila melanogaster strain (Canton S) was reared using a commercial Drosophila diet (Formula 4-24 Instant Drosophila Food, Carolina Biological, Cat#173210).
Method details
Preparation of Cas9-sgRNA RNPs
Single-guide RNAs (sgRNAs) targeting B. germanica cinnabar (GenBank: PSN36199), T. castaneum cardinal (GenPept: XP_008200769), and D. melanogaster white (GenBank: NM_057439) were synthesized as described previously (Shirai and Daimon, 2020). Briefly, annealed oligo DNA was cloned into the BsaI site of the pDR274 vector (Hwang et al., 2013). After linearization with DraI, the vector was used as a template for in vitro transcription using the T7 RiboMAX Express Large Scale RNA Production System (Promega, Cat#P1320). The synthesized sgRNAs were extracted with phenol (pH4–5):chloroform:isoamyl alcohol (125:24:1) (Sigma, Cat#77619), and then precipitated with isopropanol and dissolved in RNase-free water. For D. melanogaster white, we also purchased and used chemically synthesized sgRNAs from the Integrated DNA Technologies (IDT) (Alt-R® CRISPR-Cas9 sgRNA). Otherwise stated, commercial Cas9 protein purchased from IDT (Alt-R® S.p. Cas9 Nuclease V3, Cat#1081059), which has nuclear localization signals and a C-terminal 6-His tag (further details were not disclosed to the authors), was used in this study. Cas9 protein and sgRNAs were mixed at a molar ratio of approximately 1:2, and incubated for 10–15 min at room temperature to allow Cas9 RNP formation. In some experiments, freshly-prepared chloroquine (FUJIFILM Wako, Cat#038-17971) or saponin (Sigma, Cat#S4521) was added as an endosomal escape reagent (EER) (Chaverra-Rodriguez et al., 2018). Concentrations of Cas9 RNPs and EERs in the injection solution were adjusted with RNase-free water, without adding any other reagents (e.g., buffers or salts). The target sequences of sgRNAs are (5'- to -3′): GGTCTGGCTGTAGTCAAACA for B. germanica cinnabar sgRNA1; TTGGAGGCATGCAAAGCTCC for B. germanica cinnabar sgRNA2; GGAACAGATGAACCAAGTGA for T. castaneum cardinal sgRNA1 (Shirai and Daimon, 2020); CATTAACCAGGGCTTCGGGC for D. melanogaster white sgRNA1 (Ren et al., 2014); and AGCGACACATACCGGCGCCC for D. melanogaster white sgRNA2 (Ren et al., 2014).
Adult injection and mutant screening in Blattella germanica
Female adults carrying the ootheca were collected from a stock colony, monitored daily for ootheca drop, and were staged based on the day after the ootheca drop. The injection was performed using a glass capillary needle equipped with Femtojet 4i (Eppendorf). The females used for injection were anesthetized on ice. Approximately 4 μL of the Cas9 RNP solution containing 3.3 μg/μL Cas9 (IDT, Cat#1081059) and 1.3 μg/μL sgRNAs (a mixture of sgRNA1 and sgRNA2, Figure S1A) with or without chloroquine (2 mM) was injected into the ventral abdomen of the female adults. Injected females were individually reared in containers until the formation of the next ootheca and hatching of G0 nymphs (nymphs hatched ∼20–30 days after injection with ∼20–50 nymphs hatched from each ootheca). The eye colors of hatched G0 nymphs were examined, and all the nymphs without external phenotypes were subjected to individual genotyping. B. germanica cinnabar is an autosomal gene, as we found heterozygous males [male = XO and female = XX in B. germanica (Meisel et al., 2019)].
Genotyping of Blattella germanica
Genomic DNAs were extracted individually as described previously (Daimon et al., 2015). Genomic PCR was conducted using KOD FX Neo (TOYOBO, Cat#KFX-201). Mutations were screened by analyzing the PCR products using the heteroduplex mobility assay (HMA) using the MultiNA Microchip Electrophoresis System (MCE-202, Shimadzu). Primer sequences for HMA of B. germanica cinnabar are (5'- to -3′): GAAGGCGGATTTGATCATAGGAGC and CAATCACTTACCTCACCATCTTCTG. To determine the nucleotide sequences of mutant alleles, Sanger sequencing chromatograms were analyzed with Poly Peak Parser program (Hill et al., 2014). Primer sequences for Sanger sequencing of B. germanica cinnabar are (5'- to -3′): GGCGCACTTGAGGCAGATATG and TTCCCCTACACTTCAATGCGGG.
