Abstract
Objectives:
To fill this gap in the literature and establish a prediction model.
Methods:
This retrospective study was conducted in a Neonatal Intensive Care Unit between April 2018 and April 2019. Sixty-five sepsis cases were clinically diagnosed with early-onset neonatal sepsis (EONS) and confirmed by bacterial culture, and 91 newborns were enrolled as control. Multivariate analysis was used to analyze the data, and the predictive efficiency of the prediction model was verified using the Hosmer-Lemeshow goodness-of-fit test and receiver operating characteristic (ROC) curve.
Results:
There were differences in multiple gestation birth, cesarean section, gestational age, birth weight, Apgar scores at 1 and 5 minutes, and laboratory parameters including white blood cells, neutrophils, lymphocytes, platelets, C-reactive protein (CRP), procalcitonin (PCT), and interleukin-6 (IL-6) between the groups. Multivariable logistic analysis showed that the Apgar score at 5 minutes and CRP, PCT, and IL-6 levels remained independent significant predictors of EONS (p=0.002, p=0.020, p=0.002, p=0.021, respectively). According to multivariate logistic regression analysis, the risk prediction model was logit p=24.280–3.270 x Apgar score at 5 minutes + 1.396 x CRP + 3.409 x PCT + 0.034 x IL-6 (Lemeshow goodness-of-fit test p=0.143). Receiver operating characteristic curve analysis indicated that the area under the curve was 0.968 (95% confidence interval: 0.936–1.000, p<0.05), and the sensitivity was 90.8% and specificity was 97.8%.
Conclusion:
The Apgar score at 5 minutes and CRP, PCT, and IL-6 levels are risk factors for EONS. The risk prediction model is highly efficient in predicting EONS among preterm infants.
Keywords: bacteria profiles, early-onset neonatal sepsis, preterm newborn, risk factors
Neonatal sepsis, a systemic condition of bacterial, viral, or fungal (yeast) origin, is associated with hemodynamic changes and other clinical manifestations in the first 28 days of life. 1 It is an important cause of neonatal mortality and morbidity, particularly in preterm infants. 1,2 According to the age of onset, neonatal sepsis can be classified as either early-onset neonatal sepsis (EONS) occurring in the first 72 hours of life or late-onset neonatal sepsis occurring on or after postnatal day 4. 1 Statistically, the incidence of EONS ranges from 0.30 to 1.19/1000 live births. 3-7 Early-onset neonatal sepsis is mainly caused by an immature immune system and vertical transmission of infectious organisms from the mother, before or during birth, with nosocomial transmission being rare. 8,9 Historically, group B streptococcus (GBS) is the most common causative agent, while Escherichia coli (E. coli) is the most common cause of mortality. 10
Early detection of EONS is difficult due to clinical manifestations ranging from subtle or nonspecific signs to severe symptoms of focal or systemic disease. 1,10,11 Additionally, clinical signs associated with noninfectious causes and those of sepsis can overlap. 7, 12 Traditionally, the gold standard for confirmation of neonatal sepsis is based on bacterial culture from a normally sterile body fluid, such as, blood or cerebrospinal fluid (CSF). 1 Unfortunately, bacterial culture is hampered by its low sensitivity and long turn-around time because of maternal antibiotic therapy, small volumes of blood samples, and low or intermittent bacteremia. 13,14 The financial burden of caring for and hospitalizing the infected neonates is staggering, estimated at approximately $700 million in the United States. 15
To the best of our knowledge, many articles have recommended accurate monitoring and assessment of the risk factors, hematology profiles, and microbial profiles, which are vital for early detection, reduction of antibiotics abuse, improving prognosis, and lowering mortality in neonatal sepsis. 11,14,16,17 However, in reality, a gradual decline has occurred in the hospital admission rates for full-term newborns, but not for preterm neonates. 18 Despite the hematology profiles for recognizing neonatal sepsis is promising, many evidence shows that none of them in routine clinical setting can constantly diagnose 100% of ENOS. 19 Early-onset neonatal sepsis in preterm infants is prone to confusion with physiological changes, it remains to be seen whether the evaluation method for neonatal sepsis is still applicable. 13,20 Preterm infants have relatively weaker immune systems than full-term newborns, putting them at a higher risk of neonatal sepsis. 21 Nevertheless, there are few reports specifically on bacteria profiles and risk factors for proven EONS in preterm newborns in China. With the widespread implementation of prophylactic antibiotics for mothers, careful surveillance of the changing trend of bacterial organisms among neonates is warranted, especially among premature neonates. 22 In the present study, we retrospectively analyzed the clinical data of 65 cases of EONS, summarized the bacteria profiles and risk factors, which could help physicians identify EONS in preterm newborns early, and established a prediction model.
