SUMMARY
Nationwide prevalence and risk factors for faecal carriage of Escherichia coli O157 and O26 in cattle were assessed in a 2-year cross-sectional study at four large slaughter plants in New Zealand. Recto-anal mucosal swab samples from a total of 695 young (aged 4–7 days) calves and 895 adult cattle were collected post-slaughter and screened with real-time polymerase chain reaction (PCR) for the presence of E. coli O157 and O26 [Shiga toxin-producing E. coli (STEC) and non-STEC]. Co-infection with either serogroup of E. coli (O157 or O26) was identified as a risk factor in both calves and adult cattle for being tested real-time PCR-positive for E. coli O157 or O26. As confirmed by culture isolation and molecular analysis, the overall prevalence of STEC (STEC O157 and STEC O26 combined) was significantly higher in calves [6·0% (42/695), 95% confidence interval (CI) 4·4–8·1] than in adult cattle [1·8% (16/895), 95% CI 1·1–3·0] (P < 0·001). This study is the first of its kind in New Zealand to assess the relative importance of cattle as a reservoir of STEC O157 and O26 at a national level. Epidemiological data collected will be used in the development of a risk management strategy for STEC in New Zealand.
Key words: Emerging infections, food safety, infectious disease epidemiology, Shiga-like toxin-producing E. coli, zoonoses
INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) O157:H7 and other non-O157 STEC serogroups are recognized as important zoonotic pathogens globally. They can cause serious clinical diseases in humans such as haemorrhagic colitis and the potentially life-threatening haemolytic uraemic syndrome [1]. Ruminants, including cattle, have been identified as an important reservoir of STEC, shedding the pathogen via faeces [2, 3]. Possible sources for STEC infection in humans are STEC-shedding animals, faecally contaminated food, the environment, and infected humans (reviewed in [4]).
New Zealand's annual incidence rate of 4·1 reported STEC cases/100 000 population (2014) [5] is, along with Ireland, Denmark, Sweden [6], and Scotland [7], among the highest in the world. With STEC outbreaks being rare in New Zealand, STEC infections appear predominantly as sporadic cases or small clusters and findings from a national case-control study [8] implicate animal and environmental contact as significant exposure pathways.
New Zealand's beef and dairy production systems are pasture-based with year-round grazing, outdoor housing, and seasonal calving in winter/spring (July–October). Calves not reared for dairy replacement stock or the dairy beef market are slaughtered at a very young age (4–7 days) and exported as veal to overseas markets. Although a large number of very young calves and adult cattle (beef and dairy animals) are slaughtered in New Zealand each year (2012: ~1·7 million calves and 2·3 million adult cattle [9]), data on the prevalence of STEC in slaughter cattle are limited to a few regional studies conducted in the North Island of New Zealand [10–12]. This study was designed to investigate the prevalence and risk factors for faecal carriage of E. coli O157 and O26, including STEC O157 and O26, in slaughtered cattle nationwide, and to provide epidemiological data to be used in the development of a risk management strategy for STEC in New Zealand.
MATERIALS AND METHODS
Sample collection
Four large cattle slaughter plants, licensed to process veal and beef for export, were selected, with two plants in each of the North Island (A and B) and the South Island (C and D) of New Zealand. The plants were chosen, based on the annual number of processed cattle in 2007 and their location, to achieve a good geographical coverage in this nationwide repeated cross-sectional study. From July 2009 until June 2011, each plant was visited (1) fortnightly between July and October to collect samples from young calves (aged 4–7 days), and (2) monthly between August and July to collect samples from adult cattle. In a catchment area of a slaughter plant, calves from different farms were transported collectively and slaughtered as one mob (not grouped by farm), whereas adult cattle were transported from farms as separate mobs and slaughtered as separate ‘lines’.
Recto-anal mucosal swab (RAMS) samples were collected systematically from calves (every 20–25th carcass) and adult cattle (1–3 carcasses per line) post-slaughter on the processing chain. Samples were obtained by swabbing the recto-anal junction with firm pressure using sterile cotton-tipped swabs (Transystem®, Italy), placed in Amies transport medium provided in the swab tubes and kept refrigerated until processed within 24–48 h of collection. Sampled animal data such as breed, sex, ear tag number, carcass weight, and farm address were recorded. Cross-breeds were classified as the visually more dominant breed. The ages of adult cattle were estimated based on the number of permanent incisors [13]. Dentition was also used for grading of carcasses at the slaughter plants, classifying female adult cattle with ⩽4 and >4 permanent incisors as heifers and cows, respectively.
Laboratory methods
RAMS samples were enriched and screened with real-time polymerase chain reaction (PCR) for the presence of E. coli O157 and O26 (STEC and non-STEC), before applying culture isolation methods for E. coli on real-time PCR-positive samples. Retrieved E. coli O157 and O26 isolates were analysed for the presence of virulence genes characteristic of STEC using multiplex PCR. The methods are described in detail below.
Enrichment and direct culture plating
Each RAMS was transferred into 20 ml buffered peptone water (BPW) (Difco™, Becton, USA), mixed thoroughly, and 50 µl was directly cultured onto cefixime-tellurite sorbitol MacConkey agar (CT-SMAC; Fort Richard Laboratories, New Zealand) and cefixime-tellurite rhamnose MacConkey agar (CT-RMAC; Fort Richard Laboratories) used as selective culture media for E. coli O157 and O26, respectively. BPW broths and direct culture plates were incubated at 37 °C for 18–24 h.
Real-time PCR for detection of E. coli O157 and O26 (STEC and non-STEC)
Genomic DNA was extracted from 1 ml enriched BPW broth using 2% Chelex® beads solution (Bio-Rad, USA) and analysed for the presence of E. coli O157 and O26 using an automated real-time thermocycler (Rotor Gene 6200HRM, Corbett Research, Australia). Two separate PCR assays were performed using previously published primer sequences to detect genes encoding for serogroup-specific O-antigens of E. coli O157 (rfbEO157 [14]) and O26 (wzxO26 [14]).
