Abstract
Accurate measurement of daily infection incidence is crucial to epidemic response. However, delays in symptom onset, testing, and reporting obscure the dynamics of transmission, necessitating methods to remove the effects of stochastic delays from observed data. Existing estimators can be sensitive to model misspecification and censored observations; many analysts have instead used methods that exhibit strong bias. We develop an estimator with a regularization scheme to cope with stochastic delays, which we term the robust incidence deconvolution estimator. We compare the method to existing estimators in a simulation study, measuring accuracy in a variety of experimental conditions. We then use the method to study COVID-19 records in the United States, highlighting its stability in the face of misspecification and right censoring. To implement the robust incidence deconvolution estimator, we release incidental, a ready-to-use R implementation of our estimator that can aid ongoing efforts to monitor the COVID-19 pandemic.
Keywords: Deconvolution, COVID, Infection time series, Statistical estimation, Statistical inference
Information on the progress of an ongoing epidemic arrives with delays. New cases, hospitalizations, and deaths are reported potentially weeks after individuals are infected, which obscure the count of daily new infections. Accurate estimation of daily infections is crucial for understanding the dynamics of disease transmission, and assessing the impacts of interventions.1–3 Currently available methods for reconstructing infection curves exhibit bias and instability.
Mathematically, observations such as daily reported cases can be described as a convolution of the underlying time series of new infections with a delay distribution—the probability distribution that describes time from infection to reporting. Recovering the infections curve from delayed reports is a deconvolution operation (Figure 1). Unfortunately, deconvolution of noisy data presents an ill-posed inverse problem, in which signal and noise cannot be separated, even when the delay distribution is known perfectly.4 Ill-posedness manifests as instability in estimates, which is compounded in infection estimation by right censoring—recent infections have a smaller probability of being reported in the observation period. Instability in deconvolution problems is often addressed by regularization, which imposes structure on the signal that is recovered from noisy data.5
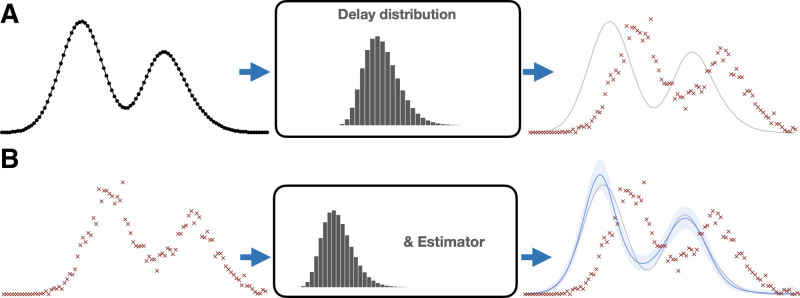
FIGURE 1.
Infection estimation overview. Top: the underlying infection time series—new infections per day—is perturbed by a delay distribution (center) that is measured with other data or assumed known. Each infection date is stochastically delayed, resulting in the reporting curve (red ×’s)—new reported cases per day. Bottom: the estimation procedure aims to undo this stochastic delay. Given observed report curve (left) we use a statistical estimator with the delay distribution to recover the underlying curve. (a) Assumed data generating process. (b) Estimation procedure.
In this work, we propose a statistically robust method to infer infection time series from delayed data, which we call the robust incidence deconvolution estimator (RIDE). This method incorporates a specific form of regularization that yields stable infection estimates, even in the presence of right censoring. In a simulation study, we compare the RIDE to existing methods, highlighting estimator accuracy and stability. As a motivating example, we use this method to study transmission dynamics of SARS-CoV-2 at state and local levels. We compare it to existing estimators on epidemic data from different regions in the United States, qualitatively showing its stability and robustness to censoring.
In our simulated and empirical examples, we compare the RIDE to two classes of existing methods. The first class, which we term reconvolution estimators, estimate the infection curve by sampling from an assumed delay distribution and shifting observed case reports backward in time—effectively, applying a convolution operation in reverse. This approach is stable and has been applied in a number of public tools for tracking the COVID-19 pandemic,6–8 but exhibits biases because it is not a deconvolution operation. The second class of estimators perform regularized deconvolution but can yield unstable estimates if regularization is inadequate or right censoring is not addressed. These include back projection (BP) (or back calculation) estimators developed to analyze the AIDS epidemic,9–14 and the Richardson-Lucy (RL) algorithm, a model-free deconvolution method that has been used to analyze influenza.15
Additionally, we make the proposed method available in an R package, incidental.
METHODS
We first give a brief overview of the statistical estimation problem, existing approaches and the RIDE. Institutional Review Board approval was not required for this research, as we only use publicly available epidemiologic data that are deidentified.