Methods
This retrospective study was conducted in the Neonatal Intensive Care Unit (NICU) at Shengjing Hospital of China Medical University, Shengjing, China between April 2018 and April 2019, as shown in Figure 1 . All newborns were premature and had a gestational age of less than 37 weeks. The need for informed consent and the ethical approval were waived because of the retrospective study design.
Figure 1.
- Strengthening the reporting of observational studies in epidemiology flow-chart.
Figure 1.
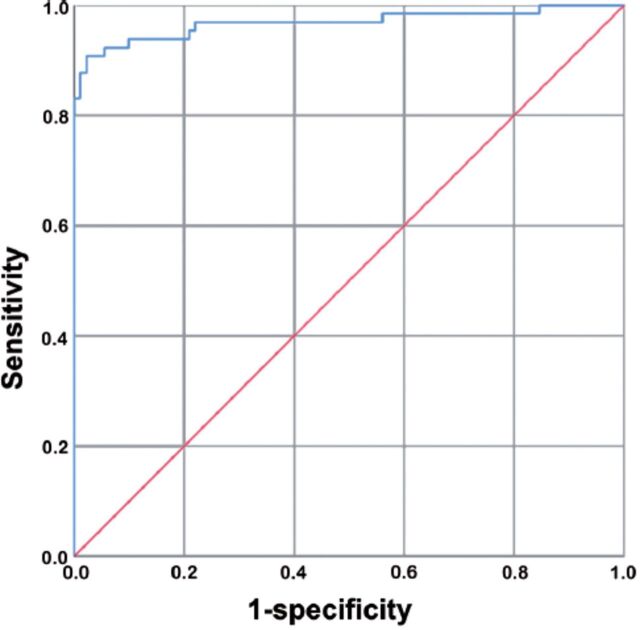
- SThe risk prediction model is able to discriminate preterm neonates with and without early-onset neonatal sepsis (EONS)(area under the curve: 0.968, 95% confidence interval: 0.936-1.000, p<0.05; sensitivity: 90.8%; specificity: 97.8%;). ROC, receiver operating characteristic curve; EONS, early-onset neonatal sepsis.
Our study was in accordance with the guidelines outlined in the Declaration of Helsinki and approved by the local Ethics Committee of Shengjing Hospital of China Medical University. All sepsis cases were consistent with the diagnostic criteria developed by international pediatric specialists in 2005. 23 Proven neonatal sepsis was present if the causative pathogen was isolated from CSF or blood. Neonates with malignancy, congenital malformations, or congenital immune diseases, and those who received immunosuppressive therapy and were treated in other hospitals for more than 3 days were excluded from the study. Patients who fulfilled these inclusion and exclusion criteria within the first 72 hours after delivery were classified as having EONS. Sixty-five sepsis patients were clinically diagnosed as having EONS, and the diagnosis was confirmed by bacterial culture.
Ninety-one newborns were enrolled as the control group. The control group was admitted to the hospital for perinatal conditions other than infection, such as hypoglycemia, very low birth weight, or intrauterine growth restriction. The inclusion criteria were as follows: neonates without signs of clinical chronic or acute infections, neonates without congenital diseases, and neonates who did not receive any antibiotic regimen.
For all neonates, the clinical profiles were collected and included the following data: gender, gestational age, mother’s age, birth weight, Apgar scores at one minute and 5 minutes, manner of delivery (natural delivery or cesarean section), and multiple gestation birth. The laboratory data included white blood cells (WBCs), neutrophils (NEUs), lymphocyte (LYMs), neutrophil-to-lymphocyte ratio (N/L), platelets (PLTs), C-reactive protein (CRP), procalcitonin (PCT), interleukin-6 (IL-6), and reticulocytes (RETs) and bacterial culture results. All blood samples were obtained within the first few hours of the onset of clinical sepsis. For the control group, the laboratory data were obtained between 6 hours and 72 hours after birth, which could reduce the effect of physiological increasing of the values. 24 Cerebrospinal fluid cultures were taken when clinically indicated. Bacterial cultures were not taken from healthy controls.
