Abstract
Simple Summary
The family Vietnamellidae (Ephemeroptera) is one of the oldest insect families in the world. However, there are still controversies about the phylogenetic relationships among Vietnamellidae, Ephemerellidae, and Teloganodidae. The mitochondrial (mt) genome can be used to discuss phylogenetic relationships and cryptic species. We sequenced and annotated three complete mt genomes of Vietnamella sinensis from three different populations, identifying a cryptic species of V. sinensis and discuss the phylogenetic relationships of Vietnamellidae. Based on the genetic distance of the whole mt genomes, the phylogenetic relationship of three populations was uncovered and the divergence time of V. sinensis QY indicated that it was a cryptic species of V. sinensis.
Abstract
Ephemeroptera (Insecta: Pterygota) are widely distributed all over the world with more than 3500 species. During the last decade, the phylogenetic relationships within Ephemeroptera have been a hot topic of research, especially regarding the phylogenetic relationships among Vietnamellidae. In this study, three mitochondrial genomes from three populations of Vienamella sinensis collected from Tonglu (V. sinensis TL), Chun’an (V. sinensis CN), and Qingyuan (V. sinensis QY) in Zhejiang Province, China were compared to discuss the potential existence of cryptic species. We also established their phylogenetic relationship by combining the mt genomes of 69 Ephemeroptera downloaded from NCBI. The mt genomes of V. sinensis TL, V. sinensis CN, and V. sinensis QY showed the same gene arrangement with lengths of 15,674 bp, 15,674 bp, and 15,610 bp, respectively. Comprehensive analyses of these three mt genomes revealed significant differences in mt genome organization, genetic distance, and divergence time. Our results showed that the specimens collected from Chun’an and Tonglu in Zhejiang Province, China belonged to V. sinensis, and the specimens collected from Qingyuan, Zhejiang Province, China were a cryptic species of V. sinensis. In maximum likelihood (ML) and Bayesian inference (BI) phylogenetic trees, the monophyly of the family Vietnamellidae was supported and Vietnamellidae has a close relationship with Ephemerellidae.
Keywords: mitochondrial genome, cryptic species, phylogenetic relationship, divergence time
1. Introduction
The mitochondrial (mt) genome is the most extensively studied genomic system in insects, and is widely used to explore phylogenetic relationships due to its characteristics of maternal inheritance and high evolutionary rate. [1]. Insect mt genomes are usually double-stranded circular molecules of 14–20 kb in length, encoding 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs), and a control region (CR; AT-rich region) [2]. Many researchers have stimulated great interest in the insects’ mt genome including testing hypotheses about microevolution, mt gene expression, population structure analysis, phylogenetic relationships, and identification of cryptic species [1,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17]. Since William Derham discovered the first cryptic species in 1718, these species are still a principal subject of several research areas and actually have a history of 300 years [18]. The study of cryptic species not only has great significance for the promotion of theories in related fields, but also is important for the quantifying biodiversity and conservation of species. Despite some controversy, most current studies define a cryptic species as one that is indistinguishable in morphology but significantly differentiated at the genetic level [19].
Ephemeroptera are generally referred to as mayflies, but have other aliases such as dayflies and fishflies [20]. Mayflies are an ancient lineage of insects, of which there are now 42 families, 400 genera, and 3500 species [21]. As early as 1988, Elliott et al. showed that Ephemeroptera play a significant role among freshwater fauna and can be used as one of the standards for water quality testing [22]. Ephemeroptera have a worldwide distribution, occurring on many continents, large islands, and archipelagos (except for Antarctica) [23,24]. Because Ephemeroptera are relatively primitive among Pterygota, considerable effort has been devoted to constructing the phylogenetic relationships within Ephemeroptera based on morphology [25,26,27], molecular evidence [28], and combined data [14,29]. The controversial points are mainly the following aspects: the phylogenetic relationship of Heptageniidae and Baetidae, and the phylogenetic relationship among Vietnamellidae, Teloganodidae, and Ephemerellidae [29,30,31].
The genus Vietnamella (Ephemeroptera: Vietnamellidae) was originally established by Tshernova in 1972, based on Vietnamella thani collected in Vietnam, and described the morphological structure of nymphs [32,33]. In 1997, McCafferty and Wang established that the subfamily Austremerellinae of the family Teloganodidae included the genus Vietnamella and Austremerella [34]. Then, this subfamily was elevated to family rank in 2000 and named Vietnamellidae [35]. In 2006, Jacobus and McCafferty moved Austremerella back to an indeterminate Austremerellidae and limited Vietnamellidae to include only one genus, Vietnamella [36]. In 2017, Hu et al. described V. dabieshanensis You and Su, 1987, V. qingyuanensis Zhou and Su, 1995, and V. guadunensis Zhou and Su, 1995 as junior synonyms of V. sinensis Hsu, 1936 [37]. As of 2021, there are six valid described Vietnamella species including V. thani Tshernova, 1972, V. ornata Tshernova, 1972, V. sinensis, V. maculosa Auychinda et al., 2020, V. nanensis Auchyinda et al., 2020, and V. chebalingensis Tong, 2020 [32,38,39,40].
The phylogenetic relationships among Vietnamellidae, Teloganodidae, and Ephemerellidae are controversial. In the early years, the most widely accepted early classification system was from McCafferty and Kluge, in which the phylogenetic relationship of Vietnamellidae was closer to Teloganodidae and more distant to Ephemerellidae [27,34]. In 2005, morphological data used by Ogden and Whiting suggested that Vietnamellidae was the sister clade to Teloganodidae, and the clade of (Vietnamellidae + Teloganodidae) formed a sister group to Ephemerellidae [28]. In 2009, Ogden et al. combined the molecular data (sRNA, IrRNA, 18S rDNA, 28S rDNA, and H3 genes) and morphological data suggesting that Vietnamellidae, Teloganodidae, and Ephemerellidae were in a parallel relationship, and the phylogenetic relationships among them had not been effectively analyzed [29]. In recent years, Li et al. [41,42,43], Zhang et al. [44], Cai et al. [12], and Gao et al. [45] reconstructed the phylogeny of Ephemeroptera based on mt genomes, all of which supported the formation of sister groups between Ephemerellidae and Vietnamellidae.
In the present study, we sequenced the mt genomes from three populations of V. sinensis from Chun’an, Tonglu, and Qingyuan in Zhejiang Province, China to discuss the phylogenetic relationship of Vietnamellidae and the cryptic species of V. sinensis.
2. Materials and Methods
2.1. Sample Collection and Morphological Identification
The Vietnamella larvae were collected by D-frame aquatic insect net from streams of Tonglu (29°79′ N, 119°68′ E), Chun’an (24°41′ N, 120°52′ E), and Qingyuan (27°61′ N, 119°06′ E), Zhejiang Province, China in July 2019. The morphological structure of Vietnamella larvae including leg, head, antenna, maxilla, labium, hypopharynx, mandible, labrum and circus were dissected, observed, and photographed under an optical SMZ-1500 stereomicroscope (Nikon, Tokyo, Japan). After morphological identification, samples were stored in 100% ethanol at Zhang’s laboratory, College of Life Science and Chemistry, Zhejiang Normal University, China. According to the description of Hu et al. [37], the three populations of Vietnamella larvae were identified as V. sinensis.
2.2. DNA Extraction, PCR Amplification, and Sequencing
Total genomic DNA from hind-leg muscle tissue of each specimen was extracted using the Ezup Column Animal Genomic DNA Purification Kit (Sangon Biotech Company, Shanghai, China). The experimental design was approved by the Animal Research Ethics Committees of Zhejiang Normal University.
Several fragments were amplified using universal primers [6]. We then used Primer Premier 5.0 to design specific primers based on sequences previously obtained from universal primers [46]. The reaction conditions for normal PCR (product length <3000 bp) and long PCR (product length >3000 bp) were performed as described in Zhang et al. [47]. After we performed electrophoresis and gel purification of PCR products, all PCR products were sequenced at Sangon Biotech Company (Shanghai, China) using bidirectional sequencing.
2.3. Mitochondrial Genome Annotation and Sequence Analyses
Manual proofreading and splicing of all nucleotide fragments were conducted using SeqMan in the DNASTAR Package v.7.1 [48]. Annotation of tRNA genes and identification of their secondary structures were identified by the online website MITOS (http://mitos.bioinf.uni-leipzig.de/index.py) (accessed on 15 September 2021) [49]. Referring to the complete mt genomes of other Ephemeroptera species, we identified the two rRNA genes (12S and 16S rRNA) using the Clustal W program of Mega 7.0 [50] and aligned the amino acid sequences of PCGs. Mega 7.0 was also used to calculate the genetic distance. We calculated the AT content and relative synonymous codon usage rate (RSCU) of the three genomes [16] using PhyloSuite v.1.2.2 [51]. With reference to invertebrate genetic codes, the nucleic acid sequences of the 13 PCGs were translated into amino acid sequences [16,52]. The three mt genomes were deposited in GenBank with accession numbers OK265109–OK265111. We used CG View online server V 1.0 to draw maps of the mt genomes (http://cgview.ca/) (accessed on 20 October 2021) [53], and separately calculated the AT and GC skews according to the following formulae: AT skew = (A − T)/(A + T) and GC skew = (G − C)/(G + C) [54].
2.4. Phylogenetic Analyses
In order to further explore the phylogenetic relationship of Vietnamellidae within Ephemeroptera, we combined the three mt genomes in this study with 69 previously sequenced Ephemeroptera mt genomes (Table S1) downloaded from NCBI including sequences from Ameletidae (1), Baetidae (4), Caenidae (5), Ephemerellidae (12), Ephemeridae (4), Heptageniidae (24), Isonychiidae (4), Leptophlebiinae (3), Polymitarcyidae (1), Potamanthidae (3), Siphlonuridae (5), Teloganodidae (2), and Vietnamellidae (3) [8,9,12,13,14,41,45,55,56,57,58,59,60,61,62].
