Abstract
The microbial capacity to degrade simple organic compounds with quaternary carbon atoms was demonstrated by enrichment and isolation of five denitrifying strains on dimethylmalonate as the sole electron donor and carbon source. Quantitative growth experiments showed a complete mineralization of dimethylmalonate. According to phylogenetic analysis of the complete 16S rRNA genes, two strains isolated from activated sewage sludge were related to the genus Paracoccus within the α-Proteobacteria (98.0 and 98.2% 16S rRNA gene similarity to Paracoccus denitrificansT), and three strains isolated from freshwater ditches were affiliated with the β-Proteobacteria (97.4 and 98.3% 16S rRNA gene similarity to Herbaspirillum seropedicaeT and Acidovorax facilisT, respectively). Most-probable-number determinations for denitrifying populations in sewage sludge yielded 4.6 × 104 dimethylmalonate-utilizing cells ml−1, representing up to 0.4% of the total culturable nitrate-reducing population.
Quaternary carbon atoms bind with all four single ς bonds to carbon atoms. This structural motif is found in many isoprenoic compounds, e.g., the monoterpenes pinene and carene, cholesterol, and carotene. The biosynthetic pathway involves an irreversible cationic polymerization of alkene bonds to newly formed carbon-carbon single bonds. As a result, quaternary carbon atoms are formed from tertiary carbon atoms (9).
The microbial degradation of compounds with quaternary C atoms has not been studied thoroughly. Already-discovered mineralization pathways in aerobic microorganisms often involve molecular oxygen as a cosubstrate. Camphor and eucalyptol feature a ketone in an α position with respect to a quaternary C-atom. This functional group enables a biological Baeyer-Villiger oxygenation to a lactone that yields, after hydrolysis, a tertiary alcohol (31). A second pathway is provided by Candida albicans sterol 14α-demethylase, a cytochrome P450 monooxygenase. This enzyme catalyzes the oxidation of a methyl group adjacent to a quaternary C atom to an aldehyde that is oxidized in a third monooxygenation reaction and is eliminated as formate. Thus, the quaternary C atom is changed via radical intermediates into a tertiary C atom with an alkene bond (27). This aldehyde lyase reaction was first described for aromatase, which catalyzes the C-19 removal of androgens, leading to the formation of estrogens (2, 32). Another pathway present in aerobic bacteria involves an oxygenation of the C-9 atom of cholesterol. The 9α-hydroxy-androsta-1,4-dien-2,17-dione formed is labile and reacts nonenzymatically to produce 9,10-seco-androsta-1,3,5(10)-trien-3-ol-9,17-dione. The spontaneous carbon-carbon cleavage obliterates a quaternary C atom and a tertiary alcohol (16).
One example of an oxygen-independent pathway is the cleavage of α-pinene oxide by a lyase. Three rings are opened in an isomerization reaction, and at the same time a quaternary C atom is transformed into a tertiary C atom (11). This rather unusual intramolecular depolymerization reaction can be perceived as the reverse of the biosynthetic pathway.
Research on the fate of quaternary carbon atoms in anoxic habitats advanced with the recent report of denitrifying β-Proteobacterium strain 72Chol, that anaerobically mineralizes cholesterol (14). In a theoretical analysis, we developed a plausible pathway for the degradation of cholesterol via carboxy-methylmalonates as possible intermediates (17). Hence, dimethylmalonate was chosen for enrichment and isolation of denitrifying bacteria to verify the existence of the physiological capacity to mineralize a small compound with a quaternary carbon atom in the absence of molecular oxygen.
MATERIALS AND METHODS
Sources of organisms.
Enrichment cultures were inoculated with activated sludge obtained from a local wastewater treatment plant (Lintel, Osterholz-Scharmbeck, Germany) or with a water-mud mixture obtained from freshwater ditches located in Bremen, Germany. Most-probable-number (MPN) counts were performed with the activated sludge. Alcaligenes defragrans strains (8), Azoarcus sp. strain 22Lin (12), and Thauera linaloolentis and Thauera terpenica strains (7) were maintained in our laboratory since their isolation.
Media and culture conditions.
Anoxic media and cultivation techniques were used in this study (33). The basic medium contained (per liter of distilled water) 0.5 g of MgSO4 · 7H2O, 0.5 g of NH4Cl, 0.5 g of KH2PO4, 0.1 g of CaCl2, and 0.85 g of NaNO3 (10 mM). After autoclaving, 2 ml of a chelated trace element mixture (1 liter of distilled water contained 2,100 mg of FeSO4 · 7H2O, 30 mg of H3BO3, 100 mg of MnCl2 · 4H2O, 190 mg of CoCl2 · 6H2O, 24 mg of NiCl2 · 6H2O, 29 mg of CuSO4 · 5H2O, 144 mg of ZnSO4 · 7H2O, 36 mg of NaMoO4 · 7H2O, and 5.2 g of EDTA, pH 6.5), 2 ml of selenite-tungstate solution (0.4 g of NaOH liter−1, 8 mg of Na2WO4 · 2H2O liter−1, and 6 mg of Na2SeO3 · 5H2O liter−1) (33), 1 ml of vitamin solution (4 mg of 4-aminobenzoic acid, 2 mg of d-(+)-biotin, 10 mg of nicotinic acid, 5 mg of calcium d-(+)-pantothenate, 15 mg of pyridoxin hydrochloride, 4 mg of folic acid, and 1 mg of lipoic acid in 100 ml of 10 mM NaH2PO4, pH 7.1), 1 ml of cyanocobalamin solution (50 mg liter−1), 1 ml of thiamine solution (10 mg of thiamine hydrochloride in 100 ml of 25 mM NaH2PO4, pH 3.4), 1 ml of riboflavin solution (2.5 mg in 100 ml of 25 mM NaH2PO4, pH 3.2) (1), and 50 ml of NaHCO3 solution (1 M) were added, and the pH was adjusted to 7.2. N2-CO2 (90:10, vol/vol) was used as the gas phase for all anoxic cultures. Electron donors were added from sterile stock solutions prior to inoculation. All incubations were performed at 28°C in the dark.