The final 20 µl PCR reaction volume for O157 contained 2x PCR buffer (Light Cycler 480 Probes Master, Roche, Germany), 50 µm SYTO® 9 dye (Invitrogen, USA), 2 µm of each primer, and 5·4 µl DNA. The amplification programme included an initial enzyme activation step at 96 °C for 5 min, which was followed by 45 cycles of denaturation at 96 °C for 15 s, annealing at 60 °C for 10 s, and extension at 72 °C for 10 s; the PCR product was detected by thermal melt from 73 °C to 82 °C at a rate of 0·1 °C/2 s. Similarly, the final 20-μl PCR reaction volume for O26 contained 2x PCR buffer (Express qPCR SuperMix, Invitrogen), 50 µm SYTO® 9 dye, 10 µm of each primer, 2·0 µl DNA, and 5·4 µl sterile water. The amplification programme included an initial enzyme activation step at 95 °C for 10 min, which was followed by 40 cycles of, 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 10 s; the PCR product was detected by thermal melt from 72 °C to 78 °C at a rate of 0·1 °C/2 s. Positive and negative template controls were included in each PCR assay.
Culture isolation of E. coli O157 and O26
Direct culture plates of real-time PCR-positive samples were scanned for suspected E. coli O157 and O26 colonies. Up to ten non-fermenting and ten fermenting colonies per culture plate were tested using serogroup-specific latex agglutination kits (E. coli O157 Latex and Serocheck O26, Oxoid, UK). Agglutination-positive E. coli O157 and O26 isolates were confirmed with real-time PCR for the presence of rfbEO157 and wzxO26 genes, respectively, as described above. Colonies on direct culture plates of real-time PCR-negative samples for E. coli O157 from 39·7% (211/532) of calf samples and 37·4% (311/832) of adult cattle samples were tested with latex agglutination for E. coli O157 to detect false-negative samples. This additional testing was not implemented on real-time PCR-negative samples for E. coli O26 because of the limited availability of the serogroup-specific agglutination kit.
If E. coli O157 and O26 isolates could not be retrieved from direct culture plates of real-time PCR-positive samples, serogroup-specific immunomagnetic beads (Dynabeads® anti-E. coli O157 and Dynabeads® EPEC/VTEC O26, Invitrogen, Norway) were used on enriched BPW broths, following the manufacturer's instructions. Bead-broth suspensions were inoculated onto CT-SMAC or CT-RMAC, incubated at 37 °C for 18–24 h, and processed as direct culture plates described above.
Molecular analysis of E. coli O157 and O26 isolates
Bacterial DNA was extracted from confirmed E. coli O157 and O26 isolates using 2% Chelex® beads solution and analysed with multiplex PCR for the presence of STEC virulence genes encoding for Shiga toxin 1 (stx1), Shiga toxin 2 (stx2), enterohaemolysin (ehxA) and intimin (eae), as described previously [15].
Statistical analysis
R software (version 2.15·2) [16] was used for all statistical analysis. χ2 and Fisher's exact tests were used to compare prevalence estimates between E. coli O157 and E. coli O26, and STEC O157 and STEC O26 in samples from calves and adult cattle.
Exposure variables were analysed using univariate and multivariate logistic regression to identify risk factors associated with a calf or adult cattle being test-positive by real-time PCR for either E. coli O157 or O26 (STEC and non-STEC). Real-time PCR results were used for this analysis because of their higher prevalence and statistical power, compared to culture results. The multivariate models were generated and evaluated for significance and goodness-of-fit as described previously [8]. To account for clustering of animals originating from the same farm, a multivariate model with ‘farm’ as a random-effect variable was built as described above, using R package ‘lme4’ [17]. The model's significance was evaluated applying likelihood ratio tests.
For consistency, univariate and multivariate logistic regression analysis was also performed on culture isolation data. The multivariate models generated based on real-time PCR data were re-run using STEC results as the binomial outcome variable; changes in exposure variables were reported.
RESULTS
Animal data
RAMS samples from a total of 695 calves and 895 adult cattle were collected. On average, RAMS samples from 27 calves (range 6–110) and 11 adult cattle (range 4–17) were collected per plant visit. The number of samples to be collected per visit was proportional to the plant's annual number of processed cattle but was also dependent on the number of animals available on each sampling day, explaining the wide range.
Calves and adult cattle originated from 496 and 513 different farms, respectively, of which 655 were dairy farms and 354 were beef farms. Over the 2-year study period, 36 dairy and seven beef farms were sampled repeatedly within the same or in the following year. The geographical distribution of farms of origin of sampled animals is shown in Figure 1 and animal data are summarized in Table 1. The majority of sampled calves were bull calves (73·7%, 512/695), dairy breeds (99·1%, 689/695), and originated from dairy farms, while adult cattle were mainly represented by steers (39·7%, 355/895), animals aged ⩽2·0 years (48·9%, 438/895), Angus breed (35·1%, 314/895), and originated from beef farms. The mean carcass weight of calves and adult cattle was 15·7 kg [95% confidence interval (CI) 9·7–21·7] and 271·2 kg (95% CI 133·9–408·4), respectively. Carcass weights were missing for 25 calves and one adult cattle due to condemnation at meat inspection.
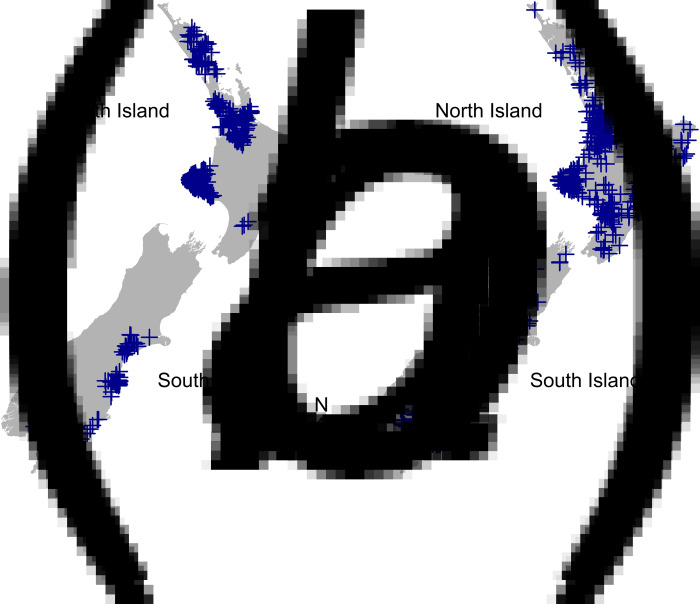
Fig. 1.