Bayesian inference (BI) and maximum likelihood (ML) phylogenetic trees were constructed based on a dataset of the nucleotide sequences of 13 PCGs. Because the third codon was saturated, all phylogenetic analyses were performed using the first and second codons. In addition, two mt genomes of Siphluriscus chinensis (HQ875717, MF352165), the most primitive species of Ephemeroptera, was selected as the outgroup species [57,62]. Our test for the heterogeneity of nucleotide sequences between taxa used AliGROOVE with the default settings [63]. In this study, the nucleotide sequences of the 13 PCGs were used for DNA alignment, which was conducted in MAFFT v.7.475 [64]. Gblock 0.91b [65] and PartionFinder version 2.2.1 [66] were employed to select the conserved regions and partitions based on Bayesian information criterion (BIC). The partition schemes and best-fit models selected for each dataset are shown in Table 1. The GTR + I + G model was used to construct ML and BI analyses. The ML analysis was performed by RaxML 8.2.0 software with rapid inference using 1000 ultrafast repetitions [67]. The BI analysis was performed by MrBayes 3.2 [68] to 10 million generations and the mean standard deviation of Bayesian split frequencies was less than 0.01. The first 25% of generations was discarded as burn in. Tracer v1.7.1 [69] was used to detect the convergence to the stationary distribution of the chains. FigTree 1.4 was used to visualize the resulting trees [70].
Table 1.
The partition schemes and best-fitting models selected. The complete names of all abbreviations are as follows: pos1: the first codon; pos2: the second codon; GTR: general time reversible; I: proportion of invariable sites; G: gamma distribution.
| Nucleotide Sequence Alignments | ||
|---|---|---|
| Subset | Subset Partitions | Best Model |
| Partition 1 | COII_pos1, COIII_pos1, Cyt b_pos1, ATP6_pos1 | GTR + I + G |
| Partition 2 | COI_pos2, Cyt b_pos2, COII_pos2, ATP6_pos2, COIII_pos2 | GTR + I + G |
| Partition 3 | ND3_pos1, ND6_pos1, ATP8_pos1, ND2_pos1 | GTR + I + G |
| Partition 4 | ND6_pos2, ATP8_pos2, ND2_pos2, ND3_pos2, ND4L_pos2 | GTR + I + G |
| Partition 5 | COI_pos1 | GTR + I + G |
| Partition 6 | ND4L _pos1, ND1_pos1, ND4_pos1, ND5_pos1 | GTR + I + G |
| Partition 7 | ND1_pos2, ND5_pos2, ND4_pos2 | GTR + I + G |
2.5. Divergence Time Estimation
This study provided fossil calibration points to accurately estimate divergence time [71]. The use of fossil evidence to estimate the divergence time can deduce other taxa without fossils [72]. In total, we included four Ephemeroptera fossils for time-calibration to access the divergence time of Ephemeroptera. The first of the calibration points was the divergence time in Siphlonuridae (159.00–160.60 Mya, an average of 159.80 Mya) [73,74], which was found for new middle Jurassic mayflies from inner Mongolia. The second calibration point was the divergence time in Atalophlebiinae of Leptophlebiidae (15.00–20.00 Mya, 17.50 Mya average) [75,76]. We used this calibration point for Choroterpides apiculate, which belongs to the subfamily Atalophlebiinae. The third calibration point was the divergence time in the genus Ephemerella of Ephemerellidae (41.30–47.80 Mya, 44.55 Mya average) [77]. The final calibration point was the divergence time in Vietnamellidae (98.17–99.41 Mya, 98.79 Mya average) [78], and this fossil was the first fossil record of Vietnamellidae from Mid-Cretaceous Burmese amber. The root age was set to 239 million years ago (the oldest age of Ephemeroptera) [79]. All fossil calibrations used the uniform model.
Based on the topology of maximum likelihood (ML) phylogenetic tree, MCMCTree [80] in the PAML v4.8 package [81] was performed to infer divergence time in Ephemeroptera. In conducting the analysis, we calculated the transition/transversion rate ratio and branch lengths first. When running MCMC, the parameters of the algorithm were set as: burn-in period = 1,000,000, sample frequency = 1000, and number of samples = 10,000. Since MCMC runs need to check convergence, they were run at least twice from different starting points. To check the convergence, we cut the time values from two output files and pasted them into an Excel spreadsheet, then used Excel to draw a scatter diagram of the time set for the first and second runs. The points should be very close to a straight line (the x = y line) [80]. Tracer v1.7.1 [69] was used to confirm the adequate mixing of the MCMC chains of the run and to check that the marginal posterior distribution value (ESS, effective sample size) were greater than 200. Finally, FigTree v1.4 was used to visualize the divergence time of every branch [70].
3. Results
3.1. Mitochondrial Genome Composition
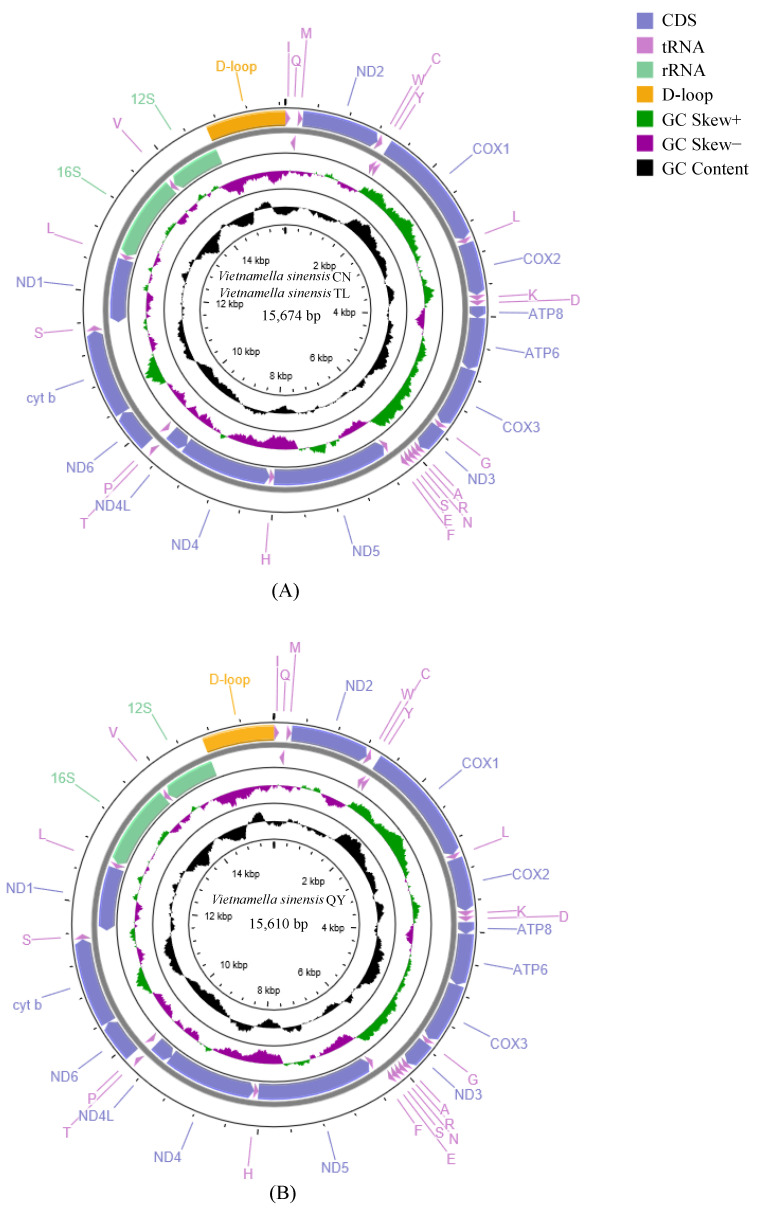
We annotated and uploaded the complete mt genome data of V. sinensis TL (OK265109), V. sinensis CN (OK265111), and V. sinensis QY (OK265110) into the GenBank database. These three mt genomes all showed double circular DNA molecules with lengths of 15,674 bp, 15,674 bp, and 15,610 bp, respectively (Figure 1A,B). The intergenic nucleotides (IGNs) of the three species ranged from 1 to 41 bp (Table S2) and the gene order of the three mt genomes were the same as those of typical insects, with a total 37 genes and an A + T rich region including 22 tRNA genes, two rRNA genes, and 13 PCGs. Of these 37 genes, 23 genes were located on the heavy (H) strand and the remaining 14 genes were on the light (L) strand (Tables S3 and S4). After subsequent analysis, V. sinensis CN and V. sinensis TL had the same length and similar characteristics, so V. sinensis CN and V. sinensis TL were collectively referred to as V. sinensis CN/TL in this paper. The nucleotide composition of the V. sinensis CN/TL mt genome was A = 32.3%, T = 38.2%, C = 17.8%, and G = 11.7%, and was very similar to V. sinensis QY, which was A = 31.9%, T = 37.6%, C = 18.4%, and G = 12.1%. There were strong A + T biases in both V. sinensis CN/TL and V. sinensis QY, 70.5% and 69.5%, respectively. According to the skew statistics, the AT skew was positive, whereas the GC skew was negative (Table 2).
Figure 1.
Circular visualization and organization of the complete mt genome of V. sinensis CN + V. sinensis TL (A) and V. sinensis QY (B). External genes on the circle are encoded by the positive strand (5′→3′) and internal genes are encoded by the negative strand (3′→5′). The second circle shows the GC skew and the third shows the GC content. GC content and GC skew are plotted as the deviation from the average value of the entire sequence. Other items are defined on the figure.
Table 2.