Enrichment and isolation of denitrifying bacteria on dimethylmalonate.
Enrichment cultures contained 330 ml of medium with 5 mM dimethylmalonate and 20 ml of sewage sludge or 250 ml of medium and 100 ml of a water-mud mixture in 0.5-liter bottles that were sealed with thick butyl rubber stoppers. Control enrichments were prepared in parallel without dimethylmalonate to account for endogeneous carbon sources of the inoculate. Overpressure due to gas formation and the amounts of nitrate and nitrite in the culture were determined regularly, and 10 mM nitrate was added when the electron acceptor was depleted. Transfer of the enrichment cultures involved an anaerobically performed serial dilution prior to inoculation to obtain an inoculation size of 1.0 × 10−6 (vol/vol) for a medium volume of 150 ml. After three passages, bacteria were isolated via repeated dilution in agar with 5 mM dimethylmalonate and 10 mM nitrate (33). Isolated colonies were pure strains according to microscopic observations of cultures grown on AC broth (Difco, Detroit, Mich.), yeast extract (0.5 g liter−1), glucose (5 mM), or pyruvate (10 mM) under fermenting or denitrifying conditions and according to the formation of homogenous colonies on oxic agar plates containing either AC broth or dimethylmalonate.
Strain maintenance and growth tests.
Isolated strains were kept in culture tubes (21 ml) with 15 ml of anoxic medium under selective growth conditions (5 mM dimethylmalonate and 10 mM nitrate). Transfer of an inoculum of 5% (vol/vol) into freshly prepared culture medium was done every second month. Grown cultures were transferred to a refrigerator and kept at 8°C until the next transfer. Growth experiments were performed in duplicate with culture tubes with an inoculum of 2% (vol/vol). Growth was monitored by turbidimetry at 660 nm, and nitrate and nitrite were analyzed in the late stationary phase. Control experiments demonstrated that the carbon sources present in the vitamin solutions and the inoculate did not support observable microbial growth.
The quantification of dimethylmalonate degradation was done by inoculating a 1- or 5-ml portion of a recently grown culture into 200 ml of medium in a 250-ml Erlenmeyer flask with a depressed sidearm for turbidimetric determinations and a threaded neck to hold the butyl rubber stopper with a screw cap against the overpressure that is build up by denitrification. The culture was regularly sampled with sterile, nitrogen-flushed syringes for analyses of organic acids, nitrite, and nitrate. Nitrogen formation was measured in cultures containing an He-CO2 (90:10, vol/vol) atmosphere. Formation of carbon dioxide was assayed in cultures that were made without bicarbonate and carbon dioxide and were buffered by potassium phosphate (20 mM, pH 7.0).
Denitrifying growth on dimethylmalonate occurred not only in oxygen-free media but also in chemically reduced media. Anoxic media were prereduced in control experiments with 4 mM ascorbate. The strains did not utilize ascorbate as a carbon and energy source. Thus, ascorbate could routinely be added to ensure anoxic conditions in the culture.
Enumeration of dimethylmalonate-utilizing denitrifying bacteria.
The size of the denitrifying population in sewage sludge was estimated with MPN dilutions in liquid medium (3). MPN counts were performed with the medium described above and either 5 mM dimethylmalonate or a mixture of fatty acids (acetate, propionate, and butyrate [2 mM each compound] and succinate, valerate, isovalerate, α-methylbutyrate, and isobutyrate [50 μM each compound]) as the sole electron donor. A 10-fold dilution was used, with three portions per dilution. The MPN culture tubes were incubated in the dark at 20°C for 18 weeks.
Chemical analyses.
Biomass formation was measured turbidimetrically at 660 nm. Cell dry weight determinations were performed as described previously (12). Qualitative measurements of nitrate consumption were performed with an indicator strip (Merck, Darmstadt, Germany). Nitrate and nitrite were determined quantitatively by high-pressure liquid chromatography (13), and gases (dinitrogen oxide, dinitrogen, and carbon dioxide) were determined by packed-column gas chromatography as described previously (14). Ammonium was measured photometrically by the indophenol method (14). Organic acids were quantified by ion exclusion chromatography on a WA1 column (7.8 by 300 mm; Sarasep, San Jose, Calif.) with a high-pressure liquid chromatography system (Sykam, Garching, Germany) equipped with a UV detector (Linear Instruments, Fremont, Calif.). Culture samples of 950 μl were acidified with 50 μl of 1 M H2SO4 and clarified by centrifugation. Subsamples of 50 μl were separated with 5 mM H2SO4 as the liquid phase at a flow rate of 0.6 ml min−1 at 35°C. Organic acids were detected at 210 nm. Net retention times for dimethylmalonate, acetate, and propionate were 5.09, 9.23, and 11.97 min, respectively.