Method Overview
Given a time series of delayed observations for T days, and a delay distribution (e.g., the distribution of time from infection to reporting) up to P days, the goal is to infer the time series of new infections The expected value of the observed data Y is a convolution of the infection time series X with the delay distribution θ; estimation of X involves the deconvolution of Y and θ. To produce an estimator that is robust to noise in Y, we propose a model-based estimator using a cubic spline16 to describe the underlying infections, X, and a Poisson likelihood to describe the observed cases. We set the degrees of freedom of the spline basis using Akaike Information Criterion (AIC).17 Additionally, we add a regularization penalty on the second difference of the spline parameters, encouraging smoothness and select regularization strength with out-of-sample log likelihood.
Finally, we include an additional adjustment for not-yet observed cases to stabilize estimates in the presence of right censoring. Cases that are observed after the current time-point T due to reporting delays are relevant for the estimation of infections for days close to T. Estimates near T rely on only a few days of observations, leading to instability or overregularization near T.
We address this issue as a missing data problem, and use a strategy similar to multiple imputation techniques18 to impute samples after T. Specifically, we sample many extrapolations of the observed time series from a random walk that encodes the assumption that the autocorrelation in the observed data, which is a direct result of the convolution of infections with the delay distribution, will remain in future observations. For each extrapolation, we condition on the simulated counts to form the incidence estimate, and average estimates across these replicates.
Right censoring corresponds to missing information in for all Those observations are, however, important for forming our estimate of for t close to T, which is where this extrapolation helps. Right censoring can be accompanied by under-ascertainment, which corresponds to incomplete counts in Yt for t close to T that might be corrected in the following days. For example, a test reported on June 1 may not enter official records and be available until a few days later, at which time it will still be reported as June 1. Right censoring produces estimator instability near T through lack of data, while under-ascertainment often produces incorrect data near T. If recording delay data are available, a runoff triangle method19 can be used as preprocessing to correct for under-ascertainment or, when used for forecasting, as an alternative to the existing extrapolation method. Since recording delay data are not widely available for COVID-19, we restrict our analysis to periods with relatively complete data.
Methodologic Details
We consider the following observation model of individual infected and reported dates. Each individual who becomes infected on day is confirmed on day and we assume that there were no infections before the initial time, denoted The date of confirmation is stochastically delayed from the date of infection In, , where Dn is a random number of days sampled from a discrete distribution with delay distribution probability vector θ. The infection curve is a time series of daily infection counts over the population, denoted where and 1 is the indicator function. The observed report curve is a time series of daily reported cases, denoted , defined analogously with Cn. Our goal is to reconstruct the infection time series X from an observed realization of counts .
These estimators consider θ fixed and known. Practically, θ can be estimated from studies or other data sources for COVID-19.20,21 We observe day-of-week and nonstationarity effects that are not captured by θ; we examine robustness to these our simulation study and case study, respectively. COVID-19 delay distribution data sources, computations, and sensitivity are given in the eAppendix (http://links.lww.com/EDE/B924).
The observed counts are related to the unobserved infection time series by a discrete convolution, which can be expressed as a matrix multiply,
| (1) |
where Pθ has a triangular structure that depends on the delay distribution θ. To deconvolve the observed signal, an estimator needs to invert Pθ. See the eAppendix (http://links.lww.com/EDE/B924) for details.
“Reconvolution” Incidence Reconstruction
A popular method for incidence estimation attempts to undo the stochastic delays by sampling from the delay distribution and subtracting the value from each observed time,7,8,22,23 effectively a convolution of the already convolved report curve. For each case n, the reconvolution estimator samples a delay and computes , aggregating these into the incidence curve estimate .
The linear relationship between X and Y makes clear the conceptual error of the reconvolution method. The reconvolution estimator has the expectation
| (2) |
which will be inconsistent in general, as . This conceptual error motivates the use of methods developed for deconvolving signals. See the eAppendix (http://links.lww.com/EDE/B924) for an in-depth discussion.
Deconvolution Estimators
Deconvolving signals is a well-studied problem in signal processing. One such deconvolution method is the RL estimator,24,25 a model-free iterative algorithm that is flexible but highly sensitive to observation noise. Nevertheless, the RL estimator has been used to reconstruct incidence curves for infectious disease.15
An alternative class of methods uses statistical models to form deconvolved incidence estimates. BP (or back calculation) methods are model-based estimators that were developed to infer HIV/AIDS infection incidence.9–13,26 BP is closely related to empirical Bayes methods for deconvolution.27 These approaches form estimates by maximizing the marginal likelihood of observed data given a model for and some form of regularization. Parameterizing the incidence time series as using smoothing splines or a step function, a model-based objective function takes the form
| (3) |
where the likelihood function varies from method to method. Previous methods have considered both multinomial12 and Poisson10 observation models, along with both model-based and post hoc methods of smoothing.26 Table summarizes various deconvolution methods in the literature.
TABLE.