Map of New Zealand showing distribution of farms of origin of (a) young calves (n = 695) and (b) adult cattle (n = 895), from which recto-anal mucosal swab samples were collected post-slaughter at four New Zealand cattle slaughter plants from July 2009 to June 2011.
Table 1.
Annual number of young calves (n = 695) and adult cattle (n = 895), from which recto-anal mucosal swab samples were collected post-slaughter at four New Zealand cattle slaughter plants (A–D) from July 2009 to June 2011, stratified by sex, estimated age, breed, farm type, and island of origin
| Categories within | Calves | Adult cattle | ||||||
|---|---|---|---|---|---|---|---|---|
| Variable | variable | 2009 | 2010 | Total | 2009 | 2010 | 2011 | Total |
| Sex | Bull (male) | 279 | 233 | 512 | 19 | 93 | 61 | 173 |
| Steer (male castrated) | — | — | — | 86 | 200 | 69 | 355 | |
| Heifer (female) | 75 | 108 | 183 | 17 | 77 | 54 | 148 | |
| Cow (female) | — | — | — | 18 | 87 | 114 | 219 | |
| Age, years (no. of permanent incisors) | ⩽2·0 (2) | — | — | — | 90 | 244 | 104 | 438 |
| 2·5 (4) | — | — | — | 20 | 84 | 58 | 162 | |
| 3·5 (6) | — | — | — | 12 | 41 | 22 | 75 | |
| ⩾4·0 (8) | — | — | — | 18 | 88 | 114 | 220 | |
| Breed | Beef | 6 | 542 | |||||
| Angus | — | 2 | 2 | 52 | 175 | 87 | 314 | |
| Hereford | 1 | 1 | 2 | 43 | 83 | 33 | 159 | |
| Charolais | — | — | — | 7 | 17 | 1 | 25 | |
| Limousin | — | — | — | — | 7 | 8 | 15 | |
| Simmental | — | — | — | 1 | 11 | 3 | 15 | |
| Other | 1 | 1 | 2 | 3 | 9 | 2 | 14 | |
| Dairy | 689 | 353 | ||||||
| Friesian | 267 | 202 | 469 | 31 | 115 | 130 | 276 | |
| Jersey | 85 | 135 | 220 | 3 | 39 | 33 | 75 | |
| Ayrshire | — | — | — | — | 1 | 1 | 2 | |
| Farm type | Beef | 5* | — | 5 | 60* | 193* | 113* | 366 |
| Dairy | 291* | 270* | 561 | 5* | 69* | 96* | 170 | |
| Island of origin | North Island | 495 | 446 | |||||
| Slaughter plant A | 205 | 80 | 285 | 22 | 111 | 79 | 212 | |
| Slaughter plant B | 73 | 137 | 210 | 30 | 120 | 84 | 234 | |
| South Island | 200 | 449 | ||||||
| Slaughter plant C | 36 | 64 | 100 | 53 | 124 | 72 | 249 | |
| Slaughter plant D | 40 | 60 | 100 | 35 | 102 | 63 | 200 | |
Number of different farms per year (excluding farms with repeated sampling of animals).
Prevalence of E. coli O157 and O26 (STEC and non-STEC) and STEC O157 and O26 at slaughter
Prevalence estimates of E. coli O157 and O26 (STEC and non-STEC) in RAMS samples from calves and adult cattle, as tested by real-time PCR and culture isolation (direct culture plating and immunomagnetic separation combined), are summarized in Table 2. In calves, the real-time PCR prevalence of E. coli O26 (STEC and non-STEC) was significantly higher than for E. coli O157 (P < 0·001), but no statistical difference was observed between E. coli serogroups in adult cattle (P = 0·717). Similarly, the culture prevalence of E. coli O26 (STEC and non-STEC) in calves was significantly higher compared to E. coli O157 (P < 0·001), but no statistically significant difference was apparent in adult cattle (P = 0·394). In contrast, however, the culture prevalence of STEC O157 in adult cattle was significantly higher than for STEC O26 (P = 0·030), but no significant difference was observed in calves (P = 0·121).
Table 2.
Prevalence estimates of E. coli O157 and O26 (STEC and non-STEC) and STEC O157 and O26 in recto-anal mucosal swab samples from young calves (n = 695) and adult cattle (n = 895) as test-positive by real-time PCR and culture isolation (direct culture plating and immunomagnetic separation combined), stratified by animal group and serogroup
| E. coli O157 and O26 (STEC and non-STEC) | STEC O157 and O26 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Real-time PCR | Culture isolation | Culture isolation | ||||||||
| Animal group | Serogroup | Prevalence % (positive/tested) | 95% CI | P | Prevalence % (positive/tested) | 95% CI | P | Prevalence % (positive/tested) | 95% CI | P |
| Calves | O157 | 23·5 (163/695) | 20·4–26·8 | <0·001 | 3·2 (22/695) | 2·0–4·8 | <0·001 | 2·3 (16/695) | 1·4–3·8 | 0·121 |
| O26 | 33·4 (232/695) | 29·9–37·0 | 8·3 (58/695) | 6·4–10·7 | 3·9 (27/695) | 2·6–5·7 | ||||
| Adult cattle | O157 | 7·0 (63/895) | 5·5–9·0 | 0·717 | 2·5 (22/895) | 1·6–3·8 | 0·394 | 1·6 (14/895) | 0·9–2·7 | 0·030* |
| O26 | 7·6 (68/895) | 6·0–9·6 | 3·2 (29/895) | 2·2–4·7 | 0·4 (4/895) | 0·1–1·2 | ||||
PCR, Polymerase chain reaction; CI, confidence interval;
P value comparing the prevalence of serogroup O157 and O26 was calculated using Fisher's exact test.