Cells were observed with a standard phase-contrast microscope (Zeiss, Oberkochen, Germany) with an oil immersion objective (100/1.6). Dense cultures were wet mounted on glass slides coated with 2% washed agar (26) and photographed by using Ortho 25 film (Agfa-Gevaert, Leverkusen, Germany).
16S rRNA gene sequence and data analysis.
In vitro amplification of the 16S rRNA gene and direct sequencing were performed as previously described (29). Sequencing reactions were performed with a Taq Dyedeoxy Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, Calif.) and were run on an Applied Biosystems 373S DNA sequencer. The obtained sequences were added to an alignment of about 5,300 homologous bacterial 16S rRNA primary structures (22), applying the aligning tool of the ARB program package (30). Phylogenetic trees were constructed by using subsets of data that included sequences from outgroup reference organisms as well as representative and genealogically related sequences of members of the alpha and beta subclasses of Proteobacteria (22). Topologies were evaluated by using distance matrix, maximum-parsimony, and maximum-likelihood methods as implemented in the ARB software to elaborate a consensus tree (21).
Whole-cell in situ hybridization.
Microbes that were grown in highly diluted MPN tubes were analyzed by whole-cell in situ hybridization for the presence of α- and β-Proteobacteria as described by Manz et al. (23). Probes EUB338 (23) and ALF968 (24) were used to detect Eubacteria and α-Proteobacteria, respectively. For β-Proteobacteria, CY3-labelled probe BET42a was used with a nonlabelled competitor probe, GAM42a (23).
Nucleotide sequences accession numbers.
The sequences determined in this work have been deposited under EMBL accession numbers AJ012067 to AJ012071.
RESULTS
Enrichment and isolation.
The capacity of nitrate-reducing microorganisms to degrade dimethylmalonate was investigated with activated sludge and a water-mud mixture from freshwater ditches. A small inoculum of the former was used to reduce the amount of endogenous electron donors. The enrichment containing 5 mM dimethylmalonate and 10 mM nitrate consumed all nitrate within 9 days and formed 30 ml of gas. After nitrate addition, the gas formation continued to a total volume of 47 ml on day 16. A control experiment without dimethylmalonate produced only 8 ml of gas in this time. The freshwater enrichment contained a significant amount of endogenous electron donors for nitrate reduction. The control consumed nearly 30 mM nitrate in 20 days and formed 89 ml of gas. Gas formation in the enrichment culture started to exceed that in the control on day 14 and accumulated to 116 ml on day 20, when 40 mM nitrate was consumed. Small inocula of these enrichment cultures (150 nl obtained by serial dilution) were transferred, resulting in a dilution of 1 ppm to select for bacteria that were probably present in larger amounts in the enrichment. Two additional passages with small inocula (1 ppm) ensured a selection for bacteria that grew well on dimethylmalonate. Isolation by agar dilution series was then successfully attempted. Two strains, B8B1 and B8B2, originated from activated sludge, and three strains, G7A1, G8A1, and G8B1, originated from the freshwater ditches.
Characterization of the isolated strains.
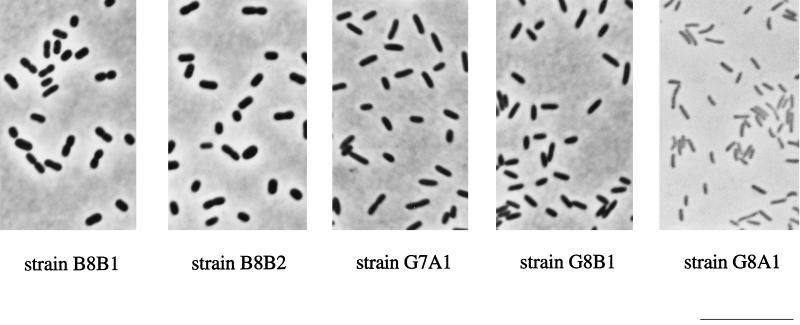
Strains B8B1 and B8B2 were nonmotile spherical cells or short rods with dimensions of 0.7 to 1.0 by 1.0 to 2.0 μm, resembling Paracoccus. Strains G7A1 and G8B1 were Pseudomonas-like motile straight rods with sizes of 0.5 to 0.7 by 1.5 to 2.5 μm. The motile strain G8A1 was a small spirillum, 0.3 to 0.5 by 1.0 to 2.0 μm in size (Fig. 1). The microorganisms grew on several organic acids, including isobutyrate. Strains G7A1 and G8B1 did not utilize methylsuccinate. Sugars were utilized only by strains B8B1 and B8B2 (Table 1).
FIG. 1.
Phase-contrast photomicrographs of new denitrifying isolates grown with dimethylmalonate (5 mM) and nitrate (10 mM). Bar, 10 μm.
TABLE 1.