Adult injection and mutant screening in Tribolium castaneum
Female adults at selected days after adult emergence, separated from males at the time of injection, were injected with approximately 0.5 μL of the Cas9 RNP solution containing 3.3 μg/μL Cas9 (IDT, Cat#1081059) and 1.3 μg/μL sgRNA, with or without saponin (100 ng/μL), as described previously (Shirai and Daimon, 2020). The injected females were grouped with males in a container with wheat flour and transferred to a new container every 24 hours to examine the relationship between the day of egg laying and the gene editing efficiencies in the hatchlings. To screen gene-edited individuals, the eye colors of the G0 insects were examined during pupal and adult stages. We also examined and compared the performance of Cas9 products from three companies additional to IDT: Sigma (Cat #CAS9PROT), FUJIFILM Wako (Cat#316-08651), and Fasmac (Cat#GE-005-S), which have a single or multiple nuclear localization signals, by targeting cardinal under the same condition (i.e., the same stage of injection and concentration of reagents). As cardinal gene locates on the X chromosome (female = XX, male = XY) (Shirai and Daimon, 2020), mutant phenotypes are not visible in heterozygous females. As we screened G0 insects based on phenotypes but not on genotypes, the GEF values for T. castaneum cardinal in this study were most likely underestimated. Primer sequences for Sanger sequencing of T. castaneum cardinal are (5'- to -3′): GGCCAAAACCGGGGCGCTTCC and CCGGAAGTTCGTGGGTACAAGCCCG (Shirai and Daimon, 2020).
Gene knock-in experiments in Tribolium castaneum
Female adults at optimized stages (i.e., 4–5 days after adult emergence) were injected as above. Injection solution contained 3.3 μg/μL Cas9 (IDT, Cat#1081059), 1.3 μg/μL sgRNA (sgRNA1 for cardinal), and ssODNs (1.6 μg/μL). ssODNs were purchased from IDT (Ultramer DNA Oligonucleotides). Injected females were allowed to lay eggs for two days, and the recovered G0 adults with both eyes whites were subjected to genotyping. For genotyping, genomic DNAs of G0 adults were individually extracted, and used as a template for PCR. PCR products were digested with HindIII and analyzed by microchip electrophoresis using the MultiNA Microchip Electrophoresis System (MCE-202, Shimadzu). Primer sequences for T. castaneum cardinal are (5'- to -3′): GTCACACATCCGGAGTGCTTTCC and GAGTTCACCCCCTGACATCGTC. To determine the nucleotide sequences of knock-in alleles, PCR products were subcloned and subjected to Sanger sequencing.
Adult injection and mutant screening in Drosophila melanogaster
Female adults at selected times after adult emergence, separated from males at the time of injection, were injected with approximately 0.5 μL of the Cas9 RNP solution containing 3.3 μg/μL Cas9 (IDT, Cat#1081059) and 1.3 μg/μL sgRNA (a mixture of sgRNA1 and sgRNA2 for white), with or without chloroquine (0.5 or 2.0 mM). The injected females were grouped with males in a vial and transferred to a new vial every 24 hours. To screen gene-edited individuals, the eye colors of the G0 insects were examined during adult stages.
Quantification and statistical analysis
Data on gene editing efficiency (GEF) of the G0 progenies (Figure S1C) were analyzed with the Mann-Whitney nonparametric U test. Statistical analyses were performed in the Prism software (Graphpad Software).