A proportion of calves (12·7%, 88/695) and adult cattle (1·7%, 15/895) were real-time PCR-positive for both E. coli (STEC and non-STEC) serogroups. The culture prevalence for both STEC serogroups was 0·1% (1/695, 95% CI 0·0–0·9) in calves and 0·2% (2/895, 95% CI 0·0%–0·9) in adult cattle. One calf and two adult cattle carried both STEC O157 and STEC O26. Taking this into consideration, the overall prevalence of STEC (STEC O157 and STEC O26 combined) was significantly higher in calves [6·0% (42/695), 95% CI 4·4–8·1] than in adult cattle [1·8% (16/895), 95% CI 1·1–3·0] (P < 0·001).
As confirmed by culture isolation, 2·8% (10/354, 95% CI 1·4–5·3) and 6·0% (39/655, 95% CI 4·3–8·1) of beef farms and dairy farms were positive for STEC O157 and/or STEC O26, respectively, resulting in 4·9% (49/1009, 95% CI 3·6–6·4) of all farms (354 beef and 655 dairy farms) being positive for STEC O157 and/or O26.
Virulence profiles of confirmed E. coli O157 and O26 isolates
A total of 44 E. coli O157 and 92 E. coli O26 isolates were retrieved and screened for the presence of virulence genes. The isolates' virulence profiles, stratified by animal group and serogroup, are summarized in Table 3. All stx-positive E. coli O157 and O26 isolates carried both ehxA and eae genes. stx1 was the only toxin gene detected in STEC O26 isolates, while stx2 was predominant in STEC O157 isolates.
Table 3.
Characteristics of 44 E. coli O157 and 92 E. coli O26 isolates retrieved from recto-anal mucosal swab samples collected from young calves (n = 695) and adult cattle (n = 895) at slaughter in New Zealand, stratified by sampled animal group and serogroup. A multiplex PCR was used to test for the presence/absence (+/–) of virulence genes encoding for enterohaemolysin (ehxA), intimin (eae), and Shiga toxins (stx1 and stx2)
| Animal group | No. of isolates | Virulence genes | ||||||
|---|---|---|---|---|---|---|---|---|
| Serogroup | Sorbitol* | Rhamnose† | ehxA | eae | stx1 | stx2 | ||
| Calves | O157 | 2 | – | + | + | + | + | |
| 14 | – | + | + | – | + | |||
| 6 | + | – | – | – | – | |||
| O26 | 27 | – | + | + | + | – | ||
| 15 | – | + | + | – | – | |||
| 3 | + | + | + | – | – | |||
| 17 | + | – | + | – | – | |||
| 1 | + | – | – | – | – | |||
| Adult cattle | O157 | 1 | – | + | + | + | + | |
| 13 | – | + | + | – | + | |||
| 8 | + | – | – | – | – | |||
| O26 | 4 | – | + | + | + | – | ||
| 7 | – | + | + | – | – | |||
| 5 | + | + | + | – | – | |||
| 13 | + | – | + | – | – | |||
Sorbitol-fermenting (+) or non-sorbitol-fermenting (–) on cefixime-tellurite sorbitol MacConkey agar.
Rhamnose-fermenting (+) or non-rhamnose-fermenting (–) on cefixime-tellurite rhamnose MacConkey agar.
Risk factors for faecal carriage of E. coli O157 and O26 (STEC and non-STEC) and STEC O157 and O26 at slaughter
Univariate logistic regression results for calves and adult cattle, stratified by serogroup of E. coli (STEC and non-STEC), are summarized in Supplementary Tables S1 and S2, respectively. Statistically significant risk factors and confounding variables in the final multivariate logistic regression models for calves and adult cattle being real-time PCR-positive for E. coli O157 or O26 (STEC and non-STEC) are presented in Table 4. ‘Slaughter plant’ and ‘presence of E. coli O26’ were identified as significant risk factors for a calf being tested real-time PCR-positive for E. coli O157 (STEC and non-STEC), with ‘sex’ and ‘carcass weight’ as confounding variables. Similarly, risk factors for a calf being test-positive for E. coli O26 by real-time PCR were ‘slaughter plant’, ‘presence of E. coli O157’, and ‘month’, while confounding variables included ‘breed’, ‘farm type’, and ‘carcass weight’. Significant risk factors for an adult animal being tested real-time PCR-positive for E. coli O157 (STEC and non-STEC) were ‘season’ and ‘presence of E. coli O26’, with ‘sex’, ‘breed’, and ‘carcass weight’ as confounders. In comparison, ‘presence of E. coli O157’ was the only risk factor for adult cattle being real-time PCR-positive for E. coli O26, with ‘age’ as the only confounding variable. Multivariate logistic regression models with ‘farm’ as a random effect were fitted on data of calves tested real-time PCR-positive for E. coli O157 and E. coli O26 (Supplementary Table S3), but could not be completed for adult cattle due to non-convergence of data.
Table 4.