Substrates for denitrifying growth of the isolated strains
| Electron donor (concn, mM) | Growtha of:
|
||||
|---|---|---|---|---|---|
| Paracoccus sp. strain B8B1 | Paracoccus sp. strain B8B2 | Acidovorax sp. strain G7A1 | Acidovorax sp. strain G8B1 | Herbaspirillum sp. strain G8A1 | |
| Acetate (10) | +++ | +++ | ++ | ++ | ++ |
| Propionate (6) | +++ | +++ | ++ | ++ | ++ |
| Butyrate (4) | +++ | +++ | ++ | ++ | ++ |
| Isobutyrate (4) | +++ | +++ | ++ | ++ | ++ |
| 2,2-Dimethylpropionate (5) | − | − | − | − | − |
| Dimethylmalonate (5) | +++ | +++ | ++ | ++ | + |
| Pyruvate (10) | + | + | + | + | ++ |
| Succinate (6) | +++ | +++ | ++ | ++ | ++ |
| Methylsuccinate (5) | +++ | +++ | − | − | ++ |
| Benzoate (2) | − | − | − | − | − |
| l-Ascorbate (10) | − | − | − | − | − |
| d-Gluconate (5) | +++ | +++ | − | − | − |
| d-Glucose (5) | +++ | +++ | − | − | − |
| d-Fructose (5) | +++ | +++ | − | − | − |
| d-Galactose (5) | +++ | +++ | − | − | − |
| d-Ribose (5) | + | + | − | − | − |
| Inositol (5) | +++ | +++ | − | − | − |
| Methanol (10) | − | − | − | − | − |
| l-Glutamate (5) | +++ | +++ | ++ | ++ | + |
| l-Arginine (5) | − | − | − | − | − |
| l-Valine (5) | ++ | ++ | − | − | − |
| l-Cysteine (5) | ++ | ++ | +++ | +++ | ++ |
| l-Phenylalanine (5) | − | − | − | − | − |
| l-Isoleucine (5) | +++ | +++ | + | ++ | + |
Bacterial growth was determined as the increase in OD at 660 nm, as follows: −, <0.030; +, >0.070; ++, >0.200, +++, >0.400.
Carbon limitation was achieved with 2 mM dimethylmalonate in the presence of 10 mM nitrate. Grown cultures of all strains contained nitrate and dinitrogen oxide but no nitrite. Nitrate-limited cultures were obtained with 5 mM dimethylmalonate and 10 mM nitrate. Nitrate and nitrite were depleted under these conditions. Nitrogen formation was observed in cultures with a helium atmosphere. Ammonium present in the medium was assimilated during denitrifying growth, excluding ammonification as a catabolic process.
Different biomass yields were observed for the strains in carbon-limited cultures and in nitrate-limited cultures; i.e., strains B8B1 and B8B2 produced an optical density (OD) at 660 nm of 0.6 on 10 mM nitrate, whereas strains G7A1, G8A1, and G8B1 produced an OD of only 0.3. Koch showed that, in the absence of pigments, cells with a volume range of 0.4 to 2 μm3 had an average dry weight/OD ratio of 371 mg (dry weight) liter−1 OD at 660 nm−1 (18). To account for the smaller size of strain G8A1 cells, we determined the dry weight formed in nitrate-limited denitrifying cultures: strain B8B2 formed 21.4 g (dry weight) mol of nitrate−1, whereas strains G8A1 and G8B1 formed only 10.3 and 10.8 g (dry weight) mol of nitrate−1, respectively.
All strains grew aerobically on nitrate-free agar plates with dimethylmalonate.
Quantification of denitrifying growth on dimethylmalonate.
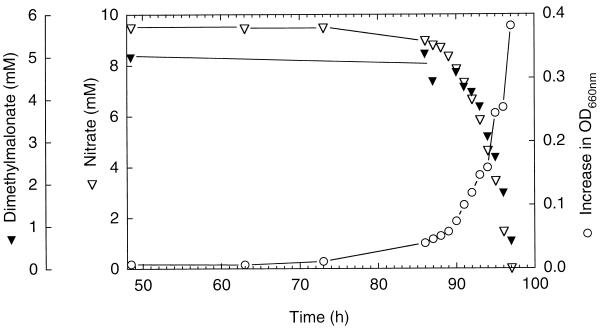
Strain B8B2 was used to determine quantitatively dimethylmalonate and nitrate consumption (Fig. 2). A biomass of 32.6 mg was formed based on a measured correlation of 427.6 mg (dry mass) liter−1 to 1 liter of culture with an OD of 1.0. According to the assimilation equation 17C5H8O4 + 2H2O → 20C4H7O3 + 5CO2, this corresponds to a consumption of 269 μmol of dimethylmalonate. Since 870 μmol of dimethylmalonate disappeared during growth, complete mineralization of the calculated amount of dissimilated dimethylmalonate (601 μmol) would provide a total of 12 mmol of electrons for denitrification. Ten millimoles of electrons is required to account for a reduction of nitrate (2 mmol consumed) to dinitrogen. In separate experiments, the nitrogen and carbon balances were analyzed. In nitrate-limited cultures grown under a helium atmosphere, nitrate consumption of 2.56 mmol of nitrogen atoms occurred with the synthesis of 160 μmol of dinitrogen oxide N and 2.13 mmol of nitrogen N. Carbon dioxide formation correlated with the amount of dimethylmalonate supplied. In grown carbon-limited cultures (2 mmol of C as dimethylmalonate), we detected 1.35 mmol of carbon dioxide; the increase in biomass represented, according to the aforementioned equation, an assimilation of 776 μmol of C. Thus, the electron, nitrogen, and carbon balances support within experimental uncertainties the complete mineralization of dimethylmalonate according to the catabolic reaction C5H8O4 + 4H+ + 4NO3− → 5CO2 + 2N2 + 6H2O.
FIG. 2.
Growth of strain B8B2 on dimethylmalonate and nitrate.
Phylogenetic affiliations of the isolated strains.