Summary of Previous Deconvolution and Back Projection Models
| Reference | Method Summary | Basis | Regularization | Extrapolation |
|---|---|---|---|---|
| Brookmeyer and Gail12 | Back projection | Step | None | Binomial model for unseen cases after last date |
| Brookmeyer11 | Back projection | Cubic spline | Second order difference squared | None |
| Liao and Brookmeyer14 | Back projection | Step | Exponentiated transform squared: | None |
| Becker et al.10 | Back projection | Step | Local smoothing of EM update | None |
| Bacchetti et al.9 | Back projection | Step | Second order difference squared | None |
| Goldstein et al.15 | Richardson-Lucy | Step | Early stopping of update algorithm | None |
Ill-posedness and Regularization
Infection time series estimation is a classic ill-posed inverse problem. Without a model, the free parameters are just identified—the number of free parameters is equal to the number of observed data points.4 Without observation noise, the convolution matrix Pθ can be inverted, and the true incidence X can be identified. With observation noise, the data alone cannot distinguish signal from noise, leading to fundamentally unstable estimation. Regularization imposes some realistic structure on the solution of the deconvolution, based on prior information, to separate out the noise. Smoothness—the belief that incidence should not vary wildly day to day—is the main property induced by regularization.
We devise a model-based estimate using a cubic spline16 to describe the underlying incidence, and a Poisson likelihood to describe the observed cases. Locations with low case counts can be prone to overfitting due to small sample size. To address this issue, we first select the spline basis degrees of freedom using AIC17 and add a squared regularization penalty on the second difference of the β parameters. To select λ, we split the observed data and use out-of-sample log likelihood. By default, we use 25% of the data to estimate out-of-sample log likelihood, and average over four random splits. The largest value of λ that gives an estimator within 2% of the highest out-of-sample likelihood is selected as a guard against overfitting. Deconvolution methods can be sensitive to loss function misspecification.9 The validation-based selection of λ allows the estimator to be robust to the types of misspecification associated with COVID-19 reporting data.
The stability of estimates in the most recent time window before T is another practical concern. In this window, the effective number of observations is small due to right censoring, leading to estimator instability. We exploit the autocorrelation structure induced by the convolution of incidence and the delay distribution by extrapolating the report curve forward in time with a random walk and condition on these simulated counts to form the incidence estimate. We first apply an Anscombe transform28 to the observed report curve to stabilize the variance and use the empirical single-lag autocorrelation to simulate random walk extrapolations centered around a first order approximation for drift. We average estimates over replicates of extrapolated random walks in a style similar to common techniques for handling missing data.18 We term this procedure the RIDE. Details are given in the eAppendix (http://links.lww.com/EDE/B924).
RESULTS
We study incidence estimator performance via a simulation study and a case study of SARS-CoV-2 infection incidence estimation in the United States.
Simulation Study
We examine the stability and reconstruction accuracy of estimators on a set of synthetic examples designed to mimic statistical issues associated with COVID-19 data: right censoring and model misspecification. We study the accuracy of various incidence estimators described herein, including reconvolution, RL, BP10,26 implemented in the surveillance package in R29 with two levels of smoothing (, and ), and our proposed method. We study the performance of the proposed RIDE under three conditions, (i) no extrapolation and a spline basis, (ii) extrapolation with a spline basis, and (iii) extrapolation with a step function basis.
We compare methods on six synthetic infection curves to study varying levels of complexity: a steep curve with a slow decay (slow decay), a symmetric curve (symmetric), a double-peaked curve representing two waves (double), a pathologic curve that has a sharp climb followed by a total drop-off in cases (stop), and two faster moving curves with Matérn covariance structure (matern and matern2). Curves are depicted in Figure 2A.
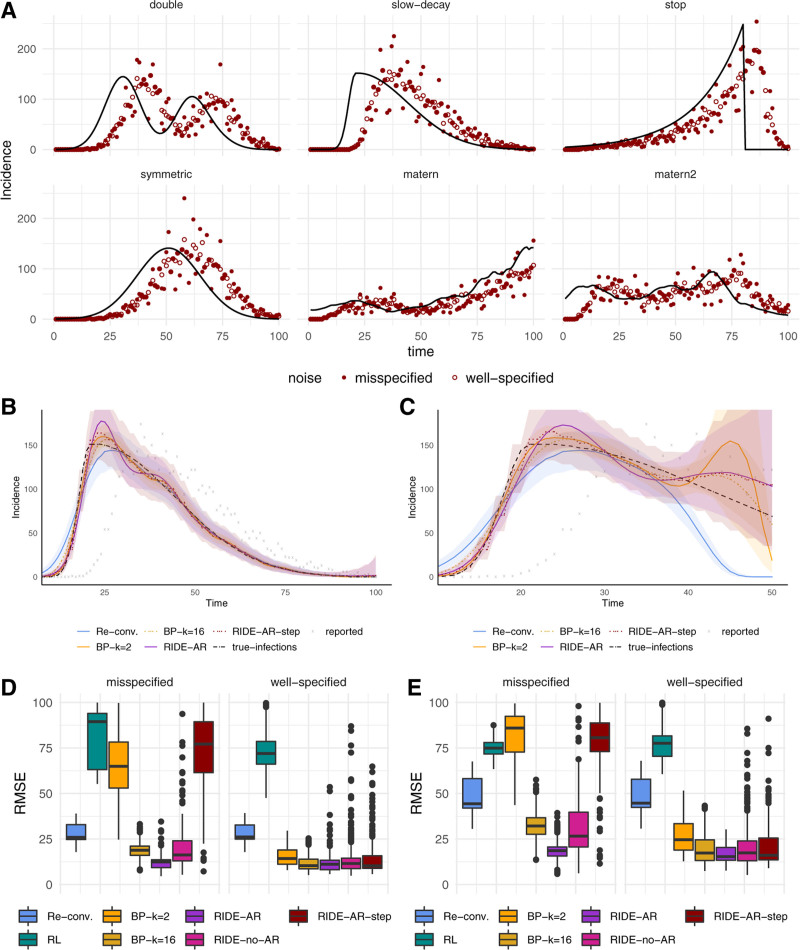
FIGURE 2.