Four multivariate logistic regression analysis models showing risk factors for a calf/adult cattle being tested real-time PCR-positive for E. coli O157/E. coli O26 (STEC and non-STEC). Due to missing carcass weights, 25 from 695 observations in calves and one from 895 observations in adult cattle were deleted
| Animal group | E. coli serogroup | Variable | Reference category | Categories within variable | Coefficient (s.e.) | OR (95% CI) | Wald test P value | Likelihood ratio test* |
|---|---|---|---|---|---|---|---|---|
| Calf† | O157 | Intercept | −1·50 (0·63) | |||||
| Slaughter plant | A | B | −0·01 (0·24) | 0·99 (0·62–1·58) | 0·965 | <0·001 | ||
| C | 0·50 (0·28) | 1·65 (0·96–2·83) | 0·070 | |||||
| D | −1·33 (0·40) | 0·26 (0·12–0·58) | 0·001 | |||||
| Presence of E. coli O26 | No | Yes | 1·22 (0·20) | 3·39 (2·28–5·02) | <0·001 | |||
| Sex | Male | Female | −0·40 (0·23) | 0·67 (0·42–1·06) | 0·089 | |||
| Carcass weight | 0·02 (0·03) | 1·02 (0·96–1·09) | 0·464 | |||||
| Calf‡ | O26 | Intercept | −1·37 (1·03) | |||||
| Slaughter plant | A | B | −1·06 (0·24) | 0·35 (0·22–0·56) | <0·001 | <0·001 | ||
| C | −0·53 (0·29) | 0·59 (0·34–1·02) | 0·061 | |||||
| D | 0·13 (0·31) | 1·13 (0·62–2·07) | 0·684 | |||||
| Month | July | Aug. | −0·67 (0·23) | 0·51 (0·33–0·79) | 0·003 | 0·004 | ||
| Sept. | −0·88 (0·31) | 0·41 (0·22–0·76) | 0·005 | |||||
| Presence of E. coli O157 | No | Yes | 1·26 (0·20) | 3·54 (2·38–5·26) | <0·001 | |||
| Breed | Friesian | Jersey | −0·27 (0·20) | 0·76 (0·51–1·13) | 0·179 | 0·366 | ||
| Other breed | 0·36 (1·04) | 1·44 (0·19–10·97) | 0·725 | |||||
| Farm type | Dairy | Beef | 1·25 (0·89) | 3·48 (0·61–19·90) | 0·162 | |||
| Carcass weight | 0·01 (0·03) | 1·01 (0·95–1·07) | 0·862 | |||||
| Adult cattle§ | O157 | Intercept | −3·92 (1·01) | |||||
| Season | Spring (Sept.–Nov.) | Summer (Dec.–Feb.) | −0·86 (0·42) | 0·42 (0·19–0·97) | 0·043 | <0·001 | ||
| Autumn (Mar.–May) | 0·48 (0·36) | 1·62 (0·79–3·30) | 0·187 | |||||
| Winter (June–Aug.) | −1·57 (0·58) | 0·21 (0·07–0·65) | 0·007 | |||||
| Presence of E. coli O26 | No | Yes | 1·72 (0·35) | 5·58 (2·80–11·13) | <0·001 | |||
| Sex | Male | Male castrated | 1·10 (0·51) | 3·00 (1·10–8·21) | 0·032 | 0·060 | ||
| Female | 0·84 (0·55) | 2·32 (0·79–6·85) | 0·126 | |||||
| Breed | Friesian | Jersey | −0·78 (0·78) | 0·46 (0·10–2·12) | 0·319 | 0·202 | ||
| Angus | 0·24 (0·39) | 1·27 (0·59–2·75) | 0·545 | |||||
| Hereford | −0·65 (0·52) | 0·52 (0·19–1·46) | 0·216 | |||||
| Other breeds | 0·49 (0·53) | 1·64 (0·58–4·62) | 0·352 | |||||
| Carcass weight | <0·01 (<0·01) | 1·00 (1·00–1·01) | 0·541 | |||||
| Adult cattle¶ | O26 | Intercept | −2·98 (0·22) | |||||
| Presence of E. coli O157 | No | Yes | 1·55 (0·33) | 4·69 (2·45–8·99) | <0·001 | |||
| Adult cattle¶ | O26 | Age (years) | ⩽2·0 | 2·5 | 0·56 (0·34) | 1·75 (0·89–3·44) | 0·102 | 0·135 |
| 3·5 | −0·06 (0·56) | 0·95 (0·32–2·82) | 0·920 | |||||
| ⩾4·0 | 0·62 (0·30) | 1·86 (1·02–3·37) | 0·042 |
s.e., Standard error; OR, odds ratio; CI, confidence interval.
P value of variable as whole.
Likelihood ratio test = 63·97 (d.f. = 6, P < 0·001) for ‘Calf-E. coli O157’ model.
Likelihood ratio test = 85·40 (d.f. = 10, P < 0·001) for ‘Calf-E. coli O26’ model.
Likelihood ratio test = 58·73 (d.f. = 11, P < 0·001) for ‘Adult cattle-E. coli O157’ model.
Likelihood ratio test = 23·18 (d.f. = 4, P < 0·001) for ‘Adult cattle-E. coli O26’ model.
Univariate logistic regression results for calves and adult cattle, stratified by STEC O157 and STEC O26, are summarized in Supplementary Tables S4 and S5, respectively. Having applied the final multivariate logistic regression models to STEC data (culture isolation), the strength of associations and significance of many variables changed due the low number of positive STEC results compared to real-time PCR data. While half of all model variables became non-significant, variables such as ‘slaughter plant’ in the ‘Calf-STEC O26’ model, ‘season’ and ‘presence of STEC O26’ in the ‘Adult cattle-STEC O157’ model, and ‘presence of STEC O157’ in the ‘Adult cattle-STEC O26’ model remained significant. Only ‘breed’ in the ‘Adult cattle-STEC O157’ model became significant when included as a single multi-category variable (P = 0·004) applied to culture data. Full details of the multivariate logistic regression models are presented in Supplementary Table S6.
DISCUSSION
This 2-year study is the first of its kind in New Zealand to provide data on the prevalence and risk factors for faecal carriage of E. coli and STEC serogroups O157 and O26 in very young calves and adult cattle at slaughter at a national level. The results showed a significantly higher overall STEC prevalence (prevalence of STEC O157 and STEC O26 combined) in calves than in adult cattle, and co-infection with either serogroup of E. coli (O157 or O26) was identified as a risk factor in both calves and adult cattle for being tested real-time PCR-positive for E. coli O157 or O26 (STEC and non-STEC). Although the animals sampled were not a random sample of slaughter cattle in New Zealand, the selection of two large slaughter plants in both North and South Island, combined with the systematic sampling of calves and adult cattle on the slaughter chains over multiple visits, resulted in animals being selected from a relatively large number of farms distributed across a wide geographical area. Based on the number of farms and the geographical coverage, the animals sampled could be considered to be broadly representative of New Zealand's slaughter cattle population.
Prevalence of STEC O157 and STEC O26
When comparing our prevalence results with previously published studies, it is essential to acknowledge variations in study designs and culture isolation methods, in addition to animal management practices and environmental factors.