Base composition of the mt genomes of the V. sinensis CN/TL, V. sinensis QY, and V. sinensis (HM067837).
| Region | Strand | V. sinensis CN/TL | V. sinensis QY | V. sinensis (HM067837) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Length (bp) | AT% | AT Skew | GC Skew | Length (bp) | AT% | AT Skew | GC Skew | Length (bp) | AT% | AT Skew | GC Skew | ||
| Whole genome | 15,674 | 70.5 | −0.083 | −0.208 | 15,610 | 69.5 | −0.083 | −0.207 | 15,761 | 70.7 | −0.092 | −0.197 | |
| PCGs | + | 6915 | 67.9 | −0.207 | −0.157 | 6915 | 66.4 | −0.208 | −0.160 | 6915 | 67.7 | −0.214 | −0.153 |
| − | 4311 | 71.6 | −0.147 | 0.289 | 4311 | 70.9 | −0.138 | 0.289 | 4308 | 71.8 | −0.144 | 0.286 | |
| tRNA | + | 910 | 71.3 | −0.005 | 0.034 | 912 | 71.5 | −0.009 | 0.031 | 915 | 71.3 | 0.002 | 0.031 |
| − | 519 | 73.6 | 0.031 | 0.314 | 521 | 74.9 | 0.046 | 0.298 | 520 | 74.8 | 0.059 | 0.313 | |
| rRNA | − | 2015 | 74.3 | 0.106 | 0.216 | 2011 | 74.2 | 0.111 | 0.215 | 2044 | 74.1 | 0.106 | 0.214 |
In both V. sinensis CN/TL and V. sinensis QY mt genomes nine genes (ND2, COI, COII, COIII, ATP6, ATP8, ND3, ND6 and Cyt b) were located on the heavy strand (H-strand), whereas the others (ND4, ND4L, ND5, and ND1) were on the light strand (L-strand) (Tables S3 and S4). Both V. sinensis CN/TL and V. sinensis QY had the same PCG lengths of 11,226 bp (Table 2). Among the three sequenced mt genomes, 13 PCGs used the typical mitochondrial start codon ATN, and 10 PCGs used typical stop codons TAA and TAG. An incomplete codon T was used for the other three PCGs (COII, ND5, and Cyt b). G-C skew values were all negative in PCGs (+), but positive in PCGs (−). Unlike G-C skew values, A-T skew values were all negative in both PCGs (−) and PCGs (+) (Table 2).
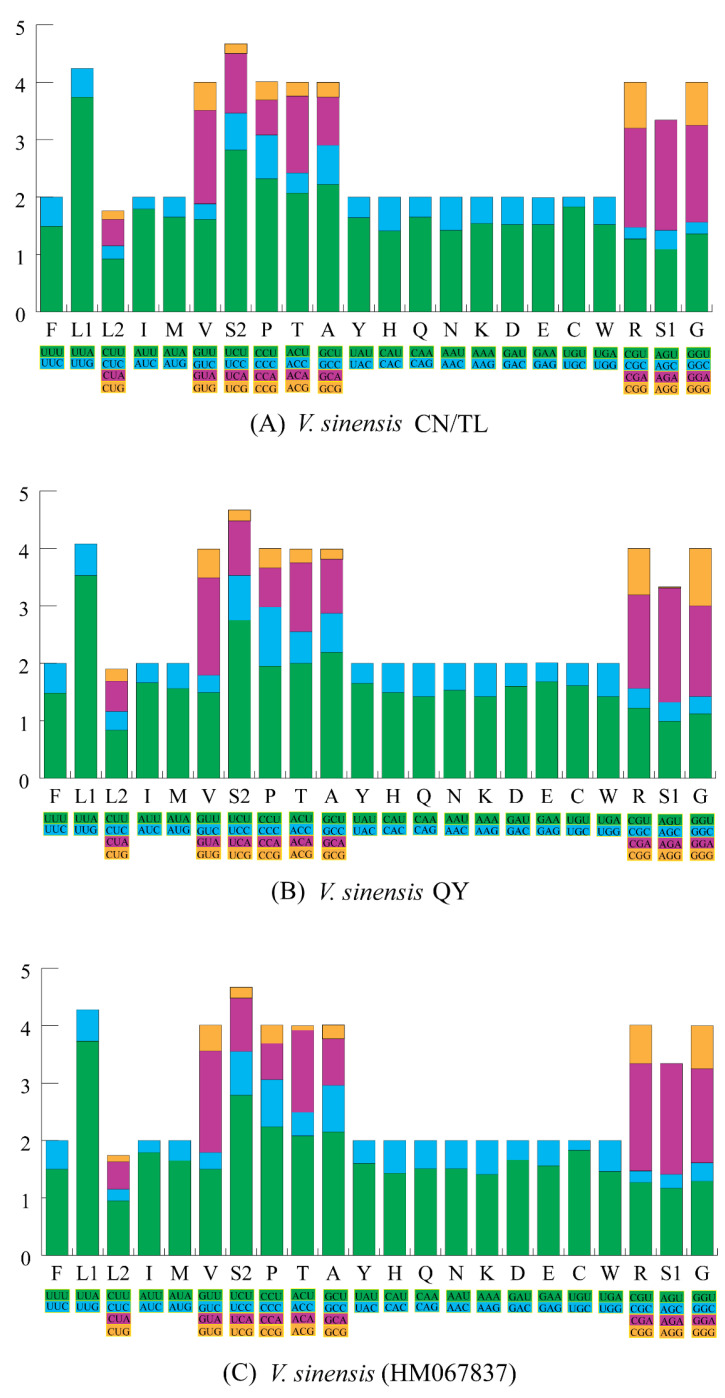
The relative synonymous codon usage (RSCU) of the three mt genomes of V. sinensis was calculated (Figure 2, Table S5). The most commonly codons in the PCGs of V. sinensis CN/TL and V. sinensis QY were UUU (Phe), UUA (Leu), and AUU (Ile), with a frequency of >230 times.
Figure 2.
The relative synonymous codon usage (RSCU) of the mt genome in V. sinensis CN/TL (A), V. sinensis QY (B), and V. sinensis (HM067837) (C).
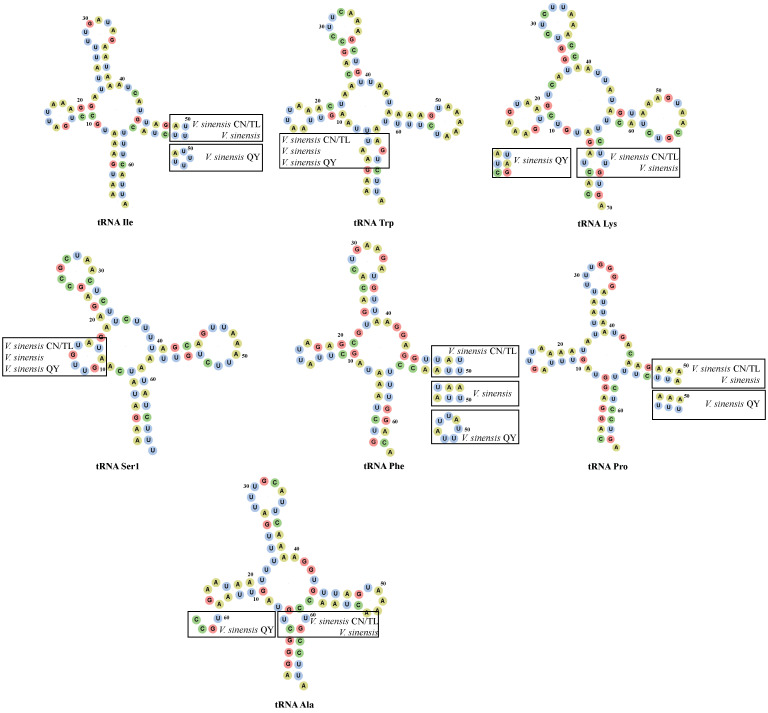
The mt genomes of V. sinensis CN/TL and V. sinensis QY possessed 22 tRNA genes. Among these genes, most tRNAs (14) were encoded on the heavy strand (H-strand), whereas eight tRNAs were encoded on the light strand (L-strand) (Tables S3 and S4). The tRNAs exhibited the classic cloverleaf secondary structure. The total tRNA sizes of V. sinensis CN/TL and V. sinensis QY were 1429 bp and 1433 bp, respectively. In both cases, the smallest tRNA was trnC with a length of 61 bp, whereas the largest tRNA was trnY with a length of 70 bp. There were differences between the tRNA secondary structures of these three mt genomes (Figure 3). The 16S rRNA gene was located between trnL and trnV with lengths of 1223 bp and 1221 bp in V. sinensis CN/TL and V. sinensis QY, respectively (Tables S3 and S4). The 12S rRNA gene was located between trnV and the CR with lengths of 792 bp and 790 bp in V. sinensis CN/TL and V. sinensis QY, respectively. The AT content of rRNA in these three mt genomes was between 74.1% and 74.3%, and the nucleotide skew was all positive for AT and GC (Table 2).
Figure 3.
Inferred different secondary structures of the tRNA genes of V. sinensis CN/TL, V. sinensis QY, and V. sinensis (HM067837).
The control region was located between the 12S rRNA and trnI genes. The length and distribution of the control region in these mt genomes of Vietnamella were relatively conservative. The length of the A + T—rich region of V. sinensis CN/TL and V. sinensis QY mt genomes was 1015 bp and 911 bp, respectively.