Synthetic experiments. (A) Six synthetic incidence curves and simulated observations with both correct and misspecified noise models. (B) Estimators on the full slow-decay data. (C) Estimators on censored slow-decay data. (D–E) Root mean squared error over all experimental settings and replicates for both correct and misspecified data for all data (D) and over the most recent 20 days of observations (E). Additional details are in the eAppendix; http://links.lww.com/EDE/B924. AR indicates autoregressive; BP, back projection; Re–conv, reconvolution; RIDE, robust incidence deconvolution estimator; RL, Richardson-Lucy; and RMSE, root mean squared error.
For each curve, we consider two additional experimental settings—different levels of right censoring (from highly censored to fully observed) and misspecification in the delay distribution (approximating processing delays in commercial testing and case reporting). For censored data, we consider observation windows with T = 40, 50, 60, 80, and 100 days. We assume a delay distribution with a mean delay of 10 days. In the misspecified setting, we mimic weekly reporting delays where on every sixth and seventh day a uniform random proportion of the cases between 30% and 50% are reported 2 days later. See the eAppendix (http://links.lww.com/EDE/B924) for a description of the synthetic data and additional results.
For each of the 60 experimental settings (six curves, five observation windows, and two noise models), we generate 80 report curves with different random seeds. For each curve, we apply each estimator and measure its accuracy with the root mean squared error between the inferred and true infection time series.
In general, we find that the model-based approaches more accurately infer the infection time series than the reconvolution and RL estimators. Figure 2B depicts typical behavior; reconvolution tends to underestimate the steep slope and the peak infection incidence. The RIDE’s regularization scheme is crucial when the model is misspecified; random walk extrapolation is key to stabilizing estimates for highly censored data. For example, BP and reconvolution dramatically under-predict the most recent infection incidence in the right-censored curve in Figure 2C.
The distribution of errors measured over all experimental settings for each estimator is presented in Figure 2D. The model-based estimators fare better in both the correctly specified and misspecified setting. BP is competitive in the well-specified setting, but in the misspecified setting accuracy suffers in comparison to our regularization scheme although both methods share the same likelihood model.
For censored data, random walk regularization stabilizes incidence reconstruction. In Figure 2E, we observe that both the reconvolution and BP methods struggle with right censoring owing to a lack of enforced smoothness. Since the incidence curve must describe unobserved cases, we include an estimate of these via extrapolation. Although this assumption may not be realistic in some instances, in smoother settings the autoregressive imputation offers a realistic assumption about the evolution of new cases. Additionally, averaging over random walk extrapolations stabilizes the uncertainty region toward the right of the curve. Coverage rates are in the eAppendix (http://links.lww.com/EDE/B924).
Case Study—SARS-CoV-2 Infection Incidence Estimation
Monitoring the COVID-19 pandemic presents numerous data challenges.2,30 Disease transmission can be studied using different sources of data, each with unique benefits and drawbacks. Data on reported cases are widely and consistently available, even at the county level in the United States; however, such data are affected by variation in testing levels across geographical regions and time periods. Hospitalization data are not affected by testing effort,7 but represent a smaller proportion of total cases, and are not widely available at the county level. Deaths incur longer delays and represent a small proportion of infections, often leading to unstable incidence estimates in locations with low population or case loads. In addition to uncertain estimates of delay, all types of observations are susceptible to random sources of error, secular changes from nonstationary patterns of surveillance, diagnosis, and treatment. There are also well-known recording delays, where case, hospitalization, and death data are often revised upward for two weeks to a month after the reporting date.
We aggregated reported COVID-19 cases, hospital admissions, and deaths for selected regions with high-quality data: Arizona, New York, Ohio, Texas, and Virginia. Hospital admission data are readily available in New York City, Arizona state, Ohio state, and within Virginia health regions. Data were collected from earliest available case records by region, from January 2020, through December, 2020. Capture occurred at least 14 days after the last date for analysis. See the eAppendix (http://links.lww.com/EDE/B924) for data sources.