Acknowledgments
We thank Masashi Tanaka, Takahiro Ohde, Takashi Koyama, Masato Kinoshita, and Jason L. Rasgon for helpful suggestions in the early stage of this project. We are grateful to three anonymous reviewers for their many constructive and insightful suggestions and comments. This research was supported by JSPS KAKENHI (nos. 20H02999 and 20K21311 to T.D.); JSPS Open Partnership Joint Research Projects (no. JPJSBP120209917 to T.D.); Spanish Ministry of Innovation and Competitiveness (PID2019-104483GB-I00 to X.B.); CSIC-Spain (2019AEP028 to M.-D.P.); and in part by Cabinet Office, Government of Japan, Cross-ministerial Moonshot Agriculture, Forestry and Fisheries Research and Development Program (no. JPJ009237 to T.D.). Y.S. is a JSPS Research Fellow with a research grant (DC1, 21J20658).
Author contributions
Conceptualization, Y.S. and T.D.; methodology, all authors; investigation, Y.S.; writing – original draft, Y.S. and T.D.; writing – review & editing, all authors; resources, Y.S. and T.D.; visualization, Y.S. and T.D.; funding acquisition, all authors; supervision, T.D.
Declaration of interests
The authors declare no competing interests.
Published: May 16, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2022.100215.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This study does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Belles X. Academic Press; 2020. Insect Metamorphosis: From Natural History to Regulation of Development and Evolution. [Google Scholar]
- Brennan M.D., Weiner A.J., Goralski T.J., Mahowald A.P. The follicle cells are a major site of vitellogenin synthesis in Drosophila melanogaster. Dev. Biol. 1982;89:225–236. doi: 10.1016/0012-1606(82)90309-8. [DOI] [PubMed] [Google Scholar]
- Chaverra-Rodriguez D., Dalla Benetta E., Heu C.C., Rasgon J.L., Ferree P.M., Akbari O.S. Germline mutagenesis of Nasonia vitripennis through ovarian delivery of CRISPR-Cas9 ribonucleoprotein. Insect Mol. Biol. 2020;29:569–577. doi: 10.1111/imb.12663. [DOI] [PubMed] [Google Scholar]
- Chaverra-Rodriguez D., Macias V.M., Hughes G.L., Pujhari S., Suzuki Y., Peterson D.R., Kim D., McKeand S., Rasgon J.L. Targeted delivery of CRISPR-Cas9 ribonucleoprotein into arthropod ovaries for heritable germline gene editing. Nat. Commun. 2018;9:3008. doi: 10.1038/s41467-018-05425-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciudad L., Piulachs M.D., Bellés X. Systemic RNAi of the cockroach vitellogenin receptor results in a phenotype similar to that of the Drosophila yolkless mutant. FEBS J. 2006;273:325–335. doi: 10.1111/j.1742-4658.2005.05066.x. [DOI] [PubMed] [Google Scholar]
- Cooper G.M., Hausman R.E. ASM Press; 2007. The Cell: A Molecular Approach. [Google Scholar]
- Cornwell P.B. Hutchinson Press; 1968. The Cockroach. Volume 1. A laboratory insect and an industrial pest. [Google Scholar]
- Daimon T., Uchibori M., Nakao H., Sezutsu H., Shinoda T. Knockout silkworms reveal a dispensable role for juvenile hormones in holometabolous life cycle. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E4226–E4235. doi: 10.1073/pnas.1506645112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermauw W., Jonckheere W., Riga M., Livadaras I., Vontas J., Van Leeuwen T. Targeted mutagenesis using CRISPR-Cas9 in the chelicerate herbivore Tetranychus urticae. Insect Biochem. Mol. Biol. 2020;120:103347. doi: 10.1016/j.ibmb.2020.103347. [DOI] [PubMed] [Google Scholar]
- Doudna J.A., Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Gantz V.M., Akbari O.S. Gene editing technologies and applications for insects. Curr. Opin. Insect Sci. 2018;28:66–72. doi: 10.1016/j.cois.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles A.F., Schinko J.B., Averof M. Efficient CRISPR-mediated gene targeting and transgene replacement in the beetle Tribolium castaneum. Development. 2015;142:2832–2839. doi: 10.1242/dev.125054. [DOI] [PubMed] [Google Scholar]