Whole blood samples were collected in an ethylenediamine tetra acetic acid anticoagulant tube and hematological parameters, such as the WBC, NEU, LYM, PLT, and RET counts, were determined by the Sysmex XN-1000 automated blood cell counter (Sysmex Corporation, Kobe, Japan). Serum concentrations of CRP were measured using nephelometry (Immage 800, Beckman-Coulter, Brea, CA, USA) according to the manufacturer’s instructions. Plasma levels of IL-6 and PCT were determined using the Roche Diagnostics Modular E170 unit (Roche Diagnostics, Mannheim, Germany) as per the manufacturer’s instructions.
Cerebrospinal fluid or 2-3 mL of venous blood was injected into the BACT/ALERT 3D culture bottles (BioMérieux, Marcy-l’Étoile, France) under sterile conditions and cultured with the BACT/ALERT 3D system (BioMérieux). When the culture result was positive, we inoculated the samples in the culture medium, identified by VITEK MS (BioMérieux), and performed antibiotic susceptibility testing using the VITEK-2 compact (BioMérieux); the clinical symptoms were also considered to determine the true or false positive result.
Statistical analysis
The data was performed with IBM SPSS Statistics for Windows, version 26.0 (IBM Corp., Armonk, N.Y., USA). Continuous variables are expressed as median (25th percentile, 75th percentile) and were compared using the non-parametric test (Wilcoxon rank sum test). Categorical variables are expressed as count (percentage) and were analyzed using the Pearson χ2 test. A logistic regression model with a backward-elimination approach was used to perform univariate analysis of all variables that exhibited a significant difference at the level of p<0.10. In the final multivariate logistic analysis, the risk prediction model was established at the same time. The adjusted odds ratio and 95% confidence interval (CI) are presented for each independent risk factor at the level of p<0.05. The Hosmer-Lemeshow goodness-of-fit test was used for model fitting evaluation, and the receiver operating characteristic (ROC) curve analysis was used to evaluate the prediction efficiency of the risk model. The prediction efficiency is expressed by the area under the curve (AUC) and 95% CI. All tests were 2-tailed, and a p-value of <0.05 was considered statistically significant.
Results
In total, 156 preterm infants were enrolled in this study and categorized into the EONS group (n=65) and non-EONS group (n=91). Baseline characteristics of preterm infants with and without EONS are compared in Table 1 . There were differences in multiple gestation birth, cesarean section, gestational age, birth weight, Apgar scores at 1 minute and 5 minutes, and WBCs, NEUs, LYMs, PLTs, CRP, PCT, and IL-6 levels between the 2 groups.
Table 1.
- Characteristics of preterm infants in the EONS and non-EONS groups. a
| Characteristic | EONS | Non-EONS | Statistical value | P-value |
|---|---|---|---|---|
| Gender | ||||
| Male | 36 (55.4) | 50 (54.9) | 0.003 | 0.957 |
| Female | 29 (44.6) | 41 (45.1) | ||
| Multiple gestation birth | ||||
| No | 45 (69.2) | 76 (83.5) | 4.447 | 0.035 |
| Yes | 20 (30.8) | 15 (16.5) | ||
| Natural delivery or cesarean section | ||||
| Natural delivery | 43 (66.2) | 76 (83.5) | 6.318 | 0.012 |
| Cesarean section | 22 (33.8) | 15 (16.5) | ||