We estimate delay distributions for infection to case, hospitalization, and death reports by composing two delay distributions: (1) time from infection to symptom onset, and (2) time from symptom onset to report. We estimate the time from infection to symptom onset by matching the quantiles of a gamma distribution to the data from31; from symptom onset to positive test report by fitting a gamma distribution to nonzero delay times from Florida for all cases through July 14, 202032; from symptom onset to hospitalization, as fitted by7 using data from33,34; and from hospitalization to death.7 Continuous distributions are discretized through rounding to estimate θ. See the eAppendix (http://links.lww.com/EDE/B924) for the full delay distribution specification.
Comparison With Existing Estimation Methods
We apply existing estimation methods and the proposed RIDE to COVID-19 case data from Queens, New York, Staten Island, New York, the Austin, Texas metro area, and the state of Arizona in Figure 3. The incidence fits are then used as inputs to the method of1 for estimating Rt. These regions were chosen for the variation in the overall number of cases, noise levels, and incidence patterns. We compare the RIDE output to three existing approaches: BP,10 RL deconvolution,15 and reconvolution.6–8,22,23 BP is fit by the backprojNP method in the surveillance package version 1.18.0 in R version 4.0.0 with parameters k = 16, eps = rep (0.005, 2), iter.max = rep (250, 2), B = –1. See the eAppendix (http://links.lww.com/EDE/B924) for a comparison of BP fits across parameter levels.
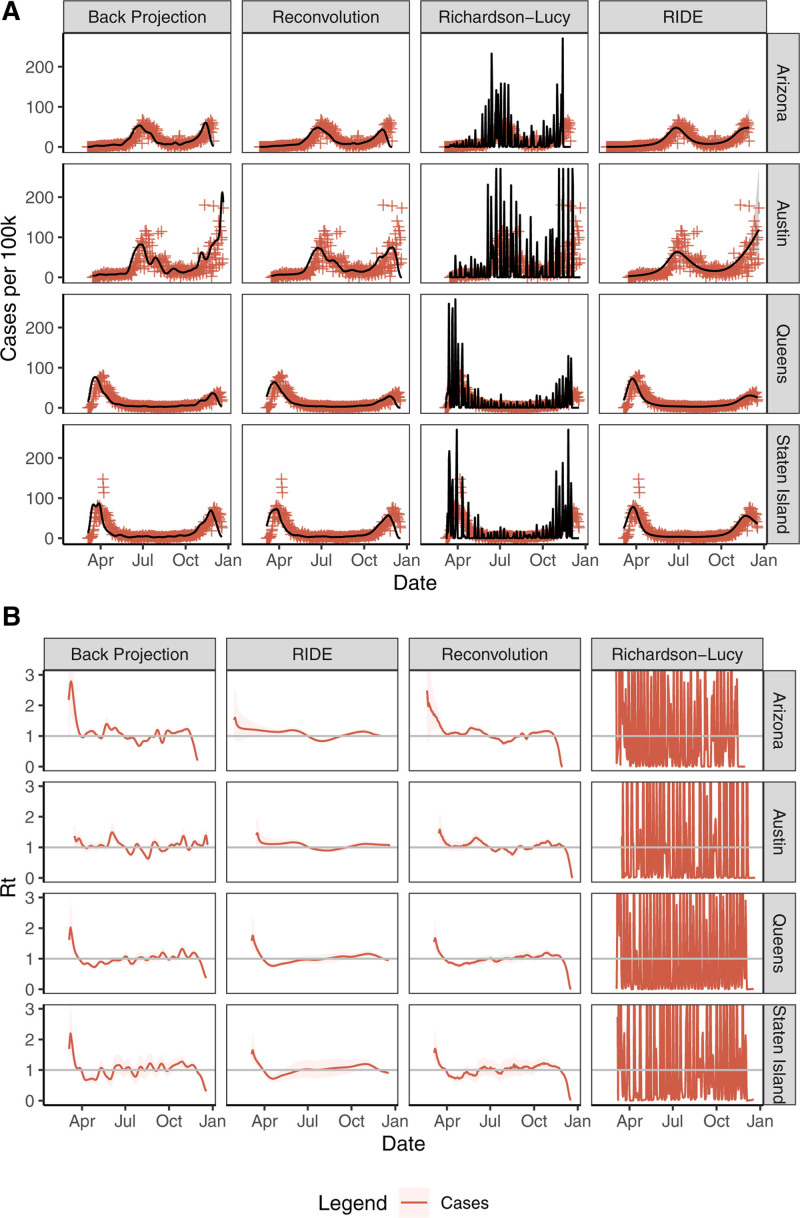
FIGURE 3.
(A) Infections incidence (solid black line), 90% credible regions (gray shaded) when available, and observed values (red plus) by data type across regions (rows) and by method (columns). (B) Estimated Rt with 90% credible regions fit using the method of1 from the incidence estimates in (A). Richardson-Lucy estimates are truncated in each panel for readability. RIDE indicates robust incidence deconvolution estimator.
The reconvolution procedure is a biased estimator and leads to oversmoothing of the infections curve and under estimating the peak. RL is sensitive to noise, leading to large oscillations in all cases due to a combination of a misspecified loss function and under-regularization. BP and reconvolution are also somewhat sensitive to noise, with moderate oscillations in the high-noise Austin region.
