SUMMARY
An outbreak of infective mastitis due to Enterococcus faecalis occurred in an intensive sheep farm in north Sardinia (Italy). E. faecalis, which is only rarely isolated from sheep milk, was unexpectedly found in 22·3% of positive samples at microbiological examination. Forty-five out of the 48 E. faecalis isolates showed the same multi-drug resistance pattern (cloxacillin, streptomycin, kanamycin, clindamycin, oxytetracycline). E. faecalis isolates were analysed by pulsed-field gel electrophoresis, and all 45 multi-drug resistant strains showed an indistinguishable macrorestiction profile, indicating their clonal origin. To our knowledge, this is the first report of an outbreak of mastitis in sheep caused by E. faecalis.
Key words: Animal pathogens, antibiotic resistance, Enterococcus, outbreaks
Infectious mastitis is one of the most important health problem affecting dairy sheep. Staphylococci represent the most common aetiological agent of intramammary infections, followed by streptococci, Gram-negative bacteria, corynebacteria and, less likely, enterococci [1]. We describe the occurrence of an unusual outbreak of sheep mastitis in a modern farm in north Sardinia (Italy) due to multi-drug resistant Enterococcus faecalis. Although enterococcal mastitis is relatively uncommon compared to mastitis caused by Staphylococcus spp. and Streptococcus spp., milk quality and udder health problems associated with enterococcal mastitis have been documented, particularly in cows [2]. Enterococci are ubiquitous Gram-positive bacteria, components of the intestinal flora of humans and animals, and can be spread in the environment through faecal contamination. Outside the host, enterococci are hardy bacteria, able to survive under a wide range of conditions. Moreover, enterococci are intrinsically resistant to a broad range of antibiotics, and are able to acquire resistance to antimicrobial agents through horizontal gene transfer [3].
The outbreak occurred in a flock of about 1200 dairy Sarda breed sheep in Sardinia, which represents the Italian region with the highest density of dairy sheep, accounting for about 3·5 million [4]. The animals were kept under intensive conditions, with no more than 2 h per day of grazing on natural pastures. The ewes were housed in a barn during the night and were mechanically milked twice a day. In August 2008 an increase in cases of mastitis accompanied by a considerable reduction in daily milk production and by a bulk tank somatic cell count of 7·8 × 106 occurred on the sheep farm, when most sheep were in the late stage of lactation. Microbiological analysis was conducted on ewes exhibiting clinical signs of acute or chronic mastitis. Milk was collected by hand milking after teat disinfection. After discarding the first streams, about 5 ml milk from both half-udders were pooled in sterile vials and immediately transported, under refrigeration, to the microbiology laboratory of the University of Sassari. Milk samples were plated on Columbia, MacConkey, bile-esculin and mannitol agar plates and incubated in aerobic conditions at 37 °C for 48 h. Of 699 milk samples tested (one from each ewe), 92 were excluded as they showed mixed bacterial flora, and 215 were considered positive. Identification of isolated bacteria was determined using both cultural, biochemical and molecular methods. As expected, most isolates were identified as staphylococci (S. aureus 23·2%, staphylococci coagulase negative 54·5%, namely S. epidermidis 32·1%, S. chromogenes 13·5%, S. xylosus 7·4%, and S. simulans 1·4%) but, surprisingly, a high number of isolates were identified as E. faecalis (n = 48, representing 22·3% of total positives). Species identification was confirmed by multiplex PCR, using primers specific for E. faecalis and E. faecium [5]. Susceptibility of enterococci to antimicrobials was determined by the automated VITEK system (bioMérieux, France), and by disk diffusion on Mueller–Hinton agar, supplemented with 5% of defibrinated sheep blood, for those antimicrobials commonly used in veterinary therapy (cloxacillin, cefazolin, amozicillin, kanamicin, and oxytetracycline) that were not included in VITEK cards.
Although E. faecalis is rarely isolated from sheep milk, with a prevalence of ⩽4% [6], in the present study we detected an unexpectedly high percentage of E. faecalis (22·3%). This is, to our knowledge, the first report of such a high number of mastitis cases due to E. faecalis occurring in sheep. Interestingly, an association was observed between the presence of mammary nodules and E. faecalis. Nodules, intended as circumscribed, solid subcutaneous or parenchymal non-ulcerated lesions, were present in 62% of E. faecalis-positive ewes and had a diameter ranging from 1 to 5 cm. These lesions were not present in sheep infected by staphylococci. Moreover, all sheep infected by enterococci showed an enlargement of supramammary lymph nodes.