| Gestational age, week | 31.6 (29.9, 33.9) | 34 (33.0, 35.1) | -4.959 | <0.001 |
| Age of mother, y | 31 (29, 34) | 31 (28, 34) | -0.197 | 0.884 |
| Birth weight, g | 1640 (1230, 1948) | 2140 (1890, 2420) | -5.932 | <0.001 |
| Apgar score at 1 min | 7 (6, 9) | 10 (9, 10) | -6.945 | <0.001 |
| Apgar score at 5 min | 10 (9, 10) | 10 (10,10) | -6.482 | <0.001 |
| WBC count, ×103/μL | 7.2 (5.1, 11.2) | 10.7 (8.7, 12.8) | -4.095 | <0.001 |
| NEU count, ×103/μL | 3.6 (1.8, 7.6) | 5.1 (3.9, 7.4) | -2.615 | 0.009 |
| LYM count, ×103/μL | 2.1 (1.35, 3.15) | 3.5 (2.7, 4.2) | -5.529 | <0.001 |
| N/L count | 1.8 (1.1, 3.8) | 1.6 (1.1, 2.2) | -1.607 | 0.108 |
| PLT count, ×103/μL | 180.0 (124.5, 231.0) | 250.0 (216.0, 285.0) | -6.005 | <0.001 |
| CRP level, mg/L | 10.4 (3.0, 27.6) | 3.1 (2.0, 3.1) | -5.67 | <0.001 |
| PCT level, ng/ml | 2.5 (0.5, 11.7) | 0.2 (0.1,0.3) | -8.476 | <0.001 |
| IL-6 level, pg/ml | 56.4 (23.0, 720.3) | 11.26 (5.3, 15.2) | -8.405 | <0.001 |
| RET count, ×106/μL | 229 (177, 270) | 242 (200, 280) | -0.727 | 0.467 |
Data are presented as n (%) or median (25th percentile, 75th percentile). EONS: early-onset neonatal sepsis, WBC: white blood cell, NEU: neutrophil, LYM: lymphocyte, N/L: neutrophil-to-lymphocyte ratio, PLT: platelet, CRP: C-reactive protein, PCT: procalcitonin, IL-6: interleukin-6, RET: reticulocyte
Univariate logistic analysis of predictors for EONS in preterm infants is shown in Table 2 . The variables in univariate logistic analysis that met the selection criterion (p<0.1) were entered into the multivariable logistic model for testing of inclusion. In multivariable logistic analysis ( Table 3 ), the Apgar score at 5 minutes and levels of CRP, PCT, and IL-6 remained independent significant predictors of EONS at a significance level of p<0.05 (p=0.002, p=0.020, p=0.002, p=0.021, respectively). We considered this model with 4 predictors as the risk prediction model.
Table 2.
- Results of univariate logistic analysis of predictors for EONS in preterm infants.
| Variable | Odds ratio (95% CI) | β oefficient | P-value |
|---|---|---|---|
| Gender | 0.982 (0.518, 1.863) | -0.018 | 0.957 |
| Multiple gestation birth | 2.252 (1.049, 4.836) | 0.812 | 0.037 |
| Natural delivery or cesarean section | 2.592 (1.218, 5.517) | 0.953 | 0.013 |
| Gestational age | 0.640 (0.535, 0.765) | -0.447 | <0.001 |
| Age of mother | 1.030 (0.951, 1.115) | 0.03 | 0.465 |
| Birth weight | 0.998 (0.997, 0.999) | -0.002 | <0.001 |
| Apgar score at 1 min | 0.397 (0.292, 0.539) | -0.924 | <0.001 |
| Apgar score at 5 min | 0.060 (0.017, 0.210) | -2.807 | <0.001 |
| WBC count | 0.956 (0.897, 1.019) | -0.045 | 0.165 |
| NEU count | 1.016 (0.947, 1.091) | 0.016 | 0.653 |
| LYM count | 0.516 (0.388, 0.687) | -0.662 | <0.001 |
| N/L | 1.334 (1.100, 1.618) | 0.288 | 0.003 |
| PLT count | 0.981 (0.974, 0.988) | -0.019 | <0.001 |
| CRP level | 1.724 (1.236, 2.405) | 0.545 | 0.001 |
| PCT level | 21.619 (5.743, 81.383) | 3.074 | <0.001 |
| IL-6 level | 1.065 (1.032, 1.099) | 0.063 | <0.001 |
| RET count | 0.999 (0.994, 1.003) | -0.001 | 0.532 |
CI: confidence interval, EONS: early-onset neonatal sepsis, BW: birth weight, WBC: white blood cell, NEU: neutrophil, LYM: lymphocyte, N/L: neutrophil-to-lymphocyte ratio, PLT: platelet, CRP: C-reactive protein, PCT: procalcitonin, IL-6: interleukin-6, RET: reticulocyte
Table 3.