BP and reconvolution are sensitive to right censoring, often lowering estimates near T to capture outliers. The RIDE is robust to noise in reporting and right censoring due to careful choice of basis, regularization, and extrapolation of observations after T. These parameters are tuned to model daily COVID-19 case report data. We note that due to its smoothing and censoring corrections, the RIDE produces Rt estimates with fewer oscillations and an absence of right-tail Rt declines when compared with the other methods.
All methods fit only as well as existing data allows. Although all COVID-19 reports include noise, there is under-ascertainment of recent reports. We tried to mitigate this by omitting recent data. However, each method responds differently to under-ascertainment. Reconvolution and BP tend to underestimate incidence at the end of the time series. The RIDE tries to correct for right censoring via extrapolation, so artificially low records can lead to a low extrapolation estimate and low incidence estimate. Recording data are not available for most COVID-19 cases, which would have allowed for under-ascertainment correction using the method of.19
Comparison of Inferences by Data Source
We next apply the RIDE to case, hospitalization, and mortality data from Arizona, Ohio, Virginia, New York City boroughs, and New York City as a whole—locations chosen for their readily available hospitalization data and relatively large number of reports. Staten Island is omitted due to a low death rate throughout the summer. Figure 4 depicts inferred daily infections that lead to reports. These values are on different scales as only a fraction of infections lead to hospitalization or death, or even reported cases. Moreover, these scaling factors can change over time as testing capacity increases and case mortality rate decreases. Nevertheless, they should have general alignment about when peaks occur and outbreak intensity. Note that the peak in estimated infections precedes the peak in reported cases, and has steeper upward and downward trajectories than the observed data time series. Additionally, uncertainty regions near the end of the time series are much larger for daily infections inferred from deaths data. This is due to the longer lag between infection and death relative to cases and hospitalizations. Reproductive numbers fitted using the method of,1 based on the inferred infection time series, are given in Figure 5.
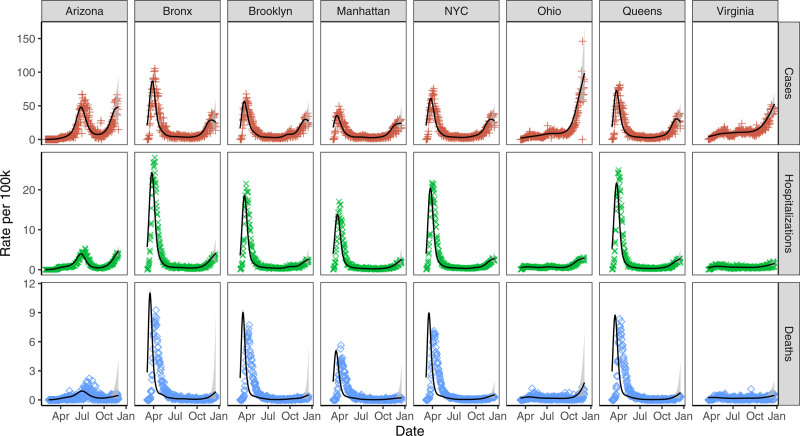
FIGURE 4.
Infections incidence (solid black line), 90% credible regions (gray shaded), and observed values by data type (red plus for cases, green cross for hospitalizations, and blue diamond for deaths) across regions.
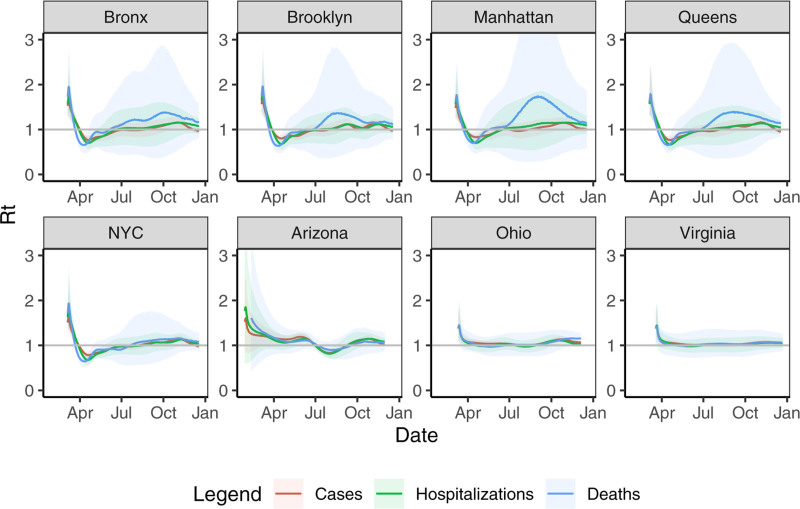
FIGURE 5.
Rt fitted on infection time series estimated by data type across regions. Solid lines are means and ribbons are 90% credible regions.