Calves
Outside of New Zealand, it is not general practice to slaughter <7-day-old calves, hence, most previous studies have investigated the prevalence of faecal carriage of STEC in calves on-farm and animals of different age groups (<24 h to <1–2 months of age), reporting prevalence estimates ranging from 0·8% to 2·2% for STEC O157 [18–20]. Due to the lack of equivalent studies, our findings can only be compared with a regional study conducted in 4- to 7-day-old calves at two slaughter plants in the lower North Island of New Zealand [10], which detected STEC O157 in 3·2% (10/309) of RAMS samples, similar to our findings (2·3%, 16/695, 95% CI 1·4–3·8). No previous surveys have been conducted in New Zealand to assess the faecal carriage of STEC O26 in calves at slaughter, but on-farm studies in Australia [20] and northern Scotland [21] also reported a higher prevalence of STEC O26 in calves compared to STEC O157 [STEC O26 ranging from 3·8% (3/79) to 93·9% (46/49); STEC O157 from 1·3% (1/79) to 8·2% (4/49), respectively].
It can be argued that the faecal carriage rates of STEC in very young calves at slaughter might be affected by transportation to, and lairage at, slaughter plants. Although impacts of transportation and lairage on faecal carriage of STEC have been studied in adult cattle overseas [22–24], very limited data are available for calves of this very young age [25]. Pseudo-vertical transmission of STEC from STEC-shedding dams to calves is a possible pathway of infection on-farm [20, 21], but other factors such as animal housing, management, and the environment could have an impact on the prevalence of STEC in calves on-farm.
Adult cattle
A number of previous studies have investigated the faecal carriage of STEC O157 and other non-O157 STEC in adult cattle at slaughter in other countries, detecting STEC O157 prevalence estimates ranging from 1·6% to 4·7% [26–29]. Their findings are similar to our results [1·6% (14/895), 95% CI 0·9–2·7] but higher compared to a previous survey conducted at a slaughter plant in the North Island of New Zealand, which observed an STEC O157 prevalence of only 0·5% (2/371) in healthy dairy cows [12]. Differences in detection and isolation methods used, season and the distribution of animals sampled (local vs. nationwide) could explain some of the observed variation of STEC O157 prevalence compared to our study.
Consistent with findings from studies outside of New Zealand [26, 27, 29], the prevalence of STEC O26 in adult cattle was lower compared to STEC O157, and only the Australian study by Barlow & Mellor [27] reported a very low STEC O26 prevalence (0·3%, 1/300) in slaughter cattle similar to our study [0·4% (4/895), 95% CI 0·1–1·2], while others reported higher prevalence estimates ranging from 1·3% to 1·5% [26, 29]. Although no previous study has investigated the faecal carriage of STEC O26 in adult cattle at slaughter in New Zealand, Cookson et al. [30] reported the isolation of STEC O26 from healthy cattle on-farm in the lower North Island (1·1%, 2/187), providing evidence of STEC O26 being prevalent in New Zealand cattle.
In general, the prevalence of STEC O157 and O26 in adult cattle was lower compared to calves and might be associated with the fully developed gastrointestinal tract of the adult cattle. Microbial fermentation of feed in the developed rumen of adult cattle produces high concentrations of volatile fatty acids, which lower the pH conditions, and are thought to inhibit the growth of STEC O157 and other pathogenic strains of E. coli [31]. These conditions and a naive gut could account therefore for a higher susceptibility for STEC colonization and thereby the higher prevalence of STEC O157 and O26 observed in calves, in addition to other external factors such as husbandry systems and animal management.
Farms
Considering the origins of calves and adult cattle sampled in this study, 6·0% (39/655) of dairy farms were positive for STEC O157 and/or STEC O26 compared to 2·8% (10/354) of beef farms. The sensitivity of the test applied at farm-level was low as only a very small number of animals were tested per farm at slaughter; hence, these values represent estimates of the minimum farm-level prevalence. No epidemiological data are available on the nationwide farm-level prevalence of STEC among New Zealand dairy and beef farms.
Risk factors for faecal carriage of E. coli O157 and O26 (STEC and non-STEC) and STEC O157 and O26 at slaughter
Calves
Plant and co-infection with either serogroup of E. coli (O157 or O26) were identified as risk factors for faecal carriage of E. coli O157 and O26 (and STEC O26) in calves at slaughter, suggesting common sources of infection for both serogroups on the farm of origin and/or during transportation to and lairage at the slaughter plant. Plant as a risk factor could indicate other risks associated with the plant's catchment area and slaughter plant-associated factors, such as animal handling or lairage management, which could affect the prevalence of faecal carriage of E. coli and STEC O157 and O26 in animals at slaughter.
Risk factors for STEC O157 infections in dairy calves were assessed in previous studies outside of New Zealand [32, 33], indicating that increased prevalence of STEC O157 on-farm is associated with age of calves and housing/farm management factors. It is general practice on New Zealand dairy farms to keep calves in groups in barns for the first week after birth, before selecting heifer calves for stock replacement and bull calves for the dairy beef market, and sending the remaining animals as calves for slaughter. Grouping calves before weaning was identified as an increased risk factor for North American dairy farms to be positive for STEC O157, compared to farms where grouping of calves was practised only after weaning [33, 34]. The use of open pails compared to nipples to feed calves was also associated with increased infection of STEC O157 in calves on Canadian dairy farms [35, 36]. Given the lack of risk factor studies on STEC infections on New Zealand dairy farms, no assumptions can be made on whether similar housing and feeding practices contribute to the level of STEC infection in calves on-farm, and consequently could have an impact on the prevalence of STEC detected in calves at slaughter.
Adult cattle
Similar to observations in calves, co-infection with either serogroup of E. coli (O157 or O26) (and similarly for serogroups of STEC) was identified as the main risk factor for faecal carriage of E. coli O157 and O26 (and STEC O157 and O26) in adult cattle, suggesting a common source of infection either on-farm or on-plant. Interestingly, season was also associated with E. coli O157 (and STEC O157) infection in adult cattle, showing a significantly lower risk in winter (autumn for STEC O157). It can be speculated whether seasonal changes in weather or feed could have an effect on the seasonality of E. coli O157/STEC O157 infection in adult cattle, or if the reduced prevalence of E. coli O157/STEC O157 in the environment over colder seasons decreases the risk of intake and colonization of E. coli O157/STEC O157 in cattle. Seasonality of STEC prevalence in cattle is well documented in studies outside of New Zealand, reporting greater faecal shedding of STEC in the warmer seasons [28, 37, 38].