- Results of multivariate logistic analysis of variables for predicting EONS in preterm infants.*
| Characteristic | Adjusted OR (95% CI) | β Coefficient | P-value |
|---|---|---|---|
| Apgar score at 5 minutes | 0.038 (0.005, 0.302) | -3.270 | 0.002 |
| CRP level | 4.039 (1.251, 13.033) | 1.396 | 0.020 |
| PCT level | 30.244 (3.349, 273.116) | 3.409 | 0.002 |
| IL-6 level | 1.034 (1.005, 1.064) | 0.034 | 0.021 |
| Intercept | 24.280 | 0.013 |
All variables identified by univariate logistic analysis (p<0.10) were tested using the backward exclusion method. In the multivariable model, the covariate with a p>0.05 was gradually removed separately from the covariate with the highest P-value. To identify other remaining potential confounders, if the effect value of any other predictor changed by 10%, all deleted variables were added separately to the multivariate model and were retained in the model. OR: odd ratio, CI: confidence interval EONS: early-onset neonatal sepsis, PLT: platelet, CRP: C-reactive protein, PCT: procalcitonin, IL-6: interleukin-6
According to multivariate logistic regression analysis, the risk prediction model was logit p=24.280–3.270 x Apgar score at 5 minutes+1.396 x CRP+3.409 x PCT+0.034 x IL-6 (p<0.05, Cox and Snell R2=0.637, Nagelkerke R2=0.857). According to the ROC curve used to evaluate the efficiency of the risk prediction model, the AUC was 0.968, (95% CI: 0.936-1.000, p<0.05), and the sensitivity and specificity were 90.8% and 97.8%, respectively (Figure 2) The result of the Hosmer-Lemeshow goodness-of-fit test used to evaluate the fitting efficiency of the equation was p=0.143, indicating that the risk prediction model was effective.
In the EONS group, 65 strains of pathogenic bacteria were detected, including 17 kinds of bacteria and 2 kinds of fungi. Twenty-one (32.3%) gram-positive bacteria strains were detected, with Staphylococcus epidermidis, Staphylococcus aureus, and Enterococcus faecium being the main strains; 41 (63.1%) gram-negative bacteria strains were detected, mainly Klebsiella pneumoniae (K. pneumoniae) and E. coli. Three strains of fungi (4.6%) were detected, and Candida albicans was the main strain, as shown in Table 4 .
Table 4.
- Distribution of the microbe profiles.
| Pathogen | Number of cases (n=65) | Proportion (%) |
|---|---|---|
| Gram-positive bacterium | ||
| Staphylococcus epidermidis | 3 | (4.6) |
| Staphylococcus aureus | 3 | (4.6) |
| Enterococcus faecium | 3 | (4.6) |
| Listeria monocytogenes | 2 | (3.1) |
| Bradylosis streptococcus | 2 | (3.1) |
| Staphylococcus hemolyticus | 2 | (3.1) |
| Streptococcus agalactiae | 2 | (3.1) |
| Streptococcus vitifolia | 1 | (1.5) |
| Staphylococcus hominis | 1 | (1.5) |
| Staphylococcus capitis | 1 | (1.5) |
| Streptococcus oralis | 1 | (1.5) |
| Gram-negative bacterium | ||
| Klebsiella pneumoniae | 23 | (35.4) |
| Escherichia coli | 14 | (21.5) |
| Enterobacter cloacae | 1 | (1.5) |
| Pseudomonas malodora | 1 | (1.5) |
| Morganella morganii | 1 | (1.5) |
| Ralstonia pili | 1 | (1.5) |
| Fungus | ||
| Candida glabrata | 2 | (3.1) |
| Candida albicans | 1 | (1.5) |
Discussion
To improve outcomes among preterm neonates with EONS, prior detection and appropriate treatment are imperative. The current mainstream diagnostic basis relies on the clinical data of maternal infection, nonspecific clinical signs of the preterm neonate, and laboratory diagnostic indicators. The selection of diagnostic indicators should be based on sensitivity, specificity, and the financial burden of EONS. In our study, with limited clinical data, multiple gestation birth, manner of delivery (natural or cesarean section), gestational age, birth weight, and Apgar score at 1 minute and 5 minutes were noted to be different between the EONS and non-EONS groups. The most common complications of multiple gestation birth were preterm birth, gestational diabetes mellitus, and premature rupture of the membrane, which facilitate the occurrence of EONS in preterm infants. Additionally, neonates from triplet pregnancies were more susceptible to being born preterm, had a lower birth weight, and needed NICU admission more often than those from twin pregnancies. 25 Although there have been reports that cesarean sections can abate the incidence of early sepsis among neonates, there still was a relatively high proportion of such cases in the EONS group. 16 This could be because most of the preterm neonates with EONS had high-risk factors, so their mothers chose cesarean sections to reduce the risk of infection as much as possible. A low gestational age and birth weight indicates the possibility of EONS. 26 Low birth weight preterm neonates have a 3-10 fold higher incidence of sepsis than full-term normal birth weight neonates, and the rate of sepsis in neonates with preterm birth at 29-33 weeks’ gestation is 11 times higher than that in full-term babies. 1 Without a doubt, preterm neonates with a low gestational age and weight at birth have relatively weaker immaturity immune systems. 21 Low Apgar scores were a high-risk factor of acquiring neonatal sepsis. The Apgar score at 5 minutes was associated with an increased risk for EONS in our multivariable logistic analysis. 27