Inferred infections from deaths consistently peaked earlier by about a week than inferred infections from hospitalizations in New York City. This may be due to nascent treatment regimes and overwhelmed hospitals in New York City during March and April. We note that Rt estimates based on case, hospitalization, and death inferences track closely in most areas. One area of disagreement is Rt estimates based on incidence inferred from deaths versus other sources in NYC boroughs during summer months. This is likely due to low New York City death rates during the late summer and early fall, where there were a daily average of 1.2 deaths in Brooklyn, 1.6 in the Bronx, 0.6 in Manhattan, 1.0 in Queens, and 4.6 in New York City overall between August 1 and September 30, 2020. In comparison, there were 10.9 (104.9) average daily hospitalizations (cases) in Brooklyn during that same period, with 6.9 (50.7) in the Bronx, 3.6 (45.5) in Manhattan, 7.9 (71.2) in Queens, and 30.7 (291.3) in New York City overall. Other regions had higher death rates during that period; there were an average of 32.1 daily deaths in Arizona, 21.6 in Ohio, and 17.0 in Virginia, which lead to more stable Rt estimates.
Relationships between the case rate, hospitalization rate, and death rate have changed between February and December. This is likely due to increases in testing, falling case mortality rates, and shifting infection demographics. Although case or death incidence curves often align well with hospitalization curves at a specific point in time, these external factors can lead to poor alignment over longer time periods.
In the eAppendix (http://links.lww.com/EDE/B924), we present additional inferences at the state and local levels, including alignment with policy interventions including stay-at-home orders and school closures, and incidence at regional versus state levels.
DISCUSSION
Infection incidence and transmission dynamics are obscured by the incubation period, testing delays, reporting delays, and time to hospitalization or death. Accurate estimation of the underlying infection time series is a pressing problem rife with challenges and complications, including observation noise, censoring, and model misspecification. Existing methods can exhibit significant bias or are sensitive to noise or missing observations in reported data. The RIDE is statistically rigorous and robust to some of the data challenges that are present with COVID-19 reported data, including high-noise levels and right censoring. Despite concerns that variation in testing effort may make hospitalizations data superior to data from all cases for monitoring infection dynamics, we found that our inferences from case data provided a reasonable proxy for those fit from hospitalization data, which are much less widely available and pose signal-to-noise ratio problems in less populous counties or regions.
One of the most promising uses for regional infections estimation is as a tool in the evaluation of public policy. Accurate reconstruction of infection time series is necessary to assess how policies influenced transmission over time, in particular when reporting is lagged and multiple interventions may have been undertaken in succession. Local SARS-CoV-2 dynamics may differ from state-level patterns, and policy decisions are often implemented to mitigate effects in the areas with the highest case loads. Only looking at state-level responses to policy decisions can blur policy effects as areas with different responses are aggregated.
There remains room to improve these estimators. As mentioned earlier, incorporation of methods to account for under-ascertainment either in the preprocessing or extrapolation phase would improve right-tail estimates. One salient aspect of the ongoing COVID-19 pandemic are day-of-week reporting effects. This source of error can be incorporated into the likelihood model with additional parameters. Additionally, reporting delays can vary by region and change over time. One potential approach for coping with such variation is to jointly model case and hospitalization data and use the relative stability of hospitalization delays to identify changes in the case reporting delay distribution. A joint approach has the potential to more efficiently use all available information, but would be limited to regions where hospitalization data are relatively available. Lastly, the delay distribution is the single-most important hyperparameter for estimating the infection time series.9 This delay may change due to reporting habits, improvements in care, or shifting demographics of the infected population, among other factors.35
Our results suggest that the method chosen to estimate infection counts influences the estimate itself and conclusions drawn. Providing a stable, accurate, and consistent way to estimate infection time series can enable more accurate characterization of real-time transmissibility (i.e., the effective reproductive number) and ultimately may help policy-makers assess the effectiveness of public health interventions at the state and local levels.
Supplementary Material
Footnotes
Supplemental digital content is available through direct URL citations in the HTML and PDF versions of this article (www.epidem.com).