Laboratory methods
The prevalence of faecal carriage of E. coli O157 and O26, and STEC O157 and STEC O26, in our study was determined by the number of test-positive samples using real-time PCR and culture isolation. When interpreting prevalence estimates based on these methods, it is important to acknowledge that other factors (e.g. selectivity of enrichment/culture media, culturability of E. coli, background microbiota) might affect the methodologies, providing false-positive or false-negative test results. For example, real-time PCR cannot differentiate DNA from viable and non-viable E. coli/STEC cells, identifying E. coli O157/O26-positive samples containing only dead cells that cannot be confirmed by culture. Confirmation by culture isolation is essential but the sensitivity of culture methods can be influenced by other factors such as choice of enrichments, selectivity of culture media, culturability of E. coli, and specificity of immunomagnetic beads. Bacterial cells are able to enter a physiological stage of being viable but non-culturable, which has been demonstrated for a large number of bacterial pathogens including E. coli and STEC [39]. Immunomagnetic beads are coated with polyclonal antibodies against surface antigens of serogroup- specific E. coli, but could cross-react with antigenically similar organisms, such as Escherichia hermannii, Salmonella O group N, or Proteus spp. [40], and provide false-negative culture results.
CONCLUSIONS
The findings of this study contribute to our understanding that STEC O157 and STEC O26 are prevalent in New Zealand slaughter cattle populations, which could represent an important source of STEC infection in humans via animal contact, environmental contamination, or contaminated food. Calves were identified as more prevalent carriers of STEC O157 and O26 than adult cattle at slaughter, and should therefore be considered as an important source of STEC likely to enter the food chain. Epidemiological data collected in this study will be used for the development of a risk management strategy for STEC and inform decision making in the meat industry to reduce the public health risks associated with the consumption of red meat produced in New Zealand. A further observational study is underway to investigate the epidemiology of STEC O157 and other non-O157 STEC serogroups on New Zealand dairy farms, including the identification of potential risk factors for STEC prevalence in very young calves on-farm.
ACKNOWLEDGEMENTS
We thank all four cattle slaughter plants and their managers for participation in this study, and their staff for providing important animal data; laboratory staff from mEpiLab, Anne Midwinter, Lynn Rogers, and Julie Collins-Emerson for technical support and assistance with processing and testing of samples.
This work was supported by the Ministry for Primary Industries, Wellington, New Zealand, and the New Zealand Meat Industry Association.
DECLARATION OF INTEREST
None.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268815003209.
click here to view supplementary material
REFERENCES
- 1.Gould LH, et al. Hemolytic uremic syndrome and death in persons with Escherichia coli O157:H7 infection, Foodborne Diseases Active Surveillance Network Sites, 2000–2006. Clinical Infectious Diseases 2009; 49: 1480–1485. [DOI] [PubMed] [Google Scholar]
- 2.Wells JG, et al. Isolation of Escherichia coli serotype O157:H7 and other Shiga-like-toxin-producing E. coli from dairy cattle. Journal of Clinical Microbiology 1991; 29: 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borczyk AA, et al. Bovine reservoir for verotoxin-producing Escherichia coli O157:H7. Lancet 1987; 1: 98. [DOI] [PubMed] [Google Scholar]
- 4.Karmali MA, Gannon V, Sargeant JM. Verocytotoxin-producing Escherichia coli (VTEC). Veterinary Microbiology 2010; 140: 360–370. [DOI] [PubMed] [Google Scholar]
- 5.Institute of Environmental Science and Research Ltd. Surveillance report – notifiable diseases in New Zealand: annual report 2014. Porirua, New Zealand, 2015. Client Report FW15010.
- 6.European Centre for Disease Prevention and Control. Annual epidemiological report 2014 – food- and waterborne diseases and zoonoses. Stockholm, Sweden: ECDC; 2014. [Google Scholar]
- 7.Locking M, et al. Surveillance report – VTEC and HUS in Scotland, 2013: enhanced surveillance, reference laboratory and clinical reporting data. HPS Weekly Report 2014; 48: 257–263. [Google Scholar]
- 8.Jaros P, et al. A prospective case-control and molecular epidemiological study of human cases of Shiga toxin-producing Escherichia coli in New Zealand. BMC Infectious Diseases 2013; 13: 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Infoshare. (http://www.stats.govt.nz/infoshare/). Accessed 17 August 2013.
- 10.Irshad H, et al. Epidemiology of Shiga toxin-producing Escherichia coli O157 in very young calves in the North Island of New Zealand. New Zealand Veterinary Journal 2012; 60: 21–26. [DOI] [PubMed] [Google Scholar]
- 11.Cookson AL, Taylor SCS, Attwood GT. The prevalence of Shiga toxin-producing Escherichia coli in cattle and sheep in the lower North Island, New Zealand. New Zealand Veterinary Journal 2006; 54: 28–33. [DOI] [PubMed] [Google Scholar]
- 12.Buncic S, Avery SM. Escherichia coli O157:H7 in healthy dairy cows. New Zealand Veterinary Journal 1997; 45: 45–48. [DOI] [PubMed] [Google Scholar]
- 13.Anon. Estimation of age by examination of the teeth. In: Kahn CM, ed. The Merck Veterinary Manual, 9th edn. Merck & Co. Inc., Whitehouse Station, NJ, USA, 2005, pp. 137–140. [Google Scholar]
- 14.Perelle S, et al. Detection by 5′-nuclease PCR of Shiga-toxin producing Escherichia coli O26, O55, O91, O103, O111, O113, O145 and O157:H7, associated with the world's most frequent clinical cases. Molecular and Cellular Probes 2004; 18: 185–192. [DOI] [PubMed] [Google Scholar]
- 15.Jaros P, et al. Geographic divergence of bovine and human Shiga toxin-producing Escherichia coli O157:H7 genotypes, New Zealand. Emerging Infectious Diseases 2014; 20: 1980–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R Core Team. R: a language and environment for statistical computing, version 2.15·2. Vienna, Austria: R Foundation for Statistical Computing, 2012.