For laboratory diagnosis, we chose WBC, NEU, LYM, N/L, PLT, CRP, PCT, IL-6, and RET as the hematology profile parameters, which constitute readily available data in hospitals and are less cost-intensive to measure. The WBC, NEU, LYM, PLT, CRP, PCT, and IL-6 values were distinct between the 2 study groups. However, these parameters may lose instant diagnostic value due to their physiological elevation in preterm neonates with EONS. Routine blood examination findings are the most basic clinical biomarkers for sepsis. 28 The WBC, NEU, and N/L values are screening laboratory indexes for diagnosis of sepsis; however, the range of these indicators vary widely within the first 72 hours in preterm infants preventing a definitive diagnosis. Further, the most commonly used biomarkers are the CRP, PCT, and IL-6 values; however, all of these parameters have shown varied sensitivity and specificity in different studies, confounding the doctor’s judgment of the diagnosis. 19,20 The CRP level shows a nonspecific physiological 3-day increase due to the stress of delivery and some other noninfectious perinatal and maternal factors making it unsuitable for the diagnosis EONS. 13 During the perinatal period, the PCT level is also affected by stress and physiological factors, which makes the PCT level less valuable within the first 12 hours after delivery. A combination of the CRP level with the PCT and IL-6 levels could increase the sensitivity of an EONS prediction model. 19,29 Similarly, our data showed that the CRP, PCT, and IL-6 levels in the risk prediction model were instrumental for the prediction efficacy. Finally, as predicted by our multivariable logistic analysis, the Apgar score at 5 minutes and CRP, PCT, and IL-6 levels remained independent significant predictors of EONS. Both the Hosmer-Lemeshow goodness-of-fit test and ROC curve analysis showed that the risk prediction model is effective. The risk prediction model suggests that pediatricians should combine CRP, PCT, and IL-6 which are easy to obtain, plus the Apgar score at 5 minutes to identify high-risk patients to begin intervention in time. Clinicians can minimize iatrogenic injury by using our risk predictive model to jointly test high performance indicators through one sample.
Because of the prophylactic use of antibiotics (intrapartum and postpartum), an improved understanding of the changing patterns of the population-level incidence of EONS and causative pathogen spectrum may be helpful in timely recognizing EONS and improving the prognosis in such patients. 7 In our study, 65 strains of causative bacteria were cultured, mainly gram-negative bacteria such as K. pneumoniae and E. coli rather than GBS. This result is consistent with that of a survey of EONS among preterm infants in China from 2015 to 2018. 30 Similarly, many systematic reviews and meta-anlysis concluded that K. pneumoniae, E. coli, and Staphylococcus aureus were the major pathogen of neonatal infection in middle-income countries. 14,31 It is suspected that gram-negative bacteria infection is strongly associated with suboptimal sterilization, unhygienic practices, vertical transmission from maternal colonization, or very early horizontal transmission in the delivery room. 30,32 Further investigation is necessary to determine the cause of these infections.
The rational administration of antibiotics can be beneficial to shorten the hospitalization period and avoid subsequent nosocomial infection. Indeed, hematology profiles and blood cultures are indispensable, especially dynamic monitoring of these indexes. White blood cells and NEU counts change dynamically during the first 12 hours after birth, and thus, continuous surveillance over 24 hours may be more informative than a single assessment. 33 Continuous PCT testing can shorten the duration of antibiotic therapy. 34 However, physicians should be aware that frequent medical procedures for obtaining blood samples can cause life-threatening complications, such as catheter-related bloodstream infections and iatrogenic anemia. 30
Study limitaitions
Our study fills the gap in the literature of bacteria profiles and risk factors for proven EONS in preterm newborns in China. This study still had some limitations. This study had a retrospective design, small sample size from a single institution, and limited clinical data that did not include parameters such as the mother’s inflammatory index and signs of labor. Thus, it is still necessary to further confirm whether the established risk prediction model has excellent predictive value for EONS in future large-sample multi-center studies.
In conclusions, the Apgar score at 5 minutes and CRP, PCT, and IL-6 levels are risk factors for EONS in preterm neonates. The risk prediction model established can effectively predict the risk of EONS and guide pediatricians in the prevention and treatment of EONS. Microbial profiles can also provide a hint on the optimal use of antibiotics.
Acknowledgment
The authors gratefully acknowledge Editage (www.editage.com) for English language editing.
Footnotes
References
- 1. Shane AL, Sánchez PJ, Stoll BJ.. Neonatal sepsis. Lancet 2017; 390: 1770–1780. [DOI] [PubMed] [Google Scholar]
- 2. Seale AC, Obiero CW, Berkley JA.. Rational development of guidelines for management of neonatal sepsis in developing countries. Curr Opin Infect Dis 2015; 28: 225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weston EJ, Pondo T, Lewis MM, Martell-Cleary P, Morin C, Jewell B, et al. The burden of invasive early-onset neonatal sepsis in the United States, 2005-2008. Pediatr Infect Dis J 2011; 30: 937–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Puopolo KM, Draper D, Wi S, Newman TB, Zupancic J, Lieberman E, et al. Estimating the probability of neonatal early-onset infection on the basis of maternal risk factors. Pediatrics 2011; 128: e1155–e1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stoll BJ, Hansen NI, Sánchez PJ, Faix RG, Poindexter BB, Van Meurs KP, et al. Early onset neonatal sepsis: the burden of group B Streptococcal and E. coli disease continues. Pediatrics 2011; 127: 817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kuhn P, Dheu C, Bolender C, Chognot D, Keller L, Demil H, et al. Incidence and distribution of pathogens in early-onset neonatal sepsis in the era of antenatal antibiotics. Paediatr Perinat Epidemiol 2010; 24: 479–487. [DOI] [PubMed] [Google Scholar]
- 7. Kuzniewicz MW, Puopolo KM, Fischer A, Walsh EM, Li S, Newman TB, et al. A quantitative, risk-based approach to the management of neonatal early-onset sepsis. JAMA Pediatr 2017; 171: 365–371. [DOI] [PubMed] [Google Scholar]
- 8. Good PI, Hooven TA.. Evaluating newborns at risk for early-onset sepsis. Pediatr Clin North Am 2019; 66: 321–331. [DOI] [PubMed] [Google Scholar]
- 9. Sgro M, Kobylianskii A, Yudin MH, Tran D, Diamandakos J, Sgro J, et al. Population-based study of early-onset neonatal sepsis in Canada. Paediatr Child Health 2019; 24: e66–e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simonsen KA, Anderson-Berry AL, Delair SF, Davies HD.. Early-onset neonatal sepsis. Clin Microbiol Rev 2014; 27: 21–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Benitz WE, Wynn JL, Polin RA.. Reappraisal of guidelines for management of neonates with suspected early-onset sepsis. The J Pediatr 2015; 166: 1070–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chauhan N, Tiwari S, Jain U.. Potential biomarkers for effective screening of neonatal sepsis infections: An overview. Microb Pathog 2017; 107: 234–242. [DOI] [PubMed] [Google Scholar]
- 13. Delanghe JR, Speeckaert MM.. Translational research and biomarkers in neonatal sepsis. Clin Chim Acta 2015; 451: 46–64. [DOI] [PubMed] [Google Scholar]
- 14. Amare D, Mela M, Dessie G.. Unfinished agenda of the neonates in developing countries: magnitude of neonatal sepsis: systematic review and meta-analysis. Heliyon 2019; 5: e02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yorita KL, Holman RC, Sejvar JJ, Steiner CA, Schonberger LB.. Infectious disease hospitalizations among infants in the United States. Pediatrics 2008; 121: 244–252. [DOI] [PubMed] [Google Scholar]
- 16. Procianoy RS, Silveira RC.. The challenges of neonatal sepsis management. Jornal de Pediatria 2020; 96: 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rafi MA, Miah MMZ, Wadood MA, Hossain MG.. Risk factors and etiology of neonatal sepsis after hospital delivery: A case-control study in a tertiary care hospital of Rajshahi, Bangladesh. PloS one 2020; 15: e0242275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lukacs SL, Schrag SJ.. Clinical sepsis in neonates and young infants, United States, 1988-2006. J Pediatr 2012; 160: 960–965.e1. [DOI] [PubMed] [Google Scholar]
- 19. Memar MY, Alizadeh N, Varshochi M, Kafil HS.. Immunologic biomarkers for diagnostic of early-onset neonatal sepsis. J Matern Fetal Neonatal Med 2019; 32: 143–153. [DOI] [PubMed] [Google Scholar]
- 20. Sharma D, Farahbakhsh N, Shastri S, Sharma P.. Biomarkers for diagnosis of neonatal sepsis: a literature review. J Matern Fetal Neonatal Med 2018; 31: 1646–1659. [DOI] [PubMed] [Google Scholar]
- 21. Collins A, Weitkamp JH, Wynn JL.. Why are preterm newborns at increased risk of infection? Arch Dis Child Fetal Neonatal Ed 2018; 103: F391–f394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sgro M, Shah PS, Campbell D, Tenuta A, Shivananda S, Lee SK.. Early-onset neonatal sepsis: rate and organism pattern between 2003 and 2008. J Perinatol 2011; 31: 794–798. [DOI] [PubMed] [Google Scholar]
- 23. Goldstein B, Giroir B, Randolph A.. International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med 2005; 6: 2–8. [DOI] [PubMed] [Google Scholar]
- 24. Subspecialty Group of Neonatology, the Society of Pediatric, Chinese Medical Association; Professional Committee of Infectious Diseases, Neonatology Society, Chinese Medical Doctor Association. [Expert consensus on the diagnosis and management of neonatal sepsis (version 2019)]. Zhonghua Er Ke Za Zhi 2019; 57: 252–257 (Article in Chinese). [DOI] [PubMed] [Google Scholar]
- 25. AlBasri SF, Shouib GM, Bajouh OS, Nasrat HA, Ahmad E, AlGreisi FM.. Maternal and neonatal outcomes in twin and triplet gestations in Western Saudi Arabia. Saudi Med J 2017; 38: 657–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stoll BJ, Puopolo KM, Hansen NI, Sánchez PJ, Bell EF, Carlo WA, et al. Early-onset neonatal sepsis 2015 to 2017, the Rise of Escherichia coli, and the Need for Novel Prevention Strategies. JAMA Pediatr 2020; 174: e200593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sorsa A. Epidemiology of neonatal sepsis and associated factors implicated: observational study at neonatal intensive care unit of Arsi University Teaching and Referral Hospital, South East Ethiopia. Ethiop J Health Sci 2019; 29: 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu C, Zhang Y, Shang Y, Fang C, He Q, Xie L.. Clinical values of common biomarkers for efficacy monitoring of antibiotics in early-onset neonatal sepsis. Transl Pediatr 2020; 9: 669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hofer N, Zacharias E, Müller W, Resch B.. An update on the use of C-reactive protein in early-onset neonatal sepsis: current insights and new tasks. Neonatology 2012; 102: 25–36. [DOI] [PubMed] [Google Scholar]
- 30. Jiang S, Hong L, Gai J, Shi J, Yang Y, Lee SK, et al. Early-onset sepsis among preterm neonates in China, 2015 to 2018. Pediatr Infect Dis J 2019; 38: 1236–1241. [DOI] [PubMed] [Google Scholar]
- 31. Zaidi AK, Thaver D, Ali SA, Khan TA.. Pathogens associated with sepsis in newborns and young infants in developing countries. Pediatr Infect Dis J 2009; 28: S10–S18. [DOI] [PubMed] [Google Scholar]
- 32. Investigators of the Delhi Neonatal Infection Study (DeNIS) collaboration. Characterisation and antimicrobial resistance of sepsis pathogens in neonates born in tertiary care centres in Delhi, India: a cohort study. Lancet 2016; 4: E752–E760. [DOI] [PubMed] [Google Scholar]
- 33. Mikhael M, Brown LS, Rosenfeld CR.. Serial neutrophil values facilitate predicting the absence of neonatal early-onset sepsis. J Pediatr 2014; 164: 522–528.e1-3. [DOI] [PubMed] [Google Scholar]
- 34. Stocker M, Fontana M, El Helou S, Wegscheider K, Berger TM.. Use of procalcitonin-guided decision-making to shorten antibiotic therapy in suspected neonatal early-onset sepsis: prospective randomized intervention trial. Neonatology 2010; 97: 165–174. [DOI] [PubMed] [Google Scholar]