- Grubbs N., Haas S., Beeman R.W., Lorenzen M.D. The ABCs of eye color in Tribolium castaneum: orthologs of the Drosophila white, scarlet, and brown genes. Genetics. 2015;199:749–759. doi: 10.1534/genetics.114.173971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M.C., Jongepier E., Robertson H.M., Arning N., Bitard-Feildel T., Chao H., Childers C.P., Dinh H., Doddapaneni H., Dugan S., et al. Hemimetabolous genomes reveal molecular basis of termite eusociality. Nat. Ecol. Evol. 2018;2:557–566. doi: 10.1038/s41559-017-0459-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heu C.C., McCullough F.M., Luan J., Rasgon J.L. CRISPR-Cas9-based genome editing in the silverleaf whitefly (Bemisia tabaci) CRISPR J. 2020;3:89–96. doi: 10.1089/crispr.2019.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J.T., Demarest B.L., Bisgrove B.W., Su Y.C., Smith M., Yost H.J. Poly peak parser: method and software for identification of unknown indels using sanger sequencing of polymerase chain reaction products. Dev. Dyn. 2014;243:1632–1636. doi: 10.1002/dvdy.24183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman J.G., Morrison P.E. Absence of female-specific protein in the hemolymph of stable fly Stomoxys calcitrans (L.)(Diptera: Muscidae) Arch. Insect Biochem. Physiol. 1986;3:205–213. doi: 10.1002/arch.940030210. [DOI] [Google Scholar]
- Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q., Sander J.D., Peterson R.T., Yeh J.R., Joung J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isasti-Sanchez J., Münz-Zeise F., Lancino M., Luschnig S. Transient opening of tricellular vertices controls paracellular transport through the follicle epithelium during Drosophila oogenesis. Dev. Cell. 2021;56:1083–1099.e5. doi: 10.1016/j.devcel.2021.03.021. [DOI] [PubMed] [Google Scholar]
- Kindle H., König R., Lanzrein B. In vitro uptake of vitellogenin by follicles of the cockroach Nauphoeta cinerea: comparison of artificial media with haemolymph media and role of vitellogenin concentration and of juvenile hormone. J. Insect Physiol. 1988;34:541–548. doi: 10.1016/0022-1910(88)90196-5. [DOI] [Google Scholar]
- Noah Koller C., Dhadialla T.S., Raikhel A.S. Selective endocytosis of vitellogenin by oocytes of the mosquito, Aedes aegypti: an in vitro study. Insect Biochem. 1989;19:693–702. doi: 10.1016/0020-1790(89)90106-6. [DOI] [Google Scholar]
- Li A., Sadasivam M., Ding J.L. Receptor-ligand interaction between vitellogenin receptor (VtgR) and vitellogenin (Vtg), implications on low density lipoprotein receptor and apolipoprotein B/E. J. Biol. Chem. 2003;278:2799–2806. doi: 10.1074/jbc.m205067200. [DOI] [PubMed] [Google Scholar]
- Linz D.M., Clark-Hachtel C.M., Borràs-Castells F., Tomoyasu Y. Larval RNA interference in the red flour beetle, Tribolium castaneum. J. Vis. Exp. 2014;92:e52059. doi: 10.3791/52059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen M.D., Brown S.J., Denell R.E., Beeman R.W. Cloning and characterization of the Tribolium castaneum eye-color genes encoding tryptophan oxygenase and kynurenine 3-monooxygenase. Genetics. 2002;160:225–234. doi: 10.1093/genetics/160.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias V.M., McKeand S., Chaverra-Rodriguez D., Hughes G.L., Fazekas A., Pujhari S., Jasinskiene N., James A.A., Rasgon J.L. Cas9-mediated gene-editing in the malaria mosquito Anopheles stephensi by ReMOT Control. G3: Genes Genom. Genet. 2020;10:1353–1360. doi: 10.1534/g3.120.401133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B.J., Vosshall L.B. How to turn an organism into a model organism in 10 ‘easy’ steps. J. Exp. Biol. 2020;223:jeb218198. doi: 10.1242/jeb.218198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J.M., Bratu D.P. Humana Press; 2015. Drosophila melanogaster Oogenesis: An Overview. [DOI] [PubMed] [Google Scholar]