3.2. Analysis of Genetic Distance
Based on the difference in organization and composition of the mt genome, the complete mt genomes of V. sinensis CN, V. sinensis TL, and V. sinensis QY was used to explore their genetic distance. Within Vietnamellidae, the genetic distance between all known species ranged from 0.1% to 21.1%, with an average of 15.58% (Table 3). The genetic distance of the two specimens from the Chun’an and Tonglu populations were similar (0.1%), and the two collection locations were about 87 km apart, suggesting that these two specimens belong to the same species. The genetic distance between V. sinensis QY and the two other species was 14.8% for V. sinensis CN and 14.9% for V. sinensis TL, respectively. Comparison of the current data with other previous reports for V. sinensis was also conducted. The calculated genetic distance between V. sinensis CN/TL and V. sp. MT-2014 (KM244655) versus between V. sinensis CN/TL and V. sp. JZ-2021 (MF352146) was 20.9% and 18.3%, respectively. However, the genetic distance between V. sinensis CN/TL and V. sinensis (HM067837) was 5.8%. The genetic difference between V. sinensis QY and V. sinensis (HM067837) was 14%, reaching the level of species. Hence, the data indicate that V. sinensis QY is a cryptic species of V. sinensis according to genetic distance.
Table 3.
The genetic distance of the complete mt genomes within Vietnamellidae.
3.3. Heterogeneous Sequences Divergence and Phylogenetic Analyses
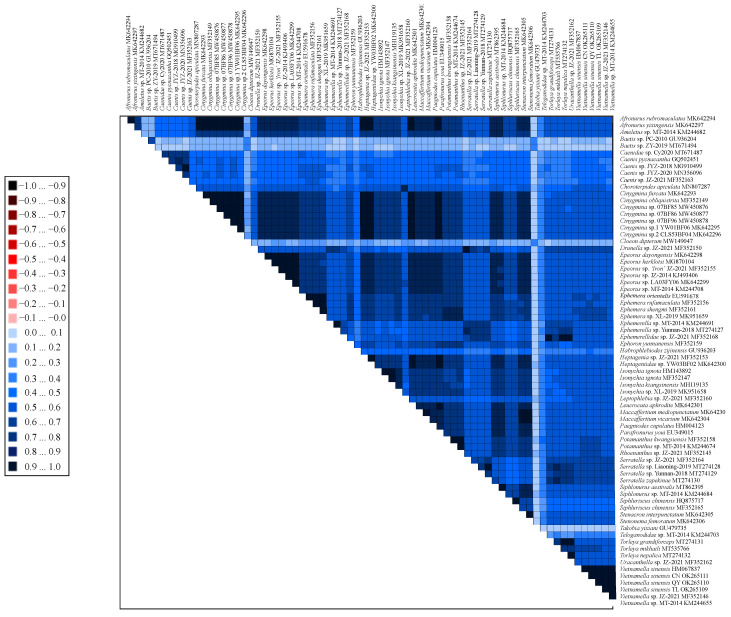
The obtained AliGROOVE matrixes helped to detect the pairwise comparisons of nucleotide datasets among all taxon comparisons that had positive similarity scores (Figure 4). Average similarity scores analyzed between sequences are represented by colored squares, and all colored squares ranged from −1 (indicating large differences in ratios to the rest of the dataset) to +1 (indicating ratios match in all other comparisons). The individual matrixes in the results revealed the degree of heterogeneity in the PCG12 matrix dataset (Figure 4). The pairwise sequence comparisons between the dataset showed a high degree of similarity, whereas the family Baetidae species showed high heterogeneity. This heterogeneity may be related to phylogenetic long-branch attraction (LBA) (Figure 5).
Figure 4.
Heterogeneous sequence divergence within two datasets of PCGs of 72 Ephemeroptera mt genomes for the PCGs matrix datasets including three codon positions of PCGs.
Figure 5.
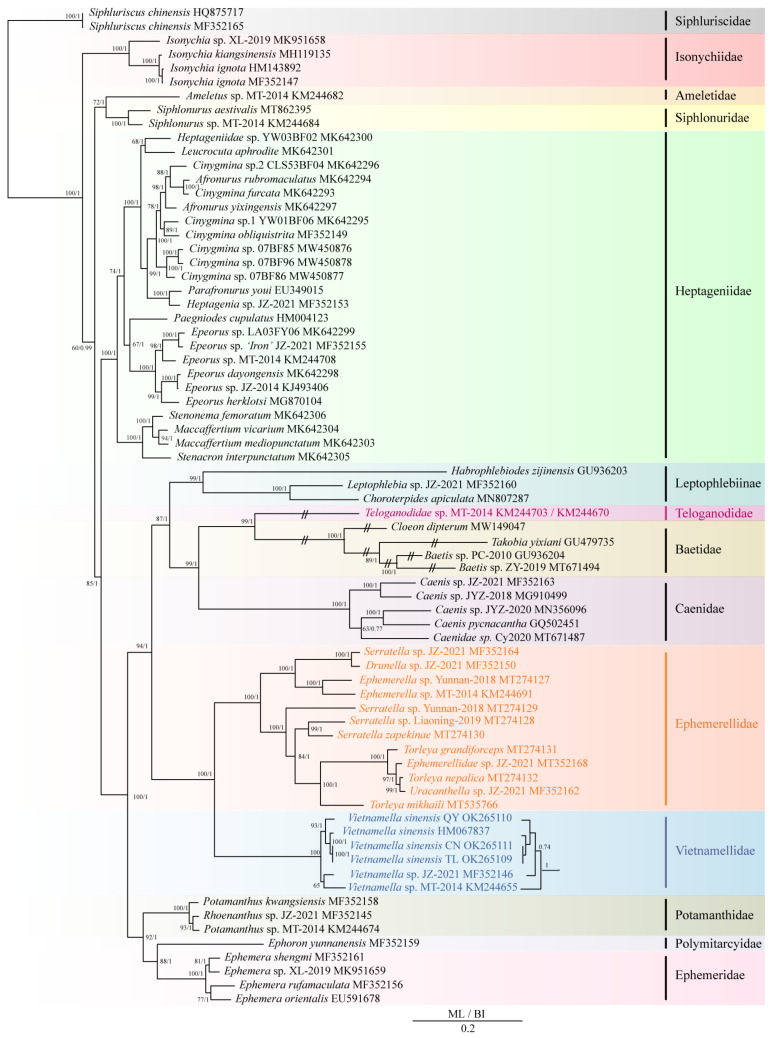
Phylogenetic tree of the relationships among 72 species of Ephemeroptera according to the nucleotide dataset of the 13 mt PCGs. Siphluriscus chinensis (HQ875717, MF352165) was used as the outgroup. The numbers above branches specify bootstrap percentages from ML (left) and posterior probabilities as determined from BI (right). The GenBank accession numbers of all species are shown in the figure. Long-branch attractions of Baetidae and Teloganodidae have been cut for aesthetics.
This study used the PCG12 matrix dataset of 72 Ephemeroptera species to analyze phylogenetic relationships, and the results showed that the monophyly of most families were supported in the phylogenetic trees. However, the presence of only one species in Ameletidae, Polymitarcyidae, and Teloganodidae limited a discussion of their monophyletic analysis (Figure 5). According to the results of phylogenetic topologies, Isonychiidae was a sister group to the other families within ingroups of Ephemeroptera. After that, Ameletidae and Siphlonuridae were found to be a sister group. Heptageniidae was a sister clade for the remaining Ephemeroptera (Heptageniidae + (((Leptophlebiidae + (Caenidae + (Teloganodidae + Baetidae))) + (Ephemerellidae + Vietnamellidae)) + (Potamanthidae + (Polymitarcyidae + Ephemeridae))). The clade of (Vietnamellidae + Ephemerellidae) was a sister clade to the clade of (Leptophlebiidae + (Caenidae + (Baetidae + Teloganodidae))).
Long-branch attraction (LBA) was observed in Teloganodidae (Teloganodidae sp. MT-2014) and Baetidae (Baetis sp. PC-2010, Baetis sp. 2 ZY-2019, Takobia yixiani and Cloeon dipterum) in both BI and ML analyses. The phylogenetic relationships between BI and ML showed roughly identical topologies, except for differences in the position of Vietnamella sp. JZ-2021 (MF352146) within Vietnamella. The ML tree showed the phylogenetic relationship of (((V. sinensis CN + V. sinensis TL) + V. sinensis) + V. sinensis QY) + (V. sp. MT-2014 + V. sp. JZ-2021). In contrast, the BI tree showed a phylogenetic relationship of (((((V. sinensis CN + V. sinensis TL) + V. sinensis) + V. sinensis QY) + V. sp. JZ-2021) + V. sp. MT-2014). Concentrating on the phylogenetic relationship of Vietnamellidae, Vietnamellidae was a sister clade to Ephemerellidae and has a distant relationship with Teloganellidae. In both BI and ML trees, the clade of V. sinensis CN and V. sinensis TL was sister clade to V. sinensis, and then were clustered together with V. sinensis QY. We realized that V. sinensis QY was a distant sister clade to V. sinensis CN and V. sinensis TL. In general, in accordance with previous phylogenetic studies based on morphological characteristics and molecular data, the monophyly of all Ephemeroptera families in this study was supported, except for Siphluriscidae. However, the monophyly of families (Ameletide, Teloganodidae and Polymitarcyidae) with only one species needs to be further explored.
3.4. Divergence Time Estimation
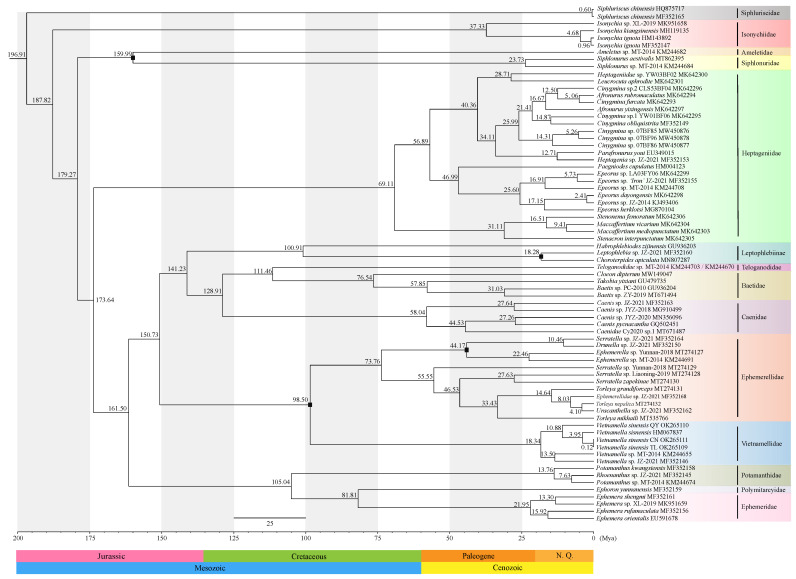
In this study, the PCG12 dataset was selected for use in divergence time estimation. The results of our divergence time analysis are shown Figure 6 and Table 4. Analysis of divergence time revealed that the diversification of Ephemeroptera occurred about 196.91 million years ago (Mya) [95% HPD (highest posterior densities), 171.18–236.55 Mya] (Figure 6). Isonychiidae originated in the Jurassic [187.82 Mya; 95% HPD, 168.38–223.63 Mya], and was a sister group to the other families within Ephemeroptera excluding Siphluriscidae. The most recent common ancestor (MRCA) of Ameletidae and Siphlonuridae also diverged in the Jurassic [159.99 Mya; 95% HPD, 159.00–161.00 Mya]. The MRCA of Heptageniidae was estimated to be at 173.64 Mya [95% HPD, 155.70–206.74 Mya]. Our results indicate that the eight currently accepted families of Ephemeridae, Polymitarcyidae, Potamanthidae, Vietnamellidae, Ephemerellidae, Caneidae, Baetidae, and Teloganodidae have similar divergence times from within the Cretaceous.
Figure 6.
Evolutionary timescale for the Ephemeroptera inferred from the PCGs dataset based on phylogenetic analyses using four fossil calibration points. Each fossil calibration point is marked with a black dot on the figure. Median ages on the chronogram are provided above nodes. A geological time scale is shown at the bottom.
Table 4.
Divergence times for nodes/clades in the Ephemeroptera based on the mt genome. All the estimates are represented in millions of years ago (Mya). “&” represents the relationship between two branches.
| Nodes/Clades | Mean Divergence Time (Mya) | 95% HPD Range (Mya) |
|---|---|---|
| Ephemeridae & Polymitarcyidae | 81.81 | 33.19~142.93 |
| (Ephemeridae + Polymitarcyidae) & Potamanthidae | 105.04 | 43.32~164.57 |
| Vietnamellidae & Ephemerellidae | 98.50 | 98.00~99.00 |
| Teloganodidae & Baetidae | 111.46 | 84.47~142.94 |
| (Teloganodidae + Baetidae) & Caenidae | 128.91 | 102.11~162.40 |
| (Teloganodidae + (Baetidae + Caenidae)) & Leptophlebiinae | 141.23 | 115.39~174.06 |
| ((Teloganodidae + (Baetidae + Caenidae)) + Leptophlebiinae) & (Vietnamellidae + Ephemerellidae) | 150.73 | 126.64~183.03 |
| (((Teloganodidae + (Baetidae + Caenidae)) + Leptophlebiinae) + (Vietnamellidae + Ephemerellidae)) & ((Ephemeridae + Polymitarcyidae) + Potamanthidae) |
161.50 | 139.21~193.76 |
| ((((Teloganodidae + (Baetidae + Caenidae)) + Leptophlebiinae) + (Vietnamellidae + Ephemerellidae)) + ((Ephemeridae + Polymitarcyidae) + Potamanthidae)) & Heptageniidae |
173.64 | 155.70~206.74 |
| Siphlonuridae & Ameletidae | 159.99 | 159.00~161.00 |
| (((((Teloganodidae + (Baetidae + Caenidae)) + Leptophlebiinae) + (Vietnamellidae + Ephemerellidae)) + ((Ephemeridae + Polymitarcyidae) + Potamanthidae)) + Heptageniidae) & (Siphlonuridae + Ameletidae) |
179.27 | 163.65~213.04 |
| ((((((Teloganodidae + (Baetidae + Caenidae)) + Leptophlebiinae) + (Vietnamellidae + Ephemerellidae)) + ((Ephemeridae + Polymitarcyidae) + Potamanthidae)) + Heptageniidae) + (Siphlonuridae + Ameletidae)) & Isonychiidae |
187.82 | 168.38~223.63 |
| (((((((Teloganodidae + (Baetidae + Caenidae)) + Leptophlebiinae) + (Vietnamellidae + Ephemerellidae)) + ((Ephemeridae + Polymitarcyidae) + Potamanthidae)) + Heptageniidae) + (Siphlonuridae + Ameletidae)) + Isonychiidae) & Siphluriscidae |
196.91 | 171.18~236.55 |
We estimated that Vietnamellidae appeared during the Cretaceous (98.50 Mya; 95% HPD, 98.00–99.00 Mya), which is supported by the first fossil record of the mayfly family Vietnamellidae from Burmese amber [78]. The MRCA of the three species in the current study and V. sinensis was estimated to be at 10.88 Mya [95% HPD, 5.11–19.87 Mya]. After V. sinensis QY was separated from the branches, then Vietnamella sinensis was separated again from V. sinensis CN and V. sinensis TL at around 3.95 Mya [95% HPD, 1.37–8.87 Mya]. Our analyses recovered a divergence between V. sinensis CN and V. sinensis TL, which is estimated to have occurred during the Neogene (0.12 Mya; 95% HPD, 0.001–0.42 Mya).
4. Discussion
4.1. Comparison of Mitochondrial Genome Composition
In 2017, Hu et al. described V. dabieshanensis, V. qingyuanensis, and V. guadunensis as junior synonyms of V. sinensis [37]. Therefore, in subsequent comparisons, V. dabieshanensis (HM067837) will be referred to as V. sinensis. As of January 2022, the National Center for Biotechnology Information (NCBI) has released three mt genomes of the Vietnamella: V. sinensis (HM067837, 15,761 bp), Vietnamella. sp. MT-2014 (KM244655, 15,043 bp), and Vietnamella. sp. JZ-2021 (MF352146, 15,043 bp). The three mt genomes of this study were shorter than the complete mt genome of V. sinensis, and the size differences of these mt genomes was mainly caused by the size of the intergenic nucleotides (IGNs) and the CR (Table S2). The intergenic nucleotides (IGNs) of V. sinensis CN/TL and V. sinensis QY ranged from 1 to 41 bp (Table S2), and these were identical with V. sinensis (HM067837). In addition, we analyzed the sizes and nucleotide compositions of V. sinensis (HM067837). The A – T content of the whole mt genome, PCGs, tRNA, and rRNA were calculated, and the results ranged from 66.4% to 71.8% (PCGs), 71.3% to 74.9% (tRNA), and 74.1% to 74.3% (rRNA), respectively (Table 2). There were strong A + T biases in both V. sinensis CN/TL and V. sinensis QY, 70.5% and 69.5%, respectively, and together, these were similar to V. sinensis (HM067837) (70.7%).
The sizes of PCGs in the three mt genomes (11,226 bp) were similar to V. sinensis (11,121 bp). We found that the start and stop codons of the 13 PCGs in both V. sinensis CN/TL and V. sinensis QY were the same as in V. sinensis (HM067837). Among the 13 PCGs, 10 PCGs used typical stop codons and three PCGs used incomplete stop codons. Incomplete stop codons are common in metazoan mt genomes [82]. We also observed that the AT content of PCGs (−) in the three sequenced mt genomes and V. sinensis (HM067837) was greater than that in PCGs (+) (Table 2). The balance between mutation, selection pressure, and genetic drift can lead to codon usage bias, so codon usage analysis is important for understanding genome evolution [83,84]. Due to the results that AT mutation bias has effects on codon usage, we found that codons with the third nucleotide being G or C in Table S5 were rarely used [85,86]. It can be seen from Table S5 that the codon count and RSCU of V. sinensis CN/TL and V. sinensis QY were relatively similar.
With few exceptions, most metazoan mt genomes contain 22 tRNA genes including two trnL (UUR and CUN) and two trnS (AGN and UCN) [87]. Among the 22 tRNA genes of the three mt genomes, the secondary structure of most tRNA genes was the normal cloverleaf model, except for trnI (V. sinensis CN/TL and V. sinensis), trnM (V. sinensis CN/TL and V. sinensis), and trnP (V. sinensis CN/TL, V. sinensis, and V. sinensis QY), which lacks the TΨC loops. Furthermore, trnS1 (V. sinensis CN/TL, V. sinensis, and V. sinensis QY) had lost the dihydrouridine (DHC) arm (Figure 3). A lack of TΨC loops or DHC arms can be found in other Ephemeroptera [57], and their translational activity was lower than the normal structures [88]. Quite a few mismatched pairs were found among the three species and V. sinensis (HM067837), and the specific mismatches are shown in Figure 3. It can be seen from the graph that the similarity of secondary structures of the tRNA genes between V. sinensis CN/TL and V. sinensis (HM067837) was higher than that of V. sinensis QY and V. sinensis (HM067837).
4.2. Phylogenetic Analyses of Vietnamellidae within Ephemeroptera
In recent years, the higher-level phylogenetic relationships within Ephemeroptera have been widely debated [8,26,27,34,89]. In most cases, Siphluriscus chinensis (HQ875717, MF352165) was primitively diverged from other mayflies within Ephemeroptera [57]. Vietnamellidae and Teloganodidae were in parallel relationship, and had a distant relationship with Ephemerellidae [27,28]. In our study, the family Vietnamellidae was strongly monophyletic in our topologies (Figure 5). Both maximum likelihood and Bayesian phylogenetic trees showed that Vietnamellidae was closely related to Ephemerellidae and had a distant relationship with Teloganellidae, which was consistent with the results of Cai et al. [12], Gao et al. [45], and Rutschmann et al. [90]. Over the last few years, many scholars did not include sequences of Teloganodidae when constructing the phylogenetic tree within Ephemeroptera, so the sister clade of Vietnamellidae and Ephemerellidae were still supported [41,42,43,44].
4.3. Identification of Cryptic Species
The intraspecific genetic distances of these three species varied between 0.1% (V. sinensis CN—V. sinensis TL) and 14.9% (V. sinensis CN/TL—V. sinensis QY), whereas the interspecific genetic distances with other Vietnamella species were very high, ranging from 18.3% (V. sinensis CN/TL—V. sp. JZ-2021) to 21.1% (V. sinensis QY—V. sp. MT-2014) (Table 3). The difference in genetic distance between V. sinensis CN/TL and V. sinensis (HM067837) published in the NCBI was 5.8%, which is between the 1% and 7% of typical insect reports [91]. However, the difference between V. sinensis QY and V. sinensis (HM067837) was 14%. Hence, we consider V. sinensis QY to be a cryptic species of V. sinensis (HM067837). Williams found that the genetic distance of Baetis rhodani in different geographic locations was 8–19% at the molecular level, and judged that some populations were cryptic species [92]. In this study, the genetic distance of V. sinensis QY reached 14%, which supports the conclusion that V. sinensis QY is a cryptic species of V. sinensis (HM067837).
Within Vietnamellidae, V. sinensis (HM067837) was a sister group to V. sinensis CN and V. sinensis TL, according to the phylogenetic topologies (Figure 5). V. sinensis QY was the earliest divergent lineage (10.88 Mya) from V. sinensis (HM067837) and was still quite closely relatively related (Figure 6). In this study, we estimated Vietnamellidae to have appeared during the Cretaceous period. After V. sinensis QY was separated from the main branches of V. sinensis at around 10.88 Mya, then V. sinensis was separated again from V. sinensis CN and V. sinensis TL at around 3.95 Mya, and the divergence time was far from V. sinensis QY. On the whole, not only the genetic distance of mt genomes, but also the phylogenetic analysis and divergence time of the three populations of Vietnamella, suggest that V. sinensis QY was a cryptic species of V. sinensis.
5. Conclusions
In this study, the complete mt genome sequences of V. sinensis CN, V. sinensis TL, and V. sinensis QY were successfully determined. The three Vietnamella mt genomes showed similar gene arrangements to V. sinensis (HM067837). BI and ML analyses indicated roughly identical topology, except for the position of Vietnamella sp. JZ-2021 (MF352146) within Vietnamella. Furthermore, our study showed that Vietnamellidae was the sister clade to Ephemerellidae, but Teloganellidae was far from Vietnamellidae and Ephemerellidae. Analysis of divergence time revealed that the diversification of Ephemeroptera occurred about 196.91 million years ago. Divergence times in most families suggested that most diversity arose during the Mesozoic era and Vietnamellidae appeared during the Cretaceous (98.50 Mya).
Overall, comprehensive comparative analysis of the mt genomes of V. sinensis CN, V. sinensis TL, and V. sinensis QY revealed that they differed significantly in various aspects such as genome composition, genetic distance, phylogenetic analyses, and divergence time estimation. The genetic distance between V. sinensis QY and V. sinensis (HM067837) reached 14%, which was much higher than that of V. sinensis CN, V. sinensis TL, and V. sinensis (HM067837), of 5.8%. In the analysis of phylogeny and divergence time estimation, V. sinensis QY first separated from V. sinensis (HM067837), V. sinensis CN, and V. sinensis TL about 10.88 Mya, and then V. sinensis (HM067837) separated from V. sinensis CN and V. sinensis TL at about 3.95 Mya. The results of this study provide evidence for the existence of cryptic species. The specific characteristics of mt genomes may be used as accessible and powerful molecular markers in the identification of cryptic species. Further research priorities can conduct additional studies of the mt genomes of Teloganodidae and Baetidae to explore long-branch attraction within Ephemeroptera.
Acknowledgments
We are grateful to Si-Si Cao, Xiao-Dong Xu, and Yi-Yang Jia for their help in these experiments.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/insects13050412/s1, Table S1: Species information used in the reconstruction of Ephemeroptera phylogeny in this study; Table S2: The size variation between V. sinensis CN/TL (CN/TL), V. sinensis QY (QY) and V. sinensis (VS) (HM067837); Table S3: Location of features in the mt genome of V. sinensis CN/TL; Table S4: Location of features in the mt genome of V. sinensis QY; Table S5: Codon numbers and relative synonymous codon usage in the protein coding genes of mt genomes of V. sinensis CN/TL (CN/TL), V. sinensis QY (QY), and V. sinensis (VS) (HM067837).
Author Contributions
Conceptualization, J.-Y.Z., Y.M., Y.T., D.-N.Y. and K.B.S.; Methodology, Y.T., L.W., J.-Y.Z. and Y.M.; Statistical analysis, D.-N.Y., S.P.G.A., Y.T. and L.W.; Investigation, J.-Y.Z., Y.M., Y.T. and S.P.G.A.; Data curation, Y.M., Y.T., L.W. and S.P.G.A.; Writing—original draft preparation, Y.T., J.-Y.Z., D.-N.Y. and K.B.S.; Writing—review and editing, Y.T., Y.M., J.-Y.Z., D.-N.Y. and K.B.S.; Maps and graphics, Y.T.; Project administration, J.-Y.Z., D.-N.Y., K.B.S. and Y.M.; Funding acquisition, J.-Y.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Natural Science Foundation of China (grant #31,370,042). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data supporting the findings of this study are openly available from the National Center for Biotechnology Information at https://www.ncbi.nlm.nih.gov (accessed on 24 September 2021). Accession numbers are: OK265109, OK265110, and OK265111.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cameron S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014;59:95–117. doi: 10.1146/annurev-ento-011613-162007. [DOI] [PubMed] [Google Scholar]
- 2.Boore J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avise J.C., Arnold J., Ball R.M., Bermingham E., Lamb T., Neigel J.E., Reeb C.A., Saunders N.C. Intraspecific phylogeography: The mitochondrial DNA bridge between population genetics and systematics. Annu. Rev. Ecol. Syst. 1987;18:489–522. doi: 10.1146/annurev.es.18.110187.002421. [DOI] [Google Scholar]
- 4.Salvato P., Simonato M., Battisti A., Negrisolo E. The complete mitochondrial genome of the bag-shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae) BMC Genom. 2008;9:331. doi: 10.1186/1471-2164-9-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao X.Y., Cai Y.Y., Yu D., Zhang J.Y. Characteristics of the complete mitochondrial genome of Suhpalacsa longialata (Neuroptera, Ascalaphidae) and its phylogenetic implications. PeerJ. 2018;6:e5914. doi: 10.7717/peerj.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J.Y., Zhang L.P., Yu D., Zheng R.Q. Complete mitochondrial genomes of Nanorana taihangnica and N. yunnanensis (Anura: Dicroglossidae) with novel gene arrangements and phylogenetic relationship of Dicroglossidae. BMC Evol. Biol. 2018;18:26. doi: 10.1186/s12862-018-1140-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu D.N., Zhang J.Y., Peng L., Zheng R.Q., Shao C. Do cryptic species exist in Hoplobatrachus rugulosus? An examination using four nuclear genes, the Cyt b gene and the complete mt genome. PLoS ONE. 2015;10:e0124825. doi: 10.1371/journal.pone.0124825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X.D., Guan J.Y., Zhang Z.Y., Cao Y.R., Cai Y.Y., Yu D.N., Zhang J.Y. Insight into the phylogenetic relationships among three subfamilies within Heptageniidae (Insecta: Ephemeroptera) along with low-temperature selection pressure analyses using mitogenomes. Insects. 2021;12:656. doi: 10.3390/insects12070656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye Q.M., Zhang S.S., Cai Y.Y., Storey K.B., Yu D.N., Zhang J.Y. The complete mitochondrial genome of Isonychia kiangsinensis (Ephemeroptera: Isonychiidae) Mitochondrial DNA Part B. 2018;3:541–542. doi: 10.1080/23802359.2018.1467233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grillo V., Jackson F., Cabaret J., Gilleard J.S. Population genetic analysis of the ovine parasitic nematode Teladorsagia circumcincta and evidence for a cryptic species. Int. J. Parasitol. 2007;37:435–447. doi: 10.1016/j.ijpara.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 11.Barber J.H.M., Zrelli S., Yanai Z., Sartori M. A reassessment of the genus Oligoneuriopsis Crass, 1947 (Ephemeroptera, Oligoneuriidae, Oligoneuriellini) ZooKeys. 2020;985:15–47. doi: 10.3897/zookeys.985.56649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai Y.Y., Gao Y.J., Zhang L.P., Yu D.N., Zhang J.Y. The mitochondrial genome of Caenis sp. (Ephemeroptera: Caenidae) and the phylogeny of Ephemeroptera in Pterygota. Mitochondrial DNA Part B. 2018;3:577–579. doi: 10.1080/23802359.2018.1467239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao S.S., Xu X.D., Jia Y.Y., Guan J.Y., Zhang J.Y. The complete mitochondrial genome of Choroterpides apiculata (Ephemeroptera: Leptophlebiidae) and its phylogenetic relationships. Mitochondrial DNA Part B. 2020;5:1159–1160. doi: 10.1080/23802359.2020.1730270. [DOI] [Google Scholar]
- 14.Xu X.D., Jia Y.Y., Cao S.S., Zhang Z.Y., Storey K.B., Yu D.N., Zhang J.Y. Six complete mitochondrial genomes of mayflies from three genera of Ephemerellidae (Insecta: Ephemeroptera) with inversion and translocation of trnI rearrangement and their phylogenetic relationships. PeerJ. 2020;8:e9740. doi: 10.7717/peerj.9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan J.Y., Zhang Z.Y., Cao Y.R., Xu X.D., Storey K.B., Yu D.N., Zhang J.Y. The complete mitochondrial genome of Choroterpes (Euthralus) yixingensis (Ephemeroptera: Leptophlebiidae) and its mitochondrial protein-coding gene expression under imidacloprid stress. Gene. 2021;800:145833. doi: 10.1016/j.gene.2021.145833. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z.Y., Guan J.Y., Cao Y.R., Dai X.Y., Yu D.N., Zhang J.Y. Mitogenome analysis of four Lamiinae species (Coleoptera: Cerambycidae) and gene expression responses by Monochamus alternatus when infected with the parasitic nematode, Bursaphelenchus mucronatus. Insects. 2021;12:453. doi: 10.3390/insects12050453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J.Y., Luu B.E., Yu D.N., Zhang L.P., Al-Alattar R., Storey K.B. The complete mitochondrial genome of Dryophytes versicolor: Phylogenetic relationship among Hylidae and mitochondrial protein-coding gene expression in response to freezing and anoxia. Int. J. Biol. Macromol. 2019;132:461–469. doi: 10.1016/j.ijbiomac.2019.03.220. [DOI] [PubMed] [Google Scholar]
- 18.Winker K. Sibling species were first recognized by William Derham (1718) Auk. 2005;122:706–707. doi: 10.1093/auk/122.2.706. [DOI] [Google Scholar]
- 19.Bickford D., Lohman D.J., Sodhi N.S., Ng P.K.L., Meier R., Winker K., Ingram K.K., Das I. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 2007;22:148–155. doi: 10.1016/j.tree.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Jacobus L.M., Macadam C.R., Sartori M. Mayflies (Ephemeroptera) and their contributions to ecosystem services. Insects. 2019;10:170. doi: 10.3390/insects10060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barber-James H.M., Gattolliat J.L., Sartori M., Hubbard M. Global diversity of mayflies (Ephemeroptera, Insecta) in freshwater. Hydrobiologia. 2008;595:339–350. doi: 10.1007/s10750-007-9028-y. [DOI] [Google Scholar]
- 22.Elliott J.M., Humpesch U.H., Macan T.T. Larvae of the British Ephemeroptera: A key with ecological notes. Sci. Publ. Freshw. Biol. Assoc. 1988;49:1–145. [Google Scholar]
- 23.Burian S.K. Ephemeroptera (Mayflies) Encycl. Inland Waters. 2009:299–314. doi: 10.1016/B978-012370626-3.00172-1. [DOI] [Google Scholar]
- 24.Ratnasingham S., Hebert P.D.N. Bold: The barcode of life data system (http://www.barcodinglife.org) Mol. Ecol. Notes. 2007;7:355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mccafferty W.P., Edmunds G.F. The higher classification of the Ephemeroptera and its evolutionary basis. Ann. Entomol. Soc. Am. 1979;72:5–12. doi: 10.1093/aesa/72.1.5. [DOI] [Google Scholar]
- 26.Mccafferty W.P. Toward a phylogenetic classification of the Ephemeroptera (Insecta): A commentary on systematics. Ann. Entomol. Soc. Am. 1991;4:343–360. doi: 10.1093/aesa/84.4.343. [DOI] [Google Scholar]
- 27.Kluge N.J. The Phylogenetic System of Ephemeroptera (the First Experience in Consistently Non-Ranking Taxonomy) Kluwer Academic Publishers; Norwell, MA, USA: 2004. 442p [Google Scholar]
- 28.Ogden T.H., Whiting M.F. Phylogeny of Ephemeroptera (mayflies) based on molecular evidence. Mol. Phylogenet. Evol. 2005;37:625–643. doi: 10.1016/j.ympev.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 29.Ogden T.H., Gattolliat J.L., Sartori M., Staniczek A.H., Soldán T., Whiting M.F. Towards a new paradigm in mayfly phylogeny (Ephemeroptera): Combined analysis of morphological and molecular data. Syst. Entomol. 2009;34:616–634. doi: 10.1111/j.1365-3113.2009.00488.x. [DOI] [Google Scholar]
- 30.Hebert P.D.N., Cywinska A., Ball S.L., de Waard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Donnell B.C., Jockusch E.L. Phylogenetic relationships of leptophlebiid mayflies as inferred by histone H3 and 28S ribosomal DNA. Syst. Entomol. 2008;33:651–667. doi: 10.1111/j.1365-3113.2008.00434.x. [DOI] [Google Scholar]
- 32.Tschernova O.A. Some new Asiatic species of mayflies (Ephemoroptera, Heptageniidae, Ephemerellidae) Entomol. Rev. (Engl. Transl.) 1972;51:604–614. [Google Scholar]
- 33.Selvakumar C., Sinha B., Vasanth M., Subramanian K.A., Sivaramakrishnan K.G. A new record of monogeneric family Vietnamellidae (Insecta: Ephemeroptera) from India. J. Asia Pac. Entomol. 2018;21:994–998. doi: 10.1016/j.aspen.2018.07.015. [DOI] [Google Scholar]
- 34.Mccafferty W.P., Wang T.Q. Phylogenetic systematics of the family Teloganodidae (Ephemeroptera: Pannota) Ann. Cape Prov. Mus. 1997;19:387–437. [Google Scholar]
- 35.Wang M. Phylogenetic systematics of the major lineages of Pannote mayflies (Ephemeroptera: Pannota) Trans. Am. Entomol. Soc. 2000;126:9–101. [Google Scholar]
- 36.Jacobus L.M., Mccafferty W.P. Reevaluation of the phylogeny of the Ephemeroptera infraorder Pannota (Furcatergalia), with adjustments to higher classification. Trans. Am. Entomol. Soc. 2006;132:81–90. [Google Scholar]
- 37.Hu Z., Ma Z.X., Luo J.Y., Zhou C.F. Redescription and commentary on the Chinese mayfly Vietnamella sinensis (Ephemeroptera: Vietnamellidae) Zootaxa. 2017;4286:381–390. doi: 10.11646/zootaxa.4286.3.5. [DOI] [Google Scholar]
- 38.You D.S., Su C.R. A new species of Vietnamella from China (Ephemeroptera: Ephemerellidae) Acta Zool. 1987;12:176–180. [Google Scholar]
- 39.Auychinda C., Jacobus L.M., Sartori M., Boonsoong B. A new species of Vietnamella Tshernova 1972 (Ephemeroptera: Vietnamellidae) from Thailand. Insects. 2020;11:554. doi: 10.3390/insects11090554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo Y., Jiang J., Wang L., Shu Z., Tong X. Vietnamella chebalingensis, a new species of the family Vietnamellidae (Ephemeroptera) from China based on morphological and molecular data. Zootaxa. 2020;4868:208–220. doi: 10.11646/zootaxa.4868.2.2. [DOI] [PubMed] [Google Scholar]
- 41.Li R., Zhang W., Ma Z.X., Zhou C.F. Novel gene rearrangement pattern in the mitochondrial genomes of Torleya mikhaili and Cincticostella fusca (Ephemeroptera: Ephemerellidae) Int. J. Biol. Macromol. 2020;165:3106–3114. doi: 10.1016/j.ijbiomac.2020.10.124. [DOI] [PubMed] [Google Scholar]
- 42.Li R., Zhang W., Ma Z., Zhou C. First complete mitogenomes of three mayflies in the genus Afronurus (Ephemeroptera: Heptageniidae) and their implications for phylogenetic reconstruction. Biologia. 2021;76:2291–2302. doi: 10.1007/s11756-021-00729-6. [DOI] [Google Scholar]
- 43.Li R., Ma Z.X., Zhou C.F. The first two complete mitochondrial genomes of Neoephemeridae (Ephemeroptera): Comparative analysis and phylogenetic implication for Furcatergalia. Genes. 2021;12:1875. doi: 10.3390/genes12121875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W., Li R., Zhou C.F. Complete mitochondrial genomes of Epeorus carinatus and E. dayongensis (Ephemeroptera: Heptageniidae): Genomic comparison and phylogenetic inference. Gene. 2021;777:145467–145475. doi: 10.1016/j.gene.2021.145467. [DOI] [PubMed] [Google Scholar]
- 45.Gao X.Y., Zhang S.S., Zhang L.P., Yu D.N., Cheng H.Y. The complete mitochondrial genome of Epeorus herklotsi (Ephemeroptera: Heptageniidae) and its phylogeny. Mitochondrial DNA Part B. 2018;3:303–304. doi: 10.1080/23802359.2018.1445482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lalitha S. Primer Premier 5. Biotech Softw. Internet Rep. 2000;1:270–272. doi: 10.1089/152791600459894. [DOI] [Google Scholar]
- 47.Zhang L.P., Cai Y.Y., Yu D.N., Storey K., Zhang J.Y. Gene characteristics of the complete mitochondrial genomes of Paratoxodera polyacantha and Toxodera hauseri (Mantodea: Toxoderidae) PeerJ. 2018;6:e4595. doi: 10.7717/peerj.4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burland T.G. Bioinformatics Methods and Protocols. Volume 132. Humana Press; Totowa, NJ, USA: 2000. DNASTAR’s Lasergene sequence analysis software; pp. 71–91. [DOI] [PubMed] [Google Scholar]
- 49.Bernt M., Donath A., Jühling F., Externbrink F., Florentz C., Fritzsch G., Pütz J., Middendorf M., Stadler P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013;69:313–319. doi: 10.1016/j.ympev.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 50.Sudhir K., Glen S., Koichiro T. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang D., Gao F., Jakovli I., Zou H., Wang G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020;20:348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- 52.Song H.J., Sheffield N.C., Cameron S.L., Miller K.B., Whiting M.F. When phylogenetic assumptions are violated: Base compositional heterogeneity and among-site rate variation in beetle mitochondrial phylogenomics. Syst. Entomol. 2010;35:429–448. doi: 10.1111/j.1365-3113.2009.00517.x. [DOI] [Google Scholar]
- 53.Grant J.R., Stothard P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:181–184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perna N.T., Kocher T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995;41:353–358. doi: 10.1007/BF01215182. [DOI] [PubMed] [Google Scholar]
- 55.Zhang J.Y., Zhou C.F., Gai Y.H., Song D.X., Zhou K.Y. The complete mitochondrial genome of Parafronurus youi (Insecta: Ephemeroptera) and phylogenetic position of the Ephemeroptera. Gene. 2008;424:18–24. doi: 10.1016/j.gene.2008.07.037. [DOI] [PubMed] [Google Scholar]
- 56.Lee E., Hong M., Kim M., Kim M.J., Park H., Kim K., Lee I., Bae C., Jin B., Kim I. The complete mitogenome sequences of the palaeopteran insects Ephemera orientalis (Ephemeroptera: Ephemeridae) and Davidius lunatus (Odonata: Gomphidae) Genome. 2009;52:810–817. doi: 10.1139/G09-055. [DOI] [PubMed] [Google Scholar]
- 57.Li D., Qin J.C., Zhou C. The phylogeny of Ephemeroptera in Pterygota revealed by the mitochondrial genome of Siphluriscus chinensis (Hexapoda: Insecta) Gene. 2014;545:132–140. doi: 10.1016/j.gene.2014.04.059. [DOI] [PubMed] [Google Scholar]
- 58.Tang M., Tan M.H., Meng G.L., Yang S.Z., Su X., Liu S.L., Song W.H., Li Y.Y., Wu Q., Zhang A.B., et al. Multiplex sequencing of pooled mitochondrial genomes-a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 2014;42:e166. doi: 10.1093/nar/gku917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song N., Li X., Yin X., Li X., Yin J., Pan P. The mitochondrial genomes of palaeopteran insects and insights into the early insect relationships. Sci. Rep. 2019;9:17765. doi: 10.1038/s41598-019-54391-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu X.D., Jia Y.Y., Dai X.Y., Ma J.L., Yu D.N. The mitochondrial genome of Caenis sp. (Ephemeroptera: Caenidae) from Fujian and the phylogeny of Caenidae within Ephemeroptera. Mitochondrial DNA Part B. 2019;5:192–193. doi: 10.1080/23802359.2019.1698986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Macher J.N., Drakou K., Papatheodoulou A., Hoorn B., Vasquez M. The mitochondrial genomes of 11 aquatic macroinvertebrate species from Cyprus. Metabarcoding Metagenom. 2020;4:91–96. doi: 10.3897/mbmg.4.58259. [DOI] [Google Scholar]
- 62.Yu D.N., Yu P.P., Zhang L.P., Storey K.B., Zhang J.Y. Increasing 28 mitogenomes of Ephemeroptera, Odonata and Plecoptera support the Chiastomyaria hypothesis with three different outgroup combinations. PeerJ. 2021;9:e11402. doi: 10.7717/peerj.11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kück P., Meid S.A., Groß C., Wäele J.W., Misof B. AliGROOVE-visualization of heterogeneous sequence divergence within multiple sequence alignments and detection of inflated branch support. BMC Bioinf. 2014;15:1–15. doi: 10.1186/1471-2105-15-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 66.Lanfear R., Calcott B., Ho S.Y.W., Guindon S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 67.Alexandros S. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;9:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Drummond A.J., Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214–221. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rambaut A. Figtree Version 1.4.0. 2012. [(accessed on 12 December 2021)]. Available online: http://tree.bio.ed.ac.uk/software/figtree/
- 71.Glazko G.V., Nei M. Estimation of divergence times for major lineages of primate species. Mol. Biol. Evol. 2003;20:424–434. doi: 10.1093/molbev/msg050. [DOI] [PubMed] [Google Scholar]
- 72.Heled J., Drummond A.J. Calibrated tree priors for relaxed phylogenetics and divergence time estimation. Syst. Biol. 2012;61:138–149. doi: 10.1093/sysbio/syr087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin Q.B., Huang D.Y. New Middle Jurassic mayfiles (Insecta: Ephemeroptera: Siphlonuridae) from inner Mongolia, China. Ann. Zool. 2008;58:521–527. doi: 10.3161/000345408X364346. [DOI] [Google Scholar]
- 74.He H.Y., Wang X.L., Zhou Z.H., Zhu R.X., Jin F., Wang F., Ding X., Boven A. 40Ar/39Ar dating of ignimbrite from Inner Mongolia, northeastern China, indicates a post-Middle Jurassic age for the overlying Daohugou Bed. Geophys. Res. Lett. 2004;31:L20609. doi: 10.1029/2004GL020792. [DOI] [Google Scholar]
- 75.Staniczek A.H., Godunko R.J., Krzeminski W. A new fossil mayfly species of the genus Borinquena Traver, 1938 (Insecta: Ephemeroptera: Leptophlebiidae: Atalophlebiinae) from Miocene Dominican amber. Ann. Zool. 2017;67:113–119. doi: 10.3161/00034541ANZ2017.67.1.013. [DOI] [Google Scholar]
- 76.Zhang W.T., Shih C.K., Shih Y.H., Ren D. A new macrolepidopteran moth (Insecta, Lepidoptera, Geometridae) in Miocene Dominican amber. Zookeys. 2020;965:73–84. doi: 10.3897/zookeys.965.54461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Staniczek A.H., Godunko R.J., Kluge N.J. Fossil record of the mayfly family Ephemerellidae (Insecta: Ephemeroptera), with description of new species and first report of Ephemerellinae from Baltic amber. J. Syst. Palaeontol. 2018;16:1319–1335. doi: 10.1080/14772019.2017.1388299. [DOI] [Google Scholar]
- 78.Godunko R.J., Martynov A.V., Staniczek A.H. First fossil record of the mayfly family Vietnamellidae (Insecta, Ephemeroptera) from Burmese amber confirms its Oriental origin and gives new insights into its evolution. Zookeys. 2021;1036:99–120. doi: 10.3897/zookeys.1036.66435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Misof B., Liu S.L., Meusemann K., Peters R.S., Donath A., Mayer C., Frandsen P.B., Ware J., Flouri T., Beutel R.G., et al. Phylogenomics resolves the timing and pattern of insect evolution. Science. 2014;346:763–767. doi: 10.1126/science.1257570. [DOI] [PubMed] [Google Scholar]
- 80.Reis M.D., Yang Z.H. MCMCTree Tutorials. 2013.
- 81.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 82.Ojala D., Montoya J., Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 83.Bachtrog D., Thornton K., Andolfatto C.P. Extensive introgression of mitochondrial DNA relative to nuclear genes in the Drosophila yakuba species group. Evolution. 2006;60:292–302. doi: 10.1111/j.0014-3820.2006.tb01107.x. [DOI] [PubMed] [Google Scholar]
- 84.Yuan Y., Kong L., Li Q. Mitogenome evidence for the existence of cryptic species in Coelomactra antiquata. Genes Genom. 2013;35:693–701. doi: 10.1007/s13258-013-0120-6. [DOI] [Google Scholar]
- 85.Powell J.R., Moriyama E.N. Evolution of codon usage bias in Drosophila. Proc. Natl. Acad. Sci. USA. 1997;94:7784–7790. doi: 10.1073/pnas.94.15.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rao Y., Wu G., Wang Z., Chai X., Nie Q., Zhang X. Mutation bias is the driving force of codon usage in the Gallus gallus genome. DNA Res. 2011;18:499–512. doi: 10.1093/dnares/dsr035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lars P., Anke B., Georg M. The complete mitochondrial genome of the onychophoran Epiperipatus biolleyi reveals a unique transfer RNA set and provides further support for the ecdysozoa hypothesis. Mol. Biol. Evol. 2008;25:42–51. doi: 10.1093/molbev/msm223. [DOI] [PubMed] [Google Scholar]
- 88.Hanada T., Suzuki T., Yokogawa T., Takemoto-Hori C., Sprinzl M., Watana B.K. Translation ability of mitochondrial tRNAsSer with unusual secondary structures in an in vitro translation system of bovine mitochondria. Genes Cells. 2001;6:1019–1030. doi: 10.1046/j.1365-2443.2001.00491.x. [DOI] [PubMed] [Google Scholar]
- 89.Kluge N.J. Phylogeny and higher classification of Ephemeroptera. Zoosyst. Ross. 1998;72:255–269. [Google Scholar]
- 90.Rutschmann S., Chen P., Zhou C.F., Monaghan M.T. Three mitochondrial genomes of early-winged insects (Ephemeroptera: Baetidae and Leptophlebiidae) Mitochondrial DNA Part B. 2021;6:2969–2971. doi: 10.1080/23802359.2021.1974966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ross K.G., Krieger M., Shoemaker D.W., Vargo E.L., Keller L. Hierarchical analysis of genetic structure in native fire ant populations: Results from three classes of molecular markers. Genetics. 1997;147:643–655. doi: 10.1093/genetics/147.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Williams H.C., Ormerod S.J., Bruford M.W. Molecular systematics and phylogeography of the cryptic species complex Baetis rhodani (Ephemeroptera, Baetidae) Mol. Phylogenet. Evol. 2006;40:370–382. doi: 10.1016/j.ympev.2006.03.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are openly available from the National Center for Biotechnology Information at https://www.ncbi.nlm.nih.gov (accessed on 24 September 2021). Accession numbers are: OK265109, OK265110, and OK265111.