All 48 E. faecalis isolates were susceptible to penicillin G, teicoplanin, amoxicillin and vancomycin. Interestingly, 45/48 E. faecalis isolates showed the same multi-drug resistance pattern (cloxacillin, streptomycin, kanamycin, clindamycin, oxytetracycline). The high degree of similarity of antibiotic resistance shown by most E. faecalis isolates suggested an outbreak of clonal origin. The potential clonal origin of the isolates was verified by macrorestriction analysis after pulsed-field gel electrophoresis (PFGE). PFGE is a highly reproducible and discriminating tool for the molecular typing of bacteria and has been successfully used for epidemiological investigations of enterococcal outbreaks and for subtyping of enterococci strains. Genomic DNA was obtained from all E. faecalis isolates and digested with the restriction enzyme SmaI [7]. PFGE was performed using the CHEF DR III apparatus (Bio-Rad, USA).
Three PFGE patterns were detected among the 48 E. faecalis isolates (see Fig. 1). Profile SS1 accounted for 43 isolates (representing 89·6% of total E. faecalis isolates) which shared also the same antibiotic resistance pattern, confirming the hypothesis of their clonal origin. The other two PFGE patterns SS2 and SS3 were identified in one and four isolates, respectively. Phenotypic and genotypic characterization of E. faecalis isolates suggests the existence of a common source of infection, most likely of faecal origin. The possible contamination of the milking machine was excluded by repeated and accurate controls. In fact, milking machines undergo periodical inspections by the Sardinian health authority, to check the functioning of mechanical parts and microbiological status; no defect or bacteriological contamination had been observed on this farm. In any case, as soon as the outbreak was detected, we performed additional microbiological tests, both on the parts of the milking machines that could represent a source of infection and on the chlorinated water used to wash the machinery. In all cases no contamination was observed.
Fig. 1.
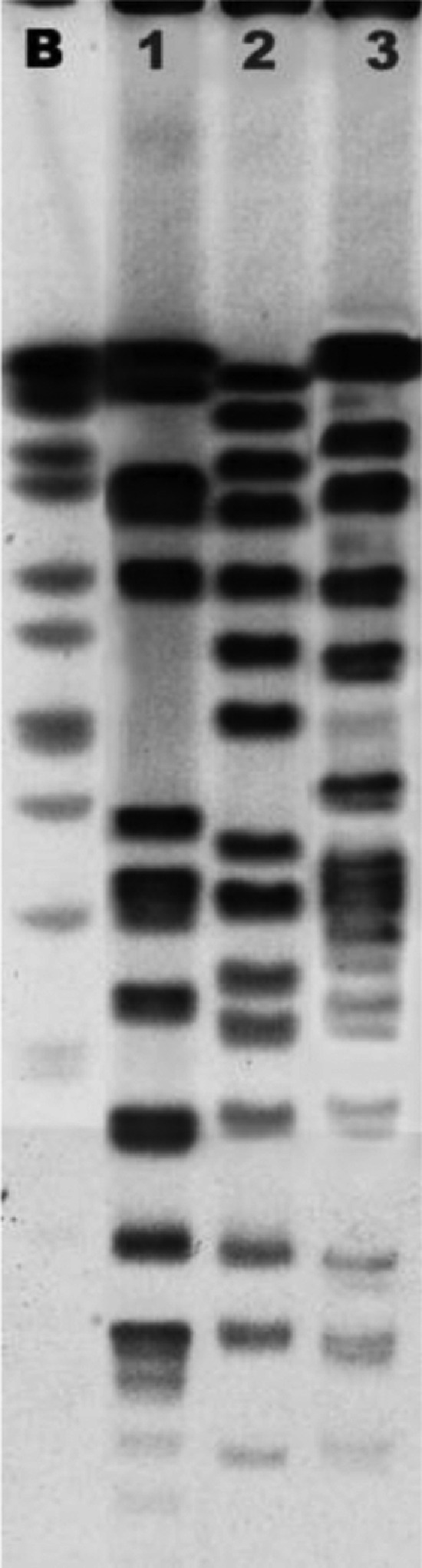
SmaI PFGE restriction patterns of representative E. faecalis isolates. B, Salmonella enterica serovar Braenderup (H9812) restricted with XbaI as reference size marker; lanes 1–3: E. faecalis pulsotypes SS1, SS2 and SS3.