- Meisel R.P., Delclos P.J., Wexler J.R. The X chromosome of the German cockroach, Blattella germanica, is homologous to a fly X chromosome despite 400 million years divergence. BMC Biol. 2019;17:100–114. doi: 10.1186/s12915-019-0721-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misof B., Liu S., Meusemann K., Peters R.S., Donath A., Mayer C., Frandsen P.B., Ware J., Flouri T., Beutel R.G., et al. Phylogenomics resolves the timing and pattern of insect evolution. Science. 2014;346:763–767. doi: 10.1126/science.1257570. [DOI] [PubMed] [Google Scholar]
- Miura T., Braendle C., Shingleton A., Sisk G., Kambhampati S., Stern D.L. A comparison of parthenogenetic and sexual embryogenesis of the pea aphid Acyrthosiphon pisum (Hemiptera: aphidoidea) J. Exp. Biol. B: Mol. Dev. Evol. 2003;295B:59–81. doi: 10.1002/jez.b.3. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Horibe T., Kinoshita M. Development of an efficient bioreactor system for delivering foreign proteins secreted from liver into eggs with a vitellogenin signal in medaka Oryzias latipes. Fish. Sci. 2019;85:677–685. doi: 10.1007/s12562-019-01320-4. [DOI] [Google Scholar]
- Parthasarathy R., Sheng Z., Sun Z., Palli S.R. Ecdysteroid regulation of ovarian growth and oocyte maturation in the red flour beetle, Tribolium castaneum. Insect Biochem. Mol. Biol. 2010;40:429–439. doi: 10.1016/j.ibmb.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual N., Cerdá X., Benito B., Tomás J., Piulachs M., Bellés X. Ovarian ecdysteroid levels and basal oöcyte development during maturation in the cockroach Blattella germanica (L.) J. Insect Physiol. 1992;38:339–348. doi: 10.1016/0022-1910(92)90058-l. [DOI] [Google Scholar]
- Posnien N., Schinko J., Grossmann D., Shippy T.D., Konopova B., Bucher G. RNAi in the red flour beetle (Tribolium) Cold Spring Harb. Protoc. 2009;2009 doi: 10.1101/pdb.prot5256. prot5256. [DOI] [PubMed] [Google Scholar]
- Quan G.X., Kim I., Komoto N., Sezutsu H., Ote M., Shimada T., Kanda T., Mita K., Kobayashi M., Tamura T. Characterization of the kynurenine 3-monooxygenase gene corresponding to the white egg 1 mutant in the silkworm Bombyx mori. Mol. Genet. Genom. 2002;267:1–9. doi: 10.1007/s00438-001-0629-2. [DOI] [PubMed] [Google Scholar]
- Raikhel A.S., Dhadialla T.S. Accumulation of yolk proteins in insect oocytes. Ann. Rev. Entomol. 1992;37:217–251. doi: 10.1146/annurev.en.37.010192.001245. [DOI] [PubMed] [Google Scholar]
- Ren X., Yang Z., Xu J., Sun J., Mao D., Hu Y., Yang S.J., Qiao H.H., Wang X., Hu Q., et al. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 2014;9:1151–1162. doi: 10.1016/j.celrep.2014.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Row S., Huang Y.-C., Deng W.-M. Developmental regulation of oocyte lipid intake through ‘patent’ follicular epithelium in Drosophila melanogaster. iScience. 2021;24:102275. doi: 10.1016/j.isci.2021.102275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai Y., Daimon T. Mutations in cardinal are responsible for the red-1 and peach eye color mutants of the red flour beetle Tribolium castaneum. Biochem. Biophys. Res. Commun. 2020;529:372–378. doi: 10.1016/j.bbrc.2020.05.214. [DOI] [PubMed] [Google Scholar]
- Tamura T., Thibert C., Royer C., Kanda T., Eappen A., Kamba M., Komoto N., Thomas J.L., Mauchamp B., Chavancy G., et al. Germline transformation of the silkworm Bombyx mori L. using a piggyBac transposon-derived vector. Nat. Biotechnol. 2000;18:81–84. doi: 10.1038/71978. [DOI] [PubMed] [Google Scholar]
- Treiblmayr K., Pascual N., Piulachs M.-D., Keller T., Belles X. Juvenile hormone titer versus juvenile hormone synthesis in female nymphs and adults of the German cockroach, Blattella germanica. J. Insect Sci. 2006;6:1–7. doi: 10.1673/031.006.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This study does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.