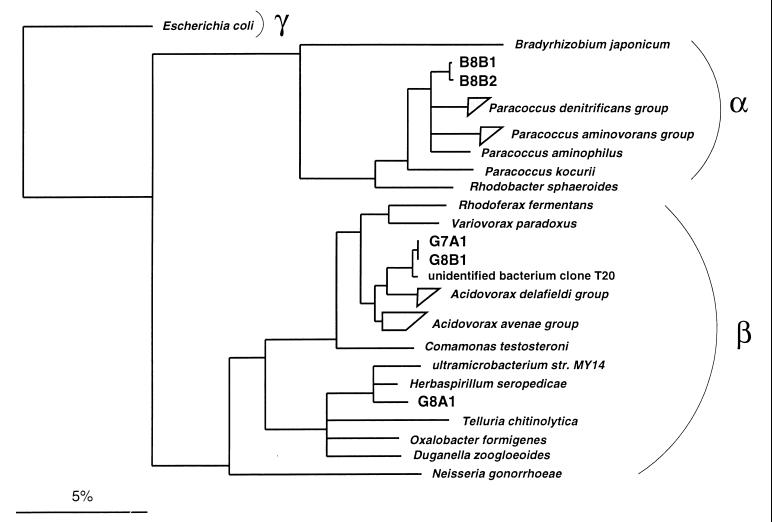
Almost-complete 16S rRNA gene sequences from the isolates were obtained by in vitro amplification and direct sequencing. The sequences of strains B8B1 and B8B2 were nearly identical, with a similarity of 99.8%, and strains G7A1 and G8B1 had identical sequences. Sequence analysis showed that both activated-sludge strains B8B1 and B8B2 were affiliated with the genus Paracoccus of the α-Proteobacteria (Fig. 3). Their closest relative was P. denitrificans, with a sequence similarity of 98.0%. Similarities of this strains to one group of Paracoccus species (P. alcaliphilus, P. aminophilus, P. aminovorans, P. thiocyanatus, and P. versutus) ranged from 96.8 to 97.5%, and those to a second group of Paracoccus species (P. alkenifer, P. marcusii, P. solventivorans, and P. kocurii) ranged from 94.9 to 95.4%. In contrast, all three ditch isolates were affiliated with the β-Proteobacteria. Strains G7A1 and G8B1 were affiliated with the genus Acidovorax, having 98.3% sequence similarity with the type strains of Acidovorax facilis. The closest relative to both ditch isolates, with a similarity of 99.3%, was the activated-sludge clone sequence T20 (28). Strain G8A1 was affiliated with the same branch as Herbaspirillum seropedicae (97.4% similarity) and a soil ultramicrobacterium, strain MY14, (97.0% similarity) within the Oxalobacter-Telluria-Duganella lineage. Based on these phylogenetic affiliations and the consistent cell morphologies, three isolated strains were selected for deposition at the German Collection of Microorganisms and Cell Cultures, Braunschweig (8a), as Acidovorax sp. strain G8B1 (DSM 12578), Herbaspirillum sp. strain G8A1 (DSM 12579), and Paracoccus sp. strain B8B2 (DSM 12584).
FIG. 3.
16S rRNA-based tree reflecting the phylogenetic relationships of (i) strains B8B1 and B8B2 and a selection of Proteobacteria from the alpha subclass and (ii) strains G7A1, G8B1, and G8A1 and a selection of Proteobacteria of the beta subclass. The tree is based on the results of a distance matrix analysis including complete or almost-complete 16S rRNA sequences from representative bacteria of the α, β, and γ subclasses (22). The topology of the tree was evaluated and corrected according to the results of distance matrix, maximum-parsimony, and maximum-likelihood analyses of various data sets. The phylogenetic positions of the analyzed strains did not differ in any of the treeing approaches. Multifurcations indicate topologies that could not be unambiguously resolved. The bar indicates 5% estimated sequence divergence. The P. denitrificans group comprises the species P. denitrificans (GenBank accession no. X69159) and P. versutus (D32243 and D32244) and Paracoccus sp. strains KL1, KS1, and KS2 (U58017, U58016, and U58015). The P. aminovorans group comprises the species P. aminovorans (D32240), P. alcaliphilus (D32238), and P. thiocyanatus (D32242). The A. delafieldii group comprises the species A. delafieldii (AF078764), A temperans (AF078766), and A. facilis (AF078765) and Acidovorax sp. strain 7078 (AF078767). The A. avenae group comprises A. anthurii (AJ007013), A. konjaci (AF078760), A. avenae subsp. avena (AF078759), A. avenae subsp. citrulli (AF078761), A. avenae subsp. cattleyae (AF078762), and Acidovorax sp. strain IMI 357678 (AF078763). Accession numbers of other bacterial 16S rRNA gene sequences are U00006 (Escherichia coli), D13429 (Bradyrhizobium japonicum), D32239 (P. aminophilus), D32241 (P. kocurii), X53855 (Rhodobacter sphaeroides), D16211 (Rhodoferax fermentans), D30793 (Variovorax paradoxus), Z93964 (unidentified bacterium clone T20), M11224 (Comamonas testosteroni), AB008503 (ultramicrobacterium strain MY14), Y10146 (H. seropedicae), X65590 (Telluria chitinolytica), U49757 (Oxalobacter formigenes), X74914 (Duganella zoogloeoides), and X07714 (Neisseria gonorrhoeae).
Population sizes of denitrifying bacteria.
Volatile fatty acids are major primary fermentation products and may as such be important carbon sources and electron donors for denitrifying bacteria in sewage plants. The population size of nitrate-reducing bacteria was determined by MPN counts with either a defined mixture of fatty acids or AC broth as an electron donor. Both measurements yielded the same population size (data not shown). Thus, we now routinely use a defined mixture of fatty acids as a carbon source for MPN counts of nitrate-reducing bacteria in order to avoid the growth of fermentative bacteria.