The progressive passage from hand milking to use of milking machines produced a general improvement in the control of infections caused by staphylococci that can be acquired during hand milking [4]. Moreover, intensive farming of sheep can cause a worsening of hygienic conditions, with a possible increase of infections due to bacteria of environmental origin, such as streptococci and enterococci. Faecal contamination of animal bedding is a primary source of bacteria such as Enterococcus and Streptococcus [8]. In fact teat ends of sheep, both during rest and rumination, are in direct contact with faecally contaminated bedding material.
When the 43 isolates showing the same genotypic and phenotypic characteristics are excluded, the percentage of enterococci isolated in this group of sheep falls from 22·3% to 2·3%, a figure comparable to those described elsewhere, confirming the hypothesis of an outbreak. Apparently, a single strain of E. faecalis found optimal environmental conditions for spreading. Such a predominance of a single strain can result from several mechanisms: it can be due to a higher virulence compared to other microorganisms, to a better adaptation to environmental conditions, but also to a selection pressure induced by the massive use of antibiotics. In fact enterococci responsible for the outbreak are multi-drug resistant, and a retrospective analysis showed extensive use of antibiotics in this flock over the previous 2 years, administered on the basis of clinical signs both by the systemic route and locally at the beginning of the dry-off period. In particular streptomycin, cloxacillin and oxytetracycline were extensively used, drugs that are ineffective against E. faecalis isolates. A few outbreaks of infective mastitis in sheep have been described previously [9–11] but, to our knowledge, this is the first report of an outbreak of mastitis in sheep caused by multi-drug resistant E. faecalis. It emphasizes the possibility of infections due to bacteria of environmental origin and the need for stricter control of antibiotic usage in the management of veterinary infections. This is particularly relevant considering the potential role of farm animals as reservoirs of antibiotic-resistant microorganisms such as E. faecalis, a leading cause of nosocomial infections and a growing public health concern.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Bergonier D, et al. Mastitis of dairy small ruminants. Veterinary Research 2003; 34: 689–716. [DOI] [PubMed] [Google Scholar]
- 2.Todhunter DA, Smith KL, Hogan JS. Environmental streptococcal intramammary infections of the bovine mammary gland. Journal of Dairy Sciences 1995; 78: 2366–2374. [DOI] [PubMed] [Google Scholar]
- 3.Hershberger E, et al. Epidemiology of antimicrobial resistance in enterococci of animal origin. Journal of Antimicrobial Chemotherapy 2005; 55: 127–130. [DOI] [PubMed] [Google Scholar]
- 4.Marogna G, et al. Clinical findings in sheep farm affected by recurrent bacterial mastitis. Small Ruminant Research 2010; 88: 119–125. [Google Scholar]
- 5.Kariyama R. Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. Journal of Clinical Microbiology 2000; 38: 3092–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mørk T, et al. Clinical mastitis in ewes; bacteriology, epidemiology and clinical features. Acta Veterinaria Scandinavica 2007; 49: 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turabelidze D, et al. Improved pulsed-field gel electrophoresis for typing vancomycin-resistant enterococci. Journal of Clinical Microbiology 2000; 38: 4242–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Contreras A, et al. The role of intramammary pathogens in dairy goats. Livestock Production Science 2003: 79: 273–283. [Google Scholar]
- 9.Berriatua E, et al. Outbreak of subclinical mastitis in a flock of dairy sheep associated with Burkholderia cepacia complex infection. Journal of Clinical Microbiology 2001; 39: 990–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Las Heras A, et al. Unusual outbreak of clinical mastitis in dairy sheep caused by Streptococcus equi subsp. zooepidemicus. Journal of Clinical Microbiology 2002; 40: 1106–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munoz M, et al. Molecular epidemiology of two Klebsiella pneumoniae mastitis outbreaks on a dairy farm in New York state. Journal of Clinical Microbiology 2007; 45: 3964–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]