REFERENCES
- 1.Cori A, Ferguson NM, Fraser C, Cauchemez S. A new framework and software to estimate time-varying reproduction numbers during epidemics. Am J Epidemiol. 2013;178:1505–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gostic KM, McGough L, Baskerville EB, et al. Practical considerations for measuring the effective reproductive number, Rt. PLoS Comput Biol. 2020;16:e1008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallinga J, Teunis P. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am J Epidemiol. 2004;160:509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Sullivan F. A statistical perspective on ill-posed inverse problems. Stat Sci. 1986;1:502–518. [Google Scholar]
- 5.Tikhonov AN, Arsenin VY. Solutions of Ill-Posed Problems. Scripta series in mathematics. V.H. Winston and Sons (distributed by Wiley); 1977. [Google Scholar]
- 6.Abbott S, Hellewell J, Thompson RN, et al. Estimating the time-varying reproduction number of sars-cov-2 using national and subnational case counts. Wellcome Open Res. 2020;5:112. [Google Scholar]
- 7.Lewnard JA, Liu VX, Jackson ML, et al. Incidence, clinical outcomes, and transmission dynamics of severe coronavirus disease 2019 in California and Washington: prospective cohort study. BMJ. 2020;369:m1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell TW, Hellewell J, Jarvis CI, et al. Estimating the infection and case fatality ratio for coronavirus disease (COVID-19) using age-adjusted data from the outbreak on the Diamond Princess cruise ship, February 2020. Euro Surveill. 2020;25:2000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bacchetti P, Segal MR, Jewell NP. Backcalculation of HIV infection rates. Stat Sci. 1993;8:82–101. [Google Scholar]
- 10.Becker NG, Watson LF, Carlin JB. A method of non-parametric back-projection and its application to AIDS data. Stat Med. 1991;10:1527–1542. [DOI] [PubMed] [Google Scholar]
- 11.Brookmeyer R. Reconstruction and future trends of the AIDS epidemic in the united states. Science. 1991;253:37–42. [DOI] [PubMed] [Google Scholar]
- 12.Brookmeyer R, Gail MH. A method for obtaining short-term projections and lower bounds on the size of the AIDS epidemic. J Am Stat Assoc. 1988;83:301–308. [Google Scholar]
- 13.Brookmeyer R, Gail MH. Minimum size of the acquired immunodeficiency syndrome (AIDS) epidemic in the United States. Lancet. 1986;328:1320–1322. [DOI] [PubMed] [Google Scholar]
- 14.Liao J, Brookmeyer R. An empirical Bayes approach to smoothing in backcalculation of HIV infection rates. Biometrics. 1995;51:579–588. [PubMed] [Google Scholar]
- 15.Goldstein E, Dushoff J, Ma J, Plotkin JB, Earn DJD, Lipsitch M. Reconstructing influenza incidence by deconvolution of daily mortality time series. Proc Natl Acad Sci U S A. 2009;106:21825–21829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green PJ, Silverman BW. Nonparametric Regression and Generalized Linear Models: a Roughness Penalty Approach. CRC Press; 1993. [Google Scholar]
- 17.Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–723. [Google Scholar]
- 18.Little RJA, Rubin DB. Statistical Analysis with Missing Data, volume 793. John Wiley & Sons; 2019. [Google Scholar]
- 19.Bastos LS, Economou T, Gomes MFC, et al. A modelling approach for correcting reporting delays in disease surveillance data. Stat Med. 2019;38:4363–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Open COVID-19 Data Working Group. Detailed Epidemiological Data from the COVID-19 Outbreak. 2020. Available at: http://virological.org. Accessed 16 June 2020.
- 21.Xu B, Gutierrez B, Mekaru S, et al. Epidemiological data from the COVID-19 outbreak, real-time case information. Sci Data. 2020;7:3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbott S, Hellewell J, Thompson RN, et al. ; CMMID COVID modeling group. Temporal variation in transmission during the COVID-19 outbreak. 2020. Available at: https://epiforecasts.io/covid/. Accessed 01 September 2020.
- 23.Systrom K, Vladeck T. Rt Covid-19. 2020. Avaiable at: https://rt.live. Accessed 16 June 2020.
- 24.Lucy LB. An iterative technique for the rectification of observed distributions. Astron J. 1974;79:745–754. [Google Scholar]
- 25.Richardson WH. Bayesian-based iterative method of image restoration. J Opt Soc Am. 1972;62:55–59. [Google Scholar]
- 26.Yip PSF, Lam KF, Xu Y, et al. Reconstruction of the infection curve for SARS epidemic in Beijing, China using a back-projection method. Commun Stat Simul Comput. 2008;37:425–433. [Google Scholar]
- 27.Efron B. Empirical Bayes deconvolution estimates. Biometrika. 2016;103:1–20. [Google Scholar]
- 28.Anscombe FJ. The transformation of Poisson, binomial and negative-binomial data. Biometrika. 1948;35:246–254. [Google Scholar]
- 29.Höhle M. Surveillance: an R package for the monitoring of infectious diseases. Comput Stat. 2007;22:571–582. [Google Scholar]
- 30.Jewell NP, Lewnard JA, Jewell BL. Caution warranted: using the Institute for Health Metrics and Evaluation model for predicting the course of the COVID-19 pandemic. Ann Intern Med. 2020;173:226–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lauer SA, Grantz KH, Bi Q, et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172:577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Florida Department of Health. Florida COVID19 case line data. 2020. Available at: https://open-fdoh.hub.arcgis.com/datasets/florida-covid19-case-line-data. Accessed 15 July 2020.
- 33.Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. J Am Med Assoc. 2020;323:1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, LItvinova M, Wang W, et al. Evolving epidemiology and transmission dynamics of coronavirus disease 2019 outside Hubei province, China: a descriptive and modelling study. Lancet Infect Dis. 2020;20:793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ali ST, Wang L, Lau EHY, et al. Serial interval of SARS-CoV-2 was shortened over time by nonpharmaceutical interventions. Science. 2020;369:1106–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.