Clarification of the wastewater at the plant in Lintel involved a limited aeration to introduce early a sequential nitrification-denitrification. MPN counts on samples from the first and third basins indicated a population of 4.6 × 104 bacteria ml−1 that were capable of denitrifying growth on dimethylmalonate. The total denitrifying population decreased from the first basin (1.1 × 108 bacteria ml−1) to the third basin (1.1 × 107 bacteria ml−1). In situ hybridization of cells was performed on the highest-diluted MPN tubes exhibiting growth. The Bacteria-specific probe EUB338 visualized 75% ± 24% of DAPI (4′,6-diamidino-2-phenylindole)-stained cells from the fatty acid-utilizing, nitrate-reducing microorganisms with a statistical population size of greater than 105 bacteria ml−1. None of the cells hybridized with the α-Proteobacteria-specific probe, but the probe for the beta subclass detected 52% ± 18% of DAPI-stained cells. Major contributions to the dimethylmalonate-degrading denitrifying subpopulation within the sludge were made by β-Proteobacteria. In MPN tubes representing greater than 103 cells ml−1, the EUB338 and BET42a probes indicated a relative abundance of 71% ± 24% of Bacteria and of 52% ± 18% of members of the beta subclass. A small contribution of α-Proteobacteria (3% of DAPI-stained cells) was found in a single MPN tube that represented a statistical population density of greater than 103 cells ml−1.
Some β-Proteobacteria that were recently isolated anaerobically on a comparable minimal medium with a single carbon and electron source and nitrate as an acceptor were tested for denitrifying growth on dimethylmalonate. Azoarcus sp. strain 22Lin and Alcaligenes defragrans 51Men and 65Phen mineralized dimethylmalonate. No utilization was observed in cultures of A. defragrans 54PinT and 62Car, Thauera linaloolentis 47LolT, and Thauera terpenica 21Mol and 58EuT.
DISCUSSION
In this study we isolated, for the first time to our knowledge, bacteria that are able to grow on dimethylmalonate as a carbon source and electron donor. The anoxic culture conditions were chosen to select for an oxygen-independent degradation pathway. In addition, aerobic growth of the isolated microorganisms on dimethylmalonate demonstrated the presence of dimethylmalonate-degrading capacities in oxygen-respiring bacteria. The observation of quantitative dimethylmalonate oxidation attests to the capacity of microorganisms to mineralize simple compounds with quaternary carbon atoms. The presence of the dimethylmalonate-degrading capacity in a medium-sized population of bacteria in sewage sludge suggests that dimethylmalonate and related disubstituted malonates may be intermediates of the microbial degradation of organic matter.
The enrichment procedure, with inocula sizes of 10−6 (vol/vol), allows the isolation of a probably abundant bacterium with efficient growth on dimethylmalonate and nitrate. The isolation of related strains B8B1 and B8B2 from sewage sludge and G7A1 and G8B1 from freshwater ditches reflects the high selection pressure during enrichment. The isolation of Paracoccus sp. strains B8B1 and B8B2 from sewage sludge corresponds to the general methylotrophic physiology of members of the genus Paracoccus (20); e.g., Paracoccus cells constituted 3.5% of the total population in a denitrifying sand filter fed with methanol (25). Denitrification is also widespread among Paracoccus species (20). However, analysis of the dimethylmalonate-degrading bacteria grown in highly diluted MPN tubes with oligodeoxynucleotide probes indicated a predominance of β-Proteobacteria. Hence, the Paracoccus strains B8B1 and B8B2 had successfully competed with the β-Proteobacteria during the enrichment. The key advantage of the nitrate-limited enrichment was probably the higher biomass yield of the Paracoccus strains. The partition of β-Proteobacteria in dimethylmalonate degradation was confirmed by the freshwater isolates and by denitrifying growth of Azoarcus sp. strain 22Lin and A. defragrans 51Men and 65Phen on dimethylmalonate.
The isolates from freshwater ditches are β-Proteobacteria related to the genera Acidovorax and Herbaspirillum. The presence of β-Proteobacteria in highly diluted MPN tubes is in agreement with in situ investigations of activated sludge that found a dominance of β-Proteobacteria (28). Curiously, a cloned 16S rRNA gene sequence obtained in that study (28), clone T20, has the highest 16S rRNA gene sequence similarity (98.3%) to the isolated strains G7A1 and G8B1. Members of Acidovorax can denitrify (34, 35), but the described H. rubrisubalbicans and H. seropedicae are only able to reduce nitrate to nitrite (4). Dinitrogen formation did not occur. Phylogenetically related ultramicrobacteria were not tested for the capacity to denitrify (15). Isolate G8A1 is the first isolate in the phylogenetic group that reduces nitrate to dinitrogen oxide and further to dinitrogen.
Two pathways for dimethylmalonate degradation can be considered. In model studies for the mechanism of the coenzyme B12-dependent methylmalonyl-coenzyme A (CoA) mutase, dimethylmalonate derivatives of organocobalamin were found to decompose spontaneously in neutral aqueous solutions. The products formed included methylsuccinate derivatives (10). A similar enzymatic rearrangement based on radical intermediates seems feasible, especially because of the utilization of methylsuccinate by strains G8A1, B8B1, and B8B2. The alternative is a decarboxylation yielding isobutyrate derivatives. During incubation with 14CO2, Mn2+, ATP, and isobutyryl-CoA, enzyme fractions of Mycobacterium sp. strain IBS-M formed two labelled acids that were identified as succinate and dimethylmalonate (19). Besides this observation, a decarboxylase entity is imaginable based on the knowledge of methylmalonyl-CoA and malonyl-CoA decarboxylases (5, 6). We will investigate these possibilities in future research.