- 17.Bates D, Maechler M, Bolker B. Linear mixed-effects models using S4 classes, version 0.999999-2. 2013.
- 18.Fernández D, et al. Short communication: characterization of Shiga toxin-producing Escherichia coli isolated from newborn, milk-fed, and growing calves in Argentina. Journal of Dairy Science 2012; 95: 5340–5343. [DOI] [PubMed] [Google Scholar]
- 19.Rugbjerg H, Nielsen EM, Andersen JS. Risk factors associated with faecal shedding of verocytotoxin-producing Escherichia coli O157 in eight known infected Danish dairy herds. Preventive Veterinary Medicine 2003; 58: 101–113. [DOI] [PubMed] [Google Scholar]
- 20.Cobbold R, Desmarchelier P. A longitudinal study of Shiga-toxigenic Escherichia coli (STEC) prevalence in three Australian dairy herds. Veterinary Microbiology 2000; 71: 125–137. [DOI] [PubMed] [Google Scholar]
- 21.Pearce MC, et al. Temporal shedding patterns and virulence factors of Escherichia coli serogroups O26, O103, O111, O145, and O157 in a cohort of beef calves and their dams. Applied and Environmental Microbiology 2004; 70: 1708–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanford K, et al. Effects of long- or short-haul transportation of slaughter heifers and cattle liner microclimate on hide contamination with Escherichia coli O157. Journal of Food Protection 2011; 74: 1605–1610. [DOI] [PubMed] [Google Scholar]
- 23.Cuesta Alonso EP, Gilliland SE, Krehbiel CR. Incidence and toxin production ability of Escherichia coli O157:H7 isolated from cattle trucks. Journal of Food Protection 2007; 70: 2383–2385. [DOI] [PubMed] [Google Scholar]
- 24.Minihan D, et al. An investigation on the effect of transport and lairage on the faecal shedding prevalence of Escherichia coli O157 in cattle. Journal of Veterinary Medicine, Series B 2003; 50: 378–382. [DOI] [PubMed] [Google Scholar]
- 25.Jaros P, et al. Population dynamics of E. coli O157 and O26 – the effect of transport and lairage on faecal shedding and carcase contamination of very young calves in New Zealand. In: Abstract Book of the 8th International Symposium on Shiga Toxin (Verocytotoxin) Producing Escherichia coli Infections. Amsterdam, The Netherlands, 2012, p 70.
- 26.Thomas KM, et al. Tracking verocytotoxigenic Escherichia coli O157, O26, O111, O103 and O145 in Irish cattle. International Journal of Food Microbiology 2012; 153: 288–296. [DOI] [PubMed] [Google Scholar]
- 27.Barlow RS, Mellor GE. Prevalence of enterohemorrhagic Escherichia coli serotypes in Australian beef cattle. Foodborne Pathogens and Disease 2010; 7: 1239–1245. [DOI] [PubMed] [Google Scholar]
- 28.Paiba GA, et al. Faecal carriage of verocytotoxin-producing Escherichia coli O157 in cattle and sheep at slaughter in Great Britain. Veterinary Record 2002; 150: 593–598. [DOI] [PubMed] [Google Scholar]
- 29.Shinagawa K, et al. Frequency of Shiga toxin-producing Escherichia coli in cattle at a breeding farm and at a slaughterhouse in Japan. Veterinary Microbiology 2000; 76: 305–309. [DOI] [PubMed] [Google Scholar]
- 30.Cookson AL, et al. Serotypes and analysis of distribution of Shiga toxin-producing Escherichia coli from cattle and sheep in the lower North Island, New Zealand. New Zealand Veterinary Journal 2006; 54: 78–84. [DOI] [PubMed] [Google Scholar]
- 31.Rasmussen MA, et al. Rumen contents as a reservoir of enterohemorrhagic Escherichia coli. FEMS Microbiology Letters 1993; 114: 79–84. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen EM, et al. Influence of age, sex and herd characteristics on the occurrence of verocytotoxin-producing Escherichia coli O157 in Danish dairy farms. Veterinary Microbiology 2002; 88: 245–257. [DOI] [PubMed] [Google Scholar]
- 33.Garber LP, et al. Risk factors for fecal shedding of Escherichia coli O157:H7 in dairy calves. Journal of the American Veterinary Medical Association 1995; 207: 46–49. [PubMed] [Google Scholar]
- 34.Shere JA, Bartlett KJ, Kaspar CW. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Applied and Environmental Microbiology 1998; 64: 1390–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson JB, et al. Risk factors for bovine infection with verocytotoxigenic Escherichia coli in Ontario, Canada. Preventive Veterinary Medicine 1993; 16: 159–170. [DOI] [PubMed] [Google Scholar]
- 36.Wilson JB, et al. Risk factors for infection with verocytotoxigenic Escherichia coli in cattle on Ontario dairy farms. Preventive Veterinary Medicine 1998; 34: 227–236. [DOI] [PubMed] [Google Scholar]
- 37.Milnes AS, et al. Factors related to the carriage of Verocytotoxigenic E. coli, Salmonella, thermophilic Campylobacter and Yersinia enterocolitica in cattle, sheep and pigs at slaughter. Epidemiology and Infection 2009; 137: 1135–1148. [DOI] [PubMed] [Google Scholar]
- 38.Fernández D, et al. Seasonal variation of Shiga toxin-encoding genes (stx) and detection of E. coli O157 in dairy cattle from Argentina. Journal of Applied Microbiology 2009; 106: 1260–1267. [DOI] [PubMed] [Google Scholar]
- 39.Oliver JD. The viable but nonculturable state in bacteria. Journal of Microbiology 2005; 43: 93–100. [PubMed] [Google Scholar]
- 40.Life Technologies™. Dynabeads® anti-E. coli O157, August 2012, Rev. No. 11 (http://tools.lifetechnologies.com/content/sfs/manuals/dynabeads_antiecoli0157_man.pdf). Accessed 24 December 2013.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268815003209.
click here to view supplementary material