Abstract
Candida albicans can cause various infections, especially in immunocompromised patients. Its ability to develop resistance to the current antifungal drugs as well as its multiple virulence factors have rendered the problem even more complicated. Thus, in the present investigation, we elucidated an in vitro and in vivo antifungal activity of Encephalartos laurentianus methanol extract (ELME) against C. albicans clinical isolates for the first time. A phytochemical identification of 64 compounds was conducted in ELME using LC-MS/MS. Interestingly, ELME exhibited antifungal activity with MIC values that ranged from 32–256 µg/mL. Furthermore, we investigated the antibiofilm activity of ELME against the biofilms formed by C. albicans isolates. ELME displayed antibiofilm activity using a crystal violet assay as it decreased the percentages of cells, moderately and strongly forming biofilms from 62.5% to 25%. Moreover, the antibiofilm impact of ELME was elucidated using SEM and fluorescent microscope. A significant reduction in the biofilm formation by C. albicans isolates was observed. In addition, we observed that ELME resulted in the downregulation of the biofilm-related tested genes (ALS1, BCR1, PLB2, and SAP5) in 37.5% of the isolates using qRT-PCR. Besides, the in vivo antifungal activity of ELME on the kidney tissues of rats infected with C. albicans was investigated using histological and immunohistochemical studies. ELME was found to protect against C. albicans induced renal damage, decrease desmin and inducible nitric oxide synthase, increase alkaline phosphatase, and increase infected rats’ survival rate. Additionally, the cytotoxicity of ELME was elucidated on Human Skin Fibroblast normal cells using MTT assay. ELME had an IC50 of 31.26 µg/mL. Thus, we can conclude that ELME might be a promising future source for antifungal compounds.
Keywords: alkaline phosphatase, desmin, fluorescent microscope, iNOs, LC-MS/MS, SEM, qRT-PCR
1. Introduction
In recent decades, Candida species have been considered a common cause of nosocomial infections, leading to high morbidity and mortality rates. Candida is capable of causing both superficial (mucocutaneous and cutaneous) and life-threatening invasive infections such as candidemia [1]. There are many species of Candida such as C. albicans, C. krusei, C. glabrata, C. parapsilosis, and C. tropicalis. However, C. albicans is the most commonly isolated species. C. albicans is a harmless commensal fungus commonly present in the oral cavity and vagina, but, upon appropriate conditions, it can be converted to be pathogenic fungus [2].
C. albicans have many virulence factors that enable them to overgrow and invade the host tissues causing different infections such as its ability to express adhesins and secretion of hydrolytic enzymes. However, the most important virulence factor is its ability to form biofilms. It is a characteristic that allows them to adhere to different surfaces, biotic and abiotic [3]. Biofilms are large communities of adjacent cells embedded in an extracellular matrix. Clinically, biofilms are difficult to be treated owing to the resistance to antimicrobials and human defences [4].
There is an increased incidence of invasive infections caused by C. albicans, especially in immuno-compromised patients such as cancer and AIDS patients. This is accompanied by the development and dissemination of resistance against most conventional antifungal agents [5]. In addition, the currently present antifungal agents suffer from many disadvantages such as the narrow spectrum of activity, low potency, high cost, and multiple adverse effects. Thus, there is an urgent need to find out and develop new therapeutics against the infections caused by C. albicans [6].
In recent decades, natural products represented a significant starting point for the therapeutic compounds that can treat various diseases. In different regions of the world, we can find that several groups of researchers have studied the biological activity of plants [7]. For example, many plants have shown significant antimicrobial activity (both in vitro and in vivo), a finding that has led to a more intense search for the antimicrobial activity of many plants [8,9]. Plants contain several active constituents that effectively control the growth of different microorganisms, including fungi [10]. There are various secondary metabolites in plants such as alkaloids, tannins, flavonoids, terpenoids, glycosides, etc., that have in vitro antimicrobial potential [11].
Encephalartos, the second biggest extant genus of the Cycadales, is restricted to Africa south of the Sahara, with roughly 66 species. The tropical regions of central and east Africa have many species, but South Africa has more than half of them. They are endemism-rich and occupy a wide range of climate regimes and habitats [12].
Encephalartos laurentianus De Wild. (Family Zamiaceae), often known as the malele or Kwango giant cycad, it is a cycad species found in northern Angola and southern Congo. It is the hugest of all the cycads. E. laurentianus was recently found to have cytotoxic, antioxidant, and antimicrobial properties [13,14].
In the current study, we aimed to investigate the antifungal activity of E. laurentianus methanol extract (ELME) against C. albicans clinical isolates, both in vitro and in vivo. In addition, the phytochemical constituents of ELME were analyzed using LC-ESI-MS/MS technique.
2. Materials and Methods
2.1. Chemicals and Plant Materials
E. laurentianus De Wild. leaves (four years old tree) were gathered from Al Abid Farms, Giza Governorate on 13 January 2017. The plant was recognized by Dr. Esraa Ammar, Faculty of Science, Tanta University. A voucher sample (PGG-003-W) was deposited at the Herbarium of the Pharmacognosy Department, Tanta University. The plant was dried for 14 days at room temperature at 25° ± 2, then for 2 days in the oven at 40°. The dried leaves were then reduced and ground to a fine powder. The powder (250 g) was extracted with methanol (2.5 L × 4 times) using the cold maceration method to yield 26.7 g of ELME. All of the other chemicals used in the current study were obtained from Merck, Kenilworth, NJ, USA.
2.2. Isolation and Identification of C. albicans Isolates
In the current study, 16 C. albicans clinical isolates were collected from different patient specimens from the clinical laboratories of Tanta University Hospitals, Egypt. The samples were cultured on sabouraud dextrose agar (SDA) plates (Oxoid, Basingstoke, UK), and they were overnight incubated at 37 °C. Then, Candida isolates were cultured on HiCrome™ Candida differential agar (Himedia, Mumbai, India) using Candida albicans (MTCC 227) as standard strain. In addition, the API 20C system (BioMérieux, Craponne, France) was used according to the manufacturer’s instructions.
2.3. Animals
A total of 75 adult male albino rats weighing 170–200 g of three months of age were used in the current work. The rats were adequately housed ventilated cages with controlled temperature (25 °C), humidity, and 12 h light/dark cycles. In addition, they were allowed free access to water and food. Strict hygiene was followed to keep a healthy medium for them. All of the animal protocols were authorized and observed via the Animal Care Committee of the Research Laboratory of Experimental Animals at the Faculty of Medicine, Menoufia University, Egypt, with approval number 2/2022OBSGN.
2.4. LC-MS/MS for Metabolite Profiling
The LC-ESI-MS/MS analysis was conducted at the Children’s Cancer Hospital’s Proteomics and Metabolomics Unit (57357). We adopted the criteria has been previously described [15,16].
Secondary metabolites of the ELME were analyzed using an ExionLC -High flow LC, Sciex®, Framingham, MA, USA UPLC analytical technique, combined with a Triple TOF. 5600+ (Sciex®) for IDA. acquisition and Analyst TF 1.7.1 (Sciex®) for LC-Triple TOF. control. After the filtration process by an in-line filter disks (0.5 μm × 3.0 mm, Phenomenex®, Torrance, CA, USA), 1 g/mL of ELME was injected into X Select HSS. T3 XP UHPLC column (2.5 μm, 2.1 × 150 mm, Waters®, Milford, MA, USA) at 40 °C, and eluted using buffer systems of 1% methanol in 5 mM ammonium formate buffer at pH 3 and pH 8 as solvent A (for positive mode MS analysis), and B (for negative mode MS analysis), respectively, and 100% of acetonitrile as solvent C at 0.3 mL/min flow rate. Gradient mobile phase mixtures were composed of 90% solvent A or B and 10% of solvent C, which were injected for 20 min, then reversed into 10% of solvent A or B to 90% of solvent C for the next 5 min and finalized by loading of the starting mixture for the last 3 min. The resultant total ion chromatogram (TIC) was used for master view peaks extraction with a signal-to-noise ratio greater than 5 (non-targeted analysis) and more than 3 feature intensities of the sample-to-blank ratio. Data interpretation was accomplished by applying a Reifycs Abf (Analysis Base File) Converter (Reifycs®, Tokyo, Japan) for Wiff file conversion and MS-DIAL 4.6 (RIKEN® Tokyo, Japan).
To identify chemicals, PeakViewTM software version 2 1 was used to compare retention duration and m/z values obtained by MS and MS2. The XIC Manager in PeakViewTM software was used to calculate peak area values. Extracted ion chromatograms (XICs) for each targeted analyte were automatically created and compared to a user-defined threshold [17].
2.5. In Vitro Antifungal Activity of ELME
2.5.1. Disc Agar Diffusion
The susceptibility of C. albicans isolates to ELME was investigated using the disk agar diffusion method [17,18]. In brief, 3–5 fungal colonies were inoculated into sabouraud dextrose broth (SDB) (Oxoid, Hampshire, UK) and overnight incubated at 37 °C. After adjusting the turbidity of the suspensions to 0.5 McFarland turbidity standard, they were spread on the surface of Muller Hinton agar (MHA) plates (Oxoid, Hampshire, UK), supplemented with 2% glucose 0.5 µg/mL methylene blue, using sterile cotton swabs. Then, sterile filter paper disks, previously loaded with ELME, were placed on the surfaces of MHA plates by sterile forceps, and the plates were incubated at 37 °C for 24 h.
2.5.2. Determination of Minimum Inhibitory Concentration (MIC)
Broth microdilution assay was utilized in the current study to determine the MIC values of ELME against the tested C. albicans isolates [18,19] in 96-well microtitration plates. In brief, fungal colonies were inoculated into SDB and incubated overnight at 37 °C in an orbital shaker (New Brunswich, Fredericton, Canada). The fungal cells were centrifuged, the pellets were rinsed twice with phosphate-buffered saline (PBS) and resuspended in Roswell Park Memorial Institute 1640 (RPMI 1640) with a concentration of 1 × 103 CFU/mL. ELME was serially diluted by adding 50 µL and 50 µL of the fungal suspensions. The concentration range of ELME was from 512 to 1 µg/mL. The plates contained positive (fungi only without ELME) and negative (RPMI 1640 only without Candida) controls, then they were incubated at 37 °C for 24 h. This test was carried out in triplicates. The lowest concentration of ELME, which inhibited the visible growth of C. albicans, was regarded as the MIC and 0.5 MIC values of ELME were used for further studies.
2.5.3. Biofilm Formation Assay
The b§iofilm formation assay was carried out as previously described [20]. Briefly, overnight C. albicans cultures were diluted with SDB and adjusted to 1 × 106 CFU/mL. Then, fungal suspensions (100 μL) were inoculated into 96-well microtitration plates and incubated at 37 °C for 48 h. The wells were gently washed with PBS, and the adherent fungal cells were fixed with 100 μL methanol (99%) for 20 min. They were then stained with crystal violet (1%) for 20 min. After that, the wells of the microtitration plates were washed using distilled water and were left to dry. The dye bound to the fungal cells was dissolved using 33% glacial acetic acid, and their optical density (OD) was recorded at 540 nm using an ELISA reader (Sunrise Tecan, Switzerland, Austria). This assay was carried out in triplicate and cut-off OD (ODc) was calculated. ODc is the mean OD of the negative control (SDB only without fungi) plus three times standard deviations (SD). C. albicans isolates were grouped into non-biofilm forming (OD ≤ ODc), weak biofilm forming (OD > ODc ≤ 2 × ODc), moderate biofilm-forming (OD > 2 × ODc ≤ 4 × ODc), and strong biofilm-forming (OD > 4 × ODc).
2.5.4. Antibiofilm Activity of ELME
The antibiofilm activity of ELME was investigated against the moderate and strong biofilm-forming C. albicans isolates before and after treatment with ELME at 0.5 MIC values using crystal violet assay as previously described [21]. The viability of the formed biofilms in the microtitration plates was assessed by counting the colony-forming units (CFU/mL). The formed biofilms were gently washed with PBS, and they were then scraped off the microtitration plate wells using pipette tips or toothpicks after adding 200 μL PBS. After that, the obtained suspensions were serially diluted, 100 μL was plated on SDA plates supplemented with chloramphenicol, incubated overnight at 37 °C, and the number of CFU/mL was counted.
-
a.
Scanning electron microscope (SEM)
As previously reported, an inspection of the biofilm morphology of C. albicans isolates by SEM was performed [22]. Briefly, glass coverslips were submerged with each C. albicans isolates (before and after treatment with ELME), and they were left for 24 h to let the isolates form biofilms. The formed biofilms were gently washed with PBS and flooded with glutaraldehyde solution (2.5%) for 24 h at 4 °C. After that, they were dehydrated by a series of ethanol with concentrations that ranged from 30% to 100%. They were then left to dry and coated with gold for inspection with SEM (Hitachi, Tokyo, Japan).
-
b.
Fluorescent microscope
Glass coverslips with C. albicans biofilms were transferred to microscopical slides and stained with a Calcofluor White stain for 2 min in a dark room. Then, biofilms were observed as soon as possible using a fluorescence microscope [23].
-
c.
Quantitative real-time polymerase chain reaction (qRT-PCR)
The impact of ELME on the gene expression of the biofilm genes (ALS1, BCR1, PLB2, and SAP5) was assessed in all of the C. albicans isolates using qRT-PCR as previously described [24]. In brief, total RNA was extracted using RNeasy mini kit (Qiagen, Hilden, Germany). Then, the extracted RNA was retrotranscribed into cDNA using SensiFAST™ cDNA kit (Bioline, London, UK). qRT-PCR was performed using SensiFAST™ SYBR green PCR master mix (Bioline, London, UK) according to the manufacturer instructions using the primers listed in Table S1. The expression of the tested genes in the fungal cells before treatment was set to be 1.
2.6. In Vivo Antifungal Activity of ELME
2.6.1. Experimental Protocol
All of the animals were exposed to immunosuppression by corticosteroid injection [25]. Then, animals were randomly divided into 5 groups as follows:
Group I (control group): 15 rats each received 1 mL saline (0.9%) by intraperitoneal (IP) injection once daily for 1 week;
Group II (C. albicans group, C6 isolate, with the concentration of 2 × 105 CFU/mL): 15 rats each received a suspension of C. albicans (0.1 mL) by intravenous (IV) injection in the tail vein once;
Group III (fluconazole treated group): 15 rats each received a suspension of C. albicans (0.1 mL) by IV injection in the tail vein once. Then, this group received 10 mg/kg fluconazole by IP injection for 1 week;
Group IV (ELME treated group, 50 mg/kg): 15 rats received a suspension of C. albicans (0.1 mL) by IV injection in the tail vein once. Then, this group received 50 mg/kg ELME by IP injection for 1 week;
Group V (ELME treated group, 100 mg/kg): 15 rats received a suspension of C. albicans (0.1 mL) by IV injection in the tail vein once. Then, this group received 100 mg/kg ELME by IP injection for 1 week;
Rats were monitored for 15 days after infection to determine the survival rate. Ether inhalation was used to anesthetize all rats to be sacrificed. Kidney tissues were obtained and processed for histological and immunohistochemical investigations. In addition, fungal tissue burden (number of CFU/g kidney tissues) was calculated.
2.6.2. Histopathological and Immunohistochemical Studies
Kidney specimens from all of the animal groups were fixed in normal saline. The specimens were processed for performing paraffin sections, 7 μm in thickness. Then, sections were stained with hematoxylin and eosin (H&E) to illustrate the histological structure, and Masson’s trichrome stain was used to detect the collagen fibers [26].
For the immunohistochemical study of desmin, a podocyte injury marker, the utilized primary monoclonal antibody was anti-desmin (Lab Vision Corp, Inc./Lab Vision, Fremont, CA, USA). A negative control was performed via replacing the primary antibody with buffer alone. Uterus smooth muscle (myometrium) was used as a positive control. The positive reaction was cytoplasmic brown color [27]. Counterstaining was performed using Mayer’s hematoxylin.
For the histochemical study, specimens from the renal cortex were fixed in cold acetone, then processed and stained using calcium phosphate according to the Gomori technique to determine the alkaline phosphatase enzyme. The histochemical activity of alkaline phosphatase in the proximal convoluted tubules was detected. The reaction product was strongly marked at the apical surface and the basal part of the cells comprising the basal infoldings with their extended tips. The positive reaction was in fine, uniform brown deposits [28].
For immunostaining of inducible nitric oxide synthase (iNOS), iNOS antibody was utilized, a polyclonal antibody (Lab Vision Corporation Laboratories, catalogue number PA1-036). Immunostaining required pretreatment by boiling for 10 min in 10 Mm citrate buffer, pH 6 for antigen retrieval, and then sections were left to cool at room temperature for 20 min. After that, the sections were incubated for one hour with the primary antibodies and immunostaining was completed using the Ultravision detection system (cat no T.P.-015- H.D.) [29].
2.6.3. Morphometric Measurements
These measurements were carried out for histological and immunohistochemical quantitative assessment. Five non-overlapping fields per animal were randomly captured by a Lieca Microscope D.M.L. B2/11888111 equipped with a Leica camera DFC450. Kidney sections were randomly selected for morphometric measurements using image analyzer software (Image J analyzer version 1.43o8, National Institutes of Health, Bethesda, MD, USA). The following parameters were measured: the glomerular basement membrane (GBM) thickness (×200), the area percentage of collagen fibers stained by Masson trichrome stain (×200) and the area percentage of desmin and iNOs positive immune staining (×400).
Histological damage of the renal tubule was assessed as the percentage of the tubules that appeared dilated, atrophied, tubules with epithelial cell vacuolated cytoplasm, and with cast formation as 0 (normal), 1 (<10%), 2 (10 to 25%), 3 (26 to 50%), 4 (51 to 75%), and 5 (>75%). H&E-stained kidney segments were utilized to examine the renal tubular damage score. Five regions of the renal tubules were randomly chosen per kidney from each of the ten rats in each group for evaluation at a magnification of ×200, and the average score was calculated [30].
2.7. Cytotoxicity MTT Assay
The MTT assay was used to determine the cytotoxicity of ELME on human skin fibroblast (HSF) normal cell line (Nawah-Scientific, Cairo, Egypt). The cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) media containing 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 100 mg/mL streptomycin. The cultures were kept at 37 °C in a humidified environment with 5% CO2. Confluent monolayers of cells were cultivated for 24 h in 96 well microtitration plates. Then, cells were cultured with varying doses of ELME in triplicate for 72 h. After that, 20 µL of 5 mg/mL MTT were gently poured into each well and incubated for 4 hours at 37 °C. The media was then removed carefully, and 150 μL of dimethyl sulfoxide (DMSO) solvent was added to dissolve the formed formazan crystals. Finally, using a microplate reader (B.M.G. Labtech, FLUOstar Omega, Ortenberg, Germany), OD values were measured at 540 nm [31].
2.8. Statistical Analysis
Data were shown as mean ± standard deviation. The differences between the tested groups were assessed using a one-way analysis of variance (ANOVA), followed by a post hoc test (Tukey). Kruskal Wallis test was used for scoring the tubular injury. In addition, Kaplan-Meier survival curve was used to calculate the survival of rats. The difference is significant at p < 0.05 using Prism version 8 (GraphPad Software, Inc., San Diego, CA, USA).
3. Results
3.1. LC-ESI-MS/MS of ELME
LC-ESI-MS/MS analysis of ELME (negative and positive mode E.S.I.) tentatively identified 64 compounds of secondary metabolites for the first time. The identified metabolites were classified as anthocyanidin-3-O-glycosides, alkaloids, aurone derivative, coumarins, flavonoid aglycone, flavonoid glycosides, phenolic, and carboxylic acid. Table 1 shows the results of LC-ESI-MS/MS analysis of ELME metabolites, and Figures S1 and S2 show the total ion chromatogram (TIC) of ELME in positive and negative mode E.S.I., respectively.
Table 1.
Phytochemical profiling of ELME by LC-MS/MS analysis (negative and positive mode E.S.I.).
| Peak NO | Identification | Error | R.T. (min.) | m/z | Adduct Ion | Formula | MS/MS |
|---|---|---|---|---|---|---|---|
| 1 | f.a Citraconic acid | −2.8 | 1.006 | 129.02 | [M − H]− | C5H6O4 | 84.99, 85.02, 129.01 |
| 2 | a Rosmarinic acid | 6.0 | 1.009 | 359.11 | [M − H]− | C18H16O8 | 181.04, 329.09, 359.07, 359.11 |
| 3 | a Malic acid | −4.7 | 1.331 | 133.10 | [M − H]− | C4H6O5 | 133.01 |
| 4 | t 3-Amino-1,2,4-triazole | −0.6 | 1.392 | 85.027 | [M + H]+ | C2H4N4 | 71.95, 85.02 |
| 5 | a 3-(4-hydroxyphenyl)prop-2-enoic acid | −0.5 | 1. 393 | 165.07 | [M + H]+ | C9H8O3 | 61.02, 85.02, 166.08 |
| 6 | s Resveratrol | 6.4 | 1.470 | 229.15 | [M + H]+ | C14H12O3 | 58.06, 60.08, 70.06, 229.15 |
| 7 | ak Harmaline | 0.6 | 1.561 | 215.05 | [M + H]+ | C13H14N2O | 72.08, 156.10, 169.13, 215.05 |
| 8 | ak Nicotinic acid | −0.9 | 1.693 | 124.03 | [M + H]+ | C6H5NO2 | 53.03, 78.03, 80.04, 124.03 |
| 9 | ak 5-Hydroxyindoleacetic acid | 0.8 | 1.3058 | 192.10 | [M + H]+ | C10H9NO3 | 148.07, 177.07, 192.10 |
| 10 | f.a Linoleic acid | 0.6 | 2.290 | 281.10 | [M + H]+ | C18H32O2 | 123.07, 151.04, 165.09, 281.05 |
| 11 | f.g Acacetin-7-O-neohesperidoside | −8.1 | 4.305 | 593.28 | [M + H]+ | C28H32O14 | 593.25 |
| 12 | f.g Kaempferol-3-Glucuronide | 10.1 | 5.108 | 461.16 | [M − H]− | C21H18O12 | 188.93, 239.09, 256.92, 324.90, 392.89, 461.15 |
| 13 | b Procyanidin B1 | 1.5 | 5.115 | 579.14 | [M + H]+ | C30H26O12 | 123.04, 127.03, 135.04, 579.14 |
| 14 | a Chlorogenic acid | 1.3 | 5.534 | 355.07 | [M + H]+ | C16H18O9 | 135.04, 163.04, 355.07 |
| 15 | co 6,7-dihydroxycoumarin | −0.4 | 5.439 | 179.03 | [M + H]+ | C9H6O4 | 51.02, 77.03 179.03 |
| 16 | f.g Isorhamnetin-3-O-rutinoside | −2.1 | 5.465 | 623.22 | [M − H]− | C28H32O16 | 315.03, 532.91, 579.15, 623.17 |
| 17 | c (+)-3 3′ 4′ 5 7-Pentahydroxyflavan | 3.6 | 5.574 | 291.08 | [M + H]+ | C15H14O6 | 111.04, 119.04, 123.04, 291.08 |
| 18 | f.g Baicalein-7-O-glucuronide | 4.2 | 5.575 | 447.16 | [M + H]+ | C21H18O11 | 125.13, 129.07, 135.04, 269.06, 273.08, 447.13 |
| 19 | f.g Luteolin-8-C-glucoside | −12.6 | 5.656 | 449.14 | [M + H]+ | C21H20O11 | 139.07, 449.12 |
| 20 | f.g Syringetin-3-O-galactoside | −5.2 | 5.693 | 507.17 | [M − H2O − H]− | C23H24O13 | 112.98, 138.02, 163.06, 218.95, 286.93, 307.10, 354.92, 507.16 |
| 21 | f.g Phlorizin | 2.7 | 5.707 | 435.22 | [M − H]− | C21H24O10 | 389.21, 433.83, 435.20, 435.23 |
| 22 | ak Dihydrocapsaicin | 3.3 | 5.974 | 308.18 | [M + H]+ | C18H29NO3 | 123.04, 131.04, 137.05, 149.05 |
| 23 | b Procyanidin B2 | 0.6 | 6.161 | 579.18 | [M + H]+ | C30H26O12 | 112.02, 579.18 |
| 24 | f.g Quercetin-4′-glucoside | −1.3 | 7.061 | 465.10 | [M + H]+ | C21H20O12 | 69.03, 115.04, 289.04, 303.04, 465.09 |
| 25 | f Myricetin | −1.5 | 7.063 | 317.06 | [M − H]− | C21H24O10 | 112.98, 155.03, 165.98, 180.96, 194.99, 274.04, 287.08, 302.07 |
| 26 | an Cyanidin-3-glucoside | 1.0 | 7.109 | 449.10 | [M]+ | C21H21O11 | 85.01, 117.02, 147.02, 201.02, 287.05, 449.10 |
| 27 | f.g Luteolin-3′,7-di-O-glucoside | −0.4 | 7.227 | 611.15 | [M + H]+ | C27H30O16 | 135.04, 148.10, 271.06, 273.06, 611.15 |
| 28 | f.g Daidzein-8-C-glucoside | −0.5 | 7.279 | 417.13 | [M + H]+ | C21H20O9 | 180.06, 399.21, 417.13 |
| 29 | f.g Isoquercitrin | −2.1 | 7.283 | 463.11 | [M − H]− | C21H20O12 | 48.17, 462.90 |
| 30 | an Petunidin-3-O-beta-glucopyranoside | 0.9 | 7.567 | 479.11 | [M]+ | C22H23O12 | 165.09, 211.12, 285.08, 299.10, 302.04, 479.11 |
| 31 | b Procyanidin C1 | 0.9 | 7.772 | 865.21 | [M-H]- | C45H38O18 | 11.46, 865.20 |
| 32 | f.g apigenin-7-O-glucoside | 3.5 | 7.891 | 433.11 | [M + H]+ | C21H20O10 | 119.04, 141.07, 148.11, 135.02, 229.04, 270.22, 433.11 |
| 33 | f.g Acacetin-7-O-rutinoside | 8.0 | 7.870 | 593.17 | [M + H]+ | C28H32O14 | 520.20, 575.32, 593.17 |
| 34 | tp Sabinene | 0.4 | 8.068 | 137.05 | [M + H]+ | C10H16 | 55.05, 66.04, 68.74, 79.05, 94.04, 137.05 |
| 35 | an Peonidine-3-O-glucoside chloride | −6.1 | 8.101 | 463.12 | [M]+ | C22H23O11 | 73.05,167.99, 197.47, 234.10, 258.04, 281.08, 286.04, 301.06, 309.47, 342.01, 399.15, 463.12 |
| 36 | co Daphnetin | −0.8 | 8.202 | 179.03 | [M + H]+ | C9H6O4 | 77.03, 104.99, 123.00, 133.02, 135.04, 151.038 |
| 37 | f.g Diosmin | −0.8 | 8.762 | 609.17 | [M + H]+ | C28H32O15 | 265.11, 303.11, 609.16 |
| 38 | au Maritimetin-6-O-glucoside | 0.8 | 8.827 | 449.16 | [M + H]+ | C21H20O11 | 74.09, 133.02,135.04, 257.04, 285.03, 360.05, 375.07, 388.05, 403.07, 417.09, 434.09, 449.12 |
| 39 | f.g Rhoifolin | 0.7 | 8.974 | 577.26 | [M − H]− | C27H30O14 | 576.81, 532.90, 269.10 |
| 40 | f.g Apigenin-6-C-glucoside -7-O-glucoside | −2.0 | 9.027 | 595.37 | [M + H]+ | C21H20O12 | 165.02, 177.06, 285.09, 303.05 |
| 41 | f 3′ 4′ 5 7-tetrahydroxyflavanone | 0.0 | 9.760 | 289.07 | [M + H]+ | C15H12O6 | 117.03, 121.06, 135.04, 139.03, 145.02, 153.01, 163.03, 179.03, 181.06, 289.07 |
| 42 | f Luteolin | 8.1 | 10.002 | 287.04 | [M + H]+ | C15H10O6 | 67.01, 77.04, 287.04 |
| 43 | an Cyanidin-3, 5-di-O-glucoside | −1.2 | 10.357 | 611.22 | [M]+ | C27H31O16 | 215.06, 266.99, 309.06, 355.06, 449.13, 594.23, 611.22 |
| 44 | f.g Luteolin-4′-O-glucoside | −3.9 | 10.370 | 449.14 | [M + H]+ | C21H20O11 | 147.04, 153.06, 167.02, 287.05, 287.10, 449.14 |
| 45 | f.g Rutin | 1.7 | 10.842 | 609.15 | [M − H]− | C27H30O16 | 609.15 |
| 46 | f Naringenin | 0.9 | 11.086 | 273.07 | [M + H]+ | C15H12O5 | 67.04, 111.08, 119.03, 125.10, 129.07, 135.01. 273.07 |
| 47 | f 3 5 7-trihydroxy-4′-methoxyflavone | 0.7 | 11.278 | 301.10 | [M + H]+ | C16H12O6 | 181.06, 215.07, 223.07, 258.08, 273.11, 301.11 |
| 48 | f 3′-Methoxy-4′,5,7-trihydroxyflavonol | 0.4 | 11.329 | 317.06 | [M + H]+ | C16H12O7 | 129.97, 137.02, 168.00, 245.04, 263.21, 274.04, 287.09, 302.04, 317.06 |
| 49 | ak Caffeine | −1.9 | 11.857 | 195.13 | [M + H]+ | C8H10N4O2 | 195.13 |
| 50 | f Formononetin | 5.9 | 12.010 | 269.07 | [M + H]+ | C16H12O4 | 137.02, 225.06, 254.06, 269.07 |
| 51 | f 4′,5-dihydroxy-7-methoxyflavone | −2.6 | 12.613 | 287.09 | [M + H]+ | C16H14O5 | 137.02, 145.08, 167.03, 175.07, 287.09 |
| 52 | f Acacetin | 1.0 | 12.776 | 285.08 | [M + H]+ | C16H12O5 | 128.06, 207.06, 241.04, 242.05, 270.05, 285.07 |
| 53 | f Apigenin | 0.8 | 12.788 | 271.09 | [M + H]+ | C15H10O5 | 65.04, 67.01, 68.99, 89.03, 109.02, 115.05, 153.02, 163.04, 253.14, 271.09 |
| 54 | f 4′,5,7-Trihydroxyflavonol | 0.9 | 14.059 | 287.09 | [M + H]+ | C15H10O6 | 67.02, 91.05, 111.04, 119.04, 124.01, 147.04, 167.03, 287.09 |
| 55 | f (+-)-Taxifolin | −0.1 | 14.290 | 305.13 | [M + H]+ | C15H12O7 | 305.13 |
| 56 | a Methyl dihydrojasmonate | −9.7 | 14.433 | 227.16 | [M + H]+ | C13H22O3 | 79.05, 95.08, 167.14, 195.14, 227.14 |
| 57 | f 3′ 4′ 5 7-tetrahydroxyflavanone | −1.4 | 14.553 | 289.18 | [M + H]+ | C15H12O6 | 271.17, 289.18 |
| 58 | f.g Apigenin 8-C-glucoside | −6.3 | 14.961 | 433.12 | [M + H]+ | C21H20O10 | 135.03, 391.09, 433.13 |
| 59 | ak Capsaicin | 0 | 15.293 | 306.20 | [M + H]+ | C18H27NO3 | 108.04, 126.02, 137.06, 153.12, 306.20 |
| 60 | f.g Quercetin-3-Arabinoside | −6.8 | 15.368 | 435.14 | [M + H]+ | C20H18O11 | 135.04, 240.04, 271.06, 389.10, 435.14 |
| 61 | an Cyanidin-3-O-(2″-O-β -xylopyranosyl-β-glucooside) | 8.6 | 17.917 | 581.13 | [M]+ | C26H29O15 | 107.04, 133.06, 135.04, 153.01, 297.07, 581.14 |
| 62 | c (-)-Epicatechin | 2.5 | 18.280 | 291.07 | [M + H]+ | C15H14O6 | 81.07, 135.05, 275.05, 291.07 |
| 63 | co Esculin | −2.2 | 18.380 | 341.19 | [M + H]+ | C15H16O9 | 112.07, 121.14, 131.04, 139.08, 161.06, 165.08, 179.12, 180.13, 287.24, 341.19 |
| 64 | f 3 3′ 4′ 5-tetrahydroxy-7-methoxyflavone | −4.7 | 20.178 | 317.11 | [M + H]+ | C16H12O7 | 105.07, 129.07, 215.18, 267.20, 299.20, 317.11 |
a: carboxylic acid or phenolic acid derivative, f.a: fatty acids, ak: alkaloid and related metabolites, an: anthocyanidin glycosides, au: aurone derivative, c: catechins co: coumarins, f: flavonoid aglycone, f.g: flavonoid glycoside, b: biflavonoids, s: stilbenes, t: triazoles tp: terpenoids.
3.1.1. Identification of Flavonoids, Biflavonoids, and Catechins
Flavonoids aglycones and their glycosides resemble most of the compounds detected in ELME. The identified flavonoids aglycones were represented by 13 compounds in positive ion mode are (41, 42, 46, 47, 48, 50, 51, 52, 53, 54, 55, 57, 64) with [M + H]+ at m/z of 289.07, 287.04, 273.07, 301.10, 317.06, 269.07, 287.09, 285.08, 271.09, 287.09, 305.13, 289.18, 317.11, respectively. Apigenin (53) was the major detected flavonoid compound in positive ion mode according to peak area measurements with [M + H]+ at m/z of 271.09 and characteristic mass fragments of m/z of 119.05 [C8H6O] + H+, 153.02 [C7H4O4] + H+, 253.14 [C15H9O4]+ followed by 3 3′ 4′ 5-tetrahydroxy-7-methoxy flavone (64) at m/z of 317.11 for [M + H]+. Myricetin (25) is the only aglycone identified in negative mode at m/z of 317.06 [M − H]−.
The flavonoids glycosides were detected in both negative and positive ion modes, these metabolites represented by 20 phytochemicals (12, 16, 20, 21, 29, 39, 45) in the negative ion mode at m/z of 461.16, 623.22, 507.17, 435.22, 463.11, 577.26, 609.15, respectively. While the remaining identified glycosides were (11, 18, 19, 20, 24, 27, 28, 32, 33, 37, 40, 44, 58, 60) in the positive ion mode at m/z of 593.28, 447.16, 449.14, 507.17, 465.10, 611.15, 417.13, 433.11, 593.17, 609.17, 595.37, 449.14, 433.12, 435.14, respectively. The major recognized glycosides were apigenin-7-O-glucoside (32) at (m/z 433.11) followed by luteolin-3′, 7-di-O-glucoside (27) at m/z 611.15 in the positive ion mode.
A total of 3 biflavonoids detected in ELME were procyanidin B1 (13) and B2 (23) at m/z 579.14 and 579.18 in positive mode, while procyanidin C1 (31) at m/z 865.21 in negative mode. Only two catechines were detected in ELME, 3 3′ 4′ 5 7-pentahydroxyflavan (17) and epicatechin (62) in the positive ion mode with [M + H]+ at m/z of 291.08 and 291.07.
3.1.2. Identification of Anthocyanidin-O-Glycosides
A total of 5 anthocyanidin glycoside derivatives were identified in ELME (26, 30, 35, 43, 61) in positive ion mode [M]+ at m/z 449.10, 479.11, 463.12, 611.22, 581.13, respectively. According to spectrum peak area, Cyanidin-3-O-2″-O-β-xylopyranosyl-β-glucopyranoside (61) was the major anthocyanidin glycoside detected in the ELME ([M]+ at m/z 581.13), followed by cyanidin-3-O-glucoside (26) [M]+ at m/z of 449.10.
3.1.3. Identification of Carboxylic or Fatty, or Phenolic Acids Derivatives
Rosmarinic acid (2) and malic acid (3) were detected in ELME in the negative ion mode [M − H] at m/z 359.11 and 133.103. At the same time, 4-hydroxyphenyl-prop-2-enoic acid (5), chlorogenic acid (14), and methyl dihydrojasmonate (56) were identified in the positive ion mode with [M + H]+ at m/z 165.07, 355.07, and 227.16. The detected fatty acids were citraconic acid (1) in the negative ion mode [M − H] at m/z 129.02 and linoleic acid (10) in the positive ion mode with [M + H]+ at m/z 281.10.
3.1.4. Identification of Alkaloids Derivatives
A total of 6 alkaloids’ derivatives were detected in the positive ion mode (7, 8, 9, 22, 49, 59) with [M + H]+ at m/z 215.05, 124.03, 192.10, 308.18, 195.13, 306.20, respectively. The highest one detected was harmaline (7) with [M + H]+ at m/z of 215.05, followed by dihydrocapsaicin (22) at m/z 308.18.
3.1.5. Identification of Coumarins Derivatives
A total of 3 coumarins were detected in the positive ion mode, and they were 6,7-dihydroxy coumarin (15), daphnetin (36), and esculin (63). The highest one detected was daphnetin with [M + H]+ at m/z of 179.03 and characteristic mass fragments of m/z 77.03 [C6H4] + H+, 104.99 [C6H3O2-2H]+, 123.00 [C6H4O3-H]+, 133.02 [C8H6O2-H]+, 135.04 [C8H6O2] + H+, 151.03 [C8H6O3] + H+.
3.1.6. Identification of Other Derivatives
Other detected compounds in the positive ion mode were 3-amino-1,2,4-triazole (m/z 85.027), resveratrol (6) at m/z 229.15, and monoterpenoids sabinene (34) m/z 137.05. Maritimetin-6-O-glucoside (38) from aurone derivatives was also detected with [M + H]+ at m/z 449.16.
3.2. In Vitro Antifungal Activity of ELME
Using a disc agar diffusion method, we performed preliminary screening of the antifungal activity of ELME against C. albicans clinical isolates. We found that ELME exhibited antifungal activity as it resulted in the formation of inhibition zones around the disks. Thus, in the following step, we determined its MIC values using broth microdilution method. ELME had MIC values that ranged from 32–256 µg/mL, as shown in Table S2.
3.3. In Vitro Antibiofilm Activity of ELME
Next, we assessed the antibiofilm activity of ELME (at 0.5 MIC values) as this virulence factor has a significant role in the development and spread of the infections caused by C. albicans. As shown in Table 2, ELME decreased the percentages of C. albicans fungal isolates that form biofilms moderately and strongly from 62.5% to 25%.
Table 2.
Effect of ELME on the biofilm-forming ability of C. albicans isolates.
| Biofilm Forming Ability | No. of Isolates before Treatment with ELME | No. of Isolates after Treatment with ELME |
|---|---|---|
| Non-biofilm forming | 4 | 6 |
| Weak biofilm-forming | 2 | 6 |
| Moderate biofilm-forming | 6 | 3 |
| Strong biofilm-forming | 4 | 1 |
3.3.1. Biofilm Viability
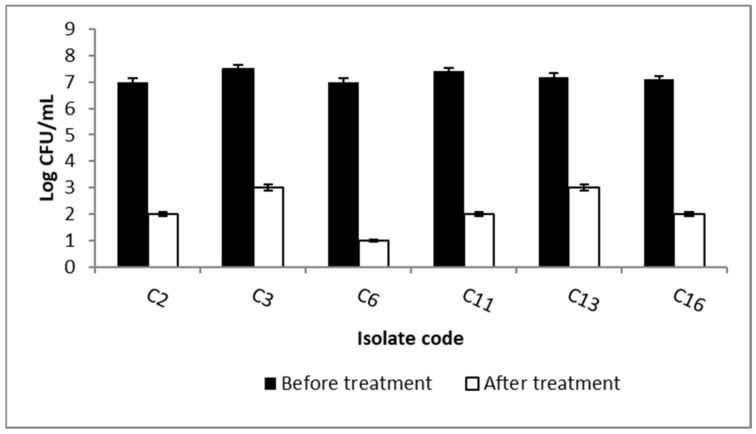
To further study the antibiofilm activity of ELME, we assessed the viability of biofilms by counting the number of CFU/mL. Interestingly, we observed a significant reduction (p < 0.05) in the viability of the formed biofilms in 37.5% of the isolates by ELME, as shown in Figure 1.
Figure 1.
Bar chart revealing a significant reduction in the count of CFU/mL of the biofilm-forming C. albicans isolates after treatment with ELME.
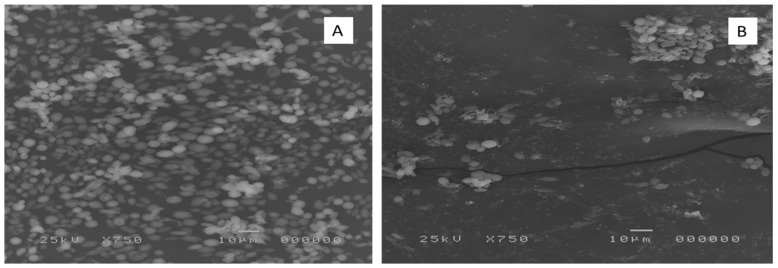
3.3.2. SEM Examination
The impact of ELME on the biofilm formed by the tested C. albicans isolates was visually confirmed using SEM. As shown in the representative example in Figure 2, SEM was used to investigate the antibiofilm activity of ELME on the isolates, which showed a decrease in their biofilm-forming ability from moderately or strongly biofilm-forming to weakly or non-biofilm forming. ELME resulted in a significant reduction in biofilm formation. This finding is consistent with the detected inhibitory effect of ELME on biofilm by crystal violet assay and the biofilm viable cells count assay.
Figure 2.
Scanning electron micrograph of the biofilm formed by a representative C. albicans isolate (C6) (A) before and (B) after treatment with ELME.
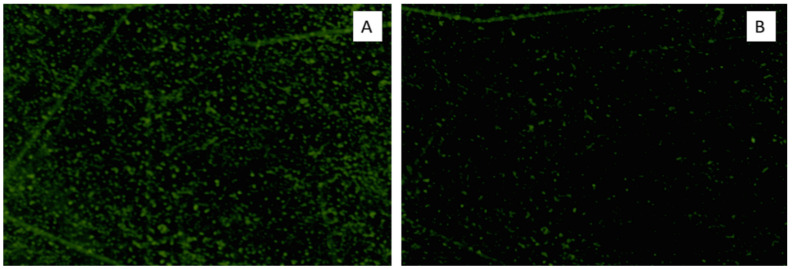
3.3.3. Fluorescent Microscope Examination
A fluorescent microscope was used to investigate the antibiofilm activity of ELME on the isolates, which showed a decrease in their biofilm-forming ability from moderately or strongly biofilm-forming to weakly or non-biofilm forming. We found that ELME exhibited a significant reduction in biofilm formation, as revealed in Figure 3. A finding that could be correlated to the results of the examination of the effect of ELME on the formed biofilms by the tested C. albicans isolates using SEM.
Figure 3.
Fluorescent microscope micrograph of the biofilm formed by a representative C. albicans isolate (C6) stained with Calcofluor White stain (A) before and (B) after treatment with ELME.
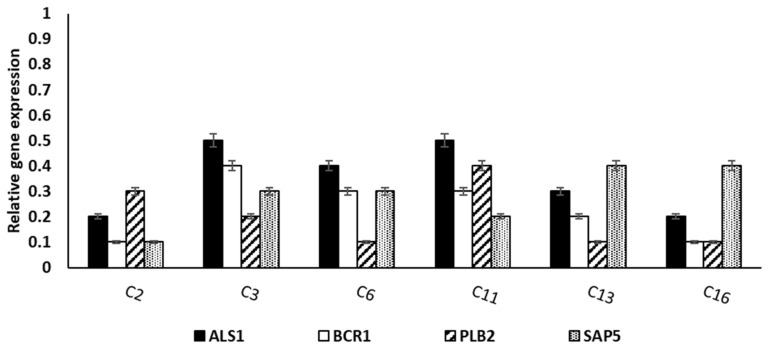
3.3.4. qRT-PCR
It was carried out to elucidate the impact of ELME on the gene expression of the biofilm genes (ALS1, BCR1, PLB2, and SAP5). Interestingly, we observed a significant reduction in the expression of the biofilm genes in 37.5% of the tested C. albicans isolates, as shown in Figure 4.
Figure 4.
Bar chart showing the significant reduction in the relative expression of the biofilm genes in 6 (37.5%) C. albicans isolates after treatment with ELME.
3.4. In Vivo Antibiofilm Activity of ELME
3.4.1. Histopathological Examination Using H&E Stain
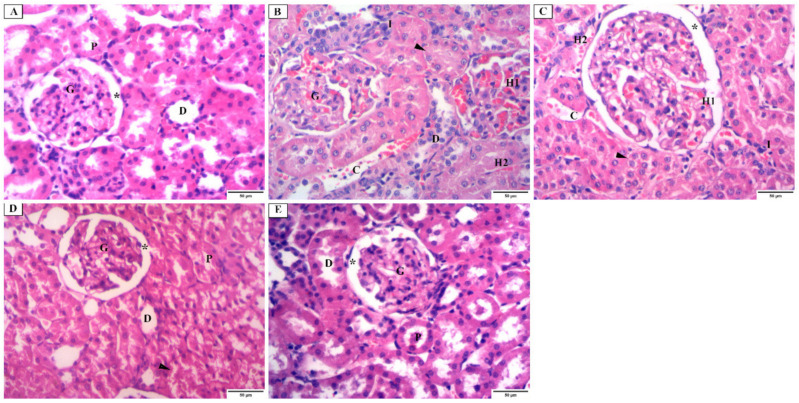
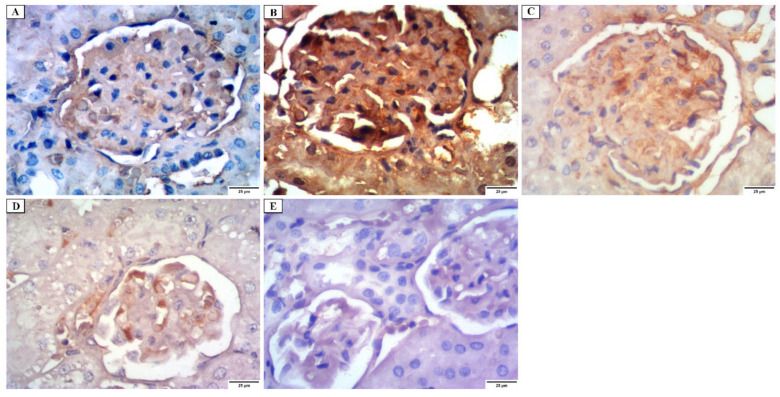
H&E-stained sections of the renal cortex of group I (control group) revealed the normal renal corpuscle, which contained glomerular capillaries and was surrounded by parietal and visceral layers of bowman’s capsule. Where bowman’s space separated the two layers, the parietal layer formed a simple squamous epithelium. Moreover, the proximal convoluted tubules (PCT) lined with pyramidal cells with deeply acidophilic cytoplasm and vesicular nuclei were observed. Distal convoluted tubules (DCT) showed a wide lumen, and its lining cells had apical nuclei and less acidophilic cytoplasm (Figure 5A). Sections of the renal cortex of group II (C. albicans group) exhibited glomerular and tubular changes. Renal glomeruli with narrow bowman’s space, intraglomerular hemorrhage, interstitial hemorrhage, cellular infiltration, and congested blood vessels were observed. The obliterated lumen and disrupted DCT were noticed (Figure 5B). Sections of the renal cortex of group III (fluconazole group) showed renal glomeruli with dilated bowman’s space and apparent partial improvement relative to the previous group. Intraglomerular hemorrhage, interstitial hemorrhage, cellular infiltration, and congested blood vessels were still seen (Figure 5C). Sections of the renal cortex of group IV (ELME treated group, 50 mg/kg) showed segmented renal glomeruli with apparently normal bowman’s space. PCT and DCT were nearly similar to the control group, but some tubules are disrupted with vacuolated cytoplasm in the tubular cells (Figure 5D). Sections of the renal cortex of group V (ELME treated group, 100 mg/kg) showed renal glomeruli, bowman’s space, PCT, and DCT are more or less similar to the control group (Figure 5E).
Figure 5.
Photomicrograph of H&E stain of the renal cortex showing: (A) the control group (group I) having a renal corpuscle with a glomerulus (G), surrounded with parietal and visceral layers of bowman’s capsule, they are separated by bowman’s space (*, proximal convoluted tubules (P) are lined with pyramidal cells which have brush border, deeply acidophilic cytoplasm, and vesicular nuclei. Distal convoluted tubules (D) have a wide lumen, and their lining cells have apical nuclei and less acidophilic cytoplasm. (B) C. albicans group (group II) having renal glomeruli (G) with narrow bowman’s space and intraglomerular hemorrhage (H1). Interstitial hemorrhage (H2), cellular infiltration (I), and congested blood vessels (C) are seen. Proximal convoluted tubules with an obliterated lumen (arrowhead) and disrupted distal convoluted tubules (D) are noticed. (C) fluconazole group (group III) having renal glomeruli with apparent dilated bowman’s space (*) and apparent partial improvement from the previous group. Intraglomerular hemorrhage (H1), interstitial hemorrhage (H2), cellular infiltration (I), and congested blood vessels (C) are still seen. Proximal convoluted tubules with the obliterated lumen (arrowhead) are still noticed. (D) ELME treated group, 50 mg/kg (group IV), having segmented renal glomeruli with apparently normal bowman’s space (*). Proximal (P) and distal (D) convoluted tubules are nearly similar to the control group, but some tubules are disrupted with vacuolated cytoplasm in the tubular cells (arrowhead). (E) ELME treated group, 100 mg/kg (group V) have renal glomerulus (G), bowman’s space (*), proximal convoluted tubules (P), distal convoluted tubules (D), more or less similar to the control group (×200).
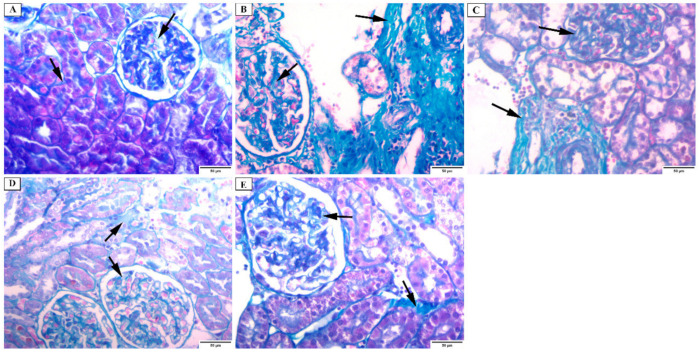
3.4.2. Masson Trichrome Stain
Masson Trichrome stained kidney sections of group I (control) showed the presence of a minimal amount of collagen fibers in the interstitium and in between the glomerular capillaries. The basal lamina was positively stained (Figure 6A). In C. albicans group (group II), collagen fibers were massively increased in the interstitium and in between the glomerular capillaries (Figure 6B). However, collagen fibers intensely increased in the fluconazole group (group III) (Figure 6C). The renal cortex of the ELME treated group, 50 mg/kg (group IV), showed a moderate increase in the collagen fibers in the interstitium and between the glomerular capillaries (Figure 6D). While renal cortex of ELME treated group, 100 mg/kg (group V) showed a mild increase in the collagen fibers in the interstitium and between the glomerular capillaries (Figure 6E).
Figure 6.
Photomicrograph of the renal cortex of Masson Trichrome stain showing: (A) group I have a minimal amount of collagen fibers in the renal interstitium and in between the glomerular capillaries (arrows). The basal lamina is positively stained. (B) group II has a massive increase in the collagen fibers in the interstitium and in between the glomerular capillaries (arrows). (C) group III has an intense increase in collagen fibers in the interstitium and between the glomerular capillaries (arrows). (D) group IV has a moderate increase in the collagen fibers in the interstitium and in between the glomerular capillaries (arrows). (E) group V has a mild increase in the collagen fibers in the interstitium and in between the glomerular capillaries (arrows) (×200).
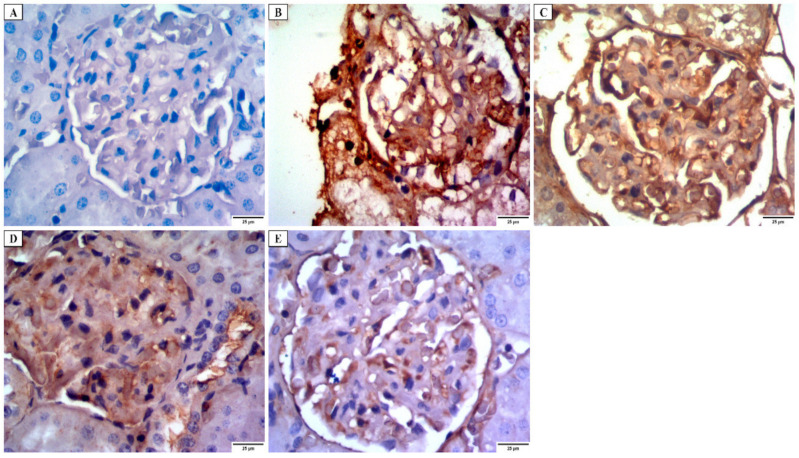
3.4.3. Immunohistochemical Investigations
-
a.
Desmin immunostaining
Kidney tissues obtained from group I showed a faint positive desmin expression in the glomerular epithelial cells (Figure 7A). Group II revealed strong positive cytoplasmic desmin immunostaining in the glomerular epithelial cells (Figure 7B). While group III revealed a moderate positive cytoplasmic desmin expression (Figure 7C). Group IV revealed a mild positive cytoplasmic desmin expression (Figure 7D). Group V showed a weak positive cytoplasmic desmin expression (Figure 7E).
Figure 7.
Photomicrograph of desmin immunostaining of the glomerular epithelial cells in the renal cortex showing: (A) group I have a faint positive cytoplasmic desmin immunostaining. (B) group II has a strong positive cytoplasmic desmin immunostaining. (C) group III has a moderately positive cytoplasmic desmin immunostaining. (D) group IV has a mild positive cytoplasmic desmin immunostaining. (E) group V has a weakly positive cytoplasmic desmin immunostaining (×400).
-
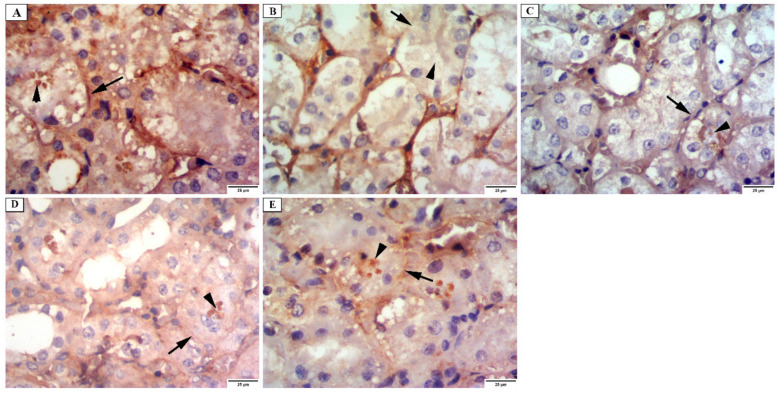
b.
Alkaline phosphatase enzyme immunostaining
Regarding the alkaline phosphatase enzyme reaction in the examined groups, group I displayed a strong positive response at the apical surfaces and the basal parts of PCT (Figure 8A). A weak reaction was observed on the examination of group II (Figure 8B). Sections of the renal cortex of group III displayed a mild positive reaction (Figure 8C). Sections of group IV showed a moderately positive reaction (Figure 8D). While sections of group V showed a strong positive reaction for alkaline phosphatase (Figure 8E).
Figure 8.
Photomicrograph of the renal cortex of alkaline phosphatase immunostaining showing: (A) group I have a strong positive reaction at the apical surfaces (arrowhead) and basal parts of the proximal convoluted tubular cells (arrow). (B) group II has a weak reaction at the apical surfaces (arrowhead) and basal parts of the proximal convoluted tubular cells (arrow). (C) group III has a mild positive reaction at the apical surfaces (arrowhead) and basal parts of the proximal convoluted tubular cells (arrow). (D) group IV has a moderate positive reaction at the apical surfaces (arrowhead) and a mild reaction in the basal parts of the proximal convoluted tubular cells (arrow). (E) group V has a strong positive reaction at the apical surfaces (arrowhead) and basal parts of the proximal convoluted tubular cells (arrow) (×400).
-
c.
iNOs immunostaining
Immunohistochemical staining for iNOs immunoreaction of the control group revealed faint positive immunoreaction in the cytoplasm of the renal tubular cells. A weak positive immunoreaction in the cytoplasm of the glomerular capillary endothelium was also observed (Figure 9A). Group II showed a strong positive reaction in the cytoplasm of the glomeruli capillary cells and the tubular cells (Figure 9B). While group III showed a moderately positive reaction in the cytoplasm of the glomeruli capillary cells and the tubular cells (Figure 9C), group IV showed a mild positive reaction in the cytoplasm of the glomeruli capillary cells and the tubular cells (Figure 9D). Group V showed a faint positive reaction in the cytoplasm of glomeruli capillary cells and the tubular cells (Figure 9E).
Figure 9.
Photomicrograph of the renal cortex of iNOs immunostaining showing: (A) group I have a weakly positive reaction at the cytoplasm of glomeruli capillary endothelium cells and a faint positive reaction in the cytoplasm of the tubular cells. (B) group II has a strong positive reaction at the cytoplasm of glomeruli capillary cells and tubular cells. (C) group III has a moderately positive reaction at the cytoplasm of glomeruli capillary cells and tubular cells. (D) group IV has a mild positive reaction at the cytoplasm of glomeruli capillary cells and tubular cells. (E) group V has a faint positive reaction at the cytoplasm of glomeruli capillary cells and tubular cells (×400).
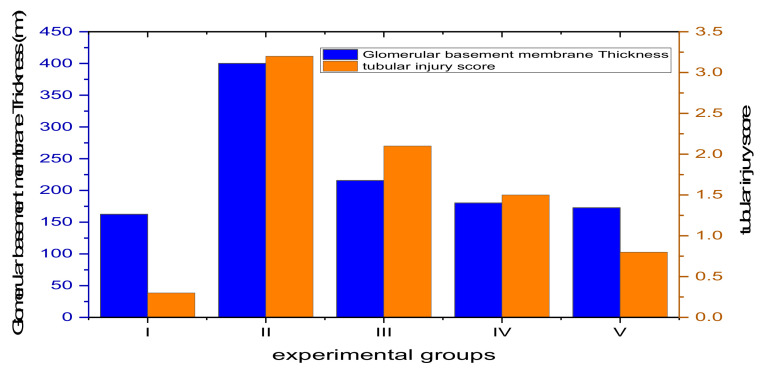
3.4.4. Morphometric Analysis
As shown in Figure 10 and Table S3, group II exhibited a highly significant increase in the thickness of the glomerular basement membrane in comparison with the control group (p ˂ 0.001). Group III displayed a substantial increase in thickness compared to the control group (p ˂ 0.05). On the other hand, groups IV and V showed a non-significant change (p > 0.05) compared to the control group. In addition, group II exhibited a significant increase (p ˂ 0.001) in the tubular injury score compared to the control group. Group III showed a substantial increase in the tubular injury score compared to the control group (p ˂ 0.05). Meanwhile, groups IV and V showed a non-significant change (p > 0.05) when compared to the control group (Figure 10).
Figure 10.
Bar chart showing the thickness of the glomerular basement membrane ((left) axis) and the tubular injury score ((right) axis) among the experimental groups.
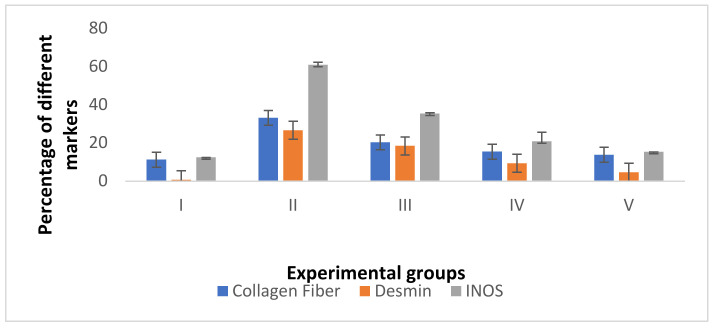
As presented in Figure 11 and Table S4, group II exhibited a highly significant increase (p ˂ 0.001) in the percentages of collagen, desmin, and iNOs when compared to the control group. However, groups III and IV showed a significant increase (p ˂ 0.05) in the percentages of desmin and iNOs compared to control. On the other side, groups IV and V exhibited a non-significant collagen change (p > 0.05) in comparison with the control group. Moreover, group V showed a non-significant desmin and iNOs increase (p > 0.05) compared to the control group.
Figure 11.
Bar chart showing the percentages of collagen fiber, desmin, and iNOs among the different groups.
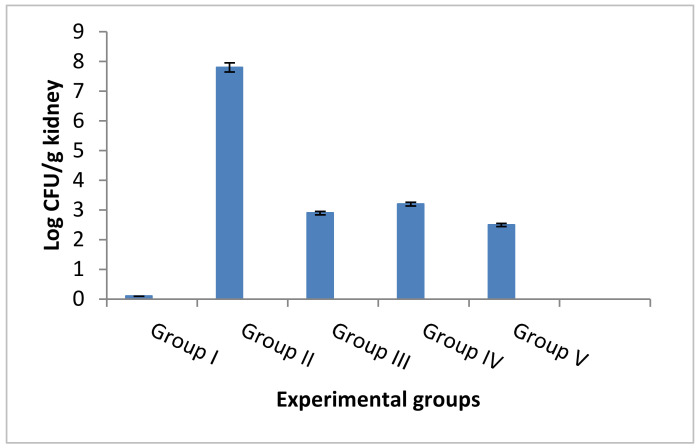
3.4.5. Counting the CFU/g and Determination of the Survival Rates
The number of CFU/g kidney tissues was counted in the tested five experimental groups. ELME resulted in a significant reduction in the number of CFU/g kidneys in the infected rats, as shown in Figure 12.
Figure 12.
Bar chart showing the number of CFU/g kidney of the rats of different experimental groups.
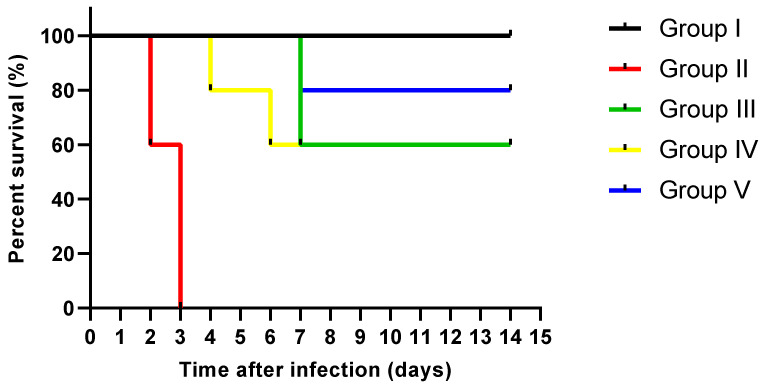
A total of 5 rats were left in each group for observing their survival rate for 14 days. As shown in Figure 13, all rats in the control group (group I) were alive till the last day of the experiment. On the other hand, two rats in C. albicans group (group II) died after two days, and three rats died after three days. In the fluconazole treated group (group III), two rats died after one week, and three remained alive until the end of the experiment. In the ELME treated group, 50 mg/kg (group IV), only 1 rat died after 4 days, and another one died after 6 days while the other rats remained alive to the end of the experiment. In ELME treated group, 100 mg/kg (group V), only 1 rat died after 7 days, and the rest were alive until the end of the experiment.
Figure 13.
Survival of rats with systemic candidiasis using Kaplan-Meier survival curve.
3.5. Cytotoxicity MTT Assay
The cytotoxicity of ELME was investigated on HSF normal cells using MTT assay. In comparison with doxorubicin as a positive control, which had an IC50 = 4.42 µg/mL, ELME had an IC50 of 31.26 µg/mL, as shown in Figure S3.
4. Discussion
Fungal infections are spreading widely on a global scale, affecting more than 1 billion people worldwide, resulting in high morbidity and mortality rates. This is in addition to high medical costs. Despite the relative availability of antifungal drugs, the fatality rates among patients infected with candidiasis are very high [32]. Thus, there is a great need for new antifungals with high activity against pathogenic fungi, particularly C. albicans. Plants are a valuable source for antimicrobials; therefore, we investigated the antifungal activity of ELME against C. albicans clinical isolates using adisc agar diffusion method as a preliminary test for screening the antimicrobial activity. Then, using broth microdilution method, ELME exhibited MIC values that ranged from 32–256 µg/mL. Several researchers worldwide have examined the antifungal activity of many plant extracts to solve the problem of the dissemination of resistance among fungal isolates [33,34,35]. Some of them have used the total plant extracts, whilst others have investigated the activity of one bioactive compound from the plant. Diba and Alizadeh [36], for example, examined the antifungal activity of Allium hirtifolium and Allium sativum plant extracts. On the other hand, Alalwan et al. [37] and Zarrinfar et al. [9] investigated the antifungal activity of curcumin against different fungi.
Candida biofilms are usually formed in the human body in mucosa or endothelium, leading to the development of candidiasis. In addition, they are associated with medical devices such as catheters. Biofilm formation by C. albicans is highly linked to resistance development to the commonly used antifungal drugs [38]. Therefore, we investigated the antibiofilm activity of ELME against the biofilms formed by C. albicans isolates. Interestingly, ELME displayed antibiofilm activity as it decreased the percentages of C. albicans isolates moderately and strongly biofilm forming from 62.5% to 25%. Herein, we chose the CFU method to assess the viability of the biofilm-forming isolates before and after treatment with ELME. It is noteworthy that the CFU method could differentiate between the living and dead cells as the viable cells only are counted with the exclusion of the dead ones and other debris [39]. The current investigation revealed that ELME resulted in a significant reduction in the viability of the formed biofilms in 37.5% of C. albicans isolates.
Unfortunately, the research scope in antifungals is not that of the antibacterial owing to the lower number of infections caused by fungi when compared to bacteria. Moreover, fungi are eukaryotes, similarly to humans, so it is important to the antifungal compound to be non-toxic to human cells. However, some studies have investigated the antifungal and antibiofilm activities of plant extracts against C. albicans, especially in oral diseases [40,41].
The impact of ELME on the biofilm morphology was elucidated using SEM and a fluorescent microscope. A significant reduction in the biofilm formation by C. albicans isolates was observed, confirming the results of crystal violet and CFU assays. Moreover, the gene expression of the genes related to biofilm formation was examined in the treated isolates. We observed that ELME resulted in the downregulation of the tested genes in 37.5% of the isolates. Biofilm formation by C. albicans is determined by many transcription factors such as ALS1, BCR1, PLB2, and SAP5 [42]. Such transcription factors have an essential function in several pathways in the fungal cells, and they could influence the adherence of C. albicans isolates [42].
Few studies investigated the in vivo antifungal activity of plant extract especially on kidney. Thus, we tried in this study to elucidate the potential protective effect of ELME against C. albicans on kidney tissues. In our in vivo experiment, C. albicans group showed damaged glomeruli and tubules accompanied by hemorrhage, cellular infiltration, and congested blood vessels. This could be explained by the finding reported by Zhao et al. [43]. They stated that yeasts pass through the vascular walls attracting neutrophilic infiltration and causing an inflammatory reaction confirmed by a highly significant response with iNOs.
Inflammation interferes with the antioxidant defense mechanism, leading to reactive oxygen species generation and renal glomeruli and tubules damage [44]. This was confirmed by morphometric and immunohistochemical results of C. albicans group, while the fluconazole treated group showed partial improvement.
The group treated with 50 mg/kg ELME showed glomeruli and tubules nearly similar to the normal control, but some tubules were disrupted. While the group treated with 100 mg/kg ELME showed glomeruli and tubules more or less similar to the control group. These findings are in harmony with morphometric and immunohistochemical results. ELME was found to have protective effects on the kidney tissues as the amount of collagen fibers in the renal interstitium and between the glomerular capillaries decreased. In addition, it increased the level of alkaline phosphatase, an enzyme having anti-inflammatory effects, and it reduced the levels of iNOs and desmin, which are highly associated with inflammation and oxidative stress [13].
LC-MS/MS of ELME identified 64 molecules belonging to several phytochemical subclasses. This investigation revealed that ELME possesses anti-inflammatory effects which may be ascribed to several active constituents present in, such as flavonoids, biflavonoids and their glycosides derivatives [45,46,47], alkaloids [48], aurone [49], coumarins [50], and phenolic acid [51,52]. ELME included various alkaloids, flavonoids, and phenolic compounds, such as harmaline, dihydrocapsaicin, luteolin, kaempferol, and isorhamnetin derivatives, which were believed to block NO generation and explain ELME’s anti-inflammatory, antioxidant, and antifungal properties [53,54,55,56].
The nature and quantity of active constituents might vary during different stages of leaves development so this could affect the activity of the plant. Moreover, it would also be interesting to evaluate the content and activity of the leaves collected in different stages of development in further studies.
Our findings align with prior research on the antimicrobial properties of certain flavonoids or flavonoid glycosides, such as myricetin [57,58], apigenin derivatives [59], luteolin derivatives [60], epicatechin [57], anthocyanidin derivatives [61,62], biflavonoids derivatives [13,14,63,64], coumarines derivatives [65,66], harmaline alkaloid [67].
5. Conclusions
In the present work, the phytoconstituents of the E. laurentianus De Wild. leaves methanol extract were determined using LC-MS/MS for the first time. Altogether, 64 compounds were identified, including anthocyanidin-3-O-glycosides, alkaloids, aurone derivative, coumarins, flavonoid aglycone, flavonoid glycoside, phenolic or organic acids.
Our results demonstrate the antifungal and antibiofilm activities of ELME against C. albicans clinical isolates in vitro using different techniques such as examination with SEM and fluorescent microscope as well as qRT-PCR In addition, the in vivo study revealed that ELEM could protect against renal cortical damage induced by C. albicans in an albino rat model using histological and immunohistochemical investigations. This could have occurred via the inhibition of oxidative stress and inflammation. Still, further studies are required to explore the exact mechanisms underlying the effects of ELEMI as an antifungal drug.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jof8050426/s1, Figure S1: Total ion chromatogram by LC-MS/MS of ELME in positive ion mode, Figure S2: Total ion chromatogram by LC-MS/MS of ELME in negative ion mode, Figure S3: MTT cytotoxicity assay of ELME against HSF normal cell line. Table S1: Sequences of primer used in qRT-PCR, Table S2: Values of MICs of ELME against the tested C. albicans clinical isolates, Table S3: Means values of the glomerular basement membrane thickness, tubular injury score in different experimental groups, Table S4: Percentages of collagen, desmin and iNOs expression in different experimental groups.
Author Contributions
Conceptualization, W.A.N. and E.E.; Data curation, M.E.-A., R.I.Y. and A.A.K.; Formal analysis, M.E.-A., M.J.A. and A.A.K.; Investigation, M.E.-A. and A.A.K.; Methodology, W.A.N., G.A., R.I.Y. and E.E.; Project administration, M.J.A.; Software, M.J.A.; Supervision, M.E.-A. and A.A.K.; Validation, M.J.A.; Visualization, G.A.; Writing—original draft, W.A.N., G.A., R.I.Y. and E.E.; Writing—review & editing, W.A.N., G.A., R.I.Y. and E.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The experiment was carried out in accordance with the criteria for the care and use of laboratory animals, which were authorized by the Research Ethical Committee (Faculty of Medicine, Menoufia University, Shebin El-Kom, Egypt), Approval No (2/2022OBSGN).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and Supplementary Material.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Talapko J., Juzbašić M., Matijević T., Pustijanac E., Bekić S., Kotris I., Škrlec I. Candida albicans—The virulence factors and clinical manifestations of infection. J. Fungi. 2021;7:79. doi: 10.3390/jof7020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh D.K., Tóth R., Gácser A. Mechanisms of pathogenic Candida species to evade the host complement attack. Front. Cell. Infect. Microbiol. 2020;10:94. doi: 10.3389/fcimb.2020.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayer F.L., Wilson D., Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4:119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kernien J.F., Snarr B.D., Sheppard D.C., Nett J.E. The interface between fungal biofilms and innate immunity. Front. Immunol. 2018;8:1968. doi: 10.3389/fimmu.2017.01968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamdy R.F., Zaoutis T.E., Seo S.K. Antifungal stewardship considerations for adults and pediatrics. Virulence. 2017;8:658–672. doi: 10.1080/21505594.2016.1226721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sousa F., Ferreira D., Reis S., Costa P. Current insights on antifungal therapy: Novel nanotechnology approaches for drug delivery systems and new drugs from natural sources. Pharmaceuticals. 2020;13:248. doi: 10.3390/ph13090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhuiyan F.R., Howlader S., Raihan T., Hasan M. Plants metabolites: Possibility of natural therapeutics against the COVID-19 pandemic. Front. Med. 2020;7:444. doi: 10.3389/fmed.2020.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azari B., Zahmatkesh Moghadam S., Zarrinfar H., Tasbandi A., Jamialahmadi T., Sahebkar A. Natural Products and Human Diseases. Springer; Berlin/Heidelberg, Germany: 2021. Antifungal Activity of Curcuminoids and Difluorinated Curcumin Against Clinical Isolates of Candida Species; pp. 123–129. [DOI] [PubMed] [Google Scholar]
- 9.Zarrinfar H., Behnam M., Hatamipour M., Sahebkar A. Pharmacological Properties of Plant-Derived Natural Products and Implications for Human Health. Springer; Berlin/Heidelberg, Germany: 2021. Antifungal Activities of Curcuminoids and Difluorinated Curcumin Against Clinical Dermatophyte Isolates; pp. 101–107. [DOI] [PubMed] [Google Scholar]
- 10.Khameneh B., Iranshahy M., Soheili V., Bazzaz B.S.F. Review on plant antimicrobials: A mechanistic viewpoint. Antimicrob. Resist. Infect. Control. 2019;8:118. doi: 10.1186/s13756-019-0559-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nigussie D., Davey G., Tufa T.B., Brewster M., Legesse B.A., Fekadu A., Makonnen E. Antibacterial and antifungal activities of Ethiopian medicinal plants: A systematic review. Front. Pharmacol. 2021;12:1327. doi: 10.3389/fphar.2021.633921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Treutlein J., Vorster P., Wink M. Molecular relationships in Encephalartos (Zamiaceae, Cycadales) based on nucleotide sequences of nuclear ITS 1&2, rbcL, and genomic ISSR fingerprinting. Plant Biol. 2005;7:79–90. doi: 10.1055/s-2004-830478. [DOI] [PubMed] [Google Scholar]
- 13.Negm W., Abo El-Seoud K., Kabbash A., El-Aasr M. Investigation of the Biological Activity Some Gymnosperm Plants Belong to Cycadales Order. J. Adv. Med. Pharm. Res. 2020;1:9–13. doi: 10.21608/jampr.2020.23512.1002. [DOI] [Google Scholar]
- 14.Negm W.A., Abo El-Seoud K.A., Kabbash A., Kassab A.A., El-Aasr M. Hepatoprotective, cytotoxic, antimicrobial and antioxidant activities of Dioon spinulosum leaves Dyer Ex Eichler and its isolated secondary metabolites. Nat. Prod. Res. 2020;35:5166–5176. doi: 10.1080/14786419.2020.1789636. [DOI] [PubMed] [Google Scholar]
- 15.Negm W.A., El-Kadem A.H., Elekhnawy E., Attallah N.G., Al-Hamoud G.A., El-Masry T.A., Zayed A. Wound-Healing Potential of Rhoifolin-Rich Fraction Isolated from Sanguisorba officinalis Roots Supported by Enhancing Re-Epithelization, Angiogenesis, Anti-Inflammatory, and Antimicrobial Effects. Pharmaceuticals. 2022;15:178. doi: 10.3390/ph15020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alotaibi B., Mokhtar F.A., El-Masry T.A., Elekhnawy E., Mostafa S.A., Abdelkader D.H., Elharty M.E., Saleh A., Negm W.A. Antimicrobial Activity of Brassica rapa L. Flowers Extract on Gastrointestinal Tract Infections and Antiulcer Potential against Indomethacin-Induced Gastric Ulcer in Rats Supported by Metabolomics Profiling. J. Inflamm. Res. 2021;14:7411. doi: 10.2147/JIR.S345780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Attallah N.G.M., Negm W.A., Elekhnawy E., Elmongy E.I., Altwaijry N., El-Haroun H., El-Masry T.A., El-Sherbeni S.A. Elucidation of Phytochemical Content of Cupressus macrocarpa Leaves: In Vitro and In Vivo Antibacterial Effect against Methicillin-Resistant Staphylococcus aureus Clinical Isolates. Antibiotics. 2021;10:890. doi: 10.3390/antibiotics10080890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing. CLSI; Wayne, PA, USA: 2017. 20th Informational Supplement; CLSI Document; [Google Scholar]
- 19.Elekhnawy E., Sonbol F., Abdelaziz A., Elbanna T. An investigation of the impact of triclosan adaptation on Proteus mirabilis clinical isolates from an Egyptian university hospital. Braz. J. Microbiol. 2021;52:927–937. doi: 10.1007/s42770-021-00485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Ganiny A.M., Kamel H.A., Yossef N.E., Mansour B., El-Baz A.M. Repurposing pantoprazole and haloperidol as efflux pump inhibitors in azole resistant clinical Candida albicans and non-albicans isolates. Saudi Pharm. J. 2022;30:245–255. doi: 10.1016/j.jsps.2022.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elekhnawy E., Negm W.A., El-Aasr M., Kamer A.A., Alqarni M., Batiha G.E.-S., Obaidullah A.J., Fawzy H.M. Histological assessment, anti-quorum sensing, and anti-biofilm activities of Dioon spinulosum extract: In vitro and in vivo approach. Sci. Rep. 2022;12:180. doi: 10.1038/s41598-021-03953-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Attallah N.G., Negm W.A., Elekhnawy E., Altwaijry N., Elmongy E.I., El-Masry T.A., Alturki E.A., Yousef D.A., Y Shoukheba M. Antibacterial Activity of Boswellia sacra Flueck. Oleoresin Extract against Porphyromonas gingivalis Periodontal Pathogen. Antibiotics. 2021;10:859. doi: 10.3390/antibiotics10070859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pu Y., Liu A., Zheng Y., Ye B. In vitro damage of Candida albicans biofilms by chitosan. Exp. Ther. Med. 2014;8:929–934. doi: 10.3892/etm.2014.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Gong H., Qi Y., Li J., Ji X., Sun J., Tian R., Bao H., Song X., Chen Q. In vitro and in vivo antifungal activities and mechanism of heteropolytungstates against Candida species. Sci. Rep. 2017;7:16942. doi: 10.1038/s41598-017-17239-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiernan J.A. Histological and histochemical methods: Theory and practice. Shock. 1999;12:479. [Google Scholar]
- 26.Pollock L., Rampling D., Greenwald S., Malone M. Desmin expression in rhabdomyosarcoma: Influence of the desmin clone and immunohistochemical method. J. Clin. Pathol. 1995;48:535–538. doi: 10.1136/jcp.48.6.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suvarna K.S., Layton C., Bancroft J.D. Bancroft’s Theory and Practice of Histological Techniques E-Book. Elsevier Health Sciences; Amsterdam, The Netherlands: 2018. [Google Scholar]
- 28.Ramos-Vara J.A., Kiupel M., Baszler T., Bliven L., Brodersen B., Chelack B., West K., Czub S., Del Piero F., Dial S. Suggested guidelines for immunohistochemical techniques in veterinary diagnostic laboratories. J. Vet. Diagn. Investig. 2008;20:393–413. doi: 10.1177/104063870802000401. [DOI] [PubMed] [Google Scholar]
- 29.Chen J., Ren J., Loo W.T., Hao L., Wang M. Lysyl oxidases expression and histopathological changes of the diabetic rat nephron. Mol. Med. Rep. 2018;17:2431–2441. doi: 10.3892/mmr.2017.8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White S. Basic & Clinical Biostatistics. 5th ed. McGraw-Hill Education; New York, NY, USA: 2020. Methods of evidence-based medicine and decision analysis. [Google Scholar]
- 31.Bongomin F., Gago S., Oladele R.O., Denning D.W. Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision. J. Fungi. 2017;3:57. doi: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Barros P.P., Rossoni R.D., Garcia M.T., de Lima Kaminski V., Loures F.V., Fuchs B.B., Mylonakis E., Junqueira J.C. The Anti-Biofilm Efficacy of Caffeic Acid Phenethyl Ester (CAPE) In Vitro and a Murine Model of Oral Candidiasis. Front. Cell. Infect. Microbiol. 2021;11:700305. doi: 10.3389/fcimb.2021.700305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonifácio B.V., Vila T.V.M., Masiero I.F., da Silva P.B., da Silva I.C., de Oliveira Lopes É., Ramos M.A.D.S., de Souza L.P., Vilegas W., Pavan F.R., et al. Antifungal Activity of a Hydroethanolic Extract from Astronium urundeuva Leaves against Candida albicans and Candida glabrata. Front. Microbiol. 2019;10:2642. doi: 10.3389/fmicb.2019.02642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma A., Singh S., Tewari R., Bhatt V.P., Sharma J., Maurya I.K. Phytochemical analysis and mode of action against Candida glabrata of Paeonia emodi extracts. J. Mycol. Med. 2018;28:443–451. doi: 10.1016/j.mycmed.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Diba A., Alizadeh F. In vitro and in vivo antifungal activity of Allium hirtifolium and Allium sativum. Avicenna J. Phytomed. 2018;8:465. [PMC free article] [PubMed] [Google Scholar]
- 36.Alalwan H., Rajendran R., Lappin D.F., Combet E., Shahzad M., Robertson D., Nile C.J., Williams C., Ramage G. The anti-adhesive effect of curcumin on Candida albicans biofilms on denture materials. Front. Microbiol. 2017;8:659. doi: 10.3389/fmicb.2017.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cavalheiro M., Teixeira M.C. Candida Biofilms: Threats, Challenges, and Promising Strategies. Front. Med. 2018;5:28. doi: 10.3389/fmed.2018.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zago C.E., Silva S., Sanitá P.V., Barbugli P.A., Dias C.M.I., Lordello V.B., Vergani C.E. Dynamics of biofilm formation and the interaction between Candida albicans and methicillin-susceptible (MSSA) and -resistant Staphylococcus aureus (MRSA) PLoS ONE. 2015;10:e0123206. doi: 10.1371/journal.pone.0123206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martins N., Petropoulos S., Ferreira I.C. Chemical composition and bioactive compounds of garlic (Allium sativum L.) as affected by pre-and post-harvest conditions: A review. Food Chem. 2016;211:41–50. doi: 10.1016/j.foodchem.2016.05.029. [DOI] [PubMed] [Google Scholar]
- 40.Abu-Darwish M., Cabral C., Gonçalves M., Cavaleiro C., Cruz M., Zulfiqar A., Khan I., Efferth T., Salgueiro L. Chemical composition and biological activities of Artemisia judaica essential oil from southern desert of Jordan. J. Ethnopharmacol. 2016;191:161–168. doi: 10.1016/j.jep.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 41.Kim D., Kim K.-Y. Hedera rhombea inhibits the biofilm formation of Candida, thereby increases the susceptibility to antifungal agent, and reduces infection. PLoS ONE. 2021;16:e0258108. doi: 10.1371/journal.pone.0258108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song Z., Papanicolaou N., Dean S., Bing Z. Localized candidiasis in kidney presented as a mass mimicking renal cell carcinoma. Case Rep. Infect. Dis. 2012;2012:953590. doi: 10.1155/2012/953590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan S.H., Younus H., Allemailem K.S., Almatroudi A., Alrumaihi F., Alruwetei A.M., Alsahli M.A., Khan A., Khan M.A. Potential of methylglyoxal-conjugated chitosan nanoparticles in treatment of fluconazole-resistant Candida albicans infection in a murine model. Int. J. Nanomed. 2020;15:3681. doi: 10.2147/IJN.S249625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ayoub I.M., Korinek M., Hwang T.-L., Chen B.-H., Chang F.-R., El-Shazly M., Singab A.N.B. Probing the antiallergic and anti-inflammatory activity of biflavonoids and dihydroflavonols from Dietes bicolor. J. Nat. Prod. 2018;81:243–253. doi: 10.1021/acs.jnatprod.7b00476. [DOI] [PubMed] [Google Scholar]
- 45.Serafini M., Peluso I., Raguzzini A. Flavonoids as anti-inflammatory agents. Proc. Nutr. Soc. 2010;69:273–278. doi: 10.1017/S002966511000162X. [DOI] [PubMed] [Google Scholar]
- 46.Maleki S.J., Crespo J.F., Cabanillas B. Anti-inflammatory effects of flavonoids. Food Chem. 2019;299:125124. doi: 10.1016/j.foodchem.2019.125124. [DOI] [PubMed] [Google Scholar]
- 47.Souto A.L., Tavares J.F., Da Silva M.S., Diniz M.D.F.F.M., Athayde-Filho D., Filgueiras P., Barbosa Filho J.M. Anti-inflammatory activity of alkaloids: An update from 2000 to 2010. Molecules. 2011;16:8515–8534. doi: 10.3390/molecules16108515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sui G., Li T., Zhang B., Wang R., Hao H., Zhou W. Recent advances on synthesis and biological activities of aurones. Bioorg. Med. Chem. 2021;29:115895. doi: 10.1016/j.bmc.2020.115895. [DOI] [PubMed] [Google Scholar]
- 49.Grover J., Jachak S.M. Coumarins as privileged scaffold for anti-inflammatory drug development. RSC Adv. 2015;5:38892–38905. doi: 10.1039/C5RA05643H. [DOI] [Google Scholar]
- 50.Su X., Zhang J., Wang H., Xu J., He J., Liu L., Zhang T., Chen R., Kang J. Phenolic acid profiling, antioxidant, and anti-inflammatory activities, and miRNA regulation in the polyphenols of 16 blueberry samples from China. Molecules. 2017;22:312. doi: 10.3390/molecules22020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu X.-W., Wei W., Yang X.-W., Zhang Y.-B., Xu W., Yang Y.-F., Zhong G.-Y., Liu H.-N., Yang S.-L. Anti-inflammatory phenolic acid esters from the roots and rhizomes of Notopterygium incisium and their permeability in the human Caco-2 monolayer cell model. Molecules. 2017;22:935. doi: 10.3390/molecules22060935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim Y., Lee J. Anti-inflammatory activity of capsaicin and dihydrocapsaicin through heme oxygenase-1 induction in raw264. 7 macrophages. J. Food Biochem. 2014;38:381–387. doi: 10.1111/jfbc.12064. [DOI] [Google Scholar]
- 53.Uddin M.J., Xu S., Crews B.C., Aleem A.M., Ghebreselasie K., Banerjee S., Marnett L.J. Harmaline analogs as substrate-selective cyclooxygenase-2 inhibitors. ACS Med. Chem. Lett. 2020;11:1881–1885. doi: 10.1021/acsmedchemlett.9b00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patil K.R., Mahajan U.B., Unger B.S., Goyal S.N., Belemkar S., Surana S.J., Ojha S., Patil C.R. Animal models of inflammation for screening of anti-inflammatory drugs: Implications for the discovery and development of phytopharmaceuticals. Int. J. Mol. Sci. 2019;20:84367. doi: 10.3390/ijms20184367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin Y.-S. Recent advances in natural antifungal flavonoids and their derivatives. Bioorg. Med. Chem. Lett. 2019;29:126589. doi: 10.1016/j.bmcl.2019.07.048. [DOI] [PubMed] [Google Scholar]
- 56.Buchmann D., Schultze N., Borchardt J., Böttcher I., Schaufler K., Guenther S. Synergistic antimicrobial activities of epigallocatechin gallate, myricetin, daidzein, gallic acid, epicatechin, 3-hydroxy-6-methoxyflavone and genistein combined with antibiotics against ESKAPE pathogens. J. Appl. Microbiol. 2022;132:949–963. doi: 10.1111/jam.15253. [DOI] [PubMed] [Google Scholar]
- 57.Chen M., Tang X., Liu T., Peng F., Zhou Q., Luo H., He M., Xue W. Antimicrobial evaluation of myricetin derivatives containing benzimidazole skeleton against plant pathogens. Fitoterapia. 2021;149:104804. doi: 10.1016/j.fitote.2020.104804. [DOI] [PubMed] [Google Scholar]
- 58.Wang M., Firrman J., Liu L., Yam K. A review on flavonoid apigenin: Dietary intake, ADME, antimicrobial effects, and interactions with human gut microbiota. BioMed Res. Int. 2019;2019:7010467. doi: 10.1155/2019/7010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qian W., Liu M., Fu Y., Zhang J., Liu W., Li J., Li X., Li Y., Wang T. Antimicrobial mechanism of luteolin against Staphylococcus aureus and Listeria monocytogenes and its antibiofilm properties. Microb. Pathog. 2020;142:104056. doi: 10.1016/j.micpath.2020.104056. [DOI] [PubMed] [Google Scholar]
- 60.Alejo-Armijo A., Glibota N., Frías M.P., Altarejos J., Gálvez A., Ortega-Morente E., Salido S. Antimicrobial and antibiofilm activities of procyanidins extracted from laurel wood against a selection of foodborne microorganisms. Int. J. Food Sci. Technol. 2017;52:679–686. doi: 10.1111/ijfs.13321. [DOI] [Google Scholar]
- 61.Oliveira H., Correia P., Bessa L.J., Guimarães M., Gameiro P., Freitas V.D., Mateus N., Cruz L., Fernandes I. Cyanidin-3-Glucoside Lipophilic Conjugates for Topical Application: Tuning the Antimicrobial Activities with Fatty Acid Chain Length. Processes. 2021;9:340. doi: 10.3390/pr9020340. [DOI] [Google Scholar]
- 62.Negm W.A., El-Aasr M., Kamer A.A., Elekhnawy E. Investigation of the Antibacterial Activity and Efflux Pump Inhibitory Effect of Cycas thouarsii R. Br. Extract against Klebsiella pneumoniae Clinical Isolates. Pharmaceuticals. 2021;14:756. doi: 10.3390/ph14080756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elmongy E.I., Negm W.A., Elekhnawy E., El-Masry T.A., Attallah N.G., Altwaijry N., Batiha G.E.-S., El-Sherbeni S.A. Antidiarrheal and Antibacterial Activities of Monterey Cypress Phytochemicals: In Vivo and In Vitro Approach. Molecules. 2022;27:346. doi: 10.3390/molecules27020346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patel J. Synthesis, characterization and antimicrobial screening of some furofused coumarines. Int. J. Adv. Res. Dev. 2018;3:1–8. [Google Scholar]
- 65.Widelski J., Luca S.V., Skiba A., Chinou I., Marcourt L., Wolfender J.-L., Skalicka-Wozniak K. Isolation and antimicrobial activity of coumarin derivatives from fruits of Peucedanum luxurians Tamamsch. Molecules. 2018;23:1222. doi: 10.3390/molecules23051222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y., Shi X., Xie X., Laster K.V., Pang M., Liu K., Hwang J., Kim D.J. Harmaline isolated from Peganum harmala suppresses growth of esophageal squamous cell carcinoma through targeting mTOR. Phytother. Res. 2021;35:6377–6388. doi: 10.1002/ptr.7289. [DOI] [PubMed] [Google Scholar]
- 67.Qaisar U. Peganum harmala: A rich source of antimicrobial agents. Int. J. Biotech Trends Technol. 2019;9:1–10. doi: 10.14445/22490183/IJBTT-V9I4P602. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article and Supplementary Material.