ACKNOWLEDGMENT
This study was supported by the Max-Planck-Gesellschaft.
ADDENDUM IN PROOF
Acidovorax sp. strains G7A1 and G8B1 belong to the recently established species Acidovorax defluvii (26a) on the basis of 16S rRNA sequences.
REFERENCES
- 1.Aeckersberg F, Bak F, Widdel F. Anaerobic oxidation of saturated hydrocarbons to CO2 by a new type of sulfate-reducing bacterium. Arch Microbiol. 1991;156:5–14. [Google Scholar]
- 2.Akhtar M, Njar V C O, Wright J N. Mechanistic studies on aromatase and related C-C bond cleaving P-450 enzymes. J Ster Biochem Mol Biol. 1993;44:375–387. doi: 10.1016/0960-0760(93)90241-n. [DOI] [PubMed] [Google Scholar]
- 3.American Public Health Association. Standard methods for the examination of water and wastewater, including bottom sediments and sludge. Washington, D.C: American Public Health Association; 1969. pp. 604–609. [Google Scholar]
- 4.Baldani J I, Pot B, Kirchhof G, Falsen E, Baldani V L D, Olivares F L, Hoste B, Kersters K, Hartmann A, Gillis M, Döbereiner J. Emended description of Herbaspirillum; inclusion of [Pseudomonas] rubrisubalbicans, a mild plant pathogen, as Herbaspirillum rubrisubalbicans comb. nov.; and classification of a group of clinical isolates (EF group 1) as Herbaspirillum species 3. Int J Syst Bacteriol. 1996;46:802–810. doi: 10.1099/00207713-46-3-802. [DOI] [PubMed] [Google Scholar]
- 5.Bott M, Pfister K, Burda P, Kalbermatter P, Woehlke G, Dimroth P. Methylmalonyl-CoA decarboxylase from Propiogenium modestum: cloning and sequencing of the structural genes and purification of the enzyme complex. Eur J Biochem. 1997;250:590–599. doi: 10.1111/j.1432-1033.1997.0590a.x. [DOI] [PubMed] [Google Scholar]
- 6.Dimroth R, Hilbi H. Enzymic and genetic basis for bacterial growth on malonate. Mol Microbiol. 1997;25:3–10. doi: 10.1046/j.1365-2958.1997.4611824.x. [DOI] [PubMed] [Google Scholar]
- 7.Foss S, Harder J. Thauera linaloolentis sp. nov. and Thauera terpenica sp. nov., isolated on oxygen-containing monoterpenes (linalool, menthol and eucalyptol) and nitrate. Syst Appl Microbiol. 1998;21:365–373. doi: 10.1016/s0723-2020(98)80046-5. [DOI] [PubMed] [Google Scholar]
- 8.Foss S, Heyen U, Harder J. Alcaligenes defragrans sp. nov., description of four strain isolated on alkenoic monoterpenes ((+)-menthene, α-pinene, 2-carene and α-phellandrene) and nitrate. Syst Appl Microbiol. 1998;21:237–244. doi: 10.1016/s0723-2020(98)80028-3. [DOI] [PubMed] [Google Scholar]
- 8a.German Collection of Microorganisms and Cell Cultures. 1 June 1999, revision date. [Online.] http://www.dsmz.de. [1 June 1999, last date accessed.]
- 9.Gibbs R A. Prenyl transfer and the enzymes of terpenoid and steroid biosynthesis. In: Sinnott M, editor. Comprehensive biological catalysis: a mechanistic reference. San Diego, Calif: Academic Press; 1998. pp. 31–118. [Google Scholar]
- 10.Grate J H, Grate J W, Schrauzer G N. Synthesis and reactions of organocobalamins relevant to the mechanism of the methylmalonyl-CoA-succinyl-CoA mutase enzyme. J Am Chem Soc. 1982;104:1588–1594. [Google Scholar]
- 11.Griffiths E T, Harries P C, Jeffcoat R, Trudgill P W. Purification and properties of α-pinene oxide lyase from Nocardia sp. strain p18.3. J Bacteriol. 1987;169:4980–4983. doi: 10.1128/jb.169.11.4980-4983.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harder J. Anaerobic degradation of cyclohexane-1,2-diol by a new Azoarcus species. Arch Microbiol. 1997;168:199–204. [Google Scholar]
- 13.Harder J, Probian C. Microbial degradation of monoterpenes in the absence of molecular oxygen. Appl Environ Microbiol. 1995;61:3804–3808. doi: 10.1128/aem.61.11.3804-3808.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harder J, Probian C. Anaerobic mineralisation of cholesterol by a novel type of denitrifying bacterium. Arch Microbiol. 1997;167:274–278. doi: 10.1007/s002030050442. [DOI] [PubMed] [Google Scholar]
- 15.Iizuka T, Yamanaka S, Nishiyama T, Hirashi A. Isolation and phylogenetic analysis of aerobic copiotrophic ultramicrobacteria from urban soil. J Gen Appl Microbiol. 1998;44:75–84. doi: 10.2323/jgam.44.75. [DOI] [PubMed] [Google Scholar]
- 16.Kieslich K. Microbial side-chain degradation of sterols. J Basic Microbiol. 1985;25:461–474. doi: 10.1002/jobm.3620250713. [DOI] [PubMed] [Google Scholar]
- 17.Kniemeyer O. Anaerober Abbau von Cholesterin durch das denitrifizierende Bakterium 72Chol. Diploma thesis. Bremen, Germany: Universität Bremen; 1998. [Google Scholar]
- 18.Koch A L. Growth measurement. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C: American Society for Microbiology; 1994. pp. 249–277. [Google Scholar]
- 19.Lafferty R M. Die an der Verwertung verzweigter Fettsäuren geknüpfte CO2-Fixierung. Zentralbl Bacteriol Parasitol Abtl 1. 1963;191:191–193. [PubMed] [Google Scholar]
- 20.Lipinski A, Reichert K, Reuter B, Spröer C, Altendorf K. Identification of bacterial isolates from biofilters as Paracoccus alkenifer sp. nov. and Paracoccus solventivorans with emended description of Paracoccus solventivorans. Int J Syst Bacteriol. 1998;48:529–536. doi: 10.1099/00207713-48-2-529. [DOI] [PubMed] [Google Scholar]
- 21.Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, Bachleitner M, Schleifer K-H. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis. 1998;19:554–568. doi: 10.1002/elps.1150190416. [DOI] [PubMed] [Google Scholar]
- 22.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 24.Neef A. Anwendung der in situ-Einzelzell-Identifizierung von Bakterien zur Populationsanalyse in komplexen mikrobiellen Biozönosen. Dissertation. Munich, Germany: Technische Universität München; 1997. [Google Scholar]
- 25.Neef A, Zaglauer A, Meier H, Amann R, Lemmer H, Schleifer K-H. Population analysis in a denitrifying sand filter: conventional and in situ identification of Paracoccus spp. in methanol-fed biofilms. Appl Environ Microbiol. 1996;62:4329–4339. doi: 10.1128/aem.62.12.4329-4339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfennig N, Wagener S. An improved method for preparing wet mounts for photomicrographs of microorganisms. J Microb Methods. 1986;4:303–306. [Google Scholar]
- 26a.Schulze R, Spring S, Amann R, Huber I, Ludwig W, Schleifer K-H, Kämpfer P. Genotypic diversity of Acidovorax strains isolated from activated sludge and description of Acidovorax defluvii sp. nov. Syst Appl Microbiol. 1999;22:205–214. doi: 10.1016/S0723-2020(99)80067-8. [DOI] [PubMed] [Google Scholar]
- 27.Shyadehi A Z, Lamb D C, Kelly S L, Kelly D E, Schunck W H, Wright J N, Corina D, Akhtar M. The mechanism of the acyl-carbon bond cleavage reaction catalyzed by recombinant sterol 14α-demethylase of Candida albicans (other names are: lanosterol 14α-demethylase, P-45014DM, and CYP51) J Biol Chem. 1996;271:12445–12450. doi: 10.1074/jbc.271.21.12445. [DOI] [PubMed] [Google Scholar]
- 28.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K-H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Springer N, Ludwig W, Amann R, Schmidt H J, Gorth H D, Schleifer K-H. Occurrence of fragmented 16S rRNA in an obligate bacterial endosymbiont of Paramecium caudatum. Proc Natl Acad Sci USA. 1993;90:9892–9895. doi: 10.1073/pnas.90.21.9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strunk, O., and W. Ludwig. 12 August 1998, revision date. [Online.] ARB: a software environment for sequence data. Department of Microbiology, Technische Universität München, Munich, Germany. http://www.mikro.biologie.tu-muenchen.de. [15 June 1999, last date accessed.]
- 31.Trudgill P W. Microbial metabolism and transformation of selected monoterpenes. In: Ratledge C, editor. Biochemistry of microbial degradation. Dordrecht, The Netherlands: Kluwer; 1994. pp. 33–61. [Google Scholar]
- 32.Vaz A D, Kessell K J, Coon M J. Aromatization of a bicyclic steroid analog, 3-oxodecalin-4-ene-10-carboxaldehyde by liver microsomal cytochrome P450 2B4. Biochemistry. 1994;33:13651–13661. doi: 10.1021/bi00250a015. [DOI] [PubMed] [Google Scholar]
- 33.Widdel F, Bak F. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. Berlin, Germany: Springer; 1992. pp. 3352–3378. [Google Scholar]
- 34.Willems A, Falsen E, Pot B, Jantzen E, Hoste B, Vandamme P, Gillis M, Kersters K, De Ley J. Acidovorax, a new genus for Pseudomonas facilis, Pseudomonas delafieldii, E. Ralsen (EF) group 13, EF group 16, and several clinical isolates, with the species Acidovorax facilis comb. nov., Acidovorax delafieldii comb. nov., and Acidovorax temperans sp. nov. Int J Syst Bacteriol. 1990;40:384–398. doi: 10.1099/00207713-40-4-384. [DOI] [PubMed] [Google Scholar]
- 35.Willems A, Goor M, Thielemann S, Gillis M, Kersters K, De Ley J. Transfer of several phytopathogenic Pseudomonas species to Acidovorax as Acidovorax avenae subsp. avenae subsp. nov., comb. nov., Acidovorax avenae subsp. citrulli, Acidovorax avenae subsp. cattleyae, and Acidovorax konjaci. Int J Syst Bacteriol. 1992;42:107–119. doi: 10.1099/00207713-42-1-107. [DOI] [PubMed] [Google